Abstract
A key developmental stage in mammalian folliculogenesis is the formation of a fluid-filled lumen (antrum) prior to ovulation. While it has long been speculated that the follicular fluid is essential for oocyte maturation and ovulation, little is known about the morphogenesis and the mechanisms driving the antrum formation and ovulation, potentially due to challenges in imaging tissue dynamics in large tissues. Misregulation of such processes leads to anovulation, a hallmark of infertility in ageing and diseases such as the polycystic ovary syndrome (PCOS). In this review, we discuss recent advances in deep tissue imaging techniques, machine learning and theoretical approaches that have been applied to study development and diseases. We propose that an integrative approach combining these techniques is essential for understanding the physics of hydraulics in follicle development and ovarian functions.
Keywords: Ovary, Folliculogenesis, Ovulation, Biophotonics, Machine learning, Tissue mechanics
Introduction
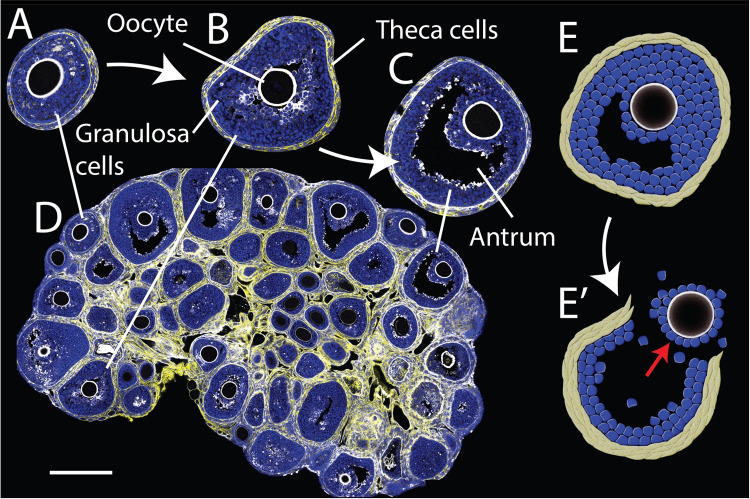
The development of functional oocytes followed by successful ovulation is a critical process in early mammalian reproduction (Biswas et al. 2022; Telfer et al. 2023). During folliculogenesis, the follicle morphology provides a rough classification of its stage of development (Fig. 1). In the secondary follicle stage, the follicle consists of an oocyte surrounded by multi-layered granulosa cells (GC) with an outer layer of elongated theca cells (TC) (Fig. 1B) (Rodgers and Irving-Rodgers 2010; Biswas et al. 2022). The formation of a fluid-filled lumen, also known as the antrum, begins as small pockets of interstitial fluids which nucleate between the GCs. As the fluid volume increases, these fluids evolve to form a singly resolved antrum showing a stereotypical “smiley” pattern in the case of mouse ovaries (Fig. 1C) (Rodgers and Irving-Rodgers 2010; Biswas et al. 2022). However, the exact mechanisms for the de novo luminogenesis during antrum development remain largely unknown. As the follicles grow further, and with the stimulation of luteinizing hormones, they rupture and release the fluid, along with the ejection of the cumulus-oocyte complex (Fig. 1D) (Converse et al. 2023; Zaniker et al. 2023; Komatsu and Masubuchi 2017; Matsuzaki 2021).
Fig. 1.
Hydraulic control of late-stage ovarian follicle development. A Early secondary follicle (100–180 m). B Secondary follicle with small pockets of fluids (180–300 m). C Pre-ovulatory follicle with fully formed “smiley-faced” antrum (300–400 m). D A mouse ovarian tissue slice with DNA (Blue), pan-collagen (yellow) and anti-Müllerian hormone (grey). Scale bar, 300 m. E-E’ Ovulation involves the rupture of the follicle wall and the release of oocyte, though the exact mechanism remains poorly understood
While extensive studies in the past have identified various key signalling pathways controlling oocyte growth, less is known about the roles of mechanical stress and biophysics in folliculogenesis (Biswas et al. 2022; Prasasya and Mayo 2019; Telfer et al. 2023). In recent years, tissue mechanics has emerged as a central regulator of various developmental processes, such as tissue folding and patterning (Matsuzaki 2021; Bevilacqua et al. 2023; Zhang and Fodor 2023; Biswas et al. 2022, 2024; Athilingam et al. 2021; Barriga et al. 2018; Fiorentino et al. 2023). In mice, mechanical stress exerted by the extracellular matrix (ECM) or the TCs surrounding the follicles have been shown to influence follicle activation and growth (Nagamatsu et al. 2019; Biswas et al. 2024). During ageing, the aged ovaries have shown significant increase in the ECM stiffness (Mara et al. 2020; Fiorentino et al. 2023), which correlates with impaired follicle and oocyte functions (Pietroforte et al. 2024) and ovulation (Umehara et al. 2022). In PCOS patients, the ovaries are characterised by multiple cystic-like follicles that fail to ovulate (Mara et al. 2020; Lee et al. 2024c; Fiorentino et al. 2023), suggesting that impaired luminogenesis and follicle rupture may contribute to developmental arrest in ovarian disease and ageing.
Indeed, in addition to cell-generated cytoskeleton forces, there has been increasing evidence that tissue-scale fluid forces can transmit long-range mechanical signals to influence morphogenesis and cellular functions during development (Chan and Hirashima 2022; Chan and Hiiragi 2020). However, how tissue hydraulics impacts mammalian follicle development has not been addressed so far. One reason for this is the challenge of studying tissue dynamics in large organs. In this review, we highlight recent advances in deep tissue imaging techniques, machine learning and theoretical models that have been increasingly applied to study complex 3D tissue dynamics in vivo. We discuss how these approaches, when applied individually or together, could deepen our basic understanding of late-stage folliculogenesis (for mechanobiology of earlier stages, see recent review (Telfer et al. 2023; Prasasya and Mayo 2019; Biswas et al. 2022)), with important clinical implications in future infertility studies and assisted reproductive technology.
Non-invasive and label-free imaging techniques to study tissue dynamics during folliculogenesis
In the study of late-stage ovarian folliculogenesis, the use of intravital imaging combined with histological analysis of fixed ovary tissues has helped to establish a general understanding of the developmental processes (Feng et al. 2018). However, the tissue dynamics of these morphological events are less well understood. To do so, direct live imaging of the spatiotemporal dynamics of cellular and fluid movements is required. Here, we discuss recent advances in the development of novel biophotonic tools that have the potential to overcome current imaging limitations and provide new mechanistic insights to our understanding of fluid antrum formation and ovulation.
Current imaging techniques utilised in ovarian studies
In the past, measurements of fluid fraction using ultrasonography have shed light on lumen expansion during follicle growth (Rodgers and Irving-Rodgers 2010). Alongside the identification of the ECM at the GC apical surface in electron micrographs, it was hypothesised that the directional secretion of large molecules, such as proteoglycans and hyaluronans, establishes an osmotic gradient against the surrounding thecal vasculature, thereby recruiting fluid for lumen growth (Clarke et al. 2006). In the study of ovulation, intravital imaging of blood vessel thickness and blood flow surrounding the ovary using multiphoton microscopy (Migone et al. 2013) and ultrasonography helped reveal the mechanism of endothelin-induced contractions of smooth muscle cells, causing basolateral invaginations prior to ovulation (Migone et al. 2016). Subsequent work using serial tissue section indirectly suggested complementary action from the inward migration of mural GCs following the surge of luteinizing hormones (Owen and Jaffe 2023). Nevertheless, ultrasonography-based intravital imaging is constrained by its poor spatial resolution of a few millimeters. In contrast, multiphoton microscopy offers better spatial resolution but is limited to a depth penetration of about 200 m in ovarian tissues (Migone et al. 2016) and has significantly slower imaging speeds. Additionally, multiphoton microscopy requires the use of fluorescent labels, which may incur phototoxic effects. While transgenic animal lines expressing fluorescent proteins are useful, such an approach is typically restricted to mice, making it difficult to translate to other mammalian species and humans. Another imaging approach is ultrastructural studies using electron microscopy. While these studies have led to the discovery of novel features such as the Call-Exner bodies during luminogenesis (Gosden et al. 1989; Van Wezel et al. 1999), they remain largely descriptive and do not inform the dynamic processes underlying luminogenesis. To address this, other label-free imaging techniques may be required, as outlined below.
Optical coherence tomography
Here, we introduce optical coherence tomography (OCT), an emerging label-free modality with intermediate depth penetration and resolution used in intravital imaging. OCT works on the basis of detecting back-scattered signals, analogous to ultrasound, except that OCT uses light rather than acoustic waves which therefore offers higher spatial resolutions of 1–15 m (Fujimoto 2003; Chow et al. 2024), with reasonable penetration depths of up to 1 mm during intravital imaging of ovarian tissues. As a label-free technique, OCT is not bound by a “fluorescence budget” as is the case for confocal microscopy, allowing for prolonged imaging without signal degradation. Typically used in ophthalmic diagnosis and research (Everett et al. 2021), OCT has recently been applied to reproductive biology (Burton et al. 2015) as an optical biopsy tool for assessing ovarian reserve and detecting metastases (Takae et al. 2017, 2018; Peters et al. 2016). Owing to the high optical contrast between different tissue components and its high-speed volumetric imaging capability, OCT has been employed to capture real-time dynamics of oocytes and pre-implantation embryo transport in the mouse oviduct, revealing uncharted location-dependent movement trajectories (Wang and Larina 2021; Umezu and Larina 2023).
Nevertheless, intravital imaging using OCT requires precise orientation of the imaged organs, and can be exacerbated by tissue motion such as muscle contraction and breathing (Burton et al. 2015). To overcome these limitations in imaging ovaries, advancement in ex vivo cultures such as 3D follicle culture (Converse et al. 2023), multifollicle tissue (Biswas et al. 2022) and slice culture of ovaries (Komatsu et al. 2018) can be utilised. Furthermore, ex vivo culture is more amenable to optical coherence microscopy (OCM), a variant of OCT that offers higher spatial resolution with reduced imaging depth (Aguirre et al. 2015). This has been demonstrated in live imaging of rapid fluid dynamics and muscle hydraulics in sea animals such as cnidarian Nematostella vectensis (Stokkermans et al. 2022) and freshwater sponge Spongilla lacustris (Ruperti et al. 2024). In a similar vein, we propose that OCM will be a powerful tool for studying antrum formation and ovulation in mammalian species, allowing for unprecedented tracking of lumen fluid growth and organisation and cellular dynamics during follicle rupture in ex vivo culture (Fig. 2H). Recently, other applications of OCM in vivo have also been explored, expanding its potential for future use in intravital imaging (Moore et al. 2019) and imaging-based biophysical measurements (see the “Biophysical control of ovulation” section).
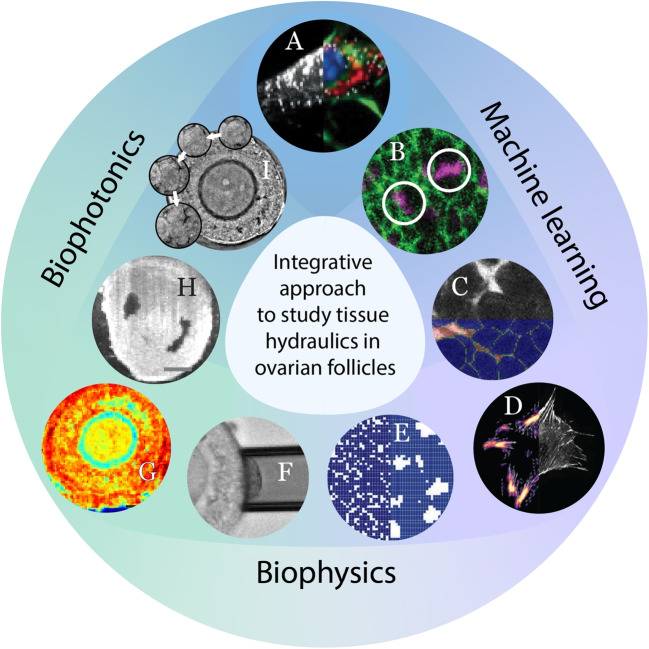
Fig. 2.
Investigating tissue hydraulics in mammalian ovarian folliculogenesis using an integrative approach that combines advanced biophotonics, deep learning and biophysical modelling. A Label-free RI imaging combined with deep learning enables visualisation of subcellular structures and dynamics with high specificity (Jo et al. 2021). B Deep learning model detects cell divisions during wound healing with high accuracy (Turley et al. 2024a). C 3D segmentation of GCs in the ovarian follicle. D Machine learning based on physics-informed neural networks allows inference of traction forces in a cell (Schmitt et al. 2024). E Computational approach to model luminogenesis in follicles based on fluid mixing-demixing transition. F Measurement of the surface tension of ovarian follicle using micropipette aspiration. G Representative “stiffness map” of an ovarian follicle imaged by Brillouin microscopy, revealing clear intrafollicular mechanical heterogeneities (Chan et al. 2021). H An image of pre-ovulatory follicle acquired with OCM. Scale bar, 100 m. I Images showing a cell undergoing division, acquired through QPI. Interstitial fluids are also visible due to its distinct RI from cellular bodies
Quantitative phase imaging
Building on existing phase measurement techniques in OCT (Nguyen et al. 2022; Park et al. 2018), quantitative phase imaging (QPI) technique was developed as another fast, label-free and non-invasive technique for biological imaging. A detailed review of the underlying principles and the different applications in biomedicine has been published (Nguyen et al. 2022; Park et al. 2018). In brief, QPI measures the change in the phase component of light, providing intrinsic optical imaging contrast. The phase change can be used to determine both the refractive index (RI) and thickness of a sample, allowing identification of subcellular structures with known RI, such as mitochondria and nuclei, thus providing greater specificity. Additionally, the RI and volume measurements can be used to estimate the dry mass of the sample (Barer 1952; Schürmann et al. 2016). In the past, QPI has been limited to the study of single cells since multiple scattering leads to reduced contrast and image quality in thick samples. Recently, the use of the phase-gradient contrast technique (Ford et al. 2012) enabled the reconstruction of quantitative phase information in thick samples, as demonstrated in dissected whole mouse brains, and human liver organoids (Ledwig and Robles 2019; Filan et al. 2024). Notably, tracking of single cells flowing through blood vessels in free-swimming zebrafish larvae exemplified QPI’s high-speed imaging capability (Kandel et al. 2019).
While complex imaging modalities often require in-house instrumentation, this challenge is mitigated by the availability of commercialised QPI systems. This had proved to be invaluable for monitoring preimplantation mouse development, revealing features used for differentiating embryos with varying developmental potentials (Lee et al. 2024b). Applied to intestinal organoids, it has successfully identified different cell types based on morphometric differences used to select viable organoids (Lee et al. 2023). In studying ovarian folliculogenesis ex vivo, QPI offers a unique approach to track GC rearrangement, proliferation and deaths (Fig. 2I). Since the lumen has a distinctly lower RI than that of cellular structures, QPI also holds promise in unravelling the precise dynamics of lumniongenesis. Direct quantification of luminal RI will also potentially inform changes in lumen composition during antral follicle development. Currently, intravital QPI imaging remains challenging due to severe light scattering in optically inhomogeneous biological tissues. However, scattering reduction techniques have successfully retrieved signals from circulating blood cells in the mesentery of live mice (Kim et al. 2016), highlighting the potential for future advancements in imaging dynamic processes in vivo.
Ideally, applying OCT to in vivo contexts will reveal true tissue dynamics in their native state, but it comes with limitations in spatial resolution and setup complexity. Alternatively, multiphoton imaging provides higher spatial detail but is constrained by reduced temporal resolution due to phototoxicity. We advocate the use of OCM and QPI to study ovarian folliculogenesis in ex vivo cultures, which should offer an optimal spatiotemporal resolution that captures the essential features of ovarian dynamics.
A general drawback of label-free imaging techniques discussed above lies in their limited cellular and molecular specificity. Nevertheless, these methods can be combined with fluorescence-based modalities (Mowla et al. 2024; Bevilacqua et al. 2023; Lee et al. 2023; Nguyen et al. 2017). Additionally, the adoption of virtual staining techniques enabled the prediction of both dye and fluorescence signals in label-free images (Fig. 2A) (Winetraub et al. 2024; Park et al. 2023a, b). This integration with the artificial intelligence (AI) approach enhances the specificity and versatility of label-free imaging, opening avenues for comprehensive cellular and molecular characterisation, a topic we will delve into in the following section.
Machine learning approaches to quantify tissue properties from large datasets
Quantitative studies of tissue dynamics in large systems can generate enormous amounts of data, particularly if the dynamics are fast. In this section, we describe several machine-learning approaches used to investigate biological processes and highlight their potential use in studying late-stage follicle development.
What is machine learning?
Machine learning comprises a set of algorithms in which a model is trained to complete repetitive sets of tasks (Hallou et al. 2021; Jones et al. 2017; Howard and Gugger 2020). Deep learning is one type of these algorithms, which are vast “artificial neural” networks with the ability to learn complex patterns from the data and solve problems which require extensive manual work (Howard and Gugger 2020). Deep learning has been particularly successful and is increasingly being used in biological and medical imaging (Culley et al. 2023; Hallou et al. 2021; Singh et al. 2020). The major advantage of using these algorithms is they often outperform other automation approaches and that once trained, the model can be applied to vast datasets with minimal or no human input. One method of training the models is by supplying a dataset of labelled examples where a human expert first completes the classification or segmentation task by hand, followed by training the machine learning model to perform a similar task on new data (Howard and Gugger 2020).
Segmenting and quantifying 3D data
One of the most common applications of deep learning is cell segmentation, which has been extensively applied to tissue morphogenesis, wound healing and tumour segmentation (Turley et al. 2024a, b; Chattopadhyay and Maitra 2022; Işin et al. 2016; McDole et al. 2018; Wang et al. 2022; Mitchell and Cislo 2023; Ichbiah et al. 2023; Boylan et al. 2024). Recently, 3D segmentation models have been used to investigate cell layers surrounding a lumen in MDCK cells (Andrés-San Román et al. 2023) and early stages of sea star embryo development (Barone et al. 2024). Following segmentation, other morphometric information such as cell volume, elongation and packing ratios can be quantified readily. The differences in tissue packing and rates of cell rearrangements can indicate changes to tissue stiffness and rheology, respectively (Lou et al. 2023; Tetley et al. 2019; Stroka and Aranda-Espinoza 2011). Without automated deep learning models, segmentation studies on this vast scale would not be feasible. Such techniques will be highly relevant to quantifying intraofollicular dynamics, where the precise segmentation of oocyte and GC size, shape and movements may lead to new insights on tissue phase transitions during folliculogenesis (see the “Potential phase transitions during antral folicle development” section).
Virtual stain of label-free data
Label-free imaging techniques, as discussed in the “Quantitative phase imaging” section, can also benefit from deep learning methods. Here, deep learning models can be used to virtually stain or segment regions of the cells such as the nucleus (Christiansen et al. 2018; Jo et al. 2021; Park et al. 2023a). This has recently been demonstrated on images of cells acquired through combined quantitative phase imaging and fluorescence microscopy (Jo et al. 2021). Using fluorescent microscopy images as the ground truth, deep learning models were trained to segment various cytoskeleton components and organelles from the quantitative phase images. This model was highly accurate and even capable of labelling other cell lines which it was not trained on Jo et al. (2021). Importantly, this implies that the model, once trained, can be extended to label cells during live imaging, where the use of dyes may be cytotoxic, or in non-transgenic organisms such as human samples. Hence, we propose that quantitative phase imaging, combined with deep learning, will provide a unique and timely approach to study the dynamics of ovarian folliculogenesis, particularly when the interstitial fluids cannot be labelled with fluorescent tags readily.
Deep learning models to detect dynamic behaviors and forces
Beyond segmenting and classifying regions of a tissue, deep learning methods can be used to detect dynamic behaviour such as cell divisions and extrusions (McDole et al. 2018; Turley et al. 2024b, a; Villars et al. 2023). The dynamics of cell divisions with daughter nuclei undergoing splitting and cytokinesis are visually distinct from the relatively stationary non-dividing cells. Therefore detection of cell divisions using deep learning models has been highly successful (McDole et al. 2018; Turley et al. 2024b, a; Villars et al. 2023). Combined with additional fluorescent markers labelling cell membrane, the detection of cell division remains robust even in noisy environments. This has been demonstrated in the case of wound healing in Drosophila (Turley et al. 2024b, a), where the presence of cell debris and immune cells did not pose a challenge for analysing cell division dynamics through deep learning. So far, deep learning analysis has been larged confined to 2D tissues. The extension of such approach to 3D tissue dynamics, such as ovarian folliculogenesis, will be necessary given the huge datasets and natural biological variation that can only be overcome with machine learning.
AI algorithms have also been used to infer cellular forces from microscopy data (Schmitt et al. 2023). Here, traction force microscopy was used as the ground truth to quantify forces generated by single cells. Based on the confocal images, the information provided by the distribution of zyxin was sufficient to train a deep-learning model to determine the traction forces. Remarkably, even though the model has not been trained on cells treated with cytoskeletal perturbations, it was able to predict the traction stress field in these conditions which matches the experimental results (Schmitt et al. 2023, 2024). Using physics-informed neural networks, these work also demonstrate the unique possibility of constructing a physical model with interpretable physics and parameters (Schmitt et al. 2024; Karniadakis et al. 2021; Colen et al. 2021). Such approach has recently been applied to Drosophila embryogenesis (Lefebvre et al. 2024), where the active stress generated by the myosin distribution is sufficient to train the neural networks to predict the tissue flow pattern driving germ-band extension. Importantly, the model can also be trained to “construct” the minimal sets of equations describing the dynamics of the myosin field (Lefebvre et al. 2024), thereby unravelling previously hidden physics underlying such processes. In ovarian follicle development, such a data-driven biophysical modelling approach may provide an exciting alternative approach to understand the underlying physical principles governing robust folliculogenesis.
Unbiased pattern detection in tissues without supervision
In addition to quantifying cellular features, deep learning methods could also inform how these properties change in time and space. Here, a certain kind of deep learning model based on unsupervised learning techniques (Zinchenko et al. 2023; Lu et al. 2019; Lafarge et al. 2019) provides an unbiased approach to distinguish the distinct morphological features from different cell types. This was recently applied to electron microscopy data from a marine annelid (P. dumerilii), where the model extracted features of both cell shape and texture using dimensionality reduction techniques (Zinchenko et al. 2023). In this “MorphoFeatures” space, clear clusters of cell types can be extracted, which demonstrates that the model has learned to distinguish individual cell types without supervision. In addition, the features extracted by the deep learning model can further reveal the unique cellular modules defining the different cell types (Zinchenko et al. 2023). In the study, different cell types were used, but similar techniques may be applied to track the different stages of development for a given cell type. In future, it will be exciting to apply such models to identify changes in GC identity during ovulation or to compare GC or oocyte features in follicles from young, old and diseased ovaries in an unbiased manner.
Theoretical approaches to study the physics of antrum formation and ovulation
In the past decades, various theoretical approaches have been developed to study tissue morphogenesis, wound healing and cancer progression (Tse et al. 2012; Jacques et al. 2023; Etournay et al. 2015; Salbreux et al. 2009; Tetley et al. 2019; Turley et al. 2022). These approaches modelled biological systems as active soft matter, where individual cells are able to convert chemical energy to execute various biological functions (Lou 2023; Marchetti et al. 2013; Fuji et al. 2022). Here, we review recent in silico work and biophysical approaches to model and measure tissue hydraulics, and propose new theoretical frameworks to investigate antrum formation and ovulation in late-stage folliculogenesis.
Computational models of luminogenesis
While the cellular dynamics in tissues have been modelled extensively, theoretical work on modelling lumen remains rather limited. Recently, a number of computational approaches have been developed to study lumen growth in development and organoids. One such approach is the phase field model, which can model cellular and lumen dynamics (Akiyama et al. 2018; Nonomura 2012; Fuji et al. 2022; Tanida et al. 2024). Phase field model has recently been applied to study the formation of MDCK cysts, pancreatic spheres and epiblasts, revealing a generic rule of the two-phase process of luminogenesis (Lu et al. 2024) characterised by lumen nucleation mediated by actin polymerization (Vasquez et al. 2021; Mukenhirn et al. 2023; Indana et al. 2024), and later expansion via osmotic gradient (Indana et al. 2024). The phase field model also reveals the interplay between cell proliferation and lumen pressure in changing organoid topology, as in the case of pancreatic spheres or branched networks (Lee et al. 2024a). Another popular approach in modelling luminogenesis is the Cellular Potts model (Graner and Glazier 1992; Hirashima et al. 2017). This technique has been applied to study organ cystogenesis (Belmonte et al. 2016; Engelberg et al. 2011) and development (Mombach et al. 2001). Recent work has extended the Cellular Potts model to include non-conservative forces arising from active cellular fluctuations (Belousov et al. 2024), which can be a novel approach to model non-equilibrium aspects of luminogenesis.
Potential phase transitions during antral folicle development
A key application of physics to the study of biological systems is the modelling of tissues as fluid- and solid-like materials, as reviewed in Lenne and Trivedi (2022). These theoretical models have predicted key cellular parameters that describe the mechanical states of tissues, and the critical points where phase transitions may occur. These parameters include cell elongation, active fluctuations and changes in extracellular space between the cells (Mongera et al. 2018; Petridou et al. 2021, 2019). An example is blastoderm spreading in early zebrafish development, where the center of the dome fluidises while the margin of the dome remains solid-like (Petridou et al. 2019). As the volume fraction of interstitial fluid increases and the cellular network breaks down, the marginal tissue becomes more fluid-like, leading to tissue flow. Here, we speculate that similar physics may apply to secondary follicles undergoing luminogenesis, where decreased GC packing may lead to increased tissue fluidisation (Biswas et al. 2022; Telfer et al. 2023). Tissue fluidization brings about another type of phase transition during antrum formation, which is the mixing-demixing transition in a fluid mixture. Here, the lumen and GCs could be considered effectively as a binary mixture of fluids, and luminogenesis may be modelled as liquid-liquid phase separation. While previous Cellular Potts model or phase field models have simulated lumen dynamics in various contexts, it is often assumed a priori that the systems exist far from criticality. Here, we hypothesise that the antrum formation in ovarian follicles may involve a phase transition from critical to supercritical states as the lumen undergoes fusion and maturation.
To test these models, it is essential to develop experimental approaches to measure and perturb tissue mechanics in follicles. Recently, 3D force sensors like polyacrylamide microbeads have been developed to measure mechanical stress distribution within cancer cell spheroids, revealing that an increased tissue pressure toward the inner core leads to inhibition of cell proliferation (Dolega et al. 2017; Taubenberger et al. 2019). Interestingly, tissue pressure has recently been shown to impact GC proliferation and follicle growth in the secondary follicle stage (Biswas et al. 2024, 2022), raising the intriguing possibility that intrafollicular pressure may impact subsequent events such as luminogenesis.
Biophysical control of ovulation
While it is well known that ovulation is triggered by hormonal signalling, the dynamics and mechanics of this process remain poorly characterised. Early studies have proposed that various mechanical factors, such as follicle volume increase, degradation of follicle wall and a build-up of hydrostatic pressure, may all be involved in ovulation (Matsuzaki 2021; Rondell 1970). However, these hypotheses remain to be tested. Here, a first characterisation of tissue mechanics during ovulation, such as changes in hydrostatic or osmotic pressure of the antrum by micropressure probes or pressure sensors may be instructive (Matousek et al. 2001; Espey and Lipner 1963; Bronson et al. 1979; Vian et al. 2023; Chan et al. 2021). Measurement of follicle wall tension or stiffness by micropipette aspiration or atomic force microscopy could also provide physical parameters to support model construction. Recently, new imaging-based tools to probe tissue stiffness have emerged. For example, optical coherence elastography (OCE), which is an extension of OCM (see the “Optical coherence tomography” section), relies on measuring local strain and stress to derive elasticity upon compressive load application (Li et al. 2021). OCE has been applied to study cancer cell metastasis, where the peripheral cells in breast cancer spheroids were shown to soften the surrounding ECM leading to invasive migration (Mowla et al. 2023). OCE has also revealed changes in mechanical properties of corpora lutea and follicles during ovarian ageing (Hepburn et al. 2024). Regardless, OCE measurements require the application of a load on the sample, which can result in changes of the sample itself. An alternative is the use of contact-free Brillouin microscopy, which measures the longitudinal modulus of tissue with submicron resolution (Prevedel et al. 2019). Recent work using Brillouin microscopy has revealed the emergence of distinct mechanical compartments within follicles during development (Fig. 2G) (Chan et al. 2021). With the recent advancement in Brillouin microscopy (Bevilacqua et al. 2023), rapid 3D live mapping of tissue material properties is now possible, paving the way for future application of such technique in studying basement membrane remodelling during ovulation.
An interesting biophysical aspect of ovulation is inspired by the physics of liquid crystals (Marchetti et al. 2013; Salbreux et al. 2009; Olenik et al. 2023; Colen et al. 2021; Popović et al. 2017). These frameworks are relevant when elongated cells align with each other and create long-range nematic order in tissues, as observed in fibroblasts, myoblasts and epithelial cells (Saw et al. 2017; Duclos et al. 2016; Sonam et al. 2022). In cases where apical-basal polarisation occurs, such polar order can further form topological defects in 3D, as observed during luminogenesis of the inner cell mass at the mouse pre-implantation stage (Guruciaga et al. 2024; Ichikawa et al. 2022). During Hydra regeneration, long, super-cellular actin bundles can form nematic order (Ravichandran et al. 2024; Maroudas-Sacks et al. 2021). The “closed” topology of this tissue implies that topological defects must be present. Interestingly, such topological defects have implications in Hydra development, where breaches only occur at these defects to relieve tissue pressure (Ravichandran et al. 2024), supporting the notion that topological defects can act as mechanical organisers during morphogenesis (Ravichandran et al. 2024). We propose that similar physics may apply to ovulation, where the spindle-like TCs may arrange into a nematic state and exhibit potential topological defects that facilitate follicle rupture during ovulation.
Conclusions and perspectives
In this review, we introduced the latest developments in microscopy, machine learning approach and theoretical models that may advance our understanding in late-stage mammalian folliculogenesis. We foresee that such techniques may lead to immediate new findings when applied to follicles grown ex vivo, but studying follicle development in vivo may pose additional challenges due to sample size and multiscale complexity. To move beyond classical histological and descriptive studies, and to have a more comprehensive and quantitative understanding of mammalian folliculogenesis, a multidisciplinary approach combining biophotonics, biophysics and machine learning is essential (Fig. 2).
Individually, the methods discussed above can provide insights but when integrated together they become more powerful. Advanced microscopy informs microscopic interactions between cells. This qualitative information can be transformed into quantified data using deep learning models, done accurately and efficiently on a large scale. Biophysical tools can be used to measure macroscopic properties of the tissue and theory can be deployed to understand how the microscopic interactions can produce macroscopic effects. Perturbations of the models can then be tested using the same quantitative tools to validate the theory. We envision this as an iterative process with advanced imaging experiments and analyses informing biophysical modelling, followed by a theoretical approach guiding further experiments and generating new hypotheses for testing. The use of novel biophotonic tools to directly infer tissue mechanics (e.g., Brillouin microscopy) also facilitates construction of biophysical models. Such an integrative framework to study 3D tissue dynamics will also be useful in the study of organoids and disease models (Beghin et al. 2022).
In addition to deepening our knowledge of female reproductive biology, a quantitative understanding of tissue hydraulics in late-stage folliculogenesis also has profound clinical implications. For example, it will be interesting to compare antrum formation and ovulation dynamics in the young and old follicles for possible biomechanical origin of infertility. Similarly, extending these approaches to ovarian disease models, such as PCOS ovaries, will shed light on the potential misregulation of tissue hydraulics in these systems.
Acknowledgements
The authors would like to acknowledge Lucie Kim (MBI, NUS) for help with graphical design. The authors thank Robert Prevedel, Tetsuya Hiraiwa, Yuting Lou, and Murat Shagirov for their valuable suggestions.
Author Contribution
This manuscript was conceived, literature search was performed, and written by JT, KWL, and CJC.
Funding
JT is supported by the Eric and Wendy Schmidt AI in Science Postdoctoral Fellowship. The Chan lab is supported by the Ministry of Education under the Research Centres of Excellence programme through the Mechanobiology Institute and the Department of Biological Sciences at the National University of Singapore, Ministry of Education Tier2 grant (T2EP30222-0026), National Research Foundation Mid-Size Grant (NRF-MSG-2023-0001) and the Bia-Echo Asia Centre for Reproductive Longevity and Equality (ACRLE) at the National University of Singapore. C.J.C. acknowledges the support of the Singaporean Teaching and Academic Research Talent Inauguration Grant (START).
Data Availability
Not applicable
Code Availability
Not applicable
Declarations
Competing of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jake Turley and Kim Whye Leong contributed equally to this work.
References
- Aguirre AD, Zhou C, Lee H-C, Ahsen OO, Fujimoto JG (2015) Optical coherence microscopy. In: Wolfgang Drexler and James G. Fujimoto, editors, Optical Coherence Tomography: Technology and Applications, pp 865–911. Springer International Publishing. ISBN 978-3-319-06419-2. URL 10.1007/978-3-319-06419-2_29 [DOI]
- Akiyama M, Nonomura M, Tero A, Kobayashi R (2018) Numerical study on spindle positioning using phase field method. Physical Biology 16(1):016005. ISSN 1478-3975. URL https://iopscience.iop.org/article/10.1088/1478-3975/aaee45 [DOI] [PubMed]
- Andres-San Roman JA, Gordillo-Vazquez C, Franco-Barranco D, Morato L, Fernandez-Espartero CH, Baonza G, Tagua A, Vicente-Munuera P, Palacios AM, Gavilán MP, Martín-Belmonte F (2023) CartoCell, a high-content pipeline for 3D image analysis, unveils cell morphology patterns in epithelia. Cell Reports Methods 3(10):100597. ISSN 26672375. URL https://linkinghub.elsevier.com/retrieve/pii/S2667237523002497 [DOI] [PMC free article] [PubMed]
- Athilingam T, Tiwari P, Toyama Y, Saunders TE (2021) Mechanics of epidermal morphogenesis in the Drosophila pupa. Seminars in Cell and Developmental Biology 120:171–180. ISSN 10963634. URL 10.1016/j.semcdb.2021.06.008. Publisher: Elsevier Ltd [DOI] [PubMed]
- Barer R (1952) Interference microscopy and mass determination. Nature 169(4296):366–367. URL 10.1038/169366b0. ISBN: 0028-0836 Publisher: Nature Publishing Group UK London [DOI] [PubMed]
- Barone V, Tagua A, Román JÁ, Hamdoun A, Garrido-García J, Lyons DC, Escudero LM (2024) Local and global changes in cell density induce reorganisation of 3D packing in a proliferating epithelium. Development. 10.1242/dev.202362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriga EH, Franze K, Charras G, Mayor R (2018) Tissue stiffening coordinates morphogenesis by triggering collective cell migration in vivo. Nature 554(7693):523–527. ISSN 0028-0836, 1476-4687. URL https://www.nature.com/articles/nature25742 [DOI] [PMC free article] [PubMed]
- Beghin A, Grenci G, Sahni G, Guo S, Rajendiran H, Delaire T, Mohamad Raffi SB, Blanc D, de Mets R, Ong HT, Galindo X, Monet A, Acharya V, Racine V, Levet F, Galland R, Sibarita JB, Viasnoff V (2022) Automated high-speed 3D imaging of organoid cultures with multi-scale phenotypic quantification. Nature Methods 19(7):881–892. ISSN 15487105. URL 10.1038/s41592-022-01508-0. Publisher: Nature Research [DOI] [PubMed]
- Belmonte JM, Clendenon SG, Oliveira GM, Swat MH, Greene EV, Jeyaraman S, Glazier JA, Bacallao RL (2016) Virtual-tissue computer simulations define the roles of cell adhesion and proliferation in the onset of kidney cystic disease. Molecular Biology of the Cell 27(22):3673–3685. ISSN 1059-1524, 1939-4586. URL https://www.molbiolcell.org/doi/10.1091/mbc.e16-01-0059 [DOI] [PMC free article] [PubMed]
- Belousov R, Savino S, Moghe P, Hiiragi T, Rondoni L, Erzberger A (2024) Poissonian cellular potts models reveal nonequilibrium kinetics of cell sorting. Phys Rev Lett 132(24):248401. ISSN 0031-9007, 1079-7114. URL https://link.aps.org/doi/10.1103/PhysRevLett.132.248401 [DOI] [PubMed]
- Bevilacqua C, Gomez JM, Fiuza UM, Chan CJ, Wang L, Hambura S, Eguren M, Ellenberg J, Diz-Muñoz A, Leptin M, Prevedel R (2023) High-resolution line-scan Brillouin microscopy for live imaging of mechanical properties during embryo development. Nat Methods 20(5):755–760. ISSN 15487105. URL 10.1038/s41592-023-01822-1. Publisher: Nature Research [DOI] [PMC free article] [PubMed]
- Biswas A, Ng BH, Prabhakaran VSO, Chan CJ (2022) Squeezing the eggs to grow: the mechanobiology of mammalian folliculogenesis. Frontiers in Cell and Developmental Biology 10. ISSN 2296634X. URL 10.3389/fcell.2022.1038107. Publisher: Frontiers Media S.A [DOI] [PMC free article] [PubMed]
- Biswas A, Lou Y, Ng BH, Tomida K, Darpe S, Wu Z, Lu TB, Bonne I, Chan CJ (2024) Theca cell mechanics and tissue pressure regulate mammalian ovarian folliculogenesis, May 2024. URL http://biorxiv.org/lookup/doi/10.1101/2024.05.06.592641
- Boylan CF, Sambo KM, Neal-Perry G, Brayboy LM (2024) Ex ovo omnia - why don’t we know more about egg quality via imaging? Biol Reprod 110(6):1201–1212, June 2024. ISSN 0006-3363, 1529-7268. URL https://academic.oup.com/biolreprod/article/110/6/1201/7676629 [DOI] [PMC free article] [PubMed]
- Bronson RA, Bryant G, Balk MW, Emanuele N (1979) Intrafollicular pressure within preovulatory follicles of the pig. Fertility and Sterility 31(2):205–213. ISSN 00150282. URL https://linkinghub.elsevier.com/retrieve/pii/S0015028216438240 [PubMed]
- Burton JC, Wang S, Stewart CA, Behringer RR, Larina IV (2015) High-resolution three-dimensional in vivo imaging of mouse oviduct using optical coherence tomography. Biomedical Optics Express 6(7):2713–2723. URL 10.1364/2FBOE.6.002713. ISBN: 2156-7085 Publisher: Optica Publishing Group [DOI] [PMC free article] [PubMed]
- Chan CJ, Hiiragi T (2020) Integration of luminal pressure and signalling in tissue self-organization. Development 147(5):dev181297. URL 10.1242/dev.181297. ISBN: 1477-9129 Publisher: The Company of Biologists Ltd [DOI] [PubMed]
- Chan CJ, Hirashima T (2022) Tissue hydraulics in reproduction. Seminars in Cell and Developmental Biology 131:124–133. ISSN 10963634. URL 10.1016/j.semcdb.2022.05.008. Publisher: Elsevier Ltd [DOI] [PubMed]
- Chan CJ, Bevilacqua C, Prevedel R (2021) Mechanical mapping of mammalian follicle development using Brillouin microscopy. Commun Biol 4(1):1133. ISSN 2399-3642. URL https://www.nature.com/articles/s42003-021-02662-5 [DOI] [PMC free article] [PubMed]
- Chattopadhyay A, Maitra M (2022) MRI-based brain tumour image detection using CNN based deep learning method. Neuroscience Informatics 2(4):100060. ISSN 27725286. URL https://linkinghub.elsevier.com/retrieve/pii/S277252862200022X
- Chow DJX, Tan TCY, Upadhya A, Lim M, Dholakia K, Dunning KR (2024) Viewing early life without labels: optical approaches for imaging the early embryo. Biology of Reproduction 110(6):1157–1174. ISSN 0006-3363, 1529-7268. URL https://academic.oup.com/biolreprod/article/110/6/1157/7655875 [DOI] [PMC free article] [PubMed]
- Christiansen EM, Yang SJ, Ando DM, Javaherian A, Skibinski G, Lipnick S, Mount E, O’neil A, Shah K, Lee AK (2018) In silico labeling: predicting fluorescent labels in unlabeled images. Cell 173(3):792–803. e19. URL 10.1016/j.cell.2018.03.040. ISBN: 0092-8674 Publisher: Elsevier [DOI] [PMC free article] [PubMed]
- Clarke HG, Hope SA, Byers S, Rodgers RJ (2006) Formation of ovarian follicular fluid may be due to the osmotic potential of large glycosaminoglycans and proteoglycans. Reproduction 132(1):119–131. URL 10.1530/rep.1.00960. ISBN: 1741-7899 Publisher: Society for Reproduction and Fertility [DOI] [PubMed]
- Colen J, Han M, Zhang R, Redford SA, Lemma LM, Morgan L, Ruijgrok PV, Adkins R, Bryant Z, Dogic Z, Gardel ML, De Pablo JJ, Vitelli V (2021) Machine learning active-nematic hydrodynamics. PNAS 118(10):e2016708118. 10.1073/pnas.2016708118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converse A, Zaniker EJ, Amargant F, Duncan FE (2023) Recapitulating folliculogenesis and oogenesis outside the body: encapsulated in vitro follicle growth. Biology of Reproduction 108(1):5–22. ISSN 0006-3363, 1529-7268. URL https://academic.oup.com/biolreprod/article/108/1/5/6711545 [DOI] [PMC free article] [PubMed]
- Culley S, Caballero AC, Burden JJ, Uhlmann V (2023) Made to measure: an introduction to quantifying microscopy data in the life sciences. Journal of Microscopy. ISSN 13652818. URL 10.1111/jmi.13208. Publisher: John Wiley and Sons Inc [DOI] [PubMed]
- Dolega ME, Delarue M, Ingremeau F, Prost J, Delon A, Cappello G (2017) Cell-like pressure sensors reveal increase of mechanical stress towards the core of multicellular spheroids under compression. Nat Commun 8(1):14056. ISSN 2041-1723. URL https://www.nature.com/articles/ncomms14056 [DOI] [PMC free article] [PubMed]
- Duclos G, Erlenkämper C, Joanny JF, Silberzan P (2016) Topological defects in confined populations of spindle-shaped cells. Nature Physics 13(1):58–62. ISSN 17452481. URL 10.1038/nphys3876. Publisher: Nature Publishing Group
- Engelberg JA, Datta A, Mostov KE, Hunt CA (2011) MDCK cystogenesis driven by cell stabilization within computational analogues. PLoS Computational Biology 7(4):e1002030, April 2011. ISSN 1553-7358. URL https://dx.plos.org/10.1371/journal.pcbi.1002030 [DOI] [PMC free article] [PubMed]
- Espey LL, Lipner H (1963) Measurements of intrafollicular pressures in the rabbit ovary. American Journal of Physiology-Legacy Content 205(6):1067–1072. ISSN 0002-9513. URL https://www.physiology.org/doi/10.1152/ajplegacy.1963.205.6.1067 [DOI] [PubMed]
- Etournay R, Popović M, Merkel M, Nandi A, Blasse C, Aigouy B, Brandl H, Myers G, Salbreux G, Jülicher F, Eaton S (2015) Interplay of cell dynamics and epithelial tension during morphogenesis of the Drosophila pupal wing. ELife 4:e07090. 10.7554/eLife.07090.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett M, Magazzeni S, Schmoll T, Kempe M (2021) Optical coherence tomography: from technology to applications in ophthalmology. Translational Biophotonics 3(1):e202000012. URL 10.1002/tbio.202000012. ISBN: 2627-1850 Publisher: Wiley Online Library [DOI]
- Feng Y, Tamadon A, Hsueh AJW (2018) Imaging the ovary. Reproductive biomedicine online 36(5):584–593. 10.1016/j.rbmo.2018.02.006. ISBN: 1472-6483 Publisher: Elsevier [DOI] [PubMed]
- Filan C, Charles S, Costa PC, Niu W, Cheng BF, Wen Z, Lu H, Robles FE (2024) Non-invasive label-free analysis pipeline for in situ characterization of differentiation in 3d brain organoid models. Research square. URL 10.21203/rs.3.rs-4049577/v1. Publisher: American Journal Experts
- Fiorentino G, Cimadomo D, Innocenti F, Soscia D, Vaiarelli A, Ubaldi FM, Gennarelli G, Garagna S, Rienzi L, Zuccotti M (2023) Biomechanical forces and signals operating in the ovary during folliculogenesis and their dysregulation: implications for fertility. Human Reproduction Update 29(1):1–23, January 2023. ISSN 14602369. URL 10.1093/humupd/dmac031. Publisher: Oxford University Press [DOI] [PubMed]
- Ford TN, Chu KK, Mertz J (2012) Phase-gradient microscopy in thick tissue with oblique back-illumination. Nature Methods 9(12):1195–1197. 10.1038/nmeth.2219. ISBN: 1548-7091 Publisher: Nature Publishing Group US New York [DOI] [PMC free article] [PubMed]
- Fuji K, Tanida S, Sano M, Nonomura M, Riveline D, Honda H, Hiraiwa T (2022) Computational approaches for simulating luminogenesis. Seminars in Cell & Developmental Biology 131:173–185. ISSN 10849521. URL https://linkinghub.elsevier.com/retrieve/pii/S108495212200180X [DOI] [PubMed]
- Fujimoto JG (2003) Optical coherence tomography for ultrahigh resolution in vivo imaging. Nat Biotechnol 21(11):1361–1367. 10.1038/nbt892. ISBN: 1546-1696 Publisher: Nature Publishing Group [DOI] [PubMed]
- Gosden RG, Brown N, Grant K (1989) Ultrastructural and histochemical investigations of call-exner bodies in rabbit graafian follicles. Reproduction 85(2):519–526. 10.1530/jrf.0.0850519. ISBN: 0022-4251 Publisher: Bioscientifica Ltd [DOI] [PubMed]
- Graner F, Glazier JA (1992) Simulation of biological cell sorting using a two-dimensional extended Potts model. Physical Review Letters 69(13). URL 10.1103/PhysRevLett.69.2013 [DOI] [PubMed]
- Guruciaga PC, Ichikawa T, Hiiragi T, Erzberger A (2024) Boundary geometry controls topological defect transitions that determine lumen nucleation in embryonic development, March 2024. URL http://arxiv.org/abs/2403.08710. arXiv:2403.08710 [cond-mat, physics:physics]
- Hallou A, Yevick HG, Dumitrascu B, Uhlmann V (2021) Deep learning for bioimage analysis in developmental biology. Development (Cambridge) 148(18). ISSN 14779129. URL 10.1242/DEV.199616. Publisher: Company of Biologists Ltd [DOI] [PMC free article] [PubMed]
- Hepburn M, Jaeschke A, Mowla A, Chan CJ, Kennedy BF (2024) Three-dimensional characterization of murine ovary elasticity using quantitative micro-elastography. In: Optical elastography and tissue biomechanics XI, pp PC128440B. SPIE, March 2024. URL 10.1117/12.3006718
- Hirashima T, Rens EG, Merks RMH (2017) Cellular Potts modeling of complex multicellular behaviors in tissue morphogenesis. Development Growth and Differentiation 59(5):329–339. ISSN 1440169X. URL 10.1111/dgd.12358. Publisher: Blackwell Publishing [DOI] [PubMed]
- Howard J, Gugger S (2020) Deep learning for coders with fastai and PyTorch. O’Reilly Media
- Ichbiah S, Delbary F, McDougall A, Dumollard R, Turlier H (2023) Embryo mechanics cartography: inference of 3D force atlases from fluorescence microscopy. Nature Methods 20(12), 1989–1999. ISSN 1548–7091:1548–7105. 10.1038/s41592-023-02084-7. URL https://www.nature.com/articles/s41592-023-02084-7 [DOI] [PMC free article] [PubMed]
- Ichikawa T, Zhang HT, Panavaite L, Erzberger A, Fabréges D, Snajder R, Wolny A, Korotkevich E , Tsuchida-Straeten N, Hufnagel L, Kreshuk A, Hiiragi T (2022) An ex vivo system to study cellular dynamics underlying mouse peri-implantation development. Developmental Cell 57(3):373–386.e9, February 2022. ISSN 15345807. URL https://linkinghub.elsevier.com/retrieve/pii/S1534580721010431 [DOI] [PMC free article] [PubMed]
- Işin A, Direkoǧlu C, Şah M (2016) Review of MRI-based brain tumor image segmentation using deep learning methods. In: Procedia computer science, vol 102, pp 317–324. Elsevier B.V., 2016. URL 10.1016/j.procs.2016.09.407. ISSN: 18770509 [DOI]
- Indana D, Zakharov A, Lim Y, Dunn AR, Bhutani N, Shenoy VB, Chaudhuri O (2024) Lumen expansion is initially driven by apical actin polymerization followed by osmotic pressure in a human epiblast model. Cell Stem Cell 31(5):640–656.e8, May 2024. ISSN 19345909. URL https://linkinghub.elsevier.com/retrieve/pii/S1934590924000973 [DOI] [PMC free article] [PubMed]
- Jacques C, Ackermann J, Bell S, Hallopeau C, Perez-Gonzalez C, Balasubramaniam L, Trepat X, Ladoux B, Maitra A , Voituriez R, Matic Vignjevic D (2023) Aging and freezing of active nematic dynamics of cancer-associated fibroblasts by fibronectin matrix remodeling. preprint, Cell Biology. URL http://biorxiv.org/lookup/doi/10.1101/2023.11.22.568216
- Jo YJ, Cho H, Park WS, Kim G, Ryu D, Kim YS, Lee M, Park S, Lee MJ, Joo H, Jo H (2021) Label-free multiplexed microtomography of endogenous subcellular dynamics using generalizable deep learning. Nature Cell Biology 23(12):1329–1337. ISSN 14764679. URL 10.1038/s41556-021-00802-x. Publisher: Nature Research [DOI] [PubMed]
- Jones W, Alasoo K, Fishman D, Parts L (2017) Computational biology: deep learning. Emerging Topics in Life Sciences 1(3):257–274. ISSN 23978562. URL 10.1042/ETLS20160025. Publisher: Portland Press Ltd [DOI] [PMC free article] [PubMed]
- Kandel ME, Hu C, Naseri Kouzehgarani G, Min E, Sullivan KM, Kong H, Li JM, Robson DN , Gillette MU, Best-Popescu C, Popescu G (2019) Epi-illumination gradient light interference microscopy for imaging opaque structures. Nature Communications 10(1):4691. ISSN 2041-1723. URL 10.1038/s41467-019-12634-3. Publisher: Nature Publishing Group [DOI] [PMC free article] [PubMed]
- Karniadakis GE, Kevrekidis IG, Lu L, Perdikaris P, Wang S, Yang L (2021) Physics-informed machine learning. Nature Reviews Physics 3(6):422–440. ISSN 2522-5820. URL https://www.nature.com/articles/s42254-021-00314-5
- Kim K, Choe K, Park I, Kim P, Park YK (2016) Holographic intravital microscopy for 2-d and 3-d imaging intact circulating blood cells in microcapillaries of live mice. Scientific Reports 6(1):33084. ISSN 2045-2322. URL 10.1038/srep33084. Publisher: Nature Publishing Group [DOI] [PMC free article] [PubMed]
- Komatsu K, Masubuchi S (2017) Observation of the dynamics of follicular development in the ovary. Reproductive Medicine and Biology 16(1):21–27. ISSN 1445-5781, 1447-0578. URL https://onlinelibrary.wiley.com/doi/10.1002/rmb2.12010 [DOI] [PMC free article] [PubMed]
- Komatsu K, Iwase A, Murase T, Masubuchi S (2018) Ovarian tissue culture to visualize phenomena in mouse ovary. Journal of Visualized Experiments 136:57794,. ISSN 1940-087X. URL https://www.jove.com/t/57794/ovarian-tissue-culture-to-visualize-phenomena-in-mouse-ovary [DOI] [PMC free article] [PubMed]
- Lafarge MW, Caicedo JC, Carpenter AE, Pluim JP, Singh S, Veta M (2019) capturing single-cell phenotypic variation via unsupervised representation learning. Proceedings of Machine Learning Research. URL https://proceedings.mlr.press/v102/lafarge19a.html [PMC free article] [PubMed]
- Ledwig P, Robles FE (2019) Epi-mode tomographic quantitative phase imaging in thick scattering samples. Biomedical Optics Express 10(7):3605–3621. 10.1364/boe.10.003605. ISBN: 2156-7085 Publisher: Optica Publishing Group [DOI] [PMC free article] [PubMed]
- Lee BH, Fuji K, Petzold H, Seymour P, Yennek S, Schewin C, Riveline D, Hiraiwa T, Sano M, Grapin-Botton A (2024a) Control of lumen geometry and topology by cell proliferation rate and pressure. bioRxiv. URL 10.1101/2024.05.29.596462 [DOI]
- Lee C, Kim G, Shin T, Lee S, Kim JY, Choi KH, Do J, Park J, Do J, Kim JH (2024) Noninvasive time-lapse 3d subcellular analysis of embryo development for machine learning-enabled prediction of blastocyst formation. bioRxiv, page 2024.05. 07.592317. URL 10.1101/2024.05.07.592317. Publisher: Cold Spring Harbor Laboratory [DOI]
- Lee S, Arffman RK, Komsi EK, Lindgren O, Kemppainen J, Kask K, Saare M, Salumets A, Piltonen TT (2024) Dynamic changes in AI-based analysis of endometrial cellular composition: analysis of PCOS and RIF endometrium. Journal of Pathology Informatics 15:100364. ISSN 21533539. URL https://linkinghub.elsevier.com/retrieve/pii/S2153353924000038 [DOI] [PMC free article] [PubMed]
- Lee MJ, Lee J, Ha J, Kim G, Kim H-J, Lee S, Koo B-K, Park YK (2023) Long-term three-dimensional high-resolution imaging of live unlabeled small intestinal organoids using low-coherence holotomography. bioRxiv, page 2023.09. 16.558039. URL 10.1101/2023.09.16.558039. Publisher: Cold Spring Harbor Laboratory [DOI] [PMC free article] [PubMed]
- Lefebvre M, Colen J, Claussen N, Brauns F, Raich M, Mitchell N, Fruchart M, Vitelli V, Streichan J (2024) Learning a conserved mechanism for early neuroectoderm morphogenesis. arXiv. URL 10.1101/2023.12.22.573058
- Lenne PF, Trivedi V (2022) Sculpting tissues by phase transitions. Nature Communications 13(1). ISSN 20411723. URL 10.1038/s41467-022-28151-9. Publisher: Nature Research [DOI] [PMC free article] [PubMed]
- Li J, Foo KY, Hepburn MS, Mowla A, Chin L, Kennedy BF (2021) Compression optical coherence elastography. In: Optical coherence elastography: imaging tissue mechanics on the micro-scale. AIP Publishing LLC. ISBN 978-0-7354-2364-0. URL 10.1063/9780735423664_007
- Lou Y (2023) Appetizer on soft matter physics concepts in mechanobiology. Development, Growth & Differentiation 65(5):234–244. ISSN 0012-1592, 1440-169X. URL 10.1111/dgd.12853 [DOI] [PMC free article] [PubMed]
- Lou Y, Rupprecht JF, Theis S, Hiraiwa T, Saunders TE (2023) Curvature-induced cell rearrangements in biological tissues. Physical Review Letters 130(10):108401. ISSN 0031-9007, 1079-7114. URL 10.1103/PhysRevLett.130.108401 [DOI] [PubMed]
- Lu AX, Kraus OZ, Cooper S, Moses AM (2019) Learning unsupervised feature representations for single cell microscopy images with paired cell inpainting. PLOS Computational Biology 15(9):e1007348. ISSN 1553-7358. URL 10.1371/journal.pcbi.1007348 [DOI] [PMC free article] [PubMed]
- Lu L, Fuji K, Guyomar T, Lieb M, Tanida S, Nonomura M, Hiraiwa T, Alcheikh Y, Yennek S, Petzold H, Martin-Lemaitre C, Grapin-Botton A, Honigmann A, Sano M, Riveline D (2024) Generic rules of lumen nucleation and fusion in epithelial organoids. URL http://biorxiv.org/lookup/doi/10.1101/2024.02.20.581158
- Mara JN, Zhou LT, Larmore M, Johnson B, Ayiku R, Amargant F, Pritchard MT, Duncan FE (2020) Ovulation and ovarian wound healing are impaired with advanced reproductive age. Aging 12(10):9686–9713. ISSN 1945-4589. URL https://www.aging-us.com/lookup/doi/10.18632/aging.103237 [DOI] [PMC free article] [PubMed]
- Marchetti MC, Joanny JF, Ramaswamy S, Liverpool TB, Prost J, Rao M, Aditi Simha R (2013) Hydrodynamics of soft active matter. Reviews of Modern Physics 85(3):1143–1189. ISSN 00346861. URL 10.1103/RevModPhys.85.1143
- Maroudas-Sacks Y, Garion L, Shani-Zerbib L, Livshits A, Braun E, Keren K (2021) Topological defects in the nematic order of actin fibres as organization centres of Hydra morphogenesis. Nature Physics 17(2):251–259. ISSN 1745-2473, 1745-2481. URL https://www.nature.com/articles/s41567-020-01083-1
- Matousek M, Carati C, Gannon B, Brännström M (2001) Novel method for intrafollicular pressure measurements in the rat ovary: increased intrafollicular pressure after hCG stimulation. Reproduction. URL 10.1530/rep.0.1210307 [DOI] [PubMed]
- Matsuzaki S (2021) Mechanobiology of the female reproductive system. Reproductive Medicine and Biology 20(4):371–401. ISSN 1445-5781, 1447-0578. URL https://onlinelibrary.wiley.com/doi/10.1002/rmb2.12404 [DOI] [PMC free article] [PubMed]
- McDole K, Guignard L, Amat F, Berger A, Malandain G, Royer LA, Turaga SC, Branson K, Keller PJ (2018) In toto imaging and reconstruction of post-implantation mouse development at the single-cell level. Cell 175(3):859–876.e33, October 2018. ISSN 10974172. URL 10.1016/j.cell.2018.09.031. Publisher: Cell Press [DOI] [PubMed]
- Migone FF, Cowan RG, Williams RM, Zipfel WR, Quirk SM (2013) Multiphoton microscopy as a tool to study ovarian vasculature in vivo. Intravital 2(1):e24334. 10.4161/intv.24334. ISBN: 2165-9087 Publisher: Taylor & Francis
- Migone FF, Cowan RG, Williams RM, Gorse KJ, Zipfel WR, Quirk SM (2016) In vivo imaging reveals an essential role of vasoconstriction in rupture of the ovarian follicle at ovulation. Proceedings of the National Academy of Sciences 113(8):2294–2299. ISSN 0027-8424, 1091-6490. 10.1073/pnas.1512304113. URL https://pnas.org/doi/full/10.1073/pnas.1512304113 [DOI] [PMC free article] [PubMed]
- Mitchell NP, Cislo DJ (2023) TubULAR: tracking in toto deformations of dynamic tissues via constrained maps. Nature Methods 20(12):1980–1988. ISSN 1548-7091, 1548-7105. URL https://www.nature.com/articles/s41592-023-02081-w [DOI] [PMC free article] [PubMed]
- Mombach JCM, De Almeida RMC, Thomas GL, Upadhyaya A, Glazier JA (2001) Bursts and cavity formation in Hydra cells aggregates: experiments and simulations. Physica A: Statistical Mechanics and its Applications 297(3-4):495–508. ISSN 03784371. URL https://linkinghub.elsevier.com/retrieve/pii/S0378437101001996
- Mongera A, Rowghanian P, Gustafson HJ, Shelton E, Kealhofer DA, Carn EK, Serwane F, Lucio AA, Giammona J, Campás O (2018) A fluid-to-solid jamming transition underlies vertebrate body axis elongation. Nature 561(7723):401–405. ISSN 14764687. URL 10.1038/s41586-018-0479-2. Publisher: Nature Publishing Group [DOI] [PMC free article] [PubMed]
- Moore EL, Wang S, Larina IV (2019) Staging mouse preimplantation development in vivo using optical coherence microscopy. Journal of Biophotonics 12(5):e201800364. 10.1002/jbio.201800364. ISBN: 1864-063X Publisher: Wiley Online Library [DOI] [PMC free article] [PubMed]
- Mowla A, Hepburn MS, Li J, Hirvonen LM, Vahala D, Amos S, Maher S, Choi YS, Kennedy BF (2023) Multimodal optical coherence microscopy, mechano-microscopy and fluorescence microscopy for three-dimensional characterization of multicellular spheroids. In: European conference on biomedical optics, pp 1263219. Optica Publishing Group. URL 10.1117/12.2670830
- Mowla A, Hepburn MS, Li J, Vahala D, Amos SE, Hirvonen LM, Sanderson RW, Wijesinghe P, Maher S, Choi YS (2024) Multimodal mechano-microscopy reveals mechanical phenotypes of breast cancer spheroids in three dimensions. bioRxiv, page 2024.04. 05.588260, April 2024. URL 10.1101/2024.04.05.588260. Publisher: Cold Spring Harbor Laboratory [DOI] [PMC free article] [PubMed]
- Mukenhirn M, Wang C-H, Guyomar T, Bovyn MJ, Staddon MF, Maraspini R, Lu L, Martin-Lemaitre C, Sano M, Hiraiwa T, Riveline D, Honigmann A (2023) Tight junctions regulate lumen morphology via hydrostatic pressure and junctional tension, May 2023. URL http://biorxiv.org/lookup/doi/10.1101/2023.05.23.541893 [DOI] [PubMed]
- Nagamatsu G, Shimamoto S, Hamazaki N, Nishimura Y, Hayashi K (2019) Mechanical stress accompanied with nuclear rotation is involved in the dormant state of mouse oocytes. Science Advances 5(6):eaav9960. ISSN 2375-2548. URL https://www.science.org/doi/10.1126/sciadv.aav9960 [DOI] [PMC free article] [PubMed]
- Nguyen TH, Kandel ME, Rubessa M, Wheeler MB, Popescu G (2017) Gradient light interference microscopy for 3d imaging of unlabeled specimens. Nature Communications 8(1):210. 10.1038/s41467-017-00190-7. ISBN: 2041-1723 Publisher: Nature Publishing Group UK London [DOI] [PMC free article] [PubMed]
- Nguyen TL, Pradeep S, Judson-Torres RL, Reed J, Teitell MA, Zangle TA (2022) Quantitative phase imaging: recent advances and expanding potential in biomedicine. ACS Nano 16(8):11516–11544. 10.1021/acsnano.1c11507. ISBN: 1936-0851 Publisher: ACS Publications [DOI] [PMC free article] [PubMed]
- Nonomura M (2012) Study on multicellular systems using a phase field model. PLoS ONE 7(4):e33501. ISSN 1932-6203. URL 10.1371/journal.pone.0033501 [DOI] [PMC free article] [PubMed]
- Olenik M, Turley J, Cross S, Weavers H, Martin P, Chenchiah IV, Liverpool TB (2023) Fluctuations of cell geometry and their nonequilibrium thermodynamics in living epithelial tissue. Physical Review E 107(1). ISSN 24700053. URL 10.1103/PhysRevE.107.014403. arXiv:2201.07154 Publisher: American Physical Society [DOI] [PubMed]
- Owen CM, Jaffe LA (2023) Luteinizing hormone stimulates ingression of mural granulosa cells within the mouse preovulatory follicle. BioRxiv. 10.1101/2023.04.21.537855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KJ, Bai B, Ryu DH, Liu T, Lee C, Luo Y, Lee MJ, Huang L, Shin J, Zhang Y, Ryu D, Li Y, Kim G, Min HS, Ozcan A, Park YK (2023a) Artificial intelligence-enabled quantitative phase imaging methods for life sciences. Nature Methods. ISSN 1548-7091. URL https://www.nature.com/articles/s41592-023-02041-4 [DOI] [PubMed]
- Park J, Shin SJ, Kim M, Kim G, Cho H, Ryu D, Ahn D, Heo JE, Jang I, Min HS (2023b) Revealing 3d cancer tissue structures using holotomography and virtual hematoxylin and eosin staining via deep learning. bioRxiv, page 2023.12. 04.569853. URL 10.1101/2023.12.04.569853. Publisher: Cold Spring Harbor Laboratory
- Park Y, Depeursinge C, Popescu G (2018) Quantitative phase imaging in biomedicine. Nature Photonics 12(10):578–589. 10.1038/s41566-018-0253-x. ISBN: 1749-4885 Publisher: Nature Publishing Group UK London
- Peters ITA, Stegehuis PL, Peek R, Boer FL, van Zwet EW, Eggermont J, Westphal JR, Kuppen PJK, Trimbos JB, Hilders CGJM (2016) Noninvasive detection of metastases and follicle density in ovarian tissue using full-field optical coherence tomography. Clinical Cancer Research 22(22):5506–5513. 10.1158/1078-0432.ccr-16-0288. ISBN: 1078-0432 Publisher: AACR [DOI] [PubMed]
- Petridou NI, Grigolon S, Salbreux G, Hannezo E, Heisenberg CP (2019) Fluidization-mediated tissue spreading by mitotic cell rounding and non-canonical Wnt signalling. Nature Cell Biology 21(2):169–178. ISSN 14764679. URL 10.1038/s41556-018-0247-4. Publisher: Nature Publishing Group [DOI] [PubMed]
- Petridou NI, Corominas-Murtra B, Heisenberg CP, Hannezo E (2021) Rigidity percolation uncovers a structural basis for embryonic tissue phase transitions. Cell 184(7):1914–1928.e19. ISSN 10974172. URL 10.1016/j.cell.2021.02.017. Publisher: Elsevier B.V [DOI] [PMC free article] [PubMed]
- Pietroforte S, Plough M, Amargant F (2024) Age-associated increased stiffness of the ovarian microenvironment impairs follicle development and oocyte quality and rapidly alters follicle gene expression, June 2024. URL http://biorxiv.org/lookup/doi/10.1101/2024.06.09.598134
- Popović M, Nandi A, Merkel M, Etournay R, Eaton S, Jülicher F, Salbreux G (2017) Active dynamics of tissue shear flow. New Journal of Physics 19(3). ISSN 13672630. URL 10.1088/1367-2630/aa5756. arXiv:1607.03304 Publisher: Institute of Physics Publishing
- Prasasya RD, Mayo KE (2019) Regulation of follicle formation and development by ovarian signaling pathways. In: The ovary, pp 23–49. Elsevier, 2019. ISBN 978-0-12-813209-8. URL https://linkinghub.elsevier.com/retrieve/pii/B9780128132098000029
- Prevedel R, Diz-Muñoz A, Ruocco G, Antonacci G (2019) Brillouin microscopy: an emerging tool for mechanobiology. Nature Methods 16(10):969–977. ISSN 15487105. URL 10.1038/s41592-019-0543-3. Publisher: Nature Publishing Group [DOI] [PubMed]
- Ravichandran Y, Vogg M, Kruse K, Pearce DJ, Roux A (2024) Topology changes of the regenerating Hydra define actin nematic defects as mechanical organizers of morphogenesis, April 2024. URL http://biorxiv.org/lookup/doi/10.1101/2024.04.07.588499
- Rodgers RJ, Irving-Rodgers HF (2010) Formation of the ovarian follicular antrum and follicular fluid. Biology of Reproduction 82(6):1021–1029. ISSN 00063363. URL 10.1095/biolreprod.109.082941 [DOI] [PubMed]
- Rondell P (1970) Biophysical aspects of ovulation. Biology of Reproduction 2(suppl_2):64–89. ISSN 0006-3363, 1529-7268. URL https://academic.oup.com/biolreprod/article/2768848/Biophysical [PubMed]
- Ruperti F, Becher I, Stokkermans A, Wang L, Marschlich N, Potel C, Maus E, Stein F, Drotleff B, Schippers KJ, Nickel M , Prevedel R, Musser JM, Savitski MM, Arendt D (2024) Molecular profiling of sponge deflation reveals an ancient relaxant-inflammatory response. Current Biology 34(2):361–375.e9. ISSN 09609822. URL https://linkinghub.elsevier.com/retrieve/pii/S0960982223016767 [DOI] [PubMed]
- Salbreux G, Prost J, Joanny JF (2009) Hydrodynamics of cellular cortical flows and the formation of contractile rings. Physical Review Letters 103(5). ISSN 00319007. URL 10.1103/PhysRevLett.103.058102 [DOI] [PubMed]
- Saw TB, Doostmohammadi A, Nier V, Kocgozlu L, Thampi S, Toyama Y, Marcq P, Lim CT, Yeomans JM, Ladoux B (2017) Topological defects in epithelia govern cell death and extrusion. Nature 544(7649):212–216. ISSN 14764687. URL 10.1038/nature21718. Publisher: Nature Publishing Group [DOI] [PMC free article] [PubMed]
- Schmitt MS, Colen J, Sala S, Devany J, Seetharaman S, Gardel ML, Oakes PW, Vitelli V (2023) Zyxin is all you need: machine learning adherent cell mechanics. arXiv. URL http://arxiv.org/abs/2303.00176. arXiv:2303.00176 [DOI] [PMC free article] [PubMed]
- Schmitt MS, Colen J, Sala S, Devany J, Seetharaman S, Caillier A, Gardel ML, Oakes PW, Vitelli V (2024) Machine learning interpretable models of cell mechanics from protein images. Cell 187(2):481–494.e24. ISSN 00928674. URL https://linkinghub.elsevier.com/retrieve/pii/S0092867423013314 [DOI] [PMC free article] [PubMed]
- Schürmann M, Scholze J, Müller P, Guck J, Chan CJ (2016) Cell nuclei have lower refractive index and mass density than cytoplasm. Journal of Biophotonics 9(10):1068–1076. 10.1002/jbio.201500273. ISBN: 1864-063X Publisher: Wiley Online Library [DOI] [PubMed]
- Singh SP, Wang L, Gupta S, Goli H, Padmanabhan P, Gulyás B (2020) 3D deep learning on medical images: a review. Sensors 20(18):5097. ISSN 1424-8220. URL https://www.mdpi.com/1424-8220/20/18/5097 [DOI] [PMC free article] [PubMed]
- Sonam S, Balasubramaniam L, Lin SZ, Ivan YMY, Pi-Jaumá I, Jebane C, Karnat M, Toyama Y, Marcq P, Prost J, Mége R-M, Rupprecht J-F, Ladoux B (2022) Mechanical stress driven by rigidity sensing governs epithelial stability. Nature Physics 19(1):132–141. ISSN 1745-2473, 1745-2481. URL https://www.nature.com/articles/s41567-022-01826-2 [DOI] [PMC free article] [PubMed]
- Stokkermans A, Chakrabarti A, Subramanian K, Wang L, Yin S, Moghe P, Steenbergen P, Mönke G, Hiiragi T, Prevedel R, Mahadevan L, Ikmi A (2022) Muscular hydraulics drive larva-polyp morphogenesis. Current Biology 32(21):4707–4718.e8. ISSN 09609822. URL https://linkinghub.elsevier.com/retrieve/pii/S0960982222013872 [DOI] [PubMed]
- Stroka KM, Aranda-Espinoza H (2011) Effects of morphology vs. cell-cell interactions on endothelial cell stiffness. Cellular and Molecular Bioengineering 4(1):9–27. ISSN 1865-5025, 1865-5033. URL http://link.springer.com/10.1007/s12195-010-0142-y [DOI] [PMC free article] [PubMed]
- Takae S, Tsukada K, Sato Y, Okamoto N, Kawahara T, Suzuki N (2017) Accuracy and safety verification of ovarian reserve assessment technique for ovarian tissue transplantation using optical coherence tomography in mice ovary. Scientific Reports 7(1):43550. 10.1038/srep43550. ISBN: 2045-2322 Publisher: Nature Publishing Group UK London [DOI] [PMC free article] [PubMed]
- Takae S, Tsukada K, Maeda I, Okamoto N, Sato Y, Kondo H, Shinya K, Motani Y, Suzuki N (2018) Preliminary human application of optical coherence tomography for quantification and localization of primordial follicles aimed at effective ovarian tissue transplantation. Journal of Assisted Reproduction and Genetics 35:627–636. 10.1007/s10815-018-1166-9. ISBN: 1058-0468 Publisher: Springer [DOI] [PMC free article] [PubMed]
- Tanida S, Fuji K, Lu L, Guyomar T, Lee BH, Honigmann A, Grapin-Botton A, Riveline D, Hiraiwa T, Nonomura M, Sano M (2024) Predicting organoid morphology through a phase field model: insights into cell division and lumenal pressure. URL http://biorxiv.org/lookup/doi/10.1101/2024.04.22.590518
- Taubenberger AV, Girardo S, Träber N, Fischer E, Kräter M, Wagner K, Kurth T, Richter I, Binner M, Hahn D, Freudenberg U, Werner C, Guck J (2019) 3D microenvironment stiffness regulates tumor spheroid growth and mechanics via p21 and ROCK. bioRxiv. URL 10.1101/586784 [DOI] [PubMed]
- Telfer EE, Grosbois J, Odey YL, Rosario R, Anderson RA (2023) Making a good egg: human oocyte health, aging, and in vitro development. Physiological Reviews 103(4):2623–2677. ISSN 15221210. URL 10.1152/physrev.00032.2022. Publisher: American Physiological Society [DOI] [PMC free article] [PubMed]
- Tetley RJ, Staddon MF, Heller D, Hoppe A, Banerjee S, Mao Y (2019) Tissue fluidity promotes epithelial wound healing. Nature Physics 15(11):1195–1203. ISSN 17452481. URL 10.1038/s41567-019-0618-1. Publisher: Nature Publishing Group [DOI] [PMC free article] [PubMed]
- Tse JM, Cheng G, Tyrrell JA, Wilcox-Adelman SA, Boucher Y, Jain RK, Munn LL (2012) Mechanical compression drives cancer cells toward invasive phenotype. Proceedings of the National Academy of Sciences 109(3):911–916. ISSN 0027-8424, 1091-6490. URL https://pnas.org/doi/full/10.1073/pnas.1118910109 [DOI] [PMC free article] [PubMed]
- Turley J, Chenchiah IV, Liverpool TB, Weavers H, Martin P (2022) iScience What good is maths in studies of wound healing? ISCIENCE 25:104778. 10.1016/j.isci [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turley J, Chenchiah IV, Martin P, Liverpool TB, Weavers H (2024a) Deep learning for rapid analysis of cell divisions in vivo during epithelial morphogenesis and repair. eLife, 12, p RP87949. 10.7554/eLife.87949.3 [DOI] [PMC free article] [PubMed]
- Turley J, Robertson F, Chenchiah IV, Liverpool TB, Weavers H, Martin P (2024b) Deep learning reveals a damage signalling hierarchy that coordinates different cell behaviours driving wound re-epithelialisation. Development, 10.1242/dev.202943 [DOI] [PMC free article] [PubMed]
- Umehara T, Winstanley YE, Andreas E, Morimoto A, Williams EJ, Smith KM, Carroll J, Febbraio MA, Shimada M, Russell DL, Robker RL (2022) Female reproductive life span is extended by targeted removal of fibrotic collagen from the mouse ovary. Science Advances, pp 4564. URL https://www.science.org. Publication Title: Sci. Adv vol 8 [DOI] [PMC free article] [PubMed]
- Umezu K, Larina IV (2023) Optical coherence tomography for dynamic investigation of mammalian reproductive processes. Molecular Reproduction and Development 90(1):3–13. 10.1002/mrd.23665. ISBN: 1040-452X Publisher: Wiley Online Library [DOI] [PMC free article] [PubMed]
- Van Wezel IL, Irving-Rodgers HF, Sado Y, Ninomiya Y, Rodgers R (1999) Ultrastructure and composition of Call-Exner bodies in bovine follicles. Springer 296. 10.1007/s004410051298 [DOI] [PubMed]
- Vasquez CG, Vachharajani VT, Garzon-Coral C, Dunn AR (2021) Physical basis for the determination of lumen shape in a simple epithelium. Nature Communications 12(1):5608. ISSN 2041-1723. URL https://www.nature.com/articles/s41467-021-25050-3 [DOI] [PMC free article] [PubMed]
- Vian A, Pochitaloff M, Yen ST, Kim S, Pollock J, Liu Y, Sletten EM, Campás O (2023) In situ quantification of osmotic pressure within living embryonic tissues. Nature Communications 14(1):7023. 10.1038/s41467-023-42024-9. ISBN: 2041-1723 Publisher: Nature Publishing Group UK London [DOI] [PMC free article] [PubMed]
- Villars A, Letort G, Valon L, Levayer R (2023) DeXtrusion: automatic recognition of epithelial cell extrusion through machine learning in vivo. Development (Cambridge) 150(13). ISSN 14779129. URL 10.1242/dev.201747. Publisher: Company of Biologists Ltd [DOI] [PMC free article] [PubMed]
- Wang A, Zhang Q, Han Y, Megason S, Hormoz S, Mosaliganti KR, Lam JKC, Li VOK (2022) A novel deep learning-based 3D cell segmentation framework for future image-based disease detection. Scientific Reports 12(1):342. ISSN 2045-2322. URL https://www.nature.com/articles/s41598-021-04048-3 [DOI] [PMC free article] [PubMed]
- Wang S, Larina IV (2021) In vivo dynamic 3d imaging of oocytes and embryos in the mouse oviduct. Cell Reports 36(2). URL 10.1016/j.celrep.2021.109382. ISBN: 2211-1247 Publisher: Elsevier [DOI] [PMC free article] [PubMed]
- Winetraub Y, Van Vleck A, Yuan E, Terem I, Zhao J, Yu C, Chan W, Do H, Shevidi S, Mao M (2024) Noninvasive virtual biopsy using micro-registered Optical Coherence Tomography (OCT) in human subjects. Science Advances 10(15):eadi5794. URL 10.1126/sciadv.adi5794. ISBN: 2375-2548 Publisher: American Association for the Advancement of Science [DOI] [PMC free article] [PubMed]
- Zaniker EJ, Babayev E, Duncan FE (2023) Common mechanisms of physiological and pathological rupture events in biology: novel insights into mammalian ovulation and beyond. Biological Reviews 98(5):1648–1667. ISSN 1464-7931, 1469-185X. URL https://onlinelibrary.wiley.com/doi/10.1111/brv.12970 [DOI] [PMC free article] [PubMed]
- Zhang Y, Fodor É (2023) Pulsating active matter. Physical Review Letters 131(23):238302. ISSN 0031–9007:1079–7114. 10.1103/PhysRevLett.131.238302 [DOI] [PubMed]
- Zinchenko V, Hugger J, Uhlmann V, Arendt D, Kreshuk A (2023) MorphoFeatures for unsupervised exploration of cell types, tissues, and organs in volume electron microscopy. eLife 12. URL 10.7554/eLife [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable
Not applicable