Abstract
Disulfidptosis, a newly identified form of regulated cell death associated with disruption of disulfide bond formation in the endoplasmic reticulum, involves the dysregulation of disulfidptosis-related genes (DRGs) that may contribute to cancer development and progression. However, the molecular mechanisms and clinical implications of DRGs in different cancer types remain poorly characterized. Therefore, in this comprehensive study, we investigated the expression, prognostic value, and functional roles of four recently identified DRGs (SLC7A11, SLC3A2, RPN1, and NCKAP1) across various cancers. Especially, in clinical samples of glioblastoma, we found that RPN1 was significantly correlated with patient survival. Through mutation landscape analysis, we identified diverse missense mutations in these DRGs, with NCKAP1 exhibiting the highest mutation frequency (5.9% in skin cutaneous melanoma). Additionally, we observed positive correlations between these DRGs and tumor stemness (DNAss DNA stemness score and RNAss RNA stemness score) as well as RNA modifications, particularly m6A modification, in several cancer types. Furthermore, high expression of SLC7A11, RPN1, and NCKAP1 was positively associated with infiltration of T-helper type 2 (Th2) cells in various cancers, while high expression of SLC7A11, SLC3A2, and RPN1 correlated with tumor mutational burden (TMB) in 10, 4, and 8 tumor types, respectively. Utilizing a protein–protein interaction network, we identified the RHO GTPases Activate WASPs and WAVEs pathway as significantly enriched, suggesting the involvement of these DRGs in cancer-related signaling pathways. Collectively, our findings provide novel insights into the molecular mechanisms and clinical implications of DRGs in pan-cancer, highlighting their potential as biomarkers and therapeutic targets for cancer treatment.
Keywords: Disulfidptosis, Disulfidptosis-related genes (DRGs), Cancer development, Prognostic markers, Tumor stemness, Therapeutic targets
Introduction
Tumors are human diseases characterized by high mortality and incidence rates worldwide [1]. One of the key factors that enable tumor cells to proliferate malignantly is the evasion of cell death, which is often influenced by metabolic reprogramming [2]. Metabolic reprogramming is a hallmark of cancer that allows cancer cells to adapt to specific nutrient or metabolic demands [3]. It also regulates various forms of cell death, such as ferroptosis [4] and cuproptosis [5]. Recently, a novel form of metabolic-related cell death, termed disulfidoptosis, has been discovered by Liu et al. [6] They showed that high expression of solute carrier family 7 member 11 (SLC7A11) in renal cancer cells leads to rapid depletion of cytoplasmic nicotinamide adenine dinucleotide phosphate (NADPH) under glucose starvation [6]. This results in the accumulation of unreduced disulfides and disulfide stress, which trigger disulfidoptosis [7, 8]. Similarly, Yan et al. reported that high expression of SLC7A11 in cancer cells, coupled with hydrogen peroxide treatment, causes the toxic buildup of intracellular cysteine and other disulfide molecules, depletion of NADPH, the collapse of the redox system, and rapid cell death, possibly through disulfide deposition [9].
Liu et al. identified four disulfidoptosis-related genes (DRGs) in their study: SLC7A11, solute carrier family 3 member 2 (SLC3A2), recombinant ribonucleoprotein 1 (RPN1), and NCK-associated protein 1 (NCKAP1) [6]. Dysregulation of DRG expression affects various cellular features, such as immune infiltration, cell cycle, and cell death. For example, inhibiting glucose-6-phosphate dehydrogenase (G6PD) significantly impairs the proliferation ability of lung cancer cells [10]. In bladder cancer, Chen et al. found that DRGs are closely associated with reduced survival rates, tumor immune microenvironment inflammation, and increased tumor mutation burden (TMB) [11]. Additionally, aberrant DRG expression in patients with kidney renal clear cell carcinoma (KIRC) [12] and thyroid carcinoma (THCA) [13] can serve as accurate prognostic models for predicting high sensitivity to immunotherapy. However, due to limited studies on disulfidoptosis, the effect of disulfidoptosis on most cancers remains elusive.
RNA modifications play a crucial role in regulating the expression of DRGs. For example, Qiu et al. demonstrated that FTO exerts its tumor-suppressive effects in papillary thyroid carcinoma (PTC) by downregulating the expression of SLC7A11 [14]. In another study, Ji et al. elucidated that METTL3 and ALKBH5 are involved in the m6A modification of SLC3A2 and SLC7A5 mRNAs, thereby facilitating their translation in uroepithelial cells undergoing 3-MC-induced transformation [15]. In this study, we conducted an integrative analysis of the four DRGs across cancers, aiming to investigate their functional significance and potential implications in tumorigenesis. We found that these genes are aberrantly upregulated in multiple cancer types and are associated with poor prognosis. Moreover, they are associated with various biological characteristics of tumors, including tumor stemness, RNA modification levels, immune cell infiltration, genomic heterogeneity, and functional signaling pathways related to tumor pathological changes. Our findings provide novel insights into the diverse and complex roles of DRGs in cancer biology and suggest potential targets for therapeutic intervention.
Methods and materials
Differential expression analysis
We downloaded the standard pan-cancer dataset TCGA (The cancer genome atlas), TARGET (TherapeuticallyApplicable Research To Generate EffectiveTreatments) and GTEx (The Genotype-Tissue Expression) (PANCAN, N = 19,131, G = 60,499) from the UCSC (https://xenabrowser.net/) database [16–18]. Gene expression data for SLC7A11, SLC3A2, RPN1, and NCKAP1 were extracted from the dataset, excluding cancer types with fewer than three samples. To ensure data comparability, a log2(x + 1) transformation was applied to each gene expression value. We performed a differential expression analysis between normal and tumor samples within 34 different cancer types by utilizing non-paired Wilcoxon Rank Sum and Signed Rank Tests implemented in R (version 4.2.2).
The prognostic value of overall survival
We collected follow-up data from TARGET via the UCSC Browser, excluding samples with follow-up times less than 30 days, and supplemented with high-quality prognostic data from a study published in Cell [19]. Cox proportional hazards regression models were built using the coxph function in R to analyze the gene expression's impact on the prognosis of overall survival in 44 cancer types. The optimal cut-off values for DRGs were calculated by the R package maxstat, and the patients were divided into two high and low groups. The Logrank test was used for statistical significance assessment in survival.
Collection of clinical tissue samples
We collected a cohort of 80 glioblastoma tissue samples, along with comprehensive clinical data including survival duration, sex, age, and other relevant variables. A glioblastoma (GBM) tissue microarray was constructed (Afantibody, AF-BraSur2201). The study using the tissue microarray was approved by the Life Sciences Ethics Committee of Changsha Yaxiang Biotechnology Co., LTD. The Ethics report is available online at yxswll.ccrl.cn. The query code is 5GQ20JNNCCA7NL.
Immunofluorescence assay
Tyramide Signal Amplification (TSA) technology (Servicebio, G1236-50 T) was utilized for the immunofluorescence (IF) assays. Following deparaffinization, antigen retrieval, and endogenous peroxidase blocking, non-specific binding was mitigated using 3% BSA. After removing the excess blocking solution, the primary antibody targeting SLC3A2 (PTG, Lot: 66,883-1-Ig) was diluted in PBS and incubated overnight at 4 °C in a humidified chamber. The HRP-labeled secondary antibody (PTG, RGAM011) was then applied to bind the primary antibody. Subsequently, TSA-555 staining buffer was introduced to the tissue sections.The procedure was repeated with the primary antibody against NCKAP1 (PTG, Lot: 12,140-1-AP), diluted in PBS, and incubated overnight at 4 °C in a humidified chamber. This was followed by application of the HRP-labeled secondary antibody (PTG, RGAR011) and TSA-488 staining buffer. Finally, the primary antibody targeting RPN1 (PTG, Lot: 12,894-1-AP) was diluted in PBS and incubated overnight at 4 °C in a humidified chamber. The HRP-labeled secondary antibody (PTG, RGAR011) was used to bind the primary antibody, and TSA-647 staining buffer was applied to the tissue sections.
Tumor stemness and DRGs expression
We obtained tumor stemness scores, calculated based on methylation features (DNAss) and mRNA features (RNAss), for each tumor from a previous study [20]. The stemness scores with gene expression data were integrated to generate a dataset of expression data for 37 cancer types. The Pearson correlation between the gene expression profiles and the stemness scores was calculated.
DRGs mutation landscape
The level 4 datasets of single nucleotide variation (SNV) and copy number variation (CNV) datasets for all TCGA samples handled by MuTect2 [21] and GISTIC [22] software, respectively, were downloaded from the Genomic Data Commons (GDC) (https://portal.gdc.cancer.gov/). The structural domain information of the DRGs protein was obtained from the maftools R package(Version 2.20.0). The mutation landscape was constructed by integrating these datasets.
RNA modification and immune cell infiltration
The expression data of DRGs and 44 genes representing three types of RNA modifications (m1A(10), m5C(13), m6A(21)) were extracted from each sample. The Spearman correlation coefficients between their expressions were computed using the corr.test function in the R package psych(Version 2.4.1). For immune cell infiltration analysis, the gene expression profiles of each tumor were mapped to GeneSymbol. Subsequently, we utilized the deconvo_xCell method from the R package IOBR(Version 1.0) [23] to re-evaluate the infiltration scores of 67 immune cell types in each tumor. Spearman's correlation coefficient between gene expression and immune cell infiltration scores was calculated across cancers.
Genomic heterogeneity and DRGs expression
The SNV information of DRGs was utilized to compute the TMB for each tumor sample using the tmb function from the R package maftools (Version 2.20.0). We assessed the Pearson correlation between the expression levels of DRGs and TMB in each specific tumor type.
Pathway enrichment analysis of DRGs-interacting protein
The protein–protein interaction network of DRGs was constructed using STRING and further visualized using Cytoscape software (version 3.9). The biological functionality of the network was analyzed using Metascape [24] (https://metascape.org/gp/index.html#/main/step1). Terms that met specific criteria (P < 0.01, count ≥ 3, enrichment factor > 1.5) were identified and grouped into clusters based on their similarities in membership.
Statistical analysis
The significance of differences between the two pairs was analyzed using the unpaired Wilcoxon rank sum and signed rank tests, and the Kruskal‒Wallis test was used to test for differences in multiple samples [25]. Unless otherwise stated, P < 0.05 was considered to indicate statistical significance.
Results
DRGs aberrantly highly expressed in multiple tumors with poor prognostic values
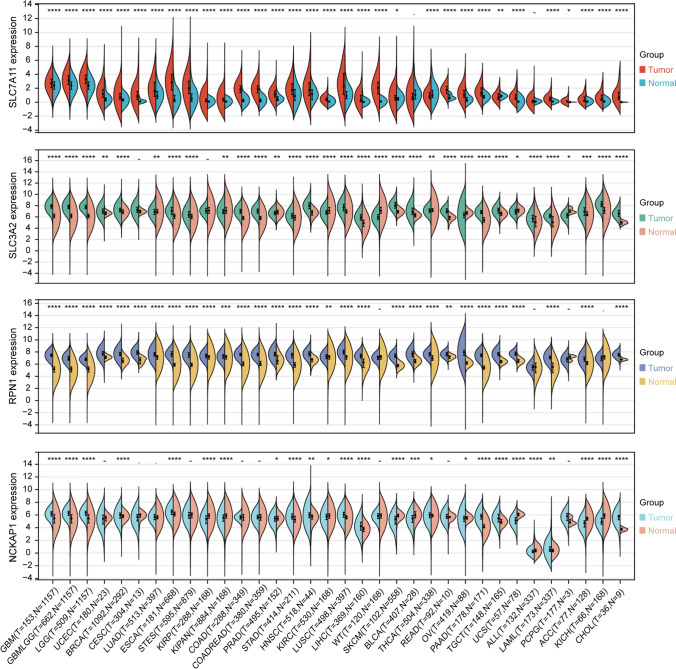
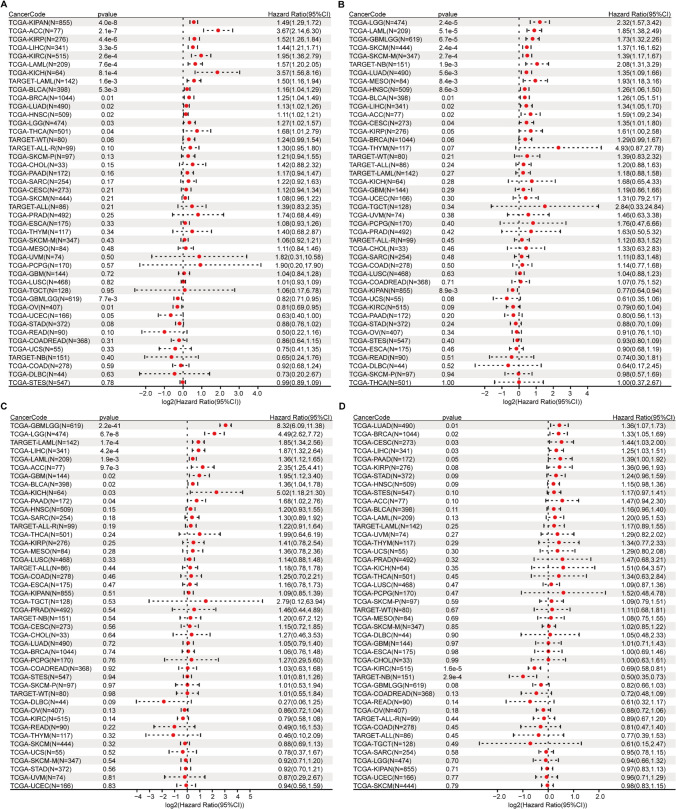
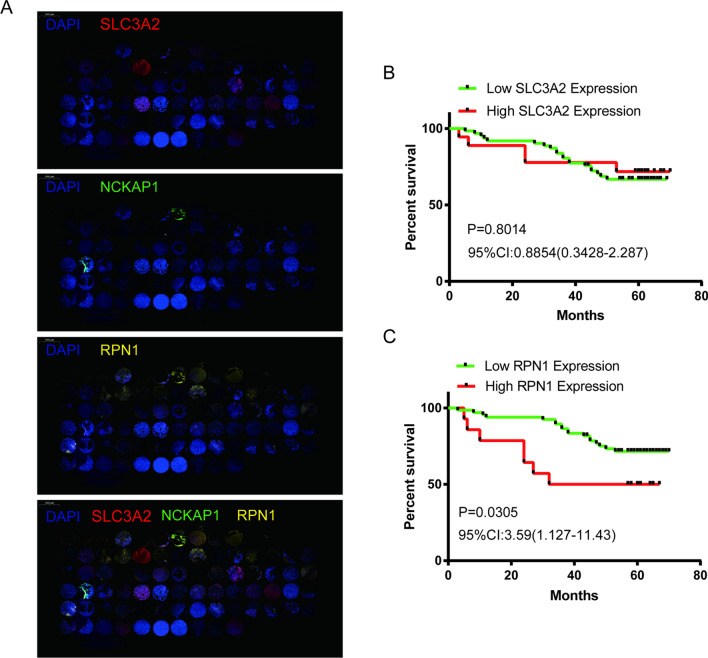
Through differential expression analysis of normal and tumor samples across 34 cancer types, we observed significant upregulation of SLC7A11, SLC3A2, RPN1, and NCKAP1 in 30, 23, 30, and 13 cancer types, respectively (Fig. 1). To validate their high expression in prognostic values across tumors, we performed Cox risk regression proportional analysis. Specifically, elevated expression of SLC7A11 and SLC3A2 was associated with poor prognosis in 14 and 23 cancer types, respectively, while elevated expression of RPN1 was associated with poor prognosis in 10 cancer types and elevated expression of NCKAP1 was associated with poor prognosis in 5 cancer types (Fig. 2). The top two tumor types showing the strongest correlation between high expression of each gene and poor prognosis were KIPAN [pan-kidney cohort (KICH + KIRC + KIRP)] (HR = 1.49) and adrenocortical carcinoma (ACC) (HR = 3.67) for SLC7A11, brain lower grade glioma (LGG) (HR = 2.32) and TCGA-LAML (acute myeloid leukemia) (HR = 1.85) for SLC3A2, glioma (GBMLGG) (HR = 8.32) and LGG (HR = 4.49) for RPN1, and lung adenocarcinoma (LUAD) (HR = 1.36) and breast invasive carcinoma (BRCA) (HR = 1.33) for NCKAP1. Further reinforcement of these findings was obtained through Kaplan–Meier survival curve analysis for these eight tumor types (Fig. 3).To further investigate the expression levels of these genes in tumor tissues, we collected 80 glioblastoma multiforme (GBM) samples and conducted immunofluorescence staining for the corresponding proteins. The immunofluorescence (IF) results demonstrated extensive expression of SLC3A2 and RPN1 in GBM tissues (Fig. 4A). Furthermore, elevated RPN1 expression was identified as a significant risk factor for GBM patients (Fig. 4C), while SLC3A2 expression was not associated with survival (Fig. 4B).
Fig. 1.
Differential expression of disulfidptosis-related genes (DRGs) in tumor versus normal samples across 34 tumor types. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, − no significance
Fig. 2.
Overall survival analysis of disulfidptosis-related genes (DRGs) across cancers using the univariate Cox regression model. A hazard ratio (HR) greater than 1 indicates that high DRGs expression is associated with an increased risk of adverse survival outcomes for patients. 95%CI, 95% confidence interval. A, SLC7A11; B, SLC3A2; C, RPN1; D, NCKAP1
Fig. 3.
Kaplan–Meier (KM) survival of disulfidptosis-related genes (DRGs) in the two tumor types with the poorest prognosis. HR hazard ratio; 95% CI 95% confidence interval. L, low expression group, H high expression group
Fig. 4.
Kaplan–Meier (KM) survival analysis of DRGs in glioblastoma multiforme (GBM) patients. HR hazard ratio; 95% CI, 95% confidence interval. A. Immunohistochemical staining of SLC3A2, RPN1, and NCKAP1 on tissue microarrays comprising 80 clinical GBM samples. B. Kaplan–Meier survival curve stratifying GBM patients based on high versus low SLC3A2 expression. C. Kaplan–Meier survival curve stratifying GBM patients based on high versus low RPN1 expression
Correlation of tumor stemness scores and DRGs expression
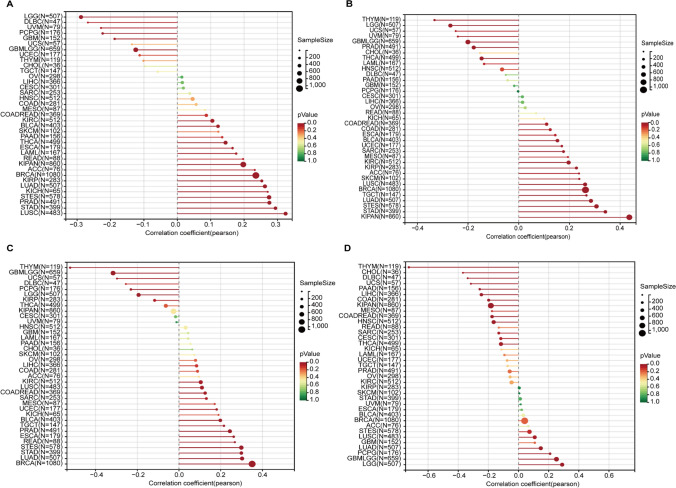
The DNAss and RNAss scores serve as quantitative measures of tumor stemness, reflecting the degree of stemness in different tumor samples. By integrating DNA and RNA-based features with DRGs expression data, we obtained tumor stemness features associated with DRGs. The DNAss results revealed significant positive correlations of SLC7A11, SLC3A2, RPN1, and NCKAP1 with 5, 10, 9, and 8 tumor types, respectively. The most correlated tumor types for these four genes were thymoma (THYM) (R = 0.32, P < 0.001), uveal melanoma (UVM) (R = 0.32, P = 0.004), GBMLGG (R = 0.47, P < 0.001), and THYM (R = 0.47, P < 0.001) (Fig. 5). Moreover, RNAss indicated a broader positive correlation of DRGs with various tumor types, particularly for SLC7A11, SLC3A2, and RPN1, which were positively correlated with 15, 16, and 14 tumor types, respectively. The tumor types most positively correlated with these three genes were lung squamous cell carcinoma (LUSC) (R = 0.32, P < 0.001), KIPAN (R = 0.44, P < 0.001), and BRCA (R = 0.35, P < 0.001) (Fig. 6).
Fig. 5.
Correlation between DNA stemness score (DNAss) and disulfidptosis-related genes (DRGs). A, SLC7A11; B, SLC3A2; C, RPN1; D, NCKAP1
Fig. 6.
Correlation between RNA stemness score (RNAss) and disulfidptosis-related genes (DRGs). A, SLC7A11; B, SLC3A2; C, RPN1; D, NCKAP1
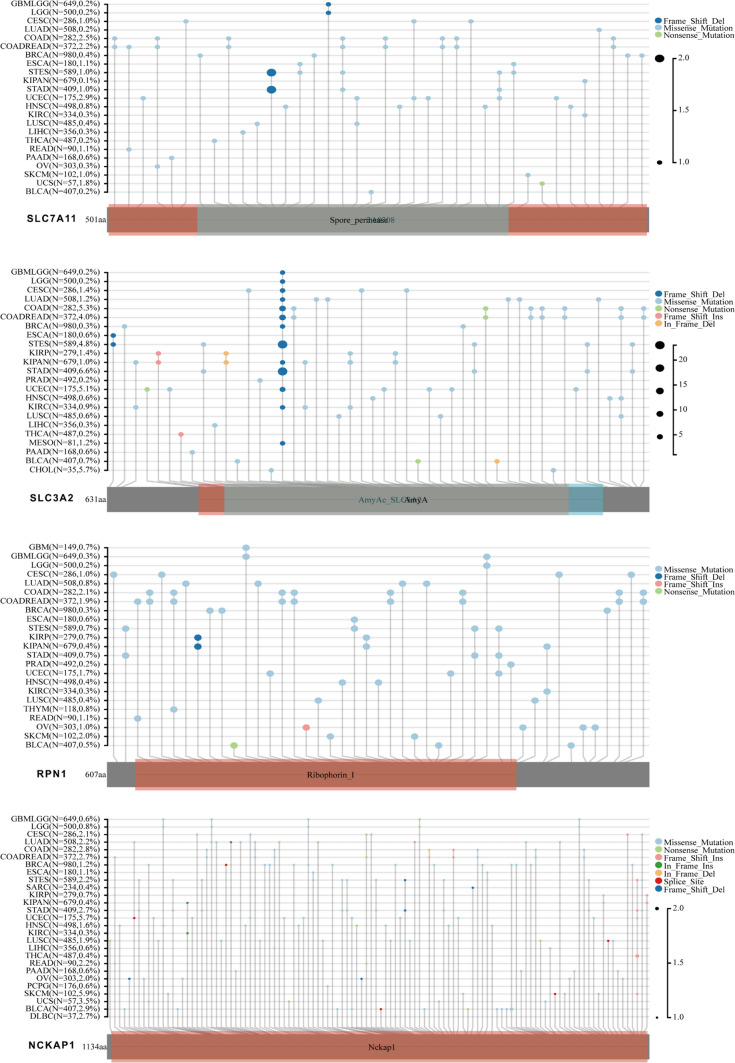
Mutational landscape of DRGs across cancers
The mutational landscape of individual genes across multiple cancer types was analyzed to elucidate their genetic alterations in pan-cancer. Through the examination of genomic data from diverse tumor samples, specific mutations occurring in these genes were identified, and their prevalence and patterns in different cancer types were evaluated. DRGs, particularly SLC3A2 and NCKAP1, exhibited a broad spectrum of mutations across various cancer types. Missense mutations spanning the entire coding sequence were the most prevalent mutation type observed across cancers. Specifically, mutations in NCKAP1 were more frequently observed across different cancers, with the highest frequency of 5.9% observed in skin cutaneous melanoma (SKCM). These findings underscore the diverse mutational landscape of DRGs and suggest their potential involvement in multiple cancer types (Fig. 7).
Fig. 7.
Mutational landscape of disulfidptosis-related genes (DRGs) across cancers
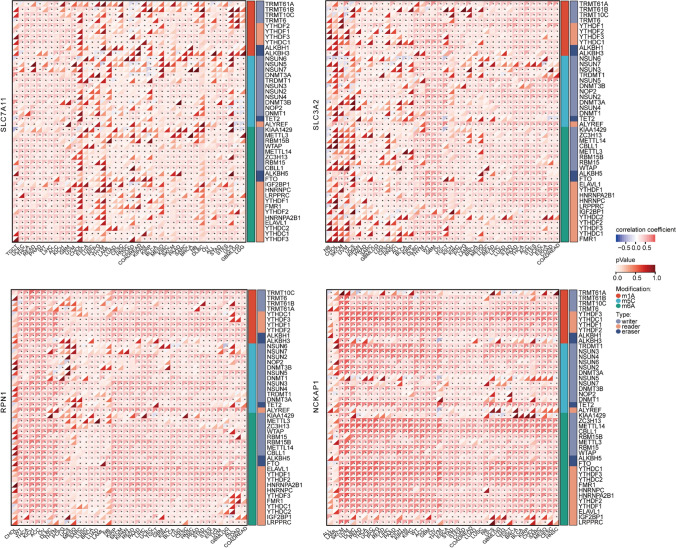
Correlation of DRGs expression with the levels of RNA modifications
Dysregulation of RNA methylation modification pathways, particularly N6-methyladenosine (m6A), is an important aspect of cancer biology [26]. Understanding the specific alterations in the expression levels of modifiers provides insights into the underlying mechanisms driving cancer development and offers potential avenues for targeted therapies. Here, we characterized the association between the expression of DRGs and the levels of 44 RNA modifications, specifically focusing on m1A, m5C, and m6A modifications. Our results revealed a widespread positive correlation between DRGs, particularly RPN1, and NCKAP1, and the three types of RNA modifications across cancers (Fig. 8). Of note, we found a significantly positive correlation between m6A modification and DRGs in multiple tumor types, particularly in the case of NCKAP1. The m6A and m1A reader modifiers showed a most significant positive correlation across cancers, such as SKCM, UVM, and THYM (Fig. 8).
Fig. 8.
Correlation between disulfidptosis-related genes (DRGs) expression and the levels of RNA modifications (m1A, m5C, and m6A) across cancers
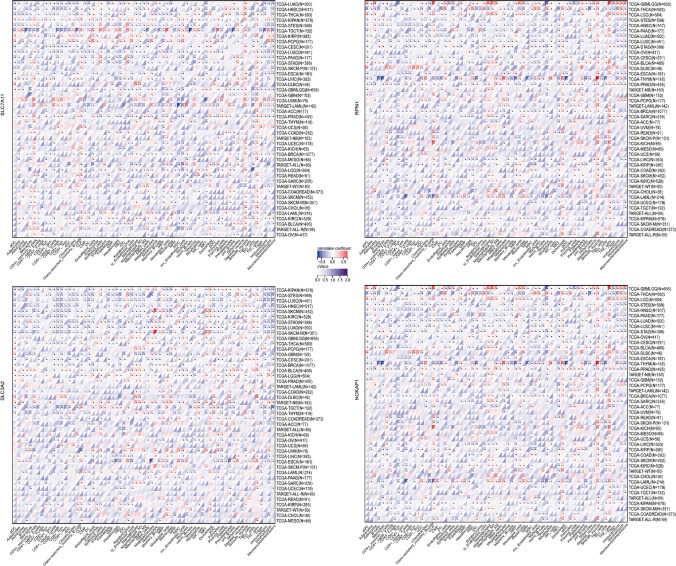
DRGs-associated tumor immune cell infiltration
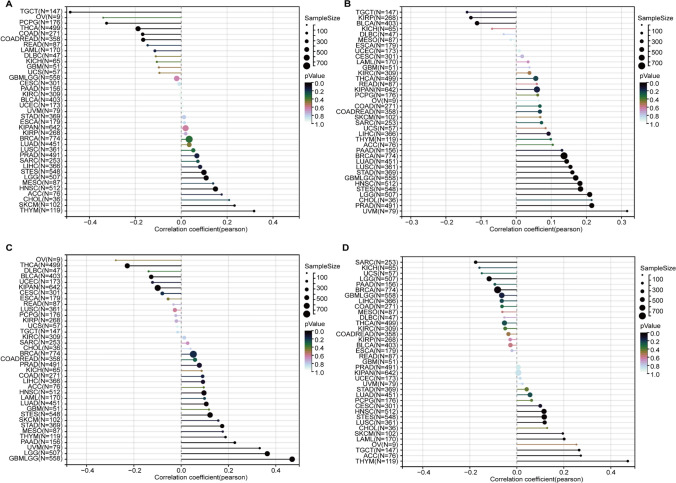
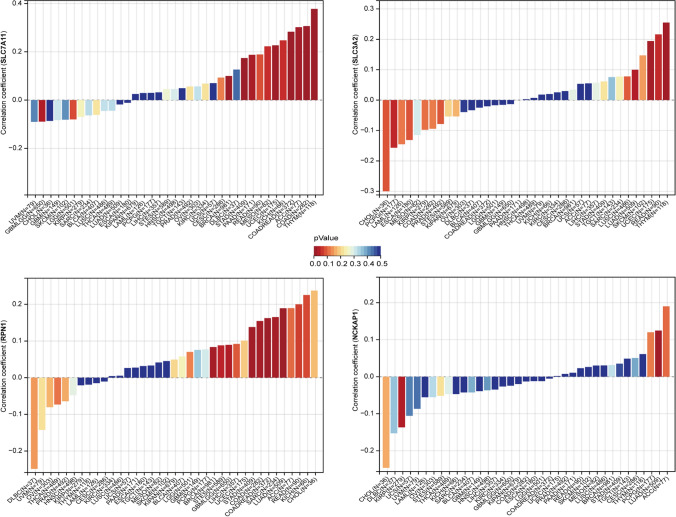
The infiltration of immune cells in tumors has a profound impact on the immune microenvironment and affects the prognosis of cancer. We assessed immune cell infiltration using the xCell method and calculated the correlation between gene expression and immune cell scores across cancers. Our findings indicate significant associations between gene expression and immune cell infiltration in multiple tumor types. The expression of SLC7A11, RPN1, and NCKAP1 showed a significant positive correlation with T-helper type 2 (Th2) cells infiltration across cancers (Fig. 9). Additionally, for GBMLGG, various immune cell infiltrates such as class-switched memory B cells and macrophages exhibited a positive correlation with the expression of RPN1 and NCKAP1 (Fig. 9).
Fig. 9.
Xcell-based analysis of immune cell infiltration across cancers
Correlation between the expression of DRGs and TMB
Tumor genomic heterogeneity influences tumor malignancy, marker expression, immunotherapy response, and prognosis. TMB, a measure of genomic instability, is associated with these aspects. High TMB indicates increased immune recognition of tumor cells, impacting tumor growth and immunotherapy response [27]. Through our analysis of the correlation between the expression of DRGs and TMB, we observed significant positive associations between SLC7A11 and TMB in 10 tumor types, including colon adenocarcinoma (COAD), COAD/rectum adenocarcinoma esophageal carcinoma (COADREAD), BRCA, stomach adenocarcinoma (STAD), uterine corpus endometrial carcinoma (UCEC), THYM, mesothelioma (MESO), pancreatic adenocarcinoma (PAAD), ACC, and kidney chromophobe (KICH) (Fig. 10). SLC3A2 exhibited a significant positive correlation with TMB in 4 tumor types [LUAD, UCEC, THYM, rectum adenocarcinoma (READ)] (Fig. 10). Furthermore, RPN1 demonstrated significant positive associations with TMB in 8 tumor types [GBMLGG, LUAD, COAD, COADREAD, BRCA, stomach and esophageal carcinoma (STES), sarcoma (SARC), STAD], while NCKAP1 displayed a positive correlation with TMB in only one tumor type LUAD (Fig. 10).
Fig. 10.
Correlation of disulfidptosis-related genes (DRGs) expression with tumor genomic heterogeneity (tumor mutation burden)
Biological function for DRGs-interacting proteins
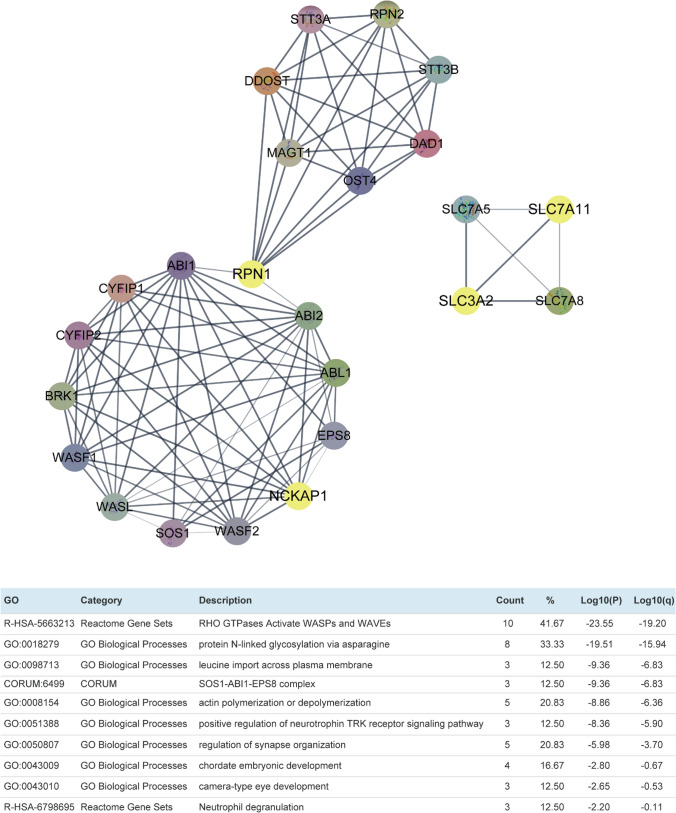
To further understand the role of DRGs in regulating tumors, we constructed a protein–protein interaction network for DRGs and performed pathway and process enrichment analysis using the online tool Metascape. The top 10 clusters identified from this analysis revealed that the proteins associated with DRGs are primarily involved in the RHO GTPases Activate WASPs and WAVEs pathway (Log10(P) = − 23.55), indicating their significant functional relevance in cancer-related signaling pathways (Fig. 11).
Fig. 11.
Protein–protein interaction network of disulfidptosis-related genes (DRG) and its pathway and process enrichment analysis. Top 10 clusters with their representative enriched terms (one per cluster). The ‘‘%’’ represents the percentage of input genes associated with the given ontology term
Discussion
Cell death is a fundamental biological process that regulates tissue homeostasis, development, and immunity. Dysregulation of cell death can lead to various diseases, such as cancer, neurodegeneration, and inflammation. Several types of regulated cell death have been identified, such as apoptosis, necroptosis, pyroptosis, ferroptosis, cuproptosis, and autophagy-dependent cell death [28]. Each type of cell death is characterized by distinct morphological, biochemical, and molecular features, and is mediated by specific signaling pathways and molecular mechanisms. Disulfidoptosis, a novel form of cell death triggered by disulfide stress, differs from other types of cell death, such as apoptosis and ferroptosis [6, 8]. Its relevance in cancer treatment is of interest, particularly considering the high expression of SLC7A11 in certain cancer types and their sensitivity to glucose deprivation [29]. Experts suggest that disulfidoptosis may serve as a potential target for metabolic cancer therapies [7]. In this study, we conducted a comprehensive analysis of the top four DRGs: SLC7A11, SLC3A2, RPN1, and NCKAP1 across cancers, aiming to investigate their functional significance and potential implications in tumorigenesis.
We observed that DRGs are significantly upregulated across various cancer types compared to normal tissues, suggesting their potential role as oncogenes. High expression levels of DRGs were associated with poor prognosis in multiple cancers, highlighting their potential as prognostic markers. Consequently, DRGs could serve as promising therapeutic targets for cancer treatment. For instance, Fang et al. demonstrated that sorafenib mitigates liver injury and extracellular matrix (ECM) accumulation in fibrotic liver while reducing SLC7A11 and GPX4 protein levels [30]. Similarly, metformin reduces the stability of SLC7A11 by inhibiting its UFMylation, which contributes to the suppression of breast cancer cell growth [31]. We also investigated the relationship between DRGs and tumor stemness scores, which reflect the presence of cancer stem cell characteristics in tumors. Our findings demonstrated a positive correlation between DRG expression and tumor stemness scores, suggesting their involvement in maintaining stemness features in multiple tumor types. These results align with previous studies highlighting the contribution of DRGs to cancer stem cell maintenance and aggressiveness [32–34].
Furthermore, we analyzed the mutational landscape of DRGs in different cancer types and identified a diverse range of genetic alterations, with missense mutations being the most prevalent. Among the DRGs examined, NCKAP1 exhibited a high frequency of mutations across various cancers, highlighting its potential role in tumorigenesis. These observations indicate that genetic alterations in DRGs contribute to the molecular heterogeneity of cancer and may impact disease progression and response to treatment. Additionally, we investigated the association between DRG expression and m6A RNA modifications, which regulate mRNA stability and translation [35]. We found a positive correlation between DRGs and m6A modifications, suggesting interactions between these genes and the m6A machinery that may impact mRNA processing and function. Gu etc. Reported that METTL3-mediated m6A modification of SLC7A11 enhances its stability and resistance to ferroptosis in hepatoblastoma (HB) [36]. In lung cancer, YTHDC2 disrupts the stability of SLC7A11 mRNA in an m6A-dependent manner by selectively binding to the m6A-modified SLC7A11 mRNA and facilitating its degradation [37]. Clinically, a considerable fraction of LUAD adenocarcinoma cases show concurrent downregulation of YTHDC2 and upregulation of SLC7A11 [38].Aboved evidences shed light on the complex post-transcriptional regulatory mechanisms involving DRGs in cancer.
Previous studies have partially elucidated the roles of specific DRGs in immune cell infiltration and genomic heterogeneity in liver hepatocellular carcinoma (LIHC)[39], bladder urothelial carcinoma (BLCA) [40], esophageal carcinoma (ESCA) [41] and kidney renal clear cell carcinoma (KIRC) [12]. Our results of immune cell infiltration demonstrated a positive correlation between DRG expression and the infiltration of Th2 cells, class-switched memory B cells, and macrophages in several cancer types, suggesting their role in modulating anti-tumor immune responses and interactions with immune cells within the tumor microenvironment. High TMB can enhance immune recognition of tumor cells by increasing the presentation of neoantigens on their surface [42]. The positive associations observed between DRGs and TMB suggest their potential involvement in DNA damage and repair pathways, further supporting their roles in tumor progression and response to immunotherapy.
Finally, we performed pathway and process enrichment analysis of proteins interacting with DRGs, revealing significant enrichment in the RHO GTPases Activate WASPs and WAVEs pathway. This pathway is known to play crucial roles in cytoskeletal dynamics, cell migration, and invasion [43], indicating the potential involvement of DRGs in tumor metastasis and progression. These functional clusters provide a foundation for future investigations into the specific mechanisms through which DRGs contribute to tumorigenesis.
Limitations inherent in this investigation include the utilization of publicly accessible datasets, which may exhibit variations in sample size, data quality, and experimental methodologies. Furthermore, the observational nature of our analyses precludes definitive establishment of causality. While we conducted a comprehensive examination of the top four DRGs across various cancer types, it's plausible that other pertinent genes were not included in our study. Additionally, our analysis focused solely on mRNA expression levels, neglecting potential post-transcriptional or post-translational regulatory processes that might influence protein function. Lastly, our findings are correlative and necessitate further experimental validation to unravel the underlying molecular mechanisms and functional implications of the observed associations.
In summary, our study highlights the aberrant expression of DRGs in multiple cancer types and their associations with poor prognosis, tumor stemness, mutational landscape, RNA modifications, immune cell infiltration, tumor mutation burden, and functional signaling pathways. These findings provide valuable insights into the diverse roles of DRGs in cancer biology and lay the groundwork for further mechanistic studies and the development of targeted therapeutic approaches.
Acknowledgements
The authors gratefully acknowledge the providers of the public database sites and software included in the paper.
Abbreviations
- ACC
Adrenocortical carcinoma
- BLCA
Bladder urothelial carcinoma
- BRCA
Breast invasive carcinoma
- CESC
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- CHOL
Cholangiocarcinoma
- COAD
Colon adenocarcinoma
- COADREAD
Colon adenocarcinoma/rectum adenocarcinoma esophageal carcinoma
- DLBC
Lymphoid neoplasm diffuse large B-cell lymphoma
- ESCA
Esophageal carcinoma
- FPPP
FFPE Pilot Phase II
- GBM
Glioblastoma multiforme
- GBMLGG
Glioma
- HNSC
Head and Neck squamous cell carcinoma
- KICH
Kidney chromophobe
- KIRC
Kidney renal clear cell carcinoma
- KIRP
Kidney renal papillary cell carcinoma
- KIPAN
Pan-kidney cohort (KICH + KIRC + KIRP)
- LAML
Acute Myeloid Leukemia
- LGG
Brain lower grade glioma
- LIHC
Liver hepatocellular carcinoma
- LUAD
Lung adenocarcinoma
- LUSC
Lung squamous cell carcinoma
- MESO
Mesothelioma
- OV
Ovarian serous cystadenocarcinoma
- PAAD
Pancreatic adenocarcinoma
- PCPG
Pheochromocytoma and paraganglioma
- PRAD
Prostate adenocarcinoma
- READ
Rectum adenocarcinoma
- SARC
Sarcoma
- STAD
Stomach adenocarcinoma
- SKCM
Skin cutaneous melanoma
- STES
Stomach and esophageal carcinoma
- TGCT
Testicular germ cell tumors
- THCA
Thyroid carcinoma
- THYM
Thymoma
- UCEC
Uterine corpus endometrial carcinoma
- UCS
Uterine Carcinosarcoma
- UVM
Uveal melanoma
- OS
Osteosarcoma
- ALL
Acute lymphoblastic leukemia
- NB
Neuroblastoma
- WT
Wilms tumor
Author contributions
ZZ: Conceptualization, Methodology, Data curation, Resources, Software, Writing-original draft. YS: Conceptualization, Formal Analysis, Project administration, Funding acquisition, Writing-review and editing.
Funding
This work was supported by the Natural Science Foundation of Ningbo [Grant Number 2021J296] and the Natural Science Foundation of the Zhejiang Province [Grant Number LQQ20H160001].
Availability of data and materials
All data supporting the findings of this study are included in this article. The datasets analysed during the current study are available in the following online repositories: https://xenabrowser.net/https://portal.gdc.cancer.gov/. https://cn.string-db.org/. https://metascape.org/gp/index.html#/main/step1
Declarations
Ethics approval and consent to participate
The study using the tissue microarray was approved by the Life Sciences Ethics Committee of Changsha Yaxiang Biotechnology Co., LTD.
Consent for publication
All authors give consent for publication of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zequn Zheng, Email: 13414057384@163.com.
Yongfei Song, Email: lhlsongyongfei@nbu.edu.cn.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12(1):31–46. [DOI] [PubMed] [Google Scholar]
- 3.Strasser A, Vaux DL. Cell death in the origin and treatment of cancer. Mol Cell. 2020;78(6):1045–54. [DOI] [PubMed] [Google Scholar]
- 4.Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji AF, Clish CB, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156(1–2):317–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang D, Chen X, Kroemer G. Cuproptosis: a copper-triggered modality of mitochondrial cell death. Cell Res. 2022;32(5):417–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X, Nie L, Zhang Y, Yan Y, Wang C, Colic M, Olszewski K, Horbath A, Chen X, Lei G, et al. Actin cytoskeleton vulnerability to disulfide stress mediates disulfidptosis. Nat Cell Biol. 2023;25(3):404–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng P, Zhou C, Ding Y, Duan S. Disulfidptosis: a new target for metabolic cancer therapy. J Experiment Clin Cancer Res CR. 2023;42(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng T, Liu Q, Xing F, Zeng C, Wang W. Disulfidptosis: a new form of programmed cell death. J Experiment Clin Cancer Res CR. 2023;42(1):137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan Y, Teng H, Hang Q, Kondiparthi L, Lei G, Horbath A, Liu X, Mao C, Wu S, Zhuang L, et al. SLC7A11 expression level dictates differential responses to oxidative stress in cancer cells. Nat Commun. 2023;14(1):3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi C, Ma J, Sun J, Wu X, Ding J. The role of molecular subtypes and immune infiltration characteristics based on disulfidptosis-associated genes in lung adenocarcinoma. Aging. 2023;15(11):5075–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen H, Yang W, Li Y, Ma L, Ji Z. Leveraging a disulfidptosis-based signature to improve the survival and drug sensitivity of bladder cancer patients. Front Immunol. 2023;14:1198878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L, Liu J, Li S, Liu X, Zheng F, Xu S, Fu B, Xiong J. Based on disulfidptosis, revealing the prognostic and immunological characteristics of renal cell carcinoma with tumor thrombus of vena cava and identifying potential therapeutic target AJAP1. J Cancer Res Clin Oncol. 2023. 10.1007/s00432-023-04877-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng Z, Zhao Q, Ding Y, Xu Y, Sun X, Chen Q, Zhang Y, Miao J, Zhu J. Identification a unique disulfidptosis classification regarding prognosis and immune landscapes in thyroid carcinoma and providing therapeutic strategies. J Cancer Res Clin Oncol. 2023;24(14):2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji FH, Fu XH, Li GQ, He Q, Qiu XG. FTO prevents thyroid cancer progression by SLC7A11 m6A methylation in a ferroptosis-dependent manner. Front Endocrinol. 2022;13: 857765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu B, Lv Y, Hu W, Huang Y, Ying X, Chen C, Zhang H, Ji W. m(6)A modification mediates SLC3A2/SLC7A5 translation in 3-methylcholanthrene-induced uroepithelial transformation. Cell Biol Toxicol. 2024;40(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blum A, Wang P, Zenklusen JC. SnapShot: TCGA-analyzed tumors. Cell. 2018;173(2):530. [DOI] [PubMed] [Google Scholar]
- 17.Li W, Shi M, Zhou P, Liu Y, Liu X, Xiao X, Zuo S, Bai Y, Sun K. Extramedullary infiltration in pediatric acute myeloid leukemia: results from the therapeutically applicable research to generate effective treatments (TARGET) initiative. Pediatr Blood Cancer. 2024;71(7): e31014. [DOI] [PubMed] [Google Scholar]
- 18.Carithers LJ, Moore HM. The genotype-tissue expression (GTEx) project. Biopreserv Biobank. 2015;13(5):307–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, Weinstein JN, Kamińska B, Huelsken J, Omberg L, Gevaert O, et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. 2018;173(2):338-354.e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC20 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng D, Ye Z, Shen R, Yu G, Wu J, Xiong Y, Zhou R, Qiu W, Huang N, Sun L, et al. IOBR: multi-omics immuno-oncology biological research to decode tumor microenvironment and signatures. Front Immunol. 2021;12: 687975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friel DC, Jeske DR. Wilcoxon rank-sum tests to detect one-sided mixture alternatives in group sequential clinical trials. Stat Methods Med Res. 2023;32(9):1711–27. [DOI] [PubMed] [Google Scholar]
- 26.Zheng Z, Song Y. Integrated analysis of the voltage-gated potassium channel-associated gene KCNH2 across cancers. BMC Bioinform. 2023;24(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sha D, Jin Z, Budczies J, Kluck K, Stenzinger A, Sinicrope FA. Tumor mutational burden as a predictive biomarker in solid tumors. Cancer Discov. 2020;10(12):1808–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tong X, Tang R, Xiao M, Xu J, Wang W, Zhang B, Liu J, Yu X, Shi S. Targeting cell death pathways for cancer therapy: recent developments in necroptosis, pyroptosis, ferroptosis, and cuproptosis research. J Hematol Oncol. 2022;15(1):174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang X, Chen W, Liu H, Liu N, Chen D, Tian D, Wang J. Research progress on SLC7A11 in the regulation of cystine/cysteine metabolism in tumors. Oncol Lett. 2022;23(2):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan S, Wei C, Liu G, Zhang L, Li J, Li L, Cai S, Fang L. Sorafenib attenuates liver fibrosis by triggering hepatic stellate cell ferroptosis via HIF-1alpha/SLC7A11 pathway. Cell Prolif. 2022;55(1): e13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, Zhou Y, Xie S, Wang J, Li Z, Chen L, Mao M, Chen C, Huang A, Chen Y, et al. Metformin induces ferroptosis by inhibiting UFMylation of SLC7A11 in breast cancer. J Exp Clin Cancer Res. 2021;40(1):206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu M, Zhang X, Zhang W, Chiou YS, Qian W, Liu X, Zhang M, Yan H, Li S, Li T, et al. Cancer stem cell regulated phenotypic plasticity protects metastasized cancer cells from ferroptosis. Nat Commun. 2022;13(1):1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polewski MD, Reveron-Thornton RF, Cherryholmes GA, Marinov GK, Aboody KS. SLC7A11 Overexpression in glioblastoma is associated with increased cancer stem cell-like properties. Stem Cells Dev. 2017;26(17):1236–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linge A, Lohaus F, Löck S, Nowak A, Gudziol V, Valentini C, von Neubeck C, Jütz M, Tinhofer I, Budach V, et al. HPV status, cancer stem cell marker expression, hypoxia gene signatures and tumour volume identify good prognosis subgroups in patients with HNSCC after primary radiochemotherapy: a multicentre retrospective study of the german cancer consortium radiation oncology group (DKTK-ROG). Radiother Oncol J Eur Soc Ther Radiol Oncol. 2016;121(3):364–73. [DOI] [PubMed] [Google Scholar]
- 35.Fish L, Navickas A, Culbertson B, Xu Y, Nguyen HCB, Zhang S, Hochman M, Okimoto R, Dill BD, Molina H, et al. Nuclear TARBP2 drives oncogenic dysregulation of RNA splicing and decay. Mol Cell. 2019;75(5):967-981.e969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu L, He J, Sun G, Huang N, Bian Z, Xu C, Zhang Y, Cui Z, Xu W, Sun F, et al. The N6-methyladenosine modification enhances ferroptosis resistance through inhibiting SLC7A11 mRNA deadenylation in hepatoblastoma. Clin Transl Med. 2022;12(5): e778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma L, Chen T, Zhang X, Miao Y, Tian X, Yu K, Xu X, Niu Y, Guo S, Zhang C, et al. The m(6)A reader YTHDC2 inhibits lung adenocarcinoma tumorigenesis by suppressing SLC7A11-dependent antioxidant function. Redox Biol. 2021;38: 101801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma L, Zhang X, Yu K, Xu X, Chen T, Shi Y, Wang Y, Qiu S, Guo S, Cui J, et al. Targeting SLC3A2 subunit of system X(C)(-) is essential for m(6)A reader YTHDC2 to be an endogenous ferroptosis inducer in lung adenocarcinoma. Free Radic Biol Med. 2021;168:25–43. [DOI] [PubMed] [Google Scholar]
- 39.Wang T, Guo K, Zhang D, Wang H, Yin J, Cui H, Wu W. Disulfidptosis classification of hepatocellular carcinoma reveals correlation with clinical prognosis and immune profile. Int Immunopharmacol. 2023;120: 110368. [DOI] [PubMed] [Google Scholar]
- 40.Zhao S, Wang L, Ding W, Ye B, Cheng C, Shao J, Liu J, Zhou H. Crosstalk of disulfidptosis-related subtypes, establishment of a prognostic signature and immune infiltration characteristics in bladder cancer based on a machine learning survival framework. Front Endocrinol. 2023;14:1180404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu F, Yuan D, Liu X, Zhuo S, Liu X, Sheng H, Sha M, Ye J, Yu H. A demonstration based on multi-omics transcriptome sequencing data revealed disulfidptosis heterogeneity within the tumor microenvironment of esophageal squamous cell carcinoma. Disc Oncol. 2023;14(1):96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miljanic M, Capasso A, Triplett TA, Eckhardt SG, Aung KL. Immune checkpoint blockade in gastrointestinal cancers: the current status and emerging paradigms. J Immunother Precision Oncol. 2020;3(1):3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moffitt L, Karimnia N, Stephens A, Bilandzic M. Therapeutic targeting of collective invasion in ovarian cancer. Int J Mol Sci. 2019. 10.3390/ijms20061466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data supporting the findings of this study are included in this article. The datasets analysed during the current study are available in the following online repositories: https://xenabrowser.net/https://portal.gdc.cancer.gov/. https://cn.string-db.org/. https://metascape.org/gp/index.html#/main/step1