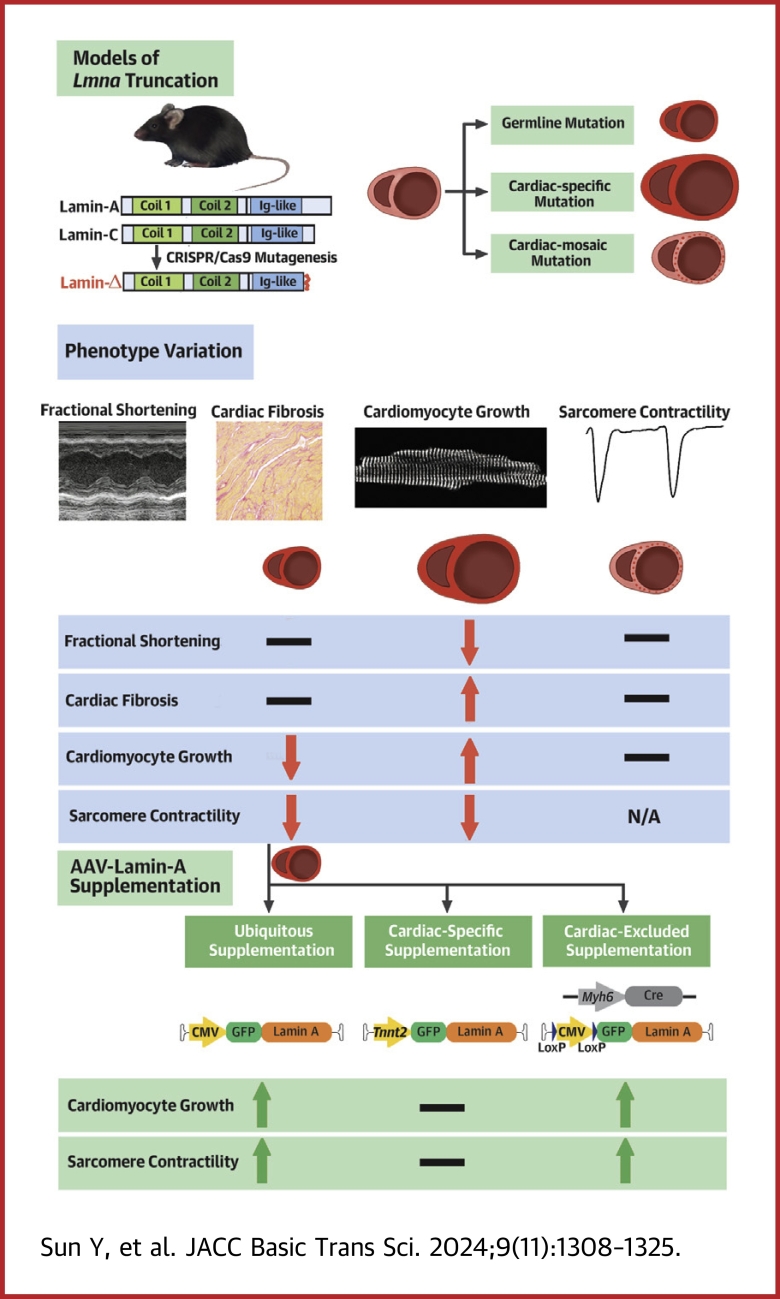
Visual Abstract
Key Words: adeno-associated virus, cardiomyocyte maturation, gene therapy, LMNA-associated cardiac defect, non-cell-autonomous
Highlights
-
•
Comparison of 3 mouse models carrying Lmna truncating mutations uncovers a non-cell-autonomous mechanism by which Lmna regulates cardiomyocyte growth and function.
-
•
Comparative analysis of 3 AAV-based lamin-A supplementation strategies elucidates noncardiomyocytes as key targets in gene therapy for Lmna-associated cardiac defects.
-
•
Genetic mosaic analysis is a crucial technique to distinguish cell-autonomous vs non-cell-autonomous effects to identify cell targets for AAV gene therapy.
Summary
The truncating mutations of LMNA are the major causes of cardiomyopathy. Here we studied 3 mouse models that carry germline, cardiomyocyte-specific, or genetic mosaic Lmna truncating mutations. Whereas the germline mutant manifested cardiac maturation defects, cardiomyocyte-specific mutation triggered pathological hypertrophy. In genetic mosaic analysis, no morphological defects were observed. Three adeno-associated virus (AAV) vectors were applied to addback lamin-A in a ubiquitous, cardiomyocyte-specific, or cardiomyocyte-excluded manner. Strikingly, only ubiquitous and cardiomyocyte-excluded AAV vectors mitigated the cardiac defects. Therefore, Lmna regulates cardiac morphology and function via a non-cell-autonomous mechanism. Noncardiomyocytes are key targets in AAV lamin-A therapy for Lmna-associated cardiac defects.
Recombinant adeno-associated viruses (AAVs) are popular gene delivery vectors for in vivo gene therapy.1 The federally approved applications of AAVs mainly involve gene supplementation therapy, in which AAVs deliver transgenes to replace or supplement endogenous genes that harbor disease-causing loss-of-function mutations.2 Myocardium is among the primary tissues that can be efficiently transduced by AAVs.3 In animal models, AAV-based gene supplementation therapy has successfully alleviated or reversed cardiac malfunctioning in an array of genetic diseases that are associated with myocardium defects, such as Danon disease,4 Barth syndrome,5 and arrhythmogenic cardiomyopathy.6 Several AAV-based clinical trials for these heart diseases are also in progress.
LMNA ranks second in prevalence among genes that cause inherited dilated cardiomyopathy.7 LMNA mutations can also lead to arrhythmogenic cardiomyopathy8 or impair cardiac hypertrophy in response to pressure overload.9 Truncating mutations, in the form of small insertions/deletions (indels), splice site mutations, or nonsense mutations, are among the major LMNA variants causing cardiomyopathy.10 The various LMNA truncating mutations share the nonsense-mediated messenger RNA (mRNA) decay mechanism that leads to cardiomyopathy due to the reduced lamin-A/C expression.11 The coding sequences of LMNA are <2 kb, which ideally fit into the 4.7 kb payload limit of AAV. Therefore, LMNA supplementation by AAV appears to be an attractive therapeutic strategy for cardiac defects that are associated with LMNA deficiency.
The gene supplementation therapy for LMNA-associated cardiomyopathy is necessary mainly because of the complicated downstream pathogenic mechanisms of LMNA mutations. LMNA deficiency is known to trigger many diverse defects in cytoskeleton organization,12 nuclear mechanics,13 signal transduction,14 DNA damage responses,15,16 chromatin organization,17 epigenetic landscape,18 and transcriptional regulation.14 Therefore, the identification of one universal therapeutic target, other than LMNA itself, to treat all these defects is very challenging. Instead, AAV-based supplementation of LMNA appears to be a more realistic approach to mitigate these disease phenotypes.
Unlike many cardiomyopathy-causing genes that are restrictively expressed in cardiomyocytes, LMNA is widely expressed in other differentiated cell types.19 Ablation of Lmna in murine and human cardiomyocytes resulted in severe cardiac dysfunction,11,20, 21, 22 leading to the general belief that the cardiomyocyte is the primary cell type that contributes to LMNA-associated heart diseases. However, accumulative evidence has started to uncover the role of noncardiomyocytes (both within and out of the heart) in LMNA-related cardiac pathogenesis.23,24 Thus, more extensive comparison of cardiomyocyte vs noncardiomyocyte would be necessary to determine whether targeting cardiomyocytes is sufficient for LMNA gene therapy.
LMNA-associated cardiac defects are often characterized by decreased systolic function, ventricular enlargement, and eccentric cardiac hypertrophy. Cardiac fractional shortening and cardiomyocyte morphology are key parameters to assess the efficacy of gene therapy for LMNA-associated cardiac defects. Unfortunately, the existing animal models carrying LMNA truncating mutations have exhibited inconsistent or even seemingly contradictory phenotypes (Table 1). For example, in an array of germline Lmna truncating mice,25, 26, 27, 28, 29, 30, 31, 32, 33 cardiomyocytes were reported to exhibit an atrophic phenotype along with whole-body growth retardation.34,35 By contrast, in mice carrying cardiomyocyte-specific Lmna loss-of-function mutations, their hearts developed elevated ventricular mass and pathological cardiomyocyte hypertrophy.20,21 Whereas multiple studies reported systolic dysfunction with decreased fractional shortening in Lmna truncating mice,25,32 other studies observed unaltered fractional shortening in mice with a similar truncating mutation30 (Table 1). Interestingly, in studies using human-induced pluripotent stem cell–derived cardiomyocytes to model LMNA-associated cardiac defects, neither atrophy nor hypertrophy phenotypes were reported, further questioning a direct, cell-autonomous role of LMNA in regulating cardiomyocyte growth and morphology. More systemic comparisons of cardiac phenotypes in distinct models are necessary to reconcile these results.
Table 1.
Summary of Mouse Models With Homozygous Lmna Truncating Mutations
| No. | Genetic Manipulation | Predicted Truncated Lamin-A/C | Death Age (wk) | Body Weight (g) |
|---|---|---|---|---|
| 1 | Exon 10 frameshift | 3-10 | <10 | |
| 2 | Exon 8-11 deletion | 5-8 | <10 | |
| 3 | Exon 4 p.R225X | 1-2 | <3 | |
| 4 | Exon 2 deletion | 2-3 | <6 | |
| 5 | Intron 1 genetrap | 2-3 | <5 | |
| 6 | Exon 2F/F;Myh6-Cre | 3-4 | <10 |
| No. | Kyphosis | Muscular Dystrophy | Lipodystrophy | Cardiac Systolic Function by LVFS | Cardiac Hypertrophy | Ref. # |
|---|---|---|---|---|---|---|
| 1 | Yes | Yes | Yes | Unaltered | Reduced | This study |
| 2 | Yes | Yes | Yes | Reduced | Reduced | 25,26,29,32,33 |
| 3 | Unknown | Unknown | Unknown | Unknown | Unknown | 27 |
| 4 | Unknown | Yes | Unknown | Reduced | Reduced | 22,28 |
| 5 | Unknown | Yes | Yes | Unaltered | Reduced | 30 |
| 6 | Unknown | Unknown | Unknown | Reduced | Increased | 20 |
LVFS = left ventricle fractional shortening.
We previously demonstrated that cardiomyocyte maturation and cardiac hypertrophy analysis can be confounded by non-cell-autonomous regulation of cardiomyocytes and the secondary effect of cardiac dysfunction.35,36 A solution to this problem is AAV-based genetic mosaics analysis. This approach harnesses low-dose AAV transduction into a small fraction of cardiomyocytes to circumvent the secondary effects.35, 36, 37, 38, 39 In this study, we generated cardiac genetic mosaic mouse models carrying Lmna truncating mutations, in comparison to the conventional germline or cardiomyocyte-specific models, to determine cell-autonomous vs non-cell-autonomous Lmna functions in cardiomyocyte growth, morphology and function. We further investigated how AAV-mediated lamin-A supplementation therapy could be complicated due to these non-cell-autonomous mechanisms.
Methods
See the Supplemental Appendix for more details about materials and methods.
Mice
All procedures involving experimental animals were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of Peking University, China (approval number LA2021332) and conformed to the Guide for the Care and Use of Laboratory Animals (8th edition, The National Academies Press, 2011) by the Association for Assessment and Accreditation of Laboratory Animal Care.
LmnaΔ/Δ mice were generated while we were generating another Lmna mutant as previously described40 at the Institute of Laboratory Animal Science, Chinese Academy of Medical Sciences, via CRISPR/Cas9-based zygotic mutagenesis. The single-guide RNA (sgRNA) sequences used in generating this allele and the genotyping primer sequences are shown in Supplemental Table 1. The RosaCas9-Tom mice were purchased from GemPharmatech (strain no. T002249). The Myh6-Cre mice were purchased from Cyagen Biosciences (strain no. C001041).
All mice were kept in a temperature-controlled room (21 ± 1 °C) with a 12-hour light/dark cycle and had free access to water and normal chow. After genotyping at postnatal day 0, neonatal LmnaΔ/Δ mice under inhalation anesthesia by isoflurane were administrated with recombinant AAVs or vehicle subcutaneously of 40 μL total volume. When necessary, adult mice were euthanized by cervical dislocation.
Adult cardiomyocyte isolation
Cardiomyocytes were isolated by retrograde perfusion.36 In brief, heparin-treated mice were anesthetized with 3% isoflurane. Hearts were extracted and cannulated onto a Langendorff perfusion apparatus. Perfusion buffer at 37 °C was first pumped into the heart to flush out blood and equilibrate the heart. Collagenase II (LS004177, Worthington) was next perfused into the heart for 8 minutes at 37 °C to dissociate cardiomyocytes. The apex was cut from the digested heart, gently dissociated into single cardiomyocytes in 10% fetal bovine serum/perfusion buffer and filtered through a 100-μm cell strainer to remove undigested tissues. To prepare for immunostaining, cardiomyocytes were cultured in Dulbecco modified Eagle medium + 10% fetal bovine serum medium for 30 minutes on a laminin-coated cover glass before fixation with 4% paraformaldehyde and permeabilization with 4% bovine serum albumin + 0.1% TritonX-100/phosphate buffered saline.
High throughput sequencing
RNA-sequencing (RNA-seq) libraries were constructed using NEBNext Ultra RNA Library Prep Kit for Illumina (E7530L, NEB). Sequencing was performed on an Illumina NovaSeq 6000 platform with 2 × 150 base pairs pair-end reads at Novogene, China. RNA-seq reads were aligned to mm10 by STAR41 and reads counts were calculated by FeatureCounts.42 DESeq243 was used to perform statistical analysis of differential gene expression. An adjusted P value <0.05 (Benjamini-Hochberg false discovery rate) was used as the cutoff to identify differentially regulated genes. Gene Ontology term analysis was performed using gene set enrichment analysis (GSEA) with ranked gene lists.44
For amplicon sequencing, genomic DNA was extracted using TIANamp Genomic DNA Kit (DP304, TIANGEN). The sgRNA-targeted loci were amplified using Taq PCR MasterMix (KT211, TIANGEN) and purified by TIANgel Purification Kit (DP219, TIANGEN). See Supplemental Table 1 for primer sequences that contain all sequences necessary for library construction. Sequencing was performed on an Illumina NovaSeq 6000 platform at Novogene. The sequencing results were processed by CRISPResso (version 2.2.14).45 The output bam files were transformed into sam files by samtools (version 1.17)46,47 and exploited to calculate the frameshifting indel rates of the target regions using a home-made python script.
AAV design and production
The human lamin-A coding sequence was acquired at Addgene (#124268) and subcloned into the AAV-Tnnt2-GFP-v239 (#165036, Addgene; control vector in this study) to build the AAV-Tnnt2-GFP-LA plasmid. Subsequently, the Tnnt2 promoter was replaced by the cytomegalovirus (CMV) promoter or the LoxP-CMV-LoxP cassette to generate AAV-CMV-GFP-LA and AAV-LoxP-CMV-LoxP-GFP-LA plasmids, respectively. These new plasmids will be available at Addgene soon after publication. CRISPR/Cas9/AAV9-mediated somatic mutagenesis (CASAAV) plasmids were produced as previously described.36 Among the 2 sgRNAs used to target Lmna, 1 sgRNA is identical to the one used to generate the LmnaΔ allele.40
AAV production was performed in house or at PackGene Biotech. We produced AAV9 as previously described.36,48 In brief, 140 μg AAV-ITR, 140 μg AAV9-Rep/Cap, and 320 μg pHelper (pAd-deltaF6, Penn Vector Core) plasmids were produced by maxiprep (DP117, TIANGEN), and triple transfected into HEK293T cells in 10 15-cm plates. Then, 60-72 hours after transfection, cells were resuspended in lysis buffer (20 mmol/L Tris pH = 8, 150 mmol/L NaCl, 1 mmol/L MgCl2, 50 μg/mL UltraNuclease (20156ES60,YEASEN)) and lysed by 2 freeze-thaw cycles. AAV in culture medium was precipitated by PEG8000 (97061-100, VMR Life Science), resuspended in lysis buffer, and pooled with cell lysates. AAV particles were next purified in an Optiprep density gradient (D1556, Sigma) by ultracentrifugation (Optima XPN-100, Beckman Coulter) with a type 70Ti rotor. The AAV were next concentrated in phosphate buffered saline with 0.001% pluronic F68 (24040032; Invitrogen) using a 100-kD filter tube (UFC910024, Thermo Fisher Scientific). AAV titer was quantified by real-time quantitative polymerase chain reaction (RT-qPCR) using a fragment of the green fluorescent protein (GFP) coding sequence to make a standard curve. See Supplemental Table 1 for primer information.
Cardiomyocyte contractility assay
Isolated ventricular cardiomyocytes were incubated in a serial concentration gradient of CaCl2 (0.375 mmol/L, 0.75 mmol/L, and 1.2 mmol/L). A IonOptix cardiomyocyte contractility system (FSI700, IonOptix) was used to measure cardiomyocyte contractility that was stimulated at 1 HZ, 10 V. The IonWizard (IonOptix) acquisition module was used to capture changes in sarcomere length. Each cardiomyocyte was recorded with 10-20 stable cycles, which were averaged by the software to reduce noise. Peak shortening (the maximal shortening of sarcomere length during contraction), fractional shortening (peak shortening divided by the diastolic sarcomere length), and the relaxation speed were automatically calculated.
Statistical analysis
Statistical analysis and plotting were performed using GraphPad Prism for Windows (version 9.5.1, GraphPad Software). Dependent on the data distribution, data were presented in figures using the mean ± SD in bar plots or median with 25th and 75th percentiles (Q1-Q3) in box/violin plots using dots to represent raw data points. Normal distribution was tested via the Shapiro–Wilk test. Comparisons were performed using Student's unpaired t-test for normally distributed data or Mann-Whitney U test for skewed data. Kaplan-Meier curves compared with log-rank test were used to evaluate survival. Categorical data were compared using the chi-square test. A P value <0.05 was considered statistically significant.
Data availability statement
Next-generation sequencing data are deposited at National Genomic Data Center under the accession code CRA011590 (6-week LmnaΔ/Δ and Lmna+/+ RNA-seq), CRA011582 (2-week Lmna+/+ RNA-seq), CRA011594 (CASAAV-treated ventricle tissue amplicon-seq), and CRA015796 (CASAAV-treated FACS-sorted cardiomyocyte amplicon-seq). AAV plasmids are available at Addgene. Other data are available on reasonable request.
Results
Germline Lmna truncation perturbs cardiomyocyte growth while maintaining normal fractional shortening
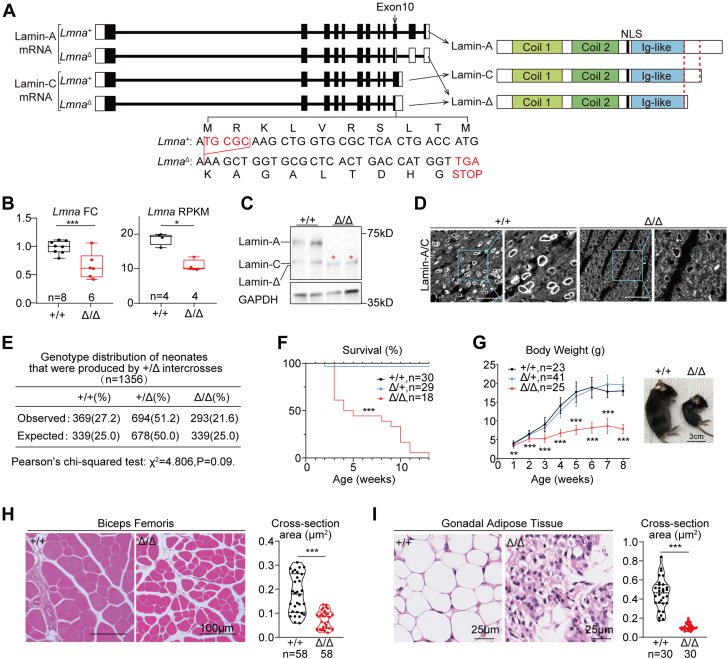
Lamin-A and lamin-C are coded by 2 Lmna transcripts that differ in mRNA splicing at exon 10 (Figure 1A). To test whether Lmna truncating mutations at the very end of the gene could still lead to dilated cardiomyopathy, a 5–base pair deletion, denoted as LmnaΔ, was introduced into exon 10 (Figure 1A) via Cas9-mediated zygotic gene editing.40 LmnaΔ was expected to shift the open reading frames for both lamin-A and lamin-C, producing a protein truncated at the C-terminus named lamin-Δ (Figure 1A). RT-qPCR and mRNA RNA-seq analysis revealed reduced Lmna mRNA level in postnatal day 14 (P14) LmnaΔ/Δ hearts (Figure 1B), suggesting nonsense-mediated mRNA decay as expected.11 Western blot validated the depletion of lamin-A/C, which was replaced by the expression of lamin-Δ (Figure 1C). Immunofluorescence analysis revealed dramatically reduced signals at the nuclear periphery in LmnaΔ/Δ hearts (Figure 1D). The reduction and truncation of lamin-A/C proteins were also validated in LmnaΔ/Δ liver and skeletal muscle (Supplemental Figure 1A).
Figure 1.
A New Mouse Model Carrying a Lmna Truncating Mutation
(A) Gene structures of wild-type and mutant Lmna variants (left) and their corresponding protein products (right). Filled boxes indicate translated regions and unfilled boxes indicate untranslated regions in exons. Mutation sites in exon 10 are enlarged to show the 5–base pair deletion and the premature stop codon. (B) Real-time quantitative polymerase chain reaction and RNA-sequencing quantification of Lmna messenger RNA (mRNA) in postnatal day 14 heart apexes. Data presented using actual data points and box plots. (C) Western blot analysis of Lmna-expressed proteins in postnatal day 14 heart apexes. Red stars indicate the truncated protein bands. (D) Immunofluorescence of Lmna-expressed proteins in heart sections. Bars = 50 μm. (E) Genotype distribution analysis of neonates. (F) Survival curve with the log-rank test between Lmna+/+ and LmnaΔ/Δ; ∗∗∗P < 0.001. (G) Growth curve presented as mean ± SD at each time point. Student’s t-test between Lmna+/+ and LmnaΔ/Δ with unpaired multiple-testing adjustment of 2-stage step-up (Benjamini, Krieger, and Yekutieli); ∗∗False discovery rate (FDR) < 0.01, ∗∗∗FDR < 0.001. A bright-field image of 6-week-old mice to the right. (H,I) Hematoxylin and eosin staining on biceps femoris sections (H) and gonadal adipose tissue sections (I) and the quantification of cross-section area. (B,H,I) Mann-Whitney U test; ∗P < 0.05, ∗∗∗P < 0.001. FC = fold change; mRNA = messenger RNA; NLS = nuclear localization sequence; RPKM = reads per kilobase per million mapped reads.
LmnaΔ/+ intercrosses reproduced Lmna+/+, LmnaΔ/+, and LmnaΔ/Δ pups following the Mendelian ratio at birth (Figure 1E). Before the 13th week after birth, all LmnaΔ/Δ animals died (Figure 1F). The body growth of Lmna+/+ and LmnaΔ/+ mice appeared indistinguishable in the first 2 months after birth, whereas LmnaΔ/Δ pups exhibited severe retardation in gaining body weight (Figure 1G). Whole-body magnetic resonance imaging demonstrated severe reduction of skeletal muscles, adipose tissues and bones in the LmnaΔ/Δ animals (Supplemental Figure 1B). The muscular dystrophy and lipodystrophy phenotypes were further validated by histological analyses (Figures 1H and 1I, Supplemental Figures 1C and 1D).
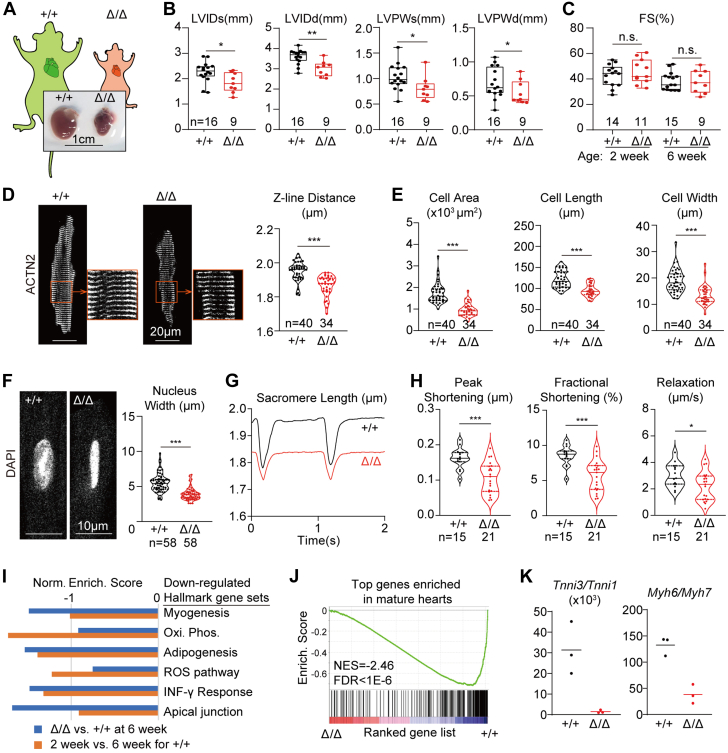
To further determine whether there are any global defects in LmnaΔ/+ mice, these mice were next examined at 6 months after birth. No body weights or heart phenotypes were observed at this age in LmnaΔ/+ mice (Supplemental Figures 1E and 1F). By contrast, the hearts of the LmnaΔ/Δ mice were significantly smaller than their control littermates (Figure 2A). Echocardiogram analysis demonstrated reduced left ventricular diameters (left ventricular internal diameter in systole [LVIDs] and left ventricular internal diameter in diastole [LVIDd]) and posterior wall thickness (left ventricular posterior wall thickness in systole [LVPWs] and left ventricular posterior wall thickness in diastole [LVPWd]) in LmnaΔ/Δ hearts (Figure 2B). However, the cardiac fractional shortening (FS) of LmnaΔ/Δ mice were comparable to control littermates at 2 and 6 weeks of age (Figure 2C). No sex influence on the hearts was found among LmnaΔ/Δ mice (Supplemental Figure 2A). In 6-week hearts, picrosirius red staining detected no changes in cardiac fibrosis (Supplemental Figure 2B). RT-qPCR and RNA-seq analysis showed little up-regulation of cardiac pathological markers Ctgf and Nppa (Supplemental Figures 2C and 2D).
Figure 2.
Cardiac Maturation Defects in LmnaΔ/Δ Mice
(A) Bright-field images of hearts extracted from 6-week-old mice. (B,C) Echocardiogram analysis of the 6-week-old mice. (D) Immunostaining of ACTN2 on isolated cardiomyocytes and quantification of the distances between adjacent Z-lines. (E) Quantification of cardiomyocyte projected cell area, cell length, and cell width. (F) DAPI (4′,6-diamidino-2-phenylindole)-stained cell nuclei and quantification of nuclear morphology. (G) Representative traces of sarcomere length changes during cardiomyocyte contraction on 1 Hz electrical stimulation. (H) Quantification of contractility parameters. (D-H) Cells from littermates were compared. (B to H) Mann-Whitney U test; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, NS. (I) Gene set enrichment analysis of the differentially expressed genes by RNA-sequencing. (J) Enrichment of cardiac mature markers among down-regulated genes in LmnaΔ/Δ ventricles. (K) Quantification of cardiomyocyte maturation isoform switching markers in RNA-sequencing data. FDR = false discovery rate; FS = fractional shortening; LVIDd = left ventricular internal diameter in diastole; LVIDs = left ventricular internal diameter in systole; LVPWd = left ventricular posterior wall in diastole; LVPWs = left ventricular systolic posterior wall in systole; NES = normalized enrichment score.
Next the LmnaΔ/Δ cardiac phenotypes were examined at the cellular level by analyzing cardiomyocytes that were isolated via Langendorff collagenase perfusion. Immunostaining of α-actinin-2 (ACTN2), a marker for sarcomere Z-lines, revealed reduced sarcomere length in the mutant cardiomyocytes (Figure 2D). These mutant cardiomyocytes appeared smaller in projected cell area, cell length, and cell width (Figure 2E). Agreeing with previous studies,49 the morphology of LmnaΔ/Δ cardiomyocyte nuclei appeared disrupted mainly because of reduced nuclear width (Figure 2F). These morphological phenotypes were repeatedly observed in multiple pairs of littermates (Supplemental Figures 2E to 2J). Interestingly, during 1 Hz electrical stimulation, LmnaΔ/Δ sarcomeres contracted to a lesser extent than control sarcomeres did, and sarcomere relaxation was also slower (Figures 2G and 2H).
To further investigate the LmnaΔ/Δ cardiac phenotypes at the gene expression level, RNA-seq differential expression analysis of 6-week Lmna+/+ vs LmnaΔ/Δ heart ventricles were performed (Supplemental Figures 2K and 2L, Supplemental Table 2). As reference data for postnatal heart maturation, we also conducted RNA-seq analysis to compared 2-week vs 6-week wild-type ventricles (Supplemental Figures 2M and 2N, Supplemental Table 3). GSEA using the hallmark gene set50 uncovered significantly reduced myogenesis, oxidative phosphorylation, and adipogenesis gene expression in the LmnaΔ/Δ group (Figure 2I). The same gene sets were also lower in the 2-week wild-type group (Figure 2I), confirming that these gene sets were relevant to cardiac maturation.34,51 We used the 2-week vs 6-week analysis to build a reference gene set of maturation markers and performed GSEA on this gene set. This analysis showed transcriptome-wide reduction of cardiac maturation genes in LmnaΔ/Δ hearts (Figure 2J). Tnni3:Tnni1 and Myh6:Myh7 ratios, which are the classic maturation metrics,34,51 were also reduced (Figure 2K). Together, these data showed that LmnaΔ/Δ cardiomyocytes exhibit transcriptional, morphologic, and functional defects in maturation, but these phenotypes were not converted into pathologic changes in cardiac FS, hypertrophy, or fibrosis.
Cardiomyocyte-specific Lmna truncation results in dilated cardiomyopathy with pathologic hypertrophy
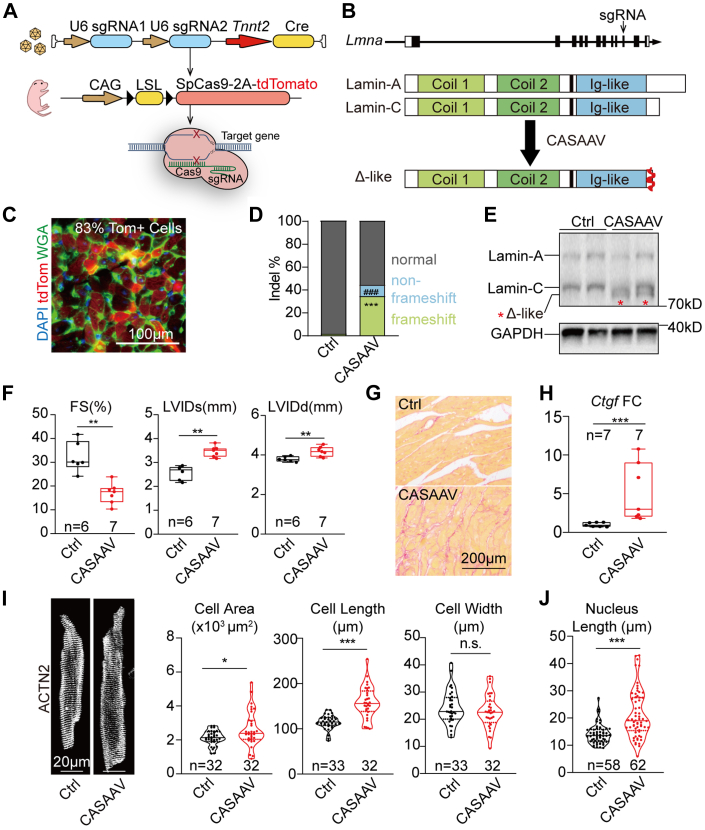
Because the LmnaΔ truncating mutation is localized at the very C-terminus of lamin-A/C, the truncated fragment might be too small to cause cardiac systolic dysfunction in LmnaΔ/Δ mice. Alternatively, the relatively normal FS of the LmnaΔ/Δ heart could originate from a compensatory effect of the whole-body growth retardation and therefore a lower demand for the heart. To test between these 2 hypotheses, cardiomyocyte-specific LmnaΔ-like mutations were created via a CASAAV system.36,48 In detail, an AAV9 vector was designed to express sgRNAs targeting Lmna exon 10 at nearly the same loci where the LmnaΔ mutation was located (Figures 3A and 3B). This vector utilized the Tnnt2 promoter (also called cardiac troponin T promoter)52 to express Cre recombinase specifically in cardiomyocytes. Therefore, when this AAV-Tnnt2-Cre-U6-Lmna-sgRNA (AAV-Lmna-sgRNA) vector was administered into RosaCAG-LSL-Cas9-tdTomato (RosaCas9-Tom) mice,48 CRISPR/Cas9-based Lmna mutagenesis would create truncating mutations specifically in cardiomyocytes that were labelled by tdTomato (Figures 3A and 3B).
Figure 3.
Cardiac Hypertrophy in Mice Undergoing CASAAV-Based Lmna Mutagenesis
(A) A diagram showing the workflow of CRISPR/Cas9/AAV9-mediated somatic mutagenesis (CASAAV). (B) The expected protein expression consequence of CASAAV. (C) A representative fluorescence image of heart sections undergoing CASAAV. TdTomato (tdTom) labels adeno-associated virus–transduced mutant cardiomyocytes. (D) Quantification of insertions/deletions (indels) by amplicon sequencing. Student’s t-test; ∗∗∗P < 0.001 (frameshifting indels), ###P < 0.001(nonframeshifting indels). (E) Western blot analysis of CASAAV-induced Lmna-expressed proteins in postnatal day 14 heart apexes. (F) Echocardiogram analysis of the 6-week-old mice. (G) Picrosirius red staining on 6-week-old heart sections. (H) Real-time quantitative polymerase chain reaction analysis of Ctgf. (I) Immunostaining of ACTN2 on isolated cardiomyocytes and quantification of cardiomyocyte morphology. (J) Nuclear shape analysis. (I,J) Cells from littermates were compared. (F, H toJ) Mann-Whitney U test; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, NS. Ctrl = control; sgRNA = single-guide RNA; WGA = wheat germ agglutinin; other abbreviations as in Figure 2.
We first applied 1 × 1014 vg/kg AAV-Lmna-sgRNA into P1 RosaCas9-Tom mice and analyzed heart samples at 6 weeks after birth. Fluorescence imaging of cardiac cryosections detected tdTomato signals in more than 80% cardiomyocytes (Figure 3C). Fluorescence-activated cell sorting (FACS) enriched tdTomato-positive cardiomyocytes (Supplemental Figure 3A) to be assessed by targeted amplicon-sequencing analyses, which confirmed the successful induction of frameshifting mutations similar to LmnaΔ (Figure 3D, Supplemental Figures 3A and 3B). The indel rates in FACS-enriched cardiomyocytes are significantly higher than in unsorted tissues, validating that CASAAV selectively edited cardiomyocytes over nonmyocytes in the heart (Supplemental Figure 3A). As compared to AAV-Tnnt2-Cre–treated control hearts, Western blot detected the expression of truncated lamin-Δ–like proteins in the CASAAV-treated hearts at the molecular weight similar to the lamin-Δ protein in the LmnaΔ/Δ hearts (Figure 3E).
By echocardiogram, CASAAV-mediated Lmna mutagenesis resulted in a significant reduction of FS in the heart accompanied with elevated LVIDd and LVIDs (Figure 3F). The body weights of the CASAAV-treated animals were slightly lower than the weights of control animals (Supplemental Figure 3C). No sex difference was detected in cardiac dysfunction among the CASAAV-treated animals (Supplemental Figure 3D). Picrosirius red–labeled cardiac interstitial fibrosis (Figure 3G) and the expression of Ctgf and Nppa were also increased (Figure 3H, Supplemental Figure 3E). CASAAV treatment increased projected cardiomyocyte area and cardiomyocyte length, whereas cell width was not affected (Figure 3I). Similarly, CASAAV-treated cell nuclei become elongated (Figure 3J). Cardiomyocyte contractility assay further validated the impaired contractile function of CASAAV-treated cells (Supplemental Figures 3F and 3G). Therefore, in sharp contrast to the normal cardiac FS and the reduced cardiomyocyte size in LmnaΔ/Δ mice, cardiomyocyte-specific Lmna truncation causes systolic dysfunction with pathological hypertrophy.
Cardiac genetic mosaic analysis uncovered Non-cell-autonomous regulation of cardiomyocyte morphology by Lmna
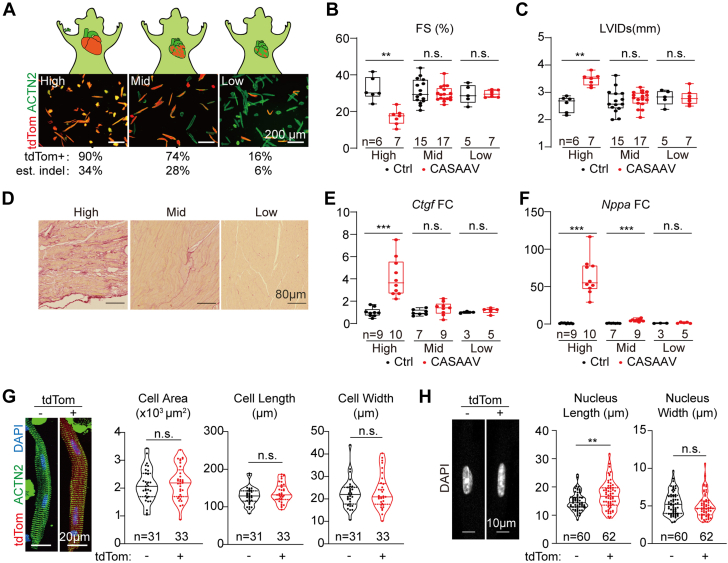
The seemingly contradictory cardiomyocyte morphology phenotypes in germline (cell atrophy) vs cardiomyocyte-specific (cell hypertrophy) Lmna mutants (Figure 2E vs Figure 3I) suggested that the impact of Lmna mutation on cardiomyocyte morphology is not cell-autonomous but is secondary to their different pathophysiological states. To test this idea, a cardiac genetic mosaic analysis36,37 was established by injecting a serial dilution of the AAV-Lmna-sgRNA vectors at high (1 × 1014 vg/kg), mid (1 × 1013 vg/kg) and low (1 × 1012 vg/kg) doses into P1 RosaCas9-Tom mice. Quantification of isolated cardiomyocytes showed ∼90%, ∼74%, and ∼16% AAV transduction rates (Figure 4A). Considering a ∼38% frameshifting mutation rate (Figure 3D), these transduction rates could be converted into ∼34%, 28%, and 6% mutation rates in cardiomyocytes.
Figure 4.
No Cardiac Morphology Changes in Lmna Cardiac Mosaics
(A) Fluorescence images of cardiomyocytes that were isolated from mice treated with serial dilutions of the CASAAV vectors. TdTomato-positive rate and estimated (est.) indel rate are labeled below the images. High, 1 × 1014 vg/kg; mid, 1 × 1013 vg/kg; low, 1 × 1012 vg/kg. (B,C) Echocardiogram analysis of the 6-week-old mice treated with diluted adeno-associated virus. (D) Picrosirius red staining on 6-week-old heart sections. (E,F) Real-time quantitative polymerase chain reaction analysis of Ctgf and Nppa. (G) Immunostaining of ACTN2 on isolated cardiomyocytes and quantification of cardiomyocyte morphology of tdTomato-positive versus -negative cells from the same heart. (H) Quantification of nuclear morphology. (B,C,E to H) Mann-Whitney U test; ∗∗P < 0.01, ∗∗∗P < 0.001, NS. Abbreviations as in Figure 1, Figure 2, Figure 3.
Echocardiography was performed to determine the impact of AAV dilution on heart functions. In the mid- and low-dose AAV-treated group, FS and LVID defects are no longer detectable (Figures 4B and 4C). The increased cardiac fibrosis and the up-regulation of Ctgf and Nppa in CASAAV-treated hearts were also eliminated in the low-dose group (Figures 4D to 4F). Therefore, in the low-dose CASAAV group, the number of mutant cardiomyocytes was so low that they were insufficient to trigger cardiac dysfunction, allowing us to separate the primary defects in cardiomyocytes from the secondary effects of pathogenesis. Based on this rationale, the phenotypes of the mutant cardiomyocytes were further analyzed in a grossly normal physiological microenvironment.
Isolated cardiomyocytes that were derived from the low-dose CASAAV-treated genetic mosaic hearts were next studied. Comparison between the tdTom-positive vs tdTom-negative cells in the same heart revealed no cardiomyocyte morphology phenotypes (Figure 4G). However, the nuclei in the tdTom-positive cardiomyocytes appeared elongated as compared to the tdTom-negative cells (Figure 4H). These data indicate that Lmna regulates cardiomyocyte morphology via a non-cell-autonomous mechanism. By contrast, nuclear shape was regulated in a cell-autonomous manner.
Global AAV-lamin-A supplementation mildly restores defects in LmnaΔ/Δ hearts
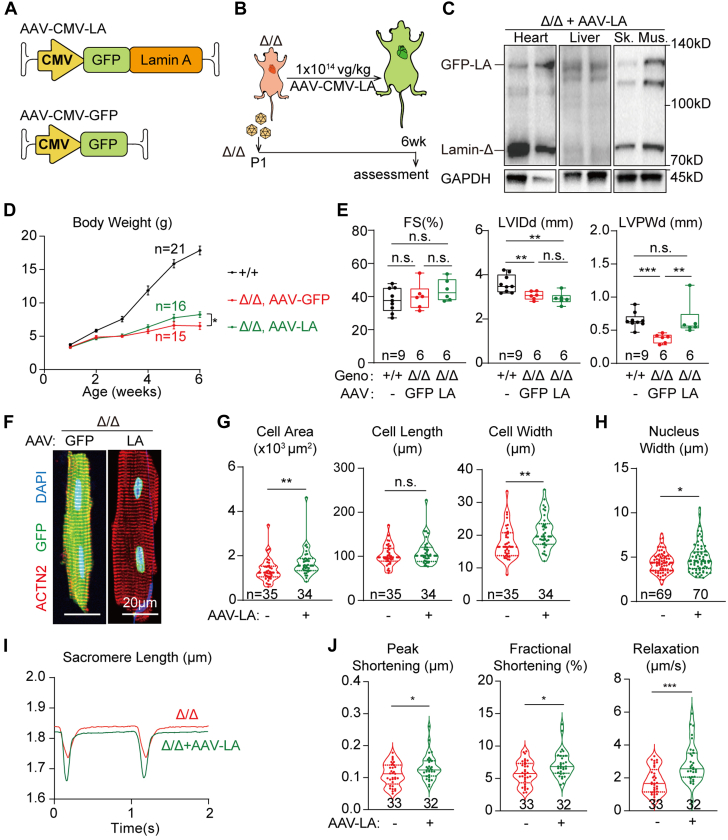
We next wondered whether AAV-based Lmna supplementation could mitigate the cardiac atrophic defects in LmnaΔ/Δ mice. An AAV9 vector was first constructed to deliver GFP-tagged lamin-A via the constitutive CMV promoter (AAV-CMV-LA) to assess its activity in wild-type mice. A single dose of 1 × 1014 vg/kg AAV-CMV-LA was subcutaneously injected at P1 to validate the transgene expression at P14 (Supplemental Figure 4A). Fluorescence imaging confirmed the nuclear lamina localization of the GFP-lamin-A proteins (Supplemental Figure 4B). Western blot detected GFP-lamin-A proteins at the level slightly lower than the endogenous lamin-A/C proteins (Supplemental Figure 4C). The GFP-lamin-A signals were mainly detected in the heart, liver, and skeletal muscles, but not brain, lung, kidney, spleen, thymus, or intestine (Supplemental Figures 4B to 4D), which agreed to the known biodistribution of AAV9 on systemic administration. Histologic analysis revealed moderate inflammatory cell infiltration into the liver after AAV-CMV-LA treatment for 6 months (Supplemental Figure 4E). However, the liver changes could not be detected at 2 weeks after AAV injection (Supplemental Figure 4F), thus the following gene therapy studies were unlikely affected by liver damages in the first several weeks after AAV administration.
Based on the above-mentioned assessment in wild-type mice, the same dose of AAV-CMV-LA was next injected into LmnaΔ/Δ mice to study the impact on cardiac defects (Figures 5A and 5B) using AAV-CMV-GFP as control vector. At 6 weeks after injection, Western blot validated GFP-lamin-A expression in the heart, liver, and skeletal muscle (Figure 5C). AAV-CMV-LA treatment moderately improved the body weight of LmnaΔ/Δ mice (Figure 5D). Echocardiogram revealed no changes in cardiac FS and LVID, but the ventricle wall thickness of LmnaΔ/Δ mice was brought to the values comparable to wild-type control mice (Figure 5E). AAV-CMV-LA treatment improved the width and the projected area of isolated LmnaΔ/Δ cardiomyocytes as compared to AAV-CMV-GFP treatment (Figures 5F and 5G). The nuclear shape phenotype was also alleviated by increasing the width of the mutant cell nuclei (Figure 5H). Cardiomyocyte contractility assay also confirmed the mitigation of cell functional defects on lamin-A supplementation (Figures 5I and 5J).
Figure 5.
Mild Rescue of Cardiac Defects via AAV-Based Lamin-A Addback
(A,B) A diagram showing the design of adeno-associated virus (AAV)-based lamin-A addback experiments. (C) Western blot analysis of 6-week-old LmnaΔ/Δ tissues that were treated with AAV-CMV-LA at postnatal day 1 (P1). (D) Growth curve of animals in the AAV-CMV-LA addback experiments. Mean ± SD. Student’s t-test for 6-week-old AAV-CMV-LA– vs AAV-CMV-GFP–treated LmnaΔ/Δ mice; ∗P < 0.05. (E) Echocardiogram analysis of the 6-week-old mice. (F,G) Immunostaining of ACTN2 on isolated cardiomyocytes and quantification of cardiomyocyte morphology. (H) Quantification of nuclear morphology. (F to H) Cells from littermates were compared. (I) Representative traces of sarcomere length changes during cardiomyocyte contraction on 1 Hz electrical stimulation. (J) Quantification of contractility parameters. (E,G,H,J) Mann-Whitney U test; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, NS. CMV = cytomegalovirus; Geno = genotype; GFP = green fluorescent protein; LA = GFP-lamin A; Sk. Mus. = skeletal muscle; other abbreviations as in Figure 2.
GFP-lamin-A-positive and -negative cardiomyocytes that were isolated from the same heart could be distinguished by fluorescence imaging (Supplemental Figure 4G), allowing a genetic mosaic analysis of the AAV-CMV-LA treatment. Counterstained with the cardiomyocyte cell membrane marker CAV3, LmnaΔ/Δ cardiomyocytes that either gained or lacked GFP-lamin-A did not show statistically significant difference in morphology (Supplemental Figures 4G and 4H). By contrast, the nuclear width phenotype was partially rescued (Supplemental Figure 4I). These results indicate that AAV-CMV-LA mildly restored cardiomyocyte morphology in LmnaΔ/Δ mice via a non-cell-autonomous mechanism, but rebuilt nuclear shape in a direct, cell-autonomous manner.
Lamin-A supplementation in nonmyocytes restores cardiomyocyte defects in LmnaΔ/Δ hearts
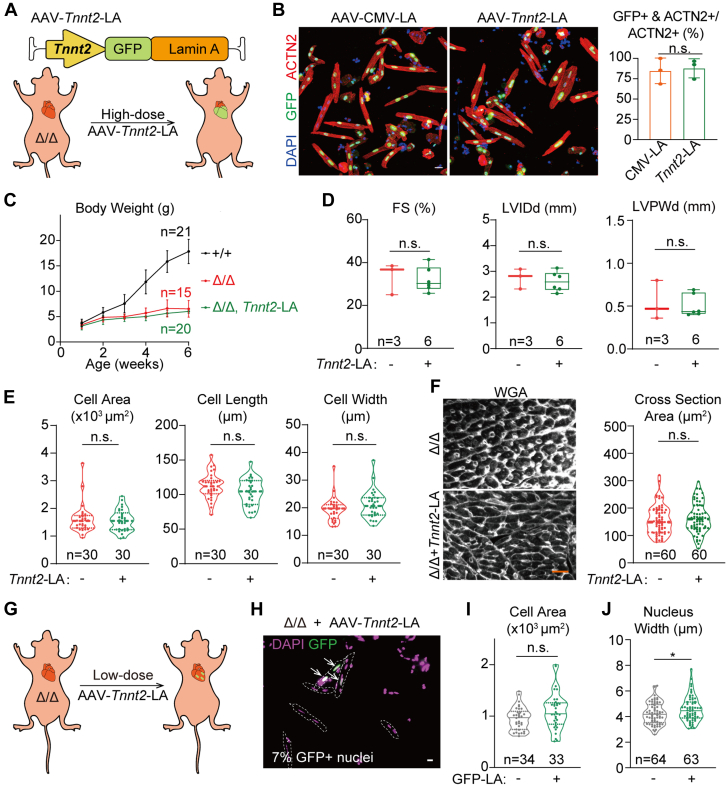
To further test the non-cell-autonomous mechanism that was implied by the above-mentioned genetic mosaic analysis, we constructed an AAV-Tnnt2-LA vector in which the Tnnt2 promoter expressed lamin-A specifically in cardiomyocytes (Figure 6A). A single dose of 1 × 1014 vg/kg AAV-Tnnt2-LA to wild-type P1 mice resulted in GFP-lamin-A expression at P14 at the level similar to endogenous lamin-A/C in the heart, but no signal was detected in the liver (Supplemental Figure 5A). When injected into LmnaΔ/Δ animals, this dose of AAV-Tnnt2-LA added back GFP-lamin-A into ∼68% nuclei in myocardium (Supplemental Figure 5B). Quantification of GFP-lamin-A-positive cardiomyocytes revealed no difference in transduction rates between AAV-Tnnt2-LA and AAV-CMV-LA treatment (Figure 6B), confirming that these 2 AAV addback experiments were comparable.
Figure 6.
No Rescue of Cardiac Morphology via Cardiomyocyte-Specific Lamin-A Addback
(A) A diagram showing the design of cardiomyocyte-specific lamin-A addback experiments. (B) Representative fluorescence images of AAV-treated cardiomyocytes and quantification of percentages of GFP-LA-–positive cardiomyocytes. Bar = 20 μm. Student’s t-test; NS. (C) Growth curve of animals in the cardiomyocyte-specific lamin-A addback experiments. (D) Echocardiogram analysis of the 6-week-old mice. (E,F) Quantification of morphology of isolated cardiomyocytes (E) or cells in heart sections (F). Bar = 20 μm. (E,F) Cells from littermates were compared. (G) A diagram showing cardiomyocyte-specific lamin-A addback in a genetic mosaic fashion. (H) Fluorescence imaging of isolated cardiomyocytes from the mosaic hearts. Cell borders was annotated with dashed lines. Arrows indicate GFP-LA–positive nuclei. Bar = 10 μm. (I,J) Quantification of cardiomyocyte (I) and nuclear (J) morphology. (D to F,I,J) Mann-Whitney U test; ∗P < 0.05, NS. Abbreviations as in Figures 2, 3, and 5.
Strikingly, comparing to control LmnaΔ/Δ animals that did not receive AAV treatment, AAV-Tnnt2-LA-treated animals did not exhibit improved body weight (Figure 6C), LVPW thickness (Figure 6D), or isolated cardiomyocyte morphology (Figure 6E). The lack of cardiac hypertrophy or other pathophysiological changes was further confirmed by histologic analysis (Figure 6F, Supplemental Figure 5C). We further administered a lower dose (5 × 1012 vg/kg) of the AAV-Tnnt2-LA vector into LmnaΔ/Δ animals and created a genetic mosaic in which only 7% nuclei were GFP-lamin-A positive (Figures 6G and 6H). Comparison between the GFP-lamin-A-positive vs -negative cells further confirmed that cardiomyocyte-specific lamin-A addback was not sufficient to rescue cardiomyocyte morphology in LmnaΔ/Δ hearts (Figure 6I), although nuclear width was significantly restored (Figure 6J).
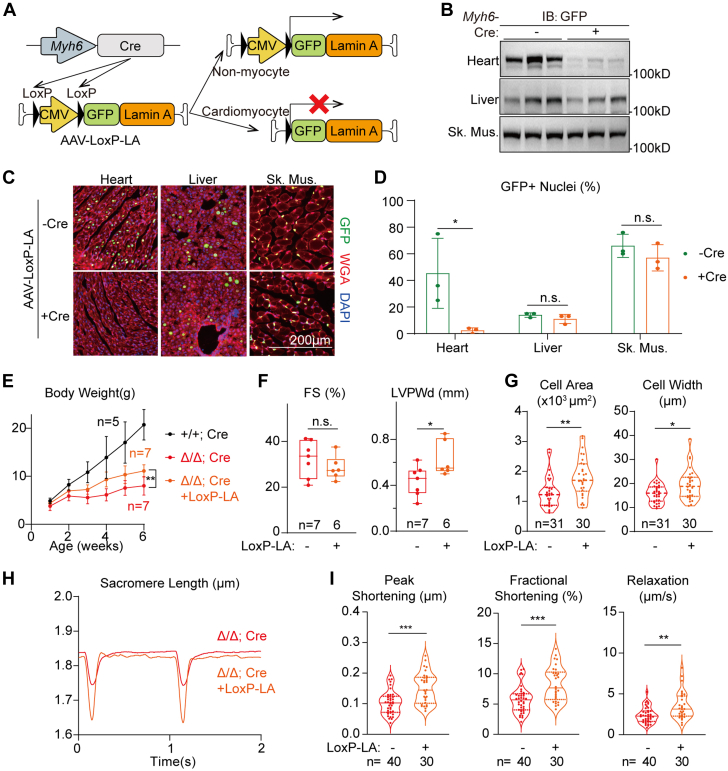
We then studied if supplementation of lamin-A into non-cardiomyocyte cell types via the same AAV serotype would be able to restore cardiomyocyte defects in LmnaΔ/Δ mice. To achieve this goal, an AAV-LoxP-CMV-LoxP-LA (AAV-LoxP-LA) vector was constructed in which the CMV promoter was flanked by LoxP sites. When this AAV was injected into mice carrying a cardiomyocyte-specific Myh6-Cre transgene, the CMV promoter would be deleted specifically in cardiomyocytes, allowing GFP-lamin-A to be expressed in AAV-transduced non-cardiomyocytes both within and out of the heart (Figure 7A). We administered 1 × 1014 vg/kg AAV-LoxP-LA into LmnaΔ/Δ;Tg(Myh6-Cre) mice at P1 and assessed transgene expression at P14. As compared to LmnaΔ/Δ mice that did not carry the Myh6-Cre allele, Western blot analysis demonstrated dramatically reduced GFP-lamin-A signal in the heart whereas the transgene expression in liver and skeletal muscles were not affected (Figure 7B, Supplemental Figure 6A). Immunofluorescence further confirmed this observation in organ sections (Figures 7C and 7D) and <4% nuclei remained GFP-lamin-A-positive, thus AAV-LoxP-LA was unlikely to rescue cardiac phenotypes by acting in the heart.
Figure 7.
Mild Rescue of Cardiac Defects via Cardiomyocyte-Excluded Lamin-A Addback
(A) A diagram showing the mechanism to achieve cardiomyocyte-excluded lamin-A addback. (B) Western blot analysis of postnatal day 14 tissues that were treated with AAV-Loxp-LA at postnatal day 1. (C) Immunostaining of AAV-Loxp-LA–treated tissues with or without the Myh6-Cre transgene. (D) Quantification of GFP-positive nuclei. (E) Growth curve in the cardiomyocyte-excluded lamin-A addback experiments. (D,E) Mean ± SD; Student’s t-test; ∗P < 0.05, ∗∗P < 0.01, NS. (F) Echocardiogram analysis of the 6-week-old mice. (G) Quantification of isolated cardiomyocyte morphology. Cells from littermates were compared. (H) Representative traces of sarcomere length changes during cardiomyocyte contraction on 1 Hz electrical stimulation. (I) Quantification of contractility parameters. (F,G,I) Mann-Whitney U test; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, NS. IB = immunoblot; other abbreviations as in Figures 2, 3, and 5.
Interestingly, AAV-LoxP-LA treatment significantly improved body weight growth and ventricular wall thickness in LmnaΔ/Δ;Tg(Myh6-Cre) mice (Figures 7E and 7F) whereas no additional cardiac fibrosis was detected (Supplemental Figure 6B). Cardiomyocyte and nuclei morphologies were both mildly restored (Figure 7G, Supplemental Figures 6C and 6D) in the LmnaΔ/Δ; Tg(Myh6-Cre) mice treated with AAV-LoxP-LA. Cell contractility tests also confirmed functional recovery of LmnaΔ/Δ; Tg(Myh6-Cre) cardiomyocytes even though AAV-LoxP-LA did not addback lamin-A in this cell type (Figures 7H and 7I). Together, these data indicate that AAV-based lamin-A supplementation mitigates LmnaΔ/Δ cardiomyocyte defects by acting in non-cardiomyocyte cell types.
Discussion
In this study, we introduced Lmna truncating mutations in 3 different forms of mouse models to understand the mechanisms by which Lmna mutations cause cardiomyocyte defects. In mice carrying the mutation throughout the body, atrophic cardiac phenotypes were observed. By contrast, cardiac-specific mutagenesis at the same genetic loci resulted in cardiac hypertrophy. These seemingly contradictory phenotypes could be reconciled via a cardiac genetic mosaic analysis, which indicated that Lmna regulated cardiomyocyte morphology via a non-cell-autonomous mechanism, depending on the pathophysiological states of the heart. The cardiac atrophic phenotype in germline mutants is secondary to global developmental defects, whereas the hypertrophic phenotype in cardiomyocyte-specific mutants is secondary to cardiac systolic dysfunction. Thus, this study highlights genetic mosaic analysis as a powerful and necessary approach to precisely define non-cell-autonomous effects in cardiac pathogenesis, circumventing secondary effects that could lead to misunderstanding of gene functions and disease mechanisms.
An array of mouse models carrying Lmna truncating mutations have been published.22,25,27, 28, 29, 30, 31,33 In Table 1, we systemically compared key phenotypes in these models together with our new LmnaΔ/Δ model. Overall, all germline mutants exhibit growth retardation and cause lethality in the first few months after birth. However, the actual body weight and death age vary greatly depending on the exact truncation sites. In our LmnaΔ/Δ model as well as the heavily studied Lmna−/− model deleted of exons 8-1125,26,29,32,33 the animals can survive to about 2 months with a body weight of ∼10 g. In these mutants, the lamin-A/C coiled coil domains and the nuclear localization signals are retained, thus the truncated proteins keep at least part of their functions in nuclear translocation and nuclear lamina polymerization. In sharp contrast, in the other models in which longer fragments including the nuclear localization signals of lamin-A/C were deleted, the resultant truncated proteins lack the ability to enter nucleus and therefore completely lose their functions in nuclear lamina. Consequently, these mice die much earlier within 1 month after birth, with body weights no more than ∼6 g.27,28,30
Although the above-mentioned protein domain analysis could explain the different phenotypes in survival and body growth, this analysis can hardly explain the inconsistent cardiac phenotypes. For example, while one model with Lmna truncated at exon 2 exhibited cardiac systolic dysfunction,22,28 the other model ablating Lmna at almost the same locus via the genetrap technique was reported to show relatively normal systolic function.30 In addition, whereas the germline Lmna exon 2–deleted mice exhibited cardiac atrophy phenotypes,22,28 cardiomyocyte-specific deletion of exon 2 using the same LmnaF/F allele resulted in pathological hypertrophy.20
These discrepant observations were recapitulated in our study comparing germline Lmna truncation vs cardiomyocyte-specific mutation at the same site. These data suggest that the cardiac phenotypes in these models are dependent on variables other than the mutation sites.
Here we propose a hypothesis that could partially reconcile these discrepancies. In germline Lmna mutants, although defective cardiac maturation leads to cardiac atrophy, this defect does not lead to cardiac dysfunction because of a compensatory effect by the retarded whole-body growth and reduced cardiac demands. By contrast, in cardiomyocyte-specific Lmna mutants, the defective cardiac function cannot meet the requirement by the normally developing body, triggering a compensatory hypertrophic response. Although this heart-body balance hypothesis nicely explained majority of the inconsistency among existing models, questions remain about why similar genotypes could lead to distinct phenotypes. Additional side-by-side comparisons of these different models are necessary to address this problem.
AAV is currently the major gene delivery vector in both basic heart research and cardiac gene therapy. AAV serotype 9 (AAV9) in combination with a Tnnt2 promoter is widely accepted as an excellent tool to achieve cardiomyocyte-specific gene transfer in vivo.52 Here we compared AAV9-CMV-LA, AAV9-Tnnt2-LA and AAV9-LoxP-LA vectors and demonstrated a non-cardiomyocyte contribution to AAV9 lamin-A supplementation therapy for Lmna-associated cardiac defects. These data indicate that it is important to extensively compare cardiomyocyte vs non-cardiomyocyte targeting when designing a cardiac gene therapy vector. This issue is particularly important when the therapeutic target is not selectively expressed in cardiomyocytes, such as in the case of LMNA.
It is important to note that lamin-A supplementation by AAV rescues nuclear morphology phenotypes in cardiomyocytes, regardless of whether lamin-A addback is targeted throughout the body or specifically in cardiomyocytes. These results agree to the well-established notion that lamin-A cell-autonomously regulates nuclear mechanics, which lays the basis for nuclear shape regulation. Interestingly, in AAV-LoxP-LA experiments when lamin-A is added back into noncardiomyocytes, nuclear morphology in cardiomyocytes is also partially rescued, uncovering a new, non-cell-autonomous layer of nuclear shape regulation by lamin-A. Overall, the distinct mechanisms by which Lmna regulates cardiomyocyte morphology vs nuclear morphology emphasize a requirement of adding back lamin-A to both nonmyocytes and myocytes in gene therapy.
Study limitations
A major limitation of this study is that the rescue effects of global LmnaΔ/Δ phenotypes are mild in all 3 AAV treatments. One likely explanation is that AAV9 adds back lamin-A mainly into heart, muscle, and liver, but not the other organs. Thus, better gene delivery vectors with broader tropisms are necessary to more globally supplement lamin-A in the germline mutants. Additionally, the therapeutic transgene in AAV could be further optimized. For example, the GFP tag could be removed to avoid potential interference of lamin-A functions. Lamin-A could also be replaced by lamin-C, which was recently suggested to exert better therapeutic effects than lamin-A.53
Another unsolved problem in this study is how to identify the key cell types that contribute to AAV-CMV-LA– or AAV-LoxP-LA–mediated mitigation of cardiac phenotypes in the germline Lmna mutants. Candidate cells include the major nonmyocytes in the heart, such as endothelial cells24 and fibroblasts,23 but the contribution of these cells was probably low due to the limited transduction efficiency by AAV. Other cell types that could potentially contribute to cardiac LMNA therapy are in nonheart organs. For example, in the AAV-LoxP-LA–based gene supplementation experiments, lamin-A was efficiently added back to the liver and skeletal muscles. Further investigation of these organs is necessary to uncover interorgan communications underlying cardiac pathogenesis and therapy (Supplemental Table 4).
Conclusions
Lmna regulates cardiomyocyte growth, morphology, and function in a non-cell-autonomous manner. Supplementation of lamin-A in noncardiomyocytes is necessary for AAV-based gene therapy for Lmna-associated cardiac defects.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE:LMNA truncating mutations frequently lead to cardiac defects in human, but their pathogenic mechanisms remain incompletely understood. In this study, we systemically compared 3 mouse models carrying Lmna truncating mutations at the same loci and uncovered the non-cell-autonomous mechanism by which Lmna regulated cardiomyocyte growth and function. This study demonstrated how systemic comparison of multiple animal models are necessary to further understand pathogenic mechanisms of the disease.
TRANSLATIONAL OUTLOOK: AAV gene therapy is emerging as a promising strategy to treat heart diseases that are associated with truncating loss-of-function mutations. Whether this strategy is suitable for LMNA-associated cardiomyopathy remains unclear. Here we showed that noncardiomyocytes in addition to cardiomyocytes are key cell types to be targeted in gene therapy for Lmna-associated heart defects. This study demonstrated how systemic comparison of multiple AAV vectors are necessary to understand the key factors that influence gene therapy for the disease.
Funding Support and Author Disclosures
This work was funded by Beijing Natural Science Foundation (7232094 to Dr Guo), the National Key R&D Program of China (2021YFF1201100 to Dr D. Zhao and 2022YFC2703100 to Drs Hu and S. Zhang), the National Natural Science Foundation of China (82222006 to Dr Guo, 82270405 to Dr Hu , 82070235 to Dr Dong, 92168113 to Dr Dong, and 32270603 to Dr Zhao), Beijing Nova Program (20220484031 to Dr Hu), the CAMS Innovation Fund for Medical Sciences (2021-I2M-5-003 to Dr Dong, 2021-I2M-1-003 to Dr S. Zhang), Haihe Laboratory of Cell Ecosystem Innovation Fund (HH22KYZX0047 to Dr Dong), and National High Level Hospital Clinical Research Funding (2022-PUMCH-D-002 and 2022-PUMCH-B-098 to Dr S. Zhang, and 2022-PUMCH-A-026 to Dr Hu). The authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
For supplemental data, methods, figures, and tables, please see the online version of this paper.
Contributor Information
Shuyang Zhang, Email: shuyangzhang103@nrdrs.org.
Yuxuan Guo, Email: guo@bjmu.edu.cn.
Appendix
References
- 1.Wang D., Tai P.W.L., Gao G.P. Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov. 2019;18(5):358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anguela X.M., High K.A. Entering the modern era of gene therapy. Annu Rev Med. 2019;70:273–288. doi: 10.1146/annurev-med-012017-043332. [DOI] [PubMed] [Google Scholar]
- 3.Rincon M.Y., VandenDriessche T., Chuah M.K. Gene therapy for cardiovascular disease: advances in vector development, targeting, and delivery for clinical translation. Cardiovasc Res. 2015;108(1):4–20. doi: 10.1093/cvr/cvv205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manso A.M., Hashem S.I., Nelson B.C., et al. Systemic AAV9.LAMP2B injection reverses metabolic and physiologic multiorgan dysfunction in a murine model of Danon disease. Sci Transl Med. 2020;12(535) doi: 10.1126/scitranslmed.aax1744. [DOI] [PubMed] [Google Scholar]
- 5.Wang S.Y., Li Y.F., Xu Y., et al. AAV gene therapy prevents and reverses heart failure in a murine knockout model of Barth syndrome. Circ Res. 2020;126(8):1024–1039. doi: 10.1161/CIRCRESAHA.119.315956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford W., Liang Y., Mataraarachchi N., et al. Plakophilin-2 gene therapy prevents arrhythmogenic right ventricular cardiomyopathy development in a novel mouse model harboring patient genetics. Faseb J. 2021;35(51) doi: 10.1096/fasebj.2021.35.S1.03193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenbaum A.N., Agre K.E., Pereira N.L. Genetics of dilated cardiomyopathy: practical implications for heart failure management. Nat Rev Cardiol. 2020;17(5):286–297. doi: 10.1038/s41569-019-0284-0. [DOI] [PubMed] [Google Scholar]
- 8.Chen L., Song J., Chen X., et al. A novel genotype-based clinicopathology classification of arrhythmogenic cardiomyopathy provides novel insights into disease progression. Eur Heart J. 2019;40(21):1690–1703. doi: 10.1093/eurheartj/ehz172. [DOI] [PubMed] [Google Scholar]
- 9.Cupesi M., Yoshioka J., Gannon J., et al. Attenuated hypertrophic response to pressure overload in a lamin A/C haploinsufficiency mouse. J Mol Cell Cardiol. 2010;48(6):1290–1297. doi: 10.1016/j.yjmcc.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Rijsingen I.A.W., Arbustini E., Elliott P.M., et al. Risk factors for malignant ventricular arrhythmias in lamin A/C mutation carriers: a European cohort study. J Am Coll Cardiol. 2012;59(5):493–500. doi: 10.1016/j.jacc.2011.08.078. [DOI] [PubMed] [Google Scholar]
- 11.Lee J., Termglinchan V., Diecke S., et al. Activation of PDGF pathway links LMNA mutation to dilated cardiomyopathy. Nature. 2019;572(7769):335–340. doi: 10.1038/s41586-019-1406-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatzifrangkeskou M., Le Dour C., Muchir A. Modulation of cytoskeleton in cardiomyopathy caused by mutations in LMNA gene. Am J Physiol Cell Physiol. 2023;324(6):C1223–C1235. doi: 10.1152/ajpcell.00471.2022. [DOI] [PubMed] [Google Scholar]
- 13.Kalukula Y., Stephens A.D., Lammerding J., Gabriele S. Mechanics and functional consequences of nuclear deformations. Nat Rev Mol Cell Biol. 2022;23(9):583–602. doi: 10.1038/s41580-022-00480-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andres V., Gonzalez J.M. Role of A-type lamins in signaling, transcription, and chromatin organization. J Cell Biol. 2009;187(7):945–957. doi: 10.1083/jcb.200904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheedipudi S.M., Asghar S., Marian A.J. Genetic ablation of the DNA damage response pathway attenuates lamin-associated dilated cardiomyopathy in mice. JACC Basic Trans Sci. 2022;7(12):1232–1245. doi: 10.1016/j.jacbts.2022.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S.N., Lombardi R., Karmouch J., et al. DNA damage response/TP53 pathway is activated and contributes to the pathogenesis of dilated cardiomyopathy associated with LMNA (lamin A/C) mutations. Circ Res. 2019;124(6):856–873. doi: 10.1161/CIRCRESAHA.118.314238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah P.P., Lv W., Rhoades J.H., et al. Pathogenic LMNA variants disrupt cardiac lamina-chromatin interactions and de-repress alternative fate genes. Cell Stem Cell. 2021;28(5):938–954.e9. doi: 10.1016/j.stem.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guenantin A.C., Jebeniani I., Leschik J., et al. Targeting the histone demethylase LSD1 prevents cardiomyopathy in a mouse model of laminopathy. J Clin Invest. 2021;131(1) doi: 10.1172/JCI136488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalo S., Kreienkamp R., Askjaer P. Hutchinson-Gilford progeria syndrome: a premature aging disease caused by LMNA gene mutations. Ageing Res Rev. 2017;33:18–29. doi: 10.1016/j.arr.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Auguste G., Rouhi L., Matkovich S.J., et al. BET bromodomain inhibition attenuates cardiac phenotype in myocyte-specific lamin A/C deficient mice. J Clin Invest. 2020;130(9):4740–4758. doi: 10.1172/JCI135922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan C.Y., Wong J.X., Chan P.S., et al. Yin yang 1 suppresses dilated cardiomyopathy and cardiac fibrosis through regulation of Bmp7 and Ctgf. Circ Res. 2019;125(9):834–846. doi: 10.1161/CIRCRESAHA.119.314794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y., Elsherbiny A., Kessler L., et al. Lamin A/C-dependent chromatin architecture safeguards naïve pluripotency to prevent aberrant cardiovascular cell fate and function. Nat Commun. 2022;13(1):6663. doi: 10.1038/s41467-022-34366-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rouhi L., Auguste G., Zhou Q., et al. Deletion of the Lmna gene in fibroblasts causes senescence-associated dilated cardiomyopathy by activating the double-stranded DNA damage response and induction of senescence-associated secretory phenotype. J Cardiovasc Aging. 2022;2(3):30. doi: 10.20517/jca.2022.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sayed N., Liu C., Ameen M., et al. Clinical trial in a dish using iPSCs shows lovastatin improves endothelial dysfunction and cellular cross-talk in LMNA cardiomyopathy. Sci Transl Med. 2020;12(554) doi: 10.1126/scitranslmed.aax9276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikolova V., Leimena C., McMahon A.C., et al. Defects in nuclear structure and function promote dilated cardiomyopathy in lamin A/C-deficient mice. J Clin Invest. 2004;113(3):357–369. doi: 10.1172/JCI19448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruan J., Liu X.G., Zheng H.L., et al. Deletion of the gene induces growth delay and serum biochemical changes in C57BL/6 mice. Asian-Australas J Anim Sci. 2014;27(1):123–130. doi: 10.5713/ajas.2013.13246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai Z.J., Lee Y.K., Lau Y.M., et al. Expression of Lmna-R225X nonsense mutation results in dilated cardiomyopathy and conduction disorders (DCM-CD) in mice: impact of exercise training. Int J Cardiol. 2020;298:85–92. doi: 10.1016/j.ijcard.2019.09.058. [DOI] [PubMed] [Google Scholar]
- 28.Kim Y., Zheng Y.X. Generation and characterization of a conditional deletion allele for Lmna in mice. Biochem Biophys Res Commun. 2013;440(1):8–13. doi: 10.1016/j.bbrc.2013.08.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sullivan T., Escalante-Alcalde D., Bhatt H., et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147(5):913–919. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubben N., Voncken J.W., Konings G., et al. Post-natal myogenic and adipogenic developmental defects and metabolic impairment upon loss of A-type lamins. Nucleus. 2011;2(3):195–207. doi: 10.4161/nucl.2.3.15731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mounkes L.C., Kozlov S., Hernandez L., Sullivan T., Stewart C.L. A progeroid syndrome in mice is caused by defects in A-type lamins. Nature. 2003;423(6937):298–301. doi: 10.1038/nature01631. [DOI] [PubMed] [Google Scholar]
- 32.Ramos F.J., Chen S.C., Garelick M.G., et al. Rapamycin reverses elevated mTORC1 signaling in lamin A/C-deficient mice, rescues cardiac and skeletal muscle function, and extends survival. Sci Transl Med. 2012;4(144) doi: 10.1126/scitranslmed.3003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jahn D., Schramm S., Schnolzer M., et al. A truncated lamin A in the Lmna(-/-) mouse line: implications for the understanding of laminopathies. Nucleus. 2012;3(5):463–474. doi: 10.4161/nucl.21676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo Y.X., Pu W.L.T. Cardiomyocyte maturation new phase in development. Circ Res. 2020;126(8):1086–1106. doi: 10.1161/CIRCRESAHA.119.315862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo Y.X., Jardin B.D., Zhou P.Z., et al. Hierarchical and stage-specific regulation of murine cardiomyocyte maturation by serum response factor. Nat Commun. 2018;9(1):3837. doi: 10.1038/s41467-018-06347-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo Y.X., VanDusen N.J., Zhang L.N., et al. Analysis of cardiac myocyte maturation using CASAAV, a platform for rapid dissection of cardiac myocyte gene function in vivo. Circ Res. 2017;120(12):1874–1888. doi: 10.1161/CIRCRESAHA.116.310283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo Y.X., Pu W.T. Genetic mosaics for greater precision in cardiovascular research. Circ Res. 2018;123(1):27–29. doi: 10.1161/CIRCRESAHA.118.313386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo Y.X., Cao Y.P., Jardin B.D., et al. Ryanodine receptor 2 (RYR2) dysfunction activates the unfolded protein response and perturbs cardiomyocyte maturation. Cardiovasc Res. 2023;119(1):221–235. doi: 10.1093/cvr/cvac077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo Y.X., Cao Y.P., Jardin B.D., et al. Sarcomeres regulate murine cardiomyocyte maturation through MRTF-SRF signaling. Proc Natl Acad Sci U S A. 2021;118(2) doi: 10.1073/pnas.2008861118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang L.Z., Sun J.H., Chen Z., et al. The LMNA p.R541C mutation causes dilated cardiomyopathy in human and mice. Int J Cardiol. 2022;363:149–158. doi: 10.1016/j.ijcard.2022.06.038. [DOI] [PubMed] [Google Scholar]
- 41.Dobin A., Davis C.A., Schlesinger F., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 43.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramanian A., Tamayo P., Mootha V.K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clement K., Rees H., Canver M.C., et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol. 2019;37(3):224–226. doi: 10.1038/s41587-019-0032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H., Handsaker B., Wysoker A., et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danecek P., Bonfield J.K., Liddle J., et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10(2):giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin J.S., Chen Z., Yang L.Z., et al. Cas9/AAV9-mediated somatic mutagenesis uncovered the cell-autonomous role of sarcoplasmic/endoplasmic reticulum calcium ATPase 2 in murine cardiomyocyte maturation. Front Cell Dev Biol. 2022;10 doi: 10.3389/fcell.2022.864516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Antoku S., Wu W., Joseph L.C., et al. ERK1/2 phosphorylation of FHOD connects signaling and nuclear positioning alternations in cardiac laminopathy. Dev Cell. 2019;51(5):602–616.e12. doi: 10.1016/j.devcel.2019.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liberzon A., Birger C., Thorvaldsdottir H., et al. The Molecular Signatures Database hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karbassi E., Fenix A., Marchiano S., et al. Cardiomyocyte maturation: advances in knowledge and implications for regenerative medicine. Nat Rev Cardiol. 2020;17(6):341–359. doi: 10.1038/s41569-019-0331-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prasad K.M.R., Xu Y., Yang Z., Acton S.T., French B.A. Robust cardiomyocyte-specific gene expression following systemic injection of AAV: in vivo gene delivery follows a Poisson distribution. Gene Ther. 2011;18(1):43–52. doi: 10.1038/gt.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan C.Y., Chan P.S., Tan H., et al. Systematic in vivo candidate evaluation uncovers therapeutic targets for LMNA dilated cardiomyopathy and risk of lamin A toxicity. J Transl Med. 2023;21(1):690. doi: 10.1186/s12967-023-04542-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Next-generation sequencing data are deposited at National Genomic Data Center under the accession code CRA011590 (6-week LmnaΔ/Δ and Lmna+/+ RNA-seq), CRA011582 (2-week Lmna+/+ RNA-seq), CRA011594 (CASAAV-treated ventricle tissue amplicon-seq), and CRA015796 (CASAAV-treated FACS-sorted cardiomyocyte amplicon-seq). AAV plasmids are available at Addgene. Other data are available on reasonable request.