Abstract
The ascomycete Saccharomyces cerevisiae exhibits alternative vegetative growth states referred to as the yeast form and the filamentous form, and it switches between the two morphologies depending on specific environmental signals. To identify molecules involved in control of morphologic differentiation, this study characterized mutant S. cerevisiae strains that exhibit filamentous growth in the absence of the normal external signals. A specific amino acid substitution in the cyclin-dependent protein kinase Cdc28 was found to cause constitutive expression of most filamentous growth characteristics. These effects include specifically modified cell polarity characteristics in addition to the defined shape and division cycle alterations typical of the filamentous form. Several other mutations affecting Cdc28 function also had specific effects on filamentous growth. Constitutive filamentous growth resulting from deletion of the protein kinase Elm1 was prevented by modification of Cdc28 such that it could not be phosphorylated on tyrosine residue 19. In addition, various mutations affecting Hsl1 or Swe1, known or presumed components of a protein kinase cascade that mediates Cdc28 phosphorylation on Y19, either prevented or enhanced filamentous growth. The data suggest that a protein kinase cascade involving Elm1, Hsl1, and Swe1 can modulate Cdc28 activity and that Cdc28 in turn exerts global effects that cause filamentous growth.
During vegetative growth, the ascomycete Saccharomyces cerevisiae is able to adopt one of two distinct morphologic forms (for a review, see reference 31), designated the yeast form and the filamentous form. Two distinct signaling regimes induce the filamentous form in wild-type cells. In the phenomenon known as pseudohyphal growth, diploid cells become filamentous when grown on solid agar medium containing a rich carbon source but limited for nitrogen (26). Haploid cells also become filamentous but in response to a different signal. This phenomenon, known as invasive growth, occurs under the surface of mature colonies on rich agar medium (55).
The filamentous form is similar in invasive and pseudohyphal growth (26, 32, 55). Filamentous-form cells are significantly elongated compared to the yeast form, and cell separation after cytokinesis is delayed. Cell polarity is altered in the filamentous form, such that a daughter cell’s initial bud is located nearly exclusively at the pole opposite the previous cytokinesis site. In contrast, in the yeast form, bud growth usually occurs adjacent to the previous site of cell division (22). Filamentous-form cells can grow invasively beneath the surface of agar media, whereas yeast-form cells grow exclusively on the surface of agar plates.
Yeast- and filamentous-form cells also differ in cell cycle control (32). In the yeast form, new daughter cells (i.e., cells that have not budded previously) expand isotropically during G1 before initiating the formation of a new bud; after reaching a critical size, the Start checkpoint is passed and bud emergence occurs (for reviews, see references 36 and 53). After passing Start, cells are able to enter mitosis and complete cell division independent of bud size. Thus, the major control of cell division cycle progression is in G1. The filamentous form differs in that buds emerge on new daughter cells very soon after cytokinesis without an intervening G1 phase (32). Thus, in filamentous-form cells, the G1 size requirement either is no longer operative or is already satisfied in the incipient new cell at the time of cytokinesis. Despite the complexity of this process, there is a simple visual indication of the difference in cell cycle progression, which is that buds emerge simultaneously in mother-daughter pairs in the filamentous form. The yeast form differs in that the mother cell usually forms a bud before the daughter cell, indicative of faster passage of Start. The apparent differences in cell division control in the filamentous form suggest involvement of the cyclin-dependent protein kinase (CDK) Cdc28, which is a central regulatory factor in determining whether S. cerevisiae passes several different cell cycle control points (for reviews, see references 23, 50, and 73).
Many factors are known to be involved in filamentous-growth signaling. These include a potential nutrient-sensing ammonia transporter (41); a trimeric G protein (33, 40); upstream regulators of a mitogen-activated protein (MAP) kinase cascade including Ras2, Ste20, and 14-3-3 proteins (26, 46, 47, 56); the Ste MAP kinase cascade; its transcription factor target, Ste12, and regulators thereof (16, 37, 43, 44, 55, 66); and other putative transcription factors (13, 24, 25, 38, 70). Despite this body of knowledge, little is known about the targets of these pathways that mediate the changes in the filamentous form, although the flocculin protein Flo11 has recently been identified as a possible candidate (39). As a potential means of identifying downstream target molecules, mutant strains were analyzed that constitutively exhibit elongated cell morphology and other specific filamentous-form characteristics independent of the signal that normally is required for morphologic differentiation in wild-type cells (8–10). One of the genes identified in this screen, ELM1, codes for a serine/threonine protein kinase (30). Deletion of ELM1 causes constitutive filamentous growth independent of the growth medium, surface contact, or cell ploidy (8), suggesting that Elm1 functions as a downstream regulator of morphologic differentiation. The targets of Elm1 that may be responsible for changes in cellular growth are not known. Another gene identified in this way, ELM7, was suggested to play a central role because of its extensive genetic interactions with other mutations affecting cell morphology (10).
Through further characterization of ELM7 and ELM1, this study describes two lines of evidence indicating that Cdc28 function is a controlling factor in the decision to grow in the filamentous or yeast form. First, a specific mutation altering Cdc28 function was identified that constitutively causes most of the filamentous-form characteristics, including cell shape and polarity in addition to cell cycle parameters. Second, several elements of the protein kinase cascade that phosphorylates Cdc28 on tyrosine 19 were found to regulate filamentous growth, both as potential downstream regulators of the effects of Elm1 and in otherwise wild-type cells exposed to the invasive growth regime. The data support the hypothesis that modulation of Cdc28 function in response to environmental signals is in large part responsible for the transition between the yeast form and the filamentous form.
MATERIALS AND METHODS
Media and strain construction.
The S. cerevisiae strains used in this study are listed in Table 1. Standard genetic methods were used for mating of S. cerevisiae strains, selection of diploids, induction of meiosis, and tetrad dissection (57). S. cerevisiae transformation was carried out by the lithium acetate procedure (3). One-step gene replacement and targeted integration of prototrophic markers into the S. cerevisiae genome were performed as described previously (58). Gene replacement was confirmed by DNA gel blot analysis and/or by the obvious appearance of the specific phenotype known to result from deletion of the gene undergoing modification (data not shown). Standard rich (YPD) and synthetic (SD) agar (2%) media were used as described previously (57). Unless otherwise noted, media contained 2% glucose as the carbon source.
TABLE 1.
S. cerevisiae strains used in this study
| Straina | Genotype | Reference, source, or derivation |
|---|---|---|
| W303 | MATa/MATα ura3/ura3 leu2/leu2 his3/his3 ade2/ade2 trp1/trp1 | 69 |
| W303-1A | MATa ura3 leu2 his3 ade2 trp1 | 69 |
| ΣΣ | MATa/MATα ura3/ura3 leu2/leu2 | L5487 × L5684b |
| DDUL | MATa/MATα ura3/ura3 leu2/leu2 his3/+ | This study |
| αDUL | MATα ura3 leu2 | This study |
| αDU | MATα ura3 | This study |
| aDUL | MATa ura3 leu2 | This study |
| aDL | MATa leu2 | This study |
| NEY2158 | MATa ura3 leu2 his3 | This study |
| NEY2159 | MATα ura3 leu2 his3 | This study |
| 104D6 | MATα ura3 elm1-1 | 10 |
| 104D576b | MATa leu2 met6 elm1-1 | 10 |
| 104DP10c | MATa ura3 his3 elm1-1 | This study |
| NEDN6d | MATα ura3 leu2 met6 elm1-1 | 10 |
| NEY1489 | MATa ura3 leu2 met6 elm1-1 | 10 |
| 127D6 | MATα ura3 cdc28-127 | 10 |
| 127D5 | MATa leu2 cdc28-127 | 10 |
| AY2287 | MATa ura3 leu2 cdc28-127 | 127D6 × aDL |
| AY2285 | MATa ura3 leu2 | 127D6 × aDL |
| AY100X | MATa ura3 leu2 CDC28-URA3 | Transformation of aDUL with p28WT-URAi |
| AY101X | MATa leu2 cdc28-127 | 127D6 × aDL |
| AY102X | MATα ura3 leu2 bud2Δ::LEU2 | Transformation of αDUL |
| AY2241 | MATα cdc28-127 bud2Δ::LEU2 leu2 | Segregant of AY102X × AY2287 |
| AY2242 | MATα cdc28-127 BUD2 leu2 | Segregant of AY102X × AY2287 |
| AY2249 | MATα ura3 leu2 clb2::LEU2 | Transformation of αDUL |
| AY2252 | MATa ura3 cdc28-127 leu2-GAL::CLB2-LEU2 | Transformation of AY2287 with YIpG2::CLB2c |
| AY2305 | MATa/MATα CDC28/cdc28::LEU2a ura3/ura3 leu2/leu2 his3/+ | Transformation of DDUL |
| AY2153 | MATα cdc28::LEU2a ura3 leu2 (p28MUT-URAc) | Segregant of AY2305 transformant |
| AY2399 | MATα ura3 leu2 his3 cdc28::LEU2a (p28Y19F-URAc) | Segregant of AY2305 transformant |
| WW28Δ/+ | MATa/MATα CDC28/cdc28::LEU2a ura3/ura3 leu2/leu2 his3/his3 trp1/trp1 ade2/ade2 | Transformation of W303 |
| Y2195 | MATα cdc28::LEU2a ura3 leu2 his3 trp1 ade2 (p28MUT-URAc) | Segregant of WW28Δ/+ transformant |
| Y2194 | MATα cdc28::LEU2a ura3 leu2 his3 trp1 ade2 (p28WT-URAc) | Segregant of WW28Δ/+ transformant |
| ΣΣ28Δ/+ | MATa/MATα CDC28/cdc28::LEU2a ura3/ura3 leu2/leu2 | Transformation of ΣΣ |
| AY2204 | MATα cdc28::LEU2a ura3 leu2 his3 trp1 ade2 (p28MUT-URAc) | Segregant of ΣΣ28Δ/+ transformant |
| NEY2170 | MATa leu2 ura3 his3 elm1::HIS3 | Transformation of NEY2158 |
| AY2451 | MATa leu2 ura3 his3 elm1::HIS3 cdc28::LEU2a (p28Y19F-URAc) | Segregant of AY2399 × NEY2170 |
| AY2452 | MATa leu2 ura3 his3 elm1::HIS3 CDC28 (p28Y19F-URAc) | Segregant of AY2399 × NEY2170 |
| NEY1494 | MATa leu2 met6 elm1-1 SEL2-1 | Mutation of 104D576b |
| NEY2018 | MATa ura3 his3 elm1-1 put3::URA3 | Transformation of 104DP10c |
| NEY2019 | MATα ura3 leu2 met6 elm1-1 bud2Δ::LEU2 | Transformation of NEDN6d |
| NEY2020 | MATa ura3 leu2 met6 elm1-1 GFA1:URA3 | Transformation of NEY1489 |
| NEY2065 | MATα ura3 elm1-1 SEL2-1 | Segregant from NEDN6d × NEY1494 |
| NEY2170 | MATa ura3 leu2 his3 elm1::HIS3 | Transformation of NEY2158 |
| NEY100X | MATα ura3 leu2 his3 sel2::LEU2 | Transformation of NEY2159 |
| NEY2299 | MATa ura3 leu2 his3 elm1::HIS3 swe1::URA3 | Transformation of NEY2170 |
| NEY2317 | MATa/MATα ura3/ura3 leu2/leu2 his3/his3 elm1::HIS3/+ sel2::LEU2/+ swe1::URA3/+ | NEY2299 × NEY100X |
| NEY2335 | MATa ura3 leu2 his3 sel2::LEU2 | Progeny of NEY2317 |
| NEY2338 | MATa ura3 leu2 his3 elm1::HIS3 | Progeny of NEY2317 |
| NEY2339 | MATα ura3 leu2 his3 sel2::LEU2 swe1::URA3 | Progeny of NEY2317 |
| NEY2340 | MATa ura3 leu2 his3 | Progeny of NEY2317 |
| NEY2341 | MATα ura3 leu2 his3 elm1::HIS3 swe1::URA3 | Progeny of NEY2317 |
| NEY2344 | MATα ura3 leu2 his3 swe1::URA3 | Progeny of NEY2317 |
| NEY2353 | MATa ura3 leu2-SEL2-1-LEU2 | Integrative transformation of aDUL with pNE33d |
All strains are related to the parental reference strain D273-10B/A1 (68) by at least five backcrosses, unless otherwise noted as being derived from W303 or Σ.
These parental strains in the Σ background were provided by C. Styles, Whitehead Institute.
Provided by D. Lew, Duke University.
Isolation and genetic analysis of suppressor mutations.
Derivatives of elm1-1 strain 104D576b that exhibited yeast-form morphology were identified by visual screening following transformation with a plasmid library. Ten of these remained in the yeast form independent of any transforming plasmid. The elongated-morphology phenotype was readily recovered in progeny of crosses to a wild-type tester strain, indicating in each instance an extragenic suppressor mutation (generically termed sel). Dominance or recessiveness was determined by mating each original isolate to elm1-1 strain 104D6. Tetrads from these diploids invariably produced two spore clones with typical yeast-form cell morphology and two with the filamentous-form characteristics typical of elm1-1 strains, indicating that the suppression phenotype is a single-gene trait. Segregants from these crosses of the genotype MATα ura3 elm1-1 sel were collected for each original isolate. These were used to determine allelism relationships of the 10 sel mutations by genetic linkage. Each segregant was mated to each original isolate (MATa leu2 elm1-1 sel). Allelism was assigned if yeast form colonies were observed exclusively in the progeny of such a cross, whereas nonallelic mutations were indicated by recovery of colonies with filamentous form characteristics typical of elm1-1 strains; at least 30 complete tetrads were examined for each cross. An allelic group of five dominant mutations defined the locus originally designated SEL2 and subsequently identified as HSL1.
Construction of plasmids containing CDC28, cdc28-127, or cdc28-Y19F.
Standard molecular biology methods were used for PCR amplification and plasmid construction (3, 61, 63). CDC28 was obtained from D. Lew (Duke University) as a 2.03-kb fragment extending from the XhoI site 342 bp upstream of the initiation codon to the PvuII site 790 bp downstream of the termination codon. This fragment contains a 2-bp stretch inserted immediately upstream of the CDC28 initiation codon that create an NdeI site not present in the native gene. This CDC28 fragment was cloned in the XhoI and SmaI sites of pRS306, pRS316, or pRS305 to construct p28WT-URAi, p28WT-URAc, and p28WT-LEUi, respectively. Plasmid p28MUT-URAc, containing the allele cdc28-127, was constructed by replacing the region between the unique EcoNI and HindIII sites of p28WT-URAc with the equivalent region from a fragment amplified by PCR from genomic DNA of a cdc28-127 haploid strain.
Plasmid p28Y19F-URAc, carrying cdc28-Y19F, was constructed as follows. Oligonucleotide primer 1022R from the noncoding strand of CDC28 contains a missense mutation that changes tyrosine codon 19 (TAC) to a phenylalanine codon (TTC). This primer was used to amplify by PCR a 126-bp region extending from 60 bp upstream of the CDC28 initiation codon to codon 22 of the open reading frame (ORF). The template for this reaction was p28WT-URAc. This small fragment was then extended by PCR to beyond the unique HindIII site within CDC28. The NdeI-HindIII fragment excised from the amplification product was used to replace the equivalent region of CDC28 in p28WT-LEUi, forming plasmid p28Y19F-LEUi. The nucleotide sequence of the entire CDC28 gene in p28Y19F-LEUi was determined, confirming the presence of the Y19F mutation. The only other changes from the wild-type sequence are a silent mutation changing codon 22 from GTA to GTT and also the mutation forming the NdeI site at the initiation codon. The XhoI-EcoNI fragment of p28Y19F-LEUi containing the Y19F mutation was used to replace the equivalent region of p28WT-URAc, forming plasmid p28Y19F-URAc.
HSL1 plasmids.
HSL1 was excised from phage clone ATCC71138 (American Type Culture Collection, Rockville, Md.) as a 7.26-kb BamHI-SstI fragment and cloned in pRS316 to form plasmid pNE30. The dominant HSL1 allele designated SEL2-1 was obtained by in vivo gap repair as follows. pNE30 was digested at the unique SunI and NruI sites within HSL1, which removes the entire ORF but leaves significant lengths of the 5′- and 3′-flanking sequences. The resulting linear fragment was used to transform the SEL2-1 strain NEY2065 to uracil prototrophy. The autonomously replicating plasmid pNE30GR was recovered from one transformant after amplification in Escherichia coli. This plasmid was shown to have been repaired from the dominant allele SEL2-1 by its ability to suppress the filamentous-growth characteristics of an elm1-1 strain. Plasmid pNE33d is a LEU2-marked integrative plasmid that was constructed by excising the BamHI-SstI fragment bearing SEL2-1 from pNE30GR and cloning it in pRS305.
Gene disruptions and tagging by integrative transformation.
Recombinant DNA fragments used to replace wild-type alleles with deletion mutations were described previously for elm1::HIS3 (8), bud2Δ::LEU2 (52), clb2::LEU2 (54), and put3::URA3 (45). The fragment used to introduce swe1::URA3 was constructed by digesting pSWE1-10g (11) with XbaI to remove the LEU2 insert from swe1::LEU2 and replacing that region with a 1.2-kb NheI fragment containing URA3 (20). The resultant plasmid is pswe1::URA3, from which a 2.5-kb disruption fragment can be excised by digestion with HindIII and BamHI. The GFA1 locus was tagged with URA3 as a selective marker by first cloning a genomic EcoRI fragment containing GFA1 (71) in pRS306, linearizing the resultant plasmid at the unique SphI site within GFA1, and then transforming strain NEY1489 to uracil prototrophy.
The deletion allele hsl1::LEU2 was constructed as follows. A 5.34-kb PvuII-SstI fragment of the HSL1 locus was excised from pNE30 and cloned in pUC118 digested with SmaI and SstI. The entire HSL1 ORF between the unique NruI and SunI sites was removed; after digestion with these two enzymes, the remaining plasmid was made blunt ended by treatment with DNA polymerase I Klenow fragment (polIK) and ligated to an NheI linker to form plasmid pNE37. The 2.2-kb XbaI fragment containing LEU2 was excised from pSWE1-10g (11) and inserted into this unique NheI site to form plasmid pNE38. Digestion of this plasmid with SstI and HindIII releases a 2.59-kb disruption fragment containing hsl1::LEU2.
The deletion allele cdc28::LEU2a was constructed as follows. The 2.03-kb XhoI-PvuII genomic DNA fragment containing CDC28 was cloned in the XhoI and SmaI sites of pBluescript SK(+). The region of CDC28 between the unique NdeI and HindIII sites (codons 1 to 247) was replaced with the LEU2 gene obtained as a 2.8-kb BglII fragment from plasmid YEp13 (12). To facilitate this cloning step, the NdeI-HindIII-cleaved CDC28 plasmid was made blunt ended by treatment with polIK and ligated to a BglII linker. The resultant plasmid, pCdc28::LEU2, provides cdc28::LEU2a as a 4.1-kb fragment excised by digestion with ApaI and BamHI.
Size measurements and estimation of cell volume.
Random fields of 30 to 50 cells were captured for analysis as digital files by using the program NIH Image; all unbudded and singly budded mother cells were measured. Cell volumes were estimated by assuming that the cell shape approximated a prolate spheroid and applying the formula V = (4/3)π(a/2)(b/2)2, where a is the length of the cell and b is the maximum width. At least 200 cells were analyzed for each genotype characterized.
Determination of bud site selection.
A segregating population was generated by crossing strain 127D6 (cdc28-127) to strain aDL (CDC28) and collecting progeny clones from complete tetrads. Wild-type and mutant siblings, along with the parental strains, were analyzed by time-lapse photography to determine directly the location of bud emergence sites and the relative timing of bud emergence on mother-daughter cell pairs. For this analysis, cells were first grown overnight on rich agar medium. Small-budded cells then were dragged to a new location on the plate by using a micromanipulator. The position of mother cells and their buds was noted. Images of the cells were captured immediately after micromanipulation and at approximately 1-h intervals thereafter. For the parental strains, at least 100 cell divisions were examined for bud site selection. For progeny clones, 32 divisions or more were characterized. A minimum of 10 independent progeny clones of each type were analyzed.
The effect of clb2::LEU2 on bud site selection was determined similarly with strain AY2249 (clb2::LEU2) and progeny of this strain crossed to strain 127D5 (cdc28-127); only progeny strains bearing the wild-type CDC28 allele were analyzed. The effect of bud2Δ::LEU2 on bud site selection in cdc28-127 strains was analyzed in progeny of a cross between strain AY102X (bud2Δ::LEU2) and strain 127D5 (cdc28-127). Seven cdc28-127 bud2Δ::LEU2 progeny and five cdc28-127 BUD2 siblings were analyzed for bud position and cell shape; distinctions in these phenotypic parameters corresponded precisely to the genotype.
RESULTS
Characterization of cdc28-127.
The mutation elm7-1, which constitutively displays multiple characteristics of the filamentous form (10) (Fig. 1A and B), was found to be an allele of CDC28. The wild-type allele of elm7-1 was cloned by complementation of the cold-sensitive growth defect also caused by the mutation. One derivative strain able to grow at 15°C after transformation of an elm7-1 mutant with a wild-type genomic DNA library also displayed the ovoid cell shape typical of yeast form cells. This strain contained plasmid p127c, which was shown to be responsible for complementation of elm7-1 by the fact that vegetative segregants lacking the plasmid regained both the abnormal cell shape and cold-sensitive growth phenotypes. Nucleotide sequence analysis revealed that the genomic insert in p127c contained CDC28. Plasmid p28WT-URAc, in which CDC28 is the only intact genetic element in the insert DNA, was introduced into elm7-1 strains and found to restore normal yeast form morphology, again in a plasmid-dependent manner (data not shown). Thus, CDC28 complements elm7-1 when present as part of a centromeric plasmid.
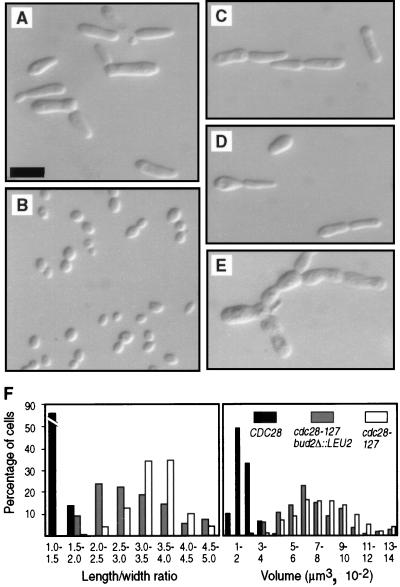
FIG. 1.
Effects of cdc28-127 on cell size and morphology. Haploid strains were grown overnight on YPD plates and photographed suspended in 1.2 M sorbitol. Bar, 10 μm. (A) Strain AY2287 (cdc28-127). (B) Strain AY2285 (CDC28). (C) Strain AY2153 (cdc28-127 reconstructed in the D273-10B/A1 background). (D) Strain AY2204 (cdc28-127 reconstructed in the Σ background). (E) Strain AY2195 (cdc28-127 reconstructed in the W303 background). (F) Distribution of length/width ratios and approximate cell volumes. The cdc28-127 and CDC28 strains analyzed were AY2287 and AY2285 (A and B, respectively); the cdc28-127 bud2Δ::LEU2 strain is AY2241 shown in Fig. 2C.
CDC28 was found to be genetically inseparable from the elm7-1 mutation. The URA3 gene was inserted as a marker into the genome adjacent to the wild-type CDC28 locus in strain AY100X. This strain was crossed to the elm7-1 mutant 127D6, and tetrads were analyzed. CDC28 (indicated by uracil prototrophy) and elm7-1 (indicated by elongated cell morphology) segregated in opposition in 30 complete tetrads, indicating tight linkage of the two genetic elements. The fact that CDC28 both complements and is tightly linked to elm7-1 indicates allelism. Accordingly, elm7-1 here is renamed cdc28-127.
The specific mutation in cdc28-127 that causes constitutive filamentous growth was found to be replacement of cysteine residue 127 by tyrosine (termed C127Y, and the mutant protein is designated Cdc28C127Y). The CDC28 locus was isolated from wild-type and cdc28-127 strains by PCR amplification of genomic DNA. Nucleotide sequence analysis of several independent clones showed that cdc28-127 differs from the wild-type allele CDC28 only by the C127Y mutation. To prove that C127Y is the causative agent of the observed phenotypes, the mutant gene was used to replace CDC28 in three different wild-type haploid genetic backgrounds, namely, D273-10B/A1, W303, and Σ. In all instances, the resultant strains displayed the morphologic and growth defects observed for the original cdc28-127 mutant strain (Fig. 1C to E). The genetic background, however, had modifying effects on the particular cell morphology and also affected the growth rate. In the D273-10B/A1 background, cdc28-127 strains grew on rich agar plates at a slightly reduced rate compared to congenic CDC28 strains; in the W303 background, the mutant strains grew significantly slower than did the wild-type control strains; and in the Σ background, the mutant strains grew extremely slowly compared to the wild type, requiring 4 to 5 days at room temperature to form a colony that could be transferred to a fresh plate (data not shown).
Filamentous growth characteristics caused by cdc28-127.
The mutation cdc28-127 causes most of the known characteristics of filamentous form cells. The following observations were made on haploid strains grown to early exponential phase in liquid cultures of rich medium (YPD), implying that filamentous growth characteristics occur in the absence of the normal requirements for diploidy, nitrogen limitation, and contact with agar medium. Cells bearing cdc28-127 were significantly elongated compared to congenic CDC28 cells (Fig. 1F). Enhanced ability to grow invasively in rich agar medium was observed previously for a cdc28-127 strain (10), and this was observed again in the cdc28-127 mutants constructed by gene replacement (data not shown). Strains containing cdc28-127 did not form large cell clumps during growth in liquid medium. Thus, the delayed cell separation phenotype characteristic of filamentous-form growth is not caused by cdc28-127.
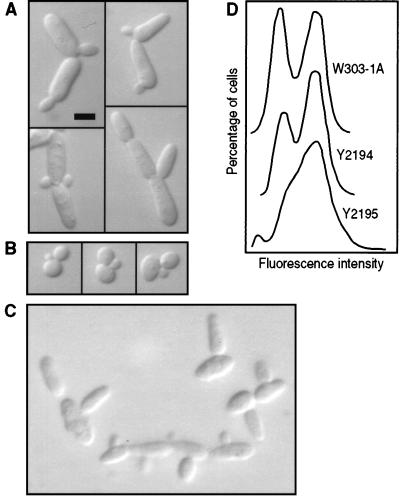
The cell division cycle characteristics of filamentous-form growth also are caused by cdc28-127. In contrast to wild-type cells, mother-daughter pairs of cdc28-127 mutants frequently contain buds very similar in size (Fig. 2A and B). Time-lapse photography commonly showed simultaneous bud emergence on mother-daughter pairs in cdc28-127 strains (10) (Fig. 3A), whereas in congenic CDC28 strains a bud almost always was visible on the mother cell prior to the daughter cell. In accordance with simultaneous bud emergence, measurement of DNA content per cell in asynchronous populations revealed that the frequency of cells in G2 was significantly increased in a cdc28-127 mutant compared to a congenic wild-type strain (Fig. 2D).
FIG. 2.
Cell cycle parameters. Bar, 5 μm. (A) Strain AY2287 (cdc28-127). (B) Strain AY2285 (CDC28). (C) Strain AY2241 (cdc28-127 bud2Δ::LEU2). Cells were grown overnight in liquid YPD medium. (D) DNA content determined by fluorescence-activated cell sorter analysis (34). The indicated strains were converted to ρ0 derivatives lacking mitochondrial DNA prior to analysis. Relevant genotypes are as follows: W303-1A, nonmutant; Y2194, cdc28::LEU2a (p28WT-URAc) (bearing CDC28); Y2195, cdc28::LEU2a (p28MUT-URAc) (bearing cdc28-127).
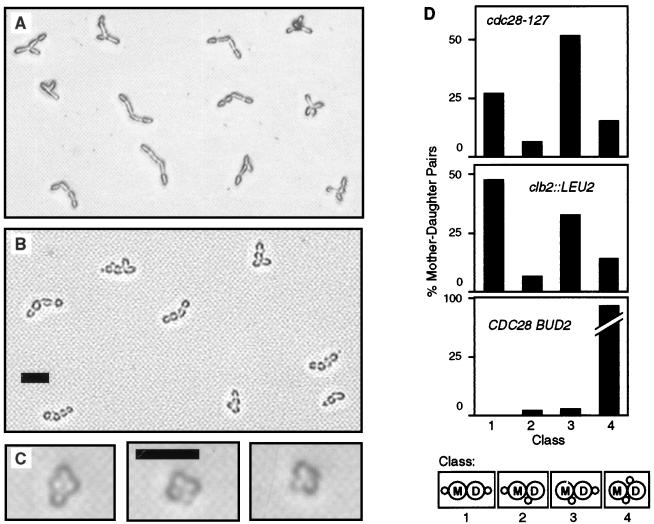
FIG. 3.
Bud site selection. (A) Strain AY2287 (cdc28-127). (B) Strain AY2249 (clb2::LEU2). (C) Strain aDUL (CDC28). Cells are photographed on the surface of YPD agar medium. Bars, 25 μm. (D) Tabulation of data shown in panels A to C. Bud site selection classes are defined at the bottom of the panel.
A possible explanation for this phenotype is that control of G1 phase is modified in cdc28-127 mutants so that cells pass Start and progress into S phase more rapidly than normal. An accelerated G1 phase, however, normally causes reduced cell size. In contrast, cdc28-127 mutant cells are significantly larger than congenic CDC28 cells (Fig. 1F). Thus, some aspects of Cdc28 activity other than activation during G1 are likely to be altered in the mutant.
The mutation cdc28-127 also affects the mechanism of bud site selection, causing haploid cells to adopt the budding pattern typically observed in diploid cells of the pseudohyphal form. Bud site selection was determined by isolating cells with small buds on solid rich medium and observing in time-lapse photography the subsequent sites of bud emergence on mother-daughter cell pairs (Fig. 3). All strains analyzed were haploids; thus, the normal budding pattern is axial (i.e., both mother and daughter cells form buds adjacent to the previous cytokinesis site). As expected, axial budding was typical of wild-type control strains. Mutant cells always formed buds at one of the two poles of the elongated cells. Axial budding was comparatively rare in the mutants, however, which instead budded with high frequency in the bipolar budding pattern (i.e., both mother and daughter cells bud at the pole opposite the cytokinesis site) or in the unipolar budding pattern (i.e., daughter’s first bud at the pole opposite the previous cytokinesis site, mother’s next bud adjacent to that site). The unipolar budding pattern predominated over the bipolar pattern in the mutant cells (Fig. 3D). This distribution of bud site selection patterns cosegregated with the cell elongation phenotype in progeny of cdc28-127/CDC28 diploid strains, indicating the alteration resulted specifically from cdc28-127 and not from an unidentified mutation present in this genetic background.
CDC28 affects bud site selection through known polarity establishment functions.
CDC28 may affect bud site selection as an upstream regulator of the “polarity establishment functions” coded for by a large group of loci including the BUD genes (for reviews, see references 14 and 18). As an initial test of this hypothesis, BUD2 was deleted in a haploid strain containing cdc28-127. Double-mutant progeny as well as cdc28-127 and bud2Δ::LEU2 single-mutant siblings were analyzed for bud site selection. As expected, bud site selection was primarily random in bud2Δ::LEU2 strains (reference 15 and data not shown). In cdc28-127 bud2Δ::LEU2 double-mutant strains, buds usually emerged from the longitudinal center of the elongated cells as opposed to the poles (Fig. 2C), a condition that was never observed in BUD2 cdc28-127 siblings. Thus, the null mutation bud2Δ::LEU2 is epistatic to cdc28-127 regarding the bud site selection phenotype, consistent with the hypothesis that filamentous-form differentiation involves a specific Cdc28-mediated modification of the site selection functions.
The BUD2 deletion mutation also affected the cell shape. The cdc28-127 bud2Δ::LEU2 double mutants were significantly less elongated than the congenic cdc28-127 BUD2 cells (Fig. 1F); this phenotype cosegregated with the bud2 mutation in the progeny of a bud2Δ::LEU2/BUD2 CDC28/cdc28-127 diploid (data not shown). The bud2Δ::LEU2 mutation did not significantly affect the enlarged volume of cdc28-127 cells (Fig. 1F). Thus, the general bud site selection functions may play an additional role in determination of cell shape in the growing bud.
Clb2 affects filamentous growth.
The G2-expressed cyclin Clb2 regulates cell shape (35), which suggests the hypothesis that Cdc28C127Y causes filamentous growth characteristics because of abnormal interaction with Clb proteins. To test this hypothesis CLB2 first was deleted in a D273-10B/A1 haploid strain. A slight degree of cell elongation resulted, although the phenotype is much less severe than that caused by cdc28-127 (Fig. 4A and B). A similar effect was observed in the haploid BF264-15DUa genetic background (54); however, a clb2/clb2 diploid in this background displayed uniformly elongated cells similar to cdc28-127 strains (35). Deletion of CLB2 in haploid cells caused another phenotype that also results from cdc28-127, specifically, alteration of the budding pattern (Fig. 3B).
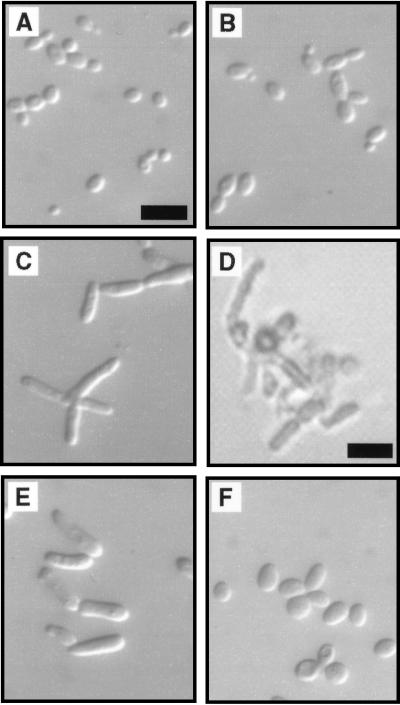
FIG. 4.
Effects of altering CLB2 expression in cdc28-127 mutant strains. (A to D) Sibling progeny from the cross of strain AY2249 (clb2::LEU2) to strain AY101X (cdc28-127). Bars, 10 μm (A) and panel (D) 25 μm. Photomicrographs are of cells grown overnight on YPD plates and suspended on a slide in 1.2 M sorbitol, except for panel D, which shows a germinating spore clone after 2 days on YPD medium, observed under an inverted microscope. (A) CDC28 CLB2. (B) CDC28 clb2::LEU2. (C) cdc28-127 CLB2. (D) cdc28-127 clb2::LEU2. The clone shown in panel D did not develop further and is representative of 15 deduced cdc28-127 clb2::LEU2 spores identified in tetratype and nonparental ditype tetrads from this cross. (E) Strain AY2252 (cdc28-127 GAL:CLB2) grown in YPD liquid medium to early log phase. (F) Strain AY2252 grown in YPGal liquid medium to early log phase.
Cells bearing cdc28-127 were shown to require Clb2 for viability, consistent with the hypothesis that Cdc28C127Y interacts abnormally with the G2-expressed cyclins. CLB2 requirement was indicated by analysis of tetrads from a cdc28-127/CDC28 CLB2/clb2::LEU2 diploid (Fig. 4A to D). From this diploid, 11 of 14 tetrads were identified as tetratypes by the presence of one cdc28-127 CLB2 progeny clone (elongated morphology, leucine auxotroph), one CDC28 CLB2 progeny clone (wild-type morphology, leucine auxotroph), and one CDC28 clb2::LEU2 progeny clone (slight cell elongation phenotype, leucine prototroph). In each instance, the fourth spore of the tetrad, predicted to carry both cdc28-127 and clb2::LEU2, germinated but failed to develop a viable clone. Two of the tetrads contained two CDC28 CLB2 progeny clones and two spores that did not form colonies; again the two inviable spores are predicted to be cdc28-127 clb2::LEU2 double mutants. The terminal phenotype of germinating clb2::LEU2 cdc28-127 spores suggests a lethal effect specifically in G2, because large buds invariably were present (Fig. 4D).
Additional evidence suggesting that Cdc28C127Y is defective in its interaction with G2 cyclins is that overexpression of CLB2 suppressed the cell elongation phenotype caused by cdc28-127. The gene fusion GAL::CLB2, in which the CLB2 coding sequence can be overexpressed from the GAL promoter, was integrated into the genome of a cdc28-127 strain. Growth of this strain on galactose caused cells to revert to the normal ovoid shape typical of the yeast form (Fig. 4E and F). The carbon source had no effect on other cdc28-127 strains in the absence of GAL::CLB2 (data not shown).
Hsl1 affects filamentous-growth characteristics.
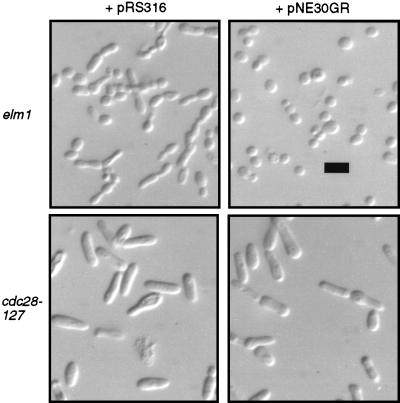
An independent line of evidence implicating CDC28 as a regulator of filamentous growth was provided by analysis of mutations that suppress the constitutive filamentous-growth phenotype induced by elm1 mutations. Five allelic, dominant mutations were obtained in the locus designated SEL2 (suppressor of elm1), each of which restores yeast-form morphology to elm1-1 strains or those bearing elm1 deletion mutations (data not shown; see Materials and Methods). SEL2 was found to be linked to ELM1 on chromosome XI, which enabled its cloning based on genetic map position. Genetic map distances from several cloned genes in the area were determined (Table 2), revealing that SEL2 is located near ORF YKL101w. This ORF was subcloned in plasmid pNE30. The dominant mutant allele SEL2-1 was rescued from the original suppressed strain for the purpose of testing whether YKL101w was in fact the locus responsible for suppression of elm1-1. The resultant gap-repaired plasmid, pNE30GR, was introduced into the parental elm1-1 strain 104D576b. All transformants were completely restored to the yeast-form morphology (Fig. 5), indicating that YKL101w and SEL2 are the same genetic element. YKL101w was previously designated HSL1 or NIK1 (42, 65), and thus SEL2 will hereafter be referred to as HSL1.
TABLE 2.
Intergenic linkage data
| Loci | Strains crossed | TT:NPD:PDa | Map distance (cM)b |
|---|---|---|---|
| HSL1-ELM1 | NEY1494 × αDU | 30:0:25 | 27.3 |
| HSL1-PUT3 | NEY2018 × NEY2065 | 11:1:3 | 56.7 |
| HSL1-BUD2 | NEY1494 × NEY2019 | 4:0:31 | 5.71 |
| HSL1-GFA1 | NEY2065 × NEY2020 | 2:0:50 | 1.92 |
Tetratype (TT), nonparental ditype (NPD), parental ditype (PD).
The map distance was calculated from the formula [50(TT + 6NPD)]/(TT + NPD + PD). cM, centimorgans.
FIG. 5.
Suppression of elongated cell morphology by SEL2-1. The elm1-1 strain NEDN6d or the cdc28-127 strain AY2287 was transformed to uracil prototrophy with pRS316 (empty vector) or pNE30GR (SEL2-1 cloned in the same vector). Transformant colonies were grown overnight on selective agar medium, suspended on a slide in 1.2 M sorbitol, and visualized with differential interference contrast optics. Bar, 10 μm.
Hsl1 is most similar to the protein kinase Nim1 from Schizosaccharomyces pombe; over a span of 304 aligned residues, 48% are identical in both proteins. The regions of high similarity are located within the protein kinase domains. HSL1 comprises a reading frame of 1,519 codons, however; therefore, there are extensive regions outside of the protein kinase domain available to provide additional functions besides this catalytic activity. Nim1 is known to promote mitosis in S. pombe by inhibiting the activity of the tyrosine kinase Wee1, which in turn phosphorylates and inhibits the CDK Cdc2 (59, 60). In S. cerevisiae, the protein kinase Swe1 functions similarly to Wee1 downstream of Hsl1 (11, 42).
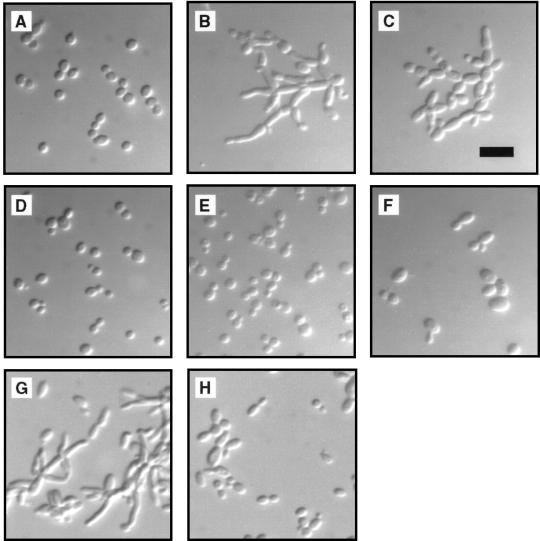
The fact that mutations of HSL1 capable of suppressing elm1-1 are dominant suggests that activation of Hsl1, predicted to result in inhibition of Swe1, is responsible for suppression of the constitutive filamentous-growth phenotype. From this hypothesis, it follows that inactivation of Hsl1, expected to result in hyperactivation of Swe1, would induce the filamentous form. Mutant strains bearing the deletion allele hsl1::LEU2 were viable and did not show any obvious growth defects. The mutant cells were significantly elongated, however, and the budding pattern was switched in haploid cells from the normal axial pattern to the bipolar pattern (Fig. 6C). Both of these phenotypes were observed during growth in rich medium. Elongated cell morphology was found previously in hsl1 mutants (42, 65), but in these strain backgrounds specific filamentous-growth characteristics were not evident.
FIG. 6.
Phosphorylation of CDC28 by Swe1 is required for mutational induction of filamentous-growth characteristics. Strains were grown overnight on YPD plates, suspended in 1.2 M sorbitol, and visualized with differential interference contrast optics. (A) Strain NEY2340 (wild type). (B) Strain NEY2338 (elm1::HIS3). (C) Strain NEY2335 (hsl1::LEU2). (D) Strain NEY2344 (swe1::URA3). (E) Strain NEY2341 (elm1::HIS3 swe1::URA3). (F) Strain NEY2339 (hsl1::LEU2 swe1::URA3). (G) Strain AY2452 (CDC28 elm1::HIS3 [p28Y19F-URAc]). (H) Strain AY2451 (cdc28::LEU2a elm1::HIS3 [p28Y19F-URAc]). Bar, 10 μm.
The Swe1-Cdc28 pathway is required for induction of filamentous-growth characteristics by elm1 or hsl1 mutations.
An hsl1::LEU2 swe1::URA3 double mutant was constructed to test the hypothesis that the morphological effects of HSL1 deletion are mediated by Swe1. The cell shape and budding-pattern phenotypes of this strain were typical of the yeast form (Fig. 6F), indicating that Swe1 is required for induction of filamentous-growth characteristics by the hsl1 mutation. An elm1::HIS3 swe1::URA3 double-mutant strain also was constructed, and again inactivation of SWE1 suppressed the constitutive filamentous-growth phenotype (Fig. 6E).
The fact that elimination of Swe1 prevents filamentous-growth characteristics induced by elm1 or hsl1 mutations suggests that Y19 phosphorylation is necessary for the morphologic phenotype. This hypothesis was tested by replacing wild-type Cdc28 in an elm1::HIS3 strain with a mutant form, Cdc28Y19F, in which the target tyrosine is replaced by a phenylalanine. Although Cdc28Y19F by itself resulted in slightly elongated cells, this phenotype was clearly distinguishable from that of elm1 CDC28 cells. The presence of Cdc28Y19F suppressed the cell elongation phenotype induced by elm1 (Fig. 6H), although apparently not as completely as did deletion of SWE1 (Fig. 6E). This result further implicates the Hsl1-Swe1-Cdc28 pathway as a controlling factor for filamentous growth, at least when it is induced by elm1 mutations. Analogous experiments showed Cdc28Y19F suppressed the cell elongation effects of hsl1 mutations (42).
The Hsl1-Swe1 pathway affects filamentous growth in otherwise wild-type cells.
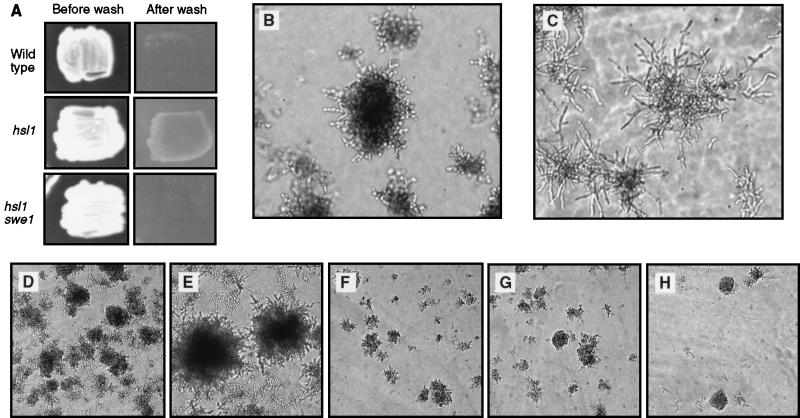
The invasive-growth response was examined in strains containing various mutations in the Hsl1-Swe1 pathway, to test whether this kinase cascade functions as a regulator of morphogenesis in response to specific external signals. Congenic strains were grown on rich medium for 2 days, at which time cells were washed off the surface of the plates to reveal the extent of invasive growth. The wild-type control strain underwent invasive growth as expected (Fig. 7A), although the response was much more extensive after 4 days of growth (data not shown). The hsl1::LEU2 mutant exhibited a much greater extent of invasive growth than did wild type cells (Fig. 7A), with a response at 2 days similar to that of the maximal wild-type response at 4 days. Examination of colonies growing inside the agar medium showed the mutants to be much more elongated than wild-type cells undergoing invasive growth (Fig. 7B and C). Enhancement of invasive growth by the hsl1 mutation was dependent on Swe1, because swe1::URA3 hsl1::LEU2 double mutants were markedly reduced in their degree of invasive growth compared to the hsl1::LEU2 single mutant (Fig. 7A, E, and H). The extent of invasive growth in the double mutant was the same as that observed in swe1 single mutants and was noticeably less than wild-type cells (Fig. 7D and G). These data are again consistent with the hypothesis that inhibition of Cdc28 by Swe1 action is required for filamentous growth and that Hsl1 activity negatively regulates Swe1. The hypothesis suggests that dominant mutations of Hsl1, originally identified as suppressors of filamentous-growth characteristics induced by elm1 mutations, also would inhibit invasive growth of otherwise wild-type cells. As predicted, the extent of invasive growth in strains bearing the dominant allele, SEL2-1, was significantly reduced compared to that in the wild-type strain (Fig. 7D and F).
FIG. 7.
Invasive growth is affected by mutations of the Hsl1-Swe1 pathway. (A) Invasive-growth assay. The indicated strains were grown for 2 days on YPD plates, and the cells were washed off of the surface of the plates. The strains are aDUL (wild type), NEY2335 (hsl1::LEU2), and NEY2339 (hsl1::LEU2 swe1::URA3). (B and C) Colonies growing underneath the agar, from the plates shown in panel A, photographed under an inverted microscope. (B) Strain aDUL (wild type). (C) Strain NEY2335 (hsl1::LEU2). (D to H) Colonies growing underneath the agar, all taken from a single YPD plate washed after 4 days of growth. In each of these panels, the photograph was taken from the densest area of invasive growth. (D) Strain NEY2340 (wild type). (E) Strain NEY2335 (hsl1::LEU2). (F) Strain NEY2353 (SEL2-1). (G) Strain NEY2344 (swe1::URA3). (H) Strain NEY2339 (hsl1::LEU2 swe1::URA3).
cdc28-127 is epistatic to mutations in the Hsl1-Swe1 pathway.
The effects of mutations in HSL1 and SWE1 regarding filamentous-growth characteristics and the invasive-growth response presumably are mediated by CDC28. This hypothesis predicts that the filamentous-growth characteristics resulting from cdc28-127 would occur independent of whether the Hsl1-Swe1 pathway was functional. To test this hypothesis, the dominant allele SEL2-1 was introduced into a cdc28-127 strain. Although this mutation clearly suppressed the filamentous-growth characteristics in an elm1-1 strain, there was no effect on the phenotype of the cdc28-127 strain (Fig. 5). In addition, SWE1 was deleted in a cdc28-127 mutant strain. The double mutant displayed constitutive filamentous-growth characteristics identical to cdc28-127 single mutants (data not shown), again indicating that the mutant Cdc28 protein is a downstream effector of filamentous-growth characteristics.
DISCUSSION
Filamentous-growth characteristics can be affected by alteration of Elm1, Hsl1, Swe1, or Cdc28.
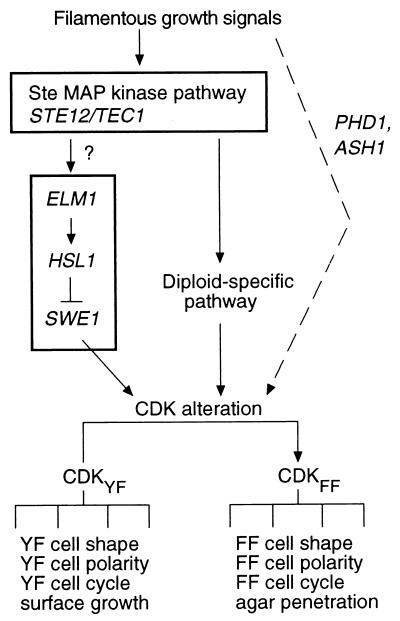
This study revealed that Cdc28C127Y simultaneously causes at least four cellular characteristics that are specific to the filamentous-growth form: simultaneous bud emergence, elongated cell shape, specifically altered bud site selection, and the ability to grow invasively in agar medium. The fact that so many specific aspects of filamentous-form cells result from this single amino acid substitution is unlikely to be coincidental. We suggest instead that wild-type cells undergoing filamentous growth in response to the normal environmental signals alter Cdc28 function in a way that is duplicated or mimicked by cdc28-127.
Identification of Cdc28 as a controlling factor in morphologic differentiation is expected from the known variations in cell cycle progression between filamentous-form and yeast-form cells (32). Kron et al. (32) proposed that a signalling pathway involving components of the Ste MAP kinase cascade is activated by environmental signals and results in inhibition of the G2 activity of the Cdc28 complex. Cdc28 activity also was known previously to control cell morphology (35). The effects of Cdc28C127Y are consistent with these studies and indicate further that a modified Cdc28 complex can also mediate the bud site selection and invasive-growth properties of the filamentous form. Thus, the Cdc28 complex is proposed to be a central downstream regulator of filamentous-form differentiation (Fig. 8). The fact that Cdc28C127Y does not cause delayed cell separation might be explained by a separate signaling pathway being responsible for this filamentous growth characteristic. Alternatively, Cdc28C127Y may not fully mimic the CDK changes that occur normally during filamentous-form differentiation.
FIG. 8.
Model for the role of the Cdc28 complex in control of filamentous growth. CDKYF and CDKFF suggest two slightly different activity states of the Cdc28 complex, which mediate different downstream effects. The Hsl1-Swe1 pathway, with Elm1 as a direct or indirect positive regulator of Hsl1, is proposed as a signal transduction module that can affect the change between CDKYF and CDKFF. At least one other means of affecting the change between CDKYF and CDKFF is proposed, because diploid cells undergo filamentous growth independent of Swe1 function. The Ste MAP kinase pathway is proposed as a possible upstream signaling module that affects the Hsl1-Swe1 pathway, although evidence of a direct connection between these modules is lacking, as noted by the question mark. The dotted line indicates the existence of possible alternative means of affecting the change between the CDKYF and CDKFF activity states, which may be mediated by other transcription factors providing morphologic differentiation functions that are partially redundant with Ste12p.
The Hsl1-Swe1 pathway also was implicated in control of filamentous growth, providing independent evidence that Cdc28 is a central regulator of this process. Although direct evidence that Hsl1 represses the activity of Swe1 by phosphorylation is not available, there is ample genetic data suggesting that these two proteins function in a hierarchical cascade (42, 65). The effects of Swe1 are dependent on phosphorylation of Y19 of Cdc28, and in this instance the enzyme-substrate relationship has been defined (11). Thus, modification of Cdc28 is most likely to be the reason that mutations of HSL1 and SWE1 can impart filamentous-growth characteristics or repress those phenotypes. In this scenario, Swe1 is needed for filamentous-form growth by inhibiting Cdc28 whereas Hsl1 is required for yeast-form growth owing to its ability to inhibit Swe1.
Figure 8 presents a model suggesting that Elm1 functions upstream in a hierarchical cascade involving Hsl1, Swe1, and Cdc28. The following facts are consistent with this model. First, phosphorylation of Cdc28 residue 19 by the Hsl1-Swe1 pathway is necessary for elm1 mutations to cause filamentous-growth characteristics. Second, dominant mutations in HSL1, expected to activate the protein, bypass the lack of Elm1. Third, deletion of HSL1 causes a morphologic phenotype similar to that of elm1 strains. These facts might also be explained by a variation of the depicted model, in which Elm1 and Hsl1 independently regulate the activity of Swe1. Consistent with either variation of the hypothesis, Hsl1 (6, 20) and Elm1 (67, 72) colocalize at the mother-bud neck.
The fact that elm1, hsl1, and cdc28-127 mutations obviate the requirement for any of the multiple aspects of the normal filamentous-growth signals implicates these proteins as downstream regulators of the response. This conclusion is consistent with the finding that ste7 and ste20 mutations do not prevent the morphological effects of ELM1 or HSL1 deletion (48). The Hsl1-Swe1 pathway could be regulated as a downstream target of the Ste MAP kinase cascade including Ste12p, although currently there is no known evidence supporting a direct connection between these two signal transduction modules.
Some insight into Hsl1 function in filamentous-growth regulation might be gained from consideration of Nim1. In S. pombe, nitrogen starvation stimulates the mating pathway and results in rapid degradation of Nim1 (7, 21, 27, 74). Loss of Nim1 results in activation of Wee1, resultant phosphorylation of the Cdc28 homolog Cdc2 on tyrosine 15, and cell cycle arrest in G2. In S. cerevisiae, hsl1 mutants are sensitive to nitrogen concentration, so that increased nitrogen source suppresses the extreme elongated morphology observed in these mutants in stationary phase (65). Collectively, these data suggest a possible scenario in which Hsl1 degradation may occur in response to nutrient deprivation, thereby eliciting a Swe1-mediated delay in the G2 phase of the cell cycle and stimulation of filamentous growth.
Two aspects of the suppression of elm1 mutations by the presence of Cdc28Y19F remain to be explained. First is the fact that the suppressive effect of the nonphosphorylatable form of Cdc28 is not as strong as that resulting from deletion of SWE1 (Fig. 6E and H). Thus, additional factors in this signaling pathway are likely to exist. Second is the fact that Cdc28Y19F was able to suppress elm1 only when it was the sole form of Cdc28 in the cell (Fig. 6G and H). The presence of normal Cdc28 apparently interferes with the ability of Cdc28Y19F to perform whatever functions are necessary to restore yeast-form growth to the elm1 mutant, although the reason for this effect currently is not known.
Possible filamentous-growth signals in nonmutant cells.
Whether the filamentous-growth signalling pathway actually uses Elm1, Hsl1, and Swe1 is an open question. Several potential mechanisms are known that might alter cell cycle control to result in a spectrum of phenotypic changes similar to that induced by the mutations investigated in this study (for reviews, see references 23, 28, 49, and 73). Any one of these, or a combination, might be used in normal filamentous-growth signaling. The fact that invasive growth is compromised by deletion of SWE1 (Fig. 7) suggests that in this instance the Hsl1-Swe1 pathway is in fact directly involved in filamentous-growth signaling.
Activity of the Hsl1-Swe1 pathway certainly cannot entirely explain filamentous-growth signaling, because swe1/swe1 diploids and Cdc28Y19F strains both respond normally to the pseudohyphal growth signals (32). To explain the fact that filamentous growth in haploids is affected by Y19 phosphorylation whereas the response in diploids does not require this cascade, we suggest that redundant mechanisms exist for filamentous-growth signaling to Cdc28 (Fig. 8). Kron et al. (32) suggested that activation of the Ste MAP kinase pathway may produce an inhibitor of the Clb-Cdc28 complex that functions independently of Y19 phosphorylation. If such an inhibitor was not present in haploids, the Swe1 pathway might be required for invasive growth but not for pseudohyphal growth in diploids. Even if such redundant pathways do exist, however, both mechanisms could be dependent on the Ste MAP kinase pathway (Fig. 8), because disruption of this signal abrogates the filamentous-growth response in both haploids and diploids (37, 55). This issue is complicated significantly by the fact that partially redundant signaling pathways seem to regulate filamentous growth, for example those involving the transcriptional regulatory proteins Ash1, Ste12, and Phd1 (13). Additional genetic and biochemical network analysis is needed to identify connections between downstream signaling modules such as the Hsl1-Swe1 pathway and upstream modules such as that comprising the Ste MAP kinase cascade.
Mutations of CLB2 constitutively cause filamentous-growth characteristics in some genetic backgrounds (1, 35), and Clb2 overexpression suppresses the morphologic effects of cdc28-127. These observations raise the possibility that decreased Cdc28-Clb activity is a general aspect of filamentous-growth signaling. Clb expression is regulated transcriptionally and by cell cycle-specific proteolytic degradation (29, 49). In addition, Cdc28-Clb function is inhibited by Sic1p (17, 62). Thus, down-regulation of Cdc28-Clb as part of filamentous-growth signalling could occur by a variety of mechanisms. Another possibility is that expression of the G1 cyclins could be altered so that the Cdc28-Cln complexes are hyperactive in filamentous-form cells compared to the yeast form. This condition is known to cause hyperpolarization of cell growth (35) and also to shorten the G1 phase (49), so that the morphologic effects of Cln hyperactivation may be similar to those of reduced Clb function. Consistent with this hypothesis, mutations of the gene GRR1, which codes for a component of a complex that promotes Cln1 and Cln2 degradation (4, 5, 64), were isolated based on their ability to cause filamentous-growth characteristics (9, 10). Furthermore, cln1 cln2 mutants are defective in filamentous growth (51). The Cln-Cdc28 and Clb-Cdc28 complexes are known to be functionally interrelated, for example by the fact that Clb2-Cdc28 inhibits transcription of CLN1 and CLN2 (2). Thus, changes in one phase of the cell cycle that result from a specific filamentous-growth signal are likely to have secondary implications throughout the cycle that contribute to morphologic differentiation.
As a last consideration regarding the mechanisms that might cause alteration of Cdc28 function so that filamentous-growth characteristics result, we expect that the changes will be subtle. Filamentous-form cells grow at nearly the same rate as do yeast-form cells (8, 32) and are tightly regulated in terms of cell size and shape. Drastic alterations in Cdc28 function would cause significant growth rate defects and frequent aberrant divisions. The uniform nature of filamentous-form cells argues that although Cdc28 activity may be altered, it still performs all essential functions, albeit with slightly different kinetics. Higher eukaryotes can regulate morphogenesis and cellular differentiation by modifying the cell cycle program (19). Further analysis of the S. cerevisiae cell division cycle and its control in filamentous growth is likely to provide significant insights into how cell division in more complex organisms can be modulated to accomplish specific functions during cellular differentiation.
ACKNOWLEDGMENTS
We thank D. Lew, M. Brandriss, H.-O. Park, C. Styles, and M. Gentzsch for supplying strains and/or plasmids.
This work was supported by National Science Foundation grants MCB-9319028 and MCB-9604247.
Footnotes
Journal paper J-18199 of the Iowa Agriculture and Home Economics Experiment Station, Ames, from project 3197.
REFERENCES
- 1.Acurio, A., S.-H. Ahn, and S. J. Kron. 1998. Personal communication.
- 2.Amon A, Tyers M, Futcher B, Nasmyth K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. doi: 10.1016/0092-8674(93)90722-3. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 4.Barral Y, Jentsch S, Mann C. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 1995;9:399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- 5.Barral Y, Mann C. Degredation des cyclines G1 et differenciation cellulaire chez Saccharomyces cerevisiae. C R Acad Sci Paris. 1995;318:43–50. [PubMed] [Google Scholar]
- 6.Barral Y, Parra M, Bidingmaier S, Snyder M. Abstracts of the 1998 Yeast Genetics and Molecular Biology Meeting. College Park, Md: Genetics Society of America; 1998. Nim1 homologs of S. cerevisiae link cell-cycle progression to proper organization of the peripheral cytoskeleton, abstr. 79; p. 84. [Google Scholar]
- 7.Belenguer P, Pelloquin L, Oustrin M L, Ducommun B. Role of the fission yeast nim1 protein kinase in the cell cycle response to nutrient signals. Biochem Biophys Res Commun. 1997;232:204–208. doi: 10.1006/bbrc.1997.6253. [DOI] [PubMed] [Google Scholar]
- 8.Blacketer M J, Koehler C M, Coats S G, Myers A M, Madaule P. Genetic control of dimorphism in Saccharomyces cerevisiae: involvement of the novel protein kinase homologue Elm1p and protein phosphatase 2A. Mol Cell Biol. 1993;13:5567–5581. doi: 10.1128/mcb.13.9.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blacketer M J, Madaule P, Myers A M. The Saccharomyces cerevisiae mutation elm4-1 facilitates pseudohyphal differentiation and interacts with a deficiency in phosphoribosylpyrophosphate synthase activity to cause constitutive pseudohyphal growth. Mol Cell Biol. 1994;14:4671–4681. doi: 10.1128/mcb.14.7.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blacketer M J, Madaule P, Myers A M. Genetic analysis of morphologic differentiation in S. cerevisiae. Genetics. 1995;140:1259–1275. doi: 10.1093/genetics/140.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Booher R N, Deshaies R J, Kirschner M W. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 1993;12:3417–3426. doi: 10.1002/j.1460-2075.1993.tb06016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broach J R, Strathern J N, Hicks J B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979;8:121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- 13.Chandarlapaty S, Errede B. Ash1, a daughter cell-specific protein, is required for pseudohyphal growth of Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:2884–2891. doi: 10.1128/mcb.18.5.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chant J. Cell polarity in yeast. Trends Genet. 1994;10:328–333. doi: 10.1016/0168-9525(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 15.Chant J, Herskowitz I. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell. 1991;65:1203–1212. doi: 10.1016/0092-8674(91)90015-q. [DOI] [PubMed] [Google Scholar]
- 16.Cook J G, Bardwell L, Kron S J, Thorner J. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 1996;10:2831–2848. doi: 10.1101/gad.10.22.2831. [DOI] [PubMed] [Google Scholar]
- 17.Donovan J D, Toyn J H, Johnson A L, Johnston L H. P40SDB25, a putative CDK inhibitor, has a role in the M/G1 transition in Saccharomyces cerevisiae. Genes Dev. 1994;8:1640–1653. doi: 10.1101/gad.8.14.1640. [DOI] [PubMed] [Google Scholar]
- 18.Drubin D G. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- 19.Edgar B A, Lehner C F. Developmental control of cell cycle regulators: a fly’s perspective. Science. 1996;274:1646–1652. doi: 10.1126/science.274.5293.1646. [DOI] [PubMed] [Google Scholar]
- 20.Edgington, N. P., and A. M. Myers. Unpublished data.
- 21.Feilotter H, Nurse P, Young P G. Genetic and molecular analysis of cdr1/nim1 in Schizosaccharomyces pombe. Genetics. 1991;127:309–318. doi: 10.1093/genetics/127.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friefelder D. Bud formation in Saccharomyces cerevisiae. J Bacteriol. 1960;80:567–568. doi: 10.1128/jb.80.4.567-568.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Futcher B. Cyclins and the wiring of the yeast cell cycle. Yeast. 1996;12:1635–1646. doi: 10.1002/(SICI)1097-0061(199612)12:16%3C1635::AID-YEA83%3E3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 24.Gavirias V, Andrianapolous A, Gimeno C J, Timberlake W E. Saccharomyces cerevisiae TEC1 is required for pseudohyphal growth. Mol Microbiol. 1996;19:1255–1263. doi: 10.1111/j.1365-2958.1996.tb02470.x. [DOI] [PubMed] [Google Scholar]
- 25.Gimeno C J, Fink G R. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol Cell Biol. 1994;14:2100–2112. doi: 10.1128/mcb.14.3.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 27.Hayles J, Nurse P. Genetics of the fission yeast Schizosaccharomyces pombe. Annu Rev Genet. 1992;26:373–402. doi: 10.1146/annurev.ge.26.120192.002105. [DOI] [PubMed] [Google Scholar]
- 28.Hoyt M A. Eliminating all obstacles: regulated proteolysis in the eukaryotic cell cycle. Cell. 1997;91:149–151. doi: 10.1016/s0092-8674(00)80396-7. [DOI] [PubMed] [Google Scholar]
- 29.King R W, Deshaies R J, Peters J-M, Kirschner M W. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 30.Koehler C M, Myers A M. Serine-threonine protein kinase activity of Elm1p, a regulator of morphologic differentiation in Saccharomyces cerevisiae. FEBS Lett. 1997;408:109–114. doi: 10.1016/s0014-5793(97)00401-8. [DOI] [PubMed] [Google Scholar]
- 31.Kron S J, Gow N A R. Budding yeast morphogenesis: signaling, cytoskeleton and cell cycle. Curr Opin Cell Biol. 1995;7:845–855. doi: 10.1016/0955-0674(95)80069-7. [DOI] [PubMed] [Google Scholar]
- 32.Kron S J, Styles C A, Fink G R. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:1003–1022. doi: 10.1091/mbc.5.9.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kübler E, Mösch H-U, Rupp S, Lisanti M P. Gpa2p, a G-protein α-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J Biol Chem. 1997;272:20321–20323. doi: 10.1074/jbc.272.33.20321. [DOI] [PubMed] [Google Scholar]
- 34.Lew D J, Marini N J, Reed S I. Different G1 cyclins control the timing of cell cycle commitment in mother and daughter cells of the budding yeast S. cerevisiae. Cell. 1992;69:317–327. doi: 10.1016/0092-8674(92)90412-6. [DOI] [PubMed] [Google Scholar]
- 35.Lew D J, Reed S I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lew D J, Weinert T, Pringle J R. Cell cycle control in Saccharomyces cerevisiae. In: Pringle J R, Broach J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. Cell cycle and cell biology. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 607–695. [Google Scholar]
- 37.Liu H, Styles C A, Fink G R. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 38.Liu H, Styles C A, Fink G R. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lo W S, Dranginis A M. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:161–171. doi: 10.1091/mbc.9.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lorenz M C, Heitman J. Yeast pseudohyphal growth is regulated by GPA2, a G protein α homolog. EMBO J. 1997;16:7008–7018. doi: 10.1093/emboj/16.23.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lorenz M C, Heitman J. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 1998;17:1236–1247. doi: 10.1093/emboj/17.5.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma X-J, Lu Q, Grunstein M. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 1996;10:1327–1340. doi: 10.1101/gad.10.11.1327. [DOI] [PubMed] [Google Scholar]
- 43.Madhani H D, Fink G R. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1316. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- 44.Madhani H D, Styles C A, Fink G R. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 45.Marczak J E, Brandriss M C. Isolation of constitutive mutants affecting the proline utilization pathway in Saccharomyces cerevisiae and molecular analysis of the PUT3 transcriptional activator. Mol Cell Biol. 1989;9:4696–4705. doi: 10.1128/mcb.9.11.4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mösch H-U, Fink G R. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics. 1997;145:671–684. doi: 10.1093/genetics/145.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mösch H-U, Roberts R L, Fink G R. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Myers, A. M. Unpublished data.
- 49.Nasmyth K. Control of the yeast cell cycle by the Cdc28 protein kinase. Curr Opin Cell Biol. 1993;5:166–179. doi: 10.1016/0955-0674(93)90099-c. [DOI] [PubMed] [Google Scholar]
- 50.Nasmyth K. At the heart of the budding yeast cell cycle. Trends Genet. 1996;12:405–412. doi: 10.1016/0168-9525(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 51.Oehlen L, Cross F R. Potential regulation of ste20 function by the Cln1-Cdc28 and Cln2-Cdc28 cyclin-dependent protein kinases. J Biol Chem. 1998;273:25089–25097. doi: 10.1074/jbc.273.39.25089. [DOI] [PubMed] [Google Scholar]
- 52.Park H-O, Chant J, Herskowitz I. BUD2 encodes a GTPase-activating protein for Bud1/Rsr1 necessary for proper bud-site selection in yeast. Nature. 1993;365:269–274. doi: 10.1038/365269a0. [DOI] [PubMed] [Google Scholar]
- 53.Pringle J R, Hartwell L H. The Saccharomyces cerevisiae cell cycle. In: Strathern J N, Jones E W, Broach J R, editors. Molecular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1981. pp. 97–142. [Google Scholar]
- 54.Richardson H E, Lew D J, Henze M, Sugimoto K, Reed S I. Cyclin-B homologs in Saccharomyces cerevisiae function in S phase and in G2. Genes Dev. 1992;6:2021–2034. doi: 10.1101/gad.6.11.2021. [DOI] [PubMed] [Google Scholar]
- 55.Roberts R L, Fink G R. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate the two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 56.Roberts R L, Mösch H-U, Fink G R. 14-3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell. 1997;89:1055–1065. doi: 10.1016/s0092-8674(00)80293-7. [DOI] [PubMed] [Google Scholar]
- 57.Rose M D, Winston F, Heiter P. Methods in yeast genetics. A laboratory course manual. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 58.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 59.Russell P, Nurse P. The mitotic inducer nim1+ functions in a regulatory network of protein kinase homologs controlling initiation of mitosis. Cell. 1987;49:569–576. doi: 10.1016/0092-8674(87)90459-4. [DOI] [PubMed] [Google Scholar]
- 60.Russell P, Nurse P. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell. 1987;49:559–567. doi: 10.1016/0092-8674(87)90458-2. [DOI] [PubMed] [Google Scholar]
- 61.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 62.Schwob E, Bohm T, Mendenhall M D, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 63.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 65.Tanaka S, Nojima H. Nik1: a Nim1-like protein kinase of S. cerevisiae interacts with the Cdc28 complex and regulates cell cycle progression. Genes Cells. 1996;1:905–921. doi: 10.1046/j.1365-2443.1996.d01-213.x. [DOI] [PubMed] [Google Scholar]
- 66.Tedford K, Kim S, So D, Stevens K, Tyers M. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr Biol. 1997;7:228–238. doi: 10.1016/s0960-9822(06)00118-7. [DOI] [PubMed] [Google Scholar]
- 67.Thomas, C., and A. M. Myers. Unpublished data.
- 68.Tzagoloff A, Akai A, Foury F. Assembly of the mitochondrial membrane system. XVI. Modified form of the ATPase proteolipid in oligomycin-resistant mutants of S. cerevisiae. FEBS Lett. 1976;65:391–396. doi: 10.1016/0014-5793(76)80154-8. [DOI] [PubMed] [Google Scholar]
- 69.Wallis J W, Chrebet G, Brodsky G, Rolfe M, Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- 70.Ward M P, Gimeno C J, Fink G R, Garrett S. SOK2 may regulate cyclic AMP-dependent protein kinase-stimulated growth and pseudohyphal development by repressing transcription. Mol Cell Biol. 1995;15:6854–6863. doi: 10.1128/mcb.15.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Watzele G, Tanner W. Cloning of the glutamine:fructose-6-phosphate amidotransferase gene from yeast. J Biol Chem. 1989;264:8753–8758. [PubMed] [Google Scholar]
- 72.Wei W, Herskowitz I. Abstracts of the 1998 Yeast Genetics and Molecular Biology Meeting. College Park, Md: Genetics Society of America; 1998. Identification of genes that are required for bud morphogenesis and the activation of G2 CLB/CDC28 activity, abstr. 314; p. 203. [Google Scholar]
- 73.Wittenberg C, Reed S I. Plugging it in: signaling circuits and the yeast cell cycle. Curr Opin Cell Biol. 1996;8:223–230. doi: 10.1016/s0955-0674(96)80069-x. [DOI] [PubMed] [Google Scholar]
- 74.Wu L, Russell P. Roles of Wee1 and Nim1 protein kinases in regulating the switch from mitotic division to sexual development in Schizosaccharomyces pombe. Mol Cell Biol. 1997;17:10–17. doi: 10.1128/mcb.17.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]