Abstract
Background
Breast cancer is one of the most common cancers in women and is closely associated with obesity. Gremlin-2 (GREM2), an antagonist for bone morphogenetic proteins (BMPs), has been considered an inhibitor of adipogenic differentiation in adipose-derived stromal/stem cells. However, the role of GREM2 in breast cancer cells remains largely unknown, and its signaling mechanism has yet to be clarified.
Methods
Bioinformatics analysis was conducted using public databases. Breast cancer cells overexpressing mock or GREM2 were used for in vitro and in vivo studies. Cell viability, colony formation, migration, and animal studies were performed to investigate the role of GREM2 in breast cancer cells. Screening of target genes affected by GREM2 overexpression in breast cancer cells was performed through RNA sequencing (RNA-seq) analysis.
Results
The expression level of GREM2 mRNA was significantly reduced in both breast cancer tissues and cell lines. Kaplan-Meier analysis showed that low expression of GREM2 and high methylation of the GREM2 promoter were each associated with poor patient survival. The low mRNA expression of GREM2 in breast cancer cells was increased by the demethylating agent decitabine. Breast cancer cells overexpressing GREM2 decreased cell proliferation when compared to control cells, both in vitro and in vivo. Through comparison of RNA-seq analysis between cell lines and tissue samples, gene ontologies that were consistently upregulated or downregulated by GREM2 in breast cancer were identified. In particular, the expression of inhibitor of DNA-binding-1 (ID1) was repressed by GREM2. BMP2 is one of the upstream regulators that increases the expression of ID1, and the expression of ID1 reduced by GREM2 was restored by overexpression of BMP2. Also, the migration ability of breast cancer cells, which had been suppressed by GREM2, was restored by BMP2 or ID1.
Conclusions
Low expression of GREM2 in breast cancer cells is associated with hypermethylation of the GREM2 promoter, which may ultimately contribute to poor patient survival. GREM2 participates in regulating the expression of various genes, including ID1, and is involved in suppressing the proliferation of breast cancer cells. This suggests that GREM2 has the potential to act as a novel tumor suppressor in breast cancer.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13058-024-01935-1.
Keywords: Gremlin-2, Breast cancer, Hypermethylation, RNA sequencing, Inhibitor of DNA-binding-1, Tumor suppressor
Background
Breast cancer is the most common malignancy in women worldwide and occurs across a variety of age groups, including women with early onset [1]. Typically, breast cancer is divided into subtypes based on the presence or absence of three receptors: estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) [2]. Although several studies have endeavored to discover novel molecular targets for each subtype of breast cancer, the range of treatments accessible to breast cancer patients remains primarily limited to those that target conventional receptors. Therefore, to propose promising treatments applicable to breast cancer, it is essential to thoroughly investigate new molecular targets for breast cancer and closely analyze their exact mechanisms.
Gremlin-2 (GREM2) is a well-known paralog of gremlin-1 (GREM1). It has a C-terminal cystine knot structure and acts as an antagonist to bone morphogenetic proteins (BMPs). Both gremlins play important roles in embryonic development, organ development, and tissue differentiation through the regulation of BMPs [3, 4]. However, studies have shown that GREM1 and GREM2 have different functions. While GREM1 has been widely reported to promote diseases like organ fibrosis [5, 6] and cancer [7–10], GREM2 has been primarily found to inhibit the differentiation of adipose [11], skin [12], or bone marrow [13]-derived stem/progenitor cells. GREM2 reduces the negative effects of inflammation after a myocardial infarction by inhibiting BMP signaling [14]. The direct role of GREM2 in cancer biology is not yet well known, but some studies suggest a tumor suppressor function of GREM2. Treatment with GREM2 protein inhibits the growth of endometrial cancer cell lines [15]. Interestingly, miR-423-5p promotes chemotherapy resistance in prostate cancer by suppressing GREM2 expression [16]. MiR-103a-3p suppresses GREM2 expression and contributes to the proliferation, migration, and invasion of colon [17] or liver [18] cancer cells. In our previous study, we demonstrated that GREM2 inhibits the proliferation and metastasis of breast cancer cells by preventing the differentiation of adipocytes. Overexpression of GREM2 in adipocytes not only inhibits adipogenic differentiation but also inhibits the expression and secretion of several adipokines, including IL-6, ultimately participating in suppressing breast cancer progression [19]. More interestingly, these functions of GREM2 are associated with the inhibition of BMP signaling [14] or the augmentation of Wnt/β-catenin [11, 19, 20] signaling.
Inhibitor of DNA-binding-1 (ID1) is a member of the helix-loop-helix (HLH) transcription factors. ID1, which lacks a DNA binding domain, forms heterodimers with other HLH transcription factors and inhibits their transcriptional activity [21]. ID1 is one of the key factors regulating cellular differentiation, lineage commitment, and development [22]. ID1 is also overexpressed in various cancers and has been found to be associated with several characteristics of cancer, such as cancer cell growth, migration, and invasion [23, 24]. In particular, ID1 is involved in cancer cell invasion [25], metastasis [25, 26], and stemness [27, 28] of cancer stem cells in breast cancer. Also, ID1 is associated with worse prognosis in breast cancer patients [29]. ID1 has been found to be significantly correlated with various signaling pathways such as BMP, K-Ras, MYC, etc [23]. Human Id1 gene contains a BMP-2-responsive element and the expression of ID1 is known to be mediated by BMP-2 and Smad1/4 [30]. ID1 cooperates with Ras to subvert the cellular senescence response and drive metastatic breast carcinoma [26]. Additionally, the MYC-ZNF148-ID1/3 axis has been reported to induce cancer stem cell characteristics in aggressive breast cancer [28].
This study demonstrates that GREM2 expression is reduced in breast cancer tissues and cells. Moreover, low levels of GREM2 mRNA expression in breast cancer cells are associated with its promoter hypermethylation and poor patient survival. Our in vitro and in vivo experiments reveal that GREM2 can inhibit the proliferation of breast cancer cells. Through RNA sequencing (RNA-seq) analysis, it was confirmed that GREM2 affects the expression of specific genes, including ID1. These results suggest that GREM2 is a novel tumor suppressor and may be a new target for breast cancer diagnosis or treatment.
Methods
Cell culture and reagents
Human breast cancer cell lines MDA-MB-453, SKBR3, and mouse breast cancer cell line MTV/TM-011 cells were originally obtained from Korean Cell Line Bank (Republic of Korea). The cells were incubated with RPMI (Corning Inc., USA) containing 10% fetal bovine serum (FBS, Thermo Fisher Scientific, USA) and 1% penicillin/streptomycin (Corning Inc.). Cells were maintained at 37 ℃ in air humidified with 5% CO2/95% air. Rabbit polyclonal GREM2 antibody was purchased from Abcam (UK). Anti-cyclin A2, anti-cyclin B1, anti-cyclin D1, anti-cyclin E2, anti-CDK2, anti-CDK4, anti-CDK6, and anti-β-actin were obtained from Cell Signaling Technology (USA).
Establishment of stable cell line
MDA-MB-453, SKBR3, and MTV-TM-011 cells were cultured in RPMI containing 10% FBS to reach 50% confluence. The next day, MDA-MB-453 and SKBR3 cells were transfected with mock or GREM2 ORF plasmid (Sino Biological. Cat#. HG10283-CY, China) using lipofectamine (Thermo Fisher Scientific) and incubated for 72 h. Stable cell lines were selected by 300 µg/ml hygromycin (InvivoGen, USA). MTV/TM-011 cell lines stably expressing either mock or Grem2 were established using a lentiviral transduction system. Briefly, mouse Grem2 lentiviral vector (EX-Mm06243-Lv122) was obtained from GeneCopoeia (USA). The lentiviruses were packaged in 293T cells using Lenti-Pac™ HIV expression packaging kit (GeneCopoeia). After 72 h transfection, the viral supernatant was collected, filtered and used for the transduction of MTV/TM-011 cells in the presence of 8 µg/ml polybrene (Merck Millipore, USA). Stable cell lines were selected by 1 µg/ml puromycin (InvivoGen).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cells using TRIzol® (Thermo Fisher Scientific). Reverse transcription of total RNA was performed using M-MLV reverse transcriptase (Enzynomics, Republic of Korea) according to the manufacturer’s protocol. Quantitative PCR (qPCR) was performed using TOP real™ qPCR 2X Pre-MIX (Enzynomics) and StepOnePlus Real-Time PCR (Thermo Fisher Scientific). ACTB or GAPDH was used as an internal reference. Primer sequences are listed in the Supplementary Table 1.
Western blot analysis
Standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis a (SDS-PAGE) and Western blotting were used to analyze the expression of various proteins. Cells were lysed in the lysis buffer (Cell Signaling Technology) containing protease inhibitors and phosphatase inhibitors (Roche, Switzerland). The quantitative protein concentration was determined by BCA Protein Assay Kit (Thermo Fisher Scientific) and equal amounts of protein were loaded on 8–12% SDS-PAGE. Proteins were transferred to polyvinylidene difluoride membrane (Merck Millipore) and subjected to immunoblotting using various antibodies overnight at 4℃, followed by further incubation with the secondary antibody (AbFrontier, Republic of Korea) at room temperature for 1 h. Visualization of protein bands was detected with Westsave Gold detection reagents (AbFrontier).
Wound healing assay
Cells were seeded in 12-well culture dishes and wounded by manually scraping the surface with a 1 ml pipette tip. The scratched surface was washed with PBS to remove cell debris. These cells were treated with medium containing 10% FBS and then cultured for 18 to 72 h. Using an inverted microscope with a 4x magnification, phase contrast images of the wound area were taken at 0 h (control) and 18–72 h after culture. Image J software was used to quantify the wound area.
Colony formation
Cells were seeded in 6-well plates at densities of 2.0 × 104 cells/well, respectively. The existing medium was replaced with medium containing 10% FBS every two days for 7 to 10 days. Cells were then fixed with methyl alcohol, stained with 0.5% crystal violet solution, and rinsed with PBS. After washing with PBS, the cells were examined under a microscope and photographs were taken in several fields. Cells stained with crystal violet were dissolved in dimethyl sulfoxide, and the absorbance was measured and quantified.
In vivo mouse model
Female BALB/c mice, 5 weeks of age (weight 18–20 ± 1–2 g) were purchased from Orient Bio Inc. (Republic of Korea). Mice were controlled in specific pathogen free conditions: 20–24 ℃, 12/12 h of dark/light cycle, 60 ± 5% humidity, and plastic cage (4 mice/cage). For the syngeneic breast cancer mouse model, MTV/TM-011-mock or MTV/TM-011-Grem2 cells were inoculated into the flanks of anesthetized mice by isoflurane inhalation. For the orthotopic breast cancer mouse model, MTV/TM-011-mock or MTV/TM-011-Grem2 cells were inoculated into the fourth mammary fat pad of anesthetized mice by isoflurane inhalation. Both the volume of the tumors and the body weight of mice were measured twice a week. At the end of the experiment, mice were euthanized by CO2 inhalation and each tumor was removed.
Immunofluorescence staining
Before staining fixed paraffin-embedded tissues, we followed the standard protocol including the steps of deparaffinization, antigen retrieval, and permeabilization. For immunofluorescence, detection of primary antibodies was done using fluorescent conjugates of Alexa Fluor® 488 antibody (Thermo Fisher Scientific) along with ProLong® Gold Antifade Reagent with DAPI (Thermo Fisher Scientific).
RNA sequencing (RNA-seq) and data analysis
Sequencing libraries were generated and sequenced by Macrogen (Republic of Korea). Total RNA was purified using RNeasy mini Kit (Qiagen, Germany). Purified RNA was randomly fragmented and synthesized into cDNA. After PCR amplification, an insert size of 200–400 bp was secured through a size selection process. The analysis results were mapped to the reference genome using the HISAT2 program, and the expression level obtained through transcript quantification of the sample was extracted as FPKM (Fragments Per Kilobase of transcript per Million mapped reads)/RPKM (Reads Per Kilobase of transcript per Million mapped reads) and TPM (Transcripts Per Kilobase Million) values. Genes or transcripts with statistically differentially expressed values of two or more groups under different conditions were selected. Additionally, functional annotation analysis was performed for these genes based on gene ontology (GO).
Statistical analysis
Data was expressed as the mean ± SD of results obtained from at least three independent experiments. Significant differences were determined by a Student’s t-test or one/two-way ANOVA. A P-value of less than 0.05 was considered to be statistically significant.
Results
The mRNA level of GREM2 is suppressed in breast cancer tissues/cells and is associated with poor survival of breast cancer patients
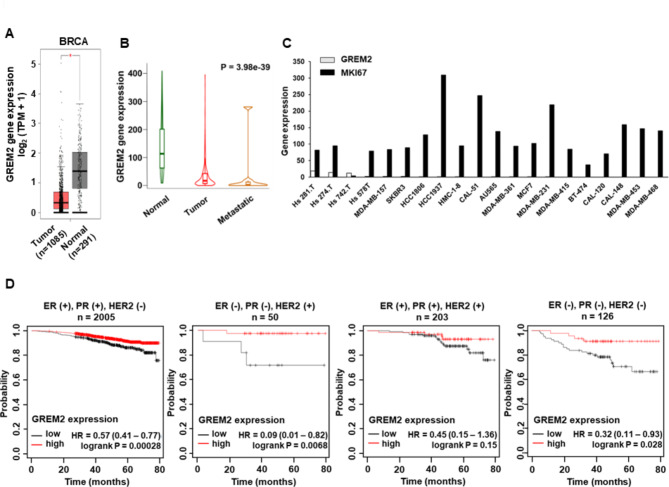
By analyzing public databases such as GEPIA and TNMplot, we discovered that the mRNA level of GREM2 was significantly suppressed in breast cancer tissues (Fig. 1A) and further reduced in metastatic tumors (Fig. 1B). The Expression Atlas database analysis revealed a significant decrease in GREM2 expression compared to MKI67, a cancer marker, in all 20 representative breast cancer cell lines (Fig. 1C). Subsequently, we investigated the correlation between GREM2 expression and the overall survival (OS) of breast cancer patients. Notably, low expression of GREM2 was associated with reduced OS in all four categories of breast cancer, although the p-value for the three positive (ER+, PR+, and HER2+, p-value = 0.15) group was not significant (Fig. 1D). The low expression of GREM2 was strongly associated with worse OS, particularly in breast cancer patients who were double negative (ER-, PR-, and HER2+) (GREM2 low vs. high expression patients: hazard ratio (HR) of survival = 0.09, 95% confidence interval (CI): 0.01–0.82, P = 0.0068) and triple negative (ER-, PR-, and HER2-) (GREM2 low vs. high expression patients: HR of survival = 0.32, 95% CI: 0.11–0.93, P = 0.028) (Fig. 1D). These findings contradict our previous results [8], which showed no significant correlation between GREM2 expression and breast cancer patient survival. In the previous study, only patient groups based on the presence or absence of ER expression were analyzed in the gene chip-based database provided by Kaplan-Meier plotter. However, the current results were based on a recently updated RNA-seq database provided by Kaplan-Meier plotter and analyzed considering the presence or absence of ER, PR, and HER2. Based on these findings, we suggest that GREM2 is significantly downregulated in breast cancer cells and that a low level of GREM2 predicts poor prognosis in breast cancer patients.
Fig. 1.
Low expression of GREM2 in breast cancer tissues/cells is associated with poor patient survival. A GREM2 expression was analyzed between TCGA breast invasive carcinoma tissues (n = 1085) and TCGA normal + GTEx normal (n = 291) tissues using GEPIA. B Expression of GREM2 in tumor (n = 1097) and metastatic tissues (n = 7) of breast cancer patients was compared with normal tissues (n = 113) using RNA-Seq TNMplot database. C The Expression Atlas database was used to analyze the mRNA levels of GREM2 and MKI67 in breast cancer cell lines. D Kaplan–Meier analysis (Kaplan-Meier Plotter) of overall survival by low or high GREM2 mRNA expression was performed in each indicated category of breast cancer patients. HR, hazard ratio
Hypermethylated GREM2 promoter is associated with poor survival of breast cancer patients
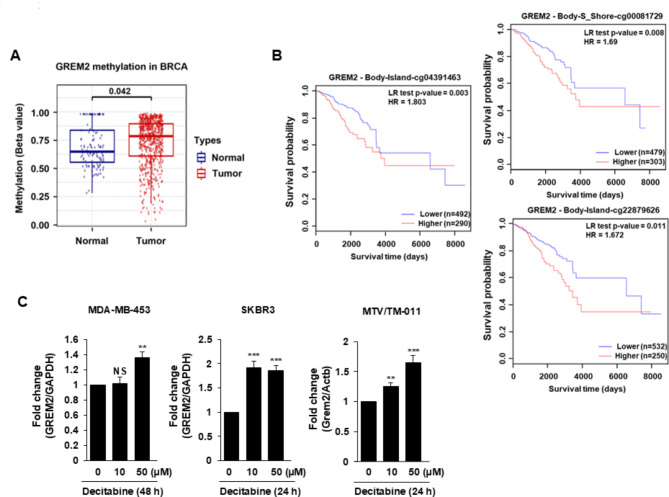
We utilized the GSCA database to investigate the potential correlation between low-level GREM2 expression in breast cancer cells and hypermethylation of the GREM2 promoter. As shown in Fig. 2A, there was a significant increase in methylation of the GREM2 promoter (cg02577267_GREM2) in breast cancer tumors compared to normal tissues. MethSurv survival analysis demonstrated a noteworthy association between higher methylation of the GREM2 promoter and poor OS in breast cancer. The Cancer Genome Atlas (TCGA) breast cancer data showed that 8 out of 23 CpGs in the GREM2 promoter showed a significant correlation between methylation levels and patient survival (Supplementary Table 2). Additionally, higher methylation of each of the eight GREM2 promoter CpGs was associated with lower patient survival (Fig. 2B, Supplementary Fig. 1). To test whether high methylation mediates low levels of GREM2 expression, three breast cancer cell lines (MDA-MB-453, SKBR3, and MTV/TM-011) were treated with demethylating agent decitabine. Decitabine significantly increased the mRNA expression of GREM2 in these cells (Fig. 2C), indicating that hypermethylation of the GREM2 promoter may be associated with transcriptional repression of GREM2 in breast cancer cells.
Fig. 2.
Hypermethylation of the GREM2 promoter is associated with poor survival in breast cancer patients. A Methylation levels of the GREM2 promoter were compared between TCGA breast invasive carcinoma patients and normal tissues in the GSCA database. B The survival rate of breast cancer patients according to the methylation level of each CpG site of GREM2 was analyzed using MethSurv. C Three breast cancer cell lines were treated with decitabine (10 or 50 µM) for 24–48 h and the mRNA level of GREM2 was quantified by qPCR analysis. Two-sided t-test. **, p < 0.01; and ***, p < 0.001. NS: not significant
GREM2 inhibits breast cancer cell proliferation in vitro and in vivo
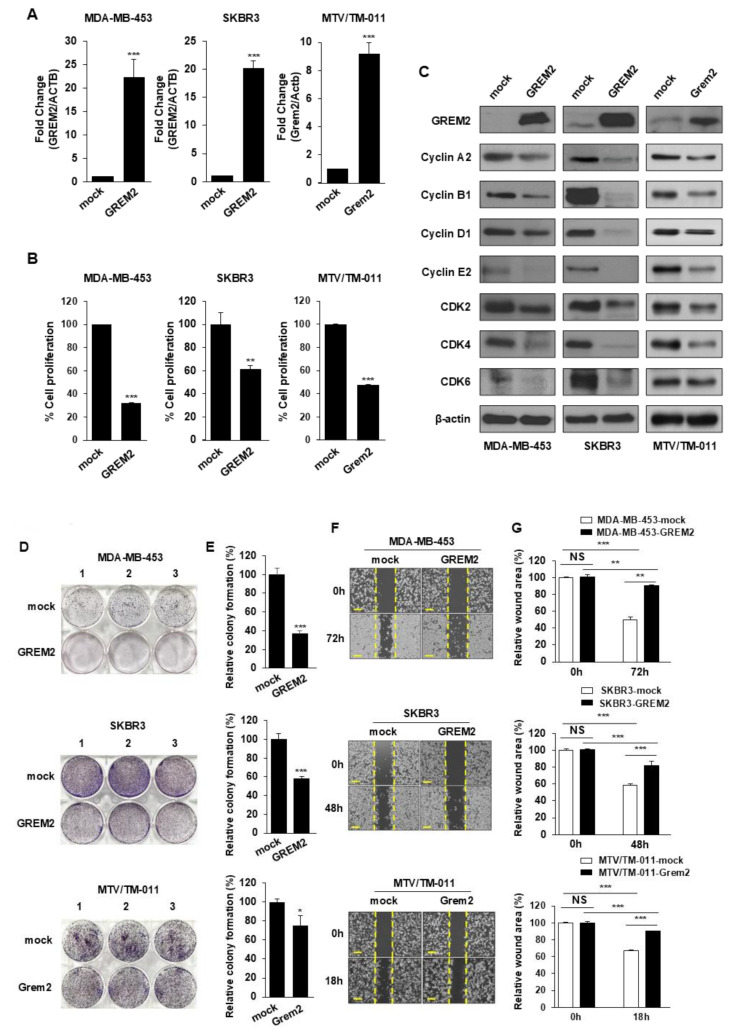
To investigate the effect of GREM2 on breast cancer cell growth, we introduced GREM2 overexpression plasmids into three different breast cancer cell lines and established stable cell lines for each. Significant increases in the mRNA (Fig. 3A) and protein (Fig. 3C) levels of GREM2 were observed in all three cell lines. The overexpression of GREM2 effectively suppressed the proliferation of breast cancer cells (Fig. 3B), as well as the expression levels of various cell cycle-related proteins when compared to the control group (Fig. 3C). Additionally, GREM2 overexpression inhibited the formation of cell colonies (Fig. 3D and E) and the migration of breast cancer cells (Fig. 3F and G). Although the ER status of MTV/TM-011 cell lines is unknown, MDA-MB-453 and SKBR3 are representative ER-negative cell lines. We further investigated the effect of GREM2 overexpression on cell proliferation in representative ER-positive cell lines, T47D (ER+/HER2-) and BT474 (ER+/HER2+). As shown in Supplementary Fig. 2, both cell viability and colony proliferation were inhibited in T47D and BT474 cell lines overexpressing GREM2 compared with the control.
Fig. 3.
Overexpression of GREM2 inhibits proliferation of breast cancer cells. A Each breast cancer cell line was established using a mock or GREM2 ORF plasmid. Expression level of GREM2 was confirmed by qPCR analysis. B Cells were seeded 96-well plates and incubated for 48 h, followed by the cell proliferation assay. C Proteins were isolated from each stable cell line, and lysates were immunoblotted with the indicated antibodies. D, E Breast cancer cells were seeded in 6-well plates and cultured for 7 to 10 days. Colony formation was analyzed and quantified using crystal violet staining. F, G Cells were seeded in 12-well plates and wounded to monitor the extent of wound healing at specified time points. Scale bar = 200 μm. A, B, E Two-sided t-test. G Two-way ANOVA. *, p < 0.05; **, p < 0.01; and ***, p < 0.001. NS: not significant
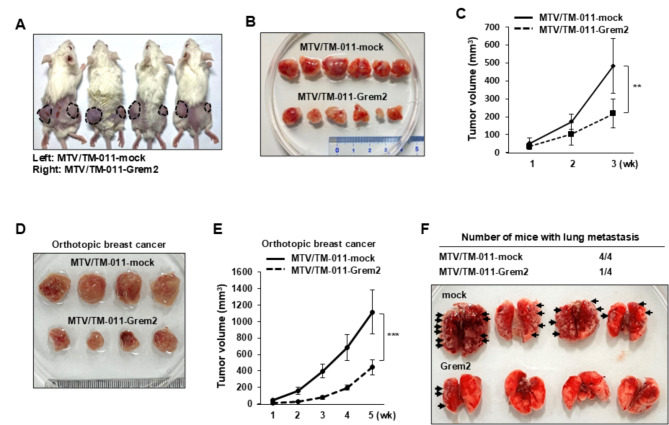
Next, to determine the effect of GREM2 on tumor growth of breast cancer cells, MTV/TM-011-mock or MTV/TM-011-Grem2 cells were injected into the left or right flank of mice, respectively. Our results showed that overexpression of GREM2 in breast cancer cells significantly reduced tumor volume compared to controls (Fig. 4A-C). Furthermore, the expressions of CDK4, cyclin B1, and cyclin D1 were reduced in Grem2-overexpressing tumors compared with control tumors (Supplementary Fig. 3A). As shown in Fig. 4D and E, mice injected with MTV/TM-011-Grem2 cells into the mammary fat pads developed smaller primary tumors than mice injected with MTV/TM-011-mock cells, resulting in a dramatic reduction in tumor volume [mean ± SD (mm3): 1116.11 ± 267.72 (MTV/TM-011-mock) vs. 443.73 ± 87.84 (MTV/TM-011-Grem2), four mice/each group]. Consistent with the growth outcome of the primary tumor, mice injected with MTV/TM-011-Grem2 had less lung metastases than mice injected with MTV/TM-011-mock (Fig. 4F). Additionally, the mRNA expression levels of Mmp2, Mmp3, Mmp9, Mmp11, and Mmp13 were significantly lower in primary breast cancer tissues of the MTV/TM-011-Grem2 group compared to the control group (Supplementary Fig. 3B). Altogether, these findings suggest that GREM2 plays an important role in suppressing the proliferation and metastasis of breast cancer cells.
Fig. 4.
GREM2 suppresses tumor growth of breast cancer cells in vivo. A-C MTV/TM-011-mock or MTV/TM-011-Grem2 cells were inoculated subcutaneously into the flanks of female nude mice (left: mock, right: Grem2, n = 6). Representative images of mice (A), dissected tumors (B), and tumor volume (C). D-F MTV/TM-011-mock or MTV/TM-011-Grem2 cells were injected into mammary fat pads of mice. Representative images of primary tumors (D), tumor volume (E), and lung metastatic foci (F). Black arrowheads indicate prominent lung metastatic foci. Two-sided t-test. **, p < 0.01 and ***, p < 0.001
Differentially expressed genes (DEGs) are identified between GREM2 overexpression group and control group in breast cancer cells
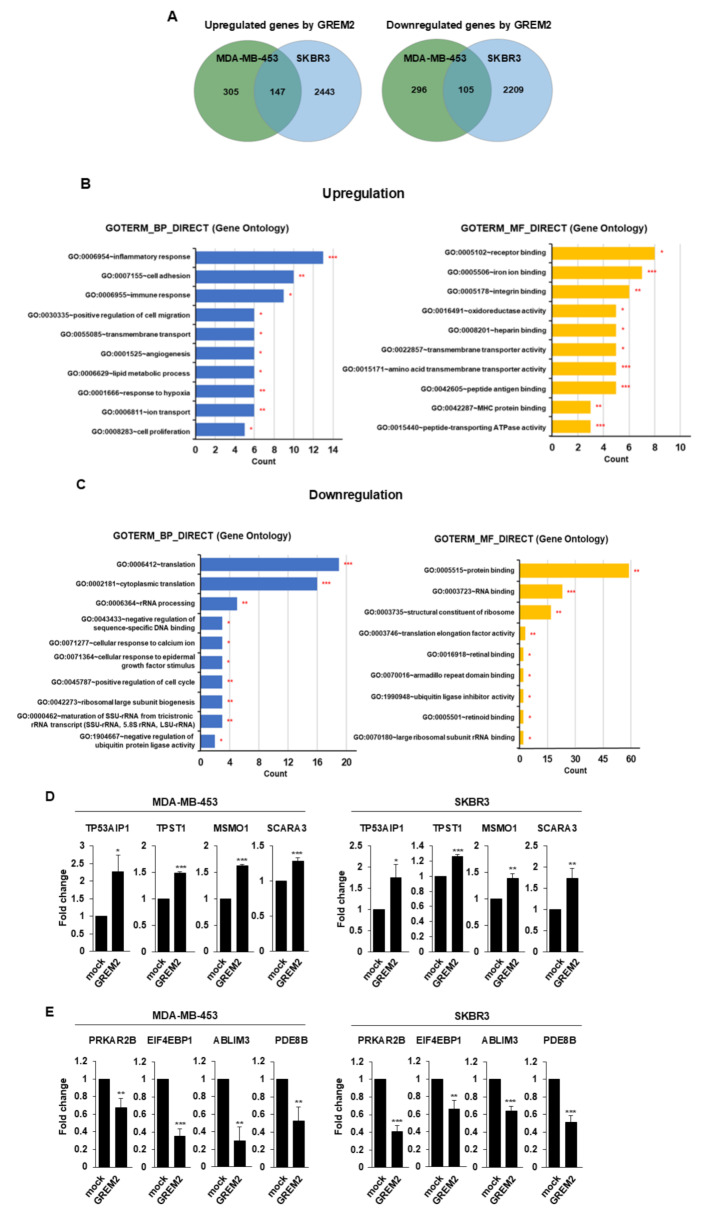
To investigate the role of GREM2 in breast cancer and its impact on tumor suppression, we identified potential target genes affected by GREM2. Gene expression levels between breast cancer cells overexpressing GREM2 (MDA-MB-453-GREM2, SKBR3-GREM2; n = 2 for each cell line) and control cells (MDA-MB-453-mock, SKBR3-mock; n = 2 for each cell line) were compared using RNA-seq analysis. Initially, we examined the differentially expressed genes (DEGs) using a fold change (FC) threshold of ≥ 1.5 and a significance level of p < 0.05 in the RNA-seq analysis. Subsequently, genes commonly upregulated or downregulated by GREM2 in both MDA-MB-453 and SKBR3 cell lines were identified. As shown in Fig. 5A and Supplementary Tables 3, 147 genes were upregulated and 105 genes were downregulated in both breast cancer cell lines by GREM2 overexpression.
Fig. 5.
GREM2 is involved in regulating the expression of various genes in breast cancer cells. A Venn diagram showing genes differentially expressed in GREM2-overexpressing breast cancer cells (MDA-MB-453-GREM2 and SKBR3-GREM2) compared to control cells (MDA-MB-453-mock and SKBR3-mock). Genes commonly increased (left) or decreased (right) by GREM2 in two breast cancer cell lines were analyzed. B, C GOs for genes up- (B) or downregulated (C) by GREM2 in breast cancer cells were analyzed using DAVID. BP, biological processes; MF, Molecular Function. D, E Some of the genes up- (D) or downregulated (E) by GREM2 were verified through qPCR analysis. Two-sided t-test. *, p < 0.05; **, p < 0.01; and ***, p < 0.001
The identified DEGs were classified based on their biological processes (BP), molecular functions (MF), and cellular components (CC) using the David database. In each category, the top 10 statistically significant GOs were selected and presented in a diagram (Fig. 5B and C, supplementary Fig. 4), ordered by gene count. For the BP category, representative GOs upregulated by GREM2 overexpression included inflammatory response, cell adhesion, and immune response. In the MF category, the representative GOs were receptor binding, iron ion binding, and integrin binding. Finally, for the CC category, the representative GOs were integral components of membrane, plasma membrane, and membrane. On the other hand, representative GOs downregulated by GREM2 overexpression included translation, cytoplasmic translation, and rRNA processing in the BP category; protein binding, RNA binding, and structural constituent of ribosome in the MF category; and cytosol, cytoplasm, and nucleus in the CC category.
Based on the RNA-seq results, qPCR analysis was further performed to verify the upregulation or downregulation of some target genes by GREM2. In GREM2-overexpressing breast cancer cells, the mRNA expression levels of the following genes were significantly increased: TP53AIP1 (p53-regulated apoptosis-inducing protein 1), TPST1 (tyrosylprotein sulfotransferase 1), MSMO1 (methylsterol monooxygenase 1), and SCARA3 (scavenger receptor class A member 3) (Fig. 5D). Conversely, the mRNA expression levels of PRKAR2B (protein kinase CAMP-dependent type II regulatory subunit beta), EIF4EBP1 (eukaryotic translation initiation factor 4E binding protein 1), ABLIM3 (actin binding LIM protein family member 3), and PDE8B (phosphodiesterase 8B) genes were significantly reduced in GREM2-overexpressing breast cancer cells compared to control cells (Fig. 5E). Additionally, the association between these genes and GREM2 was identified in the human breast tissue database. Analysis of the GTEx breast tissue database revealed that each of these genes exhibited a significant positive correlation (supplementary Fig. 5A) or negative correlation (supplementary Fig. 5B) with GREM2 expression.
ID1 is one of the potent target genes of GREM2 in breast cancer
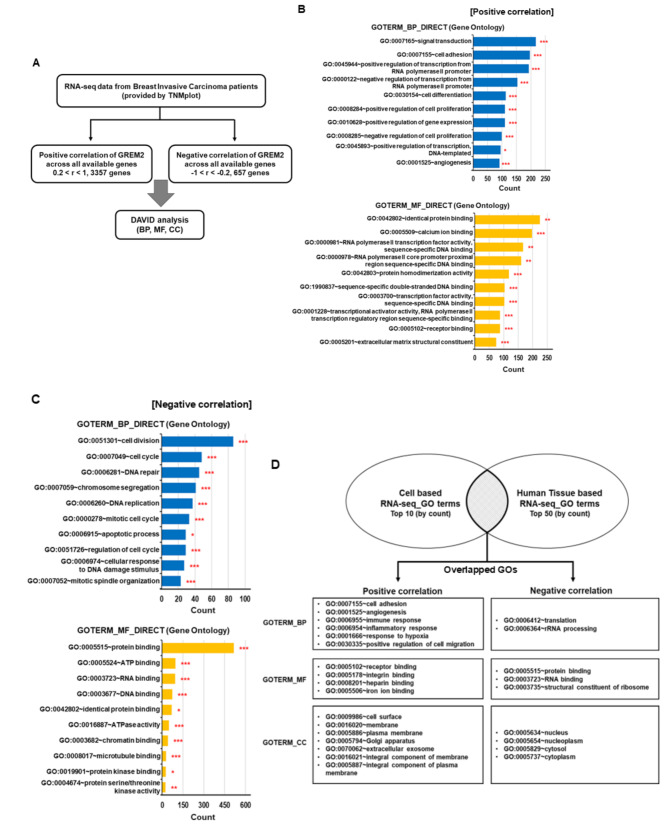
To further understand the downstream signaling pathways regulated by GREM2 in breast cancer, we conducted a comparison between the results of cell-based RNA-seq analysis (Fig. 5) and human breast tissue-based RNA-seq analysis. Using RNA-seq data from breast invasive carcinoma patients provided by TNMplot, we identified 3357 genes that showed a positive correlation with GREM2 (0.2 < r < 1.0) and 657 genes that showed a negative correlation with GREM2 (-1.0 < r < -0.2). We then performed DAVID analysis on these genes (Fig. 6A). For genes positively or negatively correlated with GREM2 in breast cancer tissues, the top 10 GO categories were identified in BP, MF, and CC (Fig. 6B and C, and supplementary Fig. 6). In each category of BP, MF, and CC, the top 10 GOs (by number of genes) from RNA-seq for GREM2 overexpressing cell lines were compared with the top 50 GOs (by number of genes) from RNA-seq for human breast cancer tissues. Interestingly, many GO pathways that showed positive or negative correlation with GREM2 overlapped between the two analyses. While a wide variety of GOs showed positive correlations with GREM2, GOs showing negative correlations with GREM2 were mainly concentrated in limited pathways associated with translation, ribosomes, and protein binding (Fig. 6D).
Fig. 6.
Specific GO pathways showing positive or negative correlation with GREM2 in breast cancer patient tissues overlap with cell line-derived RNA-seq results. A Schematic of the filtering pipeline for genes showing positive or negative correlation with GREM2 in invasive breast carcinoma patients. B, C GOs for genes showing positive- (B) or negative correlation (C) with GREM2 in breast cancer patients were analyzed using DAVID. D GO pathways that were commonly positively or negatively correlated with GREM2 in both breast cancer cell lines and invasive breast cancer patient tissues were analyzed
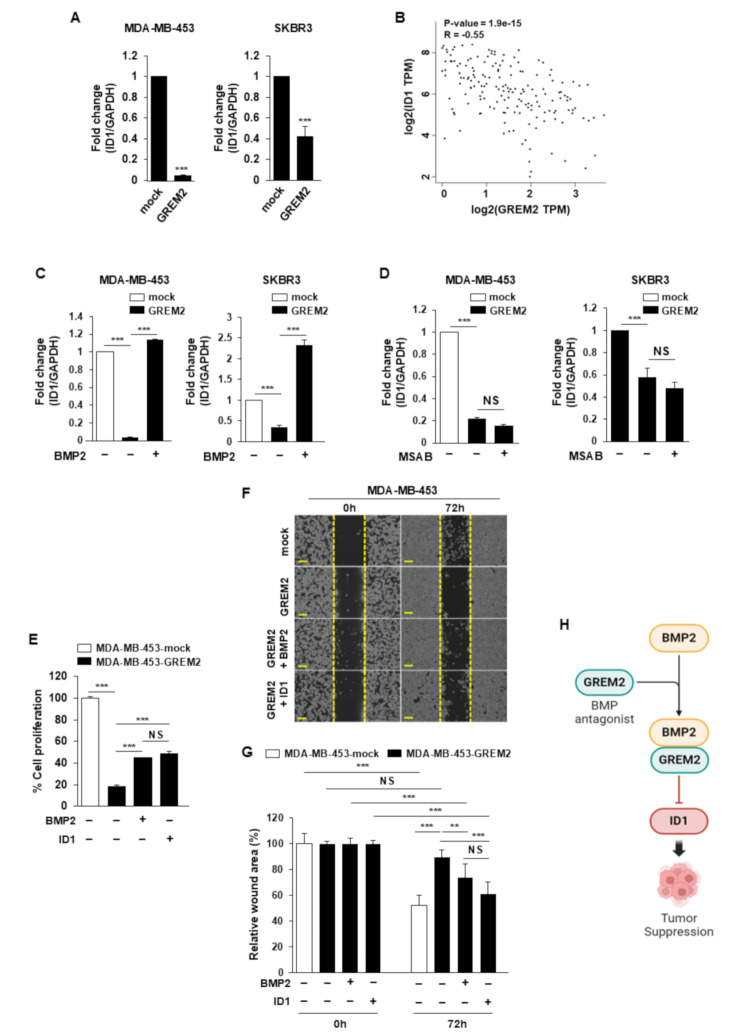
In the MF category, the GO term “protein binding” (GO:0005515) was most consistently negatively correlated with GREM2 in breast cancer cell lines and human breast cancer tissues (Fig. 6D). ID1 was one of the genes involved in “protein binding” that was significantly reduced in breast cancer cell lines overexpressing GREM2 (Supplementary Table 4). The mRNA level of ID1 was significantly decreased in two breast cancer cell lines that overexpressed GREM2 (Fig. 7A). A significant negative correlation was also identified between the mRNA expression of GREM2 and ID1 in human breast tissues (Spearman r = − 0.55, p = 1.9 × 10− 15) (Fig. 7B). Next, it was confirmed whether the decrease in ID1 expression caused by GREM2 was affected by the antagonism of GREM2 against BMP2, one of the upstream regulators of ID1. As shown in Fig. 7C, the reduced mRNA expression of ID1 in GREM2-overexpressing breast cancer cells was significantly restored by further increasing the expression of BMP2. In addition, it was confirmed whether the increase in Wnt/β-catenin signaling by GREM2 is involved in the decrease in ID1 expression. However, the expression of ID1, which was repressed by GREM2, was not affected by treatment with MSAB, a Wnt/β-catenin inhibitor (Fig. 7D). We investigated whether inhibition of ID1 or BMP2 by GREM2 was involved in the inhibition of proliferation of breast cancer cells. The decreased proliferation (Fig. 7E) and migration (Fig. 7F and G) ability of breast cancer cells by GREM2 was partially restored by increasing the expression of BMP2 or ID1. Collectively, these results demonstrate that GREM2 is a novel tumor suppressor in breast cancer that inhibits cell proliferation through negative regulation of BMP2-ID1 signaling (Fig. 7H).
Fig. 7.
GREM2 inhibits proliferation of breast cancer cells through negative regulation of BMP2-ID1. A The mRNA expression level of ID1 was analyzed through qPCR in each cell line. B The correlation between GREM2 and ID1 in GTEx human breast tissue database was investigated by using GEPIA. Correlation analysis was conducted using Spearman rank test. C Breast cancer cells overexpressing GREM2 were transfected with mock or BMP2 plasmid, and the mRNA expression level of ID1 was analyzed through qPCR. D Breast cancer cells overexpressing GREM2 were treated with or without MSAB (10 µM) for 24 h. RNA was isolated from each cell line and the mRNA expression level of ID1 was analyzed through qPCR. E-G Breast cancer cells overexpressing GREM2 were transfected with mock, BMP2, or ID1 plasmids, and then proliferation (E) or migration (F, G) analysis was performed for each cell line after 72 h. Scale bar = 200 μm. H Schematic diagram showing the proposed molecular mechanism of GREM2 in breast cancer. A Two-sided t-test. C, D, E, G Two-way ANOVA. **, p < 0.01; and ***, p < 0.001. NS: not significant
Discussion
Our previous study demonstrated that GREM2, when overexpressed in preadipocytes, not only inhibits adipocyte differentiation but also suppresses the expression of various adipokines, including IL-6. Additionally, adipocytes overexpressing GREM2 play an important role in suppressing the growth, migration, and metastasis of breast cancer cells [19]. However, GREM2, a type of cytokine, has the potential to inhibit breast cancer growth independently of adipogenic differentiation and/or adipokine inhibition. In this study, we observed a significantly lower expression of GREM2 in both breast cancer patients and cells compared to the control group. Overexpression of GREM2 in breast cancer cells led to the inhibition of cell proliferation, migration, tumor growth and lung metastasis. RNA-seq analysis showed that GREM2 may affect the regulation of various gene expressions in breast cancer cells, particularly suppressing the expression of genes involved in translation, ribosomes, and protein binding. These findings suggest that GREM2 has the potential to serve as a novel tumor suppressor in breast cancer.
Tumor suppressor genes play a crucial role in regulating various cellular functions, including cell growth, cell cycle progression, DNA repair mechanisms, and apoptosis [31]. Mutations in key tumor suppressor genes such as TP53, PTEN, and RB lead to a loss of tumor suppressor function, increasing the likelihood of developing cancer [32–34]. Mutations in BRCA1 and BRCA2 genes, which are responsible for the DNA repair system, are well-known to contribute to the development and progression of breast cancer [35, 36]. In cancer cells, the expression of tumor suppressor genes is often suppressed through hypermethylation of gene promoters. PCDH10, which acts as a tumor suppressor in various types of carcinomas, has been reported to have low mRNA expression associated with promoter hypermethylation [37, 38]. Likewise, the mRNA expression of genes such as RAI2 [39, 40], DHRS3 [41], and HOXB9 [42] is also suppressed by promoter hypermethylation in various carcinomas. Low expression of these genes is associated with increased proliferation of cancer cells and poor patient prognosis. Our results show that treatment with demethylating agent decitabine restored the suppressed expression level of GREM2 in breast cancer cells. This suggests that low levels of GREM2 expression in breast cancer cells are also likely associated with its promoter hypermethylation.
The low expression of GREM2 in breast cancer cells may be related to microRNA (miRNA) in addition to promoter hypermethylation. Recent reports have identified miRNAs that directly target GREM2 and inhibit its expression. For example, exosomal miR-423-5p secreted from cancer-related fibroblasts promotes chemotherapy resistance in prostate cancer by inhibiting GREM2 [16]. MiR-103a-3p is involved in colon cancer progression by regulating GREM2 expression [17]. Furthermore, empagliflozin could improve non-alcoholic fatty liver disease-related fibrosis by downregulating miR-34a-5p, which targets GREM2 [43]. These findings suggest the possibility that specific miRNAs may directly bind to the 3’UTR of GREM2 and inhibit its expression in breast cancer cells as well. However, more detailed studies are needed to fully understand this mechanism.
Our analysis revealed that GREM2 is involved in the upregulation of various molecular signaling pathways while downregulating pathways focused on translation, ribosomes, and especially protein binding. Some genes upregulated in breast cancer cell lines overexpressing GREM2 have been reported to have tumor suppressor functions in certain types of cancer. Among these genes, TP53AIP1 is an important gene for tumor suppressor p53-dependent apoptosis, and its overexpression promotes cell cycle arrest and apoptosis in breast cancer cells [44]. MSMO1 and SCARA3 were also genes upregulated by GREM2 and have been reported to suppress cancer progression in pancreatic cancer [45] and lung cancer [46], respectively. Meanwhile, among the protein binding-related genes significantly reduced by GREM2, PRKAR2B [47, 48], EIF4EBP1 [49–51], and ID1 [24, 52, 53] are known to be involved in the proliferation or progression of cancers. In particular, ID1 promotes proliferation and metastasis of breast cancer cells and is associated with poor prognosis of patients [25, 27, 29]. Our results indicate that reduced expression of ID1 mediated by GREM2 may ultimately contribute to the inhibition of breast cancer cell proliferation.
We propose that the up- and downregulation of various genes by GREM2 may be associated with changes in the activity of BMP or Wnt/β-catenin signaling pathway. Gremlins (GREM1 and GREM2) are representative antagonists of BMPs and are well known to directly bind to BMPs and inhibit the BMP signaling pathway [54–56]. Interestingly, ID1 is one of the target genes of BMPs, and its expression is increased by BMP2 [30, 57], BMP4 [58, 59], or BMP7 [58, 60]. In this study, the reduced expression of ID1 in GREM2-overexpressing cells was significantly restored by additional introduction of BMP2. This suggests that the reduction of ID1 expression by GREM2 may be related to the antagonism of GREM2 against BMPs in breast cancer cells. In addition, previous studies have shown that GREM2 inhibits adipogenesis in adipose-derived stromal/stem cells by activating the Wnt/β-catenin signaling pathway [11, 20]. Activation of Wnt/β-catenin signaling induced by GREM2 contributes to the reduction of IL-6 expression in adipocytes [19]. To determine whether the Wnt/β-catenin signaling increased by GREM2 is also involved in the inhibition of ID1 expression by GREM2, GREM2-overexpressing cells were treated with MSAB, a Wnt/β-catenin inhibitor. However, the expression of ID1 suppressed by GREM2 was not affected by inhibition of Wnt/β-catenin signaling, suggesting that the Wnt/β-catenin pathway is not involved in ID1 reduction by GREM2. Meanwhile, several studies have demonstrated that Wnt/β-catenin negatively regulates the C/EBP family members [61, 62]. This suggests that enhancement of Wnt/β-catenin signaling by GREM2 may reduce the expression of specific genes affected by the C/EBP pathway. Besides the C/EBP family, it is well-known that the NF-κB [63, 64] or PI3K/Akt/mTOR [65, 66] signaling pathway interacts with Wnt/β-catenin. Therefore, the expression levels of genes regulated by GREM2 in breast cancer cells may be affected not only by BMP or Wnt/β-catenin signaling but also by changes in the activity of additional kinases and/or transcription factors.
This study suggests that GREM2 may be a novel tumor suppressor gene that can suppress breast cancer proliferation, but it is still difficult to conclude whether the function of GREM2 varies depending on the type of breast cancer. Further studies are needed to elucidate the role of GREM2 and its mechanism according to the subtype of breast cancer.
Conclusion
In this study, we found that the expression of GREM2 was significantly lower in cells and tissues of breast cancer patients, and this was associated with hypermethylation of the GREM2 promoter. Additionally, low expression of GREM2 and hypermethylation of its promoter are associated with poor prognosis in breast cancer patients. RNA-seq analysis showed that GREM2 is involved in regulating the expression of specific genes in breast cancer cells and ultimately inhibits cancer cell proliferation. In particular, GREM2, a BMP antagonist, contributed to suppressing the proliferation of breast cancer cells by reducing the expression of ID1, one of the target genes of BMP2. Based on our findings, we propose that GREM2 may function as a novel tumor suppressor gene in breast cancer and potentially serve as a new diagnostic marker and therapeutic target.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
JJ and NHK are involved in study conception, collection and/or assembly of data, data analysis, and interpretation. JP, DL, MK, WG, SJ, MG, and CK are involved in collection and/or assembly of data and data analysis. YHC is involved in statistical processing and analysis. MHL, HSP, and YBE are involved in study conception and manuscript editing. SAP is involved in study conception and design, data interpretation, manuscript writing and editing, and financial support. All authors read and approved the final manuscript.
Funding
This work was supported by the Soonchunhyang University research fund, and the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2022R1F1A1074226).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
All animal experiments were performed in accordance with the guidelines for animal treatment of Soonchunhyang University. All experimental protocols in our study were conducted on protocols approved by the Institutional Animal Care and Use Committee of the Soonchunhyang University (SCH21-0014).
Consent for publication
All authors approved submission of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jiwoo Jung and Na Hui Kim contributed equally to this work.
References
- 1.Chelmow D, Pearlman MD, Young A, Bozzuto L, Dayaratna S, Jeudy M et al. Executive Summary of the Early-Onset Breast Cancer Evidence Review Conference. Obstet Gynecol. 2020;135(6):1457-78. [DOI] [PMC free article] [PubMed]
- 2.Onitilo AA, Engel JM, Greenlee RT, Mukesh BN. Breast cancer subtypes based on ER/PR and Her2 expression: comparison of clinicopathologic features and survival. Clin Med Res. 2009;7(1–2):4–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yanagita M. BMP antagonists: their roles in development and involvement in pathophysiology. Cytokine Growth Factor Rev. 2005;16(3):309–17. [DOI] [PubMed] [Google Scholar]
- 4.Ali IH, Brazil DP. Bone morphogenetic proteins and their antagonists: current and emerging clinical uses. Br J Pharmacol. 2014;171(15):3620–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller KA, Tavlaki E, Schneider M, Jorbenadze R, Geisler T, Kandolf R, et al. Gremlin-1 identifies fibrosis and predicts adverse outcome in patients with heart failure undergoing endomyocardial biopsy. J Card Fail. 2013;19(10):678–84. [DOI] [PubMed] [Google Scholar]
- 6.Church RH, Ali I, Tate M, Lavin D, Krishnakumar A, Kok HM, et al. Gremlin1 plays a key role in kidney development and renal fibrosis. Am J Physiol Ren Physiol. 2017;312(6):F1141–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim NH, Sung NJ, Youn HS, Park SA. Gremlin-1 activates Akt/STAT3 signaling, which increases the glycolysis rate in breast cancer cells. Biochem Biophys Res Commun. 2020;533(4):1378–84. [DOI] [PubMed] [Google Scholar]
- 8.Park SA, Sung NJ, Choi BJ, Kim W, Kim SH, Surh YJ. Gremlin-1 augments the oestrogen-related receptor alpha signalling through EGFR activation: implications for the progression of breast cancer. Br J Cancer. 2020;123(6):988–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sung NJ, Kim NH, Surh YJ, Park SA. Gremlin-1 promotes metastasis of breast Cancer cells by activating STAT3-MMP13 signaling pathway. Int J Mol Sci. 2020;21(23). [DOI] [PMC free article] [PubMed]
- 10.Park SA. Role of gremlin-1 in cancer. Biomedical Sci Lett. 2018;24(4):285–91. [Google Scholar]
- 11.Kawagishi-Hotta M, Hasegawa S, Igarashi T, Date Y, Ishii Y, Inoue Y, et al. Increase of gremlin 2 with age in human adipose-derived stromal/stem cells and its inhibitory effect on adipogenesis. Regen Ther. 2019;11:324–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawagishi-Hotta M, Hasegawa S, Inoue Y, Hasebe Y, Arima M, Iwata Y, et al. Gremlin 2 suppresses differentiation of stem/progenitor cells in the human skin. Regen Ther. 2021;18:191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang CL, Xiao F, Wang CD, Zhu JF, Shen C, Zuo B, et al. Gremlin2 suppression increases the BMP-2-Induced Osteogenesis of Human Bone Marrow-derived mesenchymal stem cells Via the BMP-2/Smad/Runx2 signaling pathway. J Cell Biochem. 2017;118(2):286–97. [DOI] [PubMed] [Google Scholar]
- 14.Sanders LN, Schoenhard JA, Saleh MA, Mukherjee A, Ryzhov S, McMaster WG, et al. BMP antagonist gremlin 2 limits inflammation after myocardial infarction. Circ Res. 2016;119(3):434–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsubamoto H, Sakata K, Sakane R, Inoue K, Shibahara H, Hao H, et al. Gremlin 2 is repressed in Invasive Endometrial Cancer and inhibits cell growth in Vitro. Anticancer Res. 2016;36(1):199–203. [PubMed] [Google Scholar]
- 16.Shan G, Gu J, Zhou D, Li L, Cheng W, Wang Y, et al. Cancer-associated fibroblast-secreted exosomal mir-423-5p promotes chemotherapy resistance in prostate cancer by targeting GREM2 through the TGF-beta signaling pathway. Exp Mol Med. 2020;52(11):1809–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z, Zhu X. MiR-103a-3p contributes to the progression of Colorectal Cancer by regulating GREM2 expression. Yonsei Med J. 2022;63(6):520–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma G, Chen J, Wei T, Wang J, Chen W. Inhibiting roles of FOXA2 in liver cancer cell migration and invasion by transcriptionally suppressing microRNA-103a-3p and activating the GREM2/LATS2/YAP axis. Cytotechnology. 2021;73(4):523–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung J, Kim NH, Kwon M, Park J, Lim D, Kim Y, et al. The inhibitory effect of Gremlin-2 on adipogenesis suppresses breast cancer cell growth and metastasis. Breast Cancer Res. 2023;25(1):128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Q, Tang SG, Yuan ZM. Gremlin 2 inhibits adipocyte differentiation through activation of Wnt/beta-catenin signaling. Mol Med Rep. 2015;12(4):5891–6. [DOI] [PubMed] [Google Scholar]
- 21.Wang LH, Baker NE, Proteins E, Proteins ID. Helix-Loop-Helix partners in Development and Disease. Dev Cell. 2015;35(3):269–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tzeng SF, de Vellis J. Id1, Id2, and Id3 gene expression in neural cells during development. Glia. 1998;24(4):372–81. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Z, Bo Z, Gong W, Guo Y. Inhibitor of differentiation 1 (Id1) in Cancer and Cancer Therapy. Int J Med Sci. 2020;17(8):995–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sikder HA, Devlin MK, Dunlap S, Ryu B, Alani RM. Id proteins in cell growth and tumorigenesis. Cancer Cell. 2003;3(6):525–30. [DOI] [PubMed] [Google Scholar]
- 25.Fong S, Itahana Y, Sumida T, Singh J, Coppe JP, Liu Y, et al. Id-1 as a molecular target in therapy for breast cancer cell invasion and metastasis. Proc Natl Acad Sci U S A. 2003;100(23):13543–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swarbrick A, Roy E, Allen T, Bishop JM. Id1 cooperates with oncogenic Ras to induce metastatic mammary carcinoma by subversion of the cellular senescence response. Proc Natl Acad Sci U S A. 2008;105(14):5402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teo WS, Holliday H, Karthikeyan N, Cazet AS, Roden DL, Harvey K, et al. Id proteins promote a Cancer stem cell phenotype in mouse models of Triple negative breast Cancer via negative regulation of Robo1. Front Cell Dev Biol. 2020;8:552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim M, Singh M, Lee BK, Hibbs M, Richardson K, Ellies L, et al. A MYC-ZNF148-ID1/3 regulatory axis modulating cancer stem cell traits in aggressive breast cancer. Oncogenesis. 2022;11(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Escolano M, Montoyo-Pujol YG, Ortiz-Martinez F, Ponce JJ, Delgado-Garcia S, Martin TA et al. ID1 and ID4 are biomarkers of Tumor aggressiveness and poor outcome in immunophenotypes of breast Cancer. Cancers (Basel). 2021;13(3). [DOI] [PMC free article] [PubMed]
- 30.Katagiri T, Imada M, Yanai T, Suda T, Takahashi N, Kamijo R. Identification of a BMP-responsive element in Id1, the gene for inhibition of myogenesis. Genes Cells. 2002;7(9):949–60. [DOI] [PubMed] [Google Scholar]
- 31.Sherr CJ. Principles of tumor suppression. Cell. 2004;116(2):235–46. [DOI] [PubMed] [Google Scholar]
- 32.Rivlin N, Brosh R, Oren M, Rotter V. Mutations in the p53 tumor suppressor gene: important milestones at the various steps of Tumorigenesis. Genes Cancer. 2011;2(4):466–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fusco N, Sajjadi E, Venetis K, Gaudioso G, Lopez G, Corti C et al. PTEN Alterations and Their Role in Cancer Management: Are We Making Headway on Precision Medicine? Genes (Basel). 2020;11(7). [DOI] [PMC free article] [PubMed]
- 34.MacPherson D, Sage J, Crowley D, Trumpp A, Bronson RT, Jacks T. Conditional mutation of rb causes cell cycle defects without apoptosis in the central nervous system. Mol Cell Biol. 2003;23(3):1044–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrucelli N, Daly MB, Pal T. BRCA1- and BRCA2-Associated Hereditary Breast and Ovarian Cancer. In: GeneReviews((R)). edn. Edited by Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Bean LJH, Gripp KW, Amemiya A. Seattle (WA). 1993.
- 36.Stoppa-Lyonnet D. The biological effects and clinical implications of BRCA mutations: where do we go from here? Eur J Hum Genet. 2016;24(Suppl 1):S3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu J, Cheng YY, Tao Q, Cheung KF, Lam CN, Geng H, et al. Methylation of protocadherin 10, a novel tumor suppressor, is associated with poor prognosis in patients with gastric cancer. Gastroenterology. 2009;136(2):640–51. e641. [DOI] [PubMed] [Google Scholar]
- 38.Qiu C, Bu X, Jiang Z. Protocadherin-10 acts as a tumor suppressor gene, and is frequently downregulated by promoter methylation in pancreatic cancer cells. Oncol Rep. 2016;36(1):383–9. [DOI] [PubMed] [Google Scholar]
- 39.Yan W, Wu K, Herman JG, Xu X, Yang Y, Dai G, et al. Retinoic acid-induced 2 (RAI2) is a novel tumor suppressor, and promoter region methylation of RAI2 is a poor prognostic marker in colorectal cancer. Clin Epigenetics. 2018;10:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lou X, Deng W, Shuai L, Chen Y, Xu M, Xu J, et al. RAI2 acts as a tumor suppressor with functional significance in gastric cancer. Aging. 2023;15(21):11831–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sumei S, Xiangyun K, Fenrong C, Xueguang S, Sijun H, Bin B, et al. Hypermethylation of DHRS3 as a novel tumor suppressor involved in Tumor Growth and Prognosis in Gastric Cancer. Front Cell Dev Biol. 2021;9:624871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao Y, Liu C, Wang B, Guan X, Fang L, Zhan F, et al. HOXB9 blocks cell cycle progression to inhibit pancreatic cancer cell proliferation through the DNMT1/RBL2/c-Myc axis. Cancer Lett. 2022;533:215595. [DOI] [PubMed] [Google Scholar]
- 43.Shen Y, Cheng L, Xu M, Wang W, Wan Z, Xiong H, et al. SGLT2 inhibitor empagliflozin downregulates miRNA-34a-5p and targets GREM2 to inactivate hepatic stellate cells and ameliorate non-alcoholic fatty liver disease-associated fibrosis. Metabolism. 2023;146:155657. [DOI] [PubMed] [Google Scholar]
- 44.Liang Y, Wang S, Liu J. Overexpression of tumor protein p53-regulated apoptosis-inducing protein 1 regulates proliferation and apoptosis of breast Cancer cells through the PI3K/Akt Pathway. J Breast Cancer. 2019;22(2):172–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao R, Zhang Z, Tian C, Sheng W, Dong Q, Dong M. Down-regulation of MSMO1 promotes the development and progression of pancreatic cancer. J Cancer. 2022;13(10):3013–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim J, You HJ, Youn C. SCARA3 inhibits cell proliferation and EMT through AKT signaling pathway in lung cancer. BMC Cancer. 2022;22(1):552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sha J, Han Q, Chi C, Zhu Y, Pan J, Dong B, et al. PRKAR2B promotes prostate cancer metastasis by activating Wnt/beta-catenin and inducing epithelial-mesenchymal transition. J Cell Biochem. 2018;119(9):7319–27. [DOI] [PubMed] [Google Scholar]
- 48.Xia L, Sun J, Xie S, Chi C, Zhu Y, Pan J, et al. PRKAR2B-HIF-1alpha loop promotes aerobic glycolysis and tumour growth in prostate cancer. Cell Prolif. 2020;53(11):e12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Armengol G, Rojo F, Castellvi J, Iglesias C, Cuatrecasas M, Pons B, et al. 4E-binding protein 1: a key molecular funnel factor in human cancer with clinical implications. Cancer Res. 2007;67(16):7551–5. [DOI] [PubMed] [Google Scholar]
- 50.Karlsson E, Perez-Tenorio G, Amin R, Bostner J, Skoog L, Fornander T, et al. The mTOR effectors 4EBP1 and S6K2 are frequently coexpressed, and associated with a poor prognosis and endocrine resistance in breast cancer: a retrospective study including patients from the randomised Stockholm tamoxifen trials. Breast Cancer Res. 2013;15(5):R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li F, Wang Q, Xiong X, Wang C, Liu X, Liao Z, et al. Expression of 4E-BP1 and phospho-4E-BP1 correlates with the prognosis of patients with clear cell renal carcinoma. Cancer Manag Res. 2018;10:1553–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papaspyridonos M, Matei I, Huang Y, do Rosario Andre M, Brazier-Mitouart H, Waite JC, et al. Id1 suppresses anti-tumour immune responses and promotes tumour progression by impairing myeloid cell maturation. Nat Commun. 2015;6:6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ling MT, Wang X, Zhang X, Wong YC. The multiple roles of Id-1 in cancer progression. Differentiation. 2006;74(9–10):481–7. [DOI] [PubMed] [Google Scholar]
- 54.Kisonaite M, Wang X, Hyvonen M. Structure of Gremlin-1 and analysis of its interaction with BMP-2. Biochem J. 2016;473(11):1593–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Church RH, Krishnakumar A, Urbanek A, Geschwindner S, Meneely J, Bianchi A, et al. Gremlin1 preferentially binds to bone morphogenetic protein-2 (BMP-2) and BMP-4 over BMP-7. Biochem J. 2015;466(1):55–68. [DOI] [PubMed] [Google Scholar]
- 56.Nolan K, Kattamuri C, Rankin SA, Read RJ, Zorn AM, Thompson TB. Structure of Gremlin-2 in Complex with GDF5 gives insight into DAN-Family-mediated BMP antagonism. Cell Rep. 2016;16(8):2077–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vinals F, Reiriz J, Ambrosio S, Bartrons R, Rosa JL, Ventura F. BMP-2 decreases Mash1 stability by increasing Id1 expression. EMBO J. 2004;23(17):3527–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Korchynskyi O, ten Dijke P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J Biol Chem. 2002;277(7):4883–91. [DOI] [PubMed] [Google Scholar]
- 59.Hollnagel A, Oehlmann V, Heymer J, Ruther U, Nordheim A. Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J Biol Chem. 1999;274(28):19838–45. [DOI] [PubMed] [Google Scholar]
- 60.Shaw A, Toth BB, Arianti R, Csomos I, Poliska S, Vamos A, et al. BMP7 increases UCP1-dependent and independent thermogenesis with a unique gene expression program in human neck area derived adipocytes. Pharmaceuticals (Basel). 2021;14(11). [DOI] [PMC free article] [PubMed]
- 61.de Winter TJJ, Nusse R. Running against the Wnt: How Wnt/beta-Catenin suppresses adipogenesis. Front Cell Dev Biol. 2021;9:627429. [DOI] [PMC free article] [PubMed]
- 62.Kang S, Bennett CN, Gerin I, Rapp LA, Hankenson KD, Macdougald OA. Wnt signaling stimulates osteoblastogenesis of mesenchymal precursors by suppressing CCAAT/enhancer-binding protein alpha and peroxisome proliferator-activated receptor gamma. J Biol Chem. 2007;282(19):14515–24. [DOI] [PubMed] [Google Scholar]
- 63.Ma B, Hottiger MO. Crosstalk between Wnt/beta-Catenin and NF-kappaB signaling pathway during inflammation. Front Immunol. 2016;7:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun J, Hobert ME, Duan Y, Rao AS, He TC, Chang EB, et al. Crosstalk between NF-kappaB and beta-catenin pathways in bacterial-colonized intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2005;289(1):G129–37. [DOI] [PubMed] [Google Scholar]
- 65.Prossomariti A, Piazzi G, Alquati C, Ricciardiello L. Are Wnt/beta-Catenin and PI3K/AKT/mTORC1 distinct pathways in Colorectal Cancer? Cell Mol Gastroenterol Hepatol. 2020;10(3):491–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeng H, Lu B, Zamponi R, Yang Z, Wetzel K, Loureiro J, et al. mTORC1 signaling suppresses Wnt/beta-catenin signaling through DVL-dependent regulation of wnt receptor FZD level. Proc Natl Acad Sci U S A. 2018;115(44):E10362–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.