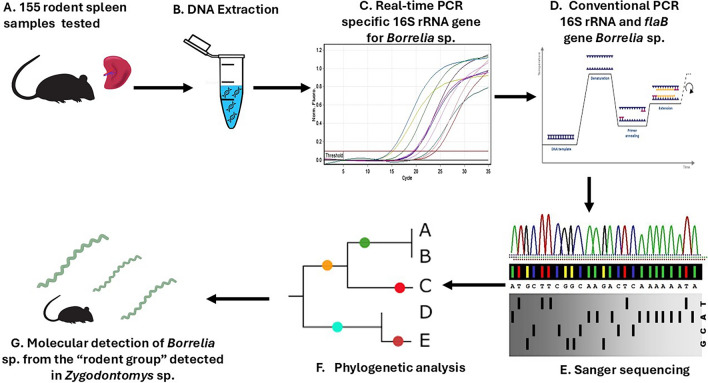
Abstract
Background
Bacteria of the genus Borrelia are agents of disease in both domestic animals and humans and pose a significant public health risk. Borrelia species have complex transmission cycles, often using rodents as vertebrate reservoir hosts. These bacteria are classified into three well-defined monophyletic groups: Borrelia burgdorferi sensu lato (Bbsl) complex, the relapsing fever (RF) group, and a third group associated with reptiles and echidnas. Moreover, a new group of Borrelia associated with rodents has recently been proposed, as these bacteria form a phylogenetic group separated from the previously mentioned groups. This study aimed to investigate the presence of DNA of Borrelia spirochetes in rodents in specific areas of the Colombian Caribbean.
Methods
A total of 155 rodent spleen samples were selected from the tissue bank. These samples were obtained in the departments of La Guajira and Córdoba (Northern Colombia). DNA extraction and specific real-time polymerase chain reaction (PCR) targeting Borrelia 16S ribosomal RNA (rRNA) gene were performed, followed by nested PCR (nPCR) on positive samples to obtain larger fragments of the 16S rRNA gene and characterize the flaB gene. Alignments of generated sequences and ortholog sequences downloaded from Genbank were performed in Clustal Omega. A phylogenetic tree was built with the maximum likelihood method in IQTREE.
Results
Spleen samples from rodents of the genera Heteromys, Mus, Necromys, Olygoryzomys, Proechymis, Rattus, Sigmodon, and Zygodontomys were processed. Overall, 6.5% (4/162) of the animals tested positive for Borrelia by real-time PCR. All quantitative PCR (qPCR)-positive samples were also positive for nPCR targeting the 16S rRNA gene, yielding fragments of 344–408 bp and 603–673 bp from two Sigmodon rodents and two Zygodontomys rodents from La Guajira and Córdoba. All samples were negative for the flaB gene. Only samples from Zygodontomys rodents presented good quality sequences. A BLASTn analysis showed a percentage of identity ranging between 98.16 and 96.06% with Borrelia sp. R57. Phylogenetic analysis revealed that sequences of the present study clustered with species of the recently proposed Borrelia “rodent group.”
Conclusions
This is the first detection of borreliae of the “rodent group” in South America. Our results reaffirm the occurrence of a group of spirochetes associated with rodents, extending its geographic distribution to the Colombian Caribbean.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13071-024-06560-7.
Keywords: Borrelia, Zygodontomys sp., Rodent reservoir hosts, Zoonosis, Tick-borne diseases
Background
The genus Borrelia includes pathogenic species that cause emerging and reemerging zoonotic diseases of significance to human and animal health [1]. The genus Borrelia is composed of three main monophyletic groups: the Borrelia burgdorferi sensu lato group (Bbsl), the relapsing fever (RF) group, and a group associated with reptiles and echidna (Tachyglossus aculeatus) hosts [2, 3]. Generally, borreliae are transmitted by ixodid (Ixodidae) and argasid (Argasidae) ticks, and one species, Borrelia recurrentis, is transmitted by the human clothing louse (Pediculus humanus humanus) [3]. Additionally, vertebrate hosts, such as bats, armadillos, monkeys, opossums, wild turkeys, deer, and squirrels, may be involved in the transmission cycle [1, 4–7]. But rodents are one of the most important vertebrate hosts for Borrelia spp. [8–10].
Indeed, many studies support this fact. In 1927, it was determined that the hosts of Borrelia of the RF group were several species of rodents [11]. Furthermore, in 1989 in the USA, B. burgdorferi was isolated from the rodent Peromyscus leucopus, which was at that time the main reservoir [12]. Subsequently, in the 1990s, Borrelia spp. of the Bbsl group were isolated in Europe in four species of rodents [13]. Furthermore, the rodent Oryzomys palustris is also a reservoir host for B. burgdorferi [14]. In South America, Thomas et al. shed light on the role of rodents as possible reservoirs of Borrelia spp. by detecting species of the Bbsl and RF group in rodents from Chile [15]. Moreover, a recent detection of those two groups of Borrelia spirochetes in rodents from Colombia, reinforces this association [16].
However, recent findings point that rodents carry spirochetes of genus Borrelia that form a consistent group that separates from Bbsl and RF groups from a phylogenetic viewpoint. In fact, a new group of borreliae associated with rodents in Australia, Spain, and the USA has been proposed and named as the “rodent group” [17–19], and adds a fresh perspective to the diversity of the genus [17]. In this study, we had access to a large number of rodent organs collected in the Colombian Caribbean and aimed to detect Borrelia DNA.
Methods
Study area and capture of rodents
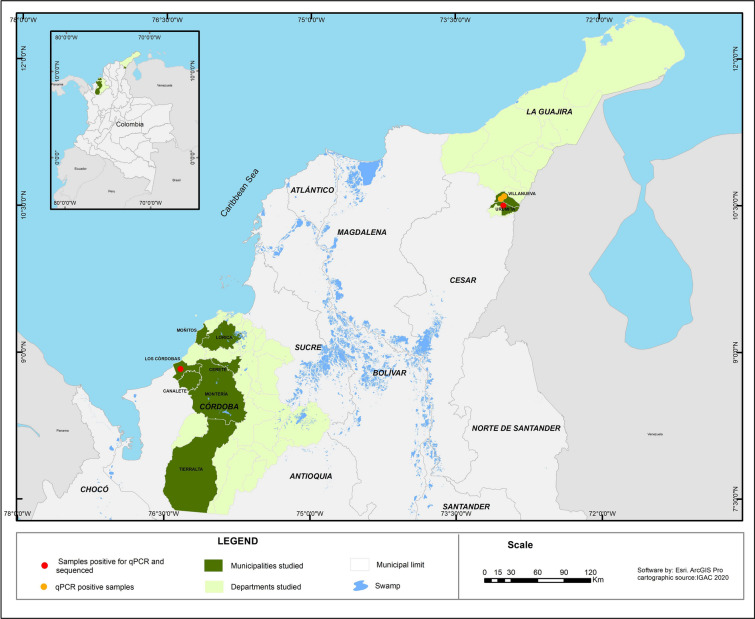
Spleen samples of rodents were obtained from the tissue bank of the Instituto de Investigaciones Biológicas del Trópico (Universidad de Córdoba). Rodent species from which spleen samples were collected were previously captured in field trips between 2011 and 2012 and 2022 and 2023 in rural and peri-urban areas of the municipalities of Los Córdobas, Montería, Tierralta, Moñitos, Cereté, and Lorica in the department of Córdoba; and rural areas of the municipalities of Villanueva and Urumita from the department of La Guajira (Additional file 1: Table S1) (Fig. 1).
Fig. 1.
Map of Colombia showing the location of the municipalities sampled in the departments of Córdoba and La Guajira. The points indicate the collection sites of qPCR-positive rodents and the sequenced samples
Molecular and phylogenetic analyses
DNA extraction from spleen was performed using the GeneJET genomic DNA purification kit (Thermo Scientific) following the manufacturer’s instructions. The DNA was quantified with a spectrophotometer. As an internal control, conventional polymerase chain reaction (PCR) targeting the mammalian β-actin gene was performed [20]. Real-time PCR (qPCR) was performed to detect Borrelia spp. using genus-specific primers (Bor16S3F, 5′-AGC CTT TAA AGC TTC GCT TGT AG-3′; Bor16S3R, 5′-GCC TCC CGT AGG AGT CTG G-3′) and a hydrolysis probe (Bor16S3P, 5′-6FAM–CCG GCC TGA GAG GGT GAA CGG BHQ-3′) that amplify a 148 bp fragment of the 16S ribosomal RNA (rRNA) gene [21]. Nested PCR (nPCR) and semi-nested PCR were implemented to positive samples to amplify three overlapping fragments of the 16S rRNA gene and two overlapping fragments of the flaB gene (Table 1) [22, 23].
Table 1.
Nested PCR primers for the detection of Borrelia sp.
| Gene | Round | Primer name | Secuencia 5′–3′ | Tm [°C] | bp |
|---|---|---|---|---|---|
| 16S-rRNA [22] | First round | FD3 [f] | AGAGTTTGATCCTGGCTTAG | 54 | 1489 |
| T50 [r] | GTTACGACTTCACCCTCCT | ||||
| Second round (hemi nested A) | FD3 [f] | AGAGTTTGATCCTGGCTTAG | 56 | 730 | |
| 16 s-1[r] | TAGAAGTTCGCCTTCGCCTCTG | ||||
| Second round (hemi nested B) | 16 s-2 [f] | TACAGGTGCTGCATGGTTGTCG | 56 | 462 | |
| T50 [r] | GTTACGACTTCACCCTCCT | ||||
| Second round (nested) | Rec4 [f] | ATGCTAGAAACTGCATGA | 54 | 520 | |
| Rec9 [r] | TCGTCTGAGTCCCCATCT | ||||
| flaB (flagellin) [23] | First round | FlaRL[f] | GCAATCATAGCCATTGCAGATTGT | 55 | 665 |
| FlaLL[r] | ACATATTCAGATGCAGACAGAGGT | ||||
| Second round A | FLaRS [f] | CTTTGATCACTTATCATTCTAATAGC | 55 | 491 | |
| FlaLL[r] | ACATATTCAGATGCAGACAGAGGT | ||||
| Second round B | FlaRL[f] | GCAATCATAGCCATTGCAGATTGT | 55 | 528 | |
| FLaLS [r] | AACAGCTGAAGAGCTTGGAATG |
Borrelia anserina genomic DNA was used as a positive control [24] and molecular grade water was used as a negative control. Sanger sequencing was performed on nPCR-positive samples. Sequences with a Phred score of 20 in Geneious (https://www.geneious.com/) were chosen and a taxonomic assessment of the sequences was performed with BLASTn [25], considering an E-value ≤ 1 × 10−5, a high Max Score and Total Score, a Query Cover > 90%, and a Per. Ident > 90%.
Sequences from the distinct groups of the Borrelia genus were downloaded from Genbank [26], to align them with the sequences of this study. Alignments were performed in Clustal Omega [27]. Phylogenetic reconstructions were performed in IQTREE with the maximum likelihood method using the TPM3 + F + G4 chosen according to BIC (Bayesian Information Criterion). We used a nucleotide substitution model with 1000 bootstraps [28]. Trees were visualized and edited with iTOL v5 [29].
Results
In total, 155 spleen samples from eight genera of rodents were analyzed; 42.6% (66/155) were of genus Rattus, 31.6% (49/155) Zygodontomys, 10.9% (17/155) Olygoryzomys, 5.1% (8/155) Mus, 3.9% (6/155) Proechymis, 1.9% (3/155) Heteromys, 1.9% (3/155) Sigmodon, and 1.9% (3/155) Necromys.
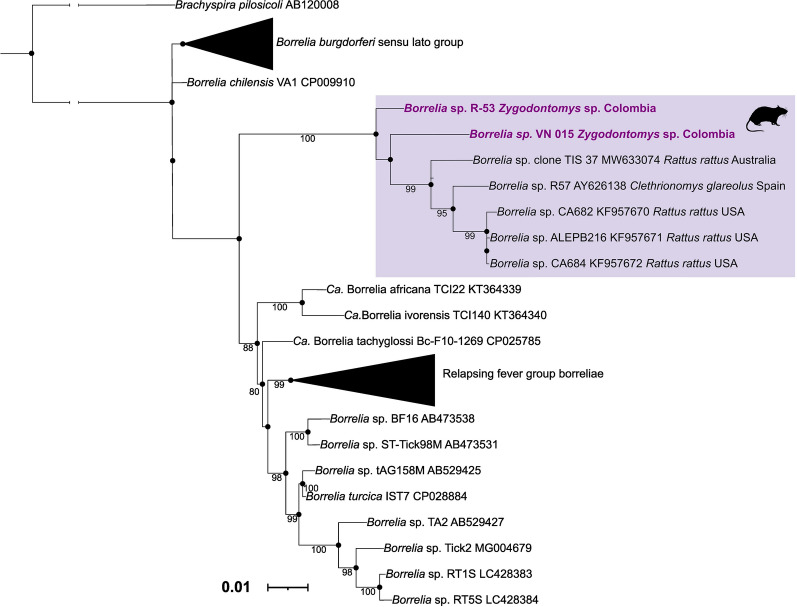
All samples were positive for the ß-actin gene PCR, demonstrating successful DNA extractions. The 6.5% (4/162), two Zygodontomys and two Sigmodon specimens, were positive for the 16S rRNA gene of Borrelia spp. by qPCR with CTs ≤ 33. The rodents were captured in the municipalities of Los Córdobas, Urumita and Villa Nueva (Table 2). Subsequent PCR protocols were positive only for the 16S rRNA gene. However, only two of the three nested reactions flanking both extremes of the 16S rRNA gene, but not overlapping, yielded amplicons of expected size (hemi nested A, heminested B). Good quality sequences were generated from samples of two rodents of the genus Zygodontomys from the departments of La Guajira and Córdoba (Fig. 1) and corresponded to sequences of 603–673 bp (heminested A) and 344–408 bp (heminested B). The BLASTn analysis of the fourth fragments showed a 98.16–96.02% of identity with Borrelia sp. R57 (GenBank accession number AY626138) (Table 2). The alignment for the 16S rRNA gene is constructed with 72 sequences, 70 downloaded from Genbank and two of our own. This alignment was adjusted to the size of the two sequences obtained (603–673 bp), products of the nPCR (hemi nested A). Phylogenetic analysis demonstrated a clustering with Borrelia sp. TIS 37 (GenBank accession number MW633074) [17], Borrelia R57 (GenBank accession number AY626138) [19], Borrelia sp. CA682 (GenBank accession number KF957670), Borrelia sp. ALEPB216 (GenBank accession number KF957671), and with Borrelia sp. CA684 (GenBank accession number KF957672) [18]. All these borreliae have been grouped in the group proposed as “rodent group” [17] (Fig. 2).
Table 2.
Rodents positive by qPCR for Borrelia spp
| Rodent code | Municipality | Department | Longitude | Latitude | Positive species by qPCR | 16S rRNA nPCR primer pairs | bp | Percentage of identity with Borrelia sp. R57 (GenBank accession number AY626138) | GenBank accession number |
|---|---|---|---|---|---|---|---|---|---|
| R-53 | Los Cordobas | Córdoba | 76°20 ′1.74"W | 8°49 ′53.1"N | Zygodontomys sp. | FD3_F/16S1_R | 673 | 96.64% | O557427 |
| 16S2_F/T50_R | 344 | 96.09% | O557428 | ||||||
| VN-15 | Urumita | La Guajira | 75° 20′ 664"W | 12° 16′ 979"N | Zygodontomys sp. | FD3_F/16S1_R | 603 | 96.02% | OR564036 |
| 16S2_F/T50_R | 408 | 98.16% | O557430 | ||||||
| VN 024 | Villa Nueva | La Guajira | 73° 02′ 261"W | 10° 35′ 563"N | Sigmodon alstoni | – | – | – | – |
| VN 025 | Villa Nueva | La Guajira | 73° 02′ 261"W | 10° 35′ 563"N | Sigmodon alstoni | – | – | – | – |
Fig. 2.
Phylogenetic analysis performed in this study. Phylogenetic tree of 16S rRNA gene constructed with 72 sequences, including two own sequences obtained in this study. The phylogenetic clade related to rodents is colored purple; the position of the detected Borrelia spp. is highlighted in bold purple. Brachyspira pilosicoli was used as an external group. The tree was built using the nucleotide substitution model TPM3 + F + G4 chosen according to BIC. The tree is drawn to scale, with the scale bar indicating nucleotide substitutions per site
Discussion
A Borrelia sp. from the “rodent group” was detected in Zygodontomys and Sigmodon in the Colombian Caribbean. Rodents are implicated as reservoirs of Borrelia species, mainly of the Bbsl group in the USA and Europe, where the bacteria are endemic [10]. However, there are reports where rodents are involved in the ecoepidemiology of the RF group [7]. Furthermore, some studies have detected species of the Borrelia genus in rodents that are in a different phylogenetic clade than the Bbsl and RF groups [17–19]. This was first demonstrated in a study conducted in Spain in 2005, where Borrelia sp. R57 was detected in rodents Apodemus sylvaticus, Clethrionomys glareolus, and Crocidura russula, with an infection rate of 8.5–12% by PCR targeting the 16S rRNA gene specific for this detected Borrelia, with primers designed by the authors. However, attempts to amplify fragments of 5S-23S, ospA, flaB, rpoB, and p66 genes were unsuccessful. In the same study, phylogenetic analyses fragments of 16S rRNA and groEL genes demonstrated that the sequences are located in a separate clade from RF and Bbsl groups [19]. These results were corroborated by a study conducted in the USA, where it was found that 43.5% of ear biopsies from R. rattus were positive for Borrelia sp. Phylogenetic analysis of the 16S rRNA gene demonstrated that the sequence from this study is related to the Spanish strain (Borrelia sp. R57).
Additionally, in 2021, a study in Australia reported a new species of Borrelia in tissue samples from nine rodents of the genus Rattus. This detection was obtained with the 16S rRNA v3–4 hypervariable region on the Illumina MiSeq, since attempts to amplify and perform Sanger sequencing of the flaB gene were unsuccessful. The identity of the Borrelia sequences showed that they were most similar to Borrelia sp. R57, as well as, phylogenetic analysis of these Australian sequences forms a separate clade, basal to the three main groups currently described. Therefore, the proposal of a new group of Borrelia associated with rodents, the “rodent group,” is suggested [17]. Our results support this hypothesis with the phylogenetic grouping of our sequences in the “rodent group” (Fig. 2). However, in all studies, there have been difficulties in amplifying other genes to confirm the taxonomic assignment of borreliae in this group.
In studies that detected Borrelia from the “rodent group”, attempts to detect this species in ticks were unsuccessful. Considering that soft and hard ticks are vectors of Borrelia species from the Bbsl group and the RF group, it is necessary to conduct studies to determine which tick species play a vectorial role in the transmission of this newly identified group.
In South America, there are few studies of Borrelia in rodents. One of the first studies was carried out in Chile by Thomas et al. in 2020, where PCR detected the presence of Borrelia of the Bbsl group in 5% (3/53) of blood samples of the rodents Oligoryzomys longicaudatus and Phyllotis xanthopygus [15].
Recently, in Colombia, Borrelia sp. related to the RF group was detected in tissues from the rodents Coendou rufescens and Microryzomys latissimus; and related to the Bbsl group in Thomasomys aureus and Mus musculus, with an infection rate of 1.9% for each group [16].
Few studies have detected Borrelia from the “rodent group” and difficulty has been generated in carrying out the molecular characterization of the “species” of this newly proposed group. Unfortunately, in our study, obtaining amplifications of the 16S-rRNA gene was only possible. This may be because this gene is highly conserved, and we are probably dealing with an atypical variant of Borrelia.
The phylogenetic analysis showed that our sequences clustered with Borrelia sp. TIS 37 (GenBank accession number MW633074) detected in Rattus rattus in Australia [17], Borrelia sp. R57 (GenBank accession number AY626138) detected in Clethrionomys glareolus in Spain [19]. (Gil et al. [19]); Borrelia sp. CA682 (GenBank accession number KF957670), Borrelia sp. ALEPB216 (GenBank accession number KF957671), and Borrelia sp. CA684 (GenBank accession number KF957672) detected in Rattus rattus in the USA [18]. The trees topology showed that our study sequences are located in a monophyletic group of borreliae associated only with rodents, basal to the RF group (Fig. 2). Unfortunately, it was only possible to obtain amplifications of the 16S rRNA gene and sequences of other genes could not be recovered to confirm the taxonomic assignment of the Borrelia detected in this study.
Our study underscores the need for further research that involves the use of multilocus genes, amplicon based whole genome sequencing with high throughput sequencing, and culturing Borrelia sp. from rodent tissues. These approaches could provide a more comprehensive understanding of the eco-epidemiology of Borrelia sp. in rodents.
Conclusions
This is the second detection of Borrelia sp. in rodents from Colombia and the first detection of Borrelia sp. in the “rodent group” in Latin America.
Furthermore, a wide geographical distribution of borreliae of the “rodent group” is demonstrated, with previous records in Australia, Spain, and the USA, and, with our results, in Colombia, being in South America for the first time.
Supplementary Information
Acknowledgements
We thank the University of Córdoba, where the project was carried out, and Marcelo Labruna for providing positive control.
Abbreviations
- Bbsl
Borrelia burgdorferi Sensu lato
- RF
Borreliae of the relapsing fever group
- qPCR
Real-time polymerase chain reaction
- nPCR
Nested polymerase chain reaction
Author contributions
Y.L., Se.Mu., Sa.Ma., and Á.A.F.M. designed the initial study; K.G., A.C., Y.L.M., and C.G. carried out the field work; Y.L., M.M., and J.R. performed DNA extraction, PCRs, and sequencing; Y.L., Se.Mu., and Á.A.F.M. implemented the phylogenetic analyzes; Y.L., Sa.Ma., and Á.A.F.M. wrote the first draft of the manuscript. All authors contributed to the interpretation and review of the data.
Funding
No funding.
Availability of data and materials
No datasets were generated or analyzed during the current study.
Declarations
Ethics approval and consent to participate
The rodents were captured under the permits of the National Authority of Environmental Licenses [ANLA], resolution no. 00914.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Oppler Z, Keeffe K, McCoy K, Brisson D. Evolutionary genetics of Borrelia. Curr Issues Mol Biol. 2021;42:97–112. 10.2177/cimb.042.097.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loh SM, Gillett A, Ryan U, Irwin P, Oskam C. Molecular characterization of ‘Candidatus Borrelia tachyglossi’ (family Spirochaetaceae) in echidna ticks, Bothriocroton concolor. Int J Syst Evol Microbiol. 2017;67:1075–80. 10.1099/ijsem.0.001929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Margos G, Henningsson AJ, Hepner S, Markowicz M, Sing A, Fingerle V. Borrelia ecology, evolution, and human disease: a mosaic of life. In: Sing A, editor. Zoonoses infect affect hum anim. Cham: Springer International Publishing; 2023. p. 1–66. [Google Scholar]
- 4.Christensen J, Fischer RJ, McCoy BN, Raffel SJ, Schwan TG. Tickborne relapsing fever, bitterroot valley, Montana, USA. Emerg Infect Dis. 2015;21:217. 10.3201/eid2102.141276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunn LH, Clark HC. Notes on relapsing fever in Panama with special reference to animal hosts. Am J Trop Med. 1933;1:201–9. 10.4269/ajtmh.1933.s1-13.201. [Google Scholar]
- 6.Muñoz-Leal S, Faccini-Martínez ÁA, Pérez-Torres J, Chala-Quintero SM, Herrera-Sepúlveda MT, Cuervo C, et al. Novel Borrelia genotypes in bats from the Macaregua Cave, Colombia. Zoonoses Public Health. 2021;68:12–8. 10.1111/zph.12789. [DOI] [PubMed] [Google Scholar]
- 7.Nieto NC, Teglas MB. Relapsing fever group Borrelia in Southern California rodents. J Med Entomol. 2014;51:1029–34. 10.1603/ME14021. [DOI] [PubMed] [Google Scholar]
- 8.Kurtenbach K, Hanincová K, Tsao JI, Margos G, Fish D, Ogden NH. Fundamental processes in the evolutionary ecology of Lyme borreliosis. Nat Rev Microbiol. 2006;4:660–9. 10.1038/nrmicro1475. [DOI] [PubMed] [Google Scholar]
- 9.Lopez J, Hovius JW, Bergstrom S. Pathogenesis of relapsing fever. Curr Issues Mol Biol. 2021;42:519–50. 10.2177/cimb.042.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steere AC, Strle F, Wormser GP, Hu LT, Branda JA, Hovius JW, et al. Lyme borreliosis. Nat Rev Dis Primers. 2016;2:1–19. 10.1038/nrdp.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicolle C, Anderson C. Etude comparative de quelques virus recurrents, pathogenes pour l’homme. Arch Inst Pasteur Tunis. 1927;16:123–206. [Google Scholar]
- 12.Mather TN, Wilson ML, Moore SI, Ribeiro JM, Spielman A. Comparing the relative potential of rodents as reservoirs of the Lyme disease spirochete (Borrelia burgdorferi). Am J Epidemiol. 1989;130:143–50. 10.1093/oxfordjournals.aje.a115306. [DOI] [PubMed] [Google Scholar]
- 13.Gorelova NB, Korenberg EI, Kovalevskii YV, Shcherbakov SV. Small mammals as reservoir hosts for Borrelia in Russia. Zentralbl Bakteriol. 1995;282:315–22. 10.1016/S0934-8840(11)80132-5. [DOI] [PubMed] [Google Scholar]
- 14.Levin M, Levine JF, Apperson CS, Norris DE, Howard PB. Reservoir competence of the rice rat (Rodentia: Cricetidae) for Borrelia burgdorferi. J Med Entomol. 1995;32:138–42. 10.1093/jmedent/32.2.138. [DOI] [PubMed] [Google Scholar]
- 15.Thomas R, Santodomingo AM, Muñoz-Leal S, Silva-De la Fuente MC, Llanos-Soto S, Salas LM, et al. Rodents as potential reservoirs for Borrelia spp. In northern Chile. Rev Bras Parasitol Vet. 2020;29:1–10. 10.1590/S1984-29612020029. [DOI] [PubMed] [Google Scholar]
- 16.Mancilla-Agrono LY, Banguero-Micolta LF, Ossa-López PA, Ramírez-Chaves HE, Castaño-Villa GJ, Rivera-Páez FA. Is Borrelia burgdorferi Sensu Stricto in South America? First molecular evidence of its presence in Colombia. Trop Med Infect Dis. 2022;7:428. 10.3390/tropicalmed7120428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egan SL, Taylor CL, Banks PB, Northover AS, Ahlstrom LA, Ryan UM, et al. The bacterial biome of ticks and their wildlife hosts at the urban–wildland interface. Microb Genom. 2021;7:1–24. 10.1099/mgen.0.000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fedorova N, Kleinjan JE, James D, Hui LT, Peeters H, Lane RS. Remarkable diversity of tick or mammalian-associated Borreliae in the metropolitan San Francisco Bay Area, California. Ticks Tick Borne Dis. 2014;5:951–61. 10.1016/j.ttbdis.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 19.Gil H, Barral M, Escudero R, García-Pérez AL, Anda P. Identification of a new Borrelia species among small mammals in areas of northern Spain where Lyme disease is endemic. Appl Environ Microbiol. 2005;71:1336–45. 10.1128/AEM.71.3.1336-1345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dean D, Rothschild J, Ruettger A, Prasad R, Sachse K. Zoonotic Chlamydiaceae species associated with trachoma, Nepal. Emerg Infect Dis. 2013;19:1948–55. 10.3201/eid1912.130656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parola P, Diatta G, Socolovschi C, Mediannikov O, Tall A, Bassene H, et al. Tick-borne relapsing fever borreliosis, rural Senegal. Emerg Infect Dis. 2011;17:883–5. 10.3201/eid1705.100573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ras N, Lascola B, Postic D, Cutler S, Rodhain F, Baranton G, et al. Phylogenesis of relapsing fever Borrelia spp. Int J Syst Bacteriol. 1996;46:859–65. 10.1099/00207713-46-4-859. [DOI] [PubMed] [Google Scholar]
- 23.Stromdahl E, Williamson P, Kollars T, Evans S, Barry R, Vince M, et al. Evidence of Borrelia lonestari DNA in Amblyomma americanum (Acari: Ixodidae) removed from humans. J Clin Microbiol. 2003;41:5557–62. 10.1128/JCM.41.12.5557-5562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ataliba AC, Resende JS, Yoshinari N, Labruna MB. Isolation and molecular characterization of a Brazilian strain of Borrelia anserina, the agent of fowl spirochaetosis. Res Vet Sci. 2007;83:145–9. 10.1016/j.rvsc.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 25.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 26.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank: update. Nucleic Acids Res. 2004;32:D23–6. 10.1093/nar/gkh045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sievers F, Higgins DG. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018;27:135–45. 10.1002/pro.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letunic I, Bork P. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–6. 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analyzed during the current study.