Abstract
Insulin-like growth factor II (IGF-II) is a major fetal growth factor. The IGF-II gene generates multiple mRNAs with different 5′ untranslated regions (5′ UTRs) that are translated in a differential manner during development. We have identified a human family of three IGF-II mRNA-binding proteins (IMPs) that exhibit multiple attachments to the 5′ UTR from the translationally regulated IGF-II leader 3 mRNA but are unable to bind to the 5′ UTR from the constitutively translated IGF-II leader 4 mRNA. IMPs contain the unique combination of two RNA recognition motifs and four hnRNP K homology domains and are homologous to the Xenopus Vera and chicken zipcode-binding proteins. IMP localizes to subcytoplasmic domains in a growth-dependent and cell-specific manner and causes a dose-dependent translational repression of IGF-II leader 3 –luciferase mRNA. Mouse IMPs are produced in a burst at embryonic day 12.5 followed by a decline towards birth, and, similar to IGF-II, IMPs are especially expressed in developing epithelia, muscle, and placenta in both mouse and human embryos. The results imply that cytoplasmic 5′ UTR-binding proteins control IGF-II biosynthesis during late mammalian development.
Specific RNA-binding proteins are emerging as regulators of cytoplasmic mRNA events such as translatability, stability, and localization. Several examples of these types of regulatory events have been reported in studies of invertebrate embryogenesis and amphibian oogenesis, in which the 3′ untranslated region (3′ UTR) has been identified as a repository of regulatory elements (reviewed in reference 35). It is anticipated that similar mechanisms operate during mammalian development, since important physiological roles for RNA-binding proteins have been discerned from deletions of the DAZ and RBM genes leading to azoospermia (9, 26) and from a point mutation in the FMR1 gene resulting in the fragile X mental retardation syndrome (7). RNA-binding proteins often contain one or more RNA-binding motif such as the RNA recognition motif (RRM) and the K homology (KH) domain (reviewed in reference 28), which may either ensure increased specificity towards a single RNA molecule or provide an ability to bind different molecules simultaneously. Moreover, solution structures of the N-terminal RRM domain of the human U1A protein in complex with its own pre-mRNA and of the first KH domain of FMR1 suggest that flexible loop regions provide discriminating binding surfaces for RNA recognition (1, 20).
Insulin-like growth factor II (IGF-II) is a fetal growth factor with auto- and paracrine modes of action. In the mouse, lack of IGF-II results in a small but apparently normal progeny (3), whereas an increased IGF-II dose is more detrimental (13, 30). In humans, increased levels of IGF-II are associated with the Beckwith-Wiedemann syndrome, which is characterized by a disproportionate overgrowth of the fetus and malformations (32). IGF-II expression is controlled by parental imprinting, since only the paternal allele is expressed in most tissues (8). However, there is no evidence that imprinting is part of a rapid temporal or spatial regulation of IGF-II. Changes in IGF-II production are likely to be executed at the posttranscriptional level, so rapid adjustments can take place at crucial developmental stages. The significance of posttranscriptional regulation of IGF-II production is reflected in an array of mRNAs transcribed from the IGF-II gene. The IGF-II gene generates overlapping primary transcripts that result in multiple mRNAs with identical coding and 3′ UTRs but distinct 5′ UTRs (31). Previously, we have shown that the major fetal mRNAs encoding preproIGF-II in the human rhabdomyosarcoma cell line RD are translated in a differential manner (22, 23). The 4.8-kb leader 4 mRNA with a 5′ UTR of 100 nucleotides is constitutively translated, whereas the abundant 6.0-kb leader 3 mRNA comprising an 1,170-nucleotide cytidine-rich (48%) 5′ UTR is stored in a 100S particle that can be translated in growing cells. Moreover, it has been demonstrated that the mouse IGF-II leader 3 mRNA homologue switches from a translated to a repressed state between embryonic day 11.5 (E11.5) and E12.5 (21). The distinct translational behavior of the two IGF-II mRNAs is likely to reflect the presence of specific trans-acting factors.
In this study, we identified and characterized a family of fetal RNA-binding proteins that exhibits high affinity and multiple attachments to the IGF-II leader 3. The RNA-binding proteins contain two RRMs and four KH domains and exhibit cell-specific and cell contact-dependent subcytoplasmic localization. Moreover, they are able to repress translation of leader 3 reporter mRNAs in vivo and are expressed in a burst at E12.5 in mice followed by a decline towards birth.
MATERIALS AND METHODS
Preparation of detergent-solubilized cytoplasmic extracts.
A frozen cell pellet of approximately 5 × 107 RD rhabdomyosarcoma cells was resuspended in 1 ml of lysis buffer (20 mM Tris-HCl [pH 8.4], 140 mM KCl, 1.5 mM MgCl2, 0.5% Nonidet P-40 [NP-40], 0.5 mM dithiothreitol [DTT]) and centrifuged at 14,000 × g for 10 min at 4°C. Glycerol was added to the supernatant at a final concentration of 5%, and the cytoplasmic extract was stored in aliquots containing 5 to 7 μg of protein per μl at −80°C.
In vitro RNA transcription.
RNA was generated by T7 RNA polymerase-directed in vitro transcription from templates inserted downstream from a T7 RNA polymerase promoter, and transcripts were purified by denaturing gel electrophoresis. In the case of full-length leader 3 RNA, purification was achieved by gel filtration in Microspin S300 spin columns (Pharmacia). Radiolabelled RNA for UV cross-linking was synthesized to a specific activity of 30 Ci of uridine/mmol by including 30 μCi of [α-32P]UTP and 1 nmol of unlabelled UTP in a 10-μl transcription reaction mixture, and 1.5 μg of Escherichia coli tRNA was added as a carrier. Radiolabelled RNA for the mobility shift assay was synthesized to a specific activity of 150 Ci of uridine/mmol.
For affinity purification, full-length biotinylated leader 3 RNA was prepared by in vitro transcription in the presence of a 1:20 molar ratio of biotin-16-UTP to UTP followed by spin column chromatography. Streptavidin-coated paramagnetic beads (0.8 mg; Promega) were washed in phosphate-buffered saline (PBS) (10 mM sodium phosphate [pH 7.2], 137 mM NaCl, 3 mM KCl) and incubated with 300 pmol of biotinylated leader 3 RNA and 30 μg of tRNA in PBS for 30 min at room temperature. Unbound RNA was removed from the leader 3 RNA-coated beads by washing with PBS.
UV cross-linking and mobility shift analysis.
In UV cross-linking assays, RD cytosolic extract containing 15 to 20 μg of protein was incubated with 100 nCi of the appropriate RNA for 25 min at room temperature in 20 mM Tris-HCl (pH 8.0)–140 mM KCl–4 mM MgCl2–0.75 mM DTT–0.1% NP-40. Samples were irradiated with 254-nm-wavelength light for 30 min at 5.4 J/cm2 on ice in a Stratalinker 1800 (Stratagene). Excess probe was removed by digestion with 80 U of RNase T1 (Amersham) at 37°C for 25 min. Samples were analyzed in 10% polyacrylamide–sodium dodecyl sulfate (SDS) gels followed by autoradiography. The leader 3-specific p69 signal was eliminated by the inclusion of 30 ng of unlabelled leader 3.
In mobility shift analyses, 2 nCi of denatured and snap-cooled RNA was incubated with recombinant IMP and 100 ng of E. coli tRNA for 10 min at 30°C in 20 mM Tris-HCl (pH 8.0)–150 mM KCl–2 mM MgCl2–0.1% Triton X-100–5% glycerol. Samples were cooled on ice and separated in 5% polyacrylamide gels run in 90 mM Tris-borate (pH 8.3)–50 mM KCl–2 mM MgCl2 followed by autoradiography.
Affinity purification and peptide sequencing of IGF-II mRNA-binding proteins (IMPs).
Detergent-solubilized cytoplasmic extracts from RD cells, prepared as described above, were centrifuged in 1-ml aliquots at 40,000 rpm for 45 min in a Beckman type 80 Ti rotor, and each pellet was resuspended in 800 μl of buffer A (20 mM Tris-HCl [pH 7.8], 20 mM KCl, 1.5 mM MgCl2, 0.05% NP-40, 5% glycerol, 0.5 mM DTT). The resuspended pellets were applied to a P11 phosphocellulose column (Whatman), and the flowthrough was collected. The ultracentrifugation and the phosphocellulose column resulted in a fivefold purification.
The salt composition and volume of the phosphocellulose column eluate were adjusted to give a final volume of 1.4 ml for each 1 ml of original cytoplasmic extract and a composition corresponding to buffer B (20 mM Tris-HCl [pH 8.0], 160 mM KCl, 4 mM MgCl2, 0.025% NP-40, 2.5% glycerol). The adjusted solution was incubated at room temperature for 15 min with 0.8 mg of leader 3-coated beads. Then, 350 U of heparin was added, and the mixture was incubated for a further 10 min. The supernatant was discarded, and the beads were washed at room temperature with 600 μl of buffer B for 30 s, with 400 μl of buffer B for 30 s, and with 200 μl of buffer B for 2 min.
The adsorbed protein was eluted from the beads by incubation in 80 μl of elution buffer (12.5 mM Tris-HCl [pH 6.8], 0.5% SDS, 2.5% glycerol) for 3 min at 44°C and subsequently in 40 μl of elution buffer for 1 min. The eluted proteins from 6 ml of cytoplasmic extract were combined and concentrated. DTT was added to a concentration of 0.1 M, and proteins were separated by polyacrylamide-SDS gel electrophoresis, followed by electroblotting onto a Problott membrane (Applied Biosystems), and stained with amido black. The predominant band with an apparent molecular mass of 67 kDa was excised and destained, and the free sites of the membrane were blocked by incubation with polyvinylpyrrolidone 30. Following digestion with trypsin for 18 h at 37°C, the resulting peptides were extracted in 10% trifluoroacetic acid and purified by high-performance liquid chromatography with a 2.1- by 150-mm C8 column (Vydac) with a gradient of acetonitrile in 0.1% trifluoroacetic acid. The peptides were analyzed by matrix-assisted laser desorption mass spectrometry (Biflex; Bruker-Franzen) and automated protein sequencing (Procise 494A; Perkin-Elmer ABD).
Isolation of cDNAs.
By using degenerate oligodeoxynucleotide primers derived from tryptic peptides 4 and 5 (see Results), three different 180-bp cDNA probes corresponding to IMP-1, IMP-2, and IMP-3 mRNAs were generated by reverse transcription (RT)-PCR on total RNA from RD cells. Subsequently, several IMP-2 and IMP-3 clones were isolated by screening a human placenta cDNA library (Clontech), one of which contained the entire reading frame of IMP-3. The IMP-2 clones from the screening contained only the 3′ half of the IMP-2 reading frame, but an expressed sequence tag (EST; H66979), obtained by comparison with the full-length IMP-3 cDNA, was found to contain the region around the IMP-2 initiation codon, and the entire IMP-2 reading frame was derived by PCR from the placenta cDNA library.
Information on the 3′ end of the IMP-1 reading frame was obtained from an EST (accession no. AA646035) containing the sequence from the codon for amino acid 132 and 350 nucleotides into the 3′ UTR. RT-PCR on RNA from RD cells using a degenerate upstream primer corresponding to amino acids 1 to 7 provided most of the remaining reading frame. The sequence around the initiation codon was obtained by inverse PCR on an IMP-1 genomic clone purchased from Genome Systems. A full-length IMP-1 reading frame was obtained by RT-PCR on RNA isolated from RD cells.
Expression and purification of recombinant IMPs.
Since recombinant N-terminal glutathione S-transferase (GST)-tagged and N- and C-terminal His6-tagged IMP-1 exhibits poor RNA binding, IMP-1, IMP-2, and IMP-3 proteins were purified by using the IMPACT system (New England Biolabs). The IMP open reading frames were inserted into NdeI- and SapI-cleaved pCYB1 vector. IMPs were expressed in E. coli BL21/DE3 cells containing pRI952 and a pCYB1-IMP plasmid. pRI952 overexpresses the tRNAs encoded by the ileA and argU genes and thereby assists translation of open reading frames containing rare isoleucine and arginine codons (10).
Cell culture and transient transfections.
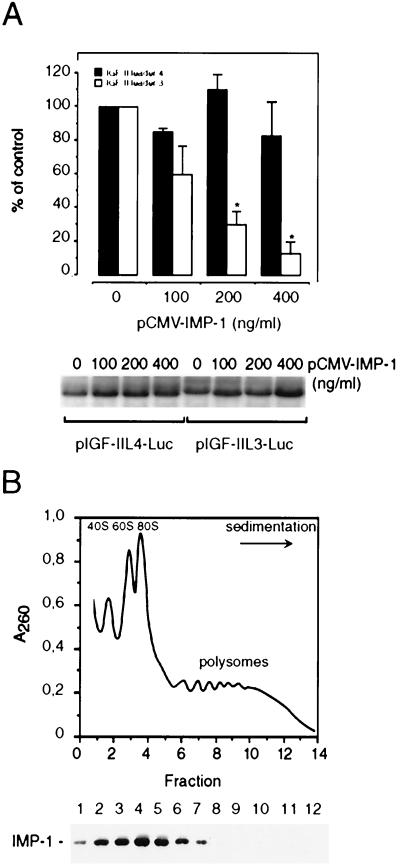
RD rhabdomyosarcoma cells and NIH 3T3 cells were obtained from the American Type Culture Collection and routinely maintained in RPMI 1640 supplemented with 10% fetal calf serum or in Dulbecco’s modified Eagle medium with 10% calf serum. Cells were transiently transfected with Lipofectamine (Life Technologies) or Superfect reagent (Qiagen) in accordance with the manufacturer’s instructions. For determination of the effect of IMP-1 and IMP-3 on translation of IGF-II leader 3- and leader 4-luciferase fusion constructs, 30,000 cells/cm2 were seeded in 24-well multidishes 24 h prior to transfection. The cells were cotransfected with 400 ng of pGL3 control vector, pGL3-IGF-II-L3, or pGL3-IGF-II-L4 per ml and from 100 to 400 ng of pCMV-IMP-1 or pCMV-IMP-3 and pCMV-β-galactosidase per ml to a total of 1,600 ng of DNA/ml. After 48 h, luciferase activity was measured. The levels of IGF-II leader 3- or IGF-II leader 4-luciferase mRNAs were determined by Northern analysis as described below. For immunocytochemistry, 30,000 cells/cm2 were seeded in 30-mm-diameter glass dishes and transfected with 200 ng of pCMV-IMP-1 or pCMV-IMP-3 per ml as described above. Staining was performed 48 h after transfection.
IGF-II leader 3 and leader 4 reporter constructs, pGL3-IGF-II-L3 and pGL3-IGF-II-L4, were generated from pGL3 vectors (Promega) by insertion of the complete 1,164-bp and 94-bp leader exons between the simian virus 40 promoter and the luciferase coding region in the BamHI and NcoI sites of the vector. The IMP-1 expression vector, pCMV-IMP-1, was generated by insertion of the cDNAs into pCMV5, which is a derivative of pCMV1 (2).
Antibodies and Western analysis.
Peptide-specific antibodies were generated against C-terminal QSNQAQARRK-OH and QSGPPQSRRK-OH sequences of IMP-1 and IMP-3, respectively, essentially as described previously (25). In Western analysis, cellular proteins were separated in a 10% polyacrylamide–SDS gel and transferred to polyvinyl difluoride Immobilon-P membranes (Millipore). After blocking, filters were incubated overnight at 4°C with anti-IMP-1 in blocking solution and with horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (IgG) in blocking solution for 1 h at room temperature. Bound antibody was detected with enhanced chemiluminescence reagents in accordance with the manufacturer’s instructions (Pierce).
Polysome isolation.
RD rhabdomyosarcoma cells (3 × 107 cells) were lysed in 500 μl containing 20 mM Tris-HCl (pH 8.5), 1.5 mM MgCl2, 140 mM KCl, 0.5 mM DTT, 0.5% NP-40, 1,000 U of RNasin (Promega) per ml, and 0.1 mM cycloheximide. The lysate was centrifuged at 10,000 × g for 10 min, and the supernatant was applied to a linear 20 to 47% sucrose gradient in 20 mM Tris-HCl (pH 8.0)–140 mM KCl–5 mM MgCl2 (22). Centrifugation was carried out at 40,000 rpm for 2 h 15 min in a Beckman type SW 41 rotor. Fractions of 1 ml were collected with concomitant measurement of the absorbance at 260 nm, followed by precipitation of sedimenting proteins in 10% trichloroacetic acid and Western analysis as described above.
Northern analysis.
Total RNA was isolated from adult NMRI mice and embryos. For staging of embryos, the appearance of the vaginal plug was considered day 0.5. RNA was isolated by the guanidinium thiocyanate method as described previously (5). RNA was denatured in glyoxal-dimethyl sulfoxide and separated in 1% agarose gels, transferred to Hybond-N membrane (Amersham), and hybridized with a 32P-labelled cDNA probe. Final washes were performed at 65°C in 0.1× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) containing 0.1% SDS. Autoradiography was done from 16 to 48 h, and the hybridization signals were visualized and quantified with a BAS 2000 bioimager (Fuji). cDNA probes for the mouse IMP-1, IMP-2, and IMP-3 mRNAs were complementary to nucleotides 680 to 1238, 50 to 650, and 680 to 1238, respectively, of the mouse DNA sequence reading frames.
Immunocytochemistry and immunohistochemistry.
Immunocytochemistry was performed as described before (24). Briefly, cells were plated on glass dishes and transfected as described above. The cells were fixed in methanol, washed, and incubated with 10% goat serum, before anti-IMP-1 or -IMP-3 was added for 60 min at 20°C in PBS with 0.5% Tween. After the cells were washed, they were incubated with fluorescein isothiocyanate-conjugated goat anti-rabbit-IgG (Sigma F-1262) for 30 min at 20°C and mounted in 90% glycerol with 0.25% 1,4-diazabicyclo[2.2.2]octane. For control stainings, the specific antibody was replaced with rabbit serum. Moreover, the peptide-specific antibodies could be preadsorbed with the peptide that was used for immunization. The percentage of cells with localized IMP was determined by microscopic examination of transfected cells in 10 consecutive visual fields (150 to 300 cells) at ×400 magnification.
Immunostaining of tissue sections from mouse embryos (E12.5 to E15.5), adult mice (8 weeks old), human fetal muscle (14 and 38 weeks), and human term placenta was performed essentially as described previously (33, 34). Cryostat sections were fixed in precooled acetone for 15 min before the antiserum (diluted 1:100) was applied to the sections and they were incubated at room temperature for 1 h. After the sections were rinsed, they were incubated with fluorescein isothiocyanate-coupled secondary antibodies (1:50; DAKO) for 30 min. Incubations with both primary and secondary antibodies were performed in 0.05 M Tris-HCl (pH 7.2), and rinses were done in 0.05 M Tris-HCl (pH 7.2) containing 0.15 M NaCl and 0.05% Tween 20. For control sections, the specific antisera were omitted or replaced with preimmune rabbit antiserum. Moreover, the inhibitory effect of simultaneously incubating the antisera with the respective peptides (20 μg/ml) was examined. The slides were mounted in buffered glycerol and examined with an Axiovert inverted microscope connected to a PentaMAX chilled charge-coupled device camera (Princeton Instruments).
RESULTS
Identification of a leader 3-specific IGF-II mRNA binding protein (IMP).
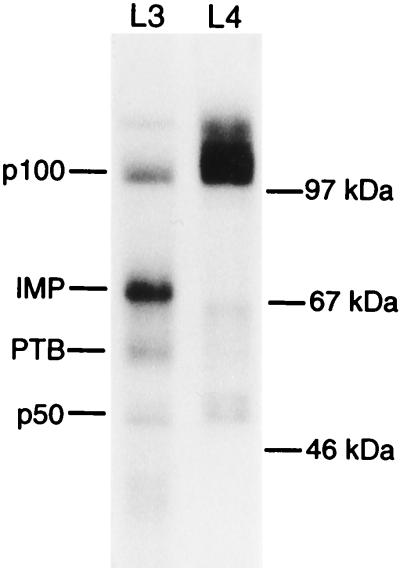
To identify a trans-acting factor that mediates the translational discrimination between IGF-II mRNAs, a UV cross-linking approach at physiological salt concentrations was employed. Randomly 32P-labelled leader 3 or leader 4 RNA was mixed with a detergent-solubilized cytoplasmic extract from the human rhabdomyosarcoma cell line RD and irradiated at 254 nm, followed by exhaustive RNase T1 digestion and SDS-polyacrylamide gel electrophoresis. The resulting autoradiograph in Fig. 1 shows that leader 3 RNA exhibits a strong cross-linking signal at 69 kDa which is absent with leader 4 RNA. Besides the 69-kDa band, cross-links at about 50 and 100 kDa originate from both leader 3 and 4 RNA, and a 60-kDa band has previously been identified as the polypyrimidine-tract binding protein (PTB) (11). The 50- and 100-kDa bands appear in cross-links with RNAs other than the IGF-II leaders (results not shown). We named the specific IGF-II leader 3 mRNA-binding protein(s) IMP.
FIG. 1.
UV cross-linking of IGF-II leaders 3 and 4 to an RD cytoplasmic extract. An autoradiograph from a UV cross-linking analysis of human IGF-II leader 3 (L3) and leader 4 (L4) in the presence of a detergent-solubilized RD cytoplasmic extract is shown. Cross-linked species and molecular size markers are indicated on the left and right, respectively.
Purification, sequencing, and cDNA cloning of IMP reveal a family of closely related RNA-binding proteins.
Since IMP can be cross-linked to IGF-II leader 3 RNA in a specific manner, IMP is a candidate for mediating the differential translatability of IGF-II mRNAs. Therefore, IMP was purified from RD cells by an RNA affinity approach, in which leader 3 RNA was biotinylated so that the RNA-protein complex could be adsorbed to streptavidin beads. A final purification in a preparative SDS-polyacrylamide gel was followed by transfer to a polyvinylidene difluoride membrane, in situ trypsin digestion of the IMP band, and sequencing of tryptic peptides as described in Materials and Methods. Based on the sequence of five tryptic fragments, (i) LYIGNLESVTPADLEK, (ii) ISYS GQFLVK, (iii) ITISSLQDLTYNPER, (iv) MVIIXXX PEAQFK, and (v) TVNELQNLTAAEVVVPR, degenerate PCR was used to obtain cDNA probes that were employed in library screening. During the course of cDNA sequencing, it became apparent that a family of at least three closely related species exists, and Fig. 2A shows an alignment of three members of this family which we have named IMP-1, IMP-2, and IMP-3. IMP-1 encompasses the five tryptic fragments, so we infer that IMP-1 is the predominant protein purified from RD cells by the RNA affinity approach.
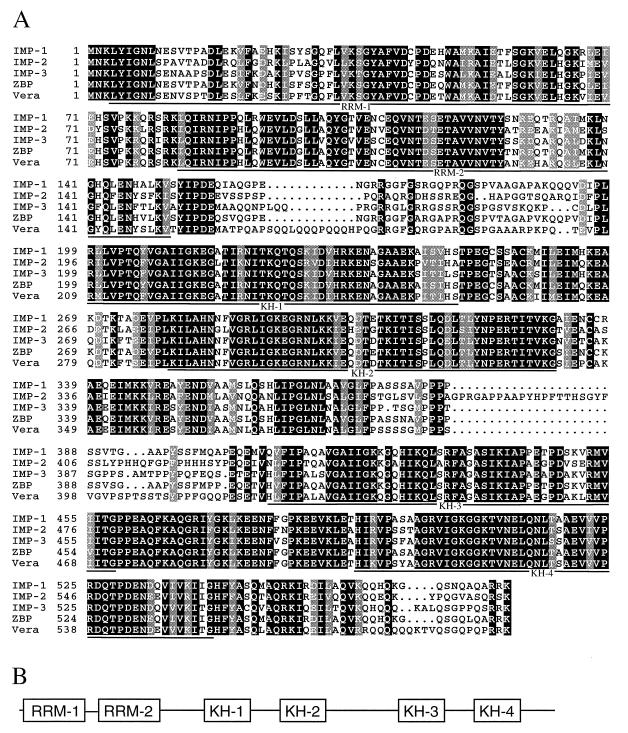
FIG. 2.
Modular structure of the human IMP family. (A) Alignment of three members of the IMP family, the zipcode-binding protein (ZBP), and the Vera protein, with black and grey boxes indicating amino acid identity and similarity, respectively. Horizontal lines below the alignment indicate the predicted two RRMs and four KH domains. (B) Schematic diagram of RRMs and KH domains indicated in panel A.
The closely related IMP family members, with expected molecular masses of 63, 66, and 64 kDa and an overall sequence identity of 59%, exhibit six characteristic RNA-binding modules, namely two RRMs (RRM-1 and -2) and four KH domains (KH-1 to -4), which are outlined in Fig. 2B. The alignment in Fig. 2A shows that the sequence similarity between members is greater for the KH domains than for the respective RRMs. There is also similarity of sequence and spacing in regions between the two RRMs and between KH domains 1 and 2 and domains 3 and 4. In contrast, regions between RRM-2 and KH domain 1 and between KH domains 2 and 3 vary considerably.
A database search reveals that IMP-1 is orthologous to the chicken zipcode-binding protein (95% identity), which has been implicated in β-actin mRNA sorting (27), and to the mouse c-myc coding region determinant-binding protein (CRD-BP;99% identity; accession no. AF061569). The IMP-3 sequence is identical to the KOC protein sequence derived from a pancreatic tumor cDNA (19), but the functional significance of the KOC protein is unknown. Moreover, IMP-3 is orthologous to the Vera protein (83% identity) in Xenopus laevis oocytes that may be involved in cytoplasmic localization of Vg1 mRNA via attachment to its 3′ UTR (12, 15).
IGF-II leader 3 RNA exhibits multiple IMP-1 attachment sites.
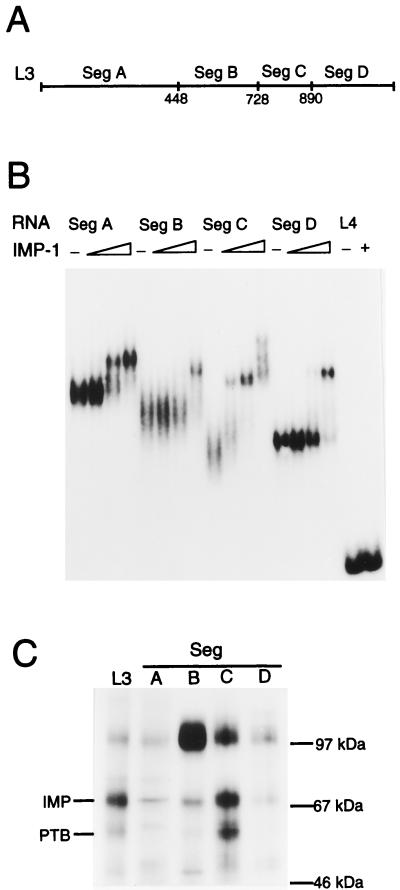
To identify IMP-1 binding sites in leader 3 RNA and estimate their apparent Kds, untagged recombinant IMP-1 was expressed in E. coli by an intein-based approach (see Materials and Methods for details), since both N- and C-terminal tagged recombinant IMP-1 exhibited poor RNA binding. The leader RNA was divided into four RNA segments (Fig. 3A) covering positions 1 to 448 (segment A), 449 to 728 (segment B), 729 to 890 (segment C), and 891 to 1164 (segment D). The four leader 3 RNA segments were subjected to mobility shift analysis, and Fig. 3B is the autoradiograph showing that all four RNA segments are able to bind to recombinant IMP-1 with Kds in the subnanomolar range. Segment C exhibits the highest affinity, with an apparent Kd of 0.1 nM, and two additional attachments appear at about 1 nM. In contrast, 10 nM IMP-1 was unable to associate with the full-length IGF-II leader 4 RNA, which implies that the mobility shifts are due to specific binding. We conclude that IGF-II leader 3 RNA contains at least six binding sites for IMP-1 and that IGF-II leader 4 RNA contains none.
FIG. 3.
Segments of IGF-II leader 3 RNA bind IMP. (A) Map of the RNA segments obtained from leader 3. Numbers below the map refer to the numbering of human IGF-II exon 5 containing 1,164 nucleotides of leader 3 RNA (31). (B) Mobility shift analysis of leader 3 RNA segments and full-length leader 4 RNA with recombinant IMP-1. In experiments with the four segments A to D, the concentrations of recombinant IMP-1 are 0, 0.07, 0.3, and 1 nM, respectively, whereas 0 and 10 nM are used in the leader 4 experiment. (C) UV cross-linking of full-length leader 3 RNA (L3) and segments A to D to a detergent-solubilized RD cytoplasmic extract. Cross-linked species and molecular size markers are indicated on the left and right, respectively.
The conclusion from the mobility shift analysis was corroborated by a UV cross-linking experiment between the four RNA segments, A to D, and an RD cytoplasmic extract. Figure 3C shows that segment C cross-links more efficiently than the other RNA segments, thus reflecting the binding affinities of recombinant IMP-1 in the mobility shift experiment. As reported previously (11), segment C also exhibits the greatest cross-linking efficiency with regard to PTB.
Since recombinant IMP-2 and IMP-3 bind to RNA segment C with an affinity similar to that exhibited by IMP-1 in mobility shift experiments (results not shown), we infer that the three members of the IMP family bind similarly to RNA segment C.
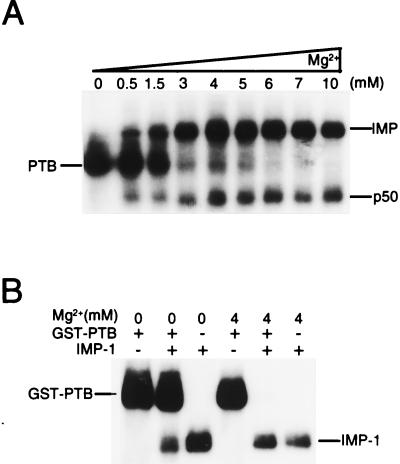
IMP-1 and PTB compete for binding to IGF-II leader 3 RNA in an Mg2+-dependent manner.
The cross-linked species that result from irradiating a mixture of leader 3 and an RD cytoplasmic extract are dependent on the Mg2+ concentration. The autoradiograph in Fig. 4A shows that PTB is the predominant species at low Mg2+ concentrations, whereas IMP dominates at higher Mg2+ concentrations in an apparently mutually exclusive manner. Since IMP-1 and PTB bind to segment C (positions 729 to 890) with high affinity, their binding sites could be overlapping. To examine this possibility, segment C was cross-linked separately to recombinant IMP-1 and GST-PTB and to a combination of both, in the absence and presence of 4 mM Mg2+ (Fig. 4B). IMP-1 and GST-PTB cross-link separately to RNA segment C at both 0 and 4 mM Mg2+, but GST-PTB is preferentially cross-linked in the absence of Mg2+ (second lane from the left) whereas IMP-1 is exclusively cross-linked at 4 mM Mg2+ (second lane from the right) when both proteins are present in equimolar amounts.
FIG. 4.
Mg2+-dependent competition of PTB and IMP. (A) UV cross-linking of full-length leader 3 RNA to a detergent-solubilized RD cytoplasmic extract in the presence of increasing Mg2+ concentrations. The major cross-linked species are indicated to the left if abundant at a low Mg2+ concentration and to the right if preferred at a high Mg2+ concentration. Moreover, the putative p50 major core protein (6) is indicated. (B) UV cross-linking of segment C to recombinant GST-PTB (8 nM) and IMP-1 (8 nM) at 0 and 4 mM Mg2+. Lanes 2 and 5 are the result of UV cross-linking in the presence of both proteins at 0 and 4 mM Mg2+, respectively.
IMP-1 inhibits translation of a leader 3 reporter mRNA in vivo.
Leader 3 reporter mRNAs cannot be translated in rabbit reticulocytes or wheat germ extracts (23), so a causal relationship between IMP-1 expression and human IGF-II leader 3 mRNA translatability was examined in vivo. NIH 3T3 cells, with their low background of endogenous IMP, were cotransfected with an expression plasmid directing IMP-1 synthesis and a firefly luciferase reporter plasmid in which leader 3 is inserted between the simian virus 40 promoter and the luciferase coding region. The equivalent experiment in which leader 3 has been replaced with leader 4 was carried out in parallel as a negative control, since Fig. 3B showed that 10 nM IMP-1 was unable to bind leader 4 in vitro. Figure 5A depicts the dose-response relationship between the amount of added expression plasmid and the obtained luciferase activity. IMP-1 could affect various transcriptional and posttranscriptional stages during formation or decay of reporter mRNAs, so the levels of leader 3- and leader 4-luciferase mRNAs were examined by Northern analysis (Fig. 5A, lower panel). Since the levels of reporter mRNAs were unaffected by the presence of IMP-1, translation of leader 3-luciferase mRNA is repressed by IMP-1 in a dose-dependent manner, whereas the translatability of leader 4-luciferase mRNA is unaffected by up to 400 ng of IMP-1 cDNA per ml.
FIG. 5.
IMP-1 inhibits translation of a chimeric IGF-II leader 3-luciferase transcript. (A) NIH 3T3 cells were transiently transfected with pIGF-IIL4-Luc or pIGF-IIL3-Luc reporter constructs and increasing concentrations of pCMV-IMP-1 as indicated. The graph depicts the dose-response relationship between the amount of pCMV-IMP-1 and the obtained luciferase activity. The results are given as a percentage of control (mean and standard error of the mean of two to four independent experiments), and the asterisks indicate statistically significant differences at a P of <0.05. A representative Northern analysis indicating the levels of pIGF-IIL4-Luc or pIGF-IIL3-Luc reporter mRNAs is shown below the bar graph. (B) Subcellular distribution of IMP-1. A detergent-solubilized RD cytoplasmic extract was applied to a linear 20 to 47% sucrose gradient. The graph shows the A260 profile of the sedimenting lysate. The positions of the 40S, 60S, 80S, and polysomal complexes are indicated. Fractions were precipitated in 10% trichloroacetic acid, and proteins were separated by SDS-polyacrylamide gel electrophoresis, followed by Western analysis with an anti-IMP-1 antibody (lower panel).
We also examined whether IMP-1 was associated with endogenous translating mRNAs or with mRNPs. A detergent-solubilized RD cytoplasmic extract was subjected to sucrose gradient ultracentrifugation, and sedimenting IMP-1 was detected by Western analysis of the gradient fractions. Figure 5B shows that IMP-1 is not polysome associated but sediments in the 40S to 150S range, implying that IMP-1 is predominantly associated with mRNPs in macromolecular assemblies. We conclude that IMP-1 is able to repress translation of a leader 3-luciferase chimeric mRNA and cosediments with untranslated mRNPs.
IMP-1 is localized in subcytoplasmic domains.
The subcellular distribution of IMP-1 was characterized immunocytochemically in transiently transfected human RD rhabdomyosarcoma cells and NIH 3T3 mouse embryo fibroblasts. Figure 6 shows the cell-specific cytoplasmic distribution of IMP-1 in RD and NIH 3T3 cells.
FIG. 6.
Subcellular localization of IMP-1. The subcellular distribution of IMP-1 was characterized immunocytochemically. RD or NIH 3T3 cells were transiently transfected with pCMV-IMP-1. After 48 h, the cells were fixed and incubated with rabbit anti-IMP-1 and stained with fluorescein isothiocyanate-conjugated anti-rabbit IgG. (a to c) Subcellular distribution of IMP-1 in RD cells; (d to f) distribution of IMP-1 in NIH 3T3 cells. (a) In RD cells, staining was observed below the plasma membrane. (b) In clustered groups of cells, IMP-1 was polarized to the free edges of the cells; (c) in myotubes, staining was distributed along the circumference of the complete myotube. (d) In NIH 3T3 cells, staining was evenly distributed in the cytoplasm in clustered cells. (e) In areas with low cell density, staining was observed in the lamellipodia of the leading edge and in the perinuclear region. (f) In completely dispersed cells, IMP-1 was found in discrete foci scattered around the nucleus, below the plasma membrane, and in the lamellipodia. Magnification, ×1,000. Arrows in panels a and d indicate nontransfected cells that served as negative controls for the immunostaining. See Materials and Methods for additional controls.
In RD cells, staining beneath the plasma membrane was invariably observed. Whereas dispersed RD cells exhibited bilateral submembranous staining (Fig. 6a), IMP-1 was polarized to the free edges of clustered cells (Fig. 6b). Moreover, RD cells have the capacity to form myotubes, in which staining was intensely distributed along the circumference of the complete myotube (Fig. 6c).
In NIH 3T3 cells, staining in areas with confluent growth-arrested cells and in areas with dispersed proliferating cells was distinct. In confluent cells, staining was evenly distributed in the cytoplasm in 93% of the cells (Fig. 6d). In areas with a low cell density, staining was located in the lamellipodia of the leading edge, in the perinuclear region, and beneath the plasma membrane in 22% of the cells (Fig. 6e). In some NIH 3T3 cells, IMP-1 was even found in discrete foci scattered around the nucleus, beneath the plasma membrane, and in the lamellipodia (Fig. 6f). If newborn calf serum was replaced with 10% fetal calf serum, localization of IMP in subcytoplasmic domains was increased to 49% of the cells in confluent areas and to 86% of the dispersed cells. We conclude that IMP-1 is localized in the cytoplasm and that the subcytoplasmic localization is cell specific and regulated by cell contact and growth. Immunocytochemical experiments with IMP-3 gave identical results (data not shown).
IMP-1, IMP-2, and IMP-3 are expressed in mammalian embryos.
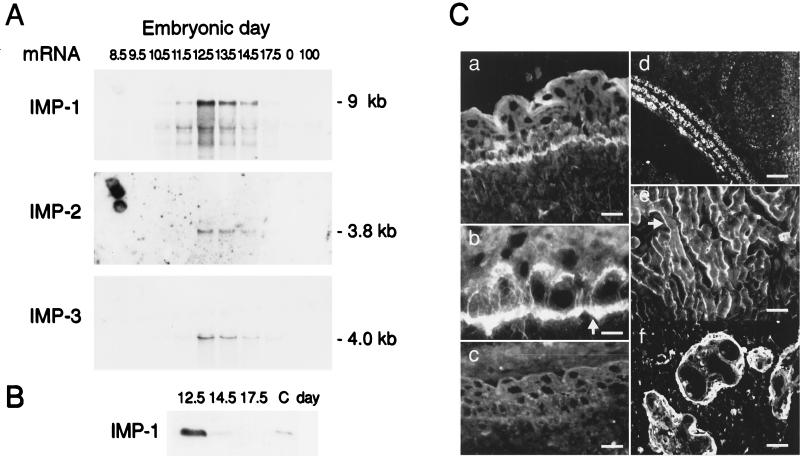
In an igf2(p−) mouse, growth deficiency becomes phenotypically manifest at E11 (3), whereas the effect of an increased IGF-II dosage is apparent at E13 (30). Moreover, the mouse IGF-II leader 3 mRNA homologue switches from a translated to a repressed state between E11.5 and E12.5 (21). Therefore, the expression of mouse homologues of IMP-1, IMP-2, and IMP-3 mRNAs was examined by Northern analysis of total RNA from different developmental stages (Fig. 7A). The results show that the three IMP mRNAs exhibit the same expression patterns in terms of timing by rising sharply to a peak at E12.5 before declining towards birth. An examination of the level of IMP-1 protein by Western analysis on days 12.5, 14.5, and 17.5 (Fig. 7B) shows a similar decline, thus demonstrating an apparent proportionality between IMP-1 mRNA and protein levels. Moreover, several ESTs reveal that the IMP family is expressed at the two-cell stage in mouse embryogenesis, indicating that there is a period of IMP expression earlier than that shown in Fig. 7A. The three IMP mRNAs could not be detected in adult mouse tissues such as the adrenal glands, brain, gut, heart, kidney, liver, lung, muscle, ovaries, spleen, stomach, and uterus (results not shown).
FIG. 7.
Expression of IMP-1, IMP-2, and IMP-3 in mouse and human embryos. (A) Expression of mouse equivalents of IMP-1, IMP-2, and IMP-3 mRNAs was examined by Northern analysis of total RNA from different embryonic (8.5 to 17.5) and postnatal (100) days as indicated. The amount of RNA in each lane is normalized to rRNA content. (B) Western analysis of IMP-1 in mouse embryos at E12.5, E14.5, and E17.5. The amounts of loaded protein in the three lanes are identical, and lane C contains a positive control sample. (C) Immunohistochemical staining of IMP-1 in mouse and human embryos. (a and b) Localization of IMP-1 in mouse epidermis at E12.5. (a) Staining is visible in the basal layer of the developing epidermis at the basal plasma cell membrane, as indicated by the arrow in panel b. (c) Parallel section where the antiserum was preadsorbed with the peptide used for immunization. (d) Localization of IMP-1 in developing mouse muscle at E15.5. (e) Localization of IMP-1 in fetal human skeletal muscle cells (38 weeks). The submembranous staining is indicated by an arrow. (f) Staining of human term placenta shows the expression of IMP-1 in the trophoblast cells. Scale bars, 11 (a), 7 (b), 11 (c), 20 (d), 11 (e), and 11 (f) μm.
The tissue distribution of IMP-1 was examined by immunohistochemical staining of frozen sections of mouse embryos (E12.5 to E15.5). Positive immunostaining was found at the basal layer of the developing epidermis of the skin, where IMP-1 was located at the basal plasma cell membrane (Fig. 7C, panels a and b). No immunostaining of the dermis layer was seen. This distinct polarized staining pattern was also seen in the developing epithelia of the lung and the intestine. Developing muscle cells exhibited prominent immunoreactivity, and IMP-1 was detected in myoblasts and myotubes in the myotome at E12.5 and later in the developing limb. Intense staining was observed during the process of myotube formation and maturation of myofibers, as illustrated by the developing panniculus carnosus muscle layer of the skin (Fig. 7C, panel d). The intensity of the muscle cell immunostaining declined shortly after birth, and no immunostaining was observed in tissues from 8-week-old mice. Finally, scattered cells in the liver exhibited positive immunoreactivity, whereas no staining was detected in the brain (data not shown).
Developing human skeletal muscle cells (14 and 38 weeks) also exhibited positive immunoreactivity (Fig. 7C, panel e). As in RD cells, IMP-1 was located beneath the plasma membrane along the circumference of myoblasts and myotubules. Finally, positive immunostaining was found in the trophoblasts of human term placenta (Fig. 7C, panel f). Moreover, an examination of dot blots containing RNA from human fetal tissues of mixed first-trimester age and from placenta (Clontech) revealed that IMP-1, IMP-2, and IMP-3 mRNAs are expressed in fetal liver, lung, kidney, thymus, and placenta. Whereas IMP-1 mRNA was expressed at moderate levels in the examined fetal tissues, IMP-2 mRNA was most abundant in fetal liver and IMP-3 mRNA was most abundant in term placenta. We conclude that IMP expression is temporally and spatially regulated.
DISCUSSION
In this study, we show that a family of embryonic RNA-binding proteins, containing two RRMs and four KH domains, associates specifically with the 5′ UTR of the human 6.0-kb IGF-II leader 3 mRNA. The RNA-binding proteins, which we have named IMPs, localize subcellularly in the cytoplasm and repress translation of a chimeric leader 3 -luciferase mRNA in vivo. Expression is developmentally regulated, and mouse IMP is expressed predominantly at E12.5 at the transition between organogenesis and fetal growth.
Previously, we have shown that the human 6.0-kb IGF-II leader 3 mRNA is stored in a 100S particle and that translation of this particular mRNA reflects the growth status of rhabdomyosarcoma RD cells (23). It is likely that at least six IMP molecules associate with the full-length leader 3, but the exact number of copies may actually be higher due to the employed segmentation approach which could sever additional attachment sites. The multiple binding sites could augment the specificity and/or strength of the biological response in a manner similar to the translational repression of oskar mRNA by bruno in Drosophila (16). The pyrimidine-rich leader 3 RNA segment C (positions 729 to 890) exhibits the highest affinity (Kd, ∼0.1 nM) towards recombinant IMPs, and it is more conserved (93%) than the translated region (87%) when human and mouse sequences are compared. The segment also binds PTB with high affinity (11) (Fig. 3C and 4), and IMP-1 and PTB exhibit mutually exclusive attachments. Regardless of the Mg2+ concentration, both proteins are able to bind to the RNA target in segment C, but their competitive abilities are strongly dependent on the Mg2+ concentration, possibly via different RNA tunings. Since PTB is predominantly a nuclear protein (14) and IMP-1 and IMP-3 are cytoplasmic, at least at the steady state, it is likely that PTB is attached to leader 3 mRNA during transcription in the nucleus. In this way, the IGF-II leader 3 mRNA may be translated by default if PTB remains attached to the leader in the cytoplasm, whereas substitution with IMP will lead to a translationally repressed form.
IMP-1 repressed translation of leader 3 mRNA dose dependently and cosedimented with untranslated mRNPs rather than being polysome associated. Although the multiple attachments of IMP-1 to the 5′ UTR imply a direct repression at the translation initiation level, the translational status may also be affected by mRNA localization. In fact, the zipcode-binding and Vera proteins have been proposed to mediate the growth-dependent localization of β-actin mRNA to the lamellipodia in chicken embryo fibroblasts and the transport of Vg1 mRNA to the vegetal pole of the X. laevis oocyte, respectively (12, 27), but data regarding localization of zipcode-binding and Vera proteins have not been reported. The discrete cytoplasmic localization of IMP in the present study complements the β-actin and Vg1 mRNA in situ hybridization data (17, 18), thus supporting a role for IMP in RNA localization.
Results from in vivo localization experiments and UV cross-linking studies suggest that an ACACCC sequence in the 54-nucleotide “zipcode” from the 3′ UTR of β-actin mRNA binds the zipcode-binding protein (27), whereas UUCAC (12) and UUUCUA (15) in the 366-nucleotide VgLE from the 3′ UTR of Vg1 mRNA have been implicated in Vera/Vg1 RNA-binding protein recognition. Segment C in leader 3 mRNA contains the putative binding motif UUCACGUUCAC, but the modular structure of this vertebrate family of RNA-binding proteins implies multiple RNA attachment sites.
IMP is expressed during fetal life in both humans and mice. The expressed sequence tags from the mouse two-cell stage indicate, in combination with our data, that IMP is expressed both in early development and from E12. The tissue-specific distribution of IMP is strikingly similar to that of IGF-II. IMP-1 is expressed at high levels in the developing muscle, in the basal layers of epidermis and other epithelia, and in trophoblasts of the placenta, whereas it is absent in the brain, similar to IGF-II (4, 29). The overlapping expression of IMP and IGF-II, in combination with the observed translational repression of the mouse IGF-II leader 3 mRNA homologue at E12.5 (21), implies that IMP participates in the physiological regulation of IGF-II production. The tissue-specific expression of IMP may adjust IGF-II production so that clonal expansion or differentiation of a particular organ is not initiated too early.
The results illustrate that mammalian cytoplasmic mRNA-binding proteins may play a major role in temporal and spatial control of gene expression during late development. IGF-II mRNA provides a powerful example of the significance of regulation by means of distinct 5′ UTRs, since the large leader 3 mRNA is controlled by IMP whereas the smaller leader 4 appears to direct constitutive translation. In this way, the production of IGF-II is maintained at a basal level, on top of which a regulated production is imposed by extracellular cues such as cell contacts and growth factors.
ACKNOWLEDGMENTS
We are grateful to Jens F. Rehfeld and Reidar Albrechtsen for help with the IMP antibodies and immunohistochemistry, respectively. Lena B. Johansson and Bente Rotbøl are thanked for technical assistance.
The research was supported by the Danish Cancer Society, the NOVO-Nordisk Foundation, and the Danish Natural Science and Medical Research Councils and their Biotek II Programme.
REFERENCES
- 1.Allain F H, Howe P W, Neuhaus D, Varani G. Structural basis of the RNA-binding specificity of human U1A protein. EMBO J. 1997;16:5764–5772. doi: 10.1093/emboj/16.18.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson S, Davis D L, Dahlback H, Jornvall H, Russell D W. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989;264:8222–8229. [PubMed] [Google Scholar]
- 3.Baker J, Liu J P, Robertson E J, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75:73–82. [PubMed] [Google Scholar]
- 4.Brice A L, Cheetham J E, Bolton V N, Hill N C, Schofield P N. Temporal changes in the expression of the insulin-like growth factor II gene associated with tissue maturation in the human fetus. Development. 1989;106:543–554. doi: 10.1242/dev.106.3.543. [DOI] [PubMed] [Google Scholar]
- 5.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 6.Davydova E K, Evdokimova V M, Ovchinnikov L P, Hershey J W. Overexpression in COS cells of p50, the major core protein associated with mRNA, results in translation inhibition. Nucleic Acids Res. 1997;25:2911–2916. doi: 10.1093/nar/25.14.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Boulle K, Verkerk A J, Reyniers E, Vits L, Hendrickx J, Van Roy B, Van den Bos F, de Graaff E, Oostra B A, Willems P J. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet. 1993;3:31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- 8.DeChiara T M, Robertson E J, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64:849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- 9.Delbridge M L, Harry J L, Toder R, O’Neill R J, Ma K, Chandley A C, Graves J A. A human candidate spermatogenesis gene, RBM1, is conserved and amplified on the marsupial Y chromosome. Nat Genet. 1997;15:131–136. doi: 10.1038/ng0297-131. [DOI] [PubMed] [Google Scholar]
- 10.Del Tito B J, Jr, Ward J M, Hodgson J, Gershater C J, Edwards H, Wysocki L A, Watson F A, Sathe G, Kane J F. Effects of a minor isoleucyl tRNA on heterologous protein translation in Escherichia coli. J Bacteriol. 1995;177:7086–7091. doi: 10.1128/jb.177.24.7086-7091.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Moor C H, Jansen M, Bonte E J, Thomas A A, Sussenbach J S, Van Den Brande J L. Proteins binding to the leader of the 6.0 kb mRNA of human insulin-like growth factor 2 influence translation. Biochem J. 1995;307:225–231. doi: 10.1042/bj3070225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deshler J O, Highett M I, Abramson T, Schnapp B J. A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr Biol. 1998;8:489–496. doi: 10.1016/s0960-9822(98)70200-3. [DOI] [PubMed] [Google Scholar]
- 13.Eggenschwiler J, Ludwig T, Fisher P, Leighton P A, Tilghman S M, Efstratiadis A. Mouse mutant embryos overexpressing IGF-II exhibit phenotypic features of the Beckwith-Wiedemann and Simpson-Golabi-Behmel syndromes. Genes Dev. 1997;11:3128–3142. doi: 10.1101/gad.11.23.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghetti A, Pinol-Roma S, Michael W M, Morandi C, Dreyfuss G. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Havin L, Git A, Elisha Z, Oberman F, Yaniv K, Schwartz S P, Standart N, Yisraeli J K. RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes Dev. 1998;12:1593–1598. doi: 10.1101/gad.12.11.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim Ha J, Kerr K, Macdonald P M. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81:403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- 17.Kislauskis E H, Li Z, Singer R H, Taneja K L. Isoform-specific 3′-untranslated sequences sort alpha-cardiac and beta-cytoplasmic actin messenger RNAs to different cytoplasmic compartments. J Cell Biol. 1993;123:165–172. doi: 10.1083/jcb.123.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melton D A. Translocation of a localized maternal mRNA to the vegetal pole of Xenopus oocytes. Nature. 1987;328:80–82. doi: 10.1038/328080a0. [DOI] [PubMed] [Google Scholar]
- 19.Mueller-Pillasch F, Lacher U, Wallrapp C, Micha A, Zimmerhackl F, Hameister H, Varga G, Friess H, Buchler M, Beger H G, Vila M R, Adler G, Gress T M. Cloning of a gene highly overexpressed in cancer coding for a novel KH-domain containing protein. Oncogene. 1997;14:2729–2733. doi: 10.1038/sj.onc.1201110. [DOI] [PubMed] [Google Scholar]
- 20.Musco G, Kharrat A, Stier G, Faternali F, Gibson T J, Nilges M, Pastore A. The solution structure of the first KH domain of FMR1, the protein responsible for the fragile X syndrome. Nat Struct Biol. 1997;4:712–716. doi: 10.1038/nsb0997-712. [DOI] [PubMed] [Google Scholar]
- 21.Newell S, Ward A, Graham C. Discriminating translation of insulin-like growth factor-II (IGF-II) during mouse embryogenesis. Mol Reprod Dev. 1994;39:249–258. doi: 10.1002/mrd.1080390302. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen F C, Gammeltoft S, Christiansen J. Translational discrimination of mRNAs coding for human insulin-like growth factor II. J Biol Chem. 1990;265:13431–13434. [PubMed] [Google Scholar]
- 23.Nielsen F C, Østergaard L, Nielsen J, Christiansen J. Growth-dependent translation of IGF-II mRNA by a rapamycin-sensitive pathway. Nature. 1995;377:358–362. doi: 10.1038/377358a0. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen F C, Wang E, Gammeltoft S. Receptor binding, endocytosis, and mitogenesis of insulin-like growth factors I and II in fetal rat brain neurons. J Neurochem. 1991;56:12–21. doi: 10.1111/j.1471-4159.1991.tb02556.x. [DOI] [PubMed] [Google Scholar]
- 25.Rehfeld J F. Radioimmunochemical analysis of neuropeptides based on general characteristics of the analyte. Trends Anal Chem. 1989;8:102–106. [Google Scholar]
- 26.Reijo R, Lee T Y, Salo P, Alagappan R, Brown L G, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O, et al. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat Genet. 1995;10:383–393. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- 27.Ross A F, Oleynikov Y, Kislauskis E H, Taneja K L, Singer R H. Characterization of a β-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siomi H, Dreyfuss G. RNA-binding proteins as regulators of gene expression. Curr Opin Genet Dev. 1997;7:345–353. doi: 10.1016/s0959-437x(97)80148-7. [DOI] [PubMed] [Google Scholar]
- 29.Stylianopoulou F, Efstratiadis A, Herbert J, Pintar J. Pattern of the insulin-like growth factor II gene expression during rat embryogenesis. Development. 1988;103:497–506. doi: 10.1242/dev.103.3.497. [DOI] [PubMed] [Google Scholar]
- 30.Sun F L, Dean W L, Kelsey G, Allen N D, Reik W. Transactivation of Igf2 in a mouse model of Beckwith-Wiedemann syndrome. Nature. 1997;389:809–815. doi: 10.1038/39797. [DOI] [PubMed] [Google Scholar]
- 31.Sussenbach J S. The gene structure of the insulin-like growth factor family. Prog Growth Factor Res. 1989;1:33–48. doi: 10.1016/0955-2235(89)90040-9. [DOI] [PubMed] [Google Scholar]
- 32.Weksberg R, Shen D R, Fei Y L, Song Q L, Squire J. Disruption of insulin-like growth factor 2 imprinting in Beckwith-Wiedemann syndrome. Nat Genet. 1993;5:143–150. doi: 10.1038/ng1093-143. [DOI] [PubMed] [Google Scholar]
- 33.Wewer U M, Iba K, Durkin M, Nielsen F C, Loechel F, Gilpin B J, Kuang W, Engvall E, Albrechtsen R. Tetranectin is a novel marker for myogenesis during embryonic development, muscle regeneration, and muscle cell differentiation in vitro. Dev Biol. 1998;200:247–259. doi: 10.1006/dbio.1998.8962. [DOI] [PubMed] [Google Scholar]
- 34.Wewer U M, Thornell L E, Loechel F, Zhang X, Durkin M E, Amano S, Burgeson R E, Engvall E, Albrechtsen R, Virtanen I. Extrasynaptic location of laminin beta 2 chain in developing and adult human skeletal muscle. Am J Pathol. 1997;151:621–631. [PMC free article] [PubMed] [Google Scholar]
- 35.Wickens M, Kimble J, Strickland S. Translational control of developmental decisions. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Vol. 30. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 411–450. [Google Scholar]