Abstract
Transcription initiation of protein-encoding genes involves the assembly of RNA polymerase II and a number of general transcription factors at the promoter. A mammalian RNA polymerase II complex containing all of the components required for promoter-specific transcription initiation can be isolated by immunopurification with a monoclonal antibody directed against the cyclin-dependent kinase CDK7, a subunit of the general transcription factor TFIIH. In vitro transcription by this immunopurified RNA polymerase II complex is effectively stimulated by thyroid embryonic factor (TEF), a basic leucine zipper transcription factor. Thus, the RNA polymerase II complex must also contain components required for activated transcription that interact with the transactivation domain of TEF. This conjecture was verified by affinity selection experiments in which the TEF transcription activation domain was used as a bait. Indeed, an RNA polymerase II complex containing all of the accessory proteins required for transcription initiation can be enriched by its affinity to recombinant proteins containing the TEF transactivation domain. These results are compatible with a mechanism by which TEF can recruit an RNA polymerase II holoenzyme to the promoter in a single step.
In prokaryotes and eukaryotes, transcription initiation can be divided into three basic steps: assembly of a closed initiation complex at the promoter, isomerization of the closed complex to the open complex, and promoter clearance (4, 11, 19, 20, 43). In principle, transcriptional regulators can affect any of these steps. For example, the Escherichia coli protein CAP (catabolite activator protein) has been shown to facilitate the binding of RNA polymerase to the promoter, isomerization, and promoter escape (4, 10, 30, 41, 43). In eukaryotes, the transactivation domain of the herpes simplex virus protein VP16 has been shown to stimulate transcription initiation, perhaps by interacting with TFIIB (18, 37), TFIIH (60), and TFIID (29). Therefore, VP16 may have a role in promoter assembly. Yankulov et al. (66) have demonstrated that the VP16 transactivation domain may also stimulate elongation, possibly by increasing the processivity of RNA polymerase II. Other activators, like the human immunodeficiency virus TAT protein, may affect still other steps (26).
Careful order-of-addition experiments with purified components of the general transcription machinery have suggested a stepwise assembly of initiation complexes in vitro. According to this model, the TATA box (or another core promoter element) is first recognized by TBP, the TATA box-binding subunit of the TFIID complex. TFIIA and TFIIB then join promoter-bound TFIID. The resulting TFIID-TFIIA-TFIIB (DAB)–promoter complex subsequently recruits RNA polymerase II and TFIIF. Finally, TFIIE and TFIIH enter the initiation complex, and isomerization can occur (2, 3, 8, 44, 45, 51, 53). A somewhat different view of initiation complex assembly has emerged with the discovery of a large multisubunit RNA polymerase II complex in yeast cells; this complex is called the holoenzyme (for reviews, see references 22, 23, 32, and 68). Such yeast holoenzyme complexes have been reported, depending on the method of isolation and analysis, to contain RNA polymerase II; SRB proteins; TFIIF, TFIIB, and TFIIH (28, 31); Sin4P, Rgr1P, and Gal11P (34); and polypeptides of the SWI-SNF complex (65). RNA polymerase II holoenzyme complexes have recently also been isolated from mammalian cells (5, 7, 39, 47, 48, 54). In three cases, such complexes have been enriched by a single affinity purification step with an immobilized CDK7 antibody (47); the immobilized elongation factors, elongin A or TF-IIS (48); or an immobilized TFIIF antibody (7). In two of these cases (47, 48), all general transcription factors required for promoter-specific initiation could be recovered. Quantitative immunoblot experiments by Pan et al. (48) revealed nearly stoichiometric amounts of the largest RNA polymerase II subunit RPB1 and TFIIB, TFIID, TFIIE, TFIIF, and TFIIH in the affinity-purified holoenzyme complex. Since all of these polypeptides coeluted in gel filtration analyses, it is likely that they are part of a large complex with a molecular mass of about 2 × 106 Da. Recently, holoenzyme complexes capable of autonomous transcription initiation have also been described for RNA polymerase I (52, 55) and RNA polymerase III (62). Evidence for the association of RNA polymerase III with its two essential initiation factors, TFIIIB and TFIIIC, in the absence of DNA had already been presented more than 10 years ago by Wingender et al. (64).
The discovery of the RNA polymerase II holoenzyme has substantially modified our view of initiation complex assembly and the way sequence-specific transcription factors participate in this process. At least at some promoters, initiation complex assembly could occur in a single step, in a way similar to the binding of bacterial RNA polymerase holoenzyme to promoters. As a consequence, transactivators may stimulate transcription simply by assisting in the recruitment of RNA polymerase II holoenzyme (for reviews, see references 22, 23, 32, and 49). In fact, genetic studies by Ptashne and coworkers demonstrated that a fortuitous contact between a promoter bound factor and a component of the RNA polymerase II holoenzyme is sufficient to efficiently activate transcription (1, 13, 16). Thus, the DNA-binding/dimerization domain of Gal4, normally inactive in transcription activation, can efficiently stimulate transcription in yeast strains expressing Gal11P, a mutant form of the holoenzyme component Gal11. Moreover, the acidic activation domain of the viral protein VP16 interacts with the RNA polymerase II holoenzyme (24). Studies by Thompson and Young (59) suggest that the yeast holoenzyme may actually be the major form of RNA polymerase II capable of transcription initiation in vivo. The yeast cup1 gene seems to be an exception, since it can be efficiently transcribed with an RNA polymerase II enzyme lacking the CTD (40).
Experimental evidence allowing the discrimination between a sequential and a single-step assembly of the RNA polymerase II initiation complex in the living cell is difficult to obtain. We therefore resorted to in vitro transcription studies to shed more light on the initiation complex assembly. In particular, we wanted to examine whether the biochemical properties of the RNA polymerase II transcription machinery is compatible with a single-step assembly mechanism. Here we show that the basic leucine zipper protein thyroid embryonic factor (TEF), a potent transactivator both in vivo (15) and in vitro (the present study), also efficiently stimulates in vitro transcription by an immunopurified RNA polymerase II holoenzyme. Moreover, the activation domain of this transcription factor binds to an RNA polymerase II complex containing all general transcription factors required for promoter-specific transcription initiation. These findings are compatible with a mechanism by which promoter-bound transcriptional activators recruit the RNA polymerase II transcription machinery in a single step, as in models proposed for prokaryotic organisms.
MATERIALS AND METHODS
Nuclear extract preparation.
Nuclear-transcription-competent extracts were prepared as described by Ossipow et al. (47). Briefly, highly purified rat liver nuclei were isolated as described by Lichtsteiner et al. (36). The nuclear pellet was resuspended at an optical density at 260 nm of 1 per ml in nuclear lysis buffer containing 10 mM HEPES (pH 7.6), 100 mM KCl, 0.15 mM spermine, 0.5 mM spermidine, 0.1 mM NaF, 0.1 mM Na3VO4, 0.1 mM EDTA, 1 mM dithiothreitol (DTT), 0.1 mM phenylmethylsulfonyl fluoride, and 10% glycerol. KCl was added to a final concentration of 0.3 M to extract soluble nonhistone proteins. After 20 min on ice, the nuclear lysate was spun at 36,000 rpm in a fixed-angle Ti60 rotor for 60 min at 0°C to sediment the insoluble chromatin components. Soluble proteins were precipitated by the addition of 0.3 g of solid (NH4)2SO4 per ml of supernatant. After 1 h on ice, the ammonium sulfate precipitate was recovered by sedimentation at 36,000 rpm in a fixed-angle Ti60 rotor for 30 min at 0°C. The precipitated proteins were resuspended in dialysis buffer (25 mM HEPES (pH 7.6), 100 mM KCl, 0.1 mM NaF, 0.1 mM Na3VO4, 0.1 mM EDTA, 0.25 mM DTT, and 10% glycerol) in 5% of the original nuclear lysate volume and dialyzed twice for 2 h against 250 volumes of the same buffer. After a 2-min centrifugation in an Eppendorf microfuge, the supernatant was divided into 100-μl aliquots, snap frozen, and kept under liquid nitrogen until use. The protein concentration was determined as described in Gorski et al. (21). Typically, 1 g of rat liver (wet weight) yields 100 μl of nuclear extract containing 8 to 12 mg of protein/ml.
RNA polymerase II holoenzyme immunopurification.
RNA polymerase II holoenzyme was immunopurified as described in Ossipow et al. (47). Briefly, per assay, 0.5 μl of ascites fluid containing a monoclonal antibody directed against human CDK7/MO15 (MO-1.1 [56]) or an irrelevant control antibody (47) was incubated with 15 μl of a suspension containing magnetic beads coated with goat immunoglobulin G (IgG) anti-mouse IgG (Dynal M450) for 4 h at 4°C. The beads were extensively washed in phosphate-buffered saline containing 0.1% Triton X-100 and incubated with 50 μl (250 to 500 μg of protein) of undiluted crude nuclear extracts for 1 h at 4°C with gentle shaking. The beads were washed three times with 400 μl of washing buffer (25 mM HEPES, pH 7.6; 50 mM KCl, 0.1 mM EDTA; 0.1 mM Na3VO4; 0.1 mM NaF; 10% glycerol; 0.1% Triton X-100).
To examine the efficacy of this purification procedure, the proteins recovered from 400 μg of liver nuclear proteins with paramagnetic beads decorated with either CDK7 or NANP control antibodies were size fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) alongside increasing amounts of input proteins (400 ng to 80 μg). Coomassie staining of these gel-separated proteins revealed that approximately 300 ng of the proteins adsorbed nonspecifically to control beads and that a somewhat higher amount adsorbed to the beads decorated with CDK7 antibodies (data not shown, but see reference 47). Since about 10 to 20% of RNA polymerase II (RPB1) and SRB7 proteins were recovered after immunoenrichment, the purification factor for the RNA polymerase II holoenzyme complex can be estimated to be approximately 100- to 200-fold.
Preparation of magnetic beads with DNA-bound TEF proteins.
First, 400 pmol of the biotinylated top strand of a 25-mer oligonucleotide encompassing the high-affinity site for TEF (and other PAR leucine zipper proteins [12]) was annealed to 400 pmol of its complementary bottom strand. The sequence of the top strand is 5′-GTTCTTGGTTACGTAATCTCCAATGGTTCTT-3′ (the TEF binding site underlined). The double-stranded oligonucleotide was bound to 200 μl (2 mg) of streptavidin-coated magnetic beads (Dynal M280) according to the manufacturer’s specifications. The decorated beads were incubated for 45 min at 4°C with 100 μg of either full-length TEF or TEF lacking the activation domain in a buffer containing 10 mM Tris (pH 7.4), 0.1 mM EDTA, 200 mM NaCl, 10 mM DTT, and 0.1% Triton X-100. The beads were washed and stored in the same buffer.
Preparation of magnetic beads with covalently coupled GST fusion proteins.
The fusion proteins glutathione S-transferase (GST)-DBP (amino acids 127 to 208) and GST-TEF (amino acids 14 to 159) were overexpressed in E. coli (strain BL21), purified, and eluted from the glutathione beads according to the manufacturer’s specifications (Pharmacia). The purified proteins were precipitated by the addition of 0.3 g of solid (NH4)2SO4 per ml and then resuspended in a buffer containing 100 mM potassium phosphate (pH 7.8) and 10 mM DTT. They were then loaded onto a 10-ml G50 size exclusion column, and the protein-containing fractions were pooled. The proteins were concentrated by centrifugation in a Centricon 30 tube to a final concentration of 8 mg/ml. Then, 1 mg of each fusion protein was covalently coupled to 166 μl (5 mg) of MPG glyceryl-porous magnetic beads (MGLY0502; CPG Inc., Lincoln Park, N.J.) in a buffer containing 100 mM potassium phosphate (pH 7.8) according to the manufacturer’s indications. The protein concentration on the beads, which was determined by including a trace of radiolabeled fusion protein in the coupling reaction, was estimated to be 120 μg/mg of beads.
Affinity enrichment of RNA polymerase II holoenzyme by adsorption to the TEF activation domain.
For each reaction, 60 μg of covalently coupled GST fusion proteins or 200 ng of full-length and truncated TEF bound to DNA-coated magnetic beads were used. The beads were incubated with 50 μl (ca. 500 μg of protein) of crude nuclear proteins for 20 min at 4°C with gentle shaking. They were then gently washed three times with 200 μl of washing buffer (25 mM HEPES, pH 7.6; 50 mM KCl; 0.1 mM EDTA; 0.1 mM Na3VO4; 0.1 mM NaF; 10% glycerol; 0.1% Triton X-100) and directly used for in vitro transcription.
The enrichment factor of the single-step affinity purification with covalently coupled GST fusion proteins was estimated to be approximately 40 to 80 by the procedure described above for the immunopurification procedure. The affinity between the TEF activation domain and the RNA polymerase II complex is several orders of magnitude lower (KD in the micromolar range [see Results]) than that expected for the interaction between the CDK7 antibody and its epitope (for the nanomolar or subnanomolar range). Therefore, a substantial amount of immobilized GST-TEF bait protein is required to retain a significant fraction of the polymerase complex. This may result in a somewhat higher contamination due to nonspecific adsorption compared to the immunopurification procedure.
In vitro transcription.
In vitro transcription reactions were performed as described by Gorski et al. (21) with either crude nuclear extract (10-μl assays) or immunopurified and affinity-purified RNA polymerase II holoenzymes immobilized on washed beads (10- to 20-μl assays). The reactions typically contained 1 μg of template DNA/10 μl and were incubated for 45 min at 37°C with gentle shaking.
Size fractionation of nuclear proteins by gel filtration.
First, 7 mg (700 μl) of nuclear extract was incubated for 20 min at 4°C with 5 mM MgCl2, 10 μM distamycin, 20 μg of ethidium bromide per ml, and 100 μg of RNase A per ml. The extract was spun at 4°C for 15 min at 15,000 rpm to remove the insoluble material, and the supernatant was loaded on a 25-ml Sepharose CL2B column. The chromatography was performed at 4°C, with a flow rate of 500 μl/min, in a buffer containing 25 mM HEPES (pH 7.6), 100 mM KCl, 5 mM MgCl2, 0.1 mM NaF, 0.1 mM Na3VO4, 0.1 mM EDTA, 0.25 mM DTT, and 10% glycerol, and 500-μl fractions were collected. For RPB1, TFIIB, TFIIE, and TBP, 20 μl of crude fractions were used for Western blot analysis. For TFIIE, TFIIH, CDK7, and SRB7, 100 μl of the fractions were trichloroacetic acid precipitated before Western blot analysis. Then 15 μl of the crude fractions was directly used in an in vitro transcription reaction, or 100 μl was concentrated sevenfold by centrifugation in a Centricon 30 tube before immunopurification with the CDK7 antibody and in vitro transcription. Western blotting was performed as described in Ossipow et al. (46, 47).
RESULTS
Recombinant TEF activates in vitro transcription by immunopurified RNA polymerase II holoenzyme.
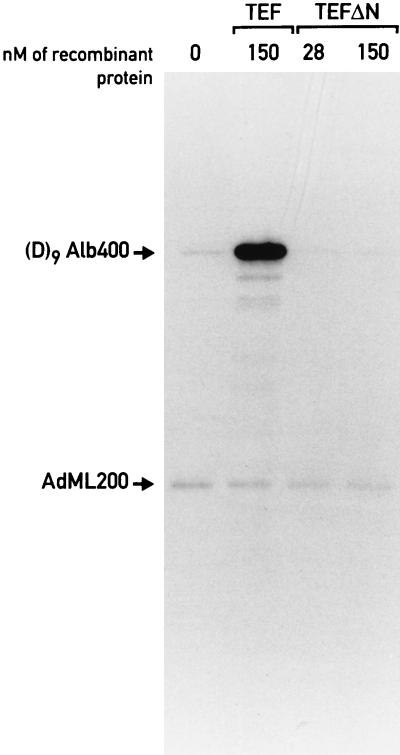
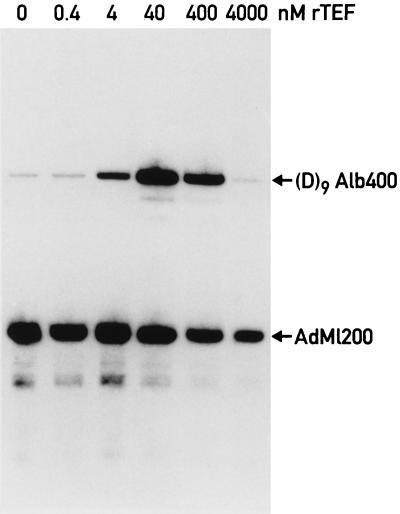
We have previously reported the isolation of an RNA polymerase II holoenzyme from rat liver nuclei by using coimmunopurification with a monoclonal antibody directed against CDK7 (MO15), the CTD kinase subunit of TFIIH (47). Our first attempts to activate transcription by this form of polymerase II with bacterially expressed C/EBPβ failed (47). However, even in unfractionated liver nuclear extracts, this transactivator stimulates transcription only moderately. We therefore searched for a more potent transcription factor and found that the leucine zipper transcription factor TEF (9) stimulates in vitro transcription very efficiently in liver nuclear extracts. In the experiment shown in Fig. 1, a promoter bearing nine TEF binding sites (D9Alb400 [38]) is strongly activated upon addition of E. coli-derived recombinant TEF. As expected, a control promoter lacking such binding sites, the adenovirus major late promoter fused to a 200-bp G-free cassette (AdML200), remains unaffected by TEF (Fig. 1, lane TEF). This activation is dependent on the TEF activation domain, since a truncated version containing only the DNA-binding domain does not stimulate transcription (Fig. 1, lane TEFΔN). Recombinant TEF also activates in vitro transcription efficiently from the albumin promoter, which contains only one TEF recognition sequence (data not shown).
FIG. 1.
Recombinant TEF is a potent transactivator of in vitro transcription in liver nuclear extracts. Two G-free cassette templates containing either the adenovirus major late promoter (AdML200, −400 to +10 [38]) or a synthetic promoter containing nine binding sites for TEF in front of the albumin core promoter (D9Alb400 [38]) were incubated with 46 μg of nuclear extract for in vitro transcription. The final concentrations of wild-type (lanes TEF) or amino-terminally truncated TEF (lanes TEFΔN) in the reactions are indicated above each lane.
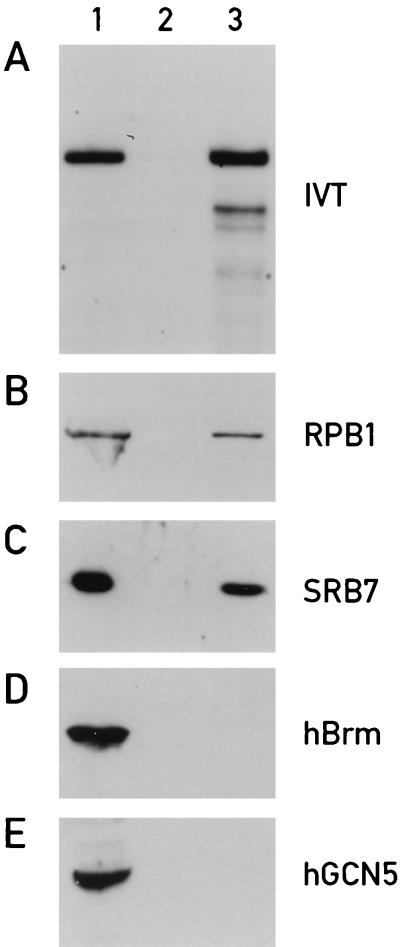
We wished to examine whether TEF can also activate transcription by an RNA polymerase II holoenzyme immunopurified from rat liver nuclei. We first assessed the quality of our holoenzyme preparation by transcription assays with the adenovirus major late promoter. Both the crude nuclear extract (Fig. 2A, lane 1) and the holoenzyme immunopurified with the CDK7 antibody (Fig. 2A, lane 3) drive efficient transcription from this viral promoter, since it contains all of the essential accessory proteins required for promoter-specific initiation (47). As expected, no transcription was detected with the material adsorbed to an irrelevant control antibody (Fig. 2A, lane 2). Immunoblots show that SRB7, a hallmark of the RNA polymerase II holoenzyme (5, 24, 39, 65), is present only in the crude nuclear extract (Fig. 2C, lane 1) and in the immunopurified holoenzyme (Fig. 2C, lane 3) but not in the control immunoprecipitation (Fig. 2C, lane 2). The same holds true for RPB1, the largest subunit of RNA polymerase II (Fig. 2B) and all of the general transcription factors (TFIIB, TFIID, TFIIE, TFIIF, and TFIIH) required for specific initiation (47). Based on the recoveries of RNA polymerase II (RPB1), SRB7, and total protein in the immunopurified preparation, we estimate that the RNA polymerase II holoenzyme complex was enriched about 100- to 200-fold during the affinity purification step (see Materials and Methods).
FIG. 2.
Analysis of immunopurified RNA polymerase II holoenzyme. (A) In vitro transcription of a template containing the adenovirus major late promoter (AdML200, −400 to +10) with 50 μg of liver nuclear extract (NE) (lane 1), proteins immunoenriched with an irrelevant antibody from 250 μg of NE (see reference 47) (lane 2), and proteins immunoenriched with CDK7 antibody from 250 μg of NE (lane 3). (B to E) Western blot analysis of immunopurified RNA polymerase II holoenzyme with antibodies against different nuclear proteins. In each experiment, 50 μg of liver nuclear proteins (lanes 1) were size fractionated by SDS-PAGE alongside proteins immunoenriched from 250 μg of liver nuclear extract with either the CDK7 antibody (lanes 3) or an antibody against an irrelevant peptide (lanes 2). The epitopes for the various antibodies used in these immunoblot experiments are indicated at the righthand side of the panels.
In contrast to RNA polymerase II and SRB7, two other nuclear proteins, Brm and GCN5, are clearly absent in the immunopurified complex (Fig. 2D and E). Brm, a component of the mammalian SWI-SNF complex (50), and GCN5, a histone acetyltransferase (61), are both thought to be involved in the modulation of chromatin structure.
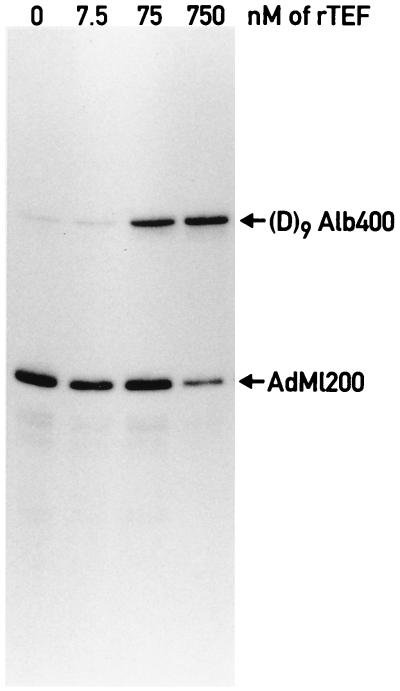
As shown in Fig. 3, the addition of bacterially expressed TEF to this immunopurified RNA polymerase II holoenzyme results in the specific stimulation of the target promoter (D9Alb400). In keeping with the results obtained with unfractionated liver nuclear extracts (Fig. 1), TEF does not affect in vitro transcription from the adenovirus major late promoter (AdML200), which is devoid of TEF recognition sequences. This demonstrates that the immunopurified holoenzyme is competent for promoter-specific activated transcription.
FIG. 3.
TEF activates transcription by immunopurified RNA polymerase II holoenzyme. Two G-free cassette templates containing either the adenovirus major late promoter (AdML200, −400 to +10) or a synthetic promoter composed of nine binding sites for TEF in front of the albumin core promoter (D9Alb400) were incubated with RNA polymerase II holoenzyme immunopurified from 500 μg of nuclear extract. The final concentration of TEF in the reactions is indicated above each lane.
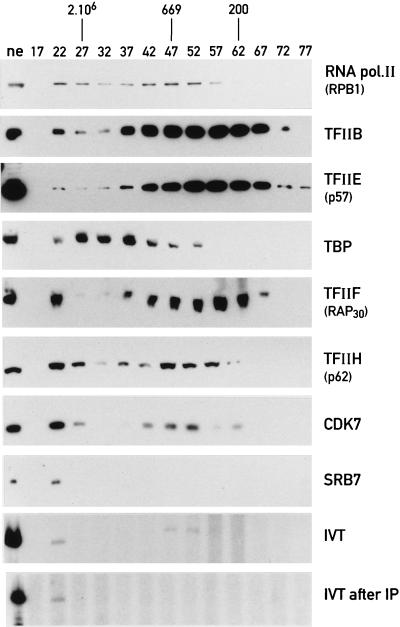
It could be argued that the CDK7 antibody immunoselects subcomplexes of the RNA polymerase II apparatus individually and that these components only assemble into a large complex during the in vitro transcription reaction. We thus wanted to test whether an RNA polymerase II complex can also be immunopurified with CDK7 antibody after size fractionation. Liver nuclear proteins were resolved on a CL2B column, after they had been treated with distamycin, ethidium bromide, and RNase A to prevent interactions of proteins with nucleic acids (6, 33). Western blot analysis of the fractions (Fig. 4) with an antibody directed against the C-terminal domain (CTD) of the largest RNA polymerase II subunit shows that the RNA polymerase II distributes in two broad peaks, one above 2 × 106 Da and one centered around 700 kDa. These presumably correspond to the RNA polymerase II holoenzyme and the core RNA polymerase II, respectively (48, 68). While a fraction of all general transcription factors comigrated with the large complex, TFIIB and the p57 subunit of TFIIE are much more abundant in fractions corresponding to lower molecular masses. Conceivably, these two factors are either present in molar excess over the other components of the RNA polymerase II holoenzyme or they dissociate from the holoenzyme during the lengthy fractionation. Consistent with the relatively tight association of TFIIF with the RNA polymerase II holoenzyme, the RAP30 subunit of TFIIF is present in significant amount in the >2 × 106-Da holoenzyme fraction. The p62 subunit of TFIIH clearly partitions into two peaks: one corresponding to the >2 × 106-Da complex and the other corresponding to ca. 700-kDa complex, probably reflecting the holo-TFIIH complex (for a review, see reference 17; see also reference 25). CDK7 distributes into three distinct peaks. The CTD kinase found in the >2 × 106-Da peak probably reflects its association with the RNA polymerase II holoenzyme, while the second peak, slightly below 700 kDa, represents holo-TFIIH, as seen for the p62 TFIIH subunit. The third and lowest-molecular-size peak corresponds to an apparent mass of roughly 200 kDa and presumably reflects the CDK7-cyclin H-MAT1 complex (14, 57). Importantly, SRB7 is found exclusively in the >2 × 106-Da holoenzyme fraction, a result in agreement with the finding that all of the SRB polypeptides of the cell are stably associated with RNA polymerase II holoenzyme (24, 31, 35, 58).
FIG. 4.
Analysis of size-fractionated liver nuclear proteins. Nuclear proteins (7 mg) were treated with distamycin, ethidium bromide, and RNase A and then size fractionated on a Sepharose CL-2B column. Every fifth fraction (lanes 17 to 77) or the unfractionated nuclear extract (lane ne) was analyzed by Western blotting with the antibodies against the polypeptides indicated at the right hand side of the figure. In vitro transcription from the adenovirus major late promoter (template AdML200, −400 to +10) with crude CL2B fractions (IVT) and after immunopurification in the CL2B fractions with the CDK7 antibody (IP).
TBP elutes in a broad peak corresponding to molecular sizes between 5 × 105 and more than 2 × 106 Da. This protein is also an important constituent of initiation factors and holoenzyme complexes for RNA polymerase I (55) and RNA polymerase III (62). Therefore, the coelution of some TBPs with the >2 × 106-Da RNA polymerase II complex cannot serve as evidence for the association of TFIID with the large RNA polymerase II complex. Nevertheless, immunopurification of this large complex with a CDK7 antibody results in the recovery of an entity competent for initiation (see below). Thus, at least a fraction of this complex must contain TBP.
In vitro transcription reactions with proteins from the various gel filtration fractions and a template containing the adenovirus major late promoter shows two peaks of activity. The major one cofractionated with the >2 × 106-Da holoenzyme complex, while the second matched the 700-kDa fractions containing RNA polymerase II and a fraction of each essential initiation factor (Fig. 4, IVT). Remarkably, after immunopurification of proteins from each fraction with the CDK7 antibody, a single peak of transcriptional activity could be detected (Fig. 4). This activity cofractionates with the >2 × 106-Da RNA polymerase II holoenzyme complex. Importantly, no detectable transcriptional activity could be immunoenriched from the fractions of around 700 kDa, despite the presence of CDK7, RNA polymerase II, and all essential general transcription factors in this fraction. These observations suggest that the affinity enrichment with the monoclonal CDK7 antibody from unfractionated nuclear extracts purifies a large >2 × 106-Da RNA polymerase II complex rather than a subset of general transcription factor complexes, each containing CDK7 kinase. Conceivably, components only present in the 2 × 106-Da complex, such as SRB proteins, are indispensable for the integrity of the RNA polymerase II holoenzyme complex.
TEF squelches transcription at high concentrations.
We observed that the addition of increasing amounts of recombinant TEF to a crude nuclear extract progressively stimulates in vitro transcription from a target promoter until it reaches a concentration of 40 nM (Fig. 5). At a TEF concentration of 400 nM, the relative activation factor is already lower than that observed at a 10-fold-lower concentration, and at 4 μM no significant stimulation of transcription can be seen (Fig. 5). The inhibitory effect of TEF at high concentrations is likely to reflect squelching, that is, competition between free and promoter-bound activator for target surfaces on the RNA polymerase II machinery (reviewed in reference 49). Since half-maximal inhibition of activated transcription is observed at TEF concentrations between 400 nM and 4 μM, the affinity (KD) of the interaction between TEF and components of the RNA polymerase II machinery can be estimated to be in the low micromolar range. Remarkably, TEF squelches transcription from the D9 target promoter much more dramatically than from the adenovirus major late promoter, indicating that the association of TEF with the transcription machinery in solution either does not attenuate or only weakly attenuates transcription initiation as such. This result also suggests that the surfaces of general transcription factors interacting with TEF are not required for transcription from the adenovirus major late promoter.
FIG. 5.
TEF squelches transcription at high concentrations. Two G-free cassette templates containing either the adenovirus major late promoter (AdML200, −400 to +10) or a synthetic promoter composed of nine binding sites for TEF in front of the albumin core promoter (D9Alb400) were incubated for in vitro transcription with 50 μg of nuclear extract. The final concentration of recombinant TEF present in the assay is indicated above each lane.
The TEF activation domain binds to the RNA polymerase II holoenzyme.
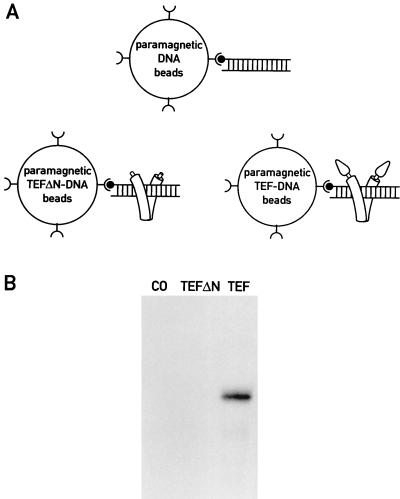
The specific squelching described in the previous section suggested that TEF interacts specifically with one or more components of the RNA polymerase II machinery in solution. Encouraged by this observation, we wanted to examine whether the activation domain of TEF can bind to an RNA polymerase II holoenzyme in the absence of promoter DNA. Towards this aim, we first immobilized biotinylated double-stranded oligonucleotides encompassing a high-affinity site for TEF (12) to streptavidin-coated magnetic beads. Full-length recombinant TEF, or an amino-terminally truncated version shown to be incompetent for transcription (Fig. 1), was then bound to the immobilized DNA. In control experiments, we have shown that these protein-DNA complexes have a half-life of much more than 1 h at 4°C (data not shown), indicating that TEF remains associated during the much shorter affinity purification procedure described below. Three sets of magnetic beads were then incubated with a transcription-competent liver nuclear transcription extract: (i) beads with the oligonucleotide alone, (ii) beads with the amino-terminally truncated TEF tethered to the oligonucleotide, and (iii) beads with full-length TEF bound to the oligonucleotide (Fig. 6A). Proteins with affinities to the beads were purified by magnetic attraction. After extensive washes, the affinity-enriched proteins were incubated with nucleoside triphosphates and a plasmid carrying the adenovirus major late promoter fused to a G-free cassette. No transcriptional activity could be recovered from beads decorated with the oligonucleotide alone or from beads with the oligonucleotide and the amino-terminally truncated version of TEF (Fig. 6B, lanes CO and TEFΔN). In contrast, incubation of nuclear proteins with beads containing the TEF binding site and full-length TEF resulted in the enrichment of an activity capable of accurate transcription initiation on the adenovirus major late promoter (Fig. 6B, lane TEF). This experiment suggests that the activation domain of TEF recruits either an RNA polymerase II holoenzyme or all individual components required for promoter-specific transcription-initiation to the beads. Although we cannot formally exclude the later possibility, the former appears more likely, in view of the evidence presented above for a large transcription-competent RNA polymerase II complex.
FIG. 6.
TEF interacts with an RNA polymerase II complex containing all components required for promoter-specific initiation. (A) Schematic representation of the different paramagnetic streptavidin beads used in the experiments shown in panel B. Double-stranded oligonucleotides encompassing a high-affinity TEF binding site (sequence given in Materials and Methods) are depicted as ladders. Biotin groups and streptavidin molecules are represented by filled circles and rounded Y-like symbols, respectively. The C-terminal DNA-binding/dimerization domains of TEF are shown as bent cylinders embracing the DNA binding sites, and the N-terminal TEF activation domains are represented as cones (see also Fig. 7). (B) Affinity selection of an RNA polymerase II complex with recombinant TEF bound to its DNA recognition site. The paramagnetic beads used in these experiments are schematically depicted in panel A. The beads were incubated with 250 μg of nuclear extract and, after being washed and after magnetic attraction, they were incubated for in vitro transcription with a G-free cassette template containing the adenovirus major late promoter (AdML200, −400 to +10 [38]). The following beads were used (see panel A): lane CO, control beads with DNA alone; lane TEFΔN, TEFΔN-DNA beads; lane TEF, TEF-DNA beads.
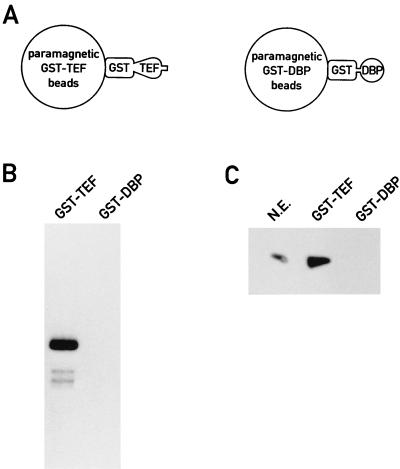
We had to examine the possibility that the double-stranded oligonucleotide encompassing the TEF binding site could have participated in the recruitment of transcriptional activity. To this end, we chose an alternative affinity enrichment approach. GST fusion proteins harboring either the activation domain of TEF (amino acids 14 to 159) or a similarly charged peptide domain without activation potential (amino acids 127 to 208 of DBP [46a]) were covalently linked to chemically activated magnetic beads (Fig. 7A). After incubation with a transcription-competent liver nuclear extract and an extensive washing, the beads were supplemented with nucleoside triphosphates and the adenovirus template. As shown in Fig. 7B, lane GST-TEF, the beads containing GST-TEF fusion protein retained an activity capable of accurate transcription initiation. In contrast, no in vitro transcripts could be observed with the magnetic beads bearing the immobilized DBP-derived polypeptide (Fig. 7B, lane GST-DBP). This experiment suggests that the TEF activation domain is able to recruit all components required for promoter-specific transcription initiation to the magnetic beads and that DNA binding or the presence of DNA is not necessary for this process.
FIG. 7.
The activation domain of TEF interacts in solution with an RNA polymerase II holoenzyme. (A) Schematic representation of the paramagnetic beads with covalently bound GST-TEF14–159 or GST-DBP127–208 fusion proteins that were used in the experiments shown in panels B and C. (B) In vitro transcription with affinity-enriched nuclear proteins. The beads schematically represented in panel A were incubated with 500 μg of liver nuclear proteins and washed. The recovered proteins were incubated for in vitro transcription with a G-free cassette template containing the adenovirus major late promoter. (C) Affinity-enriched proteins were analyzed by immunoblotting with an SRB7 antibody. Lanes: N.E., 50 μg of input nuclear extract proteins; GST-TEF, proteins recovered from 500 μg of nuclear proteins with GST-TEF magnetic beads (see panel A); GST-DBP, proteins recovered from 500 μg of nuclear proteins with GST-DBP magnetic beads (see panel A).
The affinity of the TEF activation domain with components of the RNA polymerase II holoenzyme is considerably lower than that between CDK7 and the monoclonal CDK7 antibody. It was thus not possible to affinity purify sufficient amounts of transcriptional activity from gel filtration fractions with the GST-TEF fusion protein. In order to examine whether TEF interacts with the RNA polymerase II holoenzyme, we took advantage of the observation that SRB7 is found associated exclusively with the high-molecular-weight RNA polymerase II holoenzyme complex (Fig. 4). The material enriched from nuclear extracts with magnetic beads containing either GST-TEF or GST-DBP was examined by Western blot analysis for the presence of SRB7. As shown in Fig. 7C, SRB7 was detected only in the unfractionated nuclear extract (lane N.E.) and in the material affinity enriched with the TEF activation domain (lane GST-TEF). The purification factor reached in the affinity purification with the TEF-GST fusion protein is somewhat lower than that obtained in the immunopurification with the CDK7 antibody (see above and Materials and Methods). Nevertheless, a considerable enrichment of the holoenzyme complex has been achieved. Based on the immunoblot experiment shown in Fig. 7C, we estimate that at least 10 to 20% of the SRB7 present in the input was recovered in the affinity-purified material. SDS-PAGE analysis of this preparation suggests that it contains about 0.25% of the input proteins (data not shown). Thus, the single-step affinity purification with the GST-TEF fusion protein resulted in an about 40- to 80-fold enrichment of the RNA polymerase II holoenzyme complex.
DISCUSSION
The results reported here demonstrate that the transcription factor TEF stimulates transcription in a simple reaction consisting of E. coli-derived recombinant TEF, a target promoter containing multiple TEF recognition sequences, and an immunopurified RNA polymerase II holoenzyme. The direct interaction of TEF with components of the RNA polymerase II machinery is supported by two additional findings. First, high concentrations of TEF result in specific squelching of target gene transcription in nuclear extracts (Fig. 5). This suggests that excess TEF that is not bound to promoter sites saturates its target surfaces on general transcription factors, thereby competing with promoter-bound TEF for the interaction with such components. Second, the activation domain of TEF, when immobilized on paramagnetic beads, can be used to affinity enrich all components of the RNA polymerase II transcription machinery required for promoter-specific initiation. Since SRB7, a hallmark of the large RNA polymerase II holoenzyme complex, is also among the peptides affinity enriched with TEF, it is likely that the general transcription factors are recruited to the paramagnetic beads in form of an RNA polymerase II holoenzyme. The TEF activation domain binds to this complex irrespective of whether it is tethered to the beads via binding of full-length TEF to immobilized DNA recognition sequences or via a TEF-GST fusion protein. Therefore, DNA binding of TEF is not required for its interactions with RNA polymerase II components.
The direct binding of an initiation-competent RNA polymerase II holoenzyme to the activation domain of TEF suggests a simple mechanism for transcription activation: TEF may recruit the complete transcription apparatus to the promoter in a single step. Obviously, the feasibility of such a mechanism depends on the presence of all essential initiation factors in the RNA polymerase II holoenzyme. A particularly important question is whether TFIID, the major promoter-binding component of the RNA polymerase II machinery, is associated with a fraction of the holoenzyme (see below). All authors who applied fast single-step affinity purifications have recovered some TFIID in their preparations. Thus, the presence of TBP could be demonstrated in RNA polymerase II holoenzymes immunopurified with antibodies against CDK7 kinase (47; this study), TFIIF (7), TFIIE (46a), and SRB5 (69). Likewise, TFIID was found to be associated with holoenzymes enriched by affinity chromatography on resins containing the elongation factors TFIIS or elongin A (48). In their report, Pan et al. (48) present careful stoichiometry measurements and find that about half of the affinity-enriched RNA polymerase II complexes contain TBP. It appears likely, therefore, that the failure of some authors to recover TFIID in their holoenzyme preparations is due to the lengthy and harsh purification procedures they used. In spite of the demonstration of a holoenzyme complex containing all essential initiation factors, one would expect that a fraction of RNA polymerase complexes are devoid of TFIID. According to the model proposed by Pan et al. (48), the first initiation event may be accomplished by an RNA polymerase II holoenzyme complex containing all essential initiation factors. After promoter escape has occurred, promoter-bound TFIID may serve as a landing pad for further initiations by RNA polymerases not associated with TFIID that have terminated transcription on the same (or other nearby) gene(s). In fact, many genes are transcribed in bursts of multiple rounds (42, 63). In these cases, TFIID may remain bound to the promoter during a time period, allowing repeated initiation events to occur.
We have noticed that TEF stimulates in vitro transcription from target templates with a higher amplitude with unfractionated nuclear extracts (up to 30-fold) compared to immunopurified holoenzyme (about 4-fold). Conceivably, some proteins, such as coactivators that contribute to the overall activation by TEF, are depleted during the immunoenrichment of the RNA polymerase II holoenzyme. Alternatively, repressors of basal transcription, such as NC1 or NC2/Dr1 (27, 67), may be present in nuclear extracts but absent from purified holoenzyme. The absence of such global repressors would result in a higher basal in vitro transcription with purified RNA polymerase II holoenzyme and therefore in a lower amplitude of activated transcription.
The identification of RNA polymerase II holoenzyme polypeptides that interact with the activation domain of TEF will be a major challenge. We estimate that the KD for the interaction of the TEF activation domain with the RNA polymerase II transcription machinery must be in the low micromolar range (see Results section). A KD of 10−6 M corresponds to a free energy of about −8 kcal/mol. If this binding energy were distributed over multiple TEF contacts with different components of the RNA polymerase II holoenzyme, the interaction of TEF with individual binding partners would be too weak for conventional protein-protein interaction studies. Indeed, attempts to identify TEF-interacting polypeptides of the holoenzyme by far-Western blotting techniques have failed thus far (data not shown).
The finding of holoenzyme complexes for all three forms of eukaryotic RNA polymerases (for references, see the Introduction) indicates that transcription initiation and its stimulation by sequence-specific regulatory proteins are conceptually more similar in prokaryotes and eukaryotes than hitherto assumed. Like bacterial RNA polymerase holoenzymes, all three eukaryotic RNA polymerases may occupy at least some promoters in a single step and dissociate from promoter-specific accessory factors upon promoter escape. During or after termination, the RNA polymerase core enzyme may be reassembled into an initiation competent holoenzyme, in a way similar to the transcription cycles established for bacterial RNA polymerases and sigma factors. One obvious difference between prokaryotic and eukaryotic transcription still persists, however. Most sigma factors cannot bind promoter DNA autonomously and thus have to assemble with core RNA polymerase for each initiation event. In the case of eukaryotic transcription, only the first initiation event on a given gene may require an RNA polymerase II holoenzyme containing all initiation factors. The rapidly following subsequent rounds may then be performed with incomplete RNA polymerase II complexes on already-promoter-bound initiation factors (48). The examination of such mechanisms will require novel technologies that allow multiple rounds of in vitro transcription by RNA polymerase II on the same DNA template.
ACKNOWLEDGMENTS
We are grateful to Rick Young for the antiserum against human SRB7, Erich Nigg for the CDK7 monoclonal antibody, Moshe Yaniv for hBrm antibodies, and Shelley Berger for hGCN5 antibodies. We thank Nicolas Roggli for expert preparation of the figures, and Steve Brown, Juergen Ripperger, David Gdula, Philippe Georgel, and Raphael Sandaltzopoulos for their critical reading of the manuscript.
This work was supported by the Canton of Geneva, the Swiss National Science Foundation, and a postdoctoral fellowship to V.O. from the Roche Research Foundation, Basel, Switzerland.
REFERENCES
- 1.Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 2.Buratowski S, Hahn S, Guarente L, Sharp P A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 3.Burke T W, Kadonaga J T. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- 4.Busby S, Ebright S H. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell. 1994;79:743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 5.Chao D L, Gadbois E L, Murray P J, Anderson S F, Sonu M S, Parvin J D, Young R A. A mammalian SRB protein associated with an RNA polymerase II holoenzyme. Nature. 1996;380:82–85. doi: 10.1038/380082a0. [DOI] [PubMed] [Google Scholar]
- 6.Chiang S Y, Welch J, Rauscher F J, Beerman T A. Effects of minor groove binding drugs on the interaction of TATA box binding protein and TFIIA with DNA. Biochemistry. 1994;33:7033–7040. doi: 10.1021/bi00189a003. [DOI] [PubMed] [Google Scholar]
- 7.Cho H, Maldonado E, Reinberg D. Affinity purification of a human RNA polymerase II complex using monoclonal antibodies against transcription factor IIF. J Biol Chem. 1997;272:11495–11502. doi: 10.1074/jbc.272.17.11495. [DOI] [PubMed] [Google Scholar]
- 8.Corden J L. RNA polymerase II transcription cycles. Curr Opin Genet Dev. 1993;3:213–218. doi: 10.1016/0959-437x(93)90025-k. [DOI] [PubMed] [Google Scholar]
- 9.Drolet D W, Scully K M, Simmons D M, Wegner M, Chu K T, Swanson L W, Rosenfeld M G. TEF, a transcription factor expressed specifically in the anterior pituitary during embryogenesis, defines a new class of leucine zipper proteins. Genes Dev. 1991;5:1739–1753. doi: 10.1101/gad.5.10.1739. [DOI] [PubMed] [Google Scholar]
- 10.Ebright R H, Busby S. The Escherichia coli RNA polymerase alpha subunit: structure and function. Curr Opin Genet Dev. 1995;5:197–203. doi: 10.1016/0959-437x(95)80008-5. [DOI] [PubMed] [Google Scholar]
- 11.Eick D, Wedel A, Heumann H. From initiation to elongation: comparison of transcription by prokaryotic and eukaryotic RNA polymerases. Trends Genet. 1994;10:292–296. doi: 10.1016/0168-9525(90)90013-v. [DOI] [PubMed] [Google Scholar]
- 12.Falvey E, Marcacci L, Schibler U. DNA-binding specificity of PAR and C/EBP leucine zipper proteins: a single amino acid substitution in the C/EBP DNA-binding domain confers PAR-like specificity to C/EBP. Biol Chem. 1996;377:797–809. [PubMed] [Google Scholar]
- 13.Farrell S, Simkovich N, Wu Y, Barberis A, Ptashne M. Gene activation by recruitment of the RNA polymerase II holoenzyme. Genes Dev. 1996;10:2359–2367. doi: 10.1101/gad.10.18.2359. [DOI] [PubMed] [Google Scholar]
- 14.Fisher R P, Jin P, Chamberlin H M, Morgan D O. Alternative mechanisms of CAK assembly require an assembly factor or an activating kinase. Cell. 1995;83:47–57. doi: 10.1016/0092-8674(95)90233-3. [DOI] [PubMed] [Google Scholar]
- 15.Fonjallaz P, Ossipow V, Wanner G, Schibler U. The two PAR leucine zipper proteins, TEF and DBP, display similar circadian and tissue-specific expression, but have different target promoter preferences. EMBO J. 1996;15:351–362. [PMC free article] [PubMed] [Google Scholar]
- 16.Gaudreau L, Schmid A, Blaschke D, Ptashne M, Hörz W. RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell. 1997;89:55–62. doi: 10.1016/s0092-8674(00)80182-8. [DOI] [PubMed] [Google Scholar]
- 17.Gerard M, Fischer L, Moncollin V, Chipoulet J M, Chambon P, Egly J M. Purification and interaction properties of the human RNA polymerase B(II) general transcription factor BTF2. J Biol Chem. 1991;266:20940–20945. [PubMed] [Google Scholar]
- 18.Goodrich J A, Hoey T, Thut C J, Admon A, Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 19.Goodrich J A, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 20.Goodrich J A, Cutler G, Tjian R. Contacts in context: promoter specificity and macromolecular interactions in transcription. Cell. 1996;84:825–830. doi: 10.1016/s0092-8674(00)81061-2. [DOI] [PubMed] [Google Scholar]
- 21.Gorski K, Carniero M, Schibler U. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell. 1986;47:767–776. doi: 10.1016/0092-8674(86)90519-2. [DOI] [PubMed] [Google Scholar]
- 22.Greenblatt J. RNA polymerase II holoenzyme and transcriptional regulation. Curr Opin Cell Biol. 1997;3:310–319. doi: 10.1016/s0955-0674(97)80002-6. [DOI] [PubMed] [Google Scholar]
- 23.Halle J P, Meisterernst M. Gene expression: increasing evidence for a transcriptosome. Trends Genet. 1996;12:161–163. doi: 10.1016/0168-9525(96)30035-8. [DOI] [PubMed] [Google Scholar]
- 24.Hengartner C J, Thompson C M, Zhang J, Chao D M, Liao S M, Koleske S A J, Okamura S, Young R A. Association of an activator with an RNA polymerase II holoenzyme. Genes Dev. 1995;9:897–910. doi: 10.1101/gad.9.8.897. [DOI] [PubMed] [Google Scholar]
- 25.Hoeijmakers J H, Egly J M, Vermeulen W. TFIIH: a key component in multiple DNA transactions. Curr Opin Genet Dev. 1996;1:26–33. doi: 10.1016/s0959-437x(96)90006-4. [DOI] [PubMed] [Google Scholar]
- 26.Jones K A, Peterlin B M. Control of RNA initiation and elongation at the HIV-1 promoter. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 27.Kim T K, Zhao Y, Ge H, Bernstein R, Roeder R G. TATA-binding protein residues implicated in a functional interplay between negative cofactor NC2 (Dr1) and general factors TFIIA and TFIIB. J Biol Chem. 1995;270:10976–10981. doi: 10.1074/jbc.270.18.10976. [DOI] [PubMed] [Google Scholar]
- 28.Kim Y J, Björklund S, Li Y, Sayre H M, Kornberg R D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 29.Klemm R D, Goodrich J A, Zhou S, Tjian R. Molecular cloning and expression of the 32-kDa subunit of human TFIID reveals interactions with VP16 and TFIIB that mediate transcriptional activation. Proc Natl Acad Sci USA. 1995;92:5788–5792. doi: 10.1073/pnas.92.13.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- 31.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 32.Koleske A J, Young R A. The RNA polymerase II holoenzyme and its implications for gene regulation. Trends Biochem Sci. 1995;20:113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- 33.Lai J S, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Bjorklund S, Jiang Y V, Kim Y J, Lane W S, Stillman D J, Kornberg R D. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc Natl Acad Sci USA. 1995;92:10864–10868. doi: 10.1073/pnas.92.24.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao S M, Zhang J, Jeffery D A, Koleske A J, Thompson C M, Chao D M, Viljoen M, van Vuuren H J, Young R A. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 36.Lichtsteiner S, Wuarin J, Schibler U. The interplay of DNA-binding proteins on the promoter of the mouse albumin gene. Cell. 1987;51:963–973. doi: 10.1016/0092-8674(87)90583-6. [DOI] [PubMed] [Google Scholar]
- 37.Lin Y S, Ha I, Maldonado E, Reinberg D, Green M R. Binding of general transcription factor TFIIB to an acidic activating region. Nature. 1991;353:569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- 38.Maire P, Wuarin J, Schibler U. The role of cis-acting promoter elements in tissue-specific albumin gene expression. Science. 1989;244:343–346. doi: 10.1126/science.2711183. [DOI] [PubMed] [Google Scholar]
- 39.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson W, Linn S, Reinberg D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 40.McNeil J B, Agah H, Bentley D. Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast. Genes Dev. 1998;12:2510–2521. doi: 10.1101/gad.12.16.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menendez M, Kolb A, Buc H. A new target for CRP action at the malT promoter. EMBO J. 1987;6:4227–4234. doi: 10.1002/j.1460-2075.1987.tb02771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newlands S, Levitt L K, Robinson C S, Karpf A B, Hodgson V R, Wade R P, Hardeman E C. Transcription occurs in pulses in muscle fibers. Genes Dev. 1998;12:2748–2758. doi: 10.1101/gad.12.17.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niu W, Kim Y, Tau G, Heyduk T, Ebright R H. Transcription activation at class II CAP dependent promoters: two interactions between CAP and RNA polymerase. Cell. 1996;87:1123–1134. doi: 10.1016/s0092-8674(00)81806-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohkuma Y, Roeder T G. Regulation of TFIIH ATPase and kinase activities by TFIIE during active initiation complex formation. Nature. 1994;368:160–163. doi: 10.1038/368160a0. [DOI] [PubMed] [Google Scholar]
- 45.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 46.Ossipow V, Descombes P, Schibler U. CCAAT/enhancer-binding protein mRNA is translated into multiple proteins with different transcription activation potentials. Proc Natl Acad Sci USA. 1993;90:8219–8223. doi: 10.1073/pnas.90.17.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46a.Ossipow, V., and U. Schibler. Unpublished results.
- 47.Ossipow V, Tassan J P, Nigg E A, Schibler U. A mammalian RNA polymerase II holoenzyme containing all components required for promoter-specific transcription initiation. Cell. 1995;83:137–146. doi: 10.1016/0092-8674(95)90242-2. [DOI] [PubMed] [Google Scholar]
- 48.Pan G, Aso T, Greenblatt J. Interaction of elongation factors TFIIS and elongin A with a human RNA polymerase II holoenzyme capable of promoter-specific initiation and responsive to transcriptional activators. J Biol Chem. 1997;272:24563–24571. doi: 10.1074/jbc.272.39.24563. [DOI] [PubMed] [Google Scholar]
- 49.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 50.Reyes J C, Muchardt C, Yaniv M. Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J Cell Biol. 1997;137:263–274. doi: 10.1083/jcb.137.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roeder R G. The complexities of eukaryotic transcription initiation: regulation of preinitiation complex assembly. Trends Biochem Sci. 1991;16:402–408. doi: 10.1016/0968-0004(91)90164-q. [DOI] [PubMed] [Google Scholar]
- 52.Saez-Vasquez J, Pikaard C S. Extensive purification of a putative RNA polymerase I holoenzyme from plants that accurately initiates rRNA gene transcription in vitro. Proc Natl Acad Sci USA. 1997;94:11869–11874. doi: 10.1073/pnas.94.22.11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sauer F, Tjian R. Mechanisms of transcriptional activation: differences and similarities between yeast, Drosophila, and man. Curr Opin Genet Dev. 1997;2:176–181. doi: 10.1016/s0959-437x(97)80126-8. [DOI] [PubMed] [Google Scholar]
- 54.Scully R, Anderson S F, Chao D M, Wei W, Ye L, Young R A, Livingston D M, Parvin J D. BRCA1 is a component of the RNA polymerase II holoenzyme. Proc Natl Acad Sci USA. 1997;94:5605–5610. doi: 10.1073/pnas.94.11.5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seither P, Iben S, Grummt I. Mammalian RNA polymerase I exists as a holoenzyme with associated basal transcription factors. J Mol Biol. 1998;275:43–53. doi: 10.1006/jmbi.1997.1434. [DOI] [PubMed] [Google Scholar]
- 56.Tassan J P, Schulz S J, Bartek J, Nigg E A. Cell cycle analysis of the activity, subcellular localization, and subunit composition of human CAK (CDK-activating kinase) J Cell Biol. 1994;127:467–478. doi: 10.1083/jcb.127.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tassan J P, Jaquenoud M, Fry A M, Frutiger S, Hughes G J, Nigg E A. In vitro assembly of a functional human CDK7-cyclin H complex requires MAT1, a novel 36 kDa RING finger protein. EMBO J. 1995;14:5608–5617. doi: 10.1002/j.1460-2075.1995.tb00248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson C M, Koleske A J, Chao D M, Young R A. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 59.Thompson C M, Young R A. General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci USA. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Triezenberg S J, Kingsbury R C, McKnight S L. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988;2:718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- 61.Wang L, Mizzen C, Ying C, Candau R, Barlev N, Brownell J, Allis C D, Berger S L. Histone acetyltransferase activity is conserved between yeast and human GCN5 and is required for complementation of growth and transcriptional activation. Mol Cell Biol. 1997;17:519–527. doi: 10.1128/mcb.17.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Z, Luo T, Roeder R G. Identification of an autonomously initiating RNA polymerase III holoenzyme containing a novel factor that is selectively inactivated during protein synthesis inhibition. Genes Dev. 1997;11:2371–2382. doi: 10.1101/gad.11.18.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wijgerde M, Grosveld F, Fraser P. Transcription complex stability and chromatin dynamics in vivo. Nature. 1995;377:209–213. doi: 10.1038/377209a0. [DOI] [PubMed] [Google Scholar]
- 64.Wingender E, Jahn D, Seifart K H. Association of RNA polymerase III with transcription factors in the absence of DNA. J Biol Chem. 1986;261:1409–1413. [PubMed] [Google Scholar]
- 65.Wilson C J, Chao D M, Imbalzano A M, Schnitzler G R, Kingston R E, Young R A. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:234–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 66.Yankulov K, Blau J, Purton T, Roberts S, Bentley D L. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell. 1994;77:749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 67.Yeung K, Kim S, Reinberg D. Functional dissection of a human Dr1-DRAP1 repressor complex. Mol Cell Biol. 1997;17:36–45. doi: 10.1128/mcb.17.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Young R A. RNA polymerase II. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- 69.Young, R. A. Personal communication.