Abstract
The receptor tyrosine kinase Eyk, a member of the Axl/Tyro3 subfamily, activates the STAT pathway and transforms cells when constitutively activated. Here, we compared the potentials of the intracellular domains of Eyk molecules derived from c-Eyk and v-Eyk to transform rat 3Y1 fibroblasts. The v-Eyk molecule induced higher numbers of transformants in soft agar and stronger activation of Stat3; levels of Stat1 activation by the two Eyk molecules were similar. A mutation in the sequence Y933VPL, present in c-Eyk, to the v-Eyk sequence Y933VPQ led to increased activation of Stat3 and increased transformation efficiency. However, altering another sequence, Y862VNT, present in both Eyk molecules to F862VNT markedly decreased transformation without impairing Stat3 activation. These results indicate that activation of Stat3 enhances transformation efficiency and cooperates with another pathway to induce transformation.
The oncogene v-eyk was first isolated from an acute avian retrovirus, RPL30 (38). It encodes a transmembrane receptor-type tyrosine kinase, p69gp37v-Eyk, in which the intracellular region of the putative receptor tyrosine kinase (RTK) is fused to the viral gp37 glycoprotein, leading to dimerization and activation of v-Eyk (38). The chicken proto-oncogene c-eyk, from which v-eyk is derived, codes for an RTK with a distinct extracellular region, containing two immunoglobulin-like domains and two fibronectin type III-like repeats (37). Based on its extracellular structure and the kinase domain sequence, c-Eyk is classified in the same subfamily of tyrosine kinases as Axl/Ark/UFO (36, 49; reviewed in reference 67). In addition to Axl/Ark/UFO, members of this subfamily include Sky/Rse/Tif/Brt/Tyro3 (24, 40, 43, 51), Rek (4), and Mer (31), the last of which is the closest mammalian homologue of c-Eyk. Members of this subfamily of RTKs are expressed in a variety of tissues and cell types: Tyro3 and Rek in different regions of the brain (24, 43, 51), Tyro3 in ovaries and testes (14, 45), Mer in monocytes and bone marrow cells (31), Axl in cell types of mesodermal origin such as thymic stromal cells (20, 53), and c-Eyk in adult spleen (37). Several of these RTKs are expressed during embryogenesis in different tissues (30, 37, 50). The ligands for these RTKs were identified as the anticoagulation factor protein S and the growth arrest-specific gene product Gas6 (47, 63, 68). The results of several studies addressing the physiological functions of these RTK-ligand systems have indicated roles in the growth and survival of cells (2, 28, 41) and the adhesion and migration (1, 23) of cells. In several cases, members of this subfamily have been implicated in transformation and tumor formation of a variety of cells by overexpression or constitutive activation of the RTK (36, 38, 54). v-Eyk transformation by infection with the RPL30 virus induces chicken erythroblastosis, fibrosarcomas, endotheliomas, visceral lymphomatosis, and hemorrhage (21).
We reported that chimeric forms of Eyk that are constitutively dimerized and activated can transform fibroblasts in vitro (74) without significant stimulation of the Ras/ERK (extracellular signal-regulated kinase) pathway which is characteristic for transformation by many oncogenes (60). In contrast, Eyk strongly stimulated molecules of the Jak (Janus kinase)/STAT (signal transducers and activators of transcription) pathway, namely, Stat1, Stat3, and Jak1 (74). However, most of the activation of STATs was assessed in COS cells, and transformation frequencies of Stat1 and Stat3 activation were not compared quantitatively.
Activation of the Jak/STAT pathway by cytokine receptor activation is well established (reviewed in references 16, 34, and 59). Activation of Stat3 by treatment of cells with interleukin 6 (IL-6), a ligand for the gp130 receptor, and activation of Stat1 by treatment with interferons have been well characterized. Stat3 has been implicated in proliferation of cells (13, 25), whereas Stat1 activation correlates more with immune responses and growth arrest of cells (8, 12). These notions are supported by the phenotypes of mice harboring targeted disruptions of Stat1 or Stat3. Stat1−/− mice are viable and show severe deficiencies in acute response to infections by a variety of pathogens (19, 46), whereas Stat3−/− mice die very early in embryonal development, suggesting a possible function for Stat3 activity in the proliferation of cells (64). The activation of various STAT factors by oncogenes and RTKs has been demonstrated in a growing number of cases, e.g., v-Src (73), v-Abl (15), Bcr-Abl (35), v-Fps and c-Fes (26, 48), the epidermal growth factor (EGF) receptor (55, 56), v-Eyk and c-Eyk (74), the hepatocyte growth factor/scatter factor receptor c-Met (5), v-Sis, a ligand activating the platelet-derived growth factor-receptor, and polyomavirus middle-T antigen (26). Recently, we and others have shown that in the case of v-Src, the activation of Stat3 is required for transformation (7, 66).
In our initial study (74), transient-transfection data for activated Eyk molecules in COS cells and the fact that a dominant negative Stat1, Stat1β, had a negative effect on transformation efficiency suggested that the activation of Stat1 may have been primarily responsible for transformation. In this study, we examined more extensively and quantitatively the importance of Stat1 and Stat3 activation in the transformation of rat 3Y1 fibroblasts by constitutively active c-Eyk and v-Eyk. The constitutively active receptor containing the intracellular portion of the v-Eyk molecule induced strong activation of Stat3, but not of Stat1, and resulted in a higher transformation efficiency. A single amino acid residue (glutamine 936) in the COOH-terminal tail of v-Eyk was found to be responsible for constitutive Stat3 activation and to be required for transformation. The transforming ability of c-Eyk could be converted to that of v-Eyk by changing this single residue from leucine to glutamine. Finally, we conclude from modification in another tyrosine residue in the intracellular domain of the Eyk molecules that Stat3 activation alone is not sufficient for transformation.
MATERIALS AND METHODS
Reagents.
Sources of reagents were as follows: [γ-32P]ATP (3,000 Ci/mmol), [α-32P]deoxynucleoside triphosphates (3,000 Ci/mmol), and the Renaissance immunodetection system, New England Nuclear; luciferin, Boehringer Mannheim; GammaBind G-Sepharose and glutathione-Sepharose 4B (glutathione-beads), Pharmacia; and Lipofectamine and Opti-MEM, Gibco BRL; all other reagents, Sigma.
The antibody against the COOH terminus of Eyk has been described elsewhere (38), as has an antibody against phosphotyrosine (70). Other antibodies used were anti-glutathione S-transferase (GST) epitope (Santa Cruz Biotechnology), anti-FLAG epitope (M2; Kodak), and horseradish peroxidase-conjugated anti-rabbit (Santa Cruz) and anti-mouse (Jackson Laboratories) secondary antibodies.
Cell lines.
Nontransformed 3Y1 rat fibroblasts (39) and 3Y1 cells transformed with CD8–c-Eyk, CD8–v-Eyk, or v-Src were cultured in Dulbecco’s modified Eagle medium (DMEM; Gibco BRL) supplemented with 5% calf serum (Gibco BRL). CD8–c-Eyk (74)- and CD8–v-Eyk- and v-Src-transformed cell lines (57) were described elsewhere. AD5-transformed simian virus 40 (SV40) large-T-antigen-containing 293T human embryonic kidney cells (52) were cultured in DMEM supplemented with 10% fetal bovine serum (FBS; HyClone).
Plasmid and probes.
Luciferase reporter constructs used were pTATA-tk-Luc containing a minimal thymidine kinase promoter (44), pSV40-Luc containing the SV40 promoter and enhancer (pGL2 control; Promega), and p4xM67-tk-Luc. To obtain p4xM67-tk-Luc, an oligonucleotide containing four copies of the sequence GGTTCCCGTAAATGCATCA (underline denotes the STAT-binding site [33]) was introduced in the AccI-BamHI sites of pTATA-tk-Luc upstream of the minimal promoter.
Expression plasmids for constitutively active v-Src and Ras61L (RasLeu61) were pBabe/v-src (57) and pRSV/H-ras61L (18), respectively. Expression constructs used were pEBG/erk1 (65), pRcCMV/stat3-FLAG (7), and pRcCMV/stat1-FLAG (constructed as described for pRSVstat3-FLAG except using the coding sequence of Stat1).
pMEXneo-based expression constructs for c-Eyk, CD8–c-Eyk, CD8–v-Eyk, and CD8–c-EykM have been described elsewhere (74). Point mutations in CD8-Eyk molecules were made via a site-directed mutagenesis protocol (QuickChange; Stratagene) as suggested by the manufacturer, giving rise to the constructs pMEX/CD8–c-EykL936Q (CTG to CAG, bp 2832 in c-Eyk [GenBank accession no. L21719 {37}]), pMEX/CD8–c-EykY933F (TAG to TTG, bp 2823), pMEX/CD8–c-EykY862F (TAG to TTG, bp 2610), pMEX/CD8–c-EykK609M (AAG to ATG, bp 1851), pMEX/CD8–v-EykY933F (TAG to TTG, bp 1973 in v-eyk/v-ryk [accession no. M92847 {38}]), pMEX/CD8–v-EykY862F (TAG to TTG, bp 1760), or pMEX/CD8–v-EykK609M (AAG to ATG, bp 1001). The amino acid numbers refer to the c-Eyk sequence (accession no. 438523 [37]). The construct pMEX/CD8–v-EykΔ890 was obtained by chance in an attempt to construct pMEX/CD8–v-EykY862F but with an additional thymidine introduced in the tyrosine codon (TAC-TTTC). This construct gave rise to a shortened Eyk protein which carried a Y862F frameshift followed by 28 random non-Eyk-related amino acids and a stop codon after amino acid 890. All mutations mentioned above were confirmed by sequencing of the constructs (Rockefeller University DNA Technology Center).
Oligonucleotides used for cloning or electrophoretic mobility shift assay (EMSA) were custom made by The Rockefeller University DNA Technology Center or by Genelink. The oligonucleotide M67 (upper sequence, GATCGGTTCCCGTAAAT; underline denotes STAT-binding site [37]) used in EMSAs was 32P labeled as described elsewhere (72).
Cell transfection and in vitro assays.
3Y1 fibroblasts, 293T cells, and transformed derivatives were transfected by using Lipofectamine as described by the manufacturer. The cells were either harvested for preparation of whole-cell extracts (extraction buffer consisted of 50 mM HEPES · NaOH [pH 7.4], 150 mM NaCl, 1% Triton X-100, 10% glycerol, 10 mM EDTA, 1 mM Na3VO4, 0.1 mM Na2MoO4, 1 mM phenylmethylsulfonyl fluoride, and 0.1 U of aprotinin per ml; ERK assay, Western blotting, and immunoprecipitation), cytoplasmic and nuclear extracts (74) (EMSA and related Western blot analyses), or luciferase extracts (3) or subjected to transformation assays 24 h later.
EMSA was carried out with 2 μg of nuclear extract from nontransfected (Fig. 1) or transfected (Fig. 3 to 5) cells as described previously (74), using 32P-labeled oligonucleotide M67. Western blotting of the resultant cytoplasmic extracts (20 μg) as a measure of expression of certain proteins was carried out in parallel according to standard methods (58) and instructions provided by the manufacturer of the immunodetection system.
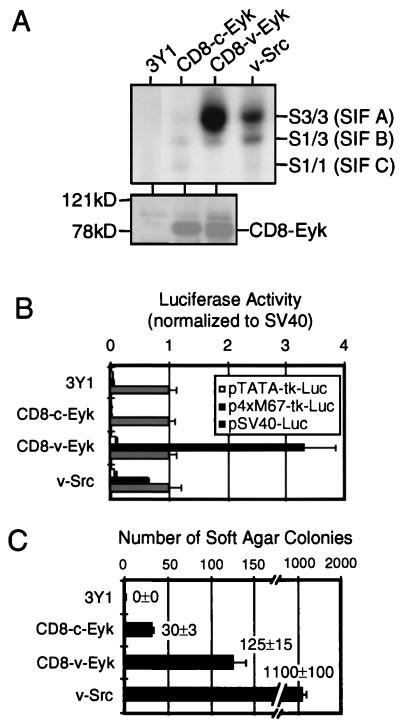
FIG. 1.
v-Eyk-transformed cells show higher binding activity primarily to Stat3 and have higher STAT-specific transcription compared to nontransformed cells. (A) Nuclear extracts (2 μg) from 3Y1 fibroblasts (3Y1) or 3Y1 cells stably transformed with CD8–c-Eyk, CD8–v-Eyk, or v-Src (0.32 × 106 cells each) were used in an EMSA with 2 ng of 32P-labeled STAT-specific oligonucleotide M67 (upper panel). The positions of oligonucleotides shifted by a homodimer of Stat3 [S3/3 factor (SIF A)], by a heterodimer of Stat3 and Stat1 [S1/3 (SIF B)], or by a homodimer of Stat1 [S1/1 (SIF C)] are indicated. Eyk expression levels were detected in cytoplasmic extracts from these cells by Western blotting (lower panel). The positions of CD8-Eyk molecules and of the molecular weight standards are indicated. (B) Cell lines (0.06 × 106 cells each) described above were transfected with 100 ng of luciferase expression vector pTATA-tk-Luc, p4xM67-tk-Luc (STAT-responsive promoter), or pSV40-Luc. Luciferase activity in 10 μg of extract was determined as described in Materials and Methods. The values were normalized with respect to that of pSV40-Luc in each cell line. Each column represents the average of duplicate samples, with the positive SD indicated as an error bar (n = 2). (C) Cells (104) from the cell lines used for panel A were plated in soft agar, and colonies were counted after 2 weeks. Each column represents the average of two samples, with the positive SD indicated as an error bar (n = 2). The original numbers are indicated beside the columns.
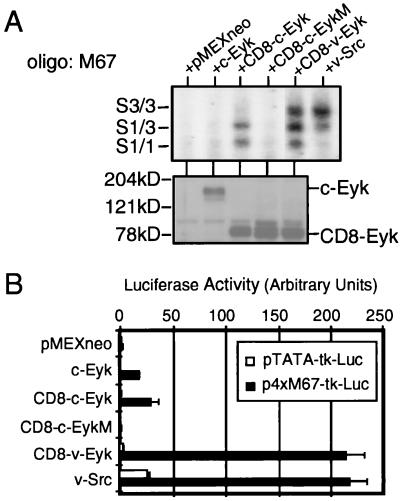
FIG. 3.
Constitutively active Eyk constructs induce Stat3 DNA binding and STAT-specific transcriptional activation to different levels. (A) 3Y1 cells were transfected without or with 2 μg of plasmids expressing the indicated proteins. Two days after transfection, the cells were harvested and nuclear extracts were prepared. DNA binding activity (upper panel) and Eyk expression levels (lower panel) were determined as described for Fig. 1A. Western blotting was performed with an Eyk-specific antibody. Oligo, oligonucleotide. (B) 3Y1 cells were transfected with 100 ng of indicated luciferase reporter construct alone or together with 50 ng of the indicated construct. Luciferase activity was determined as described for Fig. 1B. Each column represents the average of two samples, with the positive SD indicated as an error bar (n = 2).
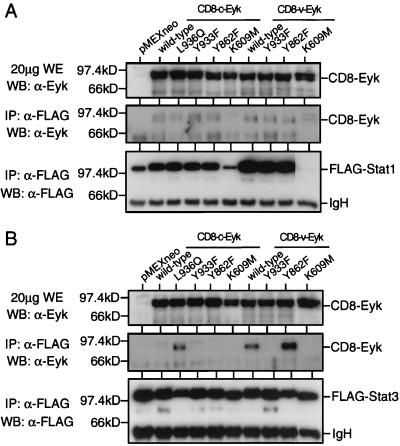
FIG. 5.
The YVPQ sequence in v-Eyk is responsible for increased Stat3 but not Stat1 binding of Eyk. (A) 293T cells were transfected with 1 μg of pRcCMV/stat1-FLAG together with 1 μg of the indicated construct. Two days after transfection, the cells were harvested and whole-cell extracts were prepared. FLAG-tagged Stat1 was immunoprecipitated (IP) from 400 μg of extract, using the FLAG-specific antibody M2 (α-FLAG). Whole-cell extracts (WE; upper panel) and immunoprecipitates (middle and lower panels) were analyzed by Western blotting (WB) for expressed or coprecipitated Eyk levels (upper and middle panel) or FLAG-tagged Stat1 levels as described for Fig. 1A. The positions of FLAG-tagged Stat1 (Stat1) and of the immunoglobulin heavy chain (IgH) are indicated. (B) Same as panel A but with FLAG-tagged Stat3 (pRcCMV/stat3-FLAG).
Luciferase reporter assays with 10 μg of extract were performed as described elsewhere (3). Results were plotted as means ± standard deviations (SD) of experiments performed in duplicate.
Immunoprecipitations were done with various amounts of whole-cell extracts with the different antibodies. Briefly, for each precipitation, 1 μg of the antibody was incubated with 20 μl of GammaBind G-Sepharose on a rocker table at 4°C for 2 h. The antibody fraction bound to Sepharose was transferred to 200 μg of whole-cell extracts (adjusted to 1 mg of total protein/ml) and incubated overnight as described above. The next day, the Sepharose beads were washed four times with extraction buffer and the bound proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting as described above.
Whole-cell extracts (100 μg/100 μl) from cells transfected with pEBG or pEBG/erk1 (expressing GST-ERK1) together with indicated constructs were precipitated with 20 μl of glutathione-beads for 1 h. The beads were washed once with extraction buffer and three times with kinase buffer (30 mM HEPES · NaOH [pH 7.4], 20 mM MgCl2, 2 mM MnCl2, 1 mM dithiothreitol). The kinase reaction was carried out for 30 min at 30°C in 20 μl of kinase buffer supplemented with 5 μg of myelin basic protein (MBP), 2 μCi of [γ-32P]ATP (3,000 Ci/mmol), and 10 μM ATP. The kinase reaction was subjected to SDS-PAGE (8% gel) and electroblotted to polyvinylidene difluoride membranes (Millipore), and the 32P-labeled MBP was detected by autoradiography.
Transformation assay.
Cells transfected with the various constructs were trypsinized, diluted in 7.5 ml of top agar (0.4% Bacto Agar in DMEM–10% FBS), and spread onto 60-mm-diameter plates containing 5 ml of 0.75% bottom agar (0.75% Bacto Agar in DMEM–10% FBS). The number of colonies of cells which were able to grow in the top agar was determined 2 to 3 weeks after plating.
RESULTS
Stable v-Eyk-transformed cells show increased Stat3 DNA binding and STAT-specific transcriptional activation.
Constitutively active forms of c-Eyk and v-Eyk activated by dimerization through linking the CD8α extracellular domain to the c-Eyk or v-Eyk internal domain are able to transform immortalized 3Y1 fibroblasts in culture. Furthermore, these dimeric receptor molecules when expressed transiently in COS cells are tyrosine phosphorylated and able to activate Stat1, Stat3, and Jak1 (74).
Full transformation of cells by v-Src is known to require activation of Stat3 (7, 66). We first compared 3Y1 cells transformed by CD8–c-Eyk and CD8–v-Eyk with v-Src-transformed cells for constitutive activation of STATs by examining Stat1/3-specific DNA binding (EMSA) in nuclear extracts. We used a probe (M67 [33]) that is known to bind both Stat1 homodimers (originally called sis-inducible factor C [SIF C] [69]) and Stat3 homodimers (SIF A) as well as the Stat1/3 heterodimers (SIF B). As found previously for NIH 3T3 fibroblasts (73), v-Src-transformed 3Y1 cells constitutively activated Stat3 as well as a lesser amount of the Stat1/3 heterodimer (Fig. 1A, upper panel). CD8–v-Eyk-transformed cells had very strong Stat3 activation, a small amount of the heterodimer, and no detectable activation of Stat1. In CD8–c-Eyk transformed cells, small amounts of all three dimers were observed. The same results were obtained in a number of different transformed 3Y1 cell clones. The nature of these complexes has been confirmed by supershift analyses using Stat1- or Stat3-specific antibodies (data not shown).
The transformed cells were examined for transcriptional activation of a reporter gene construct bearing four copies of the M67 site that is responsive to Stat1 or Stat3 activation. The CD8–v-Eyk-transformed cell line showed a significantly higher activity of the Stat1/3-responsive promoter than the v-Src-transformed cell line, correlating with the large amount of activated Stat3 in these cells. Nontransformed and CD8–c-Eyk-transformed 3Y1 cell lines did not show a Stat1/3-specific transcriptional response (Fig. 1B).
As a measure of the transformed phenotype, equal numbers of cells (104) were plated for colony formation in soft agar. v-Src-transformed cells formed 10 times as many colonies as cells transformed by CD8–v-Eyk. However, CD8–c-Eyk-transformed cells formed only about one-fourth as many colonies as CD8–v-Eyk transformed cells (Fig. 1C).
Expression of different Eyk constructs induces transformation and increased tyrosine phosphorylation but no activation of ERK1.
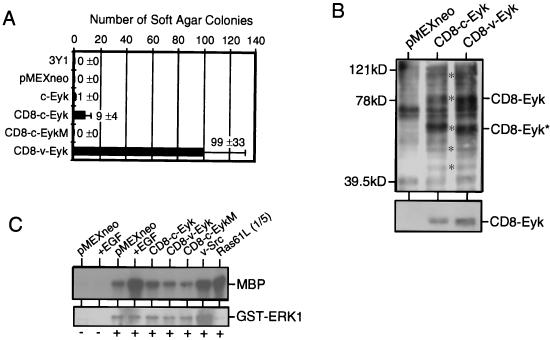
To further analyze the ability of CD8-Eyk molecules to induce transformation and to score their relative transformation efficiencies, we transfected a series of constructs into 3Y1 cells and after 3 weeks determined the number of transformed cells leading to colonies in soft agar were (Fig. 2A). The transformation assay clearly showed greater transforming ability by CD8–v-Eyk than by CD8–c-Eyk (99 ± 33 and 9 ± 4, respectively. A kinase-dead CD8–c-Eyk construct (CD8–c-EykM) included as a negative control did not lead to the formation of soft agar colonies.
FIG. 2.
Transformation efficiency and activation of different signaling events in 3Y1 cells transiently expressing different Eyk constructs. (A) 3Y1 cells (0.32 × 106) were transfected with 2 μg of the indicated constructs and 2 days later transferred to soft agar; colony numbers were determined 3 weeks later. Each column represents the average of two samples, with the positive SD indicated as an error bar (n = 2). The original numbers are indicated beside the columns. (B) 3Y1 fibroblasts were transfected with 15 μg of pMEX/neo, pMEX/CD8–c-Eyk (CD8–c-Eyk), or pMEX/CD8–v-Eyk (CD8–v-Eyk). Two days later, whole extracts were prepared and tyrosine-phosphorylated proteins (asterisks in the upper panel) and Eyk protein levels (lower panel) were detected in 20 μg of whole extract by Western blotting using a phosphotyrosine-specific antibody or an Eyk-specific antibody, respectively. (C) 3Y1 fibroblasts (0.15 × 106 cells) were transfected with 100 ng of pEBG (−) or pEBG/erk1 (+) together with 500 ng of plasmid expressing the indicated protein. Two days later, the cells were either treated with 50 ng of EGF per ml (+EGF) for 10 min or left untreated, and whole-cell extracts were prepared. GST-ERK1 was precipitated by using glutathione-beads, and kinase activity toward MBP was determined (upper panel). GST-ERK1 was detected with a GST-specific antibody (Santa Cruz) (lower panel). In the rightmost lane, one-fifth of the kinase reaction for Ras61L was loaded to avoid overexposure.
To determine why CD8–c-Eyk and CD8–v-Eyk differ in the ability to induce transformation, we studied the signaling events mediated by these two constructs in transient transfection experiments. The total tyrosine-phosphorylated proteins in CD8–c-Eyk- and CD8–v-Eyk-transfected 3Y1 cells were comparable (Fig. 2B, upper panel). The asterisks in Fig. 2B indicate phosphorylated proteins, including the expressed CD8-Eyk molecules and their degradation products and three unidentified phosphoproteins.
Activation of the Ras/ERK pathway has been shown for other members of the Axl/Tyro3 subfamily (22, 41). Results of an ERK1 assay using cells cotransfected with a construct expressing GST-ERK1 and several constructs expressing CD8-Eyk proteins, v-Src, or Ras61L is shown in Fig. 2C. Under the conditions used here, the level of ERK1 activation was not altered by CD8–c-Eyk, CD8–v-Eyk, or the kinase-dead mutant CD8–c-EykM compared to the ERK1 activity level observed in noninduced cells transfected with pMEXneo. However, ERK1 is activated in cells transfected with v-Src, Ras61L, or pMEXneo (treated with EGF). The observed activation of ERK1 by v-Src seems to be due to an increased expression level of ERK1 rather than increased activation based on the expression levels of GST-ERK1 (Fig. 2C, lower panel). Furthermore, we found no differences in ERK1/2 activation between the nontransformed and CD8-Eyk-transformed 3Y1 cell clones (data not shown).
These results together with the finding of similar levels of kinase activity of the two CD8-Eyk proteins (data not shown) indicated that the differences in transformation efficiency between CD8–c-Eyk and CD8–v-Eyk were not due to their overall ability to phosphorylate cellular proteins, their kinase activity, or their capacity to induce ERK.
Transient expression of Eyk proteins leads to differential DNA binding of Stat3 and Stat1, resulting in activation of a Stat1/3-responsive promoter construct.
Since stably transformed CD8–v-Eyk cells exhibited constitutive activation of Stat3, we determined the effects of transient transfection of CD8–c-Eyk and CD8–v-Eyk on Stat1/3-specific DNA binding and Stat1/3-specific transcriptional activation (Fig. 3). As previously shown in studies using COS cells (74), CD8–c-Eyk activates Stat1 and some Stat1/3 heterodimer, indicating that Stat3 is activated but to a lesser extent than Stat1. CD8–v-Eyk led to similar activation of Stat1 but much stronger activation of Stat3 compared to CD8–c-Eyk. A mutation in the kinase domain of Eyk in the construct CD8–c-EykM obliterated the activation of Stat1 and Stat3. As shown for v-Src-transformed 3Y1 cells (Fig. 1A, upper panel), v-Src activates Stat3 preferentially in 3Y1 cells when transiently expressed (Fig. 3A). The nature of the complexes observed in the EMSA has been confirmed by supershift analyses using Stat1- and Stat3-specific antibodies (data not shown).
Consistent with the greater activation of Stat3 by CD8–v-Eyk and v-Src than by CD8–c-Eyk, we observed a greater induction of the Stat1/3-responsive promoter construct when it was coexpressed with these molecules: about 200-fold induction by CD8–v-Eyk and v-Src but only 35-fold induction by CD8–c-Eyk (Fig. 3B).
A YVPQ sequence in the COOH terminus of CD8–v-Eyk is responsible for increased Stat3 activation and increased transformation efficiency.
The coding sequences of c-Eyk and v-Eyk differ in several regions (37). CD8–c-Eyk contains the transmembrane domain of c-Eyk including 21 intracellular amino acids, whereas CD8–v-Eyk contains the transmembrane domain of CD8α but not the first 21 intracellular amino acids of c-Eyk. This difference accounts for the slightly smaller molecular weight of CD8–v-Eyk than of CD8–c-Eyk observed in the Western blot assays using the Eyk antibody. The sequence of the remaining intracellular domains are identical between CD8–c-Eyk and CD8–v-Eyk with the exception of two point mutations. Methionine 631 in the kinase domain of c-Eyk corresponds to an isoleucine in v-Eyk, and leucine 936 in the COOH terminus of c-Eyk corresponds to glutamine in v-Eyk.
We first determined that the transmembrane domain of CD8α and the 21 intracellular amino acids of c-Eyk are not responsible for the differences in Stat3 activation and transformation (data not shown). We then tested whether the exchange of the COOH termini between CD8–c-Eyk and CD8–v-Eyk influenced the activation of Stat3 and the transformation efficiency. Interestingly, the COOH terminus of CD8–v-Eyk (amino acids 918 to 974) when linked to the NH2 terminus CD8–c-Eyk could confer Stat3 activation and high transformation capability (data not shown), suggesting that the mutation L936Q is responsible for these abilities. Amino acid 936 is juxtaposed to a potentially phosphorylated tyrosine residue at position 933, corresponding to the sequences Y933VPL in CD8–c-Eyk and Y933VPQ in CD8–v-Eyk.
To test the hypothesis that Y933VPQ in CD8–v-Eyk is responsible for Stat3 activation and higher transformation efficiency, we performed site-directed mutagenesis to obtain mutated CD8-Eyk molecules (Fig. 4A). CD8–c-EykL936Q showed strong activation of Stat3 as tested by DNA binding (Fig. 4B), STAT-responsive transcription (Fig. 4C), and increased number of soft agar colonies (Fig. 4D). CD8–v-EykY933F lost the ability to activate Stat3 and showed transformation efficiency as low as that observed for CD8–c-Eyk. As mentioned above, CD8–c-Eyk can activate Stat1 and minimally Stat3 (Fig. 3A), leading to Stat1/3-specific transcriptional activation (Fig. 3B). To further test the importance of tyrosine 933, we also introduced a Y933F mutation in CD8–c-Eyk. The construct CD8–c-EykY933F did not show any difference from the wild-type CD8–c-Eyk, suggesting that Y933 is important for Stat3 activation only in conjunction with a glutamine in the +3 position. As expected from the results with CD8–c-EykM described above, two kinase-dead constructs, CD8–c-EykK609M and CD8–v-EykK609M, neither activated the STAT pathway nor led to transformation of 3Y1 fibroblasts.
FIG. 4.
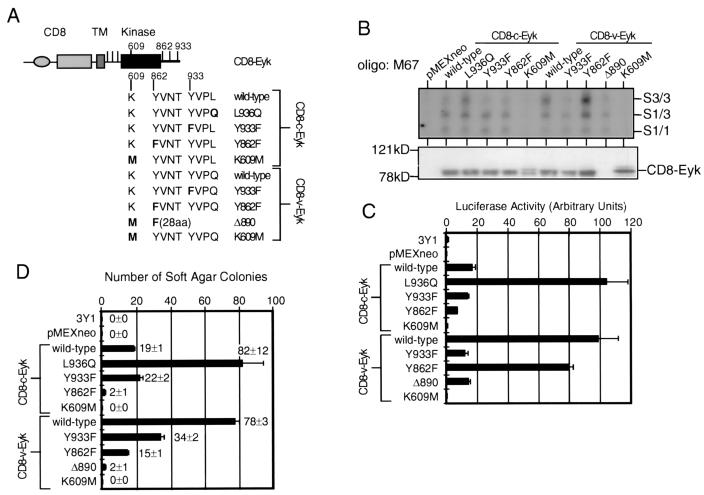
A YVPQ sequence in v-Eyk is responsible for increased DNA binding of Stat3 and transcriptional activation; tyrosine 862 is not responsible for Stat3 activation; Stat3 activation by CD8–v-Eyk and signaling events downstream of tyrosine 862 cooperate in transformation efficiency. (A) Graphic representation of the constructs used in for panels B to D. The CD8α extracellular domain (CD8), the transmembrane domain (TM), and the intracellular domain including the kinase domain (kinase) and tyrosine residues (vertical bars) are shown. Point mutations in four different sites are indicated in the context of wild-type CD8–c-Eyk and CD8–v-Eyk: leucine in CD8–c-Eyk 936 to glutamine and tyrosine 933 to phenylalanine, tyrosine 862 to phenylalanine, and lysine 609 to methionine (kinase dead) in both CD8-Eyk constructs. Δ890 denotes a COOH-terminal deletion construct of CD8–v-Eyk displaying the Y862F mutation followed by 28 unrelated amino acids (aa) and a stop codon. (B) 3Y1 cells were transfected with 2 μg of the indicated construct. Two days after transfection, the cells were harvested and nuclear and cytoplasmic extracts were prepared. EMSA using oligonucleotide M67 as the probe and Western blot analysis for Eyk expression were carried out as described for Fig. 1A. (C) 3Y1 cells were transfected with 100 ng of p4xM67-tk-Luc together without or with 50 ng of the indicated construct. Luciferase activity was determined 2 days later as described for Fig. 1B. (D) 3Y1 cells were transfected with 2 μg of the indicated construct. The transformation efficiency was determined in a soft agar assay as described for Fig. 3D.
As seen in Fig. 4B, the activation of Stat1 remained unchanged in all tested constructs excluding the kinase-dead molecules, suggesting that another region in the CD8-Eyk molecules is responsible for the activation of Stat1 and the residual activation of Stat3. In an attempt to identify this site(s), we mutated the other five tyrosines outside the kinase domain to phenylalanines in both CD8–c-Eyk and CD8–v-Eyk. All of these constructs, each containing a single amino acid substitution, behaved like their wild-type nonmutated counterparts, indicating that none of these tyrosines is required alone for the activation of Stat1 and the residual activation of Stat3 (data not shown). The mutant construct CD8–v-EykΔ890 contains a Y862F mutation, followed by 28 nonrelated amino acids and a stop codon at amino acid 890. Interestingly, this construct could still activate Stat1 and Stat1/3-responsive transcription to the same level as CD8–c-Eyk level, although the COOH terminus of CD8–v-Eyk including Y933 responsible for high Stat3 activation is missing (Fig. 4B and C). The antibody against the COOH terminus of Eyk used in Western blot analyses (Fig. 4B, lower panel) did not recognize this construct. These results indicated that the site(s) responsible for the activation of Stat1 and the residual Stat3 activation is not located in the COOH terminus and is either a tyrosine in the kinase domain or a non-tyrosine-binding site.
A YVNT sequence is required for transformation.
Two of the ten Y-to-F mutants that were generated in the CD8–c-Eyk and the CD8–v-Eyk backgrounds to identify the site responsible for Stat1 activation (see above), namely, CD8–c-EykY862F and CD8–v-EykY862F, showed interesting phenotypes. Both constructs were able to activate the STAT pathway similarly to their wild-type counterparts (Fig. 4B and C) but showed a greatly reduced ability to transform 3Y1 fibroblasts (Fig. 4D). These results indicated that activation of Stat3 by CD8–v-Eyk cooperates with another independent signaling pathway to transform 3Y1 cells in culture. Note that all transfectants analyzed in soft agar assays were also grown on plates containing selective medium. G418-resistant clone numbers were comparable between wild-type and mutant receptor constructs, indicating that there is no difference in the apoptosis in cells bearing the various receptors (data not shown).
The YVPQ sequence is required for binding of Stat3 but not of Stat1.
The sequence Y933VPQ in CD8–v-Eyk fits the determined consensus binding sequence YXPQ in the gp130 receptor for the Stat3 SH2 domain (32). Therefore, we determined whether Stat3 interacts with CD8-Eyk molecules harboring the Y933VPQ sequence.
Different mutants of CD8–c-Eyk and CD8–v-Eyk in conjunction with FLAG-tagged Stat1 (Fig. 5A) or Stat3 (Fig. 5B) were transiently expressed in 293T cells. To detect an interaction between CD8-Eyk molecules and the various STAT molecules, we immunoprecipitated the STAT molecules with the FLAG epitope-specific antibody M2 and then determined the levels of CD8-Eyk and Stat1/3 molecules in the immunoprecipitates (Fig. 5). Expression of CD8-Eyk molecules was determined in whole-cell extracts from the transfected cells (Fig. 5A and B, upper panels). All CD8-Eyk molecules including the kinase-dead mutants CD8–c-EykK609M and CD8–v-EykK609M were able to interact with Stat1 (Fig. 5A). It is noteworthy that the expression level of the Stat1 molecules is influenced by the coexpression of kinase-active CD8-Eyk molecules.
In a similar experiment using FLAG-tagged Stat3 (Fig. 5B), interaction between CD8-Eyk and Stat3 was detected only when the CD8-Eyk molecule contained tyrosine 933 and glutamine 936, i.e., in the constructs CD8–c-EykL936Q, wild-type CD8–v-Eyk, and CD8–v-EykY862F. These results show that Stat3 could specifically interact with CD8-Eyk molecules harboring the Y933VPQ sequence found originally in CD8–v-Eyk. Stat1 interacted with all CD8-Eyk constructs most likely independent of tyrosine phosphorylation. Thus, the interaction of Stat3 and Stat1 with different CD8-Eyk molecules correlated with the activation of the STAT pathway (Fig. 4B and C). Eyk molecules could not be detected when FLAG-tagged Stat3 was coexpressed with Eyk molecules which do not activate Stat3 (all CD8–c-Eyk constructs except CD8–c-EykL936Q, CD8–v-EykY933F, and CD8–v-EykK609M), indicating that the FLAG antibody did not precipitate Eyk molecules in Fig. 5 unspecifically. The additional bands observed are either Eyk degradation products (Fig. 5A and B, upper and middle panels), cross-reactivity between the anti-FLAG antibody and the horseradish peroxidase-conjugated secondary anti-rabbit antibody (Fig. 5A and B, middle panels), or Stat1 (Fig. 5A, lower panel) or Stat3 (Fig. 5B, lower panel) degradation products. None of these degradation products seem to be significant for the results presented in this study.
DISCUSSION
We have analyzed the signaling pathways downstream of activated Eyk molecules leading to transformation of 3Y1 fibroblasts and demonstrated the importance of Stat3 activation. Although Stat1 is activated by both Eyk molecules, this activation did not correlate with cellular transformation or with transcriptional activation.
The activation of Stat3 by the RTK CD8–v-Eyk presumably occurs directly after binding of Stat3 via its SH2 domain to a Y933VPQ sequence present in v-Eyk and by the subsequent tyrosine phosphorylation of Stat3 (Fig. 4 and 6). Similar sequences (YXXQ) have been shown to be important for Stat3 activation by Jak kinases in several cytokine receptors, e.g., the IL-10 receptor (71), the granulocyte colony-stimulating factor receptor (17), and gp130, the receptor activated after IL-6 treatment (32, 62). Unlike the cytokine receptors, the Stat3 docking sites for growth factor (e.g., EGF and platelet-derived growth factor) receptors which activate Stat3 had not been clearly defined. Stat3 activation by the hepatocyte growth factor/scatter factor receptor c-Met has been shown to be involved in epithelial tubule formation. A putative sequence for Stat3 binding in c-Met was suggested to be Y1356VNV, which also binds several other proteins (5). Interestingly, a similar sequence, Y862VNT, is present in Eyk, but appears not to be responsible for Stat3 activation although it is required for transformation (see below). c-Met also contains a YXXQ sequence, which raises the question of whether a synergistic effect similar to that described here for the transformation by CD8–v-Eyk is responsible for epithelial tubule formation by c-Met. It is noteworthy that v-Src does not contain a YXXQ sequence, suggesting that the activation of Stat3 by v-Src could involve another binding site for Stat3 (11).
FIG. 6.
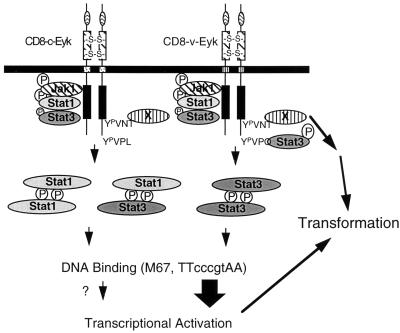
A model for signaling and transformation events downstream of CD8-Eyk. The constitutively dimerized and activated Eyk constructs activate Jak1 (74) and Stat1 and slightly activate Stat3 via an unidentified region NH2 terminal to tyrosine 862 (Fig. 4, Δ890 construct). The Stat3-binding site Y933VPQ is present only in CD8–v-Eyk and leads to increased Stat3 DNA-binding activity, 10-fold-higher activation of STAT-responsive transcription, and a 10-fold-higher number of soft agar colonies, indicating that Stat3 enhances transformation. The influence of the Stat1 homodimer and the Stat1-Stat3 heterodimer on transcriptional activation is not clear. Another protein (X) that binds to Y862VNT is activated downstream of CD8–c-Eyk and CD8–v-Eyk and leads to activation of other signaling pathways independent of Stat1 or Stat3. The activation of this pathway is required for transformation by both CD8–c-Eyk and CD8–v-Eyk.
The observed activation of Stat1 by the CD8-Eyk molecules (Fig. 3) does not require the Y933VPQ sequence; therefore, Stat1 must bind to some other sites in both c-Eyk and v-Eyk (Fig. 5). We do not know whether Stat1 activation can have a negative effect on transformation efficiency, since Stat1 activation has been previously implicated in growth arrest of cells (8, 12). In fact, comparing the transformation efficiencies of CD8–v-Eyk and v-Src (data not shown and Fig. 1), we found a significantly higher number of colonies with v-Src-expressing cells. While v-Src and CD8–v-Eyk are comparable in the ability to activate Stat3, CD8–v-Eyk also activates Stat1. Thus, it is conceivable that the activation of Stat1 by CD8–v-Eyk can decrease transformation by v-Eyk or that additional signaling pathways involved in transformation are activated by v-Src but not by CD8–v-Eyk.
Wild-type and dominant negative Stat3 molecules have been shown to be efficiently block transformation by v-Src in NIH 3T3 cells (7, 66). We tried to examine the effect of these molecules and the associated Stat1 molecules on the transformation of 3Y1 cells. Coexpression of CD8-Eyk molecules with wild-type Stat1 or Stat3 greatly increased DNA-binding activity, and coexpression of CD8-Eyk with Stat3 led to increased Stat1/3-specific transcriptional activation (data not shown). Nevertheless, neither Stat1Y-F nor Stat3Y-F molecules (7) had a dominant negative influence on the DNA binding of the endogenous STAT molecules, or transcriptional activation, or on transformation by the CD8-Eyk and the v-Src molecules. All STAT molecules in these assays were expressed to detectable levels. We do not know why these dominant negative molecules are not functional under the conditions used here.
We previously reported that Stat1β acted as a suppressor of Eyk transformation in 3Y1 cells. Stat1 and Stat1β can form heterodimers with Stat3, and eventually the dominant negative construct Stat1β blocks the activation of Stat1 and Stat3 to the same extent and leads to a reduced number of soft agar colonies because of the blockage of Stat3.
The activation of Jak1 has been shown to be important for the activation of STAT molecules. We reported previously that CD8-Eyk molecules can activate Jak1 (74). The function of this activation for the activation of Stat3 and the transformation remains to be elucidated. Interestingly, Jak1 is also activated in v-Src-transformed fibroblasts, although the importance of this activation for transformation has not been investigated (10).
We present evidence that maximal transformation frequency by CD8–v-Eyk not only is dependent on Stat3 activation but also requires the activation of another pathway activated through the Y862VNT sequence that is present in both CD8-Eyk proteins.
The amino acid sequence following the tyrosine at position 862 is VNT. The asparagine at +2 is highly conserved in the homologous sequence among all members of the Axl/Tyro3 subfamily. A YXNX motif has been reported to be a docking site for the SH2 domain of Grb2 (61). Grb2 as well as other molecules can bind to this motif in Axl (6). Since docking of Grb2 to receptors leads to the activation of the Ras/ERK pathway (42), one might expect activation of this pathway downstream of CD8-Eyk via the Y862VNT sequence. Several reports demonstrated the activation of ERK downstream of other members of the Axl subfamily in 32D cells and in NIH 3T3 fibroblasts (22, 41). The involvement of the activation of the Ras/ERK pathway for the physiological function of these receptors has not been addressed. Although we did not observe activation of ERK1 in 3Y1 cells (Fig. 2C), we found that in NIH 3T3 cells, CD8–c-Eyk and CD8–v-Eyk are able to modestly activate GST-ERK1, suggesting cell line specific regulation of this pathway (data not shown). However, increased ERK activity does not appear to occur in the transformation of 3Y1 cells by CD8-Eyk. A homologous tyrosine in c-Axl (Y821VNM) seems to be involved in the activation of phosphatidylinositol (PI) 3-kinase, S6 kinase and protein kinase B (PKB)/Akt (6, 29), and we reported that the homologous tyrosine in the c-Mer receptor, the closest mammalian homologue of c-Eyk, leads to activation of NF-κB (27). A very recent study shows that this sequence in c-Axl is required for transformation (9). We are investigating the potential role of these molecules in the signaling events downstream of Y862 in CD8-Eyk.
In conclusion, we propose the following model for the signaling events downstream of CD8–c-Eyk and CD8–v-Eyk leading to the transformation of immortalized 3Y1 fibroblasts (Fig. 6). Both molecules are able to bind and activate Jak1 (74), Stat1, and minimally Stat3 via a region residing in the kinase domain or the intracellular region N terminal to the kinase domain. The major difference between the cellular form CD8–c-Eyk and the viral form CD–v-Eyk is the strong binding of Stat3 to CD8–v-Eyk via a YVPQ sequence (Fig. 5), DNA binding, and transcriptional activation (Fig. 3). A second pathway which appeared to be important for the transformation events by both CD8-Eyk molecules involves Y862VNT. This activation seems to be responsible for the transformation events downstream of CD8–c-Eyk since a construct missing this tyrosine is virtually unable to induce transformation. In CD8–v-Eyk, this pathway synergizes with the activation of Stat3, resulting in transformation efficiency 10-fold higher than that of CD8–c-Eyk.
Our model illustrates how two independent signaling pathways cooperate in inducing transformation in 3Y1 fibroblasts. We speculate that the mutation of Y933VPL to Y933VPQ upon retroviral transduction of the RPL30 virus, harboring v-Eyk, gained a biological advantage by leading to strong activation of Stat3.
ACKNOWLEDGMENTS
We thank Inés Ibañez Tallon and Raymond Birge for discussion and critical reading of the manuscript, Sheila Harroch and all members of the Hanafusa laboratory for helpful advice, and those mentioned in the text for expression plasmids.
This work was supported by Counsel for Tobacco Research grant 4438R1 (to H.H.) and NIH grants CA44356 (to H.H.) and AI32489 and AI34420 (to J.E.D.). D.B. was supported by a fellowship from the Swiss National Foundation, and J.F.B. was supported by a Howard Hughes postdoctoral fellowship for physicians and NIH K08 grant CA67950.
REFERENCES
- 1.Bellosta P, Costa M, Lin D A, Basilico C. The receptor tyrosine kinase ARK mediates cell aggregation by homophilic binding. Mol Cell Biol. 1995;15:614–625. doi: 10.1128/mcb.15.2.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellosta P, Zhang Q, Goff S P, Basilico C. Signaling through the ARK tyrosine kinase receptor protects from apoptosis in the absence of growth stimulation. Oncogene. 1997;15:2387–2397. doi: 10.1038/sj.onc.1201419. [DOI] [PubMed] [Google Scholar]
- 3.Besser D, Presta M, Nagamine Y. Elucidation of a signaling pathway induced by FGF-2 leading to uPA gene expression in NIH 3T3 fibroblasts. Cell Growth Differ. 1995;6:1009–1017. [PubMed] [Google Scholar]
- 4.Biscardi J S, Denhez F, Buehler G F, Chesnutt D A, Baragona S C, O’Bryan J P, Der C J, Fiordalisi J J, Fults D W, Maness P F. Rek, a gene expressed in retina and brain, encodes a receptor tyrosine kinase of the Axl/Tyro3 family. J Biol Chem. 1996;271:29049–29059. doi: 10.1074/jbc.271.46.29049. [DOI] [PubMed] [Google Scholar]
- 5.Boccaccio C, Ando M, Tamagnone L, Bardelli A, Michieli P, Battistini C, Comoglio P M. Induction of epithelial tubules by growth factor HGF depends on the STAT pathway. Nature. 1998;391:285–288. doi: 10.1038/34657. [DOI] [PubMed] [Google Scholar]
- 6.Braunger J, Schleithoff L, Schulz A S, Kessler H, Lammers R, Ullrich A, Bartram C R, Janssen J W. Intracellular signaling of the Ufo/Axl receptor tyrosine kinase is mediated mainly by a multi-substrate docking-site. Oncogene. 1997;14:2619–2631. doi: 10.1038/sj.onc.1201123. [DOI] [PubMed] [Google Scholar]
- 7.Bromberg J F, Horvath C M, Besser D, Lathem W W, Darnell J E., Jr Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol. 1998;18:2553–2558. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bromberg J F, Horvath C M, Wen Z, Schreiber R D, Darnell J E., Jr Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc Natl Acad Sci USA. 1996;93:7673–7678. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burchert A, Attar E C, McCloskey P, Fridell Y W, Liu E T. Determinants for transformation induced by the Axl receptor tyrosine kinase. Oncogene. 1998;16:3177–3187. doi: 10.1038/sj.onc.1201865. [DOI] [PubMed] [Google Scholar]
- 10.Campbell G S, Yu C L, Jove R, Carter-Su C. Constitutive activation of JAK1 in Src-transformed cells. J Biol Chem. 1997;272:2591–2594. doi: 10.1074/jbc.272.5.2591. [DOI] [PubMed] [Google Scholar]
- 11.Cao X, Tay A, Guy G R, Tan Y H. Activation and association of Stat3 with Src in v-Src-transformed cell lines. Mol Cell Biol. 1996;16:1595–1603. doi: 10.1128/mcb.16.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chin Y E, Kitagawa M, Su W C, You Z H, Iwamoto Y, Fu X Y. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21 WAF1/CIP1 mediated by STAT1. Science. 1996;272:719–722. doi: 10.1126/science.272.5262.719. [DOI] [PubMed] [Google Scholar]
- 13.Cressman D E, Greenbaum L E, DeAngelis R A, Ciliberto G, Furth E E, Poli V, Taub R. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science. 1996;274:1379–1383. doi: 10.1126/science.274.5291.1379. [DOI] [PubMed] [Google Scholar]
- 14.Dai W, Pan H, Hassanain H, Gupta S L, Murphy M J., Jr Molecular cloning of a novel receptor tyrosine kinase, tif, highly expressed in human ovary and testis. Oncogene. 1994;9:975–979. [PubMed] [Google Scholar]
- 15.Danial N N, Pernis A, Rothman P B. Jak-STAT signaling induced by the v-abl oncogene. Science. 1995;269:1875–1877. doi: 10.1126/science.7569929. [DOI] [PubMed] [Google Scholar]
- 16.Darnell J E., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 17.de Koning J P, Dong F, Smith L, Schelen A M, Barge R M, van der Plas D C, Hoefsloot L H, Lowenberg B, Touw I P. The membrane-distal cytoplasmic region of human granulocyte colony-stimulating factor receptor is required for STAT3 but not STAT1 homodimer formation. Blood. 1996;87:1335–1342. [PubMed] [Google Scholar]
- 18.de Vries-Smits A M M, Burgering B M T, Leevers S J, Marshall C J, Bos J L. Involvement of p21ras in activation of extracellular signal-regulated kinase 2. Nature. 1992;357:602–604. doi: 10.1038/357602a0. [DOI] [PubMed] [Google Scholar]
- 19.Durbin J E, Hackenmiller R, Simon M C, Levy D E. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 20.Faust M, Ebensperger C, Schulz A S, Schleithoff L, Hameister H, Bartram C R, Janssen J W. The murine ufo receptor: molecular cloning, chromosomal localization and in situ expression analysis. Oncogene. 1992;7:1287–1293. [PubMed] [Google Scholar]
- 21.Fredrickson T N, Purchase H G, Burmester B R. Transmission of virus from field cases of avian lymphomatosis. III. Variation in the oncogenic spectra of passaged virus isolates. J Natl Cancer Inst Monogr. 1964;17:1–29. [Google Scholar]
- 22.Fridell Y W, Jin Y, Quilliam L A, Burchert A, McCloskey P, Spizz G, Varnum B, Der C, Liu E T. Differential activation of the Ras/extracellular-signal-regulated protein kinase pathway is responsible for the biological consequences induced by the Axl receptor tyrosine kinase. Mol Cell Biol. 1996;16:135–145. doi: 10.1128/mcb.16.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fridell Y W, Villa J, Jr, Attar E C, Liu E T. GAS6 induces Axl-mediated chemotaxis of vascular smooth muscle cells. J Biol Chem. 1998;273:7123–7126. doi: 10.1074/jbc.273.12.7123. [DOI] [PubMed] [Google Scholar]
- 24.Fujimoto J, Yamamoto T. brt, a mouse gene encoding a novel receptor-type protein-tyrosine kinase, is preferentially expressed in the brain. Oncogene. 1994;9:693–698. [PubMed] [Google Scholar]
- 25.Fukada T, Hibi M, Yamanaka Y, Takahashi-Tezuka M, Fujitani Y, Yamaguchi T, Nakajima K, Hirano T. Two signals are necessary for cell proliferation induced by a cytokine receptor gp130: involvement of STAT3 in anti-apoptosis. Immunity. 1996;5:449–460. doi: 10.1016/s1074-7613(00)80501-4. [DOI] [PubMed] [Google Scholar]
- 26.Garcia R, Yu C L, Hudnall A, Catlett R, Nelson K L, Smithgall T, Fujita D J, Ethier S P, Jove R. Constitutive activation of Stat3 in fibroblasts transformed by diverse oncoproteins and in breast carcinoma cells. Cell Growth Differ. 1997;8:1267–1276. [PubMed] [Google Scholar]
- 27.Georgescu M-M, Kirsch K H, Shishido T, Zong C, Hanafusa H. Biological effects of c-Mer receptor tyrosine kinase in hematopoietic cells depend on the Grb2 binding site in the receptor and activation of NF-κB. Mol Cell Biol. 1999;19:1171–1181. doi: 10.1128/mcb.19.2.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goruppi S, Ruaro E, Schneider C. Gas6, the ligand of Axl tyrosine kinase receptor, has mitogenic and survival activities for serum starved NIH3T3 fibroblasts. Oncogene. 1996;12:471–480. [PubMed] [Google Scholar]
- 29.Goruppi S, Ruaro E, Varnum B, Schneider C. Requirement of phosphatidylinositol 3-kinase-dependent pathway and Src for Gas6-Axl mitogenic and survival activities in NIH 3T3 fibroblasts. Mol Cell Biol. 1997;17:4442–4453. doi: 10.1128/mcb.17.8.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graham D K, Bowman G W, Dawson T L, Stanford W L, Earp H S, Snodgrass H R. Cloning and developmental expression analysis of the murine c-mer tyrosine kinase. Oncogene. 1995;10:2349–2359. [PubMed] [Google Scholar]
- 31.Graham D K, Dawson T L, Mullaney D L, Snodgrass H R, Earp H S. Cloning and mRNA expression analysis of a novel human protooncogene, c-mer. Cell Growth Differ. 1994;5:647–657. [PubMed] [Google Scholar]
- 32.Hemmann U, Gerhartz C, Heesel B, Sasse J, Kurapkat G, Grotzinger J, Wollmer A, Zhong Z, Darnell J E, Jr, Graeve L, Heinrich P C, Horn F. Differential activation of acute phase response factor/Stat3 and Stat1 via the cytoplasmic domain of the interleukin 6 signal transducer gp130. II. Src homology SH2 domains define the specificity of stat factor activation. J Biol Chem. 1996;271:12999–13007. doi: 10.1074/jbc.271.22.12999. [DOI] [PubMed] [Google Scholar]
- 33.Horvath C M, Wen Z, Darnell J E., Jr A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 1995;9:984–994. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
- 34.Ihle, J. N., T. Nosaka, W. Thierfelder, F. W. Quelle, and K. Shimoda. 1997. Jaks and Stats in cytokine signaling. Stem. Cells 15(Suppl. 1):105–112. [DOI] [PubMed]
- 35.Ilaria R L, Jr, Van Etten R A. P210 and P190BCR/ABL induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J Biol Chem. 1996;271:31704–31710. doi: 10.1074/jbc.271.49.31704. [DOI] [PubMed] [Google Scholar]
- 36.Janssen J W, Schulz A S, Steenvoorden A C, Schmidberger M, Strehl S, Ambros P F, Bartram C R. A novel putative tyrosine kinase receptor with oncogenic potential. Oncogene. 1991;6:2113–2120. [PubMed] [Google Scholar]
- 37.Jia R, Hanafusa H. The proto-oncogene of v-eyk (v-ryk) is a novel receptor-type protein tyrosine kinase with extracellular Ig/FN-III domains. J Biol Chem. 1994;269:1839–1844. [PubMed] [Google Scholar]
- 38.Jia R, Mayer B J, Hanafusa T, Hanafusa H. A novel oncogene, v-ryk, encoding a truncated receptor tyrosine kinase is transduced into the RPL30 virus without loss of viral sequences. J Virol. 1992;66:5975–5987. doi: 10.1128/jvi.66.10.5975-5987.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimura G, Itagaki A, Summers J. Rat cell line 3y1 and its virogenic polyoma- and SV40-transformed derivatives. Int J Cancer. 1975;15:694–706. doi: 10.1002/ijc.2910150419. [DOI] [PubMed] [Google Scholar]
- 40.Lai C, Gore M, Lemke G. Structure, expression, and activity of Tyro 3, a neural adhesion-related receptor tyrosine kinase. Oncogene. 1994;9:2567–2578. [PubMed] [Google Scholar]
- 41.Ling L, Kung H J. Mitogenic signals and transforming potential of Nyk, a newly identified neural cell adhesion molecule-related receptor tyrosine kinase. Mol Cell Biol. 1995;15:6582–6592. doi: 10.1128/mcb.15.12.6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lowenstein E J, Daly R J, Batzer A G, Li W, Margolis B, Lammers R, Ullrich A, Skolnik E Y, Bar-Sagi D, Schlessinger J. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell. 1992;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- 43.Mark M R, Scadden D T, Wang Z, Gu Q, Goddard A, Godowski P J. rse, a novel receptor-type tyrosine kinase with homology to Axl/Ufo, is expressed at high levels in the brain. J Biol Chem. 1994;269:10720–10728. [PubMed] [Google Scholar]
- 44.Marksitzer R, Stief A, Menoud P A, Nagamine Y. Role of LFB3 in cell-specific cAMP induction of the urokinase-type plasminogen activator gene. J Biol Chem. 1995;270:21833–21838. doi: 10.1074/jbc.270.37.21833. [DOI] [PubMed] [Google Scholar]
- 45.Matsubara N, Takahashi Y, Nishina Y, Mukouyama Y, Yanagisawa M, Watanabe T, Nakano T, Nomura K, Arita H, Nishimune Y, Obinata M, Matsui Y. A receptor tyrosine kinase, Sky, and its ligand Gas 6 are expressed in gonads and support primordial germ cell growth or survival in culture. Dev Biol. 1996;180:499–510. doi: 10.1006/dbio.1996.0323. [DOI] [PubMed] [Google Scholar]
- 46.Meraz M A, White J M, Sheehan K C, Bach E A, Rodig S J, Dighe A S, Kaplan D H, Riley J K, Greenlund A C, Campbell D, Carver-Moore K, DuBois R N, Clark R, Aguet M, Schreiber R D. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 47.Nagata K, Ohashi K, Nakano T, Arita H, Zong C, Hanafusa H, Mizuno K. Identification of the product of growth arrest-specific gene 6 as a common ligand for Axl, Sky, and Mer receptor tyrosine kinases. J Biol Chem. 1996;271:30022–30027. doi: 10.1074/jbc.271.47.30022. [DOI] [PubMed] [Google Scholar]
- 48.Nelson K L, Rogers J A, Bowman T L, Jove R, Smithgall T E. Activation of STAT3 by the c-Fes protein-tyrosine kinase. J Biol Chem. 1998;273:7072–7077. doi: 10.1074/jbc.273.12.7072. [DOI] [PubMed] [Google Scholar]
- 49.O’Bryan J P, Frye R A, Cogswell P C, Neubauer A, Kitch B, Prokop C, Espinosa R, Le Beau M M, Earp H S, Liu E T. axl, a transforming gene isolated from primary human myeloid leukemia cells, encodes a novel receptor tyrosine kinase. Mol Cell Biol. 1991;11:5016–5031. doi: 10.1128/mcb.11.10.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oelrichs R B, Reid H H, Bernard O, Ziemiecki A, Wilks A F. NYK/FLK-1: a putative receptor protein tyrosine kinase isolated from E10 embryonic neuroepithelium is expressed in endothelial cells of the developing embryo. Oncogene. 1993;8:11–18. [PubMed] [Google Scholar]
- 51.Ohashi K, Mizuno K, Kuma K, Miyata T, Nakamura T. Cloning of the cDNA for a novel receptor tyrosine kinase, Sky, predominantly expressed in brain. Oncogene. 1994;9:699–705. [PubMed] [Google Scholar]
- 52.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rinke de Wit T F, Izon D J, Revilla C, Oosterwegel M, Bakker A Q, van Ewijk W, Kruisbeek A M. Expression of tyrosine kinase gene in mouse thymic stromal cells. Int Immunol. 1996;8:1787–1795. doi: 10.1093/intimm/8.11.1787. [DOI] [PubMed] [Google Scholar]
- 54.Robinson D, He F, Pretlow T, Kung H J. A tyrosine kinase profile of prostate carcinoma. Proc Natl Acad Sci USA. 1996;93:5958–5962. doi: 10.1073/pnas.93.12.5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruff-Jamison S, Chen K, Cohen S. Induction by EGF and interferon-gamma of tyrosine phosphorylated DNA binding proteins in mouse liver nuclei. Science. 1993;261:1733–1736. doi: 10.1126/science.8378774. [DOI] [PubMed] [Google Scholar]
- 56.Ruff-Jamison S, Zhong Z, Wen Z, Chen K, Darnell J E, Jr, Cohen S. Epidermal growth factor and lipopolysaccharide activate Stat3 transcription factor in mouse liver. J Biol Chem. 1994;269:21933–21935. [PubMed] [Google Scholar]
- 57.Sabe H, Okada M, Nakagawa H, Hanafusa H. Activation of c-Src in cells bearing v-Crk and its suppression by Csk. Mol Cell Biol. 1992;12:4706–4713. doi: 10.1128/mcb.12.10.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 59.Schindler C, Darnell J E., Jr Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 60.Seger R, Krebs E G. The MAPK signaling cascade. FASEB J. 1995;9:726–735. [PubMed] [Google Scholar]
- 61.Songyang Z, Shoelson S E, Chaudhuri M, Gish G D, Pawson T, Haser W G, King F, Roberts T M, Ratnofsky S, Lechleider R J, Neel B G, Birge R B, Fajardo J E, Chou M M, Hanafusa H, Schaffhausen B, Cantley L C. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:1–20. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 62.Stahl N, Farruggella T J, Boulton T G, Zhong Z, Darnell J E, Jr, Yancopoulos G D. Choice of STATs and other substrates specified by modular tyrosine-based motifs in cytokine receptors. Science. 1995;267:1349–1353. doi: 10.1126/science.7871433. [DOI] [PubMed] [Google Scholar]
- 63.Stitt T N, Conn G, Gore M, Lai C, Bruno J, Radziejewski C, Mattsson K, Fisher J, Gies D R, Jones P F, Masiakowski P, Ryan T E, Tobkes N J, Chen D H, DiStefano P S, Long G L, Basilico C, Goldfarb M P, Lemke G, Glass D J, Yancopoulos G D. The anticoagulation factor protein S and its relative, Gas6, are ligands for the Tyro3/Axl family of receptor tyrosine kinases. The anticoagulation factor protein S and its relative, Gas6, are ligands for the Tyro 3/Axl family of receptor tyrosine kinases. Cell. 1995;80:661–670. doi: 10.1016/0092-8674(95)90520-0. [DOI] [PubMed] [Google Scholar]
- 64.Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci USA. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tanaka S, Hanafusa H. Guanine-nucleotide exchange protein C3G activates JNK1 by a ras-independent mechanism. JNK1 activation inhibited by kinase negative forms of MLK3 and DLK mixed lineage kinases. J Biol Chem. 1998;273:1281–1284. doi: 10.1074/jbc.273.3.1281. [DOI] [PubMed] [Google Scholar]
- 66.Turkson J, Bowman T, Garcia R, Caldenhoven E, de Groot R P, Jove R. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998;18:2545–2552. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van der Geer P, Hunter T, Lindberg R A. Receptor protein-tyrosine kinase and their signal transduction pathways. Annu Rev Cell Biol. 1994;10:251–337. doi: 10.1146/annurev.cb.10.110194.001343. [DOI] [PubMed] [Google Scholar]
- 68.Varnum B C, Young C, Elliott G, Garcia A, Bartley T D, Fridell Y W, Hunt R W, Trail G, Clogston C, Toso R J. Axl receptor tyrosine kinase stimulated by the vitamin K-dependent protein encoded by growth-arrest-specific gene 6. Nature. 1995;373:623–626. doi: 10.1038/373623a0. [DOI] [PubMed] [Google Scholar]
- 69.Wagner B J, Hayes T E, Hoban C J, Cochran B H. The SIF binding element confers sis/PDGF inducibility onto the c-fos promoter. EMBO J. 1990;9:4477–4484. doi: 10.1002/j.1460-2075.1990.tb07898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang J Y. Isolation of antibodies for phosphotyrosine by immunization with a v-abl oncogene-encoded protein. Mol Cell Biol. 1985;5:3640–3643. doi: 10.1128/mcb.5.12.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weber-Nordt R M, Riley J K, Greenlund A C, Moore K W, Darnell J E, Schreiber R D. Stat3 recruitment by two distinct ligand-induced, tyrosine-phosphorylated docking sites in the interleukin-10 receptor intracellular domain. J Biol Chem. 1996;271:27954–27961. doi: 10.1074/jbc.271.44.27954. [DOI] [PubMed] [Google Scholar]
- 72.Wen Z, Zhong Z, Darnell J E., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 73.Yu C L, Meyer D J, Campbell G S, Larner A C, Carter-Su C, Schwartz J, Jove R. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science. 1995;269:81–83. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 74.Zong C, Yan R, August A, Darnell J E, Jr, Hanafusa H. Unique signal transduction of Eyk: constitutive stimulation of the JAK-STAT pathway by an oncogenic receptor-type tyrosine kinase. EMBO J. 1996;15:4515–4525. [PMC free article] [PubMed] [Google Scholar]