Abstract
Ras-activated signal transduction pathways are implicated in the control of cell proliferation, differentiation, apoptosis, and tumorigenesis, but the molecular mechanisms mediating these diverse functions have yet to be fully elucidated. Conditionally active forms of Raf, v-Src, and MEK1 were used to identify changes in gene expression that participate in oncogenic transformation, as well as in normal growth control. Activation of Raf, v-Src, and MEK1 led to induced expression of c-Myc and cyclin D1. Induction of c-Myc mRNA by Raf was an immediate-early response, whereas the induction of cyclin D1 mRNA was delayed and inhibited by cycloheximide. Raf activation also resulted in the induction of an established c-Myc target gene, ornithine decarboxylase (ODC). ODC induction by Raf was mediated, in part, by tandem E-boxes contained in the first intron of the gene. Activation of the human colony-stimulating factor 1 (CSF-1) receptor in NIH 3T3 cells leads to activation of the mitogen-activated protein (MAP) kinase pathway and induced expression of c-Fos, c-Myc, and cyclin D1, leading to a potent mitogenic response. By contrast, a mutated form of this receptor fails to activate the MAP kinases or induce c-Myc and cyclin D1 expression and fails to elicit a mitogenic response. The biological significance of c-Myc and cyclin D1 induction by Raf and v-Src was confirmed by the demonstration that both of these protein kinases complemented the signaling and mitogenic defects of cells expressing this mutated form of the human CSF-1 receptor. Furthermore, the induction of c-Myc and cyclin D1 by oncogenes and growth factors was inhibited by PD098059, a specific MAP kinase kinase (MEK) inhibitor. These data suggest that the Raf/MEK/MAP kinase pathway plays an important role in the regulation of c-Myc and cyclin D1 expression in NIH 3T3 cells. The ability of oncogenes such as Raf and v-Src to regulate the expression of these proteins reveals new lines of communication between cytosolic signal transducers and the cell cycle machinery.
The Ras-activated mitogen-activated protein (MAP) kinase signaling pathway participates in the control of a variety of biological processes, including cell proliferation, migration, differentiation, and apoptosis. The importance of this pathway in metazoan development is emphasized by the fact that loss-of-function mutations of components of this pathway have severe consequences on development. Moreover, constitutively activated forms of Ras proteins are found in approximately 35% of all human cancers (13, 36, 49, 60, 63, 132).
Cycling of the Ras family of membrane-bound GTPases between GTP-bound (active) and GDP-bound (inactive) states is regulated by guanine nucleotide exchange factors and GTPase-activating proteins (25). GTP-bound Ras recruits a variety of proteins to the plasma membrane, including Ral-GDS, Rin1, p120RasGAP, p110 PI3′ kinase, and Raf (44, 46), many of which serve to propagate the signal from activated Ras throughout the cell. Among the best-characterized effectors of Ras are the Raf family of serine-specific protein kinases, which play an important role in cell proliferation, differentiation, and survival (19, 23, 62, 78, 112, 120, 122–124).
The three members of the Raf family of protein kinases, A-Raf, B-Raf, and Raf-1, share similar structural features and are activated as a consequence of the engagement of cell surface receptors by their cognate ligands (114, 125). Although the precise mechanism of Raf-1 activation is not fully elucidated, the favored model suggests that Raf-1 is recruited to the plasma membrane by direct binding to activated Ras. Membrane associated Raf-1 is then believed to be phosphorylated and thereby activated by members of the Src family of protein tyrosine kinases which have pleiotropic effects on the control of cell morphology, migration, and the cell cycle (29, 61, 65, 66, 74, 82, 93, 113, 117, 121, 126). However, there are indications that additional mechanisms for Raf-1 regulation exist (74, 130, 135). Activated Raf-1, in turn, phosphorylates to activate MAP kinase kinase (MEK), which in turn phosphorylates to activate p42 and p44 MAP kinase, also known as ERK2 and ERK1, respectively (22, 32, 53, 92). Activated MAP kinases elicit pleiotropic effects on cell function by phosphorylating a diverse array of cellular proteins, including a number of transcription factors that influence patterns of cellular gene expression (73, 77, 118).
Activated forms of Ras, Raf, and MEK can induce DNA synthesis and cellular transformation in vitro, and dominant-interfering forms of Raf and MEK can block Ras-induced mitogenesis and transformation (19, 24, 40, 50, 64, 103). In NIH 3T3 cells Raf is competent to induce DNA synthesis at low levels of activation, whereas at higher levels of activation it induces cell cycle arrest (128). To understand the molecular basis of Raf-induced cell proliferation and oncogenic transformation, we sought to identify changes in gene expression that occur after Raf activation by using conditionally active forms of these protein kinases (ΔRaf:ER) (87, 100). ΔRaf:ER proteins are rapidly activated by the addition of estrogen or its analogues to the cell culture medium, resulting in alterations in gene expression that lead to oncogenic transformation (87, 100, 128).
Considerable evidence has implicated c-Myc and cyclin D1 as important regulators of the mammalian cell cycle (5, 42, 54, 67, 136). Activation of the transcription factor c-Myc in quiescent fibroblasts induces cell cycle reentry, whereas inhibition of c-Myc function results in cell cycle arrest (8, 9, 16, 57, 127). The D-type cyclins are transcriptionally induced in the mid-G1 phase of the cell cycle and partner with cyclin-dependent kinases (CDK4 and CDK6) to elicit phosphorylation of the retinoblastoma (Rb) protein. Rb phosphorylation leads to the release of the transcription factor E2F that influences the expression of genes required for the progression of cells into S phase (107). Both c-Myc and cyclin D1 have oncogenic potential and are overexpressed in a variety of human cancers (30, 35, 43, 76, 86, 104, 111).
In this study we demonstrate the induction of the genes encoding c-Myc and cyclin D1 by the Raf/MEK/MAP kinase pathway in NIH 3T3 cells. The functional significance of the induction of these genes is substantiated by the apparent ability of Raf to complement a defective form of the human CSF-1 receptor, which fails to induce DNA synthesis in NIH 3T3 cells as a consequence of its inability to induce the expression of c-Myc and cyclin D1. Furthermore, the ability of a variety of growth factors and oncogenes to induce c-Myc and cyclin D1 is abrogated by a specific and selective inhibitor of MEK activation. Finally, since Src-family protein tyrosine kinases have been implicated in the regulation of c-Myc, we describe the ability of a conditionally active form of v-Src that activates the MAP kinase pathway leading to induced expression of c-Myc and cyclin D1, to complement the mitogenic defect of the mutated colony-stimulating factor 1 (CSF-1) receptor. These data suggest that, at least in NIH 3T3 cells, the Ras-activated MAP kinase pathway plays an important role in controlling the expression of these crucial cell cycle regulators.
MATERIALS AND METHODS
Cell culture.
NIH 3T3 cells expressing ΔRaf-1:ER, ΔB-Raf:ER, and ΔB-Raf:ER* have been described previously (87, 100, 128). Clonal populations of NIH 3T3 cells expressing either the human CSF-1 receptor or a mutated version of this receptor containing a tyrosine-to-phenylalanine mutation at amino acid 809 (CSF-1R[809F]) were a gift of Martine Roussel (97–99). NIH 3T3 cells expressing Raf-1[DD]:ER and v-Src:ER were generated by retroviral infection of the appropriate target cells as described previously (14, 100). All cells were grown in Dulbecco’s modified Eagle medium (DMEM) containing 10% (vol/vol) fetal calf serum (FCS) and rendered quiescent in either 0.5% (vol/vol) FCS, deluxe serum-free medium (DSFM), or DSFM lacking insulin, selenium, and transferrin (modified DSFM) (128). Cells expressing v-Src:ER were cultured in DMEM containing 10% (vol/vol) charcoal-stripped FCS (Gemini Bioproducts). Recombinant human CSF-1 was a gift of Steve Clark (Genetics Institute). Raf:ER proteins were activated with 4-hydroxytamoxifen (4-HT; Research Biochemicals International) or ICI 182,780 (gift of Alan Wakeling, Zeneca Pharmaceuticals). Ethanol was used as a solvent control in all experiments.
Expression of Raf-1[DD]:ER, v-Src:ER, and ΔMEK1:ER.
To construct conditionally active Raf-1[DD]:ER, DNA sequences encoding full-length human Raf-1 containing two activating point mutations (Y340D and Y341D; a gift from Debbie Morrison) were fused to the hormone binding domain of the HE14 allele of the human estrogen receptor (hbER) and cloned into the pBabepuro3 retrovirus expression vector (14, 27, 72). To generate v-Src:ER, DNA sequences encoding the Schmidt Ruppin A form of v-Src (a gift from Josh Kaplan) were fused to hbER and cloned into the pWZLblast3, pBabepuro3, or pLNCX retroviral expression vectors (45, 100). In the case of pWZLblast3 (a gift from Jay Morgenstern), expression of a bicistronic mRNA from the murine leukemia virus long terminal repeat containing the encephalomyocarditis virus internal ribosome entry site permits the expression of both v-Src:ER and the blasticidin drug resistance gene (see Fig. 6A). To generate ΔMEK1:ER, DNA sequences encoding a constitutively active form of MEK1 containing an amino terminal deletion (ΔN3) and two activating mutations (S218E and S222D) (a gift from Natalie Ahn) (64) were fused to hbER and cloned into the pBabepuro3 retroviral expression vector. Raf-1[DD], v-Src, and ΔMEK1 DNA sequences were all amplified by PCR to introduce the appropriate restriction enzyme sites for cloning (details available upon request).
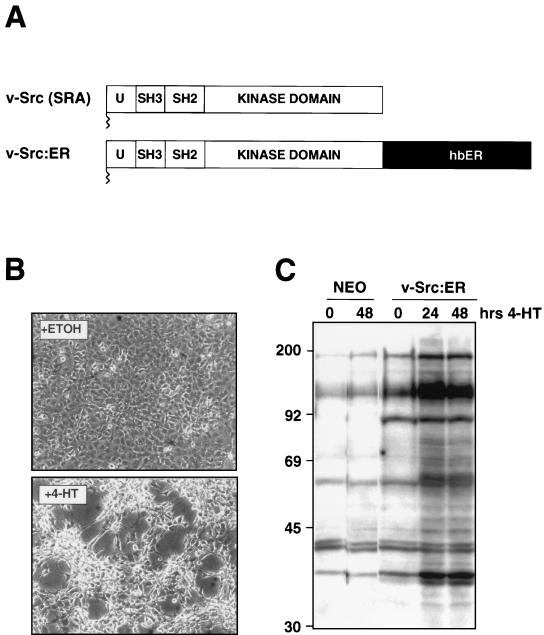
FIG. 6.
Construction of v-Src:ER. (A) Construction of v-Src:ER. DNA sequences encoding the SRA form of v-Src were fused in frame to the hormone-binding domain of the human estrogen receptor and cloned into a series of replication-defective retrovirus vectors. (B) Morphological transformation of NIH 3T3 cells after activation of v-Src:ER. NIH 3T3 cells expressing v-Src:ER were cultured in DSFM for 16 h and treated with either ethanol (solvent control) or 1 μM 4-HT (to activate v-Src:ER) for 48 h, as indicated, at which time photomicrographs were taken. (C) Tyrosine phosphorylation of cellular proteins after activation of v-Src:ER. Control NIH 3T3 cells (control cells, two left lanes) or NIH 3T3 cells expressing v-Src:ER (three right lanes) were cultured in 10% FCS and treated with 1 μM 4-HT for various lengths of time as indicated. Tyrosine phosphorylation of cellular proteins was assessed by Western blotting with the 4G10 antiphosphotyrosine monoclonal antibody.
Retrovirus stocks were prepared by the transfection of retrovirus vectors into Bosc23 cells as described previously (83, 128). Cells were infected with retrovirus stocks and cultured in neomycin-, puromycin-, or blasticidin-containing media as appropriate. After selection for drug resistance, cells were pooled and tested for expression of Raf-1[DD]:ER, v-Src:ER, and ΔMEK-1:ER by Western blotting. A clonal population of ΔMEK1:ER expressing cells was isolated by ring cloning.
Western blotting and antibody detection.
Cell extracts were prepared, quantitated, and blotted as described previously, except that 280 mM β-mercaptoethanol was used as a reducing agent and lysates were heated to 70°C prior to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Antisera used in this study were raised against c-Myc and phosphotyrosine (UBI, Inc.); p42 MAP kinase, p44 MAP kinase, and the human estrogen receptor (Santa Cruz Biotechnology); and phospho-ERK1/ERK2 (New England Biolabs). Anti-cyclin D1 antiserum was a kind gift from David Parry. Antigen-antibody complexes were visualized by using the enhanced chemiluminescence (ECL) detection system (Amersham).
Immune-complex kinase assays.
MAP kinase activity was measured by an in vitro immunoprecipitation-kinase assay by using myelin basic protein (MBP) as a substrate as described previously (100).
RNase protections.
RNase protections were conducted as described previously (68). A mouse c-Myc exon 1 riboprobe was prepared with a cDNA linearized with BamHI (a gift from Nissim Hay). This riboprobe protected a P1-initiated c-Myc transcript of approximately 400 bp and a P2-initiated transcript of approximately 350 bp. A mouse ornithine decarboxylase (ODC) cDNA (nucleotides 729 to 2357; a gift from Philip Coffino) was linearized with Eco47III, and the resulting riboprobe protected a fragment of approximately 517 bp. A full-length mouse cyclin D1 cDNA (a gift from Emma Lees) was linearized with SalI, and the resulting riboprobe protected a fragment of approximately 250 bp (nucleotides 826 to 1075). A riboprobe prepared from a plasmid containing a 120-bp fragment of human GAPDH was included in all of the hybridizations and used as a loading control. Ten micrograms of total RNA (RNeasy kit; Qiagen) or tRNA (as a negative control) were hybridized overnight at 45°C with both gene-specific and GAPDH riboprobes, treated with RNase A and RNase T1 to digest the single-stranded RNA, extracted with phenol, and ethanol precipitated, with the resulting protected fragments resolved by electrophoresis on 6% (wt/vol) acrylamide sequencing gels. Results were quantitated with a Molecular Dynamics Storm PhosphorImager.
Transient transfections.
Reporter constructs, comprising the c-Myc, ODC, or cyclin D1 promoters linked to luciferase, were transiently transfected into confluent monolayers of NIH 3T3 cells cultured in 60-mm tissue culture dishes by using either DNA-Lipofectamine complexes (Gibco-BRL) or the DEAE-dextran method (69). The mouse c-Myc promoter-luciferase construct (pGL2-myc, provided by Mike Ostrowski) was comprised of a 500-bp PvuII-SstI fragment that included approximately 140 bp upstream and 360 bp downstream of the mouse c-Myc P2 promoter. The ODC promoter-luciferase constructs (ODCΔLuc or ODCΔLucS-5A) were provided by John Cleveland and were described previously (15). The mouse cyclin D1 promoter-luciferase construct [mD1(-984)luc; provided by Gordon Peters] was comprised of 984 bp of DNA located upstream of the transcription initiation site of the mouse cyclin D1 gene. In all experiments the luciferase reporter constructs were cotransfected with pSRα–β-Gal (provided by Naoko Arai), which contains a constitutively active promoter linked to sequences encoding β-galactosidase as a control for transfection efficiency (69). After transfection the cells were split into the wells of a 24-well dish and allowed to adhere in DMEM containing 10% (vol/vol) FCS for 3 h. The medium was then replaced with DMEM containing 0.5% (vol/vol) FCS plus ethanol (solvent control), 4-HT, or ICI 182,750. Cells were harvested 36 to 48 h later in Reporter lysis buffer (Promega); luciferase activity was measured by using the luciferase assay system (Promega), and β-galactosidase activity was measured by using the Galacto-Light kit (Tropix) and quantitated with an AutoLumat LB953 luminometer (EG & G Berthold). To control for transfection efficiency, all results are expressed as the ratio of luciferase to β-galactosidase activity. In bar graphs, each bar represents the average of four samples (c-Myc experiments), three samples (cyclin D1 experiments), or six samples (ODC experiments), and error bars indicate standard deviations.
DNA synthesis assays.
Cells were grown to confluence in 96-well microtiter dishes, held at confluence for 3 days, cultured for an additional 24 h in DSFM, and then treated with either 10% (vol/vol) FCS, 300 nM CSF-1, 4-HT, or 300 nM CSF-1 plus 4-HT. Then 1 μCi of [methyl-3H]thymidine was added to each well simultaneously with the addition of growth stimuli, and the cells were incubated for a further 36 h prior to harvest with a Skatron Cell Harvester as described previously (128). Each measurement was conducted in quadruplicate, and error bars indicate standard deviations. The absence of a visible error bar indicates that the standard deviation was too small to register on the graph at the scale used.
RESULTS
Transcriptional regulation of c-Myc and cyclin D1 expression by Raf.
To determine the effects of MAP kinase activation on the expression of components of the cell cycle machinery, we explored the consequences of Raf activation on the expression of c-Myc and cyclin D1. RNA and protein samples were prepared from a pooled population of NIH 3T3 cells expressing a conditionally active form of oncogenic B-Raf (ΔB-Raf:ER*) (87) that was rendered quiescent and then treated with ICI 182,780 (ICI) to activate ΔB-Raf:ER* for different lengths of time as indicated (Fig. 1A). RNase protection analysis indicated that c-Myc mRNA was induced 4- to 15-fold after ΔB-Raf:ER* activation, with kinetics and magnitude of induction similar to those observed in response to serum stimulation (Fig. 1A and B) (37). In asynchronously growing NIH 3T3 cells, the basal level of c-Myc expression was significantly higher, such that c-Myc mRNA was induced only two- to threefold after Raf activation (data not shown). Treatment of parental NIH 3T3 cells or cells expressing a kinase-inactive form of ΔRaf-1:ER (ΔRaf301:ER) with ICI or 4-HT had no effect on the levels of c-Myc mRNA, indicating that the induction of c-Myc requires Raf kinase activity (data not shown).
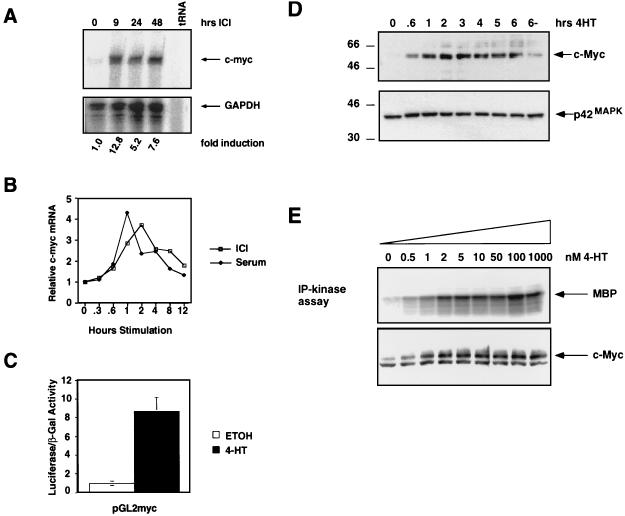
FIG. 1.
Induction of c-Myc by Raf in NIH 3T3 cells. (A) c-Myc mRNA regulation. NIH 3T3 cells expressing ΔB-Raf:ER* were cultured in 0.5% serum for 16 h, and RNA samples were prepared at different times after the addition of 1 μM ICI to activate ΔB-Raf:ER*. The expression of c-Myc (upper panel) and GAPDH (lower panel) mRNAs was detected by simultaneous RNase protection assay. The fold induction of c-Myc mRNA was quantitated by obtaining the ratio of c-Myc to GAPDH mRNAs by PhosphorImager analysis. (B) Comparison of c-Myc mRNA induction by serum versus ΔB-Raf:ER* activation. NIH 3T3 cells expressing ΔB-Raf:ER* were cultured in 0.5% serum for 16 h, and RNA samples were prepared at different times after stimulation with 10% FCS (closed diamonds) or the activation of ΔB-Raf:ER* with 1 μM ICI (open squares). c-Myc and GAPDH mRNAs were detected by RNase protection, and data were quantified by PhosphorImager analysis. Results are presented as the fold induction over the baseline level of expression. (C) Activation of the c-Myc promoter. NIH 3T3 cells expressing ΔB-Raf:ER* were transiently transfected with reporter constructs consisting of the promoter region of human c-Myc linked to luciferase (pGL2-myc) and pSRαβ-Gal as a control for transfection efficiency. Transfected cells were treated with either ethanol (solvent control, open bar) or 500 nM 4-HT (shaded bar) in 0.5% serum for 36 h, at which time the luciferase and β-galactosidase activities were measured. Results are presented as the ratios of the luciferase to β-galactosidase activities. (D) Induction of c-Myc protein expression. NIH 3T3 cells expressing ΔB-Raf:ER* were cultured in 0.5% FCS for 16 h, and cell extracts were prepared at different times after the addition of 500 nM 4-HT. The expression of p62c-Myc was detected by Western blotting with a specific antiserum (upper panel). The same Western blot was reprobed for the expression of p42 MAP kinase as a control for equal loading in each lane (lower panel). (E) Effect of increasing ΔB-Raf:ER* activity on c-Myc expression. NIH 3T3 cells expressing ΔB-Raf:ER* were cultured in modified DSFM for 24 h and stimulated with different concentrations of 4-HT, and cell extracts were prepared 4.5 h later. MAP kinase activity was measured by performing p42MAPK immune complex kinase assays with MBP as a substrate (upper panel). c-Myc expression was analyzed by Western blotting.
In order to determine if the induction of c-Myc mRNA by ΔB-Raf:ER* was mediated by an increase in c-Myc transcription, NIH 3T3 cells were transiently transfected with a reporter construct (pGL2-myc) consisting of a portion of the c-Myc promoter fused to luciferase. Activation of ΔB-Raf:ER* led to a ninefold induction of luciferase activity (Fig. 1C), indicating that Raf activation leads to transactivation of the c-Myc promoter. The induction of c-Myc mRNA in response to serum stimulation is immediate early in that it is resistant to the effects of cycloheximide, an inhibitor of protein synthesis (47). Similarly, activation of ΔB-Raf:ER* or treatment of cells with cycloheximide alone resulted in an increase in c-Myc mRNA levels (Fig. 2A, top panel). Activation of ΔB-Raf:ER* in the presence of cycloheximide led to superinduction of c-Myc mRNA, indicating that c-Myc is an immediate-early transcriptional target of Raf activation.
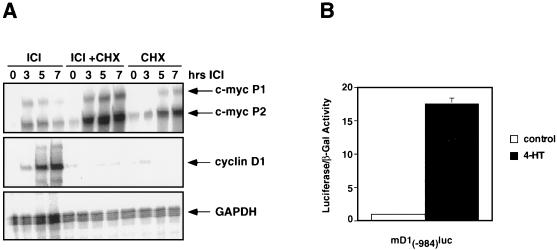
FIG. 2.
Regulation of cyclin D1 and c-Myc expression by Raf. (A) Cycloheximide sensitivity of Raf-induced cyclin D1 and c-Myc mRNA expression. NIH 3T3 cells expressing ΔB-Raf:ER* were cultured in 0.5% FCS for 24 h and were then either left untreated or treated with 25 μg of cycloheximide per ml for 1 h. These cells were then either left untreated (CHX) or treated with 1 μM ICI to activate ΔB-Raf:ER* for different lengths of time as indicated. The expression of cyclin D1, c-Myc, and GAPDH mRNAs were quantitated by RNase protection assays. (B) Cyclin D1 promoter activation. NIH 3T3 cells expressing ΔB-Raf:ER* were transiently transfected with reporter constructs consisting of the promoter region of mouse cyclin D1 linked to luciferase [mD1(-984)Luc] and pSRαβ-Gal as a control for transfection efficiency. Transfected cells were treated with either ethanol (solvent control, open bar) or 1 μM 4-HT (closed bar) for 41 h, at which time luciferase and β-galactosidase activities were measured. Results are presented as the ratios of the luciferase to the β-galactosidase activities.
Confirmation of the c-Myc mRNA analysis was sought by examining cell extracts for the expression of p62c-Myc by Western blotting. Consistent with the mRNA analysis, activation of ΔB-Raf:ER* resulted in a rapid induction of p62c-Myc that was readily detected after 40 min and was sustained at elevated levels for at least 6 h (Fig. 1D).
We have previously shown that the activity of ΔRaf:ER proteins can be titrated by varying the concentration of 4-HT added to the cell culture medium. Low levels of Raf activity (up to 2 nM 4-HT in ΔB-Raf:ER*-expressing cells) promote cell proliferation, and high levels of Raf activity (≥5 nM 4-HT) promote a G1 arrest that is mediated by p21Cip1 (128). To investigate the expression of c-Myc in response to different levels of Raf activation, we treated quiescent ΔB-Raf:ER*-expressing cells with different concentrations of 4-HT for 4.5 h and examined MAP kinase activation and c-Myc expression (Fig. 1E). Both p62c-Myc expression and MAP kinase activity were maximal at concentrations of 4-HT greater than 2 nM, with higher concentrations of 4-HT having no additional effect on MAP kinase activity or the level of p62c-Myc expression.
Cells transformed by activated Ras express elevated levels of cyclin D1, and previously we have demonstrated that activation of ΔRaf-1:ER leads to induced cyclin D1 expression (58, 128). We therefore investigated the induction of cyclin D1 mRNA by Raf. Activation of ΔB-Raf:ER* led to a sustained induction of cyclin D1 mRNA, which was maximal between 5 and 7 h after ΔB-Raf:ER* activation (Fig. 2A, middle panel). Such induction kinetics are consistent with the regulation of cyclin D1 expression after either serum stimulation or ΔB-Raf:ER* activation (128). In contrast to the induction of c-Myc mRNA by Raf, the induction of cyclin D1 mRNA was entirely abrogated by pretreatment of the cells with cycloheximide. These data are consistent with the CSF-1-mediated induction of cyclin D1 mRNA which occurs in mid-G1 and is cycloheximide sensitive (67). Given the differences in the kinetics and cycloheximide sensitivities of the c-Myc and cyclin D1 genes, it seems likely that Raf regulates these genes by different mechanisms.
In order to determine whether the induction of cyclin D1 by Raf was mediated by transcriptional activation of the cyclin D1 promoter, ΔB-Raf:ER*-expressing cells were transiently transfected with a reporter construct consisting of 984 bp of the mouse cyclin D1 promoter linked to luciferase. Activation of ΔB-Raf:ER* led to a 17-fold induction of cyclin D1 promoter activity (Fig. 2B), a finding in agreement with previous work indicating that in certain circumstances the MAP kinase pathway plays an important role in the regulation of cyclin D1 expression (56). Collectively, these data suggest that the cyclin D1 gene is a delayed-early transcriptional target of Raf activation in NIH 3T3 cells.
Induction of ODC gene expression by Raf.
The c-Myc transcription factor has been implicated in the regulation of cell proliferation, differentiation, and apoptosis (5). c-Myc acts in concert with a heterodimerization partner Max to transactivate transcription through cis-acting sequences known as E-boxes (CACGTG) (11, 12, 51, 84). A number of cellular genes have been identified as potential targets for transactivation by c-Myc, including ODC, an enzyme that catalyzes the rate-limiting step in polyamine biosynthesis which is essential for cell proliferation (7, 33, 38). If the induction of c-Myc by Raf is of functional consequence, it would be predicted that a c-Myc target gene such as ODC would be induced by Raf in an E-box-dependent manner. We therefore sought to determine whether Raf activation was sufficient to induce the expression of ODC. Activation of either ΔRaf-1:ER or ΔB-Raf:ER* in NIH 3T3 cells led to a 17- to 36-fold induction of ODC mRNA expression (Fig. 3A and data not shown). Compared to HB-EGF, which is induced with immediate-early kinetics, the induction of ODC mRNA was delayed, reaching maximum levels 4 to 12 h after Raf activation (Fig. 3A) (70). These data are consistent with a temporal requirement for intermediate events between Raf activation and ODC mRNA induction.
FIG. 3.
Induced expression of ODC after Raf activation. (A) Induction of ODC mRNA by Raf. NIH 3T3 cells expressing ΔRaf-1:ER were cultured in DMEM containing 0.5% serum for 16 h, and RNA samples were prepared at different times after the addition of 1 μM ICI. Expression of the mRNAs encoding ODC (upper panel), heparin-binding epidermal growth factor (HB-EGF, middle panel), and GAPDH (lower panel) was quantitated by using RNase protection assays. (B) ODC promoter activation. NIH 3T3 cells expressing ΔB-Raf:ER* were transiently transfected with reporter constructs consisting of the promoter region of human ODC linked to luciferase (ODCΔLuc) or a form of this promoter containing a point mutation in each of the two E-boxes located in the first intron of the gene (ODCΔLucS-5A). pSRαβ-Gal was cotransfected with the ODC reporter plasmids as a control for transfection efficiency. Transfected cells were treated with either ethanol (solvent control, open bars) or 500 nM 4-HT (shaded bars) for 40 h, at which time the luciferase and β-galactosidase activities were measured. Results are presented as the ratios of the luciferase to the β-galactosidase activities.
The transcriptional activation of ODC by c-Myc is mediated by tandem E-boxes located in the first intron of the ODC gene (7). To determine if the induction of ODC transcription by Raf requires these E-boxes, we utilized a pair of reporter constructs comprising either the wild-type ODC promoter (ODCΔLuc) or a reporter in which the E-boxes were mutated (CACGTG→CACCTG), rendering the reporter unresponsive to c-Myc (ODCΔLucS-5A) (80). Activation of ΔB-Raf:ER* led to a 3.7-fold induction of ODCΔLuc activity, whereas ODCΔLucS-5A was activated 2.3-fold (Fig. 3B). This 37% decrease in the ability of Raf to activate the ODCΔLucS-5A reporter, although modest, was highly reproducible in five separate experiments. Moreover, the residual transactivation of the ODCΔLucS-5A construct by Raf is consistent with the prevailing model of ODC transcriptional regulation that suggests that c-Myc works in concert with other regulated transcription factors to control ODC expression (1, 52, 75, 80, 81, 129). These data suggest that c-Myc binding sites are required for the full induction of ODC gene transcription in response to Raf activation.
Further support for a role for the Ras/Raf pathway in the regulation of c-Myc and ODC expression came from separate experiments with human cell lines. HCT-116 is a tumorigenic colon carcinoma cell line expressing a mutationally activated form of Ki-Ras (G13D) and elevated levels of c-Myc. Hke-3 cells are a nontumorigenic derivative of HCT-116 derived by homologous-recombination-mediated ablation of the activated Ki-Ras gene that expresses decreased levels of c-Myc (108). In these cells the wild-type ODCΔLuc reporter was transactivated 90-fold more efficiently in HCT-116 cells than in HKe-3 cells (data not shown). This is consistent with the work of others showing increased ODC gene expression in Ha-Ras-transformed fibroblasts treated with basic fibroblast growth factor (39, 41, 71) and suggest that Ras-activated signaling pathways may play a role in the regulation of c-Myc and ODC in the context of human cancer cells.
Complementation of defective CSF-1 receptor signaling by Raf.
To further address the significance of c-Myc and cyclin D1 induction by Raf, we made use of two clonal NIH 3T3 cell lines expressing either the human CSF-1 receptor (CSF-1R) or a mutated derivative containing a tyrosine-to-phenylalanine mutation at a site of ligand-dependent phosphorylation (CSF-1R[809F]) (94, 97, 98). Addition of CSF-1 to NIH 3T3 cells expressing the human CSF-1 receptor leads to the induction of c-Fos, c-Jun, c-Myc, and cyclin D1 and elicits a robust proliferative response. Although the binding of CSF-1 to the mutated CSF-1 receptor elicits tyrosine phosphorylation of the receptor and induced expression of c-Jun and c-Fos, it fails to fully induce cyclin D1 and c-Myc mRNAs, with the consequence that the cells fail to proliferate in response to CSF-1 (98). The mitogenic defect of the CSF-1R[809F] is complemented by the ectopic overexpression of either cyclin D1 or c-Myc (95, 99). Since Raf activation resulted in the induction of both c-Myc and cyclin D1, we wished to examine whether the mutated CSF-1 receptor was able to activate the Raf/MEK/MAP kinase pathway and whether Raf could functionally complement the biochemical and biological defects in the CSF-1R[809F]-expressing cells.
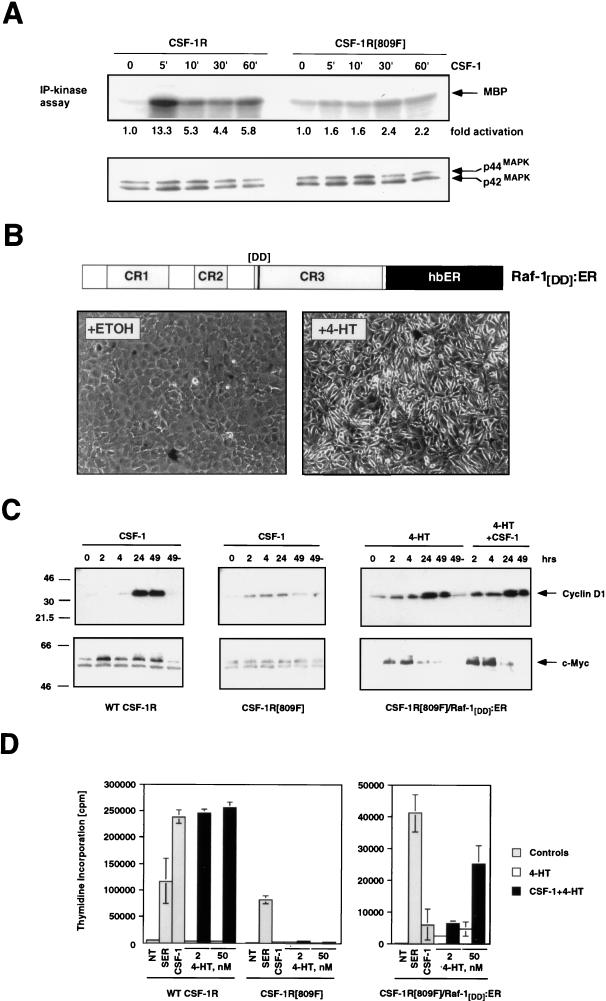
To investigate the ability of the normal and mutated CSF-1 receptors to activate the Raf/MEK/MAP kinase pathway in NIH 3T3 cells, quiescent cells of both types were treated with CSF-1 for different periods of time from 5 to 60 min, and the MAP kinase activity was measured (Fig. 4A). Activation of the wild-type CSF-1 receptor led to activation of the p42 and p44 MAP kinases that peaked after 5 min of CSF-1 addition (∼13-fold induction) and remained elevated (∼5-fold induction) for up to 60 min after CSF-1 addition. By contrast, addition of CSF-1 to cells expressing the mutated CSF-1 receptor resulted in only a twofold activation of the MAP kinases over the same time period. These data indicate that despite the apparent inability of the CSF-1R[809F] to fully activate the MAP kinase pathway, this receptor retains its ability to induce the expression of c-Fos and c-Jun (98). Such data are consistent with a requirement for MAP kinase activation for induced expression of c-Myc and cyclin D1 mRNAs.
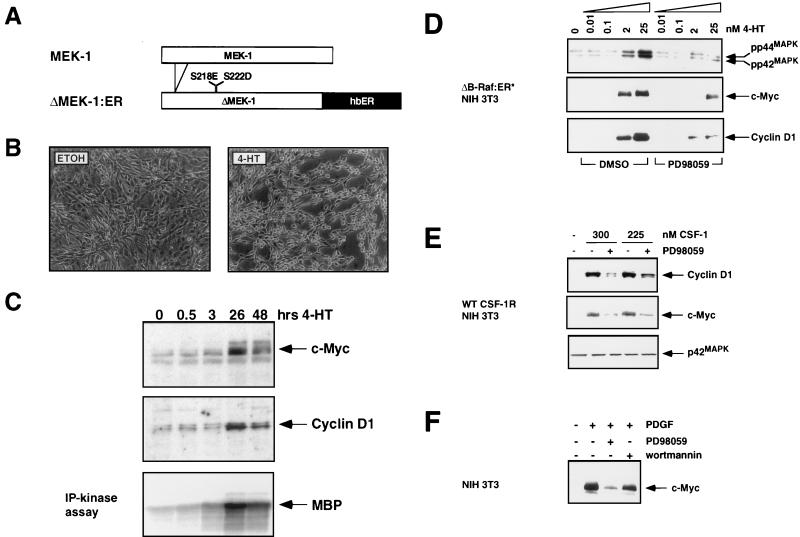
FIG. 4.
Complementation of the mitogenic and signaling defect of the CSF-1R[809F] by Raf. (A) Comparison of CSF-1-dependent MAP kinase activity in CSF-1R and CSF-1R[809F]-expressing cells. NIH 3T3 cells expressing either the wild-type human CSF-1 receptor (WT CSF-1R) or a mutated form of the receptor encoding a single tyrosine-to-phenylalanine mutation CSF-1R[809F] were cultured in DSFM for 24 h, at which time 300 nM CSF-1 was added for different periods of time as indicated. The activities of the p42 and p44 MAP kinases were measured by an immune complex kinase assay with MBP as a substrate, and the fold MAP kinase activation was quantified by PhosphorImager analysis (upper panel). Equal amounts of p42 and p44 MAP kinases in each immunoprecipitation were confirmed by Western blotting with an antiserum that recognizes p42 and p44 MAP kinases (lower panel). (B) Construction of a conditionally active form of full-length Raf-1. DNA sequences encoding a form of full-length human Raf-1 containing two activating point mutations (Y304D and Y341D) were fused in frame to sequences encoding the hormone-binding domain of the human estrogen receptor (hbER) to generate Raf-1[DD]:ER (diagram). NIH 3T3 cells infected with a replication-defective retrovirus encoding Raf-1[DD]:ER were treated with ethanol (solvent control) or 1 μM 4-HT for 48 h as indicated, at which time photomicrographs were taken. (C) Induced expression of c-Myc and cyclin D1. NIH 3T3 cells expressing either the wild-type CSF-1 receptor (left panel) or CSF-1R[809F] (middle panel) were cultured in DSFM for 40 h and treated with 300 nM CSF-1 for different periods of time as indicated. NIH 3T3 cells expressing both the CSF-1R[809F] and Raf-1[DD]:ER (right panel) were cultured in DSFM for 40 h, at which time they were treated with 50 nM 4-HT in the absence or presence of 300 nM CSF-1 for different periods of time as indicated. Expression of c-Myc and cyclin D1 was detected by Western blotting. For cyclin D1 detection, ECL exposures were all for the same length of time; for c-Myc, wild-type CSF-1R, and Raf-1[DD]:ER/CSF-1R[809F], Western blots were exposed for the same length of time but the CSF-1R[809F] Western blot was deliberately overexposed in order to detect the lower level of basal c-Myc expressed in these cells. (D) Proliferation of CSF-1R-expressing cell lines. NIH 3T3 cells expressing the wild-type CSF-1 receptor CSF-1R[809F] (left panel) or both CSF-1R[809F] and Raf-1[DD]:ER (right panel) were cultured in DSFM for 28 h and then either left untreated (NT) or treated with 10% FCS (SER), 300 nM CSF-1 (gray bars), 4-HT (2 or 50 nM, open bars), or CSF-1 plus 4-HT (solid bars). Cell proliferation was determined by measuring the incorporation of [3H]thymidine over a period of 36 h.
To determine whether Raf could functionally complement the signaling and mitogenic defects in the CSF-1R[809F]-expressing cells, we constructed a conditionally active form of full-length human Raf-1. Mutation of two tyrosine residues in full-length Raf-1 to aspartic acid (Y340D and Y341D) gives rise to a constitutively activated form of the protein that is highly transforming in NIH 3T3 cells (27). Sequences encoding this form of Raf-1 (Raf-1[DD]) were fused to the hormone-binding domain of the human estrogen receptor to generate Raf-1[DD]:ER (Fig. 4B) (14). Activation of Raf-1[DD]:ER in NIH 3T3 cells led to rapid activation of the p42 and p44 MAP kinases, induced expression of c-Myc and cyclin D1, and oncogenic transformation (Fig. 4B and data not shown). We chose to use this form of conditionally active Raf-1 because maximal activation of the p42 and p44 MAP kinases was at least threefold lower in response to Raf-1[DD]:ER activation than in the previously characterized ΔRaf-1:ER proteins that carry a deletion of the CR1 and CR2 negative regulatory regions. This allows a larger range of 4-HT concentrations in which the activation of Raf alone is insufficient to induce cell proliferation (128). In addition, since the presence of aspartic acid residues at these positions is thought to mimic phosphorylation, this form of Raf-1 may be a more accurate mimic of the protein that has been activated as a consequence of growth factor receptor engagement. NIH 3T3 cells expressing CSF-1R[809F] were infected with a retrovirus encoding Raf-1[DD]:ER, and pooled populations of cells expressing both proteins, referred to as CSF-1R[809F]/Raf-1[DD]:ER, were established.
To determine if Raf activation was able to rescue the biochemical defects in cells expressing CSF-1R[809F], cells were rendered quiescent and then stimulated with either CSF-1, 10% (vol/vol) FCS, 4-HT, or 4-HT plus CSF-1 for different lengths of time, and the expression of c-Myc and cyclin D1 proteins was assessed by Western blotting. CSF-1 treatment of cells expressing the wild-type CSF-1 receptor led to a strong induction of both cyclin D1 and c-Myc, whereas CSF-1 treatment of CSF-1R[809F]-expressing cells resulted in poor induction of both of these proteins even upon prolonged exposure of the Western blot (Fig. 4C, left and middle panels) (99). Activation of Raf-1[DD]:ER in the CSF-1R[809F]/Raf-1[DD]:ER cells either alone or in combination with CSF-1 treatment led to induction of both cyclin D1 and c-Myc (Fig. 4C, right panels). The combination of 4-HT plus CSF-1 did not appear to potentiate the kinetics of cyclin D1 or c-Myc induction by Raf-1[DD]:ER. These data indicate that activation of Raf-1[DD]:ER complemented the signaling defect of cells expressing the defective CSF-1 receptor, leading to restoration of induced c-Myc and cyclin D1 expression.
To determine if Raf-1[DD]:ER was able to complement the mitogenic defect in CSF-1R[809F]-expressing cells, cells were rendered quiescent and then treated with either CSF-1, 10% (vol/vol) FCS, 4-HT, or CSF-1 plus 4-HT, and reentry into the cell cycle was assessed by evaluating [methyl-3H]thymidine incorporation into DNA. Treatment of CSF-1R-expressing cells with either serum or CSF-1 led to induction of DNA synthesis, whereas 4-HT had no effect (Fig. 4D, left panel). Cells expressing the defective CSF-1 receptor responded to serum stimulation but not to either CSF-1 or 4-HT (Fig. 4D, left panel) (97). Quiescent CSF-1R[809F]/Raf-1[DD]:ER cells displayed a mitogenic response to serum stimulation but not to CSF-1 or activation of Raf-1[DD]:ER alone (Fig. 4D, right panel). However, concomitant activation of Raf-1[DD]:ER with CSF-1 treatment resulted in a synergistic mitogenic response that was not observed with either 4-HT or CSF-1 treatment alone. Although activation of Raf is sufficient to induce mitogenesis in NIH 3T3 cells, these experiments were conducted at levels of Raf activation (modulated by titrating the concentration of 4-HT) that were insufficient to induce DNA synthesis (Fig. 4D, right panel, open bars).
Interestingly, in multiple experiments we noted that the response of cells expressing the mutated CSF-1 receptor to concomitant treatment with CSF-1 and Raf activation was less than that observed in cells expressing the wild-type CSF-1 receptor. There are several possibilities to explain such observations. First, it may be that the phosphorylation of tyrosine-809 of the CSF-1 receptor initiates multiple signaling pathways required for cell cycle progression. Second, it is possible that the mutated CSF-1 receptor is defective in all Ras-activated signaling pathways; hence, Raf may only partially complement the loss of activated Ras. Third, such differences may be a reflection of intrinsic clonal variation between separately derived populations of NIH 3T3 cells. Finally, these results may be a reflection of the ability of Raf to inhibit cell proliferation as a consequence of the induction of p21Cip1 which could be occurring in a subpopulation of retrovirus-infected cells (128). Nonetheless, collectively, these data are consistent with a model that suggests that the defective CSF-1 receptor is unable to fully activate the MAP kinase pathway leading to c-Myc and cyclin D1 induction. Moreover, activation of Raf in conjunction with CSF-1 is sufficient to restore, at least in part, the biochemical and biological response of these cells.
MEK activation is necessary and sufficient for the induction of c-Myc and cyclin D1.
To determine whether MEK activation was either necessary or sufficient for c-Myc and cyclin D1 induction, we constructed a conditionally active form of MEK1 (ΔMEK1:ER) by fusing sequences encoding a constitutively active form of the protein to the hormone-binding domain of the human estrogen receptor as described previously (64, 128). In addition, we used a specific and selective inhibitor of MEK1 (PD098059) to address whether MEK1 activity was required for the induction of c-Myc and cyclin D1 by growth factors and oncogenes.
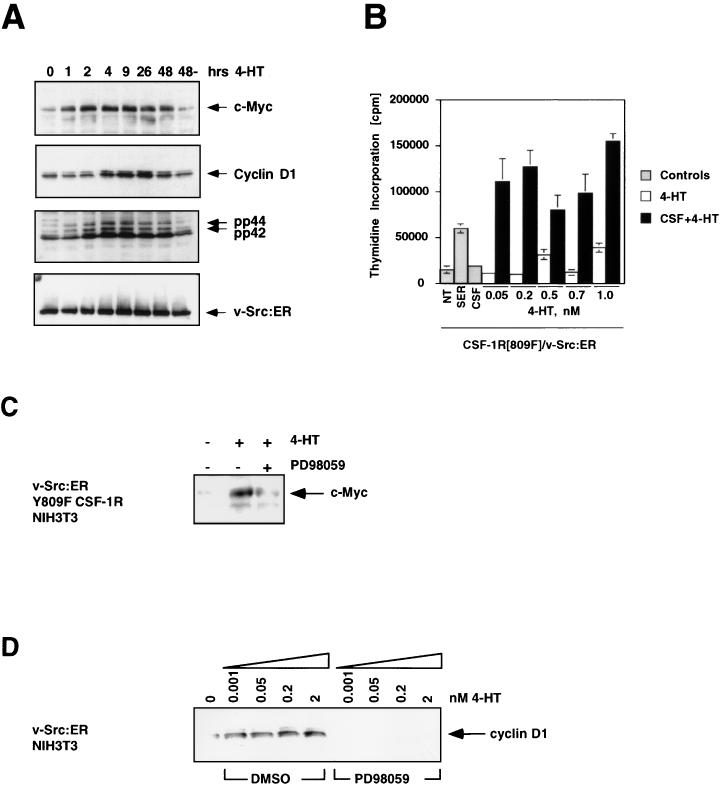
Activation of ΔMEK1:ER in NIH 3T3 cells led to activation of the p42 and p44 MAP kinases and elicited morphological transformation, although both effects occurred with slower kinetics than was observed previously with ΔRaf-1:ER (Fig. 5B and 5C and data not shown). In addition, activation of ΔMEK1:ER in quiescent NIH 3T3 cells led to the induction of c-Myc and cyclin D1 that was detected between 3 and 26 h after the activation of ΔMEK1:ER. The expression of both proteins remained elevated for at least 48 h after ΔMEK1:ER activation (Fig. 5C). The delayed induction of both proteins, compared to Raf activation, correlates with the slower activation of the MAP kinases observed in response to ΔMEK1:ER activation (Fig. 5C, lower panel). However, c-Myc and cyclin D1 were induced to the same extent by ΔMEK1:ER as by Raf. These data indicate that MEK1 activation is sufficient for the induction of c-Myc and cyclin D1 and are consistent with the ability of activated forms of MEK1 to induce DNA synthesis in NIH 3T3 cells (19, 128).
FIG. 5.
MEK activity is sufficient and necessary for the induction of c-Myc and cyclin D1. (A) Construction of ΔMEK1:ER. DNA sequences encoding a constitutively active form of MEK1 (ΔN3, S218E, and S222D) were fused to the hormone-binding domain of the HE14 allele of the human estrogen receptor and cloned into the pBabepuro3 replication-defective retroviral expression vector (64, 72). (B) Morphological transformation of NIH 3T3 cells after activation of ΔMEK1:ER. NIH 3T3 cells expressing ΔMEK1:ER were cultured in DSFM for 24 h and then treated with either ethanol (solvent control) or 1 μM 4-HT for 48 h as indicated, at which time photomicrographs were taken. (C) MAP kinase activity and expression of c-Myc and cyclin D1 after ΔMEK1:ER activation. NIH 3T3 cells expressing ΔMEK1:ER were cultured in DSFM for 24 h and then treated with 1 μM 4-HT for various lengths of time. Expression of c-Myc (top panel) and cyclin D1 (middle panel) was detected by Western blotting, and MAP kinase activity was assessed by p42 and p44 immune complex kinase assay with MBP as a substrate (lower panel). (D) Effect of inhibiting MEK activity on Raf induction of c-Myc and cyclin D1 expression. NIH 3T3 cells expressing ΔB-Raf:ER* were cultured in DSFM for 24 h and then treated with either DMSO (solvent conrol) or 100 μM PD098059 for 40 min. Different concentrations of 4-HT were then added, and cells were harvested after either 6 h (top and middle panels) or 25 h (bottom panel). The activities of the p42 and p44 MAP kinases (top panel) were measured by immune-complex kinase assay, and expression of c-Myc and cyclin D1 (middle and bottom panels) was assessed by Western blotting. (E) Effect of inhibiting MEK activity on CSF-1 induction of c-Myc and cyclin D1. NIH 3T3 cells expressing the wild-type CSF-1 receptor (WT CSF-1R) were cultured in DSFM for 20 h and then treated with either DMSO or 100 μM PD098059 for 40 min. Cells were then treated with 300 or 225 nM CSF-1 and harvested after either 1 h (middle and bottom panels) or 22.5 h (top panel). c-Myc and cyclin D1 expression (top and middle panels) was assessed by Western blotting. The c-Myc blot was reprobed with an anti-p42 MAP kinase antiserum as a control for equal loading. (F) Effect of inhibiting MEK activity on PDGF induction of c-Myc. Parental NIH 3T3 cells were cultured in DSFM for 16 h and then treated with either 100 μM PD098059, 10 nM wortmannin, or DMSO as a solvent control for 40 min. Cells were then treated with 10 ng of PDGF per ml and harvested 2 h later. c-Myc expression was assessed by Western blotting.
To address a possible requirement for MEK activity in the induction of c-Myc and cyclin D1 in response to Raf activation we pretreated NIH 3T3 cells expressing ΔB-Raf:ER* with PD098059, a specific and selective inhibitor of MEK1 activity or dimethyl sulfoxide (DMSO) as a solvent control (4, 128). Cells were then treated with increasing concentrations of 4-HT and harvested after either 6 h (to measure c-Myc expression) or 25 h (to measure cyclin D1 expression) (Fig. 5D). By 6 h after ΔB-Raf:ER* activation, p42 and p44 MAP kinase activation was readily detected with a phospho-specific anti-active MAP kinase antiserum (Fig. 5D, top panel) or by an immune-complex kinase assay (data not shown). Activation of the p42 and p44 MAP kinases was inhibited by treatment with PD098059, whereas the addition of DMSO had no effect. Treatment of cells with PD098059 inhibited the induction of both c-Myc and cyclin D1 proteins in response to ΔB-Raf:ER* activation. However, at higher levels of Raf activation (25 nM 4-HT), the inhibition of c-Myc and cyclin D1 expression by PD098059 was less profound. It is not clear if this incomplete inhibition is a consequence of only a partial blockade of MAP kinase activation in these cells or if it may reflect that, at higher levels of Raf activation, MEK-independent pathways are involved in the regulation of these genes. However, these data suggest that MEK activity is required for the full induction of c-Myc and cyclin D1 in response to Raf activation.
To determine whether MEK activity is required for the ability of the CSF-1 receptor to induce c-Myc expression, quiescent NIH 3T3 cells expressing the wild-type CSF-1 receptor were treated with different concentrations of CSF-1 (300 or 225 nM) in the absence or presence of PD098059 (Fig. 5E). Cells were harvested after either 1 or 22.5 h of CSF-1 treatment for analysis of c-Myc and cyclin D1 expression, respectively. The induction of both c-Myc and cyclin D1 proteins by CSF-1 was significantly inhibited in the presence of PD098059, indicating that MEK activity is required for the full induction of these proteins by CSF-1. These data are further consistent with a requirement for MAP kinase activation for induction of c-Myc and cyclin D1 by the CSF-1 receptor. As described above for Raf, it is not clear if the residual expression of cyclin D1 and c-Myc is due to incomplete inhibition of the MAP kinase pathway or the existence of MEK1-independent pathways leading to the control of these genes.
Finally, we addressed the requirement for MEK activity in NIH 3T3 cells for the induction of c-Myc in response to the potent mesenchymal growth factor, platelet-derived growth factor (PDGF). Quiescent NIH 3T3 cells were stimulated with PDGF in the absence or presence of PD098059. The induction of c-Myc in response to PDGF was strongly inhibited by PD098059 (Fig. 5F). Interestingly, treatment of these cells with wortmannin, an inhibitor of PI3′ kinase activity, also decreased PDGF-induced c-Myc expression, suggesting a potential role for phosphatidylinositol lipid-regulated signaling pathways in the regulation of c-Myc expression (Fig. 5F).
Complementation of defective CSF-1 receptor signaling by v-Src.
Previous experiments have indicated a potential role for the Src family of protein tyrosine kinases in the regulation of c-Myc expression in response to growth factor stimulation of mouse fibroblasts. Further, it has been suggested that a Src-dependent, Ras-independent pathway may be crucial in the regulation of c-Myc in such cells (6). We therefore sought to determine if activated Src could regulate either c-Myc or cyclin D1 expression and, additionally, if Src could complement the mitogenic defect in NIH 3T3 cells expressing the CSF-1R[809F]. A conditionally active form of v-Src was constructed by fusing sequences encoding the Schmidt Ruppin A form of v-Src to the hormone-binding domain of the human estrogen receptor as described above for Raf-1 and MEK1 (Fig. 6A). NIH 3T3 cells expressing v-Src:ER displayed hormone-dependent morphological transformation (Fig. 6B) and tyrosine phosphorylation of cellular proteins (Fig. 6C). As shown previously, treatment of NIH 3T3 cells infected with a control retrovirus (Neo) with 4-HT had no effect on the low level of tyrosine phosphorylation found in these cells and no effect on cell morphology (Fig. 6C) (100).
Activation of v-Src:ER in quiescent NIH 3T3 cells resulted in activation of the p42-p44 MAP kinases within 1 h and the induction of both c-Myc and cyclin D1 proteins within 1 and 4 h, respectively (Fig. 7A). The kinetics of c-Myc and cyclin D1 induction after v-Src:ER activation were similar to those observed in response to the activation of ΔRaf-1:ER, suggesting that v-Src:ER is able to activate some of the same changes in gene expression induced by activation of Raf and MEK. Such results are consistent with previous observations that transformation of cells by v-Src requires the activity of Ras proteins (110).
FIG. 7.
Complementation of the CSF-1R[809F] mitogenic and signaling defect by v-Src:ER. (A) Activation of the MAP kinases and induction of c-Myc and cyclin D1 by v-Src:ER. NIH 3T3 cells expressing v-Src:ER were cultured in DSFM for 2 days and treated with 500 nM 4-HT for various lengths of time. Expression of c-Myc, cyclin D1, activated p42 and p44 MAP kinases, and v-Src:ER was assessed by Western blotting. (B) Proliferation of v-Src:ER-expressing cells. NIH 3T3 cells expressing the CSF-1R[809F] and v-Src:ER were cultured in DSFM for 24 h and then either left untreated (NT) or treated with 10% FCS, 300 nM CSF-1 (gray bars), 4-HT (0.05 to 1 nM, open bars), or CSF-1 plus 4-HT (solid bars). Cell proliferation was determined by measuring the incorporation of [methyl-3H]thymidine over a period of 48 h. (C) Effect of inhibition of MEK activity on the induction of c-Myc by v-Src:ER. NIH 3T3 cells expressing CSF-1R[809F] and v-Src:ER were cultured in DSFM for 36 h, treated with 100 μM PD098059 for 1 h, and then stimulated with 50 nM 4-HT for 4 h. Expression of c-Myc was assessed by Western blotting. (D) Effect of inhibition of MEK activity on the induction of cyclin D1 by v-Src:ER. NIH 3T3 cells expressing v-Src:ER were cultured for 36 h in DSFM, treated with DMSO (solvent control) or 100 μM PD098059 for 40 min, and then stimulated with various concentrations of 4-HT (0 to 2 nM) for 24 h. Expression of cyclin D1 was assessed by Western blotting.
To address whether activation of v-Src:ER was sufficient to complement the mitogenic defect of the mutated CSF-1 receptor, we generated a population of cells expressing both the CSF-1R[809F] and v-Src:ER. These cells were rendered quiescent and then were stimulated as described in Materials and Methods. Serum stimulation of these cells led to induced DNA synthesis, whereas treatment with CSF-1 or activation of v-Src:ER alone with low concentrations of 4-HT had no effect on cell proliferation (Fig. 7B). Activation of v-Src:ER in the presence of CSF-1 resulted in a strong synergistic induction of DNA synthesis. Like Raf-1[DD]:ER, high-level activation of v-Src:ER alone results in a robust mitogenic response; hence, this experiment was performed at levels of v-Src:ER activation (modulated by varying the concentration of 4-HT) that were insufficient to induce DNA synthesis (Fig. 7B, open bars). These data are consistent with a role for Src family protein kinases in the regulation of the MAP kinase pathway leading to the induction of cyclin D1 and c-Myc. It is interesting to note that the mitogenic response of these cells to CSF-1 and v-Src:ER activation exceeded the serum response in a manner analogous to that of cells expressing the wild-type CSF-1 receptor. Such observations may reflect the fact that v-Src:ER is able to activate both Ras-dependent and Ras-independent signal transduction pathways to support cell proliferation.
To determine whether the induction of c-Myc and cyclin D1 by v-Src:ER required the activity of MEK, we assessed the effects of the MEK inhibitor PD098059 on the ability of v-Src to induce the expression of c-Myc and cyclin D1. Quiescent NIH 3T3 cells expressing CSF-1R[809F] and v-Src:ER were treated with either PD098059 or DMSO prior to the addition of 4-HT for 1 h to activate v-Src:ER. Consistent with our previous observations, the induction of c-Myc by v-Src:ER was inhibited by PD098059 (Fig. 7C). Similarly, in NIH 3T3 cells the induced expression of cyclin D1, assessed 24 h after v-Src:ER activation, was inhibited by pretreatment of the cells with PD098059 (Fig. 7D). These data suggest that v-Src requires the activity of MEK to fully induce the expression of c-Myc and cyclin D1.
DISCUSSION
Considerable effort is currently focused on forging connections between components of the cytoplasmic signal transduction machinery, transcription factors, and regulators of the cell cycle. Although the precise details remain incompletely understood, the prevailing model suggests that engagement of growth factor receptors by their cognate ligands results in activation of numerous signaling pathways that modulate gene expression leading ultimately to an appropriate biological response. Since activated growth factor receptors influence a diverse array of cytosolic signal transduction pathways, attempts are under way to link specific signal transduction pathways with the activation of particular downstream target genes.
Several lines of evidence have suggested that in mouse fibroblasts Ras regulates the expression of the Fos and Jun components of the AP-1 transcription factor, whereas a Src-dependent, Ras-independent signaling pathway mediates c-Myc expression (6, 26). First, classical experiments on oncogene cooperation indicated that ectopically introduced Ras and Myc cooperate efficiently to transform primary murine cells and to accelerate tumor formation in mice (10, 90, 105, 133). Second, MAP kinase activation was implicated in the phosphorylation of the ternary complex factor Elk-1 which binds to the serum response element in the c-Fos promoter and contributes to the activation of c-Fos transcription (85). Third, activation of the human CSF-1 receptor in NIH 3T3 cells leads to the induction of c-Fos, c-Jun, c-Myc, and cyclin D1 expression and reentry into the cell cycle. Cells expressing a mutated form of the CSF-1 receptor induce c-Fos and c-Jun but do not induce c-Myc and cyclin D1 and fail to proliferate in response to CSF-1. The fact that the CSF-1R[809F] is capable of inducing the expression of c-Fos provided circumstantial evidence for the normal functioning of the Ras-activated MAP kinase pathway. The failure to induce c-Myc and cyclin D1 was taken as evidence that a separate pathway leading to the expression of these genes was compromised by mutation. Since this mutated receptor fails to associate with c-Src and can be complemented by the ectopic overexpression of c-Myc or cyclin D1, these results provided evidence for the possible existence of a Ras-independent pathway(s) leading to the expression of these proteins (18, 97–99). Finally, microinjection studies of PDGF-induced mitogenesis in NIH 3T3 cells suggested that the effects of dominant-negative Ras were overcome by ectopic overexpression of AP-1 but not of c-Myc, whereas the effects of dominant-negative c-Src were overcome by ectopic overexpression of c-Myc but not of AP-1. These data suggested that Ras functions upstream of c-Fos and c-Jun (AP-1), whereas c-Src and its family members function upstream of c-Myc. However, the inability to conduct biochemical analyses of microinjected cells precluded a more definitive analysis of the regulation of the c-Myc and AP-1 transcription factors in these latter experiments (6).
In these studies we demonstrate a link between the activation of the Raf/MEK/MAP kinase pathway and the transcriptional activation of c-Myc and cyclin D1. The kinetics of induction of these genes by Raf is fully consistent with previously published data on their induction by serum and growth factors: Raf activation leads to the immediate-early induction of c-Myc transcription and delayed induction of cyclin D1. Evidence that the induction of c-Myc by Raf was functionally productive was inferred from the ability of Raf to induce the expression of a known c-Myc-responsive gene, ODC, in a manner that is at least partly dependent on tandem E-boxes in the first intron of the gene (7). Interestingly, of five c-Myc responsive target genes that we tested, only ODC was significantly induced after Raf activation, suggesting that there are additional factors influencing the expression of c-Myc target genes. These may include the expression or activity of the transcription factor AP-2, which has been suggested to influence c-Myc-mediated transcriptional activation (31).
The work of other investigators also supports a connection between the Ras-activated MAP kinase pathway and the expression of c-Myc. The level of c-Myc mRNA is decreased after the ablation of activated Ki-Ras in human colon carcinoma cells. In addition, c-Myc expression is upregulated in Swiss 3T3 cells transformed by oncogenic Ha-Ras (59, 108). Furthermore, it has been suggested that Raf is involved in the induction of c-Myc transcription in response to v-abl activation and growth factor stimulation (48, 137). It has also been demonstrated that Ras transformation of NIH 3T3 cells is inhibited by the expression of dominant-negative forms of c-Myc or by the inhibition of c-Myc expression (109). Finally, it is likely that regulation of c-Myc by the MAP kinase pathway is not limited to transcriptional control of c-Myc gene expression. It has been reported that MAP kinase-mediated phosphorylation of c-Myc modulates its transforming and transactivation potential and that MAP kinase and c-Myc physically associate with each other (17, 34, 88, 106). It will be important to fully characterize the transcriptional and posttranslational interactions between the MAP kinase pathway and c-Myc in order to properly understand their functional relationship. It should further be emphasized that the ability of the MAP kinase pathway to induce the expression of c-Myc and cyclin D1 in NIH 3T3 cells does not preclude an important role for other growth factor-activated signaling pathways in the expression of these proteins in fibroblasts or indeed in other cell types (102, 116).
Our studies of NIH 3T3 cells expressing the CSF-1R and CSF-1R[809F] provide additional evidence for a functional connection between the Raf/MEK/MAP kinase pathway and cyclin D1 and c-Myc. NIH 3T3 cells expressing CSF-1R[809F] are defective in their ability to activate the p42 and p44 MAP kinases in response to CSF-1 stimulation, a finding consistent with a model that places the MAP kinase pathway upstream of c-Myc and cyclin D1. Importantly, in cells expressing CSF-1R[809F], activation of the MAP kinase pathway mediated either by activated Raf-1 or v-Src complemented, at least in part, both the biochemical and the biological defects in these cells. Further evidence for a role for the MAP kinase pathway in growth factor- and oncogene-induced c-Myc and cyclin D1 expression was demonstrated by the use of a specific and selective MEK inhibitor, PD098059, which prevented the induction of c-Myc and cyclin D1 in response to both growth factors and oncogenes. Despite the strong connection between the MAP kinase pathway and the expression of c-Myc and cyclin D1 observed in these experiments, we cannot rule out the possibility that there are additional signaling pathways activated as a consequence of the phosphorylation of tyrosine-809 of the human CSF-1 receptor that participate in the regulation of these genes.
A possible resolution for some of the apparent discrepancies in the literature may reflect the mechanism of Raf-1 activation in response to growth factor stimulation. The current model suggests that activated Ras recruits Raf-1 to the plasma membrane, where it is phosphorylated by members of the Src family of protein tyrosine kinases. Phosphorylation of Raf-1 on these sites (Y340 and Y341) significantly potentiates its ability to activate the MAP kinase pathway. Indeed, the form of the conditionally active full-length Raf-1 that we used in these experiments is thought to be at least a partial mimic of the tyrosine phosphorylated form of the protein (14, 27, 65, 66). Therefore, inhibitors of both Ras and Src might be expected to interfere with the normal activation of the Raf/MEK/MAP kinase pathway and the subsequent induction of c-Myc and cyclin D1. Such a model is consistent with the observations that the activation of the wild-type CSF-1 receptor results in activation of c-Src but that the CSF-1R[809F] fails to associate with c-Src (18).
An important role for the Ras-activated MAP kinase pathway in the transmission of mitogenic signals from Src is supported by the work of others. For example, transformation by v-Src is inhibited by neutralizing anti-Ras antibodies or by overexpression of GAP (21, 79, 110). Furthermore, the Ras-activated MAP kinase pathway has been shown to be required for induction of certain Src transcriptional target genes (20, 89, 131). Finally, the phenotypic effects of overexpression of a Drosophila homologue of a Src-family kinase are inhibited by loss of Ras function (115).
Although we have not defined the cis-acting elements in either the c-Myc or cyclin D1 promoters that confer Raf responsiveness, there may be a role for Ets-family transcription factors in the transcriptional regulation of these genes. The importance of Ets transcription factors in mediating both growth factor- and oncogene-induced gene expression is illustrated by the fact that both Ets-1 and Ets-2 rescue the mitogenic defect in CSF-1R[809F]-expressing cells (96). In addition, dominant-negative forms of Ets-2 have been shown to suppress induction of c-Myc and cyclin D1 by CSF-1 and epidermal growth factor (EGF), respectively (2, 55). Dominant-negative Ets-2 has also been shown to suppress morphological transformation induced by v-Raf and CSF-1 (55, 101). Further, induction of the cyclin D1 promoter has been shown to be suppressed by the ectopic expression of dominant-negative MEK1 or MAP kinase phosphatase 1 (56). These results are consistent with the inability of the mutated CSF-1 receptor to activate the MAP kinase-mediated phosphorylation of Ets-2 in NIH 3T3 cells (28, 69, 134).
Since the CSF-1R[809F] is unable to fully activate the MAP kinase pathway and yet induces the expression of c-Fos, these data call into question the role of the MAP kinase pathway in c-Fos expression (119). In separate experiments, we have assessed the effects of Raf activation on the expression of c-Fos. Unlike c-Myc and cyclin D1, Raf activation does not mimic the effects of growth factor stimulation on c-Fos mRNA or protein expression inducing, at best, only a modest increase in c-Fos expression (17a). However, when Raf activation is combined with a second signal such as a calcium agonist, a strong synergistic induction of c-Fos expression is observed. A requirement for the MAP kinase pathway in the regulation of c-Fos was inferred from the fact that treatment of cells with PD098059 prevented c-Fos induction by serum or EGF. These data suggest that the MAP kinase pathway is required for c-Fos induction in response to mitogens but that maximal induction of c-Fos may require the cooperation of other signaling pathways. Such conclusions are supported by studies utilizing transgenic mice containing c-Fos reporter constructs, as well as by recent studies of the effects of Rho-family GTPases on c-Fos expression that indicate that c-Fos regulation is significantly more complex than earlier studies had indicated (3, 91). Clearly, future experiments must be dedicated to gaining a more precise understanding of the biochemical mechanisms underlying these transcriptional responses and the ultimate phenotypic consequences of such changes on cell physiology.
ACKNOWLEDGMENTS
We gratefully acknowledge the members of the McMahon and Lees laboratories for helpful discussions. In addition we thank Bob Eisenman and Martine Roussel for critical review of the manuscript. We thank Natalie Ahn, Naoko Arai, J. Michael Bishop, Steve Clark, John Cleveland, Simon Cook, Philip Coffino, Bob Eisenman, Gerard Evan, Nissim Hay, Josh Kaplan, John Lyons, Jay Morgenstern, Debbie Morrison, Mike Ostrowski, David Parry, Gordon Peters, Martine Roussel, Chuck Toth, and Alan Wakeling for generously providing reagents, for helpful discussions, and for communicating unpublished observations.
DNAX Research Institute is supported by the Schering Plough Corporation.
REFERENCES
- 1.Abrahamsen M S, Li R S, Dietrich-Goetz W, Morris D R. Multiple DNA elements responsible for transcriptional regulation of the ornithine decarboxylase gene by protein kinase A. J Biol Chem. 1992;267:18866–18873. [PubMed] [Google Scholar]
- 2.Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell R G. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 3.Alberts A S, Geneste O, Treisman R. Activation of SRF-regulated chromosomal templates by Rho-family GTPases requires a signal that also induces H4 hyperacetylation. Cell. 1998;92:475–487. doi: 10.1016/s0092-8674(00)80941-1. [DOI] [PubMed] [Google Scholar]
- 4.Alessi D R, Cuenda A, Cohen P, Dudley D T, Saltiel A R. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 5.Amati B, Land H. Myc-Max-Mad: a transcription factor network controlling cell cycle progression, differentiation and death. Curr Opin Genet Dev. 1994;4:102–108. doi: 10.1016/0959-437x(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 6.Barone M V, Courtneidge S A. Myc but not Fos rescue of PDGF signalling block caused by kinase-inactive Src. Nature. 1995;378:509–512. doi: 10.1038/378509a0. [DOI] [PubMed] [Google Scholar]
- 7.Bello-Fernandez C, Packham G, Cleveland J L. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci USA. 1993;90:7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett M R, Anglin S, McEwan J R, Jagoe R, Newby A C, Evan G I. Inhibition of vascular smooth muscle cell proliferation in vitro and in vivo by c-myc antisense oligodeoxynucleotides. J Clin Investig. 1994;93:820–828. doi: 10.1172/JCI117036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett M R, Evan G I, Newby A C. Deregulated expression of the c-myc oncogene abolishes inhibition of proliferation of rat vascular smooth muscle cells by serum reduction, interferon-gamma, heparin, and cyclic nucleotide analogues and induces apoptosis. Circ Res. 1994;74:525–536. doi: 10.1161/01.res.74.3.525. [DOI] [PubMed] [Google Scholar]
- 10.Birrer M J, Segal S, De Greve J S, Kaye F, Sausville E A, Minna J D. L-myc cooperates with ras to transform primary rat embryo fibroblasts. Mol Cell Biol. 1988;8:2668–2673. doi: 10.1128/mcb.8.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blackwell T K, Kretzner L, Blackwood E M, Eisenman R N, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;250:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- 12.Blackwood E M, Eisenman R N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 13.Bos J L. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 14.Bosch E, Cherwinski H, Peterson D, McMahon M. Mutations of critical amino acids affect the biological and biochemical properties of oncogenic A-Raf and Raf-1. Oncogene. 1997;15:1021–1033. doi: 10.1038/sj.onc.1201270. [DOI] [PubMed] [Google Scholar]
- 15.Brabant M, McConlogue L, van Daalen Wetters T, Coffino P. Mouse ornithine decarboxylase gene: cloning, structure, and expression. Proc Natl Acad Sci USA. 1988;85:2200–2204. doi: 10.1073/pnas.85.7.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins J F, Herman P, Schuch C, Bagby G C., Jr c-myc antisense oligonucleotides inhibit the colony-forming capacity of Colo 320 colonic carcinoma cells. J Clin Investig. 1992;89:1523–1527. doi: 10.1172/JCI115744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colman M S, Ostrowski M C. The transactivation potential of a c-Myc N-terminal region (residues 92-143) is regulated by growth factor/Ras signaling. Nucleic Acids Res. 1996;24:1971–1978. doi: 10.1093/nar/24.10.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Cook S J, Aziz N, McMahon M. The repertoire of Fos and Jun proteins expressed during the G1 phase of the cell cycle is determined by the duration of mitogen-activated protein kinase activation. Mol Cell Biol. 1999;19:330–341. doi: 10.1128/mcb.19.1.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Courtneidge S A, Dhand R, Pilat D, Twamley G M, Waterfield M D, Roussel M F. Activation of Src family kinases by colony stimulating factor-1, and their association with its receptor. EMBO J. 1993;12:943–950. doi: 10.1002/j.1460-2075.1993.tb05735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cowley S, Paterson H, Kemp P, Marshall C J. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 20.D’Arcangelo G, Halegoua S. A branched signaling pathway for nerve growth factor is revealed by Src-, Ras-, and Raf-mediated gene inductions. Mol Cell Biol. 1993;13:3146–3155. doi: 10.1128/mcb.13.6.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeClue J E, Zhang K, Redford P, Vass W C, Lowy D R. Suppression of src transformation by overexpression of full-length GTPase-activating protein (GAP) or of the GAP C terminus. Mol Cell Biol. 1991;11:2819–2825. doi: 10.1128/mcb.11.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dent P, Haser W, Haystead T A, Vincent L A, Roberts T M, Sturgill T W. Activation of mitogen-activated protein kinase kinase by v-Raf in NIH 3T3 cells and in vitro. Science. 1992;257:1404–1407. doi: 10.1126/science.1326789. [DOI] [PubMed] [Google Scholar]
- 23.Dickson B, Sprenger F, Morrison D, Hafen E. Raf functions downstream of Ras1 in the Sevenless signal transduction pathway. Nature. 1992;360:600–603. doi: 10.1038/360600a0. [DOI] [PubMed] [Google Scholar]
- 24.Downward J. Cell cycle: routine role for Ras. Curr Biol. 1997;7:R258–R260. doi: 10.1016/s0960-9822(06)00116-3. [DOI] [PubMed] [Google Scholar]
- 25.Downward J. Regulation of p21ras by GTPase activating proteins and guanine nucleotide exchange proteins. Curr Opin Genet Dev. 1992;2:13–18. doi: 10.1016/s0959-437x(05)80315-6. [DOI] [PubMed] [Google Scholar]
- 26.Eisenman R N, Cooper J A. Signal transduction. Beating a path to Myc. Nature. 1995;378:438–439. doi: 10.1038/378438a0. [DOI] [PubMed] [Google Scholar]
- 27.Fabian J R, Daar I O, Morrison D K. Critical tyrosine residues regulate the enzymatic and biological activity of Raf-1 kinase. Mol Cell Biol. 1993;13:7170–7179. doi: 10.1128/mcb.13.11.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fowles L F, Martin M L, Nelsen L, Stacey K J, Redd D, Clark Y M, Nagamine Y, McMahon M, Hume D A, Ostrowski M C. Persistent activation of mitogen-activated protein kinases p42 and p44 and ets-2 phosphorylation in response to colony-stimulating factor 1/c-fms signaling. Mol Cell Biol. 1998;18:5148–5156. doi: 10.1128/mcb.18.9.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fumagalli S, Totty N F, Hsuan J J, Courtneidge S A. A target for Src in mitosis. Nature. 1994;368:871–874. doi: 10.1038/368871a0. [DOI] [PubMed] [Google Scholar]
- 30.Garte S J. The c-myc oncogene in tumor progression. Crit Rev Oncog. 1993;4:435–449. [PubMed] [Google Scholar]
- 31.Gaubatz S, Imhof A, Dosch R, Werner O, Mitchell P, Buettner R, Eilers M. Transcriptional activation by Myc is under negative control by the transcription factor AP-2. EMBO J. 1995;14:1508–1519. doi: 10.1002/j.1460-2075.1995.tb07137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gomez N, Cohen P. Dissection of the protein kinase cascade by which nerve growth factor activates MAP kinase. Nature. 1991;353:170–173. doi: 10.1038/353170a0. [DOI] [PubMed] [Google Scholar]
- 33.Grandori C, Eisenman R N. Myc target genes. Trends Biochem Sci. 1997;22:177–181. doi: 10.1016/s0968-0004(97)01025-6. [DOI] [PubMed] [Google Scholar]
- 34.Gupta S, Davis R J. MAP kinase binds to the NH2-terminal activation domain of c-Myc. FEBS Lett. 1994;353:281–285. doi: 10.1016/0014-5793(94)01052-8. [DOI] [PubMed] [Google Scholar]
- 35.Hall M, Peters G. Genetic alterations of cyclins, cyclin-dependent kinases, and Cdk inhibitors in human cancer. Adv Cancer Res. 1996;68:67–108. doi: 10.1016/s0065-230x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- 36.Han M, Golden A, Han Y, Sternberg P W. C. elegans lin-45 raf gene participates in let-60 ras-stimulated vulval differentiation. Nature. 1993;363:133–140. doi: 10.1038/363133a0. [DOI] [PubMed] [Google Scholar]
- 37.Hann S R, Eisenman R N. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol Cell Biol. 1984;4:2486–2497. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heby O, Persson L. Molecular genetics of polyamine synthesis in eukaryotic cells. Trends Biochem Sci. 1990;15:153–158. doi: 10.1016/0968-0004(90)90216-x. [DOI] [PubMed] [Google Scholar]
- 39.Holtta E, Sistonen L, Alitalo K. The mechanisms of ornithine decarboxylase deregulation in c-Ha-ras oncogene-transformed NIH 3T3 cells. J Biol Chem. 1988;263:4500–4507. [PubMed] [Google Scholar]
- 40.Howe L R, Marshall C J. Lysophosphatidic acid stimulates mitogen-activated protein kinase activation via a G-protein-coupled pathway requiring p21ras and p74raf-1. J Biol Chem. 1993;268:20717–20720. [PubMed] [Google Scholar]
- 41.Hurta R A, Huang A, Wright J A. Basic fibroblast growth factor selectively regulates ornithine decarboxylase gene expression in malignant H-ras transformed cells. J Cell Biochem. 1996;60:572–583. doi: 10.1002/(sici)1097-4644(19960315)60:4<572::aid-jcb13>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 42.Johnson L, Greenbaum D, Cichowski K, Mercer K, Murphy E, Schmitt E, Bronson R T, Umanoff H, Edelmann W, Kucherlapati R, Jacks T. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 1997;11:2468–2481. doi: 10.1101/gad.11.19.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnston J M, Carroll W L. c-myc hypermutation in Burkitt’s lymphoma. Leuk Lymphoma. 1992;8:431–439. doi: 10.3109/10428199209051025. [DOI] [PubMed] [Google Scholar]
- 44.Joneson T, Bar-Sagi D. Ras effectors and their role in mitogenesis and oncogenesis. J Mol Med. 1997;75:587–593. doi: 10.1007/s001090050143. [DOI] [PubMed] [Google Scholar]
- 45.Kaplan J M, Mardon G, Bishop J M, Varmus H E. The first seven amino acids encoded by the v-src oncogene act as a myristylation signal: lysine 7 is a critical determinant. Mol Cell Biol. 1988;8:2435–2441. doi: 10.1128/mcb.8.6.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katz M E, McCormick F. Signal transduction from multiple Ras effectors. Curr Opin Genet Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- 47.Kelly K, Cochran B H, Stiles C D, Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983;35:603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- 48.Kerkhoff E, Houben R, Loffler S, Troppmair J, Lee J E, Rapp U R. Regulation of c-myc expression by Ras/Raf signalling. Oncogene. 1998;16:211–6. doi: 10.1038/sj.onc.1201520. [DOI] [PubMed] [Google Scholar]
- 49.Khosravi-Far R, Der C J. The Ras signal transduction pathway. Cancer Metastasis Rev. 1994;13:67–89. doi: 10.1007/BF00690419. [DOI] [PubMed] [Google Scholar]
- 50.Kolch W, Heidecker G, Lloyd P, Rapp U R. Raf-1 protein kinase is required for growth of induced NIH/3T3 cells. Nature. 1991;349:426–428. doi: 10.1038/349426a0. [DOI] [PubMed] [Google Scholar]
- 51.Kretzner L, Blackwood E M, Eisenman R N. Myc and Max proteins possess distinct transcriptional activities. Nature. 1992;359:426–429. doi: 10.1038/359426a0. [DOI] [PubMed] [Google Scholar]
- 52.Kumar A P, Mar P K, Zhao B, Montgomery R L, Kang D C, Butler A P. Regulation of rat ornithine decarboxylase promoter activity by binding of transcription factor Sp1. J Biol Chem. 1995;270:4341–4348. doi: 10.1074/jbc.270.9.4341. [DOI] [PubMed] [Google Scholar]
- 53.Kyriakis J M, App H, Zhang X F, Banerjee P, Brautigan D L, Rapp U R, Avruch J. Raf-1 activates MAP kinase-kinase. Nature. 1992;358:417–421. doi: 10.1038/358417a0. [DOI] [PubMed] [Google Scholar]
- 54.Lahti J M, Li H, Kidd V J. Elimination of cyclin D1 in vertebrate cells leads to an altered cell cycle phenotype, which is rescued by overexpression of murine cyclins D1, D2, or D3 but not by a mutant cyclin D1. J Biol Chem. 1997;272:10859–10869. doi: 10.1074/jbc.272.16.10859. [DOI] [PubMed] [Google Scholar]
- 55.Langer S J, Bortner D M, Roussel M F, Sherr C J, Ostrowski M C. Mitogenic signaling by colony-stimulating factor 1 and ras is suppressed by the ets-2 DNA-binding domain and restored by myc overexpression. Mol Cell Biol. 1992;12:5355–5362. doi: 10.1128/mcb.12.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lavoie J N, L’Allemain G, Brunet A, Muller R, Pouyssegur J. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J Biol Chem. 1996;271:20608–20616. doi: 10.1074/jbc.271.34.20608. [DOI] [PubMed] [Google Scholar]
- 57.Leonetti C, D’Agnano I, Lozupone F, Valentini A, Geiser T, Zon G, Calabretta B, Citro G C, Zupi G. Antitumor effect of c-myc antisense phosphorothioate oligodeoxynucleotides on human melanoma cells in vitro and in mice. J Natl Cancer Inst. 1996;88:419–429. doi: 10.1093/jnci/88.7.419. [DOI] [PubMed] [Google Scholar]
- 58.Li M, Lonial H, Citarella R, Lindh D, Colina L, Kramer R. Tumor inhibitory activity of anti-ras ribozymes delivered by retroviral gene transfer. Cancer Gene Ther. 1996;3:221–229. [PubMed] [Google Scholar]
- 59.Lloyd A C, Paterson H F, Morris J D, Hall A, Marshall C J. p21H-ras-induced morphological transformation and increases in c-myc expression are independent of functional protein kinase C. EMBO J. 1989;8:1099–1104. doi: 10.1002/j.1460-2075.1989.tb03479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu X, Chou T B, Williams N G, Roberts T, Perrimon N. Control of cell fate determination by p21ras/Ras1, an essential component of torso signaling in Drosophila. Genes Dev. 1993;7:621–632. doi: 10.1101/gad.7.4.621. [DOI] [PubMed] [Google Scholar]
- 61.Luttrell D K, Luttrell L M, Parsons S J. Augmented mitogenic responsiveness to epidermal growth factor in murine fibroblasts that overexpress pp60c-src. Mol Cell Biol. 1988;8:497–501. doi: 10.1128/mcb.8.1.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.MacNicol A M, Muslin A J, Williams L T. Raf-1 kinase is essential for early Xenopus development and mediates the induction of mesoderm by FGF. Cell. 1993;73:571–583. doi: 10.1016/0092-8674(93)90143-e. [DOI] [PubMed] [Google Scholar]
- 63.Mangues R, Seidman I, Pellicer A, Gordon J W. Tumorigenesis and male sterility in transgenic mice expressing a MMTV/N-ras oncogene. Oncogene. 1990;5:1491–1497. [PubMed] [Google Scholar]
- 64.Mansour S J, Matten W T, Hermann A S, Candia J M, Rong S, Fukasawa K, Vande-Woude G F, Ahn N G. Transformation of mammalian cells by constitutively active MAP kinase. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 65.Marais R, Light Y, Paterson H F, Marshall C J. Ras recruits Raf-1 to the plasma membrane for activation by tyrosine phosphorylation. EMBO J. 1995;14:3136–3145. doi: 10.1002/j.1460-2075.1995.tb07316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marais R, Light Y, Paterson H F, Mason C S, Marshall C J. Differential regulation of Raf-1, A-Raf, and B-Raf by oncogenic ras and tyrosine kinases. J Biol Chem. 1997;272:4378–4383. doi: 10.1074/jbc.272.7.4378. [DOI] [PubMed] [Google Scholar]
- 67.Matsushime H, Roussel M F, Ashmun R A, Sherr C J. Colony-stimulating factor 1 regulates novel cyclins during the G1 phase of the cell cycle. Cell. 1991;65:701–713. doi: 10.1016/0092-8674(91)90101-4. [DOI] [PubMed] [Google Scholar]
- 68.McCarthy S A, Aziz N, McMahon M. Identification of immediate-early gene targets of the Raf-1 serine/threonine protein kinase using an estradiol-dependent fusion protein, delta Raf-1:ER. Methods Mol Biol. 1997;85:137–151. doi: 10.1385/0-89603-489-5:137. [DOI] [PubMed] [Google Scholar]
- 69.McCarthy S A, Chen D, Yang B S, Garcia Ramirez J J, Cherwinski H, Chen X R, Klagsbrun M, Hauser C A, Ostrowski M C, McMahon M. Rapid phosphorylation of Ets-2 accompanies mitogen-activated protein kinase activation and the induction of heparin-binding epidermal growth factor gene expression by oncogenic Raf-1. Mol Cell Biol. 1997;17:2401–2412. doi: 10.1128/mcb.17.5.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McCarthy S A, Samuels M L, Pritchard C A, Abraham J A, McMahon M. Rapid induction of heparin-binding epidermal growth factor/diphtheria toxin receptor expression by Raf and Ras oncogenes. Genes Dev. 1995;9:1953–1964. doi: 10.1101/gad.9.16.1953. [DOI] [PubMed] [Google Scholar]
- 71.Mimori K, Mori M, Shiraishi T, Fujie T, Baba K, Kusumoto H, Haraguchi M, Ueo H, Akiyoshi T. Analysis of ornithine decarboxylase messenger ribonucleic acid expression in colorectal carcinoma. Dis Colon Rectum. 1997;40:1095–1100. doi: 10.1007/BF02050936. [DOI] [PubMed] [Google Scholar]
- 72.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moriguchi T, Gotoh Y, Nishida E. Roles of the MAP kinase cascade in vertebrates. Adv Pharmacol. 1996;36:121–137. doi: 10.1016/s1054-3589(08)60579-7. [DOI] [PubMed] [Google Scholar]
- 74.Morrison D K, Cutler R E. The complexity of Raf-1 regulation. Curr Opin Cell Biol. 1997;9:174–179. doi: 10.1016/s0955-0674(97)80060-9. [DOI] [PubMed] [Google Scholar]
- 75.Moshier J A, Osborne D L, Skunca M, Dosescu J, Gilbert J D, Fitzgerald M C, Polidori G, Wagner R L, Friezner Degen S J, Luk G D, et al. Multiple promoter elements govern expression of the human ornithine decarboxylase gene in colon carcinoma cells. Nucleic Acids Res. 1992;20:2581–2590. doi: 10.1093/nar/20.10.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nass S J, Dickson R B. Defining a role for c-Myc in breast tumorigenesis. Breast Cancer Res Treat. 1997;44:1–22. doi: 10.1023/a:1005858611585. [DOI] [PubMed] [Google Scholar]
- 77.Nishida E, Gotoh Y. The MAP kinase cascade is essential for diverse signal transduction pathways. Trends Biochem Sci. 1993;18:128–131. doi: 10.1016/0968-0004(93)90019-j. [DOI] [PubMed] [Google Scholar]
- 78.Nishida Y, Inoue Y H, Tsuda L, Adachi-Yamada T, Lim Y M, Hata M, Ha H Y, Sugiyama S. The Raf/MAP kinase cascade in cell cycle regulation and differentiation in Drosophila. Cell Struct Funct. 1996;21:437–444. doi: 10.1247/csf.21.437. [DOI] [PubMed] [Google Scholar]
- 79.Nori M, Vogel U S, Gibbs J B, Weber M J. Inhibition of v-src-induced transformation by a GTPase-activating protein. Mol Cell Biol. 1991;11:2812–2818. doi: 10.1128/mcb.11.5.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Packham G, Cleveland J L. Induction of ornithine decarboxylase by IL-3 is mediated by sequential c-Myc-independent and c-Myc-dependent pathways. Oncogene. 1997;15:1219–1232. doi: 10.1038/sj.onc.1201273. [DOI] [PubMed] [Google Scholar]
- 81.Palvimo J J, Partanen M, Janne O A. Characterization of cell-specific modulatory element in the murine ornithine decarboxylase promoter. Biochem J. 1996;316:993–998. doi: 10.1042/bj3160993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parsons J T, Parsons S J. Src family protein tyrosine kinases: cooperating with growth factor and adhesion signaling pathways. Curr Opin Cell Biol. 1997;9:187–192. doi: 10.1016/s0955-0674(97)80062-2. [DOI] [PubMed] [Google Scholar]
- 83.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Prendergast G C, Ziff E B. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991;251:186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- 85.Price M A, Hill C, Treisman R. Integration of growth factor signals at the c-fos serum response element. Philos Trans R Soc Lond B Biol Sci. 1996;351:551–559. doi: 10.1098/rstb.1996.0054. [DOI] [PubMed] [Google Scholar]
- 86.Prins J, De Vries E G, Mulder N H. The myc family of oncogenes and their presence and importance in small-cell lung carcinoma and other tumour types. Anticancer Res. 1993;13:1373–1385. [PubMed] [Google Scholar]
- 87.Pritchard C A, Samuels M L, Bosch E, McMahon M. Conditionally oncogenic forms of the A-Raf and B-Raf protein kinases display different biological and biochemical properties in NIH 3T3 cells. Mol Cell Biol. 1995;15:6430–6442. doi: 10.1128/mcb.15.11.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pulverer B J, Fisher C, Vousden K, Littlewood T, Evan G, Woodgett J R. Site-specific modulation of c-Myc cotransformation by residues phosphorylated in vivo. Oncogene. 1994;9:59–70. [PubMed] [Google Scholar]
- 89.Qureshi S A, Rim M, Bruder J, Kolch W, Rapp U, Sukhatme V P, Foster D A. An inhibitory mutant of c-Raf-1 blocks v-Src-induced activation of the Egr-1 promoter. J Biol Chem. 1991;266:20594–20597. [PubMed] [Google Scholar]
- 90.Rapp U R, Cleveland J L, Storm S M, Beck T W, Huleihel M. Transformation by raf and myc oncogenes. Princess Takamatsu Symp. 1986;17:55–74. [PubMed] [Google Scholar]
- 91.Robertson L M, Kerppola T K, Vendrell M, Luk D, Smeyne R J, Bocchiaro C, Morgan J I, Curran T. Regulation of c-fos expression in transgenic mice requires multiple interdependent transcription control elements. Neuron. 1995;14:241–252. doi: 10.1016/0896-6273(95)90282-1. [DOI] [PubMed] [Google Scholar]
- 92.Robinson M J, Cobb M H. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 93.Roche S, Fumagalli S, Courtneidge S A. Requirement for Src family protein tyrosine kinases in G2 for fibroblast cell division. Science. 1995;269:1567–1569. doi: 10.1126/science.7545311. [DOI] [PubMed] [Google Scholar]
- 94.Roussel M F. Regulation of cell cycle entry and G1 progression by CSF-1. Mol Reprod Dev. 1997;46:11–18. doi: 10.1002/(SICI)1098-2795(199701)46:1<11::AID-MRD3>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 95.Roussel M F, Cleveland J L, Shurtleff S A, Sherr C J. Myc rescue of a mutant CSF-1 receptor impaired in mitogenic signalling. Nature. 1991;353:361–363. doi: 10.1038/353361a0. [DOI] [PubMed] [Google Scholar]
- 96.Roussel M F, Davis J N, Cleveland J L, Ghysdael J, Hiebert S W. Dual control of myc expression through a single DNA binding site targeted by ets family proteins and E2F-1. Oncogene. 1994;9:405–415. [PubMed] [Google Scholar]
- 97.Roussel M F, Sherr C J. Mouse NIH 3T3 cells expressing human colony-stimulating factor 1 (CSF-1) receptors overgrow in serum-free medium containing human CSF-1 as their only growth factor. Proc Natl Acad Sci USA. 1989;86:7924–7927. doi: 10.1073/pnas.86.20.7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Roussel M F, Shurtleff S A, Downing J R, Sherr C J. A point mutation at tyrosine-809 in the human colony-stimulating factor 1 receptor impairs mitogenesis without abrogating tyrosine kinase activity, association with phosphatidylinositol 3-kinase, or induction of c-fos and junB genes. Proc Natl Acad Sci USA. 1990;87:6738–6742. doi: 10.1073/pnas.87.17.6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Roussel M F, Theodoras A M, Pagano M, Sherr C J. Rescue of defective mitogenic signaling by D-type cyclins. Proc Natl Acad Sci USA. 1995;92:6837–6841. doi: 10.1073/pnas.92.15.6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Samuels M L, Weber M J, Bishop J M, McMahon M. Conditional transformation of cells and rapid activation of the mitogen-activated protein kinase cascade by an estradiol-dependent human raf-1 protein kinase. Mol Cell Biol. 1993;13:6241–6252. doi: 10.1128/mcb.13.10.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sapi E, Flick M B, Rodov S, Kacinski B M. Ets-2 transdominant mutant abolishes anchorage-independent growth and macrophage colony-stimulating factor-stimulated invasion by BT20 breast carcinoma cells. Cancer Res. 1998;58:1027–1033. [PubMed] [Google Scholar]
- 102.Sato N, Sakamaki K, Terada N, Arai K, Miyajima A. Signal transduction by the high-affinity GM-CSF receptor: two distinct cytoplasmic regions of the common beta subunit responsible for different signaling. EMBO J. 1993;12:4181–4189. doi: 10.1002/j.1460-2075.1993.tb06102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schaap D, van der Wal J, Howe L R, Marshall C J, van Blitterswijk W J. A dominant-negative mutant of raf blocks mitogen-activated protein kinase activation by growth factors and oncogenic p21ras. J Biol Chem. 1993;268:20232–20236. [PubMed] [Google Scholar]
- 104.Schwab M. Human neuroblastoma: amplification of the N-myc oncogene and loss of a putative cancer-preventing gene on chromosome 1p. Recent Results Cancer Res. 1994;135:7–16. doi: 10.1007/978-3-642-85039-4_2. [DOI] [PubMed] [Google Scholar]
- 105.Schwartz R C, Stanton L W, Riley S C, Marcu K B, Witte O N. Synergism of v-myc and v-Ha-ras in the in vitro neoplastic progression of murine lymphoid cells. Mol Cell Biol. 1986;6:3221–3231. doi: 10.1128/mcb.6.9.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Seth A, Alvarez E, Gupta S, Davis R J. A phosphorylation site located in the NH2-terminal domain of c-Myc increases transactivation of gene expression. J Biol Chem. 1991;266:23521–23524. [PubMed] [Google Scholar]
- 107.Sherr C J. Mammalian G1 cyclins and cell cycle progression. Proc Assoc Am Physicians. 1995;107:181–186. [PubMed] [Google Scholar]
- 108.Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 1993;260:85–88. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- 109.Sklar M D, Thompson E, Welsh M J, Liebert M, Harney J, Grossman H B, Smith M, Prochownik E V. Depletion of c-myc with specific antisense sequences reverses the transformed phenotype in ras oncogene-transformed NIH 3T3 cells. Mol Cell Biol. 1991;11:3699–3710. doi: 10.1128/mcb.11.7.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Smith M R, DeGudicibus S J, Stacey D W. Requirement for c-ras proteins during viral oncogene transformation. Nature. 1986;320:540–543. doi: 10.1038/320540a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Snow E C. The role of c-myc during normal B-cell proliferation, and as B cells undergo malignant transformation. Curr Top Microbiol Immunol. 1997;224:211–220. doi: 10.1007/978-3-642-60801-8_21. [DOI] [PubMed] [Google Scholar]
- 112.Sternberg P W, Horvitz H R. Signal transduction during C. elegans vulval induction. Trends Genet. 1991;7:366–371. doi: 10.1016/0168-9525(91)90257-q. [DOI] [PubMed] [Google Scholar]
- 113.Stokoe D, McCormick F. Activation of c-Raf-1 by Ras and Src through different mechanisms: activation in vivo and in vitro. EMBO J. 1997;16:2384–2396. doi: 10.1093/emboj/16.9.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Storm S M, Brennscheidt U, Sithanandam G, Rapp U R. raf oncogenes in carcinogenesis. Crit Rev Oncog. 1990;2:1–8. [PubMed] [Google Scholar]
- 115.Takahashi F, Endo S, Kojima T, Saigo K. Regulation of cell-cell contacts in developing Drosophila eyes by Dsrc41, a new, close relative of vertebrate c-src. Genes Dev. 1996;10:1645–1656. doi: 10.1101/gad.10.13.1645. [DOI] [PubMed] [Google Scholar]
- 116.Terada K, Kaziro Y, Satoh T. Ras is not required for the interleukin 3-induced proliferation of a mouse pro-B cell line, BaF3. J Biol Chem. 1995;270:27880–27886. doi: 10.1074/jbc.270.46.27880. [DOI] [PubMed] [Google Scholar]
- 117.Thomas S M, Brugge J S. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 118.Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 119.Treisman R. Ternary complex factors: growth factor regulated transcriptional activators. Curr Opin Genet Dev. 1994;4:96–101. doi: 10.1016/0959-437x(94)90097-3. [DOI] [PubMed] [Google Scholar]
- 120.Troppmair J, Rapp U R. Apoptosis regulation by Raf, Bcl-2, and R-Ras. Recent Results Cancer Res. 1997;143:245–249. doi: 10.1007/978-3-642-60393-8_17. [DOI] [PubMed] [Google Scholar]
- 121.Twamley-Stein G M, Pepperkok R, Ansorge W, Courtneidge S A. The Src family tyrosine kinases are required for platelet-derived growth factor-mediated signal transduction in NIH 3T3 cells. Proc Natl Acad Sci USA. 1993;90:7696–7700. doi: 10.1073/pnas.90.16.7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Umbhauer M, Marshall C J, Mason C S, Old R W, Smith J C. Mesoderm induction in Xenopus caused by activation of MAP kinase. Nature. 1995;376:58–62. doi: 10.1038/376058a0. [DOI] [PubMed] [Google Scholar]
- 123.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 124.Warne P H, Viciana P R, Downward J. Direct interaction of Ras and the amino-terminal region of Raf-1 in vitro. Nature. 1993;364:352–355. doi: 10.1038/364352a0. [DOI] [PubMed] [Google Scholar]
- 125.Williams N G, Roberts T M. Signal transduction pathways involving the Raf proto-oncogene. Cancer Metastasis Rev. 1994;13:105–116. doi: 10.1007/BF00690421. [DOI] [PubMed] [Google Scholar]
- 126.Williams N G, Roberts T M, Li P. Both p21ras and pp60v-src are required, but neither alone is sufficient, to activate the Raf-1 kinase. Proc Natl Acad Sci USA. 1992;89:2922–2926. doi: 10.1073/pnas.89.7.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Williams, S. A., E. R. Gillan, E. Knoppel, J. S. Buzby, Y. Suen, and M. S. Cairo. 1997. Effects of phosphodiester and phosphorothioate antisense oligodeoxynucleotides on cell lines which overexpress c-myc: implications for the treatment of Burkitt’s lymphoma. Ann. Oncol. 8(Suppl.):25–30. [PubMed]
- 128.Woods D, Parry D, Cherwinski H, Bosch E, Lees E, McMahon M. Raf-induced proliferation or cell cycle arrest is determined by the level of Raf activity with arrest mediated by p21Cip1. Mol Cell Biol. 1997;17:5598–5611. doi: 10.1128/mcb.17.9.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wrighton C, Busslinger M. Direct transcriptional stimulation of the ornithine decarboxylase gene by Fos in PC12 cells but not in fibroblasts. Mol Cell Biol. 1993;13:4657–4669. doi: 10.1128/mcb.13.8.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xia K, Mukhopadhyay N K, Inhorn R C, Barber D L, Rose P E, Lee R S, Narsimhan R P, D’Andrea A D, Griffin J D, Roberts T M. The cytokine-activated tyrosine kinase JAK2 activates Raf-1 in a p21ras-dependent manner. Proc Natl Acad Sci USA. 1996;93:11681–11686. doi: 10.1073/pnas.93.21.11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xie W, Herschman H R. v-src induces prostaglandin synthase 2 gene expression by activation of the c-Jun N-terminal kinase and the c-Jun transcription factor. J Biol Chem. 1995;270:27622–27628. doi: 10.1074/jbc.270.46.27622. [DOI] [PubMed] [Google Scholar]
- 132.Yamauchi N, Kiessling A A, Cooper G M. The Ras/Raf signaling pathway is required for progression of mouse embryos through the two-cell stage. Mol Cell Biol. 1994;14:6655–6662. doi: 10.1128/mcb.14.10.6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yancopoulos G D, Nisen P D, Tesfaye A, Kohl N E, Goldfarb M P, Alt F W. N-myc can cooperate with ras to transform normal cells in culture. Proc Natl Acad Sci USA. 1985;82:5455–5459. doi: 10.1073/pnas.82.16.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yang B S, Hauser C A, Henkel G, Colman M S, Van Beveren C, Stacey K J, Hume D A, Maki R A, Ostrowski M C. Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activities of c-Ets1 and c-Ets2. Mol Cell Biol. 1996;16:538–547. doi: 10.1128/mcb.16.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yao B, Zhang Y, Delikat S, Mathias S, Basu S, Kolesnick R. Phosphorylation of Raf by ceramide-activated protein kinase. Nature. 1995;378:307–310. doi: 10.1038/378307a0. [DOI] [PubMed] [Google Scholar]
- 136.Zornig M, Evan G I. Cell cycle: on target with Myc. Curr Biol. 1996;6:1553–1556. doi: 10.1016/s0960-9822(02)70769-0. [DOI] [PubMed] [Google Scholar]
- 137.Zou X, Rudchenko S, Wong K, Calame K. Induction of c-myc transcription by the v-Abl tyrosine kinase requires Ras, Raf1, and cyclin-dependent kinases. Genes Dev. 1997;11:654–662. doi: 10.1101/gad.11.5.654. [DOI] [PubMed] [Google Scholar]