Abstract
Purpose
A lack of representative human disease models has limited the translation of new and more effective treatments in uveal melanoma (UM), the most common primary adult intraocular malignancy. To fill this critical need, we developed and characterized a multicenter biobank of UM patient-derived organoids (PDOs).
Methods
UM patients requiring enucleation from 2019 to 2024 donated tumor tissue for PDO generation. PDOs were cultured in Cultrex and compared to donor primary tumor using exome sequencing, RNA sequencing, and immunohistochemistry. The ability of PDOs to maintain the transformed phenotype was evaluated in an orthotopic xenograft model and monitored with fundus imaging. ATAC sequencing and drug response assays were done in a subset of PDOs to explore the feasibility of their use for mechanistic and translational studies.
Results
PDOs were successfully established in 40 of 44 cases (91%), retained clinically relevant mutations and molecular markers from the primary tumor, and displayed similar gene expression profiles and well-validated clinical prognostic markers of the disease. PDOs retained tumorigenic capacity in an in vivo model resembling human disease progression. Finally, we demonstrated that PDOs were a feasible platform to identify and evaluate novel therapeutic targets and investigate differential, personalized drug response.
Conclusions
PDO models offer a new platform with improved representation of human UM to aid in translational research for this dismal condition.
Keywords: uveal melanoma, patient-derived organoid, orthotopic xenograft, translational, epigenomics
Uveal melanoma (UM) is the most common primary intraocular malignancy of adulthood,1,2 but up to 50% of patients will ultimately develop metastatic disease.3,4 Prognosis is poor for metastatic UM, with median overall survival less than 2 years.5–8 Genomic and transcriptomic profiling of primary UM has improved prognostication,9–11 but effective targets for treatment or prevention of metastasis are still lacking. A major limitation to preclinical drug screening is a lack of human disease models representing the entire clinical and molecular spectrum of UM. Most laboratory drug studies are performed on a limited number of commercially available UM cell lines, which often differ in chromosome status and melanocyte markers compared to the tumor from which they were derived.12 Thus, there is a clear need for representative preclinical models that accurately represent UM pathobiology and drug response to develop new and more effective treatments.
Herein, we present a biobank of novel UM patient-derived organoids (PDOs) established from primary UM. PDOs are three-dimensional (3D) cultures derived from patient tissue.13 When embedded into a 3D matrix, PDOs have self-organizing capabilities and are a self-renewing resource retaining molecular and functional characteristics of the source tissue. PDOs are heterogeneous and retain genetic diversity representative of the individual tumor from which they were derived.14–19 Thus, these new models are emerging as a tool to improve translational research and propel individualized medicine. We demonstrated that UM PDOs recapitulate the characteristics of the tumor of origin, permit propagation for further mechanistic study of UM biology, and can serve as a platform to determine patient specific drug sensitivities.
Methods
Establishing PDOs: Tissue Collection
Patients seen on the Ocular Oncology Service at Mayo Clinic Rochester from July 1, 2019, through July 1, 2024, provided consent after an appropriate risks/benefits discussion regarding enrollment in the Prospective Ocular Tumor Study (POTS), which allowed collection of residual tumor tissue for laboratory specimen development from enucleated eyes with UM. When available, DecisionDx-UMSeq testing was performed on the primary tumor as part of clinical care.20 This study complied with the Health Insurance Portability and Accountability Act and adhered to the tenets of the Declaration of Helsinki. The POTS was granted Institutional Review Board (IRB) approval by Mayo Clinic. Of all study patients, those diagnosed with UM and undergoing enucleation for management of the active tumor were eligible for inclusion in the organoid biobank.
Within 30 minutes of globe removal, a fine needle aspiration biopsy (FNAB) sample was taken of the tumor using a 20-gauge needle (Fig. 1A). The aspirate was directly transferred to a 5-mL Eppendorf tube containing 1× Hanks’ Balanced Salt Solution (HBSS) without calcium, magnesium, and phenol red (21-022-CV; Corning Inc., Corning, NY, USA) for transport to the laboratory. External collaborating centers who enrolled patients under the POTS IRB deposited samples in Gibco Dulbecco's Modified Eagle Medium (DMEM; 11995065; Thermo Fisher Scientific, Waltham, MA, USA) supplement with 10% fetal bovine serum (FBS; A52567-01; Thermo Fisher Scientific) containing penicillin–streptomycin–glutamin (PSG; 10378016; Thermo Fisher Scientific), gentamicin (G1397, 10 mL; Sigma-Aldrich, St. Louis, MO, USA), and Fungizone (15290018; Thermo Fisher Scientific) for shipping to our laboratory overnight on ice.
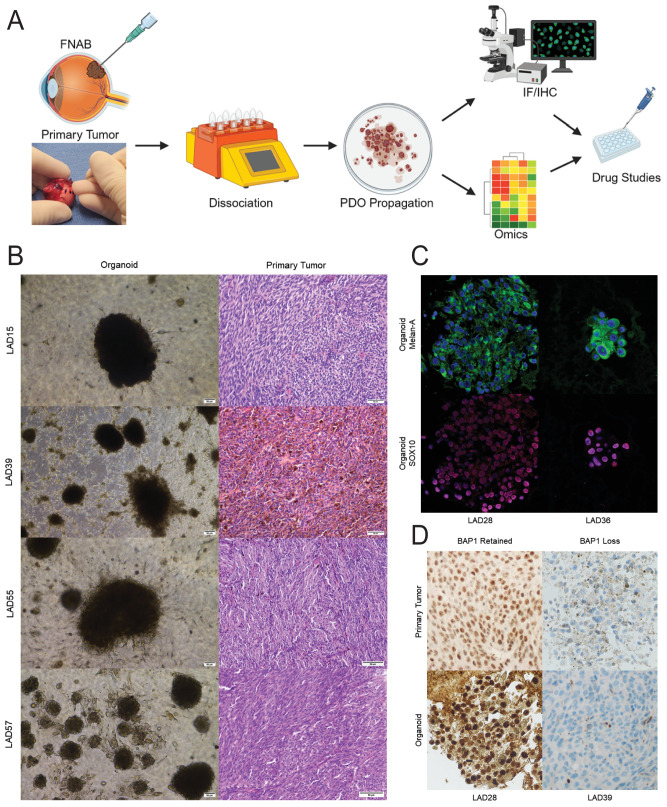
Figure 1.
Patient-derived organoids can be established from uveal melanoma tumors. (A) UM PDO pipeline. FNAB specimens are taken from the primary tumor and dissociated, and PDOs are propagated. PDOs are characterized with immunofluorescence (IF) and immunohistochemistry (IHC) to confirm melanocytic origin and similarities to the clinical sample. PDOs can be further characterized with omics analyses and are used for drug studies. The inset shows transillumination to mark the tumor in an enucleated globe followed by FNAB through the marked area to harvest tumor tissue for PDO generation. (B) H&E histopathology images of primary UM tumors from which PDOs were derived demonstrate UM of spindle (LAD15, LAD55, LAD57) or mixed (LAD39) cell type. Scale bars: 1 mm for LAD15, 100 µm for all others. Corresponding brightfield images of organoids are shown. Scale bars: 100 µm. (C) Uveal melanoma PDOs characterized by immunofluorescence with 4′,6-diamidino-2-phenylindole (DAPI) nuclear stain. PDOs have cytoplasmic staining for Melan-A (green) and nuclear staining for SOX10 (blue), confirming the melanocytic origin of the cells. (D) Uveal melanoma PDOs demonstrate histopathological features consistent with the corresponding clinical sample. The hallmark feature of nuclear BAP1 retention or loss is consistent between the clinical sample and the PDO.
Organoid Culture Initiation
Samples were centrifuged at 300g for 5 minutes at room temperature, the supernatant was aspirated, and the cell pellet was resuspended in 4 mL of HBSS and centrifuged again. Supernate was removed, and cells were resuspended in a cocktail containing 4 mL RPMI-1640 Medium (30-2001; American Type Culture Collection, Manassas, VA, USA), 1% FBS, 200 µL of Enzyme H (130-095-929; Miltenyi Biotec, Bergisch Gladbach, Germany), 100 µL of Enzyme R (130-095-929; Miltenyi Biotec), and 25 µL of Enzyme A (130-095-929; Miltenyi Biotec) and transferred to a gentleMACS C tube (130-093-237; Miltenyi Biotec). The original sample tube was washed with 700 µL of HBSS/FBS and pooled with the cells in the gentleMACS C tube. Cells were dissociated using a gentleMACS Octo Dissociator (130-096-427; Miltenyi Biotec). Dissociated cells were transferred to a 70-µM MACS SmartStrainer (130-098-462; Miltenyi Biotec) and placed in a 50-mL centrifuge tube. The MACS tube was washed with 20 mL serum-free RPMI-1640.
Cells were centrifuged at 300g for 5 minutes at room temperature. The supernatant was aspirated, and the cell pellet was resuspended in RPMI-1640 supplemented with 10% FBS, 1× Gibco Antibiotic–Antimycotic (15240062; Thermo Fisher Scientific), 1× Gibco GlutaMAX (35050061; Thermo Fisher Scientific), 10-µM Y-27632 dihydrochloride (1254; Bio-Techne/Tocris, Bristol, UK) and 5% Cultrex Reduced Growth Factor Basement Membrane Extract, Type 2, Pathclear (3533-005-02; R&D Systems, Minneapolis, MN, USA). Cells were plated in multiwell plates precoated with Cultrex and incubated at 37°C with 5% CO2 in a humidified incubator. Media were replaced every 3 to 4 days. Organoids were passaged when the growth rate approached a static state or if degradation of the Cultrex layer was observed. The detailed methodology for the PDO passaging is available in the Supplementary Materials.
Immunohistochemistry
Immunohistochemistry was done as described previously,21,22 with additional details provided in the Supplementary Materials.
Exome, RNA, and ATAC Sequencing
Exome sequencing, RNA sequencing (RNA-seq), and the assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) were done as previously described,15,23,24 with additional detail provided in the Supplementary Materials. Polymerase chain reaction (PCR) resequencing was used to confirm select clinically relevant mutations between primary tumors and PDOs as described in the Supplementary Materials.
Drug Studies
A volume-based plating Cultrex slurry method was modified by our laboratory from previously published protocols.25,26 See Supplementary Materials for details.
Orthotopic Xenograft Establishment
Animal studies were approved by the Mayo Clinic Institutional Animal Care and Use Committee, and studies were conducted in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. Athymic nude mice (Nu/J; The Jackson Laboratory, Bar Harbor, ME, USA) 9 weeks old were placed under anesthesia for tumor inoculation. Approximately 150,000 cells were concentrated in 2.5 µL of HBSS and injected suprachoroidally. A 30-gauge needle was used to make a tunnel from the limbus, extending posteriorly toward the optic nerve head. A 33-gauge blunt needle (7762-06; Hamilton Company, Reno, NV, USA) attached to a syringe (7642-01; Hamilton Company) was inserted through the tunnel, and the cell slurry was injected slowly over 10 to 20 seconds. Beginning 2 weeks after inoculation, mice were placed under anesthesia every 1 to 2 weeks for noninvasive imaging, including color fundus photography and fluorescein angiography (MICRON IV; Phoenix-Micron, Bend, OR, USA). Globes were collected at the time of euthanasia, fixed in formalin, and paraffin embedded for immunohistochemical analysis.
Data Availability
Data described in this report have been deposited in a Sequence Read Archive (BioProject accession no. PRJNA1068520).
Results
A UM PDO Model Is Feasible and Retains Stability Through Passaging
To develop models that more closely resemble primary UM, we generated a biobank of UM PDOs. A schematic of the PDO development pipeline is shown in Figure 1A. PDOs derived from primary intraocular UM were successfully established in 40 of 44 attempted cases (91%). Clinical features associated with the tumors of origin are described in the Table. Consistent with the known demographics associated with UM, patient donors were predominantly white, with similar numbers of male and females; age at the time of tumor harvest ranged from 37 to 101 years. Tumors spanning Class 1A, Class 1B, and Class 2 gene expression profiling, with both negative and positive preferentially expressed antigen in melanoma (PRAME) expression, were included. The AJCC Cancer Staging Manual, Eighth Edition,27 histopathology stage of primary tumors ranged from pT2a to pT4e. There was no apparent association between patient demographics or clinical tumor features and success of PDO generation. PDOs were also developed from externally shipped samples contributed by multicenter collaborators, and PDO generation was similarly successful as that of internally processed samples, provided sufficient tumor material was available (Supplementary Figs. S1A, S1B). As of October 2024, PDOs can be carried in culture, with more than 20 passages on some samples (Supplementary Fig. S1C).
Table.
Patient Demographics and Clinical Tumor Features for UM PDOs Derived From Primary Intraocular Tumors
| Study Number | Sex | Age (y) | Race | BAP1 Protein Expression | DecisionDx Class | PRAME | Cell Type | AJCC Histopathology T Category | Driving Mutation | Other Mutations |
|---|---|---|---|---|---|---|---|---|---|---|
| LAD3 | M | 58 | White | Lost | — | — | Epithelioid | pT4b | — | — |
| LAD11 | M | 64 | White | Lost | — | — | Mixed | pT4b | GNAQ | — |
| LAD15 | F | 48 | White | Retained | 1B | Positive | Spindle | pT3a | — | — |
| LAD17 | F | 69 | White | Lost | 2 | Positive | Epithelioid | pT4b | — | — |
| LAD24 | F | 54 | White | Lost | 2 | Positive | Mixed | pT4b | — | — |
| LAD25 | M | 56 | White | Lost | 2 | Negative | Mixed | pT3b | GNA11 | BAP1 |
| LAD26 | F | 62 | White | Lost | 2 | Negative | Mixed | pT3a | — | — |
| LAD27 | F | 101 | White | Lost | — | — | Mixed | pT4b | — | — |
| LAD28 | M | 43 | White | Retained | 1B | Negative | Spindle | pT4b | GNAQ | EIF1AX |
| LAD29 | M | 50 | White | Retained | 1B | Negative | Mixed | pT4b | GNAQ | — |
| LAD30 | F | 37 | White | Retained | 1B | Positive | Spindle | pT4b | GNA11 | SF3B1, BAP1 |
| LAD31* | M | 55 | White | Lost | 2 | Positive | Mixed | pT3a | — | — |
| LAD32 | M | 64 | White | Lost | 2 | Positive | Mixed | pT4b | GNA11 | BAP1 |
| LAD33 | M | 68 | White | Lost | 2 | Positive | Mixed | pT3b | GNA11 | BAP1 |
| LAD34 | F | 62 | White | Retained | 1B | Positive | Mixed | pT3b | — | SF3B1 |
| LAD36 | M | 52 | White | Lost | 2 | Positive | Mixed | pT3a | GNAQ | BAP1 |
| LAD37 | F | 55 | White | Lost | 2 | Positive | Epithelioid | pT4b | GNA11 | BAP1 |
| LAD38 | M | 57 | White | Retained | — | — | Spindle | pT3a | — | — |
| LAD39 | F | 62 | White | Lost | 2 | Positive | Mixed | pT4b | CYSLTR2 | BAP1 |
| LAD40 | M | 67 | White | Lost | 2 | Positive | Epithelioid | pT2a | GNA11 | BAP1 |
| LAD41 | M | 80 | White | Retained | 1B | Positive | Mixed | pT4b | GNA11 | SF3B1 |
| LAD43 | M | 66 | White | Retained | 1B | Negative | Spindle | pT4b | GNAQ | EIF1AX |
| LAD44 | M | 70 | White | Lost | 2 | Positive | Spindle | pT3b | GNA11 | BAP1 |
| LAD46 | M | 37 | White | Retained | 1B | Positive | Spindle | pT4e | GNA11 | SF3B1 |
| LAD48 | F | 64 | White | Lost | 2 | Positive | Mixed | pT3b | GNA11 | BAP1 |
| LAD49 | F | 73 | White | Lost | 2 | Positive | Mixed | pT3b | GNA11 | BAP1 |
| LAD51 | M | 72 | White | Lost | 2 | Positive | Spindle | pT4b | GNA11 | BAP1 |
| LAD52 | F | 63 | White | Retained | 1A | Positive | Mixed | pT4e | GNA11 | SF3B1 |
| LAD53 | F | 68 | White | Retained | 1A | Positive | Spindle | pT4a | GNA11 | |
| LAD55 | M | 73 | White | Lost | 2 | Positive | Spindle | pT2a | GNA11 | BAP1 |
| LAD57 | M | 47 | White | Lost | 2 | Positive | Spindle | pT3a | GNAQ | BAP1 |
| LAD58 | F | 89 | White | Lost | 2 | Positive | Mixed | pT4d | GNA11 | BAP1 |
| LAD59 | M | 72 | White | Lost | 2 | Negative | Mixed | pT4b | — | — |
| LAD60 | M | 55 | White | Lost | 2 | Positive | Mixed | pT3b | GNAQ | — |
| LAD61 | F | 81 | White | Lost | 2 | Negative | Mixed | pT3a | GNAQ | BAP1 |
| LAD62 † | M | 76 | White | — | — | — | — | — | — | — |
| CLS1 | M | 90 | White | — | — | — | Mixed | — | — | — |
| CLS2 | F | 63 | White | — | — | — | Epithelioid | pT3a | — | — |
| CLS3 | F | 64 | White | — | — | — | Mixed | pT3b | — | — |
| CLS4 | M | 61 | White | — | — | — | Mixed | pT3b | — | — |
| CLS5 | F | 48 | White | — | — | — | Mixed | pT3b | — | — |
| CLS6 | F | 60 | White | — | — | — | Mixed | pT3b | — | — |
| CLS7 | F | 64 | White | — | — | — | Epithelioid | pT4b | — | — |
| CLS8 | M | 50 | ME | — | — | — | Epithelioid | pT4b | — | — |
BAP1 status is based on immunohistochemistry analysis. Blank fields indicate that information was not available or not applicable. Italic rows failed to propagate. LAD62 was derived from a patient previously exposed to systemic immunotherapy. ME, Middle Eastern.
Recurrence after prior plaque radiotherapy.
Prior exposure to immunotherapy for treatment of metastatic disease.
Microscopically, PDOs appeared as moderate to heavily pigmented masses in culture, consisting of a mixed population of tumor cells, comparable to those seen in the primary tumor, without apparent presence of non-tumoral cell types. Corresponding clinical primary tumors were morphologically confirmed as UM by examining hematoxylin and eosin (H&E)-stained sections (Fig. 1B). The melanocytic nature of PDOs was confirmed by staining for markers commonly used for clinical histopathologic confirmation of primary tumors, Melan-A and SRY-related HMG box gene 10 (SOX10) (Fig. 1C). BRCA1-associated protein-1 (BAP1) immunohistochemistry is routinely done on clinical samples in our center due to the prognostic value of loss versus retention of nuclear BAP1 staining. PDOs were also processed for BAP1 staining and matched immunohistochemical BAP1 expression of the corresponding donor primary tumor. Supporting the goal of a diverse biobank, PDOs were generated from primary tumors with both retained and lost BAP1 expression (Fig. 1D).
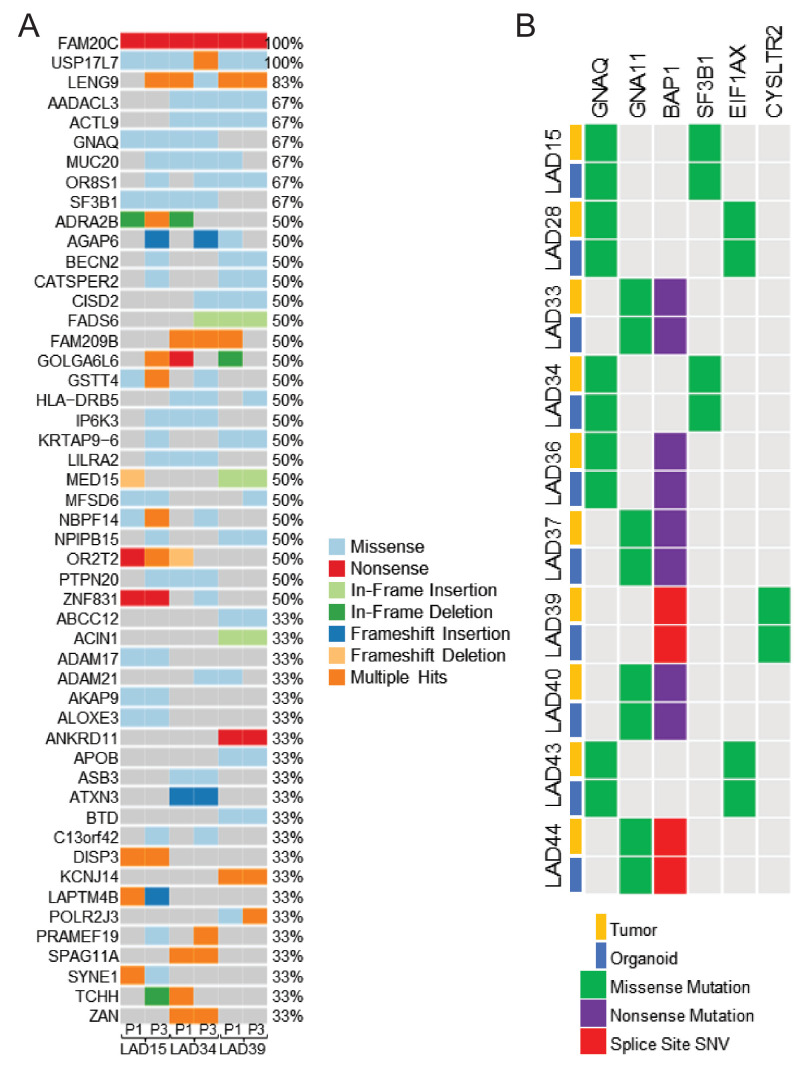
We analyzed tumor morphology and determined the stability of PDOs through passaging to support the utility of the PDO model as a renewable resource for translational UM research. Different PDOs displayed variable morphology and degrees of pigmentation, in many cases reflective of the primary tumor predominance of spindle, mixed, or epithelioid cell types (Supplementary Fig. S2A). After an initial period of establishment prior to first passage, PDO morphology remained stable with early passaging from passage 1 (P1) to P3 (Supplementary Fig. S2B). Exome sequencing of paired PDOs at P1 and P3 showed relative stability of the mutational landscape, with retention of clinically relevant UM hallmark genetic defects through the passages (Fig. 2A). Growth rates were variable, with some PDOs passaging within 1 week and others taking 2 to 3 months to passage. Passage time did not have a clear relationship with primary tumor features such as mutation status or gene expression profile.
Figure 2.
Uveal melanoma patient-derived organoids retain genomic features of the corresponding clinical tumor. (A) Oncoplot of PDOs at P1 and P3 shows relative stability and retention of key uveal melanoma hallmark mutations in GNAQ and SF3B1 through passaging. (B) Genetic comparison of clinically relevant markers between the PDO and the corresponding donor tumor (n = 10) shows retention of key driver and prognostic mutations in the PDOs. Splice site single nucleotide variants were not detected in PDOs by whole exome sequencing but were confirmed by PCR resequencing for the variant identified on clinical testing.
To determine whether organoid models retained the range of gene expression profiles seen in primary tumors, we ran RNA-seq analysis in a subset of 10 PDOs chosen for their consistent growth and passaging, with a mix of high-risk versus low-risk profiles of the corresponding primary tumor. First, we analyzed a subset of samples in pairs at P1 and P3 to determine whether UM PDOs retained stable gene expression profiles through early passaging. Data analysis of paired PDOs at P1 and P3 reported no significantly differentially expressed genes between P1 and P3 in LAD28 (BAP1 retained, low-risk) and LAD39 (BAP1 loss, high-risk), but one differentially expressed gene between P1 and P3 in LAD36. Examining a subset of genes with significant differential expression between BAP1 retained versus BAP1 loss subtypes using a false discovery rate (FDR) of 0.01 (see below), paired PDOs did not have significantly different gene expression, and same PDOs at different passages clustered together (Supplementary Fig. S3). Thus, through early passaging, PDOs retain individualized features of the UM transcriptome that are associated with tumor phenotype and metastatic risk.
PDOs Retain Clinically Relevant Features of the Primary Tumor, Cluster Into Distinct Molecular Groups Based on Proven Prognostic Markers, and Resemble Human Disease In Vivo
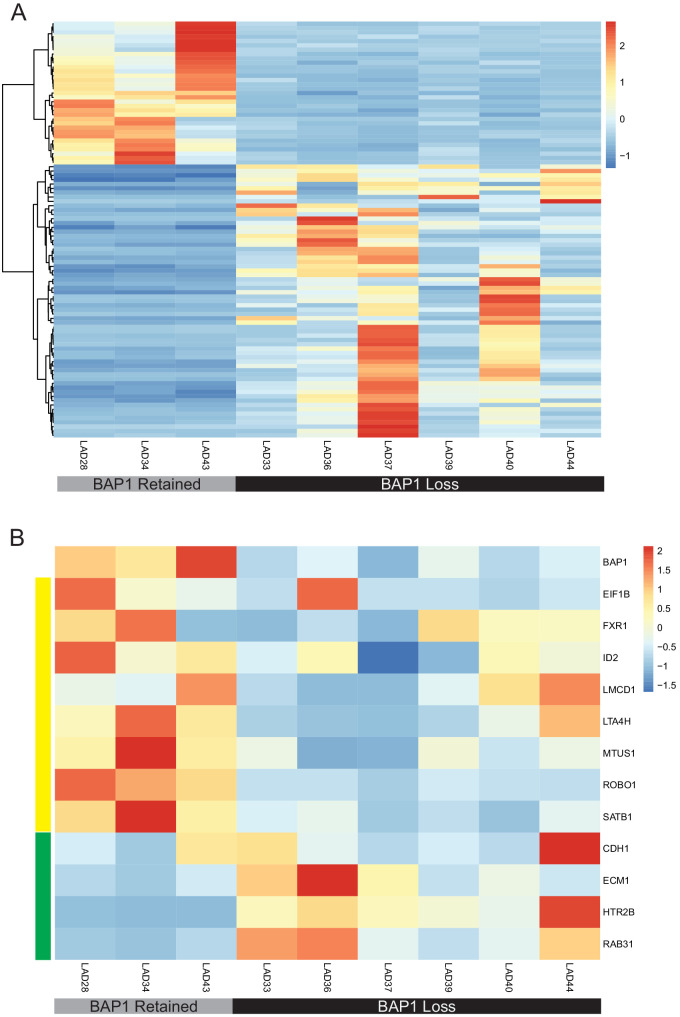
We next sought to determine how closely the PDOs clinically resembled the primary tumor. On our sequencing and marker analysis, PDOs retained key driver and prognostically important mutations detected on clinical testing of the corresponding primary tumors (Fig. 2B). Principal component analysis (PCA) comparing whole exome sequencing between passages is shown in Supplementary Figure S4. Importantly, PDOs had a mix of BAP1 status; BAP1 loss is the single most consistent alternation associated with increased risk of UM metastasis and death.9,10 Transcriptomic analysis of PDOs with retained versus lost BAP1 expression showed differential expression of 97 genes on a global level (Fig. 3A). Further analysis was done for the expression of BAP1 and 12 additional genes commonly used in a well-validated clinical prognostic gene expression profiling test.28 UM PDOs displayed similar patterns of prognostically important gene expression, stratifying into similar prognostic categories compared with the corresponding primary tumor samples, and PDOs with BAP1 loss displayed a high-risk profile, as expected (Fig. 3B). Similar patterns of gene expression in high-risk primary UM samples were also seen in analysis from The Cancer Genome Atlas (TCGA),9 altogether suggesting that UM PDOs retain gene expression profiles known to associate with metastatic risk. Correlating back to clinical data for PDOs with a high-risk profile, four of six donor patients developed metastasis, whereas there were no cases of metastasis in donors for PDOs with a low-risk profile (Supplementary Table S1).
Figure 3.
UM PDOs have differential gene expression based on BAP1 status. (A) Heatmap of z-scores for differential gene expression between BAP1-retained and BAP1-loss PDOs using DeSeq2. Differential expression determined using ±1 log fold change and P ≤ 0.1. (B) Heatmap of z-scores for gene expression of a subset of genes used on a well-validated clinical prognostic gene expression profiling test and further analyzed in TCGA samples shows that uveal melanoma PDOs simulated expression patterns seen in the clinical samples. BAP1 loss is a known poor prognostic indicator. Genes with low expression in high-risk primary UM by gene expression profiling and TCGA are denoted by the yellow panel, and those with high expression in high-risk tumors are denoted by the green panel. High-risk PDOs, defined by those with BAP1 loss, followed the expected pattern of prognostic gene expression.
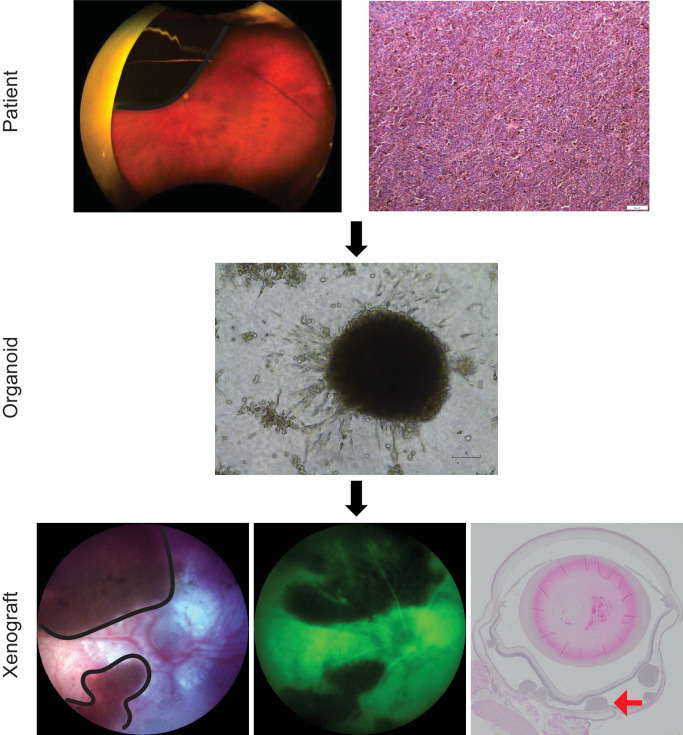
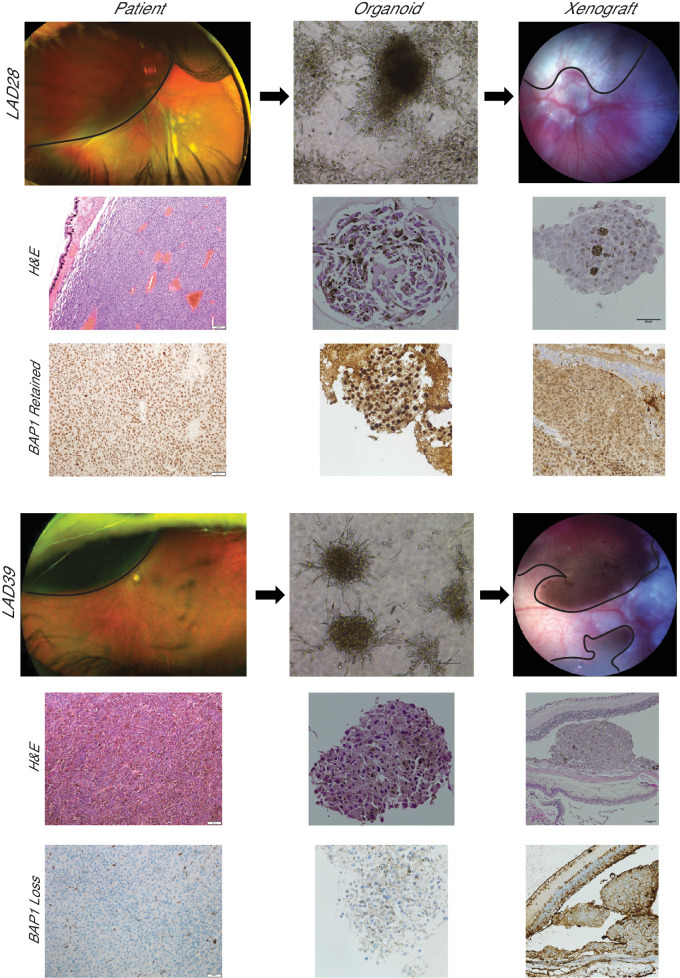
To determine the feasibility of using patient-derived samples in an in vivo model, a pilot cohort of nude mice (n = 4) were injected suprachoroidally with one of two samples, LAD28 or LAD39, chosen for their robust propagation and different metastatic risk profiles. Tumor grafting was successful in all mice. Intraocular tumors were confirmed with color fundus photography, and subretinal location was confirmed by fluorescein angiography (Fig. 4). As expected, intraocular tumors progressed over time, with increased pigmentation, clustering, and thickness (Supplementary Fig. S5). At week 12, eyes were harvested. UM tumors were seen on H&E staining, and immunohistochemistry for BAP1 revealed consistent retention or loss of BAP1 matching that of the primary tumor and PDO (Fig. 5).
Figure 4.
UM orthotopic xenografts can be generated from patient-derived material. Patients requiring enucleation presented with a unilateral, large, pigmented choroidal mass (black outline) shown on Optos pseudocolor fundus photographs. Tumor was confirmed by histopathology evaluation on H&E-stained sections. PDOs were generated from a FNAB of the primary clinical tumor. Patient-derived material was injected suprachoroidally in nude mice (n = 4) and produced viable tumor (black outline). The tumor was confirmed to be subretinal by fluorescein angiography (green image), which shows dark tumor and overlying hyperfluorescent retinal vasculature. Subretinal tumor was confirmed by H&E staining (red arrow). Representative images from LAD28 are shown.
Figure 5.
UM orthotopic xenografts generated from PDOs resemble the corresponding clinical tumor. Patients with a unilateral, large, pigmented choroidal mass (black outline) consistent with primary UM donated tumor tissue for LAD28 (BAP1 retained) and LAD39 (BAP1 loss). H&E-stained slides are shown of the primary clinical tumor (100×). BAP1 immunohistochemistry of the primary tumors of origin for LAD28 and LAD39 revealed retention or loss of nuclear BAP1 expression, respectively (200×). PDOs were generated from a FNAB of each clinical sample, and light microscopy (100×) and H&E (400×) images of LAD28 and LAD39 are shown. BAP1 immunohistochemistry of PDOs (400×) showed recapitulation of BAP1 retention or loss, matching the primary tumor. Murine orthotopic xenografts were generated from LAD28 (n = 2) and LAD39 (n = 2) via suprachoroidal injection of approximately 150,000 cells. The injected eye developed pigmented choroidal masses (black outline), simulating in vivo human pathology. H&E and nuclear BAP1 immunohistochemistry of the tumors from enucleated mouse eyes matched the primary human tumor.
PDOs Are a Suitable Model to Study Differential Drug Response and Serve as a Platform to Identify Novel Drug Targets
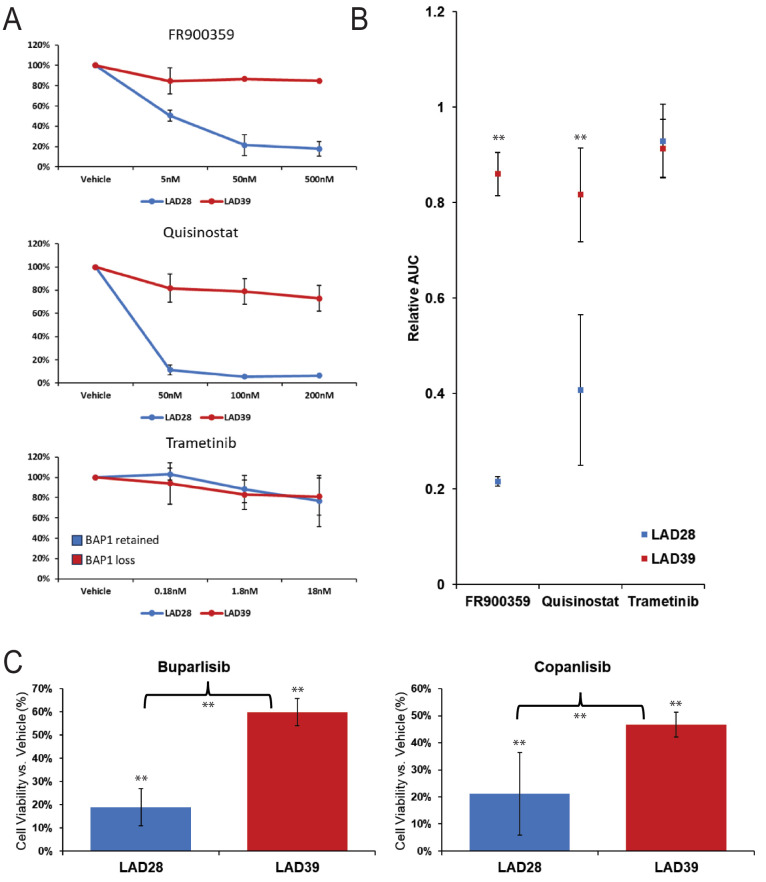
We sought to validate our in vitro PDO drug testing conditions using agents previously investigated in UM. We used a 96-well platform to test three UM-relevant drugs on two well-characterized PDOs from our biobank chosen for their reliable propagation and distinct molecular profiles (Figs. 6A, 6B). We demonstrated a dose-dependent viability reduction when PDOs were treated with the GNAQ inhibitor FR900359 and the HDAC inhibitor quisinostat in LAD28, a GNAQ-mutant PDO with retained BAP1 protein expression. LAD39, a CYSLTR2-mutant PDO with loss of BAP1 protein expression, showed treatment resistance to both drugs. In both LAD28 and LAD39, we demonstrated poor efficacy of the mitogen-activated protein kinase kinase enzyme (MEK) inhibitor trametinib, which is consistent with known poor clinical efficacy of MEK inhibitors for UM.29,30 Importantly, this is in contrast to the moderate efficacy of trametinib seen in cell culture for commercial UM cell lines.31 These results suggest that PDOs can serve as human models for exploratory drug studies, and results might simulate in vivo drug responses, without some of the false-positive results seen in a two-dimensional culture model.
Figure 6.
UM PDOs have differential drug responses based on BAP1 status. (A) Line graph showing response for UM PDOs with retained (LAD28) or lost (LAD39) BAP1 expression indicate differential responses to small molecule inhibitor therapy including FR900359, quisinostat, and trametinib. Assays were performed in triplicate with standard deviation. (B) Areas under the curve corresponding to the line graphs show significant differential drug responses to FR900359 (P = 0.003) and quisinostat (P = 0.01) with similarly poor treatment response of LAD28 and LAD39 to trametinib (P = 0.85). (C) Treatment with two different PI3K inhibitors (buparlisib and copanlisib, 5000 nm at 72 hours) showed selective treatment resistance of UM PDOs with BAP1 loss (buparlisib BAP1 retained vs. loss, P < 0.001; copanlisib BAP1 retained vs. loss, P = 0.03; all drug vs. vehicle control, P < 0.001). Data were analyzed using a two-tailed paired t-test. **Statistically significant.
To determine the suitability of UM PDOs as a platform for more in-depth mechanistic analysis, ATAC-seq was performed on six PDOs chosen from those previously analyzed with RNA-seq. PDOs were chosen with BAP1 loss (n = 3) or retained BAP1 expression (n = 3). After analysis, quality data were available from five PDOs. Differential regions of chromatin accessibility were seen in PDOs with retained BAP1 versus BAP1 loss (Supplementary Fig. S6B). We also found those differentially accessible regions were mostly located near the transcription start site (TSS) for BAP1 loss, indicating that BAP1 loss may be associated with increased chromatin accessibility near promoter regions (Supplementary Fig. S6C). We further analyzed ATAC-seq data to determine their value as a source for possible drug targets by examining chromatin accessibility in a subset of genes. We found increased chromatin accessibility in PDOs with retained BAP1 expression for phosphatidylinositol 3-kinase (PI3K) pathway-relevant components, including promotor/enhancer regions of AKT3 (1.83-fold; P < 0.001, FDR = 0.03), IGFBP7 (3.02-fold; P < 0.001, FDR < 0.001), and PIK3C2G (1.91-fold; P < 0.001, FDR = 0.03). PDOs with BAP1 loss had increased chromatin accessibility for different PI3K pathway–relevant components, including promotor or enhancer regions of Fos, a different site associated with IGFBP7 (2.12-fold; P < 0.001, FDR = 0.01), JUNB (2.52-fold; P < 0.001, FDR = 0.001), STAT3 (2.43-fold; P < 0.001, FDR = 0.003), and TEAD1 (1.80-fold; P < 0.001, FDR = 0.04) (Supplementary Fig. S7A). These findings raised our interest in the PI3K/Akt pathway. We integrated our findings with RNA-seq data. Potentially due to the small number of samples, we did not find statistically significant differences in expression of these genes after adjusting for multiple comparisons, but the data were suggestive of increased expression of IGFBP7 (P < 0.001, FDR = 0.13) and IGFBP7-AS1 (P = 0.003, FDR = 0.20) in BAP1 loss samples compared to those with retained BAP1 expression (Supplementary Fig. S7B). Larger sample sets are needed for further analysis, but previous work in other cell types suggests that IGFBP7 could be involved in PI3K inhibitor treatment response. IGFBP7 promotes cell survival through two mechanisms: (1) sustaining insulin-like growth factor 1 (IGF-1) receptor expression on the cell surface, leading to prolonged Akt activation in the presence of insulin or IGF stimulation,32 and (2) enhancing tumor-associated macrophage polarization via the FGF2/FGFR1/PI3K/Akt axis.33 To define the translational value of the epigenomics data to inform novel therapeutic responses, we investigated 72-hour responses to two different PI3K inhibitors (buparlisib and copanlisib) in LAD28 and LAD39. We found selective treatment resistance to both PI3K inhibitors (buparlisib, P < 0.001; copanlisib, P = 0.03) in the BAP1 loss PDO LAD39 compared to LAD28 with retained BAP1 expression (Fig. 6C). These PDOs are a renewable resource that can be used for in-depth epigenomic analysis allowing the identification of novel UM therapeutic sensitivities.
Discussion
UM is a devastating disease, with an approximately 50% risk of developing incurable metastasis despite successful primary tumor treatment.3,5–7 To support translational research for this dismal disease, we successfully established a multicenter biobank of UM PDOs from primary tumors that resemble the origin tumor in terms of melanocyte markers, genome, and transcriptome. Such a resource has the potential to improve successful translation of laboratory research into the clinical setting, as these models are more representative of human disease.
To date, the vast majority of in vitro UM research has been done on a limited number of commercially available cell lines.12,34,35 Many of these cell lines were established from metastases or unusual tumors, including unexpectedly aggressive disease, association with oculodermal melanocytosis, and recurrence after irradiation.12 Other cell lines were established from patient-derived xenograft models rather than the primary tumor, or, if established from the primary tumor, they differed in chromosomal status and/or melanocyte markers.12,34,35 As a result, such models may have limited translational potential to make advances for patients with typical UM tumors. Our novel PDO models offer a key advantage of retaining immunohistochemical and genomic features of the primary tumors from which they were derived. Further, the models were generated from typical, albeit larger, enucleation-requiring tumors, of which only one (LAD31) was a recurrence after prior irradiation.
We had a greater than 90% success rate for PDO generation, highlighting reliable methodology independent of tumor genetics or histological subtypes, which is especially encouraging given historically low success rates (<10%) when establishing UM cell lines.12 We were also able to establish more than 20 PDOs with loss of BAP1 protein expression that matched BAP1 loss in the primary tumor. Cells lines that lack BAP1 expression have been difficult to find even when derived from a primary tumor with BAP1 loss, possibly due to faster growing cells outcompeting those with BAP1 loss in a mixed cell population.12,34 Commercially available cell lines with BAP1 loss are also notoriously slow growing, with long doubling times,34,35 which may discourage their use in some experimental settings despite the fact that tumors with BAP1 loss are most frequently responsible for human metastatic disease.9,10 Thus, our biobank provides encouraging data for improved representation of the most aggressive UM subtypes for use in laboratory research. Ongoing immunohistochemical analyses will be important to determine if PDO BAP1 status drifts at later passages.
Although patient-derived xenograft (PDX) models have been established for UM, our PDOs offer several key advantages. Notably, our success rate for PDO generation was much higher than a reported 28% success rate when establishing 25 murine PDX models from 90 attempted samples.35,36 Furthermore, PDOs can be established within <1 to 3 months and used in a high-throughput 96-well drug screening platform with 72-hour readouts, allowing for multiple drug testing and potentially fast turnaround for drug use in the clinical setting. This is in contrast to another murine PDX study with a 38% take rate followed by up to 13 months prior to sufficient in vivo tumor growth for drug testing.37 Furthermore, these PDX models implanted tumors into the interscapular fat pads of mice, which does not accurately represent primary or metastatic UM location.37 Hepatic implantation of tumors into mice has yielded 80% or greater engraftment rates.38 Although models could be generated within less than 6 months, the authors noted that additional time would be required to generate second- or third-generation PDX models for personalized drug screening.38 Zebrafish models have been reported with up to 100% success rates when implanting material from spheroid cultures, which could improve upon the lower success rates of the reported murine PDX models.39,40 Even with the intermediary spheroid step, zebrafish models may allow drug testing within a matter of weeks rather than several months. Compared to PDX models, PDOs may be less expensive, as only cell culture facilities are required without the need for veterinary services. Both PDO and PDX models share retention of defining features of the primary tumor, so both model types may have an important role in ongoing research.35,37,38,41
PDO transcriptomic characterization has been encouraging in that PDOs retain distinct gene expression profiles. Because most of our patients with large tumors choose to have clinical prognostication by gene expression profiling,28 we were able to review the well-validated panel of genes from the clinical tests in matched organoids. We found that PDOs derived from clinically high-risk versus low-risk tumors retained gene expression profiles similar to those expected from the donor patient's prognostic test result. These high- and low-risk gene expression patterns have been reproduced in the TCGA,9 and the ability to find similar profiles even in a small cohort of PDOs suggests that we may be able to study unique clusters or subtypes of UM using these models, which could ultimately pave the way for personalized treatment strategies.
Further improving the translational value of these models, we demonstrated successful in vivo tumor growth after suprachoroidal inoculation in a nude mouse model. Historically, many animal studies of UM utilized a mouse cutaneous melanoma cell line, which would not adequately simulate UM because it is a molecularly distinct cancer from cutaneous disease.42,43 Patient-derived UM cell lines have since been implanted in immunocompromised mice, but, consistent with the scarcity and slow-growing nature of cell lines with BAP1 loss, experiments have often used BAP1-retained cell lines with or without inducible BAP1 knockdown rather than implanting human cancer cells with native BAP1 loss.42,43 We were able to show tumor growth of both a BAP1-retained and BAP1-loss sample in vivo. Importantly, immunohistochemistry showed matching BAP1 status from the primary clinical tumor to PDO to mouse. Although these results are early, these data support the representative nature of these models and suggest exciting applications for future preclinical, personalized drug studies. Although we acknowledge the PDX models mentioned above, we find it especially important to apply our PDOs in an orthotopic (primary ocular implantation) in vivo model that will ultimately allow the study of disease course simulating human UM progression. We believe the PDOs alone may be an outstanding platform but recognize that in vivo models could be needed at times for preclinical drug validation when developing novel therapeutics.
In addition to accurately representing human disease, we were able to show utility of the PDO models for in vitro drug screening. Unlike prior cell line studies that have shown moderate sensitivity to MEK inhibitors despite their ultimate clinical failure,29–31 PDOs were resistant to the MEK inhibitor trametinib, suggesting that PDOs may be less likely to yield unrealistically optimistic drug response data. Consistent with emerging techniques for other cancers, UM PDO models could be used for future high throughput drug screening and personalized drug response studies.14,15
Due to the non-renewable limited amount of material available from paraffin-embedded samples, the broad array of chromatin accessibility patterns in UM is not well described. Because these models are a renewable, living resource, we were able to analyze epigenomic data using ATAC-seq in an early cohort of PDOs. ATAC-seq analysis in UM samples has been severely limited, with one study using ATAC-seq in a single BAP1-retained cell line to investigate changes with switch/sucrose non-fermentable (SWI/SNF) inhibition.44 We were able to demonstrate chromatin accessibility differences in a small cohort of samples with BAP1 loss versus retained BAP1 expression and use this epigenomic information, integrated with RNA-seq data, to generate a hypothesis for drug targeting. A larger cohort of samples is needed to better define the global epigenomic landscape of UM, and this biobank could serve as the platform for such studies in the future. Importantly, because the PDOs can be propagated, these models will lend themselves to more in-depth study that cannot be done on fixed samples, and the three-dimensional nature of these models is crucial for accurate representation of epigenomic interactions, which are not well-represented in two-dimensional cancer cell lines.45
In conclusion, we have established a multicenter PDO biobank for UM. We achieved high success rates in PDO generation, with proof of retention of key diagnostic and prognostic features of primary tumor in these models. PDOs can be implanted for in vivo studies and show promise as a platform for in-depth epigenomic analysis, high-throughput drug screening, and personalized preclinical drug studies. UM PDO models more accurately recapitulate human disease, and we anticipate this will improve translation of future UM research.
Supplementary Material
Acknowledgments
The authors thank Jeong-Heon Lee, PhD, and Linh D. Pham from the Mayo Clinic Rochester Epigenomic Analysis Core for their assistance; Ken Walder, PhD, and Sean McGee, PhD, for their supervision as part of a PhD program through Deakin University; and Lions Gift of Sight for procurement of donor eyes for research.
Supported by the Leonard and Mary Lou Hoeft Career Development Award Fund in Ophthalmology Research; Center for Biomedical Discovery, Mayo Clinic; a grant from from the National Cancer Institute, National Institutes of Health (P30 CA015083); and a Clinical and Translational Science Award from the National Center for Advancing Translational Science (KL2 TR002379). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Disclosure: L.A. Dalvin, IDEAYA Biosciences (C); C.M. Andrews-Pfannkoch, None; D.R. Miley, None; T.L. Hogenson, None; S.A. Erickson, None; S. Malpotra, None; K.J. Anderson, None; M.E. Omer, None; L.L. Almada, None; C. Zhang, None; H. Li, None; D.R. Salomao, None; C.L. Shields, None; S.E. Lally, None; R.M. Malsch, None; J.A. Armitage, None; H.L. Holmes, None; M.F. Romero, None; M.P. Fautsch, None; S.N. Markovic, None; M.E. Fernandez-Zapico, None
References
- 1. Singh AD, Turell ME, Topham AK.. Uveal melanoma: trends in incidence, treatment, and survival. Ophthalmology. 2011; 118(9): 1881–1885. [DOI] [PubMed] [Google Scholar]
- 2. Singh AD, Bergman L, Seregard S.. Uveal melanoma: epidemiologic aspects. Ophthalmol Clin North Am. 2005; 18(1): 75–84, viii. [DOI] [PubMed] [Google Scholar]
- 3. Xu TT, Moser JC, Dalvin LA.. Uveal melanoma: laboratory advances and new frontiers in patient care. Curr Opin Ophthalmol. 2021; 32(3): 301–308. [DOI] [PubMed] [Google Scholar]
- 4. Eskelin S, Pyrhönen S, Summanen P, Hahka-Kemppinen M, Kivelä T. Tumor doubling times in metastatic malignant melanoma of the uvea: tumor progression before and after treatment. Ophthalmology. 2000; 107(8): 1443–1449. [DOI] [PubMed] [Google Scholar]
- 5. Rantala ES, Hernberg M, Kivelä TT.. Overall survival after treatment for metastatic uveal melanoma: a systematic review and meta-analysis. Melanoma Res. 2019; 29(6): 561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Orloff M, Carvajal RD, Shoushtari AN, et al.. Overall survival in patients who received checkpoint inhibitors after completing tebentafusp in a phase 3 randomized trial of first-line metastatic uveal melanoma. J Clin Oncol. 2021; 39(15_suppl): 9526. [Google Scholar]
- 7. Piperno-Neumann S, Hassel JC, Rutkowski P, et al.. Abstract CT002: phase 3 randomized trial comparing tebentafusp with investigator's choice in first line metastatic uveal melanoma. Cancer Res. 2021; 81(13 suppl): CT002. [Google Scholar]
- 8. Nathan P, Hassel JC, Rutkowski P, et al.. Overall survival benefit with tebentafusp in metastatic uveal melanoma. N Engl J Med. 2021; 385(13): 1196–1206. [DOI] [PubMed] [Google Scholar]
- 9. Robertson AG, Shih J, Yau C, et al.. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2017; 32(2): 204–220. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jager MJ, Brouwer NJ, Esmaeli B.. The Cancer Genome Atlas project: an integrated molecular view of uveal melanoma. Ophthalmology. 2018; 125(8): 1139–1142. [DOI] [PubMed] [Google Scholar]
- 11. Cai L, Paez-Escamilla M, Walter SD, et al.. Gene expression profiling and PRAME status versus tumor-node-metastasis staging for prognostication in uveal melanoma. Am J Ophthalmol. 2018; 195: 154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jager MJ, Magner JA, Ksander BR, Dubovy SR.. Uveal melanoma cell lines: where do they come from? (An American Ophthalmological Society Thesis). Trans Am Ophthalmol Soc. 2016; 114: T5. [PMC free article] [PubMed] [Google Scholar]
- 13. Tuveson D, Clevers H.. Cancer modeling meets human organoid technology. Science. 2019; 364(6444): 952–955. [DOI] [PubMed] [Google Scholar]
- 14. Drost J, Clevers H.. Organoids in cancer research. Nat Rev Cancer. 2018; 18(7): 407–418. [DOI] [PubMed] [Google Scholar]
- 15. Hogenson TL, Xie H, Phillips WJ, et al.. Culture media composition influences patient-derived organoid ability to predict therapeutic responses in gastrointestinal cancers. JCI Insight. 2022; 7(22): e158060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ou L, Liu S, Wang H, et al.. Patient-derived melanoma organoid models facilitate the assessment of immunotherapies. EBioMedicine. 2023; 92: 104614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Weeber F, van de Wetering M, Hoogstraat M, et al.. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc Natl Acad Sci USA. 2015; 112(43): 13308–13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang R, Yu Y.. Patient-derived organoids in translational oncology and drug screening. Cancer Lett. 2023; 562: 216180. [DOI] [PubMed] [Google Scholar]
- 19. Yang H, Sun L, Liu M, Mao Y.. Patient-derived organoids: a promising model for personalized cancer treatment. Gastroenterol Rep (Oxf). 2018; 6(4): 243–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alsina KM, Sholl LM, Covington KR, et al.. Analytical validation and performance of a 7-gene next-generation sequencing panel in uveal melanoma. Ocul Oncol Pathol. 2021; 7(6): 428–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chowdhury UR, Bahler CK, Hann CR, et al.. ATP-sensitive potassium (KATP) channel activation decreases intraocular pressure in the anterior chamber of the eye. Invest Ophthalmol Vis Sci. 2011; 52(9): 6435–6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dalvin LA, Fautsch MP.. Analysis of circadian rhythm gene expression with reference to diurnal pattern of intraocular pressure in mice. Invest Ophthalmol Vis Sci. 2015; 56(4): 2657–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lenkiewicz E, Malasi S, Hogenson TL, et al.. Genomic and epigenomic landscaping defines new therapeutic targets for adenosquamous carcinoma of the pancreas. Cancer Res. 2020; 80(20): 4324–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robinson JT, Thorvaldsdóttir H, Winckler W, et al.. Integrative genomics viewer. Nat Biotechnol. 2011; 29(1): 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Larsen BM, Cancino A, Shaxted JM, Salahudeen AA.. Protocol for drug screening of patient-derived tumor organoids using high-content fluorescent imaging. STAR Protoc. 2022; 3(2): 101407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Calandrini C, Drost J.. Normal and tumor-derived organoids as a drug screening platform for tumor-specific drug vulnerabilities. STAR Protoc. 2022; 3(1): 101079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kivelä TT, Simpson ER, Grossniklaus HE, et al. Uveal melanoma. In: Amin MB, Edge S, Greene F, et al. eds. AJCC Cancer Staging Manual. 8th ed. Chicago, USA: Springer International Publishing; 2017: 813–826. [Google Scholar]
- 28. Harbour JW. A prognostic test to predict the risk of metastasis in uveal melanoma based on a 15-gene expression profile. Methods Mol Biol. 2014; 1102: 427–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sacco JJ, Jackson R, Corrie P, et al.. A three-arm randomised phase II study of the MEK inhibitor selumetinib alone or in combination with paclitaxel in metastatic uveal melanoma. Eur J Cancer. 2024; 202: 114009. [DOI] [PubMed] [Google Scholar]
- 30. Steeb T, Wessely A, Ruzicka T, Heppt MV, Berking C. How to MEK the best of uveal melanoma: a systematic review on the efficacy and safety of MEK inhibitors in metastatic or unresectable uveal melanoma. Eur J Cancer. 2018; 103: 41–51. [DOI] [PubMed] [Google Scholar]
- 31. Kassumeh S, Arrow S, Kafka A, et al.. Pharmacological drug screening to inhibit uveal melanoma metastatic cells either via EGF-R, MAPK, mTOR or PI3K. Int J Ophthalmol. 2022; 15(10): 1569–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Artico LL, Ruas JS, Teixeira Júnior JR, et al.. IGFBP7 fuels the glycolytic metabolism in B-cell precursor acute lymphoblastic leukemia by sustaining activation of the IGF1R–Akt–GLUT1 axis. Int J Mol Sci. 2023; 24(11): 9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li D, Xia L, Huang P, et al.. Cancer-associated fibroblast-secreted IGFBP7 promotes gastric cancer by enhancing tumor associated macrophage infiltration via FGF2/FGFR1/PI3K/AKT axis. Cell Death Discov. 2023; 9(1): 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Amirouchene-Angelozzi N, Nemati F, Gentien D, et al.. Establishment of novel cell lines recapitulating the genetic landscape of uveal melanoma and preclinical validation of mTOR as a therapeutic target. Mol Oncol. 2014; 8(8): 1508–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Némati F, Sastre-Garau X, Laurent C, et al.. Establishment and characterization of a panel of human uveal melanoma xenografts derived from primary and/or metastatic tumors. Clin Cancer Res. 2010; 16(8): 2352–2362. [DOI] [PubMed] [Google Scholar]
- 36. Carita G, Némati F, Decaudin D.. Uveal melanoma patient-derived xenografts. Ocul Oncol Pathol. 2015; 1(3): 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nemati F, de Koning L, Gentien D, et al.. Patient derived xenografts (PDX) models as an avatar to assess personalized therapy options in uveal melanoma: a feasibility study. Curr Oncol. 2023; 30(10): 9090–9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kageyama K, Ohara M, Saito K, et al.. Establishment of an orthotopic patient-derived xenograft mouse model using uveal melanoma hepatic metastasis. J Transl Med. 2017; 15(1): 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Groenewoud A, Yin J, Gelmi MC, et al.. Patient-derived zebrafish xenografts of uveal melanoma reveal ferroptosis as a drug target. Cell Death Discov. 2023; 9(1): 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yin J, Zhao G, Kalirai H, et al.. Zebrafish patient-derived xenograft model as a preclinical platform for uveal melanoma drug discovery. Pharmaceuticals (Basel). 2023; 16(4): 598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Laurent C, Gentien D, Piperno-Neumann S, et al.. Patient-derived xenografts recapitulate molecular features of human uveal melanomas. Mol Oncol. 2013; 7(3): 625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Richards JR, Yoo JH, Shin D, Odelberg SJ.. Mouse models of uveal melanoma: strengths, weaknesses, and future directions. Pigment Cell Melanoma Res. 2020; 33(2): 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Uner OE, Gandrakota N, Azarcon CP, Grossniklaus HE.. Animal models of uveal melanoma. Ann Eye Sci. 2022; 7: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rago F, Elliott G, Li A, et al.. The discovery of SWI/SNF chromatin remodeling activity as a novel and targetable dependency in uveal melanoma. Mol Cancer Ther. 2020; 19(10): 2186–2195. [DOI] [PubMed] [Google Scholar]
- 45. Heredia-Mendez AJ, Sánchez-Sánchez G, López-Camarillo C. Reprogramming of the genome-wide DNA methylation landscape in three-dimensional cancer cell cultures. Cancers (Basel). 2023; 15(7): 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data described in this report have been deposited in a Sequence Read Archive (BioProject accession no. PRJNA1068520).