Abstract
Ribosome biogenesis plays a pivotal role in maintaining stem cell homeostasis, yet the precise regulatory mechanisms governing this process in mouse embryonic stem cells (mESCs) remain largely unknown. In this investigation, we ascertain that DEAD-box RNA helicase 10 (DDX10) is indispensable for upholding cellular homeostasis and the viability of mESCs. Positioned predominantly at the nucleolar dense fibrillar component (DFC) and granular component (GC), DDX10 predominantly binds to 45S ribosomal RNA (rRNA) and orchestrates ribosome biogenesis. Degradation of DDX10 prevents the release of U3 snoRNA from pre-rRNA, leading to perturbed pre-rRNA processing and compromised maturation of the 18S rRNA, thereby disrupting the biogenesis of the small ribosomal subunit. Moreover, DDX10 participates in the process of liquid-liquid phase separation (LLPS), which is necessary for efficient ribosome biogenesis. Notably, the NUP98-DDX10 fusion associated with acute myelocytic leukemia (AML) alters the cellular localization of DDX10 and results in loss of ability to regulate pre-rRNA processing. Collectively, this study reveals the critical role of DDX10 as a pivotal regulator of ribosome biogenesis in mESCs.
Subject terms: Pluripotency, Embryonic stem cells, RNA
Ribosome biogenesis is crucial for maintaining stem cell homeostasis. Here, authors reveal that DDX10 plays a critical role in ribosome biogenesis by regulating 18S rRNA maturation, which is vital for the proliferation and maintenance of mESCs.
Introduction
ESCs, derived from the inner cell mass (ICM) of pre-implantation blastocysts, are characterized by rapid proliferation and pluripotency, enabling them to self-renew and differentiate into diverse cell types1,2. The determination of ESC fate hinges upon intricate regulatory mechanisms that orchestrate gene expression across various tiers, encompassing chromatin, transcriptional, and post-transcriptional regulation. These regulatory processes have been extensively scrutinized in stem cells3–8. Recently, emerging evidence has underscored the pivotal role of ribosome biogenesis in upholding ESC identity9,10. ESCs exhibit robust rRNA transcription and heightened ribosome biogenesis11–14. Safeguarding chromatin integrity at actively transcribed rDNA loci shields them from epigenetic silencing, thereby promoting rRNA transcription and ribosome biogenesis, pivotal for sustaining ESC self-renewal15. Maintaining steady-state ribosome biogenesis is imperative for the maintenance of ESC homeostasis9. In ESCs, there is pronounced expression of small subunit (SSU) processome genes, ensuring efficient processing of pre-rRNA and maturation of rRNA, which are vital for preserving pluripotency. Deletion of SSU processome genes leads to diminished protein synthesis and loss of pluripotency in ESCs13. Notably, recent investigations have unveiled a dichotomy: while undifferentiated ESCs demonstrate relatively lower polysome loading compared to differentiated progeny, they still expend considerable energy to sustain an abundant ribosome pool10. In contrast, there is an increase in polysome loading, protein synthesis, and protein content that occurs during differentiation16–18. Thus, ribosome biogenesis is a linchpin for maintaining pluripotent stem cells and orchestrating their differentiation. Despite its pivotal role, the mechanisms governing ribosome biogenesis in ESCs remain unclear, warranting further exploration and study.
The nucleolus, a highly prominent and extensively studied membraneless ribonucleoprotein (RNP) entity, is a pivotal hub for ribosome biogenesis19,20. Comprising three distinct subregions – the fibrillar center (FC), the DFC, and the GC – the nucleolus plays a multifaceted role in orchestrating ribosome assembly. Within this intricate landscape, rDNA transcription occurs at the FC-DFC boundary, pre-rRNA processing takes place within the DFC, and the subsequent assembly of ribosomal subunits is executed within the GC20–22.
Ribosomal genes are transcribed by RNA polymerase I (Pol I) to produce the primary 47S rRNA precursor, which includes two external transcribed spacers (5’ETS and 3’ETS) and two internal transcribed spacers (ITS1 and ITS2) separating the mature 18S, 5.8S, and 28S rRNAs. To obtain these mature rRNAs, the transcribed spacers must be removed through a sequential series of endonucleolytic and exonucleolytic cleavages23. In mouse cells, the 47S rRNA transcript is first cleaved at site A0, generating the 46S rRNA, and then at site 6, producing the 45S rRNA. The processing of mouse 45S rRNA occurs primarily through two pathways. In pathway 1, sites A0 and 1 in the 5’ETS are successively cleaved successively to produce 43S and 41S rRNA. Subsequently, site 2c in ITS1 is cleaved to produce 20S rRNA (precursor of 18S rRNA) and 36S rRNA (precursor of 28S and 5.8S rRNA). In pathway 2, site 2c in ITS1 is firstly cleaved to produce 34S rRNA and 36S rRNA. Subsequently, sites A0 and 1 in 34S rRNA are cleaved in sequence to produce 20S rRNA. These rRNA precursors are ultimately processed into mature rRNA23,24. Ribosome biogenesis is a multi-step process underpinned by the coordinated interplay of numerous proteins and non-coding RNAs (ncRNAs). These entities collectively govern the intricate choreography of events encompassing rRNA transcription25–27, directed trafficking of nascent pre-rRNA28, and pre-rRNA processing29–31. Some small nucleolar RNAs (snoRNAs) serve as scaffolds during snoRNPs formation and base pairing with pre-rRNA to guide the directional cleavage and folding of pre-rRNA32,33, which is crucial for rRNA maturation. U3, U14, U22, U17/snR30, and snR83 affect the maturation of 18S rRNA34–39, while U8 snoRNA is essential for the accumulation of mature 5.8S and 28S rRNAs40.
DExD/H-box RNA helicases belong to the RNA-binding protein (RBP) family and are the largest consortium of RNA helicases, ubiquitously present across diverse organisms41. Their pivotal functions encompass remodeling RNA structures by harnessing the energy derived from ATP hydrolysis, thereby influencing multiple facets of cellular RNA metabolism, including transcription, splicing, ribosome biogenesis, RNA export, translation, RNA turnover, and organelle gene expression41–43. Among these, DDX10 is a constituent of the DEAD-box RNA helicases44,45. Existing literature underscores DDX10’s association with various tumors, observing abnormal expression within tumor tissues46–49. Nonetheless, the specific physiological and molecular contributions of DDX10 within ESCs remain unknown.
In this study, we unearthed that DDX10 is a crucial regulator of ribosome biogenesis and is essential for proliferation and maintenance of cell fate in mESCs. The degradation of DDX10 induces cell cycle arrest at the G1 phase while promoting apoptosis, potentially through a p53-dependent mechanism. Notably, the deficiency of DDX10 causes disruptions in pre-rRNA processing, manifesting as diminished 18S rRNA maturation and compromised ribosome biogenesis. Moreover, DDX10 interacts with the components of the SSU processome, and its absence hindering the liberation of U3 snoRNA from pre-rRNA. These insights collectively illuminate the integral role of DDX10 in maintaining proper ribosome biogenesis. Further, DDX10 undergoes LLPS, which is crucial for ribosome biogenesis. Finally, our findings show that NUP98-DDX10 fusion protein results in the loss of DDX10 function in regulating ribosome biogenesis.
Results
DDX10 is indispensable for the survival and maintenance of mESCs
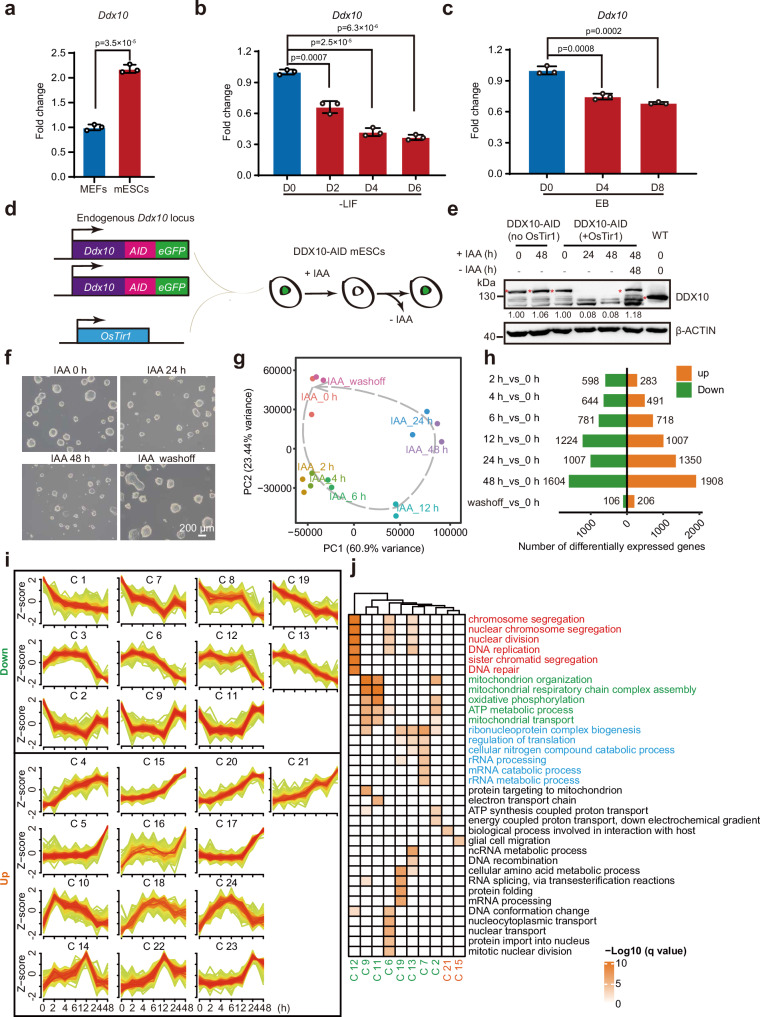
To delve into the role of DDX10 in mESCs, we initiated our investigation by assessing Ddx10 expression in mESCs and mouse embryonic fibroblasts (MEFs). Our findings unveiled heightened Ddx10 expression levels in mESCs, in contrast to MEFs (Fig. 1a). Throughout ESC differentiation, Ddx10 displayed robust expression in ESCs, which underwent rapid downregulation (Fig. 1b, c). Leveraging the auxin-inducible degron system50,51, we aimed to degrade endogenous DDX10 in mESCs. Employing the CRISPR-Cas9 genome editing technique, we introduced the AID-eGFP sequence at the stop codon of Ddx10 (Fig. 1d and Supplementary Fig. 1a). Subsequent exposure of cells to the auxin analog indole-3-acetic acid (IAA) resulted in the rapid degradation of DDX10, becoming undetectable within 2 h of IAA treatment, while regaining initial levels post IAA removal, thereby confirming the efficacy of the degradation system (Fig. 1e and Supplementary Fig. 1b, c). We observed that the protein level of DDX10 in DDX10-AID (+ OsTir1) mESCs was lower compared to wild-type mESCs (Fig. 1e). However, this reduction could not lead to significant changes in cell morphology, cell cycle, and apoptosis (Supplementary Fig. 1d-f). We observed that DDX10 degradation resulted in smaller and flattened mESC clones, with morphological recovery upon IAA withdrawal (Fig. 1f and Supplementary Fig. 1g). Scrutinizing the influence of DDX10 degradation on mESC pluripotency, we evaluated the expression of pluripotency transcription factors (OCT4, NANOG, and SOX2) revealing minimal impact due to DDX10 degradation (Supplementary Fig. 1h). Moreover, our data spotlighted that DDX10 degradation significantly impeded mESC proliferation (Supplementary Fig. 1i, j), provoking cell cycle arrest at the G1 phase (Supplementary Fig. 1k, l), while instigating apoptosis (Supplementary Fig. 1m, n).
Fig. 1. Disruption of gene expression upon acute degradation of DDX10 in mESCs.
a RT-qPCR analysis for endogenous levels of Ddx10 mRNA in MEFs and mESCs. b RT-qPCR analysis of Ddx10 expression levels following LIF withdrawal. c RT-qPCR analysis of Ddx10 expression levels during embryoid body (EB) differentiation. d Schematic illustration of the generation of DDX10-AID mESCs. e Western blot analysis of DDX10 protein levels in DDX10-AID cells with or without IAA treatment. WT: wild-type E14 mESCs. The red asterisk indicates endogenous DDX10-AID-eGFP and DDX10 proteins. β-ACTIN serves as the loading control. Experiments were repeated three times independently with similar results. f Brightfield images of DDX10-AID (+ OsTir1) mESC colonies with or without IAA treatment. Experiments were repeated three times independently with similar results. Scale bar, 200 µm. g Principal component analysis (PCA) plot displays RNA-seq data from DDX10-AID (+ OsTir1) mESCs treated with IAA at different time points or treated with IAA for 48 h followed by 48 h of washing. h Bar plots showing the number of differentially expressed genes (DEGs) upon DDX10 degradation at different time points with IAA treatment (orange: upregulated genes, green: downregulated genes). i Line chart illustrating gene expression patterns of 24 different clusters of DEGs. j Heatmap presenting gene ontology results for genes in each cluster. For (a–c) transcription levels were normalized against Gapdh. Data are presented as mean values ± SD with the indicated significance from two-sided t-test. Exact p-values are reported in the figure. n = 3 independent experiments. Source data are provided as a Source Data file.
To investigate the molecular anomalies ensuing from DDX10 degradation, we executed RNA-seq experiments on DDX10-AID mESCs, both in the presence and absence of IAA treatment. Through correlation analysis, we validated the high reproducibility among replicates. Gradual changes in gene expression patterns were discernible beginning at 2 h post-IAA treatment. Yet, the pattern closely resembled that of untreated cells at 48 h after IAA withdrawal (Fig. 1g). This temporal dynamic suggests that gene expression alterations resulting from DDX10 degradation are reversible. Compared with untreated cells, the number of differentially expressed genes (DEGs) gradually increased during IAA treatment (Fig. 1h and Supplementary Data 1). Subsequently, we clustered the DEGs at all time points into 24 groups based on their expression patterns (Fig. 1i). Notably, downregulated gene clusters predominantly associated with cell division, energy metabolism, and RNA metabolism (Fig. 1j). To further investigate the effect of DDX10 degradation on cell fate determination, we analyzed the RNA-seq data from primed (cultured in medium containing serum) and naive (cultured in medium containing 2i) mESCs52, and integrated these data with our RNA-seq data. PCA results showed that mESCs following DDX10 degradation were neither close to the primed nor to the naive mESCs (Supplementary Fig. 2a).
Then we perform Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis on the upregulated genes, which revealed significant enrichment of the p53 signaling pathway (Supplementary Fig. 2b). Concretely, genes like Mdm2 and cyclin-dependent kinase inhibitor 1 A (Cdkn1a/p21) that are associated with the p53 pathway, as well as pro-apoptotic genes Bbc3 and Pmaip1 exhibited notable upregulation post DDX10 degradation (Supplementary Fig. 2c, d). Collectively, these findings underscore that DDX10 degradation prompts cell cycle arrest and propels apoptosis by activating the p53 signaling pathway, consequently impeding mESC growth. Previous studies have shown that p53 activation can induce the transition of mESCs to 2-cell-like cells (2CLCs)53,54. Therefore, we compared our RNA-seq data with the results from previously published 2-cell data55, and observed that 2-cell specific genes, such as Zscan4b and Zscan4d, were significantly activated after 24 h of IAA treatment (Supplementary Fig. 2e, f). Together, these results indicate that DDX10 degradation promotes the transition of mESCs to 2CLCs.
DDX10 localizes to the nucleolar DFC and GC and primarily binds to 45S rRNA
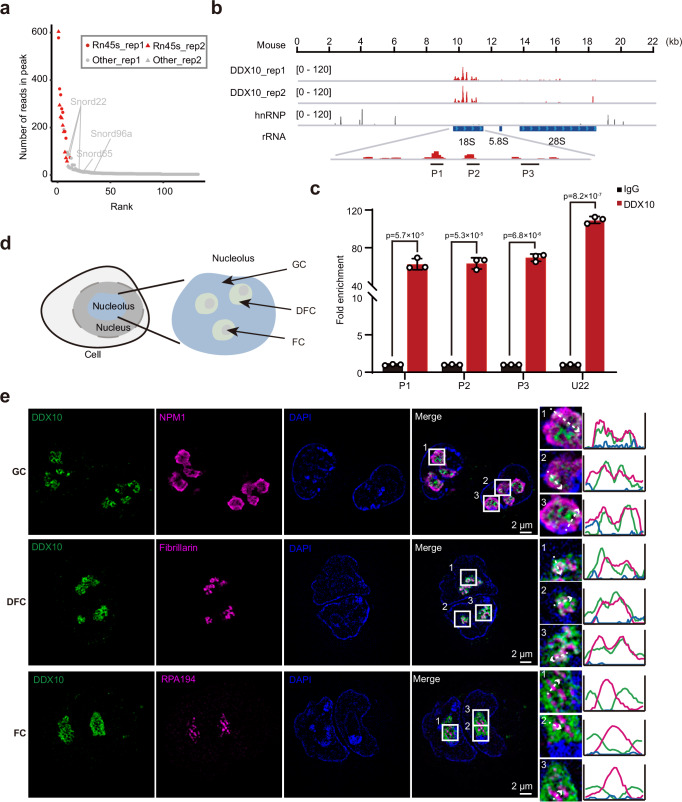
Given DDX10’s classification as an RNA binding protein, we executed crosslinking immunoprecipitation followed by high-throughput sequencing (CLIP-seq) to unveil its downstream targets in mESCs. Despite initial failures with both commercial and self-made anti-DDX10 antibodies for CLIP-seq, we engineered mESCs overexpressing FLAG-tagged DDX10, revealing nuclear localization of DDX10-FLAG (Supplementary Fig. 3a). Subsequently, we conducted CLIP-seq experiments by using anti-FLAG M2 magnetic beads to capture DDX10-bound RNAs (Supplementary Fig. 3b). Significantly, our findings illuminated that DDX10 strongly binds to 45S rRNA, with a particular preference for the 18S rRNA sequence (Fig. 2a, b). Furthermore, our data unveiled an association of DDX10 with a subset of snoRNAs, including U22 (Fig. 2a and Supplementary Data 2), recognized for guiding site-specific pre-rRNA cleavage and influencing 18S rRNA processing36. These results were further validated by RNA immunoprecipitation (RIP)-qPCR (Fig. 2c).
Fig. 2. Localization and binding preferences of DDX10 in the nucleolus.
a Scatter plot showcasing the read distribution in DDX10 peaks from CLIP-seq data. b Genomic tracks illustrating DDX10 enrichment at 45S rRNA through CLIP-seq. Control hnRNP CLIP-seq data was used. c RIP-qPCR validating DDX10 positive binding loci in FLAG-tagged DDX10 mESCs. Data are presented as mean values ± SD with the indicated significance from two-sided t-test. Exact p-values are reported in the figure. n = 3 independent experiments. d Schematic representation illustrating the nucleolus structure. e Representative SIM images of DDX10 (green), GC marker NPM1 (magenta), DFC marker Fibrillarin (magenta), and FC marker RPA194 (magenta) in mESCs. Experiments were repeated three times independently with similar results. Scale bar, 2 µm. Source data are provided as a Source Data file.
With nuclear localization of DDX10 in mind (Supplementary Fig. 3a) and its pronounced binding to 45S rRNA (Fig. 2a, b), predominantly found in the nucleolus, we conjectured that DDX10 might inhabit the nucleolus of mESCs. A verification of this notion was undertaken through immunofluorescent staining, co-staining cells with antibodies targeting DDX10, and the nucleolar marker Fibrillarin. As anticipated, DDX10 was co-localized with Fibrillarin (Supplementary Fig. 3c). The nucleolus, partitioned into three distinct subregions from interior to exterior: FC, DFC, and GC (Fig. 2d), prompted us to delve deeper into the precise nucleolar localization of DDX10. Our approach entailed antibody staining of DDX10, NPM1 (GC marker), Fibrillarin (DFC marker), and RPA194 (FC marker), followed by structured illumination microscopy (SIM) visualization. Intriguingly, our data revealed that DDX10 overlaps more significantly with Fibrillarin than with NPM1, but not with RPA194 (Fig. 2e). This observation strongly suggested DDX10’s positioning within the DFC and GC of the nucleolus, implying its involvement in pre-rRNA processing and ribosome biogenesis.
DDX10 degradation disrupts ribosome biogenesis followed by an impaired translation and disordered nucleolar structure
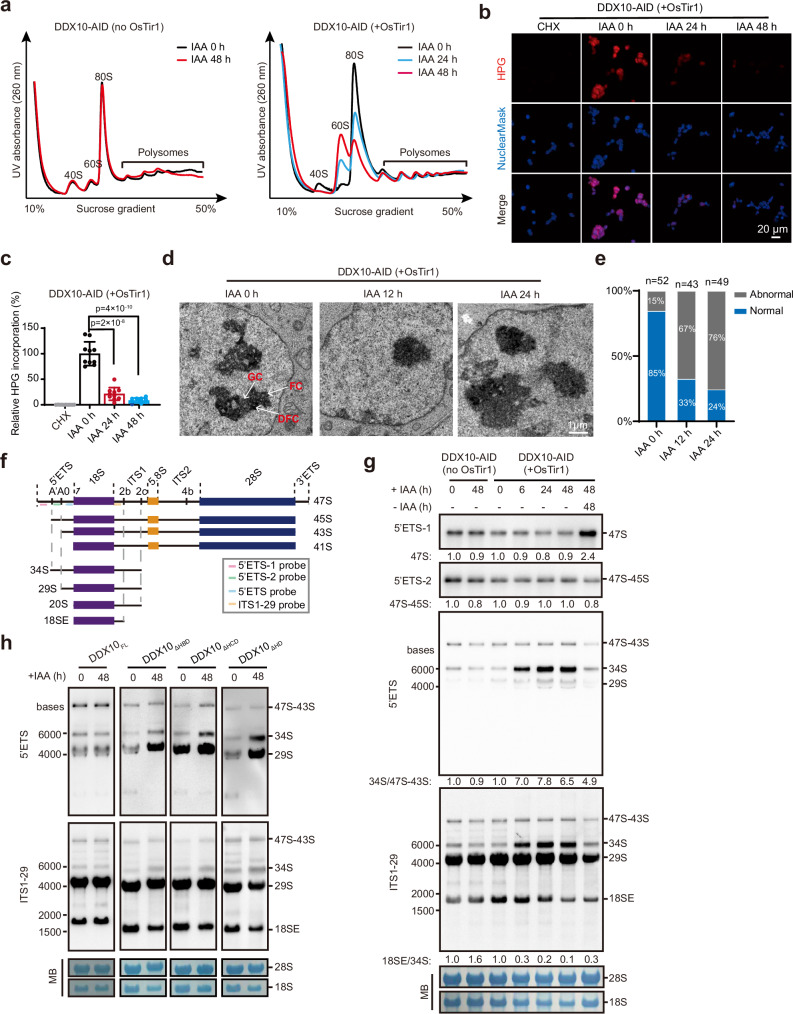
To delve deeper into the functional necessity of DDX10 in ribosome biogenesis, we executed sucrose gradient assays coupled with ribosomal fractionation to examine ribosome abundance in mESCs with and without DDX10 degradation. Strikingly, upon DDX10 degradation, a remarkable reduction in free small ribosomal subunit (40S) and ribosome (80S) content was evident, concomitant with an accumulation of free large ribosomal subunit (60S). Conversely, no discernible shifts were noted in control cells following IAA treatment (Fig. 3a). These findings definitively demonstrate that the absence of DDX10 hinders the biogenesis of the 40S ribosomal subunit, leading to an accumulation of the 60S ribosomal subunit and subsequent limits 80S ribosome assembly. Collectively, these observations provide robust evidence supporting the indispensable role of DDX10 in ribosome biogenesis.
Fig. 3. DDX10 depletion disrupts 18S rRNA maturation and small ribosomal subunit biogenesis.
a Polysome profiles of DDX10-AID (+ OsTir1) and control DDX10-AID (no OsTir1) mESCs treated with or without IAA at different time points. 40S: small ribosomal subunit, 60S: large ribosomal subunit, 80S: monosomes, and polysomes are indicated. b HPG incorporation analyzing nascent protein synthesis in DDX10-AID (+ OsTir1) mESCs at 0 h, 24 h, and 48 h after IAA treatment. Cycloheximide (CHX), a protein translation inhibitor, was used as a negative control. Experiments were repeated three times independently with similar results. Scale bar, 20 µm. c Quantification of relative HPG incorporation shown in (b), which is presented as the intensity of HPG: NuclearMask blue ratio. Data are presented as mean values ± SD with the indicated significance from two-sided t-test. Exact p-values are reported in the figure. n = 10 fields. d TEM images showing the changes in nucleolar structure following DDX10 degradation. Experiments were repeated two times independently with similar results. Scale bar, 1 µm. e Bar graphs showing the percentage of cells with normal or abnormal GC/DFC/FC structure. f Schematic illustration of mouse pre-rRNA processing with probe locations for Northern blot. The mature 18S, 5.8S, and 28S rRNAs are depicted in purple, yellow, and blue boxes, respectively. Mature rRNA sequences are flanked by 5’ETS, 3’ETS, ITS1, and ITS2. g Northern blot analysis of pre-rRNA intermediates using 5’ETS-1, 5’ETS-2, 5’ETS and ITS1-29 probes in DDX10-AID mESCs with IAA treatment (0 h, 6 h, 24 h, and 48 h) or treated with IAA for 48 h followed by 48 h of washing. MB represents methylene blue staining. Experiments were repeated three times independently with similar results. h Northern blot analysis of pre-rRNA intermediate levels in DDX10-AID (+ OsTir1) mESCs overexpressing DDX10FL, DDX10ΔHBD, DDX10ΔHCD or DDX10ΔHD at 0 h and 48 h after IAA treatment. MB represents methylene blue staining. Experiments were repeated three times independently with similar results. Source data are provided as a Source Data file.
The ribosome, which is responsible for protein synthesis, directly influences cellular protein production56,57. Then, we investigated the level of protein synthesis by assessing the translational incorporation of L-homopropargylglycine (HPG), an amino acid analog of methionine, into nascent protein. Our findings revealed that DDX10 degradation by IAA treatment for 48 h led to an ~90% reduction in HPG-labeled proteins (Fig. 3b, c, and Supplementary Fig. 3d, e). The dynamics of nucleolar architecture are intimately linked to ribosome biogenesis, and disruptions in ribosome biogenesis frequently lead to a disordered nucleolar structure56. Then we conducted transmission electron microscopy (TEM) experiments and found that DDX10 degradation resulted in abnormal structures of FC, DFC, and GC at 12 h and 24 h after IAA treatment (Fig. 3d, e). Together, these results suggest that DDX10 degradation impairs ribosome biogenesis, consequently leading to severe disruptions in both protein synthesis and nucleolar structure.
DDX10 is required for 18S rRNA maturation
The 40S ribosomal subunit is composed of mature 18S rRNA and 33 ribosomal proteins. Mature 18S rRNA is obtained by multi-step processing of 5’ETS and ITS1 in 47S rRNA23,24,58 (Fig. 3f). To decipher the influence of DDX10 on 40S subunit biogenesis, our initial examination involved assessing whether DDX10 loss impacted mature 18S rRNA levels. The outcome was a significant decrease in mature 18S rRNA upon DDX10 degradation, while 28S rRNA levels remained unaltered (Fig. 3g). Subsequently, our investigation turned to Northern blot to detect various 18S rRNA precursors using probes targeting the 5’ETS and ITS1 regions of pre-rRNA (Fig. 3f). Hybridization with these probes revealed that DDX10 degradation had no significant effect on 47S rRNA, but led to a distinct accumulation of the 34S pre-rRNA (a precursor of 18S rRNA), coupled with a noteworthy reduction in 18SE pre-rRNA (another 18S rRNA precursor) (Fig. 3g). Furthermore, we validated these findings via RT-qPCR utilizing primers specific to the cleavage sites of 5’ETS and ITS1 regions (Supplementary Fig. 3f). Overall, these results underscore the necessity of DDX10 in facilitating the processing of cleavage sites within the 5’ETS and ITS1 regions, particularly at sites A0, 1, and 2b, respectively.
To explore whether the role of DDX10 in regulating 18S rRNA processing depends on its intact helicase domain, we overexpressed HA-tagged DDX10FL (full-length DDX10), DDX10ΔHBD (DDX10 lacking helicase ATP binding domain), DDX10ΔHCD (DDX10 lacking helicase C-terminal domain), and DDX10ΔHD (DDX10 lacking both helicase ATP binding and helicase C-terminal domains) in DDX10-AID mESCs (Supplementary Fig. 3g, h). We found that DDX10FL could successfully restore the pre-rRNA processing defect caused by endogenous DDX10 degradation, while DDX10ΔHBD, DDX10ΔHCD, and DDX10ΔHD could not (Fig. 3h), indicating that the complete helicase domain of DDX10 is critical for the processing and maturation of 18S rRNA.
DDX10 interacts with the SSU processome and regulates the release of U3 snoRNA from pre-rRNA
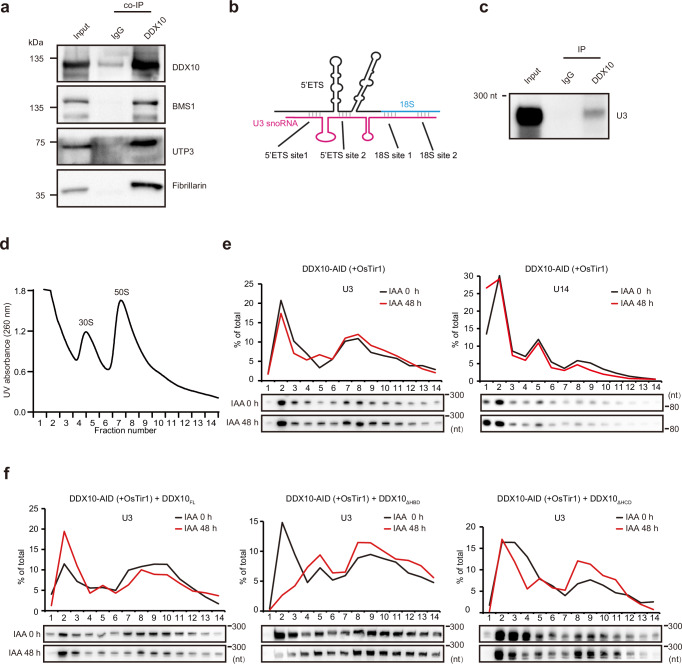
The SSU processome, also known as 90S preribosome, is an early assembly intermediate of the small ribosomal subunit. It undergoes structural changes to form the pre-40S ribosome, which is necessary for 18S rRNA maturation59,60. To investigate whether DDX10 form a complex with the SSU processome, we preformed co-immunoprecipitation (co-IP) experiments and confirmed the interactions of DDX10 with SSU processome components BMS1, UTP3, and Fibrillarin (Fig. 4a), a component of U3 snoRNPs that is positioned at the center of the SSU processome to direct pre-rRNA folding and processing61.
Fig. 4. DDX10 regulates the release of U3 snoRNA from pre-rRNA.
a Co-IP experiments confirming interactions between DDX10 and BMS1, UTP3, or Fibrillarin in FLAG-tagged DDX10 mESCs. Experiments were repeated three times independently with similar results. b Cartoon diagram illustrating binding sites among U3 snoRNA (magenta), 5’ETS (black), and 18S rRNA (blue). c RIP followed by Northern blot analyzing the interaction between DDX10 and U3 snoRNA. Experiments were repeated three times independently with similar results. d UV absorbance profiles of nuclear extracts fractionated on a 10 – 30% sucrose gradient. The peaks corresponding to 30S and 50S contain small and large ribosomal subunits, respectively, along with their precursors. Earlier preribosomal particles are shown in fractions ahead of 50S peak. e Northern blot showing the distribution of U3 and U14 snoRNAs in individual fractions of DDX10-AID (+ OsTir1) mESCs at 0 h and 48 h after IAA treatment. The data were plotted as the percentage of snoRNAs in each fraction relative to the total in all fractions. Experiments were repeated three times independently with similar results. f Northern blot analysis of the distribution of U3 snoRNA in the preribosome fraction of DDX10-AID (+ OsTir1) mESCs overexpressing DDX10FL, DDX10ΔHBD, and DDX10ΔHCD, respectively, at 0 h and 48 h after IAA treatment. Experiments were repeated two times independently with similar results. Source data are provided as a Source Data file.
Previous researches have highlighted the essential role of U3 snoRNA in pre-rRNA processing. Specifically, U3 snoRNA binds to the 5’ETS and 18S rRNA of pre-rRNA, and coordinates the early processing and folding of pre-rRNA, facilitating necessary cleavage events within the 5’ETS region35,62. However, it must be actively removed during pre-rRNA processing to complete RNA folding, which is crucial for the maturation of 18S rRNA63. Cryo-electron microscopy (Cryo-EM) structure showcased the precise location where U3 snoRNA binds to both the 5’ETS and 18S rRNA of pre-rRNA within the human SSU processome64 (Fig. 4b). Our CLIP-seq data exhibited that DDX10 binds to 18S rRNA, prompting us to investigate whether there is an interaction between DDX10 and U3 snoRNA. To this end, we conducted RIP experiments and confirmed the interaction between DDX10 and U3 snoRNA (Fig. 4c). Furthermore, in yeast, the loss of Dbp4 (a mammalian homolog of DDX10) leads to pre-rRNA processing defects by hindering the release of U14 snoRNA from pre-rRNA65,66. To unravel whether DDX10 regulates U3, U14, or both releases from pre-rRNA, we isolated preribosomes and collected the preribosome fractions using sucrose density gradient centrifugation. Subsequently, we performed Northern blot experiments, which unequivocally confirmed that DDX10 degradation inhibits the release of U3 snoRNA, but not U14 snoRNA, from preribosomes (Fig. 4d, e). This insight suggests that DDX10 loss hinders the release of U3 snoRNA from pre-rRNA, culminating in compromised pre-rRNA processing and disrupted ribosome biogenesis.
Next, we further investigated the role of the helicase domain of DDX10 in U3 snoRNA release. Our findings revealed that DDX10FL could overcome the blockage of U3 snoRNA release caused by DDX10 degradation, while DDX10ΔHBD and DDX10ΔHCD could not (Fig. 4f). These results underscore the essential involvement of both helicase ATP binding domain and helicase C-terminal domain of DDX10 in the regulation of U3 snoRNA release.
Phase separation of DDX10 regulates ribosome biogenesis
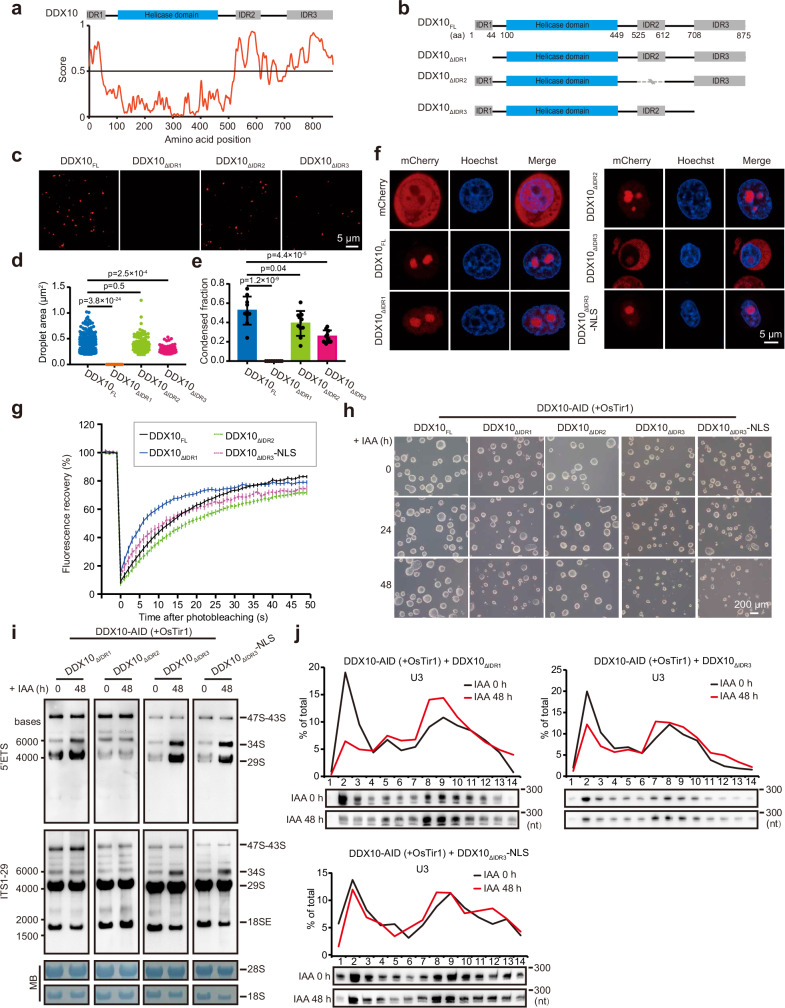
LLPS is a vital organizing principle for biomolecular condensates, intricately participating in a variety of processes encompassing RNA metabolism, ribosome biogenesis, DNA damage response, and signal transduction67–69. Of note, intrinsically disordered regions (IDRs) play a pivotal role in driving LLPS68. Given that the human DDX10 protein contains IDRs and exhibits mobility30, it follows that DDX10 could potentially partake in LLPS, a process that might regulate ribosome biogenesis.
To scrutinize the propensity of DDX10 proteins to form condensates, we embarked on an analysis of the functional domains present within the mouse DDX10 protein. This investigation revealed the presence of three IDR domains alongside a helicase domain (Fig. 5a). Subsequently, to investigate whether these IDRs could induce the condensation of DDX10, we generated both full-length and truncated versions of mCherry-tagged DDX10, denoted as DDX10FL, DDX10ΔIDR1 (DDX10 lacking N-terminal IDR domain: 1-44 amino acids), DDX10ΔIDR2 (DDX10 lacking IDR domain: 525-612 amino acids), and DDX10ΔIDR3 (DDX10 lacking C-terminal IDR domain: 708-875 amino acids) (Fig. 5b). These constructs served as our subjects for droplet formation assays in vitro through utilizing purified fusion proteins (including mCherry-tagged DDX10FL, DDX10ΔIDR1, DDX10ΔIDR2, and DDX10ΔIDR3). Our observations demonstrated that wild-type DDX10 could form droplets, while the absence of IDR1 and IDR3 disrupted DDX10 droplet formation, and the loss of IDR2 had no effect (Fig. 5c–e).
Fig. 5. DDX10 drives LLPS to modulate ribosome biogenesis.
a Top: Domain mapping of mouse DDX10 displaying a helicase domain and three intrinsically disordered regions (IDR1, IDR2, and IDR3). Bottom: Predictions of intrinsic disorder tendency of DDX10 using IUPred3 (https://iupred.elte.hu/), where scores above 0.5 indicate disorder. b Schematic representation of different truncated forms of mouse DDX10. c Droplet formation experiments evaluating 0.5 µM mCherry-tagged DDX10FL and truncated forms of DDX10: DDX10ΔIDR1, DDX10ΔIDR2, or DDX10ΔIDR3. Scale bars, 5 μm. d Droplet areas observed in panel (c). Data are presented as mean values ± SD. Significance was tested using the two-sided Mann-Whitney test. Exact p-values are reported in the figure. n = 10 fields. e Condensed fraction of DDX10-mCherry in experiments from panel (c). Data are presented as mean values ± SD with the indicated significance from two-sided t-test. Exact p-values are reported in the figure. n = 10 fields. f Live-cell images of mCherry-tagged DDX10 variants and Hoechst staining in NIH3T3 cells. Experiments were repeated three times independently with similar results. Scale bars, 5 μm. g FRAP experiments conducted on NIH3T3 cell lines. Fluorescence recovery curve obtained by bleaching a predefined spot in the nucleolus. Data are presented as mean ± SEM of n = 30 cells. h Brightfield images of DDX10-AID (+ OsTir1) mESCs overexpressing DDX10FL or different truncations treated with IAA at different time points (0 h, 24 h, and 48 h). Experiments were repeated three times independently with similar results. Scale bar, 200 µm. i Northern blot analysis of pre-rRNA intermediates in DDX10-AID (+ OsTir1) mESCs overexpressing DDX10ΔIDR1, DDX10ΔIDR2, DDX10ΔIDR3, and DDX10ΔIDR3-NLS, respectively, at 0 h and 48 h after IAA treatment. Experiments were repeated two times independently with similar results. j Northern blot analysis of the distribution of U3 snoRNA in the preribosome fraction of DDX10-AID (+ OsTir1) mESCs overexpressing DDX10ΔIDR1, DDX10ΔIDR3, and DDX10ΔIDR3-NLS, respectively, at 0 h and 48 h after IAA treatment. Experiments were repeated two times independently with similar results. Source data are provided as a Source Data file.
To further ascertain the significance of specific IDRs of DDX10 LLPS in vivo, we established a stable expression of mCherry-tagged DDX10 full-length and its various truncations in NIH3T3 cells. The results of confocal microscopy revealed distinct localization patterns and condensate formation abilities of these constructs: DDX10FL and DDX10ΔIDR2 displayed nucleolar localization and formed condensates; DDX10ΔIDR1 exhibited predominantly nucleolar localization, accompanied by dispersed signals throughout the nucleoplasm. However, its aggregation in the nucleolus was significantly lower than that of DDX10FL and DDX10ΔIDR2, exhibiting a relatively diffused distribution, and DDX10ΔIDR3 exhibited the diffused distribution in both the nucleolus and cytoplasm, with a prevalent presence in the cytoplasm (Fig. 5f). This observation led us to speculate that IDR3 of DDX10 might carry a crucial nuclear localization signal (NLS). To explore this hypothesis, we appended an NLS (Pro-Lys-Lys-Lys-Arg-Lys-Val) to the C-terminus of DDX10ΔIDR3, generating DDX10ΔIDR3-NLS (Supplementary Fig. 4a), and conducted similar experiments. The outcome demonstrated that the addition of NLS to DDX10ΔIDR3 successfully redirected it to the nucleolus, while the diffused signal was also observed in the nucleoplasm. Similar to DDX10ΔIDR1, the aggregation of DDX10ΔIDR3-NLS in the nucleolus was significantly lower than that of DDX10FL and DDX10ΔIDR2 (Fig. 5f). These in vivo results indicated that both IDR1 and IDR3 are essential for condensate formation of DDX10 in the nucleolus. Taken together, these findings suggest that both IDR1 and IDR3 are crucial for DDX10 in forming condensates both in vivo and in vitro.
In our quest to investigate the influence of IDRs on the mobility of DDX10 in vivo, we employed fluorescence recovery after photobleaching (FRAP) on the full-length and truncated versions of mCherry-tagged DDX10. Specifically, we photobleached nucleolus regions with comparable mCherry intensities and subsequently tracked the signal recovery over time (Supplementary Fig. 4b). The findings indicated that the absence of IDR2 led to decreased mobility, whereas the loss of both IDR1 and IDR3 resulted in increased mobility of DDX10 (Fig. 5g). This suggests that both IDR1 and IDR3 contribute to stabilizing DDX10 interactions, thereby facilitating condensate formation, and their deletion may accelerate the dissociation of DDX10 from RNA, leading to increased transient interactions that might affect its function. Taken together, these findings underscore the pivotal roles played by the N-terminal IDR1 and the C-terminal IDR3 in driving DDX10 phase separation.
Subsequently, our inquiry turned towards investigating the potential involvement of DDX10 phase separation in the regulation of ribosome biogenesis. To explore this, we introduced different HA-tagged DDX10 truncated variants into DDX10-AID mESCs, respectively (Supplementary Fig. 4c). Immunofluorescence staining followed by SIM imaging revealed that the localization of DDX10ΔIDR1, DDX10ΔIDR2 and DDX10ΔIDR3-NLS was mainly localized at DFC and GC of the nucleolus, which was similar to that of DDX10FL. And DDX10ΔIDR3 was present in the cytoplasm, DFC and GC of the nucleolus (Supplementary Fig. 4d). Both DDX10FL and DDX10ΔIDR2 reinstated the morphological alterations in mESC clones and compensated for the anomalies in pre-rRNA processing induced by endogenous DDX10 degradation. Conversely, DDX10ΔIDR1, DDX10ΔIDR3, and DDX10ΔIDR3-NLS failed to rescue the phenotype (Figs. 3h, 5h, i, and Supplementary Fig. 4e, f). These findings collectively indicated that the IDR1 and IDR3 are critical for DDX10 in regulating pre-rRNA processing. Moreover, DDX10FL could restore the defects of protein synthesis and disordered nucleolar structure caused by DDX10 degradation, but DDX10ΔIDR1, DDX10ΔIDR3, and DDX10ΔIDR3-NLS could not (Supplementary Fig. 4g-j).
Next, our data showed that DDX10ΔIDR1, DDX10ΔIDR3, and DDX10ΔIDR3-NLS could not restore the obstruction of U3 snoRNA release from pre-rRNA caused by DDX10 degradation (Fig. 5j), indicating that IDR1 and IDR3 of DDX10 are involved in regulating the release of U3 snoRNA. In sum, our data conclude that DDX10 phase separation plays an indispensable role in facilitating ribosome biogenesis.
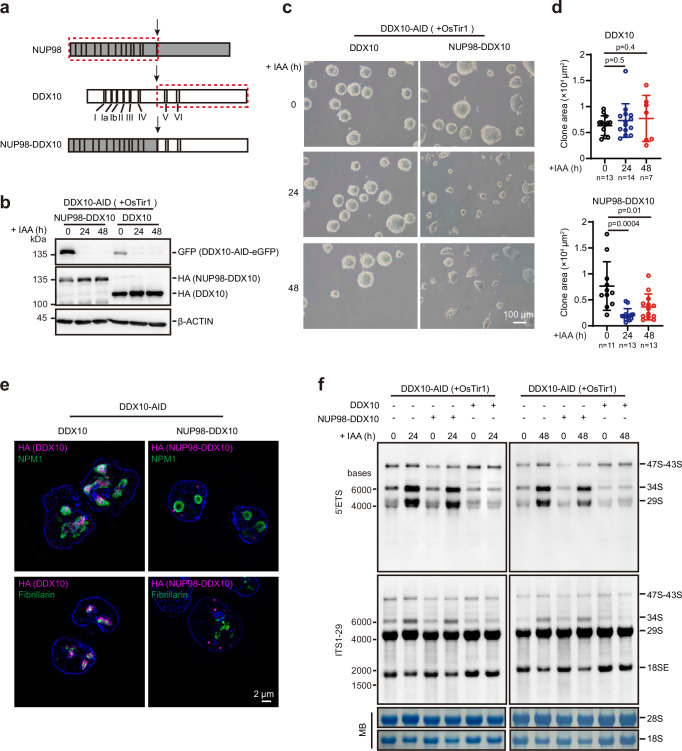
NUP98-DDX10 fusion protein lacks the normal function of DDX10 in regulating ribosome biogenesis
NUP98-DDX10 fusion arises from chromosomal abnormalities and is associated with de novo or therapy-related myeloid malignancies70,71. This fusion protein retains only two conserved motifs (V and VI) of the DDX10 protein while losing six motifs (I, Ia, Ib, II, III, and IV)72 (Fig. 6a). To further probe the potential mechanism, we attempted to obtain clinical patient samples, but encountered challenges preventing their acquisition. In lieu of this, we generated NUP98-DDX10 fusion in mESCs to explore whether the NUP98-DDX10 fusion protein retains the normal cellular function of DDX10. For this purpose, we stably expressed either HA-tagged NUP98-DDX10 or DDX10 in DDX10-depleted mESCs (Fig. 6b). As anticipated, the expression of DDX10 rescued the altered cell morphology stemming from endogenous DDX10 degradation, whereas NUP98-DDX10 could not (Fig. 6c, d), indicating that NUP98-DDX10 lacks the regulatory function of DDX10. Additionally, SIM images unveiled a punctate nuclear distribution pattern for the NUP98-DDX10 fusion protein, primarily localized in the nucleoplasm, divergent from the wild-type DDX10 localization (Fig. 6e). Furthermore, to gauge whether NUP98-DDX10 retains the ability to regulate pre-rRNA processing, we conducted Northern blot analysis and observed that DDX10 fully restored the pre-rRNA processing defect due to DDX10 degradation, while NUP98-DDX10 did not (Fig. 6f). Cumulatively, these findings affirm that the NUP98-DDX10 fusion protein fails to replicate the function of DDX10 in regulating pre-rRNA processing.
Fig. 6. NUP98-DDX10 fusion protein lacks the function in ribosome biogenesis regulation.
a Cartoon diagram illustrating the translocation of NUP98-DDX10 fusion protein, with the red dotted box representing the gene fusion segment. I-VI represent the conserved domains of DDX10. b Western blot depicting protein levels of NUP98-DDX10 and full-length DDX10, and endogenous DDX10 levels after treatment with IAA at different time points (0 h, 24 h, and 48 h). Experiments were repeated two times independently with similar results. c Brightfield images of mESCs overexpressing DDX10 or NUP98-DDX10 after DDX10 degradation with IAA treatment at different time points (0 h, 24 h, and 48 h). Scale bar, 100 µm. Experiments were repeated three times independently with similar results. d Colony areas of mESCs overexpressing DDX10 and NUP98-DDX10 after DDX10 degradation with IAA treatment at different time points in (c). The number of cells for statistical analysis is indicated in the figure. Data are presented as mean values ± SD with the indicated significance from two-sided t-test. Exact p-values are reported in the figure. e Representative SIM images of nucleolar marker proteins Fibrillarin and NPM1 (green) along with HA-tagged proteins (NUP98-DDX10-HA or DDX10-HA) (magenta) in DDX10-AID mESCs expressing NUP98-DDX10 or DDX10. Nuclei stained with Hoechst (blue). Experiments were repeated three times independently with similar results. Scale bar, 2 µm. f Northern blot analysis of pre-rRNA intermediates in DDX10-AID (+ OsTir1) mESCs overexpressing NUP98-DDX10 or DDX10, treated with IAA at different time points (0 h, 24 h, and 48 h). MB represents methylene blue staining. Experiments were repeated two times independently with similar results. Source data are provided as a Source Data file.
Discussion
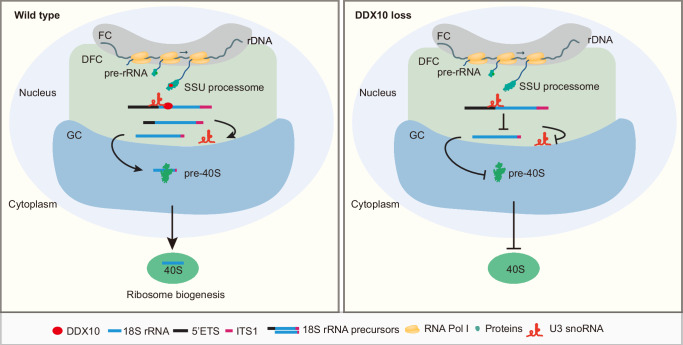
This study unveils the pivotal role of DDX10 in ribosome biogenesis and cell proliferation within mESCs. DDX10 orchestrates ribosome biogenesis by participating in critical processes, particularly the release of U3 snoRNA from pre-rRNA during the processing of 18S rRNA precursors. This regulatory mechanism ensures the accurate maturation of 18S rRNA and facilitates the biogenesis of the small ribosomal subunit (Fig. 7).
Fig. 7. Proposed model for ribosome biogenesis regulation by DDX10 in mESCs.
A schematic working model illustrating the mechanism of DDX10 in regulating ribosome biogenesis. In the presence of DDX10, it functions as a component of the SSU processome within the nucleolus, facilitating the release of U3 snoRNA from pre-rRNA. This activity ensures the maturation of 18S rRNA and the biogenesis of the 40S subunits. In contrast, in the absence of DDX10, the release of U3 snoRNA from pre-rRNA is impaired, resulting in compromised maturation of 18S rRNA and impaired biogenesis of the 40S subunits.
Ribosomes, acting as intricate protein synthesis machinery, are essential for cellular viability, growth, and proliferation56,73. When ribosome biogenesis is impaired, cells must promptly halt their cell cycle to avert incomplete growth and unprepared division56. In line with this perspective, our study illustrates that DDX10 degradation impedes the biogenesis of the small ribosomal subunit, resulting in cell cycle arrest and the downregulation of genes linked to cell division, ultimately curbing cell proliferation. Additionally, previous research has indicated that perturbation of ribosome biogenesis can undermine the maintenance of stem cell pluripotency9,15,31. Our RNA-seq data indicated that the changes in cell fate caused by DDX10 degradation are reversible. Moreover, DDX10 degradation could lead to the cell fate transition from mESCs to 2CLCs. Therefore, our study suggests that the cell fate changes caused by short-term degradation of DDX10 are reversible, but long-term degradation of DDX10 could lead to an irreversible transition of mESCs into 2CLCs.
Abnormal ribosome biogenesis may result in “nucleolar stress”, triggering cell cycle arrest in a p53-dependent manner74,75. For example, loss of DDX21 reduces endothelial ribosome biogenesis and halts the endothelial cell cycle through upregulation of p53 and p2176. In our study, we observed that DDX10 degradation activated the p53 signaling pathway. The presence of any crosstalk between DDX21 and DDX10 in regulating the p53 pathway requires further exploration. Upon disruption of ribosome biogenesis, some free and unassembled ribosomal proteins, including RPL11 and RPL5, can bind to MDM2, preventing MDM2 from exerting its ubiquitinase activity, and inhibiting MDM2-mediated P53 ubiquitination77,78. Therefore, we speculate that impaired ribosome biogenesis caused by DDX10 degradation might result in the binding of free ribosomal proteins to MDM2, which prevent MDM2 from ubiquitinating P53, thereby activating the p53 signaling pathway.
A recent study determined that DDX10 in HeLa cells localizes to a region surrounding the DFC, defined as the periphery of the dense fibrillar component (PDFC), which partially overlaps with both the DFC and GC regions30. Consistent with this, our results demonstrate that DDX10 mainly localizes to the DFC and GC regions, indicating that the specific localization of DDX10 is conserved across different species.
Our investigation uncovered that DDX10 degradation results in a decrease in the 40S ribosomal subunit, accompanied by an accumulation in the free 60S ribosomal subunit, as well as a significant reduction in the 80S ribosome. Interestingly, in yeast, Dbp4 (a mammalian DDX10 homologous protein) regulates 40S subunit production. Deletion of Dbp4 in yeast caused a pronounced reduction in free 40S ribosomal subunit and an accumulation of free 60S ribosomal subunit, while the levels of 80S ribosome remained constant65, differing from our findings in mESCs, in which we observed a reduction in both free 40S ribosomal subunit and 80S ribosome. Additionally, in yeast, the DDX10 homolog Dbp4 depletion increased the abundance of U3 and U14 snoRNAs in preribosomes, with a more robust effect on the release of U14 snoRNA from the pre-rRNA65,66. However, in our study, we determined that DDX10 is crucial for the dissociation of U3 snoRNA, but not U14 snoRNA, from pre-rRNA in mESCs (Fig. 4e). These findings suggest that the function of DDX10 in regulating ribosome biogenesis is conserved across both single-cell eukaryotes (such as yeast) and higher mammalian eukaryotes, but the molecular mechanisms are not the same. Therefore, the evolutionary variations across different species in regulating ribosome biogenesis through the same factor may involve diverse regulatory pathways.
LLPS is a process driven by weak dynamic interactions among molecules with multivalent domains or IDRs67,68. DDX10 protein contains three IDRs, two of which are critical for DDX10 droplet formation. Specifically, the loss of both the N-terminal IDR1 and the C-terminal IDR3 results in increased mobility and weakened droplet formation of DDX10. Furthermore, IDR1 was more required for the mobility and droplet formation of DDX10 compared to IDR3 (Fig. 5), indicating that IDR1 is more critical for DDX10 phase separation. However, in contrast to overexpression of Ddx10 lacking IDR1, overexpression of Ddx10 lacking IDR3 has a lowered ability to reverse the altered cell morphology and pre-rRNA processing defects resulting from endogenous DDX10 degradation. These findings suggested that IDR3 may possess additional functions, including serving as a scaffold for interactions with other proteins or nucleic acids outside of its role in phase separation.
In AML, fusion proteins linked to the NUP98 gene are frequently observed and associated with a poor prognosis79,80, and the NUP98-DDX10 fusion contributes to the development of AML70,81. However, the precise molecular mechanism involved remains undefined. In this study, we determined that NUP98-DDX10 fusion protein cannot participate in regulating ribosome biogenesis. In NUP98-DDX10 fusion-associated AML, only one allele of DDX10 gene is intact, resulting in a halved amount of DDX10 protein in these cells compared to normal cells. Consequently, cells harboring this fusion are likely more sensitive to changes in DDX10 dosage compared to healthy cells. Therefore, targeted knockout or knockdown of DDX10 in NUP98-DDX10 fusion AML cells may serve as a potential therapeutic approach for treating this type of AML.
Methods
Cell culture
mESCs were cultured on gelatin-coated plates in DMEM high-glucose media (Hyclone, SH30022.01) supplemented with 15% FBS (GIBCO, 10082147), 1 mM sodium pyruvate (GIBCO, 11360070), 1 × non-essential amino acids (GIBCO, 11140-050), 1 × GlutaMAX (GIBCO, 35050061), 0.1 mM 2-mercaptoethanol (GIBCO, 21985023), 1000 U/mL leukemia inhibitory factor (LIF) (Millipore, ESG1107), and the 2i inhibitors (3 μM CHIR99021 (Selleck, S2924) and 1 μM PD0325901 (Selleck, S1036)). NIH3T3 and HEK293T cells were maintained in DMEM high-glucose media containing 10% FBS (Natocor, SFBE). Degradation of DDX10 was induced by treating mESCs with a final concentration of 500 µM IAA (Solarbio, I8780). For the IAA wash off experiments, mESCs were initially treated with IAA for 2 days. Subsequently, the medium was replaced with culture medium without IAA, and the cells were cultured for an additional 2 days, with daily medium changes.
Plasmid constructions and lentivirus production
For the overexpression of FLAG-tagged DDX10 in mESCs, the Ddx10 fused with a Flag-tag was inserted into PiggyBac plasmid. To construct the Ddx10 donor plasmid, the AID-eGFP tag was PCR amplified from CTCF-AID-eGFP targeting vector (Addgene, 86230)51. Two homology arms (~1 kb each) around the stop codon of the Ddx10 gene were amplified by PCR from E14 genomic DNA. The homology arms and AID-eGFP tag were cloned into pEASY-Blunt plasmid (TransGen, CB101). The OsTir1 was PCR amplified from pEN114 (Addgene, 92143)51, and then cloned into pSIN-Flag lentiviral vector with a puromycin resistance gene. Ddx10-targeting sgRNAs were cloned into pX330 plasmid by annealing oligos CACCGGCCTGTAAAACAAACAAGCA and AAACTGCTTGTTTGTTTTACAGGCC. The full-length and different truncated sequences of Ddx10 were generated by PCR using mouse cDNA as the template. These truncated sequences and mCherry were cloned into pSIN-Flag lentiviral vector with a puromycin resistance gene, and HA-tagged Ddx10FL and different truncation were cloned into pSIN-Flag lentiviral vector with a BSD resistance gene. HA-tagged NUP98-DDX10 and DDX10 were generated by PCR using human cDNA as the template, then these sequences were cloned into pSIN-Flag lentiviral vector with a BSD resistance gene. Lentivirus supernatants were generated with HEK293T cells. HEK293T cells were plated and cultured overnight. The medium of a 10 cm dish was replaced with 9 mL fresh culture medium, and then co-transfected with pSIN vector containing target genes together with the packaging plasmids pMD2.G and psPAX2 using polyethyleneimine (PEI; Polysciences, 24765–2). The culture medium was refreshed 12 h after transfection. After that, the supernatant from the transfected HEK293T cells was collected 48 h post-transfection using a syringe and filtered through a 0.45 μm filter.
Generation of cell lines
To generate DDX10-AID mESCs, Ddx10 donor plasmid and sgRNA were transfected into E14 cells (ATCC, CRL-1821) using FuGENE® 6 Transfection Reagent (Promega, E2691). Single colonies were picked up and identified by PCR genotyping. Subsequently, the cells were either infected with OsTir1 lentivirus or co-transfected with pEN396 donor plasmid (Addgene, 92142) and pX330-EN1201 plasmid (Addgene, 92144) which expresses an sgRNA targeting the Tigre locus. The cells were then selected with 2 µg/mL puromycin (GIBCO, A11138-03) for 3 days. To generate overexpressing FLAG-tagged DDX10 mESCs, E14 cells were co-transfected with the PB-Ddx10-Flag-Puro vector and pBase vector, and selected with 2 µg/mL puromycin for 3 days. For phase separation experiments, NIH3T3 cells were infected with lentiviruses carrying mCherry-tagged DDX10FL, DDX10ΔIDR1, DDX10ΔIDR2, DDX10ΔIDR3 and DDX10ΔIDR3-NLS, respectively, and selected with 2 µg/mL puromycin for 3 days. To generate DDX10-AID mESCs overexpressing HA-tagged DDX10FL, DDX10ΔHBD, DDX10ΔHCD, DDX10ΔHD, DDX10ΔIDR1, DDX10ΔIDR2, DDX10ΔIDR3, DDX10ΔIDR3-NLS, NUP98-DDX10 and DDX10, cells were infected with lentiviruses and selected with 10 µg/mL blasticidin (Yeasen, 60218ES10) for 5 days. For all mESC lines, single cell clones were picked up and identified by immunoblotting.
Western blot
Protein samples were obtained with cell lysis buffer (50 mM Tris-HCl (pH7.6), 1% Triton X-100, 1 mM EDTA, 10% glycerol and 1 × protease inhibitor cocktail). The lysates were centrifuged at 12,000 × g at 4 °C for 10 min, and soluble protein was quantified. Equal amounts of total protein were separated in SDS-PAGE gel and transferred onto polyvinylidene fluoride (PVDF) membrane. The PVDF membrane was subsequently incubated with the diluted primary antibody and secondary antibodies. Antibodies are listed in Supplementary Data 3.
Quantitative RT-qPCR analysis
Total RNAs were extracted with Trizol Reagent (MRC, TR118) according to the manufacturer’s instructions. 1 µg of total RNA was reverse transcribed into cDNA using the HiScript® III RT SuperMix for qPCR (Vazyme, R323-01) or the HiScript II 1st Strand cDNA Synthesis Kit (Vazyme, R212-01) with random primers, and then analyzed by qPCR with SYBR green mixture (Genstar, A301-01) and performed on a CFX Real-Time System (Bio-Rad). The primers used for qPCR analysis are listed in Supplementary Data 4.
Immunofluorescence
Cells were seeded on glass coverslips or glass-bottom cell culture dishes. Cultured cells were fixed with 4% paraformaldehyde for 15 min at room temperature (RT). Subsequently, cells were permeabilized with 0.5% Triton X-100 in PBS for 30 min. Cells were blocked with 1% BSA (bovine serum albumin) in PBS for 1 h at RT. Primary antibodies were applied and incubated overnight at 4 °C, and then cells were incubated with the appropriate secondary antibodies in the dark for 1 h at RT. Antibodies are detailed in Supplementary Data 3. Nuclei were counterstained with DAPI (Sigma-Aldrich, D9542) or Hoechst 33342 (Sigma-Aldrich, 14533). The slides were examined using Zeiss inverted 880/900 confocal microscope, Zeiss Elyra 7 with Lattice SIM or Leica SP8 STED super-resolution microscope. Images were further processed with ZEN blue software.
Cell proliferation assay
Cell proliferation assays were performed using Cell Counting Kit-8 (CCK-8) (Selleck, B34304). Briefly, 1,500 cells were seeded in 96-well plate with 100 µL of complete medium per well. The following day, cells were treated with or without IAA. 10 µL of CCK-8 reagent was added to each well and incubated for 3 h at 37 °C. Subsequently, the absorbance at 450 nm was measured using a microplate spectrophotometer at the indicated time points after IAA treatment.
Fluorescence-activated cell sorting (FACS)
Cell cycle was conducted using Cell Cycle Detection Kit (KeyGen, KGA512). Cells were treated with IAA at different time points, harvested and fixed with 70% cold ethanol at 4 °C overnight. Subsequently, cells were incubated with propidium iodide (PI)/RNase A for 30-60 min. Cell apoptosis analysis was conducted with APC-Annexin V/PI Apoptosis Detection Kit (Bioscience, A6030L). Cells were suspended with binding buffer and incubated with Annexin V-APC and PI solution in the dark at RT for 5–15 min. FACS analysis was performed using LSR Fortessa SORP flow cytometer (BD Biosciences). FlowJo software was used for further analysis of the obtained data.
RNA-seq and bioinformatics analysis
RNAs were extracted from DDX10-AID mESCs treated with IAA at different time points. RNA libraries were generated with VAHTS Universal V6 RNA-seq Library Prep Kit for Illumina (Vazyme, NR604-01/02). Adapter sequences were removed using Trim Galore (v0.6.5) and reads < 20 nucleotides were discarded. Reads were then mapped to mouse (mm10) genome with STAR-RSEM pipeline using ENSEMBL version 79 gene annotation as reference. Differential gene expression was determined using DESeq2 (v1.32.0)82, the p-value adjusted for multiple comparisons. Genes with log2 (fold change) > 1.5 and q-value < 0.05 were considered significantly different. Gene ontology and gene set enrichment analysis (GSEA) was performed using the clusterProfiler (v4.0.0)83. Clustering of gene expression pattern was determined with Mfuzz (v2.52.0)84. RNA-seq data of mouse naive, primed ESCs (GSE226316) and preimplantation embryos (GSE66582) were analyzed using the same procedure outlined above. For analyzing 2 cell-specific genes, the significantly upregulated genes between the two-cell stage and every other stage were firstly identified, and then intersected to yield the co-upregulated genes among all comparisons. These co-upregulated genes were determined as 2 cell-specific genes.
CLIP-seq
FLAG-tagged DDX10 mESCs grown on 10 cm plate were collected and crosslinked with 400 mJ/cm2 of 254 nm UV light. Nuclei were prepared by incubating the cells in buffer A (10 mM Tris-HCl (pH7.4), 10 mM KCl, 1.5 mM MgCl2, 1 mM PMSF, 1 mM DTT, 1 × proteinase inhibitor cocktail and 40 U/mL RNase inhibitor) on ice for 15 min. Then the cells were ground with a loose pestle and centrifuged at 2000 × g at 4°C for 10 min. After centrifugation, the soluble cytoplasm was discarded and the insoluble cell nuclei were resuspended in low salt buffer (2.5 times volume of cell pellet, 20 mM Tris-HCl (pH7.4), 20 mM KCl, 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM PMSF, 1 mM DTT, 1 × proteinase inhibitor cocktail and 40 U/mL RNase Inhibitor). And then high salt buffer (0.5 times volume of low salt buffer, 20 mM Tris-HCl (pH7.4), 1.2 M KCl, 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM PMSF, 1 mM DTT, 1 × proteinase inhibitor cocktail and 40 U/mL RNase Inhibitor) was added to the nuclear lysate. The lysate was incubated on a rotator at 4 °C for 30 min, and then treated with 30 μL RQ1 DNase (Promega, M6101) at 37 °C for 3 min. The insoluble pellet was centrifuged, and the supernatant was harvested. And then the following buffer (20 mM Tris-HCl (pH7.4), 0.1% SDS and 5 mM EDTA) was added to adjust the salt concentration to 150 mM KCl. About 50 μL of FLAG-M2 magnetic beads (Sigma-Aldrich, M8823) were used for each immunoprecipitation. The beads were incubated with lysate at 4 °C overnight. The beads were washed twice with cold wash buffer (1 × PBS (tissue culture grade; without Mg2+ and Ca2+), 0.1% SDS, 0.5% sodium deoxycholate and 0.5% NP-40), followed by two times with high salt wash buffer (5 × PBS, 0.1% SDS, 0.5% NP-40 and 0.5% sodium deoxycholate) and two more times with PNK buffer (50 mM Tris-HCl (pH 7.4), 10 mM MgCl2 and 0.5% NP-40). Then the protein-RNAs complexes were partially digested with Micrococcal Nuclease (MNase, 1:1 × 103/1:5 × 105 dilution) (NEB, M0247S) at 37 °C for 10 min. The reaction was stopped by PNK-EGTA buffer (PNK buffer with 2 mM EGTA). The beads were washed once with high salt buffer, once with wash buffer and twice with PNK buffer. The beads were treated with FastAP thermosensitive alkaline phosphatase (Thermo Fisher Scientific, EF0652) at 37 °C for 20 min. After two washes with PNK-EGTA buffer and twice with PNK buffer, the pre-adenylated 3’ linker (rAppTGGAATTCTCGGGTGCCAAGG-NH2 (biotin)) was ligated with T4 RNA Ligase 2, truncated KQ (NEB, M0373S) overnight at 16 °C. The non-ligated 3’ linker was washed away with PNK buffer. The RNAs were then phosphorylated using T4 PNK (NEB, M0201S) at 37 °C for 30 min. And then the protein-RNA complexes were subjected to 4%-12% NuPAGE Bis-Tris protein gel (Invitrogen, NP0321BOX) separation and transferred to nitrocellulose membrane. The biotin-labeled RNA-protein complexes were identified by chemiluminescent nucleic acid detection module kit (Thermo Fisher Scientific, 89880). The RNAs were purified though proteinase K treatment and overnight precipitation. The purified RNAs were ligated with 5’ RNA adapter (5’-ACACGrArCrGrCrUrCrUrUrCrCrArUrCrUrNrNrNrU-3’) using T4 RNA Ligase 1 (NEB, M0204S). RNAs were reverse transcribed using Superscript III reverse transcriptase (Thermo Fisher Scientific, 18080044) with RT primer (5’-CCTTGGCACCCGAGAATTCCA-3’) and amplified for 15-18 cycles using Phusion High-Fidelity DNA Polymerase (NEB, M0530S) (DP5: 5’-ACACGACGCTCTTCCGATCT-3’; DP7: 5’-CCTTGGCACCCGAGAATTCCA-3’). Library index sequences were introduced by PCR using index primers (P5: 5’-AATGATACGGCGACCACCGAGATCTACACAACCCACACTCTTTCCCTACACGACGCTCTTCCGATCT-3’, P7: 5’-CAAGCAGAAGACGGCATACGAGATCGTGATGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA-3’) for 8-10 cycles. CLIP-seq libraries were generated and followed by high-throughput sequencing with Illumina Novaseq 6000 sequencing platform.
Analysis of CLIP-seq data
Single end reads with UMI sequences (NNNT) were selected. Fastx_clipper tool from fastx toolkit (v0.0.14) was used to remove 3’ linker sequence with parameters “-a TGGAATTCTCGGGTGCCAAGG -l 20 -n”. Fastq2collapse.pl from CLIP Tool Kit (CTK) (v1.1.3)85 was used to remove PCR duplicates, then subjected to stripBarcode.pl to remove 5’UMI sequences with parameters “-format fastq -len 4”.
Reads were mapped to mouse (mm10) genome with bwa software (v0.7.17)86. Sam files were parsed with parseAlignment.pl, tag2collapse.pl, joinWrapper.py tools from the CLIP Tool Kit (CTK)85, and then handled with tag2peak.pl to get enriched peaks, which was annotated with ChIPseeker (v1.28.3)87. BamCoverage from deeptools (v3.5.1)88 was used to generate normalized bigwig files with parameters “--normalizeUsing RPGC --effectiveGenomeSize 2494787188”. Reads were also mapped to pre-45S DNA sequence (downloaded from NCBI: X82564) with bwa software. BamCoverage was used to generate normalized bigwig files with parameters “--normalizeUsing RPGC --effectiveGenomeSize 22118”.
hnRNP CLIP data (GSE77101-GSM2044162)89 was handled as above.
Northern blot
Northern blot experiments were performed as described90. Total RNAs were extracted from mESCs treated with IAA at different times using Trizol Reagent (MRC). Biotin-labeled probes targeting 5’ETS-1, 5’ETS-2, ITS1-29, 5’ETS, U3 and U14 were synthesized by IGE biotechnology (Guangzhou). The specific sequences of the probes used are listed in Supplementary Data 4. For analysis of high-molecular-weight RNAs, 3 µg of total RNAs were resolved on agarose denaturing gels (1.3% formaldehyde/1% agarose in HEPES-triethanolamine (HT) buffer). For the analysis of the low-molecular-weight RNAs, 1.5 µg of RNAs were separated on denaturing acrylamide gels. After electrophoresis, the RNAs were transferred onto nylon membranes (GE Amersham, RPN303B). Membranes were stained with methylene blue stain solution (0.03% methylene blue (MedChemExpress, HY-14536) in 0.3 M sodium acetate (pH 5.0-5.5)), and then RNA was crosslinked onto the membrane using UV light. The membranes were prehybridized at 50 °C for 1 h in hybridization solution (5 × SSC, 5 × Denhardt’s solution and 0.5% SDS). The biotin-labeled oligonucleotide probe (20-40 pmol) was added to the hybridization solution, and the membranes were incubated at 50 °C overnight. Detection of the biotin-labeled oligonucleotide probes was performed using Chemiluminescent Nucleic Acid Detection Module Kit (Thermo Fisher Scientific, 89880) according to the manufacturer’s instructions.
Sucrose gradient fractionation of polysome
2 × 107 cells were treated with CHX (100 µg/mL) (Selleck, S7418) at 37 °C for 10 min. After treatment, the cells were lysed with lysis buffer (25 mM Tris-HCl (pH7.4), 5 mM MgCl2, 100 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 40 U/mL RNase Inhibitor, 1 × protease inhibitor cocktail, 100 µg/mL CHX and 1 mM DTT) on ice for 30 min. The cell lysates were centrifuged at 13,000 × g at 4 °C for 10 min and the supernatant was loaded onto 10% – 50% sucrose gradients. The gradients were then subjected to ultracentrifugation using a SW41 rotor at 222,228 × g at 4 °C for 3 h. The absorbance at 260 nm was recorded using a BioComp Piston Gradient Fractionator equipped with a Bio-Rad Econo UV Monitor.
Transmission electron microscopy (TEM)
The cells were cultured on glass coverslips in 24-well plates and fixed with 2.5% glutaraldehyde for 30 min at RT. Next, the cells were fixed with 1% osmium acid for 30 min and then dehydrated in a graded series of ethanol solutions (50%, 70%, 80%, 90%, 100% and 100%) for 2 min each. After dehydration, the cells were covered with resin and infiltrated at RT for 2 h. The samples were then embedded and polymerized at 40°C for 2-4 h, followed by an additional 12 h at 60 °C. After ultra-sectioning, the sections were double-stained with uranyl acetate and lead citrate. Samples were examined with a transmission electron microscope (FEI Tecnai G2 Spirit).
Quantification of nascent protein synthesis
Nascent protein synthesis in mESCs was detected using a Click-iT HPG Alexa Fluor 594 Protein Synthesis Assay Kit (Thermofisher, C10429), following the manufacturer’s instruction. Briefly, on the day of the experiment, the regular mESC culture medium was replaced by L-methionine-free DMEM medium containing 50 µM Click-iT HPG and incubated for 30 min. After incubation, the cells were washed once with 1 × PBS and fixed with 4% paraformaldehyde at RT for 15 min. Subsequently, the cells were washed twice with 3% BSA and permeabilized with 0.5% Triton X-100 in PBS for 20 min. The Click-iT reaction cocktail was added to each well and incubated at RT for 30 min. Then, the DNA was stained by HCS NuclearMask Blue Stain reagent for 30 min. Finally, the images were captured using Zeiss inverted 900 confocal microscope. For quantification, relative HPG incorporation was presented as the intensity of HPG: NuclearMask blue ratio.
Co-Immunoprecipitation (Co-IP)
Nuclear protein extracts were prepared from FLAG-tagged DDX10 mESCs. 1 mg of proteins was used for each co-IP experiment. The protein samples were incubated overnight with 2 µg of antibodies (DDX10 antibody and normal rabbit IgG). Following the incubation, 15 µL of protein A (Invitrogen, 10001D) and 15 µL of protein G (Invitrogen, 10004D) dynabeads were added to the samples and incubated at 4 °C for 4 h. After incubation, the beads were washed three times with IP wash buffer (50 mM Tris-HCl (pH7.6), 150 mM KCl, 0.1% Triton X-100, 1 mM EDTA and 1 × protease inhibitor cocktail). The bound proteins were then eluted from the beads, loaded onto SDS-PAGE gel and transferred to PVDF membrane (Merck, IPVH00010). Then the membrane was incubated with the indicated antibodies. Antibodies are detailed in Supplementary Data 3.
RNA immunoprecipitation followed by qPCR or Northern blot
2 × 107 FLAG-tagged DDX10 mESCs were lysed with lysis buffer (10 mM HEPES (pH7.0), 100 mM KCl, 5 mM MgCl2, 0.5% NP-40, 1 mM DTT, 100 U/mL RNase inhibitor and 1 × proteinase inhibitor cocktail). 2-5 µg of antibodies (DDX10 antibody and normal rabbit IgG) were added to cell lysate and incubated overnight at 4 °C. Then, protein A/G dynabeads (Invitrogen) were added to the protein-antibody complexes and incubated at 4 °C for 4 h. The beads-protein-RNA complexes were washed 5 times with RIP wash buffer (50 mM Tris-HCl (pH7.4), 1 mM MgCl2, 150 mM NaCl and 0.05% NP-40) and incubated with DNase I digestion at 37 °C for 30 min, then digested with proteinase K at 37 °C for 30 min with shaking. Finally, RNAs were extracted with Trizol Reagent and purified for qPCR or Northern blot analysis.
Preribosomes isolation
The cells were cultured in 150 mm plates, and lysed with NP-40 lysis buffer (10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.05% NP-40, 40 U/mL RNase Inhibitor, 1 × protease inhibitor cocktail) on ice for 5 min. The lysis was loaded onto the NP-40 lysis buffer containing 24% sucrose and centrifuged at 6,000 × g at 4 °C for 10 min. Then the supernatant was discarded and the nuclear precipitation was washed once with cold PBS. Nuclei were sonicated in the buffer containing 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM NaF, 2 mM EDTA, 0.05% Igepal CA-630, 1 mM DTT, 1 × protease inhibitor cocktail, and 50 U/mL RNase inhibitor and then fractionated on a 10% – 30% sucrose gradient. Following fractionation, RNAs in each fraction were extracted and analyzed by Northern blot.
Fluorescence recovery after photobleaching (FRAP)
NIH3T3 cells (overexpressing mCherry-tagged DDX10FL, DDX10ΔIDR1, DDX10ΔIDR2 and DDX10ΔIDR3-NLS, respectively) were cultured on glass-bottom cell culture dishes and seeded 12 to 24 h prior to the experiment. Cells were stained with Hoechst 33342 for 10 min at room temperature before experiments. FRAP experiments was performed using Zeiss inverted 900 confocal microscope equipped with a 63 × objective. All experimental parameters were kept constant in the independent experiments and conditions. For quantitative analysis, the average fluorescence intensity of frames captured before photobleaching was normalized to 100%. Data were analyzed using GraphPad Prism 8.
Protein purification
Ddx10FL, Ddx10ΔIDR1, Ddx10ΔIDR2 and Ddx10ΔIDR3 sequences were generated by PCR from mouse cDNA. These sequences and mCherry were cloned into a pET-28a expression plasmid. The plasmids were first transformed into BL21 (DE3) E. coli cells (TransGen). The transformed cells were grown at 37°C in LB media containing kanamycin, and then diluted 1:100 in 500 ml LB media supplemented with kanamycin. After ~4 h, the cells were grown to OD600 of ~0.6-0.8. To induce protein expression, IPTG was added to the bacterial culture at a final concentration of 0.5 µM. The cells were then induced overnight in 20°C. The cells were harvested by centrifugation at 3,000 × g at 4°C for 10 min. Cell pellet was resuspended in the buffer containing 0.5 M NaCl, 50 mM Tris-HCl (pH 7.5), 0.2% Triton X-100, 20 mM imidazole, 10% glycerol and EDTA-free protease inhibitor cocktail. The resuspended cells were lysed using Low-temperature ultra-high pressure continuous flow cell disrupter (JN-Mini Pro). The proteins were purified with Ni-NTA agarose beads and eluted from the beads using the buffer containing 0.5 M NaCl, 50 mM Tris-HCl (pH 7.5), 0.2% Triton X-100, 300 mM imidazole, 10% glycerol, 1 mM DTT and EDTA-free protease inhibitor cocktail. The eluted protein was concentrated using Amicon Ultra Centrifugal filters (Millipore). The concentration of purified proteins was determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, 23227) and stored in aliquots at -80°C.
Droplet formation
mCherry-fused DDX10 full-length and truncated proteins were thawed on ice and then centrifuged at 13,000 × g at 4°C for 10 min. The indicated protein at a final concentration of 0.5 µM was added to droplet formation buffer (5% (w/v) PEG8000, 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 10% glycerol and 1 mM DTT). The reaction mixture was incubated at RT for 1 min to allow for droplet formation, and then loaded into a custom slide chamber made from glass coverslips mounted on two parallel strips of double-sided tape on slides. Reactions were incubated for 5 min in the imaging vessel to allow droplets in solution to settle on the glass imaging surface. Phase-separated droplets were imaged by Zeiss inverted 900 confocal microscope.
Statistics and reproducibility
Data were presented as mean ± SD unless otherwise indicated in the figure legend. The number of statistical sample and experimental repeats were indicated in the figures and legends. Experimental data were plotted and analyzed using either Excel or GraphPad Prism 8, and statistical significance was determined by Student’s t-test analysis (two-tailed) for two groups, unless otherwise indicated. The exact p-values are indicated in the figures. The uncropped and unprocessed scans of all blots and gels were provided in the Source Data file and the Supplementary Information.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank Professor G Pan for helpful comments and suggestions. We thank Professor L Chen for sharing anti-RPA194 antibody with us. We thank Dr. Q Xia in assisting to construct DDX10-AID and FLAG-tagged DDX10 mESC lines together with X Wang and L Wang. This work was supported by the National Natural Science Foundation of China [31925009 to H.Y, 32430016 to H.Y, U21A20195 to H.Y], National Key R&D Program of China [2021YFA1100300 to H.Y], Guangzhou Key R&D Program [2023B03J1230 to H.Y], Guangdong Basic and Applied Basic Research Foundation [2022A1515012336 to Y.S], Science and Technology Projects in Guangzhou [202201010158 to Q.C].
Author contributions
H.Y. initiated this study, and designed the experiments. X.W. conducted most of the experiments. G.H. performed the bioinformatics analysis. L.W. assisted in the construction of DDX10-AID and FLAG-tagged DDX10 mESC lines. Ya.L. and S.Y. assisted in phase separation experiments. Yu.L., J.L., H.L., Y.F., Q.H., Q.Z., W.W., W.G., Y.S., Q.C., X.W., Xiaof.Z., Y.C., and Xiaor.Z. contributed to the work. H.Y. and X.W. wrote the manuscript. H.Y. conceived and supervised the entire study.
Peer review
Peer review information
Nature Communications thanks Miha Modic, Yangming Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The CLIP-seq and RNA-seq data reported in this paper have been deposited in the Genome Sequence Archive database in the National Genomics Data Center under the accession code GSA: CRA011147, and in the Gene Expression Omnibus under the accession code GSE232096. The dataset of this paper has been submitted to the figshare repository (10.6084/m9.figshare.27263289. Published RNA-seq datasets (GSE66582, GSE226316) and hnRNP CLIP data (GSE77101-GSM2044162 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE77101]) were used in this study. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53822-0.
References
- 1.Young, RichardA. Control of the embryonic stem cell state. Cell144, 940–954 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saba, J. A., Liakath-Ali, K., Green, R. & Watt, F. M. Translational control of stem cell function. Nat. Rev. Mol. Cell Biol.22, 671–690 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Klein, D. C. & Hainer, S. J. Chromatin regulation and dynamics in stem cells. Curr. Top. Dev. Biol.138, 1–71 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi, X. et al. RNA targets ribogenesis factor WDR43 to chromatin for transcription and pluripotency control. Mol. Cell75, 102–116.e109 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Gokbuget D., Blelloch R. Epigenetic control of transcriptional regulation in pluripotency and early differentiation. Development146, dev164772 (2019). [DOI] [PMC free article] [PubMed]
- 6.Gabut, M. et al. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell147, 132–146 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. et al. Nuclear m(6)A reader YTHDC1 regulates the scaffold function of LINE1 RNA in mouse ESCs and early embryos. Protein Cell12, 455–474 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vissers, C., Sinha, A., Ming, G. L. & Song, H. The epitranscriptome in stem cell biology and neural development. Neurobiol. Dis.146, 105139 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durand, S. et al. RSL24D1 sustains steady-state ribosome biogenesis and pluripotency translational programs in embryonic stem cells. Nat. Commun.14, 356 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabut M., Bourdelais F., Durand S. Ribosome and translational control in stem cells. Cells9, 497 (2020). [DOI] [PMC free article] [PubMed]
- 11.Woolnough, J. L., Atwood, B. L., Liu, Z., Zhao, R. & Giles, K. E. The regulation of rRNA gene transcription during directed differentiation of human embryonic stem cells. PLoS One11, e0157276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe-Susaki, K. et al. Biosynthesis of ribosomal RNA in nucleoli regulates pluripotency and differentiation ability of pluripotent stem cells. Stem Cells32, 3099–3111 (2014). [DOI] [PubMed] [Google Scholar]
- 13.You, K. T., Park, J. & Kim, V. N. Role of the small subunit processome in the maintenance of pluripotent stem cells. Genes Dev.29, 2004–2009 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu K. Quick, coordinated and authentic reprogramming of ribosome biogenesis during iPSC reprogramming. Cells9, 2484 (2020). [DOI] [PMC free article] [PubMed]
- 15.Zhang, H. et al. DEAD-box helicase 18 counteracts PRC2 to safeguard ribosomal DNA in pluripotency regulation. Cell Rep.30, 81–97.e87 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Sampath, P. et al. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell2, 448–460 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Fortier, S., MacRae, T., Bilodeau, M., Sargeant, T. & Sauvageau, G. Haploinsufficiency screen highlights two distinct groups of ribosomal protein genes essential for embryonic stem cell fate. Proc. Natl Acad. Sci. USA112, 2127–2132 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez, C. G. et al. Regulation of ribosome biogenesis and protein synthesis controls germline stem cell differentiation. Cell Stem Cell18, 276–290 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitrea D. M., Kriwacki R. W. Phase separation in biology; functional organization of a higher order. Cell Commun. Signal14, 1 (2016). [DOI] [PMC free article] [PubMed]
- 20.Lafontaine, D. L. J., Riback, J. A., Bascetin, R. & Brangwynne, C. P. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol.22, 165–182 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Cmarko, D., Smigova, J., Minichova, L. & Popov, A. Nucleolus: the ribosome factory. Histol. Histopathol.23, 1291–1298 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Boisvert, F. M., van Koningsbruggen, S., Navascues, J. & Lamond, A. I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol.8, 574–585 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Henras, A. K., Plisson-Chastang, C., O’Donohue, M. F., Chakraborty, A. & Gleizes, P. E. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA6, 225–242 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullineux, S. T. & Lafontaine, D. L. Mapping the cleavage sites on mammalian pre-rRNAs: where do we stand? Biochimie94, 1521–1532 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Calo, E. et al. RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature518, 249–253 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xing, Y. H. et al. SLERT regulates DDX21 rings associated with pol I transcription. Cell169, 664–678.e616 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Wu, M. et al. lncRNA SLERT controls phase separation of FC/DFCs to facilitate pol I transcription. Science373, 547–555 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Yao, R. W. et al. Nascent pre-rRNA sorting via phase separation drives the assembly of dense fibrillar components in the human nucleolus. Mol. Cell76, 767–783.e711 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Zhu, Q. et al. Rcl1 depletion impairs 18S pre-rRNA processing at the A1-site and up-regulates a cohort of ribosome biogenesis genes in zebrafish. Nucleic Acids Res.49, 5743–5759 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shan, L. et al. Nucleolar URB1 ensures 3’ ETS rRNA removal to prevent exosome surveillance. Nature615, 526–534 (2023). [DOI] [PubMed] [Google Scholar]
- 31.Corsini, N. S. et al. Coordinated control of mRNA and rRNA processing controls embryonic stem cell pluripotency and differentiation. Cell Stem Cell22, 543–558.e512 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Lui, L. & Lowe, T. Small nucleolar RNAs and RNA-guided post-transcriptional modification. Essays Biochem.54, 53–77 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Watkins, N. J. & Bohnsack, M. T. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA3, 397–414 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Dragon, F. et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature417, 967–970 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dutca, L. M., Gallagher, J. E. & Baserga, S. J. The initial U3 snoRNA:pre-rRNA base pairing interaction required for pre-18S rRNA folding revealed by in vivo chemical probing. Nucleic Acids Res.39, 5164–5180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tycowski, K. T., Shu, M. D. & Steitz, J. A. Requirement for intron-encoded U22 small nucleolar RNA in 18S ribosomal RNA maturation. Science266, 1558–1561 (1994). [DOI] [PubMed] [Google Scholar]
- 37.Vos T. J., Kothe U. snR30/U17 small nucleolar ribonucleoprotein: a critical player during ribosome biogenesis. Cells9, 2195 (2020). [DOI] [PMC free article] [PubMed]
- 38.Zhang, L., Wu, C., Cai, G., Chen, S. & Ye, K. Stepwise and dynamic assembly of the earliest precursors of small ribosomal subunits in yeast. Genes Dev.30, 718–732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lau, B. et al. Cms1 coordinates stepwise local 90S pre-ribosome assembly with timely snR83 release. Cell Rep.41, 111684 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peculis, B. A. & Steitz, J. A. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell73, 1233–1245 (1993). [DOI] [PubMed] [Google Scholar]
- 41.Rocak, S. & Linder, P. DEAD-box proteins: the driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol.5, 232–241 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Tanner, N. K. & Linder, P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell8, 251–262 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Linder, P. & Jankowsky, E. From unwinding to clamping - the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol.12, 505–516 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Savitsky, K. et al. A human gene (DDX10) encoding a putative DEAD-box RNA helicase at 11q22-q23. Genomics33, 199–206 (1996). [DOI] [PubMed] [Google Scholar]
- 45.Nakao, K. et al. Fusion of the nucleoporin gene, NUP98, and the putative RNA helicase gene, DDX10, by inversion 11 (p15q22) chromosome translocation in a patient with etoposide-related myelodysplastic syndrome. Intern Med.39, 412–415 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Gai, M., Bo, Q. & Qi, L. Epigenetic down-regulated DDX10 promotes cell proliferation through Akt/NF-kappaB pathway in ovarian cancer. Biochem. Biophys. Res. Commun.469, 1000–1005 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Shi, J. H. & Hao, Y. J. DDX10 overexpression predicts worse prognosis in osteosarcoma and its deletion prohibits cell activities modulated by MAPK pathway. Biochem. Biophys. Res. Commun.510, 525–529 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Quan, X. et al. DDX10 and BYSL as the potential targets of chondrosarcoma and glioma. Med. (Baltim.)100, e27669 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu, C. et al. DDX10 promotes human lung carcinoma proliferation by U3 small nucleolar ribonucleoprotein IMP4. Thorac. Cancer12, 1873–1880 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nishimura, K., Fukagawa, T., Takisawa, H., Kakimoto, T. & Kanemaki, M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods6, 917–922 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Nora, E. P. et al. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell169, 930–944.e922 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo, R. et al. TEAD2 initiates ground-state pluripotency by mediating chromatin looping. EMBO J.43, 1965–1989 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grow, E. J. et al. p53 convergently activates Dux/DUX4 in embryonic stem cells and in facioscapulohumeral muscular dystrophy cell models. Nat. Genet.53, 1207–1220 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takahiro, M., Kimura, Y., Nakano, T. & Yamaguchi, S. Ribosomal stress induces 2-cell embryo-like state transition of the mouse ESCs through p53 activation. Biochem. Biophys. Res. Commun.579, 175–180 (2021). [DOI] [PubMed] [Google Scholar]
- 55.Wu, J. et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature534, 652–657 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Turi, Z., Lacey, M., Mistrik, M. & Moudry, P. Impaired ribosome biogenesis: mechanisms and relevance to cancer and aging. Aging (Albany NY)11, 2512–2540 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bursac, S., Prodan, Y., Pullen, N., Bartek, J. & Volarevic, S. Dysregulated ribosome biogenesis reveals therapeutic liabilities in cancer. Trends Cancer7, 57–76 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Tschochner, H. & Hurt, E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol.13, 255–263 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Du, Y. et al. Cryo-EM structure of 90S small ribosomal subunit precursors in transition states. Science369, 1477–1481 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Bernstein, K. A., Gallagher, J. E., Mitchell, B. M., Granneman, S. & Baserga, S. J. The small-subunit processome is a ribosome assembly intermediate. Eukaryot. Cell3, 1619–1626 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kornprobst, M. et al. Architecture of the 90S pre-ribosome: a structural view on the birth of the Eukaryotic ribosome. Cell166, 380–393 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Marmier-Gourrier, N., Clery, A., Schlotter, F., Senty-Segault, V. & Branlant, C. A second base pair interaction between U3 small nucleolar RNA and the 5’-ETS region is required for early cleavage of the yeast pre-ribosomal RNA. Nucleic Acids Res.39, 9731–9745 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sardana, R. et al. The DEAH-box helicase Dhr1 dissociates U3 from the pre-rRNA to promote formation of the central pseudoknot. PLoS Biol.13, e1002083 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Singh, S., Vanden Broeck, A., Miller, L., Chaker-Margot, M. & Klinge, S. Nucleolar maturation of the human small subunit processome. Science373, eabj5338 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soltanieh, S. et al. DEAD-Box RNA helicase Dbp4 is required for small-subunit processome formation and function. Mol. Cell. Biol.35, 816–830 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kos, M. & Tollervey, D. The putative RNA helicase Dbp4p is required for release of the U14 snoRNA from preribosomes in saccharomyces cerevisiae. Mol. Cell20, 53–64 (2005). [DOI] [PubMed] [Google Scholar]
- 67.Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol.18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shin Y., Brangwynne C., P. Liquid phase condensation in cell physiology and disease. Science357, eaaf4382 (2017). [DOI] [PubMed]
- 69.Wheeler J. R., Matheny T., Jain S., Abrisch R., Parker., R. Distinct stages in stress granule assembly and disassembly. Elife5, e18413 (2016). [DOI] [PMC free article] [PubMed]
- 70.Gorello, P. et al. Inv(11)(p15q22)/NUP98-DDX10 fusion and isoforms in a new case of de novo acute myeloid leukemia. Cancer Genet.206, 92–96 (2013). [DOI] [PubMed] [Google Scholar]
- 71.Arai, Y. et al. The inv(11)(p15q22) chromosome translocation of De Novo and therapy-related myeloid malignancies results in fusion of the nucleoporin gene, NUP98, with the putative RNA helicase gene, DDX10. Blood89, 3936–3944 (1997). [PubMed] [Google Scholar]
- 72.Yassin, E. R., Abdul-Nabi, A. M., Takeda, A. & Yaseen, N. R. Effects of the NUP98-DDX10 oncogene on primary human CD34+ cells: role of a conserved helicase motif. Leukemia24, 1001–1011 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Derenzini, M., Montanaro, L. & Trere, D. Ribosome biogenesis and cancer. Acta Histochem.119, 190–197 (2017). [DOI] [PubMed] [Google Scholar]
- 74.Bursac, S., Brdovcak, M. C., Donati, G. & Volarevic, S. Activation of the tumor suppressor p53 upon impairment of ribosome biogenesis. Biochim. Biophys. Acta1842, 817–830 (2014). [DOI] [PubMed] [Google Scholar]
- 75.Pestov, D. G., Strezoska, Z. & Lau, L. F. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol. Cell Biol.21, 4246–4255 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koltowska, K. et al. The RNA helicase Ddx21 controls Vegfc-driven developmental lymphangiogenesis by balancing endothelial cell ribosome biogenesis and p53 function. Nat. Cell Biol.23, 1136–1147 (2021). [DOI] [PubMed] [Google Scholar]
- 77.Zhang, Y. et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell Biol.23, 8902–8912 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Han X. R. et al. CRL4(DCAF1/VprBP) E3 ubiquitin ligase controls ribosome biogenesis, cell proliferation, and development. Sci. Adv.6, eabd6078 (2020). [DOI] [PMC free article] [PubMed]
- 79.Romana, S. P. et al. NUP98 rearrangements in hematopoietic malignancies: a study of the groupe francophone de cytogenetique hematologique. Leukemia20, 696–706 (2006). [DOI] [PubMed] [Google Scholar]
- 80.Slape, C. & Aplan, P. D. The role of NUP98 gene fusions in hematologic malignancy. Leuk. Lymphoma45, 1341–1350 (2004). [DOI] [PubMed] [Google Scholar]
- 81.Morerio, C. et al. Inversion (11)(p15q22) with NUP98-DDX10 fusion gene in pediatric acute myeloid leukemia. Cancer Genet. Cytogenet.171, 122–125 (2006). [DOI] [PubMed] [Google Scholar]
- 82.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kumar, L. M EF. Mfuzz: a software package for soft clustering of microarray data. Bioinformation2, 5–7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shah, A., Qian, Y., Weyn-Vanhentenryck, S. M. & Zhang, C. CLIP Tool Kit (CTK): a flexible and robust pipeline to analyze CLIP sequencing data. Bioinformatics33, 566–567 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu, G., Wang, L. G. & He, Q. Y. ChIPseeker: an R/bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics31, 2382–2383 (2015). [DOI] [PubMed] [Google Scholar]
- 88.Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res.44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Briese, M. et al. hnRNP R and its main interactor, the noncoding RNA 7SK, coregulate the axonal transcriptome of motoneurons. Proc. Natl Acad. Sci. USA115, E2859–E2868 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang, M. & Pestov, D. G. Quantitative northern blot analysis of mammalian rRNA processing. Methods Mol. Biol.1455, 147–157 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The CLIP-seq and RNA-seq data reported in this paper have been deposited in the Genome Sequence Archive database in the National Genomics Data Center under the accession code GSA: CRA011147, and in the Gene Expression Omnibus under the accession code GSE232096. The dataset of this paper has been submitted to the figshare repository (10.6084/m9.figshare.27263289. Published RNA-seq datasets (GSE66582, GSE226316) and hnRNP CLIP data (GSE77101-GSM2044162 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE77101]) were used in this study. Source data are provided with this paper.