Abstract
Eukaryotic translation initiation factor 6 (eIF6) binds to the 60S ribosomal subunit and prevents its association with the 40S ribosomal subunit. The Saccharomyces cerevisiae gene that encodes the 245-amino-acid eIF6 (calculated Mr 25,550), designated TIF6, has been cloned and expressed in Escherichia coli. The purified recombinant protein prevents association between 40S and 60S ribosomal subunits to form 80S ribosomes. TIF6 is a single-copy gene that maps on chromosome XVI and is essential for cell growth. eIF6 expressed in yeast cells associates with free 60S ribosomal subunits but not with 80S monosomes or polysomal ribosomes, indicating that it is not a ribosomal protein. Depletion of eIF6 from yeast cells resulted in a decrease in the rate of protein synthesis, accumulation of half-mer polyribosomes, reduced levels of 60S ribosomal subunits resulting in the stoichiometric imbalance in the 40S/60S subunit ratio, and ultimately cessation of cell growth. Furthermore, lysates of yeast cells depleted of eIF6 remained active in translation of mRNAs in vitro. These results indicate that eIF6 does not act as a true translation initiation factor. Rather, the protein may be involved in the biogenesis and/or stability of 60S ribosomal subunits.
An obligatory intermediate step in initiation of protein synthesis in eukaryotic cells is the initial positioning of the 40S ribosomal subunit containing bound initiator Met-tRNAf at the AUG codon of an mRNA to form the 40S initiation complex. Subsequently a 60S ribosomal subunit joins the 40S initiation complex to form an 80S initiation complex (Met-tRNAf · 80S · mRNA) that is competent to undergo peptide bond synthesis during the elongation phase of protein synthesis (for reviews, see reference 16 and 17). Thus, the assembly of the 80S initiation complex during initiation of protein synthesis requires a cellular pool of free 40S and 60S ribosomal subunits. Two eukaryotic translation initiation factors, eIF3 and eIF6, have been implicated in maintaining a pool of free ribosomal subunits (16, 17). eIF3, a multisubunit protein complex with Mr > 500,000, binds to the 40S ribosomal subunit (3) and prevents its association with the 60S ribosomal subunit (10, 34–36). eIF6, a monomeric protein of about 26 kDa, binds to the 60S ribosomal subunit and prevents its association with the 40S ribosomal subunit (21, 26, 27, 37). This ribosomal subunit anti-association property was used originally as an assay to purify eIF6 from wheat germ (26, 27), mammalian liver (37), and rabbit reticulocyte lysates (21, 32). However, unlike eIF3, whose requirement in the formation of the 40S initiation complex is firmly established (16, 17), the requirement of eIF6 in translation of mRNAs has not been defined.
We have recently cloned a human cDNA that encodes eIF6, which is 245 amino acids long (calculated Mr, 26,558) (32). The open reading frame (ORF) of the cDNA was expressed in Escherichia coli. The purified recombinant protein exhibited biochemical properties that are similar to eIF6 isolated from mammalian cell extracts (32). When the predicted amino acid sequence of human eIF6 was used to search the yeast genomic sequence for homologous yeast eIF6 sequence, an ORF (YPR016C) that encodes a protein of 245 amino acids, with 72% identity to human eIF6, was identified in chromosome XVI of Saccharomyces cerevisiae (32).
In this paper, we describe the cloning and characterization of the yeast gene TIF6 (translation initiation factor 6), which encodes eIF6. Yeast eIF6 is a protein of 245 amino acids (calculated Mr, 25,550) with 72% identity to human eIF6 (32). Expression of eIF6 protein in E. coli confirms that the gene singled out on phylogenetic grounds (32) is indeed the structural gene of yeast eIF6. We also show that TIF6 is essential for cell growth. Additionally, we have constructed a haploid yeast strain in which a functional but a rapidly degradable form of eIF6 fusion protein was synthesized from a glucose-repressible GAL promoter. The effects of depletion of eIF6 from this strain on cell growth as well as protein synthesis in vivo and in vitro were then analyzed. The results of these experiments show that eIF6 plays a role in translation of mRNAs by maintaining the steady-state level of a pool of 60S ribosomal subunits. The protein does not function as a bona fide translation initiation factor.
MATERIALS AND METHODS
Media, growth conditions, and DNA analysis.
Yeast strains were grown at 30°C in either rich YPD medium (1% [wt/vol] yeast extract, 2% [wt/vol] Bacto Peptone, 2% [wt/vol] dextrose) or in YPGal medium, where 2% (wt/vol) galactose replaced dextrose as the carbon source. Where indicated, haploid yeast cells were also grown in synthetic complete medium (0.67% Bacto Yeast Nitrogen Base without amino acids, 0.2% amino acid mixture supplemented with nutrients required for auxotrophic deficiencies). This synthetic complete medium contained either 2% galactose (SGal) or 2% dextrose (SD) as the carbon source. For in vivo [35S]methionine incorporation, the 0.2% amino acid mixture in either the SGal or the SD medium did not contain methionine. These methionine-lacking media were designated SGal-Met and SD-Met, respectively. Yeast genetic methods were performed by standard methods (22). Standard methods were used for plasmid and genomic DNA isolation, cloning, and bacterial transformation (30). The following primers were used for PCR amplification of DNA: Yf65, 5′-CGGGATCC CATATGGCTACCAGGACTCAA (EcoRI, NdeI); yf63, 5′-GGGAATTCCTATGAGTAGGTTTCAATCAA (BamHI); Yf6G5, 5′-CGGGATCCAAGGTGCAAGATCAGAC (BamHI); yF6G3, 5′-GGGAATTCCTGCCAAGAGATACTACT (EcoRI); Yf6Ub5, 5′-GCTCTAGAATGGCTACCAGGACTCAA (XbaI); and Yf6Ub3, 5′-AACTGCAGCTATGAGTAGGTTTCAATCAA (PstI) (restriction enzyme sites are underlined).
Cloning, expression, and purification of yeast eIF6.
A 735-bp yeast genomic fragment that corresponds to the putative yeast eIF6 ORF based on sequence similarity to human eIF6 (32) was amplified by PCR with Pyrococcus DNA polymerase (Stratagene) and two primer sequences, Yf65 and Yf63, that correspond to the N-terminal and C-terminal ends of putative yeast eIF6 protein, respectively. The PCR product was digested with BamHI and EcoRI, cloned into the same sites of pBluescript SK(+) vector (Stratagene), and sequenced to ensure error-free DNA synthesis. For expression of the eIF6 protein in E. coli, the PCR product was cloned into the NdeI-EcoRI sites of the expression vector pET-5a (Novagen) to yield the recombinant plasmid pET-TIF6. The recombinant plasmid was transformed into E. coli BL21(DE3) cells, and the expression of the plasmid-encoded eIF6 protein was induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to an exponentially growing bacterial culture (2 liters). The cells were harvested by centrifugation at 3 h postinduction. The frozen cells (10 g) were suspended in 30 ml of 20 mM Tris-HCl (pH 8.5)–5 mM EDTA–100 mM KCl–10 mM 2-mercaptoethanol–0.5 mM phenylmethylsulfonyl fluoride, treated with 300 μg of lysozyme for 30 min at 4°C, and then disrupted by sonication. After centrifugation at 15,000 × g for 20 min, the supernatant was adjusted to 0.5 M KCl by the addition of 2 M KCl and centrifuged at 150,000 × g for 2 h. The postribosomal supernatant (250 mg of protein) was dialyzed against buffer A (20 mM Tris-HCl [pH 7.5], 0.2 mM EDTA, 1 mM dithiothreitol, 10% glycerol) containing 150 mM KCl and then loaded onto a 60-ml bed volume of a DEAE-cellulose column equilibrated against buffer A–150 mM KCl. After the column was washed with 300 ml of the same buffer, the bound proteins were eluted with buffer A–280 mM KCl, dialyzed against buffer A–150 mM KCl, and then applied to a 6.5-ml volume of a column of DEAE-Sephacel equilibrated against the same buffer. After the column was washed with 30 ml of buffer A–150 mM KCl, bound proteins were eluted with a 50-ml linear gradient of KCl (0.15 to 0.5 M) in buffer A. Fractions containing yeast eIF6, detected by Western blotting with anti-yeast eIF6 antibodies, were pooled, and the proteins were precipitated by addition of solid ammonium sulfate to 70% saturation. The precipitated proteins were dissolved in 1 ml of buffer A–500 mM KCl and dialyzed against the same buffer. The concentrated protein fraction (10 mg) was applied to a 33-ml bed volume column of Sephadex G-75 (superfine) equilibrated in buffer A–500 mM KCl. The column was developed with the same buffer, and fractions of 1 ml were collected. Fractions 21 to 26 containing eIF6 protein were pooled and stored in small aliquots at −70°C. The total yield was about 720 μg of purified eIF6 protein.
Disruption of the TIF6 gene in the chromosome.
The one-step gene disruption method of Rothstein (24) was used to construct the null allele of the TIF6 gene. For this purpose, the TIF6 ORF present in the vector pET-TIF6 was digested with XbaI and EcoRI and the resulting 775-bp XbaI-EcoRI fragment was cloned into the EcoRI-XbaI sites of pGEM7Zf(+) to yield the plasmid pKS60. A 1.0-kb EcoRV-SmaI fragment carrying the entire HIS3 gene was inserted into a unique EcoRV site within the TIF6 ORF (at position +362) present in pKS60 to yield the plasmid pKS601. This plasmid was linearized by digestion with SmaI and used to transform his3 homozygous W303 (a/α) cells. Stable diploid His+ transformants, designated KSY601, were selected. The disruption of one of the genomic TIF6 genes in these transformants was verified by Southern blot analysis of total DNA with a 32P-labeled 735-bp TIF6 ORF fragment of the TIF6 gene as a probe. Spores obtained from several transformants were subjected to tetrad analysis as described by Rothstein (24).
Construction of haploid yeast strains for expression of yeast eIF6.
The haploid yeast strain KSY602 (Table 1), containing the chromosomal copy of the TIF6 gene inactivated by insertion of HIS3 marker gene and harboring a centromeric expression plasmid that expresses yeast eIF6 from its own natural promoter, was constructed as follows. A 1.752-kb yeast genomic fragment containing the entire TIF6 gene was amplified from yeast genomic DNA by PCR by using a 5′ end primer, Yf6G5, and a 3′-end primer, Yf6G3. The PCR product was digested with BamHI and EcoRI and cloned into the same sites of a centromeric plasmid, pRS315 (LEU2 based) (33), to yield plasmid pRS315-TIF6. In this plasmid, the expression of eIF6 is under the control of its natural promoter present in the inserted 1.752-kb fragment. This plasmid was then used to transform the diploid strain KSY601 (a/α) (tif6::HIS3/TIF6), transformants were sporulated at 30°C, and the resulting tetrads were dissected and spores were germinated on YPD plates followed by selection on SD-His-Leu plates. Only spores containing the tif6::HIS3 gene and harboring the pRS315-TIF6 plasmid will germinate to yield His+ Leu+ colonies. The genotype of the TIF6 loci on the chromosome and on the plasmid of KSY602 was confirmed by Southern blot analysis with a 32P-labeled 735-bp TIF6 ORF DNA fragment as a probe. For expression of yeast eIF6 under the control of the galactose-inducible GAL10 promoter, we first isolated a 0.6-kb-BamHI-EcoRI fragment containing the GAL1-GAL10 promoter from plasmid pBM272 (12) and cloned it at the same sites of pRS315 to generate pTM100 (15). A 735-bp TIF6 coding region was excised from plasmid pKS60 (see above) by digestion with BamHI and XbaI and cloned into the same sites of the pTM100 vector to generate the eIF6 expression plasmid pTM100-TIF6. We also constructed a plasmid for conditional expression of a rapidly degradable form of yeast eIF6 fusion protein (see Fig. 3A). For this purpose, the coding region of TIF6 was first fused in frame to a lacI-flu segment present in plasmid pGEM-flu (20) as follows. The ORF of TIF6 was amplified by PCR with primers Yf6Ub5 and Yf6Ub3, which were flanked by XbaI and PstI, respectively, and cloned into the same sites of the plasmid pGEM-flu to make an in-frame fusion of TIF6 to the lacI-flu fragment. This plasmid was then digested with BamHI and SphI, and the resulting lacI-flu-TIF6 fragment was fused in frame to the ubiquitin gene, UBI4, under the control of GAL10 promoter, as follows. Plasmid pUB23, containing the UBI4 gene under the control of the GAL10 promoter, was cut with BamHI and EcoRI, and the resulting 2.2-kb BamHI-EcoRI fragment that contains URA3 and the upstream activation sequence of the GAL promoter (UASGal)-ubiquitin (Ub)-X (where X is the trinucleotide codon for arginine) was ligated with the BamHI-SphI fragment that encodes the lacI-flu-TIF6 gene derived from the pGEM-flu vector (see above). The resulting 3.1-kb EcoRI-SphI fragment was then cloned into the EcoRI-SphI site of the centromeric HIS3 vector pSE362 (20) to generate pUB-TIF6R (GAL10::Ub-lacI-flu-TIF6) (see Fig. 3A). For construction of a haploid yeast strain for conditional expression of ubiquitinylated eIF6 fusion protein, the haploid yeast strain KSY602, which has the disrupted chromosomal copy of TIF6 but harbors the LEU2-based centromeric pRS315-TIF6 as the complementing plasmid (see above and Table 1), was transformed with plasmid pUB-TIF6R and His+ Leu+ Ura+ transformants were selected. These transformants were then grown in SGal − His − Ura medium to promote the loss of the LEU2 plasmid, pRS315-TIF6. The newly generated yeast strain, which has the disrupted chromosomal copy of the TIF6 gene but harbors only plasmid pUB-TIF6R, was selected on appropriate SGal media. This strain, designated KSY603, expresses yeast eIF6 from GAL10 promoter initially as an N-terminal ubiquitinylated eIF6 fusion protein, which is rapidly processed in yeast cells by a deubiquitination enzyme to yield free ubiquitin and an eIF6 fusion protein having arginine (R) as the −NH2 terminal amino acid.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| W303a/α | MATa/MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 | 24 |
| W303α | MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 | 24 |
| KSY601 | MATa/MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 tif6::HIS3/TIF6 | This work |
| KSY602 | MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 tif6::HIS3 p[LEU2 TIF6] | This work |
| KSY603 | MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100 tif6::HIS3 p[URA3 GAL10::UbTIF6] | This work |
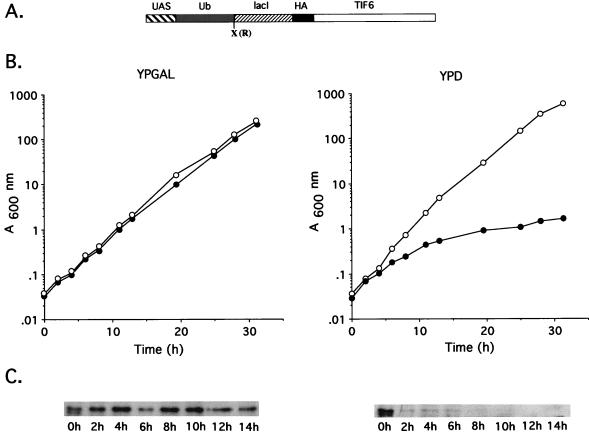
FIG. 3.
Analysis of eIF6 depletion in yeast cells and its effect on cell growth. (A) Schematic representation of the plasmid pUB-TIF6. The plasmid was constructed as described under Materials and Methods. Abbreviations: UAS, the upstream activation sequence of the GAL10 promoter; Ub, ubiquitin gene; X, the codon for arginine; lacI, a restriction fragment that encodes amino acid residues 318 to 346 of the lac repressor; HA, an epitope from the influenza virus hemagglutinin protein. (B) Exponentially growing cultures of W303α (○) and KSY603 (•) were diluted to an A600 of about 0.03 in either YPGal (galactose) medium or YPD (glucose) medium. Cell growth was monitored by measuring the A600. To keep cultures in exponential growth, they were diluted in fresh medium whenever the A600 reached 0.8 U. (C) At the indicated times following the shift from YPGal to either YPGal (left) or YPD (right), cell lysates were prepared from KSY603 as described in Materials and Methods. Approximately 50 μg of protein from each cell lysate was electrophoresed through an SDS–15% polyacrylamide gel and electrophoretically transferred to a polyvinylidene difluoride membrane. The blot was then probed with peroxidase-coupled anti-HA monoclonal antibodies to detect eIF6 fusion protein. The decrease in the amount of eIF6 observed at the 6-h time point in YPGal is due to a gel loading error.
Polysome profile analysis and isolation of ribosomal subunits.
The haploid yeast strains W303α and KSY603 were grown in YPGal or YPD medium to early logarithmic phase (absorbance at 600 nm [A600] ≈ 0.2). The cells were harvested, and 4 A600 units of cells was resuspended in 100 ml of YPGal or YPD medium and grown at 30°C. At the indicated time, 50 ml of cultures was withdrawn, treated with 50 μg of cycloheximide per ml, rapidly chilled in an ice-water bath, and then harvested. The cells were washed twice with LHB buffer (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 30 mM MgCl2, 50 μg of cycloheximide per ml). The washed cells were then suspended in 0.5 ml of LHB buffer, lysed by vortexing with an equal volume of glass beads, and then treated with an additional 0.5 ml of LHB buffer. The lysates were clarified by centrifugation at 12,000 × g for 15 min, and equivalent amounts of A254-absorbing material (approximately 10 A254 units) were layered on 11 ml of 7 to 47% (wt/vol) sucrose gradients in a low-salt buffer (10 mM Tris acetate [pH 7.0], 12 mM MgCl2, 50 mM NH4Cl) and centrifuged at 40,000 rpm for 2.5 h at 4°C in a Beckman SW41 rotor. The gradients were fractionated in an ISCO density gradient fractionator, and the A254 profile was analyzed in an ISCO UA-5 absorbance monitor.
Ribosomal subunits were isolated from mid-exponential-phase cultures of W303α (wild type) or KSY603 grown in YPD or YPGal medium. Harvested cells were washed twice with buffer B (50 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 10 mM dithiothreitol, 800 mM KCl, 0.1% Triton X-100, 0.5 mM phenylmethylsulfonyl fluoride). Cell extracts were then prepared in buffer B, as described above, and clarified by centrifugation. Approximately 2 A254 units of the supernatant was layered over an 11-ml 10 to 40% (wt/vol) sucrose gradient containing buffer B and centrifuged at 40,000 rpm for 3.5 h at 4°C in a Beckman SW41 rotor. Gradient fractionation and analysis were carried out as described above for the polysome profile analysis.
Cell-free translation.
Strain KSY603 was grown overnight at 30°C (A600 ≈ 1.0) in YPGal supplemented with adenine sulfate (0.4 mg/ml). Cells were harvested and suspended in 2 liters of YPD medium containing adenine sulfate (0.4 mg/ml) so that the initial A600 was about 0.03 unit and grown at 30°C for about 11 h, when growth was considerably inhibited and the presence of eIF6 could not be detected by immunoblot analysis of cell lysates. The cells were then harvested, cell-free translation extracts were prepared, and mRNA-dependent cell-free translation was performed with [35S]methionine as the labeled amino acid, as described by Hussain and Leibowitz (11).
Measurement of the protein synthesis rate in vivo.
The rate of protein synthesis was measured by pulse-labeling 1.0 A600 unit of yeast cells with [35S]methionine for 5 min and measuring the incorporation of 35S radioactivity into total cellular proteins as follows. Exponentially growing cultures (A600 ≈ 0.6) of W303α or KSY603 growing in YPGal were harvested, and approximately 10 A600 units of cells was resuspended in 150 ml of either YPD or YPGal medium and allowed to grow at 30°C. At the indicated times, 2 A600 units of cells was removed, centrifuged, resuspended in 300 μl of either YPGal or YPD, and labeled for 5 min with 50 μCi of [35S]methionine (Dupont-NEN) at 30°C. Following the addition of 1 ml of stop buffer (1.2 mg of unlabeled methionine per ml, 0.5 mg of cycloheximide per ml), the cells were chilled rapidly. The amount of [35S]methionine incorporated into total cellular proteins was then measured by an adaptation of the method of Kang and Hershey (13) as follows. The cells were lysed in 1.8 N NaOH containing 0.2 M 2-mercaptoethanol, and proteins were precipitated by the addition of hot 10% trichloroacetic acid. After centrifugation, the precipitate was washed twice in acetone, dissolved in 100 μl of 1% sodium dodecyl sulfate (SDS)-containing buffer, and heated at 95°C for 10 min. An aliquot of the SDS extract was counted in Aquasol for 35S radioactivity in a liquid scintillation spectrometer to determine the amount of [35S]methionine incorporated into proteins, while another aliquot containing about 100 μg of proteins was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) (15% polyacrylamide) followed by autoradiography.
Other methods.
Total and poly(A)+ RNAs of S. cerevisiae were isolated essentially as described by Rose et al. (22). Northern and Southern blot analyses were performed by standard methods (30). Immunoglobulin G antibodies specific for yeast eIF6 were isolated from specific rabbit antisera raised against purified recombinant yeast eIF6 by affinity purification with purified recombinant yeast eIF6 blotted onto aminophenyl thioether paper (Schleicher & Schuell) as an antigen as described previously (9). The procedure used for preparation of yeast cell lysates for immunoblot analysis was adapted from that described by Sachs and Davis (28). Yeast eIF6 was assayed by the ribosomal subunit anti-association assay as described for mammalian eIF6 (37). Antibodies against 60S ribosomal protein L3 (38) were kindly provided by Jonathan Warner of this institution.
RESULTS
Identification of the S. cerevisiae gene, designated TIF6, encoding eIF6.
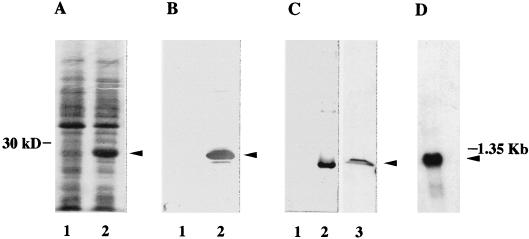
We have previously reported (32) the cloning of a human cDNA that encodes eIF6, with 245 amino acids (calculated Mr, 26,558). When we used the predicted amino acid sequence of human eIF6 to search the yeast genomic database for homologous yeast eIF6 sequence, we identified a hypothetical translation product in chromosome XVI of S. cerevisiae (YPR016C) that encodes a protein of 245 amino acids, with 72% identity to human eIF6 (32). The 735-bp YPR016C ORF was amplified by PCR from yeast genomic DNA and cloned into the E. coli expression vector pET-5a under the control of the phage T7 RNA polymerase promoter. Transformation of the resulting construct into E. coli BL21(DE3) resulted, after induction with IPTG, in the production of a polypeptide of about 26 kDa (Fig. 1A, lane 2). This polypeptide reacted specifically with the well-characterized monospecific anti-mammalian eIF6 antibodies (Fig. 1B), indicating that the expressed protein shared antigenic determinants with mammalian eIF6. When the E. coli extract was centrifuged at 10,000 × g for 10 min, a major fraction (>90%) of the expressed protein was found in the insoluble pellet fraction while only a small fraction (about 5 to 10%) was in the soluble fraction (data not shown).
FIG. 1.
Analysis of the protein and the mRNA encoded by the TIF6 gene. (A) Expression of yeast eIF6 in E. coli. Cell extracts, prepared from IPTG-induced cultures of E. coli BL21(DE3) cells harboring either the parental plasmid pET-5a (lane 1) or pET-TIF6 (lane 2), were electrophoresed in an SDS–15% polyacrylamide gel and subjected to Coomassie blue staining. The position of migration of the expressed protein of about 26 kDa is shown by an arrowhead. (B) Immunoblot analysis of the bacterially expressed 26-kDa protein with mammalian anti-eIF6 antibodies as probes. Cell extracts of IPTG-induced cultures of BL21(DE3) cells harboring either the parental vector (lane 1) or the pET-TIF6 recombinant expression plasmid (lane 2) were subjected to Western blot analysis with monospecific anti-mammalian eIF6 antibodies (32) as probes. (C) Immunoblot analysis with anti-TIF6p (eIF6) antibodies as probes. The protein samples used in lanes 1 and 2 were the same as those used in lanes 1 and 2 of panel B, while lane 3 contained 100 μg of a partially fractionated protein fraction derived from cell extracts of S. cerevisiae W303 (a/α). Immunoblot analysis was carried out with anti-yeast 26-kDa Tif6p antibodies as probes. A set of molecular weight marker proteins was run in a separate lane of each gel (data not shown). The position of eIF6 is shown by an arrowhead. (D) Northern blot analysis of mRNA expressed from the TIF6 gene. A Northern blot containing 2 μg of electrophoretically separated yeast poly(A)+ RNA was hybridized to a 32P-labeled 735-bp TIF6 ORF. The blot also contained a set of RNA size markers, of which the position of 1.35-kb RNA is indicated. The blot was analyzed by autoradiography.
The recombinant 26-kDa protein was purified initially from the insoluble pellet fraction and used as an antigen for the preparation of specific antisera in rabbits (see Materials and Methods). These polyclonal antibodies reacted specifically with the recombinant 26-kDa protein present in the E. coli lysates. (Fig. 1C, lane 2). In contrast, no immunoreactive polypeptide band was observed in E. coli cell lysates containing the parental non-recombinant pET-5a vector (lane 1). Furthermore, preimmune immunoglobulin G did not react with the 26-kDa protein (data not shown). These results indicated that these antibodies are monospecific. These monospecific antibodies were then used as a specific probe in Western blot analysis to monitor the purification of the 26-kDa protein from the soluble fraction of the bacterial cell lysates, as described in Materials and Methods. The elution profile of the recombinant protein in the Sephadex G-75 gel filtration column was identical to that of purified natural or recombinant human eIF6 (data not shown).
The biochemical properties of the recombinant 26-kDa protein with regard to its ability to interact with 60S ribosomal subunits and prevent their association with 40S ribosomal subunits at 5 mM Mg2+ concentration were investigated (Fig. 2). As reported previously (37), ribosomes washed in high-salt buffer and incubated at 1 mM Mg2+ in the absence of any protein fraction dissociated into 40S and 60S ribosomal subunits as determined by sucrose gradient centrifugation (Fig. 2A). If, however, the Mg2+ concentration of the incubation mixture was then raised to 5 mM in a second incubation, a major fraction of the subunits reassociated to form 80S ribosomes (Fig. 2B). In contrast, when the 40S and 60S ribosomal subunits were first incubated with the purified recombinant 26-kDa protein at 1 mM Mg2+ and the Mg2+ concentration was then raised to 5 mM, a major fraction of the subunits failed to reassociate to form 80S ribosomes (Fig. 2C). These results demonstrate that the recombinant 26-kDa protein can act as a ribosomal subunit anti-association factor. Thus, the 735-bp hypothetical ORF, designated YPR016C in the yeast genomic database, encodes the yeast homologue of mammalian eIF6. The derived amino acid sequence of yeast eIF6 is as predicted previously (32) based on its homology to human eIF6.
FIG. 2.
Ribosomal-subunit anti-association activity of bacterially expressed recombinant yeast eIF6. eIF6 activity was measured by the ability of the protein to prevent the association of 40S and 60S ribosomal subunits at 5 mM Mg2+ to form 80S ribosomes, as described previously (37). Three reaction mixtures, each of 100 μl and containing 20 mM Tris-HCl (pH 7.5), 100 mM KCl, 1 mM MgCl2, 1 mM dithiothreitol, and 1.0 A260 unit of Artemia salina 80S ribosomes, were prepared. Two of the reaction mixtures, A and B, contained no protein factors, while mixture C contained 1.25 μg of purified recombinant yeast eIF6 (Sephadex G-75 fraction). After incubation at 30°C for 5 min, the Mg2+ concentration of mixtures B and C was raised to 5 mM while that of mixture A was kept at 1 mM, and the incubation was continued for another 5 min at 37°C. Each reaction mixture was then chilled in an ice-water bath, loaded onto a 5-ml linear 5 to 30% (wt/vol) sucrose gradient containing 20 mM Tris-HCl (pH 7.5), 100 mM KCl, 1 mM dithiothreitol, and 5 mM MgCl2 (for mixtures B and C) or 1 mM MgCl2 (for mixture A), and centrifuged for 90 min in an SW50.1 rotor at 48,000 rpm. Each gradient was fractionated and the A254 profile was analyzed by using an UA-5 absorbance monitor. (A) No yeast eIF6 added, reaction at 1 mM Mg2+; (B) no yeast eIF6 added, reaction at 5 mM Mg2+; (C) yeast eIF6 added, reaction at 5 mM Mg2+.
To obtain evidence that TIF6 is indeed expressed in yeast cells, we carried out Western blot analysis of a fractionated yeast cell extracts with affinity-purified anti-yeast eIF6 antibodies as a probe. Two distinct immunoreactive polypeptide bands were observed (Fig. 1C, lane 3). One of the immunoreactive polypeptides migrated with the same mobility as purified recombinant yeast eIF6, while the other migrated with a slightly lower mobility. The possibility exists that the slower-migrating polypeptide was derived from the faster-migrating one by posttranslational modifications, e.g., phosphorylation.
Characterization of the TIF6 gene.
The yeast genomic database search indicated that TIF6 is a single-copy gene. This observation was confirmed by Southern blot analysis of HindIII- or EcoRI-digested yeast genomic DNA with the 32P-labeled 735-bp TIF6 ORF fragment as a probe (data not shown). TIF6 is transcribed, yielding a single-size class of RNA of about 1.2 kb (Fig. 1D).
Analysis of the nucleotide sequence of the yeast genomic DNA surrounding the TIF6 ORF indicates that the initiating ATG codon at +1, which is preceded by a translational stop codon TGA at position −12, satisfies the consensus rule of translational start sites, −3AUAAUGG+4 (the start codon is underlined) (14). There are several TATA sequences upstream of the start ATG codon which may serve as promoters for transcription of TIF6 mRNA. Based on this analysis, a 1,742-bp yeast genomic fragment that extends from 500 bp upstream of the translational start codon ATG of TIF6 ORF (position +1) to 507 bp downstream of the translation stop codon TAG (at position +735) was amplified by PCR and cloned into the LEU2-based centromeric expression plasmid pRS315. This plasmid-borne genomic fragment was sufficient to complement a null allele of TIF6, indicating that this 1,742-bp fragment can act as a transcriptional unit and contains the promoter elements of TIF6.
TIF6 is an essential gene.
To determine if TIF6 is essential for mitotic cell growth and viability, we constructed a null allele of the TIF6 gene as described in Materials and Methods. The yeast HIS3 gene was inserted into a unique EcoRV site located at position +362 of the TIF6 ORF, and the resulting construct was introduced into a diploid yeast strain, W303 (Matα/Mata), by homologous recombination. Stable His+ transformants were isolated, and the genomic DNAs of several transformants were subjected to Southern blot and PCR analyses to screen for tif6::HIS3/TIF6 heterozygous diploid yeast cells (data not shown). The resulting diploid strain, KSY601, was then sporulated for tetrad analysis. Of the 18 tetrads, 15 yielded two viable spores and 3 tetrads produced only one viable spore (data not shown). All the viable spores were His−. We conclude that TIF6 is essential for germination and/or cell growth. To determine that TIF6 was the only gene that was disrupted in strain KSY601, the strain was transformed with a centromeric LEU2-plasmid pRS315 containing the TIF6 ORF under the control of glucose-repressible GAL10 promoter, pTM100-TIF6. A Leu+ transformant was then sporulated, and a Leu+ His+ haploid strain was selected. The growth of the Leu+ His+ haploid strain (tif6::HIS3, pTM100-TIF6) was then examined on galactose- and glucose-containing plates. The strain grew on galactose plates but not on glucose plates after a 3-day incubation at 30°C, whereas W303α grew well on both (data not shown). Thus, extrachromosomal expression of TIF6 was sufficient to complement genomic disruption of the TIF6 locus. Taken together, these results show that TIF6 is an essential gene.
Construction and expression of a rapidly degradable form of eIF6 in S. cerevisiae.
To understand the effect of depletion of endogenous eIF6 on cell growth and protein synthesis in yeast cells, we constructed a haploid yeast strain, KSY603, in which the chromosomal copy of the TIF6 gene was inactivated by insertion of HIS3 marker gene and the essential eIF6 function was provided by maintenance of a centromeric plasmid harboring a conditional eIF6 expression system. In this expression system, eIF6 was expressed from a transcription unit containing a protein-destabilizing ubiquitin gene cassette consisting of GAL-UAS-UBI4-R-lacI-HA (20) fused to the NH2 terminus of the TIF6 ORF under the transcriptional control of GAL10 promoter (Fig. 3A). The eIF6 fusion protein synthesized from this construct should contain ubiquitin (Ub) at the NH2 terminus followed by an arginine residue and a 31-amino-acid segment of the lacI repressor that acts as a recognition element for ubiquitin-dependent protein degradation and finally an HA tag. Deubiquitination of the eIF6 fusion protein in yeast cells exposes arginine (R) as the NH2-terminal amino acid and should lead to rapid degradation of eIF6 protein on the basis of the N-end rule (2). As expected, strain KSY603 (GAL10::UbTIF6) grew well on plates containing galactose as the carbon source but did not form detectable colonies on plates containing glucose as the sole carbon source (data not shown). In liquid cultures containing galactose, both the wild-type W303α and strain KSY603 grew with a doubling time of about 2.5 h (Fig. 3B, left). However, when an exponentially growing culture of the KSY603 (GAL10::UbTIF6) was shifted from galactose- to glucose-containing medium to repress transcription of the TIF6 gene from the GAL10 promoter, the growth rate of KSY603 began to decrease after about 5 h and growth ceased after about 20 h (Fig. 3B, right). The growth inhibition at the earlier stages were reversible, since cells grown in glucose for about 8 h were still able to form colonies on galactose-containing plates (data not shown).
To determine whether the arrest of cell growth of KSY603 correlated with the depletion of eIF6, we carried out immunoblot analysis of the level of eIF6 protein in cell lysates following the shift from galactose to glucose medium. As shown in Fig. 3C (left), the level of eIF6 in cell lysates prepared from nonrepressed KSY603 cells remained fairly similar during the growth period. In contrast, there was a rapid decline of eIF6 fusion protein in KSY603 cells after transfer to glucose medium. After about 4 h, there was virtually no detectable eIF6 fusion protein in these cell lysates (Fig. 3C, right). Correlation of the growth of KSY603 cells in glucose-containing medium with the level of eIF6 in these cells thus clearly shows that depletion of eIF6 resulted in a growth defect in KSY603. However, even after eIF6 levels were reduced to barely detectable in 4 h, cell growth did not come to an immediate halt. Rather, there was a progressive increase in the doubling time until about 20 h, when cell growth was nearly completely arrested.
Analysis of protein synthesis in vivo upon depletion of eIF6.
The effect of depletion of eIF6 on protein synthesis in yeast cells was investigated with strain KSY603 (GAL10::UbTIF6). An exponentially growing culture of this strain was transferred from galactose- to glucose-containing medium, and the rate of protein synthesis was monitored by measuring the rate of incorporation of [35S]methionine into cellular proteins over the growth period. Figure 4A shows that after about 2 h following the shift from galactose- to glucose-containing medium, the rate of protein synthesis in this strain was reduced to nearly 50% of the initial rate, and that after about 8 h, the rate of protein synthesis was about 25% of the initial rate. This inhibition of protein synthesis in KSY603 cells following the shift from galactose- to glucose-containing medium seemed to be general, because the production of most, if not all, of the polypeptides appeared to be equally reduced (Fig. 4B). These results show that eIF6 plays a role in translation of mRNAs in yeast cells.
FIG. 4.
Inhibition of protein synthesis in eIF6-depleted cells. Exponentially growing cultures of W303α or KSY603 growing in galactose medium lacking methionine (SGal-Met medium) were harvested, and approximately 10 A260 units of cells was suspended in 150 ml of either SD-Met (glucose) or SGal-Met medium and grown at 30°C. At the indicated times, 1 A260 unit of cells from each culture was harvested and suspended in 300 μl of either SD-Met or SGal-Met medium containing 50 μCi of [35S]methionine (1175 Ci/mmol). (A) Protein synthesis rates were determined as described in Materials and Methods. The rate of protein synthesis at each time point was calculated as counts of 35S radioactivity incorporated per microgram of protein per minute. (B) Each yeast strain, as indicated, were pulse-labeled for 5 min with [35S]methionine in SD-Met medium, chased for 3 min with 1.5 mM nonradioactive methionine, and lysed. Similar amounts of total protein were separated by SDS-PAGE and autoradiographed.
To investigate whether the inhibition of protein synthesis observed in eIF6-depleted cells was due to a defect in the initiation step, we analyzed the polyribosome profile of exponentially growing cultures of wild-type W303α as well as KSY603 cells growing in galactose- or glucose-containing medium (Fig. 5). The eIF6-depleted KSY603 cells growing in glucose-containing medium showed a marked reduction in the number of large polyribosomes compared with that in wild-type W303α cells growing in glucose-containing medium (compare Fig. 5E and F with Fig. 5A and B) or KSY603 cells growing in galactose-containing medium (Fig. 5C and D). However, in contrast to cells depleted of an essential initiation factor, where a reduction in the size of polysomes is accompanied by an increase in the number of 80S ribosomes and of free ribosomal subunits, the decrease in polyribosome content in eIF6-depleted KSY603 cells was accompanied by a decrease in the number of both 80S monosomes and 60S ribosomal subunits. Furthermore, there was concomitant accumulation of half-mer polyribosomes (compare Fig. 5E and F with Fig. 5B and C).
FIG. 5.
Analysis of the polyribosome profiles of eIF6-depleted yeast cells. Exponentially growing cultures of KSY603 or W303α in YPGal medium were harvested and resuspended in either glucose (YPD) or YPGal medium such that the initial A600 of each culture was about 0.04 U. At about 5 h (E) and 10 h (B, C, and F) after the shift, 50 ml of each culture was treated with 50 μg of cycloheximide per ml and chilled cells were harvested, washed, and lysed as described in Materials and Methods. About 10 A260 units of each cell lysate was subjected to 7 to 47% (wt/vol) sucrose gradient centrifugation. Each gradient was fractionated in an ISCO gradient fractionator, and the A254 profile was analyzed in an ISCO UA-5 absorbance monitor. The positions of the half-mer polysomes are indicated by arrowheads (▾) in panels E and F.
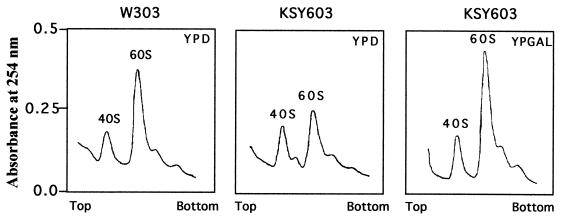
We also analyzed the relative amounts of 40S and 60S ribosomal subunits in KSY603 cells depleted of eIF6 (Fig. 6). In these cells, the level of 60S ribosomal subunits was significantly reduced relative to that of 60S subunits in KSY603 growing in YPGal or W303α shifted to YPD for 10 h (Fig. 6). Determination of the 60S-to-40S subunit A254 ratio showed that this ratio was about 2.0 for wild-type W303α or KSY603 in YPGal but decreased to about 1.3 for eIF6-depleted KSY603 in YPD.
FIG. 6.
eIF6 depletion results in a decrease in 60S subunits levels. Total ribosomes were isolated from strains W303α and KSY603 after 10 h of growth in either YPD or YPGal, dissociated into 40S and 60S ribosomal subunits, and sedimented through 15 to 40% sucrose gradients as described in Materials and Methods.
In agreement with these results, in a separate study (results not shown), we carried out an immunoblot analysis of the levels of 60S ribosomal proteins L3 and L32 and 40S ribosomal protein S12 in cell lysates following the shift of KSY603 from YPGal to YPD medium. We observed that after about 5 to 6 h there was a progressive decline in the levels of both L3 and L32 proteins in these cells with time. After about 15 h, there was no detectable L3 or L32 protein in these cell lysates. In contrast, the level of 40S ribosomal protein S12 in the cell lysates remained relatively constant during the entire growth period following the shift from galactose- to glucose-containing medium.
These results indicated that depletion of eIF6 causes a deficiency in 60S ribosomal subunits, resulting in a stoichiometric imbalance between 40S and 60S ribosomal subunits. This stoichiometric imbalance causes a subunit-joining defect, resulting in the accumulation of half-mer polysomes in eIF6-depleted cells.
Effect of depletion of eIF6 on translation of mRNAs in vitro.
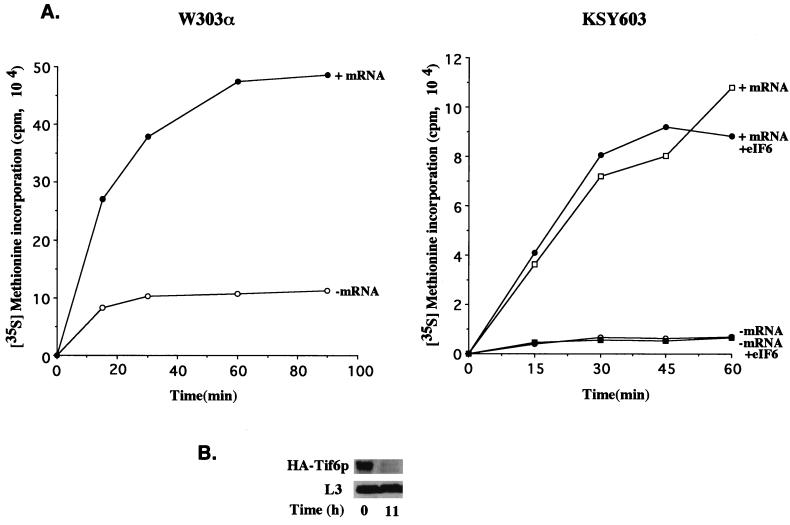
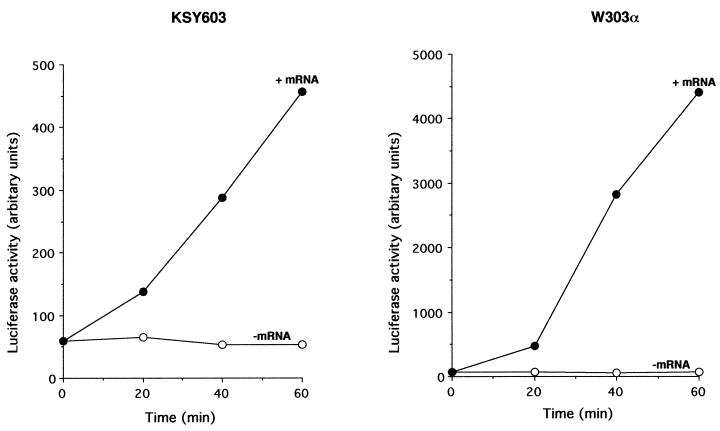
We used eIF6-depleted KSY603 cells to prepare a yeast cell-free translation system to investigate the effect of eIF6 depletion on translation of mRNAs in vitro (Fig. 7). Cell-free translation extracts prepared from exponentially growing cultures of wild-type W303α cells in glucose-containing medium were active in translation of total yeast poly(A)+ RNA without any exogenously added protein factors (Fig. 7A, left). When similar cell extracts were prepared from KSY603 (GAL10::UbTIF6) cells that were grown in glucose medium for about 11 h until eIF6 was virtually depleted from the cells (Fig. 7B), they remained active in translation of yeast poly(A)+ RNA (Fig. 7A, right). Similar results were obtained when luciferase mRNA was translated in wild-type and eIF6-depleted KSY603 extracts (Fig. 8). Additionally, analysis of the 35S-labeled translation products formed in the experiments in Fig. 7 by SDS-PAGE followed by autoradiography showed that similar polypeptides were synthesized in both the wild-type W303α and the eIF6-depleted cell extracts (data not shown). Taken together, these results show that eIF6 does not function as a translation initiation factor in vitro. If eIF6 is a bona fide translation initiation factor, its absence would have caused the cell extracts to be nearly totally inactive in translation, as was observed in eIF5-depleted cell extracts (15) or in cell extracts containing an inactive translation initiation factor, e.g., eIF4E(1) or eIF4A(5). Translation in these extracts was restored by the addition of the depleted initiation factor. It should be noted, however, that the translation efficiency of eIF6-depleted cell extracts was somewhat reduced compared to that of wild-type extracts (Fig. 7A, compare the [35S]methionine incorporation observed when comparable amounts of the two extracts were used). This is in agreement with the decrease in the rate of translation observed in eIF6-depleted cells (Fig. 4). Addition of functionally active eIF6 to eIF6-depleted lysates did not significantly increase the translation efficiency (Fig. 7A, right). Furthermore, when these extracts were supplemented with either additional 60S ribosomal subunits or 60S subunits plus eIF6, there was no effect on translation efficiency.
FIG. 7.
In vitro translation of total yeast poly(A)+ RNA in eIF6-depleted cell extracts. (A) Exponentially growing cultures of KSY603 or W303α cells in YPGal medium were harvested and suspended in glucose (YPD) medium such that the initial A600 was about 0.03 U. At about 11 h after the shift, the cells were harvested and translation cell extracts were prepared, incubated, and analyzed for [35S]methionine incorporation into proteins as described in Materials and Methods. Where indicated, 5 μg of total yeast poly(A)+ RNA (mRNA) and 0.5 μg of recombinant yeast eIF6 were added to 50 μl of reaction mixtures containing 25 μCi of [35S]methionine and all the other components required for translation including micrococcal nuclease-treated cell extracts containing approximately 150 μg of proteins. Aliquots (8 μl) from 50-μl reaction mixtures were withdrawn at the indicated times and analyzed for [35S]methionine incorporation into proteins as described in Materials and Methods. (B) At the indicated times following the shift from YPGal to YPD medium, cell lysates were prepared and 200 μg of protein from each lysate was subjected to Western blot analysis with either anti-HA monoclonal antibodies or anti-L3 antibodies as probes. It should be noted that a relatively large amount of cell lysates was analyzed in the Western blot to ensure that the lysates had no detectable levels of eIF6. Under these conditions, the level of the 60S ribosomal protein L3 was also decreased. However, no change in the level of L3 in cell lysates between 0 and 11 h was apparent in the immunoblot analysis, presumably because the amount of L3 in cell lysates analyzed was still large.
FIG. 8.
Translation of luciferase mRNA in cell extracts. Translation cell extracts were prepared from W303α (TIF6) or KSY603 (GAL10::UbTIF6) cells grown in glucose-containing medium for 11 h as described in Materials and Methods. Where indicated, 2 μg of capped luciferase mRNA was added to 50-μl reaction mixtures along with all the other components necessary for in vitro translation. The reactions were carried out at 25°C. At the indicated times, an aliquot of each reaction mixture was assayed for luciferase activity as described by Russel et al. (25).
It is not surprising that extracts prepared from eIF6-depleted KSY603 cells were rather inefficient in translation of mRNAs. In such cell extracts, concomitant with a twofold reduction in the level of total 60S ribosomal subunits, the synthesis of all cellular proteins including the proteins required for translation of mRNAs (e.g., aminoacyl-tRNA synthetases; initiation, elongation, and termination factors; and ribosomal proteins) will be significantly reduced. However, the amount of all translations components including 60S ribosomal subunits remaining was presumably still sufficient to carry out in vitro translation of exogenously added mRNAs. It should be noted that while these extracts were depleted of eIF6, a significant level of 60S ribosomal protein L3 (Tcm1p) was still present (Fig. 7B).
Association of eIF6 with free 60S ribosomal subunits in yeast cells.
Mammalian eIF6 has been shown to bind to the 60S ribosomal subunit in an approximately 1:1 ratio (37) and to prevent its association with the 40S ribosomal subunit. To investigate whether eIF6 associates with 60S ribosomal subunits in yeast cells, lysates of an exponentially growing culture of yeast strain KSY603 were subjected to sucrose gradient centrifugation. Aliquots from the gradient fractions were then analyzed by Western blotting with anti-HA antibodies to identify the location of the HA-eIF6 fusion protein (Fig. 9). A 60S ribosomal protein, L3 (Tcm1p), detected by anti-L3 antibodies (38) was used as a marker to identify the positions of 60S ribosomal proteins in the gradient (Fig. 9). As expected, 60S ribosomal protein L3 was found in the regions of the gradient containing free 60S ribosomal subunits, 80S monosomes, and polysomal ribosomes but was absent from fractions containing free 40S ribosomal subunits. In contrast, a major fraction of the expressed eIF6 fusion protein cosedimented only with free 60S ribosomal subunits but was absent from fractions containing 80S ribosomes, polysomes, or 40S ribosomal subunits (Fig. 9). The association of eIF6 with 60S ribosomal subunits is sensitive to high KCl concentration (Fig. 9). When sucrose gradient centrifugation of cell extracts was carried out with buffers containing 0.7 M KCl, eIF6 fusion protein was not associated with 60S ribosomal subunits. The protein was found in the lighter fractions (fractions 2 and 3) (Fig. 9, bottom). These results suggest that eIF6 is not a component of the 60S ribosomal particle but, rather, is a protein that associates with the 60S ribosomal subunit.
FIG. 9.
Association of eIF6 with 60S ribosomal subunits in yeast cells. An exponentially growing culture of KSY603 was harvested, the cells were washed and lysed, and the cell lysate was subjected to 5 to 30% sucrose gradient centrifugation to separate polysomes, free 80S ribosomes, and ribosomal subunits as described in the legend to Fig. 5 and Materials and Methods. Fractions from the gradient were collected, and proteins were precipitated with 10% trichloroacetic acid, analyzed by SDS-PAGE, and immunoblotted with either anti-L3 antibodies or anti-HA antibodies as probes. Fractions containing 40S and 60S subunits, 80S monosomes, and polysomes are indicated. Protein L3 was used as a marker for 60S subunit sedimentation.
DISCUSSION
It is now generally accepted that after termination of mRNA translation, 80S ribosomes are released from the polysomal complex and are then in equilibrium with their subunits. Since a pool of free ribosomal subunits is necessary for initiation of protein synthesis, a mechanism must exist for maintaining a pool of both subunits or for generating them from 80S ribosomes (16, 17). Two protein factors, eIF3 and eIF6, have been implicated as being responsible for maintaining a pool of ribosomal subunits required for initiation of protein synthesis in eukaryotic cells (17). The multisubunit initiation factor eIF3 was originally isolated from rabbit reticulocyte lysates on the basis of its near-absolute requirement for translation of globin mRNA by a protein-synthesizing system with partially purified reconstituted proteins (3, 29, 31). Purified eIF3 was shown (3) to specifically bind to the 40S ribosomal subunit in the absence of other components of translation initiation and to prevent its association with the 60S ribosomal subunit. In addition to the ribosomal subunit anti-association property, eIF3 plays a direct role at multiple steps in initiation of translation of mRNA (17). In contrast to eIF3, which was isolated based on a direct translation assay, eIF6, a 26-kDa monomeric protein, was isolated from both wheat germ (26, 27) and mammalian (21, 32, 37) cell extracts based on an in vitro assay that measured the ability of the factor to bind specifically to the 60S ribosomal subunit and prevent its association with the 40S ribosomal subunit. Because of this ribosomal-subunit anti-association property, eIF6 was thought to play a direct role in the generation of ribosomal subunits required for initiation of protein synthesis. The protein was therefore classified as a eukaryotic translation initiation factor (17), although, unlike eIF3, its role in translation of mRNA has never been demonstrated. Thus, it was important to show that the 26-kDa eIF6, purified on the basis of an in vitro partial reaction, is indeed involved in the translation of natural mRNAs before it can be regarded as a bona fide translation factor.
Data presented in this paper clearly show that as yeast cells were depleted of eIF6, the rate of protein synthesis was also inhibited. However, analysis of the polysome profiles of eIF6-depleted cells showed a reduction not only in the amounts of polysomes but also in the amounts of both 80S monosomes and free 60S ribosomal subunits. The selective reduction of total 60S ribosomal subunits with respect to 40S ribosomal subunits in eIF6-depleted cells caused a stoichiometric imbalance between 60S and 40S ribosomal subunits, resulting in the formation of half-mer polysomes. The formation of half-mer polyribosomes was presumably due to lack of joining of 60S ribosomal subunits to the stalled 43S preinitiation complexes on the 5′ UTR of mRNA templates. Such polysome-ribosome profiles are not characteristic of cells either containing an inactive translation initiation factor or depleted of a translation initiation factor. If eIF6 plays an essential role in the initiation phase of protein synthesis, its depletion from yeast cells would have caused not only a reduction in polysome content but also a simultaneous increase (not decrease) in the amounts of both 80S monosomes and free 60S and 40S ribosomal subunits, as was observed in yeast cells depleted of an essential initiation factor, e.g., eIF5 (15). Furthermore, we observed that lysates of yeast cells depleted of detectable levels of eIF6 but still containing a significant concentration of 60S ribosomal subunits were still active in translation of mRNA in vitro. Taken together, these results strongly suggest that eIF6 is not a bona fide translation initiation factor. Decreased protein synthesis rates and accumulation of half-mer polysomes observed in eIF6-depleted cells were most probably due primarily to selective reductions in the amount of 60S ribosomal subunits. It is also likely that reduction in the level of 60S ribosomal subunits will be accompanied by a decrease in the rate of synthesis of all cellular proteins, including the proteins involved in translation of mRNAs. This may explain the decrease in the rate of protein synthesis observed in vivo and in vitro in eIF6-depleted cells.
If eIF6 is not a translation factor, what essential cellular functions does it perform? More specifically, how does eIF6 maintain the steady-state level of 60S subunits in cells? Reductions in the level of 60S subunits with respect to 40S subunits and concomitant accumulation of half-mer polysomes have previously been observed for depletion or mutation of several 60S ribosomal proteins (6–8, 18, 19, 23) as well as nonribosomal proteins like Nip7p (39) and Sqt1p (7). These proteins, like eIF6, which are not constituents of 60S ribosomal subunits, were found to be associated with free 60S ribosomal subunits but not with 80S monosomes or with polysomes. The depletion of Nip7p from yeast cells has been shown to cause a significant defect in pre-rRNA processing in the nucleolus, which presumably leads to the depletion of 60S subunits (39). Sqt1p, on the other hand, appears to be involved in a late step in 60S subunit assembly or modification in the cytoplasm (7). The association of eIF6 with mature free 60S subunits (Fig. 9) suggests a role for this protein in some aspects of the late 60S subunit maturation step in the cytoplasm. On the other hand, recent immunofluorescence studies (data not shown) have indicated the presence of eIF6 also in the nucleus, supporting a role for this protein in pre-rRNA processing or ribosome assembly, as has been shown for Nip7p (39). The possibility also exists that the binding of eIF6 to free 60S ribosomal subunits in the cytoplasm is required for the stability of the 60S particle. Furthermore, although eIF6 does not act directly as a translation initiation factor, its binding to 60S ribosomal subunits may regulate the subunit-joining step during the initiation phase of protein synthesis. It has been reported (37) that 60S ribosomal subunits containing bound mammalian eIF6 are incapable of joining the 40S initiation complex to form the 80S initiation complex. Clearly, a mechanism must exist for the release of eIF6 from the 60S subunit either prior to or concomitant with the joining of the subunit to the 40S initiation complex. Further work is now directed toward understanding the cellular function(s) of eIF6.
Recently a novel β4 integrin-binding protein, designated p27BBP, has been identified through yeast two-hybrid screening (4). Sequence analysis of p27BBP revealed that the protein is identical to mammalian eIF6 reported from our laboratory (32). However, although β4 integrin is expressed mostly in epithelial cells, eIF6 is present in every cell type of higher eukaryotes (32) as well as in unicellular yeasts, where no β4 homologue has yet been found. It is likely that the primary function of eIF6 is in the biogenesis and/or maintenance of the stable population of 60S ribosomal subunits in all cell types. It may, however, perform additional functions in higher eukaryotes in coupling protein synthesis with the cellular signalling pathway through its effect on ribosome biogenesis.
ACKNOWLEDGMENTS
We are particularly grateful to Jonathan Warner and Tomohiro Matsumoto of this institution for many stimulating discussions on yeast methodologies. We are also indebted to Jonathan Warner, Stewart Shuman of Sloan Kettering Cancer Research Center, New York, and our former colleague, Jayanta Chaudhuri, now of Harvard Medical School, for critically reading the manuscript. We also acknowledge gratefully the assistance of Uttiya Basu of this laboratory for carrying out the experiment on the growth curves of yeast strains presented in Fig. 3. Finally, we thank Lyn Broccoli and Leona Connolly for their patience in the preparation of the manuscript.
This work was supported by grant GM15399 from the National Institutes of Health and by Cancer Core Support Grant P30CA 13330 from the National Cancer Institute.
Footnotes
This paper is dedicated to Jerard Hurwitz on the occasion of his 70th birthday for his enormous original scientific contributions in the field of nucleic acid biosynthesis.
REFERENCES
- 1.Altman M, Sonenberg N, Trachsel H. Translation in Saccharomyces cerevisiae: initiation factor 4E-dependent cell-free system. Mol Cell Biol. 1989;9:4467–4472. doi: 10.1128/mcb.9.10.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmair A, Varshavsky A. The degradation signal in a short-lived protein. Cell. 1989;56:1019–1032. doi: 10.1016/0092-8674(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 3.Benne R, Hershey J W B. Purification and characterization of initiation factor IF-E3 from rabbit reticulocytes. Proc Natl Acad Sci USA. 1976;73:3005–3009. doi: 10.1073/pnas.73.9.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biffo S, Sanvito F, Costa S, Preve L, Pignatelli R, Spinardi L, Marchisio P C. Isolation of a novel beta 4 integrin-binding protein (p27BBP) highly expressed in epithelial cells. J Biol Chem. 1997;272:30314–30321. doi: 10.1074/jbc.272.48.30314. [DOI] [PubMed] [Google Scholar]
- 5.Blum S, Mueller M, Schmid S R, Linder P, Trachsel H. Translation in Saccharomyces cerevisiae: initiation factor 4A-dependent cell-free system. Proc Natl Acad Sci USA. 1989;86:6043–6046. doi: 10.1073/pnas.86.16.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deshmukh M, Tsay Y-F, Paulovich A G, Woolford J L., Jr Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol Cell Biol. 1993;13:2835–2845. doi: 10.1128/mcb.13.5.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisinger D P, Dick F A, Denke E, Trumpower B L. SQT1, which encodes an essential WD domain protein of Saccharomyces cerevisiae, suppresses dominant-negative mutations of the ribosomal protein gene QSR1. Mol Cell Biol. 1997;17:5146–5155. doi: 10.1128/mcb.17.9.5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisinger D P, Dick F A, Trumpower B L. Qsr1p, a 60S ribosomal subunit protein, is required for joining of 40S and 60S subunits. Mol Cell Biol. 1997;17:5136–5145. doi: 10.1128/mcb.17.9.5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh S, Chevesich J, Maitra U. Further characterization of eukaryotic initiation factor 5 from rabbit reticulocytes. Immunochemical characterization and phosphorylation by casein kinase II. J Biol Chem. 1989;264:5134–5140. [PubMed] [Google Scholar]
- 10.Goss D J, Rounds D, Harrigan T, Woodley C L, Wahba A J. Effects of eucaryotic initiation factor 3 on eucaryotic ribosomal subunit equilibrium and kinetics. Biochemistry. 1988;27:1489–1494. doi: 10.1021/bi00405a014. [DOI] [PubMed] [Google Scholar]
- 11.Hussain I, Leibowitz M J. Translation of homologous and heterologous messenger RNAs in a yeast cell-free system. Gene. 1986;46:13–23. doi: 10.1016/0378-1119(86)90162-9. [DOI] [PubMed] [Google Scholar]
- 12.Johnston M, Davis R W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang H A, Hershey J W B. Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae. J Biol Chem. 1994;269:3934–3940. [PubMed] [Google Scholar]
- 14.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 15.Maiti T, Maitra U. Characterization of translation initiation factor 5 (eIF5) from Saccharomyces cerevisiae. Functional homology with mammalian eIF5 and the effect of depletion of eIF5 on protein synthesis in vivo and in vitro. J Biol Chem. 1997;272:18333–18340. doi: 10.1074/jbc.272.29.18333. [DOI] [PubMed] [Google Scholar]
- 16.Maitra U, Stringer E A, Chaudhuri A. Initiation factors in protein biosynthesis. Annu Rev Biochem. 1982;51:869–900. doi: 10.1146/annurev.bi.51.070182.004253. [DOI] [PubMed] [Google Scholar]
- 17.Merrick W C, Hershey J W B. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 31–69. [Google Scholar]
- 18.Moritz M, Paulovich A G, Tsay Y F, Woolford J L., Jr Depletion of yeast ribosomal proteins L16 or rp59 disrupts ribosome assembly. J Cell Biol. 1990;111:2261–2274. doi: 10.1083/jcb.111.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moritz M, Pulaski B A, Woolford J L., Jr Assembly of 60S ribosomal subunits is perturbed in temperature-sensitive yeast mutants defective in ribosomal protein L16. Mol Cell Biol. 1991;11:5681–5692. doi: 10.1128/mcb.11.11.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park E-C, Finley D, Szostak J W. A strategy for the generation of conditional mutations by protein destabilization. Proc Natl Acad Sci USA. 1992;89:1249–1252. doi: 10.1073/pnas.89.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raychaudhuri P, Stringer E A, Valenzuela D M, Maitra U. Ribosomal subunit antiassociation activity in rabbit reticulocyte lysates. Evidence for a low molecular weight ribosomal subunit antiassociation protein factor (Mr = 25,000) J Biol Chem. 1984;259:11930–11935. [PubMed] [Google Scholar]
- 22.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Rotenberg M O, Moritz M, Woolford J L., Jr Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev. 1988;2:160–172. doi: 10.1101/gad.2.2.160. [DOI] [PubMed] [Google Scholar]
- 24.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 25.Russel P J, Hambridge S J, Kirkegaard K. Direct introduction and transient expression of capped and non-capped RNA in Saccharomyces cerevisiae. Nucleic Acids Res. 1991;19:4949–4953. doi: 10.1093/nar/19.18.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russell D W, Spremulli L L. Purification and characterization of a ribosome dissociation factor (eukaryotic initiation factor 6) from wheat germ. J Biol Chem. 1979;254:8796–8800. [PubMed] [Google Scholar]
- 27.Russell D W, Spremulli L L. Mechanism of action of the wheat germ ribosome dissociation factor: interaction with the 60 S subunit. Arch Biochem Biophys. 1980;201:518–526. doi: 10.1016/0003-9861(80)90540-8. [DOI] [PubMed] [Google Scholar]
- 28.Sachs A B, Davis R W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 29.Safer B, Adams S L, Kemper W M, Berry K W, Lloyd M, Merrick W C. Purification and characterization of two initiation factors required for maximal activity of a highly fractionated globin mRNA translation system. Proc Natl Acad Sci USA. 1976;73:2584–2588. doi: 10.1073/pnas.73.8.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Schreier M H, Erni B, Staehelin T. Initiation of mammalian protein synthesis. I. Purification and characterization of seven initiation factors. J Mol Biol. 1977;116:727–753. doi: 10.1016/0022-2836(77)90268-6. [DOI] [PubMed] [Google Scholar]
- 32.Si K, Chaudhuri J, Chevesich J, Maitra U. Molecular cloning and functional expression of a human cDNA encoding translation initiation factor 6. Proc Natl Acad Sci USA. 1997;94:14285–14290. doi: 10.1073/pnas.94.26.14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas A, Goumans H, Voorma H O, Benne R. The mechanism of action of eukaryotic initiation factor 4C in protein synthesis. Eur J Biochem. 1980;107:39–45. doi: 10.1111/j.1432-1033.1980.tb04621.x. [DOI] [PubMed] [Google Scholar]
- 35.Thompson H A, Sadnik I, Scheinbuks J, Moldave K. Studies on native ribosomal subunits from rat liver. Purification and characterization of a ribosome dissociation factor. Biochemistry. 1977;16:2221–2230. doi: 10.1021/bi00629a028. [DOI] [PubMed] [Google Scholar]
- 36.Trachsel H, Staehelin T. Initiation of mammalian protein synthesis. The multiple functions of the initiation factor eIF-3. Biochim Biophys Acta. 1979;565:305–314. doi: 10.1016/0005-2787(79)90207-7. [DOI] [PubMed] [Google Scholar]
- 37.Valenzuela D M, Chaudhuri A, Maitra U. Eukaryotic ribosomal subunit anti-association activity of calf liver is contained in a single polypeptide chain protein of Mr = 25,500 (eukaryotic initiation factor 6) J Biol Chem. 1982;257:7712–7719. [PubMed] [Google Scholar]
- 38.Vilardell J, Warner J W. Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol Cell Biol. 1997;17:1959–1965. doi: 10.1128/mcb.17.4.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zanchin N I T, Roberts P, DeSilva A, Sherman F, Goldfarb D S. Saccharomyces cerevisiae Nip7p is required for efficient 60S ribosome subunit biogenesis. Mol Cell Biol. 1997;17:5001–5015. doi: 10.1128/mcb.17.9.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]