Abstract
A similarity statistic for codon usage was developed and used to compare novel gene sequences found in clinical isolates of Haemophilus influenzae with a reference set of 80 prokaryotic, eukaryotic and viral genomes. These analyses were performed to obtain an indication as to whether individual genes were Haemophilus-like in nature, or if they probably had more recently entered the H.influenzae gene pool via horizontal gene transfer from other species. The average and SD values were calculated for the similarity statistics from a study of the set of all genes in the H.influenzae Rd reference genome that encoded proteins of 100 amino acids or longer. Approximately 80% of Rd genes gave a statistic indicating that they were most like other Rd genes. Genes displaying codon usage statistics >1 SD above this range were either considered part of the highly expressed group of H.influenzae genes, or were considered of foreign origin. An alternative determinant for identifying genes of foreign origin was when the similarity statistics produced a value that was much closer to a non-H.influenzae reference organism than to any of the Haemophilus species contained in the reference set. Approximately 65% of the novel sequences identified in the H.influenzae clinical isolates displayed codon usages most similar to Haemophilus sp. The remaining novel sequences produced similarity statistics closer to one of the other reference genomes thereby suggesting that these sequences may have entered the H.influenzae gene pool more recently via horizontal transfer.
INTRODUCTION
Synonymous codons—in which >1 nt triplet encodes a particular amino acid—are an integral feature of all eukaryotic and prokaryotic genomes (1–3). Most bacterial species display unique preferences in the choice of at least some of these codons (4,5) and these preferences form a type of genomic signature that most genes of a species possess. Moreover, many bacterial species also possess a second set of genes associated with translation and basic metabolic functions that are highly expressed (6). This subset of highly expressed genes have a markedly different codon usage signature than the rest of the genome, however, they have similarities amongst themselves (7–9). Identification of genes within an organism's genome that have codon usage biases, which differ from both of the aforementioned patterns, may therefore be an indication that such sui generis genes are of foreign origin and still carry the vestiges of the codon preferences from their previous hosts (5,10). The fact that the genes making up this third group have disparate codon usage biases among themselves further supports the hypothesis that they have entered the host genome via multiple independent gene transfer events.
The selective nature of codon usage by a species offers a potential tool for the characterization of all genes within an organism, which in turn is useful in understanding the evolution of a target species, and for measuring horizontal gene flow across bacterial species. We have previously constructed a pooled genomic library from 10 clinical isolates of Haemophilus influenzae (11). Analysis of this library revealed that ∼9% of the clones contained unique sequences when compared with the H.influenzae reference strain Rd (12). Distribution studies of these novel sequences revealed that most of these unique genes were present in only a subset of the 10 strains from which the library was prepared (12). In the current study, it was our goal to characterize these novel H.influenzae genes in terms of their codon usage biases by comparing each gene against a set of reference genomes from organisms with well-established codon usage preferences. This type of genic characterization and comparison would not only provide a magnitude, but also a direction with respect to codon bias in the 59-dimensional codon space for horizontally transferred genes.
The codon adaptivity index (CAI) (4) is often used as a univariate measure of codon usage similarity to a given reference set. However, the CAI does not scale properly when used as a comparison statistic between or among different reference sets (13). An alternate, recently developed method used a statistic based on the squared difference of codon usage frequencies between the open reading frame (ORF) being tested and the chosen reference set (1). This method reliably predicts highly expressed genes among various prokaryotes (7). Such a similarity statistic could be applied to different reference sets and be directly comparable among them.
In the current study, we developed and optimized a family of statistics, based on the square of the difference of codon usage frequencies, to compare novel sequences found in our Haemophilus isolates [obtained from patients with chronic otitis media with effusion (OME)] with a reference set of 80 genomes. Included in this reference set were H.influenzae Rd, three other Haemophilus sp. and the Haemophilus phages HP1 and HP2. The codon usage for these six reference genomes formed the Haemophilus group. As a test of the method and in order for the method to be a useful tool for predicting typical Haemophilus coding patterns, a large majority of H.influenzae Rd genes were needed to select a member of the Haemophilus grouping as the closest fitting genome in terms of codon usage. The results of our investigations established the robustness of this approach and indicated that the overwhelming majority of Rd genes are more like Haemophilus genes than the genes of any of the other reference genomes.
Studies of bacterial genomes including those of several strains of H.influenzae (14–16) and the Bakaletz–Munson website at www.microbial-pathogenesis.org, have provided much of the data behind the concept of the supra-genome (17,18). The supra-genome exists only as a theoretical construct at the population level and is significantly larger than the genome of any one single strain; it is composed of (i) a common core set of genes, which define the species; and (ii) a large number of discrete contingency genes, which are distributed among the component strains in the population. Thus, each strain has a subset of the contingency genes from the population-based supra-genome (19,20) and these contingency genes are exchanged among strains of a species via horizontal gene transfer through natural competence and transformation mechanisms. In addition to intraspecies genetic exchange, evidence is mounting that bacterial genomes also evolve through horizontal gene transfer between and among species (10). Thus, although more uncommon than intraspecies gene exchange, it is likely that there exists a finite gene exchange rate across entire prokaryotic phyla and kingdoms (21–25). A method that is capable of estimating the origin of a novel gene and its duration within a given species' supra-genome would be of practical use.
METHODS
We compared the codon usage of the H.influenzae Rd genes to a set of 80 reference organisms that included eukaryotes, prokaryotes, phages and viruses. The GC content of these reference genomes spanned a wide range (24–68%). Haemophilus-like codon usage was represented by four Haemophilus strains having a GC content of 35–38% and the Haemophilus phages HP1 and HP2, both of which have a GC content of ∼40.4%. Many of the remaining 74 reference organisms were selected because they had Haemophilus-like GC contents. This was to guard against Haemophilus being chosen as a best-fit organism based upon GC content alone. The collective reference set was, however, sufficient to over-represent the 59-dimensional codon space composed of all codons except the three stop codons and the codons belonging to the single codon amino acids methionine and tryptophan.
A codon bias statistic (ɛ) was generated for each amino acid in each ORF (Equations 1, 3 and 6). Absolute codon frequencies were used rather than relative frequencies (Equation 2) (1), and an explicit amino acid usage factor (CA) was employed to compensate for differing amino acid usage between ORF and reference. Equations 1 and 2 are in fact equivalent. The amino acid usage factor was again employed in Equation 3 and explicitly optimized (Equations 4 and 5) so as to facilitate a fitting between ORF and reference in order to minimize artificial differences caused by small codon sample size. Equation 3 doesn't involve taking the absolute magnitude of the squared codon frequency differences, hence it emphasizes larger differences over many smaller differences.
| 1 |
where fi,A is the % usage of the ith codon of amino acid A in the reading frame being tested; gi,A, the % usage of the ith codon of amino acid A in the organism being tested against; and CA, the amino acid bias factor.
| 2 |
where P(A) is the % usage of amino acid A; qi,A, the relative % usage of the ith codon of amino acid A in the reading frame being tested; and ri,A, the relative % usage of the ith codon of amino acid A in the organism being tested against.
| 3 |
where fi,A is the % usage of the ith codon of amino acid A in the reading frame being tested; gi,A, the % usage of the ith codon of amino acid A in the organism being tested against; and CA, the amino acid bias factor also used as an optimization parameter.
| 4 |
| 5 |
As a final approach an amino acid ɛ statistic was obtained as a type of distance measure (Equation 6).
| 6 |
This approach does not weight each of the 59 codons equally as in Equation 1. Instead those codons associated with 2-codon amino acids have a relatively larger contribution. The reason for this approach is simply concern over small codon sample size. As an example, if it is assumed that all amino acids are used to the same extent, then a gene harboring 200 codons will have an average of five codon counts per codon for those codons associated with 2-codon amino acids, but only an average of 1.67 codon counts per codon for those codon associated with 6-codon amino acids. Hence, the codon distribution is much more reliable for 2-codon amino acids. The amino acid statistics for the 18 amino acids with more than one codon were then summed in order to produce an overall codon bias for the ORF (Equation 7).
| 7 |
Determination of codon sample size (gene length dependence) on ɛ values
The dependence of the ɛ-statistic upon gene length and subsequent codon sample size was investigated in order to obtain a gene length dependent threshold for defining high values of ɛ. Regular intervals were chosen and the average and SD for those genes in the interval were obtained and assigned to the mid-point of the interval.
Phylogenetic trees
Phylogenetic trees were generated for 10 of the novel genes identified by DNA sequence analysis of random clones from the pooled genomic library in order to obtain additional information regarding their origins. All of the trees were constructed based on the amino acid sequences and each tree was built using all of the genes available in the corresponding cluster of orthologous genes (COGs). The COG data were downloaded from the GenBank database. Sequence alignments among the members of a COG were finished using ClustalW in BioEdit 7.0 (http://www.mbio.ncsu.edu/BioEdit/). Maximum likelihood trees were constructed using PHYML 2.4.4 (http://atgc.lirmm.fr/phyml/) running on a Dell Intel XEON PC (3.06 GHz XEON, double CPU, 240 GB hard disk, 2 GB RAM). The settings used in running the PHYML program were as follows: data type: amino acid; number of data sets: 1; number of bootstraps used: 1000; substitution model: JTT; proportion of invariable sites: estimated; number of substitution rate categories: 4; gamma distribution parameter: estimated; input tree: BIONJ; optimize tree topology: yes; optimize branch lengths and rate parameters: yes. The trees were viewed by using MEGA 3.0 (http://www.megasoftware.net/). Each data set was used to make two trees: a consensus tree showing the percentage of bootstrap values that supported the tree as drawn, and a second tree showing the branch lengths proportional to the evolutionary distance. The bootstrap value for some nodes were omitted by the software due to a cut-off value set at 30%. Both trees for each gene are available at our website www.centerforgenomicsciences.org under public documents.
Software development
A java script (CODESQUARE) was written to automate the generation of the codon usage statistic for each amino acid of each ORF for each reference genome. This program is available from the CGS website (www.centerforgenomicsciences.org).
RESULTS
Reference organisms
The average codon usages of 80 reference genomes were obtained from the KAZUSA (www.kazusa.or.jp/codon/) and TIGR sites (www.tigr.org) and used to help characterize the codon usage patterns of the individual genes of the H.influenzae Rd genome via an evaluation of the ɛ statistic from Equations 1, 3 and 6. These reference genomes were also used to evaluate the probable origin of a collection of novel ORFs identified by DNA sequence analysis of random clones from a pooled genomic library prepared from 10 clinical isolates of H.influenzae recovered from chronic middle-ear infections.
The reference organisms, phage and viruses are listed in Table 1 along with their GC contents and ɛ-statistics from Equation 1, which measured the codon usage similarity of each reference organism to that of H.influenzae Rd. GC content did not impose any predictable order upon these ɛ values. The reference organisms were deliberately selected such that most had GC contents that were similar to that of the H.influenzae genome to provide an adequate test of the discriminatory power of the ɛ-statistic.
Table 1.
Characteristics of the reference organisms
| Reference organism (abbreviated) | ɛ (Equation 1) | GC% |
|---|---|---|
| H.influenzae Rd (HFRD) | 0 | 38.76 |
| H.influenzae (HF) | 7.9 | 37.45 |
| H.ducreyi (HFDY) | 12.65 | 37.15 |
| Pasteurella multocida (PM) | 12.89 | 40.74 |
| H.influenzae aegyptius (HFAY) | 15.48 | 37.15 |
| Actinobacillus actino (AA) | 16.48 | 38.99 |
| HP1 phage (HP1) | 16.51 | 40.44 |
| Listeria innocua (LI) | 17.12 | 37.79 |
| HP2 phage (HP2) | 17.32 | 40.4 |
| Staphylococcus aureus (SA) | 22.26 | 32.88 |
| Lactococcus lactis (LL) | 22.49 | 35.5 |
| enterobactphage T4 (T4) | 22.5 | 35.37 |
| Legionella pneumophila (LP) | 24.53 | 40.61 |
| Bacteriophage A118 (A118) | 25.26 | 36.33 |
| Streptococcus pneumoniae (SP) | 28.8 | 39.14 |
| Mycoplasma genitalium (MG) | 28.83 | 31.74 |
| Guillardia theta (GT) | 28.96 | 24.98 |
| Campylobacter jejuni (CJ) | 30.9 | 30.76 |
| Schizosaccharomyces pombe (SCHZ) | 33.26 | 39.8 |
| Borrelia burgdorferi (BB) | 33.29 | 29.24 |
| Ureaplasma urealyticum (UU) | 33.62 | 26.75 |
| Chlamydia muridarum (CM) | 34.24 | 40.62 |
| Candida albicans (CA) | 34.36 | 36.88 |
| Gifsy-1 (GF1) | 34.79 | 42.27 |
| Bacillus halodurans (BH) | 34.84 | 44.32 |
| Vibrio cholerae (VC) | 36.4 | 47.35 |
| Saccharomyces cerevisiae (SC) | 36.67 | 39.71 |
| Plasmodium falciparum (PF) | 36.69 | 25.87 |
| Helicobacter pylori 26695 (HP) | 36.81 | 39.56 |
| Lactobacillus helveticus (LH) | 36.91 | 38.93 |
| Salmonella enterica (SE) | 36.98 | 45.67 |
| Shigella flexneri (SF) | 37.53 | 45.49 |
| Clostridium butyricum (CB) | 38.84 | 26.25 |
| Gifsy-2 (GF2) | 39.81 | 43.88 |
| Bacillus subtilis (BS) | 40.01 | 44.32 |
| influenzae C virus (INFC) | 40.23 | 38.38 |
| Human papilomavirus 16 (HP16) | 40.8 | 38.12 |
| Yersinia pestis (YP) | 41.03 | 48.97 |
| Caenorhabditis elegans (CE) | 42.37 | 42.76 |
| Human rotavirus (strain rv5) (HR5) | 43.51 | 32.77 |
| Shigella flexneri 2a (SF2A) | 43.65 | 47.67 |
| Methanococcus jannaschii (MJ) | 43.74 | 31.85 |
| Fusobacterium nucleatum (FN) | 43.92 | 26.76 |
| FIV (FIV) | 44.07 | 36.44 |
| Arabidopsis thaliana (AT) | 45.36 | 44.41 |
| Clostridium sticklandii (CS) | 45.95 | 36.32 |
| Mycoplasma penetrans (MP) | 46.01 | 30.55 |
| bacteriophage 933W (933W) | 47.86 | 49.81 |
| Hepatitis A virus (HepA) | 47.97 | 37.15 |
| Clostridium kluyveri (CK) | 48.15 | 34.16 |
| enterobactphage Mu (Mu) | 48.22 | 52.14 |
| Mycoplasma mycoides (MM) | 51.25 | 24.64 |
| Escherichia coli K12 (K12) | 51.95 | 51.83 |
| HIV type1 (HIV1) | 52.21 | 43.33 |
| Xenopus laevis (XL) | 53.58 | 47.34 |
| Bacteriophage N15 (N15) | 53.69 | 51.39 |
| Treponema pallidum (TP) | 53.84 | 52.52 |
| influenzae A virus (HongKong) (infA) | 55.15 | 44.78 |
| Xylella fastiodosa (XF) | 55.73 | 53.78 |
| HIV type2 (HIV2) | 57.74 | 46.14 |
| Neisseria gonorrhea (NG) | 62.97 | 52.59 |
| Neisseria meningiditis (NM) | 63.68 | 53.06 |
| Plasmid R124 (R124) | 64.75 | 51.45 |
| Thermotoga maritima (TP) | 65.62 | 46.45 |
| Homo sapiens (HS) | 67.09 | 52.66 |
| Mus musculus (Mus) | 68.15 | 52.41 |
| Agrobact. sp. (Agr) | 69.52 | 56.86 |
| Klebsiella pneumoniae (KP) | 70.1 | 55.79 |
| Oryza sativa (OS) | 70.47 | 55.98 |
| Rhizobium rhizogenes (RR) | 75.35 | 57.6 |
| Drosophila melanogaster (DM) | 75.59 | 54.03 |
| Archaeoglobus fulgidus (AF) | 76.59 | 49.37 |
| Mycobacterium leprae (ML) | 79.22 | 59.9 |
| Anopheles gambiae (AG) | 86.47 | 56.93 |
| Chlorobium tepidum (CT) | 92.4 | 57.73 |
| Mesorhizobium loti (LOTI) | 99.09 | 63.24 |
| Mycobacterium tuberculosis (MT) | 101.35 | 64.48 |
| Deinococcus radiodurans (DR) | 108.1 | 67.24 |
| Pseudomonas aeruginosa (PA) | 114.7 | 66.45 |
| Caulobacter crescentus (CC) | 116.67 | 67.67 |
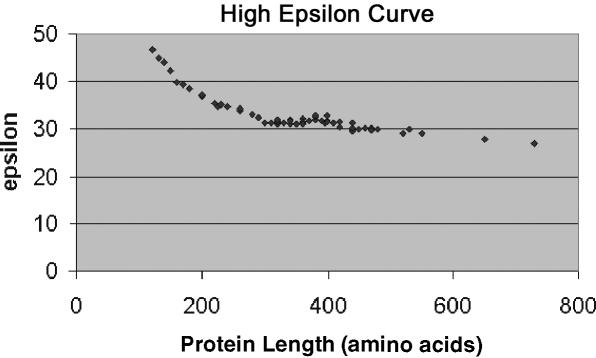
ɛ values represent the overall measure of codon usage similarity between a gene and a genome, with lower values indicative of a better fit. We calculated 80 separate ɛ values (one for each of the 80 genomes in the reference set) for each gene (N = 1538) that encoded a protein of >100 amino acids in the Rd genome. The dependence of the ɛ-statistic from Equation 1 upon gene length itself can be seen in Figure 1. The results indicate that the codon sample size of a gene imposes a significant statistical effect at gene lengths <240 amino acids. Thus, even short (<240 amino acids) Haemophilus-like genes are unlikely to obtain ɛ values as low as those seen for genes with larger codon sample sizes. This knowledge of the gene length dependence on ɛ is necessary to help identify genes within the Rd genome and our clinical isolates that are suggestive of either foreign origin or of being a member of a set of highly expressed genes. An additional interesting aspect about Figure 1 is an upwards deviation from the exponential curve between 350 and 450 amino acids. This deviation is evenmore apparent if Equations 3 or 6 are used to calculate ɛ (see Supplementary Information).
Figure 1.
Chart showing the correlation between protein length in amino acids (x-axis) and average ɛ value (y-axis).
Threshold levels and statistical choices
The choice of using the average plus SD as the threshold, for defining high ɛ genes using Equation 1, captured 110/142 genes previously classed among the highly expressed genes; 82/142 were identified using Equation 3; and using Equation 6 107/142 were identified. Equations 1, 3 and 6 identified a total of 191, 127 and 176 of 1538 Rd genes, respectively, as having high ɛ values. The ɛ-statistic in Equation 3 uses the square and not the absolute value of the codon frequency and widens the range of ɛ values obtained resulting in fewer genes above the mean ɛ value for genes of approximately the same gene length. The genes with high ɛ values based upon Equations 3 and 6 which were not identified by Equation 1 include: two ribosomal genes that are obviously part of the highly expressed set; a glycosyltransferase (HI0872) a class of genes known to be mosaic (26); and five genes associated with iron usage.
Haemophilus-like genes
The goal of this study was to find a statistic that would reliably predict if a gene was a long-standing member of the Haemophilus gene pool or not. The statistic used would only be useful if the vast majority of H.influenzae Rd genes selected a member of the Haemophilus genus as the closest fitting genome in terms of codon usage. This criterion was met, with 86% of the 635 genes encoding proteins >320 amino acids in the H.influenzae Rd genome having their closest codon usage similarity to one of the Haemophilus species (Table 2). This result was obtained regardless of whether Equations 1, 3 or 6 were used. While these methods suffered a small loss in predictive power with decreasing gene size, overall 78% of the genes encoding proteins longer than 140 amino acids selected one of the Haemophilus genomes. The method was sufficiently sensitive to discriminate the Haemophilus group from their closest relative, Pasteurella multocida (PM), in that only 6.4% of the Rd genes chose the P.multocida genome. It is important to stress that there is no learning by the program as to what is ‘Haemophilus’, rather each of the 80 reference genomes were individually tested against each Rd gene.
Table 2.
Percentage of genes best-fitting Haemophilus with optimized amino acid bias factor
| Amino acid range | Number of genes | ɛ Average | SD | % Genes selecting Rd | % Genes selecting (HF + HFAY + HFDY) | % Genes selecting (HP1 + HP2) | % Genes selecting Haemophilus group in total |
|---|---|---|---|---|---|---|---|
| 100–139 | 150 | 40.04 | 6.54 | 25.33 | 20.67 | 10 | 56 |
| 140–179 | 176 | 34.52 | 5.31 | 36.93 | 24.43 | 4.55 | 65.9 |
| 180–239 | 269 | 31.65 | 4.91 | 44.61 | 20.82 | 7.43 | 72.9 |
| 240–319 | 310 | 28.02 | 4.83 | 58.39 | 13.23 | 8.06 | 79.7 |
| 320+ | 633 | 24.36 | 5.94 | 63.19 | 15.32 | 7.42 | 85.9 |
HP1 = Haemophilus phage 1; HP2 = Haemophilus phage 2; HFAY = Haemophilus influenzae biogroup aegyptius; HFDY = Haemophilus ducreyi.
A total of 31 different non-Pasteurellaceae genomes were selected as the closest fit by at least one Rd gene using Equation 1 (Table 3). Ureaplasma urealyticum (UU) was selected the most—31 times. Approximately half of the genes (131 out of 252), which selected non-Pasteurellaceae reference genomes as most similar in codon usage, selected one of the following: Actinobacillus actinomycetemcomitans; Listeria innocua; Staphylococcus aureus; Lactococcus lactis; enterobacteriophage T4 (T4); or Legionella pneumoniae (LP). These are the six non-Haemophilus reference organisms, excluding Pasteurella, that are most similar to Rd in codon usage in this study (Table 1). Hence, the codon usage even for the majority of genes which selected non-Haemophilus reference organisms does not stray too far from the average Haemophiloid pattern. UU is, however, farther removed from Rd in its codon usage patterns than these six. We have found that the average length of a gene which selects UU is 212 amino acids whereas the average length of a gene which selects a non-Haemophilus reference organism is 260 amino acids. In addition, 21 of the 31 genes which select UU were shorter than 212 amino acids. Hence, this may indicate that a pattern of codon usage may exist for small genes within Haemophilus distinct from the typical Haemophiloid pattern. The results are similar if Equation 6 is used to generate ɛ, however, LP becomes the most selected non-Pasteurellaceae reference organism, a total of 29 times, whereas UU is selected 25 times.
Table 3.
Organisms providing the closest similarity to the 1538 Haemophilus influenzae Rd genes of length >100 amino acids
| GC content | Number of genes | H.strains | H.phage | PM | Remaindera |
|---|---|---|---|---|---|
| 25–28 | 15 | 2 | 0 | 0 | BB(2),CB(2),FN,GT(2),MM,PF,SA,UU(3) |
| 29–32 | 58 | 19 | 0 | 0 | CB(3),LL(3),MG(3),SA(3),CJ(4),T4(4),UU(7),BB(2),CM(2),LI(2),PF(2)HR5,LP |
| 33–42 | 1335 | 994 | 82 | 81 | AA(11),A118(3),BB,BH(5),BS,CN(2),CB,CJ(2),CM(2),T4(20),GF2(2),GT(6),HP(2),INFC,LH(6),LI(22),LL(17),LP(23),MG(4),MP(2),SA(19),SCHZ(2), SE,SP(3),UU(20) |
| 43–46 | 108 | 57 | 20 | 16 | AA(2),BH,BS,LH,MP,NG,SA(2),SE.SP,T4,UU,VC(2) |
| 47–50 | 22 | 0 | 13 | 2 | AA,HP,NG,VC(5) |
H. strains = Best fit to one of the four Haemophilus strains; H. phage = Best fit to Haemophilus phage HP1 or HP2; PM = Best fit to P.multocida.
aSee Table 1 for explanation of abbreviations.
Highly expressed genes
H.influenzae, like most bacteria, reserves a special codon usage pattern for a retinue of highly expressed genes. The usage patterns for these highly expressed genes are markedly different from the average H.influenzae codon usage pattern, but cluster together among themselves. As a consequence, the highly expressed genes will have unusually high ɛ values when compared to the average H.influenzae codon usage.
We have identified 191 genes that have ɛ values obtained from Equation 1 that are more than one SD above the average ɛ value for genes of equivalent length (see Figure 1). The majority of these genes (110/191) are part of the highly expressed group of genes, including ribosomal constituents, protein involved in metabolic activities and chaperonin genes that have been previously identified (8). The genes listed in Table 4 are not part of this previously published list and are therefore horizontal gene transfer candidates.
Table 4.
Characteristics of Haemophilus influenzae Rd genes displaying an ɛ value that exceeded the mean ɛ value plus one SD obtained from Equation 1
| Gene name | Gene | %GC | Length (amino acid) | ɛ | Ref. org. | % Better fit > Haemophilus | ɛ (w/r ribosomal group) |
|---|---|---|---|---|---|---|---|
| Cell envelope | |||||||
| Lipoprotein (nlpC) | HI1314 | 40 | 161 | 42.43 | HFAY | 0 | 62.3 |
| Lic-1operon protein(licA) | HI1537 | 33 | 267 | 33.45 | HFDY | 0 | 63.49 |
| Lic-1operon protein(licD) | HI1540 | 34 | 265 | 33.86 | A118 | 8.9 | 49.85 |
| Undecaprenyl-phosphate alpha-N-acetylglucosaminyl transferase(rfe) | HI1716 | 37 | 356 | 32.52 | HFRD | 0 | 69.71 |
| Cellular processes | |||||||
| Lactoylglutathionelyase(gloA) | HI0323 | 41 | 135 | 47.77 | HFRD | 0 | 53.51 |
| Competence proteinF (comF) | HI0434 | 38 | 230 | 36.22 | PM | 0.1 | 64.33 |
| Carbonic anhydrase-putative | HI1301 | 38 | 230 | 37.88 | HFRD | 0 | 55.98 |
| Conserved or predicted hypothetical | |||||||
| Conserved hypothetical protein (homol. to haloacid dehalogenase-like protein) | HI0003 | 37 | 263 | 35.27 | HFRD | 0 | 72.47 |
| H.influenzae predicted coding region HI0152 (homol. to phosphopentetheinyl transferase) | HI0152 | 35 | 236 | 35.08 | ET4 | 2.3 | 71.52 |
| H.influenzae predicted coding region HI0221.1 (homol. to IMP dehydogenase-like protein) | HI0221.1 | 44 | 162 | 43.64 | HFRD | 0 | 35.55 |
| Conserved hypothetical protein (predicted hydrolase or acyltransferase) | HI0282 | 36 | 248 | 35.42 | LP | 7.1 | 72.05 |
| Conserved hypothetical protein (homol. to transcriptional regulator) | HI0304 | 39 | 186 | 41.25 | PM | 2.5 | 68.96 |
| Conserved hypothetical protein (homol. to lysine 2,3-aminomutase) | HI0329 | 38 | 338 | 32.18 | HFRD | 0 | 66.55 |
| Conserved hypothetical protein | HI0510 | 42 | 239 | 36.98 | HP1 | 0 | 59.91 |
| Conserved hypothetical protein (homol. to pyruvate formate lyase) | HI0520 | 39 | 263 | 35.38 | PM | 1.6 | 75.17 |
| H.influenzae predicted coding region HI0554 (homol. to transposase) | HI0554 | 30 | 181 | 39.08 | UU | 4.5 | 72.21 |
| Conserved hypothetical protein | HI0638 | 38 | 205 | 37.07 | HP2 | 0 | 65.62 |
| Conserved hypothetical protein (homol. to DNA topoisomerase) | HI0656.1 | 39 | 179 | 39.7 | AA | 0.4 | 74.86 |
| Conserved hypothetical protein (4-diphosphocytidyl-2-C-methylerythritol synthase-like protein) | HI0672 | 40 | 226 | 36.26 | HFDY | 0 | 63 |
| Conserved hypothetical protein (probable pseudouridylate synthase) | HI0694 | 37 | 240 | 34.66 | HP1 | 0 | 67.36 |
| Conserved hypothetical protein (homol. to integral membrane protein) | HI0862 | 39 | 236 | 37.91 | LI | 0.5 | 43.75 |
| Conserved hypothetical protein (homol. to cytosine/adenosine deaminase) | HI0906 | 41 | 173 | 39.17 | PM | 0.3 | 72.95 |
| Conserved hypothetical protein (homol. to methylases) | HI0925 | 36 | 122 | 47.52 | HFDY | 0 | 76.27 |
| H.influenzae predicted coding region HI0983 | HI0983 | 35 | 194 | 40.33 | HFRD | 0 | 71.52 |
| H.influenzae predicted coding region HI1055 (type III restriction modification endonulase-like, strong homology between Haemophilus strains and Neisserial strains) | HI1055 | 39 | 515 | 29.75 | HP2 | 0 | 66.79 |
| H.influenzae predicted coding region HI1058 (homol. to adenine-specific DNA methylases, strong homology between Haemophilus and Neisserial strains) | HI1058 | 40 | 195 | 51.88 | GF2 | 9.9 | 88.85 |
| Conserved hypothetical protein (homology to membrane protein) | HI1073 | 38 | 125 | 49.31 | HF | 0 | 52.46 |
| Conserved hypothetical GTP-binding protein (predicted GTPase) | HI1118 | 40 | 206 | 37.32 | HP2 | 0 | 65.19 |
| Conserved hypothetical protein | HI1150 | 34 | 210 | 38.9 | HFDY | 0 | 68.94 |
| Conserved hypothetical protein (probable translation factor) | HI1198 | 39 | 207 | 36.96 | HFRD | 0 | 67.81 |
| H.influenzae predicted coding region HI1343 (probable selenocysteine lyase) | HI1343 | 39 | 239 | 36.09 | LL | 7.5 | 75.17 |
| H.influenzae predicted coding region HI1375 | HI1375 | 28 | 302 | 33.53 | HFRD | 0 | 61.53 |
| H.influenzae predicted coding region HI1498 (homol. to Mu-like phage protein gp25) | HI1498 | 47 | 139 | 44.72 | HP1 | 0 | 81.43 |
| H.influenzae predicted coding region HI1499 (homol. to Mu-like phage protein gp27) | HI1499 | 46 | 189 | 40.88 | PM | 0.1 | 71.55 |
| H.influenzae predicted coding region HI1500 (homol. to Mu-like phage protein gp28) | HI1500 | 48 | 508 | 30.69 | HP2 | 0 | 64.05 |
| H.influenzae predicted coding region HI1505 (homol. to Mu-like phage protein major head subunit) | HI1505 | 47 | 308 | 33.23 | HP1 | 0 | 53.79 |
| Conserved hypothetical protein (homol. to Mu-like phage protein gp36) | HI1508 | 47 | 141 | 45.89 | HP1 | 0 | 66.55 |
| Conserved hypothetical protein (homol. to Mu-like phage protein gp37) | HI1509 | 49 | 194 | 41.08 | VC | 6.2 | 69.66 |
| H.influenzae predicted coding region HI1518 f(homol. to Mu-like phage protein gp45) | HI1518 | 50 | 182 | 41.53 | VC | 2.9 | 74.48 |
| H.influenzae predicted coding region HI1519 (homol. to Mu-like phage protein gp46) | HI1519 | 50 | 135 | 50.11 | HP | 3.9 | 77.58 |
| H.influenzae predicted coding region HI1523 | HI1523 | 38 | 296 | 33.71 | HP2 | 0 | 57.79 |
| H.influenzae predicted coding region HI1570 (homol. to ribonuclase) | HI1570 | 42 | 170 | 47.82 | BH | 16.2 | 81.55 |
| Conserved hypothetical protein (probable 3-Deoxy-D-manno-octulosonate 8-phosphate phosphatase) | HI1679 | 43 | 180 | 38.57 | HFRD | 0 | 68.72 |
| Conserved hypothetical protein [homol. to Mn(+2) and Fe(+2) transporters] | HI1728 | 39 | 398 | 31.06 | HFRD | 0 | 61.31 |
| Conserved hypothetical protein (homol. to lactam utilization protein) | HI1729 | 39 | 258 | 34.28 | HFRD | 0 | 61.49 |
| Metabolism | |||||||
| Esterase | HI0184 | 44 | 276 | 33.45 | PM | 5 | 72.89 |
| Ferredoxin-type protein (napH) | HI0346 | 42 | 287 | 32.89 | HFRD | 0 | 62.02 |
| Urease accessory protein (ureH) | HI0535 | 44 | 262 | 36.19 | SE | 16.4 | 78.55 |
| Urease accessory protein(ureG) | HI0536 | 44 | 226 | 40.57 | VC | 5.7 | 47.87 |
| 2-Hydroxy acid dehydrogenase | HI1556 | 39 | 316 | 31.93 | HFRD | 0 | 59.98 |
| Enoyl-(acyl-carrier-protein) reductase (fabI) | HI1734 | 43 | 296 | 36.5 | HFRD | 0 | 41.78 |
| Nucleosides, nucleotides, purines, pyrimidines | |||||||
| Hydroxy ethylthiazole kinase | HI0415 | 48 | 265 | 34.79 | HP1 | 0 | 75.16 |
| Thymidylate synthetase (thyA) | HI0905 | 40 | 283 | 33.19 | HFRD | 0 | 66.13 |
| Uracil phosphoribosyl transferase (upp) | HI1228 | 41 | 209 | 36.28 | HFRD | 0 | 42.11 |
| Phosphoribosyl aminoimidazole synthetase (purM) | HI1429 | 44 | 345 | 31.19 | HP2 | 0 | 42.27 |
| Phage-like | |||||||
| Transposase (muA) | HI1478 | 48 | 686 | 30.55 | HP2 | 0 | 56.04 |
| DNA transposition protein (muB) | HI1481 | 48 | 287 | 37.29 | HP2 | 0 | 60.03 |
| E16 protein-putative | HI1488 | 42 | 184 | 41.89 | GF2 | 8.4 | 74.87 |
| Iprotein (muI) | HI1504 | 48 | 355 | 33.09 | HP2 | 0 | 56.76 |
| Sheath protein gpL (muL) | HI1511 | 48 | 487 | 30.4 | HP2 | 0 | 66.86 |
| 64 kDa virion protein (muN) | HI1515 | 46 | 455 | 31.34 | VC | 8.7 | 64.41 |
| Gprotein (muG-2) | HI1568 | 44 | 139 | 43.51 | PM | 0.1 | 60.71 |
| Regulators | |||||||
| Transcriptional regulator-putative | HI0186 | 38 | 135 | 46.22 | HP1 | 0 | 67.41 |
| Transcriptional regulatory protein | HI1476 | 43 | 240 | 37.42 | BS | 1.5 | 56.47 |
| Replication | |||||||
| Integrase/recombinase (xerD) | HI0309 | 42 | 297 | 32.44 | HFRD | 0 | 67.01 |
| Holliday junction DNA helicase (ruvB) | HI0312 | 43 | 336 | 33.63 | HFRD | 0 | 64.1 |
| RNA,tRNA modifying | |||||||
| tRNA-guanine transglycosylase (tgt) | HI0244 | 41 | 383 | 32 | HFRD | 0 | 42.85 |
| rRNAmethylase-putative | HI0766 | 39 | 161 | 40.92 | HFRD | 0 | 51.13 |
| Pseudouridylate synthase I (truA) | HI1644 | 41 | 270 | 35.53 | HFRD | 0 | 70.83 |
| Translation | |||||||
| Polypeptide deformylase (def) | HI0622 | 37 | 169 | 43.04 | HFRD | 0 | 63.76 |
| Prolyl-tRNA synthetase | HI0729 | 43 | 572 | 28.32 | HFRD | 0 | 38.02 |
| Transport | |||||||
| tonB protein | HI0251 | 40 | 271 | 35.21 | A118 | 8.3 | 57.67 |
| ABCtransporter | HI0354 | 47 | 240 | 37.97 | HP2 | 0 | 80.01 |
| ABCtransporter | HI0355 | 46 | 245 | 43.03 | BH | 0.9 | 82.72 |
| Glycerol-3-phosphatase transporter (glpT) | HI0686 | 42 | 480 | 46.66 | HFRD | 0 | 16.89 |
| Aminoacid ABCtransporter-permease protein | HI1079 | 35 | 211 | 36.96 | HFRD | 0 | 57.36 |
| Hemeexporter ATP-binding protein A (ccmA) | HI1089 | 42 | 212 | 37.11 | HFRD | 0 | 61.56 |
| Arginine ABC transporter-periplasmic-binding protein (artI) | HI1179 | 36 | 240 | 35.57 | HFRD | 0 | 54.75 |
| Ironchelatin ABC transporter | HI1472 | 44 | 352 | 30.99 | HP2 | 0 | 59.2 |
| ABC transporter-ATP-binding protein | HI1474 | 31 | 200 | 37.09 | ET4 | 9.8 | 61.34 |
| Glutamate permease (gltS) | HI1530 | 38 | 404 | 31.2 | HFDY | 0 | 37.44 |
Gene = the gene number in the annotated H.influenzae Rd genome; %GC = the percentage of GC base pairs in the gene; ɛ = the statistic derived from Equation 1; Ref. org. = The reference genome most similar in codon usage (see Table 1 for list of abbreviations); % better fit > Haemophilus = the percentage by which ɛ is lower in the most similar reference organism than Haemophilus. ɛ (w/r ribosomal group) = the ɛ value with respect to the 21 ribosomal and elongation genes of Haemophilus longer than 140 amino acids.
Phage-like genes
There are 17 genes clustered between HI1465 and HI1523 of the Rd genome that selected Haemophilus phage as most similar based upon Equation 1. Interestingly, three clinical isolates of H.influenzae do not contain this region, suggesting that this region was recently acquired by Rd. These include strain 86028 (www.microbial-pathogenesis.org) and our recently completed sequence analysis of two clinical isolates (F.Z. Hu, R. Boissy, J. Hogg, J. Gladitz, J. Hayes, B. Janto, R. Keefe, R. Preston, N. Ehrlich, J.C. Post and G.D. Ehrlich, unpublished data).
Metabolism genes
The urease accessory genes have not previously been classified as highly expressed with respect to Haemophilus (7), despite such a classification in other bacteria (7). Their high GC content certainly suggests that they are of foreign origin. We found the urease accessory genes to be amongst the few metabolism genes to select a non-Haemophilus reference organism as the most similar in codon usage (Table 4). It is conceivable that these genes may be part of a rare group of genes that are of recent foreign origin and that over time will become more typical of highly expressed genes in their codon usage (5). Their expression may be relevant to pathogenesis as ureH is up-regulated in HI during infection (27), and urease genes are present in virulent strains of Escherichia coli but absent in nonpathogenic strains (28), suggesting they may have been acquired via horizontal transfer.
Transporter genes
In contrast to the metabolic genes, there are many transporter genes with high ɛ values. The Rd transporter genes, in general, are a disparate group in terms of their codon usage; many are amongst the most well-adapted genes in the genome with respect to the average Haemophilus codon usage. The transporter genes, HI0354, HI0355 and HI1472 appear to be of foreign origin. In addition, HI1471 is a transporter gene that narrowly missed the high ɛ threshold under Equation 1, but it did make the high ɛ group using Equation 3 and it is part of the same operon with HI1472. All four of these genes have a high GC content (44–47%). HI1471 and HI1472 both selected Haemophilus phage as their best fit and they are also part of the large highly phage-like region of the Rd genome. Similarly, HI0354 and HI0355 selected non-Haemophilus reference organisms as the most similar in terms of codon usage, though with relatively high ɛ values indicating an origin outside of our set of reference genomes. Several of the transporter genes had codon usage patterns similar to the highly expressed ribosomal and elongation genes. HI0686 has not been previously classified in this category, but it had the third lowest ɛ value compared to the ribosomal group in the Rd genome.
Regulatory genes
There are few regulatory genes with high ɛ values as these genes are rarely highly expressed. HI1476 is a transcriptional regulator with a high ɛ value and it is part of the large phage-like region in Rd and HI0186 is also a transcriptional regulator which selected Haemophilus phage. A third gene, HI0304, shows protein homology to transcriptional regulators and has a high ɛ value.
Restriction/modification genes
The genes HI1055 and HI1058 have strong nucleotide homology (∼95%) to restriction/modification (R/M) genes from Neisserial strains. Neither selected Haemophilus or Neisseria as the best-fit reference genome. R/M genes can be highly mosaic (29–32) in terms of recombining sections from different organisms and so it is not surprising that the codon usage of these genes show unclear origin. Similarly, surface proteins including the lipoproteins and lic-1 operon proteins can be mosaic (33–36).
Genes selecting non-Haemophilus reference genomes
The specification of a gene length-dependent ɛ threshold (defined by Figure 1) for designating high ɛ genes is important for capturing genes of putative foreign origin, however, the majority identified are actually highly expressed genes—and not of foreign origin. An alternative way to identify genes of foreign origin is to find those that actually select non-Haemophilus genomes when run against a large set of reference genomes, i.e. give a significantly lower ɛ value for a non-Haemophilus genome.
Rd genes that displayed ɛ values based upon Equation 1 indicative of a much greater similarity to organisms other than H.influenzae are listed in Table 5. R/M and glycosyl/sialyl transferases are abundant in Table 5, however there are also several lipoproteins and outer membrane proteins in this set. The gene, however, with the largest percentage difference between the best-fitting reference genome and Haemophilus codon usage patterns is HI0687; this is the case regardless of which equation is used to calculate ɛ. This gene has homology to a drug/metabolite transporter, yet has been inserted into the middle of the Glp operon, which encodes genes involved in glycerol metabolism, further supporting the concept that this gene was acquired by horizontal transfer. The adjacent gene, HI0688, also has a very large deviation from Haemophilus-like codon usage, and like HI0687 has a very low GC content (∼25%) suggesting that these two genes were part of the same transfer event.
Table 5.
Rd genes demonstrating best-fit based upon Equation 1 (>10%) to a non-Haemophilus organism
| Gene | % GC | L (amino acids) | ɛ (Equation 1) | Gene name | Ref. org. | % better fit > Haemophilus |
|---|---|---|---|---|---|---|
| HI0916 | 35 | 198 | 32.57 | Outer membrane protein | UU | 14.5 |
| HI1407 | 38 | 448 | 20.46 | traN-related protein | LP | 14.9 |
| HI1599 | 34 | 239 | 25.15 | H.influenzae predicted coding region HI1599 | SA | 15.1 |
| HI1470 | 38 | 254 | 30.6 | Iron chelatin ABC transporter | AA | 15.2 |
| HI0855 | 42 | 116 | 43.56 | Conserved hypothetical protein (protein homol. to inner membrane protein) | INFC | 15.5 |
| HI1411 | 39 | 172 | 28.81 | Terminase-small subunit | LP | 15.8 |
| HI1412 | 36 | 174 | 27.49 | Conserved hypothetical protein (protein homol. to phage-encoded prot, possible anti-repressor) | LP | 15.8 |
| HI1070 | 43 | 1305 | 20.06 | ATP-dependent helicase (hrpa) | PM | 16 |
| HI1570 | 42 | 170 | 47.82 | H.influenzae predicted coding region HI1570 (protein homol. to ribonuclease) | BH | 16.2 |
| HI1385 | 30 | 165 | 29.38 | Ferritin (rsgA) | UU | 16.3 |
| HI0535 | 44 | 262 | 36.19 | Urease accessory protein (ureH) | SE | 16.4 |
| HI0087 | 40 | 424 | 29.21 | Threonine synthase (thrC) | BS | 16.6 |
| HI1110 | 34 | 504 | 17.68 | D-xyloseABC transporter-ATP-binding protein (xylG) | LL | 16.9 |
| HI0601 | 31 | 217 | 33.77 | DNA transformation protein (tfoX) | MG | 17.1 |
| HI1384 | 31 | 182 | 26.07 | Ferritin (rsgA) | SA | 17.1 |
| HI0724 | 32 | 186 | 28.5 | Conserved hypothetical protein | T4 | 17.6 |
| HI0011 | 41 | 135 | 34.03 | DNA polymeraseIII psi subunit (holD) | LP | 17.9 |
| HI0228 | 30 | 125 | 41.22 | H.influenzae predicted coding region HI0228 (protein homol. to lipopolysaccharide biosynthesis protein) | T4 | 18 |
| HI0977 | 30 | 191 | 29.91 | Cell filamentation protein (fic) | UU | 18.5 |
| HI1410 | 41 | 395 | 24.46 | H.influenzae predicted coding region HI1410 (bacteriophage related) | SE | 19.9 |
| HI1422 | 45 | 191 | 45 | H.influenzae predicted coding region HI1422 (protein homol. to bacteriophage anti-repressor protein) | NG | 19.9 |
| HI0802 | 39 | 327 | 31.13 | DNA-directed RNApolymerase-alpha chain (rpoA) | T4 | 20.9 |
| HI1099 | 32 | 102 | 32.67 | H.influenzae predicted coding region HI1099 | MP | 21 |
| HI1040 | 31 | 334 | 24.77 | Type II restriction enzyme | BB | 21.1 |
| HI0787 | 28 | 201 | 28.46 | H.influenzae predicted coding region HI0787 | BB | 22.7 |
| HI0358 | 42 | 215 | 34.39 | Transcriptional activator-putative | SCHZ | 24.7 |
| HI0588 | 34 | 411 | 28.55 | N-carbamyl-l-aminoacid amido hydrolase | UU | 24.7 |
| HI0872 | 30 | 471 | 27.1 | Undecaprenyl-phosphate galactose phospho transferase (rfbP) | CJ | 26.5 |
| HI1514 | 49 | 631 | 24.19 | H.influenzae predicted coding region HI1514 (protein homol. to Mu-like prophage gp42) | VC | 26.7 |
| HI1718 | 35 | 262 | 25.08 | H.influenzae predicted coding region HI1718 (protein homol. to outer membrane protein) | PF | 26.7 |
| HI0352 | 26 | 232 | 24.22 | Conserved hypothetical protein (protein homology to sialyl transferase) | UU | 27.6 |
| HI1459 | 32 | 195 | 25.09 | Putative sigma factor | T4 | 28.4 |
| HI1225 | 36 | 106 | 32.69 | Conserved hypothetical protein (homol. to translation initiation factor) | UU | 28.5 |
| HI0054 | 31 | 266 | 22.52 | Uxuoperon regulator (uxuR) | PF | 32 |
| HI0053 | 35 | 343 | 25.14 | Zinc-type alcohol dehydrogenase | CB | 37.8 |
| HI0051 | 31 | 166 | 28.76 | Conserved hypothetical transmembrane protein (homol. to transport protein) | BB | 40.5 |
| HI1647 | 44 | 291 | 25.32 | Conserved hypothetical protein (homol. to pyridoxine biosynthesis protein) | SP | 45.5 |
| HI0258 | 27 | 331 | 25.96 | Glycosyl transferase-putative | GT | 46.3 |
| HI1041 | 32 | 304 | 26.95 | Modification methylase | PF | 50.5 |
| HI0688 | 25 | 103 | 32.43 | H.influenzae predicted coding region HI0688 (homol. to type V secretory pathway adhesin AidA) | MM | 53.7 |
| HI0871 | 28 | 306 | 26.06 | H.influenzae predicted coding region HI0871 (homol. to sialyl transferase) | BB | 54 |
| HI1578 | 27 | 323 | 23.91 | Glycosyl transferase | PF | 57.4 |
| HI0512 | 30 | 259 | 21.82 | TypeII restriction endonuclease (HindIIR) | CB | 58.8 |
| HI1392 | 31 | 309 | 23.16 | Modification methylase (hindIIIM) | FN | 63.6 |
| HI1287 | 49 | 444 | 27.07 | TypeI modification enzyme (hsdM) | NG | 66.6 |
| HI1393 | 26 | 300 | 17.94 | TypeII restriction endonuclease (hindIIIR) | FN | 86.3 |
| HI0513 | 26 | 519 | 17.01 | Modification methylase (hindIIM) | CB | 89.3 |
| HI0687 | 26 | 304 | 18.13 | Conserved hypothetical protein (homol. to drug/metabolite transport protein) | CB | 99.6 |
The gene HI1647 selected Streptococcus pneumoniae as the reference genome most similar in codon usage. This is congruent with the fact that it possesses 96% amino acid identity to the pdx1 streptococcal protein. The adjacent gene (HI1648) also encodes a protein with >80% amino acid identity to a second streptococcal gene encoding a hypothetical glutamine amidotransferase, which is also contiguous with the pdx1 gene in the Streptococcal R6 genome (37). The fact that pneumococcus and H.influenzae share the human nasopharynx as an ecological niche provides a likely source for these genes.
The four contiguous genes HI0051–HI0054 all showed significantly better codon usage similarity to non-Haemophilus reference species than to Haemophilus, suggesting that this region may have been acquired en bloc. They are all low GC content genes and all select low GC content reference organisms which are closely linked to each other in terms of codon usage similarity.
The genes from HI1403 to HI1424 are part of a second locus demonstrating strong phage protein homology, but, curiously, give their lowest ɛ values primarily with the P.multocida and L.pneumoniae genomes. HI1407, HI1410–12 and HI1422 are all in Table 5. HI1405 and HI1409 did not make the cut-off for inclusion in Table 5, but they also show very good codon usage similarity to Pasteurella and Legionella genomes suggesting that this region may have been acquired en bloc.
Non-Haemophilus Rd genes identified by Equations 3 and 6
There were several genes that were identified by either Equation 3 or 6 as having ɛ values better fitting with non-Haemophilus reference organisms that were not identified by Equation 1 (Supplementary tables available on our web site). HI1407 has the largest percentage difference (64%) based upon Equation 3 and again selected L.pneumophilia. HI0618 and HI1017, both Glp family genes, gave large percentage differences (34 and 51%, respectively) based on Equation 3. Interestingly, they are not part of the Rd glp operon, which consists of the genes HI0683–HI0691. The HI0618 gene (glpG) is adjacent to glpR which has an ɛ value just below the high ɛ threshold. Both glpR and glpG are part of the major glp operon in P.multocida. The observation that HI1017 selects T4 as a reference genome under all three equations and is redundant to the glpF gene found in the major Rd glp operon makes it likely that this gene was acquired by HGT. Genes HI0507 and HI0418 which have 33 and 32% differences, respectively, based on Equation 3, and are adjacent genes in a P.multocida transport operon.
The Haemophilus Rd genome contains several sigma factor RpoE genes, only one of which (HI0628) appears to be of Haemophilus origin. Equation 3 identified HI0589 as having a much closer fit to Bacillus halodurans. Another RpoE gene, HI1459, listed in Table 5, gave a highly divergent phylogenetic tree as compared to a 16S tree, supporting its recent movement into the Rd genome.
HisH together with HisF encode the bi-enzyme complex for imidazole glycerol phosphate synthase (38). HisH (HI0472) selected A.actinomycetemcomitans by 20.8% over Haemophilus using Equation 6 and HisF (HI0474) also had an ɛ value, based upon Equation 3, that was >1 SD above the mean value for genes of its comparable length. These genes are part of the histidine biosynthesis operon, which includes eight genes (HI0468–HI0475). The ɛ values for the six other genes in this operon are low. Thus, it is possible that these two genes were acquired by horizontal transfer from a similar operon. Alternatively, they may be involved in the starvation response which has recently been suggested to utilize genes with seldom used codons (39).
HI1090, a heme exporter, has significantly better codon usage similarity to a non-Haemophilus genome using Equation 3. HI1089, adjacent to HI1090, is also a heme exporter and was identified as having a high ɛ value. The oligopeptide ABC transport operon (HI1120–HI1124) also has several genes with high ɛ values and/or, better codon usage fitting to other genomes than Rd.
Novel H.influenzae sequences
Novel (non-Rd) sequences were obtained from random sequencing of clones from a pooled genomic library prepared from 10 clinical isolates of H.influenzae (11) that had been cultured from patients with OME. All clones were subjected to DNA sequence analysis and homology searching using BLASTn and BLASTx. Only those clones containing sequences demonstrating no observable nucleotide homology to H.influenzae Rd were used in this codon analysis study. All clones were verified to have originated from one or more of the 10 clinical isolates by test amplifications of the respective genomic DNAs using primers designed from the unique DNA sequence information (12). As the study progressed and genomic data became available from additional H.influenzae strains (40) and other Haemophilus sp., we were able to demonstrate hypothetical protein homologies to one or more of these genomes for some of the novel ORFs.
Table 6 provides a list of the novel clinical H.influenzae sequences whose lowest ɛ values (obtained from Equation 1) corresponded to either the phage, HP1 or HP2. We have listed in parentheses the lowest ɛ value obtained amongst the four Haemophilus sp. for comparison with the phage values. In most cases, there is a significant difference between the phage ɛ value and the bacterial value. All of the ORFs listed in Table 6, except the hif genes and the two ATPases, also revealed amino acid sequence homology to known phage proteins. The hif cluster has previously been speculated to be of phage origin (41) and the codon usage statistic we obtained further supports this supposition. Clones 152_N2 and 4_E21 had protein identities/similarities of 85/88%, and 88/90%, respectively, to the H.influenzae strain AM30 hifD gene, notably, codon usage analysis gave high ɛ values against the Rd genome. Clone 4_E21 gave a higher ɛ value than 152_N2, and an analysis of 24 synonymous codons where the two clones differed only in the third base, showed that the 4_E21 sequence contained the less frequently used codon 16 times. The GC content of these hifD clones was also high, relative to Haemophilus, offering further support for the acquisition of this gene family from a foreign organism.
Table 6.
Novel ORFs from H.influenzae clinical isolates with ɛ values closest to the phage HP1 or HP2
| Clone no. and ORFa | ɛb | Number of amino acids | % GC | Protein homology (%ID, %Sim) | Organism with closest protein homology |
|---|---|---|---|---|---|
| 100_E23 | 22.04 (34.04) | 487 | 44 | Haemophilus phage DNA polymerase (99, 99) | H.phage HP2 |
| 103_L4 | 32.43 (35.87) | 255 | 45 | Hypoth. protein Hinf801001315 (98, 99) (underlying phage homologies) | H.influenzae 86-028NP |
| 120_O6(ORF1) | 38.88 (44.85) | 183 | 41 | Hypoth protein Hinf801001531 (94, 98) (underlying phage protein homologies) | H.influenzae 86-028NP |
| 122_N17(ORF1) | 30.32 (35.5) | 286 | 42 | ATPases of the AAA+ class (99, 100) | H.influenzae R2846 |
| 126_N4(ORF2) | 31.79 (35.75) | 279 | 39 | ATPase (AAA+ superfamily) (86, 88) | H.influenzae R2866 |
| 13_I7/135_C22(ORF2) | 31.92 (34.84) | 271 | 42 | Chromosome segregation ATPases (90, 94) (underlying phage homologies to capsid protein precursor) | HI R2866 |
| 152_N2 | 39.59 (42.83) | 213 | 46 | HifD (85, 88) | H.influenzae Str. AM30 |
| 153_I16(ORFs1,2) | 26.45 (32.12) | 295 | 43 | Phage associated baseplate assembly protein | H.influenzae 86-028NP |
| 168_P21(ORF1) | 29.64 (32.04) | 165 | 42 | Hypoth. Protein MS0093 (47, 65) (underlying phage protein homologies) | Mannheimia succiniciproducens MBEL55E |
| 32_F13 | 25.63 (31.7) | 412 | 45 | Superfamily II helicase and inactivated derivatives (49, 64) (underlying phage homologies) | H.somnus 2336 |
| 38_O23(ORF1) | 27.23 (33.93) | 187 | 43 | Hypoth. protein MS0080 (53, 69) (underlying tail fiber protein homologies) | Mannheimia succiniciproducens MBEL55E |
| 38_O23(ORF2) | 35.37 (46.4) | 235 | 49 | Hypoth protein MS0081 (51, 70) (underlying baseplate assembly homologies) | Mannheimia succiniciproducens MBEL55E |
| 4_E21(ORF1) | 28.14 (34.48) | 240 | 45 | HifC (98, 99) | H.influenzae biotype aegyptius |
| 4_E21(ORF2) | 54.19 (58.91) | 189 | 48 | HifD (88, 90) | H.influenzae Str. AM30 |
| 59_C2(ORFs1,2,3) | 30.96 (35.09) | 276 | Orf1: transcriptional regulator (82, 88). Orf2: prophage CP4-57 regulator protein AlpA. Orf3: Hypoth. Protein lpp2120 (51, 69) | Orf1: H.somnus 129PT (underlying phage homologies). Orf2: E.Coli K12. Orf3 Legionella pneumonia str Paris | |
| 67_D11(ORF1) | 28.74 (36.28) | 214 | 45 | Hypoth.protein Hflu203000157 (97, 97) (underlying phage homologies to endonuclease subunit) | H.influenzae R2866 |
| 67_D11(ORF2) | 32.46 (36.9) | 349 | 43 | Hypoth. protein Hinf801001765 (99, 100) (underlying phage homologies to capsid protein precursor) | H.influenzae 86-028NP |
| 67_D11(ORF3) | 50.71 (51.07) | 106 | 40 | Methyl accepting chemotaxis protein (97, 99) | H.influenzae 86-028NP |
| 97_H3 | 34.73 (35.05) | 327 | 41 | 2-methyl thioadenine synthetase (98, 99) | H.somnus 2336 |
| 17_D20 | 22.99 (23.52) | 392 | 44 | hypothetical protein Bucepa03004689(47, 65) (underlying phage homologies) | Burkholderia cepacia R1808 |
ɛ = codon usage bias similarity statistic; %GC = the percentage of guanine and cytosine nucleotide bases in an ORF; % ID = the percentage of amino acids from the novel ORF that are identical to the protein encoded by the paralogous reference gene; %Sim = the percentage of amino acids from the novel ORF that are similar to the protein encoded by the orthologous/paralogous reference gene.
aThe number of the ORF within the clone if the clone contained multiple ORFs.
bThe number in the parentheses is the lowest ɛ value amongst the four H.influenzae strains.
The novel ORFs that yielded their lowest ɛ values with respect to a non-Haemophilus genome are listed in Table 7. The ORF in 179_D14 displays >90% amino acid identity to the conjugal transfer protein TrbB of Ralstonia solanacearum, a pseudomonad, and has a pseudomonal-like GC content of 68%. This ORF has the largest difference in ɛ values observed between its best-fitting genome (Pseudomonas aeruginosa) and Haemophilus. The ORFs in clones 132_O3, 104_E15 and 47_C3 did not select any reference genome with a low ɛ value, but most of their protein homologies were with phage-related recombination proteins. Together these observations suggest that these ORFs may be highly mosaic genes, and that they are adapted for horizontal transfer amongst different bacteria via phage.
Table 7.
Novel ORFs from H.influenzae clinical isolates that are probably of foreign origin
| Clone no. and ORFa | Number of amino acids | % GC | Lowest ɛ (species)b | Δ%ɛ | Protein homology (%ID, %Sim) | Organism with closest protein homology |
|---|---|---|---|---|---|---|
| 179_D14 | 240 | 69 | 31.98 (PA) | 179.2 | Flp pilus assembly protein, ATPase CpaF (91, 93) | Azotobacter vinelandii |
| 132_O3 (ORF1) | 191 | 49 | 33.81 (NM) | 55.3 | Conserved hypoth. protein (52, 71) | Chromobacterium violaceum ATCC12472 |
| 151_O4 | 385 | 29 | 26.4 (BB) | 38.7 | YhbX/YhjW/YijP/YjdB family (48, 69) | N.meningiditis MC58 |
| 125_L2(ORF3) | 143 | 37 | 32.6 (GT) | 37.9 | Anaerobic decarboxylate transporter (71, 83) | Shigella flexneri 2a |
| 121_L20 | 149 | 32 | 35.66 (MM) | 32.7 | SAM-dependent methyltransferase | |
| 55_M14 | 275 | 28 | 27.21 (CJ) | 30.1 | Hydrolase (metallo-beta-lactamase) (33, 52) | H.influenzae R2846 |
| 173_G10 | 225 | 34 | 29.85 (CM) | 29.7 | Hypoth. protein Hflu20300043 (98, 98) | H.influenzae R2866 |
| 104_E15(orf1) | 180 | 41 | 35.39 (BH) | 26.4 | Hypoth. protein Hsom02001338 (61, 76) | H.somnus 129PT |
| 124_K2 | 567 | 43 | 24.64 (VC) | 23.9 | Type I restriction enzyme HsdR (70, 81) | Vibrio cholerae O1 biovar eltor Str. N16961 |
| 125_L2(ORF1) | 144 | 32 | 34.91 (GT) | 23.2 | Transcriptional regulator (LysR family) (68, 77) | Shigella flexneri 2a |
| 120_O6 (ORF 2) | 198 | 41 | 32.06 (HP) | 22.7 | Hypoth protein (73, 82) | Actinobacillus pleuropneumoniae 4074 |
| 125_L2(ORF2) | 230 | 34 | 31.14 (UU) | 21.3 | Unknown (58, 77) (putative aspartate racemase) | Shigella flexneri 2a |
| 13_D9(ORF2) | 381 | 36 | 24.64 (A118) | 19.2 | TnaB (96, 97) | H.influenzae R2866 |
| 121_J7 | 293 | 33 | 33.35 (CB) | 19.1 | DNA methylase (58, 74) | E.coli |
| 32_B2 | 198 | 33 | 22.12 (SA) | 14.6 | Hemoglobin–haptoglobin binding protein HhuA (57, 73) | H.influenzae Str. TN106 |
| 47_C3 | 306 | 41 | 29.04 (VC) | 9.8 | Recombinational DNA repair protein (99, 99) | H.influenzae 86-028NP |
| 183_E8 | 177 | 44 | 40.5 (BS) | 8.8 | Transcriptional regulator (100, 100) | H.inf 86-028NP |
| 159_B20(ORFs1,2) | 157 | 38 | 40.62 (A118) | 8.8 | HD0114 and HD0115 (40, 69) (weaker protein homologies to HI1496 and HI1495) | H.ducreyi 35000HP |
| 96_C16 | 371 | 33 | 19.82 (LL) | 7 | Restriction/modification protein HI0216 (68, 77) | H.influenzae Rd |
| 13_D9(ORF1) | 185 | 43 | 32.29 (GT) | 5.3 | TnaA (98, 98) | H.influenzae R2866 |
| 134_O6 | 270 | 39 | 30.24 (PM) | 4.6 | Fapy DNA glycosylase (98, 99) | H.influenzae R2866 |
| 112_A12(ORF1) | 250 | 34 | 23.99 (PM) | 3.9 | ADP-heptose:LPS heptosyltransferase(100, 100) | H.influenzae R2866 |
| 43_I10 | 358 | 35 | 24.28 (SA) | 2.1 | Putative glucosidase (73, 84) | Yersinia pestis C092 |
| 110_E11(ORF1) | 151 | 49 | 46.65 (HP) | 1.4 | Hypoth. protein HD1532 (68, 81) | H.ducreyi 35000HP |
| 128_C1 | 141 | 45 | 33.0 (PM) | 0.8 | Hypoth. protein Lin1719 | Listeria innoocua |
| 93_G12/117_A22 (ORF1) | 254 | 43 | 30.79 (LP) | 0.7 | Hypoth. protein Hinf8010011272 | H.influenzae 86-028NP |
| 112_A12(ORF2) | 116 | 40 | 47.52 (PM) | 0.6 | Fapy DNA glycosylase (100, 100) | H.influenzae R2866 |
aThe number of the ORF within the clone, if the clone contained multiple ORFs; ɛ = codon usage bias similarity statistic.
bThe letters in parentheses indicate the species that gave the lowest ɛ value; PA = Pseudomonas aeruginosa; NM = Neisseria meningiditis; CB = Clostridium butyricum; BH = Bacillus halodurans; MP = Mycoplasma penetrans; VC = Vibrio cholerae; CJ = Campylobacter jejuni; LL = Lactococcus lactis; T4 = enterobacteriophage T4; PM = Pasturella multocida; NG = Neisseria gonorrheae; HI = Haemophilus influenzae.
%GC = the percentage of guanine and cytosine nucleotide bases in an ORF; Δ%ɛ = the percentage difference in ɛ values between the best-fitting genome and the best-fitting Haemophilus group genome; % ID = the percentage of amino acids from the novel ORF that are identical to the protein encoded by the paralogous reference gene;% Sim = the percentage of amino acids from the novel ORF that are similar to the protein encoded by the orthologous/paralogous reference gene.
There are several presumptive R/M genes in this novel gene grouping. One of these, clone 124_K2, selected Vibrio cholerae as the closest reference organism and also demonstrated its highest protein homology to V.cholerae. In addition, clone 96_C16 encodes a restriction enzyme (RE) that shows clear evidence of a recombination event at a ggggat repeat between an Haemophilus RE gene, HI0216, and an RE gene encoded on a plasmid recovered from E.coli.
All three ORFs from clone 125_L2 are included in Table 7 and share significant amino acid homology with three contiguous genes in a Shigella flexneri pathogenicity island. ORF 2 in clone 13_D9 encodes tnaB, a tryptophan metabolism protein, and selected Guillardia theta, a eukaryotic cryptomonad, as the most similar genome among our reference set. The tryptophanase gene cluster has already been speculated to be of foreign origin (42).
Table 8 lists novel genes that demonstrated low ɛ values to Haemophilus species and strains other than Rd. The hifE entry comprised the codon counts from two separate clones, which had homology to different regions of hifE. Both had nearly complete amino acid identity to the hifE gene from H.influenzae biogroup aegyptius. The hifE gene has been documented to be highly variable amongst strains (43) and thus it is surprising that our clones' codon usage is quite Haemophilus-like. The hifE gene from the Eagan strain was analyzed for codon usage and also displayed a low ɛ value (18.24) to aegyptius. Thus, hifE appears to have assimilated much better into the Haemophilus environment than hifC and hifD, which maintain a deviant codon usage.
Table 8.
Novel ORFs from H.influenzae clinical isolates that probably represent genes that are part of the original H.influenzae gene pool
| Clone no. and ORFa | ɛ, High ɛ-thresholdb | Number of amino acids | %GC | Protein homologyc (%ID, %Sim) |
|---|---|---|---|---|
| Hb | 14.72, 26.3 | 1011 | 31 | Hemoglobin binding protein A (49, 76) |
| 166_G6(ORFs1,2,3) | 17.7, 28.5 | 567 | 39, 39, 45 | soluble lytic murein transglycosylase (94, 97): hypothetical protein Hflu2121901 (98, 99): Type IV secretory pathway, VirD4 components (99, 100) |
| 101_H6(ORFs2,3,4) | 18.85, 28.1 | 607 | 37, 36, 40 | Malata/L-lactate dehydrogenase (95, 97): permease of the major facilitator superfamily (97, 98): hypothetical protein (98,98) |
| UI | 20.42, 29.6 | 465 | 36 | Uronate isomerase (78, 79) |
| HifE | 25.83, 34.0 | 38 | HifE (94, 95) | |
| 120_C11 | 24.39, 30.9 | 284 | 37 | HMW1B (88, 93) |
| 135_I10 | 28.42, 32.3 | 291 | 38 | Las (autotransporter protein) (53, 63) |
| 36_E20 | 27.38, 30.8 | 384 | 42 | HMWA (86, 88) |
| 167_A16(ORFs2,3) | 25.35, 27.9 | 598 | 37, 36 | TPR repeat (99, 99):TPR repeat (22, 46) |
| 170_J8 | 31.18, 32.8 | 286 | 42 | HMWA (97, 99) |
aThe number of the ORF within the clone, if the clone contained multiple ORFs.
bɛ = codon usage bias similarity statistic, High ɛ threshold = Value of ɛ associated with ɛ values greater than the avg. plus SD for comparable sized genes.
cAll protein homologies are from various Haemophilus strains; % ID = the percentage of amino acids from the novel ORF that are identical to the protein encoded by the paralogous reference gene; % Sim = the percentage of amino acids from the novel ORF that are similar to the protein encoded by the orthologous/paralogous reference gene.
Two of our novel clones displayed protein homology to Rd's high molecular weight protein HMWA. Clone 170_J8 had homology to the 5′ end while clone 36_E20 had homology to the 3′ end. Though both ORFs selected Haemophilus as the reference organism most similar in codon usage, the similarity was marginal. Outer membrane proteins are known to be mosaic (33–36) and clone 36_E20 corresponds to a region that is only 90% similar between HMWA1 and HMWA2 of Rd, and itself shows only 79% similarity to HMWA.
Our clone 120_C11 encodes a protein homolog of the Haemophilus high molecular weight protein HMW1B and also shows good Haemophilus-like codon usage. This may at first seem surprising since our clones, which had protein homology to HMWA, were not strongly Haemophilus-like. However, the roles of the two proteins are very different; HMWA is an outer membrane protein responsible for adhesion while HMWB is a processing protein (44).
We identified a unique hemoglobin–haptoglobin utilization protein contig formed from several of our novel clones that produced the lowest ɛ value in the study. These genes are known to recombine frequently and to be highly mosaic among the Haemophilus species, but they appear to be well adapted to the Haemophilus genome and have probably been extant in the Haemophilus supra-genome for an extended period of time. However, one of our novel sequences, which had a 57% identity to a hemoglobin–haptoglobin protein, did have better codon usage similarity to S.aureus.
Comparison of ɛ statistics and phylogenetic tree building
As a test of the predictive value of the ɛ statistic to accurately identify the origin of novel sequences, we built phylogenetic trees for several genes to determine if the ɛ statistic followed the established 16S ribosomal phylogeny. For each of these genes, a COG was identified from the NCBI website and used to construct a maximum likelihood tree (the trees are available at www.centerforgenomicsciences.org). According to the ɛ statistic, the ORF in clone 134_O6 was predicted to have a codon usage most closely related to P.multocida and the phylogenetic tree also placed the clone closest to P.multocida with another orthologous gene from a different strain of H.influenzae being the next closest. Similarly, the Ul ORF, constructed from a three clone contig, which was predicted by the ɛ statistic to come from within the Pastuerallaceae was phylogenetically closest to several species from the enterobacteriaceae which were the most closely related species included in the COG, i.e. there were no orthologs in the database from within the Pastuerallaceae so the phylogenetic tree, to the limits of its resolution, supported the ɛ statistic results. Phylogenetic tree building also supported the ɛ statistic when it predicted that a novel gene was not from within the Haemophilus family. In the case of the ORF from 179_D14, which selected P.aeruginosa as the reference genome with the most similar codon usage, phylogenetic tree building indicated that its closest relationship was to R.solanacearum, a pseudomonad not included in our reference set of genomes. In the case of ORF2 from within clone 125_L2, the ɛ statistic suggested that the most closely related genes were from S.flexneri, and the phylogenetic tree placed the gene next to the closely related Shigella typhimurium. In this case, there was a large COG which included a P.multocida gene that was much more distant than the enterobacter genes. In the case of several clones including 97_H3, 167_A16 and 135_I10, the phylogenetic tree contained multiple gene clusters separated by very wide distances for species in the same family, thus within each cluster were found orthologs from widely divergent species according to the 16S tree. In other words, there were multiple species that had two or more orthologs within the COG that were highly divergent from one another—indicative of frequently exchanged genes. Due to the obligate requirement for multiple orthologous genes to build phylogenetic trees such testing could not be performed on the majority of the novel genes identified in this study as they are orphan genes.
DISCUSSION
Bacterial genes and genomes are highly dynamic in nature due to the fact that many contingency genes are passed between and among species via horizontal transfer methods (20,27,45–48). This movement of genes is probably responsible for the substantial differences observed in the genome size among bacterial strains of the same species (49,50). These phenomena make it difficult to develop accurate phylogenies of bacterial strains and isolates (12,17,30,51). Thus, a more useful goal is to evaluate the origins of individual genes, which we attempted in the present study by developing a general statistical method to test the relatedness of any gene in any organism against the average codon usage of the genomes of the host organism and other species. This statistic provided a measure of similarity, which was based upon the squared difference in codon usage frequencies between the gene and the average codon usage of the reference genomes (n = 80) used in the analyses. This method does not involve any learning on the part of the program and therefore introduces no bias into the system.
To test our method, we compared all of the genes (>100 amino acids in length) from the H.influenzae Rd genome against a set of 80 prokaryotic and eukaryotic reference genomes with respect to their codon usage biases. The method proved to be predictive in that 78% of all H.influenzae Rd genes >140 amino acids in length selected the Haemophilus genome as being most similar in terms of codon usage bias. Moreover, the majority of the novel sequences we identified from among our clinical isolates of H.influenzae also showed strong codon usage similarity to Haemophilus or Haemophilus phage indicating that these genes have probably been long-standing constituents of the Haemophilus supra-genome. The more commonly used CAI was not amenable for such comparisons, as it does not provide for direct comparisons across different reference genomes.
Most bacterial species reserve a set of special codon usage patterns for certain highly expressed genes and H.influenzae follows this design. These genes tend to be involved in basic metabolic and translational processes (6–9). Thus, despite their relatively poor codon usage similarities to the genome as a whole, these genes must be excluded from consideration as foreign acquisitions. In the case of the Rd genome using our statistic most of these genes still selected Haemophilus as the reference organism most similar in codon usage.
It has been suggested that there exist multiple codon use classes within a single genome based upon additional criteria including, genomic position (e.g. location of the gene on the leading or lagging strand) (52–55), expression under starvation conditions (36,56), expression for key metabolic functions (1,57) and cellular location during expression (58). Any gene fitting into one of these additional classes that was not identified as such could be misidentified as being horizontally transferred based on codon usage analyses (1). The above theoretical classes of genes would all be essential and therefore present in all strains of a species, since the novel genes that we are studying are not fixed in the H.influenzae gene pool (i.e. are present only in a subset of H.influenzae clinical strains) they would not fall into one of these categories. We did, however, identify several genes in the Rd genome, which appear to be essential genes that did not possess either Haemophilus-like codon usage patterns or Haemophilus highly expressed gene codon usage patterns. These included the DNA polymerase III ψ and δ subunit and carbonic anhydrase; it is possible that these genes belong to another, as yet unidentified class of genes.
Codon bias within a species may arise from multiple factors that affect the efficiency of mRNA translation, including the expression levels and copy number of individual tRNAs (56); the expression levels and fidelity of individual aminoacyl tRNA synthetases (57); the fidelity of the codon–anticodon pairing; and other factors such as physiological conditions and location of protein synthesis in the cell and the location of the gene on either the leading or lagging strand (1). The actual codon bias of a species appears to be an artifact of evolution, as organisms sharing similar ecological niches have distinct codon biases. However, it is not possible to rule out the possibility that the external environment may provide selective pressures that result in specific codon biases for some organisms. The fact that phage are wholly reliant on the translational machinery of the host would suggest that their codon usage patterns would be similar to the host. Indeed the HP1 and HP2 phage of Haemophilus, although having their own distinct codon bias, nevertheless are closer to Haemophilus and the other Pasteurellaceae than to any of the other reference genomes evaluated.
Codon usage patterns within a gene that are much more closely related to a non-host reference organism than to the current host strongly suggest that the gene is of foreign origin. The most likely explanation is the relatively recent acquisition of the gene via a horizontal transfer from its original host. R/M and glycosyltransferase genes accounted for 10 of the 13 genes with the largest differences between the best-fitting reference organism and Rd. Both of these groups of enzymes are large and highly mosaic in nature (7,26,29); there are >7500 R/M enzymes in the REBASE database (rebase.neb.com). This mosaicism may serve important protective and developmental purposes. R/M variation is a defense mechanism (29). Glycosyltransferases are known to be up-regulated during biofilm development (27,59,60) and may be important virulence factors for chronic infection; variability in these genes may provide H.influenzae with the capacity to present different lipopolysaccharide configurations to the host over time, so as to evade the adaptive immune response (34,35). Both of these gene classes were also characterized as having GC contents that are dissimilar to that of the Haemophilus Rd genome, thereby providing additional evidence of their foreign origin.
It is noteworthy, however, that GC content alone is insufficient to ascribe a Haemophilus-like character to a sizable proportion of foreign genes. Indeed, 31% of the genes in Table 5 displayed a Haemophilus-like GC content of 34–41%. Thus, our ɛ statistic is an independent measure of sequence relatedness not linked to GC content. The glpF and glpG genes are examples of this set.
An examination of the locations and origins of the various glp genes in Rd is highly instructive with regard to horizontal gene transfer. In P.multocida, the glpG and glpR genes are contained within the major glp operon. However, in Rd these genes are missing from the major glp operon and have been replaced with other genes. Therefore, the finding of glpG and glpR genes elsewhere in the Rd genome that do not possess the Rd standard codon usage pattern, most likely, is reflective of the fact that these genes are important to H.influenzae, and that their displacement from the major glp operon produced a strain that was at a selective disadvantage until it reacquired these genes from another source. Expression of glpR may be a necessary part of the balance between glycerol metabolism and phospholipid biosynthesis (61) and mutations in glpR can lead to cold sensitivity (61). Importantly, glpR may be expressed as a virulence factor as it has been shown to be up-regulated during otitis media infection (27).
Multiple virulence genes in H.influenzae appear to be of foreign origin, including the Hif cluster. Several of our novel sequences had protein homology to the Haemophilus Hif cluster. The codon analysis of these sequences suggested a foreign or phage lineage for these genes. This is similar to what has been observed for the pandemic forms of V.cholerae where the virulence genes are encoded by a phage (62). Thus, it is possible that pathogenic bacteria commonly evolve from commensal organisms by the acquisition of virulence genes that are transmitted by phage.
Acknowledgments
Funding to support this work and to pay the Open Access publication charges for this article was provided by a grant from the United States Department of Health and Human Services, DC02148 and Allegheny-Singer Research Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Karlin S., Mrázek J., Campbell A.M. Codon usages in different gene classes of the Escherichia coli genome. Mol. Microbiol. 1998;29:1341–1355. doi: 10.1046/j.1365-2958.1998.01008.x. [DOI] [PubMed] [Google Scholar]
- 2.Grantham R., Gautier C., Gouy M., Mercier R., Pave A. Codon catalog usage and the genome hypothesis. Nucleic Acids Res. 1980;8:R49–R62. doi: 10.1093/nar/8.1.197-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grantham R., Gautier C., Gouy M., Jacobzone M., Mercier R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981;9:R43–R74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharp P.M., Li W.H. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawrence J.G., Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- 6.Medigue C., Rouxel T., Vigier P., Henaut A., Danchin A. Evidence for horizontal gene transfer in Escherichia coli speciation. J. Mol. Biol. 1991;222:851–856. doi: 10.1016/0022-2836(91)90575-q. [DOI] [PubMed] [Google Scholar]
- 7.Karlin S., Mrázek J. Predicted highly expressed genes of diverse prokaryotic genomes. J. Bacteriol. 2000;182:5238–5250. doi: 10.1128/jb.182.18.5238-5250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karlin S., Barnett M.J., Campbell A.M., Fisher R.F., Mrazek J. Predicting gene expression levels from codon biases in alpha-proteobacterial genomes. Proc. Natl Acad. Sci. USA. 2003;100:7313–7318. doi: 10.1073/pnas.1232298100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mrazek J., Bhaya D., Grossman A.R., Karlin S. Highly expressed and alien genes of the Synechocystis genome. Nucleic Acids Res. 2001;29:1590–1601. doi: 10.1093/nar/29.7.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ochman H., Lawrence J.G., Groisman E.A. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 11.Erdos G., Sayeed S., Antalis P., Hu F.Z., Hayes J., Goodwin J., Dopico R., Post J.C., Ehrlich G.D. Development and characterization of a pooled Haemophilus influenzae genomic library for the evaluation of gene expression changes associated with mucosal biofilm formation in otitis media. Int. J. Pediatr. Otorhinolaryngol. 2003;67:749–755. doi: 10.1016/s0165-5876(03)00093-4. [DOI] [PubMed] [Google Scholar]
- 12.Shen K., Antalis P., Gladitz J., Sayeed S., Ahmed A., Yu S., Hayes J., Johnson S., Dice B., Dopico R., et al. Identification, distribution, and expression of novel (nonRd) genes in ten clinical isolates of nontypeable Haemophilus influenzae. Infect. Immun. 2005;73:3479–3491. doi: 10.1128/IAI.73.6.3479-3491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koski L.B., Morton R.A., Golding G.B. Codon bias and base composition are poor indicators of horizontally transferred gene. Mol. Biol. Evol. 2001;18:404–412. doi: 10.1093/oxfordjournals.molbev.a003816. [DOI] [PubMed] [Google Scholar]
- 14.Walker E.S., Preston R.A., Post J.C., Ehrlich G.D., Kalbfleisch J.H., Klingman K.L. Genetic diversity among strains of Moraxella catarrhalis: analysis using multiple DNA probes and a single-locus PCR-restriction fragment length polymorphism method. J. Clin. Microbiol. 1998;36:1977–1983. doi: 10.1128/jcm.36.7.1977-1983.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith H.O., Tomb J.-F., Dougherty B.A., Fleischman R.D., Venter J.C. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science. 1995;269:538–540. doi: 10.1126/science.7542802. [DOI] [PubMed] [Google Scholar]
- 16.Fleischmann R.D., Adams M.D., White O., Clayton R.A., Kirkness E.F., Kerlavage A.R., Bult C.J., Tomb J.-F., Dougherty B.A., Merrick J.M., et al. Whole-genome random sequencing and assembly of Haemophilus Influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 17.Shen K., Wang X., Post J.C., Ehrlich G.D. Molecular and translational research approaches for the study of bacterial pathogenesis in Otitis media. In: Rosenfeld R., Bluestone C.D., editors. Evidence-based Otitis Media. 2nd edn. Hamilton: B.C.Decker Inc.; 2003. pp. 91–119. [Google Scholar]
- 18.Ehrlich G.D., Hu F.Z., Post J.C. Role for biofilms in infectious disease. In: Ghannoum M., O'Toole G.A., editors. Microbial Biofilms. Washington, DC: ASM Press; 2004. pp. 332–358. [Google Scholar]
- 19.Costerton W., Veeh R., Shirtliff M., Pasmore M., Post C., Ehrlich G. The application of biofilm science to the study and control of chronic bacterial infections. J. Clin. Invest. 2003;112:1466–1477. doi: 10.1172/JCI20365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ehrlich G.D., Hu F.Z., Lin Q., Costerton J.W., Post J.C. Intelligent implants to battle biofilms. ASM News. 2004;70:127–133. [Google Scholar]
- 21.Lan R., Reeves P.R. Gene transfer is a major factor in bacterial evolution. Mol. Biol. Evol. 1996;13:47–55. doi: 10.1093/oxfordjournals.molbev.a025569. [DOI] [PubMed] [Google Scholar]
- 22.Davis J., Smith A.L., Hughes W.R., Golomb M. Evolution of an autotransporter: domain shuffling and lateral transfer from pathogenic Haemophilus to Neisseria. J. Bacteriol. 2001;183:4626–4635. doi: 10.1128/JB.183.15.4626-4635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruepp A., Graml W., Santos-Martinez M.L., Koretke K.K., Volker C., Werner H., Mewes H.W., Frishman D., Stocker S., Lupas A.N., Baumeister W. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature. 2000;407:508–513. doi: 10.1038/35035069. [DOI] [PubMed] [Google Scholar]
- 24.Nelson K.E., Clayton R.A., Gill S.A., Gwinn M.L., Dodson R.J., Haft D.H., Hickey E.K., Peterson J.D., Nelson W.C., Ketchum K.A., et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 25.Vitrescak A.G., Rodionov D.A., Mironov A.A., Gelfand M.S. Regulation of riboflavin biosynthesis genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 2002;30:3141–3151. doi: 10.1093/nar/gkf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q., Reeves P.R. Genetic variation of dTDP-l-rhamnose pathway genes in Salmonella enterica. Microbiology. 2000;146:2291–2307. doi: 10.1099/00221287-146-9-2291. [DOI] [PubMed] [Google Scholar]
- 27.Mason K.M., Munson R.S., Jr, Bakaletz L.O. Nontypeable Haemophilus influenzae gene expression induced in vivo in a chinchilla model of otitis media. Infect. Immun. 2003;71:3454–3462. doi: 10.1128/IAI.71.6.3454-3462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perna N.T., Plunkett G., III, Burland V., Mau B., Glasner J.D., Rose D.J., Mayhew G.F., Evans P.S., Gregor J., Kirkpatrick H.A., et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 29.Milkman R., Jaeger E., McBride R.D. Molecular evolution of the Escherichia coli chromosome. VI. Two regions of high effective recombination. Genetics. 2003;163:475–483. doi: 10.1093/genetics/163.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeltsch A., Pingoud A. Horizontal gene transfer contributes to the wide distribution and evolution of type II restriction-modification systems. J. Mol. Evol. 1996;42:91–96. doi: 10.1007/BF02198833. [DOI] [PubMed] [Google Scholar]
- 31.Kita K., Kawakami H., Tanaka H. Evidence for horizontal transfer of the EcoT38I restriction-modification gene to chromosomal DNA by the P2 phage and diversity of defective P2 prophages in Escherichia coli TH38 strains. J. Bacteriol. 2003;185:2296–2305. doi: 10.1128/JB.185.7.2296-2305.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29:3742–3756. doi: 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cody A.J., Field D., Feil E.J., Stringer S., Deadman M.E., Tsolaki A.G., Gratz B., Bouchet V., Goldstein R., Hood D.W., Moxon E.R. High rates of recombination in otitis media isolates of non-typeable Haemophilus influenzae. Infect. Genet. Evol. 2003;3:57–66. doi: 10.1016/s1567-1348(02)00152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patrick C.C., Kimura A., Jackson M.A., Hermanstorfer L., Hood A., McCraken G.H., Jr, Hansen E.J. Antigenic characterization of the oligosaccharide portion of the lipopolysaccharide of non-typeable Haemophilus influenzae. Infect. Immun. 1987;55:2902–2911. doi: 10.1128/iai.55.12.2902-2911.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zanze S.E., Moxon E.R. Composition of the lipopolysaccharide from different capsular serotype strains of Haemophilus influenzae. J. Gen. Microbiol. 1987;133:1443–1451. doi: 10.1099/00221287-133-6-1443. [DOI] [PubMed] [Google Scholar]
- 36.Risberg A., Masoud H., Martin A., Richards J.C., Moxon E.R., Scweda E.K.H. Structural analysis of the lipopolysaccharide oligosaccharide epitopes expressed by a capsule-deficient strain of Haemophilus influenzae Rd. Eur. J. Biochem. 1999;261:171–180. doi: 10.1046/j.1432-1327.1999.00248.x. [DOI] [PubMed] [Google Scholar]
- 37.Hoskins J., Alborn W.E., Jr, Arnold J., Blaszczak L.C., Burgett S., DeHoff B.S., Estrem S.T., Fritz L., Fu D.J., Fuller W., et al. Genome of the bacterium Streptococcus pneumoniae strain R6. J Bacteriol. 2001;183:5709–5717. doi: 10.1128/JB.183.19.5709-5717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beismann-Driemeyer S., Sterner R. Imidazole glycerol phosphate synthase from Thermotoga maritima. Quaternary structure, steady-state kinetics, and reaction mechanism of the bienzyme complex. J. Biol. Chem. 2001;276:20387–20396. doi: 10.1074/jbc.M102012200. [DOI] [PubMed] [Google Scholar]
- 39.Elf J., Nilsson D., Tenson T., Ehrenberg M. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science. 2003;300:1718–1722. doi: 10.1126/science.1083811. [DOI] [PubMed] [Google Scholar]
- 40.Munson R.S., Jr, Harrison A., Gillaspy A., Ray W.C., Carson M., Armbruster D., Gipson J., Gipson M., Johnson L., Lewis L., et al. Partial analysis of the genomes of two nontypeable Haemophilus influenzae otitis media isolates. Infect Immun. 2004;72:3002–3010. doi: 10.1128/IAI.72.5.3002-3010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mhlanga-Mutanadura T., Morlin G., Smith A.L., Eisenstark A., Golomb M. Evolution of the major pilus gene cluster of Haemophilus Influenzae. J. Bacteriol. 1998;180:4693–4703. doi: 10.1128/jb.180.17.4693-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin K., Morlin G., Smith A., Nordyke A., Eisenstark A., Golumb M. The tryptophanase gene cluster of Haemophilus influenzae type b: evidence for horizonatal gene transfer. J. Bacteriol. 1998;180:107–118. doi: 10.1128/jb.180.1.107-118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Read T.D., Satola S.W., Opdyke J.A., Farley M.M. Copy number of pilus gene clusters and variation in Haemophilus influenzae and in the hifE pilin gene. Infect. Immun. 1998;66:1622–1631. doi: 10.1128/iai.66.4.1622-1631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barenkamp S.J., St Geme J.W., III Genes encoding high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae are part of gene clusters. Infect. Immun. 1994;62:3320–3328. doi: 10.1128/iai.62.8.3320-3328.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hurtado A., Rodriquez-Valera F. Accessory DNA in the genomes of representatives of the Escherichia coli reference collection. J. Bacteriol. 1999;181:2548–2554. doi: 10.1128/jb.181.8.2548-2554.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hakenbeck R., Balmelle B., Weber B., Gardès C., Keck W., de Saizieu A. Mosaic genes and mosaic chromosomes: intra and interspecies genomic variation of Streptococcus pneumoniae. Infect. Immun. 2001;69:2477–2486. doi: 10.1128/IAI.69.4.2477-2486.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bergman N.H., Akerley B.J. Position-based scanning for comparative genomics and identification of genetic islands in Haemophilus influenzae type b. Infect. Immun. 2003;71:1098–1108. doi: 10.1128/IAI.71.3.1098-1108.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jain R., Rivera M.C., Lake J.C. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl Acad. Sci. USA. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bergthorsson U., Ochman H. Heterogeneity of genome sizes among natural isolates of Escherichia coli. J. Bacteriol. 1995;177:5784–5789. doi: 10.1128/jb.177.20.5784-5789.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berg O.G., Kurland C.G. Evolution of microbial genomes: sequence acquisition and loss. Mol. Biol. Evol. 2002;19:2265–2276. doi: 10.1093/oxfordjournals.molbev.a004050. [DOI] [PubMed] [Google Scholar]
- 51.Meats E., Feil E.J., Stringer S., Cody A.J., Goldstein R., Kroll J.S., Popovic T., Spratt B.G. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J. Clin. Microbiol. 2003;41:1623–1636. doi: 10.1128/JCM.41.4.1623-1636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lobry J.R. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 53.Kerr A.R., Peden J.F., Sharp P.M. Systematic base composition variation around the genome of Mycoplasma genitalium, but not Mycoplasma pneumoniae. Mol. Microbiol. 1997;25:1177–1179. doi: 10.1046/j.1365-2958.1997.5461902.x. [DOI] [PubMed] [Google Scholar]
- 54.Tillier E.R., Collins R.A. The contributions of replication orientation, gene direction, and signal sequences to base-composition asymmetries in bacterial genomes. J. Mol. Evol. 2000;50:249–257. doi: 10.1007/s002399910029. [DOI] [PubMed] [Google Scholar]
- 55.Daubin V., Perriere G. G+C3 structuring along the genome: a common feature in prokaryotes. Mol. Biol. Evol. 2003;20:471–483. doi: 10.1093/molbev/msg022. [DOI] [PubMed] [Google Scholar]
- 56.Kuhar I., van Putten J.P., Zgur-Bertok D., Gaastra W., Jordi B.J. Codon-usage based regulation of colicin K synthesis by the stress alarmone ppGpp. Mol. Microbiol. 2001;41:207–216. doi: 10.1046/j.1365-2958.2001.02508.x. [DOI] [PubMed] [Google Scholar]
- 57.Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E.coli translational system. J. Mol. Biol. 1981;151:389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- 58.Chiapello H., Ollivier E., Landes-Devauchelle C., Nitschke P., Risler J.-L. Codon usage as a tool to predict the cellular location of eukaryotic ribosomal proteins and aminoacyl-tRNA synthetases. Nucleic Acids Res. 1999;27:2848–2851. doi: 10.1093/nar/27.14.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y., Burne R.A. Regulation of the gtfBC and ftf genes of Streptococcus mutans in biofilms in response to pH and carbohydrate. Microbiology. 2001;147:2841–2848. doi: 10.1099/00221287-147-10-2841. [DOI] [PubMed] [Google Scholar]
- 60.Yoshida A., Kuramitsu H.K. Streptococcus mutans biofilm formation: utilization of a gtfB promoter-green fluorescent protein. (PgtfB::gfp) construct to monitor development. Microbiology. 2002;148:3385–3394. doi: 10.1099/00221287-148-11-3385. [DOI] [PubMed] [Google Scholar]
- 61.Flower A.M. SecG function and phospholipid metabolism in Escherichia coli. J. Bacteriol. 2001;183:2006–2012. doi: 10.1128/JB.183.6.2006-2012.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davis B.M., Waldor M.K. Filamentous phages linked to virulence of Vibrio cholerae. Curr. Opin. Microbiol. 2003;6:35–42. doi: 10.1016/s1369-5274(02)00005-x. [DOI] [PubMed] [Google Scholar]