Abstract
The human lbc oncogene product is a guanine nucleotide exchange factor that specifically activates the Rho small GTP binding protein, thus resulting in biologically active, GTP-bound Rho, which in turn mediates actin cytoskeletal reorganization, gene transcription, and entry into the mitotic S phase. In order to elucidate the mechanism of onco-Lbc transformation, here we report that while proto- and onco-lbc cDNAs encode identical N-terminal dbl oncogene homology (DH) and pleckstrin homology (PH) domains, proto-Lbc encodes a novel C terminus absent in the oncoprotein that includes a predicted α-helical region homologous to cyto-matrix proteins, followed by a proline-rich region. The lbc proto-oncogene maps to chromosome 15, and onco-lbc represents a fusion of the lbc proto-oncogene N terminus with a short, unrelated C-terminal sequence from chromosome 7. Both onco- and proto-Lbc can promote formation of GTP-bound Rho in vivo. Proto-Lbc transforming activity is much reduced compared to that of onco-Lbc, and a significant increase in transforming activity requires truncation of both the α-helical and proline-rich regions in the proto-Lbc C terminus. Deletion of the chromosome 7-derived C terminus of onco-Lbc does not destroy transforming activity, demonstrating that it is loss of the proto-Lbc C terminus, rather than gain of an unrelated C-terminus by onco-Lbc, that confers transforming activity. Mutations of onco-Lbc DH and PH domains demonstrate that both domains are necessary for full transforming activity. The proto-Lbc product localizes to the particulate (membrane) fraction, while the majority of the onco-Lbc product is cytosolic, and mutations of the PH domain do not affect this localization. The proto-Lbc C-terminus alone localizes predominantly to the particulate fraction, indicating that the C terminus may play a major role in the correct subcellular localization of proto-Lbc, thus providing a mechanism for regulating Lbc oncogenic potential.
The family of DH (dbl oncogene homology) domain-encoding oncogenes (8, 40) represents a unique category of transforming genes involved in cellular growth control. The DH domain is associated with guanine nucleotide exchange activation for the Rho/Rac family of small GTP binding proteins (8), resulting in the conversion of the inactive, GDP-bound form of the GTPase to the active, GTP-bound form capable of transducing signals (5, 14). In all cases, the DH domain is followed by a pleckstrin homology (PH) domain (5, 34) which can have multiple functions. Thus, these catalytic GDP-GTP exchange factors (GEFs) play a key role in regulating the Rho/Rac GTPase cycle. The Rho/Rac family of small GTPases mediates cytoskeletal reorganization (15), gene transcription (20), and cell cycle progression (36) through unique signal transduction pathways.
The 424-amino-acid Lbc oncoprotein is transforming both in vivo and in vitro and contains an N-terminal EF hand motif followed by DH and PH domains (49). We have shown that onco-Lbc activates the Rho small GTP binding protein by catalytically stimulating guanine nucleotide exchange, thereby resulting in GTP-bound Rho in vitro (55). The action of Lbc is specific for RhoA, -B, and -C (55), and the subsequent discovery that Lfc, Lsc (52, 53, 13), and P115GRF (19) also exclusively stimulate GTP exchange on Rho reveals the existence of a GEF subfamily specific for Rho. While members of this subfamily share similarity in their DH and PH domains, they otherwise encode unique domains and/or motifs, indicating that in vivo they likely serve to transduce divergent signals to their common target, the Rho GTPase. Other DH domain-encoding transforming genes such as dbl (18), tiam-1 (33), and vav (9) encode GEF activity for CDC42 and Rac GTPases. Thus, each of these cellular oncogenes is thought to regulate critical aspects of Rho/Rac GTPase function in vivo.
Much attention has focused on the Rho small GTPase that mediates actin stress fiber and focal adhesion assembly (39) in addition to gene transcription (20) and progression through the G1 phase of the cell cycle (36). As would be predicted for an in vivo activator of Rho, we have shown that microinjection of onco-Lbc into quiescent fibroblasts induces actin stress fiber and focal adhesion assembly (55), and G1 to S phase progression (37). These biological effects are identical to those reported for activated Rho (36, 39) and confirm the in vivo role of Lbc.
The precise mechanism of transformation by Rho/Rac exchange factor oncoproteins is currently poorly understood. While it is clear that activation of their target Rho GTPases is necessary for transforming activity, virtually all of the exchange factor oncoproteins are more potently transforming than activated forms of their target Rho/Rac GTPases, which are weakly transforming (15). The precise reason for this is not clear. One possibility is that the GTPases must traverse through the GTP-bound state to induce potent oncogenicity, as has been demonstrated for CDC42 (30). Additionally, in some cases, Rho/Rac GEF oncoproteins may activate multiple Rho/Rac targets coordinately, resulting in cooperative transforming activity (8). Alternatively, the exchange factors themselves have additional functions besides GTPase activation that promote oncogenicity when disrupted. The latter explanation is supported by the observation that the potently oncogenic forms of many Rho/Rac exchange factors have undergone N- or C-terminal truncation of putative regulatory motifs and/or domains (8), although the DH and PH domain cassette associated with GTPase activation is not altered.
We previously observed that the size of the onco-lbc mRNA transcript in the original tumorigenic lbc transfectants was 4 kb, while proto-lbc transcripts present in normal human tissues are at least ∼6 kb (49). This difference in transcript size indicates that onco-lbc may represent a truncated form of the lbc proto-oncogene. In order to elucidate the molecular basis for activation of the lbc oncogene, here we have analyzed its genomic structure, isolated lbc proto-oncogene cDNAs from normal tissue, and compared onco- and proto-lbc cDNA sequences. Next, the in vivo guanine nucleotide exchange factor activity of proto-Lbc was investigated and compared to that of onco-Lbc. In addition, the transforming activities of proto-Lbc and derived mutants were compared to that of onco-Lbc. Furthermore, regions necessary for onco-Lbc transforming activity were defined, and the regulatory portion of the intact proto-oncogene product that is normally responsible for inhibiting transforming potential was identified. Finally, the subcellular localizations of onco- and proto-Lbc were compared, and the roles of the PH domain and the proto-Lbc regulatory region in determining subcellular targeting were investigated.
MATERIALS AND METHODS
Genomic cloning.
Human repeat sequence was used to probe a λ EMBL4 genomic library prepared from EcoRI-digested onco-lbc transfectant DNA (49). The 7.8-kb genomic clone designated TL was restriction mapped and analyzed for transcribed sequence. Sequencing was performed with custom-made primers by using an ABI 373 automatic sequencer.
cDNA cloning.
A commercially available cDNA library prepared from normal human skeletal muscle (Clontech) was probed with the lbc oncogene 9a cDNA (49) by using stringent hybridization conditions. Two partially overlapping cDNAs were identified which together spanned 2.4 kb but lacked a 3′ stop codon in the open reading frame. To obtain further 3′ sequence, a PCR-generated 300-bp probe from the extreme 3′ sequence was used to rescreen the cDNA library. An overlapping clone was identified that extended the existing 3′ sequence by an additional 1.5 kb. A full-length lbc proto-oncogene cDNA open reading frame was generated by using Pfu polymerase and PCR to join the partial cDNAs and to provide terminal XhoI and BamHI restriction sites for subcloning into the pSRαNeo vector (48). This composite proto-lbc cDNA was sequenced in both directions to obtain correct sequence.
Chromosomal localization.
Southern blots containing human:hamster somatic cell hybrid DNA (BIOS, New Haven, Conn.) were hybridized with random-primer-labelled genomic and cDNA sequences according to the manufacturer’s methods. A mixture of proto-lbc and onco-lbc cDNAs were labelled with digoxigenin-11–dUTP by nick translation and hybridized to human metaphase chromosomes prepared from phytohemagglutinin-stimulated lymphocytes of a healthy male. Fluorescent in situ hybridization (FISH) was performed as previously described (23). Hybridization signals were detected with rhodamine-conjugated antibody to digoxigenin, localized to 4′,6-diamidino-2-phenylindole (DAPI)-banded chromosomes (0.1 μg/ml), and viewed with epifluorescence microscopy through a triple-band-pass filter set (Texas red/DAPI/FITC; ChromaTech, Inc.).
Southern and Northern blotting.
Southern blotting and hybridization was carried out according to standard procedure (43). Northern blot hybridization was carried out according to the manufacturer’s recommendation (Clontech). Probes were prepared with the Random Primed DNA Labelling Kit (Boehringer Mannheim). Filters were washed under stringent conditions.
In vivo phospholabelling and immunoprecipitation.
The wild-type RhoA cDNA sequence fused to an in-frame (six-histidine) epitope at the amino terminus was subcloned into the pMT3 vector and sequenced to ensure sequence fidelity. COS-7 cells were transfected with 0.5 to 5 μg of the desired combination of RhoA-Lbc plasmids/60-mm-diameter dish as detailed below. One day following transfection, cells were placed in serum-free medium overnight. The following morning, transfectants were incubated for 20 min in serum-containing phosphate-free minimal essential medium (Gibco BRL) and then switched to medium containing 0.25 mCi of 32PO4 (ICN)/ml/dish for 4 h. Cells were lysed in 0.5 ml. of 1% Triton X-100–1% Nonidet P-40–50 mM Tris (pH 7.5)–150 mM NaCl–10-μg/ml aprotinin–1 mM phenylmethylsulfonyl fluoride (PMSF) Hexahistidine epitope-tagged RhoA protein was isolated by a 60-min incubation of the extract with 30 μl of a 50% slurry of Ni2+-nitrilotriacetic acid agarose beads (Qiagen) preequilibrated in 50 mM NaH2PO4 (pH 8.3)–300 mM NaCl, followed by three washes in the above buffer plus 10 mM imidazole. After a final wash in phosphate-buffered saline, beads were eluted in 1 M KH2PO4 (pH 3.4) by heating at 87°C for 4 min, and the labelled nucleotides separated by thin-layer chromatography (TLC).
TLC.
Eluted samples were spotted on polyethylenemine-cellulose plates, and nucleotides were resolved by TLC in 1 M KH2PO4 (pH 3.4). The proportion of GTP-bound Rho was quantified by phosphorimager analysis, and the ratio of GDP to GTP-bound Rho was expressed as percent GTP according to the formula 100 × (GTP/GDP × 1.5 + GTP) to normalize for moles of phosphate.
COS-7 cell transfection.
COS-7 cells were maintained in Dulbecco’s modified essential medium (DMEM) (Gibco BRL) and 10% iron-supplemented calf serum (Sigma) in 6% CO2. A total of 5 × 105 cells/60-mm-diameter culture dish were transfected with 1 to 5 μg of plasmid DNA by the DEAE-dextran method (3). Four hours after transfection, the cells were subjected to a 45-s 10% dimethylsulfoxide shock. After transfection the cells were grown for 2 days, harvested, and lysed in 0.3 ml of 50 mM Tris-HCl (pH 7.5)–150 mM NaCl–1% Nonidet P-40, 1 mM PMSF–10-μg/ml aprotinin.
NIH 3T3 cell transfection, focus formation assay, and G418 selection.
For the focus formation assay, NIH 3T3 cells (D4 subclone gift of C. J. Marshall) were seeded at 1.3 × 105 cells/100-mm-diameter dish in DMEM and 10% newborn calf serum (Sigma) in 7.5% CO2. The next day, 0.025 to 1 μg of plasmid/dish was transfected with 15 μg of high-molecular-weight NIH 3T3 carrier DNA by using calcium phosphate precipitation as previously detailed (49). The following day, the precipitate was washed off with Tris-buffered saline (TBS), and the medium was replaced with DMEM and 5% lot-selected donor calf serum. Transfectants were fed with DMEM and 5% lot-selected calf serum every third day and stained with crystal violet at day 12 to 14 posttransfection, and foci were counted. Each group contained three to four dishes, and each experiment was performed at least three times. The number of foci per picomole of DNA was calculated. For G418 selection, NIH 3T3 cells were seeded at 2 × 105 cells/60-mm-diameter dish in DMEM and 10% donor calf serum (Sigma) in 7.5% CO2. The next day, 0.025 to 1 μg of plasmid/dish was transfected with 15 μg of high-molecular weight NIH 3T3 carrier DNA by using calcium phosphate precipitation as previously detailed (49). The following day, the precipitate was washed off with TBS, and the medium was replaced with DMEM and 10% donor calf serum. The next day, each dish was trypsinized and seeded into two 100-mm-diameter dishes containing DMEM and 10% donor calf serum in the presence of 1 mg of G418 sulfate (Geneticin; GIBCO BRL)/ml. After 10 days to 2 weeks, discrete colonies were visible on the dish which could be ring-cloned and expanded.
Mutagenesis.
The proto-lbc cDNA SK15 was used as template to generate the proto-lbc C-terminus truncation mutants (CT, PP, and α-HEL) and the proto-specific C terminus sequence construct (PS-1) by PCR with PFU DNA polymerase (Stratagene, La Jolla, Calif.). XhoI and BamHI sites were incorporated into the 5′ and 3′ oligonucleotides, respectively, for subcloning into the pSRαNeo vector. For all mutants, the 3′ oligonucleotide contained an in-frame octamer Flag epitope (Kodak International Biotechnology) sequence followed by a TGA stop codon at the desired position. For the PS-1 mutant, the 5′ oligonucleotide contained an in-frame GAACATG sequence to initiate translation. PCR products were agarose gel purified with GeneClean (Bio 101). cDNA was sequentially digested for ligation to BamHI- and XhoI-digested pSRαNeo vector. Mutants were fully sequenced to verify sequence fidelity. 9a2 onco-lbc cDNA (49) was used as a template to generate onco-lbc mutants. Site-directed mutagenesis was used to generate single-point mutations in the onco- and proto-lbc cDNA by the Muta-Gene Phagemid In Vitro Mutagenesis kit (Bio-Rad, Richmond, Calif.) according to the recommended procedure. The tyrosine (TAC) residue at codon 233 in the onco-lbc DH domain was changed to a phenylalanine (TTC) with the primer 5′AAAACTGGGAACTTGGTAA3′. The conserved tryptophan (TGG) residue at codon 404 in the PH domain was replaced by a leucine (TTG) residue with the primer 5′ATCTGAATCAAGCTGTTTC3′. After being sequenced to verify the mutation, mutant cDNAs were subcloned into the pSRαNeo vector. The onco-lbc PH and the DH domain deletion mutants were generated by PCR with PFU DNA polymerase according to a two-step process (25). These mutants were sequenced in their entirety to ensure sequence fidelity.
Western blotting.
Insoluble lysate material was removed by centrifugation at at 10,000 × g for 10 min. Protein content was determined by the bicinchoninic acid (BCA) assay (Pierce), and equal aliquots (50 μg total protein content) were resolved by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis. After transfer onto nitrocellulose, filters were blocked overnight at 4°C in 5% nonfat dry milk in TBS and then washed twice in TBS. Filters were probed for 1 h with anti-FLAG M2 antibody (Eastman Kodak Co., New Haven, Conn.) at a concentration of 10 μg/ml in TBS or with a 1:1,000 dilution of anti-onco-Lbc antibody in TBS containing 0.05% Tween 20 and were then washed three times for 30 min in TBS. After incubation for 1 h with anti-mouse horseradish peroxidase, filters were washed and developed with enhanced chemiluminescence reagents (Amersham).
Subcellular fractionation.
Transiently transfected COS-7 cells (four 100-mm-diameter dishes) were allowed to swell in hypotonic buffer (1 mM Tris [pH 7.5]) containing protease inhibitors (1 mM PMSF and 10-μg/ml aprotinin) on ice for 15 min. After scraping, cells were briefly pelleted, resuspended in 300 μl of hypotonic buffer, and homogenized in a Dounce homogenizer with 100 strokes. The recovered homogenate (∼200 μl) was centrifuged at 10,000 × g for 10 min to remove partially disrupted cells, and the supernatant was subjected to high-speed centrifugation at 100,000 × g for 1 h at 4°C. The resultant supernatant (∼200 μl) was collected as the S-100 soluble fraction. The P-100 particulate fraction was derived from resuspension of the pellet in 200 μl of hypotonic buffer by sonication. Comparison of the protein contents of the postnuclear supernatant versus the sum of the S and P fractions by BCA assay demonstrated minimal loss of cellular material during the fractionation procedure. Equal volumes from each fraction were analyzed by Western blotting.
RESULTS
The onco-lbc genomic structure is rearranged.
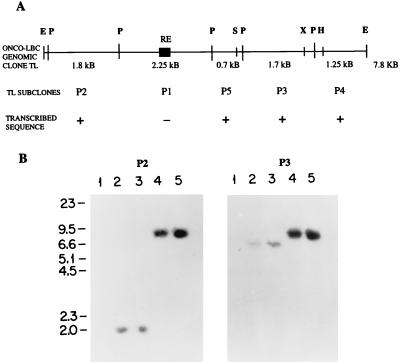
A 7.8-kb EcoRI human genomic clone designated TL was cloned from a genomic library prepared from NIH 3T3:onco-lbc transfectant cells (49). Figure 1A shows that PstI digestion of TL yields five contiguous genomic fragments, P1 to P5. P2, P3, P4, and P5 were found to encode unique lbc transcribed sequence based on the results of Northern blotting (not shown), and the P1 subclone contained a repeat element and was not used in further analyses.
FIG. 1.
Structure of the onco-lbc genomic clone TL. (A) Restriction digestion of the 7.8-kb TL clone with PstI yields five contiguous fragments P1 to P5. Subclone P1 contains a human repetitive element (RE), indicated by a solid box, and was not used in further analyses. Subclones P2 to P5 contain transcribed lbc sequence as determined by Northern blotting (results not shown) and were used for subsequent analyses. E, EcoRI; P, PstI; S, SacI; X, XbaI; H, HindIII. (B) Southern blot analysis of EcoRI-digested mouse, human, and lbc transfectant DNAs. P2, DNAs hybridized with the P2 genomic subclone; P3, DNAs hybridized with the P3 genomic subclone. Lanes: 1, NIH 3T3; 2, normal human placenta; 3, LBC patient sample; 4 and 5, secondary and tertiary, respectively, NIH 3T3:lbc transfectant DNAs.
In order to further characterize the TL genomic clone, P2 and P3 genomic subclones from opposing ends of TL were used to probe Southern blots containing EcoRI-digested DNA from murine NIH 3T3, normal human placenta, the original LBC (lymphoid blast crisis) leukemic sample, and NIH 3T3:onco-lbc transfectant cells. Figure 1B shows that, as expected, P2 and P3 subclones hybridize to a 7.8-kb EcoRI band in the NIH 3T3:lbc transfectants (lanes 4 and 5) and not to mouse DNA (lanes 1). In the normal and LBC human DNAs (lanes 2 and 3), the P2 probe detected a common 2-kb band, and the P3 probe detected a common 6.6-kb band. These data demonstrate, first, that the gross lbc gene structure in the normal human and LBC leukemic DNAs appears to be the same, and second, that transcribed portions of the onco-lbc gene are rearranged in the oncogenic NIH 3T3:lbc transfectant cells. Analysis with fragments P4 and P5 yielded results in agreement with these conclusions (not shown).
The onco-lbc cDNA is a chimera derived from fusion of the lbc proto-oncogene on chromosome 15q with unrelated chromosome 7q sequence.
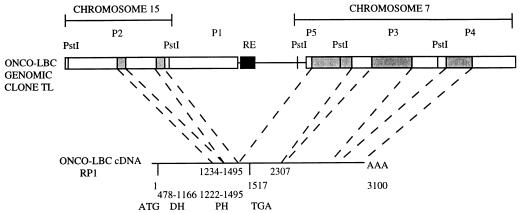
On the basis of the above results, the chromosomal localization of the onco-lbc TL genomic subclones was analyzed. Subclones P2 to P5 were used to probe Southern blots containing human:rodent somatic cell hybrid DNAs. Figure 2 presents these results in schematic form and shows that the P2 genomic subclone at one end of TL localizes to human chromosome 15, while subclones P3 to P5 from the opposite end of TL localize to human chromosome 7. For more precise analysis, the TL genomic clone was subjected to sequencing from both ends, and the sequence was compared to that of the RP1 onco-lbc cDNA. Sequenced regions are represented by the boxed areas shown in Fig. 2. The TL clone was found to encode five exons of the onco-lbc gene. The first two exons lie within the P2 subclone and encode C-terminal bases 1234 to 1495 of the onco-lbc cDNA open reading frame (49). A third exon spans the P5 and P3 subclones and encodes subsequent bases 1495 to 2306 representing onco-lbc cDNA translated (1495 to 1516) and 3′ untranslated (1517 to 2306) sequence. Two additional exons each lie within subclones P3 and P4 and encode 3′ untranslated (1517 to 2306) sequence. Two additional exons each lie within subclones P3 and P4 and encode 3′ untranslated bases 2307 to 3100 (49). In conjunction with the chromosomal localization results, these data demonstrate that the onco-lbc TL genomic clone encodes transcribed sequence derived from chromosomes 15 and 7.
FIG. 2.
Analysis of the onco-lbc genomic clone TL and cDNA. Genomic subclone P2 maps to chromosome 15, while P3, P4, and P5 map to chromosome 7. Boxed areas indicate sequenced regions. Shaded regions indicate exons encoded in onco-lbc cDNA. Bases 1234 to 1495 in the cDNA open reading frame are encoded in two exons within P2, bases 1496 to 2306 are encoded in an exon that spans P5 and P3, and bases 2307 to 3100 in the 3′ untranslated cDNA sequence are encoded in two exons each of which map to P3 and P4. ATG, start site; TGA, stop codon; AAA, poly(A) tail in the cDNA; RE, repeat element.
The TL genomic clone did not have any transforming activity in an NIH 3T3 focus formation assay either when transfected as is or when subcloned into a mammalian expression vector (results not shown), implying that it does not encode all of the onco-lbc sequences necessary for transforming activity. This is in agreement with the above findings showing that TL encodes only C-terminal exons of onco-lbc which do not include the DH domain between bases 480 and 1170 (49).
We next directly analyzed the chromosomal localizations of onco-lbc cDNA and focused on the junction around base 1495 as the possible rearrangement site. PCR-generated onco-cDNA probes were used to probe Southern blots containing rodent:human somatic cell hybrid DNAs. The results obtained (not shown) match the data for the TL genomic clone and demonstrate that onco-lbc cDNA is derived from truncation of the lbc proto-oncogene on chromosome 15 at a site corresponding to base 1495 in the cDNA sequence and subsequent fusion with unrelated sequence derived from chromosome 7, which supplies the short C terminus codons and 3′ untranslated regions (cDNA bases 1496 to 2300). Base 1495 corresponds to the end of the PH domain.
For precise chromosome mapping, 10 metaphases with single or double chromatid hybridizations were examined by FISH analysis using onco-lbc cDNA probes. The metaphases revealed hybridizations of proto-lbc at 15q24/25 and unrelated 3′ sequence at 7q36 (results not shown).
Proto-Lbc cDNA encodes an extended C terminus absent in onco-lbc.
In order to isolate the lbc proto-oncogene cDNA, a commercial oligo(dT) and random-primed human skeletal muscle cDNA library were probed with onco-lbc cDNA under stringent hybridization conditions, since we previously reported lbc mRNA expression in skeletal muscle, blood leukocytes, lung, and heart (49).
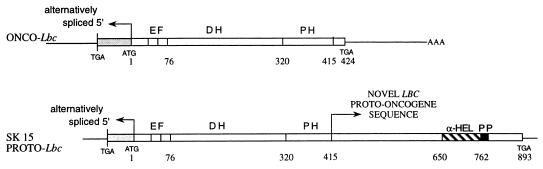
Several cDNAs ranging from 1.3 to 2.4 kb in size were isolated and sequenced. Figure 3 shows a composite of the full-length proto-lbc cDNA SK15 compared to the oncogenic form. As found for onco-lbc (46), the isolated proto-cDNAs contained variable 5′ ends which likely reflect complex splicing products in this region. These 5′ spliced cDNAs are consistent with the presence of multiple bands detected by Northern analysis of tissues (see below). Following this variable 5′ end, SK15 proto-Lbc cDNA encodes sequence identical to onco-Lbc that includes an intact EF hand motif followed by DH and PH domains which are identical to those found in onco-Lbc. In all proto-Lbc cDNAs analyzed, the PH domain is followed by an extended 1,434-bp open reading frame encoding a C terminus of 478 amino acids. This junction corresponds to base 1495 in onco-lbc. This extended C terminus is present in all proto-cDNAs isolated from both skeletal muscle and hematopoietic tissue (unpublished data) yet is entirely missing from all onco-Lbc cDNAs analyzed (49).
FIG. 3.
Schematic comparison of proto-lbc SK15 cDNA to onco-lbc cDNA. Boxed areas denote open reading frames, and single lines indicate untranslated regions. Onco- and proto-lbc cDNAs encode identical sequence until the end of the PH domain after which proto-lbc cDNA encodes a different, extended 3′ sequence. EF, hand motif; α-HEL, predicted α-helical region, PP, proline-rich sequence. Numbers underneath each schematic refer to amino acid positions.
The Lbc proto-oncogene C terminus encodes novel sequence.
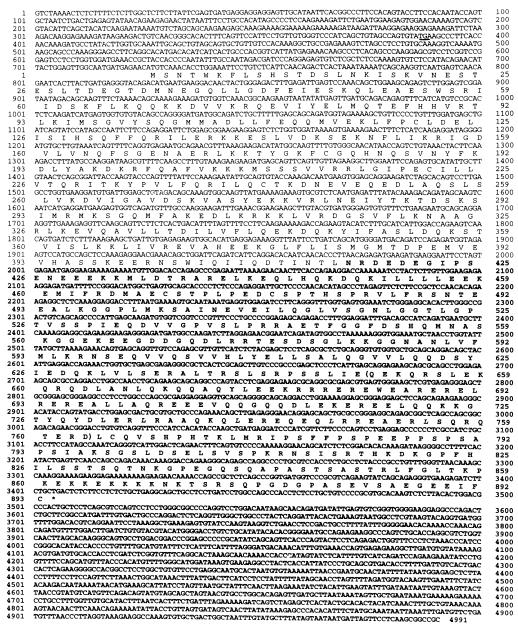
Figure 4 shows the SK15 proto-lbc cDNA sequence of 4,991 bp, although the sequence does not include complete 5′ and 3′ untranslated regions. As detailed above, the first 708 bp appear to be unique for SK15 and are likely the result of 5′ splicing. The nucleotide sequence from base 709 onward is common to all other proto-lbc cDNAs and to onco-lbc, and the in-frame ATG at nucleotide position 726 is a candidate translation initiation site of the proto-lbc product. This site is compatible with being a consensus sequence for translation initiation (24) and is in-frame with an upstream stop codon (TGA) located at nucleotide position 387. This results in a complete open reading frame of 2,679 bp with a predicted translated sequence of 893 amino acids, yielding a putative 102-kDa protein product.
FIG. 4.
Nucleotide and predicted amino acid sequences of lbc proto-oncogene SK15 cDNA. The predicted amino acid sequence is indicated in single-letter code below the nucleotide sequence. The upstream stop codon TGA is underlined. Proto-lbc-specific C-terminal sequence is shown in boldface, and the boundaries of the α-helical homology region are denoted by parentheses.
The nucleotide sequence from bases 726 to 1971 is identical between proto- and onco-lbc, and encodes 414 amino acids which contain an EF hand motif followed by DH (residues 76 to 306) and PH (residues 320 to 415) domains found in onco-Lbc (49). After nucleotide position 1972 (i.e., immediately after the TINTL amino acid sequence at the end of the PH domain), the SK15 sequence continues for an additional 1,435 bp of an open reading frame encoding 478 amino acids that is unique to proto-Lbc. This additional sequence is present in all proto-lbc cDNAs examined but is completely absent in onco-lbc. A search by using the BLAST network (2) for similarities between the translated Lbc proto-oncogene C-terminal sequence and those of known proteins revealed no overall identity or homology, and the first 235 residues appear to be unique. However, the subsequent ∼110 amino acids of the proto-specific C terminus (residues 651 to 763) are similar to those of an extensive list of known proteins, many of which are cyto-matrix associated. The highest homologies are with the human intermediate filament-associated protein trichohyalin (27) (28% identity, 57% overall homology), the human smooth muscle acto-myosin regulatory protein caldesmon (21) (20% identity, 47% homology), rat plectin (54) (22% identity, 44% homology), the chicken chromosome passenger protein class I INCENP (inner centromere protein) (31) (21% identity, 59% homology), and the human myosin beta heavy chain (4) (20% identity, 44% homology). Figure 5 shows an alignment of this region among Lbc, the two highest scoring homologous proteins, trychohyalin and caldesmon, and the derived consensus sequence. A search by using PROSITE revealed a putative leucine zipper motif (26) between residues 739 and 760. Following the α-helical region is a short proline-rich sequence, PSPEEPPSP, at residues 782 to 790.
FIG. 5.
Sequence similarities among Lbc, trychohyalin, and caldesmon. The sequences of the two proteins with the highest degrees of similarity with that of the proto-Lbc C terminus, as detected by BLAST, were aligned by using the Pileup multiple alignment program in the Genetics Computer Group package, and a consensus sequence was generated by using the Pretty program in extended GCG. The proto-Lbc residues shown (HPROTOLBC) are residues 651 to 764. HTRYCHOH, human trychohyalin residues 296 to 401; HCALDESM, human caldesmon residues 284 to 396.
Proto-lbc transcript is expressed in a wide variety of tissues.
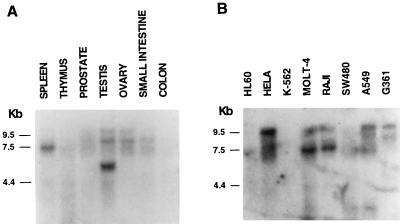
We previously reported lbc mRNA expression in human skeletal muscle, heart, and lung (49). Further Northern blot analysis using proto-lbc as a probe revealed that the spectrum of lbc expression in human tissues is wider than originally thought. Figure 6A shows high levels of variably sized lbc transcripts expressed in spleen and testis and mid-to-low levels of expression in prostate, ovary, and small intestine. Figure 6B shows lbc expression in the human cancer cell lines HeLa (epithelial), MOLT-4 (T lymphoblastic), Raji (Burkitt’s lymphoma), A549 (lung carcinoma), and G361 (melanoma) and low levels of expression in HL-60 (promyelocytic leukemia) and SW480 (colorectal adenocarcinoma). Hybridization of Northern blots with a control actin probe revealed comparable levels of RNA loading (not shown). These results indicate that lbc is expressed in myeloid and lymphoid lineages, a variety of epithelial tissues, and skeletal muscle.
FIG. 6.
Northern blot analysis of proto-lbc mRNA expression in human tissues (A) and human cancer cell lines HL-60 (promyelocytic leukemia), HeLa (cervical epitheloid carcinoma), K-562 (erythroleukemia), MOLT-4 (lymphoblastic leukemia), Raji (Burkitt’s lymphoma), SW480 (colorectal adenocarcinoma), A549 (lung carcinoma), and G361 (melanoma). (B) Northern blots were obtained from Clontech.
As reported earlier (49), the proto-lbc transcript size ranges from 6 to 9 kb with a consistent ∼7-kb transcript detected in spleen and myeloid and lymphoid cells and a larger transcript in HeLa, A549, and G361 cell lines. In contrast, testis contains a smaller-sized lbc transcript (∼5 kb) which is currently under study. The presence of multiple transcript bands of differing sizes in different tissues is thought to be due to a complex pattern of alternative splicing at the 5′ end of lbc, based on our isolation of numerous proto-lbc cDNAs with distinct 5′ sequence as represented in Fig. 3. Since the sizes of the major mRNA species of the related DH domain oncogenes lfc and lsc are 3.7 and 3.0 kb (52, 53), respectively, it is unlikely that the major bands observed with the lbc probe are due to cross-hybridization to these different gene transcripts.
Proto-Lbc can induce formation of GTP-bound Rho in vivo.
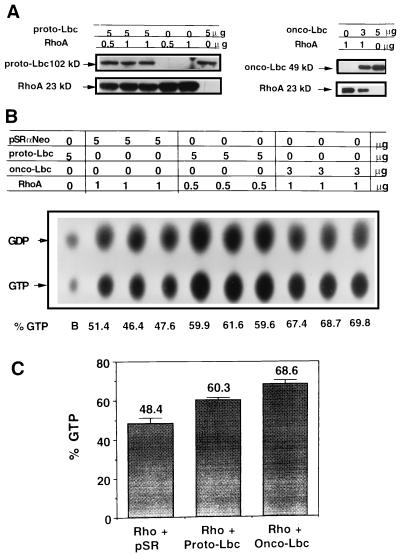
The Lbc oncoprotein has previously been shown to stimulate GTP exchange on Rho in vitro. Here, we investigated the guanine nucleotide exchange activities of onco- and proto-Lbc in vivo. Wild-type RhoA cDNA was subcloned into the simian virus 40 promoter-based vector pMT3. Since no good reagents exist for immunoprecipitating RhoA, an N-terminal (six-histidine) tag was included in the RhoA cDNA reading frame to allow purification with nickel-resin beads. Following transient transfection of COS-7 cells with 200 ng of RhoA plasmid/60-mm-diameter dish and Western blotting of cell lysates with the 26C4 anti-Rho antibody (Santa Cruz), preliminary experiments determined that substantial levels of RhoA protein could be purified with nickel-resin beads (results not shown). Lbc in vivo GEF activity was assessed by subcloning a C-terminal Flag epitope-tagged proto-Lbc cDNA into the pSRαNeo mammalian expression vector. Expression by this construct was confirmed by transient transfection into COS-7 cells followed by Western blotting of cell lysates with the M2 anti-FLAG antibody. Next, the amounts of RhoA and Lbc plasmid were titrated to determine the level of coexpression when cotransfected in COS-7 cells. Figure 7A shows Western blotting analysis of cotransfecting 5 μg of proto-Lbc and 0.5 μg of Rho plasmid and cotransfecting 3 μg of onco-Lbc and 1 μg of Rho; all constructs yield high levels of expression at these concentrations.
FIG. 7.
Onco- and proto-Lbc induce GTP exchange on RhoA. (A) Coexpression of 0 to 5 μg of proto-Lbc with 0 to 1 μg of RhoA cDNA and of 0 to 5 μg onco-Lbc with 0 to 1 μg of RhoA cDNA in COS-7 cotransfectant cells. In each case, the total amount of DNA transfected was adjusted to 6 μg by using the required amount of pSRαNeo vector. Immunoblots were probed with anti-Flag epitope antibody to detect onco- or proto-Lbc and with anti-RhoA antibody to detect RhoA. (B) Assay for onco- and proto-Lbc GTP exchange activities in vivo. Following cotransfection of COS-7 cells with the shown plasmid amounts and combinations, cells were metabolically labelled with 32PO4, and His-tagged RhoA was purified with nickel resin beads after four h. TLC separation of labelled nucleotides is shown. (C) Quantitation of the TLC results by phosphorimager. The results represent the means and SD from triplicate transfections in a representative experiment. Similar results were obtained in at least two other independent experiments.
With these concentrations, RhoA cDNA was cotransfected with either pSRαNeo empty vector, pSRαNeo:proto-Lbc, or pSRαNeo:onco-Lbc in COS-7 cells, and the effects on the level of guanine nucleotide-bound Rho were analyzed. Figure 7B shows the TLC separation results of GDP-GTP-bound Rho in the absence or presence of onco-Lbc or proto-Lbc. Figure 7C shows the quantitation of these results and demonstrates a background level of 48% ± 3% (mean ± standard deviation [SD]) GTP-bound Rho in COS-7 cells. When cotransfected with proto-lbc cDNA, an increase in the level of GTP-bound Rho to 60% ± 1% was observed. Cotransfection with onco-lbc cDNA further increased the level of GTP-bound Rho to 69% ± 2%. These results demonstrate that both onco- and proto-Lbc can promote formation of GTP-bound RhoA in vivo.
Proto-Lbc is weakly transforming.
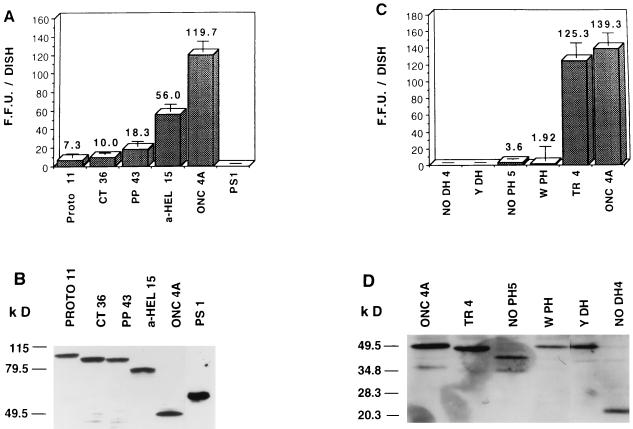
Next, the transforming activities of pSRαNeo:proto and onco-Lbc cDNAs were compared in a focus formation assay by transfection into NIH 3T3 fibroblasts. Initially, different isolates of each Flag epitope-tagged pSRαNeo construct were assessed for expression levels in COS-7 cells, and proto-Lbc clone PROTO 11 and onco-Lbc clone ONC 4A were selected for further analysis because they had comparable steady-state expression levels (results not shown). As shown in Fig. 8A, when transfected at equimolar amounts of DNA (corresponding to 80 ng of PROTO 11 and 40 ng of ONC 4A), PROTO 11 has ≤10% of the transforming activity (7 ± 4 foci/dish) of ONC 4A cDNA (120 ± 11 foci/dish). Although much reduced in number, proto-Lbc-induced foci exhibited characteristic onco-Lbc morphology (49). Figure 8B shows that PROTO 11-induced NIH 3T3 foci express a protein product of the correct predicted size of ∼102 kDa at a level comparable to that found in ONC 4A-induced NIH 3T3 foci. The Flag epitope had no significant effect on the transforming activity of Lbc, since comparison of these constructs to non-epitope-tagged versions yielded similar results (not shown). These results were in agreement with in vivo data in which nude mice were subcutaneously injected with 105 NIH 3T3:ONC 4A or NIH 3T3:PROTO 11 transfectant cells per site; onco-Lbc cDNA transfectants gave rise to 4/4 tumors per injection site in 20 days, while proto-Lbc cDNA transfectants gave rise to 1/4 tumors per injection site in 54 days (results not shown).
FIG. 8.
Transformation by proto-Lbc- and onco-Lbc-derived mutants measured by NIH 3T3 focus formation assay. (A) Transformation by proto-Lbc and its derived mutants. An equimolar amount of each plasmid was used based on 40 ng/dish of ONC 4A, except for PS-1 where a fivefold increase of plasmid was used. (C) Transformation by onco-Lbc and derived mutants. Equimolar amounts of ONC 4A and TR4 were used; for the remaining DH and PH domain mutants, a fivefold increased amount of plasmid was used. Results are derived from triplicate transfections in a representative experiment and represent the means and SD of NIH 3T3 focus numbers generated. Similar results were obtained in at least two other independent experiments. (B and D) Western blot analysis of expression of the Lbc mutants in NIH 3T3 foci. Individual NIH 3T3 foci were ring cloned and expanded for cell lysis. Cell lysates (equal protein content) were immunoblotted with anti-Flag M2 antibody. In the case of PS-1, YDH, and NODH 4 mutants, which do not generate foci, the same plasmid amounts used for the focus formation assay were used to generate G418-selected NIH 3T3 transfectant colonies; individual colonies were ring cloned and expanded for cell lysis, and lysates were immunoblotted as described above.
The transforming activity of proto-Lbc is increased by C-terminal truncation.
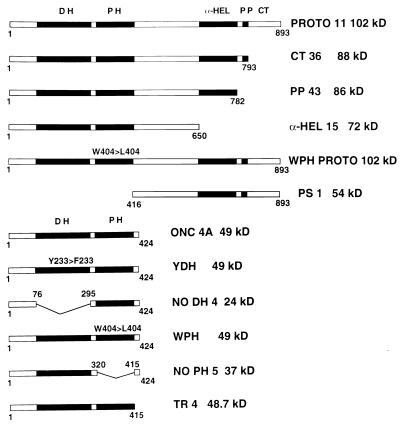
Since the proto-Lbc N terminus containing DH and PH domains is identical to that of onco-Lbc, we hypothesized that the proto-Lbc C terminus normally down-regulates Lbc transforming activity and that the potent transforming activity of the Lbc oncoprotein is due to loss of the proto-Lbc C-terminus. In order to test this hypothesis, proto-Lbc mutants representing successive truncations of the C terminus were generated each with a C-terminal Flag epitope in the pSRαNeo vector. These mutants are illustrated in Fig. 9, which shows that the CT36 (C terminus) construct lacks the final 100 residues, the PP43 (proline-rich) construct lacks the final 116 residues including the proline-rich region between residues 782 and 793, and the α-HEL 15 (α-helical) construct lacks the C-terminal 243 residues encoding the α-helical region, the PP motif, and the extreme C terminus. In addition, a construct designated PS-1 (proto specific) was generated which consists of only proto-Lbc C-terminal residues (residues 416 to 893) and lacks the DH and PH domains.
FIG. 9.
Schematic of onco-lbc and proto-lbc cDNAs and their derived mutants. α-HEL, α-helical region; PP, proline-rich motif, CT, extreme C terminus.
Dose responses of multiple cDNA ligation products of each mutant were tested in COS cell transfections to select mutant constructs at concentrations that yield comparable steady-state expression levels, and these cDNAs were used at the appropriate concentrations for focus assays. Onco-, proto-, and proto-C-terminal mutants were scored alongside each other in a NIH 3T3 cell focus formation assay to assess transforming activity at equimolar amounts with 40 ng of ONC 4A as a base, and the results of a representative experiment are shown in Fig. 8A. The CT36 mutant generated low numbers of foci (10 ± 2), which were not significantly higher than those obtained with PROTO 11 (7 ± 4). The PP43 mutant showed a very modest increase in transforming activity (18 ± 5), and the α-HEL 15 mutant had a fivefold increased transforming activity (56 ± 7) compared to that of PROTO 11. In contrast, the PS-1 construct showed absolutely no focus-forming activity, even when transfected at high concentrations (200 ng/dish). These data show that the α-HEL 15 mutant (which lacks the α-helical region, the proline-rich motif, and the extreme C terminus) is the most transforming of these mutants, although its activity is still only half that of onco-Lbc. Figure 8B shows that the CT36, PP43, and α-HEL 15 constructs express protein products of the correct predicted sizes (88, 86, and 72 kDa, respectively) at levels comparable to those of the PROTO 11 and ONC 4A constructs in NIH 3T3 foci. Although the PS-1 construct is not transforming, G418-selected NIH 3T3 colonies show a high level of expression of the correct 54-kDa product.
The chromosome 7-derived onco-Lbc C terminus is not transforming.
The results described above show that the extreme onco-Lbc C terminus after base 1495 (at the end of the PH domain) is derived from chromosome 7. We hypothesized that the primary role of this short region is to supply a premature stop codon rather than to provide transforming activity. In order to test this, an onco-Lbc cDNA mutant, designated TR4 (Figure 9), was prepared that contained a deletion of the chromosome 7-derived C-terminal nine amino acids of 9a onco-Lbc cDNA (49). When TR4 was assessed for transforming activity (125 ± 16 foci/dish), it was found to be as highly transforming as ONC 4A (139 ± 14 foci/dish) (Fig. 8C). This demonstrates that the acquisition of chromosome 7-derived sequence at the onco-Lbc tail does not confer a high level of transforming activity. The expression level of the 48.7-kDa TR4 product was comparable to that of ONC 4A in NIH 3T3 foci (Fig. 8D).
The DH and PH domains are required for onco-Lbc transformation.
In order to assess the roles of the onco-Lbc DH and PH domains in transformation, several mutants of these domains were generated (Fig. 9). For the DH domain, a construct lacking most of the domain (amino acid residues 76 to 295), designated NODH 4, was generated with a C-terminal Flag epitope; in addition, a single-point mutant (designated YDH) was prepared in which tyrosine 233 in the DH domain was conservatively altered to a phenylalanine. This tyrosine residue is part of the QRITKY sequence within the center of the DH domain that is identical in several GEFs. In addition, a PH domain deletion mutant (residues 320 to 415), termed NOPH 5, was generated from the onco-lbc cDNA template. Also, a single point mutant construct designated WPH, which altered tryptophan 404 in the PH domain to a leucine, was generated on the basis that this tryptophan is the only conserved residue in all of the PH domains (34).
The transforming activities of these mutants were assessed by transfecting fivefold increased amounts compared to that of ONC 4A (e.g., 200 ng/dish versus 40 ng/dish) in the NIH 3T3 focus formation assay. Figure 8C shows that NODH 4 was found to completely lack transforming activity, even at this high concentration. The YDH construct containing a single-point mutant in the Lbc DH domain had the same effect as deleting the entire domain, resulting in the complete loss of transforming activity. Figure 8C also shows that deletion of the onco-Lbc PH domain in the NOPH 5 construct resulted in a dramatic reduction in transforming activity (4 ± 7 foci/dish) compared to that of ONC 4A (139 ± 14 foci/dish), although trace activity was still detectable, and the foci exhibited the characteristic Lbc phenotype. The single-point mutant construct WPH also had a significantly reduced focus-forming activity (2 ± 17). Figure 8D shows that each of these mutants express the correct-sized products in NIH 3T3 transfectants. These results demonstrate that both intact DH and PH domains are required for onco-Lbc transforming ability.
Subcellular localization of different forms of Lbc.
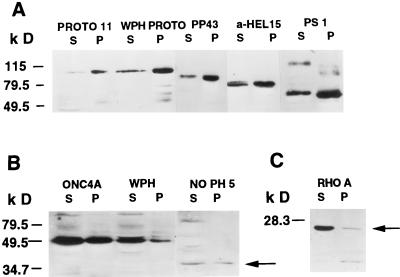
To further investigate how the transforming potential of proto-Lbc is regulated, the subcellular distributions of onco- and proto-Lbc and their derived mutants were analyzed by high-speed fractionation. Onco- and proto-Lbc and their derived mutants were transiently expressed in COS cells. Figure 10A shows that most of proto-Lbc (PROTO 11) localizes to the particulate (P) fraction. Based on reports that the PH domain of some proteins can confer membrane-lipid association (17, 22), a WPH PROTO mutant was generated where tryptophan 404 in the proto-Lbc PH domain was altered to a leucine, a mutation reported to abrogate the function of the PH domain of many proteins (8, 34). The WPH PROTO product still localized to the particulate fraction and did not show an altered distribution between fractions (Fig. 10A). Similarly, analysis of the weakly transforming proto-Lbc PP43 mutant revealed predominant localization to the P-100 fraction. In comparison, a somewhat larger proportion of the more active proto-Lbc α-HEL 15 mutant was detected in the cytosolic (S) fraction. Next, the subcellular localization of the proto-Lbc C terminus, as represented by the PS-1 construct, was analyzed and was found to localize mainly to the particulate fraction (Fig. 10A). Next, the subcellular localizations of onco-Lbc and the derived PH domain mutants were investigated. In contrast to the results obtained for proto-Lbc, Fig. 10B shows that >50% of onco-Lbc (ONC 4A) localizes to the S fraction. Both WPH and NOPH 5 onco-Lbc mutants exhibited the same localization as that of onco-Lbc (Fig. 10B). These data indicate that the Lbc PH domain does not play a primary role in conferring membrane localization. Onco-Lbc DH domain mutants did not show significant alterations in localization (results not shown). Figure 10C shows that under the same conditions, endogenous RhoA localizes primarily to the S fraction, a finding consistent with previous reports (1). Similar fractionation results were obtained for stable lbc:NIH 3T3 transfectants (results not shown). These findings indicate that (i) onco- and proto-Lbc show different subcellular localizations, (ii) the Lbc PH domain does not appear to play a major role in determining membrane localization, and (iii) the difference in localization between onco- and proto-Lbc may be attributed to the proto-Lbc C terminus.
FIG. 10.
Subcellular fractionation of proto-Lbc and onco-Lbc and their derived mutant products. (A) Proto-Lbc localizes to the P (particulate) fraction, and point mutation of the PH domain (WPH Proto) has no effect on this localization. The PP43 proto-Lbc deletion mutant product also localizes predominantly to the P fraction, while the α-HEL proto-Lbc deletion mutant shows some relocation to the S (soluble) fraction. The PS-1 product (the Proto-Lbc C terminus) localizes predominantly to the P fraction. (B) More than 50% of onco-Lbc localizes to the S fraction. A point mutation (WPH) or deletion (NOPH 5) product (indicated by arrow) of the onco-Lbc PH domain has no significant effect on onco-Lbc S or P fraction localization. (C) Endogenous or transfected RhoA (indicated by arrow) localizes predominantly to the S (soluble) fraction, as previously reported (1).
DISCUSSION
Malignant activation of several DH domain-encoding oncogenes including dbl and vav has been shown to result from molecular alterations that result in N- or C-terminal truncations, although the DH and PH domains remain intact (8). Here we describe the molecular alteration responsible for formation of the lbc oncogene. Our results demonstrate both at the genomic and cDNA levels that the lbc oncogene transcriptional unit is derived by C-terminal truncation of the lbc proto-oncogene located on chromosome 15 and subsequent fusion with unrelated sequence derived from chromosome 7. This conclusion is based on the finding that the onco-lbc genomic clone TL encodes lbc transcribed sequence which maps to chromosomes 15 and 7. Analysis of onco-lbc cDNA clones shows that the truncation site corresponds to the end of the proto-lbc PH domain. We further find that the ensuing chromosome 7-derived sequence in the onco-lbc cDNA supplies a short in-frame 3′ sequence followed by a termination codon and 3′ untranslated sequence. This chromosome 7-derived sequence does not hybridize to any of the human tissue mRNAs that we have tested by Northern blotting (results not shown). Taken together, these results indicate that the fused chromosome 7-derived sequence in the onco-lbc C terminus provides an in-frame stop codon for the truncated proto-lbc product.
The original LBC leukemia DNA whose transfection into NIH 3T3 cells yielded the lbc oncogene was classified as a lymphoid blast crisis phase of a Ph+ (Philadelphia chromosome) chronic myeloid leukemia sample, and we have previously shown that lbc is expressed in human leukemic lymphoblastic cell lines (49). Here we report that Southern blot analysis reveals no difference in the gross lbc gene structures of normal human and LBC sample DNAs. This strongly suggests (but does not prove) that the original leukemia cells did not contain the structurally altered onco-lbc gene, and hence it is likely that the genetic alteration which gave rise to onco-lbc occurred serendipitously during the course of the transfection process. This is a common event responsible for generating several transfection-derived cellular oncogenes (6). However, primary human leukemia samples can exhibit considerable cellular heterogeneity, and the conversion of chronic-phase chronic myeloid leukemia to blast phase is reported to be accompanied by acquisition of multiple poorly defined chromosomal alterations (44). Therefore, it may also be that only a small fraction of the original LBC cells contained a rearranged lbc gene not detectable by Southern blot analysis of LBC DNA. Precedent for this is provided by several reported cases in which activated ras oncogenes occur in only a fraction of the neoplastic cells (46, 50); furthermore, the in vivo assay used to detect the lbc oncogene is known to be sufficiently sensitive to detect oncogenes present in low levels in a sample (50). While PCR analysis could resolve this issue, no additional LBC sample is available for further study. In either case, for pathobiological relevance, lbc oncogene activation would be expected to occur in more than a single cancer sample, and this possibility is currently being investigated.
Isolation of lbc proto-oncogene cDNAs demonstrates that onco- and proto-lbc encode identical N termini coding for an EF hand motif and DH and PH domains. After base 1972, however, the proto-lbc open reading frame extends for an additional novel 1,434 bp not present in onco-lbc. Whether isolated from skeletal muscle tissue or from hematopoietic cells (unpublished results), we have found this proto-oncogenic C terminus to be invariant in sequence. Therefore, the oncogenic form of lbc essentially represents the N-terminal half of the proto-oncogene. Comparison of the translated sequence of the proto-specific C terminus to those of known proteins reveals an ∼110-amino-acid region (residues 651 to 763) with similarity to an extensive list of proteins, many of which are cyto-matrix associated, such as trichohyalin, plectin, caldesmon, INCENP, and myosin. In all of these proteins and in Lbc, this region is rich in the residues E/Q/R, and this likely provides the basis for the observed homology. While this region in Lbc is predicted to form an α-helical structure, it is shorter than in the homologous proteins (110 residues versus at least 300 residues). The role of this region in the known proteins appears to fall into two categories. First, this region is predicted to form a rod-like α-helical domain crucial for dimerization and higher-order assembly, such as for caldesmon (21) and myosin (4). Second, this homology region is strongly implicated in affecting protein-protein association, such as INCENP association with microtubules (31) and the association of the plectin rod domain with vimentin or lamin B (11). Therefore, this region in Lbc may play such a role. Intriguingly, a predicted α-helical region is also present in the dbl proto-oncogene product and, analogous to Lbc, is missing in the Dbl oncoprotein (42), strongly suggesting that structural and/or functional features of such domains in this family of oncogenes can normally suppress transforming activity. While in the case of Dbl this region encodes a heptad repeat motif characteristic of a coiled-coil structure (41), this does not seem to be the case for Lbc. The α-helical region of Lbc also contains a putative leucine zipper that may confer additional protein-protein association, although this motif is found in many proteins of different categories and it is far from being a specific pattern. Following the α-helical region is a proline-rich sequence (residues 782 to 790). This sequence contains a minimal PXXP core motif (P, proline; X, any amino acid) shown to provide SH3 domain binding sites (38). Therefore, this region in Lbc may be a potential SH3 binding site, and its precise role is under investigation.
Expression of the proto-lbc cDNA in NIH 3T3 and COS cells yields a protein product corresponding to a predicted size of 102 kDa that does not appear to be heavily posttranslationally modified in mammalian cells. We report here that both the onco- and proto-Lbc protein products promote formation of GTP-bound Rho in COS cells, demonstrating their GEF activity in vivo. In the absence of exogenous exchange activity, a higher percentage of exogenously expressed RhoA was found in the GTP-bound form (48%) compared to the level observed for other Ras-related small GTP binding proteins such as Ras and Ral (10 to 20%) (10, 51). This finding has been observed by others with COS-7 cells (12), but the reason is not clear. Expression of proto-Lbc increases the percentage of GTP-bound Rho by ∼30%, and expression of onco-Lbc results in an increase of 44%.
When the transforming activities of proto- versus onco-Lbc cDNAs were compared by NIH 3T3 focus formation assays, proto-Lbc was found to have ≤10% of the level of activity of onco-Lbc, even though the cDNAs were expressed at comparable levels under the control of the same promoter. While proto-Lbc-induced foci are much reduced in number, they nevertheless display characteristic Lbc morphology, indicating that relatively high-level expression of proto-Lbc by the strong promoter of the pSRαNeo vector can lead to weak transforming activity. However, loss of the proto-Lbc C terminus clearly has a major effect on amplifying transforming activity. An analogous situation is observed for Dbl where high-level expression of proto-Dbl results in weak transformation (41), although only the structurally altered oncoprotein is potently transforming (41, 42). While a considerable difference in the transforming activities of onco- and proto-Lbc is observed, the difference in GEF activities between these two forms in vivo is modest. This suggests that GEF activity levels alone may not be sufficient to account for the biological difference between onco- and proto-Lbc and that loss of C-terminal function synergizes with GEF activity to elicit potent transformation. However, detection of a potentially greater difference in GEF activities between the two forms upon measurement of endogenous GTP-Rho cannot be ruled out.
Analysis of the role of the proto-Lbc C terminus in transformation reveals that deletion of the proline-rich motif results in an ∼twofold increase in transforming activity compared to that of full-length proto-Lbc, suggesting that the proline-rich sequence may make a modest contribution to controlling transforming activity. Further truncation of the α-helical region results in a significant increase in transforming activity, resulting in ∼50% of the activity of onco-Lbc. While this demonstrates that the α-HEL 15 construct is significantly transforming, its activity is not equal to that of ONC 4A. This indicates that the remaining 235-amino-acid sequence between the PH domain and the α-helical region, which does not encode any known domains or motifs, appears to exert a significant inhibitory effect on transformation. Since an onco-Lbc cDNA mutant that contains a deletion of the chromosome 7-derived C terminus (TR4) still retains a high level of transforming activity, these findings demonstrate that it is loss of the proto-Lbc C terminus, rather than gain of unrelated sequence by the truncated proto-oncogene, that confers potent oncogenicity.
All DH domains are closely followed by a PH domain, indicating some coordinate function (5). Mutational analysis of the onco-Lbc DH domain demonstrates that an intact DH domain is necessary for Lbc transforming ability. This result is in agreement with that found for many other DH domain-encoding oncoproteins (8) and confirms our earlier findings that activation of the Rho signaling pathway by Lbc is an integral part of Lbc transformation (45, 55). In addition, we find that deletion of the entire onco-Lbc PH domain, or mutation of the conserved tryptophan at position 404 in the PH domain, significantly inhibits Lbc transformation. This illustrates the critical role of the PH domain in Lbc transformation, and the importance of the conserved W residue to PH domain function, and is consistent with the findings for other DH domain-encoding oncoproteins (8). Although we have not directly tested the GEF activities of the Lbc PH domain mutants in vitro, almost certainly they still retain exchange activity, since our earlier in vivo results obtained by using microinjection show that these mutants are still fully capable of inducing Rho-dependent actin cytoskeletal changes in fibroblasts, in contrast to onco-Lbc DH domain mutants, which retain no cytoskeletal activity (37). Taken together, these results indicate that the PH domain does not directly determine GEF activity but may regulate it in some way in vivo that is required during cellular transformation.
Within the past few years, the importance of correct intracellular targeting for oncoprotein activity has been brought to light (28). Therefore, we analyzed the subcellular localization of Lbc in order to gain more insight into its mechanism of transformation. High-speed cell fractionation analysis revealed a substantial difference between onco- and proto-Lbc localization. The proto-Lbc product was observed to localize predominantly to the particulate, membrane-associated fraction. Interestingly, similar findings have been reported for the Ras exchange factor, Ras-GRF (7), and for Tiam-1, a Rac exchange factor (47), indicating that membrane localization may be a common feature of GTP exchange factors for Ras superfamily small GTP proteins involved in cell growth control. In contrast to the proto-Lbc localization, >50% of onco-Lbc is in the soluble, cytosolic fraction. These results indicate that Lbc transforming activity may correlate with release from a membrane-associated location.
Analysis of Lbc PH domain mutants indicate that the PH domain does not play a major role in determining proto-Lbc membrane localization, although previous data indicate that it may influence onco-Lbc localization to the cytoskeleton (37). Other exchange factors such as Ras-GRF and Tiam-1 each encode an N-terminal PH domain in addition to a second PH domain that is in tandem to the DH domain in these proteins (7, 47). In both cases, the N-terminal PH domain is shown to be required for the particulate localization of these proteins (7, 47). Lbc does not contain an additional PH domain, and it may be that isolated PH domains such as those present in Ras-GRF and Tiam-1 play a different role than those of PH domains found in tandem with DH domains. Furthermore, in the case of Ras-GRF, replacement of the N-terminal PH domain with a heterologous PH domain still targets the protein to the particulate fraction but is not sufficient for Ras-GRF activation, indicating that the PH domain has an additional, critical function other than membrane localization (7). This result is in agreement with our observations reported here that the Lbc PH domain has some currently unknown critical function required for cell transformation other than membrane localization. Understanding of the considerable diversity of known PH domain ligands (17, 22, 34) is increased by the report that the PH domain of Dbl confers cytoskeletal, rather than membrane, localization (56). Additional reports on analogous PH domains of the Ras exchangers Sos (35) and Vav (16) indicate a role for phospholipid binding and signal transduction via phosphatidylinositol 3-kinase and present the possibility that the Lbc PH domain could serve a similar function.
Interestingly, the proto-Lbc C terminus alone was observed to localize predominantly to the particulate fraction. While the weakly transforming proto-Lbc PP43 mutant also strongly localizes to the particulate fraction, the more active α-HEL mutant shows some relocalization to the cytosolic fraction. This indicates that a cytosolic localization correlates with a gain in transforming activity and that the greatest cytosolic concentration occurs with the fully transforming onco-Lbc that lacks the C terminus. These results indicate that at least one function of the Lbc C terminus is to confer correct location of the proto-Lbc product required for controlling oncogenic activity. Part of this requirement may be to mediate interaction with currently unknown elements that also localize to the membrane and serve to inhibit transformation. In addition, membrane localization of proto-Lbc may serve to limit access to the physiological substrate of Lbc GEF activity, Rho. Rho localizes predominantly to the cytosol, although a small fraction is thought to cycle to and from the membrane (1), and this fraction is presumed to be the biologically active fraction. Thus, abnormal cytosolic localization of onco-Lbc could result in sustained Rho activation and consequent promotion of oncogenicity. Based on the results reported here, future experiments will address the precise identification of cellular components that interact with the C terminus and regulate proto-Lbc subcellular targeting and cell transformation.
ACKNOWLEDGMENTS
Special thanks go to Sam E. Lux for his support, Chris Marshall for NIH 3T3 cells, Lori Wirth and Christine Bogle (MBCF, Dana Farber Cancer Institute) for sequencing, and Tom Graf (MBCRR, Dana Farber Cancer Institute) for sequence analysis.
This work was funded by NIH grant CA62029 and American Cancer Society grant JFRA-524 to D.T., NIH grant HD18658 and a grant from the Beth Israel Pathology Foundation, Inc. to J.H.M.K., NIH grant GM47707 and American Cancer Society grant FRA to L.A.F., and a Human Frontiers Science Program long-term fellowship to T.U.
REFERENCES
- 1.Adamson P, Paterson H F, Hall A. Intracellular localization of the p21Rho proteins. J Cell Biol. 1992;119:617–627. doi: 10.1083/jcb.119.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, et al. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates & Wiley InterScience; 1988. [Google Scholar]
- 4.Bober E, Buchberger-Seidl A, Braun T, Singh S, Goedde H W, Arnold H H. Identification of three developmentally controlled isoforms of human myosin heavy chains. Eur J Biochem. 1990;189:55–65. doi: 10.1111/j.1432-1033.1990.tb15459.x. [DOI] [PubMed] [Google Scholar]
- 5.Boguski M S, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–653. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 6.Bos J L. The ras gene family and human carcinogenesis. Mutat Res. 1988;195:255–271. doi: 10.1016/0165-1110(88)90004-8. [DOI] [PubMed] [Google Scholar]
- 7.Buchsbaum R, Telliez J-B, Goonesekera S, Feig L A. The N-terminal pleckstrin, coiled-coil, and IQ domains of the exchange factor Ras-GRF act cooperatively to facilitate activation by calcium. Mol Cell Biol. 1996;16:4888–4896. doi: 10.1128/mcb.16.9.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerione R, Zheng Y. The Dbl family of oncogenes. Curr Opin Cell Biol. 1996;8:216–222. doi: 10.1016/s0955-0674(96)80068-8. [DOI] [PubMed] [Google Scholar]
- 9.Crespo P, Schuebel K E, Ostrom A A, Gutkind J S, Bustelo X R. Phosphotyrosine-dependent activation of Rac-1 GDP/GTP exchange by the vav proto-oncogene product. Nature. 1997;385:169–172. doi: 10.1038/385169a0. [DOI] [PubMed] [Google Scholar]
- 10.Farnsworth C L, Freshney N W, Rubin L B, Ghosh A, Greenberg M E, Feig L A. Calcium activation of Ras mediated by the neuronal exchange factor Ras-GRF. Nature. 1995;376:524–527. doi: 10.1038/376524a0. [DOI] [PubMed] [Google Scholar]
- 11.Foisner R, Traub P, Wiche G. Monoclonal antibody mappings of structural and functional plectin epitopes. J Cell Biol. 1991;112:387–394. doi: 10.1083/jcb.112.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster R, Hu K-Q, Lu Y, Nolan K M, Thissen J, Settleman J. Identification of a novel human Rho protein with unusual properties: GTPase deficiency and in vivo farnesylation. Mol Cell Biol. 1996;16:2689–2699. doi: 10.1128/mcb.16.6.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glaven J A, Whitehead I P, Nomanbhoy T, Kay R, Cerione R A. Lfc and Lsc oncoproteins represent two new guanine nucleotide exchange factors for the Rho GTP-binding protein. J Biol Chem. 1996;271:27374–27381. doi: 10.1074/jbc.271.44.27374. [DOI] [PubMed] [Google Scholar]
- 14.Hall A. Small GTP-binding proteins and the regulation of the actin cytoskeleton. Annu Rev Cell Biol. 1994;10:31–54. doi: 10.1146/annurev.cb.10.110194.000335. [DOI] [PubMed] [Google Scholar]
- 15.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 16.Han J, Luby-Phelps K, Das B, Shu X, Xia Y, Mosteller R D, Krishna U M, Falck J R, White M, Broek D. Role of substrates and products of PI 3-kinase in regulating activation of Rac-related guanosine triphosphatases by Vav. Science. 1998;279:558–560. doi: 10.1126/science.279.5350.558. [DOI] [PubMed] [Google Scholar]
- 17.Harlan J E, Hajduk P J, Yoon H S, Fesik S W. Pleckstrin homology domains bind to phosphatidylinositol 4,5-bisphosphate. Nature. 1994;371:168–170. doi: 10.1038/371168a0. [DOI] [PubMed] [Google Scholar]
- 18.Hart M J, Eva A, Evans T, Aaronson S A, Cerione R A. Catalysis of guanine nucleotide exchange on the CDC42Hs protein by the dbl oncogene product. Nature. 1991;354:311–314. doi: 10.1038/354311a0. [DOI] [PubMed] [Google Scholar]
- 19.Hart M J, Sharma S, elMasry N, Qiu R, McCabe P, Polakis P, Bollag G. Identification of a novel guanine nucleotide exchange factor for the Rho GTPase. J Biol Chem. 1996;271:25452–25458. doi: 10.1074/jbc.271.41.25452. [DOI] [PubMed] [Google Scholar]
- 20.Hill C S, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 21.Humphrey M B, Herrera-Sosa H, Gonzalez G, Lee R, Bryan J. Cloning of cDNAs encoding human caldesmons. Gene. 1992;112:197–204. doi: 10.1016/0378-1119(92)90376-z. [DOI] [PubMed] [Google Scholar]
- 22.Inglese J, Koch W J, Touhara K, Lefkowitz R J. G beta gamma interactions with PH domains and ras-MAPK signaling pathways. Trends Biochem Sci. 1995;20:151–156. doi: 10.1016/s0968-0004(00)88992-6. [DOI] [PubMed] [Google Scholar]
- 23.Knoll J H M, Lichter A. In situ hybridization to metaphase chromosomes and interphase nuclei. In: Dracopoli N C, et al., editors. Current protocols in human genetics. 1, Unit 4.3. New York, N.Y: Green-Wiley; 1994. [DOI] [PubMed] [Google Scholar]
- 24.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 25.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landschulz W H, Johnson P F, McKnight S L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 27.Lee S C, Steinert P M, Kim I G, Marekov L N, O’Keefe E J, Parry D A. The structure of human trichohyalin: potential multiple roles as a functional EF-hand-like calcium-binding protein, a cornified cell envelope precursor, and an intermediate filament-associated (cross-linking) protein. J Biol Chem. 1993;268:12164–12176. [PubMed] [Google Scholar]
- 28.Leevers S J, Paterson H F, Marshall C J. Requirement for Ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature. 1994;369:411–414. doi: 10.1038/369411a0. [DOI] [PubMed] [Google Scholar]
- 29.Lemmon M A, Ferguson K M, O’Brien R, Sigler P B, Schlessinger J. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc Natl Acad Sci USA. 1995;92:10472–10476. doi: 10.1073/pnas.92.23.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, R., S. Bagrodia, R. Cerione, and D. Manor. A novel Cdc42Hs mutant induces cellular transformation. Curr. Biol., in press. [DOI] [PubMed]
- 31.Mackay A M, Eckley D M, Chue C, Earnshaw W C. Molecular analysis of the INCENPS (inner centromere proteins): separate domains are required for association with microtubules during interphase and with the central spindle during anaphase. J Cell Biol. 1993;123:373–385. doi: 10.1083/jcb.123.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masson D, Kreis T E. Identification and molecular characterization of E-MAP-115, a novel microtubule-associated protein predominantly expressed in epithelial cells. J Cell Biol. 1993;123:357–371. doi: 10.1083/jcb.123.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michiels F, Habets G G M, Stam J C, van der Kammen R A, Collard J G. A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature. 1995;375:338–340. doi: 10.1038/375338a0. [DOI] [PubMed] [Google Scholar]
- 34.Musacchio A, Gibson T, Rice P, Thompson J, Saraste M. The PH domain: a common piece in the structural patchwork of signaling proteins. Trends Biochem Sci. 1993;18:343–348. doi: 10.1016/0968-0004(93)90071-t. [DOI] [PubMed] [Google Scholar]
- 35.Nimnual A S, Yatsula B A, Bar-Sagi D. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science. 1998;279:560–563. doi: 10.1126/science.279.5350.560. [DOI] [PubMed] [Google Scholar]
- 36.Olson M, Ashworth A, Hall A. An essential role for Rho, Rac, and CDC42 GTPases in cell cycle progression through G1. Science. 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- 37.Olson M, Sterpetti P, Nagata K, Toksoz D, Hall A. Distinct roles for DH and PH domains in the lbc oncogene. Oncogene. 1997;15:2827–2832. doi: 10.1038/sj.onc.1201594. [DOI] [PubMed] [Google Scholar]
- 38.Ren R, Mayer B J, Cicchetti P, Baltimore D. Identification of a ten-amino acid proline-rich SH3 binding site. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- 39.Ridley A J, Hall A. The small GTP-binding protein Rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 40.Ron D, Zannini M, Lewis M, Wickner R B, Hunt L T, Graziani G, Tronick S R, Aaronson A, Eva A. A region of proto-dbl essential for its transforming activity shows sequence similarity to a yeast cell cycle gene, CDC24, and the human breakpoint cluster gene, bcr. New Biol. 1991;3:372–379. [PubMed] [Google Scholar]
- 41.Ron D, Tronick S R, Aaronson S A, Eva A. Molecular cloning and characterization of the human dbl proto-oncogene: evidence that its overexpression is sufficient to transform NIH/3T3 cells. EMBO J. 1988;7:2465–2473. doi: 10.1002/j.1460-2075.1988.tb03093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ron D, Graziani G, Aaronson S A, Eva A. The N-terminal region of proto-dbl down regulates its transforming activity. Oncogene. 1989;4:1067–1072. [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Sawyers C L, Denny C T, Witte O N. Leukemia and the disruption of normal hematopoiesis. Cell. 1991;64:337–350. doi: 10.1016/0092-8674(91)90643-d. [DOI] [PubMed] [Google Scholar]
- 45.Schwartz M A, Toksoz D, Khosravi-Far R. Transformation by Rho exchange factor oncogenes is mediated by activation of an integrin-dependent pathway. EMBO J. 1996;15:6525–6530. [PMC free article] [PubMed] [Google Scholar]
- 46.Shen W P V, Aldrich T H, Venta-Perez G, Franza B R, Furth M E. Expression of normal and mutant ras proteins in human acute leukemia. Oncogene. 1987;1:157–165. [PubMed] [Google Scholar]
- 47.Stam J C, Sander E E, Michiels F, van Leeuwen F N, Kain H E T, van der Kammen R A, Collard J G. Targeting of Tiam1 to the plasma membrane requires the cooperative function of the N-terminal pleckstrin homology domain and an adjacent protein interaction domain. J Biol Chem. 1997;272:28447–28454. doi: 10.1074/jbc.272.45.28447. [DOI] [PubMed] [Google Scholar]
- 48.Takebe Y, Seiki M, Fujisawa J-I, Hoy P, Yokota K, Arai K-I, Yoshida M, Arai N. SRα promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toksoz D, Williams D A. Novel human oncogene lbc detected by transfection with distinct homology regions to signal transduction products. Oncogene. 1994;9:621–628. [PubMed] [Google Scholar]
- 50.Toksoz D, Farr C J, Marshall C J. Ras gene activation in a minor proportion of the blast population in acute myeloid leukaemia. Oncogene. 1987;1:409–413. [PubMed] [Google Scholar]
- 51.Urano T, Emkey R, Feig L A. Ral-GTPases mediate a distinct downstream signaling pathway from Ras that facilitates cellular transformation. EMBO J. 1996;15:810–816. [PMC free article] [PubMed] [Google Scholar]
- 52.Whitehead I, Kirk H, Tognon C, Trigo-Gonzalez G, Kay R. Expression cloning of lfc, a novel oncogene with structural similarities to guanine nucleotide exchange factors and to the regulatory region of protein kinase C. J Biol Chem. 1995;270:18388–18395. doi: 10.1074/jbc.270.31.18388. [DOI] [PubMed] [Google Scholar]
- 53.Whitehead I, Khosravi-Far R, Kirk H, Trigo-Gonzalez G, Der C J, Kay R. Expression cloning of lsc, a novel oncogene with structural similarities to the dbl family of guanine nucleotide exchange factors. J Biol Chem. 1996;271:18643–18650. doi: 10.1074/jbc.271.31.18643. [DOI] [PubMed] [Google Scholar]
- 54.Wiche G, Becker B, Luber K, Weitzer G, Castanon M J, Hauptmann R, Stratowa C, Stewart M. Cloning and sequencing of rat plectin indicates a 466-kDa polypeptide chain with a three-domain structure based on a central alpha-helical coiled coil. J Cell Biol. 1991;114:83–99. doi: 10.1083/jcb.114.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng Y, Olson M, Hall A, Cerione R C, Toksoz D. Direct involvement of the small GTP-binding protein Rho in lbc oncogene function. J Biol Chem. 1995;270:9031–9034. doi: 10.1074/jbc.270.16.9031. [DOI] [PubMed] [Google Scholar]
- 56.Zheng Y, Zangrilli D, Cerione R A, Eva A. The pleckstrin homology domain mediates transformation by oncogenic Dbl through specific intracellular targeting. J Biol Chem. 1996;271:19017–19020. doi: 10.1074/jbc.271.32.19017. [DOI] [PubMed] [Google Scholar]