Abstract
Background
Carbapenem-resistant Klebsiella pneumoniae (CRKP) increases the difficulty of clinical treatment of giant pandas. This study aimed to investigate the antibiotic susceptibility, antibiotic resistance genes (ARGs), mobile genetic elements (MGEs), virulence genes, and molecular epidemiology of CRKP strains isolated from giant pandas. A total of 187 nonduplicated Klebsiella pneumoniae (KP) isolates were collected from fresh feces of captive giant pandas at the Chengdu Research Base of Giant Panda Breeding. Then CRKP were isolated and identified through carbapenase Carba NP assay. Subsequently, the antimicrobial susceptibility testing and antibiotic resistance genes of CRKP isolates were studied by disk diffusion (K-B) and HT-qPCR, respectively. Then both the MGEs and virulence genes of CRKP isolates were analyzed by PCR. In addition, molecular epidemiology was analyzed among the CRKP strains using pulsed-field gel electrophoresis (PFGE) and multi-locus sequence typing (MLST).
Results
Eight strains of CRKP (4.5%) were isolated and identified among the 187 KP strains, and seven of eight CRKP strains both exhibited resistance to imipenem, while one strain showed resistance to meropenem, and one demonstrated multiple resistance; eight CRKP strains carried a large amount of ARGs, among which ampC/blaDHA, blaSHV−01, blaSHV−02, tetB−01, tetB−02, tetC−01, and tetC−02 were the most abundant. The MGEs analysis revealed the presence of intI1 in all strains, while the detection rates of other MGEs varied, and strain 24 exhibited the highest diversity of MGE species. Seven virulence genes, including wabG, uge, ycf, entB, kpn, alls, and wcaG, showed positive results with different proportions across the strains. In addition, PFGE patterns indicated a high level of genetic diversity among the CRKP strains. MLST analysis classified the strains into different sequence types (STs).
Conclusions
This study highlighted the diversity of CRKP strains isolated from giant panda feces, which exhibited varying levels of antibiotic resistance along with multiple ARGs, MGEs and virulence genes present. These findings emphasized the importance of monitoring and researching antibiotic resistance within wildlife populations to protect the health status of these conservation dependent animals.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12917-024-04377-1.
Keywords: Carbapenem-resistant Klebsiella pneumoniae, Antimicrobial resistance, Mobile genetic elements, Virulence genes, Molecular epidemiology
Background
The giant panda, an endemic species to China, serves as the flagship species for global wildlife conservation efforts [1]. Bacterial diseases have emerged as a major epidemic that poses a grave threat to the life and well-being of giant pandas. With the widespread utilization of antibiotics, bacterial resistance continues to emerge and escalate every year. The propagation of antibiotic resistance genes (ARGs) has become an emerging public health problem, and ARGs are now considered as new environmental contaminants [2–4]. ARGs could be further disseminated among antimicrobial resistant bacteria (ARB) through horizontal gene transfer (HGT) mechanisms via mobile genetic elements (MGEs) [5, 6], thereby posing an immense risk to the health of giant panda [7–9]. Gastrointestinal bacterial infections are believed to be a primary cause of mortality among giant pandas, with alterations in gut microecology leading to digestive disorders and potential disease development. The intestinal disease in giant pandas can be caused by bacterial pathogens such as Escherichia coli, Klebsiella pneumoniae (KP), Campylobacter jejuni, Arizona bacillus, Pseudomonas aeruginosa and other bacteria [10].
KP is a gram-negative, conditionally pathogenic bacillus. In captive giant pandas, KP infection has become increasingly prevalent and often occurs in conjunction with other bacteria, making it the most important pathogen to treat [11–13]. Infected pandas may develop haemorrhagic enteritis characterized by bloody stools and genitourinary bleeding characterized by haematuria, which can potentially result in fatal haemorrhagic sepsis. In addition, our previous study has identified the emergence of drug resistance in giant panda-derived KP [14].
According to our research, antimicrobials have been extensively utilized for the prevention and treatment of infectious diseases in giant pandas over the past few decades [7, 15–17]. The overuse of antibiotics is the primary factor contributing to the emergence of carbapenem-resistant Klebsiella pneumoniae (CRKP). Furthermore, HGT via MGEs such as integrons, transposons, integration-coupled elements, genomic islands and plasmids play an important role in disseminating ARGs carried by multi-drug-resistant (MDR) KP [18–20]. Integrons possess the ability to capture, transform and adapt one or more resistance gene cassettes into functionally expressed genes through self-acting gene expression systems [21]. Their association with plasmids also facilitates the transfer of these genes among different bacterial species. The three main types of MGEs associated with antimicrobial resistance are classified as type 1, type 2 and type 3 integrons [22].
Recent studies have identified a large number of ARB, ARGs and their MGEs (including integrons) in E. coli isolated from captive giant pandas [7, 15–17]. However, limited information is available regarding the prevalence of CRKPs, the diversity of ARGs and MGEs, and the correlation between antimicrobial resistance and the occurrence of integron gene cassettes in CRKP among captive giant pandas. Additionally, there is a lack of knowledge about the antimicrobial resistance profiles across different age groups of giant pandas. Therefore, the objective of this study was to analyze the antimicrobial resistance profiles of 178 KP strains collected from fecal samples obtained from captive giant pandas belonging to various age and sex groups. Furthermore, we aimed to investigate the presence of ARGs, integrative subgene cassettes and other MGEs in eight CRKP strains. These findings will provide valuable insights for guiding appropriate use of clinical antibiotics in giant pandas.
Results
Antibiotic susceptibility of CRKP isolates
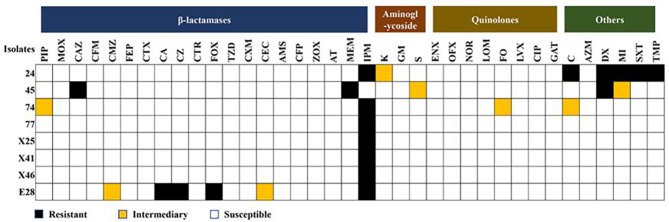
The results of Carba NP experiment showed that eight out of 178 KP isolates had yellow B solution (Fig. 1), which indicated the eight KP isolates, numbered 24, 45, 74, 77, X25, X41, X46, E28, were CRKP, with an isolation rate 4.5% (8/178). Then we further analyzed the antibiotic susceptibility of the eight CRKP isolates to 37 antibiotics, the results showed that seven of eight CRKP strains (strain 24, 74, 77, X25, X41, X46, E28) both exhibited resistance to imipenem, while one strain (strain 45) showed resistance to meropenem. Additionally, strain 24 demonstrated multiple resistance with a spectrum including imipenem, chloramphenicol, doxycycline, minocycline, compound sulfamethoxazole, and trimethoprim. Strain E28 displayed widely resistance to β-lactams such as cephalexin, cefazolin, cefoxitin, and imipenem. In addition to meropenem, strain 45 was resistant to ceftazidime and doxycycline. However, strain 74, 77, X25, X41, X46 did not exhibit any resistance except for imipenem (Fig. 2).
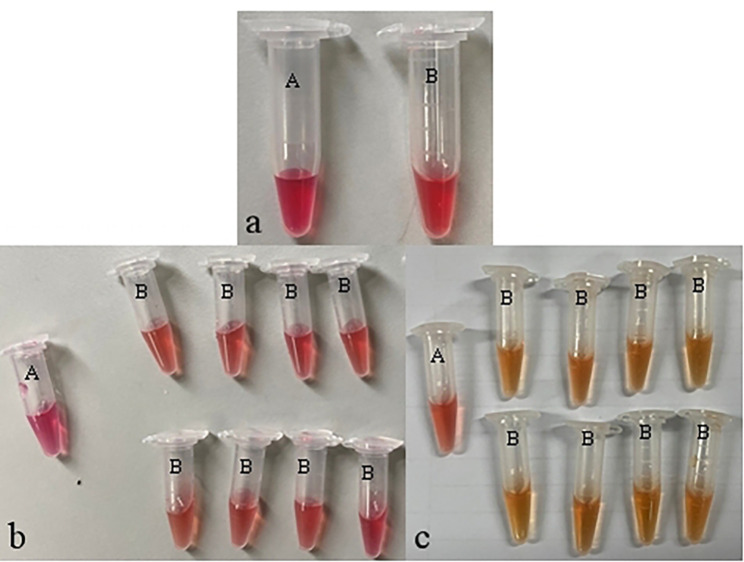
Fig. 1.
Identification results of CRKP in isolated strains a: blank control: physiological saline was added to solution B, the color did not change; b: negative result: the crude enzyme extract of the strain was added to solution B, and the color did not change, indicating that the isolated strain is a CRKP negative strain; c: positive result: The crude enzyme extract of the strain was added to the solution B, and the color changed to yellow, indicating that the isolated strain was a CRKP positive strain
Fig. 2.
The antibiotic susceptibility profiles of CRKP isolates. Columns: A total of 37 different types of antibiotics; Rows: the number of isolates in the study. PIP: piperacillin, MOX: moxalactam, CAZ: ceftazidime, CFM: cefixime, CMZ: ceftazidime, FEP: cefepime, CTX: cefotaxime, CA: cephalexin, CZ: cephazolin, CTR: ceftriaxone, FOX: cefoxitin, TZD: piperacillin/tazobactam, CXM: cefuroxime, CEC: cefaclor, AMS: ampicillin/sulbactam, CFP: cefoperazone, ZOX: ceftizoxime, AT: aztreonam, MEM: meropenem, IPM: imipenem, K: kanamycin, GM: gentamicin, S: streptomycin, ENX: enoxacin, OFX: ofloxacin, NOR: norfloxacin, LOM: lomefloxacin, FO: fleroxacin, LVX: levofloxacin, CIP: ciprofloxacin, GAT: gatifloxacin, C: chloramphenicol, AZM: azithromycin, DX: doxycycline, MI: minocycline, SXT: compound sulfamethoxazole, TMP: trimethoprim
Antibiotic resistance genes in 8 CRKP isolates
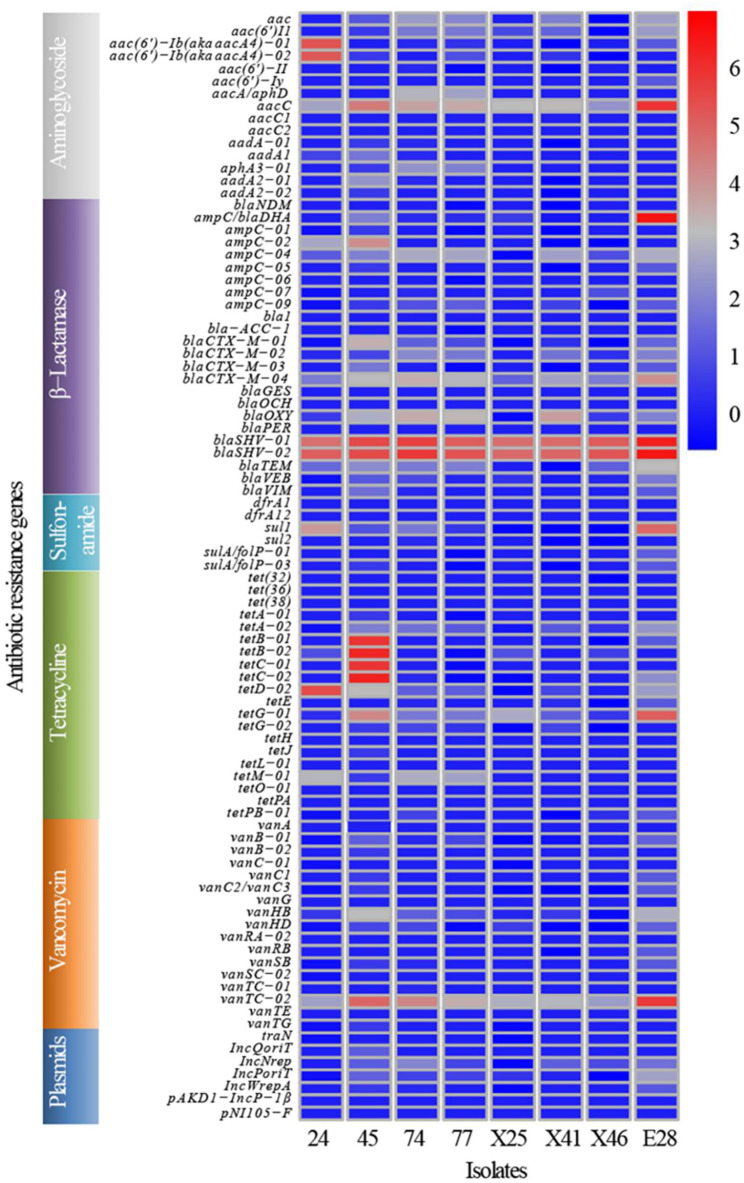
A total of 89 ARGs were assessed in eight CRKP isolates using HT-qPCR. Out of these, 47 ARGs were detected, six ARGs among which were positively present in all strains, including aacC, blaCTX−M−04, blaOXY, blaSHV−01, blaSHV−02 and vanTC−02. However, 42 ARGs such as aac(6’)−II, aac(6’)−Iy, aacC1, blaPER were not detected. Furthermore, the abundance of the identified 47 positive ARGs varied among the strains. The top ten resistance genes of abundance were: aacC, ampC/blaDHA, blaSHV−01, blaSHV−02, tetB−01, tetB−02, tetC−01, tetC−02, tetD−02, and vanTC−02. It was worth noting that ARGs of tetracycline were highly abundant in strain 45 (Fig. 3).
Fig. 3.
ARGs distribution in CRKP in giant panda. Columns: the number of isolates in the study; Rows: the 89 different types of ARGs
The virulence genes and MGEs of 8 CRKP isolates
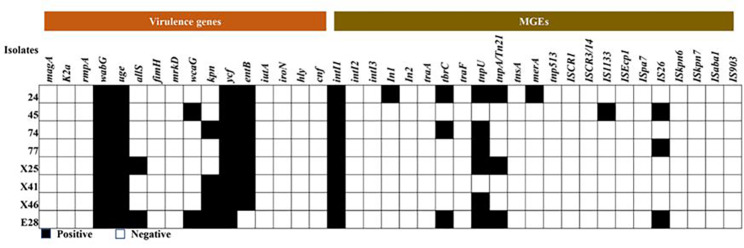
The identification of virulence-related factors is a crucial step in comprehending the molecular basis of bacterial disease. Hence, 16 virulence genes from eight isolates of CRKP were detected and analyzed in this study. Among them, seven genes tested positive, including wabG (100.0%), uge (100.0%), ycf (100.0%), entB (87.5%), kpn (50.0%), allS (25.0%), and wcaG (25.0%). In addition, strain E28 exhibited the highest number of virulence genes with a total of six virulence genes present (wagGI, uge, alls, wcaG, kpn, ycf), while other strains carried only four or five virulence genes (Fig. 4).
Fig. 4.
The detected result of virulence genes and MGEs in CRKP strains isolated from giant panda feces. Columns: the16 different types of virulence genes and 23 different types of MGEs; Rows: the number of isolates in the study
In addition, this study also included the analysis of a total 23 MGEs. The intI1 gene was detected in all eight strains, while there were significant variations in the detection rates among the other 22 MGEs. Specifically, the detection rates of In1, tbrC, tnpU, tnpA/Tn21, merA, IS1133 and IS26 were found to be 12.5%, 37.5%, 75.0%, 37.5%, 12.5%, 12.5% and 37.5% respectively. None of the remaining MGEs were detected. Furthermore, there were notable differences in the number of MGEs carried by different strains, among which strain 24 carried the most MGEs species (6/23) (Fig. 4).
The molecular epidemiology of 8 CRKP isolates
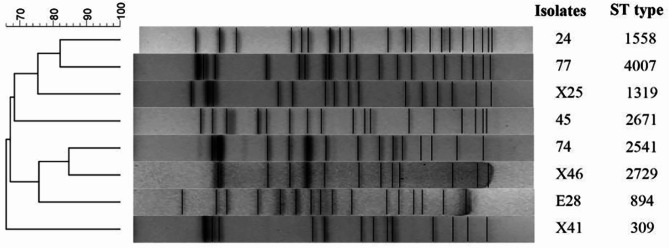
The eight CRKP isolates exhibited distinct PFGE patterns and highly diverse MLST types (Fig. 5). These CRKP strains displayed a high level of genetic diversity, with less than or equal to 84%. MLST analysis revealed that the eight CRKP strains belonged to different sequence types (ST).
Fig. 5.
Molecular epidemiology of SRKP through the analysis of PFGE and MLST The dendrogram of PFGE, generated using the BioNumerics software, illustrated the relatedness of fingerprints among the eight CRKP strains isolated from giant panda feces. The dendrogram was constructed based on the restriction patterns of XbaI-digested KP genomes, with strain numbers and ST types depicted alongside the corresponding restriction profiles
Discussion
The emergence of CRKP was initially documented in the United States, followed by subsequent reports from the France, Sweden, and Canada [23, 24]. In a hospital located in eastern China, the isolation rate of CRKP reached 41.3%, surpassing the isolation rate of 4.5% in our findings in giant pandas [25]. This disparity could potentially be attributed to the comparatively lower utilization of antibiotics for treating giant pandas, and the fact that our study isolated CRKP strains exclusively from fecal samples obtained from healthy giant pandas as opposed to clinical specimens collected in other investigations.
The antibiotic susceptibility test of CRKP isolates revealed that 87.5% of the isolates exhibited resistance to imipenem; while 25.0% showed resistance to doxycycline. Additionally, 12.5% displayed resistance to meropenem, cefazolin and chloramphenicol, indicating varying degrees of drug resistance among the eight CRKP isolates in this study, particularly towards carbapenems. Notably, the resistance of CRKP isolates to imipenem and meropenem varied across different countries and regions; for instance, in Saudi Arabia, the rates were 55.6% and 61.7%, respectively; whereas in New York, USA, they were observed at 17.0% and 20.0%; similarly in Chongqing, China, they stood at 37.2% and 30.8%, respectively [23, 24, 26, 27]. The resistance rate of the eight CRKP strains to imipenem in this study (87.5%) was significantly higher compared to that of meropenem (12.5%). The reasons for this outcome could be attributed to several factors: firstly, the utilization of antibiotics varied across different countries and regions, and the results of resistance test suggested that veterinarians may preferentially administer imipenem during clinical treatment of giant pandas; secondly, there existed disparities in the resistance mechanisms between the two CRKP strains, as meropenem targets both PBP2 and PBP3, whereas imipenem exhibits a stronger affinity towards PBP2 only [28]. Additionally, it was worth noting that all eight CRKP strains examined in this study demonstrated sensitivity towards amitrazan, kanamycin, gentamicin, ofloxacin and norfloxacin. These findings suggested potential clinical options for preventing and treating bacterial infections caused by these strains.
HT-qPCR technique enables rapid and sensitive quantification of numerous ARGs that confer resistance to nearly all major classes of antibiotics and provides a comprehensive profile of ARGs [29]. Therefore, this method was employed to perform a total of 89 ARGs in the eight CRKP strains. Out of these, 47 ARGs were detected, with six ARGs positively presented in all strains, namely aacC, blaCTX−M−04, blaOXY, blaSHV−01, blaSHV−02 and vanTC−02. The most abundance ARGs identified included ampC/blaDHA, blaSHV−01, blaSHV−02, tetB−01, tetB−02, tetC−01, tetC−02, which was consistent with the findings reported by Hu [30]. Simultaneously, the results from antimicrobial susceptibility testing revealed that the MDR isolates also exhibited significant resistance towards β-lactam antibiotics, particularly imipenem had an alarming resistance rate of 87.5%. The primary mechanism underlying KP’s β-lactam antibiotic resistance lies in its production of extended-spectrum β-lactamases encoded by extended-spectrum β-lactamase genes that disrupt the β-lactam ring structure leading to antibiotic inactivation [31]. The resistance phenotype and genotype of the isolates in this study closely corresponded with the primary mechanism of KP’s β-lactam antibiotic resistance, despite the diversity observed in both β-lactam antibiotics and genes. Given the rapid dissemination and emergence of novel ARGs among bacteria populations worldwide, it has now become imperative to monitor closely for any resistance genes carried by giant panda-associated bacteria.
MGEs play a crucial role in facilitating the transmission of ARGs within KP strains. In this study, a total of 23 MGEs were analyzed. It was observed that intI1 was present in all eight strains, while there were significant variations in the detection rates of the other 22 MGEs. The detection rates for In1, tbrC, tnpU, tnpA/Tn21, merA, IS1133, and IS26 were found to be 12.5%, 37.5%, 75%, 37.5%, 12.5%, 12.5%, and 37.5%, respectively; whereas the remaining MGEs were not detected. Interestingly, different strains exhibited varying numbers of MGE species, with strain 24 carrying the highest diversity of MGEs. The emergence of multidrug resistance in microorganisms is believed to be closely linked to MGEs due to their ability to swiftly transfer multiple ARGs [32]. The horizontal transfer of ARGs mediated by MGEs is considered as a primary mechanism driving the spread of ARGs [15]. MGEs, serving as carriers of ARGs, play a pivotal role in capturing, accumulating, and disseminating these genes. This transfer can occur both intra-strain and inter-strain within KP, thereby facilitating the rapid and widespread proliferation of antibiotic-resistant bacteria. Moreover, the horizontal transfer of ARGs contributes to the emergence of drug-resistant and multi-drug-resistant bacteria in clinical settings [33]. In this study, we found that multidrug-resistant KP strains isolated from different giant pandas carried common ARGs along with a substantial number of MGEs detected. Therefore, it is plausible to hypothesize that there might be sharing of ARGs among the giant pandas in this study. However, further investigations are required to determine the precise mechanisms underlying the sharing of ARGs.
Many types of virulence factors, including capsular polysaccharide, lipid polysaccharide, pili, and ferriferous are carried by KP and are pathogenic factors, which are encoded and expressed by virulence genes such as magA, wabG, mrk, iro respectively [58]. These virulence genes can help KP to establish immune escape and induce host sensation through different mechanisms, and the symptoms induced by different virulence genes are also different. Because of these factors the detection of the 16 common virulence genes from the eight CRKP strains was also an important part of this study. Seven genes, namely wabG (100%), uge (100%), ycf (100%), entB (87.5%), kpn (50%), allS (25%), and wcaG (25%), were found to be positive. Additionally, among the strains, E28 carried the highest number of virulence genes, with six virulence genes identified as wagGI, uge, all, wcaG, kpn, ycf, while other strains carried only four or five virulence genes, indicating a relatively high prevalence of multiple-virulence-gene-carrying strains in adult giant panda-sourced KP.
Genes associated with lipopolysaccharide (LPS) production primarily included uge and wabG, encoding UDP-galactose-4-epimerase and galactosyltransferase, respectively. These two types of virulence genes are commonly presented in a variety of virulence factors within KP strains. Studies have demonstrated that KP lacking the uge gene produces incomplete lipopolysaccharides [34]. Mutations or deletions of the wabG gene in KP can result in the absence of certain capsular antigens and hemolysins, which reduces pathogenicity in animal infection models [35]. In addition, there are several other genes potentially associated with the virulence of KP, including kpn gene involved in capsule polysaccharide synthesis, and wcaG and wagGI genes encoding extracellular toxins. Research has suggested that genes related to iron carriers, pili, and lipopolysaccharides are located on virulence plasmids in highly virulent KP strains and serve as specific molecular markers for high-virulent strains [36, 37]. Therefore, it can be inferred that wabG, uge, and ycf may potentially serve as high virulence genes. In this study, the LPS carrier genes wabG and uge exhibited a detection rate of 100%, which aligned with the findings of Zhang Wenju [38]. Conversely, unlike Han’s study [39], the fimH pilus virulence gene was not detected in KP strains from different animal sources, indicating variations in the carried virulence genes among KP isolates.
The iron carrier gene entB displayed a detection rate of 87.5%, while all urease genes had a detection rate of 25%. These results were consistent with those reported by Wang [40]. This study identified one strain of isolated bacteria carrying six virulence genes, including lipopolysaccharide carrier genes and urease genes, suggesting its potential as a highly virulent KP strain isolated from giant pandas, laying the foundation for future investigations on the pathogenicity.
The DNA fingerprinting by PFGE is considered to be the “gold standard” for typing of microorganisms due to the adequate consistency within a single assay, which is widely used in molecular epidemiology [41]. MLST technique is a well-established procedure employed for characterizing bacterial species by sequencing internal fragments of typically seven housekeeping genes [42]. Eight strains of bacteria exhibited distinct PFGE patterns, indicating a high degree of genetic diversity (≤ 84%) among these CRKP strains. Further analysis using MLST revealed that the eight CRKP strains could be classified into eight distinct STs, indicating that the molecular epidemiology was diverse among CRKP strains isolated from various giant pandas.
PCR is a molecular biology technique that can rapidly amplify specific DNA fragments in vitro. By designing gene primers with known sequences, specific gene fragments carried by bacterial samples can be directly detected, and the operation is simple and easy. However, ordinary PCR technology has disadvantages such as low detection throughput, large workload, and the need for sequencing to further analyze detection results. HT-qPCR, on the other hand, has the advantages of high throughput, sensitivity, and speed, which can perform large-scale and rapid quantitative detection of specific genes carried by bacterial strains, and provide a comprehensive overview of specific genes. This study mainly detected the ARGs, MGEs, virulence gene, and MLST molecular typing of CRKP isolated from giant pandas through HT-qPCR and ordinary PCR in the world’s largest captive population of giant pandas. The above results contribute to a comprehensive understanding of the distribution and epidemiological investigation of giant panda derived CRKP. However, both ordinary PCR and HT qPCR require bacterial culture as the basis and are limited to detecting known and limited gene sequences, making it difficult to determine the location and genetic environment of key genes in experimental strains. This poses challenges for further mechanism research and source tracing. Whole genome sequencing is the individual genome sequencing of species with unknown genome sequences, while bacteria include genetic information and frameworks of chromosomes and all plasmids. Through whole genome sequencing, we can grasp all genetic information of bacteria, including base composition, subtype types and quantities, location, and gene environment, and then compare them with their counterparts in public databases to explore antibiotic resistance and transmission mechanisms. Of course, this also requires more time and funding, and is our future research goal.
Conclusion
In this study, we investigated eight strains of CRKP isolated from giant panda fecal samples to determine their antibiotic susceptibility, ARGs, MGEs, virulence genes, and molecular epidemiology. The most abundant ARGs included ampC/blaDHA, blaSHV−01, blaSHV−02, tetB−01, tetB−02, tetC−01, and tetC−02. Analysis of MGE revealed the presence of intI1 in all strains, while other MGEs exhibited varying detection rates. Strain 24 carried the highest diversity of MGE species. Seven virulence genes including wabG, uge, ycf, entB, kpn, alls, and wcaG were detected across the strains with varying proportions. Molecular epidemiology analysis using PFGE patterns indicated a high level of genetic diversity among the CRKP strains. MLST analysis classified the strains into different STs. In conclusion, this study highlighted the remarkable diversity of CRKP strains found in giant panda feces, exhibiting varying degrees of antibiotic resistance and the presence of virulence genes and multiple ARGs which could be transmitted and shared through MGEs. These findings emphasized the critical significance of monitoring and researching antibiotic resistance in wildlife populations to safeguard the well-being of these conservation dependent animals, and we should rationally use antibiotics in the clinical treatment of wild animals such as giant pandas.
Methods
Bacterial isolates and screening for carbapenemases phenotype
One hundred seventy-eight nonduplicated KP isolates were collected from fresh feces of captive giant pandas at the Chengdu Research Base of Giant Panda Breeding (Panda Base) in Sichuan, China, between 2018 and 2019. These isolates were identified as KP by Gram staining, bacterial biochemical identification and 16 S rDNA analysis. Carba NP test was used to identify the phenotype of carbapenase of the identified KP strain [43], results were interpreted in accordance with the Clinical and Laboratory Standards Institute [CLSI], 2020 criteria [44].
Antimicrobial susceptibility testing of CRKP isolates
The antibiotic resistance testing of CRKP isolates was performed using the K-B method against a panel of antibiotics, including: piperacillin (PIP, 100 µg), moxalactam (MOX, 30 µg), ceftazidime (CAZ, 30 µg), cefixime (CFM, 5 µg), ceftazidime (CMZ, 30 µg), cefepime (FEP, 30 µg), cefotaxime (CTX, 30 µg), cephalexin (CA, 30 µg), cephazolin (CZ, 30 µg), ceftriaxone (CTR, 30 µg), cefoxitin (FOX, 30 µg), piperacillin/tazobactam (TZD, 100 µg), cefuroxime (CXM, 30 µg), cefaclor (CEC, 30 µg), ampicillin/sulbactam (AMS, 10 µg), cefoperazone (CFP, 75 µg), ceftizoxime (ZOX, 30 µg), aztreonam (AT, 30 µg), meropenem (MEM, 10 µg), imipenem (IPM, 10 µg), kanamycin (K, 30 µg), gentamicin (GM, 10 µg), streptomycin (S, 10 µg), enoxacin (ENX, 10 µg), ofloxacin (OFX, 5 µg), norfloxacin (NOR, 10 µg), lomefloxacin (LOM, 18 µg), fleroxacin (FO, 5 µg), levofloxacin (LVX, 5 µg), ciprofloxacin (CIP, 5 µg), gatifloxacin (GAT, 5 µg), chloramphenicol (C, 30 µg), azithromycin (AZM, 15 µg), doxycycline (DX, 30 µg), minocycline (MI, 30 µg), compound sulfamethoxazole (SXT, 23.75/1.25 µg), and trimethoprim (TMP, 5 µg). The antibiotic disks were purchased from Hangzhou Microbial Reagent Co., Ltd., China. E. coli ATCC25922 was used as the quality control bacterial strain. Results were interpreted as susceptible (S), intermediate (I), and resistant (R) based on the interpretative criteria of the CLSI 2020.
DNA extraction of CRKP isolates
The CRKP isolates were subjected to total genomic DNA extraction using the TIANamp Bacteria DNA Kit (Tiangen Biotech, Beijing, China) following the manufacturer’s protocol. Subsequently, the DNA samples were stored at -20 °C.
Antibiotic resistance genes (ARGs) analysis of CRKP isolates
High-throughput quantitative PCR (HT-qPCR) was performed by Yearth Biotech (Changsha, China) using the wafergen smartchip real-time PCR system to analyze the antibiotic resistance genes. A total of 89 primer sets were employed for the detection of resistance genes (Table 1.). Each sample was simultaneously replicated three times. Following a pre-denaturation step at 95 °C for 10 min, amplification was performed through 30 cycles according to the following program: denaturation at 95 °C for 30 s and annealing at 60 °C for 30 s. The obtained results were then analyzed with smartchip qPCR Software to exclude wells exhibiting multiple melting peaks or amplification efficiency beyond the range (90-110%).
Table 1.
Primers of virulence gene detection of 8 CRKP in the study
| Primer | Sequence(5’-3’) | Product size(bp) | The encoded virulence factors | related references |
|---|---|---|---|---|
| magA-F | GGTGCTCTTTACATCATTGC | 1282 | Hypermucoviscosity, capsules | [46] |
| magA-R | GCAATGGCCATTTGCGTTAG | |||
| fimH-F | TGCTGCTGGGCTGGTCGATG | 688 | fimbrial adhesins | [47] |
| fimH-R | GGGAGGGTGACGGTGACATC | |||
| uge-F | TCTTCACGCCTTCCTTCACT | 534 | Lipopolysaccharides, capsule associated genes | [48] |
| uge-R | GATCATCCGGTCTCCCTGTA | |||
| iutA-F | GGCTGGACATCATGGGAACTGG | 300 | Siderophores (iron acquisition systems) | [49] |
| iutA-R | CGTCGGGAACGGGTAGAATCG | |||
| wabG-F | ACCATCGGCCATTTGATAGA | 683 | Lipopolysaccharides, capsule associated genes | [46] |
| wabG-R | CGGACTGGCAGATCCATATC | |||
| rmpA-F | ACTGGGCTACCTCTGCTTCA | 516 | hypermucoviscosity | [46], [48] |
| rmpA-R | CTTGCATGAGCCATCTTTCA | |||
| cnf-F | AAGATGGAGTTTCCTATGCAGGAG | 498 | other virulence factors | [49] |
| cnf-R | CATTCAGAGTCCTGCCCTCATTATT | |||
| ycf-F | ATCAGCAGTCGGGTCAGC | 160 | Lipopolysaccharides, capsule associated genes | [50] |
| ycf-R | CTTCTCCAGCATTCAGCG | |||
| hly-F | AACAAGGATAAGCACTGTTCTGGCT | 1177 | other virulence factors | [49] |
| hly-R | ACCATATAAGCGGTCATTCCCGTCA | |||
| iroN-F | AAGTCAAAGCAGGGGTTGCCCG | 665 | Siderophores (iron acquisition systems) | [51] |
| iroN-R | GACGCCGACATTAAGACGCAG | |||
| k2A-F | CAACCATGGTGGTCGATTAG | 543 | encode capsules | [52] |
| k2A-R | TGGTAGCCATATCCCTTTGG | |||
| mrkD-F | TTCTGCACAGCGGTCCC | 240 | fimbrial adhesins | [53] |
| mrkD-R | GATACCCGGCGTTTTCGTTAC | |||
| kpn-F | GTATGACTCGGGGAAGATTA | 626 | fimbrial adhesins | [50] |
| kpn-R | CAGAAGCAGCCACCACACG | |||
| allS-F | CCGAAACATTACGCACCTTT | 508 | other virulence factors | [47] |
| allS-R | ATCACGAAGAGCCAGGTCAC | |||
| entB-F | ATTTCCTCAACTTCTGGGGC | 371 | Siderophores (iron acquisition systems) | [50] |
| entB-R | AGCATCGGTGGCGGTGGTCA | |||
| wcaG-F | GGTTGGKTCAGCAATCGTA | 169 | encode capsules | [48] |
| wcaG-R | ACTATTCCGCCAACTTTTGC |
Mobile genetic elements (MGEs) analysis of CRKP isolates
Twenty-three pairs of primers were used to detect MGEs in CRKP isolates by PCR, following previously described protocols [16, 17, 23, 45](Supplementary Table 1.). PCR amplification was performed in a total volume of 25 µL containing 12.5 µL of Dream Taq Green PCR Master Mix (2×), 8.5 µL ddH2O, and each forward primer and reverse primer at a concentration of 1 µL, DNA template 2 µL. Amplification was carried out under the following thermal cycling conditions: pre-denaturation at 95 °C for 5 min, followed by a total of 30 cycles consisting of denaturation at 95 °C for 30 s, annealing at the specified temperature for 30 s, extension at 72 °C for 30 s, and final extension at 72 °C for 10 min. PCR products were subsequently subjected to 1% agarose gel at 120 V, 0.5 × TAE buffer electrophoresis for 38 min.
Virulence gene analysis of CRKP isolates
Based on a number of previous investigations [46–53] related to the virulence genes of KP, four different multiplex PCR reaction mixtures were defined for the 16 virulence genes (magA-fimH-uge-iutA, wabG-rmpA-cnf-ycf, hly-iroN-K2a-mrkD, kpn-allS-entB-wcaG) and were then used to study the eight CRKP strains (Table 1). Each set consisted of a total volume of 50 µL containing 25 µL of Dream Taq Green PCR Master Mix (2×), 2 µL of total DNA, 22 µL ddH2O, and each forward primers and reverse primers (Sangon Biotech Co., Ltd., Shanghai, China) at a concentration of 0.5 µL. The amplification process was carried out with the following thermal cycling conditions: pre-denaturation at 95 °C for 5 min, followed by 30 cycles consisting of denaturation at 94 °C for 1 min, annealing at 58 °C for 1 min, extension at 72 °C for 1 min, and final elongation at 72 °C for 10 min. Finally, the PCR products were subjected to 2% agarose gel at 120 V, 1 × TAE buffer electrophoresis for 38 min.
Molecular epidemiology analysis of CRKP isolates
PFGE was conducted to investigate the molecular epidemiology of the CRKP isolates with XbaI-digested and genotyped DNA. The genomic DNA restriction patterns of the isolates were analyzed and interpreted based on the previously established criteria [54]. Additionally, in order to further assess whether clonal spread influenced the dissemination of carbapenemase-producing KP isolates in giant panda, MLST was performed by amplifying internal fragments of seven standard housekeeping loci gapA, infB, mdh, pgi, phoE, rpoB, and tonB.
Sequence and data analysis
All positive PCR products (MGEs, virulence gene and MLST) were sequenced by Sangong Biotech (Shanghai, China) in both directions. Sequences of MGEs and virulence genes were analyzed online using BLAST (http://blast.ncbi.nlm.nih.gov), and sequences of MLST were analyzed online using MLST (https://bigsdb.pasteur.fr/cgi-bin/bigsdb/bigsdb.pl?db=pubmlst_klebsiella_seqdef&page=profiles).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank the veterinary staff and keepers of the Chengdu Panda Base for collecting samples, and Cen Xin for arrangement of article data.
Abbreviations
- AMS
Ampicillin/sulbactam
- ARB
Antimicrobial Resistant Bacteria
- ARGs
Antibiotic Resistance Genes
- AT
Aztreonam
- AZM
Azithromycin
- C
Chloramphenicol
- CA
Cephalexin
- CAZ
Ceftazidime
- CEC
Cefaclor
- CFM
Cefixime
- CFP
Cefoperazone
- CIP
Ciprofloxacin
- CLSI
Clinical andLaboratory Standards Institute
- CMZ
Ceftazidime
- CRKP
Carbapenem-Resistant Klebsiella pneumoniae
- CTR
Ceftriaxone
- CTX
Cefotaxime
- CXM
Cefuroxime
- CZ
Cephazolin
- DX
Doxycycline
- ENX
Enoxacin
- FEP
Cefepime
- FO
Fleroxacin
- FOX
Cefoxitin
- GAT
Gatifloxacin
- GM
Gentamicin
- HGT
Horizontal Gene Transfer
- IPM
Imipenem
- K
Kanamycin
- K-B
Disk Diffusion
- KP
Klebsiella pneumoniae
- LOM
Lomefloxacin
- LVX
Levofloxacin
- MDR
Multi-Drug-Resistant
- MEM
Meropenem
- MGEs
Mobile Genetic Elements
- MI
Minocycline
- MLST
Multi-Locus Sequence Typing
- MOX
Moxalactam
- NOR
Norfloxacin
- OFX
Ofloxacin
- PFGE
Pulsed-Field Gel Electrophoresis
- PIP
Piperacillin
- S
Streptomycin
- SXT
Compound Sulfamethoxazole
- TMP
Trimethoprim
- TZD
Piperacillin/Tazobactam
- ZOX
Ceftizoxime
Author contributions
X Y, Xy S, R H, L L, Cj Y and Sr L contributed to conception and design of the study. Xy S played a guiding role in carrying out the experiment. X Y, M Y, Yl L and Ds Z collected samples. X Y, M Y and Y Z performed bacterial identification and isolation and related component testing. Xy S and J E. Ayala performed the statistical analysis. X Y, M Y and Xy S wrote the first draft of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This research was supported by Sichuan Science and Technology Program (2024ZYD0132, 2023NSFSC1926), the Chengdu Research Base of Giant Panda Breeding (project nos. 2021CPB-B10).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
The study, including all sample collection, received ethical approval from the Institutional Animal Care and Use Committee (IACUC) of the Chengdu Research Base of Giant Panda Breeding (No. 2018017), and the samples collected in this study were feces collected noninvasively from captive giant pandas.
Consent for publication
All authors read and approved the final manuscript and give consent for publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jia W, Yan S, He Q, et al. Giant panda Microhabitat Study in the Daxiangling Niba Mountain Corridor. Biology (Basel). 2023;12(2):165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stoll C, Sidhu JP, Tiehm A, et al. Prevalence of clinically relevant antibiotic resistance genes in surface water samples collected from Germany and Australia. Environ Sci Technol. 2012;46(17):9716–26. [DOI] [PubMed] [Google Scholar]
- 3.Pruden A, Pei RT, Storteboom H, et al. Antibiotic resistance genes as emerging contaminants: studies in northern Colorado. Environ Sci Technol. 2006;40(23):7445–50. [DOI] [PubMed] [Google Scholar]
- 4.Cooper MA, Shlaes D. Fix the antibiotics pipeline. Nature. 2011;472:32. [DOI] [PubMed] [Google Scholar]
- 5.Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3(9):679–87. [DOI] [PubMed] [Google Scholar]
- 6.Schluter A, Szczepanowski R, Puhler A, et al. Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol Rev, 2010; 31 (4), 449–77. [DOI] [PubMed]
- 7.Guo L, Long M, Huang Y, et al. Antimicrobial and disinfectant resistance of Escherichia coli isolated from giant pandas. J Appl Microbiol. 2015;119(1):55–64. [DOI] [PubMed] [Google Scholar]
- 8.Mustafa GR, Li C, Zhao S, et al. Metagenomic analysis revealed a wide distribution of antibiotic resistance genes and biosynthesis of antibiotics in the gut of giant pandas. BMC Microbiol. 2021;21(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su X, Yan X, Li Y, et al. Identification of extended-spectrum beta-lactamase (CTX-M)-producing Klebsiella pneumoniae belonging to ST37, ST290, and ST2640 in captive giant pandas. BMC Vet Res. 2022;17(1):186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiong Y, Li D, Yin W, et al. Isolation, identification and distribution of fecal microflora of giant panda in Wolong Nature Reserve. J Animal Adv. 2000;(02):165–170. (In Chinese).
- 11.Wang Q,Jiang H, Nakao Kenko, et al. A case report of Klebsiella pneumoniae hemorrhagic enteritis of giant panda. Sichuan Zool. 1998;(01): 29. (In Chinese).
- 12.Wang C, Lan J, Luo L, et al. Klebsiella pneumoniae, pathogen of infectious urogenital hematuria in giant panda. Sichuan Zool. 2006;25(1): 83–5. (In Chinese).
- 13.Wang K, Zhang H. Diagnosis of bacterial septicemia in sub-adult giant panda. Chin J vet. 2000;30(1):28-29. (In Chinese). [Google Scholar]
- 14.Xiong Y. Zhang H.Study on etiology and pathogenesis of bacterial septicemia of subadult giant panda. Chin Vet Sci.1998;028(001):7–9. (In Chinese).
- 15.Chen D, Zou W, Xie S, et al. Serotype and antimicrobial resistance of Escherichia coli isolated from feces of wild giant pandas (Ailuropoda melanoleuca) in sichuan province, China. J Wildl Dis. 2018;54(4):691–9. [DOI] [PubMed] [Google Scholar]
- 16.Zou W, Li C, Yang X, et al. Frequency of antimicrobial resistance and integron gene cassettes in Escherichia coli isolated from giant pandas (Ailuropoda melanoleuca) in China. Microb Pathog. 2018;116:173–9. [DOI] [PubMed] [Google Scholar]
- 17.Zhu Z, Pan S, Wei B, et al. High prevalence of multi-drug resistances and diversity of mobile genetic elements in Escherichia coli isolates from captive giant pandas. Ecotoxicol Environ Saf. 2020;198:110681. [DOI] [PubMed] [Google Scholar]
- 18.Chen Q, An X, Li H, et al. Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ Int. 2016;92–93:1–10. [DOI] [PubMed] [Google Scholar]
- 19.Partridge SR, Tsafnat G, Coiera E, et al. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev. 2009;33:757–84. [DOI] [PubMed] [Google Scholar]
- 20.Yan X, Su X, Ren Z, et al. High prevalence of Antimicrobial Resistance and Integron Gene cassettes in Multi-drug-resistant Klebsiella pneumoniae isolates from Captive Giant pandas (Ailuropoda melanoleuca). Front Microbiol. 2022;3:12:801292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gillings MR. Integrons: past, present, and future. Microbiol Mol Biol Rev. 2014;78(2):257–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaushik M, Kumar S, Kapoor RK, et al. Integrons in Enterobacteriaceae: diversity, distribution and epidemiology. Int J Antimicrob Agents. 2018;51:167–76. [DOI] [PubMed] [Google Scholar]
- 23.White PA, McIver CJ, Rawlinson WD. Integrons and gene cassettes in the enterobacteriaceae. Antimicrob Agents Chemother. 2001;45(9):2658–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang D, Sharma L, Dela Cruz CS, et al. Clinical epidemiology, risk factors, and control strategies of Klebsiella pneumoniae infection. Front Microbiol. 2021;12:750662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu F, Zhang L, Ji J, et al. Epidemiological and antimicrobial resistant patterns, and Molecular mechanisms of Carbapenem-resistant Klebsiella pneumoniae infections in ICU patients. Infect Drug Resist. 2023;16:2813–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al Bshabshe A, Al-Hakami A, Alshehri B, et al. Rising Klebsiella pneumoniae infections and its Expanding Drug Resistance in the Intensive Care Unit of a Tertiary Healthcare Hospital, Saudi Arabia. Cureus. 2020;12(8):e10060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaiser RM, Castanheira M, Jones RN, et al. Trends in Klebsiella pneumoniae carbapenemase-positive K. pneumoniae in US hospitals: report from the 2007–2009 sentry antimicrobial surveillance program. Diagn Microbiol Infect Dis. 2013;76(3):356–60. [DOI] [PubMed] [Google Scholar]
- 28.Zhanel GG, Wiebe R, Dilay L, et al. Comparative review of the carbapenems. Drugs. 2007;67(7):1027–52. [DOI] [PubMed] [Google Scholar]
- 29.Su J, Wei B, Ou-Yang W, et al. Antibiotic resistome and its association with bacterial communities during sewage sludge composting. Environ Sci Technol. 2015;49(12):7356–63. [DOI] [PubMed]
- 30.Hu T, Dai Q, Chen H, et al. Geographic pattern of antibiotic resistance genes in the metagenomes of the giant panda. Microb Biotechnol. 2021;14:186–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tooke CL, Hinchliffe P, Bragginton EC, et al. β-Lactamases and β-Lactamase inhibitors in the 21st Century. J Mol Biol. 2019;23(18):3472–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez JL. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ Pollut. 2009;157:2893–902. [DOI] [PubMed] [Google Scholar]
- 33.Chamosa LS, Álvarez VE, Nardelli M, et al. Lateral Antimicrobial Resistance genetic transfer is active in the open environment. Sci Rep. 2017;7(1):513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Regué M, Hita B, Piqué N, et al. A gene, uge, is essential for Klebsiella pneumoniae virulence. Infect Immun. 2004;72(1):54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cogen AL, Nizet V, Gallo RL. Skin microbiota: a source of disease or defence? Br J Dermatol. 2008;158(3):442–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russo TA, Olson R, Fang CT, et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol. 2019;57(7):e00976–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu SB, Fan L, Xia L, et al. Antimicrobial resistance and virulence genes of Klebsiella pneumoniae isolates from a tertiary hospital in Chongqing, China. Infect Drug Resist. 2017;10:111–7. [Google Scholar]
- 38.Zhang W, Liao X, Ren L et al. . Analysis of the virulence gene, drug resistance and biofilm formation of Klebsiella pneumoniae isolated from paediatric patients. PLoS ONE. 2020; 15(7): e0236283.
- 39.Han K. Analysis of virulence genes of Klebsiella pneumoniae isolated from mink pneumonia. J Anim Health Epidemic Prev. 2019;10(2):97–100. [Google Scholar]
- 40.Wang ZH, Xue WJ, Zhang YJ. Detection and drug resistance of Klebsiella pneumoniae virulence genes in respiratory tract infection. Chin J Practical Med. 2021;48(11):58–61. [Google Scholar]
- 41.Jouni Heikkinen A, von Wright I, Nikodinoska et al. An efficient Pulsed-Field Gel Electrophoresis (PFGE) method for typing autolytic Lacticaseibacillus rhamnosus strains. MethodsX. 2022; 9: 101945. [DOI] [PMC free article] [PubMed]
- 42.Jolley KA, Maiden MCJ, Bigsdb S. Sequence type analysis and recombinational tests (START). Bioinformatics. 2018;34(22):4002–4. [DOI] [PubMed] [Google Scholar]
- 43.Zhou Tingyin ZQ. Clinical Microbiology diagnosis and Diagram (Part 1) [M]. 4th Ed. Shanghai: Shanghai Science and Technology, 2017:54–6. (In Chinese).
- 44.Clinical and Laboratory Standards Institute [CLSI]. Performance standards for Antimicrobial susceptibility testing: twenty - fourth informational supplement CLSI document M100-S24. Wayne, PA: CLSI; 2020. [Google Scholar]
- 45.Lévesque C, Piché L, Larose C, et al. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39(1):185–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu WL, Ko WC, Cheng KC, et al. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis. 2006;42:1351–8. [DOI] [PubMed] [Google Scholar]
- 47.Yu WL, Ko WC, Cheng KC, et al. Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn Microbiol Infect Dis. 2008;62:1–6. [DOI] [PubMed] [Google Scholar]
- 48.Turton JF, Perry C, Elgohari S, et al. PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J Med Microbiol. 2010;9:541–7. [DOI] [PubMed] [Google Scholar]
- 49.Mamlouk K, Boutiba-Ben Boubaker I, Gautier V, et al. Emergence and outbreaks of CTX-M beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae strains in a Tunisian hospital. J Clin Microbiol. 2006;44:4049–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El Fertas-Aissani R, Messai Y, Alouache S, et al. Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens. Pathol Biol. 2013;61:209–16. [DOI] [PubMed] [Google Scholar]
- 51.Guiral E, Bosch J, Vila J, et al. Prevalence of Escherichia coli among samples collected from the genital tract in pregnant and nonpregnant women: relationship with virulence. FEMS Microbiol Lett. 2011;314:170–3. [DOI] [PubMed] [Google Scholar]
- 52.Yu WL, Fung CP, Ko WC, et al. Polymerase chain reaction analysis for detecting capsule serotypes K1 and K2 of Klebsiella pneumoniae causing abscesses of the liver and other sites. J Infect Dis. 2007;195:1235–6. [DOI] [PubMed] [Google Scholar]
- 53.Sebghati TAS, Korhonen TK, Hornick DB, et al. Characterization of the type 3 fimbrial adhesins of Klebsiella strains. Infect Immun. 1998;66:2887–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han H, Zhou H, Li H, et al. Optimization of pulse-field gel electrophoresis for subtyping of Klebsiella pneumoniae. Int J Environ Res Public Health. 2013;10(7):2720–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.