Abstract
The wild-type p53 protein exhibits a common polymorphism at amino acid 72, resulting in either a proline residue (p53Pro) or an arginine residue (p53Arg) at this position. Despite the difference that this change makes in the primary structure of the protein resulting in a difference in migration during sodium dodecyl sulfate-polyacrylamide gel electrophoresis, no differences in the biochemical or biological characteristics of these wild-type p53 variants have been reported. We have recently shown that p53Arg is significantly more susceptible than p53Pro to the degradation induced by human papillomavirus (HPV) E6 protein. Moreover, this may result in an increased susceptibility to HPV-induced tumors in homozygous p53Arg individuals. In further investigating the characteristics of these p53 variants, we now show that both forms are morphologically wild type and do not differ in their ability to bind to DNA in a sequence-specific manner. However, there are a number of differences between the p53 variants in their abilities to bind components of the transcriptional machinery, to activate transcription, to induce apoptosis, and to repress the transformation of primary cells. These observations may have implications for the development of cancers which harbor wild-type p53 sequences and possibly for the ability of such tumors to respond to therapy, depending on their p53 genotype.
The p53 gene is one of the most intensely studied human genes because of its role as a tumor suppressor gene (reviewed in reference 13). Mutations in p53 are found in over 50% of all human cancers (11), comprising more than 50 different cell and tissue types, indicating that there is a powerful selection for loss of p53 activity during tumor development. Although the fact that p53-null mice develop normally indicates that p53 is not required for normal development, these mice are susceptible to an array of spontaneous tumors in early adult life (5). The importance of p53 as a tumor suppressor is additionally demonstrated in humans with the rare autosomal dominant Li-Fraumeni syndrome, who carry heterozygous p53 mutations in the germline. Upon loss of the wild-type p53 allele, these individuals develop a variety of mesenchymal and epithelial tumors at an early age (15, 22). A number of DNA tumor virus oncoproteins also target p53, including the simian virus 40 large T, adenovirus E1b, and human papillomavirus (HPV) E6 proteins, which interact with and inactivate p53 through a variety of mechanisms (reviewed in reference 29). These interactions are at least partly responsible for the transforming activity of these viruses and are particularly important in the case of cervical cancer which is caused by high-risk oncogenic HPV types (32). Based on these key observations, p53 is considered to be the prototype tumor suppressor.
The p53 protein is normally present at low levels in the cell, but it can be upregulated by stimuli such as DNA damage, hypoxia, or the deregulated cell cycle progression caused by ectopic oncogene expression (reviewed in reference 13). The biological consequence of p53 upregulation is the induction of pathways that lead to either cell cycle arrest or apoptosis (13). At the biochemical level, p53 upregulation results in an increase in p53 sequence-specific transcriptional transactivation activity (20), resulting in the expression of a variety of genes, most notably the p21/WAF1 gene (6). The p21/WAF1 gene product is critical for the induction of cell cycle arrest in G1 through the inhibition of cyclin-dependent kinases, which are necessary for the G1/S transition. Consistent with its function as a transcriptional transactivator, p53 also associates with the TATA box binding protein (TBP) (28) and with several TFIID-associated factors (TAFs) (26). In addition to its role as an activator of transcription, it has also been reported that p53 acts as a transcriptional repressor of promoters containing a TATA element (14, 24).
The biochemical mechanism by which p53 induces apoptosis is still a matter of some controversy, since it is not clear whether p53-mediated transcription is involved in the process (3, 10). It has been reported that p53-dependent apoptosis, in response to DNA damage, is independent of the synthesis of new RNA or protein (3). Conversely it has been shown that p53 can induce the expression of the bax (17) and cd95/fas (19) genes, both of which are promoters of apoptosis.
The majority of p53 mutations found in cancer cells are missense point mutations, occurring mainly in the DNA binding domain of the protein (4). The mutant p53 proteins, differing from the wild type by only one amino acid residue, generally lose the ability to bind DNA and are thus functionally inactive. This is a clear demonstration of the strong selective pressure in tumor cells to inactivate p53 function. However, despite the justified concentration upon analysis of tumor-derived mutant p53 protein, there has been very little research, at the molecular level, on the comparative activities of the two common polymorphic variants of the wild-type p53 (16). This polymorphism arises from a single-base-pair substitution at codon 72, where either CCC encodes proline or CGC encodes arginine (16). Clearly this is a nonconservative amino acid change, and furthermore, it results in a structural change in the protein, since the p53Pro variant migrates more slowly than the p53Arg variant in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (16). Recently, we have shown that the E6 proteins from both high-risk and low-risk HPV types are able to target p53Arg more efficiently than p53Pro for ubiquitin-mediated degradation. Consistent with this observation, the majority of HPV-associated tumors so far analyzed are homozygous for the p53Arg allele (23), whereas the majority of the comparable normal population was heterozygous.
Having identified a difference in how the oncogenic HPV E6 proteins recognize these two forms of p53, we were interested in a further systematic examination of any other potential biochemical or biological differences between the two. This type of analysis is now particularly important, since recent studies have begun to define a function for this region of the p53 protein. The proline-rich domain of p53 has been shown to be required for the growth suppression activity of p53 (29), and it also plays an important role in p53-mediated apoptosis but not in cell cycle arrest (21). This polyproline region is considered to be an Src homology 3 (SH3) binding domain, and the proline at amino acid 72 constitutes one of the five PXXP SH3 binding motifs defined within this region (29). Recent studies on this region of p53 (21, 29) have considered only the p53Pro variant, and no consideration has yet been given to the p53Arg variant, which is in fact the more common wild-type variant in certain populations (2).
Given that this region is functionally important for wild-type p53 activity, an important question is whether the p53Pro and p53Arg wild-type proteins are functionally equivalent. We have therefore examined the biochemical and biological activities of these two wild-type variants of p53. Both proteins are structurally wild type, as determined by monoclonal antibody reactivity, and they exhibit similar levels of affinity for a variety of p53 DNA recognition sequences. However, there are subtle differences in their respective abilities to interact with basic elements of the transcriptional machinery, and this is reflected in differences in their transcriptional activities. In addition, there are also differences in the abilities of each form to induce apoptosis and suppress transformed cell growth. These results demonstrate that the two forms of p53 are not functionally equivalent, and this may have important implications for the management of patients with wild-type p53-containing tumors, depending on their p53 genotype.
MATERIALS AND METHODS
Cells, plasmids, and antibodies.
The p53-null 10(1) murine fibroblasts, primary baby rat kidney (BRK) cells, and Saos-2 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (GIBCO, Burlington, Canada) supplemented with 10% fetal bovine serum (FBS).
The two wild-type p53 variant cDNAs, those for p53Arg and p53Pro, initially cloned and characterized by Matlashewski et al. (16), were inserted into the SP6-driven pSP64 vector (HindIII and BamHI sites) for in vitro translation, into the pCDNA3 vector (HindIII and BamHI sites) for in vivo expression, and into the pGEX2 vector for glutathione S-transferase (GST) fusion protein production. The p53 His175 mutant (Mp53-175) was cloned into the pCDNA3 vector. The HPV type 18 (HPV-18) E6 gene within the pJ4 vector has been previously described (8) and was also cloned into the pSP64 vector (HindIII and EcoRI sites). The p53-responsive plasmids used for chloramphenicol acetyltransferase (CAT) assays were pG13CAT (12) and p53CONCAT (24). The luciferase assays were performed with the luciferase reporter plasmid pWT3L2 (kindly provided by S. Benchimol), which contains the p21 promoter-enhancer upstream of the luciferase gene. The transcription factors were expressed in vitro from the plasmids pIngTBP, pHAX-TAFII32, pHAX-TAFII70, and pHAX-TAFII250 (kindly provided by R. Tjian).
The anti-p53 PAb421 monoclonal antibody used for DNA binding assays was obtained from Cedar Lane Laboratories Limited (Toronto, Ontario, Canada). The combination of anti-p53 monoclonal antibodies used for p53 Western blot analysis included PAb1801, PAb1802, and PAb122 hybridoma supernatants (1). The anti-p53 monoclonal antibodies PAb240, PAb1620, and PAb246 used in the immunoprecipitation assays were obtained from Oncogene Research Products. The C4 antibody is a polyclonal rabbit antiserum raised against the carboxy-terminal 14 amino acids of p53.
Transfections.
Transient transfections of p53-null 10(1) murine fibroblasts were performed with the Lipofectamine system according to the manufacturer’s instructions (Gibco BRL). Briefly, cells were transfected with 1 μg of the p53 expression plasmid and 1 μg of either the CAT reporter plasmid (either pG13CAT or p53CONCAT) or the luciferase reporter plasmid pWT3L2. In the dilution CAT assay, 0.5 μg of p53CONCAT was cotransfected with between 1.0 and 0.0025 μg of the variant p53 expression plasmids and a lacZ-expressing plasmid to control for any variations in transfection efficiencies. Plasmids were incubated in the presence of 10 μl of Lipofectamine reagent for 45 min at room temperature, after which serum-free DMEM was added and the mixture was overlaid on the cells and incubated. After 6 h, the serum-free DMEM was replaced with DMEM supplemented with 10% FBS. After 48 h of incubation, cells were harvested in Nonidet P-40 (NP-40) lysis buffer (150 mM NaCl, 1.0% NP-40, 20 mM Tris [pH 8.0]), incubated on ice for 30 min, and then clarified by centrifugation at 14,000 rpm for 15 min at 4°C in an Eppendorf centrifuge. Protein concentrations were determined by the Bio-Rad protein assay, and the presence of equal amounts of p53 in the cell extracts was confirmed by Western blot analysis prior to CAT assays or luciferase assays. Cell extracts containing equal amounts of p53Arg and p53Pro proteins, as determined by Western blot analysis, were assayed for CAT or luciferase activity.
For growth suppression analysis, transfections were performed by the standard calcium phosphate precipitation method, and the cells were maintained under selection with G418 at a concentration of either 200 μg/ml (BRK cells) or 500 μg/ml (Saos-2 cells). Colonies were counted approximately 2 weeks after transfection. In the case of the BRK transfections, the colonies were pooled and then maintained as a polyclonal pool for p53 protein analysis.
CAT and luciferase assays.
CAT assays were performed essentially as previously described (8). Extracts were heated at 65°C for 10 min and centrifuged for 2 min at 14,000 rpm in an Eppendorf centrifuge. CAT assays were carried out in the presence of 5 μl of [14C]chloramphenicol (50 mCi/ml; ICN) and 5 μl of acetyl coenzyme A (33.3 mg/ml) in a final reaction volume of 100 μl at 37°C for 2 h. Samples were extracted with ethyl acetate and then analyzed by thin-layer chromatography and viewed by autoradiography. Percent CAT conversion was determined by scintillation counts of scraped thin-layer chromatography plates.
Luciferase assays were performed on cells transfected as described above with the p53 expression plasmids together with the p53-responsive luciferase reporter plasmid pWT3L2, which contains the p21 promoter and enhancer upstream from the luciferase gene. Extracts prepared as described above which contained equal amounts of plasmid-derived p53 were incubated in the presence of the luciferase substrate (Promega, Montreal, Canada) for 30 s, and the luciferase activity was determined by scintillation counting.
p53 DNA binding assays.
Three different p53 consensus sequences were used to examine p53 sequence-specific DNA binding. These sequences were p53CON (24), CON* (7), and bax (17) and were as follows: p53CON, 5′-GGA CAT GCC CGG GCA TGT CC-3′ 3′-CCT GTA CGG GCC CGT ACA GG-5′ CON*: 5′-GGG CAT GTC CGG GCA TGT CC-3′ 3′-CCC GTA CAG GCC CGT ACA GG-5′ bax: 5′-GATCTCACAAGTTAGACAAGCCTG-3′ 3′-TCTGTTCGGACCCGCACCCGATATAACAGCT-5′
p53CON and CON* were end labeled with [γ-32P]dATP (450 Ci/mmol; ICN) by using T4 polynucleotide kinase (9,700 U/ml; Pharmacia), and the bax sequence was labeled with [α-32P]dCTP by using the Klenow fragment (1,000 U/ml; Pharmacia). DNA binding assays were performed as previously described (31) with a few modifications. p53 proteins were synthesized in vitro by use of a coupled reticulocyte lysate transcription-translation system (TNT system; Promega). Reaction mixtures containing unlabeled p53 protein in the presence or absence of 300 ng of PAb421 (Cedar Lane) were preincubated at room temperature for 30 min. This reaction mixture was then added to the following cocktail containing the individual oligonucleotides: 5 μl of a 5× binding buffer (100 mM HEPES [pH 7.9], 125 mM KCl, 0.5 mM EDTA, 50% glycerol, 10 mM MgCl), 1 μl of 10 mM dithiothreitol, 1 μl of 0.5% NP-40, 200 ng of poly(dI-dC), 2 μg of bovine serum albumin, 5 ng of labeled p53 target oligonucleotide (40,000 to 100,000 cpm), and distilled water to a final volume of 25 μl. DNA binding reaction mixtures were incubated at room temperature for a further 40 min and then loaded onto a 4% nondenaturing polyacrylamide gel which had been prerun for at least 30 min at 4°C. Gels were run at 200 V for 2.5 h at 4°C, dried, and then subjected to autoradiography. A portion of in vitro-synthesized p53 was labeled with [35S]cysteine and run on an SDS–10% polyacrylamide gel, followed by autoradiography in order to verify that equal amounts of p53 proteins were synthesized in each reaction. As a negative control for DNA binding, in vitro-synthesized HPV-18 E6 protein was assayed for binding to the same oligonucleotides, and, as expected, no specific binding was obtained.
Western blot analysis.
The same NP-40 cell lysates which were used in the CAT assays were analyzed for p53 protein levels by Western blot analysis. Lysates containing equal amounts of total protein were denatured by being boiled in SDS-PAGE buffer and then resolved by SDS-PAGE. The resolved proteins were transferred onto nitrocellulose membranes and incubated overnight at 4°C in blocking solution (phosphate-buffered saline [PBS], 10% powdered milk, 0.5% Tween). Membranes were exposed to a combination of anti-p53 monoclonal antibodies (PAb1801, PAb1802, and PAb122) in PBS with 5% powdered milk for 2 h at room temperature, followed by three washes for 10 min in PBS. Membranes were then incubated with diluted antimouse antibody (1:2,000) for 2 h at 37°C, followed by three washes of 15 min in PBS. The presence of antibody was detected with an enhanced chemiluminescence (ECL) kit (Amersham Life Sciences).
To analyze the stably transfected BRK cells for p53 expression, semiconfluent monolayers were exposed to 200 J of UVC light per s. Six hours later, the monolayers were washed in PBS and extracted in extraction buffer (250 mM NaCl, 50 mM HEPES [pH 7], 0.1% NP-40, 1% aprotinin) per 100-mm-diameter dish. The protein concentration of each extract was determined with the Bio-Rad protein assay system, and equal amounts were analyzed by Western blotting. The blot was probed with a mixture of the anti-p53 monoclonal antibodies PAb1801, PAb1802, and PAb1803 and visualized with the Amersham ECL system.
GST fusion protein binding assays.
GST-p53Pro and -p53Arg fusion proteins were produced in Escherichia coli DH5α cells by IPTG (isopropyl-β-d-thiogalactopyranoside) induction of a log-phase culture for 3 h. The cell pellets were resuspended in PBS containing 0.5% Triton X-100 and sonicated for 15 s. After clarification, the supernatant was incubated overnight with glutathione-agarose at 4°C. After washing with PBS containing 0.5% Triton X-100, an aliquot of the agarose was analyzed by SDS-PAGE and Coomassie blue staining to assess levels of fusion protein binding. Aliquots of agarose containing equal amounts of fusion protein were washed with PBS and incubated with 35S-labeled in vitro-translated protein (TNT System; Promega) as indicated. After washing with PBS–0.5% NP-40, the proteins bound to the agarose were analyzed by SDS-PAGE and autoradiography.
Cell survival assays.
The cell survival assay was performed essentially as previously described (25, 30). Briefly, Saos-2 cells (105) were plated on 60-mm-diameter dishes and transfected with 2 μg of lacZ expression plasmid pCH110 and 10 μg of the p53 expression plasmids. At various time intervals, cells were fixed and stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), and live and dead cells were counted.
RESULTS
Conformational analysis of p53Pro and p53Arg.
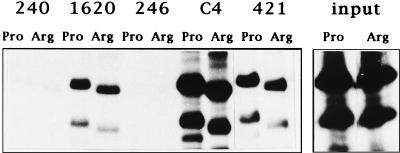
The majority of inactivating mutations in p53 result in conformational changes. Since the Pro-Arg polymorphism at codon 72 produces a major change in primary structure, as can be seen from the different mobilities of the proteins on SDS-PAGE (16), it could be argued that one or the other type is, in fact, conformationally mutant. To test this hypothesis, we made use of a range of well-characterized monoclonal antibodies to differentiate between the wild-type and mutant conformations of the p53 protein. p53Pro and p53Arg were translated in vitro and then analyzed by immunoprecipitation followed by SDS-PAGE and autoradiography; the results obtained are shown in Fig. 1. As shown by these immunoprecipitations, p53Pro migrates slower than p53Arg, and this is consistent with previous observations (16). It is clear, however, that PAb1620, which recognizes only the wild-type p53 conformation, reacted equally well with both the p53Pro and p53Arg proteins. Each protein is also equally recognized by PAb421 and by the polyclonal C4 antipeptide antiserum, both of which bind to the C-terminal domain of p53. Neither p53Pro nor p53Arg is recognized by PAb240, which is specific for an epitope exposed in mutant p53, or by PAb246, which is specific for murine p53. Thus, despite a major difference in the primary structure of the protein which results in the mobility difference in SDS-PAGE, both p53Pro and p53Arg can be considered to be conformationally indistinguishable and wild type.
FIG. 1.
p53Pro and p53Arg display similar epitopes. Immunoprecipitation analysis of in vitro-translated p53Pro and p53Arg proteins with a panel of antibodies is shown. The variants reacted equally well with the wild-type-specific PAb1620 antibody and with the carboxy-terminal-specific C4 and PAb421 antibodies. No reaction is observed with the mutant-specific PAb240 antibody or with the murine-specific PAb246 antibody. The right-hand panel shows the level of input proteins.
Comparison of the transcriptional transactivation activities of p53Pro and p53Arg.
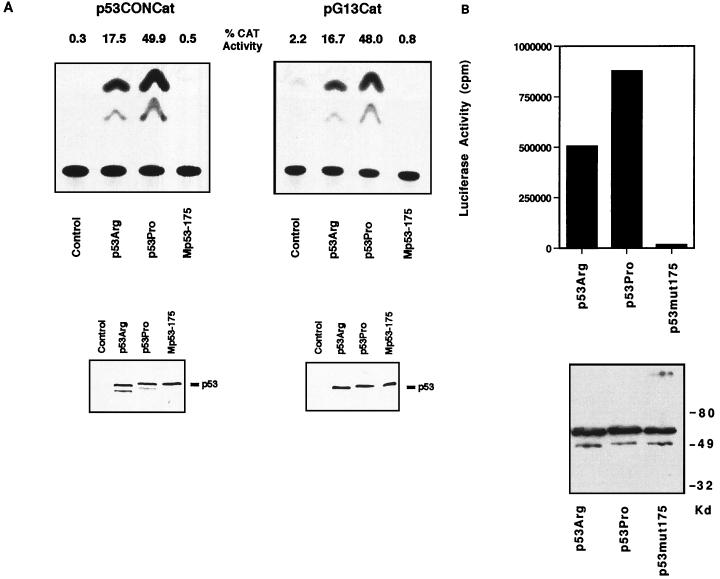
One of the major biochemical functions of p53 is sequence-specific transcriptional transactivation. Having demonstrated that both p53Pro and p53Arg retain the wild-type conformation, we were then interested in comparing the transcriptional transactivation activities of the two variants. For this analysis, plasmids expressing either p53Pro or p53Arg were transfected into p53-null murine 10(1) cells together with the pG13CAT or p53CONCAT reporter plasmid. Parallel aliquots of the cell extracts were analyzed by Western blotting to determine p53 protein levels in the transfected cells and by CAT assay to determine p53-mediated transcriptional activity. As negative controls, cells were transfected with the CAT reporter plasmids plus either empty pCDNA3 or a plasmid expressing the nonfunctional Mp53-175 mutant. As shown in Fig. 2A, under these assay conditions, p53Pro is a stronger transcriptional activator than p53Arg with both the pG13CAT and p53CONCAT reporter plasmids. Quantitation of these assays shows that p53Pro activates transcription to levels approximately twofold higher than those with p53Arg. The Western blot analysis (Fig. 2A) confirmed that equal levels of p53Pro, p53Arg, and Mp53-175 proteins were expressed in the transfected cells. When a luciferase transcriptional assay was performed under the same conditions with the p53-responsive p21 promoter linked to the luciferase reporter gene, it was likewise revealed that p53Pro induced similarly higher levels of transcription activation than p53Arg (Fig. 2B).
FIG. 2.
(A) Comparison of p53Pro and p53Arg transcriptional activation of pCONCAT and pG13CAT. The upper panels show the transcriptional activities of the wild-type p53 variants and mutant p53 protein determined by using two different reporter CAT plasmids, p53CONCAT and pG13CAT. p53-null 10(1) murine fibroblast cells were cotransfected with p53 expression plasmids or a control empty plasmid and the CAT reporter plasmid, and the CAT activity in the transfected cells was determined. p53Arg and p53Pro represent the two wild-type variants of p53, and Mp53-175 is the inactive p53 point mutant. The percent CAT conversion is also shown. The lower panels show Western blot analysis of the same cell lysates used in the CAT assay and demonstrate that equal amounts of plasmid-derived p53 protein were present in the transfected cells. (B) Comparison of p53Pro and p53Arg transcriptional activation of the of the p21 promoter by using a luciferase assay. The upper panel shows transcriptional activity of the wild-type p53 variants and mutant p53 protein determined by using the luciferase reporter plasmid pWT3L2, which contains the p53-responsive p21 promoter and enhancer. The lower panel shows Western blot analysis of the same cell lysates used in the luciferase assays and demonstrates that equal amounts of plasmid-derived p53 protein were present in the transfected cell lysates.
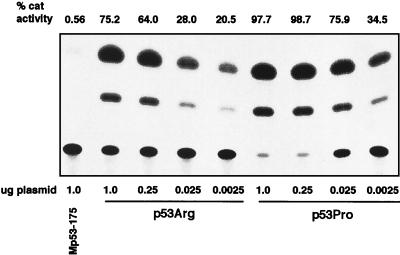
To further investigate the difference in transcriptional transactivation, we also performed a dilution assay in which decreasing amounts of variant p53 expression plasmid were cotransfected with a constant amount of the p53CONCAT reporter plasmid. As shown in Fig. 3, it was evident that at the different levels of input p53 expression plasmid, there was a consistently higher level of transcriptional activity observed with p53Pro than with p53Arg. Taken together, these data demonstrate that p53Pro is a more active transcriptional activator than p53Arg. This is the first demonstration that the two wild-type p53 variants present in the general population are not biochemically equivalent.
FIG. 3.
Comparison of p53Pro and p53Arg transcriptional activation following transfection of various concentrations of p53 variant-expressing plasmids. Between 1.0 and 0.0025 μg of p53Arg- or p53Pro-expressing plasmid was cotransfected with 5.0 μg of p53CONCAT plasmid as indicated. As a negative control for p53 activity, the p53CONCAT plasmid was cotransfected with 3 μg of the Mp53-175 plasmid. The percent CAT conversion is shown.
Affinities of p53Pro and p53Arg for enhancer DNA sequences.
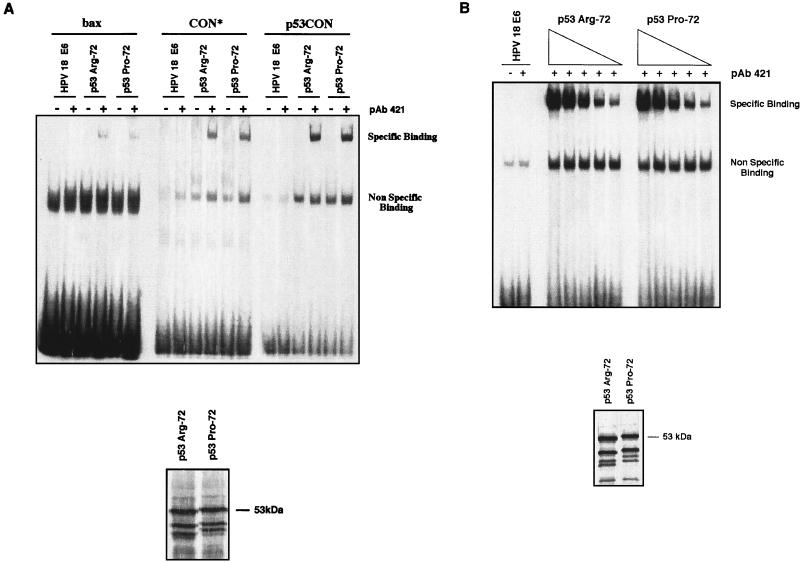
The transcriptional transactivation activity of p53 is mediated by its specific binding to p53-responsive motifs in the promoters of the relevant genes and through interactions with components of the basic transcriptional machinery. Having shown that p53Pro and p53Arg differ in their ability to transactivate certain reporter plasmids, we were interested in determining the mechanisms underlying this difference. To address this, we first analyzed the abilities of the two p53 variants to bind to three different p53-specific enhancer binding sequences. A mobility shift assay was performed with the bax, CON*, and p53CON oligonucleotides (see Materials and Methods for sequences) incubated with in vitro-translated p53Pro and p53Arg; HPV-18 E6 was also included as a negative control for binding to these oligonucleotides. The results of this assay are shown in Fig. 4A. Although both p53 variants had a higher affinity for CON* and p53CON than for the bax sequence, there was no difference between p53Pro and p53Arg in their affinities for any one sequence. As expected, HPV-18 E6 was unable to bind these sequences, nor was p53 capable of binding in the absence of the activating PAb421 antibody.
FIG. 4.
Comparison of sequence-specific DNA binding activities of p53Pro and p53Arg. (A) Comparison of the abilities of p53Arg and p53Pro to bind specific enhancer sequences (bax, CON*, and p53CON). p53 proteins and control HPV-18 E6 protein were synthesized in vitro and preincubated in the presence (+) or absence (−) of the monoclonal antibody PAb421, which activates p53 sequence-specific binding. Proteins were incubated in the presence of the labeled DNA sequence at room temperature for 40 min and then run on a nondenaturing gel. Note that the higher-molecular-weight specific binding band was obtained for the p53 variants only in the presence of PAb421, and no specific binding was obtained with the E6 protein. As shown at the bottom, a portion of the synthesized p53 proteins was also labeled and run on an SDS-polyacrylamide gel in order to ensure that the same amounts of p53 protein were synthesized and assayed in each reaction. (B) Titration of p53Pro and p53Arg binding to p53CON DNA. Decreasing amounts of unlabeled p53Arg and p53Pro proteins synthesized in vitro were preincubated in the presence of the monoclonal antibody PAb421 and then incubated in the presence of the labeled p53CON DNA sequence at room temperature for 40 min and run on a nondenaturing gel. As shown on the bottom, a portion of the synthesized p53 proteins was also labeled and run on an SDS-polyacrylamide gel in order to ensure that the same amounts of p53 protein were synthesized and assayed in each reaction.
To further compare the sequence-specific DNA binding, a titration analysis with p53Pro and p53Arg was performed. Increasing amounts of p53Pro or p53Arg were incubated in the presence of a constant amount of radiolabeled p53CON enhancer sequence oligonucleotide. It can be seen in Fig. 4B that there is no significant difference between the binding of p53Pro and p53Arg at any of the concentrations used, thus confirming that p53Pro and p53Arg have equal affinities for the p53CON sequence. This assay was repeated with both the bax and CON* sequences, with similar results (data not shown). Thus, there are no differences in the sequence-specific DNA binding activities of p53Pro and p53Arg.
Comparison of the abilities of p53Pro and p53Arg to interact with components of the basal transcriptional machinery.
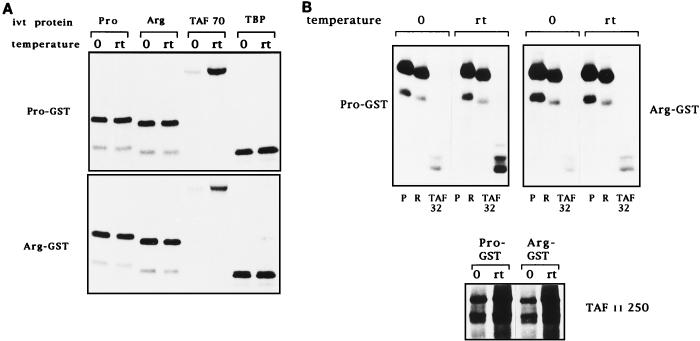
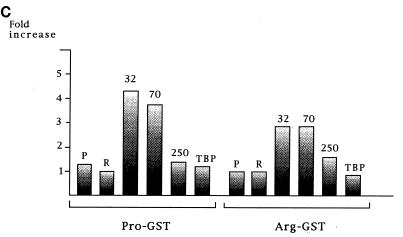
The finding that the two variants did not differ in their sequence-specific DNA binding activities suggested that the differences in transcriptional activity may be due to differences in their interactions with transcription factors involved in p53 transcriptional transactivation. Previous studies had shown that interactions with TAFII32 and TAFII70, and to some extent TBP, played a role in p53’s ability to activate transcription (26). Therefore, we next compared the abilities of p53Pro and p53Arg to interact with TAFII32, TAFII70, TAFII250, and TBP. GST-p53Pro and GST-p53Arg fusion proteins were incubated with in vitro-translated TAFII32, TAFII70, TAFII250, and TBP at either 0°C or room temperature for 1 h. For comparison, the fusion proteins were also incubated with in vitro-translated p53Pro and p53Arg, since the domains of p53 involved in dimerization lie within the carboxy-terminal domain, well away from the site of the polymorphism. After extensive washing, the bound proteins were analyzed by SDS-PAGE and autoradiography, and the results are shown in Fig. 5. Not surprisingly, p53Pro and p53Arg form hetero- and homodimers with equal efficiency at both temperatures, and both variants also bind TBP and TAFII250 with fairly equal efficiency, although the interaction with TAFII250 is clearly cold sensitive. However, significant differences in their interaction with TAFII32 and TAFII70 are seen. First, it is clear that the binding of both proteins to p53 is very weak at 0°C but that this increases dramatically at room temperature, demonstrating that the interaction between p53 and both TAFII32 and TAFII70 is also denatured by low temperature. Second, and most interestingly, it is also clear that p53Pro exhibits significantly higher levels of binding to both TAFII32 and TAFII70 than does p53Arg. The binding assay was quantitated on a Phosphorimager, and the results obtained are shown in Fig. 5C as the fold increase in binding at room temperature over binding at 0°C. These data suggest that the differences in the abilities of the two variants of p53 to activate transcription may, at least in part, be due to their differential abilities to bind to TAFII32 and TAFII70.
FIG. 5.
Interactions of p53Pro and p53Arg with transcription factors. (A) GST fusion protein pull-down assay with GST-p53Pro and GST-p53Arg bound to glutathione-agarose. In vitro-translated (ivt) 35S-labeled p53Pro, p53Arg, TAFII70, or TBP proteins were incubated with the fusion protein-agarose at either 0°C or room temperature (rt). The bound proteins were analyzed by SDS-PAGE and autoradiography. No differences in the abilities of p53Pro and p53Arg to form either hetero- or homodimers or in their abilities to bind to TBP were apparent. However, TAFII70 is bound significantly only at room temperature, and p53Pro binds more strongly than p53Arg to TAFII70. (B) The GST-fusion protein pull-down assay repeated with in vitro-translated 35S-labeled TAFII32 and TAFII250 plus p53Pro (P) and p53Arg (R) for comparison. TAFII32 binds more strongly at room temperature and binds more strongly to p53Pro than to p53Arg. TAFII250 binds to p53Pro and p53Arg with similar affinities but binds more strongly at room temperature. (C) Phosphorimager analysis of at least five GST fusion protein pull-down assays. The fold increase in binding at room temperature over binding at 0°C is shown. p53Pro binds approximately twice as strongly as p53Arg to TAFII32 and TAFII70. GST alone was also used as a negative control; in all cases, the percentage of loaded protein retained was less than 1%.
Comparison of p53Pro and p53Arg suppression of transformed cell growth.
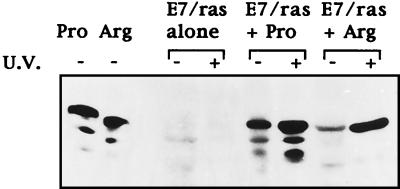
Having established that p53Pro and p53Arg differ in their respective transcriptional activities, we were next interested in comparing the biological activities of the two variants. An established assay for analyzing the growth-inhibitory effects of p53 is its ability to suppress transformed cell growth. We therefore compared the abilities of p53Pro and p53Arg to suppress the proliferation of Saos-2 cells and to suppress transformation of primary BRK cells with the HPV-16 E7-plus-EJ-ras oncogenes. Cells were transfected with the appropriate plasmid combinations, and after 2 weeks under selection, the cells were fixed and stained and the number of colonies was counted; the results obtained are shown in Tables 1 and 2. It is clear that in Saos-2 cells (Table 1), p53Pro and p53Arg suppress cell proliferation with similar levels of efficiency. In addition, colonies which do proliferate over the 2-week period cannot be further expanded, indicating that in these cells, both forms of p53 cause a complete cessation of cell proliferation. In contrast, in the suppression of E7–EJ-ras transformation assay (Table 2), differences between the two forms of p53 do appear to exist, with p53Arg being approximately twofold more active in suppressing colony formation than p53Pro. It is also clear, however, that colonies were consistently obtained in the transformation assay, regardless of whether p53Arg or p53Pro was present. We were therefore interested in determining whether these surviving BRK cells were actually expressing the transfected p53 proteins. To assess this, cell lines which had been transfected with p53Pro and p53Arg in the presence of E7 and EJ-ras were established, and the levels of p53 protein were measured following UV irradiation by Western blotting with human p53-specific monoclonal antibodies. The results obtained are shown in Fig. 6 and demonstrate that both lines continue to express the transfected p53 protein and that this can be induced following UV-induced DNA damage. These results demonstrate that these cells do contain a functional p53 protein and suggest that these surviving clones may have acquired a mutation in a gene downstream of p53 which overrides p53 function. Taken together, however, the results from the colony formation assay with BRK cells argue that p53Arg is more efficient than p53Pro in suppressing transformation by the E7 and EJ-ras oncogenes.
TABLE 1.
Suppression of growth of transformed Saos-2 cellsa
| Cell line | Mean colony no. ± SDb | % Reduction of growth |
|---|---|---|
| pCDNA3 vector | >1,000 | 0 |
| p53Pro | 239 ± 57 | 76 |
| p53Arg | 132 ± 40 | 87 |
Transfections were carried out as described in Materials and Methods.
From three separate transfections.
TABLE 2.
Suppression of E7–EJ-ras cooperation
| Cell line | Mean colony no. ± SDa | % Reduction of transformation |
|---|---|---|
| E7 + EJ-ras | 85 ± 12 | 0 |
| p53Prob | 50 ± 12 | 41 |
| p53Argb | 14 ± 4 | 83 |
From five separate transfections.
These cells were cotransfected with the p53 variants together with the EJ-ras and E7 oncogenes.
FIG. 6.
p53 induction pathways are functional in stably transfected BRK cell lines. Western blot analysis of the p53 protein in pools of colonies obtained after cotransfection of BRK cells with either p53Pro or p53Arg, with or without UV irradiation, is shown.
Comparison of p53Pro- and p53Arg-induced apoptosis.
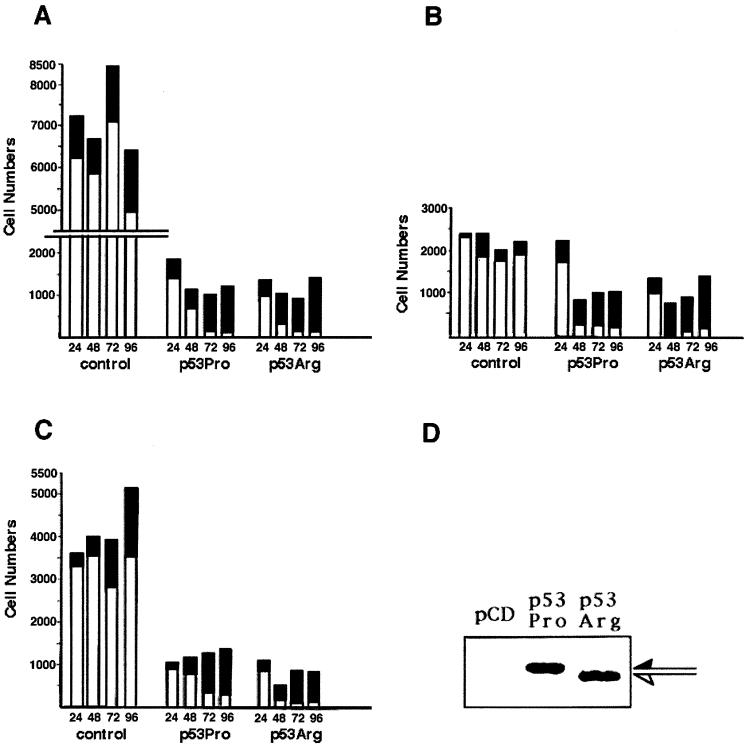
Having shown that p53Pro and p53Arg differed in their abilities to suppress the transformation of primary cells, we next determined whether there were any differences in the abilities of the two proteins to induce apoptosis. An established means of measuring apoptosis is to perform cell survival assays under conditions where the apoptotic morphology of transfected cells can be directly observed and quantitated, as previously described (25, 30). Saos-2 cells were transfected with plasmids expressing either p53Pro or p53Arg, together with a lacZ-expressing plasmid. At 24, 48, 72, and 96 h posttransfection, the cells were fixed and stained with X-Gal, and the blue cells and apoptotic bodies were counted and scored as alive or dead by morphological criteria. The results of three independent assays are shown in Fig. 7A, B, and C. It can be seen that p53Pro and p53Arg cause similar reductions in cell survival over the period of these assays; however, the kinetics of cell death are different. At the early time points there were consistently more surviving cells following the transfection with p53Pro than following that with p53Arg. In addition, at the later time points we noted a tendency for the p53Arg-containing cells to form microcolonies, whereas the p53Pro-containing cells did not. To rule out the possibility that this was due to differences in the levels of p53 in these cells, we performed a Western blot analysis 24 h following transfection of a parallel set of Saos-2 cells. As shown in Fig. 7D, there were equal levels of p53Pro and p53Arg in the transfected cells. Taken together, these results show that p53Pro and p53Arg are capable of inducing equal levels of apoptosis by 96 h posttransfection; however, the kinetics of cell death appear to be faster with p53Arg.
FIG. 7.
Comparison of the activities of p53Pro and p53Arg in inducing apoptosis. (A, B, and C) Saos-2 cells were transfected with either pCDNA3 (control vector), pCDp53Pro, or pCDp53Arg together with a lacZ-expressing plasmid. Cells were fixed at the times posttransfection indicated and stained for lacZ expression. Histograms show the number of live (white bars) and dead (black bars) blue cells present at 24, 48, 72, and 96 h posttransfection. Each panel shows an independent assay. (D) Western blot analysis of a parallel transfection harvested at 24 h confirms equivalent levels of p53Pro and p53Arg expression.
DISCUSSION
Despite the intense research into the differences between wild-type and mutant p53, there has been no study to compare the biochemical and biological activities of the structurally different wild-type variants of p53. This study was therefore initiated to address this issue, and we report several important differences between the two common wild-type p53 variants which are present in the general population. We have shown that p53Pro is a stronger inducer of transcription than p53Arg and that this appears to be related at least in part to its stronger affinity for the TAFII32 and TAFII70 transcription factors. In addition, although the two forms appear to suppress proliferation of Saos-2 cells with similar levels of efficiency, p53Arg appears to induce apoptosis in these cells with faster kinetics than p53Pro. Interestingly, p53Arg also appeared to be a more potent suppressor of the HPV-16 E7-plus-EJ-ras cotransformation of BRK cells than was the p53Pro variant. Taken together, these data argue that the common variants of wild-type p53 are neither biochemically nor biologically equivalent. These difference have potential implications both for the comparison of wild-type and mutant p53 activities and for cancer therapies based on reactivation of wild-type p53 function.
There is limited information on the potential differences which may exist between the two common polymorphic variants of wild-type p53. The first study which attempted a systematic comparison of these two variants of p53 (18) indicated that they were functionally equivalent with respect to interaction with the simian virus 40 large T antigen. Comparison of the p53Pro and p53Arg conformations determined by antibody reactivity indeed supports these conclusions. Both forms of p53 reacted equally well with the conformation-dependent antibody PAB1620, and no reactivity was detected with the PAb246 antibody, which specifically recognizes the mutant conformation of p53. However, evidence is emerging indicating that these two forms of p53 are not equivalent. The oncogenic HPV E6 proteins have been shown to preferentially target p53Arg over p53Pro for ubiquitin-mediated degradation (23), and this manifests itself in an overrepresentation of p53Arg alleles in patients with HPV-associated tumors.
The principal biological manifestations of activated p53 are the induction of growth arrest and/or apoptosis. These activities are, in part, related to its ability to activate transcription from a number of target genes. Therefore, we were interested in analyzing the activities of the p53 polymorphic variants on promoters which are known to be p53 responsive. In the first series of studies, we analyzed the effects of the two polymorphic forms of p53 in two different reporter systems, first by CAT assays with plasmids pG13CAT and pCON*CAT and second by luciferase assays with plasmid pWT3L2, which contained the p21 enhancer and promoter. In both sets of assays, p53Pro consistently activated transcription of these p53-responsive promoters to a higher level than p53Arg. Parallel Western blot analyses of transfected cells confirmed that the differences in p53-mediated transcription were not due to differences in the levels of p53 in the transfected cells but represent an intrinsic difference between the p53Pro and p53Arg forms of the protein. Therefore, p53Pro and p53Arg exhibit marked differences in their respective abilities to activate expression from a variety of p53-responsive promoters. These results may have implications for reactivation of wild-type p53 in human tumors, since one would expect that individuals expressing p53Pro would exhibit stronger transcriptional transactivation.
Having demonstrated differences in the respective transcriptional activities of the two p53 variants, we were interested in identifying the molecular basis for these observations. DNA binding assays with several p53-specific enhancer sequences revealed that the two variants of p53 bound with equal affinity to these sequences. Human p53 also interacts with a number of factors which compose the basic transcriptional machinery of the cell, in particular TAFII32, TAFII70, and TBP. The interactions with TAFII32 and TAFII70 appear to be primarily responsible for the ability of p53 to act as a transcriptional activator (26). We reasoned that differences in interactions with these proteins could provide insight into the differences in transcriptional activation. Indeed, in a series of in vitro binding assays, it was shown that p53Pro binds more strongly to TAFII70 and TAFII32 than does p53Arg. Interestingly, no difference in binding to TBP or to TAFII250 was observed. These results therefore suggest that, at least for the transcriptional transactivation function of p53, the ability of p53Pro to activate expression to a higher level than p53Arg correlates with its increased affinity for TAFII32 and TAFII70. An additional interesting point from these studies was the observation that the homodimerization of p53 and its interaction with TBP were stable at both room temperature and 0°C. In contrast, the interactions with TAFII32, TAFII70, and TAFII250 were strongly inhibited at low temperature and are therefore susceptible to cold denaturation.
Finally, we compared the abilities of the p53Pro and p53Arg proteins to inhibit cellular proliferation and induce apoptosis. In Saos-2 cells, with which colony assays were performed, p53Pro and p53Arg inhibited cell proliferation to a similar degree. However, in transient assays for apoptosis, the p53Arg variant appeared to induce apoptosis with faster kinetics that the p53Pro variant. In primary BRK cell cotransformation assays, it was also observed that p53Arg suppressed transformation to a higher degree than p53Pro. This was interesting, since previous reports had suggested that suppression of transformation by p53 correlates closely with its ability to induce apoptosis (25). Taken together, these data suggest that p53Arg may induce apoptosis more efficiently than p53Pro. Since p53Pro is a stronger transcriptional transactivator of the promoters analyzed in this study, the activation of which can be considered to be more closely associated with an induction of cell cycle arrest, studies are now under way to address the activities of these two forms of p53 on promoters more closely associated with an induction of apoptosis and also to address their effects on cell cycle.
The observations made in this study argue that the two common p53 variants are not biochemically or biologically equivalent. It is noteworthy that there should not be a selection against either form of p53 in human populations, since the onset of most cancers occurs beyond the age of reproduction. However, an interesting issue is whether the codon 72 polymorphism is maintained by any other natural selective pressure.
In conclusion, we have shown that the two common polymorphic variants of p53 are similar based on epitope characterization with monoclonal antibodies despite the fact they migrate differently in SDS-PAGE. However, the variants exhibit differences in their respective abilities to activate gene expression, and this is reflected by their different degrees of interaction with the basic components of the transcriptional machinery. Evidence is presented that the p53Arg variant induces apoptosis with faster kinetics and suppresses transformation more efficiently than the p53Pro variant. These results may suggest that the p53 genotype could affect the design of future treatments and management strategies for patients with wild-type p53-containing tumors.
ACKNOWLEDGMENTS
We thank Sam Benchimol, Bert Vogelstein, John Jenkins, and Robert Tjian for the pWT3L2, pG13CAT, pCONCAT, and TAF-expressing plasmids, respectively.
G.M. acknowledges support from the National Cancer Institute of Canada, the Cancer Society of Canada, and the Natural Sciences and Engineering Research Council of Canada. L.B. acknowledges support from the Associazione Italian per la Ricerca sul Cancro.
REFERENCES
- 1.Banks L, Matlashewski G, Crawford L. Isolation of human-p53-specific monoclonal antibodies and their use in the studies of human p53 expression. Eur J Biochem. 1986;159:529–534. doi: 10.1111/j.1432-1033.1986.tb09919.x. [DOI] [PubMed] [Google Scholar]
- 2.Beckman G, Birgander R, Sjalander A, Saha N, Holmberg P, Kiveld S, Beckman L. Is p53 polymorphism maintained by natural selection. Hum Hered. 1994;44:266–270. doi: 10.1159/000154228. [DOI] [PubMed] [Google Scholar]
- 3.Caelles C, Helmberg A, Karin M. p53-dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature. 1994;370:220–223. doi: 10.1038/370220a0. [DOI] [PubMed] [Google Scholar]
- 4.Cho Y, Gorina S, Jeffrey P, Pavletich N. Crystal structure of a p53 tumour suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 5.Donehower L, Harvey B, Slagle B, McArthur B, Montgomery C, Butel J, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 6.El-Deiry W, Tokino T, Velculescu V, Levy D, Parsons R, Trent J, Lin D, Mercer W, Kinzer K, Vogelstein B. WAF1, a potential mediator of p53 tumour suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 7.Funk W D, Pak T, Karas R, Wright W, Shay J. A transcriptionally active DNA binding site for human p53 protein complexes. Mol Cell Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu Z, Pim D, Labrecque S, Banks L, Matlashewski G. DNA damage induced p53 mediated transcription is inhibited by human papillomavirus type 18 E6. Oncogene. 1994;9:629–633. [PubMed] [Google Scholar]
- 9.Hainaut P, Milner J. Redox modulation of p53 conformation and sequence specific DNA binding in vitro. Cancer Res. 1993;53:4469–4473. [PubMed] [Google Scholar]
- 10.Haupt Y, Rowan S, Shaulian E, Vousden K, Oren M. Induction of apoptosis in Hela cells by transactivation-deficient p53. Gene Dev. 1995;9:2170–2183. doi: 10.1101/gad.9.17.2170. [DOI] [PubMed] [Google Scholar]
- 11.Hollstein M, Rice K, Greenblatt M, Soussi T, Fuchs R, Sorlie T, Hovig E, Smith-Sorensen B, Montesano R, Harris C. Database of p53 gene somatic mutations in human tumours and cell lines. Nucleic Acids Res. 1994;22:3551–3555. [PMC free article] [PubMed] [Google Scholar]
- 12.Kern S E, Pietenpol J, Thiagalingam S, Seymour A, Kinzler K, Vogelstein B. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science. 1992;256:827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- 13.Levine A. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 14.Mack D, Vartikar J, Pipas J, Laimins L. Specific repression of TATA-mediated but not initiator-mediated transcription by wild-type p53. Nature. 1993;363:281–283. doi: 10.1038/363281a0. [DOI] [PubMed] [Google Scholar]
- 15.Malkin D, Li F, Strong L, Fraumeni J, Nelson C, Kim D, Kassel J, Gryka M, Bischoff M, Tainsky M, Friend S. Germline p53 mutations in a familar syndrome of breast cancer, sarcomas and other neoplasms. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 16.Matlashewski G, Tuck S, Pim D, Lamb P, Schneider J, Crawford L. Primary structure polymorphism at amino acid residue 72 of human p53. Mol Cell Biol. 1987;7:961–963. doi: 10.1128/mcb.7.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyashita T, Reed J. Tumour suppressor p53 is a direct transcriptional activator of the human Bax gene. Cell. 1995;80:293–301. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 18.Moreau F, Matlashewski G. Molecular analysis of different allelic variants of wildtype human p53. Biochem Cell Biol. 1992;70:1014–1019. doi: 10.1139/o92-145. [DOI] [PubMed] [Google Scholar]
- 19.Owen-Schub L, Zhang W W, Cusack J, Angelo L, Santee S, Fujiwara T, Roth J, Deisseroth A, Zhang W, Kruzel E, Radinsky R. Wild-type human p53 and a temperature-sensitive mutant induce Fas/APO-1 expression. Mol Cell Biol. 1995;15:3032–3040. doi: 10.1128/mcb.15.6.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pietenpol J, Tokino T, Thiagalingam S, El-Deiry W, Kinzer K, Vogelstein B. Sequence-specific transcriptional activation is essential for growth suppression by p53. Proc Natl Acad Sci USA. 1994;91:1998–2002. doi: 10.1073/pnas.91.6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakamuro D, Sabbatini P, White E, Prendergast G. The polyproline region of p53 is required to activate apoptosis but not growth arrest. Oncogene. 1997;15:887–898. doi: 10.1038/sj.onc.1201263. [DOI] [PubMed] [Google Scholar]
- 22.Srivastava S, Zou Z, Pirolla K, Blattner W, Chang E. Germline mutations transmission of a mutated p53 gene in a family with Li-Fraumeni syndrome. Nature. 1990;348:747–749. doi: 10.1038/348747a0. [DOI] [PubMed] [Google Scholar]
- 23.Storey A, Thomas M, Kalita A, Harwood C, Gardiol D, Mantovani F, Breuer J, Leigh I, Matlashewski G, Banks L. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature. 1998;393:229–234. doi: 10.1038/30400. [DOI] [PubMed] [Google Scholar]
- 24.Subler M, Martin D, Deb S. Inhibition of viral and cellular promoters by human wild-type p53. J Virol. 1992;66:4757–4762. doi: 10.1128/jvi.66.8.4757-4762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas M, Matlashewski G, Pim D, Banks L. Induction of apoptosis by p53 is independent of its oligomeric state and can be abolished by HPV-18 E6 through ubiquitin mediated degradation. Oncogene. 1996;13:265–274. [PubMed] [Google Scholar]
- 26.Thut C, Chen J, Klemm R, Tjian R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 27.Tommasino M, Crawford L. Human papillomavirus E6 and E7: proteins which deregulate the cell cycle. Bioessays. 1995;17:509–518. doi: 10.1002/bies.950170607. [DOI] [PubMed] [Google Scholar]
- 28.Truant R, Xiao C, Ingles C, Breenblatt J. Direct interaction between the transcriptional activator domain of human p53 and the TATA box-binding protein. J Biol Chem. 1993;268:2284–2287. [PubMed] [Google Scholar]
- 29.Walker K, Levine A. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc Natl Acad Sci USA. 1996;93:15335–15340. doi: 10.1073/pnas.93.26.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Miur M, Bergeron L, Zhu H, Yuan J. Ich 1, an Ice/ced-3-related gene, encodes both positive and negative regulators of programmed cell death. Cell. 1994;78:739–750. doi: 10.1016/s0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- 31.Zhang W, Charest H, Matlashewski G. The expression of biologically active human p53 in leishmania cells: a novel eukaryotic system to produce recombinant proteins. Nucleic Acids Res. 1995;23:4073–4080. doi: 10.1093/nar/23.20.4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.zur Hausen H. Human papillomaviruses in the pathogenesis of human anogenital cancer. Virology. 1991;184:9–13. doi: 10.1016/0042-6822(91)90816-t. [DOI] [PubMed] [Google Scholar]