Abstract
Purpose
Neurochemicals of interest quantified by MRS are often composites of overlapping signals. At higher field strengths (i.e., 7T), there is better separation of these signals. As the availability of higher field strengths is increasing, it is important to re‐evaluate the separability of overlapping metabolite signals.
Methods
This study compares the ability of stimulated echo acquisition mode (STEAM‐8; TE = 8 ms), short‐TE semi‐LASER (sLASER‐34; TE = 34 ms), and long‐TE semi‐LASER (sLASER‐105; TE = 105 ms) acquisitions to separate the commonly acquired neurochemicals at 7T (Glx, consisting of glutamate and glutamine; total N‐acetyl aspartate, consisting of N‐acetyl aspartate and N‐acetylaspartylglutamate; total creatine, consisting of creatine and phosphocreatine; and total choline, consisting of choline, phosphocholine, and glycerophosphocholine).
Results
sLASER‐34 produced the lowest fit errors for most neurochemicals; however, STEAM‐8 had better within‐subject reproducibility and required fewer subjects to detect a change between groups. However, this is dependent on the neurochemical of interest.
Conclusion
We recommend short‐TE STEAM for separation of most standard neurochemicals at 7T over short‐TE or long‐TE sLASER.
Keywords: 7 T, brain, high field, magnetic resonance spectroscopy, semi‐LASER, STEAM
1. INTRODUCTION
Proton MRS is commonly used to quantify chemicals of interest in the human brain. The typically quantified neurochemicals are Glx, total N‐acetyl aspartate (tNAA), total creatine (tCr), and total choline (tCho). However, these are composites of signals from multiple neurochemicals that overlap. For example, the Glx signal consists of the overlapping signals' glutamate (Glu) and glutamine (Gln).
Higher field strength increases SNR and spectral resolution, thus potentially allowing better separation of overlapping signals. As the availability of higher field strengths (i.e., 7T) increases, it is important to re‐evaluate the separability of overlapping metabolite signals. The most commonly used acquisition protocol at 7T is stimulated echo acquisition mode (STEAM). STEAM allows for ultrashort TEs, which minimize signal modulation due to j‐coupling and T2 relaxation, desirable aspects for separating overlapping signals. 1 For example, Zhong and Ernst showed improvement of spectral quality and reduced signal loss for the coupled resonances Glu and Gln, as well as myo‐inositol at TE = 8 ms compared with TE = 30 ms. 2 Dou et al. also showed better separation of Glu and Gln signal using a short‐echo STEAM sequence (TE = 20 ms) compared with a long‐echo STEAM sequence (TE = 74 ms) at 7T. 3 However, STEAM results in a lower SNR (compared to spin echo acquisitions), because only half the signal can be obtained.
Although semi‐LASER (sLASER) is unable to achieve the very short TEs of STEAM due to the need for longer adiabatic pulses, sLASER is able to achieve higher SNR. sLASER also results in reduced chemical shift displacement and better resilience to B1 inhomogeneity, both important aspects when working at higher field strengths. 4 Furthermore, the paired adiabatic pulses in sLASER suppress j‐evolution and prolong T2 relaxation times, similar to the effects of short‐TE sequences. 5 Indeed, Marsman et al. reported that sLASER (TE = 28 ms) produces more reproducible measures of Glu than STEAM (TE = 7.5 ms) at 7T. 6 However, Okada et al. found no difference in repeatability when comparing sLASER (TE = 32 ms) with STEAM (TE = 5 ms). Additionally, although sLASER did have a higher N‐acetyl aspartate (NAA) SNR, they found that STEAM produced lower Cramér‐Rao lower bounds (CRLBs) for several metabolites. 7 Therefore, the optimal sequence and TE may depend on the metabolite of interest.
Although short TEs are typically preferred to prevent signal loss, longer TEs may sometimes be advantageous for detecting certain metabolites, particularly if optimal pulse timings are chosen to yield favorable peak detection. 8 , 9 Shorter TEs will also result in more macromolecule signal, which can complicate metabolite quantification. Furthermore, to achieve short TEs when using sLASER at 7T, special hardware modifications are sometimes needed, such as a dual transmit option, 6 which may not be available. Wong et al. looked at the signal evolution of Glu over a range of TEs from 45 ms to 225 ms and found the optimal TE to be 105 ms. 1 However, this was based on Glu only, and TEs below 45 ms were not investigated. Indeed, Najac et al. showed the highest Glu signal to be at TE = 38 ms, followed by TE = 100 ms 10 ; however, this trend of increasing signal at higher TEs was not seen for other metabolites.
The aim of this study, therefore, was to compare the ability of STEAM, short‐TE sLASER, and long‐TE sLASER acquisitions to separate the commonly acquired neurochemicals at 7T.
2. METHODS
2.1. Participants
Fourteen healthy participants between the ages of 18 and 40 years were recruited. Data were collected with approval from the local research ethics board, and written informed consent was obtained from all participants.
2.2. Data acquisition
Data were collected using a 7T Siemens Terra with a 32‐channel head coil. Each session began with a T1‐weighted image (MPRAGE, TE/TR = 1.84/4300 ms, isotropic resolution = 1 mm3) for voxel placement and segmentation. The 2.5 × 2.5 × 2.5 cm3 voxels were placed in the parietal cortex centered on the midline (Figure 1).
FIGURE 1.
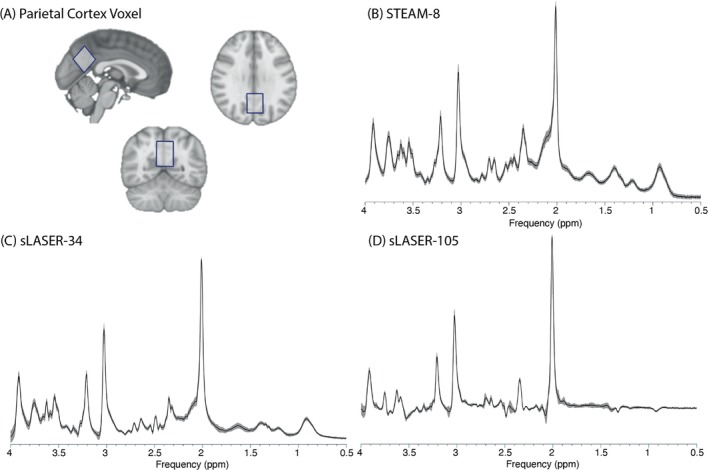
(A) Example of voxel location in the parietal cortex. Mean (black) and SD (shaded gray) of spectra acquired using (B) STEAM‐8 (stimulated echo acquisition mode, TE = 8 ms), (C) sLASER‐34 (semi‐LASER, TE = 34 ms), and (D) sLASER‐105 (semi‐LASER, TE = 105 ms).
Data were collected using three sequences: STEAM‐8 (TE/TR/TM = 8/6000/40 ms), sLASER‐34 (TE/TR = 34/5000 ms), and sLASER‐105 (TE/TR = 105/5000 ms), all with 8224 spectral points and outer‐volume suppression pulses applied in all cases. For each sequence, 64 water‐suppressed averages were acquired using VAPOR water suppression, and 8 water‐unsuppressed averages were acquired. Sequence order was counterbalanced across subjects. The STEAM sequence 11 was developed by Edward J. Auerbach and Malgorzata Marjańska, and the sLASER sequence 12 , 13 was developed by Gülin Öz and Dinesh Deelchand and provided by the University of Minnesota under a C2P agreement.
2.3. Data processing
MRS data were preprocessed using FID‐A 14 with the following steps: coil combination, removal of motion‐corrupted averages, frequency drift correction, and zero‐order phase correction. LCModel 15 was used to apply eddy current correction and quantification relative to water. The ATTH2O parameter was set to 1, to allow for correction of T2 decay of water at each TE external to LCModel.
Basis sets for quantification were simulated using FID‐A based on exact timings and RF pulses for each sequence. Basis sets included the following metabolites: alanine, aspartate, choline (Cho), glycerophosphocholine (GPC), phosphocholine (PC), creatine (Cr), phosphocreatine (PCr), γ‐aminobutyric acid, glutamate (Glu), glutamine (Gln), lactate, myo‐inositol, NAA, N‐acetylaspartylglutamate (NAAG), scyllo‐inositol, glutathione, glucose, taurine, glycine, and phosphatidylethanolamine (basis sets, including simulated macromolecules, are available at https://github.com/HarrisBrainLab/BasisSets).
Macromolecule basis sets were simulated using information obtained by parameterizing freely available 7T metabolite‐suppressed data from 6 healthy individuals (https://zenodo.org/records/3906754#.XvOWVvJ7n7e). 16 First, all six metabolite‐nulled spectra were combined into a single average macromolecule spectrum. Nine macromolecule resonances were then identified in the average macromolecule spectrum, and each macromolecule resonance was fit to a Gaussian lineshape using in‐house custom MATLAB scripts to estimate the frequency, linewidth, and relative amplitude. For each identified macromolecule resonance, a single‐proton FID‐A spin system was created, with the “scaleFactor” parameter adjusted to account for the number of protons associated with each macromolecule resonance (see Table 1; FID‐A spin systems are available at https://github.com/HarrisBrainLab/MMSim). Macromolecules were then simulated with the appropriate simulation function in FID‐A based on the parameters in Table 1, using exact timings and RF pulses as described previously. The simulated macromolecule linewidth was 6 Hz smaller than the linewidths of the parameterized spectra to account for the line broadening typically applied by LCModel, to match the narrow, simulated metabolite basis functions to the typically broader linewidths observed in in vivo data. The relative concentration of each macromolecule was constrained using the values in Table 1. Macromolecule basis sets were included for quantification of STEAM‐8 and sLASER‐34 data. At 105 ms, the macromolecule signal had decayed to approximately 0; thus, there is risk of overfitting if including macromolecule basis sets. Therefore, sLASER‐105 was quantified with and without the macromolecule basis set. All results were consistent; thus, we report sLASER‐105 quantification without macromolecule basis sets.
TABLE 1.
Values used for simulation of macromolecules.
| Macromolecule | Frequency (ppm) | No. protons a | Simulated linewidth (Hz) | LCModel constraint b |
|---|---|---|---|---|
| M0.95 | 0.95 | 3 | 32 | n/a |
| M1.24 | 1.235 | 3 | 23 | 0.41 ± 0.16 |
| M1.43 | 1.427 | 3 | 26 | 0.98 ± 0.67 |
| M1.71 | 1.705 | 2 | 31 | 1.185 ± 1.10 |
| M2.07 | 2.07 | 2 | 38 | 2.53 ± 0.81 |
| M2.30 | 2.295 | 2 | 32 | 2.00 ± 0.17 |
| M3.03 | 3.03 | 2 | 26 | 1.30 ± 0.32 |
| M3.24 | 3.24 | 2 | 48 | 0.10 ± 0.22 |
| M3.98 | 3.98 | 2 | 109 | 2.33 ± 0.72 |
Abbreviation: n/a, not applicable.
Values obtained from Cudalbu et al. 22
Concentration relative to M0.95.
T1‐weighted images were segmented using the CoRegStandAlone function from the Gannet toolbox. 17 Tissue correction was performed according to Gasparovic et al., 18 including correcting for the different TEs and T1 and T2 values for both water and metabolites (see Table S3 for T1 and T2 values used). SNR was estimated using the op_getSNR function in FID‐A, which measures the height of the NAA resonance between 1.8 and 2.2 ppm and normalizes this to the SD of the signal in a peak free region (manually set to between −1 and −2 ppm).
Consistent with the field, we used within‐subject variation as our metric of measurement precision (see Supporting Information for simulations comparing the effects of measurement precision on within‐subject and between‐subject variability). To assess repeatability, the water‐suppressed scans were split into two equal blocks of 32 averages, and the water‐unsuppressed scans were split into two equal blocks of four averages, then processed as described previously. These split scans were used for calculation of the within‐subject coefficient of variation (CV) only; all other metrics were calculated using the full 64‐average scans.
2.4. Statistical analysis
Statistical analyses were conducted using R (version 4.1.2; R Core Team [2020] https://www.R‐project.org/). 19 The analysis focused on the following combinations of metabolites: Glx, Glu + Gln; tNAA, NAA + NAAG; tCr, Cr + PCr; and tCho, Cho + PC + GPC. Due to the increasing interest in measuring glutathione (GSH) and γ‐aminobutyric acid (GABA) at high fields, results for these metabolites are reported in the Supporting Information.
Quality metrics (linewidth, SNR), metabolite levels, and CRLBs obtained from the full 64‐average scan were statistically compared using repeated‐measures analyses of variance with the “aov” function in the “stats” package. Pairwise comparisons were conducted using Bonferroni correction. CRLB was included as a measure of fit precision, as poor fitting would be more likely to result in signal being incorrectly assigned to overlapping metabolites, particularly for lower concentration metabolites.
Repeatability was assessed using the split 32‐average scans. The within‐subject CV was calculated as the within‐subject variance divided by the mean, using the RMS approach. 20 The 95% confidence intervals were calculated for the CV values using bootstrap resampling with replacement (1000 resamples). Additionally, the pairwise correlation coefficient from LCModel was used to assess the relationship between overlapping metabolites (e.g., the correlation between NAA and NAAG). A correlation closer to zero implies better separation of overlapping signal, whereas positive or negative correlations with a larger magnitude indicate poor separation, 3 and it is recommended in the LCModel manual that signal has not been adequately separated if the correlation between a pair is consistently less than −0.3.
2.5. Sample‐size calculations
Calculations were performed for each metabolite to estimate the number of participants per group that would be needed to detect a 10% change between two groups, based on the formula described by Wong et al. 1
| (1) |
where = 1.28% for 90% power; = 1.96% for 95% confidence level; = 0.1 for a 10% change; σ = the SD of each metabolite across all subjects; and = the mean of each metabolite across all subjects.
3. RESULTS
3.1. Quality metrics
There was a significant effect of sequence on NAA linewidth (p < 0.001). Pairwise comparisons showed a significant difference for all comparisons (all p < 0.001), with sLASER‐105 having the lowest NAA linewidth. There was also a significant effect of sequence on H2O linewidth (p < 0.01). Pairwise comparisons showed that sLASER‐105 had significantly lower H2O linewidth than STEAM‐8 (p = 0.02) and sLASER‐34 (p = 0.02), but there was no significant difference between STEAM‐8 and sLASER‐34 (p = 0.98). There was also a significant effect of sequence on SNR (p < 0.001). Pairwise comparisons showed that sLASER‐34 had significantly higher SNR than STEAM‐8 (p < 0.001) and sLASER‐105 (p < 0.001). There was no significant difference in SNR between STEAM‐8 and sLASER‐105 (p = 0.43, Table 2).
TABLE 2.
Mean and SDs of quality metrics for each sequence.
| NAA linewidth (Hz) | H2O linewidth (Hz) | SNR | |
|---|---|---|---|
| STEAM‐8 | 13.21 ± 1.65 | 11.73 ± 1.82 | 254.47 ± 48.23 |
| sLASER‐34 | 10.93 ± 1.42 | 11.62 ± 1.24 | 402.99 ± 42.52 |
| sLASER‐105 | 8.54 ± 1.05 | 9.89 ± 1.87 | 276.53 ± 44.90 |
| p | < 0.001 | 0.009 | < 0.001 |
Abbreviations: NAA, N‐acetyl aspartate; sLASER‐34, semi‐LASER, TE = 34 ms; sLASER‐105, semi‐LASER, TE = 105 ms; STEAM‐8, stimulated echo acquisition mode, TE = 8 ms.
3.2. Glutamate
Glu: There was a significant effect of sequence on metabolite level (F[2,38] = 178.8; p < 0.001), and pairwise comparisons showed a significant difference in metabolite levels for all three comparisons (all p < 0.001) (Figure 2A). There was also a significant effect of sequence on CRLBs (F[2,38] = 76.4; p < 0.001). Glu measured using sLASER‐105 had significantly lower CRLBs compared with Glu measured using the other sequences (Figure 2B). STEAM‐8 produced the lowest CV (Figure 2C) and required the least number of participants to detect a 10% difference (Table 3).
FIGURE 2.
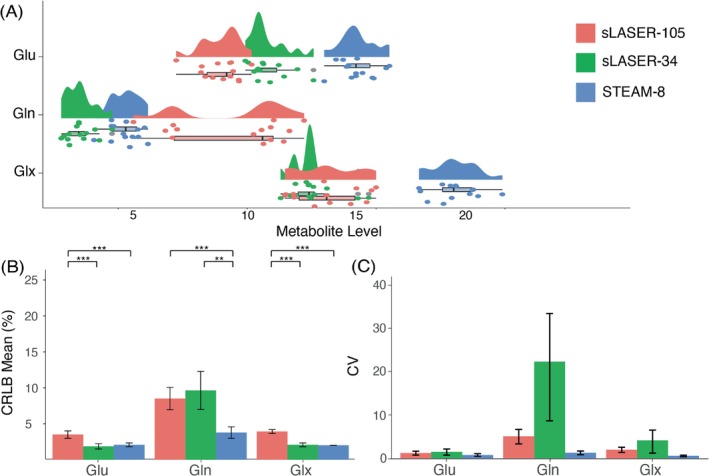
(A) Mean and spread of glutamate (Glu), glutamine (Gln), and Glu + Gln (Glx) data for each sequence. (B) Mean Cramér‐Rao lower bound (CRLB) values for Glu, Gln, and Glx, error bars represent standard deviation. (C) Coefficient of variation (CV) values for Glu, Gln, and Glx, error bars represent 95% confidence intervals, calculated using bootstrap resampling. *p < 0.05, **p < 0.01, ***p < 0.001. sLASER‐34, semi‐LASER, TE = 34 ms; sLASER‐105, semi‐LASER, TE = 105 ms; STEAM‐8, stimulated echo acquisition mode, TE = 8 ms.
TABLE 3.
Estimated number of participants per group needed to observe a 10% difference between groups for each sequence and each metabolite.
| STEAM‐8 | sLASER‐34 | sLASER‐105 | |
|---|---|---|---|
| Glu | 6 | 13 | 21 |
| Gln | 47 | 137 | 153 |
| Glx | 6 | 15 | 23 |
| NAA | 6 | 7 | 25 |
| NAAG | 21 | 76 | 174 |
| tNAA | 6 | 9 | 27 |
| Cr | 6 | 4 | 40 |
| PCr | 15 | 60 | 38 |
| tCr | 1 | 9 | 16 |
| Cho | 19 | 3342 | 901 |
| GPC | 7 | 27 | 162 |
| tCho | 7 | 22 | 47 |
Abbreviations: Cr, creatine; Glu, glutamate Gln, glutamine; Glx, Glu + Gln, GPC, glycerophosphocholine; NAA, N‐acetyl aspartate; NAAG, N‐acetylaspartylglutamate; PCr, phosphocreatine; sLASER‐34, semi‐LASER, TE = 34 ms; sLASER‐105, semi‐LASER, TE = 105 ms; STEAM‐8, stimulated echo acquisition mode, TE = 8 ms; tCho, total choline; tCr, total creatine; tNAA, total N‐acetyl aspartate.
Gln: There was a significant effect of sequence on metabolite level (F[2,38] = 77.56; p < 0.001), and pairwise comparisons showed a significant difference in metabolite levels for all three comparisons (all p < 0.001) (Figure 2A). There was also a significant effect of sequence on CRLBs (F[2,38] = 39.13; p < 0.001). STEAM‐8 produced significantly lower CRLBs compared with the other sequences (Figure 2B). STEAM‐8 produced the lowest CV (Figure 2C) and required the least number of participants to detect a 10% difference (Table 3).
Glx: There was a significant effect of sequence on metabolite level (F[2,38] = 117.2; p < 0.001), and pairwise comparisons showed a significant difference in metabolite levels for STEAM‐8 versus sLASER‐34 (p < 0.001) and STEAM‐8 versus sLASER‐105 (p < 0.001) (Figure 2A). There was also a significant effect of sequence on CRLBs (F[2,38] = 342.5; p < 0.001). STEAM‐8 produced the lowest CRLBs; however, this was not significantly different from sLASER‐34 (Figure 2B). STEAM‐8 produced the lowest CV (Figure 2C) and required the least number of participants to detect a 10% difference (Table 3).
The mean (± SD) of the correlation coefficients produced by LCModel for Glu and Gln for each sequence are as follows: STEAM‐8 = 0.02 ± 0.02, sLASER‐34 = −0.01 ± 0.05, sLASER‐105 = −0.01 ± 0.32.
3.3. N‐acetyl‐aspartate
NAA: There was a significant effect of sequence on metabolite level (F[2,38] = 36.22; p < 0.001), and pairwise comparisons showed a significant difference in metabolite levels for sLASER‐34 versus sLASER‐105 (p < 0.001) and STEAM‐8 versus sLASER‐34 (p < 0.001) (Figure 3A). There was also a significant effect of sequence on CRLBs (F[2,38] = 12.1; p < 0.001). Pairwise comparisons showed that sLASER‐34 produced significantly lower CRLBs than the other two sequences (p < 0.01) (Figure 3B). STEAM‐8 produced the lowest CV (Figure 3C). sLASER‐34 required the least number of participants to detect a 10% difference (Table 3).
FIGURE 3.
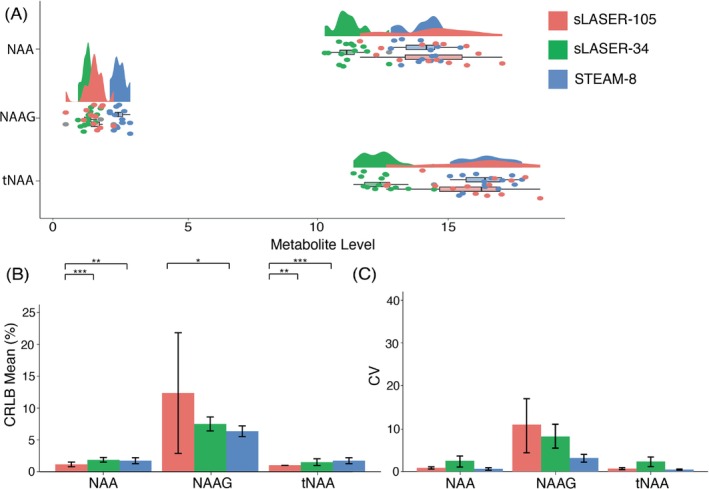
(A) Mean and spread of N‐acetyl aspartate (NAA), N‐acetylaspartylglutamate (NAAG), and total N‐acetyl aspartate (tNAA; NAA + NAAG) data for each sequence. (B) Mean Cramér‐Rao lower bound (CRLB) values for NAA, NAAG and tNAA, error bars represent standard deviation. (C) Coefficient of variation (CV) values for NAA, NAAG and tNAA, error bars represent 95% confidence intervals calculated using bootstrap resampling. *p < 0.05, **p < 0.01, ***p < 0.001. sLASER‐34, semi‐LASER, TE = 34 ms; sLASER‐105, semi‐LASER, TE = 105 ms; STEAM‐8, stimulated echo acquisition mode, TE = 8 ms.
NAAG: There was a significant effect of sequence on metabolite level (F[2,38] = 55.91; p < 0.001), and pairwise comparisons showed a significant difference in metabolite levels for STEAM‐8 versus sLASER‐34 (p < 0.001) and STEAM‐8 versus sLASER‐105 (p < 0.001) (Figure 3A). There was also a significant effect of sequence on CRLBs (F[2,38] = 5.0; p = 0.01). STEAM‐8 produced the lowest CRLBs, but these were not significantly different from sLASER‐34 (Figure 3B). STEAM‐8 produced the lowest CV (Figure 3C) and required the least number of participants to detect a 10% difference (Table 3).
tNAA: There was a significant effect of sequence on metabolite level (F[2,38] = 40.43; p < 0.001), and pairwise comparisons showed a significant difference in metabolite levels for STEAM‐8 versus sLASER‐34 (p < 0.001) and SLASER‐34 versus sLASER‐105 (p < 0.001) (Figure 3A). There was also a significant effect of sequence on CRLBs (F[2,38] = 11.28; p < 0.001). sLASER‐105 produced the lowest CRLBs; however, these were not significantly different than sLASER‐34 (Figure 3B). STEAM‐8 had the lowest CV (Figure 3C). sLASER‐34 required the least number of participants to detect a 10% difference (Table 3).
The mean (± SD) of the correlation coefficients produced by LCModel for NAA and NAAG for each sequence are as follows: STEAM‐8 = −0.04 ± 0.07, sLASER‐34 = −0.16 ± 0.10, and sLASER‐105 = −0.61 ± 0.30.
3.4. Creatine
Cr: There was a significant effect of sequence on metabolite level (F[2,38] = 86.71; p < 0.001), and pairwise comparisons showed a significant difference in metabolite levels for STEAM‐8 versus sLASER‐34 (p < 0.001) and STEAM‐8 versus sLASER‐105 (p < 0.001) (Figure 4A). There was also a significant effect of sequence on CRLBs (F[2,38] = 15.7; p < 0.001). sLASER‐34 produced significantly lower CRLBs compared with the other two sequences (Figure 4B). STEAM‐8 produced the lowest CV (Figure 4C). sLASER‐34 required the least number of participants to detect a 10% difference (Table 3).
FIGURE 4.
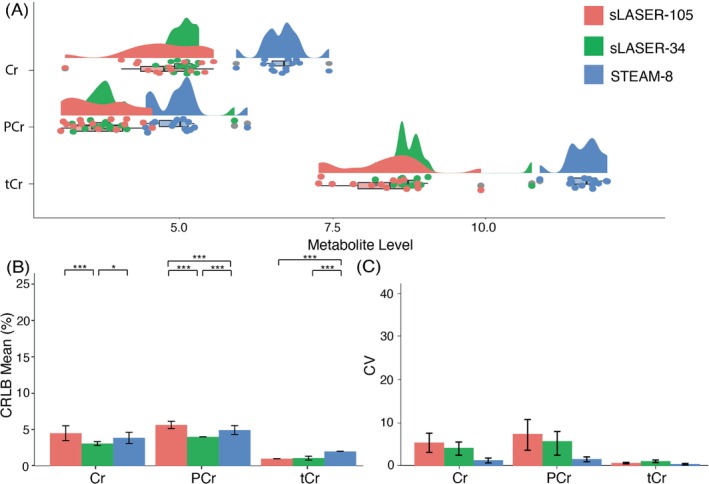
(A) Mean and spread of creatine (Cr), phosphocreatine (PCr), and total creatine (tCr; Cr + PCr) data for each sequence. (B) Mean Cramér‐Rao lower bound (CRLB) values for Cr, PCr, and tCr, error bars represent SD. (C) Coefficient of variation (CV) values for Cr, PCr, and tCr, error bars represent 95% confidence intervals calculated using bootstrap resampling. *p < 0.05, **p < 0.01, ***p < 0.001. sLASER‐34, semi‐LASER, TE = 34 ms; sLASER‐105, semi‐LASER, TE = 105 ms; STEAM‐8, stimulated echo acquisition mode, TE = 8 ms.
PCr: There was a significant effect of sequence on metabolite level (F[2,38] = 26.88; p < 0.001), and pairwise comparisons showed a significant difference in metabolite levels for STEAM‐8 versus sLASER‐34 (p < 0.001) and STEAM‐8 versus sLASER‐105 (p < 0.001) (Figure 4A). There was also a significant effect of sequence on CRLBs (F[2,38] = 47.4; p < 0.001). sLASER‐34 produced significantly lower CRLBs compared with the other two sequences (Figure 4B). STEAM‐8 produced the lowest CV (Figure 4C). STEAM‐8 required the least number of participants to detect a 10% difference (Table 3).
tCr: There was a significant effect of sequence on metabolite level (F[2,38] = 133.8; p < 0.001), and pairwise comparisons showed a significant difference in metabolite levels for STEAM‐8 versus sLASER‐34 (p < 0.001) and STEAM‐8 versus sLASER‐105 (p < 0.001) (Figure 4A). There was also a significant effect of sequence on CRLBs (F[2,38] = 178.9; p < 0.001). sLASER‐34 and sLASER‐105 produced equally low CRLBs (Figure 4B). sLASER‐105 produced the lowest CV (Figure 4C). STEAM‐8 required the least number of participants to detect a 10% difference (Table 3).
The mean (± SD) of the correlation coefficients produced by LCModel for Cr and PCr for each sequence are as follows: STEAM‐8 = −0.68 ± 0.02, sLASER‐34 = −0.71 ± 0.03, and sLASER‐105 = −0.88 ± 0.02.
3.5. Choline
Cho: There was a significant effect of sequence on metabolite level (F[2,38] = 24.0; p < 0.001), and pairwise comparisons showed a significant difference in metabolite levels for all three comparisons (all p < 0.01) (Figure 5A). There was also a significant effect of sequence on CRLBs (F[2,32] = 17.4; p < 0.001). STEAM‐8 produced the lowest CRLB, but this was not significantly lower than sLASER‐105 (Figure 5B). STEAM‐8 produced the lowest CV (Figure 5C) and required the least number of participants to detect a 10% difference (Table 3).
FIGURE 5.
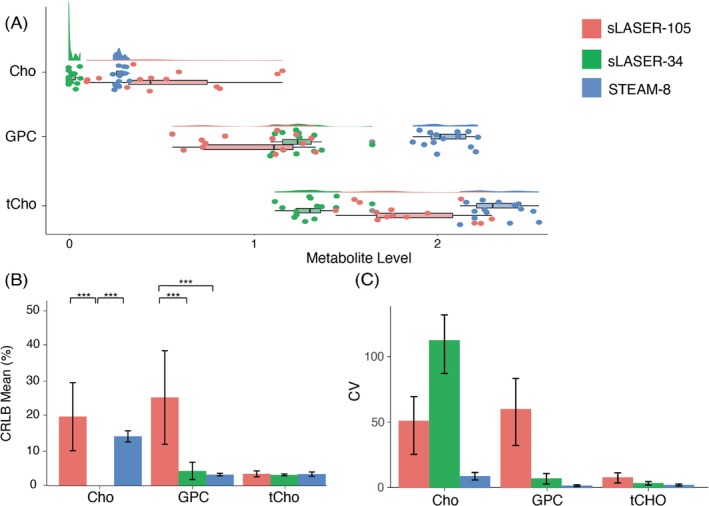
(A) Mean and spread of choline (Cho), glycerophosphocholine (GPC) and total choline (tCho; Cho + phosphocholine (PC) + GPC) data for each sequence. (B) Mean Cramér‐Rao lower bound (CRLB) values for Cho, GPC, and tCho, error bars represent SD. Mean Cho CRLB for sLASER‐34 not shown as > 500 (C) Coefficient of variation (CV) values for Cho, GPC and tCho, error bars represent 95% confidence intervals calculated using bootstrap resampling. *p < 0.05, **p < 0.01, ***p < 0.001. sLASER‐34, semi‐LASER, TE = 34 ms; sLASER‐105, semi‐LASER, TE = 105 ms; STEAM‐8, stimulated echo acquisition mode, TE = 8 ms.
PC: PC was not quantified well using any of the three sequences, and therefore will not be included for comparison.
GPC: There was a significant effect of sequence on metabolite level (F[2,38] = 112.0; p < 0.001), and pairwise comparisons showed a significant difference in metabolite levels for all three comparisons (all p < 0.01) (Figure 5A). There was also a significant effect of sequence on CRLBs (F[2,38] = 34.2; p < 0.001). STEAM‐8 produced the lowest CRLBs, but this was not significantly lower than sLASER‐34 (Figure 5B). STEAM‐8 produced the lowest CV (Figure 5C) and required the least number of participants to detect a 10% difference (Table 3).
tCho: There was a significant effect of sequence on metabolite level (F[2,38] = 95.1; p < 0.001), and pairwise comparisons showed a significant difference in metabolite levels for all three comparisons (all p < 0.001) (Figure 5A). There was no significant effect of sequence on CRLBs (F[2,38] = 1.6; p = 0.22) (Figure 5B). STEAM‐8 produced the lowest CV (Figure 5C) and required the least number of participants to detect a 10% difference (Table 3).
Due to PC not being quantified, only the correlation between Cho and GPC was assessed. The mean (± SD) of the correlation coefficients produced by LCModel for Cho and GPC for each sequence are as follows: STEAM‐8 = 0.03 ± 0.05, sLASER‐34 = −0.02 ± 0.09, and sLASER‐105 = −0.05 ± 0.27.
4. DISCUSSION
Several studies have assessed the ability of STEAM and sLASER sequences to separate Glu and Gln signal at 7T. Here we expand on previous work to include the direct comparison of three sequences (STEAM‐8, sLASER‐34, and sLASER‐105) to separate overlapping signal of the four most commonly quantified metabolites.
With the exception of creatine‐containing metabolites, there was little correlation between most overlapping metabolites, suggesting that all sequences are generally able to resolve the metabolite signals (i.e., a high correlation between two metabolites would suggest limited or inconsistent success at signal separation). The magnitude of the correlation between Cr and PCr was greater than −0.3 for all sequences, indicating they are not well separated with any sequence, despite the low fit error, and are best reported as a composite measure. NAA and NAAG measured with sLASER‐105 were also strongly correlated (−0.61 ± 0.30) and therefore should also be reported as a composite measure when using this sequence.
Based on fit error (CRLB), generally sLASER‐34 appears to perform best at quantifying the individual neurochemicals as well as quantifying the summed signals. However, this is not true for choline‐containing neurochemicals, as Cho and GPC quantified with STEAM‐8 produced the lowest fit errors. STEAM‐8 had better repeatability compared with the other sequences, and, generally, fewer participants were needed to detect a 10% change when using STEAM‐8, although again this was dependent on the neurochemical.
Interestingly, metabolites quantified using STEAM‐8 had less variation (lower CVs) than when quantified using the other two sequences. This is in contrast to Marsman et al., who found higher CVs with STEAM (TE = 7.7 ms) compared to sLASER (TE = 28 ms). 6 However, Marsman et al. compared variation across two different scan sessions rather than variation over the same session as done here. This implies that STEAM may produce more consistent results across a session but less consistent results across time. Because sLASER has a reduced chemical shift displacement and better resilience to B1 inhomogeneity, 4 it is likely less sensitive to repositioning effects.
STEAM‐8 often produced higher values compared with the other two sequences, likely due to less signal loss from the small TE. There is also evidence that apparent T2 values of metabolites may vary across sequences, 21 which may also contribute to concentration differences, although this is not expected to largely affect the values. STEAM‐8 generally also required fewer participants to detect a 10% change. This likely reflects a smaller spread of data when acquired using this sequence. As there is no ground truth when working with in vivo data, it is not possible to determine whether this reflects a more consistent measurement, or whether STEAM‐8 is not picking up on smaller differences between participants.
Wong et al. recommend sLASER‐105 for optimal Glu measurement; however, the shortest TE they included in their comparison was 45 ms. Although they observed the greatest in vivo glutamate signal at TE = 45 ms, they found more variability and larger CRLBs at this TE and suggest this to be due to variations in macromolecules. 1 We show lower variability and CRLBs using the shorter TEs, which will have the greatest macromolecule contamination. The inclusion of a simulated macromolecule baseline may have aided in controlling for this.
A limitation of this study is that only one area of the brain was assessed. Data were acquired from the parietal cortex, which generally produces good quality data. Areas of the brain where data are often of poorer quality (such as frontal areas or deep brain structures) may produce different results. Additionally, simulated macromolecule models were used to account for macromolecule signal, as opposed to directly measuring macromolecule signal in vivo. This method was chosen to reduce participant burden in the scanner and follows recommendations for accounting for macromolecule signal in metabolite spectra. 22 However, the metabolite values (and subsequently the correlation and CRLB values) may be different when using a measured macromolecule signal, partly due to the increased degrees of freedom when modeling individually simulated macromolecules. This was partly mitigated in our analysis by using fixed proportions for each macromolecule, reducing the degrees of freedom.
5. CONCLUSIONS
Overall, we show data acquired at 7T with sLASER‐34 had lower fit errors, but data acquired with STEAM‐8 had better within‐session repeatability and required fewer subjects to detect a change between groups. However, this is dependent on the neurochemical of interest. We therefore recommend short‐TE STEAM for separation of most standard neurochemicals over short‐TE or long‐TE sLASER.
Supporting information
Data S1. Supporting information.
ACKNOWLEDGMENTS
This work was undertaken thanks in part to funding from the Natural Sciences and Engineering Research Council of Canada (NSERC, RGPIN‐2020‐05917, RGPIN‐2017‐03875), a Canadian Foundation for Innovation John R Evans Leaders Fund (CFI‐JELF), and support by the Hotchkiss Brain Institute and the Alberta Children's Hospital Research Institute, University of Calgary. T.K.B is supported by One Child Every Child and Cumming School of Medicine Postdoctoral Fellowships. J.N. holds funding from the Canadian Institutes of Health Research (CIHR, PJT‐183715) and the New Frontiers Research Fund of Canada (NFRF, NFRFT 2022‐00327). A.D.H. holds a Canada Research Chair in Magnetic Resonance Spectroscopy in Brain Injury and holds funding from the CIHR (PJ‐T175085) and The Arthritis Society.
Bell TK, Goerzen D, Near J, Harris AD. Examination of methods to separate overlapping metabolites at 7T . Magn Reson Med. 2025;93:470‐480. doi: 10.1002/mrm.30293
DATA AVAILABILITY STATEMENT
Basis sets, including simulated macromolecules, are available at https://github.com/HarrisBrainLab/BasisSets. Tools to simulate macromolecules are available at https://github.com/HarrisBrainLab/MMSim. Anonymized data may be made available to qualified investigators upon request.
REFERENCES
- 1. Wong D, Schranz AL, Bartha R. Optimized in vivo brain glutamate measurement using long‐echo‐time semi‐LASER at 7 T. NMR Biomed. 2018;31:1‐13. doi: 10.1002/nbm.4002 [DOI] [PubMed] [Google Scholar]
- 2. Zhong K, Ernst T. Localized in vivo human 1H MRS at very short echo times. Magn Reson Med. 2004;52:898‐901. doi: 10.1002/mrm.20201 [DOI] [PubMed] [Google Scholar]
- 3. Dou W, Kaufmann J, Li M, Zhong K, Walter M, Speck O. The separation of Gln and Glu in STEAM: a comparison study using short and long TEs/TMs at 3 and 7 T. Magn Reson Mater Phys Biol Med. 2015;28:395‐405. doi: 10.1007/s10334-014-0479-7 [DOI] [PubMed] [Google Scholar]
- 4. Wilson M, Andronesi O, Barker PB, et al. Methodological consensus on clinical proton MRS of the brain: review and recommendations. Magn Reson Med. 2019;82:527‐550. doi: 10.1002/mrm.27742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deelchand DK, Kantarci K, Öz G. Improved localization, spectral quality, and repeatability with advanced MRS methodology in the clinical setting. Magn Reson Med. 2018;79:1241‐1250. doi: 10.1002/mrm.26788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marsman A, Boer VO, Luijten PR, et al. Detection of glutamate alterations in the human brain using 1H‐MRS: comparison of STEAM and sLASER at 7 T. Front Psych. 2017;8:60. doi: 10.3389/fpsyt.2017.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Okada T, Kuribayashi H, Kaiser LG, et al. Repeatability of proton magnetic resonance spectroscopy of the brain at 7 T: effect of scan time on semi‐localized by adiabatic selective refocusing and short‐echo time stimulated echo acquisition mode scans and their comparison. Quant Imaging Med Surg. 2021;11:9‐20. doi: 10.21037/QIMS-20-517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choi C, Ganji S, Hulsey K, et al. A comparative study of short‐ and long‐TE 1 H MRS at 3 T for in vivo detection of 2‐hydroxyglutarate in brain tumors. NMR Biomed. 2013;26:1242‐1250. doi: 10.1002/nbm.2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dobberthien BJ, Tessier AG, Yahya A. Improved resolution of glutamate, glutamine and γ‐aminobutyric acid with optimized point‐resolved spectroscopy sequence timings for their simultaneous quantification at 9.4 T. NMR Biomed. 2018;31:1‐12. doi: 10.1002/nbm.3851 [DOI] [PubMed] [Google Scholar]
- 10. Najac C, Boer VO, Kan HE, Webb AG, Ronen I. Improved detection limits of J‐coupled neurometabolites in the human brain at 7 T with a J‐refocused sLASER sequence. NMR Biomed. 2022;35:1‐16. doi: 10.1002/nbm.4801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tkáč I, Starčuk Z, Choi IY, Gruetter R. In vivo 1H NMR spectroscopy of rat brain at 1 ms echo time. Magn Reson Med. 1999;41:649‐656. doi: [DOI] [PubMed] [Google Scholar]
- 12. Deelchand DK, Berrington A, Noeske R, et al. Across‐vendor standardization of semi‐LASER for single‐voxel MRS at 3T. NMR Biomed. 2019;34;1‐11. doi: 10.1002/nbm.4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Öz G, Tkáč I. Short‐echo, single‐shot, full‐intensity proton magnetic resonance spectroscopy for neurochemical profiling at 4 T: validation in the cerebellum and brainstem. Magn Reson Med. 2011;65:901‐910. doi: 10.1002/mrm.22708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Simpson R, Devenyi GA, Jezzard P, Hennessy TJ, Near J. Advanced processing and simulation of MRS data using the FID appliance (FID‐A)—an open source, MATLAB‐based toolkit. Magn Reson Med. 2017;77:23‐33. doi: 10.1002/mrm.26091 [DOI] [PubMed] [Google Scholar]
- 15. Provencher SW. Automatic quantitation of localized in vivo 1H spectra with LCModel. NMR Biomed. 2001;14:260‐264. [DOI] [PubMed] [Google Scholar]
- 16. Považan M, Hangel G, Strasser B, et al. Mapping of brain macromolecules and their use for spectral processing of 1H‐MRSI data with an ultra‐short acquisition delay at 7T. Neuroimage. 2015;121:126‐135. https://zenodo.org/records/3906754#.XvOWVvJ7n7e doi: 10.1016/j.neuroimage.2015.07.042 [DOI] [PubMed] [Google Scholar]
- 17. Edden RAE, Puts NAJ, Harris AD, Barker PB, Evans CJ. Gannet: a batch‐processing tool for the quantitative analysis of gamma‐aminobutyric acid–edited MR spectroscopy spectra. J Magn Reson Imaging. 2014;40:1445‐1452. doi: 10.1038/jid.2014.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gasparovic C, Song T, Devier D, et al. Use of tissue water as a concentration reference for proton spectroscopic imaging. Magn Reson Med. 2006;55:1219‐1226. doi: 10.1002/mrm.20901 [DOI] [PubMed] [Google Scholar]
- 19. R Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; 2020. https://www.R‐project.org/ [Google Scholar]
- 20. Bland M. How should I calculate a within‐subject coefficient of variation? 2006. https://www‐users.york.ac.uk/∼mb55/meas/cv.htm Accessed June 13, 2024.
- 21. Deelchand DK, Auerbach EJ, Kobayashi N, Marjańska M. Transverse relaxation time constants of the five major metabolites in human brain measured in vivo using LASER and PRESS at 3 T. Magn Reson Med. 2018;79:1260‐1265. doi: 10.1002/mrm.26826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cudalbu C, Behar KL, Bhattacharyya PK, et al. Contribution of macromolecules to brain 1H MR spectra: experts' consensus recommendations. NMR Biomed. 2021;34:1‐24. doi: 10.1002/nbm.4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supporting information.
Data Availability Statement
Basis sets, including simulated macromolecules, are available at https://github.com/HarrisBrainLab/BasisSets. Tools to simulate macromolecules are available at https://github.com/HarrisBrainLab/MMSim. Anonymized data may be made available to qualified investigators upon request.


