Abstract
Background
Soda pans are unique, natural aquatic environments characterised by elevated salinity and alkalinity, creating a distinctive and often extreme geochemistry. The microbiomes of soda pans are unique, with extremophiles such as halophiles, alkaliphiles and haloalkaliphiles being important. Despite being dominated by mostly unculturable inhabitants, soda pans hold immense biotechnological potential. The application of modern “omics-based” techniques helps us better understand the ecology and true extend of the biotechnological potential of soda pan microbiomes. In this study, we used a shotgun metagenomic approach to determine the microbial diversity and functional profile of previously unexplored soda pans located in Buhera, Eastern Zimbabwe. A combination of titrimetry and inductively coupled plasma optical emission spectroscopy (ICP‒OES) was used to perform physico-chemical analysis of the soda pan water.
Results
Physicochemical analysis revealed that the Buhera soda pans are highly alkaline, with a pH range of 8.74 to 11.03, moderately saline (2.94 – 7.55 g/L), and have high carbonate (3625 mg/L) and bicarbonate ion (1325 mg/L) alkalinity. High levels of sulphate, phosphate, chloride and fluoride ions were detected. Metagenomic analysis revealed that domain Bacteria dominated the soda pan microbial community, with Pseudomonadota and Bacillota being the dominant phyla. Vibrio was shown to be the predominant genus, followed by Clostridium, Candidatus Brevefilum, Acetoanaerobium, Thioalkalivibrio and Marinilactibacillus. Archaea were also detected, albeit at a low prevalence of 1%. Functional profiling revealed that the Buhera soda pan microbiome is functionally diverse, has hydrolytic-enzyme production potential and is capable of supporting a variety of geochemical cycles.
Conclusions
The results of this pioneering study showed that despite their extreme alkalinity and moderate salinity, the Buhera soda pans harbour a taxonomically and functionally diverse microbiome dominated by bacteria. Future work will aim towards establishing the full extent of the soda pan’s biotechnological potential, with a particular emphasis on potential enzyme production.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-024-03655-0.
Keywords: Soda pan, Geochemistry, Extremophile, Haloalkaliphile, Saline-alkaline, Metagenomics, Microbial diversity
Introduction
Soda pans are unique natural aquatic environments characterized by high salinity and alkalinity [1, 2]. They have a typically alkaline pH which lies in the range 7.5 – 11.5, and highly variable salinity ranging from 1.0 to 390.0 g/L [3]. Soda pans generally contain high concentrations of carbonate and bicarbonate ions and variable quantities of other ions such as sulphate, phosphate, calcium, chloride, potassium, sodium and magnesium [1]. The actual physicochemical character of a soda pan depends on factors like climate and underlying geology. The presence of soda pans and other saline-alkaline systems has been reported worldwide, with notable cases being the East African Rift Valley [4, 5], Eastern Europe [3, 6], the Qinghai‒Tibet Plateau in China [7], the USA (California and Nevada) and the Caribbean Plateau in Canada [8].
The distinctive geochemistry and physicochemical profiles of soda pans make them an extreme microbial habitat. However, despite their extreme nature, soda pans and other saline-alkaline ecosystems have been shown to be highly productive, being capable of supporting considerable microbial taxonomic and functional diversity [9–11]. The microbial communities of soda pans are typically dominated by extremophilic bacteria and archaea, with halophiles, alkaliphiles and double extremophilic haloalkaliphiles being dominant in these ecosystems [12–14]. Different classes of microorganisms have been isolated from soda pans and lakes. These organisms include photoautotrophs, such as Cyanobacteriota, and a variety of chemolithoautotrophs like sulphur oxidisers, sulphate reducers and nitrifiers [8]. Autotrophs are primary producers that generate a significant portion of the organic matter required to support heterotrophic nutrition. The abundance of carbonate and bicarbonate ions in soda pans is believed to be the key driver of autotrophic microbial nutrition, as these ions supply the carbon needed for organic matter formation through photosynthetic and chemolithoautotrophic carbon fixation [1]. Heterotrophic bacteria and haloalkaliphilic archaea have regularly been isolated and characterized from soda pans and lakes, and these include genera such as Halomonas, Halobacillus, Halobacterium, Alkalibacillus, Salinivibrio, Desulfovibrio, Thioalkalivibrio and Haloalkalibacter [15, 16].
The ability of haloalkaliphiles to thrive under the extreme conditions of soda pans makes them potentially useful as whole-cell biocatalysts in applications such as the biodegradation of saline-alkaline industrial effluent, bioremediation of oil spills, the biosynthesis of biosurfactants and in specialist food fermentations conducted in the presence of salt [17]. Additionally, haloalkaliphilic enzymes are stable and active at high pH and salinity, making them useful in applications that include detergent formulation, paper making, starch saccharification for the production of high-fructose corn syrup, and the dehairing of salt-preserved animal skins during leather tanning [11, 18]. Several studies have reported the successful recovery of alkaline lipases, amylases and proteases from soda pans and other saline-alkaline environments [5, 19, 20]. Thus, the exploration of soda pans, among other saline-alkaline systems, provides us an opportunity to discover novel microbes that may serve as sources of useful biological resources such as whole cells, enzymes, peptides and other metabolites.
Both traditional culture-based as well as the more modern, omics-based culture-independent methods have been used to study the microbiology of soda pans and other saline-alkaline systems [21–24]. While great strides have been made in the isolation and characterization of bacteria in general and extremophiles in particular, major setbacks such as the difficulty of creating laboratory conditions for their culture remain a hindrance [9, 10]. It is estimated that up to 99.9% of extremophiles from extreme environments are uncultivable [25, 26]. This leaves only a tiny fraction of the microbial inhabitants of extreme environments at our disposal. The advent of culture-independent methods such as metagenomics has enabled the detailed exploration of the microbial communities of many previously neglected extreme environments, such as soda pans and lakes, resulting in the discovery of several novel microbial taxa [21, 27, 28].
In this study, we describe the application of a shotgun-based metagenomic approach to determine the taxonomic composition and functional profile of the microbial communities of soda pans located in Buhera in eastern Zimbabwe. Although the metagenomics of soda pans in other parts of the world, including East and North Africa, has been reported, there are no reports on the microbiomes of soda pans in Southern Africa, including Zimbabwe. This study, to the best of our knowledge, is the first to investigate the microbiomes and associated physicochemical properties of Zimbabwean soda pans, and will help improve our understanding of these previously uncharacterised microbial habitats and highlight the biotechnological potential of their unique microbiomes.
Materials and Methods
Sampling site description and sample collection
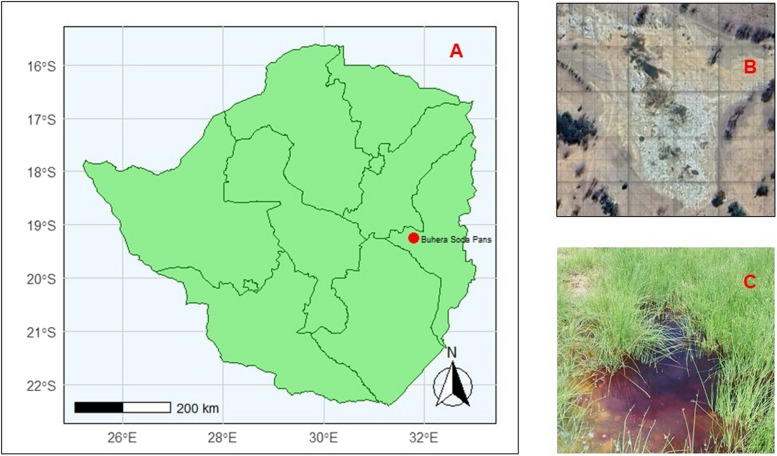
Water samples were collected at soda pans in Gombahari village (−19.26342, 31.80437), which is located in Buhera district in the Manicaland province of eastern Zimbabwe (Figure 1).
Fig. 1.
Sampling site in pictures: A Map of Zimbabwe showing the location of Buhera Soda Pans. The map was drawn using the ggplot2 package in R [29]; B Google maps image of the Buhera Soda Pans [30], C Photo showing a pool of water at the Buhera soda pans.
Sampling was conducted in October 2021 during the peak of the hot and dry season. A total of five water samples amounting to 100 mL each were aseptically collected by hand from randomly selected sampling points using the grab sampling technique into sterile glass bottles at depths of up to 30 cm from selected puddles within the soda pans. Sterility was achieved by pre-autoclaving all sampling bottles, using sterile latex gloves during sample collection and 70% ethanol as a hand sanitizer and sterilizing bottle openings. The samples were transported to the laboratory on ice, where 50 ml portions of all the samples were pooled to create one composite sample, which was then frozen at −20°C within 8 hours of collection until further processing. Approximately half of the composite sample was used for physicochemical analysis, while the remainder was used for metagenomic DNA extraction. The sample earmarked for metagenomic DNA extraction was stored for only 2 weeks before being processed for DNA extraction and sequencing.
Physicochemical analysis of the Buhera soda pan water samples
The physical and chemical properties (pH, salinity, electrical conductivity, temperature and total dissolved solids) of the water samples were measured on-site using a portable Eutech PCS Testr35 Multiparameter meter (Eutech, Singapore). In addition, the concentration of carbonate and bicarbonate ions in the sample as well as the total alkalinity were determined titrimetrically [31]. The results of this titration were used to calculate the phenolphthalein alkalinity (Eq. 1).
| 1 |
To determine the total alkalinity, 2 to 3 drops of bromocresol green indicator were added to the solution after the first titration, and the titration was continued until the solution turned yellow. The final burette reading obtained was used to calculate the total alkalinity using Eq. 2.
| 2 |
The phenolphthalein alkalinity (P) and total alkalinity (T) were used to calculate the total alkalinity, carbonate alkalinity and bicarbonate alkalinity according to methods described by the Indian Institute of Technology Kanpur [32]. Inductively coupled plasma optical emission spectroscopy (ICP‒OES) was used to determine the levels of iron, potassium, sodium, calcium, magnesium, fluoride, chloride, phosphate and sulphate in the soda pan water.
Metagenomic analysis of the Buhera soda pan water samples
In this study, a metagenomics approach was used to determine the composition and functional potential of the Buhera soda pan microbial community (Additional File 1). This involved the extraction of total metagenomic DNA, sequencing using the shotgun approach and sequence analysis on the Knowledge Base (Kbase) pipeline [33]. KBase is an integrated sequence analysis platform with an inbuilt capacity to allow the execution of a wide array of sequence analysis steps [34]. Read-based functional analysis was performed on the MG-RAST pipeline. The metagenomic workflow is described in detail in the following sections.
Metagenomic DNA extraction and sequencing
Before DNA extraction, the sample was thawed and filtered using sterile mutton cloth to get rid of excess debris and then the filtrate was centrifuged in a refrigerated centrifuge at 4 °C to collect all cells and any DNA that may have been released by lysed cells. Total metagenomic DNA was extracted from 250 ml of the sample using the ZymoBIOMICS DNA Miniprep Kit (Zymo Research, CA, USA), following manufacturer’s instructions. The concentration and quality of the extracted metagenomic DNA was determined fluorometrically (Qubit Fluorometer, Invitrogen). A total of 1 μg of metagenomic DNA was randomly fragmented using a Covaris instrument (Covaris Inc., Woburn, MA, USA), and then an Agencourt AMPure XP-Medium kit was used to select the metagenomic DNA fragments averaging 200–400 bp in size. The DNA fragments were then end-repaired using the enzyme T4 polynucleotide kinase (T4 PNK) in 100 μl of master mix containing 88 μl of 1x T4 DNA ligase buffer (New England Biolabs – NEB, Ipswich, MA, USA), 2 μl of 25 mM dNTP mix, 5 μl of 10 U/μl T4 PNK (NEB), 4 μl of 3 U/μl T4 DNA polymerase I (NEB), and 1 μl of 5 U/μl Klenow fragment of DNA polymerase I (NEB), with the reaction being incubated at room temperature for 30 minutes. Polyadenylation of the fragments was performed using 100 μl of master mix comprising 90 μl of 1x NEB buffer 2 (NEB), 5 μl of 10 mM dATP, and 5 μl of 5 U/μl Exo-Minus Klenow DNA Polymerase (NEB), and the mixture was incubated at 37°C for 30 minutes. A 1x solid-phase reversible immobilization (SPRI) clean-up was performed with elution in 100 µl of 1x NEB ligation buffer (NEB). Afterwards, 2 μl of NEB DNA Quick Ligase (NEB) and 3 μl of indexed DNA adapters (Illumina, San Diego, CA, USA) were added and mixed thoroughly. The library was amplified using a TruSeq DNA HT Sample Preparation Kit (Illumina, San Diego, CA, USA) following the TruSeq DNA Sample Preparation Guide (#15026486 Rev. C, July 2012). The PCR conditions were as follows: initial denaturation at 98°C for 30 seconds; 10 cycles of denaturation at 98°C for 10 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 30 seconds; and a final extension at 72°C for 5 minutes. PCR products were purified with an Agencourt AMPure XP-Medium Kit. The double-stranded PCR products were heat denatured and circularized by the splint oligo sequence. The single-strand circular DNA (ssCir DNA) fragments were formatted as the final library. The qualified libraries were sequenced by the BGISEQ-500 process at BGI facilities (China), in which a ssCir (single-stranded circular) DNA molecule forms a DNA nanoball (DNB) containing more than 300 copies through rolling-cycle replication. The DNBs were loaded into the patterned nanoarray using high-density DNA nanochip technology. Finally, paired-end reads 100 bp in length were obtained by combinatorial probe-anchor synthesis (cPAS).
Metagenomic sequence quality assessment and trimming
The quality of the raw sequence reads obtained from the high-throughput sequencing process was checked using the sequence quality assessment application FastQC (v0.11.9) on the KBase platform with the default settings. The QC-checked reads were trimmed using the sequence trimming application Trimmomatic (v0.36) with the following parameter settings: leading minimum quality – 3; minimum length – 36; sliding window minimum quality – 15; sliding window size – 4; trailing minimum quality – 3; and adapters – TruSeq 2-PE. The Trimmomatic software was configured to detect and remove TruSeq adapters since they were used during library creation prior to sequencing. Further quality checks of the resulting sequences were performed using FastQC. This operation produced clean reads devoid of environmental DNA, adapter sequences and low-quality bases, reads and read ends, and the clean sequences were used for further bioinformatic analysis.
Read-based taxonomic classification and community analysis
Clean unassembled QC-checked sequence reads were subjected to taxonomic classification using the Kaiju application (v1.7.3) with the following parameter settings: taxonomic level – all; reference database – RefSeq Genomes (No Euks); subsample percentage – 10%; minimum match length – 11; mode – greedy mode (allow imperfect matches); and greedy maximum mismatches – 5. Kaiju is a protein-based taxonomic classifier that allocates sequence reads to taxonomic groups such as domains, phyla, classes, orders, families, genera and species [35].
To determine species abundance and assess the adequacy of sampling and sequencing, rarefaction analysis of the sample at the species level was performed using the MG-RAST pipeline [36]. The following alpha (within sample) diversity indices were calculated in base R using species abundance data obtained from the Kaiju taxonomic classification report: Species Richness, Shannon Index (H'), Simpson’s Diversity Index (D), Simpson’s Complement (1-D), Pielou’s Evenness Index (J), Chao1 index and the abundance-based coverage estimator (ACE) metric [29]. Species Richness, ACE and Chao1 indices measure species richness in an environment, that is the total number of observed species in an environment [37]. On the other hand, Shannon, Simpson and Simpson complement indices measure both evenness and richness, and Pielou’s evenness index measures species evenness, that is how evenly spread across different species the members of a community are. Tin this study, the Shannon, Simpson, Simpson complement and Pielou’s indices were calculated based on the abundances of species with a minimum relative abundance of 0.1%, whereas Species Richness, ACE and CHAO1 were calculated based on the prevalence data of all species present.
Read-based functional profiling of the sequences
Functional annotation of the reads was performed on the MG-RAST pipeline, an end-to-end metagenomic sequence analysis platform capable of performing multiple analyses such as taxonomic classification and functional profiling of raw and assembled sequence reads [36]. MG-RAST was used to perform KEGG pathway analysis, at default settings, in order to predict the biological functions of the Buhera soda pan microbial community. Additionally, the Buhera soda pan metagenomic sequences were screened for the presence of carbohydrate-active (CAZy) enzymes in the CAZy database. The CAZy database houses structurally related domains of enzymes involved in the creation, modification or degradation of glycosidic bonds, which are often found in carbohydrate-active enzymes [38].
Results and Discussion
Buhera soda pans are highly alkaline and moderately saline
Physicochemical analysis of the soda pan water revealed that the Buhera soda pans are highly alkaline, with pH values in the range of 8.74 to 11.03 (Table 1).
Table 1.
Physicochemical characteristics of the Buhera soda pan water
| Parameter | Level |
|---|---|
| pH | 8.74 – 11.03 |
| Salinity (g/L) | 2.94 – 7.55 |
| Carbonate ion content (mg/L) | 3625.00 |
| Bicarbonate ion content (mg/L) | 1325.00 |
| Iron (mg/L) | 11.76 |
| Sodium (mg/L) | 31.23 |
| Magnesium (mg/L) | 0.002 |
| Potassium (mg/L) | 4.56 |
| Calcium (mg/L) | 7.15 |
| Fluoride (mg/L) | 71.74 |
| Chloride (mg/L) | 481.00 |
| Phosphate (mg/L) | 406.4164 |
| Sulphate (mg/L) | 1118.6089 |
The high pH of soda pans and lakes is usually due to the presence of high levels of carbonate and bicarbonate ions [13]. The ions create an alkaline pH when dissolved in water while providing a buffering effect, leading to the creation of stable alkaline conditions in the water. In fact, high levels of both carbonate and bicarbonate ions were detected in the Buhera soda pans. Apart from the high pH, the soda pans were shown to be moderately saline according to the water salinity standards of the United States Geological Service (USGS) [39]. The pH and salinity observed within the soda pans closely match findings from other soda pans and lakes around the world. A survey of soda lakes and pans in the Carpathian Basin revealed that pans have pH values in the range of 7.5–10.2 and salinities in the range of 0.7–18.9 g/L [3]. Other soda lakes with similar chemical properties include Lake Magadi in Kenya, with a salinity of 220 g/L and pH of 10.2–11.0 [2]; Lake Wadi Natrum in Egypt, with a salinity of 200–380 g/L and pH of 9.5–10.3 [14]; and Langaco Lake in the Qinghai‑Tibet Plateau in China, with a salinity of 1 g/L and pH of 8.54–8.62 [7]. In general, soda pans and lakes have alkaline pH values in the range of 8–11 and highly variable salinities, with a lower threshold of approximately 1 mg/L [1].
Buhera soda pans are also dominated by the anions sulphate, phosphate, chloride and fluoride, with sodium dominating the cations at a concentration of 31.23 mg/L (Table 1). Although the chemical composition of soda pans and lakes varies significantly, most saline-alkaline waters are dominated by the ions Ca2+, Mg2+, Na+, K+, HCO3-, CO32-, Cl- and SO42- [1]. The differential distribution of these ions in soda pan or lake water can be used in the classification of aquatic systems. According to a method for the classification of saline-alkaline waters described by Valyashko [40], a set of parameters called metamorphic coefficients (K1, K2, K3 and K4) can be used to determine whether a soda pan or lake is a carbonate, sulphate (with two subtypes; sodium sulphate or magnesium sulphate) or chloride type. The coefficient K1 (soda index) indicates carbonate type and is calculated using Eq. 3.
| 3 |
When K1>1, the water is carbonate. Using this classification system, the soda index (K1) for the Buhera soda pans was found to be 692.5, indicating that the soda pans are carbonate. In general, carbonate-type soda pans and lakes are dominated by carbonate and bicarbonate ions. Examples of such soda pans and lakes include Langaco Lake in the Qinghai‒Tibet Plateau, China [7], Lake Magadi in Kenya [5] and Lake Natron in Tanzania [41].
The physicochemical characteristics of the Buhera soda pans, as shown in this study, confirm an earlier hypothesis that these soda pans are an extremely alkaline and moderately saline environment. A high pH and moderate salinity predispose soda pans to inhabitation by extremophiles such as halophiles, alkaliphiles and haloalkaliphiles. These organisms are adaptable to extreme alkalinity and/ or salinity conditions that exist in soda pans. In general, the geochemistry of soda pans and other saline-alkaline environments plays a key role in shaping their microbial communities.
Metagenomic sequence quality
A quality assessment of the sequence reads obtained in this study revealed that the sequences were of high quality, with per-base sequence quality scores ranging from 34 to 36 and read quality scores ranging from 24 to 28 (Additional File 2). The lowest read score observed in this study was 24, which translates to a base-call accuracy probability of 0.996. Sequence quality assessment is crucial because it ensures that only high-quality sequence reads are used during sequence analysis. Raw sequence reads sometimes possess a number of aberrations, such as low-quality bases or regions, adapter sequences and environmental DNA, that may need to be removed before sequence analysis can commence.
Microbial community structure of the Buhera soda pan
The structure of the Buhera soda pan microbial community was evaluated by determining the distribution of different microbial taxa as described in sections 3.3.1 – 3.4.
The Buhera soda pans are dominated by bacteria
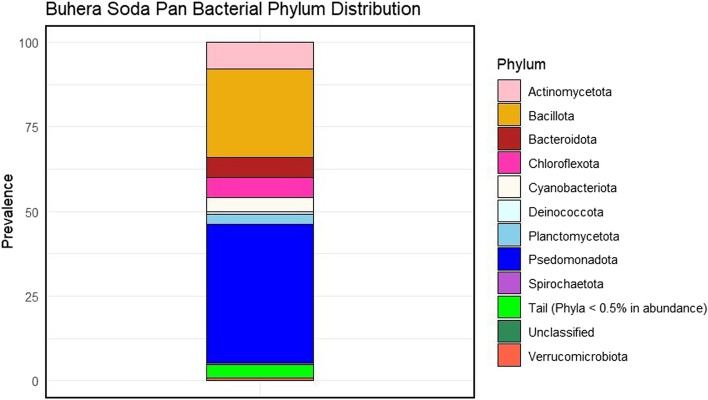
Domain analysis revealed that Bacteria dominated the Buhera soda pan microbial community, accounting for 98.5% (7,965,425 out of 8,087,434 reads evaluated) of the soda pan microbiota (Additional File 3). In addition to bacteria, small quantities of domains Archaea (0.82%), Eukaryota (0.62%) and Viruses (0.01%) were shown to be present in the soda pans. Phylum-level taxonomic analysis revealed the presence of a diverse mix of bacterial phyla in the Buhera soda pans, with at least 39 phyla being detected. Phylum Pseudomonadota (formerly known as Proteobacteria) was the dominant phylum, accounting for 41% of all bacteria present in the soda pans, followed by phylum Bacillota (formerly known as Firmicutes), which constituted 26% of the bacteria present (Figure 2). Other key bacterial phyla detected are Actinomycetota (8%), Bacteroidota (6%), Chloroflexota (6%), Cyanobacteriota (4%) and Planctomycetota (3%). Only 0.002% of bacteria from the Buhera soda pan microbial community could not be classified at the phylum level.
Fig. 2.
Taxonomic classification of the Buhera soda pans microbial community at the phylum level. Phyla with a relative abundance of at least 0.5% are shown. The stacked bar plot was created using the ggplot2 package in R [29].
The dominance of Pseudomonadota and Bacillota in many aquatic and terrestrial saline-alkaline systems, including soda pans and lakes, has been reported [3, 8, 42]. Several studies of saline-alkaline environments have shown a trend similar to findings from this study, in which Pseudomonadota tended to be the dominant bacterial phylum. For instance, in a study of the alkaline soda Langaco Lake in the Qinghai‒Tibet Plateau in China, Pseudomonadota were found to be the dominant phylum, with a relative abundance of 42.79 – 53.70% [7]. Another study covering several soda lakes in the Tibetan Plateau of China showed that the microbial communities were dominated by Pseudomonadota (28.89%), Bacteroidota (15.67%), and Bacillota (15.64%) [8]. Although the abundances may differ from one study to another, similar observations were made in this study where Pseudomonadota were shown to dominate Buhera soda pan microbial community.
Pseudomonadota are the most taxonomically and morphologically diverse bacterial phylum [43], and most of its members are flagellated and motile [44]. Pseudomonadota play several important roles in the environment such as carbon fixation through both photoautotrophy and chemolithoautotrophy, sulfur oxidation, nitrogen fixation and organic matter decomposition, thus promoting nutrient cycling within ecosystems [44]. Judging by the many roles played by Pseudomonadota in different ecosystems, their dominance in Buhera soda pans may suggest that this ecosystem is characterized by high levels of microbial activity. The presence of the photosynthetic bacterial phylum Cyanobacteriota, which constituted 4 % of the soda pan microbiome, as well as phyla like Chloroflexota, which contains several anoxygenic photosynthetic genera [45], may point to the importance of photosynthesis as a mechanism of carbon acquisition in this ecosystem. Photosynthesis in soda pans is typically supported by the presence of large quantities of carbonate and bicarbonate ions that act as a source of carbon dioxide [1]. The Buhera soda pans were shown to possess large quantities of carbonate and bicarbonate ions, a likely source of carbon for the autotrophs detected. Photosynthetic bacteria play an important role in carbon fixation and organic matter synthesis within the soda pan microbial ecosystem.
The Buhera soda pans were shown to be dominated by the following bacterial classes: Alphaproteobacteria (18%), Clostridia (16%), Gammaproteobacteria (16%), Bacilli (8%), Actinomycetia (8%), Anaerolineae (4%), Deltaproteobacteria (4%) and Planctomycetia (3%), Additional File 4. Two of the most abundant classes in the Buhera soda pans, Clostridia (mostly anaerobic) and Bacilli (mostly obligate or facultative aerobic), both belonging to phylum Bacillota, together constitute 24% of the entire Buhera soda pan bacterial community. Bacillota are a phylum of mostly gram-positive bacteria that tend to form endospores under stressful conditions, enabling their survival under extreme habitats such as soda pans [46–48]. Thus, their abundance in the Buhera soda pans may be partly explained by their ability to survive under the harsh conditions of the soda pans.
Haloalkaliphilic taxa are present in the Buhera soda pans
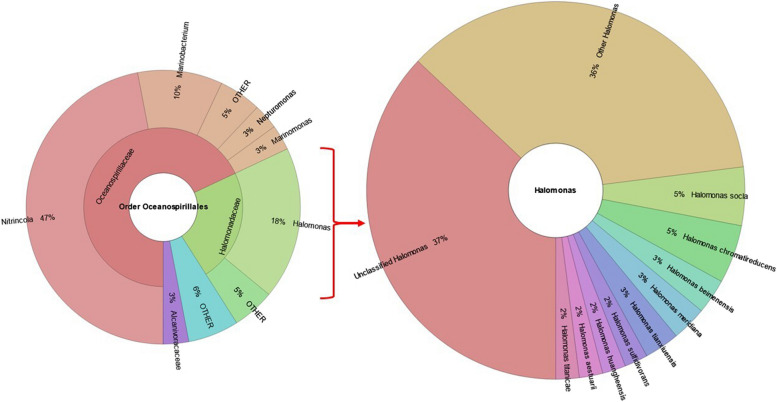
Of particular interest to this study is the discovery of the generally haloalkaliphilic order Oceanospirillales (phylum – Pseudomonadota, class – Gammaproteobacteria) in the Buhera soda pans, albeit at a modest prevalence of 1% of the entire microbial community, affirming the Buhera soda pans as a viable saline-alkaline microbial habitat. Oceanospirillales are an order of morphologically diverse bacteria, being either rod-shaped, curved or spiral, that are generally hydrolytic and perform aerobic biodegradation of a variety of organic compounds, and are either halotolerant or halophilic, having a specific requirement for elevated salt levels to sustain normal growth, and are commonly associated with marine or saline-alkaline environments such as soda pans and lakes [49]. The Oceanospirillales were shown to be important in the biodegradation of hydrocarbons during the Deep Horizon oil spill, highlighting the potential of this group of bacteria for use in the bioremediation of oil spills and other hydrocarbon pollutants under saline and alkaline conditions [50]. Although this order contains many different bacterial families, the two families that were most prevalent in the Buhera soda pans are Oceanospirillaceae (68% of Oceanospirillales found in the Buhera soda pans), and Halomonadaceae (23% of Oceanospirillales), collectively accounting for 91% of all the Oceanospirillales in the soda pans (Figure 3). Genera belonging to the Oceanospirillaceae family identified in the Buhera soda pans include Nitrincola (69%), Marinobacterium (15%), Neptunomonas (4%), Marinomonas (4%), Thalassolituus (0.8%), Bacterioplanes (0.6%) and Bermanella (0.4%). Nitrincola, the most abundant genus under order Oceanospirillales in the Buhera soda pans, is a genus of rod-shaped, Gram-negative, alkaliphilic, halotolerant and chemoorganoheterotrophic bacteria that have a specific requirement for salt for growth and are commonly isolated in soda pans and lakes [51, 52]. Meanwhile, the most dominant genus belonging to the haloalkaliphilic family Halomonadaceae was found to be Halomonas, which constituted 80% of members of this family in the Buhera soda pans (Fig. 3).
Fig. 3.
Krona charts showing the distribution of the haloalkaliphilic bacterial order Oceanospirillales and genus Halomonas in the Buhera soda pans. The Krona charts were created using Krona open-source software in excel [53].
Halomonas is a genus of halotolerant or halophilic, Gram-negative, rod-shaped bacteria that generate energy mostly through aerobic respiration and are commonly found in salty environments such as soda pans, soda lakes and marine waters [54]. Species belonging to this genus found in Buhera soda pans include, in order of abundance, Halomonas socia (5%), H. chromatireducens (5%), H. beimenensis (3%), H. meridiana (3%), H tianxiuensis (3%), H. sulfidivorans (2%), and H. aestuarii (2%). Over a third (37%) of bacteria belonging to genus Halomonas found in Buhera soda pans could not be classified at species level. Similarly, unclassified organisms were observed at all taxonomic levels in the Buhera soda pans. However, the proportion of unclassified microorganisms was found to be low at higher taxonomic levels and relatively higher at lower taxonomic levels. For instance, all organisms in this study were classified at domain level and only 0.002% could not be classified at phylum level. However, the proportion of unclassified microorganisms jumped to approximately 9% at class, 10% at genus and 20% at species level. The discovery of unclassified microbes points to the potential existence of previously undescribed microorganisms in the Buhera soda pans, highlighting the importance of this ecosystem as a habitat for novel microbiota. Future research efforts will be directed towards the discovery and possible characterisation of such novel microbes, and an evaluation of their biotechnological potential.
Vibrio and other haloalkaliphilic genera dominate the Buhera soda pans
Sequences belonging to over 1500 bacterial genera were detected in the Buhera soda pans, highlighting this habitat as an extremely rich and diverse microbial habitat. The vast majority of these genera were present in low relative abundances of less than 0.5% each, but collectively they constituted 60.9% of assigned reads. On the contrary, only 24 genera had a relative abundance of at least 0.5 % each, accounting for a combined total of 29.1% of assigned reads. Despite their often-extreme physicochemical conditions such as alkaline pH and high salinity, soda pans and lakes have been shown to harbour highly diverse microbial communities composed of different taxonomic groups and capable of mediating a wide range of biochemical transformations [9, 10]. Additionally, 10% of the reads could not be assigned to a genus, possibly suggesting the presence of novel bacterial genera in the Buhera soda pans.
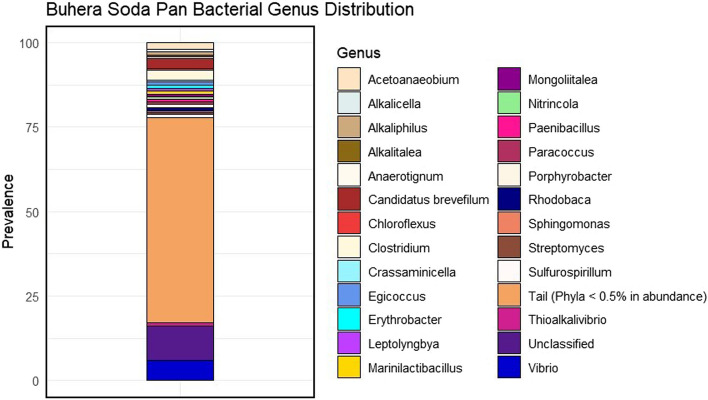
The most abundant bacterial genus in the Buhera soda pans was shown to be Vibrio, which accounted for 6% of all bacterial reads analysed (Figure 4). Other abundant genera detected include Candidatus Brevefilum (3%), Clostridium (3%), Acetoanaerobium (2%), Thioalkalivibrio (1%), Marinilactibacillus (1%), Erythrobacter (1%) and Alkaliphilus (1%).
Fig. 4.
Distribution of microbial genera in the Buhera soda pans. The stacked bar plot was created using the ggplot2 package in R [29].
The dominant genus in Buhera soda pans, Vibrio (phylum – Pseudomonadota, class – Gammaproteobacteria, order – Vibrionales, family –Vibrionaceae), is a genus of Gram-negative, motile, curved, rod-shaped, free-living bacteria that are ubiquitous in aquatic environments and have a special affinity for warm and salty marine aquatic environments [55]. The Vibrio are heterotrophic and generally halophilic, deriving carbon from the breakdown of organic compounds and obtaining energy through both aerobic respiration and fermentation [55–57]. The Vibrio genus has the largest number of species of all Bacteria [55], with 152 child taxa recognized to date [58]. At least 56 species belonging to the Vibrio genus were found in the Buhera soda pans, with the most abundant being Vibrio metshnikovii (69% of the Vibrio genus), V. cincinnatiensis (4%), V. parahaemolyticus (2%), V. cholerae (1%), V. anguillarum (0.7%) and V. vulnificus (0.5%). To date, 13 species belonging to the Vibrio genus have been shown to be associated with pathogenicity in humans [59]. Examples of pathogenic species found in the Buhera soda pans are V. parahaemolyticus, V. cholera, V. metshnikovii and V. vulnificus.
In addition to Vibrio, several other bacterial genera that are generally haloalkaliphilic and commonly associate with saline-alkaline environments were also found in Buhera soda pans. These include Alkalicella, Alkaliphilus, Alkalitalea, Thioalkalivibrio, Marinilactibacillus, Mongoliitalea, Desulfovibrio and Sulfurospirillum (Figure 4). The abundance of these haloalkaliphiles highlights the Buhera soda pans as a viable habitat for haloalkaliphilic bacteria. Some of these bacterial genera play key functions in the microbial community, a case in point being the genera Thioalkalivibrio, Desulfovibrio and Sulfurospirilum that participate in the cycling of different sulphur compounds under the saline-alkaline conditions of the soda pans, a hypothesis that can be explained by the large quantities of sulphates detected in the Buhera soda pans. A key example, Thioalkalivibrio, is an obligate genus of chemolithoautotrophic, haloalkaliphilic, sulphur-oxidising bacteria commonly inhabiting soda pans and lakes, among other saline-alkaline environments [23]. The specific members of the genus Thioalkalivibrio found in Buhera soda pans include Thioalkalivibrio paradoxus (28%), T. nitratireducens (27%), T. sulfidiphilus (6%) and T. versutus (4%).
At least 6400 bacterial species were shown to be present in the Buhera soda pans, affirming this ecosystem as a rich and diverse microbial habitat. The most abundant bacterial species was Vibrio metchnikovii, with a relative abundance of 4% of all bacteria present in the soda pans. Other abundant species found are Candidatus Brevefilum fermentans (3%), Acetoanaerobium sticklandii (2%), unclassified Marinolactibacillus sp. 15R (1%), Egicoccus halophilus (1%), Rhodobaca bogoriensis (0.9%), unclassified Leptolyngbya sp. BL0902 (0.8%) and Nitrincola iocasae (0.7%). The most prevalent bacterial species in the Buhera soda pans, Vibrio metshnikovii, is halophilic and ubiquitous in both marine and freshwater environments, and has been shown to be pathogenic to aquatic animals such as fish and to be partially zoonotic and has been reportedly isolated from seafood [59, 60].
Distribution of Archaea in the Buhera soda pans
In addition to bacteria, Archaea were also found in Buhera soda pans, although they constituted only 1% of the microbes present. The Archaea are a stand-alone domain of prokaryotic microorganisms that differ significantly from the other domain of prokaryotic microorganisms, Bacteria. Archaea are known to inhabit some of the world’s most extreme environments such as hot springs, volcanic craters, soda pans and lakes, and salt lakes [61]. While archaea generally tend to inhabit extreme environments, the relative abundance of domain Archaea in soda pans and lakes differs significantly from as low as 0.3% [12, 62] to as high as 92% [63]. The presence of Archaea in Buhera soda pans, albeit at a low prevalence, demonstrates their ability to survive under stressful conditions. The Archaea were dominated by the archaeal phylum Euryarchaeota, which accounted for 93% of the total Archaea present (Additional File 5).
The most prevalent archaeal phylum in Buhera soda pans, the Euryarchaeota, is a diverse archaeal phylum that hosts various groups of extremophiles such as methanogens, heat-loving thermophiles and salt-loving halobacteria, and commonly inhabit hot springs, volcanic craters, deep-sea geothermal vents, and soda pans and lakes [64]. The dominant genus in the archaeal phylum Euryarchaeota found in the Buhera soda pans include Methanobacterium (44% of Euryarchaeota), represented by the species M. lacus, M. congolense, M. paludis, M. formicicum and M. subterraneum. Other notable genera present are Methanosarcina (6%), Methanothermobacter (9%), Methanothrix (5%) and Methanobrevibacter (3%). Of note is the presence of archaea belonging to the genus Natronomonas in soda pans, as shown by the presence of the species Natronomonas pharaonis, N. salina, N. halophila and N. moolapensis. Although Archaea are predominantly unculturable, with most of their properties confirmed by DNA sequence analysis, some haloalkaliphilic members of the Natronomonas genus have been previously isolated. A case in point is the isolation of Natronomonas pharaonis, a polyextremophilic archaeon with an optimum pH for growth of 8.5 that is capable of surviving up to pH 11 [65].
Microbial community analysis and diversity estimation
To determine species abundance and assess the adequacy of sampling and sequencing, rarefaction analysis of the sample at the species level was performed using the MG-RAST pipeline [36], and the resulting rarefaction curve is shown in Additional File 6 (Additional Material). The rarefaction curve is a plot of the total number of distinct species annotations in a sample as a function of the number of sequences sampled. A steep slope on the left indicates that a large proportion of the species in the sampled environment has yet to be discovered. However, if the curve flattens to the right, it suggests the adequacy of the sample size, and more intensive sampling is unlikely to yield a significant number of new species. Rarefaction curves are an indicator of species abundance. The curve obtained in this study (Additional File 6) clearly shows that sufficient sequencing was performed, and near-species saturation was achieved. No reasonable prospects exist for finding a significant number of new species by sequencing more reads than were sampled in this study.
Alpha diversity analysis, performed using a number of different estimators revealed that the Buhera soda pan microbial community is highly diverse, with a high Species Richness of 6446 (Table 2).
Table 2.
Microbial diversity measures for the Buhera soda pan microbial community. All computations were performed in base R
| Diversity Index | Value |
|---|---|
| Richness | 6446 |
| Shannon | 3.81 |
| Simpson (D) | 0.04 |
| Simpson’s complement (1-D) | 0.96 |
| Pielou | 1.27 |
| ACE | 6518 |
| CHAO1 | 6534 |
While Species Richness is a commonly used diversity measure, it does not show the level of diversity in a habitat, and occasionally underestimates the actual number of species present in an environment, especially when there is a high prevalence of rare members of the community [36]. Such rare members are often missed in computations of species richness, producing lower estimates of species richness in a sample. This study produced a large number of rare members of the microbial community, creating a need to correct the observed Species Richness estimate in order to account for rare members that may have been missed. Consequently, the two richness indices, Chao1 and ACE, were used to make this correction, producing more realistic estimates of the number of species present [66]. Both ACE and Chao1 produced relatively higher estimates of species richness. The Shannon Index, also known as Shannon Entropy, provides a measure of species evenness in a sample, the Simpson index is a measure of the probability that two randomly chosen individuals present in a sample at the same time belongs to the same species, whereas the Pielou evenness index measures how evenly distributed different species are in a sample [67]. A modified Simpson index, called Simpson’s complement, provides a more direct measure of diversity and ranges from 0 (no diversity) to 1 (maximum possible diversity). In summary, all indices used in this study showed that the Buhera soda pan microbial community is very rich and highly diverse, an observation that is supported by literature affirming the fact that soda pans and lakes are highly productive and are capable of harbouring diverse microbiomes [9–11].
Read-based functional profiling of the Buhera soda pan microbial community
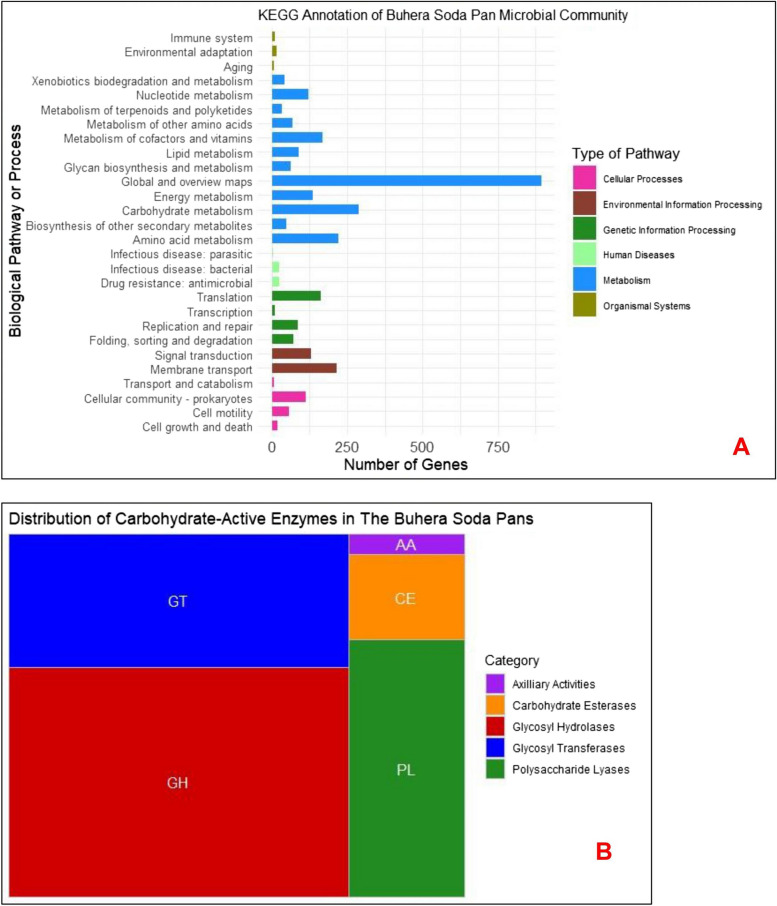
KEGG analysis of the functional profile of the microbial community of Buhera soda pan revealed that most (69.48%) of the annotated functional genes were associated with microbial metabolic processes such as amino acid biosynthesis and metabolism, carbohydrate and energy metabolism, lipid metabolism, nucleotide metabolism, secondary metabolite biosynthesis, cofactor and vitamin metabolism, and xenobiotic biodegradation and metabolism (Figure 5A).
Fig. 5.
Functional profiling of the Buhera soda pan microbial community: (A) KEGG annotation of Buhera soda pan metagenomic DNA sequences. The plot was created using the ggplot2 package in R [29]; (B) Treemap plot showing the distribution of genes containing CAZy-modules in the Buhera metagenomic sequence library. The plot was created using the ggplot2 and treemapify packages in R [29].
KEGG analysis also revealed an abundance of pathways responsible for genetic (10.60%) and environmental information processing (11.05%). Genetic information processing involves all processes related to the organization and replication of DNA, gene expression and its regulation, while environmental information processing involves pathways responsible for transmembrane transport and signal transduction. Transmembrane transport systems are key to the adaptation of haloalkaliphiles under saline-alkaline conditions, as such systems ensure efficient translocation of sodium and chloride, protons and hydroxyl ions in and out of cells as microbial cells respond to the alkaline pH and the presence of salt and in the environment [68]. In general, haloalkaliphiles pump sodium out of their cells in order to maintain an optimum intracellular solute potential. Genes for pathways associated with human disease were also detected, albeit at a low prevalence of 1.63%. These genes were most likely located in the few pathogenic bacteria and viruses detected in the soda pans.
All five classes of carbohydrate-active (CAZy) modules were detected within the Buhera soda pan bacterial functional gene pool, with glycosyl hydrolases (GHs) being the most abundant (47.3%), followed by glycosyltransferases (27.3%). These two classes of enzymes accounted for three-quarters (74.6%) of the CAZy genes detected in the microbial community (Figure 5(B)). Glycosyl hydrolases are enzymes that break glycosidic bonds in carbohydrates and include important enzymes such as amylases, cellulases and xylanases, while glycosyltransferases transfer monosaccharide units to carbohydrates during carbohydrate synthesis. Haloalkaliphilic amylases, cellulases and xylanases, among other CAZy enzymes, have potential for application in industrial processes such as detergent making, the biotreatment of saline-alkaline industrial effluent, starch saccharification, and the dehairing of animal skins during leather tanning, among other industrial applications [2, 5, 69–71]. Thus, the abundance of genes encoding carbohydrate-synthesizing and carbohydrate-hydrolyzing enzymes in Buhera soda pans demonstrates the potential of this extreme habitat to serve as a source of useful microbial enzymes with unique properties and potential for application in bioprocessing. More work needs to be done towards the recovery, characterisation and application of these enzymes. Apart from their apparent biotechnological potential, CAZy enzymes are also important in the ability of microorganisms to utilize and metabolise a wide range of carbohydrates and other carbon sources in their natural environment, allowing better colonisation of habitats and establishment of stable microbial communities.
Conclusion
This study investigated the physicochemical properties and microbial diversity of Buhera soda pans. The soda pans were shown to be highly alkaline and moderately saline, and to be of the carbonate type. The soda pans pan microbial community was shown to be highly diverse and rich, with Bacteria being the dominant domain. Bacteria belonging to phylum Pseudomonadota and Bacillota were the most abundant. Several members of the predominantly haloalkaliphilic order Oceanospirillales were detected in Buhera soda pans, represented by key halophilic/haloalkaliphilic genera such as Nitrincola, Marinobacterium, Marinomonas, Halomonas, Halotalea and Salinicola. Vibrio was the most abundant genus, accompanied by other genera such as Candidatus Brevefilum, Clostridium, Acetoanaerobium, Thioalkalivibrio, Marinilactibacillus and Alkaliphilus. A large portion of the Buhera soda pan microbial community could not be classified beyond certain taxonomic levels, signifying the potential of the Buhera soda pans as a habitat for novel microbiota. The soda pan microbial community was shown to be metabolically diverse, with the capacity to execute a wide array of important geochemical transformations. The microbiome was shown to harbour significant carbohydrate-metabolising potential, and thus could be explored for the presence of potentially useful glycosidases and other carbohydrate-active enzymes.
Supplementary Information
Additional file 1: An overview of the methods used for the collection, sequencing and analysis of metagenomic DNA from Buhera soda pans.
Additional file 2: Metagenomic sequence read statistical quality parameters.
Additional file 3: Domain-level taxonomic classification of the Buhera soda pan microbial community.
Additional file 4: Class-level taxonomic classification of the Buhera soda pans microbial community.
Additional file 5: Distribution of Archaea in the Buhera soda pans.
Additional file 6: Rarefaction analyses of the Buhera soda pan microbial community at the species level.
Acknowledgments
This work was carried out in the Biochemistry, Microbiology and Biotechnology laboratories at the National University of Science and Technology, Bulawayo, Zimbabwe. Partial geochemical analysis was performed in the Biological Sciences and Biotechnology Department, Botswana International University of Science and Technology, Palapye, Botswana. The authors would like to acknowledge the Gombahari village leadership and the local community for granting them access to the soda pans.
Authors’ contributions
N.M, N.Z, and T.N conceptualized the project, sourced project funds, and designed the methodology, N.M collected and analyzed data, N.Z and T.N supervised data collection and analysis, N.M, N.Z, and T.N interpreted results, N.M wrote the manuscript, all authors reviewed the manuscript and N.M edited the draft manuscript. All authors consent to the publishing of the manuscript.
Funding
This work was funded by the Research and Development Board (RDB) of the National University of Science and Technology, Bulawayo, Zimbabwe. Funds for metagenomic sequencing were provided by the Botswana International University of Science and Technology Research Office.
Data availability
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics, 2021) in National Genomics Data Centre (Nucleic Acid Research, 2022), China National Centre for Bioinformation/ Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA016626) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Boros E, Kolpakova M. A review of the defining chemical properties of soda lakes and pans: an assessment on a large geographic scale of Eurasian inland saline surface waters. PLoS ONE. 2018;13(8): e0202205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sorokin DY, Muntyan MS, Toshchakov SV, Korzhenkov A, Kublanov IV. Phenotypic and genomic properties of a novel deep-lineage haloalkaliphilic member of the phylum Balneolaeota from soda lakes possessing Na+-translocating proteorhodopsin. Front Microbiol. 2018;9:2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Felföldi T. Microbial communities of soda lakes and pans in the Carpathian Basin: A Review. Biologia Futura. 2020;71(4):393–404. [DOI] [PubMed] [Google Scholar]
- 4.Jeilu O, Gessesse A, Simachew A, Johansson E, Alexandersson E. Prokaryotic and eukaryotic microbial diversity from three soda lakes in the East African Rift Valley determined by amplicon sequencing. Front Microbiol. 2022;13: 999876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nyakeri EM, Mwirichia R, Boga H. Isolation and characterization of enzyme producing bacteria from Lake Magadi, an extreme soda lake in Kenya. J Microbiol Experiment. 2018;6(2):57–68. [Google Scholar]
- 6.Kaya M, Yildirim BA, Kumral M, Sasmaz A. Trace and Rare Earth Element (REE) geochemistry of recently formed stromatolites at Lake Salda, SW Turkey. Water. 2023;15(4):733. [Google Scholar]
- 7.Wang M, Zhang X, Shu Z, Wang Z, Tao Y, Lv C, Zhu D, Shen G. Bacterial and archaeal communities within the alkaline soda Langaco Lake in the Qinghai-Tibet Plateau. Annals Microbiol. 2022;72(1):33. [Google Scholar]
- 8.He Y, He L, Wang Z, Liang T, Sun S, Liu X. Salinity shapes the microbial communities in surface sediments of salt lakes on the Tibetan Plateau, China. Water. 2022;14(24):4043. [Google Scholar]
- 9.Govil T, Sharma W, Salem DR, Sani RK. Multi-Omics Approaches for Extremophilic Microbial, Genetic, and Metabolic Diversity. In: Pandey A, Sharma A, editors. Extreme Environments: Unique ecosystems - Amazing microbes. First Edition. Boca Raton: CRC Press; 2021. p. 311-329.
- 10.Sysoev M, Grötzinger SW, Renn D, Eppinger J, Rueping M, Karan R. Bioprospecting of novel extremozymes from prokaryotes—The advent of culture-independent methods. Front Microbiol. 2021;12: 630013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan TW, Lin YJ, Ou MY, Chen KB. Isolation and diversity of sediment bacteria in the hypersaline aiding lake, China. PloS ONE. 2020;15(7): e0236006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryanskaya AV, Shipova AA, Rozanov AS, Kolpakova OA, Lazareva EV, Uvarova YE, Efimov VM, Zhmodik SM, Taran OP, Goryachkovskaya TN, Peltek SE. Diversity and metabolism of microbial communities in a hypersaline lake along a geochemical gradient. Biol. 2022;11(4):605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerreiro RL, Bergier I, McGlue MM, Warren LV, de Abreu UG, Abrahão J, Assine ML. The soda lakes of Nhecolândia: A conservation opportunity for the Pantanal wetlands. Perspect Ecol Conserv. 2019;17(1):9–18. [Google Scholar]
- 14.Melton ED, Sorokin DY, Overmars L, Chertkov O, Clum A, Pillay M, Ivanova N, Shapiro N, Kyrpides NC, Woyke T, Lapidus AL. Complete genome sequence of Desulfurivibrio alkaliphilus strain AHT2 T, a haloalkaliphilic sulfidogen from Egyptian hypersaline alkaline lakes. Stand Genom Sci. 2016;11:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selim SA, El-Alfy SM, Hagagy NI, Hassanin AA, Khattab RM, El-Meleigy EA, Aziz MH, Maugeri TL. Oil-biodegradation and biosurfactant production by haloalkaliphilic Archaea isolated from Soda Lakes of the Wadi An Natrun. Egypt J Pure Appl Microbiol. 2012;6(3):1011–20. [Google Scholar]
- 16.Grant W, Gerday C, Glansdorff N. Alkaline environments and biodiversity. In: Extremophiles Volume III. 2006;21-39. EOLSS publishers.
- 17.Musikoyo EO. Distribution, Isolation and Characterization of Bacteria with Industrial Potential in Lake Nakuru. Kenya: Kenya. Master of science research project in limnology. Egerton University; 2012. p. 1–89. [Google Scholar]
- 18.Mukhtar S, Malik AK, Mehnaz S. Isolation and characterization of haloalkaliphilic bacteria isolated from the rhizosphere of Dichanthium annulatum. J Adv Res Biotechnol. 2018;3(1):1–9. [Google Scholar]
- 19.Uma G, Babu MM, Prakash VS, Nisha SJ, Citarasu T. Nature and bioprospecting of haloalkaliphilics: a review. World J Microbiol Biotechnol. 2020;36:1–3. [DOI] [PubMed] [Google Scholar]
- 20.Simair AA, Qureshi AS, Khushk I, Ali CH, Lashari S, Bhutto MA, Mangrio GS, Lu C. Production and partial characterization of α-amylase enzyme from Bacillus sp. BCC 01–50 and potential applications. BioMed Res Int. 2017;1:9173040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choure K, Parsai S, Kotoky R, Srivastava A, Tilwari A, Rai PK, Sharma A, Pandey P. Comparative metagenomic analysis of two alkaline hot springs of Madhya Pradesh, India and deciphering the extremophiles for industrial enzymes. Frontiers in Genetics. 2021;12: 643423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makhdoumi A. Bacterial diversity in south coast of the Caspian Sea: Culture-dependent and culture-independent survey. Caspian J Environ Sci.. 2018;16(3):259–69. [Google Scholar]
- 23.Ahn AC, Meier-Kolthoff JP, Overmars L, Richter M, Woyke T, Sorokin DY, Muyzer G. Genomic diversity within the haloalkaliphilic genus Thioalkalivibrio. PLoS ONE. 2017;12(3): e0173517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El Hidri D, Guesmi A, Najjari A, Cherif H, Ettoumi B, Hamdi C, Boudabous A, Cherif A. Cultivation-dependant assessment, diversity, and ecology of haloalkaliphilic bacteria in arid saline systems of southern Tunisia. BioMed Res Int. 2013;2013(1): 648141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin M, Gai Y, Guo X, Hou Y, Zeng R. Properties and applications of extremozymes from deep-sea extremophilic microorganisms: a mini review. Marine drugs. 2019;17(12):656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grötzinger SW, Karan R, Strillinger E, Bader S, Frank A, Al Rowaihi IS, Akal A, Wackerow W, Archer JA, Rueping M, Weuster-Botz D. Identification and experimental characterization of an extremophilic brine pool alcohol dehydrogenase from single amplified genomes. ACS Chem Biol. 2018;13(1):161–70. [DOI] [PubMed] [Google Scholar]
- 27.Samarasinghe SN, Wanigatunge RP, Magana-Arachchi DN. Bacterial diversity in a Sri Lankan geothermal spring assessed by culture-dependent and culture-independent approaches. Curr Microbiol. 2021;78:3439–52. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Chen F, Zeng Z, Xu M, Sun F, Yang L, Bi X, Lin Y, Gao Y, Hao H, Yi W. Advances in metagenomics and its application in environmental microorganisms. Front Microbiol. 2021;12: 766364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R Core Team. R: A language and environment for statistical computing. Foundation for Statistical Computing, Vienna, Austria. 2013.
- 30.Google. 2024. Buhera Soda Pans. Google maps. Available at: https://www.google.com/maps/place/19%C2%B015'48.3%22S+31%C2%B048'15.7%22E/@-19.26342,31.8017951,770m/data=!3m2!1e3!4b1!4m4!3m3!8m2!3d-19.26342!4d31.80437?entry=ttu&g_ep=EgoyMDI0MDkwMy4wIKXMDSoASAFQAw%3D%3D. Accessed Sept 06 2024.
- 31.Frontmatter. Environmental Laboratory Exercises for Instrumental Analysis and Environmental Chemistry. 2004. 10.1002/0471660280.fmatter.
- 32.Indian Institute of Technology Kanpur. Methods of Sampling and Test (Physical and Chemical) for Water and Wastewater, Part 23 – Alkalinity: Amendment Number 2. India: Kanpur; 2006. [Google Scholar]
- 33.Arkin AP, Cottingham RW, Henry CS, Harris NL, Stevens RL, Maslov S, Dehal P, et al. KBase: The United States department of energy systems biology knowledgebase. Nat Biotechnol. 2018;36(7):566–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chivian D, Jungbluth SP, Dehal PS, Wood-Charlson EM, Canon RS, Allen BH, Clark MM, Gu T, Land ML, Price GA, Riehl WJ. Metagenome-assembled genome extraction and analysis from microbiomes using KBase. Nat Protocols. 2023;18(1):208–38. [DOI] [PubMed] [Google Scholar]
- 35.Menzel P, Ng KL, Krogh A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nature Communications. 2016;7(1):11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J. The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC bioinformatics. 2008;9:1-8. [DOI] [PMC free article] [PubMed]
- 37.Calle ML. Statistical analysis of metagenomics data. Genomics Inform. 2019;17(1):e6. 10.5808/GI.2019.17.1.e6. [DOI] [PMC free article] [PubMed]
- 38.Drula E, Garron ML, Dogan S, Lombard V, Henrissat B, Terrapon N. The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 2022;50:D571-577. [DOI] [PMC free article] [PubMed]
- 39.USGS: Saline water and salinity. https://www.usgs.gov/special-topics/water-science-school/science/saline-water-and-salinity. Accessed 12 Mar 2024.
- 40.Valyashko MG. Basic chemical types of natural waters and the conditions producing them. Records of the Academy USSR. 1955;102:315–8. [Google Scholar]
- 41.Lameck AS, Skutai J, Boros E. Review of chemical properties of Inland soda and saline waters in East Africa (Rift Valley region). J Hydrol: Reg Stud. 2023;46: 101323. [Google Scholar]
- 42.Omeroglu E, Sudagidan M, Yurt MN, Tasbasi BB, Acar EE, Ozalp VC. Microbial community of Soda Lake Van as obtained from direct and enriched water, sediment and fish samples. Scientific Reports. 2021;11(1):18364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moon CD, Young W, Maclean PH, Cookson AL, Bermingham EN. Metagenomic insights into the roles of Proteobacteria in the gastrointestinal microbiomes of healthy dogs and cats. Microbiology Open. 2018;7(5): e00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta RS. The phylogeny of proteobacteria: relationships to other eubacterial phyla and eukaryotes. FEMS Microbiol Rev. 2000;24(4):367–402. [DOI] [PubMed] [Google Scholar]
- 45.Ward LM, Hemp J, Shih PM, McGlynn SE, Fischer WW. Evolution of phototrophy in the Chloroflexi phylum driven by horizontal gene transfer. Front Microbiol. 2018;9:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galperin MY, Yutin N, Wolf YI, Vera Alvarez R, Koonin EV. Conservation and evolution of the sporulation gene set in diverse members of the Firmicutes. J bacteriol. 2022;204(6):e00079-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galperin MY. Genome diversity of spore‐forming Firmicutes. The bacterial spore: From molecules to systems. 2016:1-18. 10.1128/microbiolspectrum.TBS-0015-2012. [DOI] [PMC free article] [PubMed]
- 48.Filippidou S, Wunderlin T, Junier T, Jeanneret N, Dorador C, Molina V, Johnson DR, Junier P. A combination of extreme environmental conditions favor the prevalence of endospore-forming firmicutes. Frontiers in Microbiology. 2016;7:1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jensen S, Duperron S, Birkeland NK, Hovland M. Intracellular Oceanospirillales bacteria inhabit gills of Acesta bivalves. FEMS Microbiol Ecol. 2010;74(3):523–33. [DOI] [PubMed] [Google Scholar]
- 50.Beyer J, Trannum HC, Bakke T, Hodson PV, Collier TK. Environmental effects of the Deepwater Horizon oil spill: a review. Marine Pollution Bull. 2016;110(1):28–51. [DOI] [PubMed] [Google Scholar]
- 51.Borsodi AK, Korponai K, Schumann P, Spröer C, Felföldi T, Márialigeti K, Szili-Kovács T, Tóth E. Nitrincola alkalilacustris sp. nov. and Nitrincola schmidtii sp. nov., alkaliphilic bacteria isolated from soda pans, and emended description of the genus Nitrincola. Int J Syst Evol Microbiol. 2017;67(12):5159–64. [DOI] [PubMed] [Google Scholar]
- 52.Joshi A, Thite S, Kulkarni G, Dhotre D, Joseph N, Venkata Ramana V, Polkade A, Shouche Y. Nitrincola alkalisediminis sp. nov., an alkaliphilic bacterium isolated from an alkaline lake. Int J Syst Evol Microbiol. 2016;66(3):1254–9. [DOI] [PubMed] [Google Scholar]
- 53.Ondov BD, Bergman NH, Phillippy AM. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics. 2011;12(385):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye JW, Chen GQ. Halomonas as a chassis. Essays in Biochemistry. 2021;65(2):393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sampaio A, Silva V, Poeta P, Aonofriesei F. Vibrio spp.: Life strategies, ecology, and risks in a changing environment. Diversity. 2022;14(2):97.
- 56.Marques PH, Prado LC, Felice AG, Rodrigues TC, Pereira UD, Jaiswal AK, Azevedo V, Oliveira CJ, Soares S. Insights into the Vibrio genus: a one health perspective from host adaptability and antibiotic resistance to in silico identification of drug targets. Antibiotics. 2022;11(10):1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang X, Lin H, Wang X, Austin B. Significance of Vibrio species in the marine organic carbon cycle—a review. Sci China Earth Sci. 2018;61:1357–68. [Google Scholar]
- 58.https://bacterio.net
- 59.Konechnyi Y, Khorkavyi Y, Ivanchuk K, Kobza I, Sękowska A, Korniychuk O. Vibrio metschnikovii: current state of knowledge and discussion of recently identified clinical case. Clin Case Reports. 2021;9(4):2236–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiao Z, Li X, Xue M, Zhang M, Liu W, Fan Y, Chen X, Chu Z, Gong F, Zeng L, Zhou Y. Vibrio metschnikovii, a potential pathogen in freshwater-cultured hybrid sturgeon. Animals. 2022;12(9):1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaban B, Ng SY, Jarrell KF. Archaeal habitats—from the extreme to the ordinary. Canadian J Microbiol. 2006;52(2):73–116. [DOI] [PubMed] [Google Scholar]
- 62.Andreote AP, Dini-Andreote F, Rigonato J, Machineski GS, Souza BC, Barbiero L, Rezende-Filho AT, Fiore MF. Contrasting the genetic patterns of microbial communities in soda lakes with and without cyanobacterial bloom. Front Microbiol. 2018;9:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang M, Xing J, Long Q, Shen G, Zhu D, Li Y. Prokaryotic Microbial Diversity Analysis and Preliminary Prediction of Metabolic Function in Salt Lakes on the Qinghai-Tibet Plateau. Water. 2024;16(3):451. [Google Scholar]
- 64.Oren A. Euryarchaeota. Encyclopedia of Life Sciences. 2019; 1–17. 10.1002/9780470015902.a0004243.pub3.
- 65.Gonzalez O, Oberwinkler T, Mansueto L, Pfeiffer F, Mendoza E, Zimmer R, Oesterhelt D. Characterization of growth and metabolism of the haloalkaliphile Natronomonas pharaonis. PLoS Comput Biol. 2010;6(6):e1000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Madison JD, LaBumbard BC, Woodhams DC. Shotgun metagenomics captures more microbial diversity than targeted 16S rRNA gene sequencing for field specimens and preserved museum specimens. Plos ONE. 2023;18(9):e0291540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pérez-Losada M, Narayanan DB, Kolbe AR, Ramos-Tapia I, Castro-Nallar E, Crandall KA, Domínguez J. Comparative analysis of metagenomics and metataxonomics for the characterization of vermicompost microbiomes. Frontiers in Microbiology. 2022;13: 854423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Banciu HL, Muntyan MS. Adaptive strategies in the double-extremophilic prokaryotes inhabiting soda lakes. Current Opinion in Microbiology. 2015;25:73–9. [DOI] [PubMed] [Google Scholar]
- 69.Ulya M, Oesman F, Iqbalsyah TM. Low molecular weight alkaline thermostable α-amylase from Geobacillus sp nov Heliyon. 2019;5(7):e02171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barakat KM, Hassan SW, Darwesh OM. Biosurfactant production by haloalkaliphilic Bacillus strains isolated from Red Sea. Egypt The Egyptian Journal of Aquatic Research. 2017;43(3):205–11. [Google Scholar]
- 71.Dahiya P, Rathi B. Characterization and application of alkaline α-amylase from Bacillus licheniformis TCC1483 as a detergent additive. International Food Research Journal. 2015;22(3):1293–7. [Google Scholar]
- 72.The Genome Sequence Archive Family. Toward Explosive Data Growth and Diverse Data Types. Genomics, Proteomics and Bioinformatics. 2021;19(4):578–83. https://doi.org/10.1016.j.gpb.2021.08.001 . PMID=34400360. [DOI] [PMC free article] [PubMed]
- 73.Database Resources of the National Genomics Data Centre. China National Centre for Bioinformation. Nucleic Acids Research. 2022;50(D1):D27–38. https://doi.org/10.1093/nar/gkab951 . PMID=34718731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: An overview of the methods used for the collection, sequencing and analysis of metagenomic DNA from Buhera soda pans.
Additional file 2: Metagenomic sequence read statistical quality parameters.
Additional file 3: Domain-level taxonomic classification of the Buhera soda pan microbial community.
Additional file 4: Class-level taxonomic classification of the Buhera soda pans microbial community.
Additional file 5: Distribution of Archaea in the Buhera soda pans.
Additional file 6: Rarefaction analyses of the Buhera soda pan microbial community at the species level.
Data Availability Statement
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics, 2021) in National Genomics Data Centre (Nucleic Acid Research, 2022), China National Centre for Bioinformation/ Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA016626) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa.