Abstract
High efficacy, selectivity and cellular targeting of therapeutic agents has been an active area of investigation for decades. Currently, most clinically approved therapeutics are small molecules or protein/antibody biologics. Targeted action of small molecule drugs remains a challenge in medicine. In addition, many diseases are considered ‘undruggable’ using standard biomacromolecules. Many of these challenges however, can be addressed using nucleic therapeutics. Nucleic acid drugs (NADs) are a new generation of gene-editing modalities characterized by their high efficiency and rapid development, which have become an active research topic in new drug development field. However, many factors, including their low stability, short half-life, high immunogenicity, tissue targeting, cellular uptake, and endosomal escape, hamper the delivery and clinical application of NADs. Scientists have used chemical modification techniques to improve the physicochemical properties of NADs. In contrast, modified NADs typically require carriers to enter target cells and reach specific intracellular locations. Multiple delivery approaches have been developed to effectively improve intracellular delivery and the in vivo bioavailability of NADs. Several NADs have entered the clinical trial recently, and some have been approved for therapeutic use in different fields. This review summarizes NADs development and evolution and introduces NADs classifications and general delivery strategies, highlighting their success in clinical applications. Additionally, this review discusses the limitations and potential future applications of NADs as gene therapy candidates.
Subject terms: Drug delivery, Drug delivery
Introduction
The central dogma of genetics posits that nucleic acids carry human genetic information and play a crucial role in life processes, such as growth, development, and reproduction. Moreover, nucleic acids can be used to modify genetic information to treat various diseases.1,2 With the advancement of the life sciences, proteomics, and genomics methods, nucleic acid drugs (NADs) have been developed to translate and regulate nucleic acid functions.3–5 These drugs can achieve long-lasting efficacy through gene repression, replacement, and editing.4,6 Many studies have shown the feasibility of NADs in disease prevention and treatment.7–9 Thus, research and development on new classes of functional NADs are gradually emerging.
NADs are a class of gene therapy agents based on DNA, RNA, or synthetic oligonucleotide analogs. They have considerable potential for clinical applications, such as treating bacterial infections, tumors, and neuromuscular diseases.10–12 However, as negatively charged biological macromolecules, NADs have difficulty in crossing cellular membranes to enter cells. Additionally, they can be easily degraded by endogenous nucleases in plasma and tissues. Furthermore, few amounts of NADs that enter cells often become trapped by endosomes and subsequently degraded by lysosomes, considerably limiting their development and application.13–15 Currently, two main strategies exist to address the application challenges of NADs. One approach is to modify the nucleic acid structure to stabilize the properties of NADs and avoid recognition by the immune system. The other approach is to use delivery systems that facilitate their passage through cell membranes and ensure their localization to specific subcellular compartments. Consequently, the modification and transformation of NADs and the development of efficient, safe, and targeted delivery systems have become the primary focus of research and development on NADs.16,17
The accelerated development of the NADs field has relied on innovations and breakthroughs in foundational technologies, such as chemical synthesis, site-specific modifications, and delivery techniques. These advancements are crucial to ensure the safety, effectiveness, targeting, and applicability of NADs.18,19 Advancements in carrier technology and delivery systems have enhanced the biological activity of NADs, which improves their cellular targeting and uptake. Thus, the concentration and bioavailability of these drugs in target tissues are increased.20 Various delivery systems for NADs have been developed, including lipid nanoparticles (LNPs), cationic polymer complexes, and ligand-mediated nucleic acid molecular targeted delivery systems based on specific receptors, peptides, and other engineered carriers.21–27 However, these systems have several drawbacks, such as nonspecific distribution, inefficient cytoplasmic delivery, and suboptimal organelle targeting. Several studies have reported that more than one strategy is needed to address the delivery challenges. Thus, combining chemical structure modifications of nucleic acids with advanced drug delivery systems could achieve enhanced therapeutic effects.
This paper outlines the history of several significant molecular biology discoveries related to NADs, tracing key milestones from initial conceptualization to clinical applications. Then, we introduce the various NAD types and their modes of action, with an overview of both approved NADs and those currently in clinical trials. Then, we discuss the challenges associated with NADs development and explore strategies for overcoming the obstacles to in vivo delivery, including chemical modifications and delivery systems. Finally, we highlight the remaining challenges for NADs development, offering references for the design and clinical application of novel NADs.
Concept and historical development of NADs
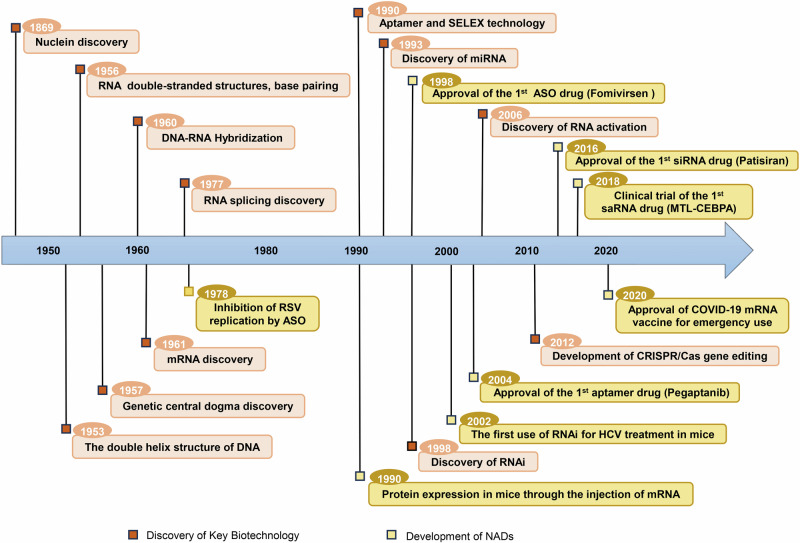
NADs development is inseparable from the major discoveries in fundamental molecular biology and the continuous observations of life activities (Fig. 1). In 1869, Friedrich Miescher discovered a new molecule called “nuclein” from white blood cells, which marked the beginning of DNA discovery.28 However, owing to the lack of advanced technologies at the time, the critical role of nucleic acids was not fully understood. The subsequent revelation of DNA’s double helix structure and the formulation of the central dogma of genetics clarified that nucleic acids are crucial participants in transmitting genetic information.1,2 Since then, researchers have understood that genetic information is encoded within nucleic acids and translated into proteins via complex mechanisms, and it plays a vital role in all life processes, such as growth, development, and reproduction.
Fig. 1.
Historical timeline of essential discoveries in fundamental molecular biology theory and critical developments in NADs therapy. The orange boxes represent major biological discoveries in nucleic acids development, including the discovery of DNA and RNA, as well as researchers’ exploration of special biological phenomena such as RNA interference, nucleic acid hybridization, and gene editing. The yellow boxes show the breakthrough progress in the clinical application of NADs based on the aforementioned biological phenomena. These include successful clinical application cases of NADs, such as the first ASO drug Fomivirsen, the first siRNA drug Patisiran, the first aptamer drug Pegaptanib, the COVID-19 mRNA vaccines, as well as clinical trials for NADs in development, such as the saRNA drug MTL-CEBPA
Additionally, there have been significant breakthroughs in NADs development including the discovery of RNA’s double-stranded structure and the phenomenon of nucleic acid hybridization.29 Early studies posited that single-stranded RNA could not form double-stranded structures. However, in 1956, Rich and Davies discovered that RNA could form double-stranded structures similar to those of DNA based on the principle of complementary base pairing. This result laid the foundation for developing RNA double-stranded drugs, including microRNAs (miRNAs) and small interfering RNAs (siRNAs).30 In 1960, Rich31 reported the phenomenon of DNA/RNA hybridization. In 1978, building on this discovery, Zamecnik and Stephenson32 used specific oligodeoxynucleotide chains that target the 35S RNA of the Rous sarcoma virus to inhibit virus replication, marking a prototype for the application of antisense oligonucleotide (ASO) drugs in disease treatment. As research on transcription and translation advanced, it was discovered that initial RNA transcripts typically require intron removal and the linkage of exons to form mature messenger RNA (mRNA), a process known as RNA splicing.33,34 RNA splicing is a crucial step in gene expression, and abnormalities in this process are the main cause of genetic variations and diseases.35,36 Dominski et al.37 found that ASOs targeting splicing sites can restore correct splicing of defective genes rather than only downregulating gene expression. This discovery provided a novel treatment strategy for diseases related to mis-splicing.
In 1998, Andrew Fire and Craig Mello reported that double-stranded RNA (dsRNA) had potent gene silencing effects in Caenorhabditis elegans,38 a discovery they termed RNA interference (RNAi). RNAi was quickly applied to inhibit the replication of the hepatitis C virus in mice,39 marking the earliest evidence of siRNA-mediated in vivo gene silencing. In 2006, they were awarded the Nobel Prize in Physiology and Medicine for their RNAi technology.40 Subsequently, researchers demonstrated that siRNA could be tailored to disrupt the expression of any pathogenic gene, propelling RNAi into the spotlight and fostering its active application in the treatment of various diseases. In 2010, the first human trial using RNAi technology was conducted to evaluate the therapeutic effect of siRNA for targeting the M2 subunit of ribonucleotide reductase in patients with melanoma.41 Since then, siRNA drugs have had issues with stability, immunogenicity, off-target effects, safety, and the delivery system. However, after advanced chemical modifications and the development of targeted delivery systems, the first siRNA drug approved by the Food and Drug Administration (FDA) in 2018 reignited interest in NADs.42
Notably, the discovery of RNA-dependent RNA polymerase and reverse transcriptase has been crucial for developing subsequent mRNA drugs.43,44 In 1984, Krieg and Melton used RNA polymerase extracted from viruses for in vitro transcription from engineered DNA templates, successfully achieving mRNA expression in cell-free systems.45 Thus, since the 1990s, in vitro-synthesized mRNA has been increasingly applied for protein replacement in preventive and therapeutic vaccines.46–48 In 2005, the discovery of pseudouridine (Ψ) modification addressed the immunogenicity issue of in vitro-synthesized mRNA,49 leading to the initiation of the first human trial of an mRNA vaccine against melanoma in 2008. In 2020, during the COVID-19 pandemic, the FDA authorized the emergency use of mRNA vaccines, providing effective measures for preventing and controlling the virus.50,51 This resulted in widespread public attention to NADs.52 In 2023, the Nobel Prize in Physiology and Medicine was awarded to Katalin Kariko and Drew Weissman for pioneering nucleoside base modification technology to decrease mRNA immunogenicity, further highlighting the critical role of chemical modification technologies in developing mRNA vaccines. This opened new clinical applications for NADs in treating human diseases.53,54
Simultaneously, discovering other types of nucleic acids and biological phenomena has further expanded the scope of NADs. Specifically, the emergence of gene-editing technology has provided a foundation for developing new therapies for genetic mutation diseases.55–57 Recently, the first gene therapy based on clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 technology has been approved for marketing,58 resulting in revolutionary changes in NADs development.
Classification and therapeutic mechanisms of NADs
NADs can be broadly divided into three categories based on their mechanisms of action. The first category includes NADs that target nucleic acids to regulate protein expression by promoting or inhibiting translation. This category primarily consists of ASOs, siRNAs, miRNAs, small activating RNAs (saRNAs), and the CRISPR/Cas system, which enables precise gene editing of genomic DNA.
The second category includes NADs that target proteins, with aptamers as the main examples. Unlike the first category, aptamers can directly and specifically bind to target proteins, functioning similarly to antibodies by providing a targeting mechanism.
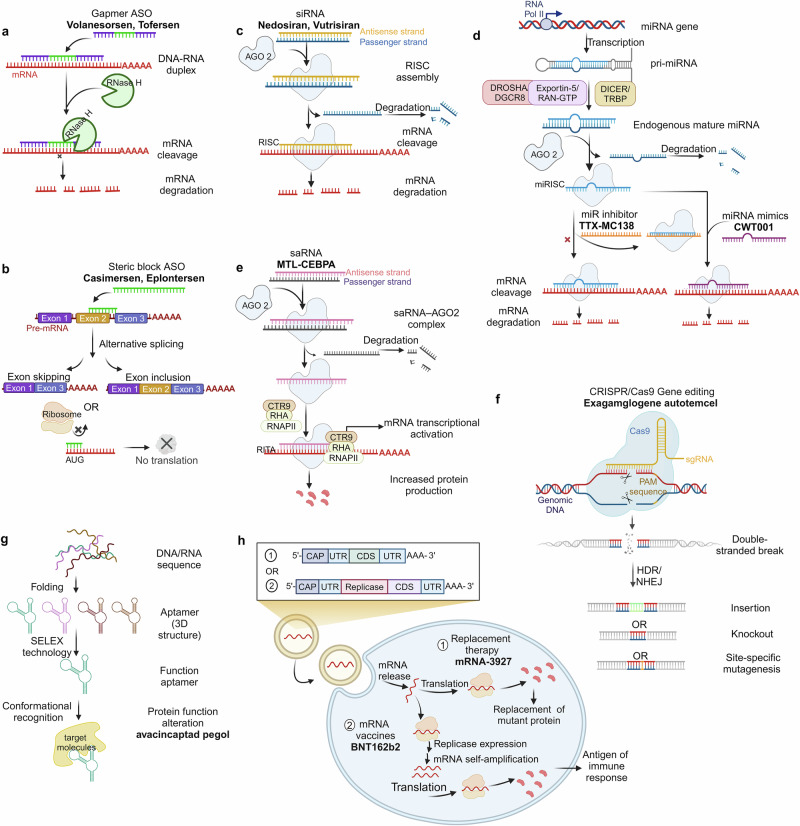
The third category includes NADs that express proteins, such as in vitro-transcribed mRNA, which can produce specific proteins in vivo to exert biological activity. This section briefly introduces the mechanisms of action of these different NAD types and highlights drugs successfully applied in clinical settings (Fig. 2).
Fig. 2.
Classification and therapeutic mechanisms of NADs. a Gapmer ASO (consisting of a DNA-based internal gap with RNA-like flanking regions) binds to target mRNA with high affinity to form an RNA-DNA duplex and participates in RNase H-mediated mRNA degradation. b Steric block ASO regulates functional target gene expression through exon skipping or exon inclusion or interrupts translation initiation by targeting and masking the AUG start codon of the target mRNA. c siRNAs form RISC with AGO2. While the passenger strand is discarded, the antisense strand binds to the target mRNA, downregulating the translation level of the target mRNA. d pri-miRNAs produced by miRNA gene transcription in the non-coding region are processed to form mature miRNAs with the help of a series of complexes (Drosha/DGCR8, Exportin-5/RAN-GTP, and Dicer/TRBP). miRNAs combine with the AGO2 to form miRISC. The activity of miRNAs can be inhibited by miRNA inhibitors that either form a complex with the mature miRNA loaded in the miRISC complex or by masking a target site via interactions with the specific transcript being targeted. e saRNAs recruit the RITA complex (including AGO2, CTR9, RHA, and RNAP II) to stimulate the initiation and extension of transcription. f CRISPR-mediated gene editing mainly uses Cas9 and sgRNA to introduce DSBs at specific positions in the genome effectively. DSBs are generally repaired by HDR or NHEJ, achieving insertion, knockout, and site-specific mutagenesis. g Aptamers screened by SELEX technology can recognize specific proteins by forming 3D structures. h Exogenous mRNAs introduced into cells undergo translation to proteins and facilitate protein function through protein replacement therapy and mRNA vaccines
NADs that target nucleic acids
ASOs
ASOs are artificially synthesized single-stranded oligonucleotide chains that regulate and target RNA’s function via specific binding according to the Watson-Crick base-pairing principle.59 The mechanisms of action of clinically used ASOs mainly include ribonuclease H (RNase H)-mediated degradation (Fig. 2a) and steric blockage mechanisms (Fig. 2b).60–62 RNase H-dependent ASOs, such as gapmers, bind to complementary mRNA, recruit RNase H to cleave mRNA, and thus block translation of the target gene. This results in the inhibition or reduction of the synthesis of the target protein. Representative drugs based on this mechanism include fomivirsen (Vitravene®), mipomersen (Kynamro®, Delisted), inotersen (Tegsedi®), volanesorsen (Waylivra®), and tofersen (Qalsody®).63–67 Because RNase H is active in the nucleus and cytoplasm, ASOs can also target other transcripts, such as long non-coding RNAs.68,69 Indeed, the pathogenesis of many diseases without clear protein targets is related to non-coding RNA, which can be used to predict the advantages of ASO in treating diseases.70,71
Additionally, ASOs can form a double-stranded structure complementary to miRNA, leading to its degradation and causing gene upregulation.72–74 Additionally, ASOs regulate transcription via steric hindrance, influencing specific splicing of pre-mRNA and selectively altering the expression of specific proteins.75,76 ASOs that employ this mechanism are splicing-switching oligonucleotides, which modulate functional target gene expression by skipping or including exons.77–79 For example, Golodirsen, an ASO drug targeting the human Duchenne muscular dystrophy (DMD) gene, was introduced in the USA in 2019 by Sarepta Therapeutics to treat DMD patients with confirmed mutations amenable to exon 53 skipping.80 Additionally, several studies have demonstrated that ASOs can disrupt translation initiation by targeting and masking the AUG start codon of the target mRNA.81 These discoveries have enabled ASOs to be applied in disease treatment via more diverse mechanisms.
siRNA
siRNA is a dsRNA molecule that is typically 19–23 base pairs long and is found naturally in various organisms or artificially synthesized.82,83 As a classical effector of RNAi, siRNA induces gene silencing by blocking mRNA translation.84,85 Unlike ASOs, siRNA-mediated gene silencing occurs via the RNA-induced silencing complex (RISC), not RNase H. Once mature siRNA enters the cell, it forms a complex with the Argonaute-2 (AGO2) protein (Fig. 2c).86,87 As the passenger strand of siRNA is degraded, the antisense strand binds to the target mRNA. It guides RISC to cleave the target sequence, thus achieving therapeutic effects by downregulating the translation of specific proteins.88–90 Based on this mechanism, researchers have designed siRNAs that target specific pathological genes to achieve specific gene silencing using RNAi.91,92 However, challenges, such as stability, specificity, and delivery obstacles, hindered progress in the early stages of development.93–96 Advancements in carrier technology and nucleic acid modification techniques have led to significant progress in overcoming these challenges, as exemplified by the first siRNA drug, patisiran (Onpattro®), which has been approved for treating hereditary transthyretin-mediated amyloidosis by degrading mRNA encoding transthyretin (TTR).97 Thus far, six siRNA drugs have received international approval. With the ongoing development of novel chemical modifications and targeted delivery systems, more siRNA drugs are anticipated to enter the market soon.
miRNA
Endogenous non-coding RNAs that have been discovered in eukaryotic organisms can act as gene regulators. Since the 1993 discovery of the first miRNA, lin-4, in the nematode C. elegans,98 miRNAs have been shown to participate in various biological functions and pathological mechanisms, including cell proliferation, differentiation, migration, disease occurrence, and progression.99 There has been extensive research on the regulatory mechanisms of miRNAs. With the assistance of complexes, such as Drosha/DGCR, Exportin-5/RAN-GTP, and Dicer/TRBP, primary miRNAs (pri-miRNAs) transcribed from the non-coding region of miRNA genes are processed to form mature miRNAs (Fig. 2d).100–105 miRNAs combine with Argonaute proteins to form the miRNA-induced silencing complex (miRISC), which can silence the target transcript via base pair complementation.106,107 Unlike siRNA-mediated gene silencing, miRNAs can simultaneously recognize and regulate the expression of multiple target mRNAs due to their low complementarity.108,109 The antisense strand of miRNA can bind to the target mRNA via complete and incomplete complementarity, leading to the cleavage of target mRNA and the inhibition of target gene expression.110,111 Contrary to the classical gene silencing mechanism, research has shown that miRNAs can interact with 3’-UTR to upregulate gene expression, indicating the complexity and diversity of miRNA regulatory mechanisms.112–114
As studies on the roles of miRNAs in diseases continue to be reported, there is anticipation for the potential use of miRNAs in different pathological processes. However, thus far, no miRNA drugs have been approved for the market.115,116 miRNA drugs’ ongoing development primarily includes two categories: miRNA mimics and miRNA inhibitors.117–121 miRNA mimics are synthetic dsRNA molecules that mimic the function of endogenous miRNAs. Similarly, miRNA inhibitors are single-stranded RNAs that are complementary to endogenous miRNAs, which can reduce the effect of gene silencing by specifically inhibiting miRNA.122,123 The miRNA drug MRG-201 (Remlarsen), developed by Viridian Therapeutics (formerly known as miRagen Therapeutics), simulates microRNA-29b to reverse regulate fibrosis, thus inhibiting fibrous proliferation in skin wounds. Clinical trials have been conducted to evaluate the efficacy and safety of MRG-201 in treating fibrotic diseases (NCT03601052).124 In addition to MRG-201, Viridian Therapeutics has several other miRNA-based drugs, including MRG-106 (Cobomarsen), which targets miR-155 to inhibit tumor development, and MRG-110, which targets microRNA-92 to promote angiogenesis.125,126 TTX-MC138 (NCT06260774 and NCT05908773), designed by TransCode Therapeutics, targets microRNA-10b in the treatment of pancreatic cancer.127 Despite the relatively slow progress in miRNA-related drug development compared with other NADs, their superior performance in treating tumors, heart failure, and diabetes indicates their promising prospects in clinical treatment.
saRNA
In addition to the classical gene silencing mechanism, the discovery of RNA activation has provided a new perspective on gene regulation. RNA activation is a gene regulation phenomenon mediated by a small dsRNA called saRNA, which targets gene promoter sequences to enhance the transcription of the target gene.128,129 After entering the cell via endocytosis, saRNA binds to AGO2. The sense strand is released and degraded, as AGO2 and the antisense strand are transported into the cell nucleus, where they bind to the promoter region (Fig. 2e). Subsequently, AGO2 recruits RNA polymerase-associated protein CTR9 homolog (part of the polymerase-associated factor 1 complex) and RNA helicase II (RHA) to form the RNA-induced transcriptional activation (RITA) complex. This complex interacts with RNA polymerase II (RNAP II) and stimulates the initiation and elongation of transcription.130,131 saRNAs are conserved in mammals and participate in the activation of various genes, such as vascular endothelial growth factor A (VEGFA), E-cadherin, progesterone receptor, and Kruppel-like factor 4.128,132–135 This conservation allows for the establishment of animal models and preclinical studies for saRNA-based therapies. Targeted activation therapy using saRNA has been validated in various disease animal models.129,136–140 However, clinical translation of saRNAs remains challenging due to the absence of an immune system in xenograft models and the complexity of drug action in the human body.141
The leading saRNA drug, MTL-CEBPA, which was developed by MiNA Therapeutics, targets the CCAAT/enhancer-binding protein alpha (CEBPA). Phase I trial results (NCT05097911) have demonstrated that MTL-CEBPA has a good safety profile for the treatment of hepatocellular carcinoma and may enhance the therapeutic effects of tyrosine kinase inhibitors by modulating immune suppression.142–144 In addition to MTL-CEBPA, MiNA Therapeutics has several saRNA candidates in preclinical development for undisclosed tumor types, metabolic diseases, and genetic disorders. RAG-01 is another saRNA candidate drug designed to treat bladder cancer by activating the expression of the human cyclin-dependent kinase inhibitor 1 A.145 Ractigen Therapeutics has submitted an Investigational New Drug application for RAG-01, and it is expected to become the second saRNA drug to enter clinical trials internationally.
CRISPR/Cas9 system
Gene editing is a genetic engineering technology that precisely modifies target genes via insertion, deletion, and site-specific mutation. The CRISPR/Cas9 system has gained widespread attention among the various genome-editing methods. As a prokaryotic adaptive immune system, CRISPR/Cas modules establish bacterial defense against exogenous nucleic acids. They have been widely used in gene editing owing to their high efficiency and accuracy.146–149 The CRISPR-mediated gene-editing system primarily involves Cas9 and single guide RNA (sgRNA) to introduce DNA double-strand breaks (DSBs) at specific positions in the genome. These DSBs are usually repaired by homologous directed repair (HDR) or nonhomologous end joining (NHEJ), achieving mutation or foreign gene insertion (Fig. 2f).150,151 CRISPR/Cas9 technology has been effective in various fields, including hematologic disorders, malignant tumors, and genetic diseases.151–153 However, this technology has several challenges, such as low delivery and HDR efficiency, off-target effects, and toxic side effects.154–157 Currently, the CRISPR/Cas9 system can enter cells for gene editing in three forms: the Cas9: sgRNA ribonucleoprotein complex, mRNA for Cas9 translation alongside a separate sgRNA, and a DNA plasmid that encodes both the Cas9 protein and sgRNA.158–160 Each method has its pros and cons. The plasmid system is relatively stable but has low cutting and editing efficiency, and continuous expression of Cas9 may increase off-target effects.161 mRNA and sgRNA are susceptible to degradation by nucleases. The Cas9/sgRNA ribonucleoprotein complex, while the most responsive mode of action, has a low off-target rate and low toxicity, but the large size of the complex complicates delivery.160 The delivery of the CRISPR/Cas9 system is a crucial issue limiting its clinical application. Like other NADs, researchers have focused on nanocarriers based on liposomes, chitosan, and other materials to achieve efficient delivery, laying a solid foundation for the clinical application of the CRISPR/Cas9 system.162–164 Exa-cel (Casgevy®) was approved in November 2023 as the first gene-editing therapeutic drug based on the CRISPR/Cas9 system for treating sickle cell disease (SCD) and transfusion-dependent β-thalassemia (TDT). Exa-cel stimulates artificial blood stem cells and progenitor cells in vitro to differentiate into red blood cells that produce high fetal hemoglobin levels.165,166 The launch of Exa-cel has filled the gap in gene-editing drugs and provided considerable motivation for developing other gene-editing therapies.
NADs that target proteins
Aptamers are single-stranded oligonucleotide molecules that have specific recognition functions obtained via iterative screening from large libraries of random oligonucleotides using the systematic evolution of ligands by exponential enrichment (SELEX) technology.167–169 Unlike other nucleic acid-based drugs, aptamers specifically recognize and bind target molecules, such as peptides, proteins, viruses, bacteria, and cells, relying on their unique three-dimensional conformation. This is similar to the conformational recognition that mediates antibody–antigen interactions and complex formation (Fig. 2g).170–173 However, compared with antibodies, aptamers have several advantages, including high thermal and physiological stability, low immunogenicity, and a wider range of target specificity. Since their first report in 1990, SELEX technology has been continuously improved,174,175 diversifying the selection and development of high-affinity aptamers.176,177 Aptamers have shown promising applications in treating cancer and ophthalmic and cardiovascular diseases (CVDs).178–180
In 2004, the FDA approved the first aptamer drug, pegaptanib (Macugen®), to treat choroidal neovascularization caused by neovascular age-related macular degeneration (AMD).181 However, Pegaptanib was withdrawn from the market owing to its poor efficacy and competition from anti-VEGF antibody drugs, such as Lucentis.64 Despite this, research on aptamers continues. In August 2023, avacincaptad pegol was approved for treating geographic atrophy (GA) secondary to dry AMD. Complex cascade overactivity is likely instrumental in AMD pathology. A crucial complement component, C5, has become a primary therapeutic target for many inflammatory diseases, including AMD.182 As a C5 inhibitor, avacincaptad pegol has been shown to slow GA progression by targeting the source of retinal cell death. Additionally, AS1411 is a candidate drug that targets nucleolin, which has been confirmed to be effective for treating renal cell carcinoma,183 glioma,184 and acute myeloid leukemia.185 In addition, aptamers are widely used in drug delivery, clinical diagnostics, and biosensing.186–188 This paper primarily focuses on their application as therapeutic drugs. Thus, we will not describe their other uses in detail further.
NADs that express proteins
mRNA, a single-stranded polynucleotide that carries genetic information, is essential for expressing encoded proteins within cells, thus exerting corresponding biological functions via these proteins. Consequently, the concept of using mRNA as NADs has been proposed. In 1990, Wolff et al.46 injected in vitro-synthesized mRNA into mouse skeletal muscle and successfully induced the expression of specific proteins. They demonstrated the feasibility of using in vitro-synthesized mRNA as an information carrier to guide somatic protein synthesis.
However, the development of mRNA-based drugs has resulted in several challenges. Unmodified mRNA can induce Toll-like receptor-mediated immune responses, leading to blocked protein synthesis. Kariko and Weissman discovered that nucleobase modifications could protect mRNA from triggering inflammatory responses.49,54 In addition, the large size of mRNA and its susceptibility to degradation by nucleases result in low cellular uptake, further limiting its application. To overcome these obstacles, researchers have developed various delivery systems, including lipids, peptides, and polymers, for mRNA delivery both in vitro and in vivo.189–191
In contrast to the previously mentioned NADs that exert therapeutic effects by directly binding mRNA or proteins, two strategies for the use of mRNA drugs have been attempted (Fig. 2h). One strategy is protein replacement therapy, which involves introducing exogenous mRNAs into cells to express functional proteins or supplement deficient ones.192,193 For example, one research group used LNPs to deliver mRNA encoding erythropoietin into mouse fetuses, thus increasing erythropoietin protein levels in the mouse bloodstream.192 Additionally, mRNA therapy has been applied to treat patients with deficiencies in essential enzyme genes, such as argininosuccinate lyase, ornithine transcarbamylase, and methylmalonyl-CoA mutase (MUT), therefore restoring enzyme levels and mitigating deficiencies.194–196 Moderna, a company dedicated to mRNA therapy, has several mRNA drugs based on enzyme replacement therapy in clinical trials. For instance, mRNA-3927, which encodes the alpha and beta subunits of the propionyl-CoA carboxylase enzyme, is designed to treat propionic acidemia. A Phase I/II trial (NCT05130437) for this indication has been initiated in pediatric patients to evaluate the long-term safety of mRNA-3927.197 Additionally, mRNA-3704 and mRNA-3705 encode MUT, which is designed to treat methylmalonic acidemia and is currently under investigation.
The second approach involves mRNA vaccines, which activate the body’s immune response to combat infectious diseases and tumors by directly translating mRNA-containing antigen proteins. Compared with traditional inactivated vaccines, mRNA vaccines have advantages such as cell-free production, high production efficiency, and low cost. Thus, they are quite promising for addressing sudden epidemic infectious diseases. During the COVID-19 pandemic, mRNA prophylactic vaccines were crucial among all candidate vaccine types. The FDA authorized the emergency use of BNT162b2, which was developed by Pfizer-BioNTech, and mRNA-1273, developed by Moderna.198,199 Moreover, the development of mRNA vaccine technology has been propelled by large-scale clinical trials of mRNA vaccines. For example, ARCov, which was jointly developed by Abogen Biosciences, the Academy of Military Medical Sciences, and Walvax Biotechnology, has addressed the issue of poor thermal stability of mRNA vaccines.200,201 Additionally, preventive vaccines against influenza viruses,202,203 respiratory syncytial virus,204 rabies virus,205 and other viruses, as well as cancer-targeted therapeutic vaccines,206–208 are being continuously researched and developed, which further reflect the application prospects of mRNA vaccines.
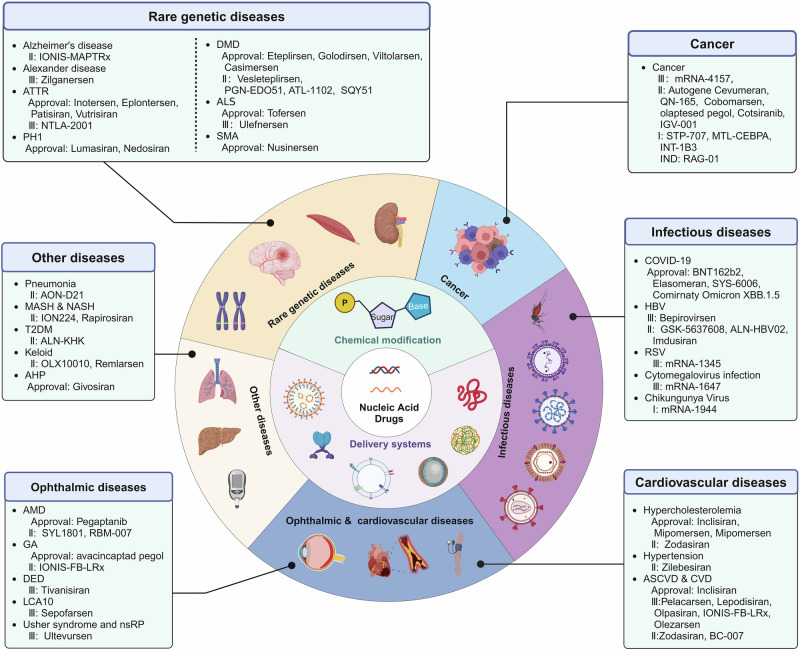
In summary, in recent decades, NADs development has undergone significant progress and achieved considerable results. More than 20 products based on ASOs, aptamers, siRNA, and mRNA have been approved for marketing to treat various rare genetic disorders (Table 1). Several companies worldwide have been active in this field, attracting substantial investment, and the market development space is expected to expand further.
Table 1.
NADs approved for clinical application
| Classification | Drug (brand name) | Company | Indication | Dose (route) | Target (organ) | Modification & delivery | Approval year | Ref(s) |
|---|---|---|---|---|---|---|---|---|
| ASO | Fomivirsen (Vitravene) | Ionis | CMV retinitis | 330 μg per eye once every 4 weeks (ITV) | CMV UL123 (eye) | PS | 1998 (delisted) | 234,578 |
| Mipomersen (Kynamro) | Ionis | HoFH | 200 mg once weekly (SC) | ApoB-100 (liver) | 2’-MOE | 2013 (delisted) | 579 | |
| Eteplirsen (Exondys 51) | Sarepta Therapeutics | DMD | 30 mg kg−1 once weekly (IV) | Exon 51 of DMD (muscle) | PMO | 2016 | 497,498 | |
| Nusinersen (Spinraza) | Ionis & Biogen | SMA | 12 mg once every 4 months (IT) | Exon 7 of SMN2 (CNS) | 2’-MOE | 2016 | 580,581 | |
| Inotersen (Tegsedi) | Ionis | hATTR | 300 mg once weekly (SC) | TTR (liver) | 2’-MOE | 2018 | 490,491 | |
| Volanesorsen (Waylivra) | Ionis | FCS | 300 mg once weekly (SC) | ApoC-III (liver) | 2’-MOE | 2019 | 66,582 | |
| Golodirsen (Vyondy 53) | Sarepta Therapeutics | DMD | 30 mg kg−1 once weekly (IV) | Exon 53 of DMD (muscle) | PMO | 2019 | 80 | |
| Viltolarsen (Viltepso) | Nippon Shinyaku | DMD | 80 mg kg−1 once weekly (IV) | Exon 53 of DMD (muscle) | PMO | 2020 | 263,583 | |
| Casimersen (Amondys 45) | Sarepta Therapeutics | DMD | 30 mg kg−1 once weekly (IV) | Exon 45 of DMD (muscle) | PMO | 2021 | 264,496 | |
| Tofersen (Qalsody) | Biogen | ALS | 100 mg once every 28 days (IT) | SOD1(CNS) | PS/ 2’-MOE | 2023 | 67,505 | |
| Eplontersen (Wainua) | Ionis & AstraZeneca | ATTRv-PN | 45 mg once monthly (SC) | TTR (liver) | LICA | 2023 | 492 | |
| siRNA | Patisiran (Onpattro) | Alnylam | hATTR | 0.3 mg kg−1 once every 3 weeks, max:30 mg (IV) | TTR (liver) | 2’-OME & LNP | 2018 | 97 |
| Givosiran (Givlaari) | Alnylam | AHP | 2.5 mg kg−1 once monthly (SC) | ALAS1(liver) | PS/ 2’-OME & GalNAc | 2019 | 584 | |
| Lumasiran (Oxlumo) | Alnylam | PH1 | 3 or 6 mg kg−1 once every 3 months (SC) | HAO1(liver) | PS/ 2’-OME & GalNAc | 2020 | 510 | |
| Inclisiran (Leqvio) | Alnylam & Novaetis | primary hypercholesterolemia | 284 mg at 0 and 3 months, and then once every 6 months (SC) | PCSK9(liver) | PS/ 2’-OME & GalNAc | 2020 | 539 | |
| Vutrisiran (Amvuttra) | Alnylam | hATTR-PN | 25 mg once every 3 months (SC) | TTR (liver) | PS/ 2’-OME & GalNAc | 2022 | 493 | |
| Nedosiran (Rivfloza) | Novo Nordisk & Dicerna | PH1 | 128 or 160 mg once monthly (SC) | LDHA (liver) | PS/ 2’-OME & GalXCTM | 2023 | 330 | |
| CRISPR/Cas | Exagamglogene autotemcel (Casgevy) | Vertex & CRISPR Therapeutics | TDT & SCD | Once (IV) | BCL11A | / | 2023 | 165,166 |
| Aptamers | Pegaptanib (Macugen) | EyeTech Pharmaceuticals & Pfizer | AMD | 0.3 mg once every 6 weeks (ITV) | VEGF-165 (eye) | pegylated | 2004 (delisted) | 181 |
| avacincaptad pegol (Izervay) | Iveric Bio | GA | 2 mg once monthly (ITV) | complement C5 (eye) | pegylated | 2023 | 585 | |
| mRNA | BNT162b2 (Comirnaty) | Pfizer & BioNTech | COVID-19 | IM | SARS-CoV-2(liver) | LNP | 2020 | 199 |
| Elasomeran (Spikevax) | Modern | COVID-19 | IM | SARS-CoV-2(liver) | LNP | 2020 | 586 | |
| mRNA-1345 (mRESVIA) | Modern | Respiratory syncytial virus infection | IM | Respiratory syncytial virus (lung) | LNP | 2024 | 587 |
Current challenges in NADs development
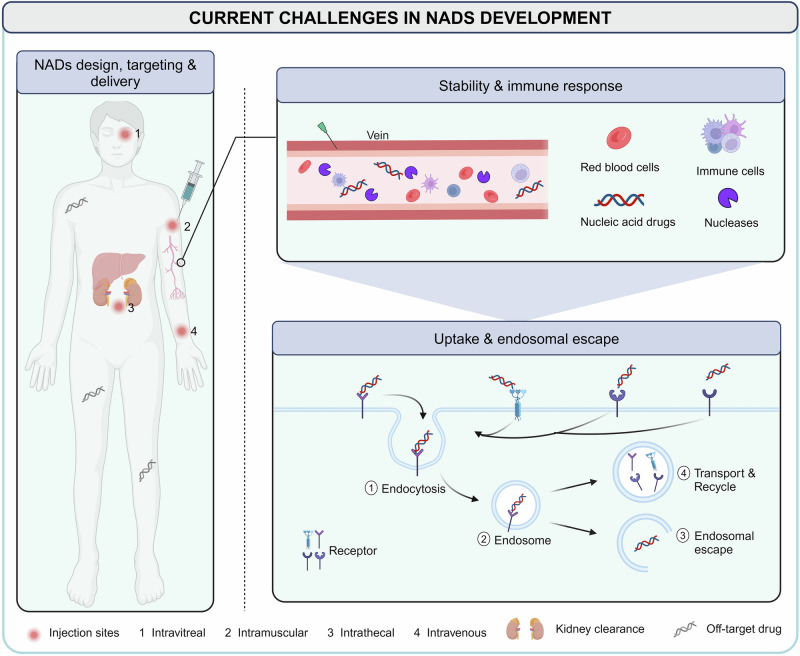
Unlike traditional small molecules and antibody drugs that exert their pharmacological effects on proteins, most NADs directly regulate gene expression, offering a broader range of targets. This is particularly valuable for addressing genes with defective proteins that are difficult to be targeted with conventional drugs, showing considerable potential in treating rare, chronic, infectious diseases and other metabolic disorders. Despite these advantages, researchers must accurately identify the genetic information related to the disease and choose the appropriate type of NADs based on the mechanism of action.209,210 For targeted NADs, including ASO and siRNA, when the relevant genetic information of the disease is determined, lead compounds can be designed for the gene sequence to avoid off-target effects during development.211 The efficacy of aptamer drugs is related to their sequence and conformation. SELEX technology has been applied to better screen specific sequences with high affinity for a target from a randomly generated single-stranded nucleic acid sequence library.212 Understanding the relationship between the mRNA sequence, structure, function, and stability for mRNA development is important to ensure the maximum functional protein output of delivered mRNA molecules.213 In recent years, the NADs sequence design process has been accelerated by the development of advanced bioinformatics tools, considerably reducing the time and costs.209,210,214,215 Nonetheless, the major obstacle in NADs development involves how to reach target cells to fully achieve therapeutic benefits (Fig. 3).13,216,217 The most relevant challenges can be summarized as follows.
Fig. 3.
Current challenges in NADs delivery. NADs are administered in many ways, such as intravitreal, intramuscular, intrathecal, and intravenous injection. For systemic delivery, NADs must first overcome renal clearance, nuclease degradation, immune system recognition, and drug off-target until reaching target tissues and organs. Subsequently, NADs successfully reach the target cells, enter the cell via endocytosis, enter the endosomes, and escape successfully to achieve the desired therapeutic effect. It is difficult for negatively charged NADs to cross the phospholipid bilayer on the surface of the cell membrane, which usually requires the help of carriers to recognize receptors or chemical modification of NADs to change properties
Pharmacokinetics, stability, and degradation
Naked nucleic acids have poor in vivo stability and can degrade in the bloodstream. They undergo either enzymatic (nucleases and RNAse) or chemical (oxidation and hydrolysis) degradation in the blood and tissue fluids or are filtered and cleared by the kidneys.218 Researchers have implemented several chemical modifications to enhance NADs stability, but almost all these methods can affect the efficacy and safety.219
Immunogenicity
Exogenous nucleic acids can be recognized as exogenous signals by pattern recognition receptors in the immune system, triggering immune responses that compromise the structural integrity and stability of the nucleic acids. Careful design and modification of NADs can mitigate immunogenicity, but this requires extensive testing.220,221
Targeting problems
Therapeutic nucleic acids often lack sufficient targeting ability in vivo. This insufficient targeting leads to low NADs concentrations at the disease site and unintended gene silencing or activation, potentially causing safety issues by requiring higher doses. Designing particular nucleic acids and conducting thorough off-target screening are critical for the efficient clinical application of NADs.
Uptake efficiency and endosomal escape
Efficient uptake of NADs into cells is difficult owing to their size, charge, and hydrophilicity. Negatively charged nucleic acids do not easily cross the negatively charged lipid bilayer of the cell membrane. Additionally, endosomal escape is a significant barrier as nucleic acids often become trapped in endosomes and degraded. After entering cells, NADs are often captured by endosomes. Only those NADs that successfully escape the endosomes can exert their therapeutic effects. Inadequate endosomal escape can result in reduced efficacy and increased off-target toxicity.222
In addition to the previous issues, NADs face manufacturing and scalability challenges. Producing NADs at scale with consistent quality requires advanced manufacturing technologies and stringent quality control measures. This results in a relatively high cost, affecting accessibility and affordability for patients.
Strategies to improve NADs performance
Because of these challenges, accurately delivering drugs to target tissues and improving patient quality of life have become the core objectives for NADs development and research. Recently, significant advancements have been made in chemical modification technologies and delivery vehicles for NADs, considerably enhancing delivery efficiency.16 Thus far, dozens of NADs have benefited from successful chemical modification and carrier delivery and have earned FDA approval. For example, the success of COVID-19 vaccines is mainly due to advancements in base modification and LNP delivery systems. Therefore, the focus of NADs research and development has shifted toward improving nucleic acid modifications and developing efficient, safe, and targeted delivery systems.
Chemical modification
Various chemical modification methods have been introduced with the continuous progress of chemical synthesis and modification technologies to provide more precise treatments.223,224 NADs can be precisely modified to improve their efficacy and stability while decreasing toxicity and immunogenicity.223,224 The most widespread modification methods include backbone, ribose, and nucleobase modifications.225–228
Backbone modification
The modification strategies used in first-generation NADs have focused on modification of the phosphate backbone, mainly using other types of groups to replace the non-bridging oxygen atoms in the phosphate backbone, such as phosphorothioate (PS), methyl phosphate, and boranophosphate.229 The most often used backbone modification method is PS modification, where the oxygen atom is replaced by sulfur.230 This modification can improve the resistance of nucleic acids to nucleases, enhance blood stability, reduce renal clearance, and prolong the circulation time of drugs in vivo by improving their binding to plasma proteins.231–233 However, it has been determined that while improving stability, PS modification can induce inflammatory responses and produce hepatotoxic effects.231 Fomivirsen, a representative drug modified by PS, was withdrawn due to its limited therapeutic effect and inflammatory reaction.234 This has inspired the exploration of new modification technologies to reduce the adverse effects associated with PS modification.
Ribose modification
Ribose modification is another common strategy that has had notable success. Changes to the 2’ position group can affect nucleic acid stability and affinity,227 with common modifications including 2’-fluoro (2’-F), 2’-O-methoxyethyl (2’-MOE), and 2’-O-methyl (2’-OMe).17,228,235 These modifications considerably increase nuclease tolerance and prolong the half-life of nucleic acids while effectively avoiding the inflammatory reactions triggered by PS modification, demonstrating a higher safety and activity. Furthermore, synergistic modifications combining PS and 2’-MOE have considerably improved the physicochemical properties and reduced NADs side effects. Dual modifications, such as 2’,4’- and 2’,5’-sugar modifications, have been employed in the development of siRNA therapeutics to further expand the potential of ribose modifications.236 Beyond specific site modifications, altering the sugar ring provides another NADs design strategy.237 A locked nucleic acid (LNA) is a nucleic acid analog with a unique bicyclic structure, where the C4’ and O2’ atoms are connected by different methylene bridges, forming a stable C3’-endo conformation.238 An LNA adheres to Watson-Crick base pairing and has a strong affinity for DNA and RNA. However, owing to its bicyclic backbone structure limitations, an LNA can only spontaneously form A-type hybrid duplexes with target nucleic acid strands.239,240 These hybrids have strong thermal stability and can activate RNase H degradation activity under specific conditions, indicating LNA’s potential as an antisense drug. Overall, LNA’s good stability, high binding specificity, and strong nuclease resistance make it advantageous for in vitro and in vivo applications.17 In contrast, unlocked nucleic acid (UNA) is a flexible RNA mimic that lacks chemical bonds between the ribose ring’s C2’ and C3’ atoms. A UNA can attach to oligonucleotide monomers to form hybrids, regulating their flexibility and thermal stability.241 A UNA supports RNase H activity, which is beneficial for antisense-based nucleic acid therapies.242 Additionally, UNA modification at the siRNA terminus improves siRNA stability and silencing efficacy while considerably reducing off-target effects, highlighting its potential in the development of new therapeutic siRNAs.243,244
Nucleobase modification
Nucleobases are essential components of nucleic acids, and changes to their structure can affect the stability, biological activity, and immunogenicity of nucleic acids.245,246 By modifying specific sites on nucleobases, the stability and affinity of nucleic acids can be greatly improved. Canonical nucleoside analogs formed from nucleobase modifications include 5-methylcytidine (m5C), 5-fluorouracil (5-FU), N7-methylguanosine (m7G), pseudouridine (Ψ), N6-methyladenosine (m6A), and 2’-deoxy-2’-fluoro-uridine (2’-FU).247–250 Modifications, such as m5C and Ψ, reduce the activity of cytokines and biomarkers in dendritic cells, helping mRNA evade the immune system.49,251,252 Yoshida et al. found that base modification could considerably reduce the hepatotoxicity of gapmer ASO,245 providing insights for developing new gapmer ASOs. Additionally, in RNAi processes, the 5’ nucleobase affects the binding activity of AGOs to siRNA, thus reducing target cleavage activity.253,254 Therefore, chemical modification at this specific position could improve siRNA binding affinity.225
Nucleic acid analogs offer new modification technologies, considerably enhancing the stability and reducing the immunogenicity of NADs in vitro and in vivo.255,256 Replacing the phosphate backbone by other group improves target affinity, nuclease resistance, and pharmacokinetic properties of nucleic acids. For peptide nucleic acids (PNAs), the sugar-phosphate backbone is replaced by a peptide backbone, which retains the specific binding ability with DNA/RNA and offers higher affinity and better stability than natural nucleic acids.257 Due to these backbone modifications, PNAs show improved resistance to nuclease and protease digestion.258 These characteristics make PNAs powerful tools for disease diagnosis and treatment.259,260 However, their poor distributions in vivo and low cellular uptake are challenges for their clinical application. Phosphorodiamidate morpholino oligonucleotides (PMOs) are another class of nucleic acid analogs with significant potential, which are characterized by a six-membered morpholine ring backbone.261 Several PMO-based NADs have been approved for treating DMD, including eteplirsen (Exondys 51®),262 golodirsen (Vyondys 53®),80 viltolarsen (Viltepso®),263 and casimersen (Amondys®).264,265 Both PMOs and PNAs are neutral nucleic acid analogs with weak binding to plasma proteins, allowing them to be easily cleared by the kidneys.266 In practice, high doses are required to maintain the therapeutic effects, which can lead to corresponding toxicities and side effects.267
Delivery systems
LNPs
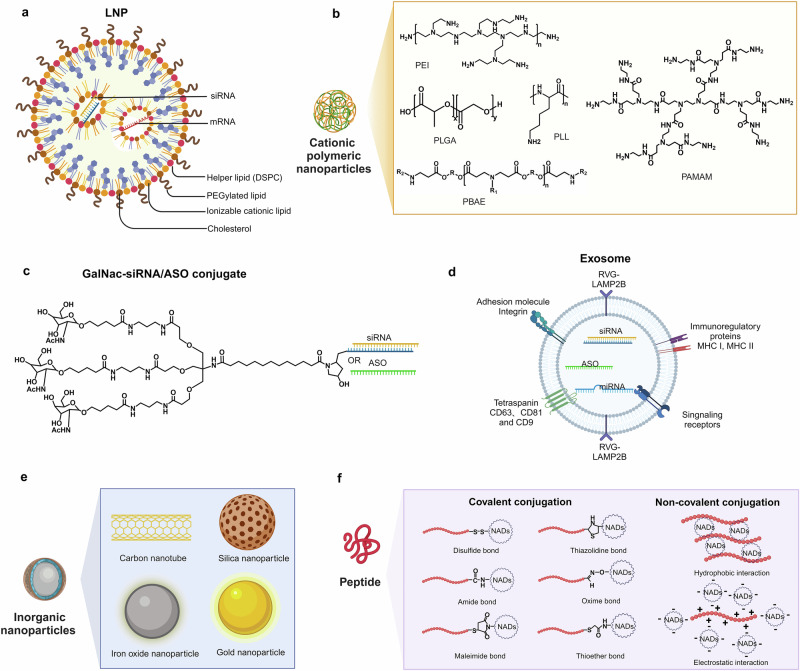
LNPs are self-assembled nanostructures with diameters of approximately 100 nm, capable of combining with negatively charged nucleic acids through electrostatic interactions. They have been widely used for NADs delivery due to their excellent compatibility with cell membranes.268,269 Classical LNPs typically include ionizable lipids (ILs) or cationic lipids (CLs), auxiliary lipids, cholesterol, and Polyethylene glycol (PEGylated) lipids (Fig. 4a).270 These components self-assemble into monodisperse nanoparticles in specific proportions through intermolecular interactions, encapsulating NADs in their core to protect them from nuclease degradation during delivery. Furthermore, modifying the surface properties of LNPs can enhance the uptake by specific cells and alter the distribution of NADs.271
Fig. 4.
Chemical structure of NADs delivery systems. a There are four types of LNPs: ILs (or CLs), auxiliary lipids, cholesterol, and PEGylated lipids. b Schematic and molecular structural formula of cationic polymeric nanoparticles. c Triantennary GalNAc moiety conjugated to siRNA or ASO. d Engineered exosome with RVG-LAMP2B displayed on the outer surface. The exosome contains therapeutic nucleic acids, such as siRNA, microRNA, and ASO. e Schematic of inorganic nanoparticles. f Peptide-assisted NADs delivery strategies. The methods of covalent conjugation include disulfide, amide, maleimide, thiazolidine, oxime, and thioether bond. The methods of non-covalent complexation include hydrophobic and electrostatic interactions
In early designs, CLs were permanently charged to bind the cell membrane and NADs effectively. However, their cytotoxicity limited their application in nanoparticle design.272,273 ILs are crucial for delivery efficacy, as they can encapsulate nucleic acids in LNPs while affecting the uptake and endosomal escape of NADs. ILs remain neutral at physiological pH, reducing the toxicity and immunogenicity of drugs. At low pH values, ILs acquire a positive charge, interacting with the negatively charged endosomal membrane and transforming its planar bilayer structure into a more hexagonal configuration. This transformation promotes the escape and release of NADs encapsulated in LNPs.274,275 1,2-Dioleyloxy-3-dimethylaminopropane (DODMA) and its analog 1,2-dioleyloxy-3-(dimethylamino) propane (DODAP) were among the first ILs used for RNA delivery.276 Continuous optimization of ILs revealed that the pKa of ILs is a critical factor in delivery efficacy. An optimal pKa value of 6.2–6.5 maximizes NADs efficacy.277 Researchers have synthesized ILs, such as DLin-KC2-DMA and DLin-MC3-DMA, with the latter used as a novel component for encapsulating nucleic acids. DLin-MC3-DMA was integral to the development of the first FDA-approved siRNA drug, Onpattro®, and has been proven effective for mRNA delivery in vivo.278,279 In addition, this technology is used in the clinically approved vaccines ALC-0315 (BNT162b2-Comirnaty®) and SM-102 (mRNA-1273-Spikevax®), where the pKa value ranges from 6.1 to 6.7.278,280 Unlike DLin-MC3-DMA, these vaccines incorporate an ester-based structure in the hydrophobic tail of lipids, enabling faster lipid clearance and improving product tolerance.281
PEGylated lipids are another crucial component of LNPs that significantly influence their size, stability, and in vivo distribution.282 However, they have encountered several challenges.283–285 PEG modification can inhibit the binding of apolipoprotein E to LNPs, a key mechanism for LNP uptake by the liver, thereby affecting liver uptake of LNPs. Moreover, repeated administration of PEGylated products may produce PEG antibodies, potentially reducing drug efficacy.286,287
1,2-Distearoyl-sn-glycero-3-phosphocholine (DSPC) and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) are common helper lipids in commercially available LNPs. DSPC, a phospholipid molecule with a choline head group and two saturated octadecyl chains, is a critical excipient in Onpattro® and COVID-19 mRNA vaccines.288 Alternatively, DOPE disrupts the endosomal membrane structure by forming a hexagonal crystalline phase, facilitating the release of encapsulated RNA from endosomes.275 Addressing the limitations of traditional phospholipid structures, Liu et al.289 developed hundreds of ionizable phospholipids called iPhos, which were capable of promoting endosomal membrane fusion and inducing hexagonal phase transitions. Cholesterol possesses a strong membrane fusion capacity, facilitating the internalization of NADs and their entry into the cytoplasm. Studies indicate that apolipoprotein E can cause cholesterol to migrate from the core of LNPs to the outer lipid layer, potentially altering the surface properties of LNPs.290,291 Additionally, Siegwart’s team introduced the fifth component, selective organ targeting, to achieve precise and universal extrahepatic-targeted mRNA delivery strategies.292
Approximately 150 molecules employ LNP technology, with nearly 75% dedicated to delivering NADs in RNA therapy, including ASOs, siRNA, and mRNA.293,294 Several new RNA therapeutics are in late-stage clinical trials, such as mRNA-1944 (NCT03829384)295 and NTLA-2001 (NCT06128629).296 Vaccines, such as BNT162b2 and mRNA-1273, have been developed based on mRNA LNP technology against COVID-19, which demonstrate the feasibility of LNPs in delivering mRNA encoding the spike protein of syndrome coronavirus 2 (SARS-CoV-2), thus eliciting an immune response against the virus.297
Cationic polymeric nanoparticles
Delivery systems for NADs based on synthetic or natural cationic polymers are complex structures formed through electrostatic interactions between polymers carrying cationic groups and NADs. Upon cell internalization via endocytosis mechanisms, these nanoparticles disrupt endosomal membranes through the proton sponge effect, facilitating the intracellular delivery of exogenous nucleic acids. Various cationic polymers, including polyethylenimine (PEI), polyamidoamine dendrimers (PAMAM), poly-L-lysine (PLL), poly-β-aminoester (PBAE), and poly-lactic-co-glycolic acid (PLGA), have been developed for this purpose (Fig. 4b).233,298 As a representative cationic polymer, PEI is widely used in non-viral gene vectors in linear PEI (LPEI) or branched PEI (BPEI).299,300 BPEI exhibits stronger gene compression ability than LPEI due to its structure containing primary, secondary, and tertiary amine groups every two carbon atoms, enhancing electrostatic interactions with nucleic acids.301 With a molecular weight of 25 kDa, PEI is considered the “gold standard” for gene transfection due to its high efficiency, but it induces significant cytotoxicity and lacks biodegradability.302,303 PLL, another widely used cationic polymer, is synthesized through ring-opening polymerization of its monomers.304 Its positively charged amino acids interact electrostatically with the phosphate backbone of NADs, enabling the delivery of RNA and DNA.305,306 Despite the biodegradability and biocompatibility of PLL, its in vivo activity remains limited due to its high toxicity, low transfection efficiency, and poor endosomal escape ability. Functional modifications, such as chloroquine, histidine, PEG, or PEI, have been incorporated into PLL to address these limitations.304,307,308 Recent advancements include PLL nanoparticles coated with hyaluronic acid shells, significantly reducing cytotoxicity, enhancing cellular uptake, and improving gene expression.305
N-Acetylgalactosamine (GalNAc)
Most approved products based on bio-conjugated delivery systems use the GalNAc-coupled delivery system developed by Alnylam Pharmaceuticals. GalNAc, a carbohydrate compound, exhibits a high affinity for the asialoglycoprotein receptor (ASGPR).309 ASGPR is an endocytic receptor notably overexpressed on the hepatocyte membrane surface.310,311 Interaction between GalNAc and ASGPR facilitates the internalization of GalNAc-bound compounds from the cell surface into endosomes via clathrin-dependent receptor-mediated endocytosis.312 As the endosome matures and pH decreases, ASGPR dissociates from the GalNAc conjugate and recycles back to the hepatocyte surface.313–315 At the same time, GalNAc is degraded, releasing NADs into the cytoplasm to initiate gene regulatory activity.
Leveraging this mechanism, Alnylam has extensively explored GalNAc conjugation with NADs, particularly siRNA, to achieve liver-specific delivery (Fig. 4c).312,316,317 Notably, the nucleic acid molecules in these conjugates are directly exposed to serum; enhancing their stability in the physiological environment is a crucial challenge. Studies have demonstrated that stability can be significantly enhanced by extensive chemical modification at the 2’ position of nucleotide sugars and by replacing the phosphodiester bond with a thiophosphate bond.318–320 Furthermore, stability can be improved without compromising drug activity by optimizing the number and modification positions of 2’-F and 2’-OME groups on both strands of double-stranded siRNA, leading to substantial efficacy improvements.319,321,322 These advancements have accelerated the shift from standard template chemistry to enhanced stability chemistry. Rapid ligand-receptor binding and efficient uptake by target cells are critical for receptor-targeted delivery systems to avoid in vivo clearance.315,323–325 Multivalent ligands show significantly enhanced binding affinity for ASGPR compared with monovalent GalNAc units, with affinity rankings in the order of tetraantennary > triantennary » biantennary » monoantennary.326 Studies by Biessen327 have suggested that a 2-nm interval between GalNAc and dendritic branch points may optimize NADs transportation efficacy. Additionally, hydrophobic linkers have been found to enhance GalNAc–ASGPR interactions.328
With the success of Alnylam in GalNAc, other companies have focused on RNA therapy and developed their delivery systems. Dicerna Pharmaceuticals has made considerable efforts to advance GalNAc delivery technology by adopting tetraantennary GalNAc conjugates (GalXC) for NADs delivery. In this conjugate, a unique four-ring structure is introduced on the passenger chain, enhancing the stability of the conjugate.310,329 Additionally, it accurately targets multiple GalXC ligands and successfully delivers siRNA to hepatocytes. Many candidates that involve this technology are currently undergoing clinical evaluation. Nedosiran,330 a siRNA targeting the silencing of lactate dehydrogenase A in hepatocytes (LDHA) with four covalently linked GalNAc, was approved by the FDA in 2023.
Exosomes
Exosomes are vesicle-like structures with a diameter of 40–160 nm released after the fusion of intracellular multivesicular bodies (MVBs) with the cell membrane.331,332 As a bridge of intercellular communication, classical exosomes have a monolayer structure, which can encapsulate bioactive substances, such as proteins, lipids, DNA, and RNA, in the core and deliver them to effector cells to play specific biological functions (Fig. 4d).333–336 They have been considered good candidates for NADs delivery.337–340 For example, Kaban et al.341 used exosomes derived from natural killer (NK) cells as a carrier of siRNA targeting BCL-2 to treat patients with estrogen receptor-positive (ER + ) breast cancer, resulting in enhanced apoptosis of breast cancer cells. Additionally, another study used plasma exosomes to deliver siRNA to T cells and monocytes and caused post-transcriptional gene silencing in recipient cells.342 Furthermore, exosomes can easily cross biological barriers, such as the blood-brain barrier (BBB), which has good application potential in extra-hepatic-targeted NADs delivery.343–345
When exosomes are used as NADs delivery vehicles, it is essential to consider the strategy of efficient drug loading. As a natural barrier of exosomes, the membrane structure of the lipid bilayer can protect NADs from external influences. Still, the existence of the membrane structure makes it difficult for exosomes to load drugs efficiently. The strategies of drug loading by exosomes are mainly divided into two categories.346–348 The first is the exogenous route, in which the NADs are directly introduced into the obtained exosomes via electroporation, co-incubation, sonication, extrusion, and freeze-thaw cycling.349–352 Although cargo uploading exogenously is simple and convenient to operate, the integrity of exosomes may be damaged during the loading process, which may affect the effect and require additional purification steps to remove the unloaded drugs.353,354 The other method is the endogenous pathway, which uses the endogenous pathway of exosome generation to indirectly improve the production of exosomes by promoting the expression of target nucleic acids and exosome secretion in productive cell lines.355 However, due to our lack of understanding of exosome biology, structure, and biogenesis, the strategy of endogenous drug loading still requires further research and optimization.
One common disadvantage of exosomes is their random movement in vivo and lack of specific targeting.355,356 The abundant lipids and membrane-bound proteins on the surface of exosomes provide binding sites for targeting ligands, such as peptides, antibodies, and aptamers. These ligands can be stably attached to the surface of exosomes through covalent bonds to enhance their targeting ability. For example, Kim et al.357 modified exosomes with transferrin receptor-binding peptide (T7 peptide), which efficiently delivered AMO-21 into glioblastoma (GBM) cells in vitro. Furthermore, in vivo delivery results have shown that T7 peptide-modified exosomes effectively reduced the level of miR-21 in tumor cells and inhibited tumor growth compared with unmodified exosomes. Another study used a single-chain variable fragment (scFv) to modify exosomes derived from human umbilical cord blood mesenchymal stem cells (MSCs).358 In addition, exosomes can be genetically engineered to express ligands on their surfaces simultaneously. The most commonly used exosome surface protein is lysosome-associated membrane protein (LAMP). The N-terminus of LAMP-2B is located on the surface of exosomes and can specifically target given sequences.359–362 After screening for cell-specific binding peptides for particular organs or tissues, such as the rabies virus glycoprotein peptide, LAMP-2B can be genetically modified to achieve targeted delivery (Fig. 4d).
Thus far, approximately 40 companies worldwide, including Codiak Biosciences, Evox Therapeutics, Tavec Pharmaceuticals, Carmine Therapeutics, Anjarium, and Micromedmark Biotech, have developed exosome-based therapies, which are expected to provide cost-effective and more accurate targeted therapies in the clinic (https://bioinformant.com/companies-developing-exosome-technologies/). However, challenges, such as large-scale production, purity, and batch homogeneity of exosomes, still limit their clinical application. Additionally, there are no regulations for the control of therapeutic drugs based on exosomes that consider safety, effectiveness, and quality control, highlighting the urgent need for standardized methods and principles to manage these molecules.363
Inorganic nanoparticles (INPs)
INPs are nanocarriers based on inorganic substances that have attracted considerable attention due to their unique electrical and optical properties, biocompatibility, and low cytotoxicity. Commonly used INPs include gold nanoparticles (AuNPs), silica nanoparticles (SiNPs), magnetic nanoparticles (MNPs), and carbon nanotubes (CNTs) (Fig. 4e).364–366 Unlike other delivery carriers, INPs possess a stable and robust structure, a large specific surface area, and tunable surface properties, allowing precise control of the drug delivery process through surface functionalization and controlled release modifications.
AuNPs are widely developed INPs with excellent biocompatibility and low toxicity. Their flexible surfaces enable nucleic acids to bind to the gold nanoparticles directly.367,368 For example, Shrestha et al.369 developed a gold nanoparticle-mediated drug delivery platform for the co-delivery of doxorubicin and polo-like kinase 1 (Plk1) siRNA, offering an adaptable and straightforward platform for studying drug-siRNA combinations in cancer treatment. Notably, NU-0129, composed of siRNA targeting the GBM oncogene Bcl2Like12 (Bcl2L12) and a gold nanoparticle core, was the first spherical nucleic acid (SNA) drug administered systemically.370 The Phase 0 clinical study (NCT03020017) in eight patients with GBM showed that NU-0129 could pass through the BBB and accumulate in tumors, reducing the abundance of BCL2L12 protein and demonstrating its potential as an innovative therapy for GBM.371
MNPs are typically made of magnetic materials, such as iron oxide or iron platinum, and are usually coated with biocompatible materials to enhance their stability and biocompatibility. Under the influence of an external magnetic field, therapeutic drugs or molecules can be loaded onto magnetic nanoparticles and targeted to precise regions in the body.372,373 The unique superparamagnetic nature, lower toxicity, and site-specific targeting capabilities of MNPs make them excellent nanocarriers for NADs delivery.374
CNTs are cylindrical structures composed of a hexagonal arrangement of sp2 hybridized carbon atoms, also known as graphene.375 CNTs can be categorized into single-walled CNTs (SWCNTs) and multi-walled CNTs (MWCNTs). SWCNTs consist of a single layer of graphene sheets rolled seamlessly into a cylindrical tube. In contrast, MWCNTs are composed of multiple graphene layers wrapped around each other in a cylindrical shape. Although CNTs have poor solubility in both water and organic media, they can be chemically modified to improve their solubility, degradation ability, and drug-loading capacity while reducing toxicity.376 This simple surface functionalization has made CNTs promising carriers for NADs delivery in various diseases.377
Moreover, SiNPs are considered excellent carriers for NADs delivery.378 Research has shown that porous silicon nanoparticles (pSiNPs) can replace commonly used viral vectors or lipid transfection reagents as novel vectors for delivering siRNA to dendritic cells.379 Luo designed pSiNPs with ASO as targeted gene and drug delivery platforms for GBM treatment. These pSiNPs penetrate the BBB monolayer in vitro and target the brain after intravenous injection in an in situ GBM mouse model. This indicates that pSiNPs and their multifunctional strategies have strong potential for cancer treatment and gene delivery research.380
Peptides
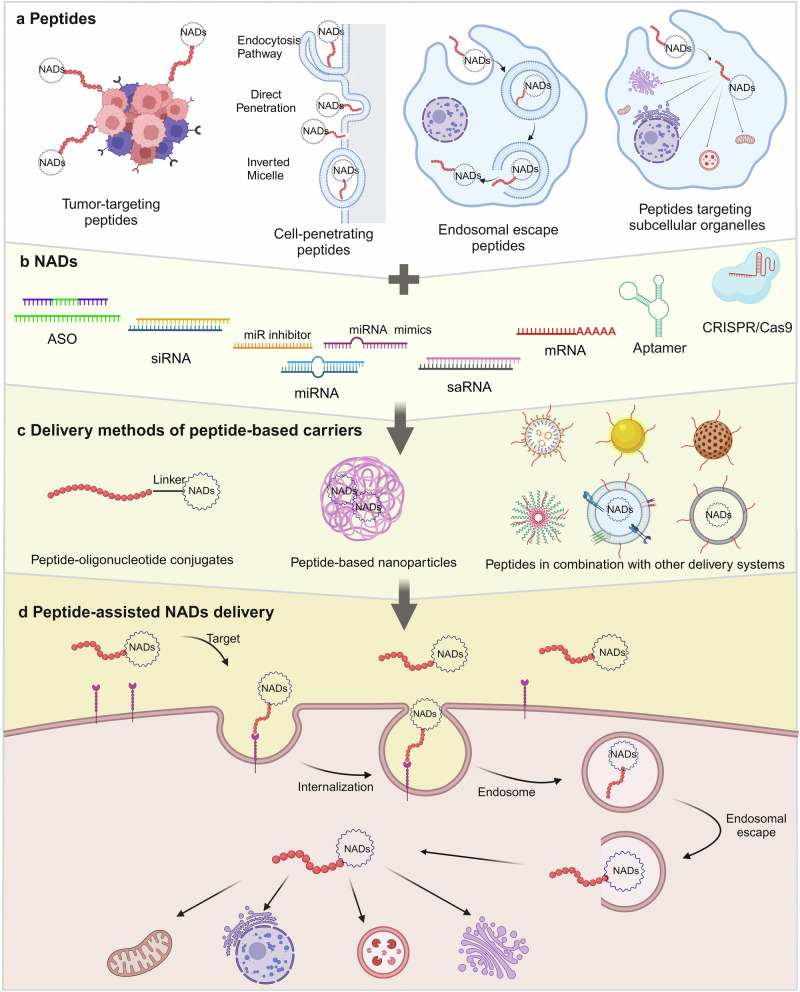
Peptides comprise fewer than 100 amino acid residues, and they are between small molecules and proteins in size. Historically, peptides have been recognized for their diverse roles as hormones, signaling molecules, carriers, and supplements.381–384 They are easily synthesized, possess relatively stable chemical properties, and exhibit a high selectivity. Due to their low immunogenicity and strong specific targeting capacity, peptides have emerged as promising carriers for selectively delivering NADs through various modalities (Fig. 4f).385,386 Peptides exhibit multiple functions, including tissue targeting,387–389 membrane penetration,390–392 endosome escape,393–395 and nuclear localization,396–398 depending on their amino acid composition. Several peptides have been developed specifically for NADs delivery based on these unique properties for cell targeting, penetration, endosomal escape, and sub-organelle targeting (Fig. 5).25,399–403
Fig. 5.
Schematic of peptide-assisted NADs delivery. a Classes of peptides facilitating the delivery of NADs across biological barriers. b Classes of NADs. c The delivery methods of peptide-based carriers include peptide–oligonucleotide conjugates, peptide-based nanoparticles, and peptides in combination with other delivery systems. d Peptides mediate the entry of NADs into cells and transfer them across the cell membrane, complete endosomal escape, and eventually release NADs in the cytoplasm, mitochondrion, nucleus, endoplasmic reticulum, lysosomes, and Golgi apparatus
Targeting peptides
Nonspecific cell binding is the leading cause of the off-target effects observed with several therapeutic molecules. The arginine-glycine-aspartic acid (RGD) sequence has been extensively applied in tumor-targeted therapy, mainly targeting αvβ3 receptors overexpressed on several tumor cells.404,405 A recent study demonstrated that siRNA conjugated to cyclic RGD (cRGD) could selectively enter cells that express αvβ3 integrin.406 Intravenously injected cRGD-siRNA molecules produced no innate immune response in mice with transplanted tumors.407 Additionally, researchers have found that incorporating siRNA into PEG-modified polylysine with cRGD can enhance gene silencing ability, improve cell uptake, inhibit glioma angiogenesis, and delay tumor progression after systemic administration in mice.408
The acidic extracellular microenvironment has become an effective disease diagnosis and treatment target.409 pH low insertion peptides (pHLIPs), derived from the C-helix of bacteriorhodopsin, can sense the pH value near the plasma membrane and deliver drugs to pathological tissues.410 In acidic environments, the elevated proton concentration increases the hydrophobicity of peptides. pHLIPs spontaneously fold to form α helices, which insert and cross the cell membrane to transport conjugated NADs into cells.191,411 In anti-tumor therapy, it has been demonstrated that pHLIPs can deliver PNAs to cancer cells due to their natural targeting abilities.412,413 For example, in a mouse model of lung adenocarcinoma, the delivery of PNAs targeting CEACAM6 using pHLIPs resulted in CEACAM6 gene silencing and tumor growth inhibition.414
Cell-penetrating peptides (CPPs)
CPPs are a short peptide class consisting of 5–30 amino acids that can cross the cell membrane directly.399 They can penetrate the cell membrane by themselves and effectively enhance the intracellular delivery of NADs, aiding their interaction with the target to achieve therapeutic goals.415–417
Despite their significant therapeutic potential, CPPs have some limitations. One major issue is their lack of selectivity, as they can carry NADs into almost all cell types. To address this, researchers have developed strategies such as local administration, targeting ligands on CPPs, or using activatable CPPs that penetrate only under specific biological stimuli.418,419 Ensuring the stability and biological activity of CPPs during the delivery process is also a challenge. Modifications can be made to the termini of existing CPPs or the peptide backbone can be adjusted, such as through peptide cyclization, to improve stability and cell permeability.420,421 Another limitation of CPPs is their limited clinical efficacy, often requiring high concentrations and doses to achieve a therapeutic effect, which can cause significant toxicity and side effects, hindering the development of clinical applications. Current research has found that inducing peptides to form multimers and using multifunctional fusion peptides can improve the delivery efficiency of CPPs to some extent.422–424
Endosomal escape peptides
Endosomal escape is a significant challenge for non-viral NADs delivery systems. Typically, after arriving at a specific cell, NADs pass through the cell membrane and reach the cytoplasm through various mechanisms, such as direct penetration or endocytic uptake.309,422 However, peptide carriers and their cargos often become trapped in acidified endosomes, preventing their entry into the cytoplasm, nucleus, and other subcellular compartments.14,425
Fusogenic peptides are short peptides that promote endosomal release by enhancing interactions with endosomal membranes.426,427 In a low pH environment, fusogenic peptides undergo pH-dependent conformational changes to form α-helices, which insert into the endosomal membrane, causing instability and decomposition of the membrane structure. This allows internalized NADs to dissociate from the fusogenic peptide delivery system and escape into the cytoplasm to exert their therapeutic effects.427 Many fusiform peptides of viral and non-viral origin, such as the INF7 family,428 L17E family,429 GALA/KALA family,430–432 and HA family,433 can disrupt endosomal membranes and increase cytoplasmic delivery. One study evaluated the ability of the H5WYG peptide to deliver NADs and promote endosomal release, confirming the elevated endosomal escape function of this clostridial peptide.434 Many studies have shown that self-assembled peptide carriers modified with fusogenic peptides can overcome intracellular delivery obstacles.435,436 For instance, the fusogenic peptide L17E was linked to peptide self-assembled disks using click chemistry. Compared with unmodified disks carrying plasmid DNA, nanodisks modified with L17E demonstrated enhanced endosomal escape and improved transfection efficiency in cell culture.429
Peptide-targeting subcellular organelles
The structural integrity and functional stability of organelles are essential for maintaining normal cellular physiological functions. Dysfunction of these organelles can lead to various diseases, including diabetes, neurodegenerative diseases, CVDs, and cancer, making them potential therapeutic targets.437–439 As mentioned earlier, the efficacy of NADs therapy largely depends on the efficient and safe delivery of NADs to their specific action sites within the body.14 After escaping from endosomes, the next challenge is transporting NADs quickly and effectively to the nucleus or specific organelles.425 Peptide targeting of subcellular organelles offers a promising strategy to guide NADs to dysfunctional organelles, thus achieving optimal therapeutic effects. This approach provides a potential method to overcome the intracellular delivery obstacles and enhance the efficacy of NAD-based therapies.
Multifunctional peptides
In previous clinical trials, using individual functional peptides for NADs delivery showed limited efficacy due to their inability to overcome various delivery obstacles.440 To address this issue, researchers have integrated different peptides or functional domains capable of overcoming transfer barriers into a single peptide-based carrier to enhance delivery efficiency.441–444 Compared with individual functional peptides, multifunctional peptide carriers considerably improve transfection efficiency and have demonstrated outstanding potential in clinical applications.445–447 For example, amino acid pairing (AAP) peptides are a novel class of self-assembled biomolecules comprising two main structural domains: an amino acid pairing domain and a cell permeability domain.448 AAP peptides possess recognition and membrane-targeting functions, facilitating gene delivery through interactions with DNA and siRNA.448
The discovery of peptide-based NADs delivery systems is accelerating by the use of various peptide combinations and high-throughput screening, providing new directions for optimizing and developing peptide carriers.449–451 For example, Li et al.452 designed a multifunctional peptide vector (PLD-R9-G-NLSW) containing the CPP R9, NLS, and 2,3-dimethylmaleic anhydride-modified PLL. This vector condenses with the pIRES-VEGF plasmid to form a complex for gene delivery. In vitro results showed that the complex considerably improved gene internalization and transfection efficiency while reducing cytotoxicity. However, researchers have discovered that the immediate coupling of individual peptides can affect the function and activity of the units. Given the likelihood that distinct peptides may interact with each other during the fusion process, the final functional characteristics of the peptides may not involve a simple integration.12,453 This phenomenon suggests that the interaction mechanism between combined peptides and the structure-activity relationship needs to be explored in more detail.
Recently, peptides have been applied as ligands to achieve delivery functions in complexes known as peptide-oligonucleotide conjugates (POCs).454,455 In this approach, oligonucleotides or their analogs are covalently linked to one or more peptide residues to form POCs. Various methods, including disulfide bonds, thioether bonds, thiol-maleimide bonds, phosphodiester bonds, and click chemistry, are used for this covalent connection.456–458 The primary advantage of covalent conjugation is that the resulting product is a single compound with defined structural and stoichiometric characteristics, aligning with the ideal properties for drug design and in vivo application.459,460 However, covalent conjugation also has drawbacks. Interactions between cationic peptides and negatively charged oligonucleotides can enhance toxicity, affect pharmacokinetic and pharmacological properties, and limit the efficacy of these therapies.461 Additionally, the interaction between anions and cations can complicate large-scale preparation and purification of these conjugates.462
The most advanced application of POCs is the delivery of charge-neutral oligonucleotides, such as PNA and PMO, using CPPs.463–465 CPP-PNA conjugates with Tat and R8 have been successfully applied to PNA delivery466,467 and showed sound therapeutic prospects in anti-virus, antibacterial, and anti-inflammatory therapy. Beyond CPPs, other functional peptides have also been exploited for PNA delivery. For instance, Soudah et al.468 used a conjugate of the cytoplasmically localized internalized peptide (CLIP6) and PNA to treat GBM cells, considerably upregulating the tumor suppressor Mnk2a and promoting cancer cell death. Kaplan et al.457 showed that pHLIP-αKu80 (γ), which was created by covalently conjugating pHLIPs with PNA targeting KU80 via disulfide bonds, could selectively reduce Ku80 expression under acidic conditions. In mice, intravenous injection of fluorescently labeled pHLIP-αKu80 (γ) targeted tumors and reduced Ku80 expression.
The development of peptide-phosphoryl diamine morpholino oligonucleotide (PPMO) conjugates has further advanced NADs delivery, particularly for neuromuscular diseases, such as spinal muscular atrophy (SMA) and DMD.469,470 A recent study demonstrated that subcutaneous injection of a CPP-DG9 and PMO conjugate improved muscle strength and innervation in mice with severe SMA, significantly extending their median survival with no apparent side effects.471 To enhance the delivery of neutrally charged oligonucleotides, Gait and Wood472 developed a series of arginine-rich CPPs named [PNA/PMO]-internalized peptides (Pip), including Pip2a, Pip2b, Pip5e, and Pip6a. Studies have shown that Pip6a-PMO effectively rescues the disease phenotype and improves the survival rate of severe SMA mice, showing strong efficacy in the central nervous system and peripheral tissues.473,474 Pip6a-PMO predicts a potent therapeutic option that combines the genetic precision of SSO with the systemic delivery efficacy of peptides.475
With technological advances, various novel POCs have demonstrated the feasibility of delivering siRNA, ASO, and miRNA in vitro.406,476 Studies have found that bivalent cRGD successfully transports VEGF receptor 2 (VEGFR2)-siRNA into tumor cells in a mouse non-small cell lung cancer xenograft model, downregulating VEGFR2 expression and significantly inhibiting cancer progression.406 Kim et al.402 designed a dual-targeting drug delivery system for miR-21 inhibitors, consisting of a PDL1-binding peptide covalently linked with an anti-miR-21 inhibitor via a click reaction. Pep-21 treatment specifically silenced target miR-21 in B16 melanoma cells and M2 macrophages, reducing tumor cell migration and inhibiting tumor progression. PepGen developed a POC drug, PGN-EDO51, using its enhanced delivery oligonucleotide platform, which skips exon 51 in the DMD gene. In Phase I clinical trials, PGN-EDO51 showed good safety, tolerability, and efficacy, and Phase II trials are ongoing.
Electrostatic or hydrophobic interactions between peptides and NADs create self-assembling peptide-based nanoparticles.477–479 These complexes protect nucleic acids from nuclease degradation and reduce the likelihood of adverse reactions.480 Using this strategy, Ryu et al.481 developed an S-R11 fusion peptide (space peptide bound to polyarginine) that forms stable nanocomposites with siRNA molecules through electrostatic attraction and hydrogen bonding, facilitating siRNA delivery to difficult-to-transfect immune cells. Sirnaomics’ histidine-lysine copolymer peptide nanoparticle (PNP) delivery platform has demonstrated clinical therapeutic potential for RNAi therapy.482 In contrast, non-covalent strategies have been used for delivering siRNA, plasmid DNA, and splicing correction oligonucleotides.483,484 The peptide p5RHH has been efficiently used to transfect siRNA and mRNA into human cartilage to reduce chondrocyte apoptosis and prevent cartilage degeneration485–487 as well as into cardiac tissue for treating abdominal aortic aneurysm to reduce the risk of aortic rupture and sudden death in mice.484
Clinical applications of NADs
Theoretically, NADs can cure any disease by allowing the selection of the correct nucleotide sequence on the target gene. Unlike conventional therapeutics, NADs induce long-lasting or curative effects due to their distinct physicochemical and biological properties. Numerous NADs have transitioned from bench to bedside and have successfully been approved for clinical trials (Fig. 6). Several NADs, including ASOs, aptamers, siRNAs, and mRNA vaccines, have been adopted as vaccines for treating rare genetic diseases, cancer, ophthalmic diseases, CVDs, and infections. Table 2 summarizes NADs successfully applied in clinical experiments for treating human diseases.
Fig. 6.
Clinical application of NADs. Several NADs have been adopted as vaccines for treating rare genetic diseases, cancer, ophthalmic diseases, cardiovascular diseases, and infection diseases, and they have shown remarkable therapeutic effects. The box summarizes the therapeutic NADs for various diseases in the clinic or on the market
Table 2.
Status of NADs-based therapeutics clinical trials
| Drug (alternate name) | Company | Classification | Indication (Phase) | Target | Modification & Delivery | Clinical Trail Number | Ref(s) |
|---|---|---|---|---|---|---|---|
| Rare genetic disorders | |||||||
| NTLA-2001 | Intellia Therapeutics | CRISPR-Cas9 | ATTR-CM (III) | TTR | LNP | NCT06128629 | 296,588 |
| Zilganersen (ION-373) | Ionis | ASO | Alexander disease (III) | GFAP mRNA | 2’-MOE | NCT04849741 | 589 |
| Fitusiran (ALN-AT3) | Sanofi | siRNA | Hemophilia A/B (III) | Serpin C1 | GalNAc | 590 | |
| Donidalorsen (IONIS-PKK-LRx) | Ionis | ASO | HAE (III) | PKK | GalNAc, 2’-MOE | NCT05392114 | 591,592 |
| Ulefnersen (ION-363) | Ionis | ASO | ALS (III) | FUS | PS, 2’-MOE | NCT04768972 | 506 |
| Cemdisiran (ALN-CC5) | Alnylam & Regeneron | siRNA | PNH, Myasthenia gravis (III), IgAN, (II) | Complement C5 | 2ʹ-OME,2’-OF & GalNAc | 593,594 | |
| Vesleteplirsen (SRP-5051) | Sarepta Therapeutics | ASO | DMD (II) | Exon 51 of DMD | PMO/ PPMO | NCT04004065 | 500 |
| PGN-EDO51 | PepGen | ASO | DMD (II) | Exon 51 of DMD | EDO | NCT06079736 | 501 |
| ATL-1102 (ISIS-107248) | Antisense Therapeutics | ASO | DMD (II) | ITGA4 | 2’-MOE | NCT05938023 | 595 |
| Zorevunersen (STK-001) | Stoke Therapeutics | ASO | Dravet Syndrome (II) | SCN1A | 2’-MOE | 596 | |
| IONIS-MAPTRx (BIIB-080) | Ionis & Biogen | ASO | Mild Alzheimer’s Disease (II) | MAPT | 2’-MOE, PS | / | |
| SQY51 | Sqy Therapeutics | ASO | DMD (I/II) | Exon 51 of DMD | tricyclo-DNA | NCT05753462 | / |
| NTLA-2002 | Intellia Therapeutics | CRISPR-Cas9 | HAE(I/II) | KLKB1 | LNP | NCT05120830 | 597 |
| mRNA-3705 | Moderna | mRNA | MMA (I/II) | MUT | LNP | 598 | |
| Cancer | |||||||
| mRNA-4157 (V-940) | Merck & Moderna | mRNA | NSCLC, Melanoma (III) | / | LNP | 599 | |
| Autogene Cevumeran (BNT-122) | BioNTech | mRNA | Colorectal, PDAC (II) | unspecified TAAs | LNP | 600 | |
| IGV-001 | Imvax | ASO | GBM (II) | IGF type 1 receptor | PS, Goldspire™ | NCT04485949 | 520 |
| Cotsiranib (STP-705) | Sirnaomics | siRNA | Keloid, BCC, hypertrophic scars, isSCC (II) | TGF-β1、COX-2 | PNP | NCT04669808 | 482 |
| QN-165 (AS1411) | Qualigen Therapeutics | Aptamer | RCC, glioma and AML (II) | Nucleolin | / | 183,184 | |
| Cobomarsen (MRG-106) | miRagen Therapeutics | miRNA | MF, CTCL, DLBCL, CLL, ATLL (I/II) | miR-155 inhibitor | LNA | 125,126 | |
| olaptesed pegol | TME Pharma | Aptamer | Tumor, GBM (I/II) | CXCL12 | / | 601 | |
| TTX-MC138 | TransCode | miRNA | advanced solid tumors (I/II) | miRNA-10b inhibitor | iron oxide nanocarrier | 127 | |
| STP-707 | Sirnaomics | siRNA | solid tumors (I) | TGF-β1、COX-2 | PNP | NCT05037149 | / |
| MTL-CEBPA | MiNA Therapeutics | saRNA | HCC (I) | CEBPA | / | NCT02716012 | 142–144 |
| RAG-01 | Ractigen Therapeutics | saRNA | NMIBC (IND) | CDKN1A | / | / | 122 |
| Ophthalmic diseases | |||||||
| Tivanisiran (SYL1001) | Sylentis, S.A. | siRNA | DED (III) | TRPV1 | / | 528 | |
| Sepofarsen (QR-110) | ProQR Therapeutics | ASO | LCA10 (III) | CEP290 | 2’-OME | NCT03913143 | 602 |
| Ultevursen (QR-421a) | ProQR Therapeutics | ASO | Usher syndrome and nsRP (II/III) | USH2A | 2’-OME | NCT05158296 | 533 |
| SYL1801 | Sylentis | siRNA | AMD (II) | NRARP | / | NCT05637255 | 526 |
| RBM-007 (APT-F2) | Ribomic | Aptamer | wet AMD (II) | FGF2 | / | 525 | |
| IONIS-FB-LRx (RO7434656) | Roche & Ionis | ASO | GA (II) | Complement Factor B | 2’-MOE & GalNAc | NCT03815825 | 603 |
| Cardiovascular diseases | |||||||
| Pelacarsen (AKCEA-APO(a)-LRx) | Novartis AG | ASO | CVD (III) | ApoA | 2’-MOE & GalNAc | 542,604 | |
| Lepodisiran | Lilly | siRNA | ASCVD (III) | ApoA | 2’-MOE, 2’-F & GalNAc | NCT06292013 | 546 |
| Olpasiran (AMG 890) | Amgen Biopharmaceuticals | siRNA | ASCVD (III) | ApoA | 2’-MOE & GalNAc | 547,548 | |
| IONIS-FB-LRx (RO7434656) | Roche & Ionis | ASO | Primary IgA Nephropathy (III) | Complement Factor B | 2’-MOE & GalNAc | NCT05797610 | / |
| Olezarsen (IONIS-APOCIII-LRx) | Akcea Therapeutics & Ionis | ASO | SHTG & FCS (III) | ApoC-III | GalNAc | 605,606 | |
| Zodasiran (ARO-ANG3) | Arrowhead Pharmaceuticals | siRNA | HoFH (II) | ANGPTL3 | 2’-OME, 2’-F & GalNAc | NCT05217667 | 607 |
| Zilebesiran (ALN-AGT01) | Alnylam | siRNA | Hypertension (II) | Angiotensinogen | 2’-MOE, 2’-F & GalNAc | 608 | |
| MRG-110 (S-95010) | miRagen Therapeutics | miRNA | Chronic heart failure (I) | miR-92a Inhibitors | LNA | NCT03603431 | 125 |
| Infection diseases | |||||||
| mRNA-1345 | Moderna | mRNA | Respiratory syncytial virus infection (III) | RSV fusion | LNP | NCT06067230 | 609 |
| mRNA-1647 | Moderna | mRNA | Cytomegalovirus infection (III) | HCMVgB & HCMVgH | LNP | NCT05085366 | 610 |
| Bepirovirsen (GSK-3228836) | GSK & Ionis | ASO | HBV (III) | HBV RNA | 2’-MOE | 559 | |
| GSK-5637608 (JNJ-73763989) | GSK | siRNA | HBV (II) | HBV RNA | GalNAc |
NCT0512359 |
560 |
| ALN-HBV02 (VIR-2218) | Alnylam & Brii Biosciences Ltd & Vir Biotechnology | siRNA | HBV (II) | HBV RNA | 2’-OME, 2’-F & GalNAc | NCT05612581 | 561 |
| Imdusiran (ARB-270729) | Arbutus Biopharma | siRNA | HBV (II) | hepatitis B surface antigen | 2’-F & GalNAc | 562 | |
| mRNA-1944 | Moderna | mRNA | Chikungunya Virus (I) | CHKV-24 | LNP | NCT03829384 | 295 |
| Other diseases | |||||||
| Sapablursen (IONIS-TMPRSS6-LRx) | Ionis | ASO | PV (II) | TMPRSS6 | 2’-MOE & GalNAc | NCT05143957 | 611 |
| CWT001 (TenoMiR) | Causeway Therapeutics | miRNA | Lateral epicondylitis (II) | microRNA-29a mimic | 2’-OME, 2’-F | NCT06192927 | / |
| ION224 | Ionis | ASO | MASH (II) | DGAT2 | 2’-MOE & GalNAc | NCT04932512 | 612 |
| Rapirosiran (ALN-HSD) | Alnylam | siRNA | NASH (II) | HSD17B13 | 2ʹ-MOE, 2’-F & GalNAc | NCT05519475 | / |
| ALN-KHK | Alnylam | siRNA | T2DM (I/II) | KHK | GalNAC | NCT05761301 | / |
| OLX10010 (BMT-101) | Olix Pharmaceuticals | siRNA | Hypertrophic scars (II) | CTGF | 2’-MOE | / | |
| Remlarsen (MRG-201) | miRagen Therapeutics | miRNA | Keloid disorder (II) | microRNA-29 mimic | / | NCT03601052 | 124 |
| AON-D21 | Aptarion Biotech | Aptamer | Community-acquired pneumonia (II) | C5a | / | NCT05962606 | 613 |
Rare genetic diseases
Transthyretin amyloidosis (ATTR) is a rare systemic disease characterized by the progressive deposition of misfolded TTR protein in the heart and peripheral nerves, leading to transthyretin amyloid polyneuropathy (ATTR-PN) and transthyretin amyloid cardiomyopathy (ATTR-CM).488,489 Several NADs have been approved for ATTR treatment, including ASOs, such as inotersen490,491 and eplontersen,492 as well as siRNAs, such as patisiran97 and vutrisiran.493 These therapies work by disrupting the relevant mRNA, thus inhibiting TTR synthesis. Additionally, researchers are exploring new methods to block TTR production via gene-editing techniques. For instance, a Phase III clinical trial evaluated the efficacy and safety of NTLA-2001, a CRISPR-Cas9-based therapy, in participants with ATTR-CM. In a previous Phase I trial, six patients treated with a single dose of NTLA-2001 showed significant reductions in serum TTR protein levels, with no serious adverse effects or liver damage reported.494
DMD is a rare genetic muscle disorder caused by mutations in the dystrophin gene, leading to the absence of the dystrophin protein. Patients typically exhibit symmetric, progressive muscle weakness and atrophy, ultimately leading to premature death due to respiratory and cardiovascular complications.495 Significant progress has been made in DMD therapeutic strategies, including NADs. Approved ASO drugs target exon 45 (casimersen),264,496 exon 51 (eteplirsen),497,498 and exon 53 (golodirsen and viltolarsen)80,263 to induce specific exon skipping in the dystrophin gene, thus delaying disease progression. Researchers have developed second-generation skipping drugs, called PPMOs, to enhance targeting and skipping activity.499 Sarepta’s SRP-5051, an ASO conjugated with a targeting peptide, has shown higher levels of dystrophin expression and exon skipping rates than eteplirsen. This drug is currently in Phase II clinical trials (NCT04004065).500 Additionally, PepGen’s PGN-EDO51, designed to skip exon 51 in the DMD gene, has shown promising results in Phase I clinical trials and is now in Phase II (NCT06079736).501
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by the dysfunction of upper and lower motor neurons.502 Approximately 10% of ALS cases are familial ALS, with various disease-causing genes identified, including superoxide dismutase 1 (SOD1), chromosome 9 open reading frame 72 (C9ORF72), and fused in sarcoma (FUS).503,504 The first NAD approved for ALS was tofersen (Qalsody®), a PS 2’-MOE ASO targeting SOD1 mutations.67,505 Ulefnersen (ION-363), an antisense therapy targeting the FUS gene, is currently in Phase III trials for FUS-ALS.506 However, clinical trials for C9ORF72-targeted therapies WVE-004 and Tadnersen (IONIS-BIIB5Rx) have been terminated due to poor efficacy.
Primary hyperoxaluria type 1 (PH1) is an autosomal recessive disorder caused by defects in hepatic glycoxidation metabolism, leading to excessive endogenous oxalate production.507 It is characterized by recurrent urinary calculi, nephrocalcinosis, and progressive renal damage. PH1 is associated with a deficiency or functional defect in alanine glyoxylate aminotransferase.508,509 Lumasiran, a siRNA drug that targets hydroxy acid oxidase 1 to reduce hepatic oxalate production, was the first drug approved for PH1.510 Nedosiran, another RNAi drug developed by Dicerna Pharmaceuticals that targets LDHA, is currently being evaluated for its efficacy in reducing oxalate production.330
SMA is a genetic neuromuscular disease caused by the deletion or mutation of the survival motor neuron 1 (SMN1) gene, leading to the loss of alpha motor neurons and progressive muscle atrophy.511 The SMN2 gene, highly similar to SMN1, produces mostly non-functional SMN protein due to differences in exon 7 splicing.512,513 Correcting exon 7 splicing in SMN2 can compensate for the lack of SMN1, producing functional SMN protein and improving symptoms.514,515 Nusinersen, a modified ASO targeting the intronic splicing silencer N1 of the SMN2 gene, promotes the production of the full-length SMN protein. Several clinical trials have confirmed the efficacy and safety of nusinersen, leading to better motor milestone responses and higher event-free survival rates.516–518
Cancer
The occurrence and progression of cancer are closely tied to the activation of oncogenes and the loss of tumor suppressor genes. NADs, which target specific genes, offer new methods for cancer treatment. For example, transforming growth factor beta 1 (TGFβ1), a multifunctional cytokine, can alter the tumor microenvironment when dysregulated, promoting angiogenesis and immune suppression, thus influencing tumor progression.482,519 Therefore, blocking TGF-β signal transduction is a critical strategy in cancer treatment.
Cotsiranib is a siRNA therapy developed by Sirnaomics that uses the PNP delivery platform, containing two active siRNAs targeting TGFβ1 and cytochrome C oxidase subunit 2 mRNA.482 A Phase II clinical trial (NCT04669808) for the in situ treatment of basal cell carcinoma showed a positive therapeutic effect and achieved 100% complete clearance. Another anticancer candidate, STP707, is currently being evaluated in a Phase I trial (NCT05037149) for safety, tolerability, and anti-tumor activity in participants with advanced/metastatic or surgically unresectable solid tumors.
In addition, tumor vaccines carrying NADs are considered feasible therapeutic approaches for most solid tumors. IGV-001 is a vaccine targeting GBM, designed based on the Imvax platform Goldspire™. Unlike traditional tumor vaccines, IGV-001 is prepared by mixing tumor tissue removed from the patient’s brain with an insulin-like growth factor 1 receptor antisense oligonucleotide (IMV-001), which inhibits tumor growth. Then, this mixture is encapsulated in biodiffusion chambers with a 0.1-μm pore size, allowing large molecules to permeate and be implanted into the patient’s abdomen.520 This aims to induce a tumor-specific immune response. In earlier clinical trials, IGV-001 demonstrated good tolerance and safety (NCT02507583).520 Imvax is conducting a randomized, multicenter, double-blind, placebo-controlled Phase IIb study (NCT04485949) to evaluate IGV-001 in patients with newly diagnosed GBM.521 Additionally, personalized mRNA neoantigen vaccines, such as mRNA-4157, encode multiple antigens to stimulate a patient-specific immune response. Clinical trials are underway for melanoma, non-small cell lung cancer, and other solid tumors. A Phase IIb trial of mRNA-4157 in combination with pembrolizumab for patients with resected high-risk melanoma showed that this combination therapy prolonged recurrence-free survival compared with pembrolizumab monotherapy and had a manageable safety profile.522 Combination therapies for melanoma and non-small cell lung cancer have entered Phase III clinical trials (NCT05933577 and NCT06077760). This is expected to be the first mRNA tumor vaccine on the market.523
Ophthalmic diseases
AMD is a complex eye disease primarily categorized into dry and wet/neovascular AMD.524 Aptamers have shown promise in AMD treatment. Pegaptanib, a VEGF antagonist with a three-dimensionally structured aptamer, was initially used to inhibit pathological neovascularization and reverse disease progression but was later removed from the market. RBM-007, an RNA aptamer designed to target fibroblast growth factor 2 (FGF2), is currently in Phase II clinical trials for treating neovascular AMD (nAMD).178,525 FGF2 promotes angiogenesis and retinal fibrosis by stimulating vascular endothelial cell proliferation and inducing VEGF secretion. Unlike traditional anti-angiogenic therapies that target VEGF, Sylentis developed SYL1801, an siRNA targeting NOTCH Regulated Ankyrin Repeat Protein (NRARP), to prevent and control nAMD progression. SYL1801 is administered as eye drops rather than via intravitreal injection, downregulating NRARP expression and inhibiting retinal neovascularization.526 Clinical trials testing various doses of SYL1801 eye drops in healthy volunteers showed good safety and tolerability. Four of the 36 patients tested experienced mild adverse events such as blepharitis, keratitis, hyperemia, and ocular irritation, all of which resolved within 72 h.527 Currently, the drug is in Phase II clinical recruitment to compare the safety and efficacy of three different doses in patients with nAMD (NCT05637255).
Dry eye disease (DED) is a common ocular condition characterized by tear film alterations, ocular inflammation, and neurosensory abnormalities.528–530 Tivanisiran (SYL1001), an unmodified siRNA eye drop developed by Sylentis, targets the transient receptor potential vanilloid 1 mRNA to alleviate ocular discomfort and pain.528 It has improved the quality of eye redness and tear film in human and animal models. It is undergoing Phase III trials to evaluate its safety in treating DED caused by dry eye syndrome (NCT05310422 and NCT04819269).
QR-110, an ASO drug in Phase III trials, was designed to address Leber congenital amaurosis type 10 caused by the p.Cys998X mutation in the centrosomal protein 290 gene.531 QR-110 binds to pre-mRNA, restoring proper splicing and producing fully functional proteins.532 Another candidate, ultevursen (QR-421a), is an ASO therapy for treating vision loss in patients with pigmentary retinopathy due to mutations in exon 13 of the USH2A gene. This drug aims to halt or reverse vision loss by restoring the function of the usherin protein through exon skipping in Usher syndrome type 2a and non-syndromic retinitis pigmentosa.533
CVDs
CVDs remain a leading cause of mortality worldwide and pose a severe public health challenge. These diseases, including atherosclerosis, myocardial infarction, cardiac hypertrophy, and heart failure, are closely linked to genetic, metabolic, and environmental factors.534 Researchers continually explore new disease targets and develop precision-targeted NADs to address CVDs. Advancements in nucleic acid chemistry have considerably improved drug stability and pharmacokinetics, offering more effective long-term treatment options for reducing the incidence and mortality of CVDs.
Genetic, epidemiological, and clinical research has indicated that lipid abnormalities, such as in low-density lipoprotein cholesterol (LDL-C), are critical factors in the occurrence and progression of CVDs.535,536 Proprotein convertase subtilisin/kexin type 9 (PCSK9) is an essential regulator of LDL-C metabolism. By reducing the number of LDL receptors (LDLR) on the surface of liver cells, PCSK9 leads to the accumulation of LDL-C, increasing the risk of CVDs.537,538 NADs targeting PCSK9 work by blocking the synthesis of PCSK9 protein and preventing LDLR degradation, thus promoting LDL-C absorption and reducing its levels in the bloodstream. Inclisiran, the world’s first siRNA drug for CVDs, is administered via subcutaneous injection and conjugated with N-acetyl galactosamine to enhance its targeted uptake by liver cells.539 Results from several Phase III clinical trials (ORION-9/10/11, NCT03397121, NCT03399370, and NCT03400800) have shown that subcutaneous injections of inclisiran every 6 months considerably reduce LDL-C levels by 48–52%, demonstrating sustained efficacy and good tolerability. Injection site reactions were more common in the inclisiran group but were generally mild and not persistent (5.0% vs. 0.7%).540,541 Inclisiran has been approved for treating various cardiovascular conditions, including atherosclerotic cardiovascular disease (ASCVD), primary hypercholesterolemia, mixed dyslipidemia, and heterozygous familial hypercholesterolemia (HeFH).
Lipoprotein(a) [Lp(a)] is a cholesterol-rich LDL-like particle formed by the covalent bonding of apolipoprotein(a) [Apo(a)] and Apo B100 via disulfide bonds. Elevated Lp(a) levels are considered independent risk factors for ASCVD, heart failure, and aortic valve stenosis.542–544 Although no targeted drug for reducing Lp(a) levels has been approved thus far, several candidates have shown promise in clinical trials.545 For example, pelacarsen, an Lp(a) ASO drug developed by Ionis in collaboration with Novartis AG, effectively lowers Lp(a) levels by selectively cleaving Apo(a) gene (LPA) mRNA. In a Phase II trial, pelacarsen reduced Lp(a) levels by 35–58% depending on the dose, with no significant differences in safety indicators compared with the placebo.542 The Phase III trial Lp(a)-HORIZON is underway, involving 8,323 patients with established ASCVD, and is expected to be completed in 2025 (NCT04023552). In addition to ASO drugs, siRNA therapies offer unique solutions for CVDs. Olpasiran and lepodisiran are GalNAc conjugates containing siRNAs that target the LPA gene, promoting RISC-mediated degradation of Apo(a) mRNA and preventing Lp(a) particle assembly in hepatocytes.546,547 Olpasiran demonstrated a dose-dependent reduction in Lp(a) levels, with a single dose of 9 mg or higher reducing Lp(a) concentrations by over 90% for several months (NCT03626662).548 Phase II trials of olpasiran [OCEAN(a)-DOSE] (NCT04270760) showed that a higher dose (75 or 225 mg) could reduce the Lp(a) level of patients by more than 95% compared with the placebo. Regarding safety, the overall incidence of AEs was similar between the dose and placebo groups. Among them, the most common AE of olpasiran was injection site reaction (primarily pain).547 The Phase III trial of olpasiran aims to compare its effects with the placebo on the risk of coronary heart disease death, myocardial infarction, and urgent coronary revascularization in ASCVD patients with elevated Lp(a), with results expected by 2026 (NCT05581303). Similarly, lepodisiran has shown well-tolerated and dose-dependent reductions in serum Lp(a) concentrations, with a Phase III trial currently recruiting participants and expected to be completed by 2029 (NCT06292013 and NCT04914546).546
Beyond lipid-lowering therapies, there are positive developments in other areas of NADs application. Zilebesiran (ALN-AGT01), which targets angiotensinogen (AGT), is being developed for hypertension treatment. It suppresses the generation of angiotensin I and II, inhibiting the renin-angiotensin-aldosterone system to reduce blood pressure.549 Phase I studies showed that a single subcutaneous dose of zilebesiran of 200 mg or more considerably decreased serum AGT levels and 24 h ambulatory blood pressure for up to 24 weeks, with only mild injection site reactions observed.550 In Phase II trials (KARDIA-1), zilebesiran reduced systolic blood pressure in all treated patients, with effects lasting 6 months, opening new possibilities for NADs use in treating hypertension and CVDs.551
Infectious diseases
The COVID-19 pandemic, caused by the severe acute respiratory SARS-CoV-2, has considerably advanced the development of mRNA vaccines. These candidate vaccines are LNP-encapsulated mRNA-based formulations that encode the full-length SARS-CoV-2 spike protein and its receptor-binding domain. They induce an immune response again st the spike protein on the virus’s surface, blocking its entry into cells and providing an antiviral effect.552,553 mRNA-1273 was one of the first COVID-19 vaccines to enter clinical trials, taking only 42 days from the publication of the virus’s genetic sequence to the production of the first batch of samples. The final analysis from its Phase III clinical trials showed a 94.1% efficacy in preventing COVID-19 illness and a 100% efficacy in preventing severe COVID-19.554 Similarly, BioNTech’s mRNA vaccine BNT162b2 demonstrated a 95% efficacy against COVID-19, with protection rates of over 94% for individuals aged 65 and older.555 These vaccines were designed, manufactured, evaluated, and brought to market in an extremely short period compared with traditional vaccine development timelines, highlighting the unique advantages of mRNA vaccines, including high safety, simple design, broad target range, and ease of scaling up production.
Chronic hepatitis B, caused by hepatitis B virus (HBV) infection, is a significant global public health problem. Current clinical treatment guidelines aim for a functional cure for CHB, which involves sustained negativity for hepatitis B surface antigen (HBsAg), undetectable HBV DNA, normal liver biochemical indicators, and improvement of liver tissue lesions after treatment cessation.556 Despite the efficacy of existing drugs, achieving a functional cure remains challenging. Bepirovirsen, developed by GlaxoSmithKline (GSK), is a 2’-MOE-modified ASO that targets all HBV RNAs, promoting RNase H-mediated RNA degradation, reducing HBV replication, and inhibiting HBsAg production.557 Additionally, bepirovirsen has immunostimulatory activity through Toll-like receptor 8, potentially helping the immune system permanently clear the virus from the bloodstream. Data from a Phase IIb clinical trial indicated that after 24 weeks of treatment, bepirovirsen can reduce levels of HBsAg and HBV DNA.558 Furthermore, another Phase II trial (B-Sure) evaluated the durability of bepirovirsen’s antiviral effect (NCT04954859). Recruitment for Phase III clinical trials to assess the efficacy and safety of bepirovirsen is also underway, with results expected in early 2026 (NCT05630807 and NCT05630820).559 Several candidate siRNA drugs, including GSK-5637608 (JNJ-73763989, GSK),560 VIR-2218 (ALN-HBV02, Alnylam & Brii Biosciences Ltd. & Vir Biotechnology),561 and imdusiran (ARB-270729, Arbutus Biopharma),562 have entered Phase II clinical trials, demonstrating a strong ability to reduce HBsAg levels. In practical clinical applications, these siRNA therapies often form combination treatments with nucleoside analogs, TLR agonists, or peginterferon, considerably enhancing the possibility of a functional cure for HBV. For example, VIR-2218, a GalNAc-siRNA conjugate that targets HBV, uses enhanced stability chemistry plus technology to stabilize the siRNA in vivo and reduce off-target effects.561 Currently, combination therapies involving VIR-2218 are in multiple Phase II trials for HBV patients, and results are eagerly awaited (NCT04856085 and NCT04412863).
Future perspectives of NADs development
Although many NADs have shown promising preclinical results, the number of NADs approved by the FDA for clinical use remains limited. The complexity of nucleic acids and their types, sizes, and mechanisms of action have made the development of NADs systematically very complex. For successful NADs design, critical factors must be considered regarding sequence design, modification, and delivery, as well as the clinical translation, medical indication, and scale production feasibility.
Sequence design of NADs
Typically, NADs are designed and screened based on the pathogenic genes of specific diseases. By leveraging existing sequence design and screening techniques, AI-assisted designs have accelerated the design and screening process, making it more accurate, and have been adapted to the needs of personalized therapy.215,563 Furthermore, patenting key modification sites in the selected target sequences is crucial to maintaining a competitive advantage.
Structure modification and delivery system
Considering the inherent physicochemical properties of NADs, the critical issue for their application is whether they can reach the target site and exert the expected therapeutic effects. Much evidence supports that it is unrealistic to address all drug delivery issues through a single modification method or a “universal” delivery vector. NADs should exhibit good stability to withstand nucleases and immune system clearance to achieve sound therapeutic effects. In addition to being delivered explicitly to the required tissues, they must reach the targeted cells, efficiently released into the cytoplasm, and exhibit in vivo biocompatibility. Initially, choosing appropriate strategies for the design of NADs is necessary based on the disease type and the required functional nucleic acid. For example, from the approved NADs, ASO drugs often do not require carriers for efficient delivery and only require appropriate modifications, such as PS and 2’ position modifications. Small NADs, such as siRNA, and large ones, such as mRNA, in addition to the necessary modifications, still require the aid of delivery vectors. siRNA typically uses nucleic acid conjugations for delivery, while mRNAs often rely on LNPs for compression and encapsulation. Furthermore, understanding the structure-activity relationship between nucleic acids and the delivery vector and different interactions (hydrophobic, electrostatic, and covalent interactions) and their effect on drug stability is crucial for developing stable and effective NADs. NADs interact with various environments during delivery, and individual chemical modifications or delivery carriers may not suffice to overcome physiological barriers. Combining nucleic acid chemical modifications with drug delivery systems is promising for better therapeutic outcomes. Moreover, researchers are developing various combined carrier-use modes to facilitate effective drug loading, precise targeting, and release. The emergence of intelligent, responsive nanocarriers may help NADs interact with complex environmental changes, responding to specific in vivo changes, such as pH, redox conditions, or external stimuli (ultrasound, light, magnetic fields, and electric fields). These carriers can improve transfection efficiency in target cells and reduce toxic side effects on normal tissues and cells.564,565 However, hybridizing multiple carriers and modifying functional ligand molecules increase the technical difficulties and manufacturing costs.
Clinical application
Safety
A serious issue in developing these therapeutics is in vivo safety. Despite the detailed understanding of the physicochemical properties of NADs, the potential immunogenicity and toxicity of carriers and related modifications may pose additional challenges to safe and effective NADs delivery.500,566 For example, early application of relatively mature PPMO technology showed that high doses of CPP-PMO conjugates caused kidney injury in rats.567 Cationic polymers, such as PLL/PEI,305,568 have induced cell apoptosis and inflammation in vivo.
Pharmacokinetic properties and adverse reactions
Advanced technologies have generated NADs complexes with different properties.569 Their chemical structure, dosage form, and administration route are the main determinants of their absorption, distribution, metabolism, and excretion, thereby affecting their efficacy and safety.570,571 For example, based on the unique mechanism of action of ASOs, the main safety issues are severe hepatotoxicity and nephrotoxicity caused by drug accumulation.64 Therefore, evaluating NADs delivery systems’ drug metabolism and pharmacokinetics is essential for clinical development. Various bioanalytical methods, such as chromatographic techniques for determining drug plasma concentrations and metabolism569,572 and imaging technology for estimating drug distribution,573 are used, although each has limitations. Low sensitivity and poor discrimination affect the accuracy of determination results,574 highlighting the importance of developing novel analytical methods. Although previous studies have used nano-fluorescent probes to detect the cellular delivery efficiency of oligonucleotide molecules,403,457,575 more pharmacokinetic data on biodistribution, immune compatibility, and toxicity are needed for clinical translation and practical therapeutic application of these systems.266,576
Pathological indications
Most diseases exhibit variability in their phenotypes, making it difficult to achieve a curative effect by targeting a single gene. Issues, such as a small patient population and unique targets, often result in a lack of attention from pharmaceutical companies. Therefore, analyzing the pathogenesis of diseases at the genetic level and developing a personalized treatment plan may offer the possibility of a permanent cure for rare and currently incurable genetic diseases. Compared with antibody drugs and small molecule drugs, NADs are specifically favorable for personalized therapies owing to their strong specificity for diverse targets. They have been explored in the development of new therapies for various diseases. Thus far, genetic diseases have been the most approved indication category for NADs. Additionally, breakthroughs in gene therapy and gene-cell combined therapy are promising for the large-scale clinical application of NADs. For instance, Casgevy®, based on the CRISPR/Cas9 gene-editing system, was the first successful attempt at disease treatment, promising a “one-time treatment for a lifelong effect.” However, the complexity of clinical applications and the difficulty of target selection in gene editing have led to long treatment cycles and high costs. Moreover, the application range of NADs is gradually expanding from rare diseases to common diseases such as chronic diseases, infectious diseases, and ophthalmic diseases. The development of curative drugs for indications with large patient populations, such as potential cures for hepatitis B, may address broader clinical needs.
Scale production
Like other drugs, the production of NADs involves multiple technical stages, including raw material collection, synthesis, separation, purification, transportation, and storage, requiring large-scale production capabilities and stringent quality control.577 Additionally, the specific requirements for sequence size, purity, and modification and delivery methods of NADs, based on different research needs, increase the demands on the production model.214 Therefore, to achieve large-scale clinical application, NADs must ensure easy production, quality control, and transportation. Moreover, as regulatory policies become more refined and the industry chain further develops, companies should seek collaboration opportunities while increasing research and development investments. Collaborations between internationally renowned enterprises in terms of sharing advanced technology and management experience can enhance research and development capabilities and production capacity.
In summary, the development of NADs has undergone a long process, with research on the mechanisms of action of different types of drugs and breakthroughs in chemical modification and delivery technologies. These have transformed NADs from conceptual to practical clinical treatment tools. However, some clinical promotion and application shortcomings still require further development. The organic combination of specific therapeutic nucleic acid molecules, targeted modifications, and functionalized delivery carriers may become vital to achieving personalized nucleic acid therapy and addressing unmet clinical needs.
Acknowledgements
This review was supported by the National Natural Science Foundation of China (82373039) and the University-Industry Collaboration Program (New Drug Targets and New Molecule Discovery and Technology 8210040001, 2020-019). We express our gratitude to Pengfei Gong, Ruipei Ouyang, and Yongjie Shi from Jiangsu Province Engineering Research Center of Synthetic Peptide Drug Discovery and Evaluation, China Pharmaceutical University, for helping revise the manuscript. Figures were created using BioRender.
Author contributions
X.S. developed the concept and wrote the paper, and C.L., S.S., and J.H. discussed, edited, and contributed to the writing. H.X. supervised and revised the paper. All authors have read and approved the article.
Competing interests
The authors declare no competing interests.
References
- 1.Zhang, S. et al. The mechanistic, diagnostic and therapeutic novel nucleic acids for hepatocellular carcinoma emerging in past score years. Brief. Bioinform.22, 1860–1883 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Smith, C. I. E. & Zain, R. Therapeutic oligonucleotides: state of the art. Annu. Rev. Pharmacol. Toxicol.59, 605–630 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Friedmann, T. & Roblin, R. Gene therapy for human genetic disease? Science175, 949–955 (1972). [DOI] [PubMed] [Google Scholar]
- 4.Vaughan, H. J., Green, J. J. & Tzeng, S. Y. Cancer-targeting nanoparticles for combinatorial nucleic acid delivery. Adv. Mater.32, e1901081 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garbo, S., Maione, R., Tripodi, M. & Battistelli, C. Next RNA therapeutics: the mine of non-coding. Int. J. Mol. Sci.23, 7471 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meng, F., Wang, J. & Yeo, Y. Nucleic acid and oligonucleotide delivery for activating innate immunity in cancer immunotherapy. J. Control. Release345, 586–600 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.To, K. K. W. & Cho, W. C. S. An overview of rational design of mRNA-based therapeutics and vaccines. Expert Opin. Drug Discov.16, 1307–1317 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Del Pozo-Rodriguez, A. et al. Gene therapy. Adv. Biochem. Eng. Biotechnol.171, 321–368 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Kumari, N. et al. Oral delivery of nucleic acid therapies for local and systemic action. Pharm. Res.40, 107–122 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maishi, N. et al. Novel antiangiogenic therapy targeting biglycan using tumor endothelial cell-specific liposomal siRNA delivery system. Cancer Sci.113, 1855–1867 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gan, L. et al. A cell-penetrating peptide enhances delivery and efficacy of phosphorodiamidate morpholino oligomers in mdx mice. Mol. Ther. Nucl. Acids30, 17–27 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan, X. et al. Redox-responsive multifunctional polypeptides conjugated with Au nanoparticles for tumor-targeting gene therapy and their 1 + 1 > 2 synergistic effects. ACS Biomater. Sci. Eng.6, 463–473 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Lehto, T., Ezzat, K., Wood, M. J. A. & El Andaloussi, S. Peptides for nucleic acid delivery. Adv. Drug Deliv. Rev.106, 172–182 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Gokirmak, T. et al. Overcoming the challenges of tissue delivery for oligonucleotide therapeutics. Trends Pharmacol. Sci.42, 588–604 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Tan, X., Jia, F., Wang, P. & Zhang, K. Nucleic acid-based drug delivery strategies. J. Control. Release323, 240–252 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sousa De Almeida, M., Rothen-Rutishauser, B., Mayer, M. & Taskova, M. Multi-functionalized heteroduplex antisense oligonucleotides for targeted intracellular delivery and gene silencing in hela cells. Biomedicines10, 2096 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao, Y., Shu, R. & Liu, J. The development and improvement of ribonucleic acid therapy strategies. Mol. Ther. Nucl. Acids26, 997–1013 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarli, S. L. & Watts, J. K. Harnessing nucleic acid technologies for human health on earth and in space. Life Sci. Space Res.35, 113–126 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scully, M. A., Sterin, E. H. & Day, E. S. Membrane-wrapped nanoparticles for nucleic acid delivery. Biomater. Sci.10, 4378–4391 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Den Berg, A. I. S., Yun, C. O., Schiffelers, R. M. & Hennink, W. E. Polymeric delivery systems for nucleic acid therapeutics: approaching the clinic. J. Control. Release331, 121–141 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Panigaj, M. et al. Therapeutic immunomodulation by rationally designed nucleic acids and nucleic acid nanoparticles. Front. Immunol.14, 1053550 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hager, S. & Wagner, E. Bioresponsive polyplexes-chemically programmed for nucleic acid delivery. Expert Opin. Drug Deliv.15, 1067–1083 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Zhang, C. et al. Modification of lipid-based nanoparticles: an efficient delivery system for nucleic acid-based immunotherapy. Molecules27, 1943 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steffens, R. C. & Wagner, E. Directing the way-receptor and chemical targeting strategies for nucleic acid delivery. Pharm. Res.40, 47–76 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evers, M. J. W. et al. Functional siRNA delivery by extracellular vesicle-liposome hybrid nanoparticles. Adv. Healthc. Mater.11, e2101202 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao, J. et al. Polyester-based nanoparticles for nucleic acid delivery. Biomater. Adv.92, 983–994 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Wang, L. et al. Improved transfer efficiency of supercharged 36 + GFP protein mediate nucleic acid delivery. Drug Deliv.29, 386–398 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thess, A. et al. Historic nucleic acids isolated by Friedrich Miescher contain RNA besides DNA. Biol. Chem.402, 1179–1185 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Varshavsky, A. Discovering the RNA double helix and hybridization. Cell127, 1295–1297 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Rich, A. & Davies, D. R. A new two stranded helical structure: polyadenylic acid and polyuridylic acid. J. Am. Chem. Soc.78, 3548–3549 (1956). [Google Scholar]
- 31.Rich, A. A hybrid helix containing both deoxyribose and ribose polynucleotides and its relation to the transfer of information between the nucleic acids. Proc. Natl Acad. Sci. USA46, 1044–1053 (1960). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zamecnik, P. C. & Stephenson, M. L. Inhibition of rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc. Natl Acad. Sci. USA75, 280–284 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berget, S. M., Moore, C. & Sharp, P. A. Spliced segments at the 5’ terminus of adenovirus 2 late mRNA. Proc. Natl Acad. Sci. USA74, 3171–3175 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suran, M. Finding the tail end: the discovery of RNA splicing. Proc. Natl Acad. Sci. USA117, 1829–1832 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, Y. I. et al. RNA splicing is a primary link between genetic variation and disease. Science352, 600–604 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian, J. et al. Aberrant RNA splicing is a primary link between genetic variation and pancreatic cancer risk. Cancer Res82, 2084–2096 (2022). [DOI] [PubMed] [Google Scholar]
- 37.Dominski, Z. & Kole, R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc. Natl Acad. Sci. USA90, 8673–8677 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in caenorhabditis elegans. Nature391, 806–811 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Mccaffrey, A. P. et al. RNA interference in adult mice. Nature418, 38–39 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Zamore, P. D. RNA interference: big applause for silencing in stockholm. Cell127, 1083–1086 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Davis, M. E. et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature464, 1067–1070 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maraganore, J. Reflections on Alnylam. Nat. Biotechnol.40, 641–650 (2022). [DOI] [PubMed] [Google Scholar]
- 43.Baltimore, D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature226, 1209–1211 (1970). [DOI] [PubMed] [Google Scholar]
- 44.Reich, E., Franklin, R. M., Shatkin, A. J. & Tatumel Action of actinomycin D on animal cells and viruses. Proc. Natl Acad. Sci. USA48, 1238–1245 (1962). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krieg, P. A. & Melton, D. A. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res12, 7057 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolff, J. A. et al. Direct gene transfer into mouse muscle in vivo. Science247, 1465–1468 (1990). [DOI] [PubMed] [Google Scholar]
- 47.Jirikowski, G. F., Sanna, P. P., Maciejewski-Lenoir, D. & Bloom, F. E. Reversal of diabetes insipidus in brattleboro rats: intrahypothalamic injection of vasopressin mRNA. Science255, 996–998 (1992). [DOI] [PubMed] [Google Scholar]
- 48.Conry, R. M. et al. Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res55, 1397–1400 (1995). [PubMed] [Google Scholar]
- 49.Kariko, K., Buckstein, M., Ni, H. & Weissman, D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity23, 165–175 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Jackson, L. A. et al. An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med.383, 1920–1931 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sahin, U. et al. COVID-19 vaccine BNT162b1 elicits human antibody and T(H)1 T cell responses. Nature586, 594–599 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Rossi, J. J. & Rossi, D. Oligonucleotides and the COVID-19 pandemic: a perspective. Nucleic Acid Ther.30, 129–132 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dolgin, E. The tangled history of mRNA vaccines. Nature597, 318–324 (2021). [DOI] [PubMed] [Google Scholar]
- 54.Kariko, K. et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther.16, 1833–1840 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gostimskaya, I. CRISPR-Cas9: a history of its discovery and ethical considerations of its use in genome editing. Biochemistry Mosc 87, 777–788 (2022). [DOI] [PMC free article] [PubMed]
- 56.Zhang, H. et al. Application of the CRISPR/Cas9-based gene editing technique in basic research, diagnosis, and therapy of cancer. Mol. Cancer20, 126 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boti, M. A. et al. Recent advances in genome-engineering strategies. Genes14, 129 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frangoul, H., Ho, T. W. & Corbacioglu, S. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. Reply. N. Engl. J. Med.384, e91 (2021). [DOI] [PubMed] [Google Scholar]
- 59.Lu, Z. G. et al. Nucleic acid drug vectors for diagnosis and treatment of brain diseases. Signal Transduct. Target Ther.8, 604–656 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsui, M. & Corey, D. R. Non-coding RNAs as drug targets. Nat. Rev. Drug Discov.16, 167–179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bennett, C. F. & Swayze, E. E. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol.50, 259–293 (2010). [DOI] [PubMed] [Google Scholar]
- 62.Dhuri, K. et al. Antisense oligonucleotides: an emerging area in drug discovery and development. J. Clin. Med.9, 2004 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stein, C. A. & Castanotto, D. FDA-approved oligonucleotide therapies in 2017. Mol. Ther.25, 1069–1075 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alhamadani, F. et al. Adverse drug reactions and toxicity of the food and drug administration-approved antisense oligonucleotide drugs. Drug Metab. Dispos.50, 879–887 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gales, L. Tegsedi (Inotersen): an antisense oligonucleotide approved for the treatment of adult patients with hereditary transthyretin amyloidosis. Pharm. (Basel)12, 78 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paik, J. & Duggan, S. Volanesorsen: first global approval. Drugs79, 1349–1354 (2019). [DOI] [PubMed] [Google Scholar]
- 67.Blair, H. A. Tofersen: first approval. Drugs83, 1039–1043 (2023). [DOI] [PubMed] [Google Scholar]
- 68.Adewunmi, O., Shen, Y., Zhang, X. H. & Rosen, J. M. Targeted inhibition of lncRNA malat1 alters the tumor immune microenvironment in preclinical syngeneic mouse models of triple-negative breast cancer. Cancer Immunol. Res.11, 1462–1479 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Amodio, N. et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia32, 1948–1957 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Esposito, R. et al. Multi-hallmark long noncoding RNA maps reveal non-small cell lung cancer vulnerabilities. Cell Genomics2, 100171 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen, Y., Li, Z., Chen, X. & Zhang, S. Long non-coding RNAs: from disease code to drug role. Acta Pharm. Sin. B11, 340–354 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.De Santi, C. et al. Precise targeting of miRNA sites restores CFTR activity in CF bronchial epithelial cells. Mol. Ther.28, 1190–1199 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sun, Q. et al. Expression and significance of miRNA-21 and BTG2 in lung cancer. Tumour Biol.34, 4017–4026 (2013). [DOI] [PubMed] [Google Scholar]
- 74.Wang, P. Y. et al. Regulating A549 cells growth by ASO inhibiting miRNA expression. Mol. Cell. Biochem.339, 163–171 (2010). [DOI] [PubMed] [Google Scholar]
- 75.Havens, M. A. & Hastings, M. L. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res44, 6549–6563 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aung-Htut, M. T. et al. Systematic approach to developing splice modulating antisense oligonucleotides. Int. J. Mol. Sci.20, 5030 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mogilevsky, M. et al. Modulation of MKNK2 alternative splicing by splice-switching oligonucleotides as a novel approach for glioblastoma treatment. Nucleic Acids Res46, 11396–11404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li, D. et al. Neurodegenerative diseases: a hotbed for splicing defects and the potential therapies. Transl. Neurodegener.10, 16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Balachandran, A. A., Raguraman, P., Rahimizadeh, K. & Veedu, R. N. Splice-switching antisense oligonucleotides targeting extra- and intracellular domains of epidermal growth factor receptor in cancer cells. Biomedicines11, 3299 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Heo, Y. A. Golodirsen: first approval. Drugs80, 329–333 (2020). [DOI] [PubMed] [Google Scholar]
- 81.Boiziau, C. et al. Inhibition of translation initiation by antisense oligonucleotides via an RNase-H independent mechanism. Nucleic Acids Res19, 1113–1119 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dong, Y., Siegwart, D. J. & Anderson, D. G. Strategies, design, and chemistry in siRNA delivery systems. Adv. Drug Deliv. Rev.144, 133–147 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hu, B. et al. Therapeutic siRNA: state of the art. Signal Transduct. Target Ther.5, 101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wittrup, A. & Lieberman, J. Knocking down disease: a progress report on siRNA therapeutics. Nat. Rev. Genet.16, 543–552 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Elbashir, S. M. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature411, 494–498 (2001). [DOI] [PubMed] [Google Scholar]
- 86.Piatek, M. J. & Werner, A. Endogenous siRNAs: regulators of internal affairs. Biochem. Soc. Trans.42, 1174–1179 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nikam, R. R. & Gore, K. R. Journey of siRNA: clinical developments and targeted delivery. Nucleic Acid Ther.28, 209–224 (2018). [DOI] [PubMed] [Google Scholar]
- 88.Rossi, J. J. & Rossi, D. J. siRNA drugs: here to stay. Mol. Ther.29, 431–432 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Robb, G. B. & Rana, T. M. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol. Cell26, 523–537 (2007). [DOI] [PubMed] [Google Scholar]
- 90.Sarisozen, C., Salzano, G. & Torchilin, V. P. Recent advances in siRNA delivery. Biomol. Concepts6, 321–341 (2015). [DOI] [PubMed] [Google Scholar]
- 91.Isazadeh, H. et al. Advances in siRNA delivery approaches in cancer therapy: challenges and opportunities. Mol. Biol. Rep.50, 9529–9543 (2023). [DOI] [PubMed] [Google Scholar]
- 92.Kurakula, H., Vaishnavi, S., Sharif, M. Y. & Ellipilli, S. Emergence of small interfering RNA-based gene drugs for various diseases. ACS Omega8, 20234–20250 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Joshi, B. H. & Pachchigar, K. P. siRNA: novel therapeutics from functional genomics. Biotechnol. Genet. Eng. Rev.30, 1–30 (2014). [DOI] [PubMed] [Google Scholar]
- 94.Ranasinghe, P., Addison, M. L., Dear, J. W. & Webb, D. J. Small interfering RNA: discovery, pharmacology and clinical development-an introductory review. Br. J. Pharmacol.180, 2697–2720 (2023). [DOI] [PubMed] [Google Scholar]
- 95.Sajid, M. I. et al. Overcoming barriers for siRNA therapeutics: from bench to bedside. Pharm. (Basel)13, 294 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gatta, A. K. et al. Strategies for improving the specificity of siRNAs for enhanced therapeutic potential. Expert Opin. Drug Discov.13, 709–725 (2018). [DOI] [PubMed] [Google Scholar]
- 97.Hoy, S. M. Patisiran: first global approval. Drugs78, 1625–1631 (2018). [DOI] [PubMed] [Google Scholar]
- 98.Lee, R. C., Feinbaum, R. L. & Ambros, V. TheC. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell75, 843–854 (1993). [DOI] [PubMed] [Google Scholar]
- 99.Bhatnagar, D., Ladhe, S. & Kumar, D. Discerning the prospects of miRNAs as a multi-target therapeutic and diagnostic for alzheimer’s disease. Mol. Neurobiol.60, 5954–5974 (2023). [DOI] [PubMed] [Google Scholar]
- 100.Gregory, R. I. et al. The microprocessor complex mediates the genesis of microRNAs. Nature432, 235–240 (2004). [DOI] [PubMed] [Google Scholar]
- 101.Han, J. et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell125, 887–901 (2006). [DOI] [PubMed] [Google Scholar]
- 102.Clancy, J. W., Zhang, Y., Sheehan, C. & D’souza-Schorey, C. An ARF6-Exportin-5 axis delivers pre-miRNA cargo to tumour microvesicles. Nat. Cell Biol.21, 856–866 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang, J. et al. XPO5 promotes primary miRNA processing independently of RanGTP. Nat. Commun.11, 1845 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li, Y. et al. The ubiquitin-specific protease USP36 associates with the microprocessor complex and regulates miRNA biogenesis by SUMOylating DGCR8. Cancer Res. Commun.3, 459–470 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wilson, R. C. et al. Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol. Cell57, 397–407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jungers, C. F. & Djuranovic, S. Modulation of miRISC-mediated gene silencing in eukaryotes. Front. Mol. Biosci.9, 832916 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Song, X., Li, Y., Cao, X. & Qi, Y. MicroRNAs and their regulatory roles in plant-environment interactions. Annu. Rev. Plant Biol.70, 489–525 (2019). [DOI] [PubMed] [Google Scholar]
- 108.Gebauer, F., Schwarzl, T., Valcarcel, J. & Hentze, M. W. RNA-binding proteins in human genetic disease. Nat. Rev. Genet.22, 185–198 (2021). [DOI] [PubMed] [Google Scholar]
- 109.Gebert, L. F. R. & Macrae, I. J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol.20, 21–37 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Naeli, P. et al. The intricate balance between microRNA-induced mRNA decay and translational repression. FEBS J.290, 2508–2524 (2023). [DOI] [PubMed] [Google Scholar]
- 111.Dalmay, T. Mechanism of miRNA-mediated repression of mRNA translation. Essays Biochem54, 29–38 (2013). [DOI] [PubMed] [Google Scholar]
- 112.Jame-Chenarboo, F., Ng, H. H., Macdonald, D. & Mahal, L. K. High-throughput analysis reveals miRNA upregulating alpha-2,6-sialic acid through direct miRNA-mRNA interactions. ACS Cent. Sci.8, 1527–1536 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Laitinen, P. et al. Nuclear microRNA-466c regulates Vegfa expression in response to hypoxia. PLoS One17, e0265948 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chipman, L. B. & Pasquinelli, A. E. miRNA targeting: growing beyond the seed. Trends Genet35, 215–222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bonneau, E. et al. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. EJIFCC30, 114–127 (2019). [PMC free article] [PubMed] [Google Scholar]
- 116.Schmidt, M. F. miRNA targeting drugs: the next blockbusters? Methods Mol. Biol.1517, 3–22 (2017). [DOI] [PubMed] [Google Scholar]
- 117.Damase, T. R. et al. The limitless future of RNA therapeutics. Front. Bioeng. Biotech.9, 628137 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim, T. & Croce, C. M. MicroRNA: trends in clinical trials of cancer diagnosis and therapy strategies. Exp. Mol. Med.55, 1314–1321 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chakraborty, C., Sharma, A. R., Sharma, G. & Lee, S. S. Therapeutic advances of miRNAs: a preclinical and clinical update. J. Adv. Res.28, 127–138 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Daige, C. L. et al. Systemic delivery of a miR34a mimic as a potential therapeutic for liver cancer. Mol. Cancer Ther.13, 2352–2360 (2014). [DOI] [PubMed] [Google Scholar]
- 121.Gambari, R., Brognara, E., Spandidos, D. A. & Fabbri, E. Targeting oncomiRNAs and mimicking tumor suppressor miRNAs: new trends in the development of miRNA therapeutic strategies in oncology (Review). Int. J. Oncol.49, 5–32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Thomson, D. W., Bracken, C. P., Szubert, J. M. & Goodall, G. J. On measuring miRNAs after transient transfection of mimics or antisense inhibitors. PLoS One8, e55214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Holjencin, C. & Jakymiw, A. MicroRNAs and their big therapeutic impacts: delivery strategies for cancer intervention. Cells11, 2332 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gallant-Behm, C. L. et al. A microRNA-29 mimic (Remlarsen) represses extracellular matrix expression and fibroplasia in the skin. J. Investig. Dermatol.139, 1073–1081 (2019). [DOI] [PubMed] [Google Scholar]
- 125.Gallant-Behm, C. L. et al. A synthetic microRNA-92a inhibitor (MRG-110) accelerates angiogenesis and wound healing in diabetic and nondiabetic wounds. Wound Repair Regen.26, 311–323 (2018). [DOI] [PubMed] [Google Scholar]
- 126.Seto, A. G. et al. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br. J. Haematol.183, 428–444 (2018). [DOI] [PubMed] [Google Scholar]
- 127.Ghosh, S. et al. The microRNA-10b targeted therapeutic, TTX-MC138, is effective in preclinical pancreatic adenocarcinoma. Cancer Res.83, 548 (2023). [Google Scholar]
- 128.Wang, X. et al. Induction of NANOG expression by targeting promoter sequence with small activating RNA antagonizes retinoic acid-induced differentiation. Biochem. J.443, 821–828 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang, C. et al. Targeted p53 activation by saRNA suppresses human bladder cancer cells growth and metastasis. J. Exp. Clin. Cancer Res.35, 53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Portnoy, V. et al. saRNA-guided Ago2 targets the RITA complex to promoters to stimulate transcription. Cell Res26, 320–335 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gregory, G. L. & Copple, I. M. Modulating the expression of tumor suppressor genes using activating oligonucleotide technologies as a therapeutic approach in cancer. Mol. Ther. Nucl. Acids31, 211–223 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Janowski, B. A. et al. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol.3, 166–173 (2007). [DOI] [PubMed] [Google Scholar]
- 133.Huang, V. et al. RNAa is conserved in mammalian cells. PLoS One5, e8848 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li, C. et al. Upregulation of E‑cadherin expression mediated by a novel dsRNA suppresses the growth and metastasis of bladder cancer cells by inhibiting beta-catenin/TCF target genes. Int. J. Oncol.52, 1815–1826 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Voutila, J. et al. Gene expression profile changes after short-activating RNA-mediated induction of endogenous pluripotency factors in human mesenchymal stem cells. Mol. Ther. Nucl. Acids1, e35 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang, M. et al. saKLK1-374 is more difficult to induce KLK1 expression in normal prostate cell lines than that in prostate cancer cell lines: Rethinking the universality of RNA activation. Biochem. Biophys. Res. Commun.643, 157–168 (2023). [DOI] [PubMed] [Google Scholar]
- 137.Li, B. & Li, C. Suppression of prostate cancer metastasis by DPYSL3-targeted saRNA. Adv. Exp. Med. Biol.983, 207–216 (2017). [DOI] [PubMed] [Google Scholar]
- 138.Yang, K. et al. Antitumor activity of small activating RNAs induced PAWR gene activation in human bladder cancer cells. Int. J. Med. Sci.18, 3039–3049 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kang, M. R. et al. Development of therapeutic dsP21-322 for cancer treatment. Adv. Exp. Med. Biol.983, 217–229 (2017). [DOI] [PubMed] [Google Scholar]
- 140.Zhang, Q. et al. p21CIP/WAF1 saRNA inhibits proliferative vitreoretinopathy in a rabbit model. PLoS One18, e0282063 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tan, C. P. et al. RNA activation-a novel approach to therapeutically upregulate gene transcription. Molecules26, 6530 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sarker, D. et al. MTL-CEBPA, a small activating RNA therapeutic upregulating C/EBP-alpha, in patients with advanced liver cancer: a first-in-human, multicenter, open-label, phase I trial. Clin. Cancer Res.26, 3936–3946 (2020). [DOI] [PubMed] [Google Scholar]
- 143.Hashimoto, A. et al. Upregulation of C/EBPalpha inhibits suppressive activity of myeloid cells and potentiates antitumor response in mice and patients with cancer. Clin. Cancer Res.27, 5961–5978 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Reebye, V. et al. Gene activation of CEBPA using saRNA: preclinical studies of the first in human saRNA drug candidate for liver cancer. Oncogene37, 3216–3228 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jarvelainen, H. et al. Preclinical development of RAG1-40-31L: a novel small activating RNA-lipid conjugate targeting tumor suppressor gene p21 for treatment of non-muscle invasive bladder cancer. J. Clin. Oncol.41, e16620 (2023). [Google Scholar]
- 146.Ishino, Y. et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol.169, 5429–5433 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science315, 1709–1712 (2007). [DOI] [PubMed] [Google Scholar]
- 148.Makarova, K. S., Wolf, Y. I. & Koonin, E. V. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res41, 4360–4377 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Charpentier, E. & Marraffini, L. A. Harnessing CRISPR-Cas9 immunity for genetic engineering. Curr. Opin. Microbiol.19, 114–119 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cox, D. B. T. et al. RNA editing with CRISPR-Cas13. Science358, 1019–1027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ghaemi, A. et al. CRISPR-cas9 genome editing delivery systems for targeted cancer therapy. Life Sci.267, 118969 (2021). [DOI] [PubMed] [Google Scholar]
- 152.Herrera-Carrillo, E., Gao, Z. & Berkhout, B. CRISPR therapy towards an HIV cure. Brief. Funct. Genomics19, 201–208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li, Y. et al. Ex vivo cell-based CRISPR/Cas9 genome editing for therapeutic applications. Biomaterials234, 119711 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol.31, 827–832 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhang, F., Wen, Y. & Guo, X. CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum. Mol. Genet.23, R40–R46 (2014). [DOI] [PubMed] [Google Scholar]
- 156.Savic, N. & Schwank, G. Advances in therapeutic CRISPR/Cas9 genome editing. Transl. Res.168, 15–21 (2016). [DOI] [PubMed] [Google Scholar]
- 157.Lino, C. A., Harper, J. C., Carney, J. P. & Timlin, J. A. Delivering CRISPR: a review of the challenges and approaches. Drug Deliv.25, 1234–1257 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang, J. Y. & Doudna, J. A. CRISPR technology: a decade of genome editing is only the beginning. Science379, eadd8643 (2023). [DOI] [PubMed] [Google Scholar]
- 159.Zhang, X. et al. Robust genome editing in adult vascular endothelium by nanoparticle delivery of CRISPR-Cas9 plasmid DNA. Cell Rep.38, 110196 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang, S., Shen, J., Li, D. & Cheng, Y. Strategies in the delivery of Cas9 ribonucleoprotein for CRISPR/Cas9 genome editing. Theranostics11, 614–648 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Taha, E. A., Lee, J. & Hotta, A. Delivery of CRISPR-Cas tools for in vivo genome editing therapy: trends and challenges. J. Control. Release342, 345–361 (2022). [DOI] [PubMed] [Google Scholar]
- 162.Chen, G. et al. A biodegradable nanocapsule delivers a Cas9 ribonucleoprotein complex for in vivo genome editing. Nat. Nanotechnol.14, 974–980 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mirjalili Mohanna, S. Z. et al. LNP-mediated delivery of CRISPR RNP for wide-spread in vivo genome editing in mouse cornea. J. Control. Release350, 401–413 (2022). [DOI] [PubMed] [Google Scholar]
- 164.Luther, D. C. et al. Delivery approaches for CRISPR/Cas9 therapeutics in vivo: advances and challenges. Expert Opin. Drug Deliv.15, 905–913 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hoy, S. M. Exagamglogene autotemcel: first approval. Mol. Diagn. Ther.28, 133–139 (2024). [DOI] [PubMed] [Google Scholar]
- 166.Philippidis, A. CASGEVY makes history as FDA approves first CRISPR/Cas9 genome edited therapy. Hum. Gene Ther.35, 1–4 (2024). [DOI] [PubMed] [Google Scholar]
- 167.Wang, T. et al. Three decades of nucleic acid aptamer technologies: lessons learned, progress and opportunities on aptamer development. Biotechnol. Adv.37, 28–50 (2019). [DOI] [PubMed] [Google Scholar]
- 168.Perret, G. & Boschetti, E. Aptamer-based affinity chromatography for protein extraction and purification. Adv. Biochem. Eng. Biotechnol.174, 93–139 (2020). [DOI] [PubMed] [Google Scholar]
- 169.Bunka, D. H., Platonova, O. & Stockley, P. G. Development of aptamer therapeutics. Curr. Opin. Pharmacol.10, 557–562 (2010). [DOI] [PubMed] [Google Scholar]
- 170.Costello, A. M. et al. Selection and characterization of vimentin-binding aptamer motifs for ovarian cancer. Molecules26, 6525 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ren, W. et al. Nanotechnology lighting the way for gene therapy in ophthalmopathy: from opportunities toward applications. Molecules28, 3500 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hermann, T. & Patel, D. J. Adaptive recognition by nucleic acid aptamers. Science287, 820–825 (2000). [DOI] [PubMed] [Google Scholar]
- 173.Nimjee, S. M., Rusconi, C. P. & Sullenger, B. A. Aptamers: an emerging class of therapeutics. Annu. Rev. Med.56, 555–583 (2005). [DOI] [PubMed] [Google Scholar]
- 174.Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science249, 505–510 (1990). [DOI] [PubMed] [Google Scholar]
- 175.Ellington, A. D. & Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature346, 818–822 (1990). [DOI] [PubMed] [Google Scholar]
- 176.Wang, Q. et al. An efficient and universal in silico screening strategy for acquisition of high-affinity aptamer and its application in analytical utility. Talanta269, 125535 (2024). [DOI] [PubMed] [Google Scholar]
- 177.Lin, Y. et al. A modified SELEX approach to identify DNA aptamers with binding specificity to the major histocompatibility complex presenting ovalbumin model antigen. RSC Adv.13, 32681–32693 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Cao, J., Zhang, F. & Xiong, W. Discovery of aptamers and the acceleration of the development of targeting research in ophthalmology. Int. J. Nanomed.18, 4421–4430 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Doherty, C., Wilbanks, B., Khatua, S. & Maher, L. J. Aptamers in neuro-oncology: an emerging therapeutic modality. Neuro Oncol.26, 38–54 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Thomas, B. J. et al. Targeting lung cancer with clinically relevant EGFR mutations using anti-EGFR RNA aptamer. Mol. Ther. Nucl. Acids34, 102046 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ng, E. W. et al. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug. Disc.5, 123–132 (2006). [DOI] [PubMed] [Google Scholar]
- 182.Kim, B. J. et al. Targeting complement components C3 and C5 for the retina: key concepts and lingering questions. Prog. Retin. Eye. Res.83, 100936 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Rosenberg, J. E. et al. A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Investig. N. Drugs32, 178–187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cheng, Y. et al. AS1411-induced growth inhibition of glioma cells by up-regulation of p53 and down-regulation of Bcl-2 and Akt1 via nucleolin. PLoS One11, e0167094 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Rizzieri, D. et al. Long-term outcomes of responders in a randomized, controlled phase II trial of aptamer AS1411 in AML. J. Clin. Oncol.28, 6557 (2010). [Google Scholar]
- 186.Ali, G. K., Algethami, F. K. & Omer, K. M. Gold single atom-based aptananozyme as an ultrasensitive and selective colorimetric probe for detection of thrombin and C-reactive protein. Mikrochim Acta191, 59 (2023). [DOI] [PubMed] [Google Scholar]
- 187.Tavassoli, M. et al. Aptamer-modified metal organic frameworks for measurement of food contaminants: a review. Microchim. Acta190, 371 (2023). [DOI] [PubMed] [Google Scholar]
- 188.Narwade, M. et al. Advanced cancer targeting using aptamer functionalized nanocarriers for site-specific cargo delivery. Biomater. Res.27, 42 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Vlatkovic, I. Non-immunotherapy application of LNP-mRNA: maximizing efficacy and safety. Biomedicines9, 530 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lee, Y. et al. Immunogenicity of lipid nanoparticles and its impact on the efficacy of mRNA vaccines and therapeutics. Exp. Mol. Med.55, 2085–2096 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ali, S. et al. Design of a new cell penetrating peptide for DNA, siRNA and mRNA delivery. J. Gene Med.24, e3401 (2022). [DOI] [PubMed] [Google Scholar]
- 192.Riley, R. S. et al. Ionizable lipid nanoparticles for in utero mRNA delivery. Sci. Adv.7, eaba1028 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Swingle, K. L. et al. Amniotic fluid stabilized lipid nanoparticles for in utero intra-amniotic mRNA delivery. J. Control. Release341, 616–633 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.An, D. et al. Systemic messenger RNA therapy as a treatment for methylmalonic acidemia. Cell Rep.21, 3548–3558 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Gurung, S. et al. mRNA therapy corrects defective glutathione metabolism and restores ureagenesis in preclinical argininosuccinic aciduria. Sci. Transl. Med.16, eadh1334 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Seker Yilmaz, B. & Gissen, P. Genetic therapy approaches for ornithine transcarbamylase deficiency. Biomedicines11, 2227 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Attarwala, H. et al. Translational pharmacokinetic/pharmacodynamic model for mRNA-3927, an investigational therapeutic for the treatment of propionic acidemia. Nucleic Acid Ther.33, 141–147 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Verbeke, R., Lentacker, I., De Smedt, S. C. & Dewitte, H. The dawn of mRNA vaccines: the COVID-19 case. J. Control. Release333, 511–520 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lamb, Y. N. BNT162b2 mRNA COVID-19 vaccine: first approval. Drugs81, 495–501 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhang, N. N. et al. A thermostable mRNA vaccine against COVID-19. Cell182, 1271–1283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Liu, X. et al. Safety and superior immunogenicity of heterologous boosting with an RBD-based SARS-CoV-2 mRNA vaccine in Chinese adults. Cell Res32, 777–780 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bahl, K. et al. Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol. Ther.30, 2874 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Feldman, R. A. et al. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine37, 3326–3334 (2019). [DOI] [PubMed] [Google Scholar]
- 204.Aliprantis, A. O. et al. A phase 1, randomized, placebo-controlled study to evaluate the safety and immunogenicity of an mRNA-based RSV prefusion F protein vaccine in healthy younger and older adults. Hum. Vaccines Immunother.17, 1248–1261 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Aldrich, C. et al. Proof-of-concept of a low-dose unmodified mRNA-based rabies vaccine formulated with lipid nanoparticles in human volunteers: a phase 1 trial. Vaccine39, 1310–1318 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lorentzen, C. L., Haanen, J. B., Met, O. & Svane, I. M. Clinical advances and ongoing trials on mRNA vaccines for cancer treatment. Lancet Oncol.23, e450–e458 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Vishweshwaraiah, Y. L. & Dokholyan, N. V. mRNA vaccines for cancer immunotherapy. Front. Immunol.13, 1029069 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wei, J. & Hui, A. M. The paradigm shift in treatment from Covid-19 to oncology with mRNA vaccines. Cancer Treat. Rev.107, 102405 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Dou, H. H. et al. An automated high-throughput fluorescence in situ hybridization (FISH) assay platform for use in the identification and optimization of siRNA-based therapeutics. SLAS Discov.26, 281–291 (2021). [DOI] [PubMed] [Google Scholar]
- 210.Sherman, M. & Contreras, L. Computational approaches in design of nucleic acid-based therapeutics. Curr. Opin. Biotech.53, 232–239 (2018). [DOI] [PubMed] [Google Scholar]
- 211.Dai, H. et al. Pancreatic cancer: nucleic acid drug discovery and targeted therapy. Front. Cell Dev. Biol.10, 855474 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kohlberger, M. & Gadermaier, G. SELEX: critical factors and optimization strategies for successful aptamer selection. Biotechnol. Appl. Bioc.69, 1771–1792 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Metkar, M., Pepin, C. S. & Moore, M. J. Tailor made: the art of therapeutic mRNA design. Nat. Rev. Drug. Disc.23, 67–83 (2024). [DOI] [PubMed] [Google Scholar]
- 214.Kawamoto, Y., Wu, Y., Takahashi, Y. & Takakura, Y. Development of nucleic acid medicines based on chemical technology. Adv. Drug Deliv. Rev.199, 114872 (2023). [DOI] [PubMed] [Google Scholar]
- 215.Leppek, K. et al. Combinatorial optimization of mRNA structure, stability, and translation for RNA-based therapeutics. Nat. Commun.13, 1536 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Jiang, X. et al. Oral delivery of nucleic acid therapeutics: challenges, strategies, and opportunities. Drug. Discov. Today28, 103507 (2023). [DOI] [PubMed] [Google Scholar]
- 217.Lu, M. et al. Overcoming pharmaceutical bottlenecks for nucleic acid drug development. Acc. Chem. Res.56, 224–236 (2023). [DOI] [PubMed] [Google Scholar]
- 218.Zhang, Z. et al. Nucleic acid-based therapy for brain cancer: challenges and strategies. J. Control. Release350, 80–92 (2022). [DOI] [PubMed] [Google Scholar]
- 219.Weng, Y. et al. The challenge and prospect of mRNA therapeutics landscape. Biotechnol. Adv.40, 107534 (2020). [DOI] [PubMed] [Google Scholar]
- 220.Kawabata, K., Takakura, Y. & Hashida, M. The fate of plasmid DNA after intravenous injection in mice: involvement of scavenger receptors in its hepatic uptake. Pharm. Res.12, 825–830 (1995). [DOI] [PubMed] [Google Scholar]
- 221.Huang, X. et al. The landscape of mRNA nanomedicine. Nat. Med.28, 2273–2287 (2022). [DOI] [PubMed] [Google Scholar]
- 222.Lechardeur, D. & Lukacs, G. L. Intracellular barriers to non-viral gene transfer. Curr. Gene Ther.2, 183–194 (2002). [DOI] [PubMed] [Google Scholar]
- 223.Eygeris, Y., Gupta, M., Kim, J. & Sahay, G. Chemistry of lipid nanoparticles for RNA delivery. Acc. Chem. Res.55, 2–12 (2022). [DOI] [PubMed] [Google Scholar]
- 224.Sasso, J. M. et al. The progress and promise of RNA medicine horizontal line an arsenal of targeted treatments. J. Med. Chem.65, 6975–7015 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Shinohara, F. et al. siRNA potency enhancement via chemical modifications of nucleotide bases at the 5’-end of the siRNA guide strand. RNA27, 163–173 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Mckenzie, L. K. et al. Recent progress in non-native nucleic acid modifications. Chem. Soc. Rev.50, 5126–5164 (2021). [DOI] [PubMed] [Google Scholar]
- 227.Crooke, S. T., Baker, B. F., Crooke, R. M. & Liang, X. H. Antisense technology: an overview and prospectus. Nat. Rev. Drug Discov.20, 427–453 (2021). [DOI] [PubMed] [Google Scholar]
- 228.Wang, J., Tian, T., Li, X. & Zhang, Y. Noncoding RNAs emerging as drugs or drug targets: their chemical modification, bio-conjugation and intracellular regulation. Molecules27, 6717 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Bege, M. & Borbas, A. The medicinal chemistry of artificial nucleic acids and therapeutic oligonucleotides. Pharm. (Basel)15, 909 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Zheng, Y. Y., Wu, Y., Begley, T. J. & Sheng, J. Sulfur modification in natural RNA and therapeutic oligonucleotides. RSC Chem. Biol.2, 990–1003 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Herkt, M. & Thum, T. Pharmacokinetics and proceedings in clinical application of nucleic acid therapeutics. Mol. Ther.29, 521–539 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Crooke, S. T., Vickers, T. A. & Liang, X. H. Phosphorothioate modified oligonucleotide-protein interactions. Nucleic Acids Res48, 5235–5253 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Vasquez, G. et al. Evaluation of phosphorus and non-phosphorus neutral oligonucleotide backbones for enhancing therapeutic index of gapmer antisense oligonucleotides. Nucleic Acid Ther.32, 40–50 (2022). [DOI] [PubMed] [Google Scholar]
- 234.Vitravene Study, G. Safety of intravitreous fomivirsen for treatment of cytomegalovirus retinitis in patients with AIDS. Am. J. Ophthalmol.133, 484–498 (2002). [DOI] [PubMed] [Google Scholar]
- 235.Chen, S. et al. Systematic evaluation of 2’-fluoro modified chimeric antisense oligonucleotide-mediated exon skipping in vitro. Sci. Rep.9, 6078 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Gangopadhyay, S. & Gore, K. R. Advances in siRNA therapeutics and synergistic effect on siRNA activity using emerging dual ribose modifications. RNA Biol.19, 452–467 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Goswami, A., Prasad, A. K., Maity, J. & Khaneja, N. Synthesis and applications of bicyclic sugar modified locked nucleic acids: a review. Nucleosides Nucleotides Nucleic Acids41, 503–529 (2022). [DOI] [PubMed] [Google Scholar]
- 238.Nielsen, K. E. et al. NMR studies of fully modified locked nucleic acid (LNA) hybrids: solution structure of an LNA:RNA hybrid and characterization of an LNA:DNA hybrid. Bioconjugate Chem.15, 449–457 (2004). [DOI] [PubMed] [Google Scholar]
- 239.Kamali, M. J. et al. Locked nucleic acid (LNA): a modern approach to cancer diagnosis and treatment. Exp. Cell Res.423, 113442 (2023). [DOI] [PubMed] [Google Scholar]
- 240.Pal, R., Deb, I., Sarzynska, J. & Lahiri, A. LNA-induced dynamic stability in a therapeutic aptamer: insights from molecular dynamics simulations. J. Biomol. Struct. Dyn.41, 2221–2230 (2023). [DOI] [PubMed] [Google Scholar]
- 241.Roxo, C. & Pasternak, A. Changes in physicochemical and anticancer properties modulated by chemically modified sugar moieties within sequence-related G-quadruplex structures. PLoS One17, e0273528 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Pasternak, A. & Wengel, J. Unlocked nucleic acid-an RNA modification with broad potential. Org. Biomol. Chem.9, 3591–3597 (2011). [DOI] [PubMed] [Google Scholar]
- 243.Werk, D. et al. Application of small interfering RNAs modified by unlocked nucleic acid (UNA) to inhibit the heart-pathogenic coxsackievirus B3. FEBS Lett.584, 591–598 (2010). [DOI] [PubMed] [Google Scholar]
- 244.Snead, N. M., Escamilla-Powers, J. R., Rossi, J. J. & Mccaffrey, A. P. 5’ unlocked nucleic acid modification improves siRNA targeting. Mol. Ther. Nucl. Acids2, e103 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Yoshida, T. et al. Identification of nucleobase chemical modifications that reduce the hepatotoxicity of gapmer antisense oligonucleotides. Nucleic Acids Res50, 7224–7234 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Svitkin, Y. V. et al. N1-methyl-pseudouridine in mRNA enhances translation through eIF2alpha-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res45, 6023–6036 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Nance, K. D. et al. Cytidine acetylation yields a hypoinflammatory synthetic messenger RNA. Cell Chem. Bio.29, 312–320 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Wang, Y. et al. Detection and application of 5-formylcytosine and 5-formyluracil in DNA. Acc. Chem. Res.52, 1016–1024 (2019). [DOI] [PubMed] [Google Scholar]
- 249.Koseki, J. et al. Theoretical analyses and experimental validation of the effects caused by the fluorinated substituent modification of DNA. Sci. Rep.10, 1138 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Sun, H., Li, K., Liu, C. & Yi, C. Regulation and functions of non-m(6)A mRNA modifications. Nat. Rev. Mol. Cell Biol.24, 714–731 (2023). [DOI] [PubMed] [Google Scholar]
- 251.Kariko, K., Muramatsu, H., Ludwig, J. & Weissman, D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res39, e142 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Andries, O. et al. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release217, 337–344 (2015). [DOI] [PubMed] [Google Scholar]
- 253.Pantazopoulou, V. I. et al. From the argonauts mythological sailors to the argonautes RNA-silencing navigators: their emerging roles in human-cell pathologies. Int. J. Mol. Sci.21, 4007 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Choung, S. et al. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem. Biophys. Res. Commun.342, 919–927 (2006). [DOI] [PubMed] [Google Scholar]
- 255.Hammond, S. M. et al. Delivery of oligonucleotide-based therapeutics: challenges and opportunities. EMBO Mol. Med.13, e13243 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Chen, C., Yang, Z. & Tang, X. Chemical modifications of nucleic acid drugs and their delivery systems for gene-based therapy. Med. Res. Rev.38, 829–869 (2018). [DOI] [PubMed] [Google Scholar]
- 257.Kuwayama, H. Peptide nucleic acid as a template for Taq DNA polymerase. Biochem. Biophys. Res. Commun.579, 76–80 (2021). [DOI] [PubMed] [Google Scholar]
- 258.Gupta, A., Mishra, A. & Puri, N. Peptide nucleic acids: advanced tools for biomedical applications. J. Biotechnol.259, 148–159 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Singh, K. R., Sridevi, P. & Singh, R. P. Potential applications of peptide nucleic acid in biomedical domain. Eng. Rep.2, e12238 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Chen, W., Dong, B., Liu, W. & Liu, Z. Recent advances in peptide nucleic acids as antibacterial agents. Curr. Med. Chem.28, 1104–1125 (2021). [DOI] [PubMed] [Google Scholar]
- 261.Maksudov, F. et al. Therapeutic phosphorodiamidate morpholino oligonucleotides: physical properties, solution structures, and folding thermodynamics. Mol. Ther. Nucl. Acids31, 631–647 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Mcdonald, C. M. et al. Open-label evaluation of eteplirsen in patients with duchenne muscular dystrophy amenable to exon 51 skipping: PROMOVI Trial. J. Neuromuscul. Dis.8, 989–1001 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Dhillon, S. Viltolarsen: first approval. Drugs80, 1027–1031 (2020). [DOI] [PubMed] [Google Scholar]
- 264.Shirley, M. Casimersen: first approval. Drugs81, 875–879 (2021). [DOI] [PubMed] [Google Scholar]
- 265.Egli, M. & Manoharan, M. Chemistry, structure and function of approved oligonucleotide therapeutics. Nucleic Acids Res51, 2529–2573 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Roberts, T. C., Langer, R. & Wood, M. J. A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov.19, 673–694 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Gait, M. J. & Agrawal, S. Introduction and history of the chemistry of nucleic acids therapeutics. Methods Mol. Biol.2434, 3–31 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Ku, S. H. et al. Chemical and structural modifications of RNAi therapeutics. Adv. Drug Deliv. Rev.104, 16–28 (2016). [DOI] [PubMed] [Google Scholar]
- 269.Zong, Y., Lin, Y., Wei, T. & Cheng, Q. Lipid nanoparticle (LNP) enables mRNA delivery for cancer therapy. Adv. Mater.35, e2303261 (2023). [DOI] [PubMed] [Google Scholar]
- 270.Jung, H. N. et al. Lipid nanoparticles for delivery of RNA therapeutics: current status and the role of in vivo imaging. Theranostics12, 7509–7531 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Hald Albertsen, C. et al. The role of lipid components in lipid nanoparticles for vaccines and gene therapy. Adv. Drug Deliv. Rev.188, 114416 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Samaridou, E., Heyes, J. & Lutwyche, P. Lipid nanoparticles for nucleic acid delivery: current perspectives. Adv. Drug Deliv. Rev.154-155, 37–63 (2020). [DOI] [PubMed] [Google Scholar]
- 273.Vhora, I. et al. Lipid-nucleic acid nanoparticles of novel ionizable lipids for systemic BMP-9 gene delivery to bone-marrow mesenchymal stem cells for osteoinduction. Int. J. Pharm.563, 324–336 (2019). [DOI] [PubMed] [Google Scholar]
- 274.Wang, C., Zhang, Y. & Dong, Y. Lipid nanoparticle-mRNA formulations for therapeutic applications. Acc. Chem. Res.54, 4283–4293 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Zhang, Y. et al. Lipids and lipid derivatives for RNA delivery. Chem. Rev.121, 12181–12277 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Heyes, J., Palmer, L., Bremner, K. & Maclachlan, I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control. Release107, 276–287 (2005). [DOI] [PubMed] [Google Scholar]
- 277.Jayaraman, M. et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed.51, 8529–8533 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Ferraresso, F. et al. Comparison of DLin-MC3-DMA and ALC-0315 for siRNA delivery to hepatocytes and hepatic stellate cells. Mol. Pharmaceutics19, 2175–2182 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Zhang, M., Sun, J., Li, M. & Jin, X. Modified mRNA-LNP vaccines confer protection against experimental DENV-2 infection in mice. Mol. Ther. Methods Clin. Dev.18, 702–712 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Escalona-Rayo, O. et al. In vitro and in vivo evaluation of clinically-approved ionizable cationic lipids shows divergent results between mRNA transfection and vaccine efficacy. Biomed. Pharmacother.165, 115065 (2023). [DOI] [PubMed] [Google Scholar]
- 281.Han, X. et al. An ionizable lipid toolbox for RNA delivery. Nat. Commun.12, 7233 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Mui, B. L. et al. Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol. Ther. Nucl. Acids2, e139 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Miao, L. et al. Synergistic lipid compositions for albumin receptor mediated delivery of mRNA to the liver. Nat. Commun.11, 2424 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Song, L. Y. et al. Characterization of the inhibitory effect of PEG-lipid conjugates on the intracellular delivery of plasmid and antisense DNA mediated by cationic lipid liposomes. Biochim. Biophys. Acta1558, 1–13 (2002). [DOI] [PubMed] [Google Scholar]
- 285.Kong, L., Campbell, F. & Kros, A. DePEGylation strategies to increase cancer nanomedicine efficacy. Nanoscale Horiz.4, 378–387 (2019). [DOI] [PubMed] [Google Scholar]
- 286.Kulkarni, J. A. et al. Lipid nanoparticle technology for clinical translation of siRNA therapeutics. Acc. Chem. Res.52, 2435–2444 (2019). [DOI] [PubMed] [Google Scholar]
- 287.Shi, D. et al. To PEGylate or not to PEGylate: immunological properties of nanomedicine’s most popular component, polyethylene glycol and its alternatives. Adv. Drug Deliv. Rev.180, 114079 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Kim, J., Eygeris, Y., Gupta, M. & Sahay, G. Self-assembled mRNA vaccines. Adv. Drug Deliv. Rev.170, 83–112 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Liu, S. et al. Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR-Cas gene editing. Nat. Mater.20, 701–710 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Paunovska, K. et al. Nanoparticles containing oxidized cholesterol deliver mRNA to the liver microenvironment at clinically relevant doses. Adv. Mater.31, e1807748 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Sebastiani, F. et al. Apolipoprotein E binding drives structural and compositional rearrangement of mRNA-containing lipid nanoparticles. ACS Nano15, 6709–6722 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Cheng, Q. et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol.15, 313–320 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Wang, Y. S. et al. mRNA-based vaccines and therapeutics: an in-depth survey of current and upcoming clinical applications. J. Biomed. Sci.30, 84 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Kenjo, E. et al. Low immunogenicity of LNP allows repeated administrations of CRISPR-Cas9 mRNA into skeletal muscle in mice. Nat. Commun.12, 7101 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.August, A. et al. A phase 1 trial of lipid-encapsulated mRNA encoding a monoclonal antibody with neutralizing activity against Chikungunya virus. Nat. Med.27, 2224–2233 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Kotit, S. Lessons from the first-in-human in vivo CRISPR/Cas9 editing of the TTR gene by NTLA-2001 trial in patients with transthyretin amyloidosis with cardiomyopathy. Glob. Cardiol. Sci. Pract.2023, e202304 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Zhang, L. et al. Effect of mRNA-LNP components of two globally-marketed COVID-19 vaccines on efficacy and stability. NPJ Vaccines8, 156 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Rai, R., Alwani, S. & Badea, I. Polymeric nanoparticles in gene therapy: new avenues of design and optimization for delivery applications. Polym. (Basel)11, 745 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Patnaik, S. & Gupta, K. C. Novel polyethylenimine-derived nanoparticles for in vivo gene delivery. Expert Opin. Drug Deliv.10, 215–228 (2013). [DOI] [PubMed] [Google Scholar]
- 300.Jiang, C. et al. Recent advances in the development of polyethylenimine-based gene vectors for safe and efficient gene delivery. Expert Opin. Drug Deliv.16, 363–376 (2019). [DOI] [PubMed] [Google Scholar]
- 301.Wang, X., Niu, D., Hu, C. & Li, P. Polyethyleneimine-based nanocarriers for gene delivery. Curr. Pharm. Des.21, 6140–6156 (2015). [DOI] [PubMed] [Google Scholar]
- 302.Zhang, Y. H. et al. Recycling gene carrier with high efficiency and low toxicity mediated by L-cystine-bridged bis(beta-cyclodextrin)s. Sci. Rep.4, 7471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Ke, X. et al. Surface-functionalized PEGylated nanoparticles deliver messenger rna to pulmonary immune cells. ACS Appl. Mater. Interfaces12, 35835–35844 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Chen, J. et al. Peptide-based and polypeptide-based gene delivery systems. Top. Curr. Chem.375, 32 (2017). [DOI] [PubMed] [Google Scholar]
- 305.Souri, M. et al. Poly-L-lysine/hyaluronan nanocarriers as a novel nanosystem for gene delivery. J. Microsc.287, 32–44 (2022). [DOI] [PubMed] [Google Scholar]
- 306.Djafari, J. et al. Study and preparation of multifunctional poly(L-Lysine)@hyaluronic acid nanopolyplexes for the effective delivery of tumor suppressive mir-34a into triple-negative breast cancer cells. Mater. (Basel)13, 5309 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Li, J. et al. Copolymer of poly(ethylene glycol) and poly(L-lysine) grafting polyethylenimine through a reducible disulfide linkage for siRNA delivery. Nanoscale6, 1732–1740 (2014). [DOI] [PubMed] [Google Scholar]
- 308.Yang, W. et al. Polymeric Micelles with pH-responsive cross-linked core enhance in vivo mrna delivery. Pharmaceutics14, 1205 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Jafari, S., Maleki Dizaj, S. & Adibkia, K. Cell-penetrating peptides and their analogues as novel nanocarriers for drug delivery. Bioimpacts5, 103–111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Debacker, A. J. et al. Delivery of oligonucleotides to the liver with GalNAc: from research to registered therapeutic drug. Mol. Ther.28, 1759–1771 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Nair, J. K. et al. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc.136, 16958–16961 (2014). [DOI] [PubMed] [Google Scholar]
- 312.Cui, H. et al. Liver-targeted delivery of oligonucleotides with N-acetylgalactosamine conjugation. ACS Omega6, 16259–16265 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Brown, C. R. et al. Investigating the pharmacodynamic durability of GalNAc-siRNA conjugates. Nucleic Acids Res48, 11827–11844 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Abdelaal, A. M. & Kasinski, A. L. Ligand-mediated delivery of RNAi-based therapeutics for the treatment of oncological diseases. NAR Cancer3, zcab030 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Huang, X., Leroux, J. C. & Castagner, B. Well-defined multivalent ligands for hepatocytes targeting via asialoglycoprotein receptor. Bioconjugate Chem.28, 283–295 (2017). [DOI] [PubMed] [Google Scholar]
- 316.Scharner, J. et al. Delivery of GalNAc-conjugated splice-switching ASOs to non-hepatic cells through ectopic expression of asialoglycoprotein receptor. Mol. Ther. Nucl. Acids16, 313–325 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Thangamani, L. et al. GalNAc-siRNA conjugates: prospective tools on the frontier of anti-viral therapeutics. Pharmacol. Res.173, 105864 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Nair, J. K. et al. Impact of enhanced metabolic stability on pharmacokinetics and pharmacodynamics of GalNAc-siRNA conjugates. Nucleic Acids Res45, 10969–10977 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Foster, D. J. et al. Advanced siRNA designs further improve in vivo performance of GalNAc-siRNA conjugates. Mol. Ther.26, 708–717 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Hassler, M. R. et al. Comparison of partially and fully chemically-modified siRNA in conjugate-mediated delivery in vivo. Nucleic Acids Res46, 2185–2196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Parmar, R. G. et al. Facile synthesis, geometry, and 2’-substituent-dependent in vivo activity of 5’-(E)- and 5’-(Z)-vinylphosphonate-modified siRNA conjugates. J. Med. Chem.61, 734–744 (2018). [DOI] [PubMed] [Google Scholar]
- 322.Janas, M. M. et al. Safety evaluation of 2’-deoxy-2’-fluoro nucleotides in GalNAc-siRNA conjugates. Nucleic Acids Res47, 3306–3320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Westerlind, U. et al. Ligands of the asialoglycoprotein receptor for targeted gene delivery, part 1: synthesis of and binding studies with biotinylated cluster glycosides containing N-acetylgalactosamine. Glycoconj. J.21, 227–241 (2004). [DOI] [PubMed] [Google Scholar]
- 324.Rensen, P. C. et al. Determination of the upper size limit for uptake and processing of ligands by the asialoglycoprotein receptor on hepatocytes in vitro and in vivo. J. Biol. Chem.276, 37577–37584 (2001). [DOI] [PubMed] [Google Scholar]
- 325.Schmidt, K. et al. Characterizing the effect of GalNAc and phosphorothioate backbone on binding of antisense oligonucleotides to the asialoglycoprotein receptor. Nucleic Acids Res45, 2294–2306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Lee, Y. C. et al. Binding of synthetic oligosaccharides to the hepatic Gal/GalNAc lectin. dependence on fine structural features. J. Biol. Chem.258, 199–202 (1983). [PubMed] [Google Scholar]
- 327.Biessen, E. A. et al. Synthesis of cluster galactosides with high affinity for the hepatic asialoglycoprotein receptor. J. Med. Chem.38, 1538–1546 (1995). [DOI] [PubMed] [Google Scholar]
- 328.Kichler, A. & Schuber, F. Versatile synthesis of bi- and tri-antennary galactose ligands: interaction with the Gal/GalNAc receptor of human hepatoma cells. Glycoconj. J.12, 275–281 (1995). [DOI] [PubMed] [Google Scholar]
- 329.Huang, Y. Preclinical and clinical advances of GalNAc-decorated nucleic acid therapeutics. Mol. Ther. Nucl. Acids6, 116–132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Syed, Y. Y. Nedosiran: first approval. Drugs83, 1729–1733 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Xu, H., Liao, C., Liang, S. & Ye, B. C. A novel peptide-equipped exosomes platform for delivery of antisense oligonucleotides. ACS Appl. Mater. Interfaces13, 10760–10767 (2021). [DOI] [PubMed] [Google Scholar]
- 332.Crescitelli, R., Lasser, C. & Lotvall, J. Isolation and characterization of extracellular vesicle subpopulations from tissues. Nat. Protoc.16, 1548–1580 (2021). [DOI] [PubMed] [Google Scholar]
- 333.Delorme-Axford, E. et al. Human placental trophoblasts confer viral resistance to recipient cells. Proc. Natl Acad. Sci. USA110, 12048–12053 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Yang, T. et al. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in danio rerio. Pharm. Res.32, 2003–2114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Su, S. A. et al. Emerging role of exosome-mediated intercellular communication in vascular remodeling. Oncotarget8, 25700–25712 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Pegtel, D. M. & Gould, S. J. Exosomes. Annu. Rev. Biochem.88, 487–514 (2019). [DOI] [PubMed] [Google Scholar]
- 337.Wang, J. J. et al. Macrophage-secreted exosomes delivering miRNA-21 inhibitor can regulate BGC-823 cell proliferation. Asian Pac. J. Cancer Prev.16, 4203–4209 (2015). [DOI] [PubMed] [Google Scholar]
- 338.Zheng, H. et al. Exosome‑encapsulated miR‑26a attenuates aldosterone‑induced tubulointerstitial fibrosis by inhibiting the CTGF/SMAD3 signaling pathway. Int. J. Mol. Med.51, 11 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Hashemi, Z. S. et al. Novel delivery of sorafenib by natural killer cell-derived exosomes-enhanced apoptosis in triple-negative breast cancer. Nanomed. (Lond.)18, 437–453 (2023). [DOI] [PubMed] [Google Scholar]
- 340.Kamerkar, S. et al. Exosome-mediated genetic reprogramming of tumor-associated macrophages by exoASO-STAT6 leads to potent monotherapy antitumor activity. Sci. Adv.8, eabj7002 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Kaban, K. et al. Therapeutic silencing of BCL-2 using NK cell-derived exosomes as a novel therapeutic approach in breast cancer. Cancers (Basel)13, 2397 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Wahlgren, J. et al. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res40, e130 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Alvarez-Erviti, L. et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol.29, 341–345 (2011). [DOI] [PubMed] [Google Scholar]
- 344.Yuan, D. et al. Macrophage exosomes as natural nanocarriers for protein delivery to inflamed brain. Biomaterials142, 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Zheng, M. et al. Harnessing exosomes for the development of brain drug delivery systems. Bioconjugate Chem.30, 994–1005 (2019). [DOI] [PubMed] [Google Scholar]
- 346.Bunggulawa, E. J. et al. Recent advancements in the use of exosomes as drug delivery systems. J. Nanobiotechnol.16, 81 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Rajput, A., Varshney, A., Bajaj, R. & Pokharkar, V. Exosomes as new generation vehicles for drug delivery: biomedical applications and future perspectives. Molecules27, 7289 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Asadirad, A. et al. Dendritic cell immunotherapy with miR-155 enriched tumor-derived exosome suppressed cancer growth and induced antitumor immune responses in murine model of colorectal cancer induced by CT26 cell line. Int. Immunopharmacol.104, 108493 (2022). [DOI] [PubMed] [Google Scholar]
- 349.Xi, X. M., Xia, S. J. & Lu, R. Drug loading techniques for exosome-based drug delivery systems. Pharmazie76, 61–67 (2021). [DOI] [PubMed] [Google Scholar]
- 350.Kim, M. S. et al. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine12, 655–664 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Han, S. et al. Delivery of anti-miRNA-221 for colorectal carcinoma therapy using modified cord blood mesenchymal stem cells-derived exosomes. Front. Mol. Biosci.8, 743013 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Luan, X. et al. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol. Sin.38, 754–763 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Rehman, F. U., Liu, Y., Zheng, M. & Shi, B. Exosomes based strategies for brain drug delivery. Biomaterials293, 121949 (2023). [DOI] [PubMed] [Google Scholar]
- 354.Johnsen, K. B. et al. Evaluation of electroporation-induced adverse effects on adipose-derived stem cell exosomes. Cytotechnology68, 2125–2138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Liang, Y., Duan, L., Lu, J. & Xia, J. Engineering exosomes for targeted drug delivery. Theranostics11, 3183–3195 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Zhang, H. et al. Exosome-mediated targeted delivery of miR-210 for angiogenic therapy after cerebral ischemia in mice. J. Nanobiotechnol.17, 29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Kim, G. et al. Systemic delivery of microRNA-21 antisense oligonucleotides to the brain using T7-peptide decorated exosomes. J. Control. Release317, 273–281 (2020). [DOI] [PubMed] [Google Scholar]
- 358.Mahati, S. et al. Delivery of miR-26a using an exosomes-based nanosystem inhibited proliferation of hepatocellular carcinoma. Front. Mol. Biosci.8, 738219 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Xu, X. et al. Exosome-mediated delivery of kartogenin for chondrogenesis of synovial fluid-derived mesenchymal stem cells and cartilage regeneration. Biomaterials269, 120539 (2021). [DOI] [PubMed] [Google Scholar]
- 360.Cooper, J. M. et al. Systemic exosomal siRNA delivery reduced alpha-synuclein aggregates in brains of transgenic mice. Mov. Disord.29, 1476–1485 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Ren, X. et al. Exosomal DNA aptamer targeting alpha-synuclein aggregates reduced neuropathological deficits in a mouse parkinson’s disease model. Mol. Ther. Nucl. Acids17, 726–740 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Fu, Z. et al. In vivo self-assembled small RNAs as a new generation of RNAi therapeutics. Cell Res31, 631–648 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Couch, Y. et al. A brief history of nearly EV-erything - the rise and rise of extracellular vesicles. J. Extracell. Vesicles10, e12144 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Erathodiyil, N. & Ying, J. Y. Functionalization of inorganic nanoparticles for bioimaging applications. Acc. Chem. Res.44, 925–935 (2011). [DOI] [PubMed] [Google Scholar]
- 365.Meena, J. et al. Inorganic nanoparticles for natural product delivery: a review. Environ. Chem. Lett.18, 2107–2118 (2020). [Google Scholar]
- 366.Luther, D. C. et al. Delivery of drugs, proteins, and nucleic acids using inorganic nanoparticles. Adv. Drug Deliv. Rev.156, 188–213 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Wang, Z. et al. Gold nanoparticle‑mediated delivery of paclitaxel and nucleic acids for cancer therapy (Review). Mol. Med. Rep.22, 4475–4484 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Graczyk, A., Pawlowska, R., Jedrzejczyk, D. & Chworos, A. Gold nanoparticles in conjunction with nucleic acids as a modern molecular system for cellular delivery. Molecules25, 204 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Shrestha, B. et al. Gold nanoparticles mediated drug-gene combinational therapy for breast cancer treatment. Int. J. Nanomed.15, 8109–8119 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Jensen, S. A. et al. Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Sci. Transl. Med.5, 209ra152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Kumthekar, P. et al. A first-in-human phase 0 clinical study of RNA interference-based spherical nucleic acids in patients with recurrent glioblastoma. Sci. Transl. Med.13, eabb3945 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Wu, K. et al. Magnetic nanoparticles in nanomedicine: a review of recent advances. Nanotechnology30, 502003 (2019). [DOI] [PubMed] [Google Scholar]
- 373.Dash, S. et al. Emerging trends in the nanomedicine applications of functionalized magnetic nanoparticles as novel therapies for acute and chronic diseases. J. Nanobiotechnol.20, 393 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Chavan, N., Dharmaraj, D., Sarap, S. & Surve, C. Magnetic nanoparticles–new era in nanotechnology. J. Drug Deliv. Sci. Technol.77, 103899 (2022). [Google Scholar]
- 375.Luo, M. et al. Delivering the promise of gene therapy with nanomedicines in treating central nervous system diseases. Adv. Sci.9, e2201740 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Rahamathulla, M. et al. Carbon nanotubes: current perspectives on diverse applications in targeted drug delivery and therapies. Mater. (Basel)14, 6707 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Ren, X. et al. Photoactivatable RNAi for cancer gene therapy triggered by near-infrared-irradiated single-walled carbon nanotubes. Int. J. Nanomed.12, 7885–7896 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Levina, A. S., Repkova, M. N., Ismagilov, Z. R. & Zarytova, V. F. Methods of the synthesis of silicon-containing nanoparticles intended for nucleic acid delivery. Eurasia. Chem.-Techno20, 177–194 (2018). [Google Scholar]
- 379.Stead, S. O. et al. siRNA gene knockdown with functionalised porous silicon nanoparticles. Transplantation104, s158 (2020). [Google Scholar]
- 380.Luo, M. et al. A multifunctional porous silicon nanocarrier for glioblastoma treatment. Mol. Pharmaceutics20, 545–560 (2023). [DOI] [PubMed] [Google Scholar]
- 381.Goyal, R. et al. Peptide-based delivery vectors with pre-defined geometrical locks. RSC Med. Chem.11, 1303–1313 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Lian, Z. & Ji, T. Functional peptide-based drug delivery systems. J. Mater. Chem. B8, 6517–6529 (2020). [DOI] [PubMed] [Google Scholar]
- 383.Jeon, B. W. et al. Recent advances in peptide signaling during arabidopsis root development. J. Exp. Bot.72, 2889–2902 (2021). [DOI] [PubMed] [Google Scholar]
- 384.Kim, J. et al. Oral supplementation of low-molecular-weight collagen peptides reduces skin wrinkles and improves biophysical properties of skin: a randomized, double-blinded, placebo-controlled study. J. Med. Food25, 1146–1154 (2022). [DOI] [PubMed] [Google Scholar]
- 385.Lindberg, J., Nilvebrant, J., Nygren, P, A. & Lehmann, F. Progress and future directions with peptide-drug conjugates for targeted cancer therapy. Molecules26, 6042 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Hao, C. et al. Renovation of old drugs’ – can peptide drug conjugates lead the post-ADC era? Aust. J. Chem.76, 318–336 (2023). [Google Scholar]
- 387.Wang, J., Tripathy, N. & Chung, E. J. Targeting and therapeutic peptide-based strategies for polycystic kidney disease. Adv. Drug Deliv. Rev.161-162, 176–189 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Feldman, K. S., Pavlou, M. P. & Zahid, M. Cardiac targeting peptide: from identification to validation to mechanism of transduction. Methods Mol. Biol.2211, 97–112 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Lu, L. et al. A novel blood-brain barrier-penetrating and vascular-targeting chimeric peptide inhibits glioma angiogenesis. Int. J. Mol. Sci.24, 8753 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Suzuki, M. et al. Characterization of the membrane penetration-enhancing peptide S19 derived from human syncytin-1 for the intracellular delivery of TAT-fused proteins. Biochem. Biophys. Res. Commun.586, 63–67 (2022). [DOI] [PubMed] [Google Scholar]
- 391.Vijakumaran, U. et al. Development of cell penetrating peptides for effective delivery of recombinant factors into target cells. Protein Pept. Lett.27, 1092–1101 (2020). [DOI] [PubMed] [Google Scholar]
- 392.Maraming, P. et al. The cationic cell-penetrating KT2 peptide promotes cell membrane defects and apoptosis with autophagy inhibition in human HCT 116 colon cancer cells. J. Cell. Physiol.234, 22116–22129 (2019). [DOI] [PubMed] [Google Scholar]
- 393.Klipp, A., Burger, M. & Leroux, J. C. Get out or die trying: peptide- and protein-based endosomal escape of RNA therapeutics. Adv. Drug Deliv. Rev.200, 115047 (2023). [DOI] [PubMed] [Google Scholar]
- 394.Guo, Y. et al. Self-assembled peptide nanoparticles with endosome escaping permits for co-drug delivery. Talanta221, 121572 (2021). [DOI] [PubMed] [Google Scholar]
- 395.Zhao, Y. et al. Engineered histidine-rich peptides enhance endosomal escape for antibody-targeted intracellular delivery of functional proteins. Angew. Chem. Int. Ed. e202304692 (2023). [DOI] [PubMed]
- 396.Lu, J. et al. Types of nuclear localization signals and mechanisms of protein import into the nucleus. Cell Commun. Signal.19, 60 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Huang, S. et al. Design of acid-activated cell-penetrating peptides with nuclear localization capacity for anticancer drug delivery. J. Pept. Sci.27, e3354 (2021). [DOI] [PubMed] [Google Scholar]
- 398.Kanazawa, T. et al. Electroporation-based ex vivo gene delivery into dendritic cells by anionic polymer-coated versatile nuclear localization signal/pDNA complex. Biol. Pharm. Bull.44, 1866–1871 (2021). [DOI] [PubMed] [Google Scholar]
- 399.Urandur, S. & Sullivan, M. O. Peptide-based vectors: a biomolecular engineering strategy for gene delivery. Annu. Rev. Chem. Biomol. Eng.14, 243–264 (2023). [DOI] [PubMed] [Google Scholar]
- 400.Varanko, A., Saha, S. & Chilkoti, A. Recent trends in protein and peptide-based biomaterials for advanced drug delivery. Adv. Drug Deliv. Rev.156, 133–187 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Falato, L., Gestin, M. & Langel, U. Cell-penetrating peptides delivering siRNAs: an overview. Methods Mol. Biol.2282, 329–352 (2021). [DOI] [PubMed] [Google Scholar]
- 402.Kim, E. H. et al. PDL1-binding peptide/anti-miRNA21 conjugate as a therapeutic modality for PD-L1(high) tumors and TAMs. J. Control. Release345, 62–74 (2022). [DOI] [PubMed] [Google Scholar]
- 403.Yang, G. et al. Improved cellular delivery of antisense oligonucleotide for miRNA-21 imaging in vivo using cell-penetrating peptide-based nanoprobes. Mol. Pharmaceutics18, 787–795 (2021). [DOI] [PubMed] [Google Scholar]
- 404.Liu, Q. et al. iRGD-modified exosomes-delivered BCL6 siRNA inhibit the progression of diffuse large B-cell lymphoma. Front. Oncol.12, 822805 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Khabazian, E. et al. Cationic liposome decorated with cyclic RGD peptide for targeted delivery of anti-STAT3 siRNA to melanoma cancer cells. J. Drug Target.30, 522–533 (2022). [DOI] [PubMed] [Google Scholar]
- 406.Liao, L. et al. A bivalent cyclic RGD-siRNA conjugate enhances the antitumor effect of apatinib via co-inhibiting VEGFR2 in non-small cell lung cancer xenografts. Drug Deliv.28, 1432–1442 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Liu, X. et al. Tumor-targeted in vivo gene silencing via systemic delivery of cRGD-conjugated siRNA. Nucleic Acids Res42, 11805–11817 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Wang, Y. et al. Tumor-targeted anti-VEGF RNAi capable of sequentially responding to intracellular microenvironments for potent systemic tumor suppression. ACS Appl. Bio Mater.3, 9145–9155 (2020). [DOI] [PubMed] [Google Scholar]
- 409.Moshnikova, A. et al. Targeting bladder urothelial carcinoma with pHLIP-ICG and inhibition of urothelial cancer cell proliferation by pHLIP-amanitin. Front Urol.2, 868919 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Zhang, M. et al. In vivo distribution and therapeutic efficacy of radioiodine-labeled pH-low insertion peptide variant 3 in a mouse model of breast cancer. Mol. Imaging2022, 7456365 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Wyatt, L. C. et al. Applications of pHLIP technology for cancer imaging and therapy. Trends Biotechnol.35, 653–664 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Dupont, M. et al. Tumor treatment by pHLIP-targeted antigen delivery. Front. Bioeng. Biotech.10, 1082290 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Visca, H. et al. pHLIP peptides target acidity in activated macrophages. Mol. Imaging Biol.24, 874–885 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Son, S. M. et al. Therapeutic effect of pHLIP-mediated CEACAM6 gene silencing in lung adenocarcinoma. Sci. Rep.9, 11607 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Luna Velez, M. V. et al. Delivery of antisense oligonucleotides for splice-correction of androgen receptor pre-mRNA in castration-resistant prostate cancer models using cell-penetrating peptides. Prostate82, 657–665 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Ervin, E. H. et al. Targeted gene silencing in human embryonic stem cells using cell-penetrating peptide PepFect 14. Stem Cell Res. Ther.10, 43 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Kurrikoff, K., Vunk, B. & Langel, U. Status update in the use of cell-penetrating peptides for the delivery of macromolecular therapeutics. Expert Opin. Biol. Ther.21, 361–370 (2021). [DOI] [PubMed] [Google Scholar]
- 418.Taniguchi, K. et al. Alpha-aminoisobutyric acid-containing amphipathic helical peptide-cyclic RGD conjugation as a potential drug delivery system for microRNA replacement therapy in vitro. Mol. Pharmaceutics16, 4542–4550 (2019). [DOI] [PubMed] [Google Scholar]
- 419.Tarvirdipour, S. et al. A self-assembling peptidic platform to boost the cellular uptake and nuclear delivery of oligonucleotides. Biomater. Sci.10, 4309–4323 (2022). [DOI] [PubMed] [Google Scholar]
- 420.Ji, K., Xiao, Y. & Zhang, W. Acid-activated nonviral peptide vector for gene delivery. J. Pept. Sci.26, e3230 (2020). [DOI] [PubMed] [Google Scholar]
- 421.Kwon, E. J., Ko, H. & Bhatia, S. N. Peptide spiders: peptide-polymer conjugates to traffic nucleic acids. Mol. Pharmaceutics17, 3633–3642 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Kim, G. C., Cheon, D. H. & Lee, Y. Challenge to overcome current limitations of cell-penetrating peptides. BBA Proteins Proteom.1869, 140604 (2021). [DOI] [PubMed] [Google Scholar]
- 423.Hadianamrei, R. & Zhao, X. Current state of the art in peptide-based gene delivery. J. Control. Release343, 600–619 (2022). [DOI] [PubMed] [Google Scholar]
- 424.Buyanova, M. et al. Discovery of a cyclic cell-penetrating peptide with improved endosomal escape and cytosolic delivery efficiency. Mol. Pharmaceutics19, 1378–1388 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Molle, L. M., Smyth, C. H., Yuen, D. & Johnston, A. P. R. Nanoparticles for vaccine and gene therapy: overcoming the barriers to nucleic acid delivery. WIREs Nanomed. Nanobiotechnol.14, e1809 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Alhakamy, N. A., Nigatu, A. S., Berkland, C. J. & Ramsey, J. D. Noncovalently associated cell-penetrating peptides for gene delivery applications. Ther. Deliv.4, 741–757 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Samec, T. et al. Peptide-based delivery of therapeutics in cancer treatment. Mater. Today Bio14, 100248 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Burks, S. R. et al. Co-encapsulating the fusogenic peptide INF7 and molecular imaging probes in liposomes increases intracellular signal and probe retention. PLoS One10, e0120982 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Feng, R., Ni, R. & Chau, Y. Fusogenic peptide modification to enhance gene delivery by peptide-DNA nano-coassemblies. Biomater. Sci.10, 5116–5120 (2022). [DOI] [PubMed] [Google Scholar]
- 430.Hagino, Y. et al. GALA-modified lipid nanoparticles for the targeted delivery of plasmid dna to the lungs. Mol. Pharmaceutics18, 878–888 (2021). [DOI] [PubMed] [Google Scholar]
- 431.Li, C., Cao, X. W., Zhao, J. & Wang, F. J. Effective therapeutic drug delivery by GALA3, an endosomal escape peptide with reduced hydrophobicity. J. Membr. Biol.253, 139–152 (2020). [DOI] [PubMed] [Google Scholar]
- 432.Miura, N. et al. A KALA-modified lipid nanoparticle containing CpG-free plasmid DNA as a potential DNA vaccine carrier for antigen presentation and as an immune-stimulative adjuvant. Nucleic Acids Res43, 1317–1331 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Dastpeyman, M. et al. Endosomal escape cell-penetrating peptides significantly enhance pharmacological effectiveness and CNS activity of systemically administered antisense oligonucleotides. Int. J. Pharm.599, 120398 (2021). [DOI] [PubMed] [Google Scholar]
- 434.Alipour, M., Hosseinkhani, S., Sheikhnejad, R. & Cheraghi, R. Nano-biomimetic carriers are implicated in mechanistic evaluation of intracellular gene delivery. Sci. Rep.7, 41507 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Samec, T. et al. Fusogenic peptide delivery of bioactive siRNAs targeting CSNK2A1 for treatment of ovarian cancer. Mol. Ther. Nucl. Acids30, 95–111 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Lu, S. et al. Multi-functional self-assembled nanoparticles for pVEGF-shRNA loading and anti-tumor targeted therapy. Int. J. Pharm.575, 118898 (2020). [DOI] [PubMed] [Google Scholar]
- 437.Luo, Y., Ma, J. & Lu, W. The significance of mitochondrial dysfunction in cancer. Int. J. Mol. Sci.21, 5598 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Yao, R. Q., Ren, C., Xia, Z. F. & Yao, Y. M. Organelle-specific autophagy in inflammatory diseases: a potential therapeutic target underlying the quality control of multiple organelles. Autophagy17, 385–401 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Machado-Oliveira, G., Ramos, C., Marques, A. R. A. & Vieira, O. V. Cell senescence, multiple organelle dysfunction and atherosclerosis. Cells9, 2146 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Hu, C., Huang, Y. & Chen, Y. Targeted modification of the cationic anticancer peptide HPRP-A1 with iRGD to improve specificity, penetration, and tumor-tissue accumulation. Mol. Pharmaceutics16, 561–572 (2019). [DOI] [PubMed] [Google Scholar]
- 441.Nakamura, M., Fujiwara, K. & Doi, N. Cytoplasmic delivery of siRNA using human-derived membrane penetration-enhancing peptide. J. Nanobiotechnol.20, 458 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Bjorge, J. D., Pang, A. & Fujita, D. J. Delivery of gene targeting siRNAs to breast cancer cells using a multifunctional peptide complex that promotes both targeted delivery and endosomal release. PLoS One12, e0180578 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Ruan, R. et al. Topical and targeted delivery of sirnas to melanoma cells using a fusion peptide carrier. Sci. Rep.6, 29159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Cerrato, C. P. et al. Intracellular delivery of therapeutic antisense oligonucleotides targeting mRNA coding mitochondrial proteins by cell-penetrating peptides. J. Mater. Chem. B8, 10825–10836 (2020). [DOI] [PubMed] [Google Scholar]
- 445.Kuang, Y. et al. Dual functional peptide-driven nanoparticles for highly efficient glioma-targeting and drug codelivery. Mol. Pharmaceutics13, 1599–1607 (2016). [DOI] [PubMed] [Google Scholar]
- 446.Bulut, S. et al. Slow release and delivery of antisense oligonucleotide drug by self-assembled peptide amphiphile nanofibers. Biomacromolecules12, 3007–3014 (2011). [DOI] [PubMed] [Google Scholar]
- 447.Nirasawa, K. et al. Development of A2G80 peptide-gene complex for targeted delivery to muscle cells. J. Control. Release329, 988–996 (2021). [DOI] [PubMed] [Google Scholar]
- 448.Jafari, M. & Chen, P. Peptide mediated siRNA delivery. Curr. Top. Med. Chem.9, 1088–1097 (2009). [DOI] [PubMed] [Google Scholar]
- 449.Yan Y. Q. et al. Localized instillation enables in vivo screening of targeting peptides using one-bead one-compound technology. ACS Nano. 10.1021/acsnano.2c09894 (2023). [DOI] [PubMed]
- 450.Paray, B. A. et al. The role of the multifunctional antimicrobial peptide melittin in gene delivery. Drug. Discov. Today26, 1053–1059 (2021). [DOI] [PubMed] [Google Scholar]
- 451.Govindarajan, S. et al. Targeting human epidermal growth factor receptor 2 by a cell-penetrating peptide-affibody bioconjugate. Biomaterials33, 2570–2582 (2012). [DOI] [PubMed] [Google Scholar]
- 452.Li, Q. et al. Multifunctional peptide-conjugated nanocarriers for pulp regeneration in a full-length human tooth root. Acta Biomater.127, 252–265 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Wan, Y., Moyle, P. M., Christie, M. P. & Toth, I. Nanosized, peptide-based multicomponent DNA delivery systems: optimization of endosome escape activity. Nanomed. (Lond.)11, 907–919 (2016). [DOI] [PubMed] [Google Scholar]
- 454.Rohira, H., Arora, A., Kaur, P. & Chugh, A. Peptide cargo administration: current state and applications. Appl. Microbiol. Biotechnol.107, 3153–3181 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Liu, Y. et al. Development and characterization of high efficacy cell-penetrating peptide via modulation of the histidine and arginine ratio for gene therapy. Mater. (Basel)14, 4674 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Tsvetkov, V. B. et al. Anticoagulant oligonucleotide-peptide conjugates: identification of thrombin aptamer conjugates with improved characteristics. Int. J. Mol. Sci.23, 3820 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Kaplan, A. R. et al. Ku80-targeted pH-sensitive peptide-PNA conjugates are tumor selective and sensitize cancer cells to ionizing radiation. Mol. Cancer Res.18, 873–882 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Dutta, K., Das, R., Medeiros, J. & Thayumanavan, S. Disulfide bridging strategies in viral and nonviral platforms for nucleic acid delivery. Biochemistry60, 966–990 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Taskova, M., Mantsiou, A. & Astakhova, K. Synthetic nucleic acid analogues in gene therapy: an update for peptide-oligonucleotide conjugates. Chembiochem18, 1671–1682 (2017). [DOI] [PubMed] [Google Scholar]
- 460.Klabenkova, K., Fokina, A. & Stetsenko, D. Chemistry of peptide-oligonucleotide conjugates: a review. Molecules26, 5420 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Cerrato, C. P., Lehto, T. & Langel, U. Peptide-based vectors: recent developments. Biomol. Concepts5, 479–488 (2014). [DOI] [PubMed] [Google Scholar]
- 462.Tomassi, S. et al. Cationic nucleopeptides as novel non-covalent carriers for the delivery of peptide nucleic acid (PNA) and RNA oligomers. Bioorg. Med. Chem.26, 2539–2550 (2018). [DOI] [PubMed] [Google Scholar]
- 463.Hansen, A. M., Shaikh, A. Y. & Franzyk, H. Facile preparation of pna-peptide conjugates with a polar maleimide-thioether linkage. Methods Mol. Biol.2105, 97–118 (2020). [DOI] [PubMed] [Google Scholar]
- 464.Schissel, C. K. et al. Cell-penetrating d-peptides retain antisense morpholino oligomer delivery activity. ACS Bio Med Chem. Au2, 150–160 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Hakata, Y. et al. Intracellular delivery of a peptide nucleic acid-based hybrid of an autophagy inducing peptide with a cell-penetrating peptide. Org. Biomol. Chem.18, 1978–1986 (2020). [DOI] [PubMed] [Google Scholar]
- 466.Linden, G. et al. Efficient antisense inhibition reveals microRNA-155 to restrain a late-myeloid inflammatory programme in primary human phagocytes. RNA Biol.18, 604–618 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Barkowsky, G. et al. Antimicrobial activity of peptide-coupled antisense peptide nucleic acids in streptococcus pneumoniae. Microbiol Spectr.10, e0049722 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Soudah, T., Mogilevsky, M., Karni, R. & Yavin, E. CLIP6-PNA-peptide conjugates: non-endosomal delivery of splice switching oligonucleotides. Bioconjugate Chem.28, 3036–3042 (2017). [DOI] [PubMed] [Google Scholar]
- 469.Sheng, L. et al. Comparison of the efficacy of MOE and PMO modifications of systemic antisense oligonucleotides in a severe SMA mouse model. Nucleic Acids Res48, 2853–2865 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Gushchina, L. V. et al. Systemic PPMO-mediated dystrophin expression in the Dup2 mouse model of duchenne muscular dystrophy. Mol. Ther. Nucl. Acids30, 479–492 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Aslesh, T. et al. DG9-conjugated morpholino rescues phenotype in SMA mice by reaching the CNS via a subcutaneous administration. JCI Insight8, e160516 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Gait, M. J. et al. Cell-penetrating peptide conjugates of steric blocking oligonucleotides as therapeutics for neuromuscular diseases from a historical perspective to current prospects of treatment. Nucleic Acid Ther.29, 1–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Hammond, S. M. et al. Systemic peptide-mediated oligonucleotide therapy improves long-term survival in spinal muscular atrophy. Proc. Natl Acad. Sci. USA113, 10962–10967 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Blain, A. M. et al. Peptide-conjugated phosphodiamidate oligomer-mediated exon skipping has benefits for cardiac function in mdx and cmah-/-mdx mouse models of duchenne muscular dystrophy. PLoS One13, e0198897 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Klein, A. F. et al. Peptide-conjugated oligonucleotides evoke long-lasting myotonic dystrophy correction in patient-derived cells and mice. J. Clin. Investig.129, 4739–4744 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Chioccioli, M. et al. A lung targeted miR-29 mimic as a therapy for pulmonary fibrosis. EBioMedicine85, 104304 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Jana, A., Narula, P., Chugh, A. & Kulshreshtha, R. Efficient delivery of anti-miR-210 using Tachyplesin, a cell penetrating peptide, for glioblastoma treatment. Int. J. Pharm.572, 118789 (2019). [DOI] [PubMed] [Google Scholar]
- 478.Schachner-Nedherer, A. L. et al. Biological activity of miRNA-27a using peptide-based drug delivery systems. Int. J. Nanomed.14, 7795–7808 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Xu, W. et al. The mirrored cationic peptide as miRNA vehicle for efficient lung cancer therapy. MedComm4, e273 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Wang, J. et al. Strategies for improving the safety and RNAi efficacy of noncovalent peptide/siRNA nanocomplexes. Adv. Colloid Interface Sci.302, 102638 (2022). [DOI] [PubMed] [Google Scholar]
- 481.Ryu, Y. C., Lee, Y. E. & Hwang, B. H. Efficient and safe small RNA delivery to macrophage using peptide-based nanocomplex. Biotechnol. Bioeng.119, 482–492 (2022). [DOI] [PubMed] [Google Scholar]
- 482.Gulley, J. L. et al. Dual inhibition of TGF-beta and PD-L1: a novel approach to cancer treatment. Mol. Oncol.16, 2117–2134 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Wu, L. P. et al. Crossing the blood-brain-barrier with nanoligand drug carriers self-assembled from a phage display peptide. Nat. Commun.10, 4635 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Yan, H. et al. Peptide-siRNA nanoparticles targeting NF-kappaB p50 mitigate experimental abdominal aortic aneurysm progression and rupture. Biomater. Adv.139, 213009 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Yan, H. et al. Induction of WNT16 via peptide-mRNA nanoparticle-based delivery maintains cartilage homeostasis. Pharmaceutics12, 73 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Jin, Y. et al. Histone demethylase JMJD3 downregulation protects against aberrant force-induced osteoarthritis through epigenetic control of NR4A1. Int. J. Oral. Sci.14, 34 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Zhou, H. F. et al. Peptide-siRNA nanocomplexes targeting NF-kappaB subunit p65 suppress nascent experimental arthritis. J. Clin. Investig.124, 4363–4374 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Ceccanti, M. & Inghilleri, M. RNA interference and neuromuscular diseases: a focus on hereditary transthyretin amyloidosis. Curr. Gene Ther.24, 6–7 (2024). [DOI] [PubMed] [Google Scholar]
- 489.Adams, D., Algalarrondo, V. & Echaniz-Laguna, A. Hereditary transthyretin amyloidosis in the era of RNA interference, antisense oligonucleotide, and CRISPR-Cas9 treatments. Blood142, 1600–1612 (2023). [DOI] [PubMed] [Google Scholar]
- 490.Keam, S. J. Inotersen: first global approval. Drugs78, 1371–1376 (2018). [DOI] [PubMed] [Google Scholar]
- 491.Benson, M. D. et al. Inotersen treatment for patients with hereditary transthyretin amyloidosis. N. Engl. J. Med.379, 22–31 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Coelho, T. et al. Eplontersen for hereditary transthyretin amyloidosis with polyneuropathy. JAMA330, 1448–1458 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Keam, S. J. Vutrisiran: first approval. Drugs82, 1419–1425 (2022). [DOI] [PubMed] [Google Scholar]
- 494.Gillmore, J. D. et al. CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med.385, 493–502 (2021). [DOI] [PubMed] [Google Scholar]
- 495.Happi Mbakam, C. & Tremblay, J. P. Gene therapy for duchenne muscular dystrophy: an update on the latest clinical developments. Expert Rev. Neurother.23, 905–920 (2023). [DOI] [PubMed] [Google Scholar]
- 496.Wagner, K. R. et al. Safety, tolerability, and pharmacokinetics of casimersen in patients with duchenne muscular dystrophy amenable to exon 45 skipping: a randomized, double-blind, placebo-controlled, dose-titration trial. Muscle Nerve64, 285–292 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Syed, Y. Y. Eteplirsen: first global approval. Drugs76, 1699–1704 (2016). [DOI] [PubMed] [Google Scholar]
- 498.Cirak, S. et al. Exon skipping and dystrophin restoration in patients with duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet378, 595–605 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Wilton-Clark, H. & Yokota, T. Recent trends in antisense therapies for duchenne muscular dystrophy. Pharmaceutics15, 778 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Sheikh, O. & Yokota, T. Pharmacology and toxicology of eteplirsen and SRP-5051 for DMD exon 51 skipping: an update. Arch. Toxicol.96, 1–9 (2022). [DOI] [PubMed] [Google Scholar]
- 501.Mellion, M. et al. PGN-EDO51, an enhanced delivery oligonucleotide (EDO) for the treatment of duchenne muscular dystrophy (DMD): results of a phase 1 study in healthy volunteers (P3-8.004). Neurology100, 4396 (2023). [Google Scholar]
- 502.Van Daele, S. H., Masrori, P., Van Damme, P. & Van Den Bosch, L. The sense of antisense therapies in ALS. Trends Mol. Med.30, 252–262 (2024). [DOI] [PubMed] [Google Scholar]
- 503.Suzuki, N., Nishiyama, A., Warita, H. & Aoki, M. Genetics of amyotrophic lateral sclerosis: seeking therapeutic targets in the era of gene therapy. J. Hum. Genet.68, 131–152 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Fang, T. et al. Gene therapy in amyotrophic lateral sclerosis. Cells11, 2066 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Miller, T. M. et al. Trial of antisense oligonucleotide tofersen for SOD1 ALS. N. Engl. J. Med.387, 1099–1110 (2022). [DOI] [PubMed] [Google Scholar]
- 506.Mccartan, R., Khorkova, O., Volmar, C. H. & Wahlestedt, C. Nucleic acid-based therapeutics for the treatment of central nervous system disorders. Front. Genet.14, 1250276 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Nieto-Romero, V. et al. Restored glyoxylate metabolism after AGXT gene correction and direct reprogramming of primary hyperoxaluria type 1 fibroblasts. iScience27, 109530 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Groothoff, J. W. et al. Clinical practice recommendations for primary hyperoxaluria: an expert consensus statement from ERKNet and OxalEurope. Nat. Rev. Nephrol.19, 194–211 (2023). [DOI] [PubMed] [Google Scholar]
- 509.Fargue, S. & Acquaviva Bourdain, C. Primary hyperoxaluria type 1: pathophysiology and genetics. Clin. Kidney J.15, i4–i8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Scott, L. J. & Keam, S. J. Lumasiran: first approval. Drugs81, 277–282 (2021). [DOI] [PubMed] [Google Scholar]
- 511.Lakhina, Y., Boulis, N. M. & Donsante, A. Current and emerging targeted therapies for spinal muscular atrophy. Expert Rev. Neurother.23, 1189–1199 (2023). [DOI] [PubMed] [Google Scholar]
- 512.Ottesen, E. W. et al. Diverse targets of SMN2-directed splicing-modulating small molecule therapeutics for spinal muscular atrophy. Nucleic Acids Res51, 5948–5980 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Gowda, V. L., Fernandez-Garcia, M. A., Jungbluth, H. & Wraige, E. New treatments in spinal muscular atrophy. Arch. Dis. Child.108, 511–517 (2023). [DOI] [PubMed] [Google Scholar]
- 514.Nishio, H. et al. Spinal muscular atrophy: the past, present, and future of diagnosis and treatment. Int. J. Mol. Sci.24, 11939 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Singh, N. N., Howell, M. D., Androphy, E. J. & Singh, R. N. How the discovery of ISS-N1 led to the first medical therapy for spinal muscular atrophy. Gene Ther.24, 520–526 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.De Vivo, D. C. et al. Nusinersen initiated in infants during the presymptomatic stage of spinal muscular atrophy: interim efficacy and safety results from the phase 2 nurture study. Neuromuscul. Disord.29, 842–856 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Montes, J. et al. Nusinersen improves walking distance and reduces fatigue in later-onset spinal muscular atrophy. Muscle Nerve60, 409–414 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Finkel, R. S. et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: final report of a phase 2, open-label, multicentre, dose-escalation study. Lancet Child Adolesc. Health5, 491–500 (2021). [DOI] [PubMed] [Google Scholar]
- 519.Neuzillet, C. et al. Targeting the TGFβ pathway for cancer therapy. Pharmacol. Ther.147, 22–31 (2015). [DOI] [PubMed] [Google Scholar]
- 520.Andrews, D. W. et al. Phase Ib clinical trial of IGV-001 for patients with newly diagnosed glioblastoma. Clin. Cancer Res.27, 1912–1922 (2021). [DOI] [PubMed] [Google Scholar]
- 521.Lee, I. Y. et al. Autologous cell immunotherapy (IGV-001) with IGF-1R antisense oligonucleotide in newly diagnosed glioblastoma patients. Future Oncol.20, 579–591 (2024). [DOI] [PubMed] [Google Scholar]
- 522.Weber, J. S. et al. Individualised neoantigen therapy mRNA-4157 (V940) plus pembrolizumab versus pembrolizumab monotherapy in resected melanoma (KEYNOTE-942): a randomised, phase 2b study. Lancet403, 632–644 (2024). [DOI] [PubMed] [Google Scholar]
- 523.Yao, R., Xie, C. & Xia, X. Recent progress in mRNA cancer vaccines. Hum. Vaccines Immunother.20, 2307187 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Nashine, S. Potential therapeutic candidates for age-related macular degeneration (AMD). Cells10, 2483 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Nakamura, Y. Multiple therapeutic applications of RBM-007, an anti-FGF2 aptamer. Cells10, 1617 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.Jimenez, A. et al. SYL1801: preclinical efficacy and safety of a sirna-based eye drops treatment for age related macular degeneration. Investig. Ophthalmol. Vis. Sci.60, 5389 (2019). [Google Scholar]
- 527.Jimenez, A. I., Ruz, V., Vargas, B. & Bleau, A. M. Phase I of SYL1801, a new siRNA delivered in eye drops for age-related macular degeneration. Investig. Ophthalmol. Vis. Sci.63, 337–F0168 (2022). [Google Scholar]
- 528.Moreno-Montanes, J., Bleau, A. M. & Jimenez, A. I. Tivanisiran, a novel siRNA for the treatment of dry eye disease. Expert Opin. Investig. Drugs27, 421–426 (2018). [DOI] [PubMed] [Google Scholar]
- 529.Valdes-Arias, D. et al. Recent United States developments in the pharmacological treatment of dry eye disease. Drugs84, 549–563 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Kuo, Y. K. et al. Dry eye disease: a review of epidemiology in taiwan, and its clinical treatment and merits. J. Clin. Med.8, 1227 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Dulla, K. et al. Splice-modulating oligonucleotide QR-110 restores CEP290 mRNA and function in human c.2991+1655A>G LCA10 models. Mol. Ther. Nucl. Acids12, 730–740 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Cideciyan, A. V. et al. Durable vision improvement after a single intravitreal treatment with antisense oligonucleotide in CEP290-LCA: replication in two eyes. Am. J. Ophthalmol. Case Rep.32, 101873 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Dulla, K. et al. Antisense oligonucleotide-based treatment of retinitis pigmentosa caused by USH2A exon 13 mutations. Mol. Ther.29, 2441–2455 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Shi, Y. et al. Epigenetic regulation in cardiovascular disease: mechanisms and advances in clinical trials. Signal Transduct. Target Ther.7, 200 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Mhaimeed, O. et al. The importance of LDL-C lowering in atherosclerotic cardiovascular disease prevention: lower for longer is better. Am. J. Prev. Cardiol.18, 100649 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Sawhney, J. P. et al. CSI clinical practice guidelines for dyslipidemia management: Executive summary. Indian Heart J.76, S6–S19 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Hummelgaard, S. et al. Targeting PCSK9 to tackle cardiovascular disease. Pharmacol. Ther.249, 108480 (2023). [DOI] [PubMed] [Google Scholar]
- 538.Ferri, N. et al. Proprotein convertase subtilisin kexin type 9 (PCSK9) secreted by cultured smooth muscle cells reduces macrophages LDLR levels. Atherosclerosis220, 381–386 (2012). [DOI] [PubMed] [Google Scholar]
- 539.Lamb, Y. N. Inclisiran: first approval. Drugs81, 389–395 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Ray, K. K. et al. Two phase 3 trials of inclisiran in patients with elevated LDL cholesterol. N. Engl. J. Med.382, 1507–1519 (2020). [DOI] [PubMed] [Google Scholar]
- 541.Ray, K. K. et al. Inclisiran and cardiovascular events: a patient-level analysis of phase III trials. Eur. Heart J.44, 129–138 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Tsimikas, S. et al. Lipoprotein(a) reduction in persons with cardiovascular disease. N. Engl. J. Med.382, 244–255 (2020). [DOI] [PubMed] [Google Scholar]
- 543.Vinci, P. et al. Lipoprotein(a) as a risk factor for cardiovascular diseases: pathophysiology and treatment perspectives. Int. J. Environ. Res. Public Health20, 6721 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Wang, S. et al. The relationship between lipoprotein(a) and risk of cardiovascular disease: a mendelian randomization analysis. Eur. J. Med. Res.27, 211 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Malick, W. A., Goonewardena, S. N., Koenig, W. & Rosenson, R. S. Clinical trial design for lipoprotein(a)-lowering therapies: JACC focus seminar 2/3. J. Am. Coll. Cardiol.81, 1633–1645 (2023). [DOI] [PubMed] [Google Scholar]
- 546.Nissen, S. E. et al. Lepodisiran, an extended-duration short interfering RNA targeting lipoprotein(a): a randomized dose-ascending clinical trial. JAMA330, 2075–2083 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.O’donoghue, M. L. et al. Small interfering RNA to reduce lipoprotein(a) in cardiovascular disease. N. Engl. J. Med.387, 1855–1864 (2022). [DOI] [PubMed] [Google Scholar]
- 548.Koren, M. J. et al. Preclinical development and phase 1 trial of a novel siRNA targeting lipoprotein(a). Nat. Med.28, 96–103 (2022). [DOI] [PubMed] [Google Scholar]
- 549.Khan, R. S. & Frishman, W. H. Zilebesiran: a promising antihypertensive therapy inhibiting angiotensinogen synthesis. Cardiol. Rev. 10.1097/CRD.0000000000000645 (2024). [DOI] [PubMed]
- 550.Desai, A. S. et al. Zilebesiran, an RNA interference therapeutic agent for hypertension. N. Engl. J. Med.389, 228–238 (2023). [DOI] [PubMed] [Google Scholar]
- 551.Bakris, G. L. et al. RNA interference with zilebesiran for mild to moderate hypertension: the KARDIA-1 randomized clinical trial. JAMA331, 740–749 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Szabo, G. T., Mahiny, A. J. & Vlatkovic, I. COVID-19 mRNA vaccines: platforms and current developments. Mol. Ther.30, 1850–1868 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Fang, E. et al. Advances in COVID-19 mRNA vaccine development. Signal Transduct. Target Ther.7, 94 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.Baden, L. R. et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med.384, 403–416 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 555.Polack, F. P. et al. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N. Engl. J. Med.383, 2603–2615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Grudda, T. et al. Integrated hepatitis B virus DNA maintains surface antigen production during antiviral treatment. J. Clin. Investig.132, e161818 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Mak, L. Y. et al. Bepirovirsen (GSK3228836) in chronic hepatitis B infection: an evaluation of phase II progress. Expert Opin. Investig. Drugs32, 971–983 (2023). [DOI] [PubMed] [Google Scholar]
- 558.Yuen, M.-F. et al. Efficacy and safety of bepirovirsen in chronic hepatitis b infection. N. Engl. J. Med.387, 1957–1968 (2022). [DOI] [PubMed] [Google Scholar]
- 559.Yuen, M. F. et al. Safety, tolerability and antiviral activity of the antisense oligonucleotide bepirovirsen in patients with chronic hepatitis B: a phase 2 randomized controlled trial. Nat. Med.27, 1725–1734 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560.Yuen, M. F. et al. Efficacy and safety of the siRNA JNJ-73763989 and the capsid assembly modulator JNJ-56136379 (bersacapavir) with nucleos(t)ide analogues for the treatment of chronic hepatitis B virus infection (REEF-1): a multicentre, double-blind, active-controlled, randomised, phase 2b trial. Lancet Gastroenterol. Hepatol.8, 790–802 (2023). [DOI] [PubMed] [Google Scholar]
- 561.Gupta, S. V. et al. Clinical and preclinical single-dose pharmacokinetics of VIR-2218, an RNAi therapeutic targeting HBV infection. Drugs RD21, 455–465 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Soriano, V. Hepatitis B gene therapy coming to age. AIDS Rev.20, 125–127 (2018). [PubMed] [Google Scholar]
- 563.Jain, S., Kaur, J., Prasad, S. & Roy, I. Nucleic acid therapeutics: a focus on the development of aptamers. Expert Opin. Drug Discov.16, 255–274 (2021). [DOI] [PubMed] [Google Scholar]
- 564.Sabir, F. et al. DNA based and stimuli-responsive smart nanocarrier for diagnosis and treatment of cancer: applications and challenges. Cancers (Basel)13, 3396 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565.Augustine, R. et al. pH-responsive polypeptide-based smart nano-carriers for theranostic applications. Molecules24, 2961 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 566.Dirin, M. & Winkler, J. Influence of diverse chemical modifications on the ADME characteristics and toxicology of antisense oligonucleotides. Expert Opin. Biol. Ther.13, 875–888 (2013). [DOI] [PubMed] [Google Scholar]
- 567.Amantana, A. et al. Pharmacokinetics, biodistribution, stability and toxicity of a cell-penetrating peptide-morpholino oligomer conjugate. Bioconjugate Chem.18, 1325–1331 (2007). [DOI] [PubMed] [Google Scholar]
- 568.Gao, X. et al. The association of autophagy with polyethylenimine-induced cytotoxicity in nephritic and hepatic cell lines. Biomaterials32, 8613–8625 (2011). [DOI] [PubMed] [Google Scholar]
- 569.Takakusa, H. et al. Drug Metabolism and pharmacokinetics of antisense oligonucleotide therapeutics: typical profiles, evaluation approaches, and points to consider compared with small molecule drugs. Nucleic Acid Ther.33, 83–94 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.Hashida, M. Role of pharmacokinetic consideration for the development of drug delivery systems: A historical overview. Adv. Drug Deliv. Rev.157, 71–82 (2020). [DOI] [PubMed] [Google Scholar]
- 571.Jiang, R. et al. Factors influencing ADME properties of therapeutic antisense oligonucleotides: physicochemical characteristics and beyond. Curr. Drug Metab.24, 536–552 (2023). [DOI] [PubMed] [Google Scholar]
- 572.Bosgra, S. et al. The pharmacokinetics of 2’-O-methyl phosphorothioate antisense oligonucleotides: experiences from developing exon skipping therapies for duchenne muscular dystrophy. Nucleic Acid Ther.29, 305–322 (2019). [DOI] [PubMed] [Google Scholar]
- 573.Gonzalez-Barriga, A. et al. Intracellular distribution and nuclear activity of antisense oligonucleotides after unassisted uptake in myoblasts and differentiated myotubes in vitro. Nucleic Acid Ther.27, 144–158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 574.Wang, L. & Ji, C. Advances in quantitative bioanalysis of oligonucleotide biomarkers and therapeutics. Bioanalysis8, 143–155 (2016). [DOI] [PubMed] [Google Scholar]
- 575.Xiao, X. et al. Multi-functional peptide-microRNA nanocomplex for targeted microRNA delivery and function imaging. Chemistry24, 2277–2285 (2018). [DOI] [PubMed] [Google Scholar]
- 576.Migliorati, J. M. et al. Absorption, distribution, metabolism, and excretion of US food and drug administration-approved antisense oligonucleotide drugs. Drug Metab. Dispos.50, 888–897 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 577.Miao, Y. et al. Current status and trends in small nucleic acid drug development: leading the future. Acta Pharm. Sin. B14, 3802–3817 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578.Perry, C. M. & Balfour, J. A. Fomivirsen. Drugs57, 375–381 (1999). [DOI] [PubMed] [Google Scholar]
- 579.Stein, E. A. et al. Apolipoprotein B synthesis inhibition with mipomersen in heterozygous familial hypercholesterolemia: results of a randomized, double-blind, placebo-controlled trial to assess efficacy and safety as add-on therapy in patients with coronary artery disease. Circulation126, 2283–2292 (2012). [DOI] [PubMed] [Google Scholar]
- 580.Hoy, S. M. Nusinersen: first global approval. Drugs77, 473–479 (2017). [DOI] [PubMed] [Google Scholar]
- 581.Finkel, R. S. et al. Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N. Engl. J. Med.377, 1723–1732 (2017). [DOI] [PubMed] [Google Scholar]
- 582.Witztum, J. L. et al. Volanesorsen and triglyceride levels in familial chylomicronemia syndrome. N. Engl. J. Med.381, 531–542 (2019). [DOI] [PubMed] [Google Scholar]
- 583.Clemens, P. R. et al. Safety, tolerability, and efficacy of viltolarsen in boys with duchenne muscular dystrophy amenable to exon 53 skipping: a phase 2 randomized clinical trial. JAMA Neurol.77, 982–991 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584.Syed, Y. Y. Givosiran: a review in acute hepatic porphyria. Drugs81, 841–848 (2021). [DOI] [PubMed] [Google Scholar]
- 585.Kang, C. Avacincaptad pegol: first approval. Drugs83, 1447–1453 (2023). [DOI] [PubMed] [Google Scholar]
- 586.Grana, C. et al. Efficacy and safety of COVID-19 vaccines. Cochrane Database Syst. Rev.12, CD015477 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 587.Britton, A. et al. Use of respiratory syncytial virus vaccines in adults aged ≥60 years: updated recommendations of the advisory committee on immunization practices - United States. Morb. Mortal. Wkly. Rep.73, 696–702 (2024). [DOI] [PubMed] [Google Scholar]
- 588.Maurer, M. S. Overview of current and emerging therapies for amyloid transthyretin cardiomyopathy. Am. J. Cardiol.185, S23–S34 (2022). [DOI] [PubMed] [Google Scholar]
- 589.Helm, J., Schöls, L. & Hauser, S. Towards personalized allele-specific antisense oligonucleotide therapies for toxic gain-of-function neurodegenerative diseases. Pharmaceutics14, 1708 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 590.Young, G. et al. Efficacy and safety of fitusiran prophylaxis in people with haemophilia A or haemophilia B with inhibitors (ATLAS-INH): a multicentre, open-label, randomised phase 3 trial. Lancet401, 1427–1437 (2023). [DOI] [PubMed] [Google Scholar]
- 591.Riedl, M. A. et al. Efficacy and safety of donidalorsen for hereditary angioedema. N. Engl. J. Med.391, 21–31 (2024). [DOI] [PubMed] [Google Scholar]
- 592.Riedl, M. A. et al. Clinical progress in hepatic targeting for novel prophylactic therapies in hereditary angioedema. J. Allergy Clin. Immunol. Pract.12, 911–918 (2024). [DOI] [PubMed] [Google Scholar]
- 593.Badri, P. et al. Pharmacokinetic and pharmacodynamic properties of cemdisiran, an RNAi therapeutic targeting complement component 5, in healthy subjects and patients with paroxysmal nocturnal hemoglobinuria. Clin. Pharmacokinet.60, 365–378 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.Caravaca-Fontan, F., Gutierrez, E., Sevillano, A. M. & Praga, M. Targeting complement in IgA nephropathy. Clin. Kidney J.16, ii28–ii39 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 595.Woodcock, I. R. et al. A phase 2 open-label study of the safety and efficacy of weekly dosing of ATL1102 in patients with non-ambulatory duchenne muscular dystrophy and pharmacology in mdx mice. PLoS One19, e0294847 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 596.Wengert, E. R. et al. Targeted augmentation of nuclear gene output (TANGO) of scn1a rescues parvalbumin interneuron excitability and reduces seizures in a mouse model of dravet syndrome. Brain Res1775, 147743 (2022). [DOI] [PubMed] [Google Scholar]
- 597.Longhurst, H. J. et al. CRISPR-Cas9 in vivo gene editing of KLKB1 for hereditary angioedema. N. Engl. J. Med.390, 432–441 (2024). [DOI] [PubMed] [Google Scholar]
- 598.Baek, R. et al. Characterizing the mechanism of action for mRNA therapeutics for the treatment of propionic acidemia, methylmalonic acidemia, and phenylketonuria. Nat. Commun.15, 3804 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599.Jeffrey, S. et al. Individualized neoantigen therapy mRNA-4157 (V940) plus pembrolizumab in resected melanoma: 3-year update from the mRNA-4157-P201 (KEYNOTE-942) trial. J. Clin. Oncol.42, LBA9512 (2024). [Google Scholar]
- 600.Rojas, L. A. et al. Personalized RNA neoantigen vaccines stimulate T cells in pancreatic cancer. Nature618, 144–150 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 601.Steurer, M. et al. Olaptesed pegol (NOX-A12) with bendamustine and rituximab: a phase IIa study in patients with relapsed/refractory chronic lymphocytic leukemia. Haematologica104, 2053–2060 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 602.Russell, S. R. et al. Intravitreal antisense oligonucleotide sepofarsen in leber congenital amaurosis type 10: a phase 1b/2 trial. Nat. Med.28, 1014–1021 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 603.Dreismann, A. K. et al. Gene targeting as a therapeutic avenue in diseases mediated by the complement alternative pathway. Immunol. Rev.313, 402–419 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 604.Tselepis, A. D. Treatment of Lp(a): is it the future or are we ready today? Curr. Atheroscler. Rep.25, 679–689 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 605.Tardif, J. C. et al. Apolipoprotein C-III reduction in subjects with moderate hypertriglyceridaemia and at high cardiovascular risk. Eur. Heart J.43, 1401–1412 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 606.Gouni-Berthold, I., Schwarz, J. & Berthold, H. K. Updates in drug treatment of severe hypertriglyceridemia. Curr. Atheroscler. Rep.25, 701–709 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607.Rosenson, R. S. et al. Zodasiran, an RNAi therapeutic targeting ANGPTL3, for mixed hyperlipidemia. N. Engl. J. Med.391, 913–925 (2024). [DOI] [PubMed] [Google Scholar]
- 608.Huang, S. A. et al. Abstract 14387: dose-related reductions in blood pressure with a RNA interference (RNAi) therapeutic targeting angiotensinogen in hypertensive patients: interim results from a first-in-human phase 1 study of ALN-AGT01. Circulation142, A14387 (2020). [Google Scholar]
- 609.Wilson, E. et al. Efficacy and safety of an mRNA-based RSV PreF vaccine in older adults. N. Engl. J. Med.389, 2233–2244 (2023). [DOI] [PubMed] [Google Scholar]
- 610.Hu, X. et al. Human cytomegalovirus mRNA-1647 vaccine candidate elicits potent and broad neutralization and higher antibody-dependent cellular cytotoxicity responses than the gB/MF59 vaccine. J. Infect. Dis.230, 455–466 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 611.Zanardi, T. A. et al. Safety, pharmacokinetic, and pharmacodynamic evaluation of a 2’-(2-methoxyethyl)-d-ribose antisense oligonucleotide-triantenarry n-acetyl-galactosamine conjugate that targets the human transmembrane protease serine 6. J. Pharmacol. Exp. Ther.377, 51–63 (2021). [DOI] [PubMed] [Google Scholar]
- 612.Prikhodko, V. A., Bezborodkina, N. N. & Okovityi, S. V. Pharmacotherapy for non-alcoholic fatty liver disease: emerging targets and drug candidates. Biomedicines10, 274 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 613.Bujko, K. et al. Signaling of the complement cleavage product anaphylatoxin C5a through C5aR (CD88) contributes to pharmacological hematopoietic stem cell mobilization. Stem Cell Rev. Rep.13, 793–800 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]