Abstract
Bcl-2-associated athanogene 3 (BAG3) plays an important function in cellular protein quality control (PQC) maintaining proteome stability. Mutations in the BAG3 gene result in cardiomyopathies. Due to its roles in cardiomyopathies and the complexity of BAG3–protein interactions, it is important to understand these protein interactions given the importance of the multifunctional cochaperone BAG3 in cardiomyocytes, using an in vitro cardiomyocyte model. The experimental assay was conducted using high pressure liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) in the human AC16 cardiomyocyte cell line with BioID technology. Proteins with BAG3-interaction were identified in all the 28 hallmark gene sets enriched in idiopathic cardiomyopathies and/or ischemic disease. Among the 24 hallmark gene sets enriched in both idiopathic cardiomyopathies and ischemic disease, 15 gene sets had at least 3 proteins with BAG3-interaction. This study highlights BAG3 protein interactions, unveiling the key gene sets affected in cardiomyopathies, which help to explain the molecular mechanisms of the cardioprotective effects of BAG3. In addition, this study also highlighted the complexity of proteins with BAG3 interactions, implying unwanted effects of BAG3.
Keywords: apoptosis, BAG3, bioID, cardiomyopathy, in vitro
1. Introduction
BAG cochaperone 3 (BAG3), also known as Bcl-2-associated athanogene 3, is a multi-functional protein, which is expressed ubiquitously in animals, and homologs have been reported in plants [1]. BAG3 was first recognized for its ability to bind to Bcl2 with subsequent inhibition of apoptosis while other studies have found that it supports a diverse array of cellular functions including autophagy [2], excitation–contraction coupling, mitochondrial function [3], and the integrity of the sarcomere [4]. BAG3 has been recognized as playing a critical role in cellular protein quality control (PQC) to maintain the health of the proteome [5]. Mutations in the BAG3 gene result in both dilated cardiomyopathy (DCM) [6] and peripartum cardiomyopathy (PPCM) [7] in a dominant inheritance model. A critical role of BAG3 in cardiomyocytes involves the maintenance of mitochondrial homeostasis that is mediated by both heat shock protein 70 (Hsp70) and the small heat shock proteins HspB6 and HspB8 [8]. BAG3 also has anti-apoptotic activity by binding Bcl-2 [9]. BAG3 has four protein binding domains, including (1) one WW (Trp–Trp) domain, binding with proteins in signal transduction processes [10], e.g., the PDZ domain containing guanine nucleotide exchange factor 2 (PDZGEF2) to promote cell adhesion [11]; (2) two IPV (Ile–Pro–Val) motifs, binding with small heat shock proteins (sHsps) HspB6/HspB8 [12]; (3) one proline-rich repeat (PXXP) region, binding with SH3 (Src homology 3) motifs, e.g., in phospholipase C-γ (PLC-γ) that also serves as an attachment site for the dynein motor transport of misfolded proteins to the peri-nuclear aggresomes [13,14]; and (4) one BAG domain, binding with Hsp70 and Bcl-2 [12,13,14].
While it is true that BAG3 is recognized for its multifunctionality and its role has been extensively studied, there is still much to uncover about its complex interactions and functions, especially in the context of different diseases. For instance, while the association of BAG3 with both cardiomyopathy and Parkinson’s disease (PD) has been reported, the genetic associations are in the opposite directions, i.e., the risk allele in DCM is protective against PD. The DCM-associated SNP rs2234962 [15,16] is in tight linkage disequilibrium with the PD-associated SNP rs72840788 (r2 = 1 in European populations) [17,18]. We advocate for a greater focus on the intricate BAG3 interaction network, as a reductionist approach may fail to capture the subtleties and multifaceted nature of BAG3’s role. Due to the complexity of BAG3–protein interactions, it is useful to gain a better understanding of the specific proteins with which BAG3 participates in its activity as a multifunctional cochaperone by using an in vitro cell model to better understand BAG3 binding with other cellular proteins. For this purpose, we performed a proteomics study using the BioID proximity-dependent biotinylation method to identify proteins that interact with BAG3, particularly those from the gene sets with expression levels correlated with cardiomyopathies. BioID is a unique technology to screen for protein interactions in living cells [19]. In addition to direct protein interactions, BioID is able to identify weak or transient interactions, as well as proteins in close proximity. This discovery experiment was carried out to identify potentially novel proteins that bind to or interact with BAG3.
2. Results
2.1. Gene Expression in Idiopathic and Ischemic Cardiomyopathies
Among the 50 hallmark gene sets, 26 showed significance in idiopathic CM, and 26 showed significances in ischemic CM (Table 1, Figure 1). From these gene sets, 24 gene sets were significant in both idiopathic and ischemic CMs, while the other 4 gene sets had significances in only idiopathic or ischemic CMs. For the latter four gene sets, the same direction trend in enrichment was observed in the other type of CM. GO, KEGG, and Reactome gene set analyses are presented in Tables S1–S3. In particular, BAG3 levels were decreased in both idiopathic (fold change = 0.646, adjusted p-value = 9.60 × 10−6) and ischemic (fold change = 0.633, adjusted p-value = 9.77 × 10−6) cardiomyopathies. WGCNA reveals that gene modules containing BAG3 are downregulated in both idiopathic and ischemic CM (Figures S1 and S2).
Table 1.
50 hallmark gene sets in idiopathic and ischemic cardiomyopathies.
| HALLMARK | SIZE | Idiopathic | NES | FDR q-val | Ischemic | NES | FDR q-val | Proteins |
|---|---|---|---|---|---|---|---|---|
| ADIPOGENESIS | 176 | case | 1.11 | 0.420844 | controls | −1.20 | 0.169207 | DBT,TST,MDH2 |
| ALLOGRAFT_REJECTION | 197 | controls | −1.05 | 0.394748 | case | 1.07 | 0.487176 | RPS9,MAP3K7 |
| ANDROGEN_RESPONSE | 98 | controls | −1.83 | 0.001006 | controls | −1.90 | 5.31 × 10−4 | SRP19,SGK1 |
| ANGIOGENESIS | 35 | case | 1.21 | 0.27238 | case | 1.24 | 0.252174 | |
| APICAL_JUNCTION | 178 | controls | −1.07 | 0.375625 | controls | −1.02 | 0.461531 | LIMA1,GTF2F1,EPB41L2,TSC1 |
| APOPTOSIS | 157 | controls | −1.51 | 0.018923 | controls | −1.40 | 0.044813 | LMNA,ANXA1,CASP1,CASP4,CASP3,PPT1,BID,BNIP3L,SPTAN1 |
| BILE_ACID_METABOLISM | 99 | case | 1.74 | 0.009496 | case | 1.62 | 0.01886 | PRDX5,ATXN1 |
| CHOLESTEROL_HOMEOSTASIS | 61 | controls | −1.32 | 0.074947 | controls | −1.28 | 0.104645 | ANXA5 |
| COAGULATION | 134 | case | 1.42 | 0.075674 | case | 1.59 | 0.018179 | ANXA1,CSRP1 |
| COMPLEMENT | 186 | controls | −1.44 | 0.028868 | controls | −1.33 | 0.0762 | CASP1,CASP4,CASP3,ANXA5,PRSS3,CSRP1 |
| DNA_REPAIR | 135 | controls | −0.93 | 0.655048 | controls | −1.01 | 0.458158 | REV3L,EDF1,CETN2,NME3,GTF2F1,RFC2,RFC3,HPRT1,TP53 |
| E2F_TARGETS | 187 | controls | −1.57 | 0.012377 | controls | −1.50 | 0.026689 | RFC2,RFC3,PSIP1,CBX5,NUP153,PNN,TMPO,STMN1,NASP,RACGAP1,TP53 |
| EPITHELIAL_MESENCHYMAL_TRANSITION | 192 | case | 1.53 | 0.031229 | case | 1.82 | 0.005753 | TAGLN |
| ESTROGEN_RESPONSE_EARLY | 198 | controls | −1.58 | 0.012051 | controls | −1.64 | 0.006863 | DHRS2,SVIL |
| ESTROGEN_RESPONSE_LATE | 199 | controls | −1.49 | 0.019649 | controls | −1.49 | 0.025326 | TST,HPRT1,DHRS2,SGK1 |
| FATTY_ACID_METABOLISM | 145 | case | 1.09 | 0.438963 | controls | −1.15 | 0.224652 | MDH2,ACADVL,UROD,MIF |
| G2M_CHECKPOINT | 195 | controls | −1.52 | 0.017902 | controls | −1.48 | 0.027784 | NCL,SFPQ,TMPO,STMN1,NASP,TOP1,KIF23,TPX2,RACGAP1 |
| GLYCOLYSIS | 186 | case | 1.02 | 0.49266 | case | 1.16 | 0.35265 | MDH2,STMN1,NASP,MIF,PGK1,PC |
| HEME_METABOLISM | 194 | case | 1.02 | 0.524825 | case | 0.98 | 0.637137 | UROD,TOP1,CAST,YPEL5,HEBP1,BNIP3L,HDGF,PC,ASNS |
| HYPOXIA | 192 | controls | −1.77 | 0.002004 | controls | −1.56 | 0.013079 | NAGK,BNIP3L,PRDX5,MIF,PGK1,FOSL2,KLF6 |
| IL2_STAT5_SIGNALING | 177 | controls | −1.50 | 0.019949 | controls | −1.45 | 0.030848 | CASP3,KLF6,PHLDA1 |
| IL6_JAK_STAT3_SIGNALING | 86 | controls | −1.84 | 8.15 × 10−4 | controls | −1.62 | 0.00786 | IRF9 |
| INFLAMMATORY_RESPONSE | 194 | controls | −1.83 | 0.001168 | controls | −1.69 | 0.004573 | KLF6 |
| INTERFERON_ALPHA_RESPONSE | 80 | case | 1.73 | 0.007606 | case | 1.78 | 0.004291 | CASP1,IRF9,TRIM21,IFI44,SP110,IFIH1 |
| INTERFERON_GAMMA_RESPONSE | 177 | case | 1.27 | 0.201025 | case | 1.41 | 0.070512 | CASP1,CASP4,CASP3,IRF9,TRIM21,IFI44,SP110,IFIH1 |
| KRAS_SIGNALING_DN | 195 | case | 0.86 | 0.829231 | case | 0.87 | 0.871536 | KMT2D,SGK1 |
| KRAS_SIGNALING_UP | 196 | controls | −1.06 | 0.38636 | case | 1.01 | 0.630874 | WDR33 |
| MITOTIC_SPINDLE | 181 | controls | −1.21 | 0.160671 | controls | −1.25 | 0.127236 | PCNT,CAPZB,PCM1,RANBP9,CD2AP,EPB41L2,TSC1,SPTAN1,KIF23,TPX2,RACGAP1,APC |
| MTORC1_SIGNALING | 193 | controls | −2.35 | 0 | controls | −2.31 | 0 | HPRT1,PGK1,PSMC2,ATP6V1D,ASNS |
| MYC_TARGETS_V1 | 189 | controls | −2.18 | 0 | controls | −2.27 | 0 | HPRT1,PGK1,HNRNPC,EEF1B2,GLO1,C1QBP,PWP1,SF3A1,SERBP1,MRPL9,HDGF,CBX3,PHB2 |
| MYC_TARGETS_V2 | 55 | controls | −2.16 | 0 | controls | −2.13 | 0 | CBX3,TCOF1 |
| MYOGENESIS | 197 | case | 1.39 | 0.080993 | case | 1.21 | 0.276254 | SVIL,SPTAN1,FHL1,TAGLN,PC |
| NOTCH_SIGNALING | 28 | case | 0.91 | 0.762965 | controls | −0.77 | 0.923318 | PPARD |
| OXIDATIVE_PHOSPHORYLATION | 194 | case | 1.82 | 0.004724 | case | 1.45 | 0.059126 | MDH2,ACADVL,ATP6V1D,PHB2,NDUFB3,OPA1,NDUFB6,NQO2,NDUFV1 |
| P53_PATHWAY | 195 | controls | −1.61 | 0.01035 | controls | −1.47 | 0.028355 | CASP1,DNTTIP2,TP53 |
| PEROXISOME | 96 | case | 1.18 | 0.309822 | case | 0.99 | 0.658104 | PRDX5,VPS4B,ATXN1 |
| PI3K_AKT_MTOR_SIGNALING | 100 | controls | −1.79 | 0.001342 | controls | −1.68 | 0.004535 | CFL1,ECSIT,PIKFYVE,MAP3K7 |
| PROTEIN_SECRETION | 95 | controls | −1.28 | 0.096338 | controls | −1.39 | 0.04544 | PPT1,SNX2,ARFGAP3,VPS4B |
| REACTIVE_OXYGEN_SPECIES_PATHWAY | 44 | controls | −1.63 | 0.008859 | controls | −1.63 | 0.007414 | PDLIM1 |
| SPERMATOGENESIS | 124 | case | 0.84 | 0.81949 | case | 0.72 | 1 | PEBP1,ZC3H14 |
| TGF_BETA_SIGNALING | 51 | controls | −1.59 | 0.011077 | controls | −1.68 | 0.005039 | APC,MAP3K7 |
| TNFA_SIGNALING_VIA_NFKB | 195 | controls | −2.50 | 0 | controls | −2.26 | 0 | NFKB2,TRIP10,SGK1,FOSL2,KLF6,PHLDA1,IFIH1 |
| UNFOLDED_PROTEIN_RESPONSE | 108 | controls | −2.20 | 0 | controls | −2.28 | 0 | PARN,BAG3,FUS,EIF4A3,DCP2,KHSRP,ASNS |
| UV_RESPONSE_DN | 140 | case | 1.09 | 0.409433 | case | 1.11 | 0.41775 | SIPA1L1,RND3,PHF3,ATXN1 |
| UV_RESPONSE_UP | 155 | controls | −1.98 | 0 | controls | −1.96 | 0 | TST,CASP3,PPT1,BID,UROD,ASNS,EIF5,CDC5L |
| WNT_BETA_CATENIN_SIGNALING | 37 | controls | −0.87 | 0.783655 | controls | −0.88 | 0.759333 | TP53,PPARD |
| XENOBIOTIC_METABOLISM | 187 | controls | −1.41 | 0.035226 | controls | −1.47 | 0.028478 | HPRT1,PC,PPARD |
| APICAL_SURFACE | 34 | controls | −0.94 | 0.655474 | controls | −1.00 | 0.471533 | |
| HEDGEHOG_SIGNALING | 36 | case | 1.03 | 0.541695 | case | 0.96 | 0.646984 | |
| PANCREAS_BETA_CELLS | 40 | controls | −0.53 | 0.998829 | case | 0.57 | 0.998143 |
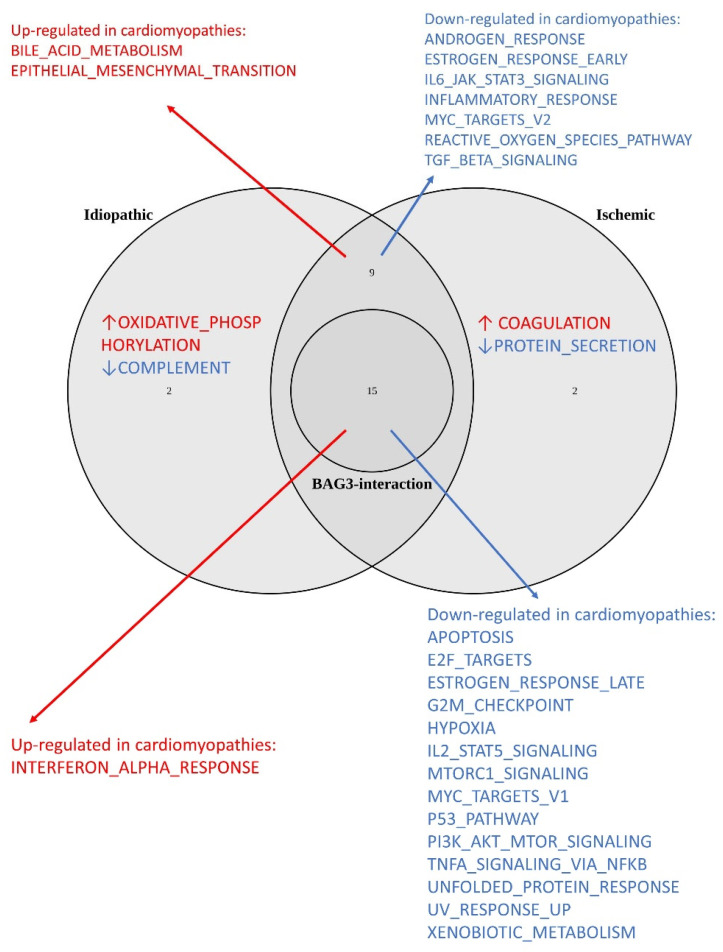
Figure 1.
Venn plot of hallmark gene sets enriched in cardiomyopathies and those with BAG3 interactions. The central circle represents 15 cardiomyopathy-related gene sets containing at least three BAG3-interacting proteins. Red arrows indicate upregulated gene sets, while blue arrows indicate downregulated gene sets.
2.2. BAG3–Protein Interactions
In total, 387 proteins were identified with significant adjusted p-values and positive BAG3–protein interactions (Table S4). Among these proteins, the BRD4, CAST, and KLF6 genes show co-expression with BAG3 in idiopathic CM based on WGCNA; the genes ARFGAP3, RBM34, BAG3, BCCIP, BID, DNTTIP2, EBNA1BP2, EIF4A3, EIF5, ENAH, FOSL2, HSPA6, PRPF4, PUS7, RND3, SFPQ, SGK1, U2AF2, and ZMYND8 show co-expression with BAG3 in ischemic CM based on WGCNA. BAG3-interaction were identified in each of the 28 hallmark gene sets enriched in the idiopathic and/or ischemic cardiomyopathies. Among the 24 hallmark gene sets enriched in both idiopathic and ischemic cardiomyopathies, 15 gene sets had at least 3 proteins with BAG3 interaction (Table 2, Figure 1).
Table 2.
Fifteen hallmark gene sets with at least thee BAG3-interacted proteins.
| Gene Set | Note | BAG3-Interacted Proteins |
|---|---|---|
| Down-regulated in cardiomyopathies | ||
| MYC_TARGETS_V1 | Signal DNA damage-induced apoptosis [20] | HPRT1,PGK1,HNRNPC,EEF1B2,GLO1,C1QBP,PWP1,SF3A1,SERBP1,MRPL9,HDGF,CBX3,PHB2 |
| E2F_TARGETS | Integrate cell cycle progression with DNA repair, replication, and G2/M checkpoints [21] | RFC2,RFC3,PSIP1,CBX5,NUP153,PNN,TMPO,STMN1,NASP,RACGAP1,TP53 |
| G2M_CHECKPOINT | Prevent DNA damaged cells from entering mitosis [22] | NCL,SFPQ,TMPO,STMN1,NASP,TOP1,KIF23,TPX2,RACGAP1 |
| P53_PATHWAY | Promote cell cycle arrest to allow DNA repair or apoptosis [23] | CASP1,DNTTIP2,TP53 |
| APOPTOSIS | Cardiomyocyte apoptosis-interruptus [24] | LMNA,ANXA1,CASP1,CASP4,CASP3,PPT1,BID,BNIP3L,SPTAN1 |
| HYPOXIA | Alter myocardial gene expression and induce cardiomyocyte apoptosis [25] | NAGK,BNIP3L,PRDX5,MIF,PGK1,FOSL2,KLF6 |
| XENOBIOTIC_METABOLISM | Oxidative stress with excessive production of reactive oxygen species (ROS) [26] | HPRT1,PC,PPARD |
| UV_RESPONSE_UP | DNA damage repair [27] | TST,CASP3,PPT1,BID,UROD,ASNS,EIF5,CDC5L |
| UNFOLDED_PROTEIN_RESPONSE | Decrease global protein synthesis, and increase refolding or degradation of misfolded proteins [28] | PARN,BAG3,FUS,EIF4A3,DCP2,KHSRP,ASNS |
| TNFA_SIGNALING_VIA_NFKB | Cardioprotective in hypoxic or ischemic myocardial injury [29] | NFKB2,TRIP10,SGK1,FOSL2,KLF6,PHLDA1,IFIH1 |
| MTORC1_SIGNALING | Regulate cell growth and metabolism [30] and mediate adaptive cardiac hypertrophy [31] | HPRT1,PGK1,PSMC2,ATP6V1D,ASNS |
| ESTROGEN_RESPONSE_LATE | Prevent apoptosis and necrosis of cardiac and endothelial cells [32] | TST,HPRT1,DHRS2,SGK1 |
| PI3K_AKT_MTOR_SIGNALING | Lead to cell proliferation [33] and inhibit cardiomyocyte apoptosis [34] | CFL1,ECSIT,PIKFYVE,MAP3K7 |
| IL2_STAT5_SIGNALING | Modulate CD4+ Th cell differentiation [35] and upregulate the expression of c-Myc, BCL-2, and BCL-x [36] | CASP3,KLF6,PHLDA1 |
| Up-regulated in cardiomyopathies | ||
| INTERFERON_ALPHA_RESPONSE | Interferon inhibits cardiac cell function in vitro [37] and interferon treatment has been shown of cardiotoxicity [38]. | CASP1,IRF9,TRIM21,IFI44,SP110,IFIH1 |
3. Discussion
This study identified 387 proteins with direct or indirect BAG3 interactions. The mRNA levels in cardiomyocytes altered in DCM, as shown in the single-cell RNA-seq study by Koenig et al. [39], are annotated in Table S4. Among these proteins, heat shock 70 kDa protein 6 (HSPA6) [40] and Hsp70-binding protein 1 (HSPBP1) [41] encode a Hsp70 protein and regulate Hsp70 function, respectively. Bcl-2-associated transcription factor 1 (BCLAF1) activates the p53 pathway and induces apoptosis [42] and may contribute to myocardial reperfusion injury [43]. BCL2/adenovirus E1B 19 kDa protein-interacting protein 3-like (BNIP3L) activates the ER and mitochondrial cell death pathways and induces cardiomyopathy [44]. In addition, we observed 15 gene sets with at least 3 BAG3-interacting proteins further supporting the important role of BAG3 in the biology of the heart (Table 2).
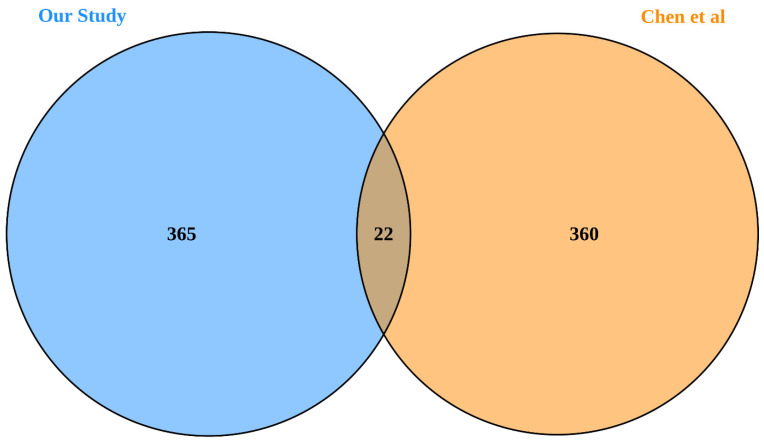
In a previous study, Chen et al. identified 382 BAG3-interacting proteins in cancer cell lines using stable isotope labeling with amino acids in cell culture (SILAC) combined with mass spectrometry (MS) [45]. Of the 387 proteins identified in our study, 22 overlapped with those reported by Chen et al. (Figure 2). Given the approximately 20,000 protein-coding genes in the human genome, this overlap is highly statistically significant and unlikely to occur by chance alone (p = 3.69 × 10−7), providing mutual validation between these two studies. Among the 360 proteins identified by Chen et al. but not replicated in our study, 250 genes were detected in heart tissue by Hannenhalli et al. [46]. In contrast, 365 of the proteins identified in our study have not been reported by Chen et al. Of these, 295 genes were detected in heart tissue by Hannenhalli et al. [46], suggesting that our cell model is more informative for understanding heart-specific gene expression.
Figure 2.
Venn plot of proteins identified in our study compared to those reported by Chen et al. [45].
3.1. Proteins with Roles in Cell Cycle
Cardiomyocytes are taken as terminally differentiated and do not proliferate after birth [47]. In a recent study, a proliferative burst in preadolescent mice was observed [48]. However, further experimental validation is needed, as this finding has not yet been replicated by other researchers. If the same phenomenon is true in humans, the highlighted cell cycle proteins in this study may be implied for early intervention for cardiomyopathies, particularly for a BAG3-based therapy. Adult cardiomyocytes also exhibit a dynamic range of cell cycle activity under various physiological and pathological conditions, e.g., in pathologic myocardial hypertrophy [49].
BAG3 interaction was identified in 31 gene proteins from four important gene sets in the cell cycle, including MYC_TARGETS_V1, E2F_TARGETS, G2M_CHECKPOINT, and P53_PATHWAY. These four gene sets were downregulated in both idiopathic and ischemic cardiomyopathies. MYC, E2F, and p53 are important regulators of cell-cycle progression [50]. We observed BAG3 interaction of 13 proteins involving MYC signaling, including target variant 1 proteins, 11 proteins in E2F signaling molecules, 9 proteins in cell cycle G2/M checkpoint, and 3 proteins in the p53 pathway. The MYC_TARGETS_V1 genes are regulated by MYC [51] and are involved in cell-cycle progression and cell proliferation [52]. c-Myc plays an essential role in signaling DNA damage-induced apoptosis through the control of the p53 tumor suppressor protein [20]. Activation of the p53 pathway promotes cell cycle arrest to allow DNA repair or apoptosis for cells with serious DNA damage [23]. The G2/M checkpoints prevent DNA damaged cells from entering mitosis [22]. Acting through the E2F targets, E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints [21].
The proteins with BAG3-interaction in these gene sets also harbor opportunities for small molecular therapies. Deficiency of the MYC target gene, phosphoglycerate kinase 1 (PGK1), from genetic mutations, may cause myopathy [53]. The protein encoded by PGK1 catalyzes the production of adenosine 5′-triphosphate (ATP) and its activity is regulated by ATP [54]. Biallelic deficiency of the MYC target gene—complement component 1 Q subcomponent-binding protein, mitochondrial (C1QBP)—causes mitochondrial respiratory-chain deficiencies and severe cardiomyopathy [55]. Copper supplementation has been shown to up-regulate the function of the C1QBP protein and may improve cardiac function [56].
3.2. Interactions with Proteins Involved in Apoptosis and Cell Damage Responses
In addition to the cell cycle proteins, BAG3 interacts with proteins in a number of gene sets downregulated in cardiomyopathies, including HALLMARK_APOPTOSIS, HALLMARK_HYPOXIA, HALLMARK_UV_RESPONSE_UP, HALLMARK_XENOBIOTIC_METABOLISM, HALLMARK_UNFOLDED_PROTEIN_RESPONSE, HALLMARK_PROTEIN_SECRETION, HALLMARK_COMPLEMENT, HALLMARK_TNFA_SIGNALING_VIA_NFKB, HALLMARK_MTORC1_SIGNALING, HALLMARK_ESTROGEN_RESPONSE_LATE, HALLMARK_PI3K_AKT_MTOR_SIGNALING, and HALLMARK_IL2_STAT5_SIGNALING. BAG3 interactions with apoptosis processes, responses to DNA damage and oxidative stress, and cell repairs were highlighted.
It has been suggested that cardiomyocyte apoptosis may contribute to myocardial reperfusion injury [57,58] and cardiomyopathies [59,60]. Currently, there is controversial evidence regarding the involvement of apoptosis in cardiomyopathies. Narula et al. [24] described an interruption in the apoptosis cascade, including nuclear fragmentation and condensation in cardiomyocytes, terming it ‘apoptosis-interruptus’. Conversely, our studies and others suggest that apoptosis plays a critical role in heart failure [61].
The gene expression data of cardiomyopathies show that the apoptosis pathway is downregulated in both idiopathic and ischemic cardiomyopathies. Cardiomyocytes with interrupted apoptosis may undergo necrosis. BAG3 binds to the Bcl-2 Homology 4 (BH4) domain of Bcl-2 to inhibit mitochondrial-dependent (intrinsic) apoptosis and, thus, protecting from cell death. Nine proteins in the APOPTOSIS gene set were identified for BAG3 interaction in this study, including three caspases CASP1, CASP3, and CASP4.
Hypoxia significantly alters myocardial gene expression, mediated by hypoxia inducible factor 1 subunit alpha (HIF1A) [62]. Hypoxia has been shown to induce dilated cardiomyopathy in chick embryos [63]. Overexpression of BAG3 has been shown to attenuate hypoxia-induced cardiomyocyte apoptosis [25]. DNA damage induces apoptosis and cardiomyopathy [27], whereas HALLMARK_UV_RESPONSE_UP genes (i.e., up-regulated genes in response to UV), play major roles in DNA damage repair [64]. Moreover, Xenobiotic toxicity causes cardiomyopathy through oxidative stress [26] resulting in dysregulated gene expression.
The unfolded protein response (UPR) decreases global protein synthesis, increases endoplasmic reticulum (ER)-associated degradation of misfolded proteins, and activates protein-folding in ER [28]. BAG3 serves as a cochaperone with Hsp70 and is involved in a wide range of protein folding processes, including the refolding of misfolded proteins [65]. The protein secretion pathway acts on protein folding, post-translational modifications (PTMs), and protein trafficking [66], and it is important in cardiac repair [67].
TNF-α [68] and complement activation [69] have been suggested as contributing factors to myocardial reperfusion injury and cardiomyopathy. However, complement activation contributes to tissue repair [70]. The genes encoding components of the complement system are downregulated in idiopathic and ischemic cardiomyopathies, which is in consistent with the recognition that long-term effects of complement inhibitors may be detrimental [71]. While NF-κB signaling may be cardioprotective in hypoxic or ischemic myocardial injury [29], it has also been shown to mediate chronic inflammation [72]. The mTORC1 signaling regulates cell growth and metabolism [30] and mediates adaptive cardiac hypertrophy [31]. Its inhibition increases overall protein degradation by the ubiquitin proteasome system [73] and may attenuate cardiac remodeling and heart failure [31]. The late estrogen response pathway may prevent apoptosis and necrosis of cardiac and endothelial cells [32]. Genes upregulated by activation of the PI3K/AKT/mTOR pathway include a major intracellular network that leads to cell proliferation [33] and inhibits cardiomyocyte apoptosis [34]. IL2_JAK_STAT5_SIGNALING genes are up-regulated by STAT5 in response to IL2 stimulation and modulate CD4+ Th cell differentiation [35], and upregulate the expression of c-Myc, BCL-2, and BCL-x further exacerbating the inflammatory response [36].
3.3. Interactions with Proteins Upregulated in Cardiomyopathies
In contrast to the above gene sets, the HALLMARK_INTERFERON_ALPHA_RESPONSE and HALLMARK_OXIDATIVE_PHOSPHORYLATION gene sets are both up regulated in cardiomyopathies. Interferon inhibits cardiac cell function in vitro [37] and interferon treatment has been shown to produce cardiotoxicity [38]. The heart is in high demand of energy by oxidative phosphorylation, where defective oxidative phosphorylation may cause cardiomyopathy [74]. Oxidative phosphorylation genes are upregulated in cardiomyopathies, which may cause overproduction of reactive oxygen species (ROS) [75]. BAG3 interactions with these upregulated genes in cardiomyopathies are interesting as this could ameliorate the cardioprotective effects of BAG3. Adjunct therapy targeting these proteins may, thus, improve the therapeutic effects of BAG3. The NADH dehydrogenases, i.e., NDUFB3 (NADH dehydrogenase 1 beta subcomplex subunit 3), NDUFB6 (NADH dehydrogenase 1 beta subcomplex subunit 6), and NDUFV1 (NADH dehydrogenase flavoprotein 1, mitochondrial), can be inhibited by metformin. The potential heart protective effects by metformin are gaining more research attention [76].
4. Research Design and Methods
Cell experimental assay: The cell model used in this study was the human AC16 cardiomyocyte cell line derived from adult human ventricular heart tissues (SCC109, Sigma-Aldrich, St. Louis, MO, USA). The cells were treated and analyzed based on five different conditions: (1) BAG3_Biotin: Cells transfected by plasmid expressing BioID-BAG3 for 48 h, then receive Biotin treatment for 16 h; (2) BioID: Cells transfected by plasmid expressing BioID vector only for 48 h, then receive Biotin treatment for 16 h; (3) BAG3 (to correct protein over-expression by BAG3): Cells transfected by plasmid expressing BioID-BAG3, without followed Biotin treatment; (4) AC16_Biotin (to correct protein-Biotin interaction): Cells receive Biotin treatment for 16 h; (5) AC16: Cells only. Three replicates were performed simultaneously for each treatment condition. Details of BAG3 construct delivered using adeno-associated virus (AAV) vector has been described in our previous study [77]. The BioID experimental assay was conducted using high pressure liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) previously described through a collaboration with the Proteomics Facility, Sanford-Burnham-Presby Medical Discovery Institute, La Jolla, CA, USA [49].
Gene expression data: Gene expression data in tissue samples from left ventricular myocardium in two disease conditions, idiopathic and ischemic cardiomyopathies, were made available by Hannenhalli et al. [46]. The study included 16 controls, 86 idiopathic, and 108 ischemic cardiomyopathies. The heart tissue was snap-frozen at time of cardiac transplantation. The gene expression assay was based on data generated using the Affymetrix Human Genome U133A Array. The data analysis was performed by the GEO2R (https://www.ncbi.nlm.nih.gov/geo/info/geo2r.html, accessed on 9 October 2024). p values were adjusted by the Benjamini and Hochberg false discovery rate method. The data are publicly available at the NCBI Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE5406, accessed on 9 October 2024).
Gene Set Enrichment Analysis (GSEA) was performed by the GSEA v4.3.2 software (Broad Institute of MIT and Harvard, MA, USA) based on the Molecular Signatures Database (MSigDB) [78] hallmark [51], Gene Ontology (GO) [79], Kyoto Encyclopedia of Genes and Genomes (KEGG) [80], and Reactome [81] gene set collections. FDR corrected p-values < 0.05 were considered statistically significant. The Weighted Gene Co-expression Network Analysis (WGCNA) analysis was performed using the WGCNA R package [82,83].
Data analysis of BAG3–Protein interactions: The BioID data were analyzed using MSstats package v4.12.1 from R Bioconductor [84]. By comparing the groups of BAG3_Biotin vs. BioID, all proteins with adjusted p-value < 0.05 by the Benjamini and Hochberg false discovery rate method were identified. The effect sizes of BAG3–protein interactions were corrected by [log2FC(BAG3_Biotin vs. BioID)]-[log2FC(AC16_Biotin vs. AC16)]-[log2FC(BAG3 vs. BioID)], i.e., BAG3–protein interactions being corrected for biotin–protein interactions and BAG3-increased protein levels.
5. Conclusions
This study utilized the relatively new technique of BioID to identify interactions between BAG3 and various gene sets. The goal was to uncover unique proteins and protein pathways that might contribute to disease when BAG3 levels are under-expressed. Such under-expression can occur due to loss-of-function mutations caused by truncations, deletions, or unique mutations secondary to single nucleotide variants. These variants may change an amino acid, insert an amino acid, or alter the reading frame, resulting in truncation. Not unexpectedly, we identified a plethora of associated proteins. In view of the small size of BAG3, the limited number of binding sites, and the somewhat focused activities of the protein, the number of intersecting proteins identified by this new technique represents an overabundance of detected proteins. However, the fact that many of the observed proteins have been associated with BAG3 in various animal studies raises the larger question of how a single protein might interact with a large number of target proteins. The fact that virtually every known protein interaction with BAG3 has been detected by the BioID assay suggests that enhancing the assay’s sensitivity might allow for a more nuanced approach to this question. Additionally, new means of discriminating interactome data, which remain proprietary, could be useful.
In conclusion, this study highlights the observed interactions of BAG3 with key gene sets that are affected in cardiomyopathies, thereby unveiling some of the molecular mechanisms involved with the cardioprotective effects of BAG3. In addition, this study also highlights the complexity of proteins with BAG3 interactions, implying unwanted effects of BAG3. Adjunct therapy to address unwanted effects of BAG3 may be indicated, such as the use of metformin to inhibit the consequences of NADH dehydrogenase activation. A limitation of this study is the AC16 cell model, which is an immortalized cell line with passages over numerous generations [85], which may not represent the actual cardiomyocytes in human heart. Although AC16 cells are derived from human ventricles, they exhibit several key limitations: (1) they do not exhibit contractile activity, (2) they are highly proliferative, and (3) the sarcomere is not organized in AC16 cells. While AC16 cells differ from primary cardiomyocytes in these aspects, they remain a widely used in vitro model for studying human cardiomyocyte-related processes due to their human origin and ability to express cardiac-specific markers. The primary objective of our study was to investigate the complex BAG3 interaction network and identify its interacting proteins, so the aforementioned limitations do not significantly impact our findings. Further study using different cell models, in particular specialized cardiomyocyte models, will help to verify the BAG3 interactions and enable investigations of the more focused phenotypic effects. Additionally, validation through co-immunoprecipitation (Co-IP) studies, although challenging to scale up and expensive for large-scale applications will be important for confirming selected candidate proteins of interest in human heart tissues and human-induced pluripotent stem cell-derived cardiomyocytes (hIPSC-CMs) to corroborate our findings and enhance their relevance to the human heart.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/1422-0067/25/20/11308/s1.
Author Contributions
Conceptualization, H.H. and A.M.F.; methodology, H.-Q.Q.; formal analysis, H.-Q.Q.; investigation, H.-Q.Q., J.-F.W. and A.R.-C.; resources, A.M.F.; data curation, H.-Q.Q. and A.R.-C.; writing—original draft preparation, H.-Q.Q.; writing—review and editing, H.H. and A.M.F.; visualization, H.-Q.Q.; supervision, H.H.; project administration, H.H. and A.M.F.; funding acquisition, H.H. and A.M.F. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was approved by the Institutional Review Board (IRB) of the Children’s Hospital of Philadelphia. Human participants and personal information are inaccessible to the research group. All human subjects or their legal guardians provided written informed consent.
Informed Consent Statement
Not applicable.
Data Availability Statement
Supporting data from this study can be obtained by emailing the corresponding author Hakon Hakonarson.
Conflicts of Interest
The authors declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article. The funding from Renovacor, Inc. (to A.M.F.) has been duly reviewed and determined to have no effect on the research.
Funding Statement
The study was supported by Institutional Development Funds from the Children’s Hospital of Philadelphia to the Center for Applied Genomics, the Neff Family Foundation, the Children’s Hospital of Philadelphia Endowed Chair in Genomic Research (to H.H.), and funding from Renovacor, Inc. (to A.M.F.).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Fagerberg L., Hallström B.M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., Habuka M., Tahmasebpoor S., Danielsson A., Edlund K., et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. MCP. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luthold C., Lambert H., Guilbert S.M., Rodrigue M.-A., Fuchs M., Varlet A.-A., Fradet-Turcotte A., Lavoie J.N. CDK1-Mediated Phosphorylation of BAG3 Promotes Mitotic Cell Shape Remodeling and the Molecular Assembly of Mitotic p62 Bodies. Cells. 2021;10:2638. doi: 10.3390/cells10102638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quintana M.T., Parry T.L., He J., Yates C.C., Sidorova T.N., Murray K.T., Bain J.R., Newgard C.B., Muehlbauer M.J., Eaton S.C. Cardiomyocyte-specific human Bcl2-associated anthanogene 3 P209L expression induces mitochondrial fragmentation, Bcl2-associated anthanogene 3 haploinsufficiency, and activates p38 signaling. Am. J. Pathol. 2016;186:1989–2007. doi: 10.1016/j.ajpath.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin T.G., Myers V.D., Dubey P., Dubey S., Perez E., Moravec C.S., Willis M.S., Feldman A.M., Kirk J.A. Cardiomyocyte contractile impairment in heart failure results from reduced BAG3-mediated sarcomeric protein turnover. Nat. Commun. 2021;12:2942. doi: 10.1038/s41467-021-23272-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stürner E., Behl C. The Role of the Multifunctional BAG3 Protein in Cellular Protein Quality Control and in Disease. Front. Mol. Neurosci. 2017;10:177. doi: 10.3389/fnmol.2017.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arimura T., Ishikawa T., Nunoda S., Kawai S., Kimura A. Dilated cardiomyopathy-associated BAG3 mutations impair Z-disc assembly and enhance sensitivity to apoptosis in cardiomyocytes. Hum. Mutat. 2011;32:1481–1491. doi: 10.1002/humu.21603. [DOI] [PubMed] [Google Scholar]
- 7.van Spaendonck-Zwarts K.Y., Posafalvi A., van den Berg M.P., Hilfiker-Kleiner D., Bollen I.A., Sliwa K., Alders M., Almomani R., van Langen I.M., van der Meer P. Titin gene mutations are common in families with both peripartum cardiomyopathy and dilated cardiomyopathy. Eur. Heart J. 2014;35:2165–2173. doi: 10.1093/eurheartj/ehu050. [DOI] [PubMed] [Google Scholar]
- 8.Ranek M.J., Stachowski M.J., Kirk J.A., Willis M.S. The role of heat shock proteins and co-chaperones in heart failure. Philos. Trans. R. Soc. B Biol. Sci. 2018;373:20160530. doi: 10.1098/rstb.2016.0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antoku K., Maser R.S., Scully W.J., Delach S.M., Johnson D.E. Isolation of Bcl-2 Binding Proteins That Exhibit Homology with BAG-1 and Suppressor of Death Domains Protein. Biochem. Biophys. Res. Commun. 2001;286:1003–1010. doi: 10.1006/bbrc.2001.5512. [DOI] [PubMed] [Google Scholar]
- 10.Macias M.J., Wiesner S., Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 2002;513:30–37. doi: 10.1016/S0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
- 11.Iwasaki M., Tanaka R., Hishiya A., Homma S., Reed J.C., Takayama S. BAG3 directly associates with guanine nucleotide exchange factor of Rap1, PDZGEF2, and regulates cell adhesion. Biochem. Biophys. Res. Commun. 2010;400:413–418. doi: 10.1016/j.bbrc.2010.08.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs M., Poirier D.J., Seguin S.J., Lambert H., Carra S., Charette S.J., Landry J. Identification of the key structural motifs involved in HspB8/HspB6–Bag3 interaction. Biochem. J. 2010;425:245–257. doi: 10.1042/BJ20090907. [DOI] [PubMed] [Google Scholar]
- 13.Doong H., Price J., Kim Y.S., Gasbarre C., Probst J., Liotta L.A., Blanchette J., Rizzo K., Kohn E. CAIR-1/BAG-3 forms an EGF-regulated ternary complex with phospholipase C-γ and Hsp70/Hsc70. Oncogene. 2000;19:4385–4395. doi: 10.1038/sj.onc.1203797. [DOI] [PubMed] [Google Scholar]
- 14.McCollum A.K., Casagrande G., Kohn E.C. Caught in the middle: The role of Bag3 in disease. Biochem. J. 2010;425:e1–e3. doi: 10.1042/BJ20091739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villard E., Perret C., Gary F., Proust C., Dilanian G., Hengstenberg C., Ruppert V., Arbustini E., Wichter T., Germain M. A genome-wide association study identifies two loci associated with heart failure due to dilated cardiomyopathy. Eur. Heart J. 2011;32:1065–1076. doi: 10.1093/eurheartj/ehr105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tadros R., Francis C., Xu X., Vermeer A.M.C., Harper A.R., Huurman R., Kelu Bisabu K., Walsh R., Hoorntje E.T., Te Rijdt W.P., et al. Shared genetic pathways contribute to risk of hypertrophic and dilated cardiomyopathies with opposite directions of effect. Nat. Genet. 2021;53:128–134. doi: 10.1038/s41588-020-00762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smeland O.B., Shadrin A., Bahrami S., Broce I., Tesli M., Frei O., Wirgenes K.V., O’Connell K.S., Krull F., Bettella F., et al. Genome-wide Association Analysis of Parkinson’s Disease and Schizophrenia Reveals Shared Genetic Architecture and Identifies Novel Risk Loci. Biol. Psychiatry. 2021;89:227–235. doi: 10.1016/j.biopsych.2020.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nalls M.A., Blauwendraat C., Vallerga C.L., Heilbron K., Bandres-Ciga S., Chang D., Tan M., Kia D.A., Noyce A.J., Xue A., et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet. Neurol. 2019;18:1091–1102. doi: 10.1016/S1474-4422(19)30320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roux K.J., Kim D.I., Burke B., May D.G. BioID: A Screen for Protein-Protein Interactions. Curr. Protoc. Protein Sci. 2018;91:19.23.11–19.23.15. doi: 10.1002/cpps.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phesse T., Myant K., Cole A., Ridgway R., Pearson H., Muncan V., Van Den Brink G., Vousden K., Sears R., Vassilev L. Endogenous c-Myc is essential for p53-induced apoptosis in response to DNA damage in vivo. Cell Death Differ. 2014;21:956–966. doi: 10.1038/cdd.2014.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zebrowski D.C., Engel F.B. The cardiomyocyte cell cycle in hypertrophy, tissue homeostasis, and regeneration. Rev. Physiol. Biochem. Pharmacol. 2013;165:67–96. doi: 10.1007/112_2013_12. [DOI] [PubMed] [Google Scholar]
- 22.Stark G.R., Taylor W.R. Analyzing the G2/M checkpoint. Methods Mol. Biol. 2004;280:51–82. doi: 10.1385/1-59259-788-2:051. [DOI] [PubMed] [Google Scholar]
- 23.Ozaki T., Nakagawara A. Role of p53 in Cell Death and Human Cancers. Cancers. 2011;3:994–1013. doi: 10.3390/cancers3010994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narula J., Kolodgie F.D., Virmani R. Apoptosis and cardiomyopathy. Curr. Opin. Cardiol. 2000;15:183–188. doi: 10.1097/00001573-200005000-00011. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J., He Z., Xiao W., Na Q., Wu T., Su K., Cui X. Overexpression of BAG3 attenuates hypoxia-induced cardiomyocyte apoptosis by inducing autophagy. Cell. Physiol. Biochem. 2016;39:491–500. doi: 10.1159/000445641. [DOI] [PubMed] [Google Scholar]
- 26.Costa V.M., Carvalho F., Duarte J.A., Bastos M.d.L., Remião F. The heart as a target for xenobiotic toxicity: The cardiac susceptibility to oxidative stress. Chem. Res. Toxicol. 2013;26:1285–1311. doi: 10.1021/tx400130v. [DOI] [PubMed] [Google Scholar]
- 27.Tsutsui H., Ide T., Kinugawa S. Mitochondrial oxidative stress, DNA damage, and heart failure. Antioxid. Redox Signal. 2006;8:1737–1744. doi: 10.1089/ars.2006.8.1737. [DOI] [PubMed] [Google Scholar]
- 28.Cao S.S., Kaufman R.J. Unfolded protein response. Curr. Biol. 2012;22:R622–R626. doi: 10.1016/j.cub.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 29.Dhingra R., Shaw J.A., Aviv Y., Kirshenbaum L.A. Dichotomous actions of NF-κB signaling pathways in heart. J. Cardiovasc. Transl. Res. 2010;3:344–354. doi: 10.1007/s12265-010-9195-5. [DOI] [PubMed] [Google Scholar]
- 30.Zou Z., Tao T., Li H., Zhu X. mTOR signaling pathway and mTOR inhibitors in cancer: Progress and challenges. Cell Biosci. 2020;10:31. doi: 10.1186/s13578-020-00396-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sciarretta S., Forte M., Frati G., Sadoshima J. New Insights Into the Role of mTOR Signaling in the Cardiovascular System. Circ. Res. 2018;122:489–505. doi: 10.1161/CIRCRESAHA.117.311147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knowlton A., Lee A. Estrogen and the cardiovascular system. Pharmacol. Ther. 2012;135:54–70. doi: 10.1016/j.pharmthera.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paplomata E., O’Regan R. The PI3K/AKT/mTOR pathway in breast cancer: Targets, trials and biomarkers. Ther. Adv. Med. Oncol. 2014;6:154–166. doi: 10.1177/1758834014530023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao H., Han X., Han X. The cardioprotection of the insulin-mediated PI3K/Akt/mTOR signaling pathway. Am. J. Cardiovasc. Drugs. 2014;14:433–442. doi: 10.1007/s40256-014-0089-9. [DOI] [PubMed] [Google Scholar]
- 35.Jones D.M., Read K.A., Oestreich K.J. Dynamic Roles for IL-2–STAT5 Signaling in Effector and Regulatory CD4+ T Cell Populations. J. Immunol. 2020;205:1721–1730. doi: 10.4049/jimmunol.2000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lord J.D., McIntosh B.C., Greenberg P.D., Nelson B.H. The IL-2 receptor promotes lymphocyte proliferation and induction of the c-myc, bcl-2, and bcl-x genes through the trans-activation domain of Stat5. J. Immunol. 2000;164:2533–2541. doi: 10.4049/jimmunol.164.5.2533. [DOI] [PubMed] [Google Scholar]
- 37.Lampidis T.J., Brouty-Boyé D. Interferon inhibits cardiac cell function in vitro. Proc. Soc. Exp. Biol. Med. 1981;166:181–185. doi: 10.3181/00379727-166-41043. [DOI] [PubMed] [Google Scholar]
- 38.Sonnenblick M., Rosin A. Cardiotoxicity of Interferon*: A Review of 44 Cases. Chest. 1991;99:557–561. doi: 10.1378/chest.99.3.557. [DOI] [PubMed] [Google Scholar]
- 39.Koenig A.L., Shchukina I., Amrute J., Andhey P.S., Zaitsev K., Lai L., Bajpai G., Bredemeyer A., Smith G., Jones C., et al. Single-cell transcriptomics reveals cell-type-specific diversification in human heart failure. Nat. Cardiovasc. Res. 2022;1:263–280. doi: 10.1038/s44161-022-00028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wisniewska M., Karlberg T., Lehtiö L., Johansson I., Kotenyova T., Moche M., Schüler H. Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B’, and HSPA5/BiP/GRP78. PLoS ONE. 2010;5:e8625. doi: 10.1371/journal.pone.0008625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shomura Y., Dragovic Z., Chang H.-C., Tzvetkov N., Young J.C., Brodsky J.L., Guerriero V., Hartl F.U., Bracher A. Regulation of Hsp70 function by HspBP1: Structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol. Cell. 2005;17:367–379. doi: 10.1016/j.molcel.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 42.Kasof G.M., Goyal L., White E. Btf, a novel death-promoting transcriptional repressor that interacts with Bcl-2-related proteins. Mol. Cell. Biol. 1999;19:4390–4404. doi: 10.1128/MCB.19.6.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y., Zhang X., Cai B., Li Y., Jiang Y., Fu X., Zhao Y., Gao H., Yang Y., Yang J. The long noncoding RNA lncCIRBIL disrupts the nuclear translocation of Bclaf1 alleviating cardiac ischemia–reperfusion injury. Nat. Commun. 2021;12:522. doi: 10.1038/s41467-020-20844-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y., Lewis W., Diwan A., Cheng E.H.-Y., Matkovich S.J., Dorn G.W. Dual autonomous mitochondrial cell death pathways are activated by Nix/BNip3L and induce cardiomyopathy. Proc. Natl. Acad. Sci. USA. 2010;107:9035–9042. doi: 10.1073/pnas.0914013107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y., Yang L.-N., Cheng L., Tu S., Guo S.-J., Le H.-Y., Xiong Q., Mo R., Li C.-Y., Jeong J.-S. Bcl2-associated athanogene 3 interactome analysis reveals a new role in modulating proteasome activity. Mol. Cell. Proteom. 2013;12:2804–2819. doi: 10.1074/mcp.M112.025882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hannenhalli S., Putt M.E., Gilmore J.M., Wang J., Parmacek M.S., Epstein J.A., Morrisey E.E., Margulies K.B., Cappola T.P. Transcriptional genomics associates FOX transcription factors with human heart failure. Circulation. 2006;114:1269–1276. doi: 10.1161/CIRCULATIONAHA.106.632430. [DOI] [PubMed] [Google Scholar]
- 47.Zhao M.-T., Ye S., Su J., Garg V. Cardiomyocyte proliferation and maturation: Two sides of the same coin for heart regeneration. Front. Cell Dev. Biol. 2020;8:594226. doi: 10.3389/fcell.2020.594226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naqvi N., Li M., Calvert J.W., Tejada T., Lambert J.P., Wu J., Kesteven S.H., Holman S.R., Matsuda T., Lovelock J.D. A proliferative burst during preadolescence establishes the final cardiomyocyte number. Cell. 2014;157:795–807. doi: 10.1016/j.cell.2014.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomar D., Thomas M., Garbincius J.F., Kolmetzky D.W., Salik O., Jadiya P., Carpenter A.C., Elrod J.W. MICU1 regulates mitochondrial cristae structure and function independent of the mitochondrial calcium uniporter channel. BioRxiv. 2019:803213. doi: 10.1161/circ.142.suppl_3.15070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patel J.H., Loboda A.P., Showe M.K., Showe L.C., McMahon S.B. Analysis of genomic targets reveals complex functions of MYC. Nat. Rev. Cancer. 2004;4:562–568. doi: 10.1038/nrc1393. [DOI] [PubMed] [Google Scholar]
- 51.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.García-Gutiérrez L., Delgado M.D., León J. MYC Oncogene Contributions to Release of Cell Cycle Brakes. Genes. 2019;10:244. doi: 10.3390/genes10030244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spiegel R., Gomez E.A., Akman H.O., Krishna S., Horovitz Y., DiMauro S. Myopathic form of phosphoglycerate kinase (PGK) deficiency: A new case and pathogenic considerations. Neuromuscul. Disord. NMD. 2009;19:207–211. doi: 10.1016/j.nmd.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 54.Singer-Sam J., Simmer R.L., Keith D.H., Shively L., Teplitz M., Itakura K., Gartler S.M., Riggs A.D. Isolation of a cDNA clone for human X-linked 3-phosphoglycerate kinase by use of a mixture of synthetic oligodeoxyribonucleotides as a detection probe. Proc Natl. Acad. Sci. USA. 1983;80:802–806. doi: 10.1073/pnas.80.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feichtinger R.G., Oláhová M., Kishita Y., Garone C., Kremer L.S., Yagi M., Uchiumi T., Jourdain A.A., Thompson K., D’Souza A.R., et al. Biallelic C1QBP Mutations Cause Severe Neonatal-, Childhood-, or Later-Onset Cardiomyopathy Associated with Combined Respiratory-Chain Deficiencies. Am. J. Hum. Genet. 2017;101:525–538. doi: 10.1016/j.ajhg.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ghosh A., Trivedi P.P., Timbalia S.A., Griffin A.T., Rahn J.J., Chan S.S., Gohil V.M. Copper supplementation restores cytochrome c oxidase assembly defect in a mitochondrial disease model of COA6 deficiency. Hum. Mol. Genet. 2014;23:3596–3606. doi: 10.1093/hmg/ddu069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoffman J.W., Jr., Gilbert T.B., Poston R.S., Silldorff E.P. Myocardial reperfusion injury: Etiology, mechanisms, and therapies. J. Extra Corpor. Technol. 2004;36:391–411. doi: 10.1051/ject/2004364391. [DOI] [PubMed] [Google Scholar]
- 58.Eefting F., Rensing B., Wigman J., Pannekoek W.J., Liu W.M., Cramer M.J., Lips D.J., Doevendans P.A. Role of apoptosis in reperfusion injury. Cardiovasc. Res. 2004;61:414–426. doi: 10.1016/j.cardiores.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 59.Takemura G., Kanoh M., Minatoguchi S., Fujiwara H. Cardiomyocyte apoptosis in the failing heart—A critical review from definition and classification of cell death. Int. J. Cardiol. 2013;167:2373–2386. doi: 10.1016/j.ijcard.2013.01.163. [DOI] [PubMed] [Google Scholar]
- 60.González A., Fortuño M.a.A., Querejeta R., Ravassa S., López B., López N., Diez J. Cardiomyocyte apoptosis in hypertensive cardiomyopathy. Cardiovasc. Res. 2003;59:549–562. doi: 10.1016/S0008-6363(03)00498-X. [DOI] [PubMed] [Google Scholar]
- 61.Feldman A.M., Gordon J., Wang J., Song J., Zhang X.-Q., Myers V.D., Tomar D., Gerhard G.S., Khalili K., Cheung J.Y. Novel BAG3 variants in African American patients with cardiomyopathy: Reduced β-adrenergic responsiveness in excitation–contraction. J. Card. Fail. 2020;26:1075–1085. doi: 10.1016/j.cardfail.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giordano F.J. Oxygen, oxidative stress, hypoxia, and heart failure. J. Clin. Investig. 2005;115:500–508. doi: 10.1172/JCI200524408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tintu A., Rouwet E., Verlohren S., Brinkmann J., Ahmad S., Crispi F., van Bilsen M., Carmeliet P., Staff A.C., Tjwa M. Hypoxia induces dilated cardiomyopathy in the chick embryo: Mechanism, intervention, and long-term consequences. PLoS ONE. 2009;4:e5155. doi: 10.1371/journal.pone.0005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Giglia-Mari G., Zotter A., Vermeulen W. DNA damage response. Cold Spring Harb. Perspect. Biol. 2011;3:a000745. doi: 10.1101/cshperspect.a000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mayer M.P., Bukau B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol. Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feizi A., Gatto F., Uhlen M., Nielsen J. Human protein secretory pathway genes are expressed in a tissue-specific pattern to match processing demands of the secretome. NPJ Syst. Biol. Appl. 2017;3:22. doi: 10.1038/s41540-017-0021-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Doroudgar S., Glembotski C.C. The cardiokine story unfolds: Ischemic stress-induced protein secretion in the heart. Trends Mol. Med. 2011;17:207–214. doi: 10.1016/j.molmed.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ceconi C., Curello S., Bachetti T., Corti A., Ferrari R. Tumor necrosis factor in congestive heart failure: A mechanism of disease for the new millennium? Prog. Cardiovasc. Dis. 1998;41:25–30. doi: 10.1016/S0033-0620(98)80028-5. [DOI] [PubMed] [Google Scholar]
- 69.Zwaka T.P., Manolov D., Özdemir C., Marx N., Kaya Z., Kochs M., Höher M., Hombach V., Torzewski J. Complement and dilated cardiomyopathy: A role of sublytic terminal complement complex-induced tumor necrosis factor-α synthesis in cardiac myocytes. Am. J. Pathol. 2002;161:449–457. doi: 10.1016/S0002-9440(10)64201-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schraufstatter I.U., Khaldoyanidi S.K., DiScipio R.G. Complement activation in the context of stem cells and tissue repair. World J. Stem Cells. 2015;7:1090. doi: 10.4252/wjsc.v7.i8.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chakraborti T., Mandal A., Mandal M., Das S., Chakraborti S. Complement activation in heart diseases: Role of oxidants. Cell. Signal. 2000;12:607–617. doi: 10.1016/S0898-6568(00)00111-X. [DOI] [PubMed] [Google Scholar]
- 72.Gordon J.W., Shaw J.A., Kirshenbaum L.A. Multiple Facets of NF-κB in the Heart. Circ. Res. 2011;108:1122–1132. doi: 10.1161/CIRCRESAHA.110.226928. [DOI] [PubMed] [Google Scholar]
- 73.Zhao J., Zhai B., Gygi S.P., Goldberg A.L. mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc. Natl. Acad. Sci. USA. 2015;112:15790–15797. doi: 10.1073/pnas.1521919112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fosslien E. Mitochondrial medicine–cardiomyopathy caused by defective oxidative phosphorylation. Ann. Clin. Lab. Sci. 2003;33:371–395. [PubMed] [Google Scholar]
- 75.Ludwig B., Bender E., Arnold S., Hüttemann M., Lee I., Kadenbach B. Cytochrome c oxidase and the regulation of oxidative phosphorylation. Chembiochem. 2001;2:392–403. doi: 10.1002/1439-7633(20010601)2:6<392::AID-CBIC392>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 76.Lal J.C., Mao C., Zhou Y., Gore-Panter S.R., Rennison J.H., Lovano B.S., Castel L., Shin J., Gillinov A.M., Smith J.D., et al. Transcriptomics-based network medicine approach identifies metformin as a repurposable drug for atrial fibrillation. Cell Rep. Med. 2022;3:100749. doi: 10.1016/j.xcrm.2022.100749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Myers V.D., Gerhard G.S., McNamara D.M., Tomar D., Madesh M., Kaniper S., Ramsey F.V., Fisher S.G., Ingersoll R.G., Kasch-Semenza L., et al. Association of Variants in BAG3 With Cardiomyopathy Outcomes in African American Individuals. JAMA Cardiol. 2018;3:929–938. doi: 10.1001/jamacardio.2018.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Consortium G.O. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kanehisa M., Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Croft D., O’kelly G., Wu G., Haw R., Gillespie M., Matthews L., Caudy M., Garapati P., Gopinath G., Jassal B. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res. 2010;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang B., Horvath S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005;4:17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 83.Langfelder P., Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Choi M., Chang C.-Y., Clough T., Broudy D., Killeen T., MacLean B., Vitek O. MSstats: An R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics. 2014;30:2524–2526. doi: 10.1093/bioinformatics/btu305. [DOI] [PubMed] [Google Scholar]
- 85.Lippi M., Stadiotti I., Pompilio G., Sommariva E. Human Cell Modeling for Cardiovascular Diseases. Int. J. Mol. Sci. 2020;21:6388. doi: 10.3390/ijms21176388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supporting data from this study can be obtained by emailing the corresponding author Hakon Hakonarson.