Abstract
The genes of the trithorax group (trxG) in Drosophila melanogaster are required to maintain the pattern of homeotic gene expression that is established early in embryogenesis by the transient expression of the segmentation genes. The precise role of each of the diverse trxG members and the functional relationships among them are not well understood. Here, we report on the isolation of the trxG gene moira (mor) and its molecular characterization. mor encodes a fruit fly homolog of the human and yeast chromatin-remodeling factors BAF170, BAF155, and SWI3. mor is widely expressed throughout development, and its 170-kDa protein product is present in many embryonic tissues. In vitro, MOR can bind to itself and it interacts with Brahma (BRM), an SWI2-SNF2 homolog, with which it is associated in embryonic nuclear extracts. The leucine zipper motif of MOR is likely to participate in self-oligomerization; the equally conserved SANT domain, for which no function is known, may be required for optimal binding to BRM. MOR thus joins BRM and Snf5-related 1 (SNR1), two known Drosophila SWI-SNF subunits that act as positive regulators of the homeotic genes. These observations provide a molecular explanation for the phenotypic and genetic relationships among several of the trxG genes by suggesting that they encode evolutionarily conserved components of a chromatin-remodeling complex.
Two classes of genes maintain regulatory decisions made during early Drosophila development by the localized expression of segmentation gene products. These are the Polycomb group (PcG) and the trithorax group (trxG) genes, which sustain, respectively, the repressed or active state of homeotic gene expression (30, 42). The trxG proteins are thought to act at many different levels of gene regulation to maintain continued and efficient expression of homeotic and other genes. While genetic tests indicate that members of this group are functionally related (see, for example, references 41 and 49), with the exception of Brahma and Snf5-related 1 (SNR1) (14), there has been no evidence that these proteins are components of the same multimeric complexes. Thus, the exact functional relationships among most members of this gene class are not yet understood.
Brahma (BRM) is a trxG protein product that is believed to increase target gene accessibility by overcoming the repressive effects of nucleosomal histones (9, 45). BRM is highly related to the Saccharomyces cerevisiae protein SNF2 (SWI2) (44), which is part of an 11-subunit, 2-MDa global regulatory complex that assists a large number of DNA-binding proteins to activate transcription of their target genes by facilitating their binding to nucleosomal sites (31, 50). Another potential fruit fly homolog of a yeast SWI-SNF component that may be involved in the regulation of homeotic gene expression is SNR1, which coisolates with BRM in a large protein complex (14). Although it was not, like most of the trxG genes, isolated in screens that were based on suppression of mutations in Polycomb, Snr1 does undergo genetic interactions with classic trxG genes (14).
To unravel the functional relationships among the different trxG members and to understand how the gene products exert regulatory control over other genes, it is necessary to obtain molecular information about those members that have not yet been cloned. One such gene is moira (mor), which was isolated in three independent screens for loci that undergo dosage-dependent interactions with Polycomb or ectopically expressed Antennapedia (29) and which exhibits many of the genetic and phenotypic characteristics of brm (6, 15, 16, 29, 44).
Here we demonstrate that mor encodes a Drosophila homolog of the S. cerevisiae SWI-SNF gene SWI3. The MOR protein sequence closely resembles those of the human SWI3-related proteins, BAF170 and BAF155 (53, 54). In accordance with its known function as a regulator of homeotic gene activity, mor is widely expressed during development and in many embryonic tissues in a spatiotemporal pattern that overlaps that of brm and Snr1. MOR is capable of forming homooligomers; optimal binding of MOR to itself is likely to require its leucine zipper motif. MOR can also bind to BRM, with which it is associated in embryonic nuclear extracts; this interaction may be mediated by the SANT domain of MOR, which is common to several proteins involved in basal or activated transcription and whose function is not known (1, 54).
Identification of MOR as an additional component of the Drosophila SWI-SNF complex provides a physical and biochemical explanation for the known functional relationship between two strong and well-characterized trxG proteins, MOR and BRM. This finding provides a conceptual framework in which to continue to analyze the role of SWI-SNF proteins in regulation of the homeotic and other developmentally regulated genes in eukaryotic organisms.
MATERIALS AND METHODS
Fruit fly culture and stocks.
Drosophila melanogaster was cultured at room temperature on standard cornmeal-yeast extract-dextrose medium or at 18°C on Instant Medium (Carolina Biological). Except for those generated in the course of this work, the mutations and transposons used are described in the online database FlyBase (20). mor region stocks and deficiencies were obtained from J. Kennison. All mor alleles were rebalanced over a TM3-ftz-lacZ balancer.
Isolation of new alleles of mor.
The original P{lacW} insertion mutagenesis was carried out with an attached-X ammunition chromosome that carries four copies of P{lacW} on each arm: C(1)RM, y1? P{lacW}5-45fD w1? P{lacW}4-5fP P{lacW}3-52d P{lacW}3-76a (24). Females carrying the attached-X ammunition chromosome and P{Δ2-3}99B were crossed with w; Sbsbd-2 Ubxbxd-1 males. F1 males with the mini-white eye color were crossed with w; Sbsbd-2 Ubxbxd-1 females. Whenever possible, homozygous insertion lines were established. F2 or F3 males were examined for changes in abdominal pigmentation or sternite bristle patterns. The insertion later designated P{lacW}89B mapped very close to Sb on the basis of segregation with the original Sbsbd-2 mutation. Mobilization of this element was accomplished by generating dysgenic males, crossing with w− females, and recovering w− Sb+ animals. Of 47 w− chromosomes recovered, 2 were lethal over Df(3R)sbd105.
Cloning of the mor region and molecular analysis of new excision alleles of mor.
The P{lacW} element allows cloning of adjacent genomic sequences by plasmid rescue (56). One isolate, pCK5.128A, obtained by EcoRI digestion of flies bearing P{lacW}89B, was labeled (random priming kit; Bethesda Research Laboratories) and used to screen a Sau3A partial genomic library of Canton-S DNA generated by using a Lambda-FIX kit (Stratagene) and high-molecular-weight genomic DNA (prepared as described in reference 34). Three independent genomic λ clones were recognized and characterized by restriction mapping and Southern analysis. Smaller genomic fragments were subcloned into pBluescript II SK (Stratagene).
The newly obtained excision mutants, mor9 and mor10, were subjected to Southern analysis with HindIII or EcoRI as described previously (12). As controls, the progenitor P{lacW}89B chromosome and a line thought to be a precise excision thereof were also analyzed. Consistent evidence of changes in both lines was obtained by using the λ probe CK128γ (Fig. 1A). When the original blot was reprobed with pP{CaSpeR} and pBluescript, there was no evidence that any portion of the P{lacW} element remained in either mor9 or mor10.
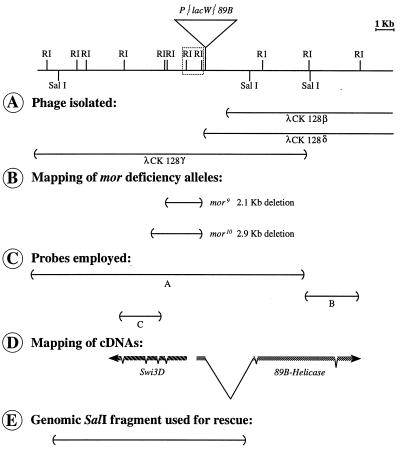
FIG. 1.
Map of the genomic region around the P{lacW}89B insertion site. The map shows the phage isolated from a Sau3A partial genomic library (A), the extent of the deletions in the new mor excision alleles (B), the probes employed in Northern analyses and for isolation of the mor cDNA (C), the map of the two mor candidate transcripts in the vicinity of the P{lacW}89B insertion site (D), and the genomic fragment used for rescue of the mor phenotype (E). In panel D, the introns indicated for 89B-Helicase are (from the 5′ to the 3′ end) about 2,800, 60, and 100 bp in length. The introns indicated for mor (Swi3D-1) are (from the 5′ to the 3′ end) 50, 63, 62, and 64 bp in length, and they are at nucleotides 1111, 1441, 2216, and 3449, respectively. The direction of transcription was determined by sequence analysis for 89B-Helicase and by the use of single-stranded probes for mor (Swi3D-1). Since the srp gene is located 10 kb to the right of 89B-Helicase and preliminary data indicate that the deficiency Df(3R)lpo4 breaks 3 to 4 kb 3′ of the mor transcript, centromere proximal is to the left.
Isolation of cDNAs and computer analyses.
To obtain the cDNA corresponding to the 4.5-kb transcript, an early embryonic cDNA library (8) was screened. One positive clone was recovered, and the cDNA insert was subcloned into pBluescript II SK. DNA, prepared by using a Wizard Miniprep kit (Promega), was sequenced at the sequencing facility of the Life Sciences Institute, Hebrew University, Jerusalem, Israel. Synthetic primers were made to allow complete sequencing of both strands. To obtain additional cDNA clones, we screened a random-primed embryonic cDNA library (27) with 5′ sequences of the one positive clone and also carried out PCR amplification (with PWO [Boehringer]) on the same library with an oligonucleotide based on the sequence of the noncoding strand of Swi3D-2 (5′ TTCAAAGCCCTGCAGCGTC 3′) and the λgt11 reverse primer. Sequences were analyzed by using the National Center for Supercomputer Applications electronic mail server and the Fasta (37), Blast (2), Gap (35), and Pileup (17) programs.
RNA preparation, Northern blotting, and determination of the direction of transcription.
Developmental Northern blotting of total nucleic acids was performed by grinding frozen samples in a 7 M urea buffer, subjecting them to phenol-chloroform extractions and ethanol precipitation, and then applying approximately 12 μg of nucleic acid from each time point to a formaldehyde-morpholinepropanesulfonic acid (MOPS) denaturing gel (12).
To determine the direction of transcription of the 4.5-kb transcript, single-stranded RNA probes were generated from subclones of the 1.3-kb EcoRI fragment to the left of the P{lacW}89B insertion, placed in both orientations into pBluescript II SK. These constructs were used to probe the total nucleic acid blots.
Southern blotting of DNA derived from fruit flies mutant in mor, and single-embryo PCR.
Genomic DNA was isolated by homogenizing 200 flies in 1.6 ml of homogenization buffer (30 mM Tris-Cl [pH 9.0], 10 mM EDTA, 100 mM NaCl, 70 g of sucrose/liter) and 0.4 ml of lysis buffer (0.5 M Tris-Cl [pH 9.0], 0.25 M EDTA, 2.5% sodium dodecyl sulfate [SDS]), incubating at 65°C for 30 min, and, after the addition of 300 μl of 8 M potassium acetate, incubating for an additional hour at 0°C. Following centrifugation, 10 μg of ethanol-precipitated DNA was used per lane. Southern blots were prehybridized at 65°C in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 1% N-lauroylsarcosine (Sigma) and 0.5 mg of herring sperm DNA (Sigma)/ml and, following addition of the probe, hybridized overnight. The washes reached a stringency of 0.1× SSC–0.5% N-lauroylsarcosine. The Swi3D probe, encompassing 3.6 kb of Swi3D cDNA (excluding the 5′-most 180 bp of Swi3D-1), was labeled by using a random primer labeling kit (Biological Industries, Beit HaEmek, Israel).
For PCR analysis of homozygous embryos, 3- to 12-h embryos laid by flies carrying mor alleles over a TM3-ftz-lacZ balancer were stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (21). DNA was amplified by using a RedHot Taq polymerase (Advanced Biotechnologies), and the amplified band was isolated for sequencing by using a QIAquick gel extraction kit (Qiagen).
P element transformation and rescue of the mor phenotype.
A genomic SalI fragment approximately 11.8 kb in length (Fig. 1E) was cloned into the XhoI site of the pP{CaSpeR-4} transformation vector (20). This fragment includes approximately 3 kb of flanking sequence at both the 5′ and 3′ ends of the 4.5-kb transcript, as well as the 5′ portion of Hel89B. Isolates with the fragment inserted in both orientations were recovered and designated pP{11.8A} and pP{11.8B}. The 4.5-kb mRNA is transcribed in the opposite direction relative to the CaSpeR mini-white gene in the pP{11.8A} construct.
y1 w1118 embryos were coinjected with pP{11.8} and pπ25.7wc in accordance with standard techniques (5, 43). Emerging adults were crossed individually to y1 w1118 flies, and transformants were identified. Two independent lines were recovered and crossed with a multiple-balancer stock; chromosomal linkage was determined, and stocks were established. Both of the transformed lines are homozygous viable and carry the P{11.8A} version of the construct. After rescue was demonstrated, the P{11.8A} transposon was renamed P{mor+11.8}. The full genotype of this construct is P{w+mC Hel89B5′ mor+t11.8 = mor+11.8}.
Antibody preparation, affinity purification, and Western blot analyses.
Antibodies were generated against MOR by inserting a 0.9-kb BamHI-EcoRI fragment into the pGEX1 expression vector (Pharmacia LKB Biotechnology Inc.), harvesting the immunogen as described previously (22), and inoculating rabbits with the immunogen in combination with Freund’s adjuvant (Sigma). The antibody was affinity purified after being allowed to bind to immunoblots containing the MOR fusion protein as described in reference 25.
For Western analysis, tissues were homogenized in a solution containing 50 mM Tris-Cl (pH 7.4), 25% glycerol, 6% β-mercaptoethanol, 4% SDS, 1 mM EDTA, protease inhibitors (1 μg of leupeptin/ml, 3 μg of aprotinin/ml, 0.06 μg of antipain/ml, 1 mM phenylmethylsulfonyl fluoride, and a 1:100 dilution of protease inhibitor cocktail P2714 [all from Sigma]). The homogenates were boiled for 5 min, and the proteins in the supernatant were separated by SDS-polyacrylamide gel electrophoresis (PAGE) on 7.5% gels. The gels were blotted onto a Hybond-C membrane (Amersham) as described previously (22). The blot was incubated for 3 h with 5.0% nonfat dry milk in 10 mM Tris-Cl (pH 7.4)–150 mM NaCl (TBS). The preimmune or anti-MOR antiserum was applied at a 1:300 dilution in TBS overnight at 4°C. Following washes in TBS and TBS–0.05% Triton X-100 and a 1-h incubation in TBS with 5% normal goat serum (Biological Industries), horseradish peroxidase-conjugated goat anti-rabbit antibodies (Jackson) were applied for 2 h at room temperature at a 1:1,500 dilution. After washes were performed as described above, the immunoreactive bands were detected by enhanced chemiluminescence (ECL; Amersham).
Immunohistochemistry and in situ hybridization.
Immunohistochemistry was carried out as described previously (22). In situ hybridizations were carried out by the method of Tautz and Pfeifle (46) with digoxigenin-labeled probes (Boehringer). An approximately 900-bp BamHI-EcoRI fragment extending from nucleotides 319 to 1234 of Swi3D-1 was inserted into pBluescript II KS to allow synthesis of RNA probes by transcription from both the T7 and T3 promoters. Following color development, the embryos were mounted in JB4 medium (Polysciences), viewed under Nomarski optics with a Zeiss Axioskop microscope, and photographed with T-MAX 100 film (Kodak).
Coimmunoprecipitation experiments.
For coimmunoprecipitations, preimmune serum or antibodies to either MOR or BRM were coupled to protein A-Sepharose beads (Pharmacia) with dimethylpimelimidate (25). The anti-BRM antibodies were obtained by immunizing rabbits with a peptide corresponding to amino acids 1617 to 1632 of BRM, coupled to keyhole limpet hemocyanin, by the use of standard procedures (25). The antibodies generated in this way recognize only one band of about 190 kDa when used in Western analysis of a crude embryo nuclear extract. Next, 15 μl of a BRM-containing embryonic nuclear fraction (see below) was added to 10 μl of beads and incubated for 2 h at 4°C in a total volume of 90 μl of HEMG (25 mM HEPES-KOH [pH 7.6], 0.1 mM EDTA, 12.5 mM MgCl2, 10% glycerol, 1.5 mM dithiothreitol, 1 mM sodium metabisulfide, 0.2 mM AEBSF, 2 mg of leupeptin/ml, and 0.7 mg of pepstatin/ml) containing 0.4 M KCl and 0.1% Triton X-100. After the incubation, the beads were washed once with a 100-fold excess volume of HEMG–0.4 M KCl–0.1% Nonidet P-40 (NP-40) and five times in HEMG–0.5 M KCl–0.01% NP-40, resuspended in SDS sample buffer, and analyzed by immunoblotting with antibodies directed against either BRM or MOR.
The BRM-containing fraction was obtained by preparing nuclear extracts from 0- to 12-h-old Drosophila embryos as previously described (52). About 400 mg of protein was further purified by heparin-agarose chromatography by standard procedures (4). The eluate of the heparin-agarose column was fractionated on an 800-ml Sephacryl S-300 gel filtration column equilibrated with HEMG–0.1 M KCl–0.01% NP-40 and developed with the same buffer (4). After a 250-ml elution volume, 10-ml fractions were collected. By Western blotting, the bulk of BRM was detected in fractions 2 to 9. Fraction 3 was used for coimmunoprecipitations.
In vitro interactions of GST fusion proteins and yeast two-hybrid analysis.
35S-labeled MOR or MOR derivatives were synthesized by using a TNT coupled rabbit reticulocyte system (Promega). To obtain MORΔNcoI and MORΔSANT, respectively, a 1,251-nucleotide NcoI fragment or a 270-nucleotide XbaI fragment was excised from the full-length mor pBluescript II SK cDNA while maintaining the same reading frame. To generate MORΔLEU, a 750-bp NcoI fragment was reinserted into MORΔNcoI. The large glutathione S-transferase (GST)–MOR fusion protein employed is encoded by a 2.1-kb EcoRI cDNA fragment placed into the pGEX1 vector. GST-MOR-NH2 is the fusion protein that was used for rabbit inoculation. To generate the GST-BRM fusion proteins, genomic fragments of brm containing no introns were PCR amplified with PWO (Boehringer) from wild-type genomic DNA, using oligonucleotide primers in which we incorporated synthetic BamHI sites. The sequences of these oligonucleotides are as follows: for the large BRM fusion protein, beginning at nucleotide 742 of the brm cDNA (44), 5′ ACCAGGGATCCAGCATGCAGGACAAC 3′; for the small BRM fusion protein, beginning at nucleotide 1625 of brm, 5′GCAAGGATCCAAGGGAGTTCCACAGAAAT 3′; and for both BRM fusion proteins, beginning at nucleotide 2263 of the noncoding strand of brm, 5′ GGCCTTGGGAATTCGATCCTTGGCATCCTCAT 3′. The large amplified fragment was cloned into pGEX3 by partial digestion with BamHI, and the small fragment was cloned into pGEX1. Both were sequenced before use. GST fusion proteins were harvested as described previously (22).
For pulldown experiments, 50 μl of a 1:1 slurry of beads with bound protein (blank beads were used to equalize all bead volumes so that the amount of bound protein was the same in each sample by Coomassie blue staining) were resuspended in 1 ml of phosphate-buffered saline–200 mM NaCl–10% bovine serum albumin (Biological Industries) for a 1-h preincubation at 4°C with gentle rocking. Labeled proteins were added, and the incubations were continued an additional 3 h. The beads were washed four times in phosphate-buffered saline with 200 or 300 mM NaCl by centrifugation at 1,000 × g for 30 s and removal of the supernatant with a 27-gauge syringe. Sample buffer was added, and labeled proteins were visualized by methyl salicylate enhancement and analyzed with a PhosphorImager (Molecular Dynamics) to determine loading variations and the amount of retention.
For yeast two-hybrid analysis, a 2.1-kb EcoRI fragment of mor cDNA was inserted into the LEXA202 (40) and pJG4-5 (23) plasmids and analysis was carried out by standard procedures (18, 19).
Nucleotide sequence accession number.
The GenBank accession number for the sequences of the mor cDNA clones isolated in this study is AF033823.
RESULTS
Isolation of new alleles of mor.
An insertion of the P transposon P{lacW} into the mor region was recovered during a screen for insertion-induced mutations resulting in abdominal phenotypes, such as the loss of male-specific pigmentation or changes in the patterns of sternite bristles. This insertion produces bristles on the sixth sternite of homozygous males; the phenotype is weak and variable. It was subsequently observed that the insertion, when homozygous, enhances the homozygous phenotype of Ubxbxd-1, a mutation of the bithorax complex. Since by recombination mapping it was determined to be immediately to the left of Sb, it appeared likely to be an insertion at or near the mor locus. However, test crosses with known mor alleles (mor5 and mor6) exhibited complementation.
The P{lacW}89B element was mobilized, and the recovered excision chromosomes were tested for viability over Df(3R)sbd105, which uncovers the 89B region. Two chromosomes that are lethal when homozygous and when placed in trans to Df(3R)sbd105 were recovered; both failed to complement the lethality of all mor alleles tested and of each other but were wild type over a serpent allele, srp3. Based on these results, the new excision alleles were designated mor9 and mor10. However, they differ from the other mor alleles in that they fail to complement the phenotype of the P{lacW}89B insertion; males of the genotype P{lacW}89B/mor9 or P{lacW}89B/mor10 display one to four small bristles on the sixth abdominal sternite.
Cloning of mor and identification of candidate transcripts.
Genomic sequences immediately adjacent to the P{lacW}89B insertion were recovered by plasmid rescue and used to screen a Canton-S genomic library. Three overlapping λ clones, spanning approximately 30 kb, were recovered (Fig. 1A). A restriction-level map was derived, and P{lacW}89B, mor9, and mor10 were placed on the map based on genomic Southern analyses (Fig. 1B). Both mor9 and mor10 are small deletions that remove the entire P{lacW} element and an additional 2 to 3 kb of genomic sequence.
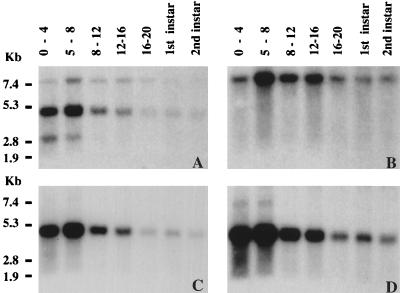
Genomic sequences around the P element insertion in P{lacW}89B were employed as probes in Northern blot analyses to identify transcripts encoded by the sequence in this vicinity. Using a large probe spanning the insertion site (labeled A in Fig. 1C), three major transcripts were visualized (Fig. 2A). One was an approximately 3-kb transcript (Fig. 2A) that is unlikely to encode mor since it exhibits limited expression in 0- to 8-h embryos. The second, an approximately 7.5-kb transcript (Fig. 2A and B) encoded by sequences to the right of the P{lacW}89B insertion, corresponds to the 89B-Helicase (Hel89B) gene that we have already described elsewhere (22). Hel89B is the Drosophila homolog of mot1 in S. cerevisiae (3, 13, 39) and of the human gene TAFII170/172 (11, 51). Because of the probable role of the Hel89B protein in anscriptional regulation of a large number of target genes (22), it was a good candidate for being encoded by the mor gene.
FIG. 2.
Developmental Northern blot analyses with genomic sequences around the P{lacW}89B insertion site reveal three transcripts. (A) When total RNA from staged embryos and larvae was probed with the probe marked A in Fig. 1C, three transcripts were visualized: an approximately 7.5-kb transcript that was present at all stages examined, an approximately 4.5-kb transcript that was expressed with a similar developmental pattern, and a ca. 3-kb transcript observed only at the earliest developmental stages. (B) When probe B was used on the blot shown in panel A, only the 7.5-kb transcript was seen. (C) When probe C was employed, again on the same blot, only the 4.5-kb transcript was seen. (D) A longer exposure of the blot in panel C reveals an additional high-molecular-weight transcript in RNA from 0- to 4- and 4- to 8-h embryos. Total RNA was isolated from 0- to 4-, 5- to 8-, 8- to 12-, 12- to 16-, and 16- to 20-h embryos and from first- and second-instar larvae. The positions of molecular size markers are shown on the left.
The third transcript, about 4.5 kb in size, is recognized by sequences to the left of the P{lacW}89B insertion (Fig. 2A and C). It, like Hel89B, is present at 0 to 4 h of embryonic development, reaches maximal levels at 4 to 8 h, and is found in decreasing amounts from 8 h until the second larval instar. Also like Hel89B, it is expressed in third-instar larvae, pupae, and adult male and female flies (data not shown). Upon longer exposure of the blot probed with the 2.3-kb genomic EcoRI fragment to the left of the P{lacW}89B insertion (labeled C in Fig. 1C), an approximately 7-kb transcript was observed in RNA derived from 0- to 8-h embryos (Fig. 2D). Because both transcripts recognized by the 2.3-kb probe are present in unfertilized eggs (data not shown), they appear to be of maternal origin. Since the gene encoding the approximately 4.5-kb transcript was unknown to us, we set out to characterize it further.
The 4.5-kb transcript encodes a Drosophila BAF170/BAF155/SWI3 homolog.
A cDNA clone corresponding to the approximately 4.5-kb transcript was obtained from an early embryonic cDNA library, using the 2.3-kb genomic EcoRI fragment (labeled C in Fig. 1C) as a probe. This cDNA, 3.8 kb in length, was sequenced, revealing a single long open reading frame followed by 210 bp of untranslated sequence. The cDNA appears to be incomplete since it lacks a poly(A) sequence at its 3′ end and there is no stop codon upstream of the first ATG at the 5′ end.
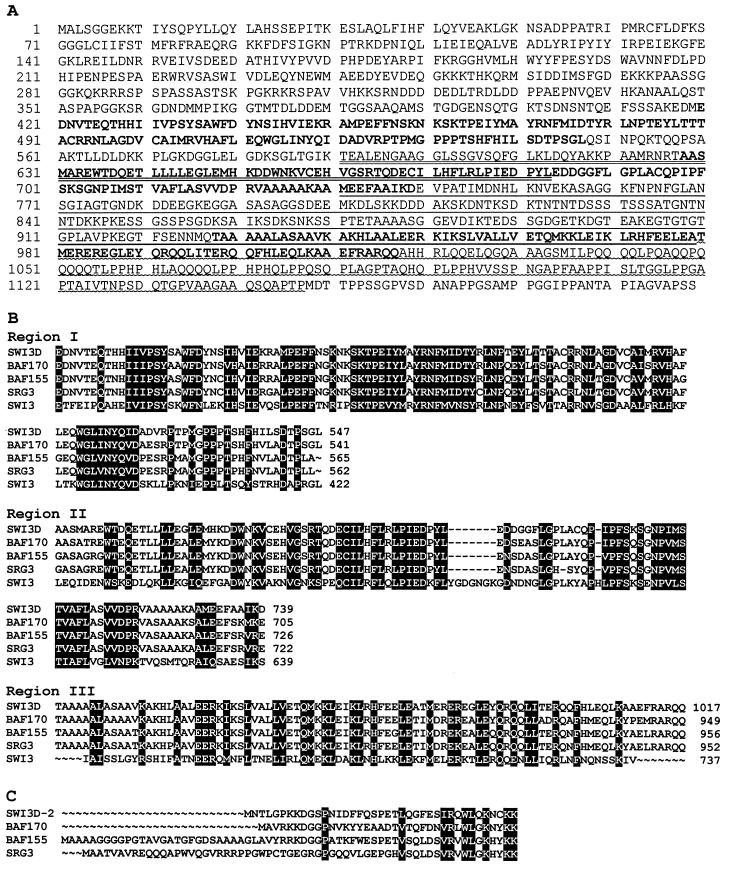
The 3.8-kb cDNA encodes a protein of 1,189 amino acids whose sequence is shown in Fig. 3A. Comparison of the predicted protein with the data bank sequences revealed that it exhibits a high degree of homology to yeast SWI3 and even greater similarity to human and mouse SWI3-related proteins. Thus, we refer to it as SWI3D. The degree of overall identity between SWI3D and the human protein BAF170 (54) is marginally higher than that between SWI3D and human BAF155 (54) (48% identity and 58% similarity versus 47% identity and 56% similarity). The SWI3D protein is as identical to the mouse SWI3 homolog, SRG3 (28), as it is to BAF155. A lower level of overall identity is exhibited to SWI3 itself (37% identity and 47% similarity).
FIG. 3.
Sequence of SWI3D protein and comparison to related proteins. (A) The amino acid sequence encoded by Swi3D-1. The regions that, with small changes, correspond to regions I to III in reference 54 are shown in boldface. The extents of the deletions in the constructs generated for the GST fusion protein interaction assay (Fig. 8) are indicated by underlining; MORΔSANT is missing the residues indicated by double underlining, MORΔNcoI lacks residues indicated with a single straight or wavy underline, and MORΔLEU lacks only the residues with a wavy underline. (B) Alignment of regions I to III of SWI3D with the same domains of BAF170, BAF155, SRG3, and SWI3. The numbers indicate the positions of the last amino acids in the regions. (C) Alignment of the sequence encoded by Swi3D-2 with the amino-terminal sequences of BAF170, BAF155, and SRG3. A stop codon is found 30 nucleotides upstream of the candidate initiator methionine. The next amino acid in the protein encoded by Swi3D-2 is tyrosine (Y, shown in position 12 of the sequence in panel A). In panels B and C, amino acids that are identical or similar in all the polypeptide sequences are shaded in black.
Alignment of the SWI3D sequence with those of BAF170, BAF155, SRG3, and SWI3 revealed that SWI3D contains all three of the conserved regions previously described in these proteins and that the level of sequence identity between SWI3D and the others is highest in these regions (Fig. 3B). SWI3D is most similar to the other proteins (especially BAF170) in region I, which is rich in prolines as well as hydrophobic and aromatic amino acids, leading to the suggestion that it may be hidden within the SWI-SNF complex (54). Region II is a tryptophan-rich domain which has been termed the SANT domain because of its presence in four proteins (SWI3, ADA2, N-CoR, and TFIIB) that participate in basal or activated transcription (1). Region III contains a leucine zipper oligomerization motif, suggesting that SWI3D may be able to bind to itself or another leucine zipper-containing protein. In addition to these conserved domains, the carboxy terminus of SWI3D is like those of BAF170, BAF155, and SRG3, very proline rich, but it is more glutamine rich than the corresponding termini of these proteins.
To isolate further-5′ regions of the Swi3D transcript, a random-primed embryonic cDNA library was probed. In this screening, an alternatively spliced cDNA, Swi3D-2 (Fig. 3C), was isolated. Swi3D-2 has a candidate initiator methionine that is preceded by a stop codon. Because of the presence of a 91-bp intron in the Swi3D-2 cDNA, it is missing the sequences which encode the first 19 amino acids of SWI3D-1 and it has a different amino-terminal sequence. Unlike that of SWI3D-1, the amino terminus of SWI3D-2 is very similar to those of BAF155 and BAF170 and a little less similar to that of SRG3 (Fig. 3C).
Analysis of DNA derived from mor mutants with Swi3D probes.
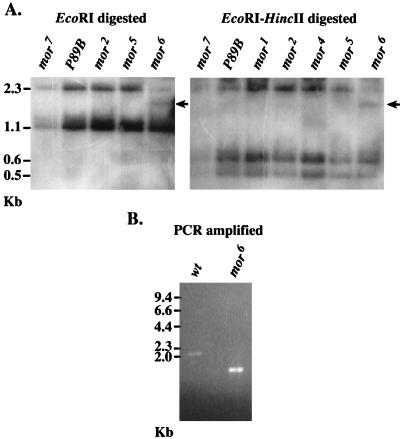
To ascribe mor function to either 89B-Helicase or Swi3D, we carried out Southern analyses of DNA extracted from heterozygous flies carrying mor alleles 1 through 6 over the same balancer chromosome. While no differences in the pattern of restriction fragments recognized by the 89B-Helicase probe were observed (data not shown), the Swi3D probe recognized one additional band in DNA extracted from flies heterozygous for the mor6 allele and digested with either EcoRI (Fig. 4A, left panel), EcoRI plus HincII (Fig. 4A, right panel), or HincII plus XbaI (data not shown). In comparison with those of the other mor alleles, the intensity of the 2.3-kb EcoRI-generated band in mor6 was reduced to the level of the same band in DNA derived from flies heterozygous for Df(3R)mor7, in which the mor locus is completely removed (7). In addition, a new, smaller, cross-hybridizing band appeared. This indicated that the 2.3-kb genomic EcoRI fragment that contains sequences of the Swi3D gene was disrupted in the mor6 allele.
FIG. 4.
(A) Southern blot analysis of genomic DNA from mor-heterozygous flies with a Swi3D probe reveals an additional cross-hybridizing band in mor6. DNA was extracted from adult flies carrying different alleles of mor over the identical balancer chromosome, restricted with either EcoRI alone (left panel) or EcoRI plus HincII (right panel), and probed with sequences representing 3.6 kb of Swi3D cDNA. The lane marked mor7 represents a haploid amount of DNA derived solely from the balancer chromosome, and the lane marked P89B serves as a control since there are no alterations in the DNA derived from heterozygous P{lacW}89B flies in the sequences recognized by the mor probe. In both panels, the lane marked mor6 has a unique cross-hybridizing band (arrow) that is not observed in DNA derived from flies carrying other mor alleles, including mor5 (which was generated in the same genetic background as mor6). In addition, the intensity of the 2.3-kb band in DNA derived from heterozygous mor6 flies is reduced relative to that of the smaller fragments and is similar to the intensity of this band in DNA derived from the Df(3R)mor7 flies. Band sizes are shown at the side in kilobases. (B) PCR amplification of genomic DNA from a single wild-type (wt) or mor6-homozygous embryo, using a pair of oligonucleotides based on sequences within the 2.3-kb genomic EcoRI fragment of Swi3D. The anticipated size of the amplified fragment obtained by using this oligonucleotide pair (one starting at bp 1670 of Swi3D and the second beginning at bp 3570) is 2.0 kb (because of the presence of two introns of 62 and 64 bp). The actual size of the amplified fragment of mor6 DNA is about 1.7 kb. Molecular size marker positions are indicated at the left.
We reasoned that the apparent reduction in size of the 2.3-kb genomic EcoRI fragment in DNA derived from heterozygous mor6 flies could be due to an internal deletion. To localize this deletion, DNA from homozygous mor6 embryos was subjected to PCR amplification with nested oligonucleotides that bracket this region. A 1.7-kb amplicon that was about 300 bp smaller than the equivalent genomic fragment amplified from wild-type DNA was obtained (Fig. 4B). Sequencing of the amplicon derived from mor6 embryos revealed that it was missing 541 nucleotides of the Swi3D-1 cDNA sequence (nucleotides 2431 to 2970) and contained an insertion of 233 bp of irrelevant DNA that did not match any known sequence. Since there are no introns in this area, these alterations caused the mor6 genomic EcoRI fragment to be 308 bp smaller than that of wild-type DNA. Thus, the mutation in mor6 constitutes a deletion of the leucine zipper motif of Swi3D (which begins at nucleotide 2790) and of the carboxyl proline- and glutamine-rich sequences that are lost due to premature closure of the translational open reading frame. This strongly suggests that Swi3D is encoded by mor and indicates that the leucine zipper domain and the carboxy-terminal sequences of Swi3D are functionally significant in vivo.
Rescue of the mor lethal phenotype with a Swi3D transgene.
A large SalI fragment, approximately 11.8 kb, that encompasses all of the genomic region known to correspond to the Swi3D transcript (Fig. 1E) was cloned into the P transformation vector pP{CaSpeR-4}. This fragment extends into the 5′ end of the transcribed region of 89B-Helicase but does not include most of the coding sequence of that gene; specifically, the essential ATPase domain (3) is not included. The resulting transformation construct, pP{mor+11.8}, was injected into y1 w1118 embryos. Two transformed lines that carry the P{mor+11.8} transposon on the second chromosome and are homozygous viable were established.
One copy of the P{mor+11.8} transgene was able to rescue all heteroallelic mor1, mor2, mor5, and mor6 combinations tested (Table 1). While in the absence of P{mor+11.8} these combinations behaved as complete lethals, in the presence of the transgene they all yielded fertile progeny of the rescued class with a frequency that did not deviate significantly from that expected assuming complete rescue (χ2 test, P > 0.05). Identical results were obtained when the rescue of mor6 was tested over the deletion Df(3R)mor7. Given the complete rescue observed for the heteroallelic combinations, the observations that homozygous combinations of these alleles were not rescued (in the cases of mor1 and mor2) or showed reduced viability and/or fertility (mor5 and mor6) is almost certainly due to the accretion of other deleterious mutations on these chromosomes. Despite this problem, a homozygous stock of P{mor+11.8}; mor6 flies has been established.
TABLE 1.
Rescue of heteroallelic mor combinations with P{mor+11.8}a
| Genotype | Viability | Fertility | Phenotype |
|---|---|---|---|
| P{mor+11.8}/+; mor1/mor6 | +b | + | + |
| P{mor+11.8}/+; mor2/mor6 | +b | + | + |
| P{mor+11.8}/+; mor5/mor6 | +b | + | + |
| P{mor+11.8}/+; mor1/mor2 | +c | NDg | + |
| P{mor+11.8}/+; mor6/Df(3R)mor7 | +c | + | +e |
| P{mor+11.8}/+; mor6/mor9 | +b | + | +e |
| P{mor+11.8}/+; mor6/mor10 | +b | + | + |
| P{mor+11.8}/+; mor9/mor10 | ±d | ± | A6, wingf |
| P{mor+11.8}/+; mor9/Df(3R)mor7 | +c | ± | A6, wingf |
+, characteristic rescued; ±, characteristic partially reserved. Several crosses were performed for each combination.
No statistically significant deviation in the observed versus expected frequency (χ2 test).
Statistical analysis was not performed.
Statistically significant reduction in the observed versus expected frequency (χ2 test).
Occasional male with one A6 sternite bristle.
A6, one to six small bristles observed in the sixth sternite of males; wing, wings were slightly smaller than those of the wild type.
ND, not determined.
The lethal P excision alleles mor9 and mor10 exhibited complete rescue in combination with mor6 but only partial rescue in combination with each other (Table 1). P{mor+11.8}; mor9/mor10 adults did survive, but at a statistically significantly lower frequency (by χ2, P < 0.005 and P < 0.025 in two different crosses). Survivors exhibited reduced fertility and had a smaller wing size. Males exhibited a weak A6 sternite bristle phenotype, similar to that of the original P{lacW}89B insertion. Rescue of mor9 over Df(3R)mor7 gave very similar results (Table 1), while P{mor+11.8}; mor9/mor9 and P{mor+11.8}; mor10/mor10 flies displayed more severe reductions in viability and fertility. Nonetheless, a homozygous P{mor+11.8}; mor9 fly stock has been established. Partial rescue of the mor9 and mor10 phenotypes by the P{mor+11.8} transgene constituted a second case in which the genetic behavior of these alleles differed from that of other mor alleles and suggested that their effects are not restricted to the mor gene (see Discussion).
We conclude that the trxG gene mor encodes SWI3D, a Drosophila homolog of proteins known to function as part of the SWI-SNF complex in yeast and mammals. In the descriptions given below, the SWI3D protein is referred to as the MOR protein. To compare the distribution of MOR with that of BRM, which is also a trxG gene product, and SNR1, which is present in a complex with BRM, and to investigate the association of MOR with BRM, we generated antibodies against MOR.
Antibodies against MOR recognize a 170-kDa nuclear protein in embryos.
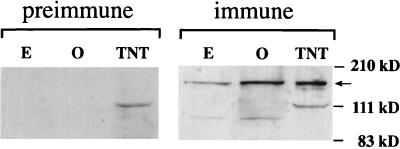
Polyclonal antibodies against a bacterial fusion protein containing amino acids 105 to 411 from the relatively nonconserved amino-terminal domain of MOR upstream of region I were raised in two rabbits. Both of the anti-MOR antisera, but neither of their preimmune controls, recognized a 170-kDa polypeptide when used in Western analysis of proteins extracted from embryos and adult ovaries (data not shown). Because both antisera also recognized additional polypeptides, one of the two anti-MOR antisera was affinity purified against the original immunogen as described in Materials and Methods. When used in Western analysis as described above, the affinity-purified immune serum specifically recognized a prevalent 170-kDa band (Fig. 5, right panel) that was not seen in blots probed with the preimmune serum (Fig. 5, left panel). The endogenous protein is of the same molecular weight as the MOR protein whose synthesis is directed in vitro by Swi3D-1, suggesting that the embryonic mor cDNA clone that we have isolated is complete or nearly so. The molecular mass of 170 kDa was larger than anticipated for MOR, but its human homologs, BAF155 and BAF170, also exhibited slower electrophoretic mobilities than predicted (54). An additional, faint band of about 100-kDa seen in the embryonic and ovarian extracts likely represented a degradation product of the 170-kDa polypeptide because it, as well as the 170-kDa band, was absent from extracts of late-stage homozygous mor6 embryos selected from among their blue (i.e., heterozygous) siblings (data not shown). In these embryos, the predicted truncated protein was also not seen, probably because of its instability when it cannot be incorporated into a multiprotein complex (see below).
FIG. 5.
Visualization of MOR by Western blot analysis of proteins extracted from embryos (E) and adult ovaries (O) and of in vitro-synthesized MOR protein (TNT) with affinity-purified anti-MOR antibodies. The anti-MOR antiserum allowed visualization of a 170-kDa protein made by in vitro transcription and translation of the mor(Swi3D-1) cDNA and an endogenous polypeptide of the same size in embryo and ovary extracts (arrow). These bands are not observed in identical Western blots treated with the preimmune serum (left panel). The smaller polypeptide seen in these extracts is likely to represent a degradation product (see text). The positions of molecular size markers are indicated at the right.
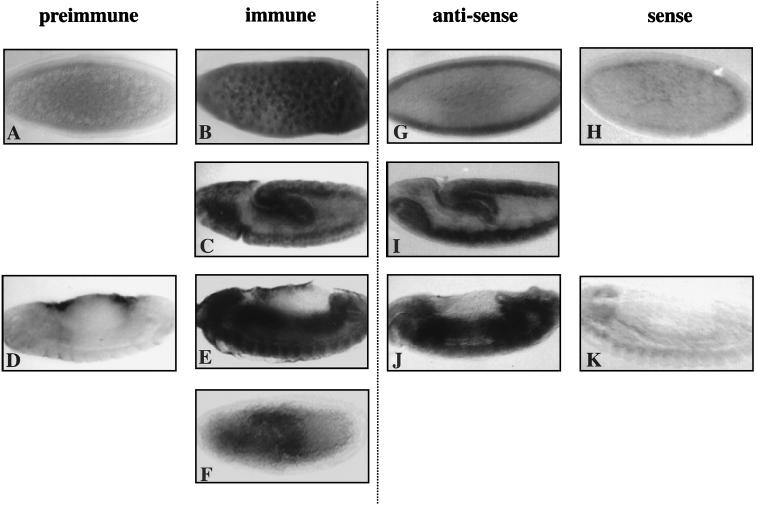
When the affinity-purified anti-MOR antibodies were used to examine the localization of MOR during embryogenesis by immunohistochemistry of whole-mount embryos, a tissue distribution exactly identical to that of mor transcripts was observed. Both the transcripts seen by in situ hybridization (Fig. 6G to K) and the protein visualized with the affinity-purified antibody (Fig. 6A to E) are ubiquitous early in development and highly expressed in the central nervous system and gut of older embryos. Using these antibodies, the subcellular localization of MOR was determined. MOR was present in all of the somatic nuclei of syncytial and cellularizing blastoderm-stage embryos (compare Fig. 6B with Fig. 6A, which was stained with affinity-purified preimmune serum) but not in the posterior pole cells (data not shown). At germ band lengthening, higher levels of MOR were seen in the developing gut (the invaginating endoderm of the posterior midgut and the stomodeum) and the ventral nerve cord (Fig. 6C). In the latter tissue, the nuclear localization of MOR was still apparent. Upon germ band retraction, MOR was preferentially enriched in the mid- and hindguts and in the ventral nerve cord (Fig. 6E; compare with control in Fig. 6D) and the brain (data not shown). At this stage, no subcellular compartmentalization could be discerned.
FIG. 6.
mor transcripts and MOR protein, ubiquitously distributed during early development, are enriched in the midgut, hindgut, and CNS of the germ-band-retracted embryo. (B, C, E, G, I, and J) In situ and immunohistochemical visualization of the distribution of mor transcripts and MOR protein at stage 4 (B and G), stage 10 (C and I), and stage 14 (E and J). (F) A Df(3R)mor7-homozygous embryo at the same stage as the embryo shown in panel C. (A, D, H, and K) Control embryos stained with affinity-purified preimmune serum (A and D) or treated with a digoxigenin-labeled sense strand probe (H and K) at stage 4 (A and H) and stage 14 (D and K). Note the nonspecific staining of the amnioserosa in panels D and E. Anterior is to the left. Staging is according to reference 26.
We examined the level of MOR immunoreactivity in embryos homozygous for Df(3R)mor7. Homozygous embryos were identified at the germ-band-extended stage, when the ftz-lacZ gene of the nonhomozygous siblings is maximally expressed and when, presumably, the effects of a possible maternal contribution are beginning to wane. In homozygous Df(3R)mor7 embryos, expression of MOR in the ventral nerve cord and brain was completely abrogated (compare Fig. 6F to the embryo of similar stage shown in Fig. 6C), further demonstrating that the anti-MOR antibodies specifically recognize the mor gene product. A small amount of residual MOR immunoreactivity may be associated with the posterior midgut and the stomodeum and may represent a persisting maternal contribution of mor.
MOR is associated with BRM in embryonic nuclear extracts.
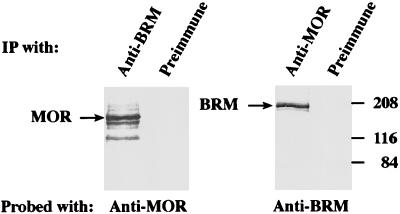
The sequence similarity of MOR to SWI3 and BAF170/155 suggests that it may function as part of an SWI-SNF complex that includes an SWI2-SNF2 homolog. To test for the association of MOR with BRM, a coimmunoprecipitation assay was used on a BRM-containing nuclear fraction obtained as described in Materials and Methods. Aliquots of this fraction were immunoprecipitated with preimmune serum or with antiserum directed against MOR or BRM. The immunoprecipitates were washed in the presence of 0.5 M KCl and detergent, resolved by SDS-PAGE, and analyzed by Western blotting with either anti-MOR or anti-BRM antiserum (Fig. 7). BRM was present in the immunoprecipitate formed with the anti-MOR antibodies, but it was not present in the preimmune-serum immunoprecipitate. Conversely, MOR was present in the immunoprecipitate formed with the anti-BRM antibodies, but it was not present in the preimmune-serum immunoprecipitate. These results indicate that the two proteins are physically complexed in embryonic nuclear extracts.
FIG. 7.
MOR coimmunoprecipitates with BRM from Drosophila embryonic nuclear extracts. A BRM-containing fraction was immunoprecipitated (IP) with either preimmune serum or polyclonal serum to MOR or BRM, as indicated above each lane. The immunoprecipitates were resolved by SDS-PAGE and analyzed by immunoblotting with either anti-MOR (left panel) or anti-BRM (right panel) antiserum. The positions of MOR, BRM, and prestained molecular mass markers (in kilodaltons) are indicated.
MOR is able to oligomerize.
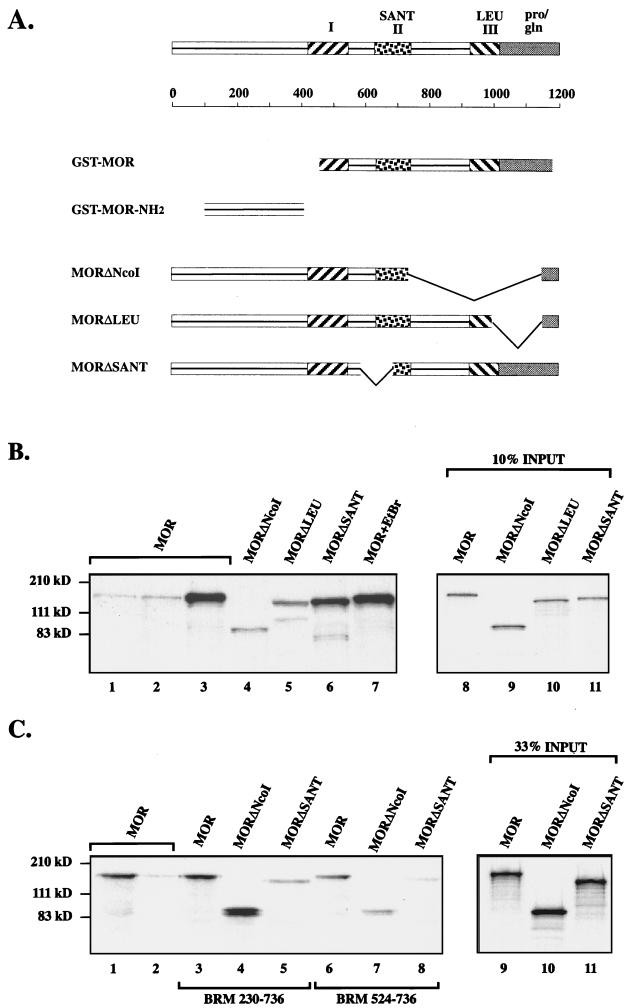
To begin to examine the protein-protein interaction capabilities of MOR, we determined whether it is able to self-associate, as might be anticipated from the presence of a candidate leucine zipper domain near its carboxyl terminus. MOR was tested by the GST fusion protein interaction assay for its ability to bind to itself. In the presence of 200 mM (data not shown) or 300 mM (Fig. 8B) NaCl, full-length labeled MOR was efficiently retained on a GST-MOR fusion protein that contained most of region I and all of regions II (the SANT domain) and III (the leucine zipper domain) (16-fold greater retention than to GST alone, as judged by PhosphorImager analysis (Fig. 8A; Fig. 8B, lane 3). In contrast, MOR bound very poorly to GST alone (Fig. 8B, lane 1) or to the GST-MOR-NH2 fusion protein that included 306 amino acids from the amino terminus of MOR (less than twofold greater retention than to GST alone) (Fig. 8A; Fig. B, lane 2). Self-association of MOR was not affected by the presence of ethidium bromide at 200 μg/ml (17-fold greater retention than in lane 1) (Fig. 8B, lane 7), indicating that DNA is not necessary for this interaction.
FIG. 8.
Protein interactions of MOR assayed by the GST fusion protein interaction assay. (A) Domains of the MOR protein, with lengths shown below in amino acids. Shown are domains included in GST-MOR constructs and the deleted derivatives of MOR that were radiolabeled and tested for their ability to bind to immobilized fusion proteins. (B) MOR-MOR interactions. Full-length MOR did not bind to GST residues alone (lane 1) or to the amino-terminal sequences of MOR in GST-MOR-NH2 (lane 2). In contrast, there was efficient retention by GST-MOR (lane 3), even in the presence of ethidium bromide (lane 7). Lanes 4 to 6 show binding of different MOR deletion derivatives to GST-MOR. Lanes 8 to 11 indicate the relative amounts of radiolabeled MOR and MOR derivatives that were incubated with the GST fusion proteins. (C) MOR-BRM interactions. Full-length MOR bound to the large BRM fusion protein (lane 3) about as well as it did to GST-MOR (lane 1). Lanes 3 to 5 compare retention of the different MOR deletion derivatives to the large BRM fusion protein, and lanes 6 to 8 compare their binding to the small BRM fusion protein. Lane 2 shows binding of full-length MOR to GST alone. Lanes 9 to 11 indicate the relative amounts of the radiolabeled proteins that were used. The positions of molecular mass markers are shown at the left (in kilodaltons).
The interaction between molecules of MOR was verified independently in the yeast two-hybrid assay. A fragment of MOR identical to that present in GST-MOR was fused both to the LexA DNA-binding domain of the LEXA202 vector (40) and to the activation domain of the pJG4-5 plasmid (23). Simultaneous transformation of S. cerevisiae by both plasmids activated the lacZ of a reporter gene containing LexA binding sites, as evidenced by the blue color of colonies on X-Gal plates, while expression of either fusion protein alone did not induce blue-colony formation (not shown).
The above observations indicate that MOR is able to self-associate and that the amino-terminal sequences of MOR cannot mediate binding between two molecules of MOR. To further localize the sequences in MOR that are required for this interaction, we labeled deleted forms of the MOR protein (Fig. 3A and 8A): MORΔNcoI, which lacks the entire highly conserved leucine zipper domain and extensive flanking sequence; MORΔLEU, which is missing 38 of 90 amino acids in the leucine zipper motif and the proline- and glutamine-rich carboxy-terminal region; and MORΔSANT, from which half (56 amino acids) of the SANT domain, as well as 37 somewhat-less-conserved residues N terminal to it, has been deleted. The MOR derivatives were tested for retention by the immobilized GST-MOR fusion protein. Removal of half of the SANT domain in MORΔSANT did not appear to affect the ability of MOR to homooligomerize (14-fold greater retention than binding of full-length MOR to GST alone) (Fig. 8B, lane 6). (Note that slightly less radiolabeled MORΔSANT was loaded onto the GST-MOR fusion protein [Fig. 8B, lane 11], thus largely accounting for the smaller amount retained.) However, self-association was severely compromised by removal of the extended leucine zipper domain in MORΔNcoI (only fourfold greater retention in lane 4 than in lane 1). Reintroduction of part of the leucine zipper motif in the MORΔLEU construct appeared to restore some of the binding activity (sevenfold greater retention) (lane 5). These data indicate that MOR is capable of forming homooligomers and that region III, the leucine zipper motif, is likely to contribute to this interaction.
MOR binds to BRM.
MOR was tested for its ability to interact with BRM, with which it is associated in embryonic nuclear extracts. In these experiments, we focused on domain II of BRM because deletion of this region causes a decrease in the size of the BRM complex, presumably due to the loss of one or several subunits (16), and because yeast two-hybrid analysis revealed an interaction between this domain of SWI2-SNF2 and the SWI3 subunit of the yeast complex (47, 48). While domain II of BRM consists of residues 549 to 610, the two GST fusion proteins that were generated for the interaction assay were more extensive and included amino acids 230 to 736 or 524 to 736. At 200 mM NaCl, labeled full-length MOR was retained on the immobilized GST-BRM fusion protein containing residues 230 to 736 approximately as well as it was retained by immobilized GST-MOR (12- and 13-fold-greater retention than of full-length MOR to GST alone) (Fig. 8C, lanes 1 and 3). In contrast, binding of MOR to beads bearing the same amount of a GST-BRM fragment containing only amino acids 524 to 736 was much less efficient (sevenfold greater retention) (lane 6). Thus, MOR is able to interact with BRM and this association can be mediated by 507 amino acids in BRM that include domain II.
To examine the role of different protein domains of MOR in binding to BRM, we tested the ability of the MOR deletion constructs to be retained by the BRM fusion proteins. MORΔNcoI was efficiently retained by GST-BRM 230–736 (15-fold greater retention in lane 4 than in lane 2), implying that the association of MOR with itself or another leucine zipper-containing protein is not necessary for its binding to BRM. This is not the case for MORΔSANT, whose binding to an equivalent amount of the large BRM fusion protein was significantly reduced relative to the binding of full-length MOR (only fourfold-greater retention in lane 5 than in the control). While MORΔNcoI was retained, albeit poorly, by the smaller BRM fusion protein (sixfold-greater retention in lane 7 than in lane 2), the level of retention of MORΔSANT fell almost to that of the negative control (twofold-greater retention than binding of the full-length protein to beads carrying GST alone) (compare lanes 8 and 2). These results suggest that the SANT domain of MOR may play a role in the association of MOR with domain II and adjacent residues of BRM.
DISCUSSION
The fruit fly BAF170/BAF155/SWI3 homolog is a member of the trxG proteins.
We have isolated a Drosophila homolog of the yeast SWI3 gene and the human BAF170 and BAF155 genes and demonstrated that it is encoded by the previously known trxG locus mor. Its identification as mor was achieved by Southern analyses of DNA derived from flies with mutations in mor, by characterization of a genomic Swi3D fragment derived from homozygous mor6 embryos, and by rescue of the mor phenotype. The origin of mor6 by γ-ray mutagenesis (29) is consistent with our observation that it encodes an altered form of mor in which 541 bp have been deleted and a 233-bp insertion of foreign DNA has occurred, leading to loss of the leucine zipper motif as well as the carboxy-terminal proline- and glutamine-rich sequences.
Genomic DNA encoding the Swi3D gene is able to completely rescue the lethal phenotype of mor alleles 1, 2, 5, and 6. In contrast, introduction of the entire neighboring 89B-Helicase cDNA into the background of the mor6 allele in combination with mor9 is completely inefficacious in rescuing the heteroallelic lethal phenotype (55). Thus, the 5′ portion of the 89B-Helicase gene included within the P{mor+11.8} construct is not mediating the observed rescue. On the other hand, the residual phenotype observed in flies carrying the mor9 and mor10 alleles after introduction of P{mor+11.8} is likely to be due to the effect of these mutations on the 89B-Helicase gene rather than failure of the transgene to completely compensate for the mor deficiency. We interpret this to be the case because the 89B-Helicase transgene appears to be able to rescue the bristle phenotype that is associated with P{lacW}89B (55). It will be interesting to test whether the introduction of both transgenes results in the complete rescue of the mor9 mor10 phenotypes.
MOR is a component of the Drosophila counterpart of the yeast SWI-SNF complex.
We have observed that the Drosophila BAF170/BAF155/SWI3 homolog, encoded by mor, is physically associated with BRM in embryonic nuclear extracts. The presence of an identical protein, named BAP155, in a highly purified 2-MDa BRM complex isolated from Drosophila embryos was very recently reported by Papoulas et al. (36). Significantly, analysis of the eight subunits of the BRM complex revealed that MOR/BAP155 is the only component, other than BRM and the previously identified SNR1 (14), that is encoded by a trxG gene. Two additional trxG proteins, ASH1 and ASH2, were found to be present in distinct high-molecular-mass complexes.
The identification of mor as a gene encoding a component of the Drosophila BRM-containing complex provides a biochemical basis for the close functional relationship between mor and brm, both strong and well-characterized trxG member. Indeed, the phenotypes resulting from mutations in each of these two genes exhibit many similarities. For example, alleles of both mor and brm were repeatedly isolated in genetic screens for dosage-dependent modifiers of dominant mutations in Pc (29); mutations in either gene affect transcription of multiple homeotic genes and the segmentation gene, engrailed; both of them are required for oogenesis; and both may be involved in cell viability of imaginal disc cells but not abdominal histoblasts (6, 7, 16, 44). More recently, mor was the only trxG gene found to interact with a dominant-negative brm transgene (36). Further analysis will be required to clarify the relationships of the trxG genes that are not components of the BRM complex to MOR and BRM.
Comparison of mor gene product distribution with those of brm and Snr1.
The expression pattern of mor overlaps with those of brm and Snr1 but is more widespread. The presence of mor transcripts in unfertilized eggs, as determined by Northern analysis, suggests that there is a maternal contribution of the mor gene product, as has been reported for brm and Snr1 (6, 14). In addition, mor, like brm and Snr1, is most highly expressed in young embryos up to 8 h of age (14, 15). Unlike those of brm and Snr1, mor transcripts are also present in adult males. Although brm transcripts were not detected in adult males, low levels of BRM protein have been reported (16).
During early embryonic development, mor transcripts and the nucleus-localized MOR protein exhibit a ubiquitous distribution like that of BRM and SNR1 (14, 16). After germ band retraction, they are observed at high levels in the central nervous system (CNS) and in the mid- and hindgut. This contrasts with SNR1, which is almost exclusively found in the ventral nerve cord and brain at the end of embryogenesis (14), and BRM, which is ubiquitously expressed and only somewhat localized to the CNS (16). Expression of mor transcripts and protein in the endodermal primordium is consistent with the midgut abnormalities described in mor mutant embryos (7) and suggests that MOR is functional in this tissue. It will be interesting to determine whether MOR is more stable in the cytoplasm, since in Df(3R)mor7 embryos what we assume to be a small amount of residual maternally derived MOR protein appears to be preferentially retained in the midgut and stomodeum, tissues in which nuclear enrichment of MOR has not been observed.
Protein interactions of MOR.
The protein-protein interactions that stabilize the SWI-SNF complex are not well understood. Some of the direct associations that have been reported are binding of TFG3/TAF30 and SNF5 (10) and an interaction between SNF11 and SNF2 (47). Because of the presence of a putative leucine zipper motif in MOR, we examined the ability of MOR to self-associate, using a GST fusion protein interaction assay and the yeast two-hybrid system. Our data indicate that MOR is able to oligomerize in vitro and in vivo in the yeast two-hybrid system. In addition to demonstrating an in vivo interaction between two molecules of MOR, our results indicate that a MOR derivative containing most of domain I, all of domains II and III, and most of the proline- and glutamine-rich tail cannot activate transcription when artificially tethered to DNA. This could be due to the absence of some essential MOR sequences or to the inability of the fruit fly protein to nucleate assembly of the entire yeast complex. By comparison, SWI2-SNF2, SNF5, and SNF6 all function as activators when targeted to a promoter as LexA fusions (32, 33).
Neither region I nor the SANT domain has to be complete for self-binding of MOR to occur. Since oligomerization is sensitive to the extent of removal of the leucine zipper domain, our results strongly suggest that the leucine zipper motif of MOR contributes to the ability of this protein to self-associate in vitro, but they do not eliminate the possibility that the carboxy-terminal region is involved.
The demonstration that MOR is able to self-associate raises the possibility that it is present in two copies in each complex, similar to BAF170 and BAF155, which are both present in each human complex (53, 54). These results support a dimer-like model for the structure of the SWI-SNF complex, with duplication of some or all subunits. Such a model has been proposed previously because the overall molecular mass of the complex is much greater than the sum of its individual components (54). It is also possible, however, that in vivo, the leucine zipper motif of MOR is involved in an association between MOR and a different leucine zipper-containing protein. In either case, the embryonic-lethal phenotype resulting from deletion of the leucine zipper and carboxy terminus in the polypeptide encoded by the mor6 allele suggests that these domains are functionally required in vivo. Robust assembly into the BRM-containing complex via interactions involving the leucine zipper motif may affect the stability of the MOR protein, since we are unable to visualize the truncated protein whose synthesis is directed by the mor6 allele. This is similar to the reduced stability of SWI3 that was noted in the absence of SWI1 and SWI2 (38) and may also derive from the failure of SWI3 to be incorporated into a complex.
We tested whether MOR, which is complexed with BRM in embryonic nuclear extracts, is able to interact with this protein in vitro. The data indicate that such an interaction can occur and that it involves domain II of BRM, as well as adjacent residues on the amino-terminal side of this domain. The motif in MOR which may mediate the interaction with BRM is the SANT domain. The possible role of the SANT domain is suggested by the significant reduction in the binding of MOR to BRM when 56 amino acids of this domain are deleted, but a role for an additional 37 non-SANT domain residues that were also removed cannot be excluded. Unlike the related DNA-binding domain in the MYB family of proteins, the SANT domain in BAF170 was not shown to have detectable DNA-binding activity in gel shift assays (54), and its function is unknown. These findings, for the first time, ascribe a possible role for the SANT domain in protein-protein interactions.
ACKNOWLEDGMENTS
We thank Daniel Jay, in whose laboratory portions of this work were carried out; James Kennison, who provided moira stocks; Yossi Markson for limitless patience in preparing the figures; Simon Greenberg, who assisted in DNA analysis; and Benny Shilo for reading the manuscript.
M.A.C. was supported by a grant from the National Science Foundation; N.B.Z. was supported by grants from the Israel Cancer Research Fund and the Israel Academy of Science.
M.A.C. and C.M. contributed equally to this work.
REFERENCES
- 1.Aasland R, Stewart A F, Gibson T. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci. 1996;21:87–88. [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Auble D T, Hansen K E, Mueller C G F, Lane W S, Thorner J, Hahn S. MOT1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 4.Austin R J, Biggin M D. Purification of the Drosophila RNA polymerase II general transcription factors. Proc Natl Acad Sci USA. 1996;93:5788–5792. doi: 10.1073/pnas.93.12.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackman R K, Sanicola L, Raftery L A, Gillevet T, Gelbart W M. An extensive 3′ cis-regulatory region directs the imaginal disk expression of decapentaplegic, a member of the TGF-β family in Drosophila. Development. 1991;111:657–665. doi: 10.1242/dev.111.3.657. [DOI] [PubMed] [Google Scholar]
- 6.Brizuela B J, Elfring L, Billard J, Tamkun J W, Kennison J A. Genetic analysis of the brahma gene of Drosophila melanogaster and polytene chromosome subdivisions 72AB. Genetics. 1994;137:803–813. doi: 10.1093/genetics/137.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brizuela B J, Kennison J A. The Drosophila homeotic gene moira regulates expression of engrailed and HOM genes in imaginal tissues. Mech Dev. 1997;65:209–220. doi: 10.1016/s0925-4773(97)00081-6. [DOI] [PubMed] [Google Scholar]
- 8.Brown N, Kafatos F C. Functional Drosophila cDNA libraries from Drosophila embryos. J Mol Biol. 1988;203:425–437. doi: 10.1016/0022-2836(88)90010-1. [DOI] [PubMed] [Google Scholar]
- 9.Burns L G, Peterson C L. Protein complexes for remodeling chromatin. Biochim Biophys Acta. 1997;1350:159–168. doi: 10.1016/s0167-4781(96)00162-5. [DOI] [PubMed] [Google Scholar]
- 10.Cairns B R, Henry N L, Kornberg R D. TFG3/TAF30/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemogenic proteins ENL and AF-9. Mol Cell Biol. 1996;16:3308–3316. doi: 10.1128/mcb.16.7.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chicca J J, II, Auble D T, Pugh B F. Cloning and biochemical characterization of TAF-170, a human homolog of yeast Mot1. Mol Cell Biol. 1998;18:1701–1710. doi: 10.1128/mcb.18.3.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crosby M A, Meyerowitz E M. Drosophila glue gene Sgs-3: sequences required for puffing and transcriptional regulation. Dev Biol. 1986;118:593–607. doi: 10.1016/0012-1606(86)90029-1. [DOI] [PubMed] [Google Scholar]
- 13.Davis J L, Kunisawa R, Thorner J. A presumptive helicase (MOT1 gene product) affects gene expression and is required for viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1879–1892. doi: 10.1128/mcb.12.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dingwall A K, Beek S J, McCallum C M, Tamkun J W, Kalpana G V, Goff S P, Scott M P. The Drosophila SNR1 and BRM proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol Biol Cell. 1995;6:777–791. doi: 10.1091/mbc.6.7.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elfring L K, Deuring R, McCallum C M, Peterson C L, Tamkun J W. Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol Cell Biol. 1994;14:2225–2234. doi: 10.1128/mcb.14.4.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elfring L K, Daniel C, Papoulas O, Deuring R, Sarte M, Moseley S, Beek S J, Waldrip W R, Daubresse G, DePace A, Kennison J A, Tamkun J W. Genetic analysis of brahma: the Drosophila homolog of the yeast chromatin remodeling factor SW12/SNF2. Genetics. 1998;148:251–265. doi: 10.1093/genetics/148.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng D F, Doolittle R F. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J Mol Evol. 1987;25:351–360. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- 18.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 19.Finley R L, Jr, Brent R. Interaction trap cloning with yeast. In: Hames D, Glover D, editors. Gene probes: a practical approach. Oxford, United Kingdom: Oxford University Press; 1995. pp. 169–203. [Google Scholar]
- 20.FlyBase. FlyBase—a Drosophila database. Nucleic Acids Res. 1998;26:85–88. doi: 10.1093/nar/26.1.85. http://flybase.bio.indiana.edu/ . [Online.] http://flybase.bio.indiana.edu/. [Updated November 1998. Last date accessed, 13 December 1998.] . [Updated November 1998. Last date accessed, 13 December 1998.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garazzo M, Christenson A C. Technical note: preparation of DNA from single embryos for PCR. Drosophila Inf Serv. 1994;75:204–205. [Google Scholar]
- 22.Goldman-Levi R, Miller C, Bogoch J, Zak N B. Expanding the MOT1 subfamily: 89B-Helicase encodes a new Drosophila melanogaster SNF-2 related protein which binds to multiple sites on polytene chromosomes. Nucleic Acids Res. 1996;24:3121–3128. doi: 10.1093/nar/24.16.3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 24.Hamilton B A, Palazzolo M J, Chang J H, Vijay Raghavan K, Mayeda C A, Whitney M A, Meyerowitz E M. Large scale screen for transposon insertions into cloned genes. Proc Natl Acad Sci USA. 1991;88:2731–2735. doi: 10.1073/pnas.88.7.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 26.Hartenstein V. Atlas of Drosophila development. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- 27.Hovemann B T. Construction of a random primed embryonic Drosophila cDNA expression library cloned into phage λ-gt11. Drosophila Inf Serv. 1991;70:250. [Google Scholar]
- 28.Jeon S H, Kang M G, Kim Y H, Jin Y H, Lee C, Chung H-Y, Kwon H, Park S D, Seong R H. A new mouse gene, SRG3, related to SWI3 of Saccharomyces cerevisiae, is required for apoptosis induced by glucocorticoids in a thymoma cell line. J Exp Med. 1997;185:1827–1836. doi: 10.1084/jem.185.10.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kennison J A, Tamkun J W. Dosage-dependent modifiers of Polycomb and Antennapedia mutations in Drosophila. Proc Natl Acad Sci USA. 1988;85:8136–8140. doi: 10.1073/pnas.85.21.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kennison J A. The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu Rev Genet. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- 31.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 32.Laurent B C, Treitel M A, Carlson M. Functional interdependence of the yeast SNF2, SNF5 and SNF6 proteins in transcriptional activation. Proc Natl Acad Sci USA. 1991;88:2687–2691. doi: 10.1073/pnas.88.7.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laurent B C, Treich I, Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- 34.Meyerowitz E M, Martin C H. Adjacent chromosomal regions can evolve at very different rates: evolution of the Drosophila 68C glue gene cluster. J Mol Evol. 1984;20:251–264. doi: 10.1007/BF02104731. [DOI] [PubMed] [Google Scholar]
- 35.Needleman S B, Wunsch C D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 36.Papoulas O, Beek S J, Moseley S L, McCallum C M, Sarte M, Shearn A, Tamkun J W. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development. 1998;125:3955–3966. doi: 10.1242/dev.125.20.3955. [DOI] [PubMed] [Google Scholar]
- 37.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peterson C L, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 39.Poon D, Campbell A M, Bai Y, Weil P A. Yeast Taf170 is encoded by MOT1 and exists in a TATA box-binding protein (TBP)–TBP-associated factor complex distinct from transcription factor IID. J Biol Chem. 1994;269:23135–23140. [PubMed] [Google Scholar]
- 40.Ruden D M, Ma J, Li Y, Wood K, Ptashne M. Generating yeast transcriptional activators containing no yeast protein sequences. Nature. 1991;350:250–252. doi: 10.1038/350250a0. [DOI] [PubMed] [Google Scholar]
- 41.Shearn A. The ash-1, ash-2 and trithorax genes of Drosophila melanogaster are functionally related. Genetics. 1989;121:517–525. doi: 10.1093/genetics/121.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon J. Locking in stable states of gene expression: transcriptional control during Drosophila development. Curr Opin Cell Biol. 1995;7:376–385. doi: 10.1016/0955-0674(95)80093-x. [DOI] [PubMed] [Google Scholar]
- 43.Spradling A, Rubin G M. Transposition of cloned P element into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 44.Tamkun J W, Deuring R, Scott M P, Kissinger M, Pattatucci A M, Kaufman T C, Kennison J A. Brahma—a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 45.Tamkun J W. The role of Brahma and related proteins in transcription and development. Curr Opin Genet Dev. 1995;5:473–477. doi: 10.1016/0959-437x(95)90051-h. [DOI] [PubMed] [Google Scholar]
- 46.Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 47.Treich I, Cairns B R, De Los Santos T, Brewster E, Carlson M. SNF11, a new component of the yeast SNF-SWI complex that interacts with a conserved region of SNF2. Mol Cell Biol. 1995;15:4240–4248. doi: 10.1128/mcb.15.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Treich I, Carlson M. Interaction of a Swi3 homolog with Sth1 provides evidence for a Swi/Snf-related complex with an essential function in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:1768–1775. doi: 10.1128/mcb.17.4.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tripoulas N A, Hersperger E, La Jeunesse D, Shearn A. Molecular genetic analysis of the Drosophila melanogaster gene absent, small or homeotic discs1 (ash1) Genetics. 1994;137:1027–1038. doi: 10.1093/genetics/137.4.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsukiyama T, Wu C. Chromatin remodeling and transcription. Curr Opin Genet Dev. 1997;7:182–191. doi: 10.1016/s0959-437x(97)80127-x. [DOI] [PubMed] [Google Scholar]
- 51.van der Knaap J A, Borst J W, van der Vliet P C, Gentz R, Timmers H T M. Cloning of the cDNA for the TATA-binding protein-associated factor II170 subunit of transcription factor B-TFIID reveals homology to global transcription regulators in yeast and Drosophila. Proc Natl Acad Sci USA. 1997;94:11827–11832. doi: 10.1073/pnas.94.22.11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wampler S L, Tyree C M, Kadonaga J T. Fractionation of the general RNA polymerase II transcription factors from Drosophila embryos. J Biol Chem. 1990;265:21223–21231. [PubMed] [Google Scholar]
- 53.Wang W, Cote J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, Workman J L, Crabtree G R. Purification and biochemical heterogeneity of the mammalian SW1-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 54.Wang W, Xue Y, Zhou S, Kuo A, Cairns B R, Crabtree G R. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 55.Watson, K. L., M. A. Crosby, and N. B. Zak. Unpublished data.
- 56.Wilson C, Pearson R K, Bellen H J, O’Kane C J, Grossniklaus U, Gehring W J. P-element-mediated enhancer detection: an efficient method for isolating and characterizing developmentally regulated genes in Drosophila. Genes Dev. 1989;3:1301–1313. doi: 10.1101/gad.3.9.1301. [DOI] [PubMed] [Google Scholar]