Abstract
Fruit color is a crucial trait for bell pepper. To investigate the mechanism of color formation, three bell pepper lines with different color (yellow, orange and red) were used as materials to conduct comprehensive targeted metabolomic and transcriptomic analyses. During the process of fruit development, 54 carotenoids metabolites were discovered, exhibiting unique accumulation patterns and notable variety specificity. The types and content of carotenoids in orange fruit (OM) were notably greater compared to the other two varieties. Red pigment (capsanthin and capsorubin) was specifically enriched in red fruit (RM), and yellow pigment (lutein and zeaxanthin) is the highest in yellow fruit (YM) and OM. Five modules positively correlated with carotenoid accumulation and one negative module was determined by weighted gene co-expression network analysis (WGCNA). Additionally, transcription factors (TFs) and hub genes related to carotenoid synthesis were predicted. By elucidating the regulation of 7 key carotenoid metabolites by 14 critical genes and 5 key TFs, we constructed a comprehensive carotenoid biosynthesis metabolic network that comprehensively explains the pigment changes observed in green and mature pepper fruit. Overall, the results not only provide important insights into carotenoid synthesis pathway, but also lay a solid base for revealing the mechanism of bell pepper color transformation.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-81005-w.
Keywords: Fruit color, Bell pepper, Metabolome analysis, Transcriptome analysis, Carotenoids
Subject terms: Plant morphogenesis, Secondary metabolism
Introduction
Capsicum annuum L., commonly known as bell pepper, is a valuable vegetable crop known for its high nutritional content and health benefits. One of the essential attributes is their fruit color, which plays a crucial role in influencing consumer preferences and breeder selection1. Mature bell peppers exhibit a diverse range of colors, including red, yellow and orange, arises from the diverse types and relative contents of carotenoids present in fruits2–6.
Carotenoids are 40-carbon isoprenoids polymers widely found in higher plants, algae, fungi and bacteria7,8. At present, more than 700 kinds of carotenoids have been discovered, mainly divided into two categories based on their chemical structure. One is type carotenes composed of carbon and hydrogen, such as α-carotene, β-carotene and (E/Z)-phytoene etc., and the other is luteins, which also contains oxygen in addition to carbon and hydrogen, such as capsanthin, neoxanthin, lutein, zeaxanthin, etc9. In green photosynthetic tissues of plants, carotenoids are involved in photosystem assembly, light-harvesting and photoprotection10–12. In non-green organs, carotenoids bring vibrant colors to plants, attracting insects and animals to participate in plant pollination and seed dissemination13. Additionally, abscisic acid (ABA) and strigolactones (SLs) is synthesized from carotenoids as precursors in plants14,15. Carotenoids are dietary sources of provitamin A, and serve as a natural antioxidant that scavenges peroxidizing free radicals and prevents the accumulation of harmful oxygen, playing a positive role in human health by reducing the incidence of many diseases9,16–19. Besides, some carotenoids are widely used in food colorants as well as cosmetic and pharmaceutical industries20,21.
In plant, isopentenyl diphosphate (IPP) and its isomer dimethylallyl pyrophosphate (DMAPP), converting to each other through IPP isomerase (IPI), are products of methylerythritol phosphate (MEP) pathway and the initial precursors of carotenoids22. The direct precursor of carotenoids is geranylgeranyl pyrophosphate (GGPP), produced by IPP and DMAPP under the catalytic action of geranylgeranyl diphosphate synthetase (GGPS)23,24. When GGPP enters the carotenoid biosynthesis pathway, the first carotenoid product, colorless 15-cis-phytoene, is produced by the rate-limiting enzyme phytoene synthase (PSY)25. Under the successive isomerization and desaturation of phytoene desaturase (PDS), ζ-carotene isomerase (Z-ISO), ζ-carotene desaturase (ZDS) and carotenoid isomerase (CRTISO), colorless phytoene is converted into red lycopene23. Due to the existence of LCYB and LCYE in plants, lycopene downstream products synthesis divided into two branches, including α- and β-carotene. Lutein is the final product of α-branch, which is transformed from α-carotene under the catalysis of P450 carotene hydroxylase CYP97A and CYP97C. As to β-branch, β-cryptoxanthin, zeaxanthin, antheraxanthin and violaxanthin are formed, respectively26,27. Under the action of neoxanthin synthase (NSY), violaxanthin is converted to neoxanthin, and it is usually the last step for the core biosynthesis pathway of carotenoids28. But in red pepper fruits, capsanthin and capsorubin are the final products, produced by capsaicin/capsorubin synthetase (CCS) catalyzing antheraxanthin and violaxanthin29.
Earlier studies perceive that three independent genes, including y, c1 and c2, control the peel color of mature pepper30, and genes encoding CCS, PRR2 and PSY1 were subsequently identified by genetic mapping techniques31–33. However, the three gene models were insufficient to explain the color variation of pepper fruits. Variation and expression of other genes in carotenoid metabolic pathway can also lead to different colors, such as PSY2, DXS, CaGLK2, BCH2, etc34–39. In higher plants, many transcription factors (TFs) also control fruit coloration by regulating carotenoid biosynthesis pathway29,40–42. In numerous plant species, MYB are known to control the synthesis and metabolism of carotenoids. For instance, in kiwifruit, MYB7 activates the promoter of AdLCY-β, which is essential for the carotenoid biosynthesis pathway43. Similarly, in the petals of Medicago truncatula, the anthocyanin-related R2R3-MYB protein WHITE PETAL1 (WP1) directly regulates the genes MtLYCe and MtLYCb expression, resulting in high accumulation of lutein and yellow petals manifestation44. In contrast, in tomatoes, SlMYB72 interacts with SlZHD17, a member of zinc-finger homeodomain TF family, to inhibit the genes SlPSY1 and SlZISO expression, thereby suppressing carotenoid synthesis45. Expanding on this theme, the phytochrome-interacting factor (PIF), a member of the bHLH TF family, downregulates carotenoid accumulation by inhibiting the expression of the rate-limiting enzyme gene PSY46. In addition, CpbHLH1 and CpbHLH2 play significant roles in regulating the transcription of CpCYC-B and CpLCY-B during the maturation of papaya fruit, thus influencing carotenoid biosynthesis47. Furthermore, the TF MADS-box also plays an important role in the carotenoid synthesis pathway. In citrus, CsMADS3 enhances carotenoid biosynthesis by interacting with the promoters of CsPSY1 and CsLCYb248. CsMADS6 also promotes the expression of PSY, PDS, and LCYb1, increasing carotenoid levels49. CsMADS5 activates carotenoid biosynthetic genes by binding to PSY, PDS, and LCYb1 promoters and interacts with CsMADS6 to form an enhancer complex50. In addition, both bZIP51 and WRKY52 have been shown to play significant roles in regulating the expression of genes within the carotenoid biosynthesis pathway, further highlighting the complexity of regulatory mechanisms governing carotenoid accumulation in plants.
In short, as a member of genus Capsicum, fruit color of bell pepper is an important breeding trait and has always been valued53. However, the complete network of bell pepper fruit color formation remain incompletely elucidated. Therefore, in the present study, taking bell peppers with yellow, orange and red fruit colors as materials, we used transcriptomics and metabolomics to detect the difference between green and mature stages, and screen metabolites and regulatory genes that cause fruit color variation. The results will reveal the genetic mechanism of fruit color change in yellow, orange and red bell peppers, and provide theoretical basis for breeding new characteristic bell pepper varieties.
Results
Metabolome analysis of different colored bell peppers
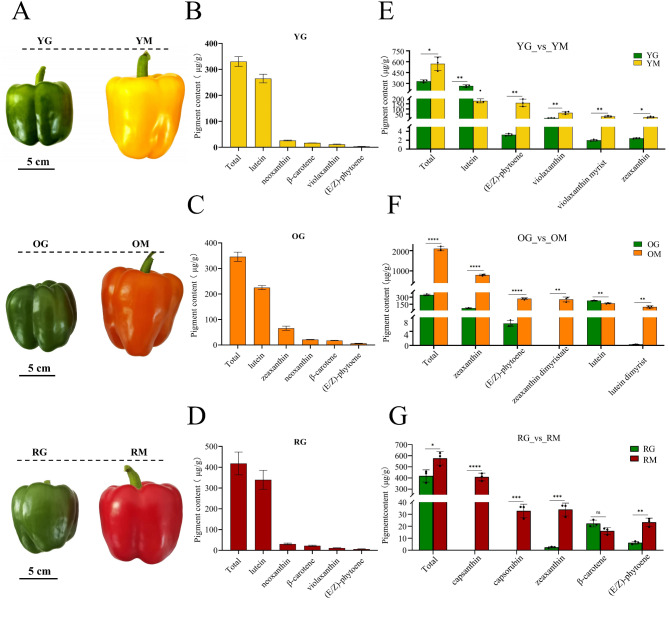
Three pepper inbred lines were used in the study, IL-Y, IL-O and IL-R (Fig. 1A), and fruit color of the three lines was green at 50 days after planting (50 DAP), the corresponding samples named YG, OG and RG. The color transformed to yellow, orange and red at 65 DAP, and the samples named YM, OM and RM, respectively. The results from metabolome analysis on samples YG, OG, RG, YM, OM and RM showed that 68 carotenoid compounds were detected, including 61 lutein (yellow pigment) and 7 carotenes (orange pigment), and among the examined carotenoids, 54 were identified in at least one sample (Table S1).
Fig. 1.
The phenotypes and carotenoid contents of three different colored bell pepper fruits. (A) The phenotype of bell pepper fruits at 50 DAP (YG, OG, and RG, green) and 65 DAP (YM, OM, and RM; yellow, orange and red). (B–D) Detection of total carotenoids and top five carotenoid pigments of IL-Y, IL-O and IL-R at green ripening stage, and the samples named YG, OG, and RG, respectively. (E–G) Detection of total carotenoids and top five carotenoid pigments of IL-Y, IL-O and IL-R at mature stage, and the samples named YM, OM, and RM, respectively, and their changes compared to green samples.
At 50 DAP, the carotenoids accumulated in the green fruits of IL-Y (YG), IL-O (OG) and IL-R (RG) were mainly lutein, β-carotene and neoxanthin, but other compounds were extremely low (Fig. 1B, C and D). With the ripening of fruits, total carotenoid contents in mature fruits of IL-Y (YM), IL-O (OM) and IL-R (RM) were 571.29 µg/g, 2098.27 µg/g and 575.62 µg/g, and it was 6.06, 1.38 and 1.73 times higher than that in YG, OG and RG, respectively. The species were also increased from 20 (YG), 18 (OG) and 17 (RG) to 39 (YM), 49 (OM) and 27 (RM) (Table S2). In YM, the main carotenoids were lutein (31.36%), (E/Z) phytoene (27.89%), violaxanthin (10.96%), violaxanthin myristate (4.55%) and zeaxanthin (3.42%) (Fig. 1E). While in OM, zeaxanthin (37.93%) was the highest, followed by (E/Z) phytoene (12.69%), zeaxanthin dimyristate (11.97%), lutein (8.02%) and lutein dimyristate (4.42%) (Fig. 1F). The accumulation of (E/Z) phytoene were both high in YM and OM, but in RM, red capsanthin was the main component of carotenoids, reaching 71.02%, which is different from YM and OM (Fig. 1G). Other carotenoids compounds varied from 0 to 5.9%. In addition, the content of lutein in RM was extremely low (0.43 µg/g), which was also different from the high content of lutein in YM and OM (Fig. 1E, F and G, Table S2).
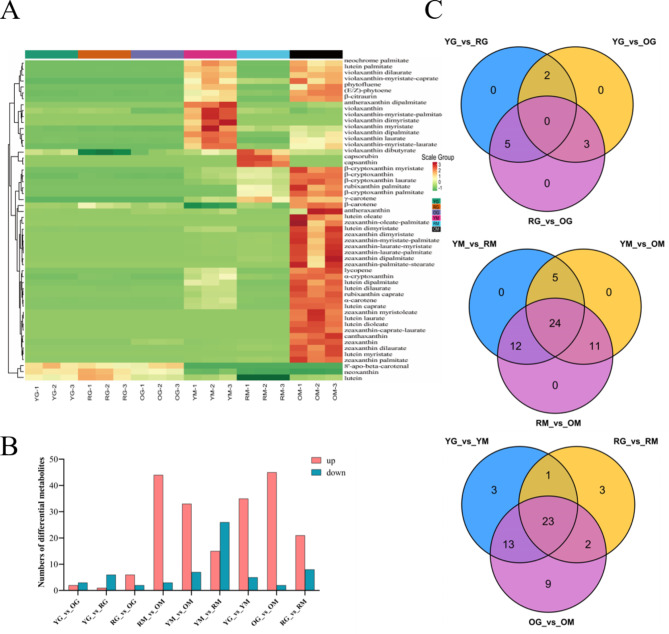
The metabolites heatmap showed significant differences among green fruits samples YG, OG and RG, and mature fruits of YM, OM and RM (Fig. 2A). Under the selection of Fold_Change (FC) ≥ 2 or ≤ 0.5, differential metabolites of three bell pepper varieties at the green ripening stage were less, ranging from 5 to 8 (Fig. 2B). At the mature stage, OM showed a higher accumulation of carotenoids compared to YM and RM. Then the differentially accumulated carotenoids (DACs) among samples were also screened (Table S3). In YM_vs_OM group, 40 DACs were identified, including 33 up-regulated and 7 down-regulated. Among the DACs, zeaxanthin and zeaxanthin-related compounds were significantly up-regulated, and the same as to orange carotenes, α-carotene and β-carotene, while violaxanthin and violaxanthin-related compounds were significantly down-regulated. Similarly in RM_vs_OM group (44 up-regulated and 3 down-regulated), zeaxanthin, zeaxanthin-related compounds, lutein-related compounds, and carotenes were also highly up-regulated, while the red pigments capsanthin and capsorubin were down-regulated. In YM_vs_RM comparison group, 41 DACs were identified, among which capsanthin and capsorubin were significantly up-regulated, while lutein and related compounds were conversely down-regulated. A total of 47, 29 and 40 DACs were identified between green and mature stages for groups OG_vs_OM, RG_vs_RM and YG_vs_YM, respectively (Fig. 2B, Table S3). Venn diagram showed 24 DACs common to the three comparison groups (Fig. 2C), detail information of DACs was showed in Table S3. The above results indicated that the unique accumulation pattern of carotenoid-related metabolites was the main reason for the color difference in bell pepper fruits.
Fig. 2.
The metabolome data analysis of three bell peppers lines. (A) Heatmap constructed based on 54 carotenoids metabolite profiles of IL-Y, IL-O and IL-R, three repeats (-1, -2 and − 3) for sample YG, OG, RG, YM, OM and RM. (B) The DACs numbers of different comparison groups. (C) The universal and specific DACs numbers of different comparison groups.
RNA-seq detection and analysis of different colored bell peppers
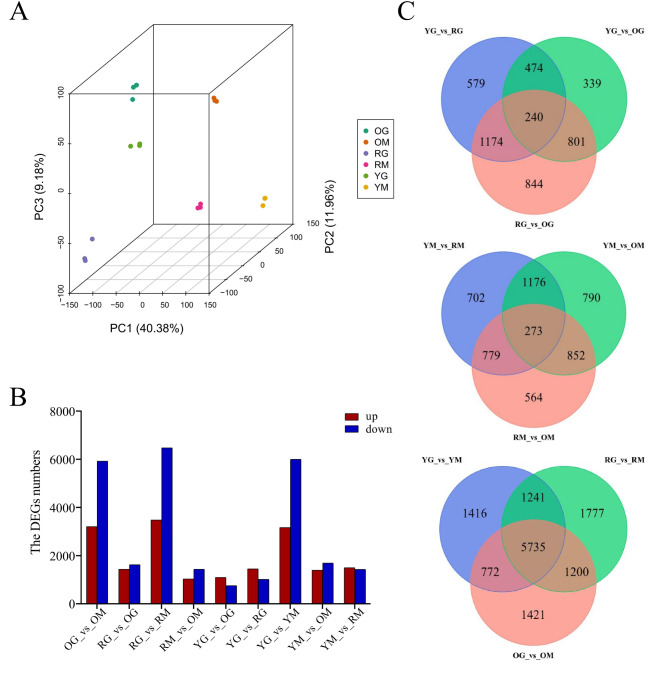
To understand the molecular regulatory network on fruit color formation of yellow, orange and red bell pepper, we performed transcriptome sequencing analysis on YG, YM, OG, OM, RG and RM with three biological replicates per sample (-1, -2, and − 3). Data analysis showed 124.82 Gb of total clean data was obtained, and clean reads per sample ranged from 41,358,224 to 52,919,370 (Table 1). The Q20 of all samples was above 97.3%, and Q30 was above 92.37%, and mean GC content was 42.53% (Table 1). The principal component analysis (PCA) score plot showed PC1, PC2 and PC3 accounted for 40.38%, 11.96% and 9.18% of the variance, respectively. Samples YG, YM, OG, OM, RG and RM were clearly separated in the analysis chart, and their three replicates were closely clustered (Fig. 3A).
Table 1.
Transcriptome sequencing data quality index.
| Sample | Raw Reads | Clean Reads | Clean Base(G) | Q20(%) | Q30(%) | GC Content(%) |
|---|---|---|---|---|---|---|
| OG-1 | 46,530,998 | 44,655,450 | 6.7 | 97.62 | 93.15 | 42.65 |
| OG-2 | 43,121,018 | 41,358,224 | 6.2 | 97.59 | 93.08 | 42.71 |
| OG-3 | 47,223,748 | 45,014,448 | 6.75 | 97.53 | 92.97 | 42.55 |
| OM-1 | 49,292,006 | 46,178,988 | 6.93 | 97.72 | 93.43 | 42.57 |
| OM-2 | 48,808,226 | 45,865,292 | 6.88 | 97.61 | 93.2 | 42.47 |
| OM-3 | 53,698,840 | 52,919,370 | 7.94 | 97.86 | 93.54 | 42.56 |
| RG-1 | 48,982,534 | 46,584,348 | 6.99 | 97.3 | 92.37 | 42.73 |
| RG-2 | 47,078,894 | 44,053,318 | 6.61 | 97.76 | 93.5 | 42.57 |
| RG-3 | 47,272,994 | 45,078,648 | 6.76 | 97.64 | 93.22 | 42.9 |
| RM-1 | 47,140,830 | 45,427,904 | 6.81 | 97.32 | 92.43 | 42.28 |
| RM-2 | 49,236,310 | 46,708,260 | 7.01 | 97.54 | 92.96 | 42.46 |
| RM-3 | 48,691,662 | 46,481,656 | 6.97 | 97.4 | 92.6 | 42.35 |
| YG-1 | 51,603,776 | 49,636,440 | 7.45 | 97.41 | 92.65 | 42.66 |
| YG-2 | 47,528,914 | 45,475,804 | 6.82 | 97.44 | 92.76 | 42.71 |
| YG-3 | 52,411,018 | 50,004,026 | 7.5 | 97.58 | 93.05 | 42.59 |
| YM-1 | 47,862,660 | 45,657,656 | 6.85 | 97.53 | 92.99 | 42.33 |
| YM-2 | 45,830,776 | 43,821,608 | 6.57 | 97.39 | 92.65 | 42.23 |
| YM-3 | 49,055,178 | 47,171,162 | 7.08 | 97.44 | 92.8 | 42.15 |
Fig. 3.
Transcriptome sequencing analyses of three bell peppers. (A) PCA score plots for all samples. (B) Numbers of up- and down-regulated DEGs detected in comparison groups. (C) Venn diagram of DEGs among different comparison groups.
Under the thresholds of log2 (FC) ≥ 1 and FDR ≤ 0.05, 14,961 differentially expressed genes (DEGs) were screened out in three bell pepper lines between green and mature stages. DEGs distribution showed in volcano plots suggested significant differences in samples YG, YM, OG, OM, RG and RM (Fig. S1). At the green ripening stage, we identified 1854, 2467 and 3059 DEGs in YG_vs_OG, YG_vs_RG and RG_vs_OG, respectively. Among them, 1099, 1452 and 1435 genes were up-regulated, while 755, 1015 and 1624 genes were conversely down-regulated. (Fig. 3B). In total, 240 common DEGs were identified at green ripening stages of yellow, orange and red fruits (Fig. 3C). At the mature stage, 2468 (1034 up- and 1434 down-regulated), 3091 (1397 up- and 1694 down-regulated) and 2930 (1500 up- and 1430 down-regulated) DEGs were screened in comparison groups RM_vs_OM, YM_vs_OM and YM_vs_RM, respectively. Venn diagrams showed that 273 DEGs were common, indicating that these DEGs may be responsible for the key differences in fruit color among the three bell peppers at mature stage(Fig. 3B and C). For two different stages of the same cultivar, we detected 3205 up- and 5923 down-regulated DEGs in OG_vs_OM, 3479 up- and 6474 down- in RG_vs_RM, and 3166 up- and 5998 down- in YG_vs_YM. Venn diagram showed 5735 common DEGs detected in all comparison groups, indicating that these DEGs might be important reasons for the color change of three bell peppers lines (Fig. 3B and C).
DEGs functional prediction
DEGs functional prediction of GO and KEGG enrichment analysis was performed to elucidate their roles in fruit color change of bell pepper (Fig. S2 and S3). All the DEGs were enriched in molecular functions, cellular components and biological processes. KEGG enrichment analysis showed the highest abundance pathway was metabolic (ko01100) and secondary metabolites biosynthesis (ko01110). Most importantly, pathway of carotenoid biosynthesis (ko00906) was significantly enriched in groups YG_vs_YM, OG_vs_OM, RG_vs_RM, RG_vs_OG, YG_vs_RG, RM_vs_OM and YM_vs_RM (Fig. S3), providing a powerful clue on genes expression in fruit coloration of bell pepper. The DEGs of group YG_vs_OG was enriched in biosynthesis pathways of phenylpropanoid (ko00940), anthocyanin (ko00942) and flavonoid (ko00941). For group RG_vs_OG, DEGs was enriched in biosynthesis of phenylpropanoid (ko00940) and anthocyanin (ko00942), and DEGs of YG_vs_RG was also significantly enriched in anthocyanin biosynthesis pathway (ko00942). The results are consistent with previous study that flavonoids are mainly synthesized in the early stage of fruit54, suggesting DEGs involved in these three pathways may be the reason for the early discoloration in bell pepper.
DEGs related to carotenoid biosynthesis
A total of 48 DEGs were screened to be related to carotenoid metabolism and assigned to pathway ko00906 (Table S4). After excluding 25 uncertain genes and low-expression genes, 23 DEGs were detected in at least one comparison group. Based on gene functional annotation, homology of these 23 genes was identified, which included 2 PSY (gene-LOC107859651, gene-LOC107868281), 1 PDS (gene-LOC107861625), 1 Z-ISO (gene-LOC107850257), 1 ZDS (gene-LOC107839468), 2 LCY-E (gene-LOC107840923, gene-LOC107852092), 1 LCYB (gene-LOC107869983), 2 BCH (gene-LOC107873401, gene-LOC107863219), 1 ZEP (gene-LOC107872926), 1 CRTISO (gene-LOC107854534), 1 CYP97 (gene-LOC107850957), 1 CCS (gene-LOC107875664), 1 NCEDs (gene-LOC107852027), 1 CCDs (gene-LOC107870081), 3 SDRS (gene-LOC107843702, gene-LOC107862477, gene-LOC107855067), 2 AAO (gene-LOC107847514, gene-LOC107847367) and 2 D27s (gene-LOC107863814, gene-LOC107875230).
In plants, phytoene is the first product of carotenoids biosynthesis pathway, which is formed by PSY catalyzing condensation of two GGPP molecules25. In our study, compared with the green ripening stage, expression of PSY1 (gene-loc107868281) was significantly increased in three bell pepper lines at coloring stage, which was corresponding to the higher content of phytoene in OM, RM and YM. The CCS gene (gene-LOC107875664) was only expressed in RM, consistent with capsaicin and capsorubin were detected only in RM. The expression level of BCH1 gene (gene-LOC107863219) increased most significantly in OG_vs_OM, RG_vs_RM, YG_vs_YM comparison groups, while the expression of BCH2 gene (gene-LOC107873401) decreased significantly, showing different expression patterns. Compared with the green ripening stage, other key genes including PDS (gene-LOC107861625), ZDS (gene-LOC107839468), Z-ISO (gene-LOC107850257), LCYB (gene-LOC107869983) and ZEP (gene-LOC107872926) also increased significantly. However, cyclase genes LCYE (gene-Loc107852092, gene-LOC107840923) and CYP97 (gene-Loc107850957) involved in α-branch synthesis decreased during maturation. In addition, downstream products of carotenoids produces precursors for abscisic acid (ABA) and strigolactone (SL)14,15. One NCED, three SDR, two AAO (related to ABA synthesis) and two D27 (related to SLs synthesis) were significantly differentially expressed in bell pepper lines at different periods (Table S4).
The direct carotenoids precursor, GGPP, is synthesized under the catalysis of GGPPS22–24. Two GGPPS (gene-LOC107843186, gene-LOC107867046) with high expression in YM, OM and RM were identified in our study, providing sufficient precursor compounds for carotenoid synthesis, which may be one of the reasons for the high carotenoid content of the three bell pepper lines at mature stage (Table S4).
DEGs analysis related to carbon and lipid metabolism
Previous studies showed positive correlation between sugar and pigment accumulation55–57. We found DEGs in groups YG_vs_YM, OG_vs_OM and RG_vs_RM were significantly enriched in carbon metabolism (ko01200) (Fig. S3). At least 80 DEGs were detected in one comparison group (Table S5). In groups YG_vs_YM, OG_vs_OM and RG_vs_RM, 32, 54 and 45 DEGs were up-regulated, while 17, 12 and 11 DEGs were down-regulated, respectively (Table S5).
In plant cells, carotenoids usually accumulate in chromoplasts, a kind of specialized storage organelles (plastids)58. In chromoplast differentiation, internal membrane remodeling is a prominent feature, and lipid substances synthesis plays an important role during the process59. In the pathway of fatty acid metabolism (ko01212), 44, 52 and 56 DEGs were detected in groups YG_vs_YM, OG_vs_OM and RG_vs_RM, respectively. Among them, 17, 22 and 25 DEGs were up-regulated, and 27, 30 and 31 were down-regulated, respectively (Table S6). In the fatty acid degradation pathway (ko00071), 17, 15, 21 DEGs were up-regulated, while 19, 15 and 22 were down-regulated in YG_vs_YM, OG_vs_OM and RG_vs_RM, respectively (Table S6). Besides, in the fatty acid biosynthesis pathway (ko00061), 7, 17 and 13 genes of YG_vs_YM, OG_vs_OM and RG_vs_RM were up-regulated, while 12, 12 and 13 genes were down-regulated (Table S6).
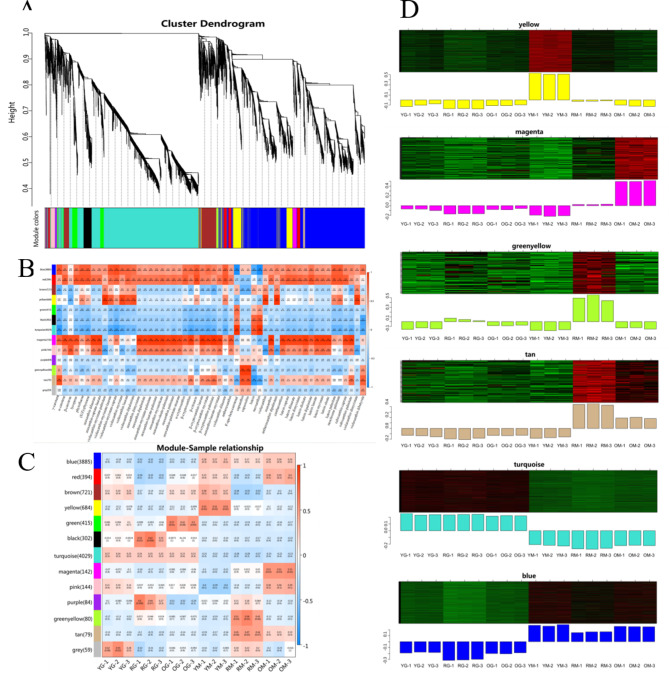
DEGs co-expression analysis
We performed weighted gene co-expression network analysis (WGCNA) using DEGs identified, and obtained 13 main branches (Fig. 4A, Table S7). Modules blue, red and magenta had a positive correlation with carotenoid metabolites, while turquoise had a negative correlation. The greenyellow module had the highest correlation with capsaicin and capsorubin, which was characteristic in RM, indicating genes in this module play more important roles in red pigment formation (Fig. 4B). Expression heat map of co-expressed genes of blue module in OM, YM and RM were much higher than OG, YG and RG, while genes of turquoise module was just the opposite, indicating that genes of the two modules played opposite roles in carotenoid synthesis (Fig. 4C). DEGs in yellow module was highly correlated with YM, while genes of magenta and greenyellow module were only highly expressed in OM and RM, showing obvious variety specificity (Fig. 4C and D). These results indicated DEGs in different modules may play different roles in carotenoid metabolites synthesis, and subsequent analysis also focuses on these six modules.
Fig. 4.
WGCNA of DEGs. (A) Hierarchical cluster tree. (B) Relationship between modules and carotenoid metabolites. (C) Relationship between modules and samples YG, OG, RG, YM, OM and RM. (D) Gene expression pattern of six modules.
Transcription factors (TFs) prediction for carotenoid metabolism regulation
Transcription factors (TFs) are also key factors in plant carotenoid biosynthesis. MYB, MAD-box, bHLH, WRKY, NAC and NY-F play important roles in regulating transcription of carotenoid metabolism related genes40–42. In blue module, 210 TFs were identified, including 16 MYB, 10 NAC, 4 WRKY, 8 MAD, 2 BHLH, 11 AP2/ERF, 9 NF-Y, and 3 HD-ZIP (Table S8). Expression abundance of 4 MYB (gene-LOC107850892, gene-LOC107867188, gene-LOC107868015 and gene-LOC107871103), 3 HD-ZIP (gene-LOC107843791, gene-LOC107862495 and gene-LOC107863056), 4 NAC (gene-LOC107842449, gene-LOC107845296, gene-LOC107847606 and gene-LOC107867776), 4 AP2/ERF (gene-LOC107855040, gene-LOC107862115, gene-LOC107867626 and gene-LOC107870958), 3 MAD (gene-LOC107847473, gene-LOC107855404 and gene-LOC107878477), 3 WRKY (gene-LOC107840352, gene-LOC107859607 and gene-LOC107872867) and 1 NF-Y (gene-LOC107861703) was higher in the colored mature fruit, indicating they may contribute more in fruit color transformation of bell pepper (Table S8). TFs such as MYB, MADS, and bZIP have been reported to regulate key genes in the carotenoid synthesis pathway, including LYC, PSY, PDS, and BCH, controlling carotenoid accumulation and influencing fruit color43–51. Notably, one MAD family member (gene-LOC107847473) was almost not expressed at the green ripening stage, but showed high expression at the mature stage with the fold change ranging from 9.18 to 15.38, indicating its pivotal role in regulating carotenoids biosynthesis in bell pepper. In the turquoise module, 269 TFs genes were identified including 25 AP2/ERF, 20 bHLH, 8 MAD, 12 MYB, 4 NAC, 8 HD-ZIP, 6 WRKY and 4 NF-Y, and detected HD-ZIP and WRKY family members were identified as negative regulators associated with carotenoid metabolism (Table S8). In the yellow module, 56 TFs genes were identified, and expression of 3 bHLH (gene-LOC107838913, gene-LOC107840824 and gene-LOC107862125) and 1 AP2/ERF (gene-LOC107864060) in YM were much higher than other samples (Table S8). The TF bHLH regulates the transcription of CYC-B and LCY-B46, promoting the yellowing of chili pepper fruit. Besides, 15, 4 and 4 TFs were predicted in red, magenta and greenyellow modules, respectively. Interestingly, one GRAS (gene-LOC107853465) was highly expressed only in greenyellow module, but no GRAS family members have been reported in carotenoids synthesis regulation, so their functions need to be further studied (Table S8).
Candidate hub genes for carotenoid synthesis in different colored bell pepper
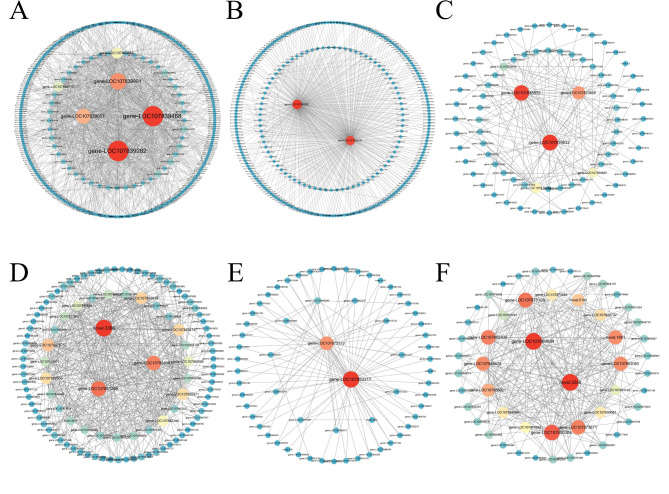
Correlation networks were constructed between genes within six key modules. In blue module, a total of 333 DEGs showed high co-expression with edge weight over 0.5, including 8 TFs, 6 carotenoid synthesis genes, 13 carbon metabolism pathway genes, and 13 genes involved in fatty acid synthesis and metabolism (Table S9), and mitochondrial carrier protein (gene-LOC107839282) and Zeta-carotene desaturase (ZDS, gene-LOC107839468) were hub genes (highly connected genes) in this module (Fig. 5A), and mitochondrial carrier protein (gene-LOC107839282) had high correlation with carotenoid synthesis genes ZDS (gene-LOC107839468), PDS (gene-LOC107861625), BCH (gene-LOC107863219), and GGPS (gene-LOC107843186), and TFs MAD (gene-LOC107879769), NAC (gene-LOC107842449 and gene-LOC107850240), indicating its key function in carotenoids synthesis. The turquoise module contained 284 co-expressed genes, including 22 TFs, 1 carotenoid decomposition gene, 3 carbon metabolism pathway genes, and 2 genes related to fatty acid synthesis and metabolism (Table S9). Hypothetical protein (gene-LOC107840002) and Ras-related protein (gene-LOC107839339) had the highest connectivity (Fig. 5B), and were both highly correlated with the carotenoid-breakdown gene CCD (gene-LOC107870081), which was negatively related to carotenoid metabolism. The yellow module contained 79 co-expressed genes, and gene-LOC107839932, putative L-ascorbate peroxidase (gene-LOC107838875) and excision repair protein (gene-LOC107870806) had the highest connectivity (Fig. 5C, Table S9). In magenta module, genes novel.3368, Chaperonin-like RBCX protein (gene-LOC107861269) and isocitrate dehydrogenase (gene-LOC107854067) had the highest connectivity (Fig. 5D, Table S9). In the red module envoloved of 63 genes, of which obg-like ATPase 1 (gene-LOC107853317) and methionine-tRNA ligase (gene-LOC107873333) showed the highest connectivity (Fig. 5E, Table S9). There were 52 genes in greenyellow module, and novel.3256 and gene-LOC107869054 were the hub genes (Fig. 5F, Table S9).
Fig. 5.
The DEGs co-expressed networks. (A) Blue module. (B) Turquoise module. (C) Yellow module. (D) Magenta module. (E) Red module. (F) Greenyellow module. DEGs with edges weight above 0.5 were displayed. The color ranges from blue to red, and the dots from small to large represent the degree of connectivity from low to high.
Metabolite and gene associations underlying color formation in bell pepper fruit
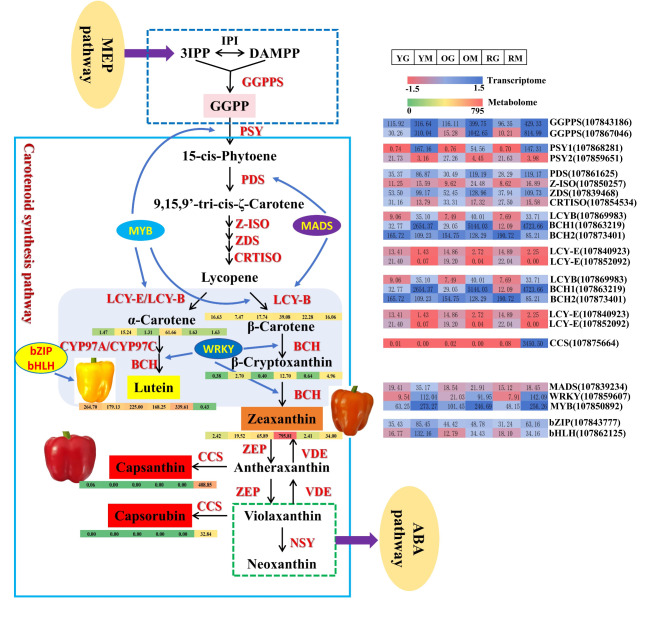
To further analyze the relationship between metabolites and related genes in carotenoid biosynthesis pathway during yellow, orange and red pepper coloration, we have established a network based on key metabolites and related genes (Fig. 6). In different pepper fruit samples undergoing color changes (YG, YM, OG, OM, RG and RM), 7 key carotenoid metabolites and 14 key biosynthetic genes were involved in the regulation. The 14 key genes are: two GGPPS genes (gene-LOC107843186 and gene-LOC107867046), PSY1 (gene-LOC107868281) and PSY2 (gene-LOC107859651), PDS (gene-LOC107861625), Z-ISO (gene-LOC107850257), ZDS (gene-LOC107839468), CRTISO (gene-LOC107854534), LCYB (gene-LOC107869983), BCH1 (gene-LOC107863219) and BCH2 (gene-LOC107873401), LCYE (gene-LOC107840923 and gene-LOC107852092), CCS (gene-LOC107875664). Among them, GGPPS, PSY1, PDS, Z-ISO, ZDS, LCYB, BCH1, and LCYE were highly expressed at mature stage (65 DAP), positively regulating pigment accumulation. In contrast, PSY2, CRTISO, and BCH2 showed higher expression in the green ripening fruit stage (50 DAP), which may be related to pigment accumulation in the green fruit.
Fig. 6.
Carotenoid synthesis pathway and key genes and metabolites regulating fruit color formation in bell pepper lines IL-Y, IL-O and IL-R. Expression of key genes and TFs (Transcriptome) and metabolites (Metabolome) were showed on scaled level among samples YG, YM, OG, OM, RG and RM.
The 7 key carotenoid metabolites are: α-carotene, β-carotene, β-cryptoxanthin, zeaxanthin, lutein, capsanthin, and capsorubin. In YM and OM samples, α-carotene content was higher, related to high expression of LCYB gene. Genes BCH2 and BCH1 promoted lutein accumulation at green ripening and mature stages, respectively, laying the foundation for pepper fruit color changes. The significantly higher accumulation of zeaxanthin in OM samples was a key factor in color transition from green to orange. Furthermore, the CCS gene was only expressed in RM samples, and it determined capsanthin and capsorubin accumulatio, leading to red color in mature RM. The distinct accumulation patterns of different metabolites are closely related to specific DEGs during fruit development, which collectively regulate the color changes in pepper fruits.
qRT-PCR analysis
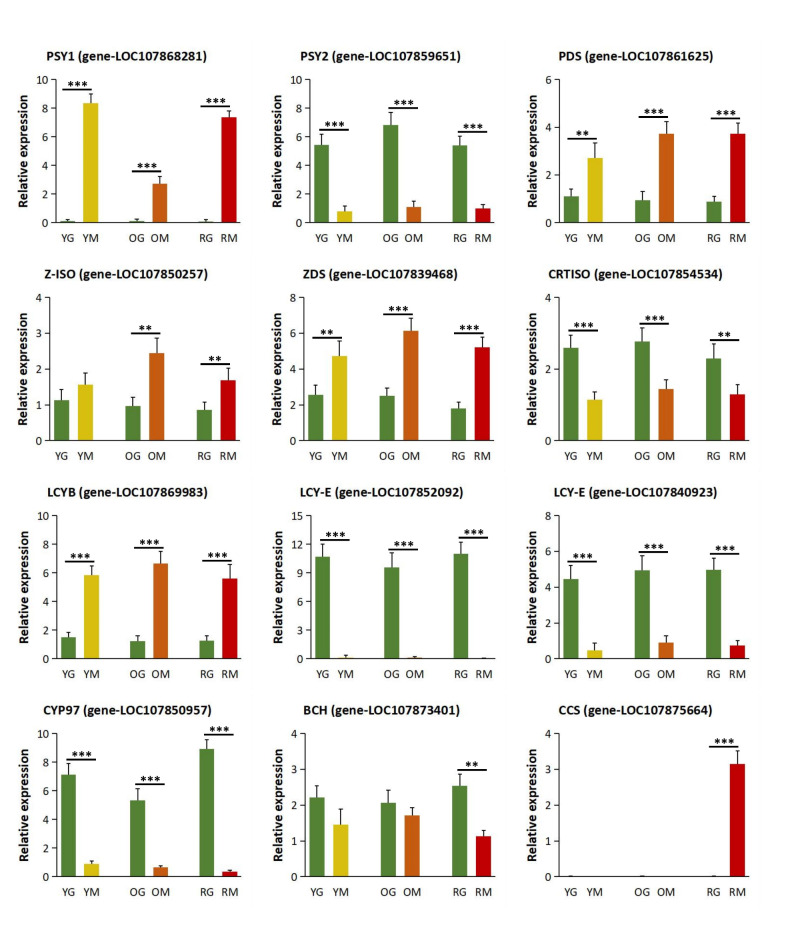
In order to confirm the accuracy of RNA-Seq data, we performed qRT-PCR experiments on the carotenoid biosynthesis pathway genes PSY1, PSY2, PDS, Z-ISO, ZDS, CRTISO, LCYB, LCY-E, CYP97, BCH, and CCS (Fig. 7). The expression levels of 12 genes were basically consistent with FPKM results, which supports the reliability of RNA-seq data in our study.
Fig. 7.
Relative expression level of 12 genes related to carotenoid biosynthesis pathway in bell pepper lines IL-Y, IL-O and IL-R. Data were presented as the average of three biological replicates of samples YG, YM, OG, OM, RG and RM, respectively. ** and *** indicated significant differences between the green and mature stage at p < 0.01 and p < 0.001 level, respectively.
Discussion
Carotenoids are natural pigments and essential fruits colorants of most horticultural crops60–64. Bell pepper contains many kinds of carotenoids with diverse fruit colors, and it is an excellent material for studying the regulation mechanism of carotenoid biosynthesis. In this study, three bell pepper lines with different fruit colors were analyzed by transcriptome and metabolome to explore the molecular mechanisms underlying the fruit color formation.
The main reason for the diversity of pepper fruit color is the variety of pigment types and contents accumulated, such as yellow, orange and red pepper, which is mainly due to the accumulation of various carotenoids4–6,65. In our study, 54 carotenoid metabolites were detected with various types enriched at different developmental stages (Table S1). Although carotenoids accumulates in fully expanded immature bell pepper fruits (50DAP), their types and content were very limited, and total carotenoid was mainly composed of lutein, β-carotene, etc. (Fig. 1B), consistent with previous study66. This suggests various carotenoids are generated mainly in late or middle developmental stages. However, in immature orange pepper OG, the content of zeaxanthin was also higher, which is worth our further exploration. With fruit ripening, (E/Z) phytoene accumulated more in OM and YM (Fig. 1F and G). As the first carotenoid product of carotenoid synthesis, (E/Z) phytoene provides precursors for the downstream carotenoid compounds synthesis. In YM and OM, yellow pigments lutein and zeaxanthin was higher, while orange pigments α-carotene and β-carotene accumulated more in OM (61.567 µg/g and 39.081 µg/g), but lower in YM (15.24 µg/g and 7.47 µg/g). The differential accumulation of α-carotene and β-carotene may be the reason for color difference between orange OM and yellow YM fruits (Table S2). Carotenoids of red RM was obviously different from that in non-red pepper fruit YM and OM. RM contains 6 carotenoids, capsanthin, capsorubin, β-carotene, zeaxanthin, (E/Z)-phytoene and violaxanthin, among which capsanthin was the highest, accounting for 71.02% of the total carotenoids (Fig. 1C), but it was not detected in OM, YM and RG, indicating that capsanthin is a unique pigment type of mature red bell pepper. In addition, the lutein content in RM was extremely low (0.43184 µg/g), which was 0.07% of the total carotenoid content (Fig. 1C, Table S2). These results suggests that different patterns of carotenoid accumulation is responsible for color differences among varieties.
At present, carotenoids biosynthetic pathway was clear. With IPP and DMAPP as precursors, various carotenoids are generated through IPI, GGPPS, PSY, PDS, ZDS, LCYB, LCYE and other enzymes (Fig. 6)23. By RNA-seq analysis, 14,961 DEGs were identified, and 48 genes were identified within carotenoid metabolic pathway (ko00906) in bell pepper genome according to KEGG enrichment analysis (Table S4). In plant plastid, three IPP and one DMAPP molecule synthesized from MEP pathway produce one GGPP, an important precursor of carotenoid biosynthesis, catalyzing by GGPPS67,68. In this study, two GGPPS genes (gene-LOC107843186 and gene-LOC107867046) were identified, with significantly increased expression at mature pepper (Table S4), thus increasing the proportion of GGPP in bell pepper fruits and changing the synthetic direction of penoids (Fig. 6). Phytoene is the first carotenoid product generated from rate-limiting reaction in carotenoid biosynthesis, GGPP condensation catalyzed by PSY25. Horticultural crops usually contain two or three PSY genes69–72. Two PSY genes were identified in our study, PSY1 (gene-LOC107868281) and PSY2 (gene-LOC107859651). Compared with the green ripening stage, PSY1 (gene-loc107868281) expression was significantly higher in mature bell pepper fruits, which was corresponding to the higher content of phytoene in YM, OM and RM (Fig. 6). The expression of PSY2 (gene-LOC107859651) decreased at maturity, indicating tissue expression preferences of PSY1 and PSY2.
Generally, PDS, ZDS, Z-ISO and CRTISO are involved in the conversion of colorless phytoene into colored carotenoids73. In our study, PDS (gene-LOC107861625) and ZDS (gene-LOC107839468) expression was significantly induced at maturity stage. Previous studies showed that photo-isomerization appears to replace CRTISO in chloroplasts74, may be responsible for why expression level of CRTISO (gene-LOC107854534) was not significantly different in comparison groups OG_vs_OM and RG_vs_RM. In plants, BCH and CYP play important roles in carotene hydroxylation75. BCH catalyzes the β-ring hydroxylation of β-carotene, and overexpression of BCH1 increased lutein content, but decreased β-carotene in Arabidopsis and kiwifruit 76–78. We found expression of BCH1 (gene-LOC107863219) was extremely high in YM and OM, consistent with high accumulation of lutein and zeaxanthin in YM and OM (Figs. 1C and 6). As to BCH2 (gene-LOC107873401), its expression decreased significantly at the mature stage. Similar results was obtained in sweet orange pulp, and CsBCH2 gene expression increased at early stage then decreased after color change39. The different expression patterns of BCH1 and BCH2 suggested their different roles in carotenoids accumulation in bell pepper fruits. In RM, BCH1 had high expression, but zeaxanthin was not accumulated much, and the possible reason may be that its downstream products were catalyzed by CCS to form capsanthin and capsorubin (Figs. 1C and 6). Previous study identified three loci controlling pepper fruit color, and CCS gene is the y loci79. Its structural variation is always the reason for color change during pepper ripening process80,81. CCS gene (gene-loc107875664) detected in our study showed high expression only in RM, consistent with the result that capsanthin and capsorubin were detected only in RM. Therefore, high expression of CCS caused red pigment accumulation and decrease of zeaxanthin, violaxanthin and antheraxanthin (Fig. 6, Table S1). Previous study showed inhibition of LCYE led to strong induction of LCYB and CCS with fruit maturity82, the same as our study, with LCYE (gene-LOC107840923 and gene-LOC107852092) expression decreasing, LCYB and CCS expression increased in mature bell pepper fruits. Therefore, antagonism between α and β branches, and decreased expression of LCYE may be the reason for lutein content decrease at mature stage (Table S1).
Taken together, we conclude that the structural genes PSY1 (gene-LOC107868281), PDS (gene-LOC107861625), ZDS (gene-LOC107839468), BCH1 (gene-LOC107863219), LCYB (gene-LOC107869983), and CCS (gene-LOC107875664) are key genes related to fruit color change in bell pepper.
TFs in plants, including MYB, bHLH, MADS, bZIP, AP2/ERF, and WRKY, play crucial roles in the regulation of carotenoid metabolism. An R-R-type MYB can directly bind to carotenoid biosynthetic genes promoter, and positively regulate capsanthin content29. In citrus, carotene genes expression was positively regulated by MADS49,50. In papaya, CpNAC1 is a positive regulator for carotenoid by interaction with CpPDS2/4 promoter83. The capsicum bHLH family may contribute to capsicum carotenoids and capsaicin biosynthesis84. In apple, MdAP2-34, a type of AP2/ERF, regulates phytoene and β-carotene accumulation85. By WCCNA analysis, blue and turquoise module were positively and negatively correlated with detected carotenoid metabolites, respectively (Fig. 5B). In blue module, 210 TFs were identified (Table S8), and 1 MADs (gene-LOC107839234), 2 AP2/ERF, 3 HD-ZIP, 1 MYB (gene-LOC107850892), 2 NAC, 1 WRKY (gene-LOC107859607) and 1 NF-Y were significantly up-regulated in YM, OM and RM (Table S8), and these TFs may be the positive regulator for carotenoid biosynthesis. In the blue module, structural genes such as ZDS (gene-LOC107839468), PDS (gene-LOC107861625), BCH (gene-LOC107863219), GGPS (gene-LOC107843186), and LCYB (gene-LOC107869983) involved in the carotenoid biosynthesis pathway were also identified (Fig. 6). These TFs and structural genes exhibit similar expression trends, jointly regulating carotenoid accumulation. It has been reported that MADS positively regulates the expression of PDS, BCH, and LCYB genes48–50, promoting carotenoid accumulation, which is consistent with our experimental results (Fig. 6). Additionally, WRKY also regulates BCH52, which aligns with our findings (Fig. 6). In turquoise module, 269 TFs were identified (Table S8), including 2 MYBs, 5 AP2/ERF, 3 HD-ZIP, 2 BHLH, 2 MAD and 1 NAC, which were significantly down-regulated, and they may be negative regulators for carotenoid biosynthesis. These results provide powerful clues for further study of carotenoid biosynthesis network.
Materials and methods
Plant materials
Three bell pepper lines, IL-Y, IL-O and IL-R, with yellow, orange and red fruit colors were used as materials (Fig. 1A). They were planted in the breeding field of Weifang University of Science and Technology (36.53°N, 118.46°E). Bell pepper fruits of IL-Y, IL-O and IL-R were sampled at 50 and 65 DAP with three replicates. After frozen immediately in liquid nitrogen, all samples were stored at -70 °C for subsequent transcriptome and metabolome analyses. Samples at 50DAP (green ripening stage) of IL-Y, IL-O and IL-R were named YG, OG and RG, and samples at 65DAP (mature period) were named YM, OM and RM.
Metabolome analysis of carotenoids
Analysis of sample preparation, metabolite extraction and identification were entrusted to Metware Biotechnology Co., Ltd. (Wuhan, China). Preparation and metabolite extraction were performed as described previously86,87. Carotenoids composition and content was detected by the ultra performance liquid chromatography (UPLC, ExionLC™ AD, https://sciex.com.cn/) system and tandem mass spectrometry (MS/MS, Applied Biosystems 6500 Triple Quadrupole, https://sciex.com.cn/) system. The multiple reaction monitoring (MRM) mode of triple quadrupole mass spectrometry was used for the quantification of carotenoid metabolite. Data acquisitions was processed by analyst 1.6.3 software (Sciex). The spectrometry data were analyzed by MultiQuant 3.0.3 software, in which the chromatographic peaks detected in different samples were integrated and corrected by referring to the retention times and peak shapes of standard compounds, ensuring the reliability of qualitative and quantitative analysis. The area of chromatographic peak represents the relative content of metabolites. Qualitative and quantitative results of tested substances was obtained using the linear equation and calculation formula. The DACs were screened under the thresholds of FC ≥ 2 or ≤ 0.5 and P-value ≤ 1.
Transcriptome analysis
Using methods described by Cao et al.88, total RNA of samples OG, OM, RG, RM, YG and YM was extracted. A total of 18 libraries were constructed with three biological replicates for each sample. RNA quality was verified by Qubit2.0 and Agilent 2100 Bioanalyzer. After qualified library inspection, Illumina HiSeq platform HiSeq 4000 (Illumina, San Diego, CA, USA) was used for sequencing. The high-quality clean reads were aligned to the reference genome (https://solgenomics.net/ftp/genomes/Capsicum_annuum/C.annuum_zunla/) using HISAT2. DEGs were screened by DESeq2 [25] with a threshold of |log2(FC)| >= 1 and FDR < 0.05. FPKM was used to evaluate gene relative expression. Gene function annotation was performed using multiple databases, including the Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology (GO), Clusters of Orthologous Groups (COG), NCBI, the non-redundant protein sequence (Nr) database, the manually annotated and reviewed protein sequence database Swiss-Prot, and the TrEMBL database which contains all the translations of EMBL nucleotide sequence entries.
WGCNA analysis
We performed WGCNA using the R package89. All DEGs and key DACs detected were selected for combined analysis. The co-expression network was visualized by free software Cytoscape.
qRT-PCR verification
Total RNA of samples YG, YM, OG, OM, RG and RM was extracted using method previously described88, respectively. The first-strand cDNA was reverse-transcribed by TIANScript IIcDNA Kit (Tiangen, China), and qRT-PCR was performed using SuperReal SYBR Green Premix kit (Tiangen, China) with three replications for each sample. Actin was used for reference, and relative gene expression was calculated by 2−∆∆Ct method. All primers used were listed in Table S10.
Conclusions
In this study, three bell pepper lines with different color (yellow, orange and red) were used as materials to conduct targeted metabolome and transcriptome analysis. The results showed that carotenoids showed different accumulation patterns during fruit development and showed significant variety specificity. The species and content of carotenoids in orange OM were significantly higher than YM and RM. Red pigment (capsanthin and capsorubin) was specifically enriched in RM, and yellow pigment (lutein and zeaxanthin) is the highest in YM and OM, and high content of orange pigment (α-carotene and β-carotene, etc.) was also accumulated in OM. Further analysis identified 27 genes involved in carotenoid biosynthesis, 9 of which expressed significantly higher at the mature stage (65DAP) than green ripening stage (50DAP), indicating their important roles in the color formation of bell pepper. Five modules positively correlated with carotenoid accumulation and one negative module were identified through WGCNA analysis, and key TFs and hub genes that may affect carotenoid content were predicted. By integrating the key DEGs and DACs, the study constructed a regulatory network for pepper fruit coloration, elucidating how the interplay between 14 key carotenoid biosynthesis genes, 5 key TFs, and 7 key metabolites collectively controlled the color changes of pepper fruits during different developmental stages. To sum up, the results not only provides important insights into carotenoid synthesis pathway, but also lays a solid data base for revealing color formation mechanism in bell pepper.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This research was funded by the Weifang Science and Technology Development Plan Project (2020GX065, 2023GX052); the Natural Science Foundation of China (32101804); the Discipline Development Project of Weifang University of Science and Technology (2021XKJS27); the High-level Talent Start-up project of Weifang University of Science and Technology (KJRC2023017).
Author contributions
Conceptualization, Q.X., Q.Z. and X.Z.; software, Q.Z., A.Z., T.H. and X.T.; validation, H.X., Q.X. and Q.Z.; investigation, D.L., Y.L., Y.Q. and F.L.; resources, Q.X.; data curation, Q.Z., A.Z., and X.Z.; writing-original draft preparation, Q.X. and Q.Z.; writing-review and editing, X.Z.; supervision, Q.X. and X.Z.; project administration, Q.X. and X.Z.; funding acquisition, Q.X. and X.Z. Q.X. and Q.Z contribute equally to the manuscript. Q.X. and Q.Z contribute equally to the manuscript. All authors have read and agreed to the published version of the manuscript.
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Qiqin Xue, Qingxia Zhang and Aiai Zhang.
References
- 1.Wahyuni, Y., Ballester, A. R., Sudarmonowati, E., Bino, R. J. & Bovy, A. G. Secondary metabolites of Capsicum species and their importance in the human diet. J. Nat. Prod.76, 783–793. 10.1021/np300898z (2013). [DOI] [PubMed] [Google Scholar]
- 2.Wahyuni, Y., Ballester, A. R., Sudarmonowati, E., Bino, R. J. & Bovy, A. G. Metabolite biodiversity in pepper (Capsicum) fruits of thirty-two diverse accessions: variation in health-related compounds and implications for breeding. Phytochemistry72, 1358–1370. 10.1016/j.phytochem.2011.03.016 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Morales-Soriano, E. et al. Carotenoid profile and basic structural indicators of native Peruvian Chili peppers. Eur. Food Res. Technol.245, 717–732. 10.1007/s00217-018-3193-2 (2018). [Google Scholar]
- 4.Tian, S. L., Li, L., Shah, S. N. M. & Gong, Z. H. The relationship between red fruit colour formation and key genes of capsanthin biosynthesis pathway in Capsicum annuum. Biol. Plant.59, 507–513. 10.1007/s10535-015-0529-7 (2015). [Google Scholar]
- 5.Guzman, I., Hamby, S., Romero, J., Bosland, P. W. & O’Connell, M. A. Variability of carotenoid biosynthesis in orange colored Capsicum spp. Plant. Sci.179, 49–59. 10.1016/j.plantsci.2010.04.014 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Azevedo-Meleiro, C. H. & Rodriguez-Amaya, D. B. Qualitative and quantitative differences in the carotenoid composition of yellow and red peppers determined by HPLC-DAD-MS. J. Sep. Sci.32, 3652–3658. 10.1002/jssc.200900311 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Botella-Pavía, P. & Rodríguez-Concepción, M. Carotenoid biotechnology in plants for nutritionally improved foods. Physiol. Plant.126, 369–381. 10.1111/j.1399-3054.2006.00632.x (2006). [Google Scholar]
- 8.Alcaíno, J., Baeza, M. & Cifuentes, V. Carotenoid distribution in Nature. Sub-Cell Biochem.79, 3–33. 10.1007/978-3-319-39126-7_1 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez-Concepcion, M. et al. A global perspective on carotenoids: metabolism, biotechnology, and benefits for nutrition and health. Prog Lipid Res.70, 62–93. 10.1016/j.plipres.2018.04.004 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Domonkos, I., Kis, M., Gombos, Z. & Ughy, B. Carotenoids, versatile components of oxygenic photosynthesis. Prog Lipid Res.52, 539–561. 10.1016/j.plipres.2013.07.001 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Nisar, N., Li, L., Lu, S., Khin, N. C. & Pogson, B. J. Carotenoid metabolism in plants. Mol. Plant.8, 68–82. 10.1016/j.molp.2014.12.007 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Niyogi, K. K. PHOTOPROTECTION REVISITED: genetic and molecular approaches. Annu. Rev. Plant. Biol.50, 333–359. 10.1146/annurev.arplant.50.1.333 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Bartley, G. E. & Scolnik, P. A. Plant carotenoids: pigments for photoprotection, visual attraction, and human health. Plant. cell.7, 1027–1038. 10.1105/tpc.7.7.1027 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreno, J. C., Mi, J., Alagoz, Y. & Al-Babili, S. Plant apocarotenoids: from retrograde signaling to interspecific communication. Plant. J.105, 351–375. 10.1111/tpj.15102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang, Y. et al. The Arabidopsis D27-LIKE1 is a cis/cis/trans-β-carotene isomerase that contributes to strigolactone biosynthesis and negatively impacts ABA level. Plant. J.113, 986–1003. 10.1111/tpj.16095 (2023). [DOI] [PubMed] [Google Scholar]
- 16.Zheng, X., Giuliano, G. & Al-Babili, S. Carotenoid biofortification in crop plants: citius, altius, fortius. Biochim. et Biophys. acta Mol. cell. Biology Lipids1865, 158664. 10.1016/j.bbalip.2020.158664 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Xavier, A. A. & Pérez-Gálvez, A. Carotenoids as a source of antioxidants in the Diet. Sub-Cell Biochem.79, 359–375. 10.1007/978-3-319-39126-7_14 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Mohd Hassan, N., Yusof, N. A., Yahaya, A. F., Rozali, M. & Othman, R. N. N. Carotenoids of Capsicum Fruits: Pigment Profile and Health-Promoting Functional Attributes. Antioxidants (Basel, Switzerland) 8, (2019). 10.3390/antiox8100469 [DOI] [PMC free article] [PubMed]
- 19.Krinsky, N. I. & Johnson, E. J. Carotenoid actions and their relation to health and disease. Mol. Aspects Med.26, 459–516. 10.1016/j.mam.2005.10.001 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Arimboor, R., Natarajan, R. B., Menon, K. R., Chandrasekhar, L. P. & Moorkoth, V. Red pepper (Capsicum annuum) carotenoids as a source of natural food colors: analysis and stability-a review. J. Food Sci. Technol.52, 1258–1271. 10.1007/s13197-014-1260-7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delgado-Vargas, F. & Paredes-Lopez, O. Natural colorants for food and nutraceutical uses. CRC Press.10.1201/9781420031713 (2002). [Google Scholar]
- 22.Pu, X., Dong, X., Li, Q., Chen, Z. & Liu, L. An update on the function and regulation of methylerythritol phosphate and mevalonate pathways and their evolutionary dynamics. J. Integr. Plant. Biol.63, 1211–1226. 10.1111/jipb.13076 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Venkatesh, J. et al. Update on the genetic and molecular regulation of the biosynthetic pathways underlying pepper fruit color and pungency. Curr. Plant. Biol. 35–36. 10.1016/j.cpb.2023.100303 (2023).
- 24.Watkins, J. L. & Pogson, B. J. Prospects for Carotenoid Biofortification Targeting Retention and Catabolism. Trends Plant. Sci.25, 501–512. 10.1016/j.tplants.2019.12.021 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Zhou, X. et al. Phytoene synthase: the key rate-limiting enzyme of Carotenoid Biosynthesis in plants. Front. Plant. Sci.13, 884720. 10.3389/fpls.2022.884720 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cazzonelli, C. I. & Pogson, B. J. Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant. Sci.15, 266–274. 10.1016/j.tplants.2010.02.003 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Lado, J., Zacarías, L. & Rodrigo, M. J. Regulation of Carotenoid Biosynthesis during Fruit Development. Sub-Cell Biochem.79, 161–198. 10.1007/978-3-319-39126-7_6 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Neuman, H., Galpaz, N., Cunningham, F. X. Jr., Zamir, D. & Hirschberg, J. The tomato mutation nxd1 reveals a gene necessary for neoxanthin biosynthesis and demonstrates that violaxanthin is a sufficient precursor for abscisic acid biosynthesis. Plant. J.78, 80–93. 10.1111/tpj.12451 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Song, J. et al. An R-R-type MYB transcription factor promotes non-climacteric pepper fruit carotenoid pigment biosynthesis. Plant. J.115, 724–741. 10.1111/tpj.16257 (2023). [DOI] [PubMed] [Google Scholar]
- 30.Hurtado-Hernandez, H. & Smith, P. G. Inheritance of mature fruit color in Capsicum annuum L. J. Hered. 76, 211–213. 10.1093/oxfordjournals.jhered.a110070 (1985).
- 31.Lefebvre, V., Kuntz, M., Camara, B. & Palloix, A. The capsanthin-capsorubin synthase gene: a candidate gene for the y locus controlling the red fruit colour in pepper. Plant. Mol. Biol.36, 785–789. 10.1023/a:1005966313415 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Jeong, H. B. et al. Candidate Gene Analysis reveals that the Fruit Color Locus C1 corresponds to PRR2 in Pepper (Capsicum frutescens). Front. Plant. Sci.11, 399. 10.3389/fpls.2020.00399 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huh, J. H. et al. A candidate gene approach identified phytoene synthase as the locus for mature fruit color in red pepper (Capsicum spp). Theor. Appl. Genet.102, 524–530. 10.1007/s001220051677 (2001). [Google Scholar]
- 34.Jang, S. J. et al. Phytoene synthase 2 can compensate for the absence of PSY1 in the control of color in Capsicum fruit. J. Exp. Bot.71, 3417–3427. 10.1093/jxb/eraa155 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeong, H. B. et al. Single-molecule real-time sequencing reveals diverse allelic variations in carotenoid biosynthetic genes in pepper (Capsicum spp). Plant. Biotechnol. J.17, 1081–1093. 10.1111/pbi.13039 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borovsky, Y. et al. Induced mutation in β-CAROTENE HYDROXYLASE results in accumulation of β-carotene and conversion of red to orange color in pepper fruit. Theor. Appl. Genet.126, 557–565. 10.1007/s00122-012-2001-9 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Lee, S. Y. et al. A mutation in Zeaxanthin epoxidase contributes to orange coloration and alters carotenoid contents in pepper fruit (Capsicum annuum). Plant. J.106, 1692–1707. 10.1111/tpj.15264 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Jang, S. et al. Investigation of genetic factors regulating chlorophyll and carotenoid biosynthesis in red pepper fruit. Front. Plant. Sci.13, 922963. 10.3389/fpls.2022.922963 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, Y. et al. Citrus β-carotene hydroxylase 2 (BCH2) participates in xanthophyll synthesis by catalyzing the hydroxylation of β-carotene and compensates for BCH1 in citrus carotenoid metabolism. Hortic. Res.10, uhac290. 10.1093/hr/uhac290 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma, X., Yu, Y. N., Jia, J. H., Li, Q. H. & Gong, Z. H. The pepper MYB transcription factor CaMYB306 accelerates fruit coloration and negatively regulates cold resistance. Sci. Hortic.295, 110892. 10.1016/j.scienta.2022.110892 (2022). [Google Scholar]
- 41.Stanley, L. & Yuan, Y. W. Transcriptional regulation of Carotenoid Biosynthesis in plants: so many regulators, so little Consensus. Front. Plant. Sci.10, 1017. 10.3389/fpls.2019.01017 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye, J. et al. Transcriptome Profiling of Tomato Fruit Development Reveals Transcription Factors Associated with ascorbic acid, Carotenoid and Flavonoid Biosynthesis. PloS One10, e0130885. 10.1371/journal.pone.0130885 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ampomah-Dwamena, C. et al. A kiwifruit (Actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New. Phytol221, 309–325. 10.1111/nph.15362 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meng, Y. et al. The MYB activator WHITE PETAL1 associates with MtTT8 and MtWD40-1 to regulate carotenoid-derived Flower pigmentation in Medicago truncatula. Plant. Cell.31, 2751–2767. 10.1105/tpc.19.00480 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi, Y. et al. SlZHD17 is involved in the control of chlorophyll and carotenoid metabolism in tomato fruit. Hortic. Res.8, 259. 10.1038/s41438-021-00696-8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toledo-Ortiz, G. et al. Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA107, 11626. 10.1073/pnas.0914428107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou, D. et al. Papaya CpbHLH1/2 regulate carotenoid biosynthesis-related genes during papaya fruit ripening. Hortic. Res.6, 80. 10.1038/s41438-019-0162-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu, K. et al. Transcription factor CsMADS3 coordinately regulates chlorophyll and carotenoid pools in Citrus hesperidium. Plant. Physiol.193, 519–536. 10.1093/plphys/kiad300 (2023). [DOI] [PubMed] [Google Scholar]
- 49.Lu, S. et al. The Citrus transcription factor CsMADS6 modulates Carotenoid Metabolism by directly regulating carotenogenic genes. Plant. Physiol.176, 2657–2676. 10.1104/pp.17.01830 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu, S. et al. A fruit ripening-associated transcription factor CsMADS5 positively regulates carotenoid biosynthesis in citrus. J. Exp. Bot.72, 3028–3043. 10.1093/jxb/erab045 (2021). [DOI] [PubMed] [Google Scholar]
- 51.Sun, Q. et al. The transcriptional regulatory module CsHB5-CsbZIP44 positively regulates abscisic acid-mediated carotenoid biosynthesis in citrus (Citrus spp). Plant. Biotechnol. J.22, 722–737. 10.1111/pbi.14219 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen, H. et al. Transcription factor CrWRKY42 coregulates chlorophyll degradation and carotenoid biosynthesis in citrus. Plant. Physiol.195, 728–744. 10.1093/plphys/kiae048 (2024). [DOI] [PubMed] [Google Scholar]
- 53.Lee, H. Y. et al. Genetic diversity and population structure analysis to construct a core collection from a large Capsicum germplasm. BMC Genet.17, 142. 10.1186/s12863-016-0452-8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu, Y. et al. Integrative analysis of metabolome and transcriptome reveals the mechanism of color formation in pepper fruit (Capsicum annuum L). Food Chem.306, 125629. 10.1016/j.foodchem.2019.125629 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Télef, N. et al. Sucrose deficiency delays lycopene accumulation in tomato fruit pericarp discs. Plant. Mol. Biol.62, 453–469. 10.1007/s11103-006-9033-y (2006). [DOI] [PubMed] [Google Scholar]
- 56.Horner, H. T. et al. Amyloplast to chromoplast conversion in developing ornamental tobacco floral nectaries provides sugar for nectar and antioxidants for protection. Am. J. Bot.94, 12–24. 10.3732/ajb.94.1.12 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Flores-Pérez, U. et al. Pleiotropic regulatory locus 1 (PRL1) integrates the regulation of sugar responses with isoprenoid metabolism in Arabidopsis. Mol. Plant.3, 101–112. 10.1093/mp/ssp100 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Sadali, N. M., Sowden, R. G., Ling, Q. & Jarvis, R. P. Differentiation of chromoplasts and other plastids in plants. Plant. Cell. Rep.38, 803–818. 10.1007/s00299-019-02420-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, Y. Q. et al. Proteomic analysis of chromoplasts from six crop species reveals insights into chromoplast function and development. J. Exp. Bot.64, 949–961. 10.1093/jxb/ers375 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu, X. et al. Organic acid and sugar components accumulation and flavor associated metabolites dynamic changes in yellow- and white-fleshed seedless loquats (Eriobotrya japonica). Food Chem. : X. 21, 101046. 10.1016/j.fochx.2023.101046 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meng, Y. et al. The MYB Activator WHITE PETAL1 Associates with MtTT8 and MtWD40-1 to Regulate Carotenoid-Derived Flower Pigmentation in Medicago truncatula. Plant cell 31, 2751–2767, (2019). 10.1105/tpc.19.00480 [DOI] [PMC free article] [PubMed]
- 62.Peng, L. et al. Integrated Metabolome and Transcriptome Analysis of Fruit Flavor and carotenoids Biosynthesis differences between mature-green and tree-ripe of cv. Golden Phoenix Mangoes (Mangifera indica L). Front. Plant. Sci.13, 816492. 10.3389/fpls.2022.816492 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou, W. et al. Analysis of carotenoid content and diversity in apricots (Prunus armeniaca L.) grown in China. Food Chem.330, 127223. 10.1016/j.foodchem.2020.127223 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Xia, Y. et al. Integrated metabolic profiling and transcriptome analysis of pigment accumulation in Lonicera japonica flower petals during colour-transition. BMC Plant. Biol.21, 98. 10.1186/s12870-021-02877-y (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kilcrease, J. et al. Correlations of carotenoid content and transcript abundances for fibrillin and carotenogenic enzymes in Capsicum annum fruit pericarp. Plant. Sci.232, 57–66. 10.1016/j.plantsci.2014.12.014 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Matsufuji, H., Ishikawa, K., Nunomura, O., Chino, M. & Takeda, M. Anti-oxidant content of different coloured sweet peppers, white, green, yellow, orange and red (Capsicum annuum L). Int. J. Food Sci. Technol.42, 1482–1488. 10.1111/j.1365-2621.2006.01368.x (2007). [Google Scholar]
- 67.Bouvier, F., Rahier, A. & Camara, B. Biogenesis, molecular regulation and function of plant isoprenoids. Prog Lipid Res.44, 357–429. 10.1016/j.plipres.2005.09.003 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Gershenzon, J. & Dudareva, N. The function of terpene natural products in the natural world. Nat. Chem. Biol.3, 408–414. 10.1038/nchembio.2007.5 (2007). [DOI] [PubMed] [Google Scholar]
- 69.Kachanovsky, D. E., Filler, S., Isaacson, T. & Hirschberg, J. Epistasis in tomato color mutations involves regulation of phytoene synthase 1 expression by cis-carotenoids. PNAS109, 19021–19026. 10.1073/pnas.1214808109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fraser, P. D., Kiano, J. W., Truesdale, M. R., Schuch, W. & Bramley, P. M. Phytoene synthase-2 enzyme activity in tomato does not contribute to carotenoid synthesis in ripening fruit. Plant. Mol. Biol.40, 687–698. 10.1023/a:1006256302570 (1999). [DOI] [PubMed] [Google Scholar]
- 71.Stauder, R. et al. Strigolactone levels in Dicot roots are determined by an ancestral symbiosis-regulated clade of the PHYTOENE SYNTHASE Gene Family. Front. Plant. Sci.9, 255. 10.3389/fpls.2018.00255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peng, G. et al. The role of 1-deoxy-d-xylulose-5-phosphate synthase and phytoene synthase gene family in citrus carotenoid accumulation. Plant. Physiol. Biochem.71, 67–76. 10.1016/j.plaphy.2013.06.031 (2013). [DOI] [PubMed] [Google Scholar]
- 73.Li, C., Li, B. & Han, X. [Advances in phytoene dehydrogenase - A review]. Wei Sheng Wu Xue bao = Acta Microbiol. Sinica56, 1680–1690 (2016). [PubMed] [Google Scholar]
- 74.Park, H., Kreunen, S. S., Cuttriss, A. J., DellaPenna, D. & Pogson, B. J. Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant. cell.14, 321–332. 10.1105/tpc.010302 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma, G. et al. Expression and functional analysis of citrus carotene hydroxylases: unravelling the xanthophyll biosynthesis in citrus fruits. BMC Plant. Biol.16, 148. 10.1186/s12870-016-0840-2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim, J. E., Cheng, K. M., Craft, N. E., Hamberger, B. & Douglas, C. J. Over-expression of Arabidopsis thaliana carotenoid hydroxylases individually and in combination with a beta-carotene ketolase provides insight into in vivo functions. Phytochemistry71, 168–178. 10.1016/j.phytochem.2009.10.011 (2010). [DOI] [PubMed] [Google Scholar]
- 77.Xia, H. et al. Characterization and functional validation of β-carotene hydroxylase AcBCH genes in Actinidia chinensis. Hortic. Res.9, uhac063. 10.1093/hr/uhac063 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.D’Ambrosio, C., Stigliani, A. L. & Giorio, G. Overexpression of CrtR-b2 (carotene beta hydroxylase 2) from S. Lycopersicum L. differentially affects xanthophyll synthesis and accumulation in transgenic tomato plants. Transgenic Res.20, 47–60. 10.1007/s11248-010-9387-4 (2011). [DOI] [PubMed] [Google Scholar]
- 79.Popovsky, S. & Paran, I. Molecular genetics of the y locus in pepper: its relation to capsanthin-capsorubin synthase and to fruit color. Theor. Appl. Genet.101, 86–89. 10.1007/s001220051453 (2000). [Google Scholar]
- 80.Bouvier, F., Hugueney, P., d’Harlingue, A., Kuntz, M. & Camara, B. Xanthophyll biosynthesis in chromoplasts: isolation and molecular cloning of an enzyme catalyzing the conversion of 5,6-epoxycarotenoid into ketocarotenoid. Plant. J.6, 45–54. 10.1046/j.1365-313x.1994.6010045.x (1994). [DOI] [PubMed] [Google Scholar]
- 81.Houlné, G., Schantz, M. L., Meyer, B., Pozueta-Romero, J. & Schantz, R. A chromoplast-specific protein in Capsicum annuum: characterization and expression of the corresponding gene. Curr. Genet.26, 524–527. 10.1007/bf00309944 (1994). [DOI] [PubMed] [Google Scholar]
- 82.Wang, Q. et al. Manipulation of Carotenoid Metabolic Flux by Lycopene Cyclization in Ripening Red Pepper (Capsicum annuum var. Conoides) fruits. J. Agric. Food Chem.67, 4300–4310. 10.1021/acs.jafc.9b00756 (2019). [DOI] [PubMed] [Google Scholar]
- 83.Fu, C. C. et al. The papaya transcription factor CpNAC1 modulates Carotenoid Biosynthesis through activating Phytoene desaturase genes CpPDS2/4 during Fruit Ripening. J. Agric. Food Chem.64, 5454–5463. 10.1021/acs.jafc.6b01020 (2016). [DOI] [PubMed] [Google Scholar]
- 84.Liu, R. et al. Genome-wide identification of the Capsicum bHLH transcription factor family: discovery of a candidate regulator involved in the regulation of species-specific bioactive metabolites. BMC Plant. Biol.21, 262. 10.1186/s12870-021-03004-7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dang, Q. et al. An apple (Malus domestica) AP2/ERF transcription factor modulates carotenoid accumulation. Hortic. Res.8, 223. 10.1038/s41438-021-00694-w (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krinsky, N. I., Mayne, S. T. & Sies, H. Carotenoids in health and disease. CRC Press. 10.1201/9780203026649 (2004).
- 87.Geyer, R., Peacock, A. D., White, D. C., Lytle, C. & Van Berkel, G. J. Atmospheric pressure chemical ionization and atmospheric pressure photoionization for simultaneous mass spectrometric analysis of microbial respiratory ubiquinones and menaquinones. J. Mass. Spectrom.39, 922–929. 10.1002/jms.670 (2004). [DOI] [PubMed] [Google Scholar]
- 88.Cao, K. et al. Transcriptome analysis reveals novel genes involved in anthocyanin biosynthesis in the flesh of peach. Plant. Physiol. Biochem.123, 94–102. 10.1016/j.plaphy.2017.12.005 (2018). [DOI] [PubMed] [Google Scholar]
- 89.Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform.9, 559. 10.1186/1471-2105-9-559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.