Abstract
A strategic research plan (SRP) serves as a compass for the patient advocacy organizations driving the therapeutic options for their rare disorder. The MED13L Foundation commissioned the SRP in 2022 through COMBINEDBrain, a consortium of patient advocacy organizations of rare neurodevelopmental disorders, working toward clinical trial readiness. The MED13L Foundation SRP is an objective evaluation of MED13L literature including clinical and basic science knowledge interwoven with an assessment of preclinical trial readiness tools necessary for achieving therapeutic interventions. Clinical evaluation is conducted through a review of the literature documenting symptoms and variant information for each individual with MED13L syndrome. Data is collated and presented as a summary, providing any unique genotype–phenotype, as applicable. Scientific literature is reviewed in the same manner, identifying areas of opportunity to expand knowledge of MED13L syndrome. Researchers and clinicians responsible for growing the understanding of MED13L syndrome are interviewed and information is shared to create an open and collaborative network. Preclinical trial readiness tools are largely framed through Food and Drug Administration guidelines for the development of therapeutics from bench to bedside. Finally, the Foundation infrastructure and community engagement are assessed providing areas of strengths and opportunities to elevate the bond formed to drive patient-centered research forward. Completed, this SRP becomes a living resource for the MED13L Foundation to set priorities, share with researchers and clinicians, and provide direction to reach their organizational goals, including therapies for their community affected by MED13L syndrome.
Keywords: advocacy led research, drug development resources, MED13L, neurodevelopmental rare disease, patient advocacy organization, research roadmap
Plain language summary
The MED13L Foundation strategic research plan
The Strategic Research Plan (SRP) serves as a guide for patient advocacy groups working to find treatments for rare disorders. The MED13L Foundation collaborated with COMBINEDBrain to create a specific SRP for MED13L Syndrome to direct clinical trial readiness. Once completed, the SRP becomes a tool for the MED13L Foundation, researchers, clinicians, and the community. It helps set priorities and guides goals. A summary of the plan, presented as a roadmap, provides a quick overview of existing tools and what else needs to be done to prepare for clinical trials. The ultimate goal is to discover therapeutics that improve the lives of those affected by MED13L Syndrome.
Background for strategic research plan
Defined as a neurodevelopmental syndrome in 2013, MED13L syndrome (MED13L) is a rare multisystem disorder. 1 While broad in phenotype, primary clinical characteristics include developmental delay, intellectual disability, recognizable facial features, mobility issues, and behavioral difficulties.1,2 Lesser prevalent symptoms but potentially more impactful, include seizures and congenital heart defects. 1 The latter served as a basis for uncovering this genetic-based disorder.3,4
First characterized in 2003, MED13L, the mediator complex subunit 13-like, was known as PROSIT240, a protein similar to THRAP2 (thyroid hormone receptor-associated protein 2), located on chromosome 12 (12q24).3,4 Initially thought to be solely responsible for severe congenital heart defects, specifically dextro-looped transposition of the great arteries (d-TGA), researchers theorized that these cardiac defects contributed to the neurodevelopmental manifestations.3,4 However, in 2013, Asadollahi, followed by Adegbola et al. in 2015, redefined the clinical presentation previously linked to congenital heart defects as primarily a neurodevelopmental phenotype driven by pathogenic variants in MED13L.1,2
Shortly after clinical recharacterization, the MED13L Foundation (Foundation) was established in 2016 by a single family as a support group for the newly diagnosed. By 2021, the Foundation refocused its mission on discovering therapeutics for those affected by MED13L syndrome. With stakeholder input, the Foundation identified several research priorities including building a collaborative clinical and scientific community, investing in drug repurposing, advancing basic scientific knowledge of MED13L, and engaging the community.
Following the 2022 inaugural Scientific and Family Meeting, the MED13L Foundation quickly assembled a diverse medical and scientific advisory board. Members included scientists with a deep knowledge base of MED13L and the mediator kinase module (MKM), as well as clinicians tied to the MED13L community or a broader background in neurogenetics. Efforts are currently underway to expand advisory board membership beyond MED13L syndrome to include experts in mitochondrial diseases, other MKM genes, common signaling pathways, and clinicians experienced in outcome measure assessment for rare diseases. These early and integral relationships fueled efforts for funding basic science experiments, as well as allowing for early interaction with the Centers for Disease Control for issuance of the coveted ICD-10 (International Classification of Diseases, 10 revision code) for MED13L syndrome (Q87.85). The success of the first in-person meeting showcased the power of togetherness and grassroot efforts to propel research forward.
The MED13L Foundation also collaborates with the France-based MED13L Syndrome Association. Both influence the local community affected by MED13L syndrome to enroll and engage in research through data collection platforms and biorepository opportunities. These partnerships also facilitate knowledge sharing and international collaboration among leading clinician-researchers in the field.
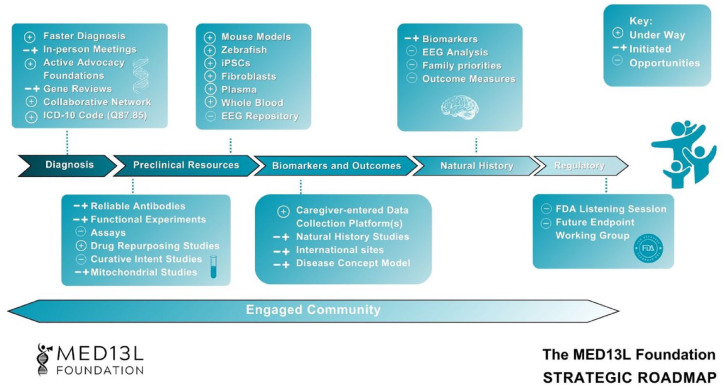
In 2023, the MED13L Foundation focused on developing a strategic research plan (SRP) to address its research priorities. The SRP evaluated the current landscape of MED13L research, reviewed published literature, and identified opportunities for collaboration and funding. This living document includes detailed reports and a summary roadmap and is regularly updated with new resources, clinical opportunities, and recommendations to propel the MED13L community toward clinical trial readiness (Figure 1).
Figure 1.
The MED13L foundation strategic roadmap.5–7 The roadmap serves as a summary of progress regarding the necessary preclinical trial tools and resources developed as part of the larger SRP. Each forward-moving arrow represents a broad category of needs, from diagnosis to regulatory tasks, essential for optimal clinical trial readiness. Each box above and below the arrows represents a set of tools and resources that align with these categories. The resources may flow across arrows and do not fully represent all that must be achieved to reach clinical trials or therapeutic development. Additionally, tools and resources may apply to many rare disorders or be specific to a particular disorder. An engaged and active community is crucial throughout the entire process. Early and intentional investment in individuals affected by MED13L syndrome is key to developing effective therapeutics.
EEG, electroencephalogram; FDA, Food and Drug Administration; ICD-10, International Classification of Diseases, 10th revision; iPSCs, induced pluripotent stem cells; SRP, strategic research plan.
About MED13L syndrome
MED13L syndrome is caused by a variety of pathogenic variants including, large gene deletions, nonsense, missense, and small genomic insertions and deletions.1,2 Variants are primarily de novo, autosomal dominant, and considered to be loss-of-function (LoF); thus, MED13L syndrome is historically considered a haploinsufficiency syndrome.1,2,8 Newer data suggests that patients harboring missense variants present with a more severe phenotype.9,10 These findings propose that select missense variants may impart a pathogenic gain-of-function (GoF) or dominant-negative effect, but the mechanism is not understood.9,10 Additionally, there are a small number of patients with full gene duplications with a reported “mild phenotype.” 1 Mosaicism has also been documented in the literature with variable phenotypes.2,11–14 Finally, it should be noted that pathogenic variants in all genes related to the MKM, except for cyclin C, are associated with neurodevelopmental disorders.2,8,9,15 Interestingly, MED12, MED13, and MED13L also present with cardiac defects in addition to developmental delay and unique musculoskeletal abnormalities.15,16
Diagnosis, incidence, and prevalence
Early identification of physical characteristics, coupled with access to more comprehensive genetic testing tools, is necessary for early and accurate diagnosis.9,10,17 Asadollahi et al. first took advantage of newer genetic technology in the mid-2000s to identify copy number variants and reframe MED13L syndrome as a neurodevelopmental disorder.1,17 Since then, MED13L syndrome has been documented as a top 10 genetic pathogenic de novo cause of intellectual delay and disability, accounting for approximately 0.5% of the population in multiple iterations of the Deciphering Developmental Disorder cohorts.18,19 This places its prevalence in line with other known autism-based neurodevelopmental disorders such as synaptic Ras GTPase-activating protein 1 (SYNGAP1), syntaxin-binding protein 1 (STXBP1), and sodium voltage-gated channel alpha subunit 2 (SCN2A).18–20 Clinicians and researchers estimate the prevalence calculated between 2 and 6 in 100,000 births.21,22 With about 100 published cases, this suggests that MED13L syndrome is significantly underdiagnosed.9,10,23 To enhance known diagnosis rates, the Foundation is focusing on increasing utilization of their ICD-10 code (Q87.85). 22 Additionally, it is postulated that missense variants, more likely to be variants of unknown significance (VUS), occur at a 2:1 ratio versus protein-truncating variants.22,24 By supporting reclassification efforts from VUS to pathogenic or likely pathogenic variants, the Foundation can also improve the known population.
MED13L syndrome clinical symptoms
The hallmark symptoms of MED13L syndrome are global developmental delay and intellectual disability, which occur in all evaluated cases.2,10,25–27 Seventy-one percent (65/92) of reported individuals are categorized as moderate intellectual disability.2,8–11,14,25,28,29 Less than 10% of cases reported are classified as mild and approximately 15% are considered severe.2,8–11,14,25,28,29 Minimal or absent speech and impaired motor capabilities dominate the presentation of global developmental delay, with 99% (81/82) and 98% (80/82) of individuals presenting with minimal/absent speech or impaired motor capabilities, respectively.2,8–11,14,25,28,29 It is postulated that hypotonia is responsible for motor dysfunction.2,8–10,25,27 Additionally, individuals present with musculoskeletal malformations, particularly in the face, hands, and feet.2,8–11,14,25,28,29 Recognizable facial features include broad prominent forehead, bulbous nasal tip, broad depressed nasal bridge, large open mouth appearance, up slanting palpebral fissures, low set ears, and jaw/chin abnormalities (micro, pro, or retrognathia).9,25 While autism or autism spectrum disorder (ASD) has contributed to top published behavioral symptoms, it should be noted that behavioral symptoms are not consistently captured with about 40% of cases making no note of behavior-based changes.2,8,10,11,13,25,30–32 Finally, visual abnormalities are reported in 31 individuals, with strabismus being a primary contributor.2,9,13,25,28,32
When it comes to behavioral symptoms, individuals with MED13L syndrome have been reported to exhibit a variety of aberrant behaviors. This includes autism and autistic traits, self-harm, tantrums, aggression, and frustration.2,8–10,13,25,28,31–33 Interestingly, individuals with MED13L syndrome are also described as having a social, friendly, and pleasant demeanor.11,27,33 Of those reported, just over 60% (38/63) of individuals are documented to have behavioral concerns.2,8–10,13,25,28,31–33 Symptoms are not well characterized or documented in the literature. There could be a potential correlation between objective magnetic resonance imaging (MRI) abnormalities and behavioral symptoms. However, an analysis is difficult to conduct due to the inconsistent collection of MRIs and behavioral phenotypes.
MRIs are potentially important for understanding pathology, symptom presentation, and perhaps biomarkers. Various MRI abnormalities are reported in 33 of 62 individuals.2,8–10,25–27,29 The most commonly reported abnormalities include ventriculomegaly (n = 9), white matter abnormalities (n = 7), myelination defects (n = 6), and corpus callosum thinning or agenesis (n = 5).2,8–10,25–27,29 Abnormalities are not mutually exclusive and do not appear to be more prevalent in any one variant type.8–10,29,33 Data linking MRI aberrations, such as corpus callosum changes, are associated with neurodevelopmental phenotypes such as cognition, speech, coordination delays, and seizures. 34 Additionally, a recent publication correlated fetal ventriculomegaly and cortical overgrowth showing a connection between ASD, including neurodevelopmental delay, focus issues, and maladaptive behaviors.35,36 While it is plausible that MRI abnormalities in MED13L publications are contributing to hallmark symptoms, there is no clear connection given the small and variable presentation noted.
Unfortunately, electroencephalograms (EEGs) are even less frequently published for MED13L syndrome. Only 9 of the 21 EEGs evaluated were reported as abnormal.2,8,26,27,32,33,37 Of the nine abnormal EEGs, five have no associated seizure symptoms.2,8,26,27,32,33,37 There is no particular repetitive pattern identified among individuals,2,8,26,27,32,33,37 and the number is too small to draw conclusions. Likewise, seizure presence is not considered a hallmark symptom of MED13L syndrome. Documented seizure presence is about 22% (15/68) in publications.2,8,10,27,32,37,38 Of these, 17 individuals with missense variants are evaluated, with 10 having seizures.10,39 Most seizure types are not characterized, though absence seizures (n = 3) are most common.8,32,38 The average age of individuals with reported seizures is 12 years.2,8,10,27,32,37,38 compared to the average age of 8 years in all reported individuals with MED13L syndrome.2,8–11,14,25–29 It is relatively unknown when seizure presence was first detected in these cases. However, if absence seizures are common and later in onset, then the subtlety of symptoms could contribute to low reporting. 40 Contributing to this knowledge base, a recent abstract from Simons Searchlight data across 28 genes described caregiver-reported data for treated seizures and severity. 41 Of 55 MED13L syndrome caregivers, treated seizures are reported in 15% and were refractory in 2%. This suggests a low occurrence rate or a later onset not captured in this study as the median age evaluated was 8 years of age. 41 Taken together, potential critical biomarkers and disease processes tied to MRIs or EEGs may present an opportunity for the Foundation to explore through consistent data collection and assessment.
Other less characterized symptoms are gastrointestinal issues,8–10,13,14,17,28,42 metabolic abnormalities, 1 and frequent childhood infections.2,9,25,32 Additionally, once considered the defining symptom of MED13L syndrome, congenital cardiac defects are present in about 20% (22/94) of individuals.3,8–10,17,26,27,31,38
While a strong genotype-phenotype correlation does not currently exist, individual missense variants located around exons 15 through 17 and exons 25 through 31 appear to show more severe motor delay, seizures, autism, and other behavioral issues.9,10,39 One publication focused on missense variants showed that of the nine individuals, there was a higher likelihood of absent speech (5/9 vs 5/21), absent ambulation (4/9 vs 1/21), seizures (5/9 vs 1/26), and autistic features (5/8 vs 5/21) compared to protein-truncating variants. 10 The reasons for the clustering of missense variants in these specific areas are not completely understood. However, extrapolating from MED13 data, it is possible that these regions experience phosphorylation dysfunction, affecting the polarity and hydrophilicity characteristics of proline, serine, and threonine residues10,39 (Table 1).
Table 1.
Summary of symptoms associated exon-specific documented missense variants in MED13L syndrome.
| Variant | Phenotype | Exon location |
|---|---|---|
| p.Pro866Leu10,39 | ID, ASD, Seizure, hypotonia, severe motor delay | 15 |
| p.Pro869Ser 10 | ID, seizures, severe motor delay | 15 |
| p.Cys1131Tyr 10 | Severe motor delay | 17 |
| p.Ser2163Leu10,39 | ID, ASD, seizures, hypotonia | 29 |
| p.Ser2177Tyr 10 | ID, ASD, seizures, hypotonia | 31 |
| p.Thr2162Met10,39 | ID, moderate speech and motor delay | 29 |
| p.Gly1899Arg 10 | ID, moderate speech and motor delay | 26 |
| p.Ser2002Leu 10 | ID, speech and motor delay, hypotonia | 27 |
ASD, autism spectrum disorder; ID, intellectual disability.
MED13L syndrome mechanism
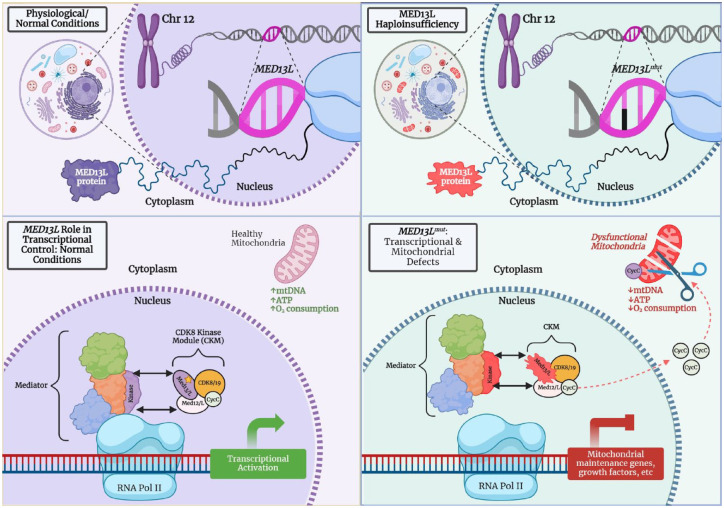
MED13L is a subunit of the MKM, which contains four subunits: CDK8/CDK19 (cyclin-dependent kinase 19), cyclin C, MED12/MED12L, and MED13/MED13L. 43 Each of the MKM subunits are present in a 1:1 ratio and the association of CDK8/CDK19, MED12/MED12L, and MED13/MED13L are mutually exclusive. 44 The larger Mediator is a 26-subunit complex that regulates RNA polymerase II transcription genome-wide. 45 Upon binding, the MKM alters the Mediator structure and function in ways that remain incompletely understood. Notably, MED13L shares roughly 50% sequence identity with its paralog MED13. Both proteins contain numerous intrinsically disordered domains, LXXLL motifs, and a medPIWi region43–45 (Figure 2).
Figure 2.
Proposed mechanism of MED13L compared to MED13L haploinsufficiency (created with BioRender). Left side: Under normal conditions, one of the functions of MED13 is to link the MKM to the Mediator Complex46,47; presumably, MED13L has a similar function given that biochemical and mass spectrometry data show Mediator association with MED13 and MED13L.48,49 MED13L has specifically been shown to regulate Wnt, FGF, and Rb/E2F pathways.44,50,51 Additionally, upon induction of stress or a loss of the nuclear-tethering of MED13L, cyclin C has been shown to exit the nucleus and interact with mitochondrial fission machinery, promoting organelle fragmentation/fission.51–53 Right side: It is hypothesized that in MED13L Haploinsufficiency Syndrome, the transcriptional process related to the mediator complex interaction with RNA polymerase II may be disrupted. Cyclin C is aberrantly released into the cytoplasm, increasing susceptibility to cell death through mitochondrial fragmentation, decreased oxygen consumption as well as decreased ATP production.50,53
FGF, fibroblast growth factor; MKM, mediator kinase module; Rb/E2F, retinoblastoma tumor suppressor; Wnt, wingless-type integration type.
While Figure 2 describes a possible mechanism for haploinsufficiency, the dominant-negative mechanism remains unconfirmed. Smol et al., as well as Hamada et al., have investigated this potential mechanism.10,39 Researchers tested this theory in a mouse model by creating customized C-terminal antibodies. Outcomes produced a similar protein compared to wild type of the p.Pro866Leu variant as well as reduced number and length of dendrites. 39 In the same study, researchers provided evidence that missense variants in the serine-rich region of the C-terminal (e.g., Ser2163Tyr) likely have both a gain and a LoF, displaying a hypomorphic state. 39 On the whole, there remains much basic biology to discover about this highly unstructured, redundant, yet seemingly unique gene. Additionally, without reliable and sensitive antibodies, confirming these functional studies remains elusive.
Treatment strategies
Treatment strategies for genetic-based diseases are contingent on several properties. The size of the gene, the function (gain, loss, and dominant-negative), the location, frequency, and variant type, human development expression, and the likeness of it to other genes. 5 This section describes potential approaches to treatment based on what is known about these factors.
Curative intent treatment strategy
Full gene replacement strategies for MED13L syndrome are limited given the size of the gene. Adeno-associated viruses (AAVs) profile have the capacity to introduce up to ~4.5 kilobases (kbs) of single-stranded DNA. 54 This approach is not feasible for MED13L syndrome given the coding sequence is over 9 kbs. 55 There are, however, different therapeutic approaches to explore where gene size is not a limitation.
The use of antisense oligonucleotides (ASOs) is a viable strategy to treat disorders of haploinsufficiency, GoF, or dominant-negative.56,57 ASOs are small, single- or double-stranded oligonucleotides that can be designed to increase stability, target binding, or improve cell uptake for GoF, LoF, or dominant-negative diseases. However, the ASO design differs by the variant’s functional consequences.56,57 Challenges for ASO design remain despite years of development experience. Barriers to ASO optimization include therapeutic window, ideal tissue target and administration, and protein dosage sensitivity to name a few.56,57 All of which are considerations for optimizing in MED13L syndrome.
A more gene-agnostic approach could be pursued for LoF variants; in particular, exploring technologies such as mRNA amplification, tNRA suppressor therapy, or readthrough.58–61 For example, molecular strategies that promote protein translation in the context of stop codons could be a viable treatment strategy for individuals with nonsense variants.60,61 Use of mRNA amplification can provide a broader approach by upregulating the wild-type copy of MED13L in those with haploinsufficiency. 58 This could be an effective strategy for over 70% of the MED13L syndrome population.
One CRISPR (clustered regularly interspaced short palindromic repeats) based therapy has already proven successful in a patient-derived fibroblast model. 53 Chang et al. applied Cas12a iCAP genome editing to a patient-derived truncating fibroblast sample, replacing exon 20. Researchers demonstrated a dramatic rescue of the MED13L cellular phenotype, including relocalization of cyclin C to the nucleus, stabilization of mitochondrial DNA, and increased ATP (Adenosine triphosphate) production and oxygen consumption. 53 While it is unknown what the impact of this rescue was on the larger transcription activation/suppression effects of the MKM dissociation mechanism, the use of this technique could provide benefits beyond mitochondrial rescue.
In addition to the use of CRISPR rescue described for mitochondrial dysfunction, cis-regulation therapy constructed as a “dead” Cas9 (CRISPR-associated protein 9) connected to a transcriptional activator could upregulate wild-type MED13L. Coined as CRISPR activation (CRISPRa), enodgenous promoters or enhancers are leveraged to restore expression in haploinsufficiency disorders, such as SCN1A (sodium voltage-gated channel alpha subunit 1) or Dravet syndrome. 62 Here researchers were able to improve epiplesy in a heterozygous Scn1a mouse model. Complimentary data to support the use for MED13L syndrome was presented in a 2022 abstract in Rennes, France. 63 Evaluating the differences in early cortical neuron development in a MED13L knock-out organoid versus a wild-type organoid showed a clear difference in the development of mature cortical neurons and functioning glutamatergic and GABAergic neurons in MED13L KO (knockout) organoids. 63 The KO model also generated increased expression to neuroretinal cells. 63 These preliminary results demonstrating neuron dysfunction in organoids provide opportunity to leverage endogenous promotors or enhancers of MED13L to rescue early developmental phenotypes in a proof-of-concept model of MED13L syndrome.62,63
Drug repurposing strategy
Current potential treatment strategies for MED13L syndrome have focused, thus far, on finding repurposed Food and Drug Administration (FDA)-approved drugs or other small molecules that impact MED13L. Comparative toxicogenomic dataset (CTD) outlines dozens of different compounds known to interact with MED13L. 64 Additionally, the MED13L Foundation has partnered with Transcripta Bio (formerly Rarebase) and the University of Alabama-Birmingham (UAB) to leverage institutional-specific technology to identify available therapeutic options that directly or indirectly interact with MED13L. University of Alabama-Birmingham uses its mediKarnren computational analysis platform to search published literature based on the following concepts65–69: (1) drugs/compounds that upregulate expression of MED13L, MED13, MED12, and MED12L, (2) preserve cyclin C in the nucleus, 53 (3) Wnt (wingless-type integration type) pathway including the following genes: WNT7A, LRP5 (low density lipoprotein related protein), FRZB (frizzled-related protein), PCDHB15 (protocadherin), PCDH17 (protocadherin), SFRP4 (secreted frizzled-related protein), FZD9 (frizzled), 38 (4) decreasing mitochondrial fission: MFN1 (mitofusion-1) (↑), MFN2 (mitofusion-2) (↑), DNM1L (Dynamin-1-like protein) (↓) 53 (Table 2). The impact and rescue of the mitochondrial mechanism is being further studied through a Foundation-issued grant to the Randy Strich Lab and Rowan University (https://med13l.org/research-hub/grant-awards/).
Table 2.
Examples of available drug compounds with proposed mechanistic impact on MED13L (direct and indirect).
| Compound | Data source | Target | Impact on MED13L |
|---|---|---|---|
| Clofibrate70,71/Bezafibrate 72 | CTD/UAB | Drp1 | Effect MED13L expression/Improves mitochondrial fission and function |
| Lithium 73 | UAB | Drp1 | Inhibition of mitochondrial fission through downregulation of Drp1 |
| Verapamil 74 | UAB | FRZB | Suppresses Wnt/β-catenin signaling, valuable in missense variants that upregulate MED13L |
| Nutraceuticals/Supplements | |||
| PSE 75 | UAB | LRP5 | The oral administration of PSE, a dietary cholesterol-lowering agent, had an effect on the expression levels of the Wnt signaling receptor |
| EPA and DHA 76 | UAB | MFN2 | Recovery of mitochondrial function by increasing Mfn2 expression |
| Ginkgo biloba 77 | UAB | Drp1 | Reduced mitochondrial fission |
The table outlines specific drugs or alternative compounds that interact with MED13L directly by upregulating or downregulating expression or through indirect mechanisms such as preservation of mitochondrial dysfunction, interaction with the Wnt pathway, or other distinct known downstream paths as outlined. CTD, comparative toxicogenomic dataset; DHA, docosahexaenoic acid; EPA, eicosapentaenoic acid; PSE, plant sterol esters; UAB, University of Alabama-Birmingham.
Drug development resources
Based on several years of ongoing guidance from the FDA,6,7,78,79 the MED13L Foundation is working to establish a toolkit for researchers, clinicians, and the community. Drug development relies on proof-of-concept studies conducted in animal and cell models of disease. 80 Patient-derived cell models, as well as animal models, are necessary for establishing disease pathology and opportunities for therapeutic rescue. Additionally, a more recent FDA guidance references the inclusion of patient/caregiver-reported outcomes to complement that of a formal Natural History Study. When available, patient/caregiver-reported outcomes should be facilitated with the use of validated assessment tools. 78
Biorepositories, cell and animal models
Cell models
The MED13L Foundation has developed a Foundation-owned biorepository in addition to a Simons Searchlight biorepository. Both repositories contain a variety of patient samples, including plasma, whole blood, serum, peripheral blood mononuclear cell, induced pluripotent stem cell, and fibroblasts. These samples are available to researchers around the world through COMBINEDBrain (https://combinedbrain.org/biorepository) or Simons Foundation Autism Research Initiative (SFARI; https://www.sfari.org/resource/ips-cells/). In total over 50 affected individuals, siblings, and parents have donated to these biorepositories.
Animal models
There are a handful of in vivo models for MED13L syndrome with published outcomes (Table 3). Utami et al. developed a Danio rerio (zebrafish) model by knocking down med13b, the zebrafish equivalent to MED13L. 38 More than 95% of embryos had underdeveloped heads, microphthalmia, and curved body axis, recapitulating the human phenotype. 38 Conclusions drawn were that MED13L is responsible for neural crest development and differentiation. This included key signaling pathways for brain, skeletal muscle, heart, and facial structure, correlating to human fibroblast growth factor (FGF) and Wnt deregulation. 38
Table 3.
Existing in vivo models for MED13L syndrome.
| Animal | Genetic model | Outcome |
|---|---|---|
| Danio rerio (zebrafish) 38 | med13b (knockdown) | Neural crest dysregulation |
| Mus musculus (mouse) 81 | Med13l N-terminal (1–668) Knockdown | MED13L nuclear and cytoplasmic neuronal co-localization |
| Mus musculus, embryonic (mouse) 39 | p.Pro866Leu p.Ser2163Leu p.Thr2162M p.Gln1922* |
Dominant-negative Hypomorphic Reduced function Loss-of-function |
| Mus musculus (mouse) 82 | Med13 + Med13l Cre-Lox alphaMHC (double KO) | Decreased ejection fraction, cardiomyopathy, increased mortality |
Additionally, Hamada et al. used a C-terminal polyclonal antibody (1998–2207 aa) to demonstrate changes in nuclear isolation in Mus musculus embryos confirming persistent cytoplasm presence in postnatally differentiated neural crest cells. 81 When repeated, MED13L protein was observed to accumulate in cytoplasmic hippocampal neurons. Hamada noticed continued co-localization of MED13L with both presynaptic and postsynaptic markers. 81 This demonstrates something fairly unique to MED13L, which is its cellular localization changes with developmental period and tissue/cell type. Additional GFP (green fluorescent protein)-tagged embryonic models were also created to characterize cortical neuron development using different patient-derived variants. 39 Using the same custom C-terminal MED13L antibody, researchers described the functional impact on developing neurons as well as changes in the localization of MED13L compared to wild type 39 (Table 3). Additional confirmation to better understand the roles of MED13L transcripts, as well as the development and validation of N-terminal antibodies in patient-derived models must be performed to further substantiate these initial experiments.
Finally, an M. musculus (mouse) model created by the Grueter Lab recently showed that both MED13 and MED13L were necessary for cardiomyocyte functionality. 82 In addition to physical cardiac deficits leading to mortality, mRNA sequencing confirmed that the Wnt pathway was upregulated, but RNA polymerase II-dependent transcription remained unchanged. 82 This confirms the necessity of both MED13 and MED13L proteins for cardiomyocyte functionality 82 (Table 3). Through a MED13L Foundation grant, the Grueter Lab at University of Iowa is expanding M. musculus models to examine the broader phenotype of MED13 and MED13L Syndromes (https://med13l.org/research-hub/grant-awards/).
Biomarkers and outcome measures
In addition to cell and animal models, it is essential for consistent measurable biomarkers. It is estimated that 20% of individuals with MED13L syndrome have seizures.2,8,10,27,32,37,38 Additionally, the literature cites about 30 individuals with abnormal MRIs.2,8–10,25–27,29 However, the rate at which these patients have disease-specific EEG or MRI features remains unclear. Disease-specific EEG signatures are often used as biomarkers for epileptic encephalopathies and could be further explored in MED13L syndrome.83,84 Similarly, MRIs have also been used in predictive endotypes of gene-based autism disorder. 36 Both research EEGs as well as prior MRIs were collected as part of the 2022 Simons-sponsored research and family conference. Analysis could provide a unique insight into MED13L syndrome.
Other molecular, metabolomic, or proteomic biomarkers are being investigated, leveraging the rich biospecimen repository. Given the transcriptional impact of MED13L on the large mediator complex, it could, in theory, provide insight into expression patterns.44,50 Additionally, a recent publication using patient-derived fibroblasts showed accelerated aging and early senescence in fibroblasts containing the MED13L variant. 23 This could indicate a larger epigenetic process and potential biomarker. The mitochondrial biogenesis mechanism also offers additional biomarker exploration.52,53
Disease phenotyping
Disease concept studies
Disease concept studies are a literature summary reviewing a specific disorder combined with personal interviews of those affected by the disorder. To undergo a formal concept study, caregivers, healthcare professionals, and educators familiar with the disorder are interviewed using open-ended questions to obtain additional details into symptoms, management, and caregiver and individual impacts of daily life. 85 To take on a formal disease concept, a draft must be completed by reviewing the available literature, compiling a list of symptoms, collating and establishing key features of the disorder, and noting ones that may be important for future tracking. 85 A formal disease concept study is underway in collaboration with the Rutgers University Genetic Counseling program and COMBINEDBrain. Preliminary information should be available by late 2024 or early 2025 (https://combinedbrain.org/conceptual-models/).
Data collection platforms
MED13L is a gene of interest for Simons Searchlight, an international online platform that studies over 150 genes causing rare neurodevelopmental disorders with known comorbid conditions of autism. Updated analyses of select surveys are provided quarterly online (https://www.simonssearchlight.org/research/what-we-study/med13l/). Similarly, the Foundation is partnering with RARE-X for patient/caregiver-entered data collection, which ensures the patient data is owned and managed by the individuals and leaders of the Foundation (https://rare-x.org/about/). The MED13L Syndrome Association is primarily using GenIDA (Genetic of Intellectual Disability and Autism Spectrum Disorders), a French-based platform to collect longitudinal caregiver-reported data. 86 GenIDA, like Simons Searchlight, is an internationally based online collection platform targeted to neurodevelopmental diseases with underlying autism. A recent publication highlighted the collaboration between The MED13L Syndrome Association and key opinion leaders, Dr. Jamal Ghoumid, Dr. Roseline Caumes, and Dr Thomas Smol. Of the 44 documented individuals with MED13L, 47% of them were French, demonstrating the need for collaboration with researchers, clinicians, and patient advocacy organizations. 86
MED13L is also a gene of interest in the Brain Gene Registry (BGR), a national repository of detailed information on individuals with variants in genes thought to be involved in intellectual and developmental disabilities. 87 The BGR collects a battery of assessments which is a composite of cognitive, adaptive, motor/sensory, autism, psychiatric, neurological, and past medical history surveys to support reclassification of VUS. The BGR will also source electronic medical record data if the affected individual is enrolled at one of the collaboration sites. 87 Given the focus on VUS, this can be particularly useful to support the reassignment of VUS in the MED13L community.
Summary
The intent of this SRP is to serve as the foundational roadmap for the path to therapeutic interventions for those affected by MED13L syndrome. Consequently, the MED13L Foundation is investing and collaborating in basic science, clinical research, and community engagement. For basic science, the Foundation is developing necessary resources for researchers, including antibodies, assays, and -omic studies for biomarkers. By providing grant funding for mitochondrial studies and mouse model development, researchers aim to establish a consistent rescuable phenotype.
Gaps identified in the roadmap (Figure 1) are prioritized for the near future. The community is engaged in providing critical biospecimens for research and providing caregiver-entered information in data collection platforms. The next phase involves identifying family priorities and holding regular convenings of families, clinicians, and researchers. This will promote long-term engagement to set the stage for a formal Natural History Study. Additionally, the Foundation is focused on collaborating worldwide to engage an international base, including The MED13L Association and other international families, to grow the community.
An SRP requires regular review and updates based on new publications, increased availability of MED13L Foundation resources, and investments in the community. The benefit for a mostly volunteer nonprofit organization in developing an SRP unique to their disorder is providing an objective framework of the tools to navigate the road to accessible therapeutic options.
Acknowledgments
None.
Footnotes
ORCID iDs: Rachel Heilmann  https://orcid.org/0009-0002-7726-4795
https://orcid.org/0009-0002-7726-4795
Anna Pfalzer  https://orcid.org/0000-0001-8317-3318
https://orcid.org/0000-0001-8317-3318
Contributor Information
Rachel Heilmann, The MED13L Foundation, PO Box 283, Barrington, NJ 08007, USA COMBINEDBrain, Brentwood, TN, USA.
Anna Pfalzer, COMBINEDBrain, Brentwood, TN, USA; Department of Neurology, Vanderbilt University, Nashville, TN, USA.
Terry Jo Bichell, COMBINEDBrain, Brentwood, TN, USA.
Ananya Terala, COMBINEDBrain, Brentwood, TN, USA.
Alicia Campbell, Department of Molecular Biology, Rowan-Virtua School of Translational Biomedical Engineering & Sciences and Virtua Health College of Medicine and Life Sciences at Rowan University, Rowan University, Glassboro, NJ, USA.
Dylan Taatjes, Department of Biochemistry, University of Colorado, Boulder, CO, USA.
Jamal Ghoumid, Department of Clinical Genetics, Lille University Hospital, Lille, France.
Chad Grueter, Department of Internal Medicine, University of Iowa, Iowa City, IA, USA.
Jennifer Bain, Departments of Neurology and Pediatrics, Columbia University Irving Medical Center, New York, NY, USA.
Randy Strich, Department of Molecular Biology, Rowan-Virtua School of Translational Biomedical Engineering & Sciences and Virtua Health College of Medicine and Life Sciences at Rowan University, Rowan University, Glassboro, NJ, USA.
Vanessa Dias, The MED13L Foundation, Barrington, NJ, USA.
Kimberly Sokorai, The MED13L Foundation, Barrington, NJ, USA.
Nicholas Seaver, The MED13L Foundation, Barrington, NJ, USA.
Kelly Sexton, The MED13L Foundation, Barrington, NJ, USA.
Kathleen Boychuck, The MED13L Foundation, Barrington, NJ, USA.
Declarations
Ethics approval and consent to participate: Not applicable.
Consent for publication: Not applicable.
Author contributions: Rachel Heilmann: Conceptualization; Writing – original draft.
Anna Pfalzer: Conceptualization; Writing – original draft.
Terry Jo Bichell: Conceptualization; Writing – review & editing.
Ananya Terala: Conceptualization; Writing – original draft.
Alicia Campbell: Conceptualization; Writing – review & editing.
Dylan Taatjes: Conceptualization; Writing – review & editing.
Jamal Ghoumid: Conceptualization; Writing – review & editing.
Chad Grueter: Conceptualization; Writing – review & editing.
Jennifer Bain: Conceptualization; Writing – review & editing.
Randy Strich: Conceptualization; Writing – review & editing.
Vanessa Dias: Conceptualization; Writing – review & editing.
Kimberly Sokorai: Conceptualization; Writing – review & editing.
Nicholas Seaver: Conceptualization; Writing – review & editing.
Kelly Sexton: Conceptualization; Writing – review & editing.
Kathleen Boychuck: Conceptualization; Writing – review & editing.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
Competing interests: The authors declare that there is no conflict of interest.
Availability of data and materials: Not applicable.
References
- 1. Asadollahi R, Oneda B, Sheth F, et al. Dosage changes of MED13L further delineate its role in congenital heart defects and intellectual disability. Eur J Hum Genet 2013; 21: 1100–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adegbola A, Musante L, Callewaert B, et al. Redefining the MED13L syndrome. Eur J Hum Genet 2015; 23: 1308–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Muncke N, Jung C, Rüdiger H, et al. Missense mutations and gene interruption in PROSIT240, a novel TRAP240-like gene, in patients with congenital heart defect (transposition of the great arteries). Circulation 2003; 108: 2843–2850. [DOI] [PubMed] [Google Scholar]
- 4. Musante L, Bartsch O, Ropers H-H, et al. cDNA cloning and characterization of the human THRAP2 gene which maps to chromosome 12q24, and its mouse ortholog Thrap2. Gene 2004; 332: 119–127. [DOI] [PubMed] [Google Scholar]
- 5. U.S. Food and Drug Administration. Rare diseases: considerations for the development of drugs and biological products [internet], https://www.fda.gov/regulatory-information/search-fda-guidance-documents/rare-diseasesconsiderations-development-drugs-andbiological-products (2023, accessed 21 February 2024).
- 6. U.S. Food and Drug Administration. FDA patient-focused drug development guidance series for enhancing the incorporation of the patient’s voice in medical product development and regulatory decision making [internet], https://www.fda.gov/drugs/development-approval-process-drugs/fda-patient-focused-drug-development-guidance-series-enhancing-incorporation-patients-voice-medical (2020, accessed 6 May 2021).
- 7. U.S. Food and Drug Administration. Rare diseases: natural history studies for drug development [internet], https://www.fda.gov/regulatory-information/search-fda-guidance-documents/rare-diseases-natural-history-studies-drug-development (2019, accessed 28 February 2024).
- 8. Asadollahi R, Zweier M, Gogoll L, et al. Genotype-phenotype evaluation of MED13L defects in the light of a novel truncating and a recurrent missense mutation. Eur J Med Genet 2017; 60: 451–464. [DOI] [PubMed] [Google Scholar]
- 9. Torring PM, Larsen MJ, Brasch-Anderson C, et al. Is MED13L related intellectual disability a recognizable syndrome? Eur J Med Genet 2019; 62: 129–136. [DOI] [PubMed] [Google Scholar]
- 10. Smol T, Petit F, Piton A, et al. MED13L-related intellectual disability: involvement of missense variants and delineation of the phenotype. Neurogenetics 2018; 19: 93–103. [DOI] [PubMed] [Google Scholar]
- 11. Codina-Solà M, Rodríguez-Santiago B, Homs A, et al. Integrated analysis of whole-exome sequencing and transcriptome profiling in males with autism spectrum disorders. Mol Autism 2015; 6: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bessenyei B, Balogh I, Mokánszki A, et al. MED13L-related intellectual disability due to paternal germinal mosaicism. Cold Spring Harb Mol Case Stud 2022; 8: a006124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carvalho LML, Costa SS, Campagnari F, et al. Two novel pathogenic variants in MED13L: one familial and one isolated case. J Intellect Disabil Res 2021; 65: 1049–1057. [DOI] [PubMed] [Google Scholar]
- 14. Yamamoto T, Shimojima K, Ondo Y, et al. MED13L haploinsufficiency syndrome: a de novo frameshift and recurrent intragenic deletions due to parental mosaicism. Am J Med Genet 2017; 173: 1264–1269. [DOI] [PubMed] [Google Scholar]
- 15. Nizon M, Laugel V, Flanigan KM, et al. Variants in MED12L, encoding a subunit of the Mediator kinase module, are responsible for intellectual disability associated with transcriptional defect. Genet Med 2019; 21: 2713–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Napoli C, Schiano C, Soricelli A. Increasing evidence of pathogenic role of the Mediator (MED) complex in the development of cardiovascular diseases. Biochimie 2019; 165: 1–8. [DOI] [PubMed] [Google Scholar]
- 17. Asadollahi R, Oneda B, Joset P, et al. The clinical significance of small copy number variants in neurodevelopmental disorders. J Med Genet 2014; 51: 677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature 2015; 519: 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wright CF, McRae JF, Clayton S, et al. Making new genetic diagnoses with old data: iterative reanalysis and reporting from genome-wide data in 1133 families with developmental disorders. Genet Med 2018; 20: 1216–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature 2017; 542: 433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. MedlinePlus. MED13L syndrome [internet] Bethesda (MD): National Library of Medicine (US), https://medlineplus.gov/genetics/condition/med13l-syndrome/ (2024, accessed 17 July 2024).
- 22. López-Rivera JA, Pérez-Palma E, Symonds J, et al. A catalogue of new incidence estimates of monogenic neurodevelopmental disorders caused by de novo variants. Brain 2020; 143: 1099–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Siavrienė E, Petraitytė G, Mikštienė V, et al. Molecular and functional characterisation of a novel intragenic 12q24.21 deletion resulting in MED13L haploinsufficiency syndrome. Medicina 2023; 59: 1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Centers for Disease Control and Prevention. ICD-10-CM: Med13L [internet], https://icd10cmtool.cdc.gov/?fy=FY2025&query=med13l (2024, accessed 24 October 2024).
- 25. Dawidziuk M, Kutkowska-Kaźmierczak A, Gawliński P, et al. The MED13L haploinsufficiency syndrome associated with de novo nonsense variant (P.GLN1981*). J Mother Child 2021; 24: 32–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yi Z, Zhang Y, Song Z, et al. Report of a de novo c.2605C > T (p.Pro869Ser) change in the MED13L gene and review of the literature for MED13L-related intellectual disability. Ital J Pediatr 2020; 46: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cafiero C, Marangi G, Orteschi D, et al. Novel de novo heterozygous loss-of-function variants in MED13L and further delineation of the MED13L haploinsufficiency syndrome. Eur J Hum Genet 2015; 23: 1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sabo A, Murdock D, Dugan S, et al. Community-based recruitment and exome sequencing indicates high diagnostic yield in adults with intellectual disability. Mol Genet Genomic Med 2020; 8: e1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gordon CT, Chopra M, Oufadem M, et al. MED13L loss-of-function variants in two patients with syndromic Pierre Robin sequence. Am J Med Genet 2018; 176: 181–186. [DOI] [PubMed] [Google Scholar]
- 30. Iossifov I, Ronemus M, Levy D, et al. De novo gene disruptions in children on the autistic spectrum. Neuron 2012; 74: 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Caro-Llopis A, Rosello M, Orellana C, et al. De novo mutations in genes of mediator complex causing syndromic intellectual disability: mediatorpathy or transcriptomopathy? Pediatr Res 2016; 80: 809–815. [DOI] [PubMed] [Google Scholar]
- 32. Mullegama SV, Jensik P, Li C, et al. Coupling clinical exome sequencing with functional characterization studies to diagnose a patient with familial Mediterranean fever and MED13L haploinsufficiency syndromes. Clin Case Rep 2017; 5: 833–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jiménez-Romero S, Carrasco-Salas P, Benítez-Burraco A. Language and cognitive impairment associated with a novel p.Cys63Arg change in the MED13L transcriptional regulator. Mol Syndromol 2018; 9: 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tsai P, Shinar S. Agenesis of the corpus callosum: what to tell expecting parents? Prenat Diagn 2023; 43: 1527–1535. [DOI] [PubMed] [Google Scholar]
- 35. Kyriakopoulou V, Davidson A, Chew A, et al. Characterisation of ASD traits among a cohort of children with isolated fetal ventriculomegaly. Nat Commun 2023; 14: 1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kundu S, Sair H, Sherr EH, et al. Discovering the gene-brain-behavior link in autism via generative machine learning. Sci Adv 2024; 10: eadl5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gilissen C. Genome sequencing identifies major causes of severe intellectual disability. Nature 2014; 511: 344–347. [DOI] [PubMed] [Google Scholar]
- 38. Utami KH, Winata CL, Hillmer AM, et al. Impaired development of neural-crest cell derived organs and intellectual disability caused by MED13L haploinsufficiency. Hum Mutat 2014; 35: 1311–1320. [DOI] [PubMed] [Google Scholar]
- 39. Hamada N, Iwamoto I, Nagata K. MED13L and its disease-associated variants influence the dendritic development of cerebral cortical neurons in the mammalian brain. J Neurochem 2023; 165(3): 334–347. [DOI] [PubMed] [Google Scholar]
- 40. Albuja AC, Ighodaro ET, Khan GQ. Absence seizure. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing, https://www.ncbi.nlm.nih.gov/books/NBK499867/ (2024, accessed 27 October 2024) [PubMed] [Google Scholar]
- 41. Kowanda M, Calakos K, Snyder LG, et al. P152: seizure severity across neurogenetic conditions in Simons Searchlight. Genet Med Open 2024; 2: 101049. [Google Scholar]
- 42. Hamdan FF, Srour M, Capo-Chichi JM, et al. De novo mutations in moderate or severe intellectual disability. PLoS Genet 2014; 10: e1004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Luyties O, Taatjes DJ. The Mediator kinase module: an interface between cell signaling and transcription. Trends Biochem Sci 2022; 47: 314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Davis MA, Larimore EA, Fissel BM, et al. The SCF–Fbw7 ubiquitin ligase degrades MED13 and MED13L and regulates CDK8 module association with Mediator. Genes Dev 2013; 27: 151–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Richter WF, Nayak S, Iwasa J, et al. The Mediator complex as a master regulator of transcription by RNA polymerase II. Nat Rev Mol Cell Biol 2022; 23: 732–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Knuesel MT, Meyer KD, Bernecky C, et al. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev 2009; 23: 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tsai K-L, Sato S, Tomomori-Sato C, et al. A conserved Mediator–CDK8 kinase module association regulates Mediator–RNA polymerase II interaction. Nat Struct Mol Biol 2013; 20: 611–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ebmeier CC, Taatjes DJ. Activator-Mediator binding regulates Mediator-cofactor interactions. Proc Natl Acad Sci U S A 2010; 107: 11283–11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sato S, Tomomori-Sato C, Parmely TJ, et al. Mediator subunits identified by multidimensional protein identification technology. Mol Cell 2004; 14(5): 685–691. [DOI] [PubMed] [Google Scholar]
- 50. Snijders Blok L, Hiatt SM, Bowling KM, et al. De novo mutations in MED13, a component of the Mediator complex, are associated with a novel neurodevelopmental disorder. Hum Genet 2018; 137: 375–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Angus SP, Nevins JR. A role for Mediator complex subunit MED13L in Rb/E2F-induced growth arrest. Oncogene 2012; 31: 4709–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stieg DC, Cooper KF, Strich R. The extent of cyclin C promoter occupancy directs changes in stress-dependent transcription. J Biol Chem 2020; 295: 16280–16291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chang K-T, Jezek J, Campbell AN, et al. Aberrant cyclin C nuclear release induces mitochondrial fragmentation and dysfunction in MED13L syndrome fibroblasts. iScience 2022; 25: 103823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Colella P, Ronzitti G, Mingozzi F. Emerging issues in AAV-mediated in vivo gene therapy. Mol Ther Methods Clin Dev 2018; 8: 87–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ensembl. Gene: MED13L (ENSG00000123066)—Summary—Homo_sapiens[internet], https://useast.ensembl.org/Homo_sapiens/Gene/Summary?db=core;g=ENSG00000123066;r=12:115957905-116277693 (2024, accessed 17 July 2024).
- 56. Gleeson JG, Bennett CF, Carroll JB, et al. Personalized antisense oligonucleotides “for free, for life”—the n-Lorem Foundation. Nat Med 2023; 29: 1302–1303. [DOI] [PubMed] [Google Scholar]
- 57. Lauffer MC, van Roon-Mom W, Aartsma-Rus A. Possibilities and limitations of antisense oligonucleotide therapies for the treatment of monogenic disorders. Commun Med (Lond) 2024; 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Torkzaban B, Kawalerski R, Coller J. Development of a Tethered mRNA Amplifier to increase protein expression. Biotechnol J 2022; 17: 2200214. [DOI] [PubMed] [Google Scholar]
- 59. Albers S, Beckert B, Matthies MC, et al. Repurposing tRNAs for nonsense suppression. Nat Commun 2021; 12: 3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Michorowska S. Ataluren—promising therapeutic premature termination codon readthrough frontrunner. Pharmaceuticals (Basel) 2021; 14: 785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dolgin E. tRNA therapeutics burst onto startup scene. Nat Biotechnol 2022; 40: 283–286. [DOI] [PubMed] [Google Scholar]
- 62. Matharu N, Ahituv N. Modulating gene regulation to treat genetic disorders. Nat Rev Drug Discov, 2020; 19: 757–775. [DOI] [PubMed] [Google Scholar]
- 63. Ghoumid J, Ziffra R, Balerdi M, et al. SS001 Caractérisation des organoïdes cérébraux KO MED13L [abstract]. Rennes, France, https://api.mycongressonline.net/api-Congress-agenda.html?record=debebeec-0fbb-1e55-6772-600ff7101225key=ab10c6a3e4ba4db1629057c3d8dca3e66653b897 (2022, accessed 24 October 2024). [Google Scholar]
- 64. Chemical Entities of Biological Interest. MED13L—chemical interactions | CTD [internet], http://ctdbase.org/detail.go?type=gene&acc=23389&view=ixn (2024, accessed 24 March 2023).
- 65. Morton K, Wang P, Bizon C, et al. ROBOKOP: an abstraction layer and user interface for knowledge graphs to support question answering. Bioinformatics 2019; 35: 5382–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bizon C, Cox S, Balhoff J, et al. ROBOKOP KG and KGB: integrated knowledge graphs from federated sources. J Chem Inf Model 2019; 59: 4968–4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fecho K, Thessen AE, Baranzini SE, et al. Progress toward a universal biomedical data translator. Clin Transl Sci 2022; 15: 1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kilicoglu H, Shin D, Fiszman M, et al. SemMedDB: a PubMed-scale repository of biomedical semantic predications. Bioinformatics 2012; 28: 3158–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Foksinska A, Crowder CM, Crouse AB, et al. The precision medicine process for treating rare disease using the artificial intelligence tool mediKanren. Front Artif Intell 5, https://www.frontiersin.org/articles/10.3389/frai.2022.910216 (2022, accessed 26 October 2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cariello NF, Romach EH, Colton HM, et al. Gene expression profiling of the PPAR-alpha agonist ciprofibrate in the cynomolgus monkey liver. Toxicol Sci 2005; 88: 250–264. [DOI] [PubMed] [Google Scholar]
- 71. Frambach SJCM. Effects of clofibrate and KH176 on life span and motor function in mitochondrial complex I-deficient mice. Biochim Biophys Acta Mol Basis Dis 2020; 1866: 1–12. [DOI] [PubMed] [Google Scholar]
- 72. Douiev L, Sheffer R, Horvath G, et al. Bezafibrate improves mitochondrial fission and function in DNM1L-deficient patient cells. Cells 2020; 9: 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wu J-H, Zhang S-H, Gao F-J, et al. RNAi screening identifies GSK3β as a regulator of DRP1 and the neuroprotection of lithium chloride against elevated pressure involved in downregulation of DRP1. Neurosci Lett 2013; 554: 99–104. [DOI] [PubMed] [Google Scholar]
- 74. Takamatsu A, Ohkawara B, Ito M, et al. Verapamil protects against cartilage degradation in osteoarthritis by inhibiting Wnt/β-catenin signaling. PLoS One 2014; 9: e92699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Borrell-Pages M, Romero JC, Badimon L. Cholesterol modulates LRP5 expression in the vessel wall. Atherosclerosis 2014; 235: 363–370. [DOI] [PubMed] [Google Scholar]
- 76. Zhang Y, Jiang L, Hu W, et al. Mitochondrial dysfunction during in vitro hepatocyte steatosis is reversed by omega-3 fatty acid-induced up-regulation of mitofusin 2. Metabolism 2011; 60: 767–775. [DOI] [PubMed] [Google Scholar]
- 77. Zhou X, Wang H-Y, Wu B, et al. Ginkgolide K attenuates neuronal injury after ischemic stroke by inhibiting mitochondrial fission and GSK-3β-dependent increases in mitochondrial membrane permeability. Oncotarget 2017; 8: 44682–44693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Deal LS, Goldsmith JC, Martin S, et al. Patient voice in rare disease drug development and endpoints. Ther Innov Regul Sci 2017; 51: 257–263. [DOI] [PubMed] [Google Scholar]
- 79. Miller KL, Mueller C, Liu G, et al. FDA orphan products clinical trial grants: assessment of outcomes and impact on rare disease product development. Orphanet J Rare Dis 2020; 15: 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Allio T. Product development under FDA’s animal rule: understanding FDA’s expectations and potential implications for Traditional Development Programs. Drug Inf J 2016; 50: 660–670. [DOI] [PubMed] [Google Scholar]
- 81. Hamada N, Iwamoto I, Nishikawa M, et al. Expression analyses of mediator complex subunit 13-like: a responsible gene for neurodevelopmental disorders during mouse brain development. Dev Neurosci 2021; 43: 43–52. [DOI] [PubMed] [Google Scholar]
- 82. Gardner RN, Oppman AM, Martins I, et al. Cardiomyocyte-specific deletion of Med13 & Med13L results in dysregulated gene expression and lethal systolic heart failure. FASEB J 2022; 36: R2395. DOI: 10.1096/fasebj.2022.36.S1.R2395 [DOI] [Google Scholar]
- 83. Bosl W, Tierney A, Tager-Flusberg H, et al. EEG complexity as a biomarker for autism spectrum disorder risk. BMC Med 2011; 9: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gülbakan B, Özgül RK, Yüzbaşıoğlu A, et al. Discovery of biomarkers in rare diseases: innovative approaches by predictive and personalized medicine. EPMA J 2016; 7: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Willgoss T, Cassater D, Connor S, et al. Measuring what matters to individuals with angelman syndrome and their families: development of a patient-centered disease concept model. Child Psychiatry Hum Dev 2021; 52(4): 654–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Burger P, Colin F, Strehle A, et al. GenIDA: an international participatory database to gain knowledge on health issues related to genetic forms of neurodevelopmental disorders. J Neural Transm 2023; 130: 459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chopra M, Savatt JM, Bingaman TI, et al. Clinical variants paired with phenotype: a rich resource for brain gene curation. Genet Med 2024; 26: 101035. [DOI] [PMC free article] [PubMed] [Google Scholar]