Summary
The escalating global prevalence of depression demands effective therapeutic strategies, with psychobiotics emerging as a promising solution. However, the molecular mechanisms governing the neurobehavioral impact of psychobiotics remain elusive. This study reveals a significant reduction in hippocampal indole-3-lactic acid (ILA) levels in depressed mice, which is ameliorated by the psychobiotic Bifidobacterium breve. In both human subjects and mice, the ILA increase in the circulatory system results from bifidobacteria supplementation. Further investigation identifies the key aromatic lactate dehydrogenase (Aldh) gene and pathway in bifidobacteria responsible for ILA production. Importantly, the antidepressant effects are nullified in the Aldh mutants compared to the wild-type strain. At the bifidobacteria species level, those with Aldh exhibit heightened antidepressant effects. Finally, this study emphasizes the antidepressant efficacy of psychobiotic-derived ILA, potentially mediated by aryl hydrocarbon receptor (AhR) signaling activation to alleviate neuroinflammation. This study unveils the molecular and genetic foundations of psychobiotics’ antidepressant effects, offering insights for microbial therapies targeting mood disorders.
Keywords: psychobiotics, indole-3-lactic acid, microbiota-gut-brain axis, depression, neuroinflammation
Graphical abstract
Highlights
-
•
ILA-producing capacity distinguishes bifidobacteria with antidepressant effects
-
•
Aldh is essential for bifidobacteria’s ILA synthesis and antidepressant efficacy
-
•
ILA’s antidepressant effects linked to AhR-mediated neuroinflammation relief
Qian et al. reveal that psychobiotic Bifidobacterium breve reverses the abnormal reduction of hippocampal indole-3-lactic acid (ILA) in depressed mice. The study highlights the role of Aldh gene in ILA production by bifidobacteria and its antidepressant effects through AhR signaling, advancing the understanding of psychobiotics in mood disorder therapies.
Introduction
Over the past few decades, the prevalence of depression has steadily risen, surpassing cardiovascular disease and cancer, making it the foremost global cause of disability.1,2 Social and psychological stressors play pivotal roles in contributing to depression, a significance accentuated during the COVID-19 pandemic, which have resulted in an upsurge in depression rates, attributed to stress from unemployment, prolonged sedentary time, and social isolation.3,4 Post-COVID-19 depression is also considered to be connected to peripheral immune inflammation triggered by viral infection, gliosis, and neuroinflammation.5 However, due to the complexity and heterogeneity of depression’s pathogenesis,6 conventional antidepressants and psychotherapy currently used in clinical practice demonstrate only limited efficacy. Evidence suggests that over one-third of patients show an insufficient response to first-line treatments.7 Classical antidepressants, rooted in the “monoamine deficiency hypothesis,” inadequately account for delayed efficacy and varied treatment responses.7,8 The deduction from the serendipitous discovery of monoamine oxidase inhibitors’ efficacy to the postulation that serotonin (5-hydroxytryptamine, 5-HT) deficiency causes depressive symptoms remains highly contentious.9,10,11 Research has established that altering brain 5-HT levels, either by reducing them in healthy volunteers or increasing them in depressed patients, is insufficient to induce or alleviate clinical depressive symptoms.10,12 Therefore, there is a pressing need to develop new antidepressant strategies that address the multifaceted nature of depression’s pathogenesis.
In recent years, attention has shifted toward the bidirectional signaling between the gut microbiota and central nervous systems, known as the microbiota-gut-brain axis.13 Some studies not only associate changes in the gut microbiota with depression but also establish causal relationships, indicating a direct impact of microbiota alterations on depressive conditions.14,15 For instance, two individuals with severe/moderate depression experienced a notable reduction in symptom severity, transitioning to a state of mild depression following a 4-week regimen of fecal microbiota transplantation (FMT) as an adjunctive therapeutic intervention; this sustained efficacy was observed in one patient for an extended period of up to 8 weeks.16 Similar effects were also observed in patients with irritable bowel syndrome, functional diarrhea, or functional constipation who underwent FMT.17 Despite the standardized management of donor screening for FMT and potential safety risks limiting the widespread application of FMT in depression, these findings underscore the intricate interplay between the gut microbiome composition and mental health outcomes.18
Alternatively, probiotics emerge as a safer and more widely applicable strategy.19,20 Coined as “psychobiotics” in 2013, these live organisms, when ingested in adequate amounts, produce health benefits in patients with psychiatric illnesses.21 Over the past decade, psychobiotics have gained academic recognition for their proven antidepressant efficacy in both animal and clinical trials.22,23,24,25 This alternative is particularly valuable for patients intolerant or unresponsive to traditional medications.25 Psychobiotics can also complement conventional drug therapies, potentially enhancing overall efficacy.20,26 Mechanisms underlying psychobiotics’ mood improvement involved increased brain-derived neurotrophic factor/serotonin levels, reduced microbiota-mediated inflammation, and vagus nerve activation.27,28,29 However, understanding the pharmacological mechanisms of psychobiotics remains challenging compared to traditional antidepressants, given the diverse strain-specific effects and limited knowledge about their common microbiological and molecular genetic features.30,31
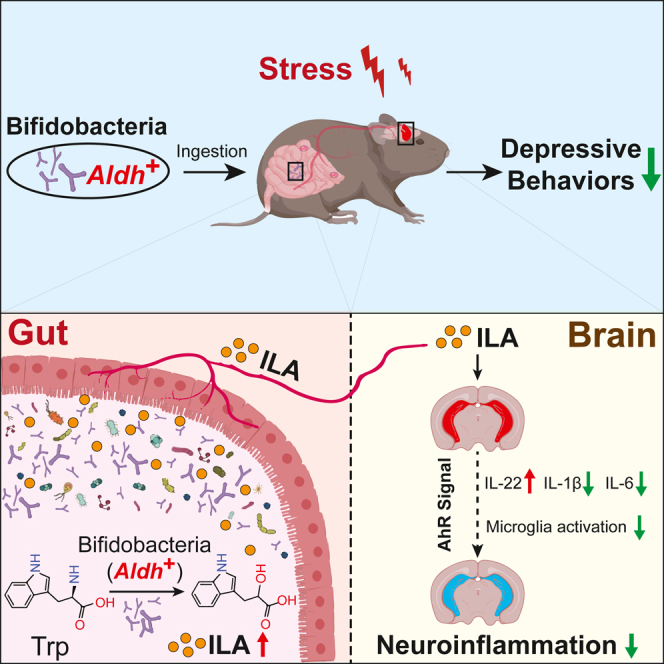
In this study, we observed a significant decrease in indole-3-lactic acid (ILA) levels in the gut and brain of chronically stressed depressive mice, while supplementation with the psychobiotic Bifidobacterium breve CCFM1025 (Bre1025) can restore these levels back to normal. To underline the critical role of ILA production for the antidepressant efficacy of Bre1025, we conducted experiments involving the insertion mutant of the aromatic lactate dehydrogenase (Aldh) gene responsible for ILA synthesis. Additionally, we investigated the psychobiotic potential of various bifidobacteria species; we confirmed that those harboring the Aldh gene exhibit psychobiotic potential. Furthermore, we suggest that antidepressant efficacy achieved by bifidobacteria-derived ILA may be attributed to the activation of the aryl hydrocarbon receptor (AhR) signaling pathway, leading to the alleviation of neuroinflammation. These findings offer insights into the therapeutic potential of psychobiotics in stress-related mood disorders.
Results
The antidepressant effects of psychobiotic Bifidobacterium breve linked to the regulation of gut ILA
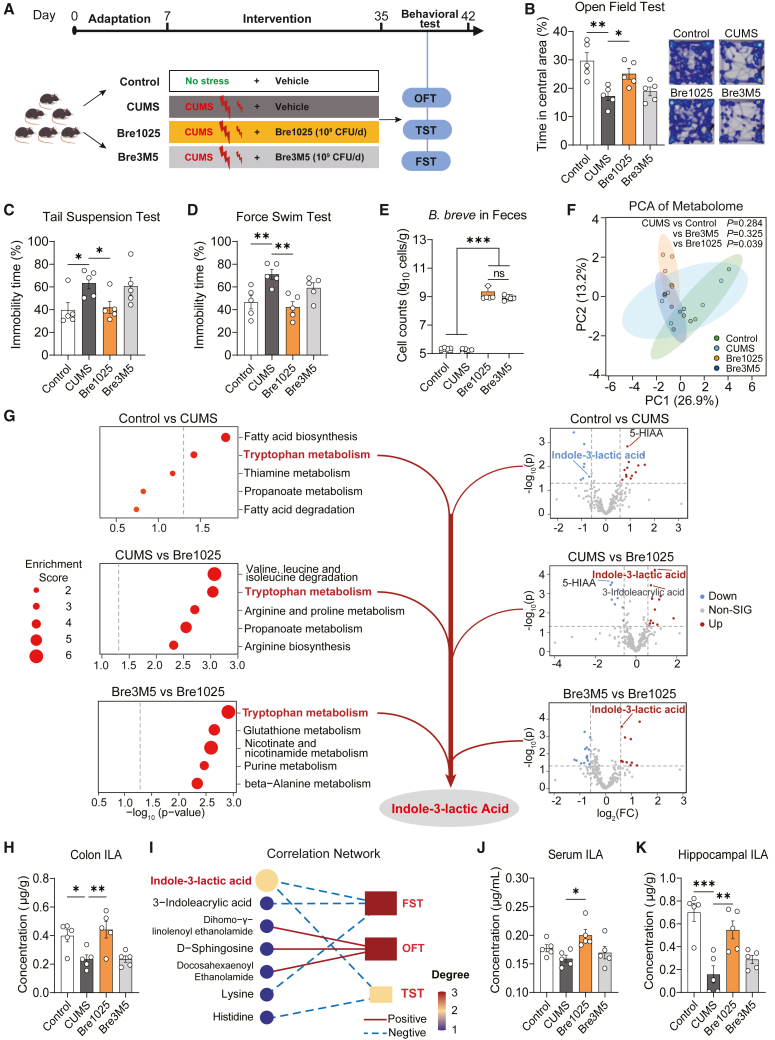
Initially, we noted significant strain-level variations in the emotional regulation capabilities of Bifidobacterium breve. Specifically, strain Bre1025 demonstrated the ability to alleviate anxiety-like and depressive-like behaviors induced by chronic unpredictable mild stress (CUMS), in contrast to the control strain, Bre3M5 (Figures 1A–1D). Notably, despite comparable colonization levels in the mouse gut (Figure 1E), Bre1025 intervention significantly altered the composition of gut microbiota metabolites (Figure 1F). Compared with the CUMS group (p < 0.05), Bre1025 intervention increased the content of ILA, indole-3-acrylic acid, and genistein, while resulting in a decrease in the content of metabolites such as 5-hydroxyindole-3-acetic acid, ornithine, and acrylic acid (Data S1). Based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathway enrichment analysis, we found that Bre1025, compared to the control strain, reversed the abnormal decrease in the metabolite ILA in the Tryptophan metabolism pathway induced by CUMS (Figure 1G). This change was further validated through targeted metabolomic detection of indole derivatives (Figure 1H). Meanwhile, we confirmed that Bre1025 exhibited significantly higher ILA production in vitro than the control strain Bre3M5 (Figure S1A). Subsequent debiased sparse partial correlation analysis highlighted ILA as the pivotal gut metabolite, demonstrating a significant negative correlation with the immobility time in the forced swimming test (FST) and tail suspension test (TST) (Figure 1I). Importantly, Bre1025 counteracted the abnormal decrease of ILA levels in the serum and hippocampus induced by CUMS (Figures 1J and 1K). Bre1025 intervention also elevated the level of the downstream indole derivative indole-3-propionic acid (IPA) production (Figure S1B), with no significant impact on other indole derivatives.
Figure 1.
Indole-3-lactic acid associated with Bre1025-mediated antidepressant effect
(A) Schematic representation of the animal experimental design. In this animal experiment, 20 mice were randomly divided into four groups (n = 5/treatment): control (blank control group), CUMS (CUMS model group), Bre1025 (B. breve Bre1025 intervention group, 1 × 10∧9 CFU/d), and Bre3M5 (B. breve Bre3M5 intervention group, 1 × 10∧9 CFU/d); OFT, open field test; TST, tail suspension test; FST, force swim test.
(B–D) Behavioral outcomes of mice assessed in different tests.
(E) Quantification of B. breve cell counts in mouse feces.
(F) Principal component analysis illustrating the metabolome of mouse gut contents.
(G) Analysis of differential metabolites in mouse gut. KEGG pathway enrichment analysis of gut metabolites are shown on the left (p < 0.05), and volcano plots of differential metabolites are shown on the right (p < 0.05 and fold change ≥1.5). 5-HIAA, 5-hydroxyindole acetic acid.
(H) Targeted and quantitative detection of indole-3-lactic acid (ILA) in mouse gut contents.
(I) Debiased sparse partial correlation analysis of gut metabolites and gut microbiota.
(J and K) Targeted quantification of ILA in mouse serum and hippocampus.
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, determined by one-way analysis of variance (ANOVA) followed by Sidak post hoc test in (B–E), (H), (J), and (K).
The potential antidepressant effects of gut microbiota-derived ILA are also supported by clinical and population studies. Previous research has found that patients with major depressive disorder (MDD) have significantly lower serum levels of indole derivatives, including indole-3-acetamide, ILA, and indole-3-aldehyde (IAld), compared to healthy controls.32 Here, we further conducted a validation experiment of serum and fecal indole derivatives in 40 subjects (patients with depression: healthy individuals = 1:1). The results revealed that patients with depression had significantly lower serum levels of ILA, indole-3-acetic acid, and IAld compared to healthy individuals, while only fecal ILA levels were significantly reduced (Figures S1C and S1D). Additionally, the results from the aforementioned animal experiments are consistent with previous data from our clinical intervention study with depressed patients using this strain, specifically showing that Bre1025 can increase ILA levels in the gut of patients with depression.20 Based on all the evidence presented, we hypothesize that ILA plays a crucial role in mediating the emotional regulation facilitated by Bre1025.
Elevated gut and serum ILA levels were attributed to the supplementation of Bre1025
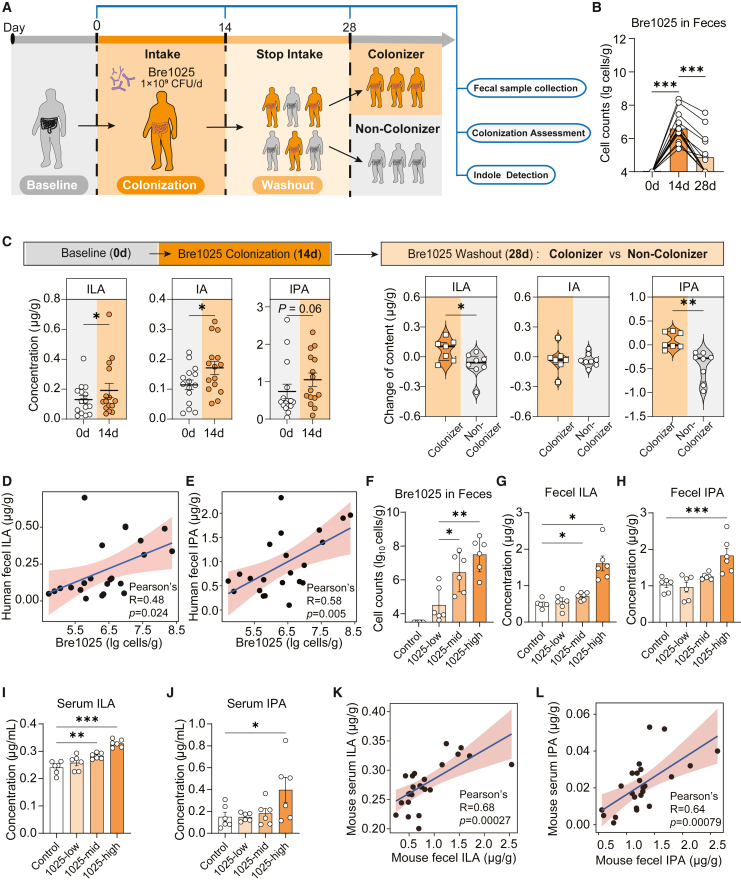
To determine whether the gut ILA content is contributed by the intake of Bre1025 rather than the indigenous microbiota, we designed strain-specific primers for Bre1025 to investigate its colonization level and assess its contribution to the gut ILA level. Healthy volunteers, upon continuous intake of Bre1025, exhibited a rapid accumulation in the gut (Figures 2A and 2B), leading to an elevation in gut ILA, as well as downstream indole acrylic acid (IA) and IPA levels (Figure 2C). In contrast, the levels of other indole derivatives from the gut microbiota significantly decreased (Figure S2A). Notably, even after discontinuation of intake for 14 days, Bre1025 was still detectable in the feces of nearly half of the volunteers (7/15) (Figure 2B), here termed as “colonizers.” Colonizers maintained stable ILA and downstream indole derivative levels after discontinuation, whereas in individuals where Bre1025 was washed out of the gut, ILA levels significantly decreased (Figures 2C and S2B). Furthermore, in individuals with the presence of Bre1025, their gut ILA and downstream IPA levels were positively correlated with the abundance of Bre1025 (Figures 2D and 2E). We subsequently analyzed the host gut microbiota structure before and after Bre1025 intake (Figures S1E–S1G). The results showed no significant changes in the β-diversity and α-diversity of the gut microbiota of healthy volunteers after 2 weeks of Bre1025 intake. However, Bre1025 intake promoted the abundance of the genera Lachnospira and Allobaculum (Figure S1H), which have been reported to be involved in the conversion of ILA to the downstream indole derivative IPA.33,34 This increase in these genera potentially explains the elevated IPA levels observed. In the mouse model, we further demonstrated that the ILA present in serum is primarily contributed by ILA produced from gut microbes. We established mice with varying doses of Bre1025 in the gut (Figure 2F), revealing a significant microbial dose-dependent ILA/IPA level trend in both the gut and serum (Figures 2G–2J). Especially noteworthy is the positive correlation between fecal and serum ILA/IPA levels observed in these mice (Figures 2K–2L). Additionally, Bre1025 ingestion did not exert significant effects on other tryptophan metabolites in either the gut or blood of mice (Figures S2C–S2J).
Figure 2.
Ingestion of Bre1025 improved host gut and serum ILA levels
(A) Schematic representation of the clinical trial experimental strategy. In this clinical trial, 15 healthy subjects were recruited for testing the colonization amount and ILA production capability of Bre1025.
(B) Quantification of Bre1025 cell counts in human feces.
(C) Content changes in indole derivatives in human feces.
(D and E) Correlation between Bre1025 biomass and ILA/IPA content in human feces.
(F) Quantification of Bre1025 cell counts in mouse feces. In this animal experiment, 24 mice were randomly divided into four groups for the quantitative analysis of intestinal and serum indole derivatives (n = 6/treatment): control (blank control group), 1025-low (low-dose Bre1025 intervention group, 1 × 10∧7 CFU/d), 1025-med (medium-dose Bre1025 intervention group, 1 × 10∧8 CFU/d), and 1025-high (high-dose Bre1025 intervention group, 1 × 10∧9 CFU/d).
(G and H) Quantification of ILA/IPA concentrations in mouse feces.
(I and J) Quantification of ILA/IPA concentrations in mouse serum.
(K) Correlation between fecal ILA content and serum ILA content in mouse feces.
(L) Correlation between fecal IPA content and serum IPA content in mouse feces.
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, determined by two-tailed paired Student’s t test in (B) and (C) and one-way ANOVA followed by Sidak post hoc test in (F–J).
Aldh is the key gene for tryptophan-dependent ILA production in Bre1025
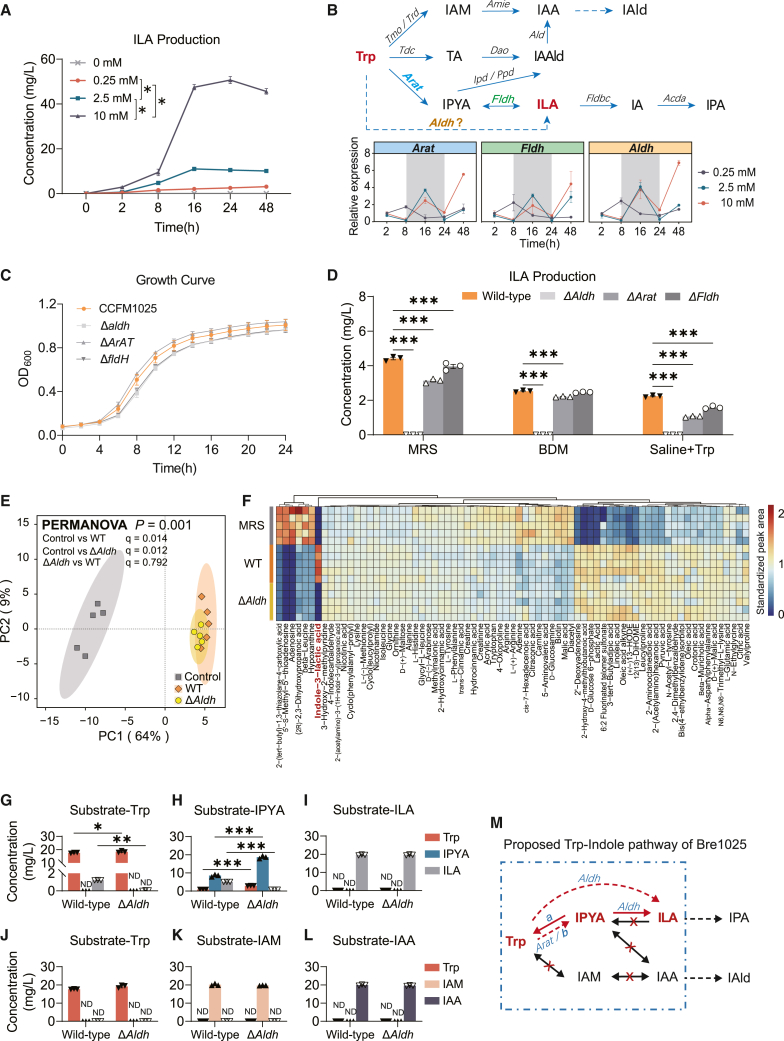
Microbial indole and its derivatives are directly or stepwise biotransformed from tryptophan (Trp).35 We assessed the ILA production ability of Bre1025 in a Bifidobacterium-defined medium (BDM, a fully defined medium) supplemented with varying concentrations of Trp. The substrate quantity did not affect the biomass accumulation of Bre1025 (Figure S3A), but it did impact ILA production, with ILA levels increasing with higher Trp concentrations (Figure 3A).
Figure 3.
Bre1025 metabolizes tryptophan to ILA through the Aldh gene
(A) ILA production by Bre1025 using tryptophan (Trp) as a substrate in a Bifidobacterium-defined medium (BDM) (n = 3/treatment).
(B) Expression of genes related to ILA production in Bre1025 with different amounts of Trp substrate. The upper pathway diagram depicts a schematic representation of how microorganisms metabolize Trp into indole derivatives. The bottom line chart displays the expression of genes in Bre1025 that are potentially involved in the microbial metabolism of Trp into ILA. IAM, indole-3-acetamide; IAA, indole-3-acetic acid; IAld, indole-3-aldehyde; TA, tryptamine; IAAld, indole-3-acetaldehyde; IPYA, indole-3-pyruvate; IA, indole acrylic acid; IPA, indole-3-propionic acid.
(C) In vitro growth curve of the Bre1025 mutant strain (n = 3/treatment). Wild-type: Bre1025 wild-type strain; Δtarget-gene: mutant strain of the Bre1025 target gene; ND, not detected.
(D) In vitro ILA production of the Bre1025 mutant strain.
(E) Principal component analysis illustrating the metabolome of the Bre1025 wild-type and mutant strains (n = 5/treatment). Control: mMRS medium supernatant.
(F) Metabolite heatmaps of Bre1025 wild-type and mutant strains (n = 3/treatment).
(G–L) Metabolism of Bre1025 in different substrates (n = 3/treatment).
(M) Proposed Trp-indole metabolism pathway of Bre1025.
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, determined by two-tailed Student’s t test in (E–J) and one-way ANOVA followed by Sidak post hoc test in (A) and (D).
Next, we examined whether genes previously reported in microorganisms to be involved in the conversion of Trp to ILA are present in the genome of Bre1025, including aromatic amino acid aminotransferase (Arat),36 phenyllactate dehydrogenase (Fldh),37 and Aldh38 (Figure 3B). However, the expression patterns of these three genes in response to changes in Trp substrate concentration were nearly identical (Figure 3B), making it challenging to distinguish their contributions to Bre1025’s ILA production. Therefore, we individually constructed insertion mutants for these three genes (Figure S3B). While the growth of the mutants in vitro was unaffected (Figure 3C), there was a significant reduction in the extracellular ILA production of Bre1025 to varying degrees (Figure 3D). Notably, only the Aldh mutant completely abolished Bre1025’s ILA production, regardless of whether it was in mMRS medium, BDM medium, or physiological saline supplemented with the substrate Trp (Figure 3D). Following that, we analyzed the metabolic composition of the Bre1025 wild-type strain and the Aldh gene mutant strain in the classic mMRS medium. The results showed no differences in metabolites between the two strains, except for ILA (Figures 3E and 3F).
Subsequently, we conducted an analysis of the Trp metabolic pathway based on both the wild-type and Aldh mutant strains. When Trp served as the substrate, the mutant failed to produce ILA, and neither the wild type nor the mutant produced indole-3-pyruvate (IPYA) (Figure 3G). This suggests that Bre1025 may either lack the enzyme necessary for converting Trp to IPYA, or the enzyme may be inactive under nutrient-deficient conditions. Additionally, when IPYA was used as the substrate, both the wild type and the mutant produced Trp, but with a significant difference in production ratios (Figure 3H). This indicates that there is an enzyme-a converting IPYA to Trp, and that Aldh is responsible for converting IPYA to ILA. Meanwhile, neither strain demonstrated the ability to reverse ILA into IPYA or Trp, nor to further convert ILA into downstream indole metabolites (Figure 3I). Furthermore, Bre1025 did not participate in another Trp-indole pathway widely reported in the intestinal microbiota (Figures 3J–3M). Based on these results, we speculate that Bre1025 has two pathways for converting Trp to ILA: one where Arat converts Trp to IPYA, followed by Aldh converting IPYA to ILA,35 and another potential pathway bypassing Arat (or another enzyme-b substituting Arat), where Aldh directly converts Trp to ILA, and lacks the ability to convert Trp and ILA into other indole metabolites (Figure 3M).
ILA production capacity is essential in enabling the antidepressant effect of Bre1025
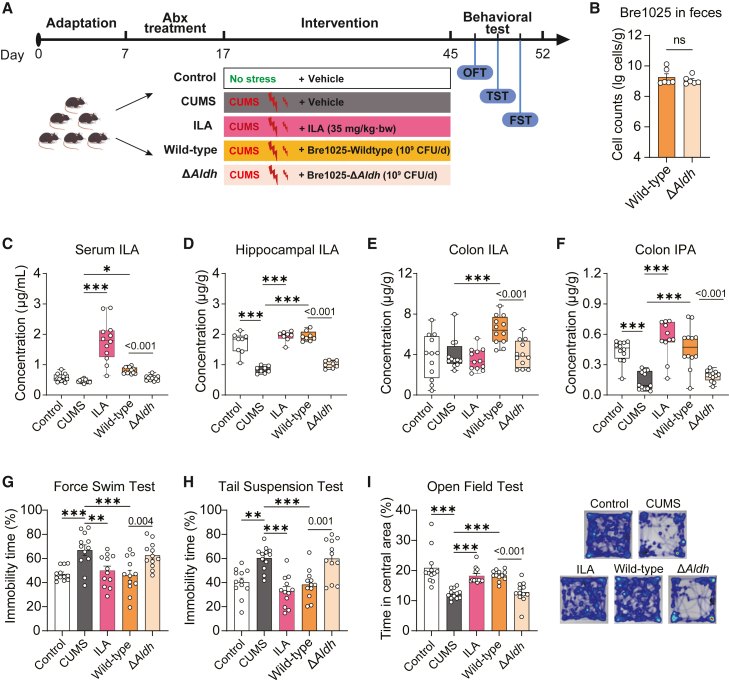
To determine the role of ILA production in Bre1025’s emotional regulation capacity, we investigated potential variations in ILA metabolism and behavioral outcomes in CUMS mice subjected to treatment with the wild-type and the Aldh mutant strain (Figure 4A). Before intervention, mice were pre-treated with antibiotics to remove the effects of ILA produced by the native gut microbiota. The findings revealed that the absence of Aldh function did not influence the accumulation of Bre1025 in the mouse intestine (Figure 4B). Nevertheless, in line with the oral intake of ILA (35 mg/kg⋅bw/day), the administration of Bre1025 counteracted the CUMS-induced reduction in serum and brain (hippocampus or prefrontal cortex) ILA levels (Figures 4C, 4D, and S4A). Notably, the loss of Aldh gene hindered the strain’s ability to regulate ILA content in the gut-blood-brain axis of mice.
Figure 4.
The production of ILA determines the antidepressant capability of Bre1025
(A) Schematic representation of the animal experimental design. In this animal experiment, 60 mice were randomly divided into five groups to evaluate the mood-regulating function of wild-type and genetically modified Bre1025 strains (n = 12/treatment): control (blank control group), CUMS (CUMS model group), ILA (oral ILA intervention group, 35 mg/kg/d), wild-type (Bre1025 intervention group, 1 × 10∧9 CFU/d), and ΔAldh (Bre1025-ΔAldh intervention group, 1 × 10∧9 CFU/d).
(B) Quantification of Bre1025 cell counts in mouse feces.
(C–E) ILA concentrations in mouse serum, hippocampus, and colon content.
(F) IPA concentration in mouse colon content.
(G–I) Behavioral outcomes of mice assessed in different tests.
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, determined by two-tailed Student’s t test in (B) and one-way ANOVA followed by Sidak post hoc test in (C–I).
It is noteworthy that orally administered ILA did not augment colonic ILA levels (Figure 4E); rather, it may be absorbed into the bloodstream before reaching the colon. Furthermore, both oral ILA and Bre1025 influenced the levels of IPA and other indole derivatives in the gut and serum (Figures 4F and S4B–S4J). Concurrently, both ILA and Bre1025 interventions significantly reduced immobility time in the FST and TST in mice, while increasing exploration time in the center zone of the open field test (Figures 4G–4I). Conversely, the Aldh mutant strain exhibited no such effects. These results underscore the essential role of ILA production in enabling Bre1025 to modulate host ILA metabolism and enhance emotional well-being.
ILA-mediated antidepressant effect extends across diverse Bifidobacterium species
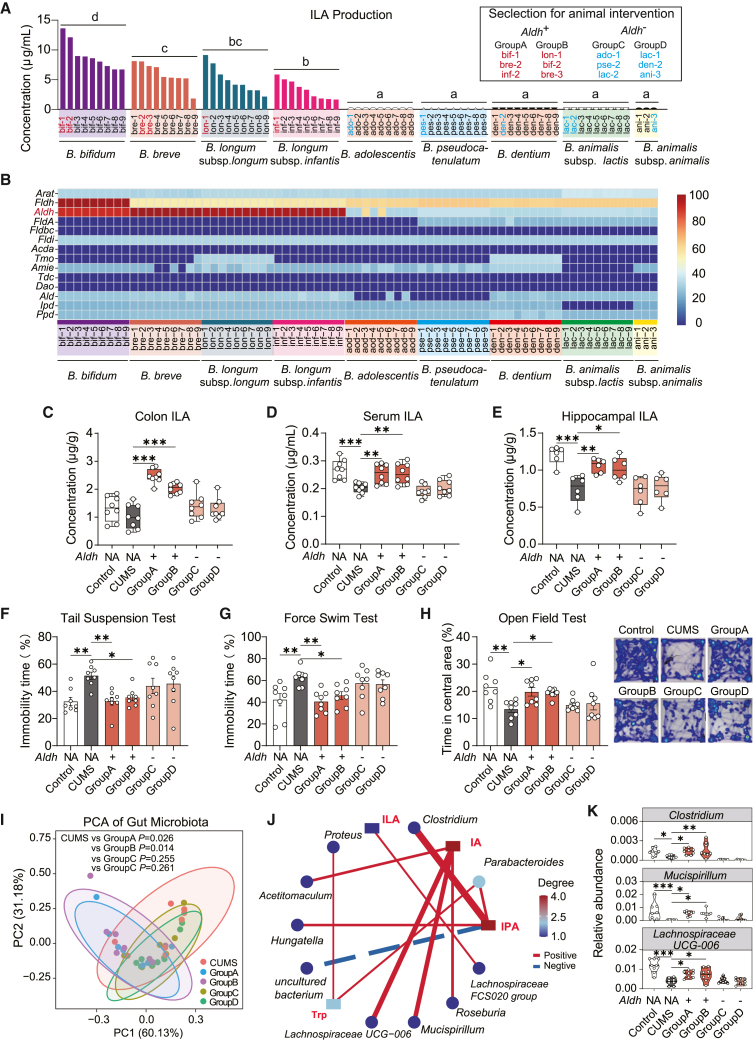
Considering that the mechanisms mentioned earlier were initially observed in only two bacterial strains, our subsequent aim was to explore their universality at the Bifidobacterium species level. Initially, we assessed the in vitro ILA production capabilities of nine different Bifidobacterium species, totaling 75 strains (using BDM). Among them, strains belonging to Bifidobacterium bifidum, B. breve, B. longum subsp. longum, and B. longum subsp. infantis exhibited ILA production, while strains from B. adolescentis, B. pseudocatenulatum, B. dentium, B. animalis subsp. lactis, and B. animalis subsp. animalis did not (Figure 5A). There were no significant differences in Trp consumption among the various Bifidobacterium species (Figure S5A). Following this, we scrutinized the genomic features related to Trp metabolism in these strains and identified that strains capable of ILA synthesis all harbor the Aldh gene. The presence of the Fldh gene may contribute to higher ILA synthesis in B. bifidum compared to other strains (Figure 5B), but further substantiation is still required.
Figure 5.
The Aldh gene distinguishes the antidepressant effects among different Bifidobacterium species
(A) Quantification of in vitro ILA production in diverse Bifidobacterium species.
(B) Identification of Trp-indole pathway genes in the genome of diverse Bifidobacterium species.
(C–E) Quantification of ILA concentrations in mouse colon content, serum, and hippocampus. In this animal experiment, 48 mice were randomly divided into six groups (n = 8/treatment): control (blank control group), CUMS (CUMS model group), and four intervention groups with Aldh+B. breve (1 × 10∧9 CFU/d): group A (bif-1, bre-2, inf-2), group B (lon-1, bif-2, bre-3), group C (ado-1, pse-2, lac-2), and group D (lac-1, den-2, ani-3).
(F–H) Behavioral outcomes of mice assessed in different tests.
(I) Principal component analysis of mouse gut microbiota.
(J) Debiased sparse partial correlation analysis of indole derivatives and gut microbiota.
(K) Abundance of gut bacteria related to the transformation of indole derivatives.
Different letters in (A), ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 in (C–H) and (K) represent significant differences between groups, as determined by one-way ANOVA followed by Sidak post hoc test.
Subsequently, we selected two groups of high-ILA-producing strains (Aldh+, group A and B) and two groups of non-ILA-producing strains (Aldh−, group C and D) from the pool of Bifidobacterium strains (Figure 5A) for intervention in the CUMS mouse model. Bifidobacteria with the Aldh gene significantly elevated ILA levels in mouse colonic contents, serum, and hippocampus (Figures 5C–5E). Moreover, these interventions alleviated anxiety- and depression-like behaviors induced by CUMS in mice (Figures 5F–5H). Bifidobacteria without the Aldh gene intervention failed to effectively regulate indole metabolism in CUMS mice (Figures 5C–5E and S5B–S5I). Moreover, an examination of the gut microbiota in all intervention groups revealed a significant alteration in the gut microbial structure in CUMS mice (Figure S5J). Although none of the four bifidobacteria intervention groups fully restored the microbial structure to that of the control group (Figures S5J–S5L), the microbial characteristics of the group with Aldh+ bifidobacteria intervention were notably distinct from those of CUMS mice (Figure 5I). The debiased sparse partial correlation analysis between microbial abundance (Data S2) and indole derivative levels indicated a positive correlation between certain genera and the levels of IA and IPA (Figures S5B and S5C), including Clostridium, Mucispirillum, and Lachnospiraceae UCG-006 (Figure 5J). Their abundance increased following the Aldh+ bifidobacteria intervention (Figure 5K). Considering that IA and IPA are downstream metabolites of ILA, these changes in microbial ecosystem structure may be a response to the elevated ILA levels.
Microbial-derived ILA alleviates neurobehavioral abnormalities by reducing neuroinflammation
In light of Bre1025’s ability to metabolize Trp, we investigated its effect on brain serotonin (5-HT) content, given its pivotal role as a neurotransmitter implicated in mood regulation and as one of the products of Trp metabolism.39,40 Although we observed that Bre1025 supplementation led to a restoration of brain 5-HT levels (Figure S6A), it did not significantly impact serum and hippocampal 5-HT precursor 5-hydroxytryptophan levels (Figures S6B and S6C). This suggests that the observed changes in brain 5-HT levels were not influenced by alterations in precursor supply within the peripheral system but may represent concurrent alterations subsequent to the mitigation of mood disorders by Bre1025 in mice.
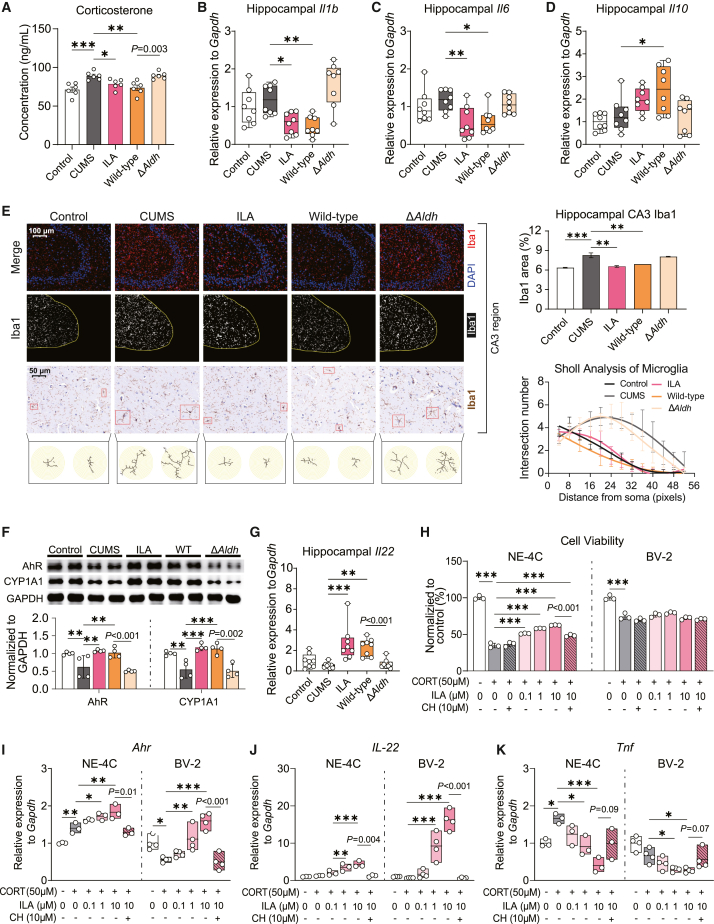
Numerous studies have highlighted that stress can trigger an upsurge in corticosterone release in mice, and prolonged high corticosterone level is associated with the inhibition of neurogenesis in neural stem cells and the promotion of neuroinflammation in the hippocampus.41,42 Compared to the Aldh- strain, Aldh+ bifidobacteria intervention effectively mitigated the aberrant rise in serum corticosterone induced by CUMS (Figures 6A and S5M). Aldh+ bifidobacteria intervention also demonstrated inhibition of the gene overexpression of pro-inflammatory cytokines interleukin-1β (IL-1β) and IL-6 in the hippocampus (Figures 6B, 6C, S5N, and S5O). Simultaneously, it enhanced the expression of the anti-inflammatory cytokine IL-10 (Figure 6D). However, the gene expression of tumor necrosis factor alpha (TNF-α) remained unaltered (Figure S6D). Immunofluorescence detection on brain slices revealed that Bre1025 intervention suppressed the abnormal activation of microglial cells in the hippocampal CA3 region and dentate gyrus, while the Aldh mutant strain’s intervention was ineffective (Figures 6E and S6E). Specifically, CUMS causes hippocampal microglia to become hyper-ramified, and intervention with ILA and its producing bacteria (Bre1025) can inhibit this abnormal change (Figure 6E). Given that Bre1025 intervention significantly increased ILA levels in the hippocampus of CUMS mice (Figure 4D), we hypothesized that the neurophysiological mechanism underlying the strain’s antidepressant effect might involve the ILA-AhR signaling pathway, known for its role in modulating inflammatory levels.43,44 As expected, the gene expression of AhR induced by ILA as its ligand, along with its downstream targets CYP1A1, CYP1A2, and CYP1B1, was upregulated under the strain intervention (Figures 6F, S5P, and S6F–I), consequently promoting the gene expression of the downstream anti-inflammatory cytokine IL-22 (Figures 6G and S5Q).
Figure 6.
Microbial-derived ILA alleviates neuroinflammation through the aryl hydrocarbon receptor
(A) Serum corticosterone concentrations in mice (n = 8/treatment).
(B–D) Expression analysis of genes associated with inflammatory factors in mouse hippocampus (n = 8/treatment).
(E) Immunofluorescence and immunohistochemical detection of Iba1 in the mouse brain (n = 3/treatment). Immunofluorescence analysis measured the percentage of Iba1 signal area relative to the total CA3 area in hippocampal. Sholl analysis was conducted on immunohistochemical slices to assess branching of labeled microglial cells, recording the intersection number every 4 pixels.
(F) Western blot analysis of aryl hydrocarbon receptor (AhR) and CYP1A1 in the hippocampus of mice (n = 4/treatment).
(G) Expression of interleukin-22 gene (Il22) in mouse hippocampus (n = 8/treatment).
(H) Protective effect of ILA intervention on the viability of NE-4C and BV-2 cells (n = 3/treatment).
(I and J) Expression of AhR (Ahr) and IL-22 (Il22) in NE-4C and BV-2 cells (n = 3/treatment).
(K) Expression of TNF-α (Tnf) in NE-4C and BV-2 cells (n = 3/treatment).
∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 indicate significant differences between groups, as determined by one-way ANOVA followed by Sidak post hoc test. P = numbers indicate significant differences between groups, as determined by two-tailed Student’s t test.
To further substantiate this hypothesis, we employed corticosterone intervention to establish an in vitro neuroinflammation model using neural stem cells (NE-4C) and microglial cells (BV-2). Corticosterone intervention significantly inhibited the viability of NE-4C cells (Figure S6J), while pre-treatment with ILA demonstrated a dose-dependent mitigation of corticosterone-induced neurotoxicity (Figures 6H and S6K). Furthermore, ILA intervention activated the AhR signaling pathway in a dose-dependent manner, which led to the promotion of gene expression of the anti-inflammatory cytokine IL-22 and inhibition of the expression of pro-inflammatory cytokines TNF-α and IL-1β (Figures 6I–6K and S6M). The use of the AhR inhibitor CH-223191 (2-methyl-2H-pyrazole-3-carboxylic acid) inhibited the activation of the AhR pathway by ILA in NE-4C cells (Figures 6I, 6J, and S6L), consequently nullifying its protective and anti-inflammatory effects on NE-4C viability (Figure 6H). While pre-treatment with ILA did not alleviate the viability damage caused by corticosterone in microglial BV-2 cells (Figure 6H), its activation of the AhR signaling pathway and regulation of inflammatory factors were consistent with the results observed in NE-4C cells (Figures 6I–6K). Therefore, we speculate that Bre1025, by elevating brain ILA levels, activates the AhR signaling pathway to maintain neuroimmune homeostasis and ultimately suppress CUMS-induced neurobehavioral abnormalities.
Discussion
Previously, several independently gathered pieces of evidence suggest that microbial ILA from the gut may play a role in regulating brain function. This is primarily evident in (1) endogenous indole derivatives in the human body that are primarily synthesized from dietary tryptophan by the gut microbiota and distributed throughout the body via the bloodstream.45 (2) Patients with MDD exhibit significantly lower serum levels of ILA compared to healthy controls. Additionally, supplemental ILA at healthy physiological levels appears to adversely affect neurobehavioral outcomes.32 (3) Indole derivatives have been shown to cross the blood-brain barrier,43,46 and their primary receptor, AhR, is expressed throughout the brain, including regions associated with emotional regulation.47 (4) Oral administration of exogenous indole compounds can activate the AhR signaling pathway in the central nervous system, influencing behavior and modulating neuroinflammation.45 Overall, direct evidence regarding the involvement of gut microbiota-derived ILA in the regulation of brain function is currently very scarce and remains in the stage of data accumulation.
Here, we have identified that the level of gut ILA serves as a core metabolic marker distinguishing normal and depressed mice (Figures 1G and 1I), and supplementation with specific bifidobacteria strains successfully reversed the reduced levels of ILA in the gut and hippocampus of depressed mice (Figures 5C–5E). Our exploration of the molecular pathway for ILA synthesis in bifidobacteria identified Aldh as the key gene responsible for ILA synthesis (Figure 3). This has been confirmed in several bifidobacteria strains, including nine different strains across three species: B. longum subsp. longum, B. longum subsp. infantis, and B. breve.48 Our study further validates that the Aldh gene is essential for ILA production across all nine commonly found human gut bifidobacteria species, encompassing a total of 75 strains. Given genetically manipulated strains to germ-free animals, we demonstrated that the mutation of Aldh in Bre1025 directly resulted in the loss of its antidepressant effects in mice (Figures 4G–4I). These results provide compelling evidence for the direct involvement of gut microbiota-derived ILA in emotional regulation and establish ILA-producing capacity as an indicator for the antidepressant effects of psychobiotic strains.
While the majority of research within the “microbiota-gut-brain axis” framework tends to focus on macroscopic effects on gut microbiota balance,49,50 we advocate for the crucial recognition of the distinct role of individual psychobiotic strains in their interactions with the host. Firstly, clarifying the pharmacological targets and mechanisms of psychobiotics can contribute to their more scientific use, including dosage selection and personalized choices, thus facilitating the translational potential of psychobiotics into clinical applications.51,52 Secondly, confirming the common molecular and genetic characteristics of strains can guide more efficient psychobiotic screening.53 As demonstrated by our widespread verification results within the Bifidobacterium genus, species with the Aldh gene and ILA synthesis ability exhibit better antidepressant potential compared to those without (Figure 5). This shift in perspective has the potential to optimize psychobiotic screening, moving away from the constraints of labor-intensive and somewhat random phenotype-based methods, such as animal behavioral experiments and psychological assessments after human consumption trials.54
Notably, Bifidobacterium species capable of ILA synthesis, including B. longum, B. breve, and B. bifidum, exhibit proficiency in utilizing human milk oligosaccharides (HMOs) and establishing dominance in the early-life gut environment.55 Since our study was conducted in adult mice, we do not propose a direct association between bifidobacteria ILA synthesis and the utilization of HMOs. Previous research has emphasized the role of these species in producing significant amounts of aromatic lactic acids, including ILA, during early life and their regulation of gut immune development through the AhR signaling pathway.38 In line with this but extending further, our results demonstrate that the immunomodulatory effects of ILA produced by gut microbiota extend to the brain. Specifically, we confirmed that ILA is directly synthesized from Trp through Aldh and is substrate dependent (Figure 3A). This discovery partially explains earlier observations, such as the occurrence of depressive symptoms under Trp-deprived diets and the overactivation of microglia in the brains of germ-free mice,45,56 which often exhibit depressive-like behavior. However, the precise interconnections between these factors still require further confirmation.
In summary, our research establishes that gut microbiota-derived ILA plays a direct role in regulating brain inflammation through the AhR signaling pathway, thereby influencing neurobehavioral outcomes. Bifidobacterium strains with the ability to synthesize ILA emerge as promising psychobiotics. These findings contribute to the expansion of our understanding of the mechanisms underlying depression and the microbiota-gut-brain axis theory. Additionally, they provide valuable insights at the molecular and genetic levels that can guide the selection and development of psychobiotic strains, as well as their future clinical applications in addressing mental disorders, including but not limited to depression.
Limitations of the study
Our study has some limitations that need to be addressed in future work. Firstly, the presence of the Aldh gene strongly distinguishes antidepressant effects among bifidobacteria primarily at the species level, rather than the strain level, since the Aldh gene is also annotated in Bre3M5, despite significant differences in ILA production between the two strains (Figure S1A). This suggests that relying solely on genomic features for psychobiotics discrimination is relatively inefficient; a combined approach considering both genomic and metabolic characteristics is essential. Nonetheless, this emerging approach proves much more efficient compared to traditional screening methods based on in vivo assessments. Additionally, we observed that B. bifidum exhibits higher ILA production capabilities compared to other species. Whether this is related to the additional presence of the Fldh gene and the superior potential in antidepressant effects warrants further evaluation. Lastly, due to constraints on animal welfare, a limited number of mice were used to explore potential differences in ILA-mediated neurobehavioral regulation between male and female individuals, despite the recognized necessity for such investigations in behavioral neuroscience studies. Future studies need to be done to address these issues.
Resource availability
Lead contact
For additional information and resource requests, please contact and they will be addressed by the lead contact, Peijun Tian (pjtian@jiangnan.edu.cn).
Materials availability
Requests for access to the materials generated in this study can be directed to the lead contact. It should be noted that this study did not produce any new unique reagents.
Data and code availability
-
•
Genome sequencing data have been deposited at SRA and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original western blot images have been deposited at Mendeley Data and are publicly available as of the date of publication. The DOI is listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact (Peijun Tian, pjtian@jiangnan.edu.cn) upon request.
-
•
This study does not include original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact (Peijun Tian, pjtian@jiangnan.edu.cn) upon request.
Acknowledgments
We appreciate the financial support from Natural Science Foundation of Jiangsu Province (BK20210456), the National Natural Science Foundation of China (no. 32201988, 32394051), the National Key R&D Program of China (2023YFC2506004), the Fundamental Research Funds for the Central Universities (JUSRP123047), and the Program of Collaborative Innovation Centre of Food Safety and Quality Control in Jiangsu Province.
Author contributions
Conceptualization, X.Q. and P.T.; investigation, X.Q., Q.L., and H. Zhu; methodology, X.Q., Y.C., and G.L.; project administration, P.T. and G.W.; data curation, X.Q. and H. Zhu; writing – original draft, X.Q. and P.T.; software, Y.C. and G.L.; formal analysis, X.Q., Q.L., and H. Zhu; visualization, X.Q., Q.L., and H. Zhu; supervision, P.T. and G.W.; validation, H. Zhang and W.C.; writing – review and editing, P.T.; funding acquisition, P.T., W.C., and G.W.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Iba1 | Beyotime | Cat# AF7143, RRID: AB_3662862 |
| Rabbit polyclonal anti-AHR | Beyotime | Cat# AF6165, RRID:AB_3662863 |
| Rabbit polyclonal anti-CYP1A1 | Beyotime | Cat# AF6642, RRID: AB_3662864 |
| GAPDH Rabbit mAb | ABclonal Technology | Cat# A19056; RRID:AB_2862549 |
| Horseradish peroxidase labeled Goat anti-Rabbit IgG | Beyotime | Cat# A0208; RRID:AB_2892644 |
| Bacterial and virus strains | ||
| Bifidobacterium breve: CCFM1025: Bre1025 | Culture Collection of Food Microorganisms (CCFM), Jiangnan University | N/A |
| B. breve: FHLJDQ3M5: Bre3M5 | CCFM, Jiangnan University | N/A |
| B. bifidum: FNXHL20M2: bif-1 | CCFM, Jiangnan University | N/A |
| B. bifidum: FHuNMY2M1: bif-2 | CCFM, Jiangnan University | N/A |
| B. bifidum: FFJND15M5: bif-3 | CCFM, Jiangnan University | N/A |
| B. bifidum: FHNXY21M6: bif-4 | CCFM, Jiangnan University | N/A |
| B. bifidum: FXJCJ22M1: bif-5 | CCFM, Jiangnan University | N/A |
| B. bifidum: FHeNJZ3M6: bif-6 | CCFM, Jiangnan University | N/A |
| B. bifidum: FXJKS43M4: bif-7 | CCFM, Jiangnan University | N/A |
| B. bifidum: FHeNJZ1M5: bif-8 | CCFM, Jiangnan University | N/A |
| B. bifidum: FGSZY50M8: bif-9 | CCFM, Jiangnan University | N/A |
| B. breve: FGZ3I2M1: bre-1 | CCFM, Jiangnan University | N/A |
| B. breve: FGZ35I2M1: bre-2 | CCFM, Jiangnan University | N/A |
| B. breve: FGZ18I2M1: bre-3 | CCFM, Jiangnan University | N/A |
| B. breve: FGZ9I2M1: bre-4 | CCFM, Jiangnan University | N/A |
| B. breve: FGZ6I2M1: bre-5 | CCFM, Jiangnan University | N/A |
| B. breve: FGZ39I2M1: bre-6 | CCFM, Jiangnan University | N/A |
| B. breve: FGZ23I2M1: bre-7 | CCFM, Jiangnan University | N/A |
| B. breve: FGZ39MM6: bre-8 | CCFM, Jiangnan University | N/A |
| B. breve: FGZ19I2M1: bre-9 | CCFM, Jiangnan University | N/A |
| B. longum subsp. longum: FHBSJZ3M1: lon-1 | CCFM, Jiangnan University | N/A |
| B. longum subsp. longum: FZJZS1M5: lon-2 | CCFM, Jiangnan University | N/A |
| B. longum subsp. longum: FHBSJZ2M3: lon-3 | CCFM, Jiangnan University | N/A |
| B. longum subsp. longum: FHBTS3M2: lon-4 | CCFM, Jiangnan University | N/A |
| B. longum subsp. longum: FHBSJZ6M1: lon-5 | CCFM, Jiangnan University | N/A |
| B. longum subsp. longum: FHBSJZ5M5: lon-6 | CCFM, Jiangnan University | N/A |
| B. longum subsp. longum: FSXBJ11M1: lon-7 | CCFM, Jiangnan University | N/A |
| B. longum subsp. longum: FGDGZ1M1: lon-8 | CCFM, Jiangnan University | N/A |
| B. longum subsp. longum: FHBSJZ1M2: lon-9 | CCFM, Jiangnan University | N/A |
| B. longum subsp. infantis: JSWX3M1: inf-1 | CCFM, Jiangnan University | N/A |
| B. longum subsp. infantis: FGZ19I2M3: inf-2 | CCFM, Jiangnan University | N/A |
| B. longum subsp. infantis: FGZ23I1M7: inf-3 | CCFM, Jiangnan University | N/A |
| B. longum subsp. infantis: FJND2M2: inf-4 | CCFM, Jiangnan University | N/A |
| B. longum subsp. infantis: JSWX25M6: inf-5 | CCFM, Jiangnan University | N/A |
| B. longum subsp. infantis: JSWX6M2: inf-6 | CCFM, Jiangnan University | N/A |
| B. longum subsp. infantis: FGZ23I1M2: inf-7 | CCFM, Jiangnan University | N/A |
| B. longum subsp. infantis: SDZC2M4: inf-8 | CCFM, Jiangnan University | N/A |
| B. longum subsp. infantis: FHeNJZ3M1: inf-9 | CCFM, Jiangnan University | N/A |
| B. adolescentis: FGSYC30M5: ado-1 | CCFM, Jiangnan University | N/A |
| B. adolescentis: FHNFQ38M3: ado-2 | CCFM, Jiangnan University | N/A |
| B. adolescentis: FHNFQ5M4: ado-3 | CCFM, Jiangnan University | N/A |
| B. adolescentis: FHNXY34M5: ado-4 | CCFM, Jiangnan University | N/A |
| B. adolescentis: FJSSZ3M10: ado-5 | CCFM, Jiangnan University | N/A |
| B. adolescentis: FNXHL20M3: ado-6 | CCFM, Jiangnan University | N/A |
| B. adolescentis: FNXHL25M2: ado-7 | CCFM, Jiangnan University | N/A |
| B. adolescentis: FXJCJ25M6: ado-8 | CCFM, Jiangnan University | N/A |
| B. adolescentis: FXJKS34M4: ado-9 | CCFM, Jiangnan University | N/A |
| B. pseudocatenulatum: FGSYC7M5: pse-1 | CCFM, Jiangnan University | N/A |
| B. pseudocatenulatum: FFJNDD5M3: pse-2 | CCFM, Jiangnan University | N/A |
| B. pseudocatenulatum: FFJNDD6M2: pse-3 | CCFM, Jiangnan University | N/A |
| B. pseudocatenulatum: FGSYC4M2: pse-4 | CCFM, Jiangnan University | N/A |
| B. pseudocatenulatum: FGSYC5M4: pse-5 | CCFM, Jiangnan University | N/A |
| B. pseudocatenulatum: FQHXN112M3: pse-6 | CCFM, Jiangnan University | N/A |
| B. pseudocatenulatum: FQHXN3M8: pse-7 | CCFM, Jiangnan University | N/A |
| B. pseudocatenulatum: FQHXN5M4: pse-8 | CCFM, Jiangnan University | N/A |
| B. pseudocatenulatum: FQHXN8M3: pse-9 | CCFM, Jiangnan University | N/A |
| B. dentium: FJSWXJ29M2: den-1 | CCFM, Jiangnan University | N/A |
| B. dentium: FGDLZ75M1: den-2 | CCFM, Jiangnan University | N/A |
| B. dentium: FGSYC1M4: den-3 | CCFM, Jiangnan University | N/A |
| B. dentium: FGZ8I1M1: den-4 | CCFM, Jiangnan University | N/A |
| B. dentium: FHuNMY10M2: den-5 | CCFM, Jiangnan University | N/A |
| B. dentium: FHuNMY7M3: den-6 | CCFM, Jiangnan University | N/A |
| B. dentium: FHuNMY9M2: den-7 | CCFM, Jiangnan University | N/A |
| B. dentium: FNMHLBE6M6: den-8 | CCFM, Jiangnan University | N/A |
| B. dentium: FWX34M7: den-9 | CCFM, Jiangnan University | N/A |
| B. animalis subsp. lactis: FNMGEL1M9: lac-1 | CCFM, Jiangnan University | N/A |
| B. animalis subsp. lactis: AHWH12M3: lac-2 | CCFM, Jiangnan University | N/A |
| B. animalis subsp. lactis: BJHD3M6: lac-3 | CCFM, Jiangnan University | N/A |
| B. animalis subsp. lactis: FNMGHHHT2M2: lac-4 | CCFM, Jiangnan University | N/A |
| B. animalis subsp. lactis: FWX37M4: lac-5 | CCFM, Jiangnan University | N/A |
| B. animalis subsp. lactis: JSYC4M3: lac-6 | CCFM, Jiangnan University | N/A |
| B. animalis subsp. lactis: NMGEL1M5: lac-7 | CCFM, Jiangnan University | N/A |
| B. animalis subsp. lactis: NMGEL2M3: lac-8 | CCFM, Jiangnan University | N/A |
| B. animalis subsp. lactis: NMGHHHT2M1: lac-9 | CCFM, Jiangnan University | N/A |
| B. animalis subsp. animalis: CGMCC1.2268: ani-1 | China General Microbiological Culture Collection Center (CGMCC) | CGMCC 1.2268 |
| B. animalis subsp. animalis: CGMCC1.3003: ani-2 | CGMCC | CGMCC 1.3003 |
| B. animalis subsp. animalis: GDMCC1.169: ani-3 | CGMCC | CGMCC 1.169 |
| Biological samples | ||
| Human stool and serum samples in cohort1 | Yixing People’s Hospital | ChiCTR2300071025 |
| Human stool samples in cohort2 | Jingjiang Chinese Medicine Hospital | ChiCTR2200057145 |
| Chemicals, peptides, and recombinant proteins | ||
| Minimum Essential Medium | Meilunbio | Cat# MA0217 |
| Dulbecco’s Modified Eagle Medium | Gibco | Cat# 11965092 |
| Fetal Bovine Serum | Gibco | Cat# A5670701 |
| Fast DNA Spin Kit for Feces | MP Biomedical | Cat# 6570200 |
| FastPure Cell/Tissue Total RNA Isolation Kit V2 | Vanzyme | Cat# DC102-01 |
| HiScript III All-in-one RT SuperMix Perfect for qPCR | Vanzyme | Cat# R333-01 |
| ChamQ Universal SYBR qPCR Master Mix | Vanzyme | Cat# Q711-02 |
| DNA gel purification Miniprep kit | Biomiga | Cat# DC3511-01 |
| Mouse Corticosterone (CORT) Enzyme-linked Immunoassay (ELISA) Kit | Senbeijia Biological Technology | Cat# SBJ-M0037 |
| FITC Immunofluorescence Detection Kit | Sangon Biotech | Cat# E670005 |
| SuperSignal West Pico PLUS Chemiluminescent Substrate | ThermoFisher | Cat# 34577 |
| Cell Counting Kit-8 solution | Beyotime | Cat# C0038 |
| RIPA Lysis Buffer | Beyotime | Cat# P0013K |
| protease phosphatase inhibitors | Beyotime | Cat# P1045 |
| BCA Protein Assay Kit | Beyotime | Cat# P0010 |
| Deposited data | ||
| Bacterial genome data | NCBI | # PRJNA1073989 |
| Original western blot images | Mendeley Data | https://doi.org/10.17632/b2xjhzsh94.1 |
| Experimental models: Cell lines | ||
| Mouse: NE-4C | ATCC | Cat# CRL-2925 |
| Mouse: BV-2 | ATCC | Cat# CRL-2467 |
| Experimental models: Organisms/strains | ||
| SPF C57BL/6J mice | Gempharmatech | Cat# N000013 |
| Oligonucleotides | ||
|
B. breve species-specific primers (BreveT2): F- GAGAAGGCTGAGGCCGT; R- GGGCAGAGAACGAACCTT |
This paper | N/A |
| Bre1025 strain-specific primers (1025T5): F- CCAATAGATTCCACATCGGTTCA; R- CCAGACCAGCCATATAATAATCCA |
This paper | N/A |
| Primers for real-time quantitative-PCR, see Data S4 | This paper | N/A |
| Software and algorithms | ||
| Ethovision version 13 | Noldus | https://www.noldus.com/ethovision-xt |
| Compound Discoverer 3.2 | Thermo Scientific | Cat# OPTON-31060 |
| SOAPdenovo v2.0.4 | BGI Genomics | http://soap.genomics.org.cn/soapdenovo.html#intro2 |
| GapCloser | BGI Genomics | https://sourceforge.net/projects/soapdenovo2/files/GapCloser/bin/r6/GapCloser-bin-v1.12-r6.tgztar |
| Prodigal v2.6.3 | Prodigal Technologies | https://github.com/hyattpd/Prodigal/wiki/introduction |
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ |
| OrthoMCL v2.0.9 | VEuPathDB Project | https://orthomcl.org/orthomcl/app |
| GraphPad Prism 8 | GraphPad Software | https://www.graphpad.com/ |
| Other | ||
| Cell chamber culture plate | NEST Biotechnology | Cat# 723001 |
| Cell culture plate | NEST Biotechnology | Cat# 701011 |
Experimental model and subject details
Animals
Male C57BL/6 mice, five weeks old and specific pathogen-free (SPF), were obtained from GemPharmatech (Shanghai, China). A total of 152 mice were used in four batches for the study: (1) 20 mice were randomly divided into four groups for non-targeted metabolomics analysis of gut metabolites: Control (blank control group), CUMS (CUMS model group), Bre1025 (B.breve Bre1025 intervention group, 1 × 10∧9 CFU/d), and Bre3M5 (B.breve Bre3M5 intervention group, 1 × 10∧9 CFU/d). (2) 24 mice were randomly divided into four groups for the quantitative analysis of intestinal and serum indole derivatives: Control (blank control group), 1025-low (low-dose Bre1025 intervention group, 1 × 10∧7 CFU/d), 1025-med (medium-dose Bre1025 intervention group, 1 × 10∧8 CFU/d), and 1025-high (high-dose Bre1025 intervention group, 1 × 10∧9 CFU/d). (3) 60 mice were randomly divided into five groups to evaluate the mood-regulating function of wild-type and genetically modified Bre1025 strains: Control (blank control group), CUMS (CUMS model group), ILA (oral ILA intervention group, 35 mg/kg/d), Wild-type (Bre1025 intervention group, 1 × 10∧9 CFU/d), and ΔAldh (Bre1025-ΔAldh intervention group, 1 × 10∧9 CFU/d). (4) 48 mice were randomly divided into six groups to evaluate the mood-regulating function of different Bifidobacterium species: Control (blank control group), CUMS (CUMS model group), and four intervention groups with Aldh+ B.breve (1 × 10∧9 CFU/d): Group A (bif-1, bre-2, inf-2), Group B (lon-1, bif-2, bre-3), Group C (ado-1, pse-2, lac-2), and Group D (lac-1, den-2, ani-3). All mice were kept in ventilated cages with a temperature range of 22°C–24°C, provided with food and water ad libitum, and maintained under a 12-h light/dark cycle. All animal handling procedures adhered to the guidelines established by the National Institutes of Health and were approved by the Animal Welfare and Ethics Committee of Jiangnan University (JN. No20210915c0901225[280], JN. No20220315c0360701[045], JN. No20220615c1041101[228], JN. No20230415c0640805[126]), Wuxi, China.
Human subjects
Cohort 1: 40 human subjects, including 20 patients with depression and 20 healthy volunteers, were recruited from Yixing People’s Hospital for the analysis of indole derivatives in stool and serum samples. Detailed subject information provided in Data S3. Inclusion criteria: a) ages between 18 and 65 years; b) body mass index (BMI) within the range of 18–25. Exclusion criteria: a) consumption of probiotics, yogurt, or other foods/medicines containing live bacteria in the past 28 days; b) use of antibiotics or other bacteriostatic drugs within the previous 56 days; c) presence of gastrointestinal disorders or other noticeable diseases. Additional inclusion criteria for patients with depression: a) clinical diagnosis of depressive disorder; b) Hospital Anxiety and Depression Scale (HADS) score ≥11. Additional exclusion criteria for patients with depression: a) other psychiatric diagnoses that meeting DSM-IV criteria, such as schizophrenia or bipolar disorder; b) behaviors like suicide or self-injury. Participants were instructed to provide stool and serum samples, with serum samples collected by 9 a.m. and fasting required after 8 p.m. the night before sampling. The trial received ethical approval from the Yixing People’s Hospital Ethics Committee (NO2023-023). Before participating in the study, all participants furnished written informed consent. The full clinical trial registration has been submitted to the Chinese Clinical Trial Registry (ChiCTR2300071025).
Cohort 2: 15 healthy subjects were recruited from Jingjiang Chinese Medicine Hospital for the clinical trial of continuous Bre1025 intake. Detailed subject information provided in Data S3. Inclusion criteria: a) ages between 18 and 65 years; b) BMI within the range of 18–25. Exclusion criteria: a) consumption of probiotics, yogurt, or other foods/medicines containing live bacteria in the past 28 days; b) use of antibiotics or other bacteriostatic drugs within the previous 56 days; c) allergies or intolerance to components within the test probiotic product; d) presence of gastrointestinal disorders or other noticeable diseases. Participants were instructed to consume freeze-dried Bre1025 powder at a dose of 1 × 10∧9 CFU per day for a period of 14 consecutive days. Stool samples were collected at three intervals: before ingestion (0days), 14 days post-ingestion (14days), and 14 days post-cessation of ingestion (28days). The trial received ethical approval from the Jingjiang Chinese Medicine Hospital Ethics Committee (EN2021078). Before participating in the study, all participants furnished written informed consent. The full clinical trial registration has been submitted to the Chinese Clinical Trial Registry (ChiCTR2200057145).
Cell culture
NE-4C and BV-2 cells, sourced from American Type Culture Collection (ATCC), were cultivated in a humidified incubator at 37°C with 5% CO2. NE-4C cells were grown in minimum essential medium (MEM, Meilunbio, Cat# MA0217), while BV-2 cells were maintained in Dulbecco’s modified Eagle medium (DMEM, Gibco, Cat# 11965092). Both media were supplemented with 10% (v/v) fetal bovine serum (FBS), 2 mM glutamine, and 1% (v/v) penicillin/streptomycin.
Bacteria culture
The Bifidobacterium spp. strains utilized in this research were sourced from the Culture Collection of Food Microorganisms (CCFM). After thawing from glycerol tubes, the strains were streaked onto modified mMRS solid media and then anaerobically cultured at 37°C for 48 h. Following incubation, individual colonies were selected and transferred into mMRS liquid medium for further cultivation, reaching the 2nd generation for subsequent experiments.
Method details
Study design and timeline
Initially, the CUMS mouse model and depression patient samples were used to screen for potential effector molecules of the psychobiotic Bre1025, leading to the discovery of ILA. Bre1025 interventions were then conducted on healthy volunteers and mice to evaluate its impact on host ILA levels. The mechanism of ILA production by Bre1025 was investigated in vitro, identifying the key gene Aldh. To confirm Bre1025’s role in mood regulation, antibiotic-treated CUMS mice were used. Additionally, the mood-regulating abilities of other high-ILA-producing Bifidobacterium species were evaluated in CUMS mouse models without antibiotics. Finally, brain tissue analysis and in vitro studies with neural stem cells and microglial cells were conducted to explore ILA’s role in alleviating neuroinflammation.
Chronic unpredictable mild stress (CUMS)
CUMS was performed as described previously.24 Mice were subjected to one or two stress stimuli daily, randomly chosen to prevent predictability and repetition. The stressors included: 1) 24-h food deprivation; 2) 24-h water deprivation with an empty bottle; 3) 24-h exposure to humid bedding; 4) 3-min tail clamping; 5) 2-h brake restraint; 6) Tilting the cage at a 45° angle for 24 h; 7) 10-min forced swimming; 8) Isolation for 24 h; 9) Continuous light exposure for 24 h; 10) Removal of bedding material for 24 h; 11) Crowding stimulation for 24 h.
Open field test
Prior to conducting the behavior tests, animals underwent a 4-h acclimatization period in the testing environment. All assessments were conducted under dim lighting conditions (60 lux) and were tracked using a video tracking system (Ethovision version 13). Animals were positioned in the center of a square arena (50 cm × 50 cm), and a 10-min monitoring period began immediately. The duration spent by the animals in the central area of the open field was recorded. To ensure consistency when transitioning between animals and prevent potential interference with subsequent test results, it is necessary to clean the inner wall and bottom surface of the open field first to remove the remaining information of the last animal (such as feces, urine, odor, etc.). Videos were analyzed digitally using EthoVision software, and the percentage of time mice spent in the central area of the open field was calculated.57
Tail suspension test
The posterior one-third of the mouse tail was fixed to the suspension bar, maintaining a distance of 20–25 cm between the tip of the nose of the animal and the floor of the device. A 6-min monitoring period began immediately. Immobility time was recorded between the third and sixth minutes after suspension of the mice using EthoVision analysis software. Immobility was a state in which an animal gives up active struggle and is completely immobile. In order to prevent test result interference, the test frame’s inside and bottom were cleaned before each test to remove traces left by the previous animal.23
Forced swim test
The swimming apparatus was filled with water to a height of 30 cm, maintained at a temperature of 24 ± 1°C. All mice underwent a 10-min adaptive swimming session 24 h before the formal experiment. During the experiment, each mouse was observed for 6 min. The immobility time was recorded between minutes 3 and 6 after the mice were placed in the water. Immobility was defined as no movement of the limbs or only slight movement of the hind limbs. In order to prevent test result interference, the swimming apparatus was cleaned, and clean water was replaced before each test to remove traces left by the previous animal.58
Quantification of target strains in fecal samples
Bre1025 was quantified in fecal DNA by quantitative PCR (qPCR) using a Bre1025 strain-specific primers (1025T5). B. breve was quantified by qPCR with species-specific primers (BreveT2). Stool samples were utilized to extract total fecal DNA employing the Fast DNA Spin Kit for Feces (MP Biomedical). Reference standards were established using blank fecal samples containing known concentrations of Bre1025 or Bre3M5 cells. The quantity of the target strain within the stool sample was determined utilizing the standard curve generated. For detailed assay and qPCR procedures, please refer to our previously published methodological article.58
Non-targeted metabolomics assay
To prepare samples, cecal contents or feces were homogenized in a methanol-water solution, followed by protein precipitation and centrifugation. The strain supernatant was homogenized in methanol. The resulting supernatant was concentrated, reconstituted, and filtered before LC-MS detection.59 Quality control (QC) samples were prepared by combining extracts from each sample, with an aqueous methanol solution used as a blank. Quality control samples were prepared by extracting and combining 5 μL from each sample, while an aqueous methanol solution was utilized as a blank sample.60 The LC-MS analysis utilized a T3 column and the target substance was separated using a gradient elution procedure with progressively increasing concentrations of the organic phase (acetonitrile). Data collection was performed in positive and negative ion modes within a range of 50–1000 m/z. Compound Discoverer 3.2 Software was employed for data extraction, filtering, metabolite identification, and statistical analysis, including enrichment analysis using MetaboAnalyst 5.0.
Targeted metabolomics of the indole derivatives
Samples, including cecal contents, feces, hippocampal tissue, serum or the strain supernatant, underwent homogenization in methanol solutions. After centrifugation, the supernatant was collected, concentrated, dried, and reconstituted. Following centrifugation and filtration, the reconstituted extract was analyzed by LC-MS. The LC-MS analysis utilized a C18 column and the target substance was separated using a gradient elution procedure with progressively increasing concentrations of the organic phase (acetonitrile). Data collection was performed in positive ion modes within a range of 50–750 m/z.57
RNA extraction and quantitative real-time PCR
RNA extraction from either Bre1025 cells or animal tissues was performed according to the kit protocol (Vanzyme, Cat#DC102-01). Subsequently, RNA samples that had been tested for concentration and purity were converted into cDNA using a reverse transcription kit (Vanzyme, Cat#R333-01). Primers used in the qPCR reactions were detailed in Data S4. Each qPCR reaction (20 μL), comprising SYBRmix (Vanzyme, Cat#Q711-02), primers (10 pmol), and 1 μL of cDNA template. qPCR protocol: 95°C (2 min); followed by 40 amplification cycles (95°C (10 s), 60°C (20 s)), and concluded with a dissolution curve spanning temperatures from 65°C to 95°C.
Construction of bacterial mutants
Insertional mutagenesis of the target gene in Bre1025 was achieved through single-crossover homologous recombination.23,61 To create the B. breve mutant plasmid pTET28 (Figure S3B), the erythromycin resistance marker in plasmid pFREM28 was replaced with the TetW sourced from B. longum H66. Primers, designed based on the target gene (refer to Data S4), were utilized to amplify a 500 bp sequence from the Bre1025 target gene domain. This sequence was then inserted into the plasmid pTET28, resulting in the generation of the recombinant plasmid. The recombinant plasmids were introduced into Bre1025 at 1800 V, following a 4-h incubation in Reinforced Clostridium Medium (RCM) preheated at 37°C, the transformed cells were plated on RCM plates containing tetracycline resistance and incubated at 37°C for 3 days. Transformants displaying tetracycline resistance were selected and further validated via PCR using primers specific to the target gene. Successfully transformed insert mutants were thereby obtained.
Bifidobacterium-defined medium (BDM)
The Bifidobacterium-defined medium (BDM) is a tailored medium formulated by adapting the growth prerequisites of Bifidobacterium, structured upon the foundation of the Lactic Acid Bacteria Defined Medium (LDM) IV.62 The specialized medium incorporates essential reagents such as vitamin solution, biotin solution, riboflavin solution, folic acid solution, nucleic acid solution, salt ion solution, and glucose solution. For a comprehensive breakdown of the exact composition of each solution, please refer to Data S5.
Identification of the genes involved in indole derivatives synthesis
The isolation strain genome sequencing was executed using the Illumina HiSeq platform. The resulting double-end reads were assembled into high-quality sequences through SOAPdenovo v2.0.4. To construct scaffolds based on read relationships and overlaps, GapCloser was employed. Prodigal v2.6.3 was utilized to translate coding sequences for subsequent alignment in identifying homologous proteins.
Microbial tryptophan metabolism related genes corresponding protein sequence is based on Enzyme Commission numbers from NCBI database (https://www.ncbi.nlm.nih.gov),63 Utilizing these sequences as reference points, a BLASTP homology analysis was conducted to pinpoint genes involved in indole derivatives synthesis within the Bifidobacterium strains utilized in this investigation. Detailed information regarding all Trp-Indole pathway genes investigated in this study is documented in Data S6.
Antibiotic (ABx) cocktail therapy
The mice underwent a 10-day course of antibiotic (ABx) cocktail treatment, followed by polyethylene glycol administration to eliminate the native intestinal microbiota.62 The antibiotic blend comprised neomycin sulfate (100 mg/kg), ampicillin sodium (100 mg/kg), metronidazole (100 mg/kg), and vancomycin hydrochloride (50 mg/kg) dissolved in a 4 mM acetic acid solution. All substances were procured from Sigma (Sigma-Aldrich) and the prepared solution was administered by gavage.
16S rRNA sequencing and data analysis
Total bacterial DNA was extracted from stool samples (MP Biomedical, Cat#6570200), followed by PCR amplification of the bacterial 16S rDNA (V3-V4 region). Amplified products were electrophoresed on a 1.5% agarose gel for 35 min. Recovered products were quantified (Biomiga, Cat#DC3511-01) and libraries of 50μL, maintaining equal mass concentrations, were constructed for sequencing on an Illumina MiSeq platform (Illumina).64 Raw sequencing data were processed using QIIME 2, with OTUs defined at a 97% nucleotide identity cutoff. Taxonomic assignments were made using the SILVA database. Evaluation of diversity involved measuring the Shannon and Simpson indices from the rarefied OTU dataset. Additionally, β-diversity was estimated via compositional data analysis employing principal component analysis (PCA) with Euclidean distance, followed by a PERMANOVA test for evaluating differences.65
Serum corticosterone detection
The serum corticosterone levels were determined using the Mouse Corticosterone (CORT) Enzyme-linked Immunoassay (ELISA) Kit (Cat# SBJ-M0037), following the manufacturer’s guidelines provided by Senbeijia Biological Technology.
Immunofluorescence staining
After cryosectioning, mouse brain tissues were affixed onto glass slides. Brain tissue antigen retrieval was performed using the FITC Immunofluorescence Detection Kit (Sangon Biotech, Cat# E670005). Subsequently, brain sections were rinsed with phosphate-buffered saline tween (PBST), followed by a 30-min blocking step. Following the incubation period, the sections underwent overnight treatment at 4°C in darkness with Rabbit polyclonal anti-Iba1 (Beyotime, Cat# AF7143) at a dilution of 1:500. Subsequent to thorough washing with PBST, the sections were exposed to FITC-labeled donkey anti-mouse IgG (1:5000 dilution) for 1 h. Following staining with 4′,6-diamidino-2-phenylindole (DAPI), the sections were sealed with coverslips. Quantitative analysis of the immunofluorescent staining images was performed using ImageJ (version 2.3.0) to assess the presence of positive signals.
Western blotting
Protein extraction from hippocampal or prefrontal samples was conducted utilizing RIPA Lysis Buffer (Beyotime, Cat# P0013K), with the addition of protease and phosphatase inhibitors to ensure integrity during the extraction process. Following extraction, protein concentrations were quantified using the BCA Protein Assay Kit (Beyotime, Cat# P0010), and the extracted proteins were subsequently denatured. Vertical electrophoresis was conducted to ensure consistent protein content, and the resolved protein bands were transferred to PVDF membranes. Following a 1-h blocking step, the PVDF membranes underwent incubation with Rabbit polyclonal anti- AHR/CYP1A1 (Beyotime, Cat# AF6165/AF6642; 1:500 dilution) at 4°C in the absence of light for 12 h. Following thorough washing with PBST, the membranes were exposed to Horseradish Peroxidase labeled Goat anti-Rabbit IgG (Beyotime, Cat# A0208; 1:5000 dilution) for 1 h. Subsequent imaging was carried out using SuperSignal West Pico PLUS Chemiluminescent Substrate (ThermoFisher, Cat# 34577), and quantitative analysis was performed using ImageJ software.
Cell viability assay
Cells were initially plated at a density of 1×104 cells/mL into 96-well cell culture plates (NEST Biotechnology) and cultured for 24 h at 37°C. Afterward, the medium was replaced with either corticosterone or ILA-containing medium for an additional 24-h incubation. Following this incubation, 10 μL of Cell Counting Kit-8 solution (Beyotime, Cat# C0038) was added to each well, and the cells were further incubated for 2 h. The absorbance was then measured at OD450, and the percentage ratio of different treatments to the blank control group was then calculated.
Quantification and statistical analysis
Statistical analysis was conducted using GraphPad Prism 8.0.1. Experimental data were presented as the mean ± SEM (standard error of the mean). Statistical significance was assessed using a two-tailed Student’s t-test for comparing two groups and a one-way analysis of variance for comparisons involving three or more groups, followed by the Sidak post hoc test. Additionally, the OmicStudio platform was used to perform debiased sparse partial correlation analysis and generate the corresponding graphs (https://www.omicstudio.cn/).
Additional resources
The two clinical studies involved in this research have been registered on the "Chinese Clinical Trial Registry." The registration IDs are ChiCTR2300071025 and ChiCTR2200057145, respectively. The access websites are as follows: https://www.chictr.org.cn/showproj.html?proj=196296; https://www.chictr.org.cn/showproj.html?proj=137056.
Published: October 24, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2024.101798.
Supplemental information
References
- 1.Ziętara K.J., Pawłowski P., Chromiak K., Orzechowska A., Michalczyk J., Orzechowska J. Multidisciplinary of depression spectrum disorders from a health care system perspective. J. Educ. Health Sport. 2022;12:206–215. [Google Scholar]
- 2.Freeman M. The World Mental Health Report: transforming mental health for all. World Psychiatr. 2022;21:391–392. doi: 10.1002/wps.21018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mazza M.G., Palladini M., Poletti S., Benedetti F. Post-COVID-19 depressive symptoms: epidemiology, pathophysiology, and pharmacological treatment. CNS Drugs. 2022;36:681–702. doi: 10.1007/s40263-022-00931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salari N., Hosseinian-Far A., Jalali R., Vaisi-Raygani A., Rasoulpoor S., Mohammadi M., Rasoulpoor S., Khaledi-Paveh B. Prevalence of stress, anxiety, depression among the general population during the COVID-19 pandemic: a systematic review and meta-analysis. Glob. Health. 2020;16:57. doi: 10.1186/s12992-020-00589-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan K., Zheng Y.B., Wang Y.J., Sun Y.K., Gong Y.M., Huang Y.T., Chen X., Liu X.X., Zhong Y., Su S.Z., et al. A systematic review and meta-analysis on prevalence of and risk factors associated with depression, anxiety and insomnia in infectious diseases, including COVID-19: a call to action. Mol. Psychiatr. 2022;27:3214–3222. doi: 10.1038/s41380-022-01638-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drevets W.C., Wittenberg G.M., Bullmore E.T., Manji H.K. Immune targets for therapeutic development in depression: towards precision medicine. Nat. Rev. Drug Discov. 2022;21:224–244. doi: 10.1038/s41573-021-00368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J.W., Suzuki K., Kavalali E.T., Monteggia L.M. Ketamine: Mechanisms and relevance to treatment of depression. Annu. Rev. Med. 2024;75:129–143. doi: 10.1146/annurev-med-051322-120608. [DOI] [PubMed] [Google Scholar]
- 8.Papp M., Cubala W.J., Swiecicki L., Newman-Tancredi A., Willner P. Perspectives for therapy of treatment-resistant depression. Br. J. Pharmacol. 2022;179:4181–4200. doi: 10.1111/bph.15596. [DOI] [PubMed] [Google Scholar]
- 9.Berman M.E., Tracy J.I., Coccaro E.F. The serotonin hypothesis of aggression revisited. Clin. Psychol. Rev. 1997;17:651–665. doi: 10.1016/s0272-7358(97)00039-1. [DOI] [PubMed] [Google Scholar]
- 10.Lacasse J.R., Leo J. Serotonin and depression: A disconnect between the advertisements and the scientific literature. PLoS Med. 2005;2:e392. doi: 10.1371/journal.pmed.0020392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warren J.B. The trouble with antidepressants: why the evidence overplays benefits and underplays risks—an essay by John B Warren. BMJ. 2020;370:m3200. doi: 10.1136/bmj.m3200. [DOI] [PubMed] [Google Scholar]
- 12.Ruhé H.G., Mason N.S., Schene A.H. Mood is indirectly related to serotonin, norepinephrine and dopamine levels in humans: A meta-analysis of monoamine depletion studies. Mol. Psychiatr. 2007;12:331–359. doi: 10.1038/sj.mp.4001949. [DOI] [PubMed] [Google Scholar]
- 13.Chakrabarti A., Geurts L., Hoyles L., Iozzo P., Kraneveld A.D., La Fata G., Miani M., Patterson E., Pot B., Shortt C., Vauzour D. The microbiota–gut–brain axis: pathways to better brain health. Perspectives on what we know, what we need to investigate and how to put knowledge into practice. Cell. Mol. Life Sci. 2022;79:80. doi: 10.1007/s00018-021-04060-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Socała K., Doboszewska U., Szopa A., Serefko A., Włodarczyk M., Zielińska A., Poleszak E., Fichna J., Wlaź P. The role of microbiota-gut-brain axis in neuropsychiatric and neurological disorders. Pharmacol. Res. 2021;172 doi: 10.1016/j.phrs.2021.105840. [DOI] [PubMed] [Google Scholar]
- 15.Li Z., Lai J., Zhang P., Ding J., Jiang J., Liu C., Huang H., Zhen H., Xi C., Sun Y., et al. Multi-omics analyses of serum metabolome, gut microbiome and brain function reveal dysregulated microbiota-gut-brain axis in bipolar depression. Mol. Psychiatr. 2022;27:4123–4135. doi: 10.1038/s41380-022-01569-9. [DOI] [PubMed] [Google Scholar]
- 16.Doll J.P.K., Vázquez-Castellanos J.F., Schaub A.-C., Schweinfurth N., Kettelhack C., Schneider E., Yamanbaeva G., Mählmann L., Brand S., Beglinger C., et al. Fecal microbiota transplantation (FMT) as an adjunctive therapy for depression—case report. Front. Psychiatr. 2022;13 doi: 10.3389/fpsyt.2022.815422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang C., Hu T., Xue X., Su X., Zhang X., Fan Y., Shen X., Dong X. Multi-omics analysis of fecal microbiota transplantation’s impact on functional constipation and comorbid depression and anxiety. BMC Microbiol. 2023;23:389. doi: 10.1186/s12866-023-03123-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlson P.E., Jr. Regulatory considerations for fecal microbiota transplantation products. Cell Host Microbe. 2020;27:173–175. doi: 10.1016/j.chom.2020.01.018. [DOI] [PubMed] [Google Scholar]
- 19.Snigdha S., Ha K., Tsai P., Dinan T.G., Bartos J.D., Shahid M. Probiotics: Potential novel therapeutics for microbiota-gut-brain axis dysfunction across gender and lifespan. Pharmacol. Ther. 2022;231 doi: 10.1016/j.pharmthera.2021.107978. [DOI] [PubMed] [Google Scholar]
- 20.Tian P., Chen Y., Zhu H., Wang L., Qian X., Zou R., Zhao J., Zhang H., Qian L., Wang Q., et al. Bifidobacterium breve CCFM1025 attenuates major depression disorder via regulating gut microbiome and tryptophan metabolism: A randomized clinical trial. Brain Behav. Immun. 2022;100:233–241. doi: 10.1016/j.bbi.2021.11.023. [DOI] [PubMed] [Google Scholar]
- 21.Dinan T.G., Stanton C., Cryan J.F. Psychobiotics: A novel class of psychotropic. Biol. Psychiatr. 2013;74:720–726. doi: 10.1016/j.biopsych.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Musazadeh V., Zarezadeh M., Faghfouri A.H., Keramati M., Jamilian P., Jamilian P., Mohagheghi A., Farnam A. Probiotics as an effective therapeutic approach in alleviating depression symptoms: an umbrella meta-analysis. Crit. Rev. Food Sci. Nutr. 2023;63:8292–8300. doi: 10.1080/10408398.2022.2051164. [DOI] [PubMed] [Google Scholar]
- 23.Qian X., Jiang J., Yang B., Zhao J., Wang G., Tian P., Chen W. Psychobiotics regulate purine metabolism to influence host emotional behavior. J. Agric. Food Chem. 2024;72:1561–1570. doi: 10.1021/acs.jafc.3c06422. [DOI] [PubMed] [Google Scholar]
- 24.Qian X., Tian P., Guo M., Yang H., Zhang H., Wang G., Chen W. Determining the emotional regulation function of Bifidobacterium breve: The role of gut metabolite regulation over colonization capability. Food Funct. 2024;15:1598–1611. doi: 10.1039/d3fo02739b. [DOI] [PubMed] [Google Scholar]
- 25.Wallace C.J.K., Milev R.V. The efficacy, safety, and tolerability of probiotics on depression: clinical results from an open-label pilot study. Front. Psychiatr. 2021;12 doi: 10.3389/fpsyt.2021.618279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson D., Letchumanan V., Thum C.C., Thurairajasingam S., Lee L.H. A microbial-based approach to mental health: The potential of probiotics in the treatment of depression. Nutrients. 2023;15:1382. doi: 10.3390/nu15061382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dehghani F., Abdollahi S., Shidfar F., Clark C.C.T., Soltani S. Probiotics supplementation and brain-derived neurotrophic factor (BDNF): A systematic review and meta-analysis of randomized controlled trials. Nutr. Neurosci. 2023;26:942–952. doi: 10.1080/1028415X.2022.2110664. [DOI] [PubMed] [Google Scholar]
- 28.Moludi J., Khedmatgozar H., Nachvak S.M., Abdollahzad H., Moradinazar M., Sadeghpour Tabaei A. The effects of co-administration of probiotics and prebiotics on chronic inflammation, and depression symptoms in patients with coronary artery diseases: a randomized clinical trial. Nutr. Neurosci. 2022;25:1659–1668. doi: 10.1080/1028415X.2021.1889451. [DOI] [PubMed] [Google Scholar]
- 29.Tyagi P., Tasleem M., Prakash S., Chouhan G. Intermingling of gut microbiota with brain: Exploring the role of probiotics in battle against depressive disorders. Food Res. Int. 2020;137 doi: 10.1016/j.foodres.2020.109489. [DOI] [PubMed] [Google Scholar]
- 30.Bi C., Guo S., Hu S., Chen J., Ye M., Liu Z. The microbiota–gut–brain axis and its modulation in the therapy of depression: Comparison of efficacy of conventional drugs and traditional Chinese medicine approaches. Pharmacol. Res. 2022;183 doi: 10.1016/j.phrs.2022.106372. [DOI] [PubMed] [Google Scholar]
- 31.Sun H., Zhao F., Liu Y., Ma T., Jin H., Quan K., Leng B., Zhao J., Yuan X., Li Z., et al. Probiotics synergized with conventional regimen in managing Parkinson’s disease. NPJ Parkinsons Dis. 2022;8:62. doi: 10.1038/s41531-022-00327-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng L., Wu H., Cai X., Zhang Y., Yu S., Hou Y., Yin Z., Yan Q., Wang Q., Sun T., et al. A Gpr35-tuned gut microbe-brain metabolic axis regulates depressive-like behavior. Cell Host Microbe. 2024;32:227–243.e6. doi: 10.1016/j.chom.2023.12.009. [DOI] [PubMed] [Google Scholar]
- 33.Vacca M., Celano G., Calabrese F.M., Portincasa P., Gobbetti M., De Angelis M. The controversial role of human gut Lachnospiraceae. Microorganisms. 2020;8:573. doi: 10.3390/microorganisms8040573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi W., Li Z., Wang W., Liu X., Wu H., Chen X., Zhou X., Zhang S. Dynamic gut microbiome-metabolome in cationic bovine serum albumin induced experimental immune-complex glomerulonephritis and effect of losartan and mycophenolate mofetil on microbiota modulation. J. Pharm. Anal. 2024;14 doi: 10.1016/j.jpha.2023.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agus A., Planchais J., Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. 2018;23:716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Z., Mu X., Cao Q., Shi Y., Hu X., Zheng H. Honeybee gut Lactobacillus modulates host learning and memory behaviors via regulating tryptophan metabolism. Nat. Commun. 2022;13:2037. doi: 10.1038/s41467-022-29760-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montgomery T.L., Eckstrom K., Lile K.H., Caldwell S., Heney E.R., Lahue K.G., D'Alessandro A., Wargo M.J., Krementsov D.N. Lactobacillus reuteri tryptophan metabolism promotes host susceptibility to CNS autoimmunity. Microbiome. 2022;10:198. doi: 10.1186/s40168-022-01408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laursen M.F., Sakanaka M., von Burg N., Mörbe U., Andersen D., Moll J.M., Pekmez C.T., Rivollier A., Michaelsen K.F., Mølgaard C., et al. Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat. Microbiol. 2021;6:1367–1382. doi: 10.1038/s41564-021-00970-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meltzer H. Serotonergic dysfunction in depression. Br. J. Psychiatr. Suppl. 1989;155:25–31. [PubMed] [Google Scholar]
- 40.Li C., Meng F., Garza J.C., Liu J., Lei Y., Kirov S.A., Guo M., Lu X.Y. Modulation of depression-related behaviors by adiponectin AdipoR1 receptors in 5-HT neurons. Mol. Psychiatr. 2021;26:4205–4220. doi: 10.1038/s41380-020-0649-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komoltsev I.G., Frankevich S.O., Shirobokova N.I., Volkova A.A., Onufriev M.V., Moiseeva J.V., Novikova M.R., Gulyaeva N.V. Neuroinflammation and neuronal loss in the hippocampus are associated with immediate posttraumatic seizures and corticosterone elevation in rats. Int. J. Mol. Sci. 2021;22:5883. doi: 10.3390/ijms22115883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim S., Yang S., Kim J., Chung K.W., Jung Y.S., Chung H.Y., Lee J. Glucocorticoid receptor down-regulation affects neural stem cell proliferation and hippocampal neurogenesis. Mol. Neurobiol. 2024;61:3198–3211. doi: 10.1007/s12035-023-03785-y. [DOI] [PubMed] [Google Scholar]
- 43.Pappolla M.A., Perry G., Fang X., Zagorski M., Sambamurti K., Poeggeler B. Indoles as essential mediators in the gut-brain axis. Their role in Alzheimer's disease. Neurobiol. Dis. 2021;156 doi: 10.1016/j.nbd.2021.105403. [DOI] [PubMed] [Google Scholar]
- 44.Cervantes-Barragan L., Chai J.N., Tianero M.D., Di Luccia B., Ahern P.P., Merriman J., Cortez V.S., Caparon M.G., Donia M.S., Gilfillan S., et al. Lactobacillus reuteri induces gut intraepithelial CD4+ CD8αα+ T cells. Science. 2017;357:806–810. doi: 10.1126/science.aah5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei G.Z., Martin K.A., Xing P.Y., Agrawal R., Whiley L., Wood T.K., Hejndorf S., Ng Y.Z., Low J.Z.Y., Rossant J., et al. Tryptophan-metabolizing gut microbes regulate adult neurogenesis via the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2021091118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin Y.T., Wu P.H., Lee H.H., Mubanga M., Chen C.S., Kuo M.C., Chiu Y.W., Kuo P.L., Hwang S.J. Indole-3 acetic acid increased risk of impaired cognitive function in patients receiving hemodialysis. Neurotoxicology. 2019;73:85–91. doi: 10.1016/j.neuro.2019.02.019. [DOI] [PubMed] [Google Scholar]
- 47.Rothhammer V., Mascanfroni I.D., Bunse L., Takenaka M.C., Kenison J.E., Mayo L., Chao C.C., Patel B., Yan R., Blain M., et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 2016;22:586–597. doi: 10.1038/nm.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yong C.C., Sakurai T., Kaneko H., Horigome A., Mitsuyama E., Nakajima A., Katoh T., Sakanaka M., Abe T., Xiao J.Z., et al. Human gut-associated Bifidobacterium species salvage exogenous indole, a uremic toxin precursor, to synthesize indole-3-lactic acid via tryptophan. Gut Microb. 2024;16 doi: 10.1080/19490976.2024.2347728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bienenstock J., Kunze W., Forsythe P. Microbiota and the gut–brain axis. Nutr. Rev. 2015;73:28–31. doi: 10.1093/nutrit/nuv019. [DOI] [PubMed] [Google Scholar]
- 50.Morais L.H., Schreiber H.L., 4th, Mazmanian S.K. The gut microbiota–brain axis in behaviour and brain disorders. Nat. Rev. Microbiol. 2021;19:241–255. doi: 10.1038/s41579-020-00460-0. [DOI] [PubMed] [Google Scholar]
- 51.Lee I.-C., Tomita S., Kleerebezem M., Bron P.A. The quest for probiotic effector molecules—unraveling strain specificity at the molecular level. Pharmacol. Res. 2013;69:61–74. doi: 10.1016/j.phrs.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 52.McFarland L.V., Evans C.T., Goldstein E.J.C. Strain-specificity and disease-specificity of probiotic efficacy: A systematic review and meta-analysis. Front. Med. 2018;5:124. doi: 10.3389/fmed.2018.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yadav R., Shukla P. An overview of advanced technologies for selection of probiotics and their expediency: a review. Crit. Rev. Food Sci. Nutr. 2017;57:3233–3242. doi: 10.1080/10408398.2015.1108957. [DOI] [PubMed] [Google Scholar]
- 54.Stenman L.K., Patterson E., Meunier J., Roman F.J., Lehtinen M.J. Strain specific stress-modulating effects of candidate probiotics: A systematic screening in a mouse model of chronic restraint stress. Behav. Brain Res. 2020;379 doi: 10.1016/j.bbr.2019.112376. [DOI] [PubMed] [Google Scholar]
- 55.Sakurai T., Odamaki T., Xiao J.Z. Production of indole-3-lactic acid by Bifidobacterium strains isolated from human infants. Microorganisms. 2019;7:340. doi: 10.3390/microorganisms7090340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lukić I., Getselter D., Koren O., Elliott E. Role of tryptophan in microbiota-induced depressive-like behavior: evidence from tryptophan depletion study. Front. Behav. Neurosci. 2019;13:123. doi: 10.3389/fnbeh.2019.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Y., Tian P., Wang Z., Pan R., Shang K., Wang G., Zhao J., Chen W. Indole acetic acid exerts anti-depressive effects on an animal model of chronic mild stress. Nutrients. 2022;14:5019. doi: 10.3390/nu14235019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qian X., Tian P., Lin G., Xu X., Wang G., Zhang H., Chen W. Detection of colonization capacity of probiotic Bifidobacterium breve CCFM1025 in the human gut. Future Microbiol. 2023;18:595–606. doi: 10.2217/fmb-2022-0131. [DOI] [PubMed] [Google Scholar]
- 59.Zhu G., Guo M., Zhao J., Zhang H., Wang G., Chen W. Integrative metabolomic characterization reveals the mediating effect of Bifidobacterium breve on amino acid metabolism in a mouse model of Alzheimer's disease. Nutrients. 2022;14:735. doi: 10.3390/nu14040735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou W., Sailani M.R., Contrepois K., Zhou Y., Ahadi S., Leopold S.R., Zhang M.J., Rao V., Avina M., Mishra T., et al. Longitudinal multi-omics of host–microbe dynamics in prediabetes. Nature. 2019;569:663–671. doi: 10.1038/s41586-019-1236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoedt E.C., Bottacini F., Cash N., Bongers R.S., van Limpt K., Ben Amor K., Knol J., MacSharry J., van Sinderen D. Broad purpose vector for site-directed insertional mutagenesis in Bifidobacterium breve. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.636822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leclercq S., Le Roy T., Furgiuele S., Coste V., Bindels L.B., Leyrolle Q., Neyrinck A.M., Quoilin C., Amadieu C., Petit G., et al. Gut microbiota-induced changes in β-hydroxybutyrate metabolism are linked to altered sociability and depression in alcohol use disorder. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108238. [DOI] [PubMed] [Google Scholar]
- 63.Pan T., Pei Z., Fang Z., Wang H., Zhu J., Zhang H., Zhao J., Chen W., Lu W. Uncovering the specificity and predictability of tryptophan metabolism in lactic acid bacteria with genomics and metabolomics. Front. Cell. Infect. Microbiol. 2023;13 doi: 10.3389/fcimb.2023.1154346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mao B., Li D., Zhao J., Liu X., Gu Z., Chen Y.Q., Zhang H., Chen W. Metagenomic insights into the effects of fructo-oligosaccharides (FOS) on the composition of fecal microbiota in mice. J. Agric. Food Chem. 2015;63:856–863. doi: 10.1021/jf505156h. [DOI] [PubMed] [Google Scholar]
- 65.Qian X., Si Q., Lin G., Zhu M., Lu J., Zhang H., Wang G., Chen W. Bifidobacterium adolescentis Is effective in relieving type 2 diabetes and may be related to its dominant core genome and gut microbiota modulation capacity. Nutrients. 2022;14:2479. doi: 10.3390/nu14122479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Genome sequencing data have been deposited at SRA and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original western blot images have been deposited at Mendeley Data and are publicly available as of the date of publication. The DOI is listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact (Peijun Tian, pjtian@jiangnan.edu.cn) upon request.
-
•
This study does not include original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact (Peijun Tian, pjtian@jiangnan.edu.cn) upon request.