Abstract
Through a computer search of the genome of the yeast Saccharomyces cerevisiae, the coding sequences of seven different box C/D antisense small nucleolar RNAs (snoRNAs) with the structural hallmarks of guides for rRNA ribose methylation have been detected clustered over a 1.4-kb tract in an inter-open reading frame region of chromosome XIII. The corresponding snoRNAs have been positively identified in yeast cells. Disruption of the nonessential snoRNA gene cluster specifically suppressed the seven cognate rRNA ribose methylations but did not result in any growth delay under the conditions of yeast culture tested. The seven snoRNAs are processed from a common polycistronic transcript synthesized from an independent promoter, similar to some plant snoRNAs but in marked contrast with their vertebrate functional homologues processed from pre-mRNA introns containing a single snoRNA. Processing of the polycistronic precursor requires nucleases also involved in rRNA processing, i.e., Rnt1p and Rat1p. After disruption of the RNT1 gene, the yeast ortholog of bacterial RNase III, production of the seven mature snoRNAs was abolished, while the polycistronic snoRNA precursor accumulated. In cells lacking functional Rat1p, an exonuclease involved in the processing of both pre-rRNA and intron-encoded snoRNAs, several processing intermediates of the polycistronic precursor accumulated. This allowed for the mapping in the precursor of the presumptive Rnt1p endonucleolytic cuts which provide entry sites for subsequent exonucleolytic trimming of the pre-snoRNAs. In line with known properties of double-stranded RNA-specific RNase III, pairs of Rnt1p cuts map next to each other on opposite strands of long double-helical stems in the secondary structure predicted for the polycistronic snoRNA precursor.
Throughout its synthesis and processing in the nucleoli of eukaryotic cells, pre-rRNA transiently associates with scores of small nucleolar RNAs (snoRNAs), only a few of which are required for the cleavages involved in formation of mature rRNA (36, 53). Cytoplasmic rRNAs of eukaryotic cells contain two prevalent types of nucleotide modification whose function is still unknown, ribose methylations and pseudouridylations, both produced posttranscriptionally on nascent pre-rRNA (34). The vast majority of snoRNAs have been recently shown to serve as guides specifying the sites of these rRNA nucleotide modifications, with each type of modification involving a distinct family of guide snoRNAs (5, 52). Ribose methylations of rRNA are guided by box C/D antisense snoRNAs which contain two short sequence motifs, box C (5′PuUGAUGA3′) and box D (5′CUGA3′), and one or occasionally two long complementarities to rRNA (11, 27, 39, 55). With each ribose-methylated nucleotide in rRNA is associated a specific box C/D antisense snoRNA, which targets the position to be methylated in pre-rRNA through transient formation of a 10- to 21-nucleotide (nt) RNA duplex at the rRNA modification site (5, 11, 27, 39, 55). Likewise, members of the other major snoRNA family, the H/ACA snoRNAs (defined by the presence of a common 3′-terminal ACA sequence) (7, 19), guide pseudouridylations through formation of a specific bipartite base-pairing with pre-rRNA around each uridine to be isomerized (for reviews, see references 20, 38, and 40).
Both families of guide snoRNAs have an unusual gene organization and a peculiar biosynthetic pathway. In vertebrates, most if not all of them are encoded in introns of protein genes and not transcribed from their own promoter in the intron but formed by processing of the host gene pre-mRNA intron (32, 36), which could provide a regulatory link between the production of the snoRNA and its host gene mRNA, all the more because most snoRNA host genes encode proteins involved in ribosome synthesis or function (36). A few intronic snoRNAs have also been detected in the yeast Saccharomyces cerevisiae (4, 45). Exonucleolytic degradation plays an essential role in the maturation of intronic box C/D and H/ACA snoRNAs, both in vertebrates (13, 14, 26, 54) and in yeast (43), with mature snoRNAs generally released by mere trimming of the debranched lariat, in line with the presence of a single snoRNA per intron. However, a minor alternative biosynthetic pathway involving prior endonucleolytic cleavages within the pre-mRNA intron can coexist in some cases, and it could become dominant when splicing efficiency is reduced (9, 10, 58). Processing of both families of intronic snoRNAs depends on cis-acting signals located within the mature snoRNA sequence, i.e., boxes C and D and the vicinal 5′- to 3′ terminal stem for the box C/D family (10, 13, 59, 62) and the conserved H/ACA motif for the second major snoRNA family (19, 20). In yeast, unlike in vertebrates, intron-encoded snoRNAs are not prevalent, probably in line with the paucity of yeast introns, and several independently transcribed S. cerevisiae modification guide snoRNAs have been reported (36). Thus, yeast U14, unlike its vertebrate ortholog, is not intron encoded (63). However, it is also produced by posttranscriptional processing, and its accumulation is dependent upon the same cis-acting signals as for processing of intronic U14 in vertebrates (23). Intriguingly, in the yeast genome the U14-coding sequence is close to that for another box C/D snoRNA, snR190, which suggests that both snoRNAs might be produced by processing of a common dicistronic transcript (63). Recent experimental findings have unambiguously confirmed this long-held hypothesis, demonstrating the involvement of both the Rat1p exonuclease and Rnt1p endonuclease in this process (16, 43). Another distinctive mode of snoRNA gene organization and expression has been identified in higher plants, with the presence of independently transcribed polycistronic snoRNA clusters, the processing of which requires endonucleolytic cleavages (30, 31).
For identifying new methylation guide snoRNAs, computer searches of genomic sequences exhibiting the structural hallmarks of box C/D antisense snoRNAs represent a powerful means which has been largely used for vertebrates (4, 39, 44, 45). In an effort to enlarge the known repertoire of S. cerevisiae snoRNAs guiding the 55 rRNA ribose methylations, we have extended this approach to the complete yeast genome and detected 15 new members of this large snoRNA family (46–48). In the present study, we have focused our attention on seven of these novel box C/D snoRNAs, snR72 to snR78 (initially termed Z2 to Z8), because of their striking clustering in a single, relatively short segment of the S. cerevisiae genome, within an inter-open reading frame (ORF) region of chromosome XIII, and have experimentally established their requirement for the cognate nonessential rRNA ribose methylations. We show that these snoRNAs result from processing of a common polycistronic snoRNA precursor, revealing in yeast the occurrence of a mode of snoRNA biosynthesis identified in plants but still undetected in metazoans. The polycistronic snoRNA transcription unit and ribosomal protein genes exhibit common cis-acting control elements, while processing of the polycistronic snoRNA precursor into individual mature snoRNAs requires endo- and exonucleases also involved in rRNA processing.
MATERIALS AND METHODS
Unless otherwise stated, all techniques for cloning and manipulating nucleic acids were performed according to standard protocols (50).
Computer search of the yeast genome.
The complete S. cerevisiae genome was searched for novel box C/D snoRNAs exhibiting structural features expected for guides of yeast rRNA ribose methylations. To also possibly identify guides for some of the 11 to 13 yet-unmapped yeast rRNA ribose methylations (34, 57), the search for rRNA complementarities was not restricted to known yeast rRNA ribose-methylated motifs but included all yeast rRNA sequences homologous to vertebrate ribose methylation sites, given that yeast and vertebrate rRNA ribose methylation patterns are largely conserved (34). Searches for a perfect 12-nt complementarity to an rRNA ribose-methylated sequence, immediately followed by the sequence NCUGA, were performed with BLAST (3) and FASTA (41) programs. Positive sequences were then screened for the presence of a box C motif within the 100 proximal upstream nucleotides and of another CUGA box D′ motif (5, 6, 56) within the 60 proximal upstream or downstream nucleotides.
Strains, media, and plasmids.
The yeast diploid strain JG1017 (MATa/MATα ade2-1/ade2-1 his4-260/his4-260 leu2-2/leu2-2 lys2-1/lys2-1 met8-1/met8-1 trp1-1/trp1-1 TYR7/tyr7-1 ura3-52/ura3-52 can1-100/can1-100 ILV1/ilv1-1) was used for preparing RNA for Northern and reverse transcription (RT) analyses and DNA for PCR amplification of the snoRNA gene cluster. The haploid strain SYNN281 (MATa ade2-101 his3-Δ200 lys2-801 trp1-Δ1 ura3-52 CAN+) and JG337.1B (MATa ade2-1 his4-260 leu2-2 lys2-1 met8-1 trp1-1 ura3-52 can1-100) were used for transformations and subsequent DNA and RNA analyses. The following strains were used to study the processing of the polycistronic transcript: an xrn1Δ strain (strain R934, kindly provided by S. Kearsey; MATa ade2-1 his3-11,15 trp1-1 ura3-52 xrn1::URA3 [43]), a rat1-1 strain (DAH18, kindly provided by C. Cole; MATa his3-Δ200 leu2-Δ1 ura3-52 rat1-1 [43]), a rat1-1 xrn1Δ strain (strain 966-1C, kindly provided by S. Kearsey; MATa xrn1::URA3 rat1-1), an rnt1Δ strain (strain SAE-52/1 [1], kindly provided by S. Abou-Elela and M. Ares; MATa his3 lys2 leu2-3,112 trp1 ura3-52 pep4 prb1 prc1 rnt1::HIS3), and an RNT1 strain (strain SAE-6, kindly provided by S. Abou-Elela and M. Ares; same as strain SAE-52/1 but transformed with plasmid pRS316-RNT1). S. cerevisiae strains were grown in rich (YPD) medium (1% yeast extract, 1% peptone, 2% glucose) or in YSD medium (0.67% yeast nitrogen base without amino acids and containing 2% glucose, supplemented with appropriate amino acids) at the temperatures specified below. Yeast was transformed by the lithium acetate method. Transformants were screened on selective plates, and the deletion of chromosomal alleles was checked by PCR. Escherichia coli TG1 [F′/supE hsdΔ5 thiΔ(lac-proAB)] or DH5α [F′ endA1 hsdr17 (rK+mK+) supE44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacIZYA-argF)U169 deoR (f80dlacΔ(lacZ)M15) grown on 2YT (1.6% Bacto tryptone, 1% Bacto yeast extract, 0.5% NaCl) or on Luria-Bertani (1% Bacto tryptone, 0.5% Bacto yeast extract, 1% NaCl) liquid or solid medium were used for all cloning procedures.
Deletion analysis of the polycistronic transcript promoter.
The following plasmids were generated. A DNA fragment consisting of the entire snoRNA gene cluster flanked by 538 bp 5′ and 216 bp 3′ was PCR amplified with oligodeoxynucleotides PC1 and PC2 (see below), which provide EcoRI and SalI restriction sites, respectively, and cloned into the pFL39 yeast centromeric shuttle vector after EcoRI/SalI digestion. The resulting plasmid was termed pΔ0. Progressive 5′ deletions of the promoter region were generated by PCR with pΔ0 as a template, the oligodeoxynucleotide 25SCm2195, and one of the following oligodeoxynucleotides: PolyRAP1.8 (generating the Δ1 deletion), PolyRAP1.7 (Δ2 deletion), Poly4 (Δ3 deletion), Poly5 (Δ4 deletion), and PC3 (Δ5 deletion). The resulting DNA fragments were digested by EcoRI and BamHI and used to replace the EcoRI/BamHI promoter fragment of pΔ0, producing plasmids pΔ1 to pΔ5. These were directly transformed into the haploid yeast strain carrying a chromosomal disruption of the snoRNA gene cluster.
RNA analyses.
Yeast cells washed with diethyl pyrocarbonate-treated H2O were ground with liquid N2. A nucleolar fraction was isolated from purified yeast nuclei through a detergent-salt extraction procedure (24). RNA was isolated by the guanidinium thiocyanate method (17) and analyzed by electrophoresis on 6 or 8% acrylamide–7 M urea gels. Capillary transfer or electrotransfer onto nylon membranes (Amersham) was carried out, followed by UV light irradiation of the membranes (Hybond-N for snoRNA expression assays and Hybond-N+ for ribose methylation assays). Northern blot hybridizations were carried out with oligodeoxynucleotide probes labeled at the 5′ ends with 32P through a 3-h incubation in 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]–1% sodium dodecyl sulfate (SDS)–5× Denhardt’s solution–150 μg of tRNA per ml at a temperature 15 to 18°C below the theoretical midpoint temperature of the hybrid. Membranes were washed twice with 0.1× SSPE–0.1% SDS or twice with 2× SSC–0.1% SDS and twice with 1× SSC–0.1% SDS before autoradiography. A TaqI/HaeIII digest of pBR322 DNA labeled with 32P served as a size marker. For identifying 5′ ends of mature snoRNAs, primer extensions were carried out in 20-μl reaction mixtures containing 15 μg of total cellular heat-denatured RNA (65°C, 5 min, in H2O) and 20 ng of 5′-end-labeled primer in the presence of 250 μM deoxynucleotide triphosphates (dNTPs) and 100 U of avian myeloblastosis virus (AMV) reverse transcriptase (Promega) for 30 min at 42°C. Sequence analysis of cDNAs was performed after purification of full-size primer extension products on denaturing polyacrylamide gels. After addition of a 3′ poly(G) tail with terminal transferase (Promega), cDNAs were amplified by PCR with a forward poly(C) primer carrying EcoRI and BamHI restriction sites and a reverse Pz2-Pz8 primer. PCR products were purified on 8% denaturing acrylamide gels and cloned into the pGEM T vector (Promega) for sequence determination (Sequenase sequencing kit; Life Sciences Co.). Primer extension experiments with RNAs extracted from the rat1-1, xrn1Δ, rat1-1/xrn1Δ, SAE-52/1, or SAE-6 strain were performed as described above except that 5 μg of each of the relevant RNAs was denatured for 4 min at 80°C and that 5 U of AMV reverse transcriptase (Promega) and 1 mM dNTPs were used.
RT-PCR experiment.
Total cellular RNA was submitted to an extensive DNase I treatment before RT with reverse primer Pz4 or Pz2 (Fig. 1). About 50 μg of RNA in 100 μl in DNase I buffer was incubated (30 min, 37°C) with 10 U of RQ1 RNase-free DNase I (Promega) and subsequently submitted to phenol-chloroform-isoamyl alcohol (50:49:1) extractions. A PCR was then carried out with one of these reverse primers and the corresponding forward primer (as depicted in Fig. 5), using the following program: 35 cycles of denaturation (30 s, 94°C), annealing (30 s, 55°C), and extension (1 min, 72°C), followed by a final extension (10 min, 72°C).
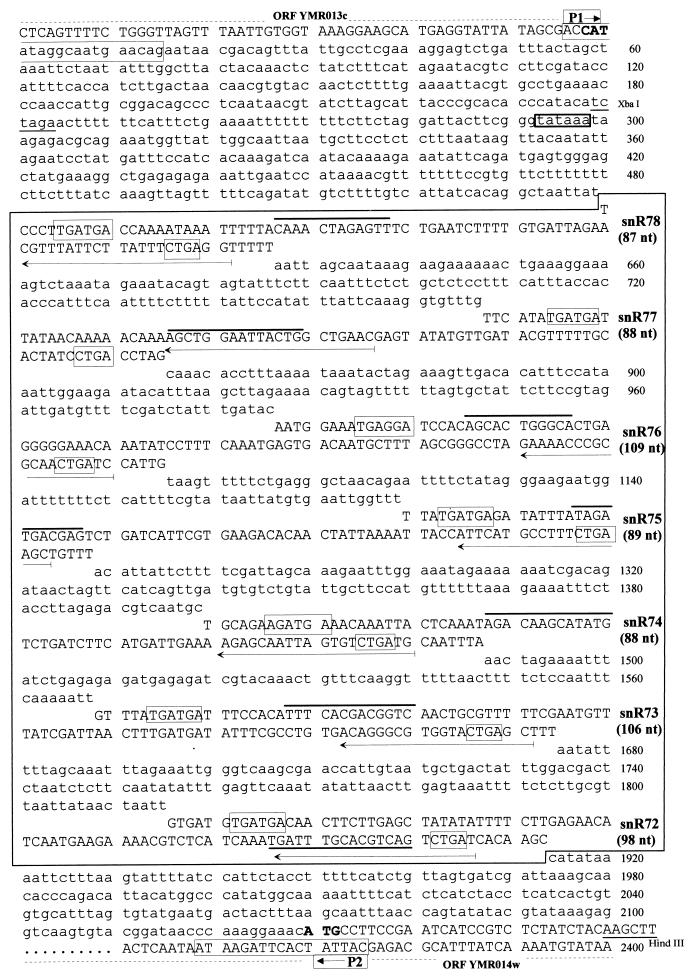
FIG. 1.
Sequence of the inter-ORF region of S. cerevisiae chromosome XIII, which spans the clustered snoRNA genes. The sequence of the 2,129-bp region separating YMR013c and YMR014w within the 3.9-kb YMR012-YMR014 inter-ORF region on the w strand is shown. The proximal sequences of flanking ORFs on opposite DNA strands, YMR013c and YMR014w, are also shown (in capital letters, with the initiation codon in boldface). The cluster of snoRNA-coding regions is boxed. Nucleotides outside snoRNA-coding regions are in lowercase letters. Within each snoRNA-coding region the C and D motifs (boxes), the 10- to 16-nt antisense elements matching sites of rRNA ribose methylation (thick overlines), and the gene-specific primers (horizontal arrows) used to delineate boundaries of mature snoRNA sequences (Fig. 2) are indicated. The locations of PCR primers P1 and P2 and restriction sites used for deleting the snoRNA gene cluster (Fig. 3) are also indicated.
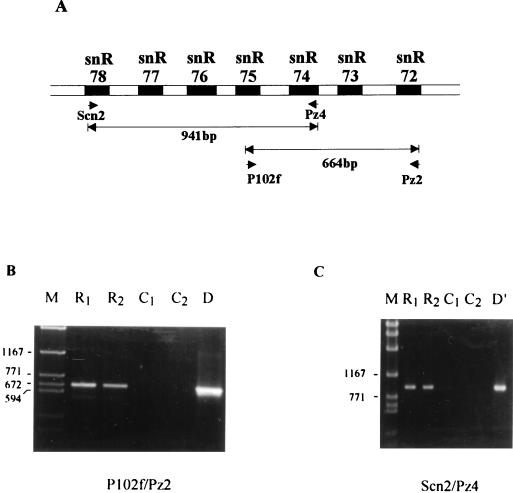
FIG. 5.
Detection of a polycistronic transcript spanning the snoRNA cluster. (A) Locations of the two pairs of primers used for RT-PCR experiments. (B and C) Products of the RT-PCR obtained with each pair of primers. Reaction products were analyzed on an 0.8% agarose gel and revealed by ethidium bromide staining. Lanes M, 2-kb molecular size marker. Lanes R1 and R2, products of two different PCRs with different concentrations of RT reaction products. Lanes C1 and C2, control reactions carried out without reverse transcriptase. Lanes D and D′, control PCR performed on yeast genomic DNA with the same pair of primers.
Detection of ribose-methylated nucleotides.
Ribose methylation was tested by RT at low dNTP concentrations (35) as follows. Five micrograms of total yeast RNA was mixed with 0.1 pmol of a gel-purified oligodeoxynucleotide labeled at the 5′ end with 32P, dried (SpeedVac), and resuspended in 20 μl of 1× RT buffer (Promega). After a heat denaturation step (90°C, 5 min), hybridization was performed at 55°C for 20 min. Primer extension with 10 U of AMV reverse transcriptase (Promega) was carried out in parallel on two aliquots (final volume, 40 μl) in the presence of either 4 μM or 1 mM dNTPs.
Immunoprecipitations.
Ten microliters of rabbit antitrimethylguanosine (anti-TMG) antibody R1131, kindly provided by R. Lührmann (33a), or 10 μl of nonimmune anti-mouse immunoglobulin Gs (IgGs) (Sigma) was incubated with gentle agitation for 1 h at 4°C with 3 mg of protein A-Sepharose (PAS) previously swollen in 150 μl of a solution containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 0.05% Nonidet P-40. PAS-IgG pellets were washed twice with 1 ml of the same buffer and then resuspended in 100 μl of the same buffer supplemented with 80 U of RNasin RNase inhibitor (Promega). Twenty micrograms of total RNA purified from either rnt1Δ cells or isogenic wild-type cells was then added to PAS-IgG samples. The RNA-IgG interaction was allowed to take place for 1 h at 4°C with gentle agitation. PAS pellets were then collected by very brief centrifugation. The supernatants were removed, and the pellets were washed five times with 1 ml of 50 mM Tris-HCl (pH 7.4)–150 mM NaCl–0.05% Nonidet P-40. RNAs were then extracted from the pellet and supernatant fractions by standard procedures.
Oligodeoxynucleotides.
Oligonucleotides were synthesized on a PerSeptive Biosystems Expedite apparatus (Y. de Préval, LBME, Toulouse, France) or obtained from the Shanghai Biochemistry Institute (Academy of Sciences, People’s Republic of China). After labeling of the 5′ ends with [32P]kinase, oligonucleotides were either directly used as probes for Northern hybridization or submitted to purification by electrophoresis on a 15% acrylamide–7 M urea gel before utilization as RT primers. Sequences of primers (termed Pz2 to Pz8) used for Northern and RT analyses of snR72 to snR78 are delineated in Fig. 1. The forward poly(C) primer used for PCR amplification of oligo(G)-tailed cDNAs for snR72 to snR78 was 5′GGAATTCGGATC163′. RT-PCR experiments (see Fig. 5) were performed with the following oligonucleotides: 5′TCCCTTGATGACCAAAATAAA3′ (Scn2), 5′CATCAGACACTAATTGCTCT3′ (Pz4), 5′CCATTCATGCCTTTCTGAAGC3′ (P102f), and 5′ATCAGACTGACGTGCTTTTC3′ (Pz2). Primers used for disrupting the snoRNA gene cluster (see Fig. 3) were as follows 5′CGTCGGTACCATATAGGCAATGAACAG3′ (P1, carrying a KpnI site) and 5′GACGGAGCTCTCGTAATAGTGAATCTTAT3′ (P2, carrying a SacI site). Ribose-methylated nucleotides in specific regions of S. cerevisiae rRNAs were assayed with the following primers: 5′TGGTTCGATTAGTCTTTCGCCC3′ (o26S-snR72), 5′CCATTGTAAGTAGTCATCC3′ (o26S-snR73), 5′TATACTTAGACATGCATGGCTTA3′ (o18S-snR74), 5′CGTTAATCCATTCATGCGCGTC3′ (o26S-snR75), 5′GCGCTTGGTTGAATTTCTTCAC3′ (o26S-snR76), 5′AACTGCAACAACTTTAATATACG3′ (o18S-snR77), and 5′GTGGGAGATACAGAGAAGTG3′ (o26S-snR78). Primers used to produce progressive 5′ deletions of the promoter of the polycistronic transcript were as follows: PC1, 5′CCCGAATTCATAGGCAATGAACAGAATAACG3′ (provides an EcoRI site); PC2, 5′CCCGTCGACGTTTCCTTTGGGTTATCCGTAC3′ (provides a SalI site); PolyRAP1.8, 5′CCCGAATTCCTCTTTTGAAAATTACGTGCC3′ (provides an EcoRI site); PolyRAP1.7, 5′CCCGAATTCTAGAACTTTTTTCATTTCTG3′ (provides an EcoRI site); Poly4, 5′CCCGAATTCCTTCTAGGATTACTTCGGG3′ (provides an EcoRI site); Poly5, 5′CCCGAATTCGAAAGGCTGAGAGAGAAATTG3′ (provides an EcoRI site); and PC3, 5′CCCGAATTCATCACAGGCTAATTATTCCCTTG3′ (provides an EcoRI site).
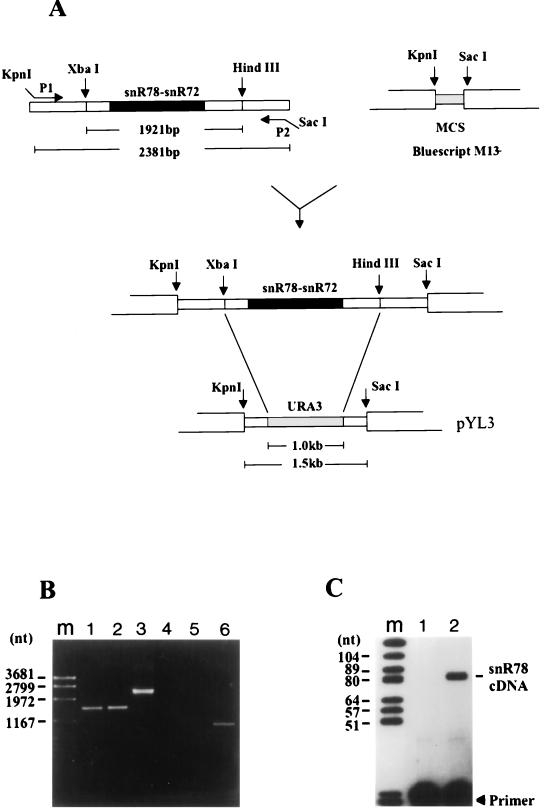
FIG. 3.
Disruption of the snoRNA gene cluster. (A) The top diagram shows PCR amplification of the 2.4-kb genomic DNA fragment of an S. cerevisiae diploid strain with primers P1 and P2 (carrying KpnI and SacI sites, respectively) and cloning of the amplified fragment into the corresponding restriction sites of E. coli plasmid Bluescript M13−. (MCS, multiple cloning site.) The bottom diagram shows how the XbaI-HindIII 1.9-kb genomic fragment containing the entire gene cluster was then replaced by the selectable marker, the URA3 gene, giving rise to plasmid pYL3, which was used to transform a haploid yeast strain after digestion with KpnI and SacI. (B) PCR analysis of URA+ transformants. DNAs from two different URA+ transformants were PCR amplified with primers P1 and P2 (lanes 1 and 2) and P1 and Pz6 (lanes 4 and 5), and PCR products were analyzed by electrophoresis on an 0.8% agarose gel followed by ethidium bromide staining. Lanes 3 and 6 are control lanes showing amplification of wild-type strain YNN281 DNA with the primer pairs P1-P2 and P1-Pz6s, respectively. Lane m contains a 2-kb molecular size marker. (C) RT analysis of RNA from a URA+ transformant. The reaction was carried out with the snR78-specific primer by using 15 μg of total RNA purified from either a URA+ transformant (lane 1) or wild-type strain YNN281 (lane 2). Lane m contains molecular size markers.
The following oligodeoxynucleotides were used in Northern or primer extension experiments with RNAs extracted from the rat1-1, xrn1Δ, rat1-1/xrn1Δ, SAE-52/1, or SAE-6 strain: snoRNA1 (hybridizing to snR78), 5′ACGTTCTAATCACAAAAG3′; 18SUm578 (hybridizing to snR77), 5′GATAGTGCAAAAACGTAT3′; 25SCm2195 (hybridizing to snR76), 5′GTGGATCCTCATTTCCAT3′; 25SCm2195.2 (hybridizing to snR76), 5′CATTTGAAAGGATATTTGTTTCC3′; 25SGm2286 (hybridizing to snR75), 5′TGGTAATTTTAATAGTTG3′; 18SAm28.2 (hybridizing to snR74), 5′CACTAATTGCTCTTTTCAATCATG3′; SnoRNA4.2 (hybridizing to snR73), 5′CGATAAACATTCGAAAAACGCAG3′; and 25SAm874 (hybridizing to snR72), 5′ATCATTTGATGAGACGTT3′.
RESULTS
Detection of a cluster of seven putative methylation guide snoRNA coding sequences on S. cerevisiae chromosome XIII.
In methylation guide snoRNAs the systematic linkage of a 10- to 21-bp antisense element with an adjacent downstream CUGA motif (6–8) provides the basis for an efficient sequence search of the 13-Mb S. cerevisiae genome for novel box C/D snoRNAs, given that positive sequences can be screened for the presence of an additional hallmark box C motif. Among the 15 novel S. cerevisiae snoRNAs eventually identified by this approach (46–48), we have focused our attention on 7 species, because their coding regions are intriguingly clustered. As shown in Fig. 1, the seven sequences, termed snR72 to snR78, are all organized in a head-to-tail fashion within a 1.4-kb segment of an inter-ORF region on the w strand of chromosome XIII. They exhibit different antisense elements corresponding to distinct 18S or 25S rRNA sequences, as detailed below, and except for box C and D motifs they are not related to each other or to any other sequence in the entire yeast genome. The same holds true for all the relatively short (85- to 141-nt) AT-rich spacers which separate the snoRNA-coding regions.
Positive identification of the snoRNAs.
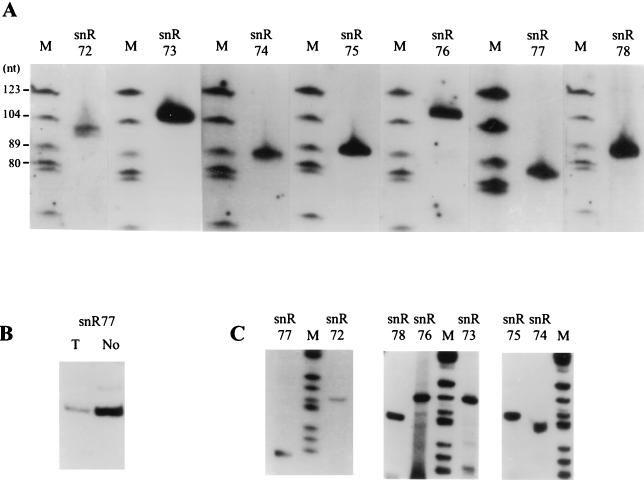
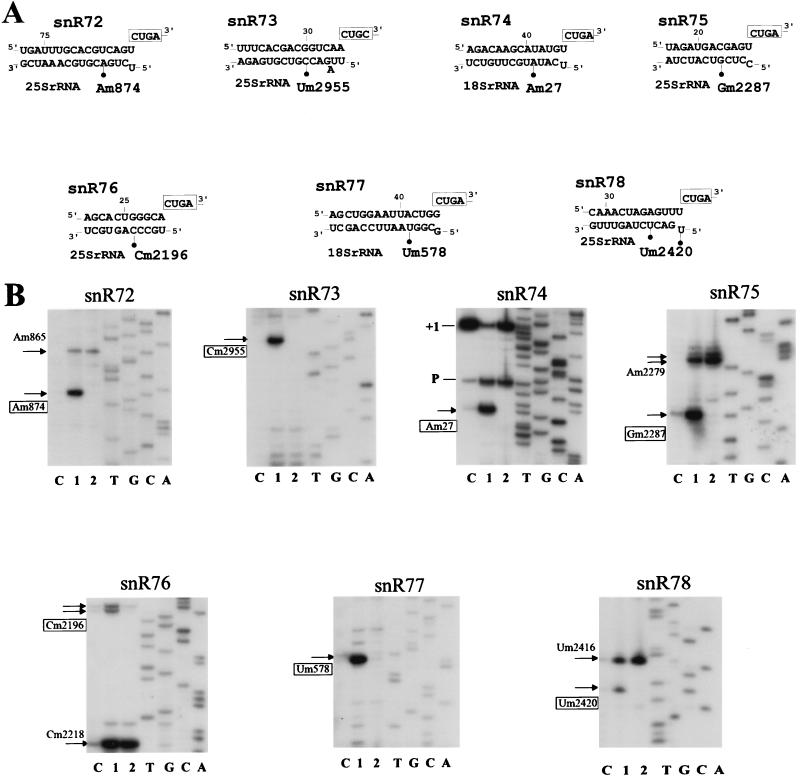
With oligonucleotide probes specific for each presumptive coding region, the seven expected RNAs were readily detected by Northern blot analysis of total yeast cellular RNA (Fig. 2A). In each case the intensity of the single band (size range, 87 to 109 nt) pointed to cellular abundances in the same order of magnitude, roughly similar to what was previously determined for intron-encoded methylation guide snoRNA U24 (45). Parallel analysis of purified yeast nucleolar RNA and total cellular RNA indicated that all these RNAs are enriched in the nucleolar fraction more than 20 times, as shown for snR77 (Fig. 2B). Mature 5′ termini of the seven RNAs were identified by primer extension with the same oligonucleotides (Fig. 2C and data not shown), followed by cloning and sequencing of the cDNA (as detailed in Materials and Methods). As expected, all cDNA sequences matched perfectly the sequences of the presumptive coding regions delineated in Fig. 1. Mature 3′ termini were inferred from RNA migration on denaturing gels (Fig. 2A). Three of the seven snoRNAs, snR72, and snR74, and snR76, have the potential to form a characteristic 5′-to-3′ terminal stem-box structure, as do about half the snoRNAs of this family (4–6).
FIG. 2.
Characterization of individual snoRNAs. (A and B) Northern analysis. (A) Total cellular RNA (15 μg) was separated on an 8% acrylamide–8 M urea gel. After electrophoretic transfer, nylon membranes were hybridized with the labeled oligonucleotide probes depicted in Fig. 1. Lanes M, size markers. (B) Parallel analysis of yeast nucleolar RNA and total cellular RNA with the snR77 oligonucleotide probe. Lane T, 15 μg of total cellular RNA. Lane No, 3 μg of RNA purified from yeast isolated nucleoli. (C) Primer extension analysis. RT was carried out with the 32P-, 5′-end-labeled oligonucleotides depicted in Fig. 1.
Disruption of the gene cluster suppresses the cognate rRNA methylations.
To test the essentiality of the clustered snoRNA genes, we replaced the ≃2.0-kb DNA fragment containing the seven snoRNA-coding regions plus vicinal flanking sequences with the URA3 marker gene (Fig. 3). After transformation of a haploid ura− strain by the disrupted allele, Ura+ colonies were selected. A PCR analysis of genomic DNA demonstrated that the seven-snoRNA gene locus was completely deleted in Ura+ cells, with the appearance of the expected ≃1.5-kb amplification product instead of the ≃2.4-kb band obtained with wild-type strain DNA when the same pair of primers was used (Fig. 3B). Disruption of the snoRNA gene cluster was further confirmed by primer extensions (Fig. 3C) and Northern blot analyses (data not shown) of Ura+ cell RNA. In both cases no positive radioactive signal was obtained with the different snoRNA-specific 5′-end-labeled oligonucleotides when probing disrupted-strain RNA, in contrast to what was observed for parent wild-type strain RNA. To our surprise, we could not detect any phenotypic difference at various growth temperatures for the Ura+ cells with a disrupted gene cluster, suggesting that the seven snoRNA genes were dispensable in yeast.
Of the seven antisense elements immediately followed by a CUGA motif found in the different snoRNA-coding regions of the gene cluster, only three, in snR74, snR77, and snR76, match previously mapped ribose-methylated nucleotides in yeast rRNAs, i.e., Am27 and Um578 in 18S rRNA and Cm2196 in 25S rRNA, respectively (34, 57). In contrast, the 11- to 15-nt antisense elements of snR72, snR73, snR75, and snR78, which are also followed by a CUGA motif and definitely exhibit the structural features of a bona fide guide sequence, did not match a known rRNA ribose-methylated nucleotide. Within all guide snoRNA-rRNA duplexes the methylated site is always at the same location, paired to the fifth nucleotide upstream from the CUGA motif (5, 6, 11, 27, 39). The positive identification of snR72, snR73, snR75, and snR78 therefore strongly suggested that 25S rRNA nucleotides A874, U2955, G2287, and U2420, respectively (Fig. 4A), were among the 11 to 13 yeast rRNA ribose methylations which remained unmapped (34, 55), and this has been recently verified experimentally (12). Using primer extension at low dNTP concentrations, which causes pauses at ribose-methylated nucleotides in the RNA template (35), we have observed that disruption of the snoRNA gene cluster results in the disappearance of the seven cognate rRNA ribose methylations (Fig. 4). Each radioactive band reflecting the presence of the cognate ribose methylation in wild-type strain RNA completely disappeared at low dNTP concentrations when disrupted-strain RNA was used (Fig. 4B, lanes 2), confirming that snR72 to snR78 are the cognate functional guides and showing that their function cannot be fulfilled in an alternative way. Conversely, the other proximal rRNA ribose methylations assayed in these experiments (Fig. 4B) were not affected by disruption of the gene cluster, as expected, indicating that most, if not all, site-specific rRNA ribose methylations are formed independently of each other and can still proceed after a substantial reduction of the snoRNA guide repertoire.
FIG. 4.
Site-specific defects in rRNA ribose methylation after disruption of the snoRNA cluster. (A) Methylation guide duplexes between snoRNAs snR72 to snR78 and yeast rRNA. Ribose methylation sites (filled circles) and box D or D′ motifs (boxes) are indicated. (B) Detection of rRNA ribose-methylated nucleotides by RT at low dNTP concentrations. Primer extensions on total cellular RNA were performed with seven appropriate oligonucleotides selected downstream from cognate sites of ribose methylation for each snoRNA of the cluster (as indicated at the top of each panel). Lanes C, control reaction at 1 mM dNTPs on wild-type strain RNA; lanes 1 and 2, primer extension at 4 μM dNTPs on wild-type strain and disrupted strain RNA, respectively. Among the sites of ribose methylation (arrows) revealed by RT pauses at low dNTP concentrations, those specifically affected in the disrupted yeast strain are indicated by boxed coordinates. In the panel for snR74, bands denoted +1 and P correspond to the 5′ end of 18S rRNA and to an RT pause at a pseudo-knot structure, respectively.
Detection of a polycistronic snoRNA transcript.
The close linkage of the seven snoRNAs, together with the short sizes of all intergenic spacers which are devoid of any known promoter sequence element, such as a TATA box, suggested that these genes might be polycistronically expressed, similar to clusters of snoRNA genes in plants (30, 31). To test this possibility we tried to detect the presence of a long polycistronic transcript spanning the gene cluster by RT-PCR. RT-PCRs were performed with two pairs of primers designed to amplify two overlapping regions which together encompass the entire cluster. In each case, the single band produced corresponded precisely to the size of the product expected for the presence of a common polycistronic transcript. Thus, with primers designed to amplify between the snR75 and snR72 genes a band of 664 bp was expected, and an amplified DNA fragment of ≃660 to 680 bp was observed (Fig. 5B). Likewise, a band of 941 bp was expected for the snR78-snR74 primer combination, and an RT-PCR product of about 950 bp was amplified (Fig. 5C). No amplification product was observed in any of the RT-PCR controls performed in the absence of reverse transcriptase (Fig. 5B and C, lanes C1 and C2). The results of the different RT-PCR experiments were therefore consistent with the existence of a common polycistronic transcript spanning the snoRNA gene cluster.
Delineation of the snoRNA gene cluster promoter.
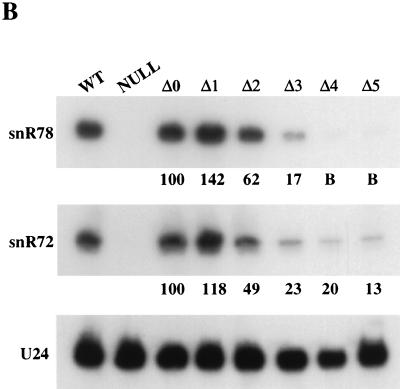
Promoter elements controlling the synthesis of the putative polycistronic transcript are most likely located between the start of the upstream ORF (YMR013c) and the start of the snR78-coding region. Intriguingly, this region contains two perfect matches to the RAP1 binding site consensus 5′(A/G)(A/C)ACCCANNCA(T/C)(T/C)3′ (reviewed in reference 21) followed by an AT-rich stretch and a TATAAA box (Fig. 6A), i.e., the elements required for full transcriptional activity of most promoters of S. cerevisiae ribosomal protein genes (61), suggesting that there might be a coordinate transcriptional control of some snoRNA and ribosomal protein genes. It is known that the latter are regulated by carbon source; their mRNA levels display a fourfold increase after cells are shifted from a medium containing a nonfermentable carbon source to a medium containing a fermentable one, and it has been suggested that this effect is mediated by Rap1p (22, 28). We therefore tested first whether the steady-state levels of snR78 are likewise affected by the carbon source. Northern analysis revealed that snR78 levels are indeed twofold higher in cells grown on glucose than in cells grown on glycerol (data not shown). We next wanted to determine whether the putative Rap1p binding sites and the T-rich sequence upstream of the snoRNA gene cluster are required for its transcription. For this purpose, the complete snoRNA gene cluster with 5′ and 3′ flanking sequences was cloned into a yeast centromeric vector, and progressive deletions in the snR78 5′ flanking region were introduced (Fig. 6A). The resulting plasmids were then transformed into a yeast strain in which the snoRNA gene cluster had been disrupted. Northern blot analysis of RNAs extracted from cells harboring the various deletion mutants revealed that (i) an 88-bp region containing the two putative Rap1p binding sites is required for full transcriptional activity, since in its absence snoRNA steady-state levels were reduced by 40 to 50%; and (ii) in the absence of this 88-bp region, the AT-rich stretch becomes essential for promoter activity, since when it was also removed, snoRNA steady-state levels dropped by 80 to 85% (compare lanes Δ2 and Δ3 in Fig. 6B). Note that snR78 and snR72 levels were similarly affected by the Δ1 to Δ3 deletions, which shows that their transcription is controlled by the same sequence elements and further supports the notion that they are cotranscribed. However, production of the 3′-most snoRNA of the polycistronic unit, snR72, was not completely abolished by the removal of the TATAAA box motif (Fig. 6B, lanes Δ4 and Δ5), in contrast to what was observed for the 5′-most snoRNA, snR78, possibly reflecting residual cryptic transcription initiation.
FIG. 6.
Deletion analysis of the snoRNA gene cluster promoter. (A) Sequence of the promoter region with the relevant promoter elements (boxed) and the 5′ ends of the various promoter deletion mutants used (bent arrows) indicated. Vertical arrows indicate the positions of the major transcription initiation sites (identified in the experiment whose results are shown in Fig. 10). (B) SnoRNA levels in the yeast strains harboring the various mutated promoters assayed by Northern blot hybridization (5 μg of total RNA per lane). Lane WT, wild-type yeast strain; lane Null, strain containing a chromosomal disruption of the snoRNA gene cluster; lanes Δ0 to Δ5, the same strain transformed with a centromeric vector bearing the entire snoRNA cluster up to the Δ0, Δ1, Δ2, Δ3, Δ4, and Δ5 5′ boundaries. The blot was hybridized with oligonucleotide probes specific for snR78, snR72, or U24 snoRNA. The percentages of snR78 and snR72 in the Δ1 to Δ5 mutants relative to the Δ0 strain, using U24 as an internal standard, are displayed below the corresponding lanes (quantifications were carried out by phosphorimager scanning). B, background.
Maturation of the snoRNA polycistronic transcript requires the exonuclease Rat1p.
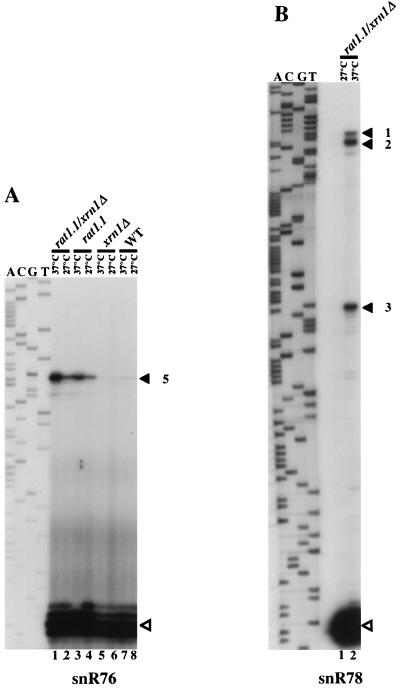
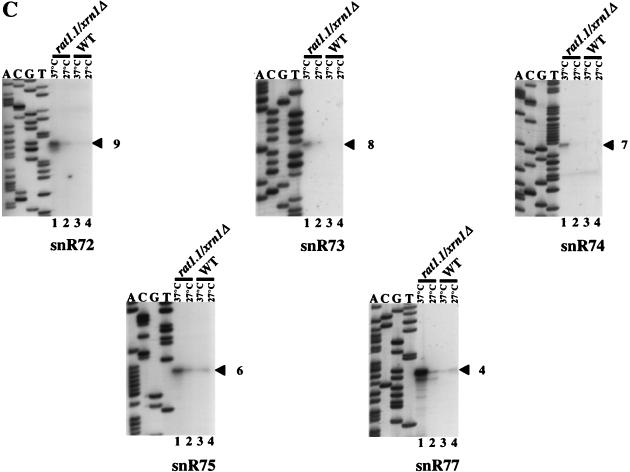
Production of mature snoRNA sequences from a polycistronic transcript could occur either by direct endonucleolytic digestion at the ends of the mature RNA sequences or by endonucleolytic cleavage within the spacers followed by 5′-to-3′ and 3′-to-5′ exonucleolytic trimming. Supporting the second scenario, analyses at nucleotide resolution show a 2- to 3-nt heterogeneity for mature snoRNA 5′ ends (Fig. 7A and data not shown). Moreover, it has recently been shown that the 5′ ends of the U14 and snR190 snoRNAs are produced by the Rat1p 5′-to-3′ exonuclease (43). We therefore tested whether the two known yeast 5′-to-3′ exoribonucleases, Rat1p and Xrn1p, are required for the 5′-end formation of snoRNAs snR72 to snR78. The RAT1 gene is essential; hence, a strain bearing a rat1-1 ts allele was used (see reference 43 for references). Primer extension experiments were performed with RNAs extracted from rat1-1 ts, xrn1::URA3-disrupted, or double mutant strains grown at the semipermissive (27°C) or nonpermissive (37°C) temperature, using oligonucleotides specific to mature snoRNA sequences (Fig. 7). With each of the six snR72- to snR77-specific primers, a single 5′-extended species relative to each mature snoRNA was detected in rat1-1 ts/xrn1::URA3 cells (Fig. 7A and C). Since the 5′-extended products must correspond to processing intermediates of the pre-snoRNA generated by endonucleolytic cuts, the primer extension experiment provided for a precise mapping of these cuts in the intergenic spacer sequence. The same extension products were also detected in the rat1-1 ts single mutant but not in xrn1::URA3 cells (Fig. 7A, compare lanes 3 and 4 to 5 and 6; also data not shown), showing that Rat1p is by far the more important of the two activities examined for 5′-end processing of snR72 to snR78. However, the increased levels of 5′-extended products in the rat1-1 ts/xrn1::URA3 double mutant strain compared to the rat1-1 ts single mutant (Fig. 7A, lanes 1 and 3, and data not shown) indicate that Xrn1p, which is exclusively cytoplasmic (25), can partially compensate for the lack of functional Rat1p. Given that the snoRNA primary transcript bears a hypermethylated cap (see below), this could point towards a cytoplasmic phase for processing of the polycistronic transcript. Primer extensions with an oligonucleotide specific to the 5′-most snoRNA in the polycistronic unit, snR78, yielded somewhat more complex results, with the detection of three major 5′-extended RNAs (Fig. 7B; see also Fig. 8A). The nature of the 3′-to-5′ exonuclease producing the 3′ ends of the snoRNAs remains to be determined.
FIG. 7.
Accumulation of 5′-extended RNAs in cells lacking functional Rat1p. (A) Primer extension products obtained with an oligonucleotide complementary to snR76 by using 5 μg of total yeast RNA extracted from the rat1-1/xrn1::URA3 strain (lanes 1 and 2), the rat1-1 strain (lanes 3 and 4), the xrn1::URA3 strain (lanes 5 and 6), and a wild-type strain (lanes 7 and 8). Lanes 1, 3, 5, and 7, strains grown for 2 h at 37°C. Lanes 2, 4, 6, and 8, strains grown at 27°C. The sequence ladder was generated with the same oligodeoxynucleotide primer. (B) Mapping of the 5′ ends of extended pre-snoRNAs with the snR78 probe. The cDNAs corresponding to the mature snoRNAs and 5′-extended pre-snoRNA intermediates are indicated by hollow and filled arrowheads, respectively, with the numbers referring to the Rnt1p cuts in the snoRNA precursor structure as depicted in Fig. 8. (C) Mapping of the 5′ ends of extended pre-snoRNAs detected with snR72- to snR75- and snR77-specific primers. Primer extensions were carried out with 5 μg of total yeast RNA extracted from the rat1-1/xrn1::URA3 strain (lanes 1 and 2) or the wild-type strain (lanes 3 and 4). Lanes 1 and 3, strains grown for 2 h at 37°C. Lanes 2 and 4, strains grown at 27°C.
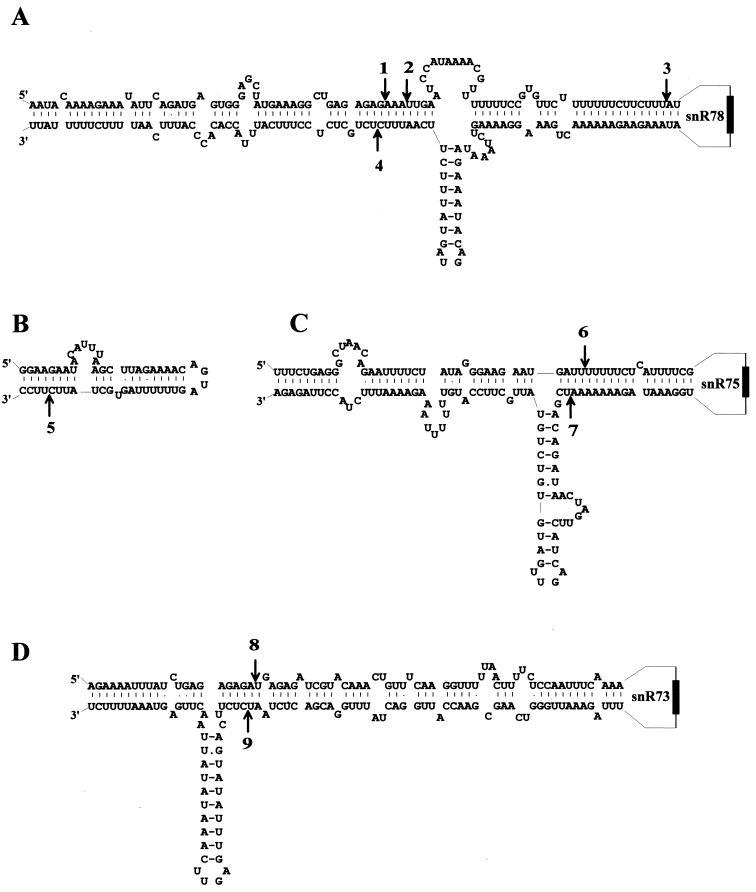
FIG. 8.
Locations of putative Rnt1p cleavage sites in potentially base-paired regions of the polycistronic snoRNA transcript. Only the parts of the polycistronic transcript structure which contain the presumptive Rnt1p cuts identified in the experiment whose results are shown in Fig. 7 are shown. Rnt1p cuts (arrows) are numbered as in Fig. 7 for products accumulating in a rat1-1 ts strain at the restrictive temperature. These cleavage sites, sufficient to free each of the snoRNAs in the primary transcript, map within interspacer pairings, involving spacers immediately upstream and downstream from snoRNAs snR78, snR75, and snR73 (A, C, and D, respectively) or within a duplex which involves only one intergenic sequence, between snR77 and snR76 (B).
Endonucleolytic cleavages of the polycistronic snoRNA transcript involve RNase III.
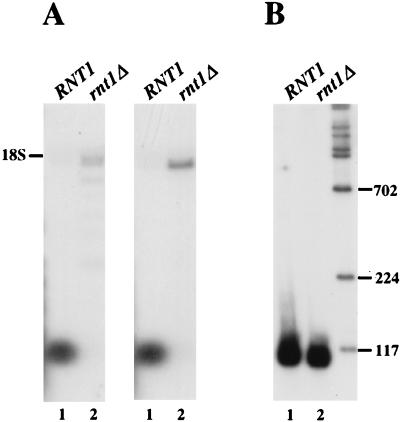
The various presumptive endonucleolytic cleavage sites mapped by primer extensions (numbered 1 to 9 in Fig. 7) are sufficient to free each of the snoRNAs in the primary transcript. A comparison of the sequences in their immediate vicinity did not reveal any obvious sequence conservation. To assess whether it is the structure rather than the sequence per se which is critical for cleavage, RNA folding predictions on a thermodynamical basis (64) were carried out for the entire snoRNA polycistronic transcript. Several very stable and extensive base-pairings, involving both interspacer and intraspacer interactions, were identified (Fig. 8). Remarkably, all mapped cuts were found within those extended double-helical structures, with six of them being grouped in pairs, found next to each other on opposite strands of the same double-helical region (Fig. 8A, C, and D). All these features pointed to RNase III (2) as the endonuclease involved. To directly test this possibility, we assessed the accumulation of snR78 and snR72 snoRNAs in a strain lacking RNase III. Northern blot analysis (Fig. 9) showed that very little mature snR78 or snR72 accumulates in the rnt1::HIS3-disrupted strain (Fig. 9A, lanes 2). Instead, a much longer RNA which comigrates with 18S rRNA accumulates. This is fully consistent with the 1.8-kb RNA being the full-length unprocessed polycistronic transcript. In addition, a series of smaller RNAs was also detected in the rnt1::HIS3-disrupted strain with the snR78 probe; these could result from premature transcription termination. Accumulation of intron-encoded snoRNA U24 (Fig. 9B) or U18 (data not shown) was not affected by the absence of RNase III, as expected.
FIG. 9.
Accumulation of the full-length polycistronic transcript in cells lacking RNase III. (A and B) Northern blot analysis of RNAs extracted from an rnt1::HIS3 strain (lanes 2) or an otherwise-isogenic wild-type strain (lanes 1) grown at 26°C. (A) RNAs were separated on a 1% agarose denaturing gel, blotted, and hybridized with probes specific for snR78 or snR72 snoRNA. (B) RNAs were separated on an 8% acrylamide gel, blotted, and hybridized with a probe specific for U24 snoRNA.
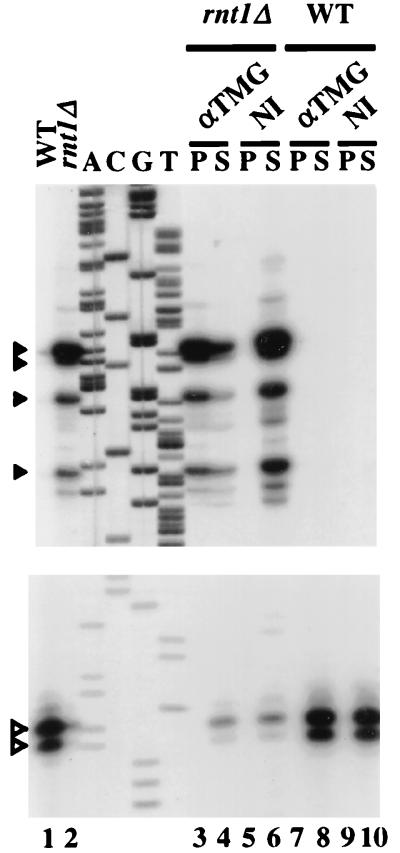
The position(s) of the 5′ end(s) of the transcript(s) accumulating in the strain lacking RNase III was determined by primer extension with an oligodeoxynucleotide hybridizing to the snR78 sequence (Fig. 10, lane 2). Four major extension products were detected (Fig. 10, lane 2) corresponding to RNA 5′ ends mapping to nucleotide positions 70, 72, 79, and 91 downstream from the TATAAA box motif (Fig. 6A). Several less abundant primer extension products could also be detected (data not shown). The fact that RNAs 5′-extended relative to the 5′ end of mature snR78 could accumulate in cells containing the wild-type Rat1p and Xrn1p exonucleases suggested the possibility that these pre-snoRNAs were protected from 5′-to-3′ exonucleolytic digestion by a modified 5′ end. We looked for the presence of a TMG cap on these 5′-extended RNAs through immunoprecipitation experiments with anti-TMG antibodies performed on RNA from cells lacking RNase III or from isogenic wild-type cells, followed by primer extensions with the anti-snR78 oligonucleotide. All the above-described 5′-extended species revealed by primer extension were exclusively detected with the immunoprecipitated RNA of the strain lacking RNase III (Fig. 10, lane 3), reflecting their carrying a 5′ TMG cap and strongly suggesting that their 5′ ends correspond to transcription initiation sites. As expected, mature snR78 snoRNA, serving as a control, was exclusively detected in the supernatant (Fig. 10, lanes 7 and 8). In conclusion, positions of the transcription initiation sites inferred from primer extension experiments performed with RNAs from cells lacking RNase III are fully consistent with the AT-rich stretch and the TATAAA motif being key promoter elements of the RNA polymerase II-transcribed polycistronic unit.
FIG. 10.
The 5′ ends of the polycistronic transcripts possess a TMG cap. Immunoprecipitations were carried out with anti-TMG antibodies (αTMG) or non-immune anti-mouse IgGs (NI) by using 20 μg of total RNA purified from cells lacking RNase III (rnt1Δ) or from isogenic wild-type cells (WT). Following interaction with the antibodies, RNAs were purified from the supernatants (S) or from the protein A-Sepharose pellets (P) and used in primer extension experiments performed with an anti-snR78 oligonucleotide. Primer extension products were resolved on a 6% polyacrylamide sequencing gel. As controls, primer extensions were also performed with 5 μg of total RNA from the rnt1::HIS3 (lane 2) and isogenic wild-type (lane 1) strains. Sequence ladders were obtained with the same oligonucleotide primers. Two identical sets of samples were loaded onto separate gels, one set being run longer to better resolve the longer extension products. Only the relevant parts of each gel are shown. Filled arrowheads point to the major long extension products, and hollow arrowheads point to cDNA of mature snR78.
DISCUSSION
Illustrating the interest in computer searches focusing on the non-protein-coding part of a genome, the identification of novel box C/D snoRNAs snR72 to snR78 represents a substantial step towards the identification of the full repertoire of snoRNAs guiding rRNA ribose methylations in the yeast S. cerevisiae. Two other large sets of novel yeast box C/D snoRNAs, also detected by computer genomic searches, have been recently included in GenBank, with 8 species, Z9 to Z16, reported by L. H. Qu (47, 48) and a series of 15 additional species identified independently (33). Together, 51 of the 55 total ribose methylations present in S. cerevisiae rRNAs (34, 57) are now assigned a cognate box C/D antisense snoRNA, strongly suggesting that all sites of rRNA ribose methylation are specified through the same snoRNA-guided process. snoRNAs snR72 to snR78 are absolutely required for the seven cognate rRNA ribose methylations, which, except for the one guided by snR75 (Gm2287 in 25S rRNA), are strongly conserved in yeast and vertebrate rRNAs (34). As far as rRNA methylation is concerned, snR73, snR74, and snR78 are functional homologues of vertebrate snoRNAs U35, U27, and U52 (reference 5 and references therein), respectively, which have unlinked coding regions. The seven cognate ribose methylations are scattered within 18S or 25S rRNA structures, and there is no evidence that these clustered snoRNAs have a more particularly related function than other methylation guide snoRNAs. Their peculiar genomic organization might therefore merely represent a simple means for producing these snoRNAs in stoichiometric amounts.
Surprisingly, the lack of the seven rRNA methylations has no detectable effect on yeast growth under the laboratory conditions examined so far, similar to what has been observed after single-gene disruption of a few methylation guide snoRNAs (references 36 and 53 and references therein), which precludes any further indication about the elusive role of these rRNA modifications in the assembly or function of eukaryotic ribosomes (6).
Common components for the biosynthesis of snoRNAs and rRNAs.
Our data conclusively show that snoRNAs snR72 to snR78 are cotranscribed and that the mature snoRNA sequences are processed from a polycistronic transcript. Taken together with the demonstration that snoRNAs snR190 and U14 are synthesized from a dicistronic unit in yeast (16, 43), these results suggest that other snoRNAs processed from polycistronic precursors might still await identification in S. cerevisiae, even if the snR72-to-snR78 gene cluster could represent a unique situation because of the high number of snoRNA cistrons involved.
Remarkably, processings of the precursor to snoRNAs snR72 to snR78 and of the snR190-U14 dicistronic transcript (16, 43) involve very similar mechanisms, with in both cases monocistronic snoRNAs released by endonucleolytic cleavage within the intergenic sequences of the precursor followed by exonucleolytic trimming. While the 3′-to-5′ exonuclease(s) catalyzing formation of the 3′ ends of mature snoRNAs remains unidentified, both snoRNA precursors are cleaved by the same endonuclease, Rnt1p, and degraded by the same 5′-to-3′ exonucleases, Rat1p and to a lesser extent Xrn1p. Interestingly, Rat1p, which functions in the nucleus, is also the major 5′ processing activity of intronic snoRNAs U18 and U24 in wild-type yeast cells, while Xrn1p, which is cytoplasmic, can partially compensate for the lack of functional Rat1p (25, 43), similar to what we have observed for the processing of the precursor of snoRNAs snR72 to snR78. This points again to the existence of redundant processing/degradation pathways involving a few enzymes with a wide range of substrate specificities. Illustrating this notion, Rat1p and Xrn1p are also responsible for the degradation of several spacer regions within pre-rRNA (43), which could provide a basis for coregulating rRNA and snoRNA processings. Likewise, endonuclease RNase III is proposed to be one of E. coli’s global regulators because of its ability to affect the expression of a large number of unrelated genes by influencing posttranscriptional control of mRNA stability or translational efficiency, in addition to its multiple roles in the processing of rRNAs, tRNAs, and other small stable RNAs or in the conversion into monocistronic mRNAs of the polycistronic transcript of the bacteriophage T7 early region (18). Paradoxically, prokaryotic RNase III is not essential for bacterial viability. Similarly, the yeast equivalent of RNase III is not strictly required for yeast growth (16). Multifunctional Rnt1p, also involved in the processing of rRNA (2) and U2 and U5 snRNAs (1, 15), seems to have a key role in snoRNA metabolism (16, 16a; also this study). It is therefore tempting to propose that the endonucleolytic activity probably required for the processing of plant polycistronic snoRNA precursors (30, 31, 51) is provided by a plant homologue of Rnt1p. Likewise, the minor, splicing-independent, endonuclease-mediated processing pathway for the maturation of a few intronic snoRNAs, both in yeast (58) and in vertebrates (9), could also involve Rnt1p (or its vertebrate homologue).
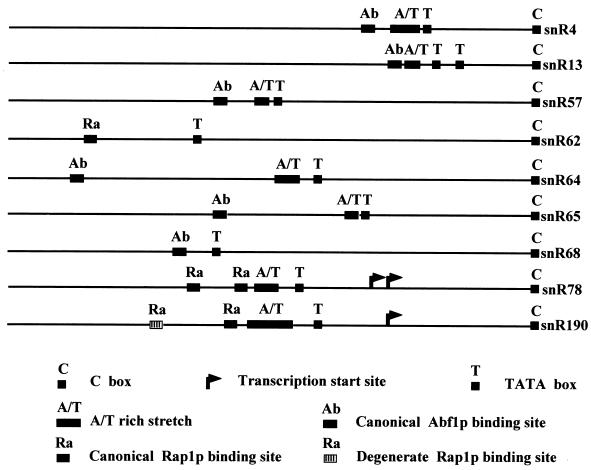
Most S. cerevisiae ribosomal protein genes contain a Rap1p or Abf1p binding site(s) followed by a T-rich sequence which together are essential for promoter activity (49, 60, 61). Deletion analysis of the polycistronic snoRNA transcript promoter showed that an 88-bp region, containing the two canonical Rap1p sites, and the AT-rich sequence upstream from the TATAAA box are sufficient for full transcriptional activity and that the latter element alone can provide 50% of the wild-type transcriptional level, as was found for the promoter of the Rp59p ribosomal protein gene (29). This suggests that coordinated production of some snoRNAs and components of cytoplasmic ribosomes might also involve common transcriptional controls, in addition to coregulated RNA processing pathways. Supporting this notion, a canonical Rap1p binding site, again closely associated with an AT-rich stretch and a TATAAA box motif, is present at a very similar location with respect to the transcription initiation site upstream of the snR190-U14 dicistronic unit. Remarkably, canonical Rap1p (or Abf1p) sites followed by an AT-rich stretch are also found upstream of several other yeast snoRNA-coding regions (Fig. 11).
FIG. 11.
Putative promoter elements found upstream of several S. cerevisiae snoRNA genes.
Recurrent themes in snoRNA gene organization and expression.
snoRNAs are produced according to a large diversity of pathways among distant eukaryotic organisms, occasionally even within a species. While most yeast snoRNAs are independently transcribed from mono-, di-, or polycistronic units, the relatively compact S. cerevisiae genome does encode a few intronic snoRNAs, U24, U18, snR38, snR39, and snR59, processed according to the vertebrate mode (4, 33, 45). The different modes of snoRNA formation are unambiguously related in their basically requiring the formation, within the snoRNA precursor, of a free entry point for a multipurpose exonuclease. Moreover, differences between these modes are blurred in a few outstanding cases. Thus, some vertebrate host genes exhibit multiple single-snoRNA-containing introns, constituting in effect snoRNA polycistronic units fused to protein-coding genes, reminiscent of the plant and yeast snoRNA polycistronic units, although the latter ones involve a splicing-independent snoRNA processing (16, 30, 31, 51; also this study). Furthermore, three host genes for mammalian intronic snoRNA do not code for proteins, exhibiting exon sequences which are not conserved between mammals, unlike the snoRNA-encoding sequences (8, 42, 56). This strongly suggests that the major if not the sole function of these genes is to express their intronic snoRNAs and that such snoRNAs have in effect a transcription unit of their own. Remarkably, one of these genes, UHG, also displays a polycistronic organization, with a cluster of seven different box C/D snoRNAs, each located in a different intron (56). Moreover, primary transcripts of the three peculiar mammalian snoRNA host genes have closely related 5′-terminal sequences, highly reminiscent of the 5′-terminal oligopyrimidine tract of ribosomal protein-coding mRNAs (37). Given the absence of protein-coding potential of the corresponding spliced RNAs, this motif is unlikely to have a role in translation and probably rather reflects the presence of promoter elements in common with ribosomal protein genes, likely providing for a coordinate expression at the transcriptional level. These three mammalian snoRNA transcription units appear thereby to be unexpectedly related to the conventional intron-encoded snoRNA units, generally hosted in genes coding for proteins directly involved in ribosome biogenesis or function (5, 6, 27, 36, 39), and also, remarkably, to the yeast independent polycistronic unit characterized in this study.
ACKNOWLEDGMENTS
We thank Guillaume Chanfreau for communicating results prior to publication. We are particularly grateful to the following persons for their gifts: M. Ares and S. Abou Elela for strains SAE-52/1 and SAE-6, S. Kearsey for strains R934 and 966-1C, C. Cole for strain DAH18, and R. Lührmann for anti-TMG antibodies. We thank A. Chambers for advice and Marlène Faubladier for experimental help and helpful discussions.
This research was supported by the National Natural Science Foundation of China (Key Project 39730300 and Fund for Distinguished Young Scholar 395250070 to L.-H.Q.), by the CNRS (Programme Physique et Chimie du Vivant 1997), the Ministère de l’Education Nationale, de l’Enseignement Supérieur et de la Recherche (MENESR, grant ACC-SV1), Université Paul Sabatier, Région Midi-Pyrénées, Association pour la Recherche sur le Cancer (ARC), and Ligue Nationale contre le Cancer. The collaboration between our labs was supported by a grant (Programme de Recherches Avancées 1998) from the Association Franco-Chinoise pour la Recherche Scientifique et Technique. A.H. was supported by a Ph.D. fellowship from MENESR.
Liang-Hu Qu and Anthony Henras contributed equally to this work.
REFERENCES
- 1.Abou-Elela S, Ares M., Jr Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J. 1998;17:3738–3746. doi: 10.1093/emboj/17.13.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abou-Elela S A, Igel H, Ares M., Jr RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Bachellerie J P, Michot B, Nicoloso M, Balakin A, Ni J, Fournier M J. Antisense snoRNAs: a family of nucleolar RNAs with long complementarities to rRNA. Trends Biochem Sci. 1995;20:261–264. doi: 10.1016/s0968-0004(00)89039-8. [DOI] [PubMed] [Google Scholar]
- 5.Bachellerie J P, Cavaillé J. Guiding ribose methylation of rRNA. Trends Biochem Sci. 1997;22:257–261. doi: 10.1016/s0968-0004(97)01057-8. [DOI] [PubMed] [Google Scholar]
- 6.Bachellerie J P, Cavaillé J. Small nucleolar RNAs guide the ribose methylations of eukaryotic rRNAs. In: Grosjean H, Benne R, editors. Modification and editing of RNA. Washington, D.C: ASM Press; 1998. pp. 255–272. [Google Scholar]
- 7.Balakin A G, Smith L, Fournier M J. The RNA world of the nucleolus: two major families of small nucleolar RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 8.Bortolin M L, Kiss T. Human U19 intron-encoded snoRNA is processed from a long primary transcript that possesses little potential for protein coding. RNA. 1998;4:445–454. [PMC free article] [PubMed] [Google Scholar]
- 9.Caffarelli E, Arese M, Santoro B, Fragapane P, Bozzoni I. In vitro study of processing of the intron-encoded U16 small nucleolar RNA in Xenopus laevis. Mol Cell Biol. 1994;14:2966–2974. doi: 10.1128/mcb.14.5.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caffarelli E, Fatica A, Prislei S, De Gregorio E, Fragapane P, Bozzoni I. Processing of the intron-incoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing and the stability of the mature snoRNA. EMBO J. 1996;15:1121–1131. [PMC free article] [PubMed] [Google Scholar]
- 11.Cavaillé J, Nicoloso M, Bachellerie J P. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- 12.Cavaillé J, Bachellerie J P. SnoRNA-guided ribose-methylation of rRNA: structural features of the guide RNA duplex influencing the extent of the reaction. Nucleic Acids Res. 1998;26:1576–1587. doi: 10.1093/nar/26.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavaillé J, Bachellerie J P. Processing of fibrillarin-associated snoRNAs from pre-mRNA introns: an exonucleolytic process exclusively directed by the common stem-box terminal structure. Biochimie (Paris) 1996;78:443–456. doi: 10.1016/0300-9084(96)84751-1. [DOI] [PubMed] [Google Scholar]
- 14.Cecconi F, Mariottini P, Amaldi F. The Xenopus intron-encoded U17 snoRNA is produced by exonucleolytic processing of its precursor in oocytes. Nucleic Acids Res. 1995;23:4670–4676. doi: 10.1093/nar/23.22.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chanfreau G, Abou-Elela S, Ares M, Jr, Guthrie C. Alternative 3′ end processing of U5 snRNA by RNase III. Genes Dev. 1997;11:2741–2751. doi: 10.1101/gad.11.20.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chanfreau G, Rotondo G, Legrain P, Jacquier A. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 1998;17:3726–3737. doi: 10.1093/emboj/17.13.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Chanfreau G, Legrain P, Jacquier A. Yeast RNase III as a key processing enzyme in nucleolar RNA metabolism. J Mol Biol. 1998;284:975–988. doi: 10.1006/jmbi.1998.2237. [DOI] [PubMed] [Google Scholar]
- 17.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 18.Court D. RNase III: a double strand processing enzyme. In: Brawerman G, Belasco J, editors. Control of messenger RNA stability. New York, N.Y: Academic Press; 1993. pp. 70–116. [Google Scholar]
- 19.Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA snoRNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 20.Ganot P, Bortolin M L, Kiss T. Site-specific pseudouridine formation in eukaryotic pre-rRNAs is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 21.Graham I R, Chambers A. Use of a selection technique to identify the diversity of binding sites for the yeast RAP1 transcription factor. Nucleic Acids Res. 1994;22:124–130. doi: 10.1093/nar/22.2.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herruer M H, Mager W H, Woudt L P, Nieuwint R T M, Waseenaar G M, Groeneveld P, Planta R J. Transcriptional control of yeast ribosomal protein synthesis during carbon-source upshift. Nucleic Acids Res. 1987;15:10133–10144. doi: 10.1093/nar/15.24.10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang G M, Jarmolowski A, Struck J C R, Fournier M J. Accumulation of U14 small nuclear RNA in Saccharomyces cerevisiae requires box C, box D, and a 5′, 3′ terminal stem. Mol Cell Biol. 1992;12:4456–4463. doi: 10.1128/mcb.12.10.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurt E C, McDowall A, Schimmang T. Nucleolar and nuclear envelope proteins of the yeast Saccharomyces cerevisiae. Eur J Cell Biol. 1988;46:554–563. [PubMed] [Google Scholar]
- 25.Johnson A W. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol Cell Biol. 1997;17:6122–6130. doi: 10.1128/mcb.17.10.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiss T, Filipowicz W. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev. 1995;9:1411–1424. doi: 10.1101/gad.9.11.1411. [DOI] [PubMed] [Google Scholar]
- 27.Kiss-Laszlo Z, Henry Y, Bachellerie J P, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 28.Klein C, Struhl K. Protein kinase A mediates growth-regulated expression of yeast ribosomal protein genes by modulating RAP1 transcriptional activity. Mol Cell Biol. 1994;14:1920–1928. doi: 10.1128/mcb.14.3.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larkin J C, Thompson J R, Woolford J L., Jr Structure and expression of the Saccharomyces cerevisiae CRY1 gene: a highly conserved ribosomal protein gene. Mol Cell Biol. 1987;7:1764–1775. doi: 10.1128/mcb.7.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leader D J, Clark G P, Watters J, Beven A F, Shaw P J, Brown J W S. Clusters of multiple different small nucleolar RNA genes in plants are expressed as and processed from polycistronic pre-snoRNAs. EMBO J. 1997;16:5742–5751. doi: 10.1093/emboj/16.18.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leader D J, Sanders J F, Waugh R, Shaw P, Brown J W. Molecular characterization of plant U14 small nucleolar RNA genes: closely linked genes are transcribed as a polycistronic U14 transcript. Nucleic Acids Res. 1994;22:5196–5203. doi: 10.1093/nar/22.24.5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leverette R D, Andrews M T, Maxwell E S. Mouse U14 snRNA is a processed intron of the cognate hsc70 heat-shock pre-messenger RNA. Cell. 1992;71:1215–1221. doi: 10.1016/s0092-8674(05)80069-8. [DOI] [PubMed] [Google Scholar]
- 33.Lowe, T. M., and S. R. Eddy. 1998. GenBank accession no. AF064263, AF064265, AF064266, AF064268, AF064269, AF064271, AF064274 to AF064277, and AF064279 to AF064283.
- 33a.Lührmann R, Appel B, Bringmann P, Rinke J, Reuter R, Rothe S, Bald R. Isolation and characterization of rabbit anti-m32,2,7 antibodies. Nucleic Acids Res. 1982;10:7103–7113. doi: 10.1093/nar/10.22.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maden B E H. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1990;39:241–301. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- 35.Maden B E H, Corbett M E, Heeney P A, Pugh K, Ajuh P M. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie (Paris) 1995;77:22–29. doi: 10.1016/0300-9084(96)88100-4. [DOI] [PubMed] [Google Scholar]
- 36.Maxwell E S, Fournier M J. The small nucleolar RNAs. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 37.Meyuhas O, Avni D, Shama S. Pages 363–388. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 38.Ni J, Tien A L, Fournier M L. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 39.Nicoloso M, Qu L-H, Michot B, Bachellerie J P. Intron-encoded, antisense small nucleolar RNAs: the characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation of rRNAs. J Mol Biol. 1996;260:178–195. doi: 10.1006/jmbi.1996.0391. [DOI] [PubMed] [Google Scholar]
- 40.Ofengand J, Fournier M J. The pseudouridine residues of rRNA: number, location, biosynthesis, and function. In: Grosjean H, Benne R, editors. Modification and editing of RNA. Washington, D.C: ASM Press; 1998. pp. 229–254. [Google Scholar]
- 41.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pelczar P, Filipowicz W. The host gene for intronic U17 small nucleolar RNAs in mammals has no protein-coding potential and is a member of the 5′-terminal oligopyrimidine gene family. Mol Cell Biol. 1998;18:4509–4518. doi: 10.1128/mcb.18.8.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petfalski E, Dandekar T, Henry Y, Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol Cell Biol. 1998;18:1181–1189. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qu L H, Nicoloso M, Michot B, Azum M C, Caizergues-Ferrer M, Renalier M H, Bachellerie J P. U21, a novel small nucleolar RNA with a 13 nt. complementarity to 28S rRNA, is encoded in an intron of ribosomal protein L5 gene in chicken and mammals. Nucleic Acids Res. 1994;22:4073–4081. doi: 10.1093/nar/22.20.4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qu L H, Henry Y, Nicoloso M, Michot B, Azum M C, Renalier M H, Caizergues-Ferrer M, Bachellerie J P. U24, a novel intron-encoded small nucleolar RNA with two 12 nt. long, phylogenetically conserved complementarities to 28S rRNA. Nucleic Acids Res. 1995;23:2669–2676. doi: 10.1093/nar/23.14.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qu, L. H. 1996. GenBank accession no. Z69294 to Z69300 and AJ010795 to AJ010801.
- 47.Qu, L. H. 1997. GenBank accession no. Z70300.
- 48.Qu, L. H. 1998. GenBank accession no. AJ002157, AJ002158, and AJ223032 to AJ223036.
- 49.Rotenberg M O, Woolford J L., Jr Tripartite upstream promoter element essential for expression of Saccharomyces cerevisiae ribosomal protein genes. Mol Cell Biol. 1986;6:674–687. doi: 10.1128/mcb.6.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 51.Shaw P J, Beven A F, Leader D J, Brown J W S. Localization and processing from a polycistronic precursor of novel snoRNAs in maize. J Cell Sci. 1998;111:2121–2128. doi: 10.1242/jcs.111.15.2121. [DOI] [PubMed] [Google Scholar]
- 52.Smith C M, Steitz J A. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 53.Tollervey D. Trans-acting factors in ribosome biogenesis. Exp Cell Res. 1996;229:226–232. doi: 10.1006/excr.1996.0364. [DOI] [PubMed] [Google Scholar]
- 54.Tycowski K T, Shu M D, Steitz J A. A small nucleolar RNA is processed from an intron of the human gene encoding ribosomal protein S3. Genes Dev. 1993;6:1120–1130. doi: 10.1101/gad.7.7a.1176. [DOI] [PubMed] [Google Scholar]
- 55.Tycowski K T, Smith C M, Shu M-D, Steitz J A. A small nucleolar RNA requirement for site-specific ribose methylation of rRNA in Xenopus. Proc Natl Acad Sci USA. 1996;93:14480–14485. doi: 10.1073/pnas.93.25.14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tycowski K T, Shu M-D, Steitz J A. A mammalian gene with introns instead of exons generating stable RNA products. Nature. 1996;379:464–466. doi: 10.1038/379464a0. [DOI] [PubMed] [Google Scholar]
- 57.Veldman G M, Klootwijk J, De Regt V C H F, Planta R J, Branlant C, Krol A, Ebel J P. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981;9:6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Villa T, Ceradini F, Presutti C, Bozzoni I. Processing of the intron-encoded U18 small nucleolar RNA in the yeast Saccharomyces cerevisiae relies on both exo- and endonucleolytic activities. Mol Cell Biol. 1998;18:3376–3383. doi: 10.1128/mcb.18.6.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watkins N, Leverette R, Xia L, Andrews M, Maxwell E S. Elements essential for processing intronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide boxes C and D. RNA. 1996;2:118–133. [PMC free article] [PubMed] [Google Scholar]
- 60.Woolford J L. The structure and biogenesis of yeast ribosomes. Adv Genet. 1991;29:63–118. doi: 10.1016/s0065-2660(08)60107-8. [DOI] [PubMed] [Google Scholar]
- 61.Woolford J L, Warner J R. The ribosome and its synthesis. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis and energetics. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 587–626. [Google Scholar]
- 62.Xia L, Watkins N J, Maxwell E S. Identification of specific nucleotide sequences and structural elements required for intronic U14 snoRNA processing. RNA. 1997;3:17–26. [PMC free article] [PubMed] [Google Scholar]
- 63.Zagorski J, Tollervey D, Fournier M J. Characterization of an SNR gene locus in Saccharomyces cerevisiae that specifies both dispensible [sic] and essential small nuclear RNAs. Mol Cell Biol. 1988;8:3282–3290. doi: 10.1128/mcb.8.8.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zuker M. Prediction of RNA secondary structure by energy minimization. Methods Mol Biol. 1994;25:267–294. doi: 10.1385/0-89603-276-0:267. [DOI] [PubMed] [Google Scholar]