Abstract
The Aspergillus nidulans transcription factor PacC, which mediates pH regulation, is proteolytically processed to a functional form in response to ambient alkaline pH. The full-length PacC form is unstable in the presence of an operational pH signal transduction pathway, due to processing to the relatively stable short functional form. We have characterized and used an extensive collection of pacC mutations, including a novel class of “neutrality-mimicking” pacC mutations having aspects of both acidity- and alkalinity-mimicking phenotypes, to investigate a number of important features of PacC processing. Analysis of mutant proteins lacking the major translation initiation residue or truncated at various distances from the C terminus showed that PacC processing does not remove N-terminal residues, indicated that processing yields slightly heterogeneous products, and delimited the most upstream processing site to residues ∼252 to 254. Faithful processing of three mutant proteins having deletions of a region including the predicted processing site(s) and of a fourth having 55 frameshifted residues following residue 238 indicated that specificity determinants reside at sequences or structural features located upstream of residue 235. Thus, the PacC protease cuts a peptide bond(s) remote from these determinants, possibly thereby resembling type I endonucleases. Downstream of the cleavage site, residues 407 to 678 are not essential for processing, but truncation at or before residue 333 largely prevents it. Ambient pH apparently regulates the accessibility of PacC to proteolytic processing. Alkalinity-mimicking mutations L259R, L266F, and L340S favor the protease-accessible conformation, whereas a protein with residues 465 to 540 deleted retains a protease-inaccessible conformation, leading to acidity mimicry. Finally, not only does processing constitute a crucial form of modulation for PacC, but there is evidence for its conservation during fungal evolution. Transgenic expression of a truncated PacC protein, which was processed in a pH-independent manner, showed that appropriate processing can occur in Saccharomyces cerevisiae.
A growing class of transcription factors is activated by the proteolytic removal of protein domains which negatively modulate their activity. These negatively acting domains can be provided in trans (i.e., by another protein in a complex) or in cis (i.e., by a region within the transcription factor’s primary translation product). Examples of the former are the p50-p52 NF-κB family and their negative regulators, the IκB proteins, which regulate human genes involved in immune and inflammatory responses (reviewed in reference 37), and their respective Drosophila homologues dorsal and cactus (reviewed in reference 3), which establish the dorsal-ventral polarity of the fly embryo and mediate the Drosophila immune response (19). Examples of the latter include the p105 precursor of NF-κB p50 (whose C-terminal moiety is homologous to a trans-acting member of the IκB family); the sterol regulatory element binding proteins (reviewed in reference 5), which activate genes for cholesterol biosynthesis, low density lipoprotein receptors, and fatty acid synthesis; the zinc finger protein Ci, which is the product of the Drosophila cubitus interruptus gene, a mediator of the Hedgehog signal (reviewed in reference 30); and the Aspergillus nidulans zinc finger protein PacC, which mediates regulation of gene expression by ambient pH (39). An extensive collection of pacC mutations, including acidity-mimicking pacC+/− loss-of-function mutations, alkalinity-mimicking pacCc gain-of-function mutations, and neutrality-mimicking pacCc/− mutations (references 6 and 39 and this work) is available, making PacC a particularly well-suited choice for investigating the proteolytic activation of eukaryotic transcription factors.
In the current model (25), the 678-residue primary translation form of PacC is activated at alkaline ambient pH by proteolytic removal of a C-terminal negatively acting domain. The resulting truncated PacC form activates transcription of genes expressed at alkaline pH through 5′-GCCARG sites in their promoters (12, 14) and prevents expression of acid-expressed genes (22, 31). PacC processing is triggered by a signal transduced by the pal gene pathway under alkaline growth conditions, resulting in an as-yet-unknown modification of the protein. This causes a conformational change in PacC, rendering it accessible to proteolytic removal of more than 400 C-terminal residues, including the negatively acting domain.
Here we address several important aspects of this model, demonstrating a precursor-to-product relationship between the two PacC forms, an obligatory requirement for a functional pal signalling pathway for proteolytic processing, and the integrity of the original N terminus in the processed form. We describe single-residue substitutions that almost certainly disrupt interactions between the N- and C-terminal moieties of PacC, resembling C-terminal truncations of PacC in their alkalinity mimicry and constitutive processing (25, 39). An acidity-mimicking deletion likely to prevent pH signal reception or its effect prevents processing. We determine that the most upstream site(s) of processing is within residues 252 to 254 or in the immediate vicinity and show that the specificity for processing resides upstream of residue 235 and thus upstream of the cleavage site(s). Finally, we show that this unusual processing reaction can occur in Saccharomyces cerevisiae, provided that the pH signalling pathway is bypassed.
MATERIALS AND METHODS
A. nidulans strains, phenotype testing, and genetic analysis.
All strains used in this work carried markers in standard use (7). Standard media, phenotype testing, and genetic procedures were used (references 1, 6, 7, and 39 and references therein). Penicillin production broth (PPB) is appropriately supplemented minimal medium (8) containing 2.5% (wt/vol) corn steep liquor, 10 mM ammonium tartrate, and the indicated carbon sources. Media were adjusted to acidic, neutral, or alkaline pH as described previously (25). For classical mutant strains, 3% (wt/vol) sucrose was present. For strains carrying gene fusions to alcAp, media (adjusted to acidic pH) contained (final concentrations) 100 mM l-threonine and 0.05% glucose for inducing conditions and 3% glucose for noninducing, repressing conditions. In all cases, except those for transient-expression experiments, cultures were grown for 24 h at 37°C.
Isolation and characterization of new pacC mutations.
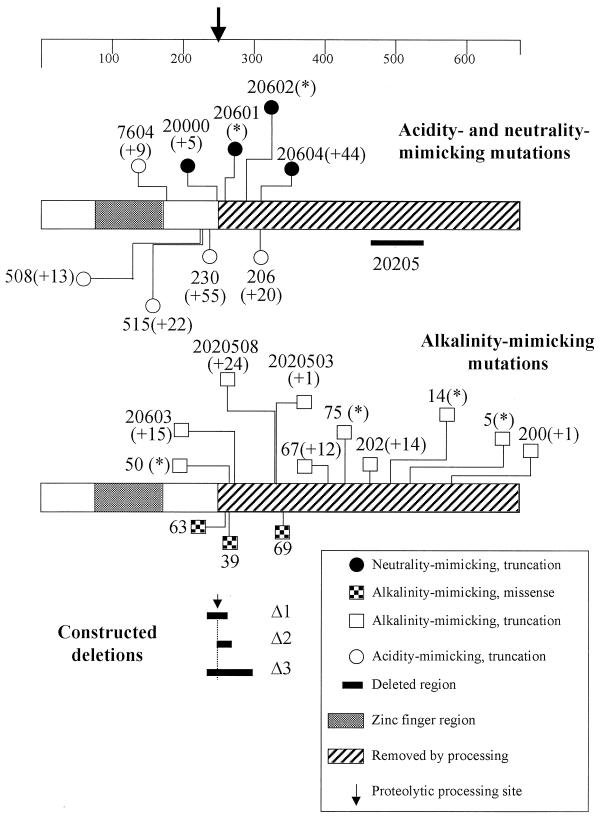
The parental strains and selection procedures used to isolate the classical pacC mutations used in this work are shown in Table 1. Their approximate positions are shown in Fig. 1. New mutations were characterized by sequencing as described previously (39). Strains carrying these mutations also carried one or more additional markers which do not affect pH regulation.
TABLE 1.
pacC mutations used in this work
| Mutation | Parental strain genotype | Mutagen | Selection method | Reference |
|---|---|---|---|---|
| pacC504 | pabaA1 pacCc5 | NQOa | Growth on 50 mM GABAb as C and N sources | 39 |
| pacC+/−7604 | inoB2 pacCc76 | UV | Resistance to neomycin (2 mg/ml) | 39 |
| pacC+/−508 and 515 | pabaA1 pacCc5 | UV | Resistance to neomycin (200 μg/ml) + 0.04% sodium deoxycholate | 39 |
| pacC+/−230 | areAr inoB2 fwA1 | UV | Growth on 5 mM GABA as N source | This work |
| pacCc20000 | yA2 gatA2 pantoB100 pacCc200 | UV | Resistance to neomycin (1 mg/ml) | This work |
| pacCc63 and 69 | pabaA1 yA2 wA3 palA1 | UV | Growth at pH 8 | This work |
| pacC+/−206 | biA1 areA1601 areAr 168 pantoB100 | UV | Growth on 5 mM GABA as N source | This work |
| pacCc/−20601, 20602, and 20604 | pacC+/−206 pantoB100 | UV | Growth at pH 8 | This work |
| pacCc20603 | pacC+/−206 pantoB100 | UV | Growth at pH 8 | This work |
| pacCc39 | biA1 pyroA4 palB7 chaA1 | NQO | Growth at pH 8 at 42°C | This work |
| pacCc50 | biA1 nis-5 | UV | Enhanced growth on nitrite as N source + 40 mM CsCl | 29 |
| pacCc2020503 and -2020508 (2020516, 2020515) | pabaA1 pacC+/−20205 pacCc202 | UV | Growth at pH 8 | This work |
| pacCc67 and 75 | inoB2 palB7 chaA1 | UV | Growth at pH 8 | This work |
| pacCc202 | pabaA1 sasA60 | Spontaneous | Resistance to GABA toxicity | 39 |
| pacC+/−20205 | pacCc202 | UV | Resistance to neomycin (1 mg/ml) | This work |
| pacCc14 | pabaA1 gatA2 palF15 fwA1 | UV | Resistance to 5 mM GABA in presence of 10 mM ammonium tartrate as N source | 39 |
| pacCc5 | biA1 | UV | Decreased staining for acid phosphatase | 11 |
NQO, 4-nitroquinoline-1-oxide.
GABA, α-aminobutyrate.
FIG. 1.
Positions of pacC mutations used in this work. The two bar diagrams represent the PacC coding sequence, with the scale at the top indicating amino acid residues. The ends of normal sequence before truncating mutations, the positions of missense mutations, and the extents of deletions are shown. Allele numbers for classical mutations are given. Their molecular descriptions are detailed in Table 3. Within parentheses is the number of additional residues added by frameshift mutations. Asterisks indicate nonsense mutations. Recombinant alleles Δ1, Δ2, and Δ3 are described in Table 2.
EMSA and protein extraction.
Electrophoretic mobility shift assay (EMSA) and protein extraction were done as described previously (25). EMSA mixtures were made with 0.3 ng (∼20,000 Cerenkov cpm) of the 32P-labelled 31-mer double-stranded oligonucleotide containing the high-affinity ipnA2 PacC binding site (12, 14, 39). Protein-DNA complexes were usually resolved in 4% polyacrylamide gels, but 8% polyacrylamide gels and longer runs were used for high resolution of complexes corresponding to proteins approximating the size of the processed form.
Western analysis.
Proteins (100 μg) were resolved in a sodium dodecyl sulfate (SDS)–11% polyacrylamide gel and were analyzed by Western blotting as described previously (25). The primary antibody (working dilution, 1:2,000) was a polyclonal antiserum raised in rats against a His-tagged PacC(5-265) protein, which was overexpressed in Escherichia coli and purified by Ni2+ affinity chromatography. The protein was denatured in a buffer containing 2% SDS and 5% β-mercaptoethanol before immunization of the animals. The secondary antibody was a peroxidase-conjugated sheep antirat antibody (working dilution, 1:2,000). Peroxidase activity was detected with the ECL system (Amersham).
Plasmids.
pALC (15) was used for expression of PacC wild-type and mutant proteins under alcAp control. This plasmid contains a functional alcAp promoter including a transcription start site separated from a downstream trpC transcription terminator by a 5′-SalI-XbaI-BamHI-SmaI-PstI-EcoRI-3′ polylinker region. Two versions of this vector, carrying as a selection marker either an argB+ allele or a frameshifted (BglII blunt-ended) argB allele (to select for integration in the argB locus), were used. Constructs derived from pALC are described in Table 2. Constructs driving expression of PacC proteins in yeast under the control of GAL1p were derivatives of Invitrogen pYES2 (see also Table 2). p[alcAp::PacC(5-678)] was constructed by subcloning a cDNA fragment containing the complete pacC coding region into pALC. This fragment was flanked by an NcoI site (converted to a BamHI site) and a second BamHI site. The NcoI site overlapped the Met5 codon. The 3′ BamHI site is located 90 bp downstream of the translation stop codon. p[alcAp::PacC(5-265)] was made by subcloning an NcoI-BstEII fragment (both cohesive ends blunted with Klenow) into the SmaI site of pGEX-2T (Pharmacia) to introduce BamHI (5′) and EcoRI (3′) flanking sites, which were used for subcloning into pALC. For construction of pPacCΔ1, a cDNA encoding an internally deleted PacC protein lacking residues 235 to 264 was reconstructed by ligating a BamHI-AccI fragment (filled in with Klenow) from the above-described pGEX-2T derivative with a BstEII-EcoRI fragment (filled in with Klenow) from the pJB2 (39) cDNA clone into pGEX-2T digested with BamHI and EcoRI. The deleted cDNA open reading frame was then subcloned as a BamHI fragment (note the presence of a second BamHI site 3′ to the translation stop codon) into pALC. Plasmid pPacCΔ2 was made by PCR amplification and in-frame rejoining of two fragments encoding residues 5 to 250 and 271 to 678, respectively. pPacCΔ3 was constructed by ligating a BamHI-AccI fragment (filled in with Klenow) and an AvaI-EcoRI fragment (filled with Klenow) in pALC digested with BamHI and EcoRI. pYES::PacC(5-265) was made by subcloning a 0.8-kb BamHI-EcoRI fragment from p[alcAp::PacC(5-678)] in pYES2. Plasmid pYES::PacC(5-678) was made by subcloning the BamHI fragment containing the complete wild-type pacC open reading frame in pYES2. Plasmid pYES::PacC(5-492) was constructed by introducing the pacCc14 nonsense mutation in pacC codon 493 of pYES::PacC(5-678). Correct in-frame joining of fragments and the absence of PCR-introduced mutations in pacC mutant versions were verified by automated DNA sequencing.
TABLE 2.
Plasmids encoding wild-type and mutant versions of PacC
| Construct name | Encoded PacC product | Host |
|---|---|---|
| p[alcAp::PacC(5-678)] | 5-678 | A. nidulans |
| p[alcAp::PacC(5-265)] | 5-265+GNSS | A. nidulans |
| pPacCΔ1 | (5-234)+C::(265-278) | A. nidulans |
| pPacCΔ2 | (5-250)::(271-678) | A. nidulans |
| pPacCΔ3 | (5-234)+S::(301-678) | A. nidulans |
| pYES::PacC(5-265) | 5-265+GNSADIHHTGGRSSMHLEGRIM | S. cerevisiae |
| pYES::PacC(5-678) | 5-678 | S. cerevisiae |
| pYES::PacC(5-492) | 5-492 | S. cerevisiae |
A. nidulans transformation.
Constructs were introduced into ΔpacC (full genotype, biA1 pabaA1 yA2 pyrG89 argB2 ΔpacC::pyr4+) and palA1 ΔpacC (full genotype, yA2 argB2 palA1 ΔpacC::pyr4+ pantoB100) recipient strains as appropriate (39, 40). Homokaryotic transformed clones were purified by repeated streaking on selective medium lacking arginine and analyzed by Southern blot hybridization. Two independent clones carrying a single-copy integration were chosen for each transforming construct and were shown to have the same phenotype.
Transient-expression experiments.
Acidic PPB cultures containing 3% (wt/vol) glucose were inoculated with 2 × 106 conidiospores/ml and incubated for 14 h at 37°C with vigorous shaking. Mycelia were harvested, washed with sterile water, and transferred to fresh acidic PPB containing 0.05% (wt/vol) glucose (a derepressing concentration) and 100 mM l-threonine (a strong inducer of alcAp), in which they were incubated for a further 6 h at 37°C. After this time, 3% (wt/vol) glucose was added to the cultures to repress alcAp transcription, and samples were taken at different time points (see Fig. 1 legend) after restoration of glucose repressing conditions. Mycelial samples were also taken immediately before and after promoter induction. Control transfer experiments showed that under such conditions 3% glucose prevented threonine induction of alcAp. Protein extracts were prepared from samples (2 to 4 g [wet weight] of mycelium) as described previously (25).
Yeast methods.
Standard methods were used for growth and maintenance of S. cerevisiae strains (33). W303-1A (38) (MATa ade2-1 his3-11,15 leu2-3,112 ura3-52 trp1-1, obtained from J. M. Gancedo) was used as the recipient for transformation by the lithium acetate procedure (17). For expression of PacC protein derivatives, cells were pregrown overnight at 30°C in 2% glucose minimal medium (lacking uracil). These primary cultures were used to inoculate secondary cultures in 2% raffinose minimal medium (lacking uracil), which were grown to an optical density at 600 nm of 0.6. At this point, 2% (wt/vol) d-galactose was added to induce the GAL1 promoter, and the cultures were further incubated for 6 h. Cells (20 ml) were collected by centrifugation, washed with water, and resuspended in twice their cell volume of lysis buffer (25 mM HEPES [pH 7.5], 50 mM KCl, 0.4 M ammonium sulfate, 5 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 μM leupeptin, and 0.6 μM pepstatin). Next, 1 to 1.5 volumes of glass beads (0.5-mm diameter) were added, and cells were lysed after vigorous vortexing (five times for 1 min each). The supernatants were decanted and collected, and glass beads and debris were washed with 0.4 ml of lysis buffer. Both supernatants were combined and centrifuged at 4°C for 60 min at 14,000 rpm in an Eppendorf microcentrifuge. The cleared supernatants (usually containing 2 mg of protein per ml) were dialyzed against lysis buffer without ammonium sulfate and assayed for PacC binding activity by EMSA, as described above, with 5 μg of protein per assay.
RESULTS
The unstable, full-length PacC form is processed to the relatively stable, short, functional form in the presence of an operational pal pathway.
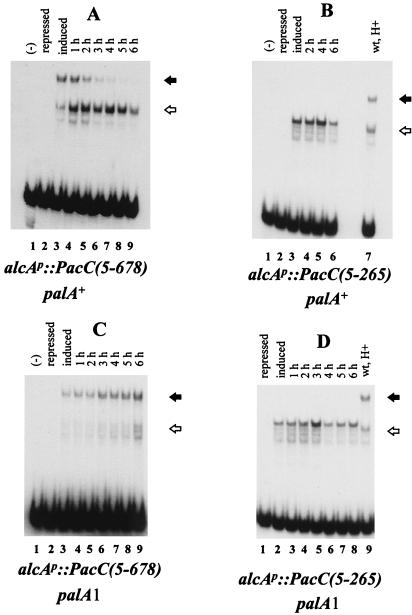
To establish a precursor-to-product relationship between the full-length and processed forms of PacC, we used transient-expression experiments, based on conditional expression (threonine inducible and glucose repressible) of proteins under the control of the alcA (alcohol dehydrogenase) promoter (28). An alcAp::PacC(5-678) construct driving expression of the full-length protein (see below) or an alcAp::PacC(5-265) construct driving expression of a truncated protein approximating the processed PacC version was introduced by transformation into a pacC null mutant background and targeted in single copy to the argB locus by homologous recombination. Neither PacC form was detectable by EMSA in protein extracts from mycelia grown under repressing conditions (Fig. 2A and B, lanes 2). Subsequent promoter induction for 6 h (after mycelial transfer) resulted in the synthesis of PacC proteins (Fig. 2A and B, lanes 3). The fate of the PacC proteins was then analyzed after the alcA promoter was switched off by addition of a repressing final concentration (3%, wt/vol) of glucose. The full-length form of PacC (low-mobility complex in Fig. 2) predominated after 6 h of induction of alcAp::PacC(5-678) (note that acidic growth conditions were used to reduce its processing), although some processed form (higher-mobility complex) was clearly visible at this point. The full-length form progressively disappeared after promoter shutdown, with less than 50% remaining after 3 h (data not shown). In contrast, the level of the processed form increased, apparently at the expense of the full-length form, not declining until 5 h after promoter shutdown (Fig. 2A). This indicates that, in the presence of a functional pal signal transduction pathway, the full-length form of PacC is significantly less stable than the processed form and that this instability results from the proteolytic processing itself. Consistent with a precursor-to-product relationship of the full-length and processed forms, the PacC(5-265) protein is markedly more stable (Fig. 2B). No reduction was evident 4 h after promoter shutdown, whereas 80% of the full-length form had been processed by this time (Fig. 2A) and only a 50% reduction was observed after 6 h. As predicted (25), the palA1 mutation, preventing pH signal transduction, stabilized the full-length form, blocking processing (Fig. 2C) but not affecting stability of the PacC(5-265) protein (Fig. 2D).
FIG. 2.
Transient-expression experiments with PacC(5-265) and PacC(5-678) proteins in the indicated genetic backgrounds. alcAp::PacC(5-678) and alcAp::PacC(5-265) constructs were transformed into ΔpacC and palA1 ΔpacC strains, and transformants having single-copy integrations of the transforming plasmids into the argB locus were identified by Southern analysis. Mycelia of the recombinant strains were grown for 14 h under repressing conditions and transferred to fresh acidic medium, with induction for alcAp, in which they were incubated for a further 6 h. After this time, a repressing concentration of glucose was added to prevent expression of the chimeric genes, and mycelial samples were harvested at the indicated time points after glucose repressing conditions were restored. Mycelial samples were also taken before (lanes labelled repressed) and after (lanes labelled induced) the 6-h induction period. Mycelial samples were used to prepare protein extracts that were assayed in EMSA experiments with a PacC target probe. Closed and open arrows indicate the protein-DNA complexes corresponding to the full-length and processed PacC forms, respectively. A control showing the complexes formed with a wild-type extract from mycelia grown under acidic conditions is shown in panels B and D. The PacC(5-265) protein used here is slightly larger than the processed PacC form (see text) but is functional in structural gene activation and repression.
Processing does not involve the N terminus of PacC.
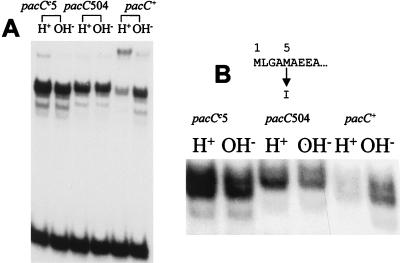
We have previously shown that PacC processing involves proteolytic removal of C-terminal residues of PacC (25), but this does not preclude the possibility of additional proteolytic removal of N-terminal residues upstream of the zinc finger region (beginning at residue 76). Two Met codons (codons 1 and 5) are present in the putative pacC coding region corresponding to the N terminus, either of which might be used for translational initiation. pacC504 (39) is a missense mutation affecting codon 5, resulting in an M5I substitution and selected as partially suppressing the alkalinity-mimicking phenotype of pacCc5. The diminution of this alkalinity-mimicking phenotype by pacC504 suggested that translational initiation occurs, at least in part, at codon 5 and that suppression results from reduced PacC synthesis (39). pacC504 does, indeed, reduce PacC levels in a pacCc5 background (Fig. 3A). Figure 3A also shows that the pacC504 mutation decreases the mobility of the complex formed by the processed form of PacC under both acidic and alkaline growth conditions. High-resolution EMSA analysis (Fig. 3B) confirmed this decreased mobility and showed that the processed PacC complex is resolved into at least two bands, which are seen with both wild-type and pacC504 extracts, excluding the possibility that they result from alternative translational initiation at Met codons 1 and 5 and suggesting C-terminal heterogeneity of the processed form. These data strongly suggest that Met5 is the major translation initiation residue and that, by mutating codon 5 to an Ile codon, pacC504 forces the use of Met codon 1 for initiation. Therefore, the major translation product contains 674 residues (not 678). However, to avoid confusion with the previous literature, we will continue numbering from Met1. In addition, the mobility shift resulting from the pacC504 mutation demonstrates that processing removes only residues C terminal to the DNA binding domain, leaving the N terminus intact.
FIG. 3.
PacC proteolytic processing does not remove N-terminal residues. (A) EMSA with mycelial extracts of wild-type, pacCc5, and pacC504 (pacCc5) strains. (B) Complexes formed by these extracts were analyzed under conditions maximizing resolution in the region of the processed-form complex. Note the presence of two predominant bands in wild-type and mutant extracts approximately corresponding to a four-residue size difference in the protein moiety, as shown by comparison of the mobilities of the pacC+ (or pacCc5) and pacC504 (pacCc5) processed-form complexes. Not only the mobility of the protein-DNA complex corresponding to the processed PacC form but also those of the misprocessing or degradation products consistently decrease due to the pacC504 (pacCc5) mutation as compared to the pacCc5 single mutant parent. Cultures were grown in PPB for 24 h at 37°C, using 3% sucrose as the main carbon source. In this and all other figures, H+ and OH− indicate acidic and alkaline growth conditions, respectively. Note that the pacCc5 strain showed pH-independent elevated PacC levels, in contrast to our previous report (25), in which we detected some pH dependence.
Certain alkalinity-mimicking missense mutations facilitate the accessibility of PacC to the processing protease.
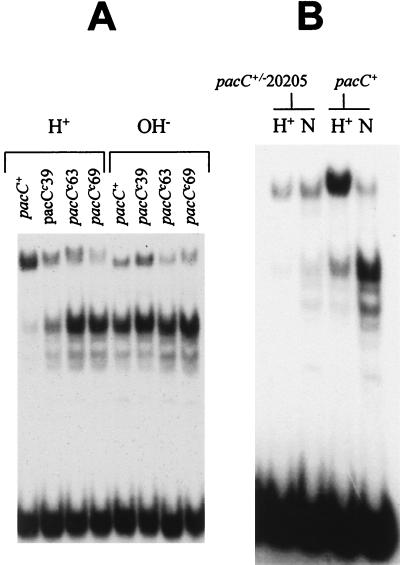
Sequencing of mutant pacC alleles revealed that three alkalinity-mimicking pacCc mutations, pacCc39, -63, and -69, are missense mutations resulting in the substitutions L266F, L259R, and L340S, respectively (Table 3). pacCc39 has a weak and thermosensitive phenotype and is only partially constitutive, as judged by its partial suppression of palB7 (in contrast to complete suppression by all other characterized pacCc mutations (1, 6). Correlating with the subtle phenotype, PacC processing in a pacCc39 strain was partially constitutive (i.e., somewhat elevated under acidic growth conditions but more so under alkaline growth conditions) (Fig. 4A). In contrast to the pH-dependent processing seen in the pacCc39 strain, PacC processing was fully constitutive in strains carrying the phenotypically more extreme pacCc63 and pacCc69 mutations (Fig. 4A), thus resembling processing in pacCc strains having a truncated PacC protein (25). The L340S (pacCc69) substitution maps at a considerable distance from the processing site (see below) and from the L266F (pacCc39) and L259R (pacCc63) substitutions. These mutations are therefore unlikely to favor protease recognition, suggesting instead that they modify the accessibility of PacC to the processing protease.
TABLE 3.
Mutant sequence changes in pacC and resulting proteins
| Genotype | Phenotype | Mutation | Mutant protein | Reference |
|---|---|---|---|---|
| pacC504 (pacC5) | ?a | G835C (C2528T) | M5I (5→523*)d | 39 |
| pacC7604 | +/− | ΔC1426 | 5→173 QSLDLATQI* | 39 |
| pacC508 | +/− | ΔC1631 | 5→224 SHRTRLTATSTTL* | 39 |
| pacC515 | +/− | 1641+A | 5→227 KPVLRQRLLRSEYRPRASPSVV* | This work |
| pacC230 | +/− | ΔT1675 | 5→238 KPAQSLTKRRMNPRSGVMMRLMSSLVTSSADNLTLIPTLPWASACSVCRTCPCLF* | This work |
| pacC20000 | c/− | 1711+C | 5→251 QEAGL* | This work |
| pacC63 | c | T1734G | L259R | This work |
| pacC20601 | c/− | G1739T | 5→260* | This work |
| pacC39 | c (ts)b | C1754T | L266F | This work |
| pacC50 | c | A1758T | 5→266* | This work |
| pacC20603 | c | ΔC1792 | 5→278 WASACSVCRTCPCLF* | This work |
| pacC20602 | c/− | C1817T | 5→287* | This work |
| pacC206 | +/− | ΔT1888; T1891C | 5→310 SLVVHMVAALTLRRHIIFHQ* | This work |
| pacC20604 (pacC206) | c/− | ΔG1913 (ΔT1888; T1891C) | 5→310 SLVVHMVAPSPCAGISSSTNEQRPNQERLDQHRPVPAANAGHNI* | This work |
| parC2020508 | c | Δ1949→1950 | 5→330 EQRPNQERLDQHRPVPAANAGHNI* | This work |
| pacC2020503 | c | Δ1959→1961; 1958+ATT | 5→333 D* | This work |
| pacC69 | c | T1977C | L340S | This work |
| pacC67 | c | ΔC218; A2182T | 5→407 ILRTPLRLALRL* | This work |
| pacC75 | c | C2250G | 5→430* | This work |
| pacC202 | c | Δ2353→2555 | 5→464 IDRPGSPFRISGRG* | 39 |
| pacC20205 (pacC202) | +/− | ΔC2577 (Δ2353→2555) | 5→464 IDRPGSPL 541→678* | This work |
| pacC14 | c | C2437A | 5→492* | 39 |
| pacC5 | c | C2528T | 5→523* | 39 |
| pacC2020515 and pacC2020516c (pacC202 pacC20205) | c | CT560T and C1932T (Δ2353→2555; Δ2557) | 5→464ID* and A323V, respectively | This work |
| pacC200 | 2690+T | 5→578 T* | 39 |
The presence of the pacCc5 mutation prevents the assessment of the phenotype of pacC504.
ts, thermosensitive phenotype.
Both mutations are present in the same revertant, which has a constitutive phenotype.
Asterisks denote stop codons.
FIG. 4.
Single-amino-acid substitutions resulting in pH-independent processing and a deletion resulting in pH insensitivity. EMSAs were performed with the indicated protein extracts. (A) PacC phenotype of single-amino-acid substitution mutations leading to alkalinity mimicry. (B) PacC phenotype of pacC+/−20205. This acidity-mimicking mutation prevents growth under alkaline conditions. N, neutral pH conditions.
An acidity-mimicking deletion mutation preventing pH signal reception.
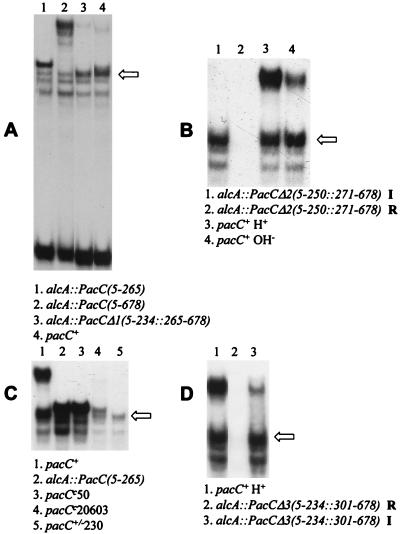
pacC+/−20205 is an extreme-acidity-mimicking loss-of-function mutation whose mutant product differs from wild-type PacC mainly through deletion of residues 465 through 540 (Table 3). Phenotypically, pacC+/−20205 strains closely resemble strains having null mutations in the palA, -B, -C, -F, and -H genes (1, 6, 10, 24) of the pH signal transduction pathway. Unlike pacC null mutants or strains having mutations such as pacC+/−206 and pacC+/−230 (see below), whose low PacC protein levels probably result, at least in part, from instability of the mutant proteins, pacC+/−20205 strains grow and conidiate normally under acidic and neutral conditions at both 25 and 37°C. The pacC+/−20205 mutation (Fig. 4B) markedly reduces PacC protein levels in extracts and largely prevents processing under neutral growth conditions (whereas wild-type PacC is nearly fully processed) (note that pacC+/−20205 strains do not grow under alkaline growth conditions). These data strongly suggest that one or more residues in the deleted region (residues 465 to 540) are involved in pH signal reception or its conformational consequences and that their absence, coupled with retention of the 138 C-terminal residues, results in insensitivity to the processing protease. If the only function absent from the mutant protein was the pH signal reception or response, an additional deletion at the C terminus should lead to pH-independent processing and alkalinity mimicry. The pacCc202 frameshift mutation (39) truncates PacC at residue 464 and results in pH-independent processing (25), strongly supporting this conclusion.
Neutrality-mimicking mutations: a new class of pacC mutations.
Selection and characterization of alkalinity- and acidity-mimicking mutations in PacC have been described previously (1, 6, 25, 39). Except for pacCc50, which truncates the protein after residue 266 (25), no mutations truncating PacC upstream of residue 464 and resulting in an alkalinity-mimicking, constitutive phenotype have been characterized (39). To characterize PacC further and define more precisely the site and requirements of processing, we sought mutations in the interval between the most-downstream characterized partial loss-of-function mutation, pacC+/−515, which truncates the protein after residue 227 (39), and codon 464 (Table 3). One technique employed was the reversion of two new extreme-acidity-mimicking loss-of-function mutations (Tables 1 and 3; Fig. 1), pacC+/−20205 (described above) and pacC+/−206. pacC+/−206 truncates the normal protein sequence after residue 310 and results in very low PacC protein levels (data not shown). Among the mutations selected in this way were alkalinity-mimicking mutations such as pacCc20603 and pacCc2020503, which truncate the normal protein sequence after residues 278 and 333, respectively (Tables 1 and 3; Fig. 1). In addition, mutations representing a new class were obtained (Tables 1 and 3; Fig. 1). These neutrality-mimicking or constitutive-derepressed (pacCc/−) mutations have a range of phenotypes but, in each case, share some aspects of the phenotypes of both alkalinity-mimicking and acidity-mimicking mutations. They do not respond to ambient pH and typically have rather high levels of both alkaline and acid phosphatases. Frequently they also lead to resistance to both molybdate (resembling pacCc mutations) and neomycin (resembling pacC+/− and pacC− mutations) toxicities. Neutrality-mimicking mutations also include pacCc/−20000, which was selected as alleviating the strongly alkalinity-mimicking phenotype of the pacCc200 mutation (39). Particularly noteworthy is pacCc/−20604, which replaces the final 12 residues of the 20-residue abnormal PacC206 C terminus by 36 residues, which are also abnormal but are encoded by a different reading frame, showing that low levels of the pacC+/−206 mutant product are possibly due to its abnormal, frameshifted C terminus.
Determination of the processing limit of PacC.
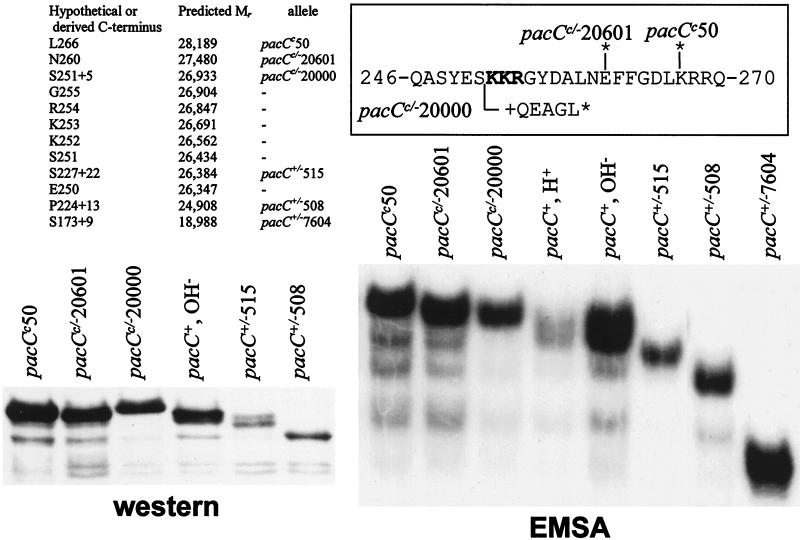
We previously proposed that the PacC proteolytic processing limit would be around residue 270 (25). We have characterized several truncating mutations by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) immunoblot analysis and high-resolution EMSA (Fig. 5) to determine this limit with greater precision. In Western analysis, the wild-type processed form, which appears as a doublet (in agreement with EMSA [Fig. 3A and 5]), showed a slight increase in mobility compared to the truncated pacCc50 and pacCc/−20601 products and a slight decrease compared to the pacC+/−515 product, indicating that the processing site would be located between residues 250 and 260 (see Fig. 5 for predictions of Mr). However, the pacCc/−20000 product showed unexpectedly reduced mobility, whereas the pacC+/−515 product showed a minor band with reduced mobility which was not observed for the pacC+/−508 protein (Fig. 5). Small differences in mobility leading to these inconsistencies might reflect differences in amino acid composition, which affects mobility because it affects SDS binding to the polypeptide (and consequently the charge/mass ratio). Basic proteins, acidic proteins, and proteins rich in proline show abnormally reduced mobility by SDS-PAGE (16), and even alterations of a single amino acid change the mobility of p21 ras in SDS-PAGE (32). Both pacCc/−20000 and pacC+/−515 are truncating mutations introducing frameshifted residues at the C termini of their products (Table 3). pacCc/−20000 removes the two basic amino acid clusters and introduces a glutamate residue in the frameshifted sequence. Its resulting ∼65 C-terminal residues (in a 252-residue protein, starting from Met5) contain no basic (but some acidic) residues and are proline rich. We suspect that one or more of these features may reduce SDS binding and consequently mobility. The pacC+/−515 protein has a 22-residue out-of-frame sequence at its C terminus, including 6 hydrophobic, 6 basic, and 3 proline residues.
FIG. 5.
Mapping of the C terminus of the processed form. The halftone on the left shows an enlarged portion of an SDS–11% PAGE immunoblot analysis showing the mobilities of different mutant truncated PacC proteins compared to that of the wild-type processed form. The right halftone is an EMSA autoradiogram showing the mobilities of complexes formed by the wild-type PacC processed form and by several mutant proteins truncated in the vicinity of the C terminus of this processed form. Culture conditions were as in Fig. 2. Acidic growth conditions were used for all mutant strains. The amino acid sequence of the region surrounding the predicted processing site is boxed, with the positions of key mutations indicated (see also Table 3). An asterisk denotes a stop codon. The table shows the predicted Mrs (starting at Met5) of hypothetical or derived (where mutations are indicated) proteins ending at the indicated positions.
Our sensitive EMSA gave much greater resolution than Western blots. In addition, it did not yield the inconsistency detected by Western blotting for the pacCc/−20000 product and resolved a single band with pacC+/−515 extracts, greatly facilitating the approximate localization of the proteolysis site. This presumably results from the fact that the net charge of the PacC-DNA complex is determined mostly by DNA phosphates, and therefore mass changes in the protein moiety are faithfully translated into mobility changes of the complex. In this high-resolution EMSA, extracts from a pacCc50 (truncating PacC after residue 266) strain revealed a prominent complex of lower mobility than the processed wild-type PacC. Wild-type PacC complex is resolved as a doublet, possibly indicating heterogeneity at the C terminus (Fig. 3 and 5). A minor band, resulting from inefficient processing of the pacCc50 protein, has wild-type mobility (Fig. 5). Therefore, the C terminus of the processed form must be upstream of residue 266. A similar result with pacCc/−20601 mutant extracts (Fig. 5) placed the proteolysis limit upstream of residue 260. In contrast, the complex formed by extracts of the pacC+/−515 strain was slightly more mobile than that with the wild-type processed form. Finally, the pacCc/−20000 strain extracts gave a complex with very slightly reduced mobility. The data in Fig. 5 enabled us to interpolate from a semilogarithmic plot of mutant protein Mr versus relative complex mobility that the most upstream C-terminal limit of the processed PacC form, corresponding to the faster band seen in the wild type, is likely to be within the basic tripeptide Lys252-Lys253-Arg254 or in its immediate vicinity.
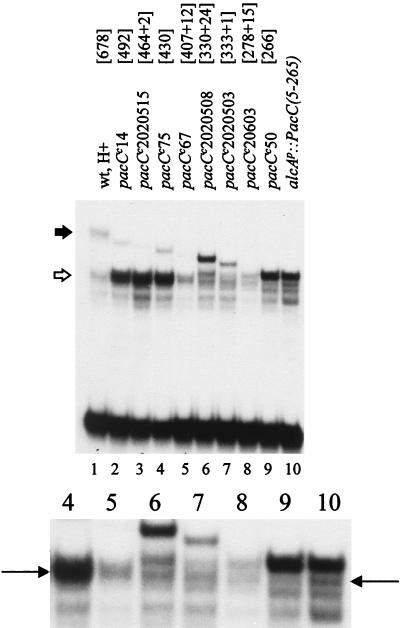
Proteolytic requirements C terminal to the processing limit.
pacCc14, a chain termination mutation which truncates PacC after residue 492 (39), results in pH-independent processing and elevated levels of processed PacC levels (Fig. 6) (25). Further C-terminal truncation (Table 3 and Fig. 1) to residue 464 (pacCc2020515) or 430 (pacCc75) had the same effect (Fig. 6). Truncation at residue 407 (pacCc67) still resulted in nearly complete processing but led to markedly reduced PacC levels (Fig. 6), in keeping with its relatively weak alkalinity-mimicking phenotype. Proteins truncated to residue 333 (pacCc2020503) or 330 (pacCc2020508) (Table 3; Fig. 1) were largely unprocessed under acidic growth conditions (Fig. 6). Extracts of strains carrying either of the latter two mutations formed, in addition to retardation complexes corresponding to (truncated) unprocessed and normally processed forms of PacC, a new complex having a mobility similar to that of the unprocessed pacCc50 (truncating after residue 266) and PacC(5-265) forms (Fig. 6). PacC in pacCc50 and alcAp::PacC(5-265) strain extracts is largely unprocessed (Fig. 5, 6, 7, and 8). PacC in extracts of a pacCc20603 strain (truncated after residue 278 but with a frameshifted 15-residue C terminus) is present at a relatively low level and processed such that the main complex is approximately equivalent in mobility to that of pacCc50. We conclude that PacC residues 407 to 678 are not required for processing but that truncating the protein at residue 333 or further upstream largely prevents it. Truncation at residue 333 or upstream also gives rise to an aberrant processing product.
FIG. 6.
Processing of mutant pacCc proteins truncated upstream of residue 492. EMSAs were performed with the indicated protein extracts. All cultures were grown under acidic conditions, using PPB with 3% sucrose, except that for lane 10, which was grown under inducing (and acidic) conditions for alcAp. Closed and open thick arrows denote the full-length and processed wild-type PacC proteins, respectively. Thin arrows in the enlarged portion of the autoradiogram indicate the position of the wild-type processed form. Numbers within brackets after mutations show the C-terminal residues of the products, with the number of additional residues added after frameshift mutations preceded by a +. See Table 3 for details.
FIG. 7.
Processing of PacC mutant proteins lacking the processing site. EMSAs of the indicated protein extracts were performed with the synthetic ipnA2 site as a probe. The arrows indicate the mobility of the complex formed by the processed PacC form. In panels A, B, and D, expression of indicated mutant PacC proteins was driven by alcAp and repressing or inducing growth conditions were used, as indicated below. (A) Data for PacC(5-234::265-678). Inducing (also acidic) growth conditions were used for alcAp. The low levels of the full-length form in lane 4 (wild type) are probably due to alkalinization resulting from threonine catabolism throughout growth. (B) Data for PacC(5-250::271-678). In lanes 1 and 2, mycelia pregrown under repressing conditions were transferred for 6 h to acidic inducing (I) or repressing (R) conditions, as indicated. (C) Data for pacC+/−230. Acidic growth conditions were used (in lane 2, alcAp inducing conditions were also used). (D) All cultures were grown under acidic conditions. In lanes 2 and 3, mycelia were pregrown under repressing conditions and transferred for 6 h to repressing (R) or inducing (I) conditions, as indicated.
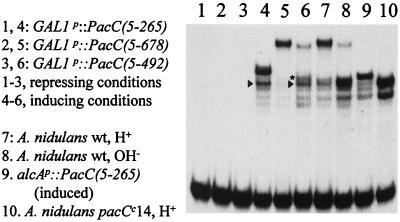
FIG. 8.
Processing of PacC in S. cerevisiae. In lanes 1 to 6, cultures of yeast strains containing the indicated constructs were grown under inducing or repressing conditions for GAL1p and used to prepare protein extracts, which were assayed by EMSA for PacC DNA binding activity. Control A. nidulans extracts were included, as indicated. Arrowheads denote the appropriately processed product in S. cerevisiae, whereas an aberrantly processed product is starred. wt, wild type.
The processing protease does not require a specific sequence at the PacC proteolysis site.
To determine whether PacC processing requires particular sequences at the proteolysis site, we constructed a mutant allele (pacCΔ1) driving expression (under alcAp control) of a PacC protein with residues 235 to 264 deleted (Table 2; Fig. 1). This 30-residue deletion resulted in constitutive processing under acidic growth conditions, giving a retardation complex indistinguishable in mobility from the processed wild-type pacC gene product (Fig. 7A). This implies that the PacC-processing protease does not require a specific target sequence (and also indicates that deletion of these 30 residues facilitates processing). To ensure that the processing of this deletion protein was not atypical, two further constructed deletion alleles plus a frameshift mutation obtained by classical genetics were analyzed for their effects on processing. Full-length PacC proteins with residues 251 to 270 (pacCΔ2 allele) or 235 to 300 (pacCΔ3 allele) deleted (Table 2; Fig. 1) resemble the pacCΔ1 product in yielding processed products indistinguishable in mobility from the wild type by EMSA (Fig. 7B and D). The acidity-mimicking pacC+/−230 frameshift mutation truncates the normal protein sequence after residue 238, which is then followed by 55 residues encoded in the −1 reading frame (Table 3; Fig. 1). pacC+/−230 results in low PacC levels, but processing appears to remove ∼40 frameshifted residues so that the retardation complex is of processed wild-type size (Fig. 7C). These results strongly suggest that the protease recognizes PacC sequences or structural features located upstream of residue 235 and cleaves the protein at a distance downstream from the recognition site.
PacC processing can occur in yeast if pH signalling is bypassed.
Homologues with regard to the DNA binding domain of PacC have been described for the yeasts S. cerevisiae and Yarrowia lipolytica (18, 36). The RIM8, -9, and -13 genes of S. cerevisiae (35) and the PAL1, -2, -3, and -4 genes of Y. lipolytica (18) are very likely to be functional homologues of the A. nidulans pal genes mediating ambient pH signal transduction (see Discussion). In particular, the RIM9- and palI-derived translation products show significant sequence similarity (9). S. cerevisiae Rim1p, whose DNA binding domain is homologous to that of PacC (39), also undergoes pH-dependent C-terminal proteolytic processing (20). This raises the question of whether the processing protease might be functionally conserved between S. cerevisiae and A. nidulans.
In A. nidulans, mutational C-terminal truncation of PacC obviates the requirement for the pal pH signal transduction pathway for processing. For example, pacCc14, an extreme-alkalinity-mimicking mutation encoding a protein truncated after residue 492 (39), results almost exclusively in the processed form of PacC irrespective of growth conditions (Fig. 6 and 8) (25). When various lengths of the pacC coding region were expressed in S. cerevisiae under GAL1p control (Fig. 8), expression of PacC(5-492) led to considerable processing, giving a major retardation band with approximately the same mobility as the processed form in A. nidulans extracts. In addition, yeast extracts contained a processed PacC form giving a slightly lower-mobility complex, approximating that formed by pacCc50 strain extracts (Fig. 8, lane 6). As in A. nidulans, PacC(5-265) is only very partially processed in S. cerevisiae (Fig. 8). Very little processing of the full-length PacC(5-678) protein occurred (Fig. 8). Taken together, these data indicate that appropriate PacC processing can occur in S. cerevisiae provided that mutational truncation has bypassed the need for the protease-sensitizing modification introduced by the pH signal transduction pathway. The presumed equivalent S. cerevisiae signal transduction process, if present, must not have been properly activated under the growth conditions used or cannot recognize the A. nidulans PacC protein (whose similarity to Rim1p is confined to the DNA binding domain).
DISCUSSION
According to the current model of pH regulation in A. nidulans (25), the full-length primary translation product is inactive. In response to alkaline ambient pH, the pal pathway mediates a modification of PacC which sensitizes it to a proteolytic activating step to yield a processed form of PacC competent in structural gene regulation. This work provides evidence for two key aspects of the model. First, relative-stability studies support a precursor-to-product relationship between the two PacC forms detected in extracts. The full-length form has a relatively short half life, as a result of its conversion to the much more stable processed form. Second, we confirm that PacC processing requires the pal signal transduction pathway. Mutational inactivation of this pathway does not affect stability of the processed form of PacC but markedly stabilizes the full-length form.
Using pacC504, which results in an M5I substitution and shifts translational initiation to codon 1, we demonstrated that proteolytic processing does not remove N-terminal residues and showed that the major translational initiation codon for PacC is AUG codon 5. The context of pacC codon 5 conforms more closely than that of codon 1 to the consensus for a strong initiation codon deduced for the A. nidulans areA gene (2). The fact that pacC504 does not lead to a loss-of-function phenotype shows that codon 1 is competent in translational initiation, even if it does not prevent leaky scanning. It is therefore possible that in the wild type a minor proportion (below current detection limits) of translation initiates at codon 1.
PacC processing is the pH-sensitive step leading to changes in gene expression in response to ambient pH. The activity of the processing protease is apparently not pH regulated, as C-terminal truncating pacCc mutations result in alkalinity mimicry and pH-independent processing (reference 25 and this work). Thus, the actual pH-regulated step is very likely to be the transition of PacC from a protease-insensitive to a protease-sensitive conformation. Under acidic growth conditions, interactions between C- and N-terminal moieties disrupted by C-terminal truncations would help maintain PacC in the protease-inaccessible conformation. Among the predictions of this model are that (i) pacC mutations outside the C terminus should also disrupt such interactions and result in pH-independent processing and a pacCc phenotype and (ii) pacC mutations rendering the protein insensitive to the alkaline pH signal or locking it in the protease-inaccessible conformation should result in a loss-of-function, acidity-mimicking phenotype. The L259R, L266F, and L340S substitutions are very likely to fall into the former class. The acidity-mimicking pacC+/−20205-containing allele is an example of the latter.
Leucine residues at positions 259, 266, and 340 are probably involved, directly or indirectly, in processing-preventing interactions with the C terminus. Constitutive (pH-independent) processing of mutant proteins with residues 235 to 264 or 251 to 270 deleted confirms the involvement of the environs of the processing site in interactions preventing processing and provides compelling evidence against the alternative interpretation that the L259R and L266F substitutions increase affinity for the processing protease (a possibility that would otherwise be formally equivalent to increased accessibility). The involvement of Leu340 in interactions with the C terminus has been established (13).
Analysis of a collection of PacC proteins mutationally truncated in the environs of the processing site(s) indicated slight heterogeneity at the C terminus of the processed form, with the most amino-proximal processing site located within or in the immediate vicinity of the basic sequence Lys252-Lys253-Arg254 and the most amino-distal limit located a further four residues downstream (Fig. 3 and 5). Processing requires a certain polypeptide chain length downstream of the proteolysis site. Truncation at residue 407 or downstream allows faithful and efficient processing. In contrast, PacC proteins ending at residue 265 or 266 (i.e., 13 or 14 residues downstream of Lys252) are inefficiently processed. Extension to residue 278 or 330 somewhat improved processing, although a significant proportion of the processed product was larger than the wild-type processed form, indicating aberrant processing. The sizes of these aberrant processing products appear to be very similar to that of an unprocessed product having Leu266 at its C terminus.
Internal deletion of the processing site(s), removing 17 residues upstream and 10 residues downstream of the Lys252-Lys253-Arg254 tripeptide, did not prevent processing, indicating a lack of specificity of the protease at the cleavage site. The processed mutant product is indistinguishable in size from the wild type and cannot result from processing at the downstream Lys267-Arg268-Arg269 tripeptide, as the resulting protein (233 to 235 residues beginning from Met5) would be detectably smaller than the wild-type processed form (248 to 250 residues). In addition, proteins with residues 251 to 270 or 235 to 300 deleted (therefore with both basic tripeptides removed) are efficiently and faithfully processed to wild-type size. This strongly suggests that the protease recognizes sequences or structural features of PacC upstream of residue 235 and cleaves at a distance downstream from the recognition sequence. Analysis of the pacC+/−230 product strongly supports this conclusion, as the 55-residue frameshifted peptide following residue 238 is appropriately cleaved, giving a processed form of approximately wild-type size. This not only confirms the lack of specificity at the cleavage site but shows that a completely different amino acid sequence can fulfil the minimum requirement for residues C terminal to the cleavage site for faithful processing.
A similar situation in which a processing protease cuts at a distance from its specificity determinants has been described for processing of the NF-κB and NF-κB2 p50 and p52 precursors, p105 (21) and p100 (4), respectively. The involvement of the ubiquitin-proteasome pathway in this reaction seems to be well established (26, 27) but raises questions of how a protein which has been ubiquitin tagged for proteasome-mediated proteolysis is not entirely degraded (27). Evidence presented by Lin and Ghosh (21) strongly suggested a two-step mechanism involving a signal-dependent endoproteolytic cleavage of p105 followed by degradation of the C-terminal moiety. They also demonstrated the presence of a context-independent 23-amino-acid signal ending 38 residues upstream of the target peptide bond which provides the cleavage specificity for the release of p50 from p105. This two-step mechanism would satisfactorily explain the partial degradation of the precursor, in which proteasome-dependent proteolysis of the released C-terminal fragment would be governed by the N-end rule (21). In common with our data for PacC, specific sequences at the cleavage site of p105 are not required. A similar conclusion was reached by Betts and Nabel (4) for processing of NF-κB2 p100.
A. nidulans PacC is a prototype of a family of filamentous fungal and yeast transcription factors having a homologous three-zinc-finger DNA binding domain and undergoing activation by proteolytic processing. S. cerevisiae Rim1p is also proteolytically activated in response to alkaline ambient pH (20). rim1 mutants show poor growth at low temperatures, altered colony morphology, inefficient sporulation, and defective invasive growth (20, 35), but the connection between these phenotypes and the absence of transduction of a pH-dependent signal through Rim1p is unclear. In contrast, it is well established that Y. lipolytica YIRim101p mediates pH regulation (18). Although proteolytic activation of YIRim101p is to be expected, based on the alkalinity-mimicking phenotype of C-terminal truncating mutations (18), it has not yet been reported.
Genes involved in transducing the ambient pH signal have been genetically identified in S. cerevisiae (RIM8, -9, and -13 [20, 35]) and Y. lipolytica (PAL1, -2, -3, and -4 [18]), and they might be isofunctional homologues of A. nidulans pal genes. Indeed, S. cerevisiae Rim9p shows 36.7% identity over 180 residues, including nearly all of the four hydrophobic, putative transmembrane segments, to A. nidulans PalI, but it lacks any sequence corresponding to the very basic, hydrophilic, C-terminal ∼400 residues of PalI (9). Sequence comparisons have also been useful in identifying putative yeast homologues of deduced pal gene products. Thus, PalB, which is likely to be a cysteine protease of the calpain family (10), shows significant similarity to the derived translation product of S. cerevisiae YMR154c in the catalytic domain and PalB homology domain (34). PalA shows 30.2% identity over 732 residues to the derived translation product of S. cerevisiae YOR275c (24). PalF shows short regions of similarity to derived translation products of S. cerevisiae YGL045w and YGL046w (23). The ability of C-terminally truncating gain-of-function mutations in RIM1 and YIRIM101 to bypass loss-of-function mutations in their respective signal transduction genes strongly suggests that the PacC model applies to its yeast homologues. However, the considerable sequence divergence in putative signal transduction homologues as well as within the transcription factors themselves outside the DNA binding domain (18, 39) provides a possible rationale for why the PacC primary translation product fails to undergo significant processing in S. cerevisiae (at least under the growth conditions tested). Very significantly, however, a mutant form of PacC truncated after residue 492, whose processing is independent of ambient pH (i.e., pal pathway) signalling, underwent processing to the correct size in S. cerevisiae, although some processing was aberrant. Thus, appropriate processing of PacC can definitely occur in yeast. An important question for the future is whether the same proteolytic activity is responsible for PacC and Rim1p processing and would thus represent a conserved proteolytic control point for transcription factors of the PacC-Rim1p family.
ACKNOWLEDGMENTS
We thank Teresa Suárez and Lynne Rainbow for discussions and Elena Reoyo for technical assistance.
We are grateful for support of the EU through Biotech contract BIO4-CT96-0535 (to M.A.P. and H.N.A.), the BBSRC through grant 60/P05893 (to H.N.A.) and research studentships (to C.V.B. and S.S.), the CICYT through grant BIO97-348 (to M.A.P.), the MEC and the Gobierno Vasco through PNFPI (to J.M.M.) and PFI (to E.D.) fellowships, respectively, and the DGICYT for a postdoctoral contract (to M.O.).
REFERENCES
- 1.Arst H N, Jr, Bignell E, Tilburn J. Two new genes involved in signalling ambient pH in Aspergillus nidulans. Mol Gen Genet. 1994;245:787–790. doi: 10.1007/BF00297286. [DOI] [PubMed] [Google Scholar]
- 2.Arst H N, Jr, Sheerins A. Translational competence, ‘leaky scanning’ and translational reinitiation in areA mRNA of Aspergillus nidulans. Mol Microbiol. 1996;19:1019–1024. doi: 10.1046/j.1365-2958.1996.470976.x. [DOI] [PubMed] [Google Scholar]
- 3.Belvin M P, Anderson K V. A conserved signaling pathway: the Drosophila toll-dorsal pathway. Annu Rev Cell Dev Biol. 1996;12:393–416. doi: 10.1146/annurev.cellbio.12.1.393. [DOI] [PubMed] [Google Scholar]
- 4.Betts J C, Nabel G J. Differential regulation of NF-κB2(p100) processing and control by amino-terminal sequences. Mol Cell Biol. 1996;16:6363–6371. doi: 10.1128/mcb.16.11.6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown M S, Goldstein J L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 6.Caddick M X, Brownlee A G, Arst H N., Jr Regulation of gene expression by pH of the growth medium in Aspergillus nidulans. Mol Gen Genet. 1986;203:346–353. doi: 10.1007/BF00333978. [DOI] [PubMed] [Google Scholar]
- 7.Clutterbuck A J. Aspergillus nidulans. In: O’Brien S J, editor. Genetic maps. Locus maps of complex genomes. 6th ed. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 3.71–3.84. [Google Scholar]
- 8.Cove D J. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta. 1966;113:51–56. doi: 10.1016/s0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- 9.Denison S H, Negrete-Urtasun S, Mingot J M, Tilburn J, Mayer W A, Goel A, Espeso E A, Peñalva M A, Arst H N., Jr Putative membrane components of signal transduction pathways for ambient pH regulation in Aspergillus and meiosis in Saccharomyces are homologous. Mol Microbiol. 1998;30:259–264. doi: 10.1046/j.1365-2958.1998.01058.x. [DOI] [PubMed] [Google Scholar]
- 10.Denison S H, Orejas M, Arst H N., Jr Signaling of ambient pH in Aspergillus involves a cysteine protease. J Biol Chem. 1995;270:28519–28522. doi: 10.1074/jbc.270.48.28519. [DOI] [PubMed] [Google Scholar]
- 11.Dorn G. Genetic analysis of the phosphatases in Aspergillus nidulans. Genet Res. 1965;6:13–26. doi: 10.1017/s0016672300003943. [DOI] [PubMed] [Google Scholar]
- 12.Espeso E A, Peñalva M A. Three binding sites for the Aspergillus nidulans PacC zinc-finger transcription factor are necessary and sufficient for regulation by ambient pH of the isopenicillin N synthase gene promoter. J Biol Chem. 1996;271:28825–28830. doi: 10.1074/jbc.271.46.28825. [DOI] [PubMed] [Google Scholar]
- 13.Espeso, E. A., T. Roncal, E. Bignell, J. Tilburn, H. N. Arst, Jr., and M. A. Peñalva. Unpublished data.
- 14.Espeso E A, Tilburn J, Sánchez-Pulido L, Brown C V, Valencia A, Arst H N, Jr, Peñalva M A. Specific DNA recognition by the Aspergillus nidulans three zinc finger transcription factor PacC. J Mol Biol. 1997;274:466–480. doi: 10.1006/jmbi.1997.1428. [DOI] [PubMed] [Google Scholar]
- 15.Fernández-Cañón J M, Peñalva M A. Overexpression of two penicillin structural genes in Aspergillus nidulans. Mol Gen Genet. 1995;246:110–118. doi: 10.1007/BF00290139. [DOI] [PubMed] [Google Scholar]
- 16.Hames B D. One-dimensional polyacrylamide gel electrophoresis. In: Hames B D, Rickwood D, editors. Gel electrophoresis of proteins. A practical approach. Oxford, United Kingdom: IRL Press; 1990. pp. 1–147. [Google Scholar]
- 17.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali ions. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambert M, Blanchin-Roland S, Le Louedec F, Lépingle A, Gaillardin C. Genetic analysis of regulatory mutants affecting synthesis of extracellular proteinases in the yeast Yarrowia lipolytica: identification of a RIM101/pacC homolog. Mol Cell Biol. 1997;17:3966–3976. doi: 10.1128/mcb.17.7.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemaitre B, Nicolas E, Michaut L, Reichhart J M, Hoffmann J A. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 20.Li W S, Mitchell A P. Proteolytic activation of RIM1p, a positive regulator of yeast sporulation and invasive growth. Genetics. 1997;145:63–73. doi: 10.1093/genetics/145.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin L, Ghosh S. A glycine-rich region in NF-κB p105 functions as a processing signal for the generation of the p50 subunit. Mol Cell Biol. 1996;16:2248–2254. doi: 10.1128/mcb.16.5.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacCabe A P, Orejas M, Pérez-González J A, Ramón D. Opposite patterns of expression of two Aspergillus nidulans xylanase genes with respect to ambient pH. J Bacteriol. 1998;180:1331–1333. doi: 10.1128/jb.180.5.1331-1333.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maccheroni W, May G S, Martinez-Rossi N M, Rossi A. The sequence of palF, an environmental pH response gene in Aspergillus nidulans. Gene. 1997;194:163–167. doi: 10.1016/s0378-1119(97)00095-4. [DOI] [PubMed] [Google Scholar]
- 24.Negrete-Urtasun S, Denison S H, Arst H N., Jr Characterization of the pH signal transduction pathway gene palA of Aspergillus nidulans and identification of possible homologs. J Bacteriol. 1997;179:1832–1835. doi: 10.1128/jb.179.5.1832-1835.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orejas M, Espeso E A, Tilburn J, Sarkar S, Arst H N, Jr, Peñalva M A. Activation of the Aspergillus PacC transcription factor in response to alkaline ambient pH requires proteolysis of the carboxy-terminal moiety. Genes Dev. 1995;9:1622–1632. doi: 10.1101/gad.9.13.1622. [DOI] [PubMed] [Google Scholar]
- 26.Orian A, Whiteside S, Israel A, Stancovski I, Schwartz A L, Ciechanover A. Ubiquitin-mediated processing of NF-κB transcriptional activator precursor p105. J Biol Chem. 1995;270:21707–21714. doi: 10.1074/jbc.270.37.21707. [DOI] [PubMed] [Google Scholar]
- 27.Palombella V J, Rando O J, Goldberg A L, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 28.Pu R T, Osmani S A. Mitotic destruction of the cell cycle regulated NIMA protein kinase of Aspergillus nidulans is required for mitotic exit. EMBO J. 1995;14:995–1003. doi: 10.1002/j.1460-2075.1995.tb07080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rand K N. Aspects of the control of nitrogen metabolism in Aspergillus nidulans. Ph.D. thesis. Cambridge, United Kingdom: University of Cambridge; 1978. [Google Scholar]
- 30.Ruiz i Altaba A. Catching a Gli-mpse of Hedgehog. Cell. 1997;90:193–196. doi: 10.1016/s0092-8674(00)80325-6. [DOI] [PubMed] [Google Scholar]
- 31.Sarkar S, Caddick M X, Bignell E, Tilburn J, Arst H N., Jr Regulation of gene expression by ambient pH in Aspergillus: genes expressed at acid pH. Biochem Soc Trans. 1996;24:360–363. doi: 10.1042/bst0240360. [DOI] [PubMed] [Google Scholar]
- 32.Seeburg P H, Colby W W, Capon D J, Goeddel D V, Levinson A D. Biological properties of human c-Ha-ras1 genes mutated at codon 12. Nature. 1984;312:71–75. doi: 10.1038/312071a0. [DOI] [PubMed] [Google Scholar]
- 33.Sherman F, Fink G R, Hicks J B. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 34.Sorimachi H, Ishiura S, Suzuki K. Structure and physiological functions of calpains. Biochem J. 1997;328:721–732. doi: 10.1042/bj3280721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su S S Y, Mitchell A P. Identification of functionally related genes that stimulate early meiotic gene expression in yeast. Genetics. 1993;133:67–77. doi: 10.1093/genetics/133.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su S S Y, Mitchell A P. Molecular characterization of the yeast meiotic regulatory gene RIM1. Nucleic Acids Res. 1993;21:3789–3797. doi: 10.1093/nar/21.16.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thanos D, Maniatis T. NF-κB: a lesson of family values. Cell. 1995;24:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 38.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 39.Tilburn J, Sarkar S, Widdick D A, Espeso E A, Orejas M, Mungroo J, Peñalva M A, Arst H N., Jr The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 1995;14:779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tilburn J, Scazzocchio C, Taylor G G, Zabicky-Zissman J H, Lockington R A, Davies R W. Transformation by integration in Aspergillus nidulans. Gene. 1983;26:205–211. doi: 10.1016/0378-1119(83)90191-9. [DOI] [PubMed] [Google Scholar]