Abstract
Digitoxin and structurally related cardiac glycoside drugs potently block activation of the TNF-α/NF-κB signaling pathway. We have hypothesized that the mechanism might be discovered by searching systematically for selective inhibitory action through the entire pathway. We report that the common action of these drugs is to block the TNF-α-dependent binding of TNF receptor 1 to TNF receptor-associated death domain. This drug action can be observed with native cells, such as HeLa, and reconstituted systems prepared in HEK293 cells. All other antiinflammatory effects of digitoxin on NF-κB and c-Jun N-terminal kinase pathways appear to follow from the blockade of this initial upstream signaling event.
Keywords: digitoxin, inflammation
The NF-κB signaling pathway is a central common regulator for the process of inflammation, and compounds that inhibit different components in the pathway are widely sought as potential therapeutics for inflammatory disorders and cancer. (1, 2). We have recently reported that cardiac glycoside drugs, such as digitoxin and oleandrin, can potently inhibit constitutive hypersecretion of the NF-κB-dependent proinflammatory cytokine IL-8 from cystic fibrosis (CF) lung epithelial cells (3). In CF cells, the active cardiac glycosides suppress constitutive phosphorylation of NF-κB pathway components, including IκB kinase (IKK)-α, inhibitor of NF-κB (IκBα), and NF-κB,p65 itself. Based on the data from CF cells, we have hypothesized that digitoxin and oleandrin may act on or upstream of the IKK complex (IKKsome). Although cardiac glycosides have a long history of use in heart failure, the structure–activity relationships clearly indicate that drug action on the NF-κB pathway in CF is clearly distinct from the cardiotonic effect on the failing heart (3). In addition, individual reports on oleandrin in TNF-α-activated HeLa cells have shown that it can block activation of NF-κB and activating protein 1 (4, 5). Finally, digitoxin itself has been reported to have cytotoxic actions on certain types of human tumor cells in vitro (6, 7). Thus, the action of cardiac glycosides on the NF-κB pathway may have general implications beyond the specific application to blocking constitutive pathway activation in CF.
Defective TNF-α/NF-κB signaling has been historically associated with the constitutive inflammatory process in CF (8, 9). We have therefore hypothesized that the mechanism of action of the active cardiac glycosides might be discovered by searching systematically for selective inhibitory action through the entire TNF-α/NF-κB pathway. In general terms, this pathway is initiated by the binding of free TNF-α to the TNF receptor (TNFR) TNFR1 on the plasma membrane (10). TNF-α-bound TNFR1 subsequently recruits the intracellular adapter protein TNFR1-associated death domain (TRADD) to form an active TNFR1/TRADD complex (11). A series of additional adapter molecules then assemble sequentially on this platform, including TNFR1-associated factor 2 (TRAF2), Fas-associated death domain (FADD), and receptor interacting protein (RIP) (12–14). This complex has been referred to as the inflammasome. At this point, the downstream signaling pathways are diverged to IKK and c-Jun N-terminal kinase (JNK) activation through distinct mitogen-activated protein kinase kinase kinases (14–22). The inactive cytosolic ternary complex of NF-κB heterodimers with IκBα becomes active when activated IKK phosphorylates IκBα. Phospho-IκBα becomes ubiquitinylated and destroyed in the proteosome (23–25). The NF-κB heterodimer is then free to translocate to the nucleus, where it can bind to κB cis-acting elements in the promoters of proinflammatory genes, such as IL-8, and drive expression (26, 27).
In this paper we have investigated the sensitivity of each step in the TNF-α/NF-κB signaling pathway to digitoxin and oleandrin. We report that the common action of these drugs is to block the upstream interaction between TNFR1 and TRADD, which follows when TNF-α binds to TNFR1. This drug action can be observed in native cells and reconstituted systems. All other effects of digitoxin and oleandrin on NF-κB and JNK pathways appear to follow from the blockade of this initial upstream signaling event. We suggest that this discovery will prove to be fundamentally important not only for understanding the mechanism of action of this important class of drugs but also as a chemical platform for development of approaches to the control of inflammatory processes in biology and medicine.
Methods
Cells, Culture Methods, Drugs, and Reagents. HeLa cells and 293T cells were obtained from the American Type Culture Collection and cultured in DMEM supplemented with 10% FBS/2 mM glutamine/100 units/ml penicillin/100 mg/ml streptomycin. Oleandrin (I) was obtained from Indofine Chemical (Hillsborough, NJ) at >98% purity. Digitoxin (II) and digoxigenin 3,12-diAc (VIII) [hereafter referred to as inactive cardiac glycoside (VIII)] at >95% purity were obtained from Sigma. Solubilized drugs were stored at 4°C. Crystalline drugs were stored in a dessicator at room temperature in the dark. Name, structures, and numbering system are as described in ref. 3. Drugs were solubilized as stock solutions in 100% ethanol and diluted for experiments to a final ethanol concentration of 0.1%.
Plasmids. Luciferase reporter plasmid pNF-κB-luc and the control plasmid pRL-CMV were purchased from Promega. Effector plasmids, Flag-TNFR1, hemagglutinin (HA)-mitogen-activated kinase/extracellular signal-regulated kinase (MEKK)3, Flag-NF-κB-inducing kinase (NIK), HA-IKKα, HA-IKKβ, and HA-NF-κB,p65 have previously been described (16, 22, 28–30). We are grateful to Michael Karin (University of California at San Diego, La Jolla) for plasmids HA-IKKα, HA-IKKβ, and HA-NF-κB,p65; Michael J. Lenardo (National Institutes of Health) for plasmid Flag-TNFR1; and Zheng-Gang Liu (National Institutes of Health) for plasmid HA-MEKK3. A sequence-verified spectinomycin-resistant clone for TRADD was subcloned by Gateway LR recombination into pDest-520 (Invitrogen) to produce a carboxy-terminal HA fusion protein.
Western Blot Analysis. Cells were collected and lysed in M2 buffer (20 mM Tris, pH 7.0/0.5% Nonidet P-40/250 mM NaCl/3 mM EDTA/3 mM EGTA/2 mM DTT/0.5 mM phenylmethylsulfonyl fluoride/20 mM β-glycerol phosphate/10 mM 4-nitrophenyl phosphate disodium salt/1 mM sodium vanadate/1 mg/ml leupeptin). Twenty micrograms of the cell lysate from each sample were fractionated by SDS/PAGE and immunoblotted. The blots were visualized with chemiluminescent substrate (Pierce).
Dual-Luciferase Reporter Assays. The luciferase reporter plasmid pNF-κB-luc and the control plasmid pRL-CMV were cotransfected into HeLa cells with FuGene 6 and incubated overnight (16 h). Thereafter, the cells were pretreated for 1 h with 100 nM oleandrin (I) or digitoxin (II) or 2,000 nM inactive cardiac glycoside (VIII). The HeLa cells were then incubated for 9 h with 30 ng/ml TNF-α. In the latter cases, cells were incubated overnight and then further treated with cardiac glycosides for 10 h. After incubation, cells were harvested and lysed immediately with 1× passive lysis buffer according to the manufacturers instructions (Promega). A volume of 20-μl cell lysate was then used for the dual-luciferase measurement by using the dual-luciferase reporter assay system (Promega).
Kinase Assays. HeLa cells were pretreated with cardiac glycosides [100 nM oleandrin (I) or digitoxin or 2,000 nM inactive cardiac glycoside (VIII)] for 6 h and then treated with 30 ng/ml TNF-α for 5 and 15 min. Cells were collected and lysed in M2 lysis buffer. IKK and JNK were allowed to bind to anti-IKKα and anti-JNK1 antibodies (Santa Cruz Biotechnology), respectively, and then immunoprecipitated with protein A agarose beads (Amersham Biosciences) The assays were performed by resuspending the agarose beads in 30 μl of kinase buffer (20 mM Hepes, pH 7.5/2 mM DTT/20 mM β-glycerol phosphate/50 μM sodium vanadate/10 mM 4-nitrophenyl phosphate disodium salt/10 mM MgCl2/20 μM ATP). The bead mixture was then incubated with 5 μCi (1 Ci = 37 GBq) [r-p33]ATP and candidate substrate for 30 min at 30°C. IKK and JNK kinase activities were determined by using 2 μg of GST-IκBα(1–54) and GST-c-Jun(1–79), respectively, as substrates. The phosphorylated proteins were then eluted with 1× loading buffer. The eluted proteins were subsequently resolved on 4–20% SDS/PAGE gels, and the gels were dried. The labeled proteins were then detected and quantitated by autoradiography (30).
Immunoprecipitation Assays. HeLa cells were pretreated with cardiac glycosides (100 nM oleandrin or digitoxin or 2,000 nM of the inactive digoxigenin 3,12-diAc) for 6 h, then incubated with 30 ng/ml TNF-α for 5 min. For the in vitro case, HA-TRADD and Flag-TNFR1 were cotransfected into 293T cells overnight and then treated with cardiac glycosides for 6 h. Cells were collected and lysed in M2 lysis buffer. The immunoprecipitation experiments were performed with protein G-Agarose beads (Invitrogen) by using anti-TNFR1 (R & D systems) or anti-Flag (Sigma) antibodies. After overnight incubation at 4°C, the beads were washed four times with M2 buffer and eluted with elution buffer (1× protein gel loading buffer). Eluted proteins were resolved by SDS/PAGE and detected by Western blot analysis.
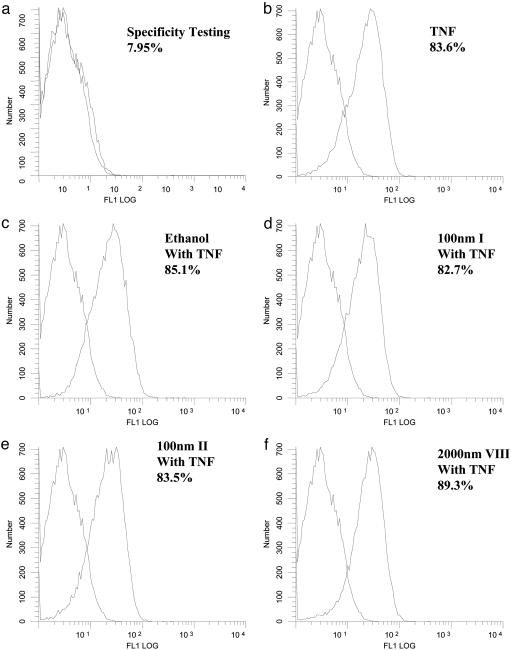
Flow Cytometry and Statistics. A 25-μl volume of washed, drug-treated cells was incubated with 10 μl of biotinylated TNF-α for 60 min at 4°C. Biotinylated soybean trypsin inhibitor was used as negative control. A 10-μl volume of avidin-FITC reagent was then added to each tube and incubated for an additional 30 min at 4°C in the dark. The cells were then washed twice with 2 ml of 1× RDF1 buffer to remove unreacted avidin-fluorescein and resuspended in 600 μl of 1× RDF1 buffer. The sample was then subjected to flow cytometric analysis by using 488-nm wavelength laser excitation (Coulter EPICS XL, Beckman Coulter). As a test of specificity, the Fc receptors on an aliquot of cells were blocked with mouse IgG. TNF-α-biotin was neutralized with anti-TNF-α antibody and then added to the cells. This process served to block nonspecific and specific binding of TNF-α-biotin to TNFR1 on the cell membrane. Experiments were performed at least three times. Statistical significance was measured by ANOVA. Differences between experimental values were said to be significant for P ≤ 0.05.
Results
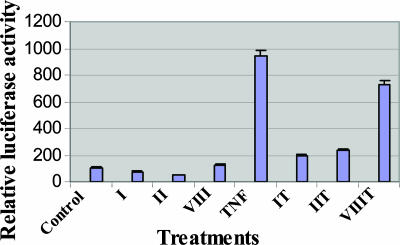
Cardiac Glycosides Inhibit NF-κB Activation. As shown in Fig. 1, when HeLa cells are transfected with a reporter gene for NF-κB, the baseline is quite low. However, this already low baseline is further suppressed by exposure to digitoxin (II) or oleandrin (I) but not by exposure to inactive cardiac glycoside (VIII). However, when the cells are incubated with TNF-α, we observe a profound activation in NF-κB reporter activity. Concurrent incubation with active cardiac glycoside drugs digitoxin (II) or oleandrin (I) reduces NF-κB activation by ≈80%. Consistently, the inactive cardiac glycoside (VIII) is relatively inactive.
Fig. 1.
Cardiac glycosides inhibit NF-κB activation. HeLa cells were cotransfected with plasmids pNF-κB-luc and pRL-CMV overnight; pretreated with 100 nM oleandrin (I), 100 nM digitoxin (II), or 2,000 nM inactive cardiac glycoside (VIII) for 1 h; and then treated with 30 ng/ml TNF-α for 10 h. Cell extracts were tested by using the dual-luciferase reporter assay system for NF-κB.
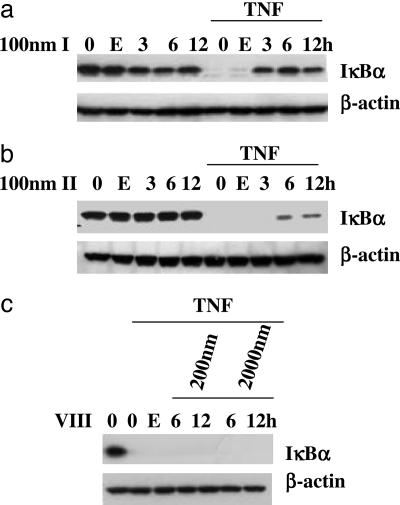
Cardiac Glycosides Prevent TNF-α-Induced Destruction of IκBα. As shown in Fig. 2a, incubation of cells with oleandrin (I) alone for 3, 6, or 12 h has little or no effect on the ambient level of IκBα. By contrast, if the cells are exposed to TNF-α, the IκBα is completely lost and activation of NF-κB can proceed. However, preincubation with oleandrin (I) for 3, 6, or 12 h before incubation with TNF-α results in virtually complete retention of IκBα in the cell. As shown in Fig. 2b, a very similar picture emerges when the cells are pretreated with digitoxin (II). The only difference is that a 3-h preincubation time is insufficient for the protective effect to be observed. The control experiment with inactive cardiac glycoside (VIII) (see Fig. 2c) indicates that when preincubated with as much as 2 μM inactive cardiac glycoside (VIII), one still observes TNF-α-dependent destruction of IκBα. Western blot analyses thus show that the drug effects on TNF-α-activated IκBα degradation consistently parallel the NF-κB reporter assays (compare Figs. 1 and 2).
Fig. 2.
Cardiac glycosides block TNF-α-induced IκBα degradation. HeLa cells were pretreated with 100 nM oleandrin (I) (a), 100 nM digitoxin (II) (b), or 2,000 nM inactive cardiac glycoside (VIII) (c) for different times, as indicated. Cells were then treated with 2 ng/ml TNF-α for 15 min. Cell extracts were tested by Western blot. E, 0.1% ethanol control; 0, no drug or carrier additions.
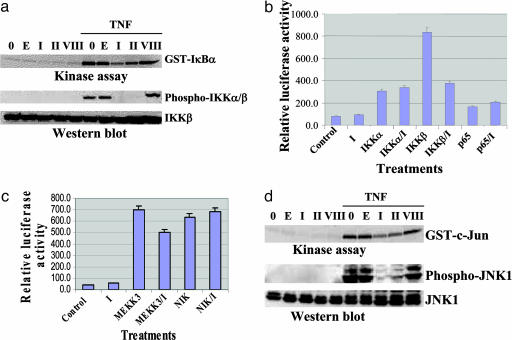
Cardiac Glycosides Affect Phosphorylation and Activation of IKK. To test whether the stabilization of IκBα by cardiac glycosides is indeed due to suppression of IκBα phosphorylation, we tested the kinase activity of IKK by incubating immunoprecipitated IKK from extracts of TNF-α-treated cells with the substrate GST–IκBα. As shown in Fig. 3a (Upper), pretreatment of cells with oleandrin (I) or digitoxin (II) substantially reduces phosphorylation of GST-IkBα. By contrast, the inactive cardiac glycoside (VIII) fails to suppress phosphorylation of the IKK substrate. We also tested the cell extracts to determine whether cardiac glycosides affect TNF-α-induced phosphorylation of IKKα/β itself. As shown in Fig. 3a (Lower), the level of IKKβ appears constant in all of the reaction mixtures. As expected, phospho-IKKα/β is readily observed when the cells are treated with TNF-α. By contrast, the active cardiac glycosides oleandrin (I) and digitoxin (II) block TNF-α-activated phosphorylation of IKKα/β. Consistently, the inactive cardiac glycoside (VIII) has no effect. Thus, it would appear that the site of cardiac glycoside action is upstream of the IKK site.
Fig. 3.
Effects of cardiac glycosides on TNF-induced IKK and JNK activation and the function of upstream or downstream kinases. (a) HeLa cells were pretreated with 100 nM oleandrin (I), 100 nM digitoxin (II), or 2,000 nM inactive cardiac glycoside (VIII) for 6 h. Cells were then treated with 30 ng/ml TNF-α for 5 min. Cell extracts were used for the IKK kinase and Western blot assays. E, 0.1% ethanol control; 0, no drug or carrier additions. (b) Reporter plasmids pNF-κB-luc and pRL-CMV were cotransfected into HeLa cells with effector plasmids HA-IKKα, HA-IKKβ, HA-p65 overnight, then treated with 100 nM oleandrin (I) for 10 h. Cell extracts were tested with the dual-luciferase reporter assay system. (c) Reporter plasmids pNF-κB-luc and pRL-CMV were cotransfected into HeLa cells with effector plasmids HA-MEKK3 and Flag-NIK overnight, then treated with 100 nM oleandrin (I) for 10 h. Cell extracts were then tested with the dual-luciferase reporter assay system. (d) HeLa cells were pretreated with 100 nM oleandrin (I), 100 nM digitoxin (II), or 2,000 nM inactive cardiac glycoside (VIII) for 6 h, then treated with 30 ng/ml TNF-α for 15 min. Cell extracts were tested by using the JNK kinase and Western blot assays.
To further test this hypothesis, we transfected cells with different constructs of the downstream IKK effectors of NF-κB signaling and examined the effects of active cardiac glycosides directly on resultant NF-κB activation. As shown in Fig. 3b, transfection of IKKα significantly activates NF-κB independently of the presence or absence of oleandrin (I). Similarly, the effect of transfection of NF-κB,p65 on NF-kB activity is independent of the presence or absence of oleandrin (I). When we transfect in IKKβ, we observe the expected profound activation of NF-κB activity. However, this activity is reduced nearly 50% in the presence of oleandrin (I) to levels seen with IKKα-induced NF-κB activity. Thus, the results for transfection with IKKα or NF-κB,p65 are consistent with an upstream action of active cardiac glycosides. The substantial cardiac glycoside-inhibited activation of IKKβ suggests the possibility of multiple sites of action for this latter kinase, including the recently identified upstream activation of Tp12 by MEKK kinase (31).
Cardiac Glycoside Action on Upstream Kinase-Driven NF-κB Activation. We next investigated the possible actions of active cardiac glycosides on NF-κB activation by MEKK3 and NIK. These kinases are considered to be upstream of the IKK complex (15, 16, 21, 22). As shown in Fig. 3c, activation of NF-κB activity by NIK is robust and is hardly affected by oleandrin (I). The activation of NF-κB by MEKK3 is also robust and is only slightly suppressed by oleandrin (I). These data therefore suggest that the site of action of the cardiac glycosides may lie at least upstream in the vicinity of the bifurcation of NF-κB and JNK signaling.
Cardiac Glycosides Block TNF-α-Driven JNK1 and c-Jun Phosphorylation. The upstream kinases that drive NF-κB activation also phosphorylate and activate the JNK and, therefore, the JNK substrate cJun (32). As shown in Fig. 3d Lower, incubation of cells with TNF-α results in the phosphorylation of JNK1. By contrast, the process is nearly completely suppressed when the cells are preincubated with oleandrin (I) or digitoxin (II). As anticipated, when preincubated with the inactive cardiac glycoside (VIII), TNF-α-driven JNK1 phosphorylation is expressed at control levels.
We proceeded to test whether cardiac glycoside-treated JNK was still active as a kinase on its physiological substrate c-Jun. Immunoprecipitates containing JNK from extracts of TNF-α-treated cells were incubated with GST-c-Jun, and the levels of phospho-GST-c-Jun were then determined by autoradiography. As shown in Fig. 3d Upper, the levels of phospho-GST-c-Jun are substantially reduced when the cells are preincubated with oleandrin (I) or digitoxin (II) but not when preincubated with the inactive cardiac glycoside (VIII), exactly consistent with phospho-JNK1. These data suggest that the site of action of oleandrin (I) and digitoxin (II) may be upstream of the bifurcation of TNF-α-dependent signals toward the JNK pathway or the NF-κB pathway.
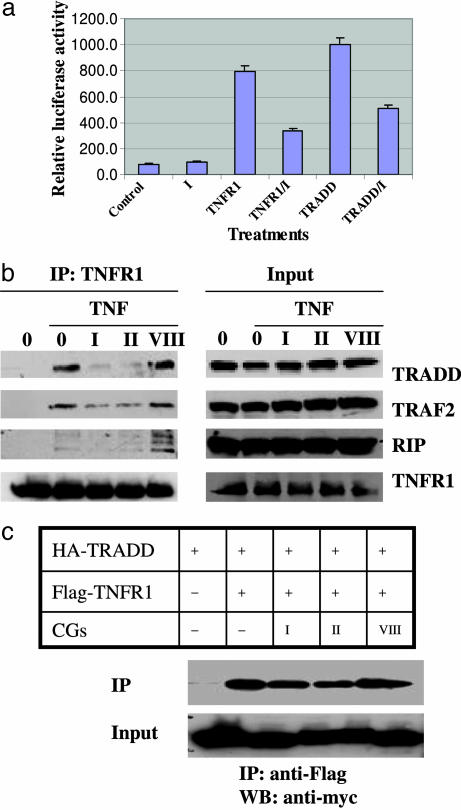
Cardiac Glycosides Block TNF-α-Dependent Recruitment of TRADD to TNFR1. The initial step leading to activation of NF-κB signaling is the formation of a complex between TNFR1 and the immediate downstream adapter molecule TRADD. To test whether active cardiac glycosides might act at this site, we transfected constructs for TNFR1 and TRADD into the cells and queried whether oleandrin (I) might interfere with signaling. As shown in Fig. 4a, the constructs for TNFR1 and TRADD increase NF-κB signaling 8- and 10-fold, respectively. By contrast, pretreatment with oleandrin (I) reduces signaling of both constructs to the NF-κB reporter by ≈50%.
Fig. 4.
Cardiac glycosides suppress TNF-α-dependent recruitment of TRADD to TNFR1. (a) Reporter plasmids pNF-κB-luc and pRL-CMV were cotransfected into HeLa cells with effector plasmids HA-TRADD and Flag-TNFR1 overnight, then treated with 100 nM oleandrin (I) for 10 h. Cell extracts were then tested with the dual-luciferase reporter assay system. (b) HeLa cells were pretreated with 100 nM oleandrin (I), 100 nM digitoxin (II), or 2,000 nM inactive cardiac glycoside (VIII) for 6 h, then treated with 30 ng/ml TNF-α for 5 min. Cell extracts were used for immunoprecipitation (IP) with anti-TNFR1 and input control. (c) The plasmids HA-TRADD and Flag-TNFR1 were cotransfected into HEK293T cells overnight and then treated with the cardiac glycosides (CGs) 100 nM oleandrin (I), 100 nM digitoxin (II), or 2,000 nM inactive cardiac glycoside (VIII) for 10 h. Cell extracts were used for immunoprecipitation experiments with anti-Flag antibody and the input control. WB, Western blot.
To directly examine the ability of cardiac glycosides to interfere with the formation of the TNFR1/TRADD complex, we performed coimmunoprecipitation experiments with anti-TNFR1. As shown in Fig. 4b (Input) TNFR1, TRADD, TRAF2, and RIP are readily identified by Western blot in extracts from untreated cells and from cells treated with drugs and/or TNF-α. Fig. 4b (IP:TNFR1) shows that when these cells are treated with TNF-α and immunoprecipitated with anti-TNFR1, TRADD, TRAF2, and RIP are also coimmunoprecipitated. However, when the cells are preincubated with oleandrin (I) or digitoxin (II), all three coimmunoprecipitants are profoundly reduced. By contrast, the inactive cardiac glycoside (VIII) fails to block the interactions.
To further validate this effect of drug action, we prepared a totally synthetic system in HEK293 cells. Our approach was to transfect the cells with Flag-tagged TNFR1 and HA-tagged TRADD. We then probed the cells for evidence of formation of Flag-TNFR1/HA-TRADD complexes. As shown in Fig. 4c, pretreatment of cells with oleandrin (I) or digitoxin (II) reduces the formation of Flag-TNFR1/HA-TRADD complexes by ≈50%. By contrast, the inactive cardiac glycoside (VIII) has no such effect. Thus, in this totally synthetic system, active cardiac glycosides can suppress the interaction between TNFR1 and TRADD independently of prior addition of TNF-α to the cell.
However, to completely exclude the possibility that the drugs might be acting by inhibiting the interaction between TNF-α itself and TNFR1, we performed a FACS analysis on HeLa cells by using biotin-labeled TNF-α and avidin-FITC. As shown in Fig. 5, the TNF-α/TNFR1 interaction is readily and reproducibly observed in cells treated with either TNF-α or TNF-α and the drug vehicle 0.1% ethanol. In addition, none of the three cardiac glycosides tested affected the TNF-α/TNFR1 interaction. These data indicate that the cardiac glycosides neither affect the density of TNFR1 molecules at the cell surface nor the binding of TNF-α to TNFR1. These four types of experiments thus validate the concept that cardiac glycosides, such as oleandrin (I) and digitoxin (II), inhibit NF-κB signaling by blocking the formation of the proximal TNF-α-dependent linkage between TNFR1 and TRADD.
Fig. 5.
Cardiac glycosides have no effect on the binding of TNF-α to TNFR1. HeLa cells were treated with 100 nM oleandrin (I), 100 nM digitoxin (II), or 2,000 nM inactive cardiac glycoside (VIII) for 6 h. The cells were then collected for staining with biotinylated TNF-α and avidin-FITC. Soybean trypsin inhibitor that had been biotinylated to the same degree was used as a negative control. Neutralized TNF-α-biotin with anti-TNF-α blocking antibody was used as a specificity control. Receptor binding activity was determined by flow cytometric analysis with 488-nm wavelength laser excitation.
Discussion
The results described in this paper give firm support to the concept that the principal mechanism by which the active cardiac glycosides oleandrin (I) and digitoxin (II) suppress the TNF-α/NF-κB signaling pathway is by blocking upstream formation of the TNF-α-dependent TNFR1/TRADD complex (see Fig. 6). The data are in complete agreement with earlier studies on CF lung epithelial cells, in which the data had been interpreted to indicate action at or upstream of the IKKsome (3). In addition, the cardiac glycoside identified as entirely inactive as an inhibitor in the CF cell system, inactive cardiac glycoside (VIII), is also inactive in the HeLa cell and HEK293 cell systems tested here. Thus, the structure–activity relationships identified previously in the CF cell system appear to be reproduced in these present studies in different human cell lines. In addition, this consistency emphasizes our previous conclusion that the suppression of NF-κB signaling is qualitatively different from the cardiotonic activity historically associated with these drugs (3, 33). The insensitivity of NIK, although consistent, must be interpreted with caution, because there is a possibility that NIK may not be involved with the TNF-α signaling pathway (34–36). Nonetheless, taken together, the logical, stepwise strategy used here to determine the site of cardiac glycoside action leaves the present study on a very firm foundation with respect to the mechanistic conclusion. We anticipate that this insight into the distinct antiinflammatory actions of cardiac glycosides may prove to be of therapeutic importance.
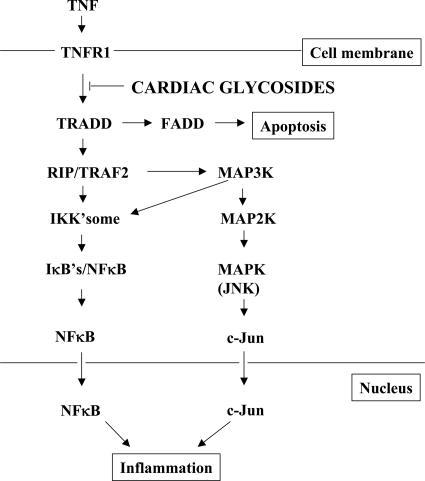
Fig. 6.
Cardiac glycosides inhibit TNF-α/NF-κB signaling by blocking recruitment of TRADD to the TNFR. The binding of TNF-α to TNFR1 initiates the TNF signal transduction pathway. TNFR1 recruits TRADD to form a complex with TNFR1. A series of additional adapter molecules then assemble sequentially on this platform, including TRAF2, RIP, and Fas-associated death domain (FADD), where they initiate downstream events leading to NF-κ B activation, JNK activation, and inflammation or apoptosis. Cardiac glycosides inhibit the TNF-α signaling pathway by blocking recruitment of TRADD to the TNFR.
The conclusion that cardiac glycosides block recruitment of TRADD to TNFR1 is based on the coincident agreement of four different types of assays. The first of these assays was to test whether the active cardiac glycosides blocked either TRADD or TNFR1 activation of NF-κB signaling in HeLa cells. We found that transfection of either construct resulted in substantial activation of NF-κB signaling, both of which were blocked by oleandrin (I). The second of these assays was an immunoprecipitation experiment with anti-TNFR1, in which the cells were treated with TNF-α. We found the now classical result (37), namely, the activation of TNFR1 results in the recruitment of TRADD, TRAF2, and RIP to TNFR1. Furthermore, the entire process was blocked by oleandrin (I) and digitoxin (II) but not the inactive cardiac glycoside (VIII). The third experiment was a totally synthetic experiment with a different human cell. We transfected HA-TRADD and Flag-TNFR1 into HEK293T cells and found that only the active cardiac glycosides blocked the coimmunoprecipitation of the HA-TRADD/Flag-TNFR1 complex. Finally, we performed a control experiment to test whether the active cardiac glycosides affected the density of TNFR1 molecules on the plasma membrane or the binding of extracellular TNF-α to membrane-bound TNFR1. We found the active drugs to be without effect on either parameter. For these logical and systematically derived reasons, we consider the mechanistic conclusions of this study to be on very solid ground.
Conclusions
The antiinflammatory effects of digitoxin on the NF-κB and JNK signaling pathways appear to follow simply from the blockade of the initial upstream recruitment event between TNF-α-activated TNFR and TRADD. The possible application of digitoxin to the therapy of inflammation in the CF airway disease is hereby based on a sound mechanistic rationale. Based on these data, we suggest the possibility that digitoxin could also make a therapeutic contribution to other inflammatory diseases for which the TNF-α/NF-κB signaling pathway is of pathophysiological importance. Finally, it has not escaped our attention that digitoxin may also be useful in addressing the proinflammatory component responsible for some cancers.
Acknowledgments
We thank Bette S. Pollard for helpful discussions. This work was supported by National Institutes of Health Grants RO1-DK53051-7 and NO1-HV-28187 (to H.B.P.) and by the Cystic Fibrosis Foundation (H.B.P.).
Author contributions: Q.Y., Y.L., M.S., and H.B.P. designed research; Q.Y., W.H., M.G., and H.C. performed research; C.J., Y.L., D.E., W.G., and J.H. contributed new reagents/analytic tools; Q.Y. and H.B.P. analyzed data; and Q.Y. and H.B.P. wrote the paper.
Abbreviations: CF, cystic fibrosis; HA, hemagglutinin; IκBα, inhibitor of NF-κB; IKK, IκB kinase; JNK, c-Jun N-terminal kinase; MEKK, mitogen-activated protein kinase/extracellular signal-regulated kinase; NIK, NF-κB-inducing kinase; TRADD, TNF-associated death domain; RIP, receptor-inhibiting protein; TRAF, TNF-associated factor.
References
- 1.Umezawa, K., Ariga, A. & Matsumoto, N. (2000) Anti-Cancer Drug Des. 15, 239-244. [PubMed] [Google Scholar]
- 2.Garg, A. & Aggarwal, B. B. (2002) Leukemia 16, 1053-1068. [DOI] [PubMed] [Google Scholar]
- 3.Srivastava, M., Eidelman, O., Zhang, J., Paweletz, C., Caohuy, H., Yang, Q. F., Jacobson, K. A., Heldman, E., Huang, W., Jozwik, C., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 7693-7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manna, S. K., Nand, K. S., Newman, R. A., Cisneros, A. & Aggarwal, B. B. (2000) Cancer Res. 60, 3838-3847. [PubMed] [Google Scholar]
- 5.Sreenivasan, Y., Sarkar, A. & Manna, S. K. (2003) Biochem. Pharmacol. 66, 2223-2239. [DOI] [PubMed] [Google Scholar]
- 6.Haux, J., Solheim, O., Isaksen, T. & Angelsen, A. (2000) Z. Onkol. 32, 11-16. [Google Scholar]
- 7.Johansson, S., Lindholm, P., Gullbo, J., Larsson, R., Bohlin, L. & Claeson, P. (2001) Anti-Cancer Drugs 12, 475-483. [DOI] [PubMed] [Google Scholar]
- 8.DiMango, E., Ratner, A. J., Bryan, R., Tabibi, S. & Prince, A. (1998) J. Clin. Invest. 101, 2598-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eidelman, O., Srivastava, M., Zhang, J., Leighton, X., Murtie, J., Jozwik, C., Jacobson, K., Weinstein, D. L., Metcalf, E. L. & Pollard, H. B. (2001) Mol. Med. 7, 523-534. [PMC free article] [PubMed] [Google Scholar]
- 10.Tartaglia, L. A. & Goeddel, D. V. (1992) Immunol. Today 13, 151-153. [DOI] [PubMed] [Google Scholar]
- 11.Hsu, H., Xiong, J. & Goeddel, D. V. (1995) Cell 81, 495-504. [DOI] [PubMed] [Google Scholar]
- 12.Hsu, H., Shu, H. B., Pan, M. G. & Goeddel, D. V. (1996) Cell 84, 299-308. [DOI] [PubMed] [Google Scholar]
- 13.Hsu, H., Huang, J., Shu, H. B., Baichwal, V. & Goeddel, D. V. (1996) Immunity 4, 387-396. [DOI] [PubMed] [Google Scholar]
- 14.Chen, G. & Goeddel, D. V. (2002) 296, 1634-1635. [DOI] [PubMed]
- 15.Woronicz, J. D., Gao, X., Cao, Z., Rothe, M. & Goeddel, D. V. (1997) Science 278, 866-869. [DOI] [PubMed] [Google Scholar]
- 16.Malinin, N. L., Boldin, M. P., Kovalenko, A. V. & Wallach, D. (1997) Nature 385, 540-544. [DOI] [PubMed] [Google Scholar]
- 17.Yeh, W. C., Shahinian, A., Speiser, D., Kraunus, J., Billlia, F., Wakeham, A., De la Pompa, J. L., Ferrick, D., Hum, B., Iscove, N., et al. (1997) Immunity 7, 715-725. [DOI] [PubMed] [Google Scholar]
- 18.Lee, F. S., Peters, R. T., Dang, L. C. & Maniatis, T. (1998) Proc. Natl. Acad. Sci. USA 95, 9319-9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishitoh, H., Saitoh, M., Mochida, Y., Takeda, K., Nakano, H., Rothe, M., Miyazono, K. & Ichijo, H. (1998) Mol. Cell 2, 389-395. [DOI] [PubMed] [Google Scholar]
- 20.Baud, V., Liu, Z.-G., Bennett, B., Suzuki, N., Xia, Y. & Karin, M. (1999) Genes Dev. 13, 1297-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao, Q. & Lee, F. S. (1999) J. Biol. Chem. 274, 8355-8358. [DOI] [PubMed] [Google Scholar]
- 22.Yang, J. H., Lin, Y., Guo, Z. J., Cheng, J. K., Huang, J. Y., Deng, L., Liao, W., Chen, Z. J., Liu, Z. G. & Su, B. (2000) Nat. Immunol. 2, 620-624. [DOI] [PubMed] [Google Scholar]
- 23.Beg, A. A. & Baldwin, A. S., Jr. (1993) Genes Dev. 7, 2064-2070. [DOI] [PubMed] [Google Scholar]
- 24.Verma, I. M., Stevenson, J. K., Schwarz, E. M., Van Antwerp, D. & Miyamoto, S. (1995) Genes Dev. 9, 2723-2735. [DOI] [PubMed] [Google Scholar]
- 25.Karin, M. & Ben-Neriah, Y. (2000) Annu. Rev. Immunol. 18, 621-630. [DOI] [PubMed] [Google Scholar]
- 26.Roebuck, K. A. (1999) J. Interferon Cytokine Res. 19, 429-438. [DOI] [PubMed] [Google Scholar]
- 27.Hoffmann, E., Dittrich-Breiholz, O., Holtmann, H. & Kracht, M. (2002) J. Leukocyte Biol. 72, 847-855. [PubMed] [Google Scholar]
- 28.Chan, F. K. M., Chun, H. J., Zheng, L. X., Siegel, R. M., Bui, K. L., Lenardo, M. J. (2000) Science 288, 2351-2353. [DOI] [PubMed] [Google Scholar]
- 29.Devin, A., Cook, A., Lin, Y., Rodriguez, Y., Kelliher, M. & Liu, Z. G. (2000) Immunity 12, 419-429. [DOI] [PubMed] [Google Scholar]
- 30.Lin, Y., Devin, A., Cook, A., Keane, M. M., Kelliher, M., Lipkowitz, S. & Liu, Z. G. (2000) Mol. Cell. Biol. 20, 6638-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waterfield, M., Jin, W., Reiley, W., Zhang, M. & Sun, S.-C. (2004) Mol. Cell. Biol. 24, 6040-6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis, R. J. (2000) Cell 103, 239-252. [DOI] [PubMed] [Google Scholar]
- 33.Kelly, R. A. & Smith, T. W. (1996) Circulation 94, 2361-2363. [DOI] [PubMed] [Google Scholar]
- 34.Matsushima, A., Kaisho, T., Rennert, P. D., Nakano, H., Kurosawa, K., Uchida, D., Takeda, K., Akira, S. & Matsumoto, M. (2001) J. Exp. Med. 193, 631-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao, G. T., Harhaj, E. W. & Sun, S. C. (2001) Mol. Cell 7, 401-409. [DOI] [PubMed] [Google Scholar]
- 36.Li, X. & Stark, G. R. (2002) Exp. Hematol. 30, 285-296. [DOI] [PubMed] [Google Scholar]
- 37.Liu, Z.-G., Hsu, H., Goeddel, D. V. & Karin, M. (1996) Cell 87, 565-576. [DOI] [PubMed] [Google Scholar]