Abstract
Background
Krüppel-like factor 4 (KLF4), a transcription factor, is involved in various biological processes. However, the role of KLF4 in regulating the intestinal epithelial barrier (IEB) in inflammatory bowel disease (IBD) and its mechanism have not been extensively studied.
Methods
KLF4 expression in IBD patients and colitis mice was analyzed using Gene Expression Omnibus(GEO) database, immunohistochemistry (IHC) and Western blot. The roles of KLF4 in IEB and colitis symptoms were verified in dextran sulfate sodium (DSS)-induced and 2,4,6-trinitrobenzenesulfonic acid (TNBS)-induced colitis model mice using an adenovirus carrying KLF4 shRNA (shKLF4-Adv). Furthermore, the influence of KLF4 on trans-epithelium electrical resistance (TEER), paracellular permeability, apical junction complex (AJC) protein expression and apoptosis was assessed in vitro and in vivo. MeRIP and RIP assays were used to verify the effects of m6A modification on KLF4 expression.
Results
KLF4 expression was significantly decreased in IBD patients and was negatively associated with inflammatory features. KLF4 deletion aggravated colitis symptoms and IEB injuries by reducing AJC protein expression and increasing apoptosis in mice with colitis. Furthermore, KLF4 transcriptionally regulated the expression of AJC proteins and inhibited apoptosis by reducing cellular ROS levels and proinflammatory cytokine expression. Moreover, we observed that METTL3/ALKBH5/YTHDF2-mediated m6A modification led to a decrease in KLF4 expression in Caco-2 cells. In addition, APTO-253, an inducer of KLF4, exhibited a synergistic effect with mesalazine on IEB function.
Conclusions
Our study demonstrated that KLF4 is a crucial regulator of IEB, suggesting that targeting KLF4 may be a promising therapeutic alternative for IBD.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-024-05514-7.
Keywords: Inflammatory bowel disease, KLF4, Intestinal epithelial barrier, Apical junction complex, Apoptosis, m6A Modification
Introduction
Inflammatory bowel disease (IBD) is a chronic relapsing gastrointestinal (GI) inflammatory condition of unknown etiology [1]. Through a comprehensive assessment encompassing clinical, biochemical, stool, endoscopic, cross-sectional imaging, and histological evaluations, IBD can be categorized into ulcerative colitis (UC) and Crohn's disease (CD) [2]. Although the specific etiology of IBD remains unclear, dysfunction of the intestinal epithelial barrier(IEB) has been implicated in its pathogenesis [3, 4].
The IEB is mainly composed of mucus and epithelial cells, which act as a dynamic interface between the luminal contents of food, commensal and pathogenic bacteria, and the gastrointestinal tract [5, 6]. Dysfunction of the IEB contributes to a variety of chronic inflammatory gastrointestinal conditions, such as IBD [6]. Epithelial cells and the intercellular apical junction complex (AJC), which consists of tight junctions (TJs) and adhesive junctions (AJs), play important roles in maintaining the IEB [7, 8]. Imparied epithelial cells and AJCs leads to increased intestinal permeability, potentially contributing to the development and exacerbation of IBD [9–12]. To restore IEB integrity and alleviate inflammation, new molecular targets must be investigated.
As a member of the Krüppel-like factor (KLF) family, KLF4 is composed of multiple functional domains, namely, a transcriptional activation domain, a DNA binding domain, and a transcriptional inhibition domain, and possesses a conserved mammalian zinc finger structure [13]. As a transcription factor, KLF4 plays a crucial role in regulating gene expression and is implicated in numerous biological processes, including embryonic development, cell differentiation, stem cell induction, neuronal regeneration, tumor formation, metastasis, and drug resistance [14]. Within the digestive tract, KLF4 is predominantly expressed in the villi and surface differentiated epithelial cells of the small intestine and colon [15–19]. Yuan Chen et al. indicated that KLF4 interacts with TXNIP and regulates the pyroptosis process in UC through the TXNIP/NLRP3 pathway [20]. KLF4 has also been implicated in the regulation of blood-tumor barrier permeability [21] and is particularly important in the final stages of goblet cell differentiation [17]. However, the function of KLF4 in IBD and the intestinal barrier and the related mechanisms have not been fully elucidated.
In the present study, we observed a decrease in KLF4 expression in the inflamed intestinal mucosa of individuals with IBD and in dextran sulfate sodium (DSS)-induced and 2,4,6-trinitrobenzenesulfonic acid (TNBS)-induced colitis model mice. KLF4 knockdown aggravated inflammation and barrier dysfunction by reducing the protein expression of AJCs and increasing the apoptosis of epithelial cells both in vivo and in vitro. Furthermore, we found that m6A modification resulted in KLF4 mRNA instability and downregulation in IBD patients. Overall, our findings reveal a novel role for KLF4 in intestinal barrier inflammation, and targeting KLF4 may be a novel strategy for treating patients with IBD.
Materials and methods
Human samples
Ethical approval for the study was granted by the Ethics Committee at the First Affiliated Hospital of Soochow University (Suzhou, China. No. 2023532), and informed consent was also obtained from all participants. Paraffin-embedded tissue specimens from UC or CD patients were collected from the First Affiliated Hospital of Soochow University. Comprehensive clinicopathological data for both UC and CD patients can be found in Additional file 1 Supplementary Table 1–2. Diagnosing CD or UC involves utilizing various methods such as clinical, biochemical, stool, endoscopic, imaging, and histological tests as recommended by European Crohn’s and Colitis Organization [ECCO] and European Society of Gastrointestinal and Abdominal Radiology [ESGAR] guidelines [2]. Ulcerative colitis disease activity and the Crohn's disease activity index (CDAI) were assessed according to established protocols [22].
Microarray data
Microarray data from GSE164985, GSE75214, GSE10616, GSE179285, GSE38713, GSE10191, GSE20881, GSE95095, GSE102133, and GSE186582 were obtained from the GEO database at http //www.ncbi.nih.gov/geo. We obtained the original data in the form of MINiML files. The analysis of gene expression was conducted utilizing the R software, specifically the ggplot2 package. The data extracted from the GEO database was normalized through log2 transformation and the microarray data was further normalized using the normalize quantiles function within the preprocessCore package in R software (version 3.4.1).
Mice and colitis models
The Ethics Committee of Soochow University (Suzhou, China) approved all animal experimental procedures (No.202311A0034). Male mice of the C57BL/6 or BALB/c strains, aged between 6 and 8 weeks, were acquired from the Laboratory Animal Center at Soochow University. A DSS-induced colitis model was established as indicated previously [23]. In brief, C57BL/6 mice were fed 2.0% DSS (#160,110, MP Biomedicals, USA) for 7 days. A TNBS-induced acute colitis model was established as previously described [23]. Following a 7-day period of skin sensitization with a 1% (wt/vol) TNBS presensitization solution, BALB/c mice were lightly anesthetized (intraperitoneal injection dosing of 80 µl/10 g body weight of ketamine/xylazine solution) and administered 2.5% (wt/vol) TNBS through a catheter inserted 4 cm into the colonic lumen. Control mice received a 100 μl dose of 50% ethanol.
In order to investigate the function of KLF4 in colitis, mice were treated with either KLF4 shRNA adenovirus (shKLF4-Adv, 1 × 1010 PFU/ml, 100 μl/mouse) or control adenovirus (control-Adv) via intracolonic administration right after receiving 2.0% DSS treatment. For TNBS-induced colitis model mice, on the day of TNBS presensitization, the first shKLF4-Adv (1 × 10^10 PFU/ml, 100 μl/mouse) or control-Adv enema was concurrently started.
In both models, mice were observed and weighed daily. Intestinal inflammation severity was evaluated with the disease activity index (DAI) according to previous descriptions [23]. The DSS-challenged mice were sacrificed at 7 days after DSS treatment, and the TNBS-treated mice were sacrificed at 3 days after 2.5% (wt/vol) TNBS treatment. The colons were removed for macroscopic examination, measuring the length of the colon, analyzing histopathology using hematoxylin–eosin (H&E) and alcian blue periodic acid–Schiff (AB-PAS) staining, and conducting cytokine analysis using RT‒qPCR.
Histology
Following treatment with 4% paraformaldehyde (#P1110, Solarbio, Beijing, China) for 24 h, the colon tissues of the mice were preserved in paraffin and cut into 5 µm slices. Following the guidelines provided by the manufacturer, H&E staining was performed using a staining kit from Beyotime in Shanghai, China. Histological scores for colitis induced by DSS and TNBS were calculated following the traditional protocol [23]. Furthermore, AB-PAS staining was carried out following the guidelines provided by the manufacturer of the product from Servicebio in Wuhan, China.
Cell culture and stimulation conditions
The colonic epithelial adenocarcinoma cell line Caco-2 was acquired from the American Tissue Culture Collection (ATCC, USA) and the colonic epithelial cell line NCM460 was purchased from Incell Corporation LLC (INCELL, USA). Caco-2 and NCM460 were grown in RPMI-1640 (Eallbio, Beijing, China) with 10% fetal bovine serum (FBS, Eallbio) and 1% penicillin‒streptomycin (Beyotime) in a humidified incubator with 5% CO2 at 37 °C. Intestinal epithelial monolayer barriers were constructed using Caco-2 cells. To investigate the role of KLF4 in intestinal epithelial barriers, Caco-2 monolayers were incubated with H2O2 (#108,597, Sigma-Aldrich, St. Louis, USA) for 4 h or TNF-α (#SRP3177, Sigma‒Aldrich)/IFN-γ (#SRP3058, Sigma‒Aldrich) mixtures for 18 h. To explore the synergistic impact of mesalazine (#HY-15027, MCE, New Jersey, USA) and KLF4 overexpression on the maintenance of intestinal epithelial barrier function, mesalazine at a concentration of 5 mmol/L and the KLF4 inducer APTO-253 (#HY-16291, MCE) at a 1 μmol/L concentration were added to Caco-2 cells and incubated for 24 h.
Cell transfection and infection
MiaoLingBio (Wuhan, China) designed and synthesized overexpression plasmids for KLF4, ALKBH5, and METTL3. GenePharma (Suzhou, China) provided commercial siRNAs targeting KLF4, ALKBH5, METTL3, YTHDF1/2/3, YTHDC1/2, and IGF2BP1/2/3 along with their corresponding controls. Caco-2 cells were grown in 6-well dishes and LipofectamineTM 3000 (#L3000001, Thermo Scientific, Waltham, USA) was utilized for transfecting Caco-2 cells with plasmids or siRNAs. GenePharma synthesized lentiviruses for KLF4 with both overexpression and knockdown capabilities, as well as their respective controls. Upon reaching 40% confluence, the Caco-2 cells were exposed to lentiviral particles with a multiplicity of infection (MOI) of 40. The effectiveness of the infection was assessed through quantitative real‑time polymerase chain reaction (qRT‒PCR) and Western blot analysis.
Immunohistochemistry (IHC)
Sections of human or mouse tissue embedded in paraffin were sliced to a thickness of 5 μm, followed by dewaxing, retrieval of antigens at high temperature, blocking of nonspecific antigens, and overnight exposure to primary antibodies at 4 °C, then incubated with secondary antibodies conjugated with HRP for 30 min at 25 °C. The antibodies utilized in the IHC assay are detailed in Supplementary Table 3. Detection was carried out using DAB substrate (#8059, CST, Danvers, USA).
Immunofluorescence (IF) staining
Paraffin-embedded sections of colon tissues were dehydrated in xylene and ethanol before being blocked with goat serum. For IF staining of Caco-2 cells, we cultured Caco-2 cells on glass coverslips the day before experimentation. The cells were treated with 4% paraformaldehyde and then exposed to 0.5% Triton X-100 for permeabilization. Afterward, the sections or cells were treated with primary antibodies overnight at 4 °C, then stained with secondary antibodies. Following staining of the nuclei with DAPI (#P0131, Beyotime), fluorescence images were captured using an immunofluorescence microscope. The antibodies utilized for immunofluorescence staining can be found in Supplementary Table 3.
Western blot
SDS lysis buffer (#P0013G, Beyotime) containing a protease and phosphatase inhibitor cocktail (#5872, CST) was used to lyse cells and tissues. The BCA protein assay kit (#P0011, Beyotime) was utilized to determine the protein concentration. The protein samples underwent SDS-PAGE and were then transferred onto PVDF membranes. After an overnight period of incubation with primary antibodies at 4 degrees Celsius, the membranes were rinsed and then treated with horseradish peroxidase (HRP)-linked secondary antibodies at 25 degrees Celsius for 2 h. The spots were observed with an improved chemiluminescence (ECL) setup.The antibodies employed in this investigation are detailed in Additional file 1 Supplementary Table 3.
qRT‒PCR
Total RNA was extracted from cells or tissues using TRIzol (#15,596,026, Thermo Scientific) according to the manufacturer’s instructions. Reverse transcription was carried out using the Transcript First Strand cDNA Synthesis Kit (#04897030001, Roche, Basel, Switzerland), and qPCR was performed using Fast Start Universal SYBR Green Master Mix (#12,239,264,001, Roche). Lithium chloride (8 M, 0.1 volume) was used to purified the samples from DSS-induced colitis model mice to prevent potential inhibition of qPCR by DSS [24]. mRNA expression was relatively quantified using the ΔΔCt technique. The control utilized was GAPDH. Three technical replicates were used in the PCR procedure. The primer sequences utilized are detailed in Additional file 1 Supplementary Table 4.
Transepithelial electrical resistance (TEER) measurement
For the TEER assay, a 24-well Transwell system (#725,101, NEST, Wuxi, China) was used. Caco-2 cells were placed in the top chamber with a concentration of 1 × 104 cells per 300 μl. Next, the lower chamber received 1.0 mL of medium. The TEER was measured on the specified day with an epithelial voltage resistance meter from Jingong Hongtai Technology Co., Ltd. in Beijing, China. TEER values were adjusted for background resistance stemming from the membrane insert and computed as Ω⋅cm2. Electrical resistance measurements were conducted until three consecutive readings were consistent.The formula used to calculate the TEER was (total resistance – blank resistance) × area.
Fluorescein isothiocyanate‑dextran permeability assay
In vivo and in vitro, intestinal permeability was evaluated using Fluorescein isothiocyanate-dextran (FD4; Sigma‒Aldrich) with a molecular weight of 4 kDa. For in vivo experiments, after forbidden food and water for 4 h, the mice were gavaged with 150 μl of FD4 solution (80 mg/mL solution). After a span of four hours, serum samples were obtained from the mice and analyzed for FD4 concentration with a TECAN Infinite F500 microplate reader configured with excitation/emission wavelengths of 485/535 nm. For the in vitro experiments, a 24-well Transwell system was used. Caco-2 cells were placed in the top chamber with a concentration of 1 × 104 cells per 300 μl. Next, the lower chamber received 1.0 mL of medium. After around 20 days of stability in TEER, Opti-MEM from Gibco in the USA was utilized to create a 2 mg/mL solution of FD4 powder. The upper transwell chamber was supplemented with FD4 solution, while the lower chamber received Opti-MEM. After incubating for 4 h, the TECAN Infinite F500 microplate reader was used to measure the fluorescence intensity emitted from the lower chamber at wavelengths of 485/535 nm.
TUNEL staining
Colon sections were subjected to TUNEL staining using a TUNEL kit (#PF00006, Proteintech, Wuhan, China) following the manufacturer's protocol. Cells positive for TUNEL staining were observed with a confocal microscope (FLUOVIEW FV3000, Olympus) and counted using ImageJ software.
Caspase3 activity assay
Caspase3 activity assays were performed using the Caspase3 Activity Assay Kit(#C1115, Beyotime). Briefly, a quantity of 10 mg of mouse colon tissue was combined with 100 μl of lysate, homogenized using a glass homogenizer on an ice bath, and subjected to lysis for a duration of 5 min. The resulting supernatants from the homogenate were collected through centrifugation at 16,000 g at 4 ºC, followed by determination of the protein concentration utilizing a Bradford protein assay kit. Subsequently, tissue lysates were incubated with Ac-DEVD-pNA (2 mM) at 37 °C for 2 h. After incubation, absorbance was read at 405 nm using a microplate reader. The enzymatic activity of caspase3 within the protein per unit weight of the sample is determined by dividing the quantity of pNA generated through catalysis in the sample by the protein concentration of the sample.
Chromatin immunoprecipitation (ChIP)
The ChIP experiment utilized a ChIP kit (Hyperactive pG-MNase CUT&RUN Assay Kit for PCR/qPCR (#HD101-01, Vazyme Biotech Co., Ltd., Nanjing, China) in accordance with the instructions provided by the manufacturer. 100,000 Caco-2 cells were gathered and exposed to ConA Beads Pro at ambient temperature. Following this, the cell-magnetic bead combination was left to incubate with primary antibodies for the entire night at a temperature of 4 °C. The pG-MNase enzyme mixture was made, flipped, and then chilled on ice. Next, the blend was introduced to the cell-magnetic bead compound and left to incubate while rotating for 1 h at 4 °C. Next, 100 μl of CaCl2 premix was introduced, followed by inverting the mixture multiple times to thoroughly blend the buffer and cell-magnetic bead complex. Next, 100 μl of stop buffer was introduced, followed by inversion of the mixture, placement in a 37 °C water bath, and an incubation period of 10–30 min. The stop buffer contained 10 pg of spike-in DNA fragments, which can be used for uniform calibration of experimental tests. FastPure gDNA Mini Columns were used for DNA extraction and purification. The qPCR utilized the purified DNA sequences for E-cadherin, Occludin, ZO-1, and Claudin-4 promoter regions as follows: E-cadherin Forward, 5ʹ-TAGAGGGTCACCGCGTCTAT-3ʹ Reverse, 5ʹ-TCACAGGTGCTTTGCAGTTC-3ʹ; Occludin Forward, 5ʹ-CCTGCTGGATGGCAACTAA-3ʹ Reverse, 5ʹ-AACGAAAGACTCCTGGGAAA-3ʹ; ZO-1 Forward, 5ʹ-TGTTTAGCAAAGCCGTCA-3ʹ Reverse, 5ʹ-CAATGCTGTCCCTCCAAC-3ʹ; and Claudin-4 Forward, 5ʹ- CTGGGGTGATGATGTCTCCAG-3ʹ Reverse, 5ʹ-CAGCTTCTCTGCTTGGCTAG-3ʹ. The fold change method was recommended for calculating the desired values, with the specific calculation procedure outlined as follows:
(1) The Ct values of the target gene and spike DNA in the treatment (anti-KLF4) group and negative control group (IgG) were determined.
(2) The ΔCt values for each group were determined by utilizing spike DNA as an internal control.
(3) Taking negative control IgG as a reference, the 2−△△Ct values of each group were calculated.
Apoptosis analysis
Apoptosis was assessed utilizing the 7-AAD/Annexin V Apoptosis Detection Kit I (#559,763, BD Bioscience, San Jose, USA). Caco-2 cells were plated in a 6-well plate at a concentration of 1 × 106 cells per well and grown until the formation of a monolayer representing the intestinal epithelial barrier. Subsequently, each cell sample was treated with 5 μl of PE-conjugated Annexin V and 5 μl of 7-AAD and incubated for 15 min at 4 °C in a light-free environment. Staining of cells was followed by flow cytometry (Beckman Coulter, California, USA). Each experimental procedure was conducted in triplicate. Annexin-V + /7-AAD- cells and Annexin-V + /7-AAD + cells were considered apoptotic cells. The data were analyzed using FlowJo statistical software (v10.62).
Determination of cellular reactive oxygen species (ROS) levels
Cellular levels of ROS were measured using a kit for detecting Reactive Oxygen Species (#S0033S, Beyotime). Caco-2 cell cultures were exposed to H2O2 (10 μmol/L) for a duration of 4 h in 6-well dishes. Cells were treated with 10 mmol/L DCFH-DA in medium without serum for 30 min at 37 °C. After washing with warm PBS, single-cell suspensions were obtained from the 6-well plates and analyzed via flow cytometry (Beckman Coulter). FlowJo statistical software (v10.62) was utilized for data analysis.
MeRIP assays
The MeRIP assay was conducted utilizing the EpiQuik™ CUT&RUN m6A RNA Enrichment (MeRIP) Kit (#P9018, Epigentek, Farmingdale, USA). Briefly, RNA fragments containing m6A were captured using a bead m6A capture antibody, followed by purification and elution of the enriched RNA. Subsequently, RT‒qPCR was used to quantify changes in the methylation of target genes, KLF4 m6A motif locations are shown in Additional File 2–2.
RNA immunoprecipitation (RIP) assay
The Magna RIP kit (#17–700, Sigma‒Aldrich) was utilized to conduct the RIP assay following the instructions provided by the manufacturer. In brief, cellular cleavage was performed on ice using RIP lysis buffer, followed by overnight incubation of whole-cell lysates with magnetic protein A/G beads and either 5 µg of normal rabbit IgG or primary antibodies (anti-METTL3, anti-METTL14, anti-WTAP, anti-FTO, anti-ALKBH5 and anti-YTHDF2) at 4 °C. Before the purification step of RNA, 50 μL of the bead suspension was taken during the last wash to test the efficiency of immunoprecipitation by Western blotting. Afterwards, the RNA–protein compound underwent ongoing eluent washing and was exposed to proteinase K at 55 °C for a duration of 30 min. The bound RNA was then extracted for qRT‒PCR analysis. The primer sequences are provided in Supplementary Table 4.
RNA stability assay
Caco-2 cells were placed in a 6-well dish and incubated for a day. Afterward, the cells were exposed to actinomycin D (10 µg/ml, #A9415, Sigma‒Aldrich) for durations of 1, 2, 3, or 4 h. RNA was isolated from the cells in preparation for future qRT‒PCR tests.
Enzyme‑linked immunosorbent assays (ELISA)
The concentration of IL-1β and IL-6 was measured using commercially-available ELISA Kits (ExCell Bio, Shanghai, China). Briefly, Caco-2 cells were cultured in a 6-well plate until a monolayer resembling the intestinal epithelial barrier was formed. After the cells were treated with TNF-α/IFN-γ mixtures for 24 h, the cell supernatant was collected via the centrifugation. The concentration of IL-1β and IL-6 in the cell supernatant was measured using the corresponding ELISA Kit following the manufacturer’s instructions.
Dual-luciferase reporter assays
A renilla luciferase plasmid and a pGL3-Basic-Luciferase reporter vector containing the promoter sequence of E-cadherin (pGL3-E-cadherin) were obtained from MiaoLingBio. KLF4 knockdown or overexpression caco-2 cells were cultured in a 24-well plate and were co-transfected with the pGL3-E-cadherin vectors (250 ng/well) and Renilla luciferase plasmid (10 ng/well). After 48 h post-transfection, the cells were lysed using passive lysis buffer, and the luciferase activity was quantified using the Dual-Luciferase Reporter Assay System kit (#E1910, Promega, Madison, USA). The Renilla luciferase activity wasserved as an internal reference.
Statistical analysis
GraphPad Prism 9 (La Jolla, CA, USA) was used to analyze all the statistical data in this study. The data are reported as the mean ± SD. Statistical analyses were conducted using unpaired two-tailed Student’s t tests, one-way ANOVA, and nonparametric Mann‒Whitney U tests. Spearman’s correlation was utilized for nonparametric correlation analysis. Significance was determined at p < 0.05. The data presented are representative of a minimum of three independent experiments.
Results
KLF4 expression is downregulated in IBD patients and colitis model mice
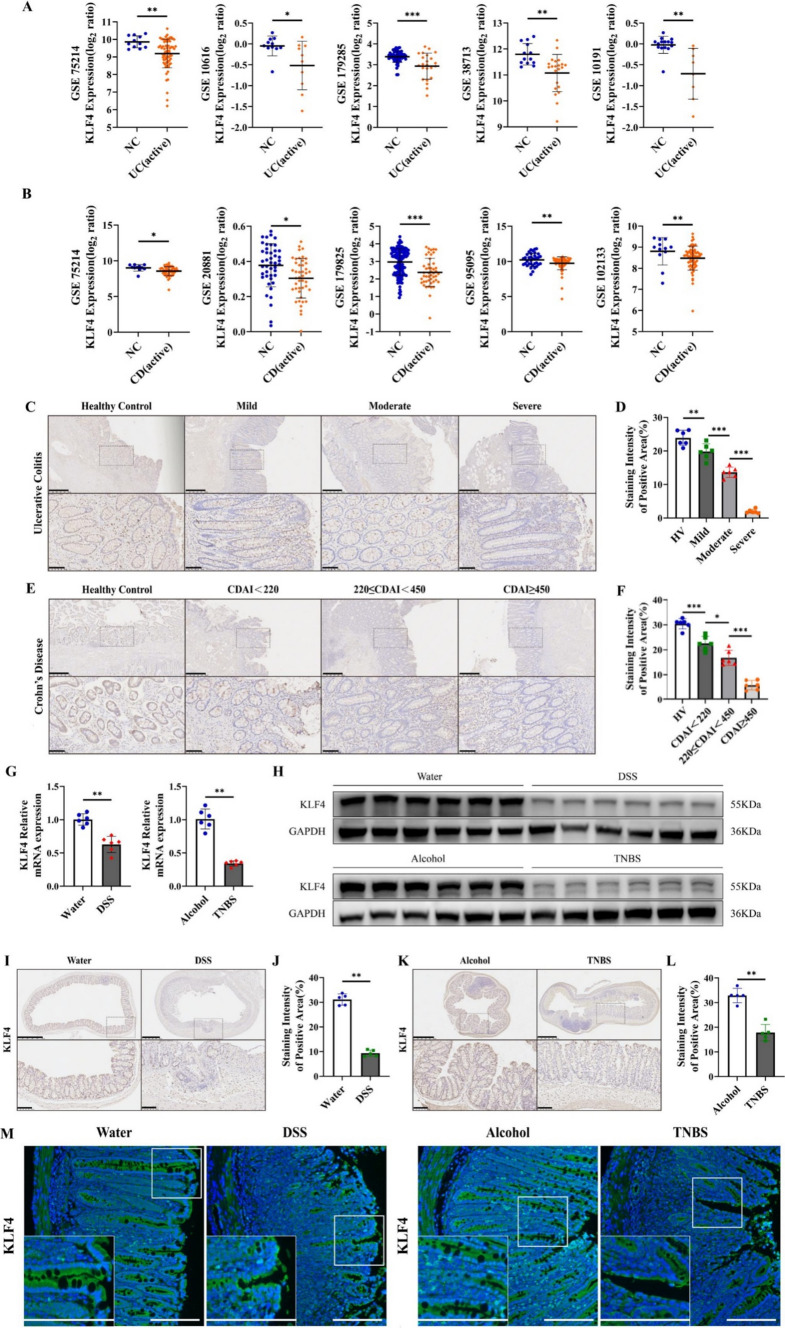
We first assessed KLF4 transcript expression in tissue specimens from IBD patients and healthy controls using the GEO database. The GSE75214 microarray dataset showed that compared with that in healthy controls, the mRNA expression of KLF4 was significantly decreased in UC and CD patients in the active phase (Fig. 1A, B). Similar results were observed in four other UC GEO datasets, GSE10616, GSE179285, GSE38713, and GSE10191, and four other CD GEO datasets, GSE20881, GSE95095, GSE102133 and GSE179825 (Fig. 1A, B). We further used scRNA-seq dataset GSE164985 to investigate KLF4 in colonic biopsy samples of CD patients. As shown in Fig. S1A, B, all cells were categorized into 6 major cell types, including epithelial cells, goblet cells, B cells, NK cells, paneth cells and T cells by using uniform manifold approximation and projection (UMAP). It was observed that KLF4 was preferentially expressed in epithelial cells and goblet cells (Fig. S1C). Moreover, the protein expression of KLF4 in IBD tissue specimens was detected by IHC. KLF4 protein expression was lower in IBD patients than in healthy controls (Fig. 1C–F and Supplementary Tables S1 and S2). Importantly, the KLF4 protein expression levels were negatively related to the disease activity of UC and the CDAI of CD, two markers of IBD disease severity [22].
Fig. 1.
KLF4 expression is downregulated in IBD patients and colitis model mice. A, B KLF4 transcript expression data were obtained from eight different GSE datasets, as indicated. All the datasets showed significantly reduced KLF4 mRNA expression in the IBD patients than in the negative controls. The UC GEO datasets used were GSE75214 [11 NC (healthy individuals), 74 active]; GSE10616 [11 NC (healthy individuals), 10 active]; GSE179285 [55 NC (healthy individuals and inactive adjacent regions), 23 active]; GSE38713 [13 NC (healthy individuals), 22 active]; and GSE10191 [15 NC (healthy individuals and inactive adjacent regions), 8 active]. The CD GEO datasets used were GSE75214 [11 NC (healthy individuals), 51 active], GSE20881 [44 NC (healthy individuals), 44 active], GSE95095 [36 NC (healthy individuals and inactive adjacent regions), 24 active], GSE102133 [12 NC (healthy individuals), 65 active] and GSE179285 [129 NC (healthy individuals and inactive adjacent regions), 47 active]. C Representative IHC images of KLF4 protein expression in colon tissues from healthy controls and patients with different stages of UC. Scale bars, 500 μm (top) and 100 μm (bottom). D The staining intensity of KLF4 in colon tissues from healthy controls and patients with different stages of UC, as determined by the staining index. (healthy control, n = 6; mild, n = 6; moderate, n = 6; severe, n = 6). E Representative IHC images of KLF4 protein expression in small intestine tissues from healthy controls and patients with different stages of CD. Scale bars, 500 μm (top) and 10 μm (bottom). F The staining intensity of KLF4 in small intestine tissues from healthy controls and patients with different stages of CD (healthy controls, n = 6; CDAI < 220, n = 6; 220 ≤ CDAI < 450, n = 6; CDAI ≥ 450, n = 6). G, H The mRNA (G) and protein (H) expression of KLF4 in DSS-induced (water, n = 6 and DSS, n = 6) and TNBS-induced (alcohol, n = 6; TNBS, n = 6) colitis mice was examined by RT‒qPCR and Western blot. I, J KLF4 protein expression in DSS-induced colitis mice was measured by IHC. Scale bars, 500 μm (top) and 100 μm (bottom). The staining intensity of KLF4 in DSS-induced colitis mice (water, n = 5; DSS, n = 5) was determined based on the IHC staining index. K, L The protein expression of KLF4 in TNBS-induced colitis mice was detected by IHC. Scale bars, 500 μm (top) and 100 μm (bottom). The staining intensity of KLF4 based on the IHC staining index in TNBS-induced colitis mice (alcohol, n = 6; TNBS, n = 6). M Representative immunofluorescence micrographs showing KLF4 (green) expression in colon biopsy specimens from DSS-induced (left) and TNBS-induced (right) colitis mice. Scale bars, 100 μm. The data are expressed as the mean ± SD. * p < 0.05, ** p < 0.01, *** p < 0.001
To further evaluate the correlation between KLF4 expression and IBD progression, colitis mouse models were established using DSS and TNBS. The results of RT-qPCR, Western blot, and IHC assays indicated a significant decrease in both mRNA and protein levels of KLF4 in mice with DSS- and TNBS-induced colitis compared to control mice (Fig. 1G–L). Moreover, the results of IF staining indicated that the KLF4 protein was mainly present in epithelial cells' membranes and cytoplasm, and the staining intensity exhibited a significant decrease in mice models of colitis induced by both DSS and TNBS (Fig. 1M). Taken together, these findings suggest that the mRNA and protein levels of KLF4 were reduced in tissue samples obtained from individuals with IBD and in mouse models of colitis, showing a negative association with the advancement of the disease.
Intestinal epithelial-specific knockdown of KLF4 exacerbates the severity of chemically induced colitis
To investigate the role of KLF4 in the progression of IBD, we utilized an adenovirus carrying KLF4 shRNA (shKLF4-AdV) to knock down the expression of KLF4 in the intestinal epithelial cells of mice (Fig. S2A). The RT‒qPCR, Western blot and IHC results showed that shKLF4-AdV markedly downregulated KLF4 expression in intestinal epithelial cells in mice (Fig. S2B–E). Furthermore, KLF4 knockdown had no influence on mouse weight or colon length (Fig. S2F–H). H&E staining analysis revealed that the deletion of KLF4 did not result in significant histological damage, including colonic epithelial injury and lamina propria lymphocyte infiltration (Fig. S2I). These data indicated that shKLF4-AdV significantly decreased KLF4 expression in intestinal epithelial cells but had no toxic effect on the mice.
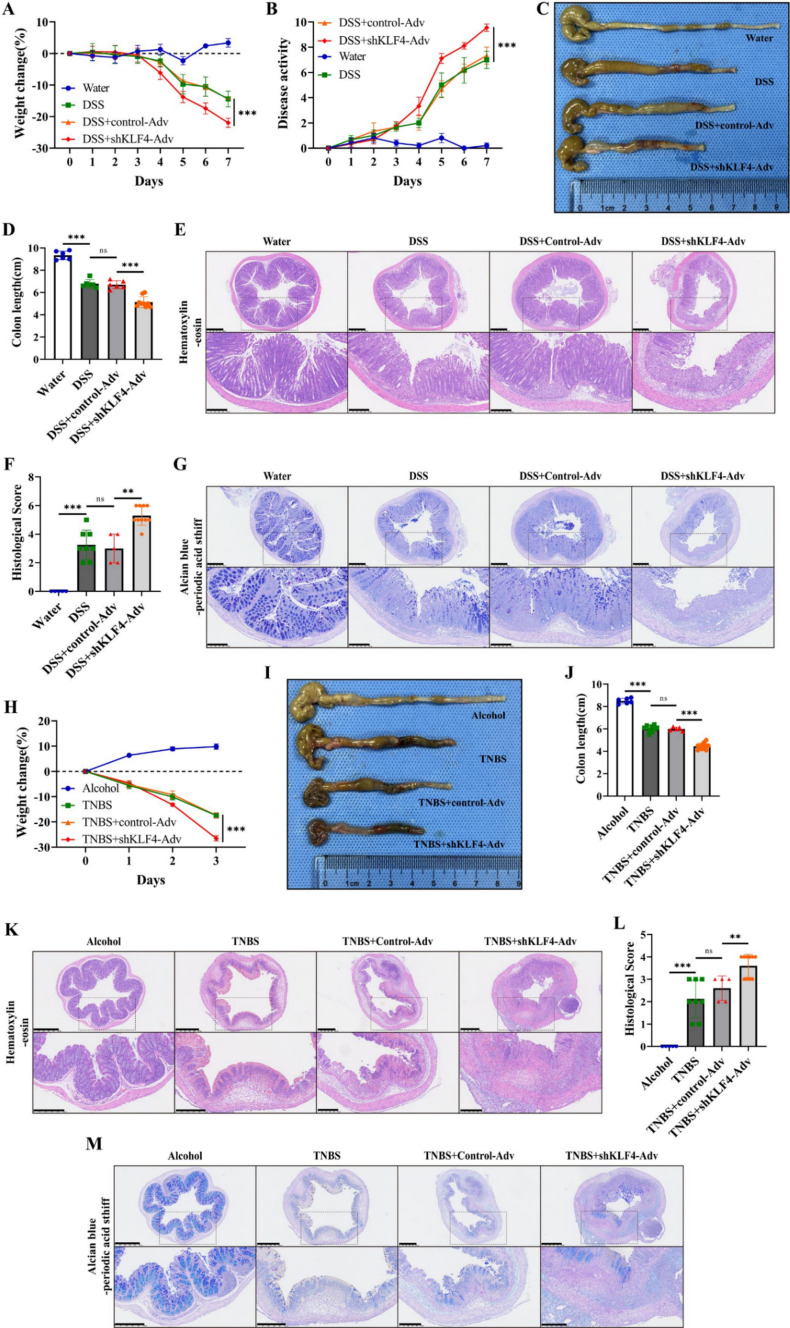
Subsequently, we investigated the role of KLF4 in experimental colitis using shKLF4-AdV. Compared with mice treated with DSS, mice subjected to co-treatment with shKLF4-AdV and DSS displayed increased disease activity, more pronounced bloody diarrhea, a more significant decrease in body weight, and shorter colons (Fig. 2A–D). Additionally, the depletion of KLF4 resulted in exacerbated histological damage, characterized by lymphocyte infiltration into the lamina propria and colonic epithelial injury, in mice subjected to DSS challenge (Fig. 2E, F). Furthermore, compared with those in the control group, mice in the DSS + shKLF4-AdV group exhibited greater mucus barrier impairment, including crypt disruption, decreased acid and neutral mucin secretion, and goblet cell depletion (Fig. 2G). In addition, KLF4 knockdown further elevated the levels of inflammation-associated factors, such as TNF-α, IL-6, and IFN-γ, in DSS-treated mice (Figure S3A). These results were further confirmed in TNBS-induced colitis (Figs. 2H–M, S3B).
Fig. 2.
KLF4 knockdown aggravates DSS- and TNBS-induced acute colitis. A, B The body weight (A) and disease activity index (DAI) (B) of DSS-challenged mice after treatment with shKLF4-Adv. C, D Representative images of mouse colons (C) and colon length (D) of DSS-challenged mice after treatment with shKLF4-Adv. E Representative images of H&E staining of colon tissues from DSS-challenged mice after treatment with shKLF4-Adv. Scale bars, 500 μm (top) and 250 μm (bottom). F Summary of the histological scores of DSS-challenged mice after treatment with shKLF4-Adv. G Representative colonic AB-PAS images of DSS-challenged mice after treatment with shKLF4-Adv. Scale bars, 500 μm (top) and 250 μm (bottom). H The body weights of TNBS-challenged mice after treatment with shKLF4-Adv. I, J Representative images of mouse colons (I) and colon length (J) of TNBS-challenged mice after treatment with shKLF4-Adv. K Representative images of H&E staining of colon tissues from TNBS-challenged mice after treatment with shKLF4-Adv. Scale bars, 500 μm (top) and 250 μm (bottom). L Summary of the histological scores of DSS-challenged mice after treatment with shKLF4-Adv. M Representative colonic AB-PAS images of TNBS-challenged mice after treatment with shKLF4-Adv. Scale bars, 500 μm (top) and 250 μm (bottom). WT + water (n = 6), WT + DSS (n = 8), control-Adv + DSS (n = 5), shKLF4-Adv + DSS (n = 10), WT + Alcohol (n = 6), WT + TNBS (n = 8), control-Adv + TNBS (n = 5), and shKLF4-Adv + TNBS (n = 10). The data are expressed as the mean ± SD. ns, not significant, ** p < 0.01, *** p < 0.001
KLF4 downregulation impairs the intestinal epithelial barrier by decreasing apical junction complex protein expression and suppressing proliferation while promoting intestinal epithelial cell apoptosis
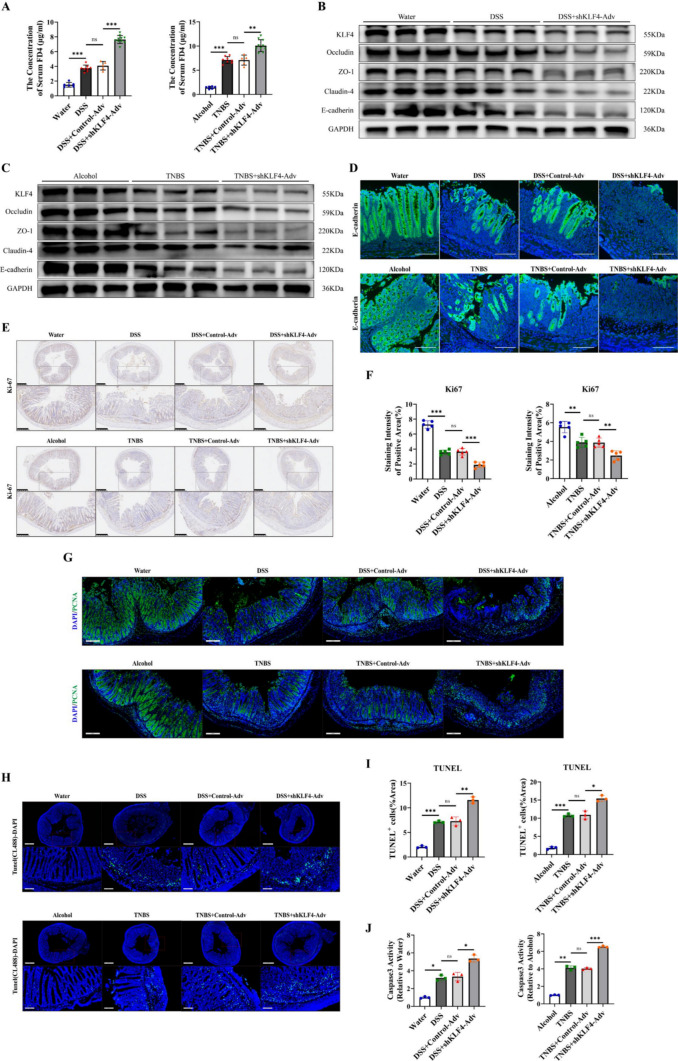
Given that intestinal epithelial barrier impairment plays a key role in the pathogenesis of IBD [25, 26], we hypothesized that severe colitis mediated by KLF4 knockdown may be associated with more pronounced impairment of the intestinal epithelial barrier. The DSS- and TNBS-treated mice displayed increased transepithelial permeability of FD4, suggesting an impairment in overall mucosal barrier function (Fig. 3A). KLF4 knockdown led to a significant increase in the serum FD4 concentration in mice challenged with DSS or TNBS (Fig. 3A). These data indicated that KLF4 knockdown aggravated transepithelial permeability in experimental colitis model mice.
Fig. 3.
KLF4 downregulation impairs intestinal epithelial barrier integrity by decreasing AJC protein expression and increasing apoptosis of IECs in DSS- and TNBS-induced colitis. A The serum concentration of FD4 in DSS- and TNBS-induced colitis mice after treatment with shKLF4-Adv. WT + water (n = 6), WT + DSS (n = 8), control-Adv + DSS (n = 5), or shKLF4-Adv + DSS (n = 10); WT + alcohol (n = 6), WT + TNBS (n = 8), control-Adv + TNBS (n = 5), or shKLF4-Adv + TNBS (n = 10). B, C The protein levels of KLF4, E-cadherin, Occludin, ZO-1 and claudin-4 in colon tissues from DSS (B)- and TNBS (C)-challenged mice after treatment with shKLF4-Adv were assessed by Western blot. D Representative immunofluorescence images of E-cadherin (green) in colon tissues from DSS- and TNBS-challenged mice after treatment with shKLF4-Adv. Scale bars, 100 μm. E, F The expression of Ki67 in colon tissues from DSS- and TNBS-challenged mice after treatment with shKLF4-Adv was detected by IHC. Scale bars, 500 μm (top) and 250 μm (bottom). The staining intensity of KLF4 based on the IHC staining index. G Representative immunofluorescence images of PCNA (green) in colon tissues from DSS- and TNBS-challenged mice after treatment with shKLF4-Adv. Scale bars, 200 μm. H Representative TUNEL images of colon tissues from DSS- and TNBS-challenged mice after treatment with shKLF4-Adv. Scale bars, 700 μm (top) and 100 μm (bottom). I Quantitative analysis of TUNEL-positive cells in colon tissues from DSS- and TNBS-challenged mice after treatment with shKLF4-Adv. J Quantitative analysis of Caspase3 activity in colon tissues from DSS- and TNBS-challenged mice after treatment with shKLF4-Adv. Data are expressed as the mean ± SD. ns, not significant, * p < 0.05, ** p < 0.01, *** p < 0.001
AJCs are crucial for maintaining the integrity and functionality of the IEB [27]. In the GSE75214 RNA microarray dataset, we found that, compared with those in healthy controls, the expression of AJC constituent proteins, including CDH1, OCLN, TJP1, CLDN4, CLDN7, CLDN8 and CLDN15, significantly decreased in IBD patients during the active phase of the disease (Fig. S4A, B). In addition, there was a positive correlation between KLF4 expression and the expression of AJC constituent proteins, such as CDH1 (encoding E-cadherin), CLDN4 (encoding claudin-4), OCLN (encoding Occludin), and TJP1 (encoding ZO-1), in UC and CD patients according to the GSE75214 microarray data (Figure S4C). Hence, we explored the roles of KLF4 in regulating the expression of E-cadherin, Occlaudin, ZO-1 and Claudin-4 in DSS-induced and TNBS-induced colitis mice. Our results revealed significantly lower levels of these proteins in the colon tissue of mice cotreated with shKLF4-Adv and DSS or TNBS (Figs. 3B, C, S4D, E). Moreover, IF staining revealed that the downregulation of KLF4 further resulted in a discontinuous distribution of E-cadherin, accompanied by a decrease in its expression, in the intestinal epithelia of mice treated with DSS and TNBS (Fig. 3D). In addition, KLF4 depletion increased the impairment of the mucus barrier, which was characterized by reduced secretion of acid and neutral mucins and depletion of goblet cells (Figure S4F, Fig. 2G, M). These findings suggest that the loss of KLF4 compromises the integrity of the IEB through the downregulation of epithelial junction proteins and reduced goblet cell secretion.
Subsequently, we conducted an examination of the impact of KLF4 depletion on the proliferation and apoptosis of intestinal epithelial cells, which are implicated in the impairment of the intestinal epithelial barrier in the context of colonic inflammation [28]. The Ki67 levels in the colon tissues of the DSS + shKLF4-Adv and TNBS + shKLF4-Adv groups were significantly lower when compared to those in the DSS and TNBS groups (Fig. 3E, F). Using PCNA staining to monitor cellular proliferation, we also noted qualitative differences in epithelial proliferation. In the colon tissues of the DSS + shKLF4-Adv and TNBS + shKLF4-Adv groups, there was a well-preserved boundary between the proliferative compartment and the differentiated epithelium, resulting in a delay in proliferation compared to the DSS and TNBS groups (Fig. 3G). Moreover, we observed greater numbers of TUNEL-positive cells in the DSS + shKLF4-Adv and TNBS + shKLF4-Adv groups (Fig. 3H, I). As shown in Fig. 3J, treatment with shKLF4-Adv significantly increased the Caspase3 activity in the colon tissues of mice challeged with DSS or TNBS. These data suggest that KLF4 deficiency can lead to impaired cell proliferation and enhanced apoptosis.
KLF4 knockdown leads to intestinal epithelial barrier damage by regulating apical junction complex proteins in vitro
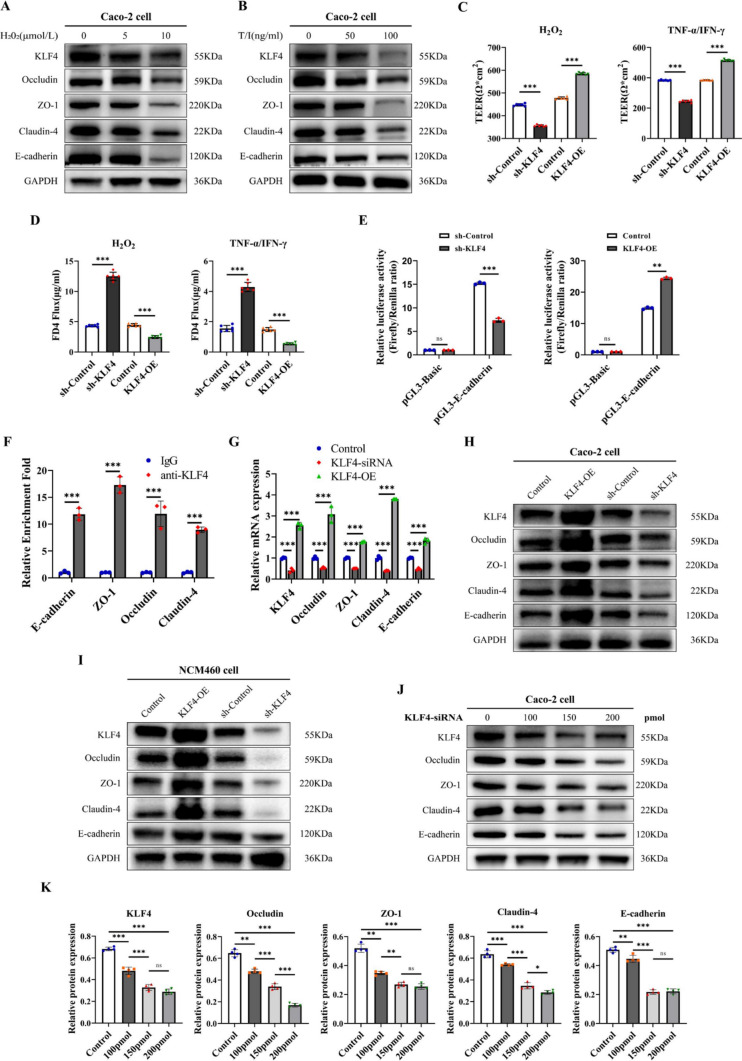
To further investigate the influence of KLF4 knockdown on the intestinal epithelial barrier, we used H2O2 or TNF-α/IFN-γ to induce damage to the intestinal epithelial barrier established by Caco-2 cells in vitro. Treatment with both H2O2 and TNF-α/IFN-γ mixtures reduced KLF4 expression in Caco-2 cells in a dose-dependent manner, and this pattern was also observed for the expression levels of AJC proteins (Figs. 4A, B, S5A, B). We found that 10 μmol/L H2O2 and 100 ng/mL TNF-α/IFN-γ were efficacious and therefore employed in subsequent in vitro experiments (Fig. S5C–G). Then, we established KLF4-overexpressing and KLF4-knockdown Caco-2 and NCM460 cell lines using a lentiviral system (Figure S5H-S5K).
Fig. 4.
KLF4 knockdown leads to intestinal epithelial barrier damage by regulating AJC proteins in vitro. A, B KLF4, Occludin, ZO-1, Claudin-4 and E-cadherin protein expression in Caco-2 cells after treatment with different doses of H2O2 (A) or TNF-α/IFN-γ (B). C, D TEER (C) and FD4 permeability (D) were measured in KLF4-overexpressing or KLF4-knockdown Caco-2 cells after stimulation with H2O2 (10 μmol/L) and TNF-α/IFN-γ (100 ng/ml). E The luciferase activity, responsive to KLF4, was measured in KLF4-overexpressing or KLF4-knockdown Caco-2 cells co-transfected with renilla luciferase plasmids and apGL3-Basic-Luciferase reporter vectors containing the promoter sequence of E-cadherin. F ChIP analysis of KLF4 binding to the promoters of E-cadherin, Occludin, ZO-1, and Claudin-4 in Caco-2 cells. The relative enrichment was calculated as the fold change in DNA abundance following pull-down of anti-KLF4 or normal rabbit IgG. G The mRNA expression levels of KLF4, Occludin, ZO-1, Claudin-4 and E-cadherin in KLF4-knockdown or KLF4-overexpressing Caco-2 cells were examined by RT‒qPCR. H, I The protein expression of KLF4, Occludin, ZO-1, Claudin-4 and E-cadherin in Caco-2 cells (H) and NCM460 cells (I) infected with sh-KLF4 lentivirus and KLF4-overexpressing lentivirus. J The protein expression of KLF4, Occludin, ZO-1, Claudin-4 and E-cadherin in Caco-2 cells treated with siKLF4 at 100 pmol, 150 pmol, or 200 pmol. K The Western blot band densities shown in Fig. 4 J were quantified using ImageJ software. The data are expressed as the mean ± SD. ns, not significant, * p < 0.05, ** p < 0.01, *** p < 0.001
The intestinal epithelial barrier function was assessed by measuring the TEER and paracellular FD4 permeability in a Caco-2 monolayer cell model. Following treatment with H2O2 or TNF-α/IFN-γ mixtures, KLF4 overexpression increased the TEER and decreased FD4 permeability compared to those in the control group (Fig. 4C, D). An opposite effect was observed when KLF4 was knocked down (Fig. 4C, D). These results indicated that KLF4 is an important regulator of the intestinal epithelial barrier in vitro.
Given that KLF4 is a transcription factor and that our data suggest that KLF4 expression is positively correlated with the expression of these AJC proteins, we wondered whether KLF4 directly binds to the promoters of these AJC proteins and promotes their transcription. The luciferase reporter gene test showed that KLF4 overexpression or knockdown could enhance or reduce the KLF4 DNA-binding activity to the promoter of E-cadherin in Caco-2 cells (Fig. 4E). ChIP assays showed that, compared with the IgG group, anti-KLF4 could enrich more E-cadherin, Occludin, ZO-1, and Claudin-4 promoter region DNA fragments in Caco-2 cells (Fig. 4F, Additional File 2–1), confirming the direct interaction of KLF4 with the promoters of these genes. Moreover, KLF4 silencing decreased, while KLF4 overexpression increased, the mRNA levels of E-cadherin, Occludin, ZO-1, and Claudin-4 in Caco-2 cells (Fig. 4G). Besides, KLF4 overexpression upregulated the protein levels of E-cadherin, Occludin, ZO-1, and Claudin-4 in Caco-2 and NCM460 cells (Fig. 4H, I). KLF4 knockdown had the opposite effect (Fig. 4H, I). Treatment with KLF4-siRNA led to a dose-dependent decrease in the expression of these AJC proteins (Fig. 4J, K). Based on these results, KLF4 promotes intestinal epithelial barrier integrity through direct regulation of AJC proteins.
KLF4 protects against intestinal epithelial cell apoptosis by reducing ROS and inflammatory cytokine levels
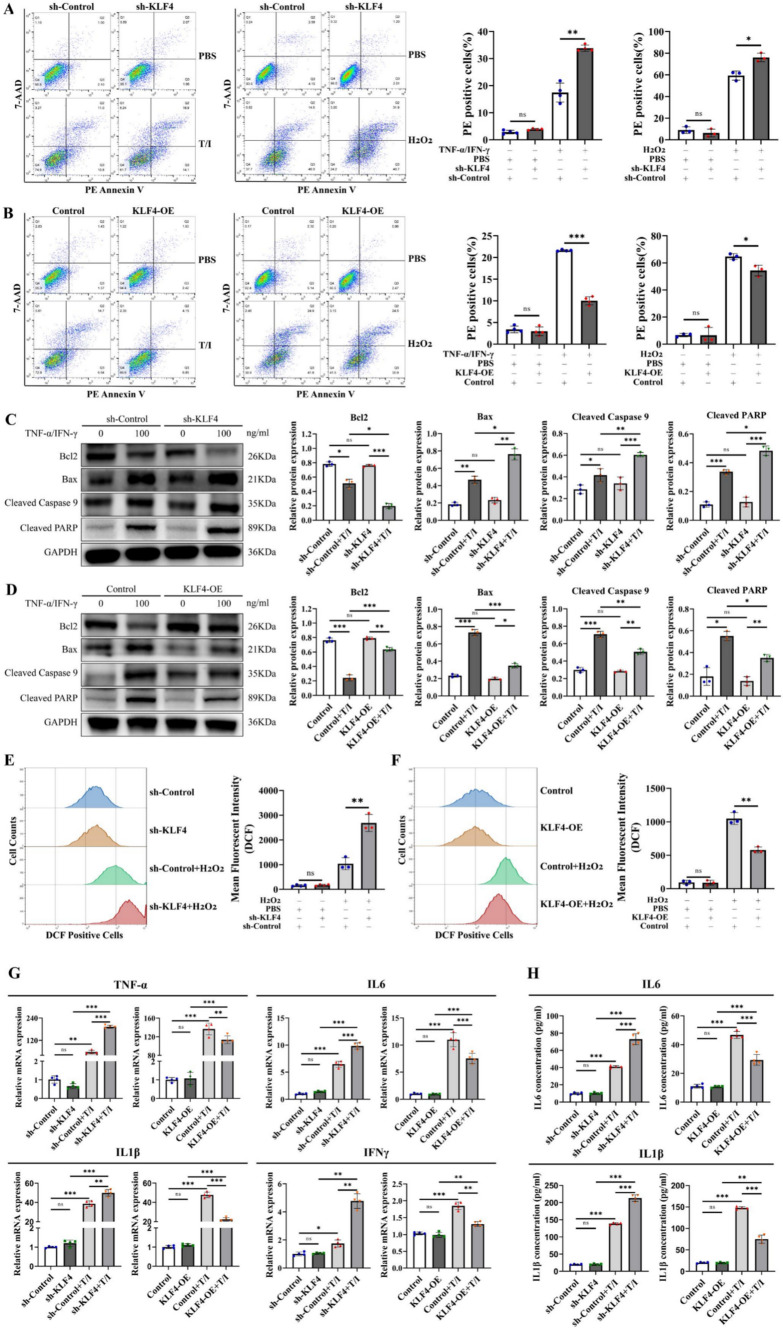
Given that intestinal epithelial cell apoptosis is also involved in the modulation of the intestinal epithelial barrier under physiological and pathological conditions [29–31], we examined whether and how KLF4 regulates intestinal epithelial cell apoptosis to protect the intestinal epithelial barrier. Treatment with H2O2 and the TNF-α/IFN-γ mixture significantly elevated the proportion of apoptotic Caco-2 cells (Fig. 5A, B), while KLF4 depletion substantially enhanced the apoptosis rate (Fig. 5A). Moreover, KLF4 overexpression reversed the promotive effect of H2O2 and the TNF-α/IFN-γ mixture on Caco-2 cell apoptosis (Fig. 5B).
Fig. 5.
KLF4 knockdown increases intestinal epithelial cell apoptosis by increasing cellular ROS and proinflammatory cytokine levels. A, B Apoptotic cells were detected using Annexin V/7-AAD double staining in KLF4-knockdown (A) or KLF4-overexpressing (B) Caco-2 cells after treatment with H2O2 (10 μmol/L) or TNF-α/IFN-γ (100 ng/mL). C, D The protein expression of Bcl2, Bax, cleaved caspase-9 and cleaved PARP in KLF4-knockdown (C) and KLF4-overexpressing (D) Caco-2 cells stimulated with TNF-α/IFN-γ (100 ng/mL). The Western blot band densities shown in (C, D) were quantified using ImageJ software. E, F The levels of intracellular ROS in KLF4-knockdown (E) or KLF4-overexpressing (F) Caco-2 cells treated with H2O2 (10 μmol/L) were determined using a DCFH-DA probe and flow cytometry. G The mRNA expression of TNF-α, IL-6, IL-1β and IFNγ in KLF4-knockdown or KLF4-overexpressing Caco-2 cells stimulated with TNF-α/IFN-γ (100 ng/ml). H The IL1β and IL-6 concentrations in the cell supernatant of KLF4-knockdown or KLF4-overexpressing Caco-2 cells stimulated with TNF-α/IFN-γ (100 ng/ml) were detected using ELISA. The data are expressed as the mean ± SD. ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001
Western blot analysis revealed that treatment with TNF-α/IFN-γ mixture reduced Bcl-2 and induced Bax, cleaved caspase-9 and cleaved PARP protein expression (Fig. 5C, D). KLF4 knockdown further decreased Bcl-2 and increased Bax, cleaved caspase-9 and cleaved PARP protein expression in Caco-2 cells after stimulation with TNF-α/IFN-γ mixture (Fig. 5C). KLF4 overexpression had the opposite effect (Fig. 5D). According to these results, KLF4 may protect intestinal barrier function by inhibiting epithelial apoptosis and regulating the Bax/Bcl2 apoptosis axis.
It has been reported that high levels of ROS contribute to the apoptosis of intestinal epithelial cells during colitis [32]. We then investigated whether KLF4-mediated inhibition of intestinal epithelial cell apoptosis was associated with intracellular ROS levels. Compared with those in the control group, KLF4 silencing increased (Fig. 5E), while KLF4 overexpression decreased (Fig. 5F) the intracellular ROS levels in Caco-2 cells after treatment with H2O2.
Another primary factor leading to the apoptosis of intestinal epithelial cells in IBD is the excessive release of inflammatory factors [33]. The mRNA expression levels of TNF-α, IL-6, IL-1β, and IFNγ were found to be downregulated in KLF4-overexpressing Caco-2 cells following treatment with the TNF-α/IFN-γ mixture, whereas they were upregulated in KLF4-knockdown cells (Fig. 5G). Moreover, ELISA assay showed that KLF4 silencing decreased, while KLF4 overexpression increased, the protein concentration of IL1β and IL-6 in the cell supernatant of Caco-2 cells treated with the TNF-α/IFN-γ mixture (Fig. 5H). These findings provide further evidence that KLF4 plays an important role in inhibiting inflammation and reducing apoptosis.
N6-methyladenosine (m6A) modification is involved in the regulation of KLF4 expression in IBD.
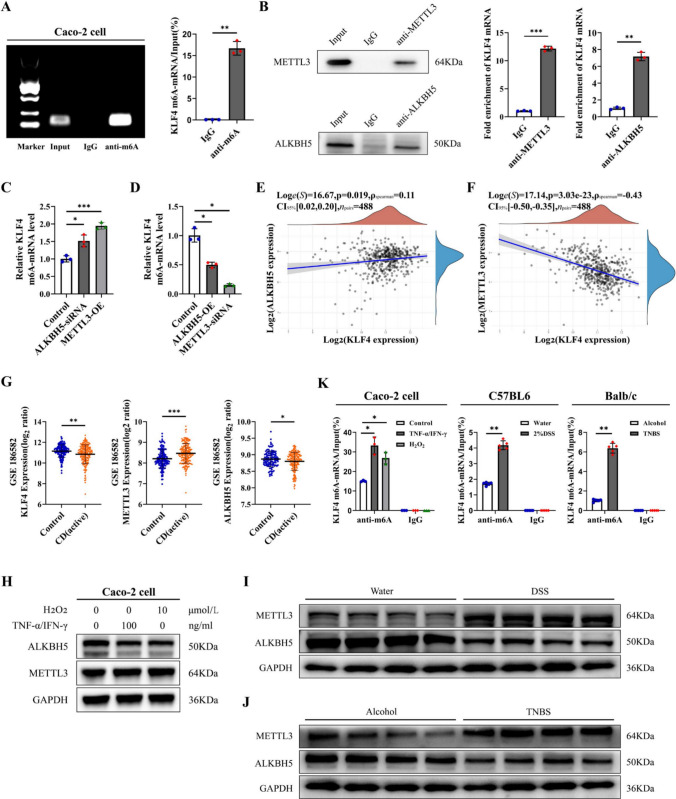
Next, we explored the reason for KLF4 downregulation in the colon tissues of IBD patients and animal models. Several studies have shown that m6A RNA modification is involved in modulating KLF4 expression in vascular endothelial and bladder cancer cells [34, 35]. Hence, we hypothesized that m6A RNA modification may regulate KLF4 expression in intestinal epithelial cells in IBD. The MeRIP-PCR assay showed that the anti-m6A antibody effectively bound to KLF4 mRNA in Caco-2 cells (Fig. 6A). To identify m6A methylation-related enzymes that bind to KLF4 mRNA, RIP-PCR assays with anti-METTL3, anti-METTL14, anti-WTAP, anti-FTO, and anti-ALKBH5 antibodies were performed. The results showed that METTL3 and ALKBH5 directly interact with KLF4 mRNA. (Fig. 6B, Figure S6A and S6B). Furthermore, MeRIP-PCR analysis revealed that depletion of ALKBH5 and overexpression of METTL3 resulted in an increase in the m6A modification of KLF4 mRNA in Caco-2 cells. In contrast, overexpression of ALKBH5 and depletion of METTL3 led to a decrease in m6A levels on KLF4 mRNA in Caco-2 cells (Fig. 6C, D).
Fig. 6.
METTL3 and ALKBH5 mediated the m6A modification of KLF4. A MeRIP-PCR analysis of m6A enrichment on the mRNA of KLF4. B RIP-qPCR analysis showed that the ALKBH5 protein and METTL3 protein bind to KLF4 mRNA. C Depletion of ALKBH5 and overexpression of METTL3 elevated m6A levels on KLF4 mRNA in Caco-2 cells. D Overexpression of ALKBH5 and knockdown of METTL3 reduced m6A levels on KLF4 mRNA in Caco-2 cells. E Spearman’s correlation between KLF4 and ALKBH5 expression in the GSE186582 dataset (n = 488). F Spearman’s correlation between KLF4 and METTL3 expression in the GSE186582 dataset (n = 488). G Transcript levels of METTL3, ALKBH5, and KLF4 in control (n = 147) and CD patient (n = 196) samples from the GSE186582 dataset. H The protein expression of METTL3 and ALKBH5 in Caco-2 cells after H2O2 (10 μmol/L) or TNF-α/IFN-γ (100 ng/ml) stimulation was detected by Western blot. I, J The protein expression of METTL3 and ALKBH5 in the colon tissues of DSS-induced (I) and TNBS-induced (J) colitis model mice. K m6A modification of KLF4 mRNA in Caco-2 cells after H2O2 (10 μmol/L) or TNF-α/IFN-γ (100 ng/ml) stimulation (left), DSS-induced colitis (middle) or TNBS-induced colitis (right). The data are expressed as the mean ± SD. * p < 0.05, ** p < 0.01, *** p < 0.001
To further investigate the associations between METTL3, ALKBH5, and KLF4 in the colonic mucosa of patients with IBD, changes in the transcript levels of METTL3, ALKBH5, and KLF4 were evaluated in the GSE186582 dataset. Spearman's correlation analysis indicated that there was a significant negative correlation between METTL3 and KLF4 expression as well as a positive correlation between ALKBH5 and KLF4 expression (Fig. 6E, F). Importantly, we found a noteworthy reduction in ALKBH5 and KLF4 expression, as well as an elevation in METTL3 expression, among individuals with IBD during the active phase in comparison to those in a healthy control group (Fig. 6G). These changes were generally consistent across 2 additional IBD datasets (GSE75214 and GSE102133, Figure S6C and S6D). Furthermore, we observed that both H2O2 and TNF-α/IFN-γ mixtures significantly decreased ALKBH5 protein expression and increased METTL3 protein expression in Caco-2 cells (Fig. 6H). Compared with those in the control group, the levels of ALKBH5 protein were reduced, whereas the expression of METTL3 protein was found to be elevated in the colon tissues of mice with colitis induced by DSS and TNBS (Fig. 6I, J). Importantly, the m6A levels on KLF4 mRNA were markedly upregulated in Caco-2 cells treated with H2O2 and TNF-α/IFN-γ, as well as in DSS-treated C57BL6 mice and TNBS-treated BALB/c mice (Fig. 6K). These data suggest that in an inflammatory milieu, the high m6A methylation level of KLF4 mRNA is caused by the high expression of METTL3 and the low expression of ALKBH5.
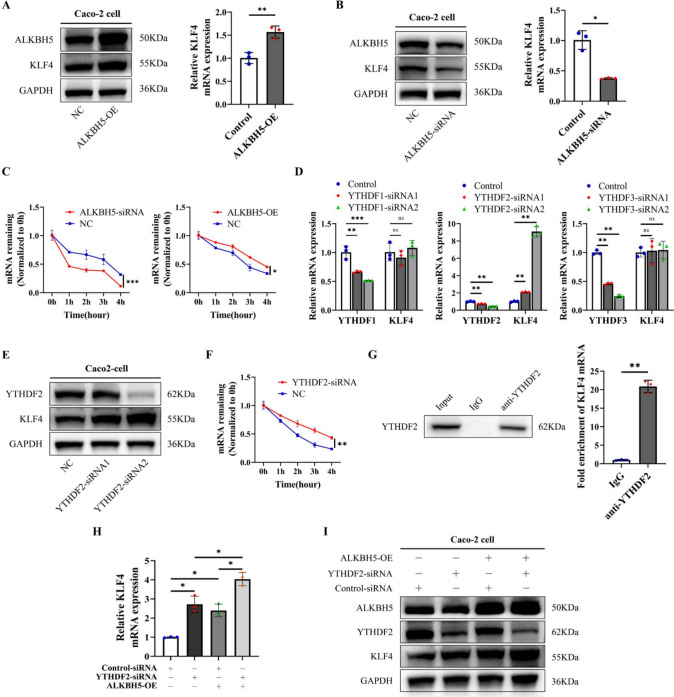
Subsequently, an investigation was conducted to determine the influence of ALKBH5 and METTL3 on KLF4 expression in Caco-2 cells. The results revealed a significant elevation in the mRNA and protein expression levels of KLF4 in Caco-2 cells overexpressing ALKBH5 and with METTL3 knockdown (Figs. 7A, S7B), whereas its expression was downregulated upon ALKBH5 knockdown and METTL3 overexpression (Figs. 7B, S7A). Considering the known impact of m6A modifications on mRNA degradation, it was postulated that m6A methylation may play a role in regulating KLF4 mRNA stability. Consistent with this hypothesis, ALKBH5 overexpression and METTL3 depletion were found to enhance the stability of KLF4 mRNA, while ALKBH5 depletion and METTL3 overexpression had the opposite effect (Figs. 7C, S7C). These results suggest that high levels of m6A methylation facilitate KLF4 mRNA degradation.
Fig. 7.
m6A modification decreased KLF4 expression via YTHDF2. A ALKBH5 overexpression elevated the expression of KLF4 in Caco-2 cells at both the mRNA and protein levels. B Knockdown of ALKBH5 reduced the expression of KLF4 in Caco-2 cells at both the mRNA and protein levels. C The mRNA stability of KLF4 in Caco-2 cells after transfection with ALKBH5 siRNAs or overexpression plasmids was measured by RT‒qPCR. D The relative mRNA levels of YTHDF1, YTHDF2, YTHDF3 and KLF4 in Caco-2 cells after transfection with YTHDF1, YTHDF2 or YTHDF3 siRNAs. E The protein expression of YTHDF2 and KLF4 in Caco-2 cells after transfection with YTHDF2 siRNAs. F The mRNA stability of KLF4 in Caco-2 cells after transfection with YTHDF2 siRNAs was measured by RT‒qPCR. G RIP-qPCR analysis showed that the YTHDF2 protein bound to KLF4 mRNA. H, I mRNA (H) and protein (I) expression levels of KLF4 in Caco-2 cells after cotransfection with YTHDF2 siRNAs and ALKBH5 overexpression plasmids. The data are expressed as the mean ± SD. ns, not significant, * p < 0.05, ** p < 0.01, *** p < 0.001
To elucidate how m6A methylation is responsible for KLF4 mRNA degradation, we suppressed the expression of m6A readers (YTHDF1/2/3, YTHDC1/2, and IGF2BP1/2/3) in Caco-2 cells. Among these, only YTHDF2 knockdown resulted in a significant increase in KLF4 mRNA expression (Fig. 7D, Figure S7D). Furthermore, YTHDF2 knockdown led to a significant increase in KLF4 protein expression (Fig. 7E). YTHDF2 depletion significantly the stability of KLF4 mRNA in Caco-2 and NCM460 cells (Figs. 7F, S7E). Moreover, the RIP assay utilizing an anti-YTHDF2 antibody validated the direct interaction between YTHDF2 and KLF4 mRNA in Caco-2 cells (Figs. 7G, S7F). We also depleted YTHDF2 in Caco-2 cells with or without ALKBH5 overexpression. The expression levels of KLF4 mRNA and protein were markedly increased in Caco-2 cells subjected to co-treatment with YTHDF2 siRNA and ALKBH5 overexpression plasmids (Fig. 7H, I). Collectively, these findings indicate that METTL3/ALKBH5/YTHDF2-mediated m6A methylation leads to a decrease in KLF4 expression in IBD.
APTO-253, which upregulates the expression of KLF4, exhibits a synergistic effect with mesalazine on the preservation of intestinal epithelial barrier function
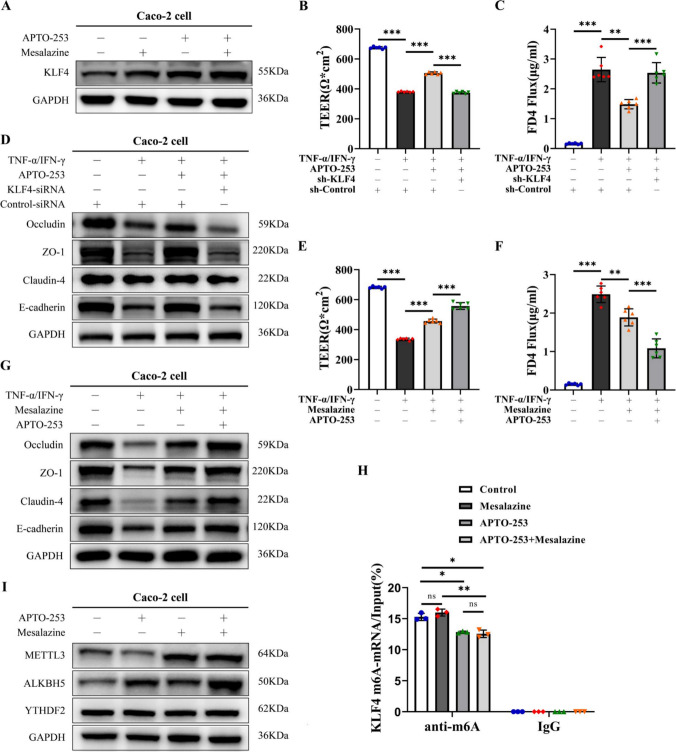
In subsequent in vitro studies, we used APTO-253, which upregulates the expression of KLF4, to evaluate the effect of KLF4 on intestinal epithelial barrier function. The results showed that APTO-253 successfully increased the protein expression of KLF4 in Caco-2 cells (Fig. 8A). Furthermore, treatment with APTO-253 significantly improved TEER and decreased FD4 permeability in Caco-2 monolayers exposed to the TNF-α/IFN-γ mixture compared to control monolayers (Fig. 8B, C). Additionally, APTO-253 treatment upregulated E-cadherin, Occludin, ZO-1, and Claudin-4 protein expression in Caco-2 cells stimulated with TNF-α/IFN-γ (Fig. 8D). Importantly, KLF4 depletion abolished the effects of APTO-253 on TEER and FD4 permeability and the expression of E-cadherin, Occludin, ZO-1, and Claudin-4 in Caco-2 cells stimulated with TNF-α/IFN-γ (Fig. 8B–D), suggesting that the protective effect of APTO-253 on the intestinal epithelial barrier is partly mediated by the upregulation of KLF4 expression.
Fig. 8.
APTO-253 exhibits a synergistic effect with mesalazine in the preservation of IEB function. A KLF4 protein expression in Caco-2 cells co-stimulated with APTO-253 and mesalazine. B, C TEER (B) and FD4 (C) permeability were measured in the KLF4 knockdown Caco-2 monolayer after costimulation with TNF-α/IFN-γ (100 ng/ml) and APTO-253. D The protein expression of Occludin, ZO-1, Claudin-4 and E-cadherin in Caco-2 cells cotreated with KLF4 siRNA, TNF-α/IFN-γ (100 ng/ml) and APTO-253. E, F TEER (E) and FD4 permeability (F) were measured in Caco-2 cell monolayers cotreated with mesalazine, TNF-α/IFN-γ (100 ng/ml) and APTO-253. G The protein expression of Occludin, ZO-1, Claudin-4 and E-cadherin in Caco-2 cells cotreated with mesalazine, TNF-α/IFN-γ (100 ng/ml) and APTO-253. H The m6A modification of KLF4 mRNA in Caco-2 cells after APTO-253 and mesalazine co-stimulation. I The protein expression of METTL3, ALKBH5 and YTHDF2 in Caco-2 cells co-treated with mesalazine and APTO-253.The data are expressed as the mean ± SD. ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001
Mesalazine, a first-line drug for IBD treatment, has been demonstrated to improve intestinal epithelial barrier function in intestinal inflammation [25, 36] and induce KLF4 expression [37] (Fig. 8A). Then, we explored the synergistic effects of APTO-253 and mesalazine on KLF4 expression and intestinal epithelial barrier function. Our study revealed that the combined administration of APTO-253 and mesalazine resulted in a significant increase in KLF4 expression (Fig. 8A). Furthermore, as depicted in Fig. 8E, F, the co-administration of APTO-253 and mesalazine led to a notable enhancement in TEER and a reduction in FD4 permeability in Caco-2 cell monolayers when exposed to TNF-α/IFN-γ stimulation, compared to treatment with APTO-253 or mesalazine alone. Moreover, we observed that the E-cadherin, Occludin, ZO-1, and Claudin-4 protein expression levels were greater in the Caco-2 cells cotreated with APTO-253 and mesalazine than in those treated with mesalazine alone (Fig. 8G).
Then, we explored whether APTO-253 and mesalazine regulated KLF4 expression via the m6A modification. As shown in Fig. 8H, APTO-253 decreased the m6A level of KLF4 mRNA in Caco-2 cells, whereas mesalazine did not significantly affect the m6A level of KLF4 (Fig. 8H). Additionally, we observed that mesalazine upregulated METTL3 and ALKBH5 expression, while APTO-253 only increased ALKBH5 expression (Fig. 8I). Neither treatment had an impact on YTHDF2 expression (Fig. 8I). Collectively, our findings suggest that the m6A modification of KLF4 mRNA plays a crucial role in the combined effects of mesalazine and APTO-253 on maintaining intestinal epithelial barrier function.
Discussion
Studies have indicated that the transcription factor KLF4 is involved in the progression of IBD. For example, KLF4 serves as a critical regulator of goblet cell differentiation and mucin production in the human colon, thereby inhibiting the development of UC [38]. In addition, KLF4 promoted the activation of alternatively activated macrophage functional subsets, which are primarily involved in tissue repair, and limited chronic inflammation in IBD [39]. Consistent with these findings, our present study showed that the expression of KLF4 was decreased in the inflamed colonic mucosa of IBD patients and colitis model mice and was negatively correlated with the inflammatory features of IBD. Although the fold differences and levels of expression of KLF4 varied among different GEO databases, the decreasing trend in KLF4 expression in IBD patients was consistent across different microarray studies. Moreover, mice with KLF4 deficiency displayed heightened vulnerability to colitis induced by 2% DSS and 2.5% TNBS. These data indicate that KLF4 may serve a protective function in the pathogenesis of IBD. In contrast, Amr M Ghaleb and colleagues observed an upregulation of KLF4 expression in the crypt zone of the colonic epithelia in mice following induction with DSS. Additionally, their study demonstrated that intestine-specific deletion of KLF4 resulted in decreased susceptibility to 3% DSS-induced colitis in mice [40]. In human UC tissues, KLF4 expression is significantly elevated, and KLF4 may regulate the pyroptosis process of intestinal epithelial cells in vitro through the TXNIP/NLRP3 pathway [20]. From these previous reports and our results, we found that inconsistencies in the expression and roles of KLF4 in IBD were associated with cell type and the degree of inflammation.
An impaired intestinal epithelial barrier is a crucial factor leading to IBD. AJC proteins are important for maintaining intestinal epithelial barrier integrity, and their dysfunction has been implicated in the development of IBD. Lijun Dong et al. reported that mannose reduced the release of the lysosomal enzyme cathepsin B, thereby enhancing tight junctions and mitigating the severity of chemically induced colitis and spontaneous colitis [25]. DDR1 knockout preserved intestinal barrier integrity by reducing the degradation of tight junction proteins in DSS-induced colitis [41]. Furthermore, TMIGD1 demonstrated a protective effect on intestinal epithelial barrier function, as indicated by increased expression of AJC proteins in cells overexpressing TMIGD1 [26]. Herein, we observed that KLF4 downregulation impaired the intestinal epithelial barrier by decreasing apical junction complex protein expression in DSS- and TNBS-induced colitis model mice. In vitro studies further demonstrated that knockdown of KLF4 led to damage to the intestinal epithelial barrier by downregulating the expression of AJC proteins. Furthermore, ChIP assays illustrated the direct binding of KLF4 to the promoters of E-cadherin, Occludin, ZO-1, and Claudin-4 in Caco-2 cells. These data indicated that KLF4 protects the intestinal epithelial barrier and suppresses the progression of IBD by transcriptionally upregulating AJC proteins in intestinal epithelial cells.
Increased intestinal epithelial cell apoptosis is also associated with the modulation of the intestinal epithelial barrier under physiological and pathological conditions [29–31]. Weiting Kuo et al. reported that the loss of Occludin in intestinal epithelial limited the severity of DSS- and TNBS-induced colitis by enhancing epithelial cell resistance to apoptosis [42]. 3-Mercaptopyruvate sulfurtransferase (MPST) deficiency elevated the apoptosis rate of intestinal epithelial cells and aggravated IBD via AKT [43]. In the present study, we found that KLF4 depletion increased, while KLF4 overexpression decreased, the apoptosis of Caco-2 cells treated with H2O2 or a TNF-α/IFN-γ mixture. Moreover, the reduction of KLF4 expression resulted in a notable increase in the percentage of apoptotic intestinal epithelial cells in mice with colitis induced by DSS and TNBS. These results indicated that KLF4 protects the intestinal epithelial barrier during IBD through the inhibition of intestinal epithelial cell apoptosis. High levels of ROS and excessive proinflammatory cytokines have been demonstrated to contribute to the apoptosis of intestinal epithelial cells during colitis [32, 33, 44, 45]. Herein, we observed that KLF4 inhibited intestinal epithelial cell apoptosis by reducing intracellular ROS levels and suppressing the production of proinflammatory cytokines such as IFNγ, IL-1β, IL-6, and TNF-α. However, the precise mechanism by which KLF4 modulates intracellular ROS and proinflammatory cytokine production is still unclear. Hence, further exploration of the mechanisms by which KLF4 regulates intracellular ROS and proinflammatory cytokine production in intestinal epithelial cells is needed.
M6A RNA modification is a widespread and abundant modification of mRNAs that plays a critical role in the regulation of various aspects of RNA metabolism, including decay, translation, localization, and splicing [46, 47]. This modification is mediated by methyltransferases (e.g., METTL3 and METTL14), demethylases (e.g., fat mass- and obesity-associated protein (FTO) and ALKBH5) and m6A binding proteins (e.g., YTHDF1/2/3, YTHDC1/2, and IGF2BP1/2/3) [48]. m6A mRNA modification plays an important role in modulating the progression of IBD [49, 50]. A study by Ting Zhang et al. noted that specific deletion of the METTL14 gene in the mouse colon resulted in colonic stem cell apoptosis, causing mucosal barrier dysfunction and severe colitis by regulating the stability of Nfkbia mRNA and modulating the NF-κB pathway [51]. Given that m6A RNA modification is involved in modulating KLF4 expression in various cells, such as vascular endothelial and bladder cancer cells [34, 35], we inferred that m6A RNA modification might modulate the expression of KLF4 in IBD. Our results revealed the presence of m6A methylation on KLF4 mRNA in Caco-2 cells. Furthermore, changes in the m6A level of KLF4 mRNA were regulated by METTL3 and ALKBH5. In addition, we observed that YTHDF2, a m6A “reader”, could directly bind to m6A-modified KLF4 mRNA and reduce its mRNA stability. Therefore, we identified that ALKBH5 removes m6A modifications and METTL3 adds m6A modifications to KLF4 mRNA, and YTHDF2 directly binds to m6A-modified KLF4 mRNA, facilitating its degradation.
We also investigated the potential synergistic effects of mesalazine and APTO-253, which upregulates the expression of KLF4, on intestinal epithelial barrier function. Mesalazine, the primary treatment for IBD, has been demonstrated to improve intestinal epithelial barrier function in intestinal inflammation [25, 36]. The current investigation demonstrates that APTO-253 exhibited a synergistic impact in maintaining the integrity of the intestinal epithelial barrier when combined with mesalazine. However, the synergistic effects on intestinal epithelial barrier function should be validated in a mouse model of colitis. This will constitute the primary focus of our research in the subsequent phase. Subsequent analysis revealed that the m6A modification of KLF4 mRNA was pivotal in mediating the collaborative effects of mesalazine and APTO-253. These findings suggest that mesalazine and the enhancement of KLF4 expression may serve as valuable targets for improving anti-inflammatory treatment strategies. Nevertheless, further investigation is required to elucidate the underlying mechanisms responsible for these synergistic effects.
Conclusions
In summary, a negative correlation exists between KLF4 expression and inflammatory characteristics of IBD (Fig. 9). m6A modification-induced KLF4 downregulation aggravated inflammation and intestinal epithelial barrier dysfunction by inhibiting the expression of AJC proteins and increasing intestinal epithelial cell apoptosis in mice with colitis. Our findings suggest that targeting KLF4 may provide insights for the development of therapeutic interventions for IBD and the optimization of anti-inflammatory treatments.
Fig. 9.
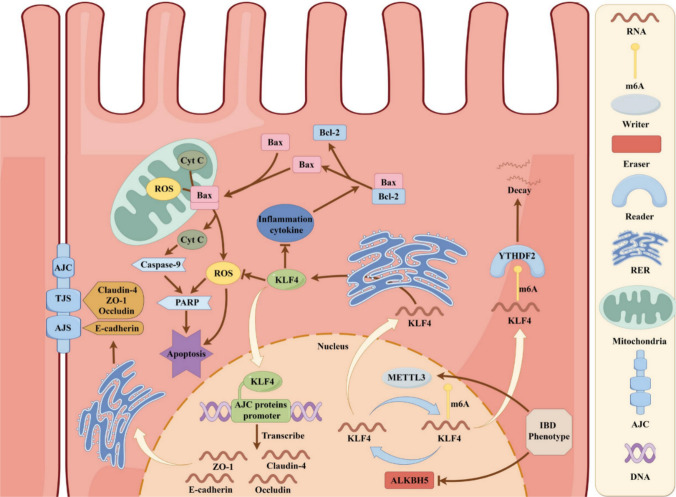
A schematic showing that KLF4 maintains intestinal epithelial barrier function and is regulated by dynamic m6A mRNA modification (By Figdraw, ID:RIPPRde807)
Supplementary Information
Below is the link to the electronic supplementary material.
Author contributions
The authors checked and approved the final manuscript. Tongguo Shi, Qinhua Xi, and Xingchao Zhu conceived and designed the experiments. Xingchao Zhu, Jiayu Wang, Jinghan Zhu, Juntao Li, Kexi Yang and Kun Wang performed most of the experiments and analyzed the data. Hongqin Yue, Huan Zhang, Kanger Shen and Xia Leng performed sample collection and clinical evaluation. The manuscript was wrote by Xingchao Zhu and revised by Tongguo Shi, Qinhua Xi.
Funding
The present work was supported by the Jiangsu Provincial Medical Key Discipline (ZDXK202246), Bo Xi Cultivation Plan of First Affiliated Hospital of Soochow University (BXQN202201).
Availability of data and material
All software used in this study were obtained from free or commercially available and listed in the “Methods” section. All data generated or analyzed during this study are included in this published article [and its Additional files].
Declarations
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethics approval and consent to participate
Ethical approval for the study was granted by the Ethics Committee at the First Affiliated Hospital of Soochow University (Suzhou, China. No. 2023532), and informed consent was also obtained from all participants. The Ethics Committee of Soochow University (Suzhou, China) approved all animal experimental procedures (No.202311A0034).
Consent for publication
All authors reviewed the results and approved this manuscript for publication.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Qinhua Xi, Email: xqhxqhxqh@126.com.
Tongguo Shi, Email: shitg@suda.edu.cn.
References
- 1.Nakase H, Uchino M, Shinzaki S, Matsuura M, Matsuoka K, Kobayashi T, Saruta M, Hirai F, Hata K, Hiraoka S et al (2021) Evidence-based clinical practice guidelines for inflammatory bowel disease 2020. J Gastroenterol 56:489–526. 10.1007/s00535-021-01784-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maaser C, Sturm A, Vavricka SR, Kucharzik T, Fiorino G, Annese V, Calabrese E, Baumgart DC, Bettenworth D, Borralho Nunes P et al (2019) ECCO-ESGAR Guideline for Diagnostic Assessment in IBD Part 1: Initial diagnosis, monitoring of known IBD, detection of complications. J Crohns Colitis 13:144–164. 10.1093/ecco-jcc/jjy113 [DOI] [PubMed] [Google Scholar]
- 3.Piovani D, Danese S, Peyrin-Biroulet L, Nikolopoulos GK, Lytras T, Bonovas S (2019) Environmental risk factors for inflammatory bowel diseases: an umbrella review of meta-analyses. Gastroenterology 157(647–659):e644. 10.1053/j.gastro.2019.04.016 [DOI] [PubMed] [Google Scholar]
- 4.Michaudel C, Sokol H (2020) The gut microbiota at the service of immunometabolism. Cell Metab 32:514–523. 10.1016/j.cmet.2020.09.004 [DOI] [PubMed] [Google Scholar]
- 5.Chen Y, Cui W, Li X, Yang H (2021) Interaction between commensal bacteria, immune response and the intestinal barrier in inflammatory bowel disease. Front Immunol 12:761981. 10.3389/fimmu.2021.761981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunleavy KA, Raffals LE, Camilleri M (2023) Intestinal Barrier Dysfunction in Inflammatory Bowel Disease: Underpinning Pathogenesis and Therapeutics. Dig Dis Sci 68:4306–4320. 10.1007/s10620-023-08122-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Hernandez V, Quiros M, Nusrat A (2017) Intestinal epithelial claudins: expression and regulation in homeostasis and inflammation. Ann N Y Acad Sci 1397:66–79. 10.1111/nyas.13360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee SH (2015) Intestinal permeability regulation by tight junction: implication on inflammatory bowel diseases. Intest Res 13:11–18. 10.5217/ir.2015.13.1.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landy J, Ronde E, English N, Clark SK, Hart AL, Knight SC, Ciclitira PJ, Al-Hassi HO (2016) Tight junctions in inflammatory bowel diseases and inflammatory bowel disease associated colorectal cancer. World J Gastroenterol 22:3117–3126. 10.3748/wjg.v22.i11.3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehandru S, Colombel JF (2021) The intestinal barrier, an arbitrator turned provocateur in IBD. Nat Rev Gastroenterol Hepatol 18:83–84. 10.1038/s41575-020-00399-w [DOI] [PubMed] [Google Scholar]
- 11.Mohanan V, Nakata T, Desch AN, Levesque C, Boroughs A, Guzman G, Cao Z, Creasey E, Yao J, Boucher G et al (2018) C1orf106 is a colitis risk gene that regulates stability of epithelial adherens junctions. Science 359:1161–1166. 10.1126/science.aan0814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muise AM, Walters TD, Glowacka WK, Griffiths AM, Ngan BY, Lan H, Xu W, Silverberg MS, Rotin D (2009) Polymorphisms in E-cadherin (CDH1) result in a mis-localised cytoplasmic protein that is associated with Crohn’s disease. Gut 58:1121–1127. 10.1136/gut.2008.175117 [DOI] [PubMed] [Google Scholar]
- 13.Wei D, Kanai M, Huang S, Xie K (2006) Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis 27:23–31. 10.1093/carcin/bgi243 [DOI] [PubMed] [Google Scholar]
- 14.Ghaleb AM, Yang VW (2017) Kruppel-like factor 4 (KLF4): what we currently know. Gene 611:27–37. 10.1016/j.gene.2017.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flandez M, Guilmeau S, Blache P, Augenlicht LH (2008) KLF4 regulation in intestinal epithelial cell maturation. Exp Cell Res 314:3712–3723. 10.1016/j.yexcr.2008.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghaleb AM, McConnell BB, Kaestner KH, Yang VW (2011) Altered intestinal epithelial homeostasis in mice with intestine-specific deletion of the Kruppel-like factor 4 gene. Dev Biol 349:310–320. 10.1016/j.ydbio.2010.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katz JP, Perreault N, Goldstein BG, Lee CS, Labosky PA, Yang VW, Kaestner KH (2002) The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development 129:2619–2628. 10.1242/dev.129.11.2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu T, Chen X, Zhang W, Li J, Xu R, Wang TC, Ai W, Liu C (2012) Kruppel-like factor 4 regulates intestinal epithelial cell morphology and polarity. PLoS ONE 7:e32492. 10.1371/journal.pone.0032492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng H, Pritchard DM, Yang X, Bennett E, Liu G, Liu C, Ai W (2009) KLF4 gene expression is inhibited by the notch signaling pathway that controls goblet cell differentiation in mouse gastrointestinal tract. Am J Physiol Gastrointest Liver Physiol 296:G490-498. 10.1152/ajpgi.90393.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Sun L, Liu H, Li J, Guo L, Wang Z (2024) KLF4 interacts with TXNIP to modulate the pyroptosis in ulcerative colitis via regulating NLRP3 signaling. Immun Inflamm Dis 12:e1199. 10.1002/iid3.1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma J, Wang P, Liu Y, Zhao L, Li Z, Xue Y (2014) Kruppel-like factor 4 regulates blood-tumor barrier permeability via ZO-1, occludin and claudin-5. J Cell Physiol 229:916–926. 10.1002/jcp.24523 [DOI] [PubMed] [Google Scholar]
- 22.Sturm A, Maaser C, Calabrese E, Annese V, Fiorino G, Kucharzik T, Vavricka SR, Verstockt B, van Rheenen P, Tolan D et al (2019) ECCO-ESGAR Guideline for Diagnostic Assessment in IBD Part 2: IBD scores and general principles and technical aspects. J Crohns Colitis 13:273–284. 10.1093/ecco-jcc/jjy114 [DOI] [PubMed] [Google Scholar]
- 23.Wirtz S, Popp V, Kindermann M, Gerlach K, Weigmann B, Fichtner-Feigl S, Neurath MF (2017) Chemically induced mouse models of acute and chronic intestinal inflammation. Nat Protoc 12:1295–1309. 10.1038/nprot.2017.044 [DOI] [PubMed] [Google Scholar]
- 24.Viennois E, Chen F, Laroui H, Baker MT, Merlin D (2013) Dextran sodium sulfate inhibits the activities of both polymerase and reverse transcriptase: lithium chloride purification, a rapid and efficient technique to purify RNA. BMC Res Notes 6:360. 10.1186/1756-0500-6-360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong L, Xie J, Wang Y, Jiang H, Chen K, Li D, Wang J, Liu Y, He J, Zhou J et al (2022) Mannose ameliorates experimental colitis by protecting intestinal barrier integrity. Nat Commun 13:4804. 10.1038/s41467-022-32505-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou L, Zhu L, Wu X, Hu S, Zhang S, Ning M, Yu J, Chen M (2023) Decreased TMIGD1 aggravates colitis and intestinal barrier dysfunction via the BANF1-NF-kappaB pathway in Crohn’s disease. BMC Med 21:287. 10.1186/s12916-023-02989-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chanez-Paredes S, Montoya-Garcia A, Castro-Ochoa KF, Garcia-Cordero J, Cedillo-Barron L, Shibayama M, Nava P, Flemming S, Schlegel N, Gautreau AM et al (2021) The Arp2/3 inhibitory protein arpin is required for intestinal epithelial barrier integrity. Front Cell Dev Biol 9:625719. 10.3389/fcell.2021.625719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo J, Xu L, Teng X, Sun M (2017) MicroRNA-7-5p regulates the proliferation and migration of intestinal epithelial cells by targeting trefoil factor 3 via inhibiting the phosphoinositide 3-kinase/Akt signalling pathway. Int J Mol Med 40:1435–1443. 10.3892/ijmm.2017.3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Citi S (2018) Intestinal barriers protect against disease. Science 359:1097–1098. 10.1126/science.aat0835 [DOI] [PubMed] [Google Scholar]
- 30.Wu WF, Tan XJ, Dai YB, Krishnan V, Warner M, Gustafsson JA (2013) Targeting estrogen receptor beta in microglia and T cells to treat experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A 110:3543–3548. 10.1073/pnas.1300313110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Looijer-van Langen M, Hotte N, Dieleman LA, Albert E, Mulder C, Madsen KL (2011) Estrogen receptor-beta signaling modulates epithelial barrier function. Am J Physiol Gastrointest Liver Physiol 300:G621-626. 10.1152/ajpgi.00274.2010 [DOI] [PubMed] [Google Scholar]
- 32.Kruidenier L, Kuiper I, Lamers CB, Verspaget HW (2003) Intestinal oxidative damage in inflammatory bowel disease: semi-quantification, localization, and association with mucosal antioxidants. J Pathol 201:28–36. 10.1002/path.1409 [DOI] [PubMed] [Google Scholar]
- 33.Weigmann B, Lehr HA, Yancopoulos G, Valenzuela D, Murphy A, Stevens S, Schmidt J, Galle PR, Rose-John S, Neurath MF (2008) The transcription factor NFATc2 controls IL-6-dependent T cell activation in experimental colitis. J Exp Med 205:2099–2110. 10.1084/jem.20072484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chien CS, Li JY, Chien Y, Wang ML, Yarmishyn AA, Tsai PH, Juan CC, Nguyen P, Cheng HM, Huo TI et al (2021) METTL3-dependent N(6)-methyladenosine RNA modification mediates the atherogenic inflammatory cascades in vascular endothelium. Proc Natl Acad Sci U S A. 10.1073/pnas.2025070118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie H, Li J, Ying Y, Yan H, Jin K, Ma X, He L, Xu X, Liu B, Wang X et al (2020) METTL3/YTHDF2 m(6) A axis promotes tumorigenesis by degrading SETD7 and KLF4 mRNAs in bladder cancer. J Cell Mol Med 24:4092–4104. 10.1111/jcmm.15063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khare V, Krnjic A, Frick A, Gmainer C, Asboth M, Jimenez K, Lang M, Baumgartner M, Evstatiev R, Gasche C (2019) Mesalamine and azathioprine modulate junctional complexes and restore epithelial barrier function in intestinal inflammation. Sci Rep 9:2842. 10.1038/s41598-019-39401-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parenti S, Montorsi L, Fantini S, Mammoli F, Gemelli C, Atene CG, Losi L, Frassineti C, Calabretta B, Tagliafico E et al (2018) KLF4 Mediates the effect of 5-ASA on the beta-catenin pathway in colon cancer cells. Cancer Prev Res (Phila) 11:503–510. 10.1158/1940-6207.CAPR-17-0382 [DOI] [PubMed] [Google Scholar]
- 38.Gersemann M, Becker S, Kubler I, Koslowski M, Wang G, Herrlinger KR, Griger J, Fritz P, Fellermann K, Schwab M et al (2009) Differences in goblet cell differentiation between Crohn’s disease and ulcerative colitis. Differentiation 77:84–94. 10.1016/j.diff.2008.09.008 [DOI] [PubMed] [Google Scholar]
- 39.Hu Q, Lyon CJ, Fletcher JK, Tang W, Wan M, Hu TY (2021) Extracellular vesicle activities regulating macrophage- and tissue-mediated injury and repair responses. Acta Pharm Sin B 11:1493–1512. 10.1016/j.apsb.2020.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghaleb AM, Laroui H, Merlin D, Yang VW (2014) Genetic deletion of Klf4 in the mouse intestinal epithelium ameliorates dextran sodium sulfate-induced colitis by modulating the NF-kappaB pathway inflammatory response. Inflamm Bowel Dis 20:811–820. 10.1097/MIB.0000000000000022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, Li Q, Xiong B, Chen H, Wang X, Zhang D (2022) Discoidin domain receptor 1(DDR1) promote intestinal barrier disruption in Ulcerative Colitis through tight junction proteins degradation and epithelium apoptosis. Pharmacol Res 183:106368. 10.1016/j.phrs.2022.106368 [DOI] [PubMed] [Google Scholar]
- 42.Kuo WT, Shen L, Zuo L, Shashikanth N, Ong M, Wu L, Zha J, Edelblum KL, Wang Y, Wang Y et al (2019) Inflammation-induced occludin downregulation limits epithelial apoptosis by suppressing caspase-3 expression. Gastroenterology 157:1323–1337. 10.1053/j.gastro.2019.07.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J, Cen L, Zhang X, Tang C, Chen Y, Zhang Y, Yu M, Lu C, Li M, Li S et al (2022) MPST deficiency promotes intestinal epithelial cell apoptosis and aggravates inflammatory bowel disease via AKT. Redox Biol 56:102469. 10.1016/j.redox.2022.102469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang R, Li R, Dai P, Li Z, Li Y, Li C (2019) Deoxynivalenol induced apoptosis and inflammation of IPEC-J2 cells by promoting ROS production. Environ Pollut 251:689–698. 10.1016/j.envpol.2019.05.026 [DOI] [PubMed] [Google Scholar]
- 45.Pott J, Maloy KJ (2018) Epithelial autophagy controls chronic colitis by reducing TNF-induced apoptosis. Autophagy 14:1460–1461. 10.1080/15548627.2018.1450021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR (2012) Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149:1635–1646. 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M et al (2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485:201–206. 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- 48.Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, Yang C, Chen Y (2021) The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther 6:74. 10.1038/s41392-020-00450-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J, Song B, Zeng Y, Xu C, Gao L, Guo Y, Liu J (2023) m6A modification in inflammatory bowel disease provides new insights into clinical applications. Biomed Pharmacother 159:114298. 10.1016/j.biopha.2023.114298 [DOI] [PubMed] [Google Scholar]
- 50.Nie K, Yi J, Yang Y, Deng M, Yang Y, Wang T, Chen X, Zhang Z, Wang X (2021) A broad m6A modification landscape in inflammatory bowel disease. Front Cell Dev Biol 9:782636. 10.3389/fcell.2021.782636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang T, Ding C, Chen H, Zhao J, Chen Z, Chen B, Mao K, Hao Y, Roulis M, Xu H et al (2022) m(6)A mRNA modification maintains colonic epithelial cell homeostasis via NF-kappaB-mediated antiapoptotic pathway. Sci Adv 8:eabl1723. 10.1126/sciadv.abl5723 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All software used in this study were obtained from free or commercially available and listed in the “Methods” section. All data generated or analyzed during this study are included in this published article [and its Additional files].