ABSTRACT
The present study was to explore relationship between serum lipid profiles and the polymorphism of rs324420 at fatty acid amide hydrolase (FAAH) gene (FAAH rs324420) and its confounders in Chinese adolescents. Serum lipids, glucose, and insulin levels were assessed using routine methods in a cohort of 708 high school students. Dietary intake was investigated by 3‐day diet records, and intakes of protein, fat, and carbohydrate were calculated. FAAH rs324420 was genotyped by polymerase chain reaction‐restriction fragment length polymorphism technique and confirmed through DNA sequencing. In the whole study population, increased HDL‐C levels were observed in FAAH rs324420 CC homozygotes than those in A allele carriers. Body mass index (BMI), gender, intake of fat, and FAAH rs324420 were predictive factors of HDL‐C levels in the whole study population. Moreover, BMI and intake of fat were predictors of HDL‐C levels in male FAAH rs324420 A allele carriers and female CC homozygotes, but only BMI was predictor of HDL‐C in female A allele carriers and male CC homozygotes. These results demonstrate that there are mutual effects of dietary fat with gender, BMI, and FAAH rs324420 on HDL‐C levels, which pave a novel way to explore the heterogeneous relationship of serum lipid profiles with diets or FAAH rs324420 and provide a new perspective of precision dietary interventions of dyslipidemia, especially in adolescents.
Keywords: fatty acid amide hydrolase, gene polymorphism, intake of fat, serum lipids
The interactions occurred between fatty acid amide hydrolase gene (FAAH) rs324420 and dietary patterns to influence serum HDL‐C levels. Meanwhile, the dietary intake of fat affected the association of FAAH rs324420 with HDL‐C, which was gender‐dependent.
1. Introduction
The prevalence of metabolic syndrome (MetS) has been increasing rapidly not only in adults but also in adolescents (Shi et al. 2022; Zhu et al. 2020). In MetS, dyslipidemia is common, which mainly manifests as the reduction of high‐density lipoprotein cholesterol (HDL‐C), as well as increase of total cholesterol (TC), low‐density lipoprotein cholesterol (LDL‐C), and triglyceride (TG) (Mach et al. 2020). Although the mechanism of dyslipidemia has not been fully elucidated yet, abnormal serum lipid profiles are believed to be resulted from combined effects of genetic, environmental, behavioral, and developmental factors (Hannon, Khan, and Teran‐Garcia 2018; Matey‐Hernandez et al. 2018).
Serum lipid profiles are intensively affected by endocannabinoid system (ECS) (de Azua and Lutz 2019), in which fatty acid amide hydrolase (FAAH) is one of the key enzymes (Cravatt and Lichtman 2003; van Egmond, Straub, and van der Stelt 2021). FAAH plays a crucial role in the catabolism of arachidonoylethanolamine (anandamide, AEA) and, to a less extent, of 2‐arachidonoylglycerol (2‐AG) (Pamplona and Takahashi 2012). An elevation of FAAH gene (FAAH) mRNA was found in the subcutaneous abdominal adipose tissue from lean individuals when compared to that from obese individuals (Engeli et al. 2014; Murdolo et al. 2007). Interestingly, despite the observed increase in blood glucose levels and enhanced TG content in plasma, skeletal muscle, adipose tissue, and liver of FAAH‐deficient mice (Tourino et al. 2010), it was also demonstrated that hepatic cholesterol synthesis was decreased, and the levels of plasma TG remained unchanged in these mice (Vaitheesvaran et al. 2012). In addition, decreased levels of plasma glucose, TG, TC, and HDL‐C were observed in the rats treated with the FAAH inhibitor URB597 (Rivera et al. 2015).
FAAH rs324420 is a single‐nucleotide polymorphism (SNP) with the substitution from C to A at nucleotide 385 (C385A) of FAAH, resulting in a missense mutation from a proline residue to threonine residue at amino acid 129 (P129T) (Sipe et al. 2002). Consequently, it has been demonstrated that individuals with FAAH rs324420 AA homozygotes exhibit only half of FAAH protein expression and enzymatic activity compared to those with the wild‐type genotype (Chiang et al. 2004). Besides, FAAH rs324420 has been found to be linked with childhood obesity (Durand et al. 2008; Muller et al. 2010) and serum lipid profiles (Ning et al. 2017; Yagin et al. 2019). A previous systematic review, encompassing 65 articles and 70 SNPs, found an association between the mutation of FAAH rs324420 and elevated TG levels in European cohorts (Doris et al. 2019). Moreover, Zeng, Li, and Huang demonstrated that the A allele carriers exhibited higher serum TG levels and lower HDL‐C levels compared with the wild type in the subjects diagnosed with MetS (Zeng, Li, and Huang 2011). However, in 70 patients who developed diabetes mellitus type 2, higher BMI, weight, waist circumference, fat mass, and insulin levels were observed in the A allele carriers, but no striking discrepancies were found in serum TG, TC, HDL‐C, and LDL‐C between allelic groups (de Luis et al. 2010). The mechanism of these heterogeneous associations between FAAH rs324420 and serum lipid profiles needs to be clarified.
Dietary fat has been one of the most persistent and challenging issues. On one hand, a high‐fat diet has long been associated with dyslipidemia, which is a contributing factor to cardiovascular disease (Forouhi et al. 2018). On the other hand, some studies demonstrated that high‐fat diet decreased the levels of serum free fatty acids (FFAs) (Shi et al. 2021) and even the risk of cardiovascular disease (Dehghan et al. 2017). The intake of fat was reported to be positively associated with the levels of TC, LDL‐C, and HDL‐C, and negatively with TG (Mente et al. 2017; Song, Song, and Song 2017). However, Jacobsen et al. failed to find such associations between fat intake and HDL‐C in adults with type 1 diabetes and type 2 diabetes (Jacobsen et al. 2020). Moreover, epidemiologic studies suggested that dyslipidemia could be resulted from interplays between genetic and environmental factors (Matey‐Hernandez et al. 2018; Stein, Ferrari, and Scolari 2019), such as dietary patterns in different races (Feeney et al. 2017; Kafyra et al. 2021; Zhang et al. 2019). Nevertheless, most of the above investigations were carried out in adults or senile populations. Much less efforts have been made in adolescents. To explain the heterogeneous relationship between diets and serum lipid profiles and further explore the prospective mechanism for the regulations of serum lipid levels, we hypothesized that there might be interactions among diets, gender, and FAAH rs324420, resulting in alterations of serum lipid profiles. Therefore, our research aimed to investigate the changes in serum lipid profiles and their relationships with dietary patterns among healthy Chinese adolescents with different genotypes of FAAH rs324420.
2. Material and Methods
2.1. Subjects
A total of 746 students were recruited through advertising in a boarding high school located in Chengdu, Sichuan Province, China. Anthropometric measurements, biochemical examinations as well as medical and dietary questionnaires were used to obtain the characteristics of the volunteers. The inclusion criteria for the students to participate in the study were understanding the procedures involved and providing written consents from the students themselves and their guardians. The criteria for exclusion were a history of cardiovascular or other chronic diseases, taking hormones or other drugs that influenced glucose and lipid metabolisms for the last 3 months, and providing questionnaires not completely finished. Finally, the analysis encompassed 708 Chinese Han volunteers with all the data and blood samples. The study was approved by the Human Research Ethics Committee of Sichuan University.
2.2. Anthropometric Measurements
Anthropometric measurements were performed from 6:30 to 8:30 a.m. before breakfast. Regular procedures were employed to measure height and weight. Body mass index (BMI) was computed by dividing the weight in kilograms by the square of the height in meters (kg/m2).
2.3. Biochemical Examinations
Blood samples were collected in veins after 12 h‐fasting. Serum levels of TG, TC, LDL‐C, HDL‐C, and glucose were analyzed by routine methods in the laboratory. Briefly, semi‐automated biochemistry analyzers were enzymatically used to measure serum TG, TC, and glucose. LDL‐C levels were assessed using the polyvinyl sulfate precipitation method with a semi‐automated biochemistry analyzer. HDL‐C levels were enzymatically determined following the precipitation of apolipoprotein B‐containing lipoproteins using phosphotungstic‐Mg2+. Insulin levels were measured using chemiluminescence immunoassays. Each variable was measured in triplicate. All measurements were conducted in triplicate, with both inter‐ and intra‐assay coefficients of variation being < 6%.
2.4. Dietary Intake
Dietary questionnaires based on 3‐day diet records were used to collect the dietary intake as described before (Zhu et al. 2014). Before starting, educations were made to instruct the students in their classroom to complete the questionnaires, which were coded confidentially. The intakes of protein, fat, and carbohydrate were calculated by the computer‐based data evaluation system that developed by the Department of Nutrition and Food Safety of the Chinese Center for Disease Control (Beijing, China) and expressed as kilocalories (Kcal). Total energy intake was the sum of the intake of protein, fat, and carbohydrate.
2.5. DNA Extraction and Genotyping
The DNAout kit (Cat No. 3671‐50, Tianze, China) was used to isolate the genomic DNA from peripheral blood leukocytes. Genotyping of FAAH rs324420 was carried out by polymerase chain reaction (PCR) using 5′‐GGAAGTGAACAAAGGGACCA‐3′ and 5′‐AGGGTCCACTCCAACAACTG‐3′ as forward and reverse primers, followed by restriction enzyme analyses. The cycling conditions for PCR were 95°C for 5 min followed by 35 cycles of 95°C for 30 s, 59°C for 30 s, and 72°C for 30 s and a final extension at 72°C for 5 min. The amplified DNA was subjected to the digestion by restriction enzyme StyI (Cat No. R3500V, New England Biolabs, USA). Two microliters of PCR products were digested at 37°C overnight with 0.6 μL of StyI enzyme in a final volume of 10 mL (Storr et al. 2009). The digested fragments were identified on gel electrophoresis of 2.5% agarose. The genotypes were identified by the resulting fragment patterns and verified by DNA sequencing. Specifically, the amplified fragments containing FAAH rs324420 site were 299 bp. As shown in Figure 1, the digestion of the PCR products with the AA genotype resulted in a single band of 299 bp fragments, the CC genotype in two bands of 228 bp and 71 bp fragments, and the CA genotype in three bands of 299 bp, 228 bp, and 71 bp fragments. These results were verified by DNA sequencing (Sangon Bioengineering Company, Shanghai, China).
FIGURE 1.
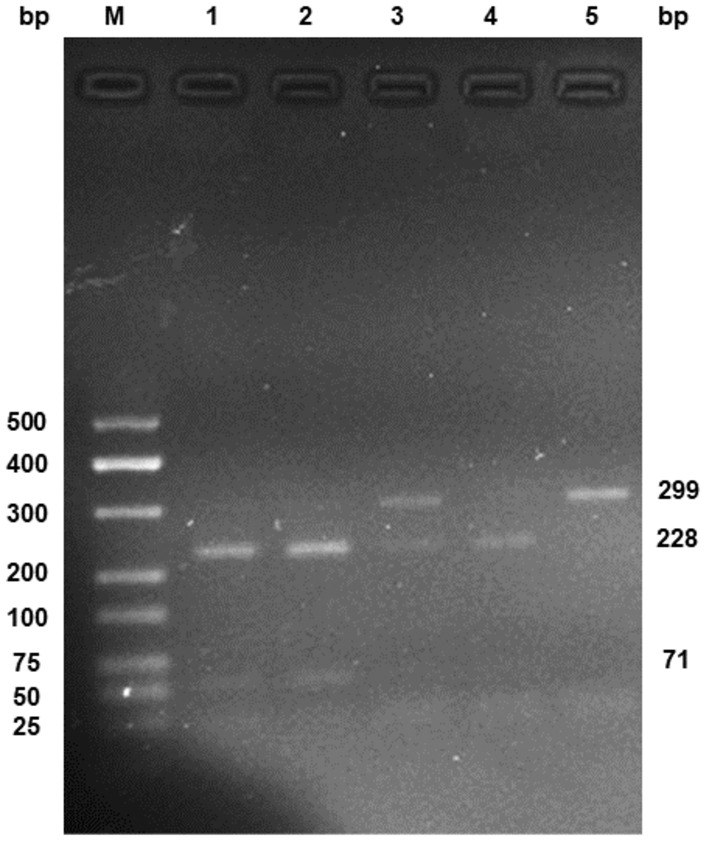
Electrophoretogram of PCR products containing FAAH rs324420 after digested by StyI. M: DNA marker; 1, 2, 4: Fragments with CC genotype; 3: Fragments with CA genotype; 5: Fragments with AA genotype.
2.6. Statistical Analyses
Data were presented as mean ± standard deviation (SD) besides its specification. Data normality was examined using Shapiro–Wilk tests. A chi‐square goodness‐of‐fit test was performed to examine whether the genotype distribution follows Hardy–Weinberg equilibrium. The chi‐square test was applied to analyze genotype and allele frequencies in individuals of different genders. Independent‐sample t‐tests were used to analyze the numerical variables of the subjects with different genotypes of FAAH rs324420. Factors influencing the levels of serum lipids, glucose, and insulin were determined by multiple linear regression analyses. The differences were judged statistically significant when p ≤ 0.05.
3. Results
3.1. Molecular Characterization of the Subjects
Genotypes and allele frequencies of FAAH rs324420 in this research are displayed in Figure 2. The genotype distribution characteristics of FAAH rs324420 agree with Hardy–Weinberg equilibrium (p = 0.549). Due to the small total number of AA genotypes, which included 15 subjects, the individuals with the CA genotype and the AA genotype were jointly defined as the A allele carriers for further analyses. There were no distinct differences in the genotype (p = 0.377), and allele (p = 0.697) frequencies were observed between the male and female subjects.
FIGURE 2.
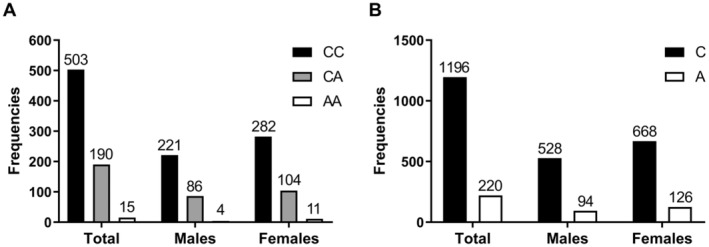
Frequencies of the genotypes and alleles of FAAH rs324420. No significant differences were observed in genotype and allele frequencies between the male and the female subjects by chi‐square tests.
3.2. Anthropometric, Biochemical, and Dietary Characteristics of the Subjects With Different FAAH rs324420 Genotypes
As indicated in Table 1, in the whole study population, only increased HDL‐C levels (p = 0.047) were observed in the CC homozygotes, when compared with those in the A allele carriers. On the contrary, there were no significant differences in the anthropometric and dietary characteristics between the CC homozygotes and the A allele carriers in the total study population.
TABLE 1.
Anthropometric, biochemical, and dietary characteristics of the adolescents with different FAAH rs324420 genotypes.
| Variables | Total (n = 708) | CC (n = 503) | AX (n = 205) |
|---|---|---|---|
| Males/females | 311/397 | 221/282 | 90/115 |
| Age, years | 16.86 ± 0.59 | 16.86 ± 0.59 | 16.85 ± 0.58 |
| BMI, kg/m2 | 20.29 ± 2.30 | 20.29 ± 2.33 | 20.29 ± 2.24 |
| TG, mmol/L | 1.12 ± 0.44 | 1.12 ± 0.46 | 1.10 ± 0.40 |
| TC, mmol/L | 3.59 ± 0.57 | 3.61 ± 0.59 | 3.53 ± 0.53 |
| LDL‐C, mmol/L | 1.67 ± 0.49 | 1.68 ± 0.50 | 1.64 ± 0.46 |
| HDL‐C, mmol/L | 1.41 ± 0.28 | 1.42 ± 0.29 | 1.38 ± 0.26 a |
| Glucose, mmol/L | 5.07 ± 0.44 | 5.06 ± 0.43 | 5.10 ± 0.46 |
| Insulin, mIU/L | 11.91 ± 5.59 | 11.73 ± 5.17 | 12.34 ± 6.49 |
| Total energy intake, 103 Kcal/day | 2.73 ± 1.45 | 2.74 ± 1.44 | 2.73 ± 1.47 |
| Intake of protein, 103 Kcal/day | 0.33 ± 0.21 | 0.33 ± 0.20 | 0.34 ± 0.24 |
| Intake of fat, 103 Kcal/day | 0.90 ± 0.61 | 0.90 ± 0.60 | 0.90 ± 0.63 |
| Intake of carbohydrate, 103 Kcal/day | 1.50 ± 0.73 | 1.51 ± 0.72 | 1.50 ± 0.74 |
Abbreviations: BMI, body mass index; FAAH, fatty acid amide hydrolase; HDL‐C, high‐density lipoprotein cholesterol; Kcal, kilocalorie; LDL‐C, low‐density lipoprotein cholesterol; TC, total cholesterol; TG, triglyceride.
p ≤ 0.05 when compared with those of the CC homozygotes (independent‐sample t‐tests).
3.3. Predictors of Serum Lipids, Glucose, and Insulin Levels
Multiple linear regression analyses were used to elucidate the factors influencing the serum levels of lipids, glucose and insulin, independent variables included BMI, age, gender, total energy intake, the intake of protein, the intake of fat, the intake of carbohydrate, and the genotype of FAAH rs324420, while the levels of serum lipids, glucose, and insulin were used as dependent variables separately. As presented in Table 2, the genotype of FAAH rs324420 was a predictor of only serum HDL‐C levels, but not serum glucose, insulin, TG, TC, and LDL‐C levels. In detail, BMI (p = 0.000), gender (p = 0.000), the intake of fat (p = 0.000), and the genotype of FAAH rs324420 (p = 0.045) were predictive factors of serum HDL‐C levels (Adjusted R 2 = 0.100).
TABLE 2.
Predictors of serum lipids, glucose, and insulin levels in the study population.
| Dependent variables | Independent variables | Adjusted R 2 | β | 95% CI | p |
|---|---|---|---|---|---|
| TG, mmol/L | BMI, kg/m2 | 0.154 | 0.045 | 0.032, 0.059 | 0.000 |
| Gender a | 0.244 | 0.183, 0.305 | 0.000 | ||
| TC, mmol/L | BMI, kg/m2 | 0.089 | 0.034 | 0.016, 0.052 | 0.000 |
| Gender a | 0.285 | 0.202, 0.368 | 0.000 | ||
| LDL‐C, mmol/L | BMI, kg/m2 | 0.063 | 0.046 | 0.031, 0.062 | 0.000 |
| Intake of carbohydrate, 103 Kcal/day | −0.102 | −0.150, −0.053 | 0.000 | ||
| HDL‐C, mmol/L | BMI, kg/m2 | 0.100 | −0.030 | −0.039, −0.022 | 0.000 |
| Gender a | 0.140 | 0.098, 0.181 | 0.000 | ||
| Intake of fat, 103 Kcal/day | 0.067 | 0.034, 0.101 | 0.000 | ||
| FAAH rs324420 b | −0.044 | −0.087, −0.001 | 0.045 | ||
| Glucose, mmol/L | Age, years | 0.070 | −0.076 | −0.129, −0.022 | 0.005 |
| Gender a | −0.114 | −0.179, −0.048 | 0.001 | ||
| Intake of protein, 103 Kcal/day | −0.336 | −0.646, −0.027 | 0.033 | ||
| Intake of fat, 103 Kcal/day | 0.236 | 0.127, 0.345 | 0.000 | ||
| Insulin, mIU/L | BMI, kg/m2 | 0.208 | 0.698 | 0.536, 0.860 | 0.000 |
| Gender a | 3.471 | 2.718, 4.224 | 0.000 |
Note: BMI, age, gender, total energy intake, intake of protein, intake of fat, intake of carbohydrate, and FAAH rs324420 were included as independent variables (multiple linear regression analysis).
Abbreviations: BMI, body mass index; CI, confidence interval; FAAH, fatty acid amide hydrolase; HDL‐C, high‐density lipoprotein cholesterol; Kcal, kilocalorie; LDL‐C, low‐density lipoprotein cholesterol; TC, total cholesterol; TG, triglyceride; β, regression coefficient.
0 = male, 1 = female.
0 = CC, 1 = AX.
3.4. Predictors of Serum HDL‐C Levels in the Subjects With Different Genders and Genotypes of FAAH rs324420
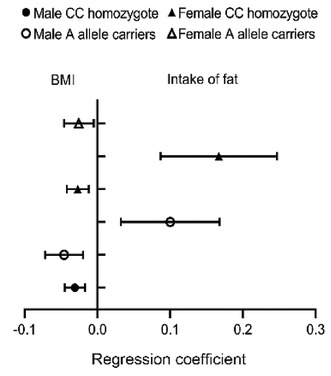
The above results showed that gender and genotype of FAAH rs324420, together with the intake of fat and BMI, had the potential to be predictors of serum HDL‐C levels. To further investigate the effect of dietary intake on HDL‐C, multiple linear regression analyses were performed in the male or the female individuals with different genotypes of FAAH rs324420 (Figure 3). Although BMI (p = 0.000 in the male CC homozygotes, p = 0.001 in the male A allele carriers, p = 0.000 in the female CC homozygotes, and p = 0.016 in the female A allele carriers) was a predictor regardless of the genders and the genotypes, the intake of fat (p = 0.004 in the male A allele carriers; p = 0.000 in the female CC homozygotes) was noticed to be a predictor of serum HDL‐C levels in the male A allele carriers (adjusted R 2 = 0.146) and the female CC homozygotes (adjusted R 2 = 0.083), but not in the female A allele carriers and the male CC homozygotes.
FIGURE 3.
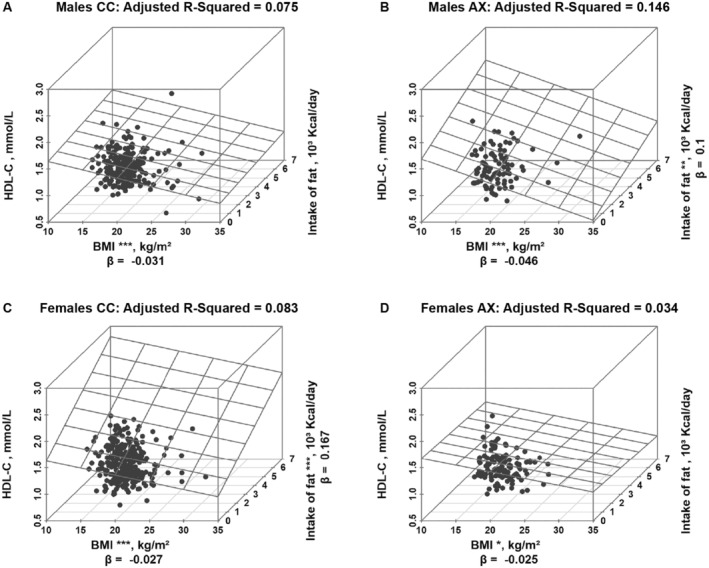
Predictors of serum HDL‐C levels in subjects with different genders and genotypes of FAAH rs324420. BMI, body mass index; FAAH, fatty acid amide hydrolase; HDL‐C, high‐density lipoprotein cholesterol; Kcal, kilocalorie; β, regression coefficient; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 (multiple linear regression analysis).
4. Discussion
Epidemiological studies have shown that there are relationships between ECS and serum lipid profiles (de Azua and Lutz 2019). FAAH, one of the key enzymes in this system (Cravatt and Lichtman 2003; van Egmond, Straub, and van der Stelt 2021), has been proven to be involved in lipid metabolism and associated with the levels of serum lipids (van Eenige et al. 2018). Therefore, exploring the above relationships may contribute to understand the regulation and its molecular mechanism of lipid metabolism and serum lipid levels, or even develop a promising strategy for the treatment of hyperlipidemia. However, there are conflicting reports about the correlation between FAAH rs324420 and serum lipid profiles (de Luis et al. 2010; Doris et al. 2019; Zeng, Li, and Huang 2011). Simultaneously, in the current research, significantly higher levels of HDL‐C were discovered in the CC homozygotes than in subjects with the A allele carriers in the whole study population (Table 1), which is inconsistent with the findings described in the previous study (de Luis et al. 2010; Zeng, Li, and Huang 2011). The endogenous cannabinoids, such as AEA and 2‐AG, have been proven to be related with food intake in both human and animal (Castonguay‐Paradis et al. 2020; van Ackern, Kuhla, and Kuhla 2021). Since FAAH is the main deactivating enzyme of AEA and 2‐AG, it may have the potential to be related to food intakes and even energy intakes of carbohydrate, fat, and protein. All the above suggested a gene–nutrient interaction to modulate serum lipid profiles.
Studies showed that the A allele of FAAH rs324420 was correlated with the significant reduction in serum TC and LDL‐C levels when experienced 3 months of hypocaloric dietary intervention (de Luis et al. 2011; Deluis et al. 2010). It was also demonstrated that the A allele carriers of FAAH rs324420 had decreased TG and TC after 6 weeks low‐fat diets (Aberle et al. 2007). However, it was also reported that the decreases in TC and LDL‐C levels after 3 months of high polyunsaturated fat dietary intervention were not associated with FAAH rs324420 (de Luis et al. 2013). In the present study, we provided the evidence that the interactions occurred between FAAH rs324420 and dietary patterns to influence serum HDL‐C levels. Meanwhile, the dietary intake of fat affected the association of FAAH rs324420 with HDL‐C, which was gender‐dependent (Table 2, Figure 3). Particularly, the intake of fat and BMI were observed to be the predictors of serum HDL‐C levels in the male A allele carriers and the female CC homozygotes, but not in the male CC homozygotes and the female A allele carriers. We could not fully explicate the opposite effects of fat intake on HDL‐C in the female versus male participants categorized by FAAH rs324420 because limited literatures could be found at the current stage. Nevertheless, sex hormones could be among the reasonable explanations. For example, FAAH was found to be a contributor to the changes in plasma lipid profiles (Rahman et al. 2021) and regulated by estrogen (Maia et al. 2017). Taken together, our results reflect the complexity of ECS itself as well as its complex interactions with the diet and gender on lipid profiles (Alexander 2015) since FAAH rs324420 was closely interacted with FAAH protein expression and enzymatic activity (Chiang et al. 2004).
The present study also existed some limitations. First, the expressions of FAAH mRNA and concentrations of FAAH protein were not measured due to the intrinsic limitation of population studies. Second, in the present research, the dietary intake was gathered by dietary questionnaires based on 3‐day diet records. The dietary habits of our study population may not be accurately reflected in the data.
5. Conclusions
The results of our research indicate that the serum level of HDL‐C is positively associated with the fat intake and negatively with BMI in the male CC homozygotes and the female A allele carriers, but only negatively with BMI in the male A allele carriers and the female students with the CC genotype of FAAH rs324420 among Chinese Han adolescents, suggesting interactions of FAAH rs324420 with diets, BMI, and genders on the levels of HDL‐C. These results may contribute to provide a new perspective of precision dietary interventions for dyslipidemia. Biological backgrounds including gender, BMI, and FAAH rs324420 need to be taken into account when diets are used to prevent or treat low HDL‐C of hyperlipidemia, MetS, and cardiovascular disease in adolescents.
Author Contributions
Yi Lin Shen: formal analysis (lead), investigation (lead), visualization (lead), writing – original draft (lead). Li Qiu: formal analysis (supporting), investigation (supporting). Jia Jing Cai: investigation (supporting), validation (supporting). Qi Wei Guo: investigation (supporting), validation (supporting). Xu Chen: investigation (supporting), validation (supporting). Guo Ming Su: investigation (supporting), validation (supporting). Jia Lin: project administration (lead), writing – review and editing (equal). Ding Zhi Fang: conceptualization (lead), data curation (lead), funding acquisition (lead), resources (lead), supervision (lead), writing – review and editing (equal).
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
The present study was supported by the Major Project of Sichuan for Science and Technology (Grant No. 2022YFH0025). Professor Ding Zhi Fang is the recipient of the grant. The authors are grateful to all the students who participated in this study.
Funding: The present study was supported by the Major Project of Sichuan for Science and Technology (Grant No. 2022YFH0025). Professor Ding Zhi Fang is the recipient of the grant.
Data Availability Statement
The interviews (datasets) generated and/or analyzed during the current study are not publicly available due to the information that could compromise the participant's privacy/consent but are available from the corresponding author upon reasonable request.
References
- Aberle, J. , Fedderwitz I., Klages N., George E., and Beil F. U.. 2007. “Genetic Variation in Two Proteins of the Endocannabinoid System and Their Influence on Body Mass Index and Metabolism Under Low Fat Diet.” Hormone and Metabolic Research 39, no. 5: 395–397. 10.1055/s-2007-977694. [DOI] [PubMed] [Google Scholar]
- Alexander, S. 2015. “The Endocannabinoid System as a Nexus of Signalling Complexity.” Pharmacological Reports 67: 3. 10.1016/j.pharep.2015.06.014. [DOI] [Google Scholar]
- Castonguay‐Paradis, S. , Lacroix S., Rochefort G., et al. 2020. “Dietary Fatty Acid Intake and Gut Microbiota Determine Circulating Endocannabinoidome Signaling Beyond the Effect of Body Fat.” Scientific Reports 10, no. 1: 15975. 10.1038/s41598-020-72861-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, K. P. , Gerber A. L., Sipe J. C., and Cravatt B. F.. 2004. “Reduced Cellular Expression and Activity of the P129T Mutant of Human Fatty Acid Amide Hydrolase: Evidence for a Link Between Defects in the Endocannabinoid System and Problem Drug Use.” Human Molecular Genetics 13, no. 18: 2113–2119. 10.1093/hmg/ddh216. [DOI] [PubMed] [Google Scholar]
- Cravatt, B. F. , and Lichtman A. H.. 2003. “Fatty Acid Amide Hydrolase: An Emerging Therapeutic Target in the Endocannabinoid System.” Current Opinion in Chemical Biology 7, no. 4: 469–475. 10.1016/s1367-5931(03)00079-6. [DOI] [PubMed] [Google Scholar]
- de Azua, I. R. , and Lutz B.. 2019. “Multiple Endocannabinoid‐Mediated Mechanisms in the Regulation of Energy Homeostasis in Brain and Peripheral Tissues.” Cellular and Molecular Life Sciences 76, no. 7: 1341–1363. 10.1007/s00018-018-2994-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Luis, D. A. , Izaola O., Aller R., de la Fuente B., and Pacheco D.. 2013. “Effects of C358A Polymorphism of the Endocannabinoid Degrading Enzyme Fatty Acid Amide Hydrolase (FAAH) on Weight Loss, Adipocytokines Levels, and Insulin Resistance After a High Polyunsaturated Fat Diet in Obese Patients.” Journal of Endocrinological Investigation 36, no. 11: 965–969. 10.1007/bf03346760. [DOI] [PubMed] [Google Scholar]
- de Luis, D. A. , Sagrado M. G., Aller R., Izaola O., and Conde R.. 2011. “Effects of C358A Missense Polymorphism of the Endocannabinoid Degrading Enzyme Fatty Acid Amide Hydrolase on Weight Loss After a Hypocaloric Diet.” Metabolism 60, no. 5: 730–734. 10.1016/j.metabol.2010.07.007. [DOI] [PubMed] [Google Scholar]
- de Luis, D. A. , Sagrado M. G., Aller R., Izaola O., Conde R., and Romero E.. 2010. “C358A Missense Polymorphism of the Endocannabinoid Degrading Enzyme Fatty Acid Amide Hydrolase (FAAH) and Insulin Resistance in Patients With Diabetes Mellitus Type 2.” Diabetes Research and Clinical Practice 88, no. 1: 76–80. 10.1016/j.diabres.2009.12.019. [DOI] [PubMed] [Google Scholar]
- Dehghan, M. , Mente A., Zhang X. H., et al. 2017. “Associations of Fats and Carbohydrate Intake With Cardiovascular Disease and Mortality in 18 Countries From Five Continents (PURE): A Prospective Cohort Study.” Lancet 390, no. 10107: 2050–2062. 10.1016/s0140-6736(17)32252-3. [DOI] [PubMed] [Google Scholar]
- Deluis, D. A. , Sagrado M. G., Aller R., Izaola O., and Conde R.. 2010. “Effects of C358A Missense Polymorphism of the Degrading Enzyme Fatty Acid Amide Hydrolase on Weight Loss, Adipocytokines, and Insulin Resistance After 2 Hypocaloric Diets.” Metabolism 59, no. 9: 1387–1392. 10.1016/j.metabol.2009.12.029. [DOI] [PubMed] [Google Scholar]
- Doris, J. M. , Millar S. A., Idris I., and O'Sullivan S. E.. 2019. “Genetic Polymorphisms of the Endocannabinoid System in Obesity and Diabetes.” Diabetes, Obesity & Metabolism 21, no. 2: 382–387. 10.1111/dom.13504. [DOI] [PubMed] [Google Scholar]
- Durand, E. , Lecoeur C., Delplanque J., et al. 2008. “Evaluating the Association of FAAH Common Gene Variation With Childhood, Adult Severe Obesity and Type 2 Diabetes in the French Population.” Obesity Facts 1, no. 6: 305–309. 10.1159/000178157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engeli, S. , Lehmann A. C., Kaminski J., et al. 2014. “Influence of Dietary Fat Intake on the Endocannabinoid System in Lean and Obese Subjects.” Obesity 22, no. 5: E70–E76. 10.1002/oby.20728. [DOI] [PubMed] [Google Scholar]
- Feeney, E. L. , O'Sullivan A., Nugent A. P., et al. 2017. “Patterns of Dairy Food Intake, Body Composition and Markers of Metabolic Health in Ireland: Results From the National Adult Nutrition Survey.” Nutrition & Diabetes 7: e243. 10.1038/nutd.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forouhi, N. G. , Krauss R. M., Taubes G., and Willett W.. 2018. “Dietary Fat and Cardiometabolic Health: Evidence, Controversies, and Consensus for Guidance.” BMJ 361: k2139. 10.1136/bmj.k2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon, B. A. , Khan N. A., and Teran‐Garcia M.. 2018. “Nutrigenetic Contributions to Dyslipidemia: A Focus on Physiologically Relevant Pathways of Lipid and Lipoprotein Metabolism.” Nutrients 10, no. 10: 1404. 10.3390/nu10101404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, S. S. , Vistisen D., Vilsboll T., Bruun J. M., and Ewers B.. 2020. “The Quality of Dietary Carbohydrate and Fat Is Associated With Better Metabolic Control in Persons With Type 1 and Type 2 Diabetes.” Nutrition Journal 19, no. 1: 125. 10.1186/s12937-020-00645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafyra, M. , Kalafati I. P., Katsareli E. A., et al. 2021. “The iMPROVE Study; Design, Dietary Patterns, and Development of a Lifestyle Index in Overweight and Obese Greek Adults.” Nutrients 13, no. 10: 3495. 10.3390/nu13103495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach, F. , Baigent C., Catapano A. L., et al. 2020. “2019 ESC/EAS Guidelines for the Management of Dyslipidaemias: Lipid Modification to Reduce Cardiovascular Risk the Task Force for the Management of Dyslipidaemias of the European Society of Cardiology (ESC) and European Atherosclerosis Society (EAS).” European Heart Journal 41, no. 1: 111–188. 10.1093/eurheartj/ehz455. [DOI] [PubMed] [Google Scholar]
- Maia, J. , Almada M., Silva A., et al. 2017. “The Endocannabinoid System Expression in the Female Reproductive Tract Is Modulated by Estrogen.” Journal of Steroid Biochemistry and Molecular Biology 174: 40–47. 10.1016/j.jsbmb.2017.07.023. [DOI] [PubMed] [Google Scholar]
- Matey‐Hernandez, M. L. , Williams F. M. K., Potter T., Valdes A. M., Spector T. D., and Menni C.. 2018. “Genetic and Microbiome Influence on Lipid Metabolism and Dyslipidemia.” Physiological Genomics 50, no. 2: 117–126. 10.1152/physiolgenomics.00053.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mente, A. , Dehghan M., Rangarajan S., et al. 2017. “Association of Dietary Nutrients With Blood Lipids and Blood Pressure in 18 Countries: A Cross‐Sectional Analysis From the PURE Study.” Lancet Diabetes and Endocrinology 5, no. 10: 774–787. 10.1016/s2213-8587(17)30283-8. [DOI] [PubMed] [Google Scholar]
- Muller, T. D. , Bronner G., Wandolski M., et al. 2010. “Mutation Screen and Association Studies for the Fatty Acid Amide Hydrolase (FAAH) Gene and Early Onset and Adult Obesity.” BMC Medical Genetics 11: 2. 10.1186/1471-2350-11-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdolo, G. , Kempf K., Hammarstedt A., Herder C., Smith U., and Jansson P. A.. 2007. “Insulin Differentially Modulates the Peripheral Endocannabinoid System in Human Subcutaneous Abdominal Adipose Tissue From Lean and Obese Individuals.” Journal of Endocrinological Investigation 30, no. 8: RC17–RC21. 10.1007/bf03347440. [DOI] [PubMed] [Google Scholar]
- Ning, T. L. , Zou Y. Y., Yang M. L., et al. 2017. “Genetic Interaction of DGAT2 and FAAH in the Development of Human Obesity.” Endocrine 56, no. 2: 366–378. 10.1007/s12020-017-1261-1. [DOI] [PubMed] [Google Scholar]
- Pamplona, F. A. , and Takahashi R. N.. 2012. “Psychopharmacology of the Endocannabinoids: Far Beyond Anandamide.” Journal of Psychopharmacology 26, no. 1: 7–22. 10.1177/0269881111405357. [DOI] [PubMed] [Google Scholar]
- Rahman, S. M. K. , Uyama T., Hussain Z., and Ueda N.. 2021. “Roles of Endocannabinoids and Endocannabinoid‐Like Molecules in Energy Homeostasis and Metabolic Regulation: A Nutritional Perspective.” Annual Review of Nutrition 41: 177–202. 10.1146/annurev-nutr-043020-090216. [DOI] [PubMed] [Google Scholar]
- Rivera, P. , Bindila L., Pastor A., et al. 2015. “Pharmacological Blockade of the Fatty Acid Amide Hydrolase (FAAH) Alters Neural Proliferation, Apoptosis and Gliosis in the Rat Hippocampus, Hypothalamus and Striatum in a Negative Energy Context.” Frontiers in Cellular Neuroscience 9: 98. 10.3389/fncel.2015.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, D. , Han T. S., Chu X., et al. 2021. “An Isocaloric Moderately High‐Fat Diet Extends Lifespan in Male Rats and Drosophila.” Cell Metabolism 33, no. 3: 581–597. 10.1016/j.cmet.2020.12.017. [DOI] [PubMed] [Google Scholar]
- Shi, J. , He L., Yu D., et al. 2022. “Prevalence and Correlates of Metabolic Syndrome and Its Components in Chinese Children and Adolescents Aged 7–17: The China National Nutrition and Health Survey of Children and Lactating Mothers From 2016–2017.” Nutrients 14, no. 16: 3348. 10.3390/nu14163348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipe, J. C. , Chiang K., Gerber A. L., Beutler E., and Cravatt B. F.. 2002. “A Missense Mutation in Human Fatty Acid Amide Hydrolase Associated With Problem Drug Use.” Proceedings of the National Academy of Sciences of the United States of America 99, no. 12: 8394–8399. 10.1073/pnas.082235799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, S. , Song W., and Song Y.. 2017. “Dietary Carbohydrate and Fat Intakes Are Differentially Associated With Lipid Abnormalities in Korean Adults.” Journal of Clinical Lipidology 11, no. 2: 338–347. 10.1016/j.jacl.2017.01.016. [DOI] [PubMed] [Google Scholar]
- Stein, R. , Ferrari F., and Scolari F.. 2019. “Genetics, Dyslipidemia, and Cardiovascular Disease: New Insights.” Current Cardiology Reports 21, no. 8: 68. 10.1007/s11886-019-1161-5. [DOI] [PubMed] [Google Scholar]
- Storr, M. , Emmerdinger D., Diegelmann J., et al. 2009. “The Role of Fatty Acid Hydrolase Gene Variants in Inflammatory Bowel Disease.” Alimentary Pharmacology & Therapeutics 29, no. 5: 542–551. 10.1111/j.1365-2036.2008.03910.x. [DOI] [PubMed] [Google Scholar]
- Tourino, C. , Oveisi F., Lockney J., Piomelli D., and Maldonado R.. 2010. “FAAH Deficiency Promotes Energy Storage and Enhances the Motivation for Food.” International Journal of Obesity 34, no. 3: 557–568. 10.1038/ijo.2009.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaitheesvaran, B. , Yang L., Hartil K., et al. 2012. “Peripheral Effects of FAAH Deficiency on Fuel and Energy Homeostasis: Role of Dysregulated Lysine Acetylation.” PLoS One 7, no. 3: e33717. 10.1371/journal.pone.0033717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ackern, I. , Kuhla A., and Kuhla B.. 2021. “A Role for Peripheral Anandamide and 2‐Arachidonoylglycerol in Short‐Term Food Intake and Orexigenic Hypothalamic Responses in a Species With Continuous Nutrient Delivery.” Nutrients 13, no. 10: 3587. 10.3390/nu13103587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eenige, R. , van der Stelt M., Rensen P. C. N., and Kooijman S.. 2018. “Regulation of Adipose Tissue Metabolism by the Endocannabinoid System.” Trends in Endocrinology and Metabolism 29, no. 5: 326–337. 10.1016/j.tem.2018.03.001. [DOI] [PubMed] [Google Scholar]
- van Egmond, N. , Straub V. M., and van der Stelt M.. 2021. “Targeting Endocannabinoid Signaling: FAAH and MAG Lipase Inhibitors.” Annual Review of Pharmacology and Toxicology 61: 441–463. 10.1146/annurev-pharmtox-030220-112741. [DOI] [PubMed] [Google Scholar]
- Yagin, N. L. , Aliasgari F., Aliasgharzadeh S., Mahdavi R., and Akbarzadeh M.. 2019. “The Influence of the Fatty Acid Amide Hydrolase 385C > A Single Nucleotide Polymorphisms on Obesity Susceptibility.” Molecular Biology Reports 46, no. 5: 5049–5055. 10.1007/s11033-019-04956-8. [DOI] [PubMed] [Google Scholar]
- Zeng, L. Y. , Li J. R., and Huang G. L.. 2011. “385 C/A Polymorphism of the Fatty Acid Amide Hydrolase Gene Is Associated With Metabolic Syndrome in the Chinese Han Population.” Archives of Medical Science 7, no. 3: 423–427. 10.5114/aoms.2011.23406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Tan S. J., Zhao A., Wang M. C., Wang P. Y., and Zhang Y. M.. 2019. “Association Between Nutrient Patterns and Serum Lipids in Chinese Adult Women: A Cross‐Sectional Study.” Nutrition & Dietetics 76, no. 2: 184–191. 10.1111/1747-0080.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X. , Lin J., Song Y., et al. 2014. “A High‐Carbohydrate Diet Lowered Blood Pressure in Healthy Chinese Male Adolescents.” Bioscience Trends 8, no. 2: 132–137. 10.5582/bst.8.132. [DOI] [PubMed] [Google Scholar]
- Zhu, Y. , Zheng H., Zou Z., et al. 2020. “Metabolic Syndrome and Related Factors in Chinese Children and Adolescents: Analysis From a Chinese National Study.” Journal of Atherosclerosis and Thrombosis 27, no. 6: 534–544. 10.5551/jat.50591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The interviews (datasets) generated and/or analyzed during the current study are not publicly available due to the information that could compromise the participant's privacy/consent but are available from the corresponding author upon reasonable request.


