Abstract
Hagfish intermediate filament (HIF) proteins, consisting of α and γ subunits, have been previously recombinantly expressed, purified, and utilized to form dry fibers with impressive mechanical properties. HIFα and HIFγ consist of three protein domains (N-termini, C-termini, and central rod domain). To begin to understand the structure–function relationship between the protein domains in fiber formation and properties in a synthetic fiber spinning system, we designed recombinant protein constructs with varying combinations of the N-terminus, central rod domain (CRD), and C-terminus for both the α and γ proteins. The constructs, for both α and γ, were expressed, purified, and spun into dry fibers, which were then tested and analyzed for mechanical and structural properties. Mechanical testing revealed that the α constructs had the highest tensile strength when both termini were removed while including either terminus improved strain and toughness compared to α CRD constructs. The γ constructs displayed improved tensile strength and elastic modulus when only the N-terminus was present. Mixing the α and γ constructs generally enhanced the mechanical properties compared to the full-length rHIFα and rHIFγ. Fourier transform infrared-attenuated total reflection (FTIR-ATR) analysis indicated that the CRD contributes more to the β-sheet content in the stretched fibers, while the termini contribute more to the α-helical/random coil regions. These findings provide valuable insights into the roles of the different protein domains in the assembly and mechanical performance of rHIF and other recombinantly expressed IF. By understanding these structure–function relationships, functionally tailored recombinant IF proteins can be designed for specific applications in biomaterials developments.
Introduction
Intermediate filaments (IF) are a family of chemically heterogeneous structural proteins that share a common tripartite domain structure consisting of a central α-helical “rod” domain (CRD) flanked by nonhelical amino-terminal “head” (N-terminus) and carboxy-terminal “tail” (C-terminus) domains.1,2 The central rod domain allows the formation of a parallel, coiled-coil dimer.3
IF proteins, such as vimentin, keratin, and fibrin, assemble into filaments in natural systems and when produced recombinantly.2,4 Previous research has primarily focused on elucidating the structure–function relationships of IF domains, with an emphasis on fiber assembly and structure. Herrmann et al. observed in their study that recombinant human and Xenopus vimentin and various truncations require the head domain for filament formation.5 The removal of the vimentin tail domain had minimal impact on vimentin assembly and filament formation.5
Research on other IF proteins, such as keratin and fibrins, has also explored the roles of protein domains in fiber assembly and structure.6,7 However, research directly correlating mechanical properties with structure by forming and testing fibers spun in a synthetic spinning process from protein constructs with varying domain compositions remains limited.
Hagfish are deep-sea jawless fish that resemble eels and possess a unique defensive mechanism.8 When threatened or bitten, they secrete a thick slime from epithelial slime glands, which clogs the gills of predators, forcing them to release the hagfish.9 Within the slime, there are threads that, when isolated from the slime, stretched, and dried, exhibit impressive mechanical properties.10 These threads are constructed from two IF proteins, denoted α and γ.11,12 In the hagfish intermediate filament (HIF), the heterodimeric proteins α and γ share this common domain architecture.13 The CRD facilitates heterodimerization between α and γ through the formation of coiled-coil structures.14 The N- and C-termini domains exhibit viscoelastic behavior, and their role in fiber assembly remains unknown.15
HIF threads have been recombinantly produced and formed.16−18 Due to high recombinant expression yields in Escherichia coli., and the impressive mechanical properties of the formed fibers, recombinant hagfish intermediate filaments (rHIF) have been proposed as an ideal protein for the production of protein-based biomaterials.17,19 It has been observed in both the naturally occurring HIF and in rHIF that the intermediate filament fibers go through an α → β transition within their central rod domains. This transition is accompanied by an increase in the mechanical properties of the synthetically formed fibers while the terminal domains remain nonhelical.16,17,19 It has been suggested that further developments in recombinant expression technologies will lead to a deeper understanding of the roles of the rHIF domains.14
With the pursuit of the production of large-scale protein-based, biodegradable fibers for a variety of applications, understanding the specific contributions of each domain to the fibers’ mechanical properties and resilience is crucial to understanding how fibers are formed and behave in a synthetic spinning system. In this project, we investigated the roles of the N-terminus, C-terminus, and CRD within a recombinant protein and nonmimetic fiber formation system to gain insights into how these domains influence dry fiber assembly, structure, and mechanical performance under stress.
The α and γ proteins are found in a mixture in nature, coexpressed in a ratio of 1:1, and evolved to fulfill a specific function.12 The evolutionary process of the proteins is unclear, and the mechanical properties presented in our study provide information regarding the properties of the three basic protein elements when spun in a synthetic fiber spinning system that does not resemble the natural process.
Early investigation of the mechanical properties of α and γ spun in a synthetic fiber spinning system revealed differences in fiber mechanical properties when spun individually.17 However, the natural occurrence of the HIF is in a 1:1 ratio, so the rHIF was also spun at a 1:1 ratio, with the proteins measured at the same proportion, to simulate the natural HIF fibers.17,18 To begin to elucidate the N- and C-termini’s functional roles in rHIF dry fiber formation, we started with the full-length constructs (rHIFα and rHIFγ(C387S)) described previously by Oliveira et al.17 Then, constructs were designed and produced with either the N-terminus removed (rHIF-C), the C-terminus removed (N-rHIF), or both termini removed (CRD-rHIF). The protein construct labeling will be simplified throughout this work to α and γ for rHIFα and rHIFγ(C387S), and the aforementioned abbreviations for inclusion or exclusion of structural elements. These new recombinant proteins were then expressed and purified as inclusion bodies, as described by Oliveira et al., with appropriate adjustments made to accommodate the different isoelectric points of these constructs.17 The dried proteins were spun into fibers following the protocol described by Bell et al., a formic-acid-solvated wet-spinning with stretches applied at two stages of fiber formation.18 These fibers were analyzed for mechanical and structural differences using tensile testing and fourier transform infrared-attenuated total reflection (FTIR-ATR) spectral analysis. By comparing the different constructs’ mechanical and structural components, the study aims to determine the contributions of the three structural elements in a synthetic spinning process.17,18
Methods
Cloning and Expression of rHIF-PC
The recombinant α and γ constructs genes were generated by polymerase chain reaction (PCR). The full-length genes (N-α-C and N-γ-C) in pET19k expressed in Oliveira et al.’s previously cited work were used as a template to generate the different constructs.17 Primers were designed (Table 1) to amplify the desired sequences and add restriction enzyme sites to allow the cloning of the specific domains while keeping the histidine tag upstream of the DNA construct.
Table 1. Primers Used in the Design of the Recombinant Constructs.
| constructs | primer |
|---|---|
| rHIFα (α) Construct Proteins | |
| N-α | forward: 5′ ccatgggccatcatcatca 3′ |
| reverse: 5′ taactagtaatacgggtttcttcgctatcc 3′ | |
| CRD-α | forward: 5′ atcctaggaaacaggatctgcagacac 3′ |
| reverse: 5′ taactagtaatacgggtttcttcgctatcc 3′ | |
| α-C | forward: 5′ atcctaggaaacaggatctgcagacac 3′ |
| reverse: 5′ ggccggatccttaactagtatagat 3′ | |
| rHIFγ(C387S) (γ) Construct Proteins | |
| N-γ | forward: 5′ taccatgggccatcatcatc 3′ |
| reverse: 5′ taactagtcaccatcagttcttgg 3′ | |
| CRD-γ | forward: 5′ tacctaggaaaaacattctggg 3′ |
| reverse: 5′ taactagtcaccatcagttcttgg 3′ | |
| γ-C | forward: 5′ tacctaggaaaaacattctggg 3′ |
| reverse: 5′ taactagtctgcagaataatg 3′ | |
The PCR products were inserted into the pET19k at AvrII (New England Biolabs R0174L) and SpeI (New England Biolabs R3133S) restriction sites for the CRD and C-termini-inclusive constructs (α-C and γ-C). The NcoI (New England Biolabs R01935) and SpeI (New England Biolabs R3133S) restriction sites were used for the N-termini-inclusive constructs (N-α and N-γ). The resulting vectors were transformed into E. coli BL21 (DE3) chemically competent cells (New England Biolabs C2527I) to produce the six recombinant hagfish protein constructs.
Expression of rHIF-PC Proteins
Initial shaker flasks, a standard tool for protein expression validation, were used to validate protein expression for each construction. As was done for the full-length proteins, all protein constructions were expressed as inclusion bodies.17 The expression of the protein constructs was then scaled up using BioFlo310 bioreactors (Eppendorf) and BioCommand software (Eppendorf). The protocol and media components were consistent with the work reported by Oliviera et al., with appropriate adjustments to the volume of components to best fit the reactor used.17 The cell mass was harvested as Oliveira et al. and Bell et al. described, with the cell mass being isolated via centrifugation before being stored frozen at −20 °C.17,18 Several small-scale purifications of 10 to 100 g of wet cell mass were performed for each construct, and the final recovery average was calculated for each construct.
Protein Purification and Verification
Cell Lysis
The cell mass was lysed in similar ratios of cell mass to lysis buffer reported by Oliveira et al., but the buffer’s pH was adjusted to reflect the isoelectric point of each construct.17 The cell mass was lysed using a Q700 sonicator (QSonica) at 50–75A for 7 min, using a cycle of 10 s on and 10 s off.
Inclusion Body Washing
The lysate was centrifuged at 10,000 rcf for 15 m at 4 °C before the washing steps began.17 The resulting inclusion body pellets were then processed through two wash buffers, a Tris-Acetate-EDTA- Isopropyl Alcohol (TAE-IPA) wash, and 50% IPA rinses.17 There are two notable changes to the method used in this work and Oliveira et al.’s work. The first is that the chemical dithiothreitol (DTT) was omitted from the wash buffer recipes, as it was omitted in the scaled-up processing described by Bell et al.18 The second is that the pH of the wash buffers was adjusted to reflect the isoelectric point of these constructs (see Table 2 in the results). The 1× TAE/50% IPA wash and the 50% IPA washes were unchanged and still served to remove nucleic acid contaminants and perform salt removal.17 The proteins were lyophilized and then crushed into a fine powder, and the final dry protein amount was recorded. The production yield was determined as grams of dry protein per kilogram of wet cell mass (g kg–1).
Table 2. Average Protein Recoveries and Purity Levels of the rHIF Constructs.
| protein construct | molecular weight (kDa) | theoretical pI | estimated protein purity (%) | purification yield (g kg–1) | references |
|---|---|---|---|---|---|
| N-α-C | 70.50 | 7.94 | 88 | 39 | (17) |
| N-α | 58.10 | 6.88 | 83 | 65 | (this work) |
| α central rod | 39.58 | 6.03 | 78 | 44 | (this work) |
| α-C | 51.61 | 6.09 | 83 | 37 | (this work) |
| N-γ-C | 66.67 | 5.52 | 91 | 45 | (17) |
| N-γ | 54.56 | 4.90 | 88 | 86 | (this work) |
| γ central rod | 40.27 | 4.57 | 78 | 107 | (this work) |
| γ-C | 52.00 | 4.78 | 86 | 59 | (this work) |
SDS-PAGE Coomassie Analysis
The dry proteins were solubilized at 0.5–1.0% w/v in 8 M urea (Sigma-Aldrich U1250) by rotating them overnight, then were mixed 1:1 v/v with 2× Laemmli Sample Buffer (Bio-Rad 1610737).17,18 These were heat treated at 100 °C for 5 min before loading on polyacrylamide gel (Invitrogen Novex Wedge Well 4–20% Tris-Glycine 1.0 mm Mini Protein Gels; ThermoFisher Scientific XP04200BOX), a dual-color protein standard (Bio-Rad) was included. The gels were allowed to run with a constant 90 V until the samples reached halfway through the gel, when the voltage was then increased to 110 V for the remainder of the gel length. The gel was then processed identically to Oliveira et al. and Bell et al., with ImageJ (NIH), used to determine protein purity based on lane and band intensity differences.17,18
Western Blot Analysis
The protein samples separated by the sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel were transferred to a poly(vinylidene fluoride) (PVDF) membrane (Bio-Rad 1620177) by electroblotting via the Mini Trans-Blot System (Bio-Rad). The transfers were accomplished using a constant amperage of 200 mA for 60 min. The membrane was then treated identically to the membranes described by Oliveira et al.17
Fiber Spinning
The dried, purified proteins were dissolved in 97% formic acid (Alfa Aesar A13285) at 15, 20, and 25% w/v concentrations. Each dope was prepared in a 4 mL glass vial (VWR 470151–622) with 2 mL of formic acid, as described previously by Bell et al.18 The vials were sealed with Parafilm and allowed to solvate for up to 24 h, after which they were transferred to microtubes for centrifugation. The supernatant was loaded into a 3 mL syringe and placed in a custom spinning machine and set up as described by Bell et al.18 Two different sets of spinning bath setups were labeled as dH2O and saltwater (SW). The dH2O setup had the coagulation bath filled with nanopure water and deionized water filling the first and second stretch baths. The SW setup had the coagulation bath and both stretch baths filled with a saltwater (SW) solution (instant ocean marine fast dissolving sea salt at 36 g/L). The fibers were extruded and removed from the coagulation bath at a rate of 15 mm s–1 and then threaded through a series of godets and stretch baths, allowing for a controlled stretching of the fibers as previously described by Bell et al.18 Each stretched section is denoted as a number followed by “X” which indicated the draw ratio compared to the speed of the previous draw ratio section speed. This means that for a 2X2X stretch, the fiber was draw-processed to 2X the initial length in the first stretch bath (deionized water or SW) and then drawn again to 2X the length in the second stretch bath (deionized water or SW) for a total of 300% draw-processed.17,18 Some of the constructs required the presence of saltwater to form fibers capable of being extracted from the bath and stretched, resulting in some construct groups not having data for the dH2O setup. Ultimately, the 20% concentration (25% for γ-CRD) spun into SW and stretched at 2X2X was used for mechanical and structural analysis comparisons due to its consistent reliability across the constructs.
Fibers Mechanical Analysis
After spinning, the fibers were allowed to air-dry on the collection spools for at least a day. The fibers were then processed similarly to Copeland et al., Jones et al., Oliviera et al., and Bell et al.17,18,20,21 The processing involved using a Motic microscope with the Motic Image Plus Program to measure the diameters of the fibers for calculation purposes. The mechanical testing occurred on an MTS system with a 10 g load cell running a tensile test at 5 mm min–1 with data collected at 120 Hz.17,18,21 The raw force and extension data were then exported to Microsoft Excel to calculate each fiber’s maximum tensile strength and strain, elastic modulus, and toughness. The calculated data was then used to calculate statistical significance using a combination of ANOVA tests and t tests to compare the constructs of each protein construct to each other at the 2X2X saltwater stretch condition for the 20% protein dopes.
Fibers Structural Analysis
Samples from the 20% saltwater spun fibers at a 2X2X stretch were sent to Arizona State University (ASU) and the United States Naval Surface Warfare Center-Panama City Division (NSWC-PCD) for Fourier transform infrared (FTIR) spectroscopy analysis. The fibers processed by ASU were done using a PerkinElmer Frontier FTIR with a Pike diamond ATR using 50 scans with a wavenumber range of 400 to 5000 cm–1. The fibers processed by NSWC-PCD utilized a Thermo Scientific Nicolet iS50 FT-IR with an iS50 ATR module with 32 scans with wavenumbers of 600 to 4000 cm–1 with an aperture setting of 100, using the OMNIC software. The FTIR data was received and deconvoluted in OriginPro2023 (OriginLab). The deconvolution process followed similar procedures described by Oliveira et al., Hu et al., Böni et al., and Zou et al.17,22−24 The raw data was normalized using the maximum of the Amide I absorbance.23 Then, the Amide I region (∼1550–1720 cm–1) was selected to go into the OriginPro software for deconvolution. The software corrected the baseline by subtracting a straight line before peak finding and fitting. Using the second derivative of the Savitzky–Golay smoothing method and a second-order polynomial and 7 points of the window, the minima of the second derivative were selected as options for the absorbances around 1650 and 1680 cm–1 wavenumbers.23 The best absorbance assignments were determined by utilizing Gaussian curves and fit parameters of a maximum of 400 iterations and a tolerance of 1 × 10–14. These best peaks maximized the R2 value while minimizing the Chi-squared value.
Results
Protein Expression, Purification, and Verification
All constructs were successfully produced, and high levels of dry protein recovered after purification consistent with previous efforts. There was some yield variability between the different constructs, likely due to differences in the OD600 at the time of induction and at the end of each run (Supporting Information).
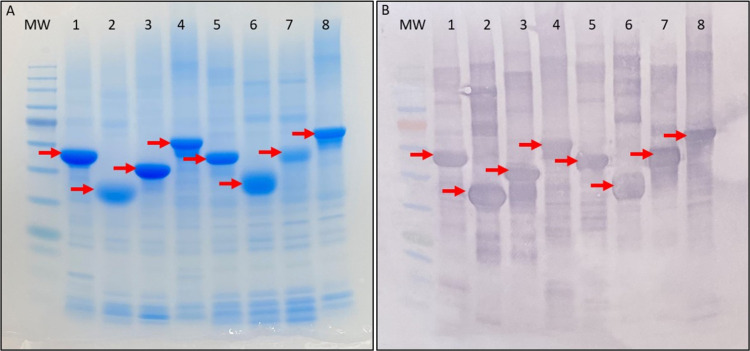
Based on ImageJ calculations, the various constructs were purified at least to the level reported by Oliviera et al., (70% pure).17 The α constructs had similar recovery levels when both of the termini were attached (N-α-C) or removed (CRD-α). However, the purity differences between N-α-C and CRD-α was a ten percent drop when the termini were removed. When only the N-terminus remained attached (N-α), the protein recovery improved with a minor impact on the relative purity (83% compared to 88% for N-α-C). The α-C had similar recovery levels as N-α-C but had similar purity as the N-α (Table 2). The N-γ-C had the lowest recovery but highest purity of the γ type proteins. Similar to α, removing both of the termini (CRD-γ) improved the yield, this time much more dramatically, but again reduced the purity by over 10%. Keeping only one terminus attached (N-γ or γ-C) resulted in similar purity levels but the N-γ had a higher recovery than the γ-C. In the SDS-PAGE Coomassie and Western Blot analyses the α constructs navigated further into the gel than the γ constructs even though the γ ones have a lower molecular weight (Figure 1), these were shown before and it is probably because of structural differences between the them.17,18
Figure 1.
Analytical protein gel and membrane of the purified constructs (indicated by the red arrows) (a) Coomassie-stained SDS-PAGE and (b) Western Blot using an antihistidine-tag antibody. Lanes are numbered from left to right as follows: MW–protein ladder; 1–N-α; 2–α central rod domain; 3–α-C; 4–N-α-C; 5–N-γ; 6–γ central rod domain; 7–γ-C; 8– N-γ-C.
Fibers Mechanical Analysis
While this study utilized an array of stretches and protein dope concentrations, this report will focus on the results of the 20% w/v dopes spun into salt water and stretched at 2X2X stretch factors. This experimental condition was chosen because of its reliability across all the constructs, with the exception of γ-CRD, which needed to be at 25% w/v to achieve the high stretch of 2X2X. This high stretch factor should reveal the maximum β-sheet formation of the recombinant proteins in this synthetic fiber formation setup. The impacts of the termini and CRD on fiber formation are best interpreted through the lens of either α or γ and not as an overarching view comparing α to γ protein constructs; the results will be described separately in the following paragraphs and Table 3 and Figure S1.
Table 3. Mechanical Properties and Structural Determination of rHIF Fibers Spun into Instant Ocean Salt Water at 2X2X Stretch Ratiosa.
| mechanical
properties |
structural composition |
||||||
|---|---|---|---|---|---|---|---|
| 20% w/v proteins at a 1:1 mixes and their components | n | toughness (MJ m–3) | tensile strength (MPa) | strain (mm mm–1) | elastic modulus (GPa) | β-sheets (%) | α-helices/random coils/turns (%) |
| N-α-C/N-γ-C | |||||||
| N-α-C | 7 | 4.04 ± 2.6b | 136 ± 41 | 0.04 ± 0.02b | 5.3 ± 0.4 | 63 | 37 |
| N-α-C/N-γ-C | 8 | 26.2 ± 17b | 126 ± 11 | 0.23 ± 0.14 | 5.1 ± 0.3 | 54 | 46 |
| N-γ-C | 9 | 1.34 ± 1.1 | 66.7 ± 17 | 0.03 ± 0.02 | 4.0 ± 0.2 | 57 | 43 |
| CRD-α/CRD-γ | |||||||
| CRD-α | 10 | 27.8 ± 12 | 184 ± 25c | 0.17 ± 0.06b | 5.0 ± 0.5 | 60 | 40 |
| CRD-α/CRD-γ | 9 | 2.25 ± 1.5b | 108 ± 27 | 0.03 ± 0.01b | 5.6 ± 1.4 | 52 | 48 |
| CRD-γ (25%) | 13 | 1.66 ± 1.0 | 93.7 ± 17b | 0.03 ± 0.01 | 4.3 ± 0.4 | 58 | 42 |
| N-α/γ-C | |||||||
| N-α | 10 | 45.0 ± 8.6 | 143 ± 19 | 0.40 ± 0.07c | 3.8 ± 0.4b | 55 | 45 |
| N-α/γ-C | 9 | 43.9 ± 9.2 | 153 ± 17 | 0.32 ± 0.07 | 5.6 ± 0.4 | 51 | 49 |
| γ-C | 10 | 1.12 ± 0.8 | 59.8 ± 11 | 0.03 ± 0.01 | 2.9 ± 0.3b | 55 | 45 |
| N-γ/α-C | |||||||
| α-C | 10 | 35.7 ± 8.4 | 130 ± 12 | 0.31 ± 0.06b | 4.4 ± 0.3b | 57 | 43 |
| N-γ/α-C | 9 | 48.1 ± 14 | 148 ± 20 | 0.37 ± 0.10 | 5.7 ± 0.6 | 50 | 50 |
| N-γ | 7 | 2.40 ± 0.9 | 122 ± 18c | 0.03 ± 0.01 | 4.8 ± 0.3c | 52 | 48 |
Here, n indicates the number of tests performed on individual fibers.
Significantly lower property values for that protein group.
Significantly higher property value for that protein group.
The α construct proteins are unable to form fibers in deionized water when both termini are removed (CRD-α) but can form robust fibers in the presence of salt water. The N-terminus of α is highly dependent on the saltwater interaction in fiber formation and can only form fibers in deionized water when the protein concentration is 25%. The γ construct proteins can form fibers in deionized water when at least one of the termini is retained but require the cation’s presence in salt water to facilitate durable fiber formation when both termini are removed (CRD-γ). This observation is similar to conclusions from research on rHIF and other IF production completed by Negishi et al, Lin et al, and Fu et al. where they found that cations provide some level of cross-linking in their synthetic fiber formation systems.10,14,25
The α protein-based groups have some notable characteristics (Tables 3 and S3), particularly in tensile strength. Here the removal of both termini (CRD-α) results in a significantly better stress value than either N-α, α-C, and N-α-C which all have relatively similar values to each other. The strain limit of the α types are all significantly different from each other. N-α has the highest strain (0.40 mm mm–1) followed by α-C but there is a drastic drop to CRD-α (0.31 to 0.17 mm mm–1 for α-C to CRD-α) and to N-α-C (0.04 mm mm–1). The α type fibers perform relatively similarly in terms of toughness, except for N-α-C which is significantly worse than the other variations. The α proteins with both termini intact (N-α-C) or both removed (CRD-α) has no impact on the elastic modulus. However, keeping just the C-terminus decreases the elastic modulus while keeping just the N-terminus further decreases the elastic modulus.
In terms of tensile strength, the N-γ (122 MPa) is the best of the γ protein types (Tables 3 and S5), followed by CRD-γ (93.7 MPa), and then a last place tie between N-γ-C and γ-C (66.7 and 59.8 MPa). The strain limit and toughness of the γ protein types are not impacted by the inclusion or removal of either the termini since all groups tested had the same average strain value (0.03 mm mm–1) and similar toughness values. Including only one terminus on the γ protein results in the two extremes for elastic modulus. The N-γ (4.8 GPa) has the highest while the γ-C (2.9 GPa) has the lowest elastic modulus value with the termini inclusive and exclusive (N-γ-C and CRD-γ, respectively) grouped in the middle.
The mix types were created by mixing equal amounts of corresponding α and γ constructs. These were the full-length constructs, the CRD constructs, and then combining N-α with γ-C and vice versa to obtain hybrid mixes with one variety of each terminus in the dope. The CRD construct mixes formed fibers in salt water and not in deionized water, potentially due to cation-based cross-linking from the salt content to form fibers robust enough for processing.10,14,25 However, we have not investigated the presence of cation-based cross-linking in this study. The hybrid mixes were not as reliant on the saltwater interaction to form fibers and could form fibers in deionized water but fared better when fiber formation occurred in saltwater. The mix types (Tables 3 and S4) were consistent in terms of tensile strength and elastic modulus performance with relatively similar stress and elastic modulus values across all varieties in this system. The worst strain value of the mix types was achieved with the CRD mix (0.03 mm mm–1), indicating that when in a combination dope, the termini are helpful for extensibility. The hybrid mixes had the best toughness (48.1 MJ m3 for N-γ/α-C and 43.9 MJ m3 for N-α/γ-C), indicating that having just one of the termini, regardless of which CRD it was attached to, improved fiber toughness. However, including both termini (N-α-C/N-γ-C) on both proteins weakened the fibers (26.2 MJ m3), though not as much as removing all the termini did (CRD-α/CRD-γ 2.25 MJ m3).
Fibers Structural Analysis
Based on the deconvolution process described, which was based on works by Oliveira et al., Hu et al., Böni et al., and Zou et al., the absorbance assignments for β-sheet and β-turn regions were around 1625 and 1683 cm–1.17,22−24 The absorbance occurring around 1652 cm–1 corresponds to random coils and turns. These curve assignments closely resemble what Böni et al. discovered when performing FTIR analysis on formic acid–based hagfish intermediate films.23 For all of the constructs, the α constructs consistently had the highest β-sheet level, with the full-length α having the highest at 62.8% β-sheet content. The CRDs of the proteins had similar amounts of β-sheet content to their corresponding full-length constructs, indicating that the termini do not contribute much to the β-sheet content of the stretched fibers in this system. The fibers of constructs with only one of the termini remaining had lower β-sheet content than their original and CRD variants (see Table 3), indicating that these domains contribute more to the α-coil/random turns than they do to the formation of β-sheet regions, similar to Fudge et al. observations.15
Discussion
The expression and purification of recombinant α and γ protein constructs yielded protein pellets with purity between 78–91% (Table 1) that was used for fiber formation and testing. Successful fiber formation of these protein domains revealed mechanical and structural properties that can build on the structure–function relationship of these proteins in a synthetic fiber manufacturing system. In the expression and purification stage, was observed a general increase in protein recovery with similar purity of the protein constructs compared to full-length rHIF (Table 2). An analysis by Francis and Page supports the increase in recovery yield of the target protein when the size of the protein is reduced.26 Additionally, Schindler et al. found that up to a 370% increase in recovery yield can be achieved by reducing the size of the construct compared to the full-length protein.27
This study focused on 20% w/v dopes spun into fibers in saltwater baths and processed at a 2X2X stretch for mechanical and structural analysis. Additional spins using 15, 20, and 25% concentrations were done using deionized water and saltwater at various stretch factors, which were also performed and are available in the Supporting Information (Tables S3, S4 and S5). The concentration, bath contents, and stretch factor were chosen because of the reliability of those factors across the different constructs. Both of the CRD constructs were able to form fibers only in salt water, probably due to cationic cross-linking from the components of the saltwater (around 400 mg L–1 Ca2+ and 1320 mg L–1 Mg2+, per Instant Ocean’s instructions).
For the synthetically formed α protein fiber types, the tensile strength of the fibers is best when neither of the termini are attached (CRD-α) with the N-α-C, N-α, and α-C having similar strength values. This observation indicates that the termini interfere with the fiber structure enough to impact the strength of the fiber. The N-terminus of α (N-α) contributes the most to fiber extensibility in this system while the C-terminus contributes in a lesser manner. However, when both the N and C-termini are attached to α (N-α-C), there are detrimental effects on the extensivity of the fiber. Additionally, the inclusion of both termini on α produces the most brittle α-type fibers in the system, suggesting that having both termini attached interferes with ideal fiber formation. The elastic modulus of the α-type fibers is impacted by the presence of the termini with softer fibers occurring when just one terminus is attached (N-α or α-C) while having both or none (N-α-C or CRD-α) having similar modulus values.
Overall, the synthetically produced γ type fibers are primarily impacted by the N-terminus for higher strength and elastic modulus while the C-terminus lowers the strength and elastic modulus while strain and toughness are not impacted by either terminus. The synthetically produced γ type protein fibers achieve the best tensile strength when only the N-terminus is attached (N-γ). The C-terminus on γ, either alone or together with the N-terminus, has a detrimental effect on the tensile strength of the fibers. The γ type fibers in this system all produced the same average strain values, indicating that the termini do not have an impact on strain for γ fibers. Additionally, the overall toughness of these fibers was not significantly impacted by the termini, indicating that they do not contribute significantly to the fiber’s toughness. Similar to the tensile strength observations, the elastic modulus is significantly higher when only the N-terminus is attached (N-γ) while the incorporation of the C-terminus softens the fibers.
Generally, when corresponding α and γ constructs are combined in a 1:1 mixture, the resulting fibers have mechanical properties that are at least in between their constituents, with some combinations and properties improving beyond their constituents. The most notable result is from a hybrid mixture that combined N-γ and α-C, resulting in an improvement of all mechanical properties compared to the corresponding constituent’s fibers. This could be from optimal interaction between the proteins that allows for improved protein alignment which was not captured in the FTIR analysis. The natural hagfish α and γ form a coiled-coil structure that could be a key component for the natural mechanical properties and could be what is happening with the N-γ and α-C hybrid combination but this cannot be confirmed with the methodologies used in this study.11,12
Removing both termini resulted in similar β-sheet/turn content as keeping both termini, indicating that the termini do not contribute significant additional β-sheet/folds in the stretched fibers in this synthetic system. This discovery remains true when looking at the recombinant proteins with one terminus removed having lower β-sheet/turn content than the CRD and original recombinant protein stretched fibers.
rHIF has been proposed as a potential candidate for the development of several novel biomaterials. Full-length rHIF has been utilized in pilot experiments, forming fibers, films, a mimetic model of Bruch’s membrane, and a three-dimensional (3D) biocompatible scaffold.10,17,23,28,29 The range of mechanical properties observed by the rHIF protein constructs, along with the high recovery yield, invites further investigation regarding the structure–function relationships of rHIF and other IF proteins, which is warranted for the development of novel protein-based biomaterials.
Conclusions
In conclusion, our study successfully demonstrated the recombinant expression, purification, and fiber formation of α and γ protein constructs, highlighting the roles of different protein domains in their structure–function relationships within our engineered spinning process. The increased protein recovery and purity observed during the purification stages align with previous findings that smaller protein sizes enhance expression yields. The lack of significant differences in β-sheet/turn content and that the mechanical properties of the CRD-only fibers were superior suggests that the termini do not contribute additional structural elements in these stretched fibers. Notably, the removal of both termini in α constructs resulted in fibers with the highest tensile strength, while the inclusion of the N-terminus in γ constructs improved both tensile strength and elastic modulus. While all the constructs studied formed fibers in saltwater, some of them were unable to form fibers in deionized water. This observation, coupled with generally improved mechanical performance of the fibers spun in saltwater over deionized water, hints that these recombinant intermediate filament constructs have some reliance on cationic cross-linking for synthetic fiber formation.10
Our results suggest that the termini play a role in the assembly process of these recombinant proteins, though their exact function in this synthetic system regarding influence on fiber assembly and structure remains unclear. These insights into protein domain functions in our synthetic system could pave the way for designing tailored biomaterials with specific mechanical properties for various applications. By investigating engineered constructs containing variations of protein domains, we gain a deeper understanding of how different protein domains contribute to fiber formation and mechanical properties in this synthetic context. Our study serves as a foundation for future research and development in the field of recombinant protein-based materials, demonstrating the potential for customizing fiber properties through protein domain engineering.
Acknowledgments
We acknowledge the use of facilities within the Eyring Materials Center at Arizona State University, supported in part by NNCI-ECCS-2025490. We also acknowledge the use of facilities within the United States Navy’s Naval Surface Warfare Center in Panama City.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.4c06950.
Amino acid sequences; bioreactor results; and full mechanical data are available in the “supplemental to recombinant HIF protein constructs reveal structure-function relationships” file (PDF)
Author Contributions
† O.W. and P.E.O. contributed equally to this work.
This research was supported by the United States Department of Defense–Navy: N6133120D0001
The authors declare no competing financial interest.
Supplementary Material
References
- Nishimura Y.; Kasahara K.; Inagaki M. Intermediate Filaments and IF-Associated Proteins: From Cell Architecture to Cell Proliferation. Proc. Jpn. Acad., Ser. B 2019, 95 (8), 479–493. 10.2183/pjab.95.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H.; Aebi U. Intermediate Filaments: Structure and Assembly. Cold Spring Harbor Perspect. Biol. 2016, 8 (11), a018242 10.1101/cshperspect.a018242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X.; Kato M.; McKnight S. L. How Do Disordered Head Domains Assist in the Assembly of Intermediate Filaments?. Curr. Opin. Cell Biol. 2023, 85, 102262 10.1016/j.ceb.2023.102262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier E. E.; Janmey P. A.. Mechanical Properties of Intermediate Filament Proteins. In Methods in Enzymology; Elsevier, 2016; Vol. 568, pp 35–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H.; Häner M.; Brettel M.; Müller S. A.; Goldie K. N.; Fedtke B.; Lustig A.; Franke W. W.; Aebi U. Structure and Assembly Properties of the Intermediate Filament Protein Vimentin: The Role of Its Head, Rod and Tail Domains. J. Mol. Biol. 1996, 264 (5), 933–953. 10.1006/jmbi.1996.0688. [DOI] [PubMed] [Google Scholar]
- Falvo M. R.; Gorkun O. V.; Lord S. T. The Molecular Origins of the Mechanical Properties of Fibrin. Biophys. Chem. 2010, 152 (1), 15–20. 10.1016/j.bpc.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F.; Li W.; Deng J.; Kan J.; Guo T.; Wang B.; Hao S. Recombinant Human Hair Keratin Nanoparticles Accelerate Dermal Wound Healing. ACS Appl. Mater. Interfaces 2019, 11 (20), 18681–18690. 10.1021/acsami.9b01725. [DOI] [PubMed] [Google Scholar]
- Zintzen V.; Roberts C. D.; Anderson M. J.; Stewart A. L.; Struthers C. D.; Harvey E. S. Hagfish Predatory Behaviour and Slime Defence Mechanism. Sci. Rep. 2011, 1 (1), 131 10.1038/srep00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudge D. S.; Levy N.; Chiu S.; Gosline J. M. Composition, Morphology and Mechanics of Hagfish Slime. J. Exp. Biol. 2005, 208 (24), 4613–4625. 10.1242/jeb.01963. [DOI] [PubMed] [Google Scholar]
- Negishi A.; Armstrong C. L.; Kreplak L.; Rheinstadter M. C.; Lim L.-T.; Gillis T. E.; Fudge D. S. The Production of Fibers and Films from Solubilized Hagfish Slime Thread Proteins. Biomacromolecules 2012, 13 (11), 3475–3482. 10.1021/bm3011837. [DOI] [PubMed] [Google Scholar]
- Spitzer R. H.; Downing S. W.; Koch E. A.; Salo W. L.; Saidel L. J. Hagfish Slime Gland Thread Cells. II. Isolation and Characterization of Intermediate Filament Components Associated with the Thread. J. Cell Biol. 1984, 98 (2), 670–677. 10.1083/jcb.98.2.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch E. A.; Spitzer R. H.; Pithawalla R. B.; Parry D. A. An Unusual Intermediate Filament Subunit from the Cytoskeletal Biopolymer Released Extracellularly into Seawater by the Primitive Hagfish (Eptatretus stouti). J. Cell Sci. 1994, 107 (11), 3133–3144. 10.1242/jcs.107.11.3133. [DOI] [PubMed] [Google Scholar]
- Schaffeld M.; Schultess J. Genes Coding for Intermediate Filament Proteins Closely Related to the Hagfish “Thread Keratins (TK)” α and γ Also Exist in Lamprey, Teleosts and Amphibians. Exp. Cell Res. 2006, 312 (9), 1447–1462. 10.1016/j.yexcr.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Fu J.; Guerette P. A.; Miserez A. Self-Assembly of Recombinant Hagfish Thread Keratins Amenable to a Strain-Induced α-Helix to β-Sheet Transition. Biomacromolecules 2015, 16 (8), 2327–2339. 10.1021/acs.biomac.5b00552. [DOI] [PubMed] [Google Scholar]
- Fudge D. S.; Gardner K. H.; Forsyth V. T.; Riekel C.; Gosline J. M. The Mechanical Properties of Hydrated Intermediate Filaments: Insights from Hagfish Slime Threads. Biophys. J. 2003, 85 (3), 2015–2027. 10.1016/S0006-3495(03)74629-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J.; Guerette P.; Pavesi A.; Horbelt N.; Lim C. T.; Harrington M.; Miserez A. Artificial Hagfish Protein Fibers with Ultra-High and Tunable Stiffness. Nanoscale 2017, 9, 12908–12915. 10.1039/C7NR02527K. [DOI] [PubMed] [Google Scholar]
- Oliveira P. E.; Chen D.; Bell B. E.; Harris T. I.; Walker C.; Zhang H.; Grob B.; Lewis R. V.; Jones J. A. The next Generation of Protein Super-Fibres: Robust Recombinant Production and Recovery of Hagfish Intermediate Filament Proteins with Fibre Spinning and Mechanical–Structural Characterizations. Microb. Biotechnol. 2021, 14 (5), 1976–1989. 10.1111/1751-7915.13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell B. E.; Burton I. K.; Arreola-Patino J.; Harris T. I.; Oliveira P.; Chen D.; Lewis R. V.; Jones J. A. Scalable Purification of Recombinant Structural Proteins, Hagfish Intermediate Filament α and γ, from Inclusion Bodies for Fiber Formation. Protein Expr. Purif. 2022, 199, 106152 10.1016/j.pep.2022.106152. [DOI] [PubMed] [Google Scholar]
- Fudge D. S.; Hillis S.; Levy N.; Gosline J. M. Hagfish Slime Threads as a Biomimetic Model for High Performance Protein Fibres. Bioinspiration Biomimetics 2010, 5 (3), 035002 10.1088/1748-3182/5/3/035002. [DOI] [PubMed] [Google Scholar]
- Copeland C. G.; Bell B. E.; Christensen C. D.; Lewis R. V. Development of a Process for the Spinning of Synthetic Spider Silk. ACS Biomater. Sci. Eng. 2015, 1 (7), 577–584. 10.1021/acsbiomaterials.5b00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. A.; Harris T. I.; Tucker C. L.; Berg K. R.; Christy S. Y.; Day B. A.; Gaztambide D. A.; Needham N. J. C.; Ruben A. L.; Oliveira P. F.; Decker R. E.; Lewis R. V. More Than Just Fibers: An Aqueous Method for the Production of Innovative Recombinant Spider Silk Protein Materials. Biomacromolecules 2015, 16 (4), 1418–1425. 10.1021/acs.biomac.5b00226. [DOI] [PubMed] [Google Scholar]
- Hu X.; Kaplan D.; Cebe P. Determining Beta-Sheet Crystallinity in Fibrous Proteins by Thermal Analysis and Infrared Spectroscopy. Macromolecules 2006, 39 (18), 6161–6170. 10.1021/ma0610109. [DOI] [Google Scholar]
- Böni L. J.; Sanchez-Ferrer A.; Widmer M.; Biviano M. D.; Mezzenga R.; Windhab E. J.; Dagastine R. R.; Fischer P. Structure and Nanomechanics of Dry and Hydrated Intermediate Filament Films and Fibers Produced from Hagfish Slime Fibers. ACS Appl. Mater. Interfaces 2018, 10 (47), 40460–40473. 10.1021/acsami.8b17166. [DOI] [PubMed] [Google Scholar]
- Zou Y.; Li Y.; Hao W.; Hu X.; Ma G. Parallel β-Sheet Fibril and Antiparallel β-Sheet Oligomer: New Insights into Amyloid Formation of Hen Egg White Lysozyme under Heat and Acidic Condition from FTIR Spectroscopy. J. Phys. Chem. B 2013, 117 (15), 4003–4013. 10.1021/jp4003559. [DOI] [PubMed] [Google Scholar]
- Lin Y.-C.; Broedersz C. P.; Rowat A. C.; Wedig T.; Herrmann H.; MacKintosh F. C.; Weitz D. A. Divalent Cations Crosslink Vimentin Intermediate Filament Tail Domains to Regulate Network Mechanics. J. Mol. Biol. 2010, 399 (4), 637–644. 10.1016/j.jmb.2010.04.054. [DOI] [PubMed] [Google Scholar]
- Francis D. M.; Page R. Strategies to Optimize Protein Expression in Escherichia coli. Curr. Protoc. Protein Sci. 2010, 61 (1), 5–24. 10.1002/0471140864.ps0524s61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler S.; Missbichler B.; Walther C.; Sponring M.; Cserjan-Puschmann M.; Auer B.; Schneider R.; Dürauer A. N(pro) Fusion Technology: On-Column Complementation to Improve Efficiency in Biopharmaceutical Production. Protein Expr. Purif. 2016, 120, 42–50. 10.1016/j.pep.2015.11.021. [DOI] [PubMed] [Google Scholar]
- Rickabaugh E.; Weatherston D.; Harris T. I.; Jones J. A.; Vargis E. Engineering a Biomimetic In Vitro Model of Bruch’s Membrane Using Hagfish Slime Intermediate Filament Proteins. ACS Biomater. Sci. Eng. 2023, 9 (8), 5051–5061. 10.1021/acsbiomaterials.3c00411. [DOI] [PubMed] [Google Scholar]
- Dastjerdi M. B.; Amini A.; Nazari M.; Cheng C.; Benson V.; Gholami A.; Ghasemi Y. Novel Versatile 3D Bio-Scaffold Made of Natural Biocompatible Hagfish Exudate for Tissue Growth and Organoid Modeling. Int. J. Biol. Macromol. 2020, 158, 894–902. 10.1016/j.ijbiomac.2020.05.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.