Abstract
Phosphatidylinositol (PI) 3-kinase is required for G1 to S phase cell cycle progression stimulated by a variety of growth factors and is implicated in the activation of several downstream effectors, including p70S6K. However, the molecular mechanisms by which PI 3-kinase is engaged in activation of the cell cycle machinery are not well understood. Here we report that the expression of a dominant negative (DN) form of either the p110α catalytic or the p85 regulatory subunit of heterodimeric PI 3-kinase strongly inhibited epidermal growth factor (EGF)-induced upregulation of cyclin D1 protein in NIH 3T3(M17) fibroblasts. The PI 3-kinase inhibitors LY294002 and wortmannin completely abrogated increases in both mRNA and protein levels of cyclin D1 and phosphorylation of pRb, inducing G1 arrest in EGF-stimulated cells. By contrast, rapamycin, which potently suppressed p70S6K activity throughout the G1 phase, had little inhibitory effect, if any, on either of these events. PI 3-kinase, but not rapamycin-sensitive pathways, was also indispensable for upregulation of cyclin D1 mRNA and protein by other mitogens in NIH 3T3 (M17) cells and in wild-type NIH 3T3 cells as well. We also found that an enforced expression of wild-type p110 was sufficient to induce cyclin D1 protein expression in growth factor-deprived NIH 3T3(M17) cells. The p110 induction of cyclin D1 in quiescent cells was strongly inhibited by coexpression of either of the PI 3-kinase DN forms, and by LY294002, but was independent of the Ras-MEK-ERK pathway. Unlike mitogen stimulation, the p110 induction of cyclin D1 was sensitive to rapamycin. These results indicate that the catalytic activity of PI 3-kinase is necessary, and could also be sufficient, for upregulation of cyclin D1, with mTOR signaling being differentially required depending upon cellular conditions.
Phosphatidylinositol (PI) 3-kinase is implicated in the receptor-mediated regulation of diverse mammalian cell functions, including insulin-stimulated glucose uptake and glycogen synthesis, exocytosis, neurite outgrowth, prevention of apoptosis, and mitogenesis (for reviews, see references 21, 25, 70, 74). Growth factor stimulation of receptor-protein tyrosine kinases rapidly activates heterodimeric isoforms of PI 3-kinase, which consist of p110 catalytic and p85 regulatory subunits (74). p85 possesses adaptor modules in its structure, among which are two SH2 regions that mediate binding to specific phosphotyrosine residues presented on either cytoplasmic region of the activated growth factor receptors or their associated substrate proteins such as insulin receptor substrate 1 (IRS-1), thereby recruiting p110 to the plasma membrane where the lipid substrates are localized. Binding of p110 via its N-terminal region to p85 in the inter-SH2 region is indispensable for its enzymatic activity (references 30, 31, and 39 and references therein), which generates the lipid second messengers 3-polyphosphoinositides (29, 70, 74, 82). In addition, p110 could directly interact with the GTP-bound active form of Ras protein (62), which interaction further contributes to membrane targeting and activation of p110.
Requirement of PI 3-kinase for mammalian cell cycle progression was first recognized by studies adopting platelet-derived growth factor (PDGF) receptor mutants that lack phosphoacceptor sites required for binding of PI 3-kinase p85 (16, 20), as well as “add back” mutants with selective restoration of these sites (73). Subsequent investigations with more specific tools confirmed these earlier observations and provided compelling evidence that PI 3-kinase is indispensable for G1 to S phase progression in response to a variety of growth factors. They include microinjection studies using inhibitory antibodies raised against p110 (60) and p85 (34) and a p85 SH2 domain peptide that also prevents the activation of p110 (34). The microinjection of these molecules inhibited DNA synthesis in mouse and rat fibroblasts stimulated by either PDGF, epidermal growth factor (EGF), basic fibroblast growth factor (bFGF), insulin-like growth factor I, and serum. Inhibitors for PI 3-kinase, LY294002 and wortmannin, have also been shown to inhibit S phase entry in a variety of cell types (14, 32, 75, 76).
The activation of PI 3-kinase is also sufficient for G1-S progression in growth factor-deprived cells, at least under certain experimental conditions. It was demonstrated for 3T3-L1 cells (24) that constitutive activation of PI 3-kinase by coexpression of the inter-SH2 region of p85 and wild-type p110 resulted in DNA synthesis to an extent that exceeded the effect of insulin, without the activation of extracellular signal-regulated protein kinase (ERK). It was also shown for CHO cells (45) that selective activation of PI 3-kinase to physiologically relevant levels was sufficient to stimulate DNA synthesis. In addition, it was reported recently that the expression of an EGF receptor mutant that caused constitutive activation of PI 3-kinase, but not GTP loading into Ras or persistent ERK activation, resulted in a reduced growth factor requirement and even anchorage-independent cell proliferation in NIH 3T3 fibroblasts (51). Indeed, recent studies provide evidence for the involvement of PI 3-kinase in the process of transformation, in addition to G1 to S progression (12, 35, 61).
PI 3-kinase activates a number of direct and indirect downstream effectors, including Akt1/protein kinase B (5, 10, 22, 23, 40, 44, 65), p70S6K (13, 14, 15, 49, 58, 80), Ca2+-independent and atypical isoforms of protein kinase C (PKC) such as PKCɛ and PKCζ (1, 50, 55, 71), a small GTPase Rac (28, 62, 72), and c-Jun N-terminal kinase (JNK) (42, 43, 54). Among others, p70S6K has been implicated as a prominent mediator of mitogenic action of PI 3-kinase (14, 19, 42, 49, 58, 69, 74). In support of this notion, it has been reported that (i) an add back mutant of PDGF receptor that restored the capacity to activate PI 3-kinase was capable of mediating activation of p70S6K (13) and DNA synthesis (73), (ii) the expression of constitutively active forms of PI 3-kinase induces the activation of p70S6K (38, 80) as well as DNA synthesis (24) in serum-deprived cells, and (iii) the inhibitors for PI 3-kinase potently suppress growth factor-stimulated activation of p70S6K and DNA synthesis (13, 14, 49). p70S6K becomes activated within minutes of growth factor stimulation through phosphorylation at multiple sites by several upstream p70S6K kinases, which include those located downstream of PI 3-kinase (57, 59, 80). In addition to the PI 3-kinase signaling pathway, the activity of the serine-threonine protein kinase mTOR (also called FRAP or RAFT), which is the mammalian target of rapamycin, is absolutely required for the activation of p70S6K (6). Thus, rapamycin blocks p70S6K activation in response to diverse mitogenic stimuli (13, 56, 58, 69). Recent investigations, including studies on truncated forms of p70S6K, provide evidence that mTOR is located in parallel to, rather than being linear downstream of, the mitogen-activated, PI 3-kinase-dependent signaling pathway (18, 19, 58, 59, 79). Very recently, it has been demonstrated that p70S6K activity is required for growth factor-responsive translational upregulation of a subset of mRNAs, termed 5′TOP mRNAs, which have a tract of oligopyrimidine at their transcriptional start site (33). In addition to p70S6K, there exists at least one more signaling molecule that is also regulated by both PI 3-kinase and mTOR: the protein synthesis initiation factor 4E (eIF4E) (4, 8, 26, 46, 77). Thus, in quiescent states eIF4E is bound to and sequestered by an eIF4E repressor, 4E-BP1 (also called PHAS-I). Growth factor stimulation induces rapid phosphorylation and inactivation of 4E-BP1, resulting in liberation of eIF4E and its subsequent incorporation into a fully functional multiprotein complex, which is required for efficient translational initiation of another subset of mRNAs that have highly structured 5′ untranslated regions (7). These findings raise the possibility that the PI 3-kinase signaling pathway and the mTOR pathway contribute to G1 phase progression through translational upregulation of crucial components of the cell cycle machinery. With regard to this point, it is interesting that, as noted previously (63), an NIH 3T3 cell line stably overexpressing eIF4E shows an elevated level of cyclin D1, which is a key player in the G1 phase progression. However, at present, it is not yet known whether mTOR or PI 3-kinase is actually involved in the induction of cyclin D1 by growth factors or other mitogens.
In the present study, we tested the possibility that the PI 3-kinase signaling pathway is linked to the cell cycle machinery through regulation of cyclin D1 expression by examining the effects of the expression of dominant negative (DN) forms of p110 and p85 as well as those of wild-type (WT) p110 and a constitutively active form of p110. We used NIH 3T3(M17) fibroblasts, in which cyclin D1 has been shown to be the only D-type cyclin upregulated by growth factors or serum and to be responsible for the phosphorylation of pRb and passage through the late G1 restriction (R) point (2, 67). We also examined the involvement of mTOR in growth factor-induced cyclin D1 expression. We demonstrate here that PI 3-kinase is absolutely necessary for mitogen induction of cyclin D1 mRNA and protein and is also sufficient, when overexpressed, for the induction of cyclin D1 protein in quiescent cells. We found that this action of PI 3-kinase is not always mediated through mTOR-dependent signaling pathways.
MATERIALS AND METHODS
Cell culture and [3H]thymidine incorporation.
NIH 3T3(M17) cells (11), a generous gift from G. M. Cooper (Harvard Medical School), were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 5% iron-enriched calf serum (Intergen) and 200 μg of geneticin (Sigma)/ml at subconfluent states. Before each experiment confluent cultures were serum deprived for 24 h in DMEM containing 0.1% bovine serum albumin (fraction V; Sigma A-8022). For induction of Ras(N17), dexamethasone (DEX) (5 × 10−7 M; Sigma) was included in the medium at this step, as previously described (11). NIH 3T3 cells were obtained from two sources, the Japanese Cancer Research Resources Bank (Tokyo, Japan) and the Riken Cell Bank (Tsukuba, Ibaraki, Japan), and were cultured in the absence of geneticin. [3H]thymidine incorporation into DNA was measured 18 h after the addition of mitogens as described previously (67), with [3H]thymidine (2 μCi/ml; DuPont-New England Nuclear) being pulse labeled during the last 1 h. Data shown are representative of at least three experiments performed in triplicate and are expressed as means ± standard errors (SE). EGF and bFGF were purchased from R&D Systems, and phorbol-12,13-dibutyrate (PDBu) was from Sigma. They were used at maximally mitogenic concentrations (30 ng/ml, 10 ng/ml, and 10−7 M, respectively). LY294002, rapamycin, and PD98059 were obtained from Carbiochem, and wortmannin was purchased from Wako Chemicals (Osaka, Japan). Unless otherwise mentioned, LY294002 (25 μM), wortmannin (300 nM), and rapamycin (30 nM) were introduced to the cells 15 min before stimulation with mitogens. In addition, wortmannin, which is labile in living cells (36), was supplemented every 3 h. As previously reported for parental NIH 3T3 cells (84), LY294002 and wortmannin at these concentrations did not cause apoptosis in mitogen-stimulated NIH 3T3(M17) cells.
Plasmids and transient transfection.
Constructions of pMIKNeop110 and pMIKNeop110EcoS, which are the expression plasmids for wild type and a DN form of the bovine PI 3-kinase p110α subunit (WTp110 and DNp110), respectively, were described elsewhere (66). pMIKNeop85ΔRV-Pvu, an expression plasmid for a DN form of the human PI 3-kinase p85 subunit (DNp85), was created by deletion of the sequence between the EcoRV and PvuII sites within the coding sequence amino acids 338 to 572, which resulted in removal of the N-terminal SH2 domain and the binding site for p110. pMIKNeoBD110 and pMIKNeoBDKD are the expression plasmids for a Myc epitope-tagged constitutively active form of p110 (BD110) which has a sequence corresponding to the p110-binding domain (BD) of p85 connected through a glycine bridge to the N terminus of full-length p110α and the kinase dead (KD) mutant form of BD110 (BDKD), respectively (31, 41). pactEF-DN-MAPK, the expression vector for a DN form of Xenopus mitogen-activated protein kinase (MAPK), was kindly donated by K. Okazaki (Kurume University Institute of Life Science, Kurume, Japan). pSV-βgal, an expression plasmid for β-galactosidase, was purchased from Promega. pCAGGS-βgal, another expression plasmid for β-galactosidase, was kindly donated by I. Saitoh (Institute of Medical Sciences, University of Tokyo, Tokyo, Japan). The plasmids were purified by two cycles of CsCl density gradient centrifugation. Cotransfections were performed in 35 mm-diameter dishes by the calcium phosphate precipitation procedure as described before (48, 67). After recovery from the transfection procedure by incubating the cells in growth medium overnight, the cells were serum deprived in the presence or absence of DEX or inhibitors as described in the legends to figures.
Immunofluorescence.
Cells were washed and fixed with 3.7% formalin in Ca2+- and Mg2+-free Dulbecco’s phosphate-buffered saline (PBS) at room temperature for 10 min. After two rinses with PBS, formalin was quenched with 50 mM glycine in PBS, followed by treatment with 0.25% Triton X-100–1% fetal calf serum in PBS for 1 h at room temperature to permeabilize membranes and to reduce nonspecific binding of antibodies. The cells were then treated with a mixture of a mouse monoclonal anti-cyclin D1 antibody (Santa Cruz) and a rabbit polyclonal anti-β-galactosidase antibody (Cappel) in PBS containing 0.25% Triton X-100–1% fetal calf serum for 1 h at room temperature, followed by detection with a mixture of fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G (IgG) (Zymed) and rhodamine-conjugated goat anti-rabbit IgG antibodies (Cappel) in PBS. This protocol gave the same results as sequential immunodetection of cyclin D1 and β-galactosidase. For detection of Myc epitope, a rabbit polyclonal antibody raised against Myc epitope (EQKLISEEDL) (Molecular and Biological Laboratory, Nagoya, Japan) was employed. For an unknown reason(s), the anti-β-galactosidase antibody which we adopted was cross-reactive for nuclear components, allowing us to identify nuclei without staining for DNA. By taking advantage of this fact, the anti-β-galactosidase antibody was routinely incorporated into immunofluorescence protocols. More than 200 cells positive for either β-galactosidase or Myc epitope were inspected per transfection, and the percentage of cyclin D1-positive cells in the transfected population was determined by using a fluorescence microscope (Olympus IX-70 inverted system microscope).
Immunoblot analysis.
Immunoblot analysis was performed as described previously, after separation of equal amounts of cellular protein by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, based upon protein contents determined in parallel cultures (67, 68). A rabbit polyclonal antibody for cyclin A and a mouse monoclonal anti-cyclin D1 antibody were purchased from UBI and Santa Cruz, respectively. A rabbit polyclonal antibody for cyclin D1 (Medical and Biological Laboratories) was also employed and the same results were obtained. The state of pRb phosphorylation was evaluated by electrophoretic mobility shift assay (85) after immunoblotting by using a mouse monoclonal anti-pRb antibody (PharMingen 14001A antibody). The activation state of p70S6K was determined by electrophoretic mobility shift assay after immunoblotting (13, 47, 49) by using a rabbit polyclonal antibody (Santa Cruz). The activation states of p44ERK1 and p42ERK2 were evaluated by electrophoretic mobility shift assay on immunoblots as described previously (48, 67).
Northern blot analysis.
The mRNA level of cyclin D1 was analyzed as described previously (67, 86). After stripping the membranes of radioactive probes, they were rehybridized with a 32P-labeled glyceraldehyde 3-phosphate dehydrogenase (GAPDH) cDNA probe, and the results were used as an internal control.
RESULTS
EGF-stimulated induction of cyclin D1 is dependent upon PI 3-kinase in NIH 3T3(M17) cells.
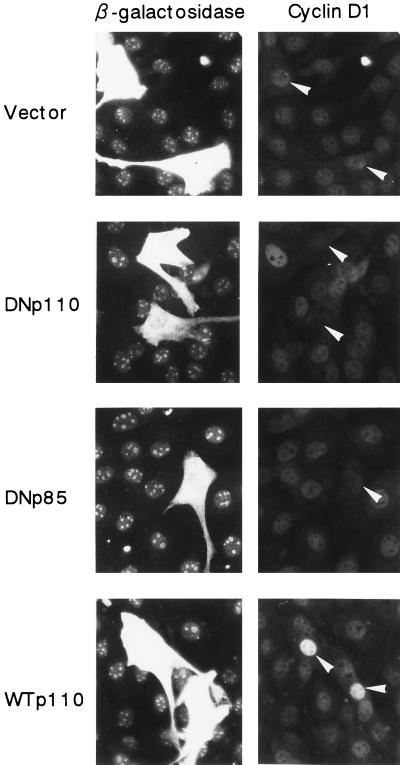
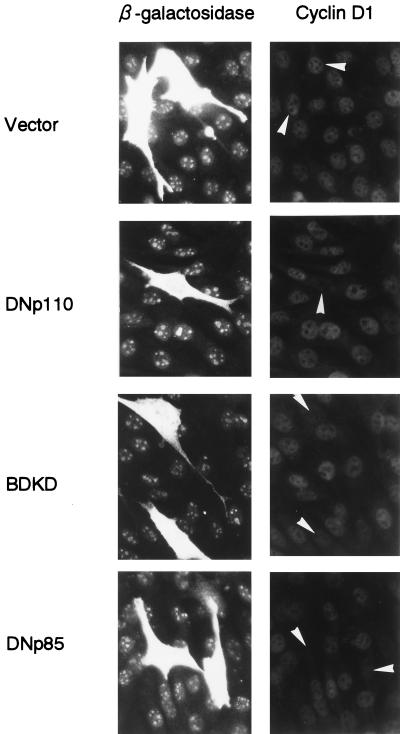
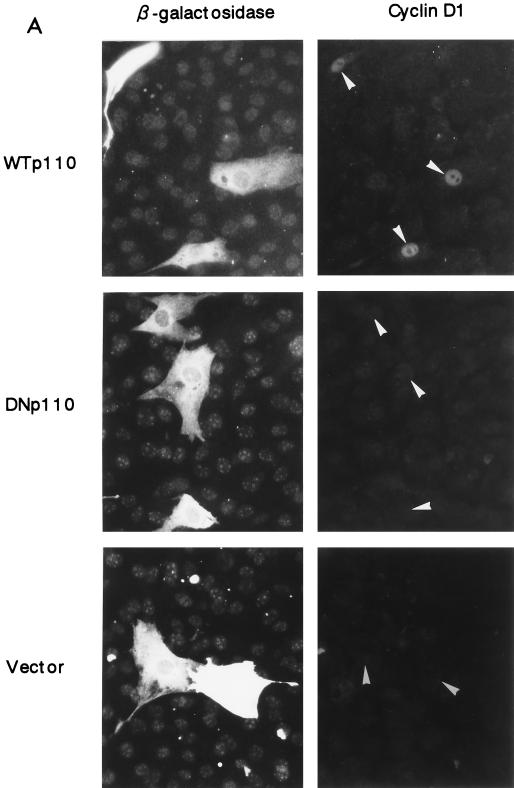
We first examined whether PI 3-kinase is required for EGF-induced expression of cyclin D1 protein. NIH 3T3(M17) cells were transfected with the expression plasmid of either DNp110, DNp85, or WTp110 together with that of β-galactosidase, which was employed as a transfection marker, and then made quiescent. After stimulation with EGF for 9 h, cyclin D1 and β-galactosidase were detected by double immunofluorescence. The expression of DNp110 and DNp85 each potently suppressed the induction of cyclin D1 protein in response to EGF (Fig. 1): the percentages of cyclin D1-positive cells in the transfected population were 0.9% ± 0.5% and 1.2% ± 0.6%, respectively, compared to 41.1% ± 1.4% in the vector control (means ± SE of three experiments). We also found that the expression of DN-MAPK(ERK) similarly inhibited cyclin D1 protein expression in EGF-stimulated NIH 3T3(M17) cells. The results indicate that PI 3-kinase and ERK are both required for upregulation of cyclin D1 protein in response to EGF. In sharp contrast, the expression of WTp110 increased the percentage of cyclin D1-positive cells (66.7% ± 2.2%) and markedly augmented the intensity of cyclin D1 staining in individual cells (Fig. 1).
FIG. 1.
Expression of DNp110 or DNp85 nearly completely abrogates EGF-stimulated induction of cyclin D1 protein expression, whereas expression of WTp110 potentiates it. Cells were transiently cotransfected with 1.5 μg each of pSV-βgal and an expression plasmid of a DN form of p110 (DNp110) or p85 (DNp85), wild-type p110 (WTp110), or an empty vector and made quiescent. Two days after transfection, the cells were stimulated with EGF (30 ng/ml) for 9 h, which corresponds to the late G1 R point. The expression of β-galactosidase (left) and cyclin D1 (right) was detected by double immunofluorescence as described in Materials and Methods. Identical fields in pairs are shown from a representative experiment. Arrowheads indicate positions of nuclei in transfected cells.
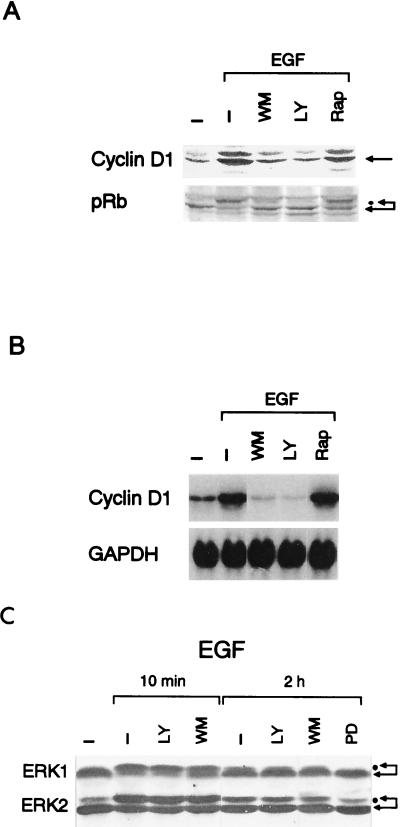
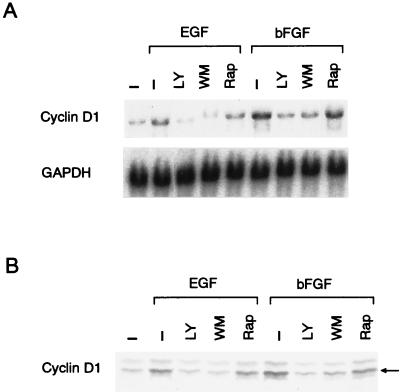
Shown in Fig. 2A are the effects of the PI 3-kinase inhibitors on the level of cyclin D1 and the extent of the phosphorylation of pRb. Both wortmannin (300 nM) and LY294002 (25 μM) nearly completely abrogated EGF-induced cyclin D1 upregulation and pRb phosphorylation. The PI 3-kinase inhibitors also abolished the EGF-stimulated increase in the cyclin D1 mRNA level (Fig. 2B). These findings agree with the results obtained with the DN forms of PI 3-kinase (Fig. 1) and indicate that PI 3-kinase is critically required for upregulation of cyclin D1 mRNA and protein in response to EGF. As reported for other cell types (14, 27, 46), the PI 3-kinase inhibitors were essentially ineffective in inhibiting EGF-stimulated activation of ERK1 and ERK2 in NIH 3T3(M17) cells (Fig. 2C), indicating that PI 3-kinase is linked to the expression of cyclin D1 through a pathway that is distinct from the ERK signaling cascade.
FIG. 2.
(A and B) The PI 3-kinase inhibitors wortmannin (WM, 300 nM) and LY294002 (LY, 25 μM) but not rapamycin (Rap, 30 nM) nearly totally abolish EGF-stimulated upregulation of cyclin D1 mRNA and protein levels and the phosphorylation of pRb. Western (A) and Northern (B) blot analyses were performed 9 h after the addition of EGF with equal amounts of total cellular protein and RNA, respectively. (C) The PI 3-kinase inhibitors are without effect on ERK activities, whereas the MEK inhibitor PD98059 (PD, 30 μM) abolishes the sustained phase of the EGF-stimulated ERK activation. The concentrations of LY and WM are the same as for panels A and B.
mTOR-p70S6K pathway is dispensable for EGF-induced upregulation of cyclin D1 or entry into S phase.
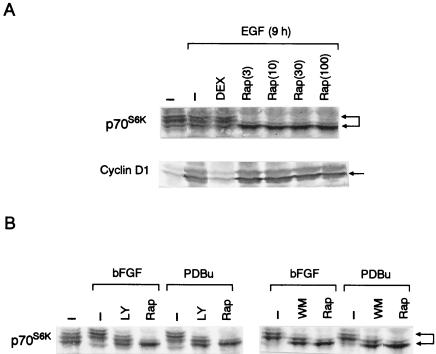
We also studied whether p70S6K is involved in EGF-stimulated, PI 3-kinase-mediated upregulation of cyclin D1, since p70S6K is widely recognized as an effector of mitogenic action of PI 3-kinase. We found that rapamycin, which is known to suppress p70S6K activity through inhibition of mTOR, slightly inhibited the EGF-stimulated increase in either cyclin D1 mRNA or protein and only marginally inhibited pRb phosphorylation (Fig. 2A and B). As shown in Fig. 3A, quiescent NIH 3T3(M17) cells showed a certain level of basal p70S6K activation as evidenced by a multiple ladder pattern on Western blots. Stimulation with EGF induced an additional, transient activation of p70S6K at 10 min (data not shown), which then returned to the basal level by 1 h and remained at this state for up to 9 h of observations. Rapamycin induced a rapid and sustained inhibition of p70S6K in EGF-stimulated cells to a level that was even much lower than the basal unstimulated level (Fig. 3A). Rapamycin at 3 nM was sufficient to cause the maximal inhibition of p70S6K, which persisted for 9 h after the addition of EGF, whereas it failed to inhibit EGF-stimulated upregulation of cyclin D1 protein even at 100 nM (Fig. 3A). As reported previously (9, 13, 14, 49), LY294002 (25 μM) was also capable of suppressing p70S6K in EGF-stimulated cells. However, the above results indicate that the inhibition of the mTOR-p70S6K pathway is not the principal mechanism for the suppression of cyclin D1 induction by the PI 3-kinase inhibitors.
FIG. 3.
(A) Rapamycin inhibits p70S6K but not the upregulation of cyclin D1 protein in EGF-stimulated cells. The cells were incubated for 9 h in the presence or absence of EGF and rapamycin at the concentrations (nanomolar) indicated in parentheses. Where indicated, DEX was introduced to cells 24 h before the addition of EGF to induce a DN Ras, Ras(N17). p70S6K and cyclin D1 were detected by Western blot analysis of identical samples. (B) Rapamycin, and the PI 3-kinase inhibitors to a lesser extent, tonically suppresses p70S6K in cells stimulated by other mitogens. The cells were growth stimulated or left unstimulated for 9 h in the presence or absence of rapamycin (30 nM), LY294002 (25 μM), or wortmannin (300 nM). Abbreviations are as defined in the legend to Fig. 2.
We also observed that the DEX induction of a DN Ras, Ras(N17), did not detectably inhibit p70S6K in EGF-stimulated cells (Fig. 3A), which is consistent with a previous report (47), yet it potently suppressed the EGF-induced increase in the level of cyclin D1 protein (Fig. 3A) (2). These results provide additional evidence that p70S6K activity and cyclin D1 upregulation could be dissociated.
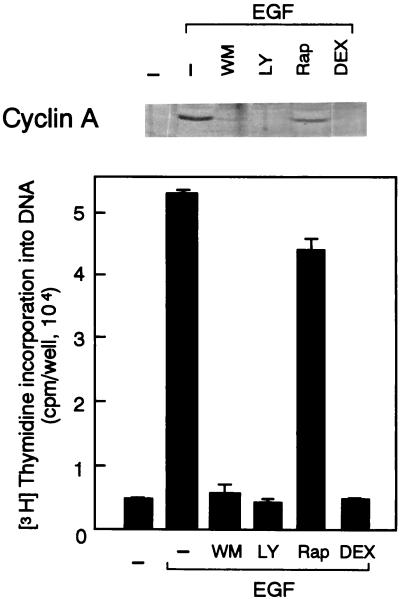
As shown in Fig. 4, either LY294002 or wortmannin, as well as the expression of Ras(N17), totally abolished the effect of EGF on DNA synthesis. By contrast, rapamycin was only marginally inhibitory. Consistent with these results, the PI 3-kinase inhibitors and the induced expression of Ras(N17), but not rapamycin, prevented the EGF induction of cyclin A, which is a hallmark of S phase entry (Fig. 4). These observations indicate that the activation of p70S6K is not critically required for either cyclin D1 expression or S phase entry in EGF-stimulated cells.
FIG. 4.
The PI 3-kinase inhibitors and the DEX induction of Ras(N17) but not rapamycin abolish DNA synthesis (lower) and the induction of cyclin A protein (upper) in response to EGF. The inhibitors and DEX were introduced to the cells as described in the legend to Fig. 3, and the cells were incubated with EGF for 18 h. Abbreviations are as defined in the legend to Fig. 2.
PI 3-kinase, but not the mTOR-p70S6K pathway, is required for upregulation of cyclin D1 by a variety of mitogens in NIH 3T3 cells.
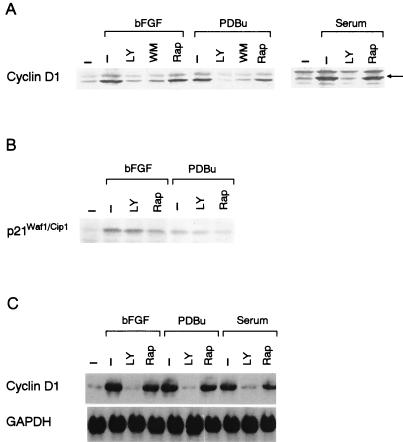
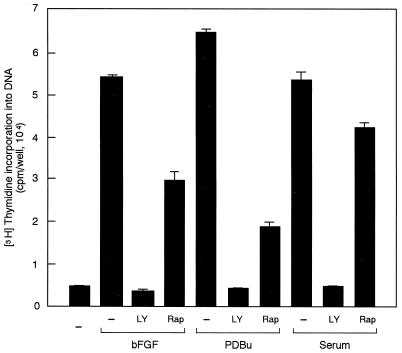
Mitogens other than EGF, including bFGF, PDBu and serum, also induced increases in the mRNA and protein levels of cyclin D1 in NIH 3T3(M17) cells (Fig. 5A and C). LY294002 and wortmannin again completely abolished the effects of these mitogens on cyclin D1 upregulation (Fig. 5A and C). By contrast, rapamycin was slightly inhibitory, if at all, despite potent and persistent inhibition of p70S6K (Fig. 3B). These mitogens, particularly bFGF and PDBu, also induced increases in the level of p21Waf1/Cip1. However, different from cyclin D1, the upregulation of p21Waf1/Cip1 was resistant to inhibition by LY294002 (Fig. 5B), demonstrating selectivity of the action of the inhibitor. As expected, LY294002, which completely prevented cyclin D1 upregulation (Fig. 5A and C), induced G1 arrest in cells stimulated by either of these mitogens (Fig. 6). In addition, rapamycin was capable of inhibiting DNA synthesis in response to bFGF and PDBu by approximately 50 and 80%, respectively (Fig. 6), implying that rapamycin has a site of action other than that for the inhibition of the cyclin D1 induction.
FIG. 5.
(A) LY294002 (LY, 25 μM) and wortmannin (WM, 300 nM), but not rapamycin (Rap, 30 nM) abrogate the effects of bFGF (10 ng/ml), PDBu (10−7 M) or serum (5% [vol/vol]) on upregulation of cyclin D1 protein. (B) bFGF- or PDBu-induced expression of p21Waf1/Cip1 is resistant to LY294002. (C) Upregulation of the cyclin D1 mRNA by bFGF, PDBu, or serum is abolished by LY294002 but not rapamycin. The cells were stimulated by either of the growth stimuli for 9 h.
FIG. 6.
LY294002 (LY, 25 μM) but not rapamycin (Rap, 30 nM) induces complete G1 arrest in cells stimulated by bFGF, PDBu, or serum.
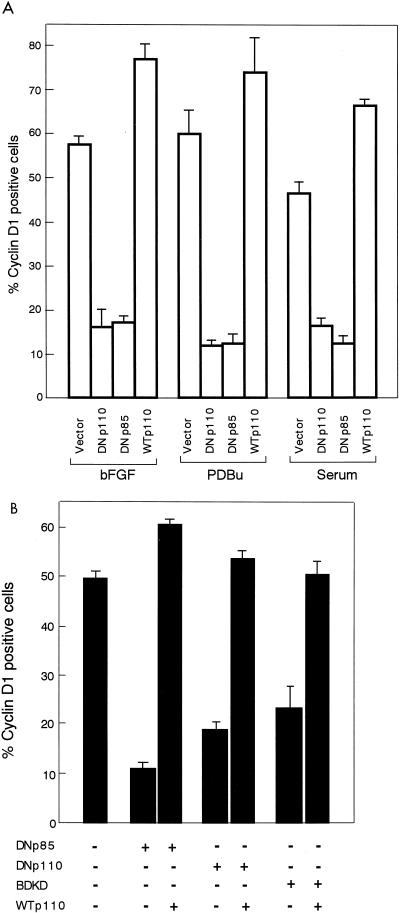
Shown in Fig. 7A are the effects of the expression of either DNp110, DNp85, or WTp110 on cyclin D1 protein induction in response to bFGF, PDBu, or serum. The expression of either of the DN forms strongly inhibited mitogen-induced expression of cyclin D1 protein. In addition, the expression of BDKD, which is a full-length, KD mutant of p110α, was also capable of inhibiting growth factor induction of cyclin D1 (Fig. 7B and 9). Conversely, the expression of WTp110 potentiated cyclin D1 upregulation by either of the mitogenic stimuli, in terms of both the percentage of cyclin D1-positive cells (Fig. 7A) and the intensity of cyclin D1 expression in individual cells (data not shown), just like the case with EGF (Fig. 1). We also found that the inhibition of cyclin D1 upregulation by the expression of either of the DN forms was counteracted by coexpression of WTp110 (Fig. 7B) or a constitutively active form of p110, BD110 (data not shown). Thus, it is concluded that PI 3-kinase participates in cyclin D1 induction by multiple growth stimuli, for the most part through a rapamycin-insensitive mechanism.
FIG. 7.
(A) Effects of the expression of DNp110, DNp85, or WTp110 on the induction of cyclin D1 protein by bFGF, PDBu, or serum. The experiments were performed as described in the legend to Fig. 1 except that the serum-deprived, transfected cells were stimulated with bFGF, PDBu, or serum for 9 h. Percentages of cyclin D1-positive cells in the transfected population were determined under a fluorescence microscope as described in Materials and Methods. (B) Coexpression of WTp110 relieves the inhibitory effect of DNp110, DNp85, or BDKD on bFGF-induction of cyclin D1. The cells were cotransfected with 1.0 μg of the expression plasmid for either of the DN forms or 1.7 μg of either pMIKNeop110 or an empty vector, together with 0.3 μg of pCAGGS-βgal, made quiescent, and then stimulated with bFGF for 9 h.
FIG. 9.
Expression of either of the DN forms of PI 3-kinase potently suppresses bFGF induction of cyclin D1 protein in wild-type NIH 3T3 cells. Cells were cotransfected with 1.5 μg each of pSV-βgal and either an empty vector or an expression plasmid for DNp110, BDKD, or DNp85, made quiescent, and then stimulated with bFGF for 9 h. The percentages of cyclin D1-positive cells in transfected population were 69.8 ± 5.1, 32.7 ± 1.4, 25.7 ± 2.2 and 20.3 ± 3.8 for vector control, DNp110, BDKD and DNp85-expressing cells, respectively (means ± SE of three determinations).
NIH 3T3(M17) cells could have a certain level of constitutive Ras(N17) expression in the absence of DEX induction, which might have altered mitogenic signalings compared to those of wild-type cells and thus have resulted in an exaggerated dependency of cyclin D1 upregulation upon PI 3-kinase. We therefore examined whether PI 3-kinase was also required for growth factor induction of cyclin D1 in wild-type NIH 3T3 fibroblasts. As shown in Fig. 8, the PI 3-kinase inhibitors, but not rapamycin, nearly completely prevented increases in the levels of cyclin D1 mRNA and protein in EGF- or bFGF-stimulated cells. In addition, expression of any of the DN forms of PI 3-kinase, including DNp110, DNp85, and BDKD, potently inhibited growth factor induction of cyclin D1 protein in wild-type NIH 3T3 cells (Fig. 9). These results indicate that PI 3-kinase is required for growth factor-stimulated cyclin D1 upregulation in wild-type NIH 3T3 fibroblasts as well.
FIG. 8.
The PI 3-kinase inhibitors but not rapamycin nearly completely inhibit growth factor-stimulated upregulation of cyclin D1 mRNA (A) and protein (B) in wild-type NIH 3T3 fibroblasts.
Expression of the WTp110 catalytic subunit of PI 3-kinase induces cyclin D1 protein in quiescent NIH 3T3(M17) fibroblasts.
We next examined the effect of overexpression of WTp110 on cyclin D1 protein induction. After transfection with the expression plasmids for WTp110 and β-galactosidase, the cells were serum deprived for 2 days. As shown in Fig. 10A and C, an enforced expression of WTp110 strongly induced nuclear cyclin D1 protein expression in more than 50% of the transfected cells, in the absence of growth factor stimulation. The expression of BD110 also caused cyclin D1 protein expression in quiescent cells (Fig. 11A). By contrast, the expression of either DNp110 or BDKD was totally ineffective in cyclin D1 induction, as was the vector control (Fig. 10A and 11A). The expression of β-galactosidase protein was not detectably affected by coexpression of either WTp110, BD110, or DN forms of p110 (Fig. 10A and 11A), indicating that they did not affect the general capacity of the cells to synthesize proteins.
FIG. 10.
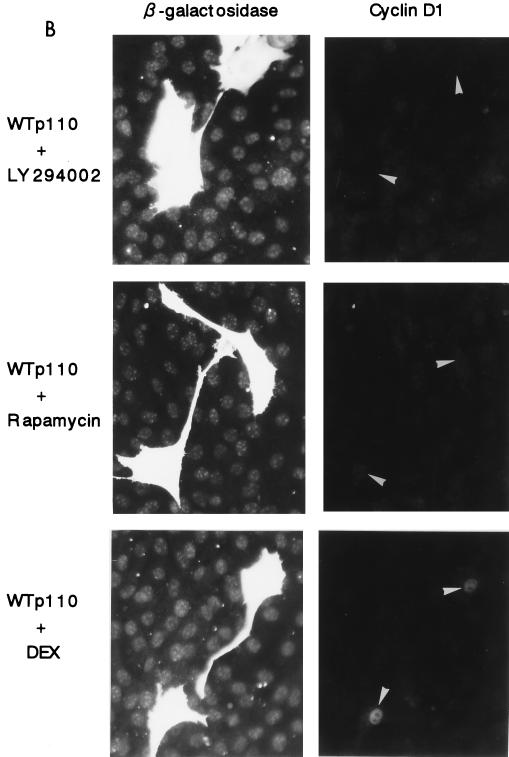
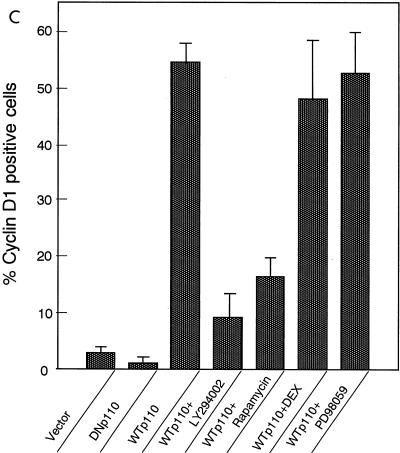
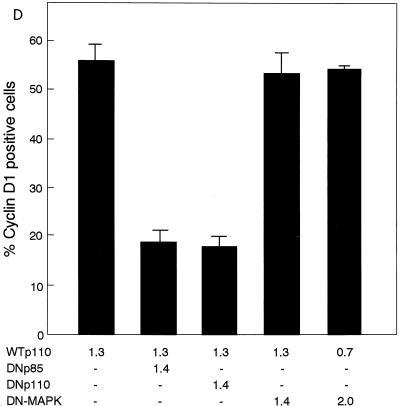
(A) Enforced expression of WTp110 but not DNp110 or an empty vector leads to the induction of cyclin D1 protein expression in serum-deprived NIH 3T3(M17) fibroblasts. Transfections were performed as described in the legend to Fig. 1, and the cells were serum deprived for 2 days. Arrowheads indicate the positions of nuclei in transfected cells. (B) The WTp110-dependent expression of cyclin D1 protein is sensitive to LY294002 (25 μM) and rapamycin (30 nM) but not DEX-induced expression of Ras(N17). (C) Percentages of cyclin D1-positive cells in the transfected population. The concentration of PD98059 was 30 μM. (D) Coexpression of either DNp110 or DNp85 but not DN-MAPK(ERK) suppresses WTp110 induction of cyclin D1 protein in quiescent cells. The cells were cotransfected with the indicated amounts (in micrograms) of expression plasmids together with 0.3 μg of pCAGGS-βgal and appropriate amounts of an empty vector so that the total amount of DNA per transfection was adjusted to 3.0 μg. The reduction in the amount of the expression plasmid for WTp110 from 1.3 to 0.7 μg by itself did not affect the percentage of cyclin D1-positive cells.
FIG. 11.
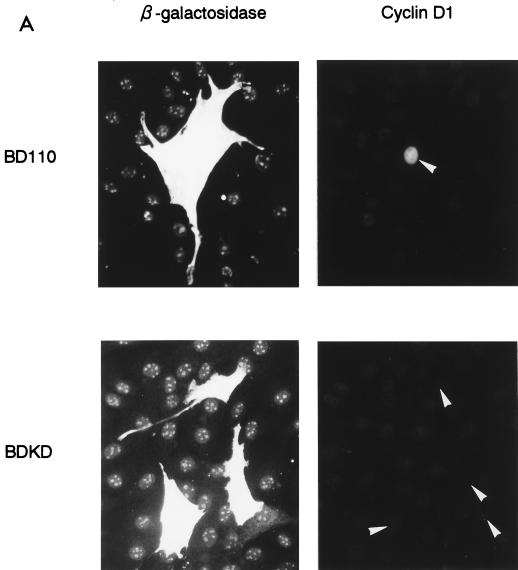
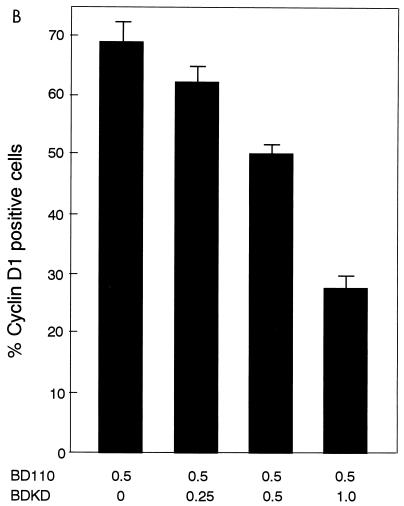
(A) Expression of BD110, a constitutively active form of PI 3-kinase p110, but not its KD mutant, BDKD, induces cyclin D1 protein expression in quiescent NIH 3T3(M17) cells. Cells were cotransfected with either of the expression plasmids (1.5 μg) and pSV-βgal (1.5 μg) and made quiescent. (B) Coexpression of BDKD dose-dependently inhibits BD110 induction of cyclin D1 protein. Cells were transfected with the indicated amounts (in micrograms) of the expression plasmids together with an appropriate amount of an empty vector so that the total amount of DNA per transfection was adjusted to 3.0 μg. The cells were made quiescent and probed for cyclin D1 and Myc epitope by double immunofluorescence.
When the cells were treated with the PI 3-kinase inhibitor LY294002 (25 μM) after the transfection with WTp110, the induction of cyclin D1 protein was strongly suppressed to less than one-fifth of nontreated cells (Fig. 10B and C). We also found that, different from mitogen-stimulated cells, WTp110 induction of cyclin D1 protein in serum-deprived cells was strongly inhibited by the addition of rapamycin after transfection (Fig. 10B and C). Neither of the inhibitors reduced the expression of β-galactosidase (Fig. 10B), indicating that their effects were selective for cyclin D1 expression.
We also tested whether coexpression of a DN form of PI 3-kinase could inhibit the ability of WTp110 to induce cyclin D1 protein in quiescent cells. As shown in Fig. 10D, the expression of either DNp85 or DNp110 potently suppressed WTp110 induction of cyclin D1 protein. Similarly, coexpression of BDKD dose-dependently inhibited BD110 induction of cyclin D1 (Fig. 11B).
We studied whether the Ras-MEK-ERK pathway was involved in the induction of cyclin D1 protein by the overexpression of WTp110. The DEX-induced expression of Ras(N17), which completely abolished the effects of EGF on both cyclin D1 upregulation and DNA synthesis (Fig. 3A and 4), failed to prevent WTp110 induction of cyclin D1 (Fig. 10B and C). Addition of the MEK inhibitor PD98059 (30 μM) was also ineffective in preventing WTp110 induction of cyclin D1 protein (Fig. 10C). This dose of PD98059 completely suppressed the sustained phase of EGF-stimulated ERK activation (Fig. 2C) and also abrogated the effect of EGF on DNA synthesis (data not shown). We also examined the effect of the expression of DN-MAPK(ERK) on WTp110 induction of cyclin D1. As shown in Fig. 10D, coexpression of DN-MAPK failed to inhibit WTp110 induction of cyclin D1, which sharply contrasts to its potent inhibitory effects observed with mitogen-stimulated cells on both the induction of cyclin D1 protein (see above) and the activation of cyclin D1 promoter-luciferase reporter activity (data not shown). These results indicate that an enforced expression of WTp110 induces cyclin D1 protein in quiescent NIH 3T3(M17) cells in a manner that is dependent upon PI 3-kinase activity and sensitive to rapamycin but independent of the Ras-MEK-ERK pathway.
DISCUSSION
During the past several years PI 3-kinase has been increasingly recognized as one of the important signaling molecules required for G1-S cell cycle progression. However, the precise molecular mechanism of PI 3-kinase-mediated mitogenic signaling has remained elusive thus far. The present study was aimed at exploring the role of PI 3-kinase in cyclin D1 induction, a critical step required for the activation of G1 cyclin-dependent kinases, which drive G1 phase progression.
The results of the present study demonstrate that the induction of cyclin D1 protein by growth factors and other mitogens is potently inhibited by the expression of DN forms of heterodimeric PI 3-kinase in NIH 3T3(M17) fibroblasts (Fig. 1 and 7). WTp110, by contrast, potentiated the mitogen induction of cyclin D1 (Fig. 1 and 7A) and counteracted the inhibition by DNp110, BDKD, or DNp85 (Fig. 7B). The inhibitors for PI 3-kinase, LY294002 and wortmannin, completely abolish mitogen-induced increases in the levels of both cyclin D1 protein and mRNA (Fig. 2A and B and 5). Similarly, the PI 3-kinase inhibitors and expression of the DN forms inhibited mitogen induction of cyclin D1 in wild-type NIH 3T3 fibroblasts as well (Fig. 8 and 9). We also demonstrate that an enforced expression of the WTp110 catalytic subunit of PI 3-kinase causes the induction of cyclin D1 protein in serum-deprived NIH 3T3(M17) fibroblasts (Fig. 10A and C). This effect of p110 is prevented by LY294002 (Figs. 10B and C) and by coexpression of either DNp110 or DNp85 (Fig. 10D). Similarly, BD110 induction of cyclin D1 protein was counteracted by coexpression of BDKD (Fig. 11B). These composite results provide evidence that the catalytic activity of PI 3-kinase is required for the induction of cyclin D1 by mitogens and is also sufficient, at least under certain conditions, for the expression of cyclin D1 protein in growth factor-deprived mammalian cells. As reported previously (2, 67), cyclin D1 is the only D-type cyclin that is upregulated by mitogens in NIH 3T3(M17) cells. Hence, PI 3-kinase is essential for the activation of D-type cyclin-dependent kinases and consequent phosphorylation of the substrate pRb, which is an absolute requirement for pRb-positive cells to pass through the late G1 R point and enter the S phase (78).
It is widely recognized that p70S6K is located downstream of PI 3-kinase and mTOR, mediating a major part of the mitogenic action of PI 3-kinase, which includes growth factor-induced translational upregulation of a subset of mRNAs. In addition, accumulating evidence in recent years indicates that mitogen-induced phosphorylation and inactivation of 4E-BP1 and resultant activation of eIF4E-dependent translation is also mediated by both PI 3-kinase and mTOR. However, the present results demonstrating that the inhibitors for PI 3-kinase, but not rapamycin, completely abrogate DNA synthesis stimulated by growth factors or serum (Fig. 4 and 6) strongly suggest that PI 3-kinase-dependent signaling other than, or in addition to, the activation of p70S6K or eIF4E plays an essential role in mediating the mitogenic effect of these growth stimuli. Our findings that rapamycin has only a limited effect, if any, on mitogen-induced cyclin D1 upregulation (Fig. 2A and B, 3A, 5A and C, and 8) despite potent suppression on both p70S6K (Fig. 3) and the phosphorylation of 4E-BP1 (data not shown) argue that neither p70S6K nor eIF4E serves as the principal mediator of the PI 3-kinase signaling that leads to cyclin D1 protein expression. In agreement with our observations, it has been reported that the overexpression of a mutant eIF4E, which was previously shown to act in a DN manner to inhibit proliferation of NIH 3T3 cells (53), does not inhibit serum-stimulated increases in the level of cyclin D1 mRNA or protein (64). The fact that the PI 3-kinase inhibitors abolish mitogen induction of cyclin D1 at the level of mRNA (Fig. 2B, 5C, and 8A) implies that the role for PI 3-kinase in the induced upregulation of cyclin D1 is not confined to translational initiation of the cyclin D1 transcript, provided that it is involved at all. In contrast to mitogen induction of cyclin D1, however, the induction of cyclin D1 protein by overexpression of WTp110 in quiescent cells is inhibited by rapamycin (Fig. 10B and C). Although the molecular basis for the discrepancy between the rapamycin sensitivities of mitogen-stimulated cells and p110-overexpressing quiescent cells is not known at present, it might be that the activation of multiple signaling pathways by mitogens would spare the requirement for the mTOR-dependent mechanisms, including p70S6K and eIF4E. In support of this notion would be an analogous observation (83) that rapamycin blocks the mitogenic effect of bombesin but not that of a combination of bombesin and insulin in Swiss 3T3 cells.
It has been demonstrated that the Ras-MEK-ERK pathway plays a pivotal role in cyclin D1 gene expression (3, 52). Indeed, we observed in the present study that the DEX induction of Ras(N17) prior to the addition of EGF prevents upregulation of cyclin D1 protein (Fig. 3A) and that the expression of a DN form of MAPK(ERK) inhibits mitogen induction of cyclin D1 protein as effectively as DNp110 and DNp85. In addition, we observed that the expression of a constitutively active form of MEK1 induced cyclin D1 protein expression in quiescent NIH 3T3(M17) cells as detected by immunofluorescence. On the other hand, we demonstrate in the present study that the induction of cyclin D1 protein by overexpression of wild-type p110 is resistant to the induced expression of Ras(N17), coexpression of DN-MAPK, or the addition of the MEK inhibitor PD98059 (Fig. 10B, C, and D). We also observed that the PI 3-kinase inhibitors abrogate mitogen-induced upregulation of cyclin D1 (Fig. 2A and B) without inhibiting ERK activation (Fig. 2C). In addition, it was reported previously that the expression in mammalian cell systems of constitutively active forms of PI 3-kinase p110 do not cause the activation of ERKs (24, 38, 41, 54). These observations indicate that the PI 3-kinase-dependent signaling leading to the induction of cyclin D1 does not involve the Ras-MEK-ERK signaling cascade. Thus, the PI 3-kinase pathway and the MEK-ERK pathway represent two independent signalings, both of which are required for cyclin D1 protein induction by such mitogens as employed in the present study, although either one of them, when overexpressed, is sufficient by itself. As previously reported for other cell types (3, 52, 81), we found that transfection of the expression plasmid of a constitutively active form of either MEK1 or Rac markedly stimulates cyclin D1 promoter-reporter activity in serum-deprived NIH 3T3(M17) cells. By contrast, the expression of WTp110 fails to do so under the same experimental condition (66a). These results suggest that the mechanism for PI 3-kinase-mediated cyclin D1 mRNA upregulation is distinct from that mediated by either the MEK-ERK pathway or Rac.
Recent reports unveil the existence of an ever-growing number of PI 3-kinase effector molecules (37). Further studies are required to identify the PI 3-kinase effector that is responsible for cyclin D1 upregulation and the exact mechanism by which this effector pathway upregulates cyclin D1 mRNA and protein.
ACKNOWLEDGMENTS
We thank R. Suzuki, M. Kato and E. Kishimoto for excellent secretarial assistance.
This work was supported by a grant-in-aid from the Japanese Ministry of Science, Education and Art.
REFERENCES
- 1.Akimoto K, Takahashi R, Moriya S, Nishioka N, Takayanagi J, Kimura K, Fukui Y, Osada S, Mizuno K, Hirai S, Kazlauslaas A, Ohno S. EGF or PDGF receptors activate atypical PKC lambda through phosphatidylinositol 3-kinase. EMBO J. 1996;15:788–798. [PMC free article] [PubMed] [Google Scholar]
- 2.Aktas H, Cai H, Cooper G M. Ras links growth factor signaling to the cell cycle machinery via regulation of cyclin D1 and the cdk inhibitor p27KIP1. Mol Cell Biol. 1997;17:3850–3857. doi: 10.1128/mcb.17.7.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell R G. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 4.Beretta L, Gingras A-C, Svitkin Y V, Hall M N, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translations. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- 5.Bos J L. A target for phosphoinositide 3-kinase: Akt/PKB. Trends Biochem Sci. 1995;20:441–442. doi: 10.1016/s0968-0004(00)89097-0. [DOI] [PubMed] [Google Scholar]
- 6.Brown E J, Beal P A, Keith C T, Chen J, Bum Shin T, Schreiber S L. Control of p70 S6 kinase by kinase activity of FRAP in vivo. Nature. 1995;377:441–446. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- 7.Brown E J, Schreiber S L. A signaling pathway to translational control. Cell. 1996;86:517–520. doi: 10.1016/s0092-8674(00)80125-7. [DOI] [PubMed] [Google Scholar]
- 8.Brunn G J, Hudson C C, Sekulic A, Willims J M, Hosoi H, Houghton P J, Lawrence J C, Jr, Abraham R T. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 9.Brunn G J, Williams J, Sabers C, Wiederrecht G, Lawrence J C, Jr, Abraham Robert T. Direct inhibition of the signaling functions of the mammalian target of rapamycin by the phosphoinositide 3-kinase inhibitors, wortmannin and LY294002. EMBO J. 1996;15:5256–5267. [PMC free article] [PubMed] [Google Scholar]
- 10.Burgering B M T, Coffer P J. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature. 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- 11.Cai H, Szeberényi J, Cooper G M. Effect of a dominant inhibitory Ha-ras mutation on mitogenic signal transduction in NIH 3T3 cells. Mol Cell Biol. 1990;10:5314–5323. doi: 10.1128/mcb.10.10.5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang H W, Aoki M, Fruman D, Anger K R, Bellacosa A, Tsichlis P N, Cantley L C, Roberts T M, Vogt P K. Transformation of chicken cells by the gene encoding the catalytic subunit of PI 3-kinase. Science. 1997;276:1848–1850. doi: 10.1126/science.276.5320.1848. [DOI] [PubMed] [Google Scholar]
- 13.Chung J, Grammer T C, Lemon K P, Kazlauskas A, Blenis J. PDGF- and insulin-dependent pp70S6K activation mediated by phosphatidylinositol-3-OH kinase. Nature. 1994;370:71–75. doi: 10.1038/370071a0. [DOI] [PubMed] [Google Scholar]
- 14.Cheatham B, Vlahos C J, Cheatham L, Wang L, Blenis J, Kahn C R. Phosphatidylinositol 3-kinase activation is required for insulin stimulation of pp70 S6 kinase, DNA synthesis, and glucose transporter translocation. Mol Cell Biol. 1994;14:4902–4911. doi: 10.1128/mcb.14.7.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou M M, Blenis J. The 70 kDa S6 kinase: regulation of a kinase with multiple roles in mitogenic signaling. Curr Opin Cell Biol. 1995;7:806–814. doi: 10.1016/0955-0674(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 16.Coughlin S R, Escobedo J A, Williams L T. Role of phosphatidylinositol kinase in PDGF receptor signal transduction. Science. 1989;243:1191–1193. doi: 10.1126/science.2466336. [DOI] [PubMed] [Google Scholar]
- 17.Cross D A E, Alessi D R, Cohen P, Andjelkovich M, Hemmings B A. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 18.Dennis P B, Pullen N, Kozma S C, Thomas G. The principal rapamycin-sensitive p70S6K phosphorylation sites, T-229 and T-389, are differentially regulated by rapamycin-insensitive kinase kinases. Mol Cell Biol. 1996;16:6242–6251. doi: 10.1128/mcb.16.11.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Downward J. Lipid-regulated kinase: some common themes at last. Science. 1998;279:673–674. doi: 10.1126/science.279.5351.673. [DOI] [PubMed] [Google Scholar]
- 20.Fantl W J, Escobedo J A, Martin G A, Turck C W, del Rosario M, McCormik F, Williams L T. Distinct phosphotyrosines on a growth factor receptor bind to specific molecules that mediate different signaling pathways. Cell. 1992;69:413–423. doi: 10.1016/0092-8674(92)90444-h. [DOI] [PubMed] [Google Scholar]
- 21.Franke T F, Kaplan D R, Cantley L C. PI3K: downstream AKTion blocks apoptosis. Cell. 1997;88:435–437. doi: 10.1016/s0092-8674(00)81883-8. [DOI] [PubMed] [Google Scholar]
- 22.Franke T F, Kaplan D R, Cantley L C, Toker A. Direct regulation of the AKT proto-oncogene product by phosphatidylinositol-3, 4-bisphosphate. Science. 1997;275:665–668. doi: 10.1126/science.275.5300.665. [DOI] [PubMed] [Google Scholar]
- 23.Franke T F, Yang S-I, Chan T O, Datta K, Kazlauskas A, Morrisin D K, Kaplan D R, Tsichlis P N. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 24.Frevert E U, Kahn B B. Differential effects of constitutively active phosphatidylinositol 3-kinase on glucose transport, glycogen synthase activity, and DNA synthesis in 3T3-L1 adipocytes. Mol Cell Biol. 1997;17:190–198. doi: 10.1128/mcb.17.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukui Y, Ihara S, Nagata S. Downstream of phosphatidylinositol-3 kinase, a multifunctional signaling molecule, and its regulation in cell responses. J Biochem (Tokyo) 1998;124:1–7. doi: 10.1093/oxfordjournals.jbchem.a022067. [DOI] [PubMed] [Google Scholar]
- 26.Gingras A-C, Kennedy S G, O’Leary M A, Sonenberg N, Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt (PKB) signaling pathway. Genes Dev. 1998;12:502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hara K, Yonezawa K, Sakaue H, Ando A, Kotani K, Kitamara T, Kitamura Y, Ueda H, Stephens L, Jackson T, Waterfield M D, Kasuga M. Phosphatidylinositol 3-kinase activity is required for insulin-stimulated glucose transport but not for RAS activation in CHO cells. Proc Natl Acad Sci USA. 1994;91:7415–7419. doi: 10.1073/pnas.91.16.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hawkins P T, Eguinoa A, Qui R-G, Stokoe D, Cooke F T, Walters R, Wennstrom S, Claesson-Welsh L, Evans T, Symons M, Stephens L. PDGF stimulates an increase in GTP-Rac via activation of phosphoinositide 3-kinase. Curr Biol. 1995;4:393–403. doi: 10.1016/s0960-9822(95)00080-7. [DOI] [PubMed] [Google Scholar]
- 29.Hawkins P T, Jackson T R, Stephens L R. Platelet-derived growth factor stimulates synthesis of PtdIns (3, 4, 5) P3 by activating a PtdIns (4, 5) P2 3-OH kinase. Nature. 1992;358:157–159. doi: 10.1038/358157a0. [DOI] [PubMed] [Google Scholar]
- 30.Hu P, Schlessinger J. Direct association of p110 beta phosphatidylinositol 3-kinase with p85 is mediated by an N-terminal fragment of p110 beta. Mol Cell Biol. 1994;14:2577–2583. doi: 10.1128/mcb.14.4.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu Q, Klippel A, Muslin A J, Fantl W J, Williams L T. Ras-dependent induction of cellular responses by constitutively active phosphatidylinositol 3-kinase. Science. 1995;268:100–102. doi: 10.1126/science.7701328. [DOI] [PubMed] [Google Scholar]
- 32.Hu S-W, Shi X-Y, Lin R Z, Hoffman B B. α1 Adrenergic receptors activate phosphatidylinositol 3-kinase in human vascular smooth muscle cells. Role in mitogenesis. J Biol Chem. 1996;271:8977–8982. doi: 10.1074/jbc.271.15.8977. [DOI] [PubMed] [Google Scholar]
- 33.Jefferies H B J, Fumagalli S, Dennis P B, Reinhard C, Pearson R B, Thomas G. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70S6K. EMBO J. 1997;16:3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jhun B H, Rose D W, Seely B L, Rameh L, Cantley L, Saltiel A R, Olefsky J M. Microinjection of the SH2 domain of the 85-kilodalton subunit of phosphatidylinositol 3-kinase inhibits insulin-induced DNA synthesis and c-fos expression. Mol Cell Biol. 1994;14:7466–7475. doi: 10.1128/mcb.14.11.7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jimenez C, Jones J R, Rodriguez-Viciana P, Gonzalez-Garcia A, Leonardo E, Wennstrom S, von Kobbe C, Toran J L, Borlado L R-, Calvo V, Copin S G, Alber J P, Gaspar M L, Diez E, Marcos M A R, Downward J, Martinez C, Mérida A I, Carreva A C. Identification and characterization of a new oncogene derived from the regulatory subunit of phosphoinositide 3-kinase. EMBO J. 1998;17:743–753. doi: 10.1093/emboj/17.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimura K, Hattori S, Kabuyama Y, Shizawa Y, Onodera K, Fukui Y. Neurite outgrowth of PC12 cells is suppressed by wortmannin, a specific inhibitor of phosphatidylinositol 3-kinases. J Biol Chem. 1994;269:18961–18967. [PubMed] [Google Scholar]
- 37.Klarlund J K, Guilherme A, Holik J J, Virbasius J V, Chawla A, Czech M P. Signaling by phosphoinositide-3, 4, 5-trisphosphate through proteins containing pleckstrin and Sec7 homology domains. Science. 1997;275:1927–1930. doi: 10.1126/science.275.5308.1927. [DOI] [PubMed] [Google Scholar]
- 38.Klippel A, Reinhard C, Kavanaugh W M, Apell G, Escobedo M-A, Williams L T. Membrane localization of phosphatidylinositol 3-kinase is sufficient to activate multiple signal-transducing kinase pathways. Mol Cell Biol. 1996;16:4117–4127. doi: 10.1128/mcb.16.8.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klippel A, Escobedo J A, Hu Q, Williams L T. A region of the 85-kilodalton (kDa) subunit of phosphatidylinositol 3-kinase binds the 110-kDa catalytic subunit in vivo. Mol Cell Biol. 1993;13:5560–5566. doi: 10.1128/mcb.13.9.5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klippel A, Kavanaugh W M, Pot D, Williams L T. A specific product of phosphatidylinositol 3-kinase directly activates the protein kinase Akt through its pleckstrin homology domain. Mol Cell Biol. 1997;17:338–344. doi: 10.1128/mcb.17.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobayashi M, Nagata S, Kita Y, Nakatsu N, Ihara S, Kaibuchi K, Kuroda S, Ui M, Iba H, Konishi H, Kikkawa U, Saitoh I, Fukui Y. Expression of a constitutively active phosphatidylinositol 3-kinase induces process formation in rat PC12 cells. Use of CrelloxP recombination system. J Biol Chem. 1997;272:16089–16092. doi: 10.1074/jbc.272.26.16089. [DOI] [PubMed] [Google Scholar]
- 42.Lane H A, Fernandez A, Lamb N J C, Thomas G. p70S6K function is essential for G1 progression. Nature. 1993;363:170–172. doi: 10.1038/363170a0. [DOI] [PubMed] [Google Scholar]
- 43.Logan S K, Falasca M, Hu P, Schlessinger J. Phosphatidylinositol 3-kinase mediates epidermal growth factor-induced activation of the c-Jun N-terminal kinase signaling pathway. Mol Cell Biol. 1997;17:5784–5790. doi: 10.1128/mcb.17.10.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marte B M, Downward J. PKB/Akt: connecting phosphoinositide 3-kinase to cell survival and beyond. Trends Biochem Sci. 1997;22:355–358. doi: 10.1016/s0968-0004(97)01097-9. [DOI] [PubMed] [Google Scholar]
- 45.McILRoy J, Chen D, Wjasow C, Michaeli T, Backer J M. Specific activation of p85-p110 phosphatidylinositol 3′-kinase stimulates DNA synthesis by ras- and p70 S6 kinase-dependent pathways. Mol Cell Biol. 1997;17:248–255. doi: 10.1128/mcb.17.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mendez R, Myers M G, Jr, White M F, Rhoads R E. Stimulation of protein synthesis, eukaryotic translation initiation factor 4E phosphorylation, and PHAS-I phosphorylation by insulin requires insulin receptor substrate 1 and phosphatidylinositol 3-kinase. Mol Cell Biol. 1996;16:2857–2864. doi: 10.1128/mcb.16.6.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ming X-F, Burgering B M T, Wennstrom S, Claesson-Welsh L, Heldin C-H, Bos J L, Kozma S C, Thomas G. Activation of p70/p85 S6 kinase by a pathway independent of p21ras. Nature. 1994;371:426–429. doi: 10.1038/371426a0. [DOI] [PubMed] [Google Scholar]
- 48.Mitsui H, Takuwa N, Kurokawa K, Exton J H, Takuwa Y. Dependence of activated Gα12-induced G1 to S phase cell cycle progression on both Ras/MAPK and Ras/Rac1/JNK cascades in NIH3T3 fibroblasts. J Biol Chem. 1997;272:4904–4910. doi: 10.1074/jbc.272.8.4904. [DOI] [PubMed] [Google Scholar]
- 49.Monfar M, Lemon K P, Grammer T C, Cheatham L, Chung J, Vlahos C J, Blenis J. Activation of pp70/85 S6 kinases in interleukin-2-responsive lymphoid cells is mediated by phosphatidylinositol 3-kinase and inhibited by cyclic AMP. Mol Cell Biol. 1995;15:326–337. doi: 10.1128/mcb.15.1.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moriya S, Kazlauskas A, Akimoto K, Hirai S, Mizuno K, Takenawa T, Fukui Y, Watanabe Y, Ozaki S, Ohno S. Platelet-derived growth factor activates protein kinase Cɛ through redundant and independent signaling pathways involving phospholipase Cγ or phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA. 1996;93:151–155. doi: 10.1073/pnas.93.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moscatello D K, Holgado-Madruga M, Emlets D R, Montgomery R B, Wong A J. Constitutive activation of phosphatidylinositol 3-kinase by a naturally occurring mutant epidermal growth factor receptor. J Biol Chem. 1998;273:200–206. doi: 10.1074/jbc.273.1.200. [DOI] [PubMed] [Google Scholar]
- 52.Lavoie J N, L’Allmain G, Brunet A, Müller R, Pouysségur J. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J Biol Chem. 1996;271:20608–20616. doi: 10.1074/jbc.271.34.20608. [DOI] [PubMed] [Google Scholar]
- 53.Lazaris-Karatzas A, Montine K S, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 54.Logan S K, Falasca M, Hu P, Schlessinger J. Phosphatidylinositol 3-kinase mediates epidermal growth factor-induced activation of the c-Jun N-terminal kinase signaling pathway. Mol Cell Biol. 1997;17:5784–5790. doi: 10.1128/mcb.17.10.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakanishi H, Brewer A, Exton J H. Activation of the zeta isozyme of protein kinase C by phosphatidylinositol 3, 4, 5-trisphosphate. J Biol Chem. 1993;268:13–16. [PubMed] [Google Scholar]
- 56.Price D J, Grove J R, Calvo V, Avruch J, Bierer B E. Rapamycin-induced inhibition of the 70-kilodalton S6 protein kinase. Science. 1992;257:973–977. doi: 10.1126/science.1380182. [DOI] [PubMed] [Google Scholar]
- 57.Price D J, Gunsalus J R, Avruch J. Insulin activates a 70-kDa S6 kinase through serine/threonine-specific phosphorylation of the enzyme polypeptide. Proc Natl Acad Sci USA. 1990;87:7944–7948. doi: 10.1073/pnas.87.20.7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Proud C G. p70 S6 kinase: an enigma with variations. Trends Biochem Sci. 1996;21:181–185. [PubMed] [Google Scholar]
- 59.Pullen N, Dennis P B, Andjelkovic M, Dufner A, Kozma S C, Hemmings B A, Thomas G. Phosphorylation and activation of p70S6K by PDK1. Science. 1998;279:707–710. doi: 10.1126/science.279.5351.707. [DOI] [PubMed] [Google Scholar]
- 60.Roche S, Koegl M, Courtneidge S A. The phosphatidylinositol 3-kinase α is required for DNA synthesis induced by some, but not all, growth factors. Proc Natl Acad Sci USA. 1994;91:9185–9189. doi: 10.1073/pnas.91.19.9185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodriguez-Viciana P, Warne P H, Khwaja A, Marte B M, Pappin D, Das P, Waterfield M D, Ridley A, Downward J. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- 62.Rodriguez-Viciana P, Warne P H, Dhand R, Vanhaesebroek B, Gout I, Fry M J, Waterfield M D, Downward J. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 63.Rosenwald I B, Lazaris-Karatzas A, Sonenberg N, Schmidts E V. Elevated levels of cyclin D1 protein in response to increased expression of eukaryotic initiation factor 4E. Mol Cell Biol. 1993;13:7358–7363. doi: 10.1128/mcb.13.12.7358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rosenwald I B, Kaspar R, Rousseau D, Gehrke L, Leboulch P, Chen J-J, Shmidt E V, Sonenberg N, London I M. Eukaryotic translation initiation factor 4E regulates expression of cyclin D1 at transcriptional and post-transcriptional levels. J Biol Chem. 1995;270:21176–21180. doi: 10.1074/jbc.270.36.21176. [DOI] [PubMed] [Google Scholar]
- 65.Stephens L, Anderson K, Stokooe D, Erdiument-Bromage H, Painter G F, Holmes A B, Gaffney P R J, Reese C B, McCormick F, Tempst P, Coadweel J, Hawkins P T. Protein kinase B kinases that mediate phosphatidylinositol 3, 4, 5-trisphosphate-dependent activation of protein kinase B. Science. 1998;279:710–714. doi: 10.1126/science.279.5351.710. [DOI] [PubMed] [Google Scholar]
- 66.Takayanagi J, Kimura K, Nishioka N, Akimoto K, Moriya S, Ohno S, Fukui Y. Dominant negative effect of the truncated p110 subunit of phosphatidylinositol-3 kinase. Biochem Mol Biol Int. 1996;39:721–728. doi: 10.1080/15216549600201801. [DOI] [PubMed] [Google Scholar]
- 66a.Takuwa, N., and Y. Kakuwa. Unpublished observations.
- 67.Takuwa N, Takuwa Y. Ras activity late in G1 phase required for p27kip1 downregulation, passage through the restriction point, and entry into S phase in growth factor-stimulated NIH 3T3 fibroblasts. Mol Cell Biol. 1997;17:5348–5358. doi: 10.1128/mcb.17.9.5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takuwa N, Zhou W, Kumada M, Takuwa Y. Ca2+-dependent stimulation of retinoblastoma gene product phosphorylation and p34cdc2 kinase activation in serum-stimulated human fibroblasts. J Biol Chem. 1993;268:138–145. [PubMed] [Google Scholar]
- 69.Thomas G, Hall M N. TOR signalling and control of cell growth. Curr Opin Cell Biol. 1997;9:782–787. doi: 10.1016/s0955-0674(97)80078-6. [DOI] [PubMed] [Google Scholar]
- 70.Toker A, Cantley L C. Signalling through the lipid products of phosphoinositide-3-OH kinase. Nature. 1997;387:673–676. doi: 10.1038/42648. [DOI] [PubMed] [Google Scholar]
- 71.Toker A, Meyer M, Reddy K K, Falck J R, Aneja R, Aneja S, Parra A, Burns D J, Ballas L M, Cantley L C. Activation of protein kinase C family members by the novel polyphosphoinositide PtdIns-3, 4-P2 and PtdIns-3, 4, 5-P3. J Biol Chem. 1994;269:32358–32367. [PubMed] [Google Scholar]
- 72.Tolias K F, Cantley L C, Carpenter L C. Rho family GTPases bind to phosphoinositide kinases. J Biol Chem. 1995;270:17656–17659. doi: 10.1074/jbc.270.30.17656. [DOI] [PubMed] [Google Scholar]
- 73.Valius M, Kazlauskas A. Phospholipase C-γ1 and phosphatidylinositol 3 kinase are the downstream mediators of the PDGF receptor’s mitogenic signal. Cell. 1993;73:321–334. doi: 10.1016/0092-8674(93)90232-f. [DOI] [PubMed] [Google Scholar]
- 74.Vanhaesebroeck B, Leevers S J, Panayotou G, Waterfield M D. Phosphoinositide 3-kinases: a conserved family of signal transducers. Trends Biochem Sci. 1997;22:267–272. doi: 10.1016/s0968-0004(97)01061-x. [DOI] [PubMed] [Google Scholar]
- 75.Vemuri G S, Rittenhouse S E. Wortmannin inhibits serum-induced activation of phosphoinositide 3-kinase and proliferation of CHRI-288 cells. Biochem Biophys Res Commun. 1994;202:1619–1623. doi: 10.1006/bbrc.1994.2118. [DOI] [PubMed] [Google Scholar]
- 76.Vlahos C J, Matter W F, Hui K Y, Brown R F. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1- benzopyran-4-one (LY294002) J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- 77.von Manteuffel S R, Dennis P B, Pullen N, Gingras A-C, Sonenberg N, Thomas G. The insulin-induced signalling pathway leading to S6 and initiation factor 4E binding protein 1 phosphorylation bifurcates at a rapamycin-sensitive point immediately upstream of p70s6k. Mol Cell Biol. 1997;17:5426–5436. doi: 10.1128/mcb.17.9.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 79.Weng Q P, Andrabi K, Kozlowski M T, Grove J R, Avruch J. Multiple independent inputs are required for activation of the p70 S6 kinase. Mol Cell Biol. 1995;15:2333–2340. doi: 10.1128/mcb.15.5.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weng Q-P, Andrabi K, Klippel A, Kazlowski M T, Williams L T, Avruch J. Phosphatidyl inositol 3-kinase signals activation of p70 S6 kinase in situ through site-specific p70 phosphorylation. Proc Natl Acad Sci USA. 1995;92:5744–5748. doi: 10.1073/pnas.92.12.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Westwick J K, Lambert Q T, Clark G J, Symons M, VanAelst L, Pestell R G, Der C J. Rac regulation of transformation, gene expression, and actin organization by multiple, PAK-independent pathways. Mol Cell Biol. 1997;17:1324–1325. doi: 10.1128/mcb.17.3.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Whitman M, Downes C P, Keeler M, Keller T, Cantley L. Type I phosphatidylinositol kinase makes a novel inositol phospholipid, phosphatidylinositol-3-phosphate. Nature. 1988;332:644–646. doi: 10.1038/332644a0. [DOI] [PubMed] [Google Scholar]
- 83.Withers D J, Seufferlein T, Mann D, Garcia B, Jones N, Rozeugurt E. Rapamycin dissociates p70S6K activation from DNA synthesis stimulated by bombesin and insulin in Swiss 3T3 cells. J Biol Chem. 1997;272:2509–2514. doi: 10.1074/jbc.272.4.2509. [DOI] [PubMed] [Google Scholar]
- 84.Yao R, Cooper G M. Growth factor-dependent survival of rodent fibroblasts requires phosphatidylinositol 3-kinase but is independent of pp70S6K activity. Oncogene. 1996;13:343–351. [PubMed] [Google Scholar]
- 85.Zhou W, Takuwa N, Kumada M, Takuwa Y. Protein kinase C-mediated bidirectional regulation of DNA synthesis, RB protein phosphorylation, and cyclin-dependent kinases in human vascular endothelial cells. J Biol Chem. 1993;268:23041–23048. [PubMed] [Google Scholar]
- 86.Zhou W, Takuwa N, Kumada M, Takuwa Y. E2F1, B-myb and selective members of cyclin/CDK subunits are targets for protein kinase C-mediated bimodal growth regulation in vascular endothelial cells. Biochem Biophys Res Commun. 1994;199:191–198. doi: 10.1006/bbrc.1994.1213. [DOI] [PubMed] [Google Scholar]