Abstract
Background
Lichen sclerosus (LS) is a chronic inflammatory cicatricial skin disease that can lead to urethral stricture or even malignant transformation and the etiology is still unknown. This study comparatively analyzed the balanopreputial swab and urine microbiota simultaneously between male patients with LS urethral stricture (LSUS) and non-LS urethral stricture (non-LSUS).
Methods
We prospectively included 31 male patients with LSUS and 30 with non-LSUS in this case–control study. Midstream urine samples and balanopreputial swabs were collected from each patient for the 16S V3-V4 hypervariable region sequencing. Operational taxonomic units were defined using a > 97% sequence similarity threshold. We compared the differences in alpha diversity, beta diversity, and microbial structure between the two groups.
Results
Whether in swab or urine samples, there was no significant difference in alpha diversity between the two groups. Swab samples showed a significant difference in beta diversity (p = 0.001). For all individuals, composition analyses showed that the most abundant phyla were Actinobacteria, Firmicutes, Proteobacteria, and Bacteroidetes in both samples. Additionally, the microbial communities of swab samples were significantly more similar to the communities of urine samples in the LSUS group (p = 0.047).
Conclusions
Microbiota showed significant variation between LSUS and non-LSUS groups, suggesting that microecological imbalance may be closely related to the occurrence of LS. Urinary irritation may be related to the unique microbiota on the genital skin of patients with LSUS.
Keywords: Lichen sclerosus, Microbiota, 16SDNA, Urethral stricture, Urine
Introduction
Lichen sclerosus (LS) is a chronic dermatosis that has a predilection for the anogenital region. In male patients, it often starts from the external genitalia and can develop into the urethra, leading to severe urethral stricture. The prior report showed that 10% of male urethral stricture was caused by LS [1]. There are currently three hypotheses related to the pathogenesis of LS to be verified: infection, autoimmunity, and chronic irritation. All the theories lead to micro-damage to the genital skin which will induce chronic inflammation and finally cause LS [2–5]. However, direct evidence of how LS occurs and develops is still lacking.
At present, major research primarily focuses on the relationship between microbiota and human health. In prior studies, microbiota exhibited a systemic impact. Disorders of the gut microbiota can cause arthritis, metabolic disease, inflammatory bowel disease, and so on. The skin microbiota is associated with the occurrence of atopic dermatitis, psoriasis, acne, and hidradenitis suppurativa. Low-level inflammation is often the result of microecological disturbances. Back to the urinary system, mounting evidence has shown that specific microbiota also colonizes the urinary tract. A variety of diseases including urinary incontinence, urinary tract infection, and interstitial cystitis have been proved to have a relationship with urinary microbiota to a certain extent [6–8]. Considering the inflammatory nature of LS, we reasonably speculate that the occurrence of LS is closely related to the microbiota.
In this research, we collected urine and balanopreputial swab samples from patients with LS urethral stricture (LSUS) and non-LS urethral stricture (non-LSUS) and compared the microbiota. We assume that differences can be found in microbial diversity and structure between the two groups.
Materials and methods
Patient selection
From 2021, patients who presented for anterior urethral stricture at the outpatient department and met the criteria were prospectively included. Including criteria were as follows: male patients with anterior urethral stricture and clinical manifestations of LS diagnosed by a professional reconstructive urologist; male patients with anterior urethral stricture caused by definite etiologies, including urethral trauma, catheterization, transurethral operation or surgery and without any suspected clinical manifestations of LS. Exclusion criteria included < 18 years old, invasive urogenital operations (< 3 months before study sample collection), history of antibiotic intaking (< 2 weeks before study sample collection), urine culture before sample collection > 100,000 colony forming unit/mL, combined with posterior urethral stricture, combined with systemic autoimmune disease, inability to urinate spontaneously through the urethra and lack of swab or urine sample. The study was approved by the ethics committee of Shanghai Sixth People's Hospital and individual consent was obtained from all the participants.
Sample collection
The samples were collected at the first visit of each included patient before any intervention. 30 mL midstream urine was collected and centrifuged at 12,000 g at 4 ℃ for 15 min [9, 10]. 1–2 mL supernatant was left to be suspended with the sediment, and the suspension was stored at −80 ℃ for 16S rDNA sequencing. Disposable sterile and DNA-free swabs were rubbed on the patients’ penile glans and inner plate of prepuce circularly 30 times. The heads of swabs were cut and stored at −80 ℃ for sequencing.
16S rDNA sequencing
Total genomic DNA was extracted using MagPure DNA LQ Kit (Magen Biotech, Guangzhou, China) following the manufacturer’s instructions. Briefly, after adding 750ul lysis buffer and mixing, the cells were lysed using Fastpreps 24 (6.5 m/s twice for 45 s) and incubated at 65 ℃ for 10 min. Samples were then centrifuged at 12,000 g for 5 min and the supernatant was collected. After using MagPure magnetic beads to bind DNA, the beads were separated on a magnetic stand and the supernatant was removed. Then, the beads were washed twice with wash buffer. Finally, the purified DNA was eluted in 100 μl of elution buffer and the concentrate was detected by NanoDrop 2000C spectrophotometer (Thermo Fisher Scientific, Waltham, USA).
The 16S rRNA gene amplicon library was generated by two consecutive PCR amplifications. In the first round, the variable region (V3-V4) of the 16S rRNA gene was amplified with universal primers 343-F and 798-R (343-F: 5'-TACGGRAGGCAGCAG −3' and 798-R: 5'-AGGGTATCTAATCCT-3') [11]. After each round of PCR, the products were detected by agarose gel electrophoresis and purified using AMPure XP magnetic beads (Beckman Coulter, California, USA). After the second round, the products were quantified using the Qubit dsDNA Assay Kit (Life Technologies, Delhi, India). Extraction and PCR negative controls were included in all steps to assess potential DNA contamination.
Equal amounts of purified amplicon were pooled for subsequent sequencing with the Illumina Mi-seq platform (Illumina, San Diego, California, USA). After obtaining the raw data, the paired-end reads were trimmed with Trimmomatic software and then assembled with FLASH software [12, 13]. Chimeric sequences were detected and removed with UCHIME [14]. The obtained clean reads were clustered to generate operational taxonomic units (OTUs) by Vsearch software with 97% similarity cutoff [15]. The representative read of each OTU was selected by the QIIME package [16] and annotated and blasted against the Silva database (Version 138) by the RDP classifier [17] with a confidence threshold of 70%.
Alpha diversity, beta diversity, community structure, and differential species of the two groups were analyzed. We used four indexes to characterize alpha diversity: Chao 1 index [18], Shannon index [19], Simpson index [20], and Good's coverage index. The microbial difference between individual patients’ swabs and urine was calculated by the Bray–Curtis distance, referred to as swab-urine distance.
Statistical analysis
Patient characteristics were compared by Student’s t-tests, Mann–Whitney U test, or Fisher’s exact test respectively for normally distributed continuous variable, non-normally distributed continuous variable, and categorical variable. Wilcoxon signed-rank test was used to compare the differential species and alpha diversity. Analysis of similarities (ANOSIM) was used to compare the difference in beta diversity which was calculated in weighted unique fraction metric (UniFrac) distance and presented by principal coordinate analysis (PCoA) between groups. The statistical difference in swab-urine distance between the two groups was tested by the Mann–Whitney U test.
Results
From 2021 to 2023, eighty-seven patients were enrolled, of which 26 patients were excluded. Nine patients were excluded due to lacking complete sample sets, five patients due to urinary infection, four patients due to a history of antibiotic use, four patients due to inability to urinate spontaneously through the urethra, two patients due to confirmed urethral squamous carcinoma and two patients due to being under 18 years of age. Finally, thirty-one patients in the LSUS group and 30 in the non-LSUS group were included in this study. Patients’ average age is 49.1 (20–88) years old. The median body mass index of the patients is 23.7 (17.1–42.3). All the included patients had suffered from the urethral stricture for more than half a year (0.5–41.0). No statistical difference was found in patients’ characteristics (Table 1).
Table 1.
The characteristics of patients in two groups
| Demographic and medical characteristics | Group | p | |
|---|---|---|---|
| LSUS | Non-LSUS | ||
| Age, mean (Year, Range) | 48.8 (27–77) | 49.4 (20–88) | 0.881 |
| BMI, median (Kg/m2, Range) | 23.7 (20.1–42.3) | 23.7 (17.1–31.6) | 0.237 |
| Course of stricture, median (Year, Range) | 6.0 (1–41) | 3.3 (0.5–29) | 0.103 |
| Urethral stricture segment, n (%) | |||
| Urethral meatus | 6 (19) | 3 (10) | 0.422 |
| Penile urethra | 14 (45) | 12 (40) | |
| Bulbar urethra | 11 (35) | 15 (50) | |
| Surgical approach, n (%) | |||
| Augmentation urethroplasty | 20 (65) | 12 (40) | N/A |
| Anastomotic urethroplasty | 0 | 11 (37) | |
| Meatotomy | 6 (19) | 2 (7) | |
| Perineal urethrostomy | 5 (16) | 0 | |
| Direct vision internal urethrotomy | 0 | 2 (7) | |
| No sugery | 0 | 3 (10) | |
LSUS lichen sclerosus urethral stricture, BMI body mass index
The urethral stricture segment was confirmed by urethrography and intraoperative findings (Table 1). The urethral meatus was affected in all the patients of the LSUS group. The stricture further affected the penile and bulbar urethra in 14 and 11 patients respectively. In the non-LSUS group, three patients had only urethral meatus stricture. Twelve and fifteen patients had penile and bulbar urethral stricture. Urethroplasty was performed on 56 patients (Table 1). Among the five patients who did not undergo urethroplasty, two underwent direct vision internal urethrotomy, one underwent cystostomy, awaiting further urethral reconstruction, and two declined surgical treatments.
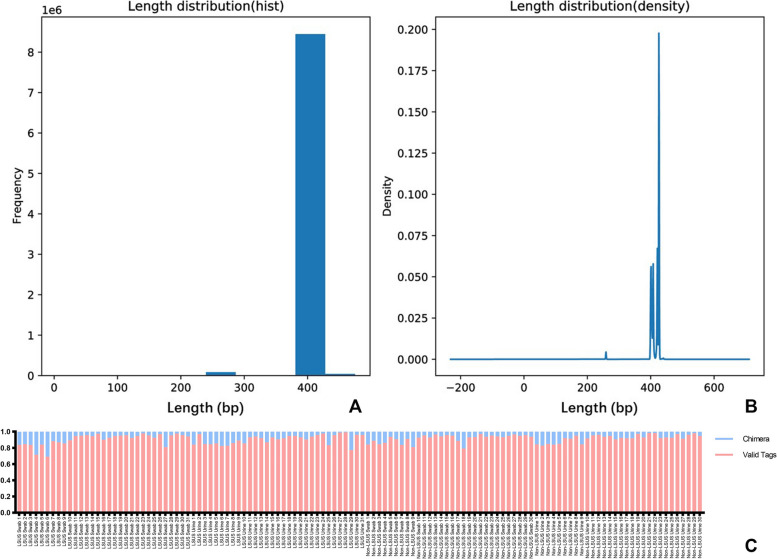
A total of 122 samples were sequenced, with an equal split between swab and urine samples. The quality assessment of 16S rDNA sequencing is shown in Fig. 1. The average length of valid tags was 385.44 ~ 425.99 bp and the number of OTUs in each sample was distributed between 141 and 3860. The median Good's coverage index of all four groups was 0.99. The median Chao 1 index, Shannon index, and Simpson index of swab and urine samples showed the approximate level between the two groups (Table 2). No significant difference was found in any of the index in alpha diversity between the LSUS patients and non-LSUS patients.
Fig. 1.
The quality assessment of 16S rDNA sequencing data. A and B show the frequency and density of sequence length distribution. The sequence length is concentrated between 400 and 450 bp. C shows the proportion of valid tags and chimeras in the sequences of each sample
Table 2.
The median of alpha diversity index in each type of samples
| Alpha diversity index | Group | |||
|---|---|---|---|---|
| LSUS-S | Non-LSUS-S | LSUS-U | Non-LSUS-U | |
| Chao 1 index | 1182.77 | 1042.98 | 1403.13 | 1231.39 |
| Shannon index | 4.89 | 4.24 | 5.55 | 5.87 |
| Simpson index | 0.88 | 0.88 | 0.91 | 0.92 |
| Good's coverage index | 0.99 | 0.99 | 0.99 | 0.99 |
LSUS lichen sclerosus urethral stricture, S swab, U urine
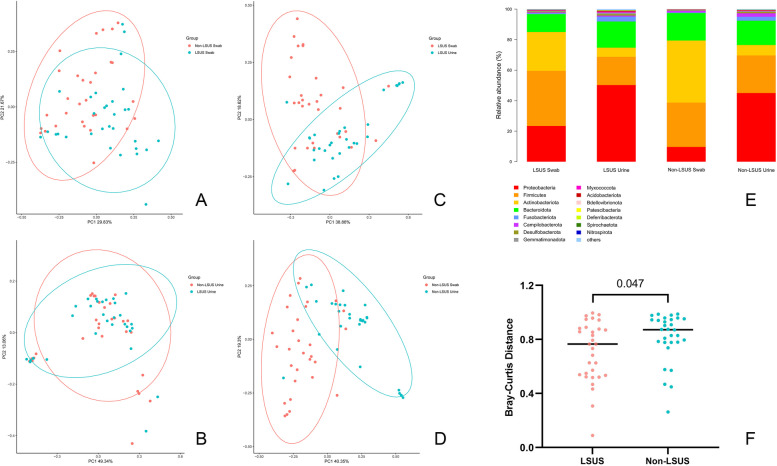
However, significant differences in beta diversity were observed in the swab samples between the LSUS and non-LSUS groups. The results of the ANOSIM showed that the R-value was 0.121 (> 0) and the p-value was 0.001, indicating that the variation between groups was significantly higher than the variation within groups in swab samples (Fig. 2A). While the R-value of the urine samples was −0.012 (p = 0.691) which denied the difference (Fig. 2B). Additionally, within the two groups, significant differences in beta diversity were observed between swab and urine samples (LSUS: R = 0.173, p = 0.001; non-LSUS: R = 0.437, p = 0.001) (Fig. 2C/D).
Fig. 2.
Results of microbiota sequencing. A/B/C/D show the beta diversity among the four types of samples. E shows the relative abundance of the top 15 bacterial phyla in four types of samples. F shows the swab-urine distance between the two groups. LSUS, lichen sclerosus urethral stricture
For all individuals, Actinobacteria, Firmicutes, Proteobacteria, and Bacteroidetes showed the highest abundance in both samples (Fig. 2E). Differences in taxonomy have been found in swab and urine samples between the LSUS and non-LSUS (Table 3). For swabs, the abundance of four phyla was significantly different, including Actinobacteriota, Proteobacteria, NB1-j, and Cyanobacteria. For urine, only RCP2-54 showed a significant difference. At the genus level, sixty-two and forty-tour genera showed statistical differences in abundance between the two groups. For swabs, Acinetobacter, Pseudomonas, and Mycobacterium were significantly more abundant in the LSUS group (p = 0.001/0.009/0.047). In contrast, Negativicoccus, Corynebacterium, Finegoldia, Peptoniphilus, and Varibaculum showed significantly decreased abundance (p = 0.001/0.002/0.002/0.002/0.002). For urine, the Pseudomonas, Cellvibrio, and Rubrobacter increased (p = 0.006/0.033/0.036), and the Ezakiella, Parvimonas, and Peptoniphilus (p = 0.003/0.017/0.048) showed a reduction in the LSUS group.
Table 3.
Number of differentially expressed taxa in different taxonomy levels
| Taxonomy | Group | |
|---|---|---|
| Swab | Urine | |
| OTUs | 306 | 224 |
| Phylum | 4 | 1 |
| Class | 10 | 3 |
| Order | 21 | 12 |
| Family | 36 | 23 |
| Genus | 62 | 44 |
| Species | 117 | 88 |
OTU Operational taxonomic unit
We also compared the microbial composition of individual swab and urine samples between the two groups. The results showed that the swab-urine distance was significantly smaller in the LSUS group, which meant the communities of swab samples were significantly more similar to the communities of urine samples in the patients of the LSUS group, compared to the non-LSUS group (Fig. 2F). Subsequently, we listed the dominant genera (relative abundance > 0.01) in the urine microbiota of the LSUS group and compared the relative abundance of these genera in the swab microbiota between the two patient groups (Table 4). Results showed that in sixteen dominant genera, thirteen genera had higher mean relative abundance in swab samples of the LSUS group. Pseudomonas, Mycoplasma, and Enterococcus showed significant differences (p = 0.009/0.023/0.031).
Table 4.
The list of dominant genera in LSUS urine samples and the differences in relative abundance of swab samples between two groups
| Dominant Genera | Relative Abundance | p | ||
|---|---|---|---|---|
| LSUS-Urine | LSUS-Swab | Non-LSUS-Swab | ||
| Escherichia-Shigella | 0.1668 | 0.0602 | 0.0237 | 0.847 |
| Ralstonia | 0.0704 | 0.0477 | 0.0074 | 0.183 |
| Prevotella | 0.0575 | 0.0475 | 0.0844 | 0.349 |
| Muribaculaceae | 0.0530 | 0.0325 | 0.0382 | 0.988 |
| Enterobacter | 0.0521 | 0.0259 | 0.0083 | 0.059 |
| Proteus | 0.0303 | 0.0059 | 0.0002 | 0.711 |
| Sneathia | 0.0276 | 0.0047 | 0.0012 | 0.351 |
| Lactobacillus | 0.0229 | 0.0482 | 0.0123 | 0.118 |
| Pseudomonas | 0.0191 | 0.0026 | 0.0012 | 0.009* |
| Bacteroides | 0.0161 | 0.0120 | 0.0169 | 0.982 |
| Mycoplasma | 0.0145 | 0.0012 | 0.0011 | 0.023* |
| Citrobacter | 0.0145 | 0.0075 | 0.0015 | 0.167 |
| Bifidobacterium | 0.0117 | 0.0113 | 0.0017 | 0.108 |
| Streptococcus | 0.0113 | 0.0260 | 0.0051 | 0.062 |
| Enterococcus | 0.0105 | 0.0024 | 0.0010 | 0.031* |
| Gardnerella | 0.0102 | 0.0026 | 0.0025 | 0.366 |
LSUS lichen sclerosus urethral stricture; * means p < 0.05
Discussion
To our knowledge, this study is the first combined comparative study of the external genital skin and urine microbiota in male urethral stricture patients with versus without LS. Studies on the relationship between microbe and pathogenesis of LS have already existed. Microorganisms including Borrelia burgdorferi [3], human papillomavirus (HPV) [21], and Human gammaherpesvirus 4 (Epstein-Barr virus, EBV) [22] were considered to be the related pathogens. LS was once suspected to be related to Borrelia burgdorferi because of the similar clinical and histological features to chronic atrophic acrodermatitis. However, the detection rates of Borrelia burgdorferi in LS varied greatly (0–100%), and there is a lack of mechanistic evidence to support it [23, 24]. HPV infection was also considered to be the cause, especially in pediatric and female patients [23]. While subsequent studies presented contradictory findings that failed to support the correlation in male patients [25, 26]. The role of EBV was not supported by strong clinical evidence either. Therefore, up to now, the pathogenesis of LS caused by a single pathogen infection has not been determined. This study focuses on the changes in local microecology rather than a single pathogen and tries to explain the pathogenesis by the imbalanced microbiota.
Alpha diversity generally refers to the mean species diversity in the biological environment, which consists of Richness (number of species in the sample) and Evenness (density of each species in the sample) [27]. From the Good’s coverage index, approximately 99% of the total species in our samples were represented by the sequencing which meant the sequencing depth was high and sufficient. Our Chao 1, Shannon, and Simpson index showed no statistical difference in the microbiota richness and diversity between the two groups which were consistent with the previous studies. Cohen et al. [28] reported that microbiota richness, evenness, and Faith’s diversity were insignificantly higher in the urine of the LSUS group than in the non-LSUS group (p = 0.076/0.36 /0.058). Watchorn et al. [29] also reported similar levels of the Shannon index in balanopreputial swab and urine samples from patients with LS and healthy controls (2.61 vs. 2.17, p = 0.2; 2.38 vs. 2.33, p = 1). Although the data from Jamil et al. [30] show a significant higher Shannon index in the urine samples of pre-operative patients with LSUS compared to non-LSUS. It remains difficult to explain the mechanism of LSUS based on changes in alpha diversity.
However, we did find significant differences in the beta diversity of swab samples between the LSUS and non-LSUS groups. It showed in different statistical methods, including Bray–Curtis distance (R = 0.125, p = 0.001), Euclidean distance (R = 0.033, p = 0.031), and weighted Unifrac distance (Table 5). These results indicate the differences in microbial existence, abundance, and evolutionary relationships of the genital skin between the two groups. The same trend of beta diversity was also revealed by Jamil et al. and Watchorn et al. They reported relatively high variations of microbiota composition in urine between preoperative LSUS and non-LSUS [30], and in balanopreputial sac between the LS and healthy control groups [29], respectively.
Table 5.
The ANOSIM results of different statistical methods for beta diversity in swab samples
| Statistical Methods | R values | p values |
|---|---|---|
| Euclidean distance | 0.033 | 0.031 |
| Bray-Curtis distance | 0.125 | 0.001 |
| Weighted Unifrac distance | 0.121 | 0.001 |
| Binary- Jaccard distance | 0.034 | 0.089 |
| Unweighted Unifrac distance | 0.013 | 0.205 |
R value > 0 indicates that the variation between the two groups is higher than the variation within each group. P value < 0.05 indicates that the difference is significant
Although there were significant variations in the beta diversity between the swab and urine samples in both groups, the variations were significantly smaller in patients with LSUS. This suggests that the microecology of the genital skin in patients with LSUS is more significantly affected by urine, which also coincides with the highly probable pathogenesis of LS: micro-incontinence. In 2002, Owen et al. proposed that genital LS is closely associated with incontinence[31]. The majority of male patients with LS reported symptoms of micro-incontinence (80–100%) [5, 32, 33]. Increasing evidences showed that the key pathogenesis of LS is the irritation of long-term, occlusive exposure of susceptible epithelium to urine [33–35]. The exposure might be from uncircumcision, trauma, urethral intervention, anatomical defects, and so forth [34]. However, it remains unclear how this exposure causes the occurrence of LS. Czajkowski et al. [33] suggested that urine might mediate the pro-inflammatory response of penile skin by stimulating the expression of pro-inflammatory cytokines genes. Our findings provide a new possibility for the interpretation of this pathogenesis: the microbial composition of genital skin in patients with LSUS changes under chronic urine irritation.
Through dominant genus analysis, we could also see the potential correlation of the microbial composition between the urine and swab samples in patients with LSUS. According to our results, three dominant genera in urine were found with significantly higher relative abundance in the swabs of the LSUS group. Pseudomonas is considered a bacterial biomarker for seborrheic dermatitis [36, 37], while it also significantly increased in corneal swabs of patients with atopic keratoconjunctivitis [38]. Mycoplasma was significantly more abundant in skin biopsies of patients with vitiligo [39] and inflamed mucosa tissue biopsies in Crohn's disease [40]. Enterococcus is a common pathogenic genus and is significantly enriched in the gut microbiota of patients with hidradenitis suppurativa or Crohn's disease [41]. However, how these differences trigger genital chronic inflammation and ultimately lead to LS still requires further study.
This research has some limitations. Firstly, the results are based on a relatively small sample size from a single center. A multicenter large sample size study is needed in the future. Secondly, due to the lack of longitudinal data, it is difficult to say whether the variation in microbiota is a cause or a consequence of LS. In addition, this study sequenced and analyzed only bacteria. Other microorganisms, such as fungi and viruses, may also have a relationship with the development of disease. Therefore, metagenome sequencing may be necessary for a more comprehensive analysis of LS-related microbiota in the future.
Despite these limitations, we mapped the microbiota of genital skin and urine in LSUS and non-LSUS and revealed the differences between the two groups in this study. Meanwhile, it provided a theoretical basis for further studies on LSUS and explored the role of microecological balance in the occurrence of LS.
Conclusion
This study analyzed the microbiota characteristics of patients with LSUS and non-LSUS and reported a significant difference in beta diversity in balanopreputial swabs between the two groups. It was also found that the variations between the swab and urine samples were significantly smaller in patients with LSUS which suggested that the micro-incontinence may be related to the change of the microbial composition on genital skin and play an important role in the pathogenesis of LSUS.
Acknowledgements
There is no acknowledgment.
Abbreviations
- LS
Lichen sclerosus
- US
Urethral stricture
- OTU
Operational taxonomic unit
- RDP
Ribosomal Database Project
- ANOSIM
Analysis of similarities
- UniFrac
Unique fraction metric
- PCoA
Principal coordinate analysis
Authors’ contributions
All authors contributed to the study conception and design. Z.Y. W, W. Y, Y.B. G, and L.J. S designed the methodology. Z.Y. W, Z.W. Y and L.J. S collected the samples and clinical information. Z.Y. W, Z.W. Y, W. Y, Y.B. G, X.J. X performed the data analysis. Z.Y. W, Z.W. Y, L.J. S performed the research and wrote first draft manuscript. The review and editing of the article were performed by all the authors. Study resource and funding were provided by L.J. S. All authors contributed to the article and approved the submitted version. All authors have read and approved the final version of the manuscript.
Funding
This project is funded by the National Natural Science Foundation of China (Grant No. 81974085), the Science and Technology Commission of Shanghai Municipality (22S31901700), the International Cooperation Fund of the Science and Technology Commission of Shanghai Municipality (No.19410741700), the Discipline Leader of Shanghai Municipal Health (No.2022XD015), and the Summit Plateau Program, Research Physician Program, Shanghai Jiao Tong University School of Medicine (20240817).
Data availability
The clinical data are available from the corresponding author upon request. The datasets generated and analysed during the current study are available in the National Center for Biotechnology Information repository (NCBI) with the project numbers PRJNA843054.
Declarations
Ethics approval and consent to participate
All procedures complied with the principles of the Declaration of Helsinki and were approved by the ethics committee of Shanghai Sixth People's Hospital (2021-KY-018(K)). Informed consent was obtained from all participants.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zeyu Wang and Zhenwei Yu contributed equally to this work.
References
- 1.Fergus KB, Lee AW, Baradaran N, Cohen AJ, Stohr BA, Erickson BA, et al. Pathophysiology, clinical manifestations, and treatment of lichen sclerosus: a systematic review. Urology. 2020;135:11–9. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Fu Q, Zhang X. The presence of human papillomavirus and Epstein-Barr virus in male Chinese lichen sclerosus patients: a single center study. Asian J Androl. 2016;18(4):650–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross SA, Sánchez JL, Taboas JO. Spirochetal forms in the dermal lesions of morphea and lichen sclerosus et atrophicus. Am J Dermatopathol. 1990;12(4):357–62. [DOI] [PubMed] [Google Scholar]
- 4.Oyama N, Chan I, Neill SM, Hamada T, South AP, Wessagowit V, et al. Autoantibodies to extracellular matrix protein 1 in lichen sclerosus. Lancet. 2003;362(9378):118–23. [DOI] [PubMed] [Google Scholar]
- 5.Bunker CB, Patel N, Shim TN. Urinary voiding symptomatology (micro-incontinence) in male genital lichen sclerosus. Acta Derm Venereol. 2013;93(2):246–8. [DOI] [PubMed] [Google Scholar]
- 6.Pearce MM, Hilt EE, Rosenfeld AB, Zilliox MJ, Thomas-White K, Fok C, et al. The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. mBio. 2014;5(4):e01283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siddiqui H, Lagesen K, Nederbragt AJ, Jeansson SL, Jakobsen KS. Alterations of microbiota in urine from women with interstitial cystitis. BMC Microbiol. 2012;12: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willner D, Low S, Steen JA, George N, Nimmo GR, Schembri MA, et al. Single clinical isolates from acute uncomplicated urinary tract infections are representative of dominant in situ populations. mBio. 2014;5(2):e01064-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whiteside SA, Razvi H, Dave S, Reid G, Burton JP. The microbiome of the urinary tract–a role beyond infection. Nat Rev Urol. 2015;12(2):81–90. [DOI] [PubMed] [Google Scholar]
- 10.Neumann CJ, Pausan MR, Haid V, Weiss EC, Kolovetsiou-Kreiner V, Amtmann B, et al. The dynamics of the female microbiome: unveiling abrupt changes of microbial domains across body sites from prepartum to postpartum phases. Microbiol Spectr. 2024;12(8): e0014724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nossa CW, Oberdorf WE, Yang L, Aas JA, Paster BJ, Desantis TZ, et al. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World J Gastroenterol. 2010;16(33):4135–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30(5):460–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4: e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chao A. Nonparametric estimation of the number of classes in a population. Scand J Statist. 1984;11:11. [Google Scholar]
- 19.Shannon CE. A mathematical theory of communication. Bell System Technical Journal. 1948;27(4):379–423. [Google Scholar]
- 20.Simpson EH. Measurement of diversity. Nature. 1949;163(4148):688-. [Google Scholar]
- 21.Lau PWY, Cook N, Andrews H, Bracka A, Myint SH. Detection of human papillomavirus types in balanitis xerotica obliterans and other penile conditions. Genitourin Med. 1995;71(4):228–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aidé S, Lattario FR, Almeida G, Do Val IC, Da Costa CM. Epstein-barr virus and human papillomavirus infection in vulvar lichen sclerosus. J Low Genit Tract Dis. 2010;14(4):319–22. [DOI] [PubMed] [Google Scholar]
- 23.Bhambhani D, Bhambhani S, Pandya NK. Penile Lichen Sclerosis: A Surgical Perspective of its Aetiology and Treatment. Cureus. 2022;14(8): e28418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fistarol SK, Itin PH. Diagnosis and treatment of lichen sclerosus: an update. Am J Clin Dermatol. 2013;14(1):27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shim TN, Harwood CA, Marsh SG, Gotch FM, Quint W, de Koning MN, et al. Immunogenetics and human papillomavirus (HPV) in male genital lichen sclerosus (MGLSc). Int J STD AIDS. 2020;31(14):1334–9. [DOI] [PubMed] [Google Scholar]
- 26.D’Hauwers KW, Depuydt CE, Bogers JJ, Noel JC, Delvenne P, Marbaix E, et al. Human papillomavirus, lichen sclerosus and penile cancer: a study in Belgium. Vaccine. 2012;30(46):6573–7. [DOI] [PubMed] [Google Scholar]
- 27.Whittaker RH. Vegetation of the siskiyou mountains Oregon and California. Ecol Monogr. 1960;30(4):279–338. [Google Scholar]
- 28.Cohen AJ, Gaither TW, Srirangapatanam S, Castellanos ER, Enriquez A, Fergus KB, et al. Synchronous genitourinary lichen sclerosus signals a distinct urinary microbiome profile in men with urethral stricture disease. World J Urol. 2021;39(2):605–11. [DOI] [PubMed] [Google Scholar]
- 29.Watchorn RE, van den Munckhof EHA, Quint KD, Eliahoo J, de Koning MNC, Quint WGV, et al. Balanopreputial sac and urine microbiota in patients with male genital lichen sclerosus. Int J Dermatol. 2021;60(2):201–7. [DOI] [PubMed] [Google Scholar]
- 30.Jamil ML, Perecman A, Sherman A, Sullivan T, Christ K, Hansma A, et al. Urinary microbiome differences between lichen sclerosus induced and non-lichen sclerosus induced urethral stricture disease. World J Urol. 2023;41(9):2495–501. [DOI] [PubMed] [Google Scholar]
- 31.Owen CM, Yell JA. Genital lichen sclerosus associated with incontinence. J Obstet Gynaecol. 2002;22(2):209–10. [DOI] [PubMed] [Google Scholar]
- 32.Panou E, Panagou E, Foley C, Kravvas G, Watchorn R, Alnajjar H, et al. Male genital lichen sclerosus associated with urological interventions and microincontinence: a case series of 21 patients. Clin Exp Dermatol. 2022;47(1):107–9. [DOI] [PubMed] [Google Scholar]
- 33.Czajkowski M, Wierzbicki P, Kotulak-Chrząszcz A, Czajkowska K, Bolcewicz M, Kłącz J, et al. The role of occlusion and micro-incontinence in the pathogenesis of penile lichen sclerosus: an observational study of pro-inflammatory cytokines’ gene expression. Int Urol Nephrol. 2022;54(4):763–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kravvas G, Shim TN, Doiron PR, Freeman A, Jameson C, Minhas S, et al. The diagnosis and management of male genital lichen sclerosus: a retrospective review of 301 patients. J Eur Acad Dermatol Venereol. 2018;32(1):91–5. [DOI] [PubMed] [Google Scholar]
- 35.Kravvas G, Muneer A, Watchorn RE, Castiglione F, Haider A, Freeman A, et al. Male genital lichen sclerosus, microincontinence and occlusion: mapping the disease across the prepuce. Clin Exp Dermatol. 2022;47(6):1124–30. [DOI] [PubMed] [Google Scholar]
- 36.Lin Q, Panchamukhi A, Li P, Shan W, Zhou H, Hou L, et al. Malassezia and Staphylococcus dominate scalp microbiome for seborrheic dermatitis. Bioprocess Biosyst Eng. 2021;44(5):965–75. [DOI] [PubMed] [Google Scholar]
- 37.Dityen K, Soonthornchai W, Kueanjinda P, Kullapanich C, Tunsakul N, Somboonna N, et al. Analysis of cutaneous bacterial microbiota of Thai patients with seborrheic dermatitis. Exp Dermatol. 2022;31(12):1949–55. [DOI] [PubMed] [Google Scholar]
- 38.Hur MS, Lee JS, Jang M, Shin HJ, Lee YW. Analysis of the conjunctival microbiome in patients with atopic keratoconjunctivitis and healthy individuals. Ann Dermatol. 2021;33(2):163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bzioueche H, Simonyté Sjödin K, West CE, Khemis A, Rocchi S, Passeron T, et al. Analysis of matched skin and gut microbiome of patients with vitiligo reveals deep skin dysbiosis: link with mitochondrial and immune changes. J Invest Dermatol. 2021;141(9):2280–90. [DOI] [PubMed] [Google Scholar]
- 40.Russo E, Cinci L, Di Gloria L, Baldi S, D’Ambrosio M, Nannini G, et al. Crohn’s disease recurrence updates: first surgery vs. surgical relapse patients display different profiles of ileal microbiota and systemic microbial-associated inflammatory factors. Front Immunol. 2022;13: 886468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cronin P, McCarthy S, Hurley C, Ghosh TS, Cooney JC, Tobin AM, et al. Comparative diet-gut microbiome analysis in Crohn’s disease and Hidradenitis suppurativa. Front Microbiol. 2023;14:1289374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The clinical data are available from the corresponding author upon request. The datasets generated and analysed during the current study are available in the National Center for Biotechnology Information repository (NCBI) with the project numbers PRJNA843054.