Abstract
The absence of functional von Hippel-Lindau (VHL) tumor suppressor gene leads to the development of neoplasias characteristic of VHL disease, including renal cell carcinoma (RCC). Here, we compared the sensitivity of RCC cells lacking VHL gene function with that of RCC cells expressing the wild-type VHL gene (wtVHL) after exposure to various stresses. While the response to most treatments was not affected by the VHL gene status, glucose deprivation was found to be much more cytotoxic for RCC cells lacking VHL gene function than for wtVHL-expressing cells. The heightened sensitivity of VHL-deficient cells was not attributed to dissimilar energy requirements or to differences in glucose uptake, but more likely reflects a lesser ability of VHL-deficient cells to handle abnormally processed proteins arising from impaired glycosylation. In support of this hypothesis, other treatments which act through different mechanisms to interfere with protein processing (i.e., tunicamycin, brefeldin A, and azetidine) were also found to be much more toxic for VHL-deficient cells. Furthermore, ubiquitination of cellular proteins was elevated in VHL-deficient cells, particularly after glucose deprivation, supporting a role for the VHL gene in ubiquitin-mediated proteolysis. Accordingly, the rate of elimination of abnormal proteins was lower in cells lacking a functional VHL gene than in wtVHL-expressing cells. Thus, pVHL appears to participate in the elimination of misprocessed proteins, such as those arising in the cell due to the unavailability of glucose or to other stresses.
Germline mutations in the von Hippel-Lindau (VHL) tumor suppressor gene predispose individuals to the development of tumors characteristic of VHL disease, including retinal angiomas, hemangioblastomas, pheochromocytomas, and renal cell carcinomas (6, 12, 20, 21, 23). In humans, the product of the VHL gene is a 213-amino-acid protein that is localized primarily in the cytoplasm (31). Although highly conserved across species, pVHL bears little similarity to other known proteins. Most studies to elucidate its function have focused on analysis of the interaction of pVHL with other cellular proteins. Thus, pVHL was found to interact with elongins B and C, the regulatory subunits of the elongin-SIII complex. Binding of elongins B and C to elongin A, the catalytic subunit, is necessary for elongin-SIII function and leads to enhanced transcriptional activity by RNA polymerase II (1); binding of pVHL to elongins B and C results in inhibition of elongin function and decreased RNA polymerase II activity in vitro (7, 26, 29). Elongin A and pVHL have not been found in the same complex, suggesting that their association with elongins B and C may be mutually exclusive. More recently, another pVHL-interacting protein, Hs-CUL-2 was identified; yeast Cul2 (also termed cdc53) is found in multiprotein complexes also, including Skp1 and cdc34, which have been proposed to participate in targeting specific proteins for ubiquitin-mediated proteolysis (24, 28, 38, 44). Based on the association between pVHL and Cul2, and the sequence similarities between Skp1 and cdc34 with elongins C and B, respectively, a role for pVHL in ubiquitin-mediated proteolysis has recently been postulated (36, 38).
The extensive vascularization of VHL tumors is likely to arise from the presence of abnormally high levels of vascular endothelial growth factor (VEGF) (49, 51, 54). Constitutive VEGF levels in tissues are normally low, but its expression is highly elevated in VHL tumors, in tissues lacking a functional VHL gene product (pVHL), and in pVHL-deficient cells cultured in vitro (13, 51, 59), with the unexpected exception of VHL knockout embryos (14). Under physiologic conditions, functional VHL downregulates expression of the VEGF gene through both inhibition of VEGF gene transcription (25, 42) and posttranscriptional destabilization of the VEGF mRNA (22, 53); the latter process has been proposed to be regulated by modulating, through proteolysis, the activity of proteins binding the VEGF mRNA (33, 34). During abnormal angiogenesis resulting in hypoxia, many tissues switch from aerobic to anaerobic metabolism by (i) activating glycolytic pathways, (ii) increasing their glucose uptake to compensate for the reduced ATP produced by glycolysis, and (iii) increasing local vascularization by stimulating angiogenesis. Indeed, there is much evidence for an overlap in the response of tissues to low oxygen availability and to glucose deprivation. For example, both hypoxia and hypoglycemia induce many common genes, such as erythropoietin and VEGF genes (15, 53); induction of VEGF gene expression by both hypoxia and hypoglycemia is mediated through stabilization of its mRNA (33, 53). In addition, the hypoxia-inducible factor 1β (HSF-1β), a transcriptional regulator of oxygen-responsive genes, was found to be required for the expression of both VEGF and the glucose-transporter GLUT-3 in hepatoma cells (40). Similarly, the transcription factor arylhydrocarbon-receptor nuclear translocator (ARNT) was found to be critical for regulating the expression of the VEGF gene and that of many glucose-responsive genes (the glucose transporter GLUT-1, aldolase, phosphofructokinase, phosphoglycerate kinase, etc.) (39). Accordingly, the impaired ability of ARNT-deficient cells to elevate the expression of these genes in response to glucose starvation or hypoxia correlated with enhanced cytotoxicity in response to these treatments (39).
In the present study, renal cell carcinoma (RCC) cells were exposed to a panel of stimuli to explore the influence of the VHL status on their responsiveness to various stresses. While the effect of most treatments was indistinguishable when cells lacking functional VHL gene expression (parental) were compared with their counterparts expressing pVHL (through stable transfection of wild-type VHL gene [wtVHL]), we observed dramatic differences with glucose deprivation: it was potently cytotoxic for parental cells, but not for cells expressing wtVHL. Studies on the mechanisms underlying these differences demonstrate a greatly diminished ability of VHL-deficient cells to survive in the presence of incompletely processed proteins, since inhibitors of protein folding and posttranslational modifications (glycosylation or Golgi processing) were also more toxic in the absence of the VHL gene. Finally, VHL-deficient cells subjected to glucose deprivation or azetidine treatment exhibited a greater accumulation of ubiquitinated proteins and a slower rate of elimination of aberrant proteins than did wtVHL-expressing cells. These findings further support the notion that wtVHL aids in the elimination of proteins that are targeted for proteolytic degradation.
MATERIALS AND METHODS
Cell culture, treatments, and transfection of bcl-2.
The human renal carcinoma cell lines UMRC6 (UMR) (18), 786-0 (cell line 786) (21), and UOK 121 (cell line 121) (13), were each stably transfected with either a vector control (parental) or a plasmid expressing wild-type VHL cDNA (wtVHL). Additionally, UMRC6 cells were also transfected with a plasmid expressing a VHL cDNA carrying a mutation in nucleotide 737, rendering a C-terminally truncated VHL protein lacking the elongin-binding site (clonal isolates XX23 and XX27). Cells were routinely cultured in Dulbecco’s modified essential medium (DMEM; Gibco BRL, Gaithersburg, Md.) supplemented with 10% fetal bovine serum (FBS; Hyclone, Logan, Utah), 100 U of penicillin per ml, and 100 μg of streptomycin (Gibco BRL) per ml and maintained in a humidified atmosphere containing 5% CO2 in air. Hypoxia treatments were carried out in a chamber receiving continuous injection of a gas mixture containing 5% CO2 and 95% N2. Under these conditions, the partial oxygen pressure reached a plateau of 10 Tor by 8 h. For the glucose deprivation experiments, cells were plated at a density of approximately 30,000, 80,000, or 200,000 to 400,000 cells per 6-well cluster plate, 60-mm dish, or 100-mm dish, respectively. Although they were cultured in glucose-free DMEM, its supplementation with 10% FBS contributed 100 mg of glucose per liter. Use of dialyzed FBS was avoided to prevent depletion of other nutrients (nucleotides, amino acids, etc.) through dialysis. The volume of glucose-free DMEM plus 10% FBS added to cells was carefully calculated so that 2 ng/cell was routinely available at the beginning of the glucose starvation period (typically about 1, 2 to 2.5, and 6 to 9 ml for 6-well cluster plates, 60-mm dishes, and 100-mm dishes, respectively). It is critical to accurately calculate the availability of glucose per cell, and not merely the glucose concentration, because glucose is not in excess. In the initial hours, the glucose contributed by the serum is depleted; the rate of depletion, and hence the toxicity, is strictly proportional to the number of cells present. Brefeldin A, tunicamycin, azetidine, neomycin, hygromycin (l-azetidine-2-carboxylic acid), glucosamine, 2-deoxyglucose, hydrogen peroxide, sodium arsenite, cycloheximide, tumor necrosis factor alpha (TNF-α), sodium phenylacetate, 4-chloro-phenylacetate, 12-O-tetradecanoylphorbol-13-acetate (TPA), and thapsigargin were from Sigma (St. Louis, Mo.). Mimosine was from Aldrich Chemical Co. (St. Louis, Mo.), and lactacystin (clasto-lactacystin β-lactone) was from Boston Biochem (Cambridge, Mass.). Drugs were added directly into the medium. For irradiation with short-wavelength UV light (UVC), cells were rinsed with phosphate-buffered saline (PBS) before irradiation, and tissue culture medium was added back to the cells immediately after irradiation. Untreated controls were subjected to mock irradiation. For the stable transfection of bcl-2, 5 × 105 cells were seeded in 100-mm plates 24 h before transfection. Then 5 μg of the pSFFV-bcl2 construct or pSFFV-neo vector control (kindly provided by G. Núñez) were transfected into cells by using standard calcium phosphate precipitation methods. Stable transformants were selected in the presence of 500 μg of neomycin (Sigma) per ml. Cell counts were performed with a hemacytometer, and crystal violet cytotoxicity assays were carried out in 96-well cluster plates, as described earlier (17).
Colony formation assays.
Cell survival was measured by a standard clonogenic assay. Cells were initially seeded at a density of 30,000 cells per well in 6-well cluster plates. At the end of each treatment, cells were trypsinized and serially diluted according to the expected surviving fraction (from 1:10 to 1:1,000,000). Plates were then returned to the incubator and cultured for an additional 10 to 12 days. The plates were fixed and stained with a crystal violet solution (10% ethanol [vol/vol], 0.1% crystal violet [wt/vol], and colonies (defined as greater than 50 cells) were counted. The surviving fraction was determined as the number of colonies divided by the dilution factor. For each treatment, plates were seeded at four different dilutions, and routinely three of these were counted. Each colony formation assay was performed at least three times.
Northern blot analysis.
Total RNA was isolated with STAT-60 (Tel-Test B, Friendswood, Tex.), and 20-μg RNA samples were denatured, size fractionated by electrophoresis in 1.2% agarose-formaldehyde gels, and transferred onto GeneScreen Plus nylon membranes (DuPont/NEN, Boston, Mass.) as described previously (16). For the detection of grp78, gadd153, hsp70, and VHL mRNAs, the corresponding cDNA inserts were excised from the plasmids p3C5grp78 (55), pCMVgadd153, pM3hsp70 (10), and pCEP4VHL, respectively, and labeled with a random primer labeling kit (Boehringer Mannheim, Indianapolis, Ind.) in the presence of [α-32P]dCTP. An oligomer complementary to the 18S rRNA (5′-ACGGTATCTGATCGTCTTCGAACC-3′) (Integrated DNA Technologies, Coralville, Iowa) was 3′ end labeled with [α-32P]dATP by terminal deoxynucleotidyl transferase (Life Technology Laboratories, Gaithersburg, Md.) and used to normalize for differences in loading and transfer among samples (data not shown). Hybridization and washes were performed according to the method of Church and Gilbert (5). Incorporation of 32P was visualized with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Western blot analysis.
Fifty-microgram samples of total cell lysates were size fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride membranes by standard techniques, and detected with the enhanced chemiluminescence (ECL) system (Amersham, Arlington Heights, Ill.). Grp78 protein was detected after incubation with the monoclonal mouse anti-human grp78 antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.), and ubiquitin was detected with a mouse monoclonal antibody (Calbiochem, San Diego, Calif.).
Assays for protein degradation.
For the analysis of protein elimination, cells were either left untreated or were treated with azetidine in methionine-free DMEM for 1 h before the addition of 50 μCi of [35S]methionine (specific activity, 1,175 Ci/mM; American Radiolabeled Chemicals, Inc., St. Louis, Mo.) per ml. After an additional 5 h, the labeling medium was removed and replaced with complete DMEM. At the appropriate times, cells were lysed in buffer containing 20 mM HEPES (pH 7.4), 50 mM β-glycerophosphate, 1% Triton X-100, 10% glycerol, 2 mM EGTA, 1 mM dithiothreitol, 10 mM sodium fluoride, 1 mM sodium orthovanadate, 2 μM leupeptin, 2 μM aprotinin, 2 μM pepstatin A, 1 mM phenylmethylsulfonyl fluoride, and 0.5 μM okadaic acid. After the determination of the protein concentration, 25-μg aliquots were size fractionated through SDS–12% PAGE, and the 35S-radiolabeled proteins were visualized with a PhosphorImager.
For the measurement of 35S-precipitable counts, lysates were prepared as described above, and 50-μg protein aliquots were precipitated in 10% trichloroacetic acid (TCA) for 30 min on ice in the presence of carrier bovine serum albumin (200 μg/ml), after which they were filtered. The filters were then rinsed and dried, and the radioactivity was measured by scintillation counting.
Measurement of glucose uptake.
Glucose measurements were taken at various intervals from the cell cultures during the course of the experiments and stored at 4°C until analysis. Glucose levels were analyzed with a Glucose Analyzer II (Beckman, Palo Alto, Calif.).
Flow cytometric analysis of cell cycle distribution and detection of DNA condensation and fragmentation by DAPI staining.
Cell cycle distribution was analyzed by flow cytometry as described earlier (27). Briefly, 1 × 106 to 2 × 106 cells were trypsinized, washed once with PBS, and fixed in 70% ethanol. Fixed cells were washed with PBS, incubated with 1 μg of RNase A per ml for 30 min at 37°C, and stained with propidium iodide (Boehringer Mannheim). The stained cells were analyzed on a FACScan flow cytometer to determine the relative DNA content. Quantitation of apoptotic cells was determined by staining with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma) at the times indicated, as described previously (37). Briefly, cells were washed three times with PBS and fixed with 4% paraformaldehyde. After being stained with DAPI for 30 min, nuclei were examined by fluorescence microscopy and apoptotic cells scored. Data are the means ± the standard deviation of three independent experiments.
RESULTS
Effect of VHL status in the response of UMR cells to stress.
In order to explore whether the VHL status influences the response of RCC cells to stresses, a survey was undertaken with a panel of stressful agents. With each treatment, the sensitivity of the RCC parental cell line UMRC6 (UMR), which lacks VHL gene function, was compared with that of UMR cells in which VHL gene function was restored through stable transfection with a wtVHL expression vector (UMR wtVHL). Stressful treatments included treatment with hydrogen peroxide, arsenite, heat shock, UVC irradiation, differentiation agents (phenylacetate and 4-Cl-phenylacetate), deprivation of nutrients (serum or glucose) or oxygen (hypoxia), and other stresses. Depending on the treatment, the toxicity was quantitated by using either direct cell counts, DAPI staining to score condensed or fragmented nuclei, or crystal violet staining of 96-well cluster plates. As shown in Table 1, the effect of each treatment ranged from undetectable to markedly cytotoxic. In most instances, we observed little difference in the response of cells lacking functional VHL gene expression (UMR parental) relative to that of their wtVHL-expressing counterparts (UMR wtVHL). Glucose deprivation, however, provided a striking exception in that cells lacking pVHL function encountered very marked cytotoxicity, while cells expressing wtVHL appeared to be protected against this treatment. Also noteworthy was the observation that similar periods of serum deprivation appeared to be more toxic for cells expressing wtVHL than for VHL gene-deficient cells, although the disparity of this response was less pronounced.
TABLE 1.
Survey of cytotoxic treatmentsa
| Treatment | Conditions
|
Relative cytotoxicity
|
||
|---|---|---|---|---|
| Dose | Time (h) | UMR parental | UMR wtVHL | |
| H2O2b | 400 μM | 24 | ++ | ++ |
| Arseniteb | 100 μM | 24 | +++++ | +++++ |
| UVCc | 20 J/m2 | 36 | ++ | ++ |
| Mimosineb | 300 μM | 24 | + | + |
| Hypoxiad | <1% O2 | 96 | ++ | ++ |
| Heat shockc | 42°C e | 24 | − | − |
| Glucose deprivationd | –f | 60 | ++++ | + |
| Cycloheximideb | 10 μg/ml | 12 | + | + |
| Phenylacetatec | 9 mM | 72 | − | − |
| 4-Cl-phenylacetatee | 3 mM | 72 | + | + |
| TPAb | 1 nM | 12 | + | + |
| TNF-αb | 10 ng/ml | 24 | − | − |
| Serum withdrawalc | –g | 72 | ++ | +++ |
UMR cells, either lacking VHL function (UMR parental) or stably expressing wild-type VHL (UMR wtVHL) were treated with various cytotoxic agents as indicated. The influence of each treatment was evaluated either by scoring DAPI-positive cells (those exhibiting condensed or fragmented nuclei), by measuring crystal violet staining of 96-well cluster plates, or by direct counts of the attached cells at the end of each treatment period (indicated in the time column), with the exception of heat shock. −, no measurable toxicity; + to ++++, relative toxicities, ranging from slight to high.
Toxicity was scored in 96-well cluster plates after staining with crystal violet.
Toxicity was scored in 6-well cluster plates after counting cells or staining nuclear DNA with DAPI.
Toxicity was scored by direct cell counts with a hemocytometer.
Cells were heat shocked for 6 h and then returned to a 37°C incubator, and toxicity was scored 24 h after the end of the heat period.
Cells were cultured in glucose-free DMEM supplemented with 10% FBS as described in Materials and Methods (about 2 mg/106 cells).
Cells were cultured in DMEM supplemented with 0.1% FBS.
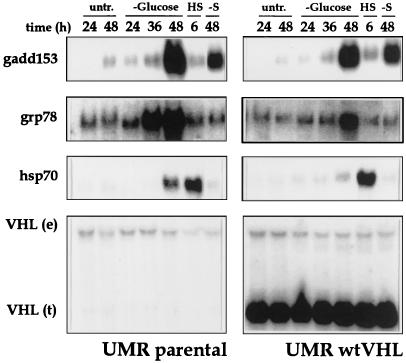
For each treatment, the expression of a number of stress-responsive genes was also monitored. Figure 1 shows representative results for three such genes: the glucose-regulated protein 78 (grp78), the growth arrest- and DNA damage-inducible 153 (gadd153), and the heat shock protein 70 (hsp70). After exposure to heat stress, glucose deprivation, or serum starvation, the expression of all three genes was elevated, although only glucose deprivation resulted in substantially higher expression of these genes in parental cells than in cells expressing wtVHL, particularly with grp78 (Fig. 1). This differential expression may contribute to or be a reflection of the different toxicities encountered by each cell line after glucose deprivation. The VHL status did not seem to substantially influence the expression of grp78, gadd153, or hsp70 when other treatments were tested (Table 2). Thus, both by direct assessment of cellular toxicity and by monitoring the expression of stress-responsive genes, the VHL status influenced the responsiveness to glucose deprivation but not to other stressful treatments tested.
FIG. 1.
Northern blot analysis of expression of stress-responsive genes. UMR cells either lacking VHL function (UMR parental) or stably expressing wild-type VHL (UMR wtVHL) were treated as indicated. Total RNA was processed as described in Materials and Methods, and representative Northern blots indicate the levels of gadd153, grp78, hsp70, and VHL mRNA. VHL (e), endogenous VHL mRNA, about 5-kb long; VHL (t), VHL transcript from the pCEP4VHL vector overexpressed in these cells, about 1-kb long; untr., untreated; −Glucose, cells cultured in glucose-free medium; −S, cells cultured in serum-free medium; HS, cells subjected to heat shock.
TABLE 2.
Relative induction of stress-responsive genes after various stressful treatmentsa
| Treatment | Conditions
|
Fold induction of stress-responsive geneb:
|
||||||
|---|---|---|---|---|---|---|---|---|
| Dose | Time (h) | gadd153
|
grp78
|
hsp70
|
||||
| P | VHL | P | VHL | P | VHL | |||
| H2O2 | 400 μM | 12 | 4 | 4 | 0.3 | 0.3 | 1 | 1 |
| Arsenite | 100 μM | 12 | 25 | 27 | 1 | 1 | 35 | 45 |
| UVC | 20 J/m2 | 12 | 2 | 2 | 0.2 | 0.3 | 0.2 | 0.1 |
| Mimosine | 300 μM | 12 | 3 | 4 | 1 | 1 | 1 | 1 |
| Heat shockc | 42°C | 6 | 2 | 2 | 1 | 1 | 11 | 12 |
| Glucose deprivation | —d | 24 | 18 | 12 | 17 | 7 | 3 | 2 |
| Cycloheximide | 10 μg/ml | 12 | 3 | 3 | 5 | 6 | 0.3 | 0.3 |
| Phenylacetate | 9 mM | 24 | 1 | 2 | 1 | 2 | 1 | 1 |
| 4-Cl-phenylacetate | 3 mM | 24 | 9 | 11 | 1 | 2 | 0.8 | 0.8 |
| TPA | 1 nM | 6 | 1 | 1 | 1 | 1 | 0.8 | 1 |
| TNF-α | 10 ng/ml | 12 | 1 | 1 | 1 | 1 | 1 | 3 |
| Serum withdrawale | −e | 24 | 7 | 9 | 1 | 1 | 1 | 1 |
UMR cells with different VHL status (parental or wtVHL) were treated with the agents indicated in Table 1, and RNA samples were prepared at the end of each treatment period (indicated in the time column), with the exception of heat shock. The expression of the stress-responsive genes grp78, gadd153, and hsp70 was studied by Northern blot analysis as described in Materials and Methods. For each treatment and gene, numbers indicate the fold induction in the abundance of that mRNA with respect to that seen in untreated controls.
Fold induction in mRNA levels relative to expression in untreated cells. P, parental cells; VHL, wtVHL-expressing cells.
Cells were heat shocked for 6 h, whereupon RNA was extracted.
Cells were cultured in glucose-free DMEM supplemented with 10% FBS, as described in Materials and Methods (about 2 mg/106 cells).
Cells were cultured in DMEM supplemented with 0.1% FBS.
wtVHL protects against stress by glucose deprivation.
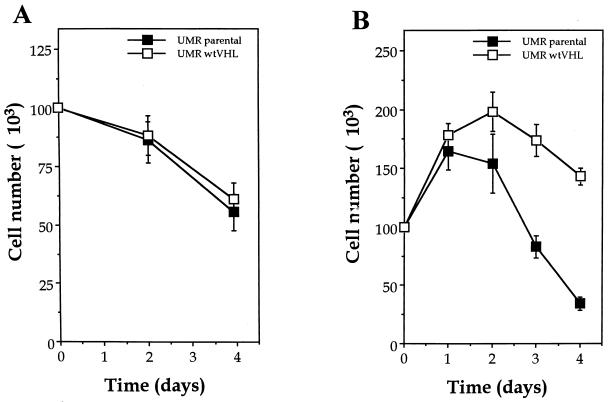
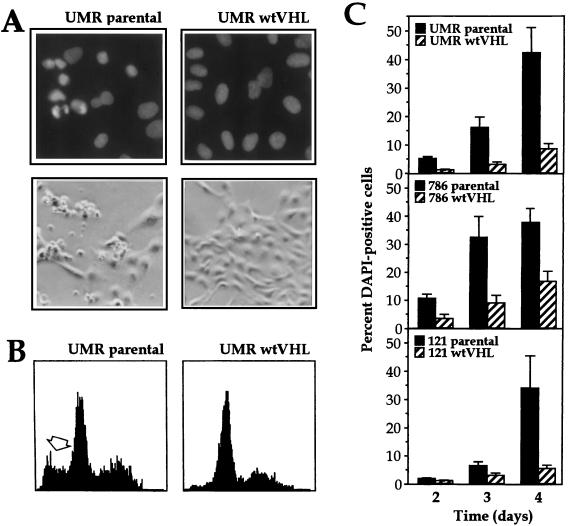
Although we did not observe VHL status-dependent differences in the sensitivity to hypoxia in our initial screen, much overlap exists in the cellular response to hypoxic and hypoglycemic stresses. Therefore, we sought to analyze more carefully the effect of functional pVHL on the sensitivity of UMR cells to glucose or oxygen deprivation. As shown in Fig. 2A, the culturing of UMR cells in hypoxic conditions was moderately toxic, but we observed no differences between parental and wtVHL-expressing cells. In contrast, the culturing of cells in glucose-free medium led to a significant loss of cells lacking functional pVHL, while it only had a modest effect on cell loss in wtVHL expressing cultures (Fig. 2B). The growth rates of untreated UMR cells were unaffected by the VHL status (not shown). A further characterization of the differences in glucose deprivation toxicity is presented in Fig. 3. Staining with the DNA dye DAPI after placement in glucose-free medium for 3 days was used to examine nuclear condensation and fragmentation. Representative results for UMR parental and UMR wtVHL are shown in Fig. 3A. The number of cells exhibiting these alterations (primarily nuclear condensation) was remarkably higher in the UMR parental population (Fig. 3A). Examination of the DNA content by fluorescence-activated cell sorter (FACS) analysis revealed the presence of a sub-G1 population of cells (Fig. 3B). Together, the results obtained by staining with DAPI and by FACS analysis suggest that glucose-depleted UMR cells undergo apoptotic cell death.
FIG. 2.
Effect of hypoxia and glucose deprivation on alterations in cell number. UMR cells were plated in 60-mm dishes and grown to a density of 100,000 cells per plate in complete medium. (A) Cells were placed in a hypoxia chamber, and cell numbers were determined at the times indicated. (B) Cell medium was replaced with 2 ml of glucose-free DMEM supplemented with 10% FBS. This contributed 100 mg of glucose (200 μg of glucose/plate, about 2 ng/cell) per ml. The number of cells per plate at each time point was determined in duplicate with a hemacytometer. Values represent the mean ± the standard error of the mean (SEM) for three independent experiments. Symbols: ■, UMR parental cells; □, UMR wtVHL cells. Untreated control plates were seeded at the same density and cultured in 2 ml of complete medium, and cell numbers indicated normal, logarithmic growth for at least 4 days (data not shown).
FIG. 3.
Effect of glucose deprivation on three pairs of RCC lines, each with a VHL-proficient and -deficient counterpart. (A) UMR cells (either parental or expressing wtVHL) were cultured in glucose-free medium for 3 days and then stained with the DNA dye DAPI. Nuclei were visualized under fluorescence microscopy (top row). Untreated control cells also displayed homogeneous DAPI staining (data not shown). Note the presence of condensed and fragmented nuclei in the parental cell population, while most nuclei of wtVHL-expressing cells are homogeneously stained. Morphological differences were also visible under light microscopy, with parental cells exhibiting distinct membrane blebbing under phase-contrast microscopy (bottom row). (B) FACS distribution of UMR cells, each with a different VHL status. Three days after culture in glucose-free medium, UMR parental and UMR wtVHL cells were subjected to FACS analysis, and the resulting histograms are shown. A sub-G1 population, characteristic of apoptosis, is indicated with an open arrow. (C) The RCC lines UMR, 786, and 121 were subjected to glucose deprivation for the times indicated and then stained with DAPI, and the condensed and/or fragmented nuclei were scored. Solid bars, parental cells; hatched bars, wtVHL-expressing cells.
Further evidence supporting the notion that the functional VHL gene is important for survival in glucose-deprived medium is shown in Fig. 4. Here, the cytotoxic influence of glucose deprivation in UMR cells was extended to include that of cells expressing, through stable transfection, a VHL cDNA with a deletion in nucleotide 737, rendering a C-terminally truncated pVHL that lacked the elongin-binding domain. The survival of 2 such clonal isolates (XX23 and XX27) was comparable to that of parental cells and substantially lower than that of wtVHL-expressing cells, thus illustrating the importance of this domain for survival in glucose-deprived medium.
FIG. 4.
Effect of glucose deprivation on UMR cells expressing C-terminally truncated pVHL. UMR cells that lacked VHL (parental), expressed wtVHL (wtVHL), or expressed a mutant VHL cDNA carrying a deletion at nucleotide position 737 that rendered a C-terminally truncated protein (clonal lines XX23 and XX27) were cultured in glucose-free medium for 4 days. At the end of the treatment period, the cultures were subjected to clonogenicity assay as described in Materials and Methods. Percentages were calculated relative to untreated, time-matched controls. Values represent the mean ± the SEM for at least three independent experiments.
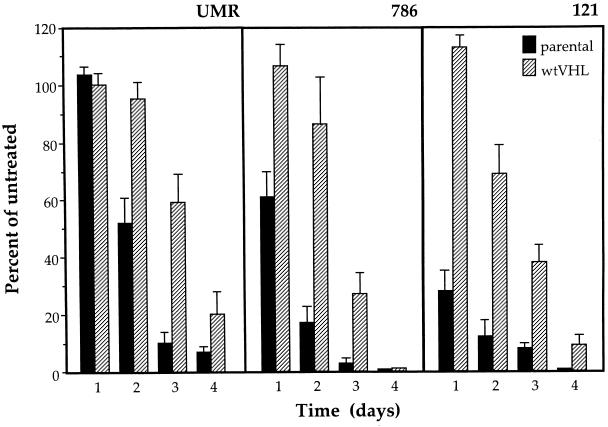
To ensure that the increased susceptibility of UMR parental cells to glucose deprivation was not due to characteristics unique to the UMR cell line, we expanded the study to include two additional RCC lines derived from other tumors deficient in pVHL function, cell lines 786 and 121 (13, 21). Cell lines 786 and 121 stably transfected with either control (parental) or wtVHL-expressing vectors were examined. As shown in Fig. 3C, the culturing of variants of all three cell lines in glucose-free medium led to time-dependent increases in the numbers of condensed and fragmented nuclei (scored as DAPI-positive cells), which occurred earlier and were more prominent in parental cells (all deficient in VHL function) compared with their wtVHL-expressing counterparts. A clonogenic assay was employed to further determine if the influence of VHL status on the sensitivity towards glucose deprivation correlated with measurable differences in a long-term survival assay. The results shown in Fig. 5 are expressed as the percentage of colonies obtained after each cell type was cultured in glucose-free medium for the times indicated relative to the number of colonies obtained from time-matched untreated controls. In all three lines tested, parental cells showed significantly lower survival rates than their wtVHL-expressing counterparts. Taken together, these findings indicate that the presence of functional VHL genes enhances survival of RCC cells subjected to glucose deprivation.
FIG. 5.
Effect of glucose deprivation on long-term survival of RCC cells each with a different VHL status. UMR, 786, and 121 cells (both parental and expressing wtVHL) were cultured in glucose-deprived medium for the times indicated, and then their long-term survival was assayed by using a clonogenic assay as described in Materials and Methods. Percentages were calculated relative to untreated, time-matched controls. In each experiment, treatments were done in duplicate, and values represent the mean ± the SEM for at least three independent experiments.
Effect of bcl-2 overexpression on cell death by glucose deprivation.
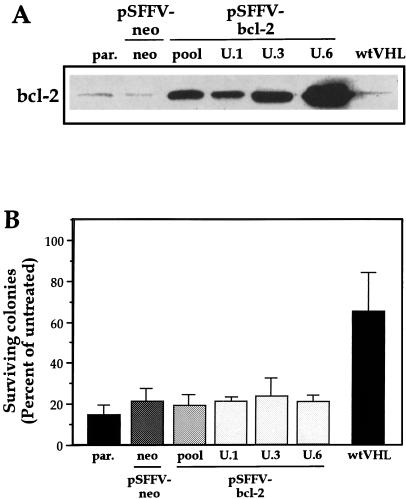
Since glucose deprivation-triggered death of RCC cells exhibited characteristics of apoptosis, we sought to determine whether this effect could be prevented by the expression of the antiapoptotic protein bcl-2 (43). To this end, UMR cells were stably transfected with a plasmid overexpressing bcl-2, which has been found to protect against apoptotic cell death triggered by a number of stresses (30, 47). Multiple clonal isolates were obtained, and the levels of bcl-2 protein expression in three of them, ranging from moderate (U.1) to high (U.6), are shown in Fig. 6A. Unexpectedly, the survival of UMR cells transfected with a vector control (neo) was virtually indistinguishable from that of any one of the bcl-2-expressing clones or the bcl-2 pool population (Fig. 6B), as assessed by colony formation assay (and by DAPI staining [data not shown]). Evidence that bcl-2 was functionally active in these transformants came from clonogenic assays after thapsigargin treatment, where overexpression of bcl-2 was found to be protective (not shown). These results indicate that bcl-2 cannot rescue UMR cells from the toxic influence of glucose deprivation.
FIG. 6.
Influence of bcl-2 overexpression in glucose deprivation toxicity. (A) Western blot analysis of bcl-2 expression in transfected UMR cells. UMR cells were transfected with the bcl-2 expression vector pSFFV-bcl-2 to obtain a pool population (pool) or isolated clones (U.1, U.3, and U.6) of bcl-2-expressing cells. neo, cells transfected with the “empty” vector control pSFFV-neo. Different levels of bcl-2 were expressed in each case. par, parental. (B) The sensitivity of each clone to the toxic influence of a 3-day glucose deprivation period was measured in a colony formation assay. In each experiment, treatments were done in duplicate, and the values represent the mean ± the SEM for at least three independent experiments.
Assessment of glucose uptake and energy availability.
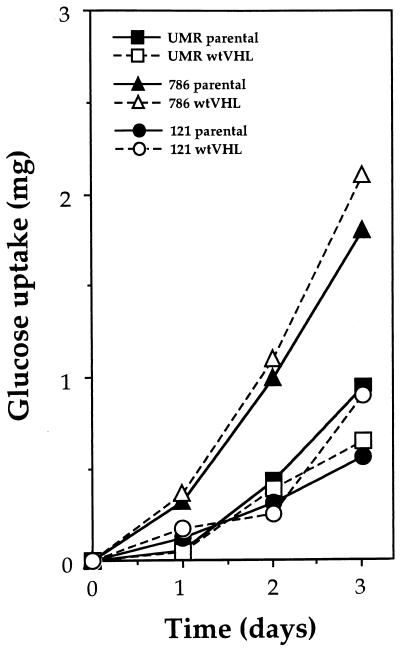
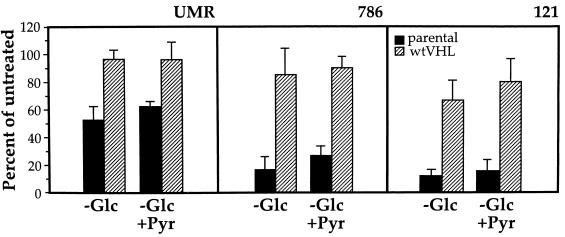
A number of hypotheses could be postulated to explain why VHL-deficient cells exhibit enhanced toxicity towards glucose deprivation. For example, glucose uptake may be different in VHL-deficient cells relative to wtVHL-expressing cells; alternatively, reduced energy availability could be more critically limiting for the VHL-deficient cells. As shown in Fig. 7, glucose uptake was virtually the same when parental (VHL-deficient) and wtVHL-expressing cells of each type were compared, although 786 cells had an overall higher rate of glucose uptake. Therefore, differences in the entry of glucose into the cell do not seem to account for the enhanced sensitivity of parental cells to glucose deprivation. To ascertain whether the sensitivity to glucose starvation was due to ATP depletion, pyruvate was added as an alternate energy source. Our earlier studies indicated that addition of 10 mM pyruvate was able to maintain cellular ATP at levels similar to control levels with glucose (4), so pyruvate was added to the glucose-depleted medium and the relative sensitivity of each cell line to each treatment condition was measured. As shown in Fig. 8, the addition of pyruvate moderately enhanced colony survival in both parental and wtVHL-expressing cells. However, it did not restore the survival of VHL-deficient cells to the level seen in their wtVHL-expressing counterparts. Thus, lower energy availability does appear to contribute somewhat to the toxic effects of glucose deprivation, but not in a VHL-specific fashion.
FIG. 7.
Glucose uptake in RCC cells. UMR, 786, and 121 cells were cultured in complete medium, and the rate of glucose uptake was determined by analyzing aliquots of medium collected at the times indicated. The glucose concentrations were determined as described in Materials and Methods.
FIG. 8.
Effect of pyruvate on the toxicity by glucose deprivation. UMR, 786, and 121 cells, plated at a density of 30,000 cells/well in 6-well cluster plates, were cultured for 48 h in glucose-free medium alone (solid bars) or supplemented with 10 mM sodium pyruvate (hatched bars). At the end of the treatment period, the cultures were subjected to clonogenicity assay as described in Materials and Methods. Percentages were calculated relative to untreated, time-matched controls. Pyruvate alone had no influence on clonogenicity relative to untreated cells. In each experiment, treatments were done in duplicate, and the values represent the mean ± the SEM for at least three independent experiments.
Impaired protein processing.
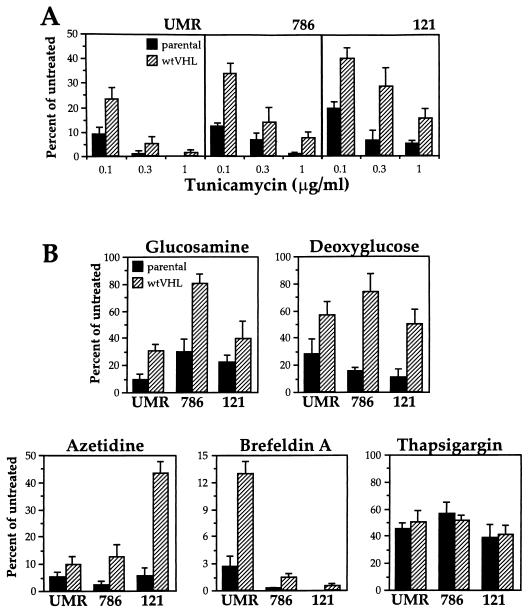
Another important consequence of glucose deprivation is impaired protein glycosylation. It is possible that such an impairment could be more damaging for cells lacking VHL function, thus explaining their greater sensitivity to glucose deprivation. To further explore this possibility, we examined whether the VHL status influenced the cellular response to treatment with other agents that affect glycosylation. First, we examined the effect of tunicamycin. As shown in Fig. 9A, tunicamycin treatment led to a dose-dependent loss in clonogenicity in all of the cell lines tested, but this treatment was much more cytotoxic for VHL-deficient cells in every case. Likewise, treatments with either glucosamine or 2-deoxyglucose, two additional agents that perturb glucose metabolism and thereby inhibit glycosylation (32, 58), were also much more cytotoxic for RCC cells lacking the VHL gene (Fig. 9B).
FIG. 9.
Effect of inhibitors of posttranslational protein processing on RCC cells. Parental (solid bars) and wtVHL-expressing (hatched bars) UMR, 786, and 121 cells, were plated at a density of 30,000 cells/well in 6-well cluster plates and cultured for 48 h in complete medium, either alone or supplemented with tunicamycin (A) or a variety of drug treatments affecting protein processing: glucosamine (10 mM), 2-deoxyglucose (10 mM), azetidine (3 mM), brefeldin A (0.3 μg/ml) and thapsigargin (3 μM) (B), with the exception of UMR cells, which were treated with 2-deoxyglucose (10 mM) for 72 h. At the end of each treatment period, cultures were subjected to clonogenic assay as described in Materials and Methods. Percentages were calculated relative to untreated, time-matched controls. Dimethyl sulfoxide alone had no influence on clonogenicity relative to untreated cells. Treatments were done in duplicate, and the values represent the mean ± the SEM for at least three independent experiments.
Among the principal consequences of impaired glycosylation is the accumulation of misfolded proteins in the cell. Therefore, we next sought to test if VHL-deficient cells had a diminished ability to tolerate, in a general sense, the presence of unprocessed proteins. To this end, we assayed the cytotoxic influence of various treatments that generate misprocessed proteins on each pair of VHL-proficient and VHL-deficient RCC lines. Brefeldin A causes the accumulation of incompletely processed proteins by preventing the shuttling of vesicles from the endoplasmic reticulum (ER) to the Golgi apparatus. This effect is mediated, at least in part, through inhibition of a guanine nucleotide-exchange protein for ADP-ribosylation factors required for this transport (19, 41). Exposure to the proline analog azetidine also results in the formation of aberrant proteins (57). Both treatments were found to be more cytotoxic for VHL-proficient than for VHL-deficient cells (Fig. 9B). Although their influence is not restricted to the ER, tunicamycin, brefeldin A, and azetidine are all considered to be ER stressors. Therefore, we examined whether cells with a different VHL status would also exhibit differential sensitivity to other ER stress agents whose mechanism of action does not work through interference with normal protein processing. Thapsigargin, which induces ER stress through perturbations in Ca2+ homeostasis (3, 56), was chosen for this purpose. As shown in Fig. 9B, thapsigargin produced similar toxicity in VHL-proficient and VHL-deficient cells. Therefore, our results suggest that the VHL gene may exert a protective influence against impaired protein processing but not against other forms of ER stress.
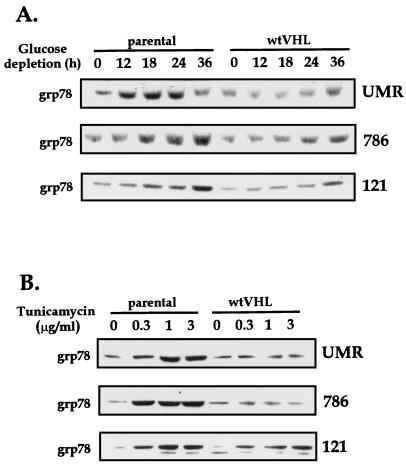
These differences in toxicity were further characterized by analyzing the expression of grp78, a marker of glucose deprivation and ER stress. As described earlier (Table 1 and Fig. 1), glucose deprivation led to a time-dependent induction of grp78 expression that was markedly attenuated in wtVHL-expressing cells (Fig. 10A); likewise, exposure to tunicamycin also led to dose-dependent elevations in grp78 expression that were more accentuated in parental cells than in wtVHL-expressing cells (Fig. 10B). Again, we believe these differences reflect the relative sensitivities of each cell line. However, other possible explanations, such as direct inhibition of the steady-state levels of grp78 mRNA by the VHL gene, cannot be ruled out at this time.
FIG. 10.
Expression of grp78 in RCC cells. Representative Western blot analysis of grp78 expression in VHL-deficient (parental) and wtVHL-expressing (wtVHL) UMR, 786, and 121 cells that were cultured in glucose-depleted medium for the times indicated (A) or in complete medium with various concentrations of tunicamycin for 12 h (B). Protein loading was the same in all lanes (data not shown).
Effect of VHL status on global ubiquitination and elimination of cellular proteins.
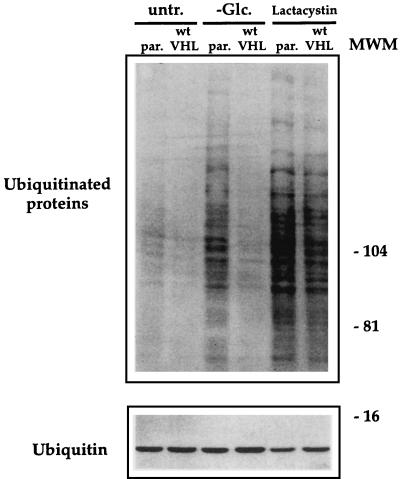
A role for pVHL in the ubiquitin-proteasome degradation pathway was recently postulated based on its association with Cul2 and elongin C (36). Cul2 and elongin C appear to be human counterparts of the yeast proteins cdc53 and Skp1, respectively, which are involved in targeting cellular proteins for degradation by the proteasome (45). In light of this proposed model for VHL function, as well as our own results showing that deletion of the elongin-binding domain is sufficient to confer sensitivity to glucose deprivation (Fig. 4), we hypothesized that we might detect differences in the extent of ubiquitination of RCC cells, depending on their VHL status. If proteins in pVHL participated in early steps, such as the recognition of substrate proteins, we would likely see less ubiquitination in VHL-deficient cells; on the other hand, if pVHL participated in subsequent steps of proteolysis, then the absence of VHL function could lead to the accumulation of more ubiquitinated proteins, as later events are impaired. The presence of ubiquitinated proteins in lysates from cell line 786 cells was determined by Western blot analysis (Fig. 11, upper panel). As shown, VHL-deficient (parental) 786 cells exhibited overall higher ubiquitination levels than did the wtVHL-expressing counterparts; these differences were further accentuated in lysates from glucose-depleted cultures (Fig. 11, upper panel). 786 cells treated with lactacystin were included in the study as positive controls. Lactacystin inhibits proteasome function and thus inhibits the de-ubiquitination of target proteins. As shown in Fig. 11, both parental and wtVHL-expressing cells had very high levels of ubiquitinated proteins after exposure to lactacystin, although it seemed to be slightly higher for parental cells. Western blot analysis of free ubiquitin (Fig. 11, lower panel) was carried out after electrophoresis through 15% SDS–polyacrylamide gels due to its small size (8.5 kDa). As shown, its expression did not change substantially with the treatments described.
FIG. 11.
Western blot analysis of ubiquitinated proteins and ubiquitin in RCC cells. (Upper panel) Parental (par.) and wtVHL-expressing 786 cells were either left untreated, subjected to glucose-deprivation for 48 h, or treated with lactacystin (10 μM) for 16 h. Whole-cell lysates were then electrophoresed in SDS–7% polyacrylamide gels and subjected to Western blot analysis to detect the presence of ubiquitin by using a monoclonal anti-ubiquitin antibody. (Lower panel) Electrophoresis in SDS–15% polyacrylamide gels was used to visualize free ubiquitin (8.5 kDa). Protein loading was the same in all lanes (not shown). MWM, molecular weight marker, indicating the size in kilodaltons.
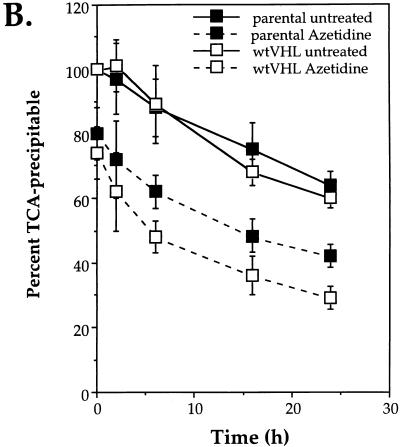
To further characterize the influence of VHL status on the fate of aberrant proteins, we studied the rate of elimination of proteins synthesized in the presence of azetidine in 786 cells either lacking or expressing wtVHL. After treatment of cells with azetidine for 6 h in the presence of [35S]methionine, both drug and [35S]methionine were removed, and the clearance of labeled proteins was monitored over the following 36 h. As shown in Fig. 12A, 35S-labeled proteins synthesized in the presence of this proline analog were eliminated more slowly in parental cells than in wtVHL-expressing cells, supporting the notion that pVHL aids in a more efficient elimination of abnormal proteins. Likewise, quantitation of TCA-precipitable counts indicated that the rate of elimination of 35S-labeled proteins was faster in the presence of wtVHL (Fig. 12B). Finally, when protein ubiquitination levels after azetidine treatment were compared, VHL-deficient cells exhibited a greater accumulation of ubiquitinated proteins over time than did cells expressing pVHL (Fig. 12C). Taken together, our results on protein ubiquitination and the elimination of abnormally processed proteins are consistent with pVHL playing a role in the process of targeted proteolysis.
FIG. 12.
Elimination of 35S-labeled proteins synthesized in the presence of amino acid analogs. (A) 786 cells either were left untreated or were pretreated with 10 mM azetidine for 1 h and incubated with [35S]methionine for an additional 5 h (as described in Materials and Methods). Then the labeling medium was removed and replaced with regular culture medium without drugs. At different times thereafter (0, 6, 18, 24, and 36 h), protein extracts from each treatment group were prepared and electrophoresed through SDS–12% polyacrylamide gels. Gels were dried, and radiolabeled proteins were visualized with a PhosphorImager. parental, VHL-deficient cells; wtVHL, cells expressing wtVHL. (B) 786 cells either were left untreated or were pretreated with 10 mM azetidine for 1 h and then incubated with [35S]methionine for an additional 5 h. At the times indicated after removal of the medium containing [35S]methionine (with or without azetidine), protein lysates were prepared, and TCA-precipitable counts were determined as described in Materials and Methods. (C) 786 cells either were left untreated or were treated with 10 mM azetidine for 6 h and then placed in medium without drugs. At different times thereafter (0, 6, 18, 24, and 36 h), protein extracts were prepared and subjected to Western blot analysis for the detection of ubiquitinated proteins as described in Materials and Methods.
DISCUSSION
Our results reported here indicate that pVHL exerts a protective influence during treatments that result in the accumulation of misprocessed proteins in RCC cells. The treatments investigated included the inhibition of protein glycosylation, the impairment of protein transport, and the perturbation of protein folding, all resulting in the elevated presence of abnormal proteins. With each treatment, RCC cells lacking VHL function exhibited marked cytotoxicity that could be relieved by restoration of VHL function. These observations are consistent with an emerging model proposed for VHL function. This model, largely based on the identification and analysis of pVHL-interacting proteins, postulates that pVHL plays a role in the targeted degradation of cellular proteins. In support of this model, pVHL has been found in complexes containing Cul2 and elongins C and B; the yeast homologs of these proteins, cdc53, cdc34, and Skp1, respectively, have been proposed to function in targeting proteins for degradation by the ubiquitin-proteasome pathway (8, 24, 35, 45). This mechanism of proteolytic breakdown involves a ubiquitin-activating enzyme (E1); a ubiquitin-conjugating enzyme (E2), which transfers ubiquitin to the protein substrate; and a ubiquitin protein ligase (E3), normally bound to the protein substrate for degradation (for reviews, see references 2, 46, and 48). Cdc53 and Cul2 appear to form part of a multiprotein complex with E3 enzymatic activity, while elongin B has a ubiquitin-like domain (11, 45). The targets of Cul2 remain to be identified, but certain RNA-binding proteins, such as those recognizing VEGF mRNA, are likely candidates. As shown by Levy et al. (33, 34), these proteins bind and stabilize the VEGF mRNA in conditions of low oxygen, thus enhancing the expression of VEGF and other hypoxia-inducible mRNAs. In VHL-deficient cells, the constitutively elevated presence of VEGF mRNA-binding proteins has been proposed to account for the heightened expression of VEGF that many groups have reported (both in cell lines and in VHL tumors) (49, 54). In the same general model, pVHL may modulate the process of transcriptional elongation through targeted proteolysis of nuclear proteins involved in RNA polymerase II elongation (7, 11, 26).
In view of our findings reported here, we propose that pVHL may play a general role in targeting cellular proteins for proteolytic elimination. As shown, VHL-deficient cells exhibit greater sensitivity to glucose deprivation, tunicamycin, brefeldin A, and azetidine than cells expressing wtVHL. We interpret this response to reflect the accumulation of a greater burden of misfolded, abnormally processed or incompletely modified proteins in VHL-deficient cells compared to VHL-proficient cells. In the simplest model, misprocessed proteins arising from these treatments would be transported back into the cytoplasm for degradation (52). In cells lacking pVHL, these proteins would accumulate and overload the cell, a situation that may become incompatible with the maintenance of cellular function and integrity. The presence of functional pVHL would aid in the elimination of proteins, possibly by facilitating their proteolytic degradation through the ubiquitin-proteasome pathway, thus preserving cellular viability. Indeed, our analysis of the ubiquitination of total cellular proteins suggests that pVHL may assist in the general elimination of cellular proteins, since VHL-deficient cells, particularly after glucose deprivation or treatment with azetidine, exhibit an elevated amount of ubiquitinated proteins. Likewise, the finding that wtVHL-expressing cells exhibit a relatively more efficient clearance of 35S-labeled proteins synthesized in the presence of azetidine is also consistent with the VHL gene playing a global role in the elimination of abnormal proteins. An alternative model can be envisioned whereby, in situations of stress, pVHL would selectively aid in the elimination of a specific subset of proteins such as, for example, certain death-promoting proteins. Yet the possibility remains that the observed differences in cytotoxicity and protein ubiquitination profiles are due to other, presently unknown, characteristics of VHL-deficient cells. However, we think that these differences are, at least in part, due to properties related to binding of pVHL to the elongin-Cul2 complex, since the expression of VHL proteins specifically lacking the elongin binding site fails to render any protection against glucose deprivation to RCC cells otherwise devoid of VHL function, as shown in Fig. 4. Discerning between these possibilities and furthering the identification of pVHL target proteins remain important areas of future investigation.
Two additional conclusions may be inferred from our analysis of general protein ubiquitination. First, although ubiquitination may also lead to protein degradation by lysosomes (46), our results suggest that the proteasome is involved, at least in part, in the process of protein elimination that is purportedly assisted by pVHL. This conclusion is based on the fact that the pattern of ubiquitinated proteins in lactacystin-treated cells resembles that seen in glucose-depleted cells and that overall the amount of ubiquitinated proteins is higher in VHL-deficient cells. Secondly, if indeed pVHL is involved in the ubiquitin-proteasome degradatory machinery, it may not be directly implicated in the recognition of substrate proteins by the elongin B/C/Cul2/pVHL complex but rather function in subsequent steps leading to its elimination. If pVHL were involved in substrate recognition, pVHL-deficient cells might have presented with less ubiquitination, particularly after glucose deprivation. On the other hand, if pVHL were involved in the ensuing phases of proteolysis, we would see an accumulation of ubiquitinated proteins in parental cells, while pVHL-expressing cells would eliminate ubiquitinated substrates more efficiently and the overall ubiquitination would be lower. Our observations presented in Fig. 11 are consistent with the latter possibility. Analysis of additional VHL mutations may provide further insight into the functional domains required for protection against glucose deprivation and the domains influencing protein ubiquitination. In this regard, our unpublished results with a variety of other VHL mutants indicate that in no case has a mutant protein engendered the same degree of protection as has wild-type VHL protein.
How does this differential responsiveness influence tumor development? The concept that pVHL, a tumor suppressor gene product, exerts protection against cell death, is somewhat counterintuitive. With a few reported exceptions (9), oncogenes are generally believed to enhance cell survival and net tumor growth, while tumor suppressor genes frequently promote cell death, thereby preventing tumor development. In VHL disease, individuals develop, in addition to RCC, a number of highly vascularized, nonmalignant tumors (retinal capillary angiomas, pheochromocytomas, and hemangioblastomas of the central nervous system). It is likely that the reportedly elevated VEGF expression in VHL-deficient cells and tumors directly contributes to their extensive vascularization. However, we propose that the absence of functional pVHL may further contribute to tumor vascularization by an additional mechanism. Based on our observations reported here, VHL-deficient cells might be rendered more vulnerable to hypoglycemia, which normally occurs during the growth of solid tumors; these focal sites of cellular death would, in turn, trigger a local inflammatory reaction, further stimulating the recruitment of angiogenic factors and promoting local vascularization. Together with the constitutively higher expression of VEGF, the resulting local angiogenesis would be further enhanced. Indeed, solid tumors have abnormal vascularization, which creates areas of poor irrigation and hence local hypoxia and hypoglycemia. Hypoxia would lead to further increases in VEGF expression, while hypoglycemia, which may be toxic for VHL-deficient cells, could lead to local cellular death, local inflammation, and increased attraction of angiogenic factors. Consequently, the combined effect of hypoxia and hypoglycemia would potently stimulate local angiogenesis. In addition, glucose depletion, as reported by other groups (50, 53), was also a potent inducer of VEGF expression in all of the RCC lines studied here (data not shown). Moreover, hypoxia accentuates the lower availability of glucose, as tissues switch from aerobic to anaerobic metabolism (a process that generates less ATP) and require higher glucose uptake. Therefore, our model proposes that in solid tumors exposure to combined hypoxic and hypoglycemic stresses leads to increased local vascularization. It is presently unclear whether this model applies only to the progression of renal cell carcinomas, which are malignant and metastatic, or also to other VHL tumors, such as retinal angiomas or angioblastomas, which are more vascular and nonmetastatic. While many important questions about VHL tumorigenesis await further investigation, our findings presented here may have important implications in the design of therapeutic strategies for the management of VHL tumors.
ACKNOWLEDGMENTS
We are grateful to J. Gnarra and W. M. Linehan for providing the UOK 121 cells, O. Iliopoulos and W. G. Kaelin for providing the 786-0 cells, and G. Núñez for the pSFFV-neo and pSFFV-bcl2 constructs. We also thank M. S. Prenger and W. Wang for their assistance with experimental procedures, D. L. Longo and K. McCullough for their critical reading of the manuscript, and S. Shack and A. Passaniti for helpful discussions.
REFERENCES
- 1.Aso T, Haque D, Barstead R J, Conaway R C, Conaway J W. The inducible elongin A elongation activation domain: structure, function and interaction with elongin BC complex. EMBO J. 1996;15:101–110. [PMC free article] [PubMed] [Google Scholar]
- 2.Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 3.Brayden D J, Hanley M R, Thastrup O, Cuthbert A W. Thapsigargin, a new calcium-dependent epithelial anion secretagogue. Br J Pharmacol. 1989;98:809–816. doi: 10.1111/j.1476-5381.1989.tb14609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson S G, Fawcett T W, Bartlett J D, Bernier M, Holbrook N J. Regulation of the C/EBP-related gene gadd153 by glucose deprivation. Mol Cell Biol. 1993;13:4736–4744. doi: 10.1128/mcb.13.8.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corless C L, Kibel A S, Iliopoulos O, Kaelin W G., Jr Immunostaining of the von Hippel-Lindau gene product in normal and neoplastic human tissues. Hum Pathol. 1997;28:459–464. doi: 10.1016/s0046-8177(97)90035-6. [DOI] [PubMed] [Google Scholar]
- 7.Duan D R, Pause A, Burgess W, Aso T, Chen D Y T, Garrett K P, Conaway R C, Conaway J W, Linehan W M, Klausner R D. Inhibition of transcriptional elongation by the VHL tumor suppressor protein. Science. 1995;269:1402–1406. doi: 10.1126/science.7660122. [DOI] [PubMed] [Google Scholar]
- 8.Feldman R M, Correll C C, Kaplan K B, Deshaies R J. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 9.Field S J, Tsai F Y, Kuo F, Zubiaga A M, Kaelin W G, Jr, Livingston D M, Orkin S H, Greenberg M E. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 10.Fornace A J, Jr, Alamo I J, Hollander M C, Lamoreaux E. Induction of heat shock protein transcripts and B2 transcripts by various stresses in Chinese hamster cells. Exp Cell Res. 1989;182:61–74. doi: 10.1016/0014-4827(89)90279-6. [DOI] [PubMed] [Google Scholar]
- 11.Garrett K P, Aso T, Bradsher J N, Foundling S I, Lane W S, Conaway R C, Conaway J W. Positive regulation of general transcription factor SIII by a tailed ubiquitin homolog. Proc Natl Acad Sci USA. 1995;92:7172–7176. doi: 10.1073/pnas.92.16.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gnarra J R, Duan D R, Weng Y, Humphrey J S, Chen D Y T, Lee S, Pause A, Dudley F, Latif F, Kuzmin I, Schmidt L, Duh F-M, Stackhouse T, Chen F, Kishida T, Wei M H, Lerman M I, Zbar B, Klausner R D, Linehan W M. Molecular cloning of the von Hippel-Lindau tumor suppressor gene and its role in renal cell carcinoma. Biochim Biophys Acta. 1996;1242:201–210. doi: 10.1016/0304-419x(95)00012-5. [DOI] [PubMed] [Google Scholar]
- 13.Gnarra J R, Zhou S, Merrill M J, Wagner J R, Krumm A, Papavassiliou E, Oldfield E H, Klausner R D, Linehan W M. Post-transcriptional regulation of vascular endothelial growth factor mRNA by the product of the VHL tumor suppressor gene. Proc Natl Acad Sci USA. 1996;93:10589–10594. doi: 10.1073/pnas.93.20.10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gnarra J R, Ward J M, Porter F D, Wagner J R, Devor D E, Grinberg A, Emmert-Buck M R, Westphal H, Klausner R D, Linehan W M. Defective placental vasculogenesis causes embryonic lethality in VHL-deficient mice. Proc Natl Acad Sci USA. 1997;94:9102–9107. doi: 10.1073/pnas.94.17.9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldberg M A, Dunning S P, Bunn H F. Regulation of the erythropoietin gene: evidence that the oxygen sensor is a heme protein. Science. 1988;242:1412–1415. doi: 10.1126/science.2849206. [DOI] [PubMed] [Google Scholar]
- 16.Gorospe M, Liu Y, Xu Q, Chrest F J, Holbrook N J. Inhibition of G1 cyclin-dependent kinase activity during prostaglandin A2-mediated growth arrest. Mol Cell Biol. 1996;16:762–70. doi: 10.1128/mcb.16.3.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorospe M, Holbrook N J. Role of p21 in prostaglandin A2-mediated cellular arrest and death. Cancer Res. 1996;56:475–479. [PubMed] [Google Scholar]
- 18.Grossman H B, Wedemeyer G, Ren L Q. Human renal carcinoma: characterization of five new cell lines. J Surg Oncol. 1985;28:237–244. doi: 10.1002/jso.2930280320. [DOI] [PubMed] [Google Scholar]
- 19.Guillemain I, Exton J H. Effects of brefeldin A on phosphatidylcholine phospholipase D and inositolphospholipid metabolism in HL-60 cells. Eur J Biochem. 1997;249:812–819. doi: 10.1111/j.1432-1033.1997.00812.x. [DOI] [PubMed] [Google Scholar]
- 20.Humphrey J S, Klausner R D, Linehan W M. Von Hippel-Lindau syndrome: hereditary cancer arising from inherited mutations of the VHL tumor suppressor gene. Cancer Treat Res. 1996;88:13–39. doi: 10.1007/978-1-4615-6343-3_2. [DOI] [PubMed] [Google Scholar]
- 21.Iliopoulos O, Kibel A, Gray S, Kaelin W G., Jr Tumor suppression by the human von Hippel-Lindau gene product. Nat Med. 1995;1:822–826. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- 22.Iliopoulos O, Levy A P, Jiang C, Kaelin W G, Jr, Goldberg M A. Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc Natl Acad Sci USA. 1996;93:10595–10599. doi: 10.1073/pnas.93.20.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iliopoulos O, Kaelin W G., Jr The molecular basis of von Hippel-Lindau disease. Mol Med. 1997;3:289–293. [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson P K. Cell cycle: cull and destroy. Curr Biol. 1996;6:1209–1212. doi: 10.1016/s0960-9822(96)00697-5. [DOI] [PubMed] [Google Scholar]
- 25.Jiang B H, Agani F, Passaniti A, Semenza G L. V-SRC induces expression of hypoxia-inducible factor 1 (HIF-1) and transcription of genes encoding vascular endothelial growth factor and enolase 1: involvement of HIF-1 in tumor progression. Cancer Res. 1997;57:5328–5335. [PubMed] [Google Scholar]
- 26.Kibel A, Iliopoulos O, DeCaprio J D, Kaelin W G., Jr Binding of the von Hippel-Lindau tumor suppressor protein to elongin B and C. Science. 1995;269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- 27.Kim Y H, Proust J J, Buchholz M J, Chrest F J, Nordin A A. Expression of the murine homologue of the cell cycle control protein p34cdc2 in T lymphocytes. J Immunol. 1992;149:17–23. [PubMed] [Google Scholar]
- 28.Kipreos E T, Lander L E, Wing J P, He W W, Hedgecock E M. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- 29.Kishida T, Stackhouse T M, Chen F, Lerman M I, Zbar B. Cellular proteins that bind the von Hippel-Lindau disease gene product: mapping of binding domains and the effect of missense mutations. Cancer Res. 1995;55:4544–4548. [PubMed] [Google Scholar]
- 30.Korsmeyer S J. Bcl-2 initiates a new category of oncogenes: regulators of cell death. Blood. 1992;80:879–886. [PubMed] [Google Scholar]
- 31.Lee S, Chen D Y T, Humphrey J S, Gnarra J R, Linehan W M, Klausner R D. Nuclear/cytoplasmic localization of the von Hippel-Lindau tumor suppressor gene product is determined by cell density. Proc Natl Acad Sci USA. 1996;93:1770–1775. doi: 10.1073/pnas.93.5.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee A S. Coordinated regulation of a set of genes by glucose and calcium ionophores in mammalian cells. Trends Biochem Sci. 1987;12:20–23. [Google Scholar]
- 33.Levy A P, Levy N S, Goldberg M A. Hypoxia-inducible protein binding to vascular endothelial growth factor mRNA and its modulation by the von Hippel-Lindau protein. J Biol Chem. 1996;271:25492–25497. doi: 10.1074/jbc.271.41.25492. [DOI] [PubMed] [Google Scholar]
- 34.Levy A P, Levy N S, Iliopoulos O, Jiang C, Kaelin W G, Jr, Goldberg M A. Regulation of vascular endothelial growth factor by hypoxia and its modulation by the von Hippel-Lindau tumor suppressor gene. Kidney Int. 1997;51:575–578. doi: 10.1038/ki.1997.82. [DOI] [PubMed] [Google Scholar]
- 35.Lisztwan J, Marti A, Sutterluty H, Gstaiger M, Wirbelauer C, Krek W. Association of human CUL-1 and ubiquitin-conjugating enzyme CDC34 with the F-box protein 45(SKP2): evidence for evolutionary conservation in the subunit composition of the CDC34-SCF pathway. EMBO J. 1998;17:368–383. doi: 10.1093/emboj/17.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lonergan K M, Iliopoulos O, Ohh M, Kamura T, Conaway R C, Conaway J W, Kaelin W G., Jr Regulation of hypoxia-inducible mRNAs by the von Hippel-Lindau tumor suppressor protein requires binding to complexes containing elongins B/C and Cul2. Mol Cell Biol. 1998;18:732–741. doi: 10.1128/mcb.18.2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lowe S W, Ruley H E, Jacks T, Housman D E. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 38.Maher E R, Kaelin W G., Jr von Hippel-Lindau disease. Medicine. 1997;76:381–391. doi: 10.1097/00005792-199711000-00001. [DOI] [PubMed] [Google Scholar]
- 39.Maltepe E, Schmidt J V, Baunoch D, Bradfield C A, Simon M C. Abnormal angiogenesis and responses to glucose and oxygen deprivation in mice lacking the protein ARNT. Nature. 1997;386:403–407. doi: 10.1038/386403a0. [DOI] [PubMed] [Google Scholar]
- 40.Maxwell P H, Dachs G U, Gleadle J M, Nicholls L G, Harris A L, Stratford I J, Hankinson O, Pugh C W, Ratcliffe P J. Hypoxia-inducible factor-1 modulates gene expression in solid tumors and influences both angiogenesis and tumor growth. Proc Natl Acad Sci USA. 1997;94:8104–8109. doi: 10.1073/pnas.94.15.8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morinaga N, Moss J, Vaughan M. Cloning and expression of a cDNA encoding a bovine brain brefeldin A-sensitive guanine nucleotide-exchange protein for ADP-ribosylation factor. Proc Natl Acad Sci USA. 1997;94:12926–12931. doi: 10.1073/pnas.94.24.12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukhopadhyay D, Knebelmann B, Cohen H T, Ananth S, Sukhatme V P. The von Hippel-Lindau tumor suppressor gene product interacts with Sp1 to repress vascular endothelial growth factor promoter activity. Mol Cell Biol. 1997;17:5629–5639. doi: 10.1128/mcb.17.9.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Núñez G, Hockenbery D, McDonnell T J, Sorensen C M, Korsmeyer S J. Bcl-2 maintains B cell memory. Nature. 1991;353:71–73. doi: 10.1038/353071a0. [DOI] [PubMed] [Google Scholar]
- 44.Pause A, Lee S, Worrell R A, Chen D Y T, Burgess W H, Linehan W M, Klausner R D. The von Hippel-Lindau tumor suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc Natl Acad Sci USA. 1997;94:2156–2161. doi: 10.1073/pnas.94.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paton E E, Willems A R, Sa D, Kuras L, Thomas D, Craig K L, Tyers M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 1998;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pickart C M. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 47.Reed J C. Bcl-2: prevention of apoptosis as a mechanism of drug resistance. Hematol Oncol Clin North Am. 1995;9:451–473. [PubMed] [Google Scholar]
- 48.Rubin D M, Finley D. Proteolysis. The proteasome: a protein-degrading organelle? Curr Biol. 1995;5:854–858. doi: 10.1016/s0960-9822(95)00172-2. [DOI] [PubMed] [Google Scholar]
- 49.Sato K, Terada K, Sugiyama T, Takahashi S, Saito M, Moriyama M, Kakinuma H, Suzuki Y, Kato M, Kato T. Frequent overexpression of vascular endothelial growth factor gene in human renal cell carcinoma. Tohoku J Exp Med. 1994;173:355–360. doi: 10.1620/tjem.173.355. [DOI] [PubMed] [Google Scholar]
- 50.Shweiki D, Neeman M, Itin A, Keshet E. Induction of vascular endothelial growth factor expression by hypoxia and by glucose deficiency in multicell spheroids: implications for tumor angiogenesis. Proc Natl Acad Sci USA. 1995;92:768–772. doi: 10.1073/pnas.92.3.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Siemeister G, Weindel K, Mohrs K, Barleon B, Martiny-Baron G, Marme D. Reversion of deregulated expression of vascular endothelial growth factor in human renal carcinoma cells by von Hippel-Lindau tumor suppressor protein. Cancer Res. 1996;56:2299–2301. [PubMed] [Google Scholar]
- 52.Sommer T, Dieter H W. Endoplasmic reticulum degradation: reverse protein flow of no return. FASEB J. 1997;11:1127. doi: 10.1096/fasebj.11.14.9409541. [DOI] [PubMed] [Google Scholar]
- 53.Stein I, Neeman M, Shweiki D, Itin A, Keshet E. Stabilization of vascular endothelial growth factor mRNA by hypoxia and hypoglycemia and coregulation with other ischemia-induced genes. Mol Cell Biol. 1995;15:5363–5368. doi: 10.1128/mcb.15.10.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takahashi A, Sasaki H, Kim S J, Tobisu K, Kakizoe T, Tsukamoto T, Kumamoto Y, Sugimura T, Terada M. Markedly increased amounts of messenger RNAs for vascular endothelial growth factor and placenta growth factor in renal cell carcinoma associated with angiogenesis. Cancer Res. 1994;54:4233–4237. [PubMed] [Google Scholar]
- 55.Ting J, Wooden S K, Kriz R, Kelleher K, Kaufman R J, Lee A S. The nucleotide sequence encoding the hamster 78-kDa glucose-regulated protein (GRP78) and its conservation between hamster and rat. Gene. 1987;55:147–52. doi: 10.1016/0378-1119(87)90258-7. [DOI] [PubMed] [Google Scholar]
- 56.Thastrup O, Dawson A P, Scharff O, Foder B, Cullen P J, Drobak B K, Bjerrum P J, Christensen S B, Hanley M R. Thapsigargin, a novel molecular probe for studying intracellular calcium release and storage. Agents Actions. 1989;27:17–23. doi: 10.1007/BF02222186. [DOI] [PubMed] [Google Scholar]
- 57.Tsai F H, Overberger C G, Zand R. Synthesis and peptide bond orientation in tetrapeptides containing l-azetidine-2-carboxylic acid and l-proline. Biopolymers. 1990;30:1039–1049. doi: 10.1002/bip.360301105. [DOI] [PubMed] [Google Scholar]
- 58.Watowich S S, Morimoto R I. Complex regulation of heat shock- and glucose-responsive genes in human cells. Mol Cell Biol. 1988;8:393–405. doi: 10.1128/mcb.8.1.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wizigmann-Voos S, Breier G, Risau W, Plate K. Up-regulation of vascular endothelial growth factor and its receptors in von Hippel-Lindau disease-associated and sporadic hemangioblastomas. Cancer Res. 1995;55:1358–1364. [PubMed] [Google Scholar]