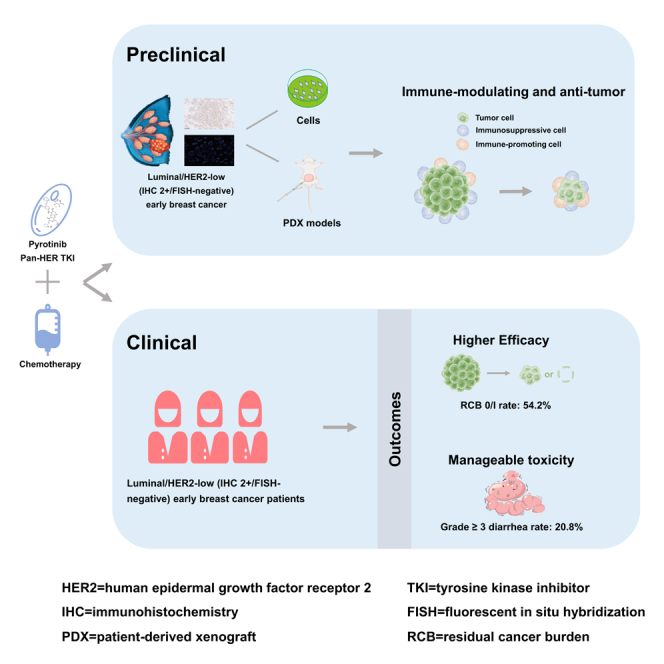
Summary
The prognosis of patients with luminal/human epidermal growth factor receptor 2 (HER2)-low early breast cancer (EBC) needs to be improved. This preclinical study and phase 2 trial (ChiCTR2100047233) aims to explore the efficacy and safety of pyrotinib (a pan-HER tyrosine kinase inhibitor) plus chemotherapy in this population. Our preclinical experiments indicate a synergistic anti-tumor effect of pyrotinib plus chemotherapy in luminal/HER2-low (immunochemistry [IHC] 2+/fluorescent in situ hybridization [FISH]-negative) breast cancer models. Furthermore, 48 women with luminal/HER2-low (IHC 2+/FISH-negative) high-risk EBC are enrolled to receive neoadjuvant pyrotinib plus chemotherapy (epirubicin-cyclophosphamide followed by docetaxel). Ultimately, 26 (54.2%; 95% confidence interval [CI] 39.2%–68.6%) patients achieve the primary endpoint (residual cancer burden [RCB] 0/I). Treatment-related adverse events of grade ≥3 occur in 21 (43.8%) patients, with the most prevalent being diarrhea (10 [20.8%]). In conclusion, neoadjuvant pyrotinib plus chemotherapy has encouraging efficacy and manageable toxicity in women with luminal/HER2-low (IHC 2+/FISH-negative) high-risk EBC. This regimen warrants to be further validated.
Keywords: early breast cancer, HER2-low, HR-positive, pyrotinib, neoadjuvant chemotherapy
Graphical abstract
Highlights
-
•
This trial evaluates the efficacy and safety of pyrotinib combined with chemotherapy
-
•
Patients with luminal/HER2-low (IHC 2+/FISH-negative) early breast cancers are included
-
•
Pyrotinib combined with chemotherapy has promising anti-tumor activity in (pre)clinical study
-
•
Pyrotinib combined with chemotherapy has a manageable toxicity profile
Gong et al. explore the efficacy and safety of pyrotinib (a pan-HER tyrosine kinase inhibitor) plus chemotherapy in women with luminal/HER2-low (immunochemistry 2+/fluorescent in situ hybridization-negative) high-risk early breast cancer. They demonstrate that this treatment regimen has promising anti-tumor activity with a manageable toxicity.
Introduction
Roughly 55% of early breast cancers (EBCs) exhibit low levels of human epidermal growth factor receptor 2 (HER2), defined as immunohistochemistry (IHC) 2+ with fluorescent in situ hybridization (FISH)-negative or IHC 1+.1 HER2-low EBC is biologically heterogeneous, with up to 90% being hormone receptor positive, falling under the category of luminal type.2,3 Significant improvements in the prognosis of patients with luminal/HER2-low EBC have been achieved through the use of chemotherapy and endocrine therapy.4 However, despite these advances, there remains an approximate 15% incidence of death within 5 years,3 which is further exacerbated among those presenting with high-risk clinical or genomic features such as TNM stage-IIB/III,3 histologic grade III,5 Ki67 ≥ 20%,6 or MammaPrint high-risk.7
In comparison to HER2-positive EBC, patients diagnosed with luminal/HER2-low EBC do not derive additional benefits from traditional anti-HER2 drugs like trastuzumab, pertuzumab, or trastuzumab-emtansine.8,9,10 Additionally, although the anti-HER2 antibody-drug conjugate (ADC) trastuzumab deruxtecan (T-DXd) has demonstrated impressive responses in HER2-low advanced breast cancer,11 its application in neoadjuvant setting for luminal/HER2-low EBC has exhibited limited tumor responsiveness, resulting in a residual cancer burden (RCB) 0/I rate of 12.1%.12 Hence, it is necessary to further explore more effective treatments for patients with luminal/HER2-low EBC.
Pyrotinib, an irreversible pan-HER tyrosine kinase inhibitor (TKI) targeting HER1, HER2, and HER4, has demonstrated excellent responses in women with HER2-positive early and advanced breast cancer.13,14,15,16 However, its effectiveness in HER2-low EBC remains uncertain. Although the I-SPY2 trial evaluated the efficacy of an irreversible pan-HER TKI neratinib in HER2-negative (IHC 0, 1+, or 2+/FISH-negative) EBC and found no additional benefit,17 it was not specifically focused on HER2-low breast cancer subgroup. Previous study reported that the antibody-dependent cellular cytotoxicity (ADCC) anti-tumor effect of anti-HER2 TKIs may be not dependent on HER2 expression and membrane localization of breast cancer cells.18 In addition to ADCC, TKIs can abrogate the immunosuppressive effects mediated by HER2 binding to STING.19 It implies that the anti-HER2 TKIs may exert anti-tumor effect through other pathway rather than blocking HER2 signaling pathway in breast cancer cells with HER2 expression.
Therefore, we conducted a preclinical study and phase 2 trial to explore the preliminary evidence on the efficacy and safety of pyrotinib plus chemotherapy in patients with luminal/HER2-low EBC, while also exploring potential biomarkers.
Results
Anti-tumor efficacy of pyrotinib in preclinical study
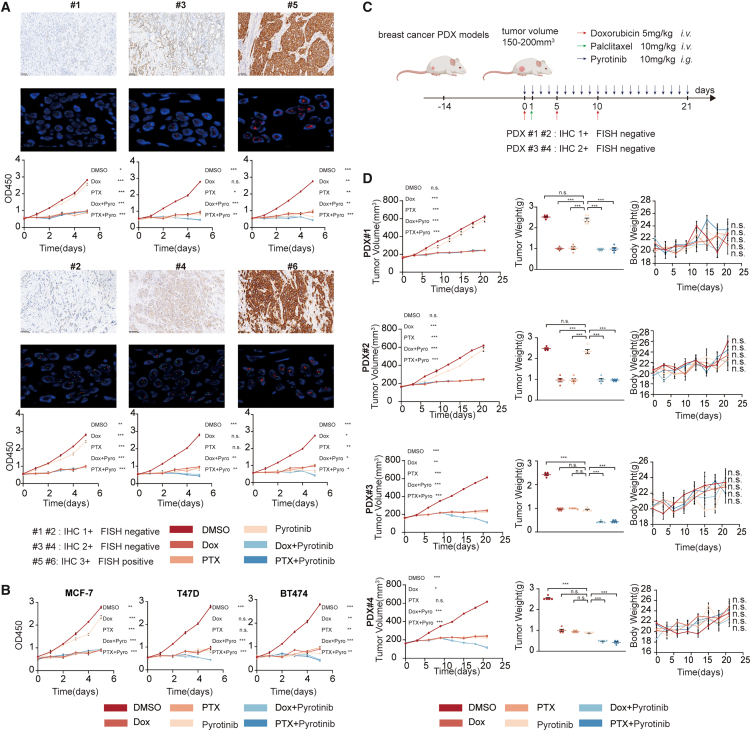
In this study, we investigated the impact of pyrotinib in luminal/HER2-low (IHC 1+), luminal/HER2-low (IHC 2+/FISH-negative), and luminal/HER2-positive (IHC 3+) breast cancer models, both in vitro and in vivo, with and without chemotherapy. Our comprehensive in vitro analysis revealed that pyrotinib monotherapy exhibited significant anti-tumor activity in luminal/HER2-low (IHC 2+/FISH-negative) and luminal/HER2-positive breast cancer cells and that this activity was further enhanced when combined with chemotherapy. However, in luminal/HER2-low (IHC 1+) breast cancer cells, neither pyrotinib alone nor in combination with chemotherapy showed substantial or synergistic anti-tumor effects (Figures 1A and 1B). We extended our investigation using patient-derived xenograft (PDX) models and found consistent results. Pyrotinib monotherapy showed pronounced anti-tumor efficacy, and the combination therapy demonstrated a synergistic anti-tumor effect in luminal/HER2-low (IHC 2+/FISH-negative) models, but not in luminal/HER2-low (IHC 1+) models (Figures 1C and 1D).
Figure 1.
The anti-tumor efficacy of pyrotinib combined with chemotherapy in different breast cancer models both in vivo and in vitro
(A) Cell Counting Kit-8 detection of primary breast cancer cells (#1–#6, both were hormone receptor positive) after treatment (0.2% DMSO, 50 nmol/L pyrotinib, 100 nmol/L Dox, 20 nmol/L PTX).
(B) Cell Counting Kit-8 detection of ER+/HER2-low MCF-7 (CCLE expression level: 2), ER+/HER2-low (CCLE expression level: 4) T47D, and ER+/HER2-positive (CCLE expression level: 7) BT474.
(C) Profile of PDX model experiments.
(D) Tumor-bearing (PDX#1–#4) mice were treated with CMC, pyrotinib, Dox, PTX, Dox + pyrotinib, and PTX + pyrotinib, n = 5. Tumor volumes, tumor weight, and body weight were measured at the indicated days.
The comparative analysis was conducted between the pyrotinib monotherapy group and each treatment group. All bar values are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. DMSO, dimethyl sulfoxide; Dox, doxorubicin; PTX, paclitaxel; CMC, carboxymethyl cellulose; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; CCLE, cancer cell line encyclopedia; PDX, patient-derived xenograft; IHC, immunohistochemistry; FISH, fluorescent in situ hybridization; SEM, standard error of the mean.
To better simulate the in vivo environment, we also developed humanized PDX breast cancer models, which confirmed that both pyrotinib monotherapy and combination therapy were more effective in luminal/HER2-low (IHC 2+/FISH-negative) and luminal/HER2-positive models compared to luminal/HER2-low (IHC 1+) models (Figures S1A and S1B). Importantly, these treatments were well tolerated in vivo, with no significant weight loss or organ damage observed (Figures S1C and S2A).
Immune microenvironment analysis in preclinical study
In our humanized breast cancer models, we analyzed the changes in the immune microenvironment following pyrotinib treatment. We found that pyrotinib upregulated infiltrating CD8+ T cells and macrophages, with a more pronounced effect in luminal/HER2-low (IHC 2+/FISH-negative) and luminal/HER2-positive models compared to Luminal/HER2-low (IHC 1+) models (Figures S2B and S3A). Notably, pyrotinib enhanced the cytotoxicity of CD8+ T cells, especially in luminal/HER2-low (IHC 2+/FISH-negative) and luminal/HER2-positive models (Figure S3B).
The aforementioned findings highlight the differential responses to pyrotinib treatment based on HER2 expression levels and emphasize the potential clinical benefit of combining pyrotinib with chemotherapy in patients with luminal/HER2-low (IHC 2+/FISH-negative) breast cancer. Therefore, we designed a single-arm clinical trial to further explore the efficacy and safety of neoadjuvant pyrotinib combined with chemotherapy in patients with luminal/HER2-low (IHC 2+/FISH-negative) high-risk EBC.
Clinical trial overview
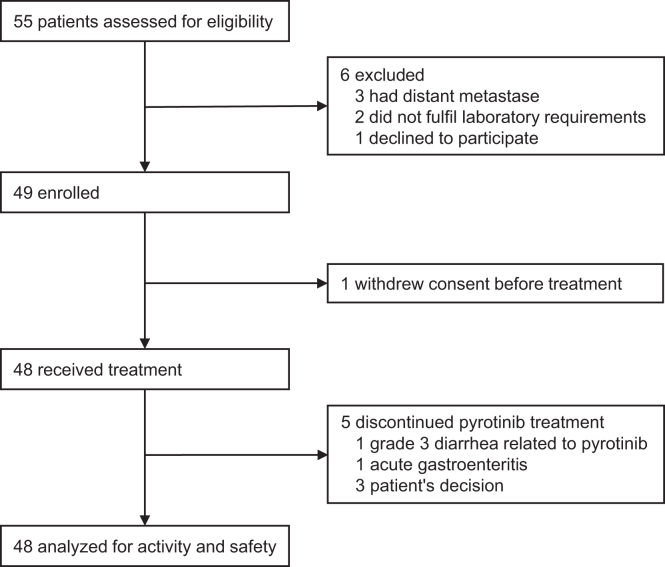
Between July 13, 2021, and March 31, 2023, a total of 55 patients were screened for eligibility. Five patients were found to be ineligible, and one patient declined to participate. Ultimately, 49 patients were enrolled in the study. However, one patient withdrew informed consent before receiving the initial neoadjuvant therapy. Consequently, the efficacy and safety of the treatment were assessed in the all-treated population (N = 48; Figure 2).
Figure 2.
The PILHLE-001 trial profile
Patients were recruited from Sun Yat-sen Memorial Hospital.
Baseline characteristics
The median age of the 48 patients was 48 years (range 28–66 years; Table 1), with 35 (72.9%) aged >40 years. Twenty-nine (60.4%) patients were premenopausal, 43 (89.6%) had cT1/2 tumors, 30 (62.5%) were lymph node-negative, 37 (77.1%) had tumors with TNM stage-II, and 39 (81.3%) had Ki67 ≥ 20%. All patients presented with clinical high-risk tumors (TNM stage-IIB/III, histologic grade III, or Ki67 ≥ 20%). The MammaPrint and BluePrint assays were conducted in 95.8% (46/48) of the patients, among whom 18 (39.1%) patients had tumors with genomic low-risk, and 28 (60.9%) had genomic high-risk. The most common subtypes are High1 luminal B (47.8%), followed by Low1 luminal A (28.3%). The detailed high-risk characteristics for each patient were shown in Table S1. Especially, among 18 patients with genomic low-risk but clinical high-risk, 5 patients had TNM stage-IIB/III, 7 had TNM stage-IIA with histologic grade III and Ki67 ≥ 20%, 2 had TNM stage-IIA with histologic grade III, and 4 had TNM stage-IIA with Ki67 ≥ 20% (40%, 50%, 30%, and 50%, respectively).
Table 1.
Baseline characteristics of enrolled patients
| Characteristic | Patients, N (%) |
|---|---|
| Age, years | |
| Median (range) | 48 (28–66) |
| ≤40 | 13 (27.1%) |
| >40 | 35 (72.9%) |
| Menopausal status | |
| Premenopausal | 29 (60.4%) |
| Postmenopausal | 19 (39.6%) |
| Clinical tumor stage | |
| T1/2 | 43 (89.6%) |
| T3/4 | 5 (10.4%) |
| Clinical nodal stage | |
| N0 | 30 (62.5%) |
| N1 | 9 (18.8%) |
| N2 | 8 (16.7%) |
| N3 | 1 (2.1%) |
| Clinical TNM stage | |
| II | 37 (77.1%) |
| III | 11 (22.9%) |
| Histological type | |
| Invasive ductal | 46 (95.8%) |
| Invasive lobular | 2 (4.2%) |
| Histological tumor grade | |
| I/II | 30 (62.5%) |
| III | 18 (37.5%) |
| Average HER2 signals/cell | |
| <4.0 | 40 (83.3%) |
| 4.0 to ≤6.0 | 8 (16.7%) |
| HER2/CEP17 ratio | |
| <1.2 (median) | 21 (43.8%) |
| ≥1.2 | 27 (56.3%) |
| ER expression | |
| 10% to <50% | 1 (2.1%) |
| ≥50% | 47 (97.9%) |
| PR expression | |
| <1% | 3 (6.3%) |
| 1% to ≤20% | 13 (27.1%) |
| >20% | 32 (66.7%) |
| Ki67 expression | |
| <20% | 9 (18.8%) |
| ≥20% | 39 (81.3%) |
| TILs | |
| ≤10% | 21 (43.8%) |
| 11% to <60% | 27 (56.3%) |
| MammaPrint/BluePrint signaturea | |
| Low2 luminal A | 5 (10.9%) |
| Low1 luminal A | 13 (28.3%) |
| High1 luminal B | 22 (47.8%) |
| High1 basal-like | 1 (2.2%) |
| High2 luminal B | 5 (10.9%) |
| Clinical or genomic riskb | |
| Clinical high-risk and genomic low-risk | 18 (39.1%) |
| Clinical and genomic high-risk | 28 (60.9%) |
Data are N (%), unless otherwise stated.
Two of 48 patients were not assessable for MammaPrint and BluePrint assay due to quality control failure.
Clinical high-risk was defined as TNM stage-IIB/III, histologic grade III, or Ki67 ≥ 20%, and genomic high-risk was defined as MammaPrint high-risk.
HER2, human epidermal growth factor receptor 2; ER, estrogen receptor; PR, progesterone receptor; TILs, tumor-infiltrating lymphocytes.
Efficacy
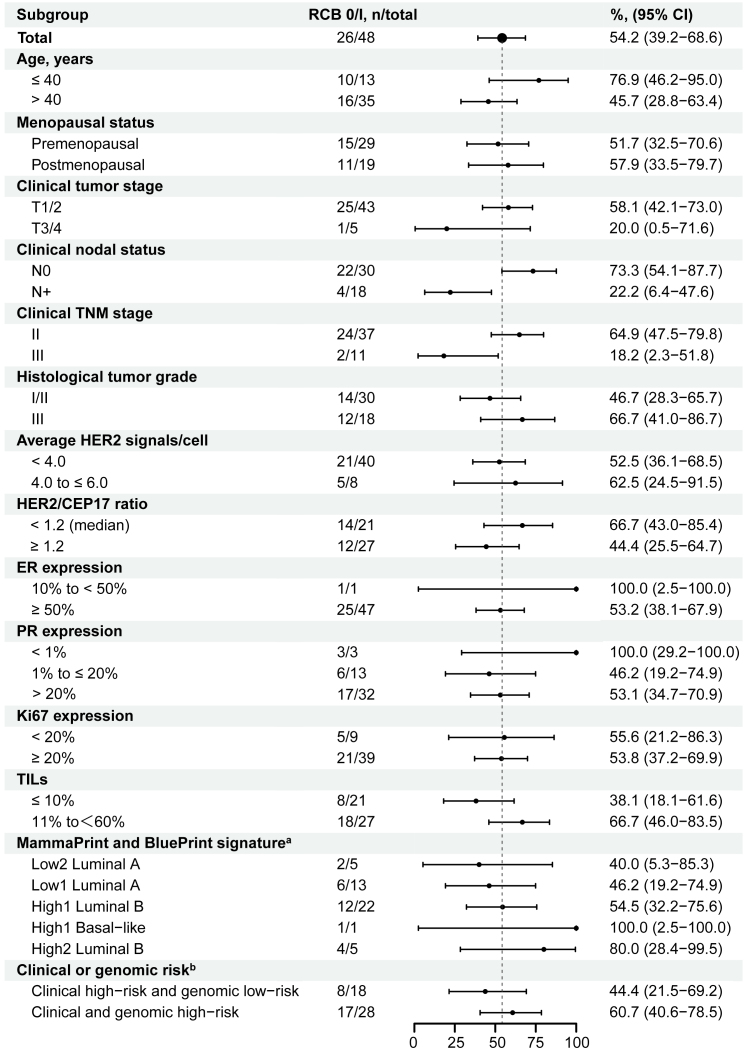
The RCB 0/I rate was 54.2% (95% confidence interval [CI] 39.2%–68.6%; N = 26; Table 2). Post hoc subgroup analyses observed that the RCB 0/I rate tended to be higher in patients with no lymph node involvement (73.3% [22/30]; Figure 3), TNM stage-II tumors (64.9% [24/37]), or intermediate tumor-infiltrating lymphocytes (TILs; 11% to <60%; 66.7% [18/27]) compared to those with lymph node-positive (22.2% [4/18]), TNM stage-III tumors (18.2% [2/11]), or low TILs (≤10%; 38.1% [8/21]), respectively. In addition, patients with both clinical high-risk and genomic high-risk tumors had a higher RCB 0/I rate compared to those with clinical high-risk but genomic low-risk (60.7% [17/28] vs. 44.4% [8/18]).
Table 2.
Primary and secondary efficacy endpoints
| Patients, N/total (%) | 95% CI | |
|---|---|---|
| Primary endpoint | ||
| RCB 0/I rate | 26/48 (54.2%) | 39.2%–68.6% |
| RCB 0 | 3/48 (6.3%) | 1.3%–17.2% |
| RCB I | 23/48 (47.9%) | 33.3%–62.8% |
| RCB II | 18/48 (37.5%) | 24.0%–52.6% |
| RCB III | 4/48 (8.3%) | 2.3%–20.0% |
| Secondary endpoints | ||
| pCR (ypT0/is, ypN0) rate | 3/48 (6.3%) | 1.3%–17.2% |
| BCS rate | 29/48 (60.4%) | 45.3%–74.2% |
| ORR by MRI (breast) at the end of cycle 2 | 22/48 (45.8%) | 31.4%–60.8% |
| Complete response | 0 | 0%–7.4% |
| Partial response | 22/48 (45.8%) | 31.4%–60.8% |
| Stable disease | 26/48 (54.2%) | 39.2%–68.6% |
| Progression disease | 0 | 0%–7.4% |
| ORR by MRI (breast) at the end of all neoadjuvant therapy | 39/48 (81.3%) | 67.4%–91.1% |
| Complete response | 3/48 (6.3%) | 1.3%–17.2% |
| Partial response | 36/48 (75.0%) | 60.4%–86.4% |
| Stable disease | 9/48 (18.8%) | 8.9%–32.6% |
| Progression disease | 0 | 0%–7.4% |
Data are N/total (%), 95% CI, unless otherwise stated. RCB, residual cancer burden; pCR, pathological complete response; BCS, breast conservation surgery; ORR, objective response rate; MRI, magnetic resonance imaging.
Figure 3.
Subgroup analyses of the RCB 0/I rate by baseline clinical-pathological factors
aTwo of 48 patients were not assessable for MammaPrint and BluePrint assay due to quality control failure.
bClinical high-risk was defined as TNM stage-IIB/III, histologic grade III, or Ki67 ≥ 20%, and genomic high-risk was defined as MammaPrint high-risk.
RCB, residual cancer burden; ER, estrogen receptor; PR, progesterone receptor; TILs, tumor-infiltrating lymphocytes.
In the prespecified secondary endpoint analyses (Table 2), three patients obtained a pathological complete response (pCR; 6.3%; 95% CI 1.3%–17.2%). The rate of breast conservation surgery was 60.4% (29/48; 95% CI 45.3%–74.2%). At the end of cycle 2 (t1), a total of 22 patients (45.8%; 95% CI 31.4%–60.8%) achieved an objective response (all partial response). At the end of all neoadjuvant therapy, 39 patients (81.3%; 95% CI 67.4%–91.1%) achieved an objective response, with 6.3% achieving a complete response and 75.0% achieving a partial response. Detailed changes in tumor size from t0 to t1 and the end of all neoadjuvant therapy are depicted in Figure S4. As of the cutoff date (Feb 01, 2024), one patient developed a disease-free survival event (liver metastasis) 21 months after the initial dose of the study drug.
Safety
Among all patients, 25 (52.1%) patients experienced grade 1 or 2 treatment-related adverse events (AEs; Table 3). The most common treatment-related AEs of any grade included diarrhea (43 [89.6%]), fatigue (35 [72.9%]), nausea (35 [72.9%]), peripheral sensory neuropathy (32 [66.7%]), alopecia (30 [62.5%]), and anemia (29 [60.4%]). Twenty-six (54.2%) patients experienced stomatitis, 22 (45.8%) experienced increased alanine aminotransferase (ALT), 19 (39.6%) experienced increased aspartate aminotransferase (AST), and 17 (35.4%) experienced hand-foot syndrome. Grade ≥3 AEs occurred in 21 (43.8%) patients, with the most prevalent being diarrhea (10 [20.8%], all grade 3), decreased lymphocyte count (5 [10.4%], all grade 3), and increased neutrophil count (grade 3: 3 [6.3%] and grade 4: 1 [2.1%]). One patient with febrile neutropenia, recognized as one serious AE (SAE) at first, was treated accordingly in the emergency outpatient department, not in inpatient department. Therefore, after discussing with the independent data monitoring committee, the occurred febrile neutropenia is not in accordance with SAE definition and may not be recognized as an SAE. Finally, one (2.1%) patient experienced SAE with increased ALT and AST.
Table 3.
Summary of treatment-related adverse events (N = 48)
| All grades, N (%) | Grade 1/2, N (%) | Grade 3, N (%) | Grade 4, N (%) | |
|---|---|---|---|---|
| Any | 48 (100.0%) | 25 (52.1%) | 21 (43.8%) | 2 (4.2%) |
| Diarrhea | 43 (89.6%) | 33 (68.8%) | 10 (20.8%) | 0 |
| Fatigue | 35 (72.9%) | 33 (68.8%) | 2 (4.2%) | 0 |
| Nausea | 35 (72.9%) | 34 (70.8%) | 1 (2.1%) | 0 |
| Peripheral sensory neuropathy | 32 (66.7%) | 32 (66.7%) | 0 | 0 |
| Alopecia | 30 (62.5%) | 30 (62.5%) | NA | NA |
| Anemia | 29 (60.4%) | 28 (58.3%) | 1 (2.1%) | 0 |
| Lymphocyte count decreased | 28 (58.3%) | 23 (47.9%) | 5 (10.4%) | 0 |
| Stomatitis | 26 (54.2%) | 26 (54.2%) | 0 | 0 |
| Alanine aminotransferase increased | 22 (45.8%) | 20 (41.7%) | 2 (4.2%) | 0 |
| White blood cell decreased | 22 (45.8%) | 19 (39.6%) | 2 (4.2%) | 1 (2.1%) |
| Neutrophil count decreaseda | 21 (43.8%) | 17 (35.4%) | 3 (6.3%) | 1 (2.1%) |
| Vomiting | 21 (43.8%) | 21 (43.8%) | 0 | 0 |
| Dizziness | 20 (41.7%) | 20 (41.7%) | 0 | 0 |
| Aspartate aminotransferase increased | 19 (39.6%) | 17 (35.4%) | 2 (4.2%) | 0 |
| Hand-foot syndrome | 17 (35.4%) | 16 (33.3%) | 1 (2.1%) | N/A |
| Stomach pain | 15 (31.3%) | 15 (31.3%) | 0 | 0 |
| Skin hyperpigmentation | 13 (27.1%) | 13 (27.1%) | 0 | 0 |
| Bone pain | 10 (20.8%) | 10 (20.8%) | 0 | 0 |
| Rash acneiform | 9 (18.8%) | 9 (18.8%) | 0 | 0 |
| Hyperhidrosis | 7 (14.6%) | 7 (14.6%) | 0 | 0 |
| Headache | 7 (14.6%) | 7 (14.6%) | 0 | 0 |
| Palpitations | 4 (8.3%) | 4 (8.3%) | 0 | 0 |
| Cough | 3 (6.3%) | 3 (6.3%) | 0 | 0 |
| Myalgia | 2 (4.2%) | 2 (4.2%) | 0 | 0 |
| Alkaline phosphatase increased | 1 (2.1%) | 1 (2.1%) | 0 | 0 |
| Serum creatinine increased | 1 (2.1%) | 1 (2.1%) | 0 | 0 |
Data are N (%), unless otherwise stated. Each patient was counted once for the highest grade of each adverse event experienced. No deaths due to adverse events were reported.
One patient had grade 3 febrile neutropenia.
Grade 3 diarrhea mainly occurred during the first cycle (7 [14.6%]) and was rarely observed thereafter (0.0%–6.3%; Figure S5). Furthermore, the majority of grade 3 diarrhea cases (75.0%) were promptly resolved within 3 days (Table S2). In total, dose reduction of pyrotinib was required in 3 (6.3%) patients due to the recurrence of grade 3 diarrhea (Table S3). Five (10.4%) patients permanently discontinued pyrotinib. Among them, 1 (2.1%) patient stopped due to grade 3 diarrhea related to pyrotinib during cycle 5, 1 (2.1%) patient discontinued due to acute gastroenteritis not related to pyrotinib during cycle 8, and 3 (6.3%) patients made the decision on their own during cycle 3 (COVID-19 outbreak), cycle 4 (intense surgery desire), and cycle 6 (intense surgery desire), respectively. Additionally, 7 (14.6%) patients required a dose reduction of chemotherapy drugs due to the decrease of neutrophil count (4 patients), increase of ALT/AST (2 patients), or hand-foot syndrome (1 patient). All AEs were reversible with symptomatic treatment, and there were no treatment-related deaths.
Magnetic resonance imaging characteristics and RCB
The associations between the RCB status and objective response status or magnetic resonance imaging (MRI) parameters were investigated. It was found that a majority of patients who achieved an objective response at t1 had an RCB 0/I status (18/22, 81.8%). This proportion was significantly higher compared to patients with stable disease (8/26, 30.8%; p < 0.001; Figure S6). The mean or median MRI parameters at t0 and the changes from t0 to t1 are presented in Table S4. Patients that achieved RCB 0/I exhibited significantly greater declines in Ktrans (median: −50.1% vs. −16.8%) and Kep (median: −42.4% vs. −13.8%) compared to those with RCB II/III (both p < 0.05).
Immune cell subpopulations and RCB
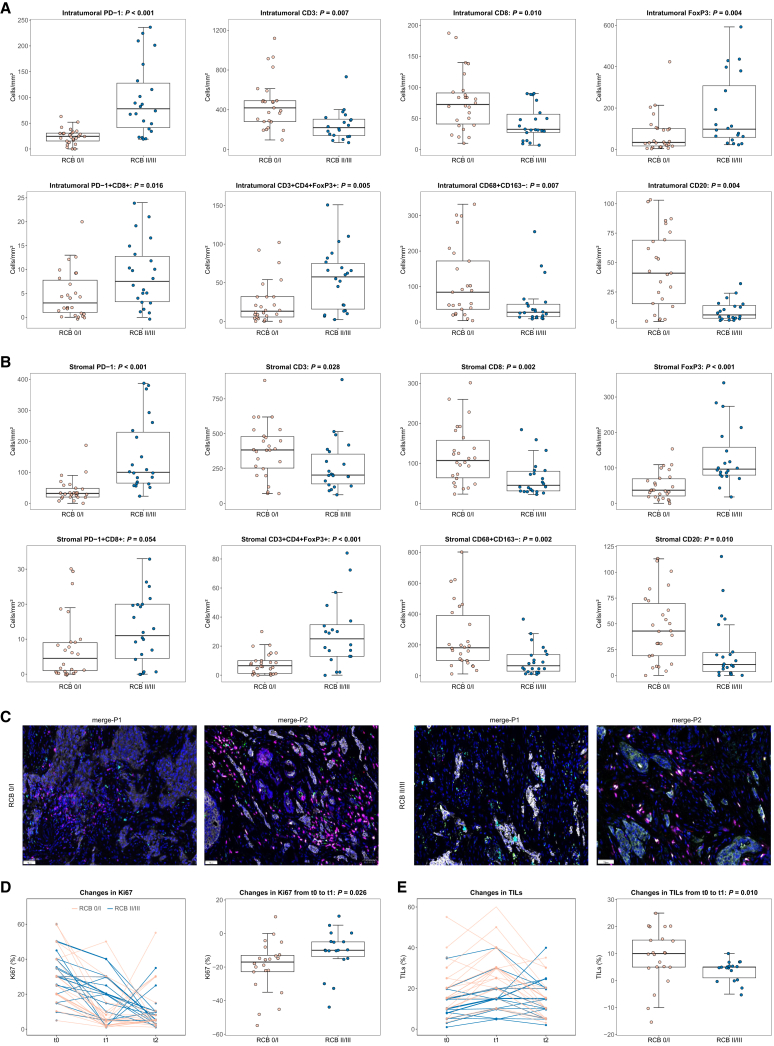
The tumor microenvironment was examined in all patients. Except for three (6.3%) patients who achieved RCB 0 at t2, the remaining patients displayed apparent infiltration of CD3 T lymphocytes and CD68 macrophages (Figures 4A, 4B, 4C, and S7). At t0, within the intratumoral compartment, patients with RCB 0/I had significantly higher levels of CD3 total T lymphocytes, CD8 cytotoxic T lymphocytes, CD68+CD163− M1 macrophages, and CD20 B lymphocytes, compared to those with RCB II/III (all p < 0.05; Figures 4A and 4B). Conversely, patients with RCB 0/I had significantly lower numbers of PD-1 cells, FoxP3 regulatory T lymphocytes, PD-1+CD8+ effector T lymphocytes, and CD3+CD4+FoxP3+ regulatory T lymphocytes (all p < 0.05). Similar results were observed in the stromal compartment, except for a non-significant difference in PD-1+CD8+ effector T lymphocytes (p = 0.054). No notable differences were found in other immune cell populations or tertiary lymphoid structures (Figure S7). Additionally, from t0 to t2, patients with RCB 0/I displayed significant increases in the density of intratumoral/stromal PD-1 cells, stromal CD3+CD4+ T lymphocytes, intratumoral/stromal FoxP3 regulatory T lymphocytes, intratumoral/stromal CD3+CD4+FoxP3+ regulatory T lymphocytes, intratumoral CD56bright natural killer cells, and intratumoral/stromal CD56dim natural killer cells, compared to those with RCB II/III (Figures S8 and S9).
Figure 4.
Baseline (t0) immune cell populations by multiplex immunofluorescence and dynamic trend of Ki67/TILs at t0, the end of cycle 2 (t1), and surgery (t2) in patients with RCB 0/I versus RCB II/III
(A) Intratumoral immune cell populationsa.
(B) Stromal immune cell populationsa.
(C) Representative pictures of multiplex immunofluorescence at t0 in patients with RCB 0/I and RCB II/III; original magnification 200×. Color code for multiplex immunofluorescence pictures of merge-P1: PD-1 in green, PD-L1 in yellow, CD8 in pink, CD68 in cyan, and CD163 in red. Color code for multiplex immunofluorescence pictures of merge-P2: CD20 in green, FoxP3 in yellow, CD3 in pink, CD56 in cyan, and CD4 in red.
(D) Ki67.
(E) TILs.
aSome points outside the range are not shown in the boxplots.
p values are from the Wilcoxon rank-sum test. RCB, residual cancer burden; TILs, tumor-infiltrating lymphocytes.
Changes in Ki67/TILs and RCB
Forty-four (91.7%) patients agreed to undergo additional core biopsies at t1, but 4 of these patients were not assessable due to the absence of invasive cells. At t2, 3 (6.3%) patients could not be evaluated because they achieved RCB 0. The dynamic trend of Ki67 and TILs from t0 to t1 and t2 is illustrated in Figures 4D and 4E. The geometric mean Ki67 was 31.4% (SD 15.3) at t0, 13.8% (13.0) at t1, and 11.2% (13.6) at t2, while TIL percentage was 16.0% (11.0) at t0, 22.3% (12.6) at t1, and 14.2% (8.9) at t2, respectively. From t0 to t1, patients with RCB 0/I displayed significantly greater declines in Ki67 (p = 0.026) and more pronounced increases in TILs (p = 0.010) compared to those with RCB II/III (Figures 4D and 4E).
Gene sequencing and RCB
Gene sequencing was conducted at t0 for quality control in 46 (95.8%) eligible patients (Figure S10). Results showed that 11 (23.9%) patients were homologous recombination deficiency (HRD) positive and 6 (13.0%) were circulating tumor DNA (ctDNA) positive. The most frequently observed altered genes in the tumor samples were PIK3CA (15 [32.6%]), GATA3 (13 [28.2%]), TP53 (11 [23.9%]), CCND1 (11 [23.9%]), and FGF19 (9 [19.6%]). Only PIK3CA was found to be significantly associated with the treatment response, as patients with the PIK3CA mutation had a lower rate of RCB 0/I (33.3%, 5/15) compared to those with the wild-type PIK3CA (64.5%, 20/31; p = 0.047). At t2, results were available for 42 (87.5%) patients (Figure S10). Four patients who were HRD positive at t0 converted to HRD negative, and two patients converted from ctDNA positive to ctDNA negative at t2. There were no significant changes in the gene mutation profile compared to t0.
Discussion
The PILHLE-001 trial evaluated the efficacy and safety of neoadjuvant pyrotinib combined with epirubicin-cyclophosphamide followed by docetaxel in women with luminal/HER2-low (IHC 2+/FISH-negative) high-risk EBC. This treatment regimen resulted in a remarkable RCB 0/I rate of 54.2% (95% CI 39.2%–68.6%) and a manageable toxicity. These results provide a rationale of pyrotinib plus chemotherapy as a promising therapeutic strategy for this specific patient population.
Patients with luminal/HER2-low EBC who achieved RCB I, had the similar long-term prognosis with those having RCB 0 following neoadjuvant treatment.20 It implies that the RCB 0/I rate may be a more suitable endpoint for evaluating the efficacy of neoadjuvant treatment in patients with luminal/HER2-low EBC compared to the pCR rate. In this study, the observed RCB 0/I rate (primary endpoint) of 54.2% was remarkably higher compared to the rates reported in trials conducted in luminal/HER2-negative (IHC 0, 1+, or 2+/FISH-negative) EBC with clinical or genomic high-risk, such as CORALLEEN trial (11.8%; risk of relapse [ROR] intermediate or high risk),21 NeoPAL trial (15.7%; ROR intermediate or high risk),22 GIADA trial (25.6% for chemotherapy plus nivolumab; clinical high-risk),23 CheckMate 7FL trial (21.3% for chemotherapy and 30.7% for chemotherapy plus nivolumab; clinical high-risk),24 I-SPY2 trial (25.0% for chemotherapy and 32.8% for chemotherapy plus targeted drugs; MammaPrint high-risk),17 and KEYNOTE-756 trial (23.7% for chemotherapy and 34.9% for chemotherapy plus pembrolizumab; clinical high-risk),25 which investigated chemotherapy or chemotherapy-based combination strategies. Previous studies have demonstrated that patients with HER2-low (IHC 1+ or 2+/FISH-negative) EBC had lower chemosensitivity compared to those with HER2 IHC 0.3 In addition, CORALLEEN and NeoPAL trials have similar characteristics with our trial (TNM stage-IIB/III: 42.6% vs. 47.2% vs. 41.7%) but reported relatively low RCB0/I rate.21,22 Therefore, although the populations in these studies may not perfectly match those in PILHLE-001 trial, it is evident that the relatively high RCB 0/I rate is primarily attributable to the combination of pyrotinib and chemotherapy, rather than differences in patient populations.
Indeed, the anti-HER2 ADC T-DXd has demonstrated a prominent anti-tumor effect in patients with HER2-low metastatic breast cancer.11 However, findings from TALENT trial indicated that T-DXd with or without anastrozole have only achieved a relatively low RCB 0/I rate (17.6% versus 6.3%) in the neoadjuvant setting in patients with luminal/HER2-low (IHC 2+/FISH-negative or IHC 1+) EBC.12 Awaitment of the final results of the TALENT trial is imperative to preliminarily assess whether T-DXd can serve as a viable alternative strategy for patients with luminal/HER2-low EBC.
The low rate of RCB 0 (pCR) in this trial can largely be attributed to the high proportion (97.9%) of patients with high estrogen receptor (ER) positive (≥50%), who generally have a lower likelihood of achieving pCR (adjusted odds ratio, 0.9 per 10% ER expression increase).3 Similar trends were observed in CheckMate 7FL trial and KEYNOTE-756 trial, where the addition of immune checkpoint inhibitors to chemotherapy resulted in a lower increase in pCR rates among patients with high ER expression compared to those with low ER expression.24,26 Future studies exploring the potential value of endocrine therapy-based combination strategies specifically in this patient population would indeed be both interesting and valuable.
In the PILHLE-001 study, although the incidence of diarrhea (any grade: 90%; grade ≥3: 21%) was relatively high, it was lower compared to previous trials of pyrotinib in combination with chemotherapy at a higher dosage of 400 mg (any grade: 95%–100%; grade ≥3: 31%–46%).13,14,15 Importantly, grade 3 diarrhea was primarily observed during the first cycle of treatment, remained at a low level thereafter, and could be rapidly reversible through the use of anti-diarrheal treatments. Incidences of pyrotinib treatment discontinuation due to safety concerns were rare (2%). The occurrence of AEs other than diarrhea was comparable to that of anthracycline and taxane-based chemotherapy.27 Overall, the combination of pyrotinib 320 mg with chemotherapy possesses a manageable toxicity for the treatment of patients with breast cancer. Additionally, in light of the growing preference for anthracycline-free regimens as a means to minimize treatment-related toxicities,28 it would be valuable to explore whether pyrotinib plus anthracycline-free regimens can offer a more favorable risk-benefit ratio.
In this trial, we explored the association between baseline immune cell profiles and treatment efficacy. Our findings revealed that immune cells, such as higher levels of cytotoxic T lymphocytes (CD8), M1 macrophages (CD68+CD163-), or B lymphocytes (CD20), were associated with better treatment response. Conversely, regulatory T lymphocytes (FoxP3+) or immune checkpoint-expressing cells (PD-1) exhibited contrasting results. These results suggest that the effectiveness of pyrotinib in combination with chemotherapy may be influenced by the anti-tumor immune response. Moreover, previous studies have indicated that anti-HER TKIs can induce immunogenic cell death, similar to the effect of chemotherapeutic agents.29,30 This further supports the notion that anti-tumor immunity plays a role in the effectiveness of this treatment regimen. Our findings differ to some extent from GIADA trial,23 which reported higher levels of regulatory T lymphocytes and immune checkpoint-expressing cells in individuals who achieved a pCR with immunotherapy. This discrepancy can be partially explained by the different mechanisms of action between immune checkpoint inhibitors and pan-HER inhibitors. In addition, research has found that anti-HER2 TKIs can abrogate the immunosuppressive effects mediated by HER2 binding to STING, which is dependent on the level of HER2 expression.3 This might explain why pyrotinib can be effective in patients with luminal/HER2-low (IHC 2+/FISH-negative) breast cancer. Further studies are needed to validate this topic in more detail, particularly in the context of the recent benefits demonstrated by the immune checkpoint inhibitors in luminal/HER2-negative high-risk EBC.24,26
Our gene sequencing analysis yielded results that aligned with previous research,31 indicating commonly altered genes in luminal/HER2-low (IHC 2+/FISH-negative) EBC. Of particular note, we observed that the presence of PIK3CA mutation at baseline was linked to a poor response to pyrotinib plus chemotherapy, mirroring findings in patients with HER2-positive EBC.32 The potential predictive role of PIK3CA status in response to pyrotinib combined with chemotherapy should be further validated.
During the early phases of treatment (at the end of cycle 2), we noted that patients with an objective response exhibited a greater pathological response compared to those with stable disease, in line with previous studies.33,34 Furthermore, we observed significant reductions in the specific kinetic parameters of MRI (transfer constant [Ktrans] and rate constant [Kep]), indicative of decreased tumor micro-vessel density and function post-treatment in patients with RCB 0/I.35 Additionally, patients who experienced greater declines in Ki67 exhibited improved therapeutic efficacy, highlighting the stronger effects of pyrotinib combined with anthracycline in inducing cell-cycle arrest in these patients.36 Consistent with GIADA trial,23 we observed more enrichment in TILs after exposure to pyrotinib plus epirubicin-cyclophosphamide in patients with RCB 0/I, suggesting a potentially more prominent immunogenic effect with the combination treatment compared to those with RCB II/III.30 Further investigation is necessary to better comprehend the underlying mechanisms driving these associations and to gain a deeper understanding of how pyrotinib plus chemotherapy exerts its actions.
Limitations of the study
The PILHLE-001 study had limitations. Firstly, the trial employed a single-arm design with a small sample size, which may introduce potential bias and limit the generalizability of the findings. Secondly, due to the short follow-up period, it is essential to assess the efficacy of combining pyrotinib with chemotherapy in a large randomized trial powered for long-term survival outcome, which is currently ongoing (NCT06144944). Lastly, the availability of immune biomarkers in the study was scarce. The related biomarkers explored based on our ongoing randomized control trial with a more comprehensive characterization of the tumor microenvironment may help physicians to apply pyrotinib-based regimen to improve the prognosis of patients with HER2-low high-risk EBC.
Conclusion
In conclusion, the combination of pyrotinib with epirubicin-cyclophosphamide followed by docetaxel demonstrated promising anti-tumor activity and a manageable toxicity in women with luminal/HER2-low (IHC 2+/FISH-negative) high-risk EBC. Several biomarkers such as baseline immune cell subpopulations, baseline PIK3CA status, early tumor response based on MRI, and changes in Ki67 or TILs during treatment might serve as potential predictors for clinical efficacy to this combination therapy. A phase 3 randomized trial is warranted to validate the clinical benefits of this combination, which is ongoing (NCT06144944).
Resource availability
Lead contact
Any further information or requests should be directed to and will be fulfilled by the lead contact, Chang Gong (gchang@mail.sysu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Participant-level clinical data are deposited to RED-Cap database (managed by the Sun Yat-sen Memorial Hospital) and can be accessed via the lead contact by reasonable request.
The raw sequencing data reported in this paper have been deposited at in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA-Human: HRA008271) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
We thank the patients and their families, nurses, radiologists, pathologists, statisticians, research staff, and members of the Independent Data Monitoring Committee who participated in this trial. We appreciate the support of Jiangsu Hengrui Pharmaceuticals, Genecast, 3D Medicines, and Burning Rock DX for this study. This work was supported by grants from the National Science and Technology Major Project of China (2023ZD0501100), National Natural Science Foundation of China (grant number 82488101, 82330056, 92159303, 81930081, 82371739, 82304120, 81872139, 82072907, 82003311, U1911204, and 51861125203), Guangdong Science and Technology Department (2020B1212060018 and 2020B1212030004), Project of The Beijing Xisike Clinical Oncology Research Foundation (Y-Roche2019/2-0078, Y-pierrefabre202102-0107), Department of Natural Resources of Guangdong Province (GDNRC[2021]51), the Science and Technology Planning Project of Guangdong Province (2023B1212060013), Guangzhou Science and Technology Program (202102010272 and 2024B01J1154), Bureau of Science and Technology of Guangzhou (20212200003), Fundamental Research Funds for the Central Universities, Sun Yat-sen University (2022005), Program for Guangdong Introducing Innovative and Enterpreneurial Teams (2019BT02Y198), High-tech, Major and Characteristic Technology Projects in Guangzhou Area (2023-2025 and 2023P-ZD14), and National Key R&D Program of China (2021YFC3001000 and 2021YFA1300602). The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of this report.
Author contributions
E.S., C.G., L.W.C.C., Y.X., Y. Zhu, Y.Y., and Q. Lin conceived and designed this study. Q. Liu improved the study design. C.G., W.Y., and Yinduo Zeng recruited and treated patients. Y.X., W.Y., J.Z., Yunjie Zeng, Z.C., J.S., Q. Lin, and Z.D. collected data. E.S., C.G., L.W.C.C., Y.X., Y. Zhu, Y.Y., Q. Liu, L.L., Q. Lin, and Z.D. contributed to the data analysis and interpretation. All authors were involved in the preparation and critically reviewed the manuscript. All authors had full access to the raw data and approved the final version of manuscript for submission. The corresponding authors had final responsibility for the decision to submit for publication.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD163 | Abcam | RRID: AB_2753196 |
| CD68 | Abcam | RRID: AB_2801637 |
| PD-1 | CST | RRID: AB_2864408 |
| PD-L1 | CST | RRID: AB_2864409 |
| CD3 | Dako | RRID: AB_2335677 |
| CD4 | Abcam | RRID: AB_2750883 |
| CD8 | Abcam | RRID: AB_2756374 |
| CD56 | Abcam | RRID: AB_2632384 |
| CD20 | Dako | RRID: AB_3075456 |
| FOXP3 | Abcam | RRID: AB_445284 |
| pan-CK | Abcam | RRID: AB_306047 |
| S100 | Abcam | RRID: AB_882426 |
| Biological samples | ||
| Tumor samples | This study | N/A |
| Critical commercial assays | ||
| QIAamp DNA Mini Kit | Qiagen | Cat#: 937236 |
| NovaSeq 6000 platform | Illumina | Cat#: 937236 |
| Deposited data | ||
| Raw sequencing data | This study | Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation (GSA-Human: HRA008271; https://ngdc.cncb.ac.cn/gsa-human) |
| Participant-level clinical data | This study | RED-Cap database (https://redcap.gzsys.org.cn:8082/redcap_v11.2.2/Design/online) managed by the Sun Yat-sen Memorial Hospital. |
| Experimental models: Cell lines | ||
| MCF-7 breast cancer cell line | ATCC | N/A |
| T47D breast cancer cell line | ATCC | N/A |
| BT474 breast cancer cell line | ATCC | N/A |
| Primary breast cancer cell lines (#1-#6) | This study | N/A |
| Experimental models: Organisms/strains | ||
| Breast cancer PDX models (#1-#4) | This study | N/A |
| Humanized breast cancer PDX models (#1-#3) | This study | N/A |
| Software and algorithms | ||
| R studio, v 4.2.2 | – | https://cran.r-project.org/https://www.rstudio.com |
| SPSS, v 26.0 | – | https://www.ibm.com/spss |
Experimental model and study participant details
Clinical trial design and patients
The PILHLE-001 (Chinese Clinical Trial registration number ChiCTR2100047233) study was a prospective, investigator-initiated, single-center, single-arm, phase II trial conducted at Sun Yat-sen Memorial Hospital. Women aged 18 years or older with newly diagnosed unilateral and primary invasive breast cancer were eligible if they had to undergo central histology assessment of core biopsies to confirm HR-positive (estrogen receptor [ER] and/or progesterone receptor [PR] ≥ 1% stained cells) and HER2-low (IHC 2+ with FISH-negative) status, according to American Society of Clinical Oncology and College of American Pathologists 2018 guideline.37 Patients were required to have a tumor size of greater than 2 cm or between 1 cm and 2 cm with histological confirmed lymph node metastasis (TNM stage-II/III). For patients with TNM stage-IIA tumor, histologic grade III or Ki67 ≥ 20% or MammaPrint high-risk were required. Patients were ineligible if they had stage-IV, occult, or bilateral breast cancer, other malignancies, serious infections, impaired liver/heart/kidney function, or clinically significant gastrointestinal abnormalities. Detailed selection criteria are available in the protocol (p15-16).
This study was approved by the ethic committee of Sun Yat-sen Memorial Hospital (2021-KY-054 and SYSKY-2022-158-02) and conducted in accordance with the Declaration of Helsinki and the Guideline for Good Clinical Practice. Each patient provided written informed consent prior to participation.
Method details
Preclinical experiments
Establishment of (humanized) patient-derived xenograft (PDX) breast cancer models
To establish the PDX models, tissues were maintained in PRI DMEM with 10% fetal bovine serum and 1% penicillin/streptomycin. Tissues were then cut into 1 × 1 × 1 mm³ pieces and rinsed with fresh PRI DMEM twice. These tissue pieces were subsequently implanted subcutaneously into the fat pad of NOD/SCID mice. When the xenografted tumor tissues reached a size of 1–2 cm³, they were harvested following the protocols mentioned earlier and transplanted into subsequent generations of NOD/SCID mice. To establish humanized PDX models, we injected 1 × 105 of CD34+hematopoietic stem cells (HSC), freshly isolated using a direct CD34+ MicroBead Kit (MiltenyiBiotec, Cologne, Germany), intravenously into 4-week-old NOD. Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice irradiated with 200cGy using 137Cs gamma irradiator. The engraftment levels of human CD45+ cells and human immune cell populations, including CD45+, CD3+, and CD8+ T cells, B cells, natural killer cells, myeloid-derived suppressor cells, and other lineage negative cells were determined in the peripheral blood, bone marrow, and spleen tissue through flow cytometry panel. Mice that had over 25% human CD45+ cells in the peripheral blood were considered humanized. The following PDX implantations were the same as mentioned above.
In vivo animal experiments
Tumor-bearing mice were randomly assigned to different groups, with each group consisting of 5 mice, when the tumors reached an average volume of 150–200 mm³. The groups included the vehicle group (0.5% carboxymethyl cellulose, CMC), the doxorubicin group at a dose of 5 mg/kg/day, the paclitaxel group at a dose of 10 mg/kg/day, the pyrotinib group at a dose of 10 mg/kg/day, and the combination group receiving doxorubicin or paclitaxel in combination with pyrotinib. Treatment was conducted for 21 days. Tumor volumes were measured twice a week using calipers (volume = L × W2/2), and mouse weights were recorded every 3 days. At the end of the treatment period, the mice were euthanized, and the tumors were collected for further analysis.
Hematoxylin and eosin (H&E) staining
After harvesting the mouse organs, tissues were fixed in 10% neutral-buffered formalin for 24 h, followed by dehydration through a graded ethanol series and clearing in xylene. The tissues were then embedded in paraffin, sectioned at 4 μm thickness, and mounted on glass slides. The sections were deparaffinized in xylene, rehydrated through graded ethanol, stained with hematoxylin for 5–10 min, differentiated in 1% acid alcohol, blued in tap water, counterstained with eosin, and finally dehydrated, cleared for histological analysis.
Flow cytometry analysis
Mononuclear cells collected from tumor tissue were first labeled for live/dead discrimination. After an incubation period of 20 min, the cells were washed with PBS and resuspended. Subsequently, the cells were labeled with antibodies targeting CD45, CD3, CD33, CD8, CD16, CD68, CD11c, and CD11b for 20 min. After another round of PBS washing and resuspension, the cells were prepared for flow cytometric analysis. Cells need to be ruptured before staining granzymeB, perforin and IFN-γ.
Cell lines and Cell Counting Kit-8 (CCK8)
Primary breast cancer cells (#1-#6) were established by our laboratory. ER+/HER2-low (expression level: 2) MCF-7, ER+/HER2-low (expression level: 4) T47D, and ER+/HER2-positive (expression level: 7) BT474 cells were obtained from American Type Culture Collection (ATCC). All cell lines were grown according to standard protocols. The CCK8 assay was performed by CCK8 assay kit manual (Dojindo, Japan). Primary BC cells (#1-#6, both were hormone receptor-positive), MCF-7, T47D, and BT474 cells were seeded in 96-well plates (3000 cells/well). CCK-8 solution was added to the cells and incubated for 2 h at 37°C. The absorbance at 450 nm was measured using a spectrophotometer.
Procedures of clinical trial
Patients in the PILHLE-001 study received pyrotinib 320 mg orally once daily, and epirubicin 90 mg/m2 plus cyclophosphamide 600 mg/m2 intravenously on day 1 for four 3-weekly cycles followed by docetaxel 100 mg/m2 intravenously on day 1 for another four 3-weekly cycles. Treatment was continued until completion of all cycles, withdrawal of consent, disease progression, death, intolerable toxicity, or other reasons as determined by the investigators. Upon completion of the above treatments, patients received surgery and subsequent therapy in accordance with the recommendations outlined in the National Comprehensive Cancer Network guideline (version 3.2021). After surgery, follow-up for survival was done every 3 months during the first 2 years and every 6 months from the third to fifth year until progression, death, loss to follow-up, or completion of the study. Detailed procedures are listed in the protocol (p26).
Data collection and biomarkers of clinical trial
At baseline (t0), we collected the demographics (including age, menopausal status, height, and weight), clinical characteristics (including tumor size and nodal status), and clinicopathological features (including ER, PR, HER2, and Ki67 expression). Breast tumor samples were collected by core needle biopsy at t0, the end of cycle 2 (t1, if consented), and surgery (t2) separately.
Stromal tumor-infiltrating lymphocytes (TILs)
Stromal TILs, defined as the percentage of mononuclear immune cell infiltrate in the stromal tissue within the borders of the invasive tumor, were evaluated by two independent pathologists who were blinded to the patient’s characteristics on H&E stained slides obtained at t0, t1, and t2, following the recommendations of the international TILs working group.38
MammaPrint/BluePrint signatures
MammaPrint/BluePrint signatures at t0 were assessed.39,40 To ensure quality control, a section of the formalin-fixed paraffin-embedded (FFPE) breast tissue was initially examined with H&E staining to confirm the presence of invasive tumor cells (≥30%) and to determine the minimum tumor surface area (10 mm2). For each tumor specimen, five to ten 5–10 μm FFPE slides were utilized.
Breast magnetic resonance imaging (MRI)
Breast MRI was conducted at t0, t1, and the end of all neoadjuvant therapy. The tumor size (at t0, t1, or the end of all neoadjuvant therapy) and related parameters (at t0 or t1) based on breast MRI were independently measured by two radiologists who were blinded to the patient’s characteristics. The tumor size was measured in the longest diameter on DCE-MRI images 90 s after the beginning of the contrast agent injection. When multifocal or multicentric lesions were identified, the maximal tumor diameters of the 2 largest lesions were added to obtain the tumor size.
Apparent diffusion coefficient (ADC; unit, ×103 mm2/s) values were measured by manually drawing a region of interesting (ROI) on the ADC map showing a hypointense tumor region while avoiding necrotic and cystic or bleeding components, referring to information from the postcontrast MRI.
The patient images were imported into a Siemens supporting workstation. DCE MRI parametric maps of quantitative and semiquantitative parameters were automatically generated after motion correction and elastic registration on the Tofts model and qualitative model, respectively, using the MRI Tissue 4D software tool (Siemens Healthcare). The individual arterial input function (AIF) was obtained from breast cancer patients scanned at different time points, and the most suitable AIF were choice based on the minimum Chi2 value on the Tofts model. Each parameter was measured with a volume of interesting (VOI). The VOI was first drawn on the influx volume transfer constant (Ktrans) derivative map and automatically generated onto the other quantitative parameter derivative maps. The VOI of each parameter derivative map encompassed the same position and range. Then, the same VOI was matched to the semiquantitative parameter derivative maps.
The quantitative parameters of DCE-MRI were as follows: Ktrans (influx volume transfer constant; unit, min−1), Kep (efflux rate constant; unit, min−1), Ve (volume fraction of the extravascular extracellular space per unit volume of tissue and is calculated as Ve = Ktrans/Kep), and iAUC (initial area under the curve for the 60 s). The semiquantitative parameters of DCE-MRI were as follows: Wash-in (the rate of contrast enhancement for contrast agent inflow; unit, min−1), Wash-out (the rate of contrast decay for contrast agent outflow; unit, min−1), and TTP (the time-to-peak enhancement after contrast agent injection; unit, min).
Immune cell subpopulations
Tumor microenvironment was characterized by multiplex immunofluorescence staining by use of the Akoya OPAL Polaris 7-Color Automation IHC kit (NEL871001KT). FFPE tissue slides were first deparaffinized in a BOND RX system (Leica Biosystems) and then incubated sequentially with primary antibodies targeting CD163 (Abcam, ab182422, 1:500), CD68 (Abcam, ab213363, 1:1000), PD-1 (CST, D4W2J, 86163S, 1:200), PD-L1 (CST, E1L3N, 13684S, 1:400), CD3 (Dako, A0452), CD4 (Abcam, ab133616, 1:100), CD8 (Abcam, ab178089, 1:100), CD56 (Abcam, ab75813, 1:100), CD20 (Dako, L26, IR604), FOXP3 (Abcam, ab20034, 1:100), and pan-CK (Abcam, ab7753, 1:100) or S100 (Abcam, ab52642, 1:200, Akoya Biosciences). This was followed by incubation with secondary antibodies and corresponding reactive Opal fluorophores. Nuclei acids were stained with DAPI. Tissue slides that were bound with primary and secondary antibodies but not fluorophores were included as negative controls to assess autofluorescence.
Multiplex stained slides were scanned using a Vectra Polaris Quantitative Pathology Imaging System (Akoya Biosciences) at 20 nm wavelength intervals from 440 nm to 780 nm with a fixed exposure time and an absolute magnification of 200×. All scans for each slide were then superimposed to obtain a single image. Multilayer images were imported to inForm v.2.4.8 (Akoya Biosciences) for quantitative image analysis. Tumor parenchyma and stroma were differentiated by pan-CK staining. The quantities (cells/mm2) of various cell populations were expressed as the number of stained cells per square millimeter.
Gene sequencing
Genomic DNA (gDNA) samples from tumor samples and whole blood control samples were analyzed at t0 and t2. FFPE tissue sections were evaluated for tumor cell content using H&E staining. Only samples with a tumor content of ≥20% were eligible for subsequent analyses. Cell-free DNA (cfDNA) was extracted using the QiAmp Circulating Nucleic Acid Kit (Qiagen) following the manufacturer’s instructions. gDNA was isolated from tumor samples or whole blood control samples using the ReliaPrep FFPE gDNA Miniprep System (Promega) or QIAamp DNA Mini Kit (Qiagen). DNA concentration was quantified using the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific) following the manufacturer’s instructions.
gDNA extracts (30–200 ng) were sheared to 250 bp fragments using an S220 focused ultrasonicator (Covaris). gDNA and cfDNA libraries were prepared using the KAPA Hyper Prep Kit (KAPA Biosystems) following the manufacturer’s protocol. cfDNA libraries were individually barcoded with unique molecular identifiers (UMI). In brief, 30–60 ng of cfDNA were subjected to end-repairing, A-tailing, and ligation with indexed adapters. The libraries were then PCR-amplified and purified for target enrichment. The concentration and size distribution of each library were determined using a Qubit 3.0 fluorometer (Thermo Fisher Scientific) and a LabChip GX Touch HT Analyzer (PerkinElmer) respectively. For targeted capture, indexed libraries were subjected to probe-based hybridization with a customized NGS panel targeting 733 cancer-related genes, where the probe baits were individually synthesized 5′ biotinylated 120 bp DNA oligonucleotides (IDT). Repetitive elements were filtered out from intronic baits according to the annotation by UCSC Genome RepeatMasker.41 The xGen Hybridization and Wash Kit (IDT) was employed for hybridization enrichment. Briefly, 500 ng indexed DNA libraries were pooled to obtain a total amount of 2 μg of DNA. The pooled DNA sample was then mixed with human cot DNA and xGen Universal Blockers-TS Mix and dried down in a SpeedVac system. The Hybridization Master Mix was added to the samples and incubated in a thermal cycler at 95°C for 10 min, before being mixed and incubated with 4 μL of probes at 65°C overnight. The target regions were captured following the manufacturer’s instructions. The concentration and fragment size distribution of the final library were determined using a Qubit 3.0 fluorometer (Thermo Fisher Scientific) and a LabChip GX Touch HT Analyzer (PerkinElmer) respectively.
The captured libraries were loaded onto a NovaSeq 6000 platform (Illumina) for 100 bp paired-end sequencing with a mean sequencing depth of ≥500× for tumor samples and ≥35000× for whole blood control samples. Raw data of paired samples (an FFPE sample and its normal tissue control) were mapped to the reference human genome hg19 using the Burrows-Wheeler Aligner.42
For gDNA, PCR duplicate reads were removed and sequence metrics were collected using Picard and SAM tools, respectively. Variant calling was performed only in the targeted regions. Somatic single nucleotide variants (SNVs) were detected using an in-house developed R package to execute a variant detection model based on binomial test. Local realignment was performed to detect indels. Variants were then filtered by their unique supporting read depth, strand bias, base quality as previously described.43 All variants were then filtered using an automated false positive filtering pipeline to ensure sensitivity and specificity at an allele frequency (AF) of ≥1%.
For cfDNA, an in-house developed software was used to generate duplex consensus sequences based on dual UMI integrated at the end of the DNA fragments. To improve specificity, especially for variants with low allele frequency in the circulating tumor DNA (ctDNA), an in-house loci specific variant detection model based on binomial test was applied. The variants were subsequently filtered by their supporting count, strand bias status, base quality, and mapping quality. In addition, variant calling was also optimized to detect variants at short tandem repeat regions.
Single-nucleotide polymorphism (SNPs) and indels were annotated by ANNOVAR against the following databases: dbSNP, 1000Genome, and ESP6500 (population frequency >0.015). Only missense, stop gain (nonsense), frameshift, and non-frameshift indel mutations were kept. Copy number variations (CNVs) and gene rearrangements were detected as described previously.43 The homologous recombination deficiency (HRD) score was counted by combining loss of heterozygosity, telomeric allelic imbalance (TAI), and large-scale state transition (LST).44,45 ctDNA-positive was defined as at least one same variant in both tumor sample and plasma sample. HRD-positive was defined as BRCA1/2 mutation or HRD score ≥ 42.46
Outcomes
The primary endpoint of this study was RCB 0/I rate, defined as the percentage of patients who are classified into RCB 0 or RCB I following neoadjuvant therapy. The primary endpoint has been amended from the pCR rate to the RCB 0/I rate in December 14, 2022 (protocol p6). During the course of the study, a multicenter pooled analysis20 demonstrated that patients with Luminal/HER2-low EBC who achieved RCB 0, equal to pCR, or RCB I had almost the same significantly improved long-term prognosis compared to those with RCB II or RCB III disease. The RCB class adds substantially to the binary assessment of pCR versus non-pCR in predicting long-term survival in this population. Therefore, the RCB 0/I rate may provide a more accurate reflection of the efficacy of neoadjuvant pyrotinib plus chemotherapy in Luminal/HER2-low EBC patients compared to the pCR rate. After thorough discussion and consideration of the updated evidence, the investigators and statisticians of the PILHLE-001 study request a revision in February 28, 2022 and achieved the approval from independent data monitoring committee (IDMC) in March 01, 2022 to modify the primary endpoint from pCR to RCB 0/I. The new ethical approval was obtained on December 14, 2022 due to the COVID-19 outbreak. The RCB score was independently evaluated by two pathologists who were blinded to the patient’s characteristics utilizing the online RCB Calculator provided by the MD Anderson Cancer Center and was classified into four levels: RCB 0 (score = 0, equivalent to pCR), RCB I (score > 0–1.36), RCB II (score >1.36–3.28), and RCB III (RCB score >3.28). Any excessive discrepancies were addressed through discussion to reach a consensus.
Secondary endpoints included the pCR rate (ypT0/is ypN0, no residual invasive tumor cells in the breast and axillary nodes, regardless of ductal carcinoma in situ), objective response rate (ORR, complete or partial response in breast according to the Response Evaluation Criteria in Solid Tumors [version 1.1], based on MRI), breast conservation surgery (BCS) rate, 5-year disease-free survival (DFS, the time from the first dose of study drug until any relapse, secondary malignancy, or any-cause death), and 5-year overall survival (OS, the time from the first dose of study drug to any-cause death). The survival outcomes are not reported in this article and will be included in a separate manuscript once more mature data are collected and analyzed.
Safety assessments included the frequency and severity of adverse events (AEs) according to the National Cancer Institute Common Terminology Criteria for Adverse Events (version 5.0). We re-verified and reorganized the original data about adverse events with independent data monitoring committee. Exploratory analyses were conducted to investigate the associations between RCB status and the objective response status at t1 or potential biomarkers (MammaPrint/BluePrint signatures, MRI parameters, immune cell subpopulations, Ki67, TILs, gene sequencing).
Quantification and statistical analysis
The sample size was recalculated based on the RCB 0/I rate. To test the null hypothesis that 15% or fewer patients (as observed previous study21,22,47,48,49) would achieve an RCB 0/I, which was not considered clinically meaningful, the planned sample size of 48 patients provided an 80% power to detect a difference of 15.0% versus 30.0% in overall pathological response at a one-sided significance level of 0.05. If more than 11 patients achieved RCB 0/I, the study treatment would be considered beneficial for these patients.
Efficacy and safety analyses were conducted in all-treated population, defined as all patients receiving at least one dose of pyrotinib. Continuous data were presented as median (range) or mean (SD), while categorical data were presented as frequency (percentage). The corresponding 95% confidence intervals (CIs) of the RCB 0/I rate, pCR rate, ORR, and BCS rate were estimated using the Clopper-Pearson method. Post-hoc analyses of the RCB 0/I rate were performed among subgroups based on baseline characteristic variables. The levels of biomarkers were compared between patients with RCB 0/I and those with RCB II/III. Student’s t test or Wilcoxon test were used to compare continuous variables, while χ2 test or Fisher’s exact test were used for comparisons of categorical variables. Statistical analyses were performed using SPSS software (version 26.0) and R studio software (version 4.2.2). Statistical significance was set at two-sided p-value <0.05.
Additional resource
Clinical trial details for the PILHLE-001 study: Chinese Clinical Trial registration number ChiCTR2100047233.
Published: November 6, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2024.101807.
Contributor Information
Chang Gong, Email: gchang@mail.sysu.edu.cn.
Louis Wing Cheong Chow, Email: lwcchow@live.hk.
Erwei Song, Email: songew@mail.sysu.edu.cn.
Supplemental information
References
- 1.Tarantino P., Hamilton E., Tolaney S.M., Cortes J., Morganti S., Ferraro E., Marra A., Viale G., Trapani D., Cardoso F., et al. HER2-Low Breast Cancer: Pathological and Clinical Landscape. J. Clin. Oncol. 2020;38:1951–1962. doi: 10.1200/jco.19.02488. [DOI] [PubMed] [Google Scholar]
- 2.Tarantino P., Viale G., Press M.F., Hu X., Penault-Llorca F., Bardia A., Batistatou A., Burstein H.J., Carey L.A., Cortes J., et al. ESMO Expert Consensus Statements (ECS) on the definition, diagnosis, and management of HER2-low breast cancer. Ann. Oncol. 2023;34:645–659. doi: 10.1016/j.annonc.2023.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Peiffer D.S., Zhao F., Chen N., Hahn O.M., Nanda R., Olopade O.I., Huo D., Howard F.M. Clinicopathologic Characteristics and Prognosis of ERBB2-Low Breast Cancer Among Patients in the National Cancer Database. JAMA Oncol. 2023;9:500–510. doi: 10.1001/jamaoncol.2022.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huppert L.A., Gumusay O., Idossa D., Rugo H.S. Systemic therapy for hormone receptor-positive/human epidermal growth factor receptor 2-negative early stage and metastatic breast cancer. CA. Cancer J. Clin. 2023;73:480–515. doi: 10.3322/caac.21777. [DOI] [PubMed] [Google Scholar]
- 5.Won H.S., Ahn J., Kim Y., Kim J.S., Song J.Y., Kim H.K., Lee J., Park H.K., Kim Y.S. Clinical significance of HER2-low expression in early breast cancer: a nationwide study from the Korean Breast Cancer Society. Breast Cancer Res. 2022;24:22. doi: 10.1186/s13058-022-01519-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Viale G., Hanlon Newell A.E., Walker E., Harlow G., Bai I., Russo L., Dell'Orto P., Maisonneuve P. Ki-67 (30-9) scoring and differentiation of Luminal A- and Luminal B-like breast cancer subtypes. Breast Cancer Res. Treat. 2019;178:451–458. doi: 10.1007/s10549-019-05402-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuadros M., Llanos A. [Validation and clinical application of MammaPrint® in patients with breast cancer] Med. Clínica. 2011;136:627–632. doi: 10.1016/j.medcli.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 8.Fehrenbacher L., Cecchini R.S., Geyer C.E., Jr., Rastogi P., Costantino J.P., Atkins J.N., Crown J.P., Polikoff J., Boileau J.F., Provencher L., et al. NSABP B-47/NRG Oncology Phase III Randomized Trial Comparing Adjuvant Chemotherapy With or Without Trastuzumab in High-Risk Invasive Breast Cancer Negative for HER2 by FISH and With IHC 1+ or 2. J. Clin. Oncol. 2020;38:444–453. doi: 10.1200/jco.19.01455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gianni L., Lladó A., Bianchi G., Cortes J., Kellokumpu-Lehtinen P.L., Cameron D.A., Miles D., Salvagni S., Wardley A., Goeminne J.C., et al. Open-label, phase II, multicenter, randomized study of the efficacy and safety of two dose levels of Pertuzumab, a human epidermal growth factor receptor 2 dimerization inhibitor, in patients with human epidermal growth factor receptor 2-negative metastatic breast cancer. J. Clin. Oncol. 2010;28:1131–1137. doi: 10.1200/jco.2009.24.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filho O.M., Viale G., Stein S., Trippa L., Yardley D.A., Mayer I.A., Abramson V.G., Arteaga C.L., Spring L.M., Waks A.G., et al. Impact of HER2 Heterogeneity on Treatment Response of Early-Stage HER2-Positive Breast Cancer: Phase II Neoadjuvant Clinical Trial of T-DM1 Combined with Pertuzumab. Cancer Discov. 2021;11:2474–2487. doi: 10.1158/2159-8290.Cd-20-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Modi S., Jacot W., Yamashita T., Sohn J., Vidal M., Tokunaga E., Tsurutani J., Ueno N.T., Prat A., Chae Y.S., et al. Trastuzumab Deruxtecan in Previously Treated HER2-Low Advanced Breast Cancer. N. Engl. J. Med. 2022;387:9–20. doi: 10.1056/NEJMoa2203690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bardia A., Hurvitz S., Press M.F., Wang L.S., McAndrew N.P., Chan D., Phan V., Villa D., Tetef M.L., Chamberlain E., et al. Abstract GS2-03: GS2-03 TRIO-US B-12 TALENT: Neoadjuvant trastuzumab deruxtecan with or without anastrozole for HER2-low, HR+ early stage breast cancer. Cancer Res. 2023;83:GS2-03. doi: 10.1158/1538-7445.SABCS22-GS2-03. [DOI] [Google Scholar]
- 13.Wu J., Jiang Z., Liu Z., Yang B., Yang H., Tang J., Wang K., Liu Y., Wang H., Fu P., et al. Neoadjuvant pyrotinib, trastuzumab, and docetaxel for HER2-positive breast cancer (PHEDRA): a double-blind, randomized phase 3 trial. BMC Med. 2022;20:498. doi: 10.1186/s12916-022-02708-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu B., Yan M., Ma F., Hu X., Feng J., Ouyang Q., Tong Z., Li H., Zhang Q., Sun T., et al. Pyrotinib plus capecitabine versus lapatinib plus capecitabine for the treatment of HER2-positive metastatic breast cancer (PHOEBE): a multicentre, open-label, randomised, controlled, phase 3 trial. Lancet Oncol. 2021;22:351–360. doi: 10.1016/s1470-2045(20)30702-6. [DOI] [PubMed] [Google Scholar]
- 15.Ma F., Yan M., Li W., Ouyang Q., Tong Z., Teng Y., Wang Y., Wang S., Geng C., Luo T., et al. Pyrotinib versus placebo in combination with trastuzumab and docetaxel as first line treatment in patients with HER2 positive metastatic breast cancer (PHILA): randomised, double blind, multicentre, phase 3 trial. BMJ (Clinical research ed.) 2023;383 doi: 10.1136/bmj-2023-076065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Y., Cao W.M., Shao X., Shi Y., Cai L., Chen W., Liu J., Shen P., Chen Y., Wang X., et al. Pyrotinib plus docetaxel as first-line treatment for HER2-positive metastatic breast cancer: the PANDORA phase II trial. Nat. Commun. 2023;14:8314. doi: 10.1038/s41467-023-44140-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Symmans W.F., Yau C., Chen Y.Y., Balassanian R., Klein M.E., Pusztai L., Nanda R., Parker B.A., Datnow B., Krings G., et al. Assessment of Residual Cancer Burden and Event-Free Survival in Neoadjuvant Treatment for High-risk Breast Cancer: An Analysis of Data From the I-SPY2 Randomized Clinical Trial. JAMA Oncol. 2021;7:1654–1663. doi: 10.1001/jamaoncol.2021.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins D.M., Madden S.F., Gaynor N., AlSultan D., Le Gal M., Eustace A.J., Gately K.A., Hughes C., Davies A.M., Mahgoub T., et al. Effects of HER Family-targeting Tyrosine Kinase Inhibitors on Antibody-dependent Cell-mediated Cytotoxicity in HER2-expressing Breast Cancer. Clin. Cancer Res. 2021;27:807–818. doi: 10.1158/1078-0432.Ccr-20-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu S., Zhang Q., Zhang F., Meng F., Liu S., Zhou R., Wu Q., Li X., Shen L., Huang J., et al. HER2 recruits AKT1 to disrupt STING signalling and suppress antiviral defence and antitumour immunity. Nat. Cell Biol. 2019;21:1027–1040. doi: 10.1038/s41556-019-0352-z. [DOI] [PubMed] [Google Scholar]
- 20.Yau C., Osdoit M., van der Noordaa M., Shad S., Wei J., de Croze D., Hamy A.-S., Laé M., Reyal F., Sonke G.S., et al. Residual cancer burden after neoadjuvant chemotherapy and long-term survival outcomes in breast cancer: a multicentre pooled analysis of 5161 patients. Lancet Oncol. 2022;23:149–160. doi: 10.1016/S1470-2045(21)00589-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prat A., Saura C., Pascual T., Hernando C., Muñoz M., Paré L., González Farré B., Fernández P.L., Galván P., Chic N., et al. Ribociclib plus letrozole versus chemotherapy for postmenopausal women with hormone receptor-positive, HER2-negative, luminal B breast cancer (CORALLEEN): an open-label, multicentre, randomised, phase 2 trial. Lancet Oncol. 2020;21:33–43. doi: 10.1016/s1470-2045(19)30786-7. [DOI] [PubMed] [Google Scholar]
- 22.Cottu P., D'Hondt V., Dureau S., Lerebours F., Desmoulins I., Heudel P.E., Duhoux F.P., Levy C., Mouret-Reynier M.A., Dalenc F., et al. Letrozole and palbociclib versus chemotherapy as neoadjuvant therapy of high-risk luminal breast cancer. Ann. Oncol. 2018;29:2334–2340. doi: 10.1093/annonc/mdy448. [DOI] [PubMed] [Google Scholar]
- 23.Dieci M.V., Guarneri V., Tosi A., Bisagni G., Musolino A., Spazzapan S., Moretti G., Vernaci G.M., Griguolo G., Giarratano T., et al. Neoadjuvant Chemotherapy and Immunotherapy in Luminal B-like Breast Cancer: Results of the Phase II GIADA Trial. Clin. Cancer Res. 2022;28:308–317. doi: 10.1158/1078-0432.Ccr-21-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loi S., Curigliano G., Salgado R., Díaz R.I.R., Delaloge S., García C.I.R., Kok M., Saura C., Harbeck N., Mittendorf E., et al. Abstract GS01-01: Biomarker Results in High-risk Estrogen Receptor Positive, Human Epidermal Growth Factor Receptor 2 Negative Primary Breast Cancer Following Neoadjuvant Chemotherapy ± Nivolumab: an Exploratory Analysis of CheckMate 7FL. Cancer Res. 2024;84:GS01. doi: 10.1158/1538-7445.SABCS23-GS01-01. [DOI] [Google Scholar]
- 25.Cardoso F., O’Shaughnessy J., McArthur H., Schmid P., Cortés J., Harbeck N., Telli M., Cescon D., Fasching P.A., Shao Z., et al. Abstract GS01-02: Phase 3 study of neoadjuvant pembrolizumab or placebo plus chemotherapy, followed by adjuvant pembrolizumab or placebo plus endocrine therapy for early-stage high-risk ER+/HER2− breast cancer: KEYNOTE-756. Cancer Res. 2024;84:GS01–GS02. doi: 10.1158/1538-7445.SABCS23-GS01-02. [DOI] [Google Scholar]
- 26.Cardoso F., McArthur H.L., Schmid P., Cortés J., Harbeck N., Telli M.L., Cescon D.W., O'Shaughnessy J., Fasching P., Shao Z., et al. LBA21 KEYNOTE-756: Phase III study of neoadjuvant pembrolizumab (pembro) or placebo (pbo) + chemotherapy (chemo), followed by adjuvant pembro or pbo + endocrine therapy (ET) for early-stage high-risk ER+/HER2– breast cancer. Ann. Oncol. 2023;34:S1260–S1261. doi: 10.1016/j.annonc.2023.10.011. [DOI] [Google Scholar]
- 27.Ejlertsen B., Tuxen M.K., Jakobsen E.H., Jensen M.B., Knoop A.S., Højris I., Ewertz M., Balslev E., Danø H., Vestlev P.M., et al. Adjuvant Cyclophosphamide and Docetaxel With or Without Epirubicin for Early TOP2A-Normal Breast Cancer: DBCG 07-READ, an Open-Label, Phase III, Randomized Trial. J. Clin. Oncol. 2017;35:2639–2646. doi: 10.1200/jco.2017.72.3494. [DOI] [PubMed] [Google Scholar]
- 28.Guarneri V., de Azambuja E. Anthracyclines in the treatment of patients with early breast cancer. ESMO open. 2022;7 doi: 10.1016/j.esmoop.2022.100461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furukawa R., Inoue H., Yoneshima Y., Tsutsumi H., Iwama E., Ikematsu Y., Ando N., Shiraishi Y., Ota K., Tanaka K., et al. Cytotoxic chemotherapeutic agents and the EGFR-TKI osimertinib induce calreticulin exposure in non-small cell lung cancer. Lung cancer (Amsterdam, Netherlands) 2021;155:144–150. doi: 10.1016/j.lungcan.2021.03.018. [DOI] [PubMed] [Google Scholar]
- 30.Hannesdóttir L., Tymoszuk P., Parajuli N., Wasmer M.H., Philipp S., Daschil N., Datta S., Koller J.B., Tripp C.H., Stoitzner P., et al. Lapatinib and doxorubicin enhance the Stat1-dependent antitumor immune response. Eur. J. Immunol. 2013;43:2718–2729. doi: 10.1002/eji.201242505. [DOI] [PubMed] [Google Scholar]
- 31.Nolan E., Lindeman G.J., Visvader J.E. Deciphering breast cancer: from biology to the clinic. Cell. 2023;186:1708–1728. doi: 10.1016/j.cell.2023.01.040. [DOI] [PubMed] [Google Scholar]
- 32.Shi Q., Xuhong J., Luo T., Ge J., Liu F., Lan Y., Chen Q., Tang P., Fan L., Chen L., et al. PIK3CA mutations are associated with pathologic complete response rate to neoadjuvant pyrotinib and trastuzumab plus chemotherapy for HER2-positive breast cancer. Br. J. Cancer. 2023;128:121–129. doi: 10.1038/s41416-022-02021-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Minckwitz G., Sinn H.P., Raab G., Loibl S., Blohmer J.U., Eidtmann H., Hilfrich J., Merkle E., Jackisch C., Costa S.D., et al. Clinical response after two cycles compared to HER2, Ki-67, p53, and bcl-2 in independently predicting a pathological complete response after preoperative chemotherapy in patients with operable carcinoma of the breast. Breast Cancer Res. 2008;10:R30. doi: 10.1186/bcr1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beresford M.J., Stott D., Makris A. Assessment of clinical response after two cycles of primary chemotherapy in breast cancer. Breast Cancer Res. Treat. 2008;109:337–342. doi: 10.1007/s10549-007-9644-2. [DOI] [PubMed] [Google Scholar]
- 35.Viallard C., Larrivée B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis. 2017;20:409–426. doi: 10.1007/s10456-017-9562-9. [DOI] [PubMed] [Google Scholar]
- 36.Wang C., Deng S., Chen J., Xu X., Hu X., Kong D., Liang G., Yuan X., Li Y., Wang X. The Synergistic Effects of Pyrotinib Combined With Adriamycin on HER2-Positive Breast Cancer. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.616443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolff A.C., Hammond M.E.H., Allison K.H., Harvey B.E., Mangu P.B., Bartlett J.M.S., Bilous M., Ellis I.O., Fitzgibbons P., Hanna W., et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J. Clin. Oncol. 2018;36:2105–2122. doi: 10.1200/jco.2018.77.8738. [DOI] [PubMed] [Google Scholar]
- 38.Salgado R., Denkert C., Demaria S., Sirtaine N., Klauschen F., Pruneri G., Wienert S., Van den Eynden G., Baehner F.L., Penault-Llorca F., et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann. Oncol. 2015;26:259–271. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cardoso F., van't Veer L.J., Bogaerts J., Slaets L., Viale G., Delaloge S., Pierga J.Y., Brain E., Causeret S., DeLorenzi M., et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N. Engl. J. Med. 2016;375:717–729. doi: 10.1056/NEJMoa1602253. [DOI] [PubMed] [Google Scholar]
- 40.Mittempergher L., Delahaye L.J., Witteveen A.T., Snel M.H., Mee S., Chan B.Y., Dreezen C., Besseling N., Luiten E.J., Annuska M Glas. Performance Characteristics of the BluePrint® Breast Cancer Diagnostic Test. Transl. Oncol. 2020;13 doi: 10.1016/j.tranon.2020.100756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su D., Zhang D., Chen K., Lu J., Wu J., Cao X., Ying L., Jin Q., Ye Y., Xie Z., et al. High performance of targeted next generation sequencing on variance detection in clinical tumor specimens in comparison with current conventional methods. J. Exp. Clin. Cancer Res. 2017;36:121. doi: 10.1186/s13046-017-0591-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abkevich V., Timms K.M., Hennessy B.T., Potter J., Carey M.S., Meyer L.A., Smith-McCune K., Broaddus R., Lu K.H., Chen J., et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br. J. Cancer. 2012;107:1776–1782. doi: 10.1038/bjc.2012.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Popova T., Manié E., Rieunier G., Caux-Moncoutier V., Tirapo C., Dubois T., Delattre O., Sigal-Zafrani B., Bollet M., Longy M., et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012;72:5454–5462. doi: 10.1158/0008-5472.Can-12-1470. [DOI] [PubMed] [Google Scholar]
- 46.Telli M.L., Timms K.M., Reid J., Hennessy B., Mills G.B., Jensen K.C., Szallasi Z., Barry W.T., Winer E.P., Tung N.M., et al. Homologous Recombination Deficiency (HRD) Score Predicts Response to Platinum-Containing Neoadjuvant Chemotherapy in Patients with Triple-Negative Breast Cancer. Clin. Cancer Res. 2016;22:3764–3773. doi: 10.1158/1078-0432.Ccr-15-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamy A.S., Darrigues L., Laas E., De Croze D., Topciu L., Lam G.T., Evrevin C., Rozette S., Laot L., Lerebours F., et al. Prognostic value of the Residual Cancer Burden index according to breast cancer subtype: Validation on a cohort of BC patients treated by neoadjuvant chemotherapy. PLoS One. 2020;15 doi: 10.1371/journal.pone.0234191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Esserman L.J., Berry D.A., Cheang M.C.U., Yau C., Perou C.M., Carey L., DeMichele A., Gray J.W., Conway-Dorsey K., Lenburg M.E., et al. Chemotherapy response and recurrence-free survival in neoadjuvant breast cancer depends on biomarker profiles: results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657) Breast Cancer Res. Treat. 2012;132:1049–1062. doi: 10.1007/s10549-011-1895-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Symmans W.F., Wei C., Gould R., Yu X., Zhang Y., Liu M., Walls A., Bousamra A., Ramineni M., Sinn B., et al. Long-Term Prognostic Risk After Neoadjuvant Chemotherapy Associated With Residual Cancer Burden and Breast Cancer Subtype. J. Clin. Oncol. 2017;35:1049–1060. doi: 10.1200/jco.2015.63.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Participant-level clinical data are deposited to RED-Cap database (managed by the Sun Yat-sen Memorial Hospital) and can be accessed via the lead contact by reasonable request.
The raw sequencing data reported in this paper have been deposited at in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA-Human: HRA008271) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.