Abstract
SWI-SNF complexes have been implicated in transcriptional regulation by chromatin remodeling. We have identified an interaction between two components of the mammalian SWI-SNF complex and cyclin E, an essential cell cycle regulatory protein required for G1/S transition. BRG1 and BAF155, mammalian homologs of yeast SWI2 and SWI3, respectively, are found in cyclin E complexes and are phosphorylated by cyclin E-associated kinase activity. In this report, we show that overexpression of BRG1 causes growth arrest and induction of senescence-associated β-galactosidase activity, which can be overcome by cyclin E. Our results suggest that cyclin E may modulate the activity of the SWI-SNF apparatus to maintain the chromatin in a transcriptionally permissive state.
Progression through the cell cycle is a tightly controlled process requiring many critical regulatory proteins (reviewed in reference 47). The cyclin-dependent kinases (cdks) and their regulatory cyclin subunits promote passage through each phase of the cell division cycle. The activation of cyclin-cdk complexes is strictly regulated both at the level of protein synthesis and destruction and by posttranslational modifications to dictate precisely when in the cell cycle each complex becomes active (28, 36).
Cyclin E is synthesized during the G1 phase of the cell cycle and binds cdk2 to become maximally active at the G1/S boundary (10, 26). Cyclin E-cdk2 complexes have been shown to play an essential and rate-limiting role in the transition between G1 and S phase (40, 44, 52, 53). The manner in which cyclin E-cdk2 promotes S-phase entry remains poorly defined, since few downstream effectors of cyclin E-cdk2 are known. One potential substrate is the protein product of the retinoblastoma tumor suppressor gene, pRb, which is also phosphorylated by the D-type cyclin-cdk complexes (9, 13, 24). However, unlike cyclin D, cyclin E remains essential in the absence of pRb, illustrating a fundamental difference between these two complexes and strongly suggesting that other key rate-limiting substrates exist for cyclin E-cdk2 (1, 33). Other targets identified more recently include SAP155, a component of the pre-mRNA splicing apparatus (46), and NPAT (58). The role of such molecules in modulating cell cycle progression has yet to be established.
SWI-SNF complexes are evolutionarily conserved and have been implicated in transcriptional regulation through remodeling of chromatin structure (reviewed in references 5 and 42). Components of the SWI-SNF apparatus are believed to bind to chromatin and relieve nucleosome-mediated repression of transcription, thus providing access to transcriptional activators (7, 21, 27, 31, 42, 45). While SWI-SNF complexes are nonessential in yeast, a second related complex, RSC, is required for yeast cell growth (3, 4, 32). In mammalian cells, SWI-SNF complexes have been implicated in hormone receptor activation and growth control (6, 11, 25, 39, 49). The ability of SWI-SNF complexes to regulate cell growth is believed to be mediated through the interaction of the human homologs of the SWI2-SNF2 protein (BRG1 and hBRM) with pRb (11, 49, 51). The mechanism by which pRb modulates BRG1 function is not known. Other modes of regulation impinging on SWI-SNF functions probably exist, but they have not been well characterized. SWI-SNF complexes have been identified with variable subunit compositions (57), and the phosphorylation states of some components of SWI-SNF complexes are altered in a cell cycle-dependent fashion (37).
To further elucidate the role of cyclin E-cdk2 in growth control and in cell cycle transitions, we looked for novel proteins that associate with cyclin E within the cell. By immunoprecipitation analysis of various cell lines using antibodies against cyclin E, we identified the presence of two components of the SWI-SNF apparatus, BAF155 and BRG1. This interaction would appear to be functionally significant, because cyclin E can abrogate the ability of BRG1 to induce growth arrest.
MATERIALS AND METHODS
Cell lines.
SW13, C33A, 293, and SAOS-2 cells were grown as monolayers, and ML-1 cells were grown as suspension cultures in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS) and nonessential amino acids. All cell lines were obtained from the American Type Culture Collection.
Antibodies.
Antibodies to cyclins D1, D2, and D3 and cdc2, cdk2, and cdk6 were raised against C-terminal peptides. Antibodies against p27 and hBRM were purchased from Transduction Labs. Antibodies against p107, p130, E2F4, and cdc25A were obtained from Santa Cruz Biotech. Antibodies to SAP155 are described elsewhere (46). HE antibodies were raised against full-length cyclin E: the HE172 epitope comprises amino acids (aa) 386 to 396, and the HE67 epitope comprises aa 366 to 381. Polyclonal antibodies against BAF155 were raised against a glutathione S-transferase (GST) fusion protein carrying aa 751 to 1105. The same antigen was used to generate monoclonal antibodies to BAF155 in BALB/c mice according to standard protocols (17), Ini1 antibodies were raised against C-terminal peptide aa residues 369 to 386. BRG1 antibodies were raised against an N-terminal GST fusion protein (residues 1 to 700). The anti-BRG1 antibody, J1, used for immunofluorescence was a kind gift of G. Crabtree (25).
Immunoprecipitations.
Cells were lysed in lysis buffer containing 5 mM HEPES (pH 7.0), 250 mM NaCl, 0.1% Nonidet P-40 (NP-40) plus protease inhibitors (Complete EDTA-free tablets, protease inhibitor cocktail; Boehringer Mannheim). Immunoprecipitations and kinase assays were performed according to standard protocols (17). For metabolic labeling, cells were starved for methionine by incubation in methionine-free DMEM (Gibco) for 30 min and subsequently labeled for 4 h with 100 μCi of [35S]methionine per ml. Two washes with ice-cold phosphate-buffered saline (PBS) were performed before cell lysis.
Covalent coupling of antibodies to protein A-Sepharose (Pharmacia) was performed with dimethyl pimelimidate (Pierce) according to the manufacturer’s instructions. The preparation of large-scale immunoprecipitations for the purification and sequencing of polypeptides is described elsewhere (46). All peptide sequencing was performed by W. Lane, Harvard Microchemistry Laboratory. Preclearing experiments were performed by three sequential rounds of antibody incubation and capture on protein A-Sepharose beads. Lysates were then spun through filters (Costar) to remove residual beads prior to immunoprecipitation.
Immunoblots.
For immunoblotting, samples were transferred to an Immobilon polyvinylidene difluoride membrane (Millipore) by Western blotting at 60 V for 6 h at 4°C. After transfer was complete, the membranes were blocked in TNT (10 mM Tris, 150 mM NaCl, 0.2% [vol/vol] Tween 20) containing 5% milk for 30 min at room temperature. The filters were probed with primary antibodies diluted to 2 μg/ml in 5% milk–TNT and incubated on a shaker for 1 h at room temperature. The filters were washed in TNT at room temperature four times for 15 min each time and probed with horseradish peroxidase-conjugated anti-rabbit or anti-mouse antibody (Amersham) diluted 1:5,000 in 5% milk–TNT and incubated on a shaker at room temperature for 1 h. The filters were washed as before and detected by enhanced chemiluminescence (ECL; Amersham).
Transfections.
For transient transfections, SW13 cells were transfected with 6 μg of total plasmid DNA, including 1 μg of pHook-1 plasmid (Invitrogen), 2 μg of pBJ5.BRG or pBJ5.BRG K798R (25), and 3 μg of cytomegalovirus (CMV) cyclin E (29) as indicated. pBSK (Stratagene) was used as the carrier. Fifteen dishes (100 by 20 mm) were transfected for each experiment with 25 μl of Lipofectamine reagent (Life Technologies, Inc.). The cells were incubated for 3 h at 37°C in a CO2 incubator. After the incubation, 5 ml of DMEM containing 20% serum and antibiotics was added. The medium was replaced with fresh complete medium 18 to 24 h after transfection. Cells were harvested with the Capture-Tec kit (Invitrogen) 72 h after transfection according to the manufacturer’s instructions. After harvesting, the cells were pelleted and lysed in lysis buffer for immunoprecipitation.
For stable transfections, SW13 cells were transfected as described above with 6 μg of total plasmid DNA, including 2 μg of pBJ5.BRG1 or pBJ5.BRG1-K798R and 3 μg of CMV cyclin E or other cyclins as indicated. pBSK was used as the carrier. Forty-eight hours posttransfection, cells were trypsinized and counted, and 107 cells were replated in the presence of puromycin (Sigma) at 2 μg/ml of complete medium for 10 days.
Crystal violet staining.
SW13 cells were transfected as described above and selected with puromycin. After 10 days of selection, cells were rinsed twice with 5 ml of PBS and fixed overnight in methanol. The methanol was removed, and the cells were stained in crystal violet solution (2% crystal violet in 20% methanol) for 5 h. The plates were rinsed with water until the background plastic was clear. The plates were inverted and dried overnight at room temperature.
BrdU staining.
Transfected SW13 cells were split into two six-well Falcon dishes 48 h after transfection. Puromycin selection of transfected cells was started on the following day (2 μg/ml of complete medium) and continued for 10 days. Cells were labeled with bromodeoxyuridine (BrdU) labeling reagent supplied in the Amersham cell proliferation kit; the labeling reagent was diluted 1:1,000 in complete medium. Cells were labeled for 18 h at 37°C in a CO2 incubator, washed briefly in PBS, and fixed in acid-ethanol (90% ethanol–5% acetic acid–5% water) for 30 min at room temperature. The cells were stained according to the manufacturer’s instructions. Cells were photographed at a magnification of ×40 on a Nikon Diaphot 300 microscope.
SA β-galactosidase staining.
SW13 cells were transfected as for BrdU staining and selected with puromycin. After 10 days of selection, cells were stained for senescence-associated (SA) β-galactosidase activity as previously described (8).
Immunofluorescence.
Cells were fixed in ice-cold methanol for 3 min at −20°C, washed in PBS, and then incubated in 0.5% NP-40–PBS for 5 min at room temperature. Following a PBS wash, samples were blocked for 1 h at room temperature in 10% normal sheep serum–0.5% Tween 20 in PBS and then incubated with primary antibody in PBS–0.5% Tween 20 (J1 [5 μg/ml]) for 1 h at room temperature. Secondary antibody (Texas red-coupled sheep anti-rabbit [Amersham]) was used at a 1:200 dilution in PBS–0.5% Tween 20, and then the mixture was incubated for 1 h at room temperature. Samples were mounted with 4′,6-diamidino-2-phenylindole (DAPI)-containing mounting medium (Vectshield; Vector Laboratories), and signals were detected by fluorescence microscopy (Axioskop; Zeiss) at ×40 magnification.
RESULTS
Cyclin E-associated proteins.
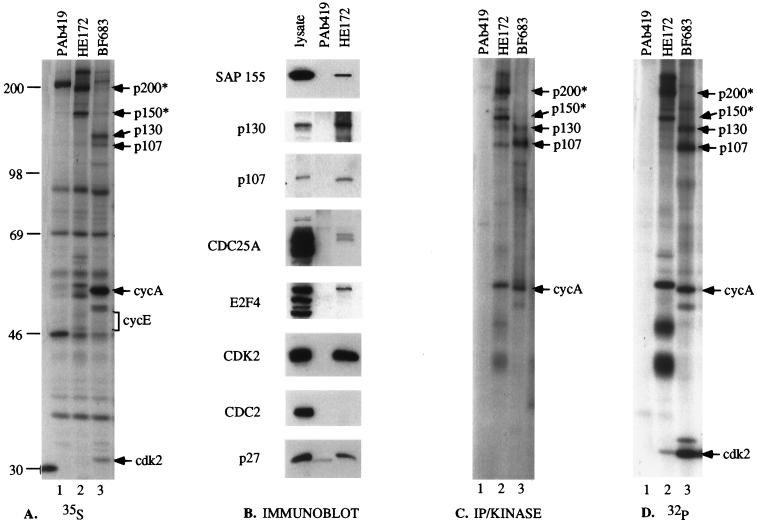
Immunoprecipitations of cyclin E were performed from [35S]methionine-labeled ML-1 cells by using a cyclin E-specific monoclonal antibody, HE172. By using this approach, we were able to identify a number of proteins specifically coprecipitating with cyclin E (Fig. 1A). Cyclin A immunoprecipitations were performed for comparison, and PAb419 immunoprecipitates were included as a negative control. The coprecipitating molecules included proteins previously established to interact with cyclin E, such as cdk2 and the pRb-related proteins p107 and p130 (Fig. 1A, lane 2), which were also present in cyclin A immunoprecipitates (Fig. 1A, lane 3) (12, 14, 29). The presence of additional proteins known to associate with cyclin E was examined by immunoblot analysis (Fig. 1B). Cyclin E was immunoprecipitated from ML-1 cell lysates, and the resulting immune complexes were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then immunoblotted with antibodies against a variety of proteins. As shown in Fig. 1B, cyclin E immune complexes from ML-1 cells contained SAP155 (46, 55), p130, p107, cdc25A (19, 22), E2F 4 (16), p27 (43), and cdk2 but not cdc2. Immunoprecipitations with a negative control antibody did not precipitate these proteins. These results demonstrate that cyclin E associates with several known cellular proteins and that complexes containing these proteins can be recognized by antibodies to cyclin E.
FIG. 1.
Association of cyclin E with cellular proteins. (A) Immunoprecipitations were performed with cyclin E monoclonal antibody (HE172), cyclin A (cycA) (BF683), or a negative control antibody (PAb419) as indicated from [35S]methionine metabolically labeled ML-1 cell lysates. (B) Cyclin E (HE172)- or control antibody (PAb419)-cross-linked beads were used to immunoprecipitate protein complexes from 1 mg of ML-1 extract. Polypeptides were separated by SDS-PAGE and analyzed by immunoblotting with antibiotics as indicated. One hundred micrograms of ML-1 extract was included as a positive control. (C) Immunoprecipitations were performed with cyclin E monoclonal antibody (HE172), cyclin A (BF683), or negative control antibody (PAb419) as indicated from ML-1 cells. Immunocomplexes were subjected to an in vitro kinase assay with 5 μCi of [γ-32P]ATP before separation by SDS-PAGE and autoradiography. (D) Immunoprecipitations were performed as described above from ML-1 cells metabolically labelled with 2 mCi of 32Pir/ml for 30 min. The identities of associated molecules are as indicated; asterisks denote unknown proteins.
Several novel proteins were also identified in the cyclin E immunoprecipitations, including two proteins with sizes of approximately 150 and 200 kDa (marked p150* and p200*, respectively, in Fig. 1A, lane 2). These molecules were not recognized under denaturing conditions, confirming that they were associated proteins and were not directly recognized by the antibody (data not shown and reference 46). To determine whether these proteins could serve as substrates for cyclin E-cdk2, we tested whether they could be phosphorylated in kinase assays in vitro and whether they existed as phosphoproteins in vivo. Immunoprecipitations were performed with lysates prepared from ML-1 cells by using antibodies against cyclin E and cyclin A and with PAb419 as a negative control. Immune complexes were then subjected to in vitro kinase assays. Phosphorylated proteins were resolved by SDS-PAGE and visualized by autoradiography. A subset of the cyclin E-associated proteins served as substrates for cyclin E-cdk2, including p107, p130, and the novel proteins p150 and p200 (Fig. 1C, lane 2). Cyclin A-associated kinase activity could also phosphorylate p107 and p130 but did not associate with or phosphorylate either p150 or p200 (Fig. 1C, lane 3). To study the in vivo phosphorylation state of the various cyclin E-associated proteins, ML-1 cells were labeled with 32Pi. Lysates were prepared and immunoprecipitated with the same antibodies as before. Both p150 and p200 were heavily phosphorylated in the cyclin E immunoprecipitates (Fig. 1D, lane 2). As expected, p107 and p130 were also found phosphorylated in both cyclin E and cyclin A immunoprecipitations, but not in control immunoprecipitations (Fig. 1D)
These experiments thus identified two novel phosphoproteins that specifically interacted with cyclin E and that were phosphorylated by kinase activity associated with cyclin E-cdk2.
p150 is BAF155, a component of the mammalian SWI-SNF complex.
To establish the identity of the cyclin E-associated phosphoprotein p150, large-scale cyclin E immunoprecipitations were performed with 2 × 109 ML-1 cells. Immune complexes were resolved by SDS-PAGE, and the protein corresponding to p150 was excised from the gel for sequencing. Mass spectroscopic sequencing of tryptic peptides yielded four peptide sequences (Fig. 2A) which were contained in a previously isolated protein, BAF155 (Fig. 2B) (57). BAF155 is a mammalian homolog of SWI3, a component of the yeast SWI-SNF complex (57).
FIG. 2.
Cyclin E associates with BAF155. (A) Peptide sequences obtained from mass spectroscopic sequencing of p150. (B) Schematic representation of BAF155. Alignments of peptides to BAF155 are shown by solid bars. The shaded regions share sequence homology with yeast SWI3 (57). (C) A SalI-NotI fragment encoding full-length BAF155 in pSPORT (Invitrogen) was in vitro translated (ivt) with T7 RNA polymerase with the TNT kit (Promega). V8 partial proteolytic mapping of in vitro-translated BAF155 protein and p150 derived from cyclin E immunoprecipitation (cycE ip) was performed according to standard protocols (46). (D) One milligram of ML-1 cell lysate was immunoprecipitated with cyclin E monoclonal antibodies from the HE series (46). PAb419 was included as a negative control. Immunoprecipitates (IP) were analyzed by immunoblotting with antibodies against BAF155 and cdk2. (E) One milligram of ML-1 (lanes 1 to 4) or U2OS (lanes 5 to 9) cell lysate was immunoprecipitated with monoclonal antibodies against cyclin E (HE172), cyclin A (BF683), and cyclin B (GNS1) and polyclonal peptide antisera against cyclins D1, D2, and D3 as indicated. PAb419 and normal rabbit serum (NR) were included as negative controls. Immunoprecipitates were separated by SDS-PAGE and immunoblotted with antibodies against BAF155. cdc2, cdk2, or cdk6.
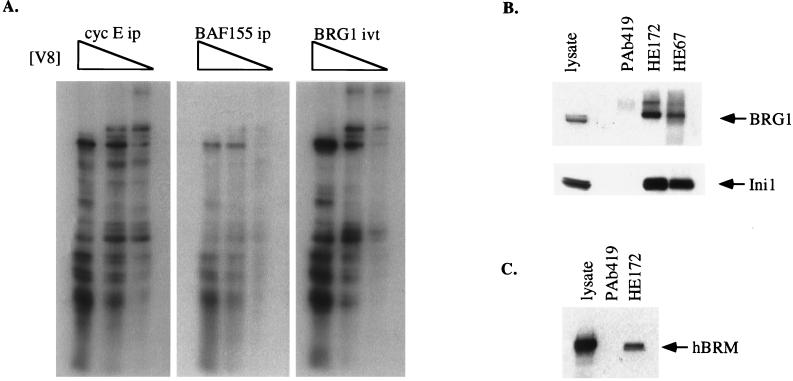
To confirm that the p150 seen in cyclin E immunoprecipitations was BAF155, we performed V8 proteolytic mapping with in vitro-translated BAF155 (Fig. 2C, left panel) and p150 isolated from a cyclin E immunoprecipitation (Fig. 2C, right panel). Both proteins yielded identical profiles, confirming that the cyclin E-associated p150 protein was BAF155. To further analyze the interaction between cyclin E and BAF155, antisera were raised against BAF155 and used in immunoblot analysis of cyclin E immunoprecipitates. Two monoclonal antibodies that recognize different epitopes of cyclin E, HE172 and HE67 (46), could both coprecipitate BAF155 (Fig. 2D). Immunoprecipitations with antibodies against cyclin A (Fig. 2E, lane 3), cyclin B1 (lane 4), and the D-type cyclins (lanes 6 to 8) did not contain detectable BAF155 (Fig. 2E, upper panel), suggesting that association with BAF155 may be specific for cyclin E.
Cyclin E associates with several SWI-SNF components.
The presence of BAF155 in cyclin E complexes raised the possibility that other components of the SWI-SNF complex may also associate with cyclin E. The novel 200-kDa protein seen in the cyclin E immunoprecipitations (p200* [Fig. 1]) has a molecular mass similar to that of BRG1, a mammalian homolog of yeast SWI2/SNF2 (25). BRG1 has DNA-dependent ATPase activity and is an integral component of the SWI-SNF complex.
V8 partial proteolytic mapping of the 200-kDa protein in cyclin E immunoprecipitations (Fig. 3A, left panel) and in vitro-translated BRG1 (Fig. 3A, right panel) showed that the two profiles were similar, hence identifying the cyclin E-associated p200 protein as BRG1. A similar 200-kDa protein seen in BAF155 immunoprecipitations was also confirmed to be BRG1 by V8 analysis (Fig. 3A, middle panel), confirming previous observations that BAF155 and BRG1 are associated in vivo (57). Immunoblot analysis with an antibody developed against BRG1 confirmed the presence of BRG1 in cyclin E immunoprecipitations with the two different cyclin E monoclonal antibodies, HE172 and HE67 (Fig. 3B, upper panel).
FIG. 3.
BRG1 is associated with the cyclin E-BAF155 complex. (A) V8 partial proteolytic mapping of in vitro-translated (ivt) BRG1 protein and p200 derived from cyclin E (cyc E) and BAF155 immunoprecipitations (ip). (B) Immunoblot analysis of cyclin E (HE172, HE67) or control (PAb419) immunoprecipitations from ML-1 cells with antibodies to BRG1 and Ini1 as indicated. One hundred micrograms of cell lysate was loaded as a positive control. (C) Immunoblot analysis of cyclin E (HE172) or control (PAb419) immunoprecipitations from ML-1 cells with antibodies to hBRM. One hundred micrograms of cell lysate was loaded as a positive control.
Another well-characterized component of the mammalian SWI-SNF complex, Ini1-hSNF5 (23), was clearly detectable in the cyclin E immunoprecipitations (Fig. 3B, lower panel). These data demonstrate that cyclin E associates with several subunits of the SWI-SNF apparatus present within a cell and suggest that the entire SWI-SNF complex may be involved. Gel filtration on Superose 6 demonstrated that the cyclin E-BAF155-containing complexes eluted well ahead of the 699-kDa thyroglobulin marker at an estimated molecular mass of over 1 MDa, consistent with the size of the SWI-SNF complexes (data not shown and reference 56). These complexes represent only a fraction of the cyclin E present within the cells. These complexes appear to be relatively stable and are resistant to stringent washing conditions with salt concentrations of up to 750 mM NaCl (data not shown).
In further analysis, we were able to demonstrate that the BRG1-related protein hBRM (39), which is believed to form distinct SWI-SNF complexes from BRG1 (56), also associates with cyclin E (Fig. 3C). This provides evidence that more than one SWI-SNF species may be recognized by cyclin E.
BRG1 promotes the association of cyclin E with BAF155.
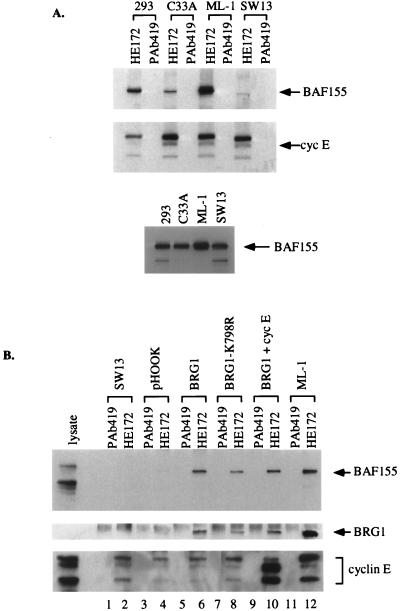
To study the interaction of cyclin E with SWI-SNF complexes in more detail, we examined several human cell lines, including two that lack components of the SWI-SNF apparatus, SW13 and C33A. SW13 is a human carcinoma cell line derived from a tumor of the adrenal cortex, which does not express detectable levels of BRG1 or hBRM (39). Although much of the SWI-SNF complex remains assembled in SW13 cells (56), and BAF155 is expressed (Fig. 4A, bottom panel), we were unable to detect any BAF155 associated with cyclin E (Fig. 4A). Experiments with C33A cells, a cervical carcinoma cell line that also does not express hBRM (39) and only expresses low levels of BRG1, revealed reduced levels of BAF155 found complexed with cyclin E (Fig. 4A, lane 3, top panel). In contrast, in 293 cells that express wild-type BRG1, coprecipitation of BAF155 with cyclin E was easily observed. These results inplied that expression of BRG1 is required for the stable association of cyclin E with the SWI-SNF complex.
FIG. 4.
BRG1 is required for the interaction between cyclin E and BAF155. (A) Immunoblot analysis of cyclin E (HE172) or control (PAb419) immunoprecipitations from cell lines as indicated, probed with antibodies against BAF155 or cyclin E. Lysates were also probed directly for BAF155 expression (lower panel). (B) SW13 cells were transiently transfected with pHook-1 (Invitrogen) alone (lanes 3 and 4) or together with pBJ5.BRG1 (lanes 5 and 6), pBJ5.BRG1-K798R (lanes 7 and 8), and pBJ5.BRG1 plus pCMV-cyclin E (lanes 9 and 10) as indicated. Transfected cells were harvested with the Capture-Tec kit (Invitrogen) 72 h after transfection. Cyclin E (HE172) or control (PAb419) immunoprecipitations were performed from 100 μg of lysate and probed with antibodies to BAF155 (upper panel), BRG1 (middle panel), and cyclin E (lower panel). Untransfected SW13 (lanes 1 and 2) and ML-1 (lanes 11 and 12) cells were included as controls.
To test this hypothesis, we asked whether introduction of BRG1 into SW13 cells would restore the stable interaction between cyclin E and BAF155. SW13 cells were transiently transfected with an expression construct for BRG1, together with a selectable cell surface marker (pHook-1; Invitrogen). Seventy-two hours posttransfection, cells were harvested and positively selected for expression of the cotransfected cell surface marker. Lysates were prepared from the selected cells and immunoprecipitated with the cyclin E monoclonal antibody, HE172, or a negative control, PAb419.
Immunoprecipitates were resolved by SDS-PAGE and immunoblotted with BAF155 antisera (Fig. 4B, upper panel). Lysates from untransfected SW13 and ML-1 cells were also immunoprecipitated to act as negative and positive controls, respectively. Ectopic expression of BRG1 was sufficient to facilitate the coprecipitation of BAF155 with cyclin E (Fig. 4B, compare lanes 4 and 6). The immunoblot was reprobed with BRG1 antisera to demonstrate the presence of BRG1 in this newly formed complex (Fig. 4B, middle panel) and with antibodies to cyclin E to show that the immunoprecipitations were equally efficient at precipitating cyclin E (Fig. 4B, lower panel). The intrinsic ATPase activity of BRG1 was not required for this association, because the catalytically inactive K798R mutant of BRG1 (39) was also able to stimulate complex formation (Fig. 4B, lane 8). Cotransfection with cyclin E did not increase the amount of cyclin E-BAF155 complexes over endogenous levels (Fig. 4B, lane 10). These results demonstrate that the expression of BRG1 facilitates the interaction of cyclin E with the SWI-SNF complex.
Cyclin E association with BRG1 does not require pRb family members.
A possible explanation for the association seen between cyclin E and the SWI-SNF complex was that it was mediated by members of the pRb family. Cyclin E binds stably to p107 and p130 (see Fig. 1 and reference 15), and since BRG1 can also interact with pRb, p107, and p130 (11, 49), it was possible that they were all contained within the same complex.
As shown previously (Fig. 4A), cyclin E was associated with BAF155 in 293 cells that do not express functional pRb, suggesting that pRb may not be an absolute requirement for the cyclin E-BAF155 complex to form. To extend this analysis, we examined SAOS-2 cells, a pRb-negative human osteosarcoma cell line. Cell lysates were prepared and immunoprecipitated with the cyclin E monoclonal antibody, HE172 (Fig. 5A, lane 1), or a control antibody, PAb419 (Fig. 5A, lane 2). Immunocomplexes were resolved by SDS-PAGE and immunoblotted with antibodies to BAF155. As shown in Fig. 5A, the association between cyclin E and BAF155 is easily detectable, confirming that pRb is dispensable for this interaction. To determine whether p107 or p130 was present in the complex, cell lysates were precleared with antibodies to p107 and p130 (Fig. 5A, lanes 4 and 6) or with normal rabbit serum as a negative control (Fig. 5A, lanes 3 and 5) prior to immunoprecipitation. Significantly, depletion of p107 and p130 did not reduce the amount of BAF155 bound to cyclin E (Fig. 5A, lane 6). Precleared lysates were immunoblotted with antibodies to p107, p130, and pRb to verify the depletion of all three proteins from these extracts (Fig. 5B). As expected, examination of cyclin E levels demonstrated that depletion of p107 and p130 led to a reduction of cyclin E levels in the SAOS-2 cell lysate (Fig. 5B, top panel), since cyclin E is found in stable complexes with p107 and p130 (29). Our findings therefore suggest that only a proportion of the cyclin E in the cell is bound to p107 and p130 and that the SWI-SNF-containing cyclin E complex is likely to be a novel and distinct complex that does not require pRb proteins for its assembly.
FIG. 5.
Association of cyclin E with SWI-SNF complex does not require the presence of pRb family member proteins. (A) Immunoblot analysis of cyclin E (HE172 [lane 1]) or control (PAb419 [lane 2]) immunoprecipitations (IP) from SAOS-2 cells. In lanes 3 to 6, lysates were precleared prior to immunoprecipitation with antibodies to p107 and p130 (lanes 4 and 6) or normal rabbit (NR) serum (lanes 3 and 5). ML-1 lysate was run as a positive control in lane 7. Blots were probed with antibodies against BAF155. (B) Extracts (100 μg) from lysates precleared with normal rabbit serum or p107 and p130 were immunoblotted with antibodies as indicated.
BRG1 induces cell cycle arrest.
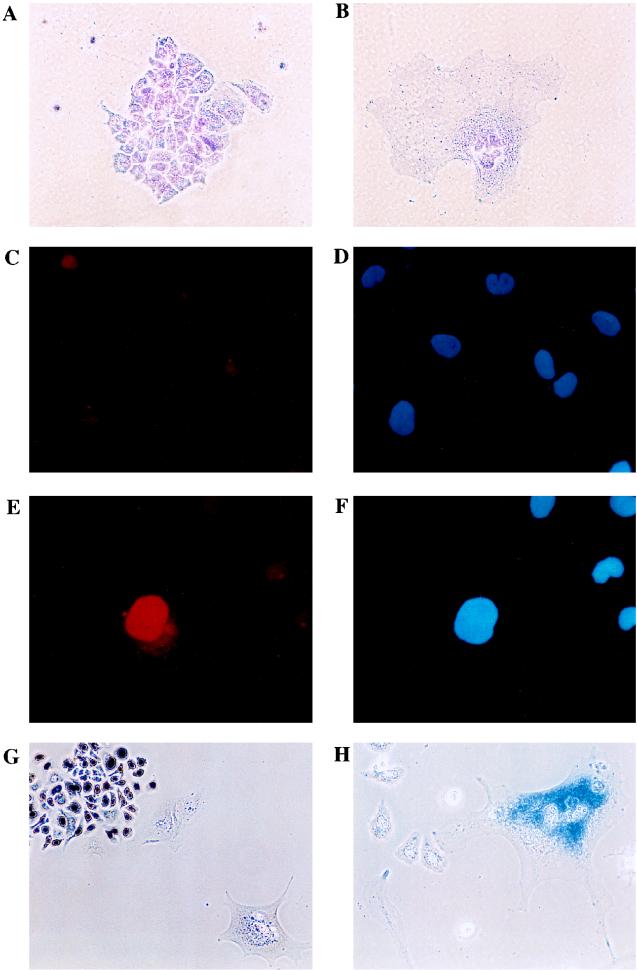
To address the functional significance of cyclin E’s interaction with the SWI-SNF complex, we examined the properties of BRG1 by using an assay that has previously been described for the analysis of BRG1 function (11, 49). When overexpressed in SW13 cells, BRG1 induces a change in cell morphology. We were interested in seeing whether we could influence the ability of BRG1 to induce such a flat-cell phenotype by the coexpression of cyclin E. SW13 cells were cotransfected with BRG1 and a puromycin resistance plasmid. Cells were grown under puromycin selection for 10 days and then stained with crystal violet, and flat cells were counted. As expected, transfection of BRG1 induced a flat-cell phenotype in a percentage of the cells (Fig. 6B and Table 1). Immunostaining with anti-BRG1 sera demonstrated that BRG1 was expressed in the flat-cell population and not in adjacent normal cells (Fig. 6E). To ascertain whether these flat cells were still proliferating, drug-selected cells were labeled with BrdU. BRG1-expressing flat cells failed to stain with BrdU, even with 18- to 24-h labeling times, while adjacent growing colonies readily incorporated BrdU (Fig. 6G), indicating that the flat cells were growth arrested. The morphological changes we observed are often associated with cellular senescence. To investigate this phenomenon further, we assayed for SA β-galactosidase activity (8). As shown in Fig. 6H, all of the flat cells stained positive for SA β-galactosidase activity, indicative of replicative senescence. Thus, introduction of BRG1 into SW13 cells was sufficient to induce growth arrest and markers of senescence.
FIG. 6.
BRG1 induces growth arrest. SW13 cells were transfected as described in Materials and Methods with 6 μg of total plasmid DNA, including 2 μg of pBJ5.BRG1. pBSK was used as the carrier. Cells were grown in the presence of puromycin for 10 days. Cells were then either fixed and stained with 2% crystal violet–20% methanol for 5 h (original magnification, ×40 [A and B]), fixed and stained with normal rabbit serum and counterstained with DAPI (original magnification, ×40) [C and D]), fixed and stained with BRG1 antibody (J1) (25) and counterstained with DAPI (original magnification, ×40 [E and F]), labelled with BrdU (original magnification, ×20 [G]), or stained for SA β-galactosidase activity as previously described (8) (original magnification, ×40 [H]).
TABLE 1.
BRG1 induction of flat-cell morphology in SW13 cells
| Plasmid(s) transfected | No. of flat cells/transfection in expta:
|
Avg % flat cells | |||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| BRG1 | 854 | 739 | 827 | 553 | 100 |
| BRG1 + cyclin E | 421 | 376 | 436 | 259 | 50 |
| Cyclin E | 121 | 67 | 73 | 22 | 9 |
| BRG1-K798R | 395 | 213 | 177 | NDb | 32 |
| BRG1-K798R + cyclin E | 193 | 147 | 116 | ND | 19 |
| BRG1 + cyclin D1 | ND | 372 | 465 | 181 | 48 |
| Cyclin D1 | ND | 118 | ND | 16 | 10 |
| Puromycin | 81 | 56 | 68 | 44 | 8 |
The numbers presented are the total number of flat cells per transfection counted 10 days after selection. Data were collected from several independent experiments (experiments 1 to 4).
ND, not determined.
Cyclin rescue of flat-cell phenotype and prevention of growth arrest.
We next tested whether cyclin E could suppress the ability of BRG1 to induce the flat-cell phenotype. SW13 cells were cotransfected with cyclin E and BRG1 and were then grown under puromycin selection for 10 days as before. The coexpression of cyclin E with BRG1 significantly reduced the frequency of flat cells by up to 50% (Table 1). The levels of BRG1 expression were not affected by the presence of cyclin E (Fig. 4B, compare lanes 6 and 10). Introduction of the ATP-binding mutant of BRG1, K798R, induced flat cells at only 32% of the frequency of wild-type BRG1 (11). However, this value was further reduced by the overexpression of cyclin E, suggesting that cyclin E affects an ATP-independent function of BRG1. We next examined whether other cyclins shared the ability with cyclin E to abrogate flat-cell production by BRG1. Cotransfection of cyclin D1 could also reduce flat-cell induction by BRG1 to a similar degree (52%). Cyclin A showed toxicity in this assay and was not evaluated further. The abilities to abrogate BRG1-induced growth arrest in this assay were therefore comparable between cyclin E and cyclin D1, although the stable association of cyclin E, but not cyclin D1, with the SWI-SNF complex suggests that cyclin E may be the more physiologically relevant effector.
DISCUSSION
Analysis of cyclin E-containing complexes in the cell demonstrated that cyclin E associated with several cellular proteins. These include its kinase partner, cdk2, and molecules that affect the activity of cyclin E-cdk2 complexes both positively (cdc25A) and negatively (p27). In addition, this approach has previously yielded the identification of novel substrates, including both p107 and p130, suggesting that cdks can form stable enzyme-substrate complexes in vivo. This stable interaction may provide a mechanism by which increased specificity and selectivity can be achieved.
In this study, we have characterized two novel cyclin E-associated proteins as components of the mammalian SWI-SNF complex. Both BAF155 and BRG1 contain cdk consensus phosphorylation sites, and both could be phosphorylated by cyclin E-cdk2-associated kinase activity in vitro. Furthermore, BRG1 and BAF155 are in a phosphorylated form in the cyclin E complex. Another component of the SWI-SNF apparatus, the Ini1-hSNF5 protein, is also present in cyclin E immunoprecipitations, which suggests that the entire SWI-SNF complex may be recognized by cyclin E. Our experiments further demonstrate an intriguing requirement for the presence of BRG1 in the SWI-SNF complex to promote the recruitment of cyclin E. This observation suggests that either BRG1 recruits some essential factor to the complex, or the SWI-SNF apparatus is somehow modified in the presence of BRG1 so that it is recognized by cyclin E. The presence of the BRG1-related molecule hBRM in cyclin E immunoprecipitations implies that at least two different SWI-SNF complexes may be targeted by cyclin E, since hBRM and BRG1 are believed to be mutually exclusive components of the SWI-SNF apparatus (56). Immunodepletion experiments suggest that the cyclin E-BAF155-BRG1 complex is distinct from those cyclin E complexes containing p107 and p130 and that neither pRb nor its related family members p107 and p130 are needed for cyclin E-BAF155-BRG1 complex assembly.
To demonstrate that the interaction of the SWI-SNF apparatus with cyclin E was functionally significant, we took advantage of a flat-cell assay established previously for BRG1 (11). BRG1 is capable of inducing a flattened-cell morphology in SW13 cells. These cells do not divide and express markers indicative of replicative senescence, namely, SA β-galactosidase activity. Both cyclin E and cyclin D1 could abrogate this property of BRG1, reducing flat-cell induction by as much as 50%, suggesting that these cyclins can modulate the activity of BRG1. The flat-cell phenotype described in this study is similar to the one observed with the introduction of pRb into SAOS-2 cells (18, 20, 35, 50). In SAOS-2 cells, pRb similarly causes growth arrest and induction of SA β-galactosidase activity, which can be rescued by overexpression of G1 cyclins (18). While a dependence on BRG1 for pRb-mediated growth suppression has not yet been shown, a mutant form of BRG1 that is defective in the ability to bind pRb can no longer induce growth arrest in SW13 cells (11). Hence, it has been proposed that pRb and BRG1 may function together to induce cell cycle arrest.
It will be important to establish the mechanism by which cyclin E suppresses flat-cell development and whether it is similar in both the SW13 and SAOS-2 assays. The ability of cyclins to rescue growth in the SAOS-2 cotransfection experiments was originally interpreted to be through pRb phosphorylation (18). However, more recent experiments show that cyclins can also rescue the growth arrest induced by a nonphosphorylatable form of pRb (30). The ability of cyclins to rescue growth in the SW13 cotransfection experiments may also be independent of the pRb family of proteins, since even in their absence, cyclin E can associate with SWI-SNF components. Furthermore, complexes between BRG1 and pRb appear unaffected by overexpression of cyclin E (data not shown). Our experiments thus raise the possibility that cyclin E may impinge on the SWI-SNF apparatus directly to revert the flat-cell phenotype. We are currently examining the phosphorylation status of various components of the SWI-SNF complex to see if they are altered with cyclin E overexpression.
A recent report by Sif et al. has demonstrated that the mitotic inactivation of the human SWI-SNF complex is caused by phosphorylation of various SWI-SNF subunits, including BRG1 (48). The identity of the kinases responsible for these regulatory phosphorylations remains unknown. These observations, taken together with our results, suggest that the SWI-SNF apparatus may be modulated both positively and negatively through the cell cycle. While mitotic phosphorylation of BRG1 may be required for chromatin-mediated transcriptional repression during mitosis, phosphorylation on different sites of BRG1 or of other SWI-SNF subunits may be required for chromatin remodeling during G1 as cells prepare for DNA synthesis. Targets for modulation may include Ini1-hSNF5, because the yeast homolog Sfh1p is phosphorylated in a cell cycle-dependent manner during G1 (4). Since Ini1-hSNF5 is present in cyclin E immunoprecipitations, it will be important to address whether it also serves as a substrate for cyclin E-cdk2. A detailed analysis of the phosphorylation status of each of the SWI-SNF components as a function of cell cycle progression, and how this affects the chromatin remodeling activity of the complex, will help to elucidate how the SWI-SNF apparatus is coordinately regulated.
The demonstration that BRG1 may induce senescence is significant, because it implies that chromatin reorganization may be important during cell cycle exit and perhaps in the maintenance of a postmitotic state. Recent data suggest that inhibition of cdk activity can induce a senescent state (2, 34). Our data are consistent with a role for cyclin E in preventing cells from exiting from the cell cycle permanently through modulation of the SWI-SNF complex. It may be significant that Ini1-hSNF5 (23, 38) has recently been shown to be mutated in aggressive cancers (54). Such observations provide further evidence that critical components of the chromatin remodeling machinery may act as growth suppressors that can be regulated by the cell cycle machinery. Overexpression of cyclin E in many human tumors may thus be a mechanism by which malignant cells escape cell cycle exit and senescence.
ACKNOWLEDGMENTS
We thank W. Wang and G. Crabtree for reagents and W. Lane at the Harvard Microchemistry Laboratory for peptide sequencing. We are grateful to W. Korver and M. McMahon for discussions and comments on the manuscript.
The DNAX Research Institute is supported by Schering-Plough Corporation.
REFERENCES
- 1.Alevizopoulos K, Vlach J, Hennecke S, Amati B. Cyclin E and c-Myc promote cell proliferation in the presence of p16INK4a and of hypophosphorylated retinoblastoma family proteins. EMBO J. 1997;16:5322–5333. doi: 10.1093/emboj/16.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown J, Wei W, Sedivy J. Bypass of senescence after disruption of p21CPI1/WAF1 gene in normal diploid human fibroblasts. Science. 1997;277:831–834. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- 3.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 4.Cao Y, Cairns B R, Kornberg R D, Laurent B C. Sfh1p, a component of a novel chromatin-remodeling complex, is required for cell cycle progression. Mol Cell Biol. 1997;17:3323–3334. doi: 10.1128/mcb.17.6.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson M, Laurent B C. The SWI/SNFI family of global transcriptional activators. Curr Opin Cell Biol. 1994;6:396–402. doi: 10.1016/0955-0674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 6.Chiba H, Muramatsu M, Nomoto A, Kato H. Two human homologues of Saccharomyces cerevisiae SWI/SNF2 complex and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic receptor. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cote J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 8.Dimri G P, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano E E, Linskens M, Rubelj I, Pereira-Smith O, Peacocke M, Campisi J. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dowdy S F, Hinds P W, Louie K, Reed S I, Arnold A, Weinberg R A. Physical interaction of the retinoblastoma protein with human D cyclins. Cell. 1993;73:499–511. doi: 10.1016/0092-8674(93)90137-f. [DOI] [PubMed] [Google Scholar]
- 10.Dulic V, Lees E, Reed S I. Association of human cyclin E with a periodic G1-S phase protein kinase. Science. 1992;257:1958–1961. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- 11.Dunaief J L, Strober B E, Guha S, Khavari P A, Alin K, Luban J, Begemann M, Crabtree G R, Goff S P. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 12.Ewen M, Faha B, Harlow E, Livingston D. Interaction of p107 with cyclin A independent of complex formation with viral oncoproteins. Science. 1992;255:85–87. doi: 10.1126/science.1532457. [DOI] [PubMed] [Google Scholar]
- 13.Ewen M E, Sluss H K, Sherr C J, Matsushime H, Kato J, Livingston D M. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell. 1993;73:487–497. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- 14.Faha B, Ewen M E, Tsai L H, Livingston D M, Harlow E. Interaction between human cyclin A and adenovirus E1A-associated p107 protein. Science. 1992;255:87–90. doi: 10.1126/science.1532458. [DOI] [PubMed] [Google Scholar]
- 15.Faha B, Harlow E, Lees E. The adenovirus E1A-associated kinase consists of cyclin E-p33cdk2 and cyclin A-p33cdk2. J Virol. 1993;67:2456–2465. doi: 10.1128/jvi.67.5.2456-2465.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ginsberg D, Vairo G, Chittenden T, Xiao Z H, Xu G, Wydner K L, DeCaprio J A, Lawrence J B, Livingston D M. E2F-4, a new member of the E2F transcription factor family, interacts with p107. Genes Dev. 1994;8:2265–2679. doi: 10.1101/gad.8.22.2665. [DOI] [PubMed] [Google Scholar]
- 17.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 18.Hinds P W, Mittnacht S, Dulic V, Arnold A, Reed S I, Weinberg R A. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 19.Hoffmann I, Draetta G, Karsenti E. Activation of the phosphatase activity of human cdc25A by a cdk2-cyclin E dependent phosphorylation at the G1/S transition. EMBO J. 1994;13:4302–4310. doi: 10.1002/j.1460-2075.1994.tb06750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang H J, Yee J K, Shew J Y, Chen P L, Bookstein R, Friedman T, Lee E Y, Lee W H. Suppression of the neoplastic phenotype by replacement of the RB gene in human cancer cells. Science. 1998;242:1563–1566. doi: 10.1126/science.3201247. [DOI] [PubMed] [Google Scholar]
- 21.Imbalzano A N, Kwon H, Green M R, Kingston R E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 22.Jinno S, Suto K, Nagata A, Igarashi M, Kanaoka Y, Nojima H, Okayama H. Cdc25A is a novel phosphatase functioning early in the cell cycle. EMBO J. 1994;13:1549–1556. doi: 10.1002/j.1460-2075.1994.tb06417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 24.Kato J, Matsushime H, Hiebert S W, Ewen M E, Sherr C J. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993;7:331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- 25.Khavari P, Peterson C, Tamkun J, Mendel D, Crabtree G. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 26.Koff A, Giordano A, Desai D, Yamashita K, Harper W, Elledge S, Nishimoto T, Morgan D, Franza R, Roberts J. Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science. 1992;257:1689–1693. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- 27.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 28.Lees E. Cyclin dependent kinase regulation. Curr Opin Cell Biol. 1995;7:773–780. doi: 10.1016/0955-0674(95)80060-3. [DOI] [PubMed] [Google Scholar]
- 29.Lees E, Faha B, Dulic V, Reed S I, Harlow E. Cyclin E/cdk2 and cyclin A/cdk2 kinases associate with p107 and E2F in a temporally distinct manner. Genes Dev. 1992;6:1874–1885. doi: 10.1101/gad.6.10.1874. [DOI] [PubMed] [Google Scholar]
- 30.Leng X, Connell-Crowley L, Goodrich D, Harper J W. S-phase entry upon ectopic expression of G1 cyclin-dependent kinases in the absence of retinoblastoma protein phosphorylation. Curr Biol. 1997;7:709–712. doi: 10.1016/s0960-9822(06)00301-0. [DOI] [PubMed] [Google Scholar]
- 31.Logie C, Peterson C L. Catalytic activity of the yeast SWI/SNF complex on reconstituted nucleosome arrays. EMBO J. 1997;16:6772–6782. doi: 10.1093/emboj/16.22.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorch Y, Cairns B R, Zhang M, Kornberg R D. Activated RSC-nucleosome complex and persistently altered form of the nucleosome. Cell. 1998;94:29–34. doi: 10.1016/s0092-8674(00)81218-0. [DOI] [PubMed] [Google Scholar]
- 33.Lukas J, Herzinger T, Hansen K, Moroni M C, Resnitzky D, Helin K, Reed S I, Bartek J. Cyclin E-induced S phase without activation of the pRb/E2F pathway. Genes Dev. 1997;11:1479–1492. doi: 10.1101/gad.11.11.1479. [DOI] [PubMed] [Google Scholar]
- 34.McConnell B B, Starborg M, Brookes S, Peters G. Inhibitors of cyclin-dependent kinases induce features of replicative senescence in early passage human diploid fibroblasts. Curr Biol. 1998;8:351–354. doi: 10.1016/s0960-9822(98)70137-x. [DOI] [PubMed] [Google Scholar]
- 35.Mittnacht S, Weinberg R A. G1/S phosphorylation of the retinoblastoma protein is associated with altered affinity for the nuclear compartment. Cell. 1991;65:381–393. doi: 10.1016/0092-8674(91)90456-9. [DOI] [PubMed] [Google Scholar]
- 36.Morgan D O. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 37.Muchardt C, Reyes J C, Bourachot B, Leguoy E, Yaniv M. The hbrm and BRG-1 proteins, components of the human SWI/SNF complex, are phosphorylated and excluded from the condensed chromosomes during mitosis. EMBO J. 1996;15:3394–3402. [PMC free article] [PubMed] [Google Scholar]
- 38.Muchardt C, Sardet C, Bourachot B, Onufryk C, Yaniv M. A human protein with homology to Saccharomyces cerevisiae SNF5 interacts with the potential helicase hbrm. Nucleic Acids Res. 1995;23:1127–1132. doi: 10.1093/nar/23.7.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohtsubo M, Theodoras A M, Schumacher J, Roberts J M, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Owen-Hughes T, Utley R T, Cote J, Peterson C L, Workman J L. Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science. 1996;273:513–516. doi: 10.1126/science.273.5274.513. [DOI] [PubMed] [Google Scholar]
- 42.Peterson C L, Tamkun J W. The SWI-SNF complex: a chromatin remodeling machine? Trends Genet. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 43.Polyak K, Lee M H, Erdjument B H, Koff A, Roberts J M, Tempst P, Massague J. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell. 1994;78:59–66. doi: 10.1016/0092-8674(94)90572-x. [DOI] [PubMed] [Google Scholar]
- 44.Resnitzky D, Gossen M, Bujard H, Reed S I. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol Cell Biol. 1994;14:1669–1679. doi: 10.1128/mcb.14.3.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnitzler G, Sif S, Kingston R E. Human SWI/SNF interconverts a nucleosome between its base and a stable remodeled state. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- 46.Seghezzi W, Chua K, Shanahan F, Gozani O, Reed R, Lees E. Cyclin E associates with components of the pre-mRNA splicing machinery in mammalian cells. Mol Cell Biol. 1998;18:4526–4536. doi: 10.1128/mcb.18.8.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherr C J. Cancer cell cycles. Science. 1996;274:1674–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 48.Sif S, Stukenburg P T, Kirschner M W, Kingston R E. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 1998;12:2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strober B E, Dunaief J L, Guha S, Goff S P. Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol Cell Biol. 1996;16:1576–1583. doi: 10.1128/mcb.16.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Templeton D J, Park S H, Lanier L, Weinberg R A. Nonfunctional mutants of the retinoblastoma protein are characterised by defects in phosphorylation, viral oncoprotein association, and nuclear tethering. Proc Natl Acad Sci USA. 1991;88:3033–3037. doi: 10.1073/pnas.88.8.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. RB and hbrm cooperate to repress the activation functions of E2F1. Proc Natl Acad Sci USA. 1997;94:11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsai L-H, Lees E, Faha B, Harlow E, Riabowol K. The cdk2 kinase is required for the G1-to-S transition in mammalian cells. Oncogene. 1993;8:1593–1602. [PubMed] [Google Scholar]
- 53.van den Heuvel S, Harlow E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- 54.Versteege I, Sevenet N, Lange J, Rousseau-Merck M-F, Ambros P, Handgretinger R, Aurias A, Delattre O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 55.Wang C, Chua K, Seghezzi W, Lees E, Gozani O, Reed R. Phosphorylation of the spliceosomal protein SAP155 is coupled with splicing catalysis. Genes Dev. 1998;12:1409–1414. doi: 10.1101/gad.12.10.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang W, Cote J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, Workman J L, Crabtree G R. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 57.Wang W, Xue Y, Zhou S, Kuo A, Cairns B, Crabtree G. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 58.Zhao J, Dynlacht B, Imai T, Hori T, Harlow E. Expression of NPAT, a novel substrate of cyclin E-CDK2, promotes S-phase entry. Genes Dev. 1998;12:456–461. doi: 10.1101/gad.12.4.456. [DOI] [PMC free article] [PubMed] [Google Scholar]