Abstract
Tyrosine kinase receptors lead to rapid activation of phosphatidylinositol 3-kinase (PI3 kinase) and the subsequent formation of phosphatidylinositides (PtdIns) 3,4-P2 and PtdIns 3,4,5-P3, which are thought to be involved in signaling for glucose transporter GLUT4 translocation, cytoskeletal rearrangement, and DNA synthesis. However, the specific role of each of these PtdIns in insulin and growth factor signaling is still mainly unknown. Therefore, we assessed, in the current study, the effect of SH2-containing inositol phosphatase (SHIP) expression on these biological effects. SHIP is a 5′ phosphatase that decreases the intracellular levels of PtdIns 3,4,5-P3. Expression of SHIP after nuclear microinjection in 3T3-L1 adipocytes inhibited insulin-induced GLUT4 translocation by 100 ± 21% (mean ± the standard error) at submaximal (3 ng/ml) and 64 ± 5% at maximal (10 ng/ml) insulin concentrations (P < 0.05 and P < 0.001, respectively). A catalytically inactive mutant of SHIP had no effect on insulin-induced GLUT4 translocation. Furthermore, SHIP also abolished GLUT4 translocation induced by a membrane-targeted catalytic subunit of PI3 kinase. In addition, insulin-, insulin-like growth factor I (IGF-I)-, and platelet-derived growth factor-induced cytoskeletal rearrangement, i.e., membrane ruffling, was significantly inhibited (78 ± 10, 64 ± 3, and 62 ± 5%, respectively; P < 0.05 for all) in 3T3-L1 adipocytes. In a rat fibroblast cell line overexpressing the human insulin receptor (HIRc-B), SHIP inhibited membrane ruffling induced by insulin and IGF-I by 76 ± 3% (P < 0.001) and 68 ± 5% (P < 0.005), respectively. However, growth factor-induced stress fiber breakdown was not affected by SHIP expression. Finally, SHIP decreased significantly growth factor-induced mitogen-activated protein kinase activation and DNA synthesis. Expression of the catalytically inactive mutant had no effect on these cellular responses. In summary, our results show that expression of SHIP inhibits insulin-induced GLUT4 translocation, growth factor-induced membrane ruffling, and DNA synthesis, indicating that PtdIns 3,4,5-P3 is the key phospholipid product mediating these biological actions.
Insulin binding stimulates tyrosine autophosphorylation of the insulin receptor and activates its intrinsic tyrosine kinase activity, leading to the phosphorylation of insulin receptor substrates and the subsequent activation of PI3 kinase (44). PI3 kinase is a heterodimer of a 110-kDa catalytic subunit (p110) and an 85-kDa regulatory subunit (p85). Once activated, it phosphorylates the D-3 position of phosphoinositides (PtdIns) (25, 44), leading to the formation of PtdIns 3,4-P2 and PtdIns 3,4,5-P3 (4, 39, 41). These PtdIns are thought to be second messengers that play a crucial role in the biologic actions of growth factors (41). However, the exact function of each of these PtdIns in hormone signaling is still unknown.
One of the major biological effects of insulin is to promote glucose uptake in muscle and fat tissue through the translocation of the glucose transporter GLUT4 to the plasma membrane (36), and PI3 kinase is both necessary (19) and sufficient (33) for this effect. Akt (PKB) is a serine-threonine kinase downstream of PI3 kinase, and overexpression of a constitutively active form of Akt leads to increased glucose uptake and GLUT4 translocation in 3T3-L1 adipocytes (28). 3′ PtdIns bind directly to Akt through its PH domain and PtdIns 3,4-P2 has been found to partially activate Akt in vitro (27), but full activation of the kinase requires Ser/Thr phosphorylation of the protein (29). Recently, an Akt kinase (PDK1) was cloned, and its activation of Akt was shown to be dependent on PtdIns 3,4,5-P3 (2, 3, 8, 38, 40). Therefore, the current data suggest that PtdIns 3,4-P2, as well as PtdIns 3,4,5-P3, play a role in insulin-induced Akt activation and GLUT4 translocation.
Growth factors such as insulin also induce actin filament rearrangement in various cell lines, leading to stress fiber breakdown and membrane ruffling (33, 34). This latter effect requires PI3 kinase activation and, in particular, PtdIns 3,4,5-P3 formation (21, 34, 43). Stress fiber formation correlates with PtdIns 4,5-P2 generation (7), and it has been suggested that stress fiber breakdown is induced by 3′ phosphorylation of 4,5-P2 induced by PI3 kinase (34). Finally, based on numerous studies encompassing different approaches, PI3 kinase activity has also been shown to be necessary for cell cycle progression (6, 11, 22, 24).
The pleiotrophic effects of PtdIns suggest that its synthesis must be highly regulated. Recently, a new family of 5′ inositol phosphatases has been described. In particular, hematopoietic cells contain an SH2 domain containing 5′ inositol phosphatase (SHIP) (26, 31, 45). It dephosphorylates 3′ PtdIns at the 5′ position and regulates the amount of PtdIns 3,4,5-P3 in the cell (31, 45). Therefore, SHIP could modulate biological effects which are dependent on the production of these PtdIns. Indeed, overexpression of SHIP in myeloid (FD-Fms) cells results in inhibition of macrophage colony-stimulating factor (M-CSF) and interleukin-3-induced cell growth (31). In addition, SHIP inhibits insulin (but not progesterone)-induced germinal vesicle breakdown when expressed in Xenopus oocytes (10).
We therefore studied the effect of expressing SHIP and a catalytically inactive mutant of SHIP (SHIPΔIP) on insulin-induced GLUT4 translocation and growth factor-induced actin filament rearrangement in 3T3-L1 adipocytes and HIRc-B fibroblasts, as well as on bromodeoxyuridine (BrdU) incorporation. Here we show that SHIP inhibits insulin-induced GLUT4 translocation, growth factor-induced membrane ruffling, and BrdU incorporation, whereas the catalytically inactive mutant had no effect. These results show that PtdIns 3,4,5-P3 plays an important role in promoting these biological effects.
MATERIALS AND METHODS
Materials.
Porcine insulin was kindly provided by Eli Lilly, Co. IGF-I was purchased from Life Technologies (Gaithersburg, Md.) and platelet-derived growth factor (PDGF) was from GIBCO BRL (Gaithersburg, Md.). Polyclonal anti-GLUT4 antibody (F349) was as described previously (18). Sheep immunoglobulin G (IgG) and fluorescein isothiocyanate (FITC)-, rhodamine-, and 7-amino-4-methylcoumarin-3-acetic acid (AMCA)-conjugated anti-mouse and anti-sheep antibodies were from Jackson Immunoresearch Laboratories, Inc. 3′-Bromo-5′-deoxyuridine (i.e., BrdU) was purchased from Amersham (Arlington Heights, Ill.), and rat anti-BrdU antibody was obtained from Accurate Scientific (Westbury, N.Y.). Anti-hemagglutinin (HA) antibody (12CA5) was purchased from Boehringer Mannheim (Indianapolis, Ind.), and anti-green fluorescent protein (GFP) antibody was from Clontech (Palo Alto, Calif.). Mouse monoclonal anti-Xpress (directed against a specific sequence of the expression product of the LacZ vector) was from Invitrogen (San Diego, Calif.). A rabbit polyclonal antibody against the dually phosphorylated form of mitogen-activated protein kinase (MAPK) was purchased from Promega (Madison, Wis.). Tetramethylrhodamine-conjugated phalloidin was from Molecular Probes, Inc. (Eugene, Oreg.); guanosine 5′-O-(3-thiotriphosphate) (GTPγS) and all other reagents were purchased from Sigma (St. Louis, Mo.).
Expression vectors.
The DNA for SHIP was cloned into a pCGN vector which contains a cytomegalovirus (CMV) promoter and has been described elsewhere (10). It has been modified in order to contain an HA (influenza HA) sequence at its NH2 terminus. Catalytically inactive SHIP was generated by deleting amino acids residues 666 to 680 (NLPSWCDRVLWKSYP) within the presumed inositol phosphatase catalytic domain (SHIPΔIP), and it has been described previously (10). It was also cloned into the mammalian expression vector pCGN and contains an HA sequence at its NH2 terminus. The p110-CAAX vector was constructed in pSG5, which contains at its carboxy terminus the sequence CKCVLS to mediate lipid modification and membrane localization of the protein, was kindly provided by Julian Downward (Imperial Cancer Research Fund, London, United Kingdom) (12). Green-Lantern (CMV-GFP) expression vector was from Life Technologies. The GFP gene is cloned into a derivative of pCVMSPORT (T7 promoter removed). The pcDNA3.1/His/LacZ control vector containing the gene for β-galactosidase was from Invitrogen. It contains a CMV promoter and an anti-Xpress antibody epitope.
Cell culture.
Rat-1 fibroblasts overexpressing wild-type human insulin receptors (HIRc-B) were maintained in Dulbecco modified Eagle medium (DMEM)–Ham F-12 (Life Technologies) supplemented with 10% fetal calf serum (FCS) and gentamicin (Gemini Bioproducts, Calabasas, Calif.), 2 mM Glutamax (Life Technologies), and 500 nM methotrexate (Sigma Chemical Co.).
3T3-L1 cells were maintained in DMEM-high glucose (GIBCO BRL) supplemented with 10% FCS and penicillin G-streptomycin (Omega Scientific) and differentiated into adipocytes as previously described (19) and then reseeded onto glass coverslips.
Microinjection.
Expression vectors were directly dissolved in microinjection buffer (5 mM sodium phosphate and 100 mM KCl, pH 7.4) to a final concentration of 0.1 mg/ml. Nuclei of single living cells were injected with a semiautomated microinjector from Eppendorf. Typically, proteins were allowed to express for 20 to 24 h after injection. Cells were then serum starved for 2 h prior to stimulation for GLUT4 detection, kept overnight for actin fiber staining, and stimulated with the appropriate ligand and fixed for staining.
GTPγS was dissolved in microinjection buffer to a final concentration of 5 mM and coinjected with sheep IgG (10 mg/ml) (to allow detection of injected cells) into the cytoplasm of serum-starved cells. Cells were fixed 30 min after microinjection.
Transient transfection of HIRc-B cells.
HIRc-B cells were grown on glass coverslips until they reached a confluency of 20 to 40% and then were transiently transfected with CMV-LacZ, SHIP, or SHIPΔIP DNA by using Superfect Reagent (Qiagen, Valencia, Calif.) as recommended by the manufacturer. After starvation for 24 or 36 h, cells were stimulated with the appropriate ligands and assessed for the presence of phospho-MAPK or BrdU incorporation.
Western blotting.
Control transfected cells or cells transfected with SHIP or SHIPΔIP were lysed in a lysis buffer containing 50 mM HEPES, 10 mM EDTA, 150 mM NaCl, 1% Triton X-100, 2 mM phenylmethylsulfonyl fluoride, 10% glycerol, 4 mM Na3VO4, 400 mM sodium fluoride, and 20 mM sodium pyrophosphate (pH 7.4) at 4°C. Then 50 μg of whole-cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (7.5% polyacrylamide) and transferred to nitrocellulose membranes in a Tris-glycine-methanol buffer. After transfer, membranes were blocked with Tris-buffered saline–5% nonfat milk (wt/vol) for 1 h at room temperature and incubated with anti-HA (1:1,000) antibody. Bound antibodies were visualized by using an enhanced chemiluminescence detection kit (Pierce).
Immunostaining and fluorescence microscopy. (i) GLUT4 protein staining.
Cells were serum starved 2 h prior to stimulation without or with insulin at 10 (1.7 nM) or 3 (0.5 nM) ng/ml for 20 min. Immunostaining of GLUT4 was performed essentially as described previously (19). The cells were fixed in 3.7% formaldehyde in phosphate-buffered saline (PBS) for 10 min at room temperature. After being washed, the cells were permeabilized and blocked with 0.1% Triton X-100 and 2% FCS in PBS for 5 to 10 minutes. Cells were then incubated overnight at 4°C with F349 (1 μg/ml, final concentration) and anti-HA or anti-GFP antibodies, as required, to allow detection of expressed protein, which were diluted in PBS with 2% FCS. After being washed with PBS for 10 min, the cells were incubated with FITC-conjugated donkey anti-rabbit (1:100), rhodamine-conjugated anti-mouse (1:100), and 7-amino-4-methylcoumarin-3-acetic acid (AMCA)-conjugated anti-sheep (1:100) antibody as needed to detect injected cells.
(ii) Actin staining.
3T3-L1 adipocytes were starved overnight before stimulation without insulin or with insulin at 100 ng/ml, insulin-like growth factor I (IGF-I) at 100 ng/ml, and PDGF at 20 ng/ml for 20 min. HIRc-B cells were starved for 36 h before stimulation with the appropriate ligands for 3 min. Cells were washed and permeabilized as described above and then incubated first with anti-HA or anti-GFP in PBS, as indicated, for 1 h at room temperature. After several washings with PBS, they were then incubated with rhodamine-phalloidin (0.125 mg/ml) to visualize the polymerized actin at the cell membrane (membrane ruffles) or stress fibers, and FITC-conjugated anti-mouse antibody was used to detect expressed protein in injected cells.
(iii) BrdU staining.
After transfection, cells were starved for 36 h and then stimulated with insulin (100 ng/ml), IGF-I (100 ng/ml), and FCS (10%) for 18 h. BrdU (1:1,000 dilution) was added to the medium for the last 6 h to allow its incorporation into newly synthesized DNA. Cells were fixed for 10 min in 3.7% formaldehyde-PBS, washed with PBS, and incubated for 1 h at room temperature with rat anti-BrdU (1:250) and either mouse anti-HA (1:250) or mouse anti-Xpress (1:2,000) antibody as needed in PBS containing 10 mM MgCl2, 20 U of DNase I, and 0.5% Nonidet P-40. Coverslips were washed with PBS and incubated for an additional hour with rhodamine anti-rat (1:100) and FITC- conjugated anti-mouse antibody (1:100) and, after being washed, were mounted on Gelvatol.
(iv) Anti-phospho-MAPK staining.
After transfection, cells were starved for 24 h and then stimulated with insulin (100 ng/ml), IGF-I (100 ng/ml), and FCS (10%) for 20 min. Cells were then washed twice with PBS, fixed for 10 min in 4% paraformaldehyde and, after two washes with PBS, permeabilized for 10 min in −20°C methanol. They were then incubated for 1 h at room temperature with rabbit anti-phospho-MAPK (1:100), mouse anti-HA (1:250), or mouse anti-Xpress (1:2,000) antibody, as needed, in cell extract obtained from unstimulated HIRc-B cells. Coverslips were washed with PBS and incubated for 30 min with rhodamine anti-mouse (1:100) and FITC anti-rabbit (1:100) antibody and, after being washed, were mounted on Gelvatol.
(v) Cell quantification.
Slides were analyzed on a Zeiss Axiophot immunofluorescence microscope (Zeiss, New York, N.Y.). The FITC- or rhodamine positive 3T3-L1 adipocytes (positive for cytoplasmic protein expression) on each coverslip were evaluated for the presence of plasma membrane-associated GLUT4. For each experiment about 30 to 50 cells were analyzed. HIRc-B cells that were positive for cytoplasmic protein expression and that displayed parallel actin fibers that colocalize with the nucleus were scored as positive for stress fibers. 3T3-L1 adipocytes or HIRc-B cells that showed actin staining at the periphery were scored as positive for membrane ruffles. HIRc-B cells positive for cytoplasmic expression of proteins from transfected plasmids, which displayed a dense nuclear staining with the anti-phospho-MAPK antibody, were scored as positive. All results are given as mean ± the standard error (SE). The observer was blinded to the experimental conditions.
(vi) Imaging.
Images were captured by using a charge-coupled device camera from Photometrics (Tucson, Ariz.) and saved with Isee software from Inovision (Durham, N.C.) to be subsequently used for making prints.
Statistics.
Statistical significance was assessed by using the Student t test for paired data.
RESULTS
Effect of SHIP on insulin-induced GLUT4 translocation.
SHIP is a 5′ phosphatase that converts PtdIns 3,4,5-P3 (PIP3) to PtdIns 3,4-P2 (10). To evaluate the role of PIP3 and SHIP in insulin-induced GLUT4 translocation, we conducted immunofluorescent staining of GLUT4 localization in 3T3-L1 adipocytes. The expression vectors for CMV-GFP (0.1 mg/ml) as a control, SHIP (0.1 mg/ml), or SHIPΔIP (0.1 mg/ml), a phosphatase-inactive mutant of SHIP, were injected into the nucleus of differentiated 3T3-L1 adipocytes. Proteins were then allowed to express for 20 to 24 h, and cells were serum starved 2 h before treatment with or without insulin. Cells were fixed and stained for GLUT4 localization. Expression of protein was confirmed with immunofluorescence detection by using either an anti-GFP antibody or an anti-HA antibody for SHIP and SHIPΔIP, combined with rhodamine anti-mouse antibody as the secondary antibody. Cells expressing SHIP and SHIPΔIP displayed similar intensities of immunofluorescent staining, indicating that the expression levels of these two proteins were comparable in our cell system (data not shown). Cells expressing the protein of interest were analyzed for GLUT4 translocation. As seen in Fig. 1A, unstimulated cells display GLUT4 staining mostly localized around the nucleus, with some staining distributed around the cytoplasm. After insulin stimulation, GLUT4 staining is seen at the plasma membrane as a clear ring, with a concomitant decrease in the cytoplasm. The cells displaying staining predominantly at the plasma membrane were scored as positive, and the results are given as a percentage of the positive cells for GLUT4 translocation, as previously described (19, 42).
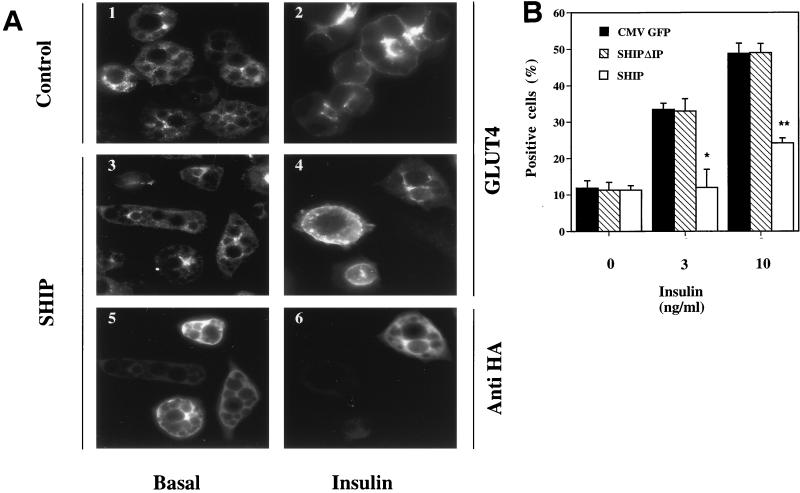
FIG. 1.
Effect of SHIP on insulin-induced GLUT4 translocation in 3T3-L1 adipocytes. 3T3-L1 adipocytes on coverslips were injected into the nucleus with CMV-GFP expression vector as a control, SHIP expression vector, or a catalytically inactive mutant of SHIP (SHIPΔIP) at a concentration of 0.1 mg/ml. Proteins were allowed to express for 20 to 24 h. Cells were then starved for 2 h in serum-free medium, stimulated without or with insulin (3 or 10 ng/ml) for 20 min, and fixed. Immunostaining was performed with rabbit polyclonal anti-GLUT4 (F349), mouse monoclonal anti-GFP, and mouse monoclonal anti-HA antibodies to allow detection of the tagged SHIP proteins. Cells positive for GFP, SHIP, or SHIPΔIP expression were counted for GLUT4 translocation. (A) Images of immunofluorescent GLUT4 staining in 3T3-L1 adipocytes. Panels 1 and 2 show typical staining in basal conditions and after insulin stimulation, respectively. Individual cells were injected into the nucleus with SHIP cDNA. Panels 5 and 6 show injected cells, and panels 3 and 4 represent GLUT4 staining of injected cells. (B) Summary data are given as bar graphs. Each bar represents mean ± SE of four to five experiments. Black bars represent values for control experiments, striped bars represent values for SHIPΔIP, and open bars represent SHIP-expressing cells. SHIP completely inhibited insulin-induced GLUT4 translocation at submaximal insulin concentrations, and by about 60% at maximal insulin concentrations (∗, P < 0.05; ∗∗, P < 0.01 versus control).
Expression of CMV-GFP had no effect on basal or insulin-stimulated GLUT4 distribution. SHIP expression did not affect basal GLUT4 localization, but it completely prevented insulin-induced GLUT4 translocation at 3 ng of insulin per ml (P < 0.05) and inhibited translocation by 64 ± 5% (mean ± SE) at 10 ng of insulin per ml (P < 0.001) (Fig. 1B). SHIPΔIP expression had no effect on basal or insulin-stimulated GLUT4 distribution (Fig. 1B).
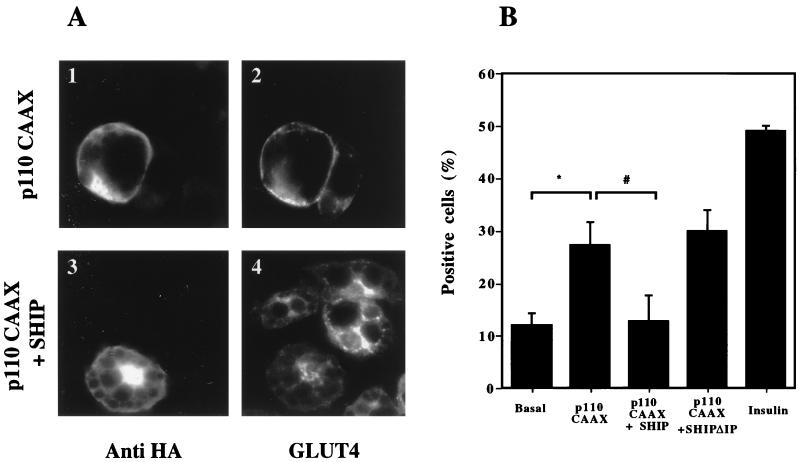
To further investigate the mechanism by which SHIP inhibits insulin-induced GLUT4 translocation, we utilized an expression vector containing the catalytic subunit of PI3 kinase with a membrane-targeting (CAAX) signal (p110-CAAX). We have recently shown that expression of this construct in 3T3-L1 adipocytes induces GLUT4 translocation to a level equal to 50% of the maximal effect of insulin (32a). p110-CAAX with CMV-GFP, p110-CAAX with SHIP, and p110-CAAX with SHIPΔIP were coinjected into the nucleus of 3T3-L1 adipocytes. After 20 to 24 h for protein expression, cells were analyzed for GLUT4 translocation as described above. As seen with the immunofluorescence staining in Fig. 2A, expression of p110-CAAX induces GLUT4 translocation to the plasma membrane in the absence of insulin. The summary data are represented as bar graphs in Fig. 2B, and p110-CAAX induced GLUT4 translocation to a level that was 50% as great as that of insulin (P < 0.05 compared to basal). Coexpression of SHIP and p110-CAAX almost completely inhibited the stimulatory effect of p110-CAAX. Immunofluorescent staining of a typical experiment is shown in Fig. 2A, and the summary data is given in Fig. 2B. Coexpression of p110-CAAX and SHIPΔIP induced the same extent of GLUT4 translocation as did p110-CAAX alone (Fig. 2B). These findings indicate that the effects of SHIP on GLUT4 translocation are mediated by a decrease in PtdIns 3,4,5-P3 due to the SHIP catalytic activity and not by the SH2 domain of SHIP (which is also contained in SHIPΔIP) or by a nonspecific effect of the protein on upstream molecules of the signaling system.
FIG. 2.
Effect of p110-CAAX and SHIP coinjection on GLUT4 translocation in 3T3-L1 adipocytes. 3T3-L1 adipocytes on coverslips were injected into the nucleus with CMV-GFP as a control, p110-CAAX with CMV-GFP, p110-CAAX with SHIP, or p110-CAAX with SHIPΔIP. All expression vectors were injected at a concentration of 0.1 mg/ml. After 20 to 24 h to allow for protein expression, cells were stimulated without or with insulin at 10 ng/ml as indicated and then fixed for staining. Cells positive for GFP or SHIP and SHIPΔIP (HA antibody) expression were scored for GLUT4 translocation. (A) Immunofluorescent GLUT4 staining. Individual cells were injected with p110-CAAX alone (1 and 2) or with p110-CAAX along with SHIP (3 and 4). The left panels show injected cells, and the right panels demonstrate GLUT4 staining. (B) Summary data are given as bar graphs. Each bar represents the mean ± SE for three experiments. p110-CAAX expression induced GLUT4 translocation, which was about half that of the insulin effect. Concomitant overexpression of SHIP inhibited the ability of p110-CAAX to induce GLUT4 translocation (∗, P < 0.05 versus basal; #, P < 0.05 versus p110-CAAX), whereas SHIPΔIP coexpression with p110-CAAX had no effect.
Effect of SHIP on GTPγS induced GLUT4 translocation in 3T3-L1 adipocytes.
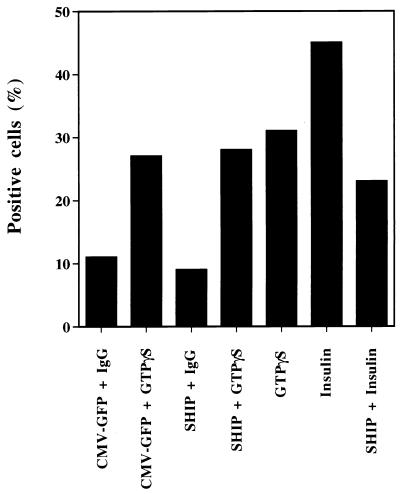
We have previously shown that microinjection of GTPγS into 3T3-L1 adipocytes leads to GLUT4 translocation (20). This effect was not inhibited by wortmannin, showing that GTPγS acts independently, or downstream, of PI3 kinase. To determine if the effect of SHIP on insulin-induced GLUT4 translocation is solely dependent on modulating phosphoinositides formed by PI3 kinase, we studied the effect of SHIP on GTPγS-induced GLUT4 translocation (Fig. 3). To accomplish this, we utilized a double-microinjection protocol in which 3T3-L1 adipocytes, on scored glass coverslips, were injected into the nucleus with either the expression vector for SHIP or CMV-GFP. Proteins were allowed to express for 20 to 24 h. After this time period, the same cells were serum starved for 2 h and reinjected into the cytoplasm with 5 mM GTPγS mixed with sheep IgG. After 30 min, the cells were fixed and immunostained for GLUT4 as described above. Cells positive for SHIP or CMV-GFP expression and cytoplasmic sheep IgG were analyzed for GLUT4 translocation. As seen in Fig. 3, cytoplasmic GTPγS injection alone induced GLUT4 translocation to about 80% of the maximal insulin effect, and this was not altered by CMV-GFP expression. Importantly, expression of SHIP inhibited insulin-induced GLUT4 translocation by about 70% but had no effect on GTPγS-stimulated GLUT4 translocation. These results demonstrate that the effects of SHIP on GLUT4 translocation are highly specific for the PI3 kinase pathway and show these effects are not related to a toxic or nonspecific effect of this protein in the cells.
FIG. 3.
Effects of SHIP on GTPγS-induced GLUT4 translocation in 3T3-L1 adipocytes. 3T3-L1 adipocytes on scored glass coverslips were injected into the nucleus with either the expression vector for SHIP or CMV-GFP. Proteins were allowed to express for 20 to 24 h. After this time period, the same cells were serum starved for 2 h and injected into the cytoplasm with 5 mM GTPγS mixed with sheep IgG. After 30 min the cells were fixed and immunostained for GLUT4 as described above. Cells positive for SHIP or CMV-GFP expression and cytoplasmic sheep IgG were counted for GLUT4 translocation. Cytoplasmic GTPγS injection alone induced GLUT4 translocation in 31% of the cells. From the cells expressing either CMV-GFP or SHIP and injected with GTPγS, 27 and 28% were positive, respectively, for GLUT4 translocation. In the same experiment, SHIP expression inhibited insulin-induced GLUT4 translocation by about 50%.
Effect of SHIP on growth factor-induced actin filament rearrangement in 3T3-L1 adipocytes.
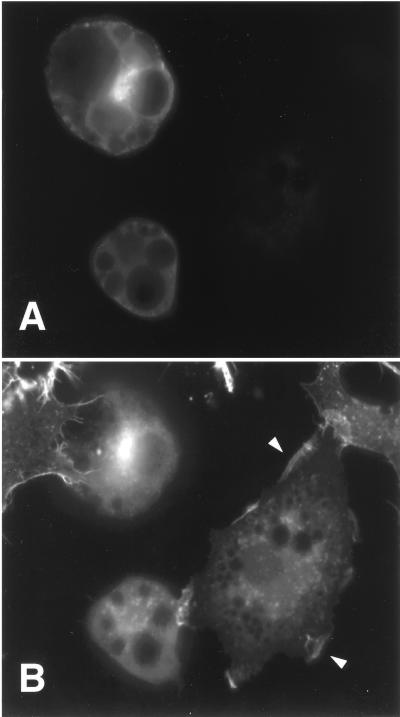
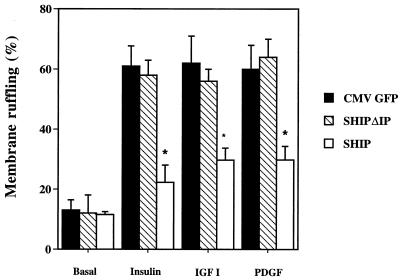
We have previously shown that growth factors, such as insulin, IGF-I, and PDGF, stimulate membrane ruffling in 3T3-L1 adipocytes and that the signaling pathway mediating this biological effect is dependent on PI3 kinase activation (33, 35). Therefore, we studied the effect of SHIP microinjection in differentiated 3T3-L1 adipocytes on growth factor-induced membrane ruffling. Expression vectors for CMV-GFP, SHIP, or SHIPΔIP, as a control, were injected directly into the cell nucleus, and encoded proteins were allowed to express for 20 to 24 h. After stimulation with the appropriate ligand, cells were fixed and stained for actin rearrangement, and expressed proteins were detected either with anti-GFP antibody or with anti-HA antibody. Figure 4A shows an example of anti-HA staining and demonstrates two cells expressing SHIP. After insulin stimulation, 3T3-L1 adipocytes display actin staining at the periphery of the cell (membrane ruffles, indicated by the arrowhead in Fig. 4B). Cells positive for protein expression were analyzed for the presence of membrane ruffles, and the results are given as the percent membrane ruffles as previously described (33). As can be seen in Fig. 4B, cells expressing SHIP have no membrane ruffles resembling unstimulated cells. Bar graphs summarizing the results for three to four independent experiments can be seen in Fig. 5. In the basal state, 13% of the cells injected with CMV-GFP exhibited membrane ruffles, and this increased to 61 ± 7 (mean ± SE), 62 ± 10, and 58 ± 8% after treatment with insulin (100 ng/ml), IGF-I (100 ng/ml), or PDGF (20 ng/ml), respectively. Nuclear microinjection of SHIP did not influence the amount of basal ruffling index, but it markedly inhibited insulin-, IGF-I-, and PDGF-induced membrane ruffling by 78 ± 10, 64 ± 3, and 62 ± 5%, respectively (all P < 0.05 compared to control) (Fig. 5). Expression of SHIPΔIP had no effect on basal or ligand induced membrane ruffling: 58 ± 5, 56 ± 4, and 64 ± 6% for insulin, IGF-I, and PDGF, respectively (all P > 0.1 compared to control) (Fig. 5).
FIG. 4.
Effects of SHIP on insulin-induced membrane ruffling in 3T3-L1 adipocytes. 3T3-L1 adipocytes on glass coverslips were injected into the nucleus with the expression vector for SHIP, and the protein was allowed to express for 24 h. After an overnight starvation, cells were then stimulated with 100 ng of insulin per ml for 20 min, fixed, and stained for expressed protein and actin filament rearrangement. (A) Photograph of 3T3-L1 adipocytes stained with anti-HA antibody demonstrating SHIP expression in the cytoplasm. (B) The same cells were photographed with an UV filter now displaying the staining for actin filament rearrangement. The two cells expressing SHIP have no membrane ruffles after insulin stimulation, whereas the uninjected cells display membrane ruffles (arrowheads).
FIG. 5.
Effects of SHIP on growth factor-induced membrane ruffling in 3T3-L1 adipocytes. 3T3-L1 adipocytes on coverslips were injected into the nucleus with expression vectors for CMV-GFP as control, SHIPΔIP, or SHIP expression vector at a concentration of 0.1 mg/ml. Proteins were allowed to express for 20 to 24 h. Cells were then starved in serum-free medium for 12 h and left either untreated or treated with insulin at 100 ng/ml, IGF-I at 100 ng/ml, or PDGF at 20 ng/ml for 15 min. Cells were then fixed and stained for actin localization with rhodamine-phalloidin. Individual adipocytes positive for GFP, SHIPΔIP, or SHIP expression were scored for the presence of membrane ruffles. Each bar represents the mean ± SE for three to four experiments. SHIP overexpression inhibited growth factor-induced membrane ruffling by about 60 to 80% (∗, P < 0.05, SHIP versus control), whereas SHIPΔIP had no effect.
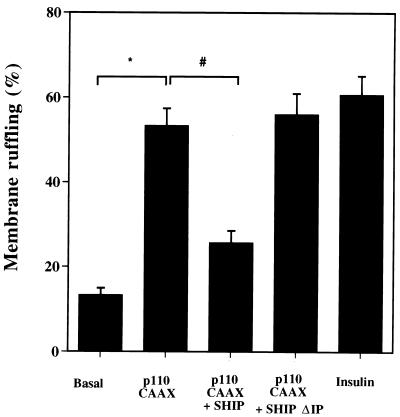
To further investigate the mechanism by which SHIP inhibits growth factor-induced membrane ruffling, we studied its effect in the context of p110-CAAX coexpression, since we have previously shown that microinjection of p110-CAAX into 3T3-L1 adipocytes induces membrane ruffling (32a). p110-CAAX expression induced membrane ruffling in the absence of ligand to about 85% the level of the insulin effect as follows: basal, 13 ± 2%, p110-CAAX 53 ± 4% (P < 0.05 compared to basal); insulin, 61 ± 4% (P < 0.05 compared to basal). SHIP coinjection with p110-CAAX significantly decreased the effect of p110-CAAX on membrane ruffling: 26 ± 3 versus 53 ± 4% (P < 0.05 compared to p110-CAAX alone), whereas coinjection of SHIPΔIP with p110-CAAX had the same effect as injection of p110-CAAX alone: 56 ± 5 versus 61 ± 4% (P > 0.1 compared to p110-CAAX alone) (Fig. 6). These results indicate that the effects of SHIP on membrane ruffling are mediated by modulating PIP3 production by PI3 kinase.
FIG. 6.
Effects of p110 CAAX and SHIP coinjection on membrane ruffling in 3T3-L1 adipocytes. 3T3-L1 adipocytes were nuclear injected with CMV-GFP as a control, p110-CAAX with CMV-GFP, p110 CAAX with SHIPΔIP, or p110-CAAX with SHIP. All of the expression vectors were injected at a concentration of 0.1 mg/ml. After 20 to 24 h to allow for protein expression and 12 h of starvation in serum-free medium, the cells were left untreated or stimulated with insulin at 100 ng/ml, as indicated, and fixed for staining. Cells positive for GFP, SHIPΔIP, and SHIP expression were scored for the presence of membrane ruffles. Each bar represents the mean ± SE for three experiments. p110-CAAX expression induced membrane ruffling in 3T3-L1 adipocytes, a finding which was similar to the insulin effect. Concomitant expression of SHIP significantly inhibited the ability of p110-CAAX to induce membrane ruffles in the absence of insulin (∗, P < 0.05 versus basal; #, P < 0.05 versus p110-CAAX). Concomintant expression of p110-CAAX and SHIPΔIP had no effect on p110-CAAX-induced GLUT4 translocation.
Effect of SHIP on actin filament rearrangement in HIRc-B cells.
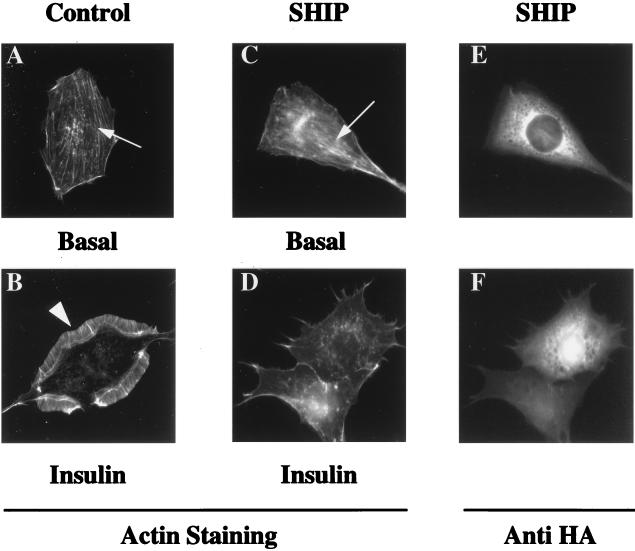
To further investigate the effect of SHIP on actin filament rearrangement, we used a Rat-1 fibroblast cell line that expresses approximately 106 human insulin receptors (HIRc-B). As visualized in Fig. 7, in the basal state, HIRc-B cells display abundant stress fibers and an absence of membrane ruffles (Fig. 7A); after ligand stimulation, the stress fibers break down, and a prominent membrane ruffling response can be easily visualized (Fig. 7B). After insulin stimulation of SHIP-expressing cells (Fig. 7C), stress fiber breakdown still proceeds normally, but the membrane ruffling response is blocked. Figure 7D shows visualization of the HA-tagged SHIP in these cells after nuclear microinjection.
FIG. 7.
Effects of SHIP overexpression on insulin-induced actin rearrangement in HIRc-B cells. Serum-starved HIRc-B cells on coverslips were injected in the nucleus with either CMV-GFP expression vector or SHIP expression vector at a concentration of 0.1 mg/ml. After 20 to 24 hours to allow for protein expression, the cells were stimulated without ligand or with insulin at 100 ng/ml for 3 min. Cells were then fixed and stained for actin filament rearrangement with rhodamine-phalloidin. (A) Photograph of actin staining in an unstimulated serum-starved cell displaying actin stress fibers in the cell cytoplasm (arrow) and the absence of membrane ruffles at the cell periphery. (B) After 3 min of insulin stimulation, a prominent membrane-ruffling response can be visualized (arrowhead), and concomitantly stress fibers have broken down. (C and D) Actin staining of cells injected into the nucleus with SHIP and stimulated without insulin (C) or with insulin for 3 min (D). Unstimulated injected cells display stress fibers as control injected cells (C) (arrowhead). After insulin stimulation, injected cells display no membrane ruffles but show stress fiber breakdown. (E and F) HA staining of the same cells as in panels C and D, respectively, showing SHIP expression in the cell cytoplasm.
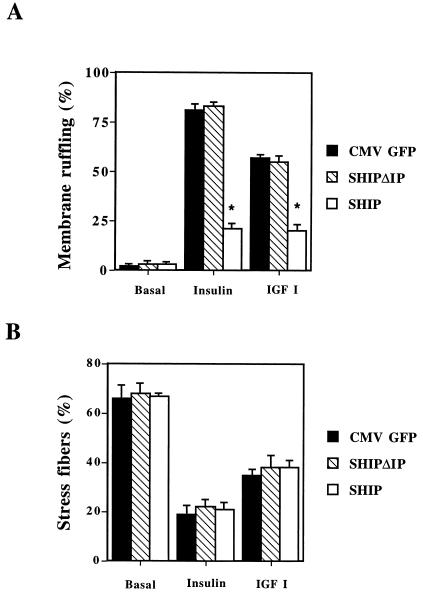
These results are quantitated in Fig. 8. In HIRc-B cells insulin stimulation leads to rapid relocalization of actin to the cell surface (membrane ruffles) in 81 ± 3% (mean ± SE) of the cells, whereas unstimulated cells display almost no membrane ruffles (2 ± 1%) (Fig. 8A). IGF-1 induces membrane ruffling to a lesser extent than insulin (57 ± 2%) (Fig. 8A), whereas PDGF has no effect (data not shown). In these cells, nuclear microinjection of SHIP dramatically inhibited insulin- and IGF-I-induced membrane ruffling by 76 ± 3 and 68 ± 5%, respectively (P < 0.005 for both) (Fig. 8A). No effect was observed on the basal appearance of the cell. Expression of SHIPΔIP had no effect on basal or ligand induced membrane ruffling in HIRc-B cells, as seen in Fig. 8A.
FIG. 8.
Effects of SHIP on growth factor-induced membrane ruffling and stress fiber breakdown in HIRc-B cells. Serum-starved HIRc-B cells were injected into the nucleus with the expression vectors for either CMV-GFP, SHIPΔIP, or SHIP, and the proteins were allowed to express for 20 to 24 h. Cells were then stimulated for 3 min with insulin (100 ng/ml), fixed, and stained for actin localization with rhodamine-phalloidin. Individual cells positive for GFP, SHIPΔIP, and SHIP expression were scored for the presence of membrane ruffles (A) or parallel actin fibers that colocalize with the nucleus (positive for stress fibers) (B). Each bar represents the mean ± SE for three to four different experiments. SHIP inhibited insulin- and IGF-I-induced membrane ruffling by about 80% (∗, P < 0.005 versus control) but had no effect on ligand-induced stress fiber breakdown, whereas SHIPΔIP did not affect ligand-induced membrane ruffling.
Serum-starved HIRc-B fibroblasts display a high content of stress fibers, as shown previously (34). As displayed in Fig. 8B, insulin and IGF-I stimulation leads to rapid breakdown of stress fibers, while expression of SHIP or SHIPΔIP had no effect on inhibiting insulin- or IGF-I-mediated breakdown.
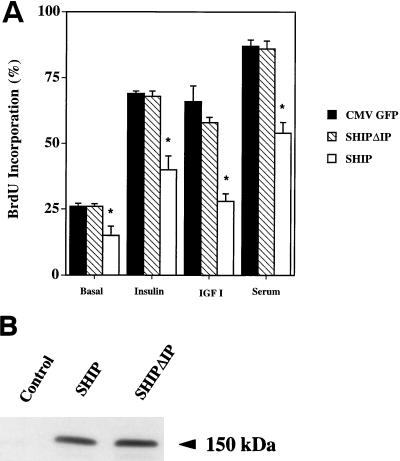
Effect of SHIP on BrdU incorporation and MAPK activation in HIRc-B cells.
In order to assess the effects of SHIP expression on ligand-induced DNA synthesis, we transiently transfected HIRc-B fibroblasts with the expression vector for either CMV-LacZ, HA-tagged SHIP, or SHIPΔIP. After 48 h, expression levels of SHIP and SHIPΔIP in whole-cell lysates were similar when assessed by Western blotting with an anti-HA antibody (Fig. 9). For BrdU incorporation, cells were starved for 24 h and stimulated for 18 h without or with insulin (100 ng/ml), IGF-I (100 ng/ml), or FCS (10%). During the last 6 h, BrdU was added to detect newly synthesized DNA. Transfected cells were detected with either an anti-Xpress or an anti-HA antibody and scored for the presence of BrdU incorporation into the nucleus. Results are given as the percent BrdU incorporation. SHIP expression slightly but significantly decreased basal BrdU incorporation (26 ± 1 versus 19 ± 2%). Insulin, IGF-1, and serum increased BrdU incorporation in control transfected cells to 69 ± 1, 66 ± 6, and 87 ± 2%, respectively (all P < 0.05 compared to basal). As seen in Fig. 9, expression of SHIP inhibited growth factor-stimulated mitogenesis by 42 ± 9, 57 ± 2, and 38 ± 10% for insulin, IGF-I, and FCS, respectively (P < 0.05 compared to control transfected cells). Expression of SHIPΔIP had no effect either on basal or growth factor-induced BrdU incorporation (Fig. 9).
FIG. 9.
Effects SHIP on growth factor-induced BrdU incorporation in HIRc-B cells. (A) HIRc-B cells were grown on glass coverslips in 12-well dishes and transiently transfected either with LacZ as a control or the expression vectors for SHIPΔIP or SHIP. Cells were starved for 24 h and then stimulated without insulin or with insulin (100 ng/ml), IGF-I (100 ng/ml), or FCS (10%) for 18 h. BrdU was added during the last 6 h of stimulation. Cells were then fixed and stained for protein expression and BrdU incorporation. Successfully transfected cells were counted for BrdU incorporation into the nucleus, and results are given as the percent BrdU incorporation. Each bar represents the mean ± SE for three separate experiments. SHIP expression slightly decreased the basal BrdU incorporation. Compared to the control, SHIP significantly inhibited insulin-, IGF-I-, and serum-stimulated BrdU incorporation by 42 ± 9, 57 ± 2, and 38 ± 10%, respectively (∗, P < 0.05 versus control). (B) HIRc-B cells were transiently transfected with the vectors for SHIP and SHIPΔIP or with no vector, and proteins were allowed to express for 48 h. Whole-cell lysates were then separated by SDS-PAGE, and membranes were blotted with an anti-HA antibody. SHIP and SHIPΔIP show similar expression levels.
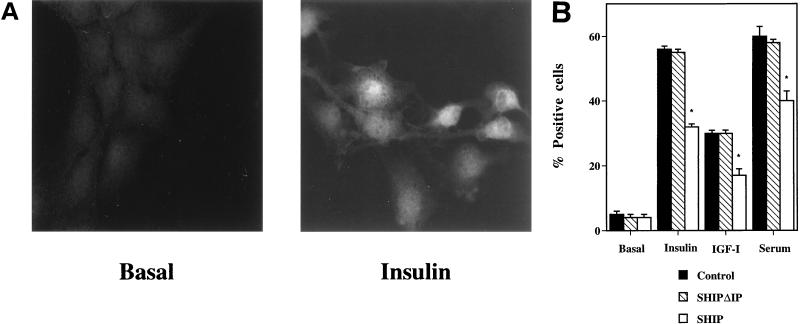
Tyrosine kinase receptors lead to mitogenesis by activating the classical pathway Grb2-Sos-Ras-Raf-MAPK. Some data suggest also that PI3 kinase is able to activate the MAPK pathway (24). To determine if inhibition of PI3 kinase signaling expression of SHIP affected MAPK signaling, we performed immunofluorescent staining on control transfected and SHIP- and SHIPΔIP-transfected cells with a specific antibody directed against the dually phosphorylated MAPK. After ligand binding, MAPK is activated by phosphorylation and relocalizes from the cytosol to the nucleus. Starved, unstimulated HIRc-B cells display almost no staining with the anti-phospho-MAPK antibody. After stimulation, bright staining in the nuclear region can be observed (Fig. 10A). Cells were transfected transiently with expression vectors for CMV-GFP, SHIP, and SHIPΔIP, stimulated with insulin, IGF-I, or serum, and then analyzed for nuclear phospho-MAPK staining. Insulin, IGF-I, and serum stimulation increased the percentage of positive cells to 55 ± 1, 30 ± 1, and 60 ± 3%, respectively, whereas after SHIP expression these responses were decreased to 32 ± 1, 17 ± 1, and 40 ± 3%, respectively (P < 0.05) (Fig. 10B). SHIPΔIP had no effect in ligand-induced MAPK phosphorylation compared to control.
FIG. 10.
Effect of SHIP on MAPK activation in HIRc-B cells. HIRc-B cells were grown on glass coverslips and transiently transfected with the expression vectors for LacZ, SHIP, or SHIPΔIP. Cells were starved for 24 h and then stimulated without or with insulin (100 ng/ml), IGF-I (100 ng/ml), or serum (10%) for 20 min. Cells were then fixed and stained for protein expression with a specific antibody directed against the dually phosphorylated MAPK. (A) Unstimulated cells display almost no staining with the anti-phospho-MAPK antibody and, after insulin stimulation, bright nuclear as well some cytoplasmatic staining can be observed. Successfully transfected cells displaying bright nuclear staining were scored as positive, and results are given as the percent positive cells. (B) Summarized data. Each bar represents the mean ± SE for three separate experiments. SHIP expression inhibited ligand-induced MAPK phosphorylation, whereas SHIPΔIP had no effect.
DISCUSSION
Growth factor-induced PI3 kinase activation by growth factors and subsequent 3′ phosphoinositide formation is required for many biological actions and, in particular, for insulin’s metabolic effects (19, 33). Our results show that nuclear microinjection of a plasmid encoding the 5′ inositol phosphatase SHIP with subsequent expression of the protein in 3T3-L1 adipocytes significantly inhibits insulin-induced or p110-CAAX (a membrane-localized form of the catalytic subunit of PI3 kinase)-induced GLUT4 translocation.
SHIP has been first described in hematopoietic cells (26, 31). It contains an SH2 domain and, in response to growth factor activation, SHIP is phosphorylated and has been shown to bind to the PTB domain of Shc (30), to Grb2 (23), and to the tyrosine phosphatase SHP2 (32), leading to translocation of the phosphatase to the plasma membrane. SHIP is a 5′ phosphatase and dephosphorylates 3′ phosphoinositides at the 5′ position. In hematopoietic cells, SHIP modulates downstream effects of growth factor action. For example, retroviral expression of SHIP in FD-Fms cells results in strong inhibition of M-CSF-stimulated cell growth (31). Furthermore, expression of SHIP in Xenopus oocytes inhibits germinal vesicle breakdown induced by insulin but not by progesterone (10). Although Xenopus oocytes do not express endogenous SHIP, the effects of SHIP expression on insulin action were directly correlated with decreased intracellular levels of PtdIns 3,4,5-P3 (10). Although SHIP has not been detected in fat cells, an insulin-sensitive 5′ phosphatase activity was found in Shc immunoprecipitates of insulin-sensitive cells (CHO-T) (16). With specific antibodies it was shown that this phosphatase activity was not due to SHIP but was possibly due to another isoform expressed in this insulin-sensitive system. Based on their results, Guilherme et al. speculated that 5′ phosphatase activity could be important in insulin signaling (16). More recently, a protein closely related to SHIP, SHIP2, has been identified (37); SHIP2 is also highly expressed in skeletal muscle (25), which is a direct target tissue for insulin-stimulated GLUT4 translocation. Based on the close similarity between SHIP and SHIP2, we believe our results accurately indicate the potential role of 5′ inositol phosphatase activity in insulin signaling.
One of the major effects of insulin is to promote GLUT4 translocation and glucose uptake in insulin-sensitive tissues, and it has been clearly demonstrated that PI3 kinase activation is both necessary and sufficient for these actions (19, 33). Our data showing that SHIP expression inhibits insulin-induced GLUT4 translocation further demonstrate the important role played by PtdIns in these actions of insulin. By using a phosphatase-inactive mutant of SHIP (10), which had no effect on insulin-induced GLUT4 translocation, we provide additional evidence that the effect of SHIP is dependent on its catalytic activity. It has been previously shown that expression of SHIP in Xenopus oocytes, a cell system where it is not endogenously expressed, inhibits insulin (but not progesterone)-induced germinal vesicle breakdown and that this effect is directly related to decreased levels of PtdIns 3,4,5-P3 in the cells in response to insulin (10). Therefore, our data clearly show that PtdIns 3,4,5-P3 is a key messenger in mediating the function of PI3 kinase for these metabolic actions. We further examined the effects of SHIP and SHIPΔIP on p110-CAAX-induced GLUT4 translocation. We have shown elsewhere that overexpression of p110-CAAX (a membrane-localized catalytic subunit of PI3 kinase) leads to GLUT4 translocation in 3T3-L1 adipocytes to a level that is about 50% that of the insulin effect (32a). Our results that concomitant expression of p110-CAAX and SHIP, but not SHIPΔIP, inhibits the p110-CAAX effect also demonstrate that the inhibition is due to decreased PtdIns 3,4,5-P3 levels and not to nonspecific binding with molecules of the signaling cascade upstream of PI3 kinase, such as the insulin receptor or the IRS proteins.
How can modulation of PtdIns have an effect on insulin-induced GLUT4 translocation? Several recent studies indicate that one effector system for PI3 kinase signaling involves a protein kinase cascade which includes the Ser-Thr kinase Akt (5, 13). Akt has been implicated in several biological responses, including GLUT4 translocation (28), glycogen synthesis, and cell survival (9, 41). The PI3 kinase dependency of insulin-induced Akt activation indicates that Akt lies downstream of PI3 kinase (5, 13). Akt contains a PH domain which binds PtdIns, a catalytic domain, and several potential Ser-Thr phosphorylation sites. Recent studies have indicated that PtdIns 3,4-P2 can bind to the PH domain and directly stimulate Akt activity (27), but additional regulatory mechanisms are required for full activation. Thus, after growth factor stimulation, Akt is phosphorylated on two sites: Thr-308 and Ser-473 (1). A new kinase, described as 3-phosphoinositide-dependent protein kinase 1 (PDK1), which phosphorylates Akt on Thr-308 and activates the enzyme, has been purified and cloned (3, 40). This kinase is itself directly activated by both PtdIns 3,4-P2 and PtdIns 3,4,5-P3 (3). The regulation of Akt in vivo is, therefore, highly complex and depends on direct activation by phospholipids and by PDK1, which is itself activated by 3′ PtdIns. These redundant mechanisms of Akt activation are consistent with our results. Complete inhibition by SHIP of insulin-stimulated GLUT4 translocation at the submaximal insulin concentration shows a critical role for PtdIns 3,4,5-P3 in this metabolic action. Indeed, as SHIP will decrease intracellular levels of PtdIns 3,4,5-P3, one would expect that Akt activation by PDK1 would be significantly decreased. At the maximal insulin concentration, we observed a 64% inhibition of insulin-induced GLUT4 translocation by SHIP, again highlighting the role of PtdIns 3,4,5-P3 as the key stimulating lipid messenger for this biologic effect. Insofar as Akt may be a mediator of PI3 kinase effects on GLUT4 translocation, the modest residual GLUT4 translocation at maximal insulin concentration in SHIP-expressing cells may represent partial activation of Akt by PtdIns 3,4-P2 or some other mechanism. Since SHIP will decrease PtdIns 3,4,5-P3 levels (10) and might also increase PtdIns 3,4-P2 levels, these results further highlight the importance of PtdIns 3,4,5-P3 as the key lipid messenger signaling to GLUT4 translocation.
To further address the specificity of the effects of SHIP expression on GLUT4 translocation, we examined its effects on GTPγS-induced GLUT4 translocation. GTPγS-induced GLUT4 translocation is independent of PI3 kinase activation, as it is not inhibited by wortmannin or by p85 SH2 GST fusion protein microinjection (20). Our data show that GTPγS-induced GLUT4 translocation was not inhibited by SHIP, again suggesting that SHIP expression inhibits insulin-induced GLUT4 translocation by inhibiting the effects of PI3 kinase upstream of GTPγS.
Actin cytoskeleton rearrangement is another biological effect exerted by insulin and other growth factors (33, 34). After ligand binding, one observes a rapid breakdown of stress fibers, followed by the appearance of membrane ruffles. Our results show that microinjection of SHIP had no effect on insulin- or IGF-I-induced stress fiber breakdown in HIRc-B cells, even though growth factor-stimulated membrane ruffling and mitogenesis were inhibited in these same cells. These results indicate that the action of PtdIns 3,4,5-P3 is not necessary for stress fiber breakdown. Indeed, previous reports suggest that stress fiber formation in different cell types is regulated by the small GTP-binding protein Rho (17). Generation of stress fibers by the Rho protein parallels its ability to stimulate the formation of 4,5-phosphorylated phosphatidylinositol (4,5-PIP2) (7). It was also suggested that 4,5-PIP2 binds directly to the focal adhesion proteins, α-actinin and vinculin, facilitating their localization at the cell surface (14, 15, 20). Dissociation of 4,5-PIP2 from these proteins upon stimulation with growth factors may lead to stress fiber breakdown (14). Since we (34) and others have shown that stress fiber breakdown is dependent on PI3 kinase, we speculate that PI3 kinase stimulates stress fiber breakdown by phosphorylating the D-3 position of 4,5-PIP2, which causes its release from focal adhesion-localized proteins. Without this anchoring effect of 4,5-PIP2, α-actinin and vinculin can no longer localize to focal adhesions, and this would lead to stress fiber breakdown. Given this possible mechanism, dephosphorylation of PtdIns 3,4,5-P3 generated by growth factor stimulation would not inhibit stress fiber breakdown; therefore, our results are fully supportive of this hypothesis.
On the other hand, several studies demonstrate that membrane ruffling is dependent on PtdIns 3,4,5-P3 formation and subsequent activation of the small GTP-binding protein Rac, even though Rac does not bind directly to PtdIns 3,4,5-P3 (43). Our current results support this view. Thus, SHIP markedly inhibited insulin- and IGF-I-induced membrane ruffling in both HIRc-B cells and 3T3-L1 adipocytes, whereas SHIPΔIP had no effect. This again shows that this effect is dependent on the phosphatase activity of SHIP and is therefore dependent on decreased levels of PtdIns 3,4,5-P3. The magnitude of the inhibition indicates that membrane ruffling is dependent on new growth factor-induced PtdIns 3,4,5-P3 synthesis.
Taken together, our results show that growth factor-induced actin reorganization, which includes stress fiber breakdown and the generation of membrane ruffles, is mediated by distinct biochemical mechanisms. Stress fiber breakdown is dependent on the activity of PI3 kinase but not on the direct positive effects of PIP3. Rather, we speculate that PI3 kinase phosphorylates PIP 4,5-P2 bound to focal adhesion proteins, causing it to dissociate from these protein structures; inducing stress fiber breakdown and expression of the 5′ phosphatase SHIP would not affect this response. On the other hand, since SHIP inhibits growth factor-induced membrane ruffling, the PIP3 generation caused by growth factor stimulation must directly participate in the formation of membrane ruffles. Furthermore, since our results show that the expression of SHIP can block one biologic effect (membrane ruffling) but not another (stress fiber breakdown) within the same cells, these results also indicate that the effects of SHIP expression are specific and not related to a toxic or nonspecific effect on the cells.
Mitogenic signaling by tyrosine kinase receptors is dependent on p21ras activation, followed by Raf activation, and then a downstream cascade of MAP kinases, leading to entrance of the cell into G1 phase. On the basis of numerous studies with different approaches, PI3 kinase activity is also known to be necessary for cell cycle progression (6, 11, 22, 24). Our results show that the expression of SHIP inhibits growth factor-induced DNA synthesis and also inhibits the activation of MAPK, indicating that generation of the lipid messenger PtdIns 3,4,5-P3 is an important event in growth factor-induced MAPK activation and subsequent mitogenic signaling.
In summary, our results show that overexpression of SHIP inhibits insulin-induced GLUT4 translocation, growth factor-induced membrane ruffling, and DNA synthesis and that its phosphatase activity is critical for this effect. These data show that PtdIns 3,4,5-P3 plays a critical role in these biological actions.
ACKNOWLEDGMENTS
This work was supported in part by NIH grant DK-33651, the VA Medical Research Service, a grant from the Schweizerische Stiftung für Medizinisch-Biologische Stipendien (to P.V.), and grants J 01287-Med and J 1584-Med from the Erwin Schrödinger Stipendium by the Austrian Fonds zur Förderung der wissenschaftlichen Forschung (to M.C.).
We thank Michael Mueckler (Washington University School of Medicine) for providing F349 anti-GLUT4 antibody.
REFERENCES
- 1.Alessi D R, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings B A. Mechanisms of activation of protein kinase B by insulin and IGF-1. EMBO. 1996;15:65541–6551. [PMC free article] [PubMed] [Google Scholar]
- 2.Alessi D R, Deak M, Casamayor A, Caudwell F B, Morrice N, Norman D G, Gaffney P, Reese C B, MacDougall C N, Harbison D, Ashworth A, Bownes M. 3-Phosphoinositide-dependent protein kinase-1 (PDK-1): structural and functional homology with the Drosophila DSTPK61 kinase. Curr Biol. 1997;7:776–789. doi: 10.1016/s0960-9822(06)00336-8. [DOI] [PubMed] [Google Scholar]
- 3.Alessi D R, James S R, Downes C P, Holmes A B, Gaffney P R J, Reese C B, Cohen P. Characterization of a 3-Phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 4.Auger K, Serunian L A, Soltoff S P, Libby P, Cantley L C. PDGF-dependent tyrosine phosphorylation stimulates production of novel polyphosphoinositides in intact cells. Cell. 1989;57:167–175. doi: 10.1016/0092-8674(89)90182-7. [DOI] [PubMed] [Google Scholar]
- 5.Burgering B M T, Coffer P J. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature. 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- 6.Cantley L C, Auger K R, Carpenter C, Duckworth B, Graziani A, Kapeller R, Soltoff S. Oncogenes and signal transduction. Cell. 1991;64:281–302. doi: 10.1016/0092-8674(91)90639-g. [DOI] [PubMed] [Google Scholar]
- 7.Chong L D, Traynor-Kaplan A, Bokoch G M, Schwartz M A. The small GTP-binding protein Rho regulates a phosphatidylinositol 4-phosphate kinase in mammalian cells. Cell. 1994;79:507–513. doi: 10.1016/0092-8674(94)90259-3. [DOI] [PubMed] [Google Scholar]
- 8.Cohen P, Alessi D R, Cross D A E. PDK1, one of the missing links in insulin signal transduction? FEBS Lett. 1997;410:3–10. doi: 10.1016/s0014-5793(97)00490-0. [DOI] [PubMed] [Google Scholar]
- 9.Cross D A E, Alessi D R, Cohen P, Andjelkovich M, Hemmings B A. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 10.Deuter-Reinhard M, Apell G, Pot D, Klippel A, Williams L T, Kavanaugh W M. SIP/SHIP inhibits Xenopus oocyte maturation induced by insulin and phosphatidylinositol 3-kinase. Mol Cell Biol. 1997;17:2559–2565. doi: 10.1128/mcb.17.5.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fantl W J, Escobedo J A, Martin G A, Turck C W, del Rosario M, McCormick F, Williams L T. Distinct phosphotyrosines on a growth factor receptor bind to specific molecules that mediate different signaling pathways. Cell. 1992;69:413–423. doi: 10.1016/0092-8674(92)90444-h. [DOI] [PubMed] [Google Scholar]
- 12.Farh L, Mitchell D A, Deschenes R J. Farnesylation and proteolysis are sequential, but distinct steps in the CaaX box modification pathway. Arch Biochem Biophys. 1995;318:113–121. doi: 10.1006/abbi.1995.1211. [DOI] [PubMed] [Google Scholar]
- 13.Franke T F, Yang S-I, Chan T O, Datta K, Kazlauskas A, Morrison D K, Kaplan D R, Tsichlis P N. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 14.Fukami K, Endo T, Imamura M, Takenawa T. α-Actinin and vinculin PIP2-binding proteins involved in signaling by tyrosine kinase. J Biol Chem. 1994;269:1518–1522. [PubMed] [Google Scholar]
- 15.Fukami K, Furuhashi K, Inagaki M, Endo T, Hatano S, Takenawa T. Requirement of phosphatidylinositol 4,5-bisphosphate for alpha-actinin function. Nature. 1992;359:150–152. doi: 10.1038/359150a0. [DOI] [PubMed] [Google Scholar]
- 16.Guilherme A, Klarlund J K, Krystal G, Czech M P. Regulation of phosphatidylinositol 3,4,5-triphosphate 5′-phosphatase activity by insulin. J Biol Chem. 1996;271:29533–29536. doi: 10.1074/jbc.271.47.29533. [DOI] [PubMed] [Google Scholar]
- 17.Hall A. Ras related proteins. Curr Opin Cell Biol. 1993;5:265–268. doi: 10.1016/0955-0674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- 18.Haney P M, Slot J W, Piper R C, James D E, Mueckler M. Intracellular targetting of the insulin-regulatable glucose transporter (GLUT4) is isoform specific and independent of cell type. J Cell Biol. 1991;114:689–699. doi: 10.1083/jcb.114.4.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haruta T, Morris A J, Rose D W, Nelson J G, Mueckler M, Olefsky J M. Insulin-stimulated GLUT4 translocation is mediated by a divergent intracellular signaling pathway. J Biol Chem. 1995;270:27991–27994. doi: 10.1074/jbc.270.47.27991. [DOI] [PubMed] [Google Scholar]
- 20.Haruta T, Morris A J, Vollenweider P, Nelson J G, Rose D W, Mueckler M, Olefsky J M. Ligand-independent GLUT4 translocation induced by guanosine 5′-O-(3-thiotriphosphate) involves tyrosine phosphorylation. Endocrinology. 1998;139:358–364. doi: 10.1210/endo.139.1.5698. [DOI] [PubMed] [Google Scholar]
- 21.Hawkins P T, Eguinoa A, Qiu R-G, Stokoe D, Cooke F T, Walters R, Wennstroem S, Claesson-Welsh L, Evans T, Symons M, Stephens L. PDGF stimulates an increase in GTP-Rac via activation of phosphoinositide 3-kinase. Curr Biol. 1995;5:393–403. doi: 10.1016/s0960-9822(95)00080-7. [DOI] [PubMed] [Google Scholar]
- 22.Hu Q, Klippel A, Muslin A J, Fantl W J, Williams L T. Ras-dependent induction of cellular responses by constitutively active phosphatidylinositol 3-kinase. Science. 1995;268:100–102. doi: 10.1126/science.7701328. [DOI] [PubMed] [Google Scholar]
- 23.Jefferson A B, Auethavekia V, Pot D A, Williams L T, Majerus P W. Signaling inositol polyphosphate-5-phosphatase. Characterization of activity and effect of GRB2 association. J Biol Chem. 1997;272:5983–5988. doi: 10.1074/jbc.272.9.5983. [DOI] [PubMed] [Google Scholar]
- 24.Jhun B H, Rose D W, Seely B L, Rameh L, Cantley L, Saltiel A R, Olefsky J M. Microinjection of the SH2 domain of the 85-kilodalton subunit of phosphatidylinositol 3-kinase inhibits insulin-induced DNA synthesis and c-fos expression. Mol Cell Biol. 1994;14:7466–7475. doi: 10.1128/mcb.14.11.7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapeller R, Cantley L C. Phosphatidylinositol 3-kinase. Bioessays. 1994;16:565–576. doi: 10.1002/bies.950160810. [DOI] [PubMed] [Google Scholar]
- 26.Kavanaugh W M, Pot D A, Chin S M, Deuter-Reinhard M, Jefferson A B, Norris F A, Masiarz F R, Cousens L S, Majerus P W, Williams L T. Multiple forms of an inositol polyphosphate 5-phosphatase form signaling complexes with Shc and Grb2. Curr Biol. 1996;6:438–445. doi: 10.1016/s0960-9822(02)00511-0. [DOI] [PubMed] [Google Scholar]
- 27.Klippel A, Kananaugh W M, Pot D, Williams L T. A specific product of phosphatidylinositol 3-kinase directly activates the protein kinase Akt through its pleckstrin homology domain. Mol Cell Biol. 1997;17:338–344. doi: 10.1128/mcb.17.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohn A D, Summers S A, Birnbaum M J, Roth R A. Expression of a constitutively active Akt Ser/Thr kinase in 3T3-L1 adipocytes stimulates glucose uptake and glucose transporter 4 translocation. J Biol Chem. 1996;271:31372–31378. doi: 10.1074/jbc.271.49.31372. [DOI] [PubMed] [Google Scholar]
- 29.Kohn A D, Takeuchi F, Roth R A. Akt, a pleckstrin homology domain containing kinase, is activated primarily by phosphorylation. J Biol Chem. 1996;271:21920–21936. doi: 10.1074/jbc.271.36.21920. [DOI] [PubMed] [Google Scholar]
- 30.Lamkin T D, Walk S F, Liu L, Damen J E, Krystal G, Ravichandran K S. Shc interaction with Src homology 2 domain containing inositol phosphatase (SHIP) in vivo requires the Shc-phosphotyrosine binding domain and two specific phosphotyrosines on SHIP. J Biol Chem. 1997;272:10396–10401. doi: 10.1074/jbc.272.16.10396. [DOI] [PubMed] [Google Scholar]
- 31.Lioubin M N, Algate P A, Tsai S, Carlberg K, Aebersold R, Rohrschneider L R. p150Ship, a signal transduction molecule with inositol polyphosphate-5-phosphatase activity. Genes Dev. 1996;10:1084–1095. doi: 10.1101/gad.10.9.1084. [DOI] [PubMed] [Google Scholar]
- 32.Liu L, Damen J E, Ware M D, Krystal G. Interleukin-3 induces the association of the inositol 5-phosphatase SHIP with SHP2. J Biol Chem. 1997;272:10998–11001. doi: 10.1074/jbc.272.17.10998. [DOI] [PubMed] [Google Scholar]
- 32a.Martin, S. S., et al. Submitted for publication.
- 33.Martin S S, Haruta T, Morris A J, Klippel A, Williams L T, Olefsky J M. Activated phosphatidylinositol 3-kinase is sufficient to mediate actin rearrangement and GLUT4 translocation in 3T3-L1 adipocytes. J Biol Chem. 1996;271:17605–17608. doi: 10.1074/jbc.271.30.17605. [DOI] [PubMed] [Google Scholar]
- 34.Martin S S, Rose D W, Saltiel A R, Klippel A, Williams L T, Olefsky J M. Phosphatidylinositol 3-kinase is necessary and sufficient for insulin-stimulated stress fiber breakdown. Endocrinology. 1996;137:5045–5054. doi: 10.1210/endo.137.11.8895379. [DOI] [PubMed] [Google Scholar]
- 35.Morris A J, Martin S S, Haruta T, Nelson J G, Vollenweider P, Gustafson T A, Mueckler M, Rose D W, Olefsky J M. Evidence for an insulin receptor substrate 1 independent insulin signaling pathway that mediates insulin-responsive glucose transporter (GLUT4) translocation. Proc Natl Acad Sci USA. 1996;93:8401–8406. doi: 10.1073/pnas.93.16.8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mueckler M. Facilitative glucose transporters. Eur J Biochem. 1993;219:713–725. doi: 10.1111/j.1432-1033.1994.tb18550.x. [DOI] [PubMed] [Google Scholar]
- 37.Peresse X, Deleu S, De Smedt F, Drayer L, Erneux C. Identification of a second SH2-domain-containing protein closely related to the phosphatidylinositol polyphosphate 5-phosphatase SHIP. Biochem Biophys Res Commun. 1997;239:697–700. doi: 10.1006/bbrc.1997.7538. [DOI] [PubMed] [Google Scholar]
- 38.Stephens L, Anderson K, Stokoe D, Erdjument-Bromage H, Painter G F, Holmes A B, Gaffney P R J, Reese C B, McCormick F, Tempst P, Coadwell J, Hawkins P T. Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-triphosphate-dependent activation of protein kinase B. Science. 1998;279:710–714. doi: 10.1126/science.279.5351.710. [DOI] [PubMed] [Google Scholar]
- 39.Stephens L R, Hughes K T, Irvine R F. Pathway of phosphatidylinositol(3,4,5)-triphosphate synthesis in activated neutrophils. Nature. 1991;351:33–39. doi: 10.1038/351033a0. [DOI] [PubMed] [Google Scholar]
- 40.Stokoe D, Stephens L R, Copeland T, Gaffney P R J, Reese C B, Painter G F, Holmes A B, McCormock F, Hawkins P T. Dual role of phosphatidylinositol-3,4,5-triphosphate in the activation of protein kinase B. Science. 1997;277:567–570. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- 41.Toker A, Cantley L C. Signalling through the lipid products of phosphoinositide-3-OH kinase. Nature. 1997;387:673–676. doi: 10.1038/42648. [DOI] [PubMed] [Google Scholar]
- 42.Vollenweider P, Martin S S, Haruta T, Morris A J, Nelson J G, Cormont M, Le Marchand-Brustel Y, Rose D W, Olefsky J M. The small guanosine triphosphate-binding protein Rab4 is involved in insulin-induced GLUT4 translocation and actin filament rearrangement in 3T3-L1 cells. Endocrinology. 1997;138:4941–4949. doi: 10.1210/endo.138.11.5493. [DOI] [PubMed] [Google Scholar]
- 43.Wennstroem S, Hawkins P, Cooke F, Hara K, Yonezawa K, Kasuga M, Jackson T, Claesson-Welsh L, Stephens L. Activation of phosphoinositide 3-kinase is required for PDGF-stimulated membrane ruffling. Curr Biol. 1994;4:385–393. doi: 10.1016/s0960-9822(00)00087-7. [DOI] [PubMed] [Google Scholar]
- 44.White M F, Kahn C R. The insulin signaling system. J Biol Chem. 1994;269:1–4. [PubMed] [Google Scholar]
- 45.Woscholski R, Parker P J. Inositol lipid 5-phosphatases-traffic signals and signal traffic. Trends Biochem Sci. 1997;22:427–431. doi: 10.1016/s0968-0004(97)01120-1. [DOI] [PubMed] [Google Scholar]