Summary
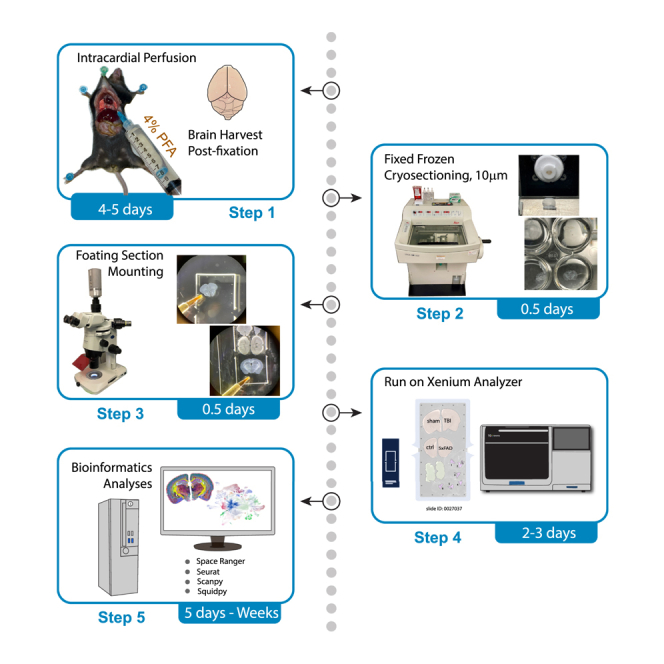
Here, we present a protocol for Xenium spatial transcriptomics studies using fixed frozen mouse brain sections. We describe steps for intracardiac perfusion, cryosectioning, and floating section mounting of brain sections, which enable runs on the Xenium analyzer and data delivery. We demonstrate that, in addition to the 10× Genomics-validated formalin-fixed paraffin-embedded (FFPE) and fresh frozen sections, fixed frozen thin brain sections are compatible with the Xenium platform and provide excellent imaging and quantification results for spatially resolved gene expression.
For complete details on the use and execution of this protocol, please refer to Ma et al.1
Subject areas: RNA-seq, molecular biology, gene expression, neuroscience, systems biology
Graphical abstract
Highlights
-
•
Steps for intracardial perfusion, brain collection, cryoprotection, and OCT embedding
-
•
Instructions for mounting fixed frozen mouse brain thin sections on Xenium slides
-
•
Fixed frozen sections allow for better sample preparation and usage of the imaging area
-
•
Fixed frozen sections provide excellent results in spatially resolved gene expressions
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Here, we present a protocol for Xenium spatial transcriptomics studies using fixed frozen mouse brain sections. We describe steps for intracardiac perfusion, cryosectioning, and floating section mounting of brain sections, which enable runs on the Xenium analyzer and data delivery. We demonstrate that, in addition to the 10× Genomics-validated formalin-fixed paraffin-embedded (FFPE) and fresh frozen sections, fixed frozen thin brain sections are compatible with the Xenium platform and provide excellent imaging and quantification results for spatially resolved gene expression.
Before you begin
Cellular functions, including gene and protein expressions, are most meaningful within the tissue spatial context in which these cells reside. While bulk and single-cell transcriptomics technologies enable the quantification of the remarkable diversity of mRNA transcripts that define cellular identity and function, these technologies often characterize dissociated cells isolated from tissue samples, resulting in the loss of the natural spatial context of molecules within cells. Recently, state-of-the-art spatial transcriptomics methods have been developed that enable multiplexed mRNA in situ detection (for a review, see You et al.1). Two primary spatial transcriptomics methods have been developed for detecting mRNA transcripts using different strategies: multiplexed fluorescent in situ hybridization (FISH) techniques such as MERFISH2,3 and SeqFISH,4,5 and in situ capture followed by high-throughput sequencing (e.g., Slide-seq6,7). A leading commercial source for these techniques is 10× Genomics, which provides the Xenium (FISH- and imaging-based) and Visium/Visium HD (in situ sequencing-based) platforms for spatially quantifying mRNA species. On the Xenium platform, FISH can be highly sensitive and specific for detecting individual RNA molecules, providing high spatial resolution and allowing for the subcellular localization of RNA molecules within a tissue section.
To resolve gene expression changes on the Xenium platform, a panel of predesigned mRNA probes must be utilized. The Xenium platform offers multiplexing capability, capable of detecting multiple RNA targets simultaneously, enabling the quantification of many genes within the same cell in a tissue section. These features are crucial for spatially variant tissues, such as the brain. The reagents and materials provided by 10× Genomics for these studies include the Xenium slide, which has defined boundaries and calibration landmarks (fiducials) that define the imaging area. Most users will mount their tissue sections inside the imaging area and send the slides to a commercial service provider or core facility for further processing. To obtain high-quality FISH-imaging data on the Xenium platform, sample preparation is critical. Currently, the Xenium platform is validated for formalin-fixed paraffin-embedded (FFPE) thin tissue sections, although fresh frozen tissue sections are also compatible with the recommended protocols. However, many neuroscience laboratories working with brain sections typically follow a protocol involving paraformaldehyde fixation, cryoprotection, cryosectioning, followed by immunohistochemistry (e.g., multichannel antibody labeling) or in situ hybridization for mRNA species (e.g., RNAscope). It is unclear whether this protocol is compatible with the 10× Genomics workflow.
Here, we report excellent results of Xenium analysis using 'fixed frozen' mouse brain sections. As a complete protocol for running tissue sections on the Xenium analyzer is beyond the scope of this protocol, we focus on the wet lab tissue section processing and mounting on Xenium slides. This approach is crucial for preparing high-quality sections without tissue folding, preserving the mRNA transcripts from degradation, and maximizing the utilization of the Xenium slide imaging areas (e.g., increasing data output and reducing cost). Finally, we provide imaging and quantification of gene expression from one dataset that demonstrates the excellent results of this tissue preparation protocol.
Institutional permissions
Appropriate precautions must be taken when handling animal specimen and fixatives. Animal studies presented here were approved by the Institutional Animal Care and Use Committee of the University of Arizona. In this study, we used C57Bl6/J mice and the 5× FAD and control littermates of 2–4 months age.
Reagents preparation and mouse brain collection
Timing: 2–3 h
Note: All reagents should be prepared from autoclaved nuclease free water. Workbench surfaces are cleaned and wiped with RNaseZap RNase decontamination solution (R2020 – Sigma-Aldrich).
The present steps aim to collect mouse brain and fix it in 4% PFA for cryosectioning.
Prepare 4% (w/v) paraformaldehyde (PFA) for a total volume of 250 mL
CRITICAL: PFA powder is very light and can be charged in the container and hard to settle. Weight the powder in a hood. Paraformaldehyde powder should always be used in a ventilated safety hood.
-
1.
Measure 125 mL of water in a beaker designated for PFA. Place the beaker on a heating pad and stir it with a stir bar.
-
2.
Weigh 10 g of PFA and add it to the beaker. Heat the mixture to approximately 65°C. Add 2.5N sodium hydroxide solution drop by drop until the PFA powder dissolves completely.
-
3.
Filter the PFA solution into a clean, designated flask using Whatman filter paper. Place the filtered solution on ice.
-
4.
Prepare 125 mL of 0.1M Phosphate Buffer (PB) by mixing 85 mL of 0.2 M Na2HPO4 (dibasic) with 40 mL of 0.2 M NaH2PO4 (monobasic).
-
5.
Mix the ∼125 mL of filtered PFA with 125 mL of PB to obtain the working 4% PFA solution. Keep the solution at 4°C. Label the bottle with the date, and do not use it if it is more than two weeks old.
4% (w/v) Paraformaldehyde (PFA)
| Reagent | Final concentration | Amount |
|---|---|---|
| Paraformaldehyde (PFA) | 1.3 mM | 10 g |
| 2.5N sodium hydroxide solution | 10 mM | 1.5 L |
| 0.2 M Na2HPO4 (dibasic) | 68 mM | 85 mL |
| 0.2 M NaH2PO4 (monobasic) | 32 mM | 40 mL |
| ddH2O | N/A | 125 mL |
| Total | N/A | ∼250 mL |
Transcardial perfusion for brain fixation
CRITICAL: Ensure that procedures involving live animals are approved by the IACUC and comply with governmental regulations.
-
6.
Anesthetize the mice with 4%–5% isoflurane in an induction chamber. Ensure the absence of tail and corneal reflexes.
-
7.
While under deep anesthesia, open the thoracic cavity to expose the heart.
-
8.
Insert a 22g perfusion needle through the left ventricle wall. Connect the needle to a 10 mL syringe containing ice-cold 0.01 M PBS mounted on a syringe pump. Set the pump speed to approximately 2 mL/min. Immediately puncture the right atrium with a needle tip.
-
9.
After the initial blood clearing, replace the PBS syringe with a 10 mL syringe containing pre-chilled, freshly made 4% PFA. Continue the perfusion for fixation for 5 min.
Brain dissection, post-fixation, cryoprotection
CRITICAL: Ensure the brain surface is not damaged by the dissecting tools. The brain surface should appear clean from blood with a uniform white color if the perfusion-fixation is done correctly.
-
10.
After perfusion-fixation, dissect the entire brain using a razor blade, a pair of forceps, and scalpel tools.
-
11.
Briefly rinse the brain in 0.01 M PBS, then transfer it to a 50 mL conical tube containing 30 mL of freshly made 4% PFA. If multiple brains are collected, use one conical tube per brain and label each tube correctly for identification.
-
12.
Keep the brain in 4% PFA for post-fixation for 10–14 h.
-
13.
Transfer the brain to 30% sucrose (in 0.01 M PBS) for 1–2 days, until the brain descends to the bottom of the tube. It is now ready for cryosectioning.
Note: The protocol assumes immediate cryosectioning after the brain is taking out of 30% sucrose solution. Alternatively, the brain can be embedded in OCT in a cryo-mold and freeze in −20°C for several weeks until more brain samples are collected for sectioning.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Experimental models: Organisms | ||
| Mouse: C57BL/6J | The Jackson Laboratory | JAX: 000664 |
| (32–50 days, wild type, male and female) | ||
| Chemicals | ||
| 1× DPBS | Gibco | 14190-144 |
| O.C.T. Compound | Sakura | 4583 |
| H&E Staining Kit (Hematoxylin and Eosin) | Abcam | ab245880 |
| Glycerol | Sigma | G5516 |
| Ethanol | Sigma | E7023 |
| Milli-Q water | Millipore | SYNSV0000 |
| NaOH | Sigma | S5881 |
| PFA | Sigma | 158127 |
| Na2HPO4 | Sigma | S5136 |
| NaH2PO4 | Sigma | S8282 |
| Sucrose | Sigma | S9378 |
| Software and algorithms | ||
| Xenium Explorer | 10× Genomics | Version 3.10 |
| R, RStudio | Open source, Posit | R- Ver 4.3, RStudio – release 2023.03.02 |
| Python | Open source, python.org | Python 3.8 |
| Other | ||
| Xenium Slide Kit | 10× Genomics | 1000659 |
| RNaseZAP | Sigma | R2020 |
| Microscope cover glass | Fisherbrand | 12541033 |
| Nickel-plated pin holder | Fine Science Tools | 26018-17 |
| Insect pins | Fine Science Tools | 26000-45 |
| Graefe forceps (Curved) | Fine Science Tools | 11051-10 |
| Graefe forceps (straight) | Fine Science Tools | 11050-10 |
| Brush | Hitobiotech | HTHS0112 |
| 6-Well plate | Falcon | 353046 |
| 50 mL centrifuge tube | VWR | 21008-178 |
| Single edge blades | MSC GEM | 62-0178 |
| Beaker 1 L (6) | Pyrex | 1000-1L |
| Glass needle | VWR | 53432-706 |
| Lint-free wipes | Texwipe | TX1010 |
| Stereo microscope | Olympus | SZX16 |
| Cryostat | Leica | CM1590 |
| Thermal cycler | Bio-Rad | C1000 |
| Low Profile Plate Insert | 10× Genomics | 3000823 |
Step-by-step method details
Timing: 6–8 h
This section describes fixed frozen thin brain cryosectioning, followed by free-floating mounting on Xenium slides. After section mounting, Xenium slides are ready for shipping to a service provider to run on Xenium Analyzer.
Cryosectioning of the mouse brain
Timing: ∼2 h
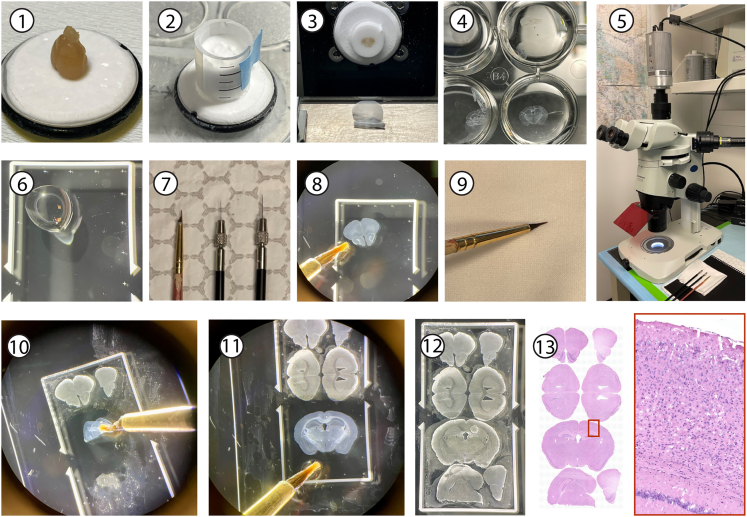
Here, we describe steps for cryosectioning of thin mouse brain sections from cryoprotected, OCT embedded mouse brains (Figure 1).
-
1.
Adjust the cryostat temperature: set the inside chamber to −20°C and the cutting head to −10°C.
-
2.
Block cut the brain sample to create a flat surface and achieve the desired orientation for mounting on the cutting head.
Note: For example, if the anterior brain region is needed, block cut the cerebellum region and mount the brain vertically.
-
3.
Add a flat layer of OCT compound to the specimen disk, and leave it in the internal cryostat chamber to solidify (step 1).
-
4.
Mount the brain to the tissue specimen disk with OCT, adding more OCT to completely embed the brain using a removable plastic container (made from a 15 mL conical tube) to form an OCT cylinder (step 2).
-
5.
After the OCT cylinder solidify, Insert the specimen disk assembly into the cryostat specimen head holder. Trim the brain under the trim mode with a section thickness of approximately 50–100 μm until the brain region of interest is reached at the coronal level (step 3).
Note: Consult the Paxinos mouse brain atlas to ensure the regions of interest are reached.
-
6.
Engage the anti-rolling device, adjust its angle, and align it with the blade cutting edge.
-
7.
Adjust the cutting thickness to 10 μm, and continue to cut a few sections to ensure they are uniform and intact.
Note: If multiple brain regions are needed, continue cutting and collecting additional sections as the desired brain regions are reached.
-
8.
Prepare a clean 6-well cell culture plate, adding 5 mL of 4°C 0.01 M PBS to each well. Using small forceps, transfer two sections to each well.
Note: The embedding OCT on the thin section should immediately dissolve, and the sections will descend to the bottom of each well.
-
9.
Rinse the sections in the 6-well plate by placing it in an ice bucket on a rocker for 5 min. Transfer the sections to a new 6-well cell culture plate containing 5 mL of 0.01 M PBS (step 4).
Figure 1.
Photomicrograph illustration of the steps involved in OCT embedding, cryosectioning, and free-floating section mounting onto Xenium slides under a dissection microscope
Floating section mounting of thin brain sections onto xenium slides
Timing: 3–5 h
Here, we describe steps for mounting free-floating thin mouse brain sections in small volumes of nuclease-free water onto Xenium slides under a stereo microscope.
CRITICAL: To make the most efficient use of the Xenium slide imaging area, increase data output, and enable multiple sample comparisons in one Xenium run, it is critical to design the layout of different samples, groupings, and brain regions on the Xenium slide. For example, instead of mounting one intact coronal section, two coronal hemisections at matched rostral-caudal levels can be mounted, one from control and the other from treated conditions. Alternatively, specific brain regions (e.g., prefrontal cortex, visual cortex, thalamus) can be dissected out if these structures are the primary interest of the study. Keep in mind there is a trade-off between sample area and the number of samples that can be mounted. This also has implications for customized downstream data analysis needs.
-
10.
Carefully plan which section or hemisection will be placed within the 10.45 mm × 22.45 mm imaging area. Sketch a diagram in a lab notebook for the layout of sections before mounting.
-
11.
Mount sections under a stereo microscope with a continuous zoom objective (1.0×, Olympus SZX-16 model), using phase contrast and dark field illumination filters (step 5).
-
12.
Clean the microscope’s glass plate with RNaseZap.
-
13.
Place the Xenium slide under the 1.0× objective. Adjust the magnification to focus on the top left corner of the sample area. Add 50 μL of nuclease-free water to form a meniscus (step 6).
-
14.
Prepare the brain section mounting tools, including a small trimmed hair brush and two dissection pin/holder assemblies. Select a whole or hemisection of a brain slice. Using a small fire-polished glass needle, pick up the section and transfer it to the drop of nuclease-free water on the Xenium slide.
Note: Transferring thin brain sections with a large area using a small paintbrush, which works well for thicker (e.g., 40 μm) sections in large liquid volumes for immunohistochemistry staining, can be very challenging for two reasons: the small liquid volume on the slide does not allow for flattening of the section using the brush, and the thin slices may easily adhere to the brush.
-
15.
Use two sharp insect pins/holder assemblies (Fine Science Tools). Insert them into the water meniscus and use the pins to guide the liquid flow to desired locations, then flattening the thin brain sections.
-
16.
Use a trimmed hair brush, insert the brush into the water meniscus, then remove the water retained by the brush by contacting a lint-free laboratory wipe. Repeat this process until all the water is removed and the slice is flattened on the Xenium slide (steps 8–9).
CRITICAL: To ensure the brain sections adhere to the slide, the water needs to be removed. This cannot be done using a vacuum suction line or pipette tips, as the sections can easily be damaged or dislocated. Instead, use a small hair brush trimmed to retain a few short hairs.
-
17.
In the empty imaging area of the Xenium slide next to the mounted slice, add another drop of PBS and repeat the above steps to mount another thin brain slice at the desired level (step 10).
CRITICAL: Ensure fiducials are not obstructed by tissue sections and tissues are not overlapping.
-
18.
Repeat these floating section mounting steps until all imaging areas of the slide are mounted with sections (steps 11–12). If desired, HE staining can be done before sending the slides for the Xenium Analyzer run (step 13).
Note: One Xenium spatial transcriptomics experiment typically uses two slides, each with a unique slide ID number. Repeat mounting of desired brain sections onto the second Xenium slide.
-
19.
After completing the mounting of one Xenium slide, take an image that covers all the mounted sections. Ensure all metadata of the sections are appropriately recorded. Insert the slide into a slide mailer, mark the slide mailer with a Sharpie marker, and store the slide at −80°C until the next step in the workflow (to be completed by a service provider).
-
20.
In a sample form, illustrate the slide tissue section layout and log all the metadata information, including Xenium slide ID, PN number, number of sections derived from different mice, treatment conditions, and annotated images of section layouts.
Xenium analyzer run and data delivery
Timing: 48–96 h to run on the Xenium analyzer, depending on the number of gene panels and the service provider. Further customized analyses of delivered data may take approximately days to weeks depending on analysis needs, computational resources, and number of samples.
-
21.
Shipping the mounted Xenium slides in frozen condition on dry ice to a service provider, e.g., the 10× Genomics Catalyst Program.
Note: Most individual labs do not have a Xenium Analyzer and therefore rely on a university core facility or a commercial service provider for the next steps, which include running the further Fixation & Permeabilization (CG000581 workflow) and in situ hybridization on a Xenium Analyzer with onboard analysis (refer to the Xenium Analyzer User Guide, CG000584). These service providers perform onboard analysis, including cell segmentation following a staining protocol that visualizes cell nuclei, content, and boundaries. They will deliver data from the Xenium Analyzer onboard analysis according to standard formats developed by 10× Genomics.
-
22.
Optional customized analysis by the end user after receiving data delivery from service provider.
Note: Customized analysis of Xenium analyzer onboard analysis files may use Linux, R, RStudio, and Python packages including Seurat, Scanpy, Squidpy etc.
Note: Users are referred to the 10× Genomics published "Getting Started with Xenium In Situ – Experiment Planning Guide" (10× Genomics - LIT000215), which is validated for fresh frozen and FFPE sections.
Comparison with fresh frozen brain section mounting
Timing: 5–7 h
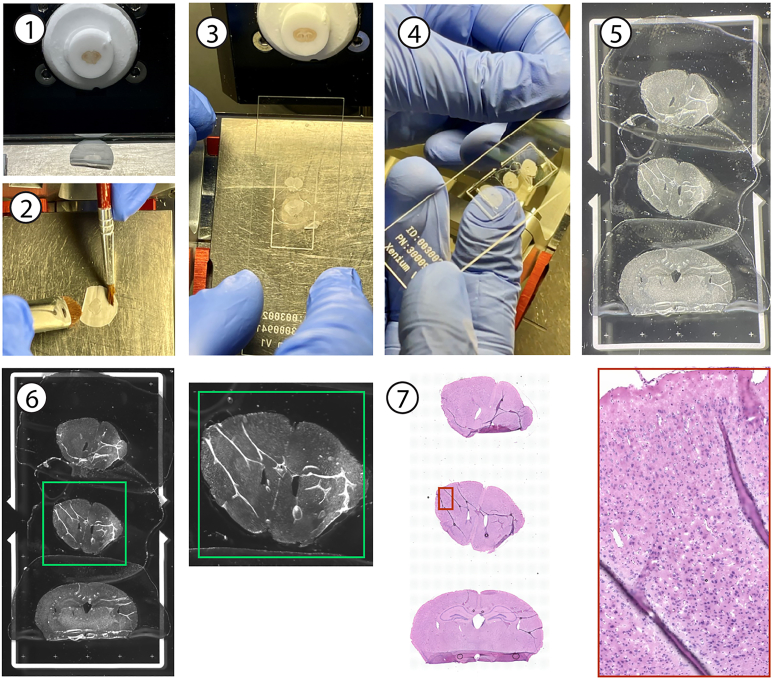
Note: Our floating mounting method for ‘fixed frozen’ thin brain sections onto Xenium slides allows mounting at room temperature (22°C–25°C) while preserving mRNA integrity for multiple rounds of high-plex hybridization of probes. Another major advantage of this approach is the ability to mount sections under a dissection microscope using very thin-tipped, rigid tools. This allows for maximum utilization of the imaging space, increasing data yield and significantly lowering experimental costs. As a comparison, we mounted another set of mouse brain sections using the 10× validated ‘fresh frozen’ method by adopting the following steps (Figure 2).
-
23.
Sacrifice mice by decapitation under deep anesthesia, and dissected the brain out.
-
24.
Quickly freeze the freshly dissected brains according to 10× Genomics literature CG000579|Rev E.
-
25.
Embed the fresh frozen brain in OCT and section the brain on a cryostat at a thickness of 10 μm. Care needs to be taken to minimize tissue rolling.
-
26.
Pick up the thin cut fresh frozen section from the cutting head and directly place onto the empty Xenium slide imaging area.
CRITICAL: Care needs be taken to avoid tissue overlapping, while maximize the number of tissue sections that can be placed in the imaging area. Despite repeated practice, obtaining a completely flat large tissue section can be extremely difficult. As such, a much larger area was left empty to avoid tissue section overlapping (Figure 2).
Figure 2.
Fresh frozen brain cryosectioning and direct pickup of sections onto a Xenium slide, following the 10× Genomics fresh frozen protocol
Note the unutilized space of the imaging area and excessive tissue curling.
Expected outcomes
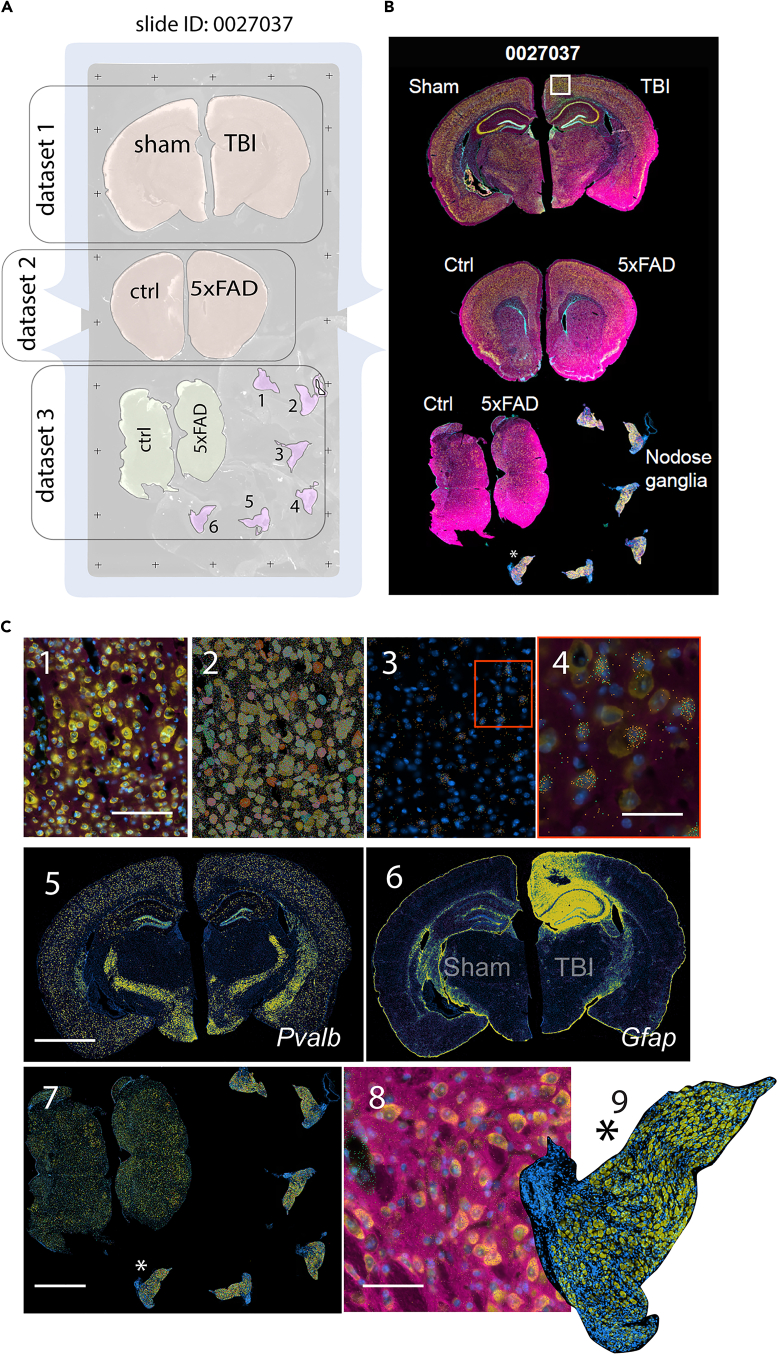
We conducted a Xenium spatial transcriptomics experiment using a predesigned 247-gene mouse brain panel of probes to demonstrate the feasibility of using fixed frozen mouse brain sections with free-floating mounting onto Xenium slides. The tissue section layout is illustrated in Figure 3A. Tissue staining, based on which cell segmentation is implemented, is shown in Figure 3B. In this slide, we mounted various brain tissue sections from different mouse strains and treatments. The analysis provided by the service provider includes data demultiplexed into three datasets.
Figure 3.
(A) Tissue section layout on an experimental Xenium slide, which generated three datasets.
(B) Overview of stained tissue on a Xenium slide.
(C) Combined staining of cell nuclei, boundaries, and contents enables excellent cell segmentation. Tissue images and overlaying transcripts can be visualized using the Xenium Explorer browser.
The first dataset consists two hemi coronal sections at the level of somatosensory cortex/mid-level of hippocampus (−2.06 mm post bregma). The right hemisection is from a mouse with mild traumatic brain injury (TBI) at the level of the direct closed head impact site; the left hemisection is from the Sham control.
The second dataset comprises two coronal hemisections at the level of the prefrontal cortex (1.34 mm anterior from bregma). The right hemisection is from a 5× FAD mouse at a pre-symptomatic age; the left hemisection is the sex- and age-matched control.
The third dataset includes two sections at the level of the brainstem (one control, one 5×FAD). It also includes six sections of nodose ganglia from wild-type mice.
In this experiment, we summarize the key findings and explore more results obtained from dataset 1, which compares the Sham-TBI hemisphere, including selected regions of interest (ROIs) such as local cortical regions under direct TBI impact and the counterpart cortical region from Sham. The following findings illustrate excellent results for these test datasets.
Tissue staining and cell segmentation
Tissue staining was performed using a combination of nucleus (DAPI), cellular content, and cell boundary markers with the Xenium Multi-Tissue Stain Mix (10× Genomics, PN 2000991), following guideline CG000749. This staining delineates cell boundaries and allows segmentation of different cells throughout the whole hemisection of the brain (Figures 3B and 3C1).
Additionally, transcript locations can be overlaid with segmented cells of different types (via unsupervised clustering, which is denoted by different cell colors) (Figure 3C2).
The locations and relative abundance of specific transcripts can be jointly visualized with nuclei staining and composite staining at higher magnification (Figure 3C3 and 3C4, for Gad1/green and Gad2/red genes; Figure 3C5, for Pvalb gene). These selected transcript spatial coordinates (x, y), when marked onto tissue images, delineate specific cell type locations and even outline the contour of these cell types (Figures 3C3 and 3C4).
Transcripts density and region of interest analysis
An overview of transcript density shows that in the local cortical region under direct impact, TBI results in increased expression of Gfap (Figure 3C6), a marker for reactive astrocytes, which is a well-reported effect of mild TBI.8,9
The staining protocol provides excellent images that allow segmentation of the brainstem and nodose ganglia, and discerning the location of selected transcripts (Figure 3C7–9).
Spatial and UMAP embedding of segmented cells
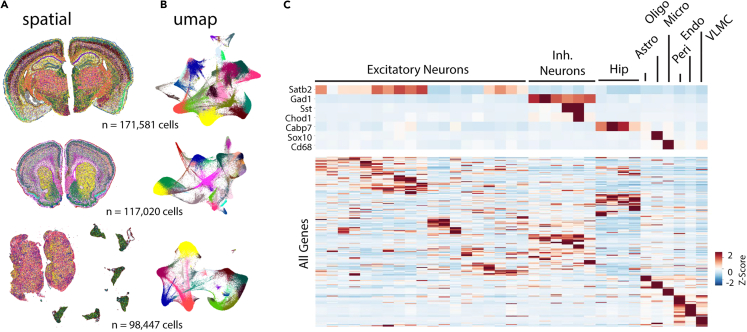
The spatial and UMAP embedding of all cells detected in these three datasets are provided in Figure 4 (4A - spatial, 4B - UMAP). Combining cells from these three datasets, supervised clustering resulted in a broad classification into various cell types, including 'Excitatory Neurons', 'Inhibitory Neurons', 'Hippocampus', 'Astrocytes', 'Oligodendrocytes', 'Microglia', 'Pericytes', 'Endothelial Cells', and 'Vascular and Leptomeningeal Cells (VLMCs)' (Figure 4C). The spatial embedding (Figure 4A) reveals the precise location of different cell types within the brain sections, showcasing the spatial organization and relationships between various cell populations. The UMAP embedding (Figure 4B) shows the clustering of cells based on their overall gene expression profiles, highlighting the distinct groups and subpopulations of cells across the different datasets.
Figure 4.
(A) Xenium analyzer onboard analysis outputs allow for unsupervised clustering of the three datasets that are visualized in spatial domain.
(B) The same three datasets visualized in UMAP embedding.
(C) Supervised clustering and label transfer identified different cell type populations based on their marker genes.
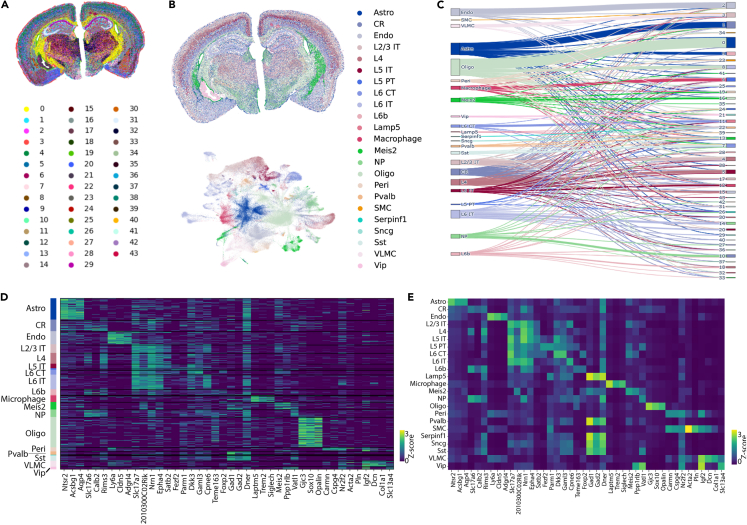
We further used the Scanpy package10 for unsupervised clustering of the Sham/TBI dataset, which includes 171,581 segmented cells. This analysis resulted in 44 distinct clusters throughout the entire brain (Figure 5A). When mapped to the Allen Brain Institute mouse cortex single-cell reference dataset,11,12 major cortical cell class labels were successfully transferred (Figure 5B). The label transfer was done using a deep generative model.13 This supervised cell type annotation identified 22 different clusters as major cell types, including astrocytes ('Astro'), endothelial cells ('Endo'), L2/3 intra-telencephalic projecting neurons ('L2/3 IT'), L5 intra-telencephalic neurons ('L5 IT'), parvalbumin-expressing interneurons ('Pvalb'), among other 17 different cell types.
Figure 5.
Additional clustering and cell type annotation using the Scanpy pipeline
(A) Unsupervised clustering resulted in 44 cell types across both hemisections.
(B) Cell type label transfer was performed using the Allen Cortex reference dataset.
(C–E) Cell cluster transitions and top marker genes for the annotated cell types are presented in the Sankey plot (C) and heatmaps (D and E).
A Sankey plot (Figure 5C) illustrates the transition of different annotated cell types and the original unsupervised clusters. Marker genes in these annotated cell types are shown in heatmaps (Figure 5D – individual cells within annotated clusters; Figure 5E – averaged gene expression levels in each cell type).
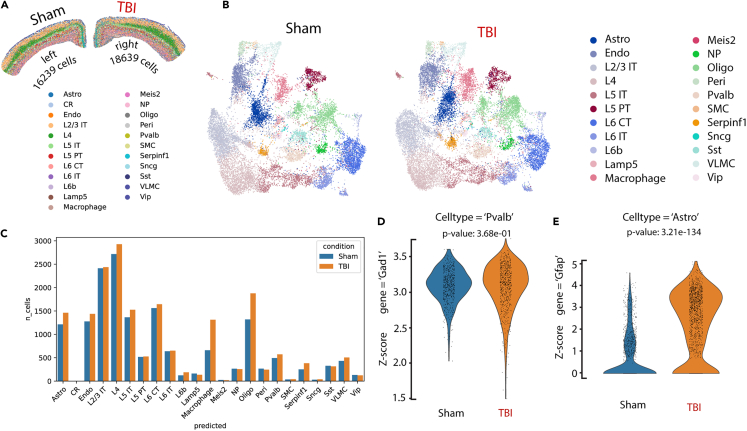
Comparison of selected gene expression from ROIs
Next, we selected cortical regions of interest (ROIs) from the Sham/TBI dataset for cell type-specific differential gene expression (DEG) analyses. Cortical regions from the TBI site, which were under direct impact, showed dramatically increased Gfap expression (Figure 3C6), while the sham side comprised anatomically matched regions (Figure 6A). After sample integration, Sham and TBI cortical neurons occupied similar UMAP spaces (Figure 6B). Annotated cell types from these two selected ROIs are plotted in Figure 6C. Comparison of selected gene expression levels between sham and TBI samples was made using violin plots with p values calculated using Mann-Whitney U test. We found that, for parvalbumin-expressing interneurons (‘Pvalb’), the Sham (Number of cells = 494) and TBI (Number of cells = 573) cortical regions have similar levels of Gad1 expression (Mann-Whitney U = 137101, z-score = −0.90, p = 0.38, Figure 6D). In comparison, Gfap gene levels were significantly higher in TBI samples in the astrocyte cluster (Sham: 1214 cells, TBI: 1461 cells, Mann-Whitney U = 414177, z-score = −23.77, p < 0.0001, Figure 6E).
Figure 6.
The Sham/TBI dataset supports spatially resolved quantitative gene expression analysis
(A) ROIs can be defined for selected cell type annotation.
(B) UMAP visualization of cells in the selected ROIs.
(C) Bar graph quantification of cell numbers in the selected ROIs.
(D) Quantitative Gad1 gene expression using violin plots in "Pvalb" cell type. p = 0.38, Mann-Whitney U test.
(E) Quantitative Gfap gene expression in the "Astro" cell type. p < 0.0001, Mann-Whitney U test.
In summary, the results from this experiment demonstrate the effectiveness of using fixed frozen mouse brain sections with free-floating mounting onto Xenium slides for the spatial transcriptomics studies. The integration of spatial and gene expression data through these advanced analyses provides a comprehensive view of the cellular architecture and the molecular landscape within the brain. This detailed classification and mapping of cell types enhance our understanding of the cellular responses to various conditions, such as TBI, and facilitate the exploration of cellular interactions and functions within the tissue context.
Limitations
The protocol was designed to test the compatibility of fixed frozen brain samples for FISH and imaging-based spatial transcriptomics studies on the Xenium platform. This complements the FFPE and fresh frozen protocols for general tissue samples that are validated by 10× Genomics. We demonstrate the feasibility and excellent data quality by adopting fixed frozen sectioning and free-floating section mounting in small volumes of water under a dissection microscope at room temperature (22°C–25°C). This should have broad applicability for the neuroscience community exploring the nascent field of spatial transcriptomics. We envision that the protocol should be compatible with the latest Xenium Prime (V2) technology.
However, there are some limitations inherent to the Xenium FISH-based technology. The protocol as presented may require fine-tuning and validation for other tissues based on their morphology, thickness, and fixation protocols. Multiple practice rounds may be necessary to achieve proficient mounting under the microscope and to maximize the usage of the imaging area and data gain on the Xenium slide.
Troubleshooting
Problem 1
The dissected mouse brain after PBS/ 4% PFA perfusion does not appear white and clean.
Potential solution
-
•
Verify correct insertion into the left ventricle during perfusion and PFA fixation.
-
•
Ensure there are no air bubbles in the syringe and tubing.
-
•
Make sure the right atrium is punctured as soon as PBS inflow/blood clearing starts.
Problem 2
During brain sectioning using a cryostat, the blade feels high resistance and tissue sections show broken stripes.
Potential solution
-
•
It is likely the cutting tissue temperature is too low.
-
•
Ensure the OCT-embedded brain is fully equilibrated to the temperature of the internal chamber of the cryostat, which should be set at −20°C.
-
•
The cutting head temperature should be set at −10°C.
Problem 3
There was excessive curling of the thin brain sections coming off the cutting blade.
Potential solution
-
•
Adjust the anti-rolling device angles and align it with the cutting edge of the blade.
-
•
Gradually adjust the device while cutting a new section.
-
•
Note that unlike cutting fresh frozen samples, curling of the fixed frozen sections is well tolerated with floating mounting methods, as the tissue section will flatten out when transferred to the cold PBS solution.
Problem 4
It is difficult to flatten out thin brain sections onto the Xenium slide.
Potential solution
-
•
Use a pair of inset pins/holders or fire-polished pulled glass needles to pick a selected brain section and transfer it to the water drop meniscus under the dissection microscope.
-
•
Avoid using a paintbrush, as it will introduce additional liquid volume and the section may easily stick to the brush.
-
•
Prior to your first Xenium run, practice tissue placement using non-experimental blocks on blank slides with hand-drawn practice frames (10.45 × 22.45 mm).
Problem 5
It is difficult to remove the water meniscus once the tissue is flattened out.
Potential solution
-
•
Use a trimmed brush to dip in the water meniscus and dab the brush on a clean, folded lint-free laboratory wipe to remove water.
-
•
Do not use a rolled Kimwipe to dry the water meniscus, as this may dry the water too quickly and draw the brain section to the Kimwipe. This will also leave lint fibers on the Xenium slides that are hard to remove.
Problem 6
What should I do after receiving the data and how do I analyze it?
Potential solution
-
•
This can be the most challenging part. Users of this protocol should have basic knowledge of spatial transcriptomics and bioinformatics data analysis.
-
•
The Xenium Analyzer guide and online resources list the next steps after receiving Xenium data, including adopting various R and Python packages for additional coded analysis.
-
•
Consult the 10× Genomics support team or a bioinformatician for customized analysis needs.
-
•
The most intuitive way to explore your Xenium data is to open the .Xenium file under the delivered data deck and browse it with Xenium Explorer. This allows visualization of images, transcripts, cell segmentations, and the export of selected ROIs as .csv files of coordinates for further analysis.
Resource availability
Lead contact
Further information and requests for resources should be directed to the lead contact, Dr. Shenfeng Qiu, sqiu@arizona.edu.
Technical contact
Technical questions on executing this protocol should be directed to and will be answered by the technical contact, Dr. Xiaokuang Ma, xiaokuangma@arizona.edu.
Materials availability
This study did not generate new unique reagents.
Data and code availability
The published article includes all details, needed resources, and datasets generated.
Acknowledgments
This study was supported by NIH grant R01MH128192 (S.Q. and D.F.), R01EY035138 (S.Q. and A.W.M.), and R21AG078700 (S.Q.), Institute of Mental Health Research (IMHR, Level 1 funding, S.Q.), Arizona Biomedical Research Commission (Investigator Award, D.F. and S.Q.), institution startup fund from The University of Arizona (S.Q.), and in part by NIH grant R01HL158596, R01HL162794, R01HL169509, and R01HL170096 and University of Arizona institution funding to Z.D.
Author contributions
X.M., J.W., C.C., D.F., and H.Z. prepared the mouse brain samples and conducted the tissue mounting and Xenium spatial transcriptomics experiments. P.C. and J.Z. analyzed data and prepared the figures. Z.D. and S.Q. conceived the study, secured funding, and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Xiaokuang Ma, Email: xiaokuangma@arizona.edu.
Zhiyu Dai, Email: z.dai@wustl.edu.
Shenfeng Qiu, Email: sqiu@arizona.edu.
References
- 1.You Y., Fu Y., Li L., Zhang Z., Jia S., Lu S., Ren W., Liu Y., Xu Y., Liu X., et al. Systematic comparison of sequencing-based spatial transcriptomic methods. Nat. Methods. 2024;21:1743–1754. doi: 10.1038/s41592-024-02325-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen K.H., Boettiger A.N., Moffitt J.R., Wang S., Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348 doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moffitt J.R., Hao J., Bambah-Mukku D., Lu T., Dulac C., Zhuang X. High-performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing. Proc. Natl. Acad. Sci. USA. 2016;113:14456–14461. doi: 10.1073/pnas.1617699113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eng C.H.L., Lawson M., Zhu Q., Dries R., Koulena N., Takei Y., Yun J., Cronin C., Karp C., Yuan G.C., Cai L. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature. 2019;568:235–239. doi: 10.1038/s41586-019-1049-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lubeck E., Coskun A.F., Zhiyentayev T., Ahmad M., Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriques S.G., Stickels R.R., Goeva A., Martin C.A., Murray E., Vanderburg C.R., Welch J., Chen L.M., Chen F., Macosko E.Z. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363:1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stickels R.R., Murray E., Kumar P., Li J., Marshall J.L., Di Bella D.J., Arlotta P., Macosko E.Z., Chen F. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat. Biotechnol. 2021;39:313–319. doi: 10.1038/s41587-020-0739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karve I.P., Taylor J.M., Crack P.J. The contribution of astrocytes and microglia to traumatic brain injury. Br. J. Pharmacol. 2016;173:692–702. doi: 10.1111/bph.13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilhelmsson U., Li L., Pekna M., Berthold C.H., Blom S., Eliasson C., Renner O., Bushong E., Ellisman M., Morgan T.E., Pekny M. Absence of glial fibrillary acidic protein and vimentin prevents hypertrophy of astrocytic processes and improves post-traumatic regeneration. J. Neurosci. 2004;24:5016–5021. doi: 10.1523/JNEUROSCI.0820-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tasic B., Yao Z., Graybuck L.T., Smith K.A., Nguyen T.N., Bertagnolli D., Goldy J., Garren E., Economo M.N., Viswanathan S., et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature. 2018;563:72–78. doi: 10.1038/s41586-018-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao Z., van Velthoven C.T.J., Nguyen T.N., Goldy J., Sedeno-Cortes A.E., Baftizadeh F., Bertagnolli D., Casper T., Chiang M., Crichton K., et al. A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell. 2021;184:3222–3241.e26. doi: 10.1016/j.cell.2021.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu C., Lopez R., Mehlman E., Regier J., Jordan M.I., Yosef N. Probabilistic harmonization and annotation of single-cell transcriptomics data with deep generative models. Mol. Syst. Biol. 2021;17 doi: 10.15252/msb.20209620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The published article includes all details, needed resources, and datasets generated.