Abstract
Background
Wilson’s disease (WD) is a hereditary disorder characterized by an abnormality in copper metabolism. Liver fibrosis, and potentially cirrhosis, induced by copper accumulation are critical factors in the pathogenesis of WD. CircRNAs exhibit high stability and play crucial roles in numerous biological processes.
Methods
RNA-seq technology was employed to conduct transcriptome sequencing of the liver from 12 homozygous (TX) mice in the model group (NL group) and 12 wild-type (WT) mice in the control group (N group). Differentially expressed circular RNAs (DE-circRNAs) were identified, and following GO and KEGG analysis, a competitive endogenous RNA (ceRNA) regulatory network was constructed. The identified DE-circRNAs were then randomly validated using RT-qPCR.
Results
Utilizing RNA sequencing (RNA-seq), the study identified 54 DE-circRNAs in TX-j mice with WD-induced liver fibrosis model, among which 19 were up-regulated and 35 were down-regulated. GO analysis revealed multiple biological processes, including single-organism process, cellular process, and metabolic process. Further pathway identification using KEGG implicated several pathways, including the HIF-1, PI3K-Akt, AMPK, FoxO, signaling pathway regulating pluripotency of stem cells, phospholipase D, mTOR, Ras, cGMP-PKG, and MAPK signaling pathway, among others. A ceRNA regulatory network was constructed with 20 circRNAs, 7 miRNAs, and 75 mRNAs as crucial core components. Additionally, RT-qPCR validation was performed on randomly selected DE-circRNAs, yielding consistent results (P<0.05).
Conclusion
The findings provide a significant molecular biology foundation for understanding the pathogenesis of liver fibrosis in WD and offer new insights for exploring potential diagnostic and therapeutic targets.
Keywords: circRNA, circRNA-miRNA-mRNA, ceRNA, Wilson’s disease, liver fibrosis
Introduction
Wilson’s Disease (WD), also referred to as Hepatolenticular degeneration (HLD), is a congenital autosomal recessive genetic disorder characterized by impaired copper ion metabolism.1 This impairment is primarily due to mutations in the ATP7B gene, which leads to dysfunction of the P-type ATPase protein. As a result, there is a reduction in copper transport capacity, causing decreased synthesis of ceruloplasmin in the patient’s plasma and an obstruction in copper excretion through the bile duct.2 The abnormal accumulation of copper can induce cellular, tissue, and organ damage and dysfunction, affecting multiple areas such as the liver, brain, kidneys, and cornea.3 The activation of hepatic stellate cells (HSCs) triggered by copper accumulation, along with the metabolic imbalance of the extracellular matrix (ECM), leads to ECM deposition in the liver and tissue structural remodeling.4 These processes contribute to liver fibrosis and, ultimately, cirrhosis, which are pivotal factors in the pathogenesis of WD. As the disease progresses, patients may encounter life-threatening liver failure.5 Consequently, anti-fibrotic therapy emerges as a crucial focus in the treatment of WD, holding significant potential for improving survival rates and prognosis.
Circular RNA (circRNA) is a type of nucleic acid molecule characterized by a closed loop RNA structure, which was first identified in plant viruses during the 1970s.6 Subsequently, Hsu et al7 reported the presence of circRNA in the cytoplasm of eukaryotic organisms. Notably, the majority of circRNAs are non-coding RNAs, with only a small fraction possessing protein-coding capabilities.8 Historically, circRNA was considered a byproduct of aberrant pre-mRNA splicing. However, with the advancement of high-throughput sequencing technology and bioinformatics engineering, circRNAs have been identified and characterized across various species.9,10 Compared to other nucleic acid molecules, the circular structure of circRNA confers resistance to cleavage and degradation by RNA proteases, demonstrating enhanced stability. The significant role of circular RNA in various biological processes, such as tissue development, cell transformation, and intercellular communication, has garnered increasing attention.11
In recent years, an increasing number of studies have elucidated the precise roles and mechanisms of circRNA in liver diseases. Recent high-throughput RNA-seq data suggested that the circAno6/miR-296-3p/TLR4 signaling axis may contribute to liver fibrosis by modulating inflammatory responses and activating hepatic stellate cells.12 Furthermore, research has revealed that circRNA IRF2 can engage with the N6-methyladenosine (m6A) reader protein YTHDF2, influencing liver fibrosis through the promotion of FOXO3 nuclear translocation.13 Wei et al14 identified genes associated with kidney injury through RNA-seq of kidney tissues from TX mice. Nevertheless, reports on potential circRNA-mediated mechanisms in the pathogenesis of liver fibrosis in WD models remain scarce. Consequently, this study employed high-throughput RNA sequencing to detect differentially expressed circular RNAs (DE-circRNAs) in the livers of TX mice and constructed a competing endogenous RNA (ceRNA) regulatory network involving circRNA-miRNA-mRNA interactions to decipher the pathological mechanisms of liver fibrosis in WD. This work aims to provide a molecular foundation for diagnosing and treating WD-induced liver fibrosis. A research design diagram based on this study was presented in Figure 1.
Figure 1.
Overall Research design Diagram.
Materials and Methods
Animal Models, Grouping, and Specimen Collection
The animal model employed in this study consisted of 4-month-old mice with toxic milk syndrome (TX-j), sourced from Jackson Laboratories. Research has demonstrated that 4-month-old TX mice exhibit distinct hallmarks of WD-induced liver fibrosis, making them the most ideal WD animal model currently available.15 Twelve homozygous (TX) mice were used as the model group (NL group), and twelve wild-type (WT) mice served as the control group (N group). RNA-seq analysis was performed on four mice from each group, while RT-qPCR analysis was conducted on eight mice. The mice were housed in independent air supply isolation cages with free access to food and water. The light/dark cycle was maintained at 12 hours, and the mice were continuously fed for 16 weeks. Death was induced through cervical dislocation to obtain mouse liver tissue. Subsequently, a portion of the liver tissue was soaked in a polyformaldehyde solution for specimen fixation, followed by routine paraffin embedding for pathological examination. The remaining mouse liver tissue was promptly sealed, placed in a liquid nitrogen box, and then transferred to a −80°C freezer for immediate freezing.
Total RNA Extraction and Sequencing
Initially, total RNA was extracted utilizing the TRIzol reagent. Subsequent to this, various reagent kits and methodologies were employed for purification and quantification to ensure the integrity and quality of the RNA. Specifically, microRNA was isolated using the MiRNeasy Mini Kit (Qiagen, Germany). This was followed by a further purification step using the RNA Clean XP Kit (Cat # A63987, Beckman Coulter, USA). Additionally, a DNase Set without RNase (Cat # 79254, Qiagen, Germany) was utilized to eliminate potential DNA contamination. The content and quality of the RNA were assessed using two advanced instruments: the NanoDrop 2000 (Thermo Fisher Scientific, USA) and the Agilent Bioanalyzer 2100 (Agilent Technologies, USA). For library construction, standardized testing and preparation were conducted in accordance with the TruSeq protocol. This step is pivotal for sequencing experiments as it influences the quality of sequencing data and the accuracy of subsequent analysis. Ultimately, transcriptome sequencing was performed on the Illumina HiSeq 6000 (Illumina, USA) instrument.
Identification and Functional Enrichment Analysis of Differential Expression of CircRNA
The study employed the CIRI (CircRNA Identification) tool16 for the identification of circRNAs. Initially, the BWA-MEM algorithm was utilized to split and align sequences. Subsequently, the resulting SAM files were analyzed for PCC (Paired Chip Clipping) and PEM (Paired End Mapping) sites. Both these methods are crucial for detecting circRNA, and by examining the information from these loci, a preliminary determination of the possible presence of circRNA can be made. During the analysis, emphasis was also placed on GT-AG splicing signals and GTAG splicing signals, which are significant features in the formation process of circRNA. Analyzing these signals further verifies the authenticity of the identified circRNA. To ensure the reliability of the identified circRNA, a dynamic programming algorithm was applied to rematch sequences with splice sites.
Gene expression was comprehensively measured using FPKM (Fragments Per Kilobase of exon model per Million mapped fragments), where an FPKM value greater than 0.5 indicates expression across all groups. Following the acquisition of FPKM expression values for genes or transcripts, differential gene expression analysis between samples/groups was conducted using the edgeR software package, and P-values were calculated. The fold change (FC) based on FPKM values is commonly referred to as log2(FC). In this experiment, a P-value of less than 0.05 and an absolute log2FoldChange value greater than 1 were used as screening criteria.
Subsequently, GO (Gene Ontology, http://geneontology.org/) and KEGG (Kyoto Encyclopedia of Genes and Genomes, http://www.kegg.jp/) were utilized to annotate and analyze the biological functions and pathways associated with the DE-circRNA.
Prediction and Annotation of circRNA Targeting miRNA and mRNA Networks
RNAhybrid (bibiserv.cebitec.uni-bielefeld.de/rnahybrid/), miRanda (www.miranda.org), and TargetScan (www.targetscan.org) software were used to predict the interactions between circRNAs, mRNAs, and miRNAs. Additionally, Cytoscape software (version 3.7.0) was employed to construct regulatory network diagrams for circRNA-miRNA, circRNA-mRNA, and ceRNA interactions, thereby illustrating the complex interplay and regulatory relationships among these targets.
Validation of Sequencing Data Using RT-qPCR
To verify the accuracy of the sequencing results, we randomly detected the expression levels of the identified DE-circRNA using RT-qPCR. After extracting the total RNA, reverse transcription reaction was performed on the sample to generate the corresponding cDNA. Next, PCR amplification was performed on each type of cDNA using specific primers, with each reaction using two microliters of cDNA template. For quantitative analysis, we used the 2-CT relative quantification method and selected β - actin as the endogenous control. Firstly, the CT values of the target and reference genes for each sample were accurately recorded. Using the standard formula ΔCT = CT (target gene) - CT (reference gene), we calculated the ΔCT values for each sample. Furthermore, by comparing the ΔCT values of the model group samples with those of the control group samples, using the formula ΔΔCT = ΔCT (model) - ΔCT (control), we evaluated the differences between the two groups. Ultimately, based on the formula 2^(-ΔΔCT), we calculated the relative expression levels of the target gene in each sample.17
Statistical Analysis
Data were statistically analyzed using SPSS 23.0 software. The Student’s t-test was employed, and a P-value of less than 0.05 was considered statistically significant. The Benjamini-Hochberg method was used for the enrichment analysis of GO and KEGG, where a P-value of less than 0.05 indicated significant enrichment.
Results
Genotyping and Pathological Observation of Liver Tissue in TX Mice
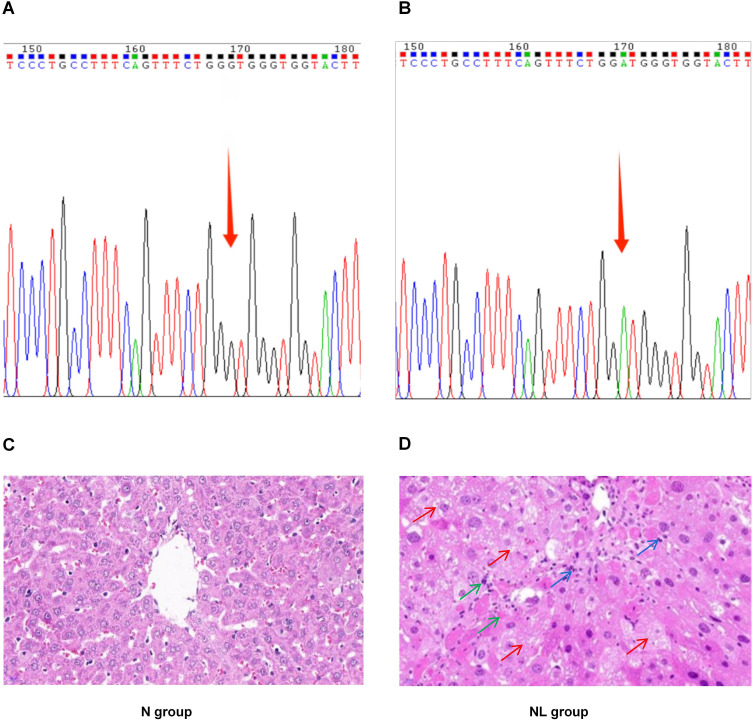
Initially, the genotype of WD model TX mice was identified. The gene testing results for the normal group mice were presented in Figure 2A, while those for the model group mice were shown in Figure 2B. Additionally, pathological evaluations were conducted on the mouse liver tissue. Following HE staining, it was observed that the liver tissue structure of the N group mice exhibited no abnormalities, with hepatic cell cords neatly arranged radiating from the central vein to the periphery. In contrast, the NL group displayed disorganized hepatic cellular arrangement, accompanied by moderate to severe edema and degeneration. The liver parenchyma presented with partial infiltration of inflammatory cells, and focal areas exhibited hepatocyte degeneration and necrosis. (Figure 2C and D).
Figure 2.
Genotyping Analysis and Histopathological Assessment of Liver Tissue in TX Mice. (A) Genetic Profile of the N Group Mice; (B) Genetic Profile of the NL Group Mice; (C) Histopathological Findings in the Liver of N Group Mice (×400); (D) Histopathological Findings in the Liver of NL Group Mice (×400). Red arrow, cellular edema and degeneration; Green arrow, cell necrosis; Blue arrow, infiltration of inflammatory cells.
Quantitative Detection of circRNA Expression
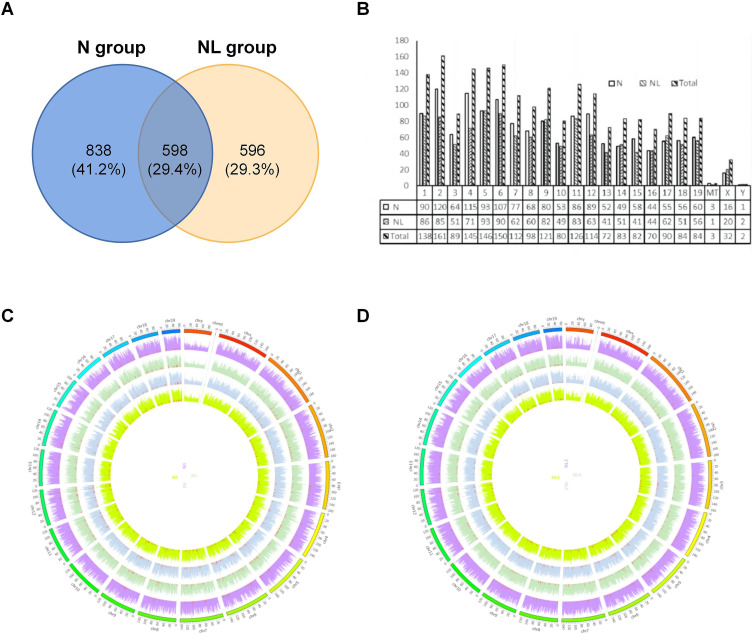
To precisely quantify the expression levels of circRNAs in the hepatic tissues of mice from groups N and NL, and to meticulously contrast and analyze the differential gene expression patterns between these two groups, we carried out an exhaustive quantitative assessment focused on circRNA expression profiles. This investigation led to the identification of 2032 circRNA transcripts. Of these, 1436 circRNAs were detected in the model group and 1194 in the control group, with an overlap of 598 circRNAs between both groups (Figure 3A). Furthermore, we analyzed the distribution of the identified circRNAs across mouse chromosomes. The 2032 circRNA transcripts were found to be distributed across all chromosomes, including ChrX and ChrY, with chromosome 2 harboring the highest number of circRNAs. All chromosomes represented in the figure (excluding ChrMT, ChrX, and ChrY) are associated with at least 50 circRNAs each (Figure 3B). As illustrated in the Circos plots presented in Figure 3C and D, the chromosome distribution of circRNA expression in two groups of mouse liver samples was depicted. In these figures, the outermost lines are segmented according to different chromosomes, while the four distinct colored rings in the center represent different samples. By analyzing the distribution of each chromosome, we can discern the specific distribution pattern of circRNA in each sample.
Figure 3.
Overall Contour of circRNA Expression Detection in TX Mouse Liver. (A) Venn diagram of the distribution of circRNA quantity in the liver of two groups; (B) Histogram of the overall distribution of circRNA chromosomes in the liver of two groups; (C) Circos diagram of chromosome distribution of circRNA expression in the liver of N group; (D) Circos diagram of chromosomal distribution of circRNA expression in the liver of NL group.
Identification of DE-circRNAs
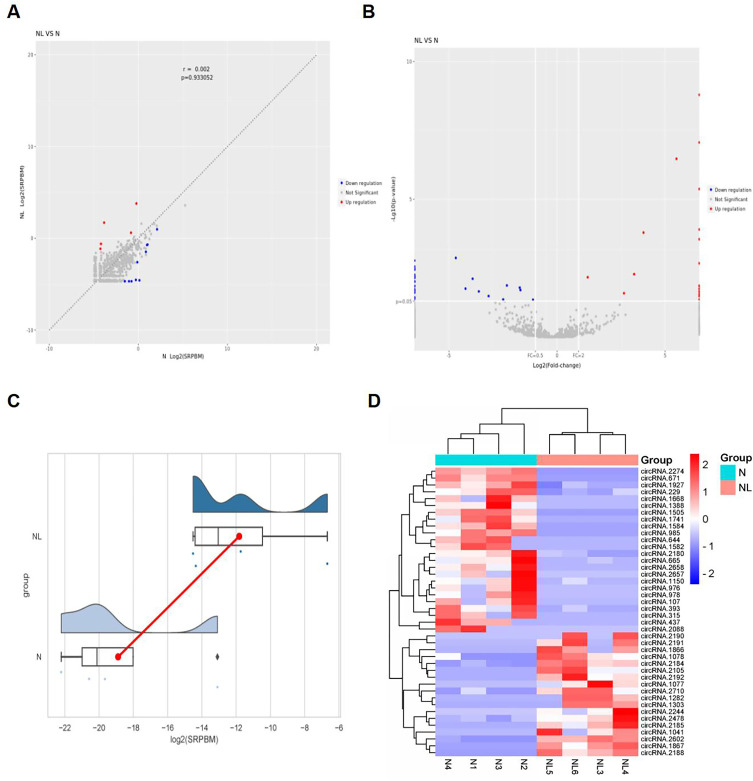
To elucidate the role of circRNA expression in liver fibrosis induced by Wilson’s disease, we conducted a comprehensive screening to identify DE-circRNAs in the liver tissues of TX mice from both the N and NL groups. Applying the screening criteria of Log2| fold change (FC) | ≥ 1 and P value < 0.05, we identified a total of 54 DE-circRNAs, comprising 19 that were up-regulated and 35 that were down-regulated. The top 10 DE-circRNAs were summarized in Table 1. The scatter plot and volcano plot were depicted in Figure 4A and B, respectively. Blue scatter points represent down-regulated DE-circRNAs, red scatter points represent up-regulated DE-circRNAs, and gray dots indicate no significant statistical significance. In Figure 4C, different colors represent the distribution of data across different groups, with the red line indicating the difference in means. The heat map shown in Figure 4D illustrates the expression levels of the identified DE-circRNAs across different samples from the two groups, with light blue representing the N group and pink representing the NL group. A redder color indicates a stronger positive correlation, while a bluer color signifies a stronger negative correlation.
Table 1.
Detailed Information on the Up-Regulated and Down-Regulated Top 10 DE-circRNAs Identified in the Liver of TX Mice
| Circrna_id | Known_circrna | Gene_id | Gene_name | Log2fc | Pvalue | Up/down |
|---|---|---|---|---|---|---|
| CircRNA.2602 | - | ENSMUSG00000093803 | Ppp2r3d | 5.533 | 3.41E-07 | UP |
| CircRNA.2710 | MMU_CIRC_0001829 | ENSMUSG00000032413 | Rasa2 | 3.577 | 0.005 | UP |
| CircRNA.1078 | - | ENSMUSG00000052942 | Glis3 | 3.107 | 0.026 | UP |
| CircRNA.2478 | - | ENSMUSG00000005534 | Insr | 1.425 | 0.007 | UP |
| CircRNA.665 | - | ENSMUSG00000055737 | Ghr | −4.688 | 0.001 | DOWN |
| CircRNA.393 | - | ENSMUSG00000047454 | Gphn | −3.908 | 0.008 | DOWN |
| CircRNA.1150 | MMU_CIRC_0000921 | ENSMUSG00000096370 | Gm21992 | −3.619 | 0.022 | DOWN |
| CircRNA.1494 | MMU_CIRC_0001017 | ENSMUSG00000052155 | Acvr2a | −3.166 | 0.033 | DOWN |
| CircRNA.97 | - | ENSMUSG00000047638 | Nr1h4 | −2.489 | 0.043 | DOWN |
| CircRNA.1927 | - | ENSMUSG00000005373 | Mlxipl | −2.317 | 0.014 | DOWN |
Figure 4.
Expression and distribution characteristics of DE-circRNA in the liver of two groups of TX mice. (A) Scatter plot of the identified DE-circRNAs; (B) Volcano map of the identified DE-circRNAs; (C) Distribution map of log2 (SRPBM) for the identified DE-circRNAs; (D) Heat map of the distribution of the identified DE-circRNAs.
GO Analysis of DE-circRNAs
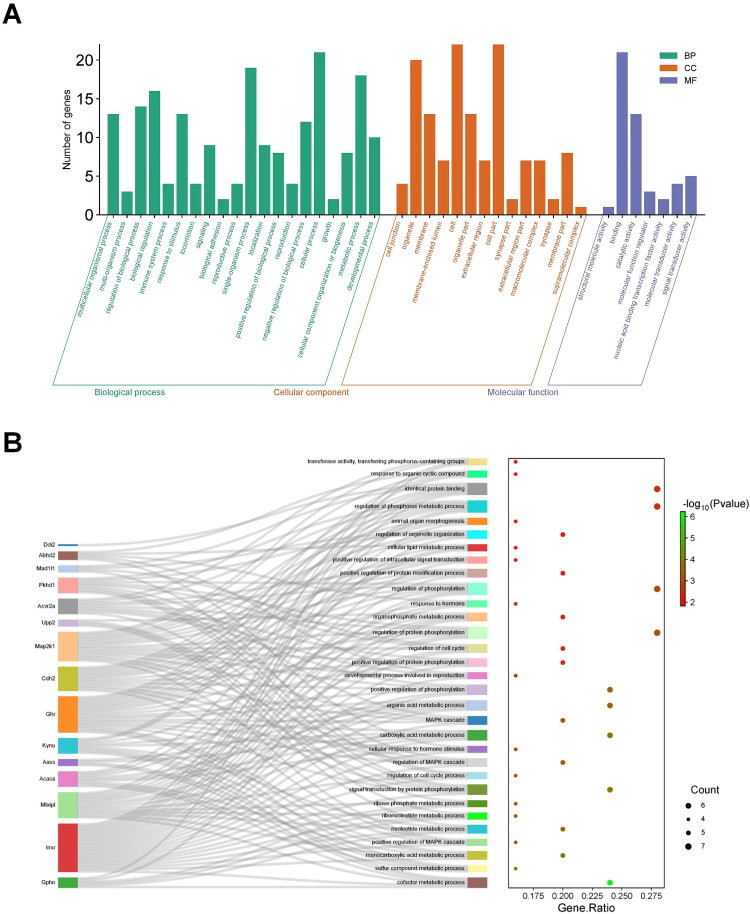
To delve deeper into the functionalities and distinctive attributes of the identified DE-circRNAs, our analysis revealed a marked enrichment in GO categories. This enrichment encompassed 20 biological process (BP), 14 cellular component (CC), and 7 molecular function (MF) items. Within the BP category, significant enrichments were detected across a diverse array of GO terms, including multicellular organismal process, multi-organism process, regulation of biological process, biological_process, immune system process, response to stimulus, locomotion, signaling, biological adhesion, reproductive process, single-organism process, localization, positive regulation of biological process, reproduction, negative regulation of biological process, cellular process, growth, cellular component organization or biogenesis, metabolic process, and developmental process. In the CC category, the identified DE-circRNAs were significantly associated with cell junction, membrane, organelle, membrane-enclosed lumen, cell, organelle part, extracellular region, cell part, synapse part, extracellular region part, macromolecular complex, synapse, membrane part, and supramolecular complex. Meanwhile, within the MF category, the enriched terms included those related to structural molecule activity, binding, catalytic activity, molecular function regulator, nucleic acid binding transcription factor activity, molecular transducer activity, and signal transducer activity (Figure 5A). The graphical representation of the interactive network between the Top 30 DE-circRNAs and their corresponding proteins was depicted in Figure 5B.
Figure 5.
GO analysis of DE-circRNA in the liver of TX mice. (A) GO classification bar chart of DE-cicRNA in the liver of TX mouse; (B) GO enriched Sangi plot of DE-cicRNA in the liver of TX mice.
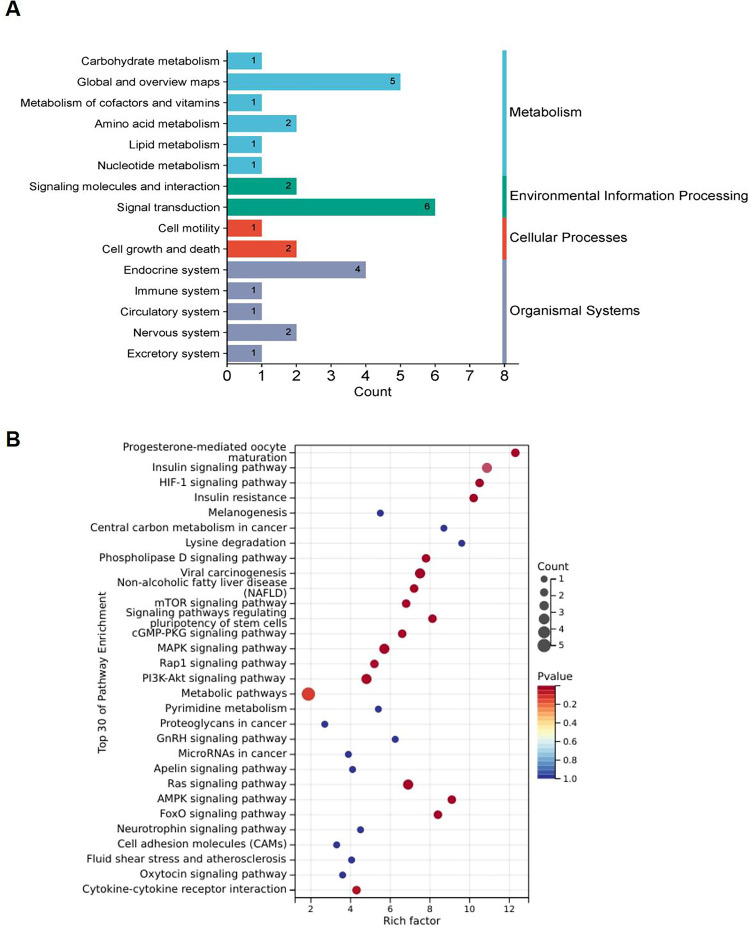
KEGG Classification and Enrichment Analysis Based on DE-circRNAs
To delineate the functional significance and biological roles of the identified DE-cicRNAs, we subjected the dataset to a KEGG pathway analysis utilizing a sophisticated cluster analyzer. The enriched pathways were classified under existing classification conditions, which include cellular processes such as cell motility, cell growth, and death; metabolic categories encompassing carbohydrate metabolism, global and overview maps, metabolism of cofactors and vitamins, amino acid metabolism, lipid metabolism, nucleotide metabolism; environmental information processing involving signaling molecules and interaction, and signal transduction; organizational systems comprising endocrine system, immune system, circulatory system, nervous system, and Excretory system (Figure 6A). In the KEGG pathway analysis, the integrated differential genes were predominantly enriched in pathways such as the HIF-1, PI3K-Akt, AMPK, FoxO, signaling pathway regulating stem cell pluripotency, phospholipase D, mTOR, Ras, cGMP-PKG, MAPK, and Rap1 signaling pathway. The top 30 KEGG enrichment pathways obtained have been visualized (Figure 6B).
Figure 6.
KEGG analysis of DE-circRNA in the liver of TX mice. (A) KEGG classification diagram of DE-circRNA in the liver of TX mice; (B) KEGG Bubble Chart of DE-circRNA Expression in the liver of TX mice.
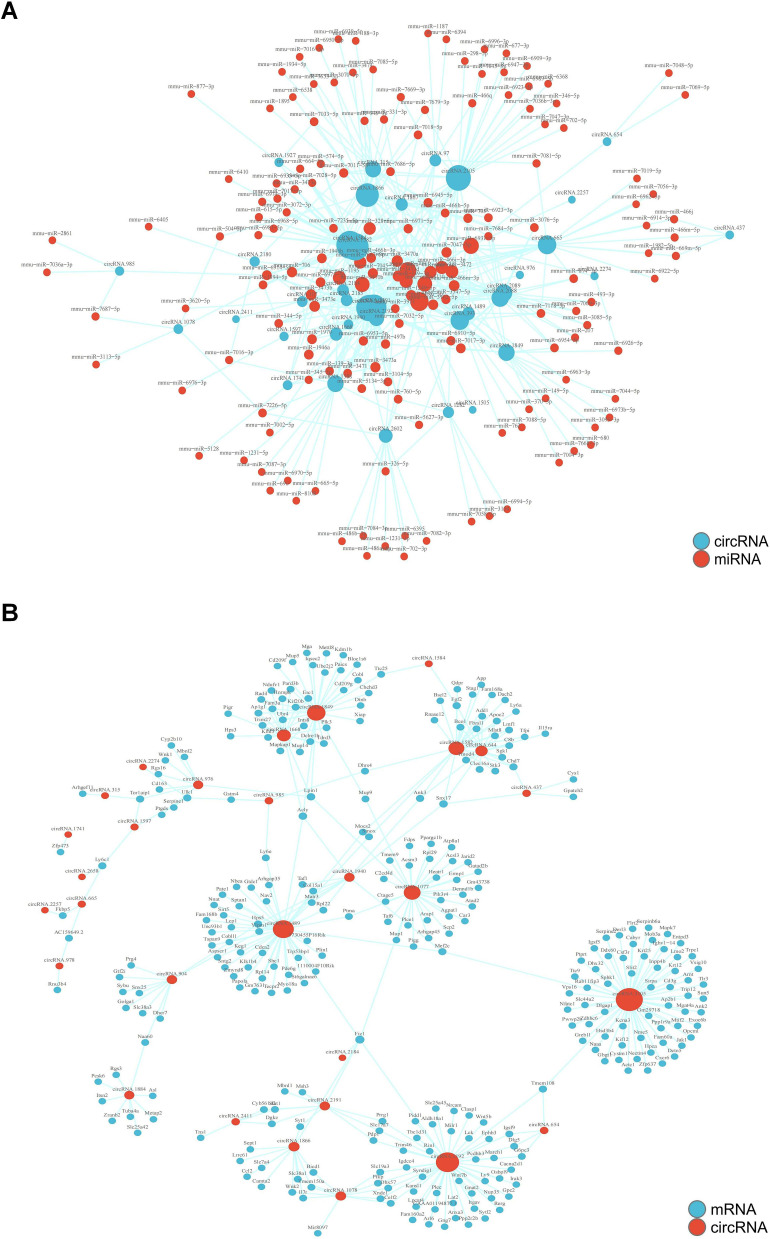
Construction of CircRNA-miRNA Interaction Network and CircRNA-mRNA Co-Expression Network
In the present study, we employed RNA-seq analysis to identify a total of 1788 miRNAs in the NL and N groups. In comparison to the N group, NL group exhibited differential expression for 182 miRNAs. By predicting the targets of these miRNAs, we were able to delineate the interactions between miRNAs and circRNAs. Subsequently, we utilized Cytoscape software to construct a comprehensive circRNA-miRNA regulatory network, highlighting the intricate interplay between these regulatory molecules. Moreover, our investigation uncovered 3379 DE-mRNA transcripts. Through the prediction of mRNA targets, we identified multiple circRNA-mRNA co-expression networks, each exhibiting varying degrees of connectivity and centered on distinct circRNAs. The interactive and co-expression relationships between circRNA-mRNA pairs were illustrated in Figures 7A and B.
Figure 7.
Interaction and co-expression network diagram of circRNA-miRNA and circRNA-mRNA in the liver of TX mice. (A) The circRNA-miRNA interaction network in the liver of TX mice; (B) The circRNA-mRNA co-expression interaction network in the liver of TX mice.
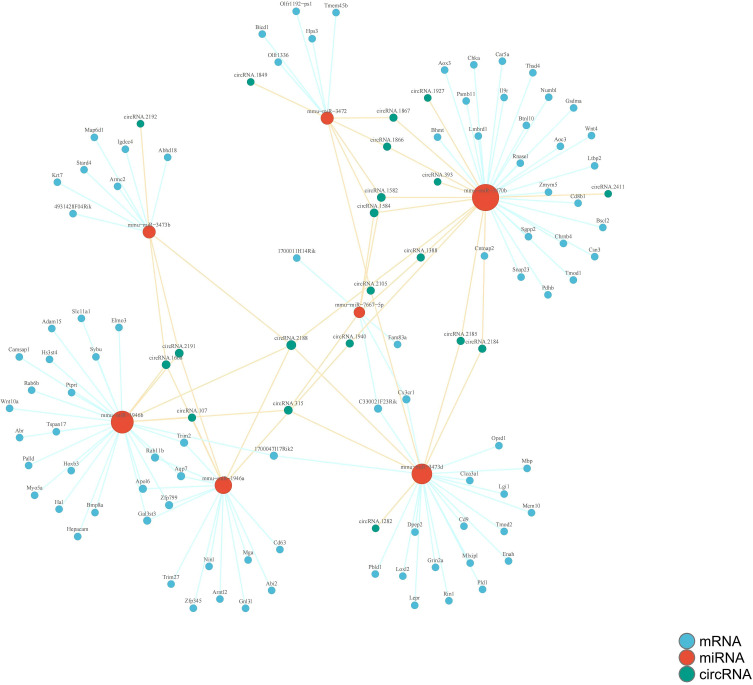
Construction of CeRNA Regulatory Network for CircRNA-miRNA-mRNA
Through the prediction of circRNA-miRNA and circRNA-mRNA interactions between two distinct groups, we elucidated a critical ceRNA regulatory network comprising 20 circRNAs, 7 miRNAs, and 75 mRNAs as pivotal nodes. The comprehensive circRNA-miRNA-mRNA interaction network diagram for the liver of TX mice was depicted in Figure 8, providing a visual representation of the intricate regulatory landscape.
Figure 8.
The circRNA-miRNA-mRNA interaction network diagram in TX mouse liver.
RT qPCR Validation
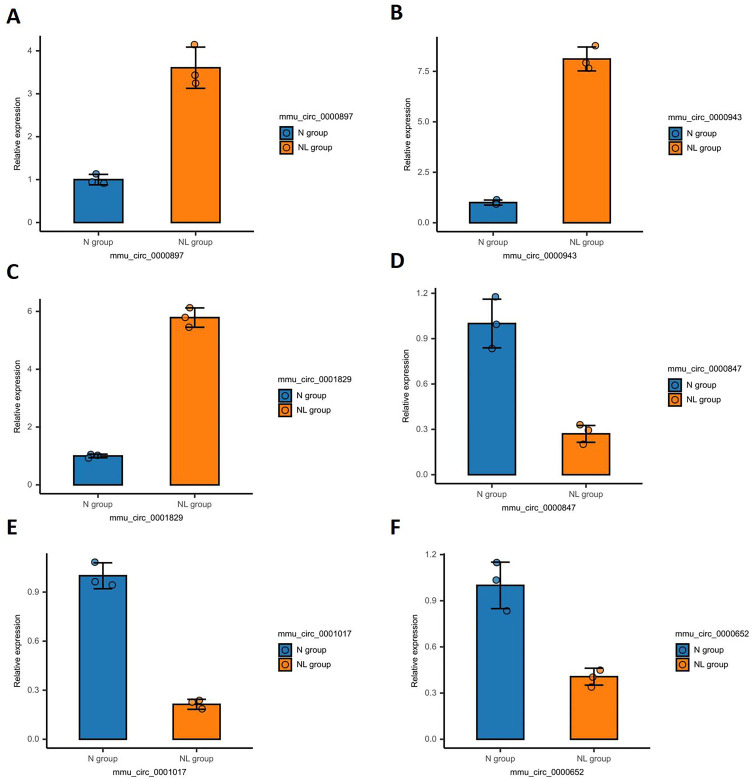
To ascertain the validity of the identified DE-circRNAs, a random validation via RT-qPCR was performed. The outcomes revealed a significant up-regulation of mmu_circ_0000897, mmu_circ_0000943, and mmu_circ_0001829, whereas a notable down-regulation was observed for mmu_circ_0000847, mmu_circ_0001017, and mmu_circ_0000652. Statistical analysis confirmed that these discrepancies were significant (P<0.05), thereby substantiating the dependability of the sequencing analysis findings (Figure 9).
Figure 9.
RT-qPCR validation of the selected DE-circRNA. (A) mmu circ 0000897; (B) mmu circ 0000943; (C) mmu circ 0001829; (D) mmu circ 0000847; (E) mmu circ 0001017; (F) mmu circ 0000652.
Discussion
The rarity of genetic features associated with WD has historically impeded comprehensive and in-depth research on the condition. Nonetheless, the advent and swift progress of high-throughput sequencing technologies in recent years have unlocked new avenues for exploring the intricate molecular and biological underpinnings of liver fibrosis instigated by WD. The expression and pivotal roles of various circRNAs in liver fibrosis have been identified and corroborated using RNA-seq technology.18 In the current investigation, we utilized transcriptome sequencing technology to screen 54 DE-circRNAs, including 19 that were up-regulated and 35 that were down-regulated within the liver tissue of TX mice. The gene functions and attributes enriched by GO mainly include single organic process, cellular process, metabolic process, cell, cell part, organelle, binding, catalytic activity, etc. Further identification of relevant pathways using KEGG involves the HIF-1, PI3K-Akt, AMPK, FoxO, signaling pathway regulating stem cell pluripotency, phospholipase D, mTOR, Ras, cGMP PKG, MAPK, Rap1 signaling pathway, etc. The circRNA-miRNA-mRNA interaction network has been established to elucidate the key mechanisms of the ceRNA ternary regulatory network. Additionally, the expression levels of six arbitrarily selected DE-circRNAs were confirmed by RT-qPCR detection, and consistent results were obtained. Therefore, the identified DE-circRNA and ceRNA networks can serve as important targets for studying WD-induced liver fibrosis, laying the foundation for further exploration of its pathological mechanisms.
The DE-circRNAs identified in this study have been validated to be implicated in glucose metabolism, lipid metabolism, and other related processes. GLIS family zinc finger 3 (GLIS3), a nuclear protein, has been found to selectively participate in the regulation of insulin gene expression.19 It is worth noting that the insulin receptor (INSR) plays a crucial role in maintaining glucose homeostasis.20 Research has shown that long-term inhibitory stress can alleviate liver fibrosis caused by carbon tetrachloride through the INSR/PI3K/AKT/AMPK signaling pathway.21 Similarly, studies have confirmed that G6pc (glucose-6-phosphatase catalytic subunit), Abhd2 (acyl-CoA binding domain containing 2), Scp2 (sterol carrier protein 2), growth hormone receptor (GHR), and MLX interacting protein-like (MLXIPL) are all associated with liver glucose and lipid metabolism.22–26
Furthermore, the regulatory roles of activin receptor type-2A (ACVR2A) and nuclear receptor subfamily 1, group H, member 4 (NR1H4) genes in liver fibrosis have been elucidated in this study. Specifically, ACVR2A is primarily responsible for mediating the biological effects of the transforming growth factor-β 1 (TGF-β1) superfamily. Research has demonstrated that downregulation of ACVR2A inhibits the activation of HSC, thereby exerting an antifibrotic effect on the liver.27 Additionally, NR1H4 plays a significant role in regulating inflammation and fibrosis. By inhibiting HSC activation, NR1H4 effectively alleviates liver fibrosis.28,29
In addition, the identified Ppp2r3d and Staphylococcal nuclease and tudor domain containing 1 (SND1) have been found to be associated with certain liver diseases. Ppp2r3d primarily originates from the serine/threonine protein phosphatase 2A (PP2A) family, which plays a key role in regulating cell division, growth, and differentiation signaling.30 Research has shown that PP2A inhibitors can participate in the progression of non-alcoholic fatty liver disease (NAFLD) by triggering miR21 activation in a NADPH oxidase 2 (NOX2)-dependent manner.31 SND1 is a conserved multifunctional protein that has been implicated in mitochondrial autophagy and specific liver pathologies.32 However, there is limited research on the role of certain DE-circRNAs identified in liver fibrosis. RASA2, as a RAS GTPase-activating protein (RasGAP), mainly inhibits the downstream RAS signaling pathway activated by T cell receptors (TCR), suggesting potential immune regulatory mechanisms.33 Gephyrin, also known as Gphn, can prevent the accumulation of neurotransmitter receptors on the postsynaptic membrane of nerve cells. This protein has been found in non-neuronal liver cells34 and is affected by serine phosphorylation when the mTOR signaling pathway is inhibited.35 The Gm21992 locus is believed to be a spontaneous transcription event that connects adjacent Rbm14 (RNA binding motif protein 14) and Rbm4 (RNA binding motif protein 4) genes.36 This study observed differential expression of Gphn and Gm21992 in the liver of WD, and their potential mechanisms need further clarification.
The roles of certain signaling pathways identified in this study have been previously reported in the pathogenesis of WD. Research has demonstrated that the activation of HIF-1 signal can improve hepatic steatosis in zebrafish ATP7B deficiency (WD model).37 The autophagy regulated by the PI3K/Akt/mTOR signaling pathway is considered a possible mechanism of traditional Chinese medicine compound intervention in WD-induced liver fibrosis.38 The research indicated that P38mapk was linked to the onset of acute liver damage in the WD model LEC (Long Evans Cinnamon) rats.39 Further elucidation of the precise mechanisms underlying the other identified signaling pathways in liver fibrosis triggered by Wilson’s disease is warranted.
In addition, this study constructed a ceRNA regulatory network based on DE-circRNAs by predicting circRNA-miRNA interactions and circRNA-mRNA co-expression relationships. This research contributes to the molecular understanding of the pathogenesis of WD-induced liver fibrosis. Moreover, by directly targeting specific nodes within the circRNA or ceRNA network, the expression of other RNA molecules (such as lncRNAs, miRNAs, etc.) in the network can be modulated. This approach restores the regulatory function of circRNA genes and rebalances the entire ceRNA network, thereby exerting a positive effect on the pathological and physiological processes associated with WD. This has significant implications for the development of treatment strategies for WD-induced liver fibrosis.
This study has several limitations. Primarily, the sample size employed is relatively small, necessitating additional research with expanded samples. While we have identified DE-circRNA in the livers of TX mice, and performed GO and KEGG analysis, constructing a competing ceRNA regulatory network, the implicated targets and pathways remain extensive and intricate. Due to limitations in our research design, we were unable to validate additional targets. Moving forward, we are committed to conducting a more in-depth investigation. Moreover, conventional transcriptome sequencing fails to provide detailed cellular data, spatial localization, or morphological information. Future studies should incorporate single-cell and spatial transcriptome sequencing to more comprehensively elucidate these mechanisms.
Conclusion
In summary, this study utilized RNA-seq to determine the differential expression of circRNAs in the liver of TX mice. We further explored the biological properties, functions, and signaling pathways of these genes through GO and KEGG analysis. Building on these findings, we constructed a ceRNA regulatory network involving circRNA, miRNA, and mRNA. This research lays the groundwork for investigating the molecular mechanisms underlying WD-induced liver fibrosis and offers novel strategies for identifying potential diagnostic and therapeutic targets.
Funding Statement
This study was funded by the National Natural Science Foundation of China (Grant No. 82274493) and Scientific Research Project of Higher Education Institutions in Anhui Province (Grant No. 2023AH050791).
Data Sharing Statement
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics Approval And Consent To Participate
The protocol of this study was in accordance with the ethical guidelines of the NIH Guide for the Care and Use of Laboratory Animals. This study was approved by The Institutional Review Committee of The Institutional Review Committee of Anhui Agricultural University (Approval No. AHAU2023043).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no conflict of interest.
References
- 1.Członkowska A, Litwin T, Dusek P, et al. Wilson disease. Nat Rev Dis Primers. 2018;4(1):21. doi: 10.1038/s41572-018-0018-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huster D, Kühne A, Bhattacharjee A, et al. Diverse functional properties of Wilson disease ATP7B variants. Gastroenterology. 2012;142(4):947–956.e5. doi: 10.1053/j.gastro.2011.12.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schilsky ML. Wilson Disease: diagnosis, Treatment, and Follow-up. Clin Liver Dis. 2017;21(4):755–767. doi: 10.1016/j.cld.2017.06.011 [DOI] [PubMed] [Google Scholar]
- 4.Parola M, Pinzani M. Liver fibrosis: pathophysiology, pathogenetic targets and clinical issues. Mol Aspects Med. 2019;65:37–55. doi: 10.1016/j.mam.2018.09.002 [DOI] [PubMed] [Google Scholar]
- 5.Gerosa C, Fanni D, Congiu T, et al. Liver pathology in Wilson’s disease: from copper overload to cirrhosis. J. Inorg Biochem. 2019;193:106–111. doi: 10.1016/j.jinorgbio.2019.01.008 [DOI] [PubMed] [Google Scholar]
- 6.Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73:3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340. [DOI] [PubMed] [Google Scholar]
- 8.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. [DOI] [PubMed] [Google Scholar]
- 11.Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–691. [DOI] [PubMed] [Google Scholar]
- 12.Li Z, Ma Y, Fan C, Jiang H. The circAno6/miR-296-3p/TLR4 signaling axis mediates the inflammatory response to induce the activation of hepatic stellate cells. Gene. 2024;920:148497. doi: 10.1016/j.gene.2024.148497 [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Zhu S, Li HD, et al. N6-methyladenosine-modified circIRF2, identified by YTHDF2, suppresses liver fibrosis via facilitating FOXO3 nuclear translocation. Int J Biol Macromol. 2023;248:125811. doi: 10.1016/j.ijbiomac.2023.125811 [DOI] [PubMed] [Google Scholar]
- 14.Wei T, Qian N, Yang W, et al. Construction of a Novel circRNA/miRNA/mRNA Regulatory Network to Explore the Potential Pathogenesis of Wilson’s Disease. Front Pharmacol. 2022;13:905513. doi: 10.3389/fphar.2022.905513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma Y, Bao Y, Wang H, et al. 1H-NMR-based metabolomics to dissect the traditional Chinese medicine promotes mesenchymal stem cell homing as intervention in liver fibrosis in mouse model of Wilson’s disease [published correction appears in. J Pharm Pharmacol. 2024;76(9):1236. doi: 10.1093/jpp/rgae036 [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Hou L, Zuo Z, et al. Comprehensive profiling of circular RNAs with nanopore sequencing and CIRI-long [published correction appears in Nat Biotechnol. Biotechnol. 2021;39(7):893. doi: 10.1038/s41587-021-00842-6 [DOI] [PubMed] [Google Scholar]
- 17.Bong D, Sohn J, Lee SV. Brief guide to RT-qPCR. Mol Cells. doi: 10.1016/j.mocell.2024.100141 [DOI] [PubMed] [Google Scholar]
- 18.Wang G, Tong J, Li Y, et al. Overview of CircRNAs Roles and Mechanisms in Liver Fibrosis. Biomolecules. 2023;13(6):940. doi: 10.3390/biom13060940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ZeRuth GT, Takeda Y, Jetten AM. The Krüppel-like protein Gli-similar 3 (Glis3) functions as a key regulator of insulin transcription. Mol Endocrinol. 2013;27(10):1692–1705. doi: 10.1210/me.2013-1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Payankaulam S, Raicu AM, Arnosti DN. Transcriptional Regulation of INSR, the Insulin Receptor Gene. Genes. 2019;10(12):984. doi: 10.3390/genes10120984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang S, Liu B, Huang L, Zhang R, An L, Liu Z. Metabolomics reveals that chronic restraint stress alleviates carbon tetrachloride-induced hepatic fibrosis through the INSR/PI3K/AKT/AMPK. Pathway J Mol Med. 2024;102(1):113–128. doi: 10.1007/s00109-023-02395-4 [DOI] [PubMed] [Google Scholar]
- 22.Guevara-Aguirre J, Guevara A, Palacios I, Pérez M, Prócel P, Terán E. GH and GHR signaling in human disease. Growth Horm IGF Res. 2018;38:34–38. doi: 10.1016/j.ghir.2017.12.006 [DOI] [PubMed] [Google Scholar]
- 23.Liu BW, Wang XY, Cao JL, et al. TDP-43 upregulates lipid metabolism modulator ABHD2 to suppress apoptosis in hepatocellular carcinoma. Commun Biol. 2022;5(1):816. doi: 10.1038/s42003-022-03788-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vujkovic M, Ramdas S, Lorenz KM, et al. A multiancestry genome-wide association study of unexplained chronic ALT elevation as a proxy for nonalcoholic fatty liver disease with histological and radiological validation. Nat Genet. 2022;54(6):761–771. doi: 10.1038/s41588-022-01078-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu C, Li H, Tang CK. Sterol carrier protein 2 in lipid metabolism and non-alcoholic fatty liver disease: pathophysiology, molecular biology, and potential clinical implications. Metabolism. 2022;131:155180. doi: 10.1016/j.metabol.2022 [DOI] [PubMed] [Google Scholar]
- 26.Youn DY, Xiaoli AM, Zong H, et al. The Mediator complex kinase module is necessary for fructose regulation of liver glycogen levels through induction of glucose-6-phosphatase catalytic subunit (G6pc). Mol. Metab. 2021;48:101227. doi: 10.1016/j.molmet.2021.101227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang J, Chen T, Xu H, et al. Echinacoside Exerts Antihepatic Fibrosis Effects in High-Fat Mice Model by Modulating the ACVR2A-Smad Pathway. Mol Nutr Food Res. 2024;68(6):e2300553. doi: 10.1002/mnfr.202300553 [DOI] [PubMed] [Google Scholar]
- 28.Al-Aqil FA, Monte MJ, Peleteiro-Vigil A, et al. Interaction of glucocorticoids with FXR/FGF19/FGF21-mediated ileum-liver crosstalk. Biochim Biophys Acta Mol Basis Dis. 2018;1864(9 Pt B):2927–2937. doi: 10.1016/j.bbadis.2018.06.003 [DOI] [PubMed] [Google Scholar]
- 29.Zhou J, Cui S, He Q, et al. SUMOylation inhibitors synergize with FXR agonists in combating liver fibrosis. Nat Commun. 2020;11(1):240. doi: 10.1038/s41467-019-14138-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fowle H, Zhao Z, Graña X. PP2A holoenzymes, substrate specificity driving cellular functions and deregulation in cancer. Adv Cancer Res. 2019;144:55–93. doi: 10.1016/bs.acr.2019.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Albadrani M, Seth RK, Sarkar S, et al. Exogenous PP2A inhibitor exacerbates the progression of nonalcoholic fatty liver disease via NOX2-dependent activation of miR21. Am J Physiol Gastrointest Liver Physiol. 2019;317(4):G408–G428. doi: 10.1152/ajpgi.00061.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang S, Zhu C, Suo C, et al. Mitochondrion-Localized SND1 Promotes Mitophagy and Liver Cancer Progression Through PGAM5. Front Oncol. 2022;12:857968. doi: 10.3389/fonc.2022.857968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Connor A, Li XC. RASA2 a new gatekeeper for TCR signaling. Am J Transplant. 2022;22(12):2715. doi: 10.1111/ajt.16684 [DOI] [PubMed] [Google Scholar]
- 34.Nawrotzki R, Islinger M, Vogel I, Völkl A, Kirsch J. Expression and subcellular distribution of gephyrin in non-neuronal tissues and cells. Histochem Cell Biol. 2012;137(4):471–482. doi: 10.1007/s00418-012-0914-7 [DOI] [PubMed] [Google Scholar]
- 35.Demirkan G, Yu K, Boylan JM, Salomon AR, Gruppuso PA. Phosphoproteomic profiling of in vivo signaling in liver by the mammalian target of rapamycin complex 1 (mTORC1). PLoS. One. 2011;6(6):e21729. doi: 10.1371/journal.pone.0021729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abuobeid R, Herrera-Marcos L, Navarro MA, et al. Dietary Erythrodiol Modifies Hepatic Transcriptome in Mice in a Sex and Dose-Dependent Way. Int J Mol Sci. 2020;21(19):7331. doi: 10.3390/ijms21197331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mi X, Li Z, Yan J, et al. Activation of HIF-1 signaling ameliorates liver steatosis in zebrafish atp7b deficiency (Wilson’s disease). models Biochim Biophys Acta Mol Basis Dis. 2020;1866(10):165842. doi: 10.1016/j.bbadis.2020.165842 [DOI] [PubMed] [Google Scholar]
- 38.Tang L, Zhao C, Zhang J, et al. Discussion on the Mechanism of Gandoufumu Decoction Attenuates Liver Damage of Wilson’s Disease by Inhibiting Autophagy through the PI3K/Akt/mTORPathway Based on Network Pharmacology and Experimental Verification. Media Inflamm. 2023;2023:3236911. doi: 10.1155/2023/3236911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kadowaki S, Meguro S, Imaizumi Y, Sakai H, Endoh D, Hayashi M. Role of p38 Mapk in development of acute hepatic injury in Long-Evans Cinnamon (LEC) rats, an animal model of human Wilson’s disease. J. Vet Med Sci. 2013;75(12):1551–1556. doi: 10.1292/jvms.13-0137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.