Abstract
Aberrant expression of the alpha-fetoprotein (AFP) gene is characteristic of a majority of hepatocellular carcinoma cases and serves as a diagnostic tumor-specific marker. By dissecting regulatory mechanisms through electromobility gel shift, transient-transfection, Western blot, and in vitro transcription analyses, we find that AFP gene expression is controlled in part by mutually exclusive binding of two trans-acting factors, p53 and hepatic nuclear factor 3 (HNF-3). HNF-3 protein activates while p53 represses AFP transcription through sequence-specific binding within the previously identified AFP developmental repressor domain. A single mutation within the DNA binding domain of p53 protein or a mutation of the p53 DNA binding element within the AFP developmental repressor eliminates p53-repressive effects in both transient-transfection and cell-free expression systems. Coexpression of p300 histone acetyltransferase, which has been shown to acetylate p53 and increase specific DNA binding, amplifies the p53-mediated repression. Western blot analysis of proteins present in developmentally staged, liver nuclear extracts reveal a one-to-one correlation between activation of p53 protein and repression of AFP during hepatic development. Induction of p53 in response to actinomycin D or hypoxic stress decreases AFP expression. Studies in fibroblast cells lacking HNF-3 further support a model for p53-mediated repression that is both passive through displacement of a tissue-specific activating factor and active in the presence of tissue-specific corepressors. This mechanism for p53-mediated repression of AFP gene expression may be active during hepatic differentiation and lost in the process of tumorigenesis.
Loss of tumor suppressor p53 function has broad-ranging effects on many cellular processes, including DNA repair, DNA replication, and cell cycle control, and is a critical step in the progression of many human cancers (reviewed in references 32, 46, and 51). p53 is frequently portrayed as an emergency response molecule, activated only under conditions of high stress or DNA damage. However, studies of cellular differentiation and p53 function (24, 55, 66, 70, 80), as well as overexpression of p53 in transgenic mice (24) or knockout of its genetic repressor MDM 2 (10), have underscored the importance of maintaining tightly regulated p53 activity during normal development.
The pleiotropic effects of p53 are most often ascribed to p53-mediated transcriptional activation of downstream target genes (reviewed in references 32, 46, and 51). However, studies of apoptosis inhibitors, including adenovirus E1B-19kD, bcl-2, and WT-1 proteins (56, 68), indicate that p53 may also control cell growth and death by means other than transcription activation. One proposed mechanism is p53-mediated repression of gene expression. For example, activation of p53 protein by UV irradiation of murine embryo-derived fibroblast cell lines downregulated transcription of genes involved in the apoptotic response of cells to stress, e.g., the MAP4 microtubule binding protein (61). As yet, the mechanism by which p53 downregulates these genes remains unclear.
Apparent inhibition of genes which lack p53-binding sites is generally due to p53 protein-protein interaction and interference with transactivators (1, 12, 19, 20, 50, 65, 78, 79, 85). However, p53 protein, when bound to its specific recognition site in DNA, can also recruit and tether adenovirus E1B 55K protein, which then serves as an active repressor of transcription (86). Recent evidence indicates that p53-mediated effects on transcription as a DNA-bound activator or repressor also may be determined by DNA sequence context. p53 protein bound to a regulatory element within the hepatitis B virus (HBV) enhancer repressed transcription in the presence of viral-enhancer-bound activators (64). Thus, the cellular fate upon p53 protein activation may be dictated in part by p53-mediated activation or the repression of specific promoters and may be modulated by context-dependent cofactors (38, 54, 69).
Functional inactivation of p53 is often a critical step in the pathway to tumorigenesis. This progression is frequently accompanied by a reversion to gene expression patterns more characteristic of fetal or undifferentiated cells. Inappropriate expression of the developmentally silenced alpha-fetoprotein (AFP) gene in adult liver occurs in 70 to 85% of all hepatocellular carcinoma (HCC) cases (reviewed in reference 75). Aberrant activation of AFP transcription provides an ideal system to study how a tightly controlled pattern of regulation established during development and differentiation is disrupted under tumorigenic conditions. HCC has been correlated with p53 gene-specific mutations and p53 protein sequestration by HBV proteins, suggesting that the loss of p53 function may be a catalyst of disease progression (reviewed in reference 73). The normal, postnatal repression of AFP is mediated through a repressor domain which lies within 1 kb of the AFP transcription start site (7, 8, 76). This domain can confer developmentally regulated repression upon a heterologous gene. It contains overlapping consensus DNA binding sites for the AFP transactivator, hepatic nuclear factor 3α (HNF-3α [2, 45]), and the tumor suppressor p53. The presence of these overlapping binding sites within a repressor domain suggested models by which formation of a multifactorial complex or the mutually exclusive binding of activator(s) and repressor(s) could direct opposing transcription patterns during hepatic development and/or tumorigenesis.
The studies described here demonstrate that p53 represses AFP transcription through site-specific DNA binding within the AFP repressor domain. p53-mediated repression of AFP gene expression acts in opposition to the hepatic activator, HNF-3α, which competes with p53 for DNA binding. Repression by p53 is further enhanced by coexpression of the histone acetyltransferase-coactivator p300, which has previously been shown to upregulate DNA binding of p53 protein to its specific response element (30). The role of p53 as a repressor of AFP transcription may be augmented by tissue-specific factors. These data suggest a novel mechanism by which the functional inactivation of p53 could potentiate aberrant activation of developmentally repressed genes during tumorigenesis.
MATERIALS AND METHODS
Cell culture and plasmids.
HepG2 cells, a human hepatocellular carcinoma cell line (AFP positive) purchased from the American Type Culture Collection, were cultured in modified Eagle medium (MEM; Gibco BRL) supplemented with 10% fetal calf serum (Gemini Bio-Products, Inc., Calabasas, Calif.). Hepa 1-6 mouse hepatoma cells (AFP positive) were grown in Dulbecco MEM (DMEM; Gibco BRL) supplemented with 10% fetal calf serum. NIH 3T3 fibroblast cells (AFP negative) were grown in DMEM with 10% bovine serum.
Plasmids encoding wild-type p53 (LTR-XA) (36), a p53 DNA-binding mutant protein (LTR-KH) (74), a p53 nuclear transport temperature-sensitive mutant protein (LTR-XV), (58) and p53 recombinant protein (pET15bp53) (39) have been previously described and were generous gifts of G. Lozano. AFP/lacZ contains 3.8 kb of DNA from the mouse AFP gene upstream of the start site, including the AFP proximal and distal promoter and enhancer I fused to the coding region of β-galactosidase (72).
The AFP template containing a mutated p53 site, AFPmut5/lacZ, was constructed by the three-step PCR method previously described (62). The entire process was performed twice to achieve the final product. The original mutated template was initially created through PCR amplification with the AFP/lacZ template and the following primers: A, 5′-CCTCCATTTTATGAGTACACTATA-3′; B, 5′-TGTCTCGAGCGGCCGCCATGTTTGCTAAGGC-3′; C, 5′-GCCTTAGCAAACATGGCGGCCGCTCGAGACA-3′; and D, 5′-CGAGGGGAAAATAGGTTGCGCG-3′. Primers A and B were used in the 5′ amplification of step 1; primers C and D were used in the 3′ amplification. Primers A and D were used in the final amplification. The PCR fragment containing the mutated site was then subcloned into a TA vector, pCR2.1, (Invitrogen) and recovered through restriction digestion and gel isolation. The fragment, containing bp −1007 to +26, was subcloned into BamHI/HindIII-digested AFP/lacZ fragment which lacked the entire AFP enhancer I region. The BamHI fragment from −3.8 to −1 kb, which consists of the AFP enhancer I region and flanking sequences, was subsequently ligated into the BamHI-digested intermediate plasmid to yield APF/lacZmutN plasmid. This plasmid was used as a template for subsequent PCR reactions in the creation of AFPmut5/lacZ with the following primers: A, 5′-CCTCCATTTTATGAGTACACTATA-3′; B, 5′-AGCAAACTCGGCTCCCGCTCAAGACAC-3′; C, 5′-GTGTCTTGAGCGGGTGCCGTGTTTGCT-3′; and D, 5′-CGAGGGGAAAATAGGTTGCGCG-3′. The fragment was generated from a second series of three-step PCRs and inserted into a TA cloning vector, pCR2.1 (Invitrogen), exactly as described above to generate the final mutated template, AFPmut5/lacZ.
The HNF-3 expression plasmid used in transient transfections contained the Rous sarcoma virus long terminal repeat as a promoter and the bovine growth hormone polyadenylation site downstream of the coding region (42). p300 (wild-type p300 expression driven by the cytomegalovirus [CMV] promoter) and pΔ300 (p300 coding sequence deleted) plasmids were provided by P. Brindle (13). Plasmids were prepared and purified by cesium chloride gradient centrifugation (67).
Transfections and reporter assays.
Cells were transfected by the calcium-phosphate method as previously described (28) with a few modifications. Cells were plated at approximately 3.0 × 105 cells per 60-mm dish 36 h before transfection. For each plate, a total of 10 to 12 μg of DNA were transfected as calcium phosphate coprecipitates and were maintained at equal concentrations in each experiment by cotransfection of control vector DNA. Cells were incubated for approximately 16 h after DNA introduction and then given a 15% glycerol shock for 3 min at 37°C. After three rinses with phosphate-buffered saline (PBS), incubation in fresh glycerol-free media continued for an additional 24 to 36 h prior to the harvesting and preparation of cell extracts. For hypoxia studies, cells were incubated for 2 h after glycerol shock, placed under hypoxic (1% oxygen) or normoxic conditions at time equal zero, and harvested at specific time points. In all studies, cells were harvested by complete medium removal, addition of 500 μl of lysis buffer (Promega, Madison, Wis.), and scraping with a rubber policeman. The protein concentration of cellular extracts was determined by the Bradford method (Bio-Rad, Melville, N.Y.).
Cellular lysates were then analyzed for β-galactosidase activity as described by Spear et al. (72). As an internal control for transfection efficiency, cells were cotransfected with pGL2 or pGL3 expression vectors (Promega) and analyzed for luciferase activity in an analytical luminescence laboratory luminometer.
In vitro transcription, cell extracts, and protein expression.
The method of Gorski et al. (25, 57) was used to prepare nuclear extracts from newborn mice aged 12 h to 4 days, mice at 2 weeks of age, and adult mice. Two grams of liver tissue from newborn and 2-week-old mice and 18 g of adult mouse liver tissue were processed. In vitro transcription extracts were prepared from human HepG2 cells as described by Dignam et al. (11) with the following minor modifications: cells were grown to 70% confluence and harvested by scraping into 1× PBS. Washed pellets were resuspended in six times the packed cell volume (PCV) with hypotonic buffer (20 mM HEPES, pH 7.9; 10 mM NaCl; 1.5 mM MgCl2; 2 mM dithiothreitol [DTT]). After cells were allowed to swell 10 min on ice, they were pelleted and resuspended in two PCVs with hypotonic buffer containing 0.05% Nonidet P-40 (NP-40) prior to Dounce homogenization (type B pestle; Wheaton). The remainder of the procedure was performed exactly as previously described (11). Protein extracts were dialyzed against two changes of 100 volumes of dialysis buffer (20 mM HEPES, pH 7.9; 50 mM KCl; 0.2 mM EDTA; 20% glycerol; 1 mM DTT; 0.2 mM phenylmethylsulfonyl fluoride [PMSF]) for 2 h. each. In vitro transcription reactions were performed as previously described (5, 16).
Purification of recombinant, full-length p53 was performed basically as described by Hupp et al. (39). BL21/DE3 cells harboring pET15bp53 were grown at 37°C to an A600 of 0.6 to 0.8 in Luria-Bertani (LB) medium (67) containing 100 μg of ampicillin per ml, at which point IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 1 mM to induce expression. After overnight incubation at 37°C, cells were harvested by centrifugation at 4°C for 15 min and resuspended in a buffer containing 10% sucrose and 50 mM HEPES (pH 8.0) to a final A600 of 150 to 200. Cells were lysed by the addition of KCl to 0.25 M, DTT to 2 mM, and lysozyme to 0.5 mg/ml with mixing. After incubation for 30 min on ice, the lysates were centrifuged. The supernatant was used as a crude source of soluble p53 protein.
Further purification of p53 was achieved as described by Gallagher and Blumenthal (22) with some modifications. After two passages through a French pressure cell, the cell lysate was slowly mixed at 4°C with 4 M ammonium sulfate (final concentration, 2 M) and allowed to incubate on ice for 30 min. Protein was pelleted by centrifugation and then resuspended in buffer B (100 mM KCl; 20% glycerol; 25 mM HEPES, pH 7.6; 0.1 mM EDTA; 2 mM DTT; 5 mM MgCl2). The ammonium sulfate-fractionated protein was dialyzed against excess buffer B. The dialyzed protein was passed over a heparin Sepharose Hi-Trap column (Pharmacia) preequilibrated in buffer B containing 50 mM KCl. Adsorbed proteins were eluted by a step gradient from 50 mM to 1 M KCl in buffer B and collected in 500-μl aliquots. Protein concentrations were determined as previously described. Peak protein fractions were determined by separation on a 12% gel by sodium dodecyl sulfate (SDS)-polyacrylamide gel and Western blot analysis (ECL; Amersham Life Science Products). Monoclonal antibody specific for the C-terminal end of p53, pAb421, was purchased from Pharmagin. Peak fractions positive for p53 were pooled and dialyzed against excess buffer B for a total of 3 h at 4°C. Proteins were stored at −100°C prior to use.
In vitro activation of recombinant p53 was achieved by casein kinase II (New England Biolabs) phosphorylation in a reaction mixture containing 20 mM Tris; HCl, pH 8.0; 50 mM KCl; 100 mM MgCl2; 1 mM ATP; and 20 mM PMSF. The reaction was incubated at 30°C for 30 min with 50 U of casein kinase II enzyme, and the activated p53 protein was utilized as described in the text.
A plasmid coding for the constitutively active human p53 protein, p53Δ30 (39), was created by PCR amplification of pET15bp53 by using the following primers: 5′-GATATACATATGGAGGAGCCGCAGTCAG-3′ and 5′-CTCGAGTGCGGCCGCGCCCTGCTCCCCCC-3′. The amplified fragment was first inserted into a TA cloning vector (Invitrogen) and then isolated through restriction enzyme digestion. The resulting DNA fragment was then inserted into a pET23b vector (Novagen), which added a histidine tag to the C terminus of the protein. BL21/DE3 cells harboring p53Δ30 were grown in LB media with 100 μg of ampicillin per ml until the optical density was between 0.6 and 0.8, at which point IPTG was added to a final concentration of 1 mM to induce expression. After an overnight induction at 37°, cells were harvested by centrifugation, and the pellets were frozen, thawed, and resuspended in 20 ml of binding buffer (5 mM imidazole; 0.5 M NaCl; 20 mM Tris-HCl, pH 7.9; 1 mM PMSF). After a 30-s sonication, cells were incubated on ice for 5 min and then pelleted by centrifugation for 15 min at 4°C. The pellets were resuspended in binding buffer containing 6 M urea and 1 mM PMSF. The cells were sonicated again and then incubated at 4° for 1 h on a rotating platform (Nutator). Supernatant and cell debris were then separated by centrifugation for 15 min. Supernatant was then passed over a nickel agarose (Qiagen) column, which had been charged with 50 mM NiSO4, and washed with binding buffer containing 6 M urea. After the supernatant had passed through, the column was washed with additional binding buffer containing 6 M urea. The column was washed with wash buffer (20 mM imidazole; 0.5 M NaCl; 20 mM Tris-HCl, pH 7.9). Proteins were then eluted with elution buffer (300 mM imidizole; 0.5M NaCl; 20 mM Tris-HCl, pH 7.8) containing 6 M urea. Eluted fractions were dialyzed against at least 100 volumes of dialysis buffer (20 mM HEPES, pH 6.5; 400 mM KCl; 1 mM MgCl2; 20% glycerol; 0.1% NP-40; 0.5 mM PMSF; 3 mM DTT) with stepwise reduction in urea concentration. Dialysis was begun in the cold room for 3 h against dialysis buffer containing 5 M urea and then for 2 h each at 4, 2, 1, and finally 0 M urea. Protein was confirmed by Western blotting analysis, frozen as aliquots in liquid nitrogen, and stored at −100°C until use.
Recombinant HNF-3α was expressed in bacteria and purified extensively by K. Stevens and K. Zaret (88). The purified HNF-3α protein used in the electrophoretic mobility shift assay (EMSA) was the generous gift of K. Zaret.
EMSAs.
The following double-stranded oligomers were used in gel retardation assays: overlapping HNF-3/p53 binding site from AFP −860 to −831 30-mer, 5′-GCCTTAGCAAACATGTCTGGACCTCTAGAC-3′ and 3′-CGGAATCGTTTGTACAGACCTGGAGATCTG-5′; consensus C/EBP site (44) 20-mer, 5′-TTCAATTGGGCAATCAGGAA-3′ and 3′-AAGTTAACCCGTTAGTCCTT-5′; consensus p53 site (15) 23-mer, 5′-TAGGCATGTCTAGGCATGTCTAAGCT-3′ and 3′-ATCCGTACAGATCCGTACAGATTCGA-5′; and consensus HNF-3 site (48) 24-mer, 5′-GATCCCCTTTATTGACTTTGACAG-3′ and 3′-GGGAAATAACTGAAACTGTCCTAG-5′.
Oligomers were radiolabeled by using [γ-32P]ATP (DuPont-NEN) and T4 polynucleotide kinase (Gibco BRL) or by [α-32P]dATP (DuPont-NEN) and Klenow fragment (Gibco BRL) filled in according to standard protocols (67). Radiolabeled, duplexed oligomers were incubated with cellular extracts in a reaction mixture containing (final concentrations) 1 mM DTT, 50 mM NaCl, 5 mM MgCl2, 20 mM EDTA, 6% glycerol, 0.1 mg of bovine serum albumin per ml, 1 μg of poly(dI)-poly(dC), and 0.025% NP-40. Incubations were carried out on ice for 30 min. In cases where antibodies or cold competitor were included for supershifts or competition, respectively, reactions were assembled, including the antibody or unlabeled oligonucleotide but without the radiolabeled probe and allowed to incubate on ice for 15 min. Similarly, when protein competition for DNA binding was analyzed, proteins were added to reactions without the radiolabeled probe and allowed to incubate on ice for 15 min. Probe was then added, and the reaction was allowed to continue for an additional 20 min. Samples were analyzed by electrophoresis at 4°C at 240 V on a 5% polyacrylamide gel (39:1, acrylamide/bis) containing 0.5× Tris-borate-EDTA (TBE), 0.05% NP-40, 5% glycerol, 1 mM EDTA, and 0.5 mM DTT in a running buffer of 0.5× TBE.
Western blot analysis and antibodies.
Western blotting was performed as previously described (9). Approximately 50 μg of total protein of newborn, 2-week-old, and adult mouse liver nuclear extracts and 30 μg of both untreated and actinomycin D-treated (59) cellular extracts were fractionated by electrophoresis on a 4.2% stacking–10% separating SDS polyacrylamide gel. Proteins were transferred to nitrocellulose (NitroBond; MicroSystem, Inc., Westborough, Mass.) overnight at 40 mA. The nitrocellulose was blocked in blocking buffer (100 mM Tris Cl, pH 7.8; 150 mM NaCl; 0.1% Tween 20; 3.0% bovine serum albumin [BSA]) and then probed with the appropriate antibody. The blot was stripped and reblocked for subsequent probing with additional antibodies as recommended by the manufacturer (Amersham), with the exception that stripping was done at 42°C for 35 to 40 min. Primary antibodies used were as follows, p53, pAb240 (Santa Cruz); p21, WAF1(Ab5) (Oncogene Research Products); alpha-fetoprotein, lyophilized anti-mouse alpha-fetoprotein serum (ICN, Irvine, Calif.). Enhanced chemiluminescence reagents and secondary antibodies were provided by the manufacturer (Amersham).
RESULTS
AFP gene expression is repressed upon p53 expression.
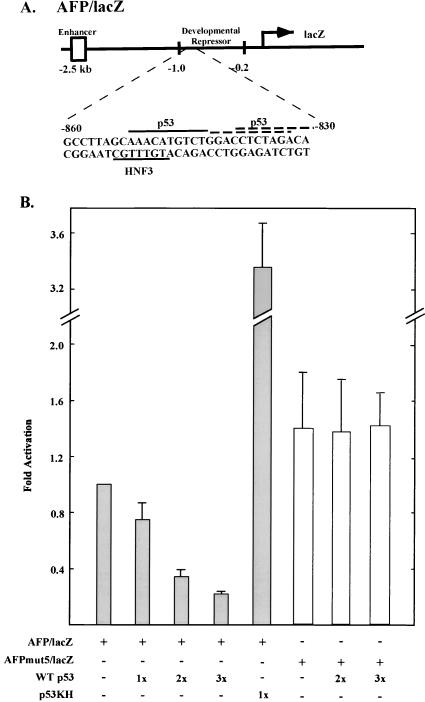
An extensive series of studies primarily using a transgenic mouse model defined a region of the mouse AFP distal promoter that confers postnatal silencing (76). Repression at the level of transcription occurs in heterologous constructs containing this regulatory element independent of the AFP gene enhancer(s) (71). In pursuing regulatory studies of aberrant AFP gene activation during tumorigenesis, we focused on potential transcription factors interacting with the developmental repressor domain. Computer-aided scanning (with Entrez/MacVector) for transcription factor consensus binding sites within this repressor domain revealed protein binding sites for the hepatic-enriched HNF-3 fetal liver activator overlapping one dimer–half-tetramer binding site of the p53 tumor suppressor protein (Fig. 1A). This dimer binding site matches the reported consensus sequence for p53 regulation, PuPuPuCA/TT/AGPyPyPy (15; 87). Directly abutting this dimer half-site is a less perfect direct repeat of the element and an additional imperfect half-site with a spacing of 3 base pairs (Fig. 1A).
FIG. 1.
AFP repression is mediated by wild-type p53 through a consensus binding site in the developmental repressor domain. (A) Schematic of overlapping p53/HNF-3 sites located between −830 and −860 bp of the AFP promoter. The solid lines represent the consensus sequence, and the dashed lines represent the imperfect p53 half-site consensus sequence. (B) Transfection results. Hepa 1-6 cells were transfected with either intact AFP/lacZ construct (3 μg/plate [shaded bars]) or a construct containing a site-specific mutation within the overlapping p53/HNF-3 binding site, AFPmut5/lacZ (3 μg/plate [open bars]). Cells were cotransfected as indicated with p53 expression vectors, either wild type at 1×, 2×, or 3× concentrations (3, 6, or 9 μg/plate, respectively) or mutated in the DNA binding domain (p53KH, 3 μg/plate). All plates were cotransfected with 1 μg of internal transfection efficiency vector pGL2 or pGL3 per plate, each of which constitutively expresses luciferase. Averaged values are expressed in the fold activation of β-galactosidase activities compared to the activity of AFP/lacZ transfected alone (set at 1×) and corrected for transfection efficiency as described in the text. Experiments were performed in duplicate or triplicate 3 to 15 times.
Transient-transfection analysis of p53 and HNF-3 coexpression with an AFP reporter gene revealed opposing regulatory functions for the two trans-acting factors (Fig. 1B and 4). Overexpression of wild-type p53 in mouse hepatoma Hepa 1-6 cells represses transfected AFP-driven lacZ expression in a dosage-dependent manner (Fig. 1B). The observed four- to fivefold repression was highly reproducible and specific to wild-type p53. Mutant p53 protein is unable to repress AFP-driven gene expression. The mutant protein p53KH encodes p53 protein harboring an amino acid substitution within the DNA binding domain, which is unable to bind DNA (74). Overexpression of p53KH results in a low-level activation of AFP/lacZ expression, pinpointing repression of AFP to normal p53 protein activity and suggesting action of the mutant protein as a dominant negative for transcription repression.
FIG. 4.
HNF-3 activation is abrogated by p53 expression. Hepa 1-6 cells were transfected with the indicated expression vectors (wild-type p53, DNA binding-deficient p53 (p53KH), and/or HNF-3) at the indicated DNA levels of 1×, 2×, or 3× concentration (3, 6, or 9 μg/plate, respectively). Reporter template AFP/lacZ (bars) was added at 3 μg/plate. Each plate was cotransfected with 1 μg of pGL2 vector (Promega) per plate to monitor and correct for transfection efficiency. Values are expressed as the fold activation above the level of AFP/lacZ activity cotransfected with carrier DNA only normalized β-galactoside activity (set at 1×). Experiments were performed in duplicate or triplicate at least three times.
In order to determine whether the observed repression of AFP by p53 was mediated by site-specific DNA binding, we mutated the p53/HNF-3 DNA binding site in our AFP reporter construct (see Material and Methods). By EMSA, the mutation generated in the AFP repressor region was found to no longer support p53 protein binding to DNA (data not shown). Transient cotransfection of an AFP/lacZ reporter containing a mutation in the p53 binding site (AFPmut5/lacZ) along with a wild-type p53 expression plasmid abolished p53-mediated repression of AFP expression (Fig. 1B). The levels of β-galactosidase expression driven by AFPmut5 remained constant and slightly elevated above the levels of the wild-type promoter in the presence or absence of cotransfected p53.
Repression of AFP gene transcription in vitro by p53 protein.
Activation of gene expression by p53 protein has been reconstituted in vitro by transcription in cell-free HeLa extracts (18, 43). We have examined whether p53 mediates a direct repression of AFP expression in a hepatoma background in the absence of protein synthesis by in vitro transcription of AFP/lacZ constructs in HepG2 human hepatoma transcription extract (Fig. 2A). Addition of recombinant p53 protein to a cell-free transcription assay of AFP/lacZ DNA has profound effects on AFP-driven expression, which are specific for the p53 DNA binding site. As amounts of recombinant p53 protein were increased, AFP/lacZ transcription is largely diminished (lanes 3 to 6, compare to lanes 2 and 7). Addition of buffer only or buffer plus carrier protein BSA had no effect on the level of transcription (lanes 2 and 7, compare to lane 1).
FIG. 2.
Direct repression of AFP transcription by p53 in vitro. (A) In vitro transcription of AFP/lacZ and AFPmut5/lacZ. All in vitro transcriptions were performed in 15 μl of whole-cell HepG2 extract and analyzed by primer extension as described elsewhere (5). Lanes 1 through 7 contain 1 μg of intact AFP/lacZ template, while lane 1 contains template only. Lane 2 has 5 μl of p53 buffer only (buffer B; see Materials and Methods) added to the reaction. Lanes 3 to 6 contain approximately 2, 6, 8, and 10 ng of p53 protein, respectively, and lane 7 contains 5 μg of BSA diluted in buffer B. (B) p53 does not repress AFP transcription by nonspecific squelching. Transcription conditions were identical to those in panel A, lanes 2 to 7, except for the use of 800 ng of AFPmut5/lacZ template along with increasing amounts of p53Δ30 protein or 1 μg of RSVcat DNA instead of the AFP/lacZ templates.
Previous in vitro transcription studies have revealed that increasing titration of recombinant p53 protein reduced p53-mediated transcription activation and repressed genes which lack a p53 binding site (37, 53). In contrast, our in vitro experiments confirm that repression of AFP is mediated through site-specific binding to DNA. The highest concentration of p53 protein used in these cell-free studies repressed AFP/lacZ transcription 78% (lane 6 compared to lane 2 or 7). In vitro transcription reactions performed in parallel with AFPmut5/lacZ template (lanes 8 to 10) lacking a recognizable p53 binding site and a p53 nonresponsive RSV-CAT gene (Fig. 2B) revealed little alteration in transcription efficiency in the presence of recombinant p53. Since expression by an AFP template with the p53 site specifically mutated, as well as RSV-CAT, was relatively unaffected by titration of p53 protein, in vitro repression of AFP transcription is not likely due to nonspecific squelching.
Developmental, postnatal repression of AFP correlates with p53 activation.
Within 2 weeks of birth, abundant AFP expression is dramatically reduced to nearly undetectable levels (75). We prepared nuclear extracts of developmentally staged hepatic tissue to determine whether AFP silencing in vivo was concomitant with activation of p53. Western blot analysis of these extracts with antibody probes specific for AFP, p53, and p21 proteins was performed (Fig. 3A). As the abundance of p53 protein increases with time of development, AFP protein becomes undetectable (lanes 1 to 3). P53 protein levels are not only increased but activated for downstream transcription as well, as illustrated by the induction of p21 protein expression at 2 weeks of age (lane 2). Previous studies of hepatic-specific p21 overexpression in a transgenic mouse model revealed a critical role for p21 in normal hepatic development, affecting postnatal decreases in cellular proliferation (83). Interestingly, p21 protein is easily detectable early in postnatal development (lane 2) but not in extracts of fully differentiated adult liver tissue (lane 3). We have not determined whether the suggestion of slower-migrating forms of p53 protein on an SDS-polyacrylamide gel, which change over time of development, represents posttranslational modification of the p53 protein.
FIG. 3.
Induction of p53 during hepatic development and in response to cellular stress results in AFP repression. (A) In vivo induction of p53 activity correlates with reduced AFP protein levels during development and in response to DNA damage. Developmentally staged mouse liver nuclear extracts (lanes 1 to 3), cellular extracts of Hepa 1-6 cells either untreated (lane 5) or treated with actinomycin D (lane 6) as described in Material and Methods were immunoblotted. Recombinant p53Δ30 protein was included as a control (lane 4). The presence of p53, AFP, and p21 proteins in each extract was revealed by sequential probing with the indicated antibody. (B) Hypoxic stress represses AFP-driven transcription. Hepa 1-6 cells were transfected with AFP/lacZ reporter template, along with control vector DNA (solid circles), wild type, or mutant p53 expression plasmids (all at 3 μg/plate). At 12 to 15 h after transfection, cells were subjected to a 15% glycerol shock and placed in either normoxic (dashed lines) or hypoxic (solid lines) 1% oxygen conditions. Cells were harvested at 0, 12, and 24 h and assayed for reporter activity. All cells were cotransfected with 1 μg of pGL2 vector (Promega) per plate to monitor and correct for transfection efficiency. Values are expressed as the fold activation above the levels of the AFP/lacZ activity cotransfected with carrier DNA only, which was set as equal to 1.0 in all cases. Values shown are the average of at least two experimental trials.
DNA damage and hypoxic stress result in reduced AFP expression.
To further address the reliability of the cell culture model and confirm the 1:1 correlation between p53 activation and AFP repression, we examined the physiological induction of p53 by using two well-established models of cellular stress. Incubation of Hepa 1-6 cells in actinomycin D as previously described (59) activated the p53 protein (lanes 5 and 6). This DNA damage-induced activation of p53 resulted in increased p21 expression and repression of endogenous AFP levels. Hepa 1-6 cells appear to express a low level of endogenous p53 that is fully capable of transactivation (and transrepression) in response to DNA damage. It is unlikely that AFP repression is the direct result of downstream p21 protein synthesis, as indicated by our in vitro transcription results showing p53-mediated repression of AFP by DNA binding.
One of the physiological responses of cells to reduced oxygen levels is the induction of p53 (26, 27). The mediators of the hypoxic signal in the activation of p53 have not been identified, but the ability of a cell’s redox state to influence p53 activity is well established (33, 43). A severalfold accumulation of wild-type p53 protein under hypoxic conditions (27), as well as activation of latent p53 pools by the redox/repair protein Ref-1, occur in vivo (43). Here we show that exposure of Hepa 1-6 hepatoma cells transfected with AFP/lacZ reporter to reduced oxygen conditions resulted in inhibition of AFP expression (Fig. 3B).
AFP repression under hypoxic conditions (Fig. 3B, solid lines) versus normoxic (dashed lines) was apparent in each experimental transfection study. In the presence of 1% oxygen (minus transfected exogenous p53 [filled circles, solid line]), AFP-driven reporter gene expression decreased over time, ending in a 3.7-fold inhibition within 24 h. Transfected cells which received exogenous p53 displayed reduced AFP expression over this time period to the same degree (3.5-fold [open squares, solid line]). Physiological induction of p53 protein activity likely resulted in additive repression of AFP with exogenously expressed p53, rather than through a cooperative potential redox activation of overexpressed, latent p53 within this time period.
Control transfection of mutant p53 unable to bind DNA (Fig. 3B, open circles solid line) slightly activated AFP gene expression early in the time-course as previously determined (Fig. 1B). Activation of AFP was then attenuated over the entire time course, a result most likely due to increased activation of endogenous p53 protein (Fig. 3B). When individual times are compared to each other, the 24-h time point samples displayed an increased activation ratio comparing p53 mut- to (minus p53)-transfected cultures at time equal zero. Thus, overexpression of mutated p53 negates repression of AFP expression and supports the model that repression is by p53 action.
Physiological p53 induction concomitant with AFP repression underscores that overexpression and squelching of transactivators of AFP are not likely causes of apparent repression. These results support the role of p53 protein as an actively repressing factor in the regulation of AFP expression, rather than as an artifactual result of p53 overexpression.
HNF-3 activates AFP expression in opposition to p53-mediated repression.
Transfection and expression of HNF-3α in hepatoma cells activates AFP-driven gene expression (Fig. 4). Activation of AFP increased from approximately 13-fold at the lowest concentration up to nearly 30-fold as greater amounts of HNF-3 expression vector were introduced. Cotransfection with a wild-type p53 expression clone markedly reduced this activation. Reduction of HNF-3 activated expression was p53 concentration dependent and relatively linear over the range analyzed. Substitution of mutant p53KH in these transfection studies reveals that repression is specific to p53 protein capable of binding to DNA. Further support for p53KH acting as a dominant-negative factor in these transfections is lent by an apparent superactivation of AFP-driven expression with HNF-3 and p53KH cotransfection. As we have shown above (Fig. 3A) that Hepa 1-6 cells express a low level of endogenous p53 that is fully capable of activation, it is likely that p53KH squelches the activity of endogenous p53 in these cells.
Both recombinant HNF-3 and p53 bind the repressor region specifically.
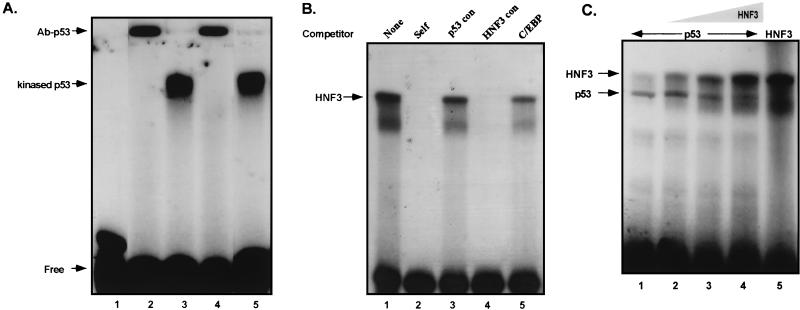
We have performed EMSA with double-stranded oligonucleotides (−860 to −830 of the mouse AFP gene; Fig. 1A) containing the overlapping HNF-3/p53 binding sites. Incubation of 32P-labeled −860/−830 oligonucleotide with recombinant p53 protein (Fig. 5A) and recombinant HNF-3 protein (Fig. 5B) reveals specific binding of the proteins to their putative sites within the AFP repressor domain. Addition of a C-terminal-specific monoclonal antibody for p53 (PAb421; Pharmingen) or in vitro modification by phosphorylation activates latent p53 tumor suppressor protein for DNA binding and is required for DNA binding of bacterially expressed p53 protein (39, 40, 60). The 1.0 M heparin-Sepharose fraction of recombinant p53 protein was activated for DNA binding by either in vitro phosphorylation by casein kinase II (lanes 3, 4, and 5) or by incubation with PAb421 antibody (lanes 2 and 4). Phosphorylation by casein kinase II did not affect the ability of p53 to interact with the C-terminal specific antibody (lane 4). Kinase-treated recombinant p53 protein was employed in further in vitro analyses as described below in order to avoid the possible influence of antibody addition on experimental outcome and interpretation.
FIG. 5.
p53 and HNF-3 bind their overlapping sites in the developmental repressor domain in a mutually exclusive manner. All EMSA reactions contained approximately 1 ng of 32P-labeled HNF-3/p53 repressor region as diagrammed in Fig 1A. (A) Purified recombinant p53 protein binds to the overlapping HNF-3/p53 sites. Lane 1 contains free probe alone. Lanes 2 to 5 contain approximately 5 ng of semipurified protein each. Lanes 2 and 4 contain approximately 40 ng of PAb421, a p53 C-terminal specific antibody. The upper arrow indicates the supershifted complex. Lanes 3, 4, and 5 contain p53 that has been phosphorylated via casein kinase II in vitro. (B) Recombinant HNF-3 protein binds this site specifically. Lanes 1 to 5 contain approximately 5 ng of purified recombinant HNF-3 protein incubated with labeled probe. Competitor oligonucleotides, p53/HNF-3 unlabeled probe, p53 consensus, HNF-3 consensus, and a nonspecific oligonucleotide were added at a 100-fold molar excess to labeled probe as indicated above the gels. (C) EMSA reactions with recombinant p53, activated by casein kinase II phosphorylation, and HNF-3 protein. Lanes 1 to 4 each contain about 150 pg of p53 protein and 0, 3, 6, and 12 ng of HNF-3 protein, respectively. Lane 5 contains 3 ng of HNF-3 protein without competing p53 protein.
Band-purified, recombinant HNF-3α protein (88) bound to the −860/−830 region of the AFP gene with high specificity (Fig. 5B). The protein-DNA complex (lane 1) was competed by a 100-fold molar excess of cold, unlabeled −860/−830 oligonucleotide (lane 2). Similarly, the HNF-3/DNA complex was competed by a consensus HNF-3 site (lane 4). Specificity was tested by competition with a 100-fold molar excess of a p53 consensus binding site oligomer (lane 3) and the C/EBP consensus binding site (lane 5). Neither of these DNA oligomers competed effectively for HNF-3 binding to the AFP −860/−830 region, demonstrating the nucleotide specificity of HNF-3 binding.
p53 and HNF-3 bind to overlapping sites in a mutually exclusive manner.
Proteins binding in close apposition to each other at composite regulatory elements can act in an additive or cooperative manner or, due to mutually exclusive binding, may direct opposing expression patterns. HNF-3α has been previously demonstrated to bind a composite HNF-3/NF1 DNA binding site and cooperate with NF1 protein in the activation of the mouse albumin enhancer (42). Since fully one-half of the 5′ p53 dimer binding site overlaps five of seven nucleotides comprising the consensus HNF-3 binding site, we examined whether p53 and HNF-3 could occupy this site simultaneously (Fig. 5C).
The specific protein-DNA complex formed by approximately 150 pg of casein kinase II-phosphorylated p53 recombinant protein is displayed in Fig. 5C (lane 1). The addition of increasing amounts of recombinant HNF-3α protein interfered with formation of the p53-DNA complex (lanes 2 to 4; 3, 6, and 12 ng, respectively). Clearly, the presence of p53 protein in these reactions is inhibitory to the binding of HNF-3α. An equivalent amount of HNF-3α protein as used in lane 5 (3 ng) bound with greatly reduced avidity in the presence of p53 protein (lane 2). An approximately 80-fold-higher molar amount of HNF-3α protein was required to completely interfere with p53 protein binding to the −860/−830 oligomer (lane 4).
The results of in vitro protein-DNA binding show (i) that recombinant p53 and HNF-3 proteins bind to the −860/−830 DNA element individually, (ii) that this binding is not cooperative or additive (i.e., no larger-molecular-weight complexes are formed with the DNA) but rather favors the binding of one factor or the other but not both, and (iii) that p53 protein displays a much higher binding affinity in competition with HNF-3α for the same AFP regulatory element.
Coexpression of p300 amplifies repression of AFP gene expression by p53.
Our experiments described thus far show that p53 mediates repression of AFP expression by a DNA sequence-specific interaction with the developmental repressor region. Further support for this mechanism is provided by the coexpression of p300 in transfection studies (Fig. 6). The acetylation of p53 protein by the cofactors CBP and p300 has recently been shown to potentiate transactivation (3, 31, 52). Acetylation modifies the C-terminal 30 amino acids of p53, the same regulatory domain modified by phosphorylation, resulting in enhancement of DNA binding ability (30). An increase in the ability of p53 to bind DNA is directly reflected in an amplified expression of genes activated by p53. Conversely, if p53 causes repression of gene expression upon DNA binding, then acetylation should increase the repression of such genes.
FIG. 6.
Coexpression of p300 increases p53 repression of AFP. All experiments contained an equal amount of transfected AFP/lacZ (3 μg/plate) with the indicated 1×, 2×, or 3× relative levels of p53 and p300 expression vectors (3, 6, or 9 μg/plate, respectively). Δp300 is the p300 expression plasmid from which the p300 coding sequence is deleted. Hepa 1-6 cells were cotransfected with 1 μg of pGL2 vector per plate to monitor and correct for transfection efficiency. Averaged values are expressed as the fold activation above the normalized AFP/lacZ β-galactosidase activity cotransfected with the carrier DNA only activity, which was set equal to 1. Experiments were performed two to four times in duplicate or triplicate.
We tested this model by transient transfection of p53, p300, and the AFP reporter constructs in hepatoma cells. At lower levels of both the p53 and p300 expression vectors (1×), repression is augmented 2.5-fold, while at higher levels of each (2×) this amplification increases to 4-fold. AFP expression drops by 85% under these conditions. Expression of either p300 or vector alone had little effect on AFP expression, further supporting a model for p53-mediated repression via binding to a specific DNA element. Our results indicate that the introduction of p300 enhances the repressive effect of p53 on AFP-driven expression.
A mechanism for p53-mediated AFP expression.
In order to further dissect the means by which p53 represses AFP gene expression, we employed a cellular background devoid of endogenous hepatic transactivators. Transient-transfection experiments were performed with NIH 3T3 fibroblast cells which do not express AFP (63). Coexpression of p53 (WT p53) and the AFP/lacZ reporter vectors in this cellular background leads to an activation rather than repression of AFP-driven expression (Fig. 7). Addition of p53KH mutant protein to transfections of AFP/lacZ in fibroblasts did not activate AFP expression (p53KH). Thus, we found that introduction of p53 into cells lacking HNF-3 and other hepatic-enriched factors leads to AFP activation rather than repression.
FIG. 7.
p53-mediated repression of AFP is hepatoma specific. NIH 3T3 cells were transfected with wild-type or mutant p53 expression vector at 1× or 2× concentrations as indicated (3 or 6 μg/plate, respectively) and/or HNF-3 expression vector (3 μg/plate), in addition to AFP/lacZ (3 μg/plate). All plates contain 1 μg of pGL2 luciferase reporter as an internal control. Averaged values are expressed as the fold activation above normalized AFP/lacZ β-galactosidase activity cotransfected with carrier DNA only. Experiments were performed in duplicate or triplicate at least twice.
Our studies indicate that AFP repression by p53 is both passive through physical exclusion of HNF-3 transactivator binding and active in transcription interference. HNF-3 addition to 3T3 fibroblast cell transfections activated AFP-driven expression 17-fold in the absence of p53, a level comparable to that seen in hepatoma transfections. The addition of p53 to HNF-3 cotransfections in NIH 3T3 cells led to a diminution of HNF-3-activated expression down to the level of activation observed with p53 alone. These results in fibroblast cells are most easily explained by the passive exclusion of the strong activator HNF-3 through (higher-affinity) binding of the weaker activator p53. Basal expression of AFP/lacZ in the absence of any cotransfected p53 or HNF-3 is comparable in both hepatoma and fibroblast cell types (data not shown).
Since p53 binds its consensus site and enhances AFP-driven expression in fibroblasts, active interference with AFP transactivation seems to be lacking. However, the ability of p53 to compete with HNF-3 protein binding within the developmental repressor appears to be maintained, resulting in an expression more closely approximating p53 activation in the absence of HNF-3. p53-mediated repression of transcription is therefore highly context dependent, suggesting that active repression requires the presence of hepatoma-specific corepressors.
DISCUSSION
A combinatorial mechanism of AFP repression.
Our studies support a model in which p53 protein represses AFP expression by site-specific binding to an overlapping p53/HNF-3 response element. Mutation of p53 protein or deletion of the target binding site abrogated repression of AFP both in cell culture and in vitro. Downregulation of AFP expression occurred under conditions of physiological induction or overexpression of p53 and was amplified by coexpression with p300. Recapitulation of AFP-specific repression in vitro indicates that regulation of AFP transcription is direct and does not require protein synthesis of downstream mediators.
Repression of transcription can be broken down into passive and active modes, though negative regulation is generally a combination of distinct mechanisms (reviewed in references 29, 34, and 49). Our studies show p53-mediated repression of AFP in hepatoma cells conforms to this generality of combinatorial regulation. Simple, passive exclusion of an activator from a common DNA binding site is supported by both in vitro DNA binding data and regulatory studies. In vitro analysis of recombinant protein binding revealed that p53 has a higher binding affinity for the overlapping HNF-3/p53 regulatory element and interferes with HNF-3 interaction at this site. p53 functions as a dose-dependent inhibitor of HNF-3-activated AFP gene expression in hepatoma transfections.
Modulation of activator or repressor binding affinities via ligand binding, heterodimer composition changes, corepressor interaction, or posttranslational modification can alter competition for a regulatory element. A role for posttranslational modification in activating p53 DNA binding and function has been demonstrated in several instances, including p53 acetylation. Expression of p300/CBP histone acetyltransferase has been shown to enhance activation of transfected p53-regulated reporters (3, 30, 31, 52). Our studies also provide support for synergy between p300 and p53 in regulation, in this case by amplification of p53-mediated repression of AFP expression in the presence of p300.
An active mode of transcription repression.
Active interference with transcription through p53 binding to the repressor site is supported by the four- to fivefold downregulation of basal AFP/lacZ expression in hepatoma cells. AFP gene expression is regulated by multiple factors binding a complex promoter and enhancer(s) (6, 21, 23, 81, 82, 89). A number of these factors are tissue specific or hepatic enriched, including HNF-3, which we show binds and activates AFP transcription from one site within the repressor region. Consensus HNF-3 binding sites also reside within the proximal promoter and enhancer. We find that p53 expression effectively silences the majority of HNF-3-mediated activation.
We are currently investigating whether this active mode of repression is due to corepressor complex formation within hepatic cells. Our studies employing cotransfection of the mutant form of p53 alongside HNF-3 show a superactivation by HNF-3 in the presence of mutant p53. One interpretation of this result is that mutation of p53 within the DNA binding domain does not interfere with corepressor-p53 interaction. These corepressor proteins could be titrated by interaction with mutant p53, resulting in higher activation by HNF-3, as well as increased expression of the AFP-driven reporter in the absence of HNF-3. p53 may be functioning in active repression of transcription by acting as a tethering agent for corepressors of transcription. This mode of action is similar to p53-mediated docking of the active repressor adenovirus E1B 55K protein, which directly downregulates genes containing p53 binding sites (86).
Tissue specificity of AFP repression.
Regulatory response to p53 activation can be determined by cell-type-specific trans-acting factors, as illustrated by DNA damage-induced cell cycle arrest versus apoptosis (38, 54, 69) or by DNA sequence context, such as the transcription control of HBV genes (64). In the presence of the HBV enhancer, p53 binds a consensus site and represses transcription independently of the cellular background. These studies suggest that HBV enhancer binding proteins and p53 bind DNA, activating transcription when the regulatory elements are uncoupled and repressing expression of the HBV X promoter when adjacent (64). AFP repression by p53 appears to be both DNA context dependent, repressing HNF-3 activation possibly by DNA binding exclusion, and cell type specific through likely interaction with corepressors of activity.
Our experiments with NIH 3T3 cells that lack endogenous hepatic transactivators recapitulate only the passive mode of AFP repression. While p53 was able to repress HNF-3-activated AFP transcription in both tissue-specific and nonspecific backgrounds, baseline AFP/lacZ transcription was activated by p53 in fibroblast transfections rather than repressed. p53-mediated activation of AFP expression in NIH 3T3 cells which lack HNF-3 argues against recruitment of ubiquitous, general repressors of transcription by p53 binding. This mode of transcription repression has been described for general transcription repressor NC2 tethering via DNA-bound AREB6 Zn-finger/homeodomain protein (41). However, our results indicate that repression of AFP may involve tissue-specific corepressors of transcription that are targeted by p53 binding within the developmental repressor domain. EMSA studies of hepatoma and adult mouse liver nuclear extracts reveal multiple protein-DNA complexes formed at the overlapping HNF-3/p53 site, which can be chased to higher-molecular-weight protein-DNA complexes with increasing concentrations of extract. The protein composition of these complexes vary between cells which do not express AFP and those that do (data not shown).
p53 activity and AFP expression.
A biological paradigm for the molecular consequences of diminished p53 function lies in AT patients. Numerous clinical studies of ataxia-telangiectasia (AT) patients since 1972 describe a nearly 1:1 correlation between the AT disorder and aberrant production of AFP protein found at high concentrations in patient serum (47, 77). This has been ascribed to abnormal tissue differentiation, since cases of HCC are not unusually high in AT patients. Whether the developmental problems of AT patients and mice with embryonic deletion of the ATM (mutated in AT) gene are part of the emerging story of p53 activity in normal development remains to be elucidated (4, 84). In certain pathways, it appears that the ATM protein acts upstream of p53 protein, although AT cells respond to extensive DNA damage with slower kinetics of apoptosis but completely lack cell cycle arrest (reviewed in references 17 and 35).
The link that we have shown between DNA damage and stress-induced p53 activity on the one hand and AFP repression on the other underscores the pleiotropic roles of p53 in development and stress response. We find that the activation of p53 occurs through increased protein abundance during a time course of hepatic development. Several studies have revealed the importance of p53 activation during spermatogenesis (66) and the differentiation of epidermal (80), kidney (24), muscle, hematopoietic (70), and neural cells (14). Mice overexpressing p53, whether by a forced overproduction of the protein (24) or through creation of MDM2 null embryos (10), do not develop normally. These studies emphasize a requirement for tight control of p53 activity during development and differentiation. Whether p53 plays a broader role in hepatic differentiation, concomitant with AFP gene repression, is unknown.
The molecular mechanisms of differentiation-coupled repression of AFP expression and tumorigenic activation of expression are complex and likely combinatorial. One regulatory mechanism revealed by these studies is the p53-mediated repression of transcription through specific DNA binding. Binding of p53 physically excludes a tissue-specific activator of AFP transcription and may tether a complex of tissue-specific corepressors which actively interfere with gene expression. Aberrant expression of AFP during tumorigenesis may require functional inactivation of the p53 protein.
ACKNOWLEDGMENTS
We are especially grateful to Jorge Bezerra for invaluable cooperation in the preparation of developmentally staged hepatic extracts, to Ling Sang for expert technical assistance, and to J. Ma, K. Fukasawa, A. Nardulli, and N. Denko for critical comments. We are indebted to G. Lozano, K. Zaret, P. Brindle, B. Spear, and B. Aronow for materials which were essential for these studies.
This work was supported by National Institutes of Health (NIH) grant GM53683 and an American Cancer Society Junior Faculty Research Award to M.C.B. Support to A.J.C. through an NIH National Research Service Award (CA73083) is also gratefully acknowledged.
REFERENCES
- 1.Agoff S N, Hou J, Linzer D I H, Wu B. Regulation of the human hsp70 promoter by p53. Science. 1993;259:84–87. doi: 10.1126/science.8418500. [DOI] [PubMed] [Google Scholar]
- 2.Ang S L, Wierda A, Wong D, Stevens K A, Cascio S, Rossant J, Zaret K S. The formation and maintenance of the definitive endoderm lineage in the mouse: involvement of HNF3/forkhead proteins. Development. 1993;119:1301–1315. doi: 10.1242/dev.119.4.1301. [DOI] [PubMed] [Google Scholar]
- 3.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 4.Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley J N, Ried T, Tagle D, Wynshaw-Boris A. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 5.Barton M C, Madani N, Emerson B M. Distal enhancer regulation by promoter derepression in topologically constrained DNA in vitro. Proc Natl Acad Sci USA. 1997;94:7257–7262. doi: 10.1073/pnas.94.14.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camper S A, Godbout R, Tilghman S M. The developmental regulation of albumin and alpha-fetoprotein gene expression. Prog Nucleic Acid Res Mol Biol. 1989;36:131–143. doi: 10.1016/s0079-6603(08)60167-1. [DOI] [PubMed] [Google Scholar]
- 7.Camper S A, Tilghman S M. Postnatal repression of the alpha-fetoprotein gene is enhancer independent. Genes Dev. 1989;3:537–546. doi: 10.1101/gad.3.4.537. [DOI] [PubMed] [Google Scholar]
- 8.Camper S A, Tilghman S M. The activation and silencing of gene transcription in the liver. Bio/Technology. 1991;16:81–87. [PubMed] [Google Scholar]
- 9.Crowe A J, Hayman M J. Post-translational modifications of the env-sea oncogene product: the role of proteolytic processing in transformation. Oncogene. 1993;8:181–189. [PubMed] [Google Scholar]
- 10.de Oca Luna R M, Wagner D S, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:206–208. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 11.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science. 1996;272:1470–1473. doi: 10.1126/science.272.5267.1470. [DOI] [PubMed] [Google Scholar]
- 13.Eckner R, Yao T P, Oldread E, Livingston D M. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 1994;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 14.Eizenberg O, Faber-Elman A, Gottlieb E, Oren M, Rotter V, Schwartz M. p53 plays a regulatory role in differentiation and apoptosis of central nervous system-associated cells. Mol Cell Biol. 1996;16:5178–5185. doi: 10.1128/mcb.16.9.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Deiry W S, Kern S E, Pietenpol J A, Kinzler K W, Vogelstein B. Definition of a consensus binding site for p53. Nat Gen. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 16.Emerson B M, Nickol J M, Fong T C. Erythroid-specific activation and derepression of the chick β-globin promoter in vitro. Cell. 1989;57:1189–1200. doi: 10.1016/0092-8674(89)90056-1. [DOI] [PubMed] [Google Scholar]
- 17.Enoch T, Norbury C. Cellular responses to DNA damage: cell-cycle checkpoints, apoptosis and the roles of p53 and ATM. Trends Biochem Sci. 1995;20:426–430. doi: 10.1016/s0968-0004(00)89093-3. [DOI] [PubMed] [Google Scholar]
- 18.Farmer G, Bargonetti J, Zhu H, Friedman P, Prywes R, Prives C. Wild-type p53 activates transcription in vitro. Nature. 1992;358:83–86. doi: 10.1038/358083a0. [DOI] [PubMed] [Google Scholar]
- 19.Farmer G, Colgan J, Nakatani Y, Manley J L, Prives C. Functional interaction between p53, the TATA-binding protein (TBP), and TBP-associated factors in vivo. Mol Cell Biol. 1996;16:4295–4304. doi: 10.1128/mcb.16.8.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farmer G, Friedlander P, Colgan J, Manley J L, Prives C. Transcriptional repression by p53 involves molecular interactions distinct from those with the TATA box binding protein. Nucleic Acids Res. 1996;24:4281–4288. doi: 10.1093/nar/24.21.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galarneau L, Pare J-F, Allard D, Hamel D, Levesque L, Tugwood J D, Green S, Belanger L. The alpha-fetoprotein locus is activated by a nuclear receptor of the Drosophila Ftz-F1 family. Mol Cell Biol. 1996;16:3853–3865. doi: 10.1128/mcb.16.7.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallagher M J, Blumenthal K M. Cloning and expression of wildtype and mutant forms of the cardiotonic polypeptide anthopleurin B. J Biol Chem. 1992;267:13958–13963. [PubMed] [Google Scholar]
- 23.Godbout R, Ingram R, Tilghman S M. Multiple regulatory elements in the intergenic region between the alpha-fetoprotein and albumin genes. Mol Cell Biol. 1986;6:477–487. doi: 10.1128/mcb.6.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Godley L A, Kopp J B, Eckhaus M, Paglino J J, Owens J, Varmus H E. Wild-type p53 transgenic mice exhibit altered differentiation of the ureteric bud and possess small kidneys. Genes Dev. 1996;10:836–850. doi: 10.1101/gad.10.7.836. [DOI] [PubMed] [Google Scholar]
- 25.Gorski K, Carneiro M, Schibler U. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell. 1986;47:767–776. doi: 10.1016/0092-8674(86)90519-2. [DOI] [PubMed] [Google Scholar]
- 26.Graeber T G, Osmanian C, Jacks T, Housman D E, Koch D J, Lowe S W, Giaccia A J. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature. 1996;379:88–91. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- 27.Graeber T G, Peterson J F, Tsai M, Monica K, Fornace A J, Jr, Giaccia A J. Hypoxia induces accumulation of p53 protein, but activation of a G1-phase checkpoint by low-oxygen conditions is independent of p53 status. Mol Cell Biol. 1994;14:6264–6277. doi: 10.1128/mcb.14.9.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–464. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 29.Gray S, Levine M. Transcriptional repression in development. Curr Opin Cell Biol. 1996;8:358–364. doi: 10.1016/s0955-0674(96)80010-x. [DOI] [PubMed] [Google Scholar]
- 30.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 31.Gu W, Shi X-L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 32.Haffner R, Oren M. Biochemical properties and biological effects of p53. Curr Opin Genet Dev. 1995;5:84–90. doi: 10.1016/s0959-437x(95)90058-6. [DOI] [PubMed] [Google Scholar]
- 33.Hainault P, Milner J. Redox modulation of p53 conformation and sequence-specific DNA binding in vitro. Cancer Res. 1993;53:4469–4473. [PubMed] [Google Scholar]
- 34.Hanna-Rose W, Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 35.Hawley R S, Friend S H. Strange bedfellows in even stranger places: the role of ATM in meiotic cells, lymphocytes, tumors, and its functional links to 53. Genes Dev. 1996;10:2383–2388. doi: 10.1101/gad.10.19.2383. [DOI] [PubMed] [Google Scholar]
- 36.Hinds P, Finlay C, Levine A J. Mutation is required to activate the p53 gene for cooperation with the ras oncogene and transformation. J Virol. 1989;63:739–746. doi: 10.1128/jvi.63.2.739-746.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horikoshi N, Maguire K, Kralli A, Maldonado E, Reinberg D, Weinmann R. Direct interaction between adenovirus E1A protein and the TATA box binding transcription factor IID. Proc Natl Acad Sci USA. 1991;88:5124–5128. doi: 10.1073/pnas.88.12.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hundley J E, Koester S K, Troyer D A, Hisenbeck S G, Subler M A, Windle J J. Increased tumor proliferation and genomic instability without decreased apoptosis in MMTV-ras mice deficient in p53. Mol Cell Biol. 1997;17:723–731. doi: 10.1128/mcb.17.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hupp T R, Meek D W, Midgley C A, Lane D P. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 40.Hupp T R, Lane D P. Allosteric activation of latent p53 tetramers. Curr Biol. 1994;4:865–875. doi: 10.1016/s0960-9822(00)00195-0. [DOI] [PubMed] [Google Scholar]
- 41.Ikeda K, Halle J-P, Stelzer G, Meisterernst M, Kawakami K. Involvement of negative cofactor NC2 in active repression by zinc finder-homeodomain transcription factor AREB6. Mol Cell Biol. 1998;18:10–18. doi: 10.1128/mcb.18.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackson D A, Rowader K E, Stevens K, Jiang C, Milos P, Zaret K S. Modulation of liver-specific transcription by interactions between hepatocyte nuclear factor 3 and nuclear factor 1 binding DNA in close apposition. Mol Cell Biol. 1993;13:2401–2410. doi: 10.1128/mcb.13.4.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jayaraman L, Kanneganti G K M, Zhu C, Curran T, Xanthoudakis S, Prives C. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev. 1997;11:558–570. doi: 10.1101/gad.11.5.558. [DOI] [PubMed] [Google Scholar]
- 44.Johnson P. Identification of C/EBP basic region residues involved in DNA sequence recognition and half-site spacing preference. Mol Cell Biol. 1993;13:6919–6930. doi: 10.1128/mcb.13.11.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson P F. Transcriptional activators in hepatocytes. Cell Growth Diff. 1990;1:47–52. [PubMed] [Google Scholar]
- 46.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 47.Kumar G K, Saadi A A, Yang S-S, McCaughey R S. Ataxia-telangiectasia and hepatocellular carcinoma. Am J Med Sci. 1979;278:157–160. doi: 10.1097/00000441-197909000-00008. [DOI] [PubMed] [Google Scholar]
- 48.Lai E, Prezioso V R, Smith E, Litvin O, Costa R H, Darnell J E. HNF-3A, a hepatocyte-enriched transcription factor of novel structure is regulated transcriptionally. Genes Dev. 1990;4:1427–1436. doi: 10.1101/gad.4.8.1427. [DOI] [PubMed] [Google Scholar]
- 49.Leng X, Cooney A J, Tsae S Y, Tsai M-J. Molecular mechanisms of COUP-TF-mediated transcriptional repression: evidence for transrepression and active repression. Mol Cell Biol. 1996;16:2332–2340. doi: 10.1128/mcb.16.5.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leveillard T, Andera L, Bissonnette N, Schaeffer L, Bracco L, Egly J-M, Wasylyk B. Functional interactions between p53 and TFIIH complex are affected by tumour-associated mutations. EMBO J. 1996;15:1615–1624. [PMC free article] [PubMed] [Google Scholar]
- 51.Levine A. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 52.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 53.Liu X, Berk A J. Reversal of in vitro p53 squelching by both TFIIB and TFIID. Mol Cell Biol. 1995;15:6474–6478. doi: 10.1128/mcb.15.11.6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ludwig R L, Bates S, Vousden K H. Differential activation of target cellular promoters by p53 mutants with impaired apoptotic function. Mol Cell Biol. 1996;16:4952–4960. doi: 10.1128/mcb.16.9.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lutzker S, Levine A J. A functionally inactive p53 protein in teratocarcinoma cells is activated by either DNA damage or cellular differentiation. Nat Med. 1996;2:804–810. doi: 10.1038/nm0796-804. [DOI] [PubMed] [Google Scholar]
- 56.Maheswaran S, Englert C, Bennet P, Heinrich G, Haber D A. The WT1 gene product stabilizes p53 and inhibits p53-mediated apoptosis. Genes Dev. 1995;9:2143–2156. doi: 10.1101/gad.9.17.2143. [DOI] [PubMed] [Google Scholar]
- 57.Maire P, Wuarin J, Schibler U. The role of cis-acting promoter elements in tissue-specific albumin gene expression. Science. 1989;244:343–346. doi: 10.1126/science.2711183. [DOI] [PubMed] [Google Scholar]
- 58.Martinez J, Georgoff I, Marinez J, Levine A J. Cellular localization and cell cycle regulation by a temperature-sensitive p53 protein. Genes Dev. 1991;5:151–159. doi: 10.1101/gad.5.2.151. [DOI] [PubMed] [Google Scholar]
- 59.McLure K G, Lee P W K. How p53 binds DNA as a tetramer. EMBO J. 1998;17:3342–3350. doi: 10.1093/emboj/17.12.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Milne D M, Palmer R H, Campbell D G, Meek D W. Phosphorylation of the p53 tumor suppressor protein at three N-terminal sites by a novel casein kinase I-like enzyme. Oncogene. 1992;7:1361–1369. [PubMed] [Google Scholar]
- 61.Murphy M, Hinman A, Levine A J. Wild-type p53 negatively regulates the expression of a microtubule-associated protein. Genes Dev. 1996;10:2971–2980. doi: 10.1101/gad.10.23.2971. [DOI] [PubMed] [Google Scholar]
- 62.Nelson R, Long G. A general method of site-specific mutagenesis using a modification of the Thermus aquaticus polymerase chain reaction. Anal Biochem. 1989;180:147–151. doi: 10.1016/0003-2697(89)90103-6. [DOI] [PubMed] [Google Scholar]
- 63.Opdecamp K, Szpirer C, Szpirer J. Major chromatin changes accompany extinction of alpha-fetoprotein gene in hepatoma x fibroblast hybrids. Somatic Cell Mol Genet. 1991;17:49–55. doi: 10.1007/BF01233204. [DOI] [PubMed] [Google Scholar]
- 64.Ori A, Zauberman A, Doitsch G, Paran N, Oren M, Shaul Y. p53 binds and represses the HBV enhancer: an adjacent enhancer element can reverse the transcription effect of p53. EMBO J. 1998;17:544–553. doi: 10.1093/emboj/17.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qi J-S, Desai-Yajnik V, Yuan Y, Samuels H H. Constitutive activation of gene expression by thyroid hormone receptor results from reversal of p53-mediated repression. Mol Cell Biol. 1997;17:7195–7297. doi: 10.1128/mcb.17.12.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rotter V, Aloni-Grinstein R, Schwartz D, Elkind N B, Simons A, Wolkowicz R, Lavigne M, Beserman P, Kapon A, Goldfiner N. Does wild-type p53 play a role in normal cell differentiation? Sem Cancer Biol. 1994;5:229–236. [PubMed] [Google Scholar]
- 67.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 68.Shen Y, Shenk T. Relief of p53-mediated transcription repression by the adenovirus E1B 19-kDa protein or the cellular bcl-2 protein. Proc Natl Acad Sci USA. 1994;91:8940–8944. doi: 10.1073/pnas.91.19.8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Soddu S, Blandino G, Scardigli R, Martinelli R, Rizzo M G, Crescenzi M, Sacchi A. Wild-type p53 induces diverse effects in 32D cells expressing different oncogenes. Mol Cell Biol. 1996;16:487–495. doi: 10.1128/mcb.16.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Soddu S, Blandino G, Scardigli R, Soen S, Marchetti A, Rizzo M G, Bossi G, Cimino L, Crescenzi M, Sacchi A. Interference with p53 protein inhibits hematopoietic and muscle differentiation. J Cell Biol. 1996;134:193–204. doi: 10.1083/jcb.134.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Spear B T. Mouse a-fetoprotein gene 5′ regulatory elements are required for postnatal regulation by raf and Rif. Mol Cell Biol. 1994;14:6497–6505. doi: 10.1128/mcb.14.10.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Spear B T, Longley T, Moulder S, Wang S L, Peterson M L. A sensitive lacZ-based expression vector for analyzing transcriptional control elements in eukaryotic cells. DNA Cell Biol. 1995;14:635–642. doi: 10.1089/dna.1995.14.635. [DOI] [PubMed] [Google Scholar]
- 73.Tabor E. Tumor suppressor genes, growth factor genes, and oncogenes in hepatitis B virus-associated hepatocellular carcinoma. J Med Virol. 1994;42:357–365. doi: 10.1002/jmv.1890420406. [DOI] [PubMed] [Google Scholar]
- 74.Tan T-H, Wallis J, Levine A J. Identification of the p53 protein domain involved in formation of the simian virus 40 large T-antigen-p53 protein complex. J Virol. 1986;59:574–583. doi: 10.1128/jvi.59.3.574-583.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tilghman S M. The structure and regulation of the alpha-fetoprotein and albumin genes. Oxf Surv Eukaryotic Genes. 1985;2:160–206. [PubMed] [Google Scholar]
- 76.Vacher J, Tilghman S M. Dominant negative regulation of the mouse alpha-fetoprotein gene in adult liver. Science. 1990;250:1732–1735. doi: 10.1126/science.1702902. [DOI] [PubMed] [Google Scholar]
- 77.Waldman T A, McIntire K R. Serum alpha fetoprotein levels in patients with ataxia telangiectasia. Lancet. 1972;2:1112–1115. doi: 10.1016/s0140-6736(72)92717-1. [DOI] [PubMed] [Google Scholar]
- 78.Wang Q, Zambetti G P, Suttle D P. Inhibition of DNA topoisomerase IIα gene expression by the p53 tumor suppressor. Mol Cell Biol. 1997;17:389–397. doi: 10.1128/mcb.17.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang X W, Forrester K, Yeh J, Feitelson M A, Gu J-R, Harris C C. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc Natl Acad Sci USA. 1994;91:2230–2234. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weinberg W C, Azzoli C G, Chapman K, Levine A J, Yuspa S H. p53-mediated transcriptional activity increases in differentiating epidermal keratinocytes in association with decreased p53 protein. Oncogene. 1995;10:2271–2279. [PubMed] [Google Scholar]
- 81.Wen P, Groupp E R, Buzard G, Crawford N, Locker J. Enhancer, repressor, and promoter specificities combine to regulate the rat α-fetoprotein Gene. DNA Cell Biol. 1991;10:525–536. doi: 10.1089/dna.1991.10.525. [DOI] [PubMed] [Google Scholar]
- 82.Wen P, Locker J. A novel hepatocytic transcription factor that binds the α-fetoprotein promoter-linked coupling element. Mol Cell Biol. 1994;14:6616–6626. doi: 10.1128/mcb.14.10.6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu H, Wade M, Krall L, Grisham J, Xiong Y, Van Dyke T. Targeted in vivo expression of the cyclin-dependent kinase inhibitor p21 halts hepatocyte cell-cycle progression, postnatal liver development, and regeneration. Genes Dev. 1996;10:245–260. doi: 10.1101/gad.10.3.245. [DOI] [PubMed] [Google Scholar]
- 84.Xu Y, Ashley T, Brainerd E E, Bronson R T, Meyn M S, Baltimore D. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 1996;10:2411–2422. doi: 10.1101/gad.10.19.2411. [DOI] [PubMed] [Google Scholar]
- 85.Yap N, Yu C-L, Cheng S-Y. Modulation of the transcriptional activity of thyroid hormone receptors by the tumor suppressor p53. Proc Natl Acad Sci USA. 1996;93:4273–4277. doi: 10.1073/pnas.93.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yew P R, Liu X, Berk A J. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 1994;8:190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]
- 87.Zambetti G P, Bargonetti J, Walker K, Prives C, Levine A J. Wild-type p53 mediates positive regulation of gene expression through a specific DNA sequence element. Genes Dev. 1992;6:1143–1152. doi: 10.1101/gad.6.7.1143. [DOI] [PubMed] [Google Scholar]
- 88.Zaret K S, Stevens K. Expression of a highly unstable and insoluble transcription factor in Escherichia coli: purification and characterization of the fork head homolog HNF3alpha. Protein Expr Purif. 1995;6:821–825. doi: 10.1006/prep.1995.0014. [DOI] [PubMed] [Google Scholar]
- 89.Zhang D-E, Hoyt P R, Papaconstantinou J. Localization of DNA protein-binding sites in the proximal and distal promoter regions of the mouse α-fetoprotein gene. J Biol Chem. 1990;265:3382–3391. [PubMed] [Google Scholar]