Summary
Starch is synthesized as insoluble, semicrystalline particles within plant chloroplast and amyloplast, which are referred to as starch grains (SGs). The size and morphology of SGs in the cereal endosperm are diverse and species–specific, representing a key determinant of the suitability of starch for industrial applications. However, the molecular mechanisms modulating SG size in cereal endosperm remain elusive. Here, we functionally characterized the rice (Oryza sativa) mutant substandard starch grain7 (ssg7), which exhibits enlarged SGs and defective endosperm development. SSG7 encodes a plant–specific DUF1001 domain‐containing protein homologous to Arabidopsis (Arabidopsis thaliana) CRUMPLED LEAF (AtCRL). SSG7 localizes to the amyloplast membrane in developing endosperm. Several lines of evidence suggest that SSG7 functions together with SSG4 and SSG6, known as two regulators essential for SG development, to control SG size, by interacting with translocon‐associated components, which unveils a molecular link between SG development and protein import. Genetically, SSG7 acts synergistically with SSG4 and appears to be functional redundancy with SSG6 in modulating SG size and endosperm development. Collectively, our findings uncover a multimeric functional protein complex involved in SG development in rice. SSG7 represents a promising target gene for the biotechnological modification of SG size, particularly for breeding programs aimed at improving starch quality.
Keywords: rice (Oryza sativa), endosperm, starch, starch grain size, translocon
Introduction
Starch provides the primary source of energy in the daily diet for both humans and animals and has extensive industrial applications (Huang et al., 2021). The endosperm of cereals accumulates substantial starch reserves in amyloplasts, where starch forms insoluble grains (starch grains [SGs]) that occupy most of the interior of amyloplasts. Despite the evolutionary conservation of the internal structure of SGs, native SGs display diverse sizes and morphologies depending on the plant species and organ (James et al., 2003; Jane et al., 1994; Matsushima et al., 2010; Seung and Smith, 2019; Zhao et al., 2018). Each amyloplast in rice (Oryza sativa) endosperm contains a single compound SG (10–20 μm in diameter) consisting of several dozen smaller non‐fusing starch granules (3–8 μm in diameter). By contrast, maize (Zea mays) and sorghum (Sorghum bicolor) possess simple and uniform sizes of SGs composed of a single granule (approximately 10 μm in diameter). Barley (Hordeum vulgare) and wheat (Triticum aestivum) contain both small (less than 10 μm) and large simple SGs (approximately 15–25 μm) that coexist in the same cell (Matsushima et al., 2010, 2014, 2016). As SG size is a key determinant of the suitability of starch for industrial applications, manipulating SG size is an important molecular target for bioengineering (Jobling, 2004; Lindeboom et al., 2004). However, the molecular mechanism that modulates SG size in the endosperm of cereals remains unclear.
Recent studies have identified several factors modulating SG size. A reduced number of enlarged starch granules was observed in chloroplasts of Arabidopsis (Arabidopsis thaliana) mutants defective in STARCH SYNTHASE4 (SS4) or its interactors, including non‐enzymatic proteins, which are involved in starch granule initiation in chloroplasts (Crumpton‐Taylor et al., 2012; Roldan et al., 2007; Seung et al., 2017, 2018; Vandromme et al., 2019). The soft starch (h) locus increases SG size in maize endosperm, which is beneficial for starch processing and improving starch properties (Gutiérrez et al., 2002; Wilson et al., 2000). In addition, the rice substandard starch grain4 (ssg4), ssg6, and enlarged starch grain1 (esg1) mutants develop enlarged compound SGs in the endosperm (Matsushima et al., 2014, 2016; Wang et al., 2021). SSG4 encodes a plastid‐localized protein containing a DUF490 domain (Matsushima et al., 2014). SSG6 encodes a plastidic membrane protein homologous to aminotransferase (Matsushima et al., 2016). ESG1 encodes a putative permease subunit of a bacterial‐type ATP‐binding cassette (ABC) lipid transporter (Wang et al., 2021). These three proteins also affect the size of chloroplasts but not the SGs within them. However, their precise modes of action and molecular mechanisms in determining SG size remain obscure. Notably, Arabidopsis TIC236, the homologue of rice SSG4, functions in chloroplast development by linking the TOC (translocon on the outer chloroplast membrane) and TIC (translocon on the inner chloroplast membrane) complexes, which orchestrate protein import in chloroplasts (Chen et al., 2018; Jarvis and López‐Juez, 2013; Richardson and Schnell, 2020).
Arabidopsis CRUMPLED LEAF (AtCRL) encodes a plastidic outer envelope membrane protein containing a DUF1001 domain that functions in cell division, cell differentiation, and plastid division (Asano et al., 2004). crl cells contain a reduced number of enlarged plastids, and crl plants display morphological abnormalities. AtCRL is homologous to the cyanobacterial phycocyanobilin T‐type (CpcT) lyases, which function in phycobilisome biogenesis by covalently attaching phycocyanobilin (PCB) to antenna proteins (Shen et al., 2006). AtCRL homologues are present in various plant species but not in yeast (Saccharomyces cerevisiae) or animals. Whether AtCRL homologues function in SG formation and endosperm development in cereals remains unclear.
In this study, we isolated and characterized the rice floury endosperm mutant ssg7, which develops enlarged SGs in starch‐accumulating tissues. SSG7 encodes a DUF1001 domain‐containing protein homologous to AtCRL. Our combined biochemical and genetic evidence suggest that SSG7 interacts with SSG4 and SSG6, and they function in SG development by associating with TOC or TIC components in rice. SSG7 represents a promising target gene for modifying SG size, particularly for breeding programs aimed at improving starch quality.
Results
Aberrant SG development in ssg7 endosperm
To dissect the molecular machinery involved in SG development in rice, we identified ssg7, a mutant with defective endosperm development, from an N‐methyl‐N‐nitrosourea‐mutagenized pool of japonica rice cultivar W017. Whereas wild‐type endosperm is translucent, the ssg7 mutant develops floury‐white endosperm (Figure 1a,b), which was apparent in developing grains beginning approximately 18 days after flowering (DAF) and in mature polished grains (Figure S1a,b). Scanning electron microscopy (SEM) showed that mature ssg7 endosperm contained spherical and loosely packed SGs, in sharp contrast to the polyhedral and tightly‐packed SGs in wild‐type endosperm (Figure 1c). Iodine‐stained semi‐thin sections of mature ssg7 endosperm clearly showed enlarged SGs, the area of which was approximately 4‐fold larger than that in the wild type (Figure 1d,e). However, the starch content was significantly lower in ssg7 grains than in the wild type (Figure 1f). We next examined the starch composition and the fine structure of amylopectin in ssg7 grains. Consistent with the declined starch accumulation, the amylose content was also significantly reduced (Figure S1c), indicating the altered starch composition in ssg7 grains. Analysis of the chain length distribution (CLD) of ssg7 amylopectin revealed an increase in the number of chains with a degree of polymerization (DP) of 6–8 branches and a decrease in the number of chains with a DP of 9–20 (except for DP13) branches (Figure S1d). The viscosity pattern of starch in ssg7 endosperm was largely similar to that of wild‐type starch but with reduced viscosity (Figure S1e), indicating that the physicochemical characteristics of the ssg7 starch were altered.
Figure 1.
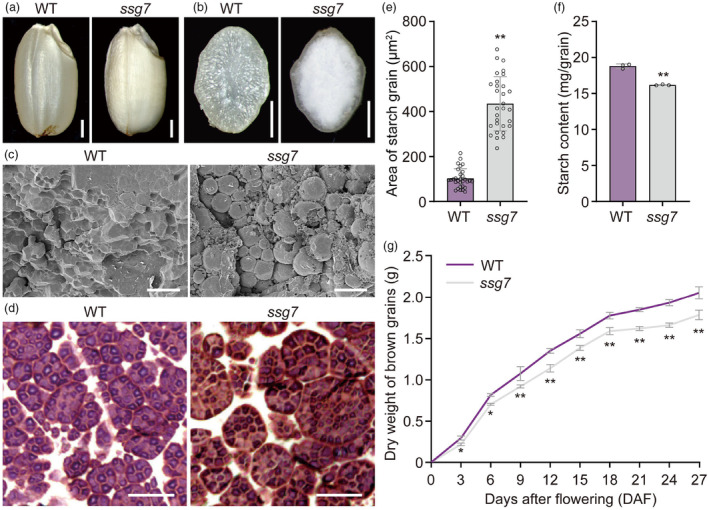
Aberrant SG development in ssg7 endosperm. (a) Representative images of wild‐type (WT) and ssg7 mature grains. Bars = 1 mm. (b) Transverse sections of the representative WT and ssg7 mature grains. Bars = 1 mm. (c) Scanning electron microscopy (SEM) images of transverse sections of the representative WT and ssg7 mature grains. Bars = 20 μm. (d) Iodine‐stained semi‐thin sections prepared from WT and ssg7 mature grains. Bars = 20 μm. (e) Quantification of the areas occupied by starch grains (SGs) in WT and ssg7 mature endosperm (n = 30). (f) Total starch contents in mature grains of WT and ssg7 (n = 3). (g) Dry substance accumulation profiles of WT and ssg7 developing grains. Data showed the weight of 100 brown grains (n = 3). Values are means ± SD. *P < 0.05, **P < 0.01 by Student's t‐test.
The ssg7 mature grains also exhibited reduced protein and lipid accumulation, indicating an important role of the SSG7 locus in storage substance accumulation (Figure S1f,g). Accordingly, the mature grain weight of ssg7 decreased by 12.78% compared to that of the wild type, which is consistent with a lower grain‐filling rate (Figure 1g; Table S1). In addition, ssg7 plants exhibited no visible differences from wild‐type plants at the vegetative stage, such as plant height and tiller number, but they exhibited a markedly reduced seed setting rate (Table S1). Collectively, the ssg7 mutation causes pleiotropic effects on reproductive development.
SSG7 plays a crucial role in modulating SG size
To investigate SG development in ssg7 endosperm, we performed cytohistological analyses of developing grains by examining periodic acid‐Schiff (PAS) and Coomassie Bright Blue (CBB) double‐stained semi‐thin sections. Enlarged SGs (stained red by PAS) and protein bodies (stained blue by CBB) were apparent in ssg7 endosperm cells at 6 DAF (Figure 2a). At 9 DAF, we observed markedly reduced numbers of even larger SGs and smaller protein bodies, together with large air spaces, in the aleurone layer and starchy endosperm cells of ssg7 (Figure 2b), which is consistent with the reduced accumulation of storage substances in the mutant (Figures 1f and S1f).
Figure 2.
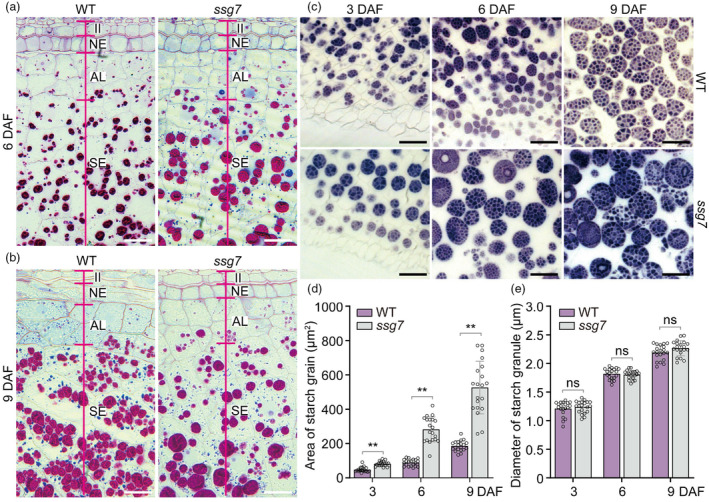
The ssg7 mutation affects starch grain development. (a, b) Light microscopy images of periodic acid‐Schiff (PAS) and Coomassie Bright Blue (CBB) double‐stained semi‐thin sections prepared from developing endosperm of the wild type (WT) and ssg7 at 6 (a) and 9 (b) DAF. DAF, days after flowering. II, inner integument; NE, nucellar epidermis; AL, aleurone layer; SE, starchy endosperm cells. Bars = 20 μm. (c) Iodine‐stained semi‐thin sections prepared from developing endosperm of WT and ssg7 at different stages. Bars = 20 μm. (d) Quantification of the areas occupied by starch grains (SGs) in semi‐thin sections of WT and ssg7 developing endosperm from 3 to 9 DAF in (c) (n = 20). Note that the quantification does not contain the very small SGs. (e) Measurement of the diameters of starch granules in WT and ssg7 developing endosperm in (c) (n = 20). Values are means ± SD. **P < 0.01 by Student's t‐test. ns, no significance.
To investigate when SG enlargement occurs in ssg7 endosperm, we analysed early‐developing grains at 3, 6, and 9 DAF. Enlarged SGs were visible as early as 3 DAF in ssg7 endosperm, occupying an area that was more than 1.7‐fold larger than that of wild‐type SGs. At 6 and 9 DAF, the area occupied by enlarged SGs was more than 2.8‐ and 3.2‐fold larger in ssg7 than the wild type, respectively (Figure 2c,d). By contrast, the size of starch granules in ssg7 was comparable to that in the wild type during endosperm development (Figure 2e). Therefore, the enlargement of SGs was due to an increased number of starch granules per SG in ssg7, suggesting that more granule initiation events likely occurred in a single amyloplast. Notably, clusters of smaller SGs coexisted with the enlarged SGs in ssg7 endosperm cells at 9 DAF (Figure 2c), as observed in the previously reported ssg6 mutant (Matsushima et al., 2016). Quantitative analysis revealed that the total numbers of SGs and granules per area were lower in ssg7 than in the wild type (Figure S2), in agreement with the reduced starch content in ssg7 (Figure 1f). In addition, the enlarged SGs in ssg7 were more spherically shaped (Figure 2c), as observed in ssg4, ssg6, and esg1 mutants (Matsushima et al., 2014, 2016; Wang et al., 2021).
As endosperm is a triploid tissue, F1 grains with different doses of the SSG7 allele can be generated by performing reciprocal crosses between the wild type and ssg7. The appearance of endosperm and the sizes of SGs in the two reciprocal F1 hybrid grains were similar to those of the wild type (Figure S3), indicating that the SSG7 allele has no apparent gene dosage effect on regulating SG size.
We also observed enlarged SGs in pollen grains and young pericarp of ssg7 (Figure S4). Unexpectedly, unlike the ssg4, ssg6, and esg1 mutants (Matsushima et al., 2014, 2016; Wang et al., 2021), ssg7 plants developed normal leaves with similar chloroplast areas compared to those of the wild type, although some chloroplasts appeared more spherical than normal (Figure S5). Altogether, these results suggest that the ssg7 mutation affects SG development in rice.
SSG7 encodes an AtCRL homologue and is generally expressed in rice
Genetic analysis indicated that the ssg7 mutation behaves as a single recessive allele (Table S2). To clone the causal gene responsible for the ssg7 phenotypes, we constructed an F2 segregating population by crossing ssg7 with the indica cultivar N22. Using 528 F2 homozygous ssg7 individuals, the ssg7 locus was delimited to a 158‐kb genomic interval flanked by the simple sequence repeat (SSR) markers RM229 and Yhg‐5 on chromosome 11 (Figure 3a). Annotation of this 158‐kb genomic region identified 17 putative open reading frames (ORFs; Figure 3a). Sequencing analysis revealed a single nucleotide substitution adjacent to the splicing donor site of the 7th intron of LOC_Os11g32160, leading to alternative splicing in ssg7 (Figure 3b,c). We detected two distinct transcripts from ssg7 grains by RT‐PCR analysis. The major transcript was found to retain 132 bp of intron 7, which was predicted to result in a frameshift and a premature stop codon, while the minor transcript corresponded to the wild‐type transcript (Figure 3b–d).
Figure 3.
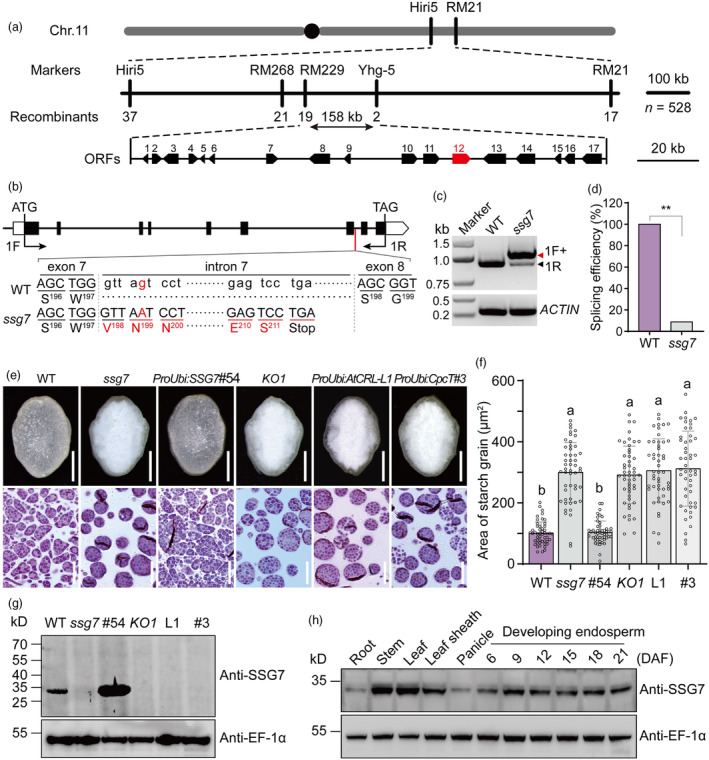
Map‐based cloning of the SSG7 gene and characterization of the SSG7 protein. (a) Fine mapping of the SSG7 locus. The SSG7 locus was narrowed to a 158‐kb interval on chromosome 11. The numbers under vertical lines and blocks at the bottom indicate the recombinant plants and candidate genes, respectively. Note that the SSG7 gene was highlighted in red. Chr, chromosome; ORFs, open reading frames. (b) Genomic structure and the mutation site of SSG7. Exons are shown as boxes, and introns as lines. The position of start (ATG) and stop (TAG) codons are indicated. A G‐to‐A substitution occurred in the 7th intron of SSG7, leading to alternative splicing in ssg7. WT, wild type. (c) Determination of SSG7 transcripts in WT and ssg7. The unspliced transcript containing the 7th intron was indicated by a red arrowhead, whereas the spliced WT‐like transcript was indicated by a black arrowhead. The primer pair (1F + 1R) in (b) was used for amplification. Rice Actin gene was used as an internal control. (d) Effect of the ssg7 mutation on splicing efficiency. RT‐PCR products from three independent experiments were subjected to sequence analysis to assess the splicing efficiency. **P < 0.01 by Student's t‐test. (e) Rescue of grain appearance (upper panels) and starch grain (SG) sizes (lower panels) of ssg7 by overexpressing SSG7 driven by the UBIQUITIN promoter, while a knockout mutant KO1 carrying a 21‐bp deletion phenocopied the ssg7 mutant. Phenotypes of AtCRL and CpcT overexpression grains in the ssg7 background were not recovered. Bars = 1 mm in upper panels, 20 μm in lower panels. (f) Quantification of the areas occupied by SGs in developing endosperm of WT, ssg7, and transgenic plants. Values are means ± SD (n = 25). P < 0.05 by Duncan's multiple range tests. Note that the quantification does not contain the very small SGs. (g) SSG7 antibodies could specifically detect endogenous SSG7 protein in total protein extracts from WT and SSG7 overexpression lines, but not in ssg7 and other transgenic plants, as indicated. EF‐1α was used as a loading control. (h) Protein accumulation profiles of SSG7 in various tissues and developing endosperm. DAF, days after flowering. EF‐1α was used as a loading control.
We conducted a complementation test by introducing the coding sequence of LOC_Os11g32160 driven by the UBIQUITIN promoter into ssg7 calli. As anticipated, transgene‐positive grains exhibited a wild‐type translucent appearance and developed normal‐sized SGs (Figures 3e,f and S6). Furthermore, we performed CRISPR/Cas9 gene editing to knock out LOC_Os11g32160. The knockout mutant KO1, with a 21‐bp deletion in LOC_Os11g32160, phenocopied ssg7, while two other mutants with frameshift mutations, named KO2 and KO3, were heterozygous at this locus, most likely due to defects in male gamete transmission (Figure S7a,b). Notably, pollen from the heterozygous KO2 plant was segregated with respect to SG morphology (Figure S7c). The segregation ratio of pollen containing simple or compound SGs was approximately 1:1 (Figure S7d), indicating a specific role of SSG7 in the compound SG formation. We raised polyclonal antibodies against SSG7, which specifically recognized a predicted ~30‐kD band in total protein extracts from developing grains of wild‐type and transgenic overexpression plants, but not the knockout mutant KO1 (Figure 3g). Little endogenous SSG7 protein was detected in ssg7, consistent with the low abundance of normal transcripts (Figures 3d,g and S8). Taken together, these results substantiate that LOC_Os11g32160 corresponds to the SSG7 gene.
SSG7 is predicted to encode a protein of 275 amino acids that harbours a putative transmembrane domain and a large DUF1001 domain (Figure S9a). Phylogenetic analysis indicated that SSG7 is a single copy gene in the rice genome that is homologous to AtCRL (Figure S9b; Asano et al., 2004). Notably, the function of SSG7 could have diverged from that of cyanobacterial CpcT lyase, as evidenced by their low sequence identity (Figure S9b,c). Supporting this notion, overexpressing AtCRL or cyanobacterial CpcT in the ssg7 background did not rescue the ssg7 mutant phenotypes (Figures 3e–g and S6). These data suggest that functional divergence has extensively occurred among AtCRL homologues during evolution.
We then used the anti‐SSG7 antibodies to analyse the accumulation pattern of SSG7 via immunoblot analysis of total protein extracts from different tissues of wild‐type plants. SSG7 was widely present in all tissues examined, with the highest accumulation in leaf and stem tissues (Figure 3h). In developing endosperm, SSG7 remained abundant until the late stage of development (Figure 3h). Together, these data suggest that SSG7 encodes the AtCRL homologue in rice and it is broadly expressed in various tissues.
SSG7 localizes to the amyloplast membrane and can form homodimers
To investigate the subcellular localization of SSG7, we separately introduced two fusion constructs into ssg7 calli: constructs harbouring SSG7‐GFP (Green fluorescent protein, fused with the C terminus of SSG7) or GFP‐SSG7 (GFP fused with the N terminus of SSG7) driven by its native promoter. The phenotypes of transgenic grains harbouring GFP‐SSG7 but not SSG7‐GFP fusion protein were restored to the wild‐type level (Figure S10), indicating that the GFP‐SSG7 fusion protein was biologically functional in vivo. Unfortunately, we failed to detect any fluorescent signals in developing endosperm, most likely due to low abundance of SSG7 in developing endosperm tissue (Figure 3h). As an alternative, we introduced GFP‐SSG7 driven by the UBIQUITIN promoter into ssg7 calli, and the results showed that the transgene‐positive grains exhibited wild‐type phenotypes (Figure S11). Given the predicted transmembrane domain in SSG7 and the defective SG development in ssg7 endosperm (Figures 1c,d, 2a–c and S9a), we reasoned that SSG7 might be an amyloplast membrane‐localized protein. As expected, GFP‐SSG7 exhibited a ring‐like localization pattern with its GFP signals surrounding the developing SGs in developing endosperm cells (Figure 4a), verifying that SSG7 may be an amyloplast membrane‐localized protein. When coexpressed with fluorescent marker proteins either for the plastid outer envelope membrane (OEP7‐DsRed) or for the inner envelope membrane (BRITTLE‐1 [BT1]‐DsRed) (Kawagoe, 2013; Yun and Kawagoe, 2010), confocal microscopy analyses revealed that GFP‐SSG7 co‐localized well with OEP7‐DsRed but not with BT1‐DsRed in developing subaleurone cells of rice endosperm (Figure 4b,c).
Figure 4.
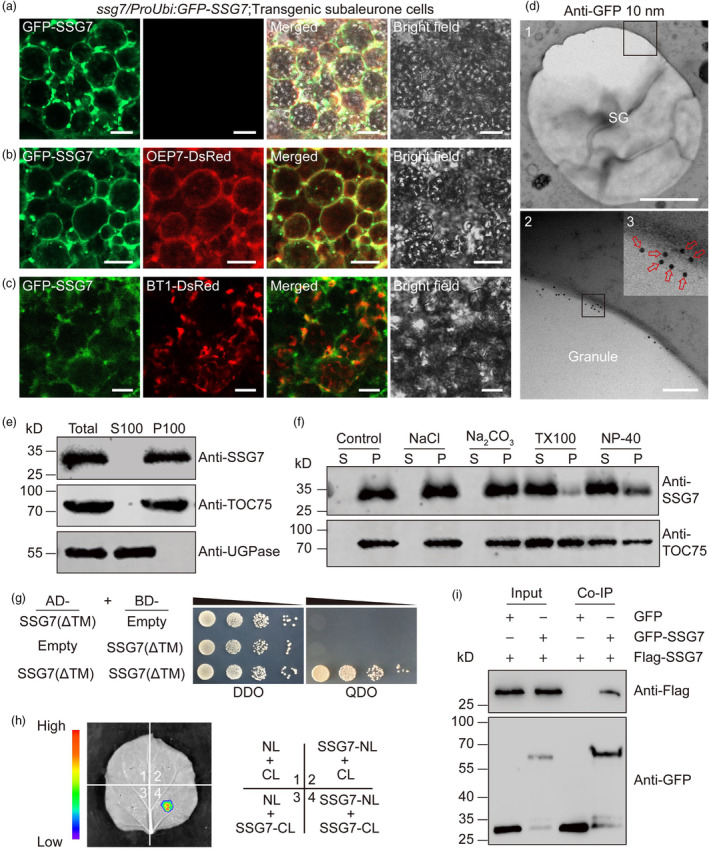
SSG7 localizes to the amyloplast membrane and can form homodimers in planta. (a) Representative confocal microscopy images showing that GFP‐SSG7 localizes to the periphery of amyloplasts and exhibits numerous ring‐like structures in developing subaleurone cells. Bars = 10 μm. (b, c) Representative confocal microscopy images showing that GFP‐SSG7 largely co‐localizes with OEP7‐DsRed (b), but not BT1‐DsRed (c) in developing subaleurone cells. Bars = 10 μm. (d) Immuno‐gold localization of GFP‐SSG7 in transgenic subaleurone cells. Ultrathin sections were prepared from HPF/FS samples of transgenic complemented GFP‐SSG7 grains, followed by hybridization with the monoclonal GFP antibody in combination with 10‐nm gold particle‐coupled secondary antibodies. The black boxes are the selected regions for magnification. Red arrows indicate the gold particles. SG, starch grain. Bars = 2 μm in panel 1; 200 nm in panel 2. (e) Subcellular fractionation assays showed that SSG7 is a membrane protein. Total protein was extracted from wild‐type developing endosperm, followed by centrifugation and fractionation into supernatant (S100, soluble fraction) and pellet (P100, membrane‐associated fraction). Both fractions were subjected to immunoblot analyses with antibodies against SSG7, an amyloplast outer envelope membrane protein TOC75, and a cytosol protein UDP‐glucose pyrophosphorylase (UGPase), respectively. (f) SSG7 is an integral membrane protein. The P100 fraction was resuspended with the different buffers and fractionated into supernatant (S) and pellet (P) fractions. Immunoblot analyses were performed with anti‐SSG7 and anti‐TOC75 antibodies, respectively. (g) Yeast two‐hybrid assay showing that SSG7 variant with a deletion of the TM domain interacts with itself. DDO, SD/‐Trp/‐Leu; QDO, SD/‐Trp/‐Leu/‐His/‐Ade. TM, putative transmembrane domain. (h) Firefly luciferase complementation imaging (LCI) assay showing that SSG7 forms a homodimer in leaf epidermal cells of N. benthamiana. NL, N terminus of LUC; CL, C terminus of LUC. (i) Co‐immunoprecipitation (Co‐IP) assay confirmed the association of SSG7 with itself. Rice protoplasts expressing GFP‐SSG7 or free GFP with Flag‐SSG7 were subjected to total extraction and IP with GFP magnetic beads followed by immunoblot analysis with anti‐GFP and anti‐Flag antibodies, respectively.
To further verify the intracellular localization of SSG7 in developing endosperm, we conducted immuno‐gold electron microscopy of ultrathin sections prepared from developing endosperm of GFP‐SSG7‐complemented lines using the primary anti‐GFP antibody in combination with 10‐nm gold particle‐coupled secondary antibodies. Consistent with our confocal microscopic observations (Figure 4a,b), the gold particles were largely detected on the surfaces of SGs (Figure 4d). Furthermore, the subcellular fractionation assay showed that SSG7 was enriched in the membrane‐associated pellet fraction (Figure 4e), and was able to be extracted from the pellet fraction after detergent treatment but not NaCl or Na2CO3 treatment (Figure 4f). Taken together, these results indicate that SSG7 is an integral membrane protein of amyloplasts.
AtCRL has been shown to be able to form homodimers in vivo and in vitro (Wang et al., 2020). To test whether rice SSG7 could form homodimers, we performed a yeast two‐hybrid assay and confirmed that SSG7 could interact with itself. Consistent with this result, a firefly luciferase complementation imaging (LCI) assay in leaf epidermal cells of Nicotiana benthamiana and an in vivo coimmunoprecipitation (Co‐IP) assay in rice protoplasts confirmed that SSG7 is able to form homodimers (Figure 4g–i).
SSG7 associates with SSG4 and SSG6
To dissect the molecular mechanism by which SSG7 modulates SG development, we performed immunoprecipitation coupled to mass spectrometry (IP‐MS) using protein extracts from developing endosperm of transgenic plants expressing either GFP‐SSG7 or free GFP. We subjected the immunoprecipitates to SDS‐PAGE, followed by CBB staining (Figure 5a) and LC–MS/MS analysis (Data S1). Intriguingly, we detected SSG4 and SSG6, known as two important regulators essential for SG development (Matsushima et al., 2014, 2016), in the GFP‐SSG7 precipitate but not in the free GFP precipitate (Figure 5b), suggesting that SSG7 forms a functional protein complex with SSG4 and SSG6 to regulate SG development. We verified the interaction between SSG7 and SSG6 by immunoblot analysis of the GFP‐SSG7 precipitate and an in vivo LCI assay in leaf epidermal cells of N. benthamiana (Figure 5c,d).
Figure 5.
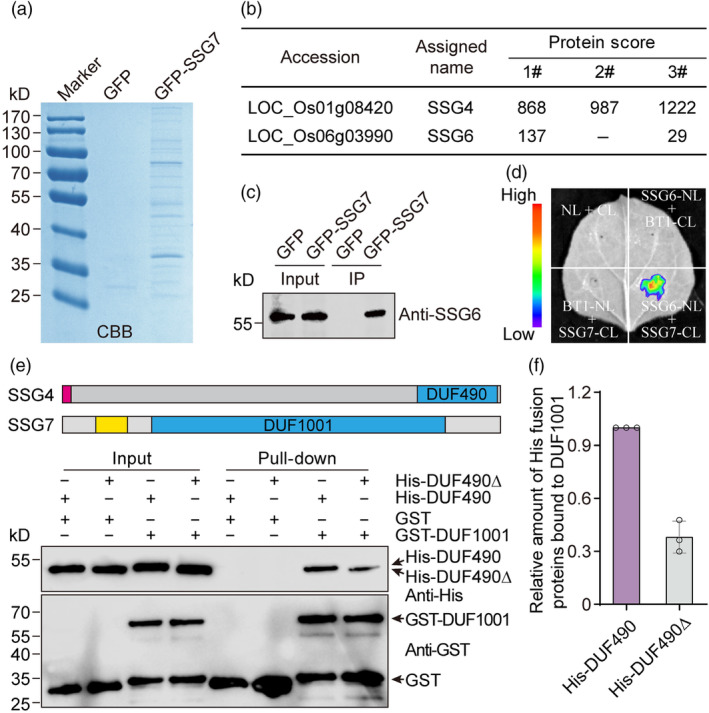
SSG7 interacts with both SSG4 and SSG6. (a) CBB‐stained SDS‐PAGE gel. Developing endosperm of transgenic rice expressing either GFP‐SSG7 or free GFP driven by the UBIQUITIN promoter was used for protein extraction. The extracts were separately incubated with GFP μMACS Microbeads and the bound proteins were subjected to SDS‐PAGE analysis. CBB, Coomassie Brilliant Blue. (b) Summary of proteins co‐precipitated with GFP‐SSG7 and identified by mass spectrometry. (c) Immunoblot analysis of the GFP‐SSG7 immunoprecipitate samples obtained from developing endosperm of transgenic rice with antibodies against SSG6. (d) Firefly luciferase complementation imaging (LCI) assay showing that SSG7 specifically associates with SSG6 in leaf epidermal cells of N. benthamiana. NL, N terminus of LUC; CL, C terminus of LUC. BT1 was used as a negative control. (e) In vitro GST pull‐down assay showing that GST‐tagged DUF1001 domain of SSG7 but not free GST can pull down His‐tagged C‐terminal DUF490 domain of SSG4. The structures of SSG4 and SSG7 proteins were shown. The DUF1001 domain of SSG7 and the C‐terminal DUF490 domain of SSG4 are highlighted in blue. The red box and yellow box represent the signal peptide of SSG4 and putative transmembrane domain of SSG7, respectively. The grey boxes represent the regions of SSG4 and SSG7 proteins containing no previously annotated domains. Note that a deletion of the C‐terminal tail of the DUF490 domain largely compromised this interaction. Black arrows indicate the corresponding recombinant proteins. DUF, domain of unknown function. (f) Quantification of the relative amounts of His fusion proteins bound to GST‐DUF1001. The intensity of pulled‐down His‐DUF490∆ was normalized by the intensity of pulled‐down His‐DUF490 using Image J software from three independent experiments.
Due to the strong inhibition of the full‐length SSG4 sequence on bacterial growth (Matsushima et al., 2014), we failed to clone the full‐length SSG4 gene. A previous study from Arabidopsis SSG4 homologue TIC236 showed that it is an integral inner envelope membrane protein, and could directly interact with the outer envelope membrane protein TOC75 via its C‐terminal DUF490 domain localized in the intermembrane space (Figure S12a; Chen et al., 2018). Interestingly, protein topology prediction showed that the C‐terminal DUF1001 domain of SSG7 might be exposed to the intermembrane space (Krogh et al., 2001). Based on these clues, we proposed that SSG7 might interact with SSG4 in a manner similar to that of Arabidopsis TOC75 (Chen et al., 2018). To verify this notion, we performed an in vitro pull‐down assay, and found that His‐DUF490 could bind to glutathione S‐transferase (GST)‐DUF1001 but not to free GST (Figure 5e). This interaction was largely compromised when the last 16 amino acid residues of the DUF490 domain were truncated (Figure 5e,f), which is in agreement with the effects of the C‐terminal tail of the DUF490 domain on protein–protein interactions (Chen et al., 2018; Selkrig et al., 2012). Collectively, these results indicate that SSG7 forms a functional protein complex with SSG4 and SSG6 to regulate SG development in rice.
SSG7 physically interacts with the translocon complex
It is reported that Arabidopsis TIC236/SSG4 functions in plastid development by linking the TOC and TIC complexes (Chen et al., 2018). We proposed that SSG4 and SSG7 might also be associated with translocons in rice. Consistent with this proposition, our LC–MS/MS assay indeed detected diverse TOC core components in the GFP‐SSG7 precipitate (Figure 6a). They included TOC159, TOC120, TOC90, TOC75, TOC64, and TOC34, with the highest protein score obtained for TOC75. Furthermore, immunoblot analyses of the GFP‐SSG7 precipitate using protein‐specific antibodies confirmed the presence of TOC34 and TOC75 (Figure 6b). A small number of peptides for TIC110, known as a core component of the TIC complex (Jackson et al., 1998), were also specifically detected in the GFP‐SSG7 precipitate (Figure 6a). In addition, immunoblot analyses showed that the protein abundances of TOC75 and TOC34 were decreased by 37.5% and 56.5% in developing ssg7 grains, respectively, compared to the wild type (Figure 6c,d). These results, combined with the finding that SSG7 interacts with SSG4 and SSG6, suggest that SSG7 functions together with SSG4 and SSG6 in regulating SG development via their association with translocons.
Figure 6.
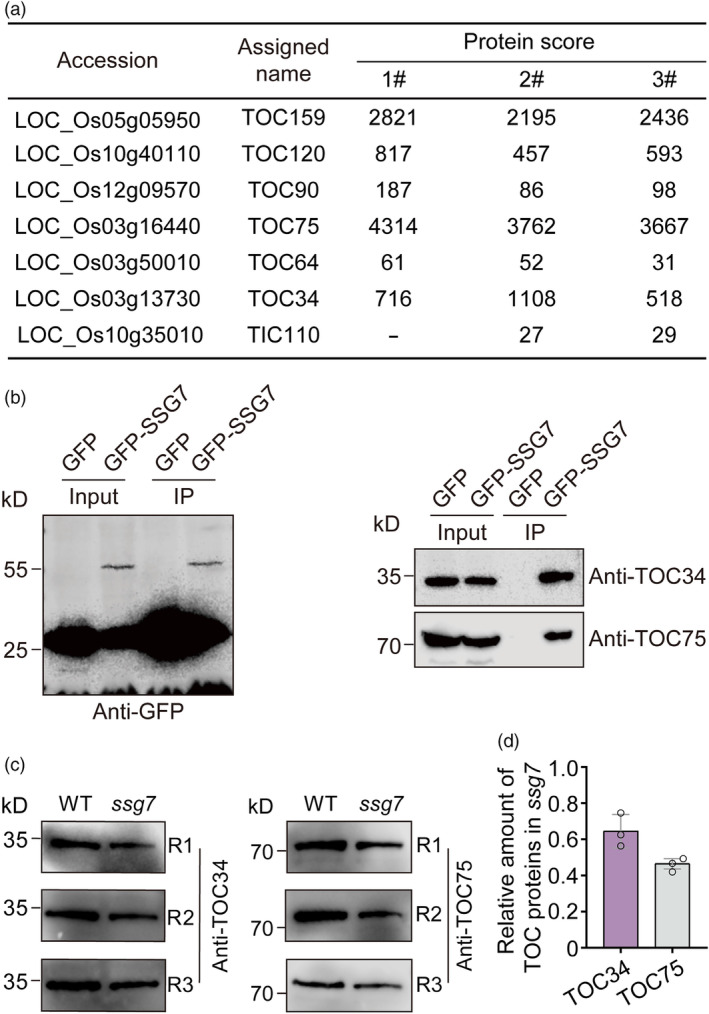
SSG7 interacts with the translocon components. (a) Summary of proteins co‐precipitated with GFP‐SSG7 and identified by mass spectrometry. Developing endosperm of transgenic rice expressing either GFP‐SSG7 or free GFP driven by the UBIQUITIN promoter was used for protein extraction. (b) Immunoblotting of the GFP‐SSG7 immunoprecipitate samples obtained from developing endosperm of transgenic rice with antibodies against translocon components. (c) Immunoblot analysis of total extracts from developing grains of the wild type (WT) and ssg7 using antibodies against TOC34 and TOC75, respectively. Equivalent volumes of total protein extracts from the same weight of grains were loaded in each lane. R1 to R3 indicate three times independent experiments. (d) Quantification of the relative amounts of translocon components in the ssg7 mutant. The intensity of translocon components in ssg7 was normalized by the corresponding intensity of those in WT using Image J software.
SSG7 genetically interacts with SSG4 and SSG6 to modulate SG development
We tried to investigate genetic relationship of SSG7 with SSG4 and SSG6. A recent study showed that a gain‐of‐function mutation of TIC236/SSG4 could rescue the chloroplast developmental defect of the Arabidopsis crl/ssg7 mutant (Fang et al., 2022). However, due to the unavailable full‐length SSG4 gene (Matsushima et al., 2014), we could not overexpress SSG4 in the ssg7 background. Alternatively, we crossed ssg4 with ssg7 in an attempt to generate a ssg4 ssg7 double mutant (Figure S12; Table S3). Unfortunately, we failed to obtain a homozygous ssg4 ssg7 double mutant, most likely due to the impaired transmission efficiency of male gametophytes (Figure 7a,b). Consistent with the phenotype observed in the KO2 line of SSG7, the segregation ratio of pollen grains containing simple SGs (resembling single mutants) versus enlarged compound SGs (likely ssg4 −/− ssg7 −/− ) was close to 1:1 in ssg4 +/− ssg7 −/− and ssg4 −/− ssg7 +/− plants (Figure 7c,d). These data suggest that SSG4 and SSG7 function synergistically to modulate SG development in rice.
Figure 7.
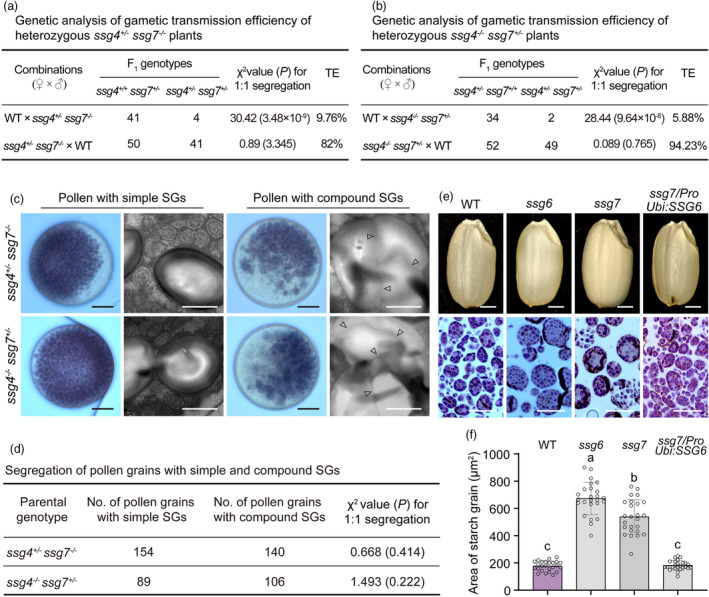
SSG7 genetically interacts with SSG4 and SSG6. (a) Genetic analysis of gametic transmission efficiency of heterozygous ssg4 +/− ssg7 −/− plants. TE, transmission efficiency; WT, wild type. (b) Genetic analysis of gametic transmission efficiency of heterozygous ssg4 −/− ssg7 +/− plants. (c) Iodine‐stained images and transmission electron microscopy (TEM) images of pollen from heterozygous ssg4 +/− ssg7 −/− and ssg4 −/− ssg7 +/− plants. Arrowheads indicate gaps between starch granules. Bars = 10 μm in iodine‐stained images, 1 μm in TEM images. SG, starch grain. (d) Segregation of pollen with simple and compound SGs from heterozygous ssg4 +/− ssg7 −/− and ssg4 −/− ssg7 +/− plants. (e, f) Appearance (e) and SG sizes (f) of WT, ssg6, ssg7, and SSG6 overexpressed grains (in the ssg7 background). P < 0.05 by Duncan's multiple range tests (n = 25). Note that the quantification does not contain the very small SGs.
We next investigated the genetic interaction between SSG7 and SSG6. Notably, when SSG6 was overexpressed in the ssg7 mutant, the transgenic grains exhibited translucent endosperm and normal‐sized SGs (Figures 7e,f and S13), suggesting that SSG6 and SSG7 share partial functional redundancy in regulating SG size in rice endosperm.
Discussion
SSG7 is particularly required for compound SG formation in rice
Starch grains size is a vital property for the industrial applications of starch, including the production of both food and non‐food products. Increased SG size could improve starch yield from maize and cassava (Manihot esculenta) during processing (Gutiérrez et al., 2002). Small SGs could be utilized as a fat substitute and for manufacturing degradable plastic film, emphasizing the importance of SG size for industrial applications (Lindeboom et al., 2004; Malinski et al., 2003). An in‐depth understanding of the molecular mechanism that governs SG size in crops is thus important.
Numerous rice mutants with altered endosperm starch synthesis have been identified and characterized. Most mutants defective in carbohydrate metabolism generally develop reduced size of SGs (Matsushima et al., 2010). By contrast, the rice ssg4, ssg6, and esg1 mutants develop enlarged chloroplasts and amyloplasts (SGs), resulting in abnormal leaf colour and chalky endosperm (Matsushima et al., 2014, 2016; Wang et al., 2021). In this study, we identified a rice ssg7 mutant containing enlarged spherical SGs in various starch‐accumulating tissues (Figures 1c,d, 2a–c and S4). Distinct from the Arabidopsis crl mutant that exhibits pleiotropic phenotypes at the vegetative stage (Asano et al., 2004), the loss of SSG7 particularly influences SG development in starch‐accumulating tissues.
During ssg7 endosperm development, a growing number of tiny SGs coexisted with enlarged SGs in the same cell, as observed in ssg6 and esg1 (Matsushima et al., 2016; Wang et al., 2021). Nevertheless, esg1 endosperm also contains small, weakly stained compound SGs, which were not observed in ssg4, ssg6, or ssg7 endosperm, suggesting that ESG1 functions in compound SG formation in a manner distinct from that of the other three SSG genes (Wang et al., 2021). The functional interaction among three SSG proteins observed in our study supports this notion (Figures 5 and 6). Furthermore, SSG4 has been proposed to control SG size in diverse starch‐accumulating tissues, like SSG7, although the requirement for SSG6 activity in modulating SG size varies in different tissues (Matsushima et al., 2016). Compared to the slightly chalky ssg6 grains, both ssg4 and ssg7 grains were highly chalky, which is consistent with the severely impaired starch biosynthesis (Figure 1f; Matsushima et al., 2014). Notably, the chain‐length distribution of amylopectin in ssg7 endosperm starch was markedly altered compared to the almost normal fine structure of amylopectin in ssg4 and ssg6 (Figure S1d; Matsushima et al., 2014, 2016). On the other hand, we could not obtain homozygous null mutants of SSG7 (Figure S7), suggesting that the ssg7 mutation in this study corresponds to a leaky mutation (Figure 3). Notably, half of the pollen grains from the heterozygous KO2 line of SSG7 contained enlarged compound SGs, although normal pollen contained simple SGs (Figure S7c,d). Altogether, these findings indicate that SSG7 has unique effects on starch biosynthesis and compound SG formation in rice.
SSG7 encodes an amyloplast membrane‐localized protein homologous to AtCRL
SSG7 encodes a DUF1001 domain‐containing protein homologous to AtCRL (Figures 3 and S9; Asano et al., 2004). The AtCRL gene was originally identified by an Arabidopsis mutant showing morphological abnormalities in many vegetative organs and developing enlarged plastids (Asano et al., 2004). Although AtCRL is homologous to the cyanobacterial CpcT lyases (Shen et al., 2006), it seems that the PCB‐binding ability of AtCRL is not functionally associated with the crl mutant phenotypes (Wang et al., 2020). Therefore, the precise molecular function of AtCRL remains elusive, despite the striking crl‐induced lesions (Li et al., 2020). The genome of the moss Physcomitrium patens harbours two homologues of AtCRL, PpCRL1, and PpCRL2, which play redundant roles in plastid division and plant growth. However, PpCRLs do not affect the orientation of the cell division plane, and AtCRL only partially rescued the defects in the plastid division of the PpCRL double knockout mutant, highlighting the divergence of CRL function in P. patens and Arabidopsis (Sugita et al., 2012).
The DUF1001 domain of SSG7 also possesses four strictly conserved amino acid residues (Figure S9c) that are required for PCB binding in AtCRL homologues (Wang et al., 2020). Consistent with the low sequence identity between SSG7 and CpcT lyase (Figure S9b,c), overexpressing CpcT failed to rescue the ssg7 phenotype (Figure 3e,f). In addition, CpcT does not function as a transporter (Shen et al., 2006), whereas AtCRL homologues in plants might have evolved other functions, including protein import (Fang et al., 2022). Moreover, SSG7 is functionally divergent from AtCRL, as evidenced by the abnormal SGs in AtCRL‐overexpressed transgenic plants (Figure 3e,f). As rice mutants defective in plastid division do not develop enlarged SGs (Yun and Kawagoe, 2009, 2010), SSG7 may not directly function in plastid division, in contrast to AtCRL and PpCRLs. Therefore, the functions of plant CRL genes have greatly diverged during evolution.
AtCRL localizes to the outer envelope membrane of chloroplasts (Asano et al., 2004). Similarly, SSG7 largely co‐localized with the amyloplast outer envelope membrane marker OEP7‐DsRed but not the inner envelope membrane marker BT1‐DsRed (Figure 4a–c). Intriguingly, only GFP‐SSG7 (but not SSG7‐GFP) rescued the ssg7 phenotype, whereas only AtCRL‐GFP or PpCRL‐GFP rescued the phenotypes of their respective mutants. In addition, SSG7 lacks a putative chloroplast transit peptide in its N‐ and C‐termini (Emanuelsson et al., 1999; Figure S9a), a characteristic of most plastidial outer envelope membrane proteins (Lee et al., 2001).
SSG7, SSG4, and SSG6 form a multivalent molecular module that regulates SG size by associating with translocons
Arabidopsis TIC236, a rice SSG4 homologue, serves as a link between the TOC and TIC complexes by interacting with TOC75 in the chloroplast intermembrane space (Chen et al., 2018). The knockdown mutant tic236‐2 develops unevenly sized chloroplasts in mesophyll cells (Fang et al., 2022). Nevertheless, the defective kernel5 (dek5) mutant of the maize SSG4 gene develops enlarged amyloplasts (SGs) and chloroplasts. The dek5 mutation decreases the amounts of TOC75 and chloroplast envelope transporters, suggesting that DEK5 functions in plastid membrane biogenesis, thereby facilitating envelope protein accumulation and metabolite transport (Zhang et al., 2019).
In our study, we verified a direct interaction between SSG4 and SSG7 in vivo and in vitro (Figure 5). SSG7 is also associated with TOC components, together with weak binding to TIC110 (Figure 6a,b; Data S1); this is similar to AtCRL, which associates with TIC236 and other translocon components (Fang et al., 2022). In addition, the ssg7 mutation markedly reduced the abundance of TOC components (Figure 6c,d). Notably, SSG4, DEK5, and TIC236 have been proposed to play a conserved role in targeting β‐barrel membrane proteins (e.g. TOC75) to the outer envelope of plastids (Zhang et al., 2019). SSG7 is also possibly involved in this process due to its direct interaction with SSG4 and TOC components (Figures 5 and 6). This may explain the reduced levels of TOC components in ssg7 (Figure 6c,d), which likely leads to defective proteins or metabolite transport, as observed in dek5 (Zhang et al., 2019). Supporting this notion, gain‐of‐function mutations of TIC236/SSG4 increase TIC236 stability and enhance the import of proteins involved in plastid division, leading to the abolishment of crl‐induced lesions (Fang et al., 2022). As we failed to clone the full‐length coding sequence of SSG4, it is challenging for us to determine whether SSG4 and SSG7 have similar genetic effects in rice. However, ssg4 and ssg7 indeed acted synergistically to modulate SG development in rice (Figure 7a–c), as previously observed for ssg4 and ssg6 (Matsushima et al., 2016).
SSG6 localizes to the outer envelope membrane of the amyloplast (Matsushima et al., 2016; Matsushima and Hisano, 2019). SSG6 likely interacts with SSG7 transiently, given that only a small amount of SSG6 peptides were detected in the GFP‐SSG7 immunoprecipitate (Figure 5b). Our data suggest that SSG4, SSG6, and SSG7 form a multi‐protein module required for SG and endosperm development. As SSG6 overexpression restored the defective SG size and endosperm development of ssg7 (Figure 7e,f), SSG6, and SSG7 may be functional redundancy in modulating SG size and endosperm development. SSG6 shares high homology with Arabidopsis aminocyclopropane‐1‐carboxylate synthase10 (ACS10) and ACS12, which are aminotransferases rather than ACS (Matsushima et al., 2016; Yamagami et al., 2003). It is unlikely that SSG7 regulates SG size by disturbing amino acid metabolism, as the loss of function of the cytosolic alanine aminotransferase OsAlaAT1 did not lead to the production of enlarged SGs in rice endosperm (Yang et al., 2015). The biological significance of SSG6 in this translocon‐associated complex is largely unknown. Further work is required to explore the molecular mechanism by which the SSG complex orchestrates SG development, amino acid metabolism, and protein import in the future.
Materials and methods
Plant materials and growth conditions
The ssg7 mutant was isolated from an N‐methyl‐N‐nitrosourea‐mutagenized pool of japonica rice (Oryza sativa) cultivar W017. Reciprocal crosses between ssg7 and W017 were used for genetic analysis. An F2 population derived from a cross between the ssg7 (japonica) with an indica cultivar N22 was used for gene mapping. The ssg4 mutant with a missense mutation derived from japonica rice cultivar W017 was crossed with ssg7 to generate the ssg4 ssg7 double mutant. The ssg6 mutant was generated using CRISPR/Cas9 technology in the W017 background. The wild type, mutants, and transgenic lines were grown in experimental fields during the normal growing seasons at the Chinese Academy of Agricultural Science, Beijing.
Physicochemical properties of the endosperm starch
Husked rice grains were processed with a polisher and ground to fine flour in a miller or directly ground into flour without polishing. The total starch content of mature grains was determined using starch assay kits (Megazyme, Wicklow, Ireland, http://www. megazyme.com). The amylose, protein, and lipid contents were measured as previously described (Kang et al., 2005). The chain‐length distribution (CLD) pattern of amylopectin was determined following the method previously described (Peng et al., 2014). The pasting properties of endosperm starch were measured with a Rapid Visco Analyser (TecMaster RVA, Perten).
Microscopy observation
Scanning electron microscopy of mature grains was performed as described previously (Yan et al., 2024). Brown rice was hand‐sectioned transversely at the mid‐section using a razor blade and coated with gold, followed by observation with a SEM5000 (CIQTEK Co., Ltd, Hefei, China).
Semi‐thin sections were prepared from developing grains as previously described (Yan et al., 2024). Briefly, developing grains were transversely cut with a razor blade, and approximately 1 mm3 blocks were taken and fixed in 0.1 M phosphate buffer (pH 7.4) containing 2.5% (v/v) glutaraldehyde at 4 °C in the dark for 12 h. Samples were dehydrated by an ethanol series [30, 50, 70, 90, and 100% (v/v)] and then embedded in LR White resin (London Resin, 14 388‐UC). After polymerization at 65 °C for 3 days, semithin sections (1 μm in thickness) were prepared using an ultramicrotome (Leica microsystems, RM2265), and cytohistological analysis was performed as described previously (Wu et al., 2016; Yan et al., 2024). Images were taken using a Nikon ECLIPSE80i microscope and the representative images were shown. The areas occupied by SGs and the diameter of starch granules were determined with Image J software (https://imagej.nih.gov/ij/).
For visualization of the subcellular localization of SSG7 in subaleurone cells of developing grains, thick sections were prepared following the method described by Ren et al. (2020), with minor modifications. Briefly, freshly dehulled developing rice grains were fixed in 5% (w/v) agarose and cut into slices (60 to 100 μm in thickness) using a Vibrating blade microtome (Leica microsystems; VT‐1200S). The fresh slices were immersed into ice‐cold phosphate buffer before observation of the fluorescence signals with a laser scanning confocal microscope (Zeiss LSM980).
Immuno‐gold electronic microscopy of developing rice grain was performed according to a previously described method (Ren et al., 2014). Developing rice grains were dehulled, followed by fixation by high‐pressure frozen/freeze substituted. After dehydration, samples were embedded in LOWICRYL HM20 resin via UV light irradiation (AFS2, Leica). Ultrathin sections (~70 nm) were prepared with a Leica EM UC7 microtome. Immunogold labelling was performed using the primary monoclonal anti‐GFP antibody in combination with 10‐nm gold‐coupled secondary antibodies. Images were taken with a transmission electron microscope (Hitachi H7700, Tokyo, Japan).
Map‐based cloning of the SSG7 gene
To map the SSG7 locus, we constructed an F2 population by crossing ssg7 (japonica) with indica cultivar N22. Individuals with floury endosperm in the F2 population were used for mapping. Molecular markers were developed based on comparisons of the sequence polymorphism between japonica cultivar Nipponbare and indica cultivar 93‐11 (Table S4).
Plasmid construction and transformation
For complementation test, full‐length coding sequence of SSG7 was amplified and inserted into the KpnI and BamHI sites of pCUBi1390 vector to generate the ProUbi:SSG7 construct. Similarly, the entire coding regions of SSG6, Arabidopsis CRL, and cyanobacteria CpcT were used to generate the ProUbi:SSG6, ProUbi:CRL, and ProUbi:CpcT constructs, respectively. The coding sequence of Synechococcus elongatus PCC 6301 CpcT (Sugita et al., 2007) was synthesized in GeneScript Biotech (Nanjing, China; https://www.genscript.com.cn/).
For the fusion complementation test, the full‐length coding sequence of SSG7 was fused to the N or C terminus of GFP in the binary vector pCAMBIA1305 driven by its native 2114‐bp promoter to produce ProSSG7:SSG7‐GFP and ProSSG7:GFP‐SSG7 constructs, respectively.
To investigate the subcellular localization of SSG7 in the developing endosperm, the full‐length coding sequence of SSG7 was inserted into the BglII site of the modified binary vector pCAMBIA1305 (hygromycin‐resistant; Ren et al., 2020) containing a GFP tag driven by the maize UBIQUITIN promoter to generate the ProUbi:GFP‐SSG7 construct. For colocalization analyses, the coding sequences of OEP7 and BT1 were separately cloned into the BamHI site of the modified binary vector pCAMBIA2300 (kanamycin‐resistant; Ren et al., 2020) containing a DsRed tag driven by the maize UBIQUITIN promoter, to generate ProUbi:OEP7‐DsRed and ProUbi:BT1‐DsRed constructs. These constructs were separately introduced into transgenic plants harbouring ProUbi:GFP‐SSG7 for colocalization analyses.
To generate a knockout construct, 20‐bp gene‐specific spacer sequences of the target genes were separately inserted into the CRISPR‐Cas9 expression vector following the method described previously (Miao et al., 2013).
Unless indicated otherwise, all constructs were separately introduced into W017 or ssg7 calli via Agrobacterium‐mediated transformation (Hiei et al., 1994). Primers are listed in Table S4.
Protein extraction and immunoblot analyses
Total protein extraction from rice seeds and immunoblot analysis were performed as described previously (Yan et al., 2024).
Subcellular fractionation
Subcellular fractionation assay was performed using the methods described previously with minor modifications (Ren et al., 2020; Wang et al., 2016). Briefly, the wild‐type developing grains (9 DAF) were dehulled and homogenized in 2‐fold volume (on a basis of fresh weight) of ice‐cold extraction buffer A (100 mm HEPS‐KOH [pH 7.5], 2 mm MgCl2, 5 mm EGTA, 300 mm sucrose, 1 mm PMSF, 1% [v/v] Triton X‐100, and proteinase inhibitor cocktail [Roche]). The homogenate was centrifuged at 200 g for 10 min at 4 °C to remove starch‐rich pellets, followed by filtration through four layers of cheesecloth, and subsequent ultracentrifugation at 100 000 g for 1 h at 4 °C to obtain the membrane‐associated pellet (P100) and soluble supernatant (S100) for immunoblot analyses using the protein‐specific antibodies.
The P100 fraction was resuspended in different solutions of buffer A (with the same volume used for initial homogenization), separately containing (1 M NaCl, 0.1 M Na2CO3 [pH 11.5], 1% [v/v] Triton X‐100, or 1% [v/v] Nonidet P‐40). After incubation for 1 h in ice, the resuspending solutions were ultracentrifugated at 100 000 g for 1 h at 4 °C to obtain the soluble supernatant (S) and insoluble pellet (P), followed by immunoblot analyses.
Immunoprecipitation and mass spectrometry (IP‐MS)
Approximately 5 g of developing grains expressing GFP‐SSG7 or free GFP were homogenized in a 1.2‐fold volume (on a basis of fresh weight) of ice‐cold extraction buffer (50 mm Tris‐MES [pH 7.5], 1 mm MgCl2, 0.5 M sucrose, 10 mm EDTA, 5 mm DTT, 0.1% [w/v] Nonidet P‐40, and proteinase inhibitor cocktail [Roche]). Starch‐rich pellets were removed by centrifugation at 200 g for 10 min at 4 °C, and the resultant supernatant was filtered through four layers of cheesecloth, followed by incubation with μMACS Microbeads conjugated to anti‐GFP (Miltenyi Biotec; 30‐091‐125) for 1 h at 4 °C. After incubation, the mixture was filtered using a μColumn that is adsorbed on a magnetic stand (Miltenyi Biotec). The μColumn was washed three times with the extraction buffer containing 0.2% (v/v) Nonidet P‐40. The bound proteins were eluted with 80 μL elution buffer (50 mm Tris–HCl [pH 6.8], 50 mm DTT, 1% [w/v] SDS, 1 mm EDTA, 0.005% [w/v] bromphenol blue and 10% [v/v] glycerol). Mass spectrometry analyses and data collection were conducted as previously described by Ren et al. (2020). Three independent experiments were performed.
In vivo co‐immunoprecipitation (Co‐IP) assay
In vivo co‐immunoprecipitation assay in rice protoplasts was performed as previously described by Chen et al. (2006) with minor modifications. Rice protoplasts were prepared from the leave sheath of 10‐day‐old seedlings. The full‐length coding sequence of SSG7 was separately cloned into pAN580 (containing a GFP tag) and pCAMBIA1300‐221‐Flag vectors (containing a Flag tag) to generate GFP‐SSG7 and Flag‐SSG7 fusion constructs. Various combinations of plasmids were transiently co‐expressed in rice protoplasts. On the second day after incubation, total protein was extracted from co‐expressed rice protoplasts using ice‐cold protein lysis buffer (50 mm Hepes [pH 7.5], 150 mm KCl, 10 mm EDTA, 1 mm DTT, 0.4% [v/v] Triton X‐100, and proteinase inhibitor cocktail [Roche]), followed by centrifugation and incubation with 20 μL of anti‐GFP mAb‐Magnetic beads (MBL, D153‐10) for 1 h at 4 °C with shaking. Before elution of the bound proteins with a reducing buffer (50 mm Tris–HCl [pH 6.8], 100 mm DTT, 2% [w/v] SDS, 3% [v/v] glycerol, and 0.005% [w/v] bromophenol blue), the beads were washed three times with extraction buffer. The immunoprecipitate samples were subjected to SDS‐PAGE and immunoblotting analyses using anti‐GFP (dilution 1 : 5000) and anti‐Flag antibodies (dilution 1 : 5000), respectively.
In vitro pull‐down assay
Partial coding sequence of SSG7 (encoding amino acids 56 to 275) was inserted into the pGEX‐4T‐2 vector at the BamHI and EcoRI sites to generate GST‐DUF1001 construct. Partial coding sequence of SSG4 (encoding amino acids 1730 to 2119 or 2135) was inserted into the pET‐30a vector at the BamHI and EcoRI sites to generate the His‐DUF490∆ and His‐DUF490 constructs, respectively. These constructs were transformed into Escherichia coli strain Rosetta (DE3) and after induction, recombinant proteins were purified using the His beads (Beaver; 70501‐100) or GST beads (Beaver; 70601‐100), following the manufacturer's instructions. Equal amounts (~2 μg) of GST and GST‐DUF1001 protein were separately incubated with 20 μL of GST beads in 1‐mL binding buffer (50 mm Tris–HCl [pH 7.5], 100 mm NaCl, 0.5% [v/v] Triton X‐100, and proteinase inhibitor cocktail [Roche]) at 4 °C for 1 h. The supernatant was collected and separately mixed with approximately 2 μg of purified His‐DUF490 or His‐DUF490∆ fusion proteins, and then incubated at 4 °C for another 2 h. After washing beads at least 5 times with the binding buffer, the bound proteins were eluted with equal volumes of SDS sample buffer and subjected to SDS‐PAGE immunodetection using anti‐GST (dilution 1 : 5000) and anti‐His (dilution 1 : 5000) antibodies. Three independent experiments were performed and similar results were obtained. The relative amounts of His fusion proteins bound to GST‐DUF1001 were calculated using Image J software (https://imagej.nih.gov/ij/).
Firefly luciferase complementation imaging (LCI) assay
Full‐length coding sequence of SSG6, SSG7 and BT1 was N terminally fused to nLUC in the pCAMBIA‐nLUC vector to generate the SSG6‐nLUC, SSG7‐nLUC, and BT1‐nLUC constructs, respectively. Full‐length coding sequence of SSG7 and BT1 was fused to cLUC in the pCAMBI‐cLUC vector to generate the SSG7‐cLUC and BT1‐cLUC constructs, respectively. Different combinations of EHA105 strains containing transformed plasmids were co‐infiltrated into leaves of N. benthaminana. After 2 days, the relative LUC activity was calculated by a Plant Molecular Imaging System in vivo as described previously by Chen et al. (2008). Three independent experiments were performed and representative images were shown.
Yeast two‐hybrid assay
The sequence encoding the SSG7 variant with a deletion of the TM domain was cloned into the pGADT7 and pGBKT7 vectors at XbaI and BamHI sites to generate AD‐SSG7(∆TM) and BD‐SSG7(∆TM) constructs, respectively. Various combinations of plasmids were cotransformed into yeast strain AH109, followed by the screening of interactions according to the manufacturer's instructions. The experiments were repeated twice independently and similar results were obtained.
Antibodies
Partial coding sequence of SSG7 (amino acids 50 to 242), SSG6 (amino acids 1 to 195), TOC159 (amino acids 1 to 150), and TOC34 (amino acids 1 to 312) were separately cloned into expression vector pET‐28a at BamHI and EcoRI sites for recombinant proteins expression. Recombinant proteins were extracted and purified using the crude extracts from transformed E. coli strain Rosetta (DE3), with the His beads (Beaver; 70501‐100). Synthetic peptides of TOC75 (C‐FERVDLEGKAK) were synthesized. The purified recombination proteins (approximately 1 mg) or synthetic peptides were injected into rabbits for polyclonal antibody production at ABclonal biotechnology (Wuhan, China; https://abclonal.com.cn/). Anti‐TIC110 antibodies were prepared as described previously (Zhu et al., 2018). Anti‐EF‐1α (Agrisera, AS10 934), anti‐GST (MBL, PM013‐7), anti‐His (MBL, D291‐7), anti‐GFP (Roche, 11 814 460 001), and anti‐Flag (Sigma, A8592) antibodies are commercially available and diluted at 1 : 5000.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: SSG7 (LOC_Os11g32160), SSG4 (LOC_Os01g08420), SSG6 (LOC_Os06g03990), TOC159 (LOC_Os05g05950), TOC120 (LOC_Os10g40110), TOC90 (LOC_Os12g09570), TOC75 (LOC_Os03g16440), TOC64 (LOC_Os03g50010), TOC34 (LOC_Os03g13730), and TIC110 (LOC_Os10g35010). Accession numbers for the proteins used in phylogenetic tree construction were listed on the tree.
Conflicts of interest
The authors declare no conflict of interest.
Author contributions
J.M.W. and W.W.Z. designed the project. H.G.Y., Y.H.W., B.L.Z., J.J., Z.J., X.Z., and Y.Z. performed the experiments. Other authors provided technical support; W.W.Z., Y.H.W., Y.L.R., and H.G.Y wrote the manuscript.
Supporting information
Data S1 Summary of proteins co‐precipitated with GFP‐SSG7 and identified by mass spectrometry.
Figure S1 ssg7 impaired endosperm development.
Figure S2 Quantification of the numbers of starch grains and starch granules.
Figure S3 The appearance of endosperm and the sizes of starch grains (SGs) of F1 grains from reciprocal crosses.
Figure S4 Starch grains (SGs) in pollen grains and pericarp cells.
Figure S5 Chloroplast morphologies in ssg7 leaves.
Figure S6 Overexpression of SSG7 driven by the UBIQUITIN promoter specifically rescued the ssg7 mutant phenotypes.
Figure S7 Characterization of SSG7 knockout mutants.
Figure S8 The expression level of SSG7 in ssg7 developing endosperm.
Figure S9 Structure and phylogenetic analyses and amino acid sequence alignment of the SSG7 protein.
Figure S10 Complementation of ssg7 by expressing GFP‐tagged SSG7 fusion protein.
Figure S11 Rescue of ssg7 mutant phenotype by overexpressing the GFP‐SSG7 fusion protein.
Figure S12 Phenotypes of the ssg4 mutant.
Figure S13 Rescue of ssg7 defective phenotype by overexpressing SSG6.
Figure S14 Assessment of the specificity of polyclonal antibodies generated in this work.
Table S1 Agronomic traits of the wild type (WT), ssg mutants, and transgenic plants.
Table S2 Genetic analysis of the ssg7 mutant.
Table S3 Segregation analysis of progenies from double heterozygous ssg4 +/‐ ssg7 +/‐ plants.
Table S4 Primers used in this study.
Acknowledgements
This work was supported by National Key R&D Program of China (2021YFF1000200), Innovation Program of Chinese Academy of Agricultural Sciences, Key R&D Program of Jiangsu Province (BE2021359), and Central Public‐Interest Scientific Institution Basal Research Fund, China (Y2021YJ18 and S2020YC05). We also gratefully thank Dr. Fan Wang and Jianan Wu from the Core Facility Platform, Chinese Academy of Agriculture Science (CAAS) for their assistance with SEM analysis.
Contributor Information
Yihua Wang, Email: yihuawang@njau.edu.cn.
Wenwei Zhang, Email: zhangww@njau.edu.cn.
Jianmin Wan, Email: wanjm@njau.edu.cn.
Data availability statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
References
- Asano, T. , Yoshioka, Y. , Kurei, S. , Sakamoto, W. , Sodmergen and Machida, Y. (2004) A mutation of the CRUMPLED LEAF gene that encodes a protein localized in the outer envelope membrane of plastids affects the pattern of cell division, cell differentiation, and plastid division in Arabidopsis. Plant J. 38, 448–459. [DOI] [PubMed] [Google Scholar]
- Chen, S.B. , Tao, L.Z. , Zeng, L.R. , Vega‐Sanchez, M.E. , Umemura, K. and Wang, G.L. (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein‐protein interactions in rice. Mol. Plant Pathol. 7, 417–427. [DOI] [PubMed] [Google Scholar]
- Chen, H. , Zou, Y. , Shang, Y. , Lin, H. , Wang, Y. , Cai, R. , Tang, X. et al. (2008) Firefly luciferase complementation imaging assay for protein‐protein interactions in plants. Plant Physiol. 146, 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.L. , Chen, L.J. , Chu, C.C. , Huang, P.K. , Wen, J.R. and Li, H.M. (2018) TIC236 links the outer and inner membrane translocons of the chloroplast. Nature 564, 125–129. [DOI] [PubMed] [Google Scholar]
- Crumpton‐Taylor, M. , Grandison, S. , Png, K.M. , Bushby, A.J. and Smith, A.M. (2012) Control of starch granule numbers in Arabidopsis chloroplasts. Plant Physiol. 158, 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O. , Nielsen, H. and Von Heijne, G. (1999) ChloroP, a neural network‐based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, J. , Li, B. , Chen, L.J. , Dogra, V. , Luo, S. , Wu, W. , Wang, P. et al. (2022) TIC236 gain‐of‐function mutations unveil the link between plastid division and plastid protein import. Proc. Natl. Acad. Sci. USA 119, e2123353119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez, O.A. , Campbell, M.R. and Glover, D.V. (2002) Starch particle volume in single‐ and double‐mutant maize endosperm genotypes involving the soft starch (h) gene. Crop Sci. 42, 355–359. [Google Scholar]
- Hiei, Y. , Ohta, S. , Komari, T. and Kumashiro, T. (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T‐DNA. Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Huang, L. , Tan, H. , Zhang, C. , Li, Q. and Liu, Q. (2021) Starch biosynthesis in cereal endosperms: an updated review over the last decade. Plant Commun. 2, 100237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, D.T. , Froehlich, J.E. and Keegstra, K. (1998) The hydrophilic domain of Tic110, an inner envelope membrane component of the chloroplastic protein translocation apparatus, faces the stromal compartment. J. Biol. Chem. 273, 16583–16588. [DOI] [PubMed] [Google Scholar]
- James, M.G. , Denyer, K. and Myers, A.M. (2003) Starch synthesis in the cereal endosperm. Curr. Opin. Plant Biol. 6, 215–222. [DOI] [PubMed] [Google Scholar]
- Jane, J.‐L. , Kasemsuwan, T. , Leas, S. , Ames, I.A. , Zobel, H. , Darien, I.L. et al. (1994) Anthology of starch granule morphology by scanning electron microscopy. Starch‐Starke 46, 121–129. [Google Scholar]
- Jarvis, P. and López‐Juez, E. (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14, 787–802. [DOI] [PubMed] [Google Scholar]
- Jobling, S. (2004) Improving starch for food and industrial applications. Curr. Opin. Plant Biol. 7, 210–218. [DOI] [PubMed] [Google Scholar]
- Kang, H.G. , Park, S. , Matsuoka, M. and An, G.H. (2005) White‐core endosperm floury endosperm‐4 in rice is generated by knockout mutations in the C4‐type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J. 42, 901–911. [DOI] [PubMed] [Google Scholar]
- Kawagoe, Y. (2013) The characteristic polyhedral, sharp‐edged shape of compound‐type starch granules in rice endosperm is achieved via the septum‐like structure of the amyloplast. J. Appl. Glycosci. 60, 29–36. [Google Scholar]
- Krogh, A. , Larsson, B. , von Heijne, G. and Sonnhammer, E.L.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Lee, Y.J. , Kim, D.H. , Kim, Y.W. and Hwang, I. (2001) Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 13, 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B.Q. , Fang, J. , Singh, R.M. , Zi, H.L. , Lv, S.S. , Liu, R.Y. , Dogra, V. et al. (2020) FATTY ACID DESATURASE5 is required to induce autoimmune responses in gigantic chloroplast mutants of Arabidopsis. Plant Cell 32, 3240–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom, N. , Chang, P.R. and Tyler, R.T. (2004) Analytical, biochemical and physicochemical aspects of starch granule size, with emphasis on small granule starches: a review. Starch‐Starke 56, 89–99. [Google Scholar]
- Malinski, E. , Daniel, J.R. , Zhang, X.X. and Whistler, R.L. (2003) Isolation of small starch granules and determination of their fat mimic characteristics. Cereal Chem. 80, 1–4. [Google Scholar]
- Matsushima, R. and Hisano, H. (2019) Imaging amyloplasts in the developing endosperm of barley and rice. Sci. Rep. 9, 3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima, R. , Maekawa, M. , Fujita, N. and Sakamoto, W. (2010) A rapid, direct observation method to isolate mutants with defects in starch grain morphology in rice. Plant Cell Physiol. 51, 728–741. [DOI] [PubMed] [Google Scholar]
- Matsushima, R. , Maekawa, M. , Kusano, M. , Kondo, H. , Fujita, N. , Kawagoe, Y. and Sakamoto, W. (2014) Amyloplast‐localized SUBSTANDARD STARCH GRAIN4 protein influences the size of starch grains in rice endosperm. Plant Physiol. 164, 623–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima, R. , Maekawa, M. , Kusano, M. , Tomita, K. , Kondo, H. , Nishimura, H. , Crofts, N. et al. (2016) Amyloplast membrane protein SUBSTANDARD STARCH GRAIN6 controls starch grain size in rice endosperm. Plant Physiol. 170, 1445–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, J. , Guo, D.S. , Zhang, J.Z. , Huang, Q.P. , Qin, G.J. , Zhang, X. , Wan, J.M. et al. (2013) Targeted mutagenesis in rice using CRISPR‐Cas system. Cell Res. 23, 1233–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, C. , Wang, Y.H. , Liu, F. , Ren, Y.L. , Zhou, K.N. , Lv, J. , Zheng, M. et al. (2014) FLOURY ENDOSPERM6 encodes a CBM48 domain‐containing protein involved in compound granule formation and starch synthesis in rice endosperm. Plant J. 77, 917–930. [DOI] [PubMed] [Google Scholar]
- Ren, Y.L. , Wang, Y.H. , Liu, F. , Zhou, K.N. , Ding, Y. , Zhou, F. , Wang, Y. et al. (2014) GLUTELIN PRECURSOR ACCUMULATION3 encodes a regulator of post‐Golgi vesicular traffic essential for vacuolar protein sorting in rice endosperm. Plant Cell 26, 410–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, Y.L. , Wang, Y.H. , Pan, T. , Wang, Y.L. , Wang, Y.F. , Gan, L. , Wei, Z.Y. et al. (2020) GPA5 encodes a Rab5a effector required for post‐Golgi trafficking of rice storage proteins. Plant Cell 32, 758–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, L.G.L. and Schnell, D.J. (2020) Origins, function, and regulation of the TOC‐TIC general protein import machinery of plastids. J. Exp. Bot. 71, 1226–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldan, I. , Wattebled, F. , Mercedes Lucas, M. , Delvalle, D. , Planchot, V. , Jimenez, S. , Perez, R. et al. (2007) The phenotype of soluble starch synthase IV defective mutants of Arabidopsis thaliana suggests a novel function of elongation enzymes in the control of starch granule formation. Plant J. 49, 492–504. [DOI] [PubMed] [Google Scholar]
- Selkrig, J. , Mosbahi, K. , Webb, C.T. , Belousoff, M.J. , Perry, A.J. , Wells, T.J. , Morris, F. et al. (2012) Discovery of an archetypal protein transport system in bacterial outer membranes. Nat. Struct. Mol. Biol. 19, 506–510. [DOI] [PubMed] [Google Scholar]
- Seung, D. and Smith, A.M. (2019) Starch granule initiation and morphogenesis‐progress in Arabidopsis and cereals. J. Exp. Bot. 70, 771–784. [DOI] [PubMed] [Google Scholar]
- Seung, D. , Boudet, J. , Monroe, J. , Schreier, T.B. , David, L.C. , Abt, M. , Lu, K.J. et al. (2017) Homologs of PROTEIN TARGETING TO STARCH control starch granule initiation in Arabidopsis leaves. Plant Cell 29, 1657–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung, D. , Schreier, T.B. , Burgy, L. , Eicke, S. and Zeeman, S.C. (2018) Two plastidial coiled‐coil proteins are essential for normal starch granule initiation in Arabidopsis. Plant Cell 30, 1523–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, G. , Saunee, N.A. , Williams, S.R. , Gallo, E.F. , Schluchter, W.M. and Bryant, D.A. (2006) Identification and characterization of a new class of bilin lyase ‐ the cpcT gene encodes a bilin lyase responsible for attachment of phycocyanobilin to CYS‐153 on the β‐subunit of phycocyanin in synechococcus sp PCC 7002. J. Biol. Chem. 281, 17768–17778. [DOI] [PubMed] [Google Scholar]
- Sugita, C. , Ogata, K. , Shikata, M. , Jikuya, H. , Takano, J. , Furumichi, M. , Kanehisa, M. et al. (2007) Complete nucleotide sequence of the freshwater unicellular cyanobacterium Synechococcus elongatus PCC 6301 chromosome: gene content and organization. Photosynth. Res. 93, 55–67. [DOI] [PubMed] [Google Scholar]
- Sugita, C. , Kato, Y. , Yoshioka, Y. , Tsurumi, N. , Iida, Y. , Machida, Y. and Sugita, M. (2012) CRUMPLED LEAF (CRL) homologs of Physcomitrella patens are involved in the complete separation of dividing plastids. Plant Cell Physiol. 53, 1124–1133. [DOI] [PubMed] [Google Scholar]
- Vandromme, C. , Spriet, C. , Dauvillée, D. , Courseaux, A. , Putaux, J.L. , Wychowski, A. , Krzewinski, F. et al. (2019) PII1: a protein involved in starch initiation that determines granule number and size in Arabidopsis chloroplast. New Phytol. 221, 356–370. [DOI] [PubMed] [Google Scholar]
- Wang, Y.H. , Liu, F. , Ren, Y.L. , Wang, Y.L. , Liu, X. , Long, W.H. , Wang, D. et al. (2016) GOLGI TRANSPORT 1B regulates protein export from the endoplasmic reticulum in rice endosperm cells. Plant Cell 28, 2850–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F. , Fang, J. , Guan, K. , Luo, S. , Dogra, V. , Li, B. , Ma, D. et al. (2020) The Arabidopsis CRUMPLED LEAF protein, a homolog of the cyanobacterial bilin lyase, retains the bilin‐binding pocket for a yet unknown function. Plant J. 104, 964–978. [DOI] [PubMed] [Google Scholar]
- Wang, R.Q. , Ren, Y.L. , Yan, H.G. , Teng, X. , Zhu, X.P. , Wang, Y.P. , Zhang, X. et al. (2021) ENLARGED STARCH GRAIN1 affects amyloplast development and starch biosynthesis in rice endosperm. Plant Sci. 305, 110831. [DOI] [PubMed] [Google Scholar]
- Wilson, J.A. , Glover, D.V. and Nyquist, W.E. (2000) Effect of dosage at the soft starch (h) locus on starch granule volume in maize. Plant Breed. 119, 177–178. [Google Scholar]
- Wu, X.B. , Liu, J.X. , Li, D.Q. and Liu, C.M. (2016) Rice caryopsis development I: dynamic changes in different cell layers. J. Integr. Plant Biol. 58, 772–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagami, T. , Tsuchisaka, A. , Yamada, K. , Haddon, W.F. , Harden, L.A. and Theologis, A. (2003) Biochemical diversity among the 1‐amino‐cyclopropane‐1‐carboxylate synthase isozymes encoded by the Arabidopsis gene family. J. Biol. Chem. 278, 49102–49112. [DOI] [PubMed] [Google Scholar]
- Yan, H. , Zhang, W. , Wang, Y. , Jin, J. , Xu, H. , Fu, Y. , Shan, Z. et al. (2024) Rice LIKE EARLY STARVATION1 cooperates with FLOURY ENDOSPERM6 to modulate starch biosynthesis and endosperm development. Plant Cell 36, 1892–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , Kim, S.R. , Lee, S.K. , Choi, H. , Jeon, J.S. and An, G. (2015) Alanine aminotransferase 1 (OsAlaAT1) plays an essential role in the regulation of starch storage in rice endosperm. Plant Sci. 240, 79–89. [DOI] [PubMed] [Google Scholar]
- Yun, M.S. and Kawagoe, Y. (2009) Amyloplast division progresses simultaneously at multiple sites in the endosperm of rice. Plant Cell Physiol. 50, 1617–1626. [DOI] [PubMed] [Google Scholar]
- Yun, M.S. and Kawagoe, Y. (2010) Septum formation in amyloplasts produces compound granules in the rice endosperm and is regulated by plastid division proteins. Plant Cell Physiol. 51, 1469–1479. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Wu, S. , Boehlein, S.K. , McCarty, D.R. , Song, G. , Walley, J.W. , Myers, A. et al. (2019) Maize defective kernel5 is a bacterial TamB homologue required for chloroplast envelope biogenesis. J. Cell Biol. 218, 2638–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, L.X. , Pan, T. , Guo, D.W. and Wei, C.X. (2018) A simple and rapid method for preparing the whole section of starchy seed to investigate the morphology and distribution of starch in different regions of seed. Plant Methods 14, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X. , Teng, X. , Wang, Y. , Hao, Y. , Jing, R. , Wang, Y. , Liu, Y. et al. (2018) FLOURY ENDOSPERM11 encoding a plastid heat shock protein 70 is essential for amyloplast development in rice. Plant Sci. 277, 89–99. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1 Summary of proteins co‐precipitated with GFP‐SSG7 and identified by mass spectrometry.
Figure S1 ssg7 impaired endosperm development.
Figure S2 Quantification of the numbers of starch grains and starch granules.
Figure S3 The appearance of endosperm and the sizes of starch grains (SGs) of F1 grains from reciprocal crosses.
Figure S4 Starch grains (SGs) in pollen grains and pericarp cells.
Figure S5 Chloroplast morphologies in ssg7 leaves.
Figure S6 Overexpression of SSG7 driven by the UBIQUITIN promoter specifically rescued the ssg7 mutant phenotypes.
Figure S7 Characterization of SSG7 knockout mutants.
Figure S8 The expression level of SSG7 in ssg7 developing endosperm.
Figure S9 Structure and phylogenetic analyses and amino acid sequence alignment of the SSG7 protein.
Figure S10 Complementation of ssg7 by expressing GFP‐tagged SSG7 fusion protein.
Figure S11 Rescue of ssg7 mutant phenotype by overexpressing the GFP‐SSG7 fusion protein.
Figure S12 Phenotypes of the ssg4 mutant.
Figure S13 Rescue of ssg7 defective phenotype by overexpressing SSG6.
Figure S14 Assessment of the specificity of polyclonal antibodies generated in this work.
Table S1 Agronomic traits of the wild type (WT), ssg mutants, and transgenic plants.
Table S2 Genetic analysis of the ssg7 mutant.
Table S3 Segregation analysis of progenies from double heterozygous ssg4 +/‐ ssg7 +/‐ plants.
Table S4 Primers used in this study.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.


