Abstract
We have identified thermosensitive mutants of five Schizosaccharomyces pombe replication proteins that have a mutator phenotype at their semipermissive temperatures. Allele-specific mutants of DNA polymerase δ (polδ) and mutants of Polα, two Polδ subunits, and ligase exhibited increased rates of deletion of sequences flanked by short direct repeats. Deletion of rad2+, which encodes a nuclease involved in processing Okazaki fragments, caused an increased rate of duplication of sequences flanked by short direct repeats. The deletion mutation rates of all the thermosensitive replication mutators decreased in a rad2Δ background, suggesting that deletion formation requires Rad2 function. The duplication mutation rate of rad2Δ was also reduced in a thermosensitive polymerase background, but not in a ligase mutator background, which suggests that formation of duplication mutations requires normal DNA polymerization. Thus, although the deletion and duplication mutator phenotypes are distinct, their mutational mechanisms are interdependent. The deletion and duplication replication mutators all exhibited decreased viability in combination with deletion of a checkpoint Rad protein, Rad26. Interestingly, deletion of Cds1, a protein kinase functioning in a checkpoint Rad-mediated reversible S-phase arrest pathway, decreased the viability and exacerbated the mutation rate only in the thermosensitive deletion replication mutators but had no effect on rad2Δ. These findings suggest that aberrant replication caused by allele-specific mutations of these replication proteins can accumulate potentially mutagenic DNA structures. The checkpoint Rad-mediated pathways monitor and signal the aberrant replication in both the deletion and duplication mutators, while Cds1 mediates recovery from aberrant replication and prevents formation of deletion mutations specifically in the thermosensitive deletion replication mutators.
Acquired genetic instability has been proposed to be an early event in tumor evolution (24, 25). The hypothesis that cancer cells exhibit a mutator phenotype or an increased rate of mutation has been validated by the finding that mutations in mismatch repair (MMR) genes lead to microsatellite instability and are the underlying cause of both hereditary nonpolyposis colon cancer (HNPCC) and sporadic tumors (reviewed in references 18, 23, and 26). In addition to MMR genes, mutations in other genes responsible for ensuring genomic stability may also generate a mutator phenotype (25). Thus, proteins that maintain genomic stability in normal cells, including those involved in DNA replication, repair, recombination, chromosomal segregation, transcription, and cell cycle control are prime proto-mutator candidates. Mutations in any of these genes may be an early event in tumorigenesis, allowing the generation of the multiple mutations observed in cancers.
In addition to microsatellite instability, another common source of mutation in human genetic diseases is deletion of sequences flanked by short direct repeats (19). For example, this type of deletion mutation has been documented in the genes responsible for retinoblastoma (5), α-, β-, and γδβ-thalassemias, ataxia telangiectasia, hemophilia B, Wilm’s tumor, breast cancer (BRCA-1 and BRCA-2), HNPCC (MLH-1 and MSH2), and xeroderma pigmentosa (10; also references 20 and 27 and references therein). In addition to human genetic diseases, similar spontaneous and ionizing radiation-induced deletion mutations have been characterized at the aprt locus of hamster cells (7, 14, 31).
Maintaining the integrity of genetic material during genome duplication requires normal and accurate DNA replication. In Saccharomyces cerevisiae, defects in three replication proteins, Pol3 (DNA polymerase δ [Polδ], Rad27 (homolog of mammalian FEN-1), and RPA (replication protein A) result in a mutator phenotype characterized by alterations of sequences flanked by short direct repeats in vivo (6, 37, 38). Strains containing thermosensitive (ts) alleles of Polδ (pol3-t) or RPA (rfa1-t29, rfa1-t6, and rfa1-t33) have a mutator phenotype exhibiting an elevated rate of deletion of sequences flanked by short direct repeats (6, 38). The pol3-t allele is thought to reduce the rate of lagging strand DNA synthesis, resulting in long stretches of single-stranded DNA (ssDNA) on the lagging strand template (12). Deletions are proposed to arise from DNA polymerase slippage over a ssDNA loop formed by slip mispairing between direct repeats (12, 38).
Cells containing a null mutation of RAD27 exhibit an increased rate of duplication of sequences flanked by short direct repeats (37). Rad27 functions as either a structure-specific 5′ flap endonuclease or exonuclease to process Okazaki fragments (16; see also references 2 and 21 and references therein). The observed duplications are thought to be initiated when strand displacement synthesis of Okazaki fragments occurs without the Rad27-dependent removal of the resulting 5′ flap (37). A subsequent ssDNA loop formed by slip mispairing of direct repeats on the 5′ flap or formation of a double-strand break (dsb) followed by mutagenic single-stranded annealing are two possible intermediates of duplication mutations (37).
These findings suggest that either the impairment or absence of a DNA replication protein could lead to a mutator phenotype that compromises genomic stability. To test this hypothesis and to further elucidate the molecular mechanisms underlying deletion and duplication mutations of sequences flanked by short direct repeats, we screened a panel of mutants of Schizosaccharomyces pombe replication proteins shown in Table 1 for a mutator phenotype. We identified specific ts alleles of polα, polδ, two polδ subunit genes, and a DNA ligase gene as mutators that generate deletions and a null mutant of rad2 (homolog of RAD27 and FEN-1) as a mutator that generates duplications of sequences flanked by short direct repeats. We characterized these mutators and found that the deletion and duplication mutation mechanisms are distinct but interdependent. Furthermore, the deletion and duplication mutators have differential requirements for the checkpoint Rad-mediated mechanisms to recover from aberrant replication.
TABLE 1.
Strains used in this study
| Mutant protein | Strain | Temp (°C)
|
Refer-ence | ||
|---|---|---|---|---|---|
| Permissive | Semipermissive | Nonpermissive | |||
| Wild-type | 972 | Not ts | |||
| FEN-1 | rad2Δ | Not ts | 30 | ||
| Polα | polαts11 | <21 | 21–25 | 30 | 3 |
| Polα | polαts13 | <21 | 21–25 | 30 | 3 |
| Polα | pol1-1 | <21 | 21–25 | 30 | 9 |
| Polδ | polδts1 | 21 | 25 | 30 | 11 |
| Polδ | polδts2 | 21 | 25 | 30 | 11 |
| Polδ | polδts3 | 25 | 30 | 33 | 11 |
| Polδ | cdc6-23 | 21 | 25 | 30 | 33 |
| Polδ | cdc6-121 | 25 | 30 | 33 | 33 |
| Polδ small subunit (p55) | cdc1-7 | 21 | 25 | 30 | 33 |
| Polδ-associated subunit (p54) | cdc27-K3 | 25 | 30 | 33 | 32 |
| Ligase | cdc17-K42 | 25 | 30 | 33 | 32 |
| Polɛ | cdc20-M10 | 30 | 33 | 36 | 32 |
| MCM2 | cdc19-P1 | 25 | 30 | 33 | 32 |
| MCM10 | cdc23-M36 | 21 | 25 | 30 | 32 |
| Orp1 | cdc30 | 21 | 25 | 30 | 13 |
| Rad26 | rad26Δ | Not ts | 1 | ||
| Cds1a | cds1Δ | Not ts | 29 | ||
| RecQ | rqh1Δ | Not tested | 35 | ||
S. cerevisiae Rad53.
MATERIALS AND METHODS
Genetic and cell biology techniques.
S. pombe 972 h− was used as the wild-type strain. All the fission yeast strains used in this study contain the wild-type ura4+ gene either at their endogenous locus or in the place of a deleted gene in the null mutants. Double mutants were constructed by standard genetic techniques (15) and were ensured to contain only one wild-type copy of the ura4+ gene in the strain. The rich medium (YE) and the minimal medium (PM) were prepared as described previously (15, 28). Unless otherwise stated, cells were grown in YES medium which is YE medium supplemented with 0.2 mg of adenine per ml and 0.5 mg (each) of uracil, leucine, and histidine per ml. Strains were generally propagated at 25°C unless otherwise stated. Conjugation and sporulation were also performed at 25°C.
Determination of permissive, semipermissive, and nonpermissive temperatures.
The permissive, semipermissive, and nonpermissive temperatures of each of the strains tested were determined by a variety of methods in rich medium (YES). The permissive temperature was defined as the condition under which cells grew at the same rate as the wild-type cells did and displayed very few, if any, elongated cells. The semipermissive condition was defined as the temperature at which a strain exhibited a lower growth rate and slightly elongated cells but still maintained a plating efficiency comparable to that observed at the permissive temperature. The plating efficiency was determined by counting the number of colonies resulting from plating 103 cells/plate on rich medium (YES). The nonpermissive temperature was determined to be the lowest temperature at which the plating efficiency of a strain was compromised (to <40%) relative to the colony-forming ability at the permissive temperature, thereby preventing the measurement of a mutation rate.
Comparison of viability between strains was assessed by spotting 5 μl of serially diluted cells onto YES plates incubated at various temperatures (21, 25, 30, 33, and 36°C). This method of assessing viability (the spot assay) is not as sensitive as measuring the plating efficiency but can detect differences of >5- to 10-fold between strains.
Mutator analysis.
Yeast strains shown in Table 1 were first grown on PM plates without uracil to select for cells harboring wild-type ura4+. After 4 days, whole colonies of ura+ cells were suspended in nonselective YES medium and grown overnight for approximately 6.5 generations. Log-phase cells were plated onto YES plates containing 1 mg of 5-fluoroorotic acid (5-FOA) per ml at a concentration of 106 cells/plate. After ∼5 days of growth, the number of FOAr colonies was assessed. Mutation rates per generation were calculated by using the formula adapted from the formula in reference 35: rate of mutation = 1 −e(1/n)lnRn/R0, where R0 and Rn are the proportions of ura+ cells 0 and n generations after removal from selection, respectively. R0 was taken as 1, since the progenitor cells for each experiment were ura+. The overall mutation rate was taken as the median value from ∼10 to 30 independent experiments per strain. The relative mutation rate was expressed as the fold induction compared to the wild-type value, which was 2.89 × 10−8 per generation. Unless otherwise mentioned, all strains were designated as having a mutator phenotype within 95% confidence intervals by a two-tailed t test (α = 0.05, β = 0.20).
To analyze the mutation spectra, genomic DNA was isolated from one FOAr colony per independent culture. The mutated ura4 gene was amplified by PCR using Vent DNA polymerase, analyzed by 1.5% agarose gel electrophoresis, and classified as having deletions, insertions, or no distinguishable size change (NDSC). The ura4 PCR products from mutants that displayed a different-sized PCR product were gel purified and sequenced to determine the nature of mutations generated. PCR primers (Anagen Technologies) used to amplify the ura4 gene in FOAr cells were P0 (aagcttagctacaaatcccac) and P8 (aacgcctaggaaaacaaacgc) at nucleotide positions 1 and 1406, respectively, of the ura4+ gene. The open reading frame of ura4+ resides between nucleotide positions 534 and 1320. In addition to primer P0, the PCR products were sequenced by using the primers P1 (tttcttaccgtattgtcctac), P2 (ggaccctatgtctgtgttatc), P3 (ggcgagggtattatacaaggc), and P4 (ggagacgggctgggacagcaa) at nucleotide positions 462, 696, 915, and 1152, respectively.
RESULTS
Identification of replication mutators.
It has been shown in S. cerevisiae that mutations of POL3, RPA, and RAD27 result in genomic instability. pol3-t, rfa1-t29, rfa1-t6, and rfa1-t33 generate deletions and a rad27 null strain generates duplications of sequences flanked by short direct repeats (6, 37, 38). To determine whether aberrant replication caused by mutations in other replication proteins could also lead to a mutator phenotype, we screened a bank of S. pombe mutants involved in DNA replication (Table 1) for an increased mutation rate. We used a forward mutation rate assay that detects mutations inactivating the ura4+ gene (FOAr mutations). Strains that had a mutation rate fivefold higher than that of the wild-type cells, which have a mutation rate of 2.89 × 10−8 per generation, were considered to have a mutator phenotype and will hereafter be referred to as mutators.
By this approach, we identified three ts alleles each of polα and polδ and one ts allele each of two subunits of polδ and a DNA ligase gene that exhibited an elevated mutation rate relative to that of wild-type cells at the semipermissive temperature (boldfaced values in Table 2). These mutants will hereafter be designated the ts replication mutators. As reported in a previous budding yeast rad27 study (37), deletion of rad2+ (S. pombe homolog of RAD27) also resulted in a mutator phenotype (Table 2). Together, the ts replication mutators and rad2Δ will be referred to as the replication mutators. Strains containing specific mutant alleles of polδ, two polδ subunit genes, a ligase gene and rad2Δ all exhibited a 9- to 22-fold-higher mutation rate than that of wild-type cells, while the mutation rates in cells containing mutant alleles of polα were almost 1 order of magnitude higher (90- to 186-fold) (Table 2).
TABLE 2.
Identification of replication mutants displaying a mutator phenotypea
| Mutant protein | Allele | Temp (°C) | Relative mutation rateb | % Mutation typec
|
||
|---|---|---|---|---|---|---|
| Del | Ins | NDSC | ||||
| Wild-type | Wild-type | 30 | 1.0 | 0 | 0 | 100 |
| Polα | polαts11 | 25 | 185.9 | 85 | 0 | 15 |
| polαts13 | 25 | 169.0 | 61 | 6 | 33 | |
| pol1-1 | 25 | 89.6 | 71 | 0 | 29 | |
| Polδ | polδts1 | 25 | 11.5 | 94 | 0 | 6 |
| polδts2 | 25 | ≤1.0 | ||||
| polδts3 | 30 | 22.3 | 0 | 0 | 100 | |
| cdc6-121 | 30 | 12.7 | 0 | 0 | 100 | |
| cdc6-23 | 25 | ≤1.0 | ||||
| Subunits of Polδ | cdc1-7 | 25 | 9.1 | 26 | 0 | 74 |
| cdc27-K3 | 30 | 10.1 | 24 | 0 | 76 | |
| Polɛ | cdc20-M10 | 33 | 2.0 | |||
| Ligase | cdc17-K42 | 30 | 13.5 | 48 | 0 | 52 |
| FEN-1 | rad2Δ | 30 | 16.5 | 5 | 37 | 58 |
| MCM2 | cdc19-P1 | 30 | ≤1.0 | |||
| MCM10 | cdc23-M36 | 25 | ≤1.0 | |||
| Orp1 | cdc30 | 25 | ≤1.0 | |||
All mutation assays were performed at the semipermissive temperature for each strain (Table 1) or at 30°C for the nonthermosensitive strains. See Materials and Methods for description of determination of the permissive, semipermissive, and nonpermissive growth conditions. Strains that had a mutation rate at least fivefold greater than the wild-type cells were designated as having a mutator phenotype, and their values are shown in boldface type.
The relative mutation rate is expressed as the fold induction relative to the wild-type mutation rate, which was 2.89 × 10−8 per generation.
Del, deletion; Ins, insertion; NDSC, no detectable size change.
As shown in Table 2, not all ts replication mutants had a mutator phenotype, as two alleles of polδ, polδts2, and cdc6-23 did not exhibit an elevated mutation rate relative to that of wild-type cells. These results suggest that the observed mutator phenotype is not replication protein specific but allele specific. Mutant alleles of other replication proteins that did not exhibit an increased mutation rate include Polɛ (cdc20-M10) and proteins in the prereplication complex such as Orp1 (cdc30), MCM2 (cdc19-P1), and MCM10 (cdc23-M36). Together, these results suggest that mutations in specific alleles of replication complex components can affect genomic stability.
Mutation spectra of the replication mutators.
We analyzed the nature of the mutations generated in the ura4 gene of the FOAr mutant cells. The size of the ura4 gene from the FOAr cells was determined by PCR amplification and agarose gel analysis as described in Materials and Methods. The wild-type ura4+ PCR product is 1,406 bp long. The mutators had either smaller, larger, or similar-sized PCR fragments compared to the wild-type ura4+ PCR product (designated Del, Ins, or NDSC in Table 2, respectively; see also Fig. 1). ura4 PCR products that were smaller than wild-type products were found in FOAr colonies derived from ts replication mutants pol1-1, polαts11, polαts13, polδts1, cdc1-7, cdc27-K3, and cdc17-K42, indicating that mutations at these specific alleles of these replication proteins caused deletions in the ura4+ gene. ura4 PCR products that were larger than wild-type products, indicative of insertion mutation events, were rarely found in those ts replication mutators that generated deletions. These mutants will hereafter be referred to as the ts deletion mutators (Table 2). The percentages of deletions versus total types of mutations generated by a ts replication mutator ranged from 24 in cdc27-K3 to 94 in polδts1. The observation that insertions, but very few deletions, were detected in FOAr rad2Δ mutant cells (Table 2) is similar to the mutation spectrum described for S. cerevisiae rad27 mutants (37).
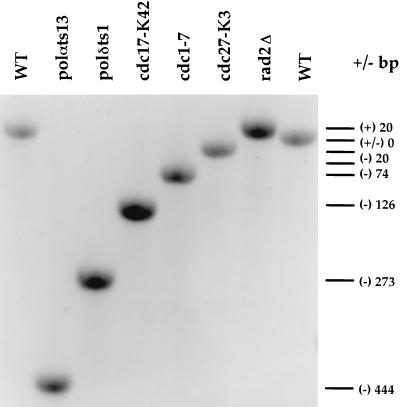
FIG. 1.
Analysis of ura4 PCR products from FOAr cells. Genomic DNA isolated from FOAr cells was amplified by PCR and analyzed by 1.5% agarose gel electrophoresis as described in Materials and Methods. The wild-type (WT) ura4 PCR product is 1,406 bp long and marked as having 0-bp alteration. The difference in size between the wild-type and other ura4 PCR products (+/− bp) is indicated to the right of the gel, with + indicating larger and − indicating smaller than the wild-type product.
In addition to deletions and insertions, some of the ura4 PCR products derived from FOAr mutators appeared to have NDSC as estimated by 1.5% agarose gel analysis. Sequence analysis of nine of these NDSC PCR products derived from each of two deletion mutators, polαts13 and cdc17-K42, showed that most of the mutations were base substitutions, and none had size changes between 3 and 17 bp (data not shown). Not all mutants displaying a mutator phenotype yielded ura4 PCR products representative of deletion or insertion mutation events. For example, polδts3 and cdc6-121 generated only NDSC ura4 PCR products (Table 2). It should be noted that in wild-type cells, only NDSC ura4 PCR products were detected. This is in agreement with the finding that base substitutions and small frameshifts (−1 and +1) are the predominant Canr mutations found in S. cerevisiae wild-type cells (37).
Sequence analysis of the smaller-than-wild-type ura4 PCR products showed a common pattern in all the observed deletions (Table 3). The pattern of deletions generated in the mutated ura4 gene in these ts replication mutators was similar to that previously described in S. cerevisiae mutators. Up to 75 bp of an artificially inserted sequence flanked by a 3- to 9-bp short direct repeat engineered in the LYS2 gene was deleted in pol3-t (38), and 8 bp to 17.7 kb of sequences flanked by short direct repeats were deleted in Canr mutants in rfa-t29 (6). In this study, we used the entire ura4+ gene as the mutagenic target reporter. The length of deleted sequence found ranged from 18 to 1,070 bp, with the median length of deleted sequence being 132 bp. The deleted sequence was flanked by short direct repeats of 4 to 12 bp (Table 3). One of the short direct repeats was always deleted, and the median size of the direct repeat involved was 7 bp. We found that 31% of deletions occurred in sequences including the initiation codon of the ura4 gene. Eleven of these deletions within the start site involved variations of the short direct repeat (aaaagcaaag) at nucleotide position 534. A 100-bp fragment including the entire direct repeat (aaaagcaaag) and a 262-bp fragment including a smaller portion of the same direct repeat (aaaagca) were deleted in five and four independent occurrences, respectively. In the remaining mutations, a 319-bp fragment including a 4-bp direct repeat (aagc) and a 100-bp fragment including a 7-bp direct repeat (aaagcaa) were deleted. It is noteworthy that deletions of sequences that inactivate the ura4 gene in FOAr cells were not limited to any specific part of the ∼790-bp open reading frame. The deletions were in a wide variety of locations, including an 18-bp deletion that is only 45 bp upstream of the ura4 termination codon. In nine instances, the direct repeats involved in the deletions were not perfectly identical, sharing all but one variant nucleotide (see short direct repeats marked by * and ** in Table 3). We found an approximately equal incidence of deletion of the first or second imperfect short direct repeat.
TABLE 3.
Spectrum of mutations observed
| Allele | Mutation typea | Repeat sequence | Repeat size (nucleotides)b | Alteration size (bp) | Positionc | Occurrenced |
|---|---|---|---|---|---|---|
| polαts13 | Del | cttagaat | 8 | 360 | 674 | 1/12 |
| Del | aaaagcaaag | 10 | 100 | 522 | 2/12 | |
| Del | aaaagca | 7 | 262 | 522 | 2/12 | |
| Del | aagc | 4 | 319 | 524 | 1/12 | |
| Del | aaaa(t/c)tgg | 7 of 8* | 74 | 690 | 1/12 | |
| Del | aatt | 4 | 138 | 679 | 1/12 | |
| Del | gtcga | 5 | 108 | 1263 | 1/12 | |
| Del | tgcttc | 6 | 444 | 431 | 1/12 | |
| Del | atc(g/a)caaatt | 9 of 10* | 80 | 811 | 1/12 | |
| Dup | tacaa | 5 | 20 | 847 | 1/12 | |
| polαts11 | Del | aaaagca | 7 | 262 | 522 | 2/9 |
| Del | tcagc | 5 | 743 | 561 | 1/9 | |
| Del | tgtttgc | 7 | 1,070 | 311 | 1/9 | |
| Del | agctggtcgt | 10 | 165 | 1079 | 2/9 | |
| Del | aaat | 4 | 138 | 679 | 1/9 | |
| Del | gacgt | 5 | 232 | 729 | 1/9 | |
| Del | tgagg | 5 | 234 | 572 | 1/9 | |
| polδts1 | Del | gctagag | 7 | 24 | 540 | 1/10 |
| Del | tggtaga(t/a)aaaa | 11 of 12* | 75 | 682 | 1/10 | |
| Del | aaaattg | 7 | 180 | 690 | 2/10 | |
| Del | aaaagcaaag | 10 | 100 | 522 | 2/10 | |
| Del | caaa | 4 | 443 | 552 | 1/10 | |
| Del | aaatcc(c/g) | 6 of 7** | 79 | 584 | 1/10 | |
| Del | cag(t/g)cg | 5 of 6** | 273 | 640 | 1/10 | |
| Del | ctttggct | 8 | 64 | 1003 | 1/10 | |
| cdc17-K42 | Del | tcctg | 5 | 75 | 1181 | 1/13 |
| Del | gtcgat | 6 | 284 | 1087 | 1/13 | |
| Del | gacgt | 6 | 232 | 729 | 1/13 | |
| Del | tggtcgt | 7 | 21 | 1226 | 2/13 | |
| Del | gaagc | 5 | 18 | 1266 | 1/13 | |
| Del | aaaagcaaag | 10 | 100 | 522 | 3/13 | |
| Del | tcaag | 5 | 126 | 715 | 1/13 | |
| Del | ctggtg | 6 | 91 | 768 | 1/13 | |
| Del | ctttggc | 7 | 64 | 1003 | 1/13 | |
| Del | tggtcgt(c/g)ga | 9 of 10** | 144 | 1082 | 1/13 | |
| cdc27-K3 | Del | cg(c/a)gg | 4 of 5* | 273 | 644 | 1/7 |
| Del | tggtcgt | 7 | 21 | 1226 | 1/7 | |
| Del | gttttctta | 9 | 333 | 460 | 1/7 | |
| Del | aaagcaa | 7 | 100 | 523 | 1/7 | |
| Del | agctggtcgt | 10 | 165 | 1079 | 1/7 | |
| Del | tcctg | 5 | 75 | 1181 | 2/7 | |
| cdc1-7 | Del | tttga(t/c) | 5 of 6** | 42 | 622 | 1/7 |
| Del | agctggtcgt | 10 | 165 | 1079 | 1/7 | |
| Del | tggtcgt | 7 | 21 | 1226 | 1/7 | |
| Del | gaagc | 5 | 18 | 1266 | 1/7 | |
| Del | tcctg | 5 | 75 | 1181 | 1/7 | |
| Del | gaaa(a/t) | 4 of 5** | 222 | 762 | 1/7 | |
| Del | ctcttt | 6 | 511 | 490 | 1/7 | |
| rad2Δ | Dup | atatc | 5 | 41 | 1172 | 1/7 |
| Dup | aaattgc | 7 | 25 | 871 | 1/7 | |
| Dup | ggct | 4 | 22 | 1137 | 1/7 | |
| Del | cttagaat | 8 | 360 | 674 | 1/7 | |
| Dup | tacaa | 5 | 20 | 847 | 2/7 | |
| Dup | attgt | 5 | 27 | 1194 | 1/7 |
Del, deletion; Dup, duplication.
One asterisk indicates that the first repeat (with variant nucleotide listed to the left of the slash) is deleted and the second repeat (with variant nucleotide listed to the right of the slash) is retained. Two asterisks indicate that the second repeat is deleted and the first repeat is retained.
The nucleotide coordinates of the wild-type genomic fragment of the ura4 gene (SPURA4) are used with the first nucleotide as nucleotide 1. The initiation codon is located at position 534.
Occurrence indicates the number of independent times the mutation was observed.
Analysis of the insertion events in rad2Δ and in the one case observed in polαts13 revealed that the pattern of insertion was similar to that described in a S. cerevisiae rad27 null mutant (37). The insertion mutation resulted in a duplication of one of the 4- to 7-bp short direct repeats and sequences flanked by the short direct repeats (Table 3). The size of the duplicated sequence ranged from 20 to 41 bp, and the median size of duplicated sequence was 22 bp. These results indicate that S. pombe rad2Δ generates the same mutator phenotype characterized by duplication of sequences flanked by short direct repeats as deletion of its S. cerevisiae homolog RAD27.
Deletion mutator phenotype is dependent upon semipermissive growth conditions.
We further characterized the replication mutators to elucidate the deletion and duplication mechanisms. Studies of an S. cerevisiae pol3-t mutant have shown an eightfold increase of the mutation rate at 30°C over that at 21°C, suggesting that Polδ has an increased mutation rate at the semipermissive temperature (38). We thus tested the effects of temperature on the mutation rates of the ts deletion replication mutators (polαts11, polαts13, polδts1, cdc1-7, cdc27-K3, and cdc17-K42), a ts replication nondeletion mutator (polδts3), and a ts replication nonmutator (polδts2) at 21, 25, or 30°C. Mutation rates of wild-type cells were similar at 25, 30, and 37°C (data not shown). The ts replication mutators polαts11, polαts13, polδts1, and cdc1-7 that displayed an elevated mutation rate relative to that of wild-type cells at 25°C had a lower mutation rate at 21°C. At 21°C, the mutation rates in polδts1 and cdc1-7 were reduced ca. five- to ninefold to near-wild-type levels, and the mutation rates in polαts11 and polαts13 were reduced from ∼180- to ∼50-fold higher than that of wild-type cells (compare Tables 2 and 6). At 25°C, cells harboring the polαts11, polαts13, polδts1, or cdc1-7 allele were slightly elongated, which is indicative of a mild replication perturbation and 25°C being the semipermissive condition for these mutants (3; also data not shown). At 21°C, cells harboring the polαts11 or polαts13 allele displayed a higher mutation rate than the wild-type cells did, indicating that 21°C is still a semipermissive temperature for these two mutants. Analogously, regardless of their ability to generate deletions, mutants such as cdc17-K42, cdc27-K3, polδts3, and cdc6-121 mutants that exhibited a mutator phenotype at 30°C had decreased mutation rates to near-wild-type levels at 25°C (compare Tables 2 and 6). Viability analysis indicated that 30°C is the semipermissive temperature of these replication mutators (Table 1). The mutation rate of a ts nonmutator (polδts2) remained at the wild-type rates at 21, 25, and 28°C (Table 2 and data not shown). Cells harboring rad2Δ are not temperature-sensitive (30), and thus, the associated mutation rate did not change in a temperature-dependent manner (data not shown). These results indicate that the observed increased mutation rate in all the ts replication mutators is dependent upon semipermissive growth conditions and confirms that the mutated replication protein is the source of the mutator phenotype.
TABLE 6.
Absence of Cds1 exacerbates deletion mutator phenotype
| Allele | Temp (°C) | Relative mutation rate in cellsa
|
|
|---|---|---|---|
| cds1+ | cds1Δ | ||
| Wild-type | 30 | 1.0 | ≤1.0 |
| polαts11 | 21 | 50.7 | SL |
| polαts13 | 21 | 52.4 | − |
| polδts1 | 21 | 1.3 | 24.3 |
| polδts2 | 25 | ≤1.0 | ≤1.0 |
| polδts3 | 25 | 2.4 | 12.2 |
| cdc6-121 | 25 | 4.4 | 16.0 |
| cdc1-7 | 21 | 2.0 | 18.6 |
| cdc27-K3 | 25 | ≤1.0 | 5.9 |
| cdc17-K42 | 25 | 2.0 | 8.1 |
| rad2Δ | 30 | 16.5 | 11.4 |
The relative mutation rate is the rate compared to that of wild-type cells. SL, synthetic lethal; −, grew too poorly to be assayed.
Formation of deletion mutations requires Rad2.
The differential mutation spectra associated with the ts replication mutators and rad2Δ suggest that the processes of generating deletions and duplications are distinct. However, the two mechanisms may be related, since proteins mutated in both pathways are directly involved in DNA replication and the sequence that is altered is always flanked by short direct repeats. To test whether the underlying mechanisms responsible for generating deletions and duplications by these replication mutators are interdependent, we constructed double mutants of each of the deletion-generating ts replication mutators in a rad2Δ background. Deletion of rad2 had no effect on the viability or restrictive temperature of all the double mutants tested (data not shown). We then compared the mutation rate of the ts replication mutators in a rad2+ or rad2Δ background at the semipermissive temperature of each respective ts replication mutator (Table 4). Surprisingly, the mutation rates of polαts11 and polαts13 decreased 4.6- to 7.1-fold, respectively, in a rad2Δ background. It was difficult to evaluate the effect of rad2Δ on the mutation rate of the other ts replication mutators within 95% confidence levels, since these ts replication mutators and rad2Δ had similar mutation rates which were 9- to 22-fold higher than that of wild-type cells (Table 4). However, agarose gel analysis of the FOAr PCR products revealed that all the ts deletion mutators in a rad2Δ background had reduced percentages of deletion mutations (Table 4). For example, in a rad2+ background, mutations generated by polδts1 were 94% deletions and 6% NDSCs (Tables 2 and 4), whereas in a rad2Δ background, deletions in polδts1 were no longer detectable (Table 4). Finding decreases in both the mutation rates in polαts11 and polαts13 and in the percentages of deletion mutations formed in all the ts deletion mutators in a rad2Δ background indicates that Rad2 is required to generate the observed deletion mutations in the ts replication mutators.
TABLE 4.
Mutation rates and types of mutations of ts replication mutators in rad2+ and rad2Δ backgroundsa
| Allele | Temp (°C) | Relative mutation ratebc
|
% Mutation type in cellscd
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Del
|
Dup
|
NDSC
|
|||||||
| rad2+ | rad2Δ | rad2+ | rad2Δ | rad2+ | rad2Δ | rad2+ | rad2Δ | ||
| Wild-type | 30 | 1.0 | 16.5 | 0 | 5 | 0 | 37 | 100 | 58 |
| polαts11 | 25 | 185.9 | 40.6 | 85 | 6 | 0 | 6 | 15 | 88 |
| polαts13 | 25 | 169.0 | 23.7 | 61 | 5 | 6 | 0 | 33 | 95 |
| polδts1 | 25 | 11.5 | 6.7 | 94 | 0 | 0 | 14 | 6 | 86 |
| polδts2 | 25 | ≤1.0 | 20.3 | ND | |||||
| polδts3 | 30 | 22.3 | 19.0 | ND | |||||
| cdc1-7 | 25 | 9.1 | 12.7 | 26 | 0 | 0 | 0 | 74 | 100 |
| cdc27-K3 | 30 | 10.1 | 13.2 | 24 | 6 | 0 | 6 | 76 | 88 |
| cdc17-K42 | 30 | 13.5 | 6.1 | 48 | 7 | 0 | 33 | 52 | 60 |
All mutation assays were performed at the semipermissive temperature (see Table 1) for each strain or at 30°C for the wild-type strain.
The relative mutation rate is the rate compared to that of wild-type cells.
Mutation rates and percentages of mutations in a rad2Δ background are shown in boldface. The remaining entries are from Table 2.
Del, deletion; Dup, duplication; NDSC, no detectable size change; ND, not determined.
Formation of duplication mutations in rad2Δ requires normal DNA polymerization, but not DNA ligation.
We also measured the duplication mutation rate of rad2Δ in all the ts replication mutator backgrounds. With the exception of the ligase mutator (cdc17-K42), the duplication mutation rate of rad2Δ was reduced in a ts replication mutator background. The percentages of duplication mutations decreased from 37 in the rad2Δ single mutant to 0 to 6 in a polαts11, polαts13, cdc1-7, or cdc27-K3 background and to 14 in a polδts1 background (Table 4). These results suggest that duplication mutations caused by rad2Δ are dependent upon normal DNA polymerization, but not upon DNA ligation. Together, these results indicate that the deletion and duplication mutation mechanisms are distinct but interdependent.
Cds1 is required for growth and viability of the ts replication mutators, but not the duplication mutator rad2Δ.
All the replication mutators described in this study involve proteins that directly function at the replication fork, suggesting that these replication mutators generate aberrant replication structures at the semipermissive temperature. We thus investigated how the cell responds to prevent cell death and mutation formation from aberrant replication(s). To this end, we analyzed these replication mutators in a cds1Δ background (Table 5). In response to hydroxyurea (HU) or DNA damage, the protein kinase Cds1 enables cells to arrest S phase reversibly through a checkpoint-Rad mediated S-phase recovery pathway (22). Cds1 is phosphorylated and activated by S-phase arrest (22). Our previous studies have shown that Cds1 protein kinase is activated in polαts11 and polαts13 mutants at the semipermissive temperature (25°C), suggesting that these mutants have aberrant replication structures that activate Cds1 (3). Furthermore, Cds1 function is required for viability of mutant cells containing polαts alleles, since cells with polαts11 or polαts13 in a cds1Δ background at 25°C are either inviable or grow poorly (3). We therefore tested the effect of deleting cds1+ in the replication mutators.
TABLE 5.
Growth of replication mutants in rad26Δ and cds1Δ backgrounds
| Allele | Temp (°C) | Mutator?a | Mutation typeb | Viability in backgroundc
|
|
|---|---|---|---|---|---|
| rad26Δ | cds1Δ | ||||
| Wild-type | 30 | No | +++ | +++ | |
| polαts-11 | 25 | Yes | Del | SL | SL |
| polαts-13 | 25 | Yes | Del | SL | − |
| polδts1 | 25 | Yes | Del | SL | +/− |
| cdc1-7 | 25 | Yes | Del | SL | +/− |
| cdc27-K3 | 30 | Yes | Del | − | +/− |
| cdc17-K42 | 30 | Yes | Del | − | − |
| polδts3 | 30 | Yes | NDSC | SL | − |
| cdc6-121 | 30 | Yes | NDSC | SL | +/− |
| rad2Δ | 30 | Yes | Dup | − | +++ |
| polδts2 | 25 | No | − | +++ | |
| cdc6-23 | 25 | No | SL | +++ | |
| cdc20-M10 | 33 | No | ND | +++ | |
| cdc19-P1 | 30 | No | ND | +++ | |
| cdc23-M36 | 25 | No | ND | +++ | |
| cdc30 | 30 | No | ND | +++ | |
Strains that had a mutation rate at least fivefold greater relative to that of wild-type cells were designated as having a mutator phenotype.
Del, deletion; Dup, duplication; NDSC, no detectable size change.
Viability was measured by spotting 5-μl amounts of serially diluted cells onto YES (rich medium) plates incubated at various temperatures (see Materials and Methods). SL, synthetic lethal at 25°C; +++, viability indistinguishable from that of either of the parental strains; +/−, ∼10- to 100-fold-lower viability of the double mutant relative to that of either of the parental strains; −, ∼100- to 1,000-fold-lower viability of the double mutant relative to that of either of the parental strains. ND, not determined.
The viability of each replication mutator in a cds1Δ background was assessed in rich medium at various temperatures, since cds1Δ single mutant were not temperature-sensitive. In a cds1Δ background, the viability of the ts replication mutators was reduced at the semipermissive temperature of the respective parental strains (Table 5). With the exception of the polαts mutants, the other ts replication mutators had normal viability in a cds1Δ background at the permissive temperature but were unable to grow at the semipermissive temperature of the parental strains. Thus, deletion of Cds1 reduced the restrictive (and semipermissive) temperatures of the ts replication mutators. In contrast, deletion of cds1+ had no effect on the viability or restrictive temperature of those ts replication mutants that did not exhibit a mutator phenotype, which included polδts2, cdc6-23, cdc20-M10, cdc19-P1, cdc23-M36, and cdc30 (Table 5). Thus, all the ts replication mutators, but not the nonmutators, required Cds1 for viability or normal growth (Table 5). These results indicate that Cds1 responds to the specific aberrant replication generated in the ts replication mutators but not the ts replication nonmutators.
Interestingly, deletion of cds1+ had no effect on the viability of rad2Δ cells (Table 5), suggesting that Cds1 does not respond to the aberrant DNA structures generated in rad2Δ cells. The specific requirement of Cds1 for the ts replication mutators but not rad2Δ suggests that the deletion and duplication mutations are processed distinctly.
Absence of Cds1 exacerbates the deletion, but not duplication, mutator phenotype.
We next analyzed the mutation rate of the ts replication mutators in a cds1Δ background. Since cds1Δ did not have a mutator phenotype (Table 6) and deletion of cds1+ reduced the semipermissive temperature of all the ts replication mutators (Table 5), analyses were performed at the permissive temperature of the respective ts replication mutators. The polαts11 cds1Δ double mutant was inviable, and the polαts13 cds1Δ double mutant grew too poorly to be assessed at 21°C. In a cds1Δ background, the mutation rates of ts deletion replication mutators, polδts1, cdc1-7, cdc27-K3, and cdc17-K42, increased 3.6- to 18.5-fold over that in a cds1+ background (Table 6). Thus, at the permissive temperature, Cds1 plays a role in preventing formation of mutations in these ts replication mutators. Similar to the effect of Cds1 on viability, deletion of cds1+ also did not have any effect on the mutation rate in the duplication mutator rad2Δ or ts nonmutator polδts2 (Table 6).
Cds1 function was also required for viability and to prevent further exacerbation of the mutator phenotype in the non-deletion-causing ts replication mutators (polδts3 and cdc6-121) (Tables 5 and 6). Neither polδts3 nor cds1Δ single mutant exhibited deletion mutations (Table 2). Interestingly, in a cds1Δ background, polδts3 was able to form deletion mutations, exhibiting 50% deletion and 50% NDSC mutations (data not shown). Thus, at the semipermissive temperature (30°C), polδts3 requires Cds1 to recover from the aberrant replication that can result in a NDSC mutator phenotype. At 25°C, the permissive temperature of polδts3, the few aberrant replication structures generated become further destabilized, resulting in deletion mutations in the absence of Cds1.
All replication mutators require checkpoint Rad proteins for viability.
Incomplete replication is monitored by the checkpoint Rad proteins either to activate Cds1 to ensure a reversible S-phase arrest (intra-S checkpoint) or to activate Chk1 and Cds1 to establish a G2 arrest (S-to-M checkpoint) (4, 22, 40). Finding that deletion of cds1+ decreased the restrictive temperature (Table 5) and enhanced the mutation rate of the ts replication mutators (Table 6) led us to test whether deletion of a Rad protein, Rad26, would similarly affect these replication mutators. Six checkpoint Rad proteins, Rad1, Rad3, Rad9, Rad17, Rad26, and Hus1, form a “guardian” complex that establishes the DNA damage and replication checkpoints, and loss of any single component results in a nonfunctional complex (1). As expected, all the ts replication mutators were either inviable or grew poorly in a rad26Δ background at the semipermissive temperature (Table 5). Similar to cds1+, deletion of rad26+ also exacerbated the mutator phenotype of two ts replication deletion mutators tested. The mutation rates of cdc27-K3 and cdc17-K42 increased ca. fourfold in a rad26Δ background at the permissive temperature (25°C) over those of the single mutants (data not shown). This suggests that aberrant replication in these two ts replication mutants is monitored through the checkpoint Rad proteins to activate Cds1 to prevent cell death and mutation formation at the semipermissive and permissive temperatures, respectively. However, the requirement for Rad26 is not restricted to the ts replication mutators. Two nonmutator Polδ mutants, polδts2 and cdc6-23, were also inviable in a rad26Δ background at their semipermissive temperatures (Table 5). This indicates that many types of replication defects are monitored through the checkpoint Rad proteins.
Deletion of rad26+, but not cds1+, reduced the growth rate (data not shown) and viability of rad2Δ cells (Table 5). This indicates that although aberrant DNA structures generated in rad2Δ cells are monitored by the checkpoint Rad proteins, they do not activate the Cds1-dependent pathway to maintain normal growth. This led us to test whether the viability of rad2Δ may be maintained by the Rqh1-dependent recovery pathway. rqh1+ encodes a putative DNA helicase with homology to the gene products of the human BLM and WRN genes and S. cerevisiae SGS1 genes (35). Genetic studies indicate that rqh1+, like BLM, functions to prevent inappropriate recombination. In response to S-phase arrest by HU or DNA damage, the checkpoint Rad proteins are thought to also activate a Rqh1-dependent process to prevent inappropriate recombination (35). The rad2Δ rqh1Δ double mutant was inviable (data not shown), suggesting that cells require Rqh1, but not Cds1, for recovery from aberrant replication structures generated in the absence of Rad2. All the ts replication mutators and some nonmutators, such as cdc6-23, cdc20-M10, and cdc23-M36, were also inviable in the rqh1Δ background at their respective semipermissive temperatures (data not shown). Thus, the requirement for Rqh1 is common, but not specific, to all the replication mutators. Unlike deletion of Cds1 or Rad26, deletion of Rqh1 did not elevate the mutation rate of the ts replication mutators at the permissive temperature (data not shown). Together, these results suggest that Rqh1 responds to the aberrant replication structures in all replication mutators, whereas Cds1 responds specifically to the aberrant replications generated in the ts replication mutators.
DISCUSSION
In this study, we have identified mutant alleles of S. pombe replication proteins that confer a mutator phenotype. Characterization of these mutators reveal the following interesting and novel findings. (i) ts alleles of polα, polδ, two polδ subunit genes, cdc1 and cdc27, and a DNA ligase gene cause aberrant replication that induces a mutator phenotype characterized by deletions of sequences flanked by short direct repeats. (ii) Generation of deletion mutations in these ts replication mutators requires a functional Rad2. (iii) Deletion of rad2+ results in an increased rate of duplication of sequences flanked by short direct repeats that requires normal DNA polymerization, but not DNA ligation. (iv) The checkpoint Rad proteins are required to prevent cell death from aberrant replication generated in the deletion and duplication mutators. (v) Cds1 is required to maintain cell viability and prevent deletion mutation formation specifically in the ts replication mutators, but not in the rad2Δ duplication mutator.
ssDNA gap initiates formation of deletion mutations in a Rad2-dependent manner.
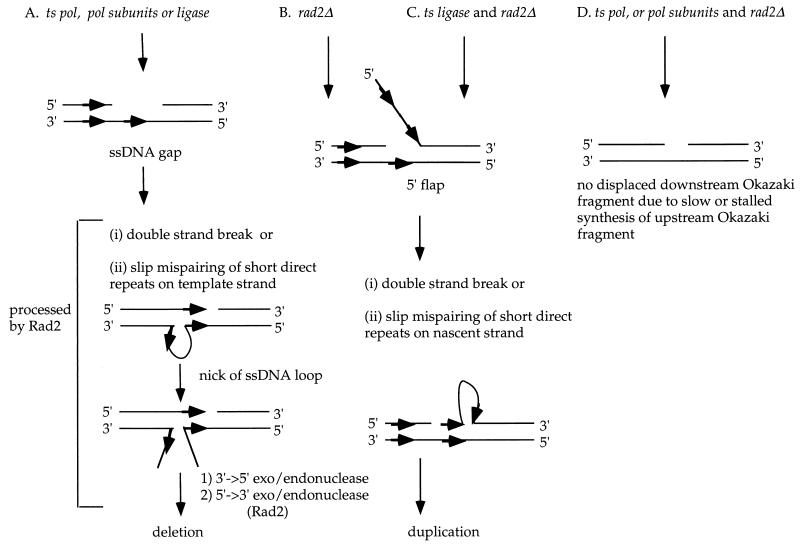
Studies of S. cerevisiae pol3-t have suggested that the observed deletion of sequences flanked by short direct repeats depends on their orientation relative to the origin of replication and are specific to either the leading or lagging strand, but not both (38). Simian virus 40 reconstituted replication studies have shown that Polα, ligase, and FEN-1 (mammalian homolog of Rad2) specifically function in lagging strand DNA synthesis, while Polδ functions in both leading and lagging strand synthesis (39). Our finding that mutant alleles of polα, polδ, two polδ subunit genes, a ligase gene and deletion of rad2 destabilize sequences flanked by short direct repeats indicates that the observed sequence alterations in these replication mutators occur predominantly during lagging strand synthesis. These mutant alleles could cause a decreased rate of lagging strand synthesis or a stalled replication fork, yielding a ssDNA gap on the template similar to the proposed mutational mechanism in S. cerevisiae pol3-t (12, 38) (Fig. 2A). The absence of ligase function results in a nick that may be resected into a ssDNA gap (Fig. 2A). Deletions are proposed to arise from DNA polymerase slippage over a ssDNA loop formed by slip mispairing of short direct repeats on the template strand (12, 38).
FIG. 2.
Proposed mechanisms for formation of deletion and duplication mutations and their interdependency in replication mutators. Two adjoining Okazaki fragments are shown, with thick arrows representing short direct repeats. (A) A ssDNA gap generated by a slow or stalled replication fork in ts DNA polymerases or by a resected nick in ts ligase mutant yields a potentially mutagenic structure that can form deletions. The ssDNA gap may initiate deletion mutation formation via a dsb or upon replication slippage over a ssDNA loop formed by slip mispairing of short direct repeats on the template strand. Following are two proposed roles (21) for Rad2 in processing a deletion mutation intermediate: (i) Rad2 may play a role in homology-dependent DNA end joining in dsb repair. After DNA ends are aligned by terminal microhomology, DNA on each strand beyond the short stretches of homology would be 3′ and 5′ flaps that would have to be cleaved off. Rad2 is the only known 5′ flap endonuclease and the only known 5′→3′-specific double-stranded DNA exonuclease activity in higher eukaryotes and thus, may be involved in resolving these structures (reviewed in reference 21). (ii) Deletion formation may involve a ssDNA loop as an intermediate and is shown in panel A. Initial nicking of the ssDNA loop by an unknown enzyme would result in 3′ and 5′ flaps that would have to be cleaved off. Rad2 may be required to remove the 5′ flaps to generate deletion mutations. (B) Duplication mutations are initiated when strand displacement synthesis of Okazaki fragments occur without Rad2-dependent removal of the resulting 5′ flap. The 5′ flap yields duplication mutations via a dsb or upon replication slippage over a ssDNA loop formed by slip mispairing of short direct repeats on the nascent strand as previously proposed (37) and described in the Discussion. (C) In a ts ligase mutant, displacement of the downstream Okazaki fragment occurs to generate a potentially mutagenic 5′ flap. The absence of Rad2 to process the displaced 5′ flap structure initiates duplication mutations as described above for panel B. (D) In a double mutant of ts polymerase and rad2Δ, a semidisabled polymerase has a reduced potential to displace a 5′ flap in the downstream Okazaki fragment. Decreased levels of displaced 5′ flap structures result in a reduced duplication mutation rate as described in the Discussion.
The deletion mutation rate of the ts replication mutators decreased in a rad2Δ background, suggesting that formation of deletion mutations is dependent upon a functional Rad2. Rad2 is a structure-specific exo- and/or endonuclease that is involved in processing Okazaki fragments (reviewed in references 2 and 21), and in S. pombe, it also functions in a recombination UV dimer endonuclease-dependent repair pathway (30). We suggest that Rad2 may be involved in the resolution of intermediates of a deletion mutation. Rad2 may be required to process the 5′ flaps generated (i) after a ssDNA loop formed by slip mispairing of direct repeats is nicked or (ii) during homology-dependent end joining (reviewed in reference 21) (Fig. 2A). Alternatively, the presence of a 5′ flap due to the absence of Rad2 may reduce either the frequency or size of the ssDNA gap on the template strand of the ts replication mutators. Thus, the absence of Rad2 would result in a reduced deletion mutation rate in the ts replication mutators (Fig. 2A).
Duplication mutations in rad2Δ are dependent upon normal DNA polymerization, but not DNA ligation.
The duplication mutations observed in S. cerevisiae rad27 mutants are proposed to be initiated by strand displacement of a 5′ flap on a downstream Okazaki fragment by synthesis of the upstream Okazaki fragment. The similarity of the duplication mutator phenotypes of S. pombe rad2Δ and S. cerevisiae rad27 mutants suggests that the source of the duplication mutations observed in rad2Δ is likely to also be initiated by a 5′ flap (Fig. 2B).
With the exception of the ligase mutator (cdc17-K42), the duplication mutation rate in rad2Δ was reduced in a ts replication mutator background (Table 4). This suggests that formation of duplications in rad2Δ cells requires wild-type replication polymerases but not DNA ligase. These results are reminiscent of a recent report showing that stimulation of microsatellite instability by a rad27 mutant was reduced in a pol3-t mutant background in S. cerevisiae (17). We suggest that the decreased duplication mutation rate of rad2Δ cells harboring a polymerase mutation is due to a reduced rate of synthesis of an upstream Okazaki fragment by the polymerase mutants. Slowed or stalled synthesis of an upstream Okazaki fragment reduces the likelihood of strand displacement of a downstream Okazaki fragment to generate a 5′ flap DNA structure that initiates duplication mutations (Fig. 2D). This would result in the observed decreased duplication mutator phenotype of rad2Δ cells in a polymerase mutant background (Fig. 2D).
Interestingly, the rate of duplication mutations generated in rad2Δ cells was not reduced in a mutant ligase background (cdc17-K42). In the absence of DNA ligase, normal synthesis of the upstream Okazaki fragment by DNA polymerases and subsequent displacement of the downstream Okazaki fragment to form a 5′ flap structure in rad2Δ cells occur. Thus, there is no effect on the duplication mutation rate of the cdc17-K42 alele in rad2Δ cells (Fig. 2C). The reductional effects of the ts polymerases, but not ts ligase, on the rates of duplication mutations in rad2Δ substantiates the hypothesis that the 5′ flap initiates the observed duplication mutations (Fig. 2B).
Cds1 responds specifically to aberrant replication induced in the ts replication mutators.
Our finding that in addition to DNA polymerases, mutations in DNA ligase could result in a mutator phenotype suggests that the observed deletion mutations are induced by aberrant replication rather than solely by polymerase slippage. The aberrant replication in rad2Δ cells that induces duplication mutations is distinct from that of the ts replication mutators. However, cells with either type of aberrant replication in a rad26Δ background are inviable (Table 5). This suggests that the checkpoint Rad proteins respond to both types of aberrant replication in the deletion and duplication mutators. The loss of viability in a rqh1Δ background indicates that the checkpoint Rad-mediated Rqh1-dependent pathway is required to prevent inappropriate recombination (35) and maintain viability from aberrant replication in the deletion and duplication mutators. However, Rqh1 does not seem to be involved in deletion mutation production because the absence of Rqh1 does not affect the mutation rate of the ts replication mutators (data not shown) and the requirement for Rqh1 is not specific to the replication mutators (data not shown).
The specific requirement of Cds1 kinase for the ts replication mutators demonstrates, for the first time, the involvement of a cell cycle checkpoint kinase in preventing the occurrence of deletion mutations and maintaining the viability of the ts replication mutators. In response to S-phase perturbation, Cds1 is activated to ensure a reversible S-phase arrest (intra-S checkpoint), thereby preventing the accumulation of unrepairable DNA lesions (22). Cds1 is thought to respond specifically to aberrant replication structures in S phase (22). Rad53 (S. cerevisiae homolog of Cds1) has been proposed to stabilize replication structures under conditions of replication inhibition and function in a DNA replication-block stress-response pathway (8). This may be accomplished by Rad53 preventing inappropriate firing of late origins by blocking recruitment of RPA to origins upon exposure to HU (36). Rad53 has also been shown to prevent early firing of late origins during undisturbed S phase (34).
At the semipermissive temperature, deletion of cds1+ reduced the viability of the ts replication mutators (Table 5). The ts replication mutator cells displayed an elongated cell morphology at the semipermissive temperature (data not shown), which is indicative of aberrant replication. This suggests that Cds1 is required to allow cells to recover from aberrant replication induced under semipermissive conditions, perhaps by preventing firing of late origins. Thus, in the absence of Cds1, cell death results from the inability to recover from a multitude of aberrant replication. This may be analogous to the primary lethal defect in a rad53 null strain being an inability to complete chromosomal replication upon exposure to HU (8). In the presence of Cds1 at the semipermissive temperature, some aberrant replication structures escape the Cds1-dependent recovery mechanism and thus result in deletion mutations. At the permissive temperature, the ts replication mutators displayed a normal cell morphology, suggesting the presence of none or very few aberrant replication structures. Under permissive conditions, deletion of cds1+ did not noticeably affect viability but did exacerbate the mutation rate of all the ts replication mutators (Table 6). Furthermore, in the absence of Cds1, deletions were generated in polδts3, which is a non-deletion-generating mutator, at the permissive temperature (Table 2 and data not shown). This suggests that at the permissive temperature, Cds1 is required to prevent formation of deletion mutations. The seemingly dual effects of Cds1 in preventing cell death and formation of deletion mutations at the semipermissive and permissive temperatures, respectively, may be explained by the cells’ response to different degrees of aberrant replication occurring at the various conditions.
In S. pombe, the protein kinases Chk1 and Cds1 appear to jointly enforce the S-to-M checkpoint (distinct from the intra-S checkpoint described above) in response to HU in a checkpoint Rad-dependent manner (4, 40). In addition, it has been reported that following exposure to HU, Cds1 phosphorylates Wee1 to establish a cell cycle checkpoint (4). Unlike Cds1, we found that effects on viability upon deletion of Chk1 or mutation of Wee1 did not correlate with the presence or absence of a mutator phenotype in the ts replication mutants (data not shown). Together, our results suggest that the roles of Cds1 in maintaining viability and preventing mutation formation are related to its function in allowing cells to recover from an aberrant replication, not by enforcing the S-to-M cell cycle checkpoint. Similarly, the lethality in rad53 null strains exposed to HU is not the result of inappropriate entry into mitosis but the inability to complete replication (8). The correlation of the deletion mutator phenotype with specific biochemical defects in replication fork progression indicates that any mutations in proteins that disrupt, destabilize, or stall replication fork progression have the potential to be mutators. Our study also predicts that mutants that require Cds1 for viability are likely to have a mutator phenotype.
Interestingly, deletion of Cds1 had no effect on either the growth or mutation rate of the duplication mutator rad2Δ (Tables 5 and 6). Thus, the Cds1-dependent pathway does not respond to a potentially mutagenic 5′ flap structure generated in the rad2Δ duplication mutator. Together, these results strongly suggest that different checkpoint Rad-mediated subpathways respond to different aberrant replication structures in order to maintain viability and prevent mutation formation.
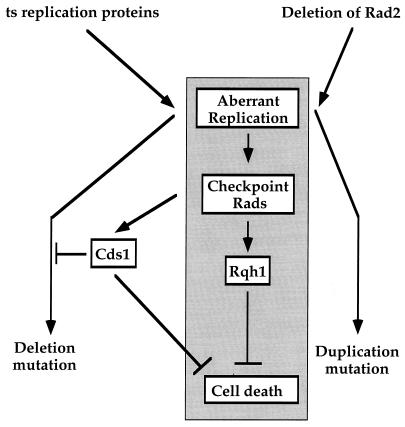
A proposed model of how deletion and duplication mutations in the replication mutators are processed by the checkpoint Rad-dependent S-phase recovery pathway is illustrated in Fig. 3.
FIG. 3.
How checkpoint Rad proteins and Cds1 respond to aberrant replication in the deletion and duplication mutators. ts replication proteins at the semipermissive temperature and deletion of Rad2 generate different types of aberrant replication structures that are monitored by checkpoint Rad proteins. Checkpoint Rad proteins activate Rqh1 to prevent inappropriate recombination and cell death from both types of aberrant replication structures. In response to prevalent levels of aberrant replication specifically in ts replication mutators at the semipermissive temperature, checkpoint Rad proteins activate the Cds1-dependent recovery subpathway to prevent cell death and formation of deletion mutations. Some aberrant replication structures escape the Cds1-dependent recovery mechanism and thus result in deletion mutations. In the presence of lower levels of aberrant replication at the permissive temperature, Cds1 prevents formation of deletion mutations in the ts replication mutators. Details of these checkpoint responses are described in the Discussion.
ACKNOWLEDGMENTS
We thank A. M. Carr for providing us with the cds1Δ and rad26Δ strains, S. Forsburg for providing most of the parental cdc strains, and T. Enoch for providing the rqh1Δ strain. We also thank Rose Borbely for excellent technical help and Alison Miyamoto and members of the Wang lab for helpful discussions and critical reading of the manuscript.
This work was supported in part by a grant from NIH (CA14835).
REFERENCES
- 1.Al-Khodairy F, Fotou E, Sheldrick K S, Griffiths D J F, Lehmann A R, Carr A M. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol Biol Cell. 1994;5:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bambara R A, Murante R S, Henricksen L A. Enzymes and reactions at the eukaryotic DNA replication fork. J Biol Chem. 1997;272:4647–4650. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- 3.Bhaumik D, Wang T S-F. Mutational effect of fission yeast Polα on cell cycle events. Mol Biol Cell. 1998;9:2107–2123. doi: 10.1091/mbc.9.8.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boddy M N, Furnari B, Mondesert O, Russell P. Replication checkpoint enforced by kinases Cds1 and Chk1. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- 5.Canning S, Dryja T P. Short direct repeats at the breakpoints of deletions of the retinoblastoma gene. Proc Natl Acad Sci USA. 1989;86:5044–5048. doi: 10.1073/pnas.86.13.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C, Umezu K, Kolodner R D. Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Mol Cell. 1998;2:9–22. doi: 10.1016/s1097-2765(00)80109-4. [DOI] [PubMed] [Google Scholar]
- 7.de Jong P J, Grosovsky A J, Glickman B W. Spectrum of spontaneous mutation at the APRT locus of Chinese hamster ovary cells: an analysis at the DNA sequence level. Proc Natl Acad Sci USA. 1988;85:3499–3503. doi: 10.1073/pnas.85.10.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desany B A, Alcasabas A A, Bachant J B, Elledge S J. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D’Urso G, Grallert B, Nurse P. DNA polymerase alpha, a component of the replication initiation complex, is essential for the checkpoint coupling S phase to mitosis in fission yeast. J Cell Sci. 1995;108:3109–3118. doi: 10.1242/jcs.108.9.3109. [DOI] [PubMed] [Google Scholar]
- 10.Efstratiadis A. The structure and evolution of the human β-globin gene family. Cell. 1980;21:653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- 11.Francesconi S, Park H, Wang T S-F. Fission yeast with DNA polymerase delta temperature-sensitive alleles exhibits cell division cycle phenotype. Nucleic Acids Res. 1993;21:3821–3828. doi: 10.1093/nar/21.16.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordenin D A, Malkova A L, Peterzen A, Kulikov V N, Pavlov Y I, Perkins E, Resnick M A. Transposon Tn5 excision in yeast: influence of DNA polymerases α, δ, and ɛ and repair genes. Proc Natl Acad Sci USA. 1992;89:3785–3789. doi: 10.1073/pnas.89.9.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grallert B, Nurse P. The ORC1 homolog orp1 in fission yeast plays a key role in regulating onset of S phase. Genes Dev. 1996;10:2644–2654. doi: 10.1101/gad.10.20.2644. [DOI] [PubMed] [Google Scholar]
- 14.Grosovsky A J, de Boer J G, de Jong P J, Drobetsky E A, Glickman B W. Base substitutions, frameshifts, and small deletions constitute ionizing radiation-induced point mutation in mammalian cells. Proc Natl Acad Sci USA. 1988;85:185–188. doi: 10.1073/pnas.85.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutz H, Heslot H, Leupold U, Loprieno N. Schizosaccharomyces pombe. In: King R C, editor. Handbook of genetics 1. Vol. 1. New York, N.Y: Plenum Press; 1974. pp. 395–446. [Google Scholar]
- 16.Harrington J J, Leiber M R. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 1994;13:1235–1246. doi: 10.1002/j.1460-2075.1994.tb06373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kokoska R J, Stefanovic L, Tran H T, Resnick M A, Gordenin D A, Petes T D. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase δ (pol3-t) Mol Cell Biol. 1998;18:2779–2788. doi: 10.1128/mcb.18.5.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 19.Krawczak M, Cooper D N. Gene deletions causing human genetic disease: mechanisms of mutagenesis and the role of the local DNA sequence environments. Hum Genet. 1991;86:425–441. doi: 10.1007/BF00194629. [DOI] [PubMed] [Google Scholar]
- 20.Krawczak M, Cooper D N. The human gene mutation database. Trends Genet. 1997;13:121–122. doi: 10.1016/s0168-9525(97)01068-8. [DOI] [PubMed] [Google Scholar]
- 21.Lieber M R. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 22.Lindsay H D, Griffiths D J F, Edwards R, Murray J M, Christensen P U, Walworth N, Carr A M. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loeb L A. Cancer cells exhibit a mutator phenotype. Adv Cancer Res. 1998;72:25–56. doi: 10.1016/s0065-230x(08)60699-5. [DOI] [PubMed] [Google Scholar]
- 24.Loeb L A. Microsatellite instability: marker of a mutator phenotype in cancer. Cancer Res. 1994;54:5059–5063. [PubMed] [Google Scholar]
- 25.Loeb L A. Mutator phenotype may be required for multi-stage carcinogenesis. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 26.Marra G, Boland C R. Hereditary nonpolyposis colorectal cancer: the syndrome, the genes and historical perspectives. J Natl Cancer Inst. 1995;87:1114–1125. doi: 10.1093/jnci/87.15.1114. [DOI] [PubMed] [Google Scholar]
- 27.Meuth M. Illegitimate recombination in mammalian cells. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. [Google Scholar]
- 28.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;94:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 29.Murakami H, Okayama H. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature. 1995;374:817–819. doi: 10.1038/374817a0. [DOI] [PubMed] [Google Scholar]
- 30.Murray J M, Tavassoli M, Al-Harithy R, Sheldrick K S, Lehmann A R, Carr A M, Watts F Z. Structural and functional conservation of the human homolog of the Schizosaccharomyces pombe rad2 gene, which is required for chromosome segregation and recovery from DNA damage. Mol Cell Biol. 1994;14:4878–4888. doi: 10.1128/mcb.14.7.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nalbantoglu J, Hartley D, Phear G, Tear G, Meuth M. Spontaneous deletion formation at the aprt locus of hamster cells: the presence of short sequence homologies and dyad symmetries at deletion termini. EMBO J. 1986;5:1199–1204. doi: 10.1002/j.1460-2075.1986.tb04347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nasmyth K, Nurse P. Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1981;182:119–124. doi: 10.1007/BF00422777. [DOI] [PubMed] [Google Scholar]
- 33.Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- 34.Santocanale S, Diffley J F X. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- 35.Stewart E, Chapman C R, Al-Khodairy F, Carr A M, Enoch T. rqh1+, a fission yeast gene related to the Bloom’s and Werner’s syndrome genes, is required for reversible S-phase arrest. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaki T, Nasmyth K. Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases. EMBO J. 1998;17:5182–5191. doi: 10.1093/emboj/17.17.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tishkoff D X, Filosi N, Gaida G M, Kolodner R D. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 38.Tran H T, Degtyareva N P, Koloteva N N, Sugino A, Masumoto H, Gordenin D A, Resnick M A. Replication slippage between distant short repeats in Saccharomyces cerevisiae depends on the direction of replication and the RAD50 and RAD52 genes. Mol Cell Biol. 1995;15:5607–5617. doi: 10.1128/mcb.15.10.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 40.Zeng Y, Forbes K C, Wu Z, Morena S, Piwnica-Worms H, Enoch T. Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature. 1998;395:507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]