Abstract
Cell death plasticity is crucial for modulating tissue homeostasis and immune responses, but our understanding of the molecular components that regulate cell death pathways to determine cell fate remains limited. Here, a CRISPR screen of acute myeloid leukemia cells identifies protein tyrosine phosphatase non-receptor type 23 (PTPN23) as essential for survival. Loss of PTPN23 activates nuclear factor-kappa B, apoptotic, necroptotic, and pyroptotic pathways by causing the accumulation of death receptors and toll-like receptors (TLRs) in endosomes. These effects are recapitulated by depletion of PTPN23 co-dependent genes in the endosomal sorting complex required for transport (ESCRT) pathway. Through proximity-dependent biotin labeling, we show that NAK-associated protein 1 interacts with PTPN23 to facilitate endosomal sorting of tumor necrosis factor receptor 1 (TNFR1), sensitizing cells to TNF-α-induced cytotoxicity. Our findings reveal PTPN23-dependent ESCRT machinery as a cell death checkpoint that regulates the spatiotemporal distribution of death receptors and TLRs to restrain multiple cell death pathways.
Subject terms: Intracellular signalling peptides and proteins, Necroptosis, Apoptosis, ESCRT, Acute myeloid leukaemia
The intricate regulation of cell death pathways remains to be understood. Here the authors identify that PTPN23 functions as a cell death checkpoint to restrain apoptosis, necroptosis and pyroptosis via regulating the endosomal sorting of death receptors and Toll-like receptors.
Introduction
Evasion of cell death is a hallmark of tumor development and a potent strategy exploited by pathogens to escape host defense. To counteract this phenomenon, cells have evolved a range of regulated cell death pathways, which serve as alternative and effective backup mechanisms. The molecular compositions and signaling cascades underlying regulated cell death pathways, including those underlying apoptosis, necroptosis, and pyroptosis, have been well-characterized1. Recent studies have shed light on the concept of plasticity, reflecting the ability of cells to adapt dynamically and switch between multiple cell death pathways, and highlighting the intricate crosstalk among them2–8. Nevertheless, the regulatory mechanisms orchestrating this plasticity in response to various environmental stimuli are not fully elucidated.
The cell survival, inflammation, and death signaling pathways are intertwined in response to the activation of immune receptors, such as death receptors that are members of the tumor necrosis factor receptor superfamily (TNFRSF), and Toll-like receptors (TLRs). The primary signaling output of immune receptor activation is often NF-κB and mitogen-activated protein kinase (MAPK)-mediated prosurvival and proinflammatory pathways. However, pathogens can inhibit these pathways using various strategies to evade the immune response2. For example, bacteria Yersinia disable NF-κB by targeting transforming growth factor beta-activated kinase 1 (TAK1)9, which is critical for maintaining cell survival through regulating RIPK1 phosphorylation status10–12. In the absence of NF-κB and TAK1 signaling, immune receptor activation can promote RIPK1 to form distinct cytosolic complexes, which comprise Fas-associated protein with death domain (FADD) and Caspase-8 (CASP8), to initiate apoptosis, or RIPK3 and MLKL, to drive necroptosis13,14. In TAK1-deficient macrophages, CASP8 can also induce pyroptosis by direct cleavage of gasdermin D (GSDMD) or by promoting inflammasome formation8,15,16. Thus, in cells where multiple death cascades are present, the intricate interplay between incoming signals is crucial for determining cell fate. Through evolutionary pressure, cells have developed diverse cell death pathways that allow them to trigger different forms of regulated cell death, thereby thwarting the efforts of pathogens to subvert these processes17. Nevertheless, it remains unclear whether evolution has also provided cells with counteracting mechanisms to limit the occurrence of multiple modes of cell death, which would ensure that cells maintain the proper balance between survival and death, preventing excessive cell death that could be detrimental to the organism.
PTPN23 is known to regulate a variety of cell-surface receptors, including EGFR18, PDGFRβ19, and integrins20, influencing various cellular processes such as proliferation, migration, and invasion in cancer21. In this study, we demonstrate that PTPN23 is essential for limiting apoptosis, necroptosis, and pyroptosis in acute myeloid leukemia (AML) cells. The depletion of PTPN23 results in the accumulation of death receptors and TLRs within the endosomal compartments, which leads to the activation of prosurvival and proinflammatory NF-κB pathway, as well as multiple cell death signaling cascades, highlighting the pivotal role of endocytic vesicles in integrating signals from diverse receptors and modulating the balance between survival and cell death signaling. We also reveal that PTPN23 functionally relies on multiple ESCRT components and that PTPN23-dependent ESCRT activity is necessary for the proper endo-lysosomal trafficking of death receptors and TLRs to attenuate downstream signaling. Moreover, we identify NAP1 as a critical PTPN23 binding partner involved in the endosomal sorting of TNFR1. Taken together, our data reveal the plasticity of myeloid cells in executing multiple cell death pathways, and underscore the importance of PTPN23-dependent ESCRT machinery as an intracellular checkpoint to restrain apoptosis, necroptosis, and pyroptosis.
Results
PTPN23 sustained cell survival in a phosphatase activity-independent manner
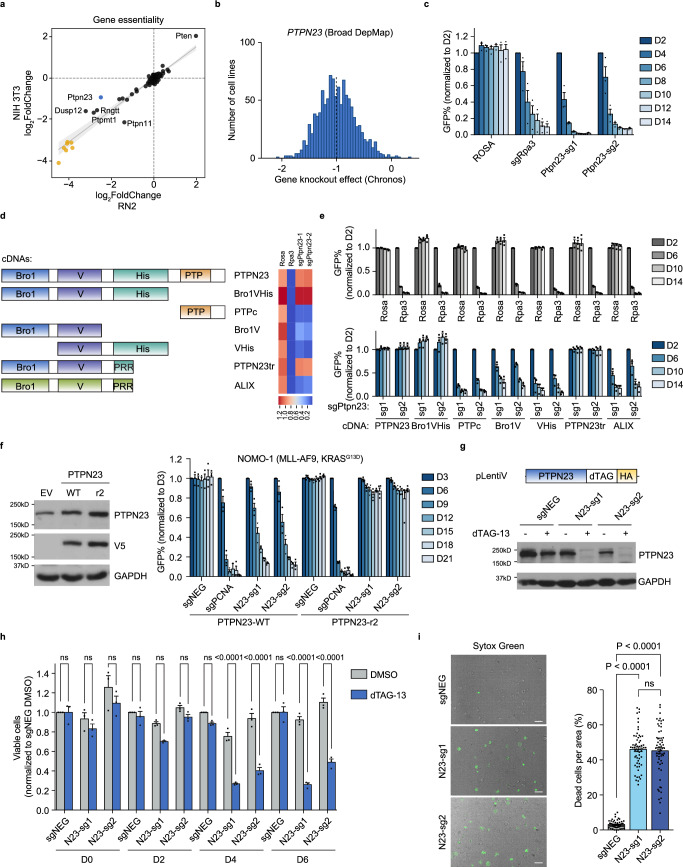
The regulation of cellular signals to determine cell fate broadly involves tyrosine phosphorylation, and accumulating evidence indicates that protein tyrosine phosphatases (PTPs) are crucial mediators of cell death and survival22–24. In light of this, we applied a domain-focused CRISPR approach25 to target the phosphatase domains of 101 murine PTP family members. We performed a dropout screen with murine RN2 (MLL-AF9; NrasG12D) AML cells26 and NIH 3T3 fibroblasts, to identify vulnerabilities preferential to AML cells. Using next-generation sequencing to measure sgRNA frequency, we identified four PTPs that may play a critical role in RN2 cells (Fig. 1a, Supplementary Data 1, 2). Among the top-scoring hits, the PTPN23 locus is frequently hemizygously deleted in various cancers27–31 and has been suggested to function as a haploinsufficient tumor suppressor18–20,32,33. However, genome-wide CRISPR screens of over 1000 cancer cell lines (DepMap)34 revealed that PTPN23 is a common essential gene in human cancer cell lines (Fig. 1b). Furthermore, homozygous deletion of Ptpn23 results in embryonic lethality in mice35. In fact, when we prolonged the culture of NIH 3T3 cells, we observed that Ptpn23 knockout also led to cell death (Supplementary Fig. 1b). Although initially, we observed a higher requirement for PTPN23 in mouse AML cells compared with NIH 3T3 fibroblast cells, the differing rates of cell depletion are likely influenced by distinct cell growth rates. Overall, our results, along with the DepMap dataset, emphasize the broad essentiality of PTPN23. Therefore, we sought to explore the molecular mechanisms underlying this essential role of PTPN23 in maintaining cell survival, which contrasts with its tumor suppressor function20,21,32,36.
Fig. 1. PTPN23 was identified to be essential for cell survival with CRISPR screening.
a Gene essentiality of mouse PTPs in RN2 cells and NIH 3T3 cells. sgRNA frequencies at Day 2 and Day 14 were determined by next-generation sequencing, with average logarithmic frequency changes calculated. Yellow dots represent pan-essential genes spiked into the screen library. b Histogram of PTPN23 gene knockout effect in 1078 cancer cell lines (DepMap 22Q2). The dashed line indicates the median score. c GFP competition growth assays performed in RN2 cells (n = 3 independent experiments). d Left, Human PTPN23 and ALIX full-length constructs and PTPN23 truncations used in rescue experiments. Right, Heatmap visualizing the complementation effects aligned with the corresponding numbers on D14 in (e). His: His domain; PRR: Proline-rich region, located at the N-terminus of the His domain. e GFP competition growth assays performed in RN2 stable cell lines expressing indicated cDNAs. Top, control sgRNAs; bottom, PTPN23 sgRNAs (n = 3 independent experiments). f Left, immunoblot of NOMO-1 cells overexpressing empty vector (EV), sgRNA-sensitive (PTPN23-WT) or sgRNA-resistant (PTPN23-r2) PTPN23 cDNA (n = 3 independent experiments). Right, GFP competition growth assays performed using NOMO-1 stable cell lines expressing either PTPN23-WT (n = 3 independent experiments) or PTPN23-r2 (n = 4 independent experiments). g Top, acute depletion of PTPN23 with dTAG system. NOMO-1 cells stably expressing sgRNA-resistant PTPN23 fused with dTAG and endogenous PTPN23 were depleted with two sgRNAs. Bottom, immunoblotting analysis of PTPN23 examined after 4-day treatment of dTAG-13 (100 nM). h CellTiter-Glo (CTG) luminescence cell viability assay measured on cells treated with either DMSO or dTAG-13. DMSO or 500 nM dTAG-13 were added to three PTPN23-dTAG NOMO-1 cell lines established in (g) (n = 3 independent experiments). i Cell death indicated by Sytox Green positivity. PTPN23-dTAG NOMO-1 cells were treated with 200 nM dTAG-13 for 4 days, and stained with Sytox Green (n = 60 fields captured, 3 independent experiments). Scale bar: 30 μm. Data are presented as mean ± SEM, statistical analysis for (h) by Two-way ANOVA, Sidak’s multiple comparisons test; (i) by One-way ANOVA, Tukey’s multiple comparisons test.
Having validated that PTPN23 is required for RN2 cell survival (Fig. 1c), we performed complementation experiments to define the essential domains (Fig. 1d). Expression of sgRNA-resistant, wild-type human PTPN23 restored the growth of Ptpn23-depleted RN2 cells (Fig. 1d, e). PTPN23 and ALIX (ALG-2-interacting protein X) are related genes in the human genome, sharing similar domain organization, with an N-terminal Bro1 domain, followed by a V domain and a Proline-rich region (PRR); however, ALIX lacks a phosphatase domain37. Through a series of PTPN23 truncations, we determined that the minimal segment capable of rescuing the lethal phenotype caused by deleting PTPN23 corresponded to the boundary of ALIX (Fig. 1d, e), suggesting that the catalytic activity of PTPN23 was dispensable for AML cell growth. The dependency upon PTPN23 was validated further in three human AML cell lines (Fig. 1f and Supplementary Fig. 1a).
To complement this genetic strategy, we employed the dTAG system to induce acute and synchronized degradation of PTPN23 in NOMO-1 human AML cells38 (Supplementary Fig. 1c). Ectopically expressed dTAG-PTPN23 fully rescued the growth disadvantages caused by the ablation of the endogenous protein, affirming that dTAG did not impair recombinant protein function (Fig. 1g and Supplementary Fig. 1d). Following dTAG-13 treatment, we observed rapid protein degradation within 90 min, and significant growth impairment after four days (Fig. 1h and Supplementary Fig. 1e), culminating in lytic cell death (Fig. 1i). In summary, both genetic and chemical-genetic approaches illustrated a PTPN23 dependency in AML cells that occurs independently of its phosphatase function.
Ablation of PTPN23 led to apoptotic, necroptotic, and pyroptotic cell death
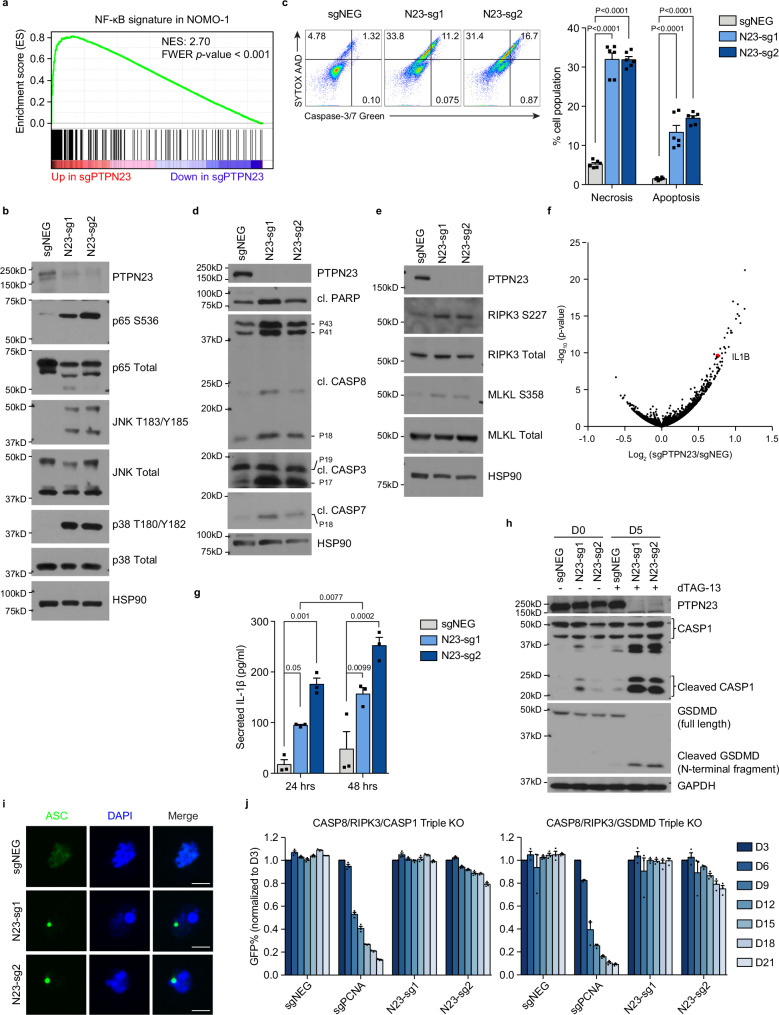
To investigate the mechanism of cell death, we performed RNA-seq analysis, following CRISPR-mediated suppression of PTPN23 in NOMO-1 cells, and used unbiased gene set enrichment analysis (GSEA) to demonstrate increased NF-κB signatures (Fig. 2a and Supplementary Fig. 2a). NF-κB activation and cell death are two opposing signaling outputs that can be elicited from activated death receptors or TLRs39–42. First, we validated NF-κB activation by immunoblotting. Upon both CRISPR knockout and dTAG-13-induced degradation of PTPN23 in NOMO-1 cells, we observed increased phosphorylation of p65 (NF-κB subunit), p38 and JNK (MAPKs) (Fig. 2b and Supplementary Fig. 2b). Considering that signaling can be diverted from NF-κB to CASP8-mediated apoptotic or RIPK3-dependent necroptotic cell death43, we tested the presence of cell death-related markers. We induced cell death by treating PTPN23-dTAG NOMO-1 cells with dTAG-13, and observed significant elevation in both necrotic (SYTOX AADvanced+/Caspase-3/7−) and apoptotic (Caspase-3/7+) cell populations in PTPN23-depleted cells (Fig. 2c). In line with this observation, depletion of PTPN23 induced extrinsic apoptosis, evident by cleaved caspases and poly(ADP ribose) polymerase (PARP) products (Fig. 2d). Furthermore, we detected the phosphorylation of RIPK3 and MLKL upon treatment with dTAG-13 (Fig. 2e), suggesting the involvement of necroptosis after PTPN23 depletion; however, double knockout of CASP8 and RIPK3 only partially rescued the lethality caused by suppression of PTPN23 (Supplementary Fig. 2c), indicating that PTPN23 deficiency triggered additional cell death modalities.
Fig. 2. PTPN23 depletion activated NF- κB and triggered apoptosis, necroptosis, and pyroptosis.
a GSEA analysis showing that NF-κB signature was upregulated upon PTPN23 depletion at post-infection day 6. Statistical significance was assessed using Family-Wise Error Rate (FWER) correction. b Immunoblot of NF-κB and MAPK signaling in NOMO-1 cells harvested 6 days after sgNEG, N23-sg1, or N23-sg2 lentiviral infection. (n = 3 independent experiments). c Left, SYTOX AADvanced and fluorogenic substrate of Caspase-3/7 stains of PTPN23-dTAG NOMO-1 cells treated with 200 nM dTAG-13 for 4 days. Right, Quantification of necrotic and apoptotic cells from negative control, PTPN23 sgRNA infected cells. Necrotic cells are SYTOX AADvanced positive and cleaved Caspase-3/7 negative; apoptotic cells are cleaved Caspase-3/7 positive (n = 6, derived from 3 independent experiments, each performed with 2 replicates). d Immunoblot analysis of cleaved forms of PARP and caspases in PTPN23-dTAG cells after 4 days of dTAG-13 (200 nM) treatment (n = 3 independent experiments). e Immunoblotting analysis of RIPK3 and MLKL in PTPN23-dTAG cells after 5 days of dTAG-13 (200 nM) treatment. (n = 3 independent experiments). f Volcano plot of RNA-seq analysis to compare transcriptional changes of sgNEG and PTPN23 sgRNA infected cells at post-infection day 6. g ELISA assay measuring the amount of secreted IL-1β in PTPN23 deficient NOMO-1 cells. PTPN23-dTAG NOMO-1 cells were treated with dTAG-13 (200 nM) for 4 days, and seeded into 96-well plates. The supernatants were then collected after 24 h and 48 h in the presence of dTAG-13 (200 nM) for ELISA analyses (n = 3 biological replicates). h Immunoblotting analysis of CASP1 and GSDMD in PTPN23-dTAG NOMO-1 cells after 5 days of dTAG-13 (200 nM) treatment (n = 3 independent experiments). Each immunoblot was reproduced three times with similar results for (b, d, e, and h). i Representative images of immunofluorescence microscopy of ASC specks in PTPN23-dTAG NOMO-1 cells. Cells were treated with 200 nM dTAG-13 for 5 days. Scale bar: 5 μm. j GFP competition growth assays performed using CASP8/RIPK3/CASP1 or CASP8/RIPK3/GSDMD triple knockout cells. Two single-cell clones derived from each genetic background were pooled together (n = 3 independent experiments). Data are presented as mean ± SEM, statistical analysis for (c) by Two-way ANOVA, Dunnett’s multiple comparisons test; (g) by Two-way ANOVA, Tukey’s multiple comparisons test.
In searching for induction of cell death-related genes, we observed that the IL-1β gene was upregulated in PTPN23-knockout cells in the aforementioned RNA-seq experiment (Fig. 2f). Upon activation of the inflammasome, mature IL-1β is secreted from pyroptotic cells following cleavage by Caspase-1 (CASP1)44, and we observed pronounced secretion of IL-1β from PTPN23-deficient cells (Fig. 2g). Furthermore, we detected cleavage of CASP1 and Gasdermin D (GSDMD), indicating activation of inflammasomes and execution of pyroptosis (Fig. 2h). The canonical inflammasome forms a macromolecular complex, known as the “ASC speck”, which can be visualized as a single micrometer-sized focus per cell44. We detected ASC specks in the PTPN23-deficient cells (Fig. 2i), consistent with triggering of pyroptotic cell death following PTPN23 ablation. In addition, we observed a high frequency of CASP8 recruitment to inflammasomes after PTPN23 depletion (Supplementary Fig. 2d), suggesting that three cell death pathways may be interconnected.
Based on our observations of the activation of caspases in the apoptotic pathway, phosphorylation of RIPK3 and MLKL in the necroptotic pathway, recruitment of CASP8 to inflammasome, and cleavage of GSDMD and CASP1 in the pyroptotic pathway, we propose that PTPN23 serves as a key regulator in limiting multiple cell death pathways. To support this hypothesis further, we established triple knockouts of CASP8−/−/RIPK3−/−/CASP1−/− and CASP8−/−/RIPK3−/−/GSDMD−/− in NOMO-1 cells. Our results showed that both cell lines were protected from cell death following sgRNA-induced depletion of PTPN23 (Fig. 2j), whereas single knockouts of CASP1 or GSDMD failed to provide complete rescue (Supplementary Fig. 2e). The protective effects observed in triple knockout cells allowed us to assess the long-term consequences of PTPN23 depletion. We performed RNA-seq analysis on CASP8−/−/RIPK3−/−/GSDMD−/− NOMO-1 cells, at Days 6, 12, and 18 following PTPN23 sgRNA infection, revealing the most pronounced alteration in the NF-κB signature (Supplementary Fig. 2f). This substantiates that the enduring consequence of PTPN23 depletion involves the sustained activation of death receptors and TLRs. Collectively, those findings suggest that PTPN23 deficiency resulted in regulated cell death in the form of apoptosis, necroptosis, and pyroptosis.
Cell death was attributed to the accumulation of death receptors and TLRs in PTPN23-deficient cells
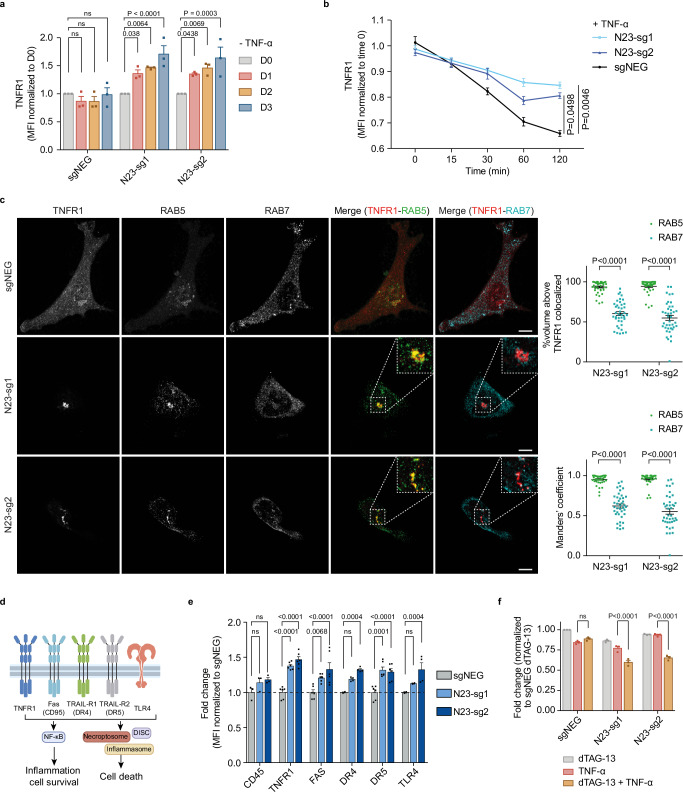
The depletion of PTPN23 resulted in an unexpected concomitant activation of prosurvival NF-κB signaling and execution of multiple cell death pathways, indicating that the balance between pro- and anti-survival was affected. Given the function of PTPN23 in receptor trafficking and lysosomal degradation18,20,45, we investigated receptors implicated in apoptotic, necroptotic, or pyroptotic signaling, such as death receptors, and TLR4, which are potent activators of interlinked NF-κB and cell death signaling39,46–48. We validated the rapid internalization and subsequent downregulation of endogenous TNFR1, the prototypic death receptor, determining a half-life (t1/2) of 67 ± 5.7 min (Supplementary Fig. 3b). Notably, treating NOMO-1 cells with cycloheximide for 4 h substantially reduced TNFR1 levels, whereas the presence of Bafilomycin A1 (Baf-A1), a lysosomal inhibitor, attenuated the degradation rate, confirming a lysosomal-dependent degradation mechanism (Supplementary Fig. 3c). Subsequently, we depleted PTPN23 protein in NOMO-1 cells using the dTAG system and measured the abundance of TNFR1 using flow cytometry. We observed elevated TNFR1 levels in PTPN23-depleted cells in comparison to controls, suggesting that ligand-independent degradation of TNFR1 was impaired in PTPN23-deficient cells (Fig. 3a, Supplementary Fig. 3a). Additionally, we observed a significantly slower degradation rate of TNFR1 in PTPN23-deficient cells compared to PTPN23-expressing cells (Supplementary Fig. 3d). Furthermore, we examined ligand-dependent regulation of TNFR1 by treating cells with dTAG-13 to degrade exogenous PTPN23, and then stimulated with TNF-α. In the negative control cells, total TNFR1 levels gradually decreased, indicating ligand-dependent receptor internalization and degradation, whereas in PTPN23-knockout cells a substantial reduction in the rate of total TNFR1 degradation was observed (Fig. 3b). The elevated abundance of TNFR1 in PTPN23-knockout cells was consistent with our observations that these cells displayed a prolonged duration and higher amplitude of NF-κB and MAPK signaling in response to TNF-α treatment (Supplementary Fig. 3e). Therefore, genetic ablation of PTPN23 extended the duration of the TNFR1 signal in both a TNF-α-independent and TNF-α-dependent manner.
Fig. 3. PTPN23 controlled the spatiotemporal regulation of TNFR1.
a Quantitation of fold changes in endogenous TNFR1 in the presence or absence of PTPN23 normalized to negative control at day 0 (n = 3 independent experiments). PTPN23-dTAG NOMO-1 cells were treated with 200 nM dTAG-13 for indicated periods. b Flow cytometry analysis depicting total TNFR1 levels in PTPN23-dTAG NOMO-1 cells stimulated with 200 ng/ml FLAG-TNF-α for the indicated time. Prior to stimulation, cells were treated with 200 nM dTAG-13 for 2 days to degrade PTPN23-dTAG recombinant protein (n = 4 independent experiments). c Left, representative immunofluorescence images of TNFR1, RAB5 and RAB7 in the modified HeLa cells. Hela cells were engineered with tetracycline-responsive element (TRE)-Cas9, and then a vector containing TNFR1-mCherry and either negative control or PTPN23 sgRNAs was ectopically expressed. Cells were treated with 1 μg/ml doxycycline (Dox) for 5 days and depletion of PTPN23 was examined (Supplementary Fig. 3f). Insets are higher magnification views of dashed rectangular regions. Images are shown as 3D reconstructions from confocal image stacks. Scale bar: 10 μm. Top right, quantification of volume percentage of TNFR1 colocalized with RAB5 or RAB7 in PTPN23-deficient cells. Bottom right, Manders’ colocalization coefficients of TNFR1-RAB5 and TNFR1-RAB7 (n = 40 per group, 2 independent experiments). d Death receptors and TLR4 subjected to examination. Created with Adobe Illustrator and BioRender.com (License reference: https://BioRender.com/a17y329). e Fold changes of death receptors and TLR4 in PTPN23-dTAG NOMO-1 cells after 3 days of 200 nM dTAG-13 treatment measured by flow cytometry (n = 3 biological replicates for CD45, n = 4 for DR4 and TLR4, n = 6 for TNFR1, FAS and DR5). f Cytotoxicity of TNF-α in PTPN23-dTAG NOMO-1 cells. Cells were incubated with 1 μM dTAG-13 or 100 ng/ml TNF-α as indicated for 48 h, and cell viability was measured using CTG assay (n = 3 independent experiments). Data are presented as mean ± SEM. Statistical analysis for (a, b, e, and f) by two-way ANOVA, Tukey’s multiple comparisons test; (c) by two-way ANOVA, Sidak multiple comparisons test.
In addition to studying the temporal dynamics of TNFR1, we sought to investigate its spatial distribution in the absence of PTPN23. For this purpose, we established an inducible knockout system in Hela cells (Supplementary Fig. 3f). Following depletion of PTPN23 (Supplementary Fig. 3f), we tracked TNFR1 subcellular localization using confocal microscopy. Compared to negative control sgRNA-infected cells, PTPN23-deficient cells exhibited discrete TNFR1-mCherry puncta in the cytoplasm, which colocalized with RAB5 to a greater extent than RAB7 (Fig. 3c); this suggests that TNFR1 trafficking from early to late endosomes was compromised in the PTPN23-knockout cells. This spatial redistribution of TNFR1 resembled the phenotype observed upon knockdown of ESCRT components, including Tsg101, Vps28, UBAP1, and CHMP4B49, supporting an ESCRT-mediated function of PTPN23.
Multiple death receptors (DRs) are known to play important roles in regulated cell death46. Our RNA-seq analysis showed that DR3 mRNA expression was below the detection limit in NOMO-1 cells, therefore, we used flow cytometry to measure the level of the other four death receptors, and TLR4, following the depletion of PTPN23 (Fig. 3d). PTPN23 sgRNA-transfected cell lines showed marked elevation of TNFR1, Fas, DR4, DR5, and TLR4, whereas CD45 (PTPRC) and transferrin receptor (TFRC) did not change significantly (Fig. 3e, Supplementary Fig. 3g). Furthermore, PTPN23 depletion rendered previously resistant NOMO-1 cells more susceptible to TNF-α-induced cell death (Fig. 3f), and similar sensitization effects were observed when PTPN23-deficient cells were stimulated with LPS (Supplementary Fig. 3h). This indicates that PTPN23 affects the balance between NF-κB and cell death signaling, which dictates the cell death threshold. Collectively, these data suggest that PTPN23 functions in regulating death receptors and TLRs in space and time, to restrain their downstream cell death signaling.
PTPN23 engaged ESCRT machinery through its V domain interacting with UBAP1
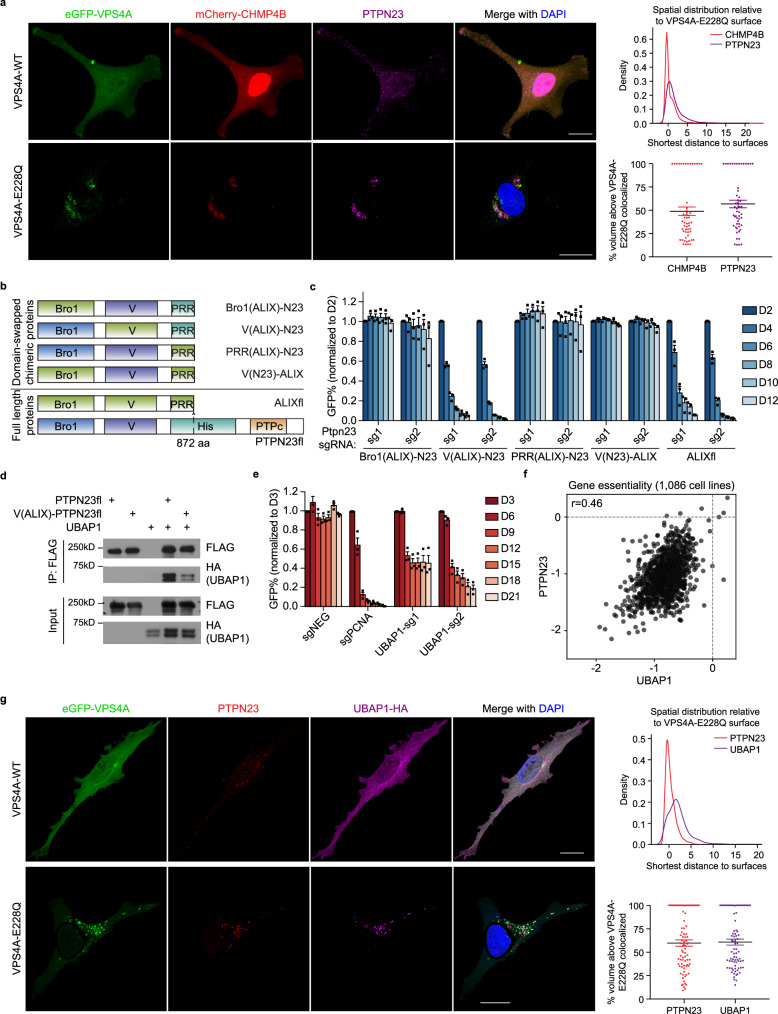
To demonstrate the integral role of PTPN23 within the ESCRT machinery and its involvement in the dynamic assembly and disassembly of ESCRT, we examined the coordination between PTPN23 and CHMP4B, a core ESCRT-III component. We used dominant-negative form of AAA ATPase VPS4, VPS4A-E228Q, which cannot hydrolyze ATP and leads to the formation of characteristic puncta in the cytoplasm50,51 and performed fluorescence microscopy to examine the distribution dynamics of PTPN23 and CHMP4B. In the VPS4A-WT-expressing cells, both PTPN23 and CHMP4B exhibited a diffuse cytoplasmic distribution. In contrast, following overexpression of VPS4A-E228Q, both proteins appeared in the vicinity of VPS4A-E228Q puncta, as measured by the distance between CHMP4B or PTPN23 and VPS4A-E228Q (Fig. 4a and Supplementary Fig. 4a). This suggests that, as with the core ESCRT-III subunit CHMP4B, the subcellular localization of PTPN23 coincided with VPS4A-E228Q, consistent with PTPN23 functioning in the ESCRT pathway.
Fig. 4. V domain of PTPN23 is indispensable for sustaining cell survival via interaction with UBAP1.
a Left, representative immunofluorescence images of HeLa cells expressing eGFP-VPS4A (WT or E228Q), mCherry-CHMP4B, and PTPN23. VPS4A (WT or E228Q) cDNAs were transiently expressed in HeLa cells stably expressing mCherry-CHMP4B. Immunofluorescence staining was performed after 36 h of VPS4A transfection. Scale bar: 20 μm. Top right, the density plot of the shortest distance between CHMP4B or PTPN23 spots to eGFP-VPS4A-E228Q surfaces. Bottom right, volume percentages of CHMP4B or PTPN23 spots colocalized above the VPS4A-E228Q surface. The percentage of volume colocalized between PTPN23 and CHMP4B is 60.6 ± 2.3 (mean ± SEM). A total of 54 cells were analyzed, in 3 independent experiments. b Schematic depiction of domain-swapping experiments. c GFP competition growth assays measuring PTPN23 dependency on RN2 cell lines overexpressing indicated chimeric proteins (n = 3 independent experiments). Data are presented as mean ± SEM. d Immunoprecipitation assay to measure the binding between UBAP1 and WT PTPN23 or ALIX V domain-swapped PTPN23 (n = 3 independent experiments). e GFP competition growth assay of UBAP1 knockout in NOMO-1 cells (n = 3 independent experiments). Data are presented as mean ± SEM. f Scatter plot of PTPN23 versus UBAP1 gene knockout effect in 1086 cancer cell lines using DepMap 22Q2 Public dataset from Broad DepMap project. g Left, representative immunofluorescence images of eGFP-VPS4A (WT or E228Q), PTPN23 and UBAP1 in HeLa cells. Scale bar: 20 μm. Top right, density plot of the shortest distance from PTPN23 or UBAP1 to VPS4A-E228Q. Bottom right, volume percentage of PTPN23 or UBAP1 spots colocalized above the VPS4A-E228Q surface. The percentage of volume colocalized between PTPN23 and UBAP1 is 46.9 ± 3.8 (mean ± SEM). A total of 80 cells were analyzed, in 3 independent experiments.
We sought to identify the components of ESCRT that coordinate with PTPN23 to promote the sorting of TNFR1. Two Bro1 domain-containing proteins, ALIX and PTPN23, have been shown to function in the ESCRT pathway; however, depletion of ALIX had no impact on AML cell growth and survival (Supplementary Fig. 4b). The minimal segment of PTPN23 that rescued the growth defects caused by PTPN23 knockout consists of the domains within the protein that correspond with those in ALIX (Fig. 1e). This allowed us to use a domain-swapping strategy to interrogate the functional region of PTPN23 that is uniquely responsible for AML cell survival. We constructed chimeric proteins with ALIX Bro1/V/PRR domains swapped into the PTPN23tr (truncated PTPN23 corresponding to ALIX boundary) backbone. The chimera containing the ALIX Bro1 domain or PRR segment completely compensated for the function of PTPN23; however, the chimeric protein with the V domain from ALIX failed to rescue the cell growth defects caused by PTPN23 knockout. Reciprocally, although ALIX could not rescue the lethal phenotype caused by Ptpn23 knockout, the chimeric ALIX containing the V domain from PTPN23 gained the capacity to rescue (Fig. 4b, c). These results illustrate that the V domain supports a unique and non-redundant function of PTPN23 in AML cell survival.
We performed site-directed mutagenesis to abrogate each of the binding pockets in PTPN23 that have been reported to associate with ESCRT components18,45,52 and assessed the ability of each mutant to rescue the lethality caused by PTPN23 sgRNAs. Consistent with the domain-swapping experiments, the PTPN23-F678D mutation located in the V domain impaired rescue, whereas mutations in Bro1 or PRR were able to compensate for PTPN23 function, confirming the importance of the V domain (Supplementary Fig. 4c). UBAP1, an ESCRT-I component, has been identified as a binding partner of the PTPN23 V domain52,53. We confirmed the interaction between the PTPN23 V domain and UBAP1 by co-immunoprecipitation. UBAP1 bound to PTPN23 WT, but not chimeric PTPN23 containing the V domain from ALIX (Fig. 4d). Similar to PTPN23 knockout, homozygous deletion of UBAP1 is lethal (International Mouse Phenotyping Consortium), and sgRNA-induced UBAP1 depletion impaired the viability of NOMO-1 cells (Fig. 4e). In addition, we analyzed the DepMap database34 to examine the effect of UBAP1 knockout across a large panel of cell lines, and we observed a high co-dependency between PTPN23 and UBAP1 (Fig. 4f). Co-dependency denotes a situation in which the knockout of two genes independently elicits similar lethal effects across the examined cell lines, suggesting that they regulate related biological processes54. In this case, our data indicate that PTPN23 and UBAP1 may function together in a common protein complex or pathway.
To examine the involvement of PTPN23 and UBAP1 in the ESCRT pathway, particularly in regulating death receptor signaling, we treated control and UBAP1-knockout NOMO-1 cells with TNF-α. Similar to the effects of PTPN23 depletion, UBAP1-deficient cells exhibited a higher amplitude and longer duration of phosphorylation of p65 and p38 (Supplementary Fig. 4d). Moreover, we observed a redistribution of PTPN23 and UBAP1 upon overexpression of VPS4A-E228Q, providing further evidence of their concordant function in the ESCRT pathway (Fig. 4g and Supplementary Fig. 4e). Taken together, these data suggest that PTPN23, through its V domain, engaged the ESCRT machinery via UBAP1, which conferred specificity between the effects of PTPN23 and ALIX in regulating cell death.
ESCRT machinery restrained cell death and promoted the degradation of death receptors and TLRs
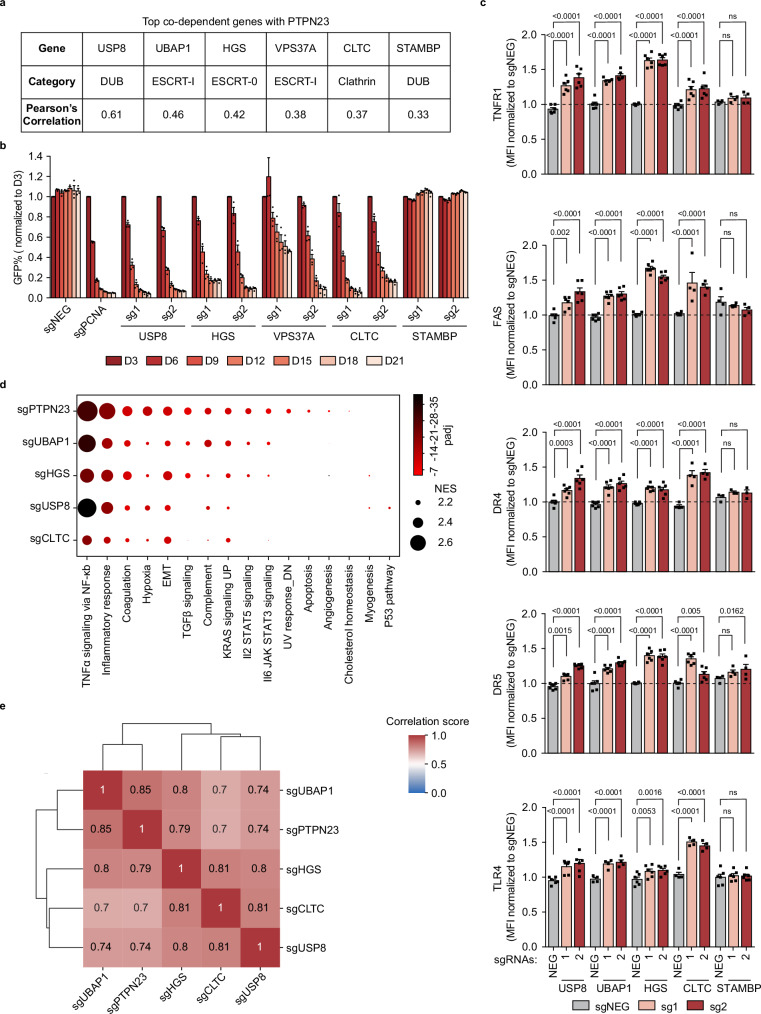
Upon mining the DepMap database, we noted that the top PTPN23 co-dependent genes were all involved in the ESCRT pathway (Fig. 5a). Therefore, we extended our study of the cell death mechanism to encompass these co-dependent genes and asked whether they shared a common mechanism with PTPN23 to sustain cell survival. USP8 and STAMBP are two endosomal deubiquitinating (DUB) enzymes, which remove ubiquitin chains from cargo proteins to regulate the balance between receptor lysosomal degradation and recycling to plasma membrane55–58. UBAP159 and VPS37A60 are ESCRT-I components, whereas HGS is an ESCRT-0 component61. CLTC, the clathrin heavy chain, is involved in the recruitment of ESCRT-0 onto endosomes62,63. These PTPN23 co-dependent genes are early-acting members of the ESCRT pathway, implying that cargo selection prior to commitment to lysosomal degradation plays an important role in ESCRT-mediated cell death. We showed that other than STAMBP, which exhibited a lineage-specific requirement in upper digestive cancers64, these were all required for NOMO-1 survival (Fig. 5b and Supplementary Fig. 5b).
Fig. 5. ESCRT machinery is essential to limit cell death mediated by death receptors and TLRs.
a The list of PTPN23’s top co-dependent genes with correlation factor >0.3 from DepMap Portal (22Q4 Public dataset). b GFP competition growth assay of PTPN23’s co-dependent gene depletion in NOMO-1 cells (n = 3 independent experiments). c Flow cytometric measurements of death receptors and TLR4 in NOMO-1 cells. Receptor levels were measured at day 7 post-infection with sgRNAs against corresponding ESCRT components. The number of biological replicates (n) for each gene knockout (KO) and receptor measurement is as follows: USP8 KO (n = 6), UBAP1 KO (n = 6, except for TLR4 measurement, n = 4), HGS KO (n = 6), CLTC KO (n = 4, except for TNFR1 and DR5 measurements, n = 6), and STAMBP KO (n = 4, except for DR4 measurement, n = 3, and TLR4 measurement, n = 6). d Dot plot of enriched pathways with normalized enrichment score (NES) > 2 derived from GSEA analysis. The sizes of the dots correspond the NES, and the colors of the dots represent the -log10 (padj). The NES is calculated by normalizing the raw enrichment score to account for differences in gene set sizes. The adjusted p-value (padj) is computed to control the false discovery rate when comparing multiple datasets. EMT: epithelial-mesenchymal transition. e Global correlation matrix of gene expression profile changes between samples. The log2 fold changes were calculated for each gene to compare the knockout and negative control samples. Heatmap represents Pearson’s correlation coefficient for log2 (fold changes) across five genes. NOMO-1 cells were transfected with two independent sgRNAs targeting the indicated gene, harvested at day 5 post-infection for USP8 and day 6 for remaining samples (n = 3 biological replicates). Data are presented as mean ± SEM. Statistical analysis for (c) by two-way ANOVA, Dunnett’s multiple comparisons test.
To investigate whether ESCRT components were involved in receptor trafficking and degradation, we examined the effects of depleting individual co-dependent genes on the level of death receptors and TLR4 in NOMO-1 cells. We showed that similar to the depletion of PTPN23, sgRNAs targeting USP8, UBAP1, HGS, and CLTC all caused receptor accumulation, indicating that disruption of these ESCRT components impaired trafficking and degradation of death receptors and TLRs (Fig. 5c). By contrast, STAMBP depletion did not lead to receptor accumulation, which is in agreement with its dispensability for NOMO-1 cell survival (Fig. 5c). These results suggest that multiple ESCRT components and associated proteins function to promote degradation of death receptors and TLRs, which is necessary for the maintenance of cell survival.
To validate further the correlation between PTPN23 and ESCRT pathway members, we ablated PTPN23 and its co-dependent genes in NOMO-1 cells, followed by RNA-seq analysis to compare changes in the mRNA profile. Our analyses revealed that the NF-κB-induced transcriptional program was the most enriched gene signature (Fig. 5d and Supplementary Fig. 5a), consistent with the observed accumulation of death receptors and TLRs (Fig. 5c). Moreover, the overall transcriptional alterations displayed high correlations between PTPN23 and its co-dependent genes (Fig. 5e). Collectively, our data reveal that the impact of disruption of PTPN23 and its co-dependent genes was mostly indistinguishable in triggering cell death, accumulation of death receptors and TLRs, and transcriptomic profiles. Thus, our study has uncovered the crucial role of PTPN23 and multiple members in the ESCRT pathway in limiting cell death mediated by death receptors and TLRs.
The BioID interactome of PTPN23 revealed NAP1 as a potential adapter protein to TNFR1 signaling complex
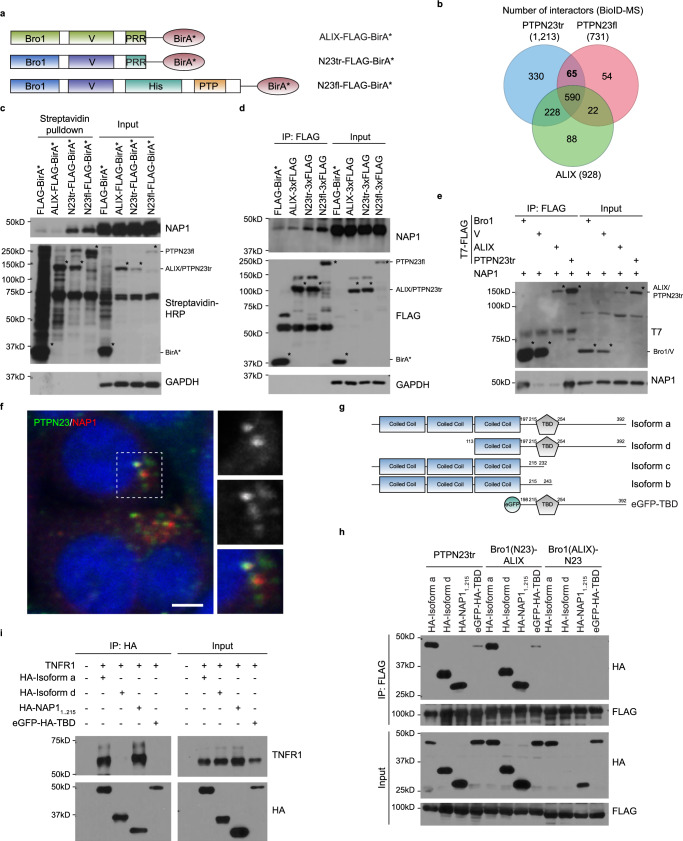
PTPN23 plays a critical role in the degradation of death receptors and TLRs, thus, to identify additional components of this pathway, we employed a proximity-dependent biotin identification (BioID) approach in AML cells. We established NOMO-1 cell lines that stably express ALIX and PTPN23 constructs in which the BirA* biotin ligase was fused to the C-terminus of each recombinant protein (Fig. 6a and Supplementary Fig. 6a). The BirA*-tagged recombinant proteins biotinylate endogenous proximal interacting proteins, which were subsequently isolated using streptavidin pulldown and identified through mass spectrometry (MS)65. We aimed to identify PTPN23-specific interactors that were distinct from those of ALIX, by ranking the spectral counts based on the fold change of PTPN23 relative to ALIX in the presence of biotin (Fig. 6b, Supplementary Data 3). The top 8 candidates in Table 1, which included known binding partners of PTPN23, such as ZFYVE9 (SARA) and ZFYVE16 (Endofin)66, were selected based on the average fold change of protein enrichment in PTPN23tr- and PTPN23fl-overexpressing cell lines versus the ALIX-overexpressing cell line, employing a threshold of 2 for inclusion. Importantly, we also identified AZI2 (5-azacytidine induced 2, also known as NAP1 (NF-κB-activating kinase-Associated Protein 1)), which has been implicated in TNFR1 signaling complex via its binding to TBK167.
Fig. 6. PTPN23 directly interacts with NAP1 via the Bro1 domain.
a NOMO-1 cell lines that constitutively express ALIX-FLAG-BirA*, PTPN23tr-FLAG-BirA*, or PTPN23fl-FLAG-BirA* used in BioID-MS experiment. b Venn diagram showing the intersection and difference of PTPN23 and ALIX interactors. Three independent experiments were performed and included only proteins detected in all three repeats. The targeted binding group was the 65-membered PTPN23tr+/PTPN23fl+/ALIX− group. c Biotinylation of various BirA* tagged recombinant proteins in NOMO-1 cells. NOMO-1 cell lines stably expressing indicated constructs were treated with 50 μM Biotin for 24 h, followed by streptavidin pulldown. Asterisks mark the self-labeling of BirA* tagged recombinant proteins. d Immunoprecipitation assays of endogenous NAP1 in NOMO-1 stable cell lines expressing indicated constructs. Asterisks mark the Flag-tagged recombinant proteins. e Co-immunoprecipitation of NAP1 and indicated PTPN23 domains or ALIX using proteins purified from E. coli. Each Immunoblots was reproduced three times with similar results for (c–e). f Immunostaining of HA-NAP1-overexpressing HeLa cells with PTPN23 (green) and HA (red) antibodies. Nuclei were counterstained with DAPI (blue). %volume of NAP1 above threshold colocalized 45.7 ± 12.4 (mean ± s.d.). Data were derived from 43 total cells captured in z-stacks across 2 independent transfection experiments; scale bar: 5 μm. g Schematic representation of NAP1 isoforms and constructs. h Co-immunoprecipitation assays of NAP1 constructs and truncated PTPN23 or PTPN23/ALIX chimeras. The plasmids expressing indicated proteins were transfected into HEK 293 T cells. Cells were harvested 48 h after transfection. (n = 3 independent experiments). i Co-immunoprecipitation assays of NAP1 isoforms constructs and TNFR1 in HEK 293 T cells. Cells were harvested 36 h after transfection. (n = 3 independent experiments).
Table 1.
Candidate PTPN23 interactome protein list
| Protein Name | Control-biotin | Control+biotin | ALIX-biotin | ALIX+biotin | PTPN23tr-biotin | PTPN23tr+biotin | PTPN23fl-biotin | PTPN23fl+biotin |
|---|---|---|---|---|---|---|---|---|
| ZFYVE9 | 0 | 0 | 0 | 0 | 3.0 | 7.3 | 2.3 | 10.2 |
| ZFYVE16 | 7.1 | 2.0 | 0 | 1.2 | 20.5 | 47.8 | 43.9 | 89.2 |
| RABEP1 | 0.8 | 0 | 0.3 | 0.3 | 2.4 | 7.4 | 0.3 | 3.3 |
| AZI2 | 0 | 0 | 0 | 0.9 | 4.5 | 12.5 | 3.3 | 10.4 |
| KIFC1 | 1.9 | 2.6 | 0.9 | 0.3 | 1.1 | 2.6 | 2.3 | 3.3 |
| CC2D1A | 0.3 | 0.0 | 0.6 | 3.4 | 5.9 | 15.9 | 13.4 | 38.7 |
| CALCOCO2 | 0.3 | 0 | 0.3 | 1.2 | 1.8 | 6.2 | 1.0 | 2.8 |
| GSR | 1.6 | 1.3 | 4.6 | 1.8 | 2.7 | 6.8 | 1.3 | 3.1 |
Identification of binding partners of PTPN23 pulled down from the cytoplasmic fraction of indicated stable cell lines by mass spectrometry analyses. The list was ranked based on the average fold change of protein enrichment in PTPN23tr and PTPN23fl-overexpressing cell lines compared to ALIX overexpressing cell line, with a cutoff value of 2. Only proteins with a fold change greater than 2 in the presence of biotin compared to its absence were included in the table.
We validated the BioID results using NOMO-1 cell lines that overexpressed various BirA* fusion proteins. We observed that both truncated and full-length PTPN23 biotinylated endogenous NAP1, which was not detected with ALIX or BirA* alone, confirming a specific interaction between NAP1 and PTPN23 (Fig. 6c). Furthermore, we observed that FLAG-tagged truncated and full-length PTPN23 co-immunoprecipitated with NAP1, whereas this was not observed with FLAG-tagged ALIX or BirA*, providing further support for a specific physical interaction between PTPN23 and NAP1 (Fig. 6d). To explore whether the interaction was direct, we used purified proteins and demonstrated that PTPN23 bound directly to the Bro1 domain of PTPN23 (Fig. 6e). Furthermore, we performed additional experiments wherein we swapped the Bro1 domain from PTPN23 to ALIX, resulting in the chimeric ALIX gaining the ability to interact with NAP1. Nonetheless, this chimeric protein’s interaction with NAP1 was less efficient than truncated PTPN23 (Supplementary Fig 6b,c). Following overexpression in Hela cells, immunostaining revealed a punctate distribution pattern of NAP1 (Fig. 6f). The partial colocalization of PTPN23 and NAP1 indicates potential differences in their protein levels or the involvement of additional regulatory mechanisms. Collectively, these data support a direct binding of NAP1 to PTPN23, suggesting a functional collaboration within distinct subcellular regions.
There are four isoforms of NAP1, with the longest isoform consisting of three coiled-coil domains and a TBK1 binding domain (TBD) (Fig. 6g). To refine further the interaction site between PTPN23 and NAP1, we evaluated their association by co-immunoprecipitation and observed interaction through the third coiled-coil domain of NAP1. Moreover, we confirmed this pattern of interaction using an ALIX chimeric protein containing the Bro1 domain from PTPN23. In contrast, no interaction was detected with a PTPN23tr chimeric protein containing the Bro1 domain from ALIX (Fig. 6h). In addition, we mapped the interaction between NAP1 and TNFR1 to the first two coiled-coil domains of NAP1 (Fig. 6i). In summary, our data indicate that the first two coiled-coil domains of NAP1 are responsible for interaction with TNFR1, and the third coiled-coil domain mediates direct binding to the Bro1 domain of PTPN23. This prompted us to hypothesize that NAP1 functions as an adapter protein to promote the sorting of TNFR1 signaling complex, and recruits PTPN23-dependent ESCRT machinery to direct TNFR1 into the endo-lysosomal pathway.
NAP1 functions as an adapter protein that modulates TNFR1 signaling
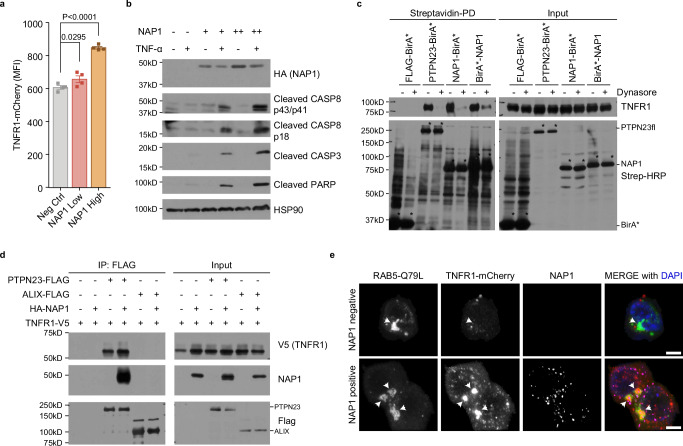
To test the hypothesis that NAP1 functions as a sorting adapter protein for TNFR1, we measured receptor levels in response to increasing amounts of NAP1. Upon transient expression of NAP1, we observed a dose-dependent increase in total TNFR1-mCherry levels (Fig. 7a, Supplementary Fig. 7a). This coincided with increased sensitivity to TNF-α-induced apoptosis, as illustrated by enhanced cleavage of CASP8, CASP3, and PARP (Fig. 7b). Additionally, depletion of NAP1 in NOMO-1 cells led to reduced activation of NF-κB and RIPK3 (Supplementary Fig. 7b), further supporting the role of NAP1 in TNFR1 signaling.
Fig. 7. NAP1 acts as an adapter protein in the regulation of TNFR1 signaling.
a Flow cytometric analysis of TNFR1-mCherry. The HEK 293 T cells stably expressing TNFR1-mCherry were transiently transfected with two doses of NAP1 constructs. Data are presented as mean ± SEM, n = 4 individual experiments. Statistics were determined by two-tailed, paired Welch’s t-test. b Immunoblotting analysis of cleaved CASP3, CASP8, and PARP in HEK 293 T cells. HEK 293 T cells were transiently transfected with increasing amounts of NAP1 plasmids. TNF-α (100 ng/ml) was added to fresh culture media 6 h post-transfection. Cells were harvested 24 h after TNF-α stimulation (n = 3 independent experiments). c Proximity labeling of TNFR1 by PTPN23-BirA* or NAP1-BirA* in the presence or absence of dynasore. HEK 293 T cells were transiently transfected with TNFR1 and Flag-BirA*, PTPN23-BirA*, NAP1-BirA*, or BirA*-NAP1. Biotin (50 μM) and dynasore (50 μM) were added into fresh media 6 h post-transfection, and incubated for 24 h. Asterisks mark the self-labeling of BirA* tagged recombinant proteins. d Co-immunoprecipitation of TNFR1 and NAP1 by FLAG-tagged PTPN23 or ALIX in HEK 293 T cells. Each immunoblot was reproduced three times with similar results for (c, d). e Representative confocal images of TNFR1-mCherry and eGFP-RAB5-Q79L in the presence or absence of NAP1. The HEK 293 T stably expressing TNFR1-mCherry cell line was transiently transfected with RAB5-Q79L and NAP1 plasmids. Cells were subjected to immunofluorescence staining 24 h after transfection. Scale bar: 5 μm.
The observed accumulation of TNFR1 indicated a delay in its degradation, which could arise from either impaired internalization at the plasma membrane or a subsequent defect in cargo sorting to the endosome and lysosome. To distinguish between these two possibilities, we transiently co-expressed BirA*-tagged PTPN23 or NAP1 constructs with TNFR1 in HEK 293 T cells. Consistent with immunoprecipitation assays, PTPN23 and NAP1 fusion proteins, but not BirA* alone, biotinylated TNFR1. Furthermore, treatment with dynasore68, to inhibit dynamin-mediated endocytosis resulted in a loss of TNFR1 biotinylation by PTPN23-BirA* and NAP1-BirA* fusion proteins, suggesting that NAP1 and PTPN23 interacted with TNFR1 downstream of endocytic internalization (Fig. 7c).
To investigate the effect of NAP1 on TNFR1 degradation, we co-expressed TNFR1 and FLAG-tagged PTPN23 or ALIX in HEK 293 T cells, both in the presence and absence of HA-tagged NAP1. Surprisingly, overexpression of HA-NAP1 led to an increase of TNFR1 co-immunoprecipitated with PTPN23 (Fig. 7d), indicating that NAP1 was unlikely to impair ESCRT-mediated degradation. To explore the potential role of NAP1 in the endosomal sorting of TNFR1, we used a constitutively active Rab5-Q79L mutant, which enhances the homotypic fusion of early endosomes to form enlarged endosomes, to allow for improved visualization69. In NAP1-overexpressing cells, although NAP1 displayed a different cellular distribution compared to Rab5-Q79L, TNFR1 was found highly enriched in large, ring-like, Rab5-Q79L-positive early endosomal structures, suggesting that NAP1 may promote the endosomal sorting of TNFR1 (Fig. 7e and Supplementary 7c).
PTPN23 is crucial for the survival of bone marrow-derived macrophages
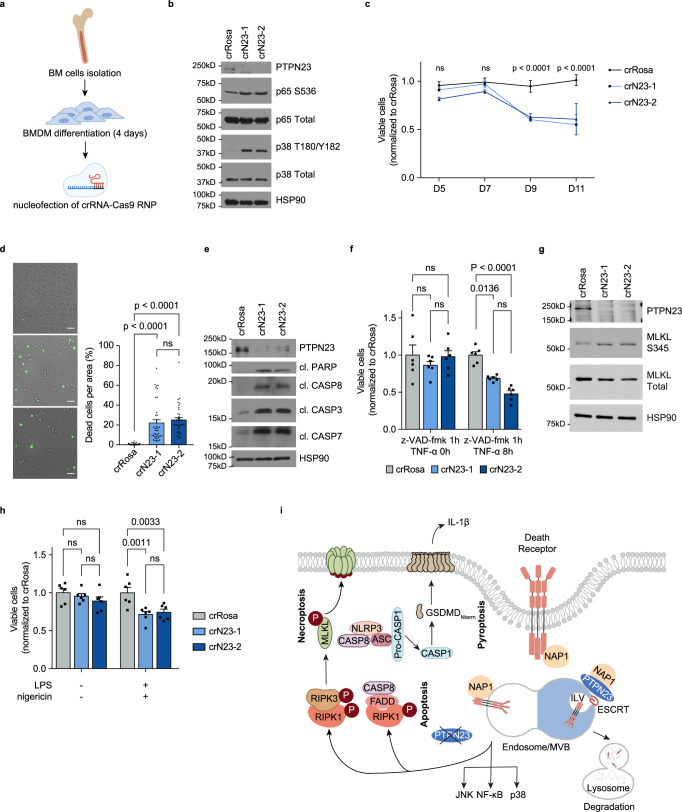
Considering the lethality of Ptpn23 homozygous deletion in a mouse model35, we sought to investigate the role of PTPN23 in normal cell survival using bone marrow-derived macrophages (BMDMs) to demonstrate effects beyond cancer cells. We induced the differentiation of BMDMs from hematopoietic progenitors by culture of total bone marrow in the presence of macrophage colony-stimulating factor (M-CSF). Following 4 days of differentiation, we utilized Cas9-ribonuclear protein (RNP) electroporation (nucleofection) to disrupt PTPN23 (Fig. 8a). Consistent with our observations in AML cells, depletion of PTPN23 in mouse BMDMs led to activation of NF-κB and MAPK signaling, suggesting the activation of death receptors (Fig. 8b). Prolonged culture of PTPN23-depleted BMDMs for 11 days revealed a significant decline in viable cells after day 9, as assessed by the CTG assay (Fig. 8c). Additional staining with Sytox Green after 9 days of nucleofection indicated an increased presence of lytic cell death (Fig. 8d).
Fig. 8. Impact of PTPN23 depletion on multiple cell death modalities in BMDM cells.
a Workflow for crRNA-Cas9-RNP-mediated knockout of Ptpn23 in mouse BMDM cells. The CRISPR RNAs (crRNAs) targeting Ptpn23 were consistent with those employed in the mouse AML validation study. Created with Adobe Illustrator and BioRender.com (License reference: https://BioRender.com/p57w564). b Immunoblotting analysis of NF-κB and MAPK pathway in BMDM cells harvested 6 days post-nucleofection with crRosa, crN23-1, or crN23-2 (n = 3 mice). c CTG luminescence cell viability assay in WT or Ptpn23 knockout BMDM cells (n = 5 mice). Statistical analysis was conducted via two-way ANOVA followed by Tukey’s multiple comparisons test. d Left, Cell death measured by Sytox Green staining in cells cultured for 9 days post-nucleofection (n = 40 fields from 4 independent experiments). Scale bar: 40 μm. Right, Quantification of Sytox Green positivity. Data are mean ± SEM, analyzed using one-way ANOVA followed by Tukey’s test. e Immunoblot of apoptosis markers in BMDMs treated with 50 ng/ml TNF-α for 4 h, 6-7 days post-nucleofection (n = 3 mice). f CTG assay in WT and Ptpn23-knockout BMDMs pretreated with 40 μM z-VAD-fmk for 1 h, followed by treatment with 1 μg/ml TNF-α for the indicated time, 6 days post-nucleofection. g Immunoblot of MLKL in BMDMs 7 days post-nucleofection with crRosa or crN23, treated with 40 μM z-VAD-fmk for 1 h, followed by 1 μg/ml TNF-α for 4 h (n = 2 independent experiments, each using cells from three mice). Immunoblots in (b, e, and g) showed consistent results across three, three, and two independent experiments, respectively, with representative blots presented. h CTG assay in WT and Ptpn23-knockout BMDMs primed with 100 ng/ml LPS for 4 h, followed by 2.5 μM nigericin for 4 h, 6 days post-nucleofection. Data for (f, h) are mean ± SEM from six replicates derived from three mice, analyzed using two-way ANOVA followed by Tukey’s multiple comparisons tests. i Proposed model: PTPN23 collaborates with NAP1 to facilitate receptor sorting. On the right-hand side, representing the normal condition, this process involves the engagement of ESCRT, leading to the lysosomal degradation of death receptors. On the left-hand side, loss of PTPN23 results in the prolonged accumulation of death receptors within the endosomal compartment, triggering the activation of multiple cell death pathways. Created with Adobe Illustrator and BioRender.com (License reference: https://BioRender.com/x06g968).
To elucidate the modality of cell death, we induced apoptosis, necroptosis, or pyroptosis in BMDMs that were nucleofected with Ptpn23 CRISPR RNAs at 6 or 7 days, the time points when significant cell death was not observed in controls. Compared to the Rosa control, PTPN23-depleted cells exhibited increased sensitivity to TNF-α treatment, and displayed elevated markers for apoptosis, including cleaved PARP, CASP8, CASP3, and CASP7 (Fig. 8e). Upon treatment with z-VAD-fmk and TNF-α, increased cell death and phosphorylation of MLKL was observed, suggesting that PTPN23 loss sensitized BMDMs to necroptosis (Fig. 8f, g). Moreover, LPS priming followed by nigericin treatment significantly increased cell death in PTPN23-deficient BMDMs, indicating an enhanced sensitivity to pyroptotic stimuli (Fig. 8h). In summary, our study underscores that PTPN23 plays a pivotal role in restraining multiple cell death modalities in primary myeloid cells, thereby indicating its broader relevance beyond transformed cancer cell contexts.
Discussion
The proper functioning of the PTPN23-dependent ESCRT machinery is essential for preventing the accumulation of death receptors and TLRs in the endosomal compartments. Our study supports a model in which PTPN23 functions in concert with NAP1, and potentially other sorting adapters, to facilitate receptor sorting. This process is followed by engagement with the ESCRT complex to promote the degradation of death receptors and TLRs. Loss of PTPN23 leads to the accrual of death receptors and TLRs in the endosomal compartment, culminating in ligand-independent activation and extended duration of activation, ultimately triggering apoptotic, necroptotic, and pyroptotic pathways (Fig. 8i). These findings suggest that PTPN23-dependent ESCRT machinery serves as an intrinsic cell death checkpoint, highlighting the multifaceted nature of cell death and the critical role of the spatiotemporal distribution of death receptors and TLRs in modulating signal output.
Previous research has indicated that the selective transmission of signals leading to cell survival or death is regulated by death receptor internalization and endosomal trafficking70,71. Upon ligand binding at the plasma membrane, death receptors activate the NF-κB and MAPK pathways72. Following this activation, receptors are internalized and transported to the endosomal compartment, which is necessary to induce cell death signaling14,73,74. However, our study reveals that the spatiotemporal distribution of receptors is crucial for the organization of signals derived from various death receptors and TLRs, which collectively activate NF-κB, apoptosis, necroptosis, and pyroptosis. The confinement of receptors to endosomal membranes may contribute to the formation of receptor clusters to activate downstream signaling75–77. Therefore, our findings provide insights into a spatiotemporal regulatory mechanism, whereby PTPN23-dependent ESCRT machinery controls the amount of death receptors and TLRs over time to modulate the duration and amplification of NF-κB and MAPK signaling, and the sensitivity threshold to cell death. Furthermore, the endosomal compartmentalization of death receptors and TLRs resulting from the disruption of the ESCRT pathway tilts the balance towards cell death pathways and away from pro-survival NF-κB signaling.
An intriguing aspect of our study is the fact that these effects of PTPN23 are independent of its intrinsic catalytic activity. We conducted a comprehensive characterization of domain truncation and domain-swapping constructs to elucidate the importance of the non-enzymatic domains of PTPN23 in its essential function. Our results indicate that the V domain is critical for the interaction with UBAP1 to engage ESCRT machinery. This is consistent with the requirement for UBAP1 to prevent endosomal accumulation of cytokine receptors and to limit downstream NF-κB signaling49. It also provides a mechanism through which PTPN23 and ALIX execute non-redundant functions. PTPN23 and ALIX share some common interactors; however, only the V domain of PTPN23 associated with UBAP152. In addition, we demonstrate that TNFR1 sorting adapter protein NAP1 binds directly to the Bro1 domain of PTPN23 but not the Bro1 domain of ALIX, underscoring the distinct roles of PTPN23 and ALIX in the TNFR1 pathway. Ubiquitination has been suggested to serve as a signal for the recognition of substrates targeted for degradation78. In the present study, we propose that interactions between adapter proteins, such as NAP1, and receptors may further contribute to receptor sorting and degradation. Collectively, these findings underlie the biochemical basis for the non-redundant function of PTPN23 and ALIX in promoting cargo lysosomal degradation. Specifically, the Bro1 domain of PTPN23 interacts with NAP1 and potential additional adapters to regulate the endosomal sorting of death receptors and TLRs, which is followed by engagement of ESCRT machinery via the V domain to promote receptor lysosomal degradation.
Our study demonstrates the co-dependency between PTPN23 and multiple subunits in the ESCRT pathway, including endosomal DUB, Clathrin heavy chain, ESCRT-0, and -I. While the late-acting ESCRT-III subcomplex is well-known for its role in cell survival through facilitating cytokinetic abscission79 and repairing various membrane damage70,80–84, the role of early-acting ESCRT (ESCRT-0 and -I) components in regulating cell death is not fully understood. Our findings show that cells that are deficient in PTPN23-co-dependent ESCRT components exhibit impaired cell survival, accumulation of death receptors and TLRs, and transcriptional program changes. These observations suggest that early-acting ESCRT components contribute to the maintenance of cell survival through a mechanism distinct from that of late-acting ESCRT components, thereby expanding the functional repertoire of the evolutionarily conserved ESCRT pathway. Importantly, our study sheds light on the involvement of PTPN23 in early-acting ESCRT, providing insights into its function within this regulatory network.
We show that loss of PTPN23 results in the activation of CASP3/7/8 and CASP1, phosphorylation of RIPK3 and MLKL, formation of inflammasomes, and recruitment of CASP8 into inflammasomes. These observations indicate that cells coordinate signals emitted from various death receptors and/or TLRs to control cell death modality. The PANoptosome is an emerging concept that features the assembly of key molecules from pyroptosis, apoptosis, and necroptosis (PANoptosis) into a single complex through protein-protein interaction2,8,15,85–87; however, it remains unclear whether the absence of PTPN23 induces all three cell death pathways concurrently, or whether mediators released from dying cells trigger a different cell death pathway in surrounding cells. Notably, necroptosis and pyroptosis are proinflammatory forms of cell death, which may potentiate anti-tumor immunity within the tumor microenvironment88–91. In this context, cell death is not the ultimate outcome, but rather the initiation of an immune response17. The PTPN23 gene is located in the human 3p21.3 chromosomal region, a tumor suppressor cluster that frequently exhibits hemizygous loss in kidney, lung, breast, and cervical tumors27,28,30. Cancer cells with deletion of a single allele of PTPN23 may be reliant on the retained allele for survival and could represent a cancer-specific vulnerability to PTPN23 inhibition. Future studies will further investigate whether interference with PTPN23 or ESCRT components has therapeutic benefits.
Methods
Mice
All mouse experiments were conducted under protocol 21-18-15-6 approved by Cold Spring Harbor Laboratory Institutional Animal Care and Use Committee (IACUC). Wild-type C57BL/6 J mice were obtained from Jackson Laboratory. All mice were aged 8–12 weeks and acclimated for at least 1 week in the mouse facility.
Cell culture and cell lines
NIH 3T3, NOMO-1, MV4-11, Kasumi-1, HEK 293 T, and HeLa cells were obtained from the American Type Culture Collection (ATCC). Platinum-E (Plat-E) was purchased from Cell Biolabs, Inc. The generation of cancer cell lines with Cas9 was performed as previously described25,92. NOMO-1, MV4-11, and murine RN2 (MLL-AF9/NrasG12D AML) cells26 were cultured in RPMI 1640 supplemented with 10% FBS. KASUMI-1 cells were cultured in RPMI 1640 supplemented with 20% FBS. HEK 293 T and HeLa cells were maintained in DMEM with 10% FBS. Murine NIH 3T3 cells were cultured in DMEM supplemented with 10% FCS. Plat-E cells were maintained in DMEM supplemented with 10% FBS, 1 μg/mL puromycin, and 10 μg/mL blasticidin. 100 U/ml penicillin/streptomycin was added to all culture media. Cells were grown in a 37 °C humidified incubator with 5% CO2, and experiments were conducted within 1–2 months after thawing cryopreserved vials. All cell lines were subjected to be tested for mycoplasma using a MycoAlert Mycoplasma Detection kit (Lonza).
Transfection and viral infection of cells
For lentivirus generation, HEK 293 T cells were seeded 16–24 h before transfection in a 10 cm plate and grown into 90–100% confluency. 10 μg of indicated lentiviral constructs, the envelope plasmid 5 μg VSV-G, and packaging plasmid 7.5 μg psPAX2 were transfected by polyethylenimine (PEI). For retrovirus generation, Plat-E cells were seeded into a 10 cm plate 16–24 h prior to transfection. 10 μg of indicated retroviral constructs, 1.66 μg pCL-Eco, and 1.33 μg VSV-G were mixed with PEI to transfect cells. Cell culture media were replaced with fresh media after 6 h of transfection. Viral supernatants were harvested every 12 h 2–3 times, pooled together, and passed through a 0.45 μm filter. Lentivirus-containing media were added to targeted cells in the presence of polybrene (8 μg/ml) before cells were subjected to spin infection (2000 rpm, 40 min, RT). Infected cells were either selected with antibiotics based on the selection marker on the plasmids (5–10 μg/ml Puromycin, 400 μg/ml G418, 200 μg/ml Hygromycin B, or 10 μg/ml Blasticidin) or sorted using FACS. The single guide RNA sequences used in this study are presented in Supplementary Data 4.
Construction of domain-focused sgRNA library and pooled CRISPR screen
The list of murine protein tyrosine phosphatases was generated by searching mouse orthologs of PTPs in the human genome93. The catalytic domain information was retrieved from the NCBI GenBank database with annotation of either protein tyrosine phosphatase or dual-specificity phosphatase. The exon sequences corresponding to PTP catalytic domains were then used to design sgRNAs, and ~6 different sgRNAs with low off-target effects were designed94. The pooled library containing phosphatase domain targeting and control sgRNAs (Supplementary Data 1) was synthesized in duplicates or triplicates on an array platform (Twist Bioscience) and then cloned into the LRG2.1 vector25 with Gibson Assembly kit (NEB).
The pooled sgRNA library was used to produce lentivirus, and the multiplicity of infection (MOI) was set to ~0.3 for infection of RN2-Cas9+ and NIH-3T3-Cas9+. The number of infected cells was over 1000 times of the number of sgRNAs in the library to ensure enough representation of each sgRNAs. Cells were harvested on day 2 post-infection and day 14 post-infection and cell pellets were stored in a −80 °C freezer. Genomic DNA was extracted using QIAamp DNA mini kit (QIAGEN, 51304) following the manufacturer’s instructions.
Genomic DNA fragment containing sgRNA insertion was amplified by PCR using primer pair: TCTTGTGGAAAGGACGAAACACCG and TCTACTATTCTTTCCCCTGCACTGT. The PCR products were subjected to end-repair to generate blunt ends using T4 DNA polymerase (NEB M0203L), DNA polymerase I (NEB M0210L) and T4 polynucleotide kinase (NEB M0201L). The 3′ A-overhang was added to the end-repaired DNA using Klenow Fragment (3′→ 5′ exo-) (NEB M0212L). The diversity-increased custom barcodes were ligated with T4 DNA ligase (NEB Quick ligation kit M2200L) and followed by pre-capture PCR. Barcoded libraries were sequenced using the MiSeq instrument (Illumina) with MiSeq Reagent Kit v2 (Illumina MS-102-2002)25.
Pair-end reads were de-multiplexed and mapped to reference sgRNA library, with zero mismatch allowed. The read counts for each sgRNA were calculated and normalized to total read counts. The average log2(fold change) for each gene was calculated by averaging the log2(sgRNA abundance in the day 2 post-infection/ sgRNA abundance in the day 14 post-infection) of all the sgRNAs targeting a given gene (Supplementary Data 2).
GFP competition growth assay
Lentiviral vector co-expressing sgRNA and an eGFP reporter were used to transduce cells. The titer of lentivirus was determined to achieve a 30–60% GFP-positive population at D2 or D3. The GFP percentages were measured every 2 days for murine AML RN2 cells, and every 3 days for all the human AML cells using Guava easyCyte Flow Cytometer (Millipore).
Immunoblotting
Cells were washed with cold PBS and lysed in RIPA buffer, with freshly added cOmplete EDTA-free protease inhibitors (Roche, 11836153001) and Halt phosphatase inhibitors (Thermo Fisher Scientific, 78428). Equally loaded proteins were separated using 8–12% of protoGel (National Diagnostics, EC-890) with Tris-glycine-SDS running buffer. Proteins were transferred onto 0.2 μm Amersham nitrocellulose membranes (Cytiva). Membranes were blocked in 5% BSA in Tris-buffered saline Tween-20. Proteins were detected using antibodies as indicated. Details of the antibodies used for immunoblotting are provided in Supplementary Data 5.
Degradation of PTPN23 protein using the dTAG system
The full-length PTPN23 gene was fused with degrader FKBPF36V (dTAG) at C-terminus, and then chimeric protein was exogenously expressed in the NOMO-1 cells. Next, the endogenous PTPN23 alleles were disrupted with two distinct gRNAs, N23-sg1 and N23-sg2. Cell lines were established by single colony expansion from sgNEG, N23-sg1, and N23-sg2 infected cells, with criteria that dTAG-13 (100 nM) can induce recombinant protein degradation within 90 min, and endogenous PTPN23 was completely abolished (except for sgNEG control).
CellTiter-Glo luminescence cell viability assay
Cells were seeded at 10,000–50,000 cells per well in duplicates into 96-well plates, and treated as indicated. CellTiter-Glo 2.0 reagents (Promega, G9242) were subsequently added to plates according to the manufacturer’s instructions. Luminescence was measured using a microplate reader, and the viability was calculated via normalization to the negative control cells.
Sytox green staining
Negative control gRNAs and two PTPN23 sgRNAs infected PTPN23-dTAG overexpressed NOMO-1 cells were treated with dTAG-13 (200 nM) for 4 days. Cells were pelleted and then suspended in RPMI at a concentration of 500,000 cells/ml. Cells were stained with 2.5 μM cell-impermeable dye, Sytox Green nucleic acid stain (Thermo Fisher Scientific, S7020), and incubated for 30 min at room temperature. 50,000 cells were plated onto a poly-L-lysine coated 8-well μ-slide (Ibidi). The Sytox Green staining was imaged using a Spinning disk confocal fluorescence microscope, and GFP and bright field channels were used to capture Sytox Green positive cells and total cells. Images were acquired within 2 h of staining, during which Sytox Green treatment did not affect cell viability. The percentage of dead cells was determined by manually counting the positively stained cells/total cells.
RNA sequencing and data analysis
Total RNAs were isolated and purified with TRIzol reagent (Invitrogen) following the manufacturer’s protocol. A cDNA library was prepared with the Illumina TruSeq sample prep kit v2 following the manufacturer’s protocol with 1 μg total RNA. The library was sequenced on an Illumina NextSeq with single-end 76 bp reads. Sequencing reads were mapped to the human reference genome (GRCh38/hg38) using Hisat2 2.1.0 with default settings. Differentially expressed genes were analyzed using counts from HTSeq-count (version 0.13.5). Genes with at least 10 reads were then fed into DESeq2 with masking structural RNAs and non-coding RNAs to calculate fold changes. For pre-ranked gene set enrichment analysis (GSEA), genes were ranked by their log2(fold-change) normalized to negative controls. RNA-seq data have been deposited in GEO under accession number GSE272123.
Caspase-3/7 flow cytometry assay
PTPN23-dTAG cells were treated with 200 nM dTAG-13 for 4 days. Cells were harvested, and the concentration of each sample was adjusted to 5 × 106 cells/mL in RPMI medium. The staining was performed according to the manufacturer’s instructions.
IL−1β ELISA
PTPN23-dTAG cells were treated with dTAG-13 for 4 days and then were seeded into a 96-well plate. For 24 h treatment, cells were seeded at a density of 200,000 per well in the presence of 200 nM dTAG-13, for 48 h treatment, cells were seeded at a density of 100,000 per well in the presence of 200 nM dTAG-13. The plate was centrifugated, and the supernatants were subject to IL-1β ELISA kit (R & D Systems, DLB50) detection.
Confocal immunofluorescent microscopy
For NOMO-1 cells, cells were harvested and fixed in 4% PFA on ice for 10 min. Cells were then washed with cold PBS, blocked, and permeabilized in 5% donkey serum, and 0.1% Triton X-100 in PBS for 1 h at room temperature. The primary antibodies indicated in figure legends were used to stain overnight at 4 °C. Next day, cells were washed with PBS, incubated with secondary antibodies and DAPI (Abcam, ab228549) for 1 h at room temperature. Slides were washed with PBS and filled with ibidi Mounting Medium (ibidi, 50001) before imaging.
Confocal microscopy and image analysis
Images were acquired using a Zeiss LSM 780 laser scanning confocal microscope. 3D z-stacks were recorded with 63x/1.4NA Plan-Apochromat oil immersion objective lens, full cell depth with optimized z interval step size were applied, and all 3D z-stacks were recorded with the same laser power and detector gain settings. The image stacks were then imported into Imaris 3D/4D Visualization and Analysis Software (Version 9.9.1, Oxford Instrument) for volumetric reconstruction and colocalization analysis. The Surface function in Imaris was used to create a 3D surface around NAP1, TNFR1, and RAB5 signals. The surface threshold was set using manual thresholding to identify two classes of voxels with or without NAP1. The 3D object-based colocalization was processed with the same intensity thresholding parameters to ensure consistency in intensity data measurements both within and between datasets. The Spot function in Imaris was used to measure the shortest distance from CHMP4B, PTPN23 or UBAP1 to VPS4A-E228Q. We applied the same thresholding parameters and distance filters to identify and distinguish CHMP4B, PTPN23 or UBAP1 to VPS4A-E228Q populations.
Immunoprecipitation (IP)
For examination of the interaction between proteins, cells were transfected with responding constructs, and cultured for 24 h to allow protein overexpression. Cells were then washed with cold PBS, and lysed in IP buffer (1% NP-40, 150 mM NaCl, 20 mM HEPES, pH 7.5, 1 mM EDTA, freshly supplemented with protease inhibitor) at 4 °C for 1 h. Subsequently, the lysates were centrifuged at 13,500 rpm for 20 min and the Flag-tagged proteins were immunoprecipitated using Anti-FLAG M2 Magnetic Beads (M8823, Sigma) overnight at 4 °C. The following day, the beads were washed three times with IP buffer, and proteins were eluted by boiling in 2X SDS loading buffer. Samples were analyzed by Immunoblotting.
Flow cytometry
For ligand-independent receptor degradation examination, NOMO-1 cells overexpressing PTPN23-dTAG were treated with dTAG-13 for indicated days. 2,000,000 cells were collected, and fixed with 4% PFA on ice for 8 min. Cells were washed once with cold PBS and then blocked and permeabilized with 5% donkey serum, and 0.1% Triton X-100 in PBS for 1 h. Cells were then transferred into a U-bottom 96-well plate, and incubated with 100 μl primary antibodies for overnight. On the second day, cells were washed with cold PBS, incubated with appropriate secondary antibodies, or fluorophore-conjugated primary antibodies for 1 h at room temperature and then subjected to flow cytometry analysis.
For TNFα-dependent receptor degradation assessment, NOMO-1 cells overexpressing PTPN23-dTAG were treated with dTAG-13 for 2 days to deplete PTPN23. Cells were added with 100 ng/ml TNFα and incubated in the cold room for 2 h with rotation. Cells were then transferred into a 37 °C incubator for indicated time to allow receptor internalization and degradation. Subsequently, cells were washed with cold PBS and centrifuged at 4 °C to remove all the media, and followed by flow cytometry sample preparation as described in ligand-independent. Details of the antibodies used for confocal imaging or flow cytometry are provided in Supplementary Data 5.
Proximity-dependent biotin identification (BioID)
ALIX-BirA*, N23tr-BirA*, and N23fl-BirA* were stably overexpressed in NOMO-1 cells. Three independent experiments were conducted for each condition. Cells in each 150 mm plate were pretreated with 78 μl BioLock biotin-blocking solution (IBA, 2-0205-050) for 48 h. Cells in the experimental groups were then treated with 50 μM biotin diluted in RPMI-1640 with 10% FBS for 24 h. At the end of labeling, cells were washed 4 times with cold PBS to remove free biotin. Cells were lysed with Lysis Buffer (LB) [50 mM HEPES-KOH, (pH 7.5), 140 mM NaCl, 1 mM EDTA, 10% (vol/vol) glycerol, 0.5% (vol/vol) NP-40, 0.25% (vol/vol) Triton X-100, 1X freshly added cOmpleteTM EDTA-free protease inhibitors (Roche, 11836153001)]. Cell lysates were rotated in the Protein LoBind Tubes (Eppendorf, 0030108116) at 4 °C for 15 min, and clarified by centrifugation at 2000 × g for 5 min at 4 °C. The supernatants were collected into new tubes, and Triton X-100 was added to reach a final concentration of 0.5%. The cytoplasmic fractions of cell lysates were rotated at 4 °C for 15 min, and clarified by centrifugation at 20,000 × g for 20 min. The supernatants were transferred to new tubes and added Dynabeads™ MyOne™ Streptavidin C1 (ThermoFisher, Cat #65001). After overnight rotation at 4 °C, the beads were washed once with SDS-Wash buffer (25 mM Tris-HCl, pH 7.4, 2% SDS), twice with RIPA (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP40, 0.1% SDS, 0.4% sodium deoxycholate), once with TNNE buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% NP40), and three times with 50 mM ammonium bicarbonate (ABC buffer), pH 8.0.
Trypsin digestion
Beads were resuspended in 70 μl of 50 mM ABC buffer with 1 μg trypsin (Promega, V511A) added and digested at 37 °C overnight with agitation. On the next day, an additional 0.5 μg of trypsin was added to each sample, and incubated for 4 h at 37 °C with agitation. Supernatants were collected into a new tube, beads were washed twice with mass spectrometry grade water, and supernatants were pooled with the peptide supernatants.
Mass spectrometry
Peptides were dried using vacuum centrifugation and reconstituted in 30 μl of 2% acetonitrile: 0.1% formic acid, and then injected onto a self-pack PicoFritTM 75 μm analytical column with an 8 μm emitter (New Objective, Woburn, MA) packed to 25 cm with ReproSil-Pur C18-AQ, 1.9 μM material (Dr. Maish GmbH). A total of 1 μl of the resulting peptidic solution was used for liquid chromatography (LC)-MS/MS analysis. LC-MS/MS was performed using an Orbitrap Fusion Lumos mass spectrometer (Thermo Scientific) coupled to an EASY-nLC 1200 system (Thermo Scientific). Peptides were separated using a changing ratio of solvent A (3% ACN/0.1% FA) to solvent B (90% ACN/0.1% FA) with the following steps at a flow rate of 200 nL/min: 2% B to 6% B over 1 min, 6% B to 30% B over 84 min, 30% B to 60% B over 9 min, 60% B to 90% B over 1 min, held at 90% B for 5 min, 90% B to 50% B over 1 min and then flow rate was increased to 500 nL/min as 50% B was held for 9 min.
Full-scan mass spectrum (Res = 60,000; 400–1600 m/z) was followed by MS/MS using the “Top Speed” method for selection. High-energy collisional dissociation (HCD) was used with the normalized collision energy set to 35 for fragmentation, the isolation width set to 1.2 and a duration of 10 s set for the dynamic exclusion with an exclusion mass width of 10 ppm.
Raw data were analyzed using Mascot version 2.6.2, where they were searched against the human UniProt database (20181009) with the following settings: Fragment Tolerance: 0.30 Da (Monoisotopic); Parent Tolerance: 20 PPM (Monoisotopic); Variable Modifications: +16 on M (Oxidation); Digestion Enzyme: stricttrypsin; Max Missed Cleavages: 2. Label-free quantification was used to quantify the MS data. The data were analyzed using Scaffold version 4.8.9, with the following settings: Modification Metadata Set: 2334 modifications; Protein Grouping Strategy: Experiment-wide grouping with protein cluster analysis; Peptide Thresholds: 69.0% minimum; Protein Thresholds: 13.0% minimum and 2 peptides minimum. To ensure high-confidence identifications, peptides were filtered at 0.1% (Decoy) and proteins were filtered at 0.7% (Decoy) false discovery rate. Proteins identified with less than two peptides were filtered out. Quantitative values were normalized to total spectra, and data are provided in Supplementary Data 3.
Protein purification
The His-tagged Bro1, V domain of PTPN23, PTPN23tr, ALIX, and NAP1 were cloned into the pET21b vector, and purified from BL21 E. coli culture. Cultures were incubated at 37 °C with shaking at 180 rpm until OD (600 nm) reached 0.7. The flasks were then cooled down to 18 °C and 0.5 mM IPTG (Roche, 11411446001) was added to induce protein expression overnight. Cells were harvested by centrifugation at 3500 rpm at 4 °C for 15 min.
Cell pellets were resuspended in Binding buffer (50 mM HEPES, pH 7.5; 500 mM NaCl; 5% Glycerol; 5 mM Imidazole; supplemented with 1X cOmpleteTM EDTA-free protease inhibitors). Samples were subsequently sonicated on ice for 2 min with 5 s on and 5 s off at 20–40% amplitude, and cleared by ultracentrifugation (35,000 × g for 1 h at 4 °C). The supernatants were mixed with Ni-NTA beads (Cytiva, 17531806), and loaded onto gravity columns. Beads were washed with 10 column volume (CV) of wash buffer 1 (50 mM HEPES, pH 7.5; 500 mM NaCl; 5% Glycerol; 30 mM Imidazole), followed by 10 CV of wash buffer 2 (50 mM HEPES, pH 7.5; 500 mM NaCl; 5% Glycerol; 50 mM Imidazole), followed by 10 CV of wash buffer 3 (50 mM HEPES, pH 7.5; 500 mM NaCl; 5% Glycerol; 70 mM Imidazole). Proteins were eluted with 5 CV of Elution buffer (50 mM HEPES, pH 7.5; 500 mM NaCl; 5% Glycerol; 250 mM Imidazole). An equal volume of glycerol was added to each protein eluent, flash frozen in liquid N2 and proteins were stored at −80 °C.
Bone marrow-derived macrophage (BMDM) cultures
Bone marrow cells were aseptically flushed from the femurs and tibias of mice. The cells were then induced to differentiate into BMDMs through culturing in RPMI-1640 medium, supplemented with 20% heat-inactivated FBS, 2 mM GlutaMAX (35050061, Gibco), 1 mM Sodium Pyruvate (11360070, Gibco), and 1% penicillin and streptomycin. Additionally, freshly added components included 0.055 mM β-Mercaptoethanol, and 10 ng/ml M-CSF (Prospec CYT-439). BMDMs were seeded at a density of 2 × 106 cells/well in non-tissue culture-treated 6 well plates.
Gene editing via nucleofection of Cas9-RNP in mouse BMDMs
Negative control CRISPR RNA (crRNA) and two distinct crRNAs against mouse PTPN23 were synthesized by Integrated DNA Technologies (IDT). The crRNAs were annealed to transactivating crRNA (tracrRNA) in equimolar concentration using a PCR machine with gradual cooling steps: 95 °C, 5 min; 90 °C, 1 min; 85 °C, 1 min; 55 °C, 5 min; 25 °C, 10 min. To prepare Cas9-RNP complex, the annealed crRNA/tracrRNA duplexes were mixed with Cas9 (1081059, IDT, Alt-R™ S.p. Cas9 Nuclease V3) at a 3:1 molar ratio. The mixture was incubated at room temperature for 20 min. Meanwhile, BMDMs were harvested after four days of cell culture using Detachin (T100100, Genlantis) to release them from plates. Following a PBS wash and cell counting, 106 cells per reaction were resuspended in 20 μl of P3 primary nucleofection solution (V4XP-3032, P3 Primary Cell 4D-Nucleofector™ X Kit S, Lonza). The cells/P3 nucleofection solution was then combined with each Cas9-RNP complex, gently pipetted to ensure thorough mixing without introducing bubbles. The cell/RNP mix was immediately loaded into the nucleofector cassette strip and inserted into the Lonza 4D-Nucleofector. Nucleofection was performed with Buffer P3, CM-137 condition. Following nucleofection, the cassette strip was removed, and 180 μl of prewarmed medium was added to each cassette well. The medium/cell/RNP mix was transferred to non-tissue culture-treated 6-well plates, and the cells were cultured according to the aforementioned conditions95.
Statistical analysis
All data are presented as mean ± standard error of the mean (SEM), and statistical analyses were performed with Prism v7 software (GraphPad Software). Comparisons between two groups were performed using unpaired or paired two-tailed t-tests as indicated in figure legends. Comparisons for more than two groups were analyzed using one-way or two-way ANOVA followed by a post-doc test as indicated in figure legends.
Reporting summary
Further information is available in the Nature Portfolio Reporting Summary linked to this article.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We express our gratitude to Dr. Christopher Bonham for his insightful discussions and suggestions throughout the course of this study. Special thanks to Xiao Han for her dedicated assistance in the collection and culture of BMDM, and to Dr. Mikala Egeblad for her support and guidance. N.K.T. is the Caryl Boies Professor of Cancer Research at Cold Spring Harbor Laboratory. Research in the Tonks lab was supported by NIH grant CA53840, the CSHL Cancer Centre Support Grant CA45508, the Robertson Research Fund of CSHL, the Don Monti Memorial Research Foundation, the Hansen Foundation, and the Simons Foundation. Additional funding was provided to C.R.V. by the Pershing Square Sohn Cancer Research Alliance, National Institutes of Health grants CA013106 and CA242919, Department of Defense grant W81XWH1910317, and the Cold Spring Harbor Laboratory and Northwell Health Affiliation. These studies utilized the Functional Genomics, Next Generation Sequencing, Flow Cytometry, Mass Spectrometry, and Microscopy Shared Resources of the CSHL Cancer Center. Figures 3d, 8a, 8i in this manuscript were created using BioRender.com and Adobe Illustrator.
Author contributions
Conceptualization, D.S. and N.K.T.; investigation, D.S., Y.C., Z.Q., X.S.W., and K.R.; resources, K.C.; formal analysis, D.S., X.S.W., K.R., T.W., O.E.D., and D.P.; writing-original draft, D.S. and N.K.T.; writing-review & editing, Y.C., Z.Q., X.S.W., K.R., T.W., O.E.D., K.C., D.P., and C.R.V.; supervision, D.P., C.R.V., and N.K.T.; funding acquisition, C.R.V., and N.K.T.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The FASTQ files generated in CRISPR screen have been deposited in the Sequence Read Archive (SRA) database under accession number PRJNA1187288. The raw RNA-seq data generated in this study have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE272123. The raw mass spectrometry data have been deposited to the ProteomeXchange Consortium via the iProX repository96,97 with the dataset identifier PXD056836. The sgRNA sequences used in the CRISPR screen are provided in Supplementary Data 1, and sgRNA sequences used in the validation study are provided in Supplementary Data 4. Supplementary Data 2 contains the average log2 frequency changes of sgRNAs targeting the same gene at Days 2 and 14 post-sgRNA transduction. Proteomic data analyzed using Scaffold software are available in Supplementary Data 3. Supplementary Data 5 provides a list of antibodies used for immunoblotting, confocal imaging, and flow cytometry. All other relevant data are included within the main article, Supplementary Information, or Source Data files. Source data are provided with this paper.
Competing interests
N.K.T. is a member of the Scientific Advisory Board of DepYmed Inc. and Anavo Therapeutics. C.R.V. has received consulting fees from Flare Therapeutics, Roivant Sciences, and C4 Therapeutics; has served on the advisory boards of KSQ Therapeutics, Syros Pharmaceuticals, and Treeline Biosciences; has received research funding from Boehringer-Ingelheim and Treeline Biosciences; and owns a stock option from Treeline Biosciences. The other authors declare that they have no conflicts of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54749-2.
References
- 1.Galluzzi, L. et al. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ.25, 486–541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malireddi, R. S., Kesavardhana, S. & Kanneganti, T.-D. ZBP1 and TAK1: master regulators of NLRP3 inflammasome/pyroptosis, apoptosis, and necroptosis (PAN-optosis). Front. Cell. Infect. Microbiol.9, 406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fritsch, M. et al. Caspase-8 is the molecular switch for apoptosis, necroptosis and pyroptosis. Nature575, 683–687 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Newton, K. et al. Activity of caspase-8 determines plasticity between cell death pathways. Nature575, 679–682 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Gurung, P. et al. FADD and caspase-8 mediate priming and activation of the canonical and noncanonical Nlrp3 inflammasomes. J. Immunol.192, 1835–1846 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuriakose, T. & Kanneganti, T.-D. ZBP1: innate sensor regulating cell death and inflammation. Trends Immunol.39, 123–134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng, M., Karki, R., Vogel, P. & Kanneganti, T.-D. Caspase-6 is a key regulator of innate immunity, inflammasome activation, and host defense. Cell181, 674–687.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orning, P. et al. Pathogen blockade of TAK1 triggers caspase-8–dependent cleavage of gasdermin D and cell death. Science362, 1064–1069 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukherjee, S. et al. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science312, 1211–1214 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Dondelinger, Y. et al. NF-κB-independent role of IKKα/IKKβ in preventing RIPK1 kinase-dependent apoptotic and necroptotic cell death during TNF signaling. Mol. Cell60, 63–76 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Haas, T. L. et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol. Cell36, 831–844 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Jaco, I. et al. MK2 phosphorylates RIPK1 to prevent TNF-induced cell death. Mol. Cell66, 698–710.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun, L. et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell148, 213–227 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Schneider-Brachert, W., Heigl, U. & Ehrenschwender, M. Membrane trafficking of death receptors: implications on signalling. Int. J. Mol. Sci.14, 14475–14503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malireddi, R. et al. Innate immune priming in the absence of TAK1 drives RIPK1 kinase activity–independent pyroptosis, apoptosis, necroptosis, and inflammatory disease. J. Exp. Med.217, jem.20191644 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malireddi, R. S. et al. TAK1 restricts spontaneous NLRP3 activation and cell death to control myeloid proliferation. J. Exp. Med.215, 1023–1034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Legrand, A. J., Konstantinou, M., Goode, E. F. & Meier, P. The diversification of cell death and immunity: memento mori. Mol. cell76, 232–242 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Doyotte, A., Mironov, A., McKenzie, E. & Woodman, P. The Bro1-related protein HD-PTP/PTPN23 is required for endosomal cargo sorting and multivesicular body morphogenesis. Proc. Natl Acad. Sci.105, 6308–6313 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma, H. et al. Histidine-domain-containing protein tyrosine phosphatase regulates platelet-derived growth factor receptor intracellular sorting and degradation. Cell. Signal.27, 2209–2219 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Manteghi, S. et al. Haploinsufficiency of the ESCRT component HD-PTP predisposes to cancer. Cell Rep.15, 1893–1900 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Gingras, M.-C., Kazan, J. M. & Pause, A. Role of ESCRT component HD-PTP/PTPN23 in cancer. Biochem. Soc. Trans.45, 845–854 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Suzuki, A. et al. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr. Biol.8, 1169–1178 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Chan, G. et al. Essential role for Ptpn11 in survival of hematopoietic stem and progenitor cells. Blood, J. Am. Soc. Hematol.117, 4253–4261 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marsh, H. N. et al. SHP-1 negatively regulates neuronal survival by functioning as a TrkA phosphatase. J. Cell Biol.163, 999–1010 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi, J. et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat. Biotechnol.33, 661–667 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuber, J. et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat. Biotechnol.29, 79–83 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji, L., Minna, J. D. & Roth, J. A. 3p21. 3 tumor suppressor cluster: prospects for translational applications. Future Oncol.1, 79–92 (2005). [DOI] [PubMed]
- 28.Hesson, L., Cooper, W. & Latif, F. Evaluation of the 3p21. 3 tumour-suppressor gene cluster. Oncogene26, 7283–7301 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Imreh, S., Klein, G. & Zabarovsky, E. R. Search for unknown tumor‐antagonizing genes. Genes Chromosomes Cancer38, 307–321 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Senchenko, V. N. et al. Discovery of frequent homozygous deletions in chromosome 3p21. 3 LUCA and AP20 regions in renal, lung and breast carcinomas. Oncogene23, 5719–5728 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Braga, E. et al. Critical tumor‐suppressor gene regions on chromosome 3P in major human epithelial malignancies: allelotyping and quantitative real‐time PCR. Int. J. Cancer100, 534–541 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Zhang, S. et al. Suppression of protein tyrosine phosphatase N23 predisposes to breast tumorigenesis via activation of FYN kinase. Genes Dev.31, 1939–1957 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kharitidi, D. et al. Interplay of endosomal pH and ligand occupancy in integrin α5β1 ubiquitination, endocytic sorting, and cell migration. Cell Rep.13, 599–609 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Tsherniak, A. et al. Defining a cancer dependency map. Cell170, 564–576.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gingras, M.-C. et al. Expression analysis and essential role of the putative tyrosine phosphatase His-domain-containing protein tyrosine phosphatase (HD-PTP). Int. J. Dev. Biol.53, 1069–1074 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Lin, G., Aranda, V., Muthuswamy, S. K. & Tonks, N. K. Identification of PTPN23 as a novel regulator of cell invasion in mammary epithelial cells from a loss-of-function screen of the ‘PTP-ome’. Genes Dev.25, 1412–1425 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bissig, C. & Gruenberg, J. ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell Biol.24, 19–25 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Nabet, B. et al. The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol.14, 431–441 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He, S., Liang, Y., Shao, F. & Wang, X. Toll-like receptors activate programmed necrosis in macrophages through a receptor-interacting kinase-3–mediated pathway. Proc. Natl Acad. Sci.108, 20054–20059 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaiser, W. J. et al. Toll-like receptor 3-mediated necrosis via TRIF, RIP3, and MLKL. J. Biol. Chem.288, 31268–31279 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peltzer, N., Darding, M. & Walczak, H. Holding RIPK1 on the ubiquitin leash in TNFR1 signaling. Trends Cell Biol.26, 445–461 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Annibaldi, A. & Meier, P. Checkpoints in TNF-induced cell death: implications in inflammation and cancer. Trends Mol. Med.24, 49–65 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Legler, D. F., Micheau, O., Doucey, M.-A., Tschopp, J. & Bron, C. Recruitment of TNF receptor 1 to lipid rafts is essential for TNFα-mediated NF-κB activation. Immunity18, 655–664 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Sharma, D. & Kanneganti, T.-D. The cell biology of inflammasomes: mechanisms of inflammasome activation and regulation. J. Cell Biol.213, 617–629 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ali, N. et al. Recruitment of UBPY and ESCRT exchange drive HD-PTP-dependent sorting of EGFR to the MVB. Curr. Biol.23, 453–461 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Annibaldi, A. & Walczak, H. Death receptors and their ligands in inflammatory disease and cancer. Cold Spring Harb. Perspect. Biol.12, a036384 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaiser, W. J. & Offermann, M. K. Apoptosis induced by the toll-like receptor adaptor TRIF is dependent on its receptor interacting protein homotypic interaction motif. J. Immunol.174, 4942–4952 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Bauernfeind, F. G. et al. Cutting edge: NF-κB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol.183, 787–791 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mamińska, A. et al. ESCRT proteins restrict constitutive NF-κB signaling by trafficking cytokine receptors. Sci. Signal.9, ra8–ra8 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Fujita, H. et al. A dominant negative form of the AAA ATPase SKD1/VPS4 impairs membrane trafficking out of endosomal/lysosomal compartments: class E vps phenotype in mammalian cells. J. Cell Sci.116, 401–414 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Adell, M. A. Y. et al. Coordinated binding of Vps4 to ESCRT-III drives membrane neck constriction during MVB vesicle formation. J. Cell Biol.205, 33–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gahloth, D. et al. Structural basis for selective interaction between the ESCRT regulator HD-PTP and UBAP1. Structure24, 2115–2126 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Agromayor, M. et al. The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a SOUBA domain. Structure20, 414–428 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pan, J. et al. Interrogation of mammalian protein complex structure, function, and membership using genome-scale fitness screens. Cell Syst.6, 555–568.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Komada, M. Controlling receptor downregulation by ubiquitination and deubiquitination. Curr. Drug Discov. Technol.5, 78–84 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Clague, M. J., Liu, H. & Urbé, S. Governance of endocytic trafficking and signaling by reversible ubiquitylation. Dev. cell23, 457–467 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Mizuno, E., Kobayashi, K., Yamamoto, A., Kitamura, N. & Komada, M. A deubiquitinating enzyme UBPY regulates the level of protein ubiquitination on endosomes. Traffic7, 1017–1031 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Clague, M. J. & Urbé, S. Endocytosis: the DUB version. Trends Cell Biol.16, 551–559 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Stefani, F. et al. UBAP1 is a component of an endosome-specific ESCRT-I complex that is essential for MVB sorting. Curr. Biol.21, 1245–1250 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Stuchell, M. D. et al. The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding. J. Biol. Chem.279, 36059–36071 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Ren, X. & Hurley, J. H. VHS domains of ESCRT‐0 cooperate in high‐avidity binding to polyubiquitinated cargo. EMBO J.29, 1045–1054 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Christ, L., Raiborg, C., Wenzel, E. M., Campsteijn, C. & Stenmark, H. Cellular functions and molecular mechanisms of the ESCRT membrane-scission machinery. Trends Biochem. Sci.42, 42–56 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Raposo, G., Tenza, D., Murphy, D. M., Berson, J. F. & Marks, M. S. Distinct protein sorting and localization to premelanosomes, melanosomes, and lysosomes in pigmented melanocytic cells. J. Cell Biol.152, 809–824 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doherty, L. M. et al. Integrating multi-omics data reveals function and therapeutic potential of deubiquitinating enzymes. Elife11, e72879 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roux, K. J., Kim, D. I., Raida, M. & Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol.196, 801–810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gahloth, D. et al. Structural basis for specific interaction of TGFβ signaling regulators SARA/Endofin with HD-PTP. Structure25, 1011–1024. e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lafont, E. et al. TBK1 and IKKε prevent TNF-induced cell death by RIPK1 phosphorylation. Nat. Cell Biol.20, 1389–1399 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Macia, E. et al. Dynasore, a cell-permeable inhibitor of dynamin. Dev. cell10, 839–850 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Sun, Q., Westphal, W., Wong, K. N., Tan, I. & Zhong, Q. Rubicon controls endosome maturation as a Rab7 effector. Proc. Natl Acad. Sci.107, 19338–19343 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Radulovic, M. et al. ESCRT‐mediated lysosome repair precedes lysophagy and promotes cell survival. EMBO J.37, e99753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schütze, S., Tchikov, V. & Schneider-Brachert, W. Regulation of TNFR1 and CD95 signalling by receptor compartmentalization. Nat. Rev. Mol. Cell Biol.9, 655–662 (2008). [DOI] [PubMed] [Google Scholar]
- 72.Schneider-Brachert, W. et al. Compartmentalization of TNF receptor 1 signaling: internalized TNF receptosomes as death signaling vesicles. Immunity21, 415–428 (2004). [DOI] [PubMed] [Google Scholar]
- 73.Lee, K. H. et al. The role of receptor internalization in CD95 signaling. EMBO J.25, 1009–1023 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mazurek, N. et al. Cell-surface galectin-3 confers resistance to TRAIL by impeding trafficking of death receptors in metastatic colon adenocarcinoma cells. Cell Death Differ.19, 523–533 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vanamee, É. S. & Faustman, D. L. Structural principles of tumor necrosis factor superfamily signaling. Sci. Signal.11, eaao4910 (2018). [DOI] [PubMed] [Google Scholar]
- 76.Karathanasis, C. et al. Single-molecule imaging reveals the oligomeric state of functional TNFα-induced plasma membrane TNFR1 clusters in cells. Sci. Signal.13, eaax5647 (2020). [DOI] [PubMed] [Google Scholar]
- 77.Pan, L. et al. Higher-order clustering of the transmembrane anchor of DR5 drives signaling. Cell176, 1477–1489. e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Raiborg, C. & Stenmark, H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature458, 445–452 (2009). [DOI] [PubMed] [Google Scholar]
- 79.Vietri, M., Radulovic, M. & Stenmark, H. The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol.21, 25–42 (2020). [DOI] [PubMed] [Google Scholar]
- 80.Gong, Y.-N. et al. ESCRT-III acts downstream of MLKL to regulate necroptotic cell death and its consequences. Cell169, 286–300. e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rühl, S. et al. ESCRT-dependent membrane repair negatively regulates pyroptosis downstream of GSDMD activation. Science362, 956–960 (2018). [DOI] [PubMed] [Google Scholar]
- 82.Dai, E., Meng, L., Kang, R., Wang, X. & Tang, D. ESCRT-III–dependent membrane repair blocks ferroptosis. Biochem. Biophys. Res. Commun.522, 415–421 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ritter, A. T. et al. ESCRT-mediated membrane repair protects tumor-derived cells against T cell attack. Science376, 377–382 (2022). [DOI] [PubMed] [Google Scholar]
- 84.Raab, M. et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science352, 359–362 (2016). [DOI] [PubMed] [Google Scholar]
- 85.Wang, Y. & Kanneganti, T.-D. From pyroptosis, apoptosis and necroptosis to PANoptosis: a mechanistic compendium of programmed cell death pathways. Comput. Struct. Biotechnol. J.19, 4641–4657 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kuriakose, T. et al. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci. Immunol.1, aag2045 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee, S. et al. AIM2 forms a complex with pyrin and ZBP1 to drive PANoptosis and host defence. Nature597, 415–419 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou, J. et al. Immunogenic cell death in cancer therapy: Present and emerging inducers. J. Cell. Mol. Med.23, 4854–4865 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yatim, N. et al. RIPK1 and NF-κB signaling in dying cells determines cross-priming of CD8+ T cells. Science350, 328–334 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Snyder, A. G. et al. Intratumoral activation of the necroptotic pathway components RIPK1 and RIPK3 potentiates antitumor immunity. Sci. Immunol.4, eaaw2004 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang, Q. et al. A bioorthogonal system reveals antitumour immune function of pyroptosis. Nature579, 421–426 (2020). [DOI] [PubMed] [Google Scholar]
- 92.Tarumoto, Y. et al. LKB1, salt-inducible kinases, and MEF2C are linked dependencies in acute myeloid leukemia. Mol. Cell69, 1017–1027.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Alonso, A. et al. Protein tyrosine phosphatases in the human genome. Cell117, 699–711 (2004). [DOI] [PubMed] [Google Scholar]
- 94.Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol.31, 827–832 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Freund, E. C. et al. Efficient gene knockout in primary human and murine myeloid cells by non-viral delivery of CRISPR-Cas9. J. Exp. Med.217, e20191692 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ma, J. et al. iProX: an integrated proteome resource. Nucleic Acids Res.47, D1211–D1217 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen, T. et al. iProX in 2021: connecting proteomics data sharing with big data. Nucleic Acids Res.50, D1522–D1527 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The FASTQ files generated in CRISPR screen have been deposited in the Sequence Read Archive (SRA) database under accession number PRJNA1187288. The raw RNA-seq data generated in this study have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE272123. The raw mass spectrometry data have been deposited to the ProteomeXchange Consortium via the iProX repository96,97 with the dataset identifier PXD056836. The sgRNA sequences used in the CRISPR screen are provided in Supplementary Data 1, and sgRNA sequences used in the validation study are provided in Supplementary Data 4. Supplementary Data 2 contains the average log2 frequency changes of sgRNAs targeting the same gene at Days 2 and 14 post-sgRNA transduction. Proteomic data analyzed using Scaffold software are available in Supplementary Data 3. Supplementary Data 5 provides a list of antibodies used for immunoblotting, confocal imaging, and flow cytometry. All other relevant data are included within the main article, Supplementary Information, or Source Data files. Source data are provided with this paper.