Abstract
Fine particulate matter (PM2.5) exposure can induce immune system pathology via epigenetic modification, affecting pregnancy outcomes. Our study investigated the association between PM2.5 exposure and immune response, as well as epigenetic changes using high-dimensional epigenetic landscape profiling using cytometry by time-of-flight (EpiTOF) at the single cell. We found statistically significant associations between PM2.5 exposure and levels of certain cytokines [interleukin-1RA (IL-1RA), IL-8/CXCL8, IL-18, and IL-27)] and histone posttranslational modifications (HPTMs) in immune cells (HPTMs: H3K9ac, H3K23ac, H3K27ac, H2BK120ub, H4K20me1/3, and H3K9me1/2) among pregnant and nonpregnant women. The cord blood of neonates with high maternal PM2.5 exposure showed lower IL-27 than those with low exposure. Furthermore, PM2.5 exposure affects the co-modification profiles of cytokines between pregnant women and their neonates, along with HPTMs in each immune cell type between pregnant and nonpregnant women. These modifications in specific histones and cytokines could indicate the toxicological mechanism of PM2.5 exposure in inflammation, inflammasome pathway, and pregnancy complications.
PM2.5 exposure differentially alters cytokine and histone modification profiles in pregnant versus nonpregnant women.
INTRODUCTION
Exposure to ambient air pollution (AAP) has been shown to increase health risks by triggering oxidative stress and inflammation (1). Exposure may alter immune cell profiles (2), elevate levels of proinflammatory response across immune cell types (3), and lead to disorders linked with immune system dysfunction (4). Pregnant women are especially susceptible to AAP because increased respiration rates and the higher oxygen demand for the developing fetus lead to greater inhalation and circulation of pollutants (5, 6). Increased susceptibility to adverse health effects of AAP may affect maternal and prenatal outcomes, including preeclampsia, low birth weight, and preterm birth (6). Furthermore, prenatal exposure is known to influence neurodevelopment and metabolic dysfunction during early childhood (7, 8).
Exposure to AAP during pregnancy can lead to cytokine dysregulation and T cell polarization associated with epigenetic modifications (1, 2, 9). Aberrant expression of maternal cytokines can be related to fetal development failure and pregnancy complications (10, 11). Limited but recent studies suggest the possible association between maternal environmental exposure and cord blood (CB) cytokines (11, 12). Therefore, it is important to understand the effects of prenatal AAP exposure on the association between maternal and neonatal responses, which have been underexplored.
Recent studies indicate that AAP can affect gene expression through epigenetic modifications in histone modification that regulates DNA-templated biological processes, including transcription, replication, and repair (13). Histone posttranslational modifications (HPTMs) also play a crucial role in maternal and fetal immune health by involving embryo implantation and placenta development (14). In particular, our recent review found that the epigenetic mechanisms have a larger impact when exposed to AAP in the first trimester of pregnancy (6). However, these studies cannot differentiate the affected cells, which is important in identifying specific targets to prevent and treat disease. Considering heterogeneity in epigenetic modifications linked to immune cellular responses, our approach to studying single cell–specific epigenetic changes in immune cells offers a distinct advantage to heretofore-used methods. Moreover, studies on the association between AAP and cell-specific histone changes during pregnancy have never been reported.
In previous studies, our group demonstrated that exposure to AAP during pregnancy is linked to a negative impact on T helper subsets involved in inflammatory responses. In addition, we and other researchers have recently reviewed the literature on the harmful effects of AAP during pregnancy on maternal, fetal, and neonatal health (2, 6, 9, 15). Thus, we hypothesized that exposure to elevated PM2.5 (fine particulate matter ≤2.5 μm in diameter) during pregnancy might affect immune responses and molecular signaling pathways. Here, we studied the immunological responses and HPTMs at the single-cell level in pregnant women exposed to PM2.5, analyzing molecular profiling of blood features and identifying aberrant features associated with PM2.5 exposure during the second trimester. Our study provides the first comprehensive analysis of the impact of AAP exposure on immune single-cell phenotypes and epigenetic alterations. This will enable the identification of specific markers for the treatment and prevention of diseases associated with AAP exposure during pregnancy.
RESULTS
Study participants were collected from Fresno and the Bay area in California
Table 1 presents characteristics of study samples from pregnant women (n = 168) living in Fresno, California, at approximately 20 weeks of gestation (range: 18 to 25 weeks) and age-matched nonpregnant women residing in Fresno or the Bay area in California (n = 151) (fig. S1). Most enrolled subjects were Hispanic for pregnant women (71.9%) while non-Hispanic white for nonpregnant women (44.8%).
Table 1. Demographic characteristics of the study participants.
Values for age are years in median ± SD and those for ethnicity and history of clinical variables are in percentage.
| Types of participants | Characteristics | Luminex | EpiTOF |
|---|---|---|---|
| Pregnant women | Sample size (n) | 167 | 36 |
| Age (median, years) | 28.7 ± 6.1 | 28.5 ± 6.2 | |
| Race and ethnicity* | |||
| Hispanic (%) | 71.9 | 72.2 | |
| Non-Hispanic white (%) | 10.2 | 13.9 | |
| Non-Hispanic Black (%) | 7.2 | 11.1 | |
| Non-Hispanic Asian or Pacific Islander (%) | 10.2 | 2.8 | |
| Non-Hispanic other races (%) | 0.5 | 0 | |
| Current smoker (%) | 0 | 0 | |
| SDI score (median)‡ | 98.0 | 98.0 | |
| History of asthma† (%) | 22.0 | 16.7 | |
| Nonpregnant women | Sample size (n) | 145 | 20 |
| Age (median, years) | 27.1 ± 7.7 | 24.1 ± 6.8 | |
| Race and ethnicity* | |||
| Hispanic (%) | 14.5 | 30.0 | |
| Non-Hispanic white (%) | 44.8 | 10.0 | |
| Non-Hispanic Black (%) | 3.4 | 50.0 | |
| Non-Hispanic Asian or Pacific Islander (%) | 24.8 | 10.0 | |
| Non-Hispanic other races (%) | 12.4 | 0 | |
| Current smoking (%) | 3.5 | 5.0 | |
| SDI score (median)‡ | 51.0 | 89.0 | |
| History of asthma† (%) | 20.0 | 15.0 | |
| Neonates | Sample size (n) | 33 | |
| Mother’s ethnicity* | |||
| Hispanic (%) | 81.8 | ||
| Mother’s age (median, years) | 29.67 ± 5.8 | ||
| Prenatal smoke exposure (%) | 0 | ||
| Mother with gestational diabetes | |||
| Yes (%) | 12.1 | ||
| No (%) | 69.7 | ||
| Missing (%) | 18.2 |
*Participants used “Hispanic or Latino” to answer questions about both their race and ethnicity. For the analysis, individuals who identified their race or ethnicity as “Hispanic or Latino” or “Mexican American” were categorized as “Hispanic.”
†History of asthma is self-reported asthma status.
‡SDI score is Social Deprivation Index (SDI), a composite score (0-100) showing deprivation level in a geographic area. SDI scores at the ZCTA level were matched to the participant’s home address. A higher score indicates greater social deprivation.
Average PM2.5 exposure was calculated for each participant over 1 week, 3, 6, and 9 months preceding blood collection (see details in Materials Methods section, “Air pollution exposure estimation and analysis;” table S1). The low-concentration exposure group was defined as having an average PM2.5 exposure below 12 μg/m3, the 2012 Environmental Protection Agency (EPA) National Ambient Air Quality Primary Standard for PM2.5 levels (16). Figure 1 presents the schematic overview of the workflow applied to our study samples. An 80-plex custom Luminex assay with an EMD Millipore human cytokine panel was used to quantify 80 cytokines from the plasma (see details in Materials and Methods section, “Luminex”). Epigenetic landscape profiling by cytometry by time-of-flight [EpiTOF, Cheung et al. (17)] was performed per our previously published methods using samples from a subset of 36 pregnant and 20 nonpregnant women at the same time points matched for age. In addition, CB samples (n = 33) were collected from a subset of pregnant women for whom CB was available at birth. Average PM2.5 exposure for CB was calculated at 1 week, one, two, and three trimesters preceding CB collection (table S1).
Fig. 1. Schematic overview of the workflow.
Pregnant status modified the association between PM2.5 and IL-1RA, IL-8, IL-18, and MIF and was significantly associated with 12 of 80 cytokines
To explore how exposure to PM2.5 affects cytokines differently in pregnant and nonpregnant women, we assessed the expression of 80 proteins while considering the pregnant status, level of PM2.5 exposure, and their interaction using a linear regression. For PM2.5 exposure, we primarily focused on the 3-month window, which our previous studies identified as having the strongest effects on immune and epigenetic changes (2, 9, 18). For sensitivity analysis, we also examined 1-week, 6-month, and 9-month exposure periods. We also adjusted for age, ethnicity, asthma status, social deprivation index (SDI) at zip code level, and batch effect, which were identified as potential confounding in our preliminary analysis.
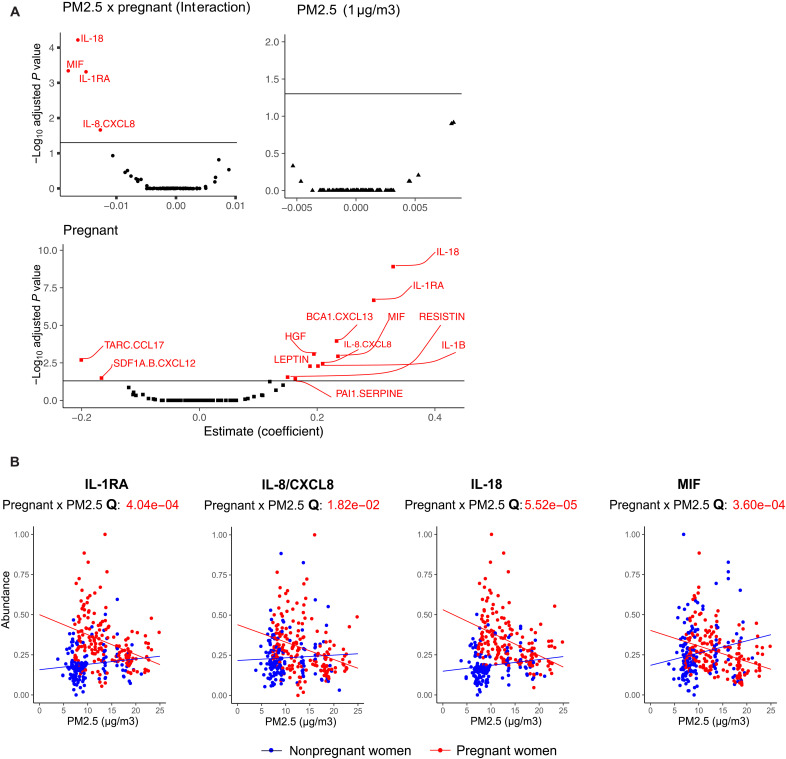
Figure 2A illustrates how pregnancy status and PM2.5 exposure, separately and in combination, influence each of the 80 cytokine levels. After applying statistical corrections for multiple testing [false discovery rate (FDR) < 0.05], 12 cytokines significantly increased during pregnancy. No cytokines were exclusively influenced by PM2.5 exposure alone. However, four cytokines demonstrated significant interaction effects, suggesting a complex relationship between pregnancy and PM2.5 exposure. Figure 2B delves into these four proteins by pregnancy status. In pregnant women, the expression levels of four markers that are normally increased in pregnancy, interleukin-1RA (IL-1RA) (Q = 4.04 × 10−4), IL-8/CXCL8 (Q = 1.82 × 10−0.2), IL-18 (Q = 5.52 × 10−5), and migration inhibitory factor (MIF) (Q = 3.60 × 10−4), were negatively associated with the level of PM2.5 exposure, indicating a potentially impactful interaction between PM2.5 exposure and pregnancy on these cytokines. Our sensitivity analysis, using different PM2.5 exposure windows, confirmed that the 3-month exposure window showed the strongest association with cytokine levels (fig. S2). We found that long-term PM2.5 exposure significantly affected the IL-7 (6 months: Q = 0.020; 9 months: Q = 0.034) and hepatocyte growth factor (HGF) (9 months: Q = 0.016) based on pregnant status.
Fig. 2. Pregnancy modifies the association between PM2.5 and four cytokines (IL-1RA, IL-8, IL-18, and MIF) and significantly affects 12 of 80 cytokines.
(A) Volcano plots summarize the beta (β) coefficients and −log transformed adjusted P values from multivariable linear regression adjusted for potential confounding such as age, ethnicity, asthma status, zip code–level SDI, and batch effect (see Materials and Methods for details). Multiple testing corrections were performed by using FDR ≥ 0.05. Significant cytokines were labeled red. A negative β coefficient of interaction between pregnant and PM2.5 status can be interpreted as the relationship between PM2,5 and cytokines getting weaker in pregnant women compared to nonpregnant women. β greater than 0 indicates a positive association. For PM2.5, it means that for every 1-unit (μg/m3) increase in PM2.5, the expression of markers will increase by the β coefficient value. For pregnant status, the expression of markers will increase by the beta coefficient value among pregnant women compared to nonpregnant women. (B) Scatter plots of PM2.5 concentration versus the levels for IL-1RA, IL-8/CXCL8, IL-18, and MIF, which had a significant interaction effect. Points and lines on the scatter plot are colored by pregnant status (pregnant women in red and nonpregnant women in blue). Lines shown are fitted within the pregnant status. FDR-adjusted P values (Q value) of the statistical interaction term between pregnant status and PM2.5 levels are noted below the cytokine name.
CB from neonates with high maternal PM2.5 exposure is likely to have lower IL-27 than that from those with lower maternal PM2.5 exposure
We examined cytokines in the CB of 33 neonates born to mothers exposed to varying levels of PM2.5 (fig. S3). Using multivariable regression, we evaluated CB cytokine in relation to PM2.5 exposure levels, considering the corresponding maternal cytokine levels, mother’s age, and ethnicity. We assumed that CB cytokine levels reflect corresponding maternal cytokine levels, and this relationship varies on the basis of the extent of PM2.5 exposure. Because of the small sample size, we dichotomized exposure variables into low- and high-exposure groups using average PM2.5 values of 1 week before CB collection and for the first, second, and third trimesters based on the EPA standard (see details in the method section). The primary focus was the average exposure during the third trimester, which is comparable to a 3-month exposure window before CB collection.
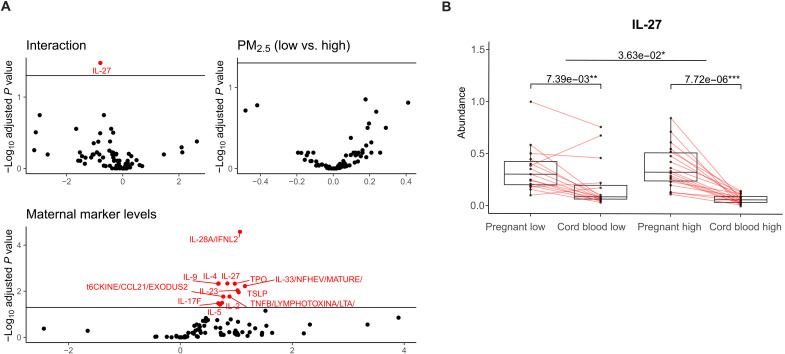
Our analysis of 80 cytokines in neonates’ CB found third-trimester exposure window had the strongest impact on CB cytokines (Fig. 3A and fig. S3, A to C). We found a significant positive correlation between 13 cytokines in CB and their corresponding maternal cytokine levels. We did not find any cytokines solely affected by PM2.5 for each trimester-specific exposure window. To check the robustness of the analysis, a sensitivity analysis (fig. S3D) using a different cutoff point, using the 25th percentile, showed similar results to the main findings in Fig. 3A. For the interaction term, we found that IL-27 was significantly lower in the high maternal exposure group compared to the low exposure group, holding the mothers’ IL-27 levels constant as shown in Fig. 3B.
Fig. 3. Impact of prenatal PM2.5 exposure during third trimester on neonatal-maternal cytokine association, especially IL-27.
(A) Volcano plots summarize the β coefficients and statistical significance for the association between maternal marker levels and CB markers (see Materials and Methods). Multiple testing corrections were performed by using FDR at 0.05. A positive β coefficient can be interpreted that 1-U increase in maternal cytokine will increase the cytokines level in CB by β estimates. A β coefficient of PM2.5 (binary indicator; low versus high) can be interpreted that cytokine levels in CB will change by β among high maternal PM2.5 exposure compared to the low exposure group. A β coefficient of interaction between maternal cytokine levels and PM2.5 status can be interpreted as the relation between maternal cytokine and corresponding cytokine in CB getting weaker in CB from high maternal PM2.5 exposure compared to those with low maternal PM2.5 exposure. (B) Box plots for levels of IL-27 in pregnant women and CB. The exposure group is defined on the basis of the mother’s exposure status during the 3 months before the blood collection, categorized as low or high. Points are samples in each group and matched pregnant women and CB are connected by a line. Q values of the differences within exposure status and between pregnant and nonpregnant women are noted with significant levels. Q values below 0.05 get one star (*); below 0.01, two stars (**); and below 0.001, three stars (***).
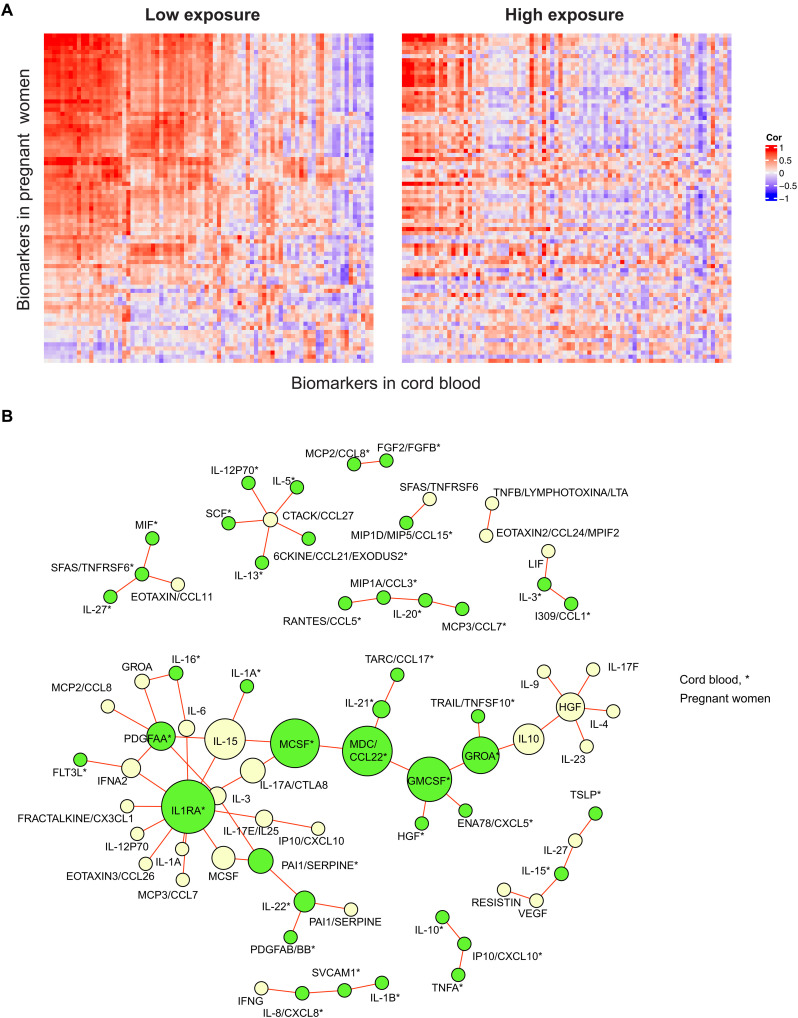
Figure 4A highlights the coexpression profiles of cytokines in maternal-neonatal pairs under low and high PM2.5 exposure. The exposure group was defined using the third-trimester exposure window, which corresponds to approximately 3 months before the CB collection. Under high PM2.5 exposure, we observed a loss of strong correlations among maternal-neonatal pairs compared to the low exposure situation. Figure 4B specifically presents strong cytokines coexpression pairs that have opposite directions under low and high exposure (see Materials and Methods). Top three cytokines among pregnant women with the highest betweenness centrality (BC) were IL-1RA (BC:382), macrophage-derived chemokine (MDC)/CCL22 (BC:347), and macrophage colony-stimulating factor (M-CSF) (BC:338). These cytokines have been linked to maternal-fetal immune tolerance, placental development, and pregnancy complications (19–21). The findings suggest that exposures to high PM2.5 levels will likely dysregulate the maternal-fetal immune responses compared to a healthy state.
Fig. 4. Different coexpression patterns in maternal-neonatal pairs between low versus high exposure.
(A) The Pearson correlation matrixes for PM2.5 exposure level (low versus high during the third trimester) show correlations between maternal and neonatal cytokines. Red and blue indicate direct and reverse correlations, respectively. (B) A coexpression differential network was created to compare cytokines that have different coexpression patterns by PM2.5 exposure levels (low versus high) during the third trimester. The edges in the network indicate that the markers are strongly coexpressed, but the direction of coexpression is opposite in low and high exposure. An asterisk (*) next to the marker name indicates that the marker is from CB. The nodes, which represent the cytokines, were colored according to sample type, with pregnant women in beige and CB in green. The size of the nodes is based on the BC, which measures the centrality based on shortest paths.
Exposure to PM2.5 alters the epigenetic profiles in the immune cells of pregnant women compared to nonpregnant women
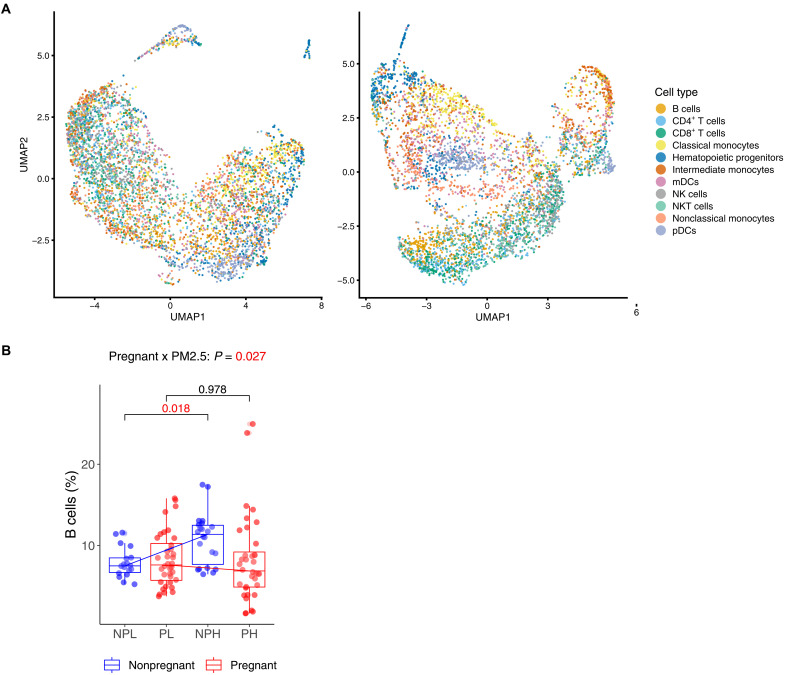
We used an EpiTOF previously published by our group members, to profile 38 targeted HPTMs, at the single-cell level (17). Previously, we characterized and validated more than 150 commercial antibodies to create two panels, each of them consisting of 18 acetylation and 20 methylation-based modifications (table S2) (17). EpiTOF enables us to measure global per-cell levels of 20 methylation and 18 acetylation histone modifications in 11 specific immune cell types (see Materials and Methods), including hematopoietic progenitor cells (HPCs), plasmacytoid dendritic cells (pDCs), myeloid DCs (mDCs), natural killer (NK) cells, NKT cells, B cells, CD4 T cells, CD8 T cells, classical monocytes (cMOs), intermediate monocytes (iMOs), and non-cMOs (ncMOs). UMAP (Uniform Manifold Approximation and Projection) analysis, using all measured cell surface markers as feature inputs (table S3) in the acetylation and methylation panels, shows that the immune cell clusters were expected for both panels (fig. S4). In addition, single-cell UMAP analysis of histone acetylation and methylation showed that histone modifications mostly separated myeloid and lymphoid-lineage cells in both panels (Fig. 5A).
Fig. 5. Impact of PM2.5 on immune cell type proportion depending on pregnancy status.
(A) Single-cell UMAP visualizations of histone acetylation and methylation, colored by cell type identity. (B) Frequency of immune cell subtypes in low and high PM2.5 exposure groups of pregnant and nonpregnant women. The y axis represents the frequency of cells as a percentage of a total number of 11 immune cells population, and the x axis shows the groups categorized by pregnant and PM2.5 exposure states: NPL (nonpregnant low), NPH (nonpregnant high), PL (pregnant low), and PH (pregnant high). The box plot is colored by pregnant status (red, pregnant; blue, nonpregnant). Lines present the changes in cell proportions between low and high exposures within each pregnant status. P values (P) of the interaction term between pregnant status and PM2.5 exposure are noted above graph.
Exposure to PM2.5 significantly changes the cell type proportion of B cells and ncMOs
Both normal pregnancy and exposure to AAP are known to modulate immune cell profiles, yet understanding immune response changes associated with AAP exposure in pregnant women remains limited. We investigated whether changes in the composition of 11 immune cells due to PM2.5 exposure varied by pregnancy status using a mixed-effect regression (see details in Materials and Methods). Figure 5B shows the differentiated impact of PM2.5 exposure on immune cell proportion levels, specifically B cells and ncMOs, modified by pregnancy status. For nonpregnant women, the high PM2.5 exposure group had a 3.6% (P = 0.02) higher proportion of B cells and a 0.86% (P = 0.01) lower proportion of ncMOs compared to those in the low exposure group. Conversely, among pregnant women, the high PM2.5 exposure did not significantly alter cell proportions.
Differential posttranslational epigenetic modifications were associated with PM2.5 exposure
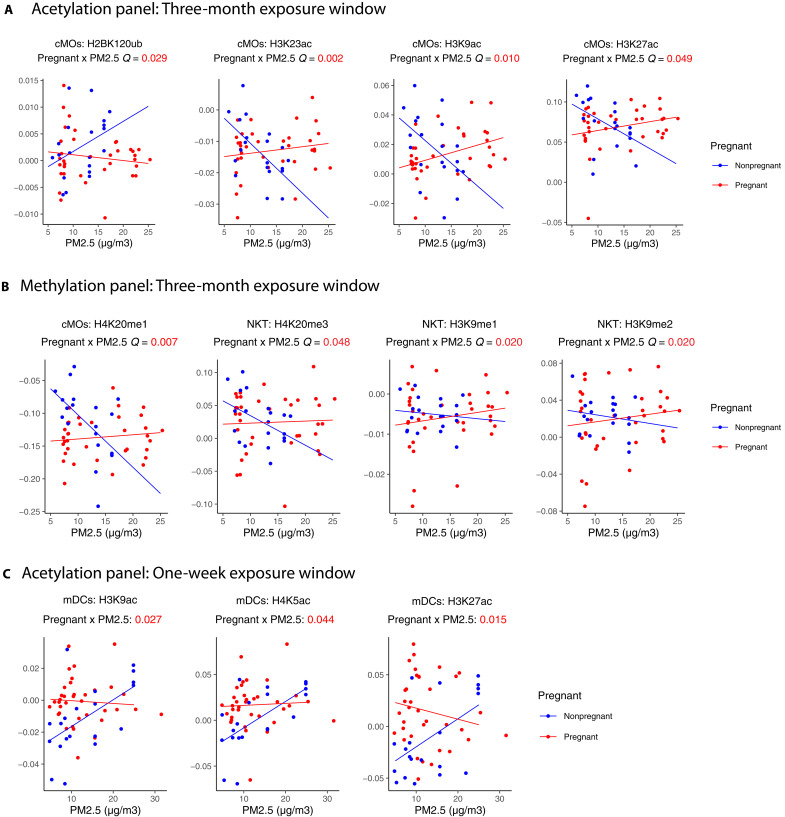
In a multivariable mixed-effect regression using median values of HPTMs at each cell type as the dependent variable, we demonstrated an association between the PM2.5 exposure and the abundance of specific HPTMs in immune cell types differing between pregnant and nonpregnant women. Heatmaps illustrate the main effect of PM2.5, pregnancy, and their interaction from the mixed-effect regression for each acetylation and methylation panel by 1 week, 3 months, 6 months, and 9 months before blood collection (figs. S5 and S6). As in our previous study, a 3-month exposure window showed the strongest results in both panels. Significant interaction effects were observed in H3K9ac (3 months: β = 0.004, Q = 0.010; 6 months: β = 0.006, Q = 0.037), H3K23ac (3 months: β = 0.002, Q = 0.002; 6 months: β = 0.003, Q = 0.019), H3K27ac (3 months: β = 0.008, Q = 0.029), H2AK119ub (3 months: β = 0.004, Q = 0.049), and H4K20me1(3 months: β = 0.008, Q = 0.007; 6 months: β = 0.012, Q = 0.017; 9 months: β = 0.018, Q = 0.035) in cMOs; H3K9me1 (3 months: β = 0.002, Q = 0.020), H3k9me2 (3 months: β = 0.007, Q = 0.020), and H4k20me3 (3 months: β = 0.006, Q = 0.048) in NKT cells; and H3K9ac (1 week: β = −0.002, Q = 0.027), H3K27ac (1 week: β = −0.004, Q = 0.15), and H4K5ac 1 week: β = −0.003, Q = 0.0.44) in mDCs. Our findings indicate that the correlation between PM2.5 exposure and HPTMs in specific immune cell types varies between pregnant statuses.
Figure 6 depicts the changes in HPTM abundance by PM2.5 exposure windows in both groups for HPTMs with a significant interaction effect. For nonpregnant women, a general trend of decreasing abundance was observed with increasing 3-month average PM2.5 exposure, notably for H3K9ac, H3K23ac, H3K27ac, and H4K20me1 in cMOs and H4K20me3, H3K9me1, and H3K9me2 in NKT cells. Conversely, H2BK120ub in cMOs increased with high exposure in nonpregnant women. We also observed a positive association between the average weekly PM2.5 before blood collection and H3K9ac, H4K5ac, and H3K27ac in mDCs. These differences described in nonpregnant women were not observed in pregnant women, indicating a dominant effect of pregnancy modifying the influence of air pollution in specific immune cells.
Fig. 6. An association between PM2.5 exposure and H2AK119Ub, H3K9ac, H3K23ac, H3K27ac, H4K5ac, H3K9me1/2 and H4k20me1/3 in pregnant women versus nonpregnant women across different exposure windows and cell types.
Scatter plots of (A) 3-month window PM2.5 concentration versus the levels for H2BK120ub, H3K23ac, H3K9ac, and H3K27ac in cMOs; (B) 3-month window PM2.5 concentration versus the levels for H4K20me1, H4K20me3, H3K9me1 and H3K9me2 in cMOs; and (C) 1-week window PM2.5 concentration versus the levels for H3K23ac, H3K9ac, H4K5ac, and H3K27ac in mDCs, which had a significant interaction effect. Points and lines on the scatter plot are colored by pregnant status (pregnant women in red and nonpregnant women in blue). Lines shown are fitted within the pregnant status. FDR-adjusted P values (Q value) of the statistical interaction term between pregnant status and PM2.5 levels are noted below the cytokine name. Descriptions for the abbreviations of histone modifications can be found in table S1.
Altered HPTM co-modification profiles during pregnancy are due to PM2.5 exposure level
Understanding the co-modification of HPTMs is crucial to comprehend the regulation of HPTM function in specific cell types under given conditions. One recent study by Ganesan et al. (22) showed direct associations between HPTMs changing 30 days after vaccination, yet no investigation has explored whether exposure to PM2.5 during pregnancy alters the co-modification of HPTMs. In this study, we examined the impact of PM2.5 on the co-modification profile of HPTMs to identify changes in epigenetic regulations in pregnant women. We used a 3-month exposure window, which shows the strongest effect on epigenetic changes in previous analyses. The methods and parameters used in the co-modification profile construction are detailed in Materials and Methods (see “EpiTOF: Network preservation analysis”).
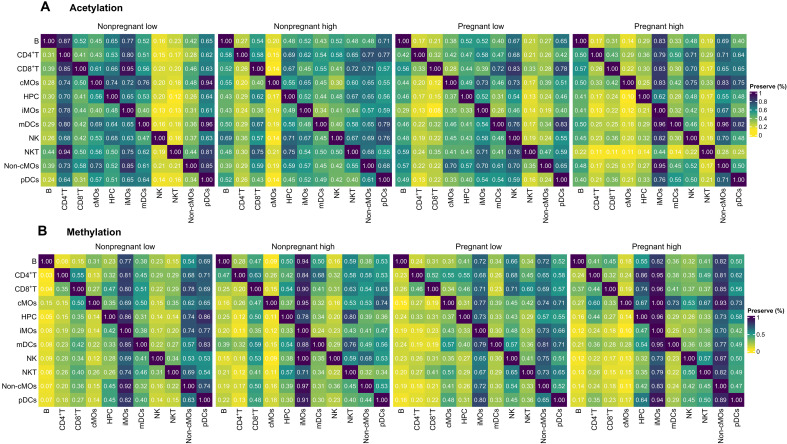
First, we calculated the preservation score, indicating the proportion of HPTM co-modifications found in one cell type that is preserved in another (see details in Materials and Methods). Heatmaps represent co-modification profiles differing across immune cell types and groups (Fig. 7). Figure 7A displays the preservation score for histone acetylation, and Fig. 7B depicts the preservation score for histone methylation across pregnant low-exposure (PL), pregnant high-exposure (PH), nonpregnant low-exposure (NPL), and non-pregnant high-exposure (NPH) groups. Heatmaps suggest altered acetylation- and methylation-associated regulatory profiles of immune cells under air pollution. Overall, immune cells displayed lower preservation scores in pregnant women, indicating less shared co-modification profiles with other immune cells, meaning, unique profiles. For instance, Fig. 7B shows lower preservation scores of immune cells with cMOs, indicating unique co-modification profiles. Conversely, iMOs or ncMOs show high preservation scores, meaning, overlapping profiles with other immune cells. Our results suggest pregnancy and PM2.5 exposure status are associated with immune system dysregulation, affecting HPTM co-modifications in immune cell types.
Fig. 7. PM2.5 exposure affects the similarity of immune cell type–specific HPTM interactions.
Module similarity is based on the proportion of HPTM interactions found in the row immune cells with strong evidence of preservation in the column immune cells. Heatmap (A) represents the acetylation HPTMs and (B) shows the methylation HPTMs. Note that the preservation scores should be read horizontally. Figure 7 summarizes the proportion of significant and strong (rho > 0.5) comethylation and coacetylation profiles predicted in each cell type (row) that are preserved in the other cell types (column). Preservation scores are expressed as % preserved co-modifications and are color-coded from yellow (low) to blue (high).
In the next step, we aim for a comprehensive understanding of potentially associated pathology by analyzing changes in biological pathways and their networks across all immune cell type groups. We identified HPTM co-modification networks using the NPL group as a reference, identifying each of three distinct modules, highly connected groups of HPTMs that have common correlation patterns, for acetylation and methylation panels. We tested whether the modules were preserved between the low and high groups in pregnant and nonpregnant women, respectively (details in Materials and Methods). Table 2 presents P values from the module preservation test between groups, with P values below 0.05 considered preserved. We found the co-modulate of acetylation module (AM) 3 (H3K18ac, H2BK5ac, H3S10ph, H4K5ac, and H3K9ac) and methylation module (MM) 3 (H3K27me1, Rme2sym, Rme2asy, and H3K27me2) were consistent between low and high exposure in both nonpregnant and pregnant women. In addition, MM2 (H3K36me1, H3K36me2, H4K20me2, and H4K20me3) is preserved regardless of PM2.5 exposure level (high versus low) in pregnant women but not in nonpregnant women.
Table 2. HPTM modules and model preservation.
Modules within networks are groups of HPTMs that are tightly connected or coexpressed. To check whether the network topology of each module is preserved between groups, we run a permutation test. We considered a module to be preserved between two groups if a permutation test P value <0.05. Descriptions for the abbreviations of histone modifications can be found in table S1.
| Epigenetic marker | Nonpregnant low versus high | Pregnant low versus high | ||
|---|---|---|---|---|
| (P value) | (P value) | |||
| Acetylation | Module 1 | H2BK120ub, CrotonylK, H3R2cit | 0.236 | 0.292 |
| Module 2 | H4K16ac, H3K23ac, H3K56ac, AcetylK, gammaH2AX | 0.433 | 0.315 | |
| Module 3 | H3K18ac, H2BK5ac, H3S10ph, H4K5ac, H3K9ac | 0.049 | 0.039 | |
| Methylation | Module 1 | H3K4me3, H2A.Z, Rme1, H3K4me2, H3K9me1, CENPA | 0.708 | 0.716 |
| Module 2 | H3K36me1, H3K36me2 H4K20me2, H4K20me3 | 0.282 | 0.030 | |
| Module 3 | H3K27me1, Rme2sym, Rme2asy, H3K27me2 | 0.035 | 0.026 |
DISCUSSION
Our study investigated the impact of PM2.5 exposure on the maternal immune programs, which are crucial for protecting the woman and fetus during pregnancy, including anti-inflammatory immune responses fostering maternal-fetal tolerance. Proinflammatory cytokine responses during pregnancy are associated with adverse birth outcomes, potentially exacerbated by environmental stressors like AAP (6). Furthermore, histone modifications are key epigenetic regulators of cytokine expression (23). This study has uncovered the associations among PM2.5 pollutants, pregnancy, offspring, and maternal immune response using cytokines and HPTMs, and the impact of AAP on maternal-neonatal immune response associations. Our data show that maternal PM2.5 exposure can alter circulating cell frequencies and the abundance and co-modification of cytokines. In addition, PM2.5 was associated with the modulation of HPTMs in pregnant women compared to nonpregnant women. Evidence presented here suggests that maternal exposure to PM2.5 may also affect CB levels of cytokines and distributions of specific immune cells. In addition, we identified the 3-month exposure window before the maternal blood or CB collection had the strongest impact on immune response and epigenetic changes compared to 1-week, 6-month, and 9-month exposure windows.
PM2.5 exposure in pregnant women during the 3 months leading up to the second trimester (18 to 25 weeks) was associated with a significant decrease of four cytokines (IL-RA, IL-8/CXCL8, IL-18, and MIF). Consistent with prior studies, nonpregnant women exposed to high PM2.5 have elevated trends in IL-RA, IL-8/CXCL8, IL-18, and MIF (24–26). While the studies on the effect of PM2.5 exposure on cytokine levels during pregnancy are scarce, previously, our group reported that exposure to AAP is likely to increase the methylation levels at specific CPG sites in the IL-4, IL-10, and interferon-γ genes in pregnant women, suggesting insights into potential mechanism behind the cytokine dysregulation (9). In addition, previous research on maternal levels of cytokines during pregnancy has reported that aberrant levels of IL-RA, IL-8/CXCL8, IL-18, and MIF during pregnancy are linked to complications such as preeclampsia and preterm birth (20, 27–30). Our sensitivity analysis using the different exposure windows also showed consistent outcomes. We observed that the 9-month window was associated with a decrease in IL-7, a cytokine linked to pregnancy loss, similar to recent findings (31).
Since there are limited data on the association between prenatal AAP exposure and fetal inflammation in humans, we conducted high-dimensional immunophenotyping in matched CB pairs with pregnant women in our cohort. Our findings suggest that prenatal exposure to high PM2.5 influences neonatal immune systems. Specifically, we found that pregnant women with high PM2.5 exposure had elevated levels of IL-27, while their newborns had lower levels of IL-27 in CB. IL-27, known to increase considerably in CB dendritic cells upon Toll-like receptor ligation to support neonatal immune response (32), was found to be lower in neonates with high prenatal exposure.
A successful pregnancy requires a healthy maternal immune system to promote communication between the mother and fetus while maintaining tolerance toward the fetus and transferring passive immunity to the offspring. However, we found that PM2.5 exposure can disrupt the normal relationship between cytokines in the mother and fetus, potentially leading to adverse pregnancy outcomes due to the over- or underexpression of specific markers. To maintain a healthy pregnancy, maternal and neonatal immune systems may regulate cytokine levels in response to PM2.5, potentially causing a lack of or weakened correlations between maternal-neonatal pairs under high PM2.5 exposure, as in Fig. 4A. However, it is important to acknowledge that the sample size of pregnant women and CB pairs in this study is small, which may result in limited statistical power.
Identifying how PM2.5 exposure alters cytokines can help us understand the biological mechanism by which AAP exposure affects immune responses during pregnancy. Cytokines often interact cooperatively within and across biological pathways rather than solely affecting the immune system. Differential co-modification network analysis provides valuable information in identifying the important cytokines related to PM2.5 exposure in pregnancy. Our results suggest that highly connected hub markers, including IL-1RA, MDC/CCL22, and M-CSF from CB, might play a critical immune regulatory role in the setting of high prenatal PM2.5 exposure. Aberrant levels of IL-1RA in CB are associated with neonatal morbidity and adverse outcomes (20, 25). A recent study found that parental AAP exposure affects CCL22, a T helper 2–associated chemokine, levels shortly after birth (33). Furthermore, CB levels of the CCL22 are known to be associated with total immunoglobulin E or allergic sensitization risk in asthma later in life (25, 34). Prenatal AAP exposure is likely to elevate levels of M-CSF, which play an essential role in placenta maintenance and lead to preeclampsia (35).
Using a unique and innovative technique to examine epigenetic changes at the single-cell level, we conducted EpiTOF to investigate histone modification at the single-cell level to understand molecular regulatory mechanisms potentially altered by air pollution during pregnancy. In nonpregnant women exposed to high PM2.5 levels, we observed a decrease in the circulating percentage of ncMOs and an increase in B cells compared to women exposed to low PM2.5 levels, consistent with previous studies indicating decreased monocyte counts and impaired phagocytosis and oxidative burst associated with PM exposure (36, 37). We did not observe the same trend in pregnant women. We propose that this finding in pregnant women may reflect the maternal immune system’s efforts to maintain a healthy pregnancy characterized by lower levels of B cells (38) and ncMOS (39).
We observed four histone acetylation sites—H3K23ac, H3K27ac, H3K9ac, and H2AK119Ub—in cMOs decreased with increased PM2.5 exposure during the 3-month window before blood collection in nonpregnant women, while no significant changes in pregnant women. H3K9ac is known to increase after short-term exposure to PM (40, 41) but is also known to be reduced in response to DNA damage (42). Our findings align with these patterns as we also found that PM2.5 exposure during the week before blood collection was positively associated with H3K9ac in mDCs. H3K23ac and H2AK119Ub are, in general, understudied, including how they may be affected by AAP and pregnancy. In pregnant women, H3K23ac remained unchanged regardless of PM2.5 exposure, while a negative association was found in nonpregnant women. According to a recent animal study, utero alcohol exposure may lead to H3K23ac hypoacetylation in the fetal brain (43). Recent studies link H3K23ac to cancer, learning and memory impairment, and obstructive sleep apnea (44–46). Studies have shown that healthy placentas and placentas with fetal growth restriction have different H3K27ac profiles (47). In addition, a recent study found that H3K27ac profiles are affected by air pollution. However, there are no studies that have considered the effects of pregnancy and air pollution together on pregnancies (48). On the basis of previous studies, we can infer the potential impact of PM2.5 on pregnant outcomes. However, further research is necessary to fully understand the implications of this altered response compared to nonpregnant women.
We also found that four histone methylation sites, H4K20me1 in cMOs and H4K20me3 and H3K9me1/2 in NKT cells, were significantly lower in nonpregnant women with high exposure but not in pregnant women. Decreases in H4K20me1 are known to be related to DNA damage accumulation (49). The knockdown of poly(adenosine diphosphate–ribose) polymerase 1 (PARP1) increases SET8 protein levels, which mediate the regulation of H4K20 monomethylation, leading to aberrant H4K20me1.(50) Our previous work found that PARP1 can be affected by wildfire smoke, often containing PM (51). Further, a study on polybrominated diphenyl ethers exposure showed that decabromodiphenyl ether (BDE-209), often found in PM2.5, lowers the levels of SETD9/H4K20me1 and activates the upstream signaling of DNA damage response (DDR) (Mre11/Rad50/NBS1) (52). PR-SET7–mediated H4K20me1/3 is critical in suppressing endogenous retroviruses at trophoblasts to prevent double-stranded RNA stress, excessive interferon response, and necrosis. Moreover, dysregulation of H4K20me1 can result in recurrent miscarriage (53). While no study has investigated the impact of PM2.5 exposure on H4K20me1/3 during pregnancy, it may be reasonable to assume that pregnant women’s H4K20me1 levels remain stable to protect their babies. In addition, studies on the H3K9me1/2 in the context of AAP and pregnancy are limited; a recent study suggested that oxidative stress in the intrauterine environment is likely to induce aberrant H3K9me1/2 levels in placental trophoblasts in preeclampsia (54). Therefore, AAP exposure in mothers might trigger oxidative stress, leading to abnormal histone levels.
This study uses a network approach to understand the co-modification between HPTMs, which is not captured with a traditional approach. We found that cross-immune cell HPTM co-modification profiles varied depending on pregnancy and exposure status, suggesting that AAP exposure may influence chromatin structure and gene expression differently based on pregnancy status. Notably, monocytes exhibit intriguing characteristics in this regard. Our previous study found that PM2.5 contributes to trained immunity in monocytes (55). Thus, the differences in co-modification by PM2.5 exposure level in monocytes might relate to trained immunity, which relies on histone modification and is induced by epigenetic reprogramming (56).
We further test whether HPTM modules were preserved across PM2.5 exposure levels. AM3, related to the opening and relaxation of chromatin structure to promote active transcription, was preserved between low- and high-exposure groups. AM1, related to transcriptional regulation, and AM2, related to DDR, were not. AAP exposures are known to cause changes in the transcriptomic profile due to epigenetic changes and DDR, which may affect the interactions of HPTMs in those modules. Among MMs, MM3, related to transcriptional regulation, was preserved regardless of exposure and pregnancy status. In addition, MM2, related to DNA repair, was only preserved in pregnant women. Histone modifications are important for gene expression and epigenetic regulation. Our results indicate that PM2.5 exposure–induced alterations in gene expression and epigenetic regulation can vary by pregnant status. However, further research is required to explore how changes in these interactions between HPTMs can affect immune response during pregnancy.
One of the strengths of our research is the inclusion of age-matched, nonpregnant participants as controls, allowing us to study how AAP affects the maternal immune system while considering natural pregnancy-related changes in cytokine and HPTM levels. Our analyses identified several previously unidentified associations between maternal AAP exposures and cytokine and HPTM levels, using 3-month exposure periods based on our previous studies (2, 9, 18). Our sensitivity analysis, using another exposure window, also showed that the 3-month period that is the most relevant to immune and epigenetic changes. However, future research should investigate the impact of cumulative or multiple pollutants exposure, which may affect the mechanism of exposure. Prior air pollution health studies typically include adults over a broad range of demographics and lack appropriate controls. Here, we specifically investigated the second trimester, often characterized by a predominant anti-inflammatory maternal immune response crucial for maintaining a healthy pregnancy. Moreover, analysis of matched CB plasma of neonates born to pregnant women in our study enabled us to analyze cytokine expressions as a function of maternal exposure to air pollution. Using a custom human 80-plex assay, we were able to perform simultaneous and comprehensive quantification of 80 cytokines in this study. In addition, this study investigated previously unreported associations of immune cell–specific histone (methylation and acetylation) levels with AAP exposure during pregnancy. Last, our use of the network analysis provided insights into the regulatory mechanisms underlying changes in the interactions between cytokines or HPTMs due to AAP exposure, which was not captured by standard approaches.
Our study recognizes some limitations and challenges in interpreting results. First, estimated PM2.5 exposure was based on residence-based inverse distance–weighted interpolation from the EPA air quality monitoring stations. Although we did validate our data with the high-resolution data (57), we did not compare other studies using different modeling methods for exposure assessment. In addition, the lack of personal exposure data and information on potential confounders or modifiers, such as outdoor activity, individual-level socioeconomic status (SES), and PM2.5 chemical compositions, may introduce a bias or misclassification. Blood draws occurred across different seasons but using a 3-month exposure window rather than a single time point led us to conclude that simply using the four seasons to control for seasonality does not accurately reflect the seasonal variations in environmental exposure levels. To address the limited availability of individual-level SES data, we use the area-level SES, which is recommended and commonly used in the AAP studies (58, 59). Another limitation is that pregnant women were from Fresno, while nonpregnant women were from Fresno or the Bay Area in California, which may lead to potential selection bias. However, we did not find distinct patterns in cytokine or histone modification levels by geographic location, and we control potential confounding in our statistical analysis to mitigate this difference. However, caution is warranted when interpreting the results. Other limitations include a lack of adjustment for clinical information of pregnant women and birth outcomes, which could contribute to cytokine levels of CB and potentially confound results. In addition, maternal blood samples were collected at a single mid-pregnancy time point, limiting the generalization of conclusions to other pregnancy periods regarding inflammatory cytokines and HPTMs in response to AAP exposures. EpiTOF offers insights into global epigenetic states at a single-cell level, but it lacks locus-specific information. Integrating EpiTOF with single-cell assay for transposase-accessible chromatin (ATAC) sequencing and/or cellular indexing of transcriptomes and epitopes by sequencing/single-cell RNA sequencing in future studies could enhance understanding of epigenetic processes due to air pollution by identifying HPTM combinations associated with chromatin accessibility or transcriptome profiles in a given cell type during different immunological states. Despite its limitations, EpiTOF is a powerful tool that provides multiplexed analysis of chromatin modification in single cells and offers single-cell resolution and multilayered information. In conclusion, we found that exposure to PM2.5 was significantly associated with cytokines and HPTM responses in pregnancy, and this association could be due to a competing effect between exposure and the maternal immune response, which is altered during pregnancy to potentially prevent adverse pregnancy outcomes.
MATERIALS AND METHODS
Sample and data collection
Blood samples were collected from pregnant women (n = 168), approximately 20 weeks into their pregnancy, living in Fresno, California, a city with high air pollution. Neonatal samples (n = 33) were also collected from a subset of the pregnant women. Blood samples from age-matched nonpregnant women (n = 151), a control group in the analyses, were collected from Fresno and the Bay Area in California. None of the homes used wood burning, and the participants had infections or other acute or chronic medical conditions or were on medications that could modify the immune system. In addition, no pregnant women were currently smoking.
An 80-plex Luminex assay with an EMD Millipore human cytokine panel was used to quantify cytokine levels in the plasma samples of the pregnant women, nonpregnant women, and CB plasma of the neonates that match the pregnant women. We used a subset of samples from the pregnant women (n = 36) and nonpregnant women (n = 20) for EpiTOF. We quantified 14 surface markers and 38 posttranslational modifications of histone tails (HPTMs) in the plasma samples of pregnant and nonpregnant women.
Guidelines and ethical statement
All participants in this study gave written informed consent for the protocol that was approved by Stanford University’s Institutional Review Board (Ethics approval number 28263). This study was conducted following the Declaration of Helsinki, which covers informed consent, privacy and confidentiality, ethical considerations, and assessment of any risks. This study followed all relevant guidelines of clinical research, including clinical value, scientific validity, fair subject selection, favorable risk-benefit ratio, independent review, informed consent, and respect for potential and enrolled participants.
Air pollution exposure estimation and analysis
Average air pollutant exposures were calculated for each participant over a 3-month period before the blood sample was collected. The 3-month exposure windows before the blood draw were selected on the basis of our prior research because this time frame is associated with the strongest effect on immune and epigenetic changes (2, 9, 18). The 3-month exposure window before CB collection corresponded to the exposure period during the third trimester of pregnancy. To calculate each trimester, we used the date of the CB collection and the number of weeks of gestation at the time of the mother’s blood collection. For the sensitivity analysis, we included the exposure window of 1 week, 6 months, and 9 months before the blood draw to calculate the average PM2.5 exposure. The CB analysis used the exposure window of 1 week before a CB collection date, as well as the first and second trimesters. We modeled outdoor residential air pollution exposure to PM2.5 using air quality data collected at outdoor monitors from the US EPA Air Quality System, applying inverse distance–squared weighting. EPA’s Air Quality System offered provided hourly, quality-assured ambient pollutant concentrations from on-site monitoring stations, with daily summary PM2.5 Federal Reference Methods/Federal Equivalent Methods Mass (88101) data available from 2010 through 2020, covering the time window of our study. First, we checked the participants’ residential addresses (street addresses, cities, zip codes, and states) obtained through a self-report survey and consent form. For the pregnant women study, we additionally collected residential address history with residential periods. For individuals with missing information or incorrect addresses in the database, we cross-checked the information using their hard copy consent forms from clinic visits. Participants with missing information were excluded (n = 9) from our study. We primarily used street addresses to convert their residential locations into latitude and longitude using the R package ggmap and the Google Application Programming Interface key. If the geographic location did not convert to latitude and longitude due to incorrect information (e.g., incorrect zip code), we manually verified them using the map. Furthermore, we confirmed that no residential mobility occurred during the 3 months leading up to blood sample collection. For interpolation, we considered up to four closest stations within 50 km of the participant’s address, excluding stations near roads. We have excluded near-road sites because the near-road data are only representative of conditions in a narrow band along major roadways/freeways. In addition, we have excluded the exceptional event data from the monitor PM2.5 values, which were flagged by the monitor report. If the nearest monitor is within 0.25 km, we only use data from that monitor. Most of our study population lives within 10 km of the EPA stations (fig. S1A). We calculated the distance between a participant and stations and calculated the weighted concentration using PM2.5 measurement from the closest station. In addition, we verified our estimates with the high spatiotemporal resolution PM2.5 data (10-km grid, 2000 to 2016) from Di et al. (57, 60) using our study samples that overlapped with their time period. The Pearson correlation indicated that our estimates are consistent (ρ = 0.97) (fig. S1B).
Blood processing
We extracted human peripheral blood mononuclear cells (PBMCs) and plasma from the collected blood sample using a Ficoll procedure, allowing rapid and efficient isolation of mononuclear cells and storing them in liquid nitrogen and at −80°C per published techniques (61).
Luminex
Luminex assay
Luminex-EMD Millipore Human 80 Plex kits: This assay was performed by the Human Immune Monitoring Center at Stanford University. Kits were purchased from EMD Millipore Corporation, Burlington, MA, and run according to the manufacturer’s recommendations with the following modifications: H80 kits included three panels: Panel 1 was Milliplex HCYTA-60 K-PX48. Panel 2 was Milliplex HCP2MAG-62 K-PX23. Panel 3 included the Milliplex HSP1MAG-63 K-06 and HADCYMAG-61 K-03 (resistin, leptin, and HGF) to generate a nine-plex. The assay setup followed the recommended protocol: Briefly, samples were diluted 3-fold (panels 1 and 2) and 10-fold (panel 3). Twenty-five microliters of the diluted sample was mixed with antibody-linked magnetic beads in a 96-well plate and incubated overnight at 4°C with shaking. Cold and room temperature incubation steps were performed on an orbital shaker at 500 to 600 rpm. The plates were washed twice with wash buffer in a BioTek ELx405 washer (BioTek Instruments, Winooski, VT). Following 1 hour of incubation at room temperature with a biotinylated detection antibody, streptavidin-phycoerythrin was added for 30 min with shaking. The plates were washed as described above, and phosphate-buffered saline (PBS) was added to wells for reading in the Luminex FlexMap3D Instrument with a lower bound of 50 beads per sample per cytokine. Each sample was measured in a single well. Custom Assay Chex control beads were purchased and added to all wells (Radix BioSolutions, Georgetown, Texas). The wells with a bead count <50 were flagged, and data with a bead count <20 were excluded.
PCA and ComBat
To identify the potential batch and confounder effect, we used principal components analysis (PCA) to determine the significant sources of variation. We included age, batch, race/ethnicity (Hispanic white, non-Hispanic white, and non-Hispanic others), whether having asthma (yes, no, and do not know), PM2.5 exposure levels (μg/m3), zip code level SDI score, and pregnancy states. We found batch effects account for a significant source of variation in the data along with race/ethnicity, age, asthma, and SDI score. To adjust the batch effect in the data, the ComBat was applied using the sva package in R. After implementing the ComBat, the PCA results showed that the batch effect was removed. Age, race/ethnicity, asthma status, and SDI score were included as confounders in the model. (fig. S5A).
Multivariable linear regression analysis
To investigate the association between PM2.5 levels measured 3 months before the blood draw and the 80 cytokine and chemokine marker levels in pregnant women compared to nonpregnant women, we used multivariable linear models at the person-level that included pregnant status, (pregnant versus nonpregnant) PM2.5 (μg/m3, continuous), the interaction between pregnant and PM2.5, age (continuous), ethnicity (Hispanic versus not Hispanic), age, asthma status (yes versus no), and zip code level SDI score. Because of a lack of individual-level SES, we used the SDI score, a composite measure of area-level deprivation based on seven demographic characteristics, collected in the American Community Survey: percent living in poverty, percent with less than 12 years of education, percent single-parent households, the percentage living in rented housing units, the percentage living in the overcrowded housing unit, percent of households without a car, and percentage nonemployed adults under 65 years of age (62). Moss et al. (58) suggested that area-level SES measured at smaller geographic units can be the best option when the individual-level SES is not available. We additionally used the 1 week, 6 months, and 9 months to calculate the average PM2.5 levels for the sensitivity analysis.
Next, to investigate that the impact of PM2.5 levels during the third trimester affects the correlation between immune biomarkers in CB and pregnant women, we used a multivariable regression model for 33 pairs of pregnant women and CB samples. The level of cytokine in CB is a function of the level of matching cytokines in CB’s mother, PM2.5 exposure level (low versus high; the cutoff point is 12 μg/m3), the interaction between the mother’s matching cytokine level and PM2.5, mother’s age, ethnicity (Hispanic versus not Hispanic), asthma status, and SDI score. Because of the small sample size, we dichotomized PM2.5 exposure using the 2012 EPA National Ambient Air Quality Standards for PM2.5, which is set at 12 μg/m3 to protect public health. We categorize the low-exposure group as below the 12 μg/m3 cutoff point, although this level does not indicate as safe to AAP. We hypothesized that the exposure level affects immunity passed from mother to baby by including interaction terms between exposure and maternal expression markers during pregnancy. We additionally used the 1 week, first trimester, and second trimester to calculate the average PM2.5 levels for the sensitivity analysis. We also used an alternative cutoff point (12 μg/m3) for the EPA standard to check the robustness of the main results of the CB analysis: We used the 25th quartile (11.14 μg/m3) of exposure levels in the third trimester of pregnancy to separate the low- and high-exposure groups.
For both models, we used FDR correction to correct for multiple testing. We used a Q value to denote the adjusted P value across all regression models, and Q < 0.05 was considered statistically significant.
Differential coexpression network analysis
To investigate the cytokines interactions between maternal-neonatal pairs exposed to low and high PM2.5, we used differential coexpression network analysis following the methodology outlined in Pettersen and Almaas (63). We first calculated the Pearson correlation between each pair of cytokines separately for the low- and high-exposure groups. We then used the csdR package [v.1.4.0] to calculate the differentiated scores by comparing the value of the mean correlation and SDs from the low- and high-exposure groups. By using
Conserved, Specific, and Differentiated approach, we calculated the differentiated (D) score, representing coexpression in both low and high exposures, but with the opposite sign, to estimate the differentiated effect of PM2.5 on maternal-neonatal coexpression. To identify the cytokine pairs with the highest value for D score, we used the cutoff point using an inverse of the common importance level P = 0.01. Using the list of markers, we generated a network that shows the strong interactions present in both low- and high-exposure groups but with opposite signs. The size of the nodes in the network indicates the number of connections between cytokines.
EpiTOF
Detailed protocols for EpiTOF sections, including lanthanide labeling of antibodies, mass cytometry, immune cell population definitions and data preprocessing, fluorescence-activated cell sorting (FACS) and Western blotting, and data processing and normalization are described in Cheung et al. (17). We followed these methods precisely in our study. Here, we provide a brief summary of the protocol.
Lanthanide labeling of antibodies for mass cytometry
Antibodies were conjugated using lanthanides from tables S3 and S4 using the MAXPAR antibody labeling kit (Fluidigm). Tris-(2-Carboxyethyl)phosphine, Hydrochloride (TCEP) (Thermo Fisher Scientific) was added to create sulfhydryl groups for maleimide-mediated conjugation of metal-chelating polymers. Conjugated antibodies were diluted in antibody stabilizing solution (Boca Scientific) with sodium azide (Sigma-Aldrich) for storage.
Mass cytometry (sample processing, staining, barcoding, and data collection)
Briefly, cryopreserved PBMCs were thawed and incubated in RPMI 1640 media with 10% fetal bovine serum at 37°C for 1 hour. After adding 10 μM cisplatin for viability staining, the cells were quenched with cytometry by time of flight (CyTOF) buffer and centrifuged. They were then stained with lanthanide-labeled antibodies against immunophenotypic markers, washed with CyTOF buffer, fixed in 1.6% paraformaldehyde, centrifuged for post-fixation, permeabilized with methanol, and processed for mass-tag sample barcoding. Intracellular staining was performed overnight in CyTOF buffer, followed by staining with 250 nM 191/193Ir DNA intercalator. The cells were washed, filtered, and analyzed on the CyTOF2 in Stanford Shared FACS Facility. Raw data were concatenated and normalized using calibration beads for down streaming processing.
Immune cell population definitions and data preprocessing
Raw data were preprocessed using FlowJo (FlowJo LLC) to identify cellular events through palladium-based mass tags and to isolate specific immune cell populations using immunophenotypic markers. Single-cell data for different immune cell subtypes in individual subjects were exported from FlowJo for downstream computational analyses.
FACS and Western blotting
Fresh PBMCs or cells recovered from cryopreservation were resuspended in PBS and incubated for viability staining, quenched with CyTOF buffer, centrifuged, and resuspended in CyTOF buffer containing antibodies against immunophenotypic markers. Cells were sorted on the basis of markers for cell types (table S.3). FACS was performed on sorters in Stanford Shared FACS Facility. Sorted cells were collected, centrifuged, and resuspended in 5× SDS sample buffer, and sonicated to obtain homogenized lysates for Western blot analysis.
Data processing and normalization
Single-cell chromatin data were normalized to basal levels of histone proteins to account for variations in mark levels due to changes in global amounts of chromatin. We used a multivariable linear regression model for each chromatin mark across all cell types and subjects. The normalized chromatin score is the residual of the regression. Normally distributed residuals suggest that the model fits the data. For each chromatin mark, we calculated the mean level of the mark in each cell type across all subjects.
UMAP analysis
For the UMAP analysis using the cell surface markers listed in table S3, a maximum of 5000 cells for each cell type were subsampled to ensure consistency as the proportions of immune cell types in PBMCs vary. For the cell type–specific UMAP, either 5000 cells or the total number of cells of that specific cell type were used, with the epigenetic modification markers shown in table S3. The analyses were performed separately for the acetylation and methylation panels. All analyses were conducted using the uwot package (v.0.1.10).
Principal components analysis
PCA was used to determine sources of significant variation and detect potential batch and/or confounding effects. We included age, batch, race/ethnicity (Hispanic white, non-Hispanic white, and non-Hispanic others), whether having asthma (yes, no), PM2.5 exposure levels (microgram per cubic meter), pregnancy status, and SDI score. (fig. S7B).
Mixed-effect regression analysis
To evaluate the association between PM2.5 levels measured 3 months before the blood draw and (i) the cell type proportion and (ii) HPTM levels, a multivariable mixed-effect model was performed. Cell proportion was computed as the proportion of each immune cell type for every individual. The association between cell proportion and PM2.5 exposure during pregnancy was estimated, considering pregnant status, PM2.5 exposure status (low versus high), the interaction term of these two factors, age, asthma history, ethnicity, and random effects for batch and panels. We reported an unadjusted P value, and P < 0.05 was considered statistically significant.
The association between the cell-specific HPTM levels and PM2.5 across pregnant status was measured using the mixed-effect model, including pregnant status, PM2.5 exposure status (microgram per cubic meter), the interaction term of these two factors, age, asthma history, and random effects for batch. We included age and asthma history, as these were the most significant factors affecting epigenetic changes. We used the average PM2.5 levels for 1 week, 6 months, and 9 months before the blood collection for the sensitivity analysis. To correct for multiple testing, we use FDR correction. We used a Q value to denote the adjusted P value across all models, and Q < 0.05 was considered statistically significant.
Network preservation analysis
Initially, we prepared the dataset by converting the single-cell level data into sample-wise data. We calculated the median value of HPTM for each sample and immune cell type, subsequently grouping the samples on the basis of pregnant status and PM2.5 exposure levels into four datasets: PL, PH, NPL, and NPH groups. Using these datasets, we conducted edge and module preservation analysis.
For edge preservation, we followed Ritchie et al. (2016) to examine whether the discovered co-modification profile within an immune cell type was preserved across other immune cells (64). We constructed the Pearson correlation matrices from each dataset, retaining only strong and significant correlations between pairs of HPTMs (rho > 0.5 and P value <0.05). These matrices were converted into network matrices using the graph.adjacency function from igraph R packages (v.1.5.1). Total edge counts were then calculated for each cell type and group. To test whether the co-modification profiles within one immune cell type were preserved in others, we applied the intersection function to calculate shared edge numbers. By comparing the total edge and shared edge, we were able to calculate the proportion of preserved edges across different cells. For example, if B cells and CD4+ T cells shared five edges of B cells’ total 20 edges, then 25% of co-modifications in B cells were preserved in CD4+ T cells.
Regarding module preservation, we first identified co-modification histone sets with similar expression patterns using the mclust (v.6.0.1) R package for model-based clustering. The PL dataset served as the reference group for model-based clustering, resulting in three distinct modules for each acetylation and methylation panel. With modules from the PL dataset, we used NetRep, a rapid and computationally efficient method with a permutation approach, to test the module preservation across different groups (PL, PH, NPL, and NPH). NetRep analysis involved calculating Pearson correlation matrixes and network edge matrixes for each group. For the network edge matrixes, we only considered the strong (rho > 0.5) and significant (P value < 0.05) correlation between HPTMs. We tested the impact of PM2.5 exposure on module preservation by comparing (i) PL and PH and (ii) NPL and NPH. Therefore, we tested whether each module is preserved between low and high PM2.5 exposure groups by estimating permutation P values with 10,000 permutations per test. In our study, module preservation was determined whether permutation test P values below 0.05. More details on the permutation test from NetRep can be found in the Ritchie et al. paper (64).
Acknowledgments
We would like to thank the UCSF-Fresno research team (G. Aguilar,C. Bonilla, K. Corona, C. Cortez, A. Lopez, C. Orozco, and J. Blaauw) for work in conducting the clinical visits; B. Nguyen and K. Nabaglo for assistance with blood and urine analyses in the Holland laboratory; B. Thapa, N. Myren, and B. MacDonald for general assistance with study management; and all of the participating children and families for their patience and dedication to the study.
Funding: This work was supported by the National Institutes of Environmental Health Science grant R01ES032253 (K.C.N); National Heart, Lung, and Blood Institute grant R01HL081521; National Institutes of Health/ Environmental Protection Agency grants EPA R834596/NIEHS P01ES022849 and EPA RD835435/NIEHS P20ES018173; and SNP Center for Asthma and Allergy Research at Stanford University.
Author contributions: All the other authors reviewed the draft and approved the final manuscript. Conceptualization: Y.S.J., J.A., J.W.H., S.K.H., J.B., E.N., T.D.B., P.J.U., A.R.W., R.D., H.M., M.P. (now M.M.J.), and K.C.N. Methodology: Y.S.J., J.A., A.K., J.W.H., E.Y., R.A., F.L., L.L., K.H., P.J.U., T.D.B., Y.R.H., P.K., H.T.M., M.P. (now M.M.J.), and K.C.N. Investigation: J.A., J.W.H., S.C., E.Y., R.A., K.H., Y.R.H., M.P. (now M.M.J.), and K.C.N. Visualization: Y.S.J., R.A., H.T.M., and K.C.N. Supervision: J.A., A.K., J.W.H., S.K.H., P.J.U., Y.R.H., H.T.M., M.P. (now M.M.J.), and K.C.N. Writing—original draft: Y.S.J., J.A., A.K., J.W.H., R.D., and K.C.N. Writing—review and editing: Y.S.J., J.A., A.K., J.W.H., E.Y., L.L., J.B., PJ.U., P.K., R.A., T.D.B., N.A., A.R.W., H.T.M., and K.C.N. Resources: J.W.H., S.C., E.Y., S.K.H., P.J.U., H.T.M., Y.R.H., and K.C.N. Funding acquisition: S.K.H., J.B., T.D.B., and K.C.N. Validation: J.W.H., E.Y., R.A., S.K.H., A.K., and Y.R.H. Formal analysis: Y.S.J., A.K., J.W.H., S.C., E.Y., P.K., N.A., and K.C.N. Project administration: J.A., A.K., J.W.H., J.B., P.J.U., S.K.H., P.K., R.A., Y.R.H., and K.C.N. Data curation: Y.S.J., J.W.H., L.L., and K.C.N. Software: Y.S.J., J.W.H., F.L., and L.L.
Competing interests: The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Tables S1 to S4
REFERENCES AND NOTES
- 1.Glencross D. A., Ho T.-R., Camiña N., Hawrylowicz C. M., Pfeffer P. E., Air pollution and its effects on the immune system. Free Radic. Biol. Med. 151, 56–68 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Prunicki M., Cauwenberghs N., Lee J., Zhou X., Movassagh H., Noth E., Lurmann F., Hammond S. K., Balmes J. R., Desai M., Wu J. C., Nadeau K. C., Air pollution exposure is linked with methylation of immunoregulatory genes, altered immune cell profiles, and increased blood pressure in children. Sci. Rep. 11, 4067 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H., Ryu M. H., Orach J., Yuen A., Lau K. S. K., Yeung C., Wen W., Carlsten C., Acute air pollution exposure increases TET enzymes in human PBMCs. J. Allergy Clin. Immunol. 150, 477–488.e9 (2022). [DOI] [PubMed] [Google Scholar]
- 4.Thangavel P., Park D., Lee Y.-C., Recent insights into particulate matter (PM2.5)-mediated toxicity in humans: An overview. Int. J. Environ. Res. Public Health 19, 7511 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kannan S., Misra D. P., Dvonch J. T., Krishnakumar A., Exposures to airborne particulate matter and adverse perinatal outcomes: A biologically plausible mechanistic framework for exploring potential effect modification by nutrition. Environ. Health Perspect. 114, 1636–1642 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aguilera J., Konvinse K., Lee A., Maecker H., Prunicki M., Mahalingaiah S., Sampath V., Utz P. J., Yang E., Nadeau K. C., Air pollution and pregnancy. Semin. Perinatol. 47, 151838 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holzhausen E. A., Shen N., Chalifour B., Tran V., Li Z., Sarnat J. A., Chang H. H., Jones D. P., Goran M. I., Liang D., Alderete T. L., Longitudinal profiles of the fecal metabolome during the first 2 years of life. Sci. Rep. 13, 1886 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgan Z. E. M., Bailey M. J., Trifonova D. I., Naik N. C., Patterson W. B., Lurmann F. W., Chang H. H., Peterson B. S., Goran M. I., Alderete T. L., Prenatal exposure to ambient air pollution is associated with neurodevelopmental outcomes at 2 years of age. Environ. Health 22, 11 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aguilera J., Han X., Cao S., Balmes J., Lurmann F., Tyner T., Lutzker L., Noth E., Hammond S. K., Sampath V., Burt T., Utz P. J., Khatri P., Aghaeepour N., Maecker H., Prunicki M., Nadeau K., Increases in ambient air pollutants during pregnancy are linked to increases in methylation of IL4, IL10, and IFNγ. Clin. Epigenetics 14, 40 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yockey L. J., Iwasaki A., Interferons and proinflammatory cytokines in pregnancy and fetal development. Immunity 49, 397–412 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hahn J., Gold D. R., Coull B. A., McCormick M. C., Finn P. W., Perkins D. L., Rifas Shiman S. L., Oken E., Kubzansky L. D., Air pollution, neonatal immune responses, and potential joint effects of maternal depression. Int. J. Environ. Res. Public Health 18, 5062 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X., Tong J., Liang C., Wang X., Ma Y., Tao S., Liu M., Wang Y., Liu J., Yan S., Gao G., Wu X., Huang K., Cao Y., Tao F., Trimester-specific effects of maternal exposure to single and mixed metals on cord serum inflammatory cytokines levels: A prospective birth cohort study. Sci. Total Environ. 895, 165086 (2023). [DOI] [PubMed] [Google Scholar]
- 13.Mukherjee S., Dasgupta S., Mishra P. K., Chaudhury K., Air pollution-induced epigenetic changes: Disease development and a possible link with hypersensitivity pneumonitis. Environ. Sci. Pollut. Res. Int. 28, 55981–56002 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrawus M., Sharvit L., Atzmon G., Epigenetics and pregnancy: Conditional snapshot or rolling event. Int. J. Mol. Sci. 23, 12698 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leong M., Karr C. J., Shah S. I., Brumberg H. L., Before the first breath: Why ambient air pollution and climate change should matter to neonatal-perinatal providers. J. Perinatol. 43, 1059–1066 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.US EPA, National Ambient Air Quality Standards (NAAQS) for PM. (n.d.). https://www.epa.gov/pm-pollution/national-ambient-air-quality-standards-naaqs-pm.
- 17.Cheung P., Vallania F., Warsinske H. C., Donato M., Schaffert S., Chang S. E., Dvorak M., Dekker C. L., Davis M. M., Utz P. J., Khatri P., Kuo A. J., Single-cell chromatin modification profiling reveals increased epigenetic variations with aging. Cell 173, 1385–1397.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prunicki M., Stell L., Dinakarpandian D., de Planell-Saguer M., Lucas R. W., Hammond S. K., Balmes J. R., Zhou X., Paglino T., Sabatti C., Miller R. L., Nadeau K. C., Exposure to NO2, CO, and PM2.5 is linked to regional DNA methylation differences in asthma. Clin. Epigenetics 10, 2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keith J. C., Pijnenborg R., Luyten C., Spitz B., Schaub R., Van Assche F. A., Maternal serum levels of macrophage colony-stimulating factor are associated with adverse pregnancy outcome. Eur. J. Obstet. Gynecol. Reprod. Biol. 89, 19–25 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Ruiz R. J., Jallo N., Murphey C., Marti C. N., Godbold E., Pickler R. H., Second trimester maternal plasma levels of cytokines IL-1Ra, Il-6 and IL-10 and preterm birth. J. Perinatol. 32, 483–490 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao Y., Xu X.-H., Jin L., Macrophage polarization in physiological and pathological pregnancy. Front. Immunol. 10, 792 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganesan A., Dermadi D., Kalesinskas L., Donato M., Sowers R., Utz P. J., Khatri P., Inferring direction of associations between histone modifications using a neural processes-based framework. iScience 26, 105756 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munro S. K., Balakrishnan B., Lissaman A. C., Gujral P., Ponnampalam A. P., Cytokines and pregnancy: Potential regulation by histone deacetylases. Mol. Reprod. Bev. 88, 321–337 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Zou W., Wang X., Hong W., He F., Hu J., Sheng Q., Zhu T., Ran P., PM2.5 induces the expression of inflammatory cytokines via the Wnt5a/Ror2 pathway in human bronchial epithelial cells. Int. J. Chron. Obstruct. Pulmon. Dis. 15, 2653–2662 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calderon-Garciduenas L., Cross J., Franco-Lira M., Aragon-Flores M., Kavanaugh M., Torres-Jardon R., Chao C., Thompson C., Chang J., Zhu H., D’Angiulli A., Brain immune interactions and air pollution: Macrophage inhibitory factor (MIF), prion cellular protein (PrPC), Interleukin-6 (IL-6), interleukin 1 receptor antagonist (IL-1Ra), and interleukin-2 (IL-2) in cerebrospinal fluid and MIF in serum differentiate urban children exposed to severe vs. low air pollution. Front. Neurosci. 7, 183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucht S., Hennig F., Moebus S., Führer-Sakel D., Herder C., Jöckel K.-H., Hoffmann B., Air pollution and diabetes-related biomarkers in non-diabetic adults: A pathway to impaired glucose metabolism? Environ. Int. 124, 370–392 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Sun L., Mao D., Cai Y., Tan W., Hao Y., Li L., Liu W., Association between higher expression of interleukin-8 (IL-8) and haplotype −353A/−251A/+678T of IL-8 gene with preeclampsia: A case–control study. Medicine 95, e5537 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang X., Huang H., Dong M., Yao Q., Wang H., Serum and placental interleukin-18 are elevated in preeclampsia. J. Reprod. Immunol. 65, 77–87 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Todros T., Paulesu L., Cardaropoli S., Rolfo A., Masturzo B., Ermini L., Romagnoli R., Ietta F., Role of the macrophage migration inhibitory factor in the pathophysiology of pre-eclampsia. Int. J. Mol. Sci. 22, 1823 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jahantigh D., Ghazaey Zidanloo S., Forghani F., Doroudian M., IL-27 variants might be genetic risk factors for preeclampsia: Based on genetic polymorphisms, haplotypes and in silico approach. Mol. Biol. Rep. 47, 7929–7940 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Yount C. S., Scheible K., Thurston S. W., Qiu X., Ge Y., Hopke P. K., Lin Y., Miller R. K., Murphy S. K., Brunner J., Barrett E., O’Connor T. G., Zhang J., Rich D. Q., Short term air pollution exposure during pregnancy and associations with maternal immune markers. Environ. Res. 260, 119639 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Birkholz J., Doganci A., Darstein C., Gehring S., Zepp F., Meyer C. U., IL-27 improves migrational and antiviral potential of CB dendritic cells. Hum. Immunol. 75, 584–591 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Pedersen C.-E. T., Eliasen A. U., Ketzel M., Brandt J., Loft S., Frohn L. M., Khan J., Brix S., Rasmussen M. A., Stokholm J., Chawes B., Morin A., Ober C., Bisgaard H., Pedersen M., Bønnelykke K., Prenatal exposure to ambient air pollution is associated with early life immune perturbations. J. Allergy Clin. Immunol. 151, 212–221 (2023). [DOI] [PubMed] [Google Scholar]
- 34.Følsgaard N. V., Chawes B. L. K., Bønnelykke K., Jenmalm M. C., Bisgaard H., Cord blood Th2-related chemokine CCL22 levels associate with elevated total-IgE during preschool age. Clin. Exp. Allergy 42, 1596–1603 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Hayashi M., Numaguchi M., Ohkubo N., Yaoi Y., Blood macrophage colony-stimulating factor and thrombin-antithrombin III complex concentrations in pregnancy and preeclampsia. Am. J. Med. Sci. 315, 251–257 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Li Z., Li X., Song H., Tao B., Qiu B., Tian D., Zhan M., Wu Z., Wu J., Zhang Q., Wang J., Effects of short-term ambient PM2.5 exposure on the blood cell count and hemoglobin concentration among 82,431 people in eastern China. Sci. Total Environ. 776, 146046 (2021). [Google Scholar]
- 37.Becker S., Decreased Cd11b expression, phagocytosis, and oxidative burst in urban particulate pollution-exposed human monocytes and alveolar macrophages. J. Toxicol. Environ. Health 55, 455–477 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Lima J., Martins C., Leandro M. J., Nunes G., Sousa M.-J., Branco J. C., Borrego L.-M., Characterization of B cells in healthy pregnant women from late pregnancy to post-partum: A prospective observational study. BMC Pregnancy Childbirth 16, 139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al-ofi E., Coffelt S. B., Anumba D. O., Monocyte subpopulations from pre-eclamptic patients are abnormally skewed and exhibit exaggerated responses to toll-like receptor ligands. PLOS ONE 7, e42217 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng Y., Sanchez-Guerra M., Zhang Z., Joyce B. T., Zhong J., Kresovich J. K., Liu L., Zhang W., Gao T., Chang D., Osorio-Yanez C., Carmona J. J., Wang S., McCracken J. P., Zhang X., Chervona Y., Díaz A., Bertazzi P. A., Koutrakis P., Kang C.-M., Schwartz J., Baccarelli A. A., Hou L., Traffic-derived particulate matter exposure and histone H3 modification: A repeated measures study. Environ. Res. 153, 112–119 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding R., Jin Y., Liu X., Ye H., Zhu Z., Zhang Y., Wang T., Xu Y., Dose- and time- effect responses of DNA methylation and histone H3K9 acetylation changes induced by traffic-related air pollution. Sci. Rep. 7, 43737 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tjeertes J. V., Miller K. M., Jackson S. P., Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 28, 1878–1889 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rychlik K. A., Secrest J. R., Lau C., Pulczinski J., Zamora M. L., Leal J., Langley R., Myatt L. G., Raju M., Chang R. C.-A., Li Y., Golding M. C., Rodrigues-Hoffmann A., Molina M. J., Zhang R., Johnson N. M., In utero ultrafine particulate matter exposure causes offspring pulmonary immunosuppression. Proc. Natl. Acad. Sci. U.S.A. 116, 3443–3448 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li K.-L., Zhang L., Yang X.-M., Fang Q., Yin X.-F., Wei H.-M., Zhou T., Li Y.-B., Chen X.-L., Tang F., Li Y.-H., Chang J.-F., Li W., Sun F., Histone acetyltransferase CBP-related H3K23 acetylation contributes to courtship learning in Drosophila. BMC Dev. Biol. 18, 20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klein B. J., Jang S. M., Lachance C., Mi W., Lyu J., Sakuraba S., Krajewski K., Wang W. W., Sidoli S., Liu J., Zhang Y., Wang X., Warfield B. M., Kueh A. J., Voss A. K., Thomas T., Garcia B. A., Liu W. R., Strahl B. D., Kono H., Li W., Shi X., Côté J., Kutateladze T. G., Histone H3K23-specific acetylation by MORF is coupled to H3K14 acylation. Nat. Commun. 10, 4724 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y.-C., Hsu P.-Y., Chin C.-H., Hsiao C.-C., Liou C.-W., Wang T.-Y., Lin Y.-Y., Lee C.-P., Lin H.-C., Lin M.-C., Su M.-C., H3K23/H3K36 hypoacetylation and HDAC1 up-regulation are associated with adverse consequences in obstructive sleep apnea patients. Sci. Rep. 11, 20697 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu C., Xu J., Chen Y., Guo X., Zheng Y., Wang Q., Chen Y., Ni Y., Zhu Y., Joyce B. T., Baccarelli A., Deng F., Zhang W., Hou L., Characterization of genome-wide H3K27ac profiles reveals a distinct PM2.5-associated histone modification signature. Environ. Health 14, 65 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paauw N. D., Lely A. T., Joles J. A., Franx A., Nikkels P. G., Mokry M., van Rijn B. B., H3K27 acetylation and gene expression analysis reveals differences in placental chromatin activity in fetal growth restriction. Clin. Epigenetics 10, 85 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shikata D., Yamamoto T., Honda S., Ikeda S., Minami N., H4K20 monomethylation inhibition causes loss of genomic integrity in mouse preimplantation embryos. J. Reprod. Dev. 66, 411–419 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Estève P.-O., Sen S., Vishnu U. S., Ruse C., Chin H. G., Pradhan S., Poly ADP-ribosylation of SET8 leads to aberrant H4K20 methylation in mammalian nuclear genome. Commun. Biol. 5, 1–18 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aguilera J., Kaushik A., Cauwenberghs N., Heider A., Ogulur I., Yazici D., Smith E., Alkotob S., Prunicki M., Akdis C. A., Nadeau K. C., Granzymes, IL-16, and poly(ADP-ribose) polymerase 1 increase during wildfire smoke exposure. J. Allergy Clin. Immunol. Glob. 2, 100093 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X., Zhang Y., Gao L., Yang X., Zhou G., Sang Y., Xue J., Shi Z., Sun Z., Zhou X., BDE-209 induced spermatogenesis disorder by inhibiting SETD8/H4K20me1 related histone methylation in mice. Sci. Total Environ. 864, 161162 (2023). [DOI] [PubMed] [Google Scholar]
- 53.Zhou X., Xu Y., Ren S., Yang N., Sun Y., Yang Q., Zhang Y., Cai H., Deng W., Chen J., Chen D., Cao B., Qi H., Wang H., Lu J., Trophoblast PR-SET7 dysfunction induces viral mimicry response and necroptosis associated with recurrent miscarriage. Proc. Natl. Acad. Sci. U.S.A. 120, e2216206120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gu Y., Cooper D., Lewis D. F., Zoorob D., Wang Y., Oxidative stress contributes to hypermethylation of Histone H3 lysine 9 in placental trophoblasts from preeclamptic pregnancies. Front. Endocrinol. (Lausanne) 15, 1371220 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Movassagh H., Prunicki M., Kaushik A., Zhou X., Dunham D., Smith E. M., He Z., Aleman Muench G. R., Shi M., Weimer A. K., Cao S., Andorf S., Feizi A., Snyder M. P., Soroosh P., Mellins E. D., Nadeau K. C., Proinflammatory polarization of monocytes by particulate air pollutants is mediated by induction of trained immunity in pediatric asthma. Allergy 78, 1922–1933 (2023). [DOI] [PubMed] [Google Scholar]
- 56.Ochando J., Mulder W. J. M., Madsen J. C., Netea M. G., Duivenvoorden R., Trained immunity — Basic concepts and contributions to immunopathology. Nat. Rev. Nephrol. 19, 23–37 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Di Q., Amini H., Shi L., Kloog I., Silvern R., Kelly J., Sabath M. B., Choirat C., Koutrakis P., Lyapustin A., Wang Y., Mickley L. J., Schwartz J., An ensemble-based model of PM2.5 concentration across the contiguous United States with high spatiotemporal resolution. Environ. Int. 130, 104909 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moss J. L., Johnson N. J., Yu M., Altekruse S. F., Cronin K. A., Comparisons of individual- and area-level socioeconomic status as proxies for individual-level measures: Evidence from the Mortality Disparities in American Communities study. Popul. Health Metr. 19, 1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hajat A., MacLehose R. F., Rosofsky A., Walker K. D., Clougherty J. E., Confounding by socioeconomic status in epidemiological studies of air pollution and health: Challenges and opportunities. Environ. Health Perspect. 129, 065001 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Q. Di, Y. Wei, A. Shtein, X. Xing, E. Castro, H. Amini, C. Hultquist, L. Shi, I. Kloog, R. Silvern, J. Kelly, M. B. Sabath, C. Choirat, P. Koutrakis, A. Lyapustin, Y. Wang, L. J. Mickley, Y. Daouk, J. Schwartz, Daily and Annual PM2.5 Concentrations for the Contiguous United States, 1-km Grids, Version 1.10 (2000–2016), Palisades, NY: NASA Socioeconomic Data and Applications Center (SEDAC) (2024); 10.7927/G2N9-CA10. [DOI]
- 61.Fuss I. J., Kanof M. E., Smith P. D., Zola H., Isolation of whole mononuclear cells from peripheral blood and cord blood. Curr. Protoc. Immunol. 85.1, 7.1.1–7.1.8 (2009). [DOI] [PubMed] [Google Scholar]
- 62.Butler D. C., Petterson S., Phillips R. L., Bazemore A. W., Measures of social deprivation that predict health care access and need within a rational area of primary care service delivery. Health Serv. Res. 48, 539–559 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pettersen J. P., Almaas E., csdR, An R package for differential co-expression analysis. BMC Bioinformatics 23, 79 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ritchie S. C., Watts S., Fearnley L. G., Holt K. E., Abraham G., Inouye M., A scalable permutation approach reveals replication and preservation patterns of network modules in large datasets. Cell Syst. 3, 71–82 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 to S4