Abstract
Background
Members of the interferon regulatory factor (IRF) family are transcriptional regulators that play vital roles in the inflammatory response of macrophages. IRF1, IRF3, and IRF9 regulate the expression of immune-responsive gene 1 (IRG1) in macrophages. However, the role of IRF2 in the inflammatory response of macrophages remains somewhat contradictory. The regulatory relationship between IRF2 and IRG1 and their role in macrophages remain unclear. This study aimed to explore the role of IRF2 and IRG1 in macrophages.
Methods
Overexpression plasmids of IRF2 (IRF2-pcDNA3.1) and silencing siRNA targeting IRF2 and IRG1 (si-IRF2, si-IRG1) were constructed and transfected into RAW264.7 cells respectively. Subsequently, cells were treated with LPS and IFN-γ for 24 h. The expression of IRF2, IRG1, iNOS, IL-6, and BCL-xl was detected using Western blotting and qRT-PCR. Cell viability, migration and apoptosis were determined by CCK-8, transwell and flow cytometry. The IRG1 promoter region was cloned into the pGL3-basic plasmid. A dual-luciferase reporter assay was performed to verify the regulatory relationship between IRF2 and IRG1.
Results
IRF2 overexpression inhibited the expression of IL-6 and iNOS, cell migration, and apoptosis and elevated the expression of BCL-xl, IRG1, and cell viability of LPS- and IFN-γ-induced macrophages. IRF2 silencing had an opposite effect. IRF2 activated the promoter activity of IRG1. The inhibitory effects on LPS- and IFN-γ-induced proinflammatory responses, cell migration, apoptosis, and enhancing effects on the cell viability of over-expressed IRF2 were reversed by IRG1 silencing.
Conclusion
IRF2 promoted the promoter activity of IRG1 and regulated directly the expression of IRG1. IRF2 inhibited LPS and IFN-γ-induced pro-inflammatory responses, cell migration and apoptosis, enhanced cell viability in macrophages through regulating IRG1. IRF2 affected LPS- and IFN-γ-induced pro-inflammatory responses, cell viability, migration and apoptosis of macrophages by regulating IRG1.
Keywords: IRF2, IRG1, macrophages, pro-inflammatory responses, apoptosis
Introduction
In the initial stage of the host inflammatory response, macrophages, pivotal members of immune cells, mainly exhibit a pro-inflammatory phenotype (M1 polarization), whereas in the process of inflammation resolution, macrophages present a dominant anti-inflammatory phenotype (M2 polarization).1–6 Subsequently, macrophages can further affect the local immune response, regulate the infection of pathogens in the body and the immune response of tumors through synergistic effects with various cytokines, participate in immune regulation, tissue repair, and remodeling, and play a crucial role in inflammation, the occurrence and development of diseases, and the repair of tissues and organs.7–11 Interferon regulatory factor (IRFs) protein family members, including IRF1-9, are a class of transcriptional regulators that modulate the host cellular response to interferon (IFN), bacterial, and viral infections and regulate the expression of downstream genes by binding to DNA. Available studies have shown that IRF proteins are involved in immunomodulation, IFN transcription, cell differentiation and apoptosis, signal transduction, viral defense, and the regulation of several diseases.12–18
Members of the IRF family have different roles in the inflammatory response of macrophage.19 IRF1 synergistically enhances the expression of pro-inflammatory cytokines with NF-κB, ultimately leading to the M1 polarization of macrophages.20 Simultaneously, IRF1 binds to the IL-4 promoter and acts as a repressor of IL-4, thereby inhibiting M2 macrophage polarization.21 Mouse models of spinal cord injury pretreated with low-dose LPS showed elevated IRF3 expression in brain microglia, resulting in upregulation of IL-10, causing macrophage polarization into the M2 phenotype.22 IRF4 is a key transcription factor mediate M2 polarization of macrophages.23,24 The expression of related genes (Arg-1, FIZZ1, Ym1) of M2 polarization in macrophages also depends on the presence of IRF4.25 IRF4 and IRF5 competitively bind to MyD88, thereby promoting the transformation of macrophages into M2 phenotypes.26,27 IRF5 is activated in macrophages through the TLR4-MyD88 pathway, thereby causing macrophages to polarize into the M1 phenotype.28 IRF8 plays a key role in monocyte maturation and macrophage differentiation into pro-inflammatory M1 phenotypes. In LPS-stimulated macrophages, IRF8 interacts with IRF1 and AP1 to induce the expression of proinflammatory cytokines, causing macrophages to exhibit a proinflammatory M1 phenotype.29 Under the stimulation of type I and type II IFN, IRF9 combines with STAT1 and STAT2 to form ternary complexes, inducing the expression of pro-inflammatory cytokines and causing M1 polarization of macrophages.30 Studies have shown that IRF2 is degraded in the early stage of LPS-induced macrophages (0–3 hours) through a proteasome-mediated pathway, is subsequently restored through trans-activation (3–6 hours), and exerts anti-inflammatory effects by regulating glycolytic-dependent genes.31 In addition, studies have shown that IRF2 inhibits macrophage polarization into the M2 anti-inflammatory phenotype by binding to the promoter region of IL-4.32 Therefore, the role of IRF2 in the inflammatory response of macrophages is somewhat contradictory and complex.
Immune response gene 1 (IRG1) is a multi-functional regulator of inflammation and infection, is highly expressed in mammalian inflammatory processes, and plays an important role in immunity and diseases.33 Abnormal IRG1 expression leads to immune disorders, tumor progression, and neurodegenerative diseases.34 Studies have suggested that IRF3 can regulate the expression of IRG1 through a synergistic action with JUN, thus playing an important role in lethal innate immunity.35 MiRNA93 can promote M2 polarization of macrophages in ischemic muscle by decreasing the expression of IRF9, and IRF9 regulates the expression of IRG1, thus promoting the formation of blood vessels and restoration of perfusion in peripheral artery disease.36 IRF1 silencing obviously lowered the expression of IRG1 in macrophages.37 These studies suggest a regulatory relationship between IRF3, IRF9, IRF1, and IRG1. However, the specific mechanisms require further investigation. However, the regulatory relationship between IRF2 and IRG1 remains to be elucidated. Therefore, our study aimed to explore the effects of IRF2 on macrophage inflammation, cell viability, migration, and apoptosis, to determine whether there is a regulatory relationship between IRF2 and IRG1, and whether the effect of IRF2 on macrophage function is realized by regulating the expression of IRG1, in order to provide an experimental and theoretical basis for the diagnosis and treatment of clinically related diseases.
Materials and Methods
Cell Culture, Plasmid and siRNA Transfection, and Drug Administration
RAW264.7 cells were purchased from FuHeng Biology and cultured in DMEM containing 10% fetal bovine serum (FBS) (VivaCell) at 37 °C in a humidified atmosphere with 5% CO2. The full-length coding sequence of IRF2 was cloned into pcDNA3.1. si-IRF2 and si-IRG1 were designed and synthesized to silence IRF2 and IRG1 genes (GenePharma). The overexpressed plasmid and silencing RNA were transfected into RAW264.7, using Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen). After transfection for 24 h, cells were incubated with LPS (400 ng/mL, Sangon) and IFN-γ (20 ng/mL, Solarbio) for another 24 h. The siRNA sequences used in this study are shown in Table 1.
Table 1.
The siRNA Sequences Used in This Study
| Name | Sequence (5′–3′) | Sequence (5′–3′) |
|---|---|---|
| si-IRF2 | GGGCUAAAGUGGCUGAACATT | UGUUCAGCCACUUUAGCCCTT |
| si-IRG1 | CACGGUGGAAAGCCUUAUATT | UAUAAGGCUUUCCACCGUGTT |
| si-NC | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
Western Blot Analysis
Proteins from each group were extracted using RIPA lysis buffer. The concentration of each protein group was determined using the BCA protein assay kit (Beyotime). Subsequently, the protein samples were electrophoresed by SDS-PAGE and transferred to a PVDF membrane (0.45 µm, Millipore). The membrane was then blocked with a protein-free rapid blocking buffer (EpiZyme) for 30 min at room temperature and immunoblotted with primary antibodies (GAPDH, IRF2, IRG1, iNOS, IL-6, and BCL-xl (Proteintech)) for 16 h at 4 °C. The membrane was washed five times with TBST buffer solution and then incubated with the secondary antibody to detect immunoreactive proteins by enhanced chemiluminescence (Proteintech).
Quantitative Real-Time qRT-PCR
RNA was isolated from the samples of each group using the TRIZOL reagent and reverse-transcribed into cDNA. qPCR amplification was performed using the SYBR Green reagent. Primer pairs used in this study are listed in Table 2. Gene expression levels were determined by 2-ΔΔCT, as previously described.38
Table 2.
Primers Used for qRT- PCR
| Name | Sense (5′–3′) | Antisense (5′–3′) |
|---|---|---|
| IRF2 | TAGCCCATTACCCACGCCTCTC | CTTCTCCTCCTCCTCCTCCAAGATG |
| IRG1 | AACTACTCCTGCCATCCACTCCTG | TCCTCTTGCTCCTCCGAATGATACC |
| IL6 | CTTCTTGGGACTGATGCTGGTGAC | TCTGTTGGGAGTGGTATCCTCTGTG |
| iNOS | ATCTTGGAGCGAGTTGTGGATTGTC | TAGGTGAGGGCTTGGCTGAGTG |
CCK‑8 Assay
Cell viability in each group was determined the CCK‑8 assay. Briefly, RAW264.7 cells were seeded in 96‑well plates at a density of 5*104 cells/well. CCK‑8 reagent (Vazyme) was added and the cells were incubated for 3 h. The absorbance of the cell solutions was read at 450 nm using a microplate reader (Thermo Scientific), following the manufacturer’s instructions.
Transwell Assay
RAW264.7 cells were diluted to 1.5×105 cells in serum-free medium and grown in a Transwell chamber (8.0 μm, Corning, USA) which was added complete medium in the lower chamber. After incubation at 37 °C with 5% CO2 for 48 h, the cells were fixed with 4% paraformaldehyde and stained with a 0.1% crystal violet solution. The cells remaining on the upper surface of the membrane were removed and the cells on the lower surface of the membrane were the migrated cells. The cell migration was observed and photographed using an inverted microscope.
Flow Cytometry
RAW264.7 cells were seeded into 6-well plates, collected when the cell density reached 80%, and subsequently incubated with Annexin V-FITC and PI staining solution to detect apoptosis using the Annexin V-FITC/PI Apoptosis Detection Kit (Vazyme).
Dual‑luciferase Reporter Assay
Promoter region of IRG1 was cloned into the pGL3-basic plasmid. IRF2-pCDNA3.1, pCDNA3.1 empty, IRG1-pGL3, or pGL3 empty and pRL-SV40 were co-transfected into RAW264.7 cells. Forty-eight hours after successful co-transfection, firefly and Renilla luciferase activities were determined using a dual-luciferase reporter assay kit (Beyotime) following the manufacturer’s protocols.
Statistical Analysis
Data were analyzed using SPSS software (version 17.0) and expressed as the mean ± SD of at least three independent experiments. The differences between the groups were evaluated using the Student’s t-test and one-way analysis of variance (ANOVA). P<0.05 was regarded as statistically significant.
Results
IRF2 Inhibited LPS and IFN-γ-Induced Pro-Inflammatory Responses of Macrophages
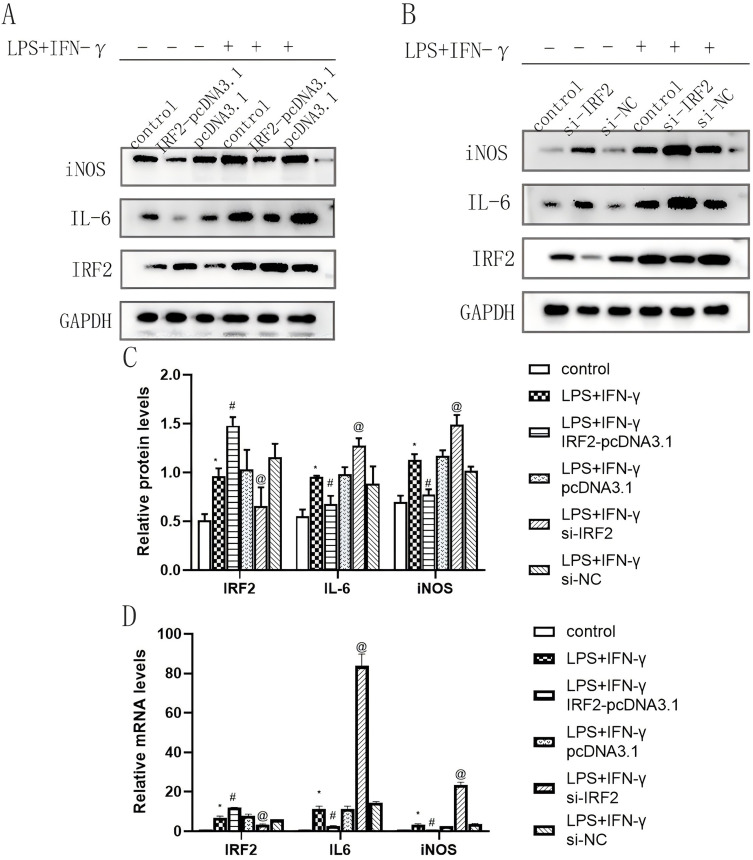
To determine the expression of IRF2 in the inflammatory response of macrophages, RAW264.7 cells were treated with LPS and IFN-γ for 24 h, and the expression of IL-6 and iNOS protein and mRNA levels were verified by Western blotting and qRT-PCR. The results showed that the expression of iNOS and IL-6 protein and mRNA was significantly elevated after 24 h of LPS and IFN-γ treatment, indicating that the establishment of the inflammation model was successful (Figure 1A, C and D). To explore the influence of IRF2 on the inflammatory process in macrophages, overexpressed plasmids and siRNAs of IRF2 were transfected into RAW264.7 cells respectively. Results of qRT-PCR and Western blotting showed that the expression levels of IRF2 protein and mRNA in the IRF2-pCDNA3.1 group were higher than those in the control group (Figure 1A, C and D, Figure S1). In contrast, IRF2 protein and mRNA levels in the si-IRF2 group were lower than those in the control group (Figure 1B, C, and D, Figure S1), indicating that transfection was successful. IRF2 overexpression significantly repressed the expression of iNOS and IL-6 (Figure 1A, C and D), whereas IRF2 silencing had the opposite effect (Figure 1B, C and D), suggesting that IRF2 may be a negative regulator of the inflammatory response of macrophages.
Figure 1.
Effects of IRF2 overexpression and silencing on the protein and mRNA expression levels of IRF2, iNOS and IL-6 in LPS+IFN-γ-induced macrophages. Representative Western blot bands of IRF2, iNOS and IL-6 (A and B); Relative protein levels of IRF2, iNOS and IL-6 were normalized to that of the GAPDH internal control (C); Relative mRNA levels of IRF2, iNOS and IL-6 (D); Mean±standard error of the mean values is presented for each group. *: LPS+IFN-γ group compared with control group, P < 0.05; #: LPS+IFN-γ+IRF2-pcDNA3.1 group compared with LPS+IFN-γ group and LPS+IFN-γ+pcDNA3.1 group, P < 0.05; @: LPS+IFN-γ+si-IRF2 group compared with LPS+IFN-γ group and LPS+IFN-γ+si-NC group, P < 0.05. P < 0.05 by one-way analyse of variance (ANOVA). Control group was defined as RAW264.7 cells without LPS+IFN-γ treated group.
IRF2 Elevated Cell Viability of Macrophages
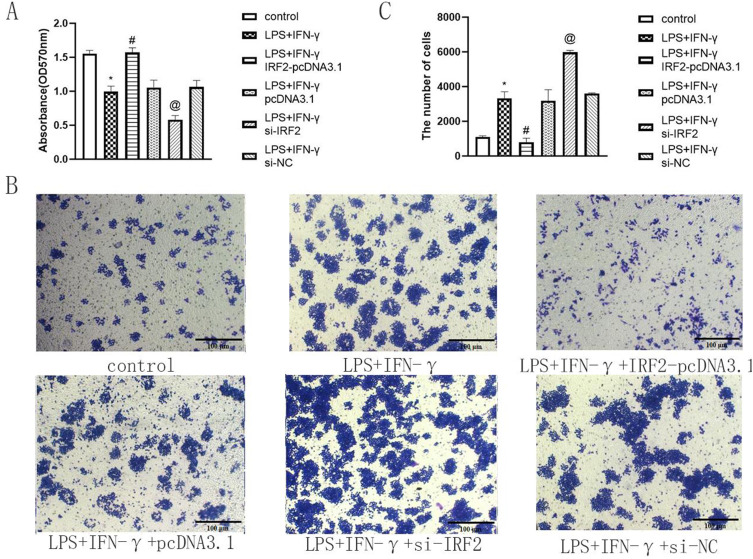
CCK8 assay was performed to explore the effect of IRF2 on macrophage viability. Our observations showed that after 24 h of treatment with LPS and IFN-γ, the viability of RAW264.7 cells reduced. Overexpression of IRF2 elevated the viability of RAW264.7, whereas IRF2 silencing had a negative effect (Figure 2A). This suggests that IRF2 increases the viability of macrophages.
Figure 2.
IRF2 influenced cell viability and migration in LPS+IFN-γ induced macrophages. Relative cell viability of different group (A) and representative images of transwell assay in each group (B) when IRF2 overexpression and silencing. Relative migration rate in different groups (RAW264.7 cells) (C); Mean±standard error of the mean values is presented for each group. *: LPS+IFN-γ group compared with control group, P < 0.05; #: LPS+IFN-γ+IRF2-pcDNA3.1 group compared with LPS+IFN-γ group and LPS+IFN-γ+pcDNA3.1 group, P < 0.05; @: LPS+IFN-γ+si-IRF2 group compared with LPS+IFN-γ group and LPS+IFN-γ+si-NC group, P < 0.05. P < 0.05 by one-way analyse of variance (ANOVA). Control group was defined as RAW264.7 cells without LPS+IFN-γ treated group. Scale bar, 100 μm.
IRF2 Inhibited Migration of Macrophages
Transwell assay was performed to determine the influence of IRF2 on macrophage migration. The data revealed that after a 24-hour treatment with LPS and IFN-γ, an increase in the number of cells passing through the chamber compared with control group exhibited. There was a reduction in the number of cells traversing the chamber in the LPS+IFN-γ+IRF2-pcDNA3.1 group compared to LPS+IFN-γ group and LPS+IFN-γ+pcDNA3.1 group. Conversely, the LPS+IFN-γ+si-IRF2 group presented more cells acrossing the chamber compared with LPS+IFN-γ group and LPS+IFN-γ+si-NC group. (Figure 2B and C). This suggested that IRF2 repressed macrophage migration.
IRF2 Suppressed Apoptosis of Macrophages
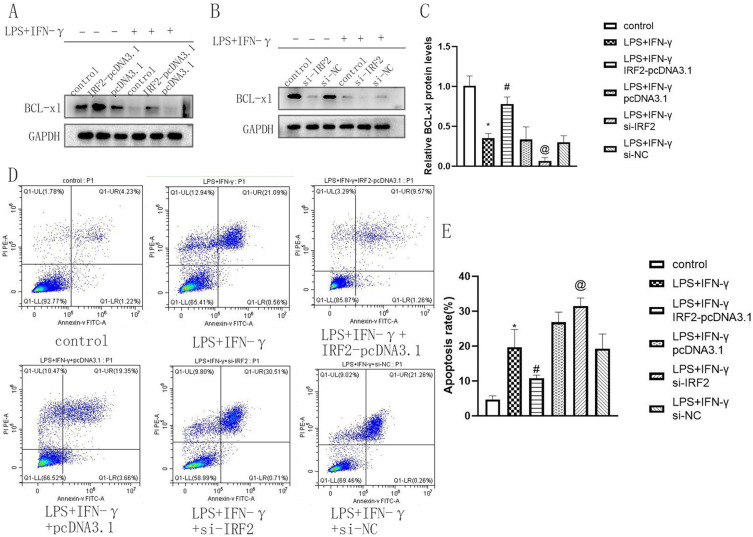
To determine the effect of IRF2 on macrophage apoptosis, the expression level of the anti-apoptotic protein BCL-xl and the cell apoptosis rate were detected by Western blotting and flow cytometry assays. After 24 h of treatment with LPS and IFN-γ, BCL-xl levels were significantly reduced compared to those in the control group (Figure 3A and C). When IRF2 was overexpressed, BCL-xl expression significantly improved (Figure 3A and C). IRF2 silencing suppressed BCL-xl expression (Figure 3B and C). Flow cytometric analysis showed that macrophage apoptosis increased after 24 h of LPS and IFN-γ treatment. IRF2 overexpression reduced macrophage apoptosis, whereas IRF2 silencing induced macrophage apoptosis (Figure 3D and E, Figure S2). This indicated that IRF2 suppressed macrophage apoptosis.
Figure 3.
The influences of IRF2 on cell apoptosis in LPS+IFN-γ induced macrophages. Representative Western blot bands of BCL-xl (A, B) and relative protein levels of BCL-xl were normalized to that of the GAPDH internal control (C); Representative images of flow cytometry in each group (D). Relative apoptosis rate in different groups (E); Mean±standard error of the mean values is presented for each group. *: LPS+IFN-γ group compared with control group, P < 0.05; #: LPS+IFN-γ+IRF2-pcDNA3.1 group compared with LPS+IFN-γ group and LPS+IFN-γ+pcDNA3.1 group, P < 0.05; @: LPS+IFN-γ+si-IRF2 group compared with LPS+IFN-γ group and LPS+IFN-γ+si-NC group, P < 0.05. P < 0.05 by one-way analyse of variance (ANOVA). Control group was defined as RAW264.7 cells without LPS+IFN-γ treated group.
IRG1 Was a Direct Target of IRF2
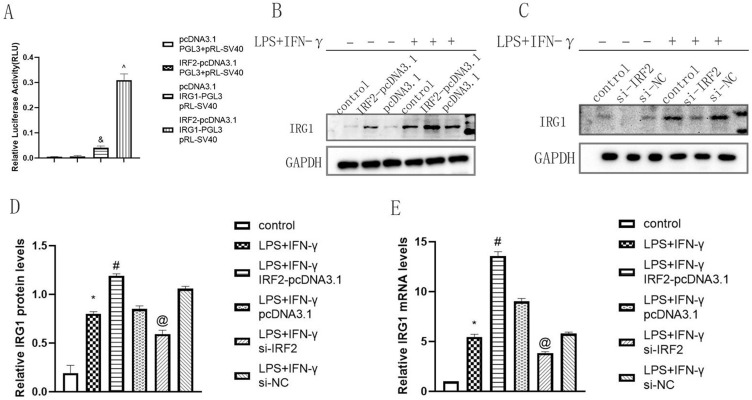
First, we performed Dual-Luciferase reporter assay to verify the transcriptional regulatory relationship between IRF2 and IRG1. The results showed that the luciferase activity of the IRF2-pcDNA3.1+IRG1-PGL3+pRL-SV40 group was significantly higher than that of the other groups, demonstrating that IRF2 positively regulated the promoter activity of IRG1 (Figure 4A). IRG1 expression was determined using Western blotting and qRT-PCR. The findings indicated that after transfection with IRF2-pcDNA3.1, the protein and mRNA levels of IRG1 were increased, whereas the expression level of IRG1 was significantly decreased after transfection with si-IRF2 (Figure 4B–E). These findings indicated that IRG1 was a direct target of IRF2.
Figure 4.
IRG1 was a direct target of IRF2. The luciferase activity of IRG1 promoter (A), Representative Western blot bands of IRG1 (B and C) and relative protein and mRNA levels of IRG1 were normalized to that of the GAPDH internal control (D and E) when IRF2 was overexpressed and silencing; Mean±standard error of the mean values is presented for each group. and: pcCDNA3.1+IRG1-PGL3+pRL-SV40 group compared with pcDNA3.1+PGL3+pRL-SV40 group and IRF2-pcDNA3.1+PGL3+pRL-SV40 group, P < 0.05; ^: IRF2-pcDNA3.1+IRG1-PGL3+pRL-SV40 group compared with pCDNA3.1+IRG1-PGL3+pRL-SV40 group, P < 0.05; *: LPS+IFN-γ group compared with control group, P < 0.05; #: LPS+IFN-γ+IRF2-pcDNA3.1 group compared with LPS+IFN-γ group and LPS+IFN-γ+pcDNA3.1 group, P < 0.05; @: LPS+IFN-γ+si-IRF2 group compared with LPS+IFN-γ group and LPS+IFN-γ+si-NC group, P < 0.05. P < 0.05 by one-way analyse of variance (ANOVA). Control group was defined as RAW264.7 cells without LPS+IFN-γ treated group.
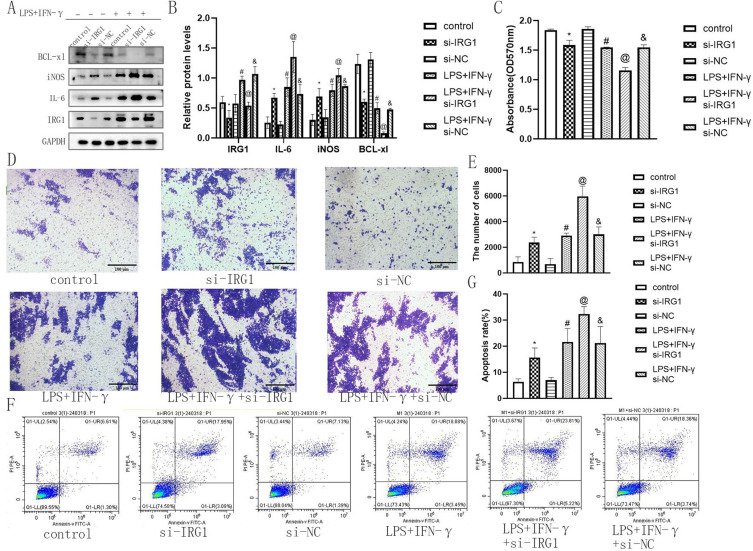
IRG1 Affected the Inflammatory Response, Cell Viability, Migration and Apoptosis of Macrophages
To determine the function of IRG1 in the inflammatory response, cell viability, migration, and apoptosis of macrophages, si-IRG1 was designed and transformed into RAW264.7 cells. Western blotting and qRT-PCR results showed that IRG1 silencing decreased the expression of IRG1 (Figure 5A and B, Figure S3), suggesting that IRG1 siRNA was transfected successfully. IRG1 silencing enhanced the expression of iNOS and IL-6 and lowered the expression of BCL-xl (Figure 5A and B), indicating that IRG1 might act as a negative regulator of the inflammatory response and apoptosis of macrophages. In addition, CCK-8, Transwell, and flow cytometry results indicated that IRG1 silencing inhibited cell viability and promoted cell migration and apoptosis (Figure 5C–G). These findings indicated that IRG1 affected the inflammatory response, cell viability, migration, and apoptosis of macrophages.
Figure 5.
IRG1 affected the inflammatory response, cell viability, migration and apoptosis of macrophages. Representative Western blot bands of IRG1, iNOS, IL-6 and BCL-xl (A) and relative protein levels of IRG1, iNOS, IL-6 and BCL-xl were normalized to that of the GAPDH internal control (B) when IRG1 was silencing. Relative cell viability rate of different group (C) and representative images of transwell assay (D) and Relative migration rate in each group (E), Representative images of flow cytometry (F) and apoptosis rate in each group (G) when IRG1 was silencing; Mean±standard error of the mean values is presented for each group. *: si-IRG1 group compared with control group and si-NC group, P < 0.05; #: LPS+IFN-γ group compared with control group and si-NC group, P< 0.05; @: LPS+IFN-γ+si-IRG1 group compared with LPS+IFN-γ group and LPS+IFN-γ+si-NC group, P < 0.05; and: LPS+IFN-γ+si-NC group compared with control group and si-NC group, P < 0.05. P < 0.05 by one-way analyse of variance (ANOVA). Control group was defined as RAW264.7 cells without LPS+IFN-γ treated group. Scale bar, 100 μm.
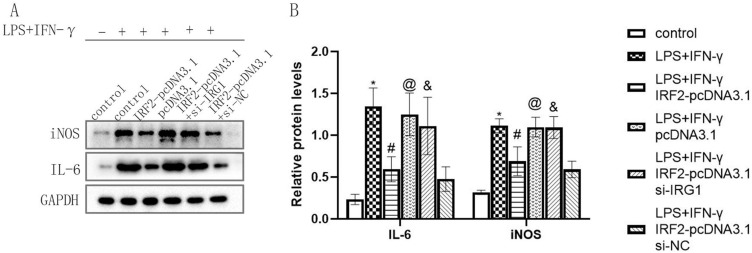
IRF2 Influenced the Inflammatory Response, Cell Viability, Migration and Apoptosis of Macrophages by Regulating IRG1
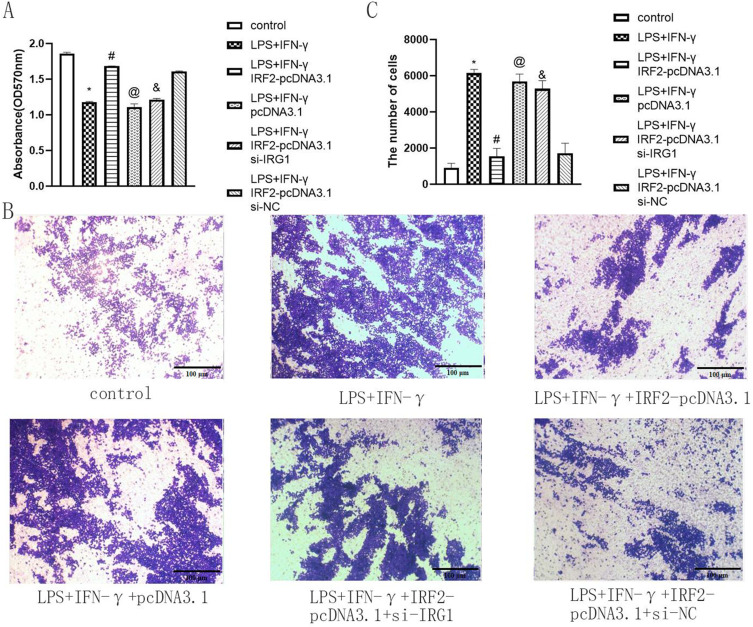
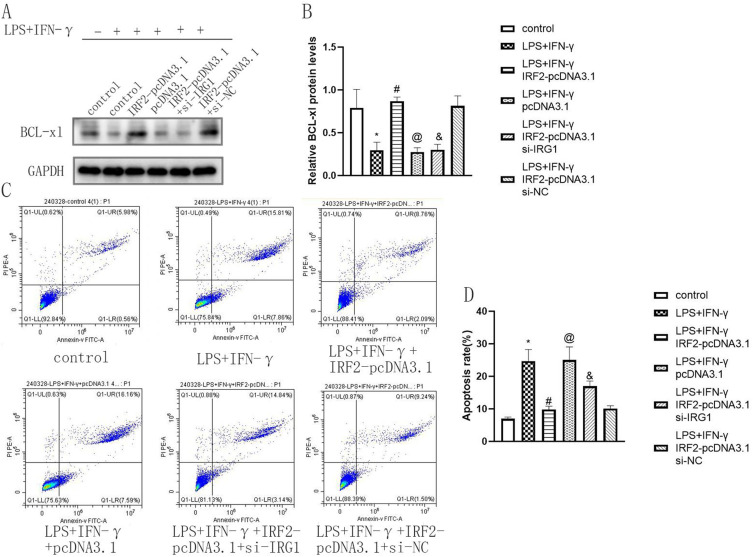
To determine the effect of IRF2 on the physiological function of macrophages by regulating IRG1, IRF2-pcDNA3.1, and si-IRG1 were co-transfected into RAW264.7 cells and treated by LPS+IFN-γ for 24 hours. si-IRG1 weakened the enhancing effect of IRF2 overexpression on IRG1 expression and the inhibitory effect of IRF2 overexpression on iNOS and IL-6 expression (Figure 6A and B). This indicated that IRF2 affected the inflammatory response of macrophages by targeting IRG1. Subsequently, the effect of IRF2 on macrophage viability via the regulation of IRG1 was investigated. The results of the CCK-8 assay showed that si-IRG1 reduced the enhancing effect of IRF2 overexpression on macrophage viability (Figure 7A). This suggested that IRF2 influenced macrophage viability by regulating IRG1 expression. Moreover, the results of the transwell assay revealed that si-IRG1 lowered the inhibitory effect of IRF2 overexpression on cell migration of RAW264.7 cells (Figure 7B and C). The influence of IRF2 on macrophage apoptosis via the regulation of IRG1 was confirmed. Western blotting and flow cytometry assays showed that silencing of IRG1 reduced the enhanced effect of IRF2 overexpression on BCL-xl expression (Figure 8A and B) and the inhibitory effect of IRF2 overexpression on macrophage apoptosis (Figure 8C and D). This demonstrated that IRF2 caused a difference in macrophage apoptosis by regulating IRG1.
Figure 6.
IRF2 affected the inflammatory response of macrophages by target regulating IRG1. Representative Western blot bands of iNOS and IL-6 (A) and relative protein levels of iNOS and IL-6 were normalized to that of the GAPDH internal control (B); Mean±standard error of the mean values is presented for each group. *: LPS+IFN-γ group compared with control group, P < 0.05; #: LPS+IFN-γ+IRF2-pcDNA3.1 group compared with LPS+IFN-γ group and LPS+IFN-γ+pcDNA3.1 group, P < 0.05; @: LPS+IFN-γ+pcDNA3.1 group compared with LPS+IFN-γ+IRF2-pcDNA3.1 group, P < 0.05; &: LPS+IFN-γ+IRF2-pcDNA3.1+si-IRG1 group compared with LPS+IFN-γ+IRF2-pcDAN3.1 group and LPS+IFN-γ+IRF2-pcDAN3.1+si-NC group, P < 0.05. P < 0.05 by one-way analyse of variance (ANOVA). Control group was defined as RAW264.7 cells without LPS+IFN-γ treated group.
Figure 7.
IRF2 affected cell viability and migration of macrophages by regulating IRG1. Relative cell viability of different group (A), representative images of transwell assay (B) and relative migration rate in different groups (C); Mean±standard error of the mean values is presented for each group. *: LPS+IFN-γ group compared with control group, P < 0.05; #: LPS+IFN-γ+IRF2-pcDNA3.1 group compared with LPS+IFN-γ group and LPS+IFN-γ+pcDNA3.1 group, P < 0.05; @: LPS+IFN-γ+pcDNA3.1 group compared with LPS+IFN-γ+IRF2-pcDNA3.1 group, P < 0.05; and: LPS+IFN-γ+IRF2-pcDNA3.1+si-IRG1 group compared with LPS+IFN-γ+IRF2-pcDAN3.1 group and LPS+IFN-γ+IRF2-pcDAN3.1+si-NC group, P < 0.05. P < 0.05 by one-way analyse of variance (ANOVA). Control group was defined as RAW264.7 cells without LPS+IFN-γ treated group. Scale bar, 100 μm.
Figure 8.
IRF2 affected the apoptosis of macrophages by regulating IRG1. Representative Western blot bands of BCl-xl (A), relative protein levels of BCl-xl were normalized to that of the GAPDH internal control (B), representative images of flow cytometry in each group (C) and relative apoptosis rate in different groups (D); Mean±standard error of the mean values is presented for each group. *: LPS+IFN-γ group compared with control group, P < 0.05; #: LPS+IFN-γ+IRF2-pcDNA3.1 group compared with LPS+IFN-γ group and LPS+IFN-γ+pcDNA3.1 group, P < 0.05; @: LPS+IFN-γ+pcDNA3.1 group compared with LPS+IFN-γ+IRF2-pcDNA3.1 group, P < 0.05; and: LPS+IFN-γ+IRF2-pcDNA3.1+si-IRG1 group compared with LPS+IFN-γ+IRF2-pcDAN3.1 group and LPS+IFN-γ+IRF2-pcDAN3.1+si-NC group, P < 0.05. P < 0.05 by one-way analyse of variance (ANOVA). Control group was defined as RAW264.7 cells without LPS+IFN-γ treated group.
Discussion
It is widely recognized that the polarization of macrophage M1/M2 balance during inflammation or injury determines the outcome of organ function. When an infection or inflammation is severe enough to affect an organ, macrophages initially adopt the M1 phenotype, secreting pro-inflammatory cytokines such as TNF-α, IL-1β, IL-12, and IL-23 in response to stimulation. However, if the M1 stage persists, it may lead to tissue damage. Therefore, M2 macrophages secrete large amounts of IL-10 and TGF-β to inhibit inflammation, promote tissue repair, remodeling, angiogenesis, and maintain homeostasis.1 Macrophages are involved in multiple organ systems, and they not only promote inflammation and wound healing, but are also involved in fibrosis, anti-inflammatory responses, anti-fibrosis, and promote dissolution and tissue regeneration.39,40 For example, recent studies have shown that although genetic destiny mapping studies have confirmed that most macrophages in the adult heart are derived from yolk sacs and fetal progenitor cells, the macrophages that dominate the early inflammatory response after heart tissue injury are CCR2+ monocyte derived macrophages.41 On the contrary, resident cardiac macrophages, which originate from embryonic sources, are pivotal cells that facilitate recovery.42 These studies have demonstrated that these resident tissue macrophages stimulate cardiomyocyte proliferation and blood vessel development. This implies that macrophages derived from embryonic tissue and those from CCR2+ monocytes have distinct roles in the process of heart repair. Therefore, macrophages play an important role in human and animal disease models.
Members of the IRFs family have been proven to be multi-functional in regulating immune responses and to play a strong role in the physiological environment.29,30 During inflammation, IRF family proteins perform important functions by regulating the pro-inflammatory and anti-inflammatory processes of immune cells, especially macrophages. IRF1, IRF5, IRF8, and IRF9 participate in mediating pro-inflammatory effects (M1 polarization) of macrophages, whereas IRF3 and IRF4 participate in mediating anti-inflammatory effects (M2 polarization) of macrophages.20,28 However, IRF2 exhibits unique characteristics that differ from those of the other IRF proteins. On the one hand, IRF2 supports the production of IFN-γ, IL-1, and IL-12 induced by LPS and inhibits macrophage polarization into the M2 phenotype through binding to the promoter region of IL-4, showing a pro-inflammatory effect.43 In contrast, IRF2 inhibited LPS-induced TNF-α expression and exerted anti-inflammatory effects.31 Therefore, the effects of IRF2 on macrophages are somewhat contradictory and complex.
In our study, the findings indicated that overexpression of IRF2 inhibited the expression of IL-6 and iNOS, cell migration, and apoptosis, and elevated the expression of BCL-xl and cell viability in RAW264.7 cells treated by LPS and IFN-γ (treated 24 hours). In contrast, silencing of IRF2 enhanced the expression of IL-6 and iNOS, cell migration, and apoptosis, decreased the expression of BCL-xl, and decreased the viability of macrophages (Figures 1–3). These results indicated that IRF2 affects the inflammatory response, cell viability, migration, and apoptosis of macrophages.
IRG1 is a multi-functional regulator of inflammation and infection.33 IRG1-encoded cis-aconite decarboxylase produces itaconate via cis-decarboxylation.44 Itaconate is an organic compound that inhibits isocitrate lyase, a key enzyme that diverts glyoxylic acid, and is necessary for bacterial growth under certain conditions. Related research has indicated that the TET-family DNA dioxygenase (TET) is the main target of itaconate in blocking LPS-induced gene expression, including genes controlled by the NF-κB and STAT signaling pathways. Itaconate alleviates LPS-induced acute pulmonary edema and lung and liver injury. This study has identified itaconate as an immunomodulatory metabolite that selectively targets the TET enzyme to suppress inflammatory responses.44 Another study has shown that itaconate and its derivatives have anti-inflammatory effects in preclinical models of sepsis, viral infection, psoriasis, gout, ischemia/reperfusion injury, and pulmonary fibrosis, suggesting that itaconic acid-based therapies may be useful for a range of inflammatory diseases.33,45 A recent study demonstrates that a cell‐permeable derivative of itaconate can reduce the prolonged inflammatory response by activating the transcription factor Nrf2.46 Considering that IRG1/itaconate pathway triggers an Nrf2-mediated antioxidative response in hepatocytes, thereby protecting the liver from I/R injury.47 The IRG1 pathway observed in macrophages may also play a role in this protective mechanism. Furthermore, lentivirus-encoded Irg1 shRNA significantly reduced peritoneal tumors and led to decreased ROS production and ROS-mediated MAPK activation in tumor cells.48 As reflected in the literature, IRG1 and members of the IRFs family are inextricably linked. siRNA-mediated silencing of IRF1 in macrophages sharply reduced IRG1 expression at both the gene and protein levels, which was associated with reduced itaconic acid production.37 Overexpressed IRF9 elevated the mRNA level of IRG1 in macrophages.36 This suggests that some IRF family members regulate IRG1 expression, and some IRF family members may play a pivotal role in inflammation, the onset and progression of diseases, as well as in the repair of tissues and organs by modulating the expression of IRG1. However, it is unclear whether IRF2 has a regulatory relationship with IRG1, and whether it plays a role in inflammatory responses and other biological processes in macrophages. Therefore, further experiments were conducted to explore these mechanisms. In order to verify the targeted regulatory relationship between IRF2 and IRG1, dual-luciferase reporter assay was used for verification. The promoter of IRG1 was cloned into PGL3 plasmid and transfected into RAW264.7 cells for dual-luciferase reporter assay. The experimental results indicated that IRF2 could activate the promoter activity of IRG1 (Figure 4A). And overexpression of IRF2 enhanced the expression of IRG1, while silence of IRF2 inhibited the expression of IRG1 (Figure 4B–E). This indicated that IRF2 had a positive regulatory effect on IRG1.
In addition, IRG1 silencing by si-IRG1 increased the expression of iNOS and IL-6 and decreased the expression of BCL-xl (Figure 5A, B), suggesting that IRG1 might function as a negative regulator of the inflammatory response and apoptosis of macrophages. Furthermore, IRG1 silencing suppressed cell viability and enhanced cell migration and apoptosis (Figure 5C–G). These findings demonstrated that IRG1 affected the inflammatory response, cell viability, migration, and apoptosis of macrophages. Moreover, after co-transfection of IRF2-pcDNA3.1 and si-RG1 into macrophages, it was found that the inhibitory effects on inflammation, cell migration, apoptosis, and enhancing effects on the cell viability of IRF2 over-expression were reversed by IRG silencing. Previous studied had showed that IRG1/itaconate pathway functioned as an suppressor of inflammatory responses by modulating NF-κB, STAT and Nrf2 signaling,44–46 suggesting that IRF2 might inhibit inflammation responses by regulating the IRG1/itaconate pathway. Subsequent investigation on the regulation of the IRF2 and IRG1/itaconate pathway might further elucidate the relevant mechanisms. Our above observations indicated that there was a positive regulatory relationship between IRF2 and IRG1, and that IRF2 affected the inflammatory responses, cell viability, migration, and apoptosis of macrophages by directly regulating IRG1.
Conclusion
In summary, these findings suggested that IRF2 could repress the inflammatory response, cell migration, apoptosis and improve cell viability of macrophages. IRG1 was a regulatory target of IRF2. IRF2 caused differences in inflammation, cell migration, apoptosis, and macrophage viability by regulating IRG1. Therefore, the IRF2/IRG1 axis is expected to provide new clues for clinical macrophage-related inflammatory diseases and offer a new direction for the better clinical diagnosis and treatment of related diseases.
Funding Statement
This work was supported by the Excellent Young Talents Support Program of Anhui Universities (Grant No. gxyq2021171), Research Fund of Anhui Institute of Translational Medicine (Grant No. 2023zhyx-C63), and Natural Science Research Project of Colleges and Universities of Anhui Province (Grant No. 2024AH050800, 2024AH040107).
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors have no relevant financial or non-financial interests to disclose.
References
- 1.Wynn TA, Vannella KM. Macrophages in tissue repair, regeneration, and fibrosis. Immunity. 2016;44(3):450–462. doi: 10.1016/j.immuni.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ginhoux F, Schultze JL, Murray PJ, Ochando J, Biswas SK. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat Immunol. 2016;17(1):34–40. doi: 10.1038/ni.3324 [DOI] [PubMed] [Google Scholar]
- 3.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496(7446):445–455. doi: 10.1038/nature12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Epelman S, Lavine KJ, Randolph GJ. Origin and functions of tissue macrophages. Immunity. 2014;41(1):21–35. doi: 10.1016/j.immuni.2014.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oishi Y, Manabe I. Macrophages in inflammation, repair and regeneration. Int Immunol. 2018;30(11):511–528. doi: 10.1093/intimm/dxy054 [DOI] [PubMed] [Google Scholar]
- 6.Chazaud B. Macrophages: supportive cells for tissue repair and regeneration. Immunobiology. 2014;219(3):172–178. doi: 10.1016/j.imbio.2013.09.001 [DOI] [PubMed] [Google Scholar]
- 7.Italiani P, Boraschi D. From monocytes to M1/M2 macrophages: phenotypical vs functional differentiation. Front Immunol. 2014;5:514. doi: 10.3389/fimmu.2014.00514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viola A, Munari F, Sánchez-Rodríguez R, Scolaro T, Castegna A. The metabolic signature of macrophage responses. Front Immunol. 2019;10:10. doi: 10.3389/fimmu.2019.01462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray P, Allen J, Biswas S, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41(1):14–20. doi: 10.1016/j.immuni.2014.06.008.r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8(12):958–969. doi: 10.1038/nri2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32(5):593–604. doi: 10.1016/j.immuni.2010.05.007 [DOI] [PubMed] [Google Scholar]
- 12.Ikushima H, Negishi H, Taniguchi T. The IRF family transcription factors at the interface of innate and adaptive immune responses. Cold Spring Harb Symp Quant Biol. 2013;78:105–116. doi: 10.1101/sqb.2013.78.020321 [DOI] [PubMed] [Google Scholar]
- 13.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623 [DOI] [PubMed] [Google Scholar]
- 14.Tailor P, Tamura T, Ozato K. IRF family proteins and type I interferon induction in dendritic cells. Cell Res. 2006;16(2):134–140. doi: 10.1038/sj.cr.7310018 [DOI] [PubMed] [Google Scholar]
- 15.Barnes B, Lubyova B, Pitha PM. On the role of IRF in host defense. J Interferon Cytokine Res. 2002;22(1):59–71.12. doi: 10.1089/107999002753452665 [DOI] [PubMed] [Google Scholar]
- 16.Savitsky D, Tamura T, Yanai H, Taniguchi T. Regulation of immunity and oncogenesis by the IRF transcription factor family. Cancer Immunol Immunother. 2010;59(4):489–510.13. doi: 10.1007/s00262-009-0804-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400 [DOI] [PubMed] [Google Scholar]
- 18.Jefferies CA. Regulating IRFs in IFN driven disease. Front Immunol. 2019;10:325. doi: 10.3389/fimmu.2019.00325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chistiakov DA, Myasoedova VA, Revin VV, Orekhov AN, Bobryshev YV. The impact of interferon-regulatory factors to macrophage differentiation and polarization into M1 and M2. Immunobiology. 2018;223(1):101–111. doi: 10.1016/j.imbio.2017.10.005 [DOI] [PubMed] [Google Scholar]
- 20.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92(1):5–8. doi: 10.1016/s0092-8674(00)80893-4 [DOI] [PubMed] [Google Scholar]
- 21.Elser B, Lohoff M, Kock S, Giaisi M, Kirchhoff S, Krammer PH. IFN-gamma Represses IL-4 Expression via IRF-1 and IRF-2. Immunity. 2002;17(6):703–712. doi: 10.1016/s1074-7613(02)00471-5 [DOI] [PubMed] [Google Scholar]
- 22.Osman N, Omar SZ, Bolong J, Dsilva JL, Shaffril HAM. Lipopolysaccharide preconditioning facilitates M2 activation of resident microglia after spinal cord injury. J Neurosci Res. 2015;92(11):1647–1658. doi: 10.1002/jnr.23448 [DOI] [PubMed] [Google Scholar]
- 23.El Chartouni C, Schwarzfischer L, Rehli M. Interleukin-4 induced interferon regulatory factor (Irf) 4 participates in the regulation of alternative macrophage priming. Immunobiology. 2010;215(9–10):821–825. doi: 10.1016/j.imbio.2010.05.031 [DOI] [PubMed] [Google Scholar]
- 24.Tarassishin L, Suh HS, Lee SC. Interferon regulatory factor 3 plays an anti-inflammatory role in microglia by activating the PI3K/Akt pathway. J Neuroinflammation. 2011;8:187. doi: 10.1186/1742-2094-8-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Satoh T, Takeuchi O, Vandenbon A, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11(10):936–944. doi: 10.1038/ni.1920 [DOI] [PubMed] [Google Scholar]
- 26.Negishi H, Fujita Y, Yanai H, et al. Evidence for licensing of IFN-γ-induced IFN regulatory factor 1 transcription factor by MyD88 in toll-like receptor-dependent gene induction program. Proc Natl Acad Sci USA. 2006;103(41):15136–15141. doi: 10.1073/pnas.0607181103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang SC, Smith AM, Everts B, et al. Metabolic reprogramming mediated by the mTORC2-IRF4 signaling axis is essential for macrophage alternative activation. Immunity. 2016;45(4):817–830. doi: 10.1016/j.immuni.2016.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takaoka A, Yanai H, Kondo S, et al. Integral role of IRF-5 in the gene induction programme activated by toll-like receptors. Nature. 2005;434(7030):243–249. doi: 10.1038/nature03308 [DOI] [PubMed] [Google Scholar]
- 29.Mancino A, Termanini A, Barozzi I, et al. A dual cis-regulatory code links IRF8 to constitutive and inducible gene expression in macrophages. Genes Dev. 2015;29(4):394–408. doi: 10.1101/gad.257592.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kessler DS, Veals SA, Fu XY, Levy DE. Interferon-alpha regulates nuclear translocation and DNA-binding affinity of ISGF3, a multimeric transcriptional activator. Genes Dev. 1990;4(10):1753–1765. doi: 10.1101/gad.4.10.1753 [DOI] [PubMed] [Google Scholar]
- 31.Cui H, Banerjee S, Guo S, Xie N, Liu G. IFN regulatory factor 2 inhibits expression of glycolytic genes and lipopolysaccharide-induced proinflammatory responses in macrophages. J Immunol. 2018;200(9):3218–3230. doi: 10.4049/jimmunol.1701571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fehr T, Schoedon G, Odermatt B, et al. Crucial role of interferon consensus sequence binding protein, but neither of interferon regulatory factor 1 nor of nitric oxide synthesis for protection against murine listeriosis. J Exp Med. 1997;185(5):921–931. doi: 10.1084/jem.185.5.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michelucci A, Cordes T, Ghelfi J, et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc Natl Acad Sci USA. 2013;110(19):7820–7825. doi: 10.1073/pnas.1218599110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu R, Chen F, Wang N, Tang D, Kang R. ACOD1 in immunometabolism and disease. Cell Mol Immunol. 2020;7(8):822–833. doi: 10.1038/s41423-020-0489-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen F, Wu R, Liu J, Kang R, Li J, Tang D. The STING1-MYD88 complex drives ACOD1/IRG1 expression and function in lethal innate immunity. iScience. 2022;25(7):104561. doi: 10.1016/j.isci.2022.104561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ganta VC, Choi MH, Kutateladze A, Fox TE, Farber CR, Annex BH. A MicroRNA93-interferon regulatory factor-9-immunoresponsive Gene-1-itaconic acid pathway modulates M2-like macrophage polarization to revascularize ischemic muscle. Circulation. 2017;135(24):2403–2425. doi: 10.1161/CIRCULATIONAHA.116.025490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tallam A, Perumal TM, Antony PM, et al. Gene regulatory network inference of immunoresponsive Gene 1 (IRG1) identifies interferon regulatory factor 1 (IRF1) as its transcriptional regulator in mammalian macrophages. PLoS One. 2016;11(2):e0149050. doi: 10.1371/journal.pone.0149050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor SC, Nadeau K, Abbasi M, Lachance C, Nguyen M, Fenrich J. The ultimate qPCR experiment: producing publication quality, reproducible data the first time. Trends Biotechnol. 2019;37:761–774. doi: 10.1016/j.tibtech.2018.12.002 [DOI] [PubMed] [Google Scholar]
- 39.Kluth DC. Pro-resolution properties of macrophages in renal injury. Kidney Int. 2007;72(3):234–236. doi: 10.1038/sj.ki.5002332 [DOI] [PubMed] [Google Scholar]
- 40.Mitchell S, Thomas G, Harvey K, et al. Lipoxins, aspirin-triggered epi-lipoxins, lipoxin stable analogues, and the resolution of inflammation: stimulation of macrophage phagocytosis of apoptotic neutrophils in vivo. J Am Soc Nephrol. 2002;13:2497–2507. doi: 10.1097/01.asn.0000032417.73640.72 [DOI] [PubMed] [Google Scholar]
- 41.Epelman S, Lavine KJ, Beaudin AE, et al. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity. 2014;40:91–104. doi: 10.1016/j.immuni.2013.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lavine KJ, Epelman S, Uchida K, et al. Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proc Natl Acad Sci USA. 2014;111:16029–16034. doi: 10.1073/pnas.1406508111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cuesta N, Salkowski CA, Thomas KE, Vogel SN. Regulation of lipopolysaccharide sensitivity by IFN regulatory factor-2. J Immunol. 2003;170(11):5739–5747. doi: 10.4049/jimmunol.170.11.5739 [DOI] [PubMed] [Google Scholar]
- 44.Chen LL, Morcelle C, Cheng ZL, et al. Itaconate inhibits TET DNA dioxygenases to dampen inflammatory responses. Nat Cell Biol. 2022;24(3):353–363. doi: 10.1038/s41556-022-00853-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peace CG, LA O. The role of itaconate in host defense and inflammation. J Clin Invest. 2022;132(2):e148548. doi: 10.1172/JCI148548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mills EL, Ryan DG, Prag HA, et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature. 2018;556(7699):113–117. doi: 10.1038/nature25986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yi Z, Deng M, Scott MJ, et al. Immune-responsive gene 1/Itaconate activates nuclear factor erythroid 2-related factor 2 in hepatocytes to protect against liver ischemia-reperfusion injury. Hepatology. 2020;72(4):1394–1411. doi: 10.1002/hep.31147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiss JM, Davies LC, Karwan M, et al. Itaconic acid mediates crosstalk between macrophage metabolism and peritoneal tumors. J Clin Invest. 2018;128(9):3794–3805. doi: 10.1172/JCI99169 [DOI] [PMC free article] [PubMed] [Google Scholar]