Abstract
In recent years, the development of novel chemistry routes for the synthesis of organic compounds has attracted special attention. 2,3-Diaminophenazine (DAP), a derivative of Phenazine, is a large group of nitrogen-containing heterocyclic compound with diverse chemical structure and various biological activities, such as antibacterial, antimicrobial, anti-inflammatory, and anticancer activities. Phenazine is a fluorescent molecule with wide range of biological properties. Therefore, a novel chemical methodology is required for effective synthesis of this product. Numerous oxidants can easily oxidize ortho-phenylenediamine (OPD) to create luminous DAP. This article discusses a simple, sustainable, and safe way to synthesize DAP using water as a green solvent and Jeanbandyite as a catalyst. Mass spectrometry, 1H-NMR and 13C-NMR were used to characterize the molecule, and the catalytic efficacy of Jeanbandyite was assessed. The cellular uptake and cytotoxicity of DAP were investigated to determine whether DAP can be used as a bioprobe in bioapplications. Finally, DAP binding to DNA was methodically performed and confirmed using molecular docking.
Keywords: Catalysis, Cellular internalization, Green chemistry, Jeanbandyite, Phenazine
Subject terms: Chemistry, Materials science
Introduction
Phenazine and its derivatives are electron-deficient π-systems containing a pyrazine ring with two annulated benzene rings1. The electronic properties and positions of the functional groups determine the differences in the chemical and physical characteristics of the Phenazine derivatives. Due to its rich organic chemistry and strong luminescence properties, 2,3-diaminophenazine (DAP), a derivative of Phenazine, has garnered much attention in the field of chemistry because of its fascinating features. A wide range of biological activities, such as antibacterial, neuroprotective, antiparasitic, anti-inflammatory, anticancer, and antitumor effects makes the derivatives useful as medications2–6. Their luminous properties are particularly intriguing because they can be used as organic semiconductors, photosensors, lactose- and glucose-sensitive, and electroluminescent devices7–9. Therefore, pharmacologists and chemists both are interested in the development of Phenazines derivatives.
The general approach for the synthesis of Phenazine involves the condensation reaction of nitrobenzene and aniline in the presence of an alkali medium10. Synthetic strategies of environmental friendliness, use of less hazardous substances, minimized waste using catalytic reactions, and low cost are the preliminary targets of organic synthesis. Numerous limitations of different synthetic techniques have been reported, such as controlling the reaction, hazardous metal contamination, and use of toxic solvents. These drawbacks have inhibited the use of Phenazine derivatives as theranostic molecules. Therefore, there is an urgent need to develop synthetic strategies that use less toxic solvents or materials, green catalysts, and which are environment friendly11,12.
In recent years, catalysts have attracted the interest of researchers owing to their wide range of application13,14. Among two types of catalysts heterogeneous catalysts are preferred over homogenous catalysts because they are easy to separate from the mixture; therefore, the chance of metal contamination is very low, which makes heterogeneous catalyst superior15. Separation techniques from the reaction mixture involve various methods such as filtration, centrifugation, vacuum distillation, and magnetic separation, among which magnetic separation is a fast, simple, and efficient process. Thus, a heterogeneous catalyst with superparamagnetic characteristics can be easily separated from the reaction mixture using an external magnet, thereby potentially increasing the value of the catalyst.
Perovskite materials are currently one of the most useful materials, with the general formula ABX316. In recent years, many perovskites have been used as catalysts in organic reactions because of their structural flexibility and controllable physiochemical properties17–19. Among the perovskite family, Jeanbandyite which is a hydroxide perovskite with superparamagnetic properties, was identified in Bolivia and has not yet been well explored. A cost-effective synthesis technique would arise from the use of magnetic Jeanbandyite, which would make it easy to replenish the catalyst by simply applying an external magnetic field20.
Numerous chemical compounds with fluorescent properties have been investigated for their internalization into cells21. As DAP is luminous, we envisioned that it could penetrate cells for cell imaging, making it a crucial tool for drug administration and other cellular applications. Since organic compounds may be used to treat cancer or tumors, their interactions with DNA have drawn much research attention. Numerous chemical compounds have been observed to interact with DNA either through weak interactions, such as π–π stacking and ionic interactions, or intercalation22. Therefore, in this study, a sustainable and facile synthesis strategy for DAP was reported using a hydroxide perovskite, and the cellular uptake, cytotoxicity, and DNA binding activity of the synthesized compound were determined by fluorescence quenching experiments, which were further validated by molecular docking studies (Figs. 1 and 2).
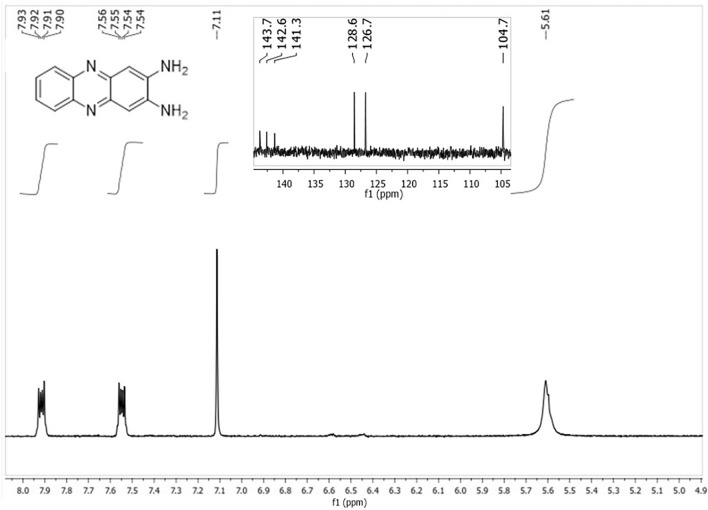
Fig. 1.
1H-NMR and 13C-NMR spectra (inset) of DAP.
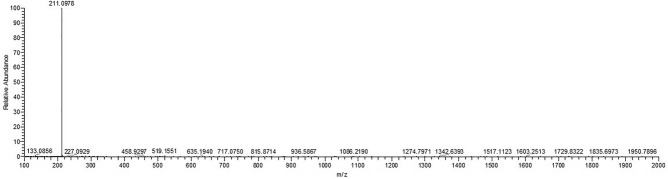
Fig. 2.
Mass spectra of DAP.
Results and discussions
XRD was used to determine the structure of the synthesized catalyst. At room temperature, the XRD pattern of FeSnO(OH)5 Jeanbandyite (Fig. S1a) was obtained within the (2θ) range of 10–90°. The XRD pattern of the studied sample indicates the existence of the Jeanbandyite iron-tin-oxide-hydroxide Fe3+Sn4+O(OH)5 with the cubic space group Pn-3 (ICDD# 01-074-1745), revealing that this compound adopts a cubic crystal lattice20. Figure S1b depicts the FT-IR spectra of Jeanbandyite, from which various distinguishing bands were observed. The hydroxyl (–OH) group is responsible for the broad band near 3082 cm−1, while the O–H bending vibrations of adsorbed H2O are responsible for the peak at 1570 cm−1, the Sn–OH bending vibrations are responsible for the sharp peak observed at 1186 cm−1, and the Fe–OH formed by the coordination of iron ions and H2O is responsible for the peak at 797 cm−123. Moreover, the peak at 569 cm−1 is attributed to the stretching modes of Sn–O and Fe–O24. As a result, the FT-IR data provided more evidence for the formation of Jeanbandyite. One of the primary features of the synthesized compound is their magnetic behavior. To measure the magnetic moment, a vibrating sample magnetometer (VSM) analysis was performed at 300 K and in the range of − 5 to + 5 T (Fig. S1c). The saturation magnetization (Ms) and retentivity (Mr) were 15.62 emu/gm and 0.6527 emu/gm respectively. The superparamagnetic behavior of Jeanbandyite was validated by the lack of any hysteresis loop (negligible hysteresis and squareness ratio (Mr/Ms) of approximately 0.041)25. The morphology of the sample was investigated by SEM (Fig. S1d) and TEM (Fig. S1e), and it is evident from these techniques that the compound has a cubic grain morphology with an average grain size of approximately 58 nm. This observation was further corroborated by the STEM images (Fig. S1f). These results are in line with those of previously published studies that demonstrated the cubic shape of Jeanbandyite24,26,27.
Characterization of DAP
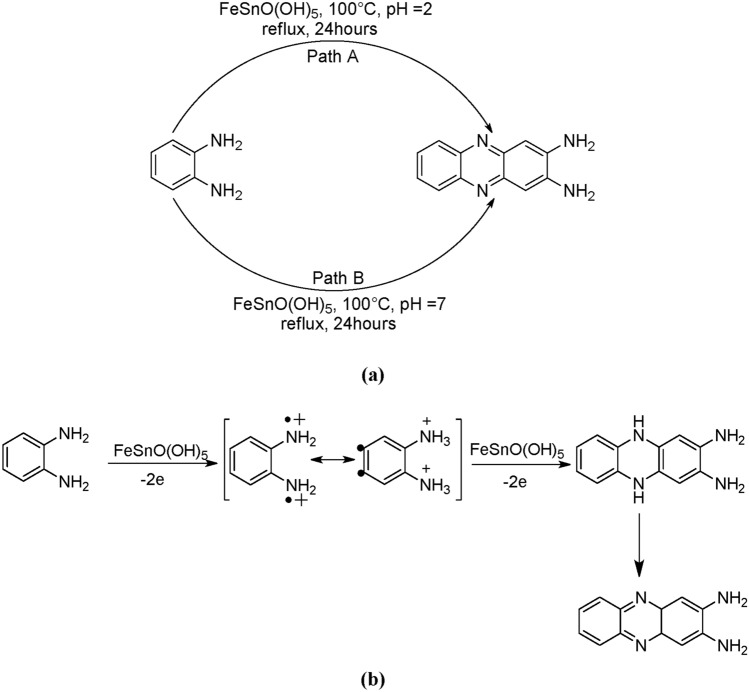
DAP was synthesized at two different pH values (2 and 7), with a maximum yield at pH ~ 2 compared to that at pH ~ 7 (24%), having purity more than 95% (Fig. S2). Which indicates that pH plays an important role in DAP synthesis (Scheme 1a) (Figs. 1, 2) .
Scheme 1.
(a) Synthetic route and (b) Plausible mechanism for DAP formation.
Catalytic recyclability
An important aspect that must be addressed for the reaction using the Jeanbandyite catalyst is the possibility of catalyst recycling. In the best case, it is possible to recover and reutilize the solid catalyst several times before it is completely deactivated. Consequently, the reutilization of the Jeanbandyite catalyst was investigated for the synthesis of DAP from OPD under standard conditions, with 100 mg of OPD and 24 mg of the catalyst, at pH ~ 2 (i.e., its maximum performance). At the end of each run, the catalyst was collected from the reaction mixture by using an external magnet and carefully washed with water, methanol, and ethyl acetate. The recovered catalyst was dried overnight under reduced pressure at room temperature and was reutilized for subsequent experiments. The experimental results indicate that the catalyst could be reused four times during the reaction. After the third cycle, the % of yield decreased significantly, which may be due to the marginal loss of the catalyst. This indicates that after the third cycle, the effectiveness of the catalyst decreased (Fig. 3).
Fig. 3.
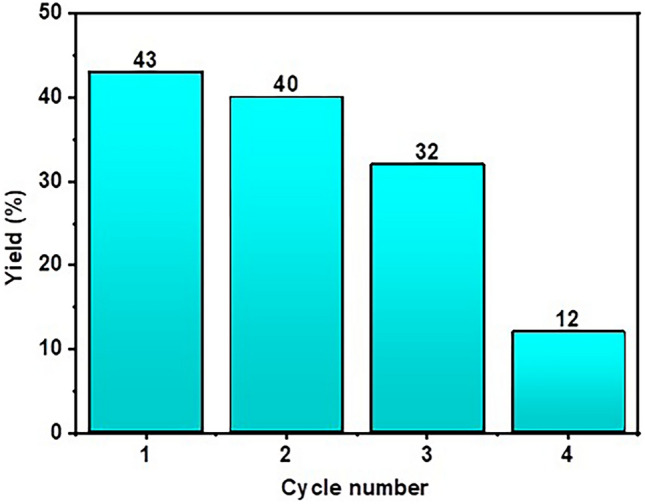
Catalytic recyclability studies (% of yield vs. cycle number).
Comparison with other methods
There has been reported of DAP synthesis employing a variety of catalysts, including metal complexes, nanoparticles, and inorganic salts (Table S1). The DAP has been synthesized in the reported approach employing metal complexes such as [Cu(L)(NO3)2], [Cu(L)Br2], [Fe(L)Cl2]Cl and [Fe(L)(NO3)2]NO328,29. Inorganic salts like FeCl3, CuCl2 and other nanoparticles such as CdFe2O4 / TiO2 have also been utilized in the production of DAP30–32. All the documented methods are effective, albeit with some drawbacks. The primary issue with the most widely reported approach is the catalyst recyclability. This is a key component of the recyclable catalysts. Nonetheless, the majority of catalysts have been used only once because of various factors, such as solubility or difficulty in separation. However, in the described procedure, the superparamagnetic Jeanbandyite catalyst, which is easily separated from the reaction mixture, is recycled four times, improving its catalytic activity in DAP synthesis. Another important consideration was the use of solvents. Methanol and other organic solvents were used in most of the reported procedures. However, in this study, water was used as the solvent, making the procedure less harmful and greener than that previously reported.
Mechanism
Based on the process of oxidative polymerization from monomers to polymers, at least two steps (Scheme 1b) may be involved in the formation of DAP molecules: (1) oxidation of OPD molecules by ferric ions to form radical cations or semiquinone radicals, and (2) dimerization or coupling of the radical cations to form DAP molecules. The DAP thus formed was rich in π-bonding and proton donor and acceptor sites. It is reasonable to assume that these functional sites would cause hydrogen bonds and intermolecular π–π interactions, causing DAP molecules to self-assemble. There are two steps in the process that result in the production of structures from OPD, as illustrated in Scheme 1b. During the initial phase, DAP is produced when Jeanbandyite provides Fe3+ to oxidize OPD. Using π – π and hydrogen bonding interactions, DAP molecules undergo self-assembly in the second stage to form a dimeric structure33. Previously, numerous oxidants, such as silver oxide, lead (IV) oxide, ferric chloride, cupric chloride, perchlorate, and cobalt perchlorate, have catalyzed the oxidation to the formation of DAP34. But there are few reports where heterogenous catalysts been utilized for DAP synthesis. Here, FeSnO(OH)5, having vacant orbital and oxidizing property, is acting in the same manner, helping the oxidation of OPD to formation of DAP.
Cellular internalization study
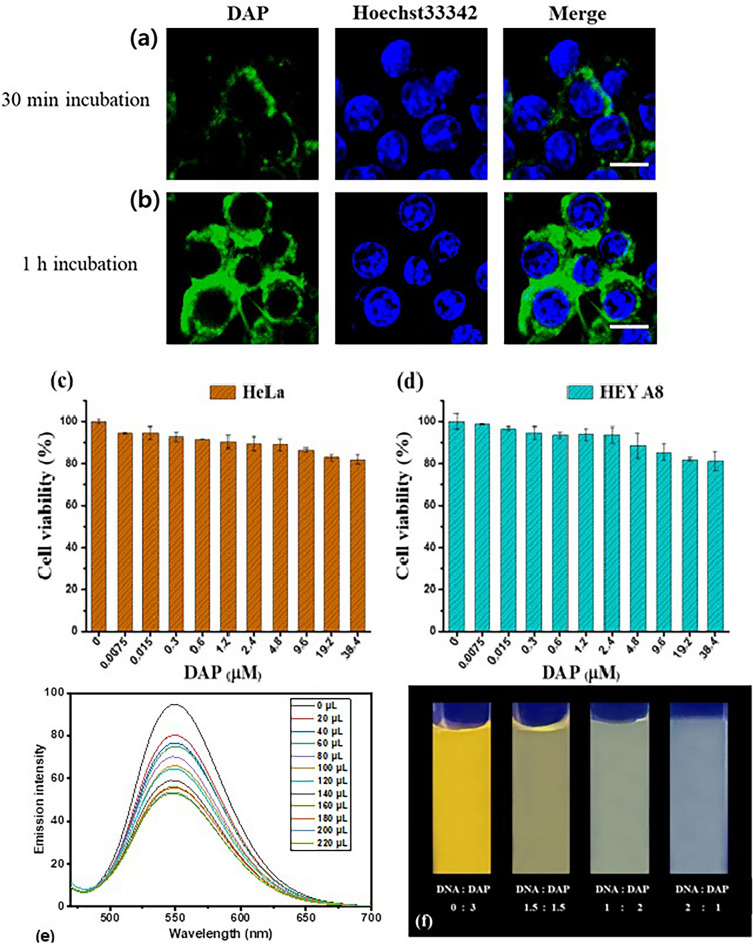
The cellular uptake of DAP was investigated using confocal laser scanning microscopy. HeLa cells were incubated with 10 µM DAP for either 30 min or 1 h. The cells were then washed three times with fresh cell medium and Hoechst33342 was added to stain the nuclei. After further incubation for 5 min, the cells were examined using confocal microscopy. As shown in Fig. 4a, green fluorescence was observed in the cytoplasm, indicating cellular uptake of DAP. Cell nuclei are displayed as blue fluorescence from Hoechst33342. However, the green fluorescence appeared dim, indicating 30 min incubation time was not enough for the compound to diffuse fully into the cytoplasm. After 1 h of incubation (Fig. 4b), the compound diffused well into the cytoplasm, but not into the cell nucleus. This result suggests that the compound can internalize rapidly into the cytoplasm of HeLa cells within 30 min at 10 µM and diffuse to most of the cytoplasm but not the nucleus within 1 h.
Fig. 4.
Cellular internalization of DAP in HeLa cells. The green fluorescence was emitted from DAP, and blue from Hoechst33342. (a) Incubation time of 30 min (b) 1 h (scale bar: 10 µm); Cytotoxicity measurement by MTT assay. (c) HeLa and (d) HEY A8 cells were treated with DAP for 24 h; (e) Fluorescence emission spectra of DAP (0.002 M) in the presence of increasing [DNA] (1.24 × 10–6–1.15 × 10–5 M). (f) Change in color of DAP in presence and absence of DNA.
Cytotoxicity assay
To assess the potential of DAP as a genuine biomolecular probe, we examined the cytotoxicity of DAP following the exposure of cells to different concentrations of DAP. After 24 h of incubation with DAP in HeLa (human cervical cancer) and HEY A8 (human ovarian cancer) cell lines, the cells were analyzed using MTT assay to determine cell viability (Hela and HEYA8 cells were kindly provided from Samsung Advanced Institute for Health Sciences & Technology, Sungkyunkwan University School of Medicine, Seoul, Republic of Korea). As shown in Fig. 4c,d, HeLa cells showed a cell viability of more than 82% and HEY A8 cell viability of more than 81%, suggesting that DAP has low toxicity towards both cell lines. Taken together, DAP is cell membrane-permeable and shows rapid cellular uptake with negligible cytotoxicity, indicating that DAP can be utilized as a useful tool in applications related to bioimaging.
DNA binding
Fluorescence spectroscopy is one of the most frequently employed techniques for investigating interactions between small ligand molecules and DNA. Molecular fluorescence has several advantages over other methods, including selectivity, wide linear concentration range, and high sensitivity. Compounds with low-energy π–π transition states containing aromatic functional groups exhibit the strongest and most potent fluorescence35,36. The mechanism of the interaction between the compound and SS-DNA was determined using fluorescence quenching studies. The fluorescence spectra of the phenazine derivatives are shown in Fig. 4e. The fluorescence intensity decreased significantly upon addition of DNA because of the interaction of the compound with DNA. The binding constant Kb was calculated to be 3.2 × 103 M−1. Quenching of the emission of compounds is due to photoelectron transfer from the nitrogenous base (guanine) to the excited states of the compounds. Hence, it is conventional for compounds to bind to DNA via electrostatic interactions or groove binding. The interaction between DNA and DAP is shown in Fig. 4f, where the color of the compound changes gradually with the addition of DNA, providing evidence supporting the DNA-binding property of DAP37.
Thermal denaturation
Thermal denaturation was conducted to validate the fluorescence titration data. As temperature increased, the hydrogen bond energy between the two DNA helices decreased. Thus, heat energy modifies the structural makeup of the DNA. Double-stranded DNA is denatured o its single-stranded form by hydrogen bond breakage at higher temperatures. The temperature at which 50% of molecules are denatured is known as melting temperature ("Tm"). Through stacking interactions, intercalation of compounds stabilizes DNA’s double helix structure of DNA and raises the Tm value38,39. The melting temperature (Tm) of SS-DNA was found to be 61.4 °C but increased to 64.5 °C in the presence of DAP. This result supports an interaction between DAP and SS-DNA.
Computational study
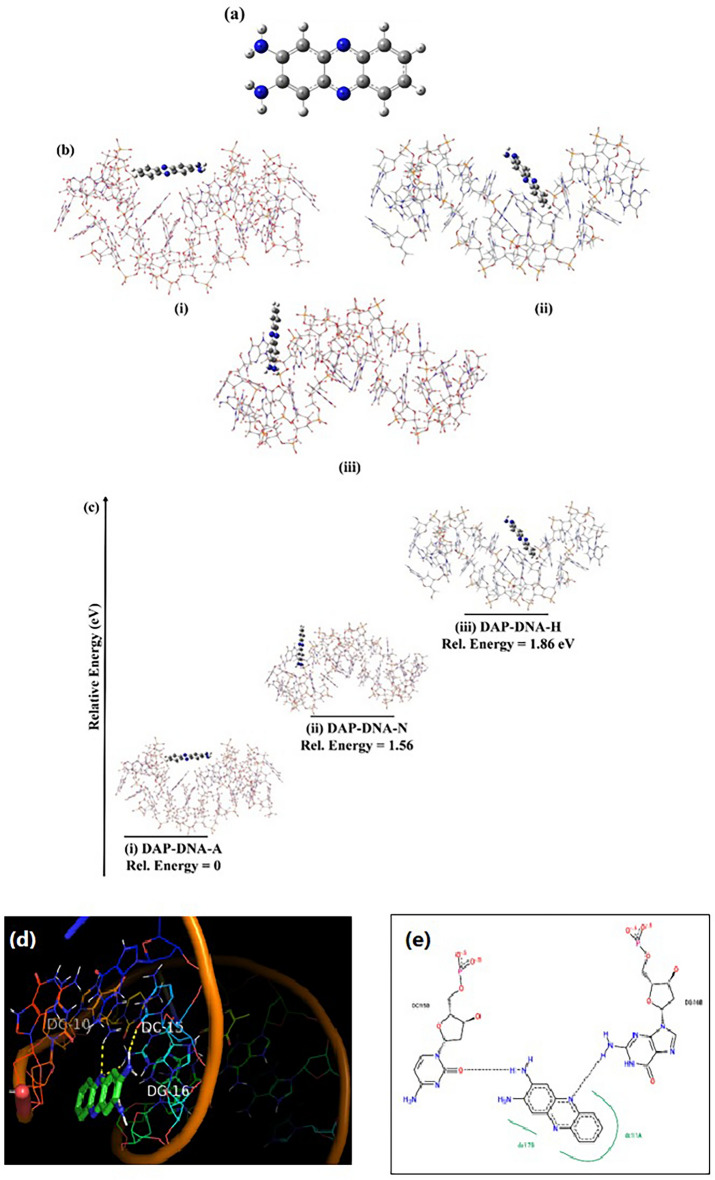
The ground state optimized structure of DAP is shown in Fig. 5a and the DAP can interact with the DNA at different sites, these various optimized interaction sites (DAP-DNA complexes) are shown in Fig. 5b. Based on the extent of their interactions, the binding energies of these complex systems were calculated using the following formula:
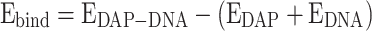 |
1 |
Fig. 5.
(a) Ground State optimized structure of DAP (b) The Orientations of various DAP-DNA complexes (i) DAP-DNA-A, (ii) DAP-DNA-N, (iii) DAP-DNA-H (c) Relative energies of the PDA-DNA complexes in various orientations. As observed in the figure, the stability order is (i) DAP-DNA-A > (ii) DAP-DNA-N > (iii) DAP-DNA-H.; (d) 3D and (e) 2D H-bonded interaction of the DAP with DNA.
If the 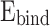 is negative, the structure of the DAP-DNA is stable. In this context, we studied the interaction between DAP and DNA as follows: (i) DAP is maintained in such a manner that it can interact with the bases and phosphates of SS-DNA (DAP-DNA-A). (ii) In another orientation, the terminal amine group is intercalated between the strands (DAP-DNA-N); and (iii) in the third orientation, the other side is intercalated between the strands (DAP-DNA-H). The binding energies of these complex systems are shown in Fig. 5c. Our calculations revealed that the DAP-DNA-A configuration exhibited the highest stability.
is negative, the structure of the DAP-DNA is stable. In this context, we studied the interaction between DAP and DNA as follows: (i) DAP is maintained in such a manner that it can interact with the bases and phosphates of SS-DNA (DAP-DNA-A). (ii) In another orientation, the terminal amine group is intercalated between the strands (DAP-DNA-N); and (iii) in the third orientation, the other side is intercalated between the strands (DAP-DNA-H). The binding energies of these complex systems are shown in Fig. 5c. Our calculations revealed that the DAP-DNA-A configuration exhibited the highest stability.
Molecular docking
The molecular docking technique helps to understand the non-covalent drug-DNA interactions of small molecules in the targeted region of DNA for rational drug design and discovery. The minor groove of DNA is preferred over the major groove because of its low steric hindrance, electrostatic factors, and its narrow shape. Molecular docking studies were performed to determine binding affinities of phenazine towards DNA. Molecular docking studies in ADV revealed that the designed compound showed sufficient binding affinity towards DNA (− 7.3 kcal/mol). The 3D and 2D interactions are shown in Fig. 5d,e, which prove that DAP has a good binding affinity with DNA.
Conclusion
Jeanbandyite was synthesized and characterized using SEM, TEM, STEM, FT-IR, XRD and VSM. The characterization results indicated that the Jeanbandyite had a square shape and exhibited superparamagnetic behavior. The synthesis of DAP from OPD using Jeanbandyite in an environmentally sustainable way is discussed in this article. Here, Jeanbandyite catalyzes the production of DAP with a good yield at pH = 2 and 24 h in an aqueous medium, the reaction occurred in neutral media also. The Jeanbandyite catalyst also offers good recyclability up to four times, and the recycling process becomes easier owing to its magnetic character. Mass spectroscopy, 1H-NMR, 13C-NMR, and other techniques were used to characterize the DAP. DAP showed efficient cellular uptake and low cytotoxicity in cells, suggesting its potential use as a biomolecular probe for various biomedical applications. Fluorescence measurements were performed to analyze the DNA-binding properties of DAP. These results demonstrated the superior DNA-binding capacity of the compound, which was further supported by a molecular docking investigation.
Methods
Synthesis of Jeanbandyite
In 20 mL of deionized water (DW), 3.24 gm of anhydrous FeCl3 and 2.26 gm of SnCl2 were mixed. To complete the precipitation reaction, 5.5 gm of NaOH in 80 mL of DW was added to the mixture. The mixture was then agitated for an additional two hours at 15 °C in air (final pH of ~ 13). After stopping the reaction, centrifugation was performed to obtain the final precipitate, which was then washed three times with deionized water. After separation, the brown magnetic material was collected and allowed to dry overnight in a vacuum desiccator20.
Synthesis of DAP
DAP was synthesized under two different conditions: a neutral medium (pH = 7) (path A) and an acidic medium (pH = 2) (path B), as described in Scheme 1. OPD (9.25 mmol) and FeSnO(OH)5 (0.86 mmol) dissolved in a minimum amount of water in a round-bottom flask was refluxed in a silicon oil bath at 100 °C for 24 h under different pH conditions [pH = 2(Path A); pH = 7(Path B)] (Scheme 1a). After the reaction was completed by TLC monitoring, the reaction mixture was cooled and 10% sodium hydroxide (NaOH) solution was slowly added with constant stirring until the mixture became alkaline to litmus. The reaction mixture was then quenched with water and extracted three times with ethyl acetate. The combined organic layers were dried over Na2SO4, filtered, and concentrated using a rotary distillation apparatus, leaving the crude product, which was purified by the decantation method. In this method, a minimum amount of hexane was added and the solution was sonicated. The mixture was centrifuged for 10 min to collect the solid precipitate (desired product) and the hexane portion (containing unwanted product) separately. The centrifugation process was repeated thrice, and the solid part was dried. The decantation method was monitored by TLC.
Brown solid, Yield = 43%; Rf = 0.42(Methanol: Chloroform = 1:9).
1H NMR (400 MHz): δ(ppm): 5.61 (s, 4 H, NH2); 7.11 (s, 2H); 7.55 (m, 2H); 7.92 (m, 2H). 13C NMR (100 MHz): δ(ppm): 143.7(Ar–C), 142.6(Ar–C), 141.3(Ar–C), 128.6(Ar–C),126.7 (Ar–C), 104.7 (Ar–C). The 1H-NMR and 13C-NMR spectra were similar to the previously reported one (Fig. 1)33. HRMS-ESI+ (m/z): calcd. For C12H10N4, [M + H]+ 211.0969; found: 211.0978 (Fig. 2). These characterization techniques provide sufficient information for the formation of DAP using Jeanbandyite as a catalyst.
Catalytic recyclability
The feasibility of reusing the Jeanbandyite catalyst was ascertained after completion of the reaction. An external magnet made it simple to remove the catalyst from the reaction mixture, and any leftover product was first cleaned with methanol and water. The catalyst was then used under the same conditions for the next reaction cycle. After each cycle, isolated yield% were calculated, higher yields indicate higher activity and lower values indicate poor activity.
Confocal microscopy experiment
HeLa cells were seeded in a cover glass (35 mm)-bottomed dish at a concentration of 2 × 103 cells/dish. After 24 h incubation and removal of the cell medium, the cells were treated with DAP in serum-free cell media at a concentration of 10 μM. Following incubation for either 30 min and 4 h at 37 °C, the cells were washed three times with PBS. Next, 2 ml of Hoechst33342 (8.1 μM) was added and incubated for 5 min. Confocal laser scanning microscopy (Carl Zeiss LSM 710) using an oil immersion lens (NA 1.30, 100X) was used to observe fluorescence in the cells. DAP and Hoechst were illuminated at 488 and 405 nm, respectively. Fluorescence was observed at 500–525 nm, and 440–460 nm, respectively.
MTT assay
HeLa cells were seeded in 96-well plates containing 10% FBS (Welgene) and 0.1% penicillin–streptomycin (Hyclone) in DMEM. After 24 h of culturing at 37 °C in a CO2 incubator, the cell media was removed and washed with 200 μL of PBS. In serum-free media at 37 °C, the cells were treated with different concentrations of DAP from 0 to 38.4 μM. Then, to investigate the cytotoxicity, the treated cells were incubated for 48 h and then rinsed with cold PBS before being added with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) dissolved in media. Four hours after the media was switched to DMSO, the formazan dye was quantified using a microplate reader to assess the absorbance at 540 nm. Non-treated cells were used as controls. The cell viability was determined by comparing the fluorescence intensities of the experimental and control wells. The percentage viability of cells was estimated as the ratio of the fluorescence intensity in the experimental wells to that in the control well. All experiments were performed in triplicate.
DNA binding study
Fluorescence quenching study
SS-DNA binding experiments with DAP were carried out in a 10 mM Tris-HCl/50 mM NaCl (pH 7.5) buffer solution using fluorescence quenching study. The quality of SS-DNA (salmon sperm DNA) was tested before the binding experiment by measuring the absorbance ratio at 260 and 280 nm in Tris-HCl-NaCl buffer solution. This ratio was 1.81, suggesting that the DNA was suitable and pure. The SS-DNA concentration was calculated using the absorbance at 260 nm and a molar extinction value € of 6600 M−1 cm−1, and an absorption titration experiment was carried out using 20 μL aliquots of DNA solution added sequentially to a fixed amount of metal complex. For each sample the mixed solutions were incubated for 2 min before the fluorescence spectra were recorded (excitation wavelength 400 nm, emission wavelength 420–800 nm)37,40. The binding constant (Kb of the SS-DNA-DAP complexes was calculated using the following equation41.
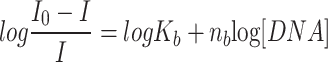 |
2 |
where Kb is the binding constant and nb is the number of binding sites. I and Io are the fluorescence intensities in the presence and absence of the DNA, respectively.
Thermal denaturation
The thermal stability of the DNA in the presence of DAP was measured as a function of temperature. For a fixed [DAP]/[DNA] ratio, the temperature change in absorbance at 260 nm was monitored from 30 to 90 °C at a scan rate of 1 °C/min. Consequently, the temperature at which DNA melts (Tm) was measured36.
Computational study
To determine the interaction between DAP and DNA at the molecular level, further computational quantum chemical calculations were performed. To do so, we employed Density Functional Theoretical (DFT) method in combination with the 6–31 G + (d) basis set to optimize the ground state structures of DAP. The PBE functional was used to compute the exchange–correlation energy. On the other hand, the DNA structure is obtained from PDB ID 1BNA42. However, to compute the geometry of the large DNA structure, we applied molecular mechanical methods using a universal force field. Furthermore, the Own N-layered Integrated molecular Orbital and Molecular mechanics (ONIOM) scheme43–45 was adopted to reveal the binding of the ground state optimized DAP with the optimized SS-DNA structure. Unless otherwise mentioned, all the calculations were performed using the g16 program suit46.
Molecular docking
Receptor preparation
The X-ray crystal structure of DNA (PDB ID: 1BNA) was retrieved from the RCSB Protein Data Bank47. After importing the crystal structure into AutoDock Tools 1.5.6, the heteroatoms and water molecules were eliminated, polar hydrogen atoms were inserted, and the Kollman charge was computed. Ultimately, the protein was saved in the pdbqt36,48.
Ligand preparation
Ligands were converted into PDB format using the OpenBabel software49. Furthermore, charges were allocated, torsional angles were changed, the ligands were optimized using the MGL tool, and finally, the ligands were converted into the pdbqt format50.
Docking procedure
Molecular testing of all compound libraries was performed using AutoDock Vina (ADV) software51. The protein was assumed to be rigid during docking, whereas the ligands were assumed to be flexible. A grid box with a size of 24 × 30 × 46 Å3 with coordinates of center_x = 15.393, y = 21.256, and z = 9.613 during docking exhaustiveness was set at 50. Using Pymol software, the top docking pose of the ADV output file was visualized52.
Supplementary Information
Acknowledgements
G.B, R.M, and S.S want to acknowledge UGC-DAE Consortium for Scientific Research for supporting this research (Sanction Order No. CRS/2022-23/02/818, dated 15-05-2023, PFMS Ref. No. C032393527418). SB acknowledges NIT Sikkim for computational support. This work was supported by the Technology Innovation Program (RS-2024-00400101, Multi-omics-based ovarian cancer undruggable target discovery and PROTAC drug delivery development) funded by the Ministry of Trade, Industry & Energy (MOTIE, Korea). This work was also supported by Soonchunhyang University research fund.
Author contributions
S.S.: Conceptualization, Formal analysis, Methodology, Writing—original draft. S.B.: Formal analysis, Methodology, Writing—original draft. S.P. and P.S. Formal analysis, Methodology. T.W.K., N.G., H.J.K., J.S.R., and S.J.: Formal analysis. B.S.: Formal analysis, Characterization. R.R., R.M. and A.S.: formal analysis, characterization. S.B.: Formal analysis, Writing—original draft. J.I.: Conceptualization, supervision, investigation, writing, reviewing and editing. G.B.: Conceptualization, formal analysis, supervision, investigation, writing, reviewing and editing.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sumantra Bhattacharya, Email: sumantrabhattacharya1@gmail.com.
Jungkyun Im, Email: jkim5279@sch.ac.kr.
Goutam Biswas, Email: goutam@cbpbu.ac.in.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-81330-0.
References
- 1.Che, Y. et al. Synthetic strategies of phenazine derivatives: A review. J. Heterocycl. Chem.59, 969–996 (2022). [Google Scholar]
- 2.Makgatho, E. M. & Mbajiorgu, E. F. In vitro investigation of clofazimine analogues for antiplasmodial, cytotoxic and pro-oxidative activities. Afr. Health Sci.17, 191 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pachón, O. G. et al. Antitumoral effect of phenazine N 5, N 10 -dioxide derivatives on caco-2 cells. Chem. Res. Toxicol.21, 1578–1585 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Krishnaiah, M. et al. Synthesis, biological evaluation, and metabolic stability of phenazine derivatives as antibacterial agents. Eur. J. Med. Chem.143, 936–947 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Huigens, R. W., Brummel, B. R., Tenneti, S., Garrison, A. T. & Xiao, T. Pyrazine and phenazine heterocycles: Platforms for total synthesis and drug discovery. Molecules27, 1112 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Serafim, B., Bernardino, A. R., Freitas, F. & Torres, C. A. V. Recent developments in the biological activities, bioproduction, and applications of pseudomonas spp. Phenazines. Molecules28, 1368 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanekar, D. et al. Fluorescent Indolo[2,3-b]Quinoxalin-2-Yl)(Phenyl)Methanone dyes: Photophysical, Aie activity, electrochemical, and theoretical studies. https://www.researchsquare.com/article/rs-1195228/v1 (2021) 10.21203/rs.3.rs-1195228/v1.
- 8.Pauliukaite, R., Ghica, M. E., Barsan, M. M. & Brett, C. M. A. Phenazines and polyphenazines in electrochemical sensors and biosensors. Anal. Lett.43, 1588–1608 (2010). [Google Scholar]
- 9.Romadina, E. I., Akkuratov, A. V., Simoska, O. & Stevenson, K. J. Phenazine-based compound as a universal water-soluble anolyte material for the redox flow batteries. Batteries8, 288 (2022). [Google Scholar]
- 10.Pachter, I. J. & Kloetzel, M. C. The Wohl-Aue reaction. I. Structure of benzo [a] phenazine oxides and syntheses of 1,6-dimethoxyphenazine and 1,6-dichlorophenazine1. J. Am. Chem. Soc.73, 4958–4961 (1951). [Google Scholar]
- 11.Rubab, L. et al. Green chemistry in organic synthesis: Recent update on green catalytic approaches in synthesis of 1,2,4-thiadiazoles. Catalysts12, 1329 (2022). [Google Scholar]
- 12.Nanda, B., Sailaja, M., Mohapatra, P., Pradhan, R. K. & Nanda, B. B. Green solvents: A suitable alternative for sustainable chemistry. Mater. Today Proc.47, 1234–1240 (2021). [Google Scholar]
- 13.Xu, C., Paone, E., Rodríguez-Padrón, D., Luque, R. & Mauriello, F. Reductive catalytic routes towards sustainable production of hydrogen, fuels and chemicals from biomass derived polyols. Renew. Sustain. Energy Rev.127, 109852 (2020). [Google Scholar]
- 14.Das, C. et al. Green synthesis, characterization and application of natural product coated magnetite nanoparticles for wastewater treatment. Nanomaterials10, 1615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang, D. & Astruc, D. Fast-growing field of magnetically recyclable nanocatalysts. Chem. Rev.114, 6949–6985 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Wang, K., Yang, D., Wu, C., Sanghadasa, M. & Priya, S. Recent progress in fundamental understanding of halide perovskite semiconductors. Prog. Mater. Sci.106, 100580 (2019). [Google Scholar]
- 17.Yuan, Y. et al. Stereoselective C−C oxidative coupling reactions photocatalyzed by zwitterionic ligand capped CsPbBr3 perovskite quantum dots. Angew. Chem. Int. Ed.59, 22563–22569 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Wu, W.-B., Wong, Y.-C., Tan, Z.-K. & Wu, J. Photo-induced thiol coupling and C–H activation using nanocrystalline lead-halide perovskite catalysts. Catal. Sci. Technol.8, 4257–4263 (2018). [Google Scholar]
- 19.Wang, C. et al. Photothermal Suzuki coupling over a metal halide perovskite/Pd nanocube composite catalyst. ACS Appl. Mater. Interfaces14, 17185–17194 (2022). [DOI] [PubMed] [Google Scholar]
- 20.El Hachmi, A. et al. Structural, morphological, magnetic and optical properties of Jeanbandyite prepared by the co-precipitation method. Mater. Today Commun.34, 105358 (2023). [Google Scholar]
- 21.Meher, N., Bidkar, A. P., Barman, D., Ghosh, S. S. & Iyer, P. K. A conformational tweak for enhanced cellular internalization, photobleaching resistance and prolonged imaging efficacy. Chem. Commun.56, 14861–14864 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Ebadi, A., Najafi, Z., Pakdel-yeganeh, H., Dastan, D. & Chehardoli, G. Design, synthesis, molecular modeling and DNA-binding studies of new barbituric acid derivatives. J. Iran. Chem. Soc.19, 3887–3898 (2022). [Google Scholar]
- 23.Dong, S. et al. Crystal structure and photocatalytic properties of perovskite MSn(OH)6 (M = Cu and Zn) composites with d10–d10 configuration. Appl. Surf. Sci.463, 659–667 (2019). [Google Scholar]
- 24.Wu, J., Sun, J., Jiao, Y., Hu, W. & Xu, J. Preparation of submicrometer mesoporous iron-tin oxide hydroxide and its flame-retardant and smoke-suppressant properties on flexible poly(vinyl chloride). J. Appl. Polym. Sci.135, 46218 (2018). [Google Scholar]
- 25.Lavanya Rathi, P. & Deepa, S. Structural, magnetic, thermal and optical properties of Sn2+ cation doped magnetite nanoparticles. Ceram. Int.46, 2969–2978 (2020). [Google Scholar]
- 26.Betterton, J., Green, D. I., Jewson, C., Spratt, J. & Tandy, P. The composition and structure of jeanbandyite and natanite. Mineral. Mag.62, 707–712 (1998). [Google Scholar]
- 27.Zhao, H., Wu, J., Hu, W.-D., Jiao, Y.-H. & Xu, J.-Z. The block combustion, high char residues and smoke restrain effect of nano-FeSnO(OH)5 on polyvinyl chloride. J. Therm. Anal. Calorim.137, 1255–1265 (2019). [Google Scholar]
- 28.Khattar, R., Yadav, A. & Mathur, P. Copper(II) complexes as catalyst for the aerobic oxidation of o-phenylenediamine to 2,3-diaminophenazine. Spectrochim. Acta. A. Mol. Biomol. Spectrosc.142, 375–381 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Tyagi, N. & Mathur, P. Iron(III) complexes of bis (benzimidazol-2-yl) methyl) thiophene-2,5-dicarboxamide: Synthesis, spectral and oxidation of o-phenylenediamine. Spectrochim. Acta. A. Mol. Biomol. Spectrosc.96, 759–767 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Ragab, S. S., Badawy, A. A. & El Nazer, H. A. A green approach to the synthesis of 2,3-diaminophenazine using a photocatalytic system of CdFe2O4/TiO2 nanoparticles. J. Chin. Chem. Soc.66, 719–724 (2019). [Google Scholar]
- 31.Mei, L., Tai, L. S., Tao, F. H., Jie, S. & Rong, L. Q. A novel synthesis of 2,3-diaminophenazine. Res. Chem. Intermed.38, 499–505 (2012). [Google Scholar]
- 32.El Bahnasawy, A. A. & El Ahwany, M. F. Synthesis of some pyrido-imidazophenazine derivatives. Phosphorus Sulfur Silicon Relat. Elem.182, 1937–1944 (2007). [Google Scholar]
- 33.He, D., Wu, Y. & Xu, B.-Q. Formation of 2,3-diaminophenazines and their self-assembly into nanobelts in aqueous medium. Eur. Polym. J.43, 3703–3709 (2007). [Google Scholar]
- 34.Doyle, R. P., Kruger, P. E., Mackie, P. R. & Nieuwenhuyzen, M. Phenazine-2,3-diamine. Acta Crystallogr. C57, 104–105 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Sirajuddin, M., Ali, S. & Badshah, A. Drug–DNA interactions and their study by UV–Visible, fluorescence spectroscopies and cyclic voltametry. J. Photochem. Photobiol. B124, 1–19 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Sen, S. et al. Anticancer, antibacterial, antioxidant, and DNA-binding study of metal-phenalenyl complexes. Bioinorg. Chem. Appl.2022, 1–15 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niu, S. Y. & Zhang, S. S. Electrochemical and spectroscopic studies on the interaction between DNA and the product of enzyme-catalyzed reaction of OPD-H2O2-HRP. Bull Korean Chem Soc25, 829 (2004). [Google Scholar]
- 38.Selvakumar, B., Rajendiran, V., Uma Maheswari, P., Stoeckli-Evans, H. & Palaniandavar, M. Structures, spectra, and DNA-binding properties of mixed ligand copper(II) complexes of iminodiacetic acid: The novel role of diimine co-ligands on DNA conformation and hydrolytic and oxidative double strand DNA cleavage. J. Inorg. Biochem.100, 316–330 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Neyhart, G. A. et al. Binding and kinetics studies of oxidation of DNA by oxoruthenium(IV). J. Am. Chem. Soc.115, 4423–4428 (1993). [Google Scholar]
- 40.Das, T. et al. Phenazine virulence factor binding to extracellular DNA is important for Pseudomonas aeruginosa biofilm formation. Sci. Rep.5, 8398 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Almarhoon, Z. M., Al-Onazi, W. A., Alothman, A. A., Al-Mohaimeed, A. M. & Al-Farraj, E. S. Synthesis, DNA Binding, and molecular docking studies of dimethylaminobenzaldehyde-based bioactive Schiff bases. J. Chem.2019, 1–14 (2019). [Google Scholar]
- 42.Dickerson, R. E. & Drew, H. R. Kinematic model for B-DNA. Proc. Natl. Acad. Sci.78, 7318–7322 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung, L. W. et al. The ONIOM method and its applications. Chem. Rev.115, 5678–5796 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Dapprich, S., Komáromi, I., Byun, K. S., Morokuma, K. & Frisch, M. J. A new ONIOM implementation in Gaussian98. Part I. The calculation of energies, gradients, vibrational frequencies and electric field derivatives. J. Mol. Struct. Theochem.461–462, 1–21 (1999). [Google Scholar]
- 45.Vreven, T., Morokuma, K., Farkas, Ö., Schlegel, H. B. & Frisch, M. J. Geometry optimization with QM/MM, ONIOM, and other combined methods. I. Microiterations and constraints. J. Comput. Chem.24, 760–769 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Gaussian 16, Revision C.01, Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., Scalmani, G., Barone, V., Petersson, G. A., Nakatsuji, H., Li, X., Caricato, M., Marenich, A. V., Bloino, J., Janesko, B. G., Gomperts, R., Mennucci, B., Hratchian, H. P., Ortiz, J. V., Izmaylov, A. F., Sonnenberg, J. L., Williams-Young, D., Ding, F., Lipparini, F., Egidi, F., Goings, J., Peng, B., Petrone, A., Henderson, T., Ranasinghe, D., Zakrzewski, V. G., Gao, J., Rega, N., Zheng, G., Liang, W., Hada, M., Ehara, M., Toyota, K., Fukuda, R., Hasegawa, J., Ishida, M., Nakajima, T., Honda, Y., Kitao, O., Nakai, H., Vreven, T., Throssell, K., Montgomery, J. A., Jr., Peralta, J. E., Ogliaro, F., Bearpark, M. J., Heyd, J. J., Brothers, E. N., Kudin, K. N., Staroverov, V. N., Keith, T. A., Kobayashi, R., Normand, J., Raghavachari, K., Rendell, A. P., Burant, J. C., Iyengar, S. S., Tomasi, J., Cossi, M., Millam, J. M., Klene, M., Adamo, C., Cammi, R., Ochterski, J. W., Martin, R. L., Morokuma, K., Farkas, O., Foresman, J. B. & Fox, D. J. Gaussian, Inc., Wallingford CT, (2016).
- 47.Binda, C. et al. Binding of rasagiline-related inhibitors to human monoamine oxidases: A kinetic and crystallographic analysis. J. Med. Chem.48, 8148–8154 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morris, G. M. et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem.30, 2785–2791 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Boyle, N. M. et al. Open babel: An open chemical toolbox. J. Cheminf.3, 33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sen, S. et al. Experimental and theoretical study on supramolecular encapsulation of molnupiravir by Cucurbit[7]uril: A potential formulating agent for COVID-19. J. Mol. Liq.395, 123877. 10.1016/j.molliq.2023.123877 (2023). [Google Scholar]
- 51.Trott, O. & Olson, A. J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. NA-NA10.1002/jcc.21334 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roy, S. et al. Design, synthesis and molecular docking studies of 5-fluoro 1-aryl/alkyl sulfonyl benzimidazole derivatives for treatment of Parkinson’s disease. Phosphorus Sulfur Silicon Relat. Elem.198, 336–344 (2023). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.