Abstract
Recent studies have unveiled disrupted metabolism in the progression of cleft palate (CP), a congenital anomaly characterized by defective fusion of facial structures. Nonetheless, the precise composition of this disrupted metabolism remains elusive, prompting us to identify these components and elucidate primary metabolic irregularities contributing to CP pathogenesis. We established a murine CP model by retinoic acid (RA) treatment and analyzed control and RA-treated embryonic palatal tissues by LC-MS-based proteomic approach. We identified 220 significantly upregulated and 224 significantly downregulated proteins. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis revealed that these differentially expressed proteins (DEPs) were involved in translation, ribosome assembly, mitochondrial function, mRNA binding, as well as key metabolic pathways like oxidative phosphorylation (OXPHOS), glycolysis/gluconeogenesis, and amino acid biosynthesis. These findings suggest that dysregulated ribosome-related pathways and disrupted metabolism play a critical role in CP development. Protein-protein interaction analysis using the STRING database revealed a tightly connected network of DEPs. Furthermore, we identified the top 10 hub proteins in CP using the Cytohubba plugin in Cytoscape. These hub proteins, including RPL8, RPS11, ALB, PA2G4, RPL23, RPS6, CCT7, EGFR, HSPD1, and RPS28, are potentially key regulators of CP pathogenesis. In conclusion, our comprehensive proteomic analysis provides insights into the molecular alterations associated with RA-induced CP in Kun Ming mice. These findings suggest potential therapeutic targets and pathways to understand and prevent congenital craniofacial anomalies.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12920-024-02054-8.
Keywords: Cleft palate, Retinoic acid, Proteomic, Metabolic pathway, Gene Ontology, Kyoto Encyclopedia of Genes and Genomes
Introduction
Cleft palate (CP) is one of the most prevalent congenital birth defects in the craniofacial region, affecting approximately one in every thousand newborns worldwide [1, 2]. Its multifaceted origins have been reported to intertwine with genetic background and environmental factors [3, 4], yet the comprehensive understanding of CP’s etiology and pathogenesis remains elusive.
The formation of the secondary palate involves the growth, horizontal elevation, and fusion of the palatal shelves. In mice, palate specification occurs at embryonic gestation day (E) 11.5, and palatogenesis initiates around E12.5, with the palatal shelves emerging from the lateral sides of the maxillary processes. From E12.5 to E13.5, the palatal shelves grow downward vertically alongside the tongue, then elevate to a horizontal position by E14.0. After this elevation, the bilateral palatal shelves progress toward the midline and fuse to form the complete palate between E14.5 and E16.5 [5, 6].
Retinoic acid (RA), a major metabolic form of vitamin A in the body, holds a pivotal role in organ morphology, cell proliferation, and differentiation [7–9]. Researchers have successfully established CP animal models with RA [10]. Notably, studies have demonstrated that an excess of RA can induce CP in mice [11].
Building upon this CP model, comprehensive analyses of differentially expressed genes and proteins have been undertaken through integrated omics approaches. For example, the integration of genomic and transcriptomic data identified nine genes as potential biomarkers for CP [12], while proteomic data illustrated the correlation between peroxiredoxin-1 protein and CP development [13]. Additionally, recent studies have implicated the involvement of abnormal metabolism in CP. For instance, increased levels of unsaturated triglycerides were observed in RA-induced CP [14], and modulation of lipid metabolic defects has shown promise in rescuing CP in Tgfbr2 mutant mice [15]. Elevated lipid concentration was also identified in dexamethasone-induced CP [16]. Moreover, glycolysis has emerged as a key regulator of palate development [17], and reduced carbon metabolism has been associated with a high risk of CP formation [18]. Despite these advancements, the involvement of other metabolic pathways, such as oxidative phosphorylation, in palatal development has been largely unexplored.
In this study, we explored the existence of common differentially expressed proteins (DEPs) in RA-induced CP compared with normal control during the palatal process growth period at E13.5, using proteomics technology-specifically, isobaric tags for relative and absolute quantitation [19]. This investigation aims to provide valuable insights for future studies elucidating the mechanisms contributing to CP formation.
Materials and methods
Animal and sample preparation
Eight-week-old Kun Ming mice were housed in a temperature-controlled room (22–25 °C, 45% humidity) in the Center Laboratory Animal Sciences of Shantou University Medical College. Female mice were paired with fertile male mice overnight in a 2:1 females-to-male ratio. The day of vaginal plug observation was designated as E0.5. Six inseminated females were randomly assigned to two groups. At E10.5, the treatment group received oral administration of RA (Sigma, MO, USA) dissolved in sesame oil at a dosage of 70 mg/kg by gavage. For hematoxylin and eosin (H&E) staining, the embryos were collected at E13.5, E14.5, or E15.5 and then fixed with 4% paraformaldehyde overnight at 4 °C. Subsequently, the fixed embryos were subjected to paraffin embedding and sectioning, followed by H&E staining.
For proteomic analysis, pregnant female mice were euthanized and E13.5 embryos were collected. The palatal tissues were dissected by microsurgery. Palatal tissues of embryos from each pregnant female were pooled together, forming one replicate. Three pairs of control and RA-treated replicates were frozen and stored in liquid nitrogen for further analysis. All Kun Ming mice used in this study were placed in a euthanasia chamber and then 100% carbon dioxide gas was introduced at a flow rate of 30–70% of the chamber volume per minute. When there was no corneal reflex, no breathing, and no heartbeat for more than 5 min, the mice were judged dead. Every effort is made to minimize animal suffering.
The frozen embryonic palatal tissues were thawed slowly at 4 °C and homogenized in 200 µL ddH2O. Subsequently, 240 µL of precooled methanol was added to the samples, and they were mixed, followed by the addition of 800 µL of methyl tert-butyl ether (MTBE). The samples were then centrifuged at 14,000 × g and 10 °C for 15 min. The upper organic phase was collected and dried using nitrogen gas. For mass spectrometry (MS) resolution, 200 µL of a 90% isopropanol/acetonitrile solution was added to the dried samples. To prepare quality control (QC) samples, 10 µL from each sample was taken and mixed. The supernatant was sampled and analyzed by centrifugation at 14,000 ×g and 10 °C for 15 min. All animal experiments are compliant with the ARRIVE guidelines and are carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory animals (NIH Publications No. 8023, revised 1978), and the procedures conducted in this study were approved by the Animal Ethics Committee of Shantou University Medical College (Approval number: SUMC2020-295).
Proteomics analysis
A total of 100 µg of proteins from each sample solution were combined with triethylammonium bicarbonate (TEAB) containing 0.1% SDS, followed by incubation with 1 µL of trypsin (40 ng/µL) at 37 °C for 4 h. A second trypsin addition at the same ratio extended the digestion for 8 h continuously. Each sample was digested with 1.25 µL trypsin (40 ng/µL). The resulting digestion solution was frozen, re-dissolved with TEAB (water: TEAB = 1: 1), and subjected to incubation with iTRAQ® Reagent-8PlexMultiplex Kit (AB Sciex Inc., USA) at 25 °C. Each labeled reagent tube received 70 µL of isopropanol, was vortexed for 1 min, and then centrifuged to collect at the tube’s bottom. NP-13-1, NP-13-2, and NP-13-3 were used for the control group, while RP-13-1, RP-13-2, and RP-13-3 were used for the RA-induced group. NP stands for normal palate while RP stands for RA-induced palate. The labeled samples were mixed, centrifuged at 25 °C for 2 h, and subsequently lyophilized.
Identification of DEPs
The DEPs between normal and RA-induced palatal tissues were identified with the criterion of |log2FC| > 0 and P < 0.05. The volcano plot was visualized using the R package “ggplot2”. The top 20 DEPs were visualized using heatmap, accomplished by the R package “Pheatmap”.
Functional enrichment analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) database enrichment analysis were utilized to annotate the function of DEPs [20]. This analysis was conducted using the R packages, including “clusterProfiler”, “org.Mm.eg.db”, and “enrichplot“ [21].
Western blot
Palatal tissues were lysed with RIPA lysis buffer containing protease inhibitor (P0013K, Beyotime, China), with homogenization on ice. After centrifugation, the supernatants were collected, and their protein concentration was determined by a BCA kit (P0012, Beyotime). Samples containing 25 µg protein were separated by 10% SDS-PAGE and then transferred to 0.22 μm PVDF membrane (Millipore, USA). Next, the membrane was blocked by 5% non-fat milk for 1 h at room temperature (RT), followed by incubation with primary antibodies at 4 °C overnight. After washing with TBST (TBS + 0.1% Tween 20), the membrane was incubated with HRP-conjugated goat anti-rabbit IgG (H + L) (1:3000, A0208, Beyotime) at RT for 1 h. Finally, the membrane was washed with TBST again, and the signal was visualized using an ECL kit and documented using a Bio-Rad ChemiDoc XRS + machine. The primary antibody information is listed as follows: anti-NU5M (1:1000, 55410-1-AP, Proteintech, China), anti-PIGT (1:1000, 16906-1-AP, Proteintech), anti-MGAT1 (1:1000, 15103-1-AP, Proteintech), and anti-GAPDH (1:1000, 10494-1-AP, Proteintech). The band intensity was quantified by Image J (NIH, USA). Palatal tissues from three pairs of control and RA-treated embryos at E13.5 were assayed. Unpaired two-tailed Student’s t-test was used to evaluate the significance.
Protein-protein Interaction (PPI) analysis
The PPI of DEPs was first analyzed in the STRING database (https://cn.string-db.org/). The PPI among the top 10 hub proteins was further calculated by cytoHubba plugins [22] in Cytoscape software (version 3.9.1).
Statistical analysis
R software (v.4.0.5, https://www.r-project.org/) was used to perform the statistical analyses. Student’s t-test was employed to analyze the significant changes of proteins between RA-induced and control palatal tissues. Significance levels were denoted as * for P < 0.05, ** for P < 0.01, and *** for P < 0.001.
Results
RA-induced CP in Kun Ming Mouse
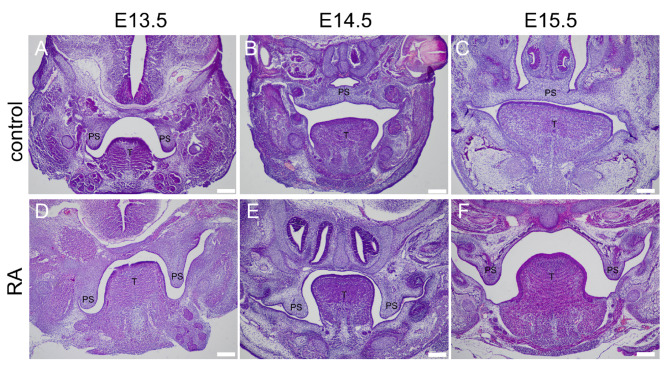
Among the embryos exposed to RA, 18 out of 20 exhibited CP phenotype, and 2 embryos died before analysis. While none of the 22 embryos in the control group showed such malformation. H&E staining revealed that control animals demonstrated normal palatal frame development, elevation, and fusion from E13.5 to E15.5 (Fig. 1A-C). In contrast, the RA treatment group showed a failure in proper elevation and fusion of the palatal frame (Fig. 1D-F).
Fig. 1.
H&E staining of palatal sections of control and RA-induced embryos. A-C: Control embryonic head at E13.5, E14.5, E15.5. D-F: RA-exposed embryonic head at E13.5, E14.5, E15.5. PS represents palatal shelves, and T represents togue. Control and RA-exposed PS structures are comparable at E13.5. However, while the control PS structures fuse at E14.5 and E15.5, RA-exposed embryos exhibit CP. Scale bars: 42 μm
Identification of DEPs in CP
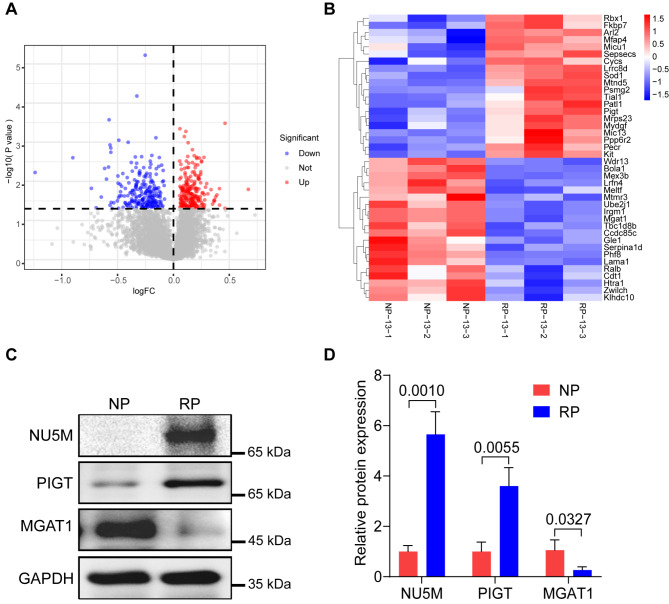
To gain a comprehensive understanding of significantly expressed proteins in RA-induced CP, proteins with |log2(Fold Change)| > 0 and P value < 0.05 were identified as DEPs between the RA-treated and control groups. The results showed that there are 220 significantly upregulated proteins and 224 significantly downregulated proteins (Fig. 2A and Supplemental Table 1). A heatmap was generated to visually represent the top 20 upregulated and top 20 downregulated DEPs (Fig. 2B). To verify the accuracy of the proteomic results, we performed Western blot assays to test the expression of a few randomly selected DEPs associated with metabolism including NU5M, PIGT, and MGAT1. The results confirmed the overexpression of NU5M and PIGT and the reduced expression of MGAT1 in RA-induced palatal tissues compared to untreated palatal tissues (Fig. 2C and D).
Fig. 2.
Identification of DEPs in RA-induced palatal tissues. (A) The volcano plot was utilized to indicate the DEPs. Red dots represent the significant upregulated proteins, while blue dots represent the significant downregulated proteins. (B) The top 20 upregulated proteins and top 20 downregulated proteins were visualized by heatmap. Control samples: NP-13-1, NP-13-2, NP-13-3; RA-induced palatal samples: RP-13-1, RP-13-2, RP-13-3. (C) Western blot data show that the expression of NU5M and PIGT is upregulated, while MGAT1 expression is decreased in RA-induced palatal tissues compared to control palatal tissues. (D) Quantification result of the Western blot data. The experiment was repeated three times independently. Unpaired two-tailed Student’s t-test was used to evaluate the significance
Signaling pathway enrichment of DEPs in CP
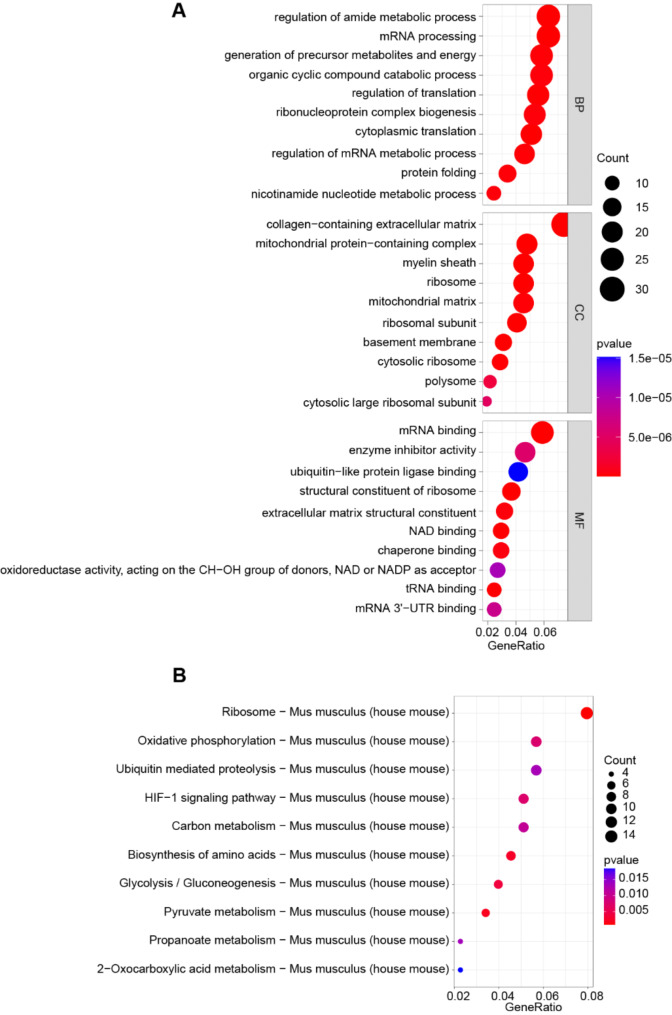
To further explore the potential signaling pathways involved in CP, we employed GO and KEGG enrichment analysis with a significance filter of P < 0.05. GO Biological Process (BP) revealed enrichment in translation, including mRNA processing and regulation of translation. Cellular Component (CC) illustrated enrichment in ribosome and mitochondrial matrix, while Molecular Function (MF) analysis indicated enrichment in mRNA binding, ubiquitin-like protein ligase binding, and structural constituent of ribosome (Fig. 3A). For a deeper insight into the key pathways of DEPs involved, we performed the KEGG analysis, revealing enrichment in pathways such as ribosome, oxidative phosphorylation (OXPHOS), HIF-1, glycolysis/gluconeogenesis, and biosynthesis of amino acid (Fig. 3B). These findings indicate that ribosome-related pathways are dysregulated in CP tissues. Moreover, disruptions in metabolism may occur within CP tissues, affecting amino acids, OXPHOS, and glycolysis.
Fig. 3.
GO and KEGG analysis of DEPs. (A) GO BP, CC, and MF analysis of 444 DEPs. (B) KEGG enrichment results of 444 DEPs in CP
Identification of hub proteins in CP
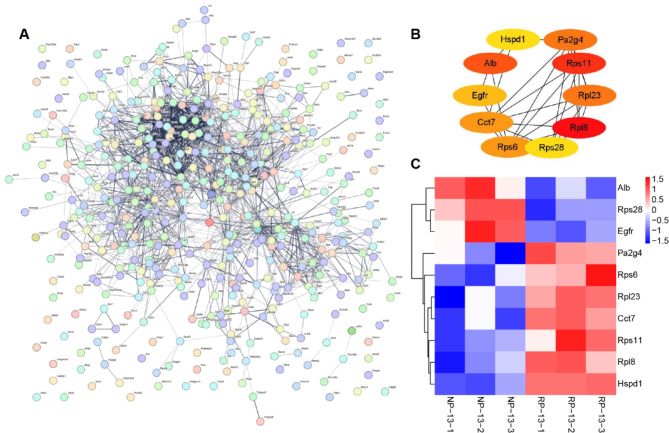
To comprehensively understand PPIs in CP, we employed the STRING database for network analysis of DEPs. As shown in Fig. 4A, the DEPS exhibits complicated interactions. To figure out the PPIs among the core DEPs, we utilized the Cytohubba plugin in Cytoscape software, the results revealed PPIs among the top 10 hub proteins: Rpl8, Rps11, Alb, Pa2g4, Rpl23, Rps6, Cct7, Egfr, Hspd1, and RPS28 (Fig. 4B). Among these, RPS11 exhibited the highest expression, while ALB displayed the lowest expression in RA-induced palate (Fig. 4C).
Fig. 4.
Identification of hub proteins by PPI. (A) The PPI network of all DEPs. (B) The PPI network of the top 10 hub proteins. (C) Expression of the top 10 hub proteins was visualized by heatmap. Red and blue colors represent upregulated and downregulated expression of the indicated DEPs, respectively
Discussion
In this study, we conducted a comprehensive proteomic analysis to gain insights into RA-induced CP formation at the vertical growth period of the mouse palatal shelves. We identified a total of 220 significantly upregulated and 224 significantly downregulated proteins in RA-induced CP. These DEPs likely play a pivotal role in the etiology of CP. Notably, many of these proteins are involved in crucial cellular processes, including translation, ribosome assembly, mitochondrial function, and mRNA binding. This observation aligns with the finding of reduced mitochondria in dexamethasone-induced CP [23]. The dysregulation observed in these processes suggests that RA exposure disrupts the normal cellular activities crucial for proper palatal development.
The GO and KEGG enrichment analyses provide deeper insights into the biological processes and pathways affected by the DEPs. The GO analysis highlights significant enrichment in translation-related processes, encompassing mRNA processing, and regulation of translation. KEGG analysis also underscores the involvement of the dysregulated metabolism of amino acids and other metabolites in CP formation. This notion is supported by the finding that CP patients have abnormal levels of plasma metabolites compared to normal subjects [24]. Moreover, phosphoglucomutase 1 deficiency, which impacts glycolysis/glycogen metabolism/protein glycosylation, can also cause CP [25]. Therefore, our finding hints at the likelihood that RA-induced CP entails aberrant metabolic processes, with potential ramifications for palatal morphogenesis.
The identification of hub proteins through PPI analysis is crucial for elucidating the principal regulators within CP. Enumerating the top 10 hub proteins, including Rpl8, Rps11, Alb, Pa2g4, Rpl23, Rps6, Cct7, Egfr, Hspd1, and RPS28, provides a foundational platform for unraveling the intricate network of PPI pertinent to CP. Among these, RPL8 and RPS28 are associated with Diamond–Blackfan anemia, a condition featuring CP [26, 27]. CCT7 is also a candidate gene for increased CP risk [28]. EGFR is required for normal palate closure [29]. The function of other hub proteins in CP formation remains to be determined.
In summary, these proteins emerge as potential focal points in the dysregulation of cellular processes contributing to CP development. Therefore, our findings suggest that the occurrence of CP may be attributed to disturbed metabolic processes, potentially leading to abnormal behaviors of embryonic palatal cells.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Author contributions
W. Z. wrote the manuscript, L. C. and A. M. plotted Figs. 1 and 2, W. J., M. X. and X. B. plotted Figs. 3 and 4, J. Z. and S. T. analyzed the results. All the authors reviewed and approved the manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (82071101), Guangdong Basic and Applied Basic Research Foundation (2019A1515011857, 2021A1515011142, 2023A1515012343, 2022A1515220099), 2020 Li Ka Shing Foundation Cross-Disciplinary Research Grant (2020LKSFG18B, 2020LKSFG02E), Guangdong University Innovation Team Project (2021KCXTD047), Provincial science and technology innovation strategy special project funding program (200114165897946, 210714106901245, STKJ202209067, STKJ2023004), 2022 Shantou University Graduated Student Innovation Program (Shandafa[2022]207).
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and/or its supplementary materials, and from the corresponding authors upon reasonable request.
Declarations
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Animal ethics declaration and consent to participate
This study was conducted following the guidelines and approved by the Animal Ethics Committee of Shantou University Medical College.
Experimental statement
All experimental protocols of this study have been approved by Shantou University Medical College, and the experimental operation is carried out in accordance with relevant standards and regulations of Shantou University Medical College.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Evenson KR, Mowla S, Olshan AF, Shaw GM, Ailes EC, Reefhuis J, Joshi N, Desrosiers TA. Maternal physical activity, sitting, and risk of non-cardiac birth defects. Pediatr Res. 2024;95(1):334–41. [DOI] [PMC free article] [PubMed]
- 2.Xing Y, Zhang W, Wan X, Hong Z, Zhao H, Liang W, Shi L, Chen J, Zhong X, Zhou J, et al. Association between an Interferon Regulatory Factor 6 gene polymorphism and nonsyndromic cleft palate risk. Genet Test Mol Biomarkers. 2019;23(9):652–63. [DOI] [PubMed] [Google Scholar]
- 3.Deņisova A, Pilmane M, Kažoka D. Antimicrobial peptides and interleukins in Cleft Soft Palate. Children (Basel). 2023;10(7):1162. [DOI] [PMC free article] [PubMed]
- 4.Zhang W, Tan L, Xing Y, Zhao H, Shi L, Zhou J, Lang X, Cai J, Tang S. Association between SATB2 gene polymorphism and cleft palate only risk in eastern Guangdong population and a meta-analysis. Cell Mol Biol (Noisy-le-grand). 2018;64(14):101–7. [PubMed] [Google Scholar]
- 5.Li Q, Ding J. Gene expression analysis reveals that formation of the mouse anterior secondary palate involves recruitment of cells from the posterior side. Int J Dev Biol. 2007;51(2):167–72. [DOI] [PubMed] [Google Scholar]
- 6.Li C, Lan Y, Jiang R. Molecular and Cellular mechanisms of Palate Development. J Dent Res. 2017;96(11):1184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W, Kirsch T. Retinoic acid stimulates annexin-mediated growth plate chondrocyte mineralization. J Cell Biol. 2002;157(6):1061–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song C, Li T, Zhang C, Li S, Lu S, Zou Y. RA-induced prominence-specific response resulted in distinctive regulation of wnt and osteogenesis. Life Sci Alliance. 2023;6(10):e202302013. [DOI] [PMC free article] [PubMed]
- 9.Campero-Basaldua C, Herrera-Gamboa J, Bernal-Ramírez J, Lopez-Moran S, Luévano-Martínez L-A, Alves-Figueiredo H, Guerrero G, García-Rivas G, Treviño V. The retinoic acid response is a minor component of the cardiac phenotype in H9c2 myoblast differentiation. BMC Genomics. 2023;24(1):431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Chen J, Wang X, Guo C, Peng X, Liu Y, Li T, Du J. Novel investigations in retinoic-acid-induced cleft palate about the gut microbiome of pregnant mice. Front Cell Infect Microbiol. 2022;12:1042779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu X, Gao J, Liao Y, Tang S, Lu F. Retinoic acid alters the proliferation and survival of the epithelium and mesenchyme and suppresses Wnt/β-catenin signaling in developing cleft palate. Cell Death Dis. 2013;4(10):e898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan F, Dai Y, Iwata J, Zhao Z, Jia P. An integrative, genomic, transcriptomic and network-assisted study to identify genes associated with human cleft lip with or without cleft palate. BMC Med Genomics. 2020;13(Suppl 5):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan X, Liu L, Pu Y, Zhang X, He X, Fu Y. 2,3,7,8-Tetrachlorodibenzo-p-dioxin induces a proteomic pattern that defines cleft palate formation in mice. Food Chem Toxicol. 2012;50(7):2270–4. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Zhao H, Chen J, Zhong X, Zeng W, Zhang B, Qi K, Li Z, Zhou J, Shi L, et al. A LCMS-based untargeted lipidomics analysis of cleft palate in mouse. Mech Dev. 2020;162:103609. [DOI] [PubMed] [Google Scholar]
- 15.Iwata J, Suzuki A, Pelikan RC, Ho T-V, Sanchez-Lara PA, Chai Y. Modulation of lipid metabolic defects rescues cleft palate in Tgfbr2 mutant mice. Hum Mol Genet. 2014;23(1):182–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xing Y, Zhang W, Zhao H, Shen Z, Liang W, Zhou J, Shi L, Chen J, Zhong X, Tang S. Multi–organ assessment via a 9.4–Tesla MRS evaluation of metabolites during the embryonic development of cleft palate induced by dexamethasone. Mol Med Rep. 2019;20(4):3326–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng X, Chen J, Wang Y, Wang X, Zhao X, Zheng X, Wang Z, Yuan D, Du J. Osteogenic microenvironment affects palatal development through glycolysis. Differentiation. 2023;133:1–11. [DOI] [PubMed]
- 18.Inostroza V, Salamanca C, Recabarren AS, Pantoja R, Leiva N, Pardo R, Suazo J. Maternal genotypes of folate/one-carbon metabolism gene variants and nonsyndromic cleft lip with or without cleft palate risk in Chile. Eur J Oral Sci. 2021;129(5):e12813. [DOI] [PubMed] [Google Scholar]
- 19.Zhang M, Wang D, Xu X, Xu W, Zhou G. iTRAQ-based proteomic analysis of duck muscle related to lipid oxidation. Poult Sci. 2021;100(4):101029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innov (Camb). 2021;2(3):100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lan S-J, Yang X-G, Chen Z, Yang T-Y, Xiang C-H, Zhang D, Li Y-X, Rong L. Role of GATA-6 and bone morphogenetic Protein-2 in Dexamethasone-Induced Cleft Palate formation in Institute of Cancer Research Mice. J Craniofac Surg. 2016;27(6):1600–5. [DOI] [PubMed] [Google Scholar]
- 24.Lei HG, Hong L, Kun SJ, Hai YX, Dong WY, Ke Z, Ping X, Hao C. A preliminary investigation of NSCL/P Plasma and urine in Guizhou Province in China using NMR-Based metabonomics. Cleft Palate Craniofac J. 2013;50(5):603–9. [DOI] [PubMed] [Google Scholar]
- 25.Altassan R, Radenkovic S, Edmondson AC, Barone R, Brasil S, Cechova A, Coman D, Donoghue S, Falkenstein K, Ferreira V, et al. International consensus guidelines for phosphoglucomutase 1 deficiency (PGM1-CDG): diagnosis, follow-up, and management. J Inherit Metab Dis. 2021;44(1):148–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lebaron S, O’Donohue MF, Smith SC, Engleman KL, Juusola J, Safina NP, Thiffault I, Saunders CJ, Gleizes PE. Functionally impaired RPL8 variants associated with Diamond-Blackfan anemia and a Diamond-Blackfan anemia-like phenotype. Hum Mutat. 2022;43(3):389–402. [DOI] [PubMed] [Google Scholar]
- 27.Gripp KW, Curry C, Olney AH, Sandoval C, Fisher J, Chong JX, Pilchman L, Sahraoui R, Stabley DL, Sol-Church K. Diamond-Blackfan anemia with mandibulofacial dystostosis is heterogeneous, including the novel DBA genes TSR2 and RPS28. Am J Med Genet A. 2014;164A(9):2240–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gowans LJJ, Comnick CL, Mossey PA, Eshete MA, Adeyemo WL, Naicker T, Awotoye WA, Petrin A, Adeleke C, Donkor P, et al. Genome-wide scan for parent-of-origin effects in a sub-saharan African cohort with nonsyndromic cleft lip and/or Cleft Palate (CL/P). Cleft Palate Craniofac J. 2022;59(7):841–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miettinen PJ, Chin JR, Shum L, Slavkin HC, Shuler CF, Derynck R, Werb Z. Epidermal growth factor receptor function is necessary for normal craniofacial development and palate closure. Nat Genet. 1999;22(1):69–73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and/or its supplementary materials, and from the corresponding authors upon reasonable request.