Abstract
Hematopoietic progenitor kinase 1 (HPK1), a mammalian Ste20-related protein kinase, is an upstream activator of c-Jun N-terminal kinase (JNK). In order to further characterize the HPK1-mediated JNK signaling cascade, we searched for HPK1-interacting proteins that could regulate HPK1. We found that HPK1 interacted with Crk and CrkL adaptor proteins in vitro and in vivo and that the proline-rich motifs within HPK1 were involved in the differential interaction of HPK1 with the Crk proteins and Grb2. Crk and CrkL not only activated HPK1 but also synergized with HPK1 in the activation of JNK. The HPK1 mutant (HPK1-PR), which encodes the proline-rich region alone, blocked JNK activation by Crk and CrkL. Dominant-negative mutants of HPK1 downstream effectors, including MEKK1, TAK1, and SEK1, also inhibited Crk-induced JNK activation. These results suggest that the Crk proteins serve as upstream regulators of HPK1. We further observed that the HPK1 mutant HPK1-KD(M46), which encodes the kinase domain with a point mutation at lysine-46, and HPK1-PR blocked interleukin-2 (IL-2) induction in Jurkat T cells, suggesting that HPK1 signaling plays a critical role in IL-2 induction. Interestingly, HPK1 phosphorylated Crk and CrkL, mainly on serine and threonine residues in vitro. Taken together, our findings demonstrate the functional interaction of HPK1 with Crk and CrkL, reveal the downstream pathways of Crk- and CrkL-induced JNK activation, and highlight a potential role of HPK1 in T-cell activation.
The c-Jun N-terminal kinase (JNK)/stress-activated protein kinase (SAPK) signaling cascade is critical for cells to transmit extracellular stimuli into the nucleus, thereby leading to appropriate cellular responses such as proliferation, differentiation, and apoptosis (16, 28, 62). JNK is activated by numerous extracellular stimuli, including mitogens, epidermal and transforming growth factors (EGF and TGF) (29, 60), inflammatory cytokines (tumor necrosis factor alpha and interleukin-1 [IL-1]) (30, 47), and environmental stresses (osmotic shock, UV and gamma irradiation) (6, 9, 14). However, most of the signaling pathways that link extracellular stimuli to JNK have not been fully characterized. Identification of the upstream regulators of JNK and delineation of the link between extracellular stimuli and JNK via these upstream regulators are necessary to define the entire JNK signaling cascade.
Previous works have identified a number of upstream activators of JNK. MKK4, also known as SEK1/JNKK1, was the first kinase identified as an immediate upstream activator of JNK (10, 20, 40), and recently another immediate upstream activator of JNK was cloned and designated as MKK7/JNKK2 (57, 63, 64). MKK4 is a direct target of MEKK1, a member of the MAP kinase kinase kinases (MAPKKKs) (20, 40). Other members of the MAPKKK family with the ability to activate JNK include TGF-β-activated kinase 1 (TAK1), tumor progression locus-2 (Tpl-2), and mixed lineage kinase-3 (MLK-3) (39, 56, 60). Furthermore, several Ste20-related protein kinases which activate JNK through MEKK1 or other MAPKKKs have been identified as MAPKKKKs, including germinal center kinase (GCK), hematopoietic progenitor kinase 1 (HPK1), HPK1/GCK-like kinase (HGK), Nck interacting kinase (NIK), GCK-like kinase (GLK), and the kinase homologous to SPS1/ST20 (KHS) (11, 15, 36, 50, 58, 65). Novel upstream activators of the JNK signaling cascade are still emerging. Some of the upstream activators not only participate in the JNK signaling cascade but may also mediate other MAPK signaling cascades such as the ERK and p38-MAPK cascades.
HPK1, a mammalian Ste20-related protein kinase, was recently cloned (15, 17). HPK1 is predominantly expressed in hematopoietic organs and cells, suggesting that it plays an important role in regulating the JNK signaling cascade in blood cells. It has been demonstrated that transient expression of HPK1 activates JNK via the signaling pathway MEKK1→SEK1→JNK but does not stimulate the Erk or p38 signaling pathways (15, 17). Data from our laboratory also indicate that TAK1, like MEKK1, is a downstream effector for HPK1-induced JNK activation (60). HPK1 is a component in a scaffold complex formed by JNK interacting protein-1 (JIP-1) with other components, including MLK3, MKK7, and JNK (61). Unlike Ste20 and PAK, HPK1 does not interact with the small GTPases Rac1 and Cdc42. Thus, the extracellular stimuli and upstream regulators of HPK1 are still unknown. To further characterize the HPK1-mediated JNK signaling pathway, we focused on identifying HPK1-interacting proteins. HPK1, a serine-threonine protein kinase, contains a Ste20-like kinase domain in the N terminus, four proline-rich motifs in the middle region, and a distal C-terminal regulatory domain like those of NIK, GLK, and GCK (50). It is known that proline-rich motifs interact with Src-homology 3 (SH3) domains (7, 35, 66). Thus, the four proline-rich motifs of HPK1 are potential binding sites for SH3 domain-containing proteins. In comparing the proline-rich motifs of HPK1 with several SH3 domain-binding consensus motifs recognized by known SH3 domain-containing proteins (48), we found that some of the HPK1 proline-rich motifs match the Crk and Grb2 N-SH3-binding consensus motifs. Therefore, SH2/SH3 adapter proteins, such as Crk, CrkL, Grb2, and Nck, are potential HPK1-interacting proteins.
SH2/SH3 adapter proteins are known to mediate the formation of multiprotein complexes that allow signals to transmit to downstream effectors that may regulate various cellular responses (7, 35, 65). The Crk proteins, originally identified as cellular homologues of the v-Crk oncogene product, are SH2 and SH3 domain-containing adapter proteins (13a, 26, 27). SH2 domains recognize a phosphorylated tyrosine residue within a certain sequence context, while SH3 domains recognize certain proline-rich motifs with a specific helical structure (7, 35, 65). Two forms of the c-Crk protein, Crk-I and Crk-II, result from alternative RNA splicing. Crk-I consists of one SH2 domain and one SH3 domain, and the longer Crk-II consists of one SH2 domain and two tandem SH3 domains (25, 37). CrkL, cloned from chronic myelogenous leukemia cells, has 60% overall homology to Crk-II (55). The biological roles of the Crk proteins are unclear. Previous findings indicate that the overexpression of v-Crk or Crk-I induces the morphologic transformation of fibroblasts but that Crk-II does not (25, 37). Transient expression of v-Crk induces JNK activation in 293T cells through C3G, a guanine nucleotide exchange protein for Rap1 (53). CrkL has also been shown to activate the Ras and JNK signaling pathways in cells (44). To further define the Crk- and CrkL-mediated signaling pathways, we investigated whether Crk and CrkL transmit upstream signals to HPK1. Here we report that HPK1 bound the SH2/SH3 adaptor proteins Crk and CrkL, which acted as upstream regulators to couple signals to the HPK1-mediated JNK signaling cascade.
MATERIALS AND METHODS
Plasmids.
pCIneo-FLAG-tagged vectors of wild-type HPK1 and various HPK1 mutants, including a kinase-dead mutant (HPK1-M46) made by substituting lysine 46 with methionine, the HPK1 kinase domain alone (HPK1-KD; amino acids 1 to 291), and the HPK1 carboxyl domain alone (HPK1-CD; amino acids 292 to 833) were described previously as pCI-HPK1, pCI-HPK1-M46, pCI-HPK1-KD, and pCI-HPK1-CD, respectively (15). pCR3.1-HA-tagged vectors of HPK1 mutants, including the HPK1 proline-rich region (HPK1-PR; amino acids 288 to 482), the HPK1 distal regulatory region (HPK1-DR; amino acids 484 to 833), and the kinase domain of HPK1-M46 [HPK1-KD(M46); amino acids 1 to 291], were generated by amplifying the indicated regions by PCR. The PCR products were then cloned into pCR3.1 vector (Invitrogen) by the TA cloning method. Hemagglutin (HA)-tagged JNK was described previously (11). pUna3-FL-MEKK-KR, pEF-TAK1-K63W, and pEBG-SEK1-AL were the dominant-negative mutant constructs as described previously (15, 17, 60). pEBB-Crk-II and pEBB-Crk-II-R38K (51) were generously provided by B. J. Mayer (The Children’s Hospital, Harvard Medical School, Boston, Mass.). pSRαMSVtkNeo retrovirus vector containing CrkL cDNA (44) was kindly provided by C. L. Sawyers (University of California, Los Angeles, Los Angeles, Calif.). GST-Crk and GST-CrkL were described previously (13, 18). GST-Grb2, GST-Nck, and GST-p85 SH3 domain (50) were kindly provided by E. Y. Skolnik (New York University Medical Center, New York, N.Y.). GST-c-Jun (1-79) and GST-Rac1 were described previously (15, 17).
Cell culture and transfections.
293T and COS-1 cells were gifts from M. C.-T. Hu (Amgen, Inc., Thousand Oaks, Calif.) and L.-Y. Yu-Lee (Baylor College of Medicine, Houston, TX), respectively. Both 293T and COS-1 cells were grown in Dulbecco modified Eagle’s medium with 10% fetal bovine serum (FBS). 293T cells (2 × 105 cells per well) were plated in a six-well plate the day before transfection. Transfections were then performed by the calcium phosphate precipitation protocol (Specialty Media, Inc.) with various plasmids as indicated in the figures. After 40 h, the cells were harvested and lysed in lysis buffer (150 mM NaCl; 20 mM HEPES, pH 7.4; 2 mM EGTA; 50 mM β-glycerophosophate; 1% Triton X-100; 10% glycerol; 500 μM phenylmethylsulfonyl fluoride [PMSF], 5 μg of leupeptin per ml, 3 μg of aprotinin per ml). COS-1 cells were transfected by the same method with various plasmids as indicated. Jurkat T cells were maintained in RPMI 1460 medium supplemented with 10% FBS. The cells were then lysed in lysis buffer as described above. Cell lysates were utilized for binding assays and kinase assays.
Antibodies, immunoprecipitation, and immunoblot analysis.
Anti-HPK1 antibodies were generated against the HPK1 peptide (amino acids 341 to 366). Other antibodies include anti-HA monoclonal antibodies (12CA5; Boehringer Mannheim); anti-FLAG monoclonal antibodies (M2; Kodak); anti-Crk, anti-CrkL, and anti-EGF receptor antibodies (Santa Cruz Biotechnology). Immunoprecipitation and Western blot analysis were performed as described previously (15). Anti-HPK1 and anti-HA immunocomplexes were recovered by using protein-A beads (Sigma). Anti-FLAG immunocomplexes were recovered by using protein-G beads (Santa Cruz Biotechnology).
Preparation of GST fusion proteins.
The various GST fusion proteins were prepared according to manufacturer’s instructions (Pharmarcia Biotech, Inc.). Briefly, bacterial cultures were grown in 2× YT-G medium (16 mg of tryptone per ml, 10 mg of yeast extract per ml, 85.56 mM NaCl, 2% glucose, and 100 μg of ampicillin per ml) to an A600 of 0.6 to 0.8 at 30°C. IPTG (isopropyl-β-d-thiogalactopyranoside; 5 mM) was added to the cultures for 3 h to induce the expression of fusion proteins. The cells were harvested and sonicated in bacterial lysis buffer (10 mM phosphate, 30 mM NaCl, 0.25% Tween 20, 10 mM β-mercaptoethanol, 10 mM EDTA, 500 μM PMSF, 5 μg of leupeptin per ml, 3 μg of aprotinin per ml). Cell lysates were centrifuged at 12,000 × g for 30 min. The fusion proteins were purified by using glutathione-Sepharose beads.
Preparation of in vitro-translated proteins.
Plasmids indicated in the figures were utilized to synthesize [35S]methionine-labeled proteins by an in vitro transcription and translation method according to the manufacturer’s instructions (Promega Biotech, Inc.). Plasmids (1 μg/reaction) were added to a reaction mixture, including 25 μl of rabbit reticulocyte lysate, 2 μl of reaction buffer, 1 μl of T7 RNA polymerase, 1 μl of amino acid mixture minus methionine (1 mM), 4 μl of [35S]methionine (10 mCi/ml), ribonuclease inhibitor (40 U/μl), and nuclease-free H2O to a final volume of 50 μl. The reactions were incubated at 30°C for 90 min. The in vitro-translated proteins were then used for in vitro binding assays. The interactions of in vitro-translated proteins with GST fusion proteins were directly examined by autoradiography.
In vitro binding assay and competition binding assay.
The various GST fusion proteins were generated as described above and were coupled to glutathione-Sepharose beads. Jurkat T-cell lysates or in vitro-translated proteins were incubated with the various GST fusion proteins in 500 μl of lysis buffer for 1 to 2 h at 4°C. Interacting complexes were washed three times with radioimmunoprecipitation assay buffer (25 mM Tris, pH 7.5; 150 mM NaCl, 1 mM EDTA, 0.5% deoxycholate, 1% Nonidet P-40, 500 μM PMSF, 5 μg of leupeptin per ml, 3 μg of aprotinin per ml) and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The interaction of HPK1 with specific GST fusion proteins was analyzed by immunoblotting with an anti-HPK1 antibody. Four wild-type HPK1 proline-rich peptides and two mutant peptides of HPK1 second proline-rich motif were synthesized and utilized for competition binding assays (see Fig. 3A). To test the ability of individual HPK1 proline-rich peptides to block the interaction of HPK1 with adaptor proteins, [35S]methionine-labeled HPK1-CD was incubated with immobilized GST fusion proteins in the presence of wild-type or mutant HPK1 proline-rich peptides (1 mM) in 200 μl of lysis buffer for 1 to 2 h at 42°C. After an extensive washing, the bound [35S]methionine-labeled HPK1-CD was examined by autoradiography.
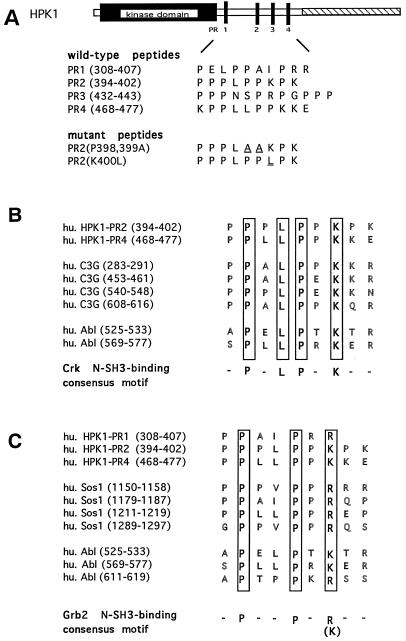
FIG. 3.
Sequence comparison of the HPK1 proline-rich motifs with the Crk and Grb2 N-SH3-binding consensus motifs. (A) Amino acid sequences of the four wild-type HPK1 proline-rich motifs (PR1, PR2, PR3, and PR4) and the two mutants generated from the second proline-rich motif, PR2(P398, 399A) and PR2(K400L) are shown. The amino acid positions of each motif are indicated in parentheses. For the two PR2 mutants, the mutated residues are underlined. The four wild-type peptides and the two mutant peptides were synthesized and utilized for competition binding assays. (B) The second and fourth proline-rich motifs of HPK1 showed the same Crk N-SH3-binding consensus motifs (-P-x-L-P-x-K-) as the known Crk N-SH3-binding sites of C3G and c-Abl. (C) The first, second, and fourth proline-rich motifs of HPK1 also showed the same Grb2 N-SH3-binding consensus motif (-P-x-x-P-x-R/K-) as the known Grb2 N-SH3-binding sites of Sos1 and c-Abl.
Immunocomplex kinase assay.
Assays for JNK activity were performed as described previously (15). In brief, HA-tagged JNK1 from 50 μg of transfected 293T cell lysates was immunoprecipitated by using an anti-HA monoclonal antibody and protein A-agarose beads in 500 μl of lysis buffer. After being washed twice with lysis buffer, twice with LiCl buffer (500 mM LiCl; 100 mM Tris-Cl, pH 7.6; 0.1% Triton X-100), and twice with kinase buffer (20 mM MOPS [morpholine propanesulfonic acid], pH 7.2; 2 mM EGTA; 10 mM MgCl2; 0.1% Triton X-100), the immunoprecipitates were incubated with 30 μl of kinase buffer containing 2 μg of GST–c-Jun (1-79), 20 μM unlabeled ATP, and 20 μCi of [γ-32P]ATP. The reactions were incubated at 30°C for 30 min and terminated by boiling the samples in 6× loading buffer, and the final products were resolved by SDS-PAGE (15%). HPK1 activity was measured like the assay for JNK activity. Jurkat cells and 293T cells transiently expressing HPK1 were immunoprecipitated by using an anti-HPK1 antibody or an anti-FLAG monoclonal antibody, and kinase assays were performed with myelin basic protein (MBP), GST-Crk, or GST-CrkL as a substrate where indicated.
Phosphoamino acid analysis.
The phosphorylated proteins obtained from the immunocomplex assays were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The spots containing phosphorylated proteins were excised and hydrolyzed in 6 N HCl for 1 h at 110°C. The supernatants were lyophilized and then dissolved in 10 μl of pH 2.5 buffer (5.9% glacial acetic acid, 0.8% formic acid, 0.3% pyridine, 0.3 mM EDTA). The phosphoamino acids were resolved in one dimension with a thin-layer plate in pH 2.5 buffer at 20 mA for 90 min. The phosphoamino acid residues were detected by autoradiography.
CAT assay.
Plasmids were mixed with 6 μl of DMRIE-C lipid transfection reagent (GIBCO-BRL) in 1 ml of Opti-Mem I (GIBCO-BRL) for 45 min at room temperature. Jurkat T antigen cells (2 × 106) were added to the mixture and incubated at 37°C. After 5 h, 2 ml of RPMI with 15% FBS was added to the cells. At 34 h after the RPMI was added, cells were stimulated for 8 h with phytohemagglutinin (PHA; 1 μg/ml), phorbol myristate acetate (PMA; 10 ng/ml), and ionomycin (1 μM). The cells were collected, washed twice in phosphate- buffered saline, suspended 250 μl of TE buffer (250 mM Tris-HCl, 5 mM EDTA), and lysed by three freeze-thaw cycles. Chloramphenicol acetyltransferase (CAT) assays were performed as described previously (19).
RESULTS
HPK1 interacts with Crk and CrkL adapter proteins in vitro and in vivo.
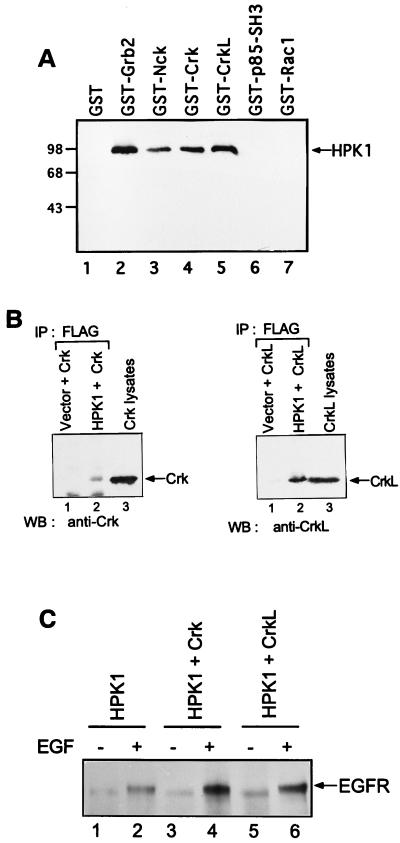
To identify HPK1-interacting proteins, we utilized an in vitro binding assay to test several SH3 domain-containing proteins, including Grb2, Nck, Crk, CrkL, and the p85 SH3 domain of PI3 kinase, as well as the small GTPase Rac1 as a non-SH3 containing control protein. Jurkat T-cell lysates containing endogenous HPK1 were incubated with the various GST fusion proteins described above. Association of HPK1 with specific GST fusion proteins was determined by immunoblotting with an anti-HPK1 antibody. We found that the GST adapter proteins GST-Grb2, GST-Nck, GST-Crk, and GST-CrkL associated with HPK1, whereas GST alone, the GST-p85 SH3 domain, and GST-Rac1 did not pull down any detectable amounts of HPK1 (Fig. 1A). Recently, Grb2 and Nck were shown to link tyrosine kinases to HPK1 (1). However, the Crk C-SH3 domain alone does not interact with HPK1 (1). Our results here support this previous finding that HPK1 bound Grb2 and Nck (1) and also demonstrate the interaction of HPK1 with the full-length Crk and CrkL in vitro. Next, we determined whether HPK1 could associate with Crk and CrkL in mammalian cells. FLAG-tagged HPK1 was cotransfected with Crk or CrkL into COS-1 cells. Anti-HPK1 immunoprecipitates from the transfected COS-1 cells showed the association with Crk and CrkL, respectively (Fig. 1B), indicating that HPK1 formed complexes with Crk and CrkL in vivo. Since Crk is known to recognize the activated EGF receptor through its SH2 domain (2, 22), we further examined whether Crk and CrkL could recruit HPK1 to EGF receptor upon EGF stimulation. We found that HPK1 associated with EGF receptor upon EGF stimulation. Transient expression of HPK1 with Crk and CrkL strongly enhanced the association of HPK1 with EGF receptor in response to EGF (Fig. 1C), suggesting that the Crk proteins play a role in the recruitment of HPK1 to the EGF receptor.
FIG. 1.
The adapter proteins Crk and CrkL interact with HPK1 in vitro and in vivo. (A) Various GST-fusion proteins as indicated and the GST protein alone were first immobilized on glutathione-agarose beads. Jurkat T-cell lysate (5 × 106) was incubated with the immobilized GST fusion proteins for 1 to 2 h at 4°C. The interacting complexes were resolved by SDS-PAGE (8%). The association of HPK1 with specific GST fusion proteins was examined by immunoblotting with an anti-HPK1 antibody. (B) Crk (15 μg) and CrkL (15 μg) were transfected into COS-1 cells (100-mm culture dish) in combination with FLAG-tagged HPK1 (10 μg) or pCI-neo vector (10 μg). Cell lysates were immunoprecipitated with anti-FLAG monoclonal antibodies and then separated by SDS-PAGE (10%). Coprecipitated Crk and CrkL were examined by immunoblotting with anti-Crk and anti-CrkL antibodies, respectively. Crk- and CrkL-transfected 293T cell lysates were included as positive controls for immunoblotting. (C) Transfections were performed as described in Fig. 1B. Transfected cells were treated with serum deprivation or EGF as indicated. Cell lysates were immunoprecipitated by using anti-FLAG monoclonal antibodies and immunoblotted by using an anti-EGF receptor antibody.
The HPK1 proline-rich region is responsible for the HPK1 interaction with Crk and CrkL.
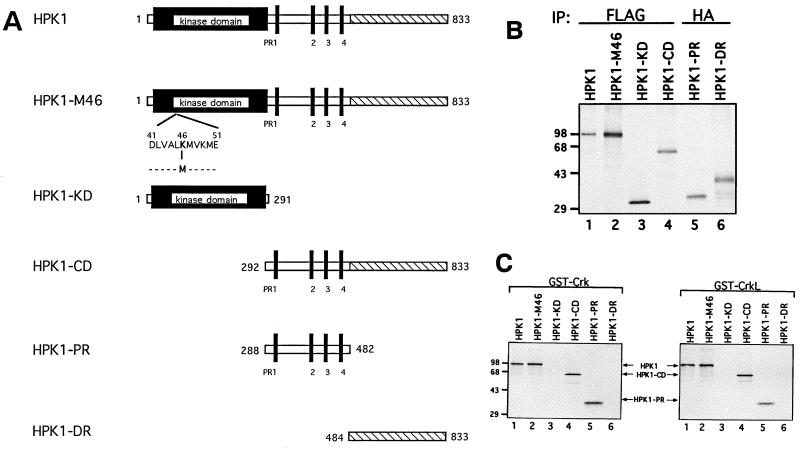
To determine the region(s) of HPK1 responsible for its interaction with Crk and CrkL, a variety of [35S]methionine-labeled HPK1 proteins (Fig. 2A), including wild-type HPK1, a kinase-dead mutant (HPK1-M46), the HPK1 kinase domain (HPK1-KD), the HPK1 carboxyl domain (HPK1-CD), the HPK1 proline-rich region (HPK1-PR), and the HPK1 distal region (HPK1-DR), were generated by an in vitro transcription and translation method. The in vitro-translated products were examined by immunoprecipitation prior to the binding assay (Fig. 2B). Results showed that full-length HPK1, HPK1-M46, and the HPK1 carboxyl domain (HPK1-CD) bound Crk and CrkL (Fig. 2C). The HPK1 proline-rich region (HPK1-PR) alone was sufficient for binding to Crk and CrkL, whereas the HPK1 kinase domain (HPK1-KD) and the HPK1 distal region (HPK1-DR), which lack proline-rich motifs, did not bind Crk or CrkL. These data reveal that the HPK1 proline-rich region, which contains four proline-rich motifs, mediates the interaction of HPK1 with Crk and CrkL.
FIG. 2.
The HPK1 proline-rich region is essential for HPK1 interaction with Crk and CrkL. (A) A schematic diagram displays various HPK1 constructs, including wild-type HPK1, a kinase-dead mutant (HPK1-M46), the kinase domain (HPK1-KD; amino acids 1 to 291), the carboxyl domain (HPK1-CD; amino acids 292 to 833), the proline-rich region (HPK1-PR; amino acids 288 to 482), and the distal regulatory region (HPK1-DR; amino acids 484 to 833). HPK1 is composed of the N-terminal kinase domain (shown as the solid box), four proline-rich motifs (shown as solid vertical bars) in the middle region, and the distal regulatory region (shown as the hatched area) in the C terminus. (B) The generation of in vitro-translated wild-type HPK1 and its mutants was examined by immunoprecipitation with anti-FLAG and anti-HA monoclonal antibodies, as indicated. (C) A variety of [35S]methionine-labeled wild-type HPK1 and its mutants were used to determine the region of HPK1 critical for binding to Crk and CrkL. [35S]methionine-labeled wild-type HPK1 and its mutants (20 μl/sample) were incubated with immobilized GST-Crk or GST-CrkL. The interacting complexes were resolved by SDS-PAGE (10%) and examined by autoradiography.
The HPK1 proline-rich motifs mediate the differential interaction of HPK1 with the Crk proteins and Grb2.
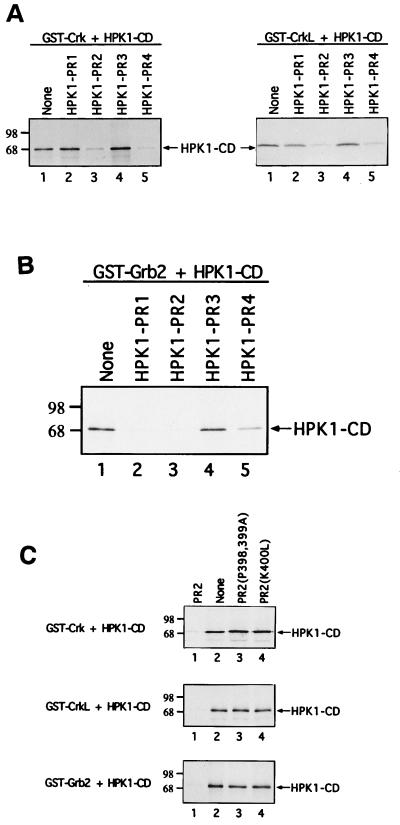
Signaling molecules such as c-Abl and C3G, a guanine nucleotide exchange factor for the small GTPase Rap1, recognize the Crk N-SH3 domain via their proline-rich motifs (12, 38, 52). Comparison of the Crk N-SH3-binding consensus motif (-P-x-L-P-x-K-) with the four proline-rich motifs of HPK1 revealed that the second and fourth proline-rich motifs of HPK1 contain the consensus motif necessary for recognition by the Crk N-SH3 domain (Fig. 3B). Therefore, we investigated whether the individual HPK1 proline-rich motifs are involved in the interaction of HPK1 with Crk and CrkL. Four wild-type HPK1 proline-rich peptides, termed PR1, PR2, PR3, and PR4, and two mutant peptides of the second proline-rich motif, PR2(P398, 399A) and PR2(K400L), were synthesized for this study (Fig. 3A). Competition binding assays were performed by adding individual HPK1 proline-rich peptides to Crk–HPK1-CD and CrkL–HPK1-CD complexes to test whether the proline-rich peptides could block the interaction of HPK1-CD with Crk and CrkL. The HPK1 second (PR2) and fourth (PR4) proline-rich peptides, which contain the Crk N-SH3-binding consensus motif (-P-x-L-P-x-K-), blocked the interaction of HPK1-CD with Crk and CrkL (Fig. 4A), suggesting that both the second and fourth proline-rich motifs may be involved in the recognition of Crk and CrkL by HPK1. Since the fourth proline-rich motif is not conserved between human and mouse HPK1 (15, 17), it is unlikely to play an essential role.
FIG. 4.
The HPK1 proline-rich peptides differentially block the interaction of HPK1 with SH2/SH3 adapter proteins. Competition binding assays were performed to determine the ability of individual HPK1 proline-rich peptides to block the binding of HPK1-CD to adapter proteins. (A) GST-Crk and GST-CrkL were first immobilized on beads. Four wild-type HPK1 proline-rich peptides (shown in Fig. 3A) were added to block the interaction of [35S]methionine-labeled HPK1-CD with GST-Crk and with GST-CrkL, respectively. The amount of bound [35S]methionine-labeled HPK1-CD from the samples was shown by autoradiography and compared with that of the controls, which do not contain HPK1 proline-rich peptides. (B) The same assays were utilized to examine the blocking effect of HPK1 proline-rich peptides on the Grb2–HPK1-CD complex. (C) The HPK1 second proline-rich peptide and its two mutants, PR2(P398, 399A) and PR2(K400L), were examined for their ability to block the interaction of HPK1-CD with Crk, CrkL, or Grb2.
Because the HPK1 first, second, and fourth proline-rich motifs also match the Grb2 N-SH3-binding consensus motif (-P-x-x-P-x-R/K-) (Fig. 3C), we examined the ability of HPK1 proline-rich peptides to block the formation of Grb2/HPK1-CD complexes. The first and second proline-rich peptides efficiently blocked the interaction of HPK1-CD with Grb2 (Fig. 4B); however, the fourth proline-rich peptide had at best a marginal blocking effect on the Grb2–HPK1-CD complex. Thus, the HPK1 proline-rich peptides showed the differential blocking effects on the Crk–HPK1-CD complex and the Grb2–HPK1-CD complex. To ensure that the typical -P-x-L-P-x-K- motif is required for the recognition of the Crk N-SH3 domain, two mutant HPK1 second proline-rich peptides PR2(P398, 399A) and PR2(K400L) were analyzed in competition binding assays. PR2(P398, 399A) was designed to disrupt the typical -P-x-L-P-x-K- motif by changing proline residues at positions 398 and 399 to alanine, and PR2(K400L) was designed by switching the critical lysine residue at position 400 to leucine. Both PR2(P398, 399A) and PR2(K400L) failed to block the interaction of HPK1-CD with Crk and CrkL, while the same concentration of the wild-type PR2 peptide effectively blocked these interactions (Fig. 4C). A similar result was also observed in the case of the Grb2–HPK1-CD complex (Fig. 4C). Thus, the blocking effect of HPK1 proline-rich peptides is highly selective and is not due to nonspecific interference or the large amount of peptides used in these reactions. Together, our results indicate that the proline-rich motifs within HPK1 are not only critical for the recognition of SH2/SH3 adapter proteins but may also be implicated in the differential binding of HPK1 to these adapter proteins or other SH3 domain-containing proteins.
Crk and CrkL activate HPK1 and synergize with HPK1 in the activation of JNK.
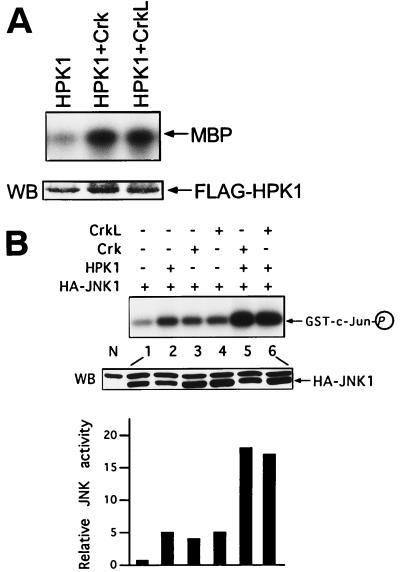
After determining that HPK1 interacted with Crk and CrkL, we addressed the functional relevance of these interactions. We first investigated whether Crk and CrkL could regulate HPK1 activity. Immunocomplex kinase assays were performed to determine HPK1 activity on 293T cells transfected with HPK1 alone or in the presence of various constructs, including Crk and CrkL. We found that Crk and CrkL activated HPK1 in 293T cells (Fig. 5A), indicating that the Crk proteins not only physically associate with HPK1 but also regulate HPK1 activity. Since HPK1 and the Crk proteins are upstream activators of JNK, we examined whether Crk and CrkL could affect HPK1-induced JNK activation. Transient expression of HPK1, Crk, or CrkL alone induced a 4- to 5-fold JNK activation in 293T cells, whereas HPK1, in the presence of Crk or CrkL, significantly induced JNK activation 15-fold over the basal activity (Fig. 5B), suggesting that the Crk proteins cooperate with HPK1 to activate JNK synergistically. Based on these observations, it is likely that the Crk proteins and HPK1 are located in the same JNK signaling cascade.
FIG. 5.
Crk and CrkL activate HPK1 and synergize with HPK1 in the activation of JNK. (A) 293T cells were transfected with HPK1 alone or with HPK1 plus Crk and CrkL. The total amount of plasmids in each sample was equalized with the pCI-neo vector. Cell lysates were immunoprecipitated with anti-FLAG antibodies, and equal amounts of HPK1 were used for immunocomplex kinase assays. The expression of HPK1 was examined by Western blotting with an anti-FLAG antibody. (B) HA-JNK1 was transfected into 293T cells alone (lane 1) or in combination with 2 μg each of the plasmids, as indicated (lanes 2 to 6). The expression of JNK in transfected cells was examined by Western blotting (WB) with an anti-HA antibody, and a nontransfected cell lysate (N) was included as a negative control. The bottom bands in the WB panel were HA-JNK. JNK activity was determined by immunocomplex kinase assays with an anti-HA antibody. GST–c-Jun (1-79) was utilized as a substrate for HA-JNK1. The relative JNK kinase activity was measured by densitometer.
The HPK1 proline-rich region mutant and dominant-negative mutants of MEKK1, TAK, and SEK1 inhibit JNK activation by the Crk proteins.
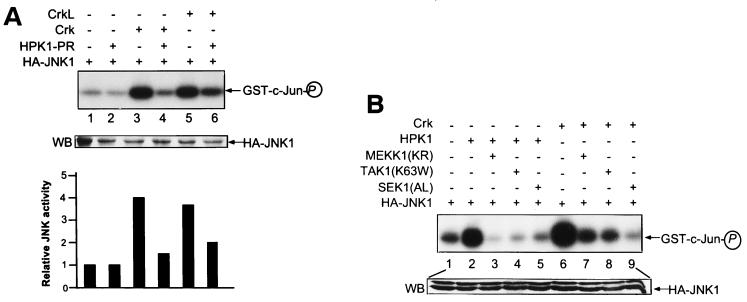
To determine whether Crk and CrkL act as upstream regulators of HPK1 in the JNK signaling cascade, we tested whether the HPK1 mutants had an effect on JNK activation by the Crk proteins. We observed that HPK1-PR, an HPK1 mutant encoding the proline-rich region only, blocked JNK activation by Crk and CrkL (Fig. 6A). Meanwhile, we tested if the dominant-negative mutants of HPK1 downstream effectors, including MEKK1-KR, TAK1-K63W, and SEK1-AL, could block Crk-induced JNK activation. We found that these mutants blocked HPK1-induced JNK activation as shown previously (15, 60) and also inhibited Crk-induced JNK activation effectively, suggesting that Crk and HPK1 shared common downstream effectors for JNK activation (Fig. 6B). The results from the binding assays and kinase assays indicate that the Crk proteins function as upstream regulators of the HPK1-mediated JNK signaling cascade.
FIG. 6.
HPK1-PR and dominant-negative mutants of MEKK1, TAK1, and SEK1 inhibit activation of JNK by the Crk proteins. (A) 293T cells were transfected with HA-JNK1 alone or in combination with 3 μg each of the indicated constructs. JNK activity was then measured by immunocomplex kinase assay as described previously (Fig. 5B). (B) The mutants of HPK1 downstream effectors, including MEKK1-KR, TAK1-K63W, and SEK1-AL, were used to block JNK activation induced by HPK1 and Crk. 293T cells were transfected with HA-JNK1 alone or with various combinations of plasmids as indicated. Similar transfections and kinase assays were performed to determine the JNK activity.
Inhibition of IL-2 induction by the HPK1 and Crk mutants.
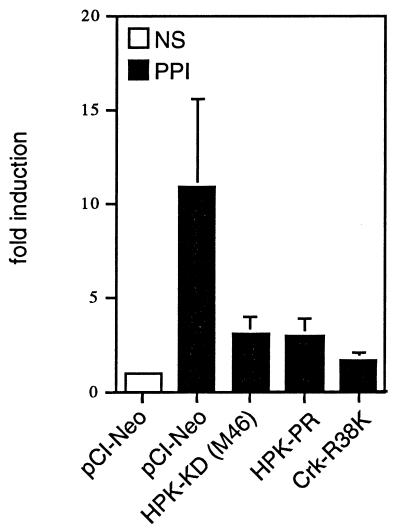
HPK1 is an upstream activator of the transcription factor AP1, which is involved in IL-2 induction, and is expressed predominantly in hematopoietic cells. Thus, we examined whether HPK1 played a role in IL-2 induction in T cells. To that end, we conducted CAT reporter assays to determine the effect of different HPK1 mutants on IL-2 promoter activation. Jurkat T cells were transfected with an IL-2–CAT reporter in the presence of an empty vector or various HPK1 mutants, and then the transfected cells were stimulated with T-cell stimuli (PHA plus PMA and ionomycin). Our result indicated that HPK1-KD(M46), an HPK1 mutant encoding the kinase-dead domain of HPK1-M46, and HPK1-PR inhibited IL-2 induction in Jurkat T cells (Fig. 7). Since Crk potentially acts as an upstream regulator of HPK1, we also examined the effect of a Crk SH2 domain mutant (Crk-R38K) on IL-2 induction. We found that Crk-R38K, which is unable to bind tyrosine-phosphorylated proteins, also blocked IL-2 induction (Fig. 7). This result indicates that the HPK1 signaling may be involved in the regulation of IL-2 promoter after T-cell stimulation.
FIG. 7.
The HPK1 mutants and a Crk mutant block IL-2 promoter induction by T-cell stimuli. Jurkat T cells were transfected with 2 μg of the IL-2–CAT reporter construct with 10 μg of the indicated constructs. Where indicated, the cells were stimulated with PHA (1 μg/ml), PMA (10 ng/ml), and ionomycin (1 μM) 30 h posttransfection and collected 8 h later. CAT activities were measured as described in Materials and Methods. The fold induction is relative to nonstimulated cells transfected with empty vector.
HPK1 phosphorylates Crk and CrkL mainly on serine and threonine residues in vitro.
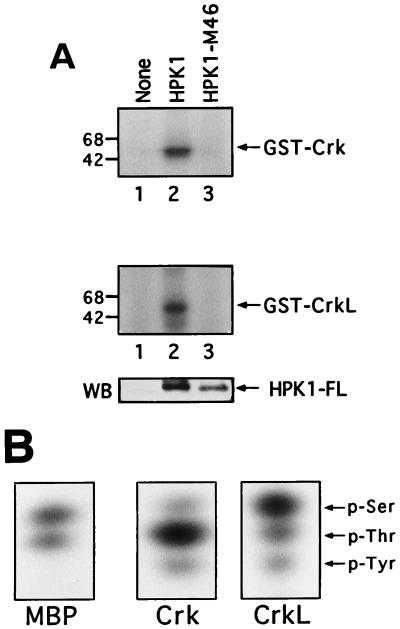
Previous studies have shown that c-Abl bound the N-SH3 domain of Crk and phosphorylated Crk on tyrosine 221 (Y221) (12) and that CrkL was associated with BCR-Abl and was tyrosine-phosphorylated in BCR-Abl-expressing cells (32, 34, 54). Because HPK1, like c-Abl, recognized the Crk N-SH3 domain, we examined whether HPK1 could phosphorylate Crk and CrkL. Immunocomplex kinase assays were used to test HPK1 phosphorylation of Crk and CrkL. We first observed that both GST-Crk and GST-CrkL, but not GST alone, were phosphorylated by anti-HPK1 immunoprecipitates from Jurkat T-cell lysates (data not shown), suggesting that Crk and CrkL are substrates for HPK1. We further demonstrated that Crk and CrkL were phosphorylated by anti-HPK1 immunoprecipitates from 293T cells transfected with HPK1 but not with HPK1-M46, indicating that the HPK1 phosphorylation of Crk and CrkL is highly specific (Fig. 8A). Consequently, we were interested in determining the phosphorylated residues on the Crk and CrkL by phosphoamino acid analysis. HPK1 phosphorylated MBP on serine and threonine residues (Fig. 8B) as shown in previous work (15). We next found that HPK1 phosphorylated Crk mainly on threonine and weakly on serine and tyrosine, whereas HPK1 phosphorylated CrkL mainly on serine and weakly on threonine and tyrosine (Fig. 8B). This is the first evidence that the Crk proteins are phosphorylated by a protein kinase on serine and threonine residues. The tyrosine phosphorylation of the Crk proteins is most likely induced by an HPK1-associated tyrosine kinase (e.g., c-Abl) rather than by HPK1 itself. More studies are needed to determine both the HPK1 phosphorylation sites on Crk and CrkL and the functional significance of these phosphorylations.
FIG. 8.
HPK1 phosphorylates Crk and CrkL on serine and threonine in vitro. (A) pCI-neo vector, HPK1, and HPK1-M46 were transfected into 293T cells. Cell lysates were immunoprecipitated by using an anti-HPK1 antibody, and then immunocomplex kinase assays were performed by using GST-Crk and GST-CrkL as substrates for HPK1. HPK1-FL represented wild-type HPK1 and HPK1-M46. (B) Phosphorylated MBP, GST-Crk, and GST-CrkL were obtained from immunocomplex kinase assays. Phosphorylated residues of these proteins were analyzed by phosphoamino acid analysis.
DISCUSSION
We have investigated the potential interaction of HPK1 with several SH2/SH3 adapter proteins, including Grb2, Nck, Crk, and CrkL. Here we provide evidence from binding assays and kinase assays to demonstrate the functional interaction of HPK1 with Crk and CrkL. We showed that fusion proteins GST-Crk and GST-CrkL pulled down HPK1 from Jurkat T-cell lysates and that HPK1 formed complexes with Crk and CrkL in mammalian cells. Also, the Crk proteins enhanced the recruitment of HPK1 to the EGF receptor, supporting the previous report that SH2/SH3 adapter proteins link surface receptors to HPK1 (1). Although the expression of HPK1 is mainly restricted to hematopoietic cells, SH2/SH3 adapter proteins such as Grb2, Crk, and CrkL are ubiquitously expressed in a wide range of tissues and cells, including T cells, B cells, hematopoietic progenitor cells, and other cells (25,37). Based on the previous evidence and our own data, we speculate that HPK1 may form complexes with Crk and CrkL in hematopoietic cells constitutively or upon extracellular stimulation. The proline-rich motifs of HPK1 mediated the complex formation between HPK1 and various SH2/SH3 adapter proteins (Fig. 4). Particularly, the second proline-rich motif containing the Crk N-SH3-binding consensus sequence (-P-x-L-P-x-K-) was critical for the binding of HPK1 to Crk and CrkL, whereas the first and second proline-rich motifs containing the Grb2 N-SH3-binding consensus sequence (-P-x-x-P-x-R/K-) were implicated in the binding of HPK1 to Grb2. The third proline-rich motif did not play a role in the interaction of HPK1 with the SH2/SH3 adapter proteins tested; however, it may be involved in the association of HPK1 with other SH3 domain-containing proteins. Since the fourth proline-rich motif is not conserved between human and mouse HPK1, it may not play a critical functional role for HPK1. Consistent with previous findings (18), our data showed that lysine-400 in the HPK1 second proline-rich motif is critical for the recognition of Crk and CrkL by HPK1 (Fig. 4C).
SH2/SH3 adapter proteins, which themselves have no enzymatic activity, are signaling modulators to form multiple protein complexes selectively and thereby to transmit upstream signals to downstream targets. SH2/SH3 adapter proteins usually localize close to the cell membrane, and some are even directly associated with surface receptors or the cytoskeleton (24). For instance, Grb2 and Crk are recruited to activated EGF receptors via their SH2 domains (2). Crk is also recruited indirectly to the nerve growth factor receptor through Shc (23) and to integrin through Paxillin, a constituent of focal adhesions (5, 42). Thus, SH2/SH3 adapter proteins play their roles in the very upstream or initial events during signal transduction. According to the features of adapter proteins and our data, it is likely that the differential interaction of HPK1 with various SH2/SH3 adapter proteins provides a mechanism for HPK1 to integrate different upstream signals to the JNK signaling cascade. A similar phenomenon may occur in other mammalian Ste20-related MAPKKKKs containing the proline-rich motifs, such as NIK, GLK, and KHS. It will be of interest to dissect the HPK1-mediated JNK signaling pathways by using specific HPK1 proline-rich motif mutants and the related adapter mutants. This approach will be useful in characterizing parallel JNK signaling cascades mediated by other MAPKKKKs.
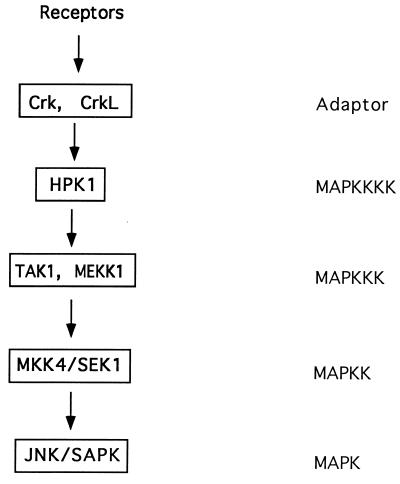
Through kinase assays, we have shown that the Crk proteins directly activated HPK1 and also cooperated with HPK1 to activate JNK synergistically (Fig. 5). These results indicated that the Crk proteins not only physically interacted with HPK1 but also functionally regulated HPK1-mediated JNK activation. A previous study demonstrated that C3G, a Crk-interacting protein, mediates the downstream signaling of v-Crk-induced JNK activation (53). Like C3G, HPK1 bound Crk via recognition of the Crk N-SH3 domain (Fig. 3B), suggesting that HPK1 and C3G may be at the same level of the parallel Crk-mediated JNK signaling pathways. We further demonstrated that expression of HPK1-PR, the region responsible for binding to Crk and CrkL, blocked JNK activation by Crk and CrkL and that the downstream effectors of HPK1, such as MEKK1, TAK1, and SEK1, were also involved in Crk-induced JNK activation (Fig. 6). Although it is possible that the overexpression of HPK1-PR may compete with endogenous C3G for Crk binding, the evidence from the kinase assays (Fig. 5 and 6) strongly suggests that HPK1 is one of the Crk downstream effectors for the Crk-mediated signaling pathways. Therefore, we propose the model for the HPK1-mediated JNK signaling cascade shown in Fig. 9. HPK1 receives the upstream signals through its differential interaction with various SH2/SH3 adapter proteins and subsequently transmits these signals to downstream targets (MEKK1 and TAK1) via its kinase domain or distal regulatory region. Afterwards, the signals are transmitted to a target further downstream (SEK1), thereby leading to JNK activation. Although HPK1 was shown to associate EGF receptor in transfected COS-1 cells, the surface receptors, which recruit the Crk-HPK1 and CrkL-HPK1 complexes in hematopoietic cells, are yet to be determined. By identifying the surface receptors that activate HPK1, we may elucidate the entire HPK1-mediated JNK signaling cascade from the cell membrane to the nucleus.
FIG. 9.
The proposed model for the HPK1-mediated JNK signaling cascade. The adapter proteins Crk and CrkL couple surface receptors to HPK1. Subsequently, HPK1 transmits the upstream signals to downstream MAPKKKs, such as MEKK1 and TAK1, which transmit the signals to further downstream SEK1 for JNK activation.
Optimal IL-2 induction, a central event of T-cell activation, requires two major signals triggered by costimulation of the T-cell receptor (TCR) and CD28 or mimicked by incubating T cells with phorbol ester (i.e., PMA) and Ca2+ ionophore (8, 43). Early studies have indicated that AP1 binds the IL-2 promoter (31, 46) and forms a complex with NF-AT to induce IL-2 activation (33, 59). Another study also showed that JNK is involved in the integration of the costimulatory signals CD3 plus CD28 or PMA plus Ca2+ ionophore in T cells (49). However, the response of JNK to Ca2+ ionophore is T cell specific, suggesting that a cell type-specific component plays a role in this event (49). As a hematopoietic upstream activator of JNK and AP1, HPK1 may be involved in IL-2 induction in T cells. By CAT reporter assays, we found that the HPK1 mutants, HPK1-KD(M46) and HPK1-PR, blocked IL-2 activation by T-cell stimuli (PHA, PMA, and ionomycin) in Jurkat T cells (Fig. 7). Inhibition of IL-2 induction by HPK1-PR may be due to the interruption of upstream signals mediated by the SH2/SH3 adapter proteins. The SH2/SH3 adapter proteins, Crk and Grb2, are known to form distinct signaling complexes during TCR stimulation (3, 4, 21, 41), suggesting that these adapter proteins are involved in the TCR signaling pathway. Grb2 is also shown to couple upstream tyrosine kinases such as v-Src to HPK1 (1). In addition, we also found that Crk-R38K, a Crk SH2 domain mutant, blocked IL-2 induction. These observations further support the idea that HPK1-PR may block the transmission of upstream signals to the IL-2 promoter. The mechanism by which HPK1-KD(M46) inhibits of IL-2 induction is unclear. One possible explanation is that HPK1-KD(M46) may compete with wild-type HPK1 for the binding of the downstream targets (MAPKKKs), which are essential for JNK and AP1 activation. More studies are needed to define the role of HPK1 in IL-2 activation. Future studies will examine whether T-cell costimulatory signals, TCR plus CD28 or PMA plus Ca2+ ionophore, can activate HPK1.
The phosphorylation of Crk and CrkL by HPK1 further supported the notion that HPK1 bound Crk and CrkL. Interestingly, our data from phosphoamino acid analysis revealed that Crk and CrkL were phosphorylated mainly on serine and threonine (Fig. 8B). We noticed that phosphorylated Crk and CrkL also contained some phosphotyrosine residues. Since HPK1 is a serine-threonine kinase, HPK1 is unlikely responsible for this tyrosine phosphorylation. The c-Abl tyrosine kinase was previously shown to bind the HPK1 proline-rich motifs via its SH3 domain (17). Thus, these tyrosine phosphorylations are most likely due to the coimmunoprecipitation of c-Abl with HPK1. Previous evidence indicated that c-Abl binds the Crk N-SH3 domain and phosphorylates Crk on tyrosine 221 (Y221). This phosphorylated Y221 then provides a binding site for the Crk SH2 domain to form an intramolecular interaction resulting in an uncomplexed Crk molecule (12). Similarly, tyrosine residue 207 (Y207) in CrkL has been identified to function as a negative regulatory site (45). Although HPK1 is unlikely to phosphorylate Crk and CrkL on Y221 and Y207, serine and threonine phosphorylation of the Crk proteins by HPK1 might still play a role in the feedback regulation of the Crk proteins. Future studies will focus on determining the phosphorylation sites of Crk and CrkL induced by HPK1 and the functional significance of these phosphorylations. These studies will provide critical insights for understanding the Crk- and CrkL-mediated cellular responses. It will be also interesting to investigate whether HPK1, like c-Abl and C3G, plays a role in cell transformation induced by the Crk proteins.
ACKNOWLEDGMENTS
We thank M. C.-T. Hu, B. J. Mayer, P. Polverino, C. L. Sawyers, E. Y. Skolnik, and L.-Y. Yu-Lee for generous gifts of materials as described in experimental procedures. We also thank Kathie Mihindukulasuriya for critical reading of the manuscript, Susan Lee and Roshi Afshar for technical assistance, Mary Lowe for secretarial assistance, and members of the Tan laboratory for discussions.
W. Oehrl and S. M. Feller are supported by SFB465 of the Deutsche Forschungsgemeinschaft. T.-H. Tan is a Scholar of the Leukemia Society of America and is supported by the NIH grants RO1-AI42532 and RO1-AI38649.
REFERENCES
- 1.Anafi M, Kiefer F, Gish G D, Mbamalu G, Iscove N N, Pawson T. SH2/SH3 adaptor proteins can link tyrosine kinases to a Ste20-related protein kinase, HPK1. J Biol Chem. 1997;272:27804–27811. doi: 10.1074/jbc.272.44.27804. [DOI] [PubMed] [Google Scholar]
- 2.Birge R B, Fajardo J E, Mayer B J, Hanafusa H. Tyrosine-phosphorylated epidermal growth factor receptor and cellular p130 provide high affinity binding substrates to analyze Crk-phosphotyrosine-dependent interactions in vitro. J Biol Chem. 1992;267:10588–10595. [PubMed] [Google Scholar]
- 3.Boussiotis V A, Freeman G J, Berezovskaya A, Barber D L, Nadler L M. Maintenance of human T cell anergy: blocking of IL-2 gene transcription by activated Rap1. Science. 1997;278:124–128. doi: 10.1126/science.278.5335.124. [DOI] [PubMed] [Google Scholar]
- 4.Buday L, Khwaja A, Sipeki S, Farago A, Downward J. Interactions of Cbl with two adapter proteins, Grb2 and Crk, upon T cell activation. J Biol Chem. 1996;271:6159–6163. doi: 10.1074/jbc.271.11.6159. [DOI] [PubMed] [Google Scholar]
- 5.Burridge K, Turner C E, Romer L H. Tyrosine phosphorylation of paxillin and pp125FAK accompanies cell adhesion to extracellular matrix: a role in cytoskeletal assembly. J Cell Biol. 1992;119:893–903. doi: 10.1083/jcb.119.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y-R, Wang X, Templeton D, Davis R J, Tan T-H. The role of c-Jun N-terminal kinase (JNK) in apoptosis induced by ultraviolet C and γ radiation: duration of JNK activation may determine cell death and proliferation. J Biol Chem. 1996;271:31929–31936. doi: 10.1074/jbc.271.50.31929. [DOI] [PubMed] [Google Scholar]
- 7.Cohen G B, Ren R, Baltimore D. Modular binding domains in signal transduction proteins. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 8.Crabtree G R. Contingent genetic regulatory events in T lymphocyte activation. Science. 1989;243:355–361. doi: 10.1126/science.2783497. [DOI] [PubMed] [Google Scholar]
- 9.Derijard B, Hibi M, Wu I-H, Barrett T, Su B, Deng T, Karin M, Davis R J. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 10.Derijard B, Raingeaud J, Barrett T, Wu I-H, Han J, Ulevitch R J, Davis R J. Independent human MAP kinase signal transduction pathways defined by MEK and MKK isoforms. Science. 1995;267:682–685. doi: 10.1126/science.7839144. [DOI] [PubMed] [Google Scholar]
- 11.Diener K, Wang X S, Chen C, Meyer C F, Bray J, Zukowski M, Tan T-H, Yao Z. Activation of the c-Jun N-terminal kinase pathway by a novel protein kinase related to human germinal center kinase. Proc Natl Acad Sci USA. 1997;94:9687–9692. doi: 10.1073/pnas.94.18.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feller S M, Knudsen B, Hanafusa H. c-Abl kinase regulates the protein binding activity of c-Crk. EMBO J. 1994;13:2341–2351. doi: 10.1002/j.1460-2075.1994.tb06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feller S M, Knudsen B, Hanafusa H. Cellular proteins binding to the first homology 3 (SH3) domain of the proto-oncogene product c-Crk indicate Crk-specific signaling pathways. Oncogene. 1995;10:1465–1473. [PubMed] [Google Scholar]
- 13a.Feller SM, Posern G, Voss J, Kardinal C, Sakkab D, Zheng J, Knudsen B. Physiological signals and oncogenesis mediated through Crk family adaptor proteins. J Cell Physiol. 1998;177:535–552. doi: 10.1002/(SICI)1097-4652(199812)177:4<535::AID-JCP5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 14.Galcheva-Gargova Z, Derijard B, Wu I-H, Davis R. An osmosensing signal transduction pathway in mammalian cells. Science. 1994;265:806–808. doi: 10.1126/science.8047888. [DOI] [PubMed] [Google Scholar]
- 15.Hu M C-T, Qiu W R, Wang X, Meyer C F, Tan T-H. Human HPK1, a novel human hematopoietic progenitor kinase that activates the JNK/SAPK kinase cascade. Genes Dev. 1996;10:2251–2264. doi: 10.1101/gad.10.18.2251. [DOI] [PubMed] [Google Scholar]
- 16.Ip Y T, Davis R J. Signal transduction by the c-Jun N-terminal kinase (JNK)—from inflammation to development. Curr Opin Cell Biol. 1998;10:205–219. doi: 10.1016/s0955-0674(98)80143-9. [DOI] [PubMed] [Google Scholar]
- 17.Kiefer F, Tibbles L, Anafi A M, Janssen A, Zanke B W, Lassam N, Pawson T, Woodgett J R, Iscove N N. HPK1, a hematopoietic protein kinase activating the SAPK/JNK pathway. EMBO J. 1996;15:7013–7025. [PMC free article] [PubMed] [Google Scholar]
- 18.Knudsen B S, Zheng J, Feller S M, Mayer J P, Burrell S K, Cowburn D, Hanafusa H. Affinity and specificity requirements for the first Src homology 3 domain of the Crk proteins. EMBO J. 1995;14:2191–2198. doi: 10.1002/j.1460-2075.1995.tb07213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai J-H, Hovath G, Subleski J, Bruder J, Ghosh P, Tan T-H. RelA is a potent transcriptional activator of the CD28 response element within the interleukin-2 promoter. Mol Cell Biol. 1995;15:4260–4271. doi: 10.1128/mcb.15.8.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin A, Minden A, Martinetto H, Claret F-X, Lange-Carter C, Mercurio F, Johnson G L, Karin M. Identification of a dual specificity kinase that activates the Jun kinases and p38-Mpk2. Science. 1995;268:286–290. doi: 10.1126/science.7716521. [DOI] [PubMed] [Google Scholar]
- 21.Marengere L E, Mirtsos C, Kozieradzki I, Veillette A, Mak T W, Penninger J M. Proto-oncoprotein Vav interacts with c-Cbl in activated thymocytes and peripheral T cells. J Immunol. 1997;159:70–76. [PubMed] [Google Scholar]
- 22.Margolis B, Silvennoinen O, Comoglio F, Roonprapunt C, Skolnik E, Ullrich A, Schlessinger J. High-efficiency expression/cloning of epidermal growth factor-receptor-binding proteins with Src homology 2 domains. Proc Natl Acad Sci USA. 1992;89:8894–8898. doi: 10.1073/pnas.89.19.8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuda M, Hashimoto Y, Muroya K, Hasegawa H, Kurata T, Tanaka S, Nakamura S, Hattori S. CRK protein binds to two guanine nucleotide-releasing proteins for the Ras family and modulates nerve growth factor-induced activation of Ras in PC12 cells. Mol Cell Biol. 1994;14:5495–5500. doi: 10.1128/mcb.14.8.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuda M, Kurata T. Emerging components of the Crk oncogene product: the first identified adaptor protein. Cell Signalling. 1996;8:335–340. doi: 10.1016/0898-6568(96)00067-8. [DOI] [PubMed] [Google Scholar]
- 25.Matsuda M, Tanaka S, Nagata S, Kojima A, Kurata T, Shibuya M. Two species of human CRK cDNA encode proteins with distinct biological activities. Mol Cell Biol. 1992;12:3482–3489. doi: 10.1128/mcb.12.8.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayer B J, Hamaguchi M, Hanafusa H. A novel viral oncogene with structural similarity to phospholipase C. Nature. 1988;332:272–275. doi: 10.1038/332272a0. [DOI] [PubMed] [Google Scholar]
- 27.Mayer B J, Hanafusa H. Association of the v-crk oncogene product with phosphotyrosine-containing proteins and protein kinase activity. Proc Natl Acad Sci USA. 1990;87:2638–2642. doi: 10.1073/pnas.87.7.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minden A, Karin M. Regulation and function of the JNK subgroup of MAP kinases. Biochim Biophys Acta. 1997;1333:85–104. doi: 10.1016/s0304-419x(97)00018-8. [DOI] [PubMed] [Google Scholar]
- 29.Minden A, Lin A, Claret F-X, Abo A, Karin M. Selective activation of the JNK signaling cascade and c-Jun transcription activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 30.Minden A, Lin A, Smeal T, Derijard B, Cobb M, Davis R, Karin M. c-Jun N-terminal phosphorylation correlates with activation of the JNK subgroup but not the ERK subgroup of mitogen-activated protein kinases. Mol Cell Biol. 1994;14:6683–6688. doi: 10.1128/mcb.14.10.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muegge K, Williams T M, Kant J, Karin M, Chiu R, Schmidt A, Siebenlist U, Young H A, Durum S K. Interleukin-1 costimulatory activity on the interleukin-2 promoter via AP-1. Science. 1989;246:249–251. doi: 10.1126/science.2799385. [DOI] [PubMed] [Google Scholar]
- 32.Nichols G, Raines M, Vera J, Lacomis L, Tempst P, Golde D. Identification of CRKL as the constitutively phosphorylated 39-kD tyrosine phosphoprotein in chronic myelogenous leukemia cells. Blood. 1994;84:2912–2918. [PubMed] [Google Scholar]
- 33.Northrop J P, Ullman K S, Crabtree G R. Characterization of the nuclear and cytoplasmic components of the lymphoid-specific nuclear factor of activated T cells (NF-AT) complex. J Biol Chem. 1993;268:2917–2923. [PubMed] [Google Scholar]
- 34.Oda T, Heaney C, Hagopian J, Okuda K, Griffin J, Druker B. Crk1 is the major tyrosine-phosphorylated protein in neutrophils from patients with chronic myelogenous leukemia. J Biol Chem. 1994;269:22925–22928. [PubMed] [Google Scholar]
- 35.Pawson T. Protein modules and signalling networks. Nature. 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 36.Pombo C M, Kehrl J H, Irma S, Woodgett J R, Force T, Kyriakis J M. Activation of the SAPK pathway by the human STE20 homogoeu germinal centre kinase. Nature. 1995;377:750–754. doi: 10.1038/377750a0. [DOI] [PubMed] [Google Scholar]
- 37.Reichman C, Mayer B, Keshav S, Hanafusa H. The product of the cellular crk gene consists primarily of SH2 and SH3 regions. Cell Growth Differ. 1992;3:451–460. [PubMed] [Google Scholar]
- 38.Ren R, Ye Z S, Baltimore D. Abl protein-tyrosine kinase selects the Crk adapter as a substrate using SH3-binding sites. Genes Dev. 1994;8:783–795. doi: 10.1101/gad.8.7.783. [DOI] [PubMed] [Google Scholar]
- 39.Salmeron A, Ahmad T B, Carlile G W, Pappin D, Narsimhan R P, Ley S C. Activation of MEK-1 and SEK-1 by Tpl-2 proto-oncogene, a novel MAP kinase kinase kinase. EMBO J. 1996;15:817–826. [PMC free article] [PubMed] [Google Scholar]
- 40.Sanchez I, Hughes R T, Mayer B J, Yee K, Woodgett J R, Avruch J, Kyriakis J M, Zon L I. Role of SAPK/ERK kinase-1 in the stress-activated pathway regulating transcription factor c-Jun. Nature. 1994;372:794–798. doi: 10.1038/372794a0. [DOI] [PubMed] [Google Scholar]
- 41.Sawasdikosol S, Chang J H, Pratt J C, Wolf G, Shoelson S E, Burakoff S J. Tyrosine-phosphorylated Cbl binds to Crk after T cell activation. J Immunol. 1996;157:110–116. [PubMed] [Google Scholar]
- 42.Schaller M D, Parsons J T. pp125FAK-dependent tyrosine phosphorylation of paxillin creates a high-affinity binding site for Crk. Mol Cell Biol. 1995;15:2635–2645. doi: 10.1128/mcb.15.5.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwartz R H. Costimulation of T lymphocytes: the role of CD28, CTLA-4, and B7/BB1 in interleukin-2 production and immunotherapy. Cell. 1992;71:1065–1068. doi: 10.1016/s0092-8674(05)80055-8. [DOI] [PubMed] [Google Scholar]
- 44.Senechal K, Halpern J, Sawyers C L. The CRKL adaptor protein transforms fibroblasts and functions in transformation by the BCR-ABL oncogene. J Biol Chem. 1996;271:23255–23261. doi: 10.1074/jbc.271.38.23255. [DOI] [PubMed] [Google Scholar]
- 45.Senechal K, Heaney C, Druker B, Sawyers C L. Structural requirements for function of the Crk1 adapter protein in fibroblasts and hematopoietic cells. Mol Cell Biol. 1998;18:5082–5090. doi: 10.1128/mcb.18.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Serfling E, Barthelmas R, Pfeuffer I, Schenk B, Zarius S, Swoboda R, Mercurio F, Karin M. Ubiquitous and lymphocyte-specific factors are involved in the induction of the mouse interleukin 2 gene in T lymphocytes. EMBO J. 1989;8:465–473. doi: 10.1002/j.1460-2075.1989.tb03399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sluss H K, Barrett T, Derijard B, Davis R J. Signal transduction by tumor necrosis factor mediated by JNK protein kinases. Mol Cell Biol. 1994;14:8376–8384. doi: 10.1128/mcb.14.12.8376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sparks A B, Rider J E, Hoffman N G, Fowlkes D M, Quilliam L A, Kay B K. Distinct ligand preferences of Src homology 3 domains from Src, Yes, Abl, Cortactin, p53bp2, PLCγ, Crk, and Grb2. Proc Natl Acad Sci USA. 1996;93:1540–1544. doi: 10.1073/pnas.93.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su B, Jacinto E, Hibi M, Kallunki T, Karin M, Ben-Neriah Y. JNK is involved in siganl integration during costimulation of T lymphocytes. Cell. 1994;77:727–736. doi: 10.1016/0092-8674(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 50.Su Y-C, Han J, Xu S, Cobb M, Skolnik E Y. NIK is a new Ste20-related kinase that binds NCK and MEKK1 and activates the SAPK/JNK cascade via a conserved regulatory domain. EMBO J. 1997;16:1279–1290. doi: 10.1093/emboj/16.6.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanaka M, Gupta R, Mayer B J. Differential inhibition of signaling pathways by dominant-negative SH2/SH3 adapter proteins. Mol Cell Biol. 1995;15:6829–6837. doi: 10.1128/mcb.15.12.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanaka S, Morishita T, Hashimoto Y, Hattori S, Nakamura S, Shibuya M, Matuoka K, Takenawa T, Kurata T, Nagashima K. C3G, a guanine nucleotide-releasing protein expressed ubiquitously, binds to the Src homology 3 domains of CRK and GRB2/ASH proteins. Proc Natl Acad Sci USA. 1994;91:3443–3447. doi: 10.1073/pnas.91.8.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanaka S, Ouchi T, Hanafusa H. Downstream of Crk adaptor signaling pathways: activation of Jun kinase by v-Crk through the guanine nucleotide exchange protein C3G. Proc Natl Acad Sci USA. 1997;94:2356–2361. doi: 10.1073/pnas.94.6.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.ten Hoeve J, Kaartinen V, Fioretos T, Voncken J, Heisterkamp N, Groffen J. Cellular interactions of CRKL, and SH2-SH3 adaptor protein. Cancer Res. 1994;54:2563–2567. [PubMed] [Google Scholar]
- 55.ten Hoeve J, Morris C, Heisterkamp N, Groffen J. Isolation and chromosomal localization of CRKL, a human crk-like gene. Oncogene. 1993;8:2469–2474. [PubMed] [Google Scholar]
- 56.Teramoto H, Coso O A, Miyata H, Igishi T, Miki T, Gutkind J S. Signaling from the small GTP-binding proteins Rac1 and Cdc42 to the c-Jun N-terminal kinase/stress-activated protein kinase pathway: a role for mixed lineage kinase 3/protein-tyrosine kinase 1, a novel member of the mixed lineage kinase family. J Biol Chem. 1996;271:27225–27228. doi: 10.1074/jbc.271.44.27225. [DOI] [PubMed] [Google Scholar]
- 57.Tournier C, Whitmarsh A J, Vavanagh J, Barret T, Davis R J. Mitogen-activated protein kinase kinase 7 is an activator of the c-Jun NH2-terminal kinase. Proc Natl Acad Sci USA. 1997;94:7337–7342. doi: 10.1073/pnas.94.14.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tung RM, Blenis J. A novel human SPS1/STE20 homologue, KHS, activates Jun N-terminal kinase. Oncogene. 1997;14:653–659. doi: 10.1038/sj.onc.1200877. [DOI] [PubMed] [Google Scholar]
- 59.Ullman K S, Northrop J P, Admon A, Crabtree G R. Jun family members are controlled by a calcium-regulated, cyclosporin A-sensitive signaling pathway in activated T lymphocytes. Genes Dev. 1993;7:188–196. doi: 10.1101/gad.7.2.188. [DOI] [PubMed] [Google Scholar]
- 60.Wang W, Zhou G, Hu M C-T, Yao Z, Tan T-H. Activation of the hematopoietic progenitor kinase-1 (HPK1)-dependent, stress-activated c-Jun N-terminal kinase (JNK) pathway by transforming growth factor β (TGF-β)-activated kinase (TAK1), a kinase mediator of TGF-β signal transduction. J Biol Chem. 1997;272:22771–22775. doi: 10.1074/jbc.272.36.22771. [DOI] [PubMed] [Google Scholar]
- 61.Whitmarsh A J, Cavanagh J, Tournier C, Yasuda J, Davis R J. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;281:1671–1674. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 62.Whitmarsh A J, Davis R J. Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J Mol Med. 1996;74:589–607. doi: 10.1007/s001090050063. [DOI] [PubMed] [Google Scholar]
- 63.Wu Z, Wu J, Jacinto E, Karin M. Molecular cloning and characterization of human JNKK2, a novel Jun NH2-terminal kinase-specific kinase. Mol Cell Biol. 1997;17:7407–7416. doi: 10.1128/mcb.17.12.7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yao Z, Diener K, Wang X S, Zukowski M, Matsumoto G, Zhou G, Tibbles L A, Sasaki T, Nishina H, Tan T-H, Woodgett J, Penninger J M. Activation of stress-activated protein kinase/c-Jun N-terminal protein by a novel mitogen-activated protein kinase kinase (MKK7) J Biol Chem. 1997;272:32378–32383. doi: 10.1074/jbc.272.51.32378. [DOI] [PubMed] [Google Scholar]
- 65.Yao, Z., G. Zhou, X.S. Wang, A. Brown, K. Diener, H. Gan, and T.-H. Tan. A novel human STE20-related protein, HGK, that specifically activates the JNK signaling pathway. J. Biol. Chem., in press. [DOI] [PubMed]
- 66.Yu H, Chen J K, Feng S, Dalgarno D C, Brauer A W, Schreiber S L. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]