Abstract
Olaparib (AZD2281) is used as a first-line maintenance treatment for patients with ovarian cancer (OC) with a breast cancer susceptibility gene (BRCA) mutation. Fatty acid binding protein 4 (FABP4) may serve an important role in cancer, but its role in olaparib-treated OC with a BRCA mutation requires further clarification. To explore the function of FABP4 and enhance the efficacy of AZD2281 in OC, cell counting kit-8, cell apoptosis, cell cycle, colony formation, cell transfection, western blotting, reverse transcription-quantitative polymerase chain reaction, chromatin immunoprecipitation, seahorse and reactive oxygen species assays were performed. In the present study, AZD2281 significantly promoted cell apoptosis, and inhibited cell cycle progression and colony formation in COV362 cells. In addition, AZD2281 significantly upregulated the levels of CCAAT enhancer binding protein α (CEBPα), peroxisome proliferator activated receptor γ (PPARγ) and FABP4. AZD2281 markedly promoted fold enrichment of CEBPα in the promoters of PPARγ and FABP4. Furthermore, FABP4 overexpression significantly decreased cell apoptosis and promoted cell cycle progression and colony formation. In contrast, FABP4 knockdown demonstrated the opposite effects. In addition, FABP4 significantly regulated levels of reactive oxygen species, adenosine triphosphate, aerobic glycolysis, basal respiration rate and fatty acid oxidation. The combination of AZD2281 with the FABP4 inhibitor BM S309403 further significantly increased cell apoptosis and decreased colony formation. In conclusion, the findings of the present study demonstrated that AZD2281 significantly enhanced FABP4 expression, leading to diminished antitumor efficacy in OC cells with a BRCA mutation by regulating CEBPα-PPARγ. Conversely, the combination of AZD2281 and FABP4 inhibitor BM S309403 demonstrated heightened antitumor effectiveness, presenting a promising therapeutic strategy for treating patients with OC with a BRCA mutation.
Keywords: ovarian cancer, olaparib, FABP4, BM S309403, BRCA1 mutation
Introduction
Ovarian cancer (OC) is one of the deadliest diseases in women, which threatens their health (1). With a 5-year survival rate of 47%, patients with OC grapple with challenges such as chemotherapy resistance, recurrence, metastasis and associated complications (2). Notably, 15–22% of OCs are caused by a breast cancer susceptibility gene (BRCA) mutation (3–5). Women with pathogenic mutations in BRCA are 10–30 times more likely to develop OC than those without such mutations (6,7).
Poly (ADP-ribose) polymerase (PARP)-1 mostly exists in the nucleus of eukaryotic cells and catalyzes the transfer of ADP-ribose to target proteins (8). PARP-1 inhibitors constitute a novel molecularly targeted therapy and are used to treat breast cancer and OC (9–13). Olaparib, a targeted PARP inhibitor, is used as a first-line treatment for patients with OC with a BRCA mutation (14–17). Studies indicate that maintenance treatment with olaparib enhances progression-free survival (PFS) in patients with platinum-sensitive recurrent advanced OC with a BRCA mutation (18). Moreover, it demonstrates notable efficacy in improving PFS for individuals with newly diagnosed advanced OC harboring BRCA1/2 mutations (15,19). However, several patients with OC have shown no response to treatment with olaparib. Long-term treatment with olaparib can lead to the development of resistance in OC (20–22). To address resistance to olaparib, ongoing research is focused on understanding the underlying mechanisms and developing a novel combinatorial therapy for OC with a BRCA mutation (23).
Fatty acid binding protein 4 (FABP4) binds to long-chain fatty acids and regulates the uptake, transport and metabolism of fatty acids (24,25). Previous studies have reported that FABP4 serves an important role in several cancers, including breast (26,27), hepatic (28), colorectal (29,30) cervical (31) and gastric (32) cancers. FABP4 expression is upregulated in recurrent breast cancer and is associated with a worse prognosis in patients with breast cancer (26). Moreover, it modulates the import and metabolism of fatty acids in these patients (27). FABP4 also promotes hepatocarcinogenesis by regulating inflammatory pathways (28). In addition, it serves as a key determinant of the metastatic potential of OC and mediates carboplatin resistance (33,34). One study indicated that PARP-1 inhibition promotes CCAAT enhancer binding protein (cEBP)β and FABP4 expression and regulates the adipogenic transcriptional program (35). Therefore, it is necessary to determine the efficacy of a FABP4 inhibitor and the combinatorial efficacy of olaparib and FABP4 inhibitors for treating patients with OC with a BRCA1 mutation.
In the present study, to explore the function of FABP4 and enhance the efficacy of AZD2281 in OC, cell counting kit-8, cell apoptosis, cell cycle, colony formation, cell transfection, western blotting, reverse transcription-quantitative polymerase chain reaction, chromatin immunoprecipitation, seahorse and reactive oxygen species assays were performed. In addition, the combination of AZD2281 with the FABP4 inhibitor BM S309403 were performed to enhance the efficacy of AZD2281 in OC.
Materials and methods
Cell culture and transfection
COV362 and OVCAR5 cell lines were purchased from Shanghai Xuanya Biotechnology Co., Ltd., and were cultured in Dulbecco's Modified Eagle Medium containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and penicillin/streptomycin (100 U/ml) in an incubator with 5% CO2 at 37°C. For overexpression experiments, cells were transfected with plasmid vectors, and for knockdown experiments, cells were transfected with shRNA targeting FABP4. The negative control for overexpression transfection was an empty plasmid vector, while the negative control for knockdown transfection was a scrambled shRNA.
Transfections were performed using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) for plasmid DNA and Lipofectamine RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.) for shRNA according to the manufacturer's instructions. Briefly, cells were seeded into 24-well plates at a density of 1×105 cells/well and cultured overnight until reaching 50–60% confluence. For transfection, 0.5 µg of plasmid DNA or 50 nM shRNA was mixed with 1.5 µl of the appropriate transfection reagent in serum-free medium and incubated at room temperature for 10–20 min. The mixture was then added to the cells, which were incubated at 37°C for 48 h before subsequent experiments.
FABP4 overexpression (OE) was performed using a PcDNA3.1 plasmid (Shanghai Yasai Biotechnology Co., LTD.; cat. no. AVB010021), and the following sequences: FABP4-Forward (F): 5′-CTCGGATCCGCCACCATGTGTGATGCTTTTGTAGG-3′ and FABP4-Reverse (R): 5′-CCCTCTAGACTCGAGTTATGCTCTCTCATAAACTC-3′ [Asia-Vector Biotechnology (Shanghai) Co., Ltd.]. Knockdown of FABP4 expression with shRNA was performed using a PGIPZ plasmid. The sequences for FABP4 shRNA were as follows: FABP4-shRNA forward: 5′-TGCTGTTGACAGTGAGCGACTCACTGCAGATGACAGGAAAGTCAAATAGTGAAGCCACAGATGTATTTGACTTTCCTGTCATCTGCAGTGAGGTGCCTACTGCCTCGGA-3′ and FABP4-shRNA reverse: 5′-TCCGAGGCAGTAGGCACCTCACTGCAGATGACAGGAAAGTCAAATACATCTGTGGCTTCACTATTTGACTTTCCTGTCATCTGCAGTGAGTCGCTCACTGTCAACAGCA-3′ [Asia-Vector Biotechnology (Shanghai) Co., Ltd.]. The negative control shRNA (NC shRNA) sequence was 5′-CCTAAGGTTAAGTCGCCCT-3′ [Asia-Vector Biotechnology (Shanghai) Co., Ltd.]. Cells were transfected with 0.5 µg FABP4-SH, 0.5 µg FABP4-OE or 0.5 µg negative control plasmids (Asia-Vector Biotechnology) using the FuGENE® HD Transfection Reagent (cat. no. E2311; Promega Corporation), according to the protocol described in previous studies (36,37). In brief, 1×105 cells/well were seeded into 24-well culture plates overnight at 37°C. A mixture of 50 µl serum-free medium and 0.5 µg plasmid was gently incubated at room temperature for 5 min. Additionally, 2 µl FuGENE HD Transfection Reagent dissolved in 50 µl serum-free medium was gently mixed and incubated at room temperature for 5 min. Subsequently, the plasmid and liposome solution were gently mixed and incubated at room temperature for 15 min. Finally, the mixed solution was added to the cells in the 24-well plate for 6 h at 37°C. The mixed solution was then discarded and the COV362 cells were re-cultured using normal medium (DMSO; BBI Solutions) for 48 h at 37°C and collected for further experiments.
Cell Counting Kit-8 assay
Cells (COV362 and OVCAR5) were seeded onto 96-well plates at a density of 5,000 cells/well and cultured for 12, 24 and 48 h with or without AZD2281 (400 nm; Selleck Chemicals) treatment at 37°C for 48 h. Subsequently, the culture medium was replaced with a fresh medium and 10 µl of the Cell Counting Kit-8 reagent (BBI Solutions) was added to each well. The cells were incubated at 37°C for 1 h. Finally, the absorbance of cells at 450 nm was measured using a microplate reader.
Cell apoptosis assay
Cells (COV362 and OVCAR5) were seeded in a 12-well plate (3×105/well) for 24 h, and treated with or without 400 nM AZD2281 or 500 nM BM S309403 (Selleck Chemicals) at 37°C for 0, 12, 24 or 48 h. They were then collected using centrifugation at 300 × g for 5 min at 4°C, washed twice with phosphate-buffered saline (PBS), and resuspended by adding 250 µl of binding buffer. Subsequently, 100 µl cell suspension (1×106 cells/ml), 5 µl Annexin V-FITC and 10 µl propidium iodide were added to a 5-ml flow cytometer tube. After which, 400 µl binding buffer was added and mixed, and the samples were tested within 1 h. For the flow cytometry analysis, cells were treated at room temperature using the Annexin V Apoptosis Detection Kit (Beyotime Institute of Biotechnology). Red blood cell lysis was not performed. After staining, the cells were immediately analyzed using a Thermo Attune NXT flow cytometer, with the analysis conducted within 1 h. For intracellular staining, fixation and permeabilization were not necessary as surface staining was performed. The Annexin V-FITC and PI labeling was sufficient for the detection of apoptotic and necrotic cells. No additional cytokine/chemokine treatments were applied in this experiment. The antibodies used included Annexin V-FITC (Beyotime Institute of Biotechnology; cat. no. C1062) and propidium iodide (PI; Beyotime Institute of Biotechnology; cat. no. E607306). Flow cytometry was performed on a Thermo Attune NXT, and data were analyzed using Attune™ NxT software (version 3.1.2, Thermo Fisher Scientific, Inc.).
Cell cycle assay
Cells (COV362 and OVCAR5) were seeded in a 12-well plate (1×106/well) for 24 h at 37°C, treated with or without 400 nM AZD2281 or 500 nM BM S309403 at 37°C for 0, 12, 24 or 48 h, and then harvested and centrifuged at ~150 × g for 5 min at room temperature. The supernatant was then discarded. The cells were fixed with 70% ethanol at 4°C for 1 h and then washed and precipitated with PBS. After adding a cell staining solution, the cells were resuspended. Finally, cells sample was treated with 500 µl propyl iodide dye solution, gently mixed using a pipette and incubated in the dark at 37°C for 30 min. Upon completion, the samples were analyzed using flow cytometry. For apoptosis detection, Annexin V-FITC/PI staining was performed using the Annexin V-FITC Apoptosis Detection Kit (Beyotime Institute of Biotechnology; cat. no. C1062), with Annexin V-FITC detected in one channel and PI in a separate channel. For cell cycle analysis, the Cell Cycle Analysis Kit (Beyotime Institute of Biotechnology; cat. no. C1052) was used to detect a single channel. The data were collected and stored for further analysis.
Colony formation assay
Cells (COV362 and OVCAR5) were cultured in 6-well plates at a density of 200 cells/well, treated with or without 400 nM AZD2281 or 500 nM BM S309403 at 37°C for 0, 12, 24 or 48 h, and then fixed with 4% paraformaldehyde at 37°C for 30 min. Subsequently, they were stained with 1% crystal violet at 37°C for 10 min, observed and imaged using an inverted fluorescence microscope. The number of colonies with >50 cells in each group was counted.
Reverse transcription-quantitative (q)PCR
Total RNA was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. For cDNA synthesis, 1 µg of total RNA was reverse transcribed using the SuperScript™ III First-Strand Synthesis System (Invitrogen; Thermo Fisher Scientific, Inc., cat. no. 18080-051) according to the manufacturer's instructions. The reverse transcription reaction was carried out at 25°C for 10 min, followed by 50°C for 50 min, and the reaction was terminated by heating at 85°C for 5 min. Total RNA was extracted from the cells (COV362 and OVCAR5) using the TRIzol method (Invitrogen; Thermo Fisher Scientific, Inc.; cat. no. 15596-018). Gene expression was assessed using the AceQ™ Universal SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd.; cat. no. Q511-03). The thermocycling conditions for the qPCR were as follows: An initial denaturation at 95°C for 5 min, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. The final stage included 95°C for 15 s, 60°C for 60 s and 95°C for 15 s. Gene expression levels were normalized to GAPDH, and the relative quantification of gene expression was performed using the 2−ΔΔCq method (38). The analysis was conducted on a QuantStudio™ 5 Real-Time PCR System (Thermo Fisher Scientific, Inc.). The primers used for the PCR are listed in Table SI.
Western blotting
Cells (COV362 and OVCAR5) were collected, and the protein concentration was determined using western blotting. Proteins were separated and transferred onto membranes, followed by their incubation with 5% milk at room temperature for 1 h. The protein extraction buffer used was RIPA from Beyotime Institute of Biotechnology. For protein determination, the BCA protein concentration determination kit was utilized. Each lane was loaded with 20 µg of protein, and a 12% gel was used for electrophoresis. The proteins were transferred to a PVDF membrane. The membranes were incubated at 4°C overnight with the following antibodies: Anti-FABP4 (1:1,000; cat. no. ab92501; Abcam), anti-Snail (1:1,000; cat. no. 3879T; CST Biological Reagents Co., Ltd.), anti-Slug (1:1,000; cat. no. ab27568; Abcam), anti-B-cell lymphoma 2 (BCL2; 1:1,000; cat. no. AB1722; MilliporeSigma), anti-BRCA1 (1:1,000; cat. no. 22362-1-AP; Proteintech Group, Inc.), anti-BRCA2 (1:1,000; cat. no. 29450-1-AP; Proteintech Group, Inc.), anti-cEBPα (1:1,000; cat. no. 18311-1-AP; Proteintech Group, Inc.), anti-peroxisome proliferator activated receptor γ (PPARγ; 1:1,000; cat. no. 16643-1-AP; Proteintech Group, Inc.) and anti-GAPDH (1:20,000; cat. no. 5174; CST Biological Reagents Co., Ltd.). The membranes were then incubated with the secondary antibodies at room temperature for 1 h. Specifically, anti-mouse IgG (HRP-conjugated; CST Biological Reagents Co., Ltd., cat. no. 7076) and anti-rabbit IgG (HRP-conjugated; CST Biological Reagents Co., Ltd., cat. no. 7074) were both used at a dilution of 1:5,000. Protein levels were then assessed using the Tanon 5200a Chemiluminescence Imaging System (Shanghai Tianneng Technology Co., Ltd.).
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed using a ChIP kit (cat. no. ab500; Abcam). COV362 Cells were washed with PBS and collected using centrifugation at 300 × g at 4°C for 30 sec. The supernatant was then discarded. Subsequently, the precipitate was lysed with 400 µl of 1% SDS lysate, mixed and incubated for 10 min at 4°C. The supernatant was then collected and diluted with 0.6 ml dilution buffer containing phenylmethylsulfonyl fluoride. Finally, the diluted solution was washed with TE and was equally divided into each tube. Samples were incubated at 4°C overnight with anti-cEBPα antibodies (2 µg; cat. no. 18311-1-AP; Proteintech Group, Inc.) and normal IgG antibodies (cat. no. IgG 30000-0-AP; Proteintech Group, Inc.; 1 ug/ml). Later, agarose A or G (Beyotime Institute of Biotechnology; cat. no. P2055; 20 ul beads per set) was added and centrifuged at ~1,000 × g for 5 min at 4°C, and the supernatant was discarded completely. Finally, DNA was purified using a PCR purification kit (Beijing Kangrun Chengye Biotechnology, Co., Ltd.; cat. no. A304). The primers used in this experiment are provided in Table SII.
Determination of reactive oxygen species (ROS) level
Dichloro-dihydro-fluorescein diacetate was added to COV362 cells at a final concentration of 10 µM and incubated for 30 min at 37°C. The cells were then washed thrice and imaged under a fluorescence microscope. The cells were digested with trypsin, neutralized and collected in Eppendorf tubes. The cells in each group were harvested and detected using flow cytometry.
Adenosine triphosphate (ATP) assay
ATP standard solution was diluted with ATP test lysate to achieve an appropriate concentration gradient. Subsequently, 100 ml ATP test working solution was added to the tube, followed by incubation at room temperature for 3–5 min. Subsequently, 20 ml sample of COV362 cells with or without FABP4 overexpression or FABP4 knockdown was added to the test tube and mixed rapidly. Finally, the absorbance was measured using a luminometer (Promega Corporation).
Seahorse assay
Hydration solution (180 µl) was added to the lower layer of the XF96 extracellular flux assay kit (cat. no. Q29216; Agilent Technologies, Inc.) and the XF cell mito stress test kit (cat. no. 103015-100; Agilent Technologies, Inc.), both of which were hydrated overnight in an incubator without CO2 at 37°C. After incubation, the COV362 cells were washed twice with hippocampal XF basic medium (cat. no. 102353; Agilent Technologies, Inc.), and 175 µl hippocampal XF basic medium was added to each well, followed by incubation for 1 h at 37°C. Finally, the cells were assessed using the Seahorse XF24 instrument (Agilent Technologies, Inc.).
Statistical analysis
Data are presented as mean ± standard deviation. Statistical tests were performed using SPSS version 24.0 (IBM Corp.). One-way ANOVA was used to analyze data from the following experiments: Cell apoptosis, colony formation, mRNA expression, fold enrichments of cEBPα in the FABP4 or PPARγ promoter, FABP4 overexpression and knockdown, the half-maximal inhibitory concentration (IC50) of AZD2281, the levels of ROS and ATP, extracellular acidification rate, basal respiration and fatty acid oxidation at different time points or different groups. Two-way ANOVA was used to compare cell cycle analysis between different time groups or different groups and cell cycle phases (G1, S and G2), and also to compare the gene expression between different groups and different genes. Dunnett's or Tukey's multiple comparison tests were subsequently used. P<0.05 was considered to indicate a statistically significant difference.
Results
AZD2281 promotes cell apoptosis and inhibits colony formation in COV362 cells
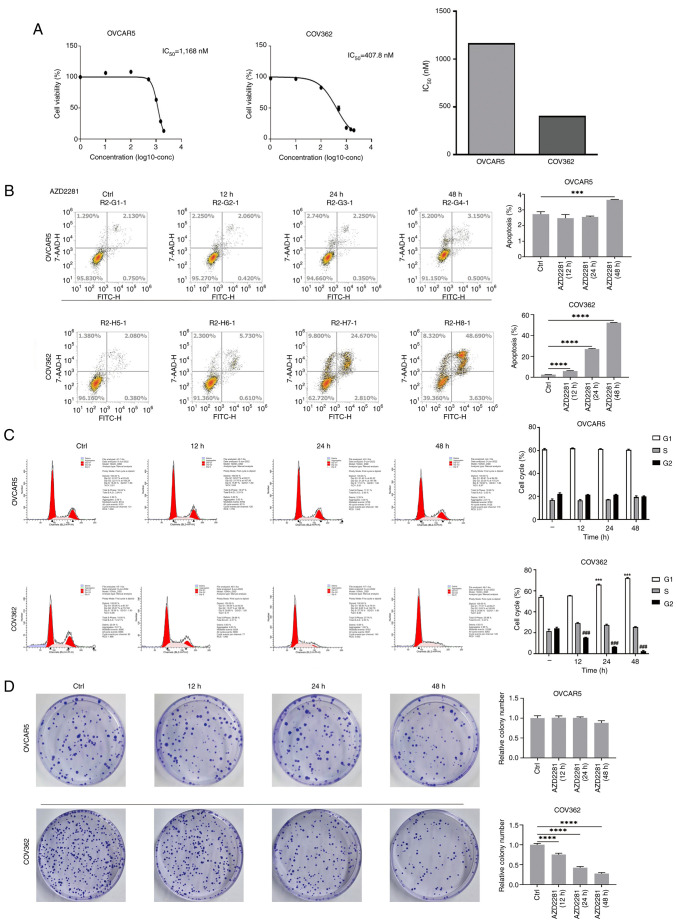
COV362 cells contain a BRCA1 mutation, whereas OVCAR5 cells contain the wild-type BRCA1; therefore, COV362 and OVCAR5 cells were selected for further analyses. The results revealed that the IC50 of AZD2281 in OVCAR5 cells was 1,168 nM, whereas in COV362 cells it was 407.8 nM, indicating that BRCA1-mutant cells are more sensitive to AZD2281 than BRCA1-wild-type cells (Fig. 1A). Moreover, compared with the control group, AZD2281 treatment significantly increased apoptosis in OVCAR5 cells at 48 h, whereas in COV362 cells, apoptosis was significantly increased at 12 h, which gradually increased with time after AZD2281 treatment (Fig. 1B). In addition, compared with the control group, AZD2281 significantly reduced cell cycle progression and colony formation in COV362 cells, but not in OVCAR5 cells (Fig. 1C and D). These results indicate that AZD2281 inhibits the function of OC cells with a BRCA1 mutation.
Figure 1.
AZD2281 promotes apoptosis and inhibits colony formation in COV362 cells. (A) IC50 of AZD2281 in OVCAR5 and COV362 cells were assessed. (B) Cell apoptosis at different time points (ctrl, 12, 24 and 48 h) after treatment with 400 nM AZD2281 in OVCAR5 and COV362 cells. ***P<0.001; ****P<0.0001. (C) Cell cycle progression at different time points (0, 12, 24 and 48 h) after treatment with 400 nM AZD2281 and in different cell cycle phases (G1, S, and G2) in OVCAR5 and COV362 cells. ***P<0.001 vs. Ctrl (G1); ###P<0.001 vs. Ctrl (G2). (D) Colony formation at different time points (ctrl, 12, 24 and 48 h) after treatment with 400 nM AZD2281 in OVCAR5 and COV362 cells. ****P<0.0001. IC50, half-maximal inhibitory concentration; Ctrl, control.
AZD2281 upregulates FABP4 expression
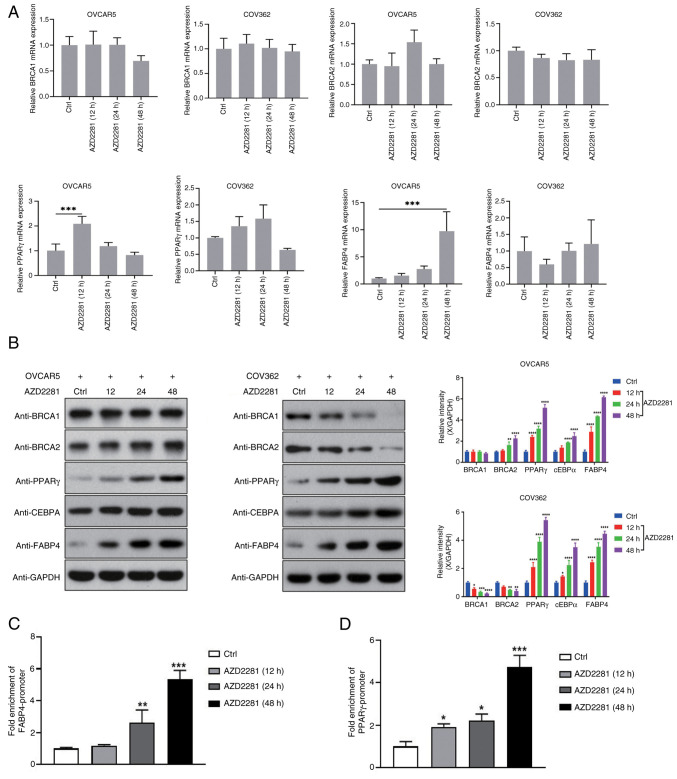
Previous studies have reported that PARP-1 inhibition upregulates levels of cEBPβ and PPARγ (35,39), whereas cEBPβ and PPARγ could regulate FABP4 expression (40,41). Therefore, the present study assessed their expression after AZD2281 treatment. The results revealed that compared with the control group, AZD2281 did not significantly regulate the mRNA levels of BRCA1 and BRCA2, but increased the mRNA levels of PPARγ after AZD2281 treatment for 12 h, and also increased the mRNA level of FABP4 after AZD2281 treatment for 48 h (Fig. 2A); whereas, AZD2281 significantly upregulated cEBPα, PPARγ and FABP4 protein expression, in COV362 and OVCAR5 cells (Fig. 2B). In addition, compared with the control group, AZD2281 significantly downregulated BRCA1 and BRCA2 expression in COV362 cells, but not in OVCAR5 cells (Fig. 2B). Furthermore, the results of the ChIP assay indicated that compared with the control group, AZD2281 treatment significantly promoted the binding of cEBPα to the promoters of PPARγ and FABP4 (Fig. 2C and D), suggesting that AZD2281 upregulates FABP4 expression by modulating cEBPα expression.
Figure 2.
AZD2281 upregulates FABP4 expression. (A) mRNA expression of BRCA1, BRCA2, PPARγ and FABP4 at different time points (ctrl, 12, 24 and 48 h) after treatment with 400 nM AZD2281 in OVCAR5 and COV362 cells. ***P<0.001. (B) Protein expression of BRCA1, BRCA2, PPARγ, cEBPα and FABP4 at different time points (ctrl, 12, 24 and 48 h) after treatment with 400 nM AZD2281 in OVCAR5 and COV362 cells. *P<0.05 vs. Ctrl;**P<0.01 vs. Ctrl;***P<0.001 vs. Ctrl; ****P<0.0001 vs. Ctrl. Fold enrichments of cEBPα in the (C) FABP4 and (D) PPARγ promoters at different time points (ctrl, 12, 24 and 48 h) after treatment with 400 nM AZD2281. *P<0.05 vs. Ctrl; **P<0.01 vs. Ctrl; ***P<0.001 vs. Ctrl; ***P<0.001 vs. Ctrl. FABP4, fatty acid binding protein 4; BRCA, breast cancer susceptibility gene; PPARγ, peroxisome proliferator activated receptor γ; cEBPα, CCAAT enhancer binding protein α; ChIP, chromatin immunoprecipitation; Ctrl, control.
FABP4 regulates the function of OC in COV362 cells
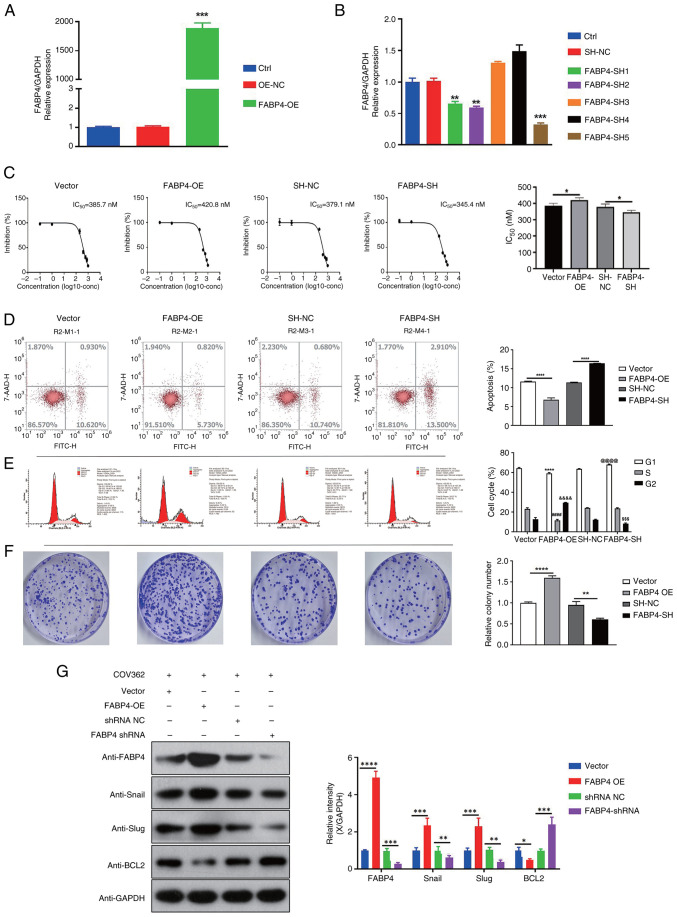
To assess the function of FABP4 in OC cells with a BRCA1 mutation, FABP4 overexpression and knockdown were performed in COV362 cells (Fig. 3A and B). Due to its superior silencing efficacy, FABP4-SH5 was chosen to use in the subsequent experiments. Compared with the vector group, FABP4 overexpression significantly increased the IC50 of AZD2281 in COV362 cells. Moreover, compared with the SH-NC group, FABP4 knockdown significantly decreased the IC50 of AZD2281 in COV362 cells (Fig. 3C). In addition, compared with the vector group, FABP4 overexpression significantly reduced apoptosis and promoted cell cycle progression and colony formation, whereas compared with the SH-NC group, FABP4 knockdown significantly increased apoptosis and reduced cell cycle progression and colony formation in COV362 cells (Fig. 3D-F). Furthermore, compared with the vector group, FABP4 overexpression significantly upregulated the expression of Snail, Slug and BCL2, whereas compared with the SH-NC group, FABP4 knockdown significantly downregulated the expression of the three proteins in COV362 cells (Fig. 3G).
Figure 3.
FABP4 regulates the function of COV362 cells in OC. (A) FABP4 expression in different groups (ctrl, OE-NC and FABP4-OE) after FABP4 overexpression. ***P<0.001 vs. OE-NC. (B) FABP4 expression in different groups (ctrl, SH-NC, FABP4-SH1, FABP4-SH2, FABP4-SH3, FABP4-SH4 and FABP4-SH5) after FABP4 knockdown. **P<0.01 vs. SH-NC; ***P<0.001 vs. SH-NC. (C) IC50 of AZD2281 in different groups (vector, FABP4-OE, SH-NC and FABP4-SH) after FABP4 overexpression and knockdown. *P<0.05. (D) Cell apoptosis in different groups (vector, FABP4-OE, SH-NC and FABP4-SH) after FABP4 overexpression and knockdown. ****P<0.0001. (E) Cell cycle progression between different groups (vector, FABP4-OE, SH-NC and FABP4-SH) and cell cycle phases (G1, S and G2). ****P<0.0001 vs. Vector (G1); ####P<0.0001 vs. Vector (S); &&&&P<0.0001 vs. Vector (G2); @@@@P<0.0001 vs. SH-NC (G1); $$$P<0.001 vs. SH-NC (G2). (F) Colony formation in different groups (vector, FABP4-OE, SH-NC and FABP4-SH) after FABP4 overexpression and knockdown. **P<0.01; ****P<0.0001. (G) Gene expression of Snail, Slug and BCL2 in different groups (vector, FABP4-OE, SH-NC, and FABP4-SH). *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. FABP4, fatty acid binding protein 4; OC, ovarian cancer; BCL2, B-cell lymphoma 2; OE, overexpression; NC, negative control; SH/sh, short hairpin; ctrl, control; IC50, half-maximal inhibitory concentration.
FABP4 regulates mitochondrial function
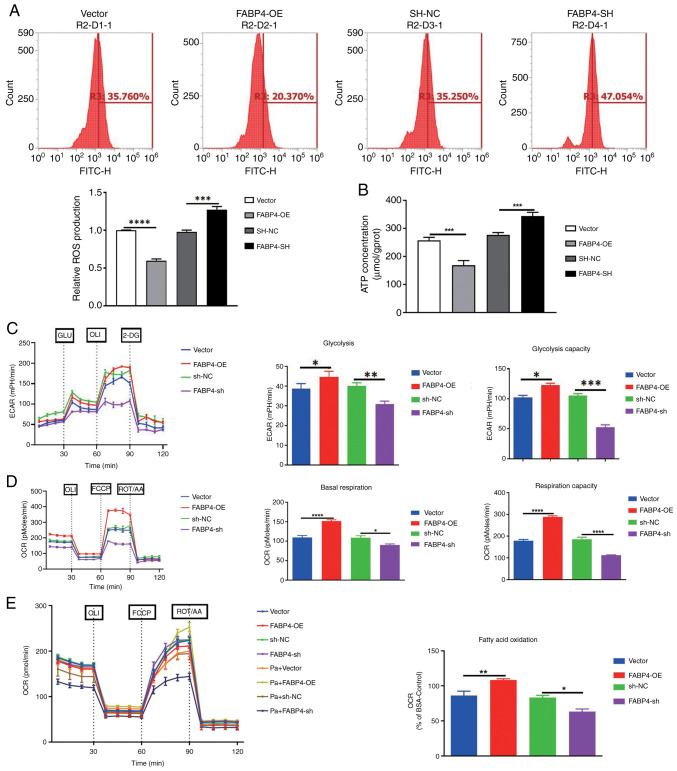
A previous study reported that FABP4 could regulate mitochondrial membrane homeostasis and mitochondrial function (42). Therefore, the present study assessed the effects of FABP4 on mitochondrial function. The results revealed that compared with the vector group, FABP4 overexpression significantly reduced ROS and ATP levels (Fig. 4A and B). Further results demonstrated that FABP4 overexpression significantly enhanced aerobic glycolysis, basal respiration rate and fatty acid oxidation, promoting mitochondrial function and providing energy for OC growth (Fig. 4C-E). FABP4 knockdown also significantly increased ROS and ATP levels (Fig. 4A and B) and significantly reduced glycolysis, basal respiration rate and fatty acid oxidation (Fig. 4C-E). Whereas compared with the SH-NC group, These results indicate that FABP4 regulates the progression of OC by modulating mitochondrial function.
Figure 4.
FABP4 regulates mitochondrial function. (A) Levels of ROS, (B) ATP, (C) ECAR, (D) basal respiration and (E) fatty acid oxidation in different groups (vector, FABP4-OE, SH-NC, FABP4-SH, Pa + Vector, Pa + FABP4-OE, Pa + SH-NC, and Pa + FABP4-SH) after FABP4 overexpression and knockdown, as well as after Pa treatment. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. FABP4, fatty acid binding protein 4; ROS, reactive oxygen species; ATP, adenosine triphosphate; ECAR, extracellular acidification rate; OCR, oxygen consumption rate; OE, overexpression; NC, negative control; SH/sh, short hairpin; Pa, palmitic acid; GLU, glucose; OLI, oligomycin; FCCP, Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone; ROT/AA, Rotenone + Antimycin A; 2-DG, 2-deoxy glucose.
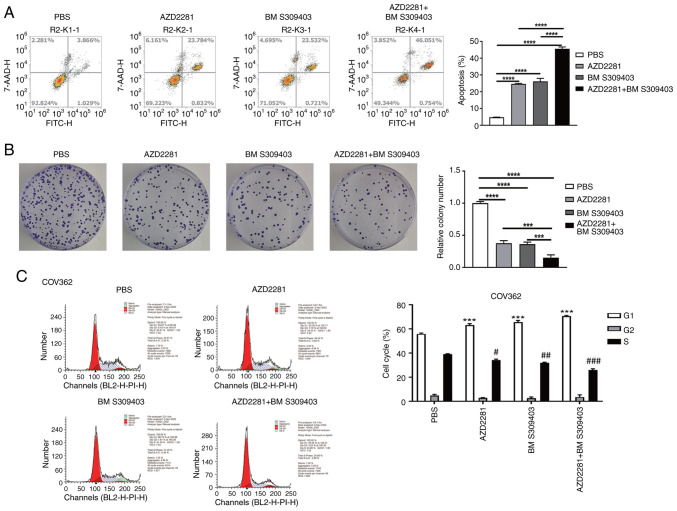
Combinatorial efficacy of AZD2281 and BM S309403 in BRCA1-mutant OC cells
Compared with the PBS group, AZD2281 and BM S309403 (a FABP4 inhibitor) both significantly promoted apoptosis (Fig. 5A) and significantly inhibited colony formation (Fig. 5B) and cell cycle progression (Fig. 5C) in COV362 cells. Compared with AZD2281 or BM S309403 alone, their combination further significantly promoted cell apoptosis (Fig. 5A) and inhibited colony formation (Fig. 5B) and cell cycle progression (Fig. 5C), indicating the combinatorial antitumor efficacy of olaparib and a FABP4 inhibitor for treating OC with a BRCA1 mutation.
Figure 5.
Combinatorial efficacy of AZD2281 and BM S309403 in breast cancer susceptibility gene 1-mutant ovarian cancer cells. (A) Cell apoptosis and (B) colony formation in different groups (PBS, AZD2281, BM S309403 and AZD2281 + BM S309403). ***P<0.001; ****P<0.0001. (C) Cell cycle progression in different groups (PBS, AZD2281, BM S309403 and AZD2281 + BM S309403) and cell cycle phases (G1, S and G2). ***P<0.001 vs. PBS (G1); #P<0.05 vs. PBS (S); ##P<0.01 vs. PBS (S); ###P<0.001 vs. PBS (S).
Discussion
The results of the present study revealed that AZD2281 (olaparib) significantly promoted apoptosis and inhibited cell cycle progression and colony formation in COV362 cells, which contain a BRCA1 mutation. However, AZD2281 also significantly upregulated FABP4 expression in COV362 cells. FABP4 overexpression inhibited cell apoptosis and promoted cell cycle progression and colony formation, whereas FABP4 knockdown demonstrated the opposite effects. Further analyses demonstrated that FABP4 overexpression reduced ROS and ATP levels and enhanced aerobic glycolysis, basal respiration rate and fatty acid oxidation, whereas FABP4 knockdown demonstrated the opposite effects. Furthermore, the combination of AZD2281 and the FABP4 inhibitor BM S309403 significantly promoted cell apoptosis and inhibited colony formation, indicating the combinatorial efficacy of olaparib and FABP4 inhibitor for treating OC with a BRCA1 mutation (Fig. 6).
Figure 6.
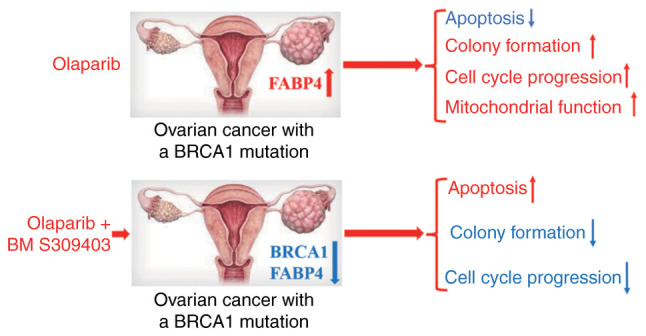
AZD2281 (olaparib) markedly inhibits the progression of ovarian cancer cells with a BRCA1 mutation. Furthermore, AZD2281 increases the protein level of FABP4, which regulates the progression of ovarian cancer by mediating function of mitochondria. Finally, the combination of AZD2281 with a FABP4 inhibitor increases the antitumor efficacy in ovarian cancer cells with a BRCA1 mutation, providing a potential new treatment strategy for patients with ovarian cancer with a BRCA mutation. BRCA1, breast cancer susceptibility gene 1; FABP4, fatty acid binding protein 4.
Previous studies have demonstrated that the incidence of OC is higher among individuals with a BRCA mutation (43,44) and that BRCA mutations are the main mutations in patients with OC (45). Olaparib has been used as a maintenance therapy for patients with newly diagnosed advanced OC and a BRCA mutation, but not for patients with OC and wild-type BRCA (14,16,17). The present study demonstrated that olaparib (AZD2281) significantly promoted apoptosis and inhibited cell cycle progression and colony formation in COV362 cells; however, no effect was observed in OVCAR5 cells.
PARP-1 regulates cEBPβ expression via PARylation modification (35,39,46), whereas cEBPβ and cEBPα (an isoform of cEBPβ) promote the activities of FABP4 and PPARγ, ultimately regulating adipogenesis (35,47,48). A previous study reported that FABP4 promoted metastasis in OC, whereas FABP4 knockout reduced it (33,34). BM S309403, a small-molecule inhibitor of FABP4, has been reported to reduce the tumor burden and increase the sensitivity of OC cells to carboplatin (33), indicating that FABP4 may serve an important role in the progression of OC. In the present study, although AZD2281 inhibited the progression of OC cells with a BRCA1 mutation, it also significantly upregulated cEBPα, PPARγ and FABP4 expression. A previous study reported that olaparib was a pan-agonist of PPARγ (49). We hypothesize that olaparib may directly modulate PPAR expression and potentially exert other functions with temporal effects. In a previous study, FABP4 contributed to poor prognosis and regulated the metastasis and metabolism of OC cells (34). The present study demonstrated that FABP4 could regulate the progression of OC cells with a BRCA1 mutation. However, olaparib significantly promoted FABP4 expression, which is associated with resistance to olaparib in patients with OC and a BRCA1 mutation (50).
Furthermore, BM S309403, a FABP4 inhibitor, has been reported to inhibit the progression of OC (33). The results of the present study revealed that BM S309403 also promoted cell apoptosis and inhibited cell cycle progression and colony formation. The combination of AZD2281 and BM S309403 further promoted cell apoptosis and inhibited colony formation. Previous studies have reported that the combination of olaparib and bevacizumab could serve as a first-line maintenance therapy in patients with advanced OC (16,51,52). Olaparib alone has limited efficacy in treating OC. However, drug combinations have been shown to improve cure rates. The FDA approval of the Olaparib-bevacizumab combination as a first-line treatment highlights the benefits of such approaches (53). This further supports the view that combining Olaparib with BM S309403 may offer a more effective treatment for OC. Therefore, the findings of the present study suggest a potential new treatment strategy for patients with OC with a BRCA1 mutation.
One challenge for treating OC is the development of drug resistance, as cancer cells can adapt and develop mechanisms to evade treatment, leading to the emergence of drug-resistant phenotypes and reduced treatment efficacy over time (54,55). Whilst the initial results with olaparib (AZD2281) and FABP4 inhibitors may be promising in inhibiting tumor growth, the specificity of treatment poses another challenge. Targeting specific mutations like BRCA1 or BRCA2 may be effective in certain patient subsets but not universally applicable to all patients with OC. Certain individuals may harbor different genetic mutations or molecular profiles that make them less responsive to the proposed treatment regimen. Therefore, developing new combination therapies that target different mutations could offer a more comprehensive approach to benefit a wider range of patients.
The present study has certain limitations. First, further determination of the optimal concentration and mode of administration of FABP4 inhibitors in the clinical treatment of BRCA1-mutant patients with OC is warranted. Pharmacokinetic trials and exploration of several administration modes could provide valuable insights into achieving the appropriate concentration and mode of FABP4 inhibitors. Additionally, the validation of the antitumor effects of AZD2281 and FABP4 inhibitors in combination with BRCA1-mutant OC using multiple cell models and animal models should be performed. Utilizing organoids and patient-derived xenograft models may offer enhanced benefits for observing the efficacy of combination therapy. Third, in previous studies, it has been reported that olaparib (AZD2281) has the potential to modulate the progression of OC in BRCA2-mutant OC cells (56,57). The present study also demonstrated that AZD2281 significantly decreased BRCA2 expression. However, whether AZD2281 also influences FABP4 expression in BRCA2-mutant cell lines remains uncertain. Subsequent research should delve deeper into this aspect for further clarification. Finally, treatment with AZD2281 did not alter mRNA expression levels but exhibited regulation of protein expression in the present study. We hypothesize that these protein changes may be governed by influencing post-transcriptional modifications. The precise mechanism underlying this phenomenon requires elucidation through additional investigations in future studies.
In summary, the present study demonstrated that olaparib (AZD2281) significantly inhibited the progression of OC cells with a BRCA1 mutation. However, olaparib also significantly upregulated FABP4 expression in COV362 cells, which contain a BRCA1 mutation. Furthermore, FABP4 regulated the progression of OC by modulating mitochondrial function. Finally, the combination of AZD2281 and a FABP4 inhibitor (BM S309403) significantly increased its antitumor efficacy, thus providing a potential new treatment strategy for patients with OC with a BRCA1 mutation.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- OC
ovarian cancer
- BRCA
breast cancer susceptibility gene
- FABP4
fatty acid binding protein 4
- PARP-1
poly (ADP-ribose) polymerase 1
- PFS
progression-free survival
- SH
short-hairpin
- PBS
phosphate-buffered saline
- OE
overexpression
- qPCR
quantitative polymerase chain reaction
- cEBP
CCAAT enhancer binding protein
- PPARγ
peroxisome proliferator activated receptor γ
- ChIP
chromatin immunoprecipitation
- ROS
reactive oxygen species
- BCL2
B-cell lymphoma 2
- ATP
adenosine triphosphate
Funding Statement
The present work was supported by the National Natural Science Foundation of China (grant nos. 82173238 and 81872507), Harbin Medical University Cancer Hospital Nn10 Project (grant no. Nn10py2017-01], the Key Projects of Haiyan Fund (grant no. JJZD2017-01) and the Key Projects of Heilongjiang Natural Science Foundation (grant no. ZD2020H007).
Availability of data and materials
The data generated in the present study may be requested from the corresponding author.
Authors' contributions
LH, GL and WH contributed to the conceptualization and methodology. HM collected the data. YX analyzed the data. LH and GL confirm the authenticity of all the raw data. LH and GL contributed to manuscript writing and reviewing. All authors reviewed the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A, Siegel RL. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68:284–296. doi: 10.3322/caac.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harter P, Johnson T, Berton-Rigaud D, Park SY, Friedlander M, Del Campo JM, Shimada M, Forget F, Mirza MR, Colombo N, et al. BRCA1/2 mutations associated with progression-free survival in ovarian cancer patients in the AGO-OVAR 16 study. Gynecol. Oncol. 2016;140:443–449. doi: 10.1016/j.ygyno.2015.12.027. [DOI] [PubMed] [Google Scholar]
- 4.Tewari KS, Burger RA, Enserro D, Norquist BM, Swisher EM, Brady MF, Bookman MA, Fleming GF, Huang H, Homesley HD, et al. Final Overall Survival of a Randomized Trial of Bevacizumab for Primary Treatment of Ovarian Cancer. J Clin Oncol. 2019;37:2317–2328. doi: 10.1200/JCO.19.01009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sunar V, Korkmaz V, Topcu V, Cavdarli B, Arik Z, Ozdal B, Ustun YE. Frequency of germline BRCA1/2 mutations and association with clinicopathological characteristics in Turkish women with epithelial ovarian cancer. Asia Pac J Clin Oncol. 2022;18:84–92. doi: 10.1111/ajco.13520. [DOI] [PubMed] [Google Scholar]
- 6.Daly MB, Pal T, Berry MP, Buys SS, Dickson P, Domchek SM, Elkhanany A, Friedman S, Goggins M, Hutton ML, et al. Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2021;19:77–102. doi: 10.6004/jnccn.2021.0001. [DOI] [PubMed] [Google Scholar]
- 7.Walker M, Jacobson M, Sobel M. Management of ovarian cancer risk in women with BRCA1/2 pathogenic variants. CMAJ. 2019;191:E886–E893. doi: 10.1503/cmaj.190281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langelier MF, Planck JL, Roy S, Pascal JM. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012;336:728–732. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zimmer AS, Gillard M, Lipkowitz S, Lee JM. Update on PARP inhibitors in breast cancer. Curr Treat Options Oncol. 2018;19:21. doi: 10.1007/s11864-018-0540-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Layman RM, Arun B. PARP inhibitors in triple-negative breast cancer including those With BRCA Mutations. Cancer J. 2021;27:67–75. doi: 10.1097/PPO.0000000000000499. [DOI] [PubMed] [Google Scholar]
- 11.Tung NM, Zakalik D, Somerfield MR, Hereditary Breast Cancer Guideline Expert Panel Adjuvant PARP Inhibitors in Patients With High-Risk Early-Stage HER2-Negative Breast Cancer and Germline BRCA Mutations: ASCO Hereditary Breast Cancer Guideline Rapid Recommendation Update. J Clin Oncol. 2021;39:2959–2961. doi: 10.1200/JCO.21.01532. [DOI] [PubMed] [Google Scholar]
- 12.Mirza MR, Coleman RL, Gonzalez-Martin A, Moore KN, Colombo N, Ray-Coquard I, Pignata S. The forefront of ovarian cancer therapy: Update on PARP inhibitors. Ann Oncol. 2020;31:1148–1159. doi: 10.1016/j.annonc.2020.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Hong T, Lei G, Chen X, Li H, Zhang X, Wu N, Zhao Y, Zhang Y, Wang J. PARP inhibition promotes ferroptosis via repressing SLC7A11 and synergizes with ferroptosis inducers in BRCA-proficient ovarian cancer. Redox Biol. 2021;42:101928. doi: 10.1016/j.redox.2021.101928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banerjee S, Moore KN, Colombo N, Scambia G, Kim BG, Oaknin A, Friedlander M, Lisyanskaya A, Floquet A, Leary A, et al. Maintenance olaparib for patients with newly diagnosed advanced ovarian cancer and a BRCA mutation (SOLO1/GOG 3004): 5-year follow-up of a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2021;22:1721–1731. doi: 10.1016/S1470-2045(21)00531-3. [DOI] [PubMed] [Google Scholar]
- 15.Friedlander M, Moore KN, Colombo N, Scambia G, Kim BG, Oaknin A, Lisyanskaya A, Sonke GS, Gourley C, Banerjee S, et al. Patient-centred outcomes and effect of disease progression on health status in patients with newly diagnosed advanced ovarian cancer and a BRCA mutation receiving maintenance olaparib or placebo (SOLO1): A randomised, phase 3 trial. Lancet Oncol. 2021;22:632–642. doi: 10.1016/S1470-2045(21)00098-X. [DOI] [PubMed] [Google Scholar]
- 16.Ray-Coquard I, Pautier P, Pignata S, Pérol D, González-Martín A, Berger R, Fujiwara K, Vergote I, Colombo N, Mäenpää J, et al. Olaparib plus Bevacizumab as First-Line Maintenance in Ovarian Cancer. N. Engl J Med. 2019;381:2416–2428. doi: 10.1056/NEJMoa1911361. [DOI] [PubMed] [Google Scholar]
- 17.Pujade-Lauraine E, Ledermann JA, Selle F, Gebski V, Penson RT, Oza AM, Korach J, Huzarski T, Poveda A, Pignata S, et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): A double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18:1274–1284. doi: 10.1016/S1470-2045(17)30469-2. [DOI] [PubMed] [Google Scholar]
- 18.Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, Scott CL, Meier W, Shapira-Frommer R, Safra T, et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: A preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014;15:852–861. doi: 10.1016/S1470-2045(14)70228-1. [DOI] [PubMed] [Google Scholar]
- 19.DiSilvestro P, Colombo N, Scambia G, Kim BG, Oaknin A, Friedlander M, Lisyanskaya A, Floquet A, Leary A, Sonke GS, et al. Efficacy of maintenance olaparib for patients with newly diagnosed advanced ovarian cancer with a BRCA Mutation: Subgroup analysis findings from the SOLO1 Trial. J Clin Oncol. 2020;38:3528–3537. doi: 10.1200/JCO.20.00799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noordermeer SM, van Attikum H. PARP Inhibitor Resistance: A Tug-of-War in BRCA-Mutated Cells. Trends Cell Biol. 2019;29:820–834. doi: 10.1016/j.tcb.2019.07.008. [DOI] [PubMed] [Google Scholar]
- 21.Dias MP, Moser SC, Ganesan S, Jonkers J. Understanding and overcoming resistance to PARP inhibitors in cancer therapy. Nat Rev Clin Oncol. 2021;18:773–791. doi: 10.1038/s41571-021-00532-x. [DOI] [PubMed] [Google Scholar]
- 22.Jiang X, Li X, Li W, Bai H, Zhang Z. PARP inhibitors in ovarian cancer: Sensitivity prediction and resistance mechanisms. J Cell Mol Med. 2019;23:2303–2313. doi: 10.1111/jcmm.14133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang N, Yang Y, Jin D, Zhang Z, Shen K, Yang J, Chen H, Zhao X, Yang L, Lu H. PARP inhibitor resistance in breast and gynecological cancer: Resistance mechanisms and combination therapy strategies. Front Pharmacol. 2022;13:967633. doi: 10.3389/fphar.2022.967633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang PB, Hou PP, Liu FY, Hong WB, Chen HZ, Sun XY, Li P, Zhang Y, Ju CY, Luo LJ, et al. Blocking PPARү interaction facilitates Nur77 interdiction of fatty acid uptake and suppresses breast cancer progression. Proc Natl Acad Sci USA. 2020;117:27412–27422. doi: 10.1073/pnas.2002997117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jabs M, Rose AJ, Lehmann LH, Taylor J, Moll I, Sijmonsma TP, Herberich SE, Sauer SW, Poschet G, Federico G, et al. Inhibition of endothelial notch signaling impairs fatty acid transport and leads to metabolic and vascular remodeling of the adult Heart. Circulation. 2018;137:2592–2608. doi: 10.1161/CIRCULATIONAHA.117.029733. [DOI] [PubMed] [Google Scholar]
- 26.Luis G, Godfroid A, Nishiumi S, Cimino J, Blacher S, Maquoi E, Wery C, Collignon A, Longuespée R, Montero-Ruiz L, et al. Tumor resistance to ferroptosis driven by Stearoyl-CoA Desaturase-1 (SCD1) in cancer cells and Fatty Acid Biding Protein-4 (FABP4) in tumor microenvironment promote tumor recurrence. Redox Biol. 2021;43:102006. doi: 10.1016/j.redox.2021.102006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gyamfi J, Yeo JH, Kwon D, Min BS, Cha YJ, Koo JS, Jeong J, Lee J, Choi J. Interaction between CD36 and FABP4 modulates adipocyte-induced fatty acid import and metabolism in breast cancer. NPJ Breast Cancer. 2021;7:129. doi: 10.1038/s41523-021-00324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H, Deng Q, Ni T, Liu Y, Lu L, Dai H, Wang H, Yang W. Targeted Inhibition of LPL/FABP4/CPT1 fatty acid metabolic axis can effectively prevent the progression of nonalcoholic steatohepatitis to liver cancer. Int J Biol Sci. 2021;17:4207–4222. doi: 10.7150/ijbs.64714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian W, Zhang W, Zhang Y, Zhu T, Hua Y, Li H, Zhang Q, Xia M. FABP4 promotes invasion and metastasis of colon cancer by regulating fatty acid transport. Cancer Cell Int. 2020;20:512. doi: 10.1186/s12935-020-01582-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Zhao X, Deng L, Li X, Wang G, Li Y, Chen M. High expression of FABP4 and FABP6 in patients with colorectal cancer. World J Surg Oncol. 2019;17:171. doi: 10.1186/s12957-019-1714-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li G, Wu Q, Gong L, Xu X, Cai J, Xu L, Zeng Y, He X, Wang Z. FABP4 is an independent risk factor for lymph node metastasis and poor prognosis in patients with cervical cancer. Cancer Cell Int. 2021;21:568. doi: 10.1186/s12935-021-02273-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen QY, Huang XB, Zhao YJ, Wang HG, Wang JB, Liu LC, Wang LQ, Zhong Q, Xie JW, Lin JX, et al. The peroxisome proliferator-activated receptor agonist rosiglitazone specifically represses tumour metastatic potential in chromatin inaccessibility-mediated FABP4-deficient gastric cancer. Theranostics. 2022;12:1904–1920. doi: 10.7150/thno.66814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukherjee A, Chiang CY, Daifotis HA, Nieman KM, Fahrmann JF, Lastra RR, Romero IL, Fiehn O, Lengyel E. Adipocyte-Induced FABP4 expression in ovarian cancer cells promotes metastasis and mediates carboplatin resistance. Cancer Res. 2020;80:1748–1761. doi: 10.1158/0008-5472.CAN-19-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gharpure KM, Pradeep S, Sans M, Rupaimoole R, Ivan C, Wu SY, Bayraktar E, Nagaraja AS, Mangala LS, Zhang X, Haemmerle M, et al. FABP4 as a key determinant of metastatic potential of ovarian cancer. Nat Commun. 2018;9:2923. doi: 10.1038/s41467-018-04987-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo X, Ryu KW, Kim DS, Nandu T, Medina CJ, Gupte R, Gibson BA, Soccio RE, Yu Y, Gupta RK, Kraus WL. PARP-1 Controls the Adipogenic Transcriptional Program by PARylating C/EBPβ and modulating its transcriptional activity. Mol Cell. 2017;65:260–271. doi: 10.1016/j.molcel.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan J, Liu F, Xiao X, Xu R, Dai L, Zhu M, Xu H, Xu Y, Zhao A, Zhou W, et al. METTL3 promotes colorectal carcinoma progression by regulating the m6A-CRB3-Hippo axis. J Exp Clin Cancer Res. 2022;41:19. doi: 10.1186/s13046-021-02227-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dang Y, Xu R, Pan J, Xiao X, Zhang S, Zhou W, Xu Y, Ji G. Dynamic changes in DNA methylation and hydroxymethylation revealed the transformation of advanced adenoma into colorectal carcinoma. Clin Transl Med. 2023;13:e1202. doi: 10.1002/ctm2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Ryu KW, Nandu T, Kim J, Challa S, DeBerardinis RJ, Kraus WL. Metabolic regulation of transcription through compartmentalized NAD(+) biosynthesis. Science. 2018;360:eaan5780. doi: 10.1126/science.aan5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su X, Jin M, Xu C, Gao Y, Yang Y, Qi H, Zhang Q, Yang X, Ya W, Zhang Y, Yang R. FABP4 in Paneth cells regulates antimicrobial protein expression to reprogram gut microbiota. Gut Microbes. 2022;14:2139978. doi: 10.1080/19490976.2022.2139978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moreno-Vedia J, Girona J, Ibarretxe D, Masana L, Rodriguez-Calvo R. Unveiling the role of the fatty acid binding protein 4 in the metabolic-associated fatty liver disease. Biomedicines. 2022;10:197. doi: 10.3390/biomedicines10010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang M, Shu W, Zhai X, Yang X, Zhou H, Pan B, Li C, Lu D, Cai J, Zheng S, et al. Integrated multi-omic analysis identifies fatty acid binding protein 4 as a biomarker and therapeutic target of ischemia-reperfusion injury in steatotic liver transplantation. Cell Mol Life Sci. 2024;81:83. doi: 10.1007/s00018-023-05110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sekine M, Nishino K, Enomoto T. Differences in ovarian and other cancers risks by population and BRCA Mutation Location. Genes (Basel) 2021;12:1050. doi: 10.3390/genes12071050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schouten PC, Richters L, Vis DJ, Kommoss S, van Dijk E, Ernst C, Kluin RJC, Marmé F, Lips EH, Schmidt S, et al. Ovarian Cancer-Specific BRCA-like Copy-number aberration classifiers detect mutations associated with homologous recombination deficiency in the AGO-TR1 Trial. Clin Cancer Res. 2021;27:6559–6569. doi: 10.1158/1078-0432.CCR-21-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samuel D, Diaz-Barbe A, Pinto A, Schlumbrecht M, George S. Hereditary Ovarian Carcinoma: Cancer Pathogenesis Looking beyond BRCA1 and BRCA2. Cells. 2022;11:539. doi: 10.3390/cells11030539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vidakovic M, Dinic S, Grdovic N, Mihailović M, Uskoković A, Quesada P, Poznanović G. Regulation of rat haptoglobin gene expression is coordinated by the nuclear matrix. J Cell Biochem. 2009;107:1205–1221. doi: 10.1002/jcb.22225. [DOI] [PubMed] [Google Scholar]
- 47.Gui L, Raza SHA, Ma B, Easa AA, Althobaiti F, Shukry M, Alotaibi MA, Al Hazani TMI, Dawood MAO, Khan R, et al. CEBPβ binding directly to the promoter region drives CEBPa transcription and improves FABP4 transcriptional activity in adipose tissue of yak (Bos grunniens) Res Vet Sci. 2021;141:174–179. doi: 10.1016/j.rvsc.2021.10.018. [DOI] [PubMed] [Google Scholar]
- 48.Regassa A, Park KW, Kim WK. Phenamil enhances the adipogenic differentiation of hen preadipocytes. Cell Biol Int. 2016;40:1123–1128. doi: 10.1002/cbin.10651. [DOI] [PubMed] [Google Scholar]
- 49.Mandal SK, Puri S, Kumar BK, Muzaffar-Ur-Rehman M, Sharma PK, Sankaranarayanan M, Deepa PR. Targeting lipid-sensing nuclear receptors PPAR (α, ү, β/δ): HTVS and molecular docking/dynamics analysis of pharmacological ligands as potential pan-PPAR agonists. Mol Divers. 2024;28:1423–1438. doi: 10.1007/s11030-023-10666-y. [DOI] [PubMed] [Google Scholar]
- 50.Baczewska M, Bojczuk K, Kołakowski A, Dobroch J, Guzik P, Knapp P. Obesity and energy substrate transporters in ovarian cancer-review. Molecules. 2021;26:1659. doi: 10.3390/molecules26061659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arora S, Balasubramaniam S, Zhang H, Berman T, Narayan P, Suzman D, Bloomquist E, Tang S, Gong Y, Sridhara R, et al. FDA Approval Summary: Olaparib monotherapy or in combination with bevacizumab for the maintenance treatment of patients with advanced ovarian cancer. Oncologist. 2021;26:e164–e172. doi: 10.1002/onco.13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gonzalez-Martin A, Desauw C, Heitz F, Cropet C, Gargiulo P, Berger R, Ochi H, Vergote I, Colombo N, Mirza MR, et al. Maintenance olaparib plus bevacizumab in patients with newly diagnosed advanced high-grade ovarian cancer: Main analysis of second progression-free survival in the phase III PAOLA-1/ENGOT-ov25 trial. Eur J Cancer. 2022;174:221–231. doi: 10.1016/j.ejca.2022.07.022. [DOI] [PubMed] [Google Scholar]
- 53.Moore K, Colombo N, Scambia G, Kim BG, Oaknin A, Friedlander M, Lisyanskaya A, Floquet A, Leary A, Sonke GS, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2018;379:2495–2505. doi: 10.1056/NEJMoa1810858. [DOI] [PubMed] [Google Scholar]
- 54.Nikolaou M, Pavlopoulou A, Georgakilas AG, Kyrodimos E. The challenge of drug resistance in cancer treatment: A current overview. Clin Exp Metastasis. 2018;35:309–318. doi: 10.1007/s10585-018-9903-0. [DOI] [PubMed] [Google Scholar]
- 55.Wang X, Zhang H, Chen X. Drug resistance and combating drug resistance in cancer. Cancer Drug Resist. 2019;2:141–160. doi: 10.20517/cdr.2019.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Biegala L, Gajek A, Marczak A, Rogalska A. Olaparib-Resistant BRCA2(MUT) Ovarian Cancer Cells with Restored BRCA2 Abrogate Olaparib-Induced DNA Damage and G2/M Arrest Controlled by the ATR/CHK1 Pathway for Survival. Cells. 2023;12:1038. doi: 10.3390/cells12071038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Biegala L, Gajek A, Szymczak-Pajor I, Marczak A, Sliwinska A, Rogalska A. Targeted inhibition of the ATR/CHK1 pathway overcomes resistance to olaparib and dysregulates DNA damage response protein expression in BRCA2(MUT) ovarian cancer cells. Sci Rep. 2023;13:22659. doi: 10.1038/s41598-023-50151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in the present study may be requested from the corresponding author.