Abstract
Protein nuclear import is generally mediated by basic nuclear localization signals (NLSs) that serve as targets for the importin α (Imp α) NLS receptor. Imp α is in turn bound by importin β (Imp β), which targets the resultant protein complex to the nucleus. Here, we report that the arginine-rich NLS sequences present in the human immunodeficiency virus type 1 regulatory proteins Tat and Rev fail to interact with Imp α and instead bind directly to Imp β. Using in vitro nuclear import assays, we demonstrate that Imp α is entirely dispensable for Tat and Rev nuclear import. In contrast, Imp β proved both sufficient and necessary, in that other β-like import factors, such as transportin, were unable to support Tat or Rev nuclear import. Using in vitro competition assays, it was demonstrated that the target sites on Imp β for Imp α, Tat, and Rev binding either are identical or at least overlap. The interaction of Tat and Rev with Imp β is also similar to Imp α binding in that it is inhibited by RanGTP but not RanGDP, a finding that may in part explain why the interaction of the Rev nuclear RNA export factor with target RNA species is efficient in the cell nucleus yet is released in the cytoplasm. Together, these studies define a novel class of arginine-rich NLS sequences that are direct targets for Imp β and that therefore function independently of Imp α.
The majority of nuclear proteins are targeted to the nucleus by basic, generally lysine-rich nuclear localization signals (NLSs) that serve as binding sites for an NLS receptor termed importin α (Imp α) or karyopherin α (reviewed in references 29 and 44). Imp α in turn interacts with a second import factor, termed importin β (Imp β) or karyopherin β1, that mediates docking of the resultant ternary complex to the cytoplasmic face of the nuclear pore complex (NPC) via a direct interaction with specific nucleoporins (5, 16, 32, 38). The subsequent translocation of this heterotrimer through the NPC remains poorly understood but is known to require energy and may be mediated by additional Imp β-nucleoporin interactions (39, 45). Once the heterotrimer reaches the nuclear face of the NPC, the GTP-bound form of the Ran GTPase directly binds to Imp β, resulting in the release of Imp α and the NLS protein into the nucleoplasm (15, 25, 33). Ran, which is found in the GDP-bound form in the cytoplasm and in the GTP-bound form in the nucleus, is therefore a major determinant of the directionality of nuclear import and may also provide a source of energy (21, 31, 36, 45). Once the NLS protein is released, both Imp α and Imp β are separately recycled back to the cytoplasm, where they can then participate in additional rounds of nuclear import.
Human immunodeficiency virus type 1 (HIV-1) encodes two essential regulatory proteins that are both active in the cell nucleus (reviewed in references 8 and 11). The Tat protein is an unusual transcriptional transactivator that dramatically enhances the processivity of transcription directed by the viral long terminal repeat promoter element. Tat function involves a direct interaction between Tat and an RNA target site, termed TAR, that is mediated by an arginine-rich RNA binding motif (ARM) that also functions as the Tat NLS (18, 40). The Rev protein, while equally critical for HIV-1 replication, acts posttranscriptionally to induce the sequence-specific nuclear export of late HIV-1 mRNA species. This RNA export activity requires a direct interaction between Rev and a specific RNA target sequence present in these RNAs, termed the RRE. Rev binding to the RRE is, in turn, mediated by an ARM sequence that is somewhat similar to the ARM present in Tat (Fig. 1), although the RNA sequence specificities of these two motifs are distinct. The Rev ARM also shares the ability of the Tat ARM to function as an effective NLS (3, 6, 17, 27).
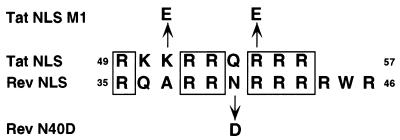
FIG. 1.
Sequence of the Tat and Rev NLS. A potential alignment of the 9-amino-acid Tat NLS with the somewhat larger Rev NLS is shown. The Tat M1 mutant encodes glutamic acid residues in place of lysine 51 and arginine 55 while the Rev N40D mutation substitutes aspartic acid for asparagine 40, as indicated. The Rev M6 mutant has been previously described (27, 28) and bears an aspartic acid and a leucine residue in place of arginines 41 to 44.
Because the NLS sequences present in Tat and Rev are arginine rich, and therefore basic, it might be assumed that they would mediate nuclear import via the same import pathway utilized by the prototypic basic NLSs found in simian virus 40 (SV40) large T antigen and nucleoplasmin (9, 23). However, several reports have shed doubt on this hypothesis. Thus, Efthymiadis et al. (10) have reported that the Tat NLS is able to mediate nuclear import in vitro in the absence of both Imp α and Imp β, that nuclear import of a Tat NLS substrate is not inhibited by an excess of an SV40 T antigen NLS peptide, and, finally, that the Tat NLS fails to bind to either Imp α or Imp β. These data were interpreted to suggest that the Tat NLS functioned via an entirely novel nuclear import pathway. In the case of HIV-1 Rev, Fankhauser et al. (12) have proposed that the nucleolar protein B23 binds to the Rev basic domain and mediates its nuclear import. In contrast, Henderson and Percipalle (20) have reported that the Rev NLS can bind directly to Imp β and that this interaction is inhibited by added Imp α, thus suggesting that Imp α and Rev compete for overlapping binding sites on Imp β. While these authors also reported a Rev-Imp α interaction, this was not dependent on a functional Rev NLS and was therefore viewed as nonspecific. However, because this latter article did not report any nuclear import assays, it remained unclear whether Imp β was indeed sufficient or even necessary for Rev nuclear import.
While the reports described above might suggest that the Tat and Rev NLSs are functionally distinct, the sequence similarity between the Tat and Rev NLS-ARM sequences (Fig. 1) seemed more consistent with the hypothesis that these NLSs utilize the same nuclear import pathway. Here, we demonstrate that both the Tat and the Rev NLS directly interact with Imp β but not Imp α in vitro and further demonstrate that Imp β is both necessary and sufficient for the nuclear import of both Tat and Rev into isolated nuclei. These studies therefore demonstrate the existence of a novel class of basic NLS sequences that function independently of Imp α.
MATERIALS AND METHODS
Bacterial expression plasmids.
Plasmids for the expression in Escherichia coli of the recombinant glutathione S-transferase (GST) fusion proteins GST-SV40 large T antigen NLS (T NLS), GST-Imp β, GST-Imp α, GST-IBB (Imp β binding domain of Imp α), GST-M9, GST-Ran, GST-RanQ69L, GST-p10/NTF2, GST-Rev, and GST-RevM6 have been described previously (13, 28, 42). Plasmid pMBP-Trn, encoding the human transportin protein fused to maltose binding protein (MBP), has also been described (13, 42). Plasmid pGST-RevN40D was made by site-directed mutagenesis, using pGST-Rev as a template, by the Quick change method (Stratagene) changing the codon at amino acid position 40 from AAT (asparagine) to GAC (aspartic acid). Plasmid pGST-R5QR4 was constructed by annealing two complementary oligonucleotides encoding the amino acid sequence NH2-RRRRRQRRRR-COOH, bearing 5′-BamHI and 3′-XhoI overhanging ends, and ligating into BamHI- and XhoI-digested pGex5X-1. Plasmid pGST-TatNLS, encoding the Tat amino acid sequence 49-RKKRRQRRRAHQ-60, was constructed similarly to pGST-R5QR4. Plasmid pGST-TatM1 is similar to pGST-TatNLS, except that oligonucleotides encoding the amino acid sequence 49-RKeRRQeRRAHQ-60 were used.
Recombinant protein expression and purification.
GST-Rev, GST-RevM6, and GST-RevN40D proteins were purified from E. coli BL21 cells containing the relevant plasmids. Bacteria were grown overnight to saturation and then diluted 1:10 in fresh media for growth at 30°C for 3 h prior to induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 5 h. The proteins were then purified on glutathione-Sepharose 4B beads (Pharmacia Biotech, Inc.), dialyzed into storage buffer A (100 mM NaCl, 10 mM HEPES [pH 7.4], 1 mM dithiothreitol [DTT], 10% glycerol), and frozen in aliquots at −80°C. For RNA mobility shift assays, the GST-Rev protein was further purified over a Q-Sepharose Fast Flow column (Pharmacia Biotech, Inc.), eluted in 500 mM NaCl, dialyzed into storage buffer B (50 mM NaCl, 10 mM KCl, 10 mM HEPES [pH 7.4], 10% glycerol, 2 mM DTT), and frozen in single-use aliquots at −80°C. The GST-IBB, GST-T NLS, GST-R5QR4, GST-M9, GST-TatNLS, and GST-TatM1 fusion proteins were purified by the standard batch method recommended by the glutathione-Sepharose bead supplier (Pharmacia Biotech, Inc.), dialyzed into storage buffer A, and frozen at −80°C.
MBP-transportin, Imp β, Imp α (Rch1), Ran, RanQ69L, and p10/NTF2 were expressed, purified, cleaved, and stored as described previously (42). For protein binding experiments to the Tat NLS and Rev protein, the recombinant Imp β protein was further purified by affinity chromatography on a column of GST-IBB protein covalently linked to Affi-Gel active ester agarose (Affi-10; Bio-Rad Laboratories) at a 5-mg/ml concentration.
For peptide competition experiments, the GST-T NLS, GST-IBB, GST-TatNLS, and GST-Rev proteins were expressed in bacteria and purified by the standard batch procedure onto glutathione-Sepharose 4B beads (Pharmacia Biotech, Inc.). The glutathione-Sepharose resin-bound fusion proteins were quantitated by the Bradford assay reagent (Bio-Rad Laboratories) and cleaved by the appropriate protease (factor Xa or thrombin) at 25°C for 12 h. For final quantitation, 80% efficiency of cleavage by proteases was assumed. The supernatants were treated with 1,5 Dansyl-Glu-Gly-Arg chloromethyl ketone HCl (Calbiochem) to inactivate the protease and were then aliquoted and frozen at −80°C. The integrity of all recombinant proteins used in this study was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Protein affinity chromatography.
Purified proteins (GST-Tat or GST-Rev) were coupled at 4- to 8-mg/ml concentrations onto Affi-Gel 10 active ester agarose beads (Bio-Rad Laboratories) in coupling buffer (500 mM NaCl, 10 mM HEPES [pH 7.4], 10% glycerol). Columns of 10-μl bed volume were constructed in 100-μl borosilicate pipettes and equilibrated with 50 column volumes of AC buffer (50 mM NaCl, 10 mM HEPES [pH 7.4], 0.1 mM DTT, 10% glycerol) injected with a 100-μl Hamilton syringe. Approximately 2 μg of Imp β or Imp α was loaded onto the columns in 25 μl of AC buffer, and another 25 μl of AC buffer was then added and collected to constitute the flow-through fraction. The columns were then washed with 3 column volumes of AC buffer, and bound proteins were eluted with 50 μl of 800 mM MgCl2. The entire flow-through and eluate (bound) fractions were analyzed by SDS–12% PAGE (Ready gels; Bio-Rad Laboratories) and visualized by Coomassie blue R-250 (Life Technologies, Inc.) staining.
RanGTP release experiments.
For the RanGTP and -GDP release experiment, 5 μg of Imp β was incubated with ∼20 μg of GST-TatNLS protein in a total volume of 50 μl of AC buffer. Next, ∼50 μl of equilibrated glutathione-Sepharose 4B beads (Pharmacia Biotech, Inc.) was added, and the reactions were spun at 250 × g for 1 min. A total of 100 μl of Ran buffer (20 mM NaHPO4, 50 mM NaCl, 1 mM Mg acetate) was then added to the beads, which were then incubated for 30 min at 25°C in the presence of buffer alone or in buffer with RanGTP or RanGDP. The Q69L mutant of Ran, which is unable to hydrolyze bound nucleotides (2), was used for these release experiments. RanQ69L (5 μg) and 2 mM GTP or GDP were premixed in Ran binding buffer prior to addition to the Ran binding buffer-bead mixture. The reactions were then spun at 250 × g for 1 min, the supernatant was discarded, and 50 μl of 1% SDS sample loading buffer was added. The reactions were then spun at 500 × g for 1 min, and the supernatant was analyzed by SDS–12% PAGE and visualized by Coomassie blue R-250 staining.
In vitro nuclear uptake assays.
In vitro nuclear uptake assays were performed using digitonin-permeabilized HeLa cells, as previously described (1, 42). Recombinant purified GST-Rev, GST-Tat, and GST-T NLS fusion moieties were fluorescein labeled (FLOUS; Boehringer Mannheim) and used at 2 μM final concentrations. The import factor proteins Imp β, Imp α, MBP-transportin, Ran, and p10/NTF2 were used at 2 μM final concentrations. For competition assays, fluorescein-labeled proteins were used at a 500 nM final concentration, and rabbit reticulocyte lysate (Promega) was used as cytosol. Competing peptides were used at an ∼60-fold molar excess (∼30 μM). Competition assays were performed in a total volume of 70 μl for 20 min at 18°C. Images were digitally captured with a Leica DMRB fluorescence microscope and converted to 8-bit gray scale with Adobe Photoshop 4.0 software.
Rev-RRE mobility shift assays.
The GST-Rev mobility shift assay on the HIV-1 RRE was performed essentially as previously described (28), with the exception that 0.5% Triton X-100 was added to the binding buffer and a different nonspecific RNA competitor (4 μg of 16S rRNA; Boehringer Mannheim) was used. Thirty nanograms of GST-Rev, 500 ng of Imp β or transportin, and 1 μg of RanQ69L were used per reaction. RanQ69L protein was preincubated with 1 mM GTP or GDP in the presence of 0.1 mM MgCl2 prior to addition. All proteins were added simultaneously to the 32P-CTP-labeled RRE probe and incubated for 20 min on ice before loading into a 6% Tris-glycine–5% glycerol (acrylamide/bis ratio, 80:1) amino-gel. The gel was run at 180 V at 25°C and was visualized by autoradiography after drying.
RESULTS
To examine whether either Imp α or Imp β is required for the nuclear import of HIV-1 Tat or Rev, we performed in vitro nuclear uptake assays using suspended, digitonin-permeabilized HeLa cells, as previously described (42). The permeabilization procedure used, which involves digitonin treatment followed by the isolation of intact nuclei on a sucrose cushion (42), results in a preparation of nuclei with only limited residual cytoplasm. In comparison to cells that have been digitonin permeabilized on coverslips, this procedure has the advantage that cytoplasmic import factors are more effectively depleted but has the disadvantage that lack of nuclear import gives a lack of any detectable signal, rather than a cytoplasmic halo (1, 35, 42). The import substrates used were FLUOS-labeled proteins consisting of GST fused to full-length wild-type or mutant (M6 or N40D) Rev, to wild-type or M1 mutant forms of the Tat NLS (Tat amino acids 49 to 60), or to the SV40 large T antigen NLS (T NLS). The T NLS is known to be dependent on both Imp β and Imp α for nuclear import (38).
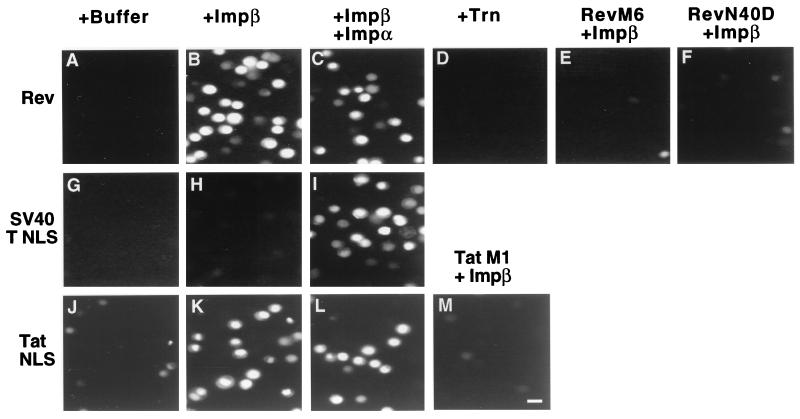
Nuclear import reactions were performed in a buffer containing recombinant human Ran and p10/NTF2, as well as a source of energy, as previously described (42). As shown in Fig. 2, the GST-Rev, GST-TatNLS, and GST-T NLS substrates all failed to effectively enter nuclei when incubated in buffer alone, i.e., in the absence of both Imp β and Imp α (panels A, G, and J). However, in the presence of recombinant Imp β, both the GST-Rev and the GST-TatNLS substrates displayed readily observable nuclear import (panels B and K), although the GST-T NLS substrate was still not detectably imported (panel H). Further addition of recombinant Imp α induced GST-T NLS import as expected (panel I) but did not appreciably inhibit or enhance nuclear import of either the GST-Rev or the GST-TatNLS substrate (panels C and L). The nuclear import of GST-TatNLS and GST-Rev in the absence of added Imp α was specific in that mutations known to inactivate Rev NLS function (Rev M6 and Rev N40D) or Tat NLS function (TatM1) in vivo (17, 18, 27) also blocked Imp β-dependent import in vitro (panels E, F, and M). This import was also specific for Imp β in that the related nuclear import factor transportin (4, 13, 37) was not able to mediate the nuclear import of these substrates (panel D). We therefore conclude that Imp β is able to mediate the in vitro nuclear import of substrates containing either the Rev NLS or the Tat NLS in the absence of functionally detectable levels of Imp α.
FIG. 2.
In vitro nuclear import assays. Permeabilized HeLa cells were prepared as previously described (13, 42) and incubated in a buffer containing recombinant human Ran and p10/NTF2 as well as a source of energy. Import substrates consisted of wild-type (GST-Rev, GST-T NLS, GST-TatNLS) or mutant (GST-M6, GST-N40D, GST-TatM1) fusion proteins labeled with FLUOS. Recombinant purified import factors (Imp β, Imp α, Trn) were added as indicated. After incubation at 30°C for 15 min, the nuclei were fixed and then analyzed for nuclear import of the fluorescent substrate using a fluorescence microscope. Bar in panel M ≈ 20 μm.
The Rev and Tat NLS both directly bind Imp β.
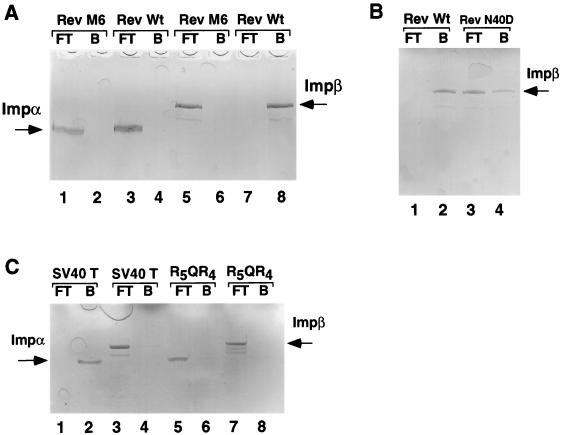
The observation that Imp β alone can mediate both Rev NLS and Tat NLS function in vitro (Fig. 2) implies a direct interaction between these NLSs and Imp β. Initially, we examined whether the Rev NLS can bind to either Imp β or Imp α in vitro, using an affinity chromatography assay. A direct and specific interaction between Rev and Imp β was indeed observed (Fig. 3A, lanes 7 and 8, and Fig. 3B, lanes 1 and 2), which was inhibited by both the M6 NLS mutation (Fig. 3A, lanes 5 and 6) and the less severe N40D NLS mutation (Fig. 3B, lanes 3 and 4). No interaction between Imp α and Rev (Fig. 3A, lanes 3 and 4) under conditions where the SV40 T NLS bound Imp α effectively (Fig. 3C, lanes 1 and 2) was detected. Finally, a recombinant protein consisting of GST fused to a highly arginine-rich sequence (R5QR4) failed to bind to either Imp α or Imp β in vitro (Fig. 3C, lanes 5 to 8) and also failed to import in vitro in the presence of Imp β plus or minus Imp α (data not shown). Therefore, this simple basic sequence is not sufficient for Imp β binding even though its charge is comparable to that of the Rev NLS (Fig. 1). We conclude that the interaction between Rev and Imp β is both direct and specific.
FIG. 3.
Direct binding of Imp β by the Rev NLS. The indicated purified wild-type (Rev, TNLS) or mutant (Rev M6, Rev M40D, R5QR4) GST fusion proteins were coupled to agarose beads and used to construct micro-affinity columns that were then loaded with ∼2 μg of recombinant Imp β or Imp α. Proteins present in the column flow-through (FT) fraction were collected, and bound (B) proteins were eluted from the column by using 800 mM MgCl2. The FT and B fractions were then subjected to analysis by SDS-PAGE.
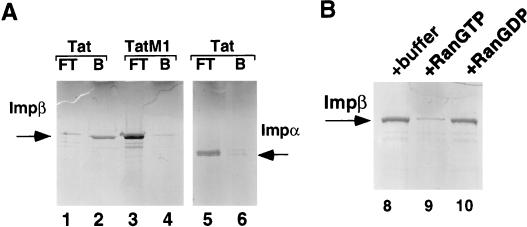
As shown in Fig. 4, we further extended these data by examining the interaction of the Tat NLS with Imp β and Imp α. As may be observed, wild-type Tat, but not the Tat M1 mutant, proved able to directly bind to Imp β (Fig. 4, lanes 1 to 4) but not to Imp α (lanes 5 and 6). Therefore, the Tat NLS shares the ability of the Rev NLS to directly interact with Imp β in vitro.
FIG. 4.
The interaction of the Tat NLS with Imp β is inhibited by Ran GTP. (A) Micro-affinity chromatography, using columns bearing the wild-type GST-TatNLS or mutant GST-TatM1 fusion proteins, was performed as described in the legend to Fig. 3. (B) Glutathione-Sepharose beads bearing the wild-type GST-TatNLS protein were loaded with recombinant Imp β and then incubated in buffer alone or with added RanGTP or RanGDP. After washing, the residual bound Imp β was released with SDS and analyzed by SDS-PAGE.
The Imp β-Tat NLS interaction is inhibited by RanGTP.
As described in the introduction, Imp β and related import factors, such as transportin, bind import substrates in the cytoplasm where RanGTP levels are low and then release these substrates, including Imp α, into the nucleoplasm when they encounter nuclear RanGTP (21, 29, 33, 44). If the interaction between the Tat NLS and Imp β indeed leads to the productive nuclear import of Tat, then this interaction should also be released by RanGTP but not by RanGDP. As shown in Fig. 4B, RanGTP indeed proved to be an effective inhibitor of the Imp β-Tat NLS interaction while added RanGDP had no detectable effect. Similar data showing inhibition of Rev NLS binding to Imp β were previously reported (20) and have been confirmed in this laboratory (data not shown).
Tat, Rev, and Imp α bind to overlapping sites on Imp β.
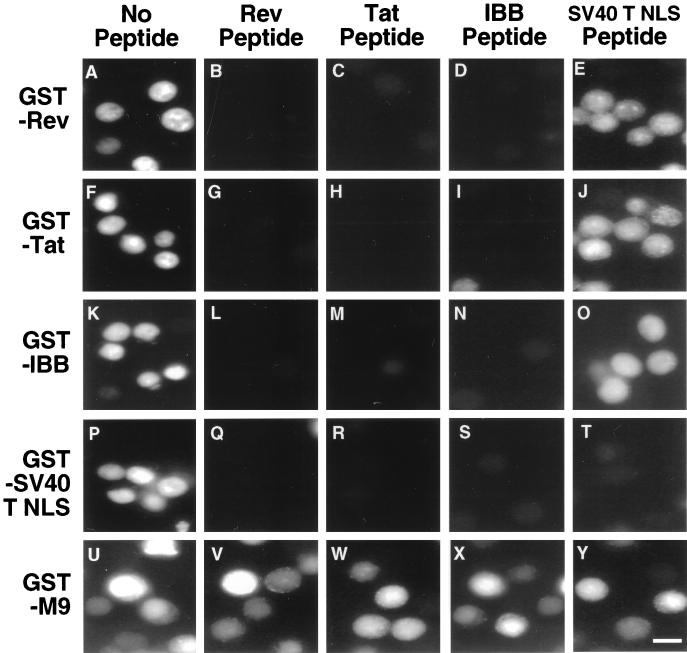
The interaction between Imp β and Imp α is mediated by a short sequence, termed the Imp β binding or IBB domain, located near the Imp α amino terminus (14, 46). Importantly, the IBB domain can function as an effective Imp β-dependent but Imp α-independent NLS both in vitro and in vivo. Because the interaction between Imp β and the Rev NLS or the Tat NLS is inhibited by RanGTP, a property that is also shared by the Imp β-IBB interaction (21), it appeared possible that these three NLS signals might bind to the same, or at least overlapping, regions on Imp β. To examine this question, we asked whether nuclear import of GST-Rev, GST-Tat NLS, or GST-IBB would be specifically inhibited by an ∼60-fold molar excess of a Rev NLS, Tat NLS, or IBB peptide. Relevant controls included the GST-T NLS substrate, import of which is Imp α dependent and should therefore be inhibited by the IBB peptide (27), and an SV40 T NLS peptide, which should only inhibit GST-T NLS import. A final control is the GST-M9 substrate, which is imported into the nucleus by the distinct transportin import factor (4, 13, 37) and should therefore be unaffected by all four peptides. These in vitro nuclear uptake experiments were performed using isolated HeLa cell nuclei with reticulocyte lysate as a source of native import factors.
As shown in Fig. 5, the Rev, Tat, and IBB peptides inhibited import of all substrates except for GST-M9, while the T NLS peptide, as predicted, only inhibited the import of the cognate GST-T NLS substrate. The specificity of the inhibition shown in Fig. 5 therefore strongly suggests that the interaction of Rev, Tat, and Imp α (IBB) with Imp β is mediated by identical, or at least overlapping, Imp β sequences and that these three protein-protein interactions are therefore mutually incompatible. While the observed inhibition of Tat and Rev nuclear import by added IBB peptide (Fig. 5) might appear to contradict the earlier finding that added full-length Imp α has no evident effect on Tat and Rev import (Fig. 2), it should be recalled that the IBB peptide was added at an ∼60-fold molar excess over Imp β, while the full-length Imp α was added at an equimolar level.
FIG. 5.
Competitive in vitro nuclear import assays. Nuclear import assays were performed using permeabilized HeLa cells in the presence of rabbit reticulocyte lysate as a source of native nuclear import factors. The FLUOS-labeled substrates analyzed in this assay are given at left. Competitor peptides were added at an ∼60-fold molar excess and are listed at the top of the figure. Nuclear uptake assays were performed for 20 min at 18°C and were then analyzed as described in the legend to Fig. 2. Bar in panel Y ≈ 20 μm.
Imp β can inhibit the Rev-RRE interaction.
While Tat remains in the cell nucleus after import, Rev continuously shuttles between the nucleus and cytoplasm in the process of mediating HIV-1 late mRNA export from the nucleus (8, 11, 30). It has, however, remained unclear why the interaction of Rev with the RRE RNA, which is mediated by an ARM sequence extensively overlapping with the Rev NLS (3, 28), is efficient in the cell nucleus yet is released in the cell cytoplasm. One possible explanation for this compartmentalization is that Imp β might inhibit the Rev-RRE interaction by competing with the RRE for Rev binding, a process that should only occur in the cytoplasm where RanGTP levels are low (21, 25).
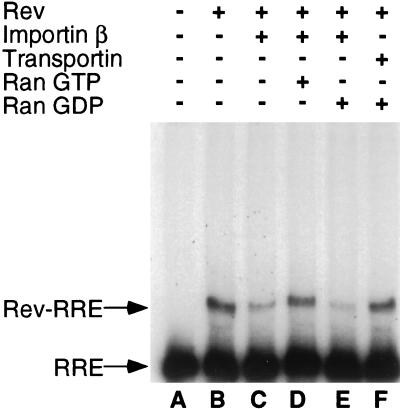
To examine this question, we performed an RNA gel shift analysis using a radiolabeled HIV-1 RRE RNA probe and recombinant Rev and Ran proteins. As shown in Fig. 6, and reported by many groups in the past (19, 24, 28, 47), Rev is able to form a specific complex with the RRE RNA probe in vitro, resulting in the appearance of a retarded RNA-protein complex. Interestingly, while addition of Imp β results in the inhibition of this interaction, Rev-RRE binding can be rescued by the further addition of RanGTP, but not of RanGDP (Fig. 6). No inhibition of the Rev-RRE interaction was observed upon addition of an equivalent level of the Imp β-like import factor transportin. We therefore conclude that Imp β indeed has the potential to specifically inhibit the Rev-RRE interaction in the cytoplasm, but not the nuclei, of HIV-1 infected cells.
FIG. 6.
Imp β inhibits RNA binding by HIV-1 Rev. The indicated electrophoretic mobility shift assay was performed essentially as previously described (28) using recombinant HIV-1 Rev protein and a radiolabeled HIV-1 RRE RNA probe. Recombinant Imp β or transportin was added to the RNA binding assays visualized in lanes C to F. In addition, RanGTP (lane D) or RanGDP (lanes E and F) was added as indicated.
DISCUSSION
As described in more detail in the introduction, nuclear import of lysine-rich NLS sequences, such as the SV40 T NLS, requires an indirect interaction between the NLS and the Imp β nuclear import factor that is mediated by the Imp α NLS receptor (29, 44). The role of Imp α in the process of nuclear import therefore appears to be solely that of targeting the NLS to Imp β. Several lines of evidence have suggested that NLS sequences that directly interact with Imp β, and that therefore would be Imp α independent, might exist. First, the short IBB motif present in Imp α, which is necessary and sufficient for Imp β binding, has itself been shown to function as an Imp β-dependent, Imp α-independent NLS when attached to a substrate protein (14, 46). Second, the NLS found in the yeast nucleocytoplasmic shuttle protein Nab2 has been shown to mediate protein nuclear import via a direct interaction with Imp β when expressed in mammalian cells (42). Third, it is now apparent that Imp β is unusual, and possibly unique, in relying on the Imp α adaptor protein to recognize target NLS sequences. In contrast, other Imp β-related nuclear import factors, such as transportin, are now known to bind to their cognate NLS sequences directly (4, 13, 37).
In this article, we present evidence demonstrating that both the Tat NLS and the Rev NLS in fact function as direct Imp β-dependent NLSs. Specifically, we have shown that the in vitro nuclear import of substrates bearing the Tat and Rev NLS is dependent on Imp β but independent of Imp α under conditions where SV40 T NLS import requires both added proteins (Fig. 2). We have further demonstrated a highly specific, direct interaction between Imp β, but not Imp α, and both the Tat NLS and the Rev NLS under conditions where the SV40 T NLS binds Imp α but not Imp β (Fig. 3 and 4A). The interaction between Imp β and the Tat NLS was found to be potently inhibited by RanGTP, but not RanGDP, as predicted for a functionally relevant Imp β interaction (Fig. 4B) (21). Finally, we have demonstrated that the Tat NLS, the Rev NLS and the Imp α IBB motif can all act as competitive inhibitors of all forms of Imp β- but not transportin-dependent nuclear import, thus strongly suggesting that these proteins bind to an identical or overlapping surface on Imp β (Fig. 5). Collectively, these data identify a novel class of Imp α-independent basic NLS sequences that are functionally distinct from Imp α-dependent NLSs, such as the monopartite SV40 T NLS and the bipartite nucleoplasmin NLS (9, 23).
The data presented in this manuscript partly contradict the report of Efthymiadis et al. (10), which noted that Tat NLS function is both Imp α and Imp β independent. This earlier result may reflect the use of the SV40 T NLS as a control for in vitro factor dependence. If the permeabilized cells used by these workers retained Imp β activity but lacked functional Imp α, then it would appear that the Tat NLS differed from the SV40 T NLS in being factor independent. However, our data both explain and reproduce (Fig. 5) this group’s finding that Tat NLS function is not inhibited by an excess of an SV40 T NLS peptide. In addition, our data confirm the observation by Henderson and Percipalle (20) that the Rev NLS can directly bind Imp β and now show, for the first time, that Rev NLS function is indeed independent of Imp α. Finally, we have not observed any evidence in support of the hypothesis of Fankhauser et al. (12) that Rev NLS function is mediated by the B23 protein. While it remains formally possible that B23 could affect Rev nuclear import, it is clearly not essential (Fig. 2).
At least six NLS sequences able to directly interact with Imp β have now been reported, namely, the Tat and Rev NLSs (Fig. 1), the Imp α IBB, the Rex NLS, and the NLSs found in the T-cell protein tyrosine phosphatase and in the yeast protein Nab2 (14, 35, 41, 42, 46). Inspection of these six sequences reveals that they are all arginine rich. Thus, the ∼40-amino-acid IBB motif conserved in all six human Imp α homologs contains nine conserved arginine residues, four of which are sequentially arranged (14, 46), in a manner reminiscent of the Tat and Rev NLSs (Fig. 1), while the ∼18-amino-acid Rex NLS contains seven arginine residues (35). Finally, the Nab2 NLS contains eight arginine residues, located over a 31-amino-acid core sequence, and mutation of two of these has been shown to result in a loss of Nab2 NLS function (42). In contrast, basic NLS sequences of the Imp α-dependent type are lysine rich and generally contain at least two, and more often three or more, lysine residues (7, 9, 23, 29). While the number of examples of Imp β-dependent, Imp α-independent NLSs remains too small for confident prediction, it nevertheless seems possible that basic NLSs may be subdivided into at least two types, a more common, lysine-rich, Imp α-dependent class and a less common, arginine-rich, Imp α-independent class. The recently reported (7) structure of a complex of Imp α with the SV40 T NLS provides a molecular explanation for the requirement for lysine residues in Imp α-dependent NLSs, as these have been found to make critical hydrophobic contacts with Imp α that could not be formed by arginine residues.
Immediately prior to submission of this article, Jäkel and Görlich (22) reported that several ribosomal proteins, including particularly rpL23a, contained NLSs that could function as Imp β-dependent, Imp α-independent NLSs. Interestingly, the rpL23a NLS was mapped to a 32-amino-acid sequence that contained eight arginine residues. Nevertheless, the rpL23a NLS appears to be functionally distinct from the NLSs present in the Tat and Rev proteins. Specifically, the rpL23a NLS was reported to bind to a region on Imp β that is distinct from the Imp α IBB binding site. In contrast, we observed that the IBB effectively competed the in vitro nuclear import of both the Tat NLS and the Rev NLS, and vice versa (Fig. 5), thus strongly suggesting that the Tat and Rev NLS bind to a site on Imp β that overlaps the IBB site. Secondly, the rpL23a NLS was reported to function as a target not only for Imp β but also for transportin and two other Imp β-like factors, termed RanBP5 and RanBP7. In contrast, we did not observe any nuclear import of a Rev NLS substrate into isolated nuclei in the presence of recombinant transportin (Fig. 2), even though we have previously shown that transportin is fully able to mediate nuclear import of substrates bearing the cognate M9 NLS under these assay conditions (13, 42). In addition, the finding that an IBB peptide can entirely block nuclear import of both Rev and Tat NLS substrates in vitro in the presence of an added cytoplasmic extract that can fully support M9 NLS nuclear import (Fig. 5) demonstrates that Imp β is necessary, as well as sufficient (Fig. 2), for Tat and Rev NLS function. The data of Jäkel and Görlich (22) would therefore seem to imply the existence of a third class of basic NLSs that differ from the Tat and Rev NLS, as well as from the IBB and Rex NLS (14, 35, 46), in terms of both their binding site on Imp β and their ability to also use other nuclear import factors, such as transportin.
An unresolved question is why Tat and Rev would evolve arginine-rich, Imp α-independent NLSs rather than the more common Imp α-dependent NLS. Two possibilities suggest themselves. Firstly, both Tat and Rev are short proteins (86 and 116 amino acids, respectively) that are expressed by a virus containing a small genome (8, 11). Both proteins are also RNA binding proteins that contain ARMs. It is therefore possible that these proteins have simply evolved to also use the preexisting ARMs as NLS sequences rather than acquire an additional, lysine-rich NLS that would require an increase in both protein size and complexity. On the other hand, it is also known that the various forms of Imp α (at least five distinct variants are encoded in the human genome) are expressed at widely divergent levels in different tissues and also display very different affinities for distinct NLS sequences (26, 34, 43). Potential difficulties in achieving efficient nuclear import in all of the various tissues infected by HIV-1 in vivo could therefore be avoided by targeting Imp β directly, rather than relying on one or more forms of Imp α as an intermediary.
While the reasons why Tat and Rev contain an unusual Imp α-independent NLS are presently unclear, this finding does have potential implications for the cytoplasmic release of the RRE containing RNAs that are exported from the nucleus by the Rev protein. Specifically, as shown in Fig. 6, Imp β can effectively inhibit the Rev-RRE interaction, but only in the absence of RanGTP. Because RanGTP is found at high levels in the cell nucleus but only at low levels in the cytoplasm (21, 25, 29), this finding provides a potential mechanism to explain why the Rev-RRE interaction is efficient in the nucleus, where RanGTP would prevent the Rev-Imp β interaction, but is released in the cytoplasm, where little RanGTP is found. This may therefore represent another example of the known critical role of Ran in determining the directionality of nucleocytoplasmic transport pathways.
REFERENCES
- 1.Adam S A, Sterne-Marr R E, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bischoff F R, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H. RanGAP1 induces GTPase activity of nuclear ras-related Ran. Proc Natl Acad Sci USA. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Böhnlein E, Berger J, Hauber J. Functional mapping of the human immunodeficiency virus type 1 Rev RNA binding domain: new insights into the domain structure of Rev and Rex. J Virol. 1991;65:7051–7055. doi: 10.1128/jvi.65.12.7051-7055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonifaci N, Moroianu J, Radu A, Blobel G. Karyopherin β2 mediates nuclear import of a mRNA binding protein. Proc Natl Acad Sci USA. 1997;94:5055–5060. doi: 10.1073/pnas.94.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chi N C, Adam E J H, Adam S A. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cochrane A W, Perkins A, Rosen C A. Identification of sequences important in the nucleolar localization of human immunodeficiency virus Rev: relevance of nucleolar localization to function. J Virol. 1990;64:881–885. doi: 10.1128/jvi.64.2.881-885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 8.Cullen B R. HIV-1 auxiliary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 9.Dingwall C, Sharnick S V, Laskey R A. A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell. 1982;30:449–458. doi: 10.1016/0092-8674(82)90242-2. [DOI] [PubMed] [Google Scholar]
- 10.Efthymiadis A, Briggs L J, Jans D A. The HIV-1 Tat nuclear localization sequence confers novel nuclear import properties. J Biol Chem. 1998;273:1623–1628. doi: 10.1074/jbc.273.3.1623. [DOI] [PubMed] [Google Scholar]
- 11.Emerman M, Malim M H. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science. 1998;280:1880–1884. doi: 10.1126/science.280.5371.1880. [DOI] [PubMed] [Google Scholar]
- 12.Fankhauser C, Izaurralde E, Adachi Y, Wingfield P, Laemmli U K. Specific complex of human immunodeficiency virus type 1 Rev and nucleolar B23 proteins: dissociation by the Rev response element. Mol Cell Biol. 1991;11:2567–2575. doi: 10.1128/mcb.11.5.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fridell R A, Truant R, Thorne L, Benson R E, Cullen B R. Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-β. J Cell Sci. 1997;110:1325–1331. doi: 10.1242/jcs.110.11.1325. [DOI] [PubMed] [Google Scholar]
- 14.Görlich D, Henklein P, Laskey R A, Hartmann E. A 41 amino acid motif in importin-α confers binding to importin-β and hence transit into the nucleus. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- 15.Görlich D, Panté N, Kutay U, Aebi U, Bischoff F R. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 16.Görlich D, Vogel F, Mills A D, Hartmann E, Laskey R A. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- 17.Hammerschmid M, Palmeri D, Ruhl M, Jaksche H, Weichselbraun I, Böhnlein E, Malim M H, Hauber J. Scanning mutagenesis of the arginine-rich region of the human immunodeficiency virus type 1 Rev transactivator. J Virol. 1994;68:7329–7335. doi: 10.1128/jvi.68.11.7329-7335.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hauber J, Malim M H, Cullen B R. Mutational analysis of the conserved basic domain of human immunodeficiency virus Tat protein. J Virol. 1989;63:1181–1187. doi: 10.1128/jvi.63.3.1181-1187.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heaphy S, Dingwall C, Ernberg I, Gait M J, Green S M, Karn J, Lowe A D, Singh M, Skinner M A. HIV-1 regulator of virion expression (Rev) protein binds to an RNA stem-loop structure located within the Rev response element region. Cell. 1990;60:685–693. doi: 10.1016/0092-8674(90)90671-z. [DOI] [PubMed] [Google Scholar]
- 20.Henderson B R, Percipalle P. Interactions between HIV Rev and nuclear import and export factors: the Rev nuclear localisation signal mediates specific binding to human importin-β. J Mol Biol. 1997;274:693–707. doi: 10.1006/jmbi.1997.1420. [DOI] [PubMed] [Google Scholar]
- 21.Izaurralde E, Kutay U, von Kobbe C, Mattaj I W, Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jäkel S, Görlich D. Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalderon D, Roberts B L, Richardson W D, Smith A E. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 24.Kjems J, Brown M, Chang D D, Sharp P A. Structural analysis of the interaction between the human immunodeficiency virus Rev protein and the Rev response element. Proc Natl Acad Sci USA. 1991;88:683–687. doi: 10.1073/pnas.88.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koepp D M, Silver P A. A GTPase controlling nuclear trafficking: running the right way or walking RANdomly? Cell. 1996;87:1–4. doi: 10.1016/s0092-8674(00)81315-x. [DOI] [PubMed] [Google Scholar]
- 26.Köhler M, Ansieau S, Prehn S, Leutz A, Haller H, Hartmann E. Cloning of two novel human importin-α subunits and analysis of the expression pattern of the importin-α protein family. FEBS Lett. 1997;417:104–108. doi: 10.1016/s0014-5793(97)01265-9. [DOI] [PubMed] [Google Scholar]
- 27.Malim M H, Böhnlein S, Hauber J, Cullen B R. Functional dissection of the HIV-1 Rev transactivator: derivation of a transdominant repressor of Rev function. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 28.Malim M H, Cullen B R. HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral RRE: implications for HIV-1 latency. Cell. 1991;65:241–248. doi: 10.1016/0092-8674(91)90158-u. [DOI] [PubMed] [Google Scholar]
- 29.Mattaj I W, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;167:256–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 30.Meyer B E, Malim M H. The HIV-1 Rev transactivator shuttles between the nucleus and the cytoplasm. Genes Dev. 1994;8:1538–1547. doi: 10.1101/gad.8.13.1538. [DOI] [PubMed] [Google Scholar]
- 31.Moore M S, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 32.Moroianu J, Blobel G, Radu A. Previously identified protein of uncertain function is karyopherin α and together with karyopherin β docks import substrate at nuclear pore complexes. Proc Natl Acad Sci USA. 1995;92:2008–2011. doi: 10.1073/pnas.92.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moroianu J, Blobel G, Radu A. Nuclear protein import: Ran-GTP dissociates the karyopherin αβ heterodimer by displacing α from an overlapping binding site on β. Proc Natl Acad Sci USA. 1996;93:7059–7062. doi: 10.1073/pnas.93.14.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nadler S G, Tritschler D, Haffar O K, Blake J, Bruce A G, Cleaveland J S. Differential expression and sequence-specific interaction of karyopherin α with nuclear localization sequences. J Biol Chem. 1997;272:4310–4315. doi: 10.1074/jbc.272.7.4310. [DOI] [PubMed] [Google Scholar]
- 35.Palmeri D, Malim M H. Importin β can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin α. Mol Cell Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paschal B M, Delphin C, Gerace L. Nucleotide-specific interaction of Ran/TC4 with nuclear transport factors NTF2 and p97. Proc Natl Acad Sci USA. 1996;93:7679–7683. doi: 10.1073/pnas.93.15.7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pollard V W, Michael W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 38.Radu A, Blobel G, Moore M S. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 40.Siomi H, Shida H, Maki M, Hatanaka M. Effects of a highly basic region of human immunodeficiency virus Tat protein on nucleolar localization. J Virol. 1990;64:1803–1807. doi: 10.1128/jvi.64.4.1803-1807.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tiganis T, Flint A J, Adam S A, Tonks N K. Association of the T-cell protein tyrosine phosphatase with nuclear import factor p97. J Biol Chem. 1997;272:21548–21557. doi: 10.1074/jbc.272.34.21548. [DOI] [PubMed] [Google Scholar]
- 42.Truant R, Fridell R A, Benson R E, Bogerd H, Cullen B R. Identification and functional characterization of a novel nuclear localization signal present in the yeast Nab2 poly(A)+ RNA binding protein. Mol Cell Biol. 1998;18:1449–1458. doi: 10.1128/mcb.18.3.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuji L, Takumi T, Imamoto N, Yoneda Y. Identification of novel homologues of mouse importin α, the α subunit of the nuclear pore-targeting complex, and their tissue-specific expression. FEBS Lett. 1997;416:30–34. doi: 10.1016/s0014-5793(97)01092-2. [DOI] [PubMed] [Google Scholar]
- 44.Weis K. Importins and exportins: how to get in and out of the nucleus. Trends Biochem Sci. 1998;23:185–189. doi: 10.1016/s0968-0004(98)01204-3. [DOI] [PubMed] [Google Scholar]
- 45.Weis K, Dingwall C, Lamond A I. Characterization of the nuclear protein import mechanism using Ran mutants with altered nucleotide binding specificities. EMBO J. 1996;15:7120–7128. [PMC free article] [PubMed] [Google Scholar]
- 46.Weis K, Ryder U, Lamond A I. The conserved amino-terminal domain of hSRP1α is essential for nuclear protein import. EMBO J. 1996;15:1818–1825. [PMC free article] [PubMed] [Google Scholar]
- 47.Zapp M L, Green M R. Sequence-specific RNA binding by the HIV-1 Rev protein. Nature. 1989;342:714–716. doi: 10.1038/342714a0. [DOI] [PubMed] [Google Scholar]